Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
84results about How to "Polymorphism rich" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
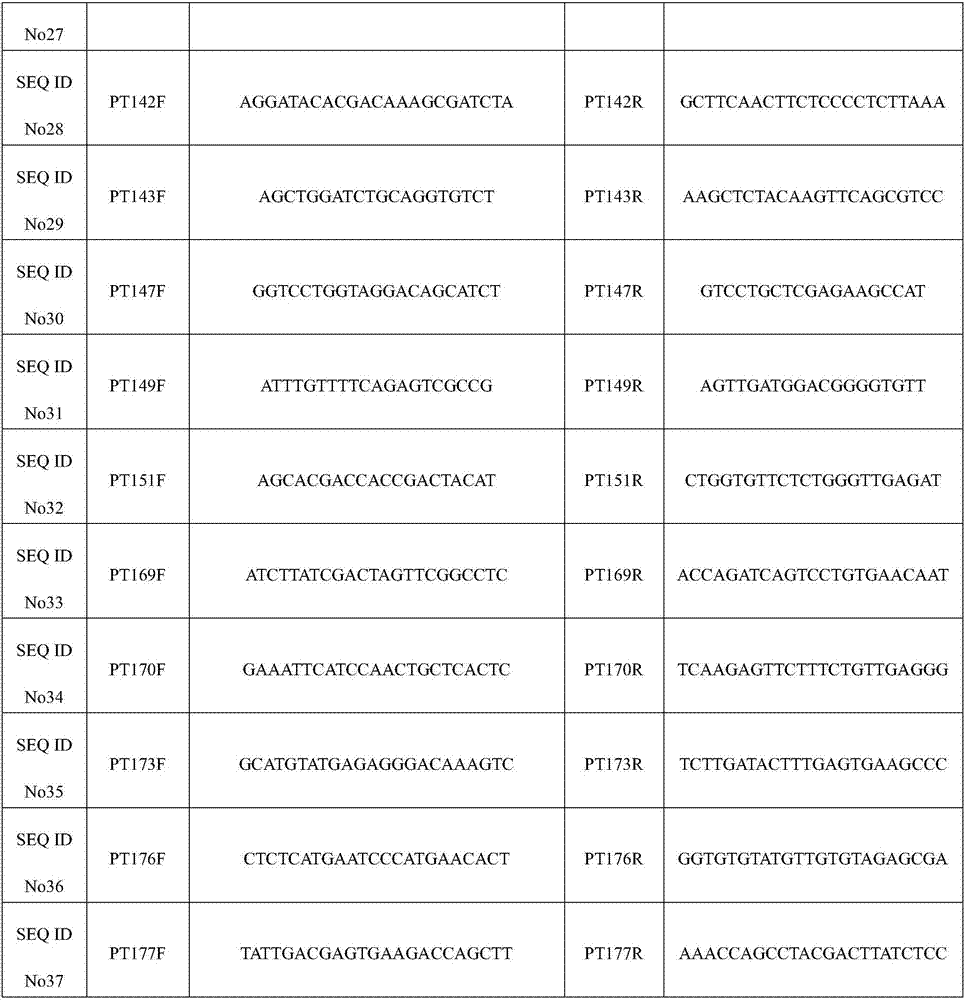
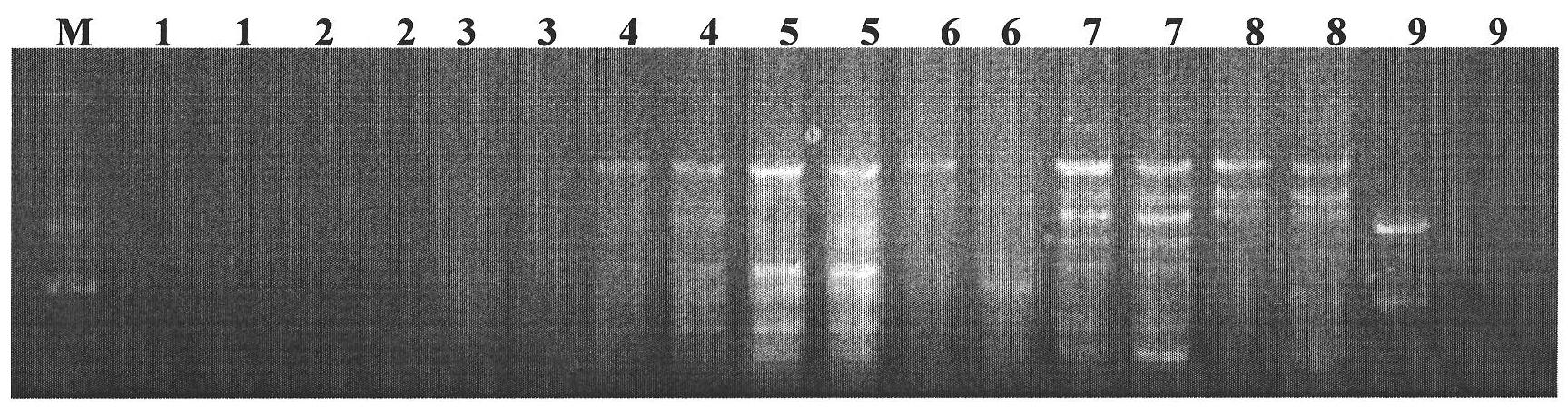
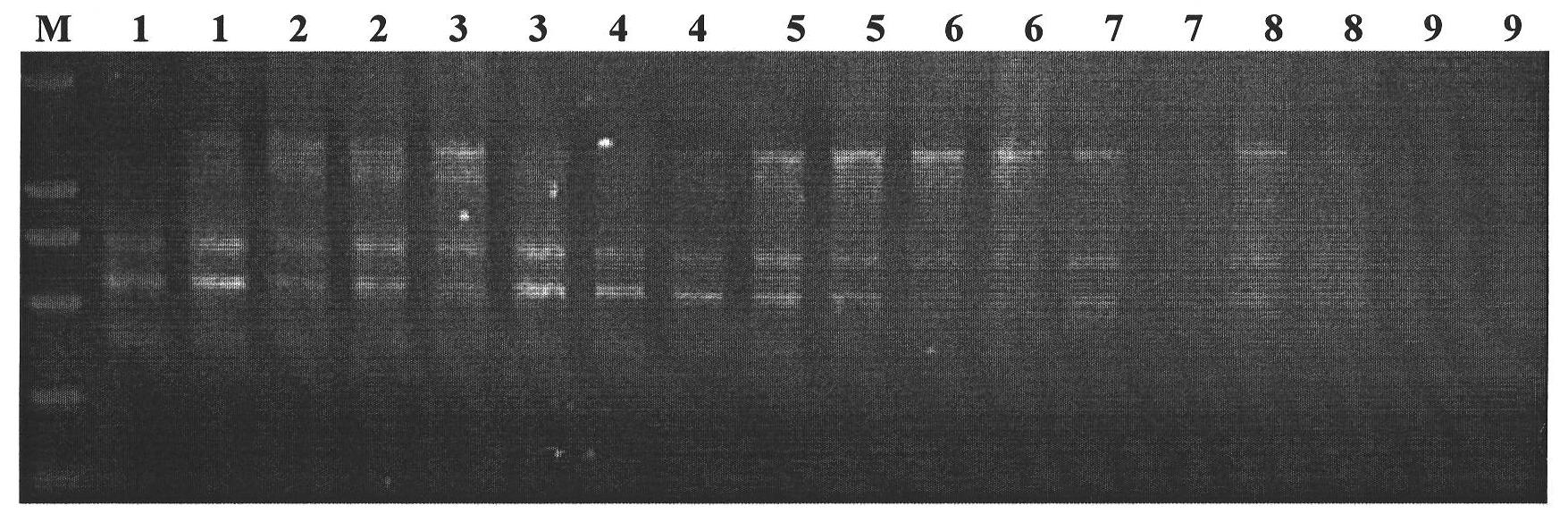
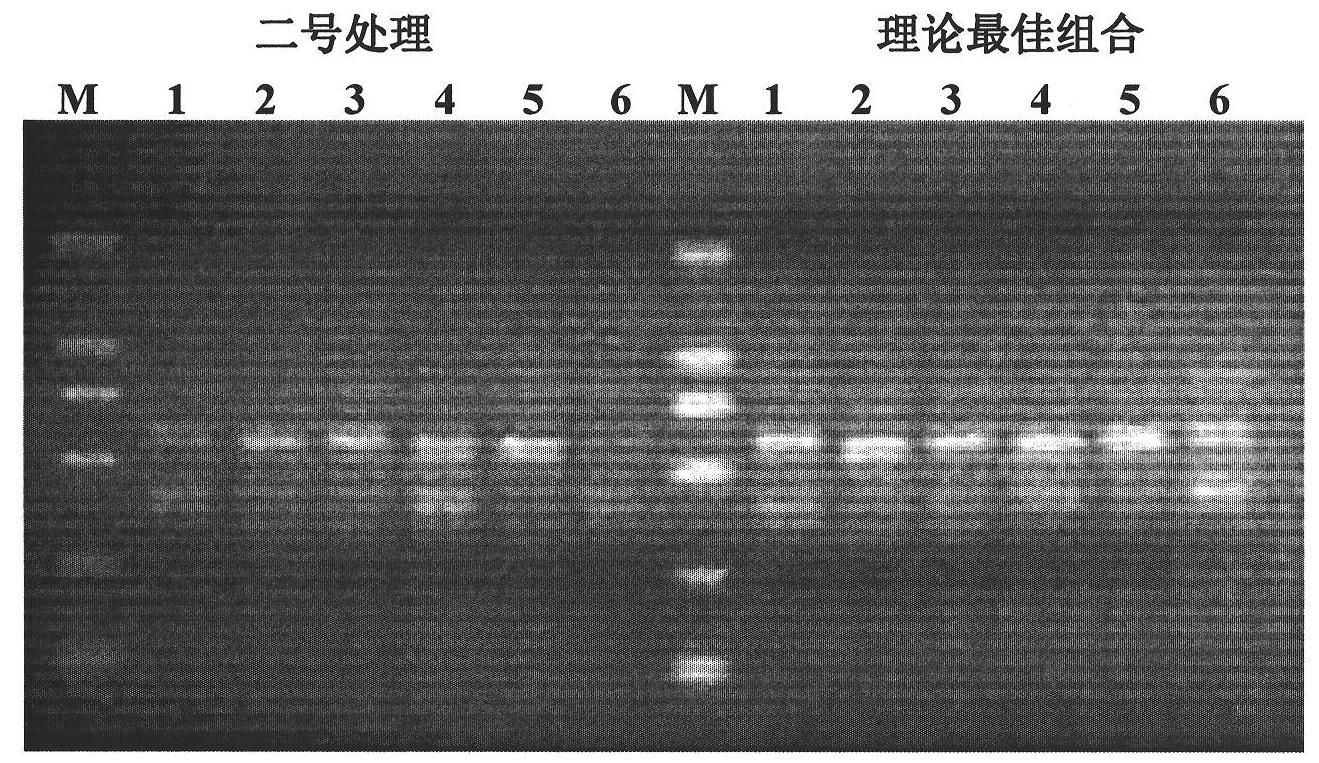
SSR (Simple Sequence Repeat) core primer group developed based on whole genome sequence of foxtail millet and application of SSR core primer group
ActiveCN103060318AImproved geneticsEvenly distributedMicrobiological testing/measurementDNA preparationAgricultural scienceGenetic diversity
The invention discloses an SSR (Simple Sequence Repeat) core primer group developed based on whole genome sequence of foxtail millet and an application of the SSR core primer group, and belongs to the technical field of molecular biology. The core primer group comprises 30 pairs of primers, wherein nucleotide sequences are represented by SEQ ID NO. 1-60. The primer has advantages of clear electrophoretic band and rich polymorphism, and is uniformly distributed and stable in amplification. The invention also provides the application of the SSR core primer group in identifying the genetic diversity and variety of the foxtail millet. The primer group can be used for precisely and quickly identifying the variety of the foxtail millet and precisely reflecting a genetic relationship among the varieties of the foxtail millet.
