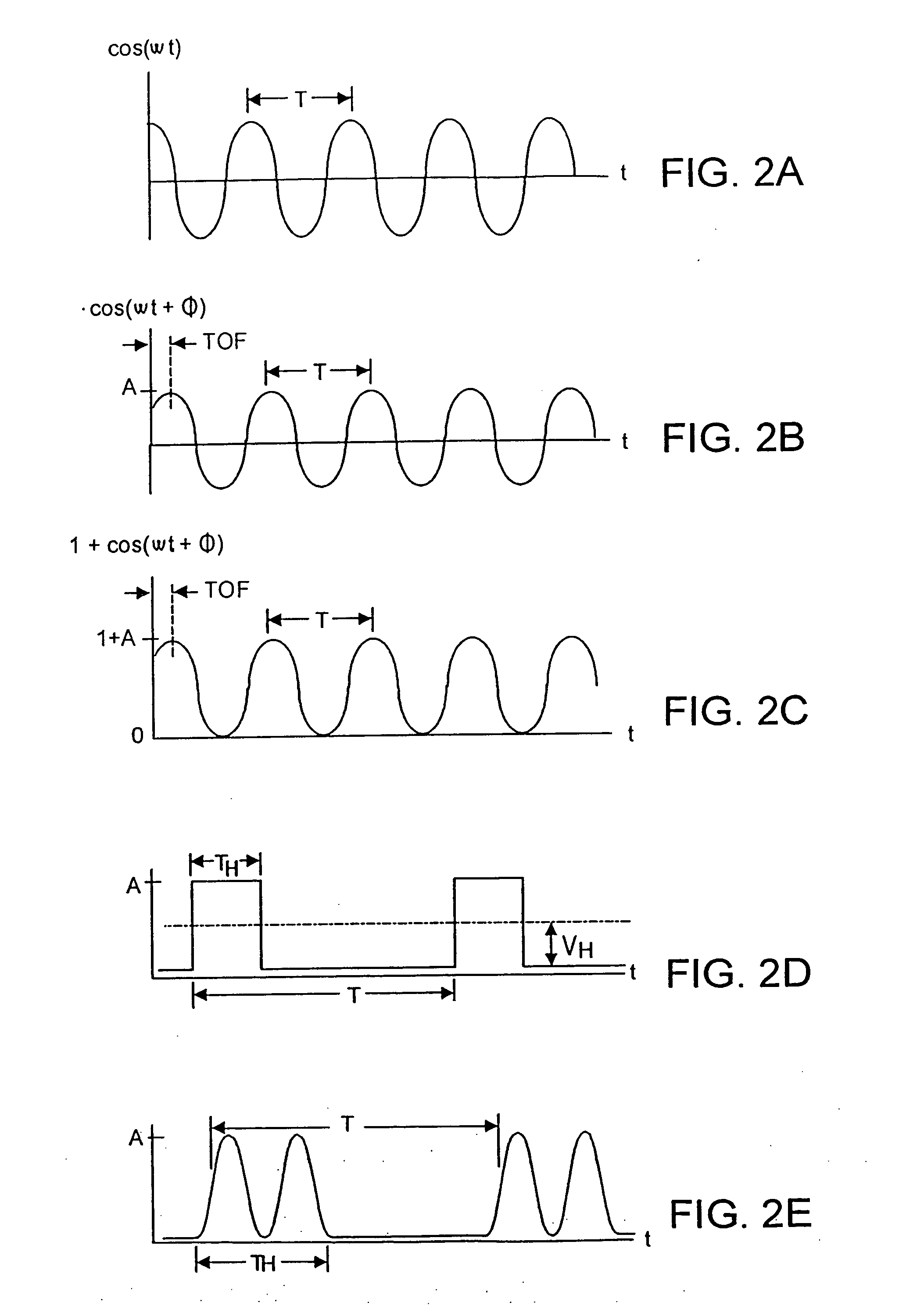
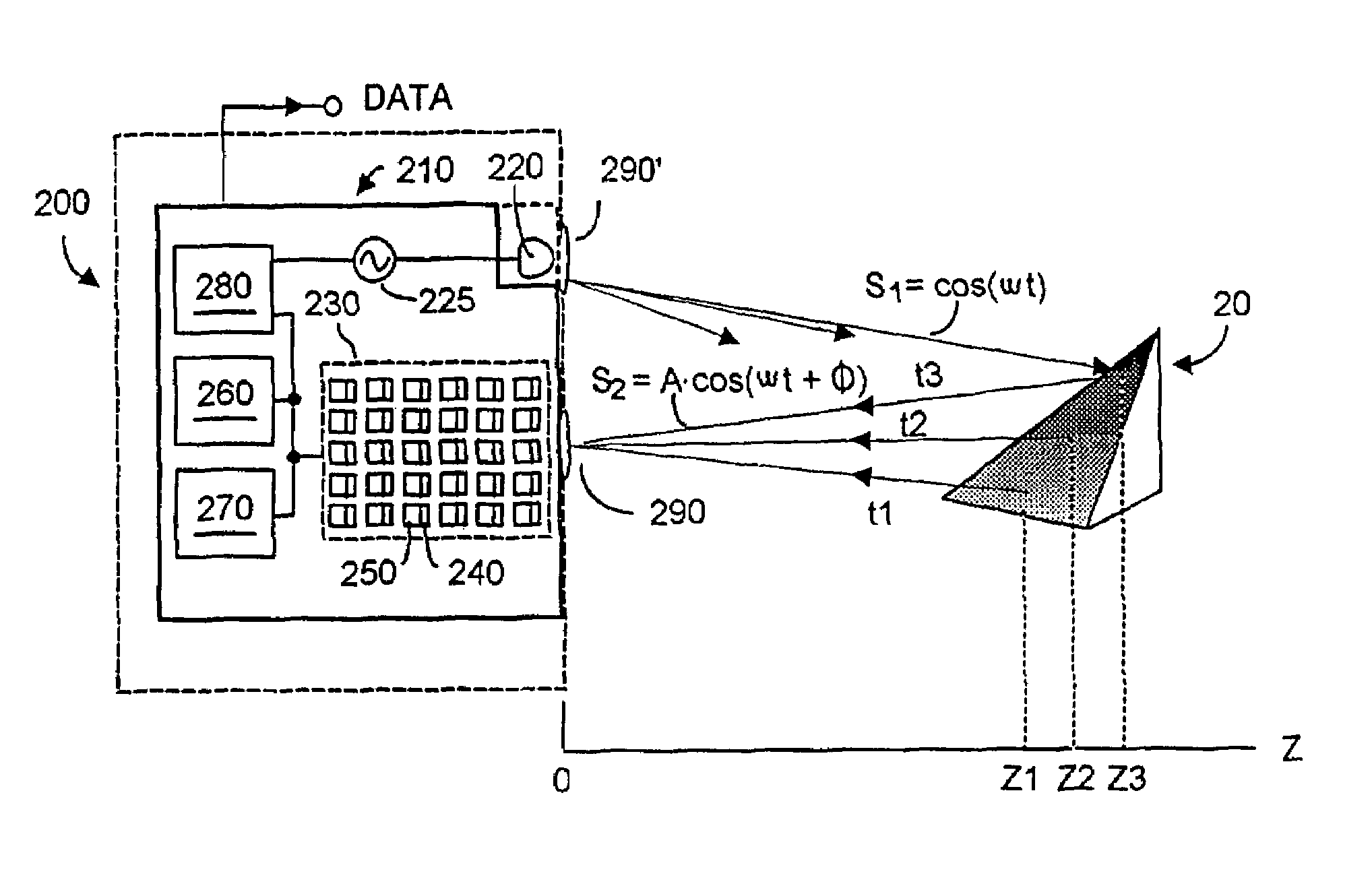
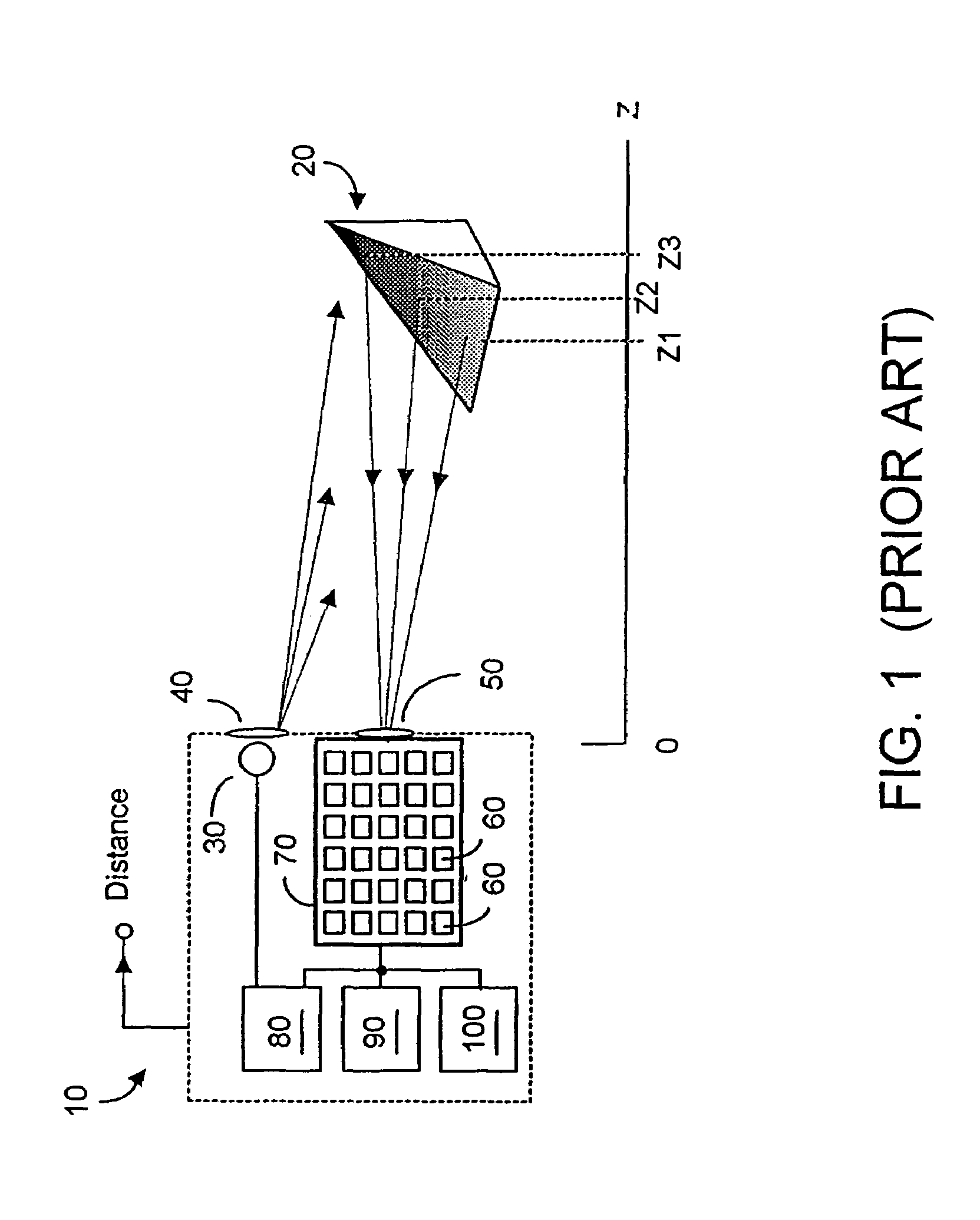
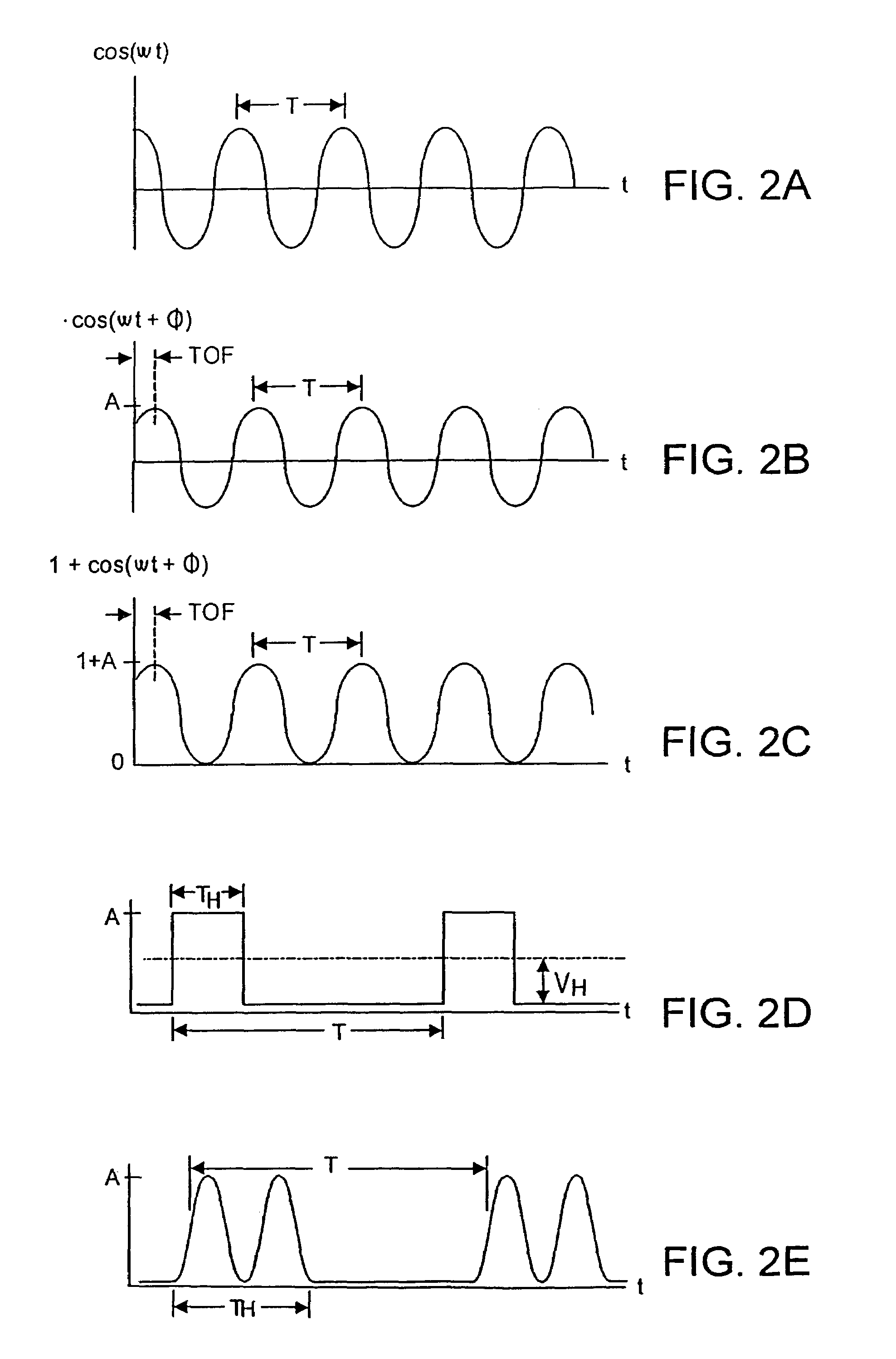
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
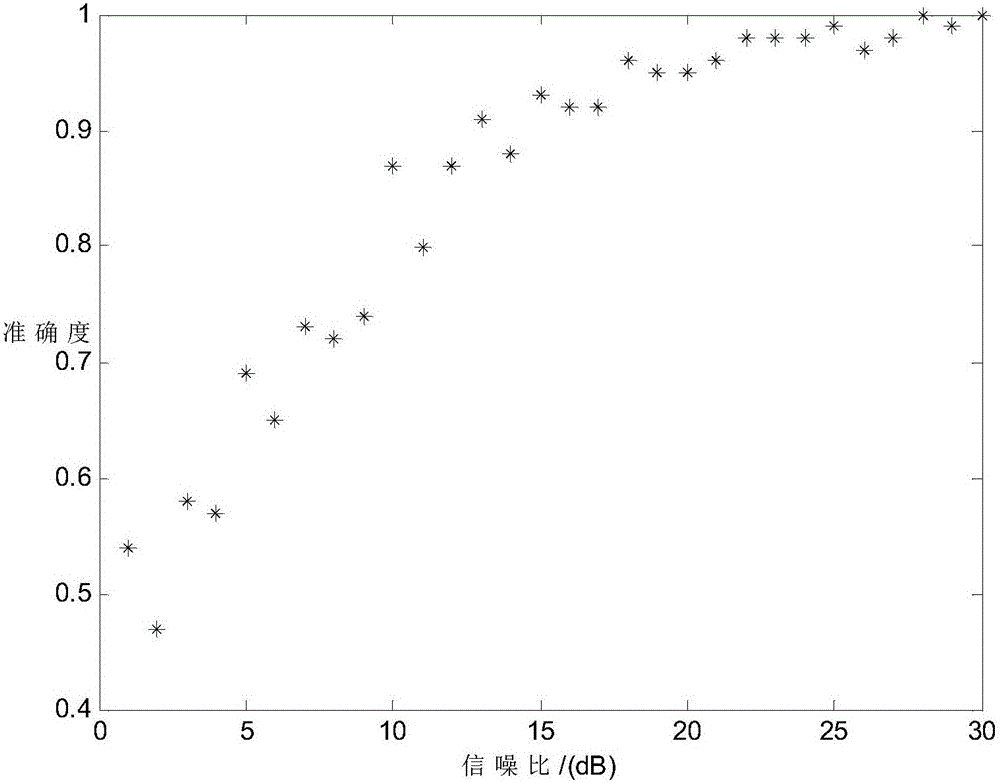
207results about How to "Reduce aliasing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
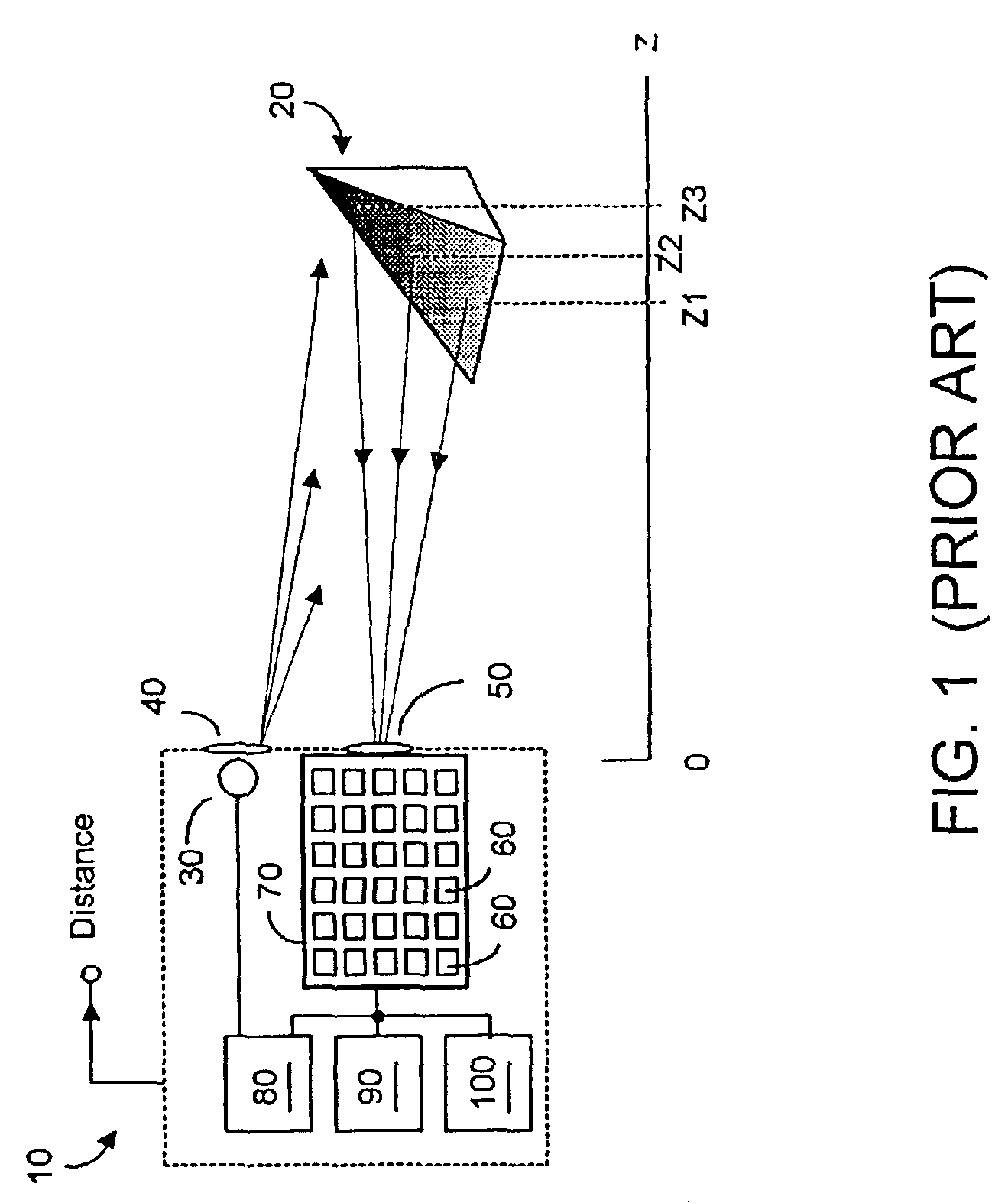
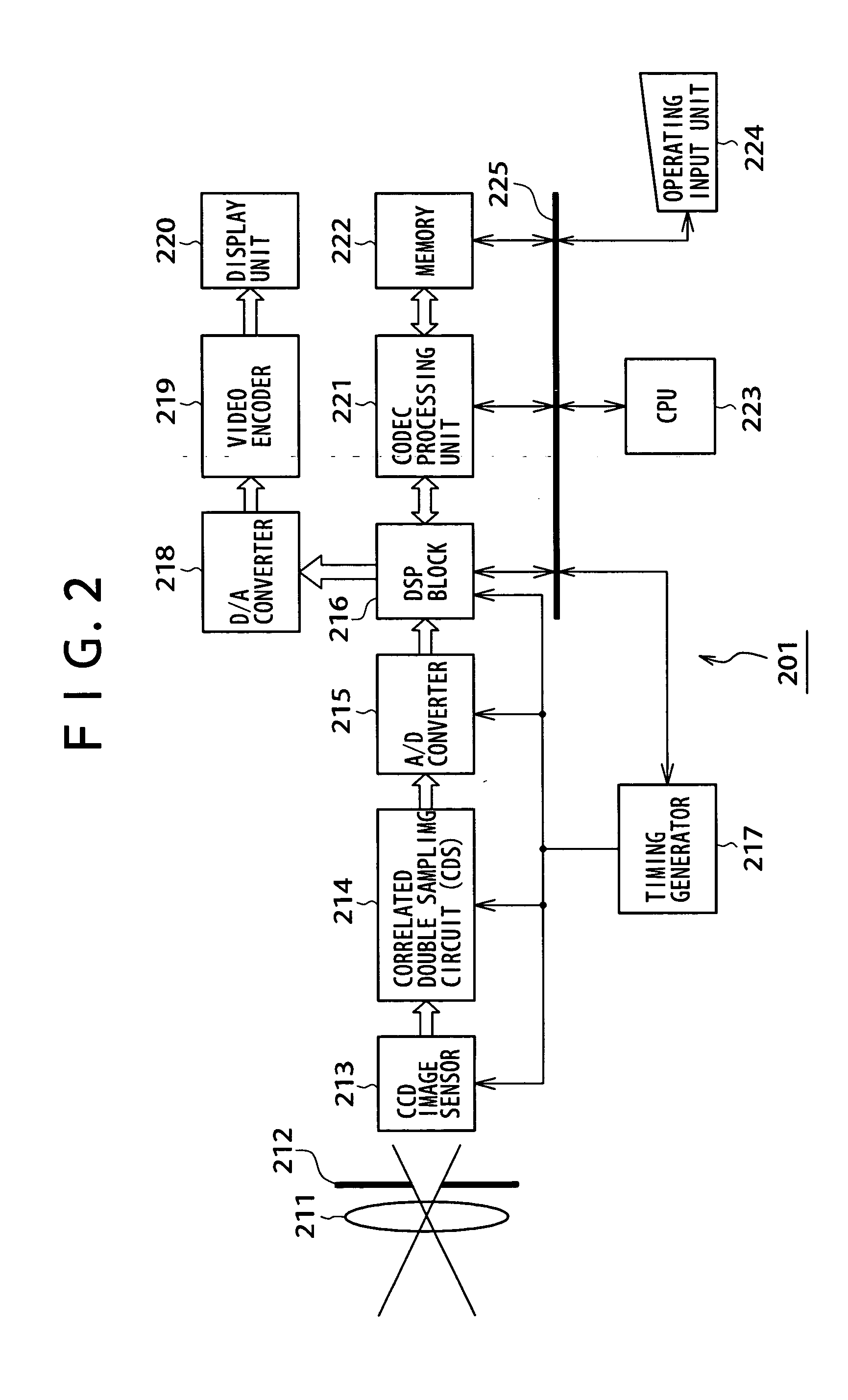
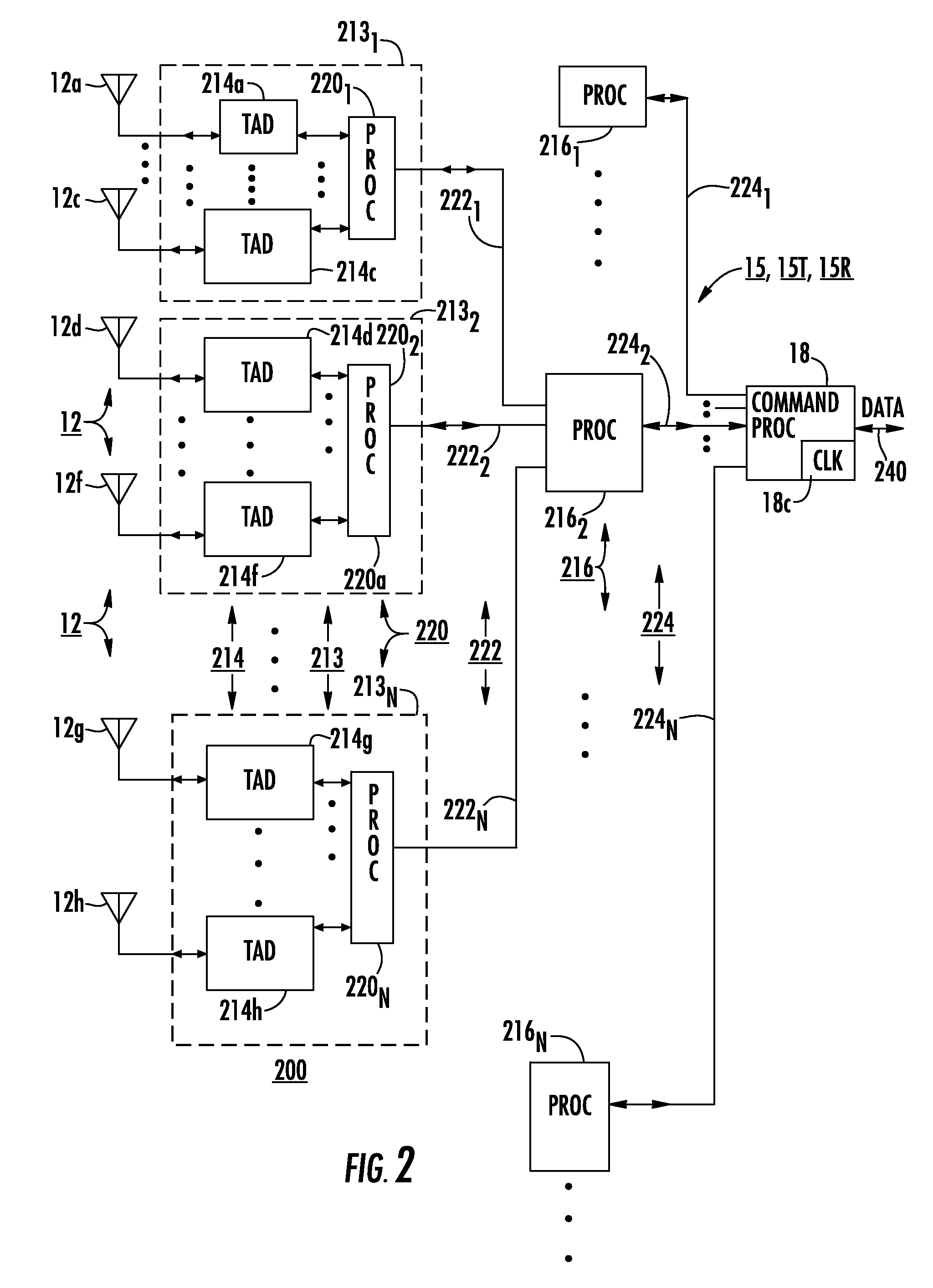
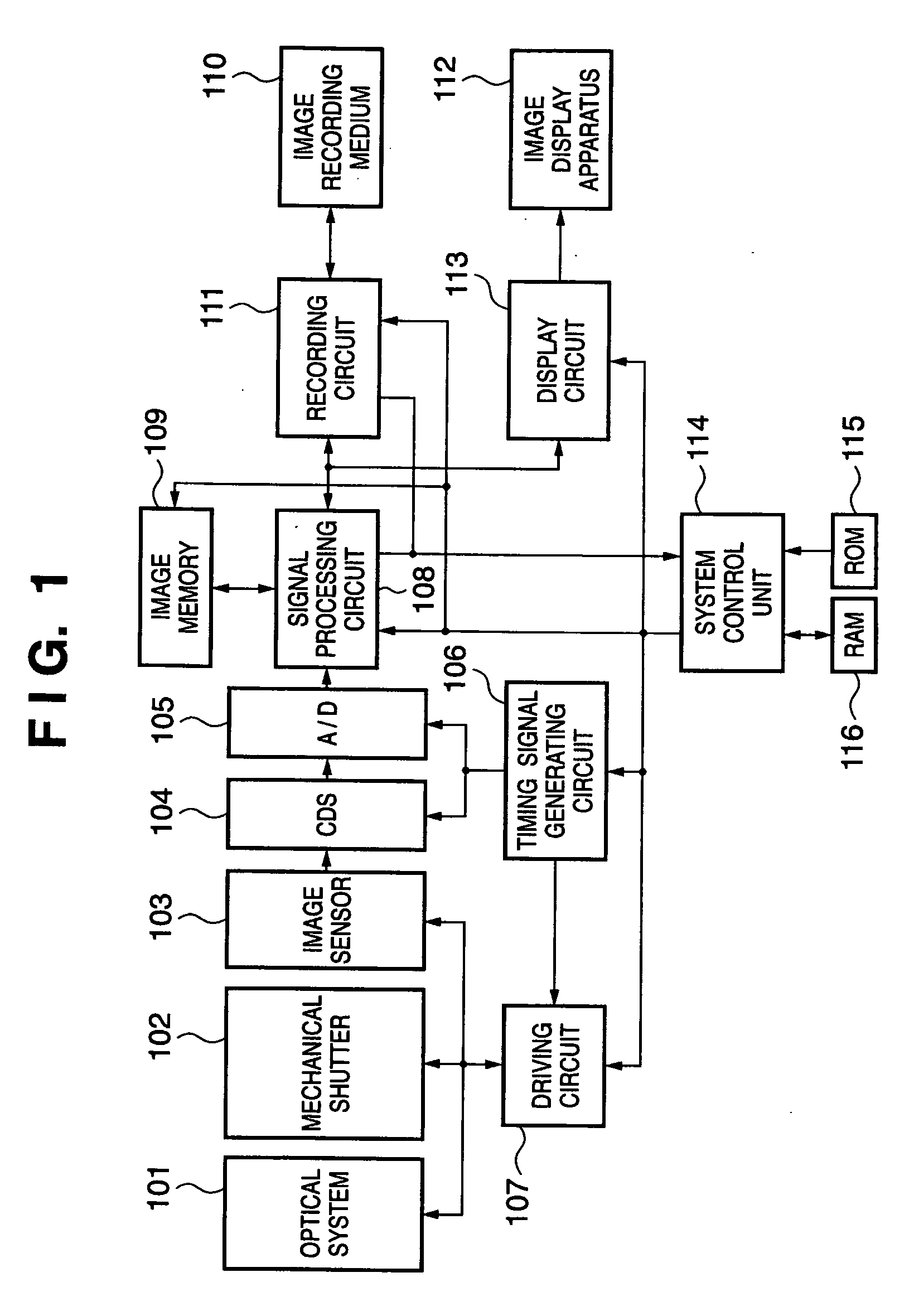
Methods and devices for charge management for three-dimensional sensing
InactiveUS6906793B2Minimal overheadEffective coloringTelevision system detailsOptical rangefindersCMOSHigh frequency modulation
Structures and methods for three-dimensional image sensing using high frequency modulation includes CMOS-implementable sensor structures using differential charge transfer, including such sensors enabling rapid horizontal and slower vertical dimension local charge collection. Wavelength response of such sensors can be altered dynamically by varying gate potentials. Methods for producing such sensor structures on conventional CMOS fabrication facilities include use of “rich” instructions to command the fabrication process to optimize image sensor rather than digital or analog ICs. One detector structure has closely spaced-apart, elongated finger-like structures that rapidly collect charge in the spaced-apart direction and then move collected charge less rapidly in the elongated direction. Detector response is substantially independent of the collection rate in the elongated direction.
