Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
157 results about "Actin beta" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Beta-actin (human gene and protein symbol ACTB/ACTB) is one of six different actin isoforms which have been identified in humans. This is one of the two nonmuscle cytoskeletal actins. Actins are highly conserved proteins that are involved in cell motility, structure and integrity.
Kit for quickly detecting polymorphism of human CYP2C19 gene and using method of kit
ActiveCN104212904AAvoid the disadvantages of difficult judgment of resultsNo human errorMicrobiological testing/measurementFluorescenceActin beta
The invention belongs to the gene detecting technology in clinical detecting technologies of the bio-medical field and particularly relates to a kit for quickly detecting polymorphism of a human CYP2C19 gene and a using method of the kit. The kit comprises three pairs of primers, three probes, six PNA (pentose nucleic acid) sequences, a pair of beta-Actin interior label primers and an interior label probe, and further comprises a PCR (polymerase chain reaction) buffer solution of Mg<2+>, a dNTP mixture, Taq enzyme and ddH2O. The kit can be utilized to carry out qualitative detection on a polymorphic site of the CYP2C19 gene and carry out two-tube PNA-PCR fluorescent amplified reaction according to the SNP site and can carry out result judgment according to a condition whether a two-tube fluorescence curve is started or not without producing personal errors, so that a false positive rate and a false negative rate are low. And moreover, the kit can be used to realize high-flux detection easily and is capable of clinical use.
Owner:WUHAN HEALTHCHART BIOLOGICAL TECH
Fluorescent quantitative PCR kit for detecting alpha-globin gene deletion
InactiveCN102146476AImprove featuresGood repeatabilityMicrobiological testing/measurementFluorescence/phosphorescenceFluorescenceThalassemia
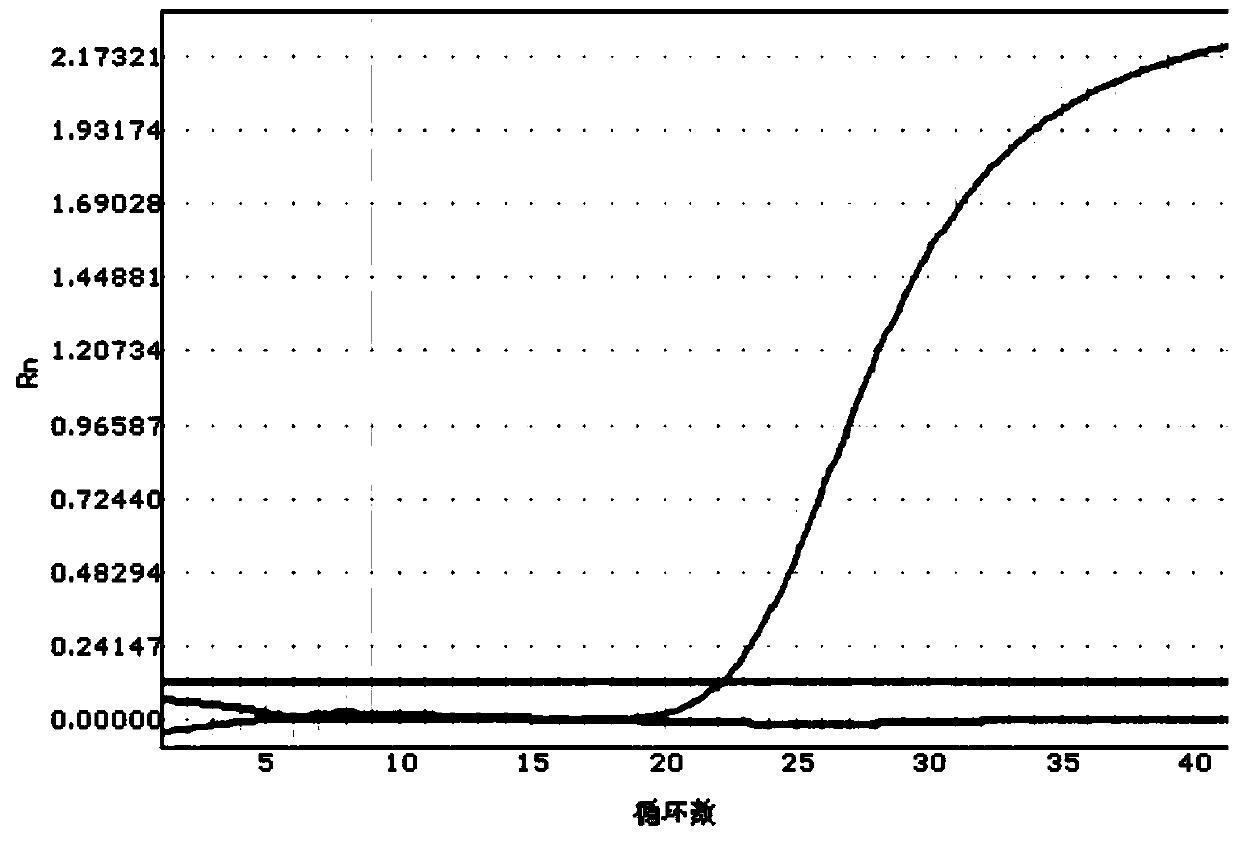
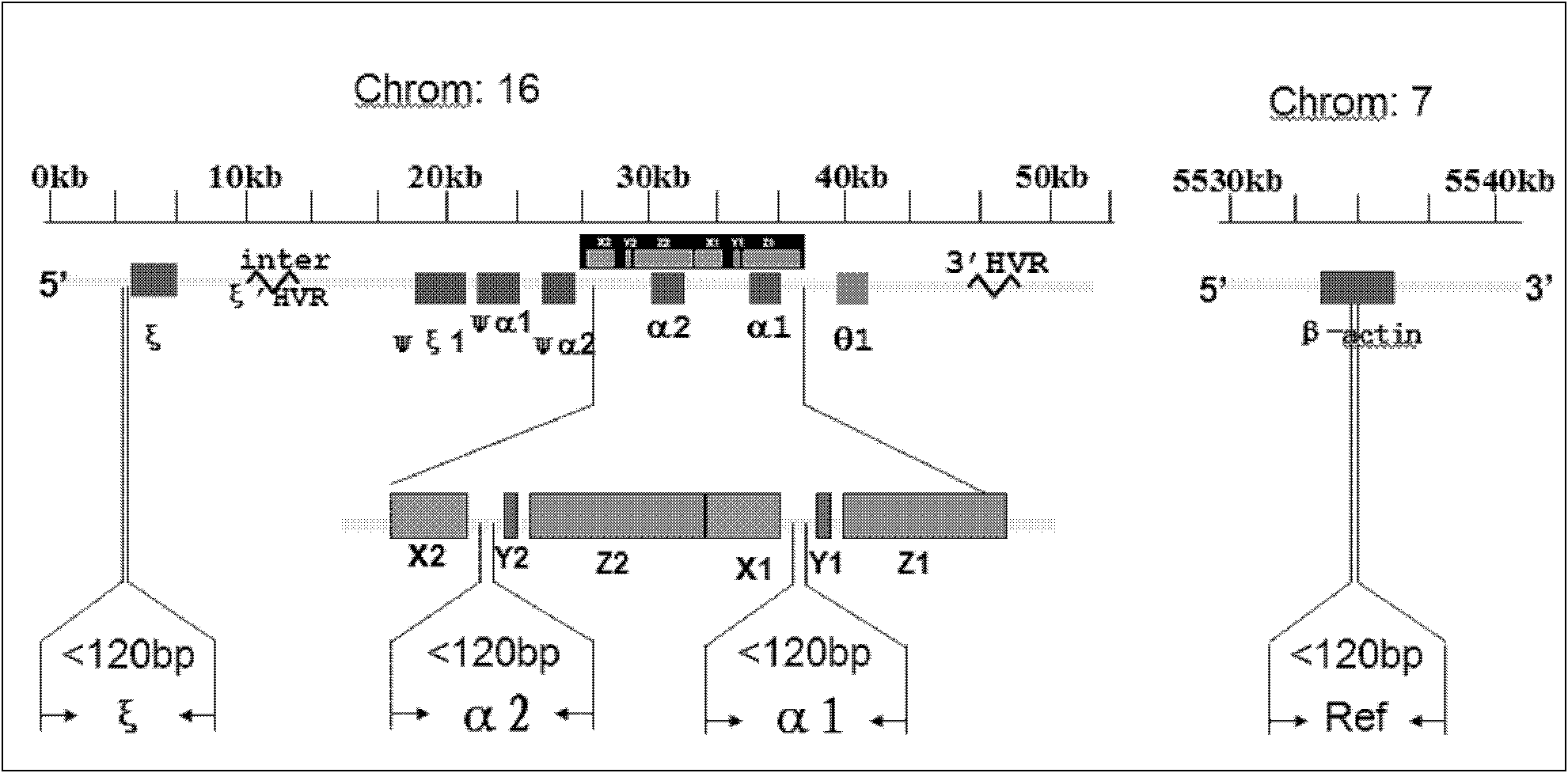
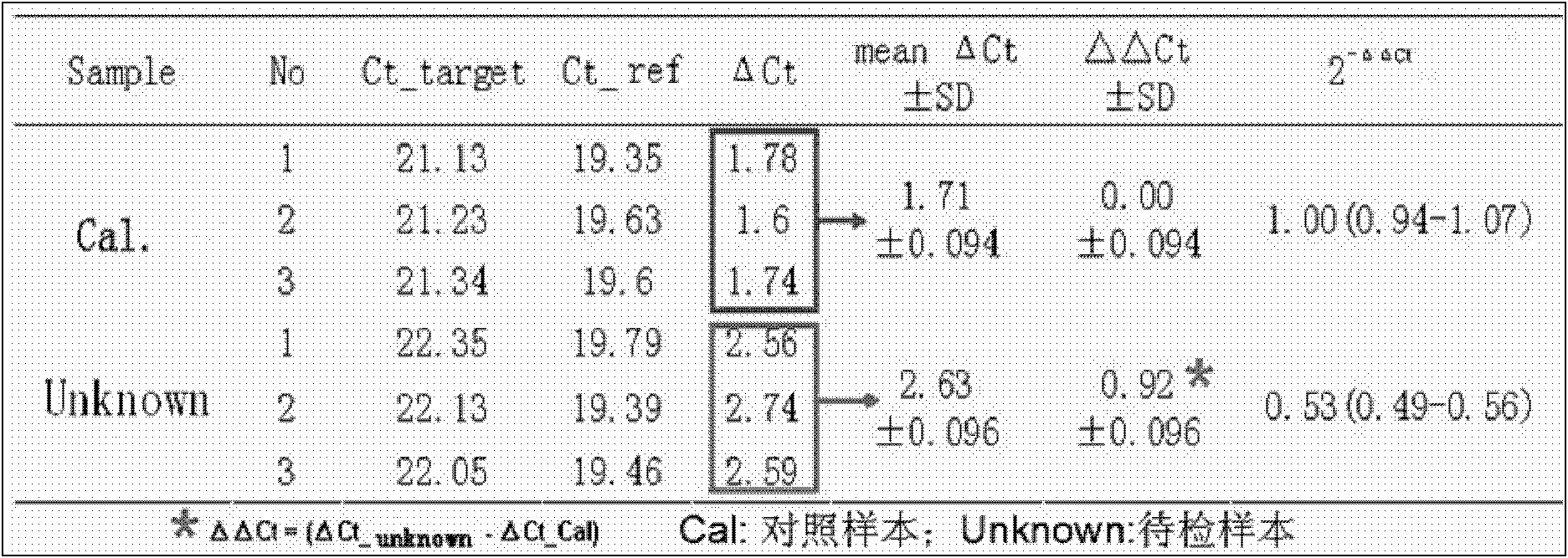
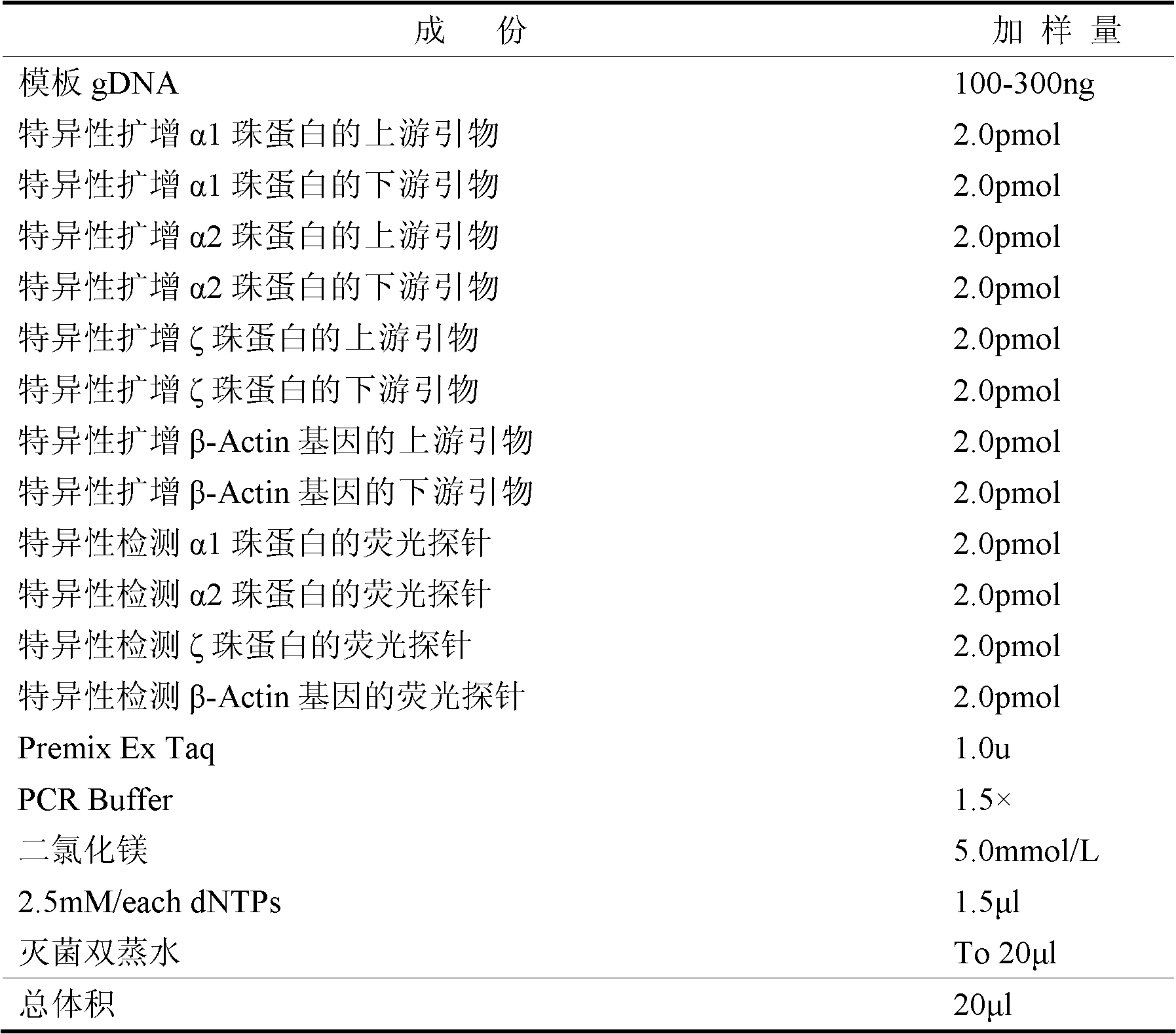
The invention belongs to the field of biochemistry, and provides a fluorescent quantitative polymerase chain reaction (PCR) kit for detecting alpha-globin gene deletion. The kit consists of a pair of specific amplified xi globin primers, a pair of specific amplified alpha1 globin primers, a pair of specific amplified alpha2 globin primers, a pair of specific amplified beta-Actin gene primers, a fluorescent probe for specifically detecting xi globin, a fluorescent probe for specifically detecting alpha1 globin, a fluorescent probe for specifically detecting alpha2 globin, a fluorescent probe for specifically detecting beta-Actin genes, a DNA polymerase and the like. The kit has good sensitivity and accuracy for detecting depletion alpha-thalassemia, has good repeatability and stability, and is totally suitable for clinical detection of alpha-thalassemia.
Owner:SOUTHERN MEDICAL UNIVERSITY
Reagent for detecting Notch signal path as well as PCR (Polymerase Chain Reaction) detecting method and application thereof
