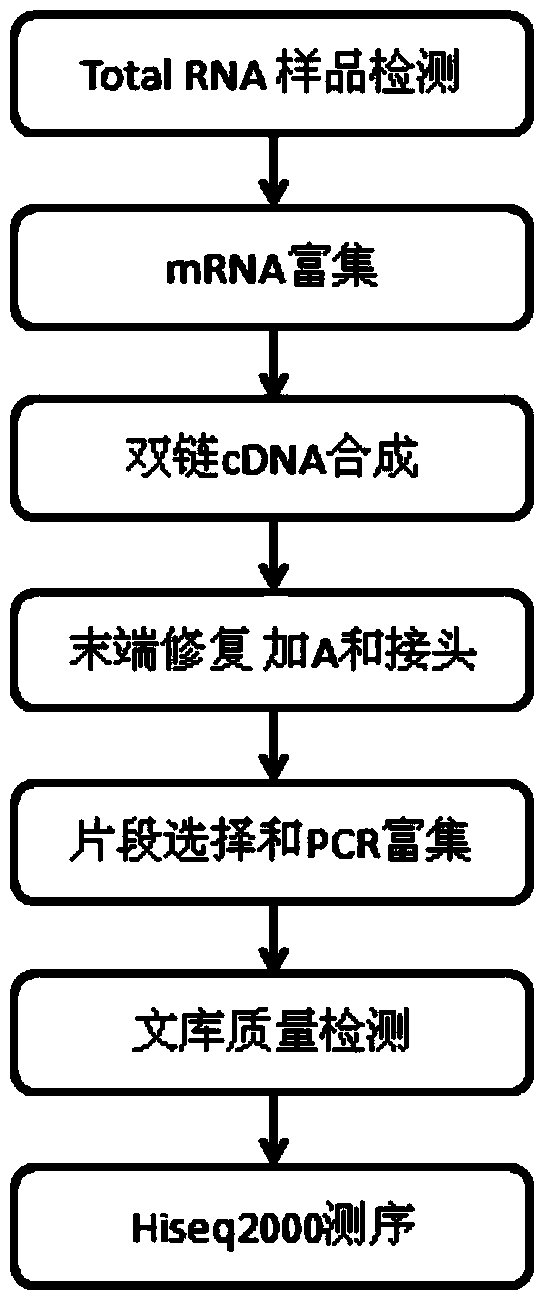
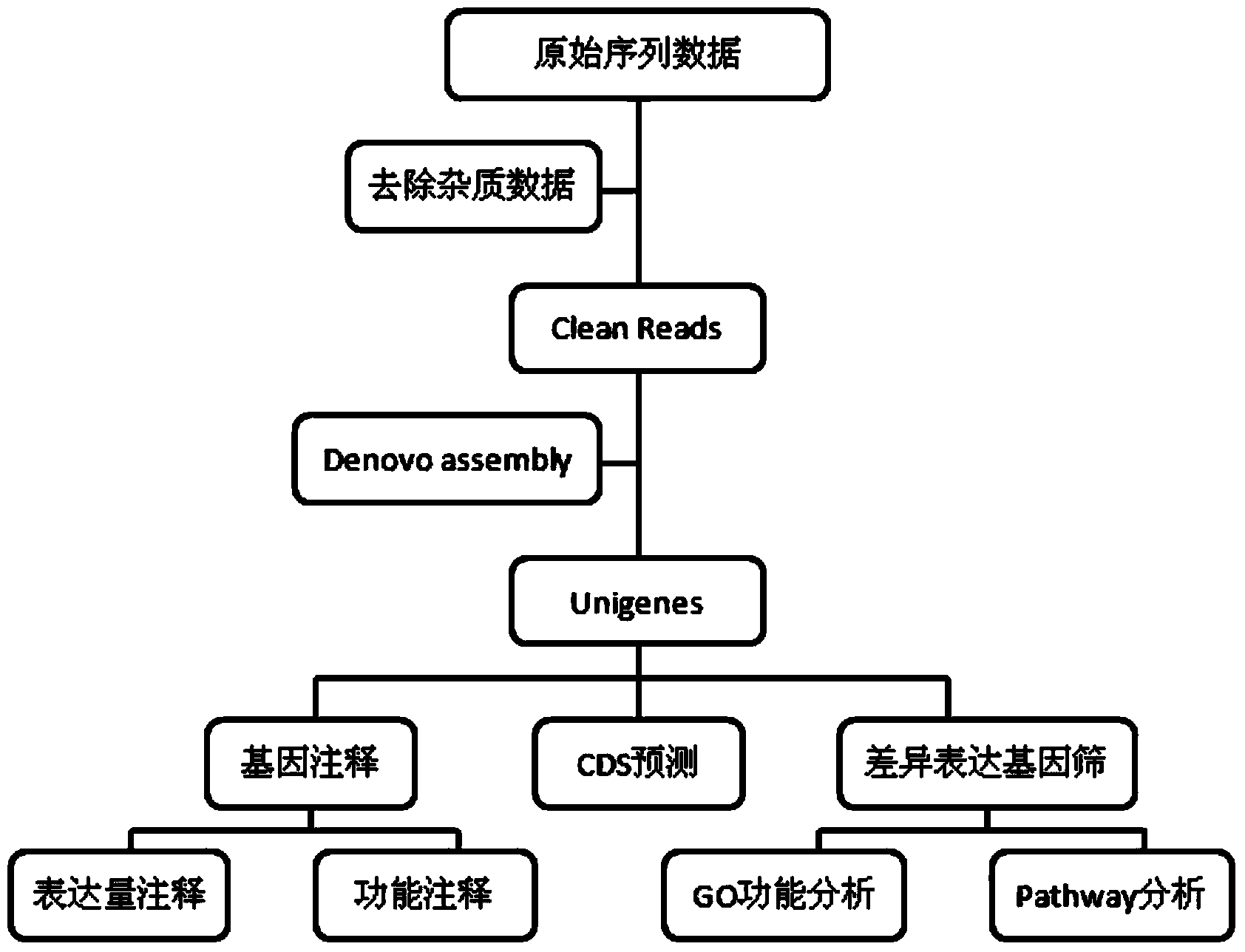
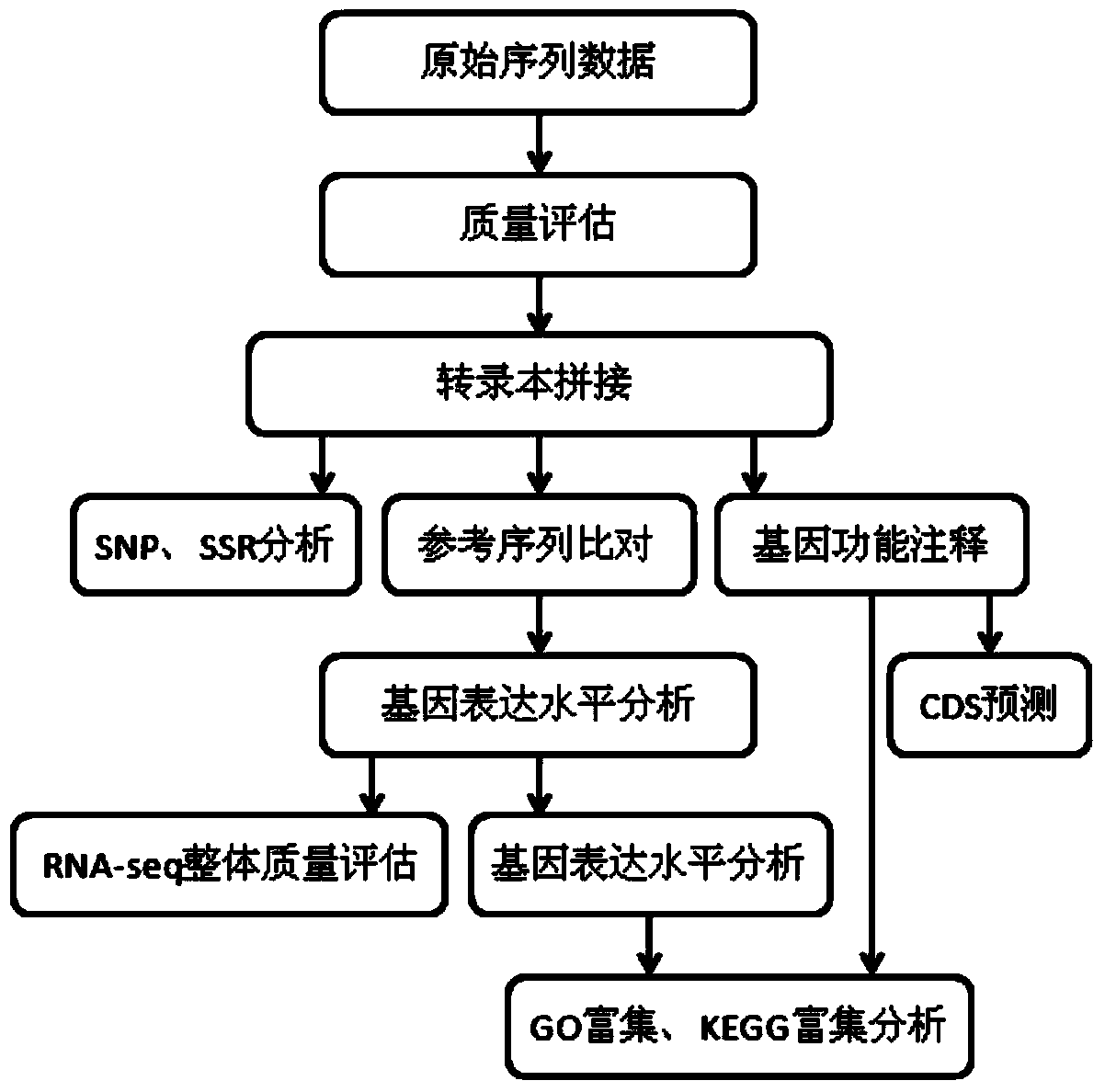
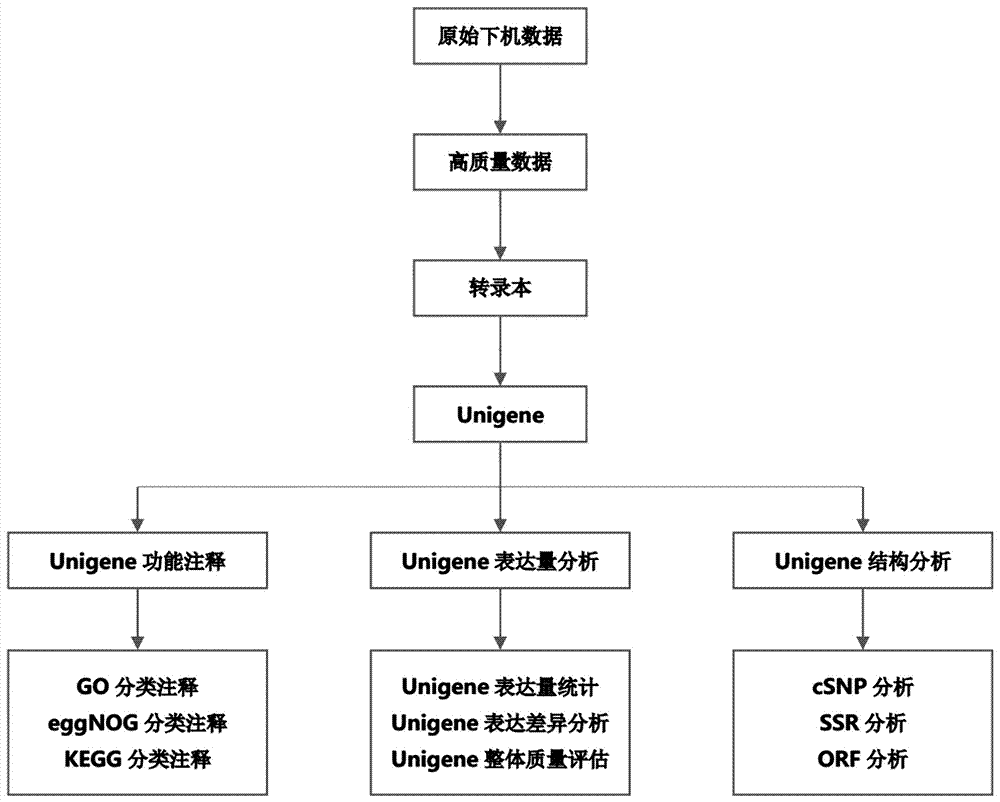
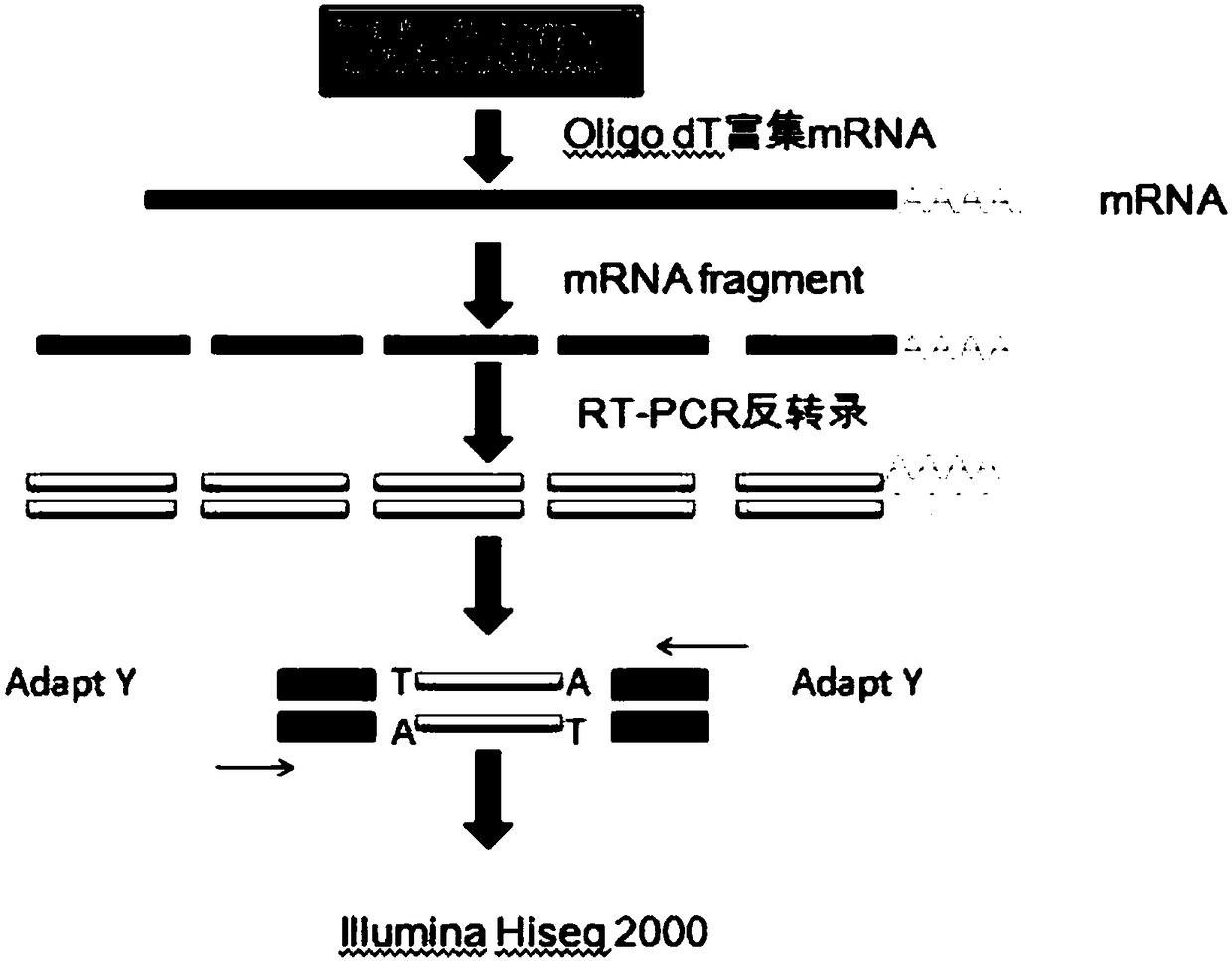
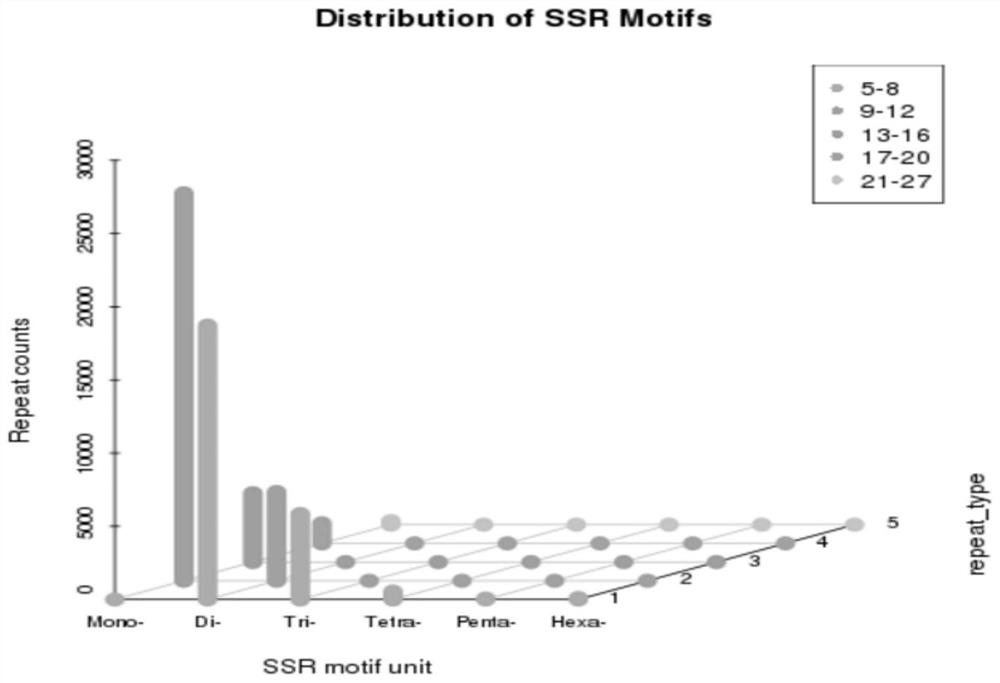
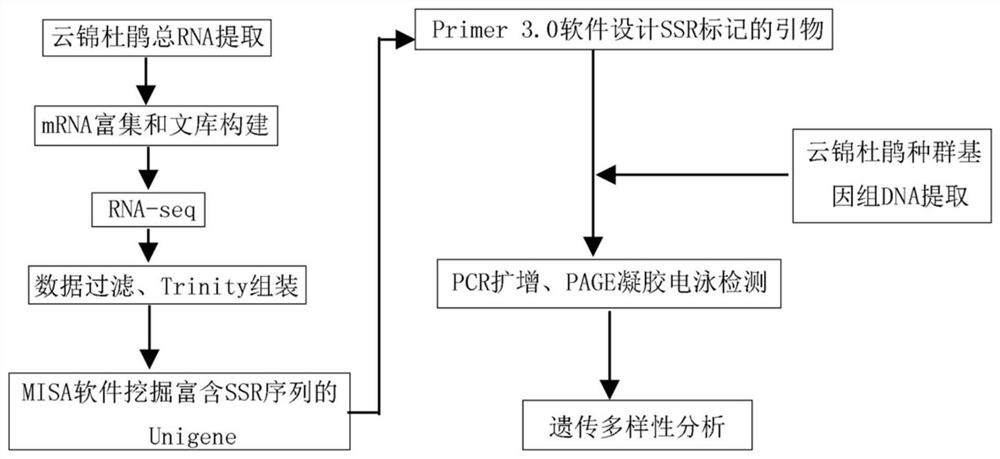
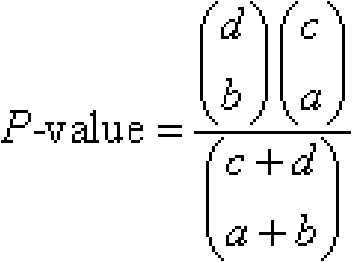Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
44 results about "UniGene" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
UniGene is a NCBI database of the transcriptome and thus, despite the name, not primarily a database for genes. Each entry is a set of transcripts that appear to stem from the same transcription locus (i.e. gene or expressed pseudogene). Information on protein similarities, gene expression, cDNA clones, and genomic location is included with each entry.
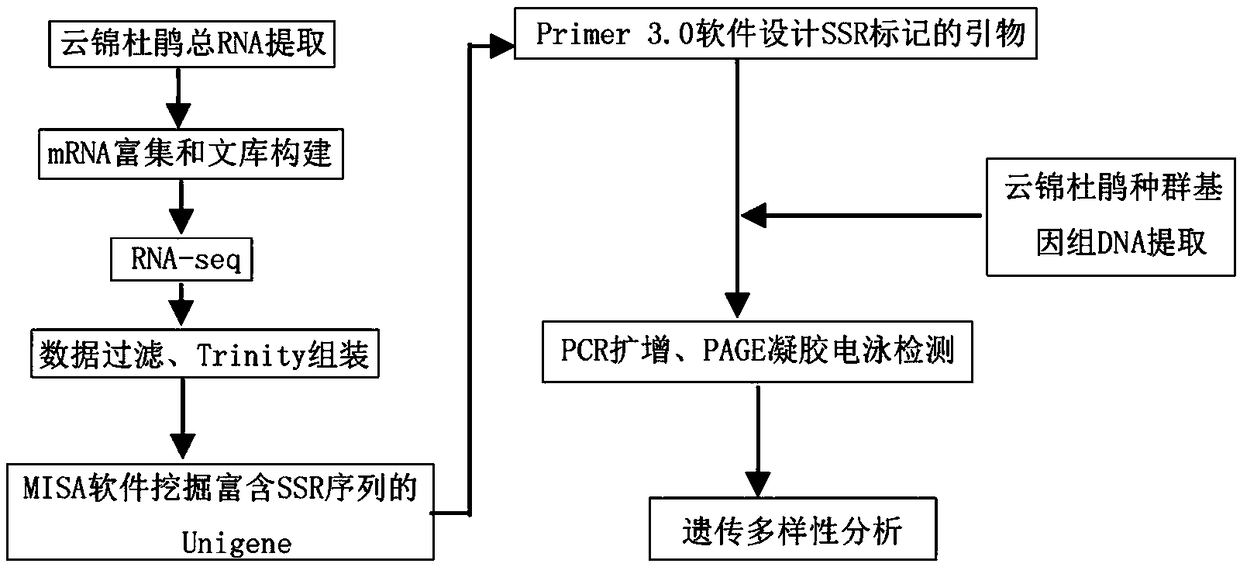
Method for developing mung bean simple sequence repeat (SSR) primer based on transcriptome sequencing
InactiveCN103642912AImprove design success rateSimple designMicrobiological testing/measurementDNA/RNA fragmentationSequence databaseTranscriptome Sequencing
The invention provides a method for developing a mung bean simple sequence repeat (SSR) primer based on transcriptome sequencing. The method comprises the following steps: obtaining a set of mung bean genome-wide transcription, and forming a sequence database; splicing sequencing sequences into a transcriptome by Trinity; taking the longest transcript in each gene as Unigene; carrying out bioinformatics analysis of a Unigene sequence; carrying out SSR detection on the Unigene by adopting MISA1.0; carrying out SSR primer design by using a Primer 3, and carrying out SSR primer polymorphism identification. 