Owner:MICROSOFT TECH LICENSING LLC
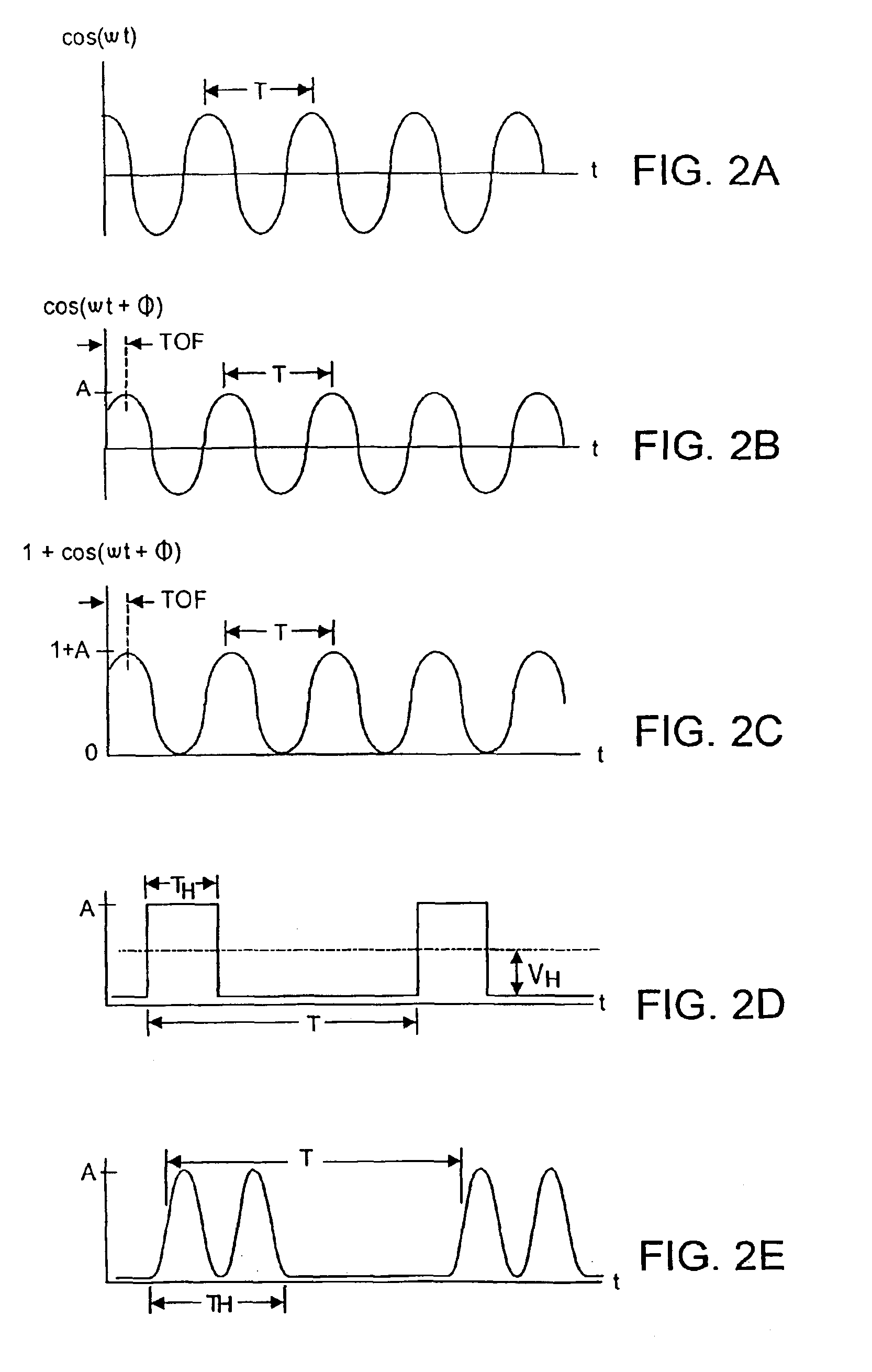
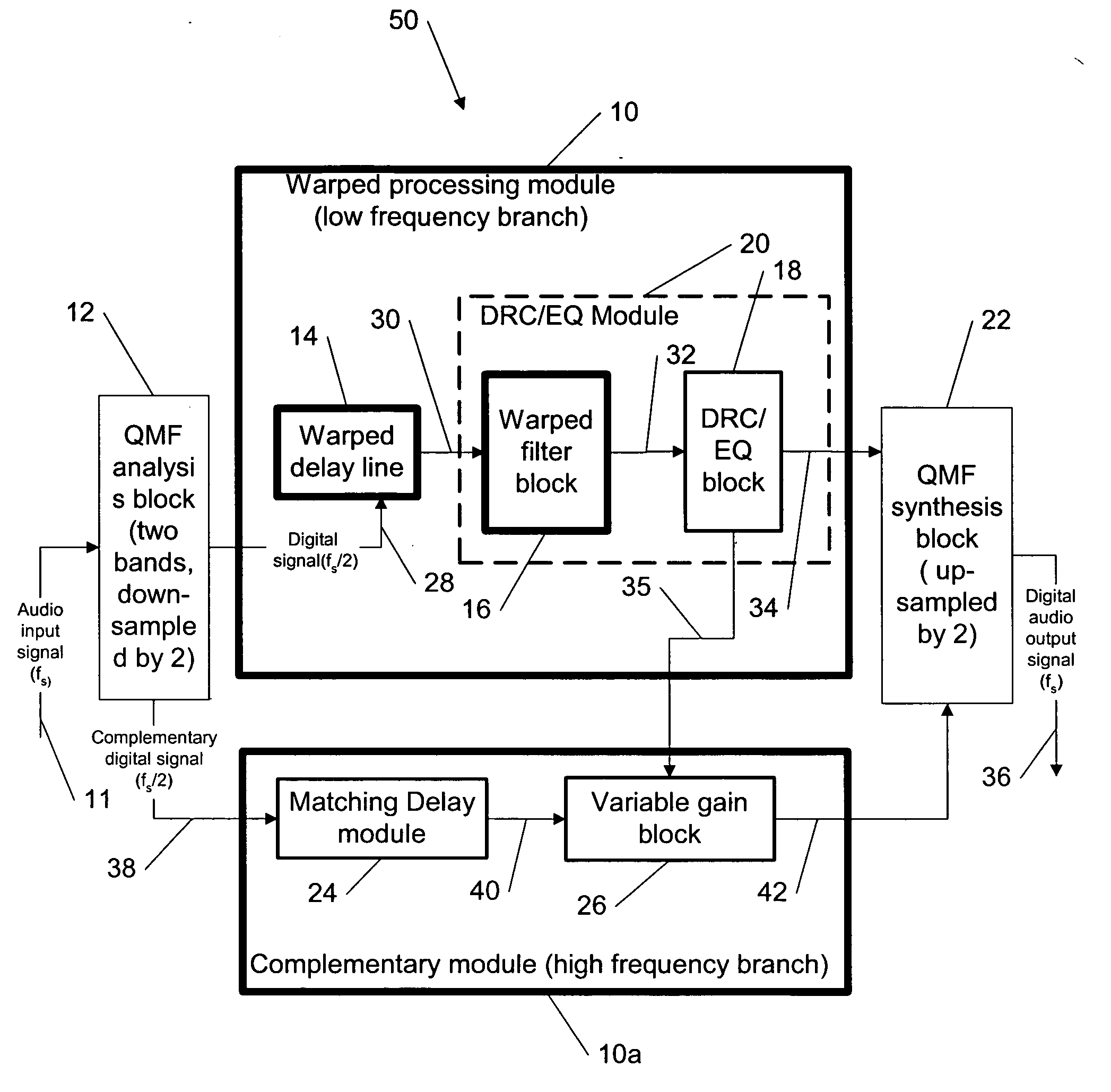
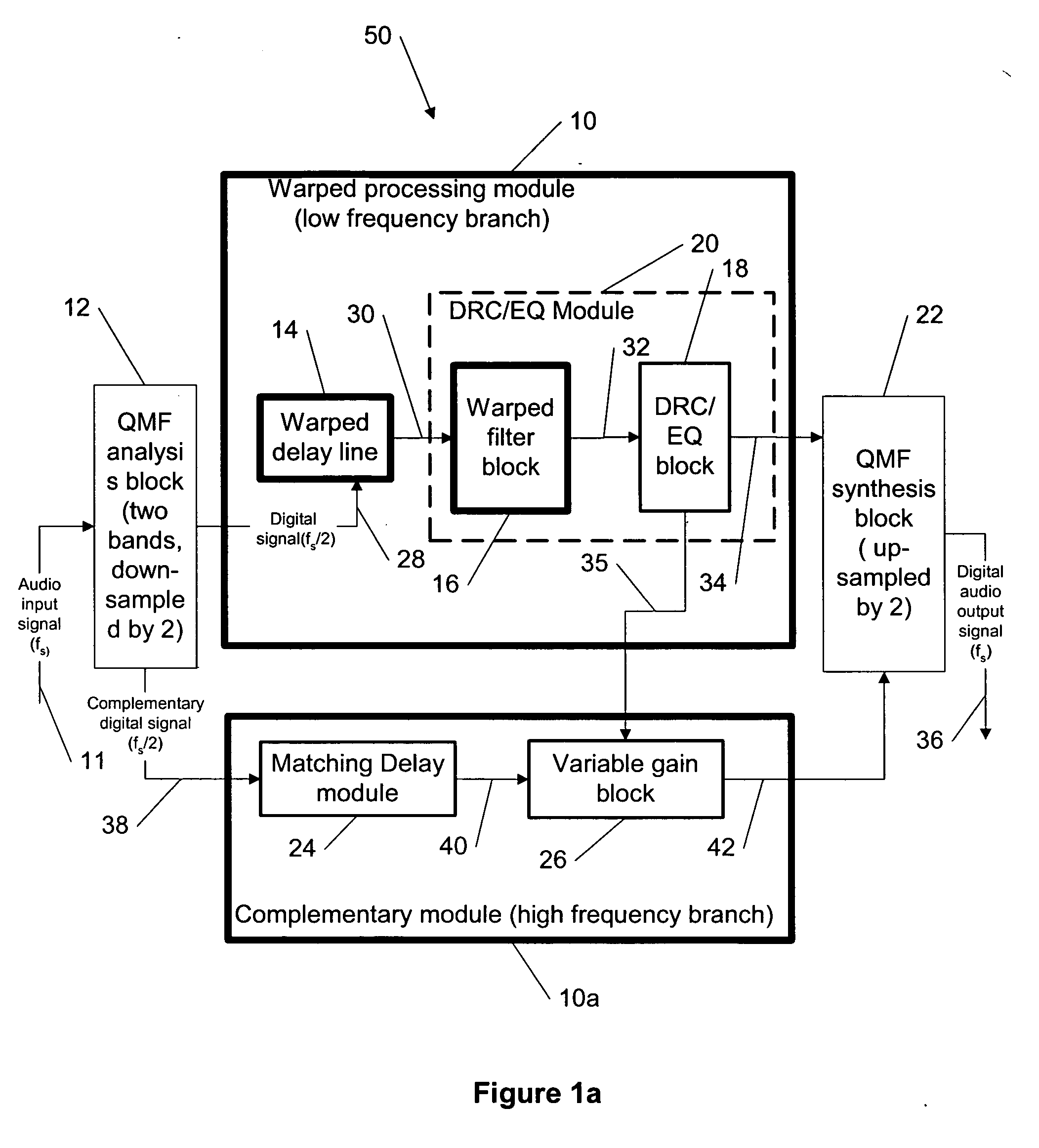
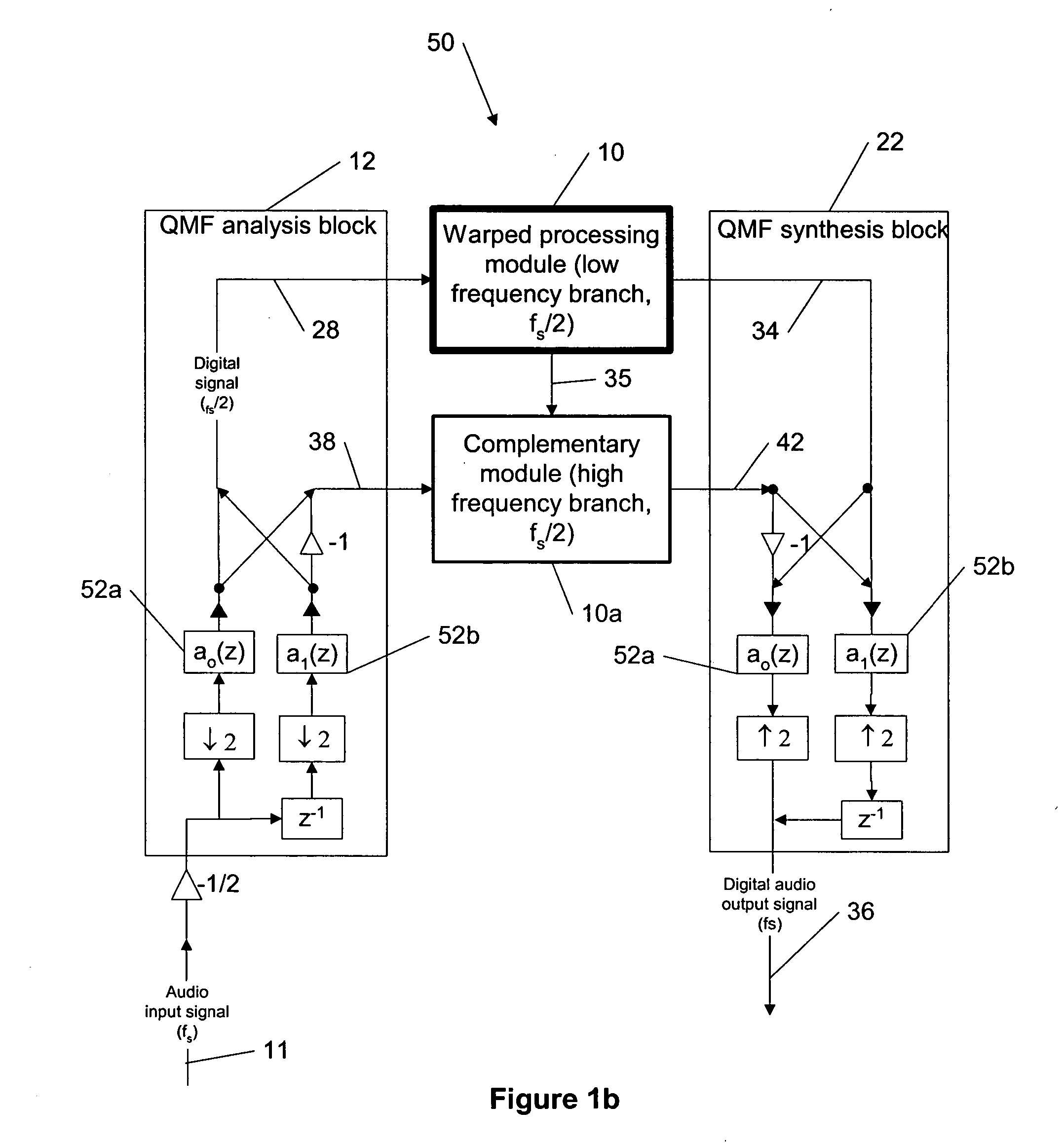
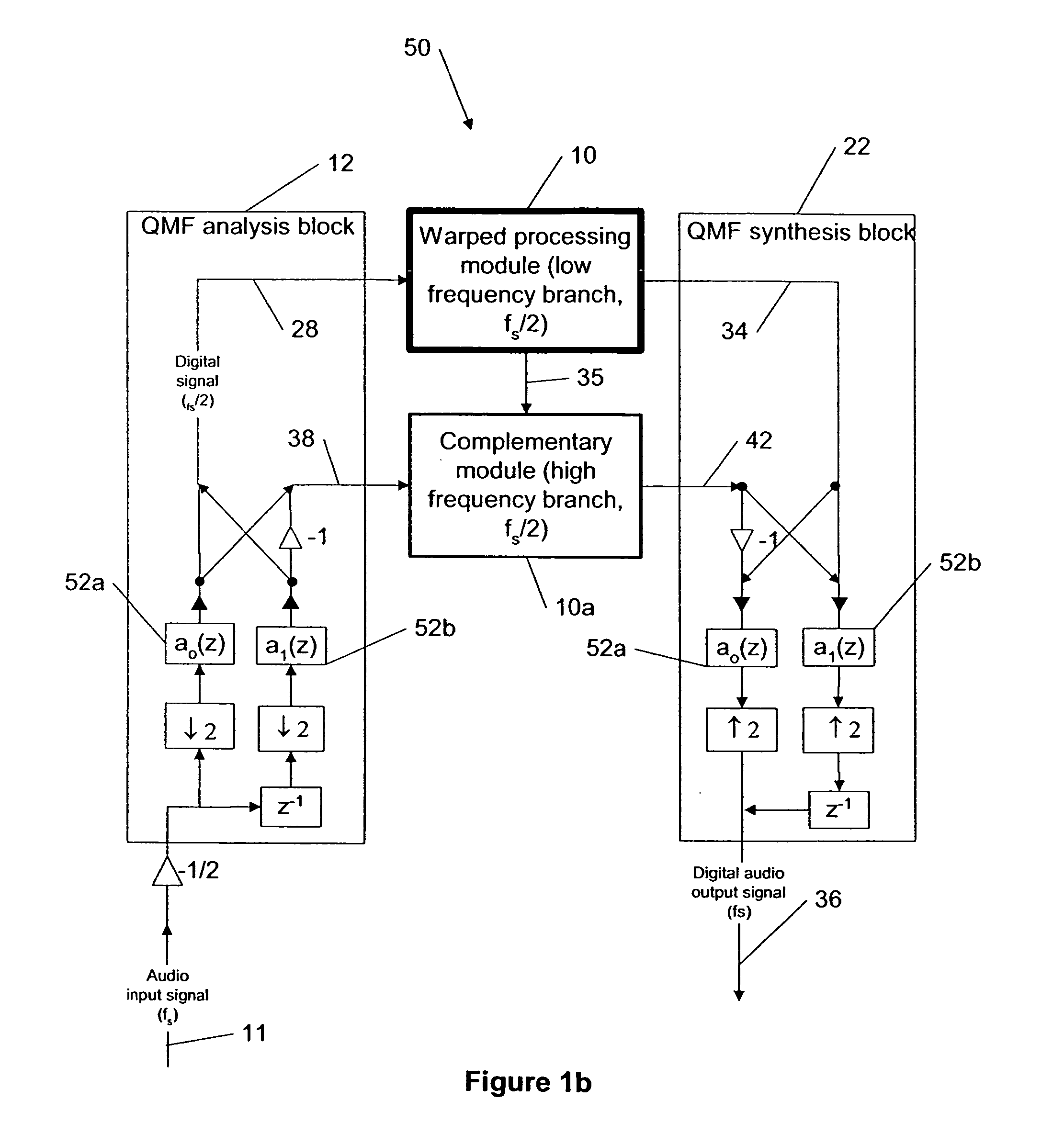
Dynamic range control and equalization of digital audio using warped processing
ActiveUS20050249272A1Reduce processing loadComputationally efficientMultiple-port networksDelay line applicationsFrequency bandLoudness
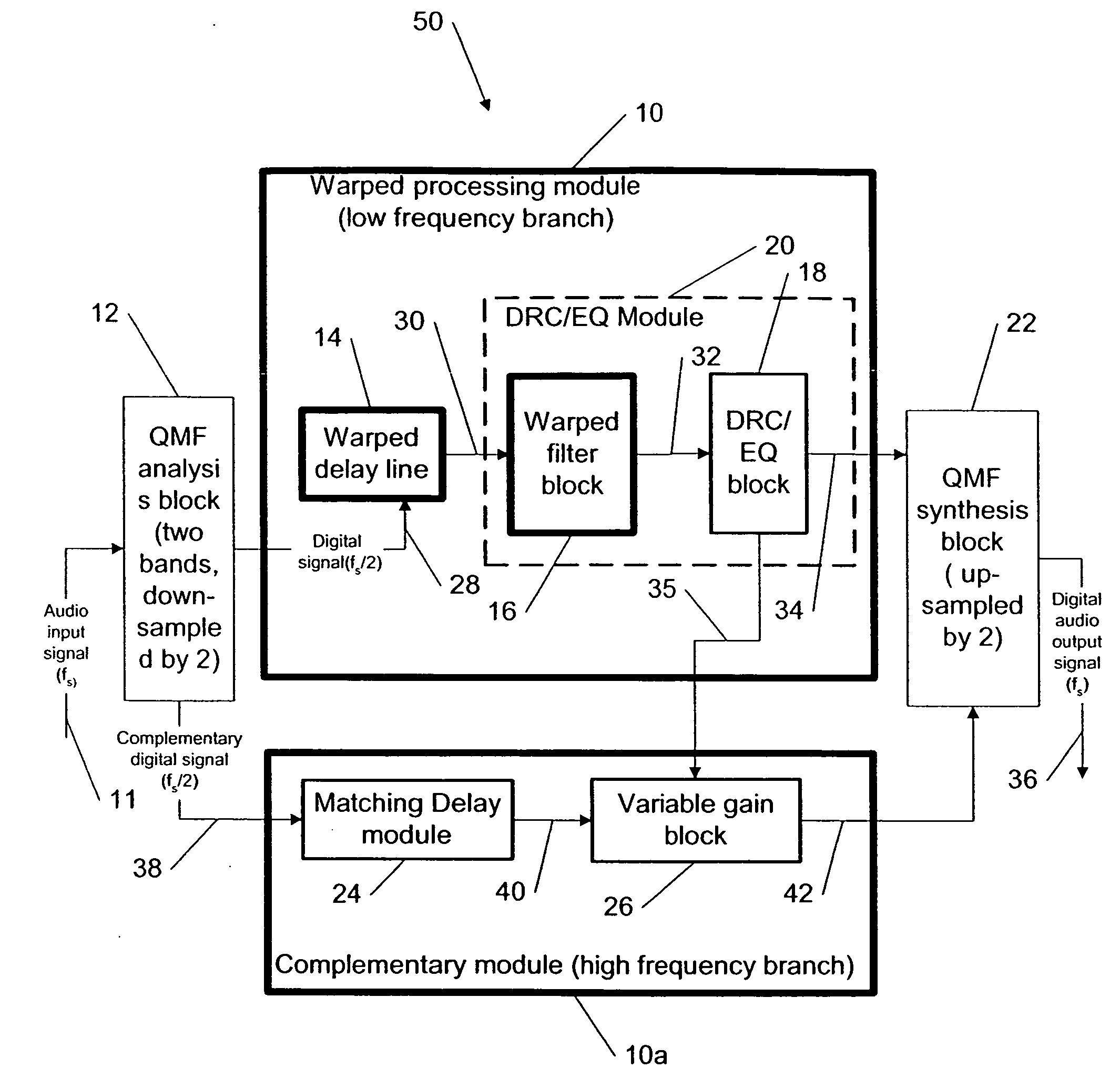
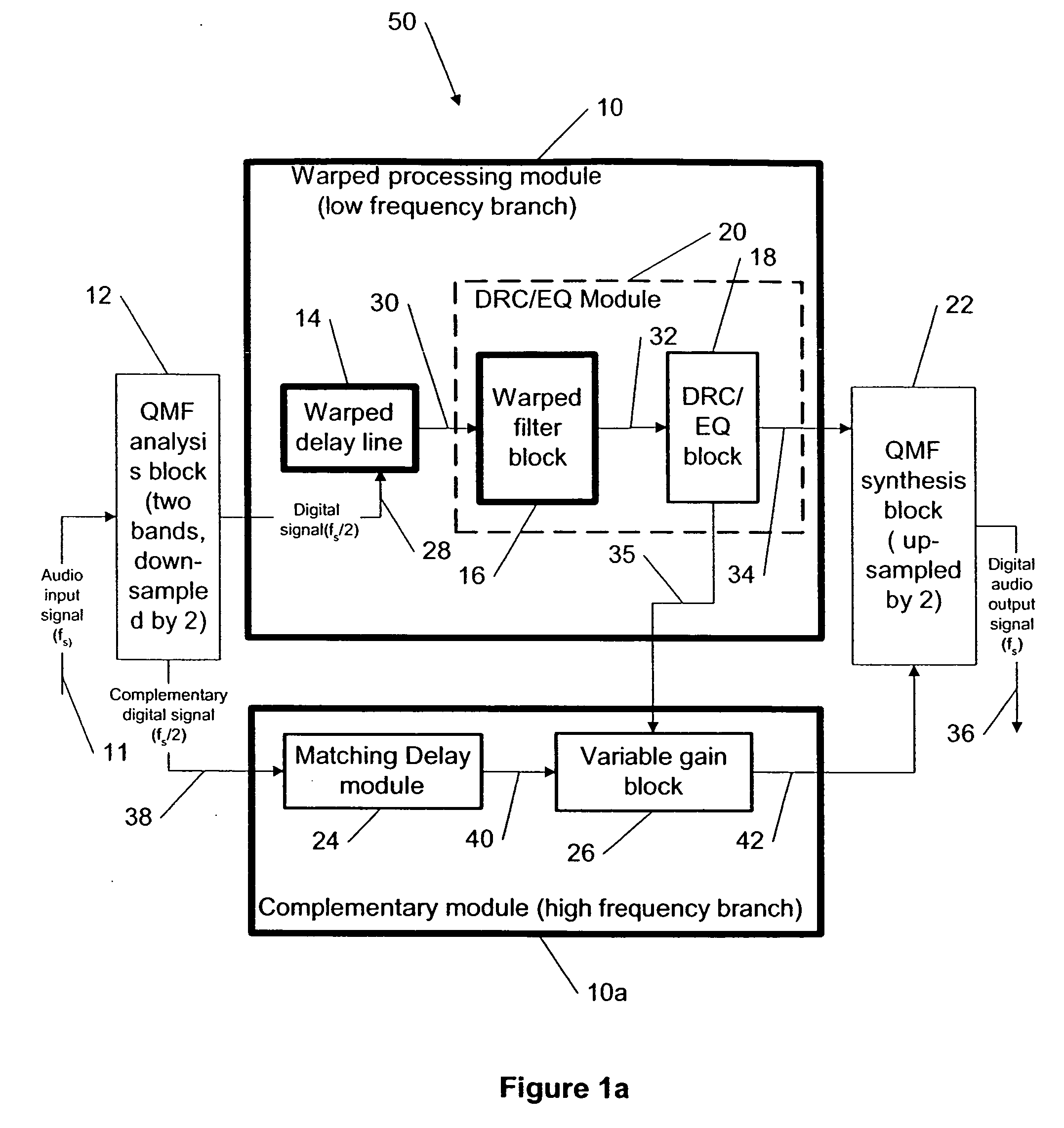
This invention describes a method for adjusting the loudness and the spectral content of digital audio signals in a real-time using warped spectral filtering. A warped processing module modifies a spectral content of a digital audio signal with a set of gains for a plurality of non-linearly-scaled frequency bands determined by a warping factor λ of a warped delay line. Warped delay line signals, generated by the warped delay line, are processed by a warped filter block containing multiple warped finite impulse response filters, e.g., Mth band filters, using individual warped spectral filtering in said plurality of the non-linearly-scaled frequency bands, which is