The invention provides a reagent for detecting a Notch signal path as well as a PCR (Polymerase Chain Reaction) detecting method and application thereof. The detecting reagent comprises PCR reaction primers for detecting an ADAM10 gene, an ADAM17 gene, an AES gene, a CBL gene, a CCND1gene, a CD44 gene, and the like, and primers of corresponding internal-reference regulatory genes GAPDH (Reduced Glyceraldehyde-Phosphate Dehydrogenase), ACTB (Beta-Actin) and B2M (Beta-2-Microglobulin). The invention provides the reagent for detecting core molecule changes of the NOTCH signal channel; by virtue of the detecting reagent, genes associated with the NOTCH channel can be quickly detected on transcriptional level by virtue of real-time fluorescent quantitative PCR; on this basis, core molecules and a mechanism of action of a tumor NOTCH signal channel are analyzed and researched, so that an NOTCH regulatory channel of tumor-associated medicaments is quickly and accurately found, and therefore, a powerful tool is provided for screening anti-tumor medicaments, discussing the mechanism of a novel targeted medicament, and the like.
Owner:HOSPITAL ATTACHED TO QINGDAO UNIV
Diagnosis Of Neurodegenerative Diseases
InactiveUS20080286263A1Improve trustNervous disorderElectrolysis componentsHemoglobin Beta ChainApolipoprotein C-II
The invention relates to a method of diagnosis of Huntington's Disease in a diagnostic sample of a valid body tissue taken from a human subject, which comprises detecting an altered concentration of a protein in the diagnostic sample, compared with a sample of a control human subject, the protein being selected from: Swiss Prot accession number: Protein name; P10909: Clusterin precursor; P00738: Haptoglobin precursor; P01009: Alpha-1-antitrypsin precursor; P01024: Complement C3 precursor; P01620: 1 g kappa chain V-III region; P01834: 1 g kappa chain C region P01842: 1 g lambda chain C regions; P01857: 1 g gamma-1 chain C region; P01859: Ig gamma-2 chain C region; P01876: 1 g alpha-1 chain C region P02647: Apolipoprotein A-I precursor; P02649: Apolipoprotein E precursor; P02652: Apolipoprotein A-II precursor; P02655: Apolipoprotein C-II precursor; P02656: Apolipoprotein C-II precursor P02671: Fibrinogen alpha / alpha-E chain precursor; P02763: Alpha-1-acid glycoprotein 1 precursor; P02766: Transthyretin precursor; P02768: Serum albumin precursor; P02787: Serotransferrin precursor; P04196: Histidine-rich glycoprotein precursor; P06727: Apolipoprotein A-IV precursor; P19652: Alpha-1-acid glycoprotein 2 precursor; P68871 / P02042: Hemoglobin beta chain / Hemoglobin delta chain; P60709: Beta actin.
Owner:ELECTROPHORETICS LTD
Pseudovirions and preparation method and application thereof
InactiveCN105200018ANon-infectiousImprove stabilityMicrobiological testing/measurementInactivation/attenuationNucleic acid detectionRibonuclease
The invention discloses pseudovirions; a capsid protein of MS2 bacteriophage is wrapped with a DNA-protein complex of a detection sequence formed by series connection of three base fragments of influenza A virus, influenza B virus and beta-actin successively. The invention also provides a preparation method of the pseudovirions and an application of the pseudovirions as quality control products for detecting the influenza A virus and the hepatitis B virus. The preparation method of the pseudovirions is simple and is easy to operate; the obtained pseudovirus is high in purity, can be used as the quality control products in an influenza A virus and influenza B virus nucleic acid detection kit, and has the characteristics of no infectivity, good stability, ribonuclease resistance and the like.