13134 pairs of SSR primers are successfully designed by application of the method; 50 pairs of primers are randomly selected to verify 8 parts of mung bean deoxyribonucleic acids (DNAs) from different countries, wherein 32 pairs of polymorphic primers are formed in all; the mung bean materials with different geographical origins can be distinguished by using the 32 pairs of SSR primers. The method disclosed by the invention is convenient, fast and accurate, and low in cost, and a new thought is provided for development of the mung bean SSR primer.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
Lathyrus quinquenervius EST-SSR primer group developed based on transcriptome sequencing, method and application
InactiveCN107345256ALow costMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceSequence database
The invention provides a lathyrus quinquenervius EST-SSR primer group developed based on transcriptome sequencing, a method and application. The method comprises the steps of obtaining a set of all root, stem and leaf transcripts of two lathyrus quinquenervius varieties of different flower colors at a seedling stage to form an original sequence database; removing low-quality sequencing data with a joint and forming a high-quality filtered sequence database; splicing high-quality filtered sequences, reducing the transcripts and carrying out BLAST comparison on the transcripts and a reference protein database NR, reserving the optimal comparison result and determining the maximal sequence as a Unigene representative sequence; carrying out SSR locus scanning in Unigene by using MISA 1.0; and carrying out SSR primer design by adopting primer 3.0, screening SSR primers and carrying out fingerprint map construction on 43 parts of lathyrus quinquenervius materials. The lathyrus quinquenervius EST-SSR primer group is convenient, fast, accurate, low in cost, and a new thought is provided for the development of lathyrus quinquenervius EST-SSR primers and germplasm resources identification.
