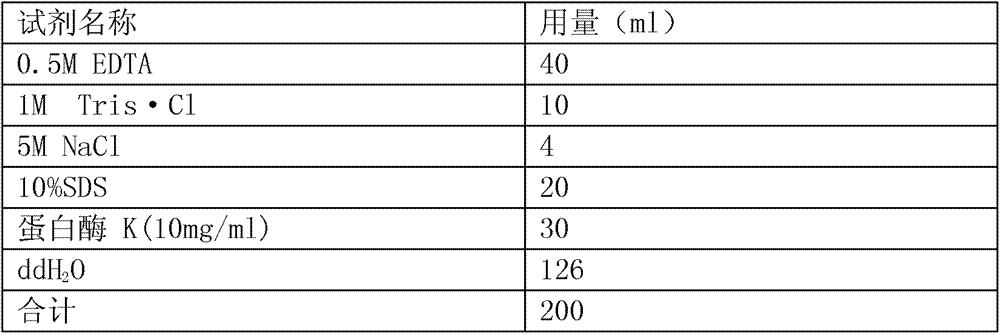
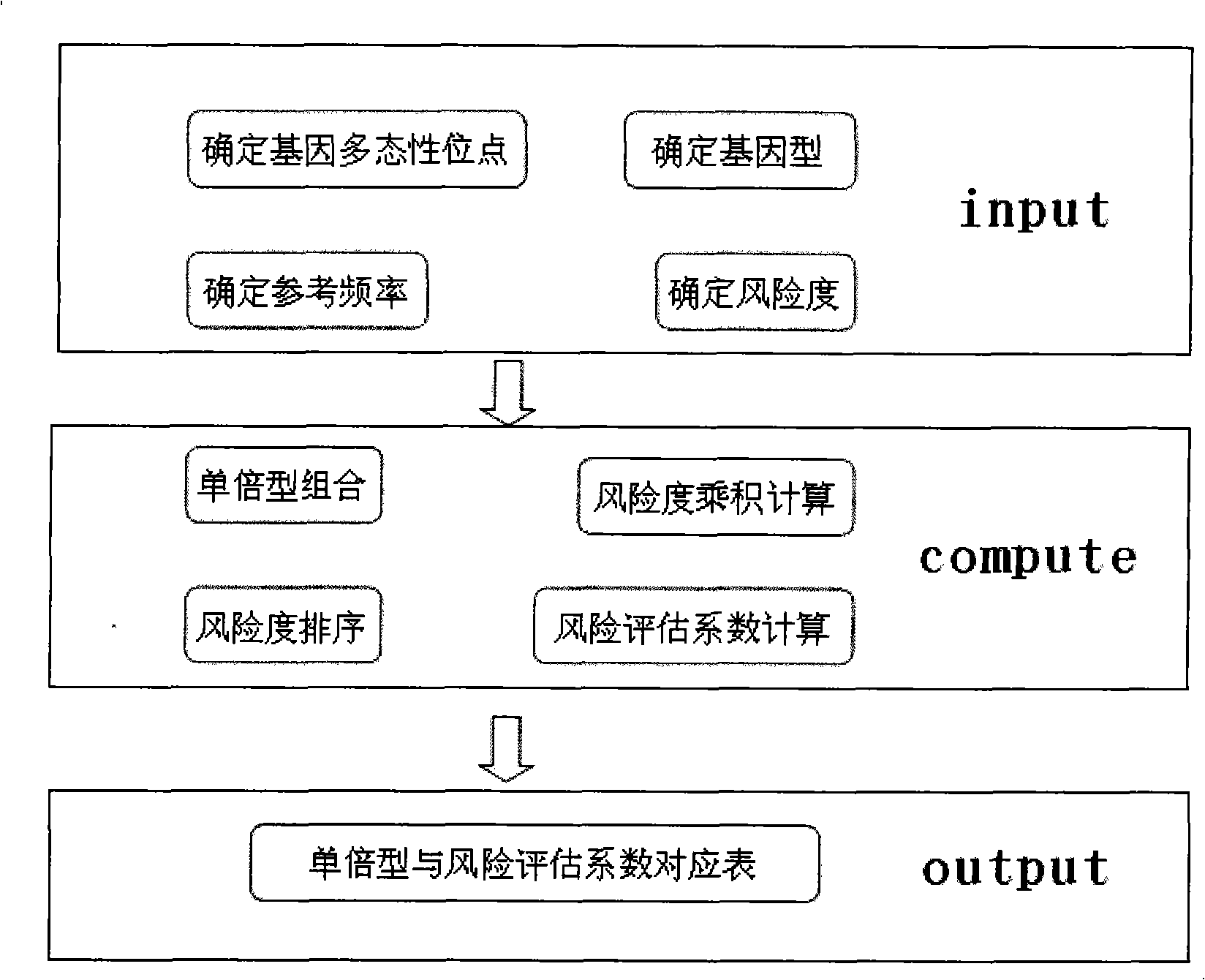
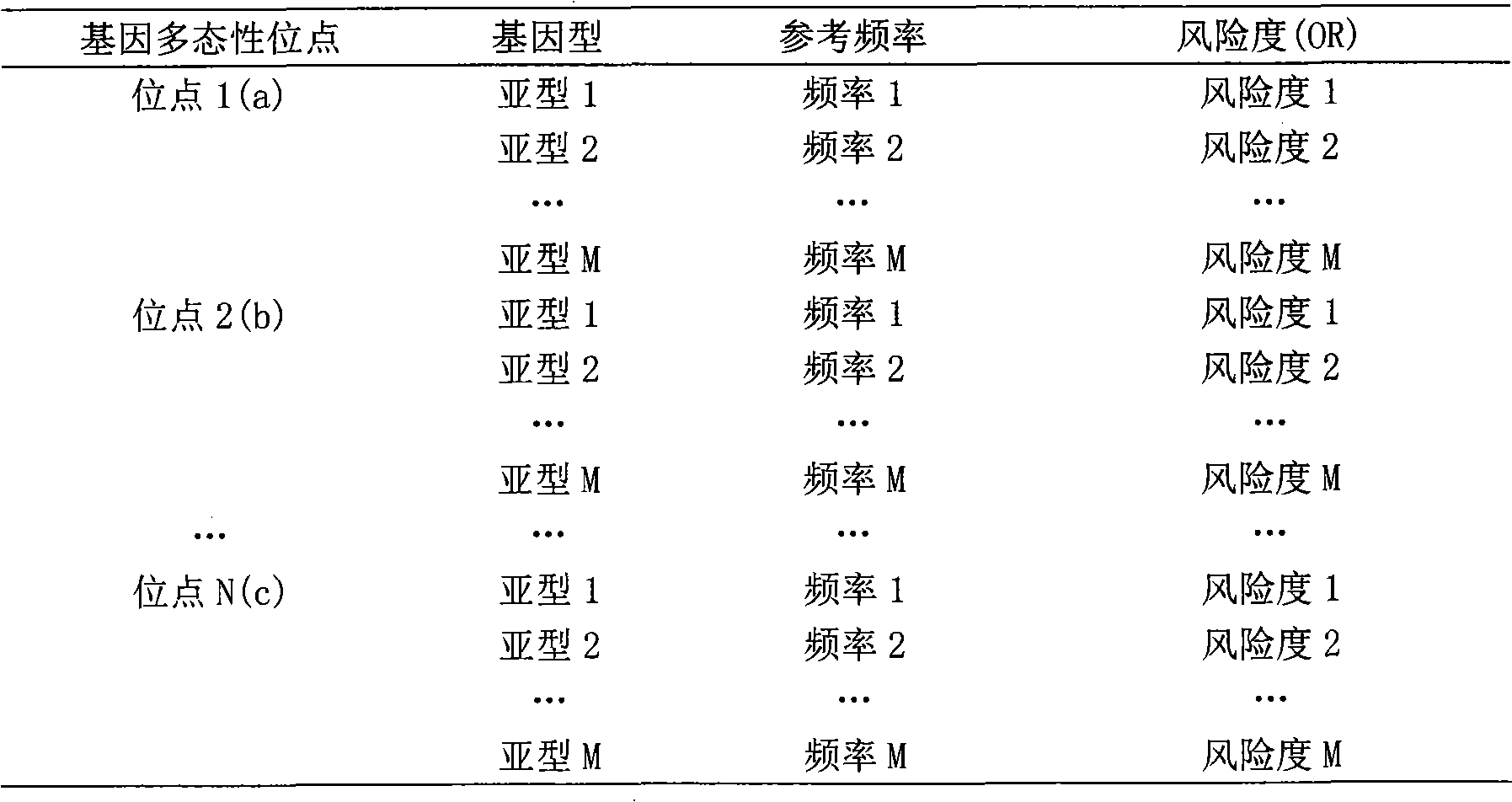
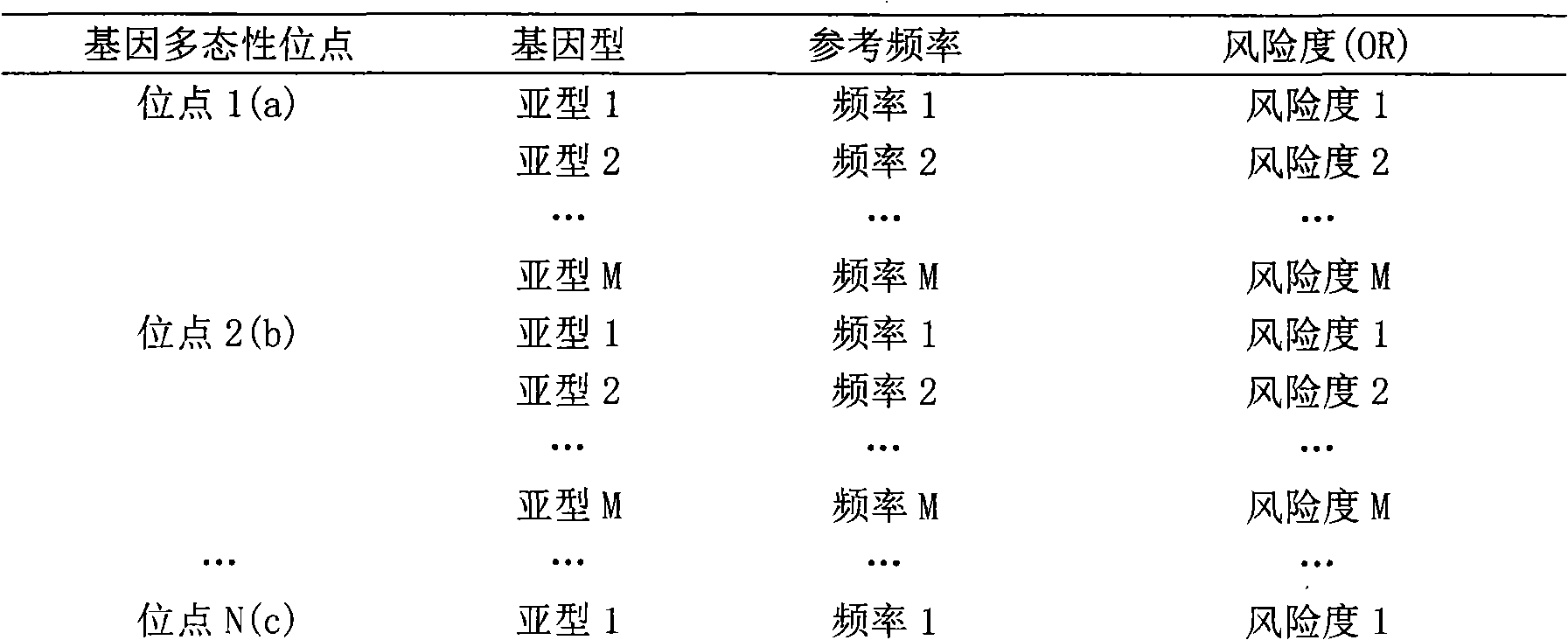
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
511 results about "Polyphenism" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
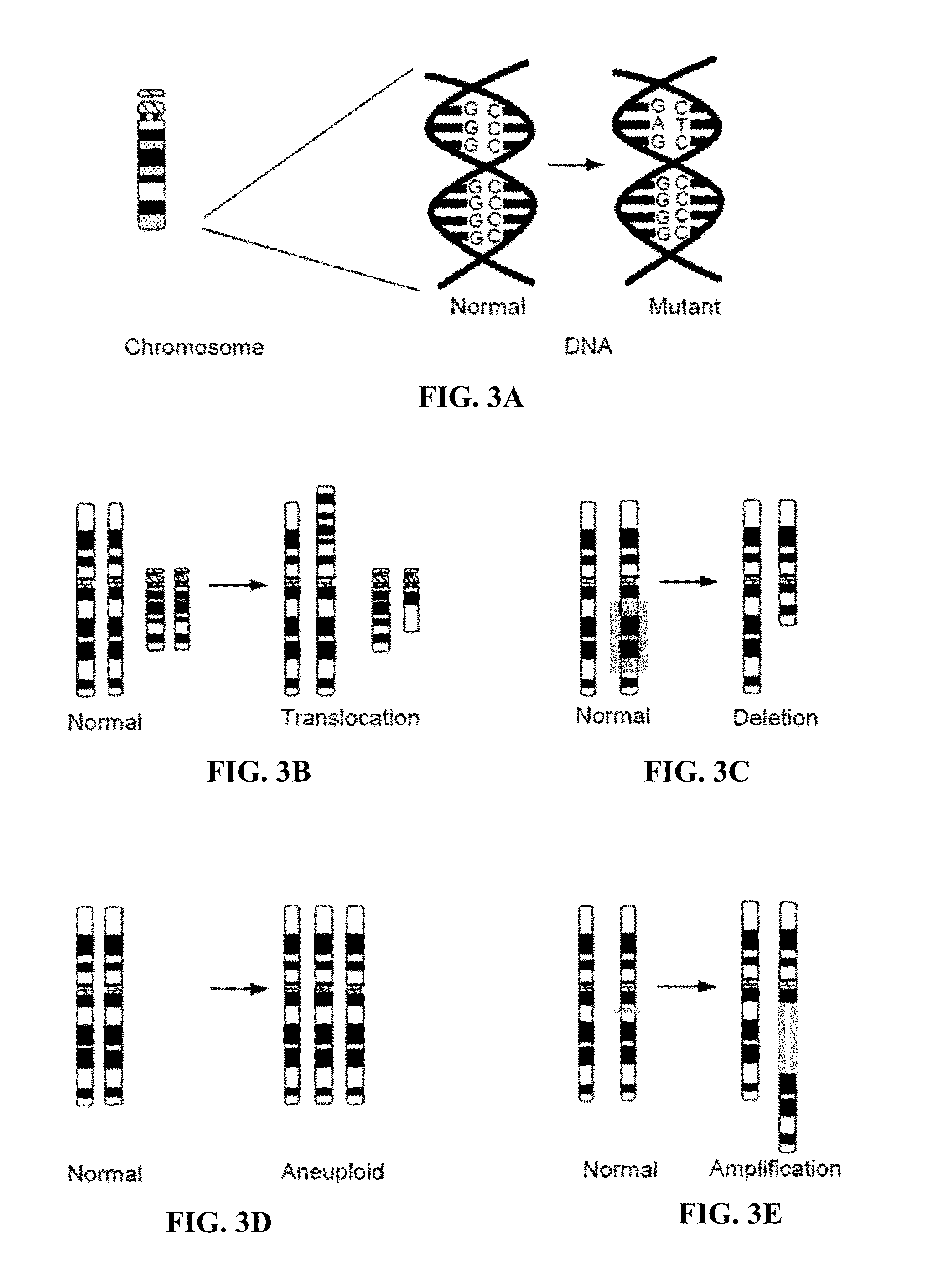
A polyphenic trait is a trait for which multiple, discrete phenotypes can arise from a single genotype as a result of differing environmental conditions. It is therefore a special case of phenotypic plasticity.
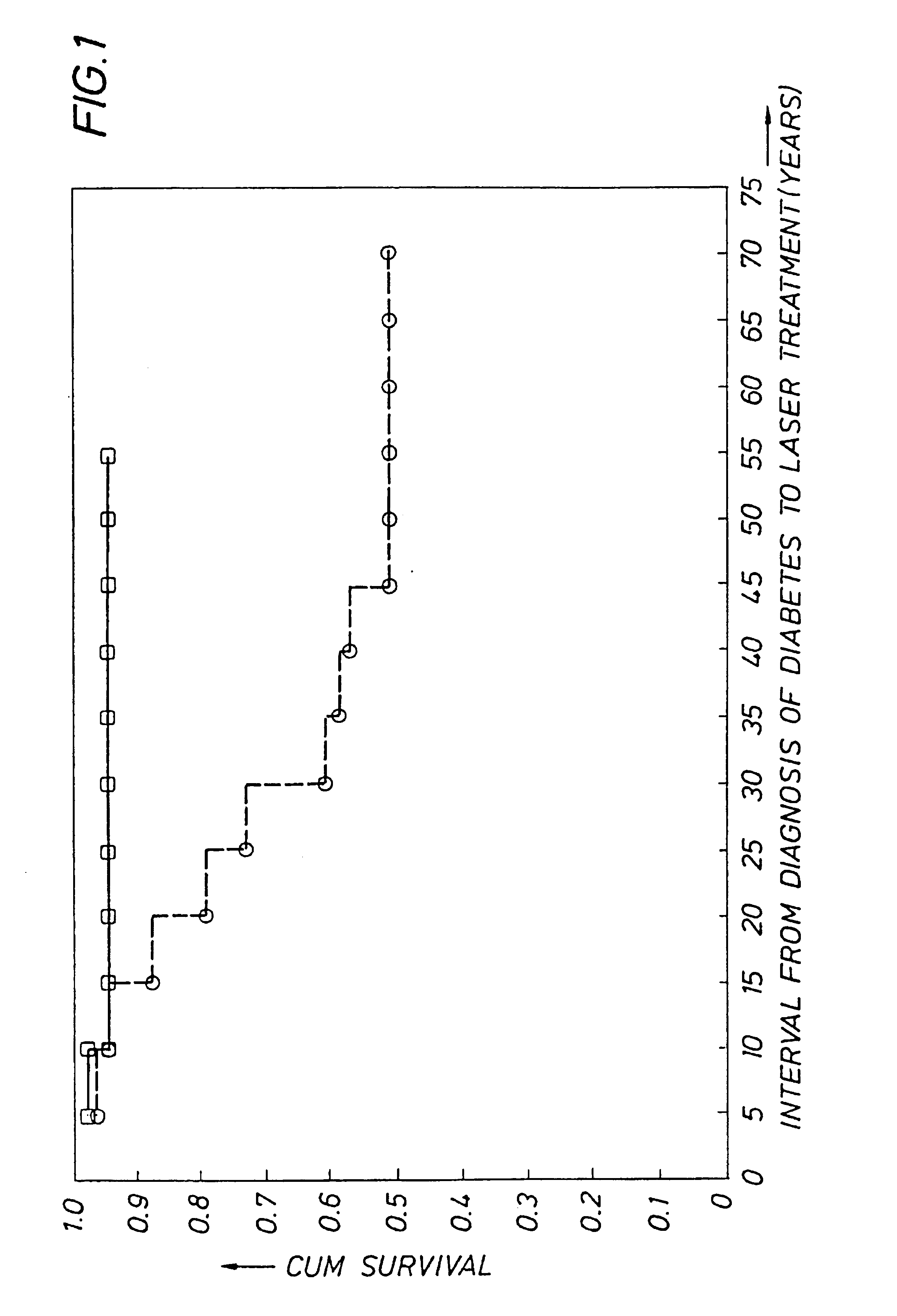
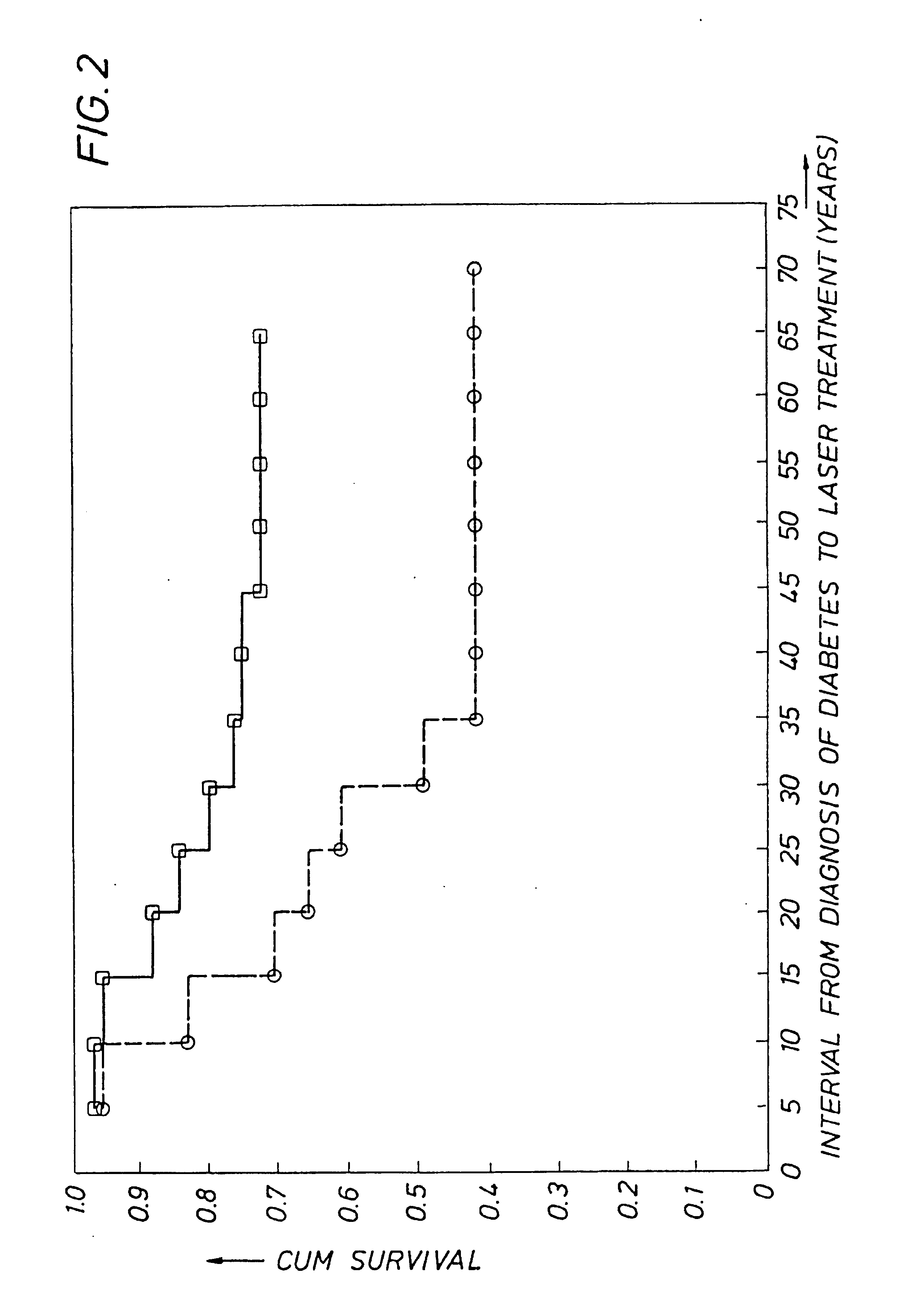
Detecting genetic predisposition to sight-threatening diabetic retinopathy
InactiveUS6713253B1Reduce and prevent occurrencePredict riskSugar derivativesMicrobiological testing/measurementDiabetes retinopathyGenomic DNA
A method and kit for predicting increased risk of sight-threatening diabetic retinopathy which includes isolating genomic DNA from a sample from a diabetic patient. The genetic polymorphism pattern for the genes IL-1A, IL-1B and IL-1RN is then identified in the DNA. The identified pattern is compared with control patterns of known polymorphisms, and patients expressing a genetic polymorphism pattern associated with increased risk of sight-threatening diabetic retinopathy are identified.
Owner:INTERLEUKIN GENETICS
Evaluating genetic disorders
The present invention relates to genetic analysis and evaluation utilizing copy-number variants or polymorphisms. The methods utilize array comparative genomic hybridization and PCR assays to identify the significance of copy number variations in a human, non-human animal, and plant subject or subject group.
Owner:POPULATION BIO INC
Systems and methods for identifying polymorphisms
InactiveUS20150356243A1Convenient treatmentGood estimateHealth-index calculationProteomicsDiseasePhenotypic trait
Owner:MULTIMODAL IMAGING SERVICES CORP +1
Identification and mapping of single nucleotide polymorphisms in the human genome
The invention relates to the role of genes in human diseases. More particularly, the invention relates to compositions and methods for identifying genes that are involved in human disease conditions. The invention provides identification and mapping of a very large number of SNPs throughout the entire human genome. This contribution allows scientists to isolate and identify genes that are relevant to the prevention, causation, or treatment of human disease conditions.
