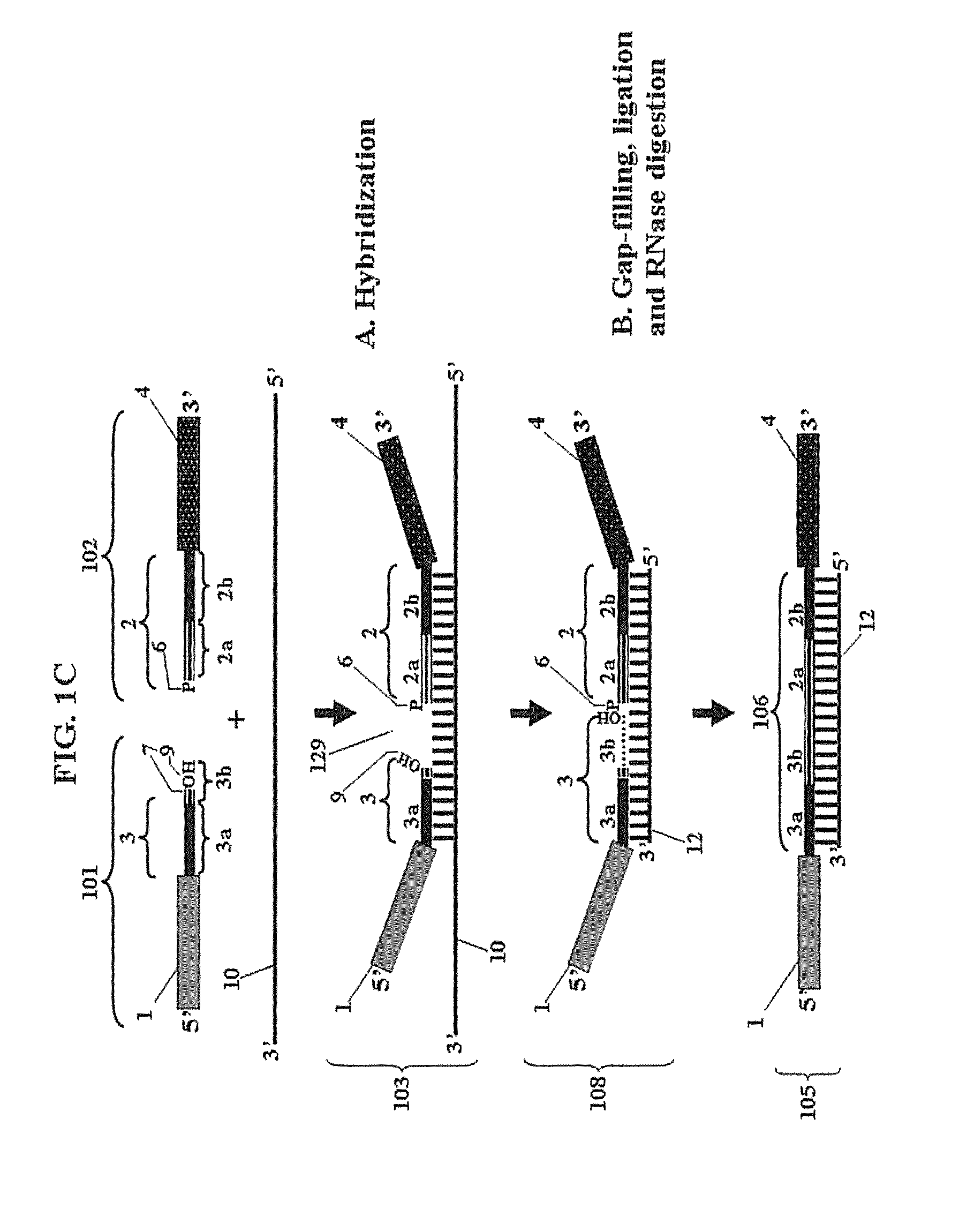
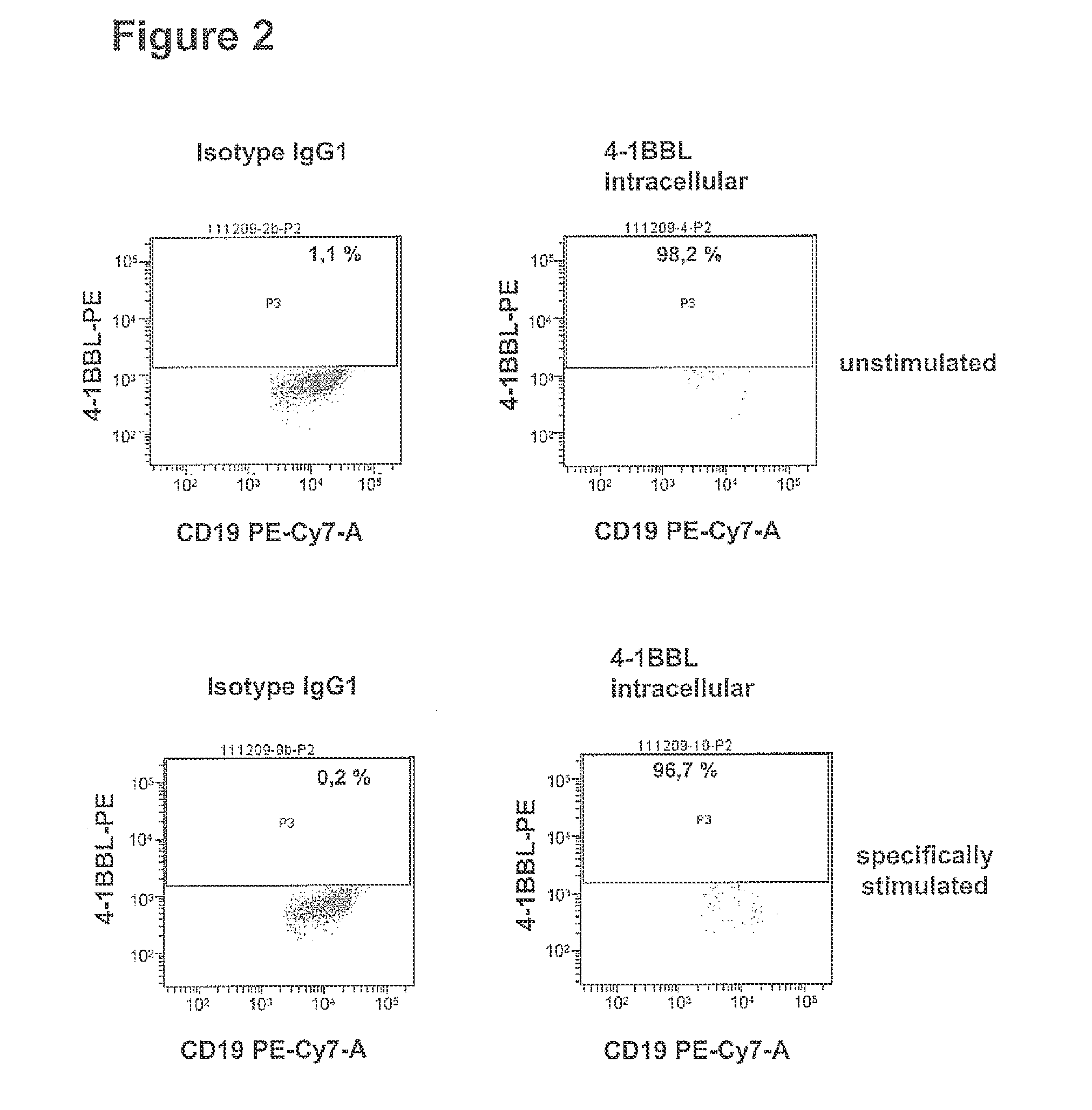
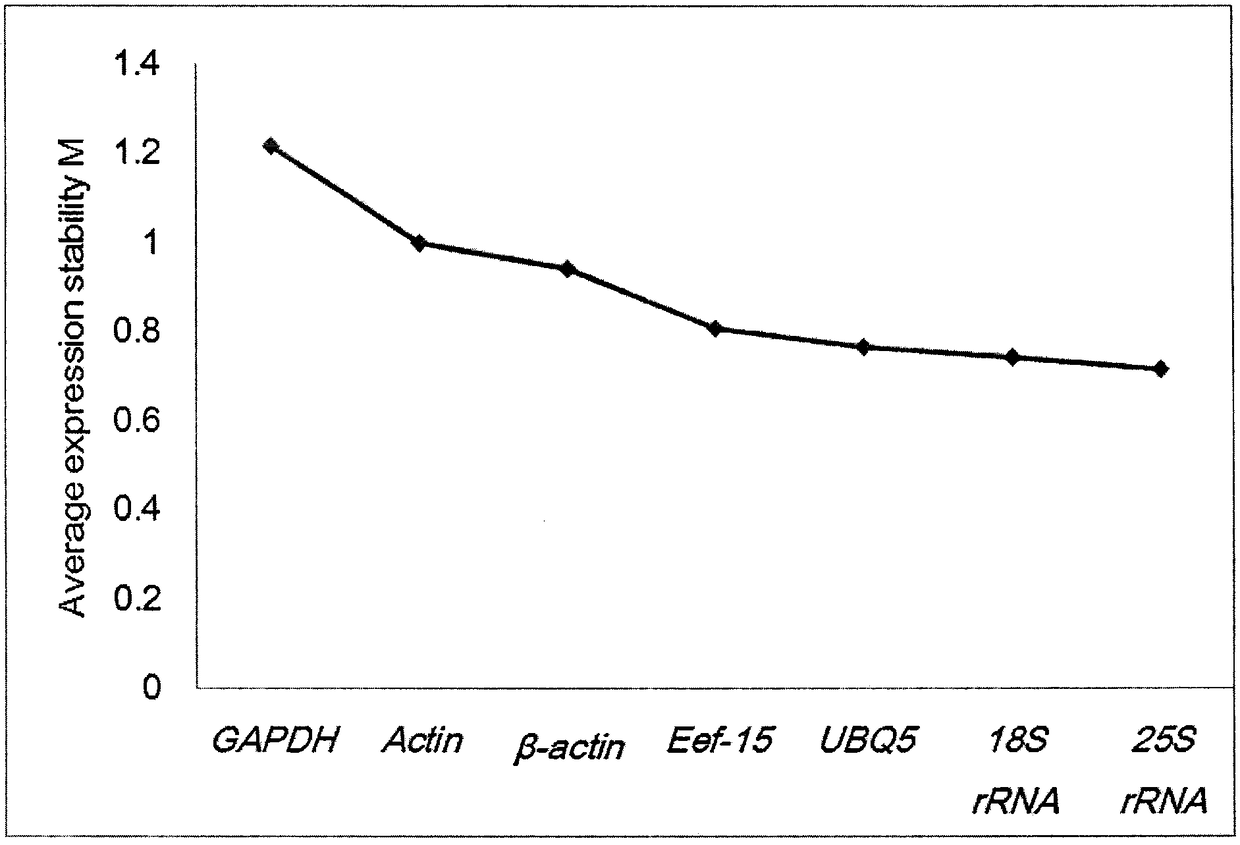
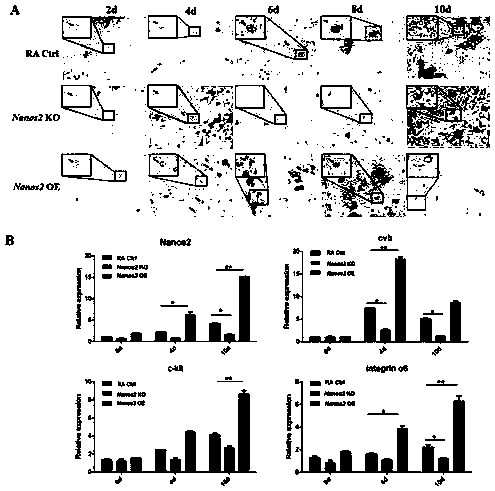
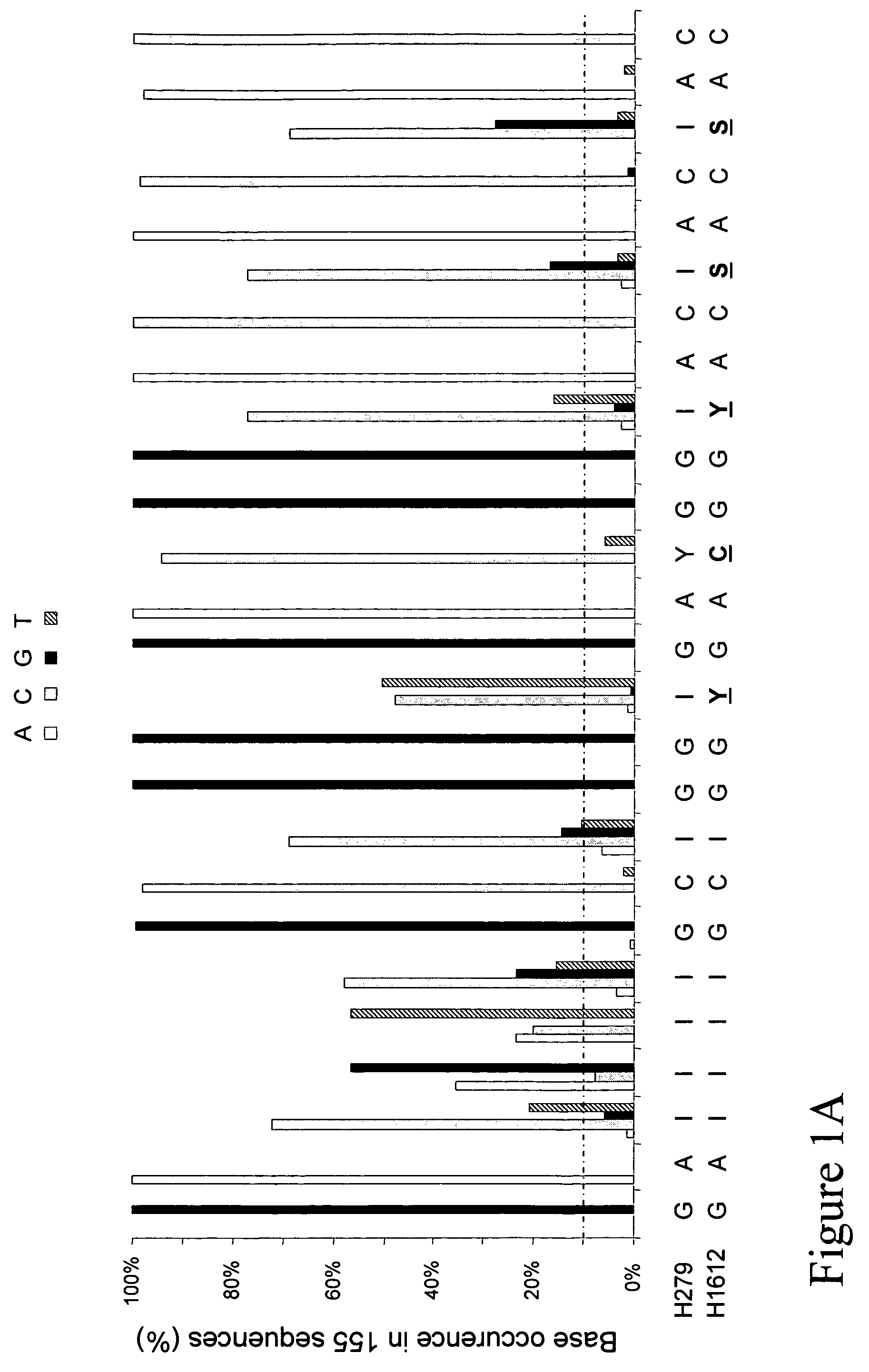
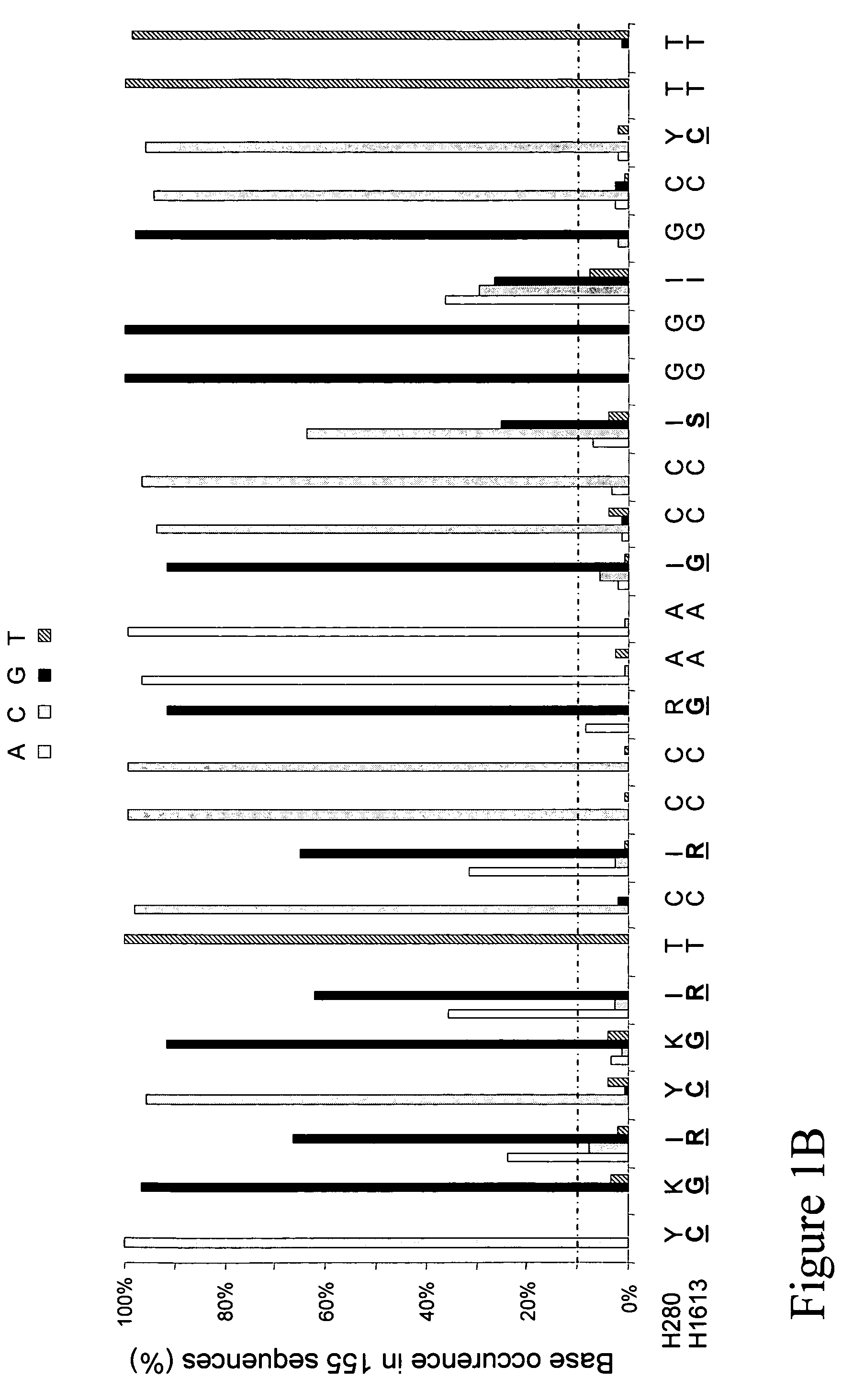
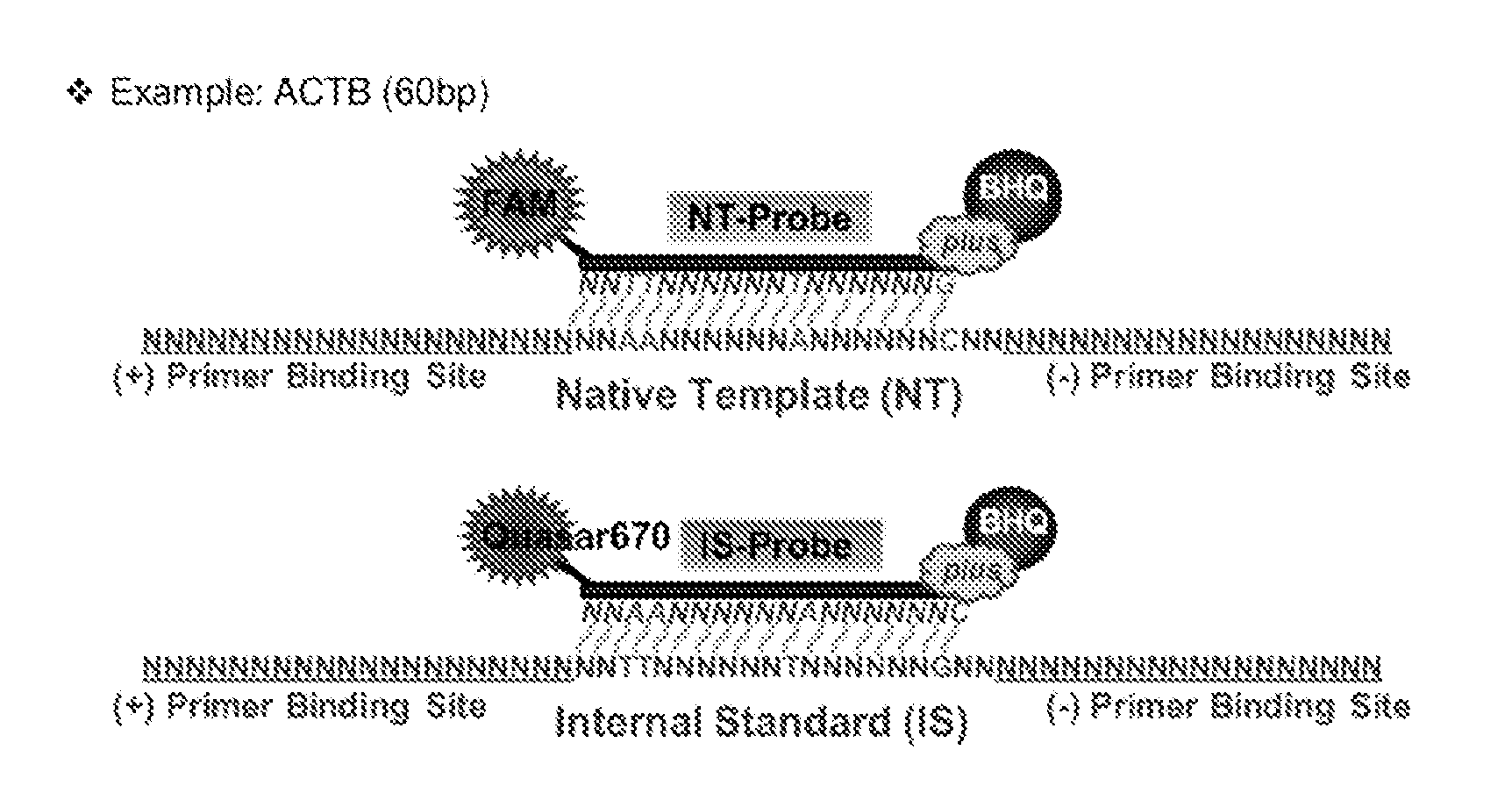
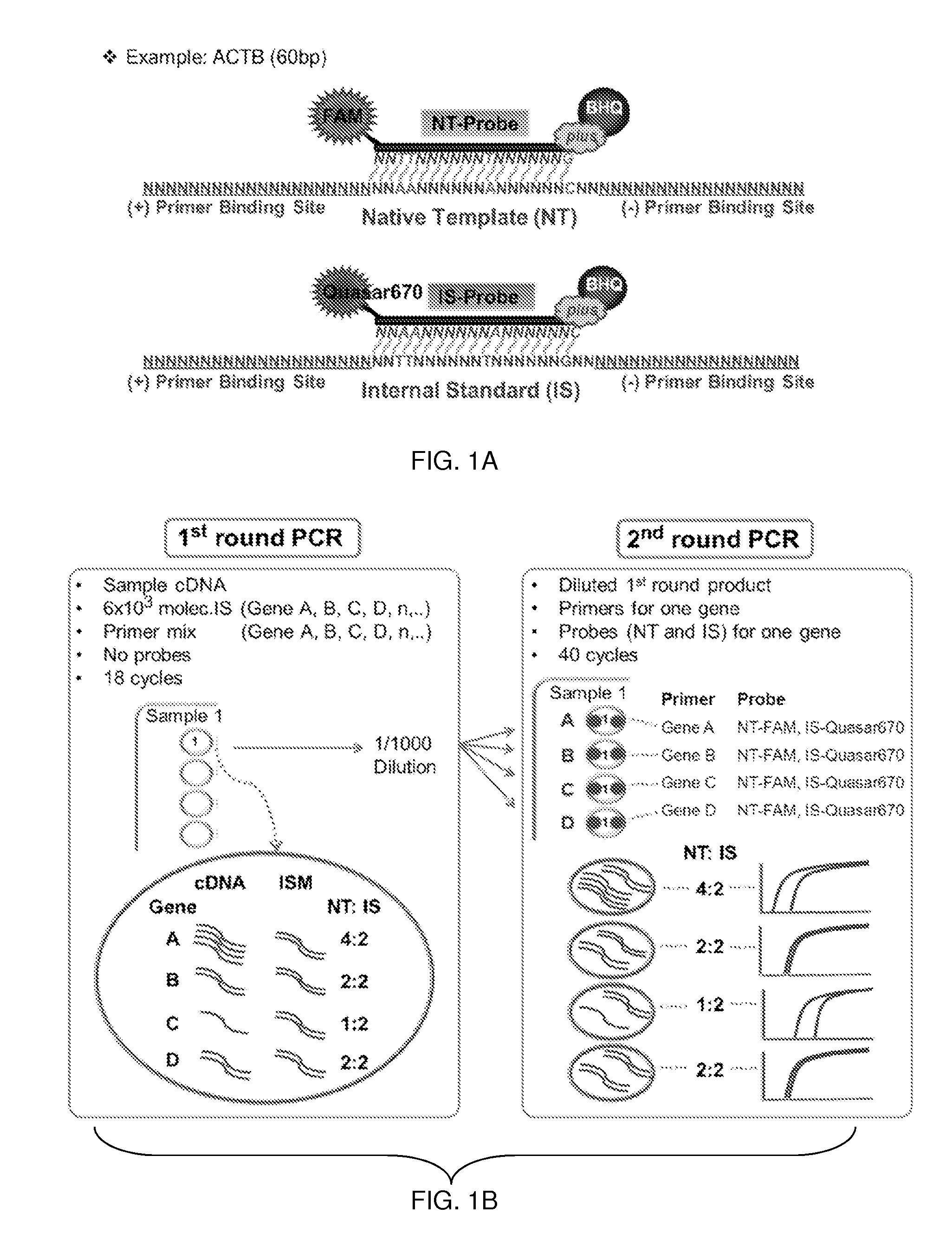
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
32 results about "Quantitative Real Time PCR" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Use Of Genes As Molecular Markers In Diagnosis Of Schizophrenia And Diagnostic Kit For The Same
InactiveUS20080274455A1Sugar derivativesMicrobiological testing/measurementQuantitative Real Time PCRScreening method
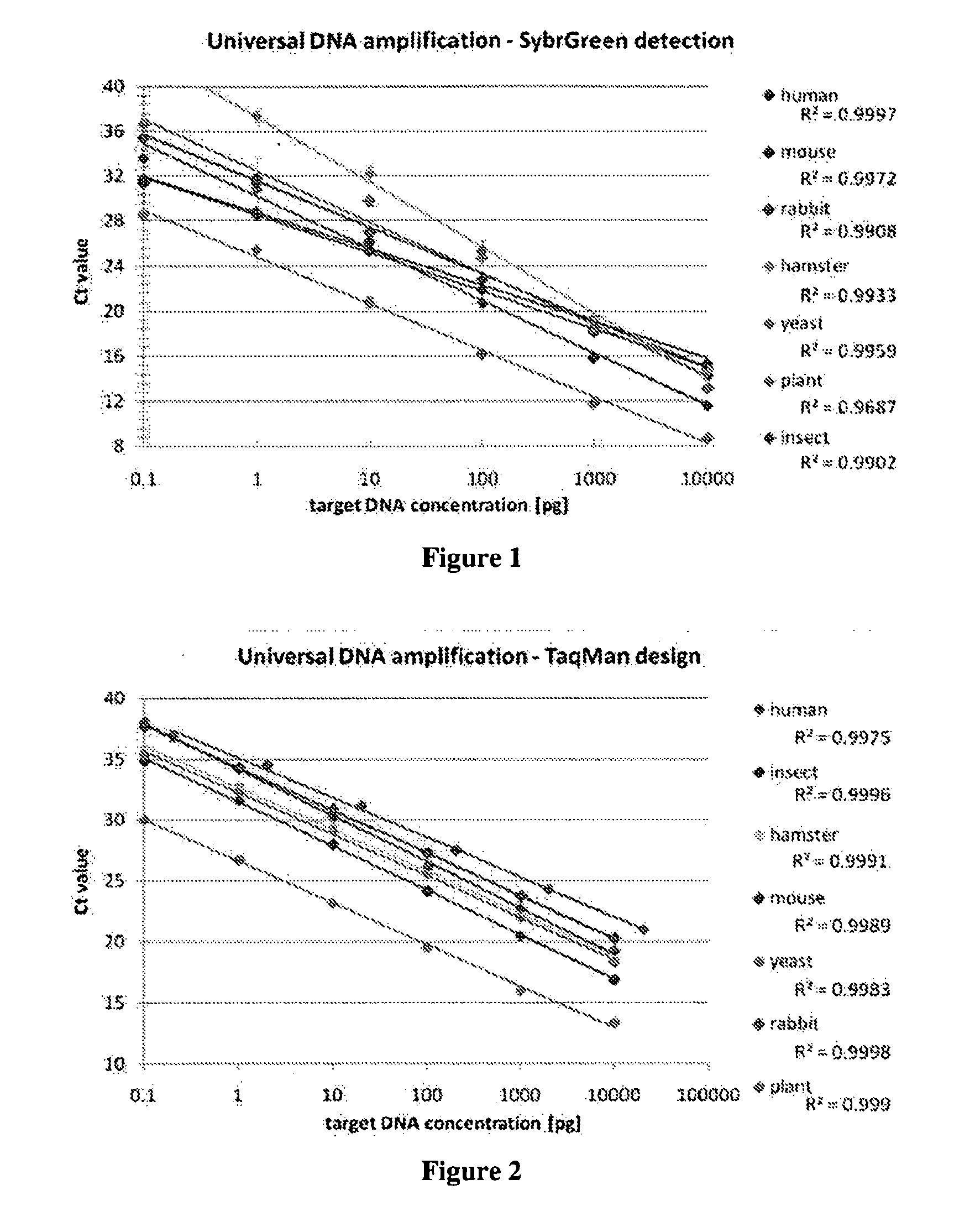
Drug-naive and drug-free schizophrenic PBL were screened to identify additional markers that are differentially expressed compared to healthy individuals using microarray and quantitative real-time PCR (QRT-PCR) techniques. Genes for dopamine D2 receptor (DRD2) and inwardly rectifying potassium channel (Kir2.3) were found to be overexpressed in microarray analysis. Increased mRNA levels were confirmed by QRT-PCR using SybrGreen method and dual labeled TaqMan probes.The invention relates to a method for diagnosing schizophrenia in a subject comprising assessing the level or the expression level of at least one of the following genes or proteins: Kir2.3 or DRD2 or a gene encoding Kir2.3 or DRD2. The invention further relates to agents and uses thereof, said agents specifically binding to said proteins or nucleic acids encoding them, diagnostic kits and screening methods.Use of both molecular markers allow prediction of schizophrenia and help to follow efficiency of drugs in therapy in order to provide a more tailored medication for schizophrenic patients.
Owner:THE BIOLOGICAL RES CENT OF THE HUNGARIAN ACAD OF SCI
Method for detecting and quantifying perchlorate-reducing bacteria
InactiveUS20090258349A1Enhanced hybridizationMicrobiological testing/measurementQuantitative Real Time PCRMicrobiology
The present invention provides methods, compositions, and kits for detecting and quantitating perchlorate-reducing bacteria in samples using reagents that hybridize to and allow amplification of the pcrA gene. The invention includes a quantitative real-time PCR assay for amplification of the pcrA gene which may be used to detect and quantitate perchlorate-reducing bacteria in samples.