Owner:山西省农业科学院农作物品种资源研究所 +1
Preparation method and application for Miscanthus Genic-SSR mark
InactiveCN102424826ASimple methodImprove efficiencyMicrobiological testing/measurementDNA preparationCritical energyTotal rna
The invention discloses a preparation method and application for a Miscanthus Genic-SSR mark. The method comprises the following steps: A, acquiring Miscanthus transcriptomic sequence, that is, selecting young and tender Miscanthus leaves, extracting total RNA respectively so as to obtain transcriptome Reads, removing the pollution of carrier RNA sequence and carrying out splicing so as to obtainthe Miscanthus transcriptomic sequence; B, identifying SSR sites in Miscanthus ESR sequence, that is, searching SSR sites in obtained non-redundant Unigene-EST sequence by using SSR retrieval software, with repeats being 2 to 6 bases, so as to obtain a series of Unigene sequences containing information about the SSR sites; C, designing Miscanthus Genic-SSR primers, that is, using primer design software to design primers according to the obtained Unigene sequences containing the SSR sites so as to obtain a series of the Miscanthus Genic-SSR primers. According to the invention, the method is simple, has high efficiency and is simple to operate; a huge amount of marks are obtained; EST sequence obtained through transcriptomic sequencing lays a foundation for genetic research, molecular improvement and the like of the critical energy plant Chinese silvergrass.
Owner:湖北光芒能源植物有限公司
Rhododendron fortunei SSR primer pair based on transcriptome sequencing development, screening method and application
ActiveCN109468405AFast wayAccurate methodMicrobiological testing/measurementDNA/RNA fragmentationSequence analysisGenetic diversity
The invention discloses a rhododendron fortunei SSR primer pair based on transcriptome sequencing development, a screening method and application, and belongs to the technical field of biology. The primers are obtained on the basis of transcriptome sequencing and sequence analysis, on the basis of transcriptome sequencing, a large number of data are screened, a unigene sequence which is rich in SSR sites is acquired and SSR labeled primers are designed, and the barriers of a small number of rhododendron fortunei SSR molecular markers and low development efficiency at present are overcome. By arhododendron fortunei group, the effectiveness of the SSR primers is verified, and a foundation is laid for genetic research such as rhododendron fortunei genetic diversity research, genetic linkagemap, QTL positioning and molecular marker-assisted breeding.
Owner:HUANGGANG NORMAL UNIV
Method for developing meconopsis SSR (simple sequence repeat) primers on basis of transcriptome sequences
PendingCN106834510ASolve the problem of few SSR primersAdd raw dataMicrobiological testing/measurementAgricultural scienceMeconopsis
The invention relates to a method for developing meconopsis SSR (simple sequence repeat) primers on the basis of transcriptome sequences. The method includes particular steps of (1), constructing transcriptome libraries; (2), carrying out sequencing, splicing sequenced sequences to obtain a complete transcriptome by the aid of software Trinity, utilizing each transcript with the longest genes as Unigene and carrying out bioinformatics analysis on Unigene sequences; (3), carrying out SSR detection on the Unigene by the aid of software MISA1.0; (4), designing SSR primers by the aid of software Primer3 and identifying the polymorphism of the primers. The method has the advantages that raw data for developing the primers can be greatly increased by the aid of the obtained transcriptome sequences; SSR markers can be developed on a large scale in bioinformatics ways, and accordingly the development efficiency can be greatly improved; the primers with the SSR markers are novel markers which are stably available, and can be directly used for related research on meconopsis plants, and accordingly the problem of scarceness of meconopsis SSR primers can be solved.
Owner:SOUTHWEST FORESTRY UNIVERSITY
Specific primers and screening method for EST-SSR (Express Sequence Tag-Simple Sequence Repeat) marker of torreya grandis transcriptome sequence
ActiveCN107287288AMicrobiological testing/measurementDNA/RNA fragmentationGerm plasmGenetic diversity
The invention belongs to the field of molecular biology DNA (Deoxyribonucleic Acid) marker technology and application, and particularly relates to specific primers and a screening method for an EST-SSR (Express Sequence Tag-Simple Sequence Repeat) marker of a torreya grandis transcriptome sequence. The screening method comprises the following steps: firstly, assembling fine torreya grandis seed kernel and circular torreya grandis seed kernel transcriptome data by adopting Trinity software to obtain a transcript sequence; secondly, carrying out SSR site research on Unigene with 1kb or above obtained by screening by using an MISA (MIcroSAtellite identification tool) program, and carrying out screening and separating on 2 to 6 base repeat units and repeat times according to microsatellite fragments with repeat times of 9, 8, 5, 5 and 5 in sequence, thereby obtaining an ESTs sequence with microsatellite repeat; thirdly, designing the primers by using a Primer 3 program; finally, comprehensively evaluating repeatability, stability and polymorphism of the EST-SSR marker to obtain the EST-SSR marker. The specific primers disclosed by the invention can be used for conveniently and quickly analyzing germ plasm resources of different types and genetic diversity of interspecific and intraspecific torreya grandis.
Owner:ZHEJIANG FORESTRY UNIVERSITY +1
Construction method of Plectropomus microsatellite DNA molecular markers
ActiveCN104357547AEfficient methodSimple methodMicrobiological testing/measurementTranscriptome SequencingMetapopulation