followed by a conventional processing by a dynamic range control / equalization block. The present invention describes another innovation, that is embedding the warped processing module in a two-channel quadrature mirror filter (QMF) bank for improving processing efficiency at high sample rates.
Owner:BEIJING XIAOMI MOBILE SOFTWARE CO LTD
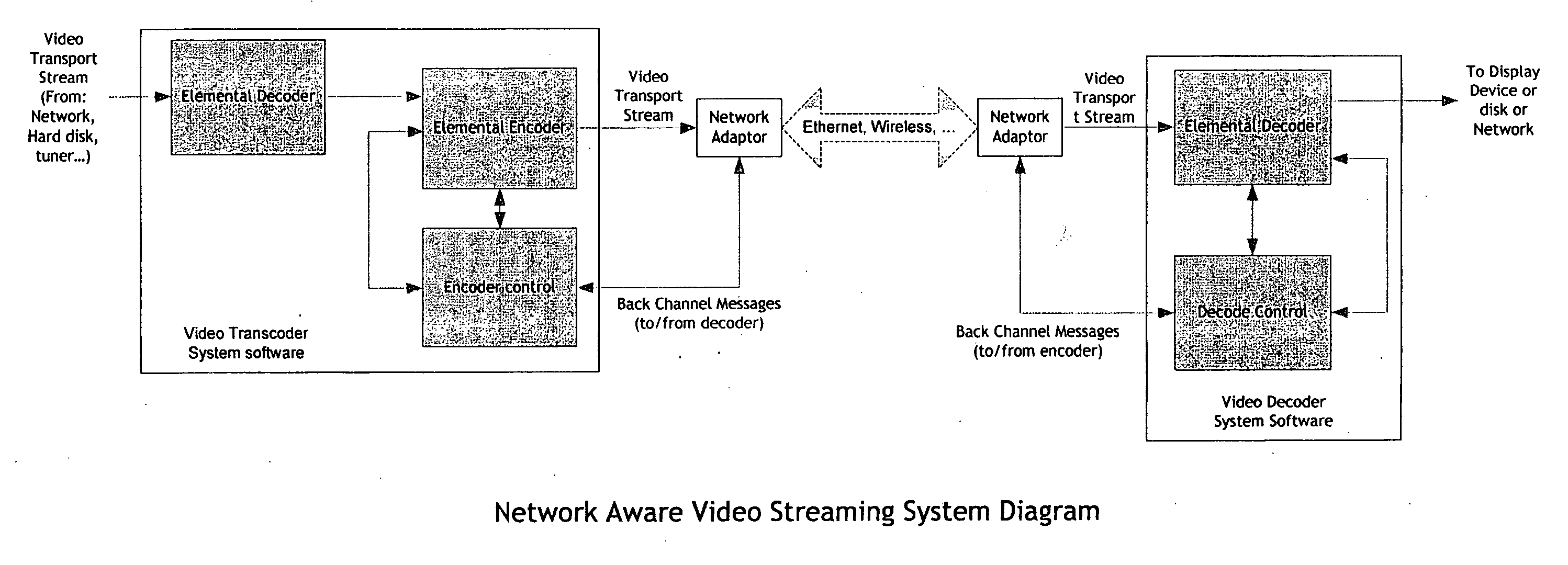
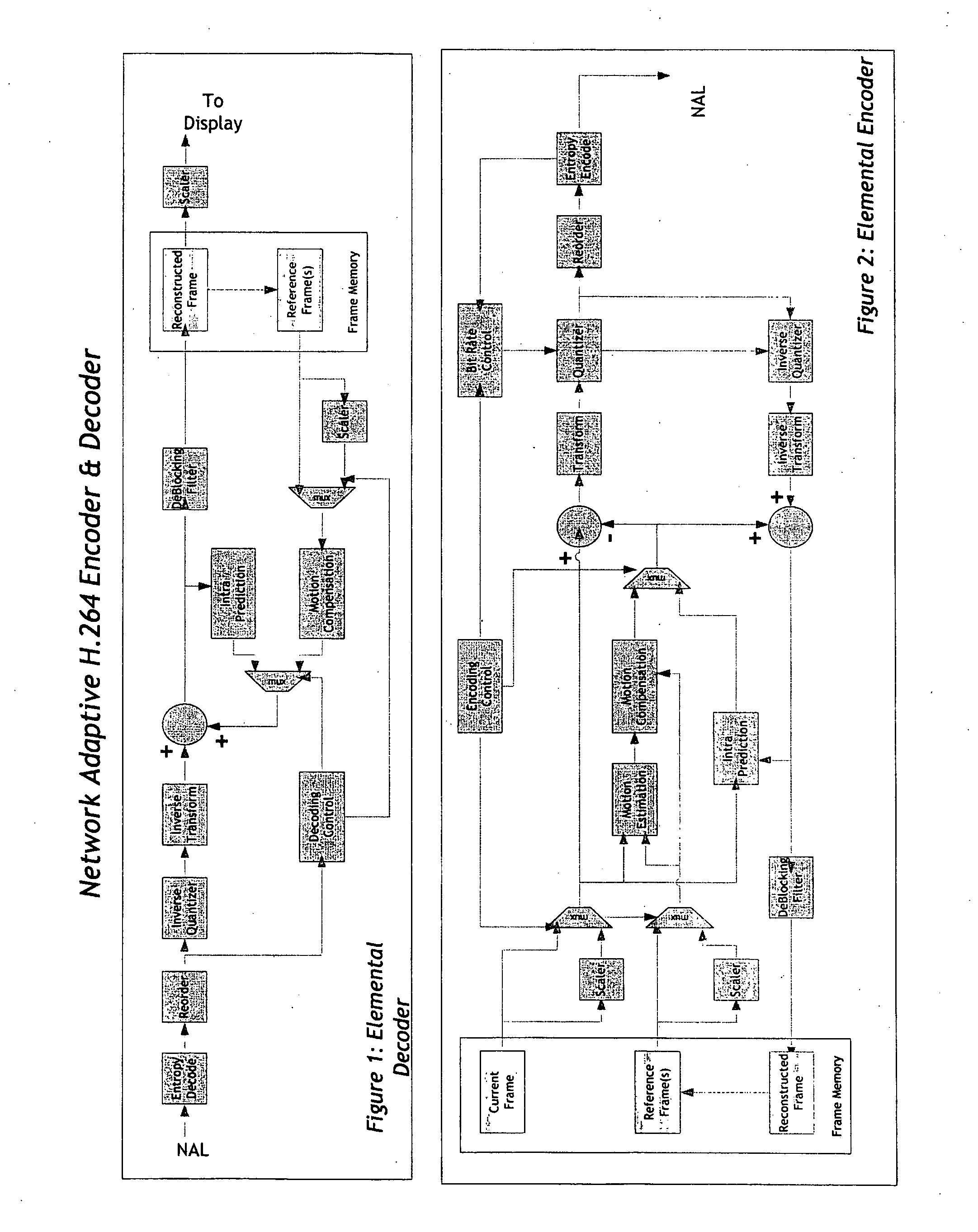
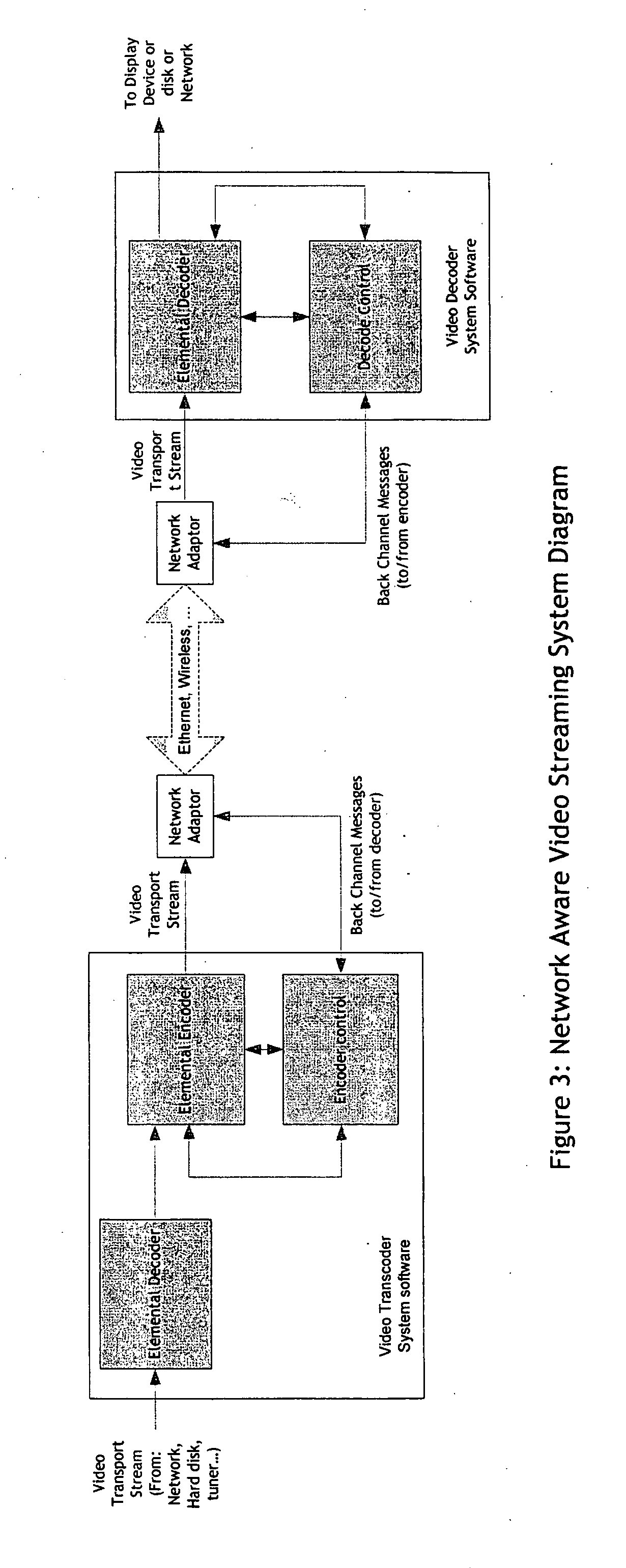
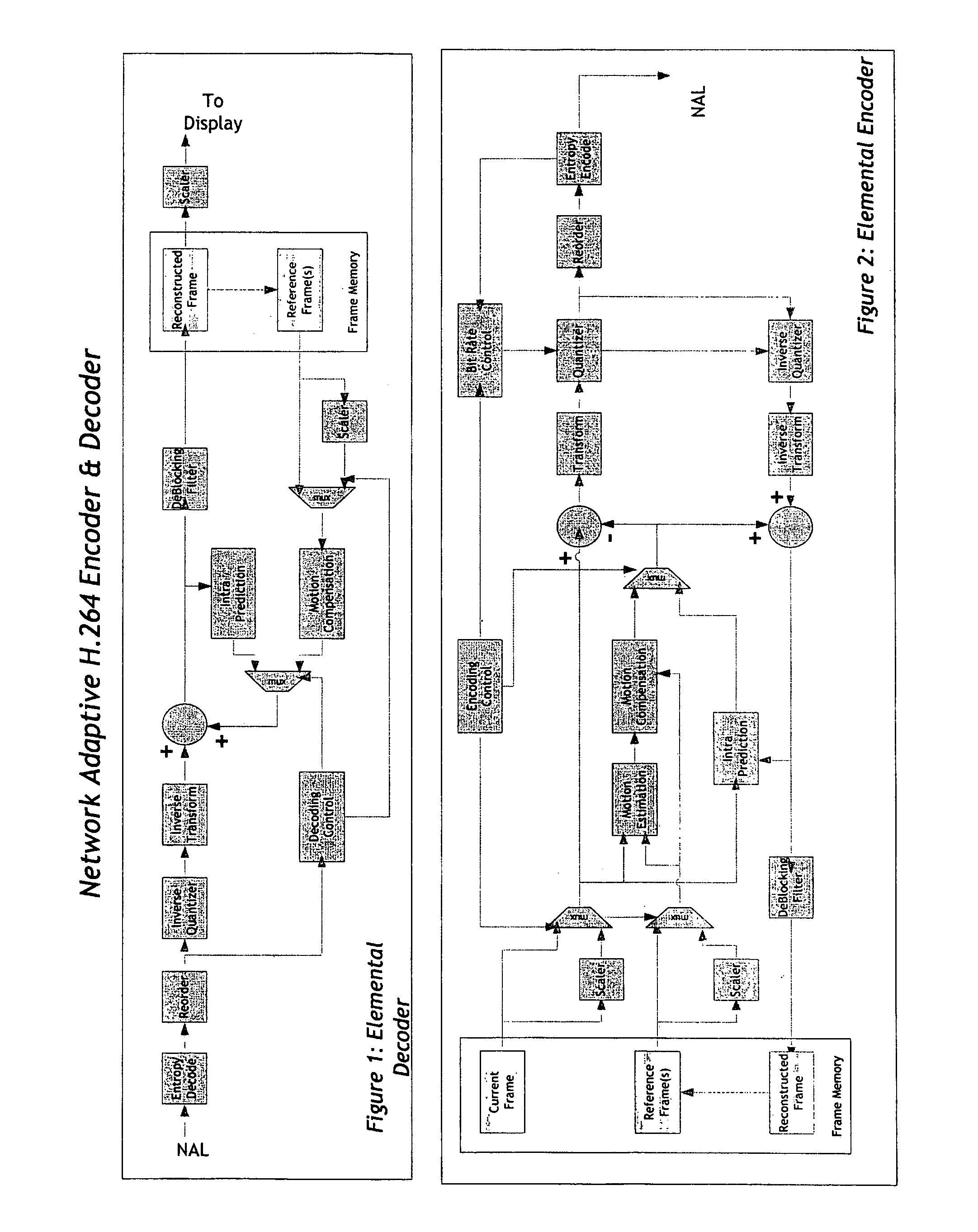
Real-time network adaptive digital video encoding/decoding
ActiveUS20080084927A1Lowering scale factorLow data ratePulse modulation television signal transmissionPicture reproducers using cathode ray tubesDigital videoImaging quality
A method for real time video transmission over networks with varying bandwidth is described. Image quality is maintained even under degrading network performance conditions through the use of image scaling in conjunction with block based motion compensated video coding (MPEG2 / 4, H.264, et. Al.). The ability to quickly switch resolutions without decreasing reference frame correlation is shown enabling a fast switch to reduce the required bandwidth for stable image quality.
