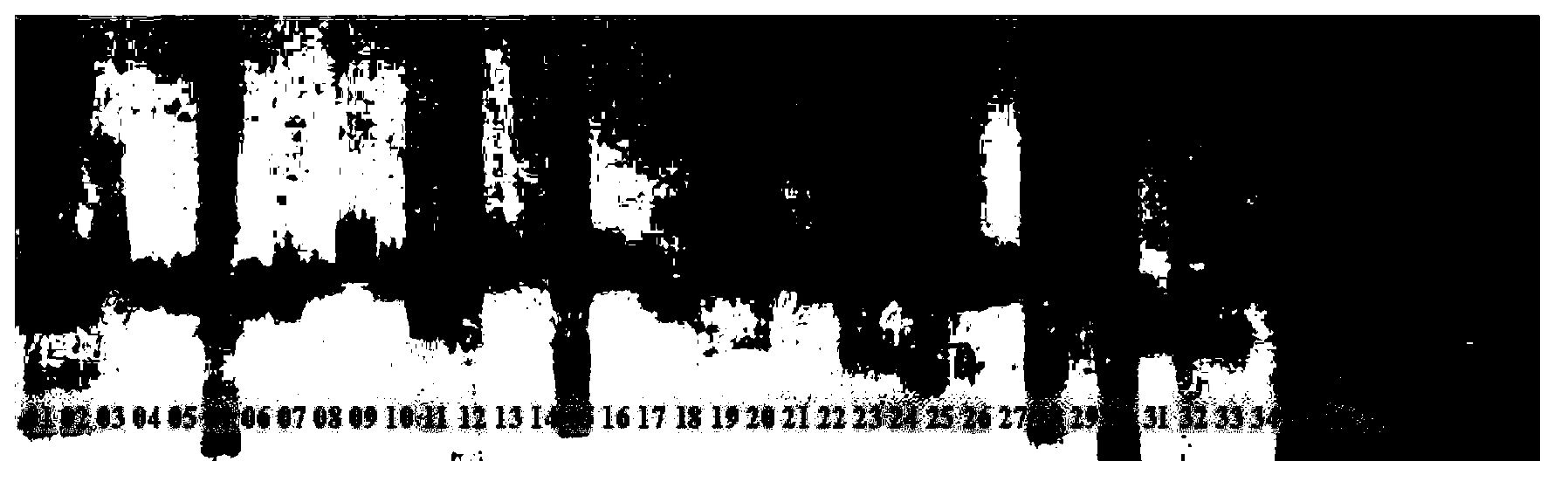SSR (Simple Sequence Repeat) core primer group developed based on whole genome sequence of foxtail millet and application of SSR core primer group
A genome sequence and core primer technology, applied in the field of molecular biology, can solve the problem of the small number of SSR markers, achieve the effects of enriching polymorphism, promoting the improvement of millet genetic breeding level, and stable amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1: Application of SSR Marker of Millet Genome in Variety Identification
[0023] 1. Extraction of millet genomic DNA
[0024] (1) Material
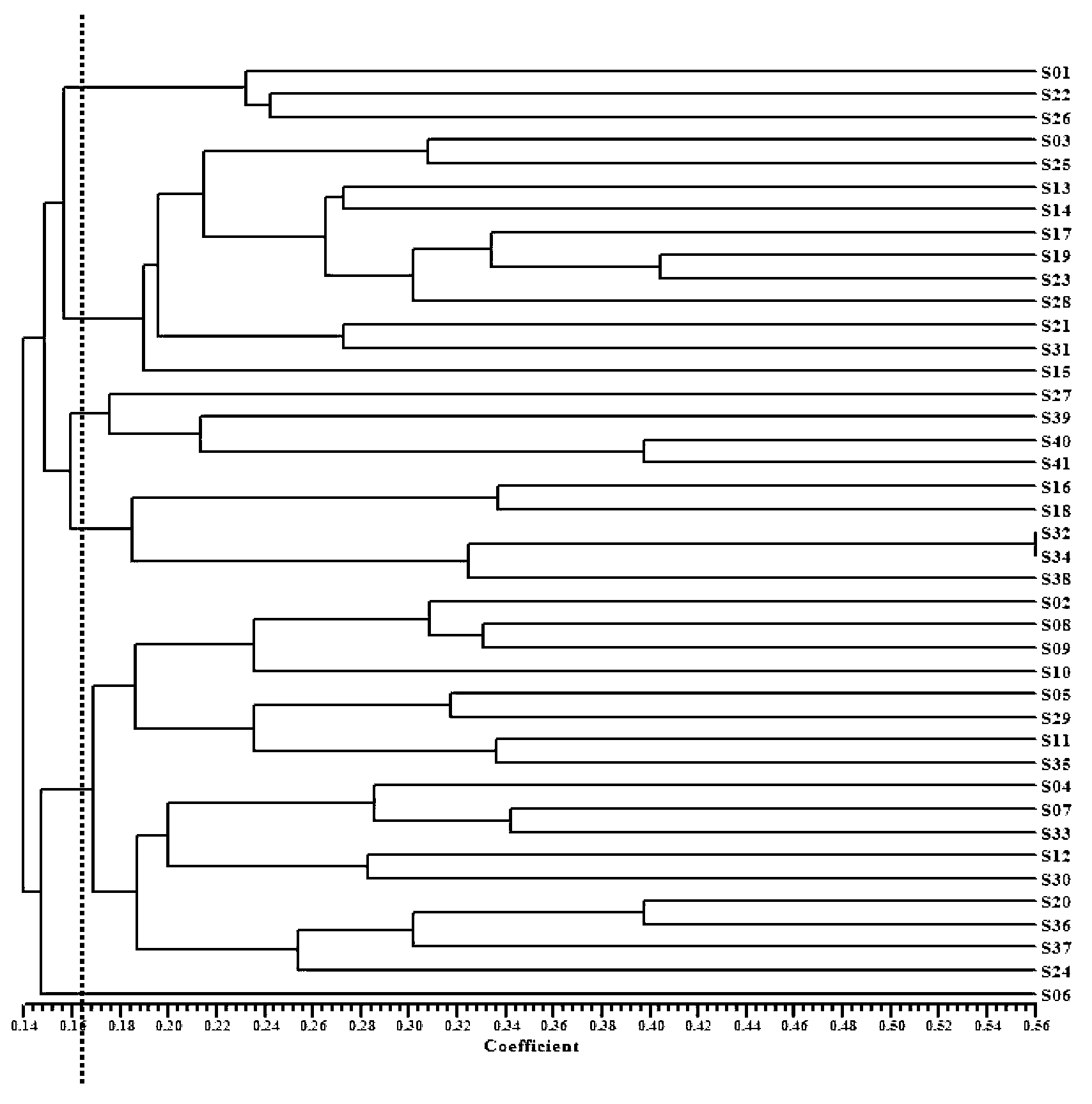
[0025] 41 representative local varieties and bred varieties were selected from the core germplasm materials of millet, covering 6 ecological regions in the Northeast Plain, North China Plain, South of Huaihe River, Loess Plateau, Inner Mongolia Plateau and Northwest Inland (see Table 1) , Used for the evaluation of millet primers and variety identification research.
[0026] Table 1 Information of materials used in the identification of millet varieties in the present invention
[0027]
[0028]
[0029]
[0030] (2) CTAB method to extract genomic DNA
[0031] The modified CTAB method is used to extract the DNA of millet, the specific steps are as follows:
[0032] 1) Sprouting and cultivating the tested millet materials, and extracting DNA when two leaves and one heart stage.
[0033] 2) Cut an appropriate amount of leaves, place them...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com