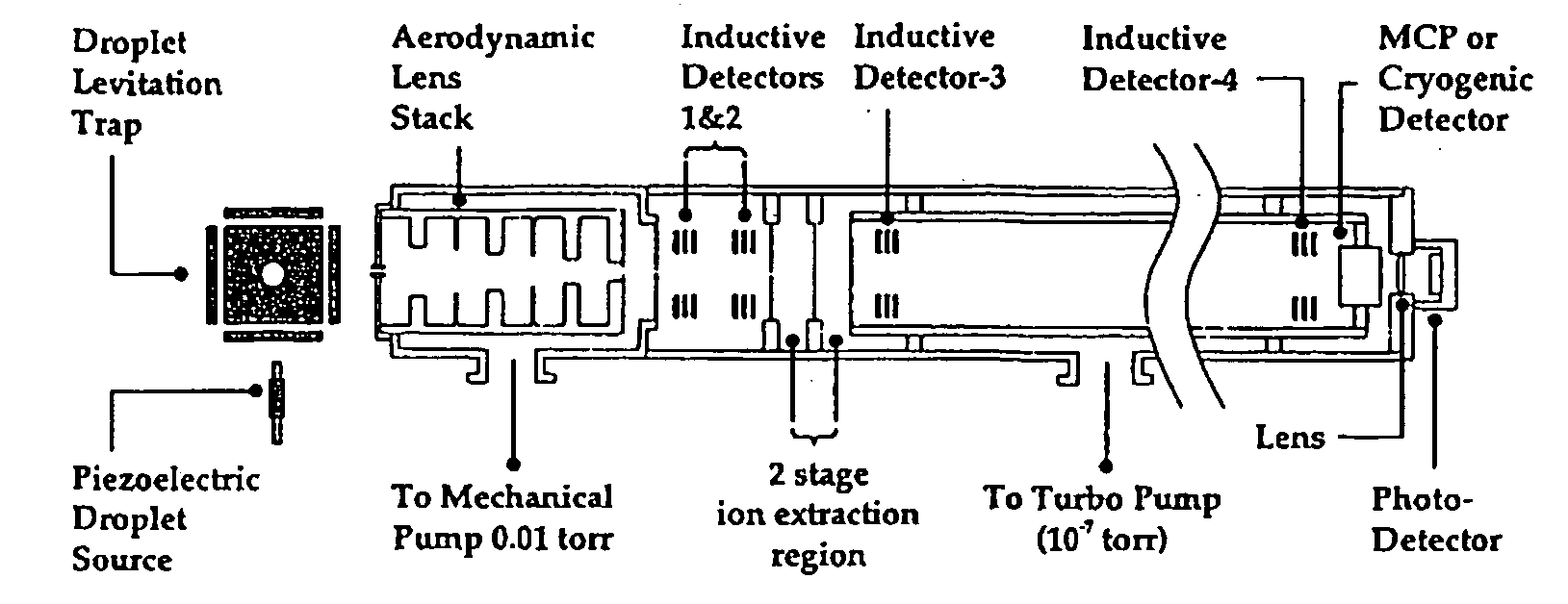
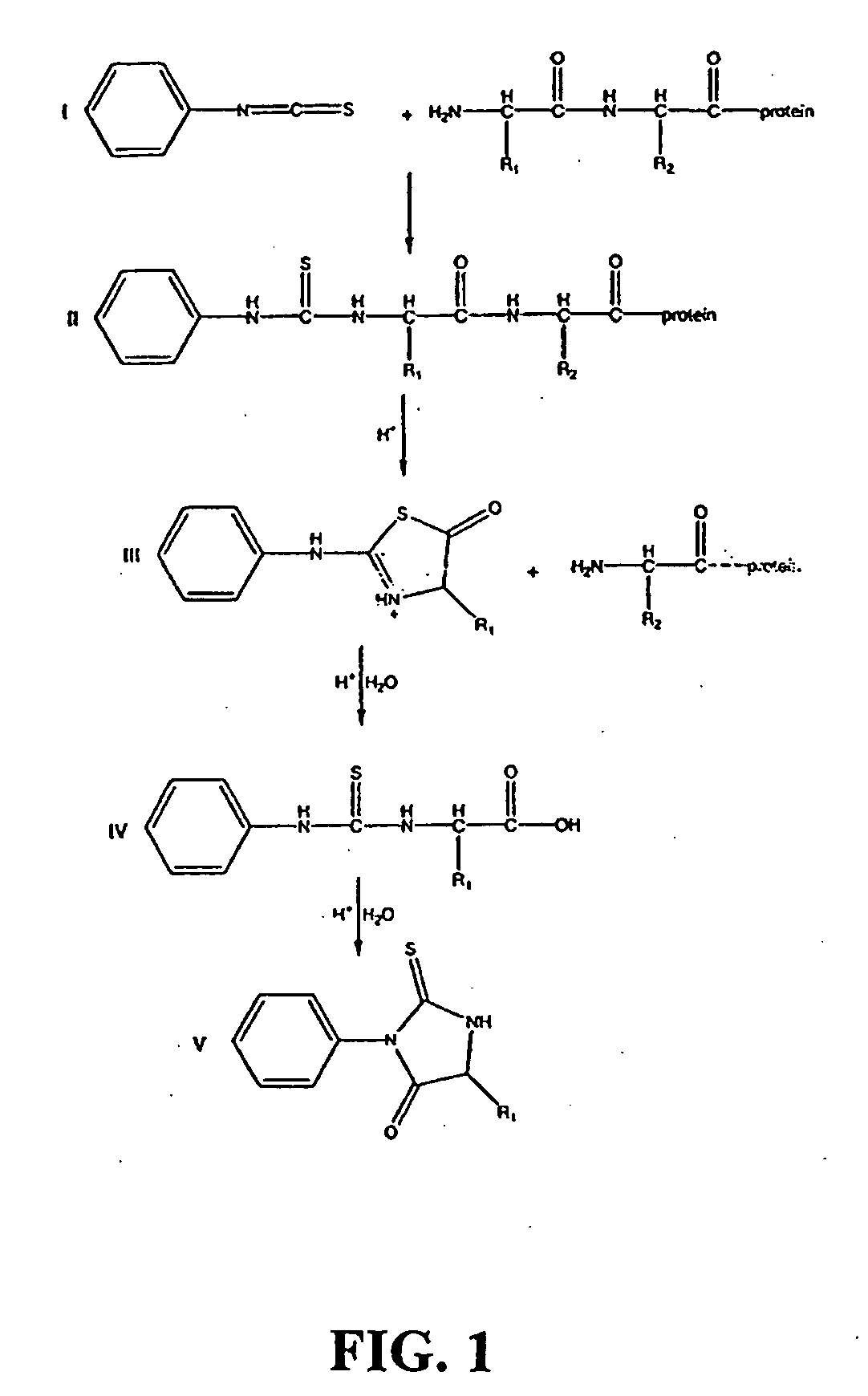
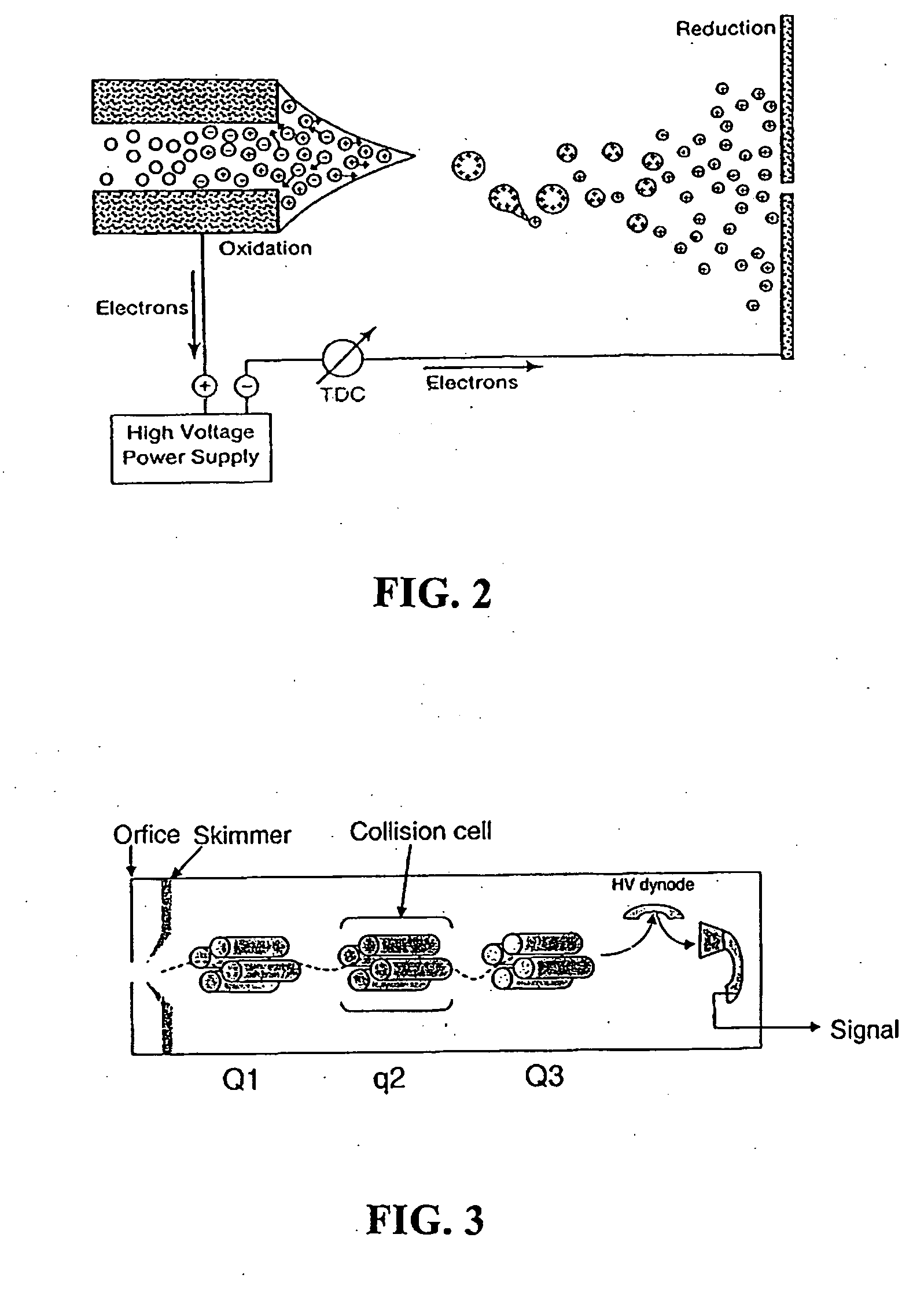
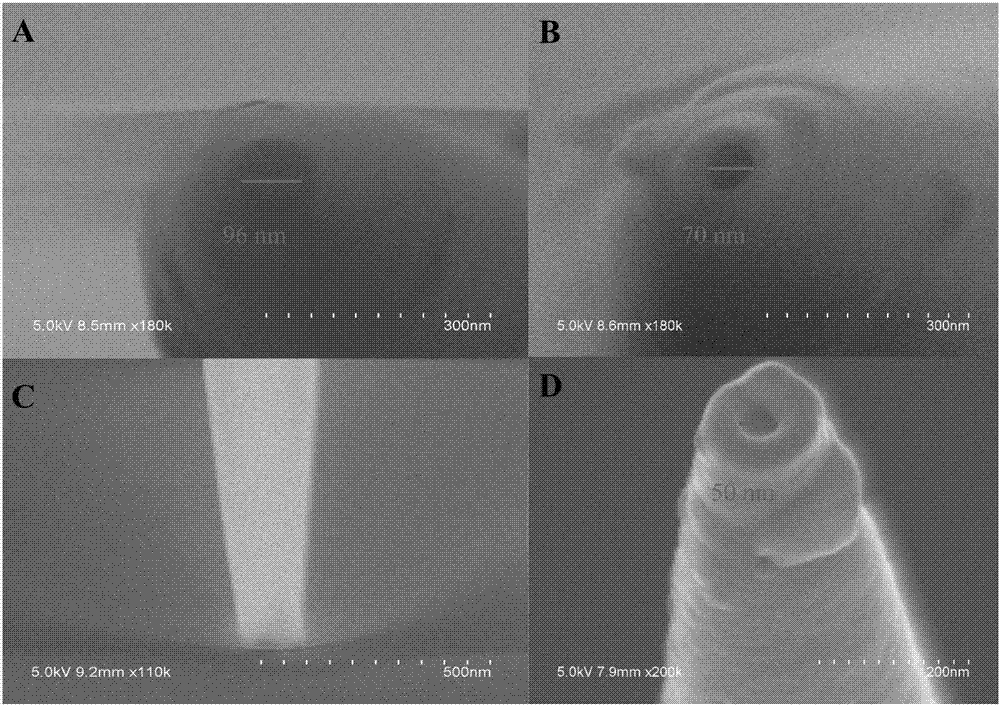
