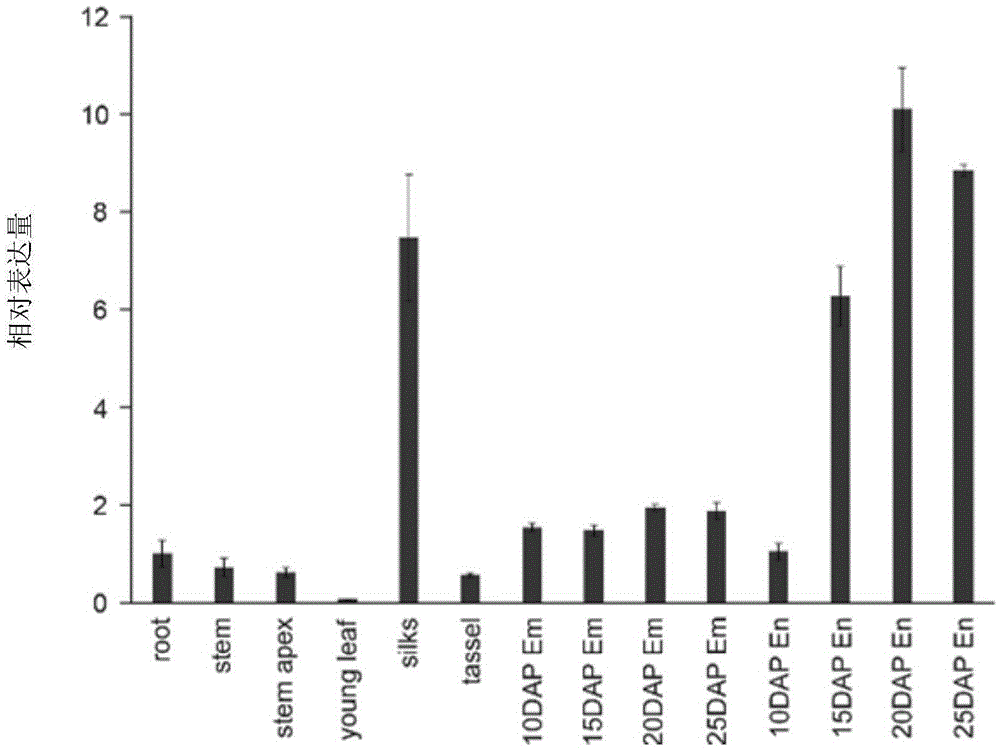
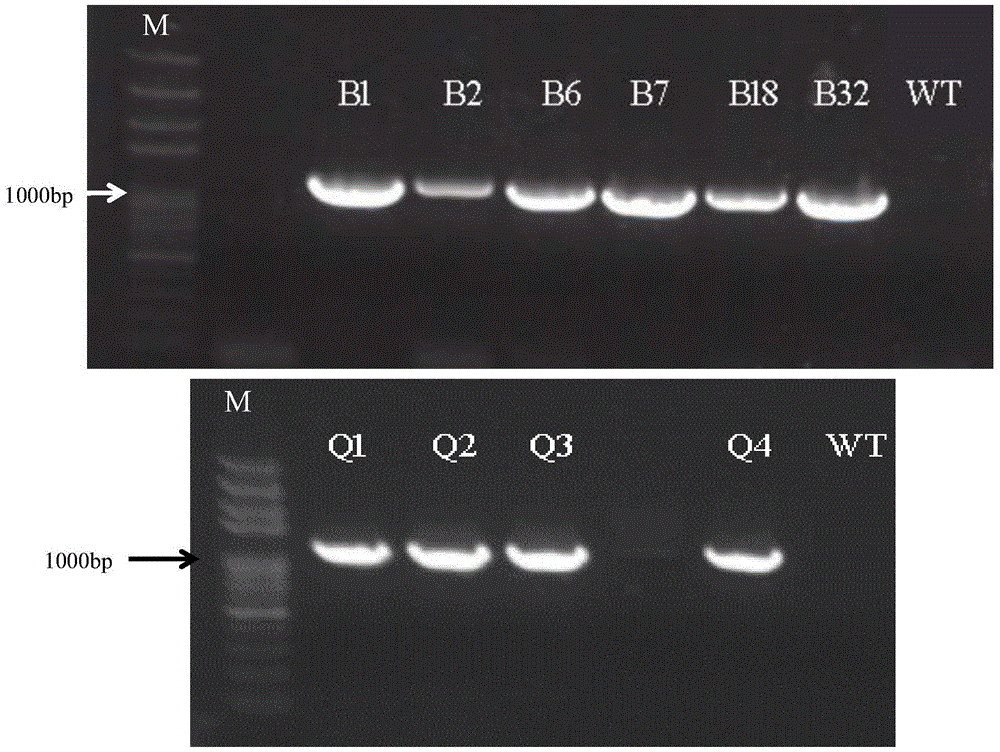
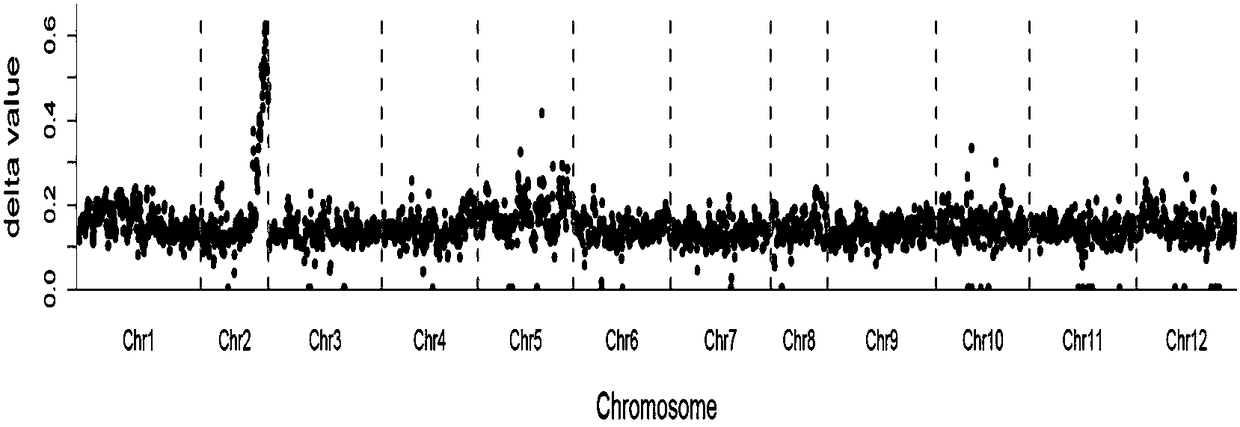
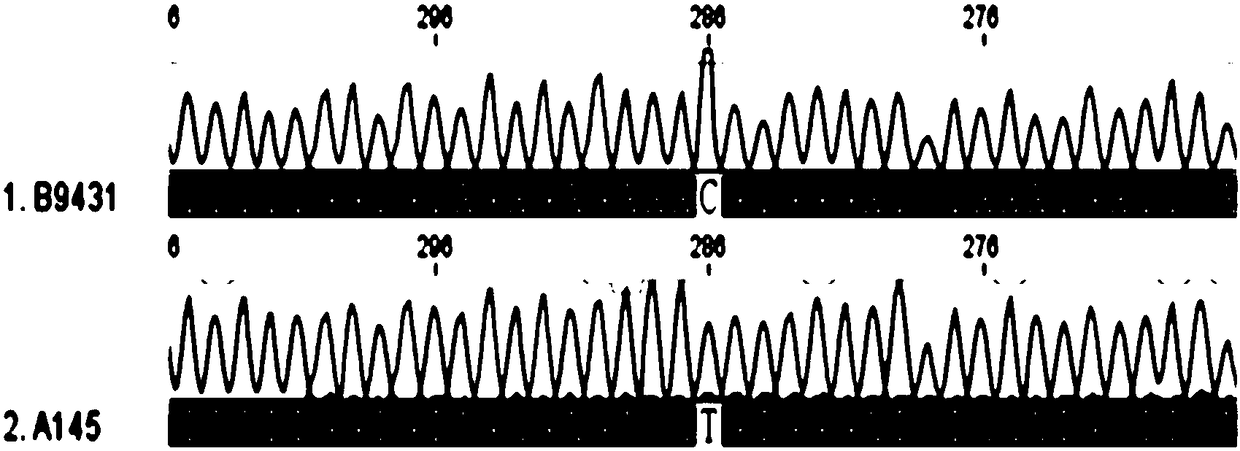
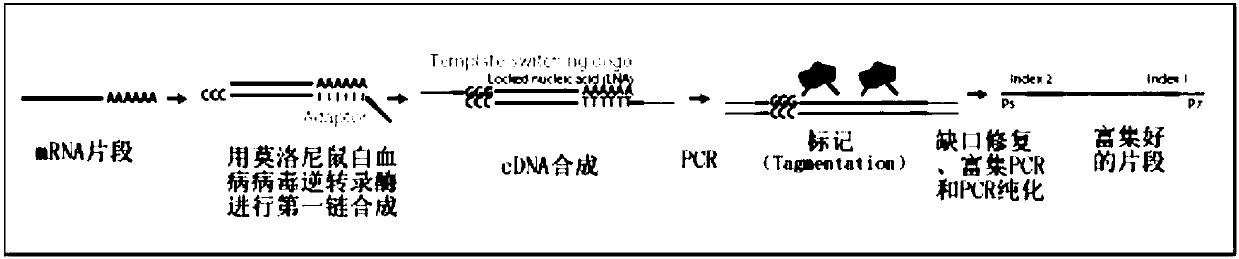
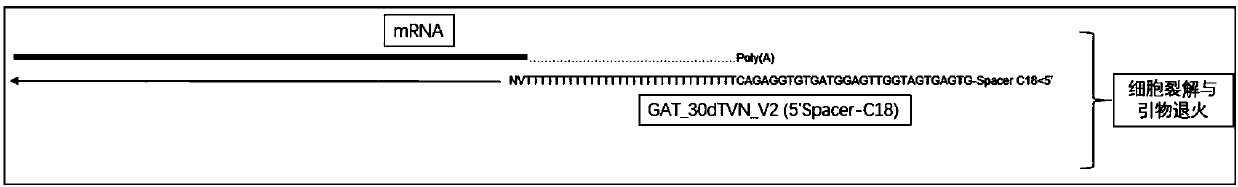
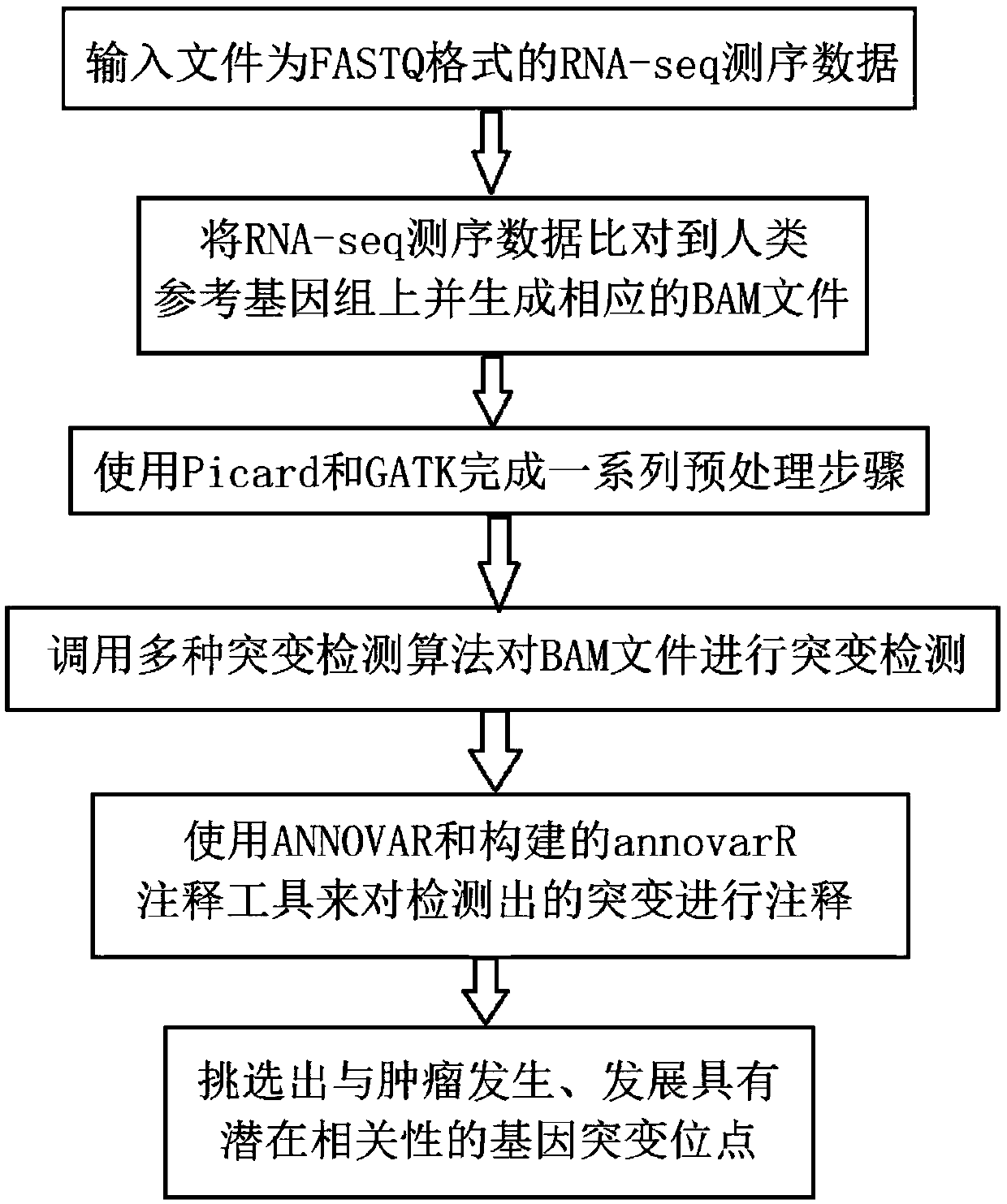
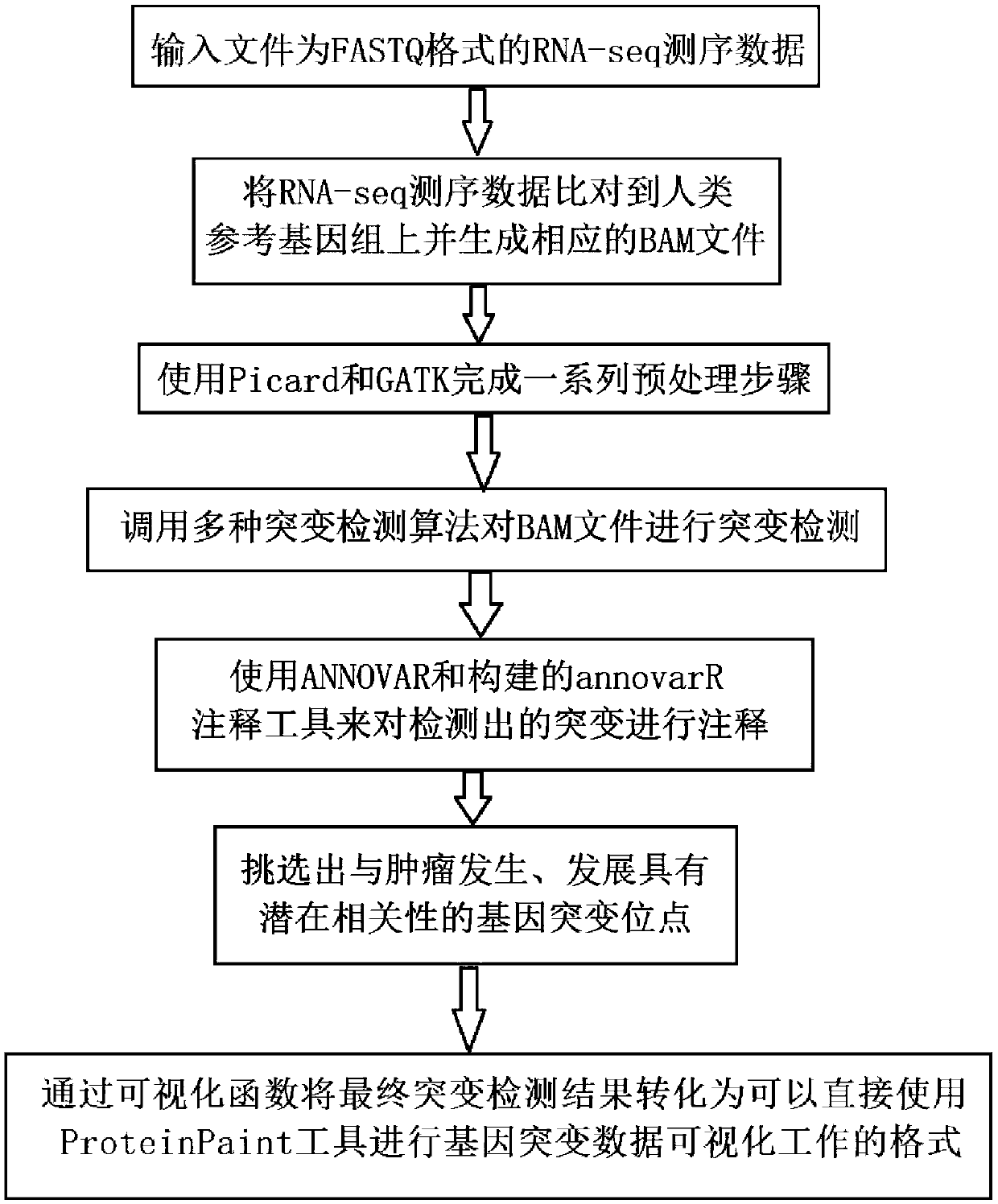
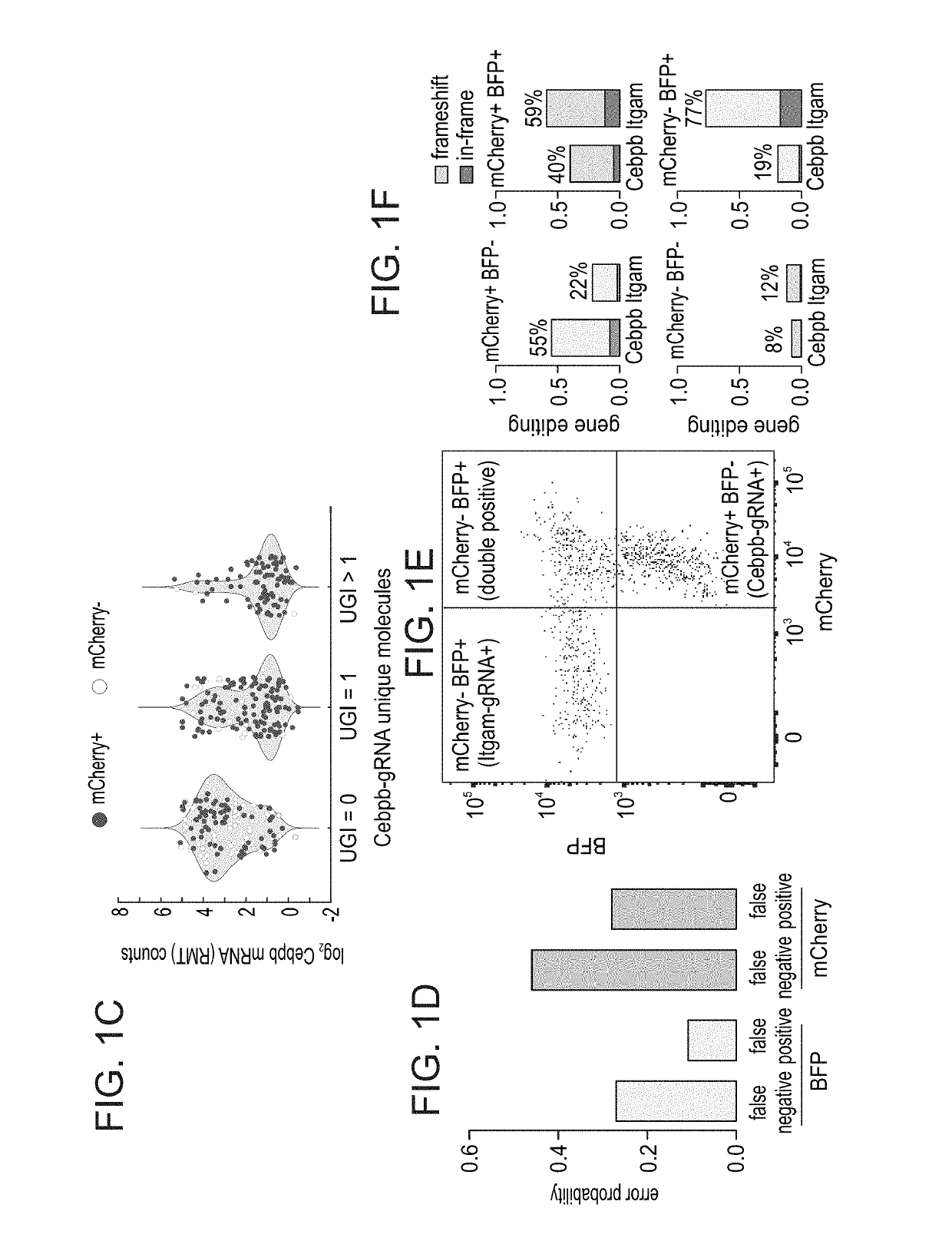
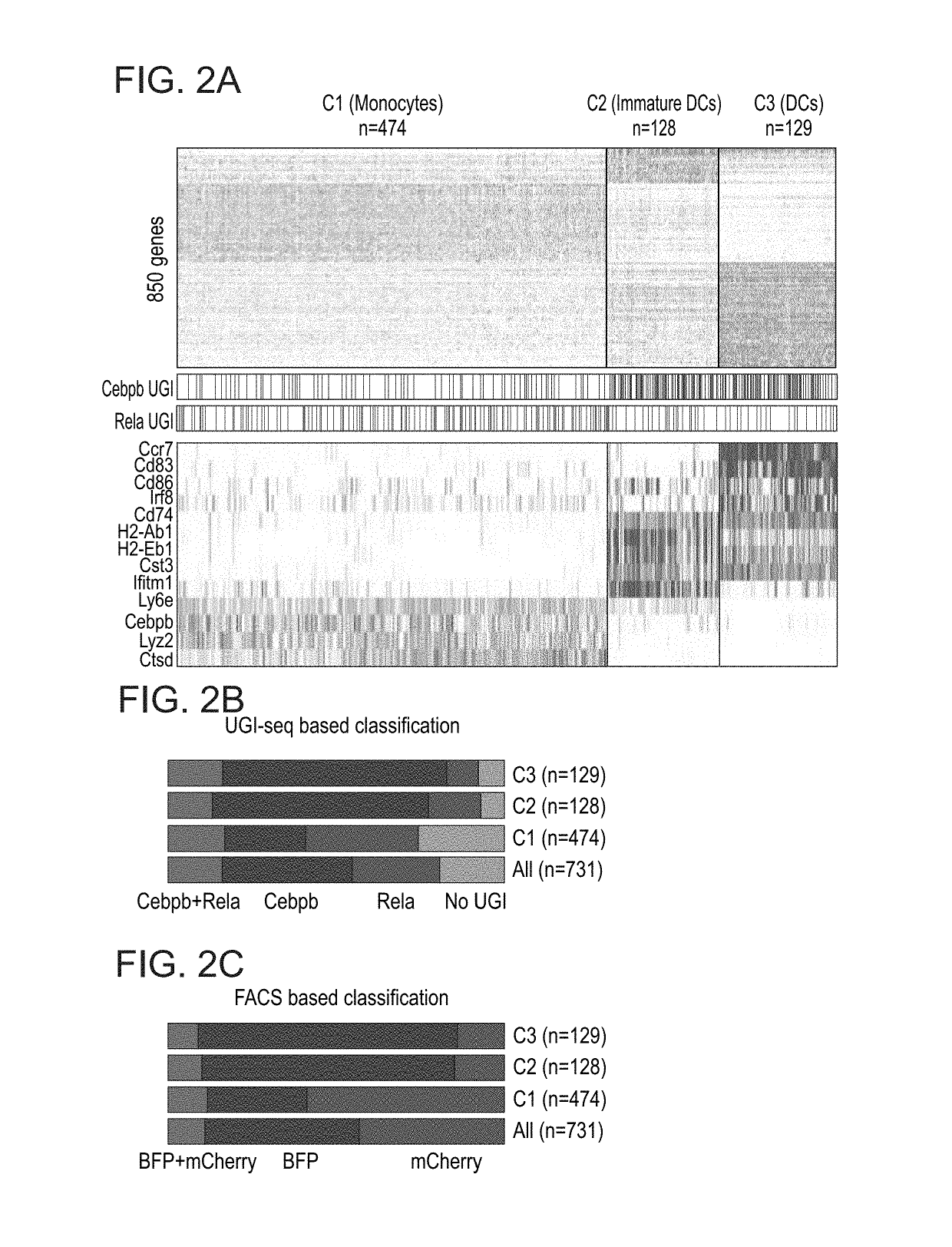
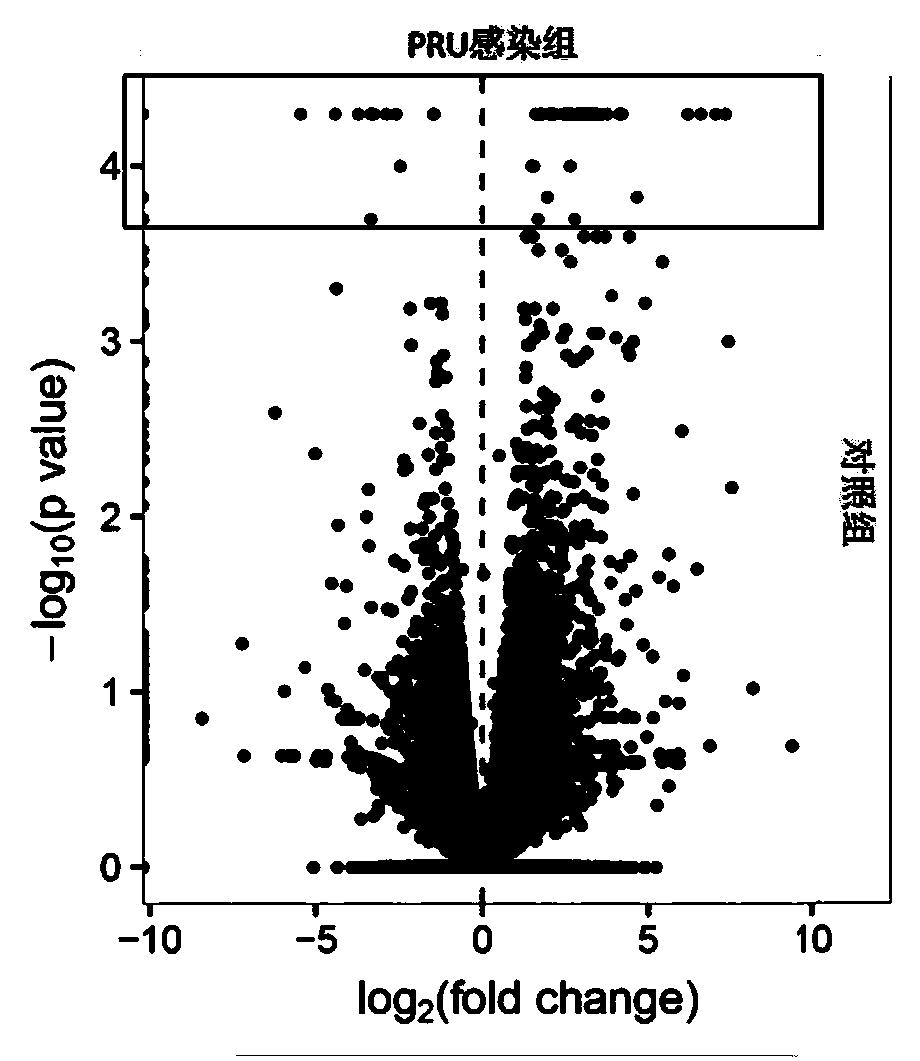
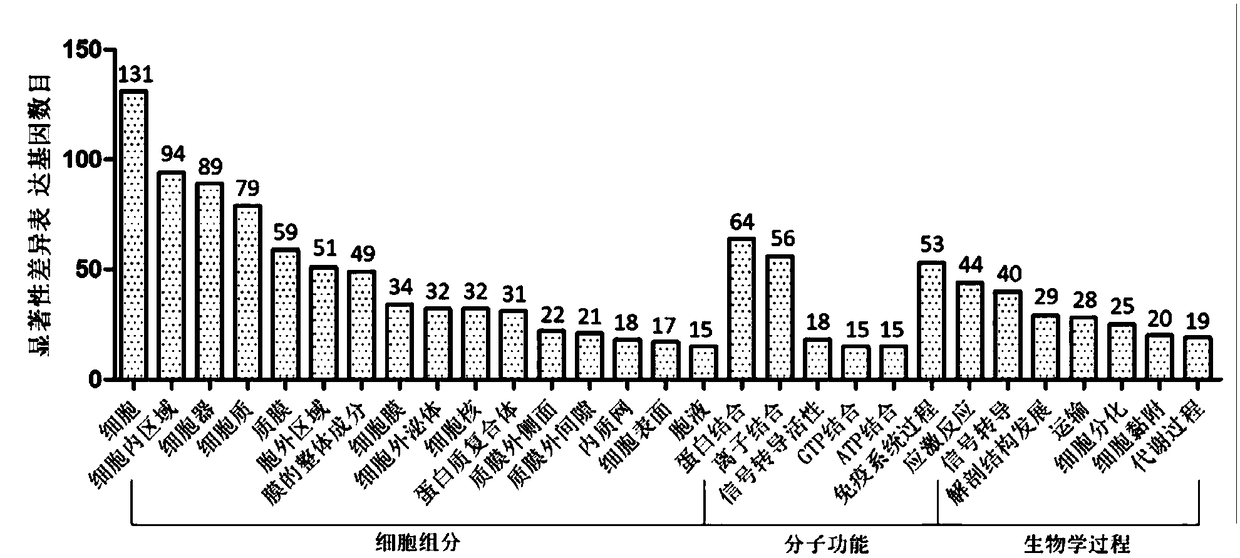
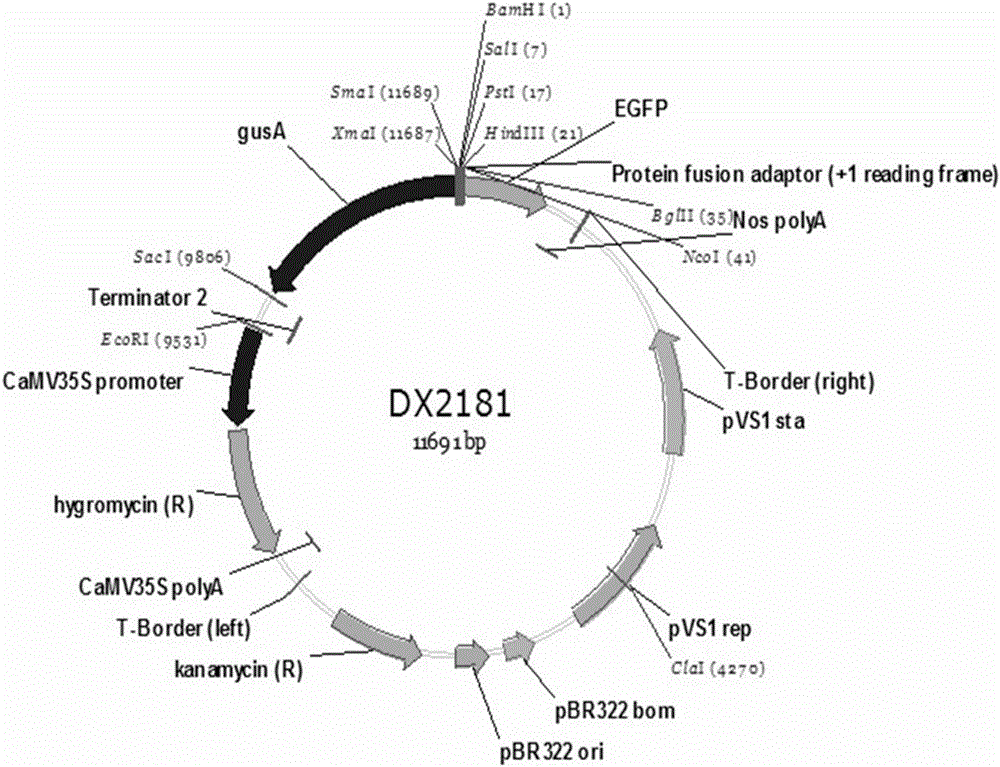
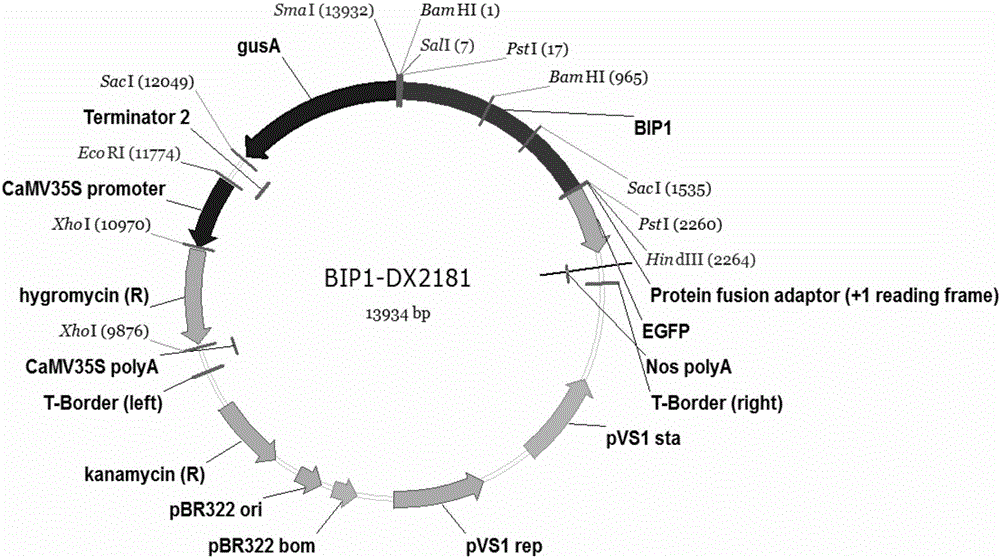
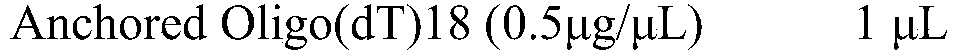Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
39 results about "RNA-Seq" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
RNA-Seq uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a given moment, analyzing the continuously changing cellular transcriptome. Specifically, RNA-Seq facilitates the ability to look at alternative gene spliced transcripts, post-transcriptional modifications, gene fusion, mutations/SNPs and changes in gene expression over time, or differences in