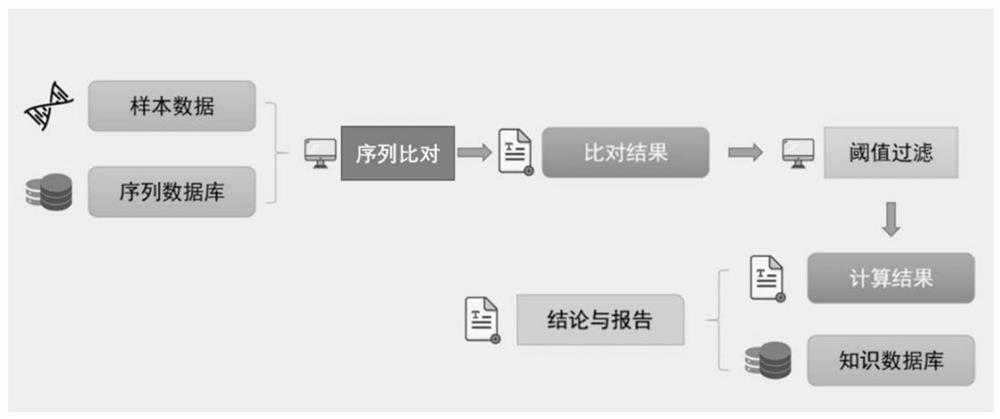
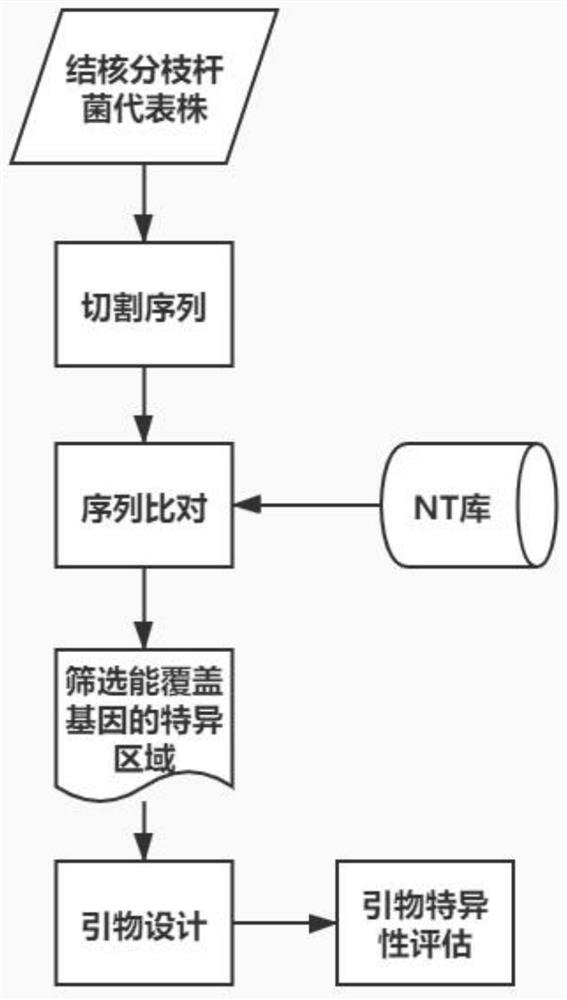
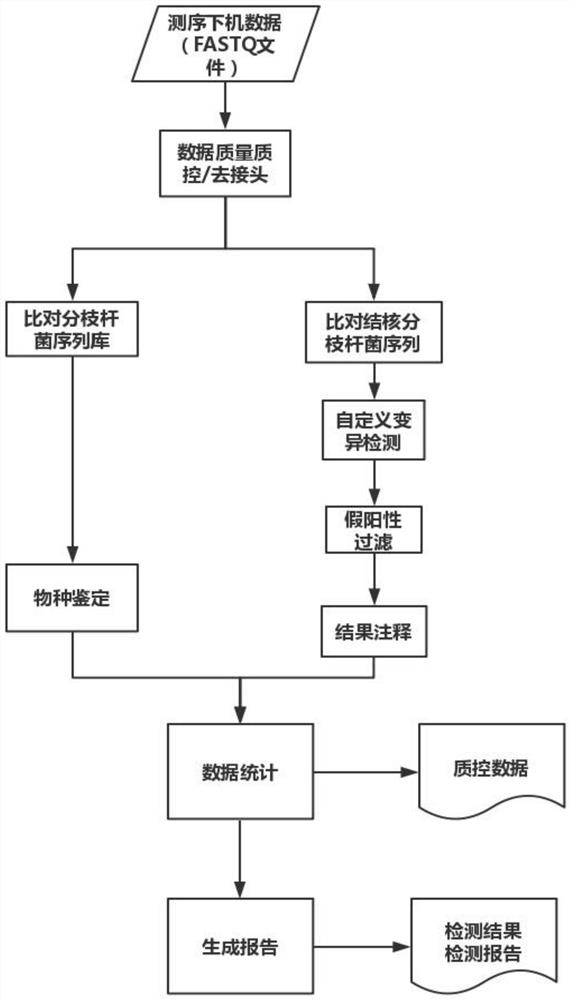
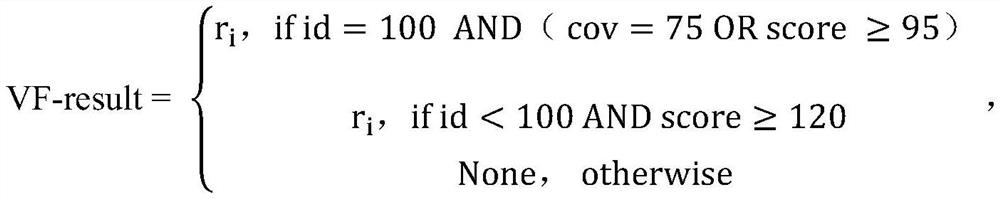Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
79 results about "Genetic Databases" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Animal genetic and health profile database management
InactiveUS7134995B2Facilitate and restrict flowImproving outcomeSurgeryNutrition controlVoice communicationGenetic Databases
A system, method, and apparatus is provided for computerized management of the databases relating to phenotypic health assessment and genomic mapping and genetic screening of animals. Multiple remote users can access a computer network connected with a central database processing resource for managing the data There is appropriate security and payment of appropriate fees for accessing and retrieving data Users may input data relating to animal health, lifespan, and genetic background, and obtain reports relating to phenotypic health assessment of a particular animal subject or animal group, and genotypic characteristics of a particular subject or animal group to which this subject belongs. The central computer database processing resource stores phenotypic data and genotypic data relating to animals and analyzes their relationship according to predetermined criteria. The input of data and reporting of data is preferably electronic, or through fax or voice communication. The analysis of the health assessment database and genetic database can be automatic and / or in part manually interpreted by experts related to the central database processing resources.
Owner:HEMOPET
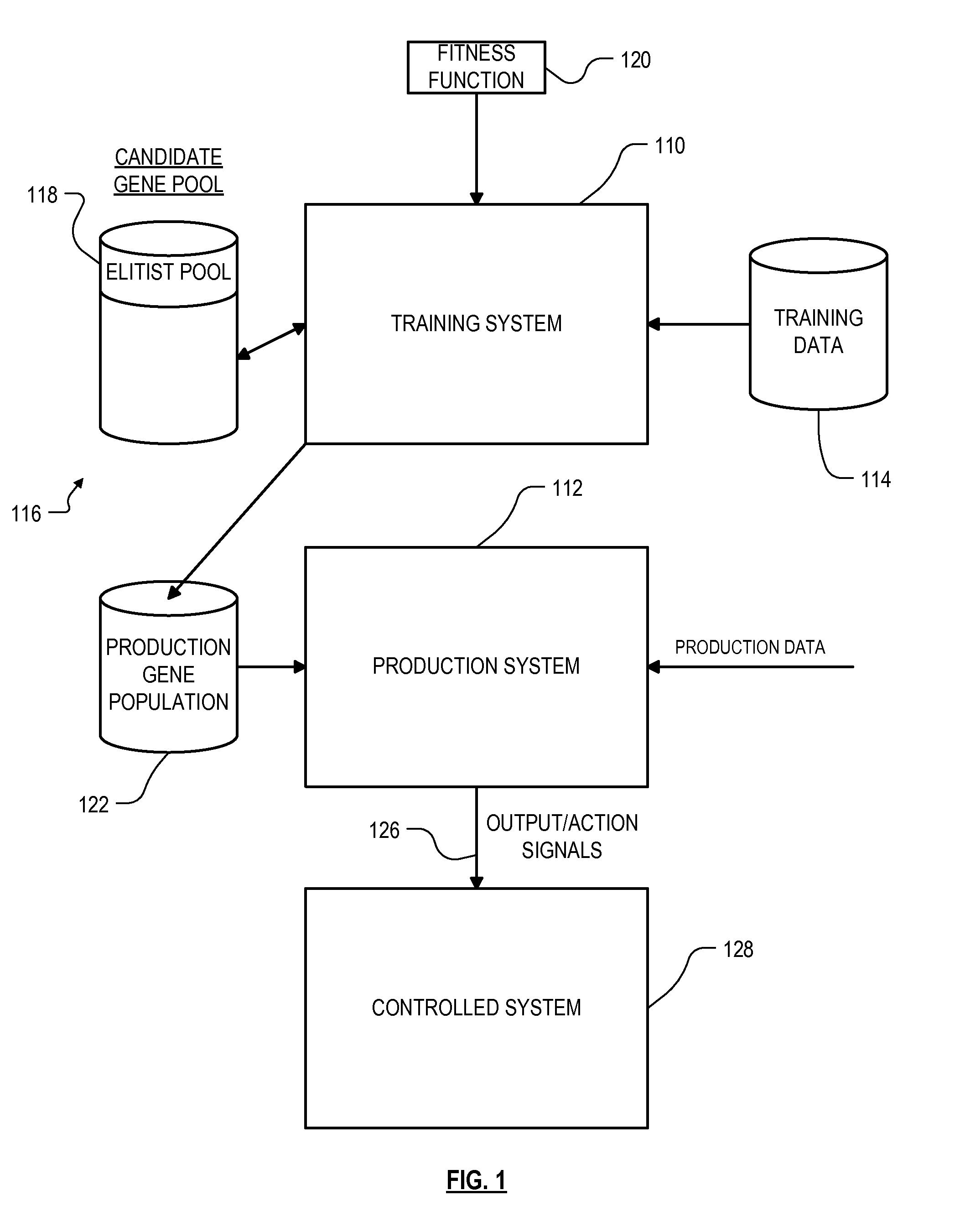
Data mining technique with experience-layered gene pool
ActiveUS8909570B1Well formedDigital computer detailsMachine learningEvolutionary data miningCandidate Gene Association Study
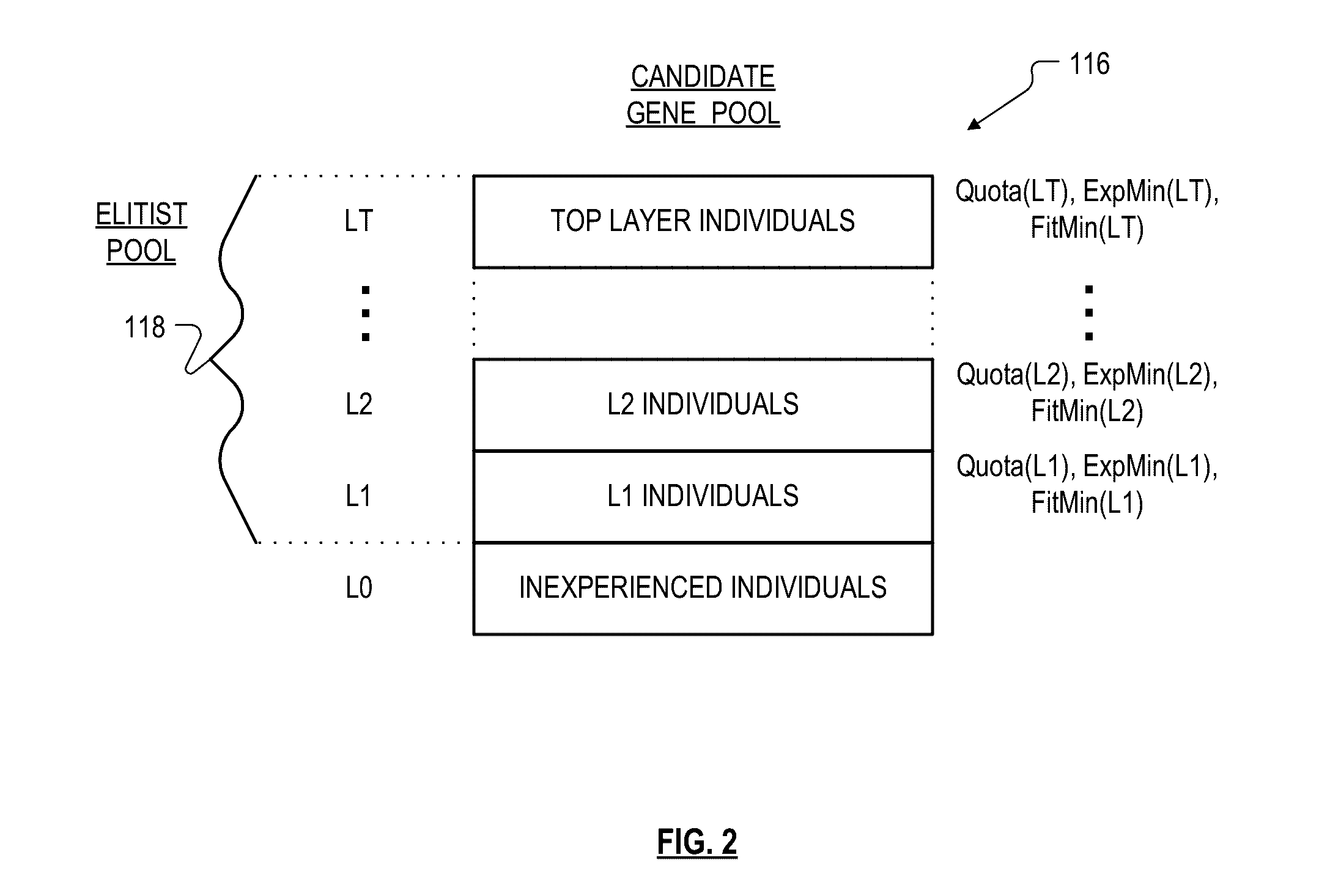
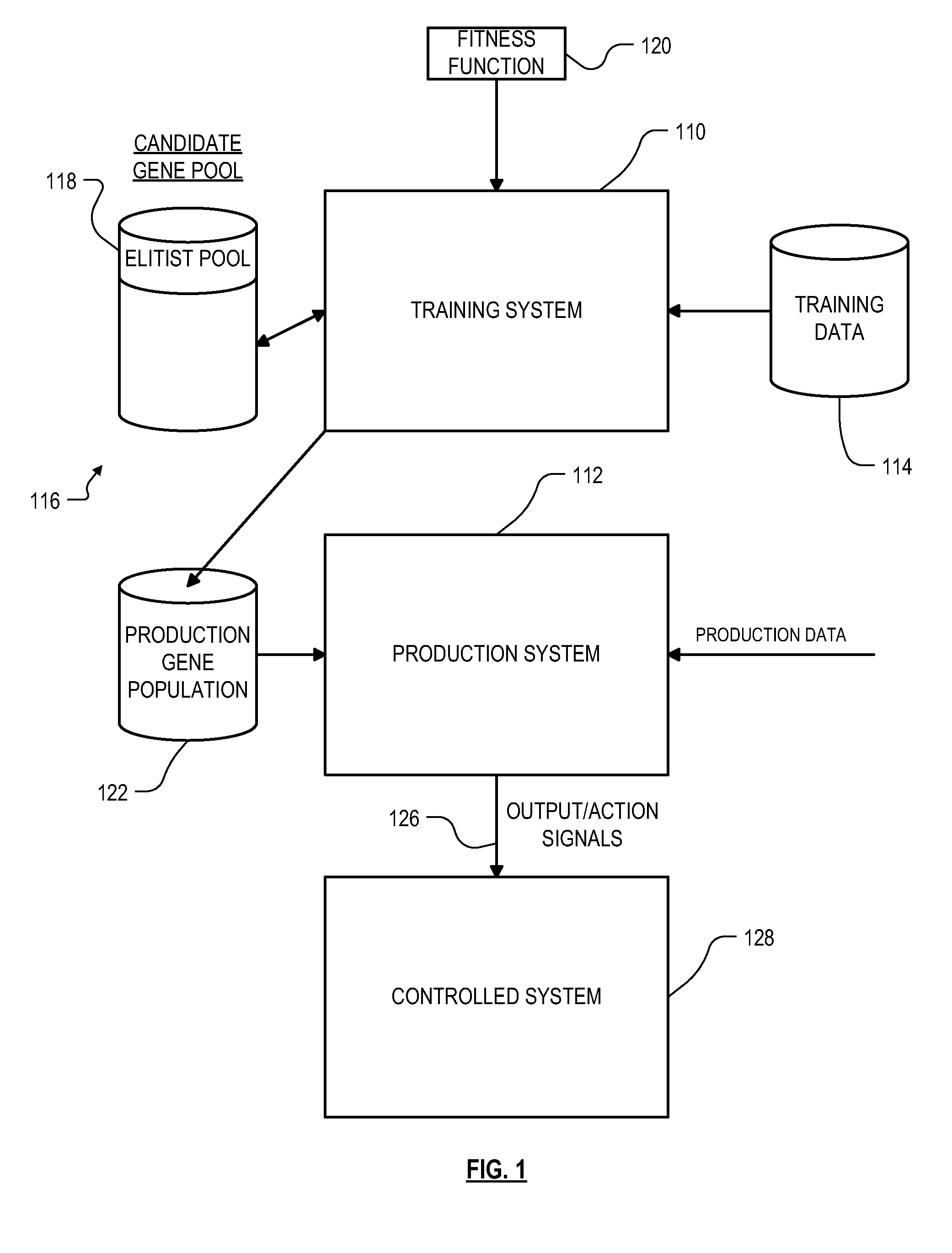
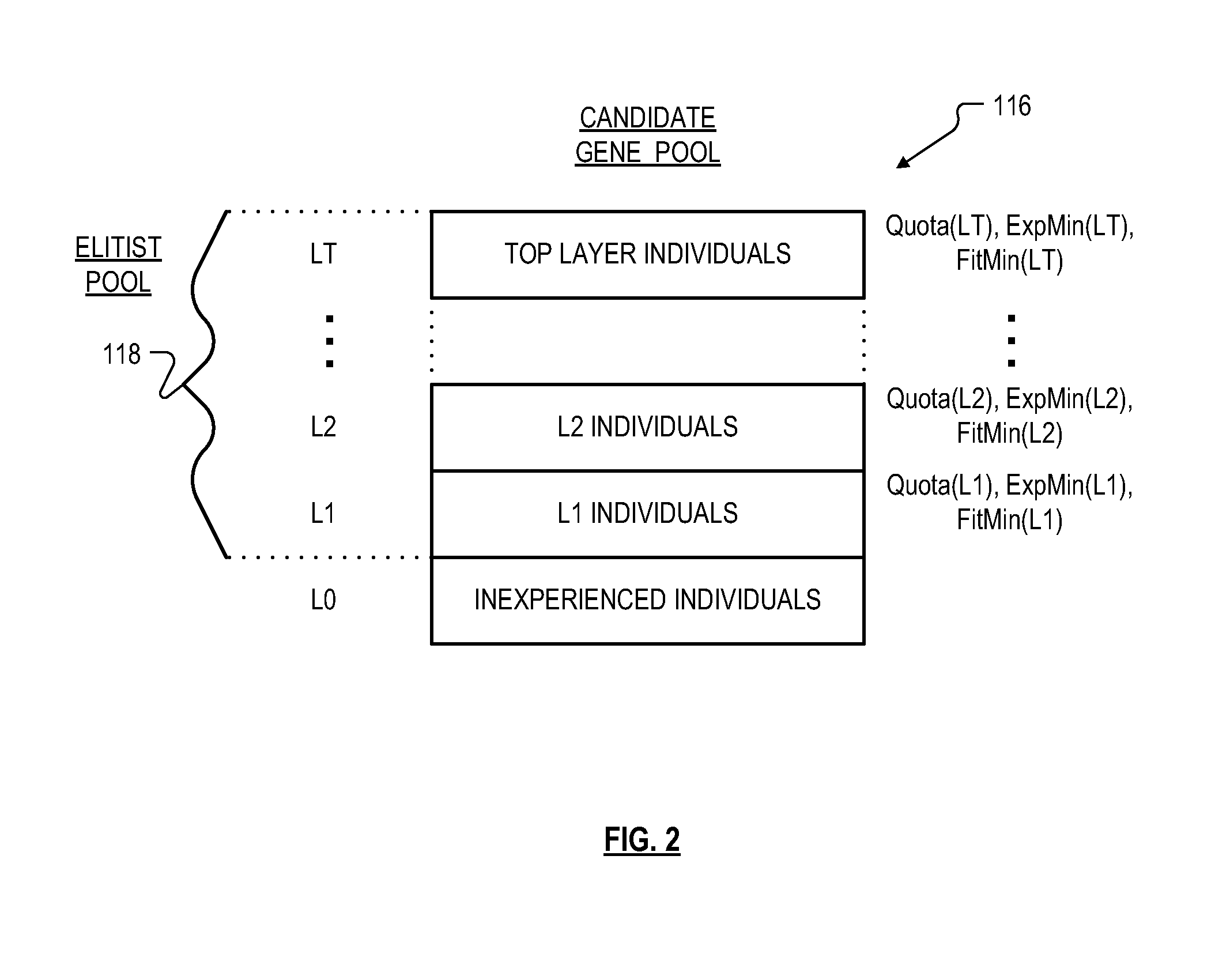
Roughly described, a computer-implemented evolutionary data mining system includes a memory storing a candidate gene database in which each candidate individual has a respective fitness estimate; a gene pool processor which tests individuals from the candidate gene pool on training data and updates the fitness estimate associated with the individuals in dependence upon the tests; and a gene harvesting module providing for deployment selected ones of the individuals from the gene pool, wherein the gene pool processor includes a competition module which selects individuals for discarding from the gene pool in dependence upon both their updated fitness estimate and their testing experience level. Preferably the gene database has an elitist pool containing multiple experience layers, and the competition module causes individuals to compete only with other individuals in their same experience layer.