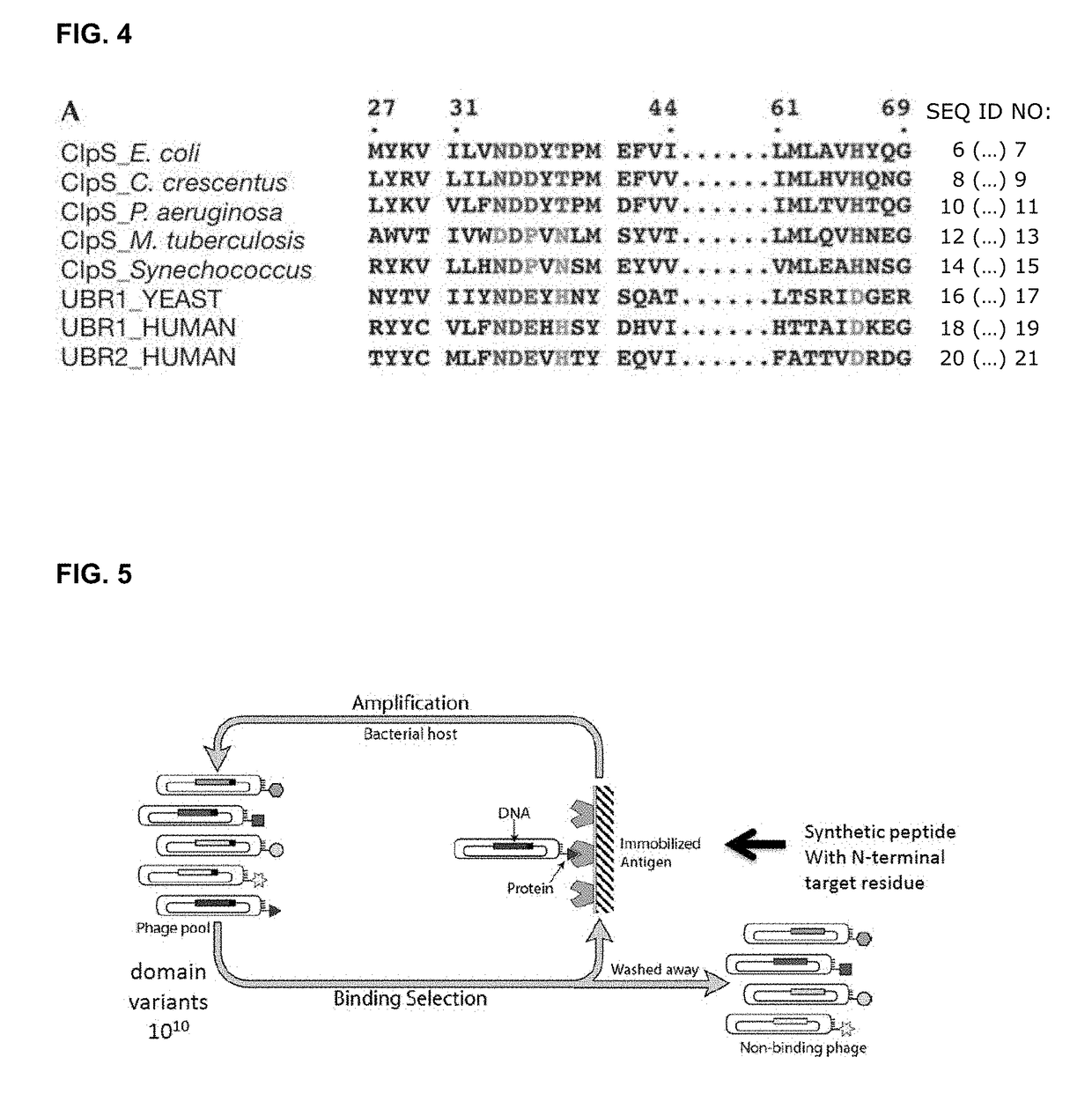
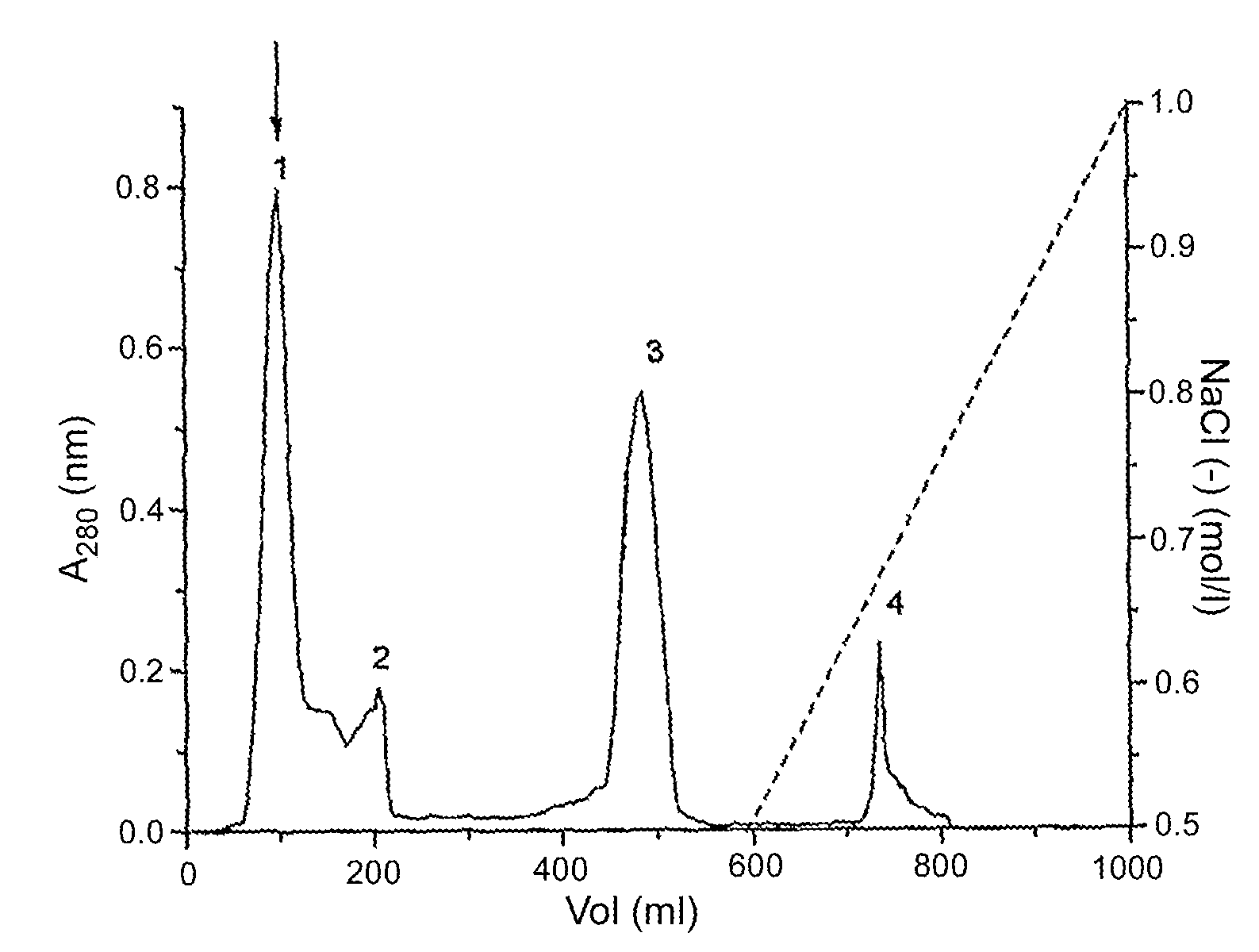
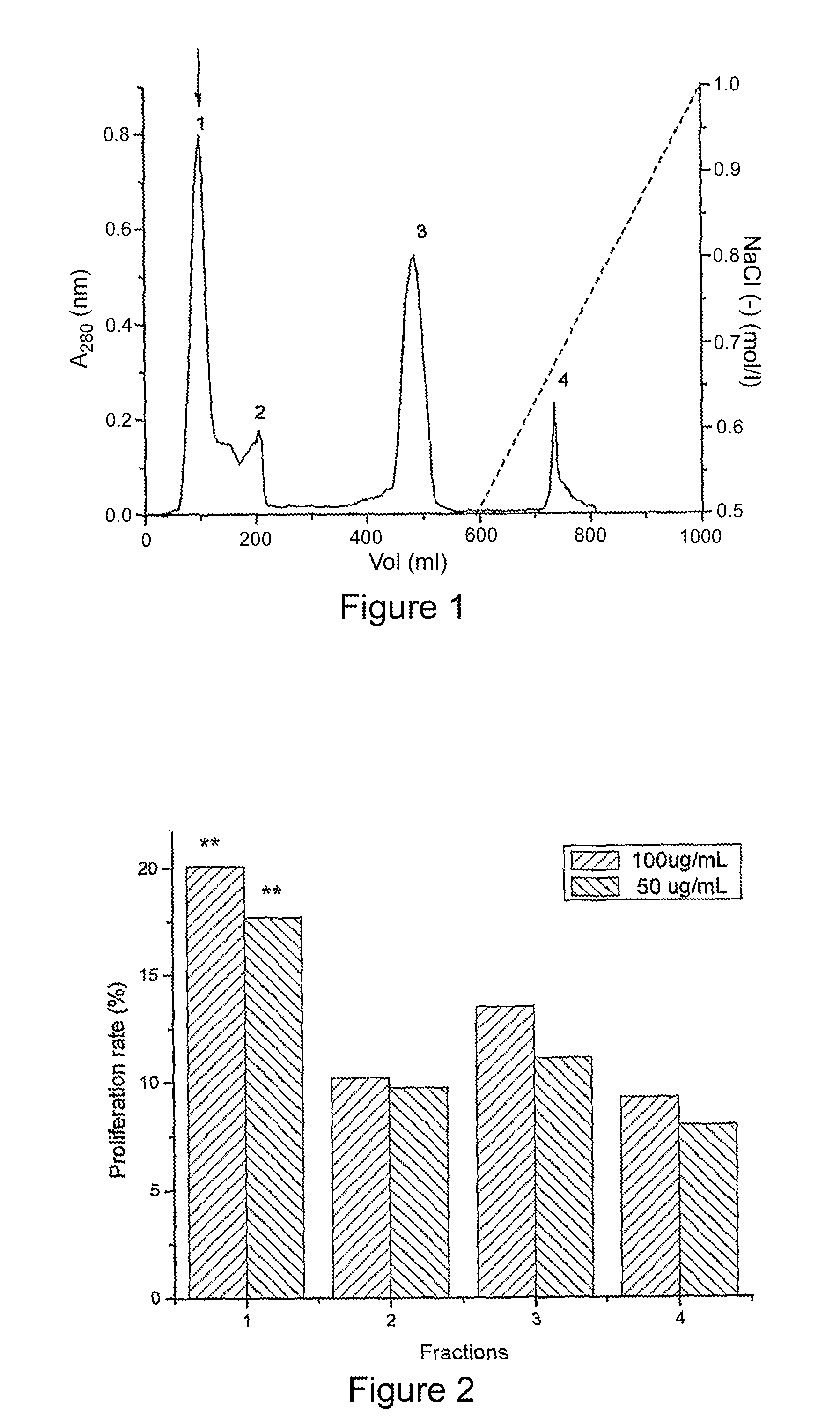
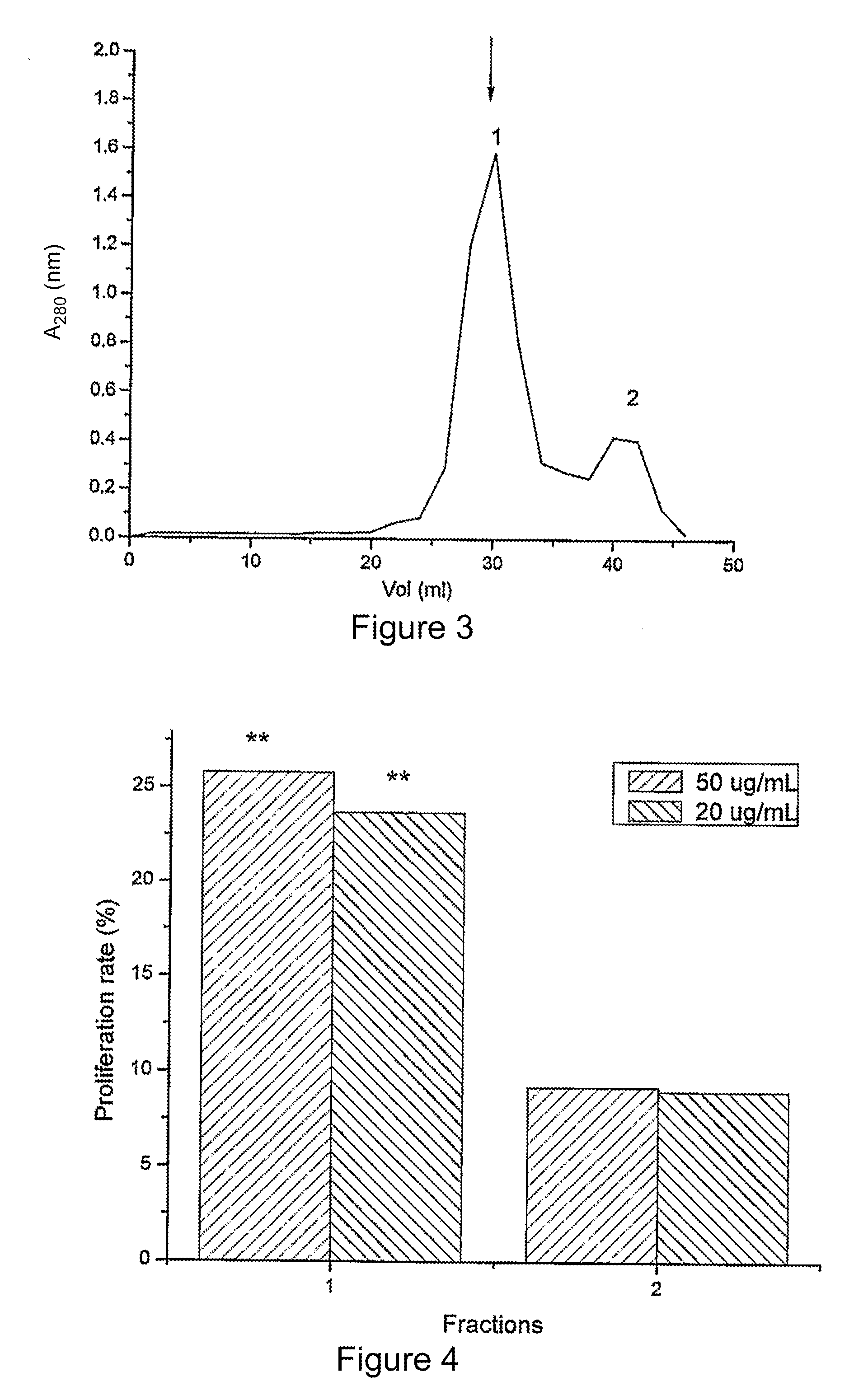