gene expression in different groups or treatments. In addition to mRNA transcripts, RNA-Seq can look at different populations of RNA to include total RNA, small RNA, such as miRNA, tRNA, and ribosomal profiling. RNA-Seq can also be used to determine exon/intron boundaries and verify or amend previously annotated 5' and 3' gene boundaries. Recent advances in RNA-Seq include single cell sequencing and in situ sequencing of fixed tissue.
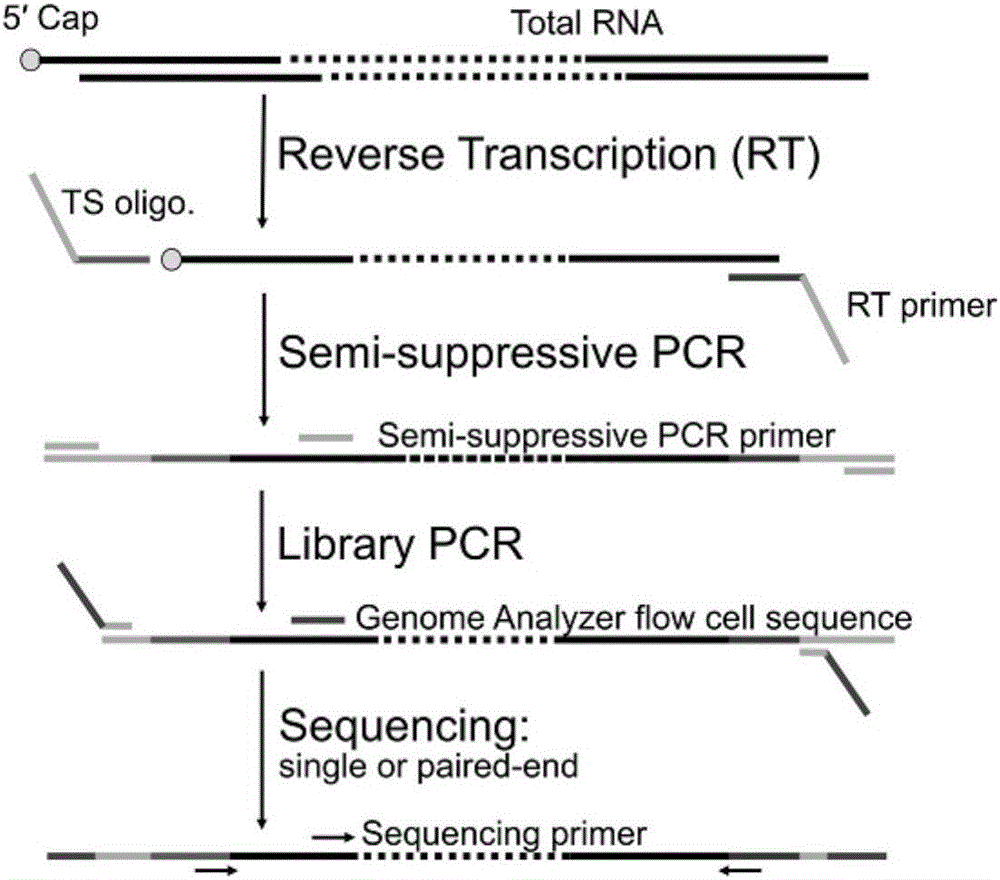
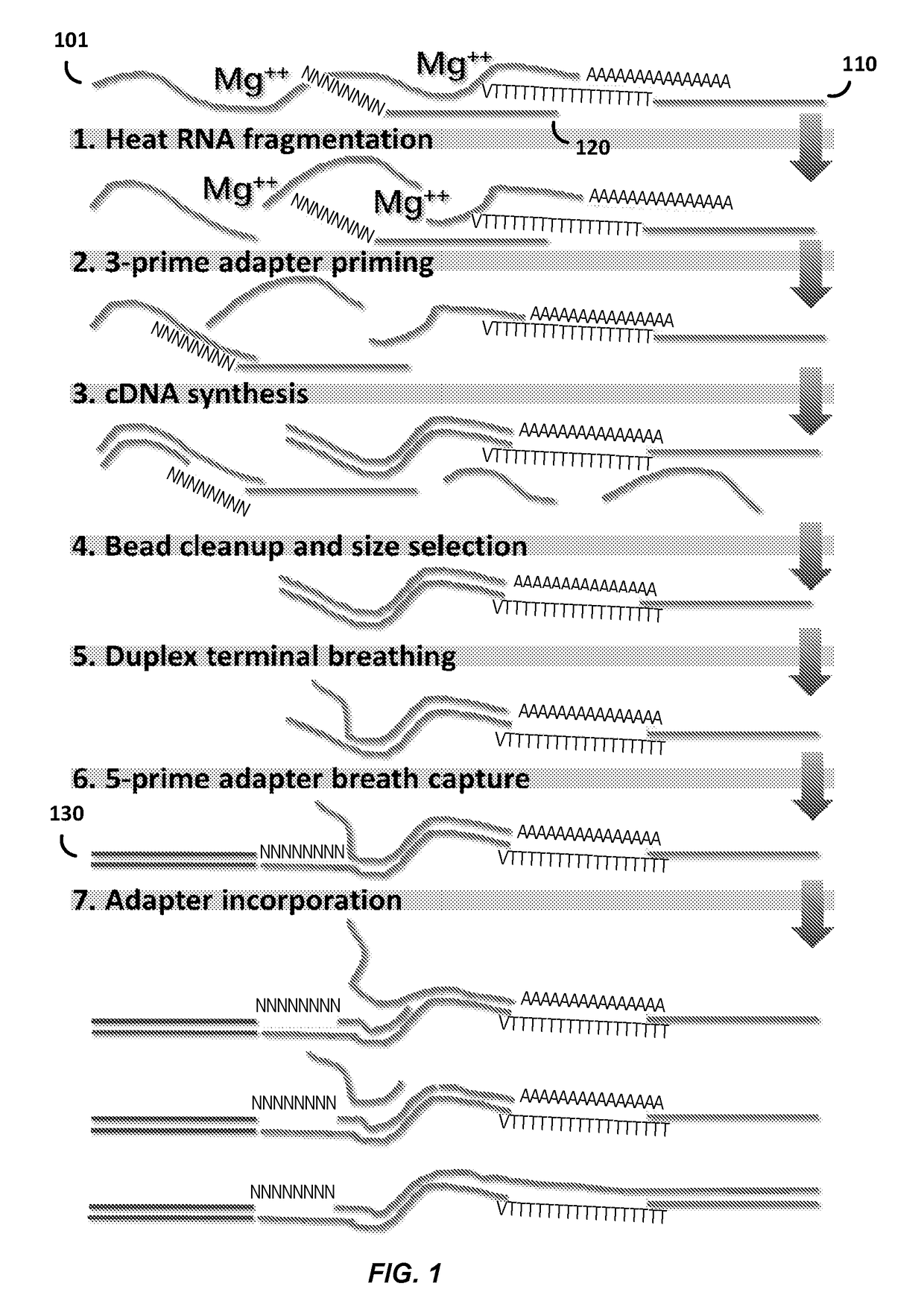
High-throughput rna-seq
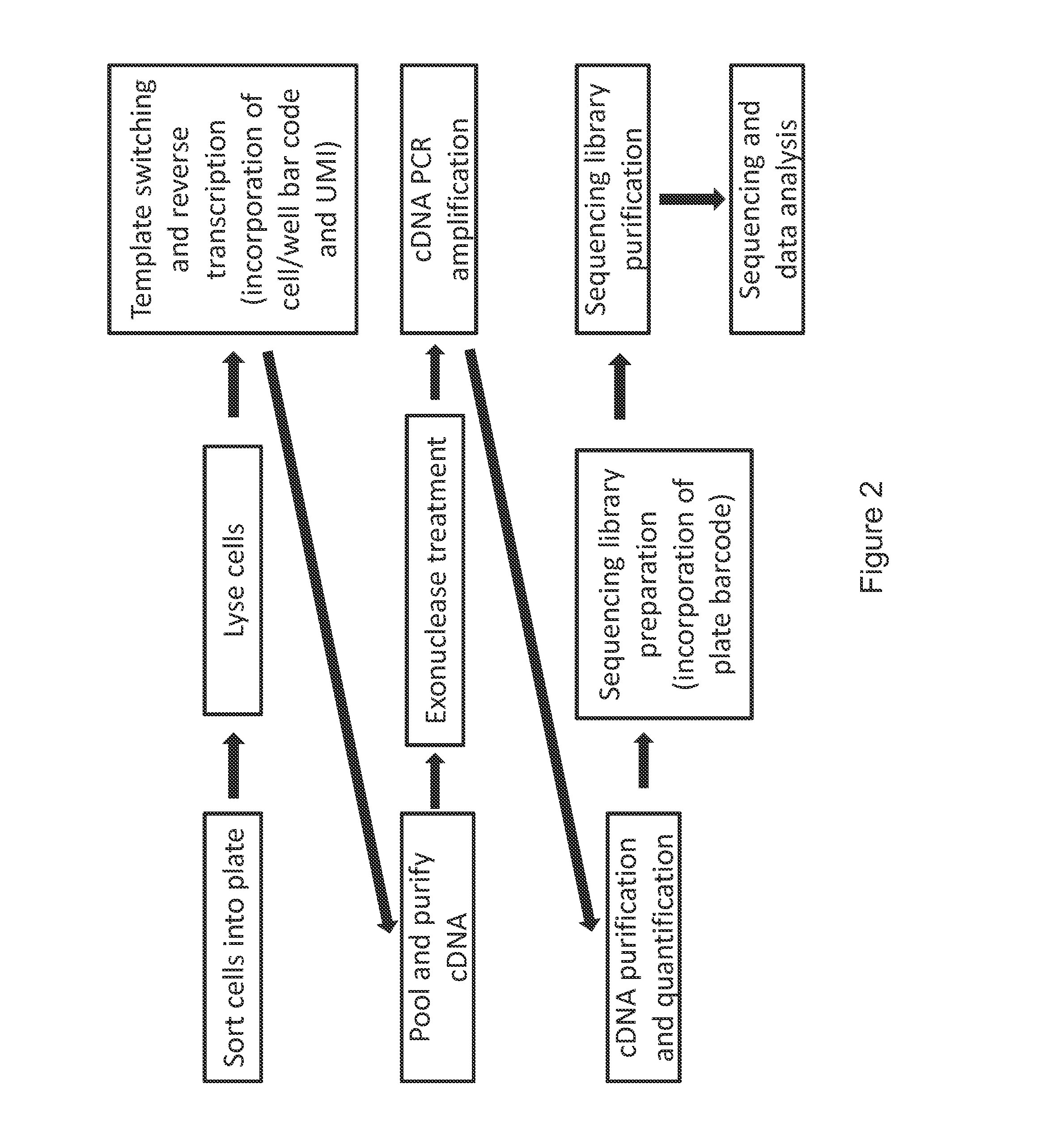
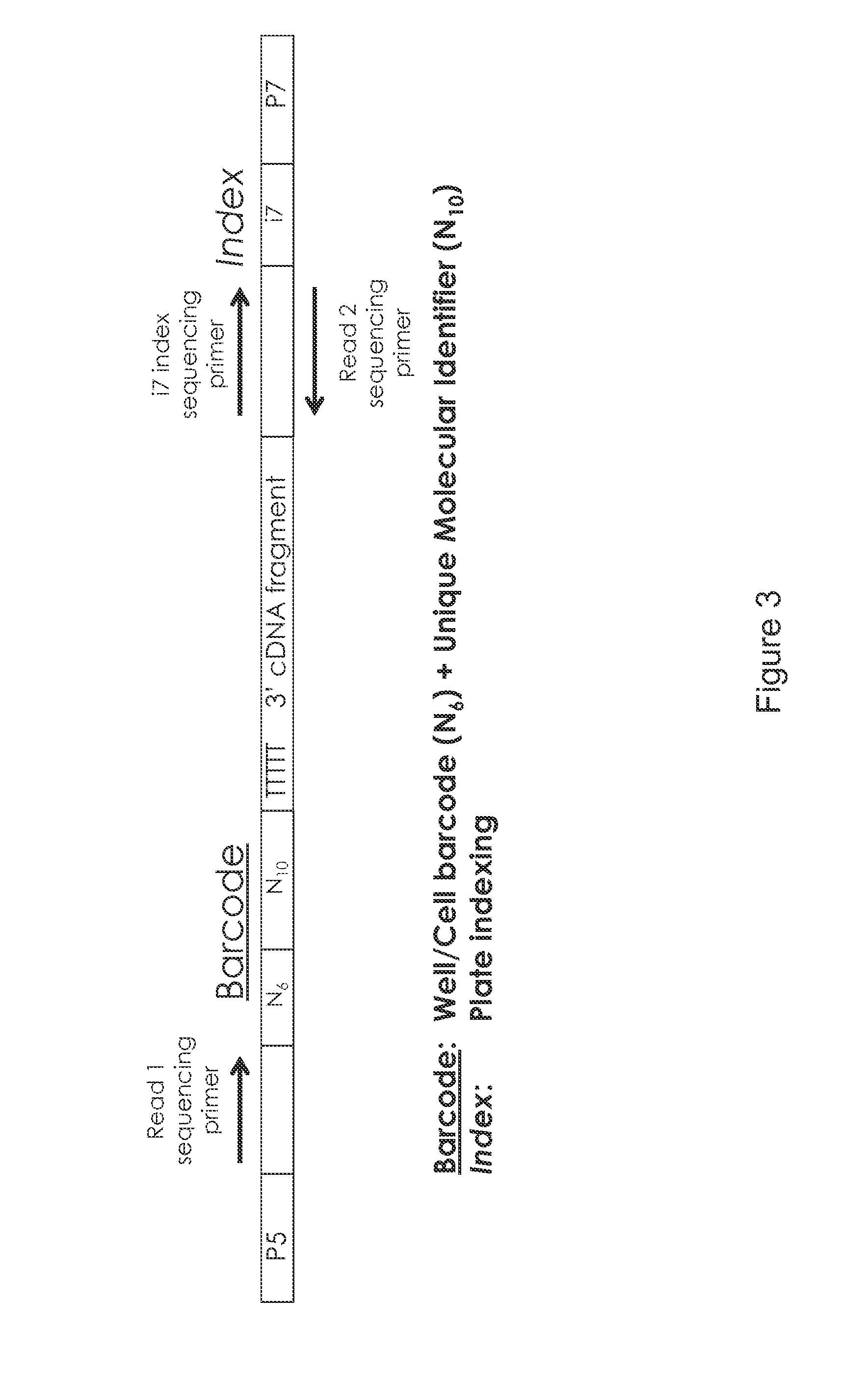
The present invention relates generally to methods for single-cell nucleic acid profiling, and nucleic acids useful in those methods. For example, it concerns using barcode sequences to track individual nucleic acids at single-cell resolution, utilizing template switching and sequencing reactions to generate the nucleic acid profiles. These methods and compositions are also applicable to other starting materials, such as cell and tissue lysates or extracted / purified RNA.