Owner:CROP RES INST SHANDONG ACAD OF AGRI SCI
Method for quickly identifying purity of watermelon variety Zhentian 1217 by using EST-SSR (expressed sequence tag-simple sequence repeat) molecular marker
The invention relates to the field of molecular markers, and provides a method for quickly identifying purity of a watermelon variety Zhentian 1217 by using an EST-SSR (expressed sequence tag-simple sequence repeat) molecular marker. According to the method, a specially designed EST-SSR primer is adopted, a genome DNA of the Zhentian 1217 is used as a template to perform amplification, and a specific band of an amplification result is used for detecting the variety purity of the Zhentian 1217 to be detected. By adopting the method, the identification can be performed at any period of the growth of a Zhentian 1217 plant, and hybrid seeds can be differentiated from female parent selfed seeds and male parent selfed seeds; and the method is high in accuracy and broad in application popularization prospect, and provides a scientific basis for carrying out watermelon seed preparation and selling high-quality seeds timely.
Owner:华盛农业集团股份有限公司
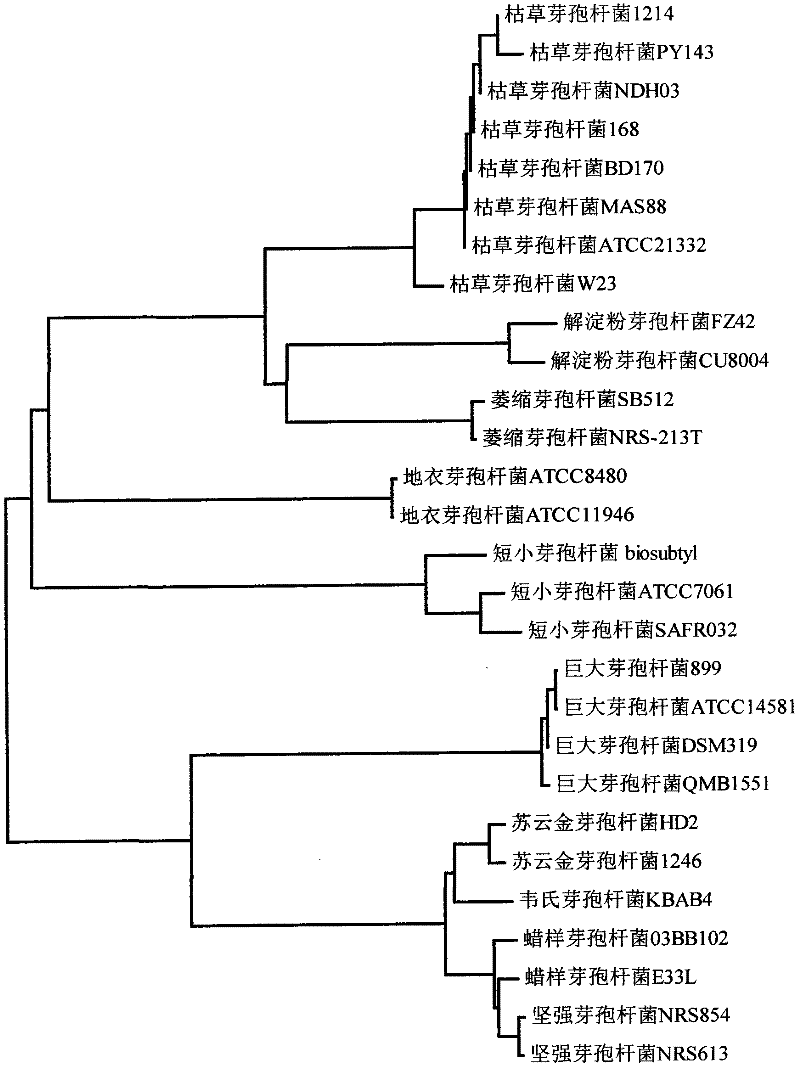
Universal primers for identifying bacillus and classification method using same
ActiveCN102226216APolymorphism richThe classification result is accurateMicrobiological testing/measurementDNA/RNA fragmentationClassification methodsSubspecies
The invention discloses universal primers for identifying bacillus. The universal primers comprise a forward degenerate primer composed of a nucleotide sequence shown as SEQID NO:1 and a reverse degenerate primer composed of a nucleotide sequence shown as SEQID NO:2. The invention also discloses a method for classifying and identifying bacillus, which comprises the following steps: utilizing the universal primers to perform PCR (Polymerase Chain Reaction) amplification; sequencing the PCR amplification product; utilizing sequence comparison software to perform sequence comparison in a Genbank data base; and acquiring the variety of a known strain having the highest sequence similarity, namely the variety of the strain. All varieties of bacillus can be classified by the universal primers, and the classification result is accurate and reliable; and the universal primers can be used for classifying bacillus on a variety level and can be also used for classifying bacillus on a subvariety level. Besides, the classification and identification method is simple and has a high identification speed.
Owner:南京科绿丰科技有限公司
Traceablility SNP marker in pig GIGYF2 gene and detection method thereof
InactiveCN102485892AReduce distribution variancePolymorphism richMicrobiological testing/measurementDNA/RNA fragmentationGenomic DNADNA fragmentation
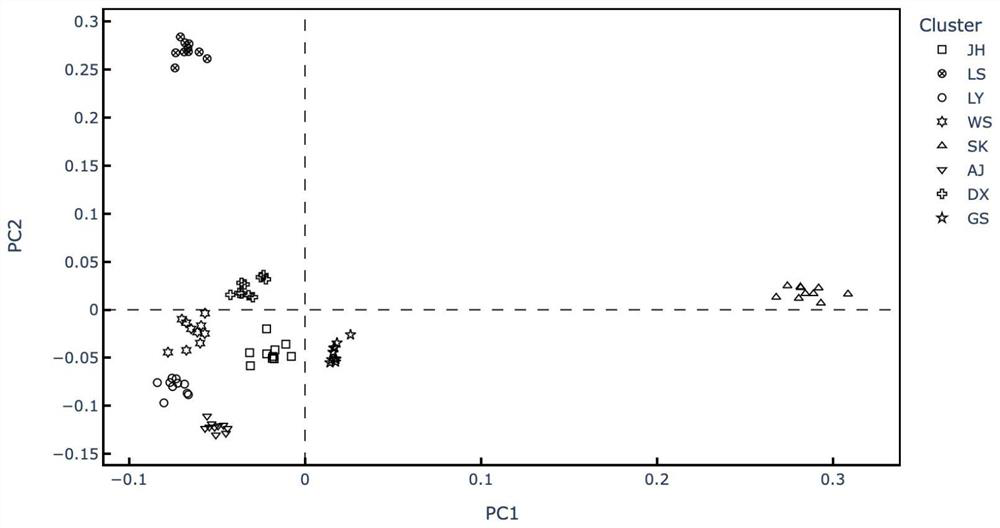
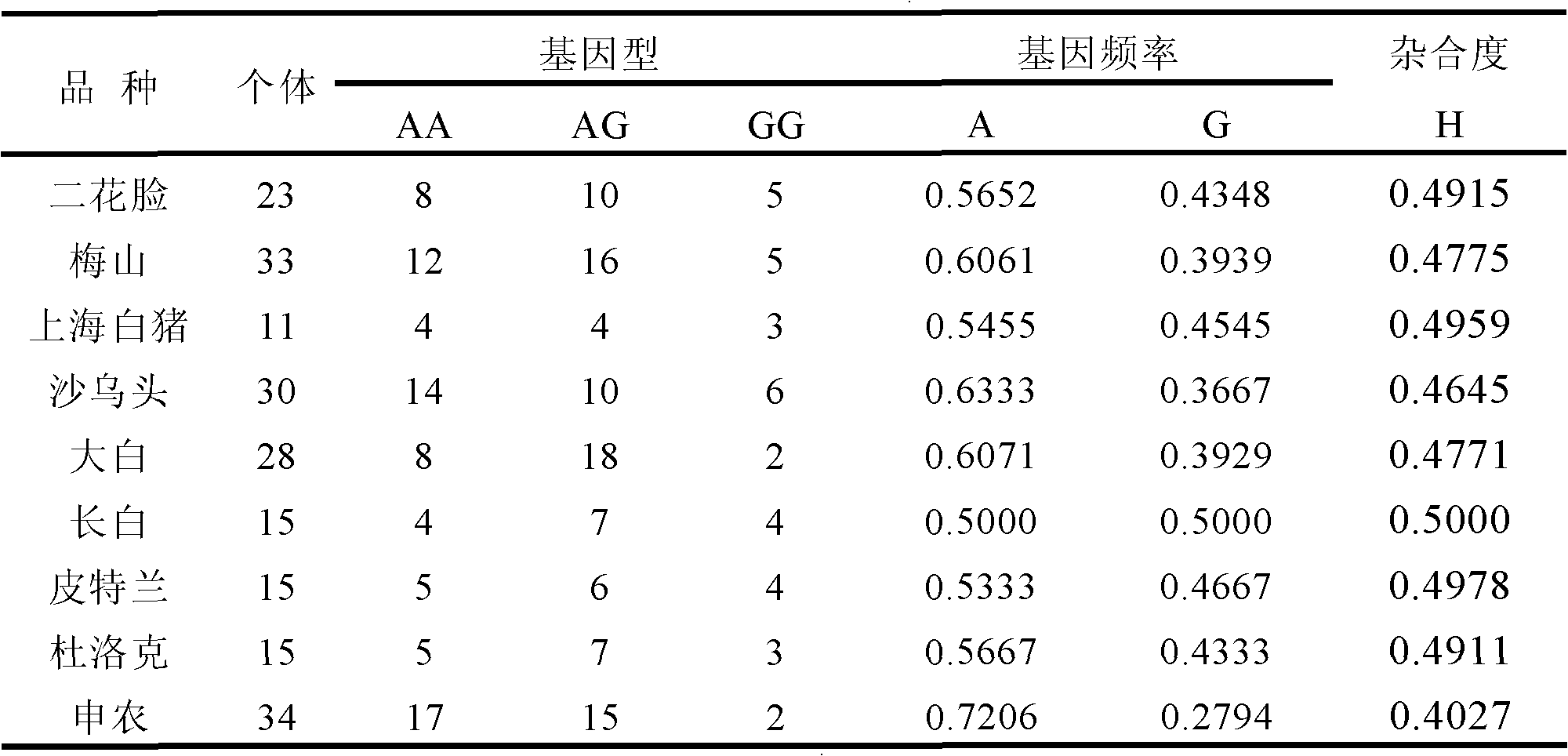
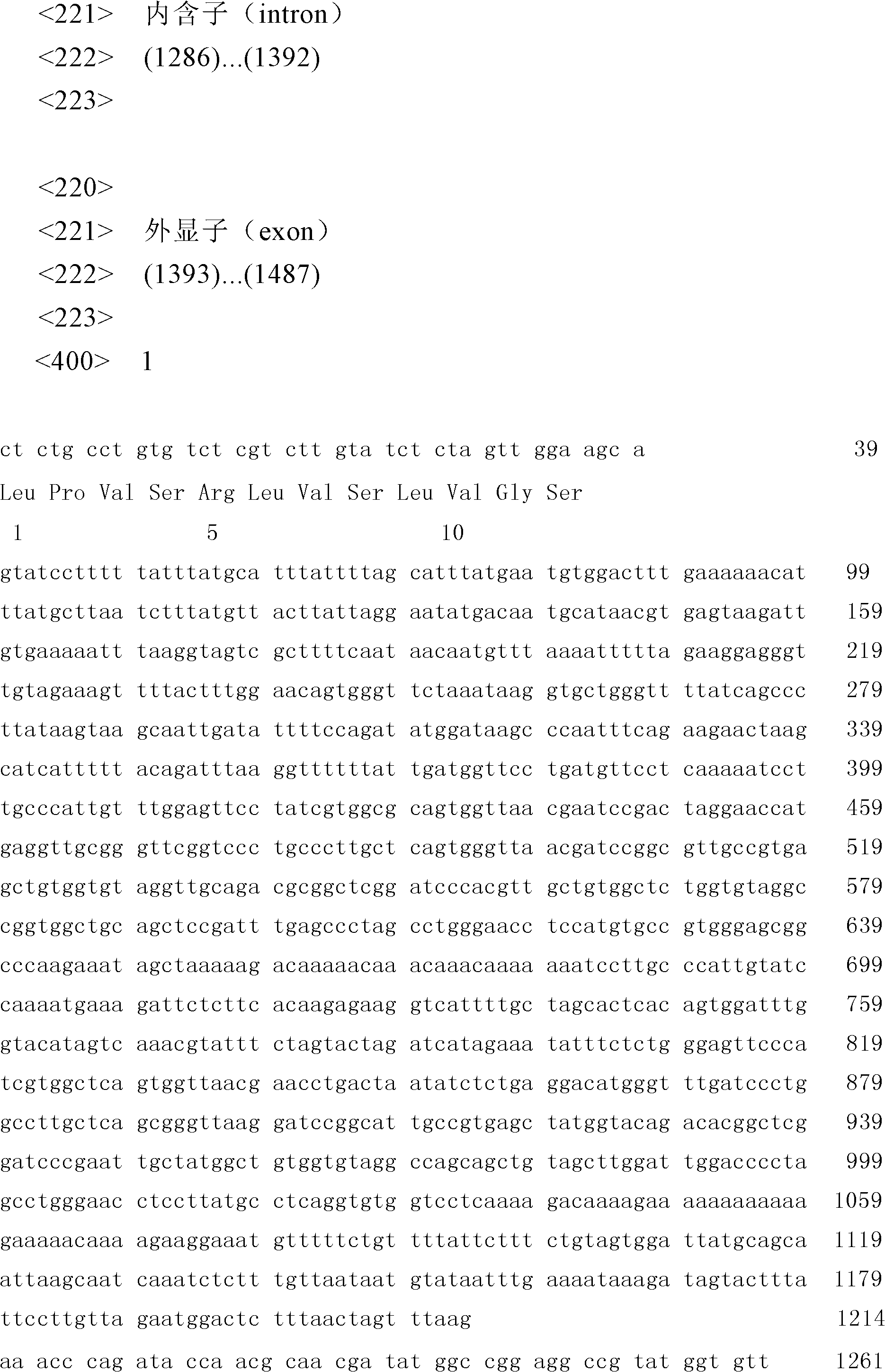
The invention relates to a traceablility SNP marker in a pig GIGYF2 gene and a detection method thereof. The SNP marker which is positioned in a genomic DNA fragment in the pig GIGYF2 gene is obtained through DNA pool sequencing. The genomic DNA fragment in the pig GIGYF2 gene, which is represented by SEQ ID NO.1 and comprises bps with the number of 452, comprises parts of twenty-three introns, and there is a base mutation from 187A to 187C at the 187th bit. The distribution case of the SNP marker of the invention in allelic genes in a test colony comprising ten pig kinds or lines is analyzed, and results show that frequencies of the marker in the allelic genes of different kinds or lines are close, so the polymorphism is abundant, there is a small difference among the frequencies of the allelic genes of different kinds or lines, and heterozygosities are greater than 0.3, so a case that the SNP marker of the invention can be used for pig product DNA traceablility is preliminarily determined.
Owner:SHANGHAI ACAD OF AGRI SCI
Paphiopedilum Henryanum EST-SSR (expressed sequence tag-simple sequence repeat) marker primers and their development method and application
ActiveCN107365874APolymorphism richGood repeatabilityMicrobiological testing/measurementDNA preparationAgricultural scienceSubgenus
The invention discloses an EST-SSR (expressed sequence tag-simple sequence repeat) marker primer group developed based on Paphiopedilum henryanum transcriptome sequences; the group includes 34 pairs of primers having nucleotide sequences shown as in SEQ ID NO. 1 to SEQ ID NO. 68. The invention also discloses an agent comprising the Paphiopedilum henryanum EST-SSR marker primers. The invention further discloses a method of developing the Paphiopedilum henryanum EST-SSR marker primers. The Paphiopedilum henryanum EST-SSR marker primers provided herein have the advantages such as rich polymorphism, good repeatability, good amplification stability and good static convenience, and are applicable to genetic diversity analysis and molecular mark assisted breeding of germplasm resources of Paphiopedilum henryanum and other species in Paphiopedilum, especially subgenus Paphiopedilum, as well as to molecular research relating to plants in Paphiopedilum.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