Owner:ELEMENTAL TECH LLC
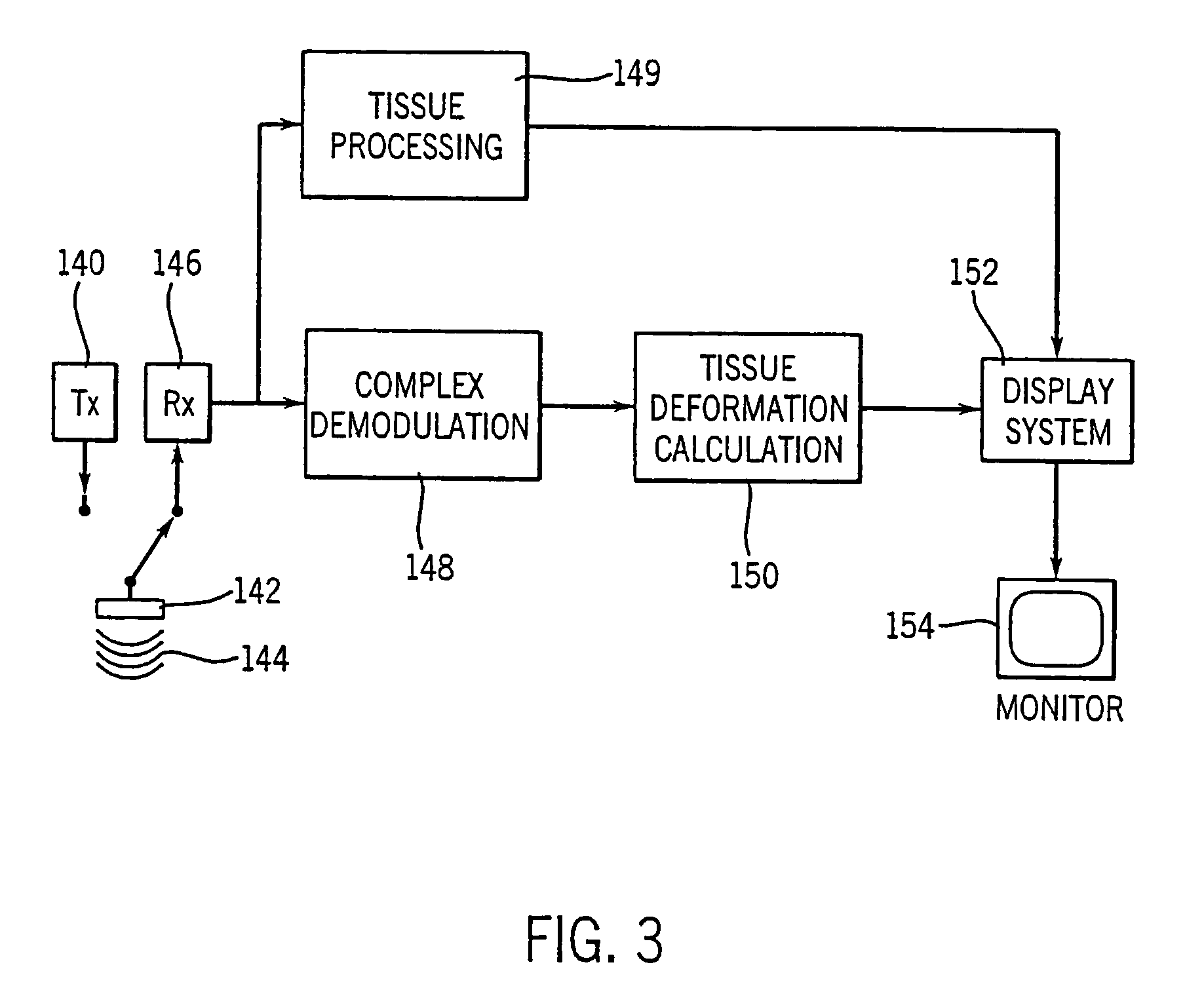
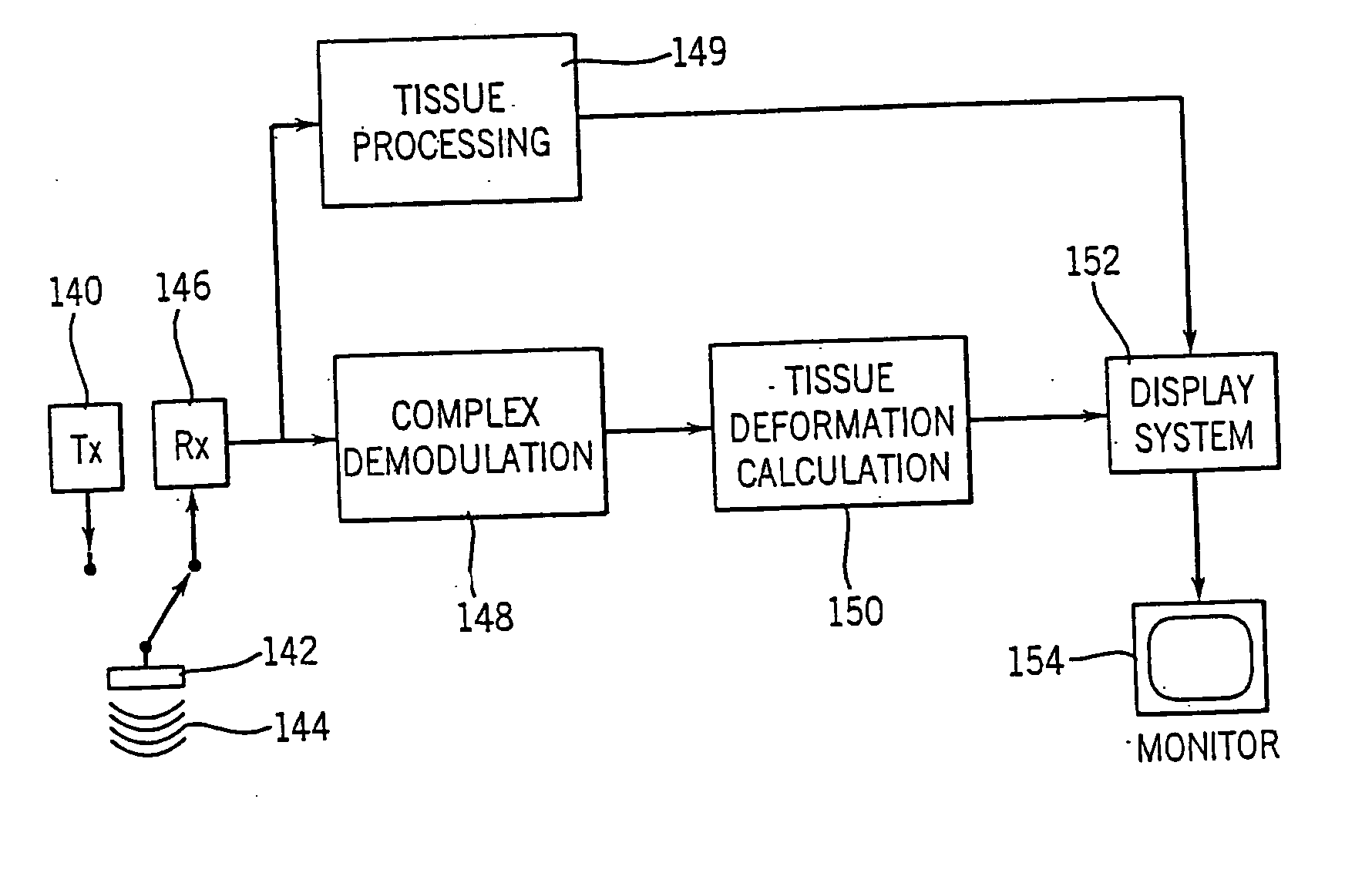
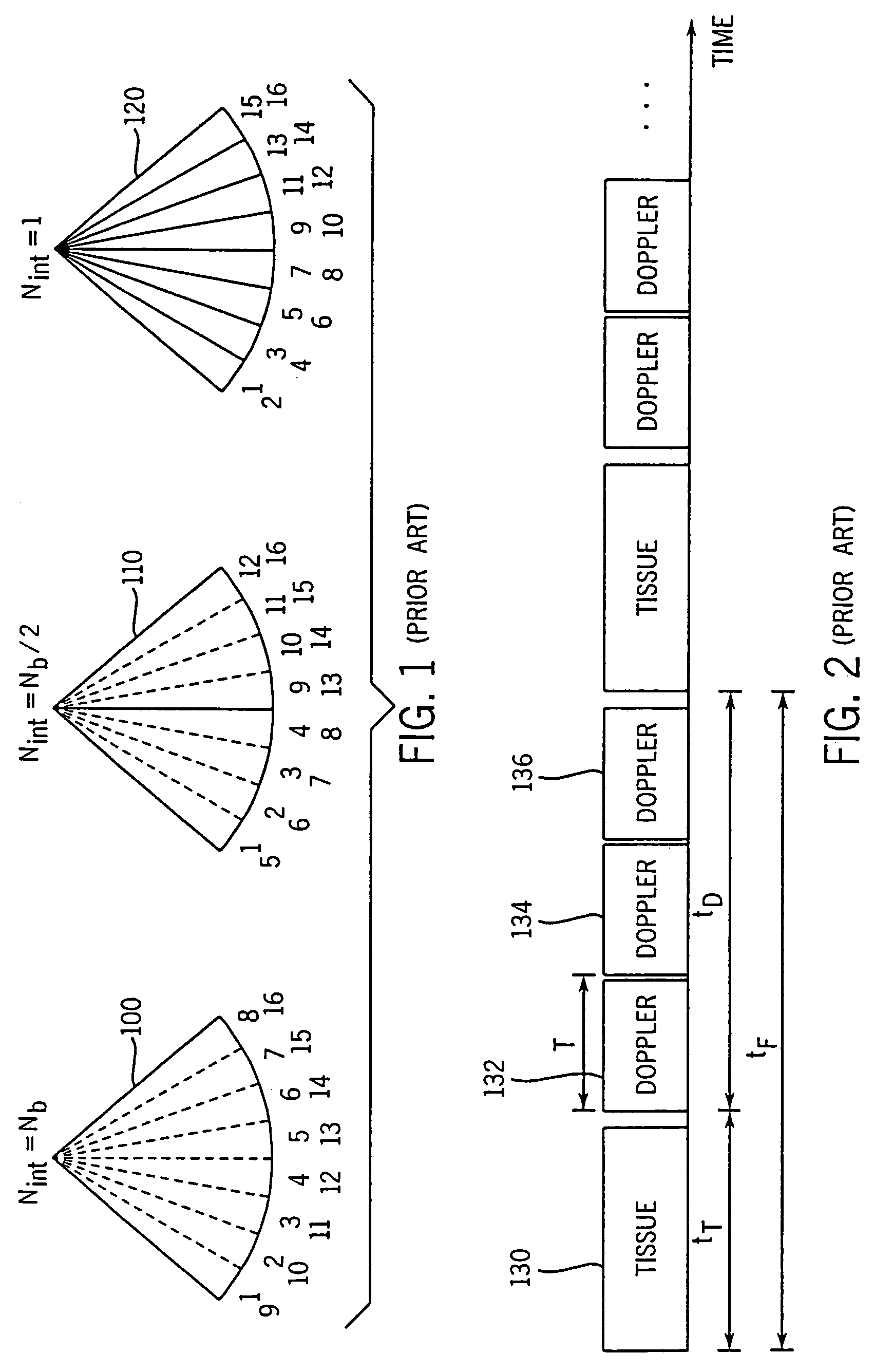
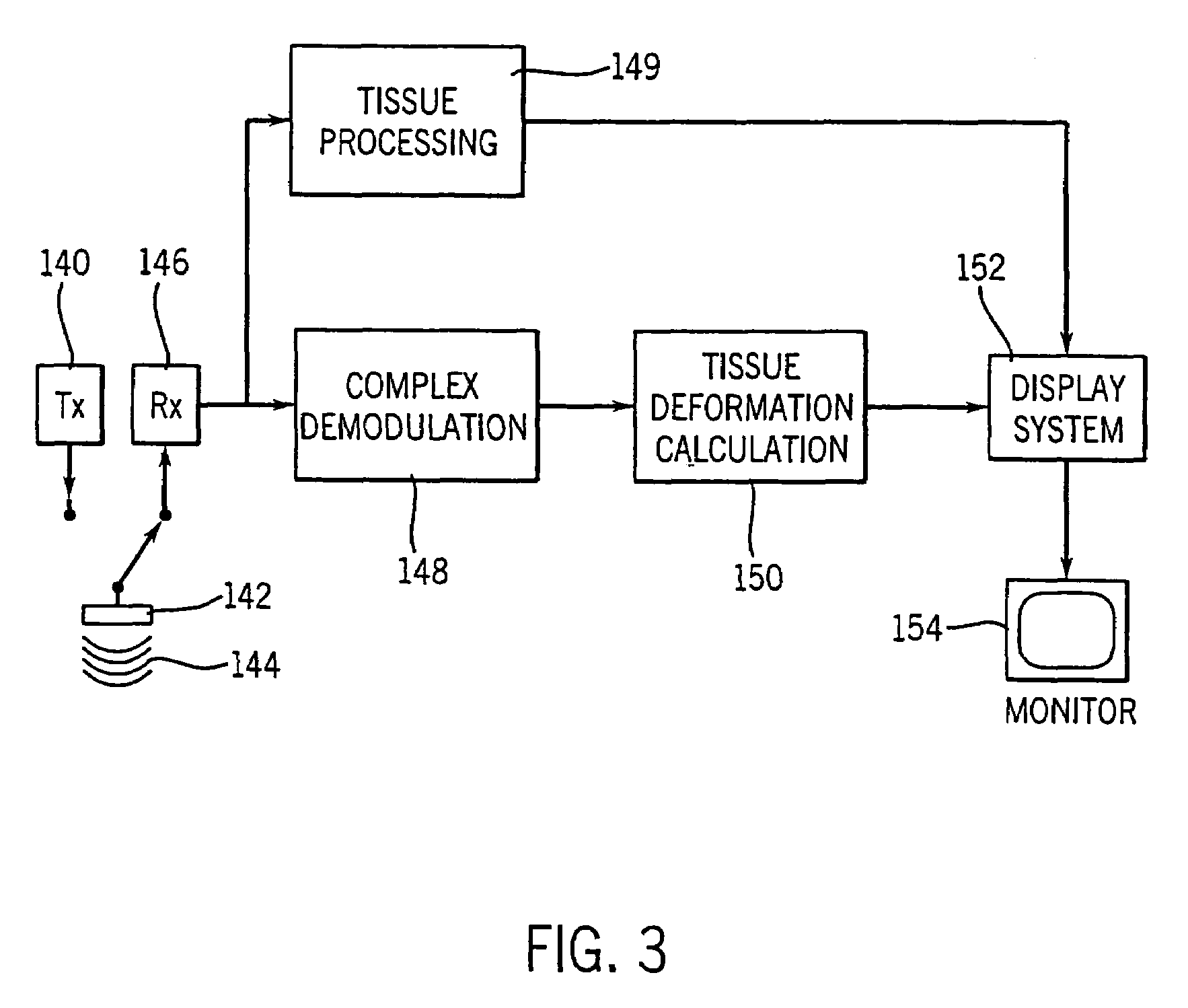
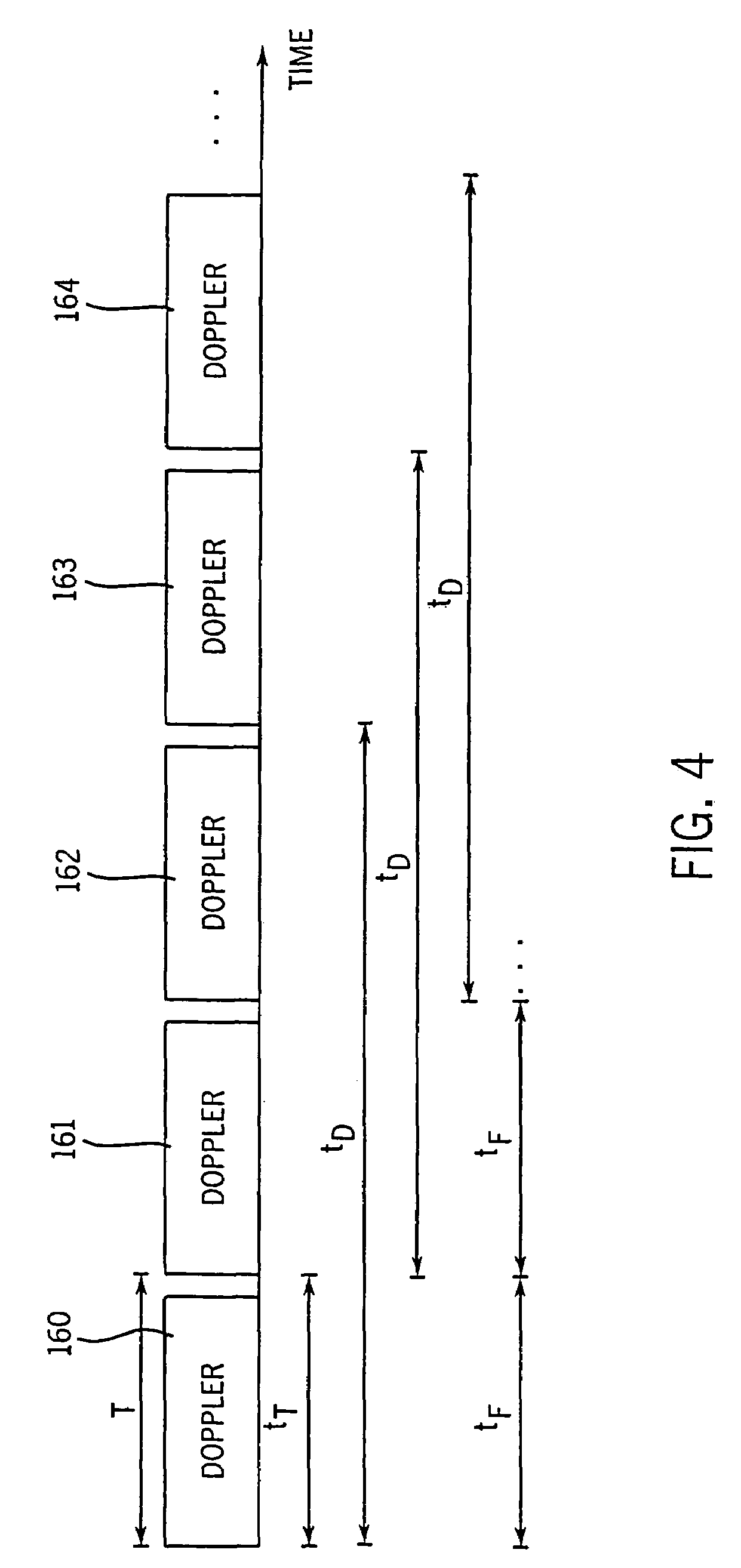
Method and apparatus for providing real-time calculation and display of tissue deformation in ultrasound imaging
InactiveUS7077807B2Reduce impactReduce aliasingElectrocardiographyOrgan movement/changes detectionUltrasound imagingSonification
An ultrasound system and method for calculation and display of tissue deformation parameters are disclosed. A method to estimate a strain rate in any direction, not necessarily along the ultrasound beam, based on tissue velocity data from a small region of interest around a sample volume is disclosed. Quantitative tissue deformation parameters, such as tissue velocity, tissue velocity integrals, strain rate and / or strain, may be presented as functions of time and / or spatial position for applications such as stress echo. For example, strain rate or strain values for three different stress levels may be plotted together with respect to time over a cardiac cycle.
Owner:G E VINGMED ULTRASOUND
Image processing apparatus and image processing method, and program
InactiveUS20050088550A1Good reproducibilityReduce aliasingTelevision system detailsTelevision system scanning detailsColor imageImaging processing
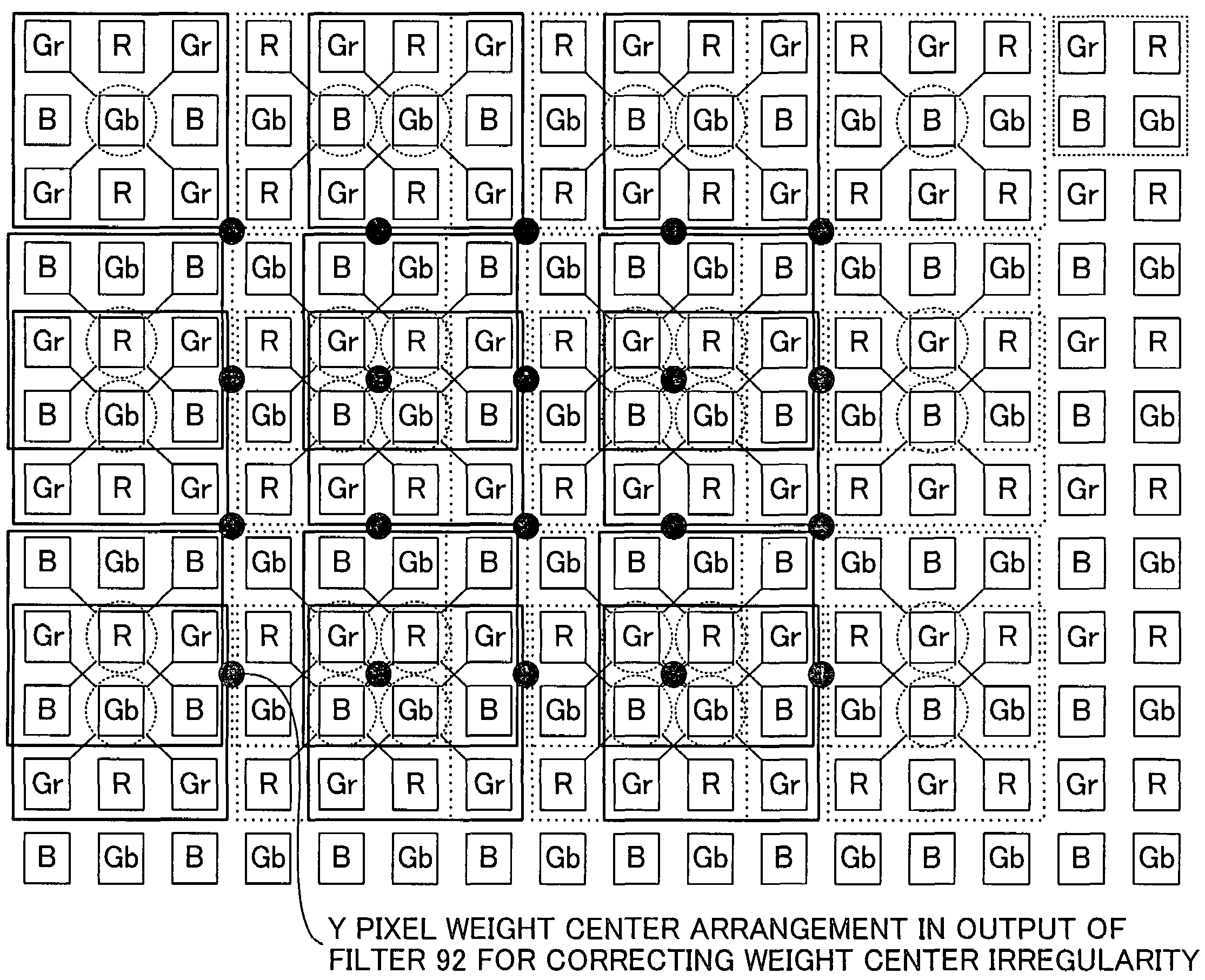
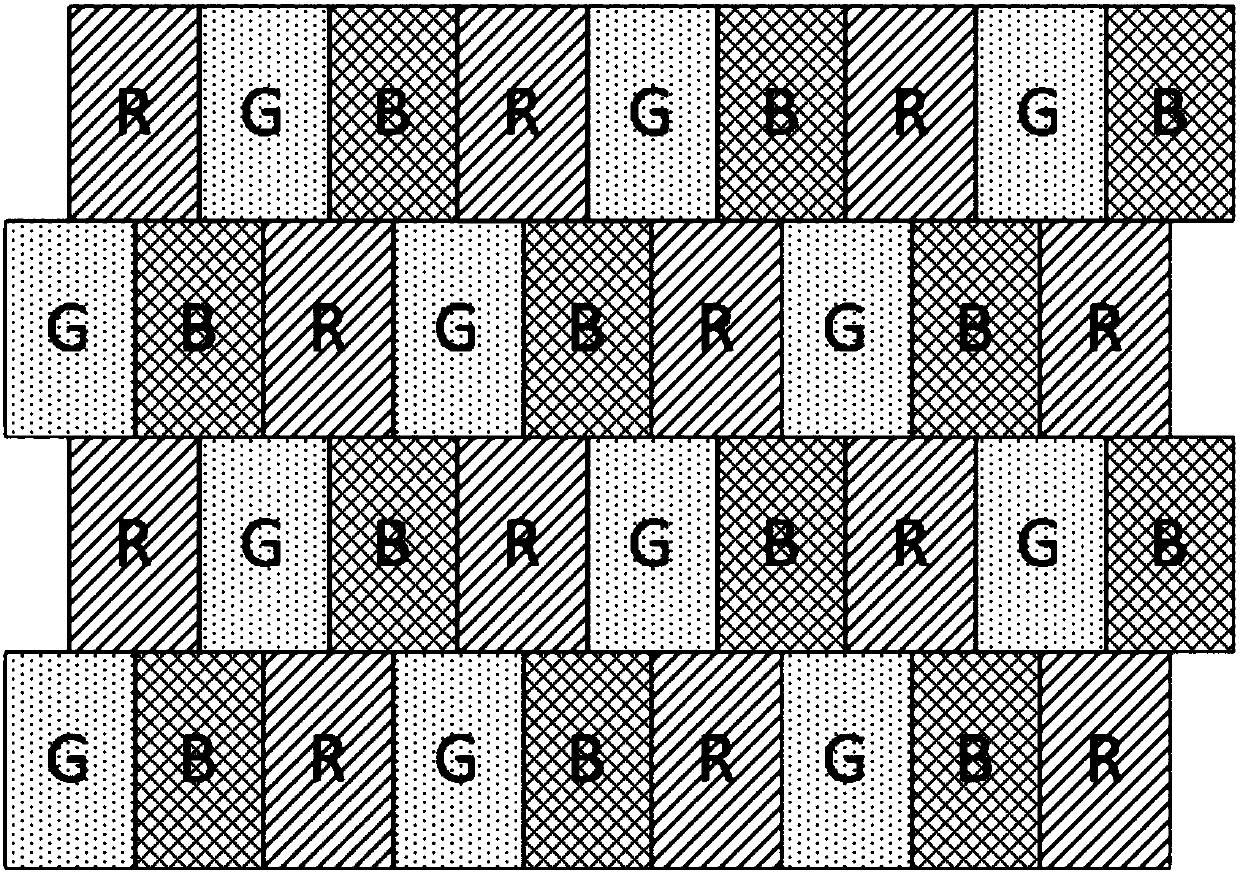
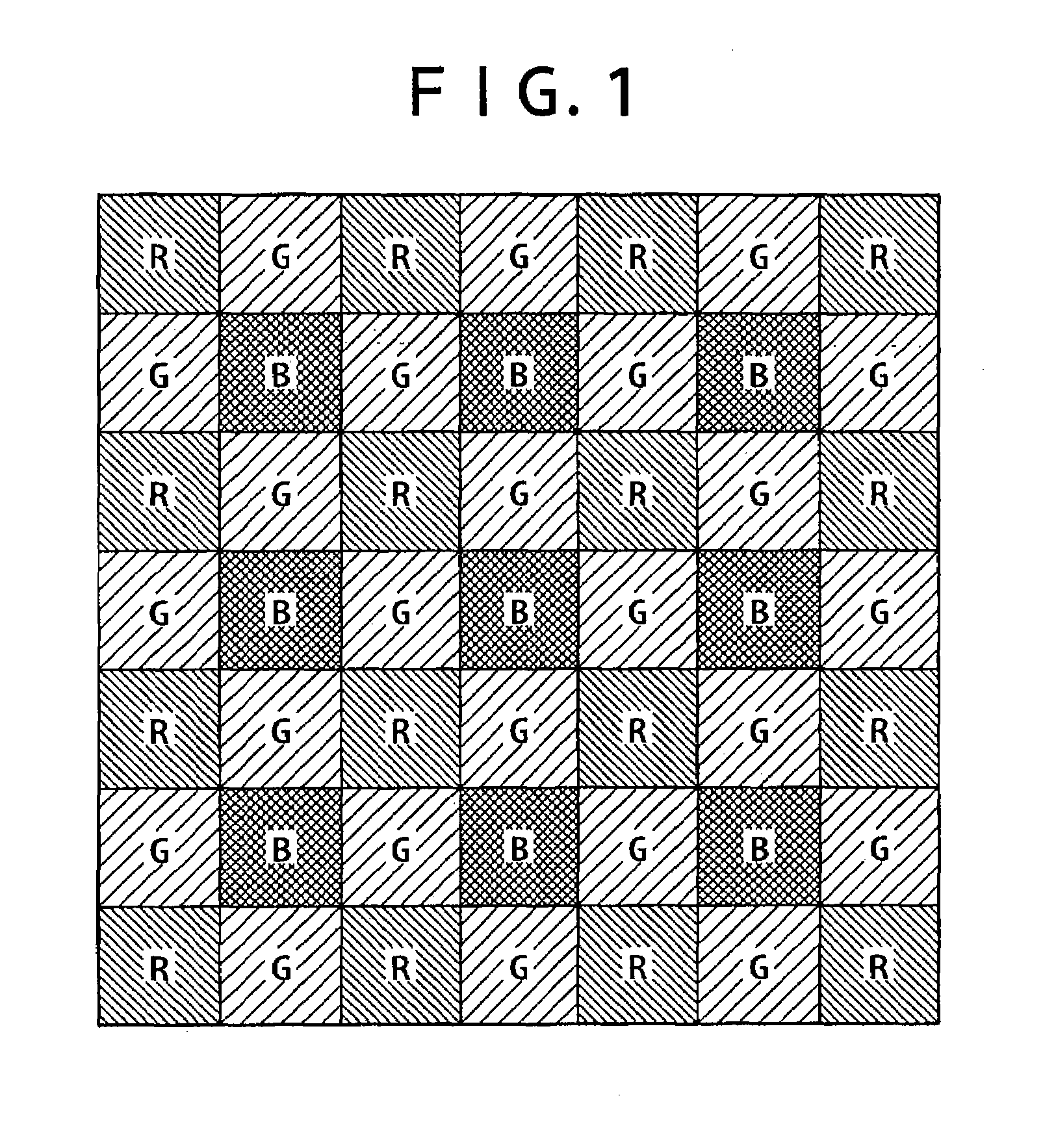
An image processing apparatus is provided which includes: an image obtaining unit for obtaining a mosaic image by an image sensor having at least four kinds of filters with different spectral sensitivities, one of the at least four kinds of filters being used for each pixel; and an image processing unit for generating a color image such that intensity information at a position of each pixel, the intensity information corresponding to a number of colors determined by the spectral sensitivities of the filters, is computed for all pixels, from the mosaic image obtained by the image obtaining unit. In a color filter arrangement of the image sensor of the image obtaining unit, filters possessed by pixels arranged in a checkered manner in half of all the pixels have at least some spectral characteristics having a strong correlation with the spectral characteristic of luminance. The image processing unit calculates estimated intensity values of colors possessed by the pixels arranged in the checkered manner at each pixel and calculates a luminance value of each pixel on a basis of the estimated intensity values.