Owner:COGNIZANT TECH SOLUTIONS U S CORP
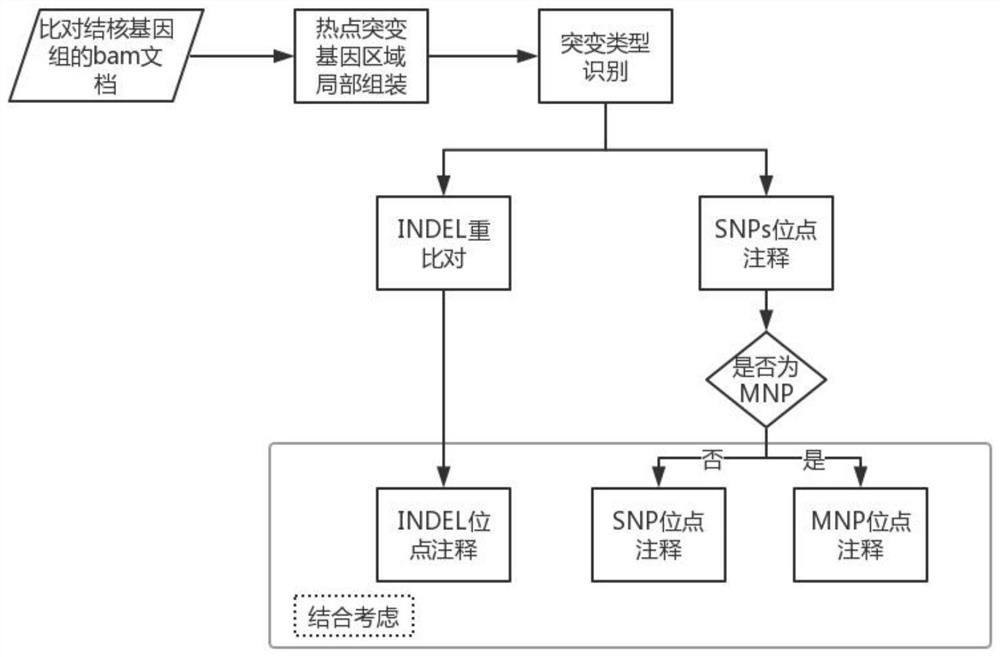
Comprehensive genetic analysis method of susceptibility of complex diseases
InactiveCN101845501AImprove graspReduce genetic analysis timeMicrobiological testing/measurementSpecial data processing applicationsCalculation errorGenetic Databases
The invention relates to a comprehensive genetic analysis method of susceptibility of complex diseases, which comprises the following steps: 1) establishing related genetic databases of complex diseases and determining related detection sites; 2) carrying out genotyping for SNPs sites within individual whole genome by using the Affymetrix6.0 chip technology to obtain the corresponding genotype of each SNP site; 3) exporting determined disease-related SNP site typing results from 900,000 SNP site detection results of a 6.0 chip, and calculating a CGR value according to the genotyping results; and 4) carrying out particular and deep genetic analysis for increased-risk diseases and high-risk diseases according to the calculated disease-related CGR value. The invention can improve the certainty of the susceptibility predication of complex diseases of Chinese Han population, shortens genetic analysis time and prevents calculation errors caused by manual calculation.
Owner:孟涛 +1
Data Mining Technique With Experience-layered Gene Pool
ActiveUS20160283563A1Digital data information retrievalMachine learningEvolutionary data miningCandidate Gene Association Study
Roughly described, a computer-implemented evolutionary data mining system includes a memory storing a candidate gene database in which each candidate individual has a respective fitness estimate; a gene pool processor which tests individuals from the candidate gene pool on training data and updates the fitness estimate associated with the individuals in dependence upon the tests; and a gene harvesting module providing for deployment selected ones of the individuals from the gene pool, wherein the gene pool processor includes a competition module which selects individuals for discarding from the gene pool in dependence upon both their updated fitness estimate and their testing experience level. Preferably the gene database has an elitist pool containing multiple experience layers, and the competition module causes individuals to compete only with other individuals in their same experience layer.
Owner:COGNIZANT TECH SOLUTIONS U S CORP
Rational selection of putative peptides from identified nucleotide or peptide sequences
InactiveUS20020059032A1Facilitates pharmacological studyBiological testingSpecial data processing applicationsNucleotideGenetic database
The present invention relates to a method for identifying putative peptides from nucleotides or peptide sequences of unknown function such as both nucleic acid and peptide precursors of a peptide comprising an amidated C-terminal end and, more particularly, to a method wherein putative peptide precursors are identified from a genetic database.
Owner:SOC DE CONSEILS DE RECH & DAPPLICATIONS SCI SAS
Method for obtaining characterization parameters of diabetes mellitus and complications
InactiveCN102930135AEfficient integrationAutomate processingSpecial data processing applicationsDiabetes mellitusSusceptibility gene
The invention relates to a method for obtaining characterization parameters of diabetes mellitus and complications. The method comprises the following steps of: 1) establishing a diabetes mellitus and complications susceptibility gene database; 2) establishing a personal diabetes mellitus and complications characterization parameters analysis platform according to the database; 3) analyzing personal diabetes mellitus and complications characterization parameters based on the analysis platform; and 4) outputting the personal diabetes mellitus and complications characterization parameters. The invention provides the method for obtaining the characterization parameters of the diabetes mellitus and complications, so that obtained characterization information is accurate, the information is obtained in time, and the foundation is laid for follow-up treatment work.
Owner:西安时代基因健康科技股份有限公司
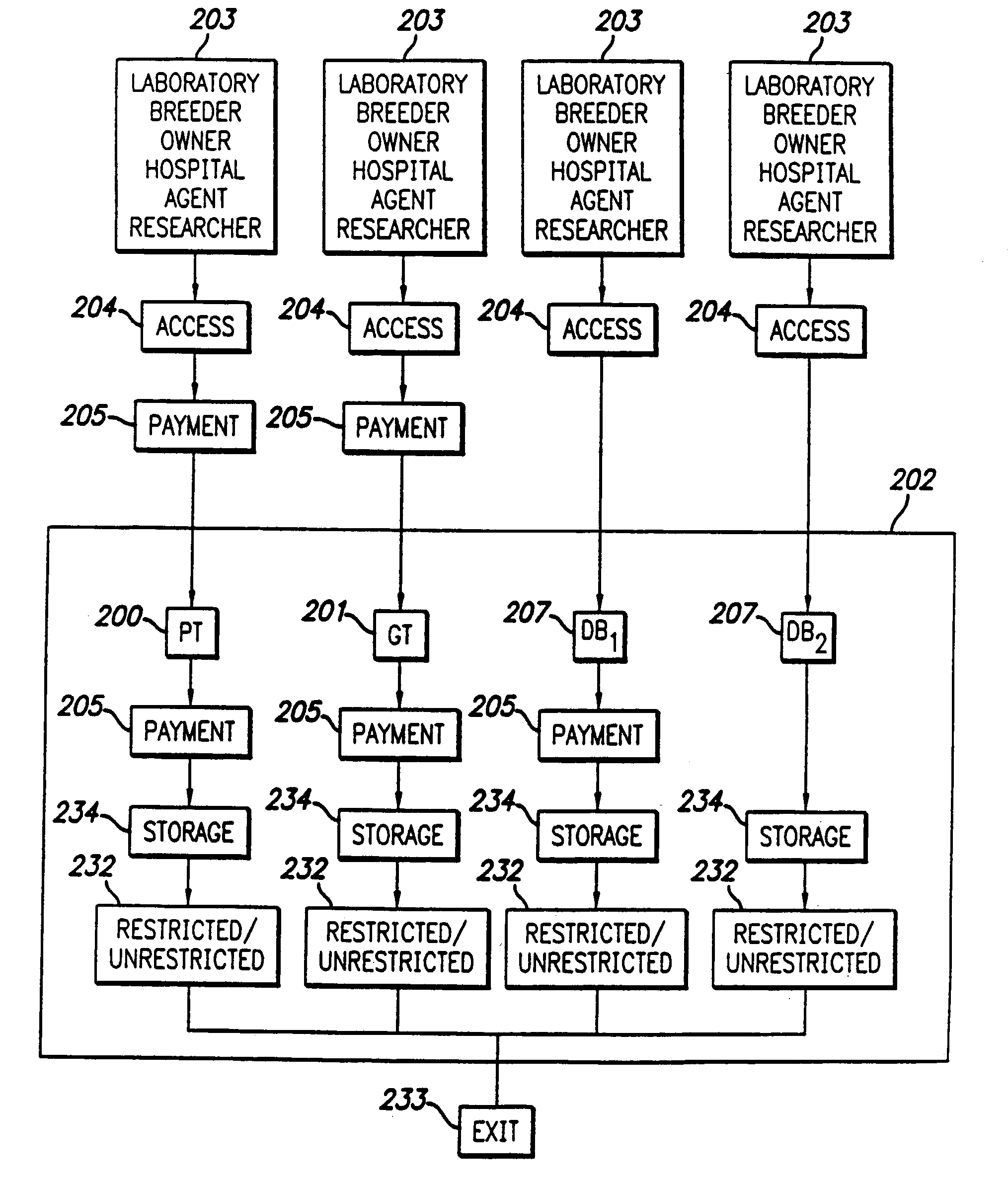
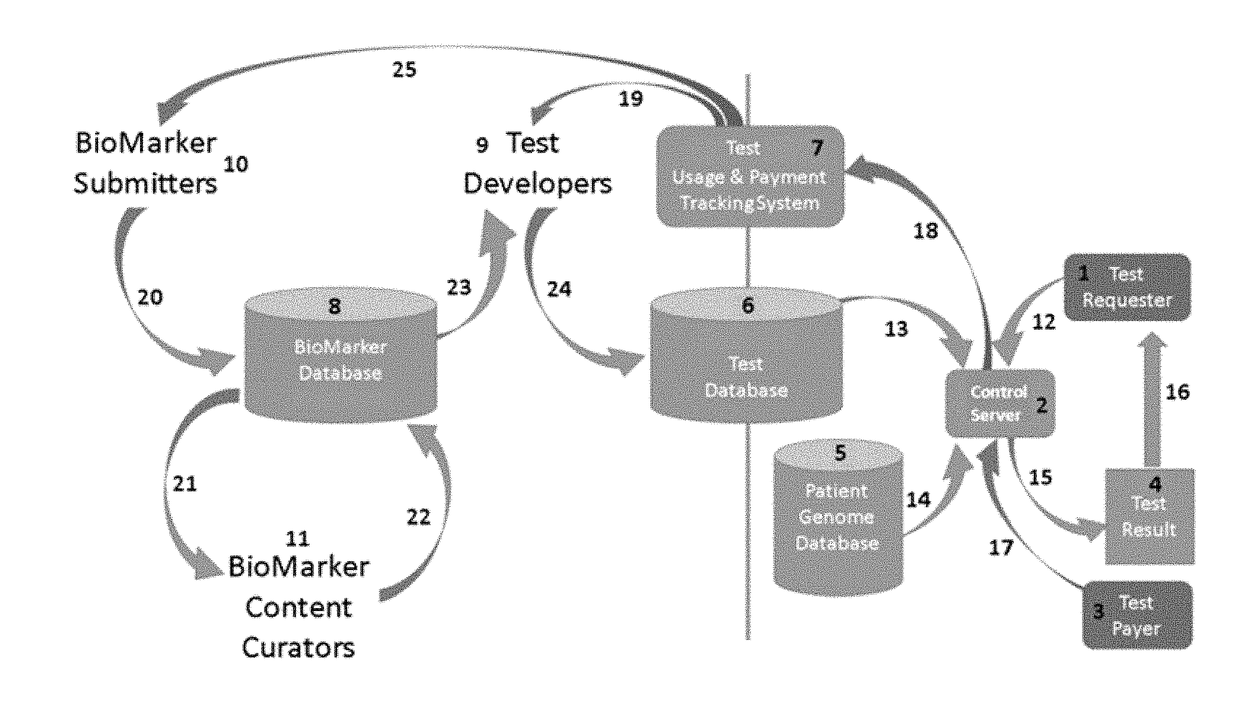
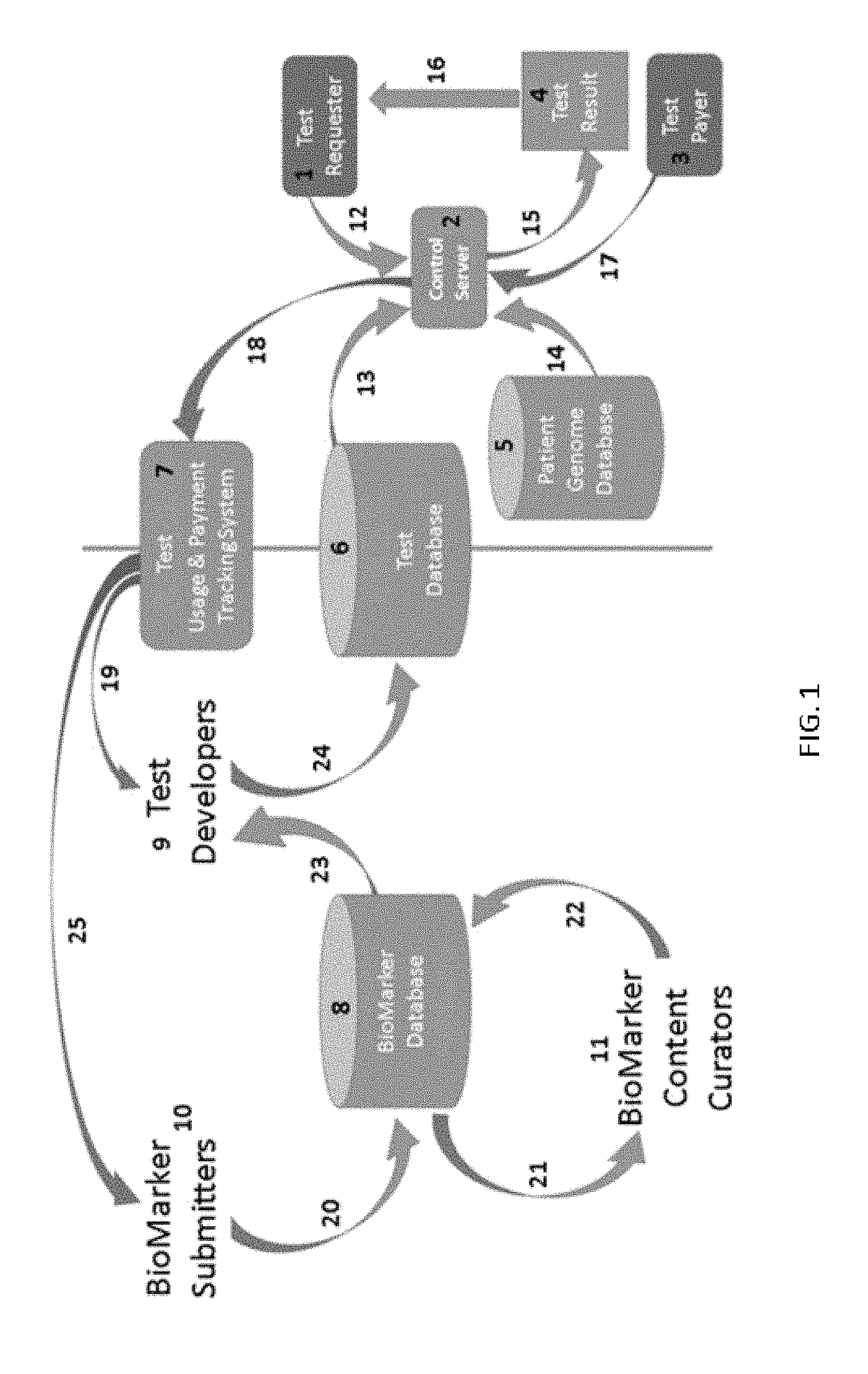
Curated genetic database for in silico testing, licensing and payment
InactiveUS20190026425A1Medical data miningEncryption apparatus with shift registers/memoriesGenetic DatabasesNew genetics
This disclosure relates to methods and systems for a curated genetic variant database and systems and methods for submitting new genetic tests based on the information in the curated database. The methods and systems of the invention further provide for a single curated variant database that allows curation of genetic variants while protecting the proprietary nature of the information submitted to the database. The system and methods also provide for submission of new genetic tests based on genetic variants, conducting genetic tests, and for determining payments to submitters and test developers based on the genetic tests.