Method for sifting macrobrachium nipponensis microsatellite mark with magnetic bead concentration method
InactiveCN101403013APolymorphism richImprove reliabilityMicrobiological testing/measurementGerm plasmEnrichment methods
The invention provides a method for screening microsatellite markers of Macrobrachium nipponensis by a magnetic beads enrichment method, including the following steps: the extraction of the genomic DNA of the Macrobrachium nipponensis; the hybridization of a probe and a target segment; the enrichment of magnetic beads; positive sequencing clone and the amplification of the genomic DNA according to primers which are related to the flanking sequences of the microsatellite. As the invention combines the magnetic beads enrichment and PRC amplification of bacterial liquid to carry out second-time screening, no isotope is needed. The method has the advantages of safety and high efficiency and low cost, short cycle, high reliability and the like. At the same time, in the enrichment process, the magnetic beads are repeatedly washed for a plurality of times, which can clean and remove microsatellite sequences which are not firmly adhered because of less repetition times. Therefore, the obtained microsatellite sequences are longer and are repeated for more times. The microsatellite sequences which are more frequently repeated are considered to have rich diversity at interspecies and intraspecies and can be widely applied to the diversity identification of germ plasm, the building of genetic maps, and QTL. Therefore, the method is one which screens the microsatellite markers of Macrobrachium nipponensis and can be easily popularized.
Owner:FRESHWATER FISHERIES RES CENT OF CHINESE ACAD OF FISHERY SCI
InDel molecular marker interlocked with gossypium harknessii male sterile recovery gene and application
ActiveCN108728571ASpeed up breedingGuaranteed purityMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceCytoplasm
The invention discloses an InDel molecular marker interlocked with a gossypium harknessii male sterile recovery gene and application. The molecular marker is 5'-TAGAAGACTGGA-3', and is located at a position, which is distant from an initiation codon by 1247 bp in a promoter region of the recovery series gene Gh_D05G3392, and an InDel molecular marker primer is as shown in SEQID NO. 1-2. By the InDel molecular marker primer, whether recovery genes in materials exist or not and are homozygous or not is effectively identified by operations such as simple PCR and polyacrylamide gel electrophoresis, therefore, breeding of a field cotton cytoplasmic male sterile new recovery series material is accelerated, and seed purity of cotton recovery series is ensured.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Pepper hybrid seed purity testing EST-SSR (expressed sequence tag-simple sequence repeat) molecular marker and application thereof
ActiveCN103642906AReduce demandQuick checkMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceHybrid seed
The invention relates to the field of molecular markers, particularly relates to a pepper hybrid seed purity testing EST-SSR (expressed sequence tag-simple sequence repeat) molecular marker and application thereof, and discloses the pepper hybrid seed purity testing EST-SSR (expressed sequence tag-simple sequence repeat) molecular marker HE4 applied to 'hot No 1' hybrid seed purity testing.
Owner:TROPICAL CORP STRAIN RESOURCE INST CHINESE ACAD OF TROPICAL AGRI SCI
Method for rapidly identifying hot pepper species and golden pepper purity degree by using EST-SSR molecular markers
ActiveCN104928396AQuick checkLower quality requirementsMicrobiological testing/measurementHot peppersRepeatability
The invention relates to the field of molecular markers and provides a method for rapidly identifying hot pepper species and golden pepper purity degrees by using the EST-SSR molecular markers. The method adopts the specially designed EST-SSR primers, augmentation is conducted with the leaf genome DNA of golden pepper, the species purity degree of the golden pepper to be detected is tested through the specific bands of the augmentation result, by the adoption of the method, the identification can be conducted at any period during the growth of golden pepper, and the coeno-species can be separated from female parent selfed seeds and male parent selfed seeds; the requirement on DNA quality is not high, and the needing amount is less; the cost is low, the operation is simple; the repeatability and the stability are good; the fastness and the high efficiency are achieved; the accuracy is high, the application and the popularization prospect is wide, and scientific basis is provided for the development of pepper seed production and good seeds and the in-time selling.
