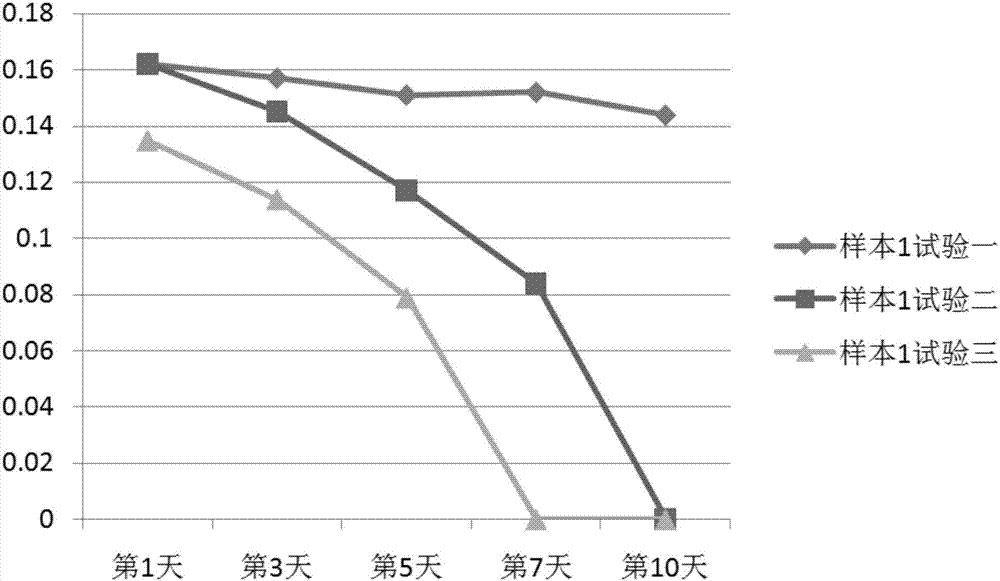
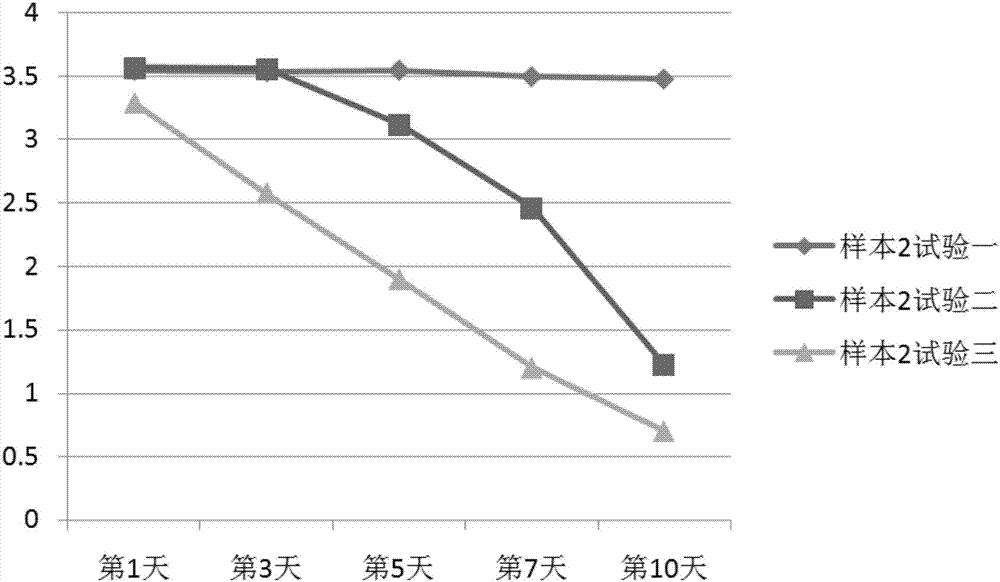
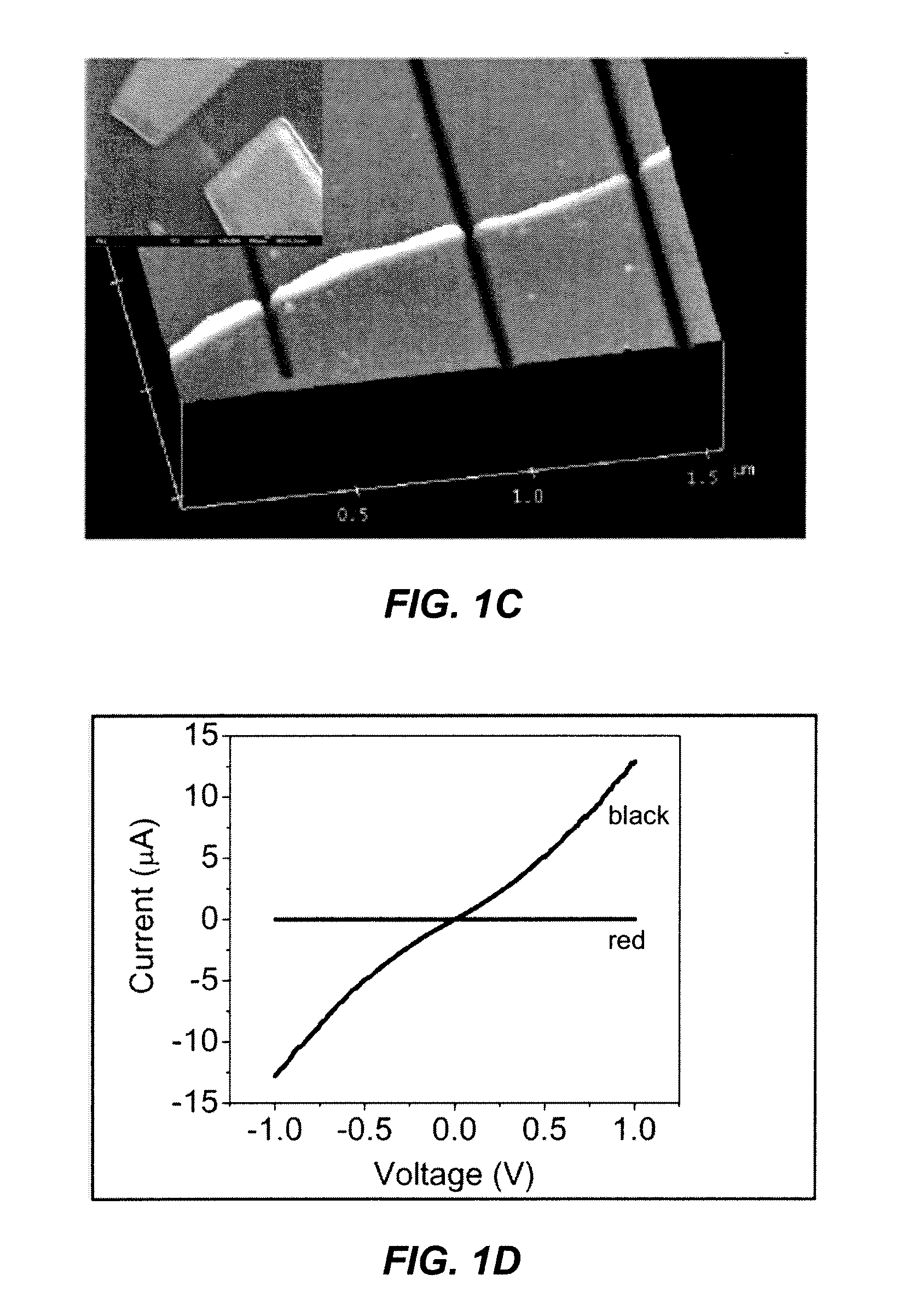
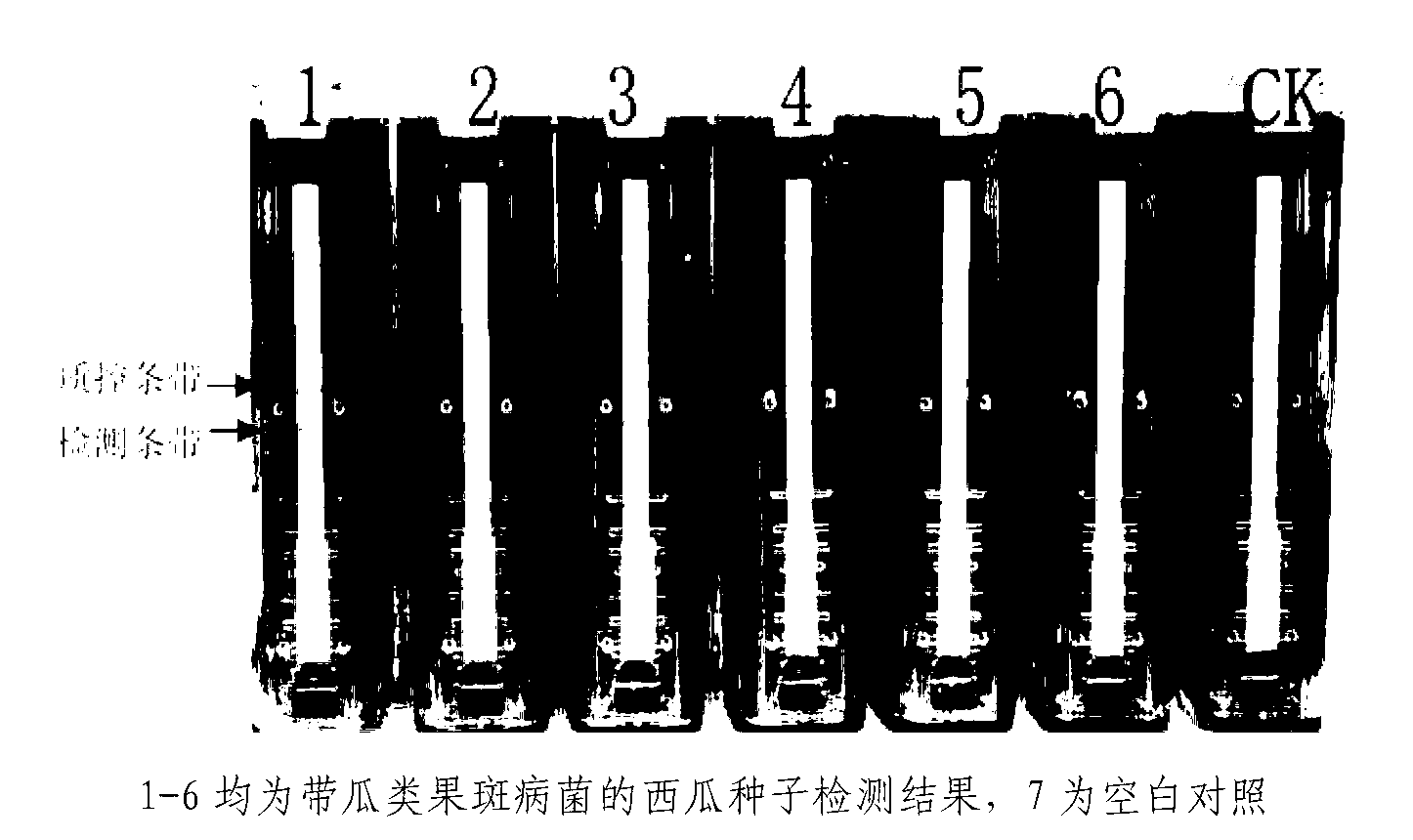
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
431 results about "Dna detection" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
DNA-Based Methods. The most common technique for detecting a specific DNA sequence that comprises a given biotech product is the polymerase chain reaction (PCR). This technique can be qualitative to indicate the presence or absence of a sequence or quantitative to determine the amount of DNA from a biotechnology-derived crop present in a sample.