Owner:SONY CORP
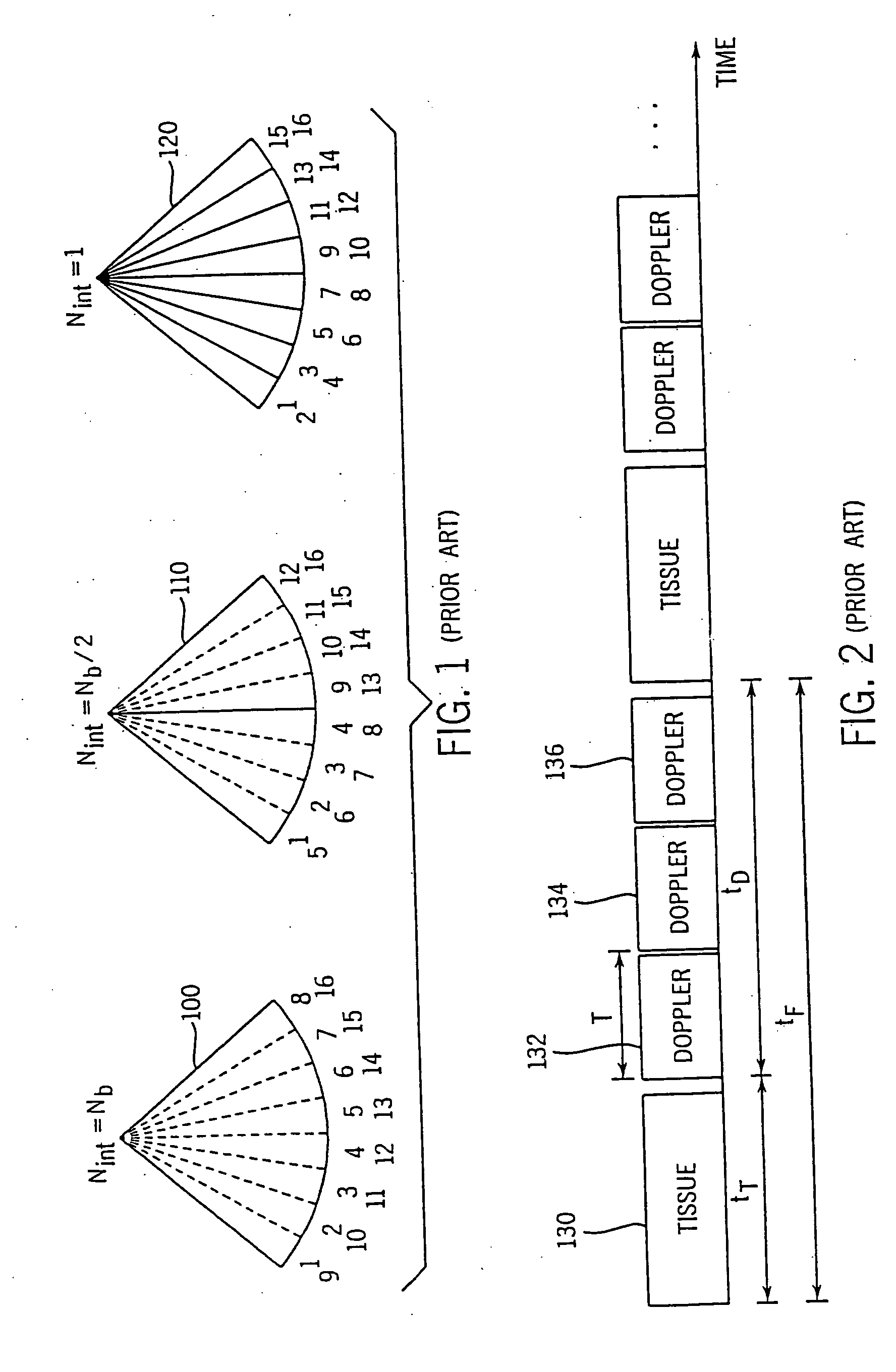
Method and apparatus for providing real-time calculation and display of tissue deformation in ultrasound imaging
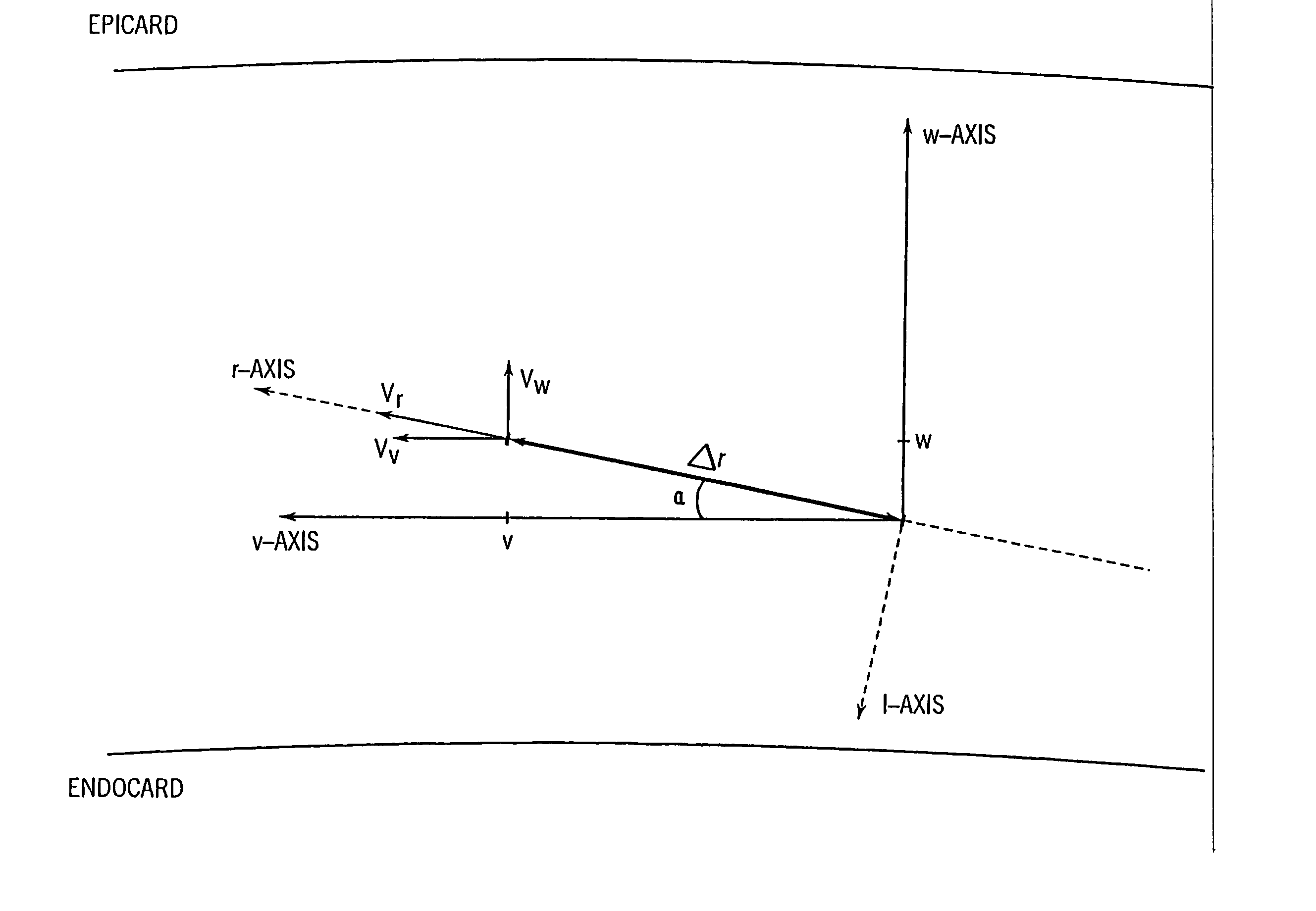
InactiveUS20050203390A1Reduce aliasingMaintain spatial resolutionElectrocardiographyOrgan movement/changes detectionRelative displacementImage resolution
An ultrasound system and method for calculation and display of tissue deformation parameters are disclosed. An ultrasound acquisition technique that allows a high frame rate in tissue velocity imaging or strain rate imaging is employed. With this acquisition technique the same ultrasound pulses are used for the tissue image and the Doppler based image. A sliding window technique is used for processing. The tissue deformation parameter strain is also determined by an accumulation of strain rate estimates for consecutive frames over an interval. The interval may be a triggered interval generated by, for example, an R-wave in an ECG trace. The strain calculation may be improved by moving the sample volume from which the strain rate is accumulated from frame-to-frame according to the relative displacement of the tissue within the original sample volume. The relative displacement of the tissue is determined by the instantaneous tissue velocity of the sample volume. An estimation of strain rate based upon a spatial derivative of tissue velocity is improved by adaptively varying the spatial offset, dr. The spatial offset, dr, can be maximized to cover the entire tissue segment (e.g., heart wall width) while still keeping both of the sample volumes at each end of the offset within the tissue segment. This may be accomplished by determining whether various parameters (e.g., grayscale value, absolute power estimate, magnitude of the autocorrelation function with unity temporal lag and / or magnitude of strain correlation) of the sample volumes within in the spatial offset are above a given threshold. Strain rate may be estimated using a generalized strain rate estimator that is based on a weighted sum of two-sample strain rate estimators with different spatial offsets. The weights are proportional to the magnitude of the strain rate correlation estimate for each spatial offset, and thus reduce the effect of noisy, i.e. poorly correlated, samples. An improved signal correlation estimator that uses a spatial lag in addition to the usual temporal lag is disclosed. The spatial lag is found from the tissue velocity. The improved signal correlation estimator can be utilized both in the estimation of strain rate and tissue velocity. Tissue velocity may be estimated in a manner that reduces aliasing while maintaining spatial resolution. Three copies of a received ultrasound signal are bandpass filtered at three center frequencies. The middle of the three center frequencies is centered at the second harmonic of the ultrasound signal. A reference tissue velocity is estimated from the two signals filtered at the outside center frequencies. The reference tissue velocity is used to choose a tissue velocity from a number of tissue velocities estimated from the signal centered at the second harmonic. A method to estimate the strain rate in any direction, not necessarily along the ultrasound beam, based on tissue velocity data from a small region of interest around a sample volume is disclosed. Quantitative tissue deformation parameters, such as tissue velocity, tissue velocity integrals, strain rate and / or strain, may be presented as functions of time and / or spatial position for applications such as stress echo. For example, strain rate or strain values for three different stress levels may be plotted together with respect to time over a cardiac cycle. Parameters which are derived from strain rate or strain velocity, such as peak systolic wall thickening percentage, may be plotted with respect to various stress levels,
Owner:G E VINGMED ULTRASOUND
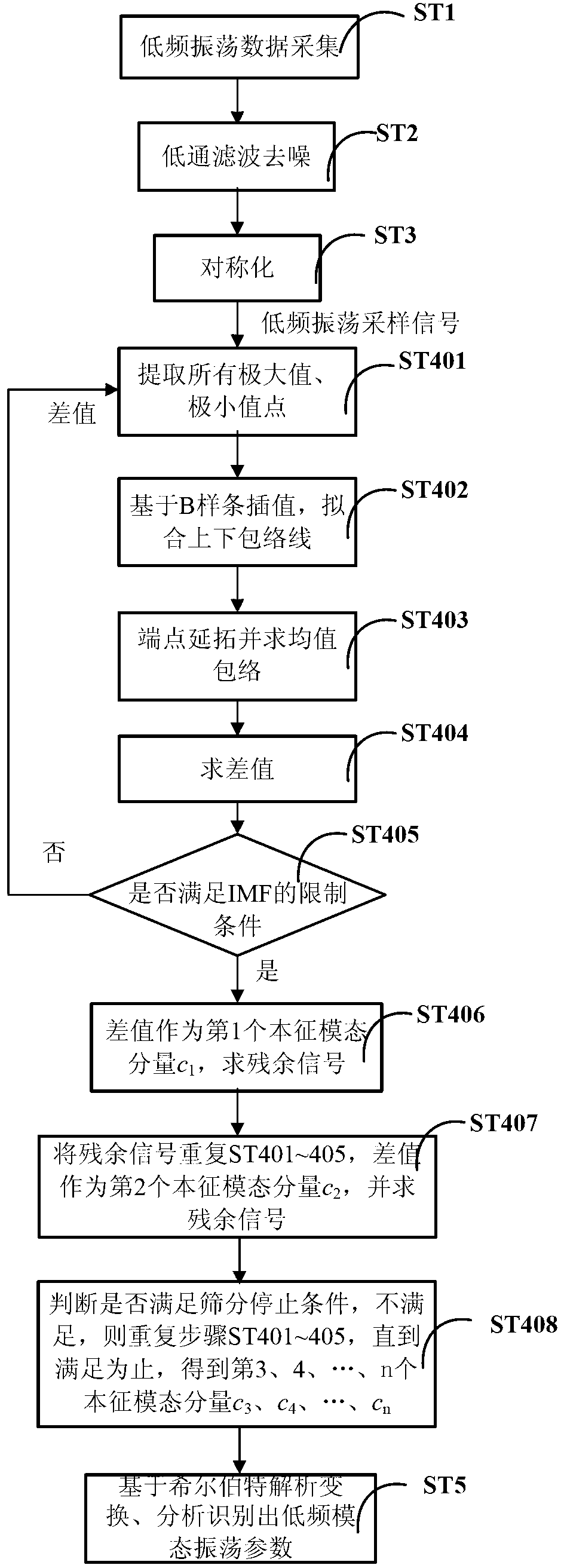
Electric system low-frequency oscillation detection method
InactiveCN102937668AReduce aliasingReduce damping lossSpectral/fourier analysisScreening algorithmEngineering
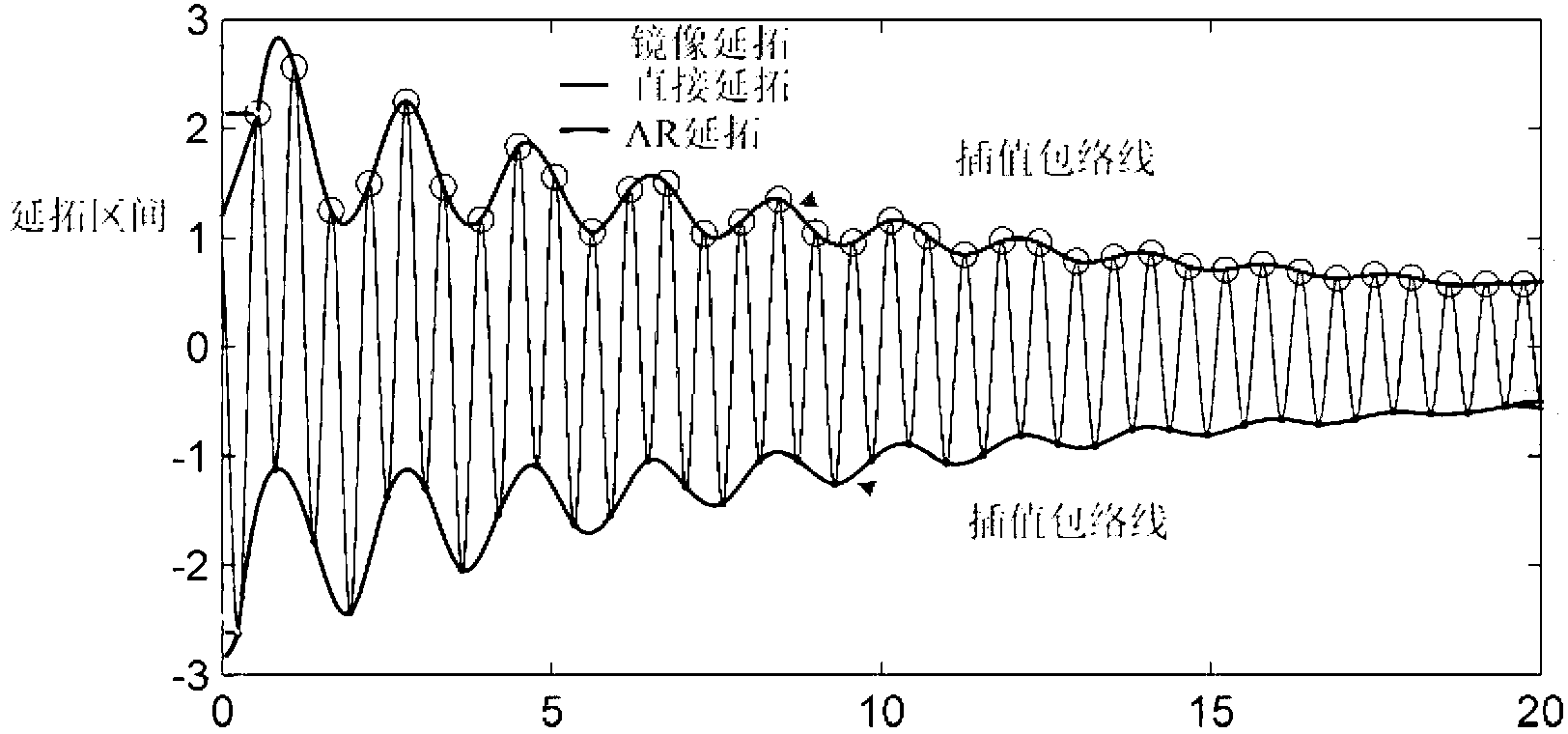
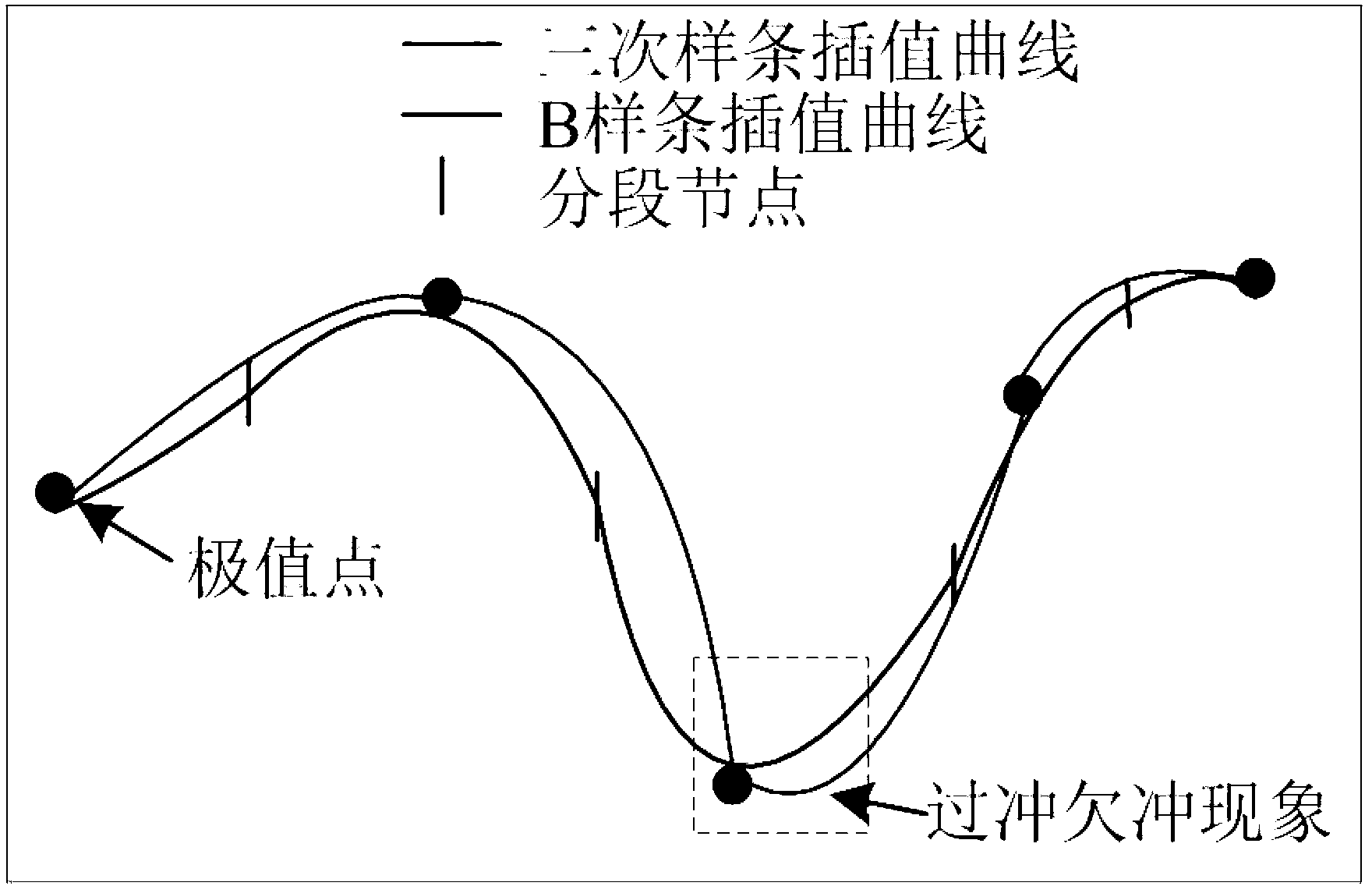
The invention discloses an electric system low-frequency oscillation detection method. On the basis of the existing nonlinear and non-stable signal empirical mode decomposition algorithm, the improved empirical mode screening algorithm enables the electric system low-frequency oscillation detection method to adapt to low-frequency oscillation signal processing, decomposed eigen mode components are combined with low-frequency oscillation mode physical expression meaning, and accordingly all low-frequency oscillation mode parameters of low-frequency oscillation sampling signals are recognized. The electric system low-frequency oscillation detection method reduces influence of the endpoint effect through improved mixed endpoint forecast, removes overshoot and undershoot by adopting triple B spline interpolation, simultaneously effectively reduces mode aliasing degree, analyzes and detects aliasing mode parameters through refined complex wavelets, controls algorithm screening depth through the improved energy elementary error screening stopping condition, reduces damping loss of mode components, and accordingly effectively improves low-frequency oscillation mode characteristic parameter detection accuracy.
Owner:UNIV OF ELECTRONICS SCI & TECH OF CHINA
Dynamic range control and equalization of digital audio using warped processing
ActiveUS20100010651A1Reduce aliasingComputationally efficientDelay line applicationsDigital technique networkFinite impulse responseFrequency spectrum
This invention describes a method for adjusting the loudness and the spectral content of digital audio signals in a real-time using warped spectral filtering. A warped processing module modifies a spectral content of a digital audio signal with a set of gains for a plurality of non-linearly-scaled frequency bands determined by a warping factor λ of a warped delay line. Warped delay line signals, generated by the warped delay line, are processed by a warped filter block containing multiple warped finite impulse response filters, e.g., Mth band filters, using individual warped spectral filtering in said plurality of the non-linearly-scaled frequency bands, which is followed by a conventional processing by a dynamic range control / equalization block. The present invention describes another innovation, that is embedding the warped processing module in a two-channel quadrature mirror filter (QMF) bank for improving processing efficiency at high sample rates.
Owner:BEIJING XIAOMI MOBILE SOFTWARE CO LTD
Method and apparatus of M/r imaging with coil calibration data acquisition
InactiveUS7064547B1High calibration signalFull FOV coverageMagnetic measurementsElectric/magnetic detectionImage resolutionData acquisition
A system and method for MR imaging with coil sensitivity or calibration data acquisition for reducing wrapping or aliasing artifacts is disclosed. Low resolution MR data representative of coil sensitivity of a coil arrangement within an FOV is acquired prior to application of an imaging data acquisition scan and is used to reduce wrapping or aliasing artifacts in a reconstructed image.
Owner:GENERAL ELECTRIC CO
Method and apparatus for providing real-time calculation and display of tissue deformation in ultrasound imaging
InactiveUS7261694B2Reduce impactReduce aliasingElectrocardiographyOrgan movement/changes detectionUltrasound imagingSonification
An ultrasound system and method for calculation and display of tissue deformation parameters are disclosed. The tissue deformation parameter strain is determined by an accumulation of strain rate estimates for consecutive frames over an interval. The interval may be a triggered interval generated by, for example, an R-wave in an ECG trace. Three quantitative tissue deformation parameters, such as tissue velocity, tissue velocity integrals, strain rate and / or strain, may be presented as functions of time and / or spatial position for applications such as stress echo. For example, strain rate or strain values for three different stress levels may be plotted together with respect to time over a cardiac cycle. Parameters which are derived from strain rate or strain velocity, such as peak systolic wall thickening percentage, may be plotted with respect to various stress levels.
Owner:G E VINGMED ULTRASOUND
Spectral imaging method and spectrum imaging instrument of snapshot-type high throughput
ActiveCN102944305AImprove throughputPixel Count ReductionRadiation pyrometrySpectrum investigationMicro lens arrayPrimary mirror
The invention relates to a spectral imaging method and a spectrum imaging instrument of a snapshot-type high throughput, which aim at realizing a picture-type photograph. The spectrum instrument comprises a linear gradient filter array and an optical-field imaging mechanism, and the optical-field imaging mechanism comprises a primary mirror, a micro lens array and a detector. The filter array is arranged on an aperture diaphragm between lenses of the primary mirror, the filter array consists of a plurality of filters which have identical width and are arranged at intervals, and the filter is a linear gradient filter band with continuous spectrum. The micro lens array is arranged on an imaging surface of a front-mounted optical imaging system, and the detector is arranged on a focus plane of the micro lens array. The spectrum imaging method is characterized in that light in different directions transmitted or reflected by a target is imaged on one micro lens after being modulated by the primary mirror and the filter array, the light is dispersed by the micro lens onto a picture element of the detector to form a subimage, and finally three-dimensional spectrum image data is obtained. The complete spectrum image information of the target can be obtained through the photograph in one step, and the spectral imaging method and the spectrum imaging instrument can be used for monitoring and tracking a rapid variable or movable target.