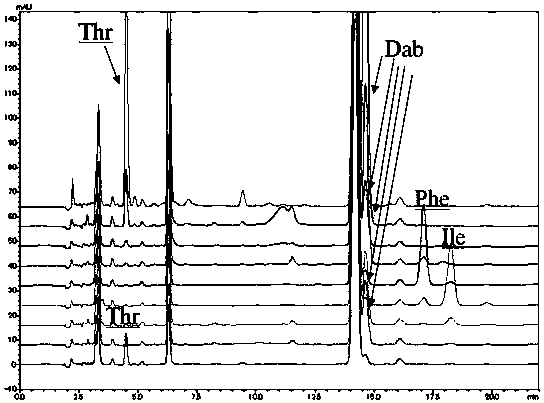
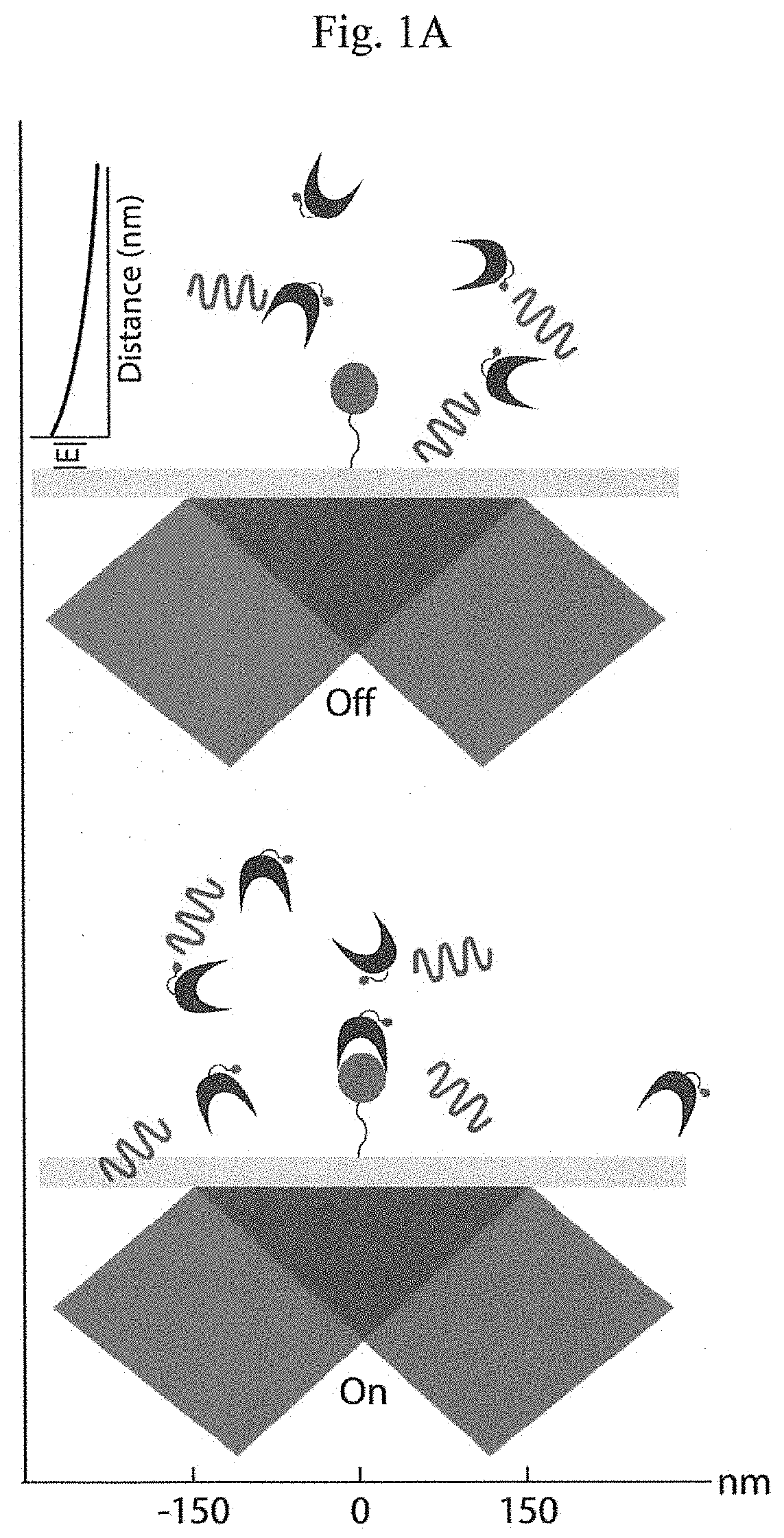
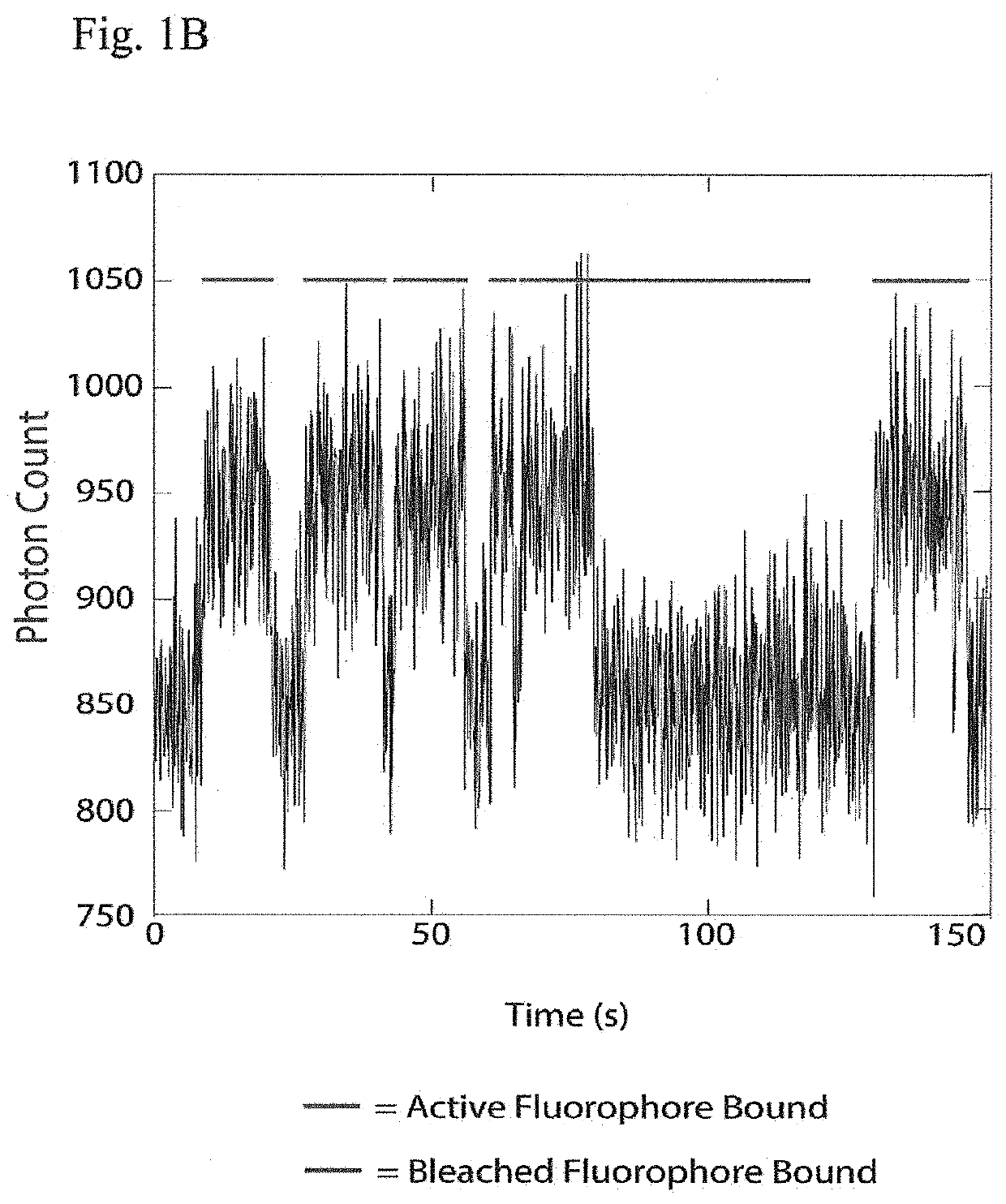
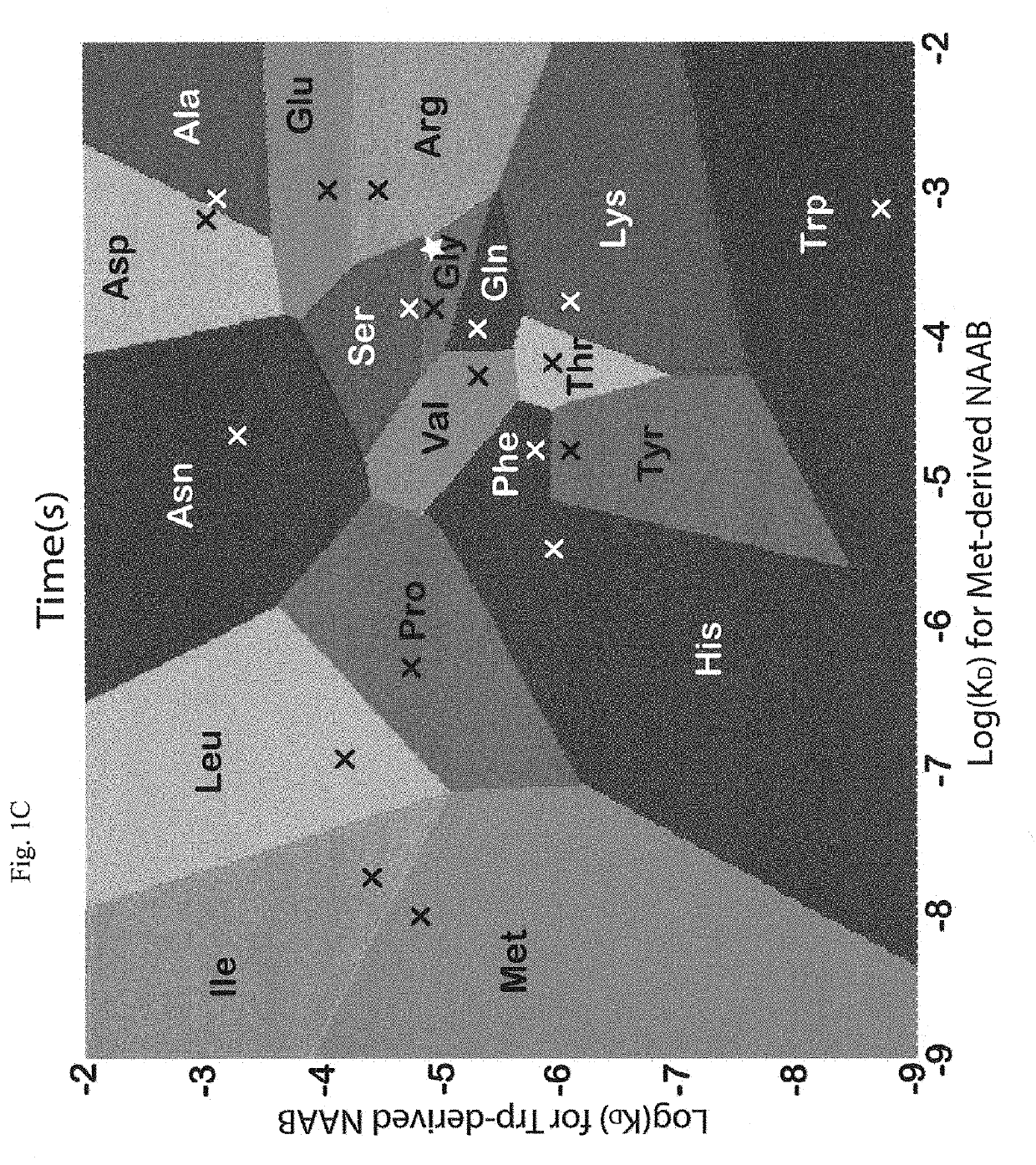