Owner:SNP CONSORTIUM THE
SSR markers lined with major gene of cotton fiber strength
InactiveCN101613761AImprove selection efficiencyGood effectMicrobiological testing/measurementFermentationAgricultural scienceA-DNA
The invention discloser SSR markers lined with major genes of cotton fiber strength, which is obtained by the following steps: generating F2 and F2:3 populations by using a cotton search 41 line sGK9708 selected from cultivated varieties of gossypium hirsutum and a high quality line 0-153 of gossypium hirsutum as parents; allowing generation within the family of the F2:3 population to self cross till the F2:6 generation, performing within-family individual selection of the F2:6 generation once, and planting two generation till F6:8; performing polymorphism screening of the parents by using SSR primers and creating an RIL population linkage map; and performing the multi-environment major QTL screening of the cotton fiber strength to screen 6 QTLs of a cotton fiber strength character from line 0-153, wherein 5 QTLs are multi-environment stable QTLs and are FS1 linkage marker NAU2119330, FS2 linkage markers BNL2572125, BNL1064110 and DPL0874210, FS4 linkage markers are NAU1048250 and NAU2627350, and FS5 linkage markers BNL1421200 and NAU2730450. The SSR markers lined with the major genes of the cotton fiber strength are screen from high fiber quality materials and used as molecular markers to perform early auxiliary selection on a DNA level to improve the selection efficiency of the cotton fiber strength.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Genemap of the human genes associated with crohn's disease
InactiveUS20090181380A1Microbiological testing/measurementAnalogue computers for chemical processesGenomicsLinkage Disequilibrium Mapping
The present invention relates to the selection of a set of polymorphism makers for use in genome wide association studies based on linkage disequilibrium mapping. In particular, the invention relates to the fields of pharmacogenomics, diagnostics, patient therapy and the use of genetic haplotype information to predict an individual's susceptibility to IBD (ex: Chrohn's disease) and / or their response to a particular drug or drugs.
Owner:BELOUCHI ABDELMAJID +12
Molecule marking method for pig backfat thickness property