Owner:BEIHANG UNIV
Digital processing radar system
InactiveUS7492313B1Reduce aliasingSuppress signalAntennasRadio wave reradiation/reflectionDigital dataRadar systems
A radar system array antenna includes plural elements. Each element is coupled to a port of a transmit-receive device. Digital data representing the characteristics of the radar signal is applied to a monolithic IC combination DAC / ADC, which converts the digital data into analog transmit signal. The analog transmit signal is applied to a monolithic microwave IC transceiver, which may be monolithic with the DAC / ADC. The transceiver upconverts the transmit signal. The upconverted transmit signal is applied to a transmit port of the transmit-receive device and transmitted. Receive RF signal is coupled from each antenna element through the transmit-receive device to a receive signal port of the transceiver. The transceiver downconverts the receive signal to produce received downconverted signal. The downconverted signal is coupled to the ADC part of the DAC / ADC, which converts to receive digital signal. Digital processing processes the receive digital signal to produce the return target information.
Owner:LOCKHEED MARTIN CORP
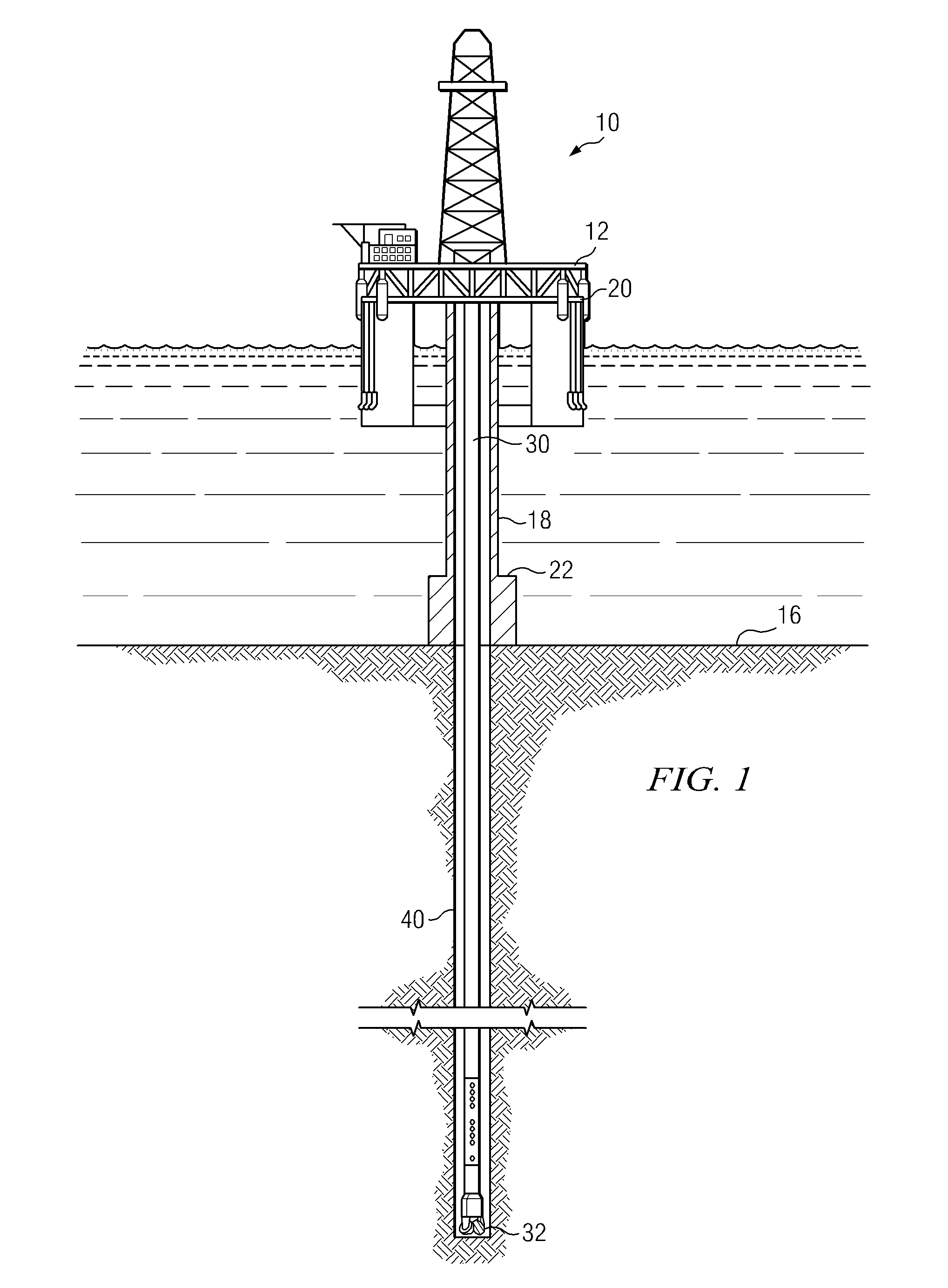
Downhole Sonic Logging Tool Including Irregularly Spaced Receivers
InactiveUS20110188345A1Reduces (or even substantially eliminates) aliasingReducing aliasing effectSeismology for water-loggingAcousticsMechanical engineering
A downhole acoustic measurement tool includes at least one transmitter longitudinally spaced apart from a non-uniformly spaced longitudinal array of acoustic receivers. The array has a non-uniform spacing such that a first spacing between a first pair of consecutive acoustic receivers in the array is not equal to a second spacing between a second pair of consecutive acoustic receivers in the array. Non-uniform spacing of the receivers in the array reduces aliasing when the received waveforms are processed, for example, to obtain semblance data.
Owner:SCHLUMBERGER TECH CORP
Methods and devices for charge management for three-dimensional and color sensing
InactiveUS20050156121A1Reduce peak powerImprove resolution accuracyOptical rangefindersTelevision system scanning detailsCMOSHigh frequency modulation
Structures and methods for three-dimensional image sensing using high frequency modulation includes CMOS-implementable sensor structures using differential charge transfer, including such sensors enabling rapid horizontal and slower vertical dimension local charge collection. Wavelength response of such sensors can be altered dynamically by varying gate potentials. Methods for producing such sensor structures on conventional CMOS fabrication facilities include use of “rich” instructions to command the fabrication process to optimize image sensor rather than digital or analog ICs. One detector structure has closely spaced-apart, elongated finger-like structures that rapidly collect charge in the spaced-apart direction and then move collected charge less rapidly in the elongated direction. Detector response is substantially independent of the collection rate in the elongated direction.
Owner:MICROSOFT TECH LICENSING LLC
Method enabling a standard CMOS fab to produce an IC to sense three-dimensional information using augmented rules creating mask patterns not otherwise expressible with existing fab rules
InactiveUS7464351B2Reduce peak powerImprove resolution accuracyOptical rangefindersTelevision system scanning detailsCMOSEngineering
CMOS implementable three-dimensional silicon sensors are fabricated using a standard fab but using augmented rules that create mask patterns not expressible with existing fab rules. Standard fab rules are not optimized to produce high quality three-dimensional silicon sensors. Accordingly, the normal set of rules does not permit creating the fab mask patterns necessary for high performance such sensors. However, the present invention can use the fab standard mask set with a rich set of fab instructions to express mask patterns from the mask set that would not otherwise be expressible. The resultant method enables high quality silicon sensors for three-dimensional sensing to be readily mass produced from a standard fab.
Owner:MICROSOFT TECH LICENSING LLC
Real-time network adaptive digital video encoding/decoding
ActiveUS8250618B2Reduce scaleLow data ratePicture reproducers using cathode ray tubesPicture reproducers with optical-mechanical scanningDigital videoImage resolution
A method for real time video transmission over networks with varying bandwidth is described. Image quality is maintained even under degrading network performance conditions through the use of image scaling in conjunction with block based motion compensated video coding (MPEG2 / 4, H.264, et. Al.). The ability to quickly switch resolutions without decreasing reference frame correlation is shown enabling a fast switch to reduce the required bandwidth for stable image quality.
Owner:ELEMENTAL TECH LLC
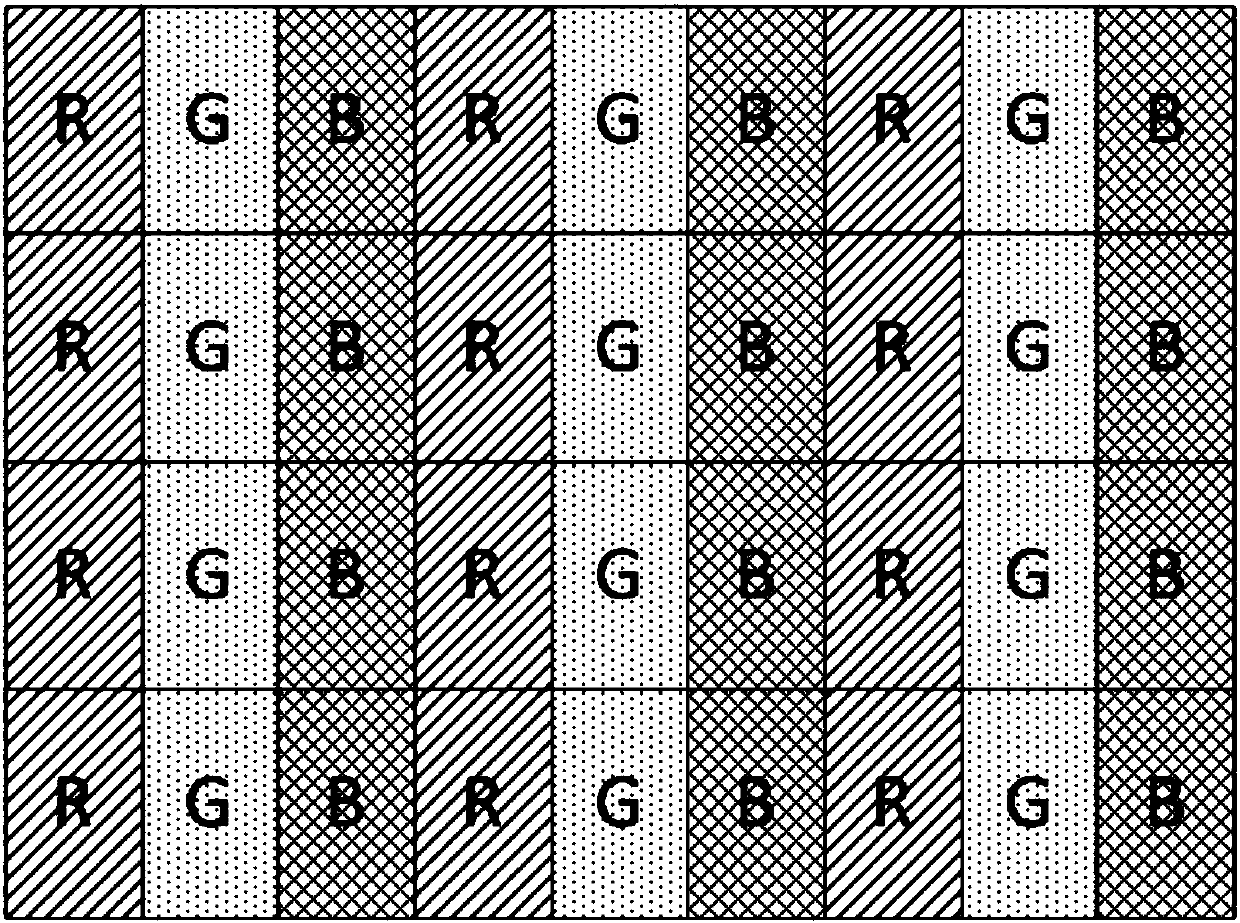
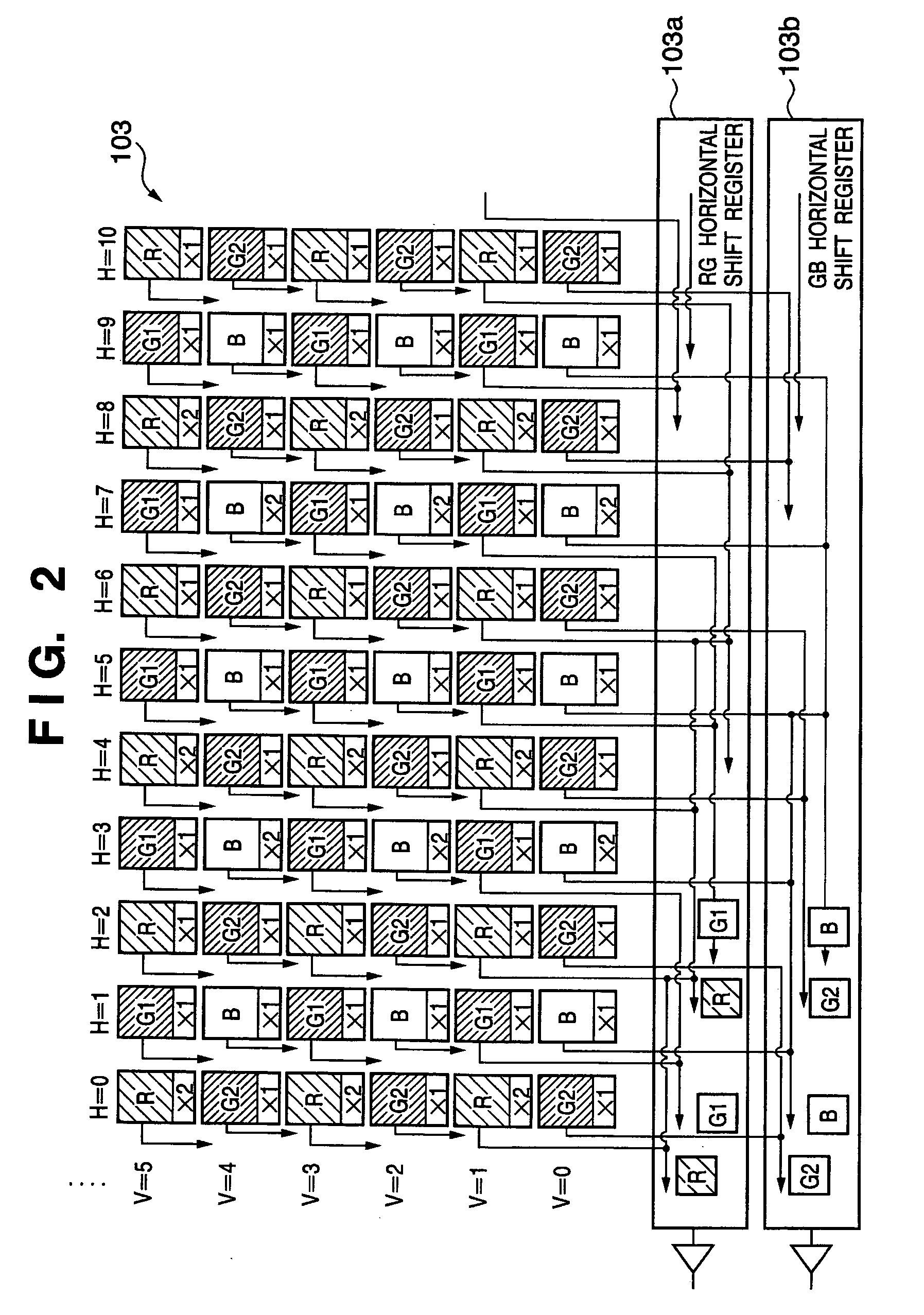
Vertical color filter sensor group array that emulates a pattern of single-layer sensors with efficient use of each sensor group's sensors
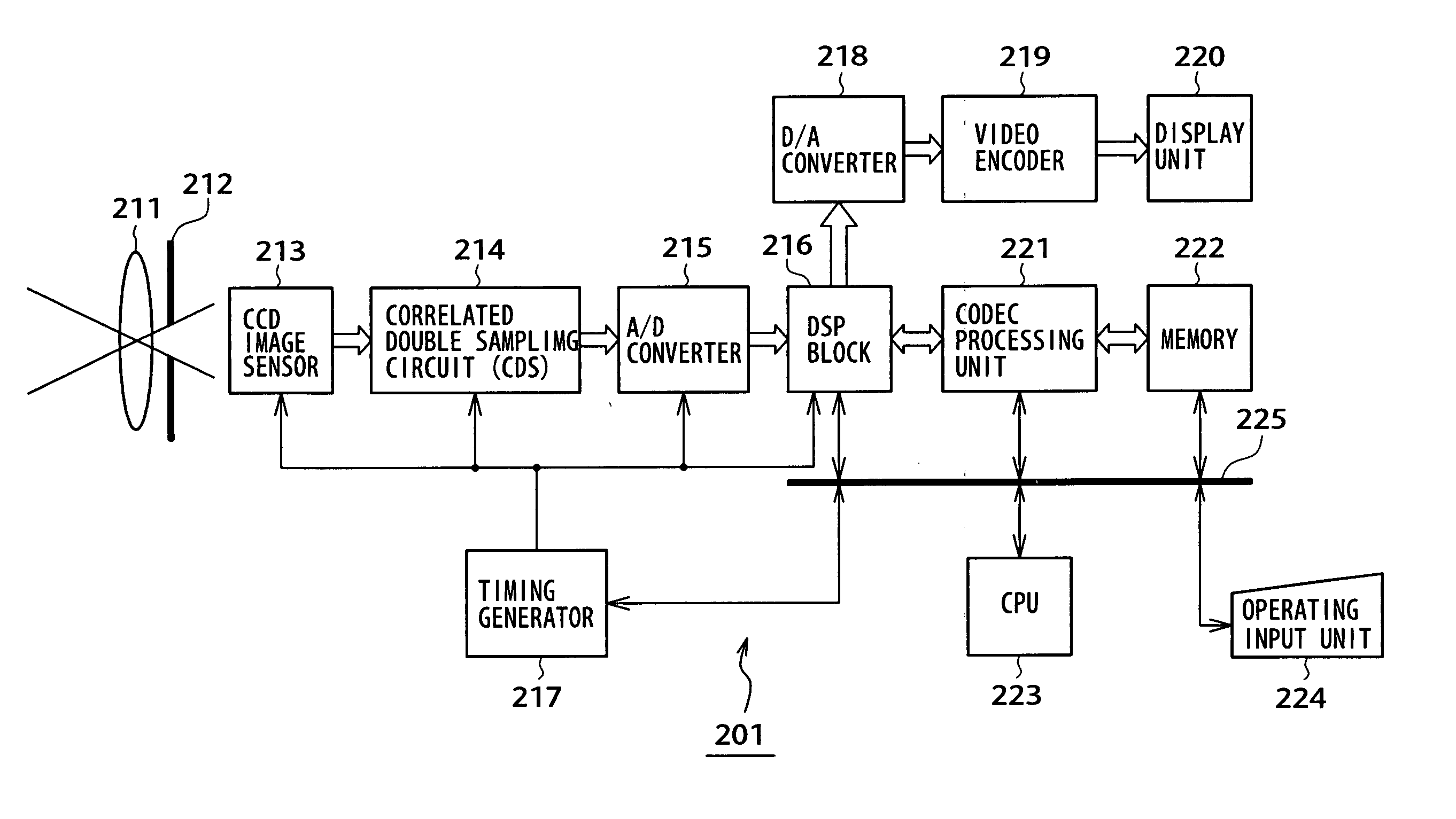
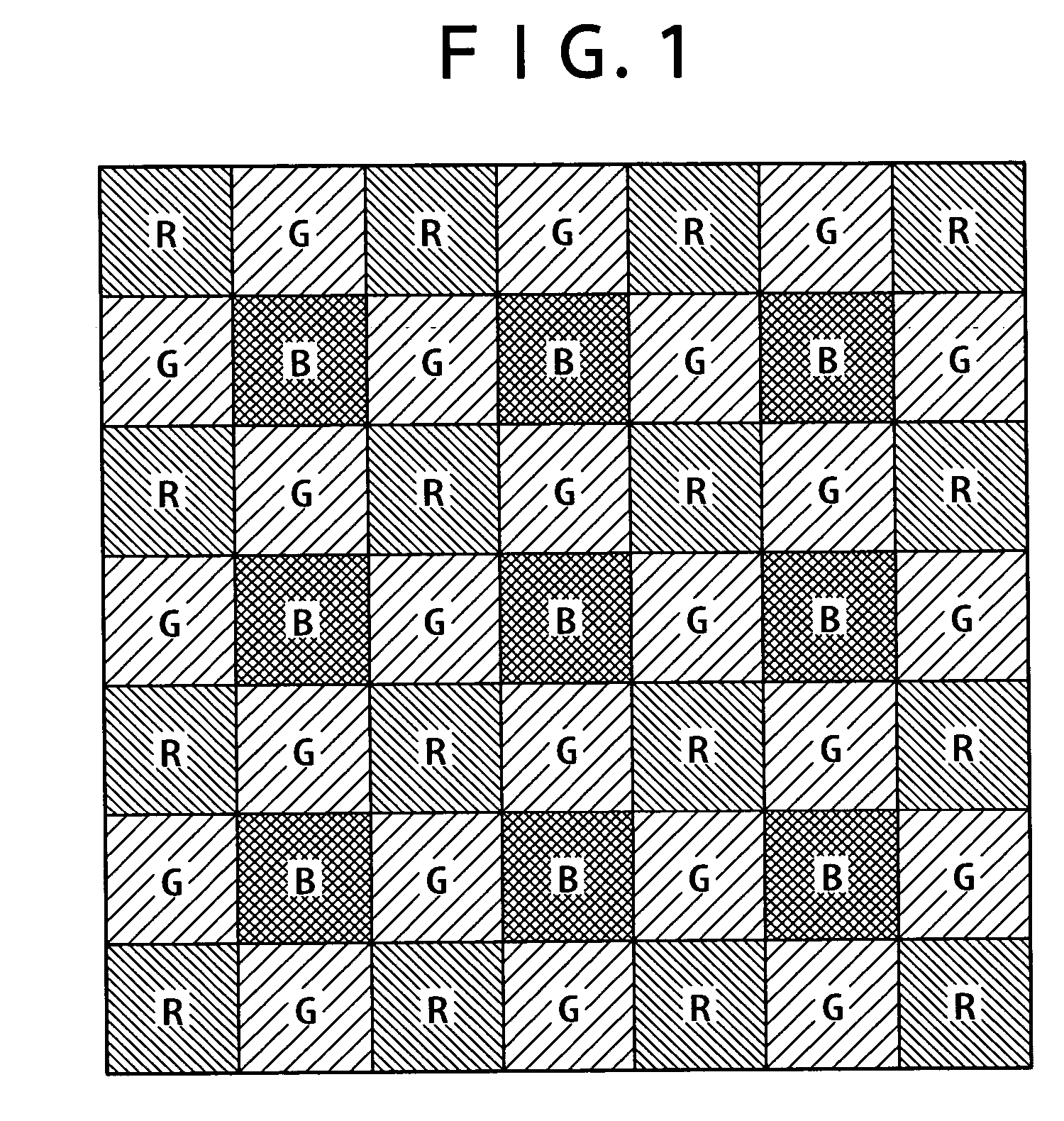
InactiveUS6998660B2High resolutionEasy to integrateSolid-state devicesRadiation controlled devicesSensor arrayMultiple sensor
An array of vertical color filter (VCF) sensor groups, each VCF sensor group including at least two vertically stacked, photosensitive sensors. Preferably, the array is fabricated, or the readout circuitry is configured (or has a state in which it is configured), to combine the outputs of sensors of multiple sensor groups such that the array emulates a conventional array of single-layer sensors arranged in a Bayer pattern or other single-layer sensor pattern, and such that the outputs of at least substantially all of the sensors of each of the VCF sensor groups are utilized to emulate the array of single-layer sensors.
Owner:FOVEON
Method, computer readable medium and apparatus for converting color image resolution
InactiveUS20050008258A1Reduce aliasingHigh resolutionGeometric image transformationVisual presentation using printersColor imageImage resolution
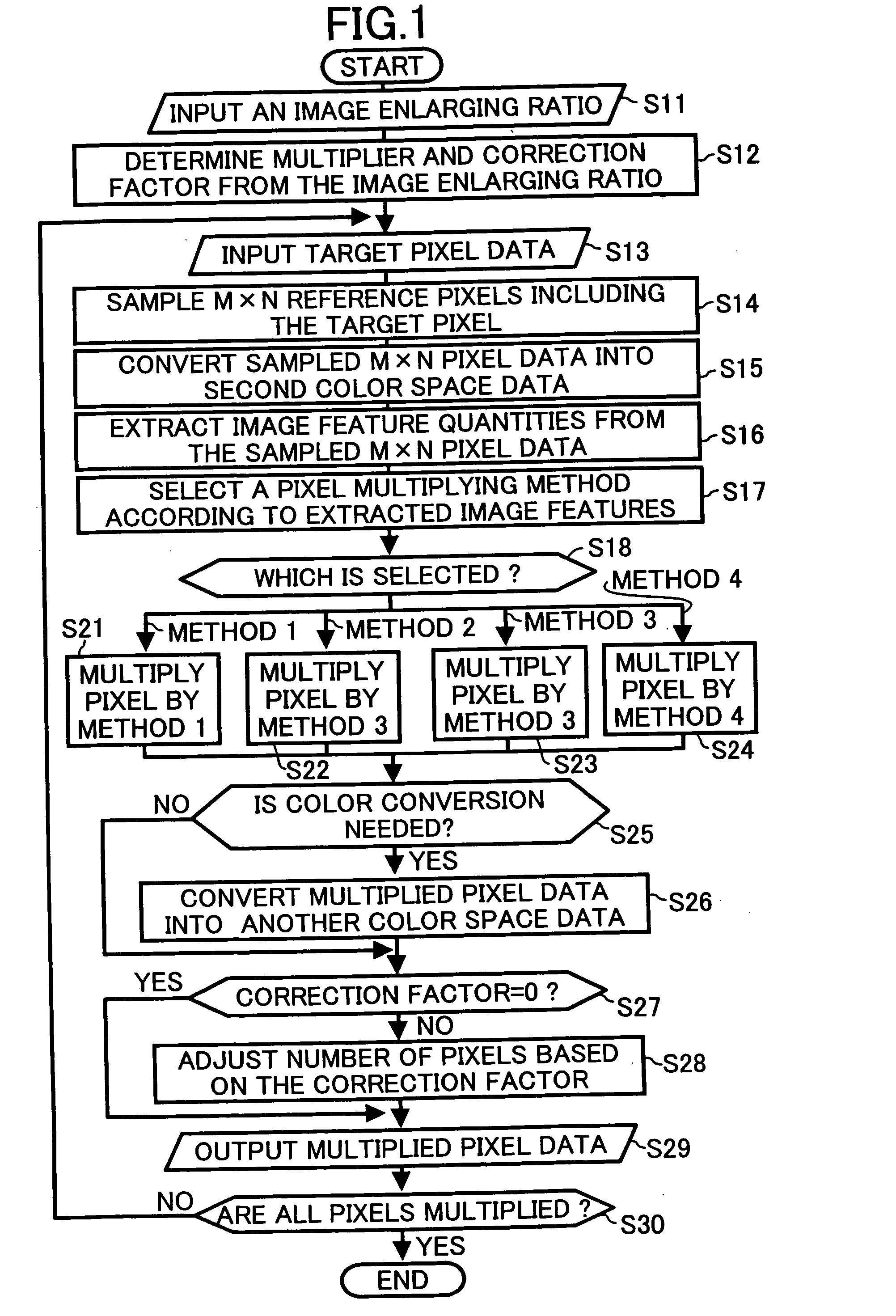
A method, computer readable medium and apparatus for converting color image resolution including inputting an image enlarging ratio, inputting target pixel data of an image in a first color space to be enlarged in a sequence, sampling reference pixels including the target pixel and pixels at least one of which links to the target pixel, and converting the reference pixel data into a second color space data. Other functions include extracting image feature quantities from the reference pixel data, selecting one of a plurality of pixel multiplying methods according to the extracted image feature quantities, multiplying the target pixel by the selected image multiplying method with an integer value based on the input image enlarging ratio, and outputting pixel data that have been generated by the target pixel multiplying step in a sequence.