Owner:RGT UNIV OF CALIFORNIA
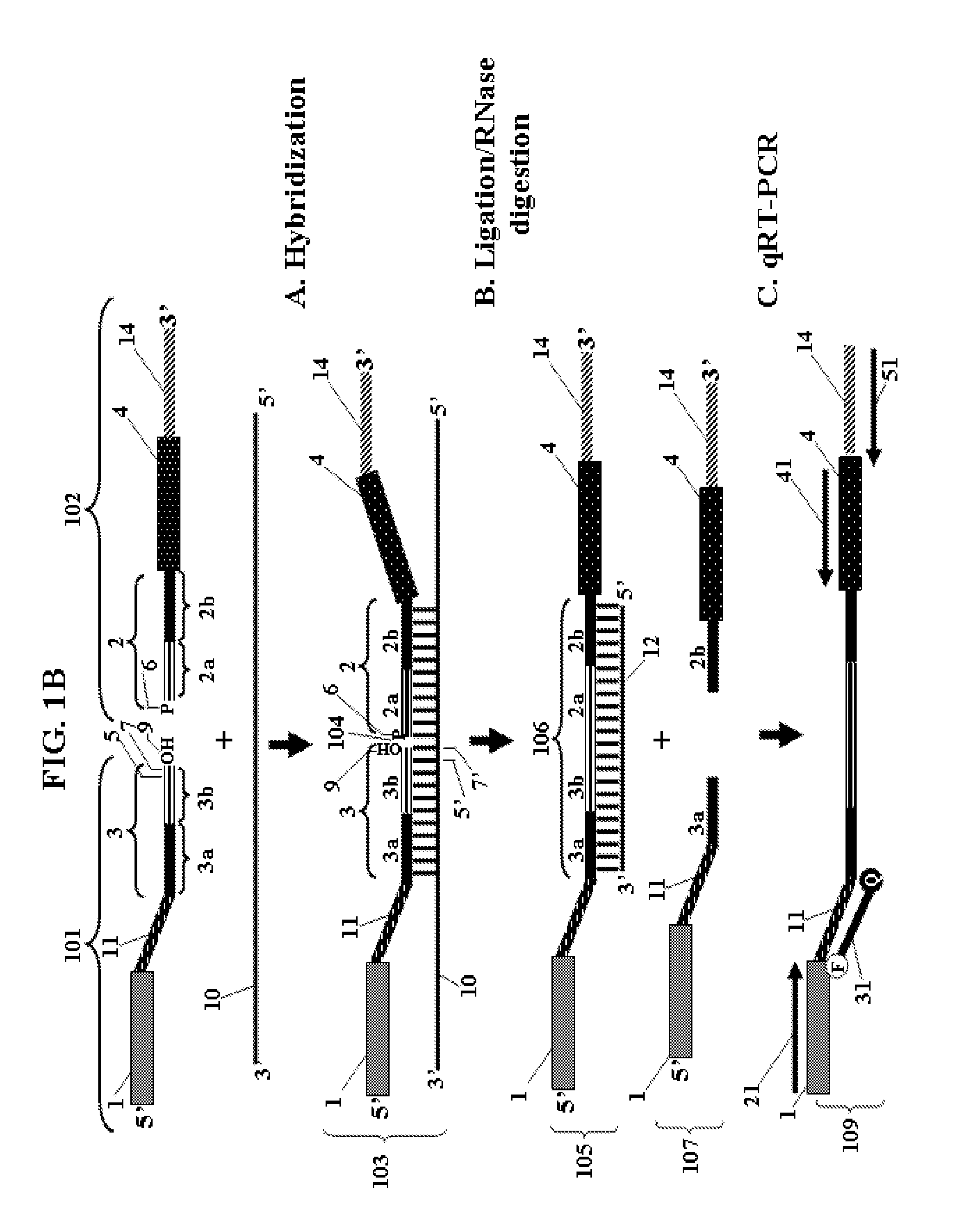
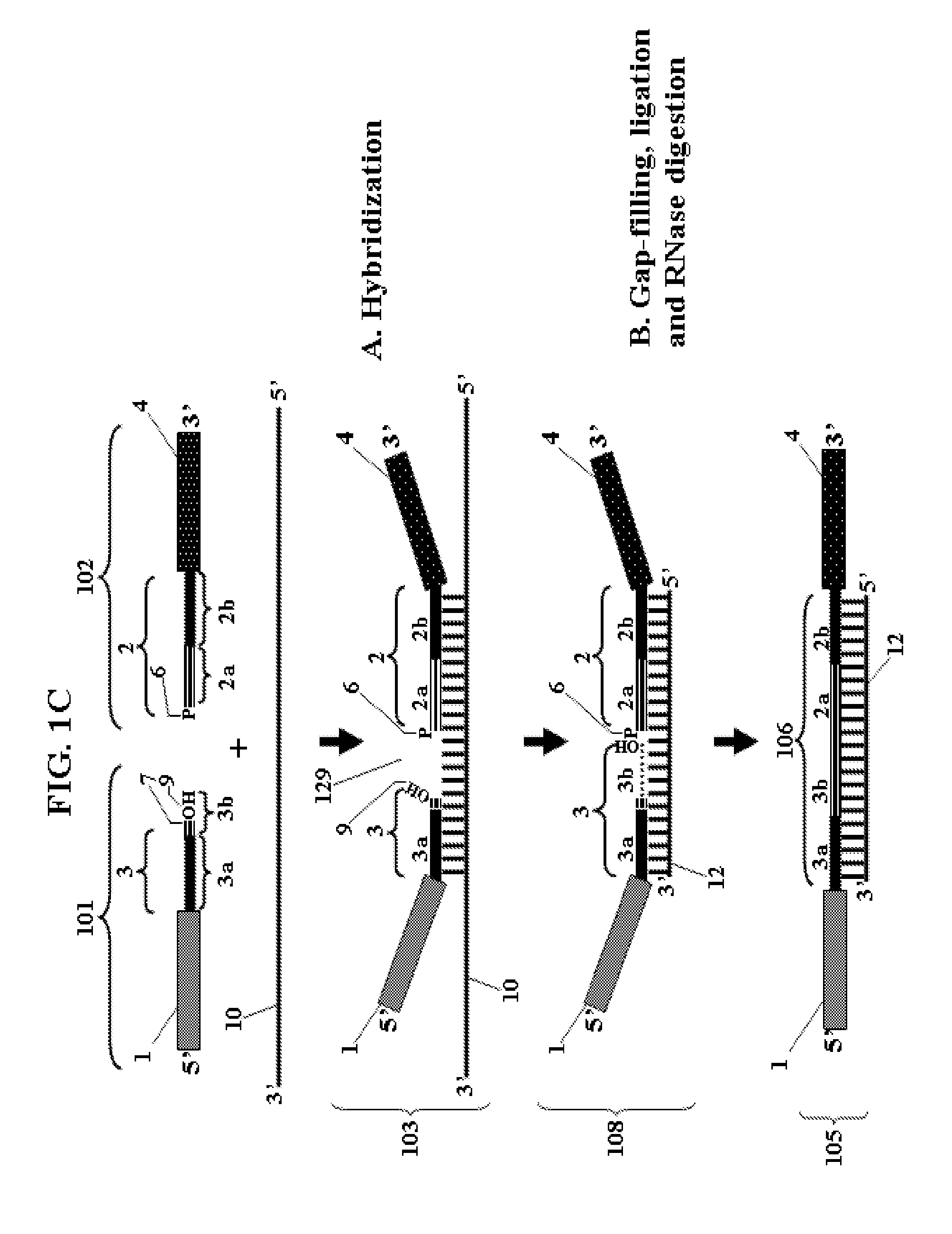
Chimeric oligonucleotides for ligation-enhanced nucleic acid detection, methods and compositions therefor
ActiveUS8008010B1Sugar derivativesMicrobiological testing/measurementQuantitative Real Time PCRCross-link
Ligation-enhanced nucleic acid detection assay embodiments for detection of RNA or DNA are described. The assay embodiments rely on ligation of chimeric oligonucleotide probes to generate a template for amplification and detection. The assay embodiments are substantially independent of the fidelity of a polymerase for copying compromised nucleic acid. Very little background amplification is observed and as few as 1000 copies of target nucleic acid can be detected. Method embodiments are particularly adept for detection of RNA from compromised samples such as formalin-fixed and paraffin-embedded samples. Heavily degraded and cross-linked nucleic acids of compromised samples, in which classic quantitative real time PCR assays typically fail to adequately amplify signal, can be reliably detected and quantified.
Owner:APPL BIOSYSTEMS INC
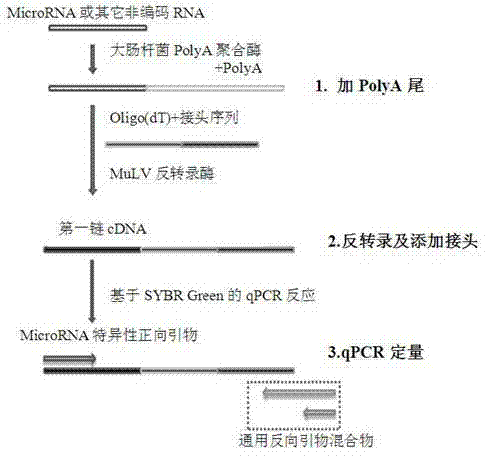
Quantitative detection method for micro RNA (Ribose Nucleic Acid)
ActiveCN102676677AImprove accuracyIncreased sensitivityMicrobiological testing/measurementFluorescence/phosphorescenceQuantitative Real Time PCRTotal rna
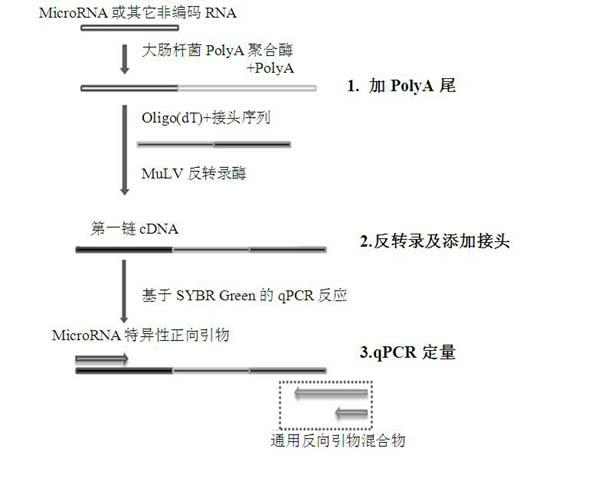
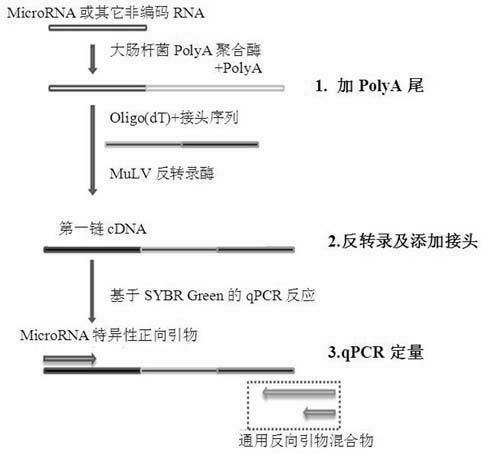
The invention discloses a quantitative detection method for micro RNA (Ribose Nucleic Acid), and the method comprises the following steps of: (1) extracting total RNA of a sample; (2) treating the total RNA by poly A polymerase, and adding a section of poly A tail at a 3' end of the mi RNA; (3) carrying out inverse transcription for oligo (dT) inverse transcription primer with joint sequence at a5' end, and adding a section of joint at the obtained first-chain c DNA (Deoxyribonucleic Acid); and (4) enabling sequence specificity of the mi RNA to be tested and general reverse primer complementary with the joint sequence to carry out quantitative real-time PCR (Polymerase Chain Reaction) amplification, wherein the general reverse primer is mixture of two types of reverse primers with different length. By means of the method disclosed by the invention, quantitative detection and analysis for mi RNA expression can be achieved, and the specificity, sensitivity and repeatability of the existing detection method can be greatly improved.
Owner:天津旷博同生生物技术有限公司
Quantification of residual host cell DNA by real-time quantitative PCR
The present invention relates to a method for quantifying residual host cell genomic DNA in recombinant protein biologics using quantitative real time PCR.
Owner:CONFARMA FRANCE
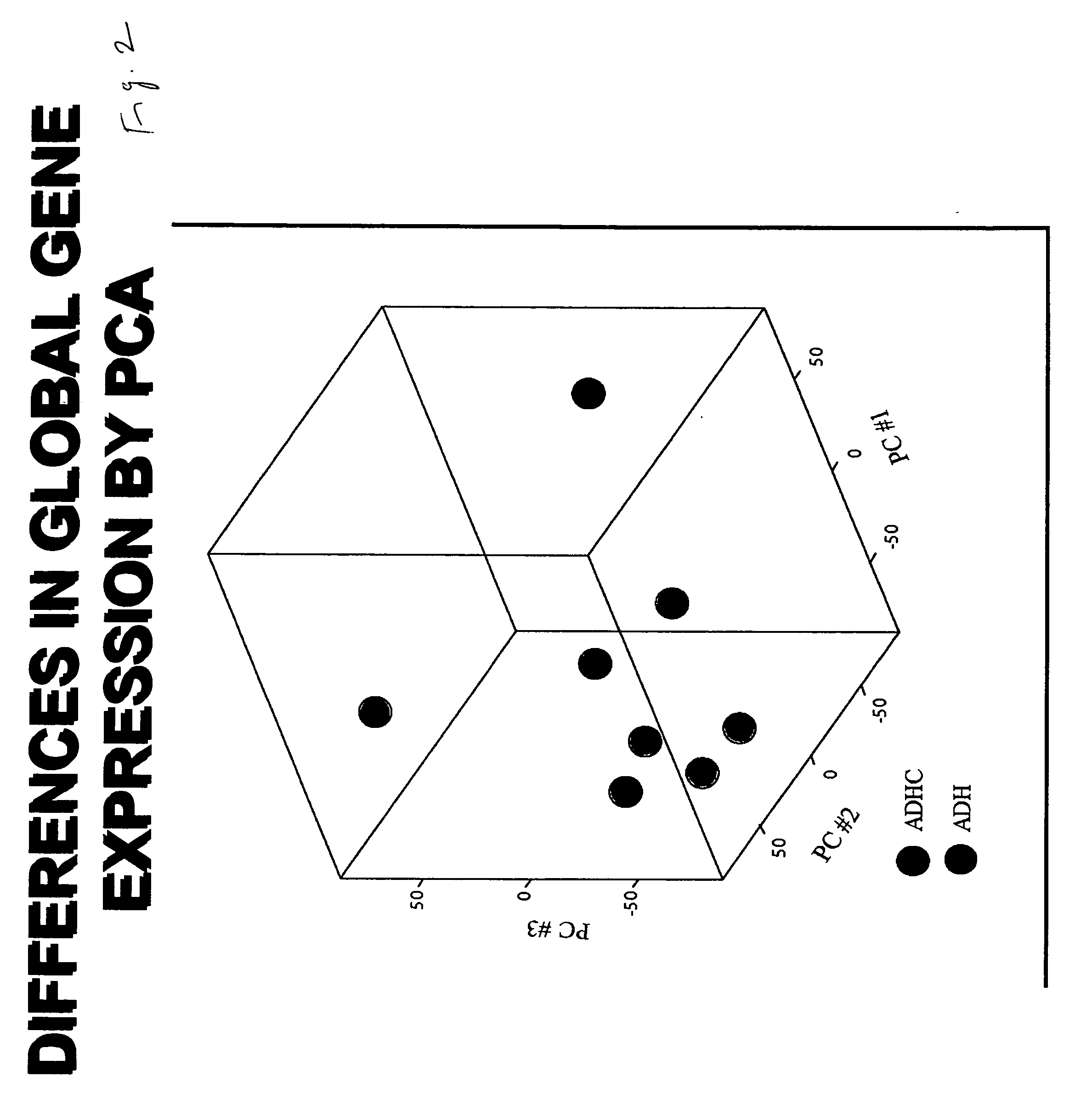
Molecular Markers that predict breast cancer development
InactiveUS20070254286A1Prevent hyperprolific conditionMicrobiological testing/measurementBiological material analysisDiseaseQuantitative Real Time PCR
A number of selected genes / gene products; Application of selected genes / gene products at mRNA or protein levels either singly or in combination; Application of selected genes / gene products at mRNA levels by any of the methods such as: Northern blotting, or reverse transcription and conventional PCR, or reverse transcription and quantitative real-time PCR or gene expression micro-arrays; Application of selected genes / gene products at protein levels by either Western Blotting, or immunohistochemistry, or ELISA or functional assays or gel electrophoretic separation followed by spectroscopic identification (proteamics); Application of selected genes / gene products at peptide levels derived from proteins and spectroscopic methods of identification; Detection of a hyperproliferative condition, a precancerous condition, a predisposition to develop hyperproliferative condition or cancer by applying any one of the selected genes either singly or in combination in breast tissue, breast fluid, breast cells, blood or any other tissues or cells of a mammal; Application of selected genes either singly or in combination with others for designing molecular therapeutic drugs to treat a hyperproliferative condition, a precancerous condition, or predisposition to develop hyperproliferative condition or cancer of the breast or any other tissue of a mammal; Application of selected genes either singly or in combination with others for following up of a therapeutic treatment to a hyperproliferative condition, or precancerous condition, or predisposition to a hyperproliferative condition, or cancer, of the breast or any other tissue of a mammal; Application of selected genes either singly or in combination with others for screening of therapeutic drugs to a hyperproliferative condition, or precancerous condition, or predisposition to a hyperproliferative condition, or cancer, of the breast or any other tissue of a mammal; and Application of selected genes either singly or in combination with others for designing vaccines to prevent a hyperproliferative condition, or precancerous condition, or predisposition to a hyperproliferative condition, or cancer, of the breast or any other tissue of a mammal.