InactiveCN103497994ASolve the problems of high blindness and low selection accuracyShorten the generation intervalMicrobiological testing/measurementBOARAcyl coenzyme
The invention relates to a molecule marking method for pig backfat thickness property. The method comprises: extraction of pig genome DNA, design of a long-chain acyl-coenzyme Asynthetase 1 (ACSL1) gene seventh exon primer, amplification in vitro and genotype detection. For the first time, it is discovered that the polymorphism of the long-chain acyl-coenzyme Asynthetase 1 (ACSL1) gene seventeenth exon has significant correlation with the pig backfat thickness, and a method for detecting the restriction fragment polymorphism at mutation sites of the seventeenth exon is established. The molecule marking method is applicable to auxiliary selection on backfat thickness property during breeding of pigs, helps to realize early-stage breeding selection of boars, and even helps to accurately select breeding just when pigs are born, so that the generation interval is shortened, and the selection progress of the backfat thickness property is accelerated; and the method is simple in operation, condition requirements during a polymerase chain reaction are low, the length of amplified fragments is relatively short (292 bp), amplification is relatively easy, and the method helps to improve amplification efficiency and accuracy of genotype determining.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY MEDICINE ANHUI ACAD OF AGRI SCI
Comprehensive evaluation method of polygenic diseases genetic risk
The invention relates to a method for performing multi-gene disease inheritance risk comprehensive assessment to an individual. The method comprises the following steps that: inheritance risk factors of a special multi-gene disease in a special crowd are screened; the risk degree of each risk factor to the special multi-gene disease is determined; and resources such as NCBI, HapMap databases and so on, are utilized to determine the frequencies of gene types of polymorphic gene type loci in the special crowd. The method is characterized in that: an assessment model is established, and the risk rank of the multi-gene disease of the individual is assessed from the genetic angle. The method is a comprehensive, objective, easily-operated, and intellectualized risk assessment method.