Owner:GUANGDONG HECIN SCI INC
Real-time fluorescence quantitative PCR (polymerase chain reaction) kit for quantitatively detecting TRECs and KRECs genes and application thereof
ActiveCN103173533AEasy accessEasy to storeMicrobiological testing/measurementFluorescence/phosphorescenceT cellBiology
The invention relates to a real-time fluorescence quantitative PCR (polymerase chain reaction) kit for quantitatively detecting TRECs and KRECs genes and application thereof. The kit comprises a PCR system based on nested PCR technology and a real-time fluorescence quantitative PCR system based on real-time fluorescence PCR technology, wherein the nested PCR system comprises forward and reverse primers for TRECs, KRECs and beta-actin genes; and the real-time fluorescence quantitative PCR system comprises forward and reverse primers and specific fluorescence probes for TRECs, KRECs and beta-actin genes. The kit can be used for quickly screening the T-cell level and B-cell level of a neonatal immune system, and has the advantages of high sensitivity, high stability and excellent reproducibility. The method is suitable for combined quantitative detection of TRECs and KRECs, can be used for screening functions of the neonatal immune system, and has practical clinical application value.
Owner:无锡联合利康临床检验所有限公司
Primer pair, kit and method for methylation detection of stomach cancer related genes Reprimo and RNF180
InactiveCN107904313AEasy to useReliable test resultsMicrobiological testing/measurementDNA/RNA fragmentationForward primerRibonucleoside
The invention relates to a primer pair for methylation detection of stomach cancer related genes Reprimo and RNF180. The primer pair comprises forward primers, reverse primers and detection probes ofReprimo, RNF180 and beta-Actin. The invention relates to the kit for methylation detection of the stomach cancer related genes Reprimo and RNF180. The kit comprises a PCR (polymerase chain reaction) solution of the primers and the probes, and the PCR solution comprises the forward primers, the reverse primers, the detection probes, a 10*PCR buffer, dNTP (deoxy-ribonucleoside triphosphate), Mg<2+>,nuclease-free water and an Ex Taq enzyme. The invention further relates to the method for methylation detection of the stomach cancer related genes Reprimo and RNF180. The kit and the detection method thereof have the advantages of accurate detection result, high sensitivity, high flux, high specificity and short detection time.
Owner:韩林志
Kit for quickly detecting polymorphism of human VKORC1 and CYP2CP genes and using method of kit
ActiveCN104293920ANo human errorLow false positiveMicrobiological testing/measurementVKORC1Fluorescence
The invention belongs to the gene detecting technology in the clinical detecting technology in the bio-medical field and particularly relates to a kit for quickly detecting polymorphism of human VKORC1 and CYP2CP genes and a using method of the kit. The kit comprises three pairs of primers, three probes, six PNA sequences, 1 pair of beta-Actin interior label primers and one interior label probe, further comprises Mg<2+> PCR (polymerase chain reaction) buffer liquor, a dNTP mixture, Tag enzyme and ddH2O. The kit disclosed by the invention can be used for performing qualitative detection on polymorphic sites of the VKORC1 and CYP2C9 genes, performing two-tube PNA-PCR fluorescent amplification reaction according to an SNP site and performing result judgment according to the condition whether a two-tube fluorescent curve is formed or not without producing manual errors, so that the false positive rate and the false negative rate are low. The kit disclosed by the invention can easily realize high-flux detection and is capable of satisfying clinical use very well.
Owner:WUHAN HEALTHCHART BIOLOGICAL TECH
Huaman plasma DNA quantitative analyser
InactiveCN1811387AAccurate quantitative detectionOvercoming Extraction EfficiencyMicrobiological testing/measurementFluorescence/phosphorescenceFluorescenceTherapeutic effect
The present invention relates to a quantitative detection analysis method of human plasma DNA level. Said method includes the following steps: selecting artificially-synthetic exogenous DNA series whose extraction efficiency is identical to or similar to that of human plasma DNA, but they are non-homologous, selecting housekeeping gene beta actin as determination gene, inserting artificially-synthetic exogenous DNA into carrier pMD18T and placing it in the bacteria to make culture, making blue and white spot screening, colony PCR identification, enzyme-cutting into linearity, placing the above-mentioned material into plasma, adopting double fluorescent quantitative gene amplification, fluorescent groups are respectively FAM and HEX, extracting known exogenous positive plasmid recovery efficiency, using computation formula to make computation so as to obtain the quantity of plasma DNA.
Owner:潘世扬
Methylation gene combination, primer and probe combination and kit for cervical cancer methylation detection and using method of kit
PendingCN110452984AReduce randomnessReduce protein expressionMicrobiological testing/measurementDNA/RNA fragmentationReference genesHigh flux
The invention provides a methylation gene combination, primer and probe combination and kit for cervical cancer methylation detection and a using method of the kit, and belongs to the technical fieldof medical gene detection. Based on a real-time fluorescent PCR technology, specific methylation detection primers and probes in the promoter regions of FAM19A4, has-miR-124-2 and ZNF671 genes are designed and prepared into a PCR reaction solution to amplify a sample DNA after vulcanization, the quality of the sample is controlled by means of an internal reference gene beta-actin, and whether or not the sample is methylated is determined. Compared with the conventional method, the method has the advantages of fast speed, high flux and good sensitivity and specificity, and plays an important role in the early diagnosis and prognosis of cervical cancer.
Owner:韩林志
Detection reagent kit for methylation of septin 9 genes in human peripheral blood circulation tumor DNA
ActiveCN105331727AEasy to detectEasy to operateMicrobiological testing/measurementCpG siteOperability
Owner:湖南宏雅基因技术有限公司
Group B streptococcus fluorescent quantitative PCR detection kit
InactiveCN107557443AImprove detection accuracyHigh sensitivityMicrobiological testing/measurementMicroorganism based processesNucleotideTrue positive rate
The invention belongs to the technical field of bacteria detection, and more specifically discloses a group B streptococcus fluorescent quantitative PCR detection kit. The group B streptococcus fluorescent quantitative PCR detection kit comprises a group of group B streptococcus fluorescent quantitative PCR detection primers and probes; the primers and probes comprise GBS amplification primer pairGBS-F and GBS-R, GBS detection probe GBS-P, internal reference primer pair beta-actin-F and beta-actin-R, and internal reference probe beta-actin-P; the nucleotide sequences of GBS-F, GBS-R, GBS-P, beta-actin-F, beta-actin-R, and beta-actin-P are represented by SEQ ID NO:1-6 respectively. The group B streptococcus fluorescent quantitative PCR detection kit can be used for detecting GBS in samplewith complex composition, and detecting single copy templates, is wider in linearity range, is capable of avoiding false positive results caused by amplification products pollution, and is high in sensitivity and specificity.
Owner:GUANGZHOU SAGENE BIOTECH
Real time fluorescent PCR method for detecting aquatic product food allergen gene
InactiveCN101434991AHigh sensitivityShort detection timeMicrobiological testing/measurementFluorescence/phosphorescenceTotal rnaTest sample
The invention discloses a real-time fluorescent PCR method for testing the allergen genes of aquatic food, comprising the following steps: total RNA of samples is extracted; the allergen genes Par and TM of aquatic food, and internal standard gene Beta-actin are respectively tested by SYBR Green or TaqMan probe real-time fluorescent PCR technology; and result analysis is carried out to the tested samples according to the amplification kinetic curve and the Ct value. As the real-time fluorescent PCR technology is used for testing the allergen genes Par and TM of aquatic food, compared with other methods, the method has the advantages of high sensitivity, short testing time, available quantitative analysis, and the like, and can be used in the qualitative screening, quantitative analysis and confirmation identification of allergen genes in unknown species or mixed samples of aquatic food.