Owner:SUZUKI HIROAKI +1
Solid-state color imaging apparatus capable of reducing false signals with regard to both luminance and chrominance
ActiveUS7349016B2Reduce the number of pixelsIncrease speedTelevision system detailsTelevision system scanning detailsLow-pass filterImaging equipment
A pixel area of a megapixel solid-state color imaging device is divided into unit areas for pixel adding and all the pixels for the same color are added together in each unit area. Accordingly, the percentage of utilized pixels is raised to 100% and aliasing noise to low frequencies in a high-frequency video signal is greatly suppressed by a spatial LPF (low pass filter) effect of the pixel addition in the area units.
Owner:PANASONIC SEMICON SOLUTIONS CO LTD
Device and Method for Processing a Real Subband Signal for Reducing Aliasing Effects
ActiveUS20100013987A1Great filter lengthReduce aliasingMultiple-port networksGain controlDiscrete-time signalComputer science
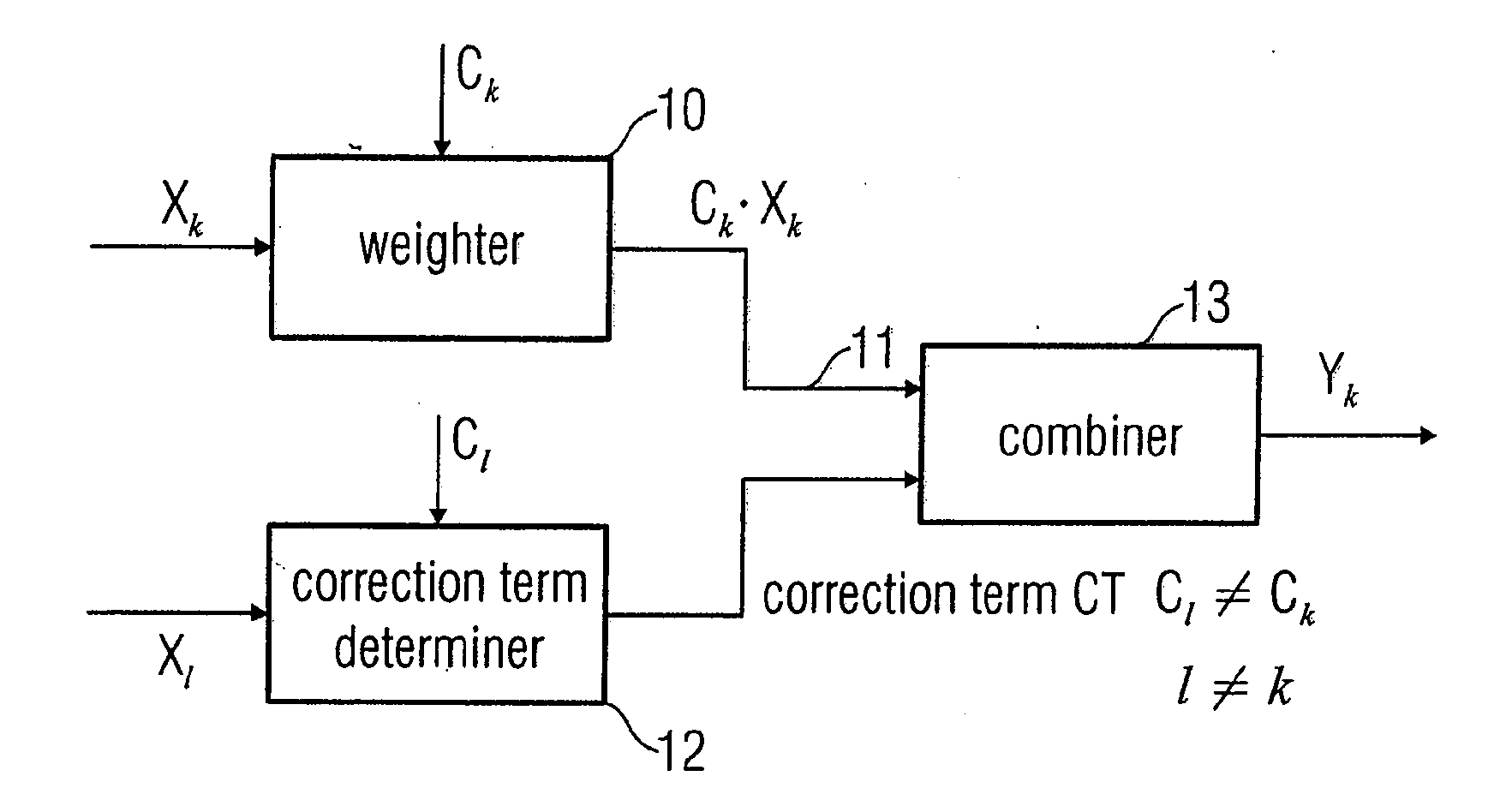
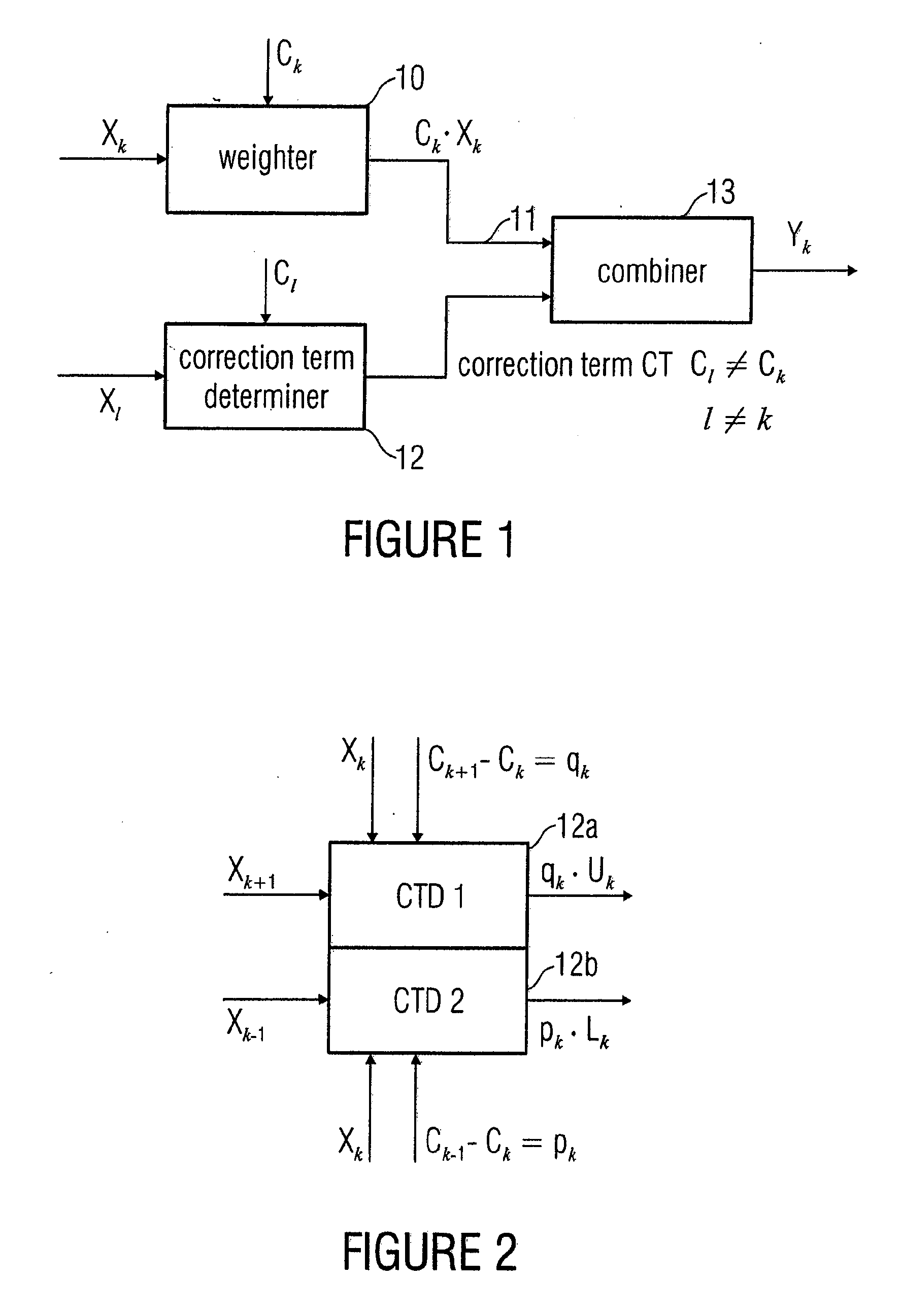
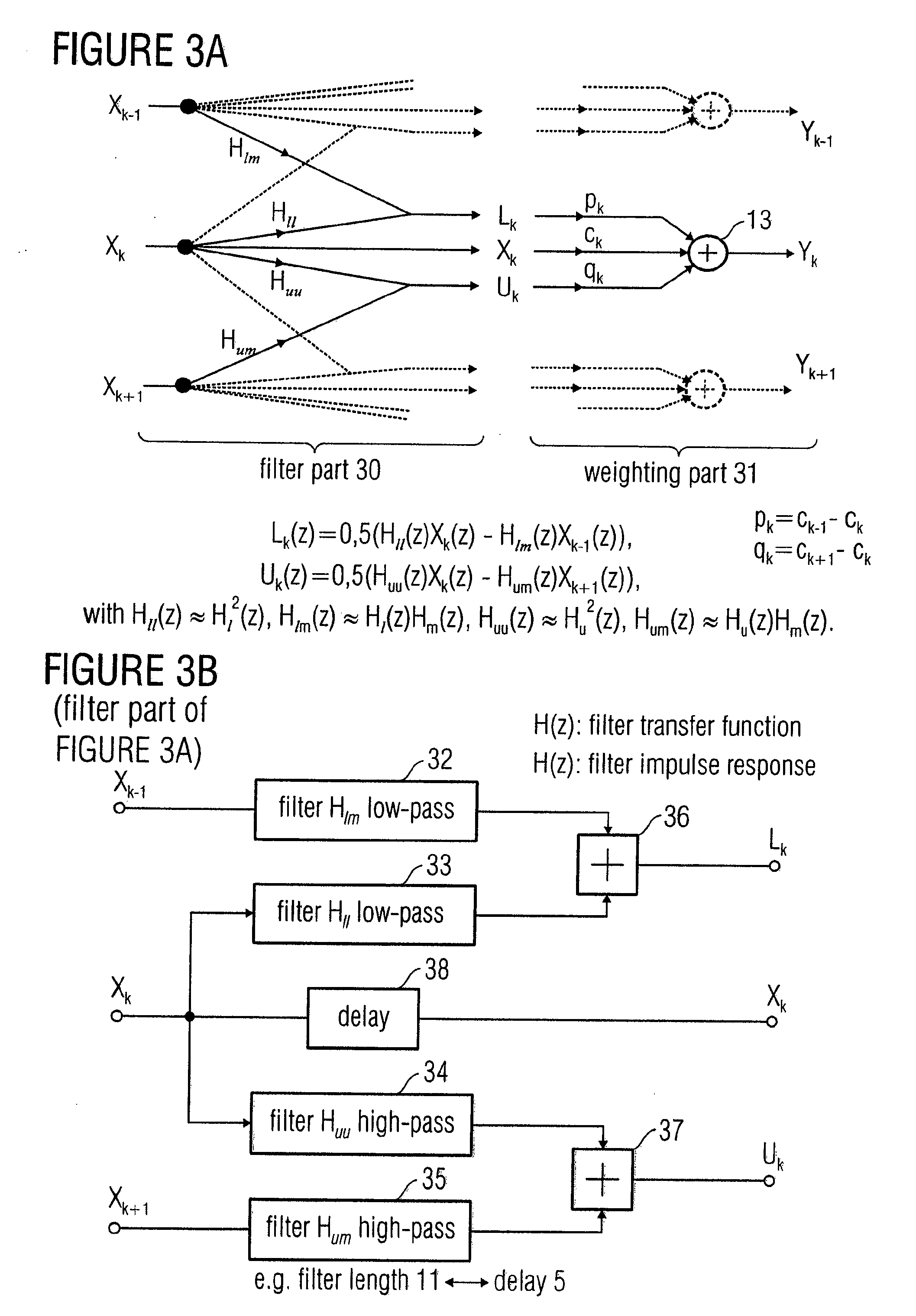
In order to process a subband signal of a plurality of real subband signals which are a representation of a real discrete-time signal generated by an analysis filter bank, a weighter for weighting a subband signal by a weighting factor determined for the subband signal is provided to obtain a weighted subband signal. In addition, a correction term is calculated by a correction term determiner, the correction term determiner being implemented to calculate the correction term using at least one other subband signal and using another weighting factor provided for the other subband signal, the two weighting factors differing. The correction term is then combined with the weighted subband signal to obtain a corrected subband signal, resulting in reduced aliasing, even if subband signals are weighted to a different extent.
Owner:FRAUNHOFER GESELLSCHAFT ZUR FOERDERUNG DER ANGEWANDTEN FORSCHUNG EV
Solid-state imaging apparatus, imaging system and driving method for solid-state imaging apparatus
ActiveUS20100060754A1Reduce aliasingHigh speedTelevision system detailsTelevision system scanning detailsEngineeringImage system
A solid-state imaging apparatus, an imaging system and a driving method for the solid-state imaging apparatus that can reduce jaggy while increasing speed of operation for reading out signals are provided. The driving method includes a first step of storing one or more signals from the plurality of pixels in each of the plurality of first holding units; a second step of adding the signals from the plurality of pixels stored in the plurality of first holding units; and a third step of outputting the signal stored in the second holding unit, such that at least a part of a period of the first step is overlapped with a period of the third step.
Owner:CANON KK
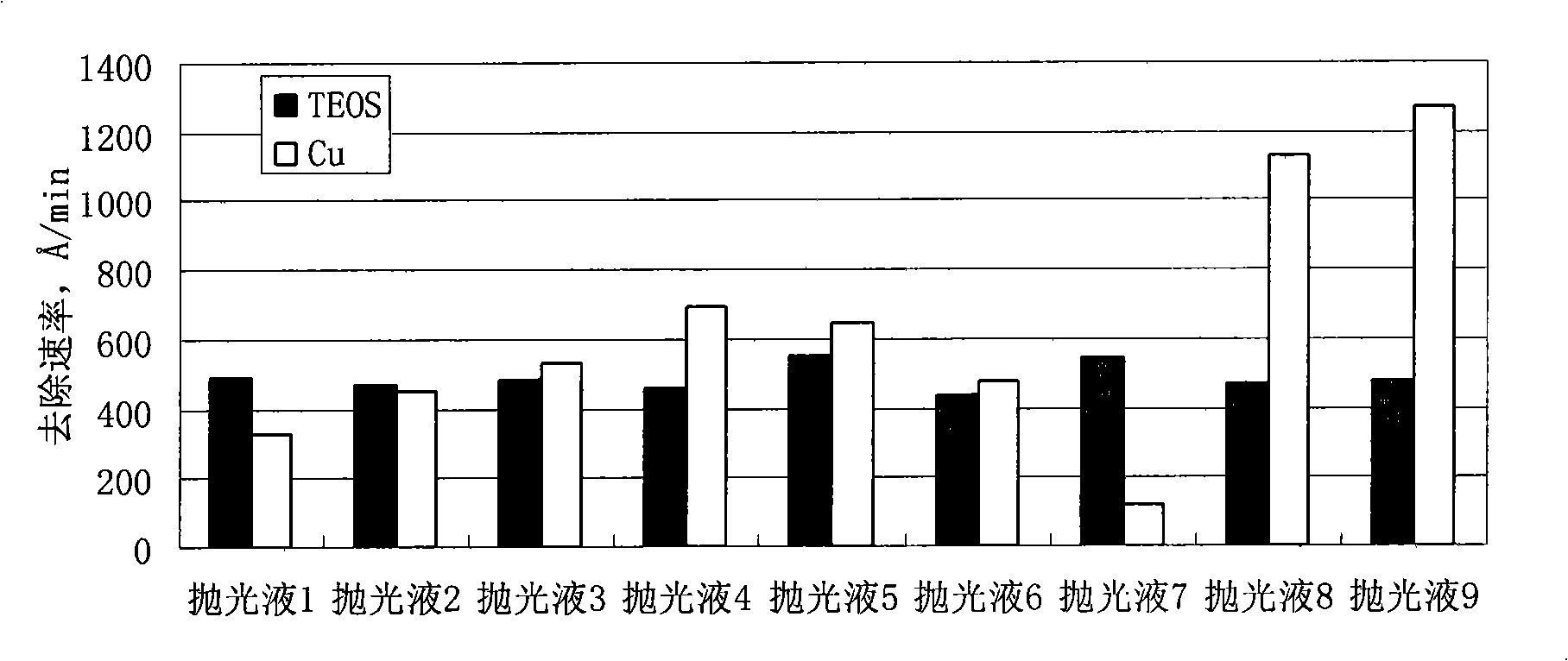
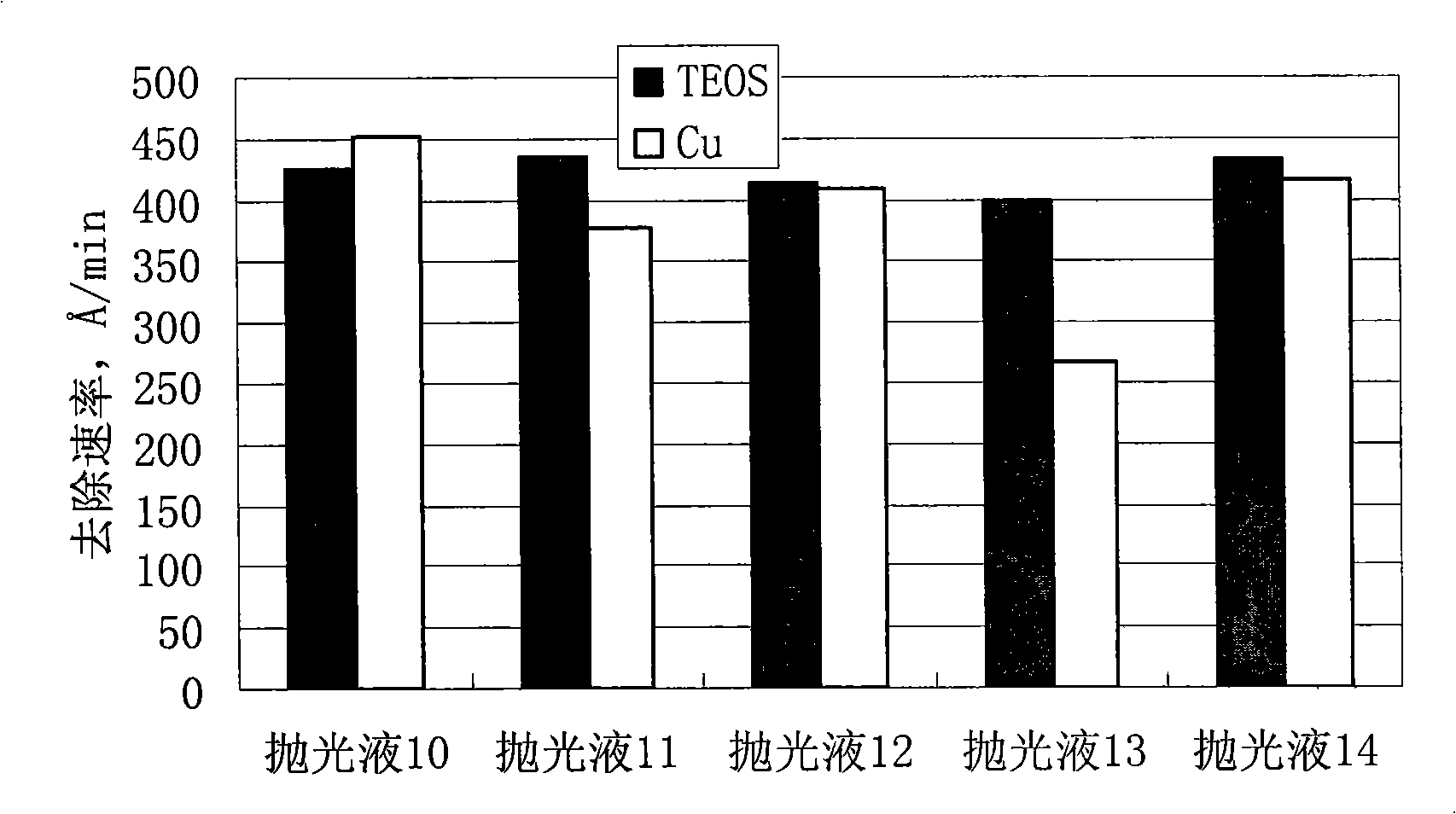
Polishing solution for polishing low dielectric material
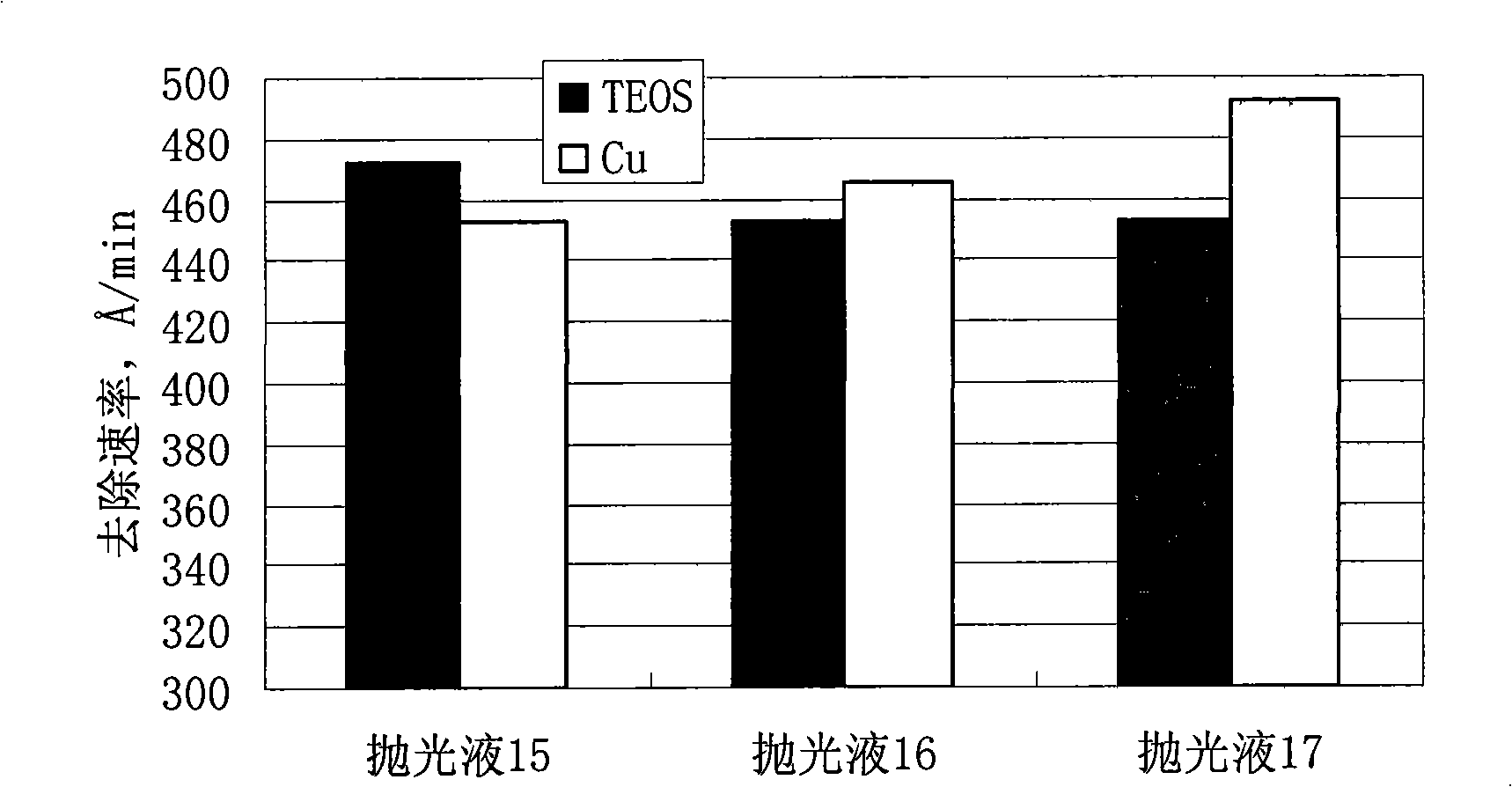
InactiveCN101407699AImprove protectionImprove electrical performancePolishing compositions with abrasivesElectrical performanceThin line
The invention discloses polishing liquid for polishing low dielectric materials, which contains silicon dioxide and water; and the polishing liquid is characterized in that the polishing liquid further contains a mixed corrosion inhibitor. The polishing liquid can enhance the protection of a Cu thin line, significantly reduce the defects of saw teeth, dents, scratches, erosion, scratches, and the like, and improve the electrical performance of a polishing chip.