Owner:上海中优医药高科技股份有限公司
PRKAG3 alleles and use of the same as genetic markers for reproductive and meat quality traits
InactiveUS6919177B2Improve selection accuracyImprove eating qualitySugar derivativesMicrobiological testing/measurementLitterAllele
Disclosed herein are genetic markers for animal meat quality and reproductive efficiency, methods for identifying such markers, and methods of screening animals to determine those more likely to produce larger litters and / or better meat quality and preferably selecting those animals for future breeding purposes. The markers are based upon the presence or absence of certain polymorphisms in the PRKAG3 gene.
Owner:PIG IMPROVEMENT UK
Identification and mapping of single nucleotide polymorphisms in the human genome
The invention relates to the role of genes in human diseases. More particularly, the invention relates to compositions and methods for identifying genes that are involved in human disease conditions. The invention provides identification and mapping of a very large number of SNPs throughout the entire human genome. This contribution allows scientists to isolate and identify genes that are relevant to the prevention, causation, or treatment of human disease conditions.
Owner:SNP CONSORTIUM
Sudden cardiac death mutant gene detection kit
ActiveCN104561310AChange bad habitsAchieve the purpose of preventionMicrobiological testing/measurementHigh risk populationsCvd risk
The invention relates to the field of molecular biology and medical science, and in particularly relates to a sudden cardiac death mutant gene detection kit which is high in accuracy and good in predictability. The kit is used for performing polymorphism detection on 12 SNP sites of 8 major genes related to sudden cardiac death; three forward and reverse specific primers are respectively adopted for each SNP site by combining the characteristics of specific allelic gene PCR (polymerase chain reaction) and temperature gradient descent PCR; and the mutation of the 12 SNP sites can be simultaneously determined by virtue of program amplification of the temperature gradient descent PCR and agarose gel electrophoretic analysis. The detection kit provided by the invention is strong in specificity, high in detection rate, high in efficiency and low in cost, and can be used for screening high-risk population of sudden cardiac death, evaluating the risk degree of having the sudden cardiac death for a detected patient, and making clear pathogenic factors of patient sudden death from a gene level, thereby providing a new way of preventing, diagnosing and treating clinical sudden death.
Owner:谢怡
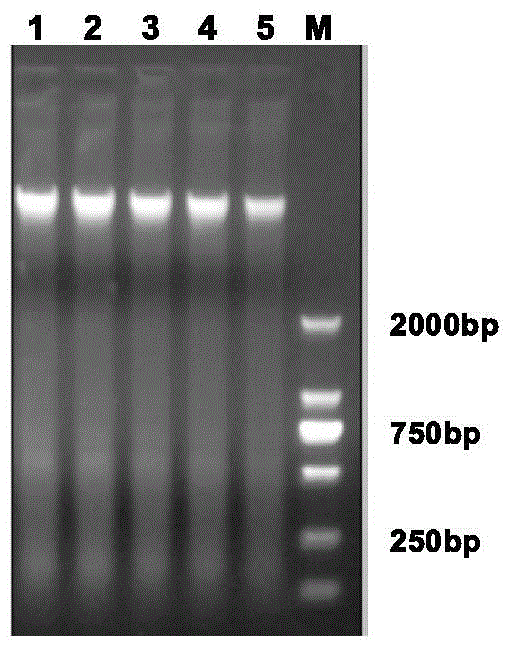
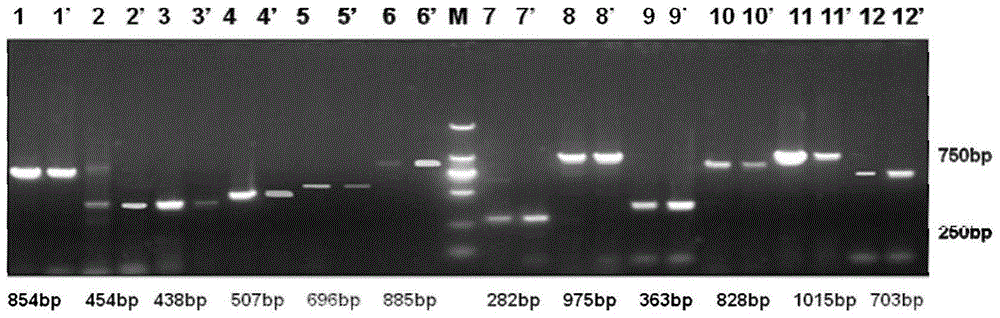
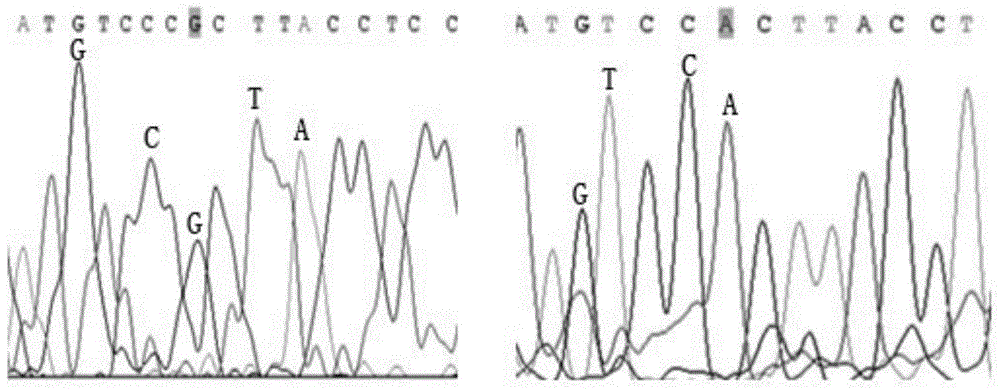
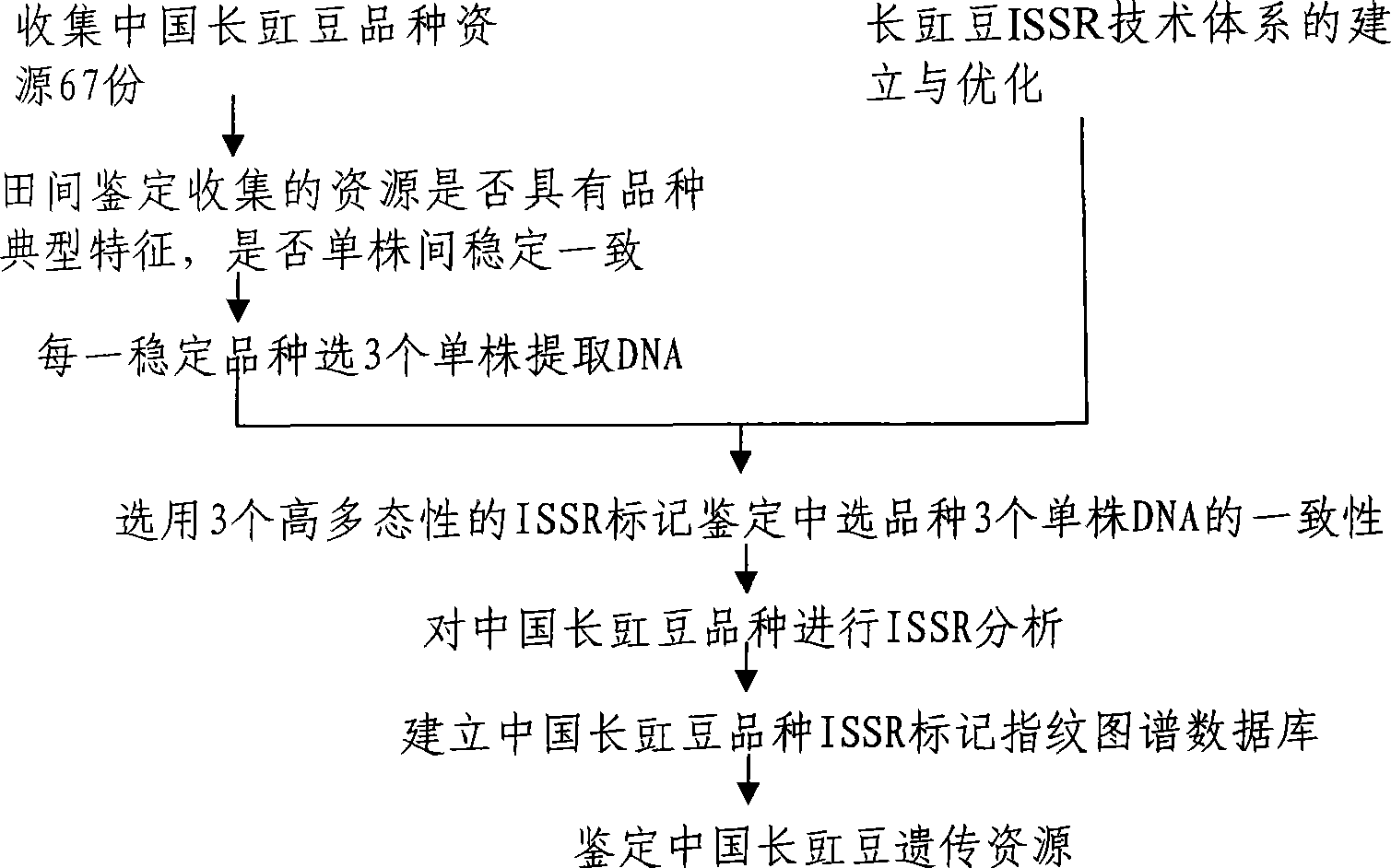
Method for constructing China asparagus bean genetic resource database based on ISSR molecular marker and uses thereof
InactiveCN101436229AValid identificationRapid identificationMicrobiological testing/measurementSpecial data processing applicationsDependabilityBiology
The invention discloses an ISSR molecular marking based method for constructing a Chinese asparagus bean generic resource database and application thereof. The invention also utilizes the data base to identify the trueness of a variety; according to field forms, the consistency and typicality among individual plants of the variety are identified, separated and atypical materials are given up, and DNA of the individual plant of the consistent and typical variety is picked up; by utilizing three ISSR markings with high polymorphism, the consistency of DNA level among the individual plants is primarily selected, and the variety and individual plant constructing a fingerprint are finally determined; and by utilizing the finally selected variety and individual plant, the ISSR analysis of the Chinese asparagus bean is made to construct a fingerprint database. The invention overcomes the defects existing in the construction of the prior generic resource database, such as separation among different individual plants of the variety and impurity on the DNA level, the reliability and accuracy of the Chinese asparagus bean ISSR fingerprint data base constructed by the method are greatly improved, and the planting varieties of the Chinese asparagus bean can be effectively and conveniently identified and distinguished.
