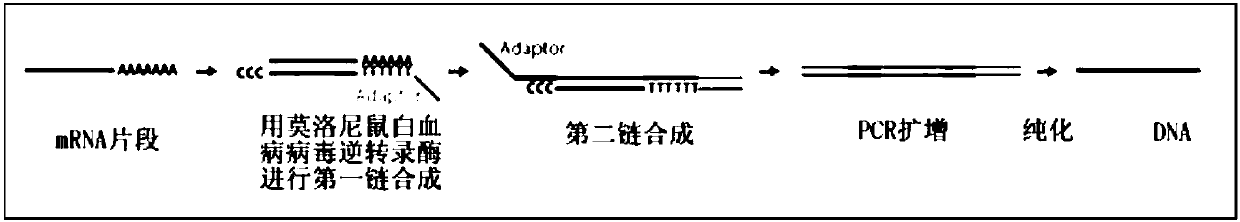
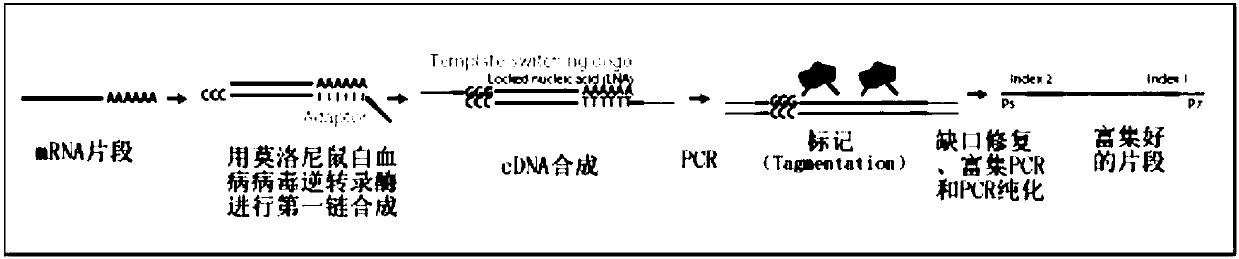
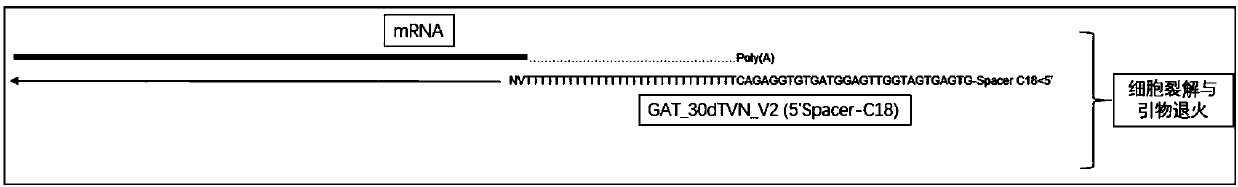
Single cell mRNA reverse transcription and amplification method
A reverse transcription, single-cell technology, applied in the field of genome sequence analysis and biology, it can solve the problems of low amplification efficiency, biased transcript length, and inability to efficiently transcribe sequences, so as to expand the scope of application and avoid 3' bias. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Example 1 Obtaining single cell (or trace cell) total RNA
[0062] 1. Provide an aliquot of cells growing adherently on a cell culture flask (with medium). In this method, human lymphocyte GM12878 was selected as the material for subsequent experiments.
[0063] 2. Gently pour out the medium in the cell culture flask described in step 1 and discard it. Add 5mL 1x PBS (the following PBS refers to 1×PBS buffer) buffer, wash twice, then pour out the washed liquid gently and discard it. Dilute trypsin with ultrapure water to a concentration of 0.25%, add 3mL into the culture bottle, rotate gently to make the liquid evenly stick to the inner wall of the bottle, and treat at 37°C for 3 minutes. After the incubation, hold the bottle mouth and use the wrist Gently shake the liquid in the bottle with force, suck out the liquid with a pipette gun and place it in a 15mL centrifuge tube for later use.
[0064] 3. Connect the centrifuge tube with the liquid obtained in step 2, pu...
Embodiment 2
[0070] Example 2 reverse transcription to obtain double-stranded full-length cDNA
[0071] 1. Pipette 1.7*N (N is the number of reactions, pay attention to the addition of 10% pipetting loss) μL of reverse transcriptase mixture (component name: RT Enzyme Mix) and add it to the RT Buffer obtained in step 8 of Example 1 , mixed by hand, and centrifuged to form a mixture labeled "RT Mix".
[0072] Wherein said reverse transcriptase mixture contains reverse transcriptase, reverse transcriptase protective agent, BSA, RNase inhibitor.
[0073] The reverse transcription linker is in *GTGAGTGATGGTTGAGGTAGTGTGGAGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT (SEQ ID NO: 2, * means blocked by C18 carbon skeleton).
[0074] The template switching oligonucleotide is
[0075] #GTGAGTGATGGTTGAGGTAGTGTGGAGNNNNNNNrGrGLNA-G (SEQ ID NO: 3, # means NH 2 -C6 closed).
[0076] 2. Add 15 μL of "RT Mix" to the cell lysate sample obtained in step 8 of Example 1, place it on ice immediately after centrifugation...
Embodiment 4
[0089] Example 4 Single Cell mRNA Reverse Transcription and Amplified Product (cDNA) Fragment Interruption
[0090] The following are the steps of Covaris M220DNAShearing:
[0091] Power-on inspection
[0092] 1. Check whether the computer fixed on the top of the machine is properly connected to the line of the machine.
[0093] 2. Confirm that the Drip Tray is placed under the machine.
[0094] 3. Insert the operation tube holder (Tube Holder)
[0095] water bath setup
[0096] 1. Pull up the sliding weight (Sliding Weight) on the top of the tube holder (Tube Holder) and rotate it 90°C.
[0097] 2. Use a random water bottle (wash bottle) to add about 15ml of distilled water or deionized water to the center of the bracket. It is advisable that the water level reaches or exceeds the "RUN" marker.
[0098] sample placement
[0099] 1. Pull up the sliding weight (Sliding Weight) on the top of the tube holder (Tube Holder) and rotate it 90°C.
[0100] 2. Put in the sample t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com