Owner:JIMEI UNIV
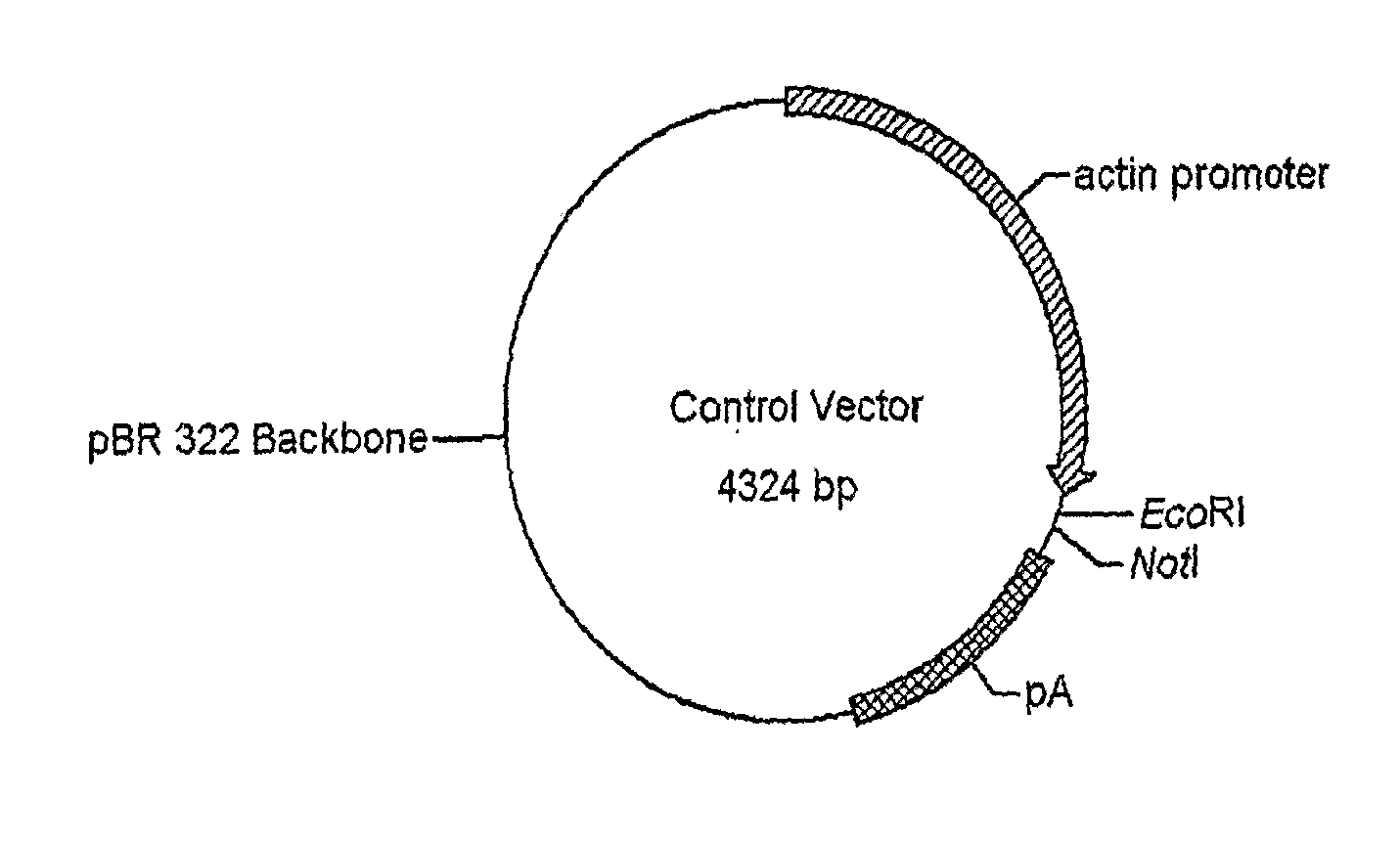
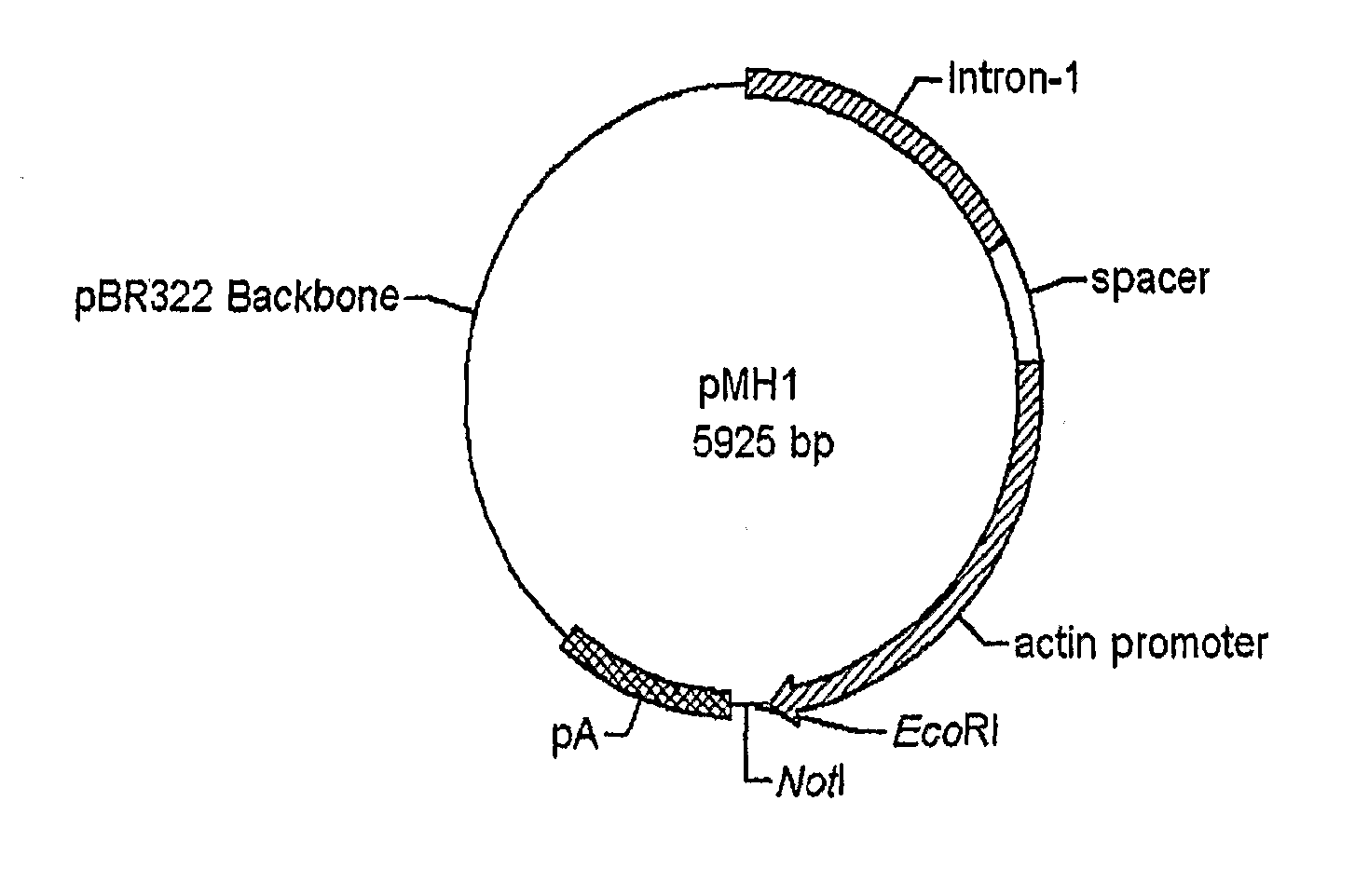
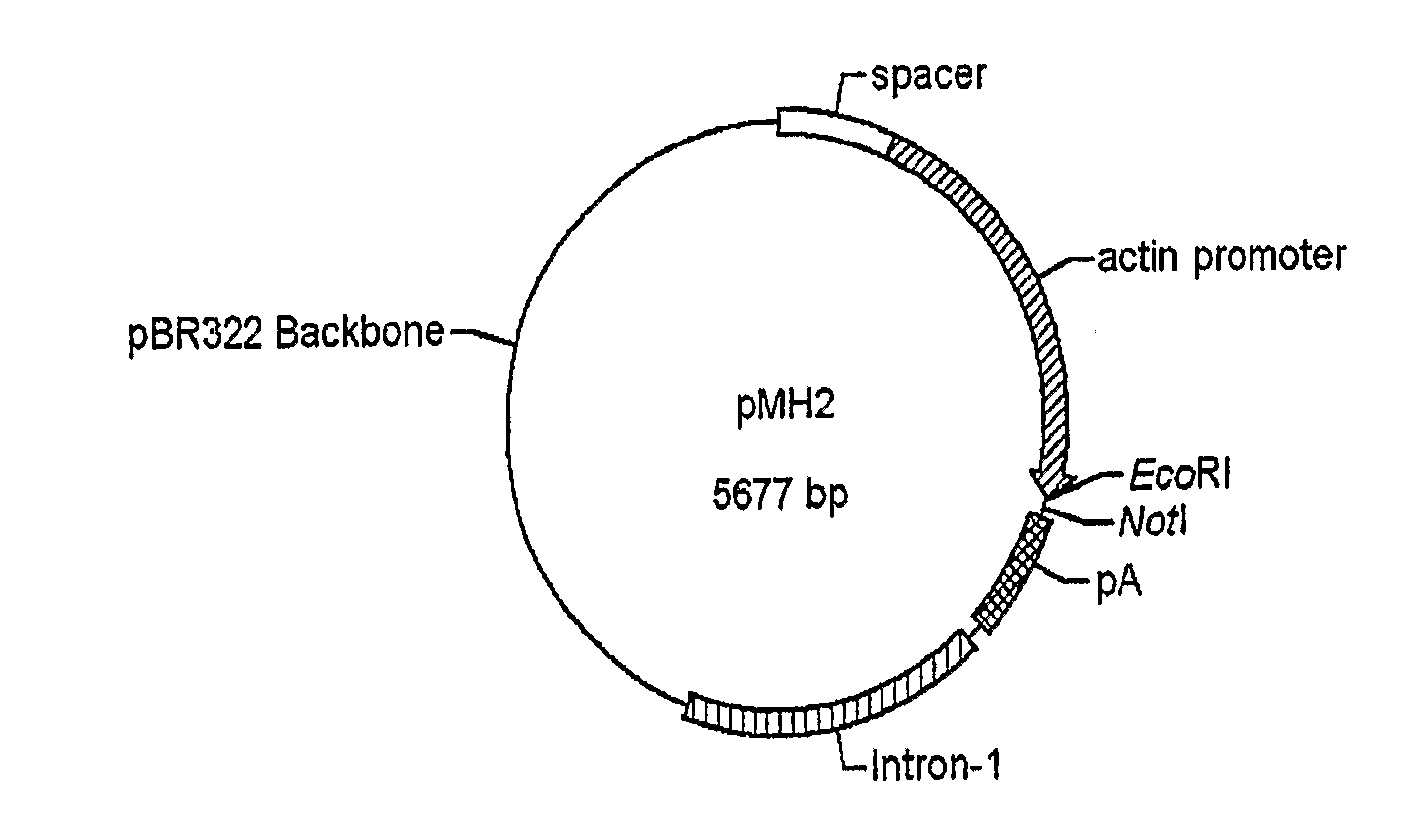
Use of chick beta actin gene intron-1
A method to use chick beta actin gene intron-1 or functional equivalent as a gene expression enhancer eiement or a gene expression 'hot spot' sequence for constructing or reconstructing a mammalian expression vector for extremely high expression of recombinant proteins is disclosed. Composition of a set of extremely strong gene expression vectors is also disclosed.