Owner:THE BROAD INST INC +1
Method for obtaining gene information and functional genes from species without genome reference sequence
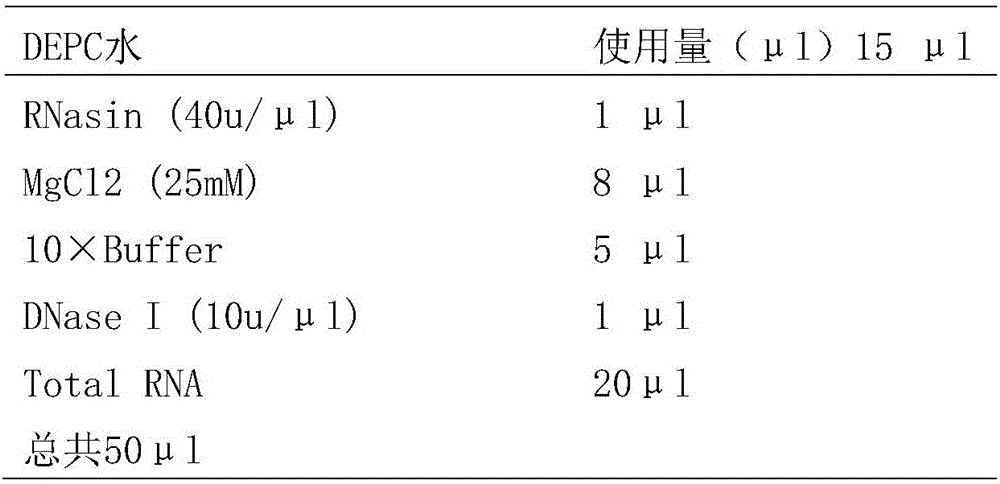
The invention relates to a method for obtaining gene information and functional genes from species without genome reference sequences. The invention also discloses a method for obtaining the gene expression profile of the Asian corn borer. The invention also discloses a method for obtaining transcriptome information and functional genes of the Asian corn borer. The present invention combines DGE-tag technology with RNA-seq technology for the first time to obtain gene expression period, expression level, corresponding metabolic pathway and gene function information from species without genome reference sequence. The method is convenient and fast , accurate and low cost.
Owner:SHANGHAI INST OF BIOLOGICAL SCI CHINESE ACAD OF SCI
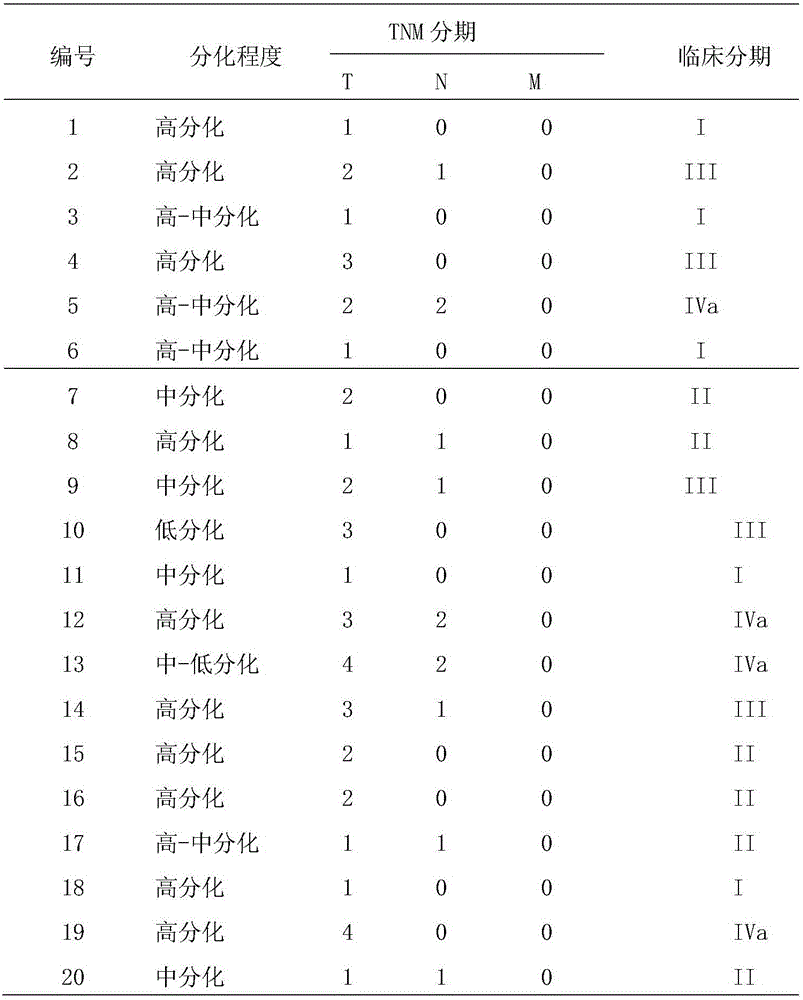
Novel target gene for diagnosing and treating tongue squamous carcinoma and application thereof
ActiveCN105779618AHigh selectivitySimplify the process of quantitative detectionOrganic active ingredientsGenetic material ingredientsDiseaseCandidate Gene Association Study
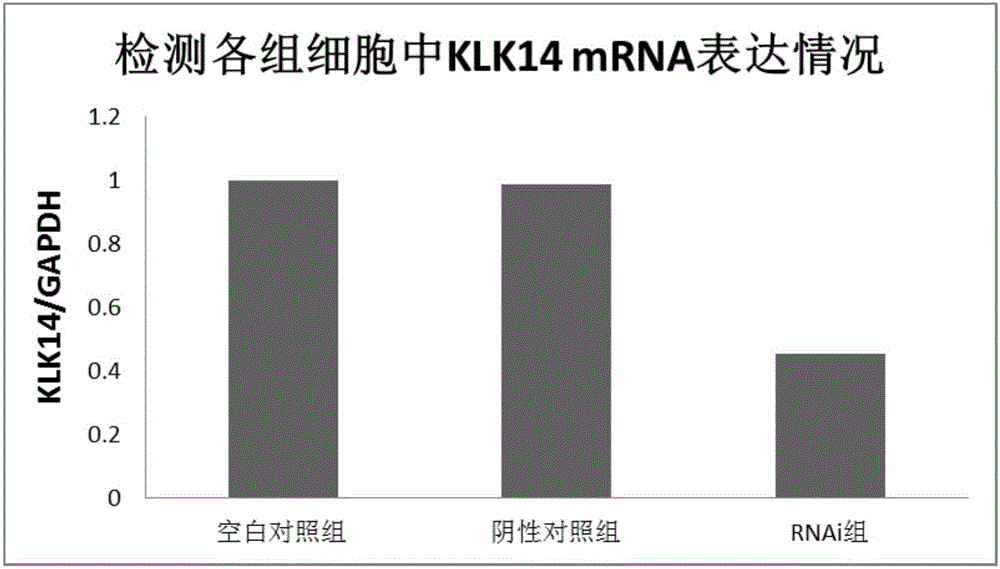
The invention provides a novel target gene for diagnosing and treating tongue squamous carcinoma and application thereof and particularly relates to application of a KLK14 gene and an expression product thereof to diagnosis and treatment of tongue squamous carcinoma. To research the occurrence and development mechanisms of tongue squamous carcinoma, search for an effective molecular target gene for diagnosing and treating tongue squamous carcinoma, promote early diagnosis, prevention and treatment of the disease and lower the death rate of tongue cancer, firstly, RNA-seq sequencing is utilized to detect differential expression genes of tongue squamous carcinoma, cancer branch and normal oral mucosa; secondly, a Real-time PCR technology is utilized to verify the sequencing result; then, an interference technology is utilized, and expression of the candidate gene KLK14 in tongue squamous carcinoma cells SCC15 is silenced. By means of the novel target gene for diagnosing and treating tongue squamous carcinoma and application thereof, an experimental foundation is laid for clinical application of the KLK14 gene to tongue squamous carcinoma, and a new target gene and theoretical basis are provided for early diagnosis and treatment of tongue squamous carcinoma.
Owner:THE SECOND XIANGYA HOSPITAL OF CENT SOUTH UNIV
Sequential sequencing
InactiveUS20140274738A1Microbiological testing/measurementLibrary member identificationNucleic acid sequencingExon
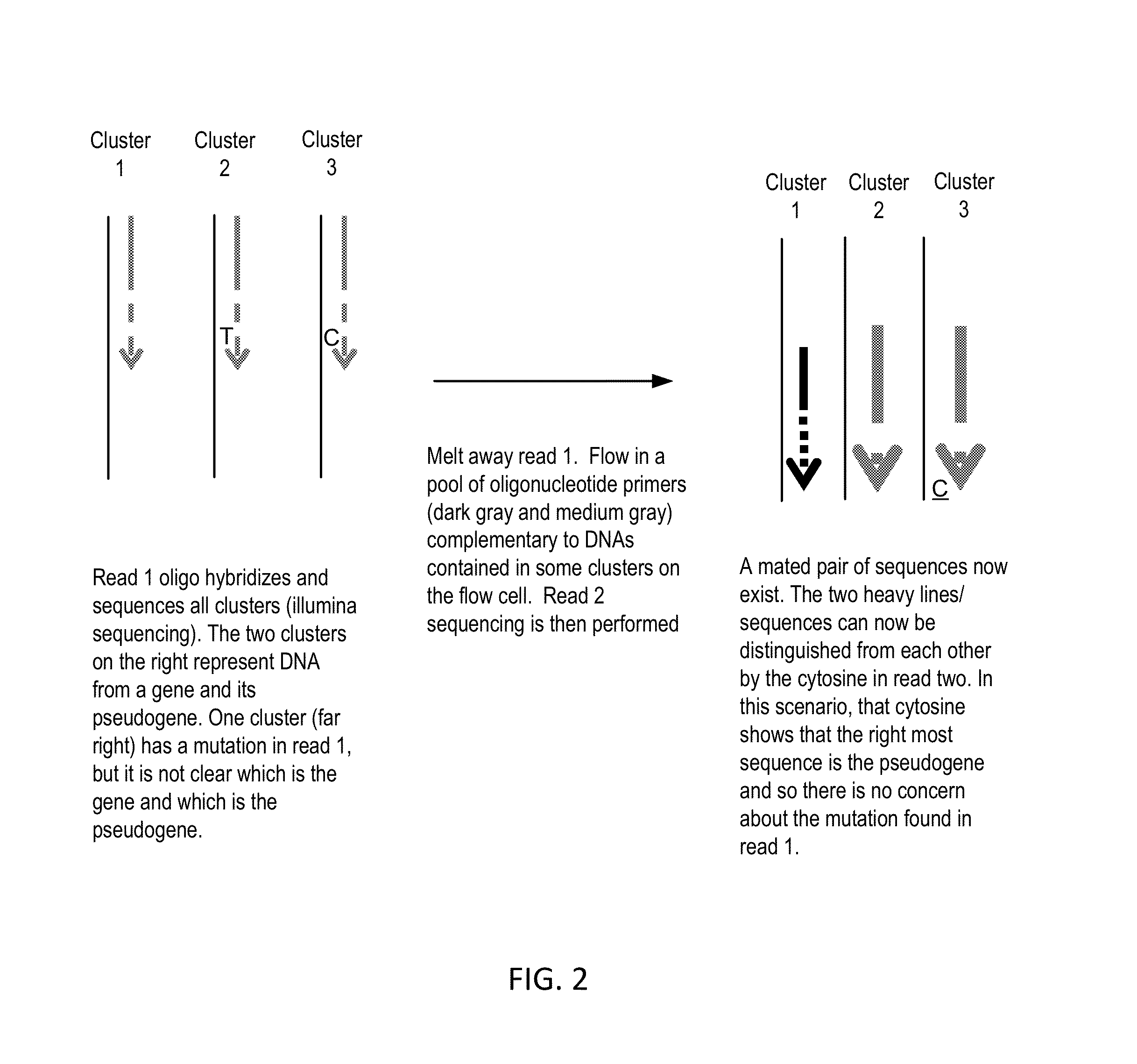
The present invention provides improved methods, compositions and kits for short read next generation sequencing (NGS). The methods, compositions and kits of the present invention enable phasing of two or more nucleic acid sequences in a sample, i.e. determining whether the nucleic acid sequences (typically comprising regions of sequence variation) are located on the same chromosome and / or the same chromosomal fragment. Phasing information is obtained by performing multiple, successive sequencing reactions from the same immobilized nucleic acid template. The methods, compositions and kits provided herein are useful, for example, for haplotyping, SNP phasing, or for determining downstream exons in RNA-seq.
Owner:NUGEN TECH
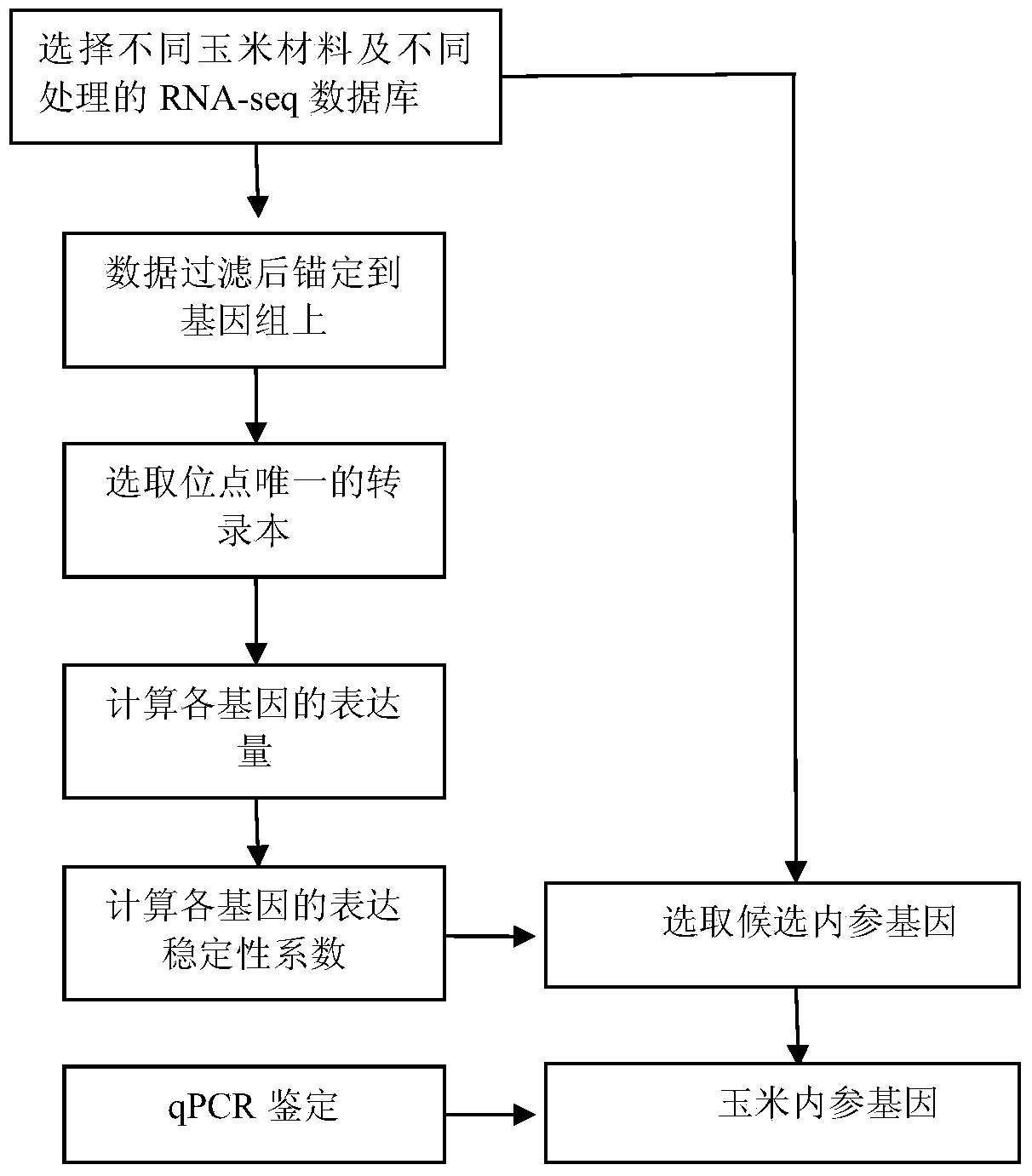
Method for high-throughput screening of maize reference gene
InactiveCN104131107AImprove selection efficiencyReduce experimental workloadMicrobiological testing/measurementReference genesHigh-Throughput Screening Methods
The invention discloses a method for high-throughput screening of a maize reference gene. The method comprises the following steps: by using a high-throughput transcriptome sequencing (RNA-seq) database which is sourced from different maize inbred lines with high diversity, different tissues and different developmental periods and is constructed after different stress treatment, analyzing the expression conditions of a whole transcriptome massively; screening a gene of which the expression is the most constant in the different developmental periods and under different conditions as the reference gene, and providing an accurate comparison for the study of transcriptome expression. The method disclosed by the invention is used for analyzing all the expressed genes of the whole transcriptome by using data of the whole transcriptome, is used for systematically screening the maize reference gene, and is more accurate and reliable compared with a conventional screening method.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