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
55 results about "Protein Sequence Determination" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
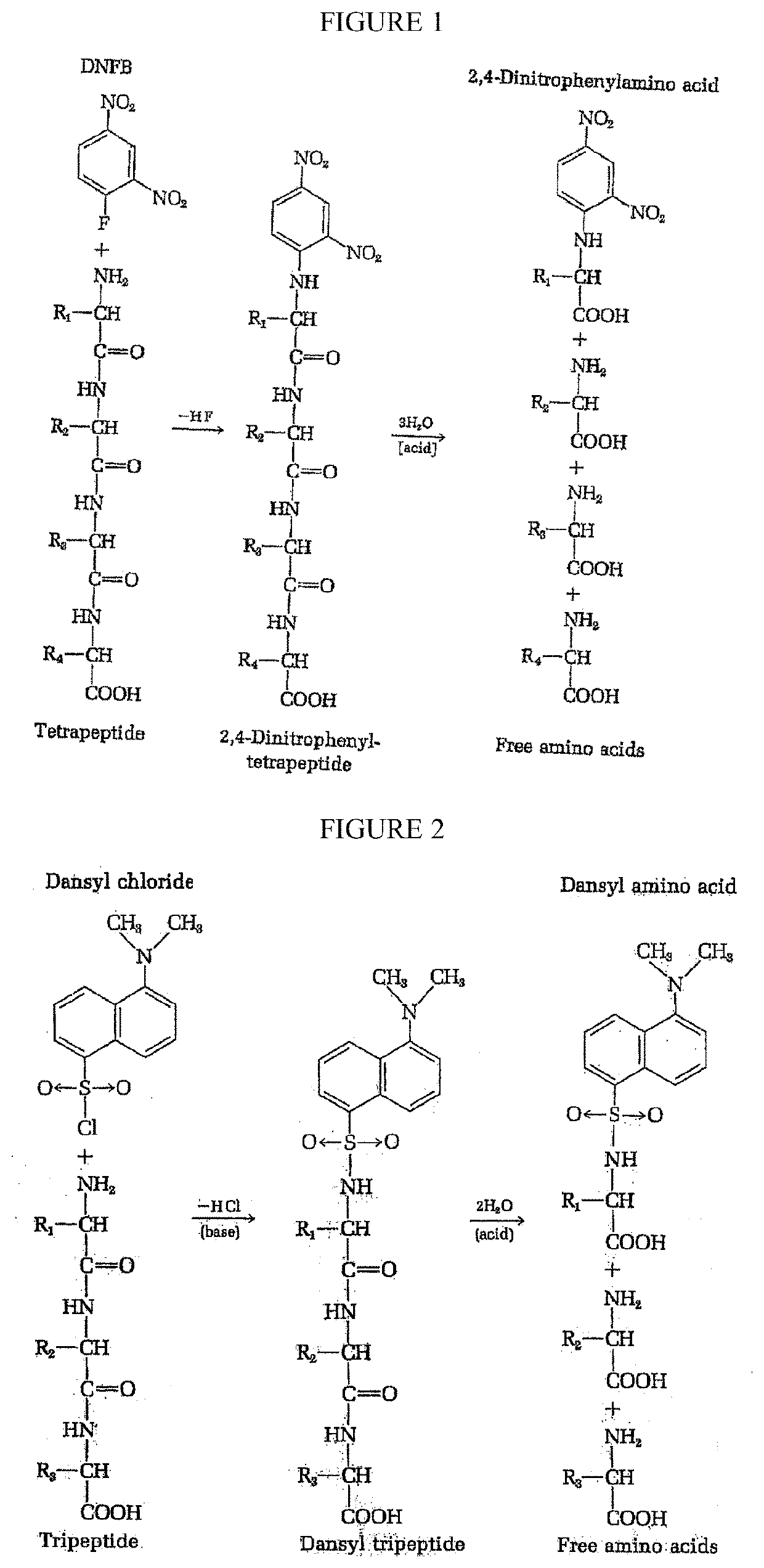
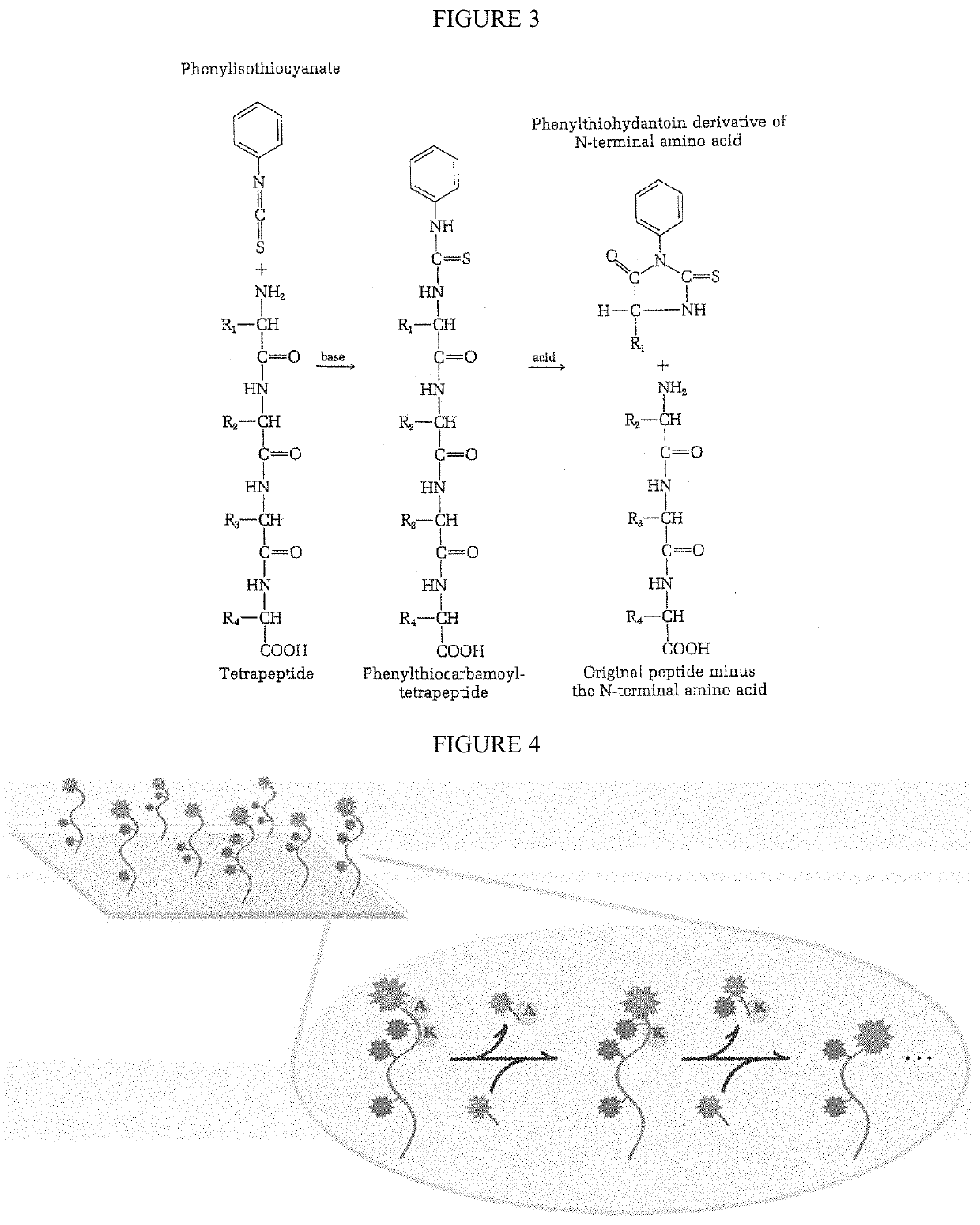
Protein sequencing is the practical process of determining the amino acid sequence of all or part of a protein or peptide. This may serve to identify the protein or characterize its post-translational modifications.