The invention discloses a construction method of Plectropomus microsatellite DNA molecular markers. According to the method, Unigene containing Plectropomus microsatellite repeat sequences is firstly screened out by transcriptome sequencing. Then microsatellite sites are detected in the Unigene sequences. Specific primers of microsatellite markers are designed according to the sequences of both ends of the microsatellite sites and are used for detecting polymorphism of the microsatellite sites. The microsatellite markers with abundant polymorphism of Plectropomus are exploited, and seven microsatellite DNA molecular markers of Plectropomus are verified further. The Plectropomus microsatellite DNA molecular markers provided by the invention can be applied to population genetic structure and genetic breeding study of Plectropomus population.
Owner:SUN YAT SEN UNIV
Method for developing simple sequence repeats molecular markers of genome of ammopiptanthus mongolicus (Maxim.) Chengf. plant
ActiveCN106282330AImprove efficiencyShorten the timeMicrobiological testing/measurementGenetic diversityShort read
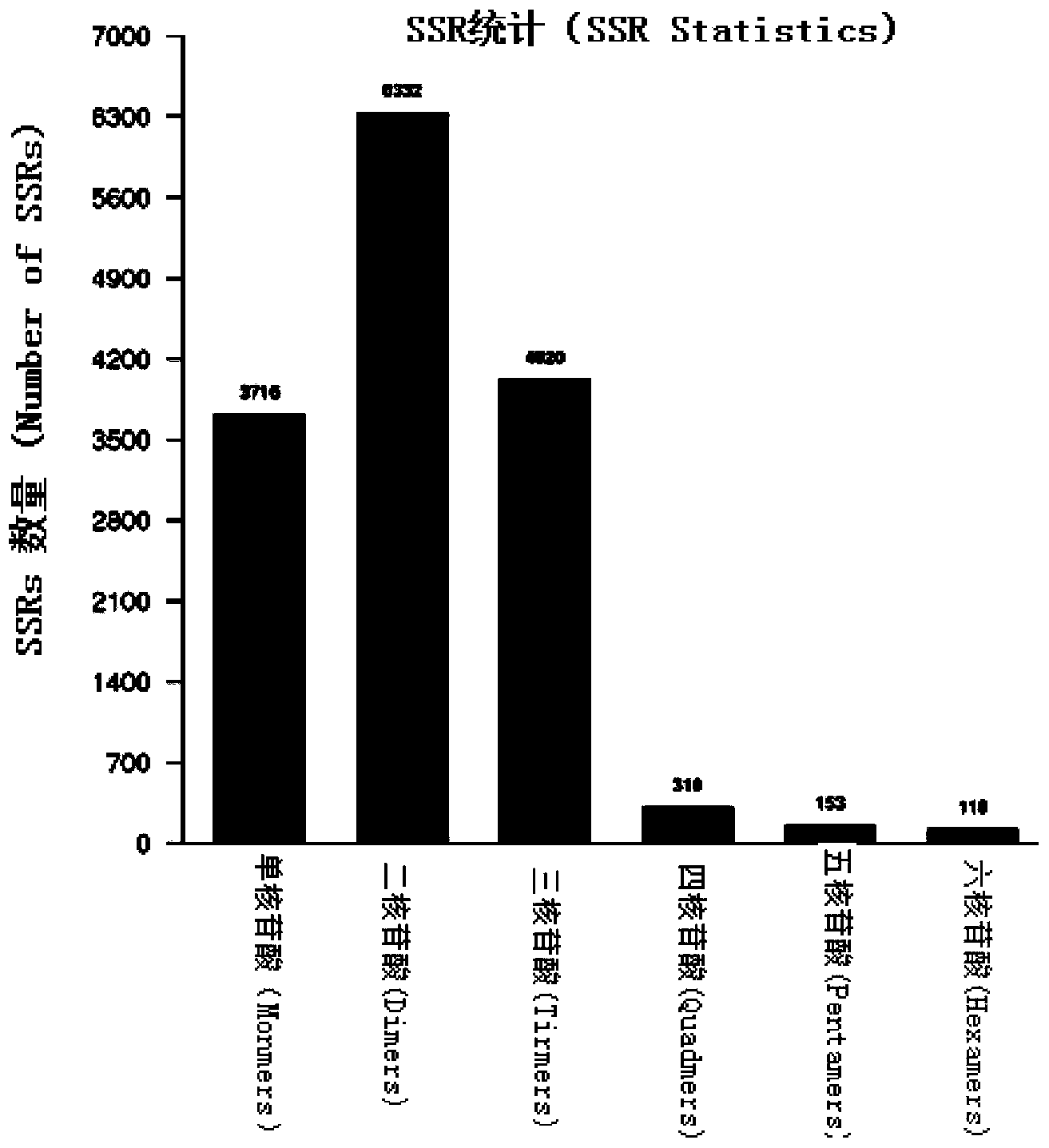
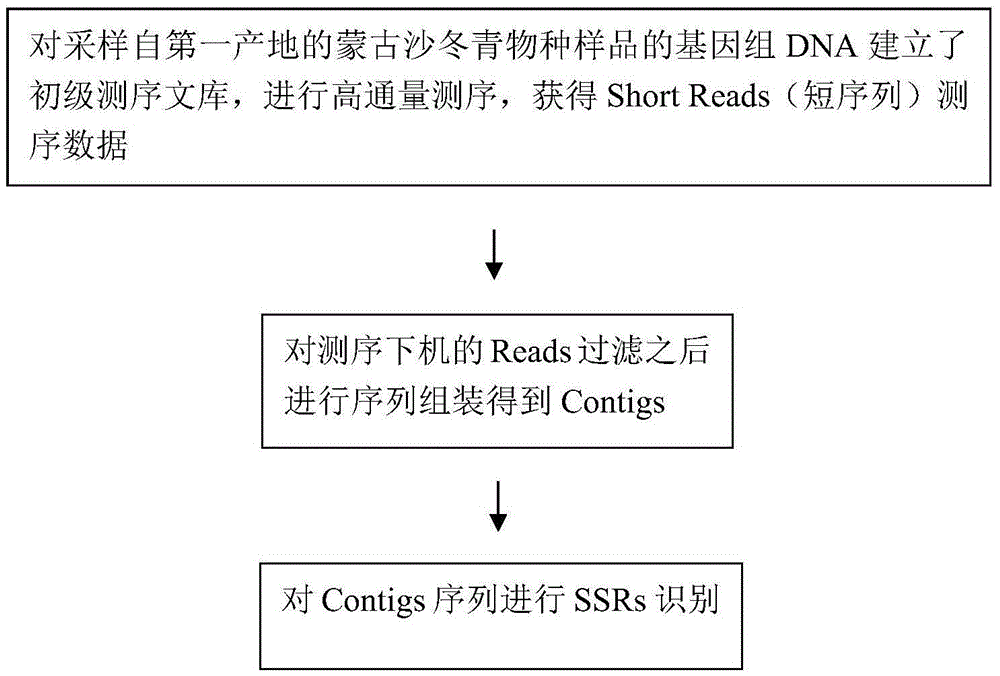
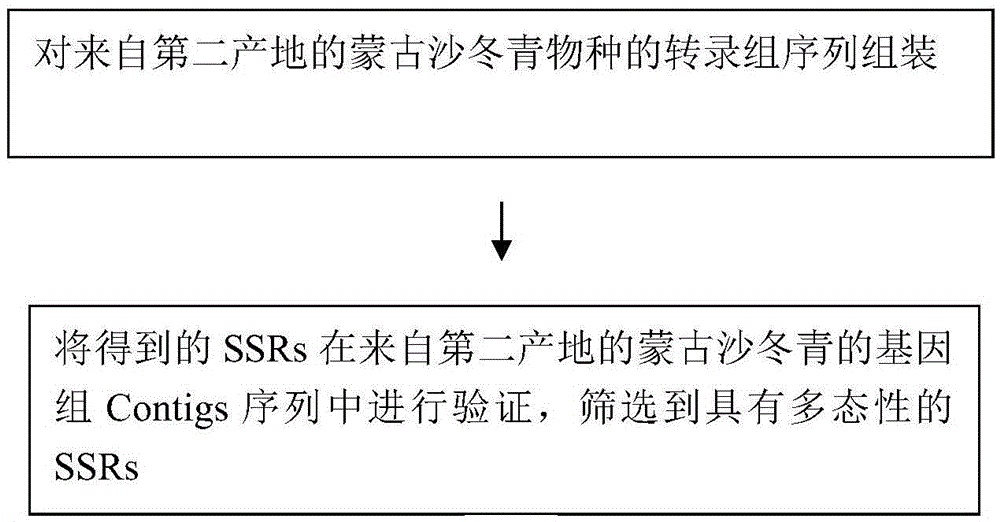
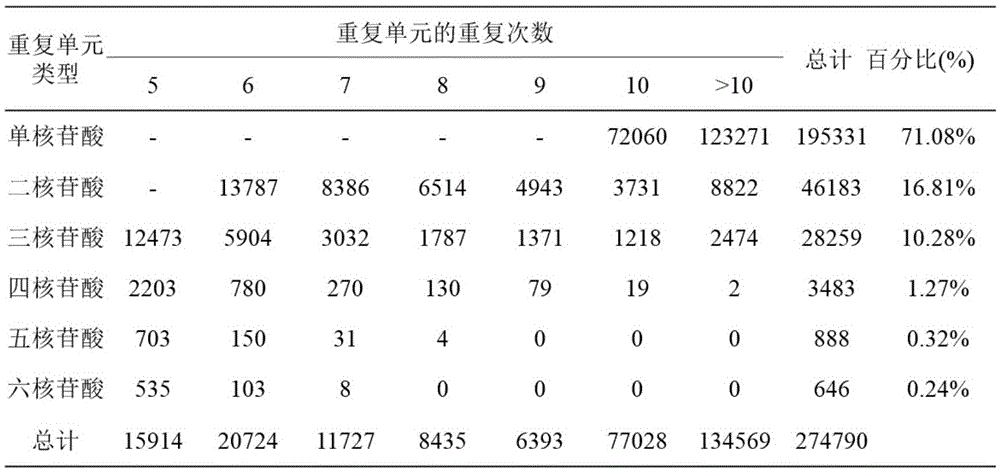
The invention relates to a method for developing SSRs (simple sequence repeats) molecular markers of genome of an ammopiptanthus mongolicus (Maxim.) Chengf. plant, wherein the method comprises the following steps: (1) establishing a primary sequencing library for genome DNA of a species sample sampled from a first production area, and conducting high-throughput sequencing so as to obtain Short Reads sequencing data; (2) filtering Reads obtained from sequencing and conducting sequence assembling so as to obtain Contigs; (3) conducting SSRs identification on Contigs sequences; and (4) verifying the SSRs in a Unigene sequence of a same species derived from a second production area, and conducting screening so as to obtain the SSRs having polymorphism. The high-throughput SSRs molecular marker discovering method provided by the invention is applicable to such researches as genetic map construction, QTL (quantitative trait loci) location, genetic diversity analysis and the like of the plant species.
Owner:SHENZHEN RES INST THE CHINESE UNIV OF HONG KONG
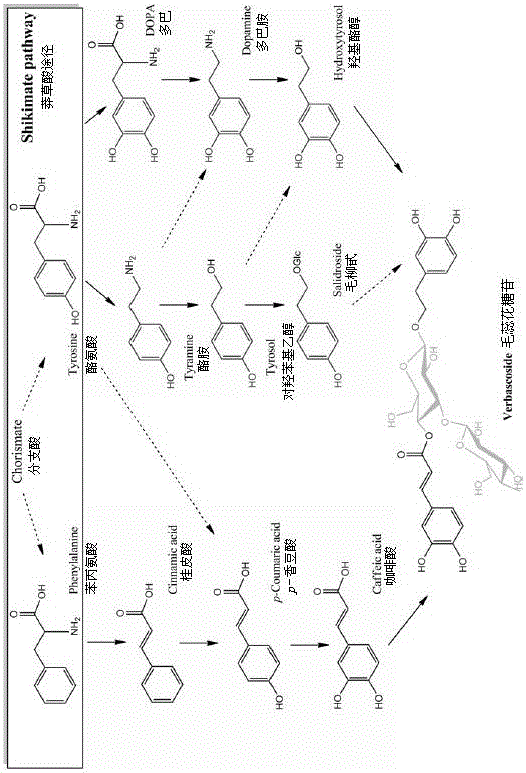
Verbascoside biosynthesis pathway and verbascoside biosynthesis enzyme related genes
InactiveCN106498009AIncreased glycoside contentIncrease contentOxidoreductasesLigasesTotal rnaMetabolism pathway
The invention discloses a verbascoside biosynthesis pathway and verbascoside biosynthesis enzyme related genes. Tuberous roots of rehmannia at six different development stages serve as materials, overall RNA (ribonucleic acid) is extracted and detected, combined transcriptome sequencing is performed by an Illumina HiSeq2500 platform to obtain 149.8 million reads, and head assembly is performed to obtain 96961 Unigenes sequences. The Unigenes sequences are compared with a KEGG (Kyoto encyclopedia of genes and genomes) database to perform metabolism pathway analysis and obtain 280 KEGG pathways, a metabolism pathway is compared with a supposed verbascoside biosynthesis pathway, and 5 candidate KEGG pathways are screened. The supposed verbascoside biosynthesis pathway is compared with the screened 5 candidate KEGG pathways, 19 gene coding enzyme catalysis verbascoside biosynthesis pathways are built, the Unigenes sequences and the database are subjected to Blast X comparison, and ESTscan software functions are added to obtain 13 candidate verbascoside biosynthesis enzyme related genes. A foundation is laid for illuminating the complete verbascoside biosynthesis pathway and producing verbascoside by secondary metabolism engineering or synthetic biology.
Owner:HENAN NORMAL UNIV
Enterolobium cyclocarpum root transcriptome database, fusion protein, soaking system and silencing system
ActiveCN107506616AReduction in knot formationReduce infection rateConnective tissue peptidesProteomicsNicotiana tabacumCandidate Gene Association Study
The invention belongs to the field of bioinformatic analysis technology and discloses an enterolobium cyclocarpum root transcriptome database, fusion protein, a soaking system and a silencing system. The database is composed of more than 38,000 Unigenes sequences, and bioinformatic analysis is utilized to predict potential RNAi target candidate genes of more than 230 enterolobium cyclocarpum root-knot nematodes. A prokaryotic expression technology is utilized to obtain ME-MAPK1, ME-COL1 and MeHsp70 recombinant protein, and a foundation is laid for protein characteristic analysis; the M.enterolobii in-vitro RNAi soaking system and the VIGSRNAi in-vivo silencing system with a tobacco rattle virus (TRV) carrier being a medium are successfully constructed, and Me-mapk1, Me-crt1 and Me-cbp1 gene down-regulated expression is induced; and a necessary molecular foundation is laid for M.enterolobii prevention and treatment.
Owner:INST OF PLANT PROTECTION HAINAN ACADEMY OF AGRI SCI
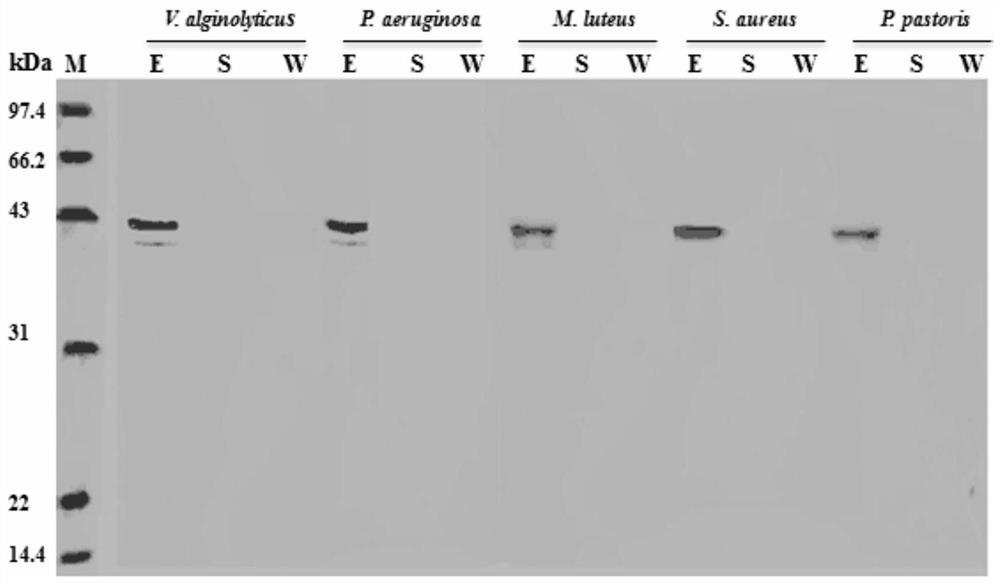
C-type lectin PtCLec1 gene of portunus trituberculatus and encoded protein and application thereof
ActiveCN110343703AGood effectEnhanced inhibitory effectAntibacterial agentsPeptide/protein ingredientsPichia pastorisAgglutination
The invention belongs to the technical field of molecular biology, and particularly relates to a C-type lectin PtCLec1 gene of portunus trituberculatus and an encoded protein and application thereof.Through unigene obtained by transcriptome sequencing and the RAC technology, amplification is conducted in the portunus trituberculatus to obtain PtCLec1 gene cDNA, and the recombinant protein PtCLec1has obvious bacteriostasis, bacterium binding and bacterium agglutination activity. The recombinant protein PtCLec1 has an obvious inhibition effect on gram negative bacteria and gram-positive bacteria. The recombinant protein PtCLec1 has obvious binding activity to vibrio alginolyticus, vibrio parahaemolyticus, pseudomonas aeruginosa, micrococcus luteus and pichia pastoris. In the presence of Ca<2+>, the recombinant protein PtCLec1 has an obvious agglutination effect on vibrio alginolyticus, vibrio parahaemolyticus, pseudomonas aeruginosa, staphylococcus aureus, micrococcus luteus and pichiapastoris.