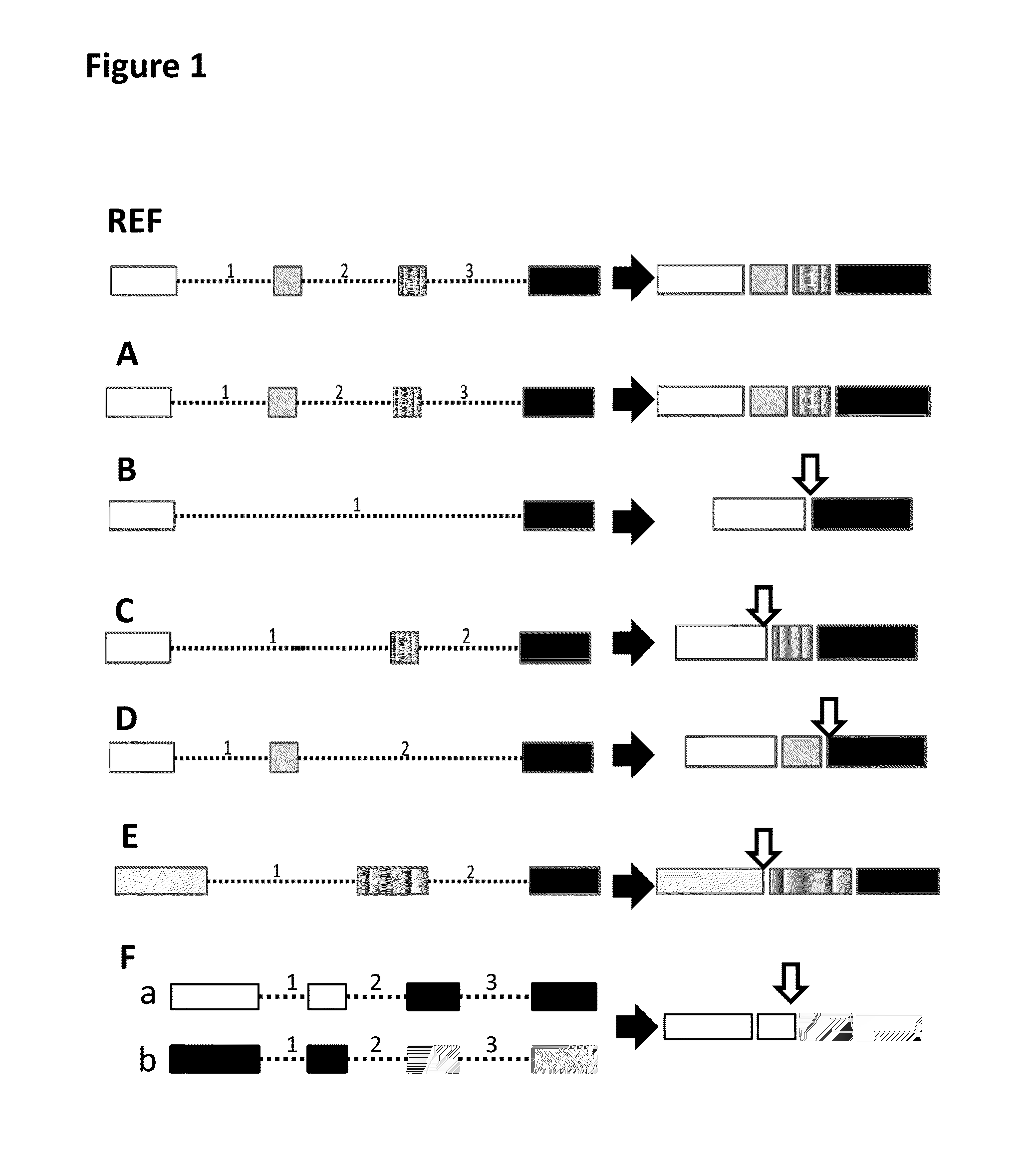
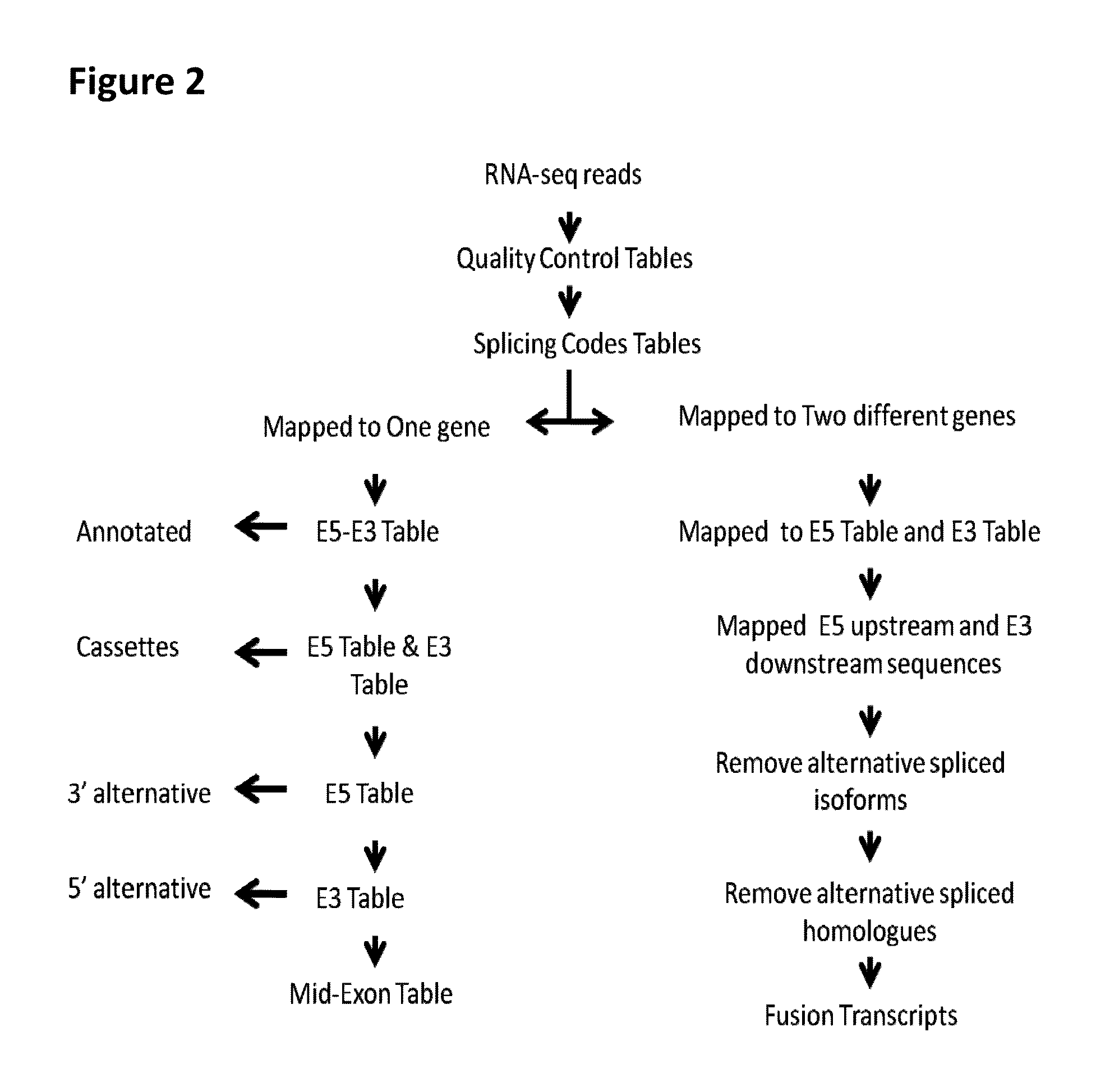
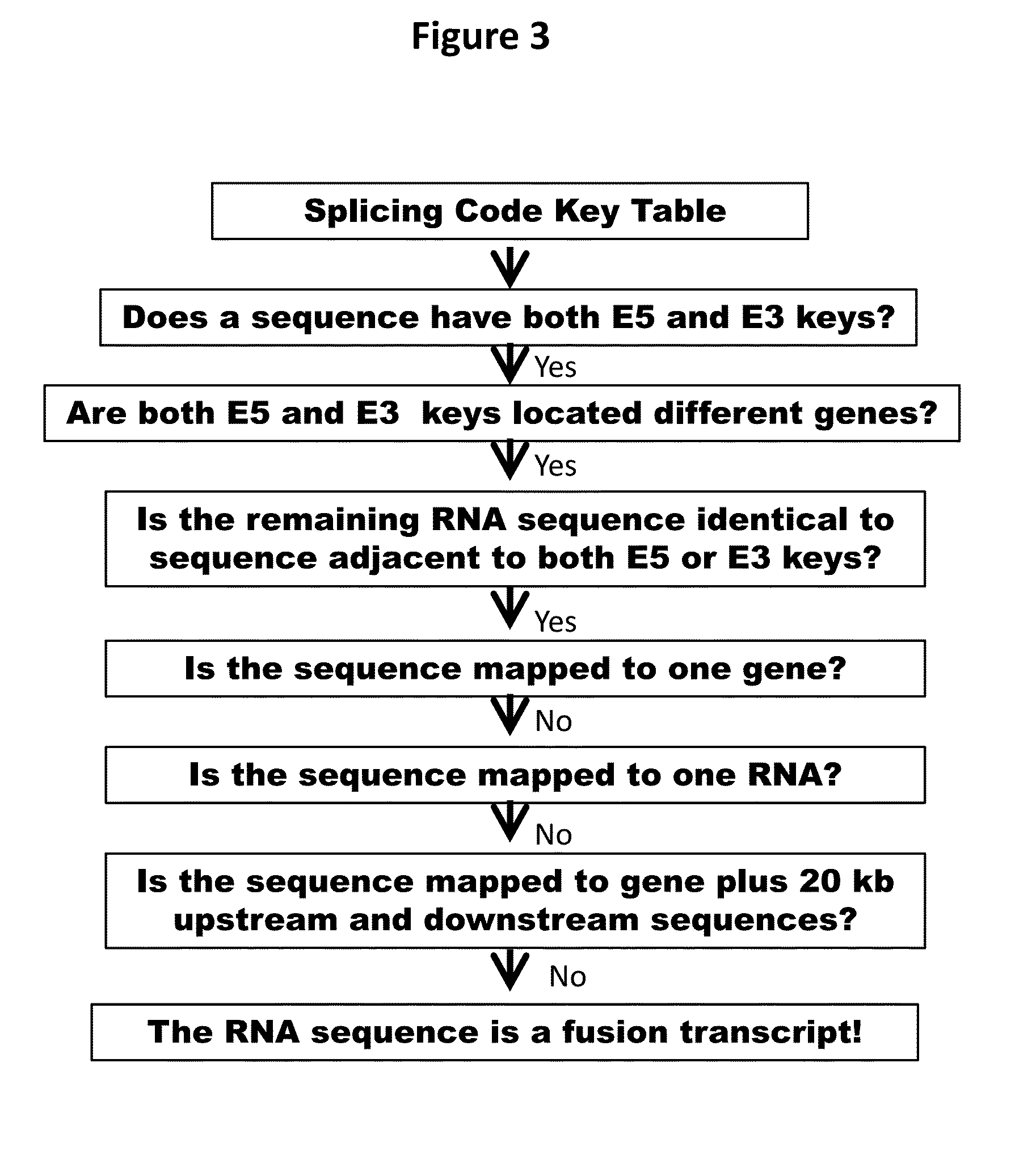
Fusion transcript detection methods and fusion transcripts identified thereby
InactiveUS20160078168A1Simple and accurate and fast computationMicrobiological testing/measurementSequence analysisHuman cancerCancers diagnosis
This present disclosure generally relates to compositions and methods for cancer diagnosis, research and therapy, including but not limited to, cancer markers. In particular, the present disclosure provides a computerized method for detecting fusion transcripts from RNA-seq data and provides the fusion transcripts identified thereby in human cancers. Compositions and methods for identifying the fusion transcripts are also provided.
Owner:SPLICINGCODES COM
Wheat BSR-Seq gene locating method
ActiveCN106202995ALow costHigh precisionSequence analysisSpecial data processing applicationsReference genome sequenceAllele frequency
The invention discloses a wheat BSR-Seq gene locating method. The wheat BSR-Seq gene locating method comprises the following steps of: constructing and sequencing mixing pools, performing quality variation mining, screening a transcript tightly linked to a target gene, developing and locating a molecular marker, etc. The next-generation transcriptome sequencing technology (transcriptome sequencing, RNA-Seq) and a mixing pool technology (Bulked Segregant Analysis BSA) are combined; a wheat sequencing draft sequence is used as a reference sequence at first; then, a lot of high-quality SNP heritable variations on the transcript is mined at high throughput by adopting the next-generation sequencing technology; the allele frequency is precisely calculated in combination with the mixing pool technology, so that the transcript tightly linked to the target gene possibly can be rapidly screened out; false positive is precisely checked and controlled through Fish; the method is independent of a reference genomic sequence, low in cost, rapid and high in precision; the wheat gene locating efficiency and precision are increased; the wheat polymorphic molecular marker developing cost is reduced; therefore, the wheat gene fine locating working time is reduced to several months from several years; the locating precision is reduced to a fraction or 0 cM from several cM; and the fine locating cost is reduced to several thousands from several ten thousands.
Owner:北京麦美瑞生物科技有限公司
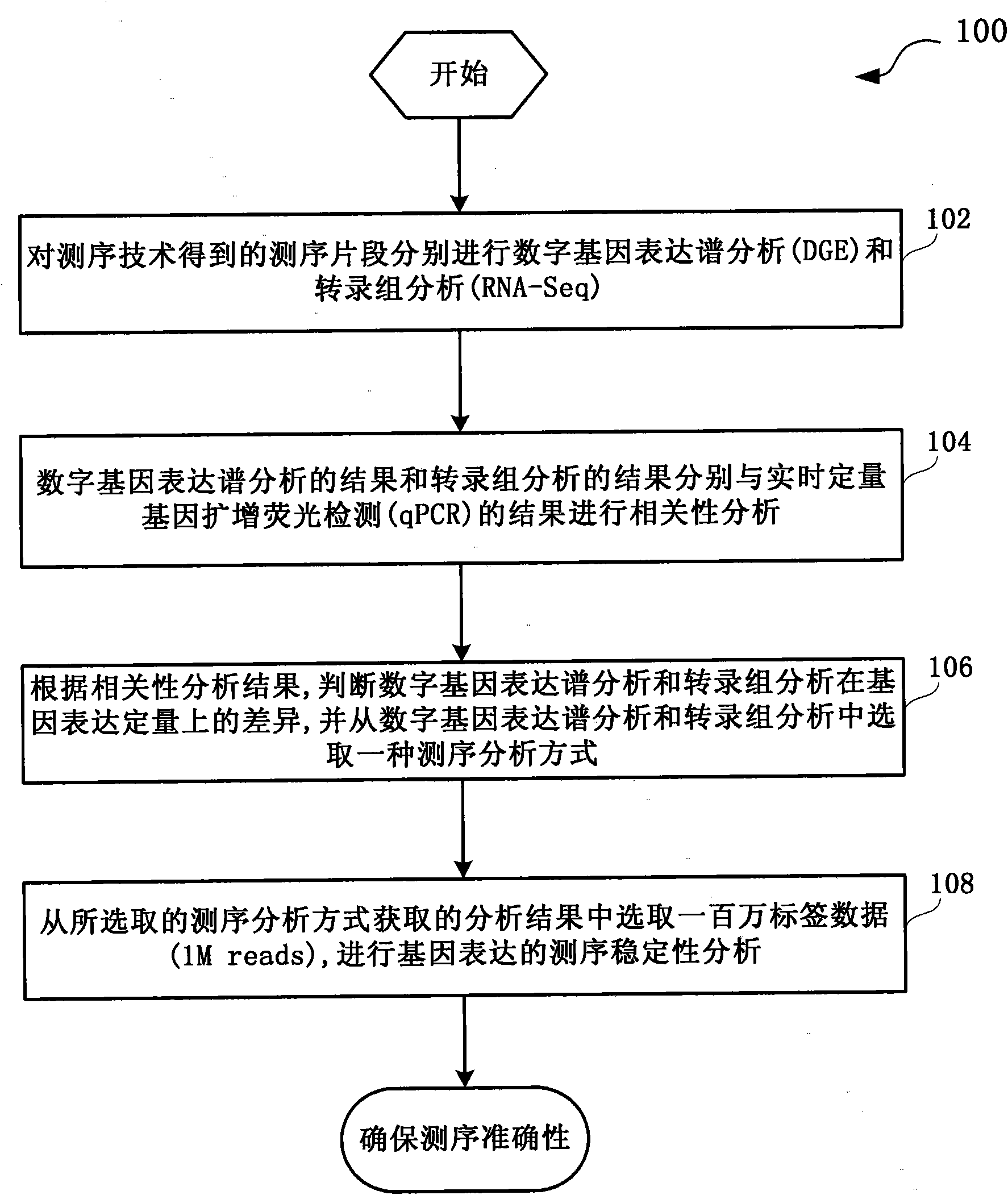
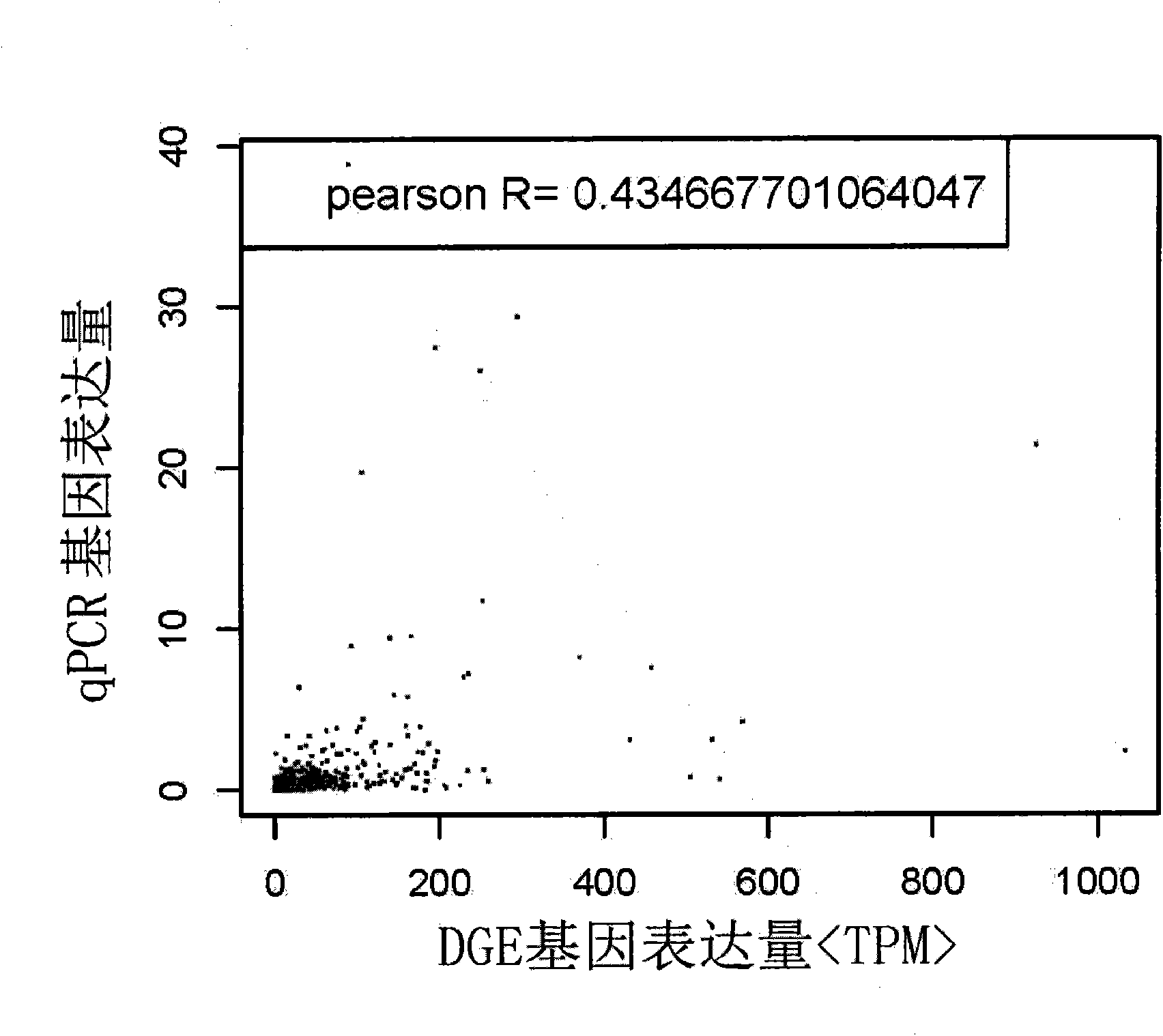
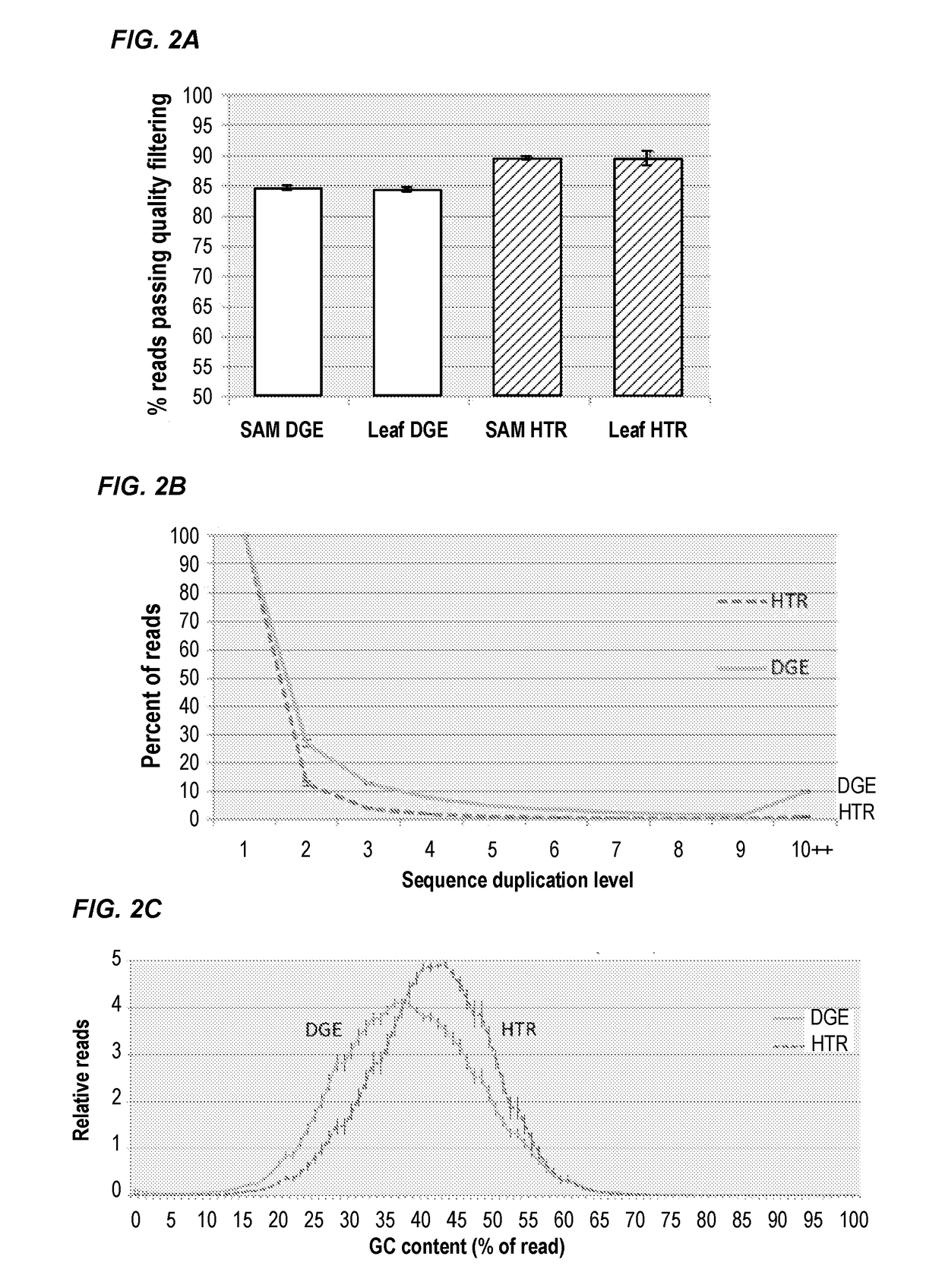
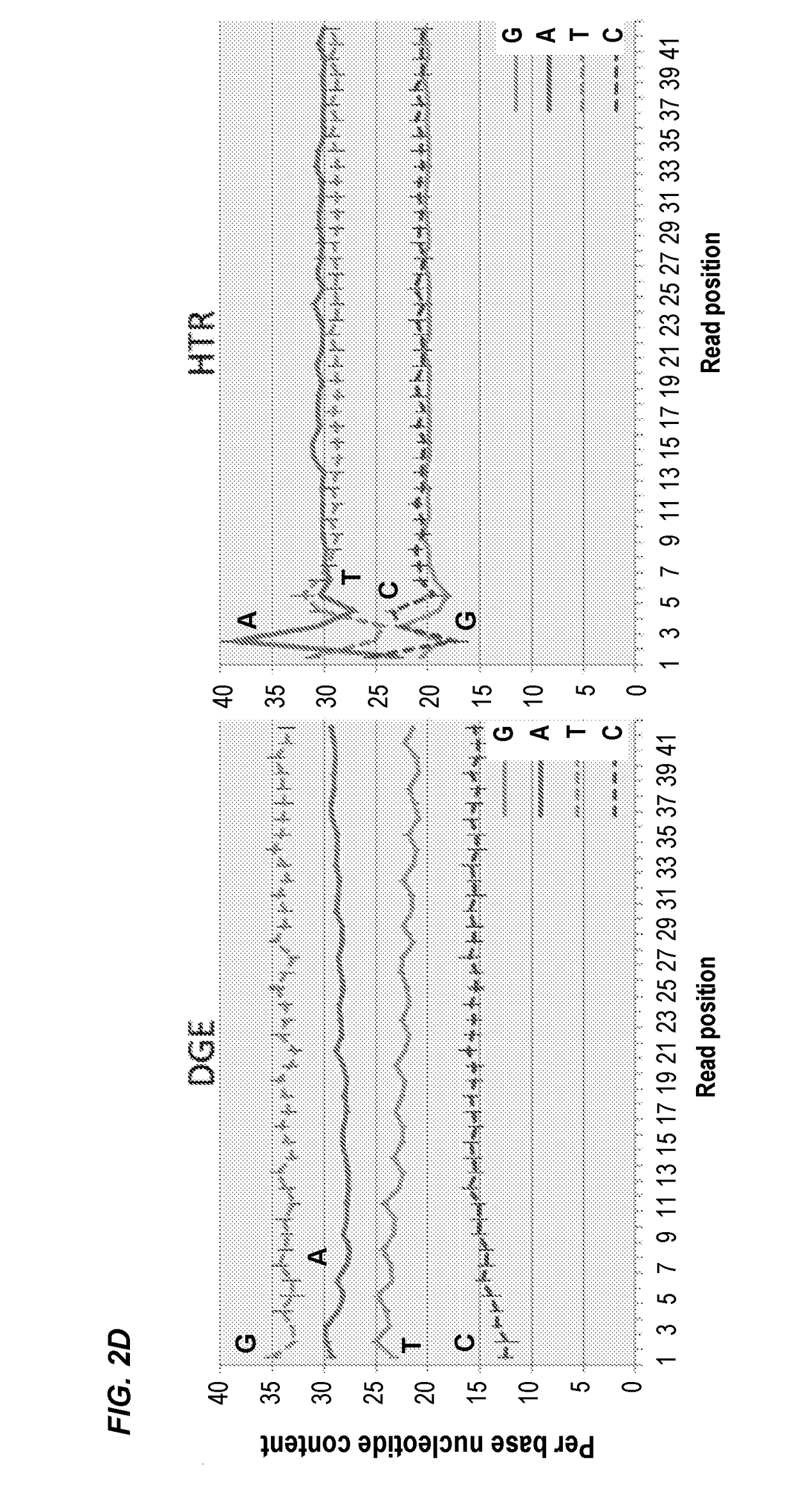
RNA (Ribonucleic Acid) sequencing quality control method and device relating to gene expression
InactiveCN101914619AImprove reliabilityTrue reflection accuracyMicrobiological testing/measurementHybridisationSequence analysisQuality control
The invention discloses RNA (Ribonucleic Acid) sequencing quality control method and device relating to gene expression. The method comprises the following steps of: respectively carrying out DGE (Digital Gene Expression) analysis and RNA-Seq (Ribonucleic Acid-Sequence) analysis on a sequencing segment obtained through a sequencing technology; respectively carrying out correlation analysis between a DGE analysis result and a qPCR (quantitative Polymerase Chain Reaction) result and between a transcriptome analysis result and the qPCR result; judging the difference of DGE analysis and the transcriptome analysis in gene expression quantification according to a correlation analysis result and selecting a sequencing analysis mode from the DGE analysis and the transcriptome analysis; and selecting 1M of reads from analysis results acquired from the selected sequencing analysis mode to analyze the sequencing stability of the gene expression. By carrying out the correlation analysis and the comprehensive estimation of the gene segment, the invention selects a gene expression analysis method with higher reliability, ensures the accuracy of the sequencing work and provides a quality control scheme for the production stability.