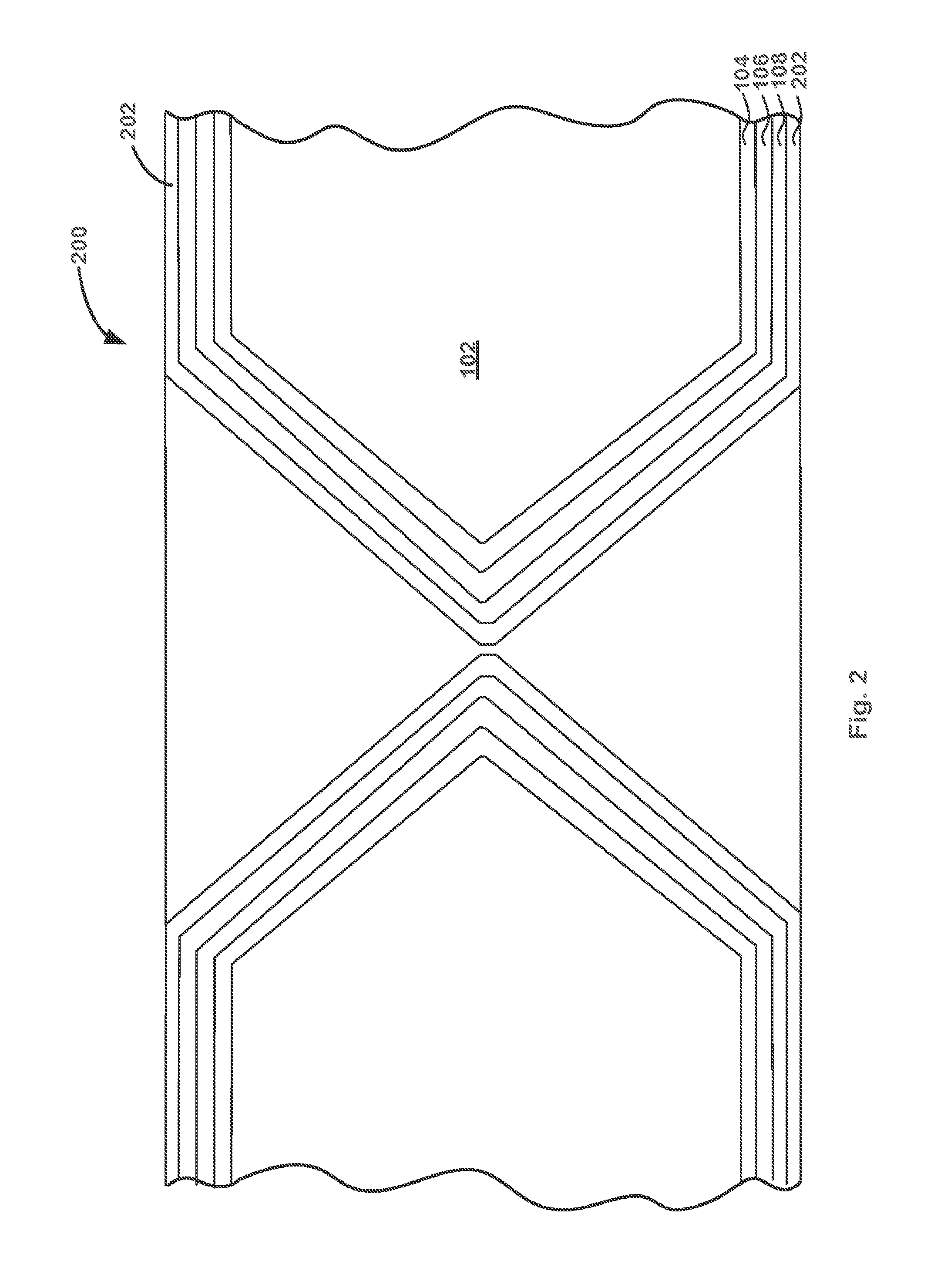
Method and system for rapid biomolecular recognition of amino acids and protein sequencing
InactiveUS6846638B2Low costHigh sensitivityBioreactor/fermenter