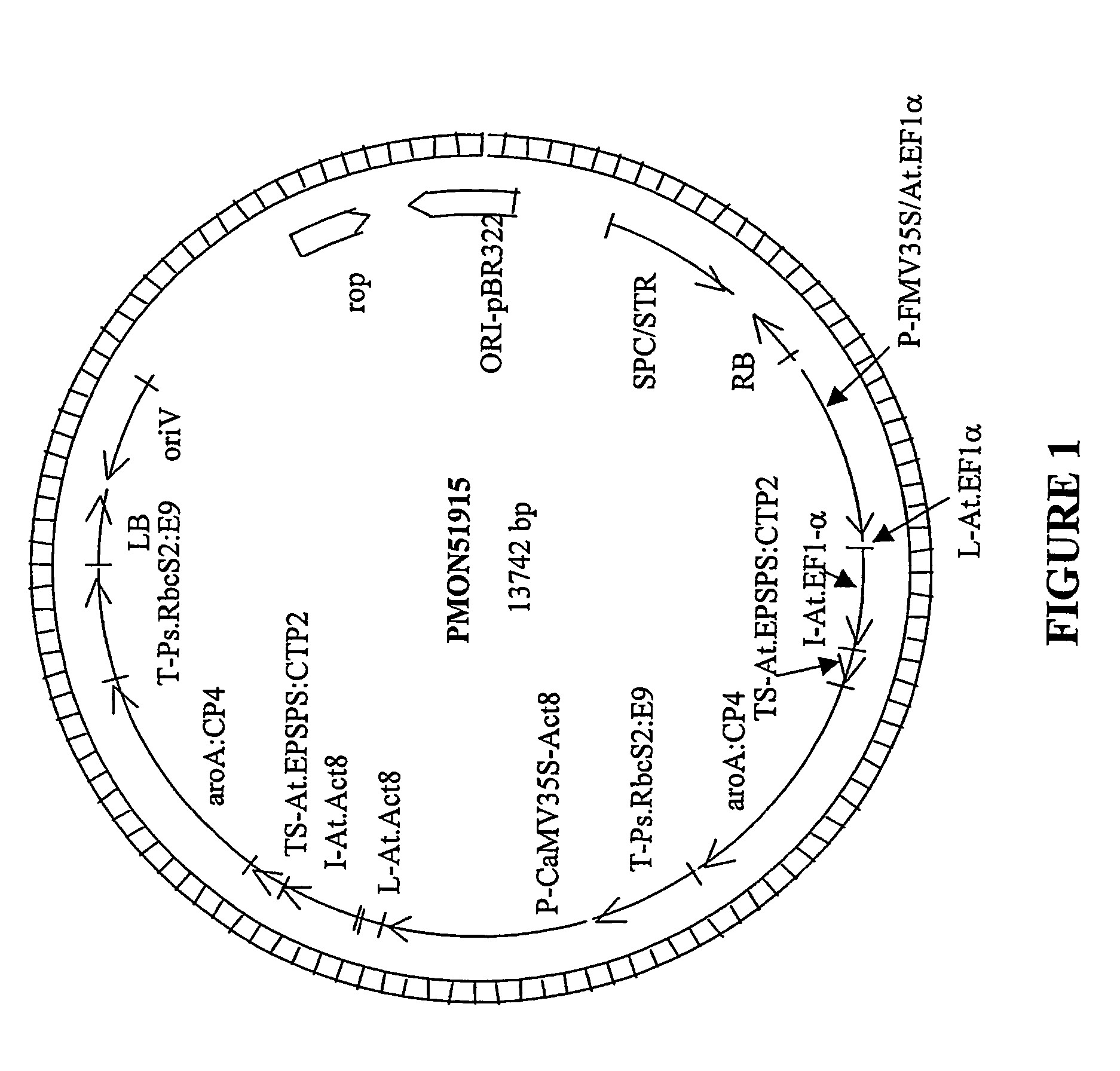
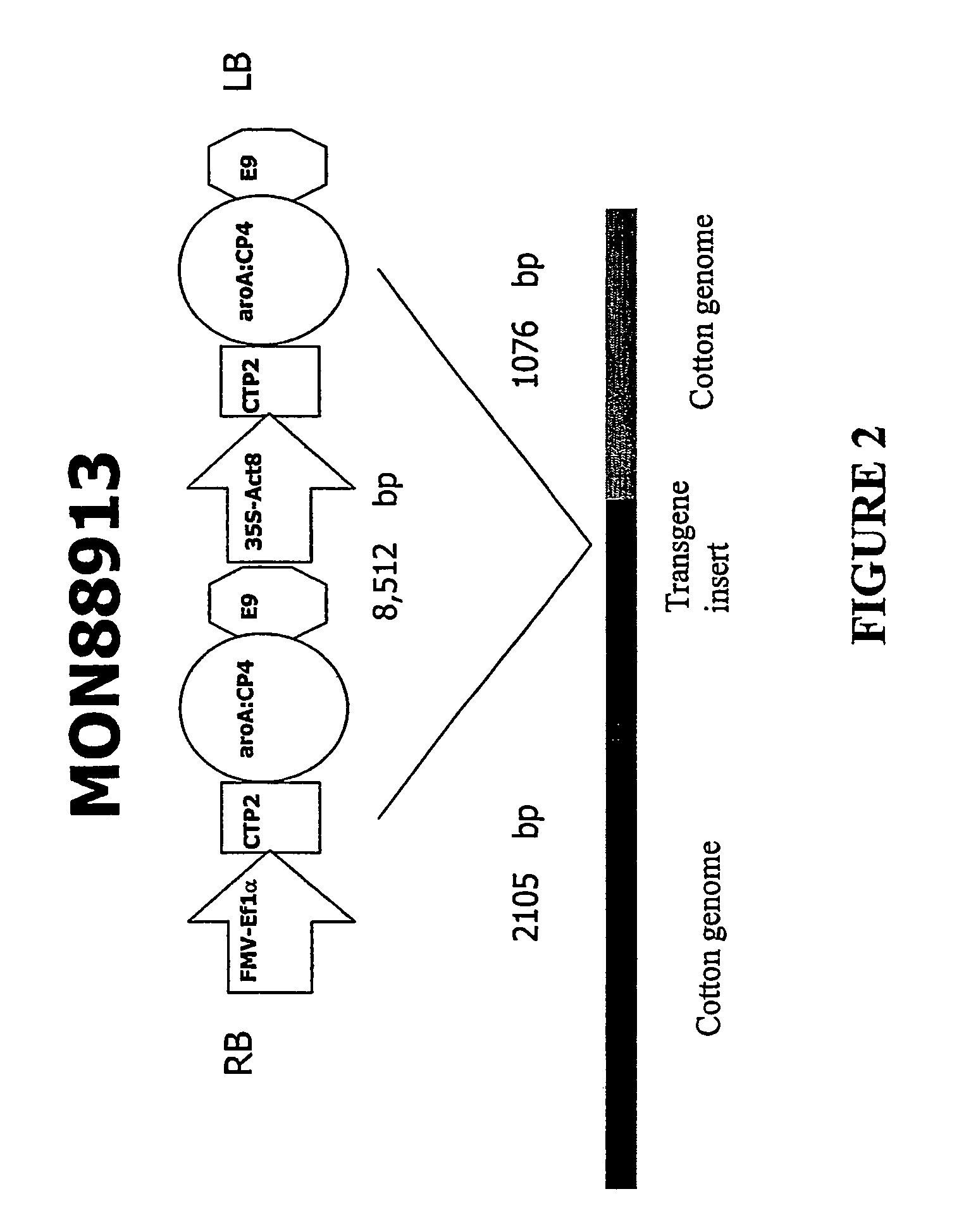
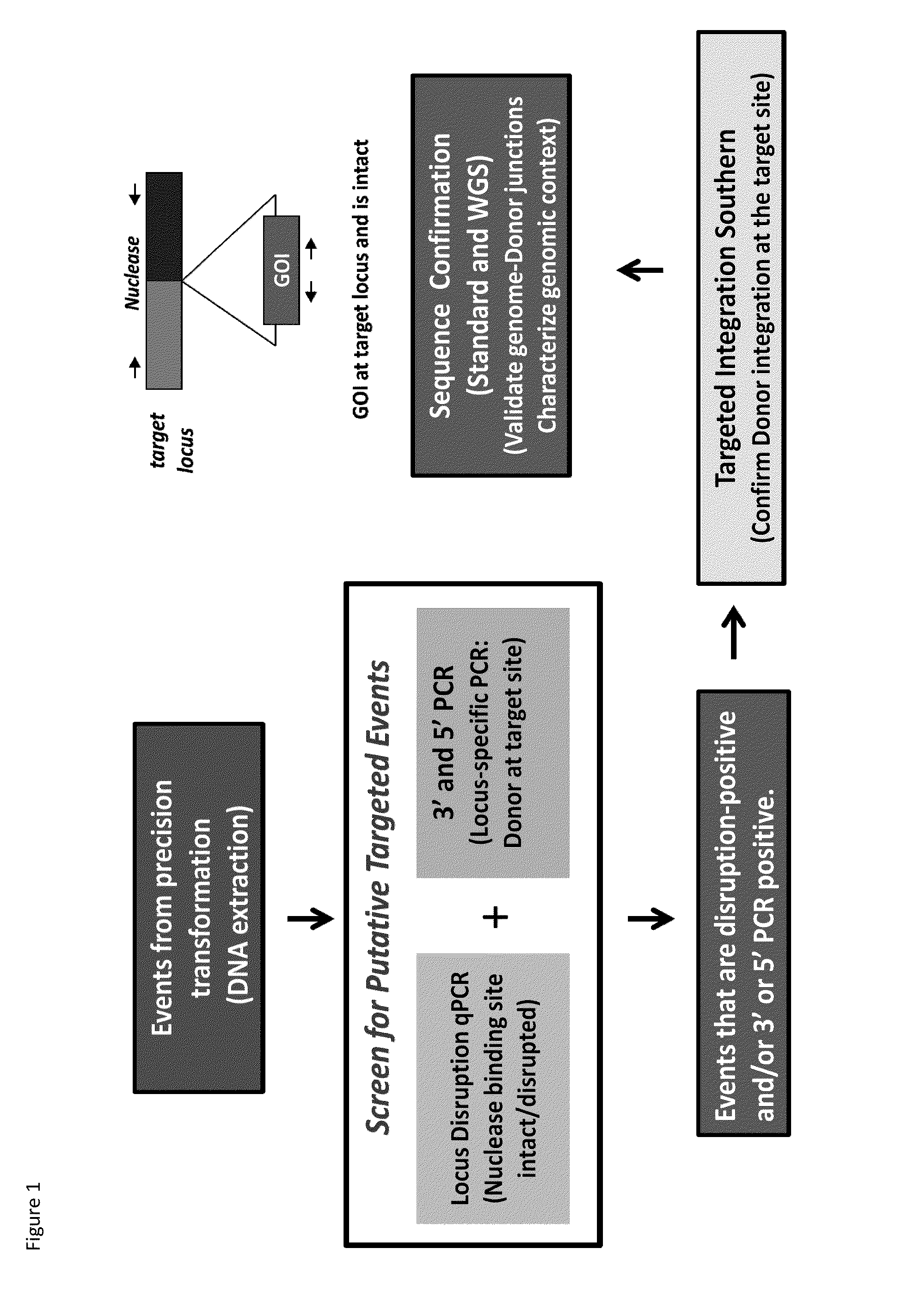
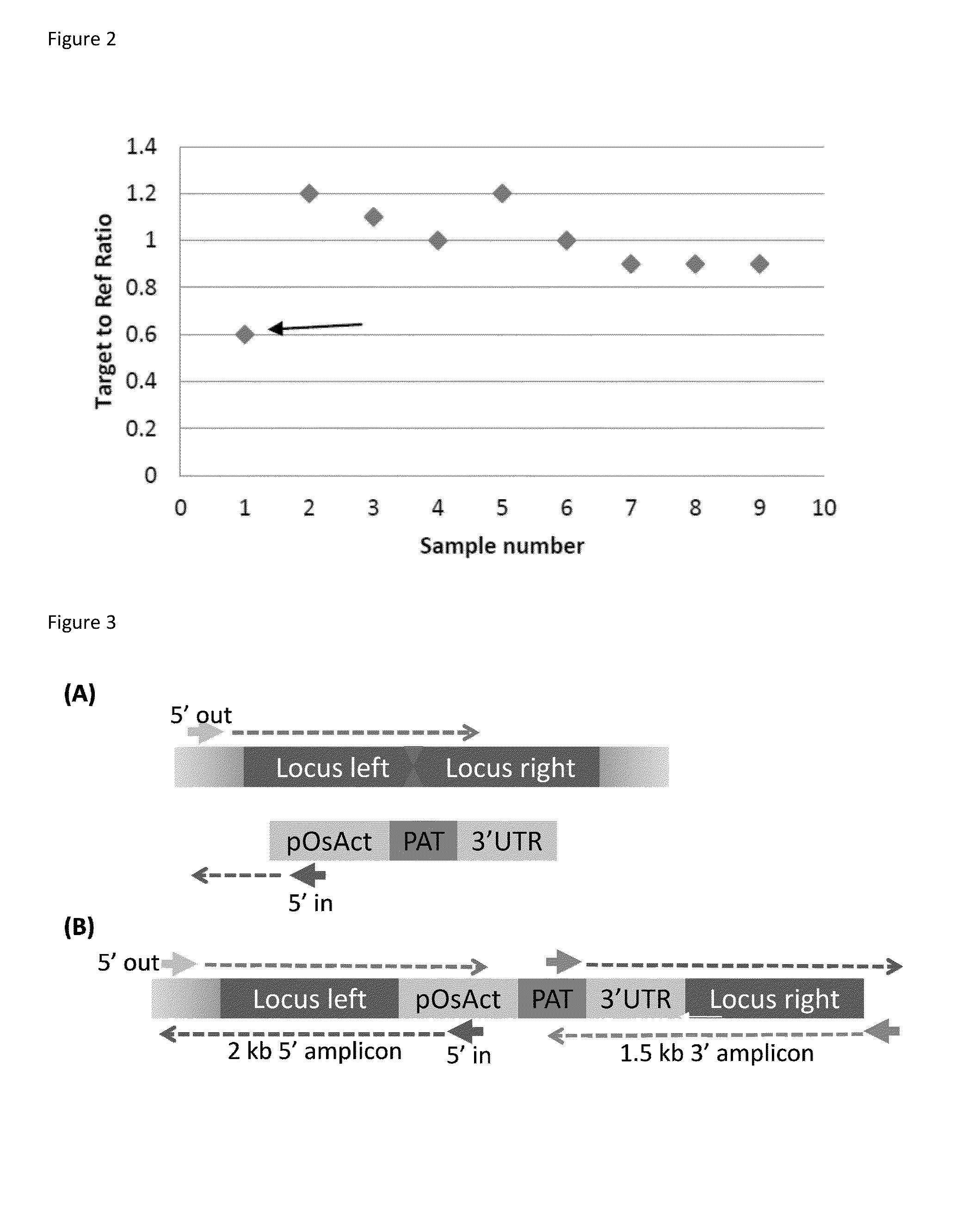
DNA detection methods for site specific nuclease activity
The present disclosure provides methods for detecting and identifying plant events that contain precision targeted genomic loci, and plants and plant cells comprising such targeted genomic loci. The method can be deployed as a high throughput process utilized for screening the intactness or disruption of a targeted genomic loci and optionally for detecting a donor DNA polynucleotide insertion at the targeted genomic loci. The methods are readily applicable for the identification of plant events produced via a targeting method which results from the use of a site specific nuclease.
Owner:CORTEVA AGRISCIENCE LLC
Multilayered microfluidic DNA analysis system and method
A multilayered microfluidic DNA analysis system includes a cell lysis chamber, a DNA separation chamber, a DNA amplification chamber, and a DNA detection system. The multilayered microfluidic DNA analysis system is provided as a substantially monolithic structure formed from a plurality of green-sheet layers sintered together. The substantially monolithic structure has defined therein a means for heating the DNA amplification chamber and a means for cooling the DNA amplification chamber. The means for heating and means for cooling operate to cycle the temperature of the DNA amplification chamber as required for performing a DNA amplification process, such as PCR.
Owner:GOOGLE TECH HLDG LLC
Nanotube sensor devices for DNA detection
InactiveUS20070178477A1Bioreactor/fermenter combinationsBiological substance pretreatmentsDNA SolutionsSingle strand dna
A nanotube device is configured as an electronic sensor for a target DNA sequence. A film of nanotubes is deposited over electrodes on a substrate. A solution of single-strand DNA is prepared so as to be complementary to a target DNA sequence. The DNA solution is deposited over the electrodes, dried, and removed from the substrate except in a region between the electrodes. The resulting structure includes strands of the desired DNA sequence in direct contact with nanotubes between opposing electrodes, to form a sensor that is electrically responsive to the presence of target DNA strands. Alternative assay embodiments are described which employ linker groups to attach ssDNA probes to the nanotube sensor device.