Owner:JIANGHAN UNIVERSITY
SNP (single nucleotide polymorphism) marker related to melon powdery mildew resistance and application of SNP marker
ActiveCN105803071AImprove breeding efficiencyEasy to breedMicrobiological testing/measurementPlant genotype modificationAgricultural scienceMolecular breeding
The invention discloses an SNP (single nucleotide polymorphism) marker related to melon powdery mildew resistance and an application of the SNP marker. The application provided by the invention is the application of SNP at a following SNP site in a melon genome or a substance for detecting SNP of the following SNP site in the melon genome in identification or assistant identification of the melon powdery mildew resistance; the SNP site in the melon genome corresponds to the 23rd site in a nucleotide sequence shown in sequence 8 in a sequence table; nucleotide at the SNP site is C or G. The SNP marker provides technical support for selective breeding and molecular breeding of melon powdery mildew resistant varieties, and has significant application value in selective breeding of melon powdery mildew resistant varieties.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
Pepper plants with improved disease resistance
ActiveUS20180171353A1Vector-based foreign material introductionPlant genotype modificationDiseaseGermplasm
The present disclosure provides pepper plants exhibiting resistance to Phytophthora capsici. Such plants may comprise novel introgressed chromosomal regions associated with disease resistance. In certain aspects, compositions, including novel polymorphic markers and methods for producing, breeding, identifying, and selecting plants or germplasm with a disease resistance phenotype are provided.
Owner:SEMINIS VEGETABLE SEEDS
EST-SSR core primer group developed on basis of transcriptome sequence of towel gourd and application thereof
ActiveCN104004833APolymorphism richStable amplificationMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGourd
The invention discloses an EST-SSR core primer group developed on the basis of the transcriptome sequence of towel gourd. The EST-SSR core primer group comprises 50 pairs of primers with nucleotide sequences as shown in SEQ ID No. 1-100 in a sequence table. The 50 pairs of EST-SSR core primers have the advantages of abundant polymorphism, stable amplification, good repeatability, convenience in statistics, etc.; meanwhile, since the primers are originated from the expressed gene of towel gourd and can reflect variation of functional sequences in a genome, so germplasm genetic diversity of towel gourd can be better distinguished. The core primer group can be used in fields like identification of the variety of towel gourd, genetic genealogical analysis of varieties and analysis of germplasm genetic diversity, is beneficial for protection of legal interests of breeders, producers and consumers and promotes sound development of genetic breeding and production of towel gourd.
Owner:INST OF VEGETABLES GUANGDONG PROV ACAD OF AGRI SCI
Authentication method of cattle CAPN1 gene as longisimus dorsi tenderness molecular marker and application
InactiveCN101624598AMicrobiological testing/measurementGenetic engineeringAgricultural sciencePolymorphism analysis
The invention belongs to the technical field of animal gene engineering, relating to clone of cattle longisimus dorsi tenderness related CAPN1 gene segment and an application thereof in cattle marker assisting selection. The obtained CAPN1 gene segment has the nucleotide sequence length of 520bp and comprises full fourteenth exons. A base mutation from A to G exists at the 166bp of the segment, which results in the generation of PsuI-RFLP restriction enzyme polymorphism. The mutation site is taken as a molecular marker, the restriction enzyme PsuI is used for detecting the mutation site, and association analysis can be carried out on the detection result and the cattle longisimus dorsi tenderness. Analysis result shows that striking difference exists among the longisimus dorsi tenderness of different genotype individuals, thus providing a theoretical basis for the seed selection and breeding of cattle. The invention is characterized by obtaining the CAPN1 gene part segment related to the cattle longisimus dorsi tenderness and providing the molecular marker and the application for the marker assisting selection of cattle by using a PCR-RFLP method to carry out polymorphism analysis on the segment.
Owner:JILIN UNIV
Molecular marker for identifying genetic gender of nibea albiflora and application thereof
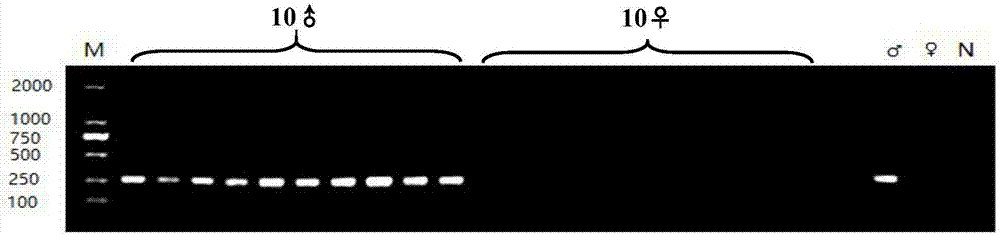
ActiveCN107326077AImprove aquaculture profitabilityRapid identificationMicrobiological testing/measurementDNA/RNA fragmentationEconomic benefitsNucleotide sequencing
The invention discloses a molecular marker for identifying the genetic gender of nibea albiflora and application thereof. The molecular marker shows insertion / deletion length polymorphism of a nucleotide sequence shown as SEQ ID NO: 1; an individual having the nucleotide sequence shown as SEQ ID NO: 1 is a female nibea albiflora, and an individual without the nucleotide sequence shown as SEQ ID NO: 1 is a male nibea albiflora. The invention further discloses a primer pair, a kit and a method for detecting the molecular marker. The molecular marker can simply, conveniently, rapidly and stably identify the genetic genders of different individuals in each colony of the nibea albiflora, so that development of a unisexual breeding technology of the nibea albiflora and development of unisexual breeding of the nibea albiflora are facilitated, the breeding efficiency is further improved and nibea albiflora breeding economic benefits are increased; meanwhile, nibea albiflora gender determining mechanism and other relevant scientific researches are benefited.
Owner:JIMEI UNIV
Kiwi fruit InDel molecular marker and screening method and application thereof
ActiveCN106498075APrimer variant stabilitySpeed up the breeding processMicrobiological testing/measurementDNA/RNA fragmentationResearch ObjectActinidia
The invention discloses a kiwi fruit InDel molecular marker and a screening method and application thereof. Two different cold-proof Actinidia arguta genome DNAs are used as research subjects, resequencing of four variety genes is carried out, InDel primers are designed according to resequencing data, and PCR (polymerase chain reaction) amplification, agarose electrophoresis and non-denaturing polyacrylamide gel electrophoresis are carried out; polymorphism detection is carried in Actinidia arguta to screen out 14 pairs of InDel primers, the 14 pairs of InDel primers are applicable to the genetic diversity analysis for Actinidia chinensis, Actinidia deliciosa and the like and are also applicable to the researches, such as hybrid offspring authenticity identification, genetic map construction, and molecule-assisted breeding. The kiwi fruit InDel primers according to the embodiment of the invention has good mutation stability and low detection difficulty, allow InDel insertion / loss of large fragments, and allows agarose analysis, with steps that may be simplified.
Owner:ZHENGZHOU FRUIT RES INST CHINESE ACADEMY OF AGRI SCI
Microsatellite DNA markers for Trionyx sinensis
The invention discloses microsatellite DNA markers for Trionyx sinensis, which is characterized in that an enriched library of microsatellites (CA)n, (AAC)n, (ACAG)n and (GATA)n from the Trionyx sinensis is constructed, and positive clones of microsatellite sequences are screened and sequenced to obtain 63 clones containing microsatellite repetitive sequences, thus determining 15 microsatellite markers with rich polymorphism, namely, PSW01, PSW02, PSW03, PSW04, PSW05, PSW06, PSW07, PSW08, PSW09, PSW10, PSW11, PSW12, PSW13, PSW14 and PSW15. The microsatellite DNA markers for the Trionyx sinensis of the invention can be applied to researching genetic diversity and paternity test for the Trionyx sinensis in different geographical populations, have the advantage of good repeatability, and are reliable and effective molecular markers.
Owner:ANHUI NORMAL UNIV
Molecular marker of brassica napus anti-clubroot gene and application of molecular marker to anti-clubroot breeding
ActiveCN105063033AImprove breeding efficiencyShorten the breeding processMicrobiological testing/measurementDNA/RNA fragmentationRe sequencingAgricultural science
The invention discloses a molecular marker of a brassica napus anti-clubroot gene PbBa8.1 and a method for applying the molecular marker to anti-clubroot breeding through re-sequencing. The DNA molecular marker, capable of distinguishing anti-clubroot brassica napus and brassica napus being not resistant to clubroot, is obtained by conducting polymorphic analysis on sequences, around a PbBa8.1 locus, of two parents, and carrying out primer development design by utilizing the flanking sequences of an Indel locus. Primers, namely F A08-300: 5'-GTAGTGCGGGCCACAAAAT-3' and R A08-300: 5'-CACAATGGAGTGTTGAAATTCACT-3', aimed at the molecular marker are designed, and can be used for improving the anti-clubroot gene breeding speed and identification efficiency. The method is easy to carry out, high in repeatability, operability and identification speed, and low in detection cost, time consumption and labor cost, and has the advantages that the workload of selecting an anti-clubroot singleplant through phenotype identification in a field is greatly reduced.