Owner:华盛农业集团股份有限公司
Method for monitoring Siniperca chuatsi germplasms by virtue of microsatellite genetic markers
PendingCN106222271AHigh amplification efficiencyClear bandData processing applicationsMicrobiological testing/measurementGermplasmPolymorphic microsatellites
The invention discloses a method for monitoring Siniperca chuatsi germplasms by virtue of microsatellite genetic marker. The method comprises genome DNA extraction and detection, microsatellite sequence search and analysis, polymorphic microsatellite primer selection and optimization, polymorphic microsatellite detection and monitoring of different geographic siniperca chuatsi populations and hereditary features thereof. Microsatellite loci selection and polymorphic microsatellite marker development and verification are performed on a large number of EST generated by siniperca chuatsi transcriptome sequencing, so that 15 EST-SSR polymorphic microsatellite markers are developed, and different geographic siniperca chuatsi populations and hereditary features thereof can be monitored; the limitations of fuzzy genetic background and less molecular marker amount in existing germplasm detection, selective breeding and germplasm improvement research of siniperca chuatsi can be broken, and a basis is provided for germplasm detection, selective breeding and germplasm improvement of siniperca chuatsi, so that germplasm detection, auxiliary genetic breeding and genomic research processes of siniperca chuatsi molecular markers are promoted.
Owner:SUZHOU UNIV
SSR (Simple Sequence repeats) primer group based on Fraxinus Velutina Torr. transcriptome sequencing information development and application of primer group in germplasm identification
InactiveCN104046697APolymorphism richGood repeatabilityMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceNucleotide
The invention discloses an SSR (Simple Sequence repeats) primer group based on Fraxinus Velutina Torr. transcriptome sequencing information development and an application of the primer group in germplasm identification, belonging to the technical field of molecular biology. The SSR primer group comprises 42 pairs of primers, wherein the nucleotide sequences of the first pair to the forty-second pair of primers are shown in SEQ ID No.1 to SEQ ID No.84. Experiments verify that the SSR primer group disclosed by the invention has the advantages of rich polymorphism, good repeatability, clear electrophoretic band and the like. The SSR primer group can be effectively used for researches on germplasm identification, DNA fingerprint establishment and the like. A germplasm identification method is sensitive and reliable, can be used for rapidly and accurately realizing and finishing distinguishment and identification of Fraxinus chinensis germplasm, and has important significances in identification and intellectual property protection of Fraxinus chinensis germplasm resources and genetic breeding of genetic breeding molecules.
Owner:CHINA AGRI UNIV +1
DNA (Deoxyribonucleic Acid) molecular identification method for Luchuan pigs and meat products thereof
ActiveCN103215365AEasy to operatePolymorphism richMicrobiological testing/measurementBiotechnologyMolecular identification
The invention relates to a DNA (Deoxyribonucleic Acid) molecular identification method for Luchuan pigs and meat products thereof. According to the method, primers used in ISSR-PCR (Inter-Simple Sequence Repeat-Polymerase Chain Reaction) are WM14, WM12 and WM29, and nucleotide sequences of the primers are 5'-CTCCTCCTCGC-3', 5'-CACCACCACGC-3' and 5'-CTCCTCCTCAT-3'; and finally, seven electrophoretograms of the Luchuan pigs and the meat products thereof are drawn, a DNA fingerprint atlas of the Luchuan pigs and the meat products thereof is established, and the electrophoretograms and the DNA fingerprint atlas can serve as a basis for the distinction and identification of the Luchuan pigs and the meat products thereof. The method has the advantages that a reliable DNA molecular identification technique is adopted, so that the differences among the Luchuan pigs, the meat products of the Luchuan pigs, Luchuan pig hybrids, meat products of the Luchuan pig hybrids, Bama miniature pigs, meat products of the Bama miniature pigs, Huanjiang miniature pigs and meat products of the Huanjiang miniature pigs are identified quickly and accurately, and the development of advantaged brand pigs, namely the Luchuan pigs, is promoted.
Owner:GUANGXI UNIV
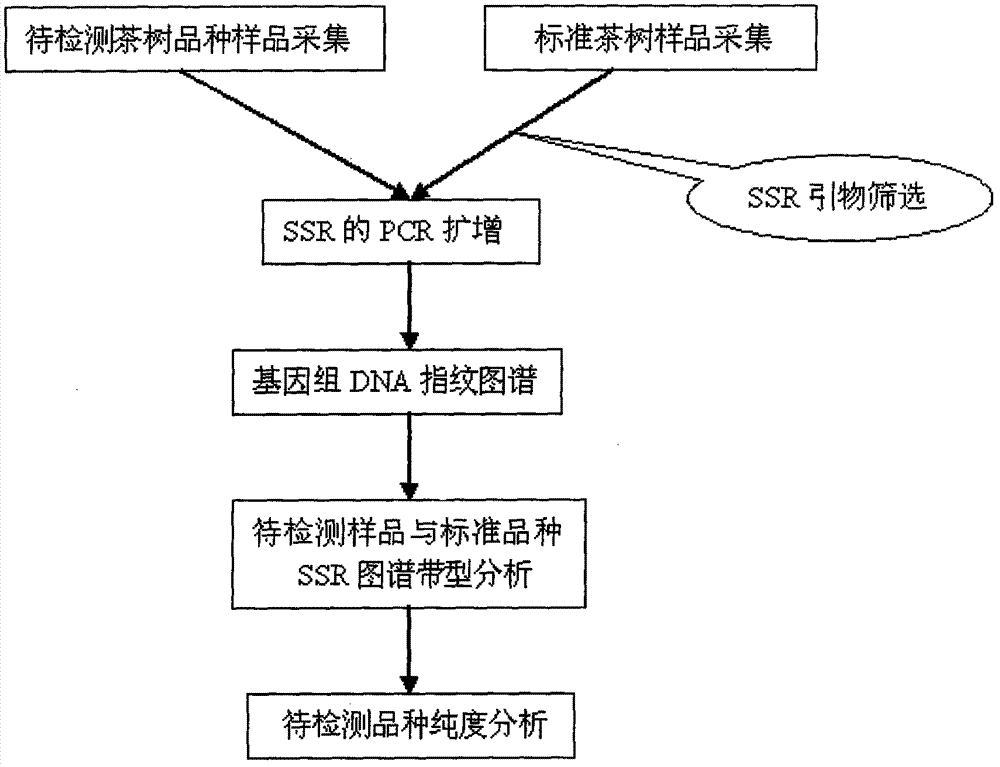
Molecular marking method for identifying tea clonal variety of tea trees
InactiveCN102776271ASimplified extraction procedureOmit purificationMicrobiological testing/measurementA-DNABud
Belonging to the field of biotechnologies, the invention provides a molecular marking method for identifying a tea clonal variety of tea trees. The method includes the following steps of: 1) putting 1.0g of fresh and tender buds and leaves of a variety to be tested into a mortar, adding liquid nitrogen and grinding the mixture into powder, adding a CTAB buffer solution, conducting water bathing at a temperature of 65DEG C for 30-60 minutes, then adding a DNA extracting solution to obtain the genome DNA of the tea tree variety to be tested; 2) taking the genome DNA extracted in step 1) as a template, using SSR (simple sequence repeat) primers for PCR amplification, and subjecting the amplification product to 10% native polyacrylamide gel electrophoresis, thus obtaining an electropherogram of the variety to be tested; and 3) comparing the electropherogram of the variety to be tested with a standard tea tree variety electropherogram obtained according to the step 1) and step 2), and if the variety to be tested and the standard variety are consistent in band composition, determining the variety to be tested and the standard variety as an identical variety, thus obtaining the purity of the variety to be tested. Compared with other methods, the invention can reflect the genetic background and genetic relationship of tea tree varieties more truly. And the technology provided in the invention can be used as a scientific basis for identifying the authenticity of the tea tree varieties.
Owner:刘本英
Chenopodium quinoa willd EST-SSR molecular marker, development method of chenopodium quinoa willd EST-SSR molecular marker and application of chenopodium quinoa willd EST-SSR molecular marker
ActiveCN105087574AIncrease success ratePolymorphism richMicrobiological testing/measurementDNA/RNA fragmentationForward primerBio informatics
The invention relates to the technical field of molecular markers, in particular to a chenopodium quinoa willd EST-SSR (Expressed Sequence Tag-Simple Sequence Repeat) molecular marker, a development method of the chenopodium quinoa willd EST-SSR molecular marker and application of the chenopodium quinoa willd EST-SSR molecular marker. A group of chenopodium quinoa willd EST-SSR molecular markers comprises forward primers and reverse primers corresponding to 66 sites. The development method of the chenopodium quinoa willd EST-SSR molecular marker comprises the following steps of: 1, obtaining RNA-seq and EST sequences from an NCBI (National Center of Biotechnology Information) database; 2, using bioinformatics software to process, assemble and splice sequences into unigenes, and performing SSR site searching on the unigenes to obtain the EST-SSR; and 3, designing EST-SSR primer amplification chenopodium quinoa willd genome DNA for verifying the EST-SSR, and using the verified EST-SSR molecular marker to realize application of genetic diversity analysis in chenopodiaceae germplasm. The EST-SSR molecular marker developed on the basis of a public database has the advantages of high efficiency, high success rate, rich polymorphism and high universality.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
SSR core primer set based on Pennisetum purpureum Schum transcriptome sequence development and its application
InactiveCN106591302AA large amountPolymorphism richMicrobiological testing/measurementDNA/RNA fragmentationPennisetum purpureumAgricultural science
The invention discloses a SSR core primer set based on Pennisetum purpureum Schum transcriptome sequence development and its application. The SSR core primer set comprises 50 pairs of primers and the primers have nucleotide sequences shown in SEQUENCE ID NO. 1 to 100 in the sequence table. The invention also relates to a use of the SSR core primer set in Pennisetum purpureum Schum genetic diversity analysis and Pennisetum purpureum Schum germplasm genetic genealogy analysis. The 50 pairs of the SSR primers are derived from Pennisetum purpureum Schum spire transcriptome sequences. Compared with the existing Chinese pennisetum common SSR primers, the SSR core primer set has the advantages of high polymorphism, stable amplification, good repeatability and statistics convenience. The primers are successfully applied to Pennisetum purpureum Schum genetic diversity analysis and Pennisetum purpureum Schum germplasm genetic genealogy analysis, increase the amount of SSR primers used by Pennisetum purpureum Schum, can be effectively used for Pennisetum purpureum Schum genetic diversity analysis, identification of variety, fingerprinting construction and molecular marker-assisted breeding, and other Chinese pennisetum related sturdy. The SSR core primer set is suitable for promotion and application in the field of biotechnology.