Owner:NANOMIX
Microelectronic sensor device for DNA detection
ActiveCN101466848AMicrobiological testing/measurementMaterial analysis by optical meansFluorescenceBiological target
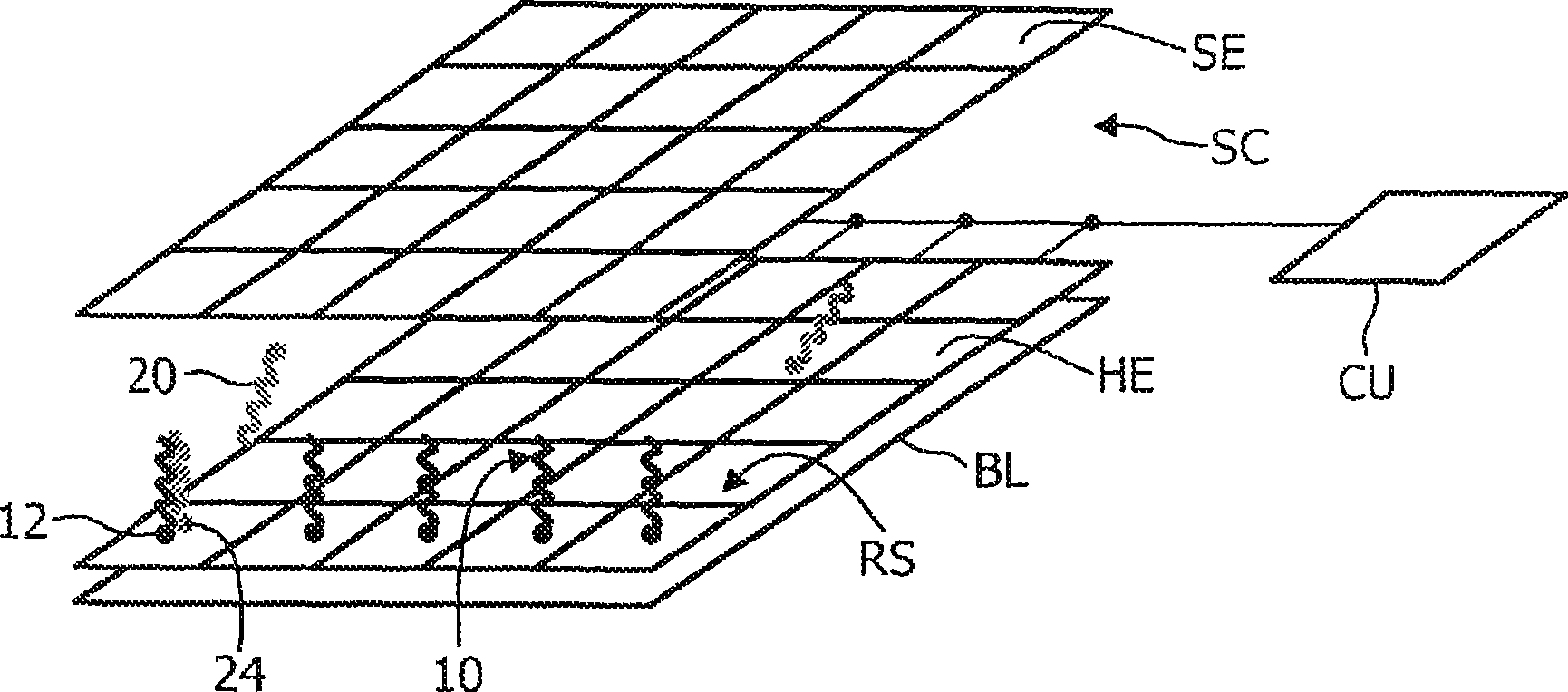
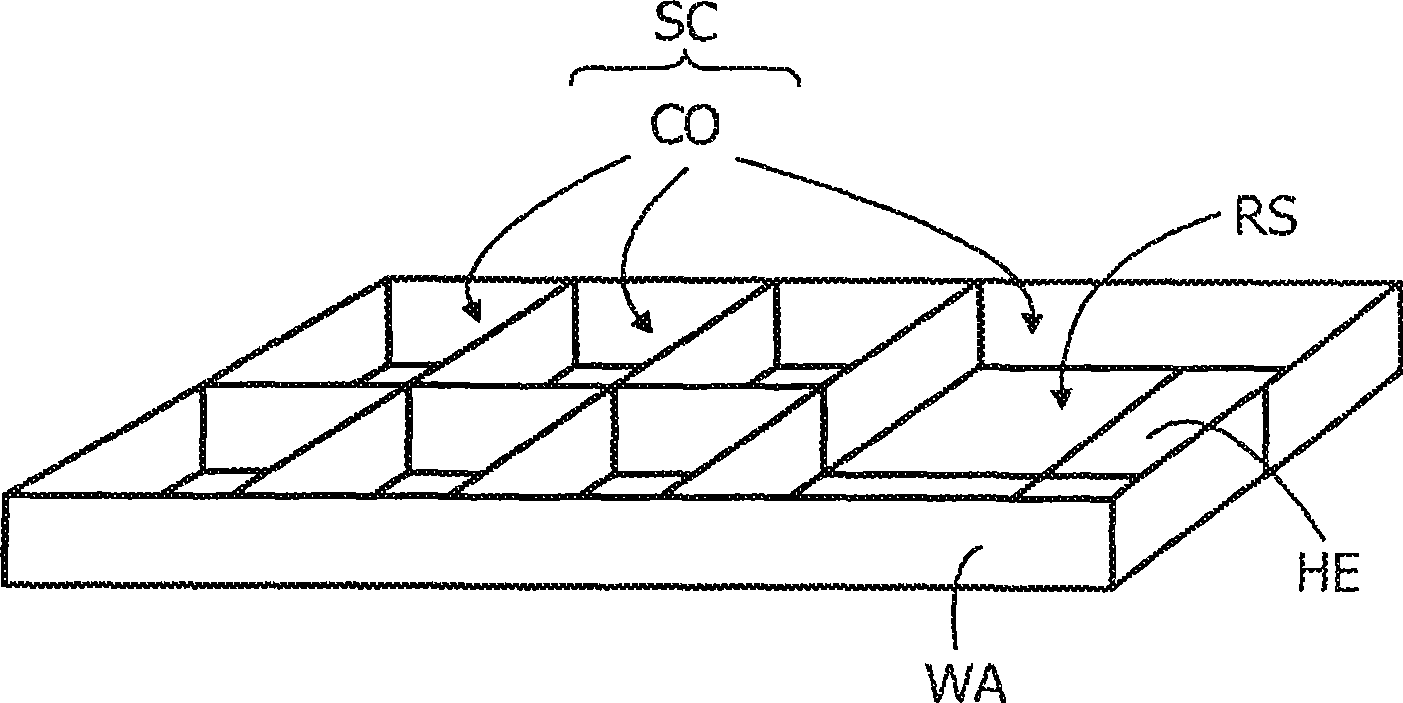
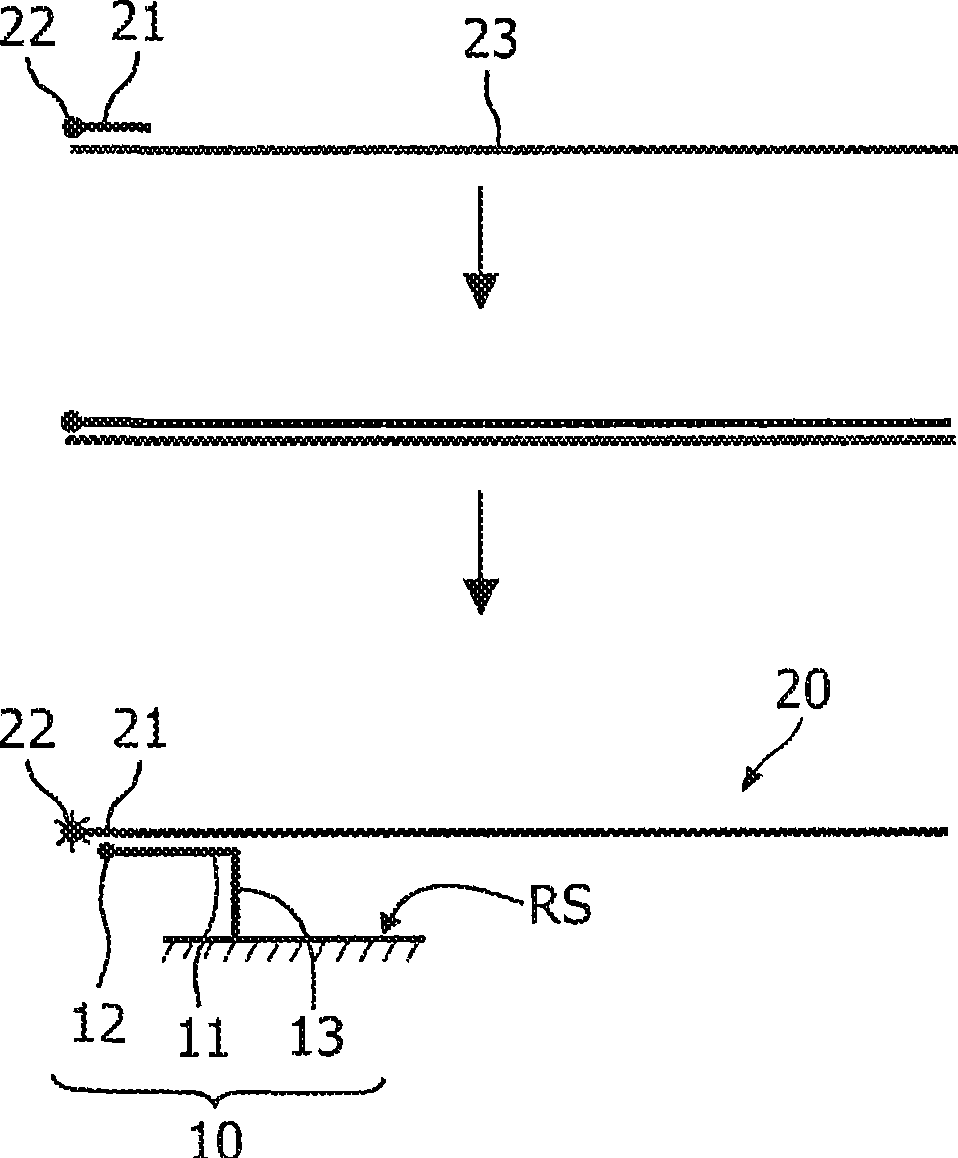
The invention relates to a microelectronic sensor device and a method for the investigation of biological target substances (20), for example oligonucleotides like DNA fragments. In one embodiment, the device comprises a reaction surface (RS) to which target specific reactants (10) are attached and which lies between a sample chamber (SC) and an array of selectively controllable heating elements (HE). The temperature profile in the sample chamber (SC) can be controlled as desired to provide for example conditions for a PCR and / or for a controlled melting of hybridizations. The reactant (10) and / or the target substance (20) comprises a label (12) with an observable property, like fluorescence, that changes if the target substance (20) is bound to the reactant (10), said property being detected by an array of sensor elements, for example photosensors (SE). The fluorescence of the label (12) may preferably be transferred by FRET to a different fluorescent label (22) or quenched if the target substance (20) is bound.
Owner:KONINKLIJKE PHILIPS ELECTRONICS NV
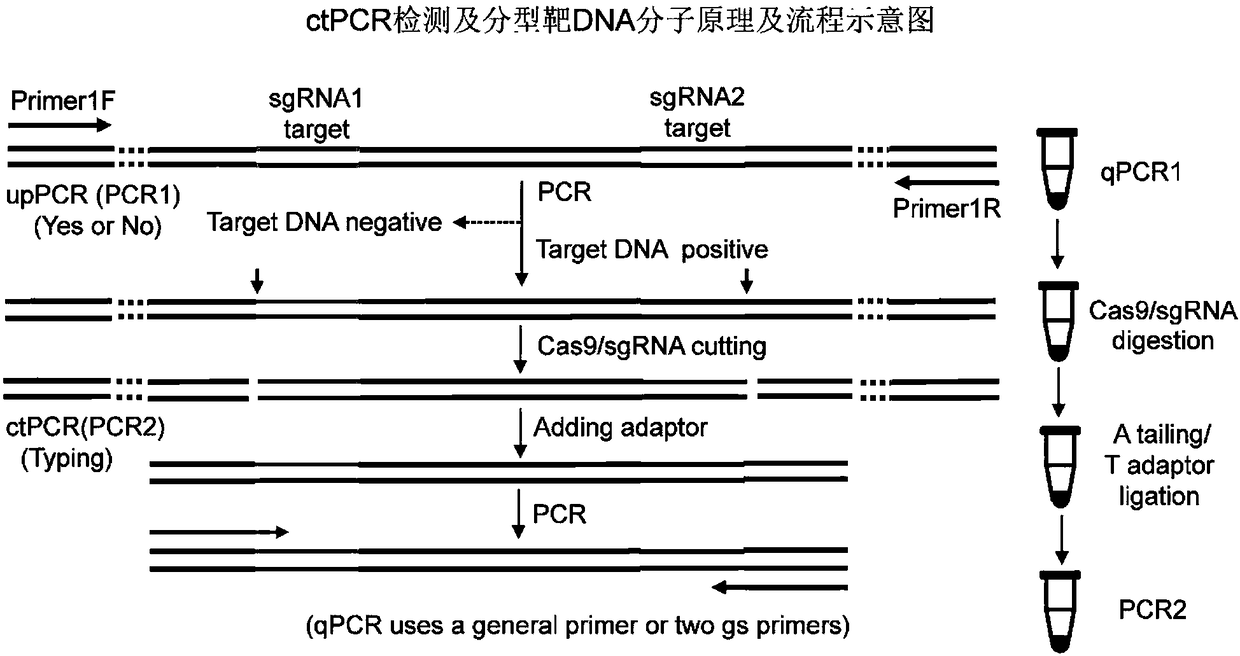
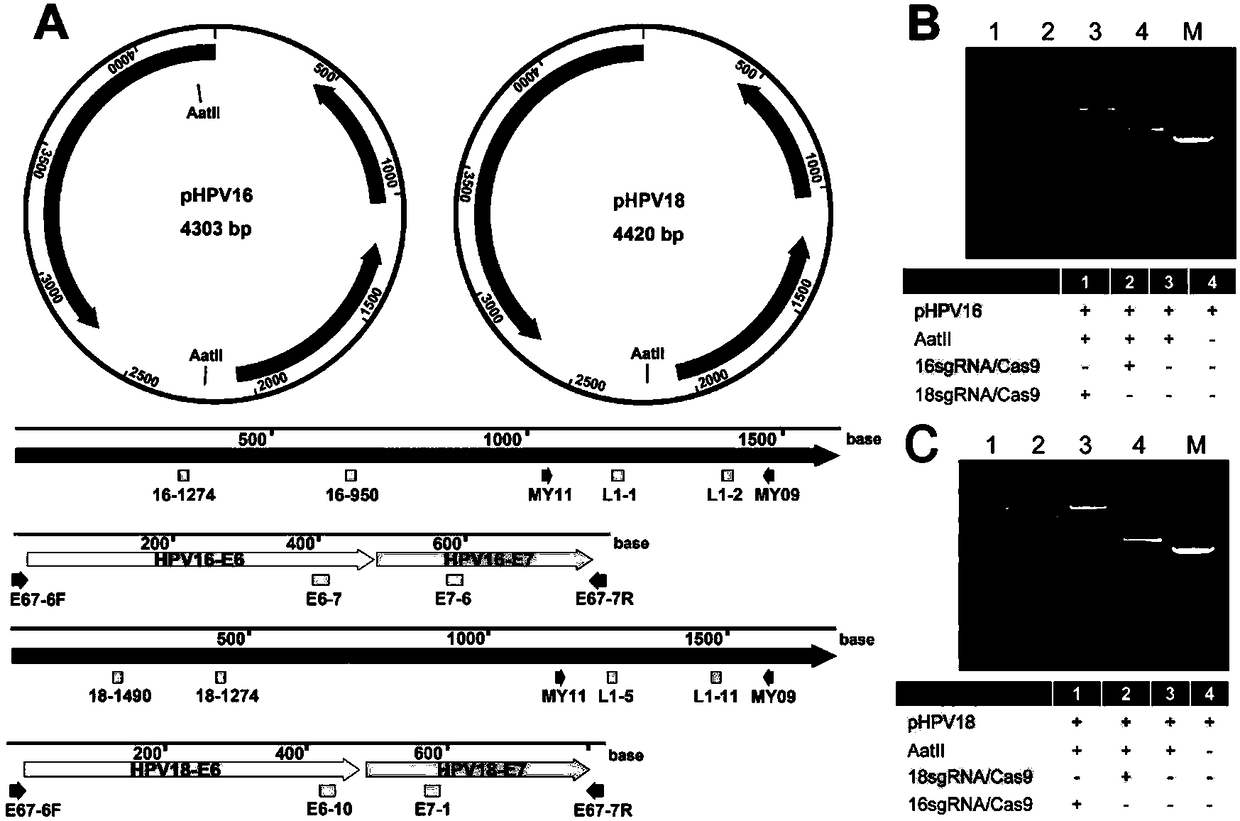
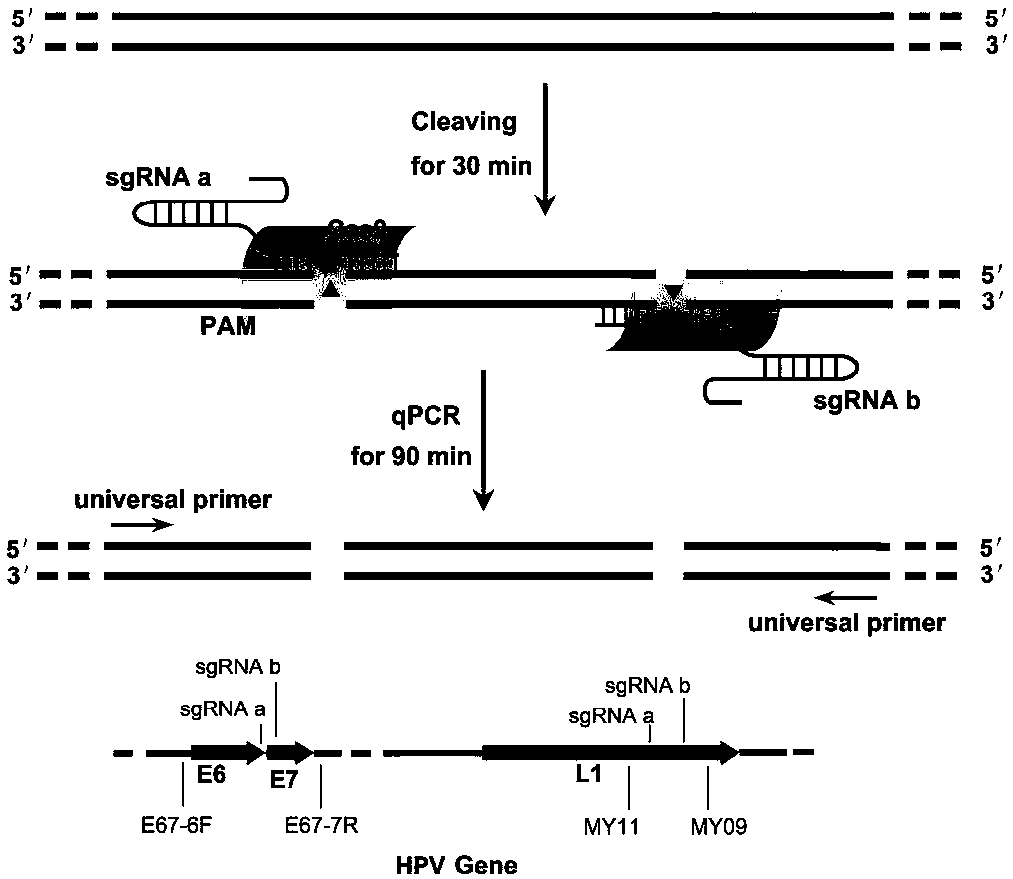
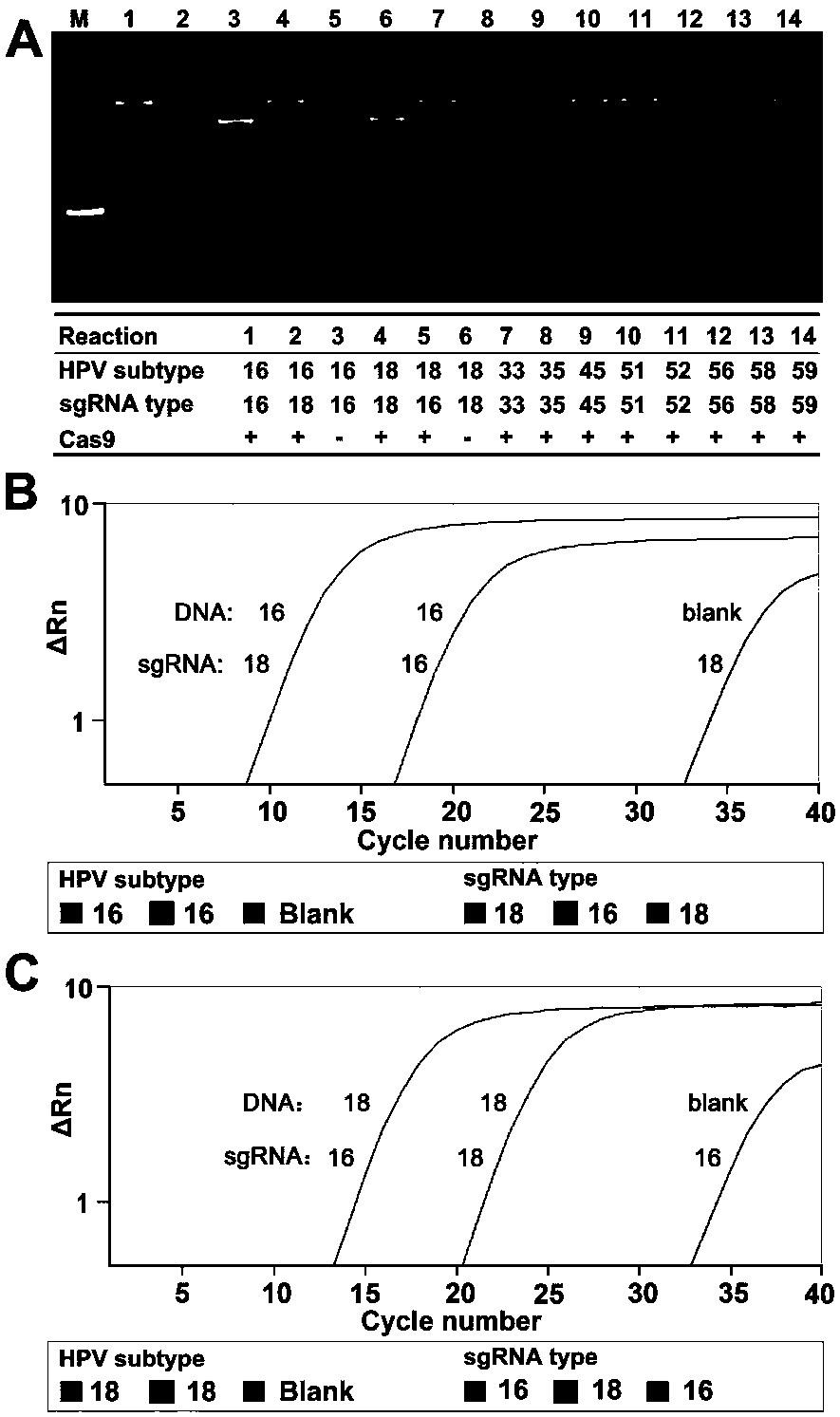
DNA detection and analysis method based on Cas9 nuclease and application thereof
ActiveCN108192956AImprove featuresHigh sensitivityMicrobiological testing/measurementNucleic acid detectionTyping
The invention provides a DNA detection and analysis method based on Cas9 nuclease and application thereof. The method comprises the following three steps: (1) carrying out PCR amplification on targetDNAs; (2) treating a PCR amplification product by using a CAT method; and (3) carrying out PCR amplification with DNAs treated by using the CAT method as a template. The method can successfully detectL1 and E6 / E7 genes of HPV16 and HPV18 in human cervical carcinoma cells. According to the invention, the method successfully avoids current key bottleneck problems in nucleic acid hybridization, designing of specific PCR primers and the like in the fields of nucleic acid detection and typing.
Owner:SOUTHEAST UNIV