Owner:QINGDAO HUINUODE BIOLOGICAL TECH CO LTD
GLP-1 gene delivery for the treatment of type 2 diabetes
ActiveUS20030220274A1Rapid clearanceEasy to controlSugar derivativesPeptide/protein ingredientsGene deliveryGlucose polymers
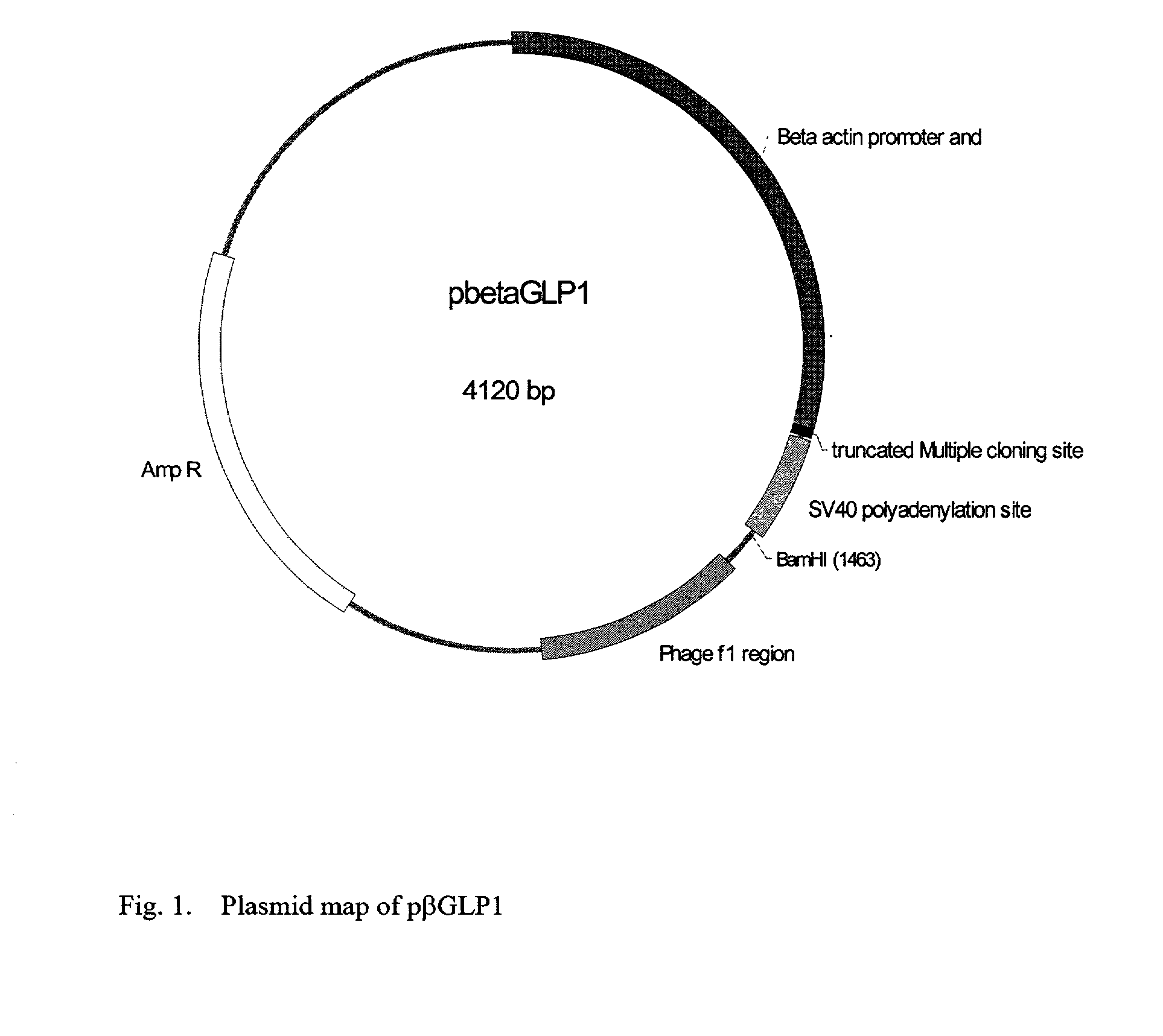
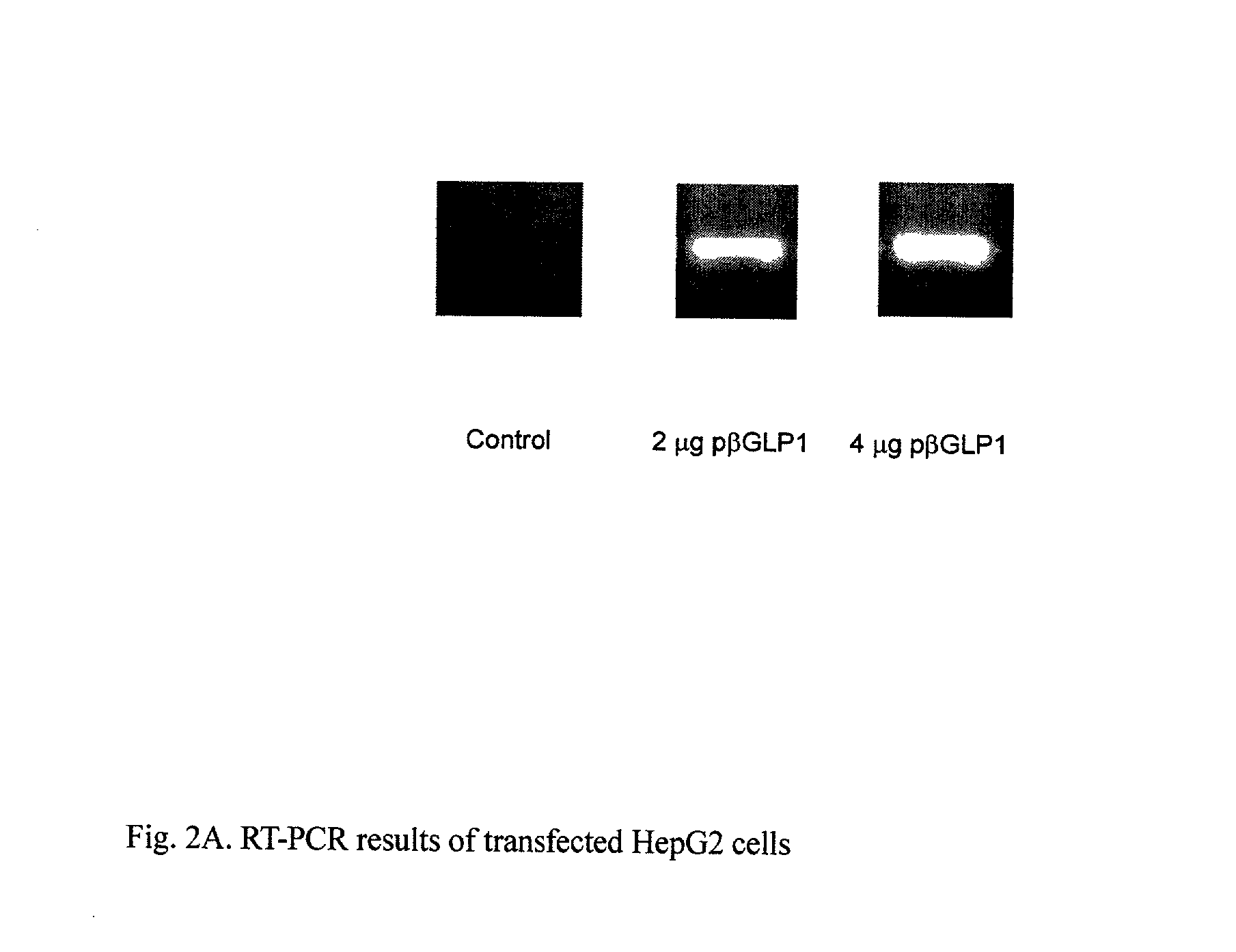
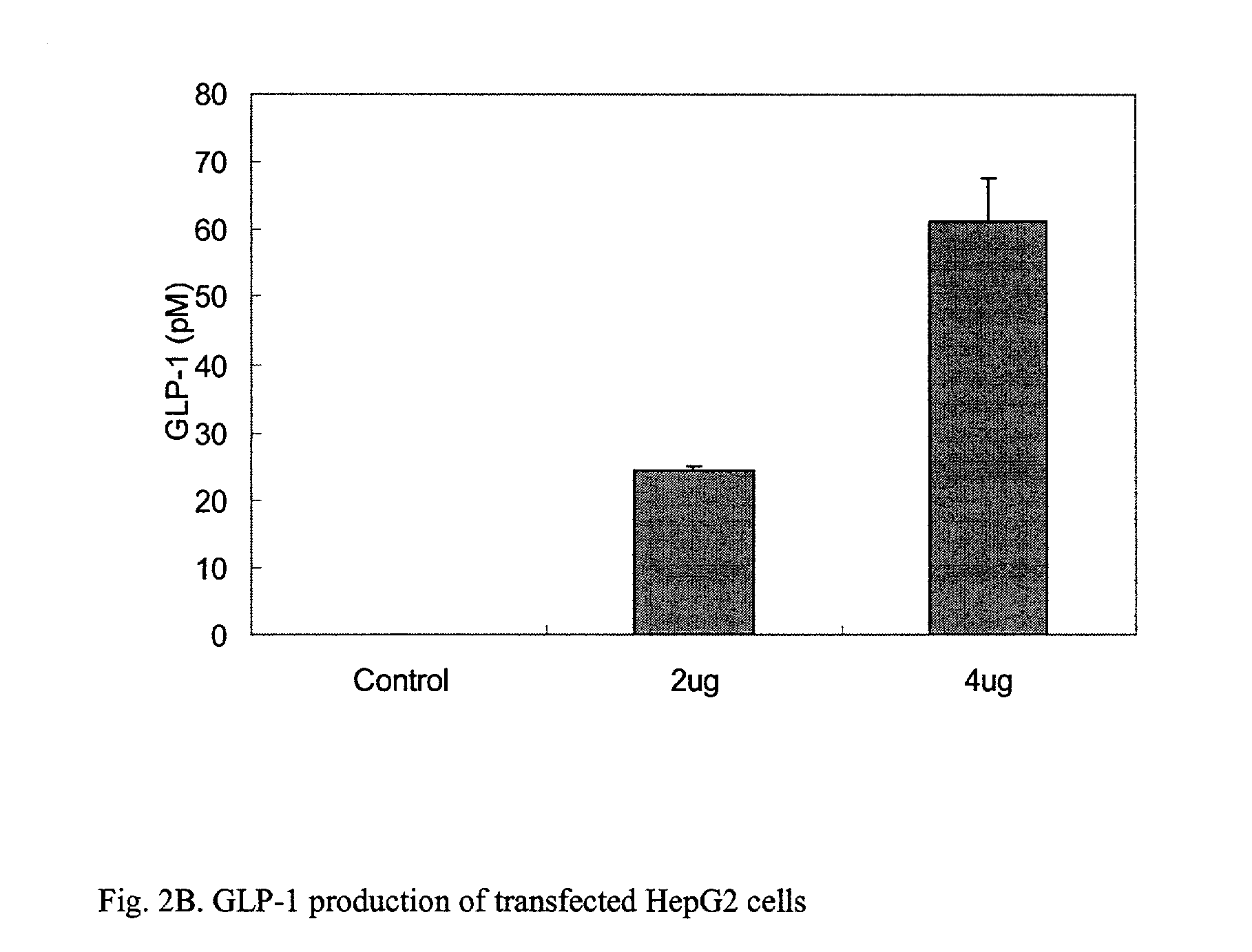
This patent discloses compositions and methods of use thereof to normalize the blood glucose levels of patients with type 2 diabetes. It relates particularly to a plasmid comprising a chicken beta actin promoter and enhancer; a modified GLP-1 (7-37) cDNA (pbetaGLP1), carrying a furin cleavage site, which is constructed and delivered into a cell for the expression of active GLP-1.