Owner:SILBIOTECH
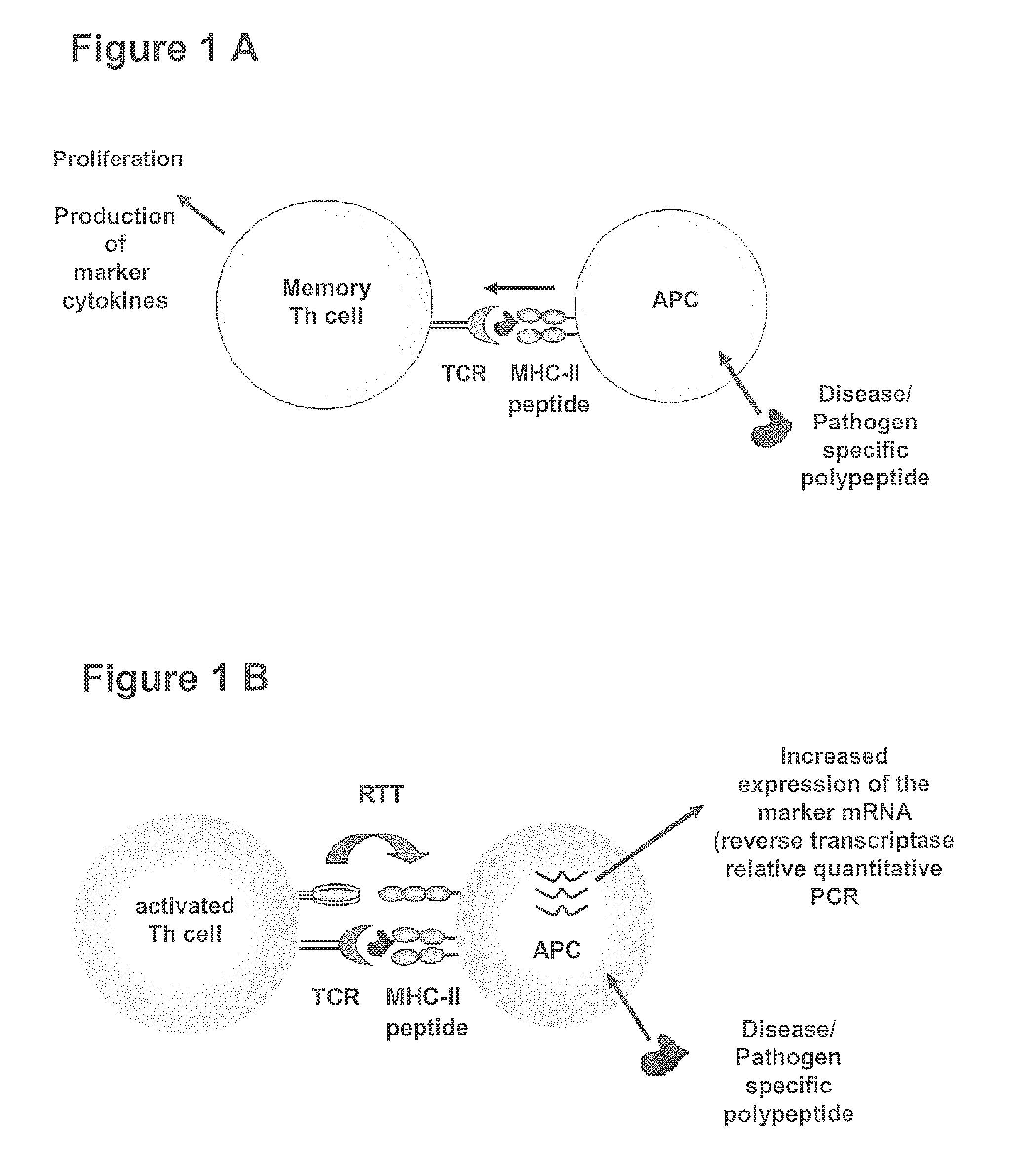
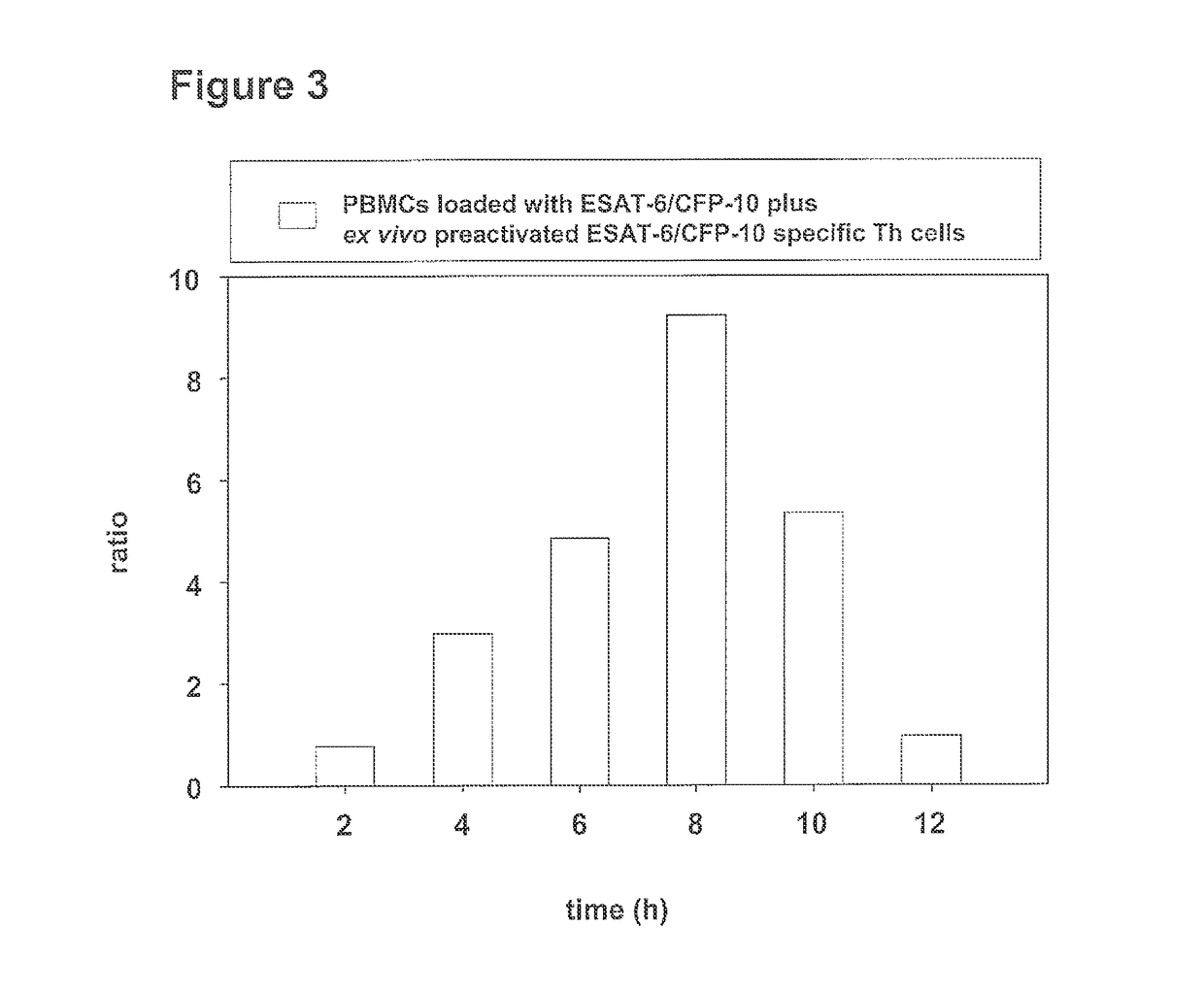
METHOD FOR DETECTION, DIFFERENTIATION AND QUANTIFICATION OF T CELL POPULATIONS BY WAY OF REVERSE TRANSCRIPTION QUANTITATIVE REAL TIME PCR (RT-qPCR) TECHNOLOGY
ActiveUS20130288229A1Microbiological testing/measurementMaterial analysisQuantitative Real Time PCRAntigen
The present invention relates to a method for detection, differentiation and quantification of T cell populations, comprising the following steps a) contacting a first aliquot of a body fluid of an individual with at least one antigen, wherein the body fluid contains antigen presenting cells (APC) and T cells, b) incubating the first aliquot with at least one antigen for a certain period of time, c) detection and differentiation of the T cell population by detecting in the first aliquot and in a second aliquot of the body fluid of the individual, which has not been incubated with the at least one antigen, at least a first marker of the APC induced by T cells in a specific T cell population using reverse transcription quantitative real time-time polymerase chain reaction (RT-qPCR), and d) detection and quantification of the T cell population by determining the ratio of the detected marker of the APC of the first aliquot to the second aliquot as well as a kit for performing the method.
Owner:LOPHIUS BIOSCI
Selection method of reference genes in quantitative real-time PCR analysis of jerusalem artichoke
InactiveCN108588191AStable expressionMicrobiological testing/measurementQuantitative Real Time PCRReference genes
The invention discloses a selection method of reference genes in quantitative real-time PCR analysis of jerusalem artichoke, and relates to the field of quantitative PCR. The method comprises the following steps: taking different tissues (radicles, immature stems, leaves, stem blocks and petals) of the jerusalem artichoke as materials, and carrying out expression analysis of the reference genes of18S ribosomal RNA gene (18S rRNA), transcription elongation factor gene (Ef-1a), actin gene (Actin and beta-actin), 3-glyceraldehyde phosphate dehydrogenase (GAPDH), 25S ribosomal RNA gene (25S rRNA)and poly-ubiquitin enzyme gene (UBQ)7 by using a q PCR technology; carrying out statistical assessment on the obtained data and analyzing expression change of all housekeeping genes by utilizing GeNorm and NormFinder software, so as to screen out relatively-stable genes as the reference genes of the jerusalem artichoke, which are used for studying the gene dosage changes of the jerusalem artichoke.
Owner:青海大学农林科学院
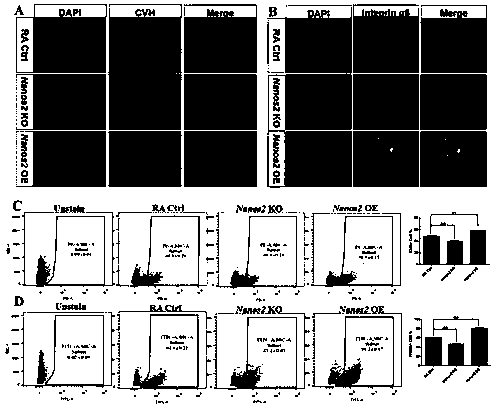
Method for function verification of Nanos2 in differentiation of chicken ESCs into male germ cells
InactiveCN109022484AHigh differentiation efficiencyImprove production efficiencyHydrolasesStable introduction of DNAQuantitative Real Time PCRPlant Germ Cells
