Class I sequence based typing of HLA-A, -B, and -C alleles by direct DNA sequencing
a technology of hla-a, -b, and -c alleles, applied in the field of direct dna sequencing of hlaa,b, andc alleles, can solve the problems of high level of allelic diversity, mhc polymorphism exerting immunological burden on the transplanted host, and a formidable task of hla class i typing at the nucleic acid level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example i
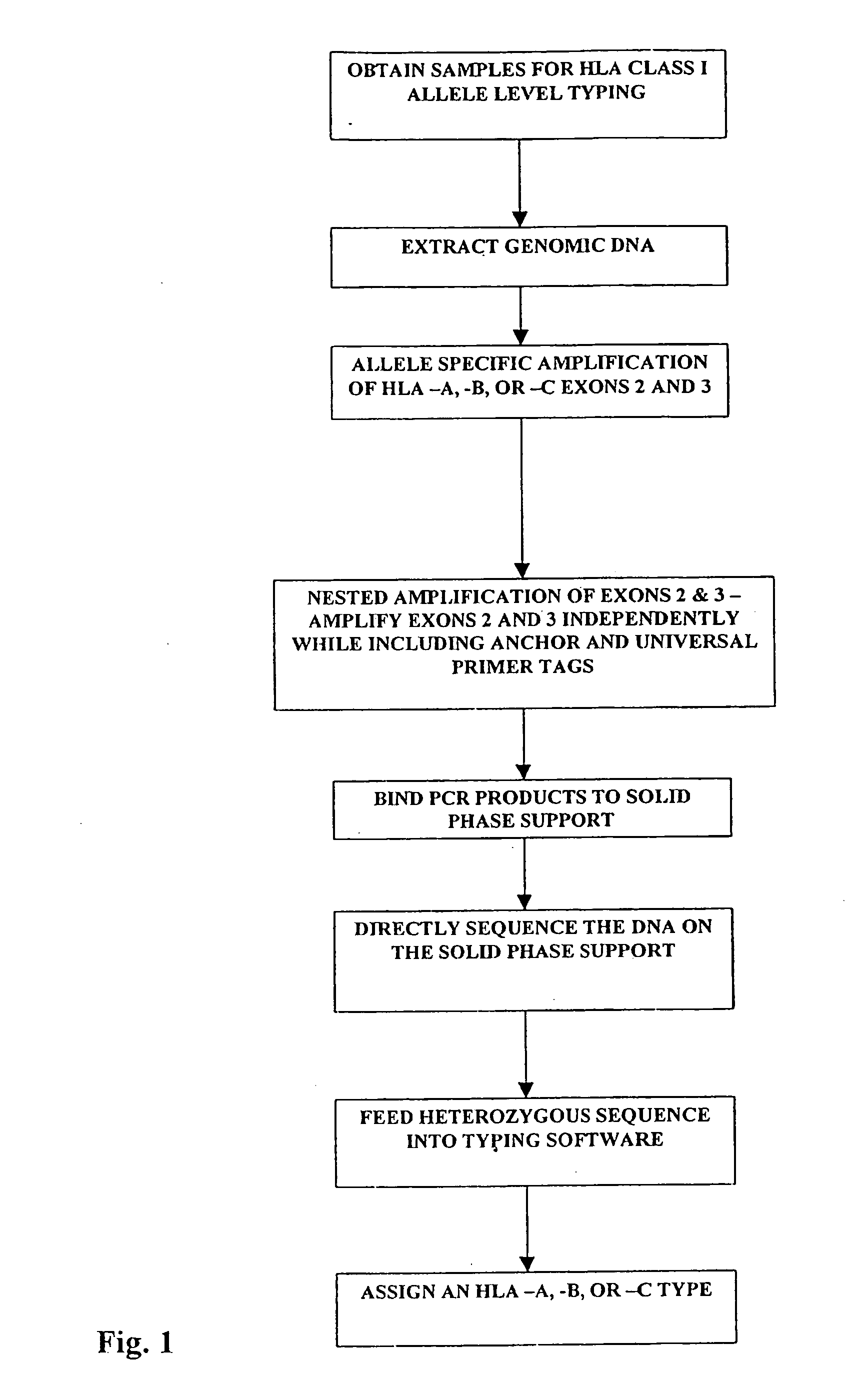
[0036] Once a sample is obtained, the first step is to treat the tissue sample so as to obtain nucleic acids for amplification. The method herein described can be performed on whole blood, tumor cells, sperm, hair follicles, or any other nucleated tissue sample.