Owner:BGI TECH SOLUTIONS
Application of maize gene ZmGFT1 in increasing folic acid content in plants
ActiveCN105647942AAlleviate hidden hungerMicrobiological testing/measurementTransferasesBiotechnologyNutrition
The invention provides application of maize gene ZmGFT1 in increasing folic acid content in plants. 5-Formyltetrahydrofolate content in 513 selfing line materials of a maize natural variant group is measured, the influence of group structure and genetic relationship of the natural variant group is estimated in connection with genotype data mined from RNA-Seq data, gene GFT1 having significant influence on the 5-formyltetrahydrofolate content and located on maize 5th chromosome is located by means of genome-wide association study, and the gene GFT1 encodes glutamate formiminotransferase 1. The invention also provides an SNP marker associated with maize folic acid content and its application. The maize gene ZmGFT1 and discovery of the key SNP site therein having significant influence on the 5-formyltetrahydrofolate content not only provide theoretical basis for studying folic acid metabolism in maize and other crops, but also enable the hidden hunger problem of human in terms of folic acid nutrition to be solved through the increase in the folic acid content of transgenic plants.
Owner:THE INST OF BIOTECHNOLOGY OF THE CHINESE ACAD OF AGRI SCI
Chili maturity SNP (Single Nucleotide Polymorphism) molecular marker and application thereof
ActiveCN108486276AAccurate identificationAchieve early identification of cotyledonsMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceCotyledon plant
The invention discloses a chili maturity SNP (Single Nucleotide Polymorphism) molecular marker and the application thereof. The chili maturity SNP molecular marker comprises a sequence of SEQ ID NO.1as shown in the specification. By using a BSR-Seq (Bulked Segregant RNA-Seq) technique, a chromosome site controlling first flower nodes can be rapidly identified, and the chili maturity SNP molecularmarker is specifically developed for the site. By adopting the molecular marker and an amplification primer thereof, and with specific restriction endonuclease treatment, restriction endonuclease chilies can be rapidly and accurately screened at a cotyledon stage, molecular marker auxiliary selection upon chili maturity breeding can be achieved, the breeding selection efficiency can be improved,and the molecular marker has significant application values in chili maturity breeding.
Owner:VEGETABLE & FLOWER INST JIANGXI ACADEMY OF AGRI SCI
Analytical method for excavating key lncRNA in process of differentiating chick embryo stem cells into male germ cells
PendingCN105838791AShort cycleImprove efficiencyMicrobiological testing/measurementTotal rnaPlant Germ Cells
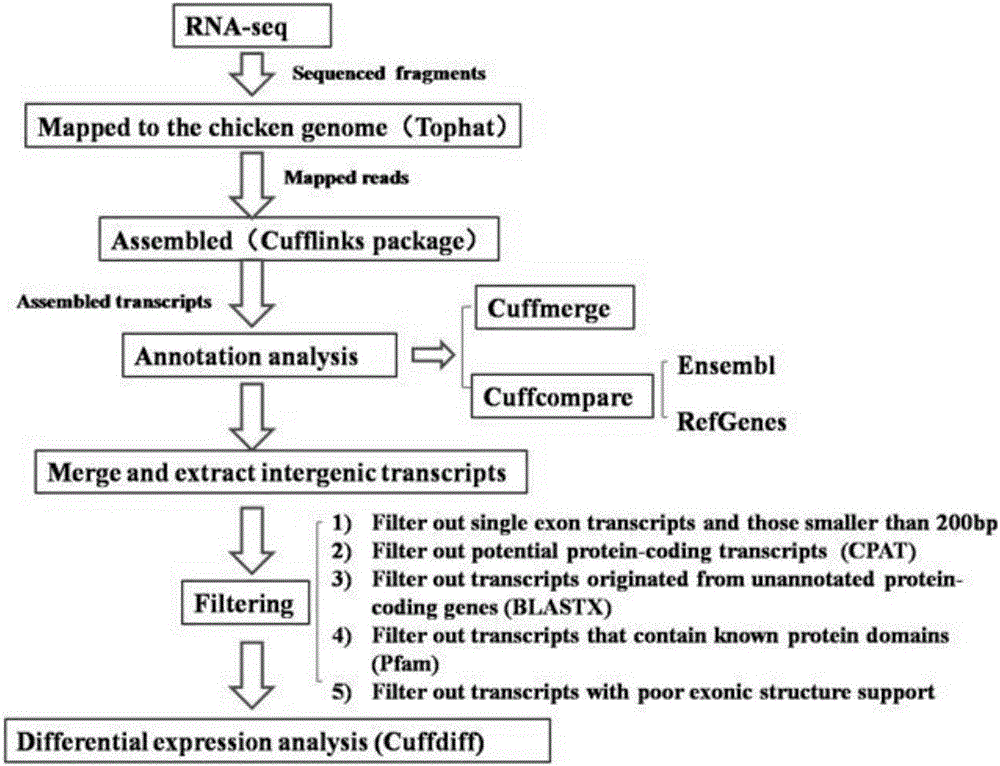
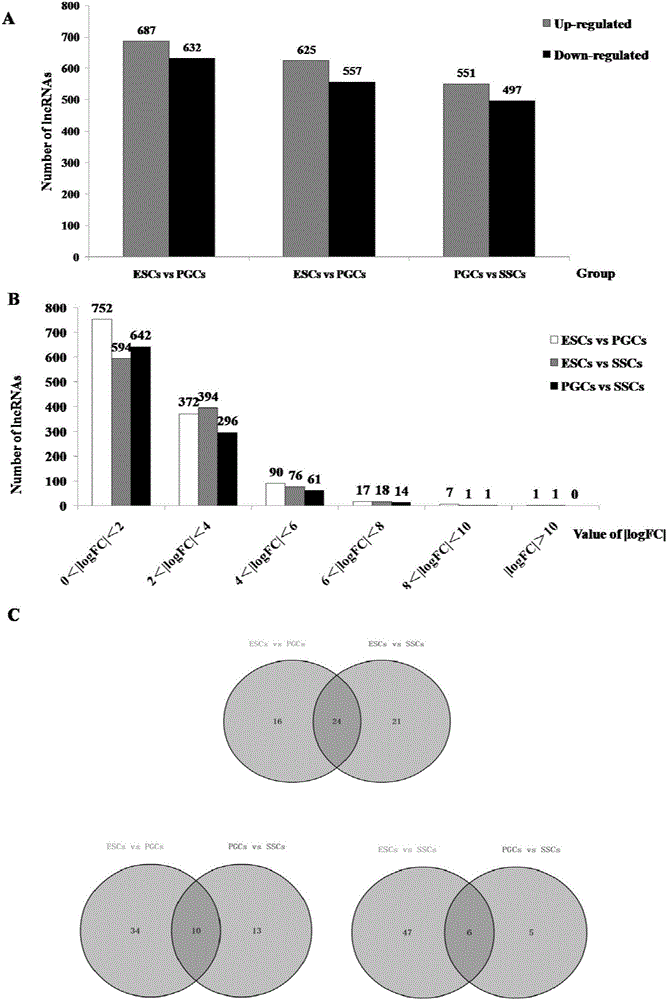
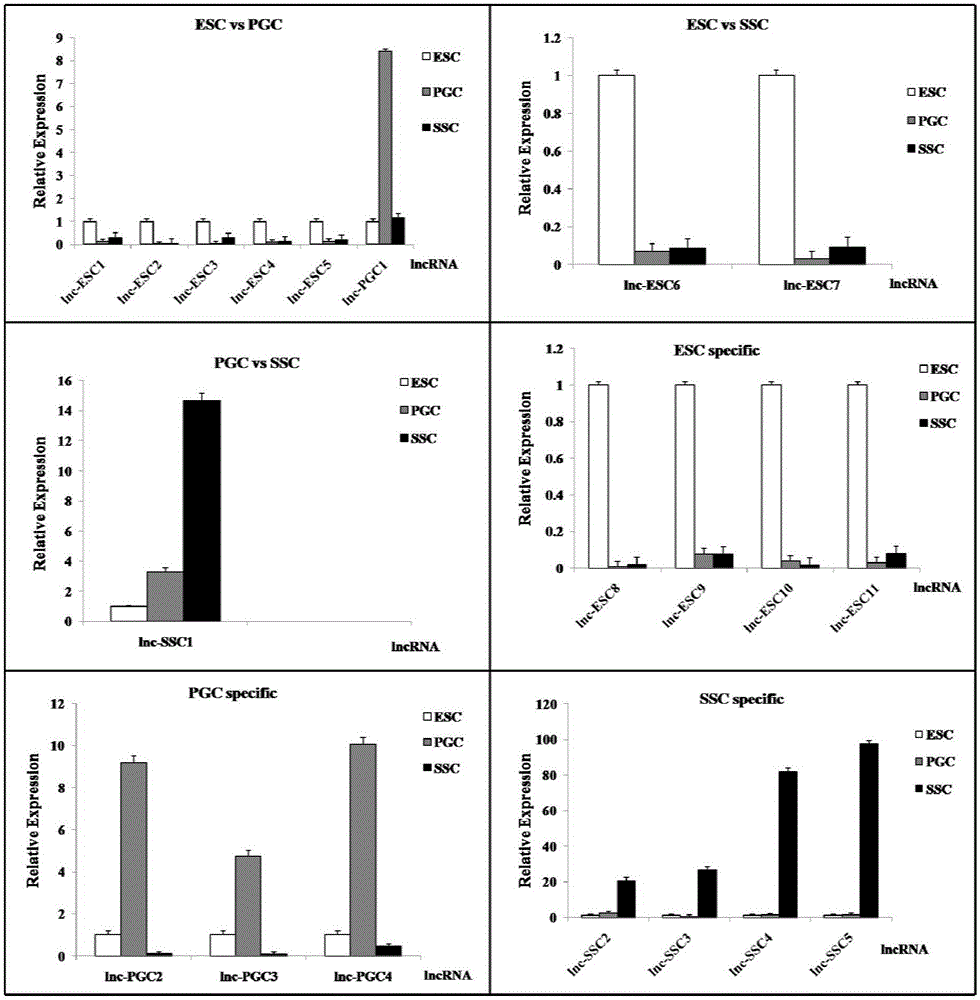
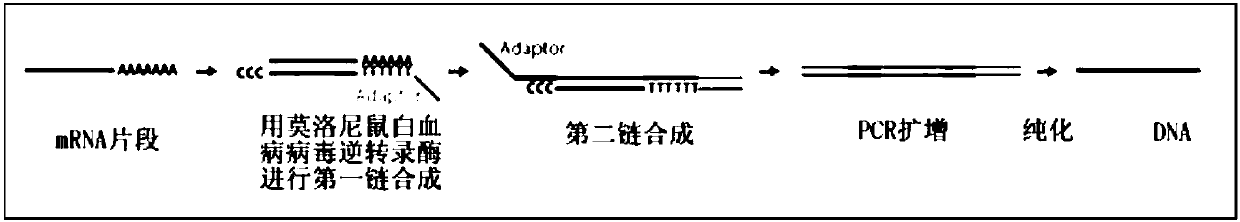
The invention discloses an analytical method for excavating key lncRNA in a process of differentiating chick embryo stem cells into male germ cells. The method comprises the following steps: firstly, separating chick ESCs, PGCs and SSCs from an embryonic disc of a fresh fertilized egg of a Rugao yellow chicken, a germinal ridge of the 19th stage (72h) and a testicle of the 18th day, performing sex determination by using a PCR amplification method according to a design specificity of a CDH1 gene, sorting antibody labeled ESCs, PGCs and SSCs, extracting cell total RNA from positive cells, detecting the quality of RNA after purification, and performing RNA-Seq sequencing after passing the detection; screening a transcript of assembled RNA from a sequencing result; separating chicken ESCs, PGCs and SSCs, extracting total RNA, and reversely transcribing the total RNA into cDNA; performing predictive analysis of Trans and Cis target genes on candidate lncRNA, searching a GO function item of the lncRNA by virtue of a DAVID database, annotating functions of the lncRNA by virtue of GO analysis, and performing functional analysis on the lncRNA; and drawing an interaction network diagram of the relationship between the key lncRNA, a target gene thereof and related signal channels.
Owner:YANGZHOU UNIV
Single cell mRNA reverse transcription and amplification method
InactiveCN107893100AGet transcription directionAvoid pollutionMicrobiological testing/measurementSequence analysisTotal rnaFull length cdna
The invention belongs to the field of transcriptome analysis, and in particular relates to a single cell mRNA reverse transcription and amplification method. 20 to 500 ng of high-quality full-length double-stranded cDNA can be amplified from 1 to 2,000 cells or 10 pg to 20 ng of extracted eukaryotic total RNA as a start by the method within 5-6h. The success rate of reverse transcription and amplification can reach 95% or above, and the amplified full-length cDNA can seamlessly connected with a mainstream sequencing platform. The method is a powerful tool for studying gene expression at the single cell level and greatly expands the application range of RNA-Seq. The cDNA reversely-transcribed and amplified by the method can be used for expression analysis of a very small amount of sample (single cell), early embryonic development studies, tumor cell heterogeneity studies, immune cell population studies, and stem cell differentiation studies.
Owner:XUKANG MEDICAL SCI & TECH (SUZHOU) CO LTD
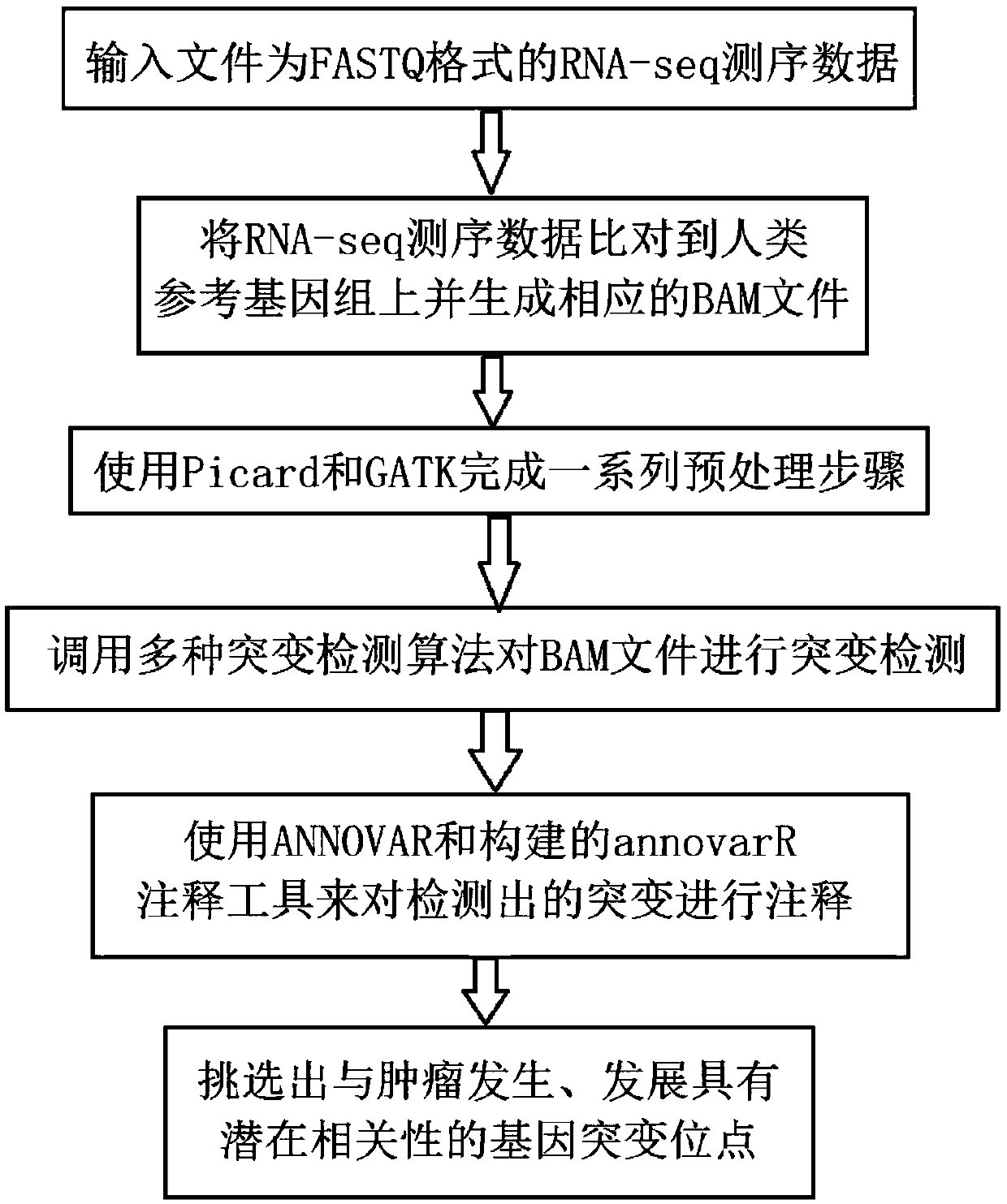
Tumor gene mutation detection method and system based on RNA-seq data
The invention discloses a tumor gene mutation detection method based on RNA-seq data. The method comprises the following steps that gene mutations for different high-incidence tumors and diseases arecollected and sorted so as to build a high-priority mutation detection gene list and a mutation site database; gene mutations currently having specific targeted or chemotherapeutic drug action sites and gene database are collected and sorted; a gene mutation annotation database based on RNA-seq is built; quality control, gene sequence alignment and gene mutation detection algorithms used for RNA-seq mutation detection are collected and sorted; and a set of complete analysis process from initial RNA-seq sequencing data to final mutation detection result is built. The invention further disclosesa tumor gene mutation detection system based on RNA-seq data. By using the integrated multiple databases, people can more rapidly find the known gene mutation sites related to tumorigenesis through the detection analysis process.