combinationsBiological substance pretreatmentsProtein Sequence DeterminationDigestion
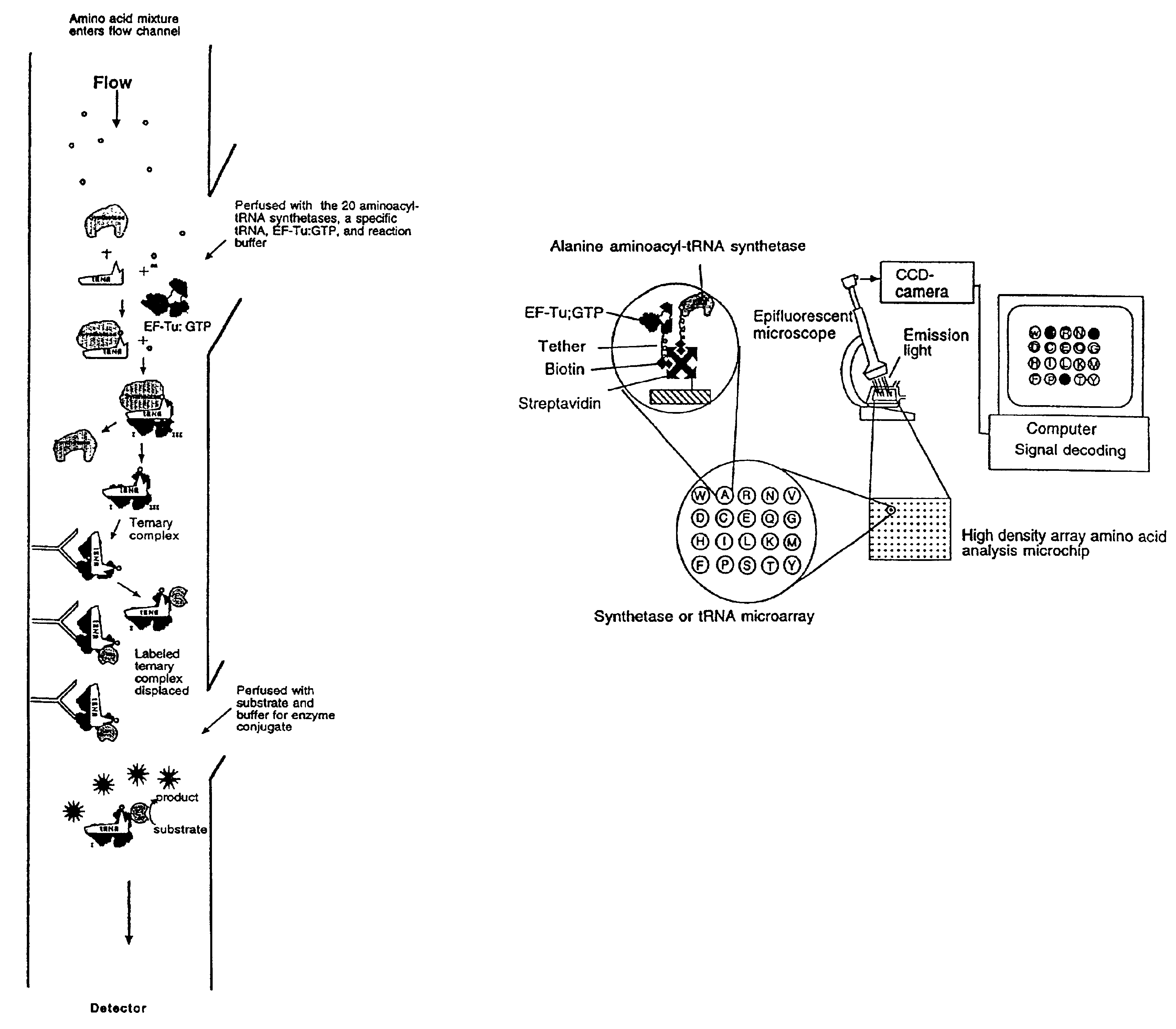
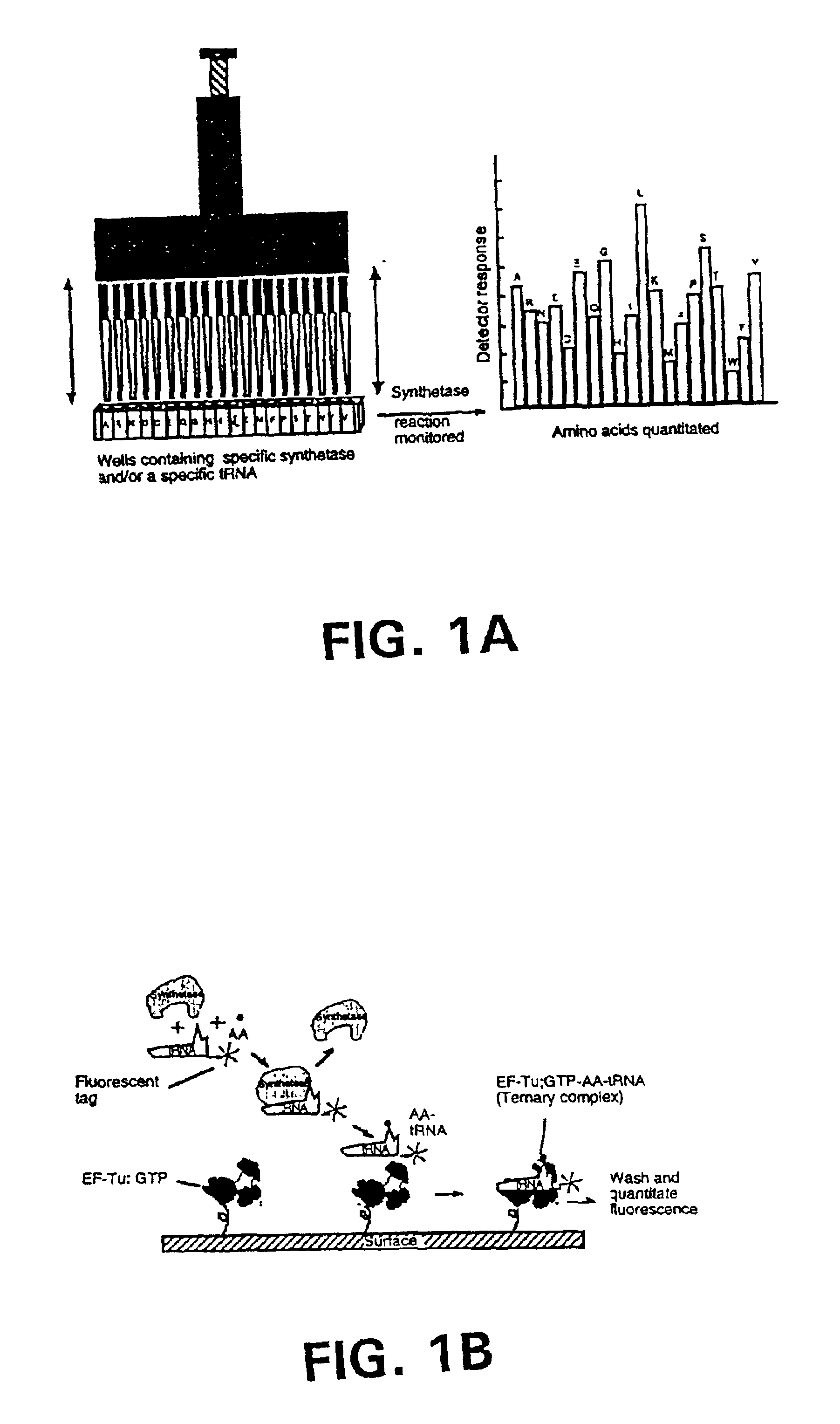
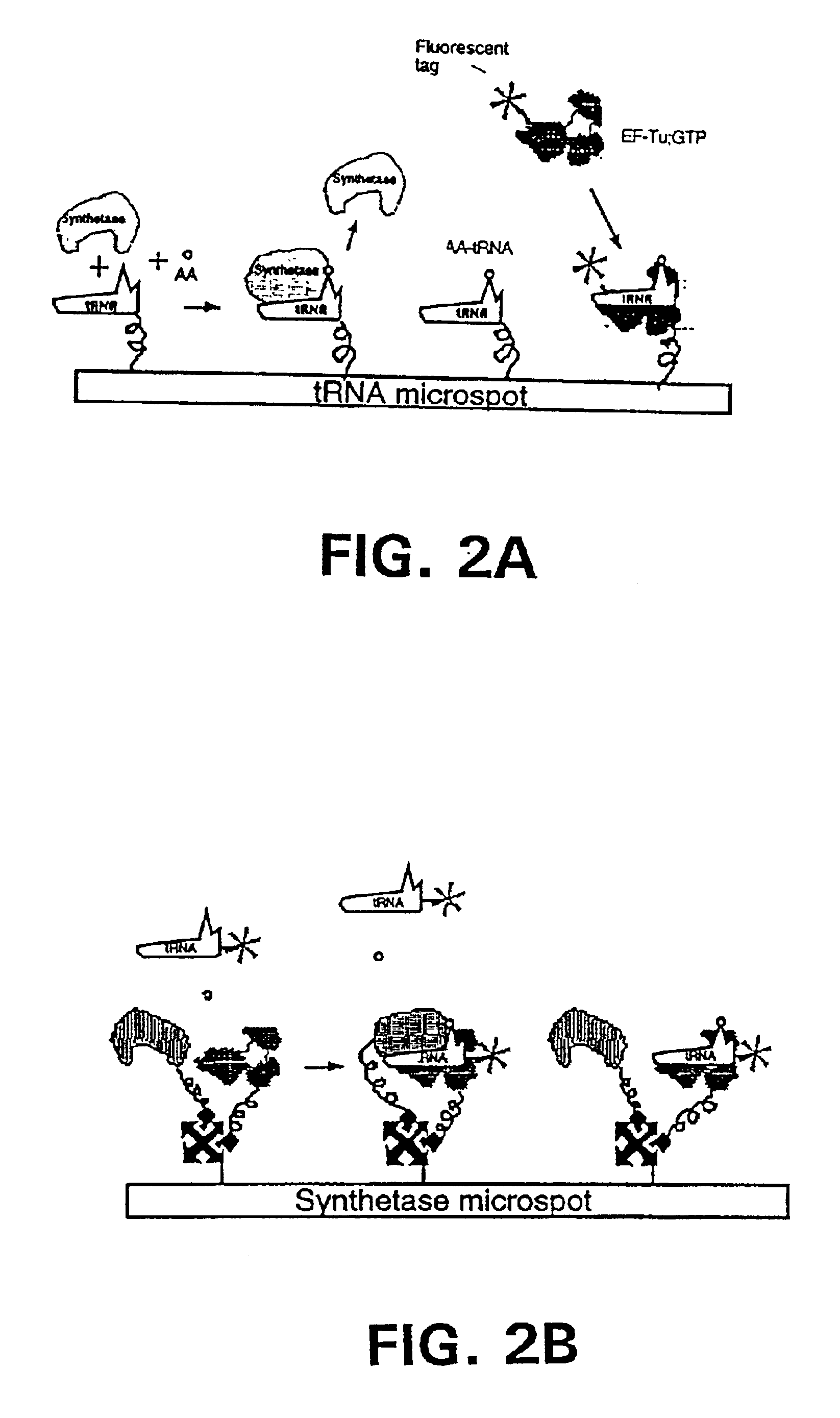