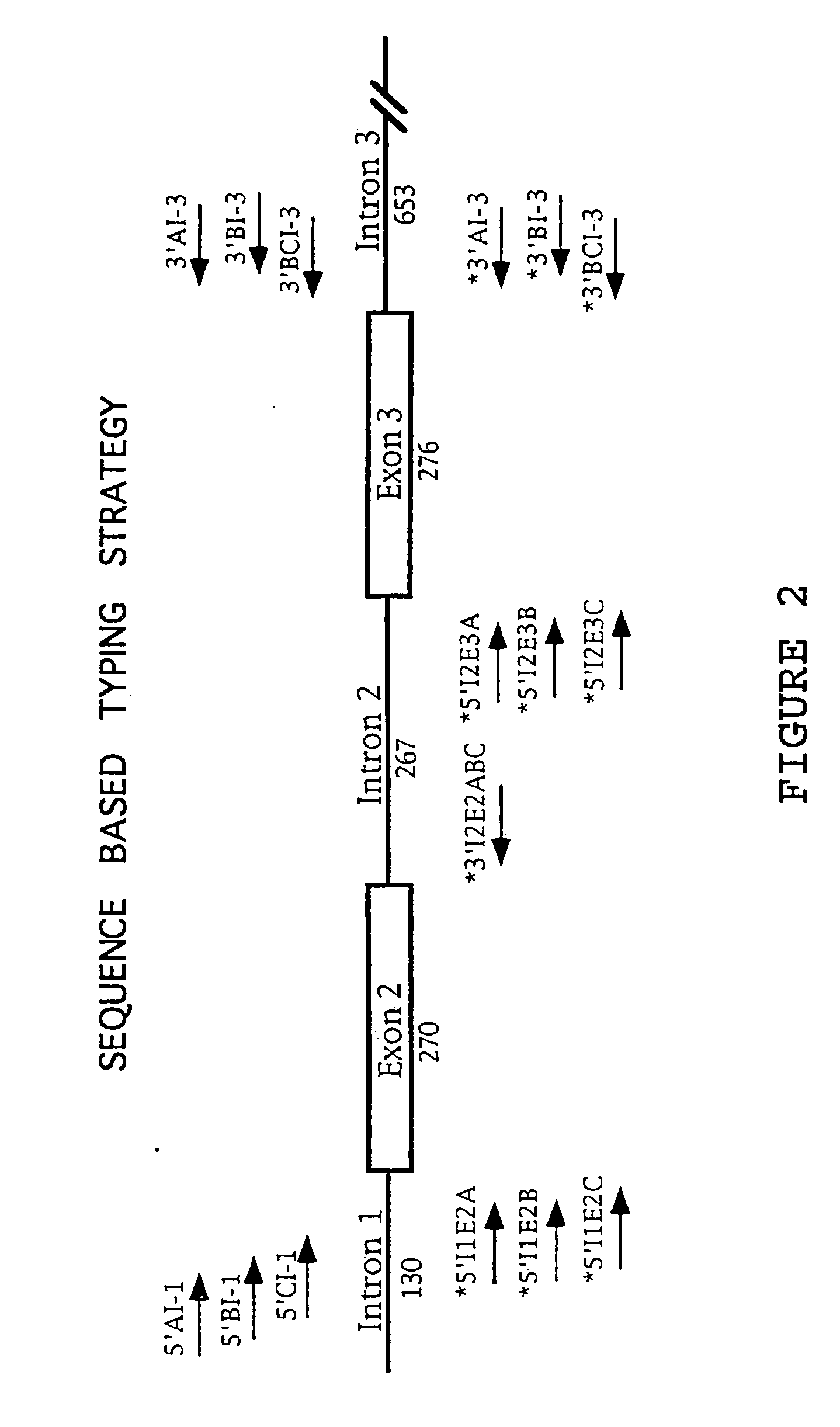
[0037] A. Locus Specific Amplification of HLA-A, -B, -C Alleles and of Exons 2 and 3
[0038] Genomic DNA was extracted from 200 μL of whole blood using the Qiagen DNA extraction kit otherwise known as a “QIAamp blood kit” according to the supplied protocol. Exons 2 and 3 of HLA class I-A, -B, and -C loci were PCR amplified from 500 ng of genomic DNA using 20 picomoles of HLA class I locus specific primers located within introns 1 and 3 (Table 1).
[0039] The primers listed in Table 1 correspond to the bases which are the same across the introns and are indicated as a single base (A, C, G, T), while bases which are variable across the introns are indicated by a code for alternative bases. In general, it will be advantageous to ...
example 2
[0076] We first obtained cells for category two typing. The cells we received required HLA-A, -B, and -C nucleotide sequence analysis. After thawing and resuspending the cells in RPMI 1640 supplemented with 15% fetal calf serum, 10 U / ml penicillin, 10 mg / ml streptomycin, and 2 mM L-glutamine, the cells were counted and cell viability was determined using trypan exclusion. Each sample contained approximately 5×106 total cells, however, viability varied significantly between the three samples. A summary of the cells received and their work-up is shown in Table 3.
TABLE 3# of CellsAllelesPre-Test Sample #CategoryReceivedViabilitySequenced0121-3981-2HLA-A5 × 106N.D.0121-2817-9HLA-B5 × 106N.D.0121-5337-5HLA-C5 × 106>90%Cw*0802,*020220104-8698-32a; HLA-A5 × 106>90%N.D.0104-9545-52b; HLA-B5 × 106>90%B*1510,*5001
[0077] As can be seen from Table 3, samples 3981-2 and 2817-9 were not viable.