Owner:RUIJIN HOSPITAL AFFILIATED TO SHANGHAI JIAO TONG UNIV SCHOOL OF MEDICINE
Long-chain non-coding RNA and application thereof
ActiveCN109897853APrevent proliferationPromote apoptosisNervous disorderAntipyreticNon-coding RNAGene
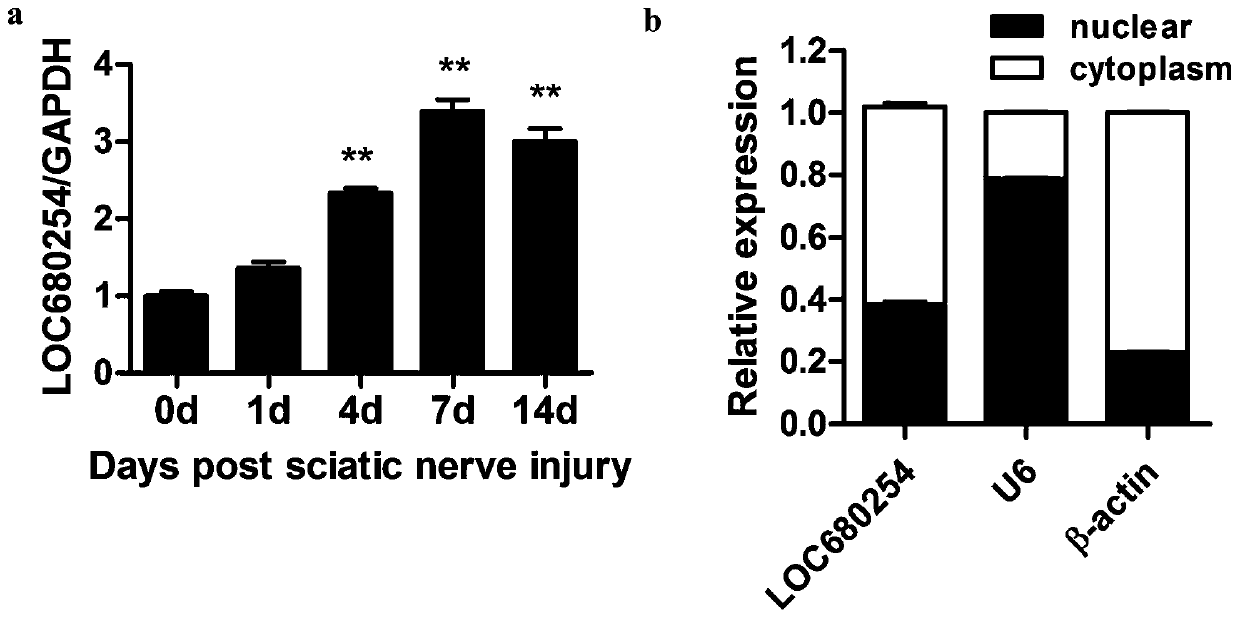
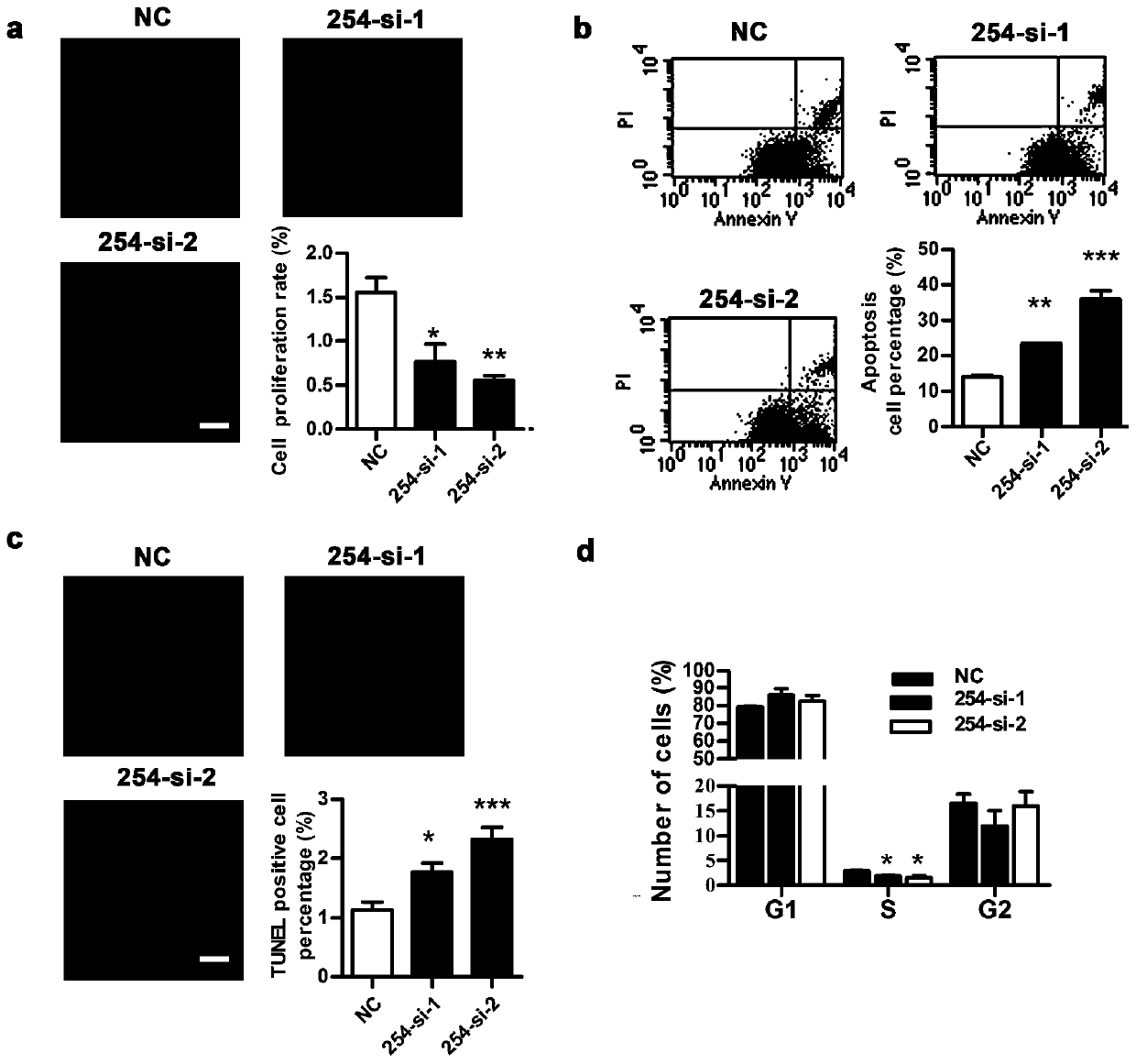
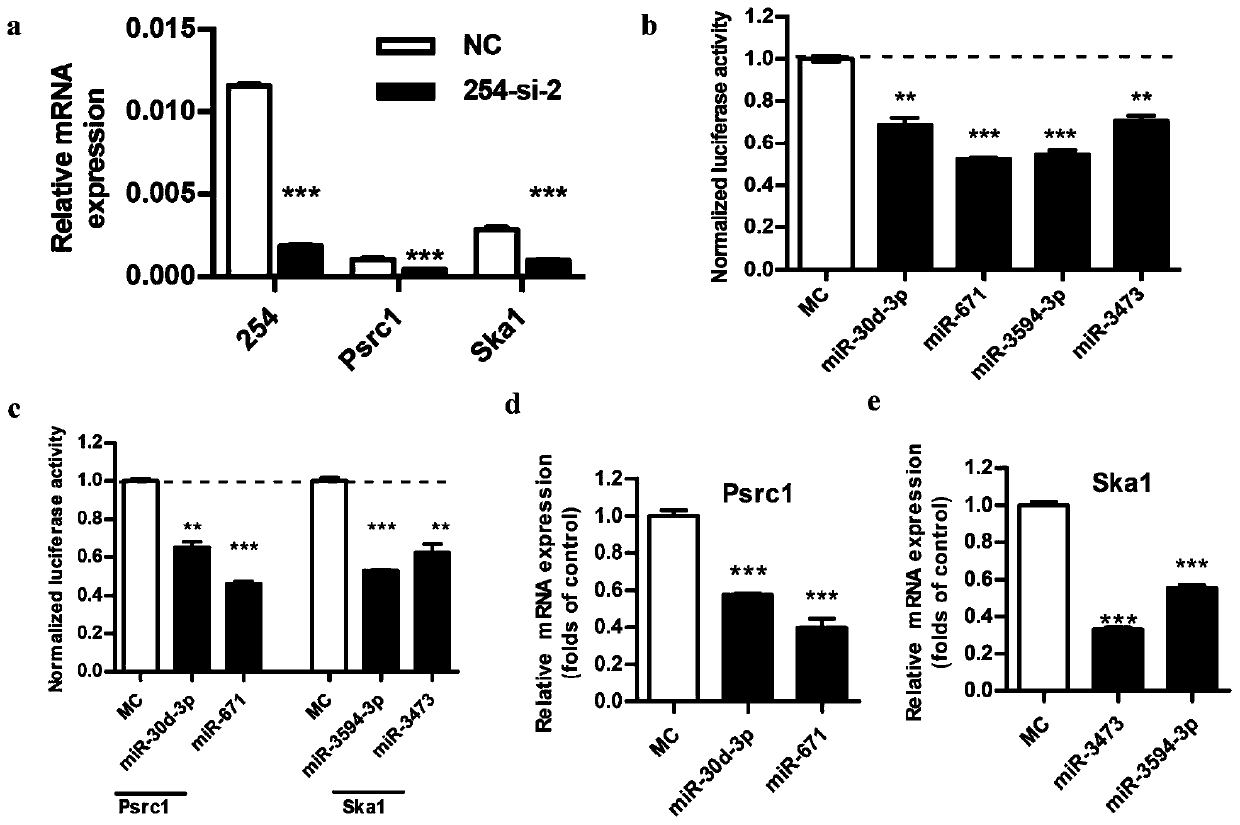
The invention relates to long-chain non-coding RNA and an application thereof. LncRNA LOC680254 related to cell proliferation and cell cycle control is obtained by analyzing different LncRNAs in different time point samples after sciatic nerve injury of SD rats by use of RNA-seq and can be used as miRNA molecular sponge to specifically bind miR-30d-3p, miR-671, miR-3594-3p or miR-3473 so as to resist miRNA functions and promote expression of Psrc1 and Ska1, and cell proliferation is promoted.
Owner:NANTONG UNIVERSITY
Method for constructing mRNA 5'-termnal information library
ActiveCN105734052APromote enrichmentShorten the lengthMicrobiological testing/measurementLibrary creationGenomicsStart site
The invention belongs to the technical field of genomics, and particularly relate to a method for simplifying CAGE-seq library establishing.The method comprises the steps that mRNA and lncRNA containing cap structures are enriched, a 5'-termnal connector is specifically added, after fragmentation processing, reverse transcription is carried out, and 5'-terminal information is further enriched.According to the whole method, an existing mature RNA-seq kit of the gnomegen company is mixed.The method is easier to operate and high in success rate, the 5'-terminal information can be specifically enriched, the method is more helpful for verifying the transcriptional start site of mRNA in a whole cell, and thus acquiring accurate promoter information and 5'UTR information.The method is a powerful mode for studying gene expressions and is suitable for gene regulatory network studies.
Owner:武汉生命之美科技有限公司
Screening method and application of target gene SSR molecular markers of acanthopanax senticosus
ActiveCN108754018AOvercome uncertaintyOvercoming the disadvantages of blindnessMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationAgricultural science
The invention discloses a screening method and application of target gene SSR molecular markers of acanthopanax senticosus. The screening method specifically includes the following steps that 1, totalRNAs of all kinds of tissue of acanthopanax senticosus are extracted separately; 2, RNA-Seq sequencing is conducted after reverse transcription of the total RNAs is conducted, and Trinity software isused for conducting unigene assembly; 3, unigenes related to synthetic accumulation of acanthopanax senticosus saponins, SOD and the like are screened out, and MicroSatellite software is used for screening SSR sites on the unigenes; 4, Primer3.0 software is used for designing primers, and the primers are synthesized; 5, the extracted genomic DNAs serve as a template for PCR amplification; 6, PCRproducts are subjected to electrophoresis detection, and target gene SSR primers are developed. By means of the method and application, the candidate SSR molecular markers related to target traits areefficiently and more specifically developed, and effective markers are provided for molecular identification of excellent acanthopanax senticosus germplasm resources. An important meaning is achievedfor conducting early identification and speeding up a breeding process of the excellent acanthopanax senticosus germplasm resources.
Owner:DALIAN NATIONALITIES UNIVERSITY
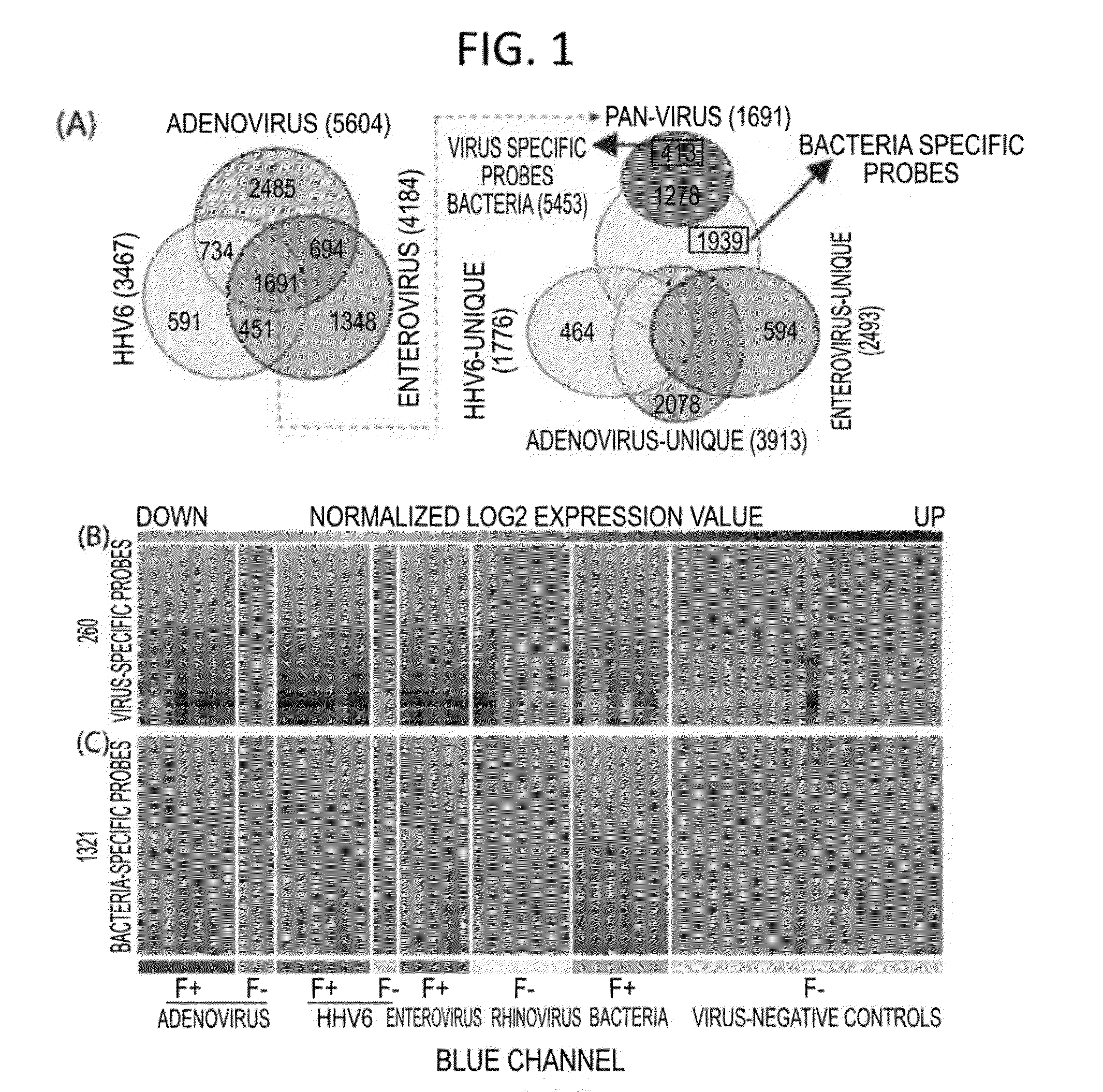
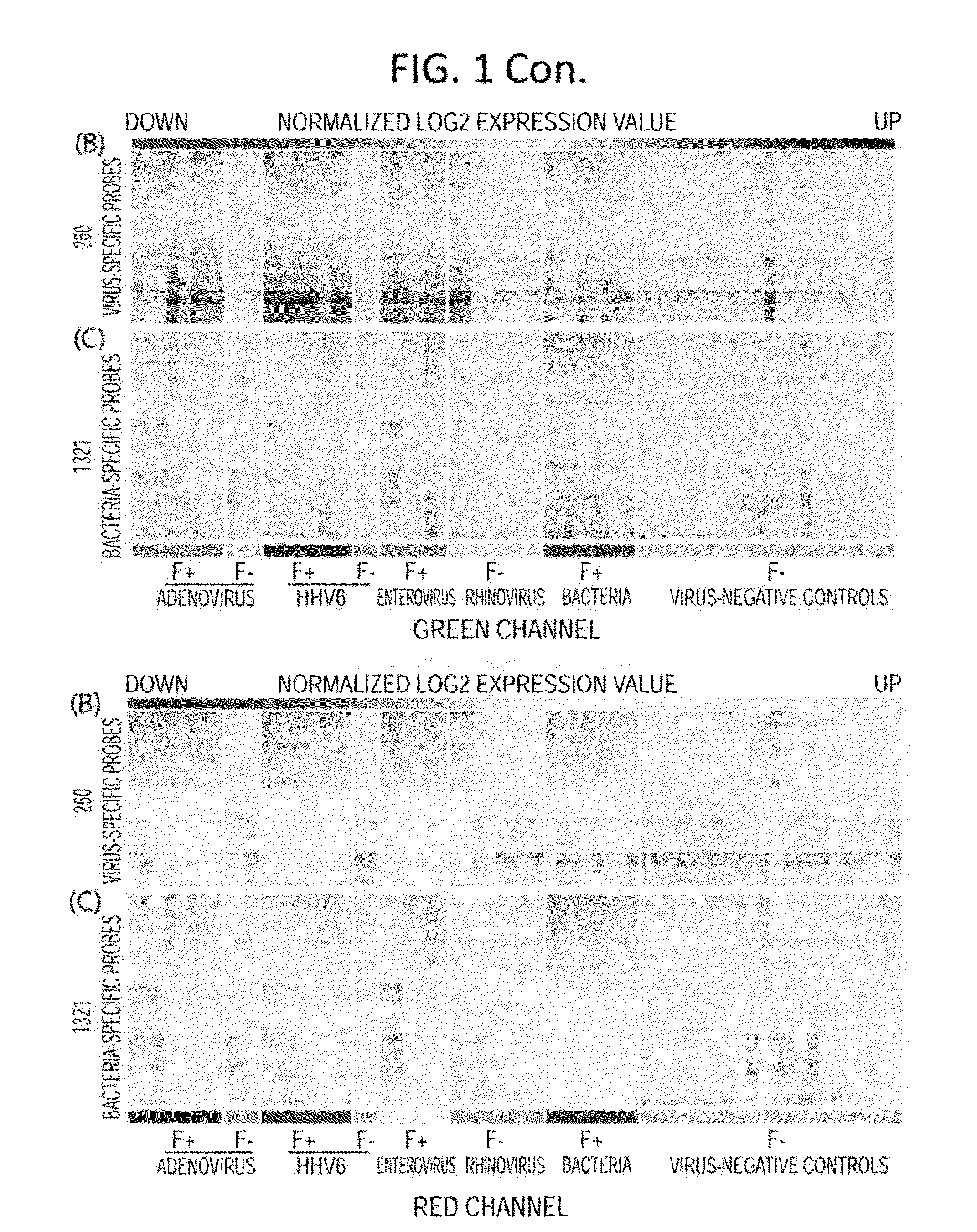
Diagnostic methods for infectious disease using endogenous gene expression
Disclosed are methods of diagnosis of a pathogen-associated disease. These methods comprise: providing a biological sample from a human subject; determining presence, absence and / or quantity of a bacterial pathogen, a viral pathogen, or a combination thereof, by a pathogen culture, a serum antibody detection test, a pathogen antigen detection test, a pathogen DNA and / or RNA detection test, or a combination thereof; determining in the sample, expression levels of at least one endogenous gene in which aberrant expression levels are associated with infection with a pathogen, by a microarray hybridization assay, RNA-seq assay, polymerase chain reaction assay, a LAMP assay, a ligase chain reaction assay, a Southern, Northern, or Western blot assay, an ELISA or a combination thereof. The subject can be diagnosed with the disease if the subject comprises the pathogen and an aberrant level of expression of an endogenous gene.
Owner:WASHINGTON UNIV IN SAINT LOUIS
Sub-population detection and quantization of receptor-ligand states for characterizing inter-cellular communication and intratumoral heterogeneity
A system for characterizing intercellular communication and heterogeneity in cancer tumors, and more particularly a method for detecting sub-populations and receptor-ligand states for providing predictive information in relation to cancer and cancer treatment is disclosed. The system comprises the steps of obtaining from a NGS sequencer, single-cell RNA-seq for a plurality of cells within a tumor, correlation with a plurality of data sets from a curated gene list of receptor-ligand pairs, normalizing their transcript abundance data, assigning states (e.g. 0,1,2,3) to each curated receptor-ligand pair in each cell (e.g. depending on {L:R}={0:0, 0:1, 1:0, 1:1}), thereby forming a matrix of receptor-ligand states, extracting sub-groups from the matrix that are not invariant and applying unsupervised clustering methods to identifying sub-clusters, identifying sub-populations within the set based on pair-wise distances between individual cells and similarity of cellular transcriptomes, identifying expressed ligands and receptors across the sub-populations, cross-referencing against the curated set of receptor-ligand pairs and providing a visually display the results by a mapping module for the clinician. The method can be used to study intercellular communication to elicit the etiology of diseases, and can be used to measure the disruption of intercellular communication to diagnose similarly disrupted disease patterns across patients.
Owner:KONINKLJIJKE PHILIPS NV
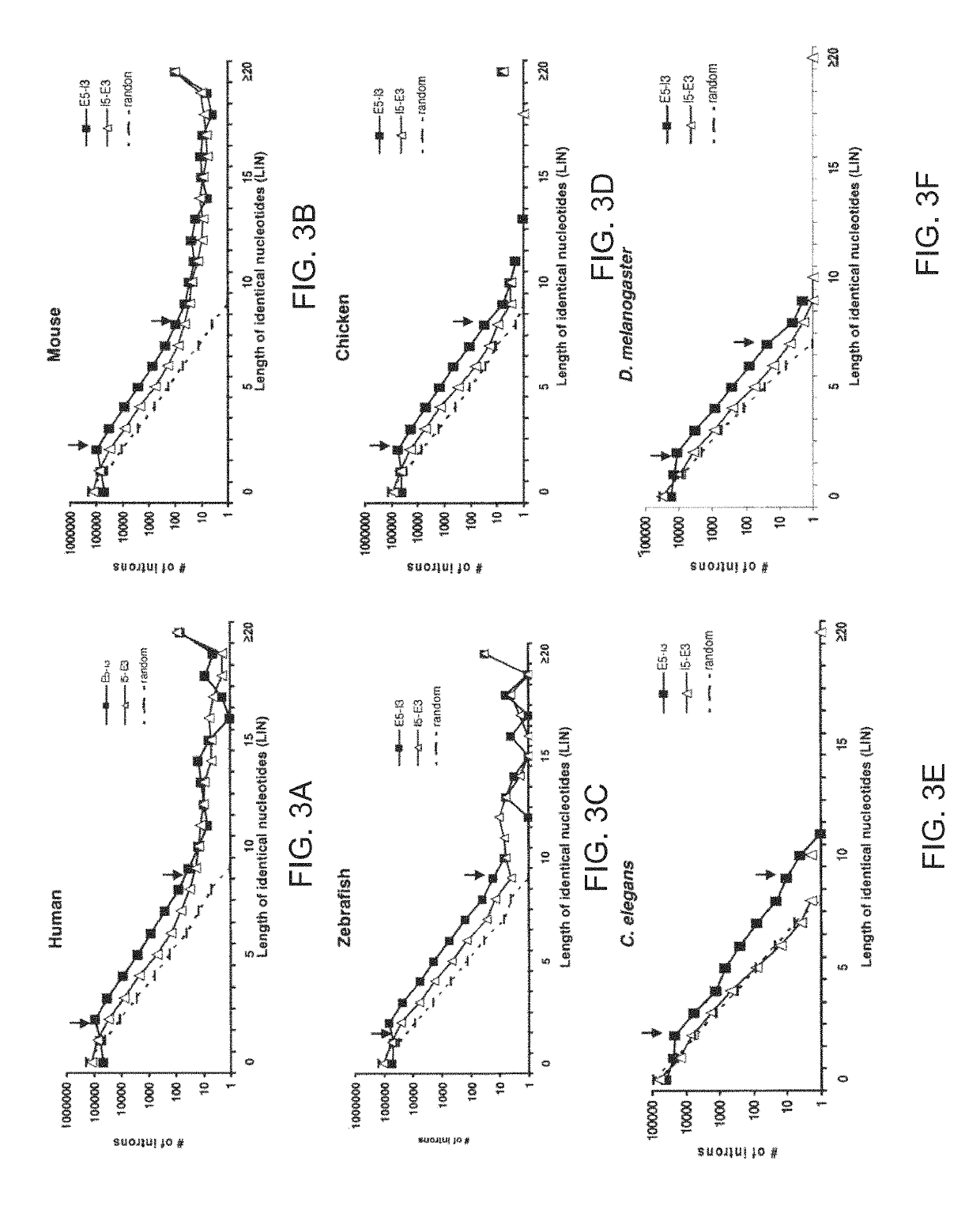
System and method for analyzing splicing codes of spliceosomal introns
ActiveUS10446261B1Lower Level RequirementsMicrobiological testing/measurementProteomicsDiseaseGenomic DNA
A system and method for analyzing splicing codes of spliceosomal introns is disclosed. One embodiment comprises methods of identifying introns and exons in genomic DNA or pre-mRNA sequences by locating characteristic markers in splicing junctions by computation and / or manually. Exon sequences predicted by computation can be verified and characterized by employing standard amplification methods, such as comparative genomic, RNA-seq, next-generation sequencing, RT-PCR. DNA / RNA / oligo, electrophoretic or protein chip technologies. If a given sample is verified, its polypeptide can be translated based on genetic codons. Its functions can be deduced based on its characteristics, computation predictions and related knowledge databases. These data can be used to compare databases which correlate the characterized intron or exon or gene to characterized diseases or genetic mutations. Isoforms can be detected and analyzed at mRNA and protein levels alone and with other isoforms predicted by computation, characterized by experiments and stored in existing databases.