Owner:YOUGENE
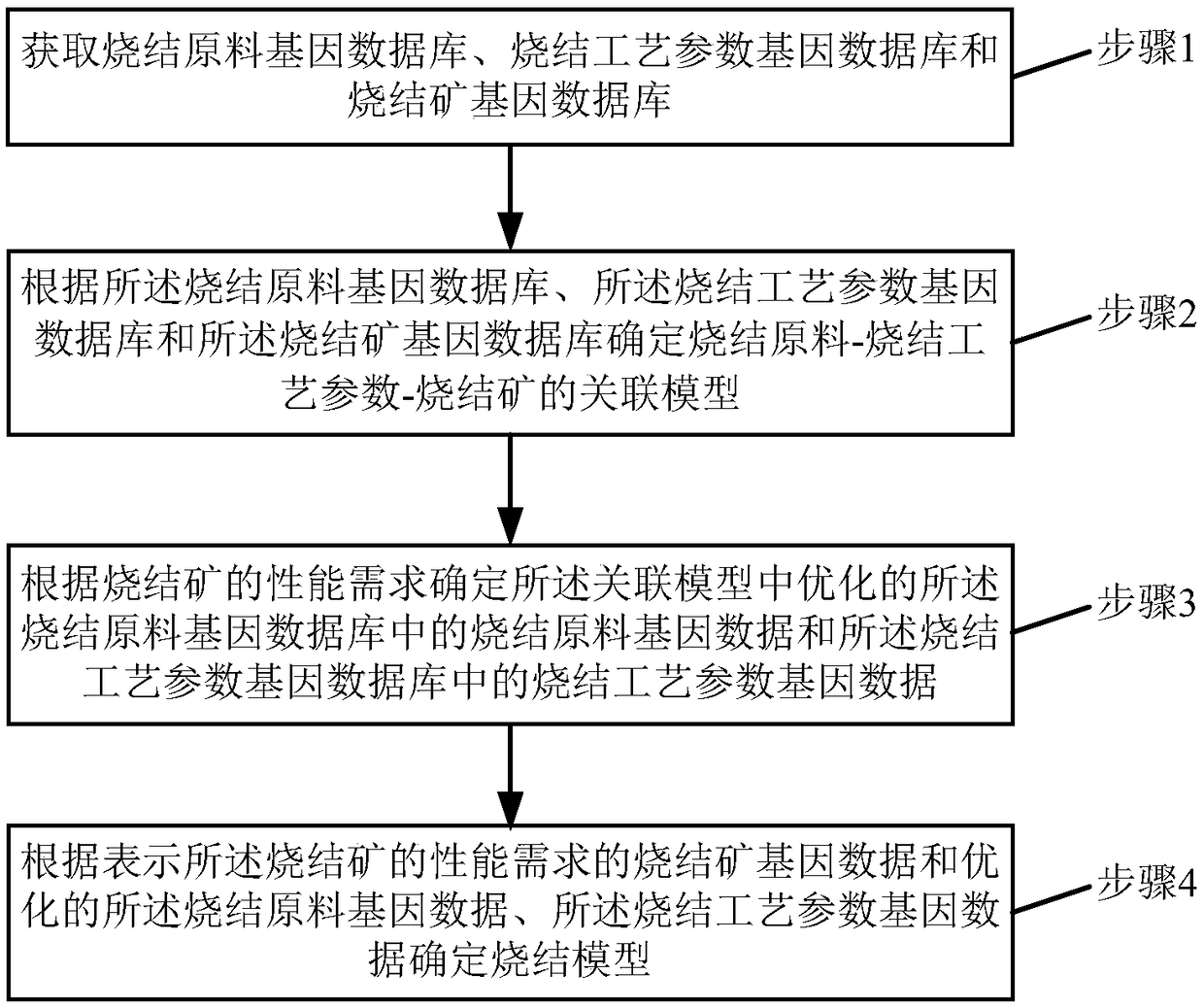
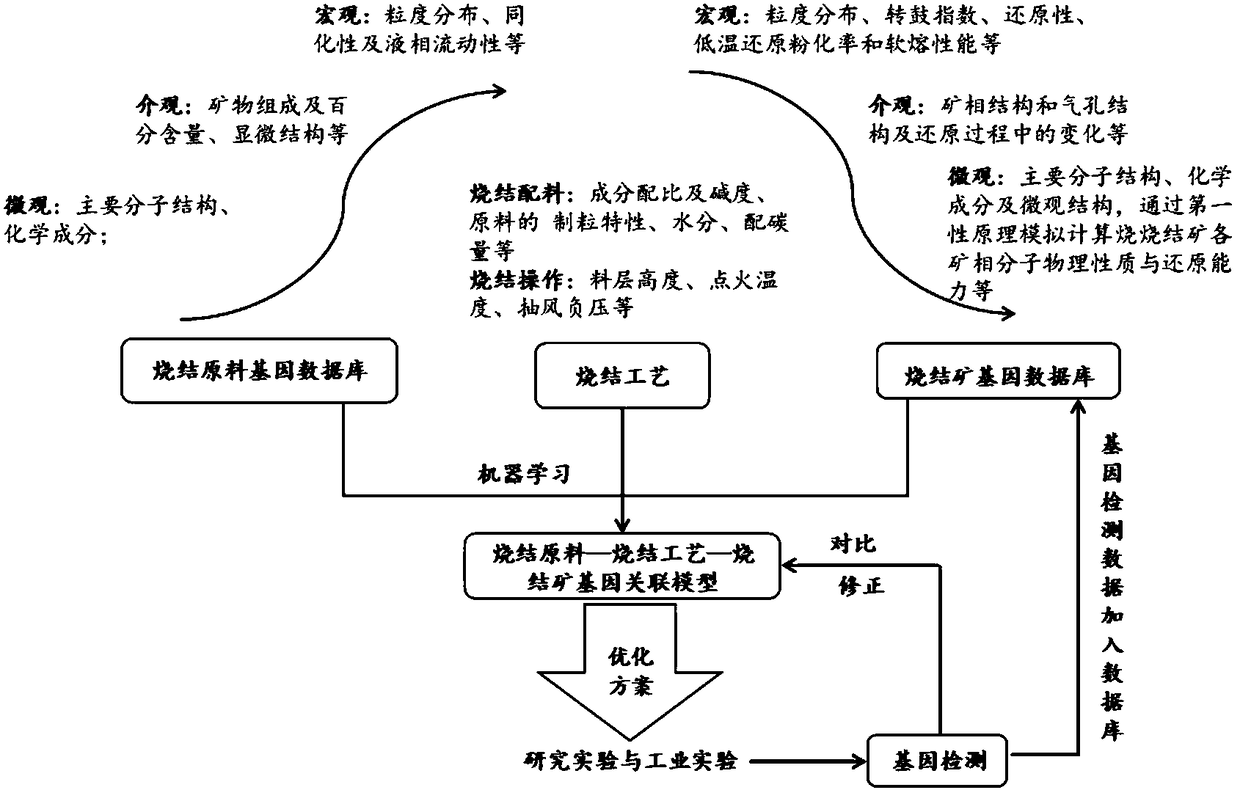
Sintered ore sintering model determination method and system based on material genes
InactiveCN108536997AEfficient integrationGuarantee the quality of sinterMolecular entity identificationCheminformatics data warehousingAssociation modelMaterials science
The invention relates to a sintered ore sintering model determination method and system based on material genes. The method includes the steps: acquiring a sintering raw material gene database, a sintering process parameter gene database and a sintered ore gene database; determining a sintering raw material, sintering process parameter and sintered ore association model according to the sinteringraw material gene database, the sintering process parameter gene database and the sintered ore gene database; determining optimized sintering raw material gene data in the sintering raw material genedatabase and sintering process parameter gene data in the sintering process parameter gene database in the association model according to performance requirements of sintered ores; determining a sintering model according to sintered ore gene data representing the performance requirements of the sintered ores, the optimized sintering raw material gene data and the sintering process parameter gene data. The optimal sintering process can be rapidly set according to the performance requirements of the sintered ores, and the quality of the sintered ores is ensured.
Owner:WUHAN UNIV OF SCI & TECH
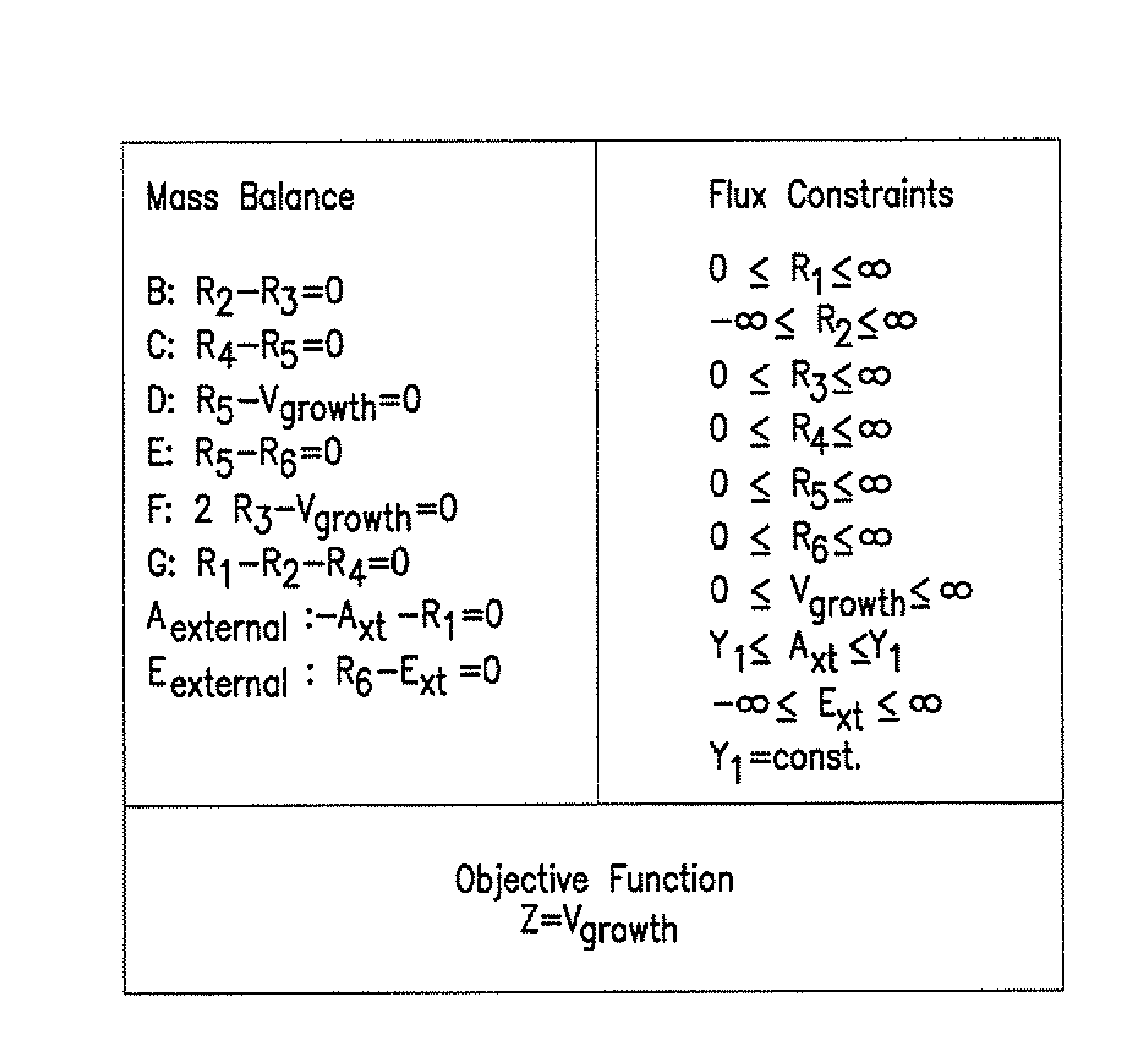
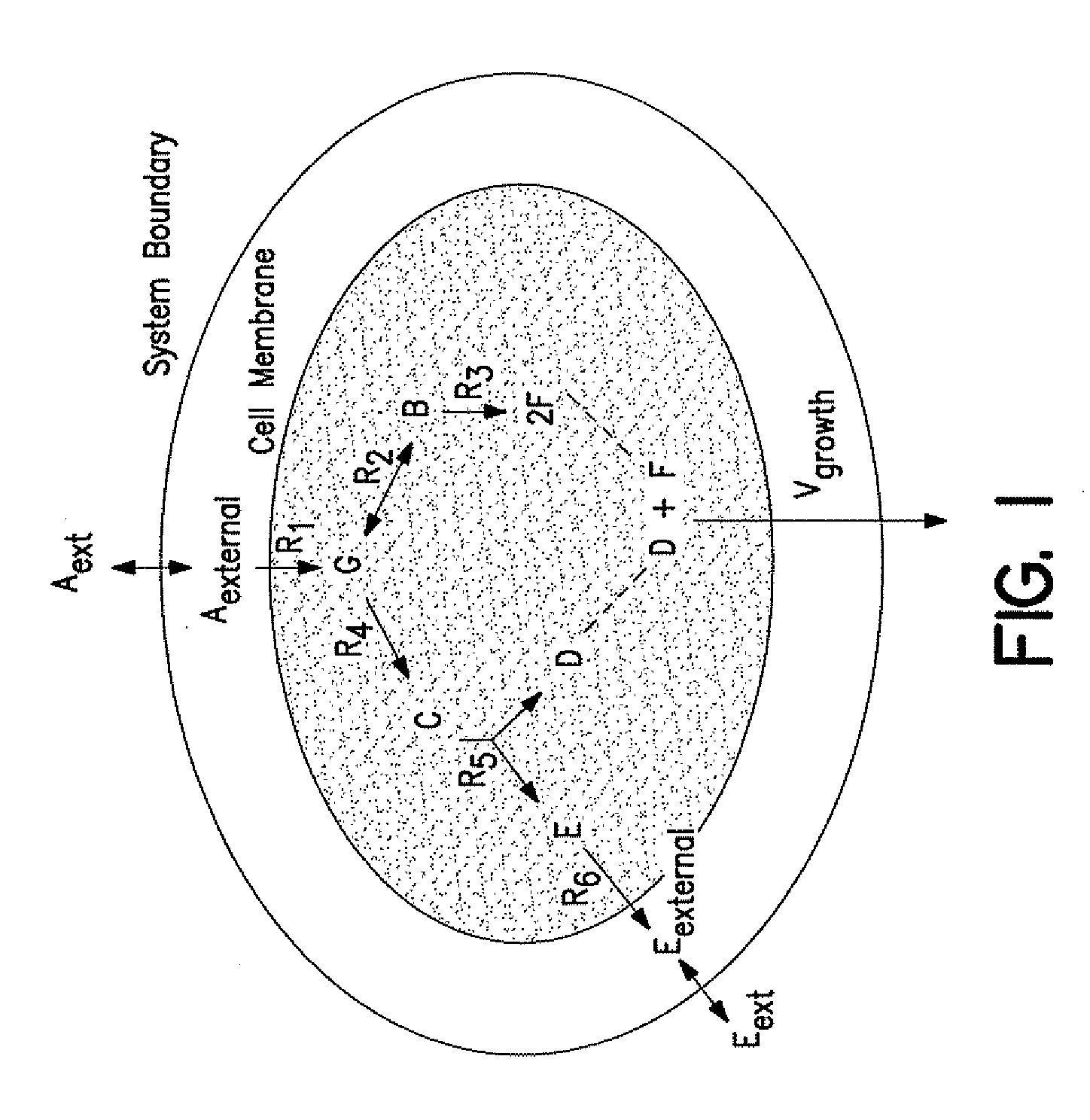
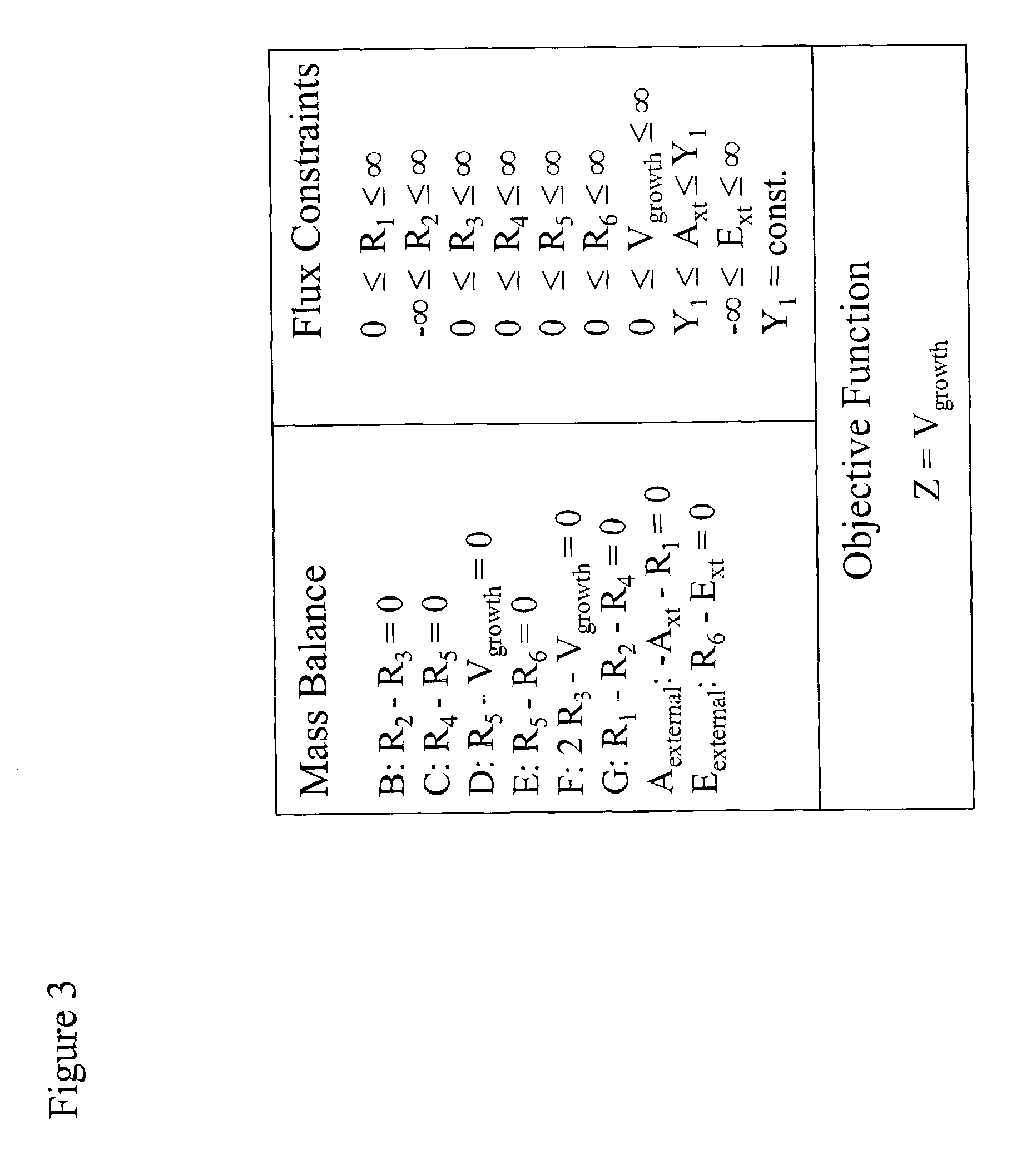
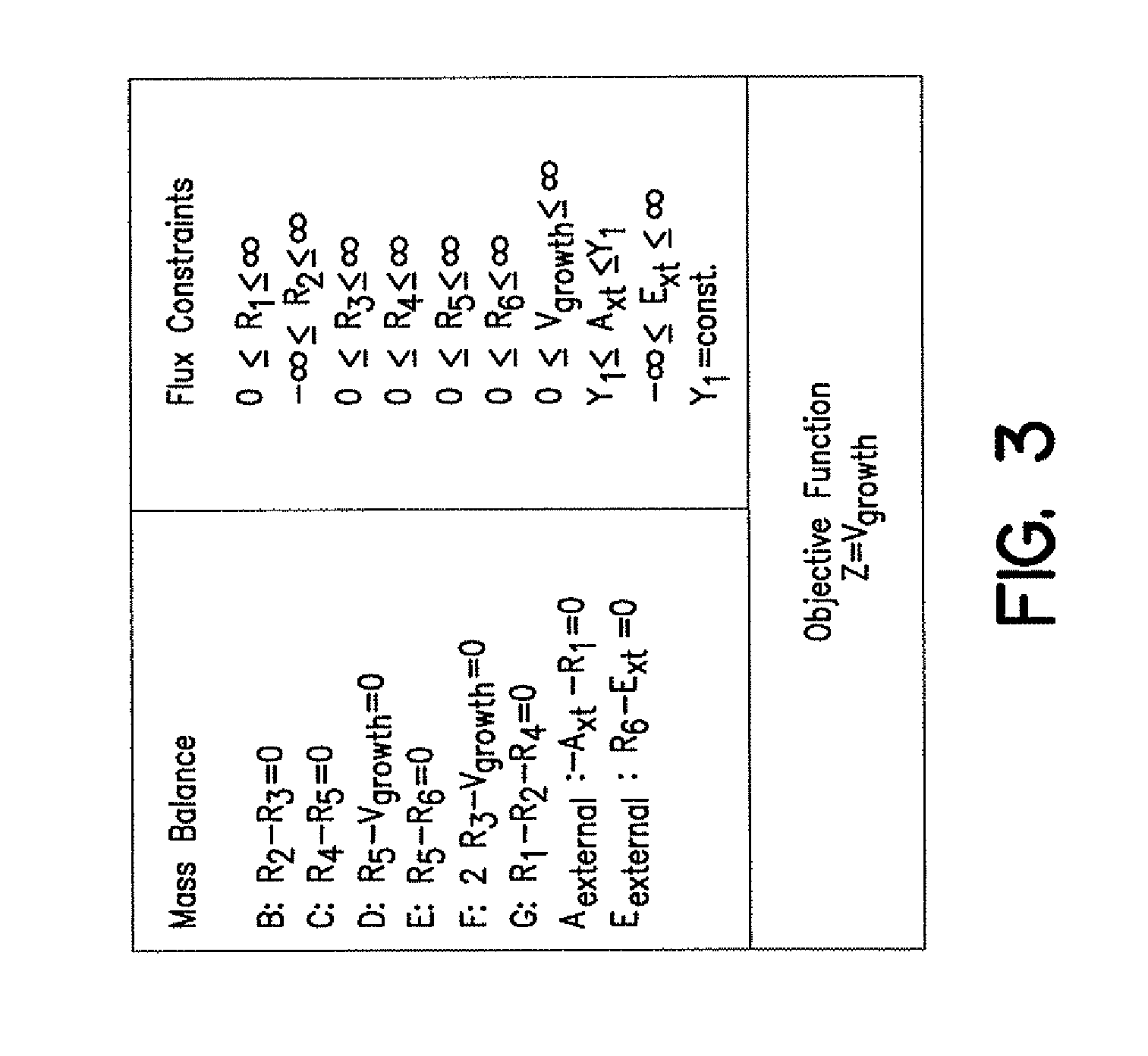
Compositions and Methods for Modeling Saccharomyces cerevisiae Metabolism
The invention provides an in silico model for determining a S. cerevisiae physiological function. The model includes a data structure relating a plurality of S. cerevisiae reactants to a plurality of S. cerevisiae reactions, a constraint set for the plurality of S. cerevisiae reactions, and commands for determining a distribution of flux through the reactions that is predictive of a S. cerevisiae physiological function. A model of the invention can further include a gene database containing information characterizing the associated gene or genes. The invention further provides methods for making an in silico S. cerevisiae model and methods for determining a S. cerevisiae physiological function using a model of the invention.
Owner:RGT UNIV OF CALIFORNIA
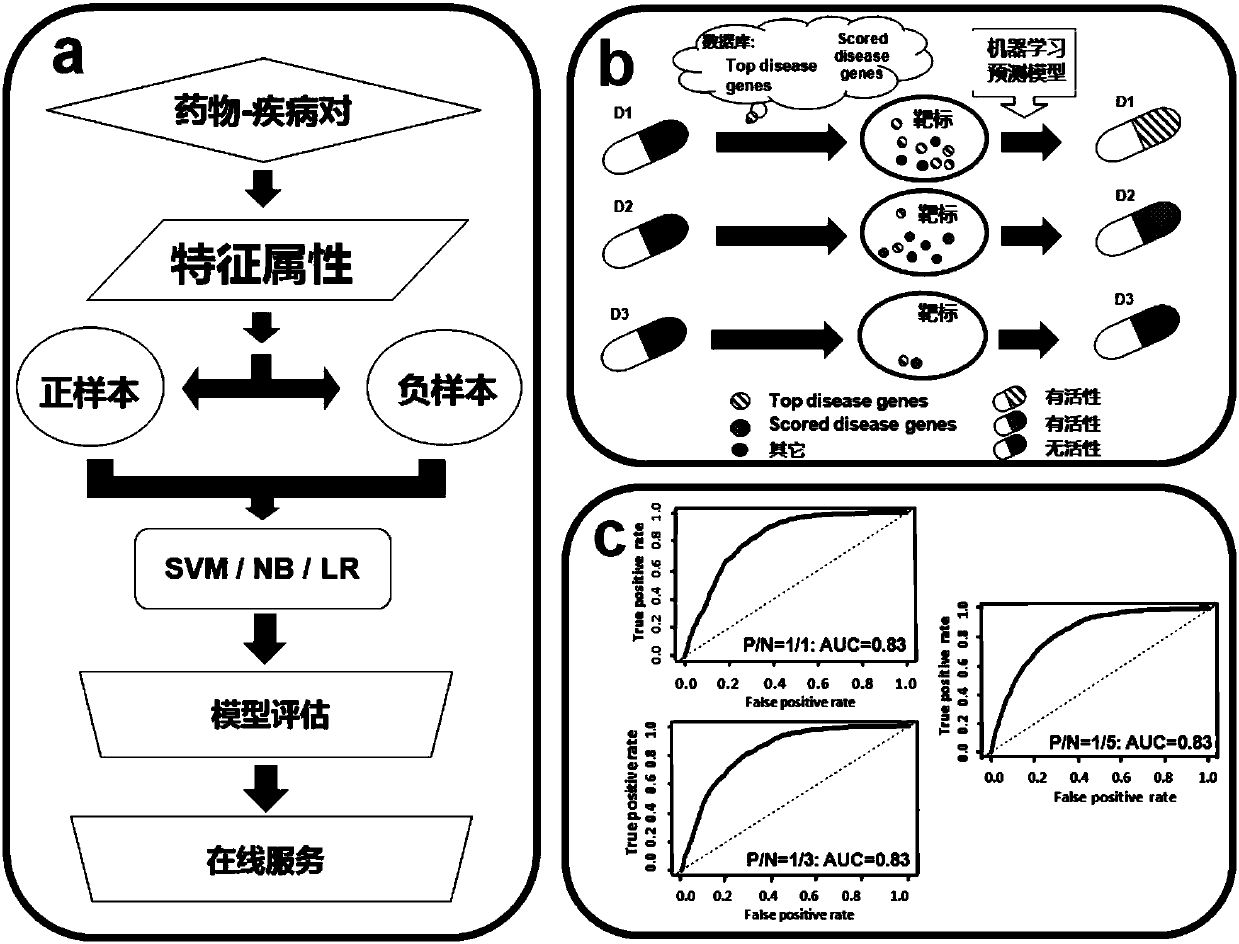