Owner:HUAZHONG AGRI UNIV
SNP locus related to growth characteristics of patinopecten yessoensis and detection and application thereof
InactiveCN103740729AHigh trait valueAvoid crossbreedingMicrobiological testing/measurementClimate change adaptationInsulin-like growth factor bindingNucleotide sequencing
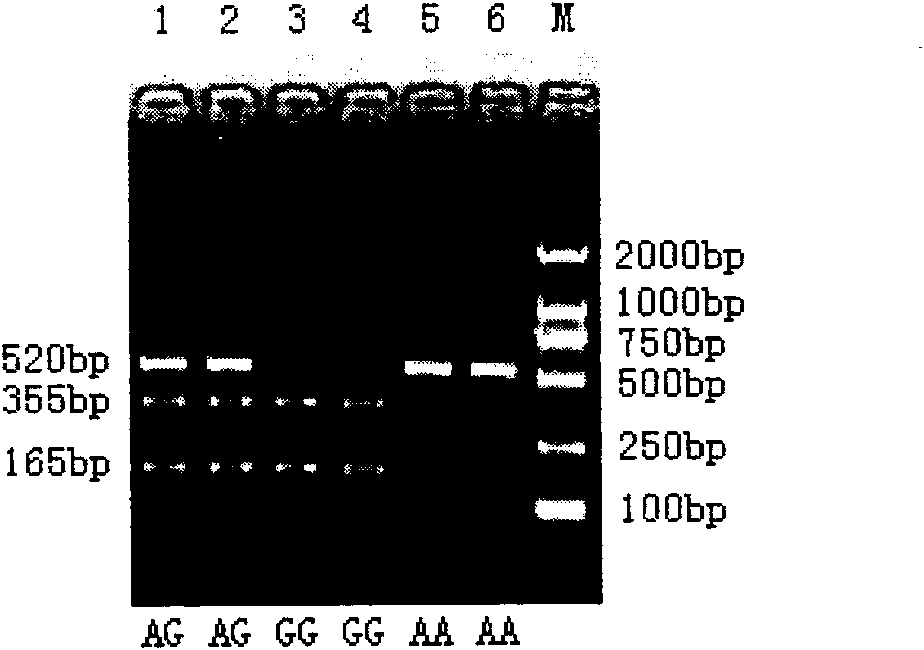
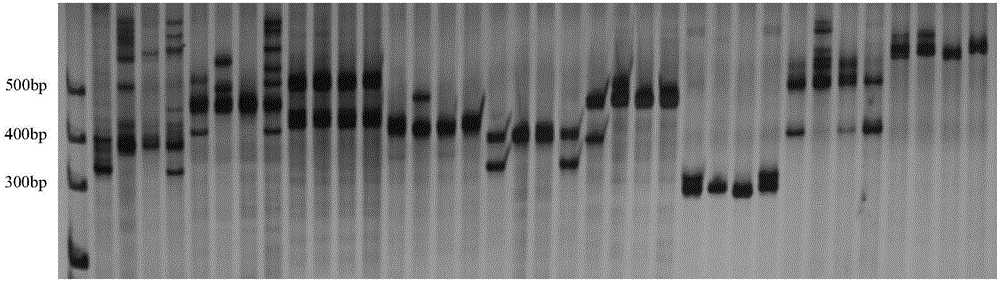
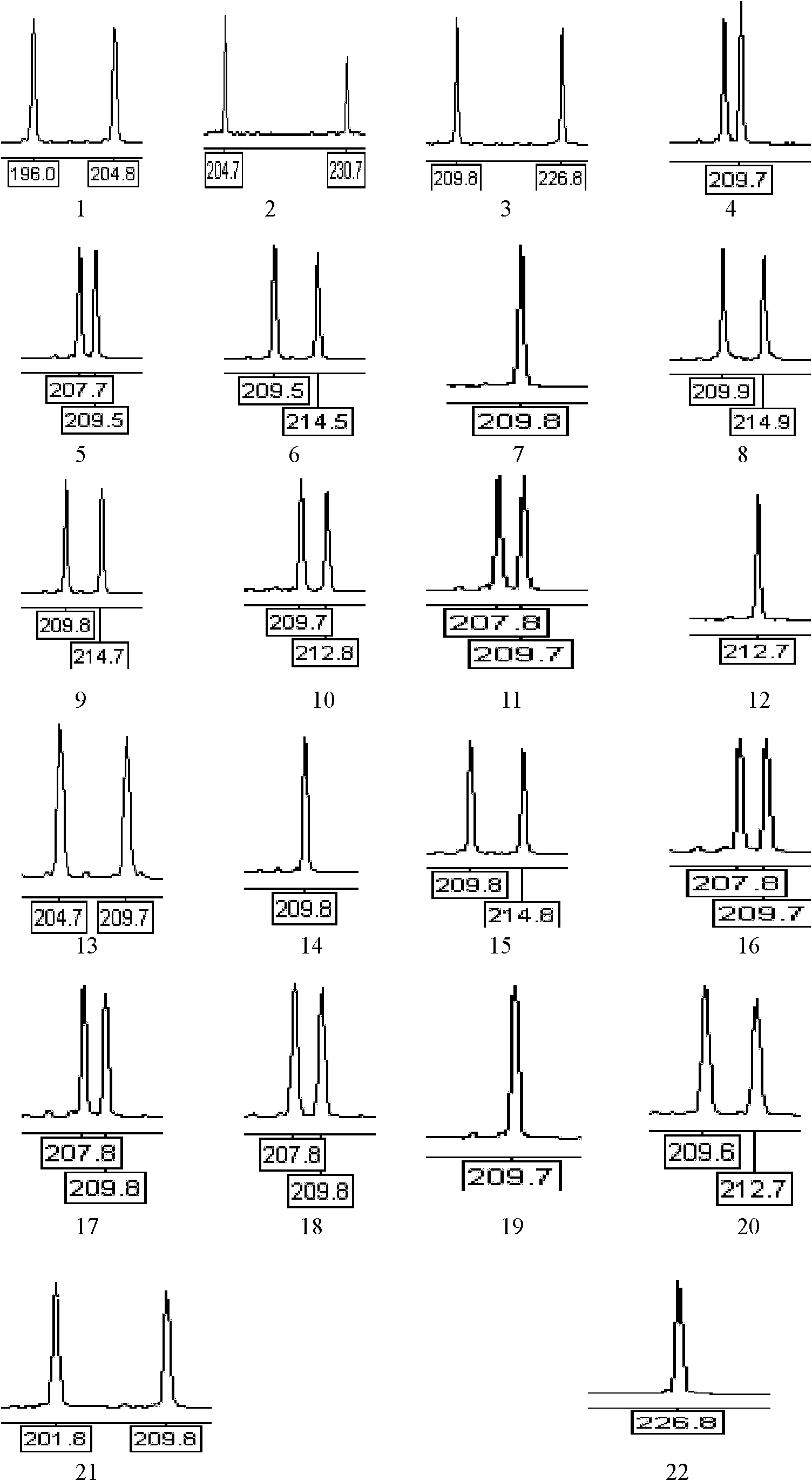
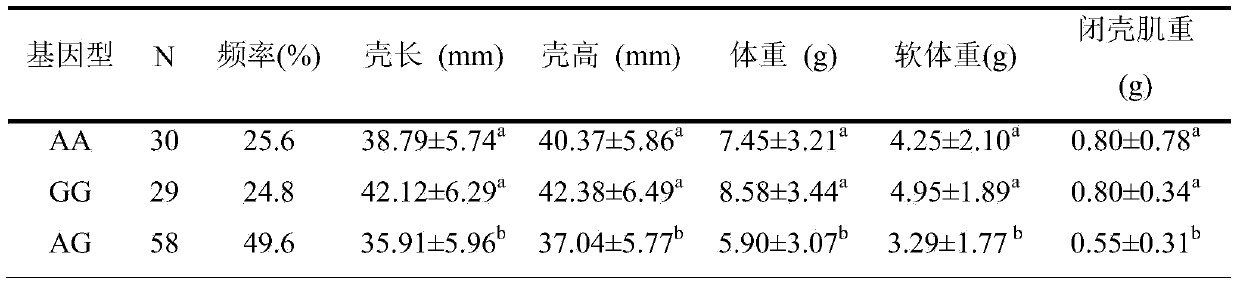
The invention provides an SNP locus related to growth characteristics of patinopecten yessoensis. The locus is the 1054 site of IGFBP (Insulin Like Growth Factor Binding Proteins) genes of which the nucleotide sequence is shown as SEQ ID NO:2 and has the base of A or G. The invention also provides a probe for detecting the SNP locus and a parting primer. The transcription sequence of the IGFBP genes in the patinopecten yessoensis is subjected to sequencing and Clustal comparison, and three SNP loci are screened. The site polymorphism is detected in the patinopecten yessoensis group by using a high resolution melting curve technology, and correlation between the site genotype frequency and the growth characteristics of patinopecten yessoensis is analyzed. The locus C.1054A>G is obviously related to important growth characteristics such as the height, shell length, weight, soft weight and adductor muscle weight of the patinopecten yessoensis, the growth characteristics of the AG type individuals are obviously lower than those of AA and GG type individuals (P is less than 0.05), and the characteristic value of the GG type is the highest. Therefore, individuals of which the genotype is GG on the locus can be preferentially selected during production and are used as parents for performing high-yield patinopecten yessoensis breeding or performing large-scale breeding.