Owner:CLSN LAB
Human red cell ABO blood type antigen multi-PCR genotyping method and kit
PendingCN108624663ADetection site simplificationLow costMicrobiological testing/measurementDNA/RNA fragmentationAntigenMultiplex pcrs
Owner:SHANGHAI BLOOD CENT
Lentiviral gene transfer vector, preparation method and application thereof
InactiveCN101705246AIncrease productionIncrease nuclear inputViruses/bacteriophagesVector-based foreign material introductionLarge fragmentWild type
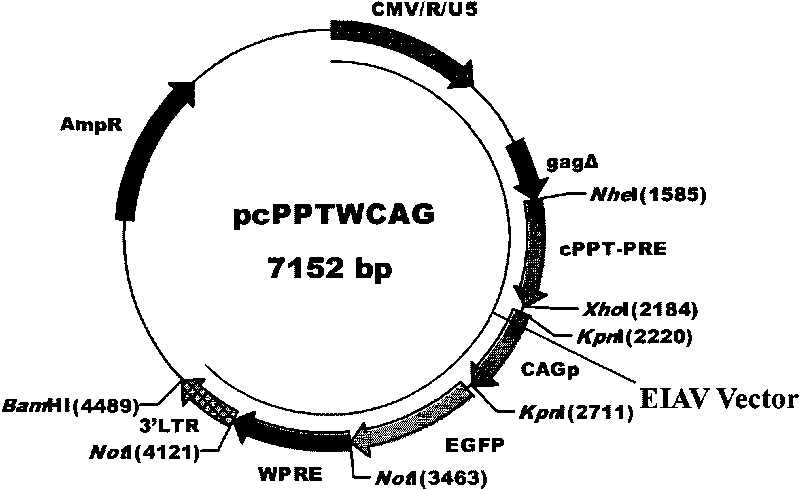
The invention discloses an equine infectious anemia virus (EIAV) gene transfer vector, a constructed method and an application thereof. The vector of the invention comprises a CMV / R / U5 promoter, a 5' non-translated region homing sequence, a partial 5' gag gene coding region sequence, a central polypurine sequence, an Rev reaction element of EIAV, a CMVIE / chicken beta-actin promoter, a reporter gene, WPRE of WHBV, a polyclone locus, a poly (A) signal before EIAV 3'LTR, complete 3'LTR and partial sequence of eukaryon expression vector. The vector of the invention possesses common advantages of lentiviral vector, lacks auxiliary genes of all the wild-type virus, can carry one or a plurality of selected genes to transmit to target cells, can insert with target DNA sequence with large fragments and can lead the foreign genes and reporter genes to express at a high level in the cells.
Owner:HARBIN VETERINARY RES INST CHINESE ACADEMY OF AGRI SCI
Parkin gene mutation relating to Parkinson's disease and its detection method
InactiveCN1664098AAccurate detectionQuick checkSugar derivativesMicrobiological testing/measurementNucleotideIntein
The invention relates to the gene mutation relevant to the paralysis agitans and the detecting method, comprising: extracting DNA sample; designing primer; expanding with PCR; analyzing nucleic acid by means of dHPLC technology; testing the gene order of the sample which is dHPLC positive reaction to definite the genetype; carrying out beta-actin comparison for the sample of repeated extanding negative reaction to eliminate extron deletion mutation. The parkin gene mutation is in the codon promoter of the parkin gene, the second basic group in the exon1, ATG mutated to ACG; on the intersection point of thefifth introne and sixth extron in the exon6, AG mutated to AC; in the sixth extron, the 727th basic group, G mutated to A; in the exon9, the 1016th basic group, C mutated to T; in the position of ring finger intermediate region, in the Znf C6HC zinc region, GCG mutated to CGTC. By applying the gene mutation and the detection method, the detection for the mutation site of the parkin fene is high-effective and low-cost, and the diagnose to the potential PD patient can be made in an early stage.
Owner:XUANWU HOSPITAL OF CAPITAL UNIV OF MEDICAL SCI
RNA (Ribonucleic Acid) circEPSTI1 (Epithelial-stromal Interaction 1) and application thereof in triple negative breast cancer
The invention relates to the technical field of breast cancer diagnosis, and specifically discloses RNA (Ribonucleic Acid) circEPSTI1 (Epithelial-stromal Interaction 1) and application thereof in triple negative breast cancer. The application of the RNA circEPSTI1 in the triple negative breast cancer comprises the following steps: firstly, collecting a to-be-detected suspected triple negative breast cancer tissue sample, extracting total RNA, specifically and reversely transcribing the cyclic RNA circEPSTI1 (hsa_circRNA_000479) into cDNA (Complementary Deoxyribonucleic Acid) by using the total RNA as a template; carrying out real-time quantitative PCR (Polymerase Chain Reaction) amplification by using a specific PCR primer; using beta-actin as a reference gene, thus obtaining a relative quantitative deltaCT value of circEPSTI1; reminding that the circEPSTI1 is in positive expression when deltaCT is smaller than or equal to 5.52. According to the RNA circEPSTI1 and the application thereof in the triple negative breast cancer, disclosed by the invention, the RNA circEPSTI1 is applied to prognostic prediction of a triple negative breast cancer patient through relatively and quantitatively detecting the expression situation of the circEPSTI1 in the suspected triple negative breast cancer tissue sample.
Owner:SUN YAT SEN UNIV CANCER CENT
Real-time fluorescent quantitative PCR (polymerase chain reaction) kit for one-step quantitative detection of TRECs and KRECs genes and its application
InactiveCN105274230AEasy accessEasy to transportMicrobiological testing/measurementFluorescenceT cell
The invention relates to a real-time fluorescent quantitative PCR (polymerase chain reaction) kit for one-step quantitative detection of TRECs and KRECs genes and its application. The kit comprises a real-time fluorescent quantitative PCR reaction system based on the real-time fluorescent PCR technique. The real-time fluorescent quantitative PCR reaction system comprises forward and reverse primers specific to TRECs, KRECs and beta-actin genes, and a specific fluorescent probe. The kit allows quick joint screening of neonatal immune system T-cell level and B-cell level, has high sensitivity and stability and provides excellent reproducibility, and this method is applicable to the joint quantitative detection of TRECs and KRECs and to functional screening of the neonatal immune system and is worthy of practical clinical application.
Owner:SHANGHAI ADVANCED CLINICAL LAB SCI
Method for preparing a sterile transgenic fish
InactiveUS20050257281A1Easy to operateSolve the real problemPisciculture and aquariaFermentationCommon carpGonadotropin-releasing hormone-III
The present application disclosed a method for preparing a sterile transgenic fish, comprising constructing antisense RNA expression vector of salmon-type gonadotropin-releasing hormone, introducing the recombinant DNA fragment into carp oosperm by microinjection, and screening the sterile transgenic fish by Polymerase Chain Reaction and radioimmunoassay, wherein the expression vector comprising a promoter of carp beta actin (β-actin) gene, a complementary DNA fragment of antisense salmon-type gonadotropin-releasing hormone (sGnRH) gene from carp with 323 bp as a target gene comprising sGnRH decapeptide, the coding region of gonadotropin-releasing hormone associated peptide and 3′ non-coding sequence, and 3′ flanking sequence of grass carp growth hormone gene as a stop sequence. The method of the present invention is easy and convenient for operation which provides a basis for providing a technical platform of general applicability for solving the hereditary and ecological safety problems of transgenic fishes.