Owner:SICHUAN AGRI UNIV
Rapid method for identifying truth of maize variety
InactiveCN101469347AHigh quality and stableShorten germination timeMicrobiological testing/measurementAgricultural scienceRapid identification
The invention relates to a rapid identification method for truth of corn varieties. In the method, an embryo of a single dry seed is adopted to extract DNA of a genome of a sample to be detected, and the method is time-saving and ensures that the extracted genome has high and steady quality; 20 pairs of SSR molecular marker core primers are used, have rich polymorphism and steady banding patterns, and are evenly distributed on 10 chromosomes; and the method also adopts an optimized TouchDown PCR amplification program. The rapid identification method for the truth of the corn varieties has low design cost for the primers, is simple in operation and time-saving, can conveniently and quickly identify the authenticity of the corn varieties, and is particularly suitable for material identification of a large amount of samples.
Owner:BEIJING KINGS NOWER SEED S&T
Sacalia bealei microsatellite marked specific primer and detection method
InactiveCN103627807AEfficient amplificationPolymorphism richMicrobiological testing/measurementDNA/RNA fragmentationPopulation geneticsGenetic diversity
The invention discloses a Sacalia bealei microsatellite marked specific primer and a detection method. The detection method comprises the following steps: performing polymerase chain reaction (PCR) amplification on microsatellite site sequence by taking Sacalia bealei genome DNA as a template and using the microsatellite marked specific primer; performing non-denaturing polyacrylamide gel electrophoresis detection on the PCR amplification products; and performing polymorphic analysis on microsatellite sites according to the detection result. The Sacalia bealei microsatellite marked specific primer and the detection method thereof, provided by the invention, can be applied in the fields of genetic diversity level evaluation, population genetic structure analysis and relationship identification of Sacalia bealei.