Owner:BIOTAILOR
NF-kappaB dual-luciferase report cell capable of stably expressing pig derived TLR8 receptor gene, and construction method of NF-kappaB dual-luciferase report cell
InactiveCN107794245ARich researchReliable resultsGenetically modified cellsPeptidesHigh-Throughput Screening MethodsRenilla luciferase
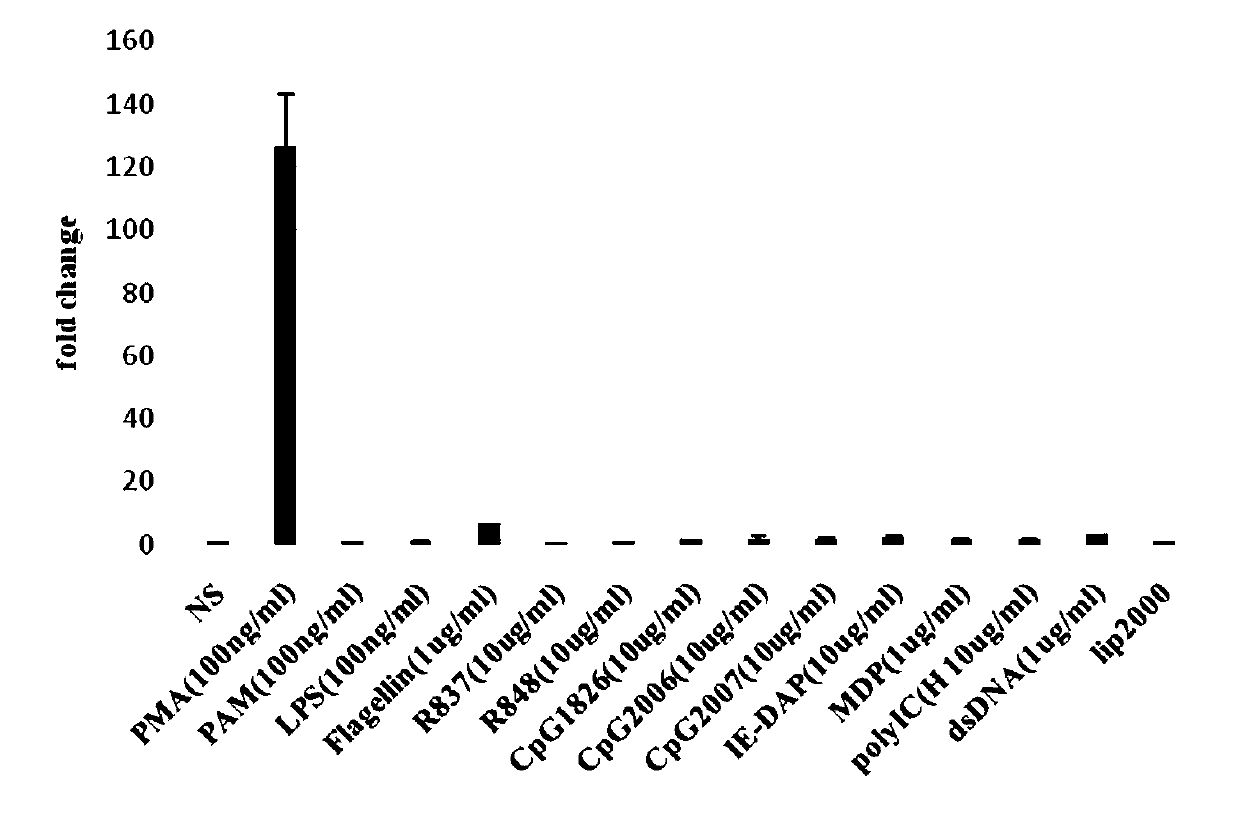
The invention belongs to the field of biological technology, and in particular relates to an NF-kappaB dual-luciferase report cell line capable of stably expressing a pig derived TLR8 receptor gene, and a construction method of the NF-kappaB dual-luciferase report cell line. The cell line is an HEK293-NF-kappaB cell, in which a pTLR8 gene is transferred; and the HEK293-NF-kappaB cell is an HEK293cell, in which NF-kappaB luciferase and endodontic luciferase are introduced. The invention provides a stable cell line, which can be used for researching pTLR8 activated downstream NF-kappaB promoterreport gene detection. The invention also provides a biological material for an RNA-Seq technology to perform high-throughput screening on differential expression genes in the stable cell under the bTLR8 silent and activated states, and lays the foundation and a platform for further researching the species specificity of TLR8.
Owner:YANGZHOU UNIV
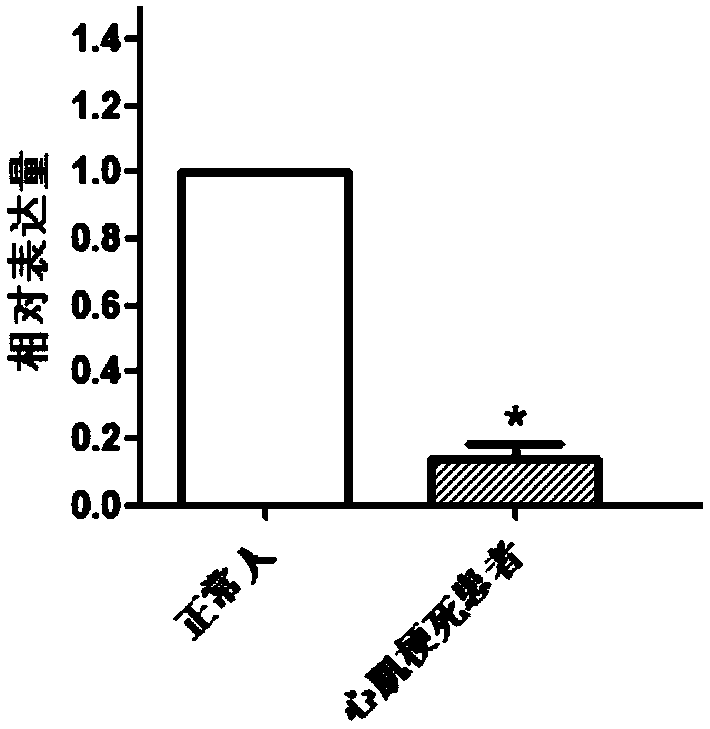
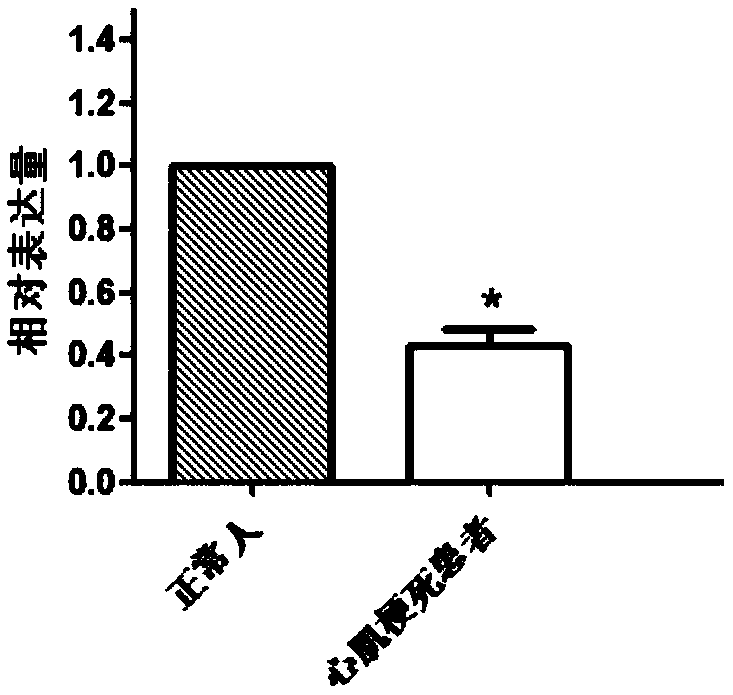
DOCK10 gene for early diagnosis of myocardial infarction
InactiveCN108796068AAchieve early diagnosisReduce mortalityMicrobiological testing/measurementDisease diagnosisDiagnosis earlyBiomarker (petroleum)
The invention discloses a myocardial infarction diagnosis related gene. Genes in differential expression between a myocardial infarction patient and a normal person are screened through RNA-seq, and according to large sample QPCR verification, contents of a DOCK10 gene and DOCK10 protein in blood of the myocardial infarction patient are lower than those in blood of the normal person. Therefore, the DOCK10 gene and the DOCK10 protein can be used as biomarkers for diagnosis of myocardial infarction. By research results, a noninvasive method is provided for clinical early diagnosis of myocardialinfarction and suitable for clinical popularization.
Owner:QINGDAO MEDINTELL BIOMEDICAL CO LTD
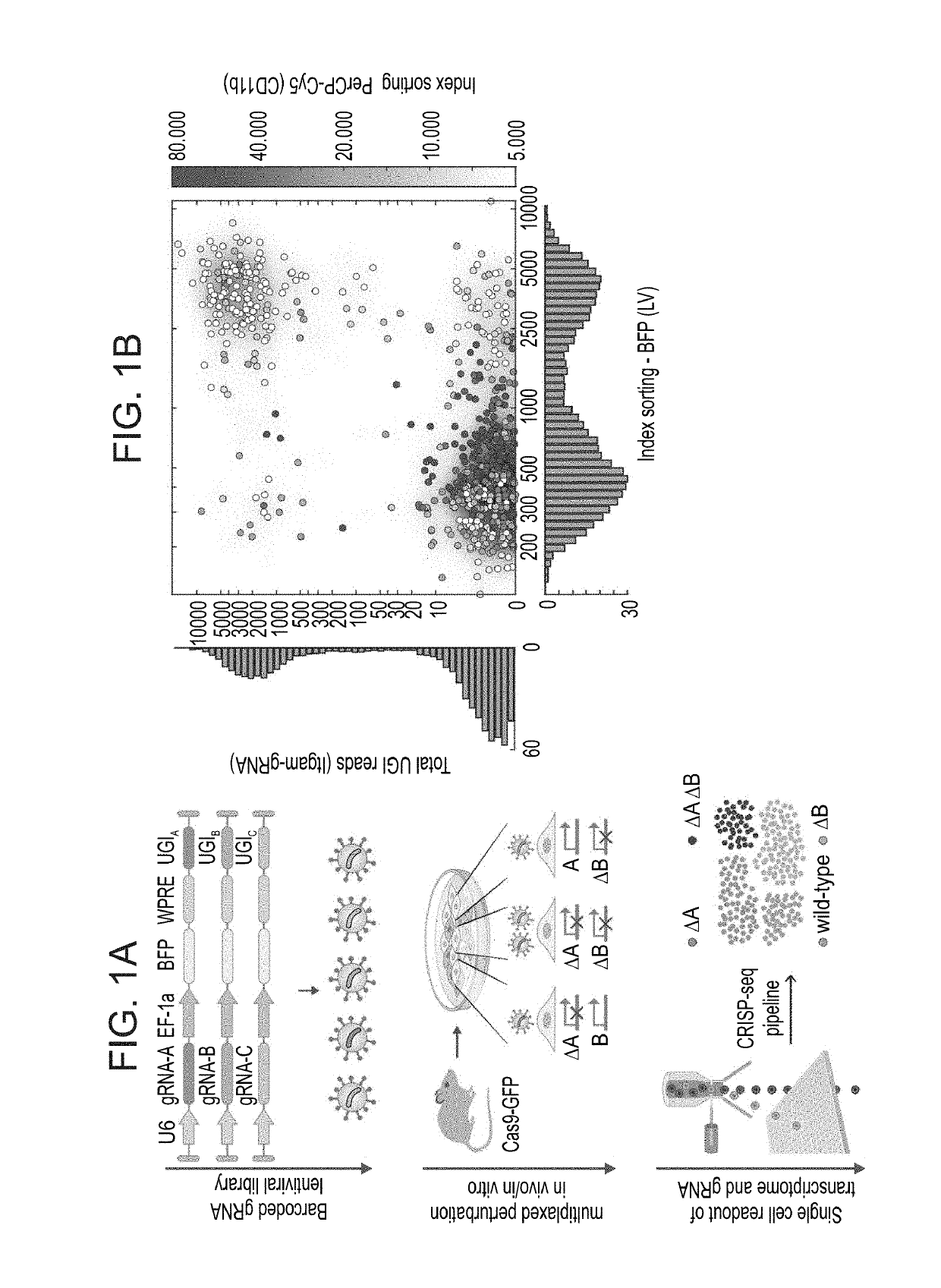
Crisp-seq, an integrated method for massively parallel single cell rna-seq and crispr pooled screens
An expression construct is disclosed which comprises:(i) a DNA sequence which encodes at least one guide RNA (gRNA) operatively linked to a transcriptional regulatory sequence so as to allow expression of the gRNA in a target cell;(ii) a barcode sequence for identification of the at least one gRNA operatively linked to a transcriptional regulatory sequence so as to allow expression of the barcode sequence in the target cell.
Owner:YEDA RES & DEV CO LTD
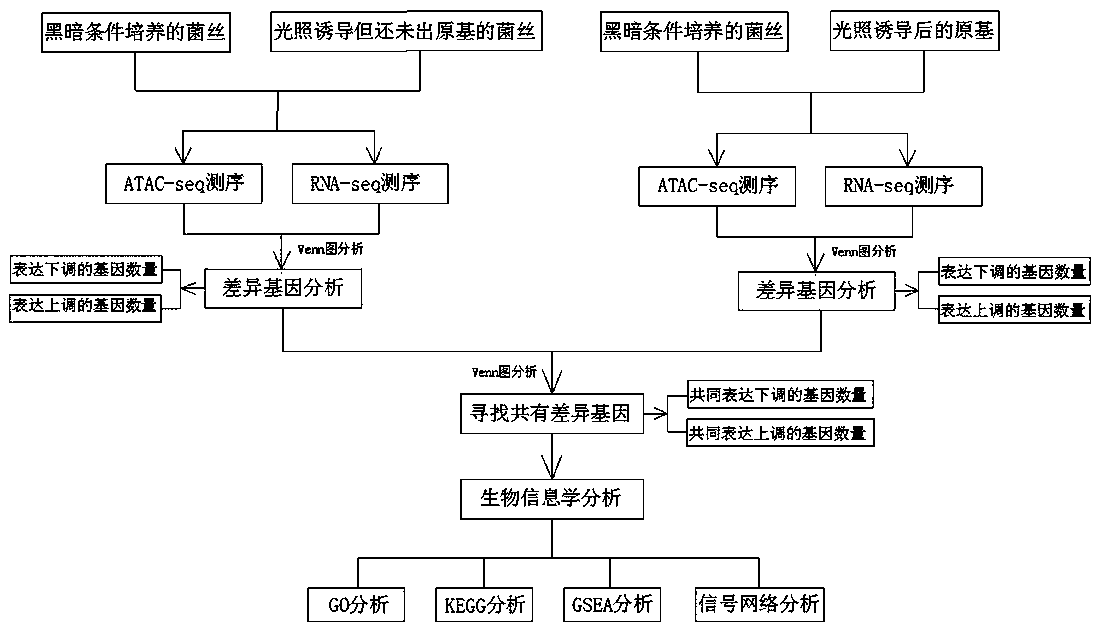
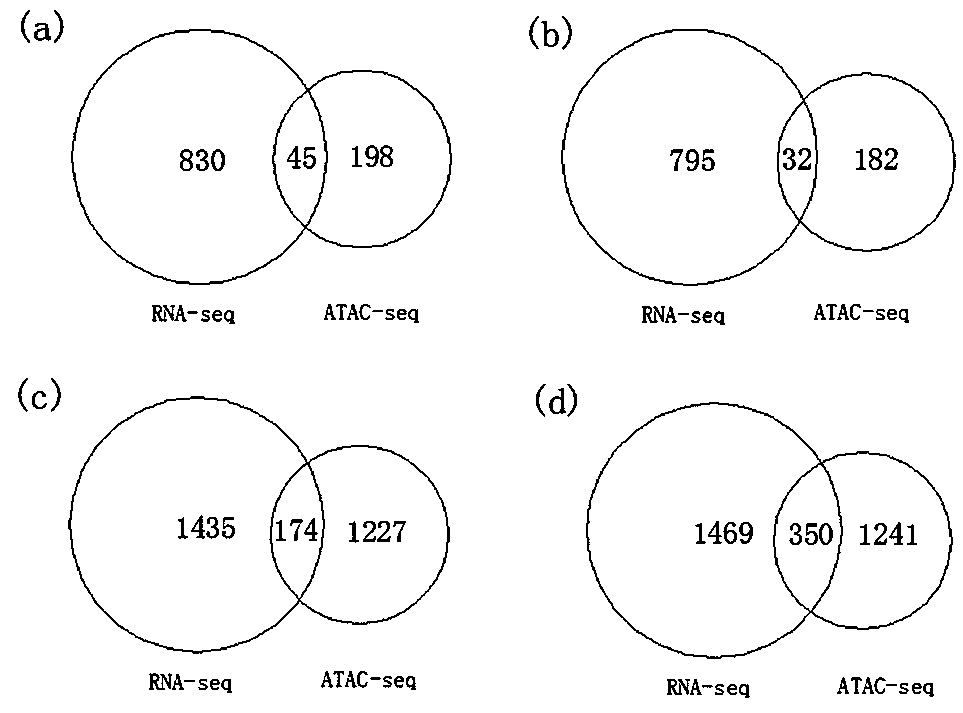
Method for screening functional genes of edible and medicinal fungi by combining ATAC-seq with RNA-seq
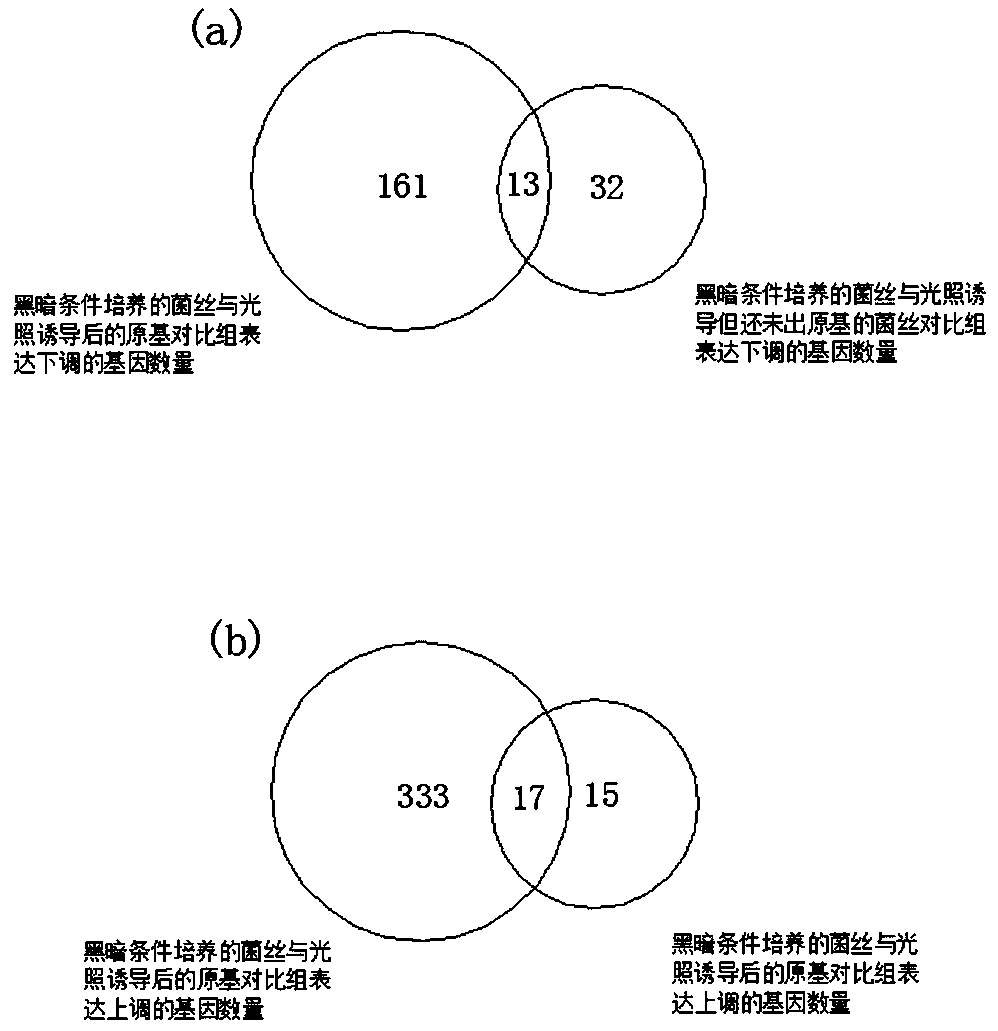
PendingCN109837335AImprove efficiencyImprove featuresMicrobiological testing/measurementBiotechnologyPrimordium
The invention discloses a method for screening functional genes of edible and medicinal fungi by combining ATAC-seq with RNA-seq. The method comprises the following steps: testing mycelia of the edible and medicinal fungi at a mycelium culture stage before and after induction of environmental factors and an open chromatin area of primordium after primordium differentiation by induction of environmental factors with an ATAC-seq method and analyzing coding genes of the open area, testing mycelia of the edible and medicinal fungi at a mycelium culture stage and an mRNA sequence after primordium differentiation with an RNA-seq method, searching common differential genes of gene sequences obtained by the ATAC-seq method and the RNA-seq method by use of a Venn map, and finally, searching key functional gene sequences influencing primordium differentiation of the edible and medicinal fungi in the common differential genes by biological analysis.
Owner:INST OF EDIBLE FUNGI FUJIAN ACAD OF AGRI SCI
NF-kB dual-luciferase reporter cell for stably expressing swine TLR5 receptor genes and construction method
InactiveCN107760653AMicroorganism based processesPeptidesBiotechnologyHigh-Throughput Screening Methods
The invention belongs to the technical field of biology and in particular relates to an NF-kB dual-luciferase reporter cell for stably expressing swine TLR5 receptor genes and a construction method thereof. A cell line refers to HEK293-NF-kB cells in which the swine TLR5 receptor genes are transferred, and the HEK293-NF-kB cells refer to HEK293 cells in which NF-kB firefly luciferase and renilla luciferase are transferred. The invention provides the stable cell line used in detection for researching pTLR5 activated downstream NF-kB promoter report genes. The invention also provides a biological material for performing high-throughput screening on differentially expressed genes in stable cells in quiesced and activated states in an RNA-Seq technology, and lays a foundation and platform forfurther researching species specificity of TLR5.