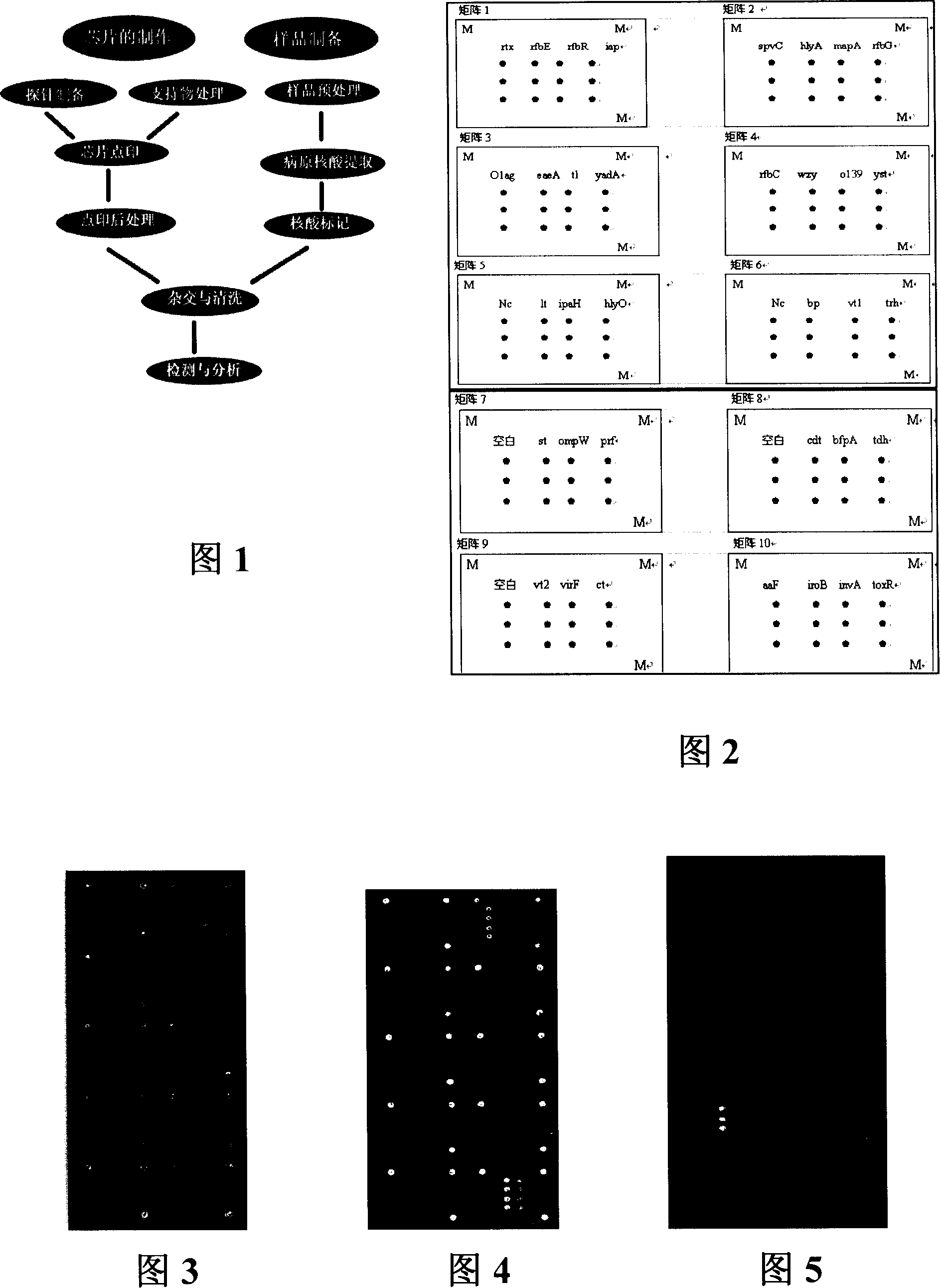
Pathogenic microorganism DNA detecting chip and preparation method and application thereof
InactiveCN101113476ASensitive hybridization detectionAccurate readingMicrobiological testing/measurementAgainst vector-borne diseasesESCHERICHIA COLI ANTIGENNucleic Acid Probes
The invention relates to a pathogenic microorganism DNA detection chip and the chip comprises a carrier and a nucleic acid probe on the carrier. The nucleic acid probe is provided with a target detection probe used for detecting target gene of pathogenic microorganism. The pathogenic microorganism comprises but not limits to vibrio cholerae, pathogenic escherichia coli, campylobacter jejuni, Yersinia enterocolitica, parahemolytic vibrio, salmonella, and shigella and Listeria monocytogenes. The invention also relates to a preparation method of the pathogenic microorganism DNA detection chip and provides applications of the pathogenic microorganism DNA detection chip in detection of pathogenic microorganism. Also a pathogenic microorganism DNA detection chip kit is provided.
Owner:ICDC CHINA CDC
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) based PCR (Polymerase Chain Reaction) typing method and application thereof
ActiveCN108486234ASimplified typesQuick checkMicrobiological testing/measurementSpecific detectionTyping methods
The invention discloses a CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) based PCR (Polymerase Chain Reaction) typing method and the application thereof. The method is a method forperforming DNA detection and typing. The method comprises the following steps: adding a Cas9 protein and sgRNA for targeting target DNA into a conventional PCR reaction system, adding a short-term thermostatic incubation program before the PCR reaction program, and starting a conventional PCR amplification program. The CRISPR based PCR typing method disclosed by the invention adopts a homogeneousdetection technology, and detection can be completed by a PCR amplification step only. By utilizing a specific recognition cutting characteristic of the CRISPR technology on the DNA, the target DNA can be simply, homogeneously, rapidly and sensitively subjected to specific detection and typing, and the method is a novel DNA detection method with high specificity and sensitivity. According to themethod in the invention, human HPV DNA in clinical samples can be successfully detected.