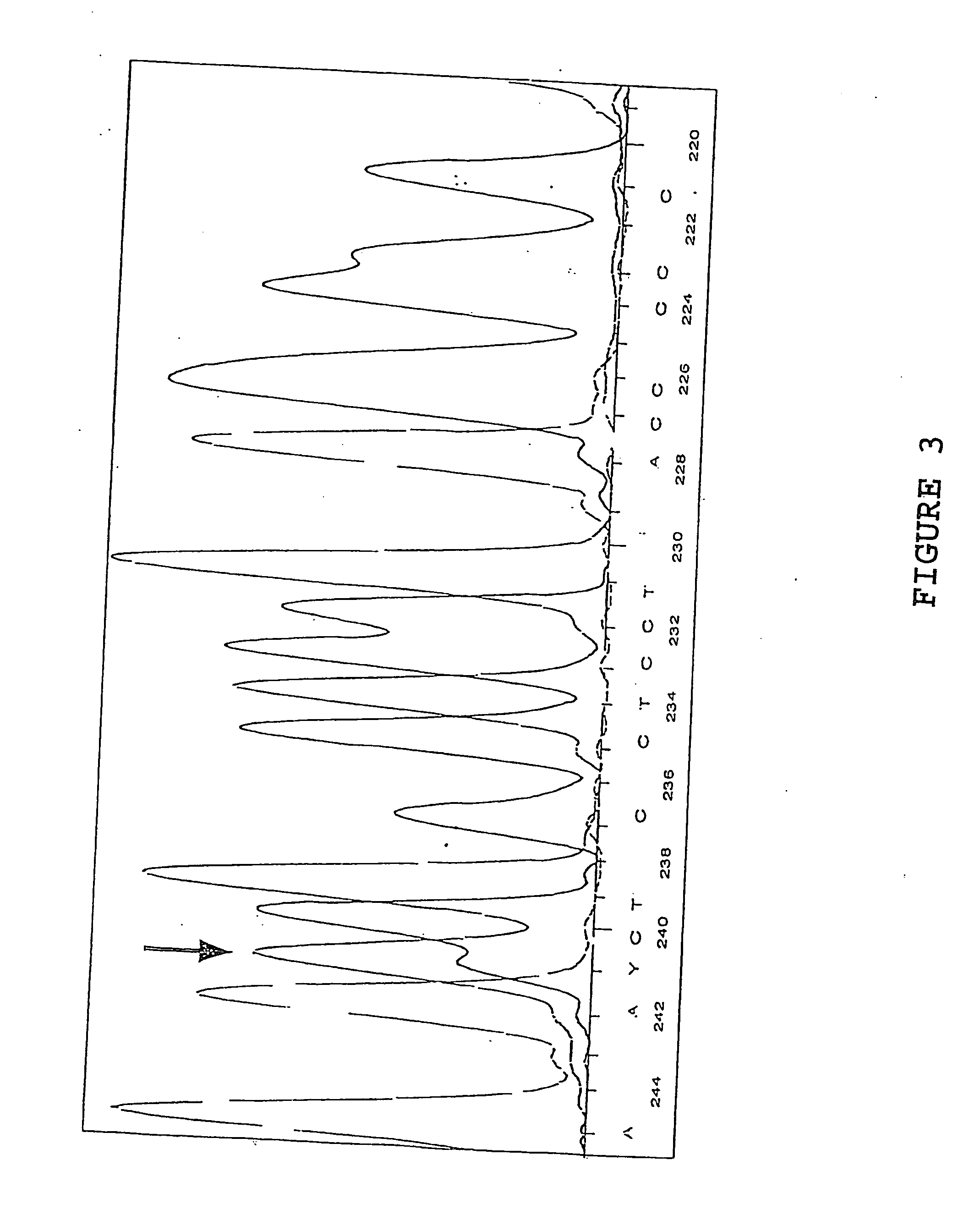
[0078] Sample 0121-5337-5 was sequenced according to the method hereinabove described and it was dete...
example 3
[0079] Samples 1548 through 1579 (as shown in Table 4) were received as frozen transformed lymphocytes excepting samples 1551, 1552 and 1553 which were delivered as granulocytes. Once these samples were assigned their respective run numbers processing began. DNA extraction using the QIAMP 96 Spin Blood kit was carried out yielding sufficiently good quality DNA at a suitable concentration for primary PCR amplification. 25 μl primary PCR reactions were set up and were checked by loading 5 μl of the amplicon on a 2% agarose gel run at 200 volts for twenty minutes.
[0080] Once it was established that the primary PCR had produced a suitable product this was then diluted 1:200 with distilled water ready for the secondary PCR reactions. Each dilution was then used in the four secondary PCR reactions with the resulting amplicons again being checked on a 2% agarose gel. The secondary PCR products then underwent solid phase sequencing reactions using the Amersham Pharmacia Autoload SPS kit an...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Volume | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com