Drug activity prediction method and application thereof
ActiveCN107731309AImprove development success rateLow costMedical data miningMolecular designRelational databaseMedicine
The invention discloses a drug activity prediction method and application thereof. The method comprises the following beneficial effects: step one, targets of human on-market or under-study drugs andtreatment activity information are collected by inquiring drug target interaction database information; step two, searching for a plurality of virulence gene databases, collecting disease associationgenes, and according to activity rates of drugs corresponding to the disease association genes, different assignment values of disease association genes of different database sources are given; step three, feature attributes of the drug targets and the disease association genes are built; step four, a machine learning prediction model is constructed; step five, a model prediction result is evaluated; and step six, a drug with activity to a specific disease is predicted. According to the invention, the drug activity prediction method can be used as a GPS in the drug discovery field; disease association genes can be identified efficiently; the effective guidance is provided for prediction, research and development of active drugs; and a novel method and idea is provided for the drug discovery field in future.
Owner:武汉百药联科科技有限公司 +1
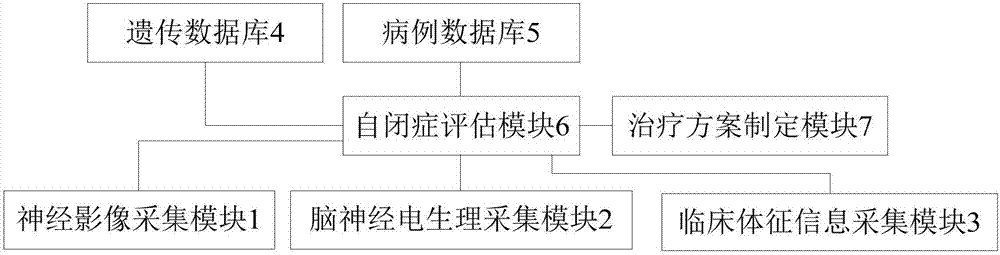
Autism diagnosis and treatment system
InactiveCN107887027AImprove the effect of diagnosis and treatmentGood treatment effectHealth-index calculationMedical automated diagnosisTreatment effectGenetic database
The present invention discloses an autism diagnosis and treatment system provided by the embodiment of the invention. The system comprises a neuroimaging collection module, a cranial nerve electrophysiologic collection module, a clinical sign information collection module, a genetic database, a case database and an autism diagnostic assessment module. The autism diagnostic assessment module can combine genetic information and autism cases to determine whether a patient makes a definite diagnosis of the autism or not according to neuroimaging, cranial nerve electrophysiologic signals and clinical sign information, and a diagnosis result is obtained. According to the embodiment of the invention, remote automatic diagnosis and treatment and diagnosis definition can be achieved, a customized treatment rehabilitation scheme is made, and therefore, a diagnosis and treatment effect is improved.