Owner:INST OF AQUATIC LIFE ACAD SINICA
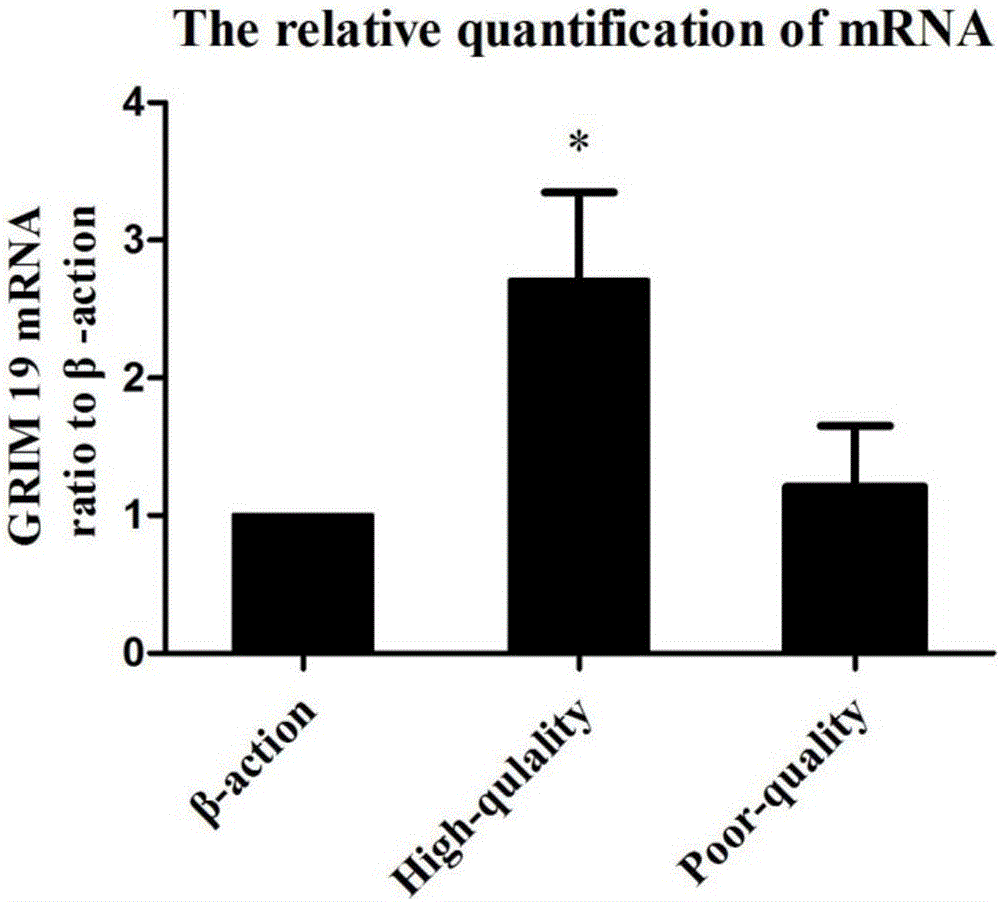
Method for evaluating developmental potential of oocyte by utilizing relative expression of GRIM19 in granulosa cell
ActiveCN105861709ANon-invasive injuryReduce stimulationMicrobiological testing/measurementDNA/RNA fragmentationNumerical rangeGranulosa cell
The invention discloses a method for evaluating developmental potential of an oocyte by utilizing the relative expression of GRIM-19 in a granulosa cell. When the relative expression of the GRIM-19 in the granulosa cell of the oocyte is 0.16-0.35, the obtained oocyte is judged to be high in developmental potential; when the relative expression of the GRIM-19 is 0-0.12, the obtained oocyte is judged to be low in developmental potential; a beta-actin gene is selected for internal reference. The method disclosed by the invention is a more objective, accurate and quantizable oocyte quality evaluation method, is noninvasive without repeatedly taking out an incubator for observation, so that the adverse environmental stimulation is reduced. Due to being quantizable, the method disclosed by the invention has a specific numerical range, is easy to operate and solves the problem that due to artificial subjective factors, the judgment standards are not unified.
Owner:SHANDONG UNIV QILU HOSPITAL
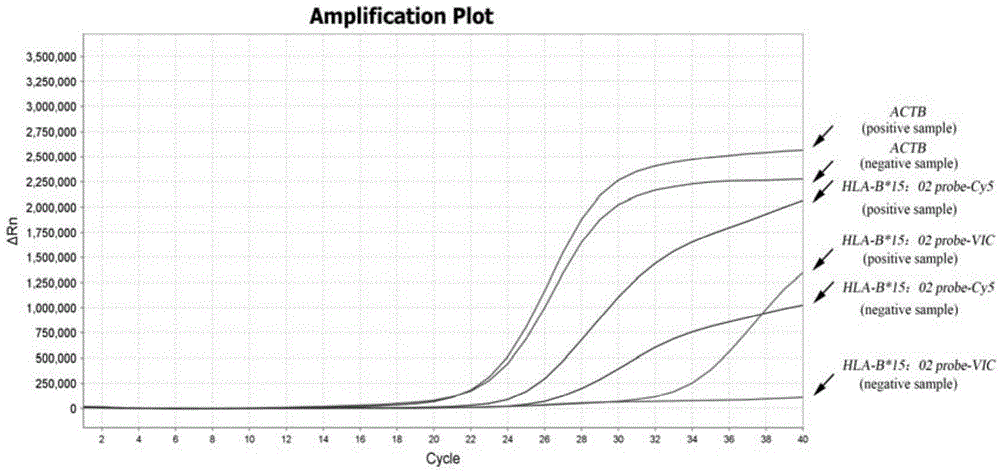
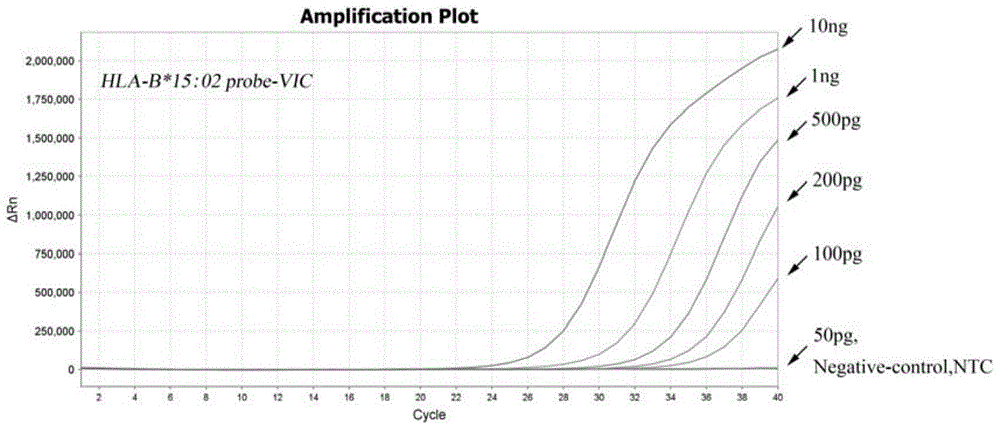
Multiplex real-time fluorescent PCR (polymerase chain reaction) method for detecting HLA-B*15:02 alleles
ActiveCN104830852AShorten the timeAvoid secondary pollutionMicrobiological testing/measurementDNA/RNA fragmentationFluorescenceHLA-B
The invention discloses a primer-probe combination of two pairs of high specific amplification HLA-B*15:02 alleles designed on the basis of using a TaqMan probe detection method. On the basis, by using a primer and probe of a reference gene beta-Actin, specific primers and probes of two pairs of target genes and the primer and probe of the reference gene are added into a same pipe to carry out a multiplex fluorescent PCR, and then results are analyzed by using a fluorescent amplification curve. The method disclosed by the invention has the characteristics of high specificity, flexibility, rapidness, high flux, no pollution, high resolution, capability of carrying out real-time monitoring on a reaction process, and the like, and can be applied to the detection of HLA-B*15:02 alleles of whole-genome DNA samples in human peripheral blood and saliva.
Owner:SHANXI LIFEGEN
PCR technology-based quail sex identification method
ActiveCN106244689AReduce the amount of breedingReduce the cost of farmingMicrobiological testing/measurementPcr ctppBiology
The invention discloses a PCR technology-based quail sex identification method, and belongs to the technical field of biological detection identification. Two pairs of PCR primers used for PCR identification comprise a pair of micro-satellite primers and a pair of internal control gene (beta-actin) primers. The PCR identification method comprises the following steps: carrying out PCR amplification by adopting the quail sex identification PCR primers with the blood sample genome DNA of a young quail to be detected as a template, detecting the above obtained PCR amplification product by adopting agarose gel electrophoresis, judging the young quail to be detected as a male quail when the amplification product only has a 259 bp specific amplification band, and judging the young quail to be detected as a female quail when the amplification product has a 259 bp specific amplification band and a 114 bp specific amplification band. The method has the advantages of simplicity and convenience in operation, realization of sex discrimination in at the hatching time, rapidness in discrimination, accurate result, and avoiding of the disadvantages of long time, high breeding cost and high error rate of traditional methods.