Owner:INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Screening method and application of target gene SSR molecular markers of acanthopanax senticosus
ActiveCN108754018AOvercome uncertaintyOvercoming the disadvantages of blindnessMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationAgricultural science
The invention discloses a screening method and application of target gene SSR molecular markers of acanthopanax senticosus. The screening method specifically includes the following steps that 1, totalRNAs of all kinds of tissue of acanthopanax senticosus are extracted separately; 2, RNA-Seq sequencing is conducted after reverse transcription of the total RNAs is conducted, and Trinity software isused for conducting unigene assembly; 3, unigenes related to synthetic accumulation of acanthopanax senticosus saponins, SOD and the like are screened out, and MicroSatellite software is used for screening SSR sites on the unigenes; 4, Primer3.0 software is used for designing primers, and the primers are synthesized; 5, the extracted genomic DNAs serve as a template for PCR amplification; 6, PCRproducts are subjected to electrophoresis detection, and target gene SSR primers are developed. By means of the method and application, the candidate SSR molecular markers related to target traits areefficiently and more specifically developed, and effective markers are provided for molecular identification of excellent acanthopanax senticosus germplasm resources. An important meaning is achievedfor conducting early identification and speeding up a breeding process of the excellent acanthopanax senticosus germplasm resources.
Owner:DALIAN NATIONALITIES UNIVERSITY
Method for developing SSR primers of Blumea balsamifera based on transcriptome sequencing
ActiveCN108192893AImprove design success rateSimple methodMicrobiological testing/measurementDNA/RNA fragmentationSequence databaseTranscriptome Sequencing
The invention provides a method for developing SSR primers of Blumea balsamifera based on transcriptome sequencing. The method comprises: obtaining a Blumea balsamifera genome transcription set, and forming a sequence database; splicing the sequenced sequences by using Trinity to form a transcriptome, and taking the longest transcripts in each gene as Unigene; carrying out Unigene sequence bioinformatics analysis; carrying out SSR detection on the Unigene by using MISA1.0; and carrying out SSR primer design by using Primer3, and carrying out SSR primer polymorphism identification. According tothe present invention, with the method, 17979 pairs of the SSR primers are successfully designed, the 30 pairs of the primers related to the metabolic pathway of the active ingredient are screened, the DNA of 9 Blumea balsamifera with different sources are verified, 27 pairs of the polymorphic primers are determined, and the plant materials Blumea balsamifera with different geographical sources can be distinguished with the 27 pairs of the SSR primers; and the method provides the new idea for the development of the SSR primers of Blumea balsamifera.
Owner:TROPICAL CORP STRAIN RESOURCE INST CHINESE ACAD OF TROPICAL AGRI SCI
Portunus trituberculatus C-type lectin PtCLec2 gene and encoded protein thereof and application of encoded protein
ActiveCN110317813ACohesive activityGood effectAntibacterial agentsPeptide/protein ingredientsAgglutinationPortunus trituberculatus
The invention belongs to the technical field of molecular biology, and particularly discloses a portunus trituberculatus C-type lectin PtCLec2 gene and an encoded protein thereof and application of the encoded protein. A RACE technology and a unigene obtained by adopting transcriptome sequencing are adopted to acquire cDNA of the PtCLec2 gene from portunus trituberculatus through amplification, and it is found that a recombinant PtCLec2 gene has significant activity of bacterium inhibition, somatic agglutination and bacterium removal. The recombinant PtCLec2 gene has significant inhibiting effects on Gram-negative bacteria (vibrio alginolyticus and pseudomonas aeruginosa) and Gram-positive bacteria (staphylococcus aureus and micrococcus luteus). In the presence of Ca<2+>, the recombinant PtCLec2 gene has significant agglutination effects on vibrio alginolyticus, pseudomonas aeruginosa, staphylococcus aureus and micrococcus luteus. Meanwhile, the recombinant PtCLec2 gene has a removal effect on vibrio alginolyticus.
Owner:INST OF OCEANOLOGY - CHINESE ACAD OF SCI
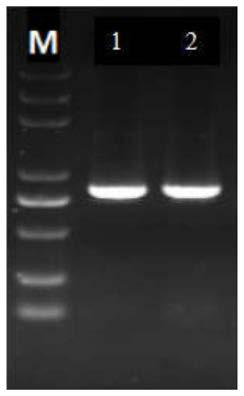
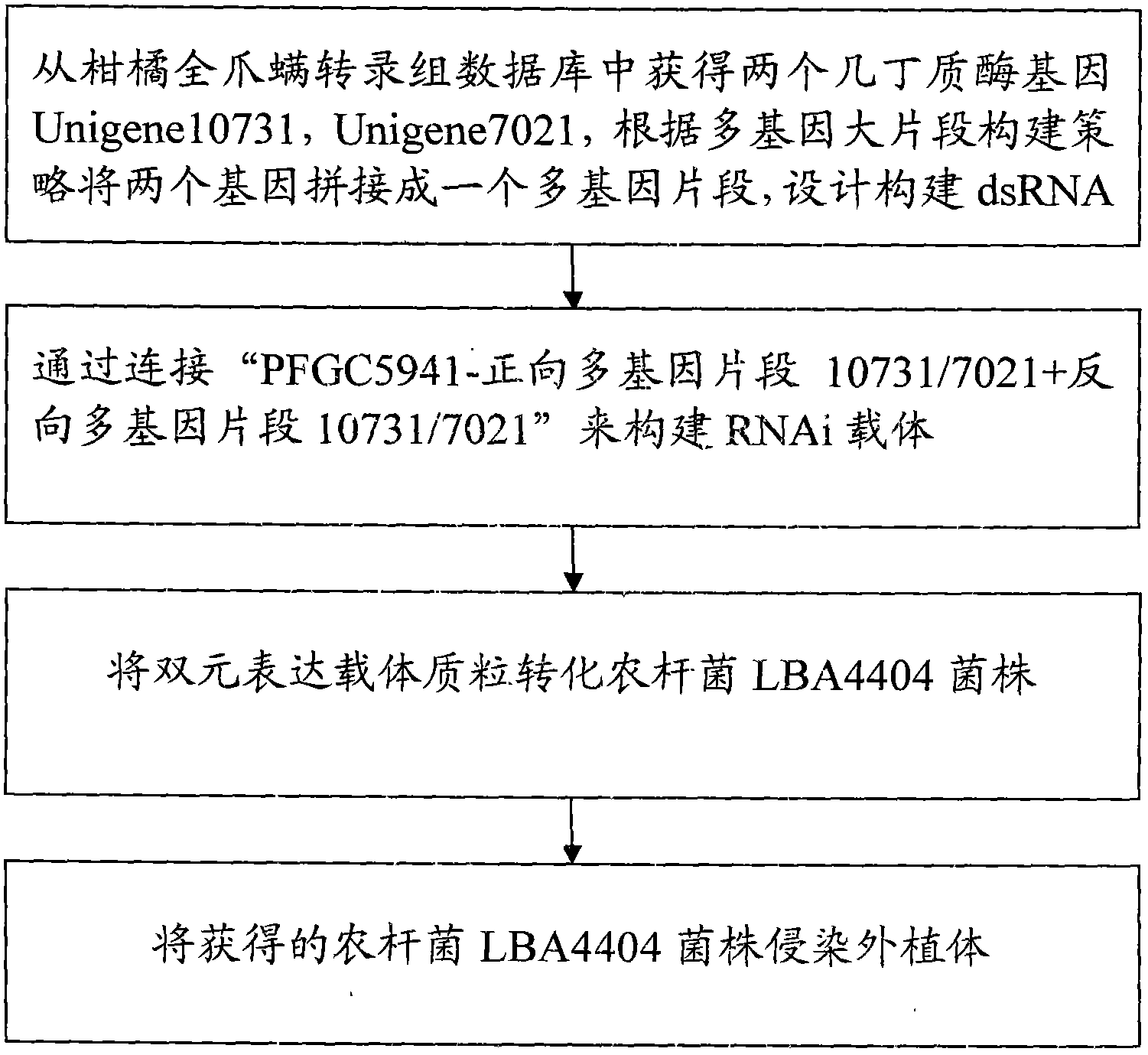
Preparation method of panonychus citri me gregor chitin gene-based double-target transgenetic vector
InactiveCN103014059AReduce usageVector-based foreign material introductionAngiosperms/flowering plantsConserved sequenceIntein
The invention discloses an application method of 2 chitinase genes. Two chitinase genes Unigene10731 and Unigene 7021 are identified from an orange panonychus citri me gregor transcriptome database and are subjected to authenticity testing; the two selected genes are compared on NCBI (National Center for Biotechnology Information) to find a conserved sequence; and the two genes are jointed into a multi-gene segment according to the multi-gene large-segment construction strategy, and the Unigene10731 and the Unigene 7021 are jointed together to form a multi-gene; a primer containing a restriction enzyme cutting site is designed, and forward and reverse segments of the multi-gene chitinase order are sequentially jointed into two terminals of the intron of the vector, so that an RNAi (ribonucleic acid interfere) interference vector 'PFGC+Ug10731 / Ug7021' with a reverse complementary hairpin structure can be formed, and the vector is reprinted into agrobacterium to infect an orange nursery stock.
Owner:CITRUS RES INST SOUTHWEST UNIV
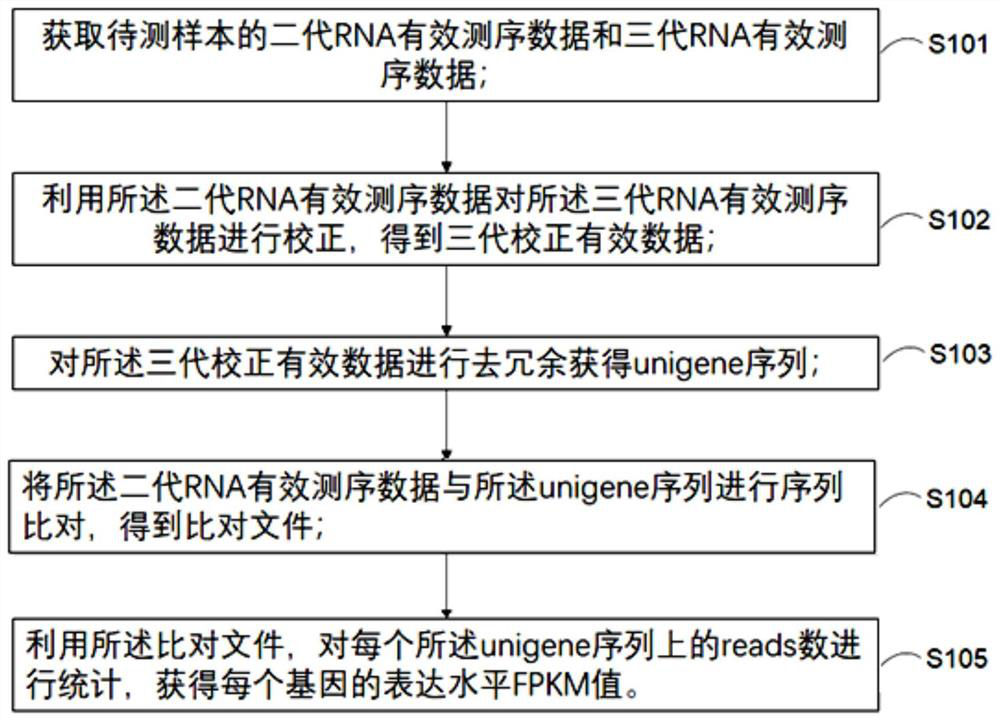
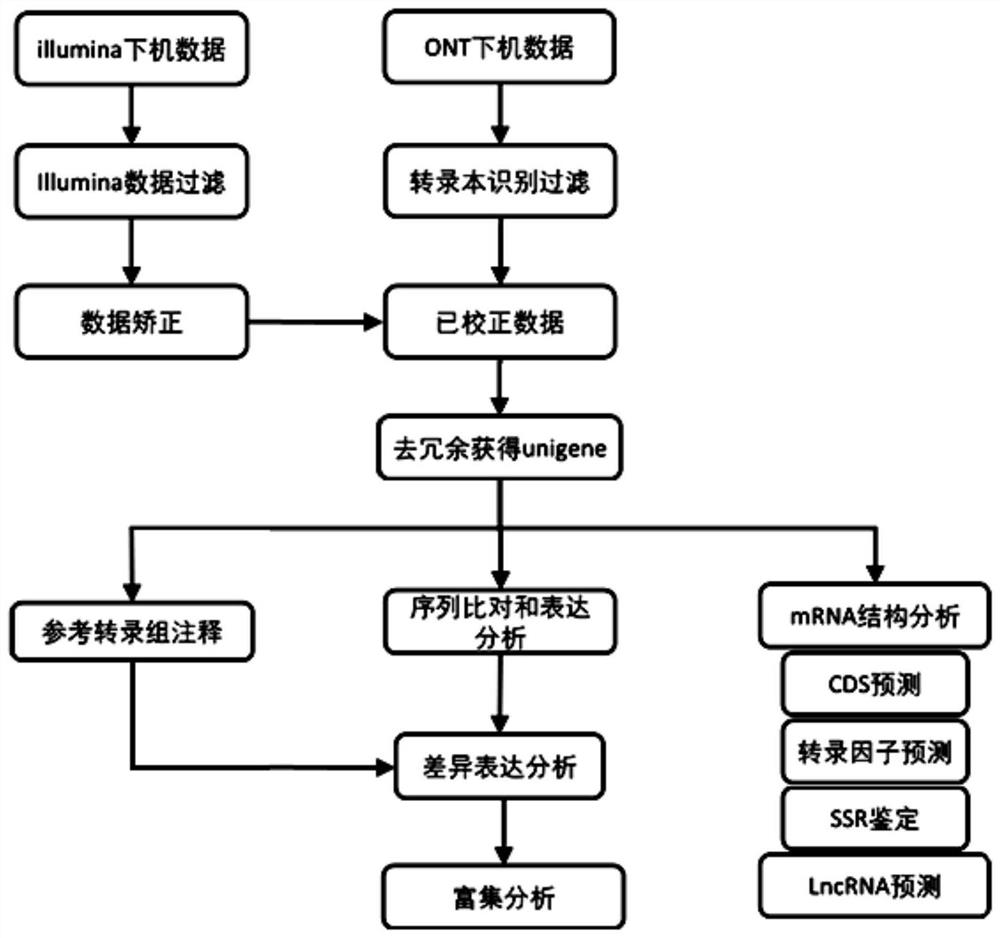
Transcriptome analysis method and system without reference genome sequence
The invention provides a transcriptome analysis method and system without a reference genome sequence. The analysis method comprises the following steps: acquiring second-generation RNA effective sequencing data and third-generation RNA effective sequencing data of a to-be-detected sample; correcting the third-generation RNA effective sequencing data by utilizing the second-generation RNA effective sequencing data; redundancy elimination is carried out on the valid data after the third-generation correction to obtain a unigene sequence; performing sequence comparison on the second-generation RNA effective sequencing data and the unigene sequences, and performing statistics on the reads number on each unigene sequence by utilizing a comparison file to obtain an expression level FPKM value of each gene. The analysis method integrates the advantages of a second-generation sequencing technology and a third-generation sequencing technology, and solves the problem that no transcriptome of areference-free genome sequence is analyzed by utilizing third-generation sequencing data at present.