Owner:广州优涵信息技术有限公司
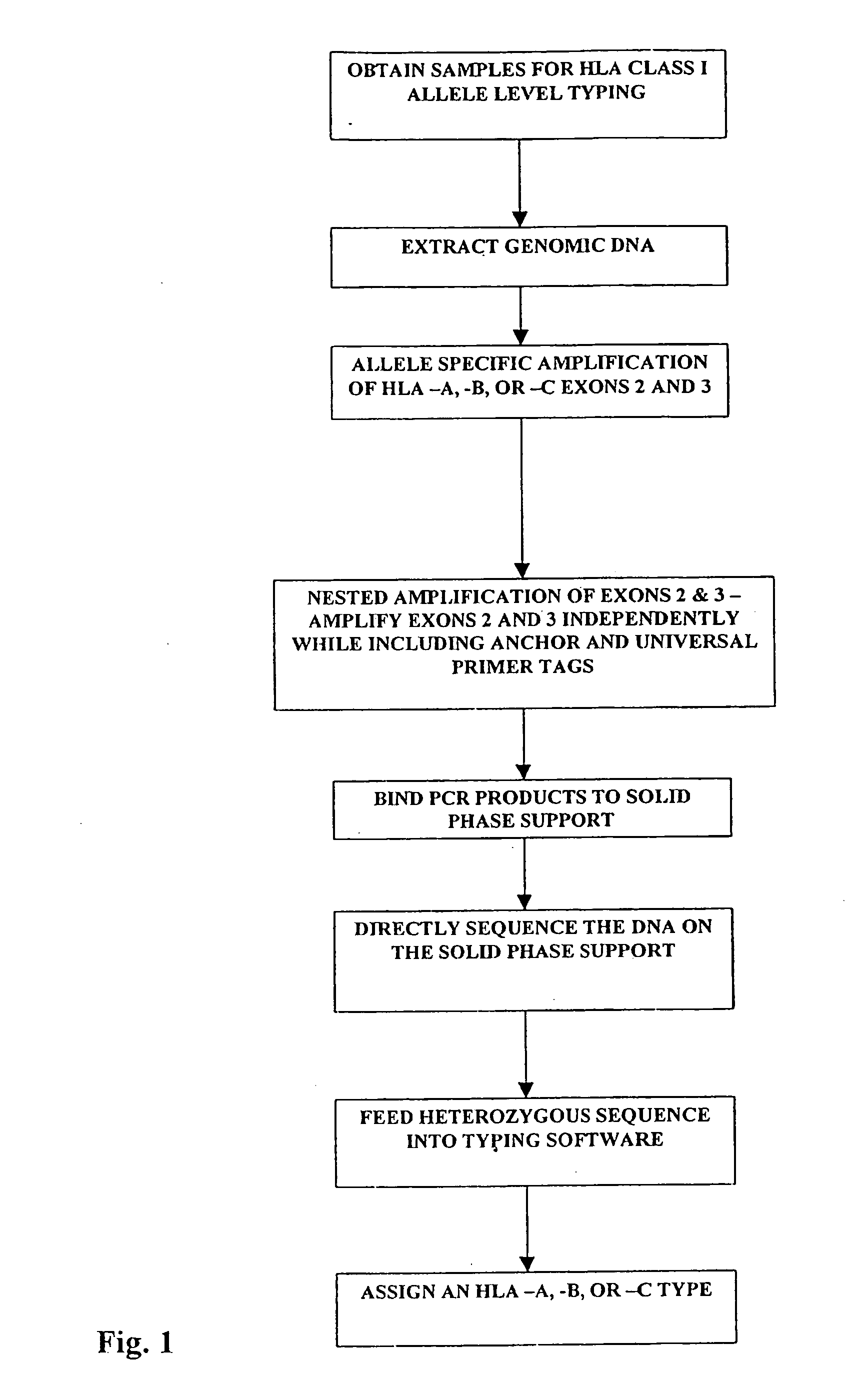
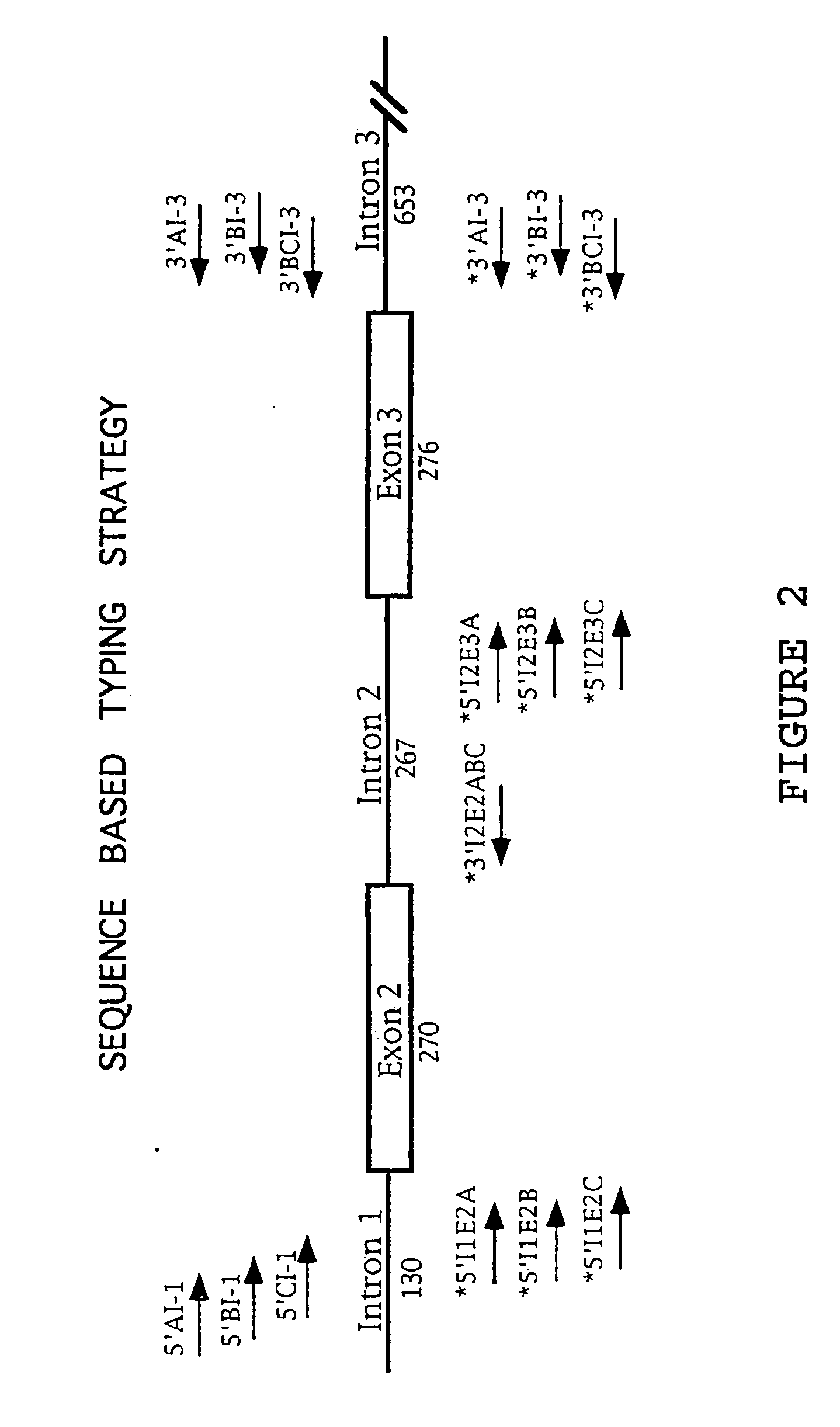
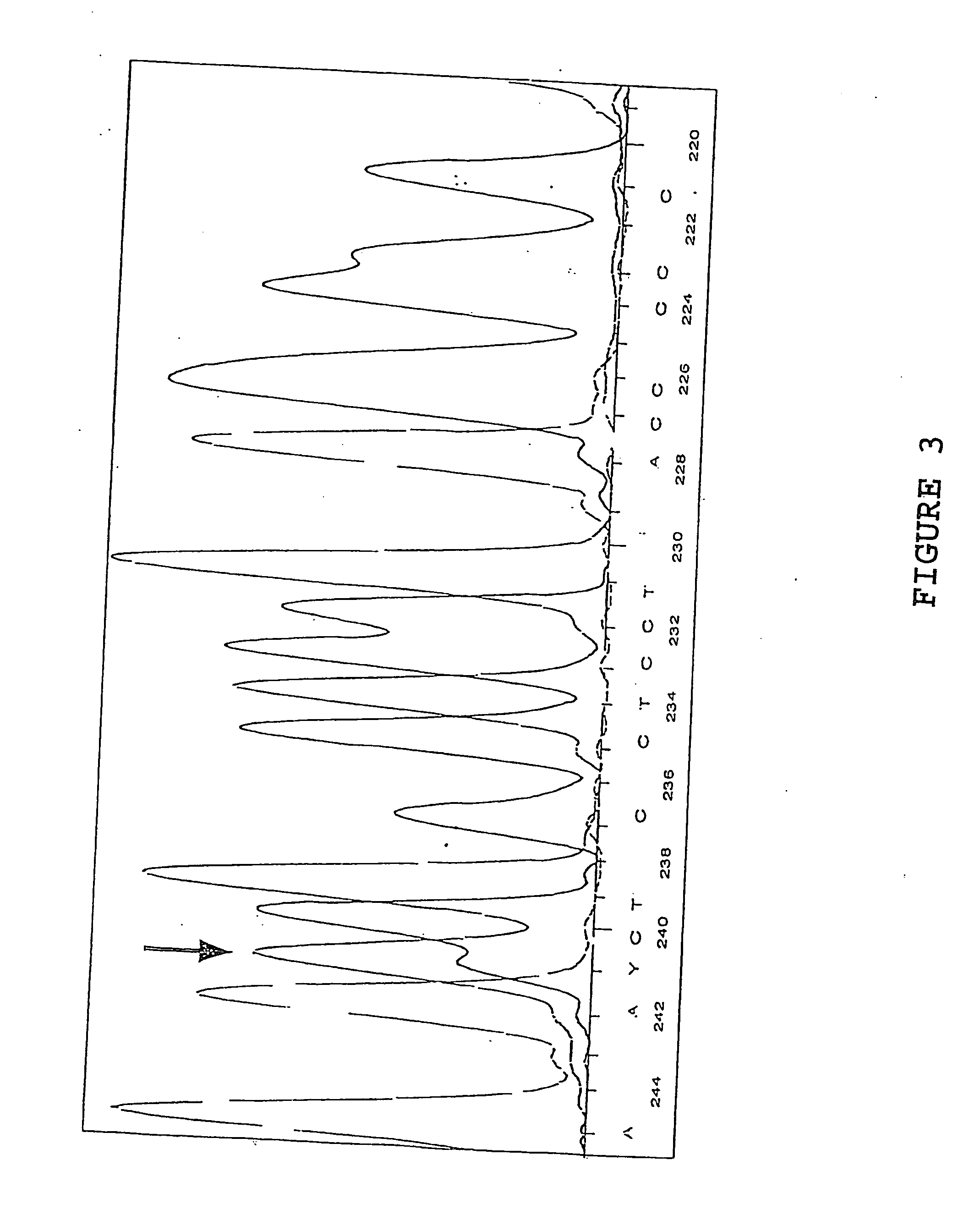
Class I sequence based typing of HLA-A, -B, and -C alleles by direct DNA sequencing
InactiveUS20070128629A1Microbiological testing/measurementFermentationTissue sampleDirect sequencing
There is provided a method for directly typing or sequencing HLA-A, -B, or -C alleles from a tissue sample wherein exons 2 and 3 of the HLA-A, -B, or -C alleles from the sample are amplified together in a locus specific manner and then separated out and individually amplified in a locus specific manner. After the two amplifications, the amplified exons are directly sequenced, the sequences are recombined, and a comparison is made between the derived HLA allele sequence and an HLA allele database, thereby giving an exact HLA-A, -B, or -C type for the sample being tested.
Owner:HILDEBRAND WILLIAM H +3
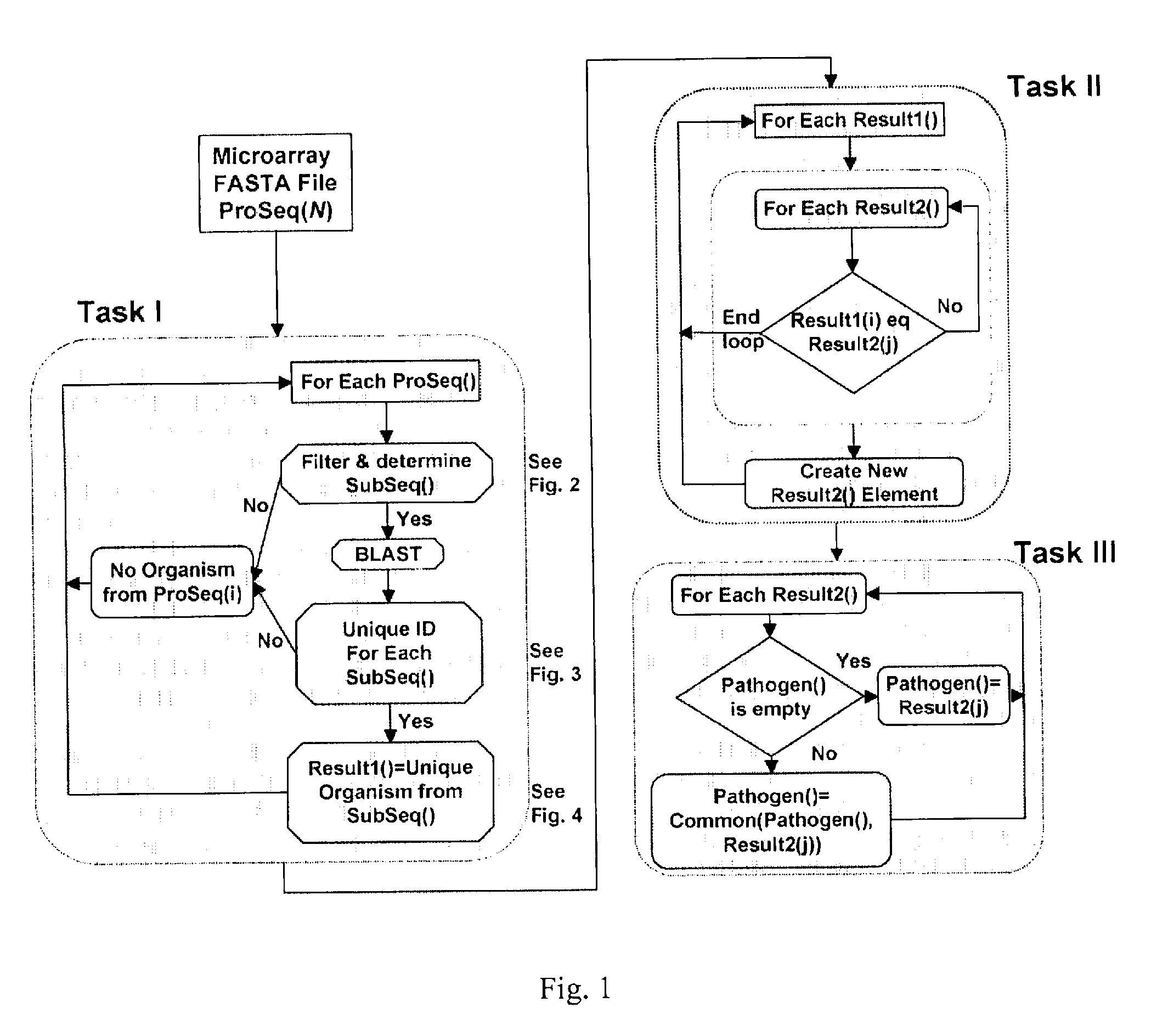
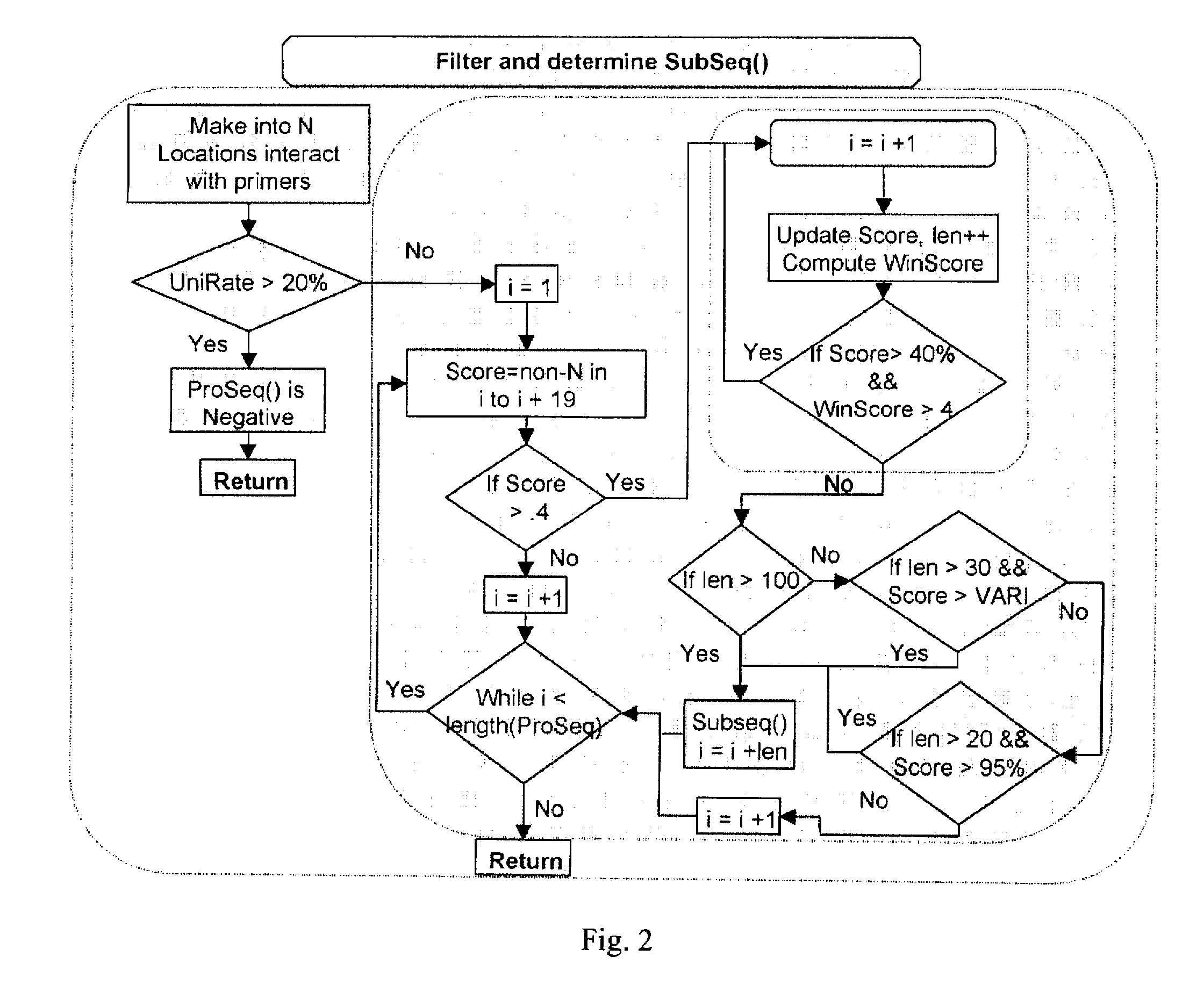
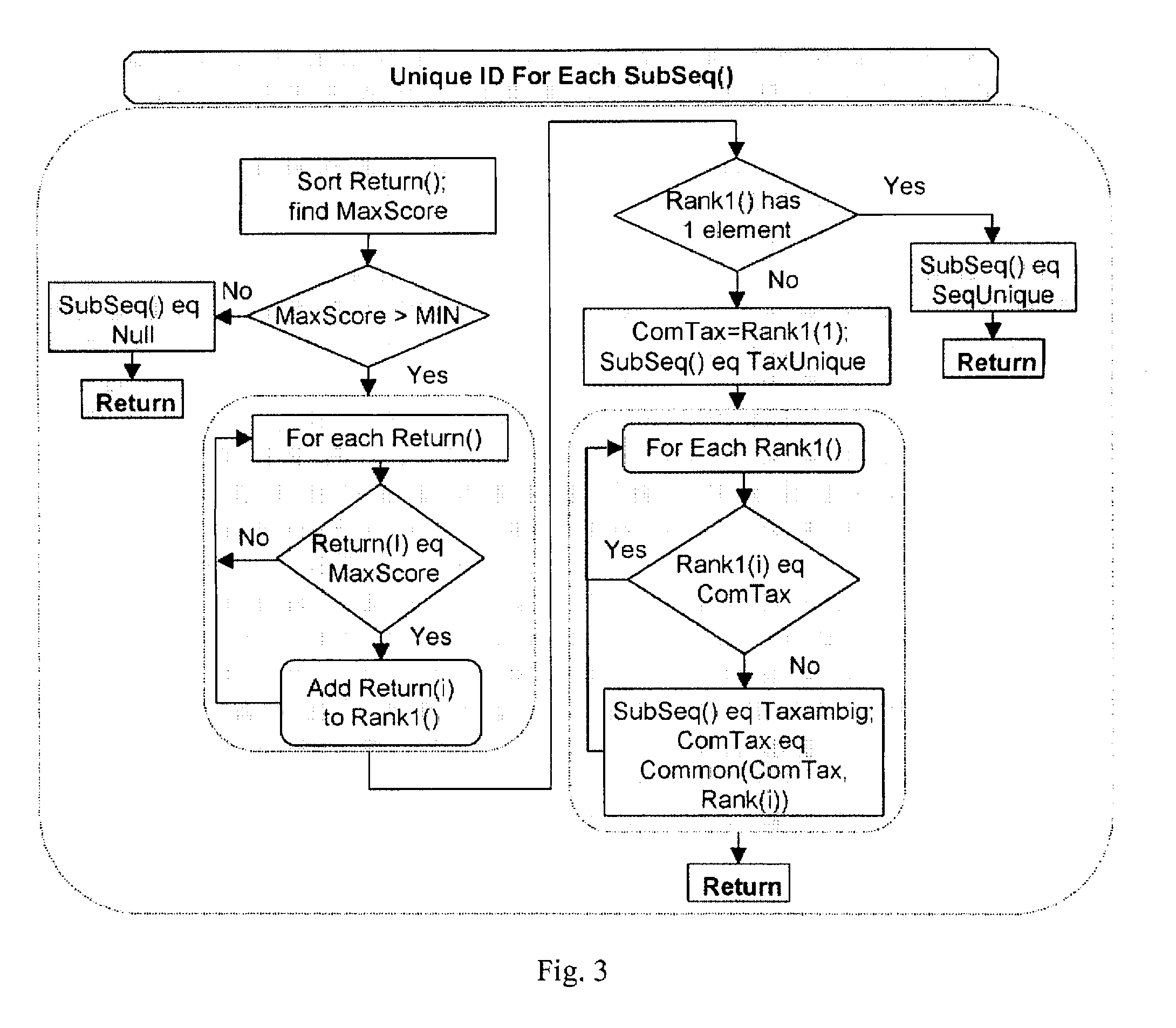
Computer-implemented biological sequence identifier system and method
InactiveUS20070059728A1Bioreactor/fermenter combinationsBiological substance pretreatmentsBase callingGenetic database
A method of: submitting reference sequences to a taxonomic database to produce taxonomic results; and reporting a taxonomic identification based on the taxonomic results. The reference sequences are the output of genetic database queries that return a score for each reference sequence. A method for processing a biological sequence obtained from an assay by: converting base calls located in a predetermined list of positions within the biological sequence to N; and determining the ratio of single nucleotide polymorphisms in the biological sequence relative to a reference sequence. Each entry in the predetermined list of positions represents the capability of a substance hybridizing to a microarray used to generate the biological sequence. The substance is not the nucleic acid of a target pathogen.