Owner:INST OF ZOOLOGY GUANGDONG ACAD OF SCI
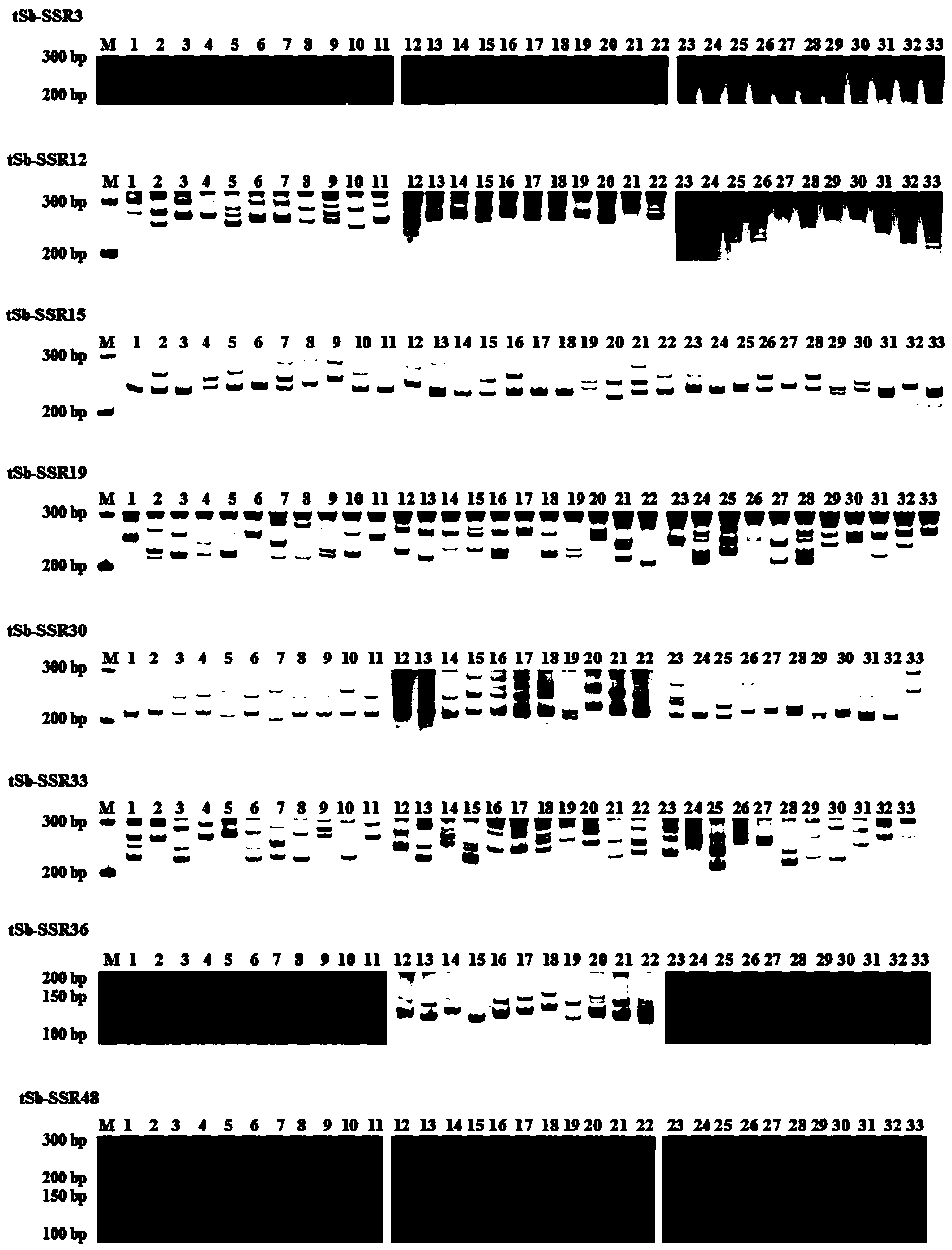
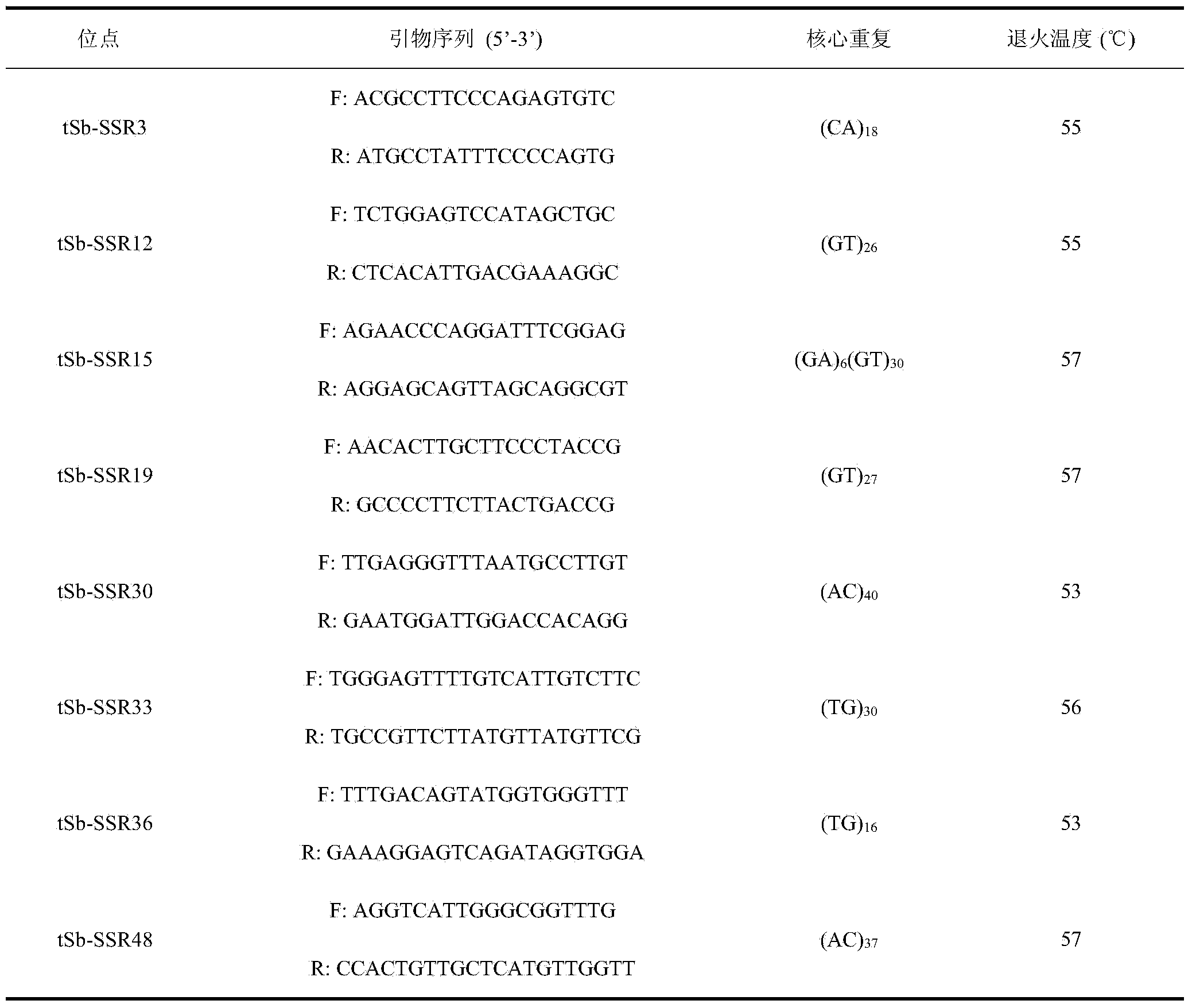
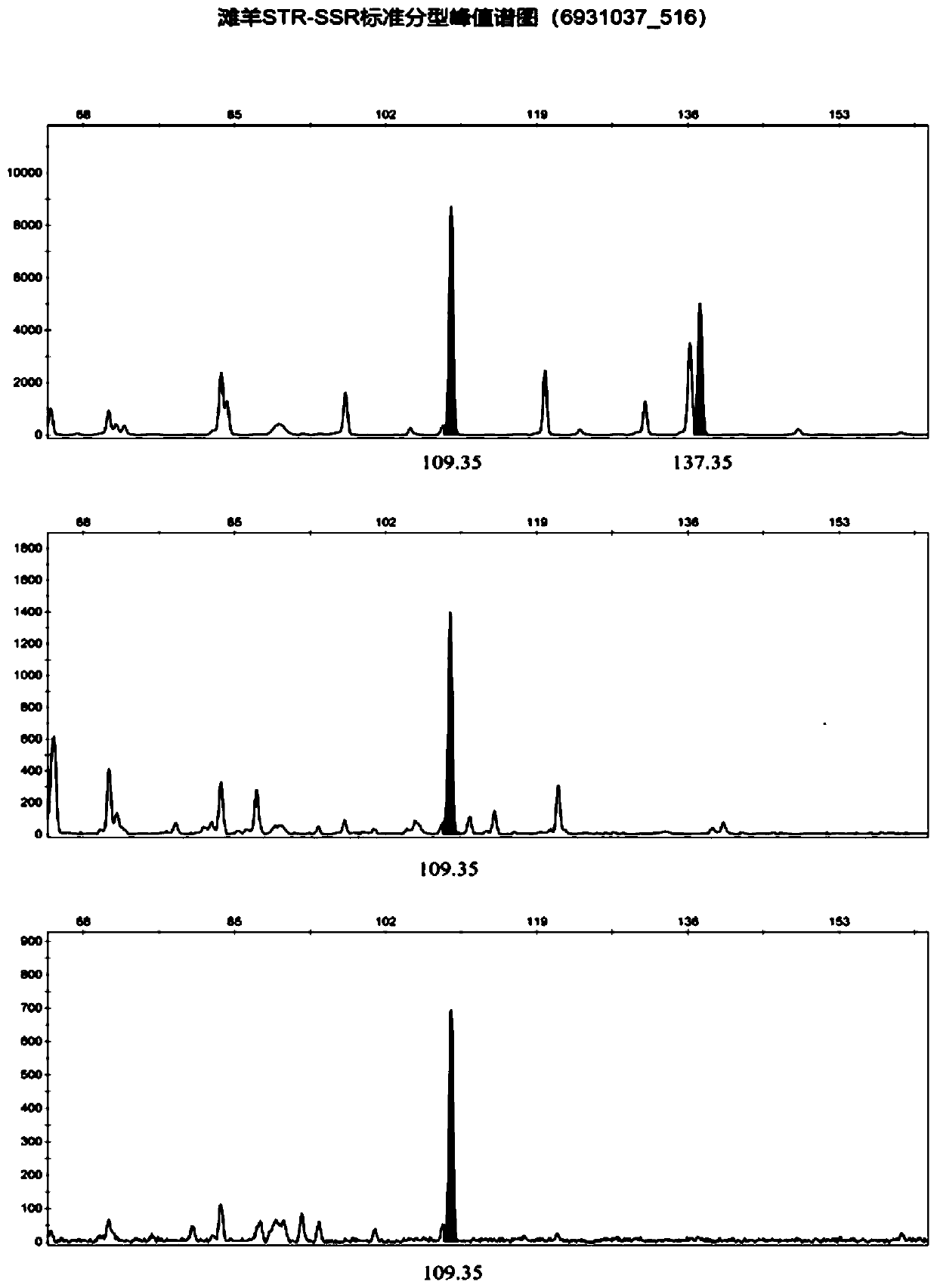
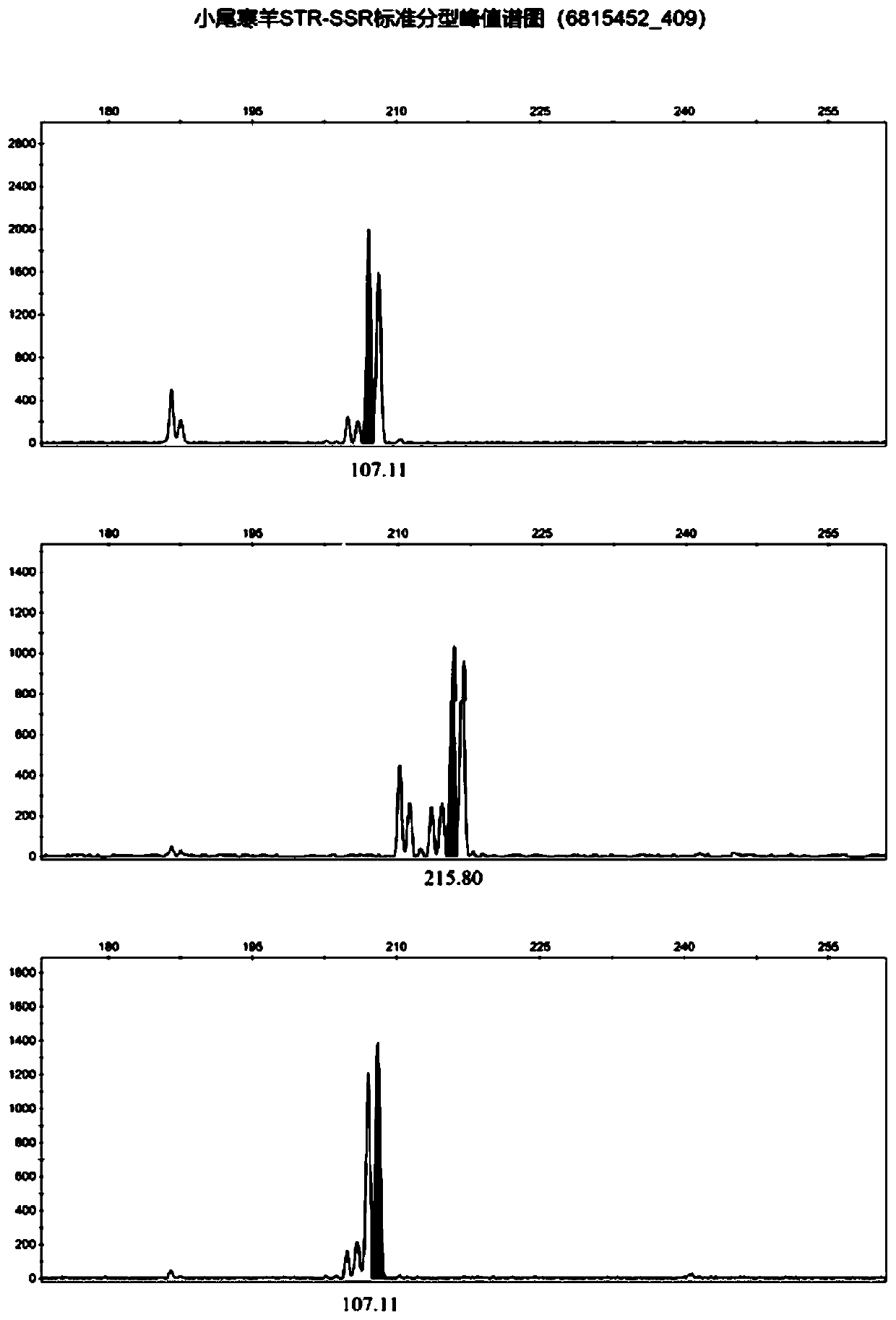
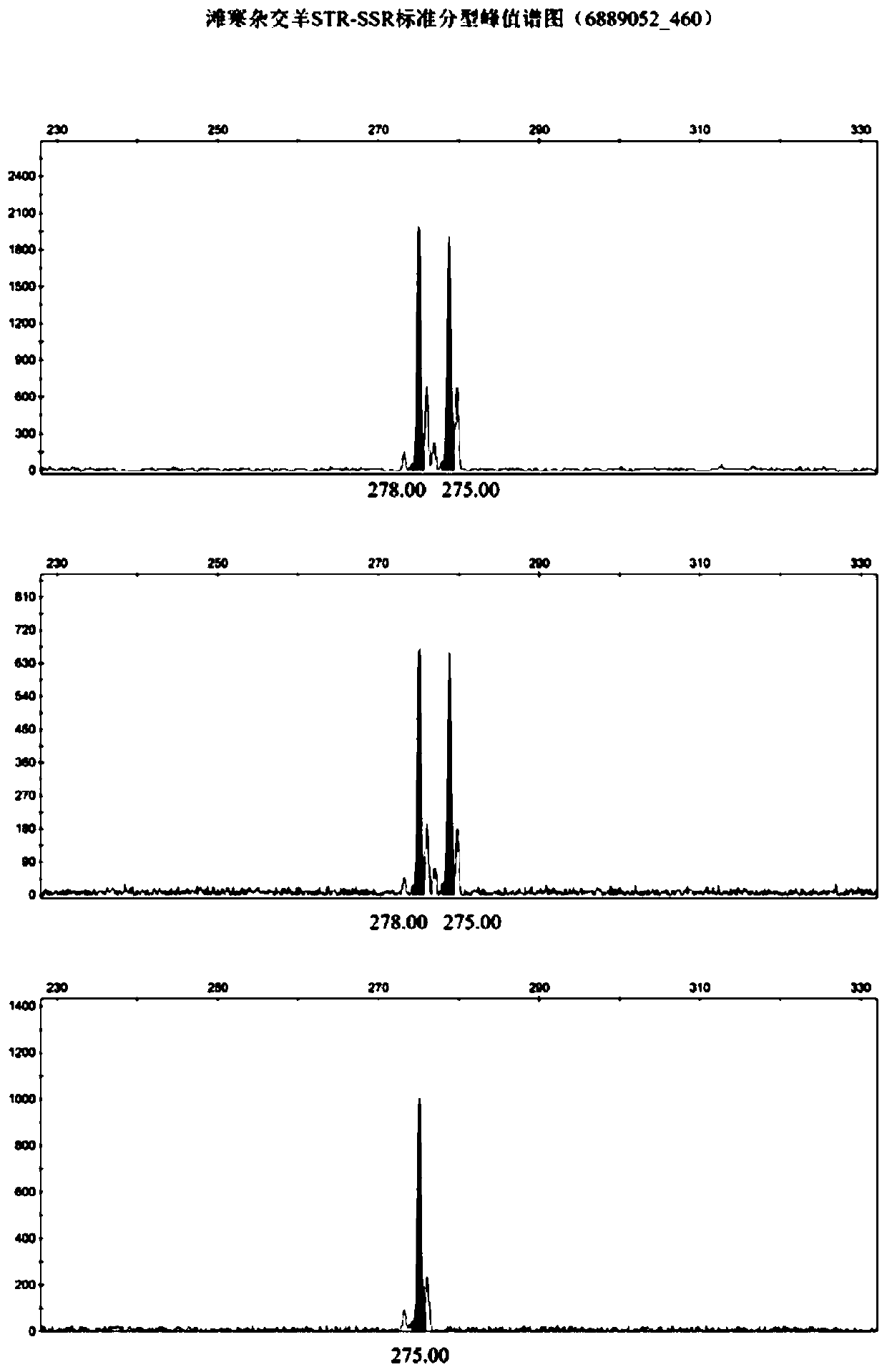
Affinis sheep meat detection method based on species specificity STR-SS
PendingCN110004218APromote healthy and sustainable developmentShort timeMicrobiological testing/measurementGenetic engineeringCapillary electrophoresisFluorescence
The invention belongs to the technical field of food detection. The detection method comprises the following specific steps that DNA of a standard sheep meat sample and DNA of a to-be-detected sheep meat sample are extracted, the sheep meat sample DNA concentration of the standard sample DNA and the sheep meat sample DNA concentration of the to-be-detected sample DNA are detected respectively, theSSR gene sequence of the standard sample DNA and the SSR gene sequence of the to-be-detected sample DNA are subjected to fluorescence labeling, PCR amplification is conducted by using primers designed by the SSR sequences, the PCR amplification result is detected through agarose gel electrophoresis, the SSR gene sequence type of the standard sample and the SSR gene sequence type of the to-be-detected sample are detected through capillary electrophoresis, an STR-SSR standard type peak spectrogram is built. The detection method is s used for specifically identifying Tan sheep, small tailed Hansheep and Tan-Han crossbred sheep, the method takes short time, the operation is simple, and the specificity is high. The detection results are compared by using the peak spectrogram, the detection results can be mutually identified, the method can be used for precisely identifying the three kinds of affinis sheep species which cannot be identified morphologically, and sustainable and healthy development of the Tan sheep industry and consumer rights and interests are facilitated.