Owner:TIANJIN MODERN INNOVATIVE TCM TECH CO LTD
Antifungal protein and usage thereof
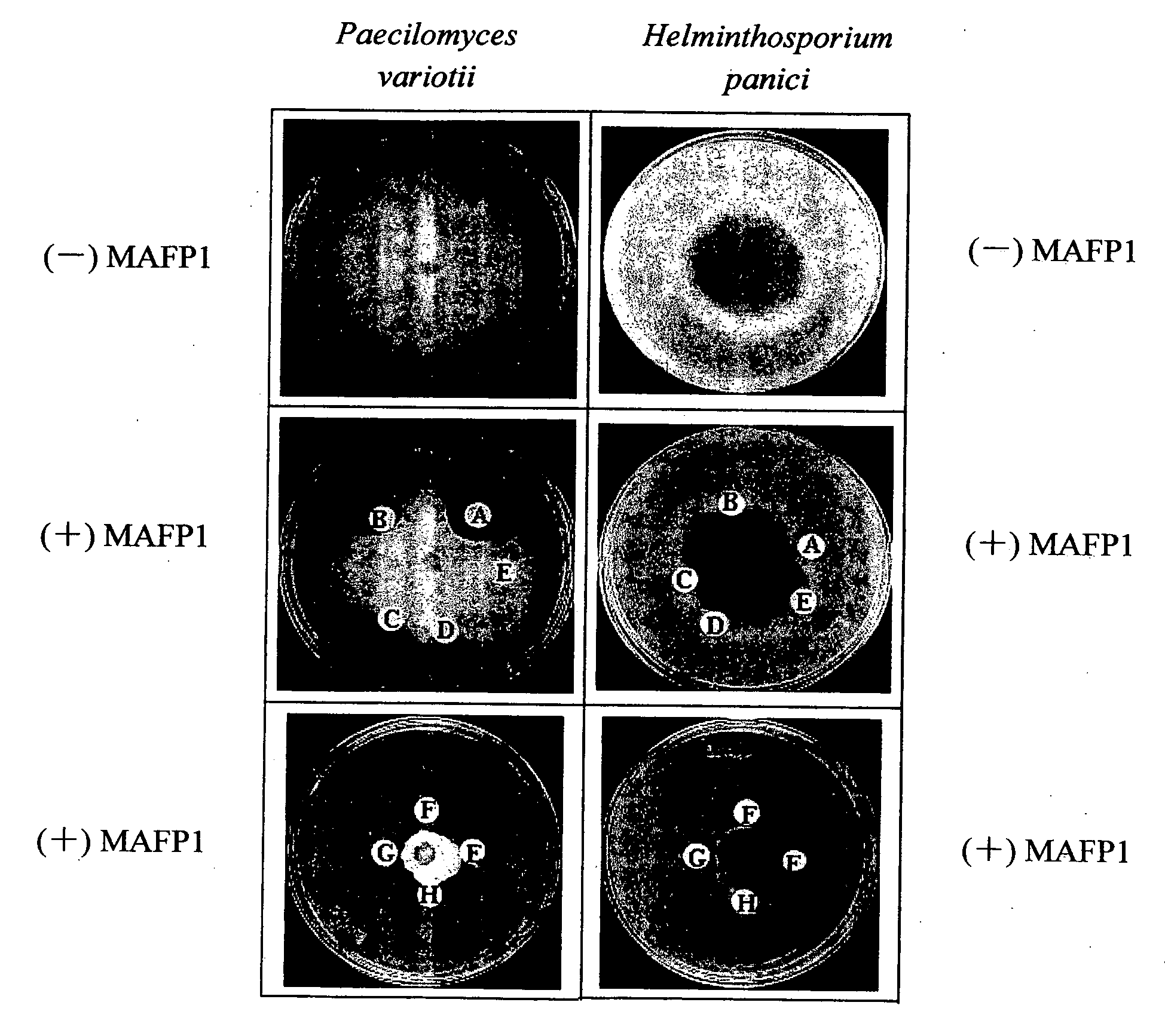
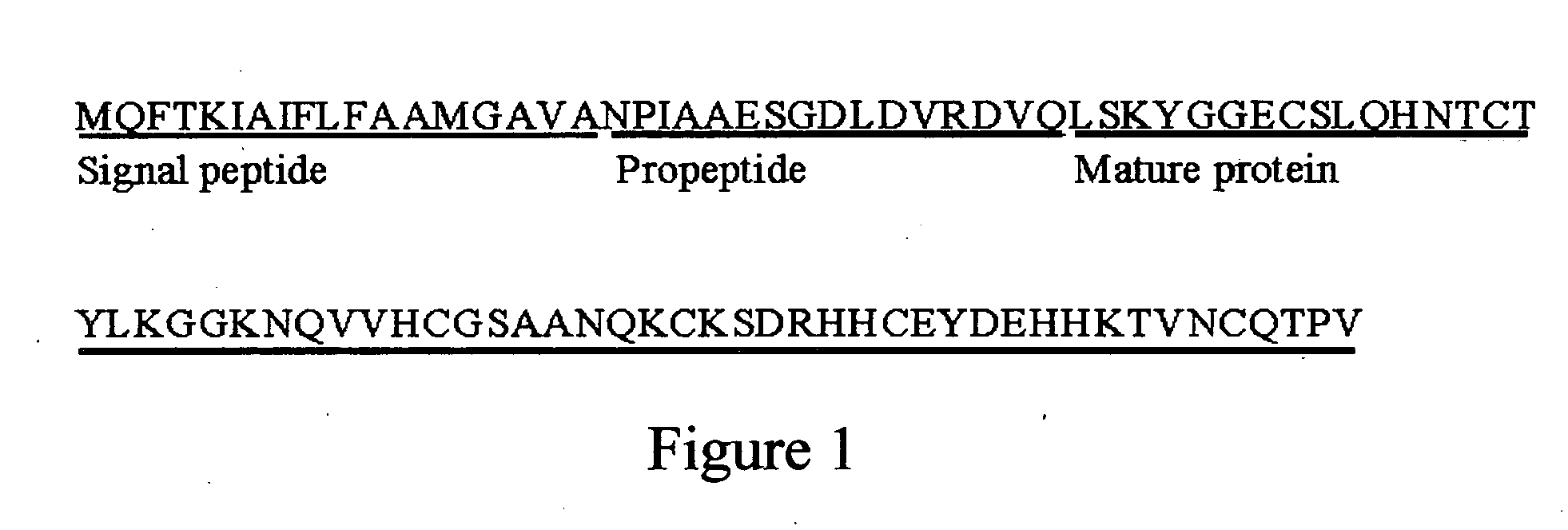
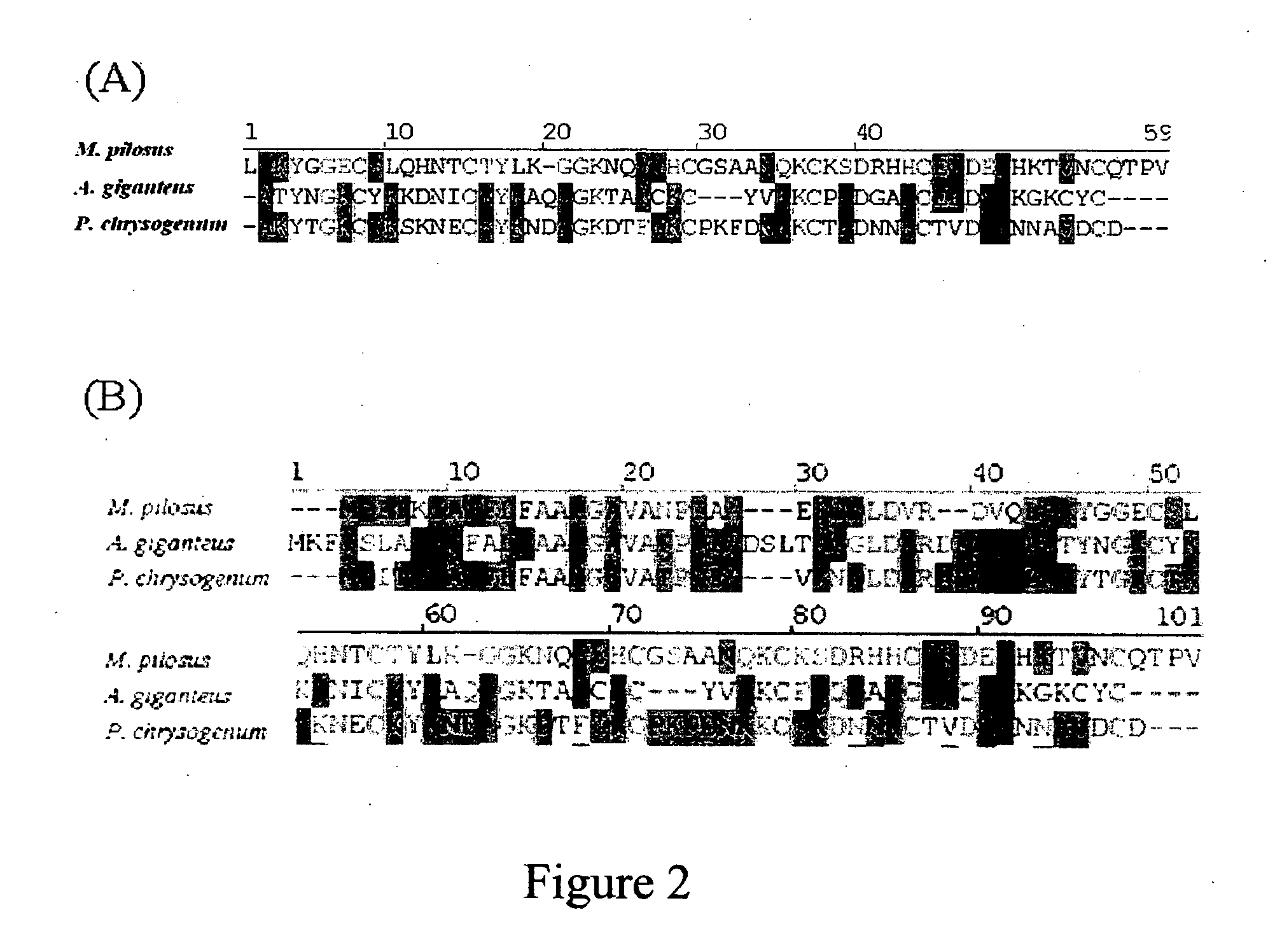
The present invention relates to an antifungal protein gene and cDNA sequence thereof, which is obtained by mining the whole genome sequences of Monascus pilosus BCRC 38072 and the unigene database. The gene can encode an antifungal protein MAFP1. A purified protein obtained from M. pilosus culture broth having molecular weight of about 7 kDa is identified as MAFP1 by N-terminal protein sequencing and comparative analysis. The purified MAFP1 protein can inhibit the growth of pathogens such as Paecilomyces variotii BCRC 33174 and Helminthosporium panici BCRC 35004. In addition, it is found by PCR test that the gene of this antifungal protein exists in other Monascus species such as M. Barkeri, M. floridanus, M. lunisporas, M. pilosus, M. ruber and the like. It is also been proved that the mafp1 gene and cDNA thereof in four Monascus strains, M. pilosus (BCRC 38072, BCRC 38093 and BCRC 31502) and M. ruber BCRC 31533, have the same DNA sequences.