The invention belongs to the field of animal breeding and cell biology, and particularly relates to a method for function verification of Nanos2 in differentiation of chicken ESCs into male germ cells, and the method includes detection of the expression level of the Nanos2 in different chicken tissues, Nanos2 clone and construction of pcDNA3.0-Nanos2 vector, Nanos2 knockout vector construction andknockout activity verification, in-vitro Nanos2 function verification, in-vivo Nanos2 function verification, Quantitative Real Time PCR (quantitative RT-PCR) verification, cell morphology observationand cellular immunochemical detection, flow cell analysis, paraffin section and PAS staining. Compared with the prior art, the function verification of the Nanos2 can be accomplished relatively quickly at the chicken germ cell level. The method is simple and easy to perform, has high repeatability and can be performed in an ordinary laboratory. The method greatly enhances the efficiency of in-vitro induction differentiation of the chicken ESCs into the male germ cells. The method greatly improves the efficiency of in-vivo induction generation of chicken PGCs and SSCs.
Owner:YANGZHOU UNIV
Method for providing information for diagnosing cancer using quantitative real-time PCR and kit for diagnosing cancer for the same
InactiveUS20120244531A1Strong specificityEasy to findMicrobiological testing/measurementQuantitative Real Time PCRReal-Time PCRs
Owner:M&D
Chimeric Oligonucleotides for Ligation-Enhanced Nucleic Acid Detection, Methods and Compositions Therefor
InactiveUS20120065105A1Nucleotide librariesMicrobiological testing/measurementQuantitative Real Time PCRCross-link
Owner:LIFE TECH CORP
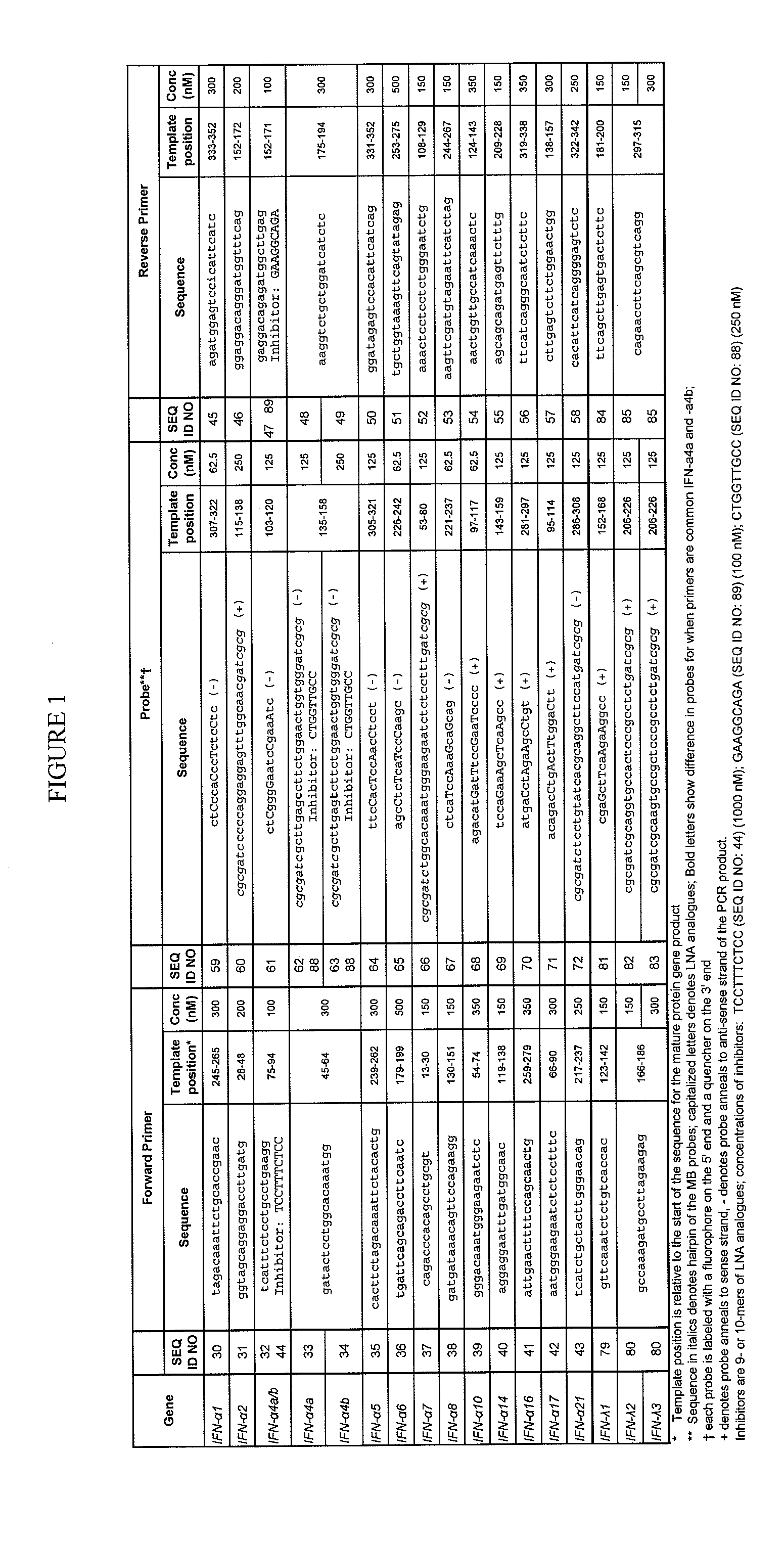
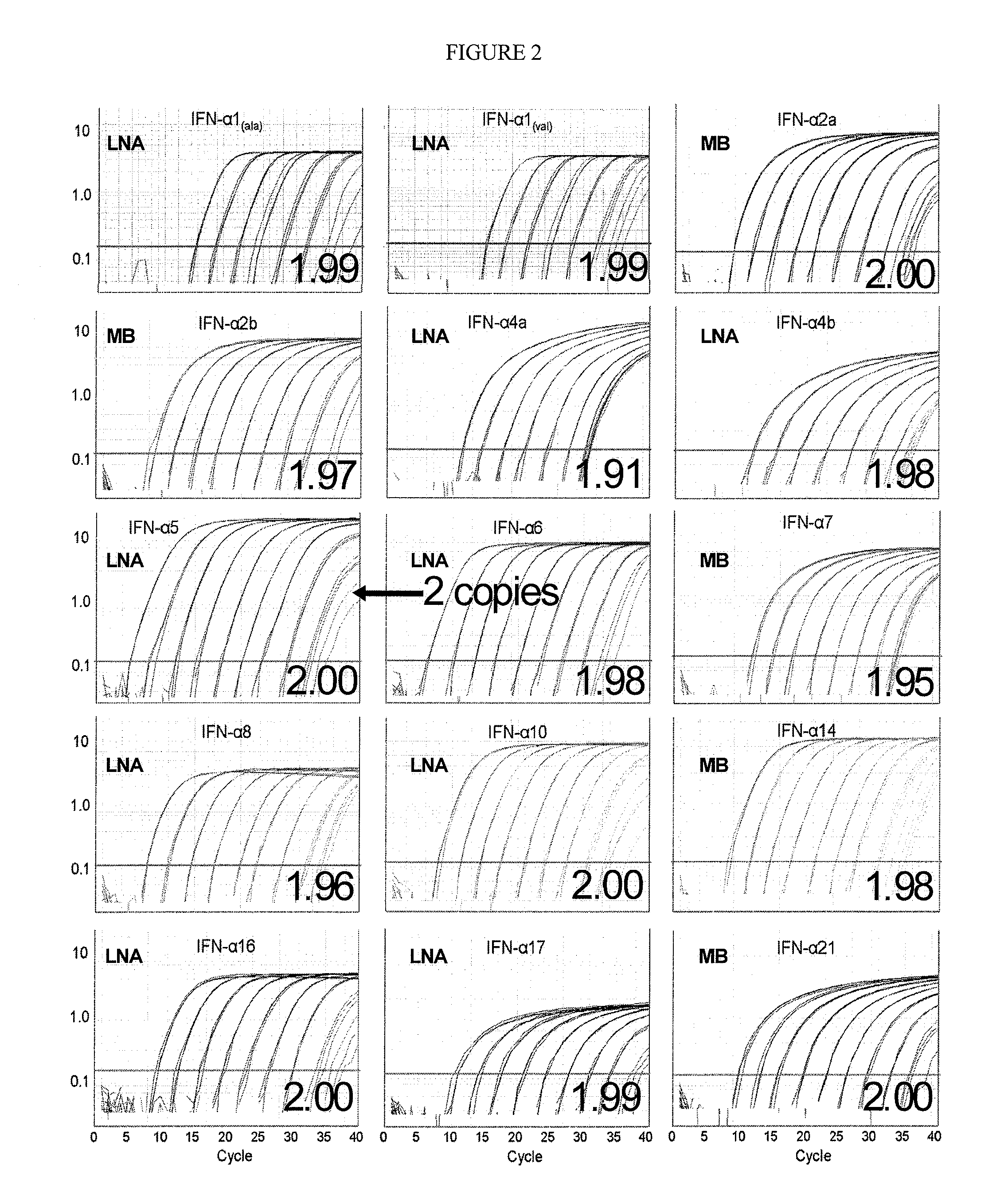
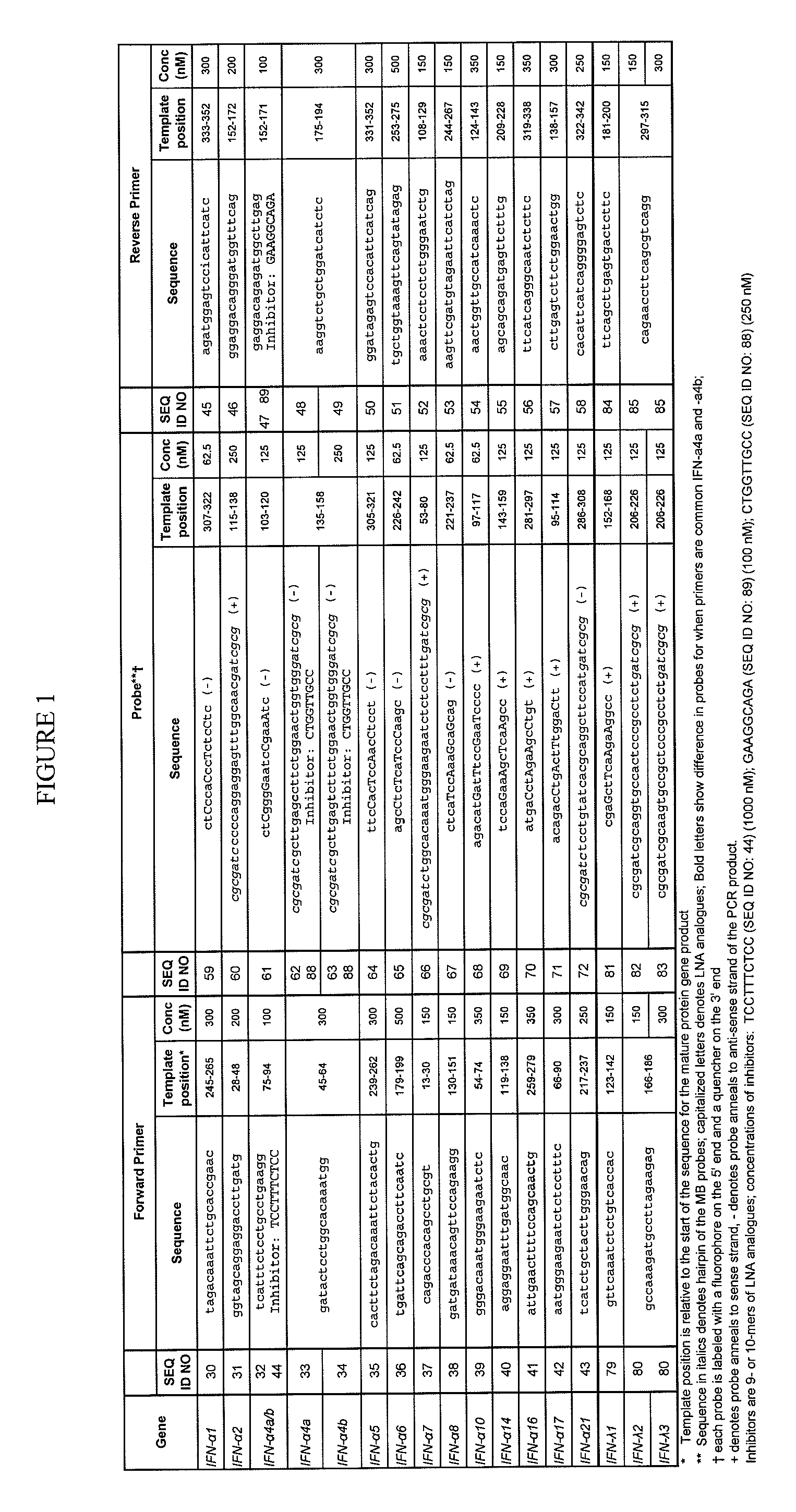
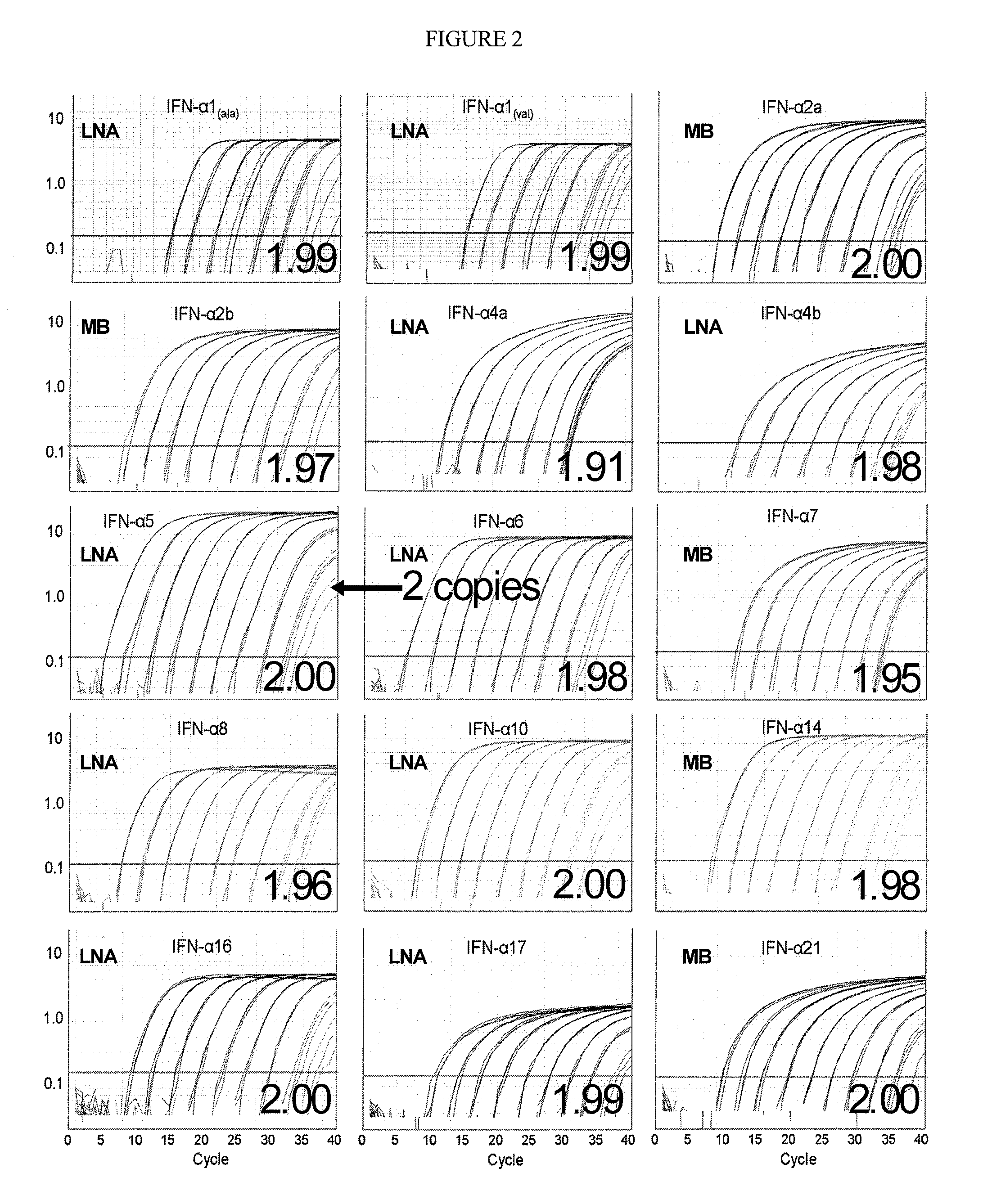
Compositions for detecting human interferon-alpha subtypes and methods of use
ActiveUS20110311973A1Good correlationGreat amplificationSugar derivativesMicrobiological testing/measurementQuantitative Real Time PCRTumor therapy
The invention provides highly sensitive, specific and efficient quantitative real-time PCR compositions, methods and assay kits to detect at least one IFN subtype and / or IFN subtype allotypic variants. Primer / probe sets complementary to the coding sequence of an IFN subtype of interest avoid spurious detection of degraded mRNA and enhances the correlation between the IFN subtype that is measured by the assays of the invention and the protein that is actually expressed. The invention also provides methods for designing primers and methods of using the compositions and assay kits. The compositions, kits, and methods of the invention may be used, for example, to monitor vaccine efficacy, autoimmune disease, chronic infections, or tumor therapy.
Owner:UNITED STATES OF AMERICA
Strong PCR primers and primer cocktails
InactiveUS7507535B2Efficient amplificationAccurate representationSugar derivativesMicrobiological testing/measurementQuantitative Real Time PCRMammal
The invention disclosed relates to a PCR primer pair for amplication of chaperonin-60 (cpn60) targets having high G+C content and to a PCR primer “cocktail” to improve the representation of diverse sequences in chaperonin-60 based PCR product libraries derived from complex templates. In previous cpn60-based and 16S rDNA-based studies of mammalian intestinal microbiota, it has been observed that some classes of organisms such as the Actinobacteria, which are known through culture-based studies to