Owner:SOUTHEAST UNIV
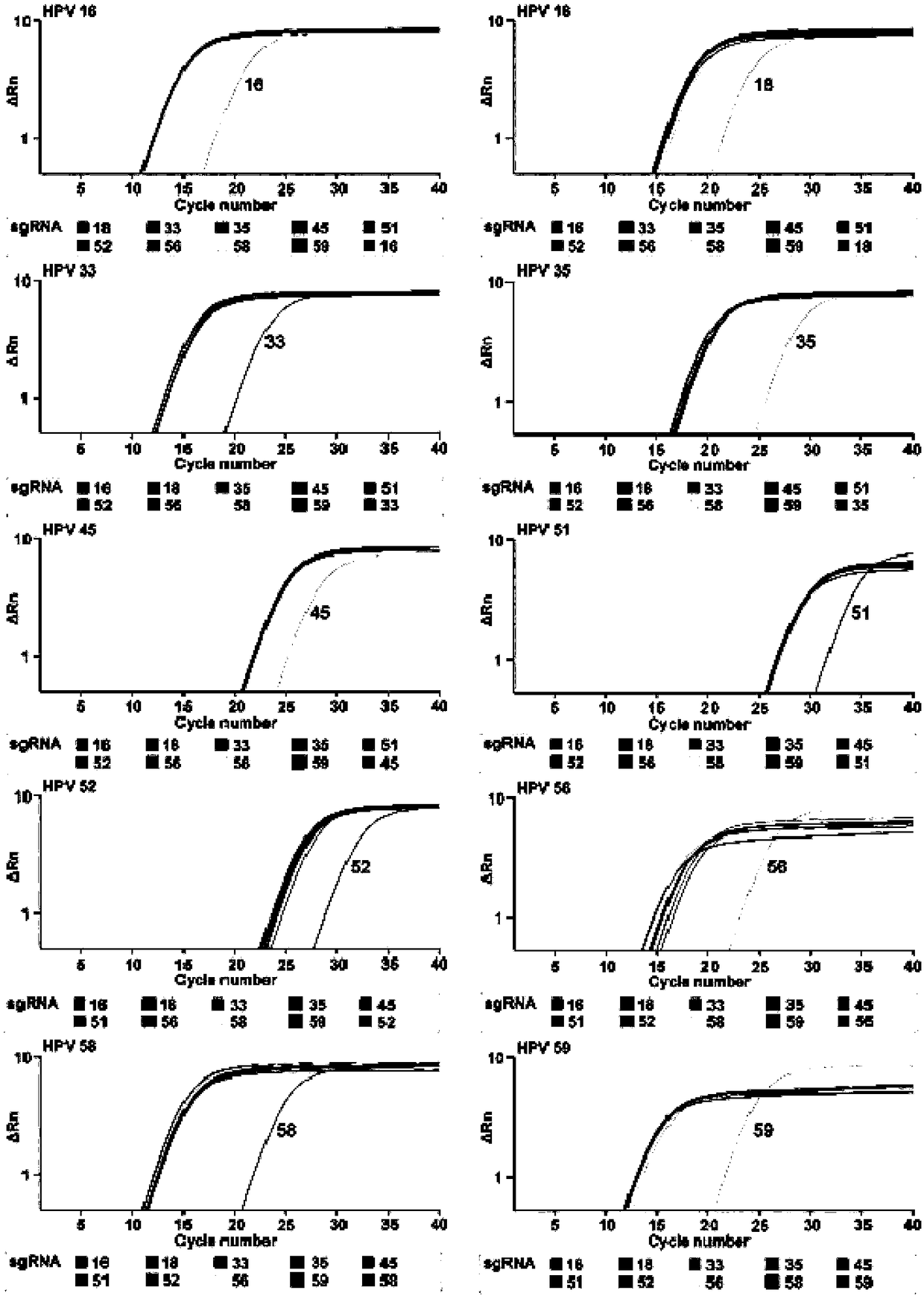
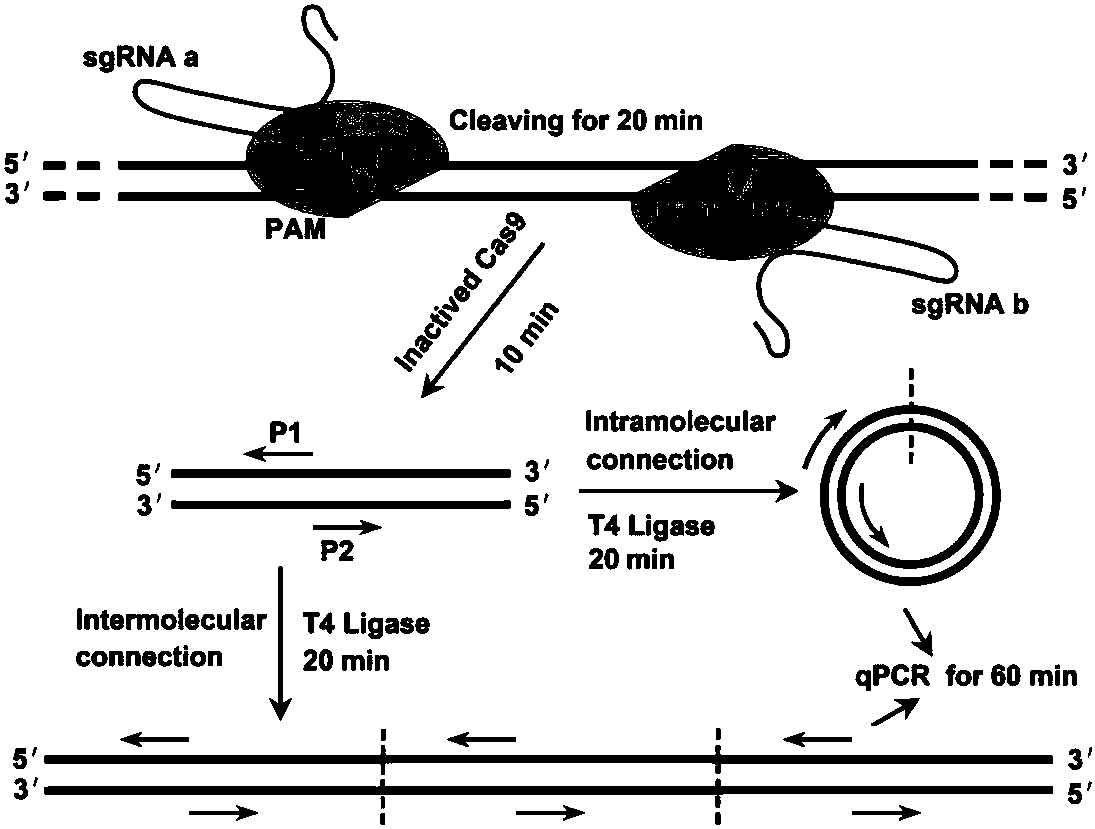
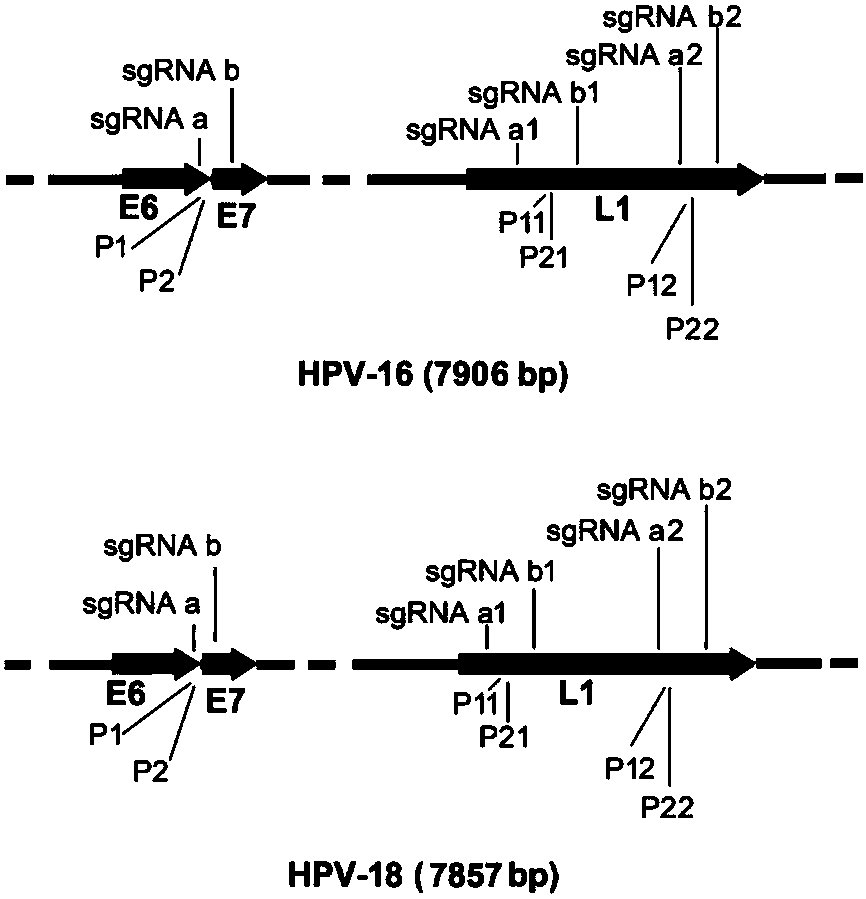
CRISPR (clustered regularly interspaced short palindromic repeat) based DNA detecting and typing method and application thereof
ActiveCN107828874ASimple detectabilitySimplified typesMicrobiological testing/measurementNucleic acid detectionSpecific detection
The invention discloses a CRISPR (clustered regularly interspaced short palindromic repeat) based DNA detecting and typing method and an application thereof. The method comprises following steps: (1),target DNA is subjected to PCR (polymerase chain reaction) amplification by a pair of universal primers; (2), the amplified target DNA is cut by Cas9 / sgRNA; (3), the cut target DNA is linked with DNAligase; (4), the linked target DNA is subjected to PCR amplification. According to the method, the target DNA can be subjected to specific detection and typing simply, rapidly and sensitively according to the specific identification cutting characteristics of the CRISPR technology on DNA, the method is one novel DNA detection method with high specificity and sensitivity, and the critical bottleneck problems about nucleic acid hybridization, specific PCR primer design and the like in current nucleic acid detection and typing fields are solved successfully. With the application of the method, L1 and E6 / E7 genes of HPV16 and HPV18 in human cervical cancer cells are detected successfully.
Owner:SOUTHEAST UNIV
Nano-gold signal probe for DNA detection, production method and method for detecting DNA
The invention discloses a nano-gold signal probe for DNA detection and the preparation method thereof, as well as a DNA detection method; the nano-gold signal probe is that the surface thereof is simultaneously assembled with proteins which can generate chromogenic or fluorescence signals and a sulfhydryl-modified signal probe DNA nano-gold-particle solution. A solid phase carrier is utilized in the DNA detection, and a sandwich complex of the solid phase carrier-target DNA-nano-gold signal probe is formed by DNA hybridization. As the surface of one nano-gold particle can be connected with a plurality of proteins which can generate chromogenic or fluorescence signals, one protein which can generate chromogenic or fluorescence signals, especially an enzyme catalyst, can catalyze a plurality of substrates to have chemical reactions, so as to amplify reaction signals, and further to significantly improve detection sensitivity. Therefore, the nano-gold signal probe, the preparation method and the DNA detection method have important significances in the biological detection, the early diagnosis and the treatment of diseases.
Owner:SHANGHAI INST OF APPLIED PHYSICS - CHINESE ACAD OF SCI
Probe and method for DNA detection
InactiveUS20110212540A1Enhanced fluorescence emissionSugar derivativesMicrobiological testing/measurementHybridization probeFluorescence
A hybridization probe containing two linear strands of DNA lights up upon hybridization to a target DNA using silver nanoclusters that have been templated onto one of the DNA strands. Hybridization induces proximity between the nanoclusters on one strand and an overhang on the other strand, which results in enhanced fluorescence emission from the nanoclusters.
Owner:TRIAD NAT SECURITY LLC
Luminol direct bonded nano gold nucleic acid analyzing probe and application thereof
InactiveCN102021226AOvercome operabilityOvercoming time consumingMicrobiological testing/measurementChemiluminescene/bioluminescenceSingle strand dnaChloroauric acid
The invention discloses a luminol direct bonded nano gold nucleic acid analyzing probe and a novel chemiluminescence analysis method. The invention provides the luminol direct bonded nano gold nucleic acid analyzing probe, a chemiluminescence analysis method based on the luminol direct bonded nano gold nucleic acid analyzing probe and a kit for constructing the analyzing method; the analyzing probe comprises nucleic acid which is directly bonded by luminol and marked by nano gold; and luminol direct bonded nano gold is obtained by reducing chloroauric acid at one step by luminol. The chemiluminescence analysis method based on the nucleic acid analyzing probe of the invention has the advantages of high sensitivity (for example, specific sequence single-chain DNA detection limit can reach 1.9*10<-16>mol / L), wide linear range, high repeatability, simple operation, low cost, and the like, can be used for the detection of DNA, RNA and adapter corresponding ligands in various samples and has wide application prospect in the fields of clinical diagnosis and treatment, pharmaceutical analysis, food safety detection, environmental monitoring, and the like.
Owner:UNIV OF SCI & TECH OF CHINA
Micro fluidic chip apparatus by integrating continuous flow PCR and capillary electrophoresis function
InactiveCN102899238AEasy to integrateReduce miniaturizationBioreactor/fermenter combinationsBiological substance pretreatmentsContinuous flowBiology
The invention discloses a micro fluidic chip apparatus by integrating a continuous flow PCR and a capillary electrophoresis function, which comprises a continuous flow type PCR chip and a capillary electrophoresis chip, and is characterized in that an output channel of the continuous flow type PCR chip is directly connected in a sample pool of the capillary electrophoresis chip, a sample introduction channel and a separating channel are provided on the capillary electrophoresis chip, the sample introduction channel and the separating channel present an intersect shape, the top of the sample introduction channel is the sample pool and a sample waste liquid pool, and the top of the separating channel is a buffer liquid pool and a waste liquid pool. The apparatus provided by the invention can enhance the automation degree and whole process operation speed for DNA detection, the operation step is reduced, and the apparatus is convenient for miniaturization and portable performance of the apparatus.