Owner:FOOD IND RES & DEV INST
Antifungal protein and usage thereof
Owner:FOOD IND RES & DEV INST
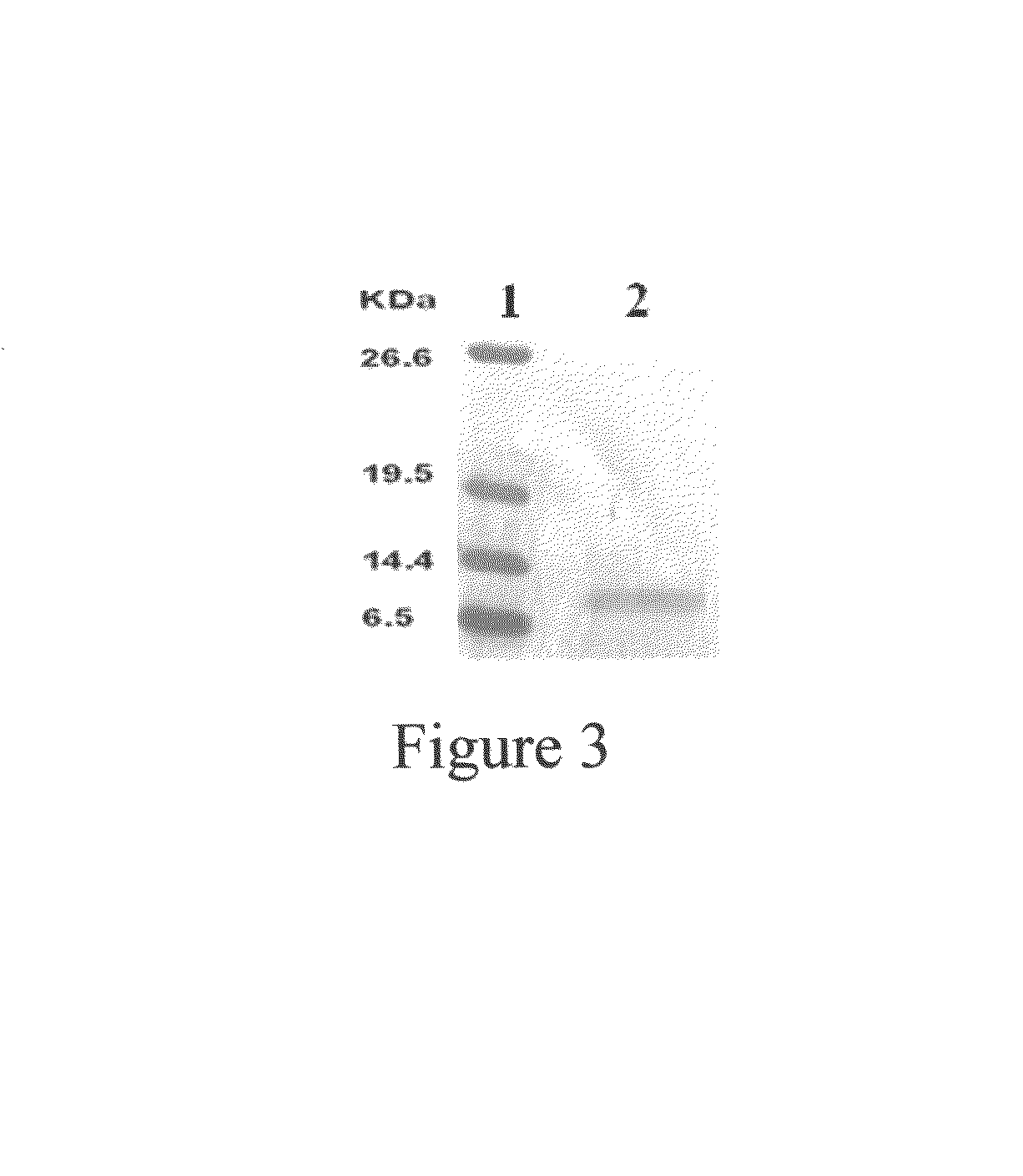
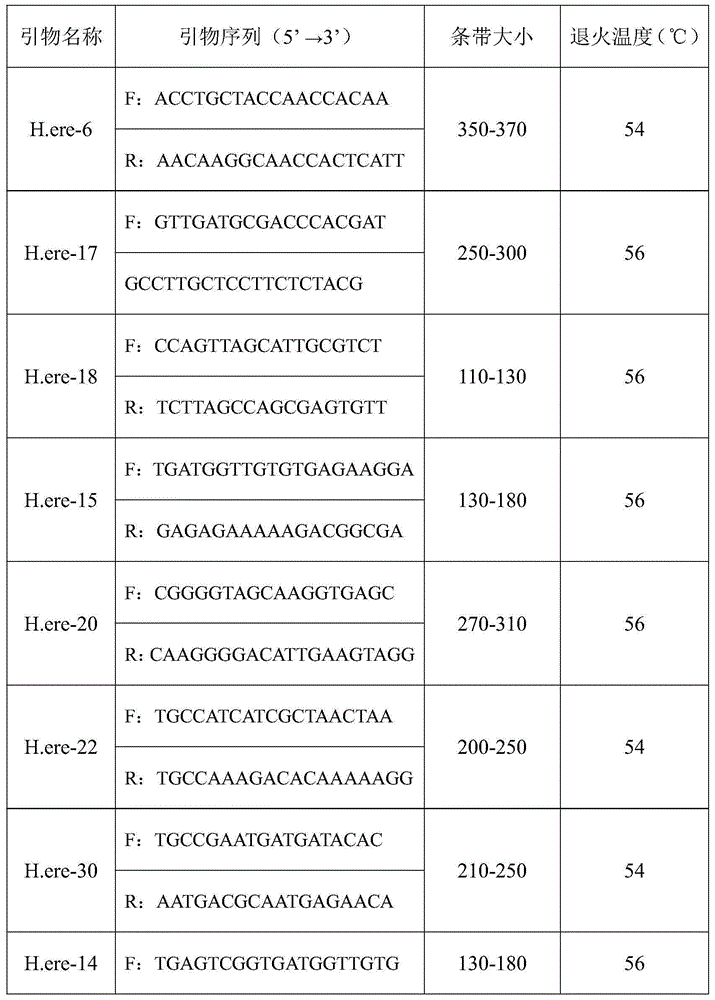
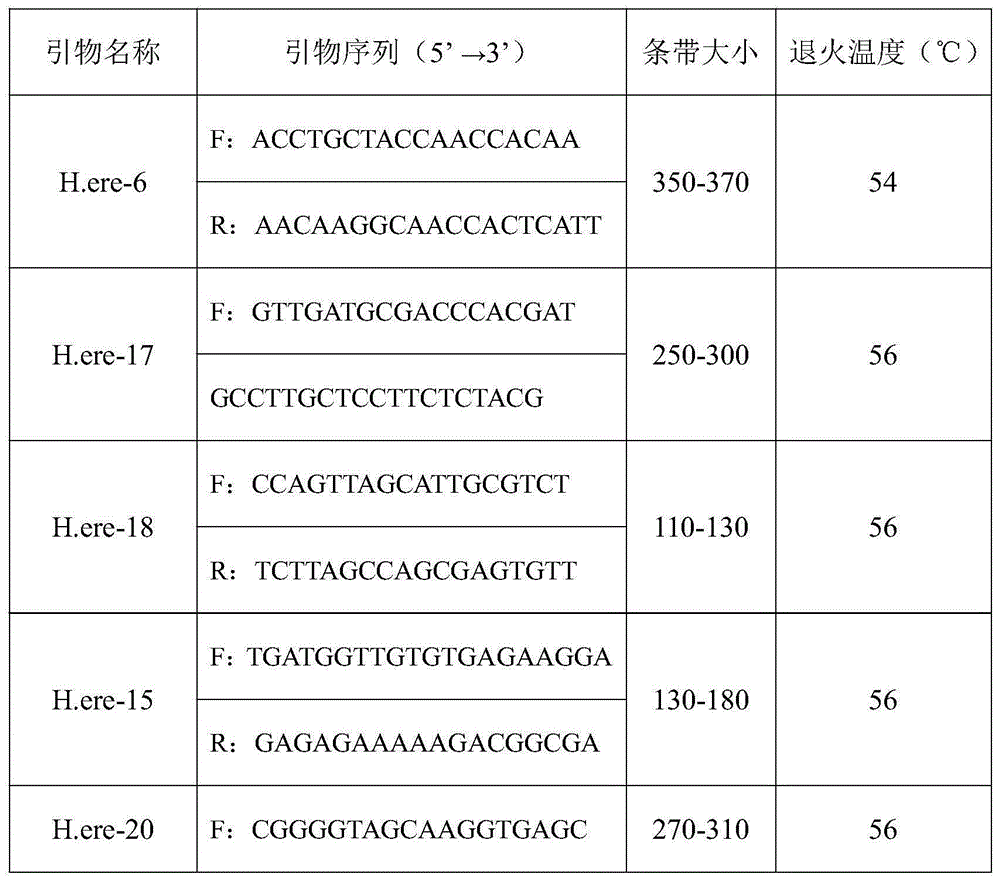
Hippocampus erectus microsatellite markers and screening method thereof
ActiveCN105177142AEfficient build methodEasy to buildMicrobiological testing/measurementDNA/RNA fragmentationDNA paternity testingScreening method
The invention belongs to the technical field of fish breeding, and discloses Hippocampus erectus microsatellite markers and a screening method thereof. The Hippocampus erectus microsatellite markers are disclosed as SEQ.ID.NO:1-9. The screening method comprises the following steps: 1) carrying out transcriptome sequencing screening to obtain Hippocampus erectus Unigenes; 2) detecting microsatellite sites in the Unigenes sequence; 3) designing specific primers according to sequences on both ends of the microsatellite site; and 4) carrying out PCR (polymerase chain reaction) amplification and detecting the polymorphism, thereby obtaining the 9 microsatellite markers. The invention discloses the Hippocampus-erectus-related microsatellite markers and specific primers thereof for the first time. The Hippocampus erectus microsatellite markers can be used for researching Hippocampus erectus population genetic structure, genetic breeding, identification in disputed paternity and the like.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Gene excavating method based on EST database and UniGene database
InactiveCN101661536AMining results are accurateEfficient miningMicrobiological testing/measurementSpecial data processing applicationsExpression genePcr ctpp
The invention provides a gene excavating method based on EST database and UniGene database, relating to the field of applied bioinformatics. The method overcomes the problem that characters-correlative induce gene can not be exactly excavated in the existing gene excavating method. The method comprises the steps: digitizing the EST expression quantity of expression gene UniGene transcriptome in the UniGene database with an EST sequence in the EST database; building hypergeometric distribution test; adjusting a hypergeometric distribution test value P-value of the differential expression of theexpression gene UniGene transcriptome by combining with an FDR method; screening error state response gene; and verifying the response gene is the characters-correlative induce gene with a RT-PCR technology. The method can be used for excavating the characters-correlative induce gene in the process of the disease occurrence of human beings, the growing development regulation of animals and plants, the disease regulation of the animals and plants, the adversity stress of the animals and plants, etc.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
SSR marker for detecting bruchid-resistant variety of broad beans and application of SSR marker
ActiveCN114032321AGood choiceEfficient identification of soybean weed resistance traitsMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyNucleotide
The invention provides an SSR marker for detecting a bruchid-resistant variety of broad beans and application of the SSR marker. According to the invention, a PacBio three-generation full-length sequencing technology is combined with an RNA-seq method to carry out whole genome sequencing onbroad bean varieties, software MISA is combined to search Unigene of a transcriptome to obtain a specific broad bean genome sequence containing an SSR core motif, primers are designed according to the SSR core motif, and are used for amplifying an SSR marker, the nucleotide sequence of the upstream primer is shown as SEQ ID NO.1, and the nucleotide sequence of the downstream primer is as shown in SEQ ID NO. 2; the SSR marker primer pair is used for amplifying a broad bean genome template, a PCR amplification product is subjected to polyacrylamide gel electrophoresis, the banding pattern of the PCR product is detected, and the broad bean material for identifying and detecting is the bruchid-resistant material; and the method for detecting the bruchid resistance of broad beans is reliable, simple, convenient and practical, and has an important application prospect in broad bean germplasm resource identification and molecular marker-assisted breeding selection.