Owner:YANGZHOU UNIV
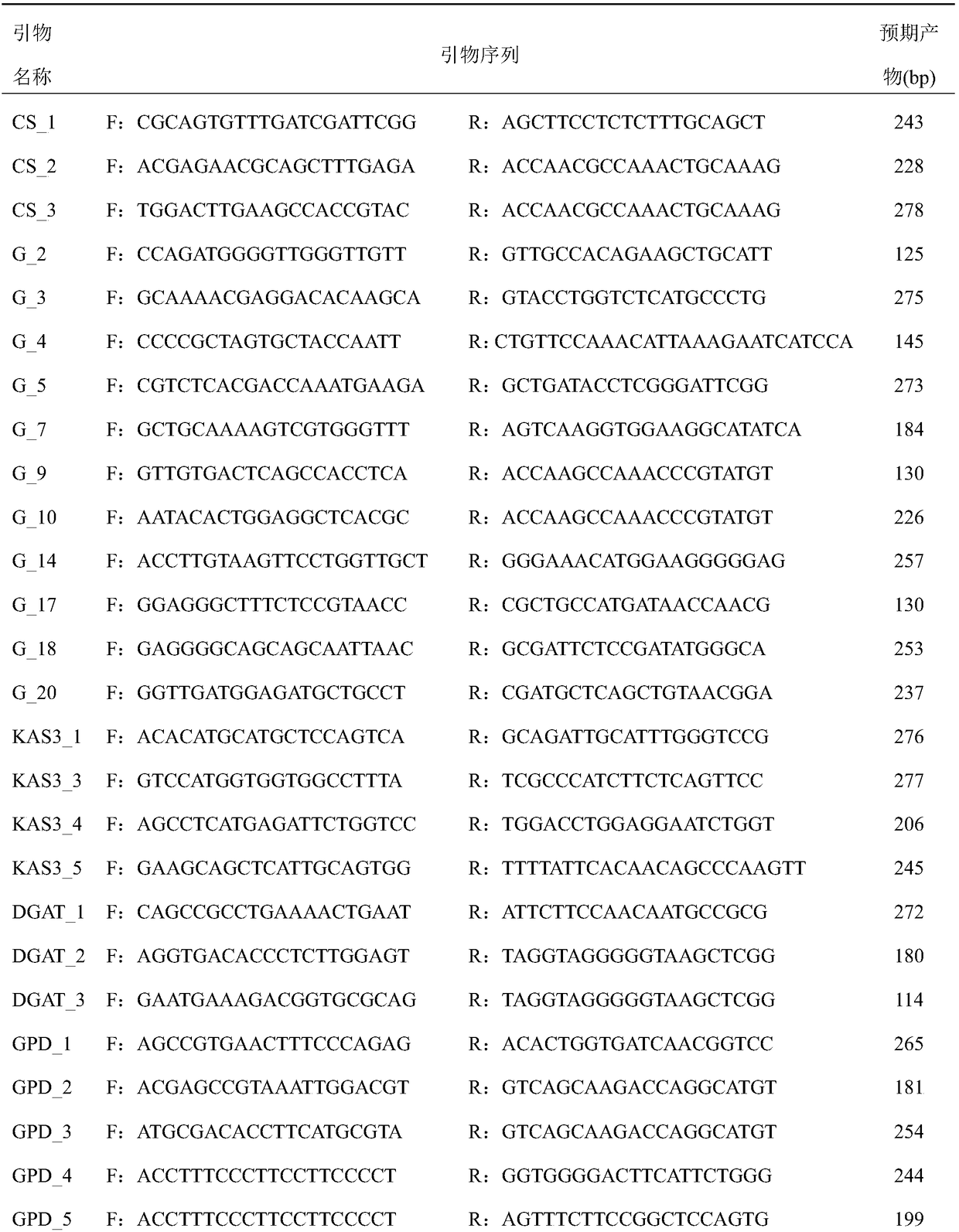
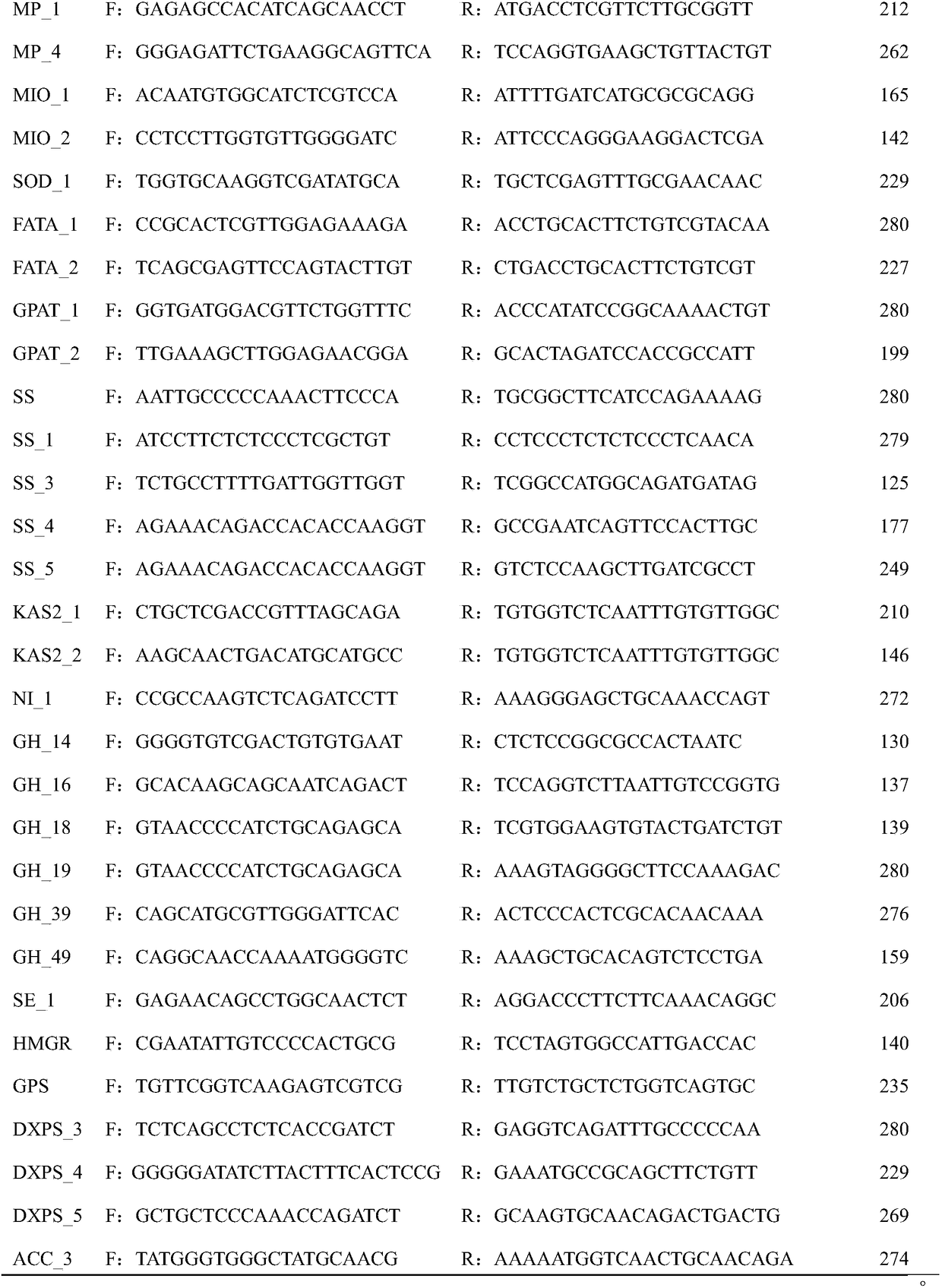
Lentil KASP marker developed based on RNA-Seq and application thereof
ActiveCN109929947AImprove conversion rateEasy to detectMicrobiological testing/measurementDNA/RNA fragmentationForward primerGenetics
Owner:SHANDONG CROP GERMPLASM CENT +1
NF-kB dual-luciferase reporting cell line for stably expressing human TLR8 receptor gene and construction method thereof
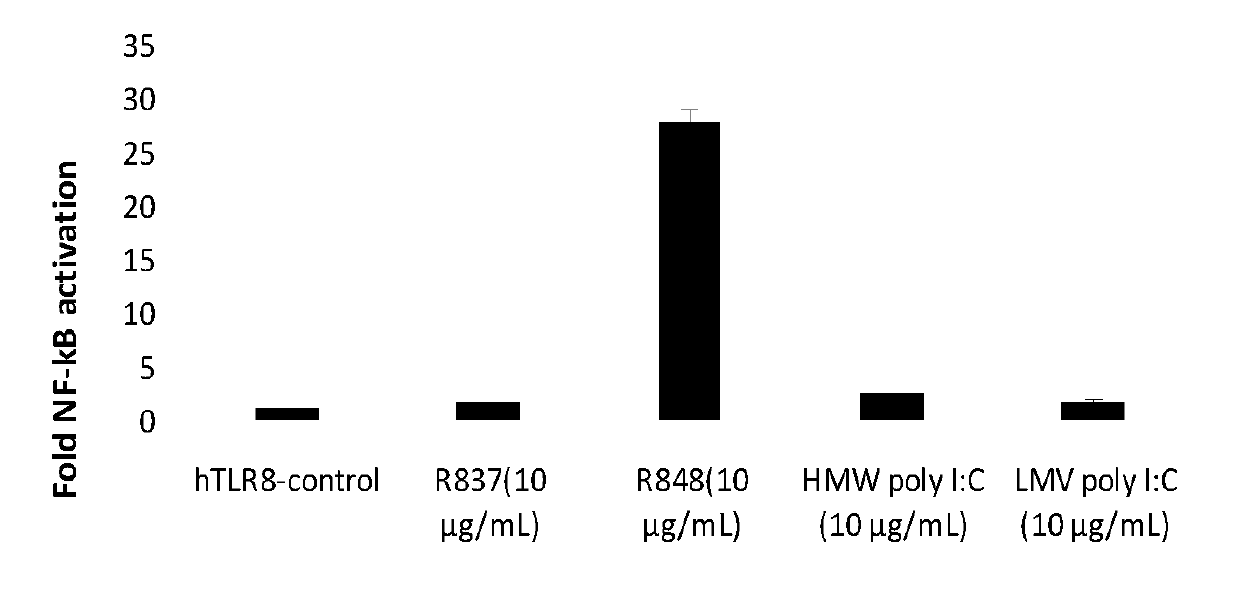
InactiveCN107828733ACorrect and avoid errorsReliable resultsGenetically modified cellsMicroorganism based processesHigh-Throughput Screening MethodsRenilla luciferase
The invention belongs to the technical field of biology and particularly relates to an NF-kB dual-luciferase reporting cell line for stably expressing human TLR8 receptor gene and a construction method thereof. The cell line is HEK293-NF-kB cells with transgenosis of the human TLR8 gene, and the HEK293-NF-kB cells are HEK293 cells with introduction of NF-kB luciferase and internal reference ranilla luciferase. The NF-kB dual-luciferase reporting cell line and the construction method also provide a biological material for studying the action of TLR8 ligands in tumor treatment and utilizing an RNA-Seq technology to screen genes for stabilizing differential expression in the cells under the silent and activated states of hTLR8 by high flux, and lays foundation and platform for further studying the species specificity of the TLR8.
Owner:YANGZHOU UNIV
Compositions and methods for constructing strand specific cdna libraries
Provided herein are compositions, kits and methods for the production of strand-specific cDNA libraries. The compositions, kits and methods utilize properties of double stranded polynucleotides, such as RNA-cDNA duplexes to capture and incorporate a novel sequencing adapter. The methods are useful transcriptome profiling by massive parallel sequence, such as full-length RNA sequencing (RNA-Seq) and 3′ tag digital gene expression (DGE).
Owner:RGT UNIV OF CALIFORNIA
Identification of toxoplasma gondii chronic infection male mouse genital system target gene and clinical application thereof
ActiveCN108060220AFast and effective transcriptomic researchMicrobiological testing/measurementSequence analysisRNA extractionMouse Epididymis
The invention provides a method for applying an RNA-seq technology to research toxoplasma gondii chronic infection mouse epididymis tissue differential expression genes. The method comprises the following steps of (1) extracting RNA of mouse epididymis tissues; (2) carrying out library construction and sequencing for the RNA obtained in the step (1); (3) carrying out bioinformatics analysis on a sequencing result of the step (2), obtaining the toxoplasma gondii chronic infection mouse epididymis tissue differential expression genes, and screening and identifying reproduction-related genes. Themethod for the toxoplasma gondii chronic infection mouse epididymis tissue differential expression genes provided by the invention can be used for quickly and effectively researching a gene expression difference, and identifying whether an epididymis is infected with toxoplasma gondii or not through using a qPCR method for detecting a screened target gene, has an important theoretical value and an application prospect on improving reproductive potential of male animals and animal reproduction rate, and is beneficial to quickly positioning detection targets and prevention and cure drug targetswith research values or commercial application values by adopting the genes as molecular markers for identifying whether reproductive systems of the male animals are infected with the toxoplasma gondii or not.
Owner:SOUTH CHINA AGRI UNIV
Isolation and cloning and function analysis of endogenous bidirectional expression promoter of rice
ActiveCN106676107AHigh-efficiency bidirectional promoter resourcesMicrobiological testing/measurementDNA/RNA fragmentationPlant genetic engineeringProtein activity
The invention belongs to the field of plant genetic engineering, and relates to isolation and cloning and function analysis of an endogenous bidirectional expression promoter of rice. A candidate bidirectional promoter BIP1 is selected from a rice genome by combining RNA-seq data in MSU and RAP databases and expression profile chip data in a CREP database, and an experiment proves that the candidate bidirectional promoter BIP1 has bidirectional expression activity; 5' and 3' deletion analysis is carried out, a bidirectional expression control section and a section for controlling the basic expression activity in 5' and 3' directions are identified; and then the conservatism of the BIP1 in gramineous plants is analyzed by using means of bioinformatics. The invention discloses a screening and isolation and cloning method of the bidirectional promoter, a building process of a transformation vector, a genetic transformation process of the rice, a GUS histochemical binding method of a transformation plant, a GUS protein activity detection method, a GFP histological analysis method, a GFP quantitative analysis method and a conservatism analysis method of the bidirectional promoter.
Owner:HUAZHONG AGRI UNIV
A new lncRNA extracted from porcine intramuscular adipose tissue and its application
ActiveCN107904242BSpeed up entryMicrobiological testing/measurementBiological testingBiotechnologyIntramuscular fat
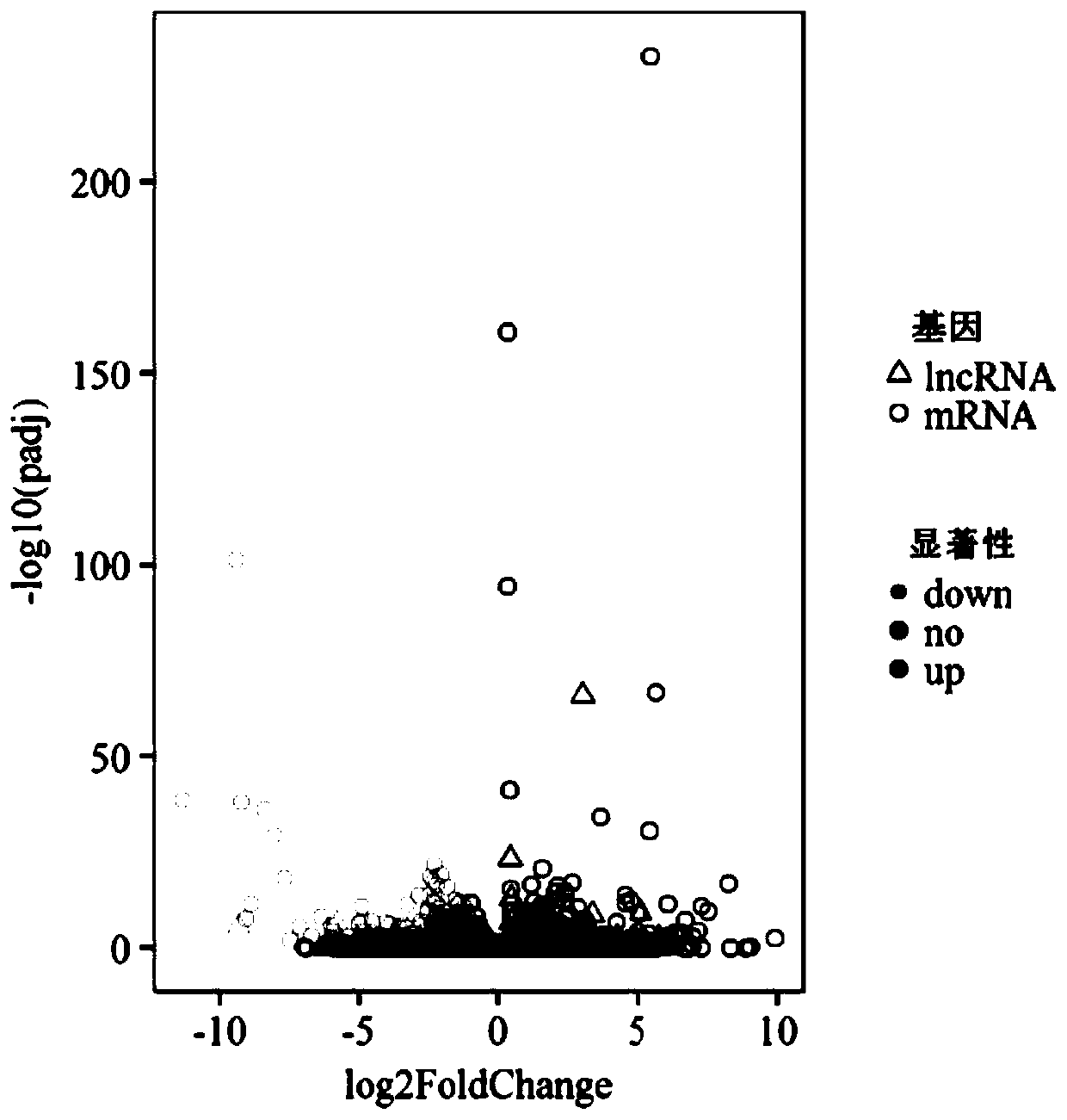
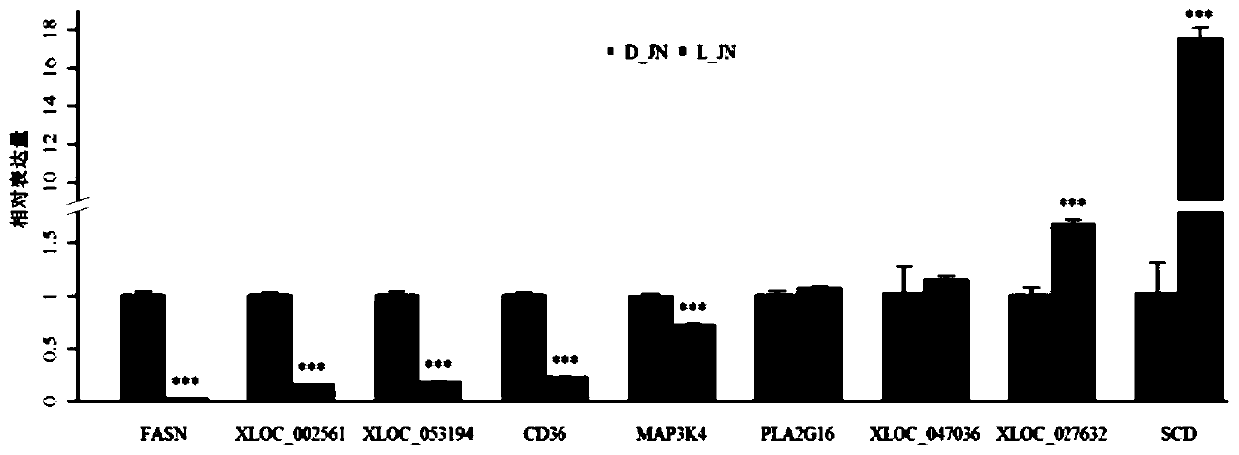
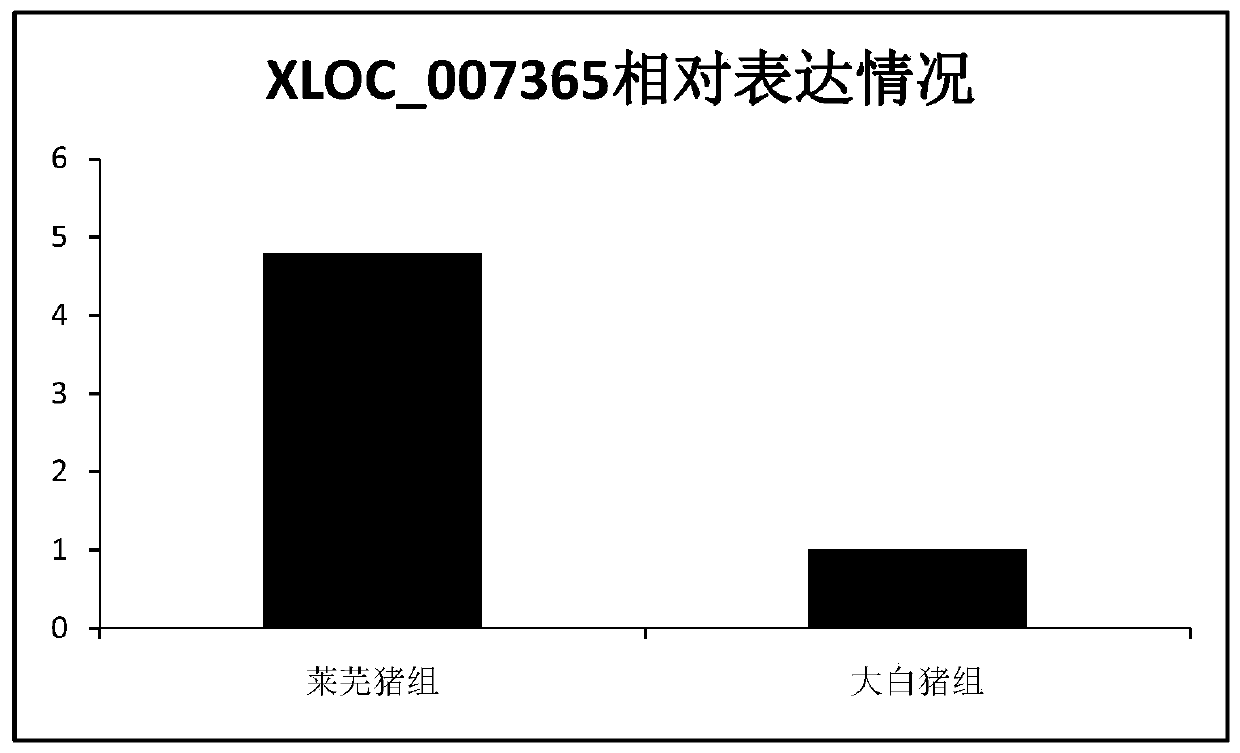
The invention discloses new lncRNA extracted from pig intramuscular fat tissue and application of the new lncRNA. A RNA-seq technology and a bioinformatics method are adopted to compare the gene expression profiles of the intramuscular fat tissues of a large white pig and a Laiwu pig, and the key differentially expressed lncRNA related to adipogenic differentiation and lipid metabolism are identified and screened out. Molecular verification experiments indicate that the lncRNA and the target genes PLEKHG3, SERPINB7, SPTLC3 and TMOD2 thereof are all closely related to the pig intramuscular fat.The invention lays a theoretical basis for the pig lipidosis mechanism study and livestock fat tissue lncRNA study, and has an important application prospect in molecular breeding of livestock.
Owner:INST OF ANIMAL SCI OF CHINESE ACAD OF AGRI SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com