Owner:UNITED STATES OF AMERICA
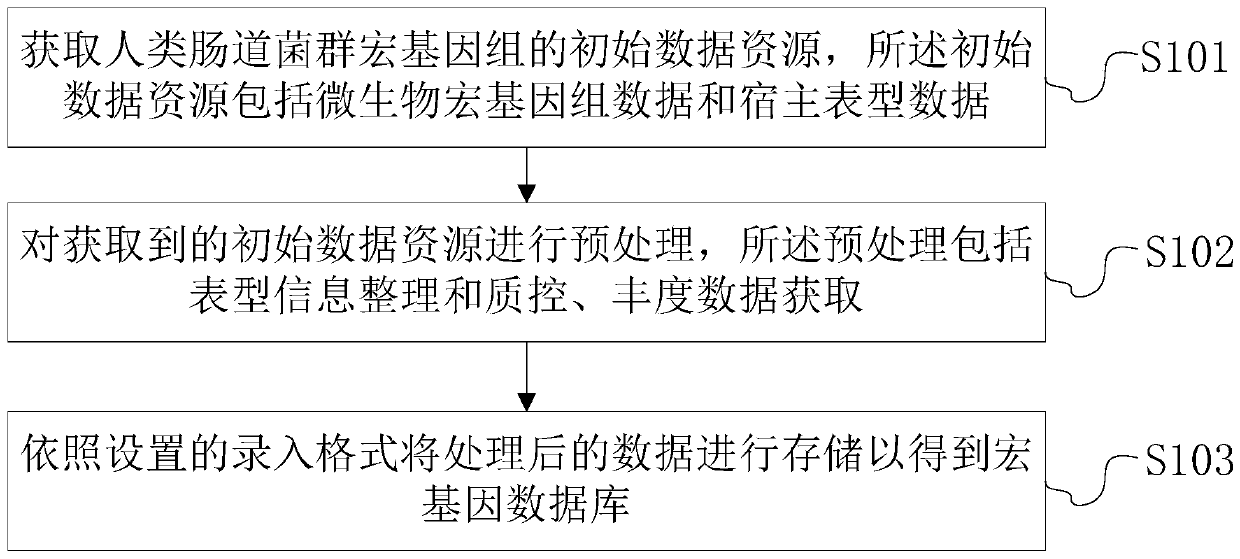
Intestinal flora metagenome database construction method and device and intestinal flora metagenome database analysis method and device
The invention discloses an intestinal flora metagenome database construction method which comprises the following steps: acquiring initial data resources of a human intestinal flora metagenome, the initial data resources comprising microbial metagenome data and host phenotype data; preprocessing the acquired initial data resources, wherein the preprocessing comprises phenotypic information arrangement and quality control and abundance data acquisition; and storing the processed data according to a set entry format to obtain a metagene database. The invention further discloses an analysis method and system based on the intestinal flora metagenome database, and a storage medium. According to the intestinal flora metagenome database construction method, screening and unified standard processing are carried out on original data, so that obtained database information is accurate, reliable and standard, and information transmission, database management and database data interaction are facilitated.
Owner:康美华大基因技术有限公司
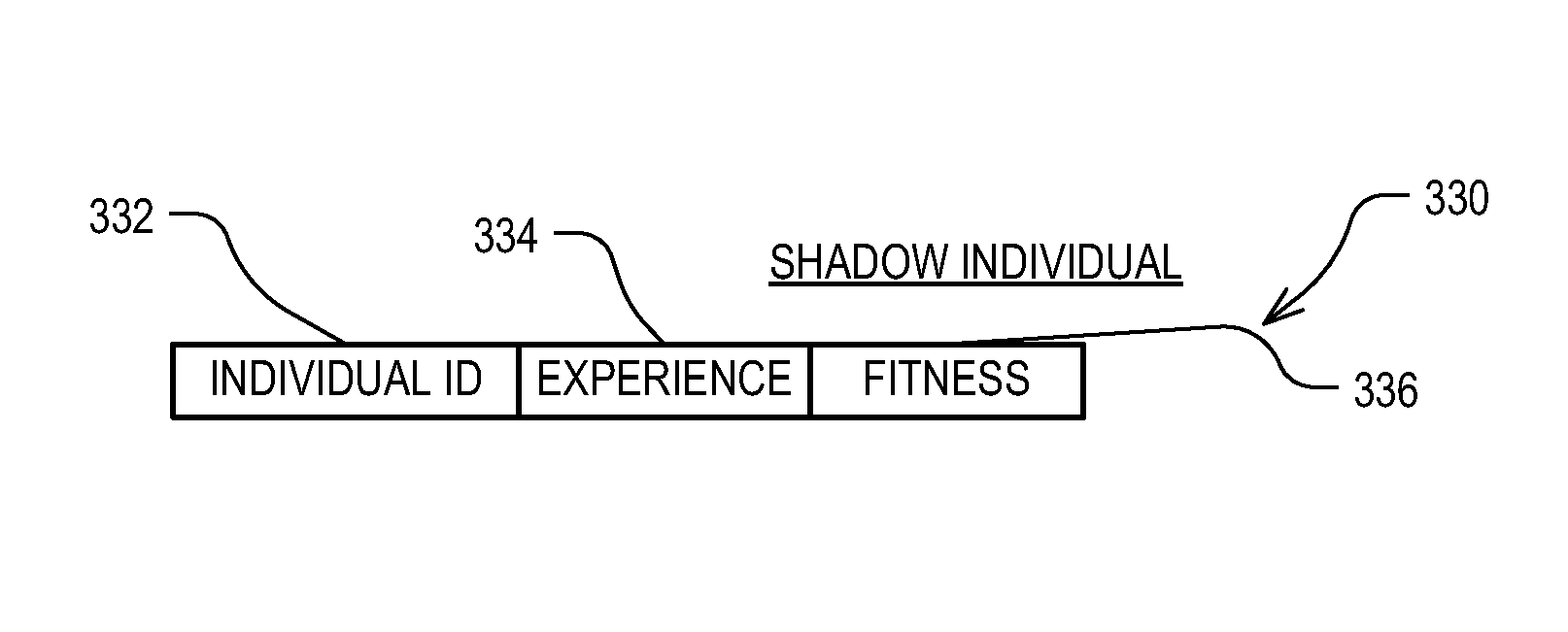
Data mining technique with shadow individuals
ActiveUS9256837B1Waste of scarceWell formedMachine learningSpecial data processing applicationsEvolutionary data miningCandidate Gene Association Study
Roughly described, a computer-implemented evolutionary data mining system includes a memory storing a candidate gene database containing active and shadow individuals; a gene pool processor which tests only active individuals on training data and updates their fitness estimates; a competition module which selects individuals (both active and shadow) for discarding from the gene pool in dependence upon both their updated fitness estimate and their testing experience level; and a gene harvesting module providing for deployment selected ones of the individuals from the gene pool. The gene database has an experience layered elitist pool, and individuals to compete only with other individuals in their same layer. Shadow individuals are created in each layer for active individuals that survive all competition with the layer before their testing experience exceeds the testing experience range for the layer.
Owner:COGNIZANT TECH SOLUTIONS U S CORP
Articles of manufacture and methods for modeling Saccharomyces cerevisiae metabolism
The invention provides an in silico model for determining a S. cerevisiae physiological function. The model includes a data structure relating a plurality of S. cerevisiae reactants to a plurality of S. cerevisiae reactions, a constraint set for the plurality of S. cerevisiae reactions, and commands for determining a distribution of flux through the reactions that is predictive of a S. cerevisiae physiological function. A model of the invention can further include a gene database containing information characterizing the associated gene or genes. The invention further provides methods for making an in silico S. cerevisiae model and methods for determining a S. cerevisiae physiological function using a model of the invention.
Owner:RGT UNIV OF CALIFORNIA
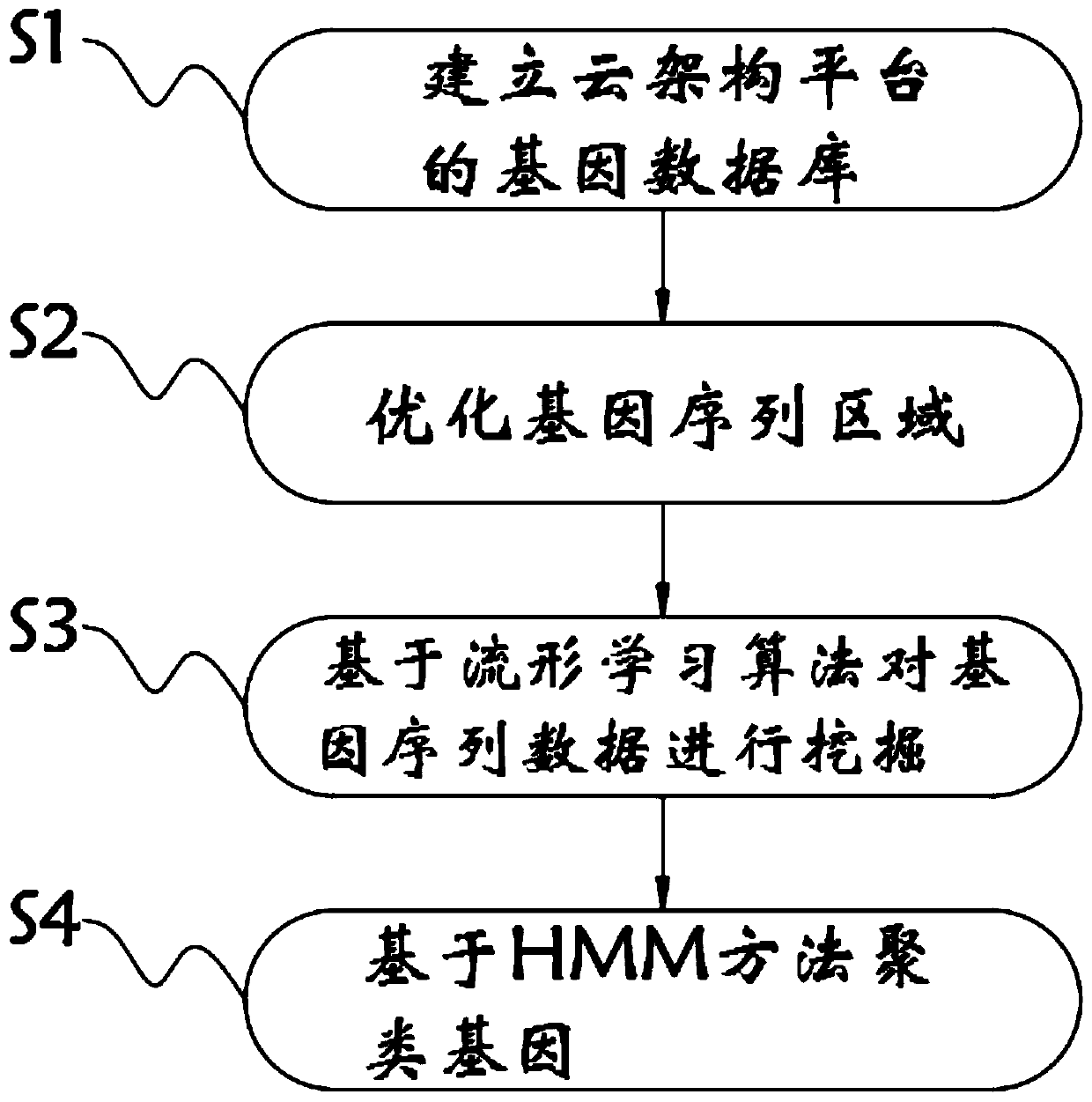
Gene data analysis method based on cloud architecture platform
InactiveCN110648723AImprove data processing efficiencyAchieve contrastBiostatisticsSequence analysisGenetic DatabasesEngineering
The invention relates to the technical field of gene data analysis, in particular to a gene data analysis method based on a cloud architecture platform. The method comprises the following steps: establishing a gene database of the cloud architecture platform, and storing gene sequence data in the gene database by taking the cloud architecture platform as a framework; optimizing the gene sequence region, and carrying out global and local optimization on the gene sequence region; and mining the gene sequence data based on a manifold learning algorithm. The gene data analysis method based on thecloud architecture platform takes the cloud architecture platform as the framework, and makes the gene sequence data stored in the gene database, so that big data which is no longer single is formed,and the data processing efficiency is improved; comparison and splicing of gene sequences are realized through a global optimization module and a local optimization module; and the gene sequence datais mined based on the manifold learning algorithm, so that important data in the sequence data is extracted conveniently.
Owner:南京江北新区创新投资基金管理有限公司
Determination of one section vagina bacillus Gardner DNA sequence (3) specificity and its application
InactiveCN1493698AFast wayAccurate methodSugar derivativesMicrobiological testing/measurementOrganismBiomedical technology
The specificity and PCR amplification primer of a viginal Gardener's bacillus DNA sequence (3) fragment are determined by searching and analyzing the data of gene database. Said sequence with 196 bp in length is obtained by PCR amplification and clone of said viginal Gardener's bacillus infected specimen. It can be used to diagnose the viginal Gardener's bacillus with high speed and correctness and low cost.
Owner:昆明寰基生物芯片开发有限公司
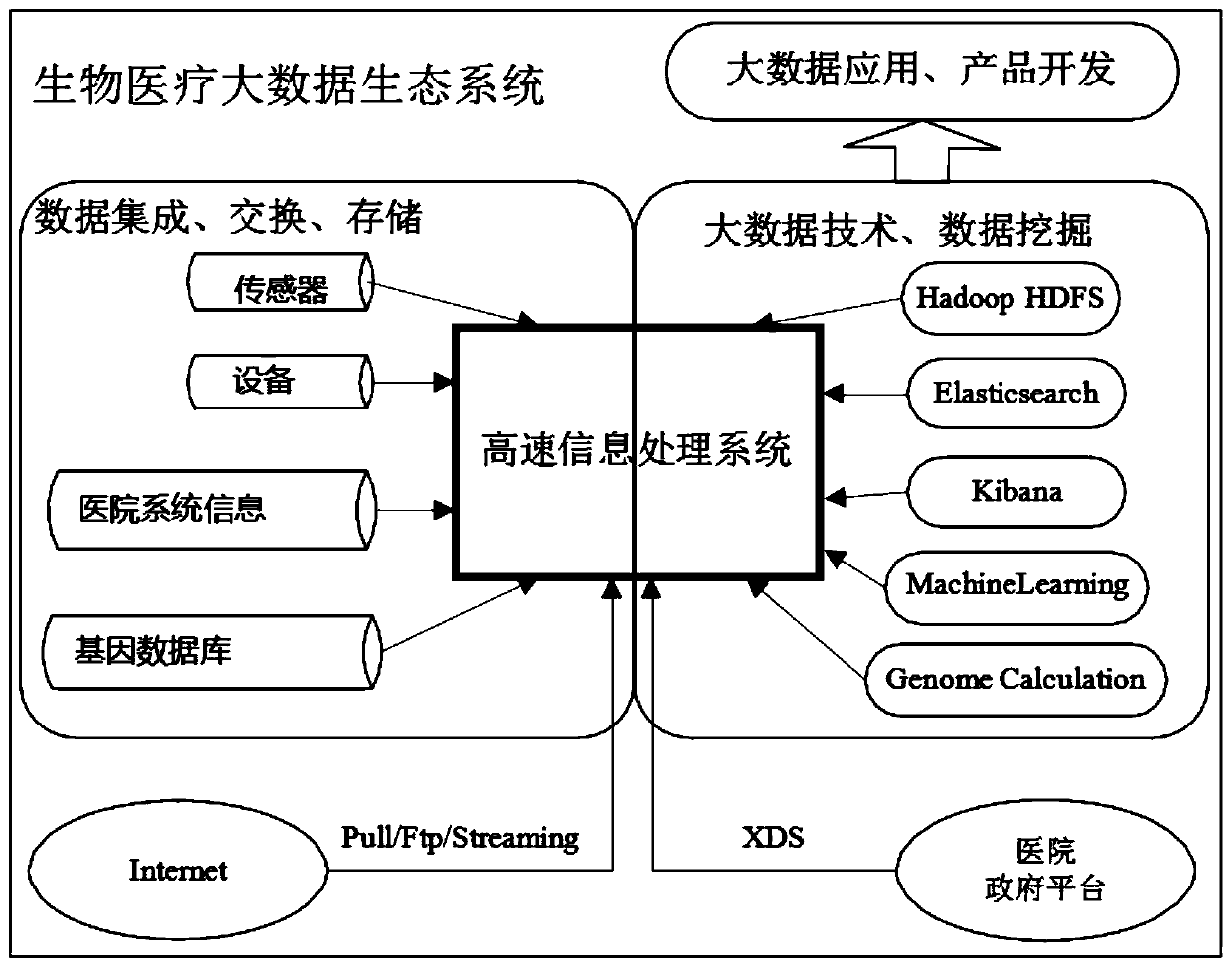
Biomedical high-speed information processing and analyzing system based on big data technology
PendingCN111324671ARealize free searchPrevent theftDatabase distribution/replicationRelational databasesInformation processingRelational database
The invention discloses a biomedical high-speed information processing and analyzing system based on big data technology, which comprises a hospital government platform, a data integration module anda big data technology module, and is characterized in that the data integration module comprises hospital system information, gene database information, sensor acquisition information and equipment acquisition information based on the hospital government platform; according to the invention, distributed storage of the relational database and the NoSQL database is realized; a multilayer data modelof the data warehouse can be constructed; according to the method, historical case data can be searched freely by utilizing the powerful function of an Elasticsearch search engine, private data and common data can be stored separately through a hierarchical data model, multiple backups of the data can be carried out, important data is prevented from being lost, and the method has perfect data warehouse and database function support capacity.