Owner:NINGXIA UNIVERSITY
Method for quickly identifying purity of cucumber rootstock Heli II by use of molecular marker
ActiveCN106755387AQuick checkReduce incubation timeMicrobiological testing/measurementGrowth plantHybrid seed
The invention relates to the field of molecular markers and provides a method for quickly identifying purity of cucumber rootstock Heli II by use of a molecular marker. According to the method, a specially designed EST-SSR primer is adopted, and the genome DNA of Heli II serves as a template for amplification, and the purity of the F1-generation seeds of the to-be-detected cucumber rootstock Heli II is detected according to the specific band of the amplification result; by adopting the method, the plant growth of cucumber rootstock Heli II can be identified in any stage, and the hybrid seeds can be distinguished from the female-parent selfing seeds and male-parent selfing seeds; and moreover, the method has high accuracy and broad prospect in application and promotion and provides scientific basis for rootstock seed production and timely selling of improved seeds.
Owner:华盛农业集团股份有限公司
SNP marker used for traceability in pig SFTPC gene and detection method thereof
InactiveCN102477424APolymorphism richSmall differences in allele frequency distributionMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceSFTPC gene
The invention relates to a SNP marker used for traceability in a pig SFTPC gene and a detection method thereof. The SNP molecular marker is obtained through DNA pool sequencing, and has a sequence as shown in SEQ ID NO 1; a base is mutated from 639A to 639T at the 639th base of the SNP molecular marker, and the coded amino acid sequence is shown in SEQ ID NO 2. In a test group including 11 pig varieties or strains, the allele distribution condition of the SNP molecular marker in the test group is analyzed, and the allele frequencies of the molecular marker in different varieties or strains are found to be approximate; the polymorphism is abundant; the allele frequency distribution difference between varieties or strains is fewer; the heterozygosity is more than 0.3; and the SNP marker is preliminary determined to be applicable to DNA traceablility for pork products.
Owner:SHANGHAI ACAD OF AGRI SCI
Primer set of puccinia recondita tritici EST-SSR molecular marker, detection method and application thereof
InactiveCN107058602AAmplification results are stablePolymorphism richMicrobiological testing/measurementDNA/RNA fragmentationPuccinia reconditaAgricultural science
Relating to the field of plant protection, the invention specifically discloses a primer set of a puccinia recondita tritici EST-SSR molecular marker, a detection method and application thereof. The primer set of the Puccinia recondita tritici EST-SSR molecular marker includes primers shown as nucleotide sequences SEQ ID No:1-37. The detection method for puccinia recondita tritici genetic diversity employs the primer set to analyze the puccinia recondita tritici diversity and the population genetic structure, has the characteristics of simple operation, convenient detection, stable PCR amplification result, clear electrophoretic band and rich polymorphism, plays an important role in puccinia recondita tritici toxicity monitoring and genetic diversity analysis.
Owner:HEBEI AGRICULTURAL UNIV.
Buchloe dactyloides ISSR-PCR molecular marker system
InactiveCN101985655APolymorphism richClear backgroundMicrobiological testing/measurementEnzymeMolecular breeding
The invention belongs to the technical field of buchloe dactyloides molecular breeding, and particularly relates to an optimal buchloe dactyloides ISSR-PCR (inter-simple sequence repeat-polymerase chain reaction) reaction system. The invention provides the following ISSR-PCR reaction system that: each 25muL of reaction system contains 1*PCR buffer solution, 0.3mM of dNTPs, 1.0U of Taq enzyme, 0.6muM of any one of primers shown as SEQ ID 1-7, 2.0mM of Mg2+ and 50ng of DNA template; and the reaction mixed solution is amplified by the following procedures of: pre-denaturing for 3 minutes at 94 DEG C, denaturing for 30 seconds at 94 DEG C, annealing for 30 seconds at 50 to 60 DEG C, stretching for 2 minutes at 72 DEG C, circulating for 35 times, and stretching for 4 minutes at 72 DEG C. The buchloe dactyloides ISSR-PCR provided by the invention has good stability and high definition, and increases the richness of strips.
Owner:CHINA AGRI UNIV
Primer set for hybrid cymbidium tortisepalum EST (expressed sequence tags)-SSR (simple sequence repeats) labeling and screening method
PendingCN110699480AConducive to genetic diversity analysis of germplasm resourcesAssisted BreedingMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyNucleotide
The invention discloses a primer set for hybrid cymbidium tortisepalum EST (expressed sequence tags)-SSR (simple sequence repeats) labeling. The primer set includes 4 pairs of primers, and the nucleotide sequences of the primers are as shown in the sequence tables SED ID NO.1-SED ID NO.8. The invention further discloses a screening method of the primer set for hybrid cymbidium tortisepalum EST-SSRlabeling. The primer set for hybrid cymbidium tortisepalum EST-SSR labeling obtained by the screening method has the advantages of rich polymorphisms, stable amplification and convenient statistics,and can be used for genetic diversity analysis of germplasm resources of hybrid cymbidium tortisepalum plant varieties, molecular marker-assisted breeding and related molecular research.
Owner:GUANGXI SUBTROPICAL CROPS RES INST GUANGXI SUBTROPICAL AGRI PROD PROCESSING RES INST
Rhizoctonia solani kuha SSR mark, as well as preparation method and application thereof
ActiveCN106399508AEfficient enrichmentRapid enrichmentMicrobiological testing/measurementMicroorganism based processesRestriction enzyme digestionMagnetic bead
The invention discloses a rhizoctonia solani kuha SSR mark, as well as a preparation method and application thereof, and belongs to the technical field of molecular organism. The rhizoctonia solani kuha SSR mark comprises 8 groups of primer pairs, and the preparation method comprises the following steps: firstly, extracting genome DNA of rhizoctonia solani kuha; secondly, obtaining a DNA fragment through restriction enzyme digestion total NDA; thirdly, ensuring that a biotin labeled probe is cross-fertilized with a target fragment; fourthly, enriching a DNA fragment containing an SSR sequence through a magnetic bead; fifthly, amplifying and purifying the DNA fragment containing the SSR sequence; sixthly, performing clone sequencing on the DNA fragment; seventhly, designing an SSR primer through an SSR flanking sequence, which is used for amplifying a microsatellite fragment of a locus. The rhizoctonia solani kuha SSR mark is high in primer specificity, stable in amplification, simple in operation, short in period and low in cost, meanwhile has the advantages of rich codominant inheritance and polymorphism, and can be applied to analysis of the genetic diversity of rhizoctonia solani kuha.
Owner:CHINA NAT RICE RES INST
EST-SSR marker primer developed based on platycladus orientalis transcriptome sequence and application of EST-SSR marker primer
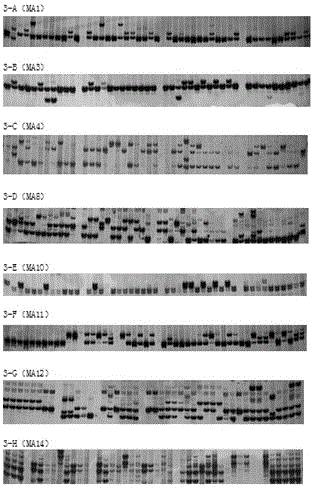
ActiveCN114317800APolymorphism richStable amplificationMicrobiological testing/measurementDNA/RNA fragmentationGenetic diversityGel electrophoresis