Owner:张影频
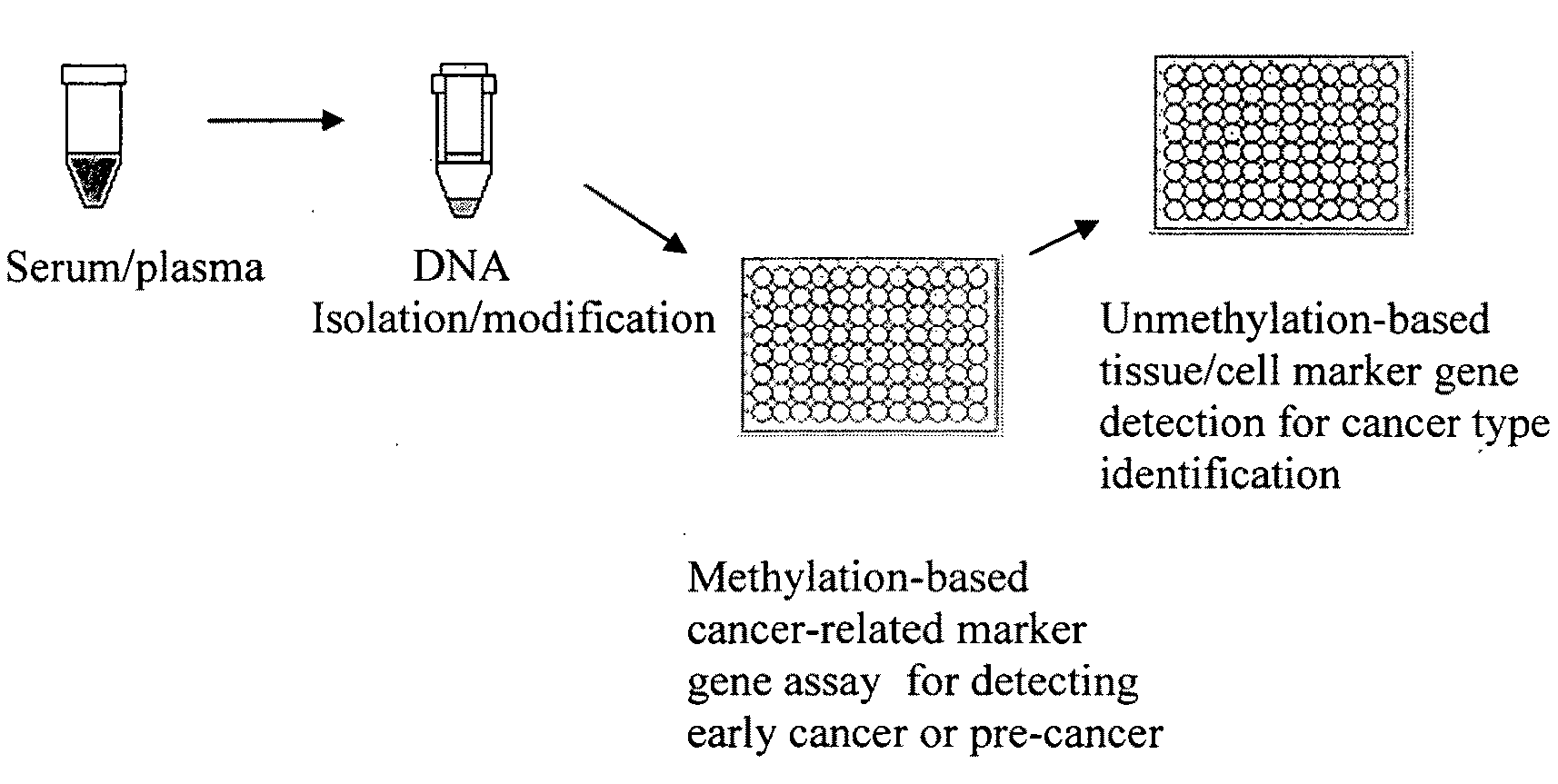
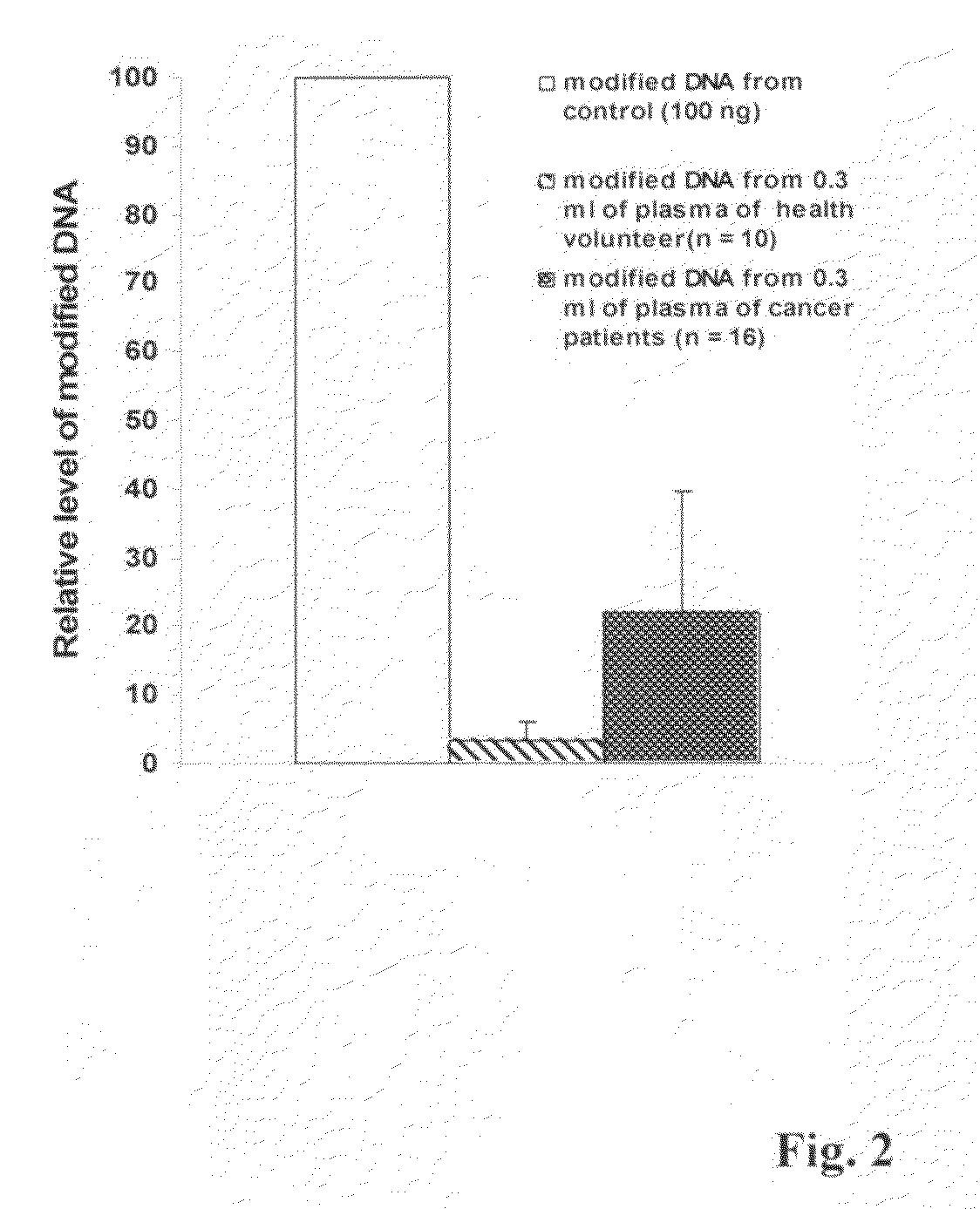
Method and kit for detection of early cancer or pre-cancer using blood and body fluids
Owner:LI WEIWEI +1
High resolution DNA detection methods and devices
InactiveUS20050287589A1Bioreactor/fermenter combinationsBiological substance pretreatmentsOligonucleotideElectrical current
The present invention provides methods and devices for detecting a target nucleic acid molecule. A set of oligonucleotide probes integrated into an electric circuit, where the oligonucleotide probes are positioned such that they can not come into contact with one another, are contacted with a sample. If the sample contains a target nucleic acid molecule, one which has sequences complimentary to both probes, the target nucleic acid molecule can bridge the gap between the probes. The resulting bridge can then carry electrical current between the two probes, indicating the presence of the target nucleic acid molecule.
Owner:CONNOLLY DENNIS MICHAEL
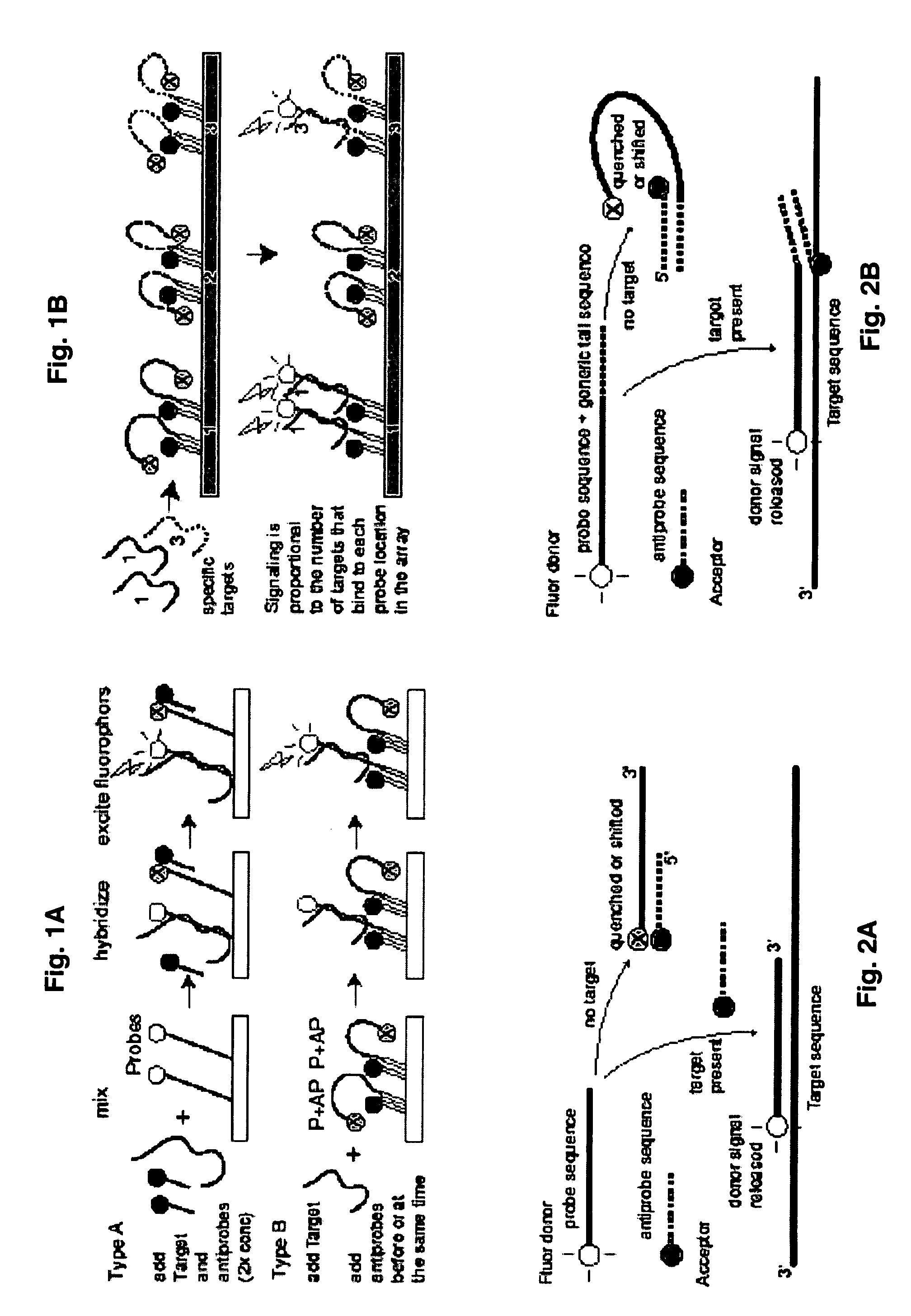
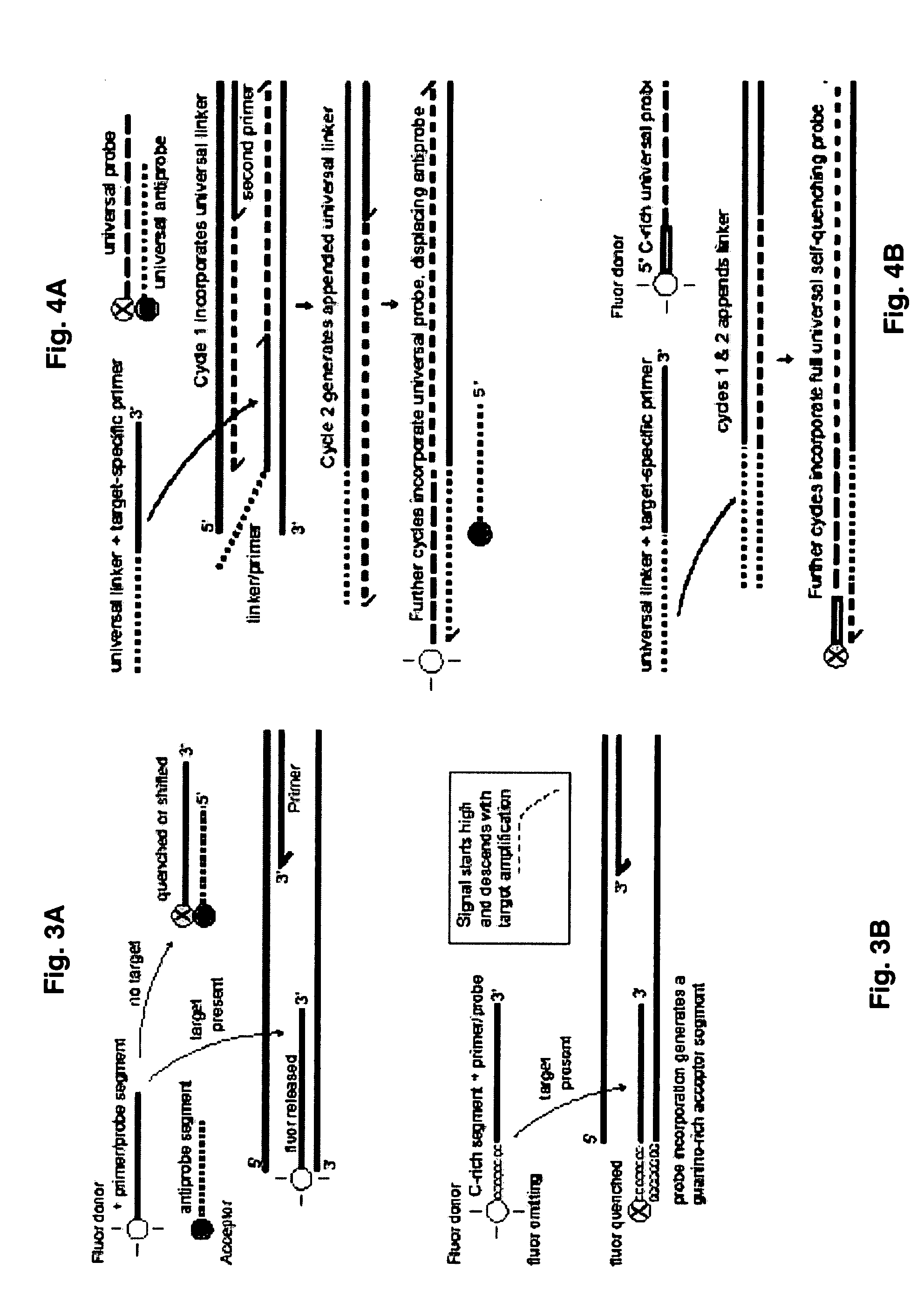
Probe-antiprobe compositions and methods for DNA or RNA detection
ActiveUS20090209434A1Avoiding preventing detectionSimple compositionSugar derivativesMicrobiological testing/measurementSubspeciesDrug resistant mutants
The invention provides novel compositions and methods for detecting unlabeled nucleic acid targets using labeled polynucleotide probes and partially complementary antiprobes. The interaction of probes, antiprobes and targets result in signaling changes that indicate target frequency. This novel detection mechanism is called a DNA detection switch, and it enable end-point detection, microarray detection and real-time PCR detection of a variety of nucleic acid targets including microbial species and subspecies, drug resistant mutants, and pathogenic strains.