Owner:苏州工业园区洛加大先进技术研究院
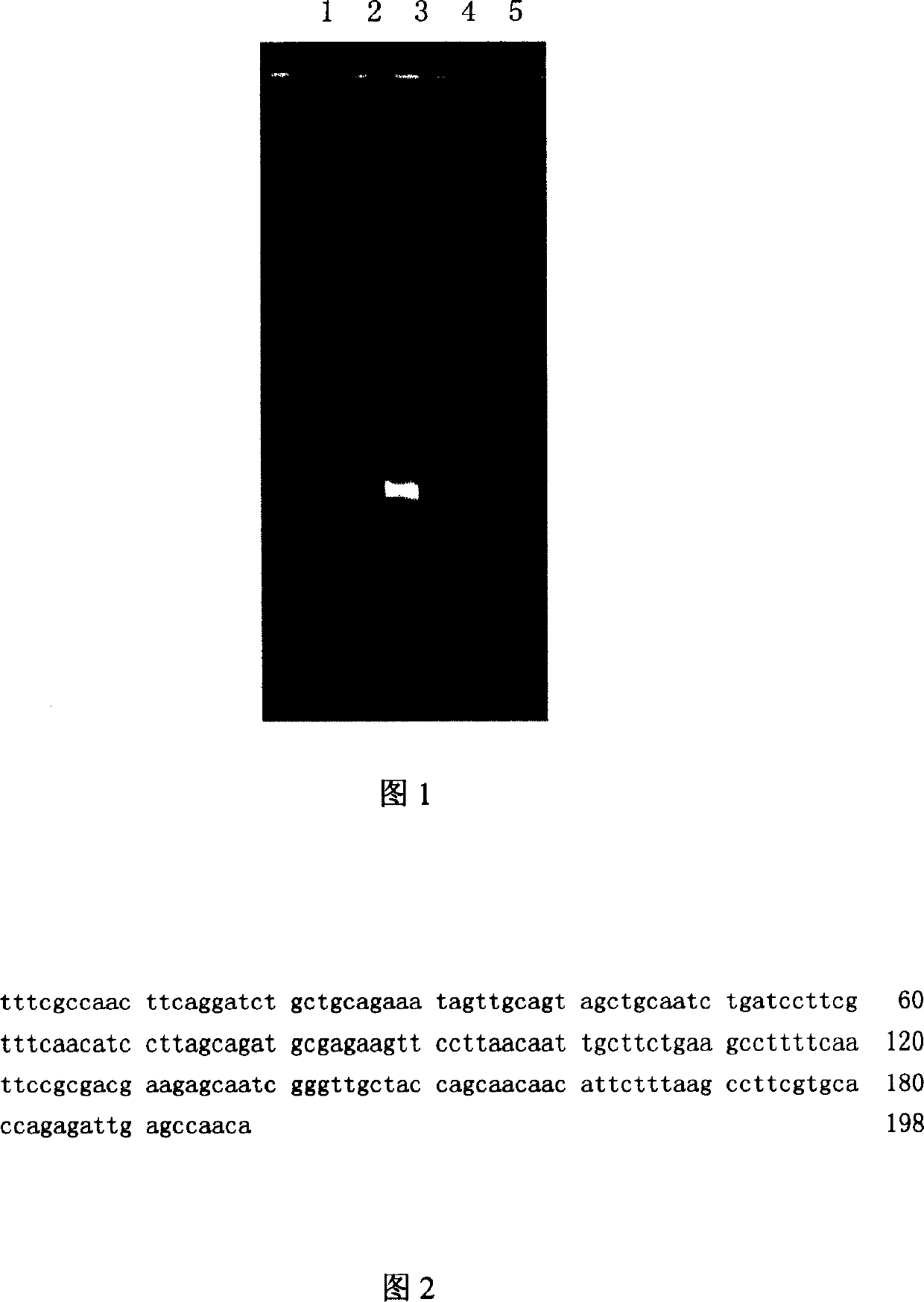
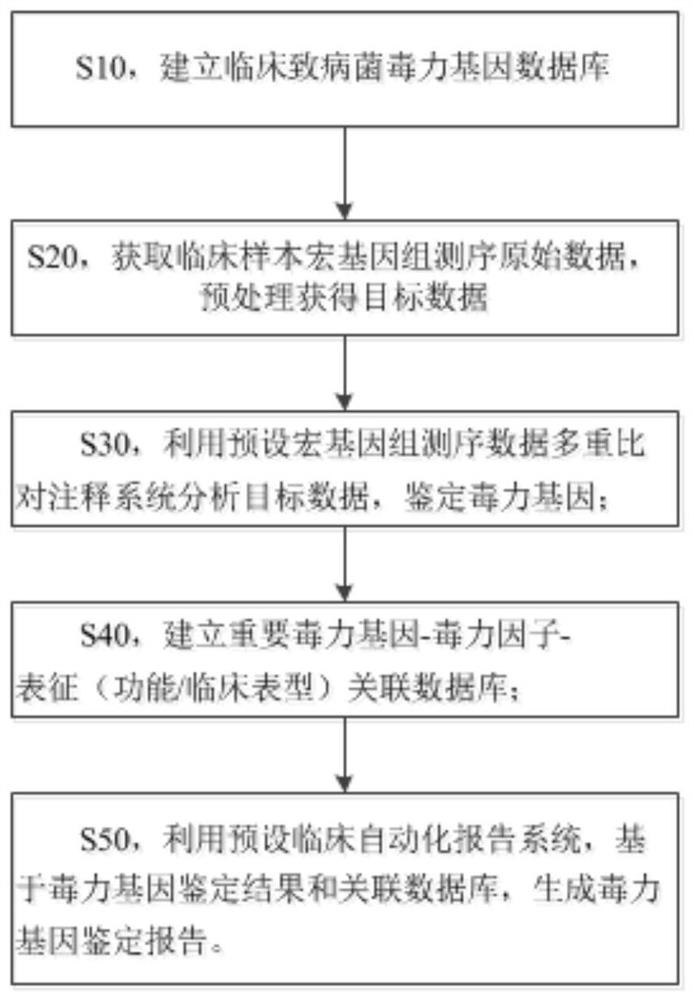
Metagenome-based clinical important pathogenic bacterium virulence gene detection method and system
ActiveCN113223618ATimely diagnosisTimely therapeuticBiostatisticsSequence analysisGenomic sequencingCerebrospinal fluid
The invention discloses a metagenome-based clinical important pathogenic bacterium virulence gene detection method and system. The method comprises the following steps: s10, establishing a clinical pathogenic bacterium virulence gene database; s20, acquiring clinical sample metagenome sequencing original data, and preprocessing the clinical sample metagenome sequencing original data to obtain target data; s30, analyzing the target data by using a preset metagenome sequencing data multiple comparison annotation system, and identifying virulence genes; s40, establishing an important virulence gene-virulence factor-characterization (function / clinical phenotype) associated database; s50, generating a virulence gene identification report based on the virulence gene identification result and the association database by using a preset clinical automatic report system. The system can perform virulence gene identification on metagenome sequencing data of clinical infection samples of different types (cerebrospinal fluid and the like), and multiple important virulence genes of multiple pathogenic bacteria in the samples are identified at a time. The system has good sensitivity and accuracy and helps doctors to perform diagnosis, treatment and prognosis in time.
Owner:HUGOBIOTECH BEIJING CO LTD +2
Mycobacterium tuberculosis multi-line drug resistance gene identification method and device
PendingCN113621716AImprove detection accuracyHelp identifyMicrobiological testing/measurementProteomicsGenomic sequencingGenetic Databases
The invention relates to a mycobacterium tuberculosis multi-line drug resistance gene identification method and device. The method comprises the following steps: acquiring a whole genome sequence obtained by metagenome sequencing of a sample or a sequence of a mycobacterium tuberculosis drug-resistant target region obtained on the basis of targeted amplification sequencing, and carrying out data quality control; comparing the sequence subjected to the data quality control to a mycobacterium tuberculosis genome, sequencing and screening out a comparison result meeting the quality requirement; carrying out local assembly and variation detection on the sequences of the gene regions where the variation sites of which the comparison positions are located in the mycobacterium tuberculosis drug resistance gene database are located; annotating a variation detection result to the mycobacterium tuberculosis drug resistance gene database according to the positions and mutation types of variation sites on the genome; and outputting a detection result of each drug resistance related mutation sites in the mycobacterium tuberculosis drug resistance gene database based on an annotation result. According to the invention, multiple drugs and corresponding drug resistance sites can be covered in one detection, and the detection precision is high.
Owner:深圳华大因源医药科技有限公司 +2
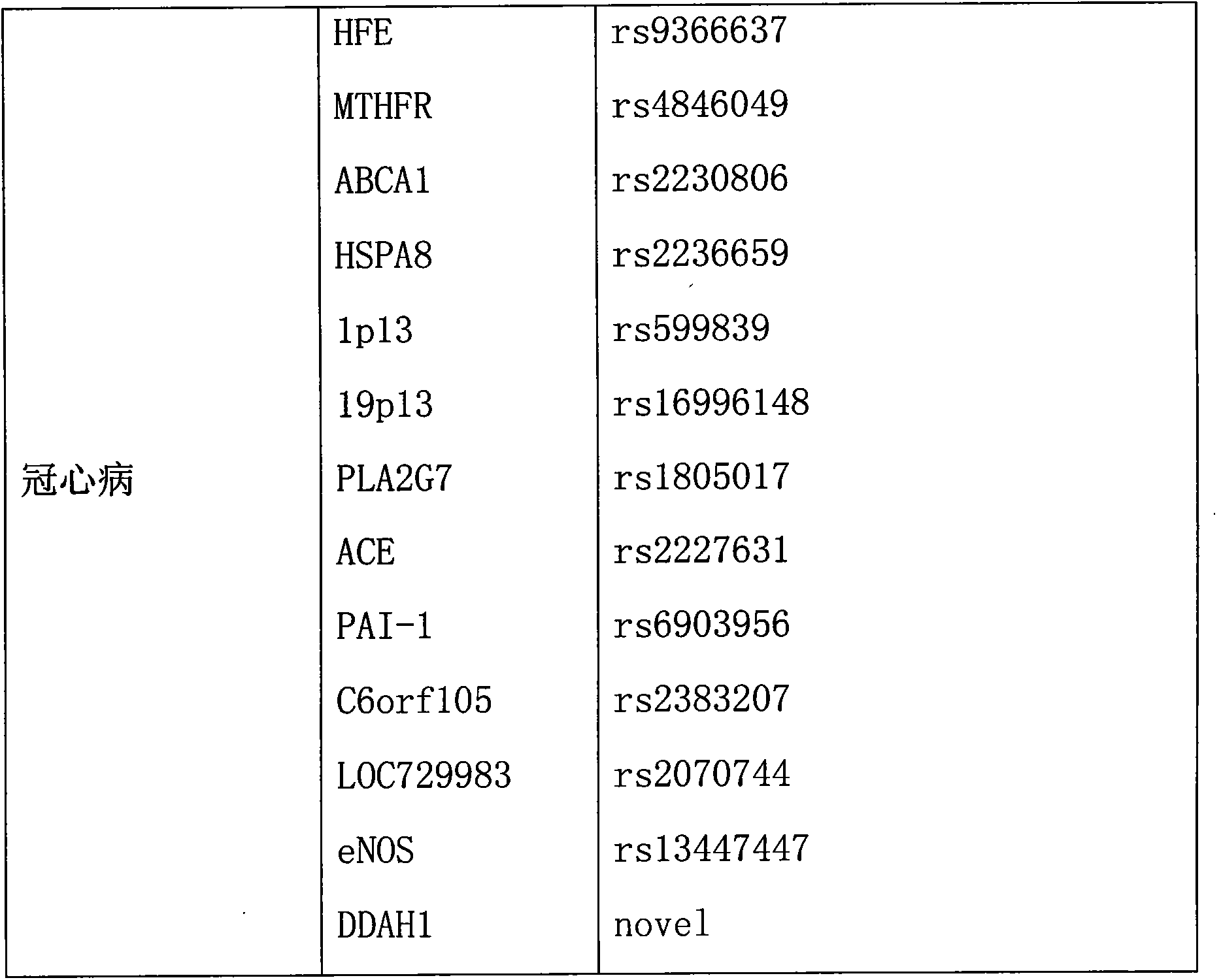
Method for acquiring characterization parameters of coronary disease
InactiveCN104050344AEfficient integrationAutomate processingSpecial data processing applicationsParameter analysisCoronary heart disease
The invention discloses a method for acquiring characterization parameters of a coronary disease. The method comprises the following steps: 1) establishing a coronary disease susceptibility gene database; 2), establishing a characterization parameter analysis platform of an individual coronary disease according to the coronary disease susceptibility gene database established in the step 1); 3) analyzing the characterization parameters of the individual coronary disease on the basis of the analysis platform established in the step 2); 4) outputting the characterization parameters of the individual coronary disease. The method disclosed by the invention is capable of accurately acquiring characterization information in time and laying a foundation for sequential treatment work.
Owner:西安时代基因健康科技股份有限公司
Curated genetic database for in silico testing, licensing and payment
Disclosed are systems and methods for managing and using a curated genetic biomarker database. The systems and methods allow for submission and curation of genetic biomarkers, the development of new genetic tests based on the submitted biomarkers, conducting the genetic tests, and accounting for payments due to biomarker submitters and test developers for use of the genetic tests.
Owner:YOUGENE
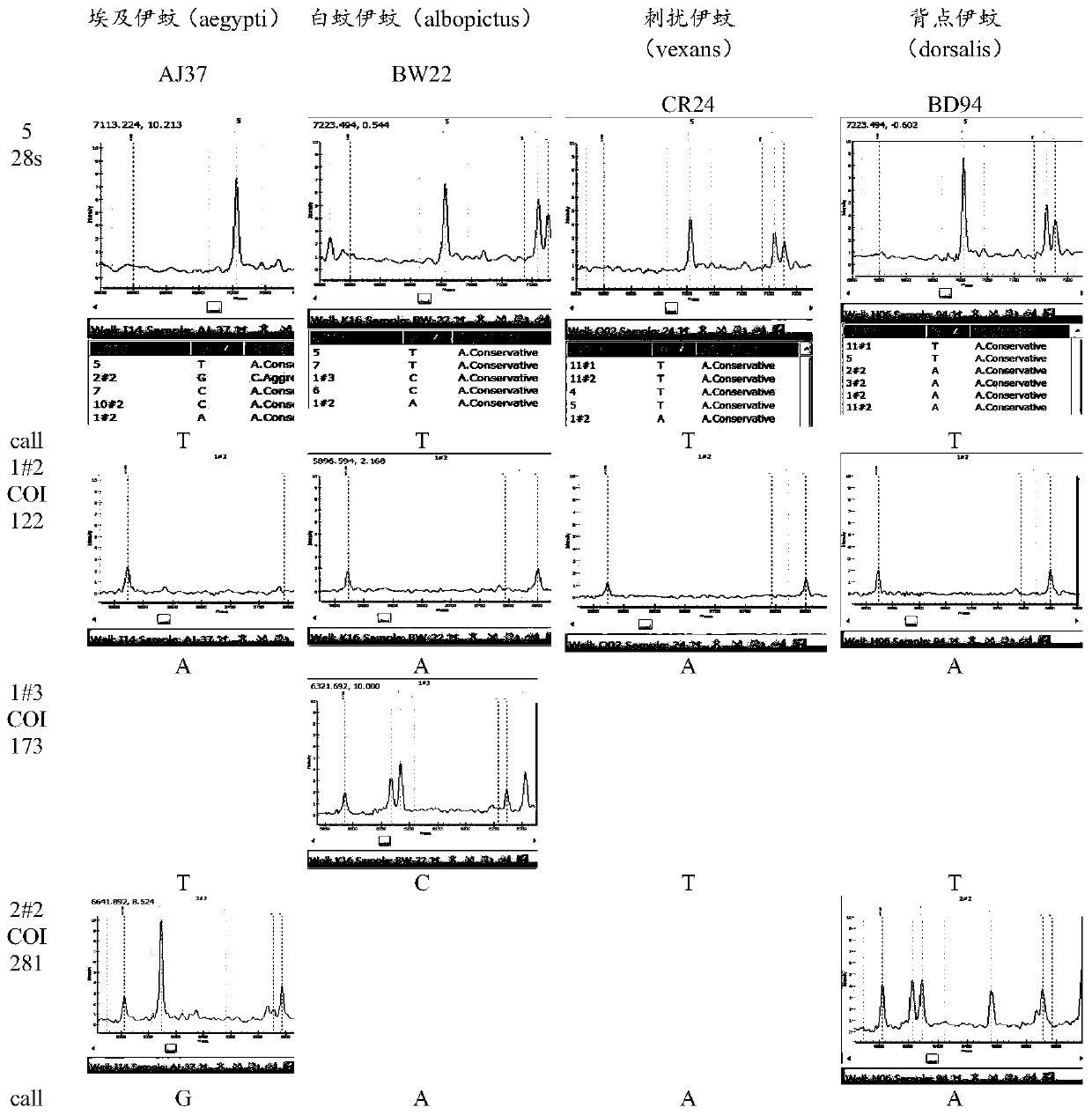
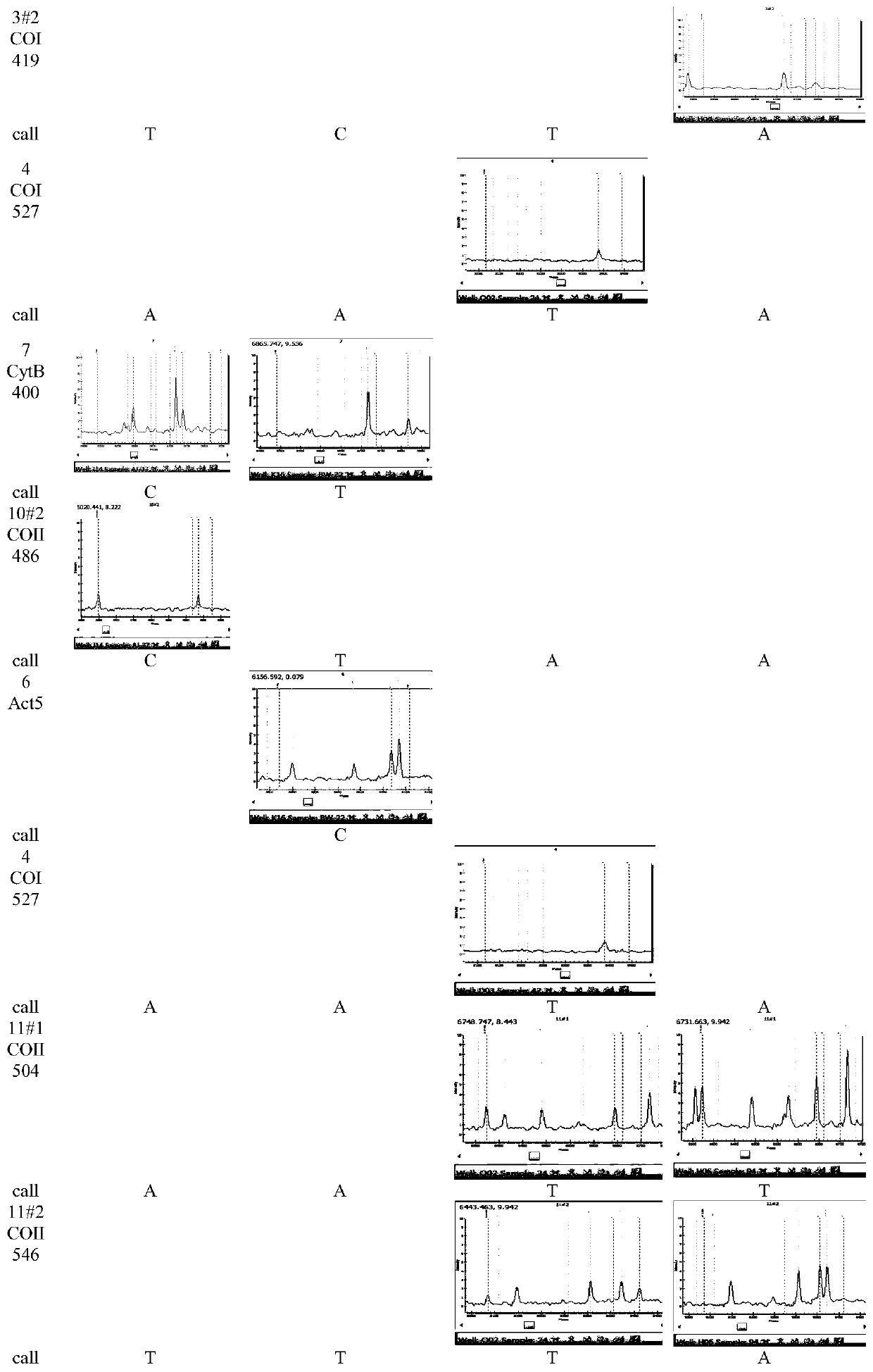
Method for identifying varieties of aedes based on PCR-MS (polymerase chain reaction-mass spectrum) scanning
The invention discloses a method for identifying varieties of aedes based on PCR-MS (polymerase chain reaction-mass spectrum) scanning. The method comprises the following steps: constructing an aedesvariety identification gene database (COI, COII, 28s, CytB400); constructing an aedes variety identification DNA (deoxyribonucleic acid) two-dimensional code, designing multiple PCR-MS primers, and performing multiple PCR-MS scanning; and performing analysis on thousands of gene sequences of nine aedes varieties, so as to obtain specific SNP (single nucleotide polymorphism) loci of each aedes variety. According to the loci, specific SNP loci of two or more genes of each variety are selected, meanwhile, specific and non-specific loci of other varieties are referred and treated as an exclusive method, analysis software is designed to recognize basic groups of multiple loci, and varieties of aedes can be confirmed, so that the accuracy in identifying aedes varieties can be improved, and detection omission can be avoided.