Owner:OCEAN UNIV OF CHINA
Single nucleotide polymorphisms as genetic markers for childhood leukemia
InactiveUS20100092959A1Reduce riskIncreased riskMicrobiological testing/measurementIncreased riskChildhood leukemia
The present invention is directed to a panel of single nucleotide polymorphisms (SNPs) in specific genes that serve as biomarkers for sex-specific childhood leukemia risk. There is provided herein methods and reagents for assessing the specific SNPs in those genes. The method useful in applying these SNPs in predicting an increased risk or a decreased risk for childhood leukemia for males and females is also disclosed.
Owner:MEDICAL DIAGNOSTIC LAB
Cucumber SNP marker and detection methods thereof
InactiveCN101591708AShorten development timeReduce verification timeMicrobiological testing/measurementDNA/RNA fragmentationF1 generationCucumber family
The invention relates to a cucumber SNP marker and detection methods thereof in the technical field of plant gene engineering. A method for identifying cucumber SNP locus comprises the following steps: selecting homozygous cucumber parents, and performing sequencing to determine candidate SNP locus; hybridizing to obtain F1 generation single plants, and performing sequencing; and detecting a peak shape chart of a F1 generation sequencing result to determine candidate locus with heterozygous peaks. A method for determining SNP marked polymorphic single plants in F2 generation separation group of cucumber comprises the following steps: selecting homozygous cucumber parents, and obtaining the F2 generation separation group; selecting recessive phenotype single plants in the F2 generation, mixing DNAs of the single plants in equal quantity, and construct a recessive gene pool; performing amplification and sequencing by an SNP primer, and detecting a peak shape chart of a sequencing result; and respectively performing amplification and sequencing on single plants in the recessive gene pool by the SNP primer, and detecting a peak shape chart of a sequencing result. The cucumber SNP marker has a sequence shown as SEQ ID NO:1. The invention shortens the development and testing time of the SNP marker, and reduces the detection cost of the marker.
Owner:SHANGHAI JIAO TONG UNIV
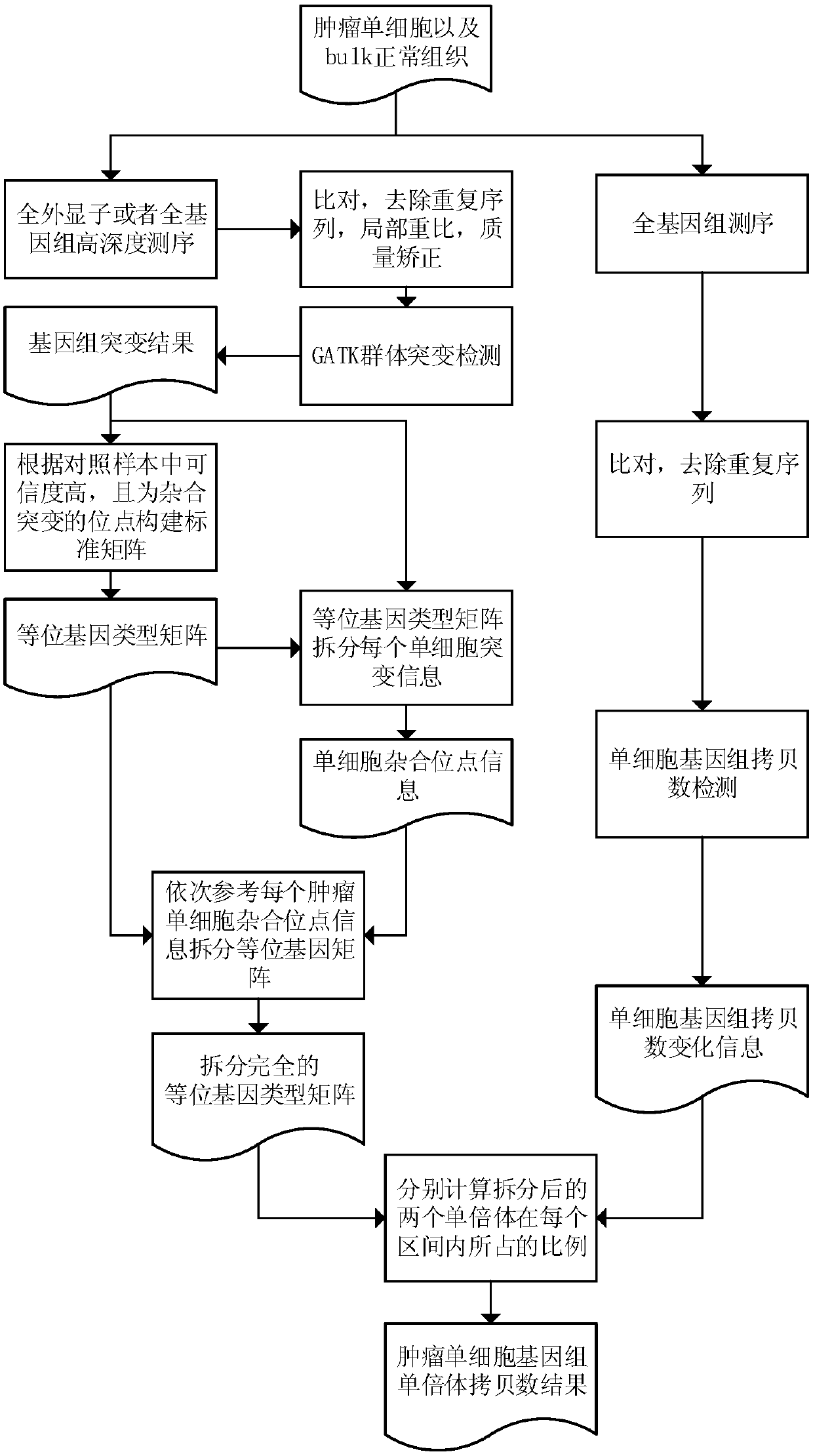
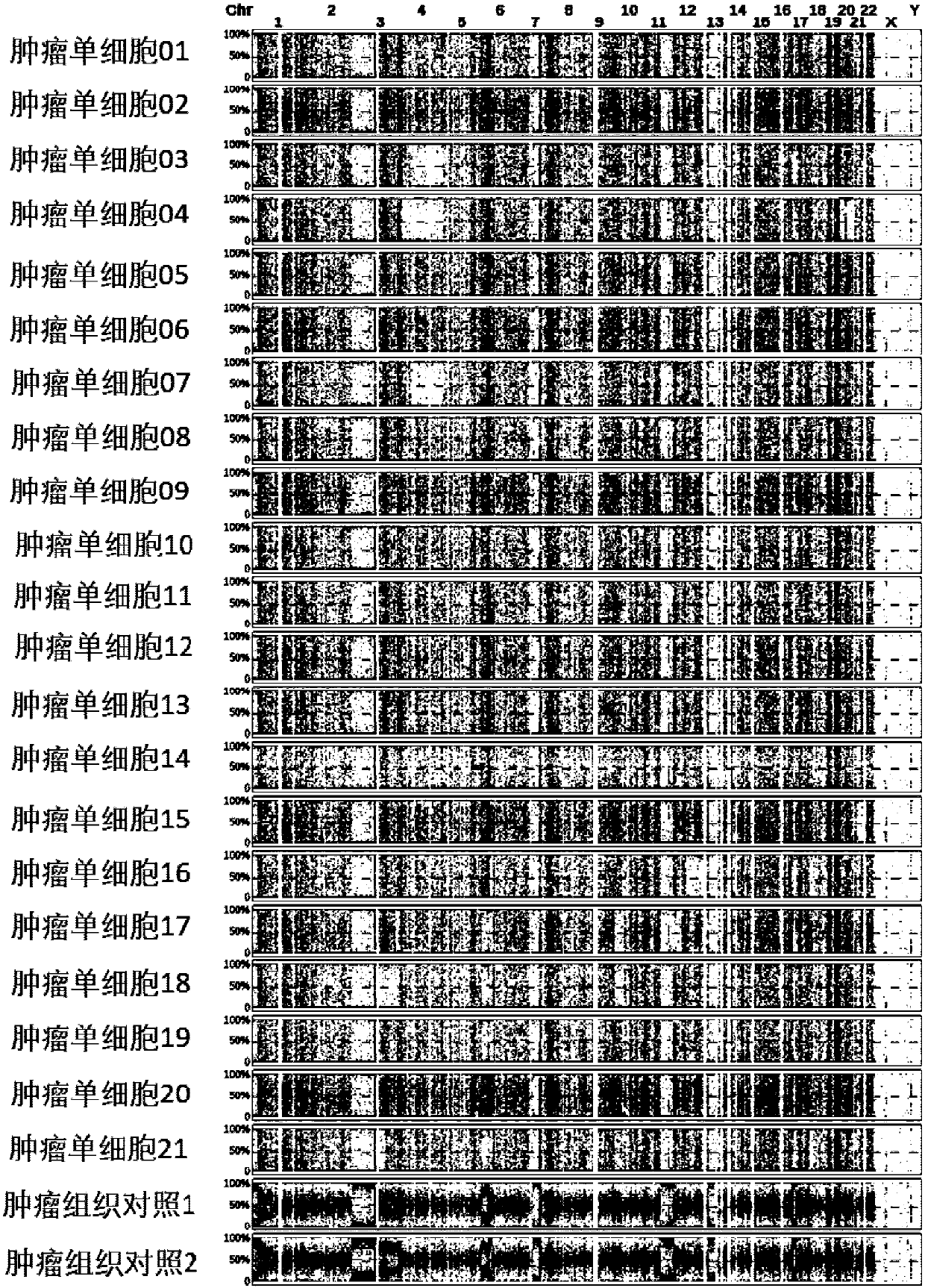
Method for detecting haploid copy number variation of tumor unicell genome
The invention discloses a method for detecting haploid copy number variation of tumor unicell genome. The method combines the population polymorphism site information of the tumor unicell and the change information of the genome copy number to analyze an abnormal region of the tumor unicell genome allele copy number, Compared with simple genome copy number changes or somatic mutations, one more dimension is increased, different unicell alleles can be effectively distinguished to from a zone with proportion abnormity, which has important significance for describe the heterogeneity of tumor genome and tumor genome evolution information.