be present in large numbers in these environments, are underrepresented or even absent from PCR product libraries. Using library sequence data and reference cpn60 sequence data from cpnDB, the chaperonin sequence database, we designed a pair of PCR primers which can be used alone for higher G+C content targets and, when used in combination with a previously developed degenerate, universal cpn60 primer pair, improve the representation of complex templates with high G+C content. We have validated these primers using a combination of traditional and quantitative real-time PCR on both artificially constructed complex templates and biological samples. The development and optimization of this primer cocktail represents a significant advance in our ability to generate cpn60 PCR product libraries which more closely represent the biodiversity in complex microbial communities.
Owner:NAT RES COUNCIL OF CANADA
Materials and Methods for Quality-Controlled Two-Color RT-QPCR Diagnostic Testing of Formalin Fixed Embedded and/or Fresh-Frozen Samples
InactiveUS20150225798A1Nucleotide librariesMicrobiological testing/measurementQuantitative Real Time PCRFluorescence
Described herein are materials, methods, and kits enabling accurate and reproducible two-color reverse-transcription real-time quantitative PCR (RT-qPCR) for quality-controlled molecular diagnostic testing of samples that may contain degraded RNA. In certain aspects described herein are materials, methods, and kits for use in the molecular diagnostic testing of lung cancer in FFPE samples and / or fresh-frozen samples. Also described herein are materials and methods to control for inter-experimental variation occurring during two-color RT-qPCR amplification arising from variation in fluorescence specific activity, use of different thermocyclers, and inter-laboratory differences.
Owner:UNIVERSITY OF TOLEDO
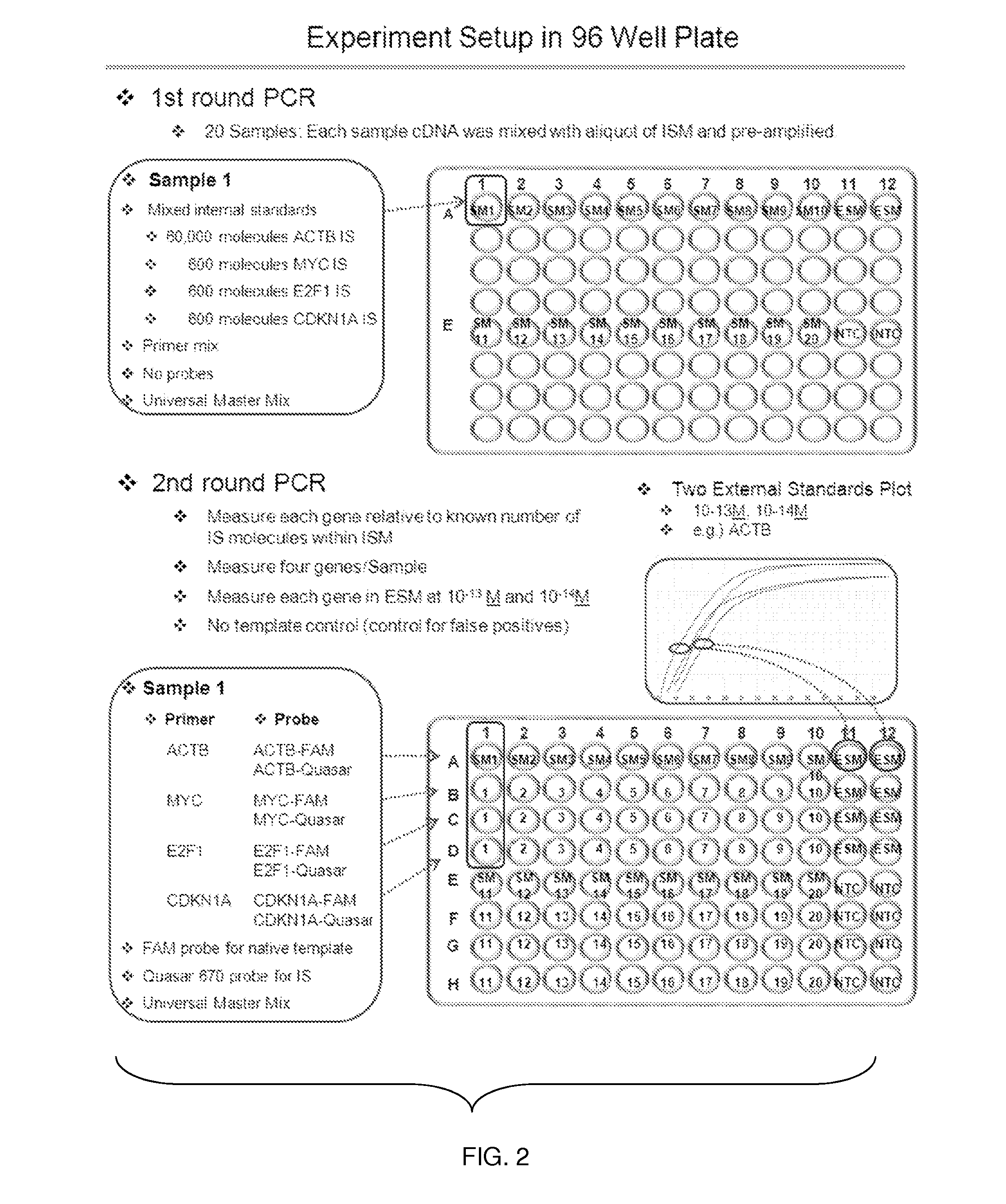
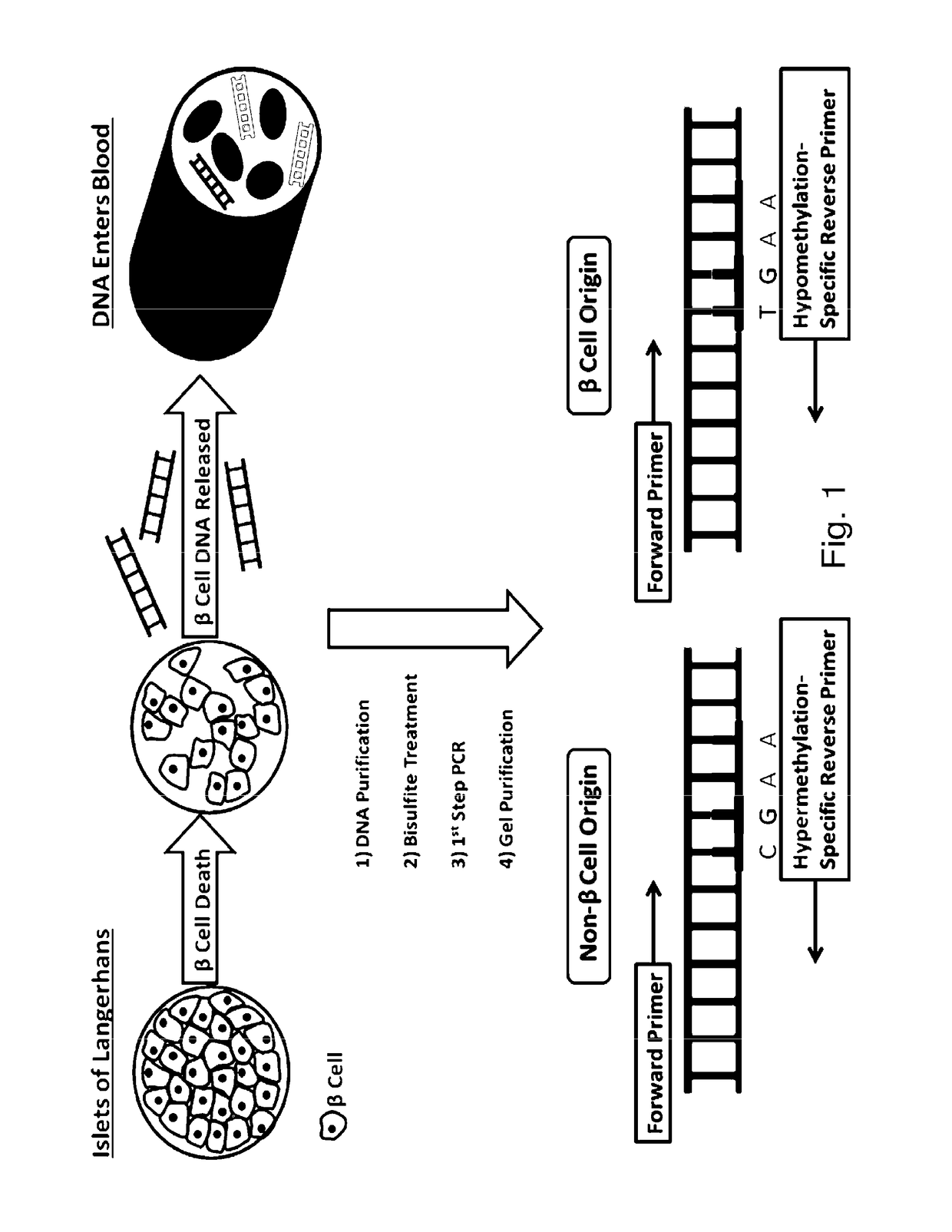
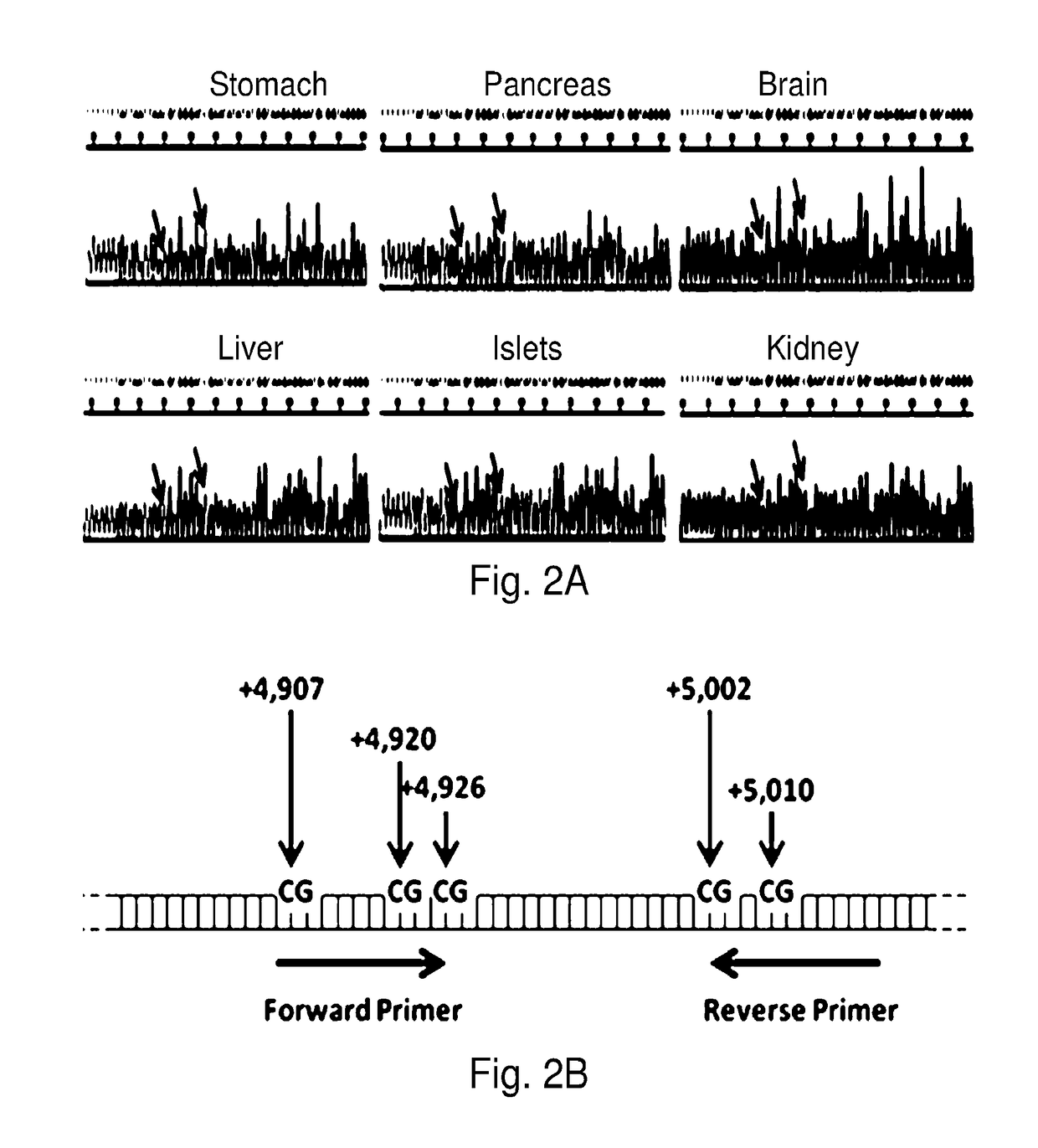
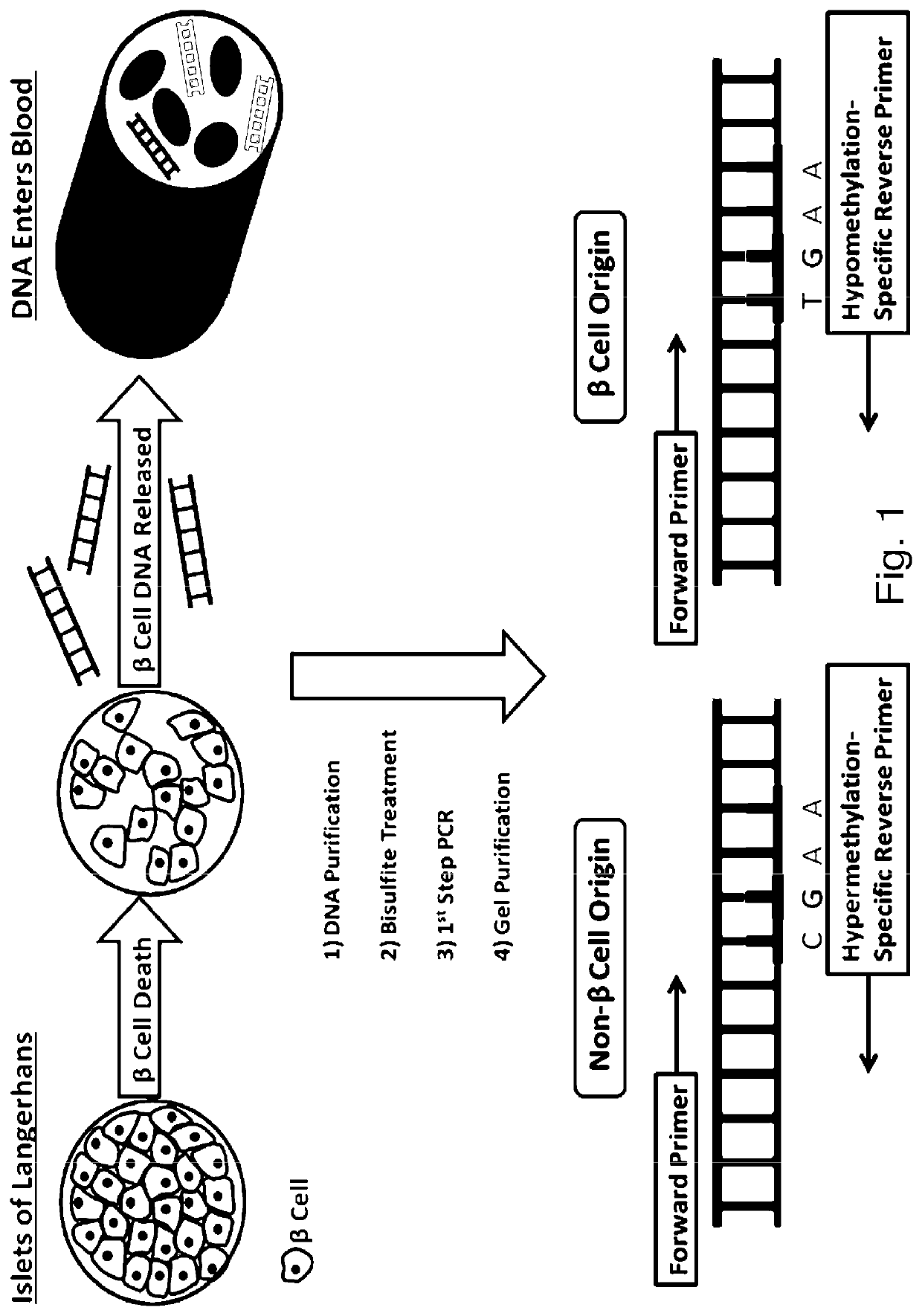
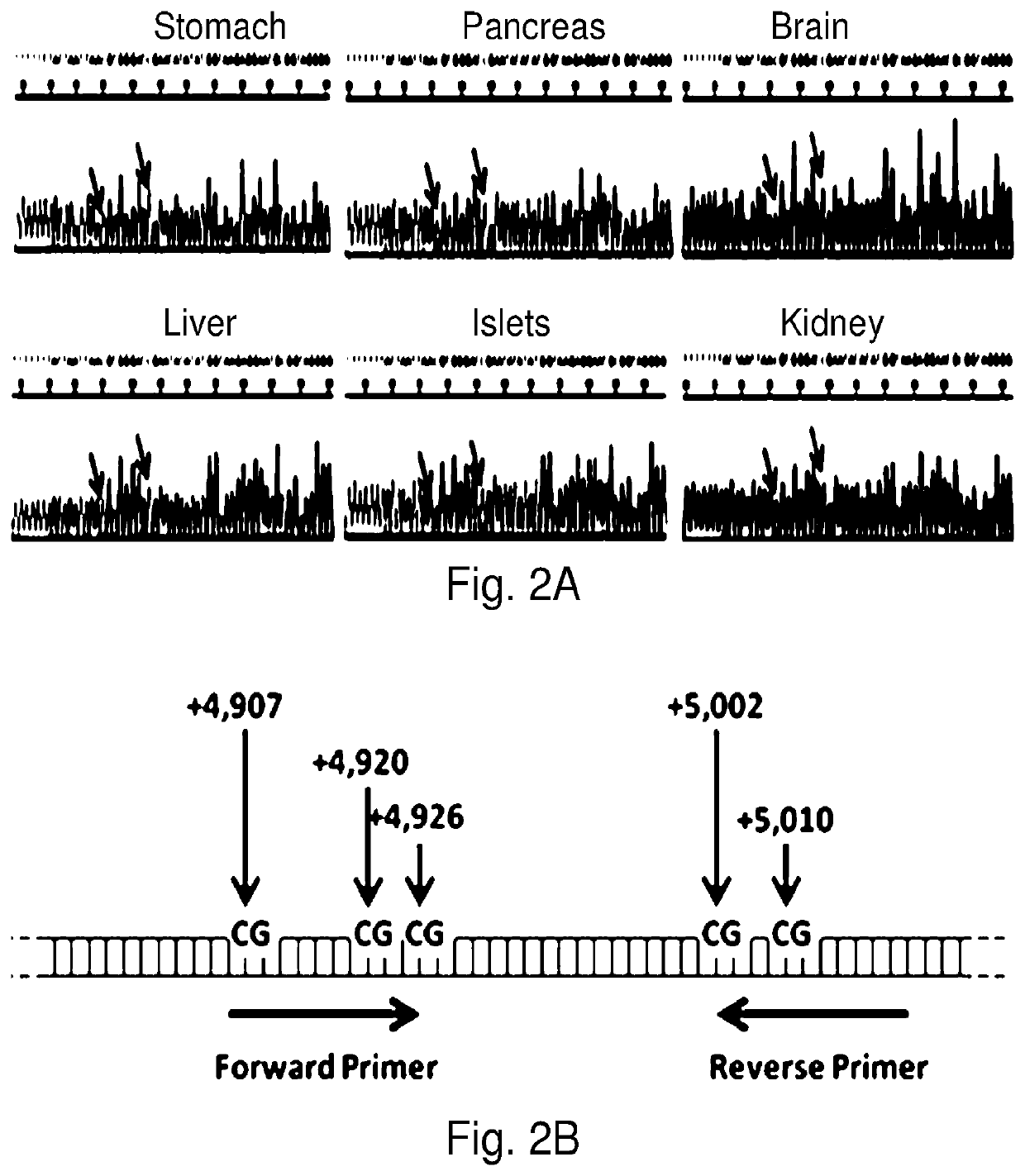
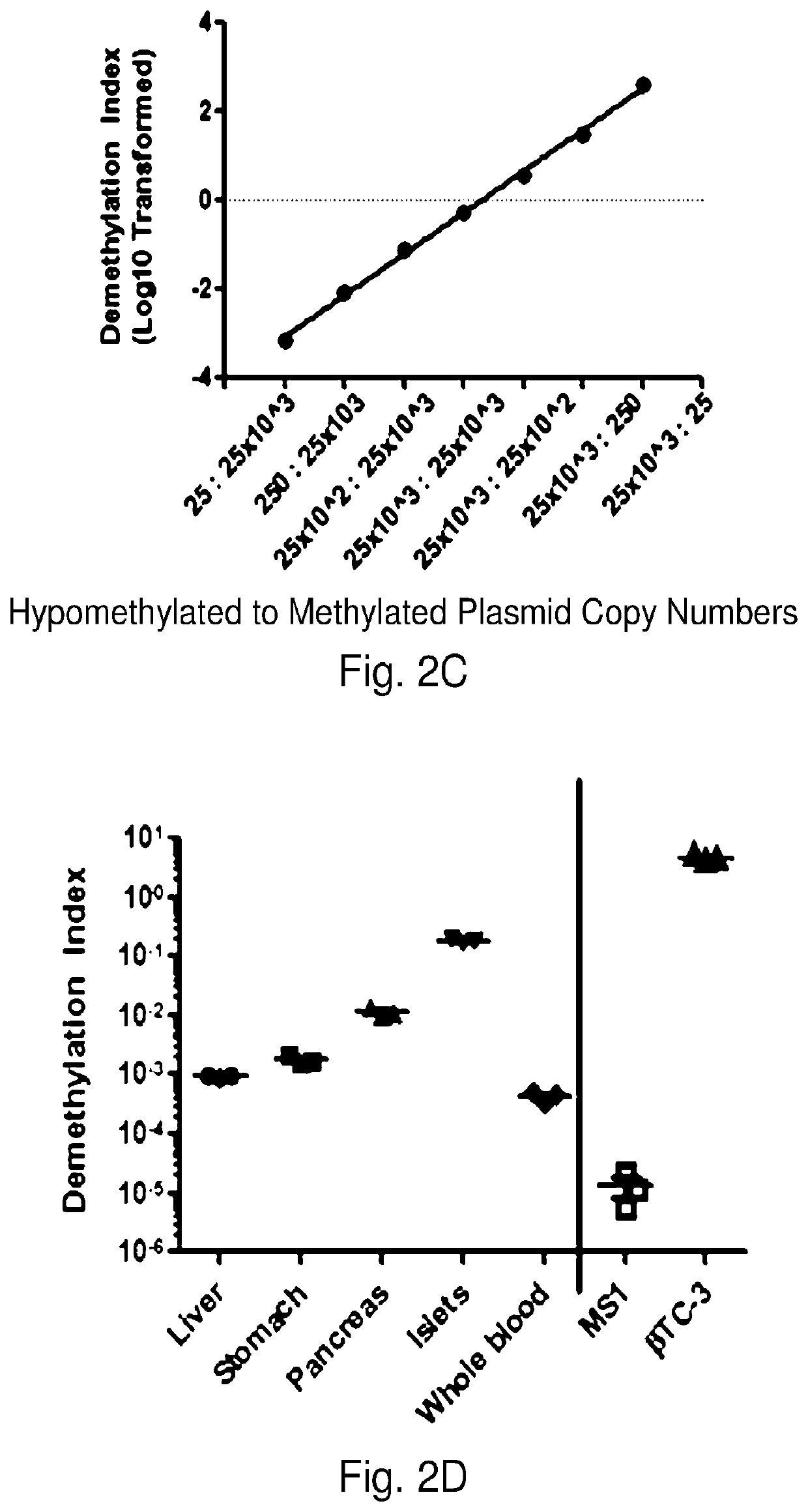
System, method and kit for analysis of circulating differentially methylated DNA as a biomarker of beta-cell loss
ActiveUS20180208985A1Reducing and preventing degradationInhibitory activityMicrobiological testing/measurementCell specificQuantitative Real Time PCR
β-cell loss in In Type 1 diabetes is typically undetected until the development of hyperglycemia, at which point β-cell mass is significantly reduced. Methylation sensitive quantitative real time PCR (qRTPCR) of demethylated circulating free β-cell specific DNA can be used as a biomarker of β-cell death. Such DNA includes insulin gene and amylin gene DNA. Detection may be by determination of a gene demethylation index. Methylated and demethylated DNA may be distinguished by bisulfite treatment and use of specific PCR primers or probes to detect the different bisulfite treatment products. Detection of demethylated circulating free amylin DNA is useful in identifying β-cell death. The amylin DNA may be used in conjunction with other β-cell specific genes, such as insulin, to provide a multi-gene approach towards the detection of β-cell loss.
