The ssr primer set developed based on eggplant transcriptome sequencing data and its application
A transcriptome sequencing and primer set technology, applied in the field of molecular biology, can solve the problems of low polymorphism of SSR markers and few SSR markers, and achieve the effects of clear amplification bands, stable amplification and rich polymorphisms.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0044] The invention is further illustrated in connection with the examples, but is not limited thereto.
[0045] First, the source of transcription group data
[0046] The eggplant transcript data is derived from the inventors in 2019 to perform ILLUMINA high-throughput depth sequencing of eggplant in 2019.
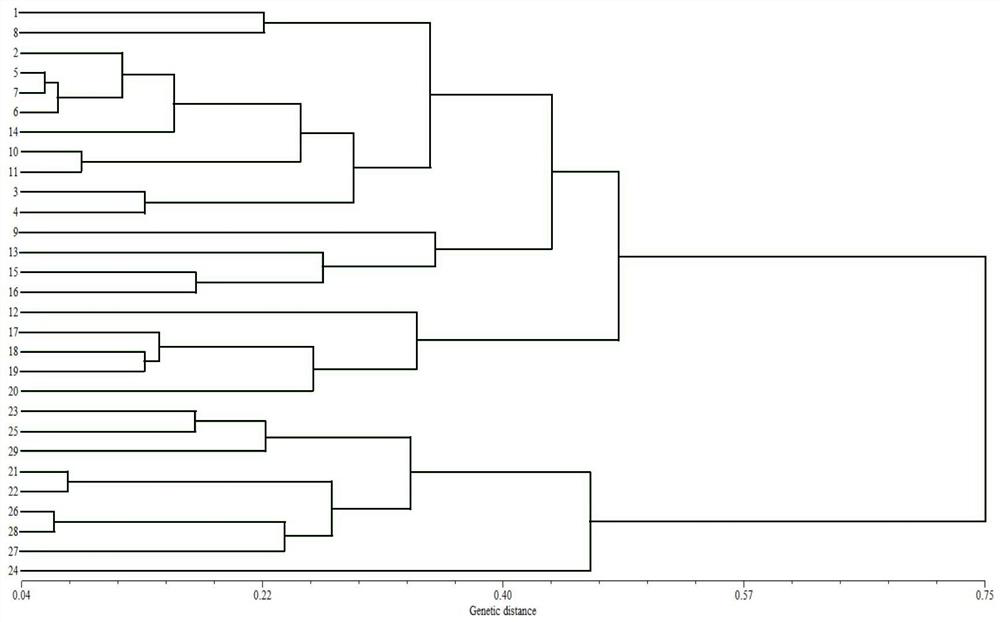
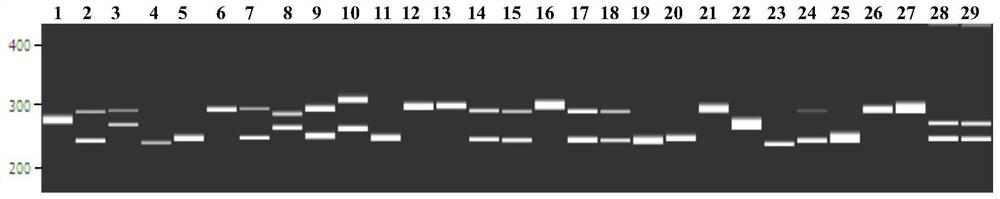
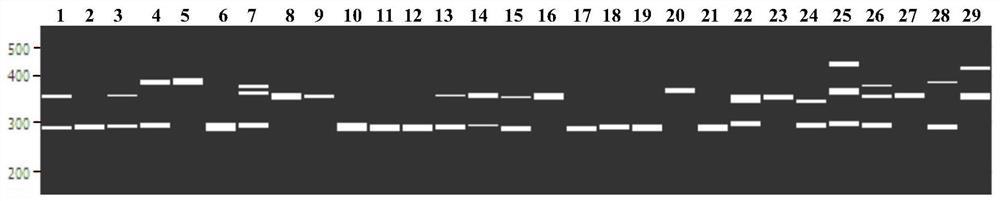
[0047] Second, the transcription group SSR site identification and SSR primer design
[0048] Use software Misa to search for SSR sites in the eggplant transcription group, search criteria: repeating unit length 2 to 6 bp, dinucleotide, tri-nucleotide, tetrapliocide, pentuclearide and hexa nucleotide least The number of repetitions is 6, 5, 5, 4, and 4 times respectively. Primer 3.0 software The sequence before and after the SSR repeating unit was used to evaluate 3 pairs of primers per SSR. The primer sequence length is 18 to 25 bp, the primer is too short to ensure the uniqueness of the sequence, and the primer may be hybridized with the error pair of sequences, reducing s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com