The invention discloses an EST-SSR (expressed sequence tag-simple sequence repeat) marker primer developed based on a platycladus orientalis transcriptome sequence and application of the EST According to the EST-SSR marker primer, an SSR sequence is obtained on the basis of platycladus orientalis transcriptome sequencing data, a primer is designed, and the stably-amplified, efficient and polymorphic microsatellite marker primer is obtained through a triple screening method of agarose gel electrophoresis, polyacrylamide gel electrophoresis and capillary electrophoresis screening. The effectiveness of the developed EST-SSR marker primer in the aspects of genetic diversity analysis and genetic relationship identification is verified by utilizing different varieties and clone of platycladus orientalis, and the EST-SSR marker primer can be applied to genetic diversity analysis, genetic relationship and variety identification, molecular marker-assisted selective breeding, genetic relationship identification and genetic relationship identification of platycladus orientalis plants. And the research fields of new plant variety protection and the like.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
Construction method for high-density genetic map of scylla paramamosain
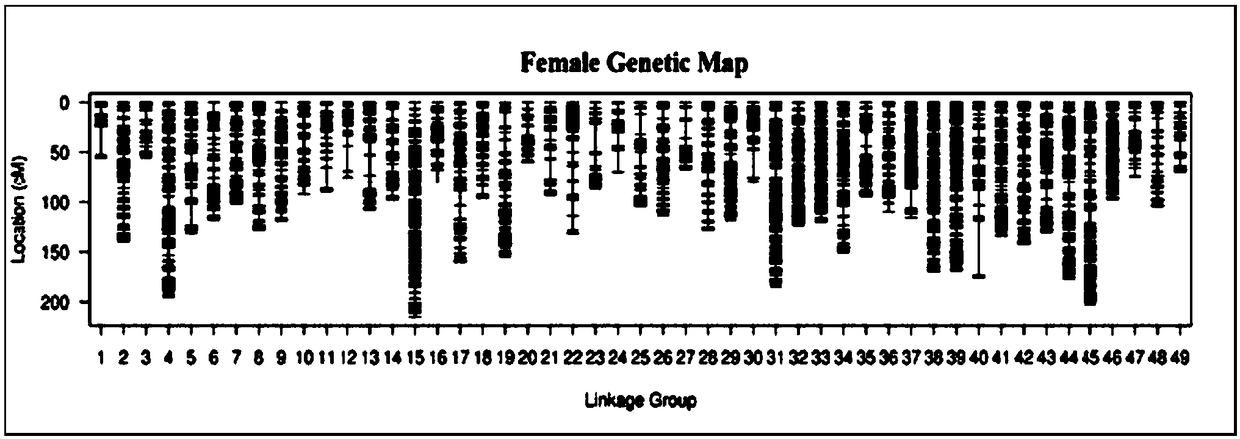
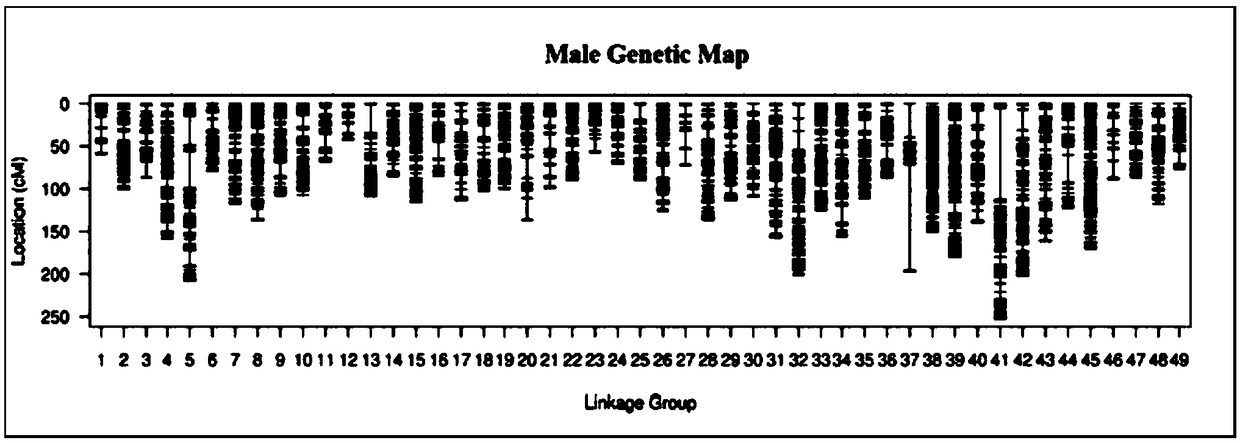
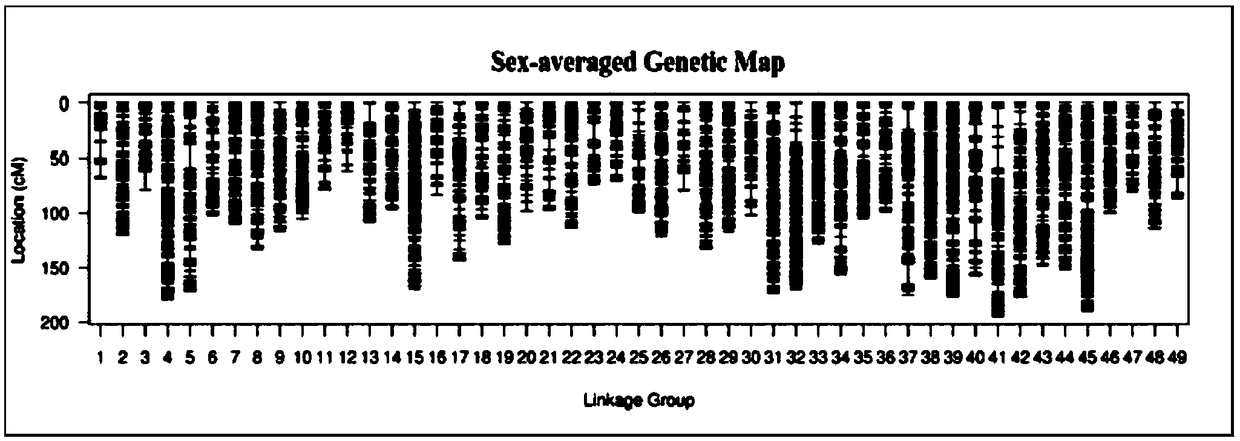
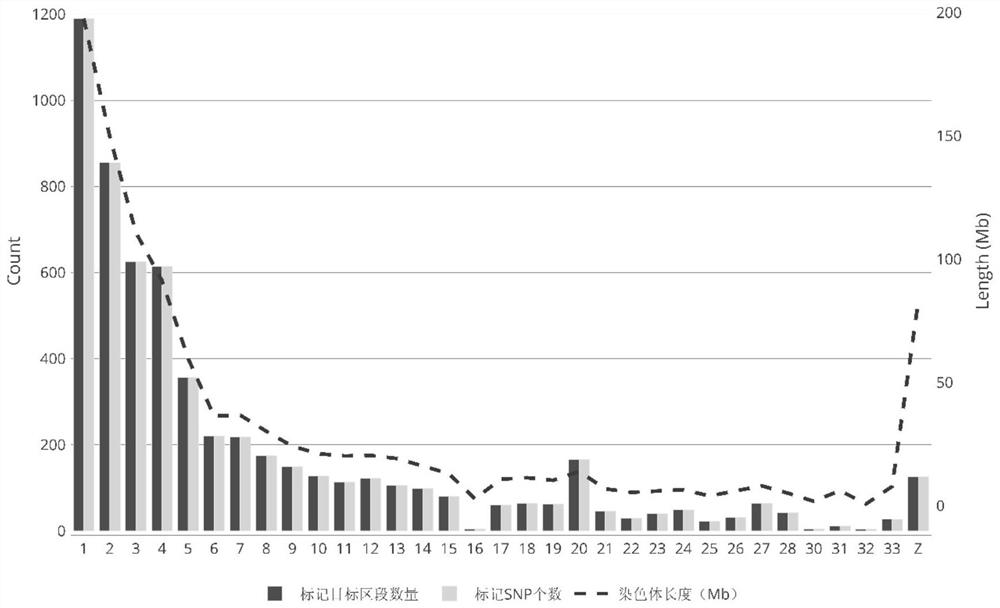
ActiveCN109486961AEasy constructionGuaranteed build stabilityMicrobiological testing/measurementClimate change adaptationBiotechnologyScylla paramamosain
The invention relates to a construction method for a high-density genetic map of scylla paramamosain. The construction method for the high-density genetic map of the scylla paramamosain comprises thefollowing four steps: constructing a mapping population, constructing a simplified genomic library and carrying out high-throughput sequencing, carrying out genetic typing and constructing a high-density genetic map. The genotype of a male parent is reversely ratiocinated by the genotype of a female parent and the genotype of an offspring, then genetic typing is carried out, therefore, the geneticmap is constructed, a new thinking and method are provided for construction of genetic maps of animal and plant families which only have single parent or difficultly have double parents simultaneously in a sample material, construction of the genetic map is facilitated, a research course of genetic background is pushed, and the construction method has good application prospect in the field of molecular breeding. A parting technology based on a SNP label is adopted, the construction method has the advantages of numerous labels, abundant polymorphism, genetic stability and the like, and the obtained genetic map is high in density and good in quality, and can be used for QTL positioning of follow-up important economic character and assisted assembly of genome.
Owner:SHANTOU UNIV
Chicken low-density SNP liquid phase chip based on targeted capture sequencing and application thereof
ActiveCN114480673ALow parting costGood value for moneyMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyBroiler
The invention discloses a chicken low-density SNP liquid phase chip based on targeted capture sequencing and application of the chicken low-density SNP liquid phase chip, the chip contains 5912 SNP loci, 2582 SNP loci obviously associated with economic traits of slaughter type chicken breeds are derived from whole genome re-sequencing data of 33 representative local chicken breeds in China and related researches, and the SNP loci have the advantages that the SNP loci are high in specificity and high in specificity; the method is suitable for resource evaluation and genetic identification of local chicken varieties and application to economic character genetic improvement of slaughter type broiler chicken varieties. Meanwhile, according to the chicken low-density liquid-phase chip, the target SNP sites can be adjusted by directly increasing or decreasing the probes, and compared with a solid-phase chip, the chicken low-density liquid-phase chip has better flexibility.
Owner:JIANGSU INST OF POULTRY SCI
SNP molecular maker in pig PSMA1 gene for traceability and detection method thereof
InactiveCN102399775APolymorphism richSmall differences in allele frequency distributionMicrobiological testing/measurementDNA/RNA fragmentationAmino acidA-DNA
The invention relates to an SNP molecular maker in a pig PSMA1 gene for traceability and a detection method thereof. The SNP molecular maker is obtained by DNA pooling and sequencing and is represented by SEQ ID NO.3, and an amino acid sequence encoded by the SNP molecular maker is represented by SEQ ID NO.4. The SNP molecular maker is in a DNA fragment of the pig PSMA1 gene, wherein, the DNA fragment of the pig PSMA1 gene is represented by SEQ ID NO.1, and an amino acid sequence encoded by the DNA fragment of the pig PSMA1 gene is represented by SEQ ID No.2. The analysis of the allelic distribution of the SNP molecular maker in a group of trials containing 9 pig species / strains shows that allelic frequencies of the SNP molecular maker in different species or strains are approximate, the polymorphism is high, allelic frequency distribution difference between species or strains is small, and the heterozygosis degree is all larger than 0.3, thus the SNP molecular maker disclosed herein can be used for DNA technology for pork traceability.
Owner:SHANGHAI ACAD OF AGRI SCI
SSR primer group for analyzing Hevea brasiliensis (Willd. ex A. Juss.) Muell. Arg. powdery mildew colony genetic structure, and application of SSR primer group
ActiveCN113755632AGenetic Diversity RevealedConvenient statisticsMicrobiological testing/measurementMicroorganism based processesBiotechnologyNucleotide
The invention discloses an SSR primer group for analyzing a Hevea brasiliensis (Willd. ex A. Juss.) Muell. Arg. powdery mildew colony genetic structure, and application of the SSR primer group. The SSR primer group for analyzing the Hevea brasiliensis (Willd. ex A. Juss.) Muell. Arg. powdery mildew colony genetic structure comprises 16 pairs of primers, wherein the nucleotide sequences of the primers are as shown in SEQ ID NO.17-48. The invention also provides application of the primer group in the Hevea brasiliensis (Willd. ex A. Juss.) Muell. Arg. powdery mildew colony genetic structure analysis. The primer group has the characteristics of being good in specificity, stable in amplification, rich in polymorphism, good in repeatability, convenient in counting and the like, and a new technology and a new means are provided for research on the Hevea brasiliensis (Willd. ex A. Juss.) Muell. Arg. powdery mildew colony genetic structure.
Owner:ENVIRONMENT & PLANT PROTECTION INST CHINESE ACADEMY OF TROPICAL AGRI SCI
SSR (Simple Sequence Repeat) core primer group developed based on whole genome sequence of foxtail millet and application of SSR core primer group
ActiveCN103060318BImproved geneticsEvenly distributedMicrobiological testing/measurementDNA preparationAgricultural scienceGenetic diversity
The invention discloses an SSR (Simple Sequence Repeat) core primer group developed based on whole genome sequence of foxtail millet and an application of the SSR core primer group, and belongs to the technical field of molecular biology. The core primer group comprises 30 pairs of primers, wherein nucleotide sequences are represented by SEQ ID NO. 1-60. The primer has advantages of clear electrophoretic band and rich polymorphism, and is uniformly distributed and stable in amplification. The invention also provides the application of the SSR core primer group in identifying the genetic diversity and variety of the foxtail millet. The primer group can be used for precisely and quickly identifying the variety of the foxtail millet and precisely reflecting a genetic relationship among the varieties of the foxtail millet.
Owner:CROP RES INST SHANDONG ACAD OF AGRI SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com