Owner:INST OF ANIMAL SCI & VETERINARY HUBEI ACADEMY OF AGRI SCI
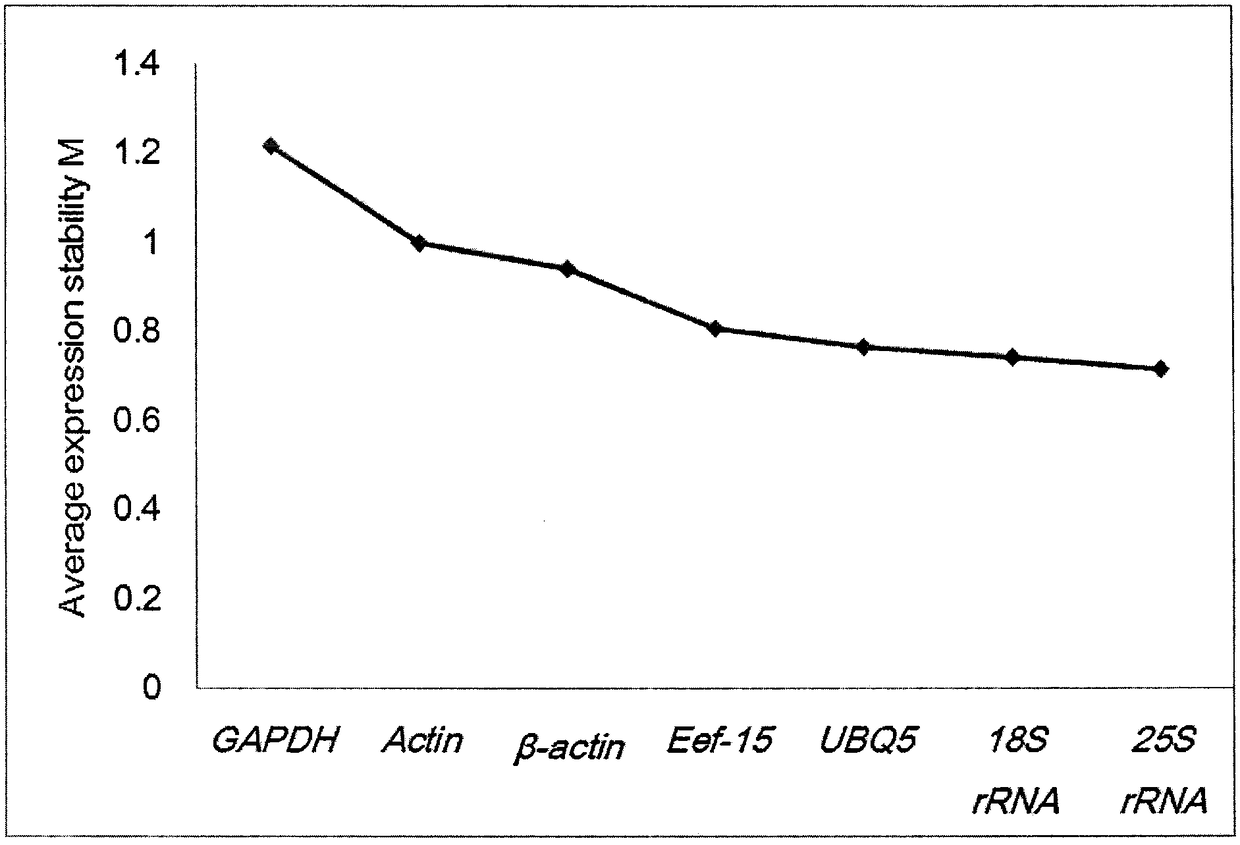
Selection method of reference genes in quantitative real-time PCR analysis of jerusalem artichoke
InactiveCN108588191AStable expressionMicrobiological testing/measurementQuantitative Real Time PCRReference genes
The invention discloses a selection method of reference genes in quantitative real-time PCR analysis of jerusalem artichoke, and relates to the field of quantitative PCR. The method comprises the following steps: taking different tissues (radicles, immature stems, leaves, stem blocks and petals) of the jerusalem artichoke as materials, and carrying out expression analysis of the reference genes of18S ribosomal RNA gene (18S rRNA), transcription elongation factor gene (Ef-1a), actin gene (Actin and beta-actin), 3-glyceraldehyde phosphate dehydrogenase (GAPDH), 25S ribosomal RNA gene (25S rRNA)and poly-ubiquitin enzyme gene (UBQ)7 by using a q PCR technology; carrying out statistical assessment on the obtained data and analyzing expression change of all housekeeping genes by utilizing GeNorm and NormFinder software, so as to screen out relatively-stable genes as the reference genes of the jerusalem artichoke, which are used for studying the gene dosage changes of the jerusalem artichoke.
Owner:青海大学农林科学院
Primer pair, kit and method for colorectal-cancer related gene Septin9 methylation detection
InactiveCN108048570AEasy to useReliable test resultsMicrobiological testing/measurementDNA/RNA fragmentationForward primerEnzyme
The invention relates to a primer pair for colorectal-cancer related gene Septin9 methylation detection. The primer pair comprises a Septin9 forward primer, a Septin9 reverse primer, a Septin9 detection probe, a beta-Actin forward primer, a beta-Actin reverse primer and a beta-Actin detection probe. The invention relates to a kit for colorectal-cancer related gene Septin9 methylation detection. The kit comprises PCR reaction liquid comprising the primers and the probes, and the PCR reaction liquid comprises the forward primers, the reverse primers, the detection probes, 10*PCR buffer, dNTP, nuclease-free water and Ex Taq enzyme. The invention also relates to a method for colorectal-cancer related gene Septin9 methylation detection. The kit and the detection method have the advantages of being accurate in detection result, high in detection flux, good in specificity and sensitivity, short in detection time, convenient to use and capable of effectively meeting the clinical requirements.
Owner:韩林志
Method for identifying drought resistance of highland barley by utilizing semi-quantitative PCR method of HVA1 gene
The invention discloses a method for identifying the drought resistance of highland barley by utilizing a semi-quantitative PCR (Polymerase Chain Reaction) method of an HVA1 gene, which comprises the following steps: (1) carrying out stress treatment for 4 days by utilizing 0-30 percent of PEG (Polyethyleneglycol) on the two-leaf stage of the highland barley; (2) carrying out total RNA (Ribose Nucleic Acid) extraction and reverse transcription on a blade by adopting a total RNA extraction reagent kit and a reverse transcription reagent kit from Shanghai Sangon; (3) amplifying a target gene HVA1 through taking a barley actin (Beta-Actin) gene as a reference gene; (4) carrying out quadratic function fitting on a relative expression map of the obtained (Beta-Actin) gene by adopting Gel-pro software; and (5) calculating the highest relative expression quantity of the HVA1 gene or the corresponding PEG concentration according to a fitting curve, and if the numerical value is larger, a highland barley variety with strong drought resistance is determined.
Owner:QINGHAI UNIVERSITY
Use of chick beta actin gene intron-1
A method to use chick beta actin gene intron-1 or functional equivalent as a gene expression enhancer element or a gene expression “hot spot” sequence for constructing or reconstructing a mammalian expression vector for extremely high expression of recombinant proteins is disclosed. Composition of a set of extremely strong gene expression vectors is also disclosed.
Owner:AMPROTEIN CORP
Real-time fluorescence quantitative PCR (polymerase chain reaction) kit for quantitatively detecting TRECs gene and application thereof
The invention relates to a real-time fluorescence quantitative PCR (polymerase chain reaction) kit for quantitatively detecting TRECs gene and application thereof. The kit comprises a PCR system based on nested PCR technology and a real-time fluorescence quantitative PCR system based on real-time fluorescence PCR technology, wherein the nested PCR system comprises forward and reverse primers for TRECs and beta-actin genes; and the real-time fluorescence quantitative PCR system comprises forward and reverse primers and specific fluorescence probes for TRECs and beta-actin genes. The kit can be used for quickly screening the T-cell level of a neonatal immune system, and has the advantages of high sensitivity, high stability and excellent reproducibility. The method is suitable for quantitative detection of TRECs, can be used for screening functions of the neonatal immune system, and has practical clinical application value.
Owner:无锡联合利康临床检验所有限公司
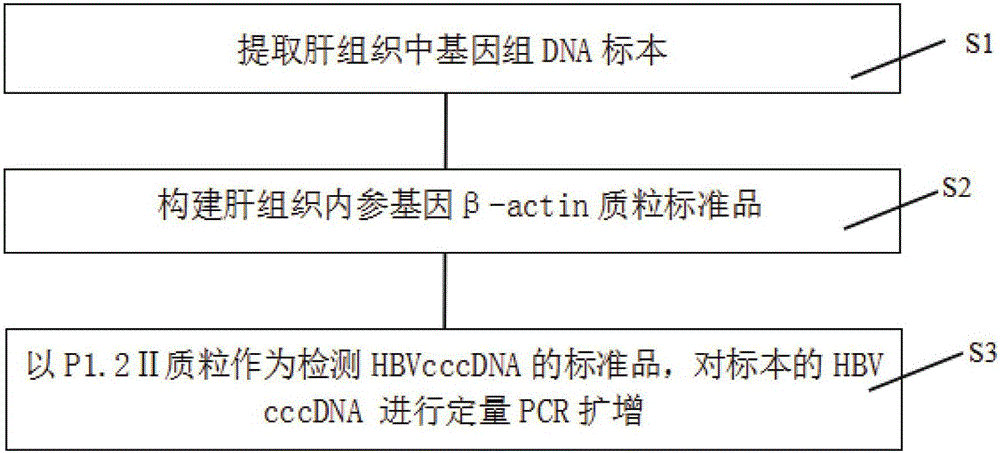
Quantitative PCR detection method for hepatic tissue HBVcccDNA
InactiveCN105755151AStrong specificityIncreased sensitivityMicrobiological testing/measurementTotal rnaElectrophoresis
The invention relates to a quantitative PCR detection method for hepatic tissue HBVcccDNA. The method comprises the following steps: extracting a genome DNA specimen in hepatic tissues; establishing a beta-actin plasmid standard; taking total RNA of adult peripheral blood cells as a template, performing inverse transcription to synthesize cDNA; performing PCR amplification on corresponding cDNA target fragments of human beta-actin genes by taking the cDNA as a template, establishing to form recombinant plasmid by taking pMD18-T as a carrier, performing electrophoresis and sequencing identification, and making a beta-actin standard curve by using fluorescent quantitative PCR. According to the method disclosed by the invention, HBVcccDNA and beta-actin in the hepatic tissues can be subjected to sensitive and specific amplification, quantification of the HBVcccDNA in hepatic cells is realized, and the method can be used for detection and application of HBVcccDNA in the clinical hepatic tissue and hepatic cells.
Owner:陈延平
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com