Owner:HAINAN UNIVERSITY
Method for obtaining transcriptome and functional genes of blumea balsamifera
PendingCN109423490AHigh content of active ingredientsMicrobiological testing/measurementPlant peptidesPoly-A RNATotal rna
The invention provides a method for obtaining transcriptome and functional genes of blumea balsamifera. The method comprises (a) extracting the total RNA(ribonucleic acid) of blumea balsamifera, separating out mRNA (messenger RNA) with polyA (polyadenylation) at the 3' end, randomly breaking the mRNA, performing inverse transcription to synthesize double-stranded cDNA (complementary DNA); (b) sequencing an obtained sequence; (c) splicing and assembling a sequencing result to obtain Unigene and determining the orientation of the Unigene; (d) performing bioinformatics analysis on an obtained gene transcript to obtain the transcriptome and the function genes of blumea balsamifera. The method for obtaining the transcriptome and the functional genes of blumea balsamifera can help obtain 48197273 pieces of sequence information and 100341 Unigenes, involving 60477 pieces of information including RNA-seq names, sequence length and expression quantity, COG (cluster of ortholog genes) predication, COG functional annotations, KEGG (Kyoto encyclopedia of genes and genomes) annotations, KEGG-pathway and GO (gene ontology) annotations, and 37283 pieces of information protein function annotationsfor performing CDS (coding sequence) prediction on the obtained sequence information.
Owner:TROPICAL CORP STRAIN RESOURCE INST CHINESE ACAD OF TROPICAL AGRI SCI
A method for developing elderberry ssr primers based on transcriptome sequencing
ActiveCN108998561BGood polymorphismMicrobiological testing/measurementDNA/RNA fragmentationTranscriptome SequencingSambucus nigra flower
The present invention provides a method for developing elderberry SSR primers based on transcriptome sequencing, the method comprising: S1. Constructing an elderberry transcriptome library and sequencing, splicing and assembling the sequencing results into a complete transcriptome, taking each The longest transcript in the gene is used as Unigene, and the mixed Unigenes library is obtained by splicing; S2. Perform SSR detection on the mixed Unigenes library in step S1; S3. Design SSR primers and identify polymorphisms of the SSR primers. A total of 64,148 pairs of SSR-specific primers were designed in the present invention, and the optimal system of western elderberry fluorescent SSR-PCR was obtained through the experimental optimization scheme. Among the 100 pairs of fluorescently labeled SSR primers synthesized, 25 pairs of primers had good polymorphism. The method for developing elderberry SSR primers based on transcriptome sequencing in the present invention is fast and accurate.
Owner:SHANDONG FOREST SCI RES INST
A kind of screening method and application of ssr molecular marker of Acanthopanax target gene
ActiveCN108754018BMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationTotal rna
The invention discloses a screening method and application of the SSR molecular marker of the target gene of Acanthopanax. The specific steps of the screening method are: 1) extracting the total RNA of each tissue of Acanthopanax anticana; 2) performing RNA- Seq sequencing, Trinity software for unigene assembly; 3) Screen out unigenes related to the synthesis and accumulation of eleutherosides, SOD, etc., and MicroSatellite software screens the SSR sites on these unigenes; 4) Primer3.0 software designs primers and synthesizes them; 5) Genomic DNA is used as a template for PCR amplification; 6) PCR products are detected by electrophoresis, and SSR primers for the target gene are developed. The invention efficiently and more targetedly develops candidate SSR molecular markers related to target traits, and provides effective markers for molecular identification of excellent Acanthopanax germplasm resources. It is of great significance to the early identification of excellent Acanthopanax germplasm resources and to speed up the breeding process.
Owner:DALIAN NATIONALITIES UNIVERSITY
The ssr primer pair, screening method and application of rhododendron yunnanensis developed based on transcriptome sequencing
ActiveCN109468405BFast wayAccurate methodMicrobiological testing/measurementDNA/RNA fragmentationSequence analysisGenetics
The invention discloses a pair of SSR primers for Rhododendron versicolor developed based on transcriptome sequencing and a screening method and application. Belongs to the field of biotechnology. The primers are obtained based on transcriptome sequencing and sequence analysis. On the basis of transcriptome sequencing, a large amount of data is screened to obtain unigene sequences rich in SSR sites and design primers for SSR markers, which overcomes the current SSR molecular The obstacles of small number of markers and low development efficiency. The validity of the SSR primers was verified by Rhododendron versicolor population, which laid a foundation for the genetic diversity research, genetic linkage map structure, QTL mapping, molecular marker-assisted breeding and other genetic research of Rhododendron versicolor.
Owner:HUANGGANG NORMAL UNIV
A kind of est-ssr marker specific primer and screening method for the transcriptome sequence of Torreya torreya
The invention belongs to the field of molecular biology DNA (Deoxyribonucleic Acid) marker technology and application, and particularly relates to specific primers and a screening method for an EST-SSR (Express Sequence Tag-Simple Sequence Repeat) marker of a torreya grandis transcriptome sequence. The screening method comprises the following steps: firstly, assembling fine torreya grandis seed kernel and circular torreya grandis seed kernel transcriptome data by adopting Trinity software to obtain a transcript sequence; secondly, carrying out SSR site research on Unigene with 1kb or above obtained by screening by using an MISA (MIcroSAtellite identification tool) program, and carrying out screening and separating on 2 to 6 base repeat units and repeat times according to microsatellite fragments with repeat times of 9, 8, 5, 5 and 5 in sequence, thereby obtaining an ESTs sequence with microsatellite repeat; thirdly, designing the primers by using a Primer 3 program; finally, comprehensively evaluating repeatability, stability and polymorphism of the EST-SSR marker to obtain the EST-SSR marker. The specific primers disclosed by the invention can be used for conveniently and quickly analyzing germ plasm resources of different types and genetic diversity of interspecific and intraspecific torreya grandis.
Owner:ZHEJIANG FORESTRY UNIVERSITY +1
Method of developing elderberry SSR primer on the basis of transcriptome sequencing
ActiveCN108998561AThe method is fast and accurateGood polymorphismMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceFluorescence
The invention provides a method of developing elderberry SSR primer on the basis of transcriptome sequencing. The method includes: S1) establishing and sequencing elderberry transcriptome library andsplicing and jointing sequencing results to form a complete transcriptome, and with the longest transcript in every gene being Unigene, splicing the Unigenes to form a blend Unigenes library; S2) performing SSR detection to the blend Unigenes library in the step S1); S3) performing SSR primer design and identifying the polymorphism of the SSR primer. In the invention, 64148 pairs of SSR specific primers are designed, wherein through experimenal optimization, a fluorescent SSR-PCR optimum system of golden elder is acquired; among the synthesized 100 pairs of the fluorescence label SSR primers,25 pairs of primers show excellent polymorphism. The method of developing elderberry SSR primer on the basis of transcriptome sequencing is quick and accurate.
Owner:SHANDONG FOREST SCI RES INST
Syringa oblata chalcone synthase gene and application thereof
The invention relates to the technical field of biology, and concretely relates to a syringa oblate chalcone synthase gene. According to the invention, an Unigene sequence fragment of chalcone synthase gene (SoCHS) obtaining by syringa oblate transcriptome sequencing is taken as a template, a SoCHS cDNA full length sequence is obtained; based on bioinformatics analysis on a structure and coding protein, a molecular biology technology is used, an overexpression and YFP positioning expression carrier of the gene is constructed, through agrobacterium mediation, arabidopis thaliana and tobacco are converted, the protein characteristic is analyzed and the function is verified; and a RT-PCR technology is used for analyzing the tissue specific expression mode of the gene. A molecule mechanism formed by SoCHS-participated pattern control is researched and revealed.
Owner:BEIJING UNIV OF AGRI
Mannose-binding lectin ptmbl gene of Portunus trituratus and its encoded protein and application
ActiveCN109797155BEnhanced inhibitory effectAntibacterial agentsAntimycoticsImmunologic preparationFeed additive
The invention belongs to the technical field of molecular biology, and specifically relates to a PtMBL gene of Portunus trituberculatus mannose-binding lectin, its encoded protein and its application. The present invention uses the unigene and RACE technology obtained by transcriptome sequencing to amplify the PtMBL gene cDNA from Portunus trituberculatus, and finds that the recombinant PtMBL protein has significant bacteriostasis, bacterium binding and bacterium agglutination activities. The recombinant protein PtMBL has obvious inhibitory effects on Gram-negative bacteria (Vibrio alginolyticus, Pseudomonas aeruginosa) and Gram-positive bacteria (Staphylococcus aureus, Micrococcus luteus), and the minimum inhibitory concentrations are 0.81~1.61μM, 0.81μM, 0.81~1.61μM, 3.22~6.44μM. The recombinant protein PtMBL has obvious binding activity to Vibrio alginolyticus, Pseudomonas aeruginosa, Staphylococcus aureus, Micrococcus luteus and Pichia pastoris. In addition, in Ca 2+ In the presence of the recombinant protein PtMBL, it has obvious agglutination effect on Vibrio alginolyticus, Pseudomonas aeruginosa, Staphylococcus aureus, Micrococcus luteus and Pichia pastoris. The invention lays a foundation for the disease prevention and treatment of portunus trituberculatus, and has potential application value in the development of antibacterial drugs, bacterial agglutination preparations, new immune preparations, feed additive production and the like.
Owner:INST OF OCEANOLOGY - CHINESE ACAD OF SCI
The Manshan Red SSR primer based on the transcription group sequencing method and screening method and application
ActiveCN109536632BFast wayAccurate methodMicrobiological testing/measurementDNA/RNA fragmentationSequence analysisGenetic diversity
The invention discloses a pair of SSR primers developed based on transcriptome sequencing and a screening method and application. Belongs to the field of biotechnology. The primers are obtained based on transcriptome sequencing and sequence analysis. On the basis of transcriptome sequencing, a large amount of data is screened to obtain unigene sequences rich in SSR sites and design primers for EST‑SSR markers, which overcomes the current problems The low number and low development efficiency of red SSR molecular markers are obstacles. The validity of the EST-SSR primers was verified by the Mangospermum population, which laid a foundation for the genetic diversity research, genetic linkage map structure, QTL mapping, molecular marker-assisted breeding and other genetic studies of Mangosteen.
Owner:HUANGGANG NORMAL UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com