Owner:ANJI MICROELECTRONICS (SHANGHAI) CO LTD
Method and device for mura compensation of display panel
ActiveCN108538264AImprove the compensation effectSmooth transitionStatic indicating devicesPattern recognitionCompensation effect
The present invention provides a method and a device for mura compensation of a display panel. The method comprises the steps of: taking one pixel selected in each compensation area as one compensation pixel to measure and store compensation data corresponding to each compensation pixel, calculating the compensation data of other pixels except the compensation pixels in each compensation area according to the compensation data corresponding to each compensation pixel and a preset bicubic interpolation algorithm, and employing the bicubic interpolation algorithm to calculate the compensation data of other pixels except the compensation pixels in each compensation area. Compared to traditional bilinear interpolation, the image transition is smoother, sawtooth is reduced, the mura compensation effect is improved, and the image quality is improved.
Owner:TCL CHINA STAR OPTOELECTRONICS TECH CO LTD
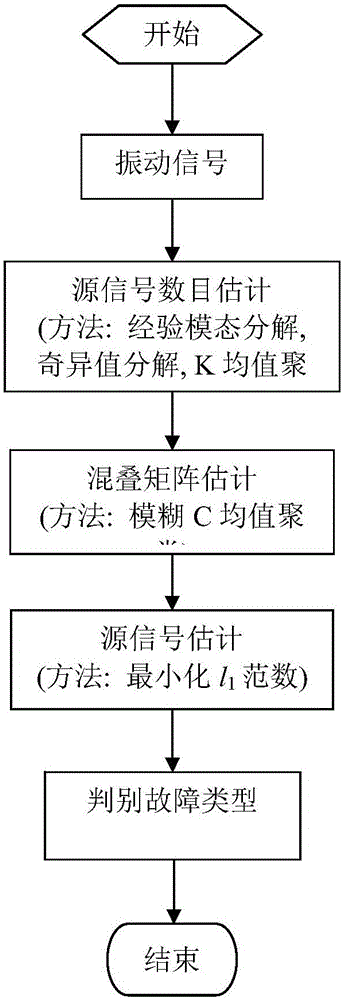
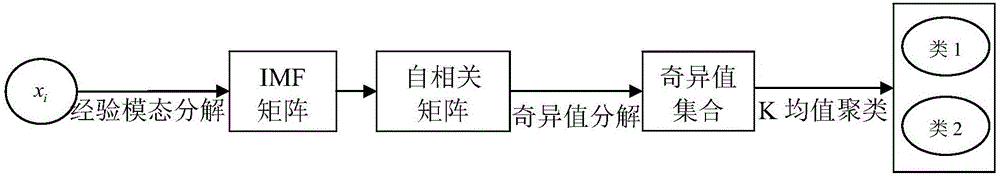
Wind turbine generator gearbox fault diagnosis method based on vibration signal blind source separation and sparse component analysis
ActiveCN106096562AReduce aliasingReduce maintenance costsData processing applicationsCharacter and pattern recognitionSingular value decompositionElectricity
The invention discloses a wind turbine generator gearbox fault diagnosis method based on vibration signal blind source separation and sparse component analysis. The wind turbine generator gearbox fault diagnosis method mainly comprises three algorithms, which are an empirical mode decomposition (EMD), singular value decomposition (SVD) and K-means clustering (K-Means) based source signal number estimation algorithm, a fuzzy C-means clustering (FCM) based aliasing matrix estimation method, and a minimized l1-norm based source signal estimation and diagnosis algorithm. An idea of blind source separation (BSS) and sparse component analysis (SCA) is introduced into the whole algorithm process. The wind turbine generation gearbox fault diagnosis method takes simulated signals and actual vibration signals as a test object, detailed algorithm description is provided, and the effectiveness of the algorithms in the aspects of signal processing and fault analysis is verified through a series of experiments.
Owner:ZHEJIANG UNIV
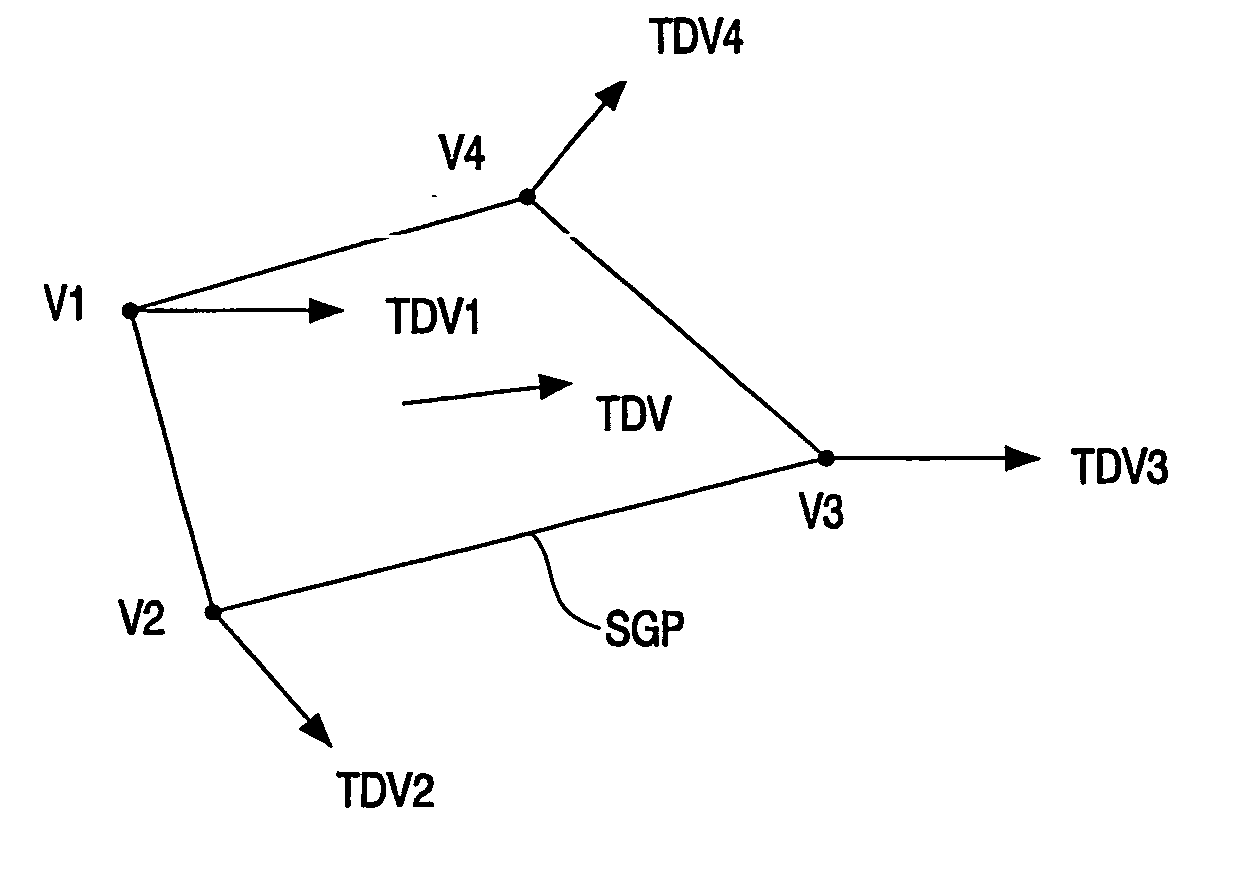
Generation of motion blur
InactiveUS20070120858A1Acceptable motion blurReduce aliasingAnimation3D-image renderingGraphicsGraphic system
In a method of generating motion blur in a 3D-graphics system, geometrical information (GI) defining a shape of a graphics primitive (GP) is received (RSS; RTS) from a 3D-application. A displacement vector (SDV; TDV) defining a direction of motion of the graphics primitive (GP) is also received from the 3D-application or is determined from the geometrical information. The graphics primitive (GP) is sampled (RSS; RTS) in the direction indicated by the displacement vector to obtain input samples (RPi), and an one dimensional spatial filtering (ODF) is performed on the input samples (RPi) to obtain temporal prefiltering.
Owner:NXP BV
Pixel structure, driving method thereof, display panel and display device
ActiveCN107621716ATake advantage of luminosityIncrease display resolutionStatic indicating devicesNon-linear opticsImage resolutionComputer science
The invention discloses a pixel structure, a driving method thereof, a display panel and a display device. Each pixel group is divided into three columns of pixel areas, the length of sub-pixels in the first-column pixel area is greater than that of each sub-pixel in the second-column pixel area and the third-column pixel area in the column directions, the colors of any two adjacent sub-pixels inthe column and row directions in each pixel group are different, and therefore high display resolution is achieved by utilizing low physical resolution of the pixel structure. According to the pixel structure, the sawtooth and particle senses during display can be reduced because of the arrangement mode, the luminous performance of the sub-pixels different in color is fully utilized, and thereforean image is displayed more evenly.
Owner:WUHAN TIANMA MICRO ELECTRONICS CO LTD
Image processing apparatus and image processing method, and program
InactiveUS7548264B2Good reproducibilityReduce aliasingTelevision system detailsColor signal processing circuitsColor imageImaging processing
An image processing apparatus is provided which includes: an image obtaining unit for obtaining a mosaic image by an image sensor having at least four kinds of filters with different spectral sensitivities, one of the at least four kinds of filters being used for each pixel; and an image processing unit for generating a color image such that intensity information at a position of each pixel, the intensity information corresponding to a number of colors determined by the spectral sensitivities of the filters, is computed for all pixels, from the mosaic image obtained by the image obtaining unit. In a color filter arrangement of the image sensor of the image obtaining unit, filters possessed by pixels arranged in a checkered manner in half of all the pixels have at least some spectral characteristics having a strong correlation with the spectral characteristic of luminance. The image processing unit calculates estimated intensity values of colors possessed by the pixels arranged in the checkered manner at each pixel and calculates a luminance value of each pixel on a basis of the estimated intensity values.
Owner:SONY CORP
Image capturing apparatus, image sensor, and image processing method
InactiveUS20060244841A1Reduce deteriorationReduce aliasingTelevision system detailsTelevision system scanning detailsImaging processingImaging quality
This invention is directed to reduce a deterioration in image quality after thinning operation when pixels are read out upon thinning. An image sensor is designed such that different gains are applied on a pixel basis. After periodical gains are applied to the respective pixels, the pixels of the same colors are added and averaged to obtain a low-pass filter effect before thinning processing for the pixels in the sensor, thereby preventing the occurrence of aliasing (moiré) due to thinning.
Owner:CANON KK
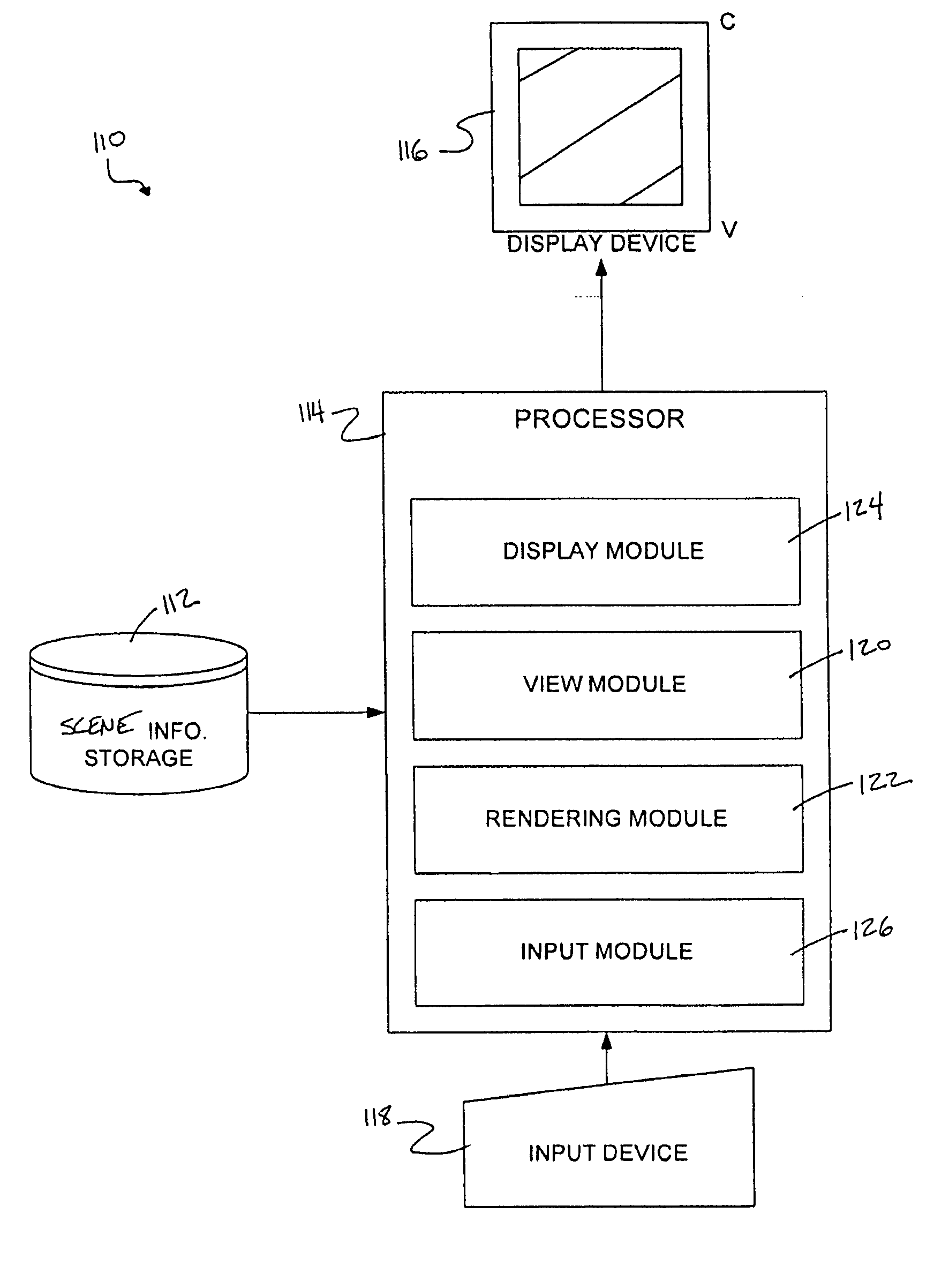
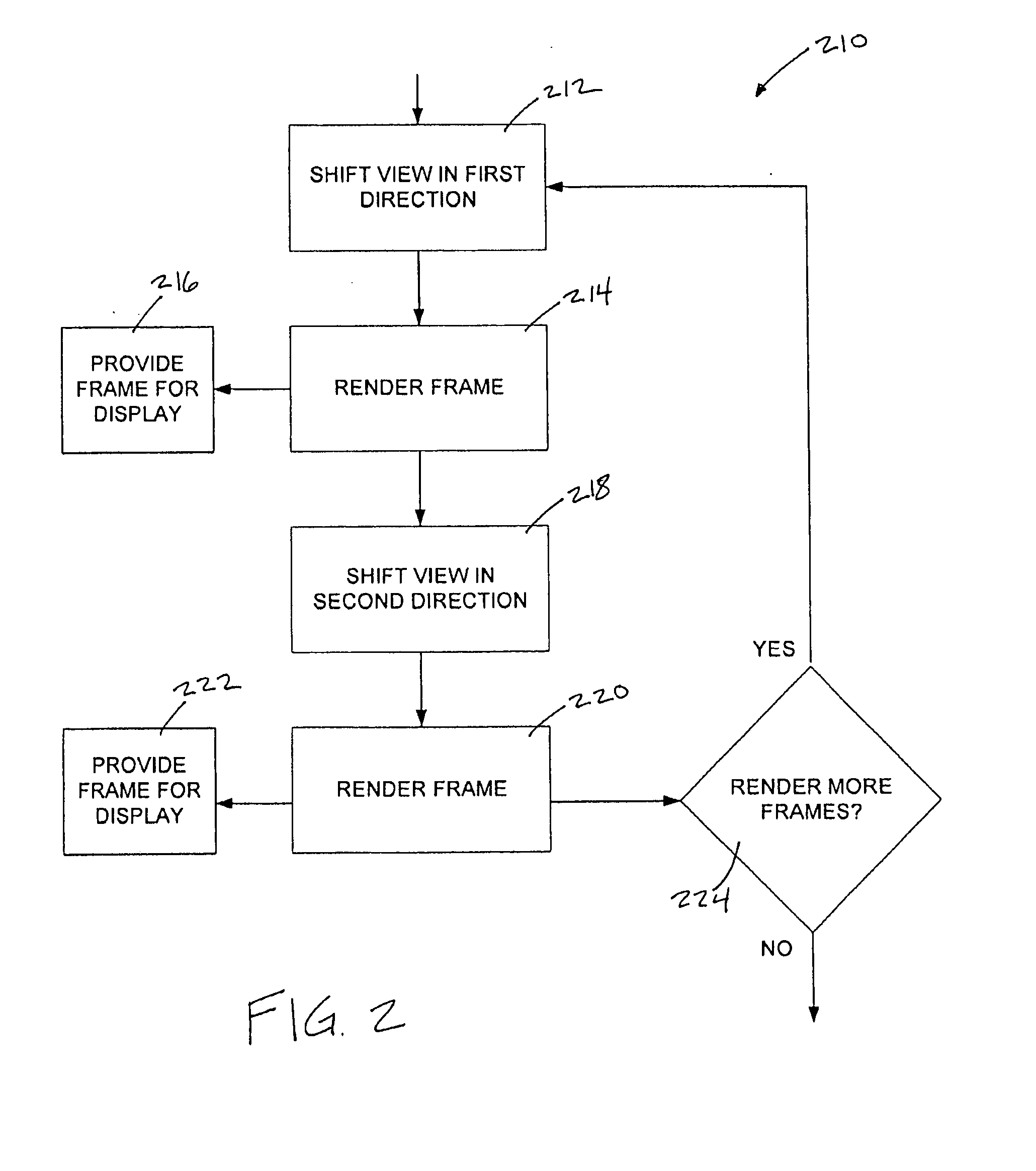
System and method of temporal anti-aliasing
InactiveUS20070153023A1Increase throughputReduce aliasingDetails involving antialiasingCathode-ray tube indicatorsAnti-aliasingColor transformation
A system and method for temporally anti-aliasing a display of a set of frames rendered from scene information without significantly increasing an amount of processing power used in rendering the frames. In some embodiments of the invention, a view used to render the frames from the scene information may be modulated between renderings of successive frames. This modulation of the view may temporally blend pixel colors displayed in pixels at or near color transitions in the frames by causing the pixel colors of these pixels to fluctuate between frames, thereby reducing aliasing present at these transitions as the human eye observing the display of the frames innately averages the fluctuating pixel colors.
Owner:COMP ASSOC THINK INC
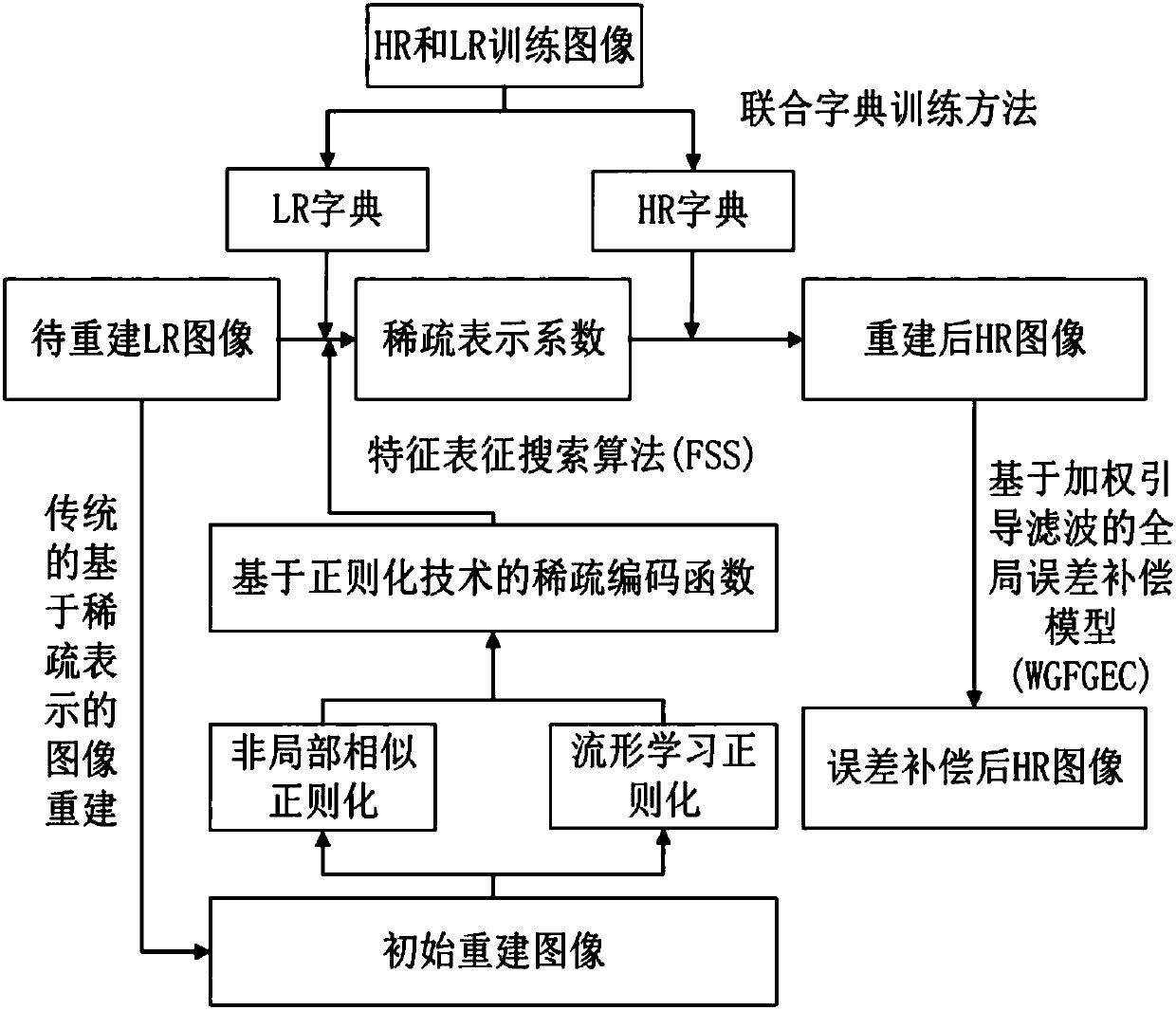
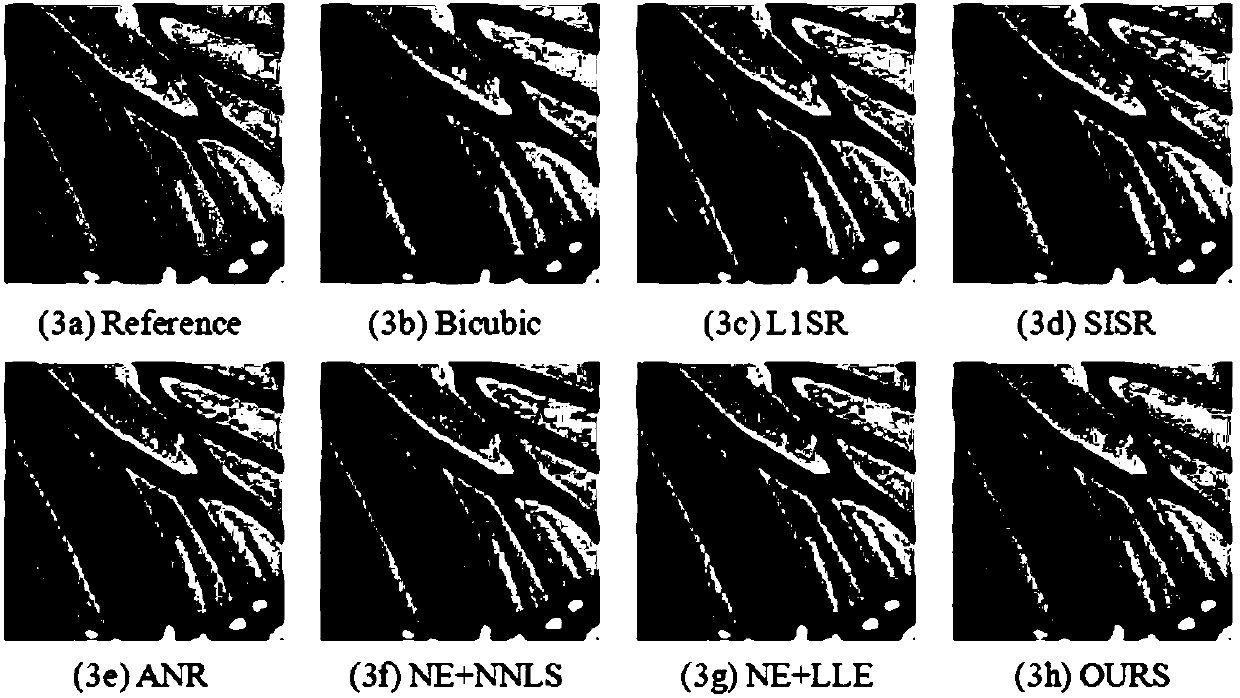
Image super-resolution method based on sparse regularization technology and weighted guidance filtering
ActiveCN107610049AImprove edge detailImprove visual effectsImage enhancementGeometric image transformationReconstruction errorSparse regularization
The present invention discloses an image super-resolution method based on the sparse regularization technology and the weighted guidance filtering. The method is characterized in that: a new sparse coding objective function is constructed by combining the non-local similarity of the image and the manifold learning theory, so that on one hand, similar image blocks are searched in the initial reconstruction image to construct the no-local similarity regularization term, and non-local redundancy of the image is obtained to maintain the edge information, and on the other hand, the local linear embedding method is used to construct the manifold learning regularization term, and the prior knowledge of the structure of the image is obtained to enhance the structure information; and the global error compensation model of the weighted guidance filtering is used to carry out error compensation on the reconstructed high-resolution image to obtain the image with a smaller reconstruction error andhigher quality.
Owner:HUAQIAO UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com