Owner:GENETAG TECH
Method for detecting embryonic chromosome abnormality by virtue of blastochyle free DNA
InactiveCN104450923AProbability of small developmental abnormalitiesSimple and fast operationMicrobiological testing/measurementFragment sizeEmbryo
The invention relates to a method for detecting embryonic chromosome abnormality by virtue of blastochyle free DNA. The method comprises the following steps: acquiring blastochyle free DNA, detecting the blastochyle DNA, carrying out whole genome amplification of the free DNA, analyzing a product of the whole genome amplification, implementing fragmenting treatment on genome DNA, carrying out quantitative analysis and fragment size analysis on fragmented target DNA, constructing a library, sequencing by virtue of a computer and analyzing biological information. By virtue of high-throughput sequencing, the method disclosed by the invention can be used for overcoming shortcomings of a conventional DNA analysis method which is merely used for researching partial region of a single cell genome, and is capable of completely analyzing the genetic information of the single cell genome; the method is simple and convenient to operate, time-saving and efficient; meanwhile, by using the blastochyle free DNA as a detection sample, the method is convenient and safe to sample, so that the probability of later embryonic development abnormality is reduced and embryo is protected from being influenced in later development.
Owner:SUZHOU BASECARE MEDICAL DEVICE CO LTD
SNP typing method and kit
ActiveCN103937896AImprove throughputAvoid mutual interferenceMicrobiological testing/measurementDNA preparationGenotypeNucleic acid
The invention relates to the technical field of DNA detection, and provides an SNP typing method and a kit. The method comprises the following steps: A, fixing a nucleic acid segment containing a to-be-tested SNP site on a solid-phase carrier in an addressable manner; B, sequencing the nucleic acid segment containing the to-be-tested SNP site fixed on the solid-phase carrier, so as to determine the genetype of the to-be-tested SNP site on the nucleic acid segment, wherein the nucleic acid segment containing the to-be-tested SNP site is the product obtained by non-single molecule amplification. The kit comprises a specific primer pair and a solid-phase carrier, wherein the specific primer pair is used for obtaining the nucleic acid segment containing the to-be-tested SNP site by non-single molecule amplification; the solid-phase carrier is used for fixing the nucleic acid segment containing the to-be-tested SNP site in the addressable manner. By adopting the method and the kit disclosed by the invention, mutual interference between different probes can be avoided, and the flux for detecting the SNP site is greatly improved.
Owner:SHENGZHEN CHINA GENE TECH COMPANY
ApoE gene primer group, detection kit and detection method
ActiveCN105886608ARaise the ratioImprove detection accuracyMicrobiological testing/measurementDNA/RNA fragmentationApolipoprotein e4Agricultural science
The invention relates to the technical field of DNA (Deoxyribonucleic Acid) detection, and provides an ApoE (Apolipoprotein E) gene primer group, a detection kit and a detection method, wherein the ApoE gene primer group comprises a primer group for detecting an rs429358 site and a primer group for detecting an rs7412 site; an upstream primer in the primer group for detecting the rs429358 site has any one sequence in SEQ ID NO: 1-4; a downstream primer in the primer group for detecting the rs429358 site has any one sequence in SEQ ID NO: 5-8; an upstream primer in the primer group for detecting the rs7412 site has any one sequence in SEQ ID NO: 9-12; and a downstream primer in the primer group for detecting the rs7412 site has any one sequence in SEQ ID NO: 13-16. The primer group, the kit and the method provided by the invention have the advantage that the correct signal proportion in the ApoE gene detection process can be improved.
Owner:WUHAN CONSIDERIN GENE & HEALTH TECH CO LTD
Tortoise shell DNA detection kit and identification method
InactiveCN103255220AMultiplex PCR identification method is simpleReliable test resultsMicrobiological testing/measurementMultiplex pcrsBiology
The invention relates to the technical field of the identification of a Chinese medicine, and specifically relates to a tortoise shell DNA detection kit and identification method. The tortoise shell DNA detection kit comprises the following four parts: sample pretreatment fluid and decalcifying solution, a mitochondria DNA extraction system, a PCR reaction system and a result observing system. The tortoise shell DNA identification method comprises the following five steps: detection sample pretreatment, mitochondria DNA extraction, PCR primer design and synthesis, PCR reaction establishment and result determination. The determination standards are that if 100bp and 400bp strips simultaneously appear, the tortoise shell is an authentic tortoise shell, and if only one or no one appears, the tortoise shell is a fake tortoise shell. The multiple identification method provided by the invention has simple, quick, and reliable in the detection results and can accurately and simultaneously identify the specificities of the authentic tortoise shell and the fake tortoise shell.
Owner:JILIN LEIBO TECH
HPA allelic gene typing detection reagent kit
InactiveCN101845520AImprove objectivityImprove accuracyMicrobiological testing/measurementFluorescence/phosphorescenceHigh fluxGene type
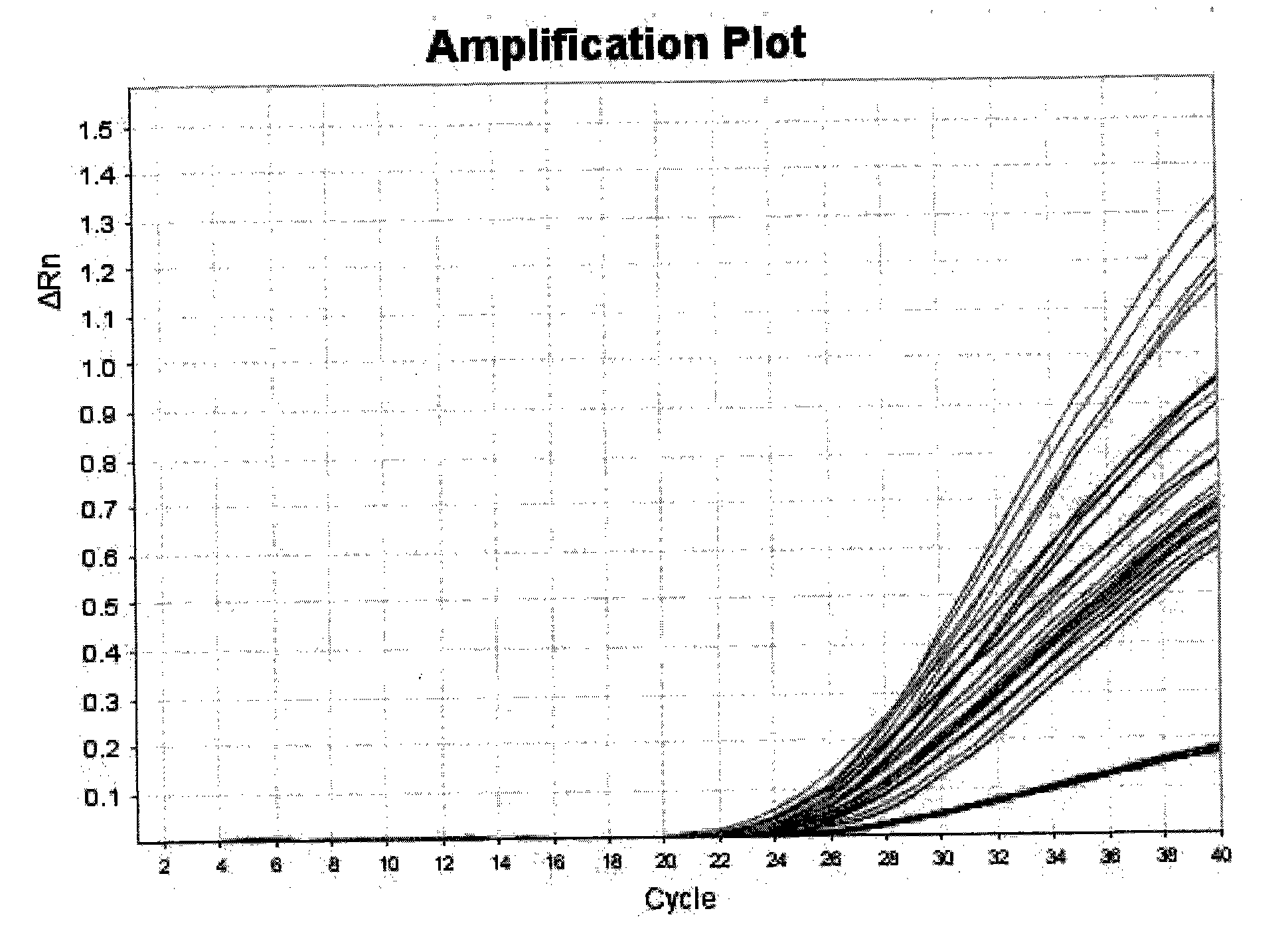
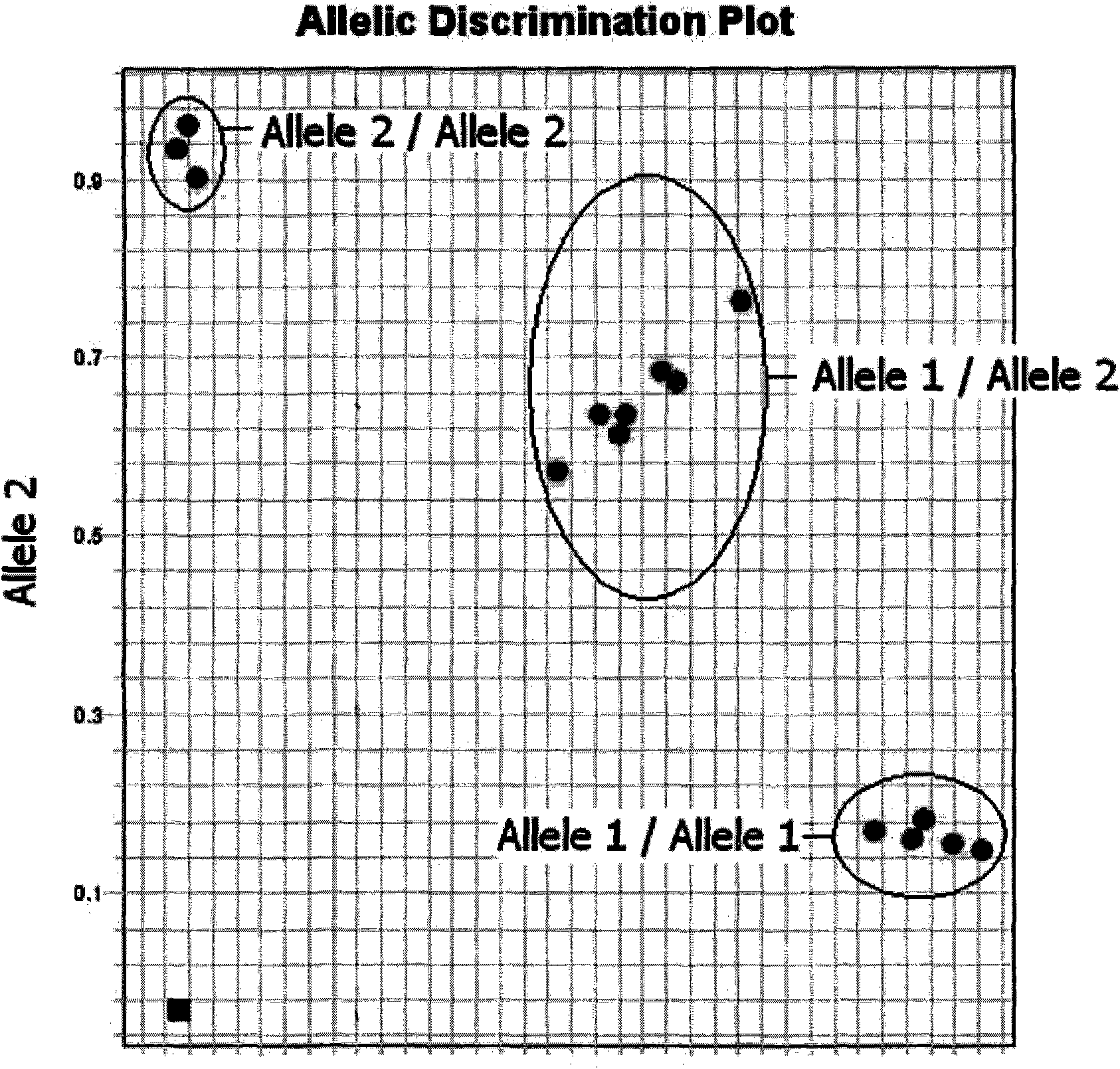
The invention belongs to the technical field of biology, in particular to an HPA allelic gene typing detection reagent kit. The reagent kit of the invention comprises a primer and an MGB-probe. The reagent kit can judge the gene type of the allelic gene according to the observation of the difference of two kinds of fluorescence curves and the distribution of scattergrams after the amplification, so the HPA allelic gene typing detection can be completed. The reagent kit of the invention can meet the experiment requirement of high flux, and the whole typing experiment can be completed in two hours under the condition of the existence of proper DNA detection materials. The problem of fast systemic typing on HPA-1 to 5 and 15 on people groups in China can be solved, at the same time, the condition of homozygotes and heterozygotes can be identified, the typing results can be directly obtained, and the invention has the characteristics of flexibility, stability, accuracy and high efficiency.
Owner:上海血液生物医药有限责任公司 +1
Method for determining contents of biological marker and DNA through direct-reading portable glucometer
A method for determining the contents of a biological marker and DNA through a direct-reading portable glucometer is characterized in that a glucose molecule embedded carrier liposome, hollow metal ball, porous carbon ball, porous silicon ball or polymer is introduced into the surface of a sensor as a composite marker through a sandwich immunization reaction or a DNA reaction, so the detection of the biological marker or DNA is converted into the detection of a final product glucose, the contents of the biological marker and DNA for detection are in direct proportion to the content of the embedded glucose according to the sandwich immunization analysis principle, and the contents of the biological marker and DNA can be obtained by detecting the concentration of glucose through the glucometer. The glucometer is developed into a portable and universal biological marker and DNA tester, and the glucometer detection technology based biological marker and DNA analysis new method is constructed, and can be used for the real-time, online, simple and sensitive determination of various biological markers and DNA. An apparatus used in the method is simple, so the analysis cost is low.