Owner:NYU WINTHROP HOSPITAL
Method of detecting and quantifying coccidioides species
ActiveUS9404161B2High sensitivityGood suitSugar derivativesMicrobiological testing/measurementQuantitative Real Time PCRMicrobiology
Owner:TRANSLATIONAL GENOMICS RESEARCH INSTITUTE
Strong PCR primers and primer cocktails
InactiveUS20070072206A1Efficient amplificationAccurate representationSugar derivativesMicrobiological testing/measurementChaperonin 60Quantitative Real Time PCR
The invention disclosed relates to a PCR primer pair for amplication of chaperonin-60 (cpn60) targets having high G+C content and to a PCR primer “cocktail” to improve the representation of diverse sequences in chaperonin-60 based PCR product libraries derived from complex templates. In previous cpn60-based and 16S rDNA-based studies of mammalian intestinal microbiota, it has been observed that some classes of organisms such as the Actinobacteria, which are known through culture-based studies to be present in large numbers in these environments, are underrepresented or even absent from PCR product libraries. Using library sequence data and reference cpn60 sequence data from cpnDB, the chaperonin sequence database, we designed a pair of PCR primers which can be used alone for higher G+C content targets and, when used in combination with a previously developed degenerate, universal cpn60 primer pair, improve the representation of complex templates with high G+C content. We have validated these primers using a combination of traditional and quantitative real-time PCR on both artificially constructed complex templates and biological samples. The development and optimization of this primer cocktail represents a significant advance in our ability to generate cpn60 PCR product libraries which more closely represent the biodiversity in complex microbial communities.
Owner:NAT RES COUNCIL OF CANADA
High-sensitivity BHV-2 (bovine herpes virus 2) quantitative real-time PCR (polymerase chain reaction) detection method and kit
InactiveCN106521038AStrong specificityHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationQuantitative Real Time PCRDNA fragmentation
The invention provides a kit and a method for detecting BHV-2 (bovine herpes virus 2) and particularly discloses a high-sensitivity BHV-2 quantitative real-time PCR (polymerase chain reaction) detection method and a kit. The kit comprises a primer group and a fluorescent probe for detecting the BHV-2; the primer group comprises four primers for nested amplification of specific DNA fragments of the BHV-2. With the adoption of the detection method, nested amplification and quantitative real-time PCR can be completed in one tube through one-step amplification by the aid of the four primers and the fluorescent probe. With the adoption of the BHV-2 quantitative real-time PCR detection method and the kit with high sensitivity, specificity and accuracy, quick detection of the BHV-2 can be realized, the kit is very convenient to apply, and on the basis of the advantages, the kit is suitable to be popularized and applied in livestock breeding units, animal inspection and quarantine departments and the like and has wide application prospect.
Owner:BEIJING INST OF GENOMICS CHINESE ACAD OF SCI CHINA NAT CENT FOR BIOINFORMATION
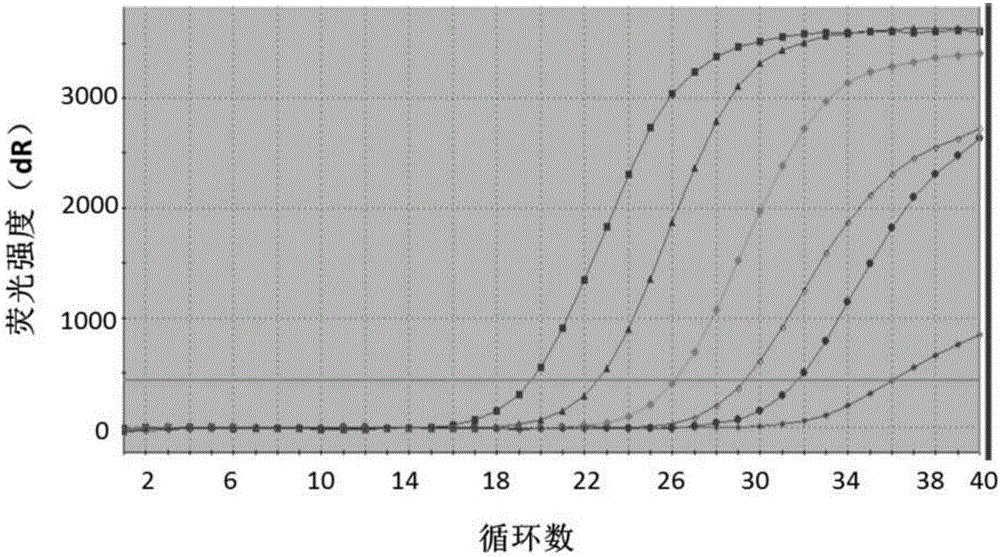
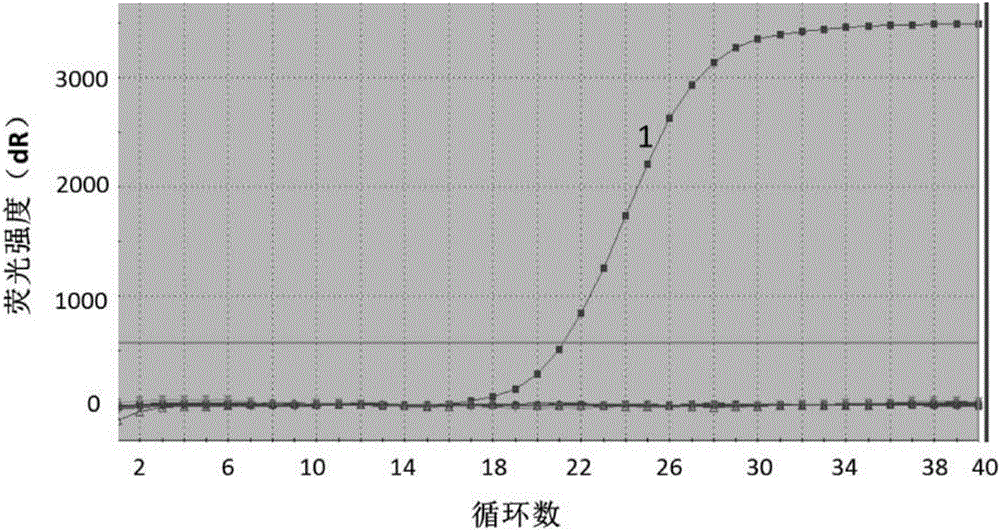
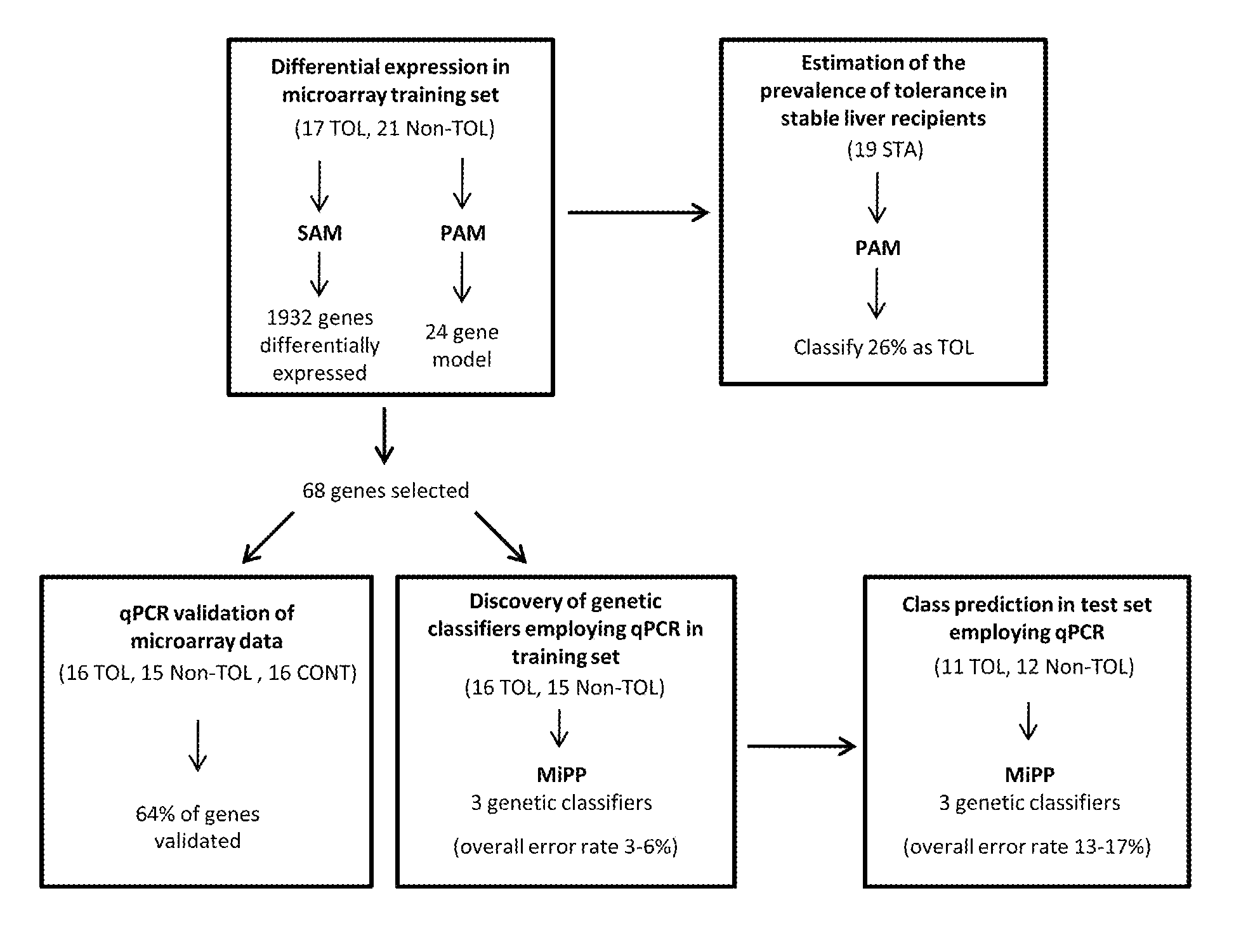
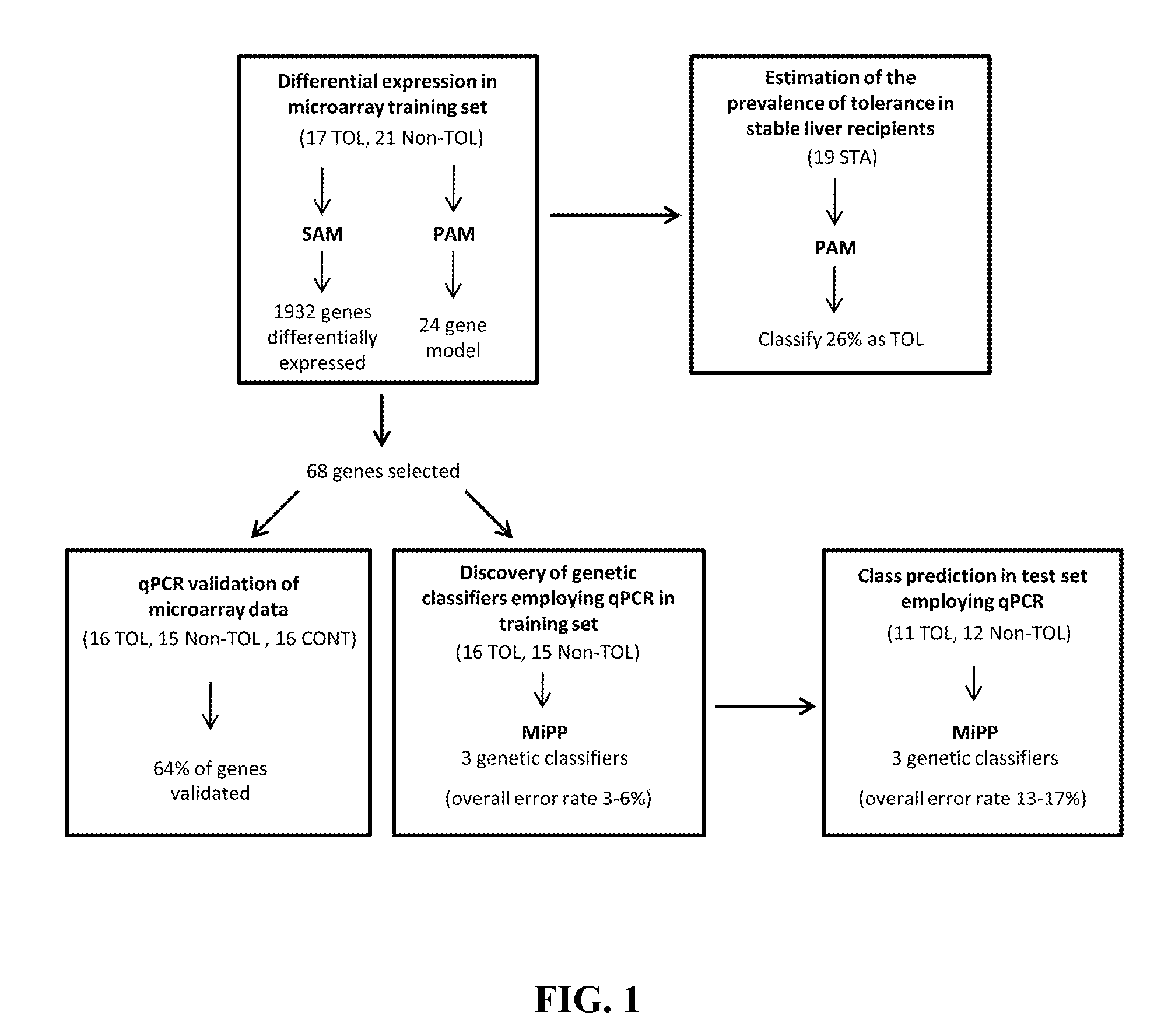
In vitro diagnosis/prognosis method and kit for assessment of tolerance in liver transplantation
InactiveUS20110130303A1Improve diagnostic accuracyImprove accuracyBioreactor/fermenter combinationsPeptide librariesQuantitative Real Time PCRLiver transplant recipient
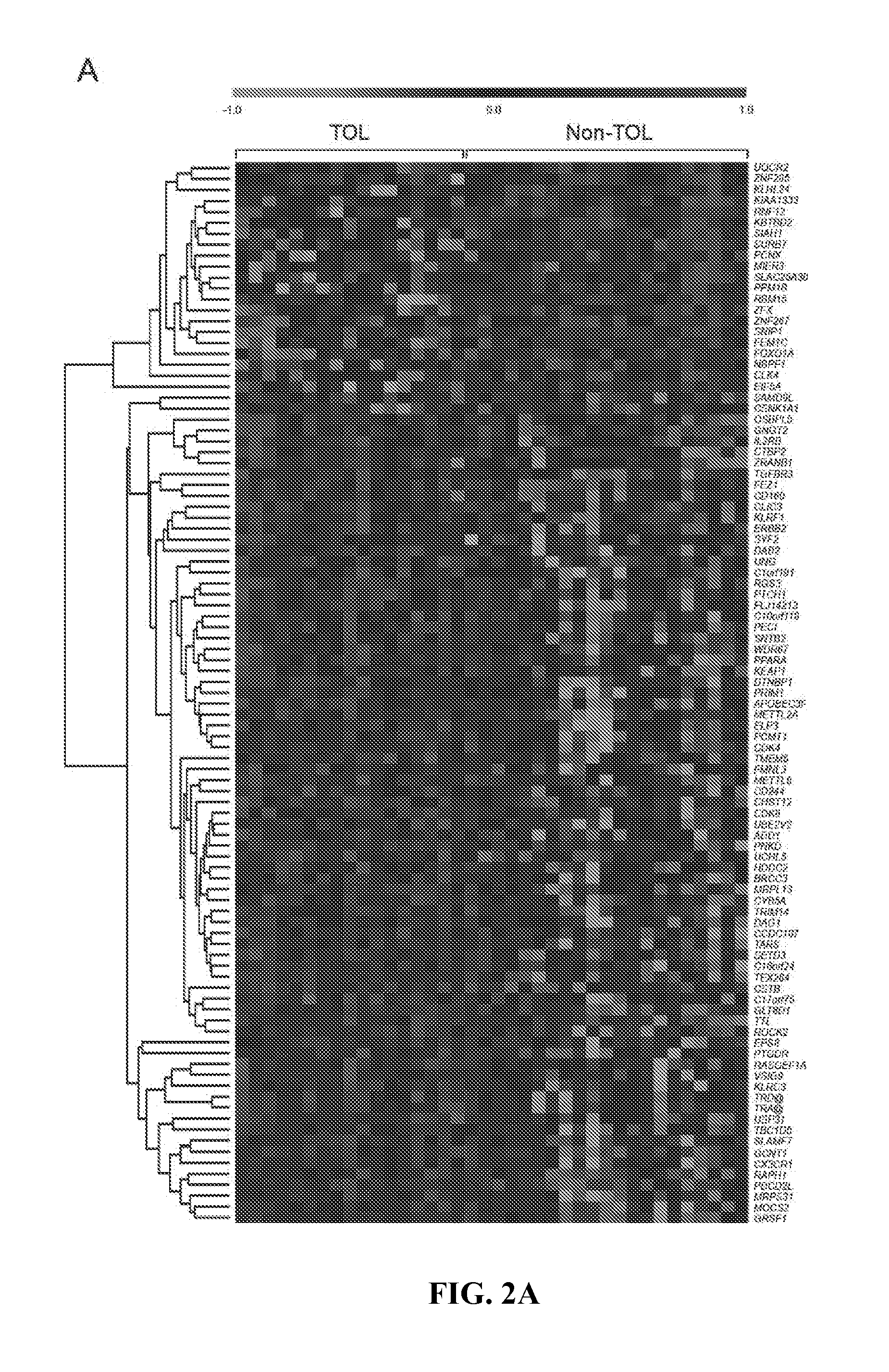
In vitro diagnosis / prognosis method and kit, for assessment of tolerance in liver transplantation. The present invention refers to the study of peripheral blood transcriptional patterns from 80 liver transplant recipients and 16 non-transplanted healthy individuals employing either oligonucleotide microarrays and / or quantitative real-time PCR to design a clinically applicable molecular test. This has resulted in the discovery and validation of several gene signatures comprising a modest number of genes capable of identifying tolerant and non-tolerant recipients with high accuracy. The marker genes are KLRF1, SLAMF7, NKG7, IL2RB, KLRB1, FANCG, GNPTAB, CLIC3, PSMD14, ALG8, CX3CR1, RGS 3. Multiple peripheral blood lymphocyte subsets contribute to the tolerance-associated transcriptional patterns with NK and γdelta T cells exerting a predominant influence. The invention concludes that transcriptional profiling of peripheral blood can be employed to identify liver transplant recipients who can discontinue immunosuppressive therapy and that innate immune cells are likely to play a major role in the maintenance of operational tolerance in liver transplantation.
