Triple fluorescent quantitative PCR detection primers and kit for identifying African swine fever wild strain and gene deletion strain
A detection kit and gene deletion technology, which is applied in the determination/inspection of microorganisms, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problems of application limitations, achieve rapid detection process, wide detection range, and detection reaction sensitivity high effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
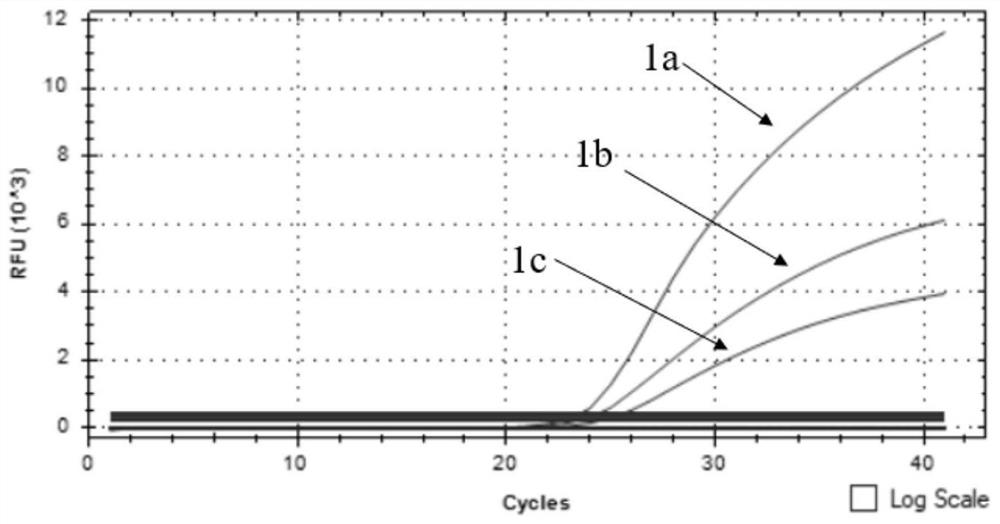
[0049] The design of embodiment 1 specific primer and probe
[0050] According to the B646L, EGFP and mCherry gene sequences published by GenBank, specific primers and probes for detecting B646L, EGFP and mCherry genes were designed in their conserved regions. The nucleotide sequences and amplified fragment sizes of the specific primers are shown in Table 1. The nucleotide sequences of the specific probes are shown in Table 2, and both the primers and the probes were synthesized by Yingwei Jieji. Wherein, since the RFP gene and the mCherry gene have high homology, both genes can be detected by the specific primers and probes designed for the mCherry gene in the present invention.
[0051] Table 1. Nucleotide sequences and amplified fragment sizes of specific primers
[0052]
[0053]
[0054] Table 2. Nucleotide sequences of specific probes
[0055]
Embodiment 2 3
[0056] Embodiment 2 Triple fluorescent quantitative PCR detection kit and detection method
[0057] 1. Triple fluorescence quantitative PCR detection kit
[0058] A triple fluorescent quantitative PCR detection kit for detecting African swine fever wild strains and gene deletion strains, comprising the following components:
[0059] (1) the specific primer and probe of embodiment 1;
[0060] (2) AceQ Universal U + Probe Master Mix V2, ddH 2 O (negative control);
[0061] (3) Positive plasmid: The positive plasmid was used as the positive control, and the positive plasmid was the recombinant plasmid vector pUC57-EGFP, pMD18-B646L and pUC57-mCherry, all of which were stored in the Department of Infectious Diseases, School of Veterinary Medicine, South China Agricultural University.
[0062] 2. Triple fluorescent quantitative PCR detection method
[0063] (1) Sample nucleic acid extraction
[0064] Use the Axygen nucleic acid extraction kit from Corning Biotechnology Co., L...
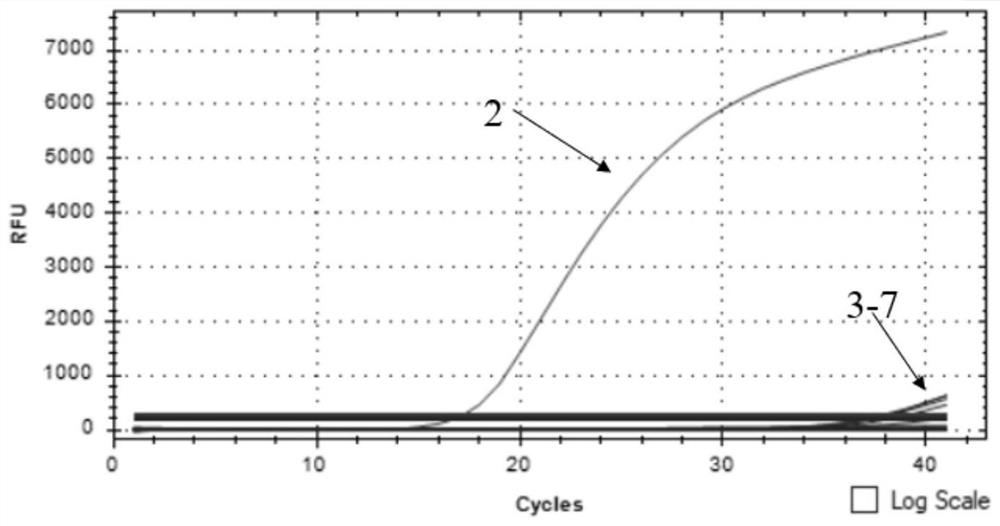
Embodiment 3 3
[0078] The clinical sample detection of embodiment 3 triple fluorescence quantitative PCR
[0079] 1. Clinical sample testing
[0080] A total of 849 clinical samples were collected from pig farms in Guangdong Province, of which 513 were pig anticoagulant blood, 256 were pig mouth swabs, 35 were pig tonsils, and 45 were pig lymph nodes; among them, 15 were positive for African swine fever virus Samples, nucleic acid extracted from 2 African swine fever-affected pigs (from the case announced by the Ministry of Agriculture and Rural Affairs in Huangpu District, Guangzhou City), were prepared and preserved by the National African Swine Fever Regional Laboratory (Guangzhou) (because the gene deletion strain The vaccine has not yet been successfully marketed, so it is currently only tested on clinical samples infected with wild African swine fever virus). These 849 clinical samples were detected using the detection kit and detection method of Example 2.
[0081] 2. Test results ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com