Owner:HUAZHONG NORMAL UNIV
Method for preparing biosensor based on silicon nanowires and application of biosensor to detecting DNA
InactiveCN102072931AImprove conductivityGood biocompatibilityNanosensorsMaterial electrochemical variablesBiocompatibility TestingIntegrated circuit
The invention discloses a method for preparing a biosensor based on silicon nanowires and application of the biosensor to detecting DNA. The method is characterized by preparing the silicon nanowires by a wet chemical method, modifying gold nanoparticles on the silicon nanowires via a silane coupling agent and grafting a probe DNA on the silicon nanowires through binding of chemical bonds betweenthe gold nanoparticles and the DNA to prepare a sensor probe. The sensor is applied to detecting the unknown DNA sequences in the target solution to be detected. The detecting results are mainly obtained by analyzing the data measured by cyclic voltammetry. The invention has the following advantages: (1) mass production can be carried out through simple microprocessing technology, the cost is lowand the microprocessing technic is compatible with the large scale integration technology; (2) the biosensor mainly utilizes the specificity and biocompatibility among the silicon nanowires, the goldnanoparticles and the DNA, is easy to realize and has wide applicability; and (3) the biosensor is simple and convenient to manufacture, has good repeatability and high sensibility, is easy to realize microminiaturization and can realize real-time monitoring.
Owner:EAST CHINA NORMAL UNIV
Turtle shell DNA detection kit and identification method
InactiveCN103074433AMultiplex PCR identification method is simpleRapid multiplex PCR identification methodMicrobiological testing/measurementBioinformaticsDna identification
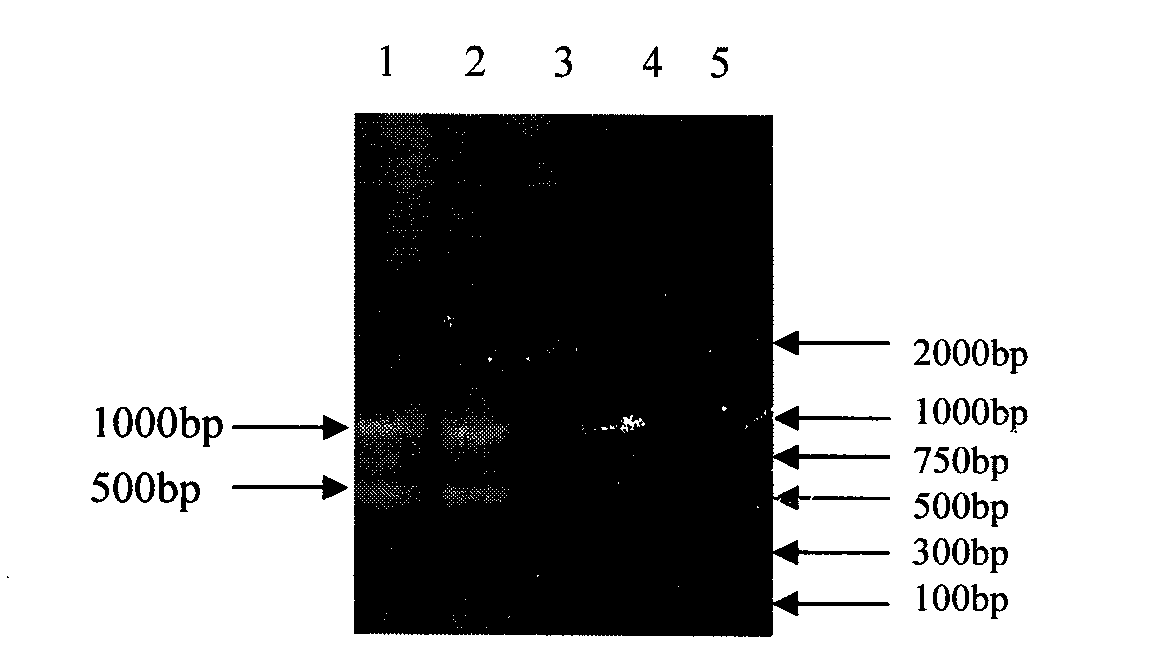
The invention relates to the technical field of traditional Chinese medicine identification, in particular to a turtle shell DNA (deoxyribonucleic acid) detection kit and an identification method. The turtle shell DNA detection kit comprises a sample pretreatment liquid, a decalcification liquid, a mitochondrial DNA extraction system, a PCR (polymerase chain reaction) system and a result observation system. The turtle shell DNA identification method comprises the steps of pretreating a detection sample, extracting mitochondrial DNA, conducting PCR primer design and synthesis, establishing PCR, and judging a result. A judgment standard is that a turtle shell is authentic if 1000bp and 500bp bands appear simultaneously, and the turtle shell is fake if only one or no band appears. The established multi-PCR identification method has the advantages of simplicity, convenience, quickness, reliable detection result and the like, and can differentiate and identify specificities of the turtle shell and the fake turtle shell accurately and simultaneously.
Owner:JILIN LEIBO TECH
Urine preservation reagent and application thereof
InactiveCN107881213AEffective protectionReduce the impactMicrobiological testing/measurementArginineDiazolidinyl urea
The invention discloses a urine preservation reagent and an application thereof. The urine preservation reagent comprises a nucleic acid buffer, a metal ion chelating agent, an antiseptic, a cell fixing agent, and a formaldehyde quenching agent; a nucleic acid buffer is Tris-HCl, a working pH value is 7-8, the metal ion chelating agent is EDTA, the antiseptic is citric acid and / or sodium citrate,the cell fixing agent is selected from at least one of paraformaldehyde, diazolidinyl urea, imidazolidinyl urea, and diethyl urea; and the formaldehyde quenching agent is selected from at least one ofglycine, lysine, ethene diamine and arginine. The urine preservation reagent can effectively preserve free DNA in urine, degradation can be avoided, and the urine preservation reagent establishes a base for subsequently detecting the free DNA; fragmentation of cells in the urine is prevented, a genome DNA background is reduced, and free DNA detection result accuracy is guaranteed. The urine preservation reagent is the novel reagent for preserving a urine sample, and provides the guarantee for clinical detection of the urine free DNA.
Owner:GENETALKS BIO TECH CHANGSHA CO LTD
Nanoscale DNA detection system using species-specific and/or disease- specific probes for rapid identification
InactiveUS20100101956A1Increase probabilityAccurate detectionElectrostatic separatorsSludge treatmentNanowireCarbon nanotube
A method and system for detecting a DNA strand using carbon nanotubes or nanowires. A specific single strand of template DNA serves as a probe for its complementary strand in a solution containing DNA segments to be tested. The single-stranded sequence-specific DNA probe segment, whose ends are modified with amine, is attached between two carbon nanotubes / nanowires. When complementary strands representing DNA segments under test are brought near the probe strands, a dielectrophoresis (DEP) field may enhance the probability of selective hybridization between the complimentary target DNA and probe DNA. A change in electrical conductance in the probe strand occurs upon hybridization of the complementary target DNA with the single probe strand. This conductance change may be measured using the two carbon nanotubes or nano-dimensional electrodes. By exploiting nano-dimensional electrodes and single strand probe DNA, the proposed system is capable of accurately detecting a single molecule of DNA.
Owner:FLORIDA INTERNATIONAL UNIVERSITY
Primers and method for cross primer isothermal amplification detection of acidovorax citrulli
The invention provides primers and a method for cross primer isothermal amplification detection of acidovorax citrulli. Nucleotide sequences of the primers areACLF3:5'-GGCTAACTACGTGCCAGC-3'ACLB3:5'-ACGCATTTCACTGCTACA-3'ACLBIP:5'-CAGATGTGAAATCCCCGGGCTCTGCCGTACTCCAGCGAT-3'ACDF5b1:5'-GCAAGCGTTAATCGGAATTACT-3'ACDF5f2:5'-CAACCTGGGAACTGCATTTGT-3', wherein the end 5 of the primer ACDF5b1 is labeled with biotin, and the end 5 of the primer ACDF5f2 is labeled with FITC (Fluorescein Isothiocyanate). The invention further provides a cross primer isothermal amplification technology for detecting the acidovorax citrulli, wherein with total DNA of a sample or a bacterial liquid as a template, isothermal amplification is carried out by using the primers; and after the reaction is finished, a result is judged directly by using a sealed colloidal gold-labeled DNA detecting device. The primers and the method provided by the invention have the advantages of good specificity, high accuracy, high sensitivity as well as simplicity, convenience and rapidness in operation and provides assurance for import / export safety.
Owner:CHINESE ACAD OF INSPECTION & QUARANTINE +1
Process for improving DNA detecting sensitivity
InactiveCN1464070AIncrease the degree of fixationImprove adsorption capacityMicrobiological testing/measurementGold particlesSingle strand
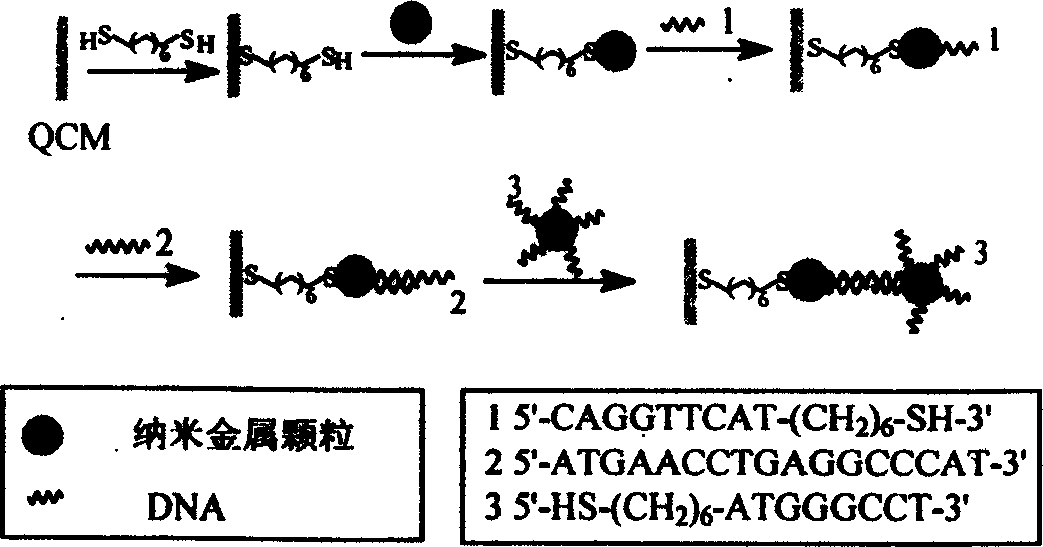
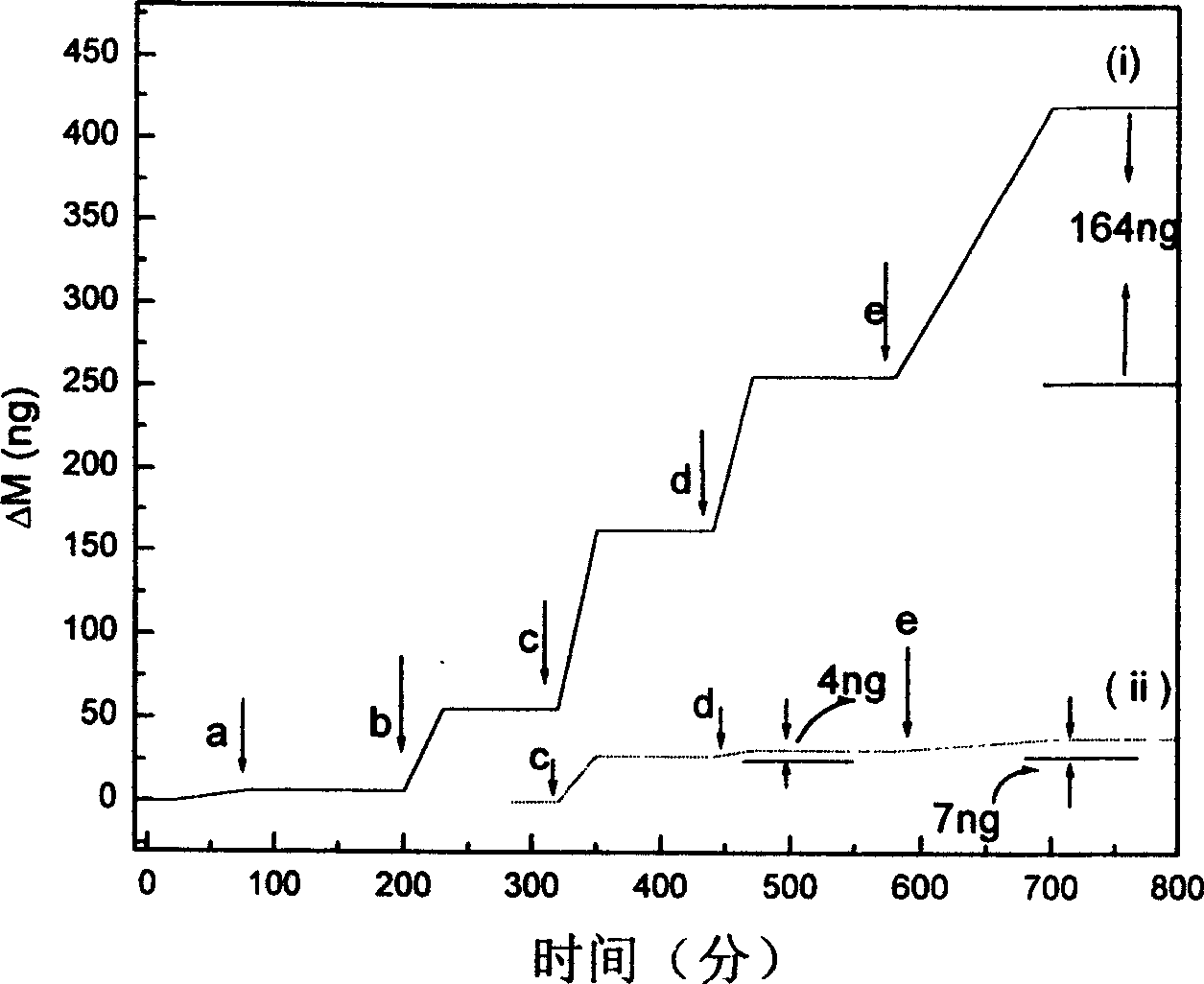
The present invention relates to one kind of method of detecting DNA crossbreeding and mispairing in raised sensitivity based on DNA base pairing principle. Nano gold particles in different sizes are used separately as the surfactant of quartz crystal microbalance and distinguishing amplifier as DNA detector. By means of the decoration of the metal surface of the quartz crystal microbalance with double-functional radical connecting agent, nano metal particles are fixed to the quartz crystal microbalance. Single-strand DNA probe is then fixed onto the nano metal decorated surface, and the DNA probe and the target DNA single strand to be detected are made to crossbred. The relatively longer target DNA has one part complementary with the probe DNA via being fixed onto the quartz crystal microbalance and the other part paired with the complementary DNA single strand connected to nano metal particles to raise the detection sensitivity to 10E(-16).
Owner:INST OF CHEM CHINESE ACAD OF SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com