Owner:CHINESE ACAD OF INSPECTION & QUARANTINE
Genetic database system and method
InactiveUS20140236607A1High riskPayment schemes/modelsPathological referencesEpigenomeGenetic database
A system and method for performing an analysis of a subject's genome, transcriptome and / or epigenome is described. The identities of a predetermined number of, such as ten or more, characteristics in the subject's genome, transcriptome and / or epigenome which are associated with one or more particular phenotype(s) or an increased risk of a particular phenotype are determined by performing an analytical technique upon a biological sample obtained from the subject. The amounts of any royalties or fees required for determination of the identities of one or more of the ten or more characteristics in the subject's genome, transcriptome and / or epigenome are determined via a computer. The amount of a fee charged for a determination of the identities of the ten or more characteristics in the subject's genome, transcriptome and / or epigenome are adjusted via a computer to reflect the amount of the determined royalties or fees and / or the amount of a fee for a future treatment of the subject. A database accessed during the method is also described.
Owner:ALEXANDRE LAURENT
Universal primer for identifying pathogenic species of potato scabs and detection method
ActiveCN106957923AAvoiding the Drawbacks of Amplifying False PositivesHigh resolutionMicrobiological testing/measurementMicroorganism based processesFragment sizeImage resolution
The invention provides a universal primer for identifying pathogenic species of potato scabs and a detection method, which relates to a detection primer and a PCR (polymerase chain reaction) amplification method. The invention aims at solving the problems of low identification resolution ratio, low identification efficiency and poor identification accuracy of the pathogenic species of the potato scabs. The sequences of the primer are gyrBF2:5'-TCTGCACGGYGTSGGYGTCTC-3' and gyrBR2:5'-TTGCGGGAGTTGAGGTAC-3'. The detection method comprises the following steps: extracting genomic DNA of a bacterial strain, performing PCR amplification, sequencing products with fragment size of 400bp, comparing a sequencing result and a gene database and finally determining the pathogenic spices. The universal primer has comparatively high identification resolution ratio between spices, high identification efficiency and high repeatability. A detected sample can be pure bacterial strain DNA, and potato tuber tissue DNA after being infected by a single pathogenic bacterium serves as a template for PCR amplification and sequencing.
Owner:黑龙江省农业科学院植物脱毒苗木研究所
Data structures and methods for modeling Saccharomyces cerevisiae metabolism
The invention provides an in silico model for determining a S. cerevisiae physiological function. The model includes a data structure relating a plurality of S. cerevisiae reactants to a plurality of S. cerevisiae reactions, a constraint set for the plurality of S. cerevisiae reactions, and commands for determining a distribution of flux through the reactions that is predictive of a S. cerevisiae physiological function. A model of the invention can further include a gene database containing information characterizing the associated gene or genes. The invention further provides methods for making an in silico S. cerevisiae model and methods for determining a S. cerevisiae physiological function using a model of the invention.
Owner:RGT UNIV OF CALIFORNIA
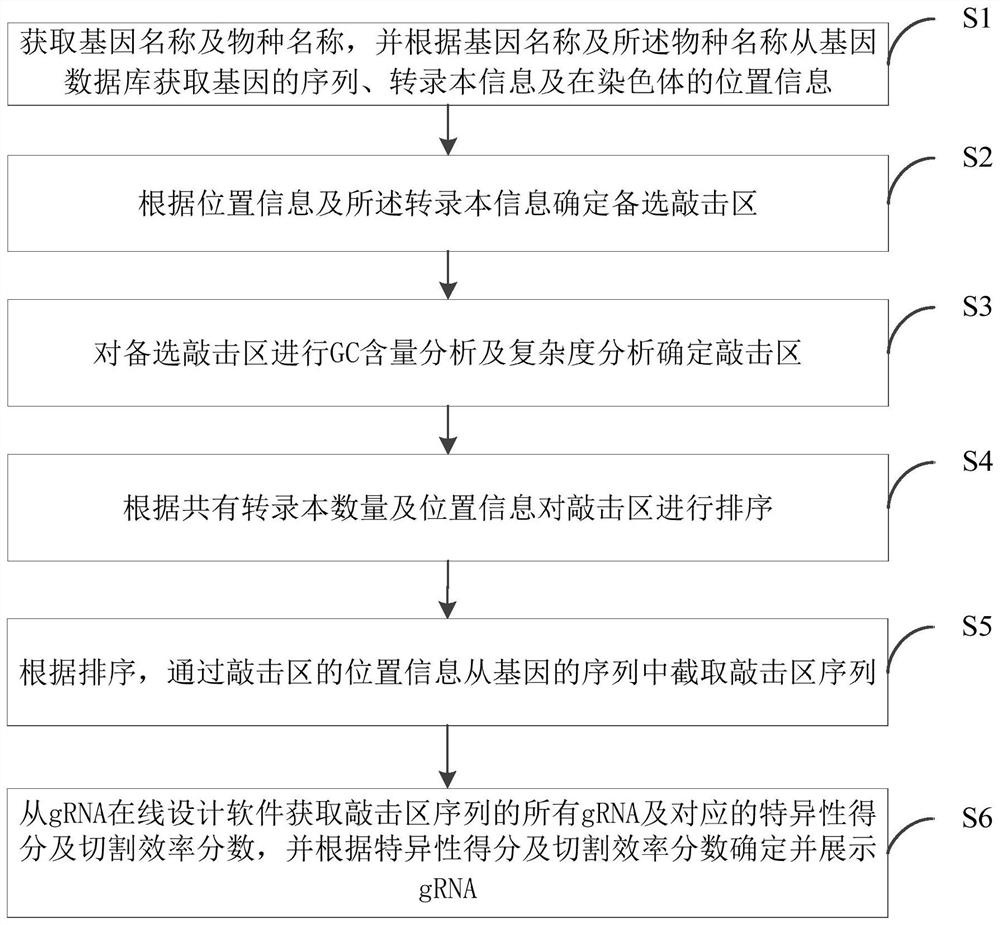
Automatic screening method, system and device for gene editing sites and storage medium
PendingCN112614541AStrong specificityImprove featuresProteomicsGenomicsGenome editingGenetic Databases
The invention discloses an automatic screening method, system and device for gene editing sites and a storage medium. The automatic screening method comprises the steps: obtaining a gene name and a species name, and obtaining a gene sequence, transcript information and chromosome position information from a gene database; determining an alternative knocking area according to the position information and the transcript information; carrying out GC content and complexity analysis on the alternative knocking areas to determine knocking areas; sequencing the knock areas according to the quantity and the position information of the common transcripts; intercepting a knock region sequence from the gene sequence through the position information of the knock region according to the sequence; and obtaining all gRNAs of the knocking region sequence and corresponding specific scores and cutting efficiency scores from gRNA online design software, and determining and displaying editing sites of the gRNAs according to the specific scores and the cutting efficiency scores. According to the embodiment of the invention, the gene editing sites with good knockout effect, strong specificity and high cutting efficiency can be automatically and efficiently screened, and the method can be widely applied to the technical field of gene editing.
Owner:广州源井生物科技有限公司
Construction method and system of microbial gene database
ActiveCN114121167AQuick searchQualitatively accurateBioinformaticsInstrumentsGenetic DatabasesGenomic data
Owner:SHENZHEN AIMIGENE TECH CO LTD
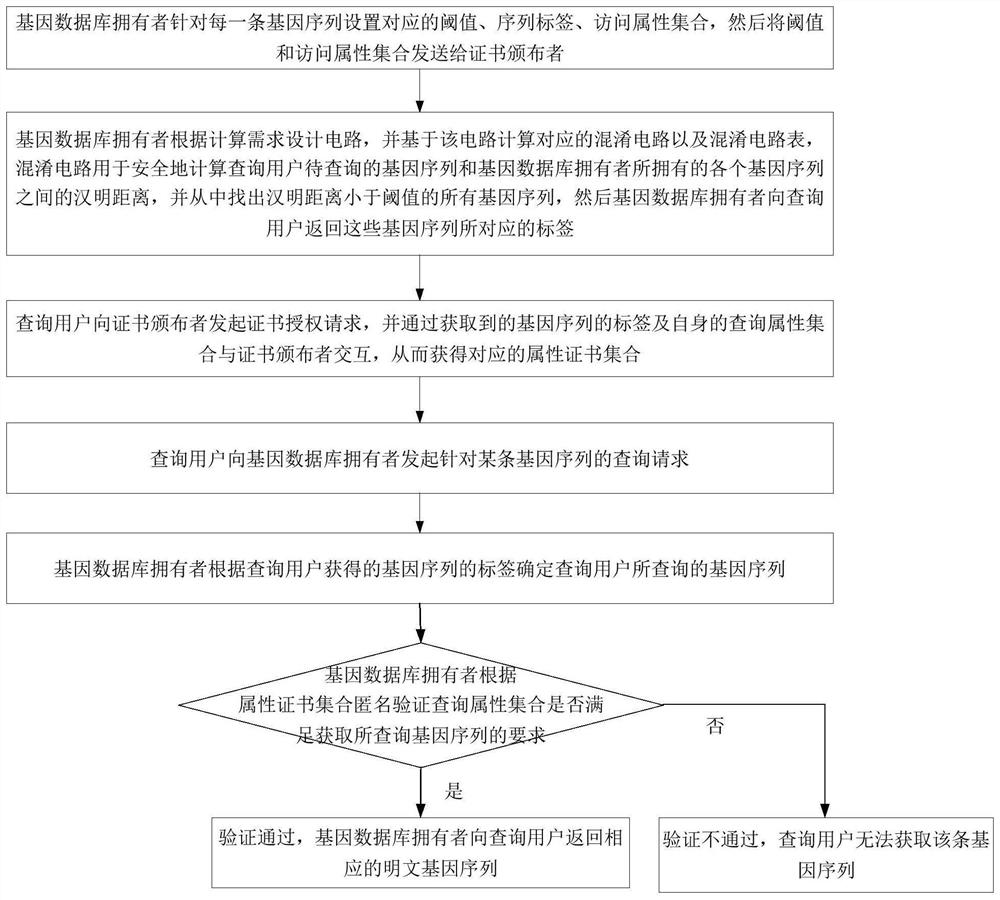
Gene sequence security comparison method and system supporting multi-attribute anonymous authentication
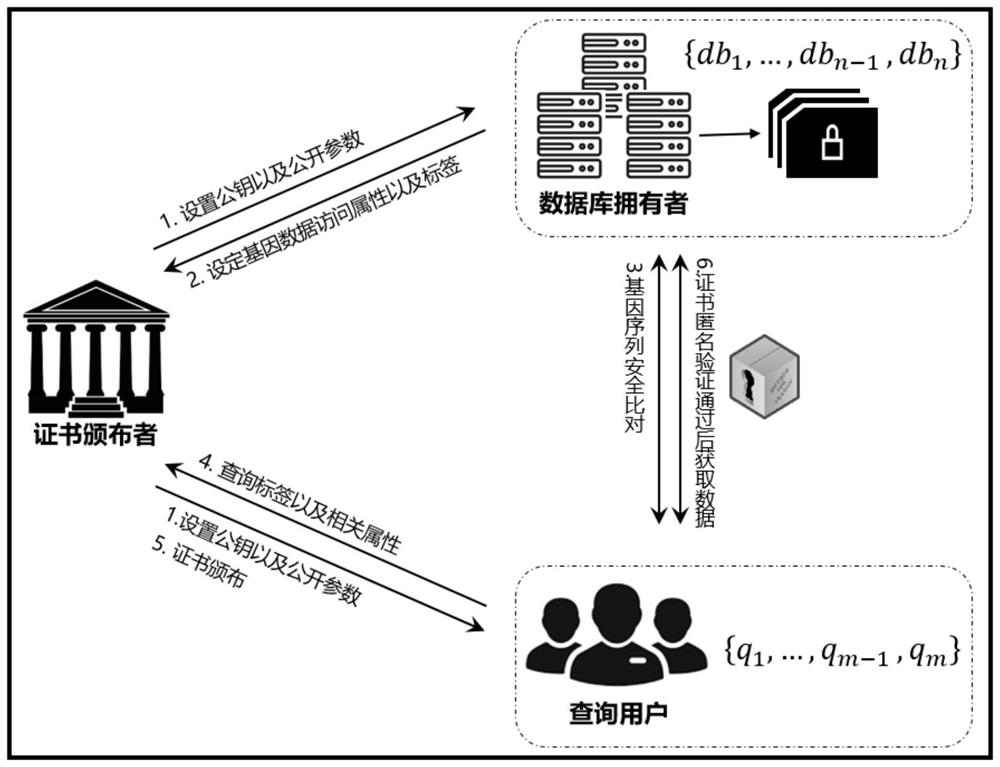
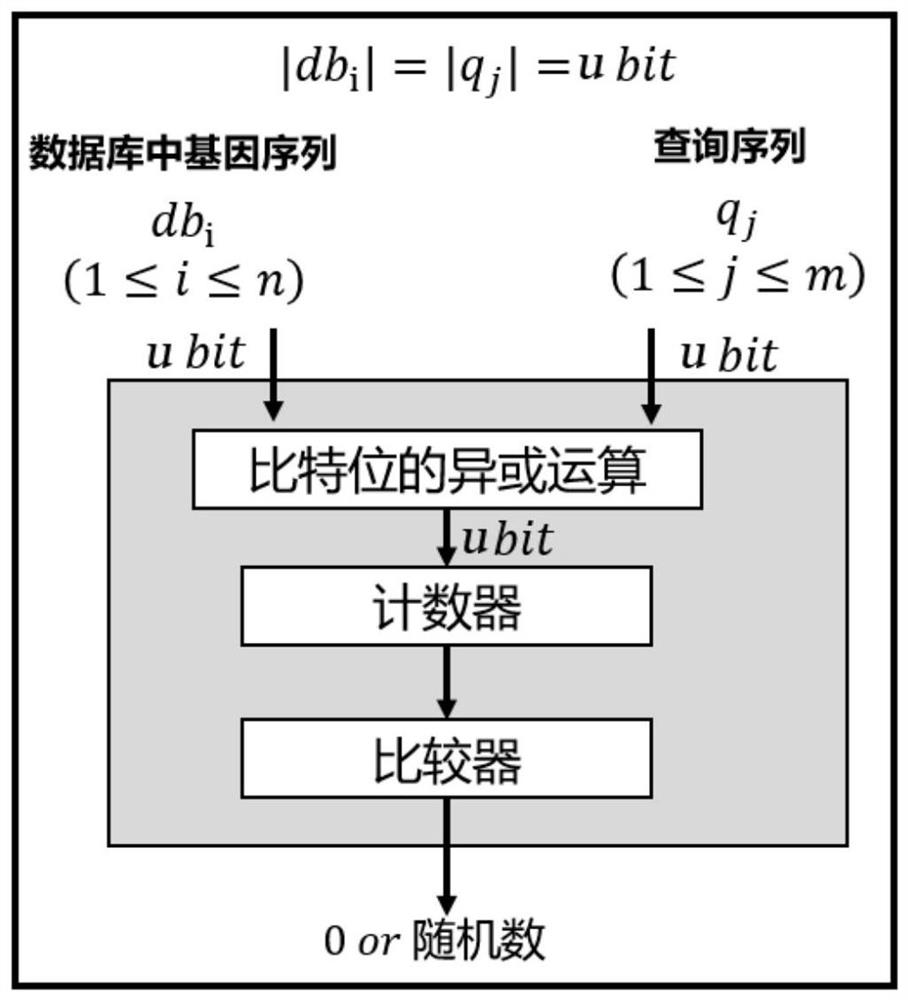
ActiveCN112614545AGuaranteed confidentialityIntegrity guaranteedDigital data protectionSequence analysisGenetic DatabasesAnonymous authentication
The invention discloses a gene sequence security comparison method and system supporting multi-attribute anonymous authentication; the method comprises the steps: firstly enabling a gene database owner to transmit a threshold value and an access attribute set of a gene sequence to a certificate issuer, and then designing a confusion circuit according to a calculation demand, so as to find out a gene sequence which can be queried by a query user from the confusion circuit; returning labels corresponding to the gene sequences to the query user; initiating, by a query user, a certificate authorization request to a certificate issuer and interacts with the certificate issuer, and after an attribute certificate set is obtained, initiating the query request for a certain gene sequence to a gene database owner; anonymously verifying, by the gene database owner, the attribute certificate of the query user, determining whether the query attribute set of the query user meets the acquisition requirement or not, and returning a corresponding plaintext gene sequence to the query user under the condition that the query attribute set meets the acquisition requirement. According to the invention, the security, privacy, high efficiency and controllability of data query can be realized.
Owner:JINAN UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com