Owner:PEKING UNIV
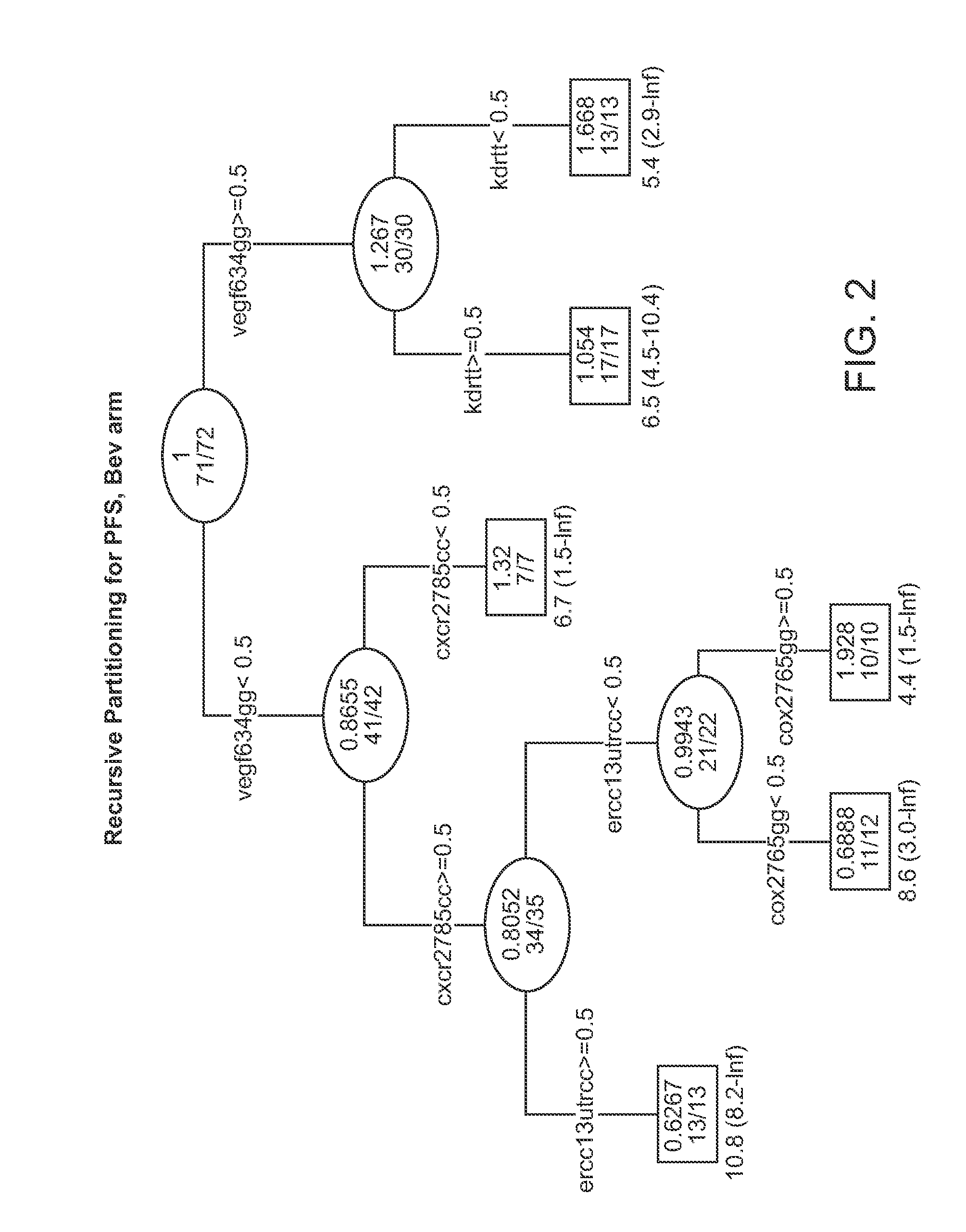
Genetic variants in angiogenesis pathway associated with clinical outcome
InactiveUS20120100134A1Heavy metal active ingredientsMicrobiological testing/measurementGenotype determinationMedicine
The invention provides methods for determining the clinical outcomes for treatment with various treatment regimens available to cancer patients based on genotypes of the patients for genetic polymorphism markers. The invention also provides kits for making the determination.
Owner:UNIV OF SOUTHERN CALIFORNIA
Porcine NTF3 promoter region SNP as molecular marker for boar breeding traits and applications of porcine NTF3 promoter region SNP
ActiveCN107022604AMicrobiological testing/measurementDNA/RNA fragmentationMotilityCorrelation analysis
The invention belongs to the technical field of porcine molecular marker screening, and in particular relates to porcine NTF3 promoter region SNP as a molecular marker for the boar breeding traits and applications of the porcine NTF3 promoter region SNP. The molecular marker is cloned from the 5' flank promoter fragment of the porcine NTF3 gene, the nucleotide sequences of the molecular marker are shown as SEQ ID NO:1 and SEQ NO:2, allele mutation of A / G and G / A exists at the 92nd basic group site of the sequence shown as SEQ ID NO:1 and SEQ NO:2, and the allele mutation causes the polymorphism of MvaI-RFLP. The marker is utilized for carrying out correlation analysis on the boar breeding traits of large white pigs and duroc pigs. The breeding traits comprise the boar sperm motility and the sperm density trait. The invention further discloses a typing detection method of the marker, and therefore, a novel marker is provided for selecting the boar breeding traits. The marker can be used for evaluating the boar breeding traits and researching the genetic improvement on the breeding boar breeding performance.
Owner:HUAZHONG AGRI UNIV
Molecular marker linked with bruchid resistance gene in mung bean
InactiveCN101358232AAssisted breeding selection with clear goalsShorten the timeMicrobiological testing/measurementPlant genotype modificationResistant genesAgricultural science
The invention relates to a molecular marker for mungbean weevil resistance gene linkage, which is used for screening mungbean weevil resistant genes. V2709, VC1973 (zhonglu No. 1) and the individual DNA of F2 separation population thereof are extracted by the CTAB method. 63 RAPD molecular markers, 100 pairs of SSR molecular markers and 28 pairs of STS molecular markers are adopted to carry out the PCR product polymorphism screening on the resistant bulk and the susceptible bulk of the two parents and the F2 progeny, and then each of F2 generation individuals is screened out the weevil resistance gene linkage marker. A RAPD primer, OPC-06 and a STS primer are found to link with a weevil resistance genes The molecular marker can target the weevil resistance gene, and the selection efficiency of weevil resistance breeding is greatly improved.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
Molecular marking method for indicating and identifying litter size in pigs by MMP23 gene
InactiveCN101440399AGood choiceMicrobiological testing/measurementFermentationNucleotideLong segment
The invention aims to provide a detection method for searching the genetic mutation sites of porcine MMP23 and the genetic polymorphism, namely, the molecular marking method for speculating and identifying the litter size with MMP23. The invention provides the sequence of genomic nucleotides with a length of 482bp including the exons 2, 3 and 4 and the introns 2 and 3 of the porcine MMP23 gene of large white sows, Meishan pigs and Qingping pigs, and provides the single nucleotide polymorphism (SNP) of at two sites situating in the interior of the proliferated 482bp-long segment. The method provided by the invention can be used for developing diagnostic procedures or kits used for selecting the pigs carrying the advantageous alleles in breeding programs with better selective outcome.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
Prepn process and application of sea island cotton EST SSR marker
The present invention belongs to the field of cotton breeding technology, and is especially the preparing technology of sea island cotton EST-SSR primer sequence and its application in cotton genetic diversity evaluation, molecular genetic linkage construction and TQL location of important cotton characters. The preparation process includes the following steps: establishing cDNA library of sea island cotton strain Pima3-79 fiber development and selecting EST of partial bases; designing SSR lateral wing primer; taking total DNA with plant leaf; amplifying with the designed primer; calculating the polymorphism information content of each pair of primers and performing cluster and main coordinate analysis of taken total DNA; and integrating the amplified EST-SSR to corresponding linkage group and performing TQL location of important cotton characters. The preparation results are also given.
Owner:HUAZHONG AGRI UNIV
Kit for detecting disease-causing genic mutation of neural tube defect of neonatus and application thereof
InactiveCN102181569AReduce morbidityQuick and easy detectionMicrobiological testing/measurementFluorescence/phosphorescenceDiseasePrenatal diagnosis
The invention discloses a kit for detecting whether mutation occurs at a relevant single nucleotide polymorphism (SNP) locus on a disease-causing gene of a neural tube defect of a neonatus. The kit mainly comprises a specific primer pair and a specific fluorescent probe pair which are used for detecting a No.rs1801133 SNP locus polymorphism genotype and a No.rs1801131 SNP locus polymorphism genotype on a methylenetetrahydrofolate reductase (MTHFR) gene and a No.rs1801394 SNP locus polymorphism genotype on a methionine synthase reductase (MTRR) gene, and the conventional fluorescent quantitative PCR reaction reagent. The kit is used for detecting a mutant of the disease-causing gene of the neural tube defect of the neonatus, can be used for quickly and conveniently searching a carrier with the disease-causing gene, can be applied to antenatal diagnosis and timely treatment, and reduces the morbidity of the neutral tube defect of the neonatus so as to fulfill the aims of promoting good prenatal and postnatal care.
Owner:XINBAXIANG SHANGHAI MOLECULAR MEDICAL TECH SHANGHAI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com