Owner:INST DINVESTIGACIONS BIOMEDIQUES AUGUST PI I SUNYER IDIBAPS
Method of detecting and quantifying coccidioides species
ActiveUS20140011693A1High sensitivityGood suitBioreactor/fermenter combinationsBiological substance pretreatmentsQuantitative Real Time PCRMicrobiology
Owner:TRANSLATIONAL GENOMICS RESEARCH INSTITUTE
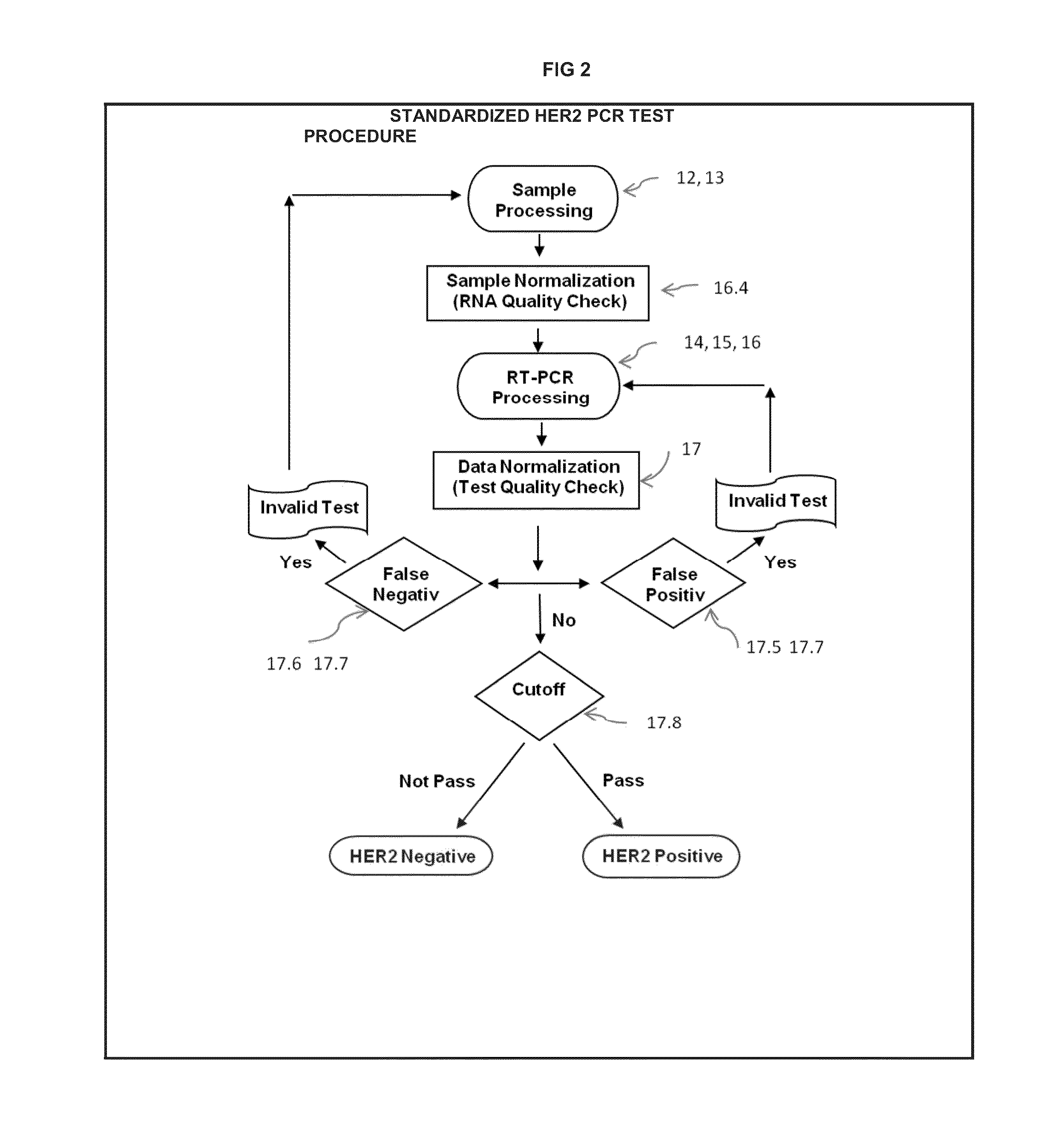
Cutoff Point Delta Ct. Method for HER2 PCR Testing in Breast Cancer
ActiveUS20140335525A1Heart valvesMicrobiological testing/measurementQuantitative Real Time PCRReference genes
The present invention is related to an improved method for HER2 gene test by using quantitative real-time PCR (Polymerase Chain Reaction) technique. Our invention streamlines test process, and incorporates quality control for each major step, including sample, reagent, operation, and data report. We eliminate the need for reference genes which is hard to standardize in HER2 PCR test. We develop a cutoff reference point by using the statistical mean of tumor tissue population, and adopt a simplified scoring scheme for evaluation of HER2 status. Our invention produces consistent result across machines and labs, and has proven to be clinically successful in HER2 test.
Owner:GENETICS DEV
Compositions for detecting human interferon-alpha subtypes and methods of use
ActiveUS9193995B2Good correlationGreat amplificationSugar derivativesMicrobiological testing/measurementQuantitative Real Time PCRTumor therapy
Owner:UNITED STATES OF AMERICA
Quantitative detection method for micro RNA (Ribose Nucleic Acid)
ActiveCN102676677BImprove accuracyIncreased sensitivityMicrobiological testing/measurementFluorescence/phosphorescenceQuantitative Real Time PCRTotal rna
The invention discloses a quantitative detection method for micro RNA (Ribose Nucleic Acid), and the method comprises the following steps of: (1) extracting total RNA of a sample; (2) treating the total RNA by poly A polymerase, and adding a section of poly A tail at a 3' end of the mi RNA; (3) carrying out inverse transcription for oligo (dT) inverse transcription primer with joint sequence at a5' end, and adding a section of joint at the obtained first-chain c DNA (Deoxyribonucleic Acid); and (4) enabling sequence specificity of the mi RNA to be tested and general reverse primer complementary with the joint sequence to carry out quantitative real-time PCR (Polymerase Chain Reaction) amplification, wherein the general reverse primer is mixture of two types of reverse primers with different length. By means of the method disclosed by the invention, quantitative detection and analysis for mi RNA expression can be achieved, and the specificity, sensitivity and repeatability of the existing detection method can be greatly improved.
Owner:天津旷博同生生物技术有限公司
System, method and kit for analysis of circulating differentially methylated DNA as a biomarker of beta-cell loss
ActiveUS10858706B2Improve information availableAccelerated deathMicrobiological testing/measurementQuantitative Real Time PCRCell mass
Owner:NYU WINTHROP HOSPITAL
A highly sensitive bhv-2 real-time fluorescent quantitative PCR detection method and kit
InactiveCN106521038BHigh sensitivityStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationQuantitative Real Time PCRFluorescence
The invention provides a kit and a method for detecting BHV-2 (bovine herpes virus 2) and particularly discloses a high-sensitivity BHV-2 quantitative real-time PCR (polymerase chain reaction) detection method and a kit. The kit comprises a primer group and a fluorescent probe for detecting the BHV-2; the primer group comprises four primers for nested amplification of specific DNA fragments of the BHV-2. With the adoption of the detection method, nested amplification and quantitative real-time PCR can be completed in one tube through one-step amplification by the aid of the four primers and the fluorescent probe. With the adoption of the BHV-2 quantitative real-time PCR detection method and the kit with high sensitivity, specificity and accuracy, quick detection of the BHV-2 can be realized, the kit is very convenient to apply, and on the basis of the advantages, the kit is suitable to be popularized and applied in livestock breeding units, animal inspection and quarantine departments and the like and has wide application prospect.
Owner:BEIJING INST OF GENOMICS CHINESE ACAD OF SCI CHINA NAT CENT FOR BIOINFORMATION
Method of detecting coccidioides species
ActiveUS20160326603A1Lack of sensitivityLoad accuratelyMicrobiological testing/measurementQuantitative Real Time PCRMicrobiology
Owner:ARIZONA BOARD OF REGENTS ACTING FOR & ON BEHALF OF NORTHERN ARIZONA UNIV +1
Method of detecting Coccidioides species
ActiveUS9970066B2Sugar derivativesMicrobiological testing/measurementQuantitative Real Time PCRPcr assay
Owner:ARIZONA BOARD OF REGENTS ACTING FOR & ON BEHALF OF NORTHERN ARIZONA UNIV +1
Method for detection, differentiation and quantification of T cell populations by way of reverse transcription quantitative real time PCR (RT-qPCR) technology
The present invention relates to a method for detection, differentiation and quantification of T cell populations, comprising the following steps a) contacting a first aliquot of a body fluid of an individual with at least one antigen, wherein the body fluid contains antigen presenting cells (APC) and T cells, b) incubating the first aliquot with at least one antigen for a certain period of time, c) detection and differentiation of the T cell population by detecting in the first aliquot and in a second aliquot of the body fluid of the individual, which has not been incubated with the at least one antigen, at least a first marker of the APC induced by T cells in a specific T cell population using reverse transcription quantitative real time-time polymerase chain reaction (RT-qPCR), and d) detection and quantification of the T cell population by determining the ratio of the detected marker of the APC of the first aliquot to the second aliquot as well as a kit for performing the method.
Owner:LOPHIUS BIOSCI
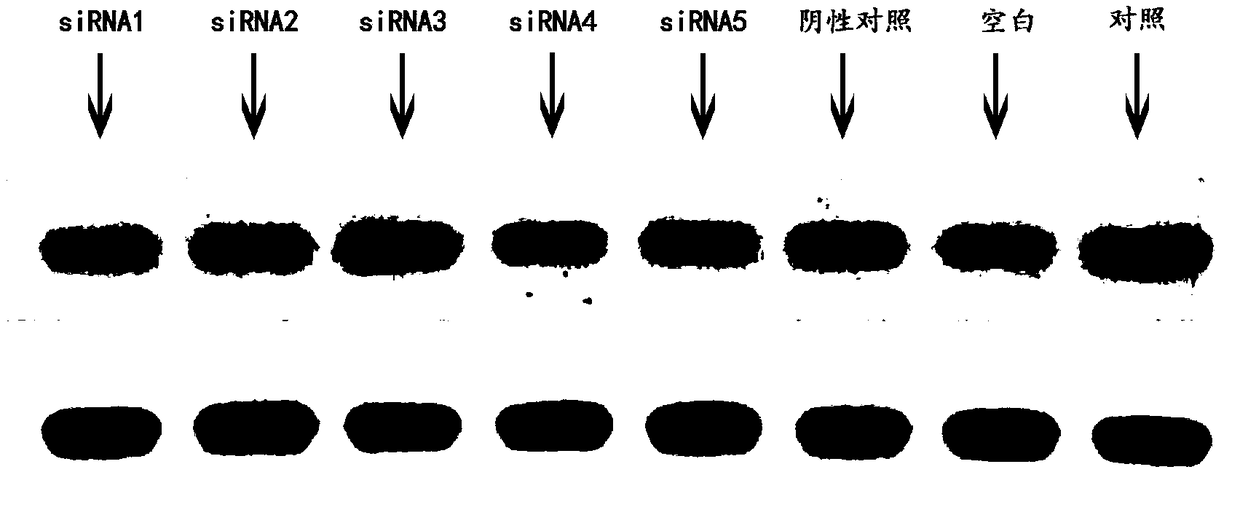
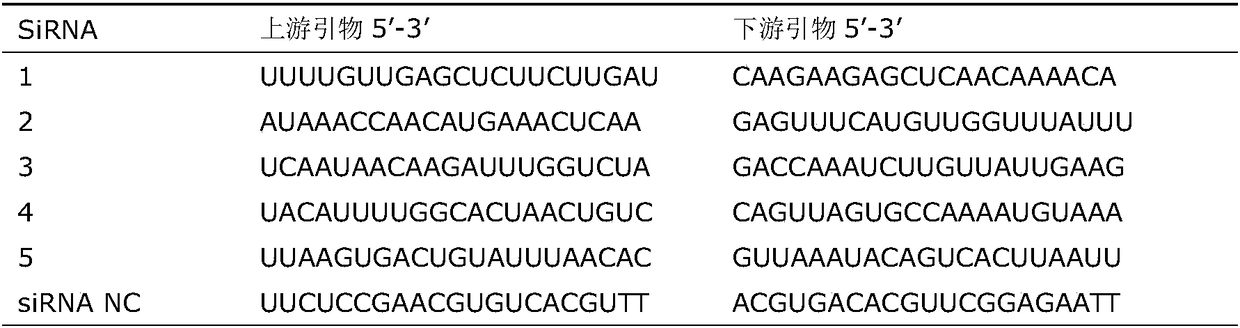
siRNA capable of inhibiting expression of HSP90 gene in breast cancer cells
InactiveCN108486106AMicrobiological testing/measurementDNA/RNA fragmentationQuantitative Real Time PCRFluorescence
The invention discloses a siRNA capable of inhibiting the expression of an HSP90 gene in breast cancer cells. The siRNA is constructed by synthesizing interference sequences directed at the human Hsp90alpha gene and designed by siRNA design software after cell culture of breast cancer cells. The invention also provides a method for constructing the siRNA. The method comprises the following steps:subjecting the MDA-MB-231 cell line of human breast cancer cells to cell culture; designing five pairs of interference sequences directed at the human Hsp90alpha gene by using an siRNA designing tool,and carrying out synthesizing; detecting the expression of the Hsp90alpha gene in the breast cancer cells through the fluorescence quantitative real-time PCR technology; and transfecting the breast cancer cells with the designed siRNA, and carrying out a western-blot test so as to detect the expression situation of a Hsp90 gene protein. According to the invention, after MDA-MB-231 breast cancer cells are transfected by the siRNA constructed by using the method provided by the invention, the mRNA expression level and protein expression level of the MDA-MB-231 cells are significantly decreased,so the siRNA can effectively inhibit the expression of the Hsp90alpha gene in the MDA-MB-231 breast cancer cells, and has certain scientific values and clinical values for treatment of breast cancer.
Owner:华子昂
Qpcr-based method to assess t cell function
InactiveUS20170058353A1Cell from functioningMicrobiological testing/measurementBlood/immune system cellsQuantitative Real Time PCRActivation cells
The invention provides a method of determining T cell function in a subject in need of immunotherapy comprising:i) providing a blood sample comprising a population of T cells from the subject;ii) activating the T cells in the sample; andiii) assaying an expression level of one or more T cell activation markers using quantitative real time PCR (qPCR) after activating the T cells in the sample.
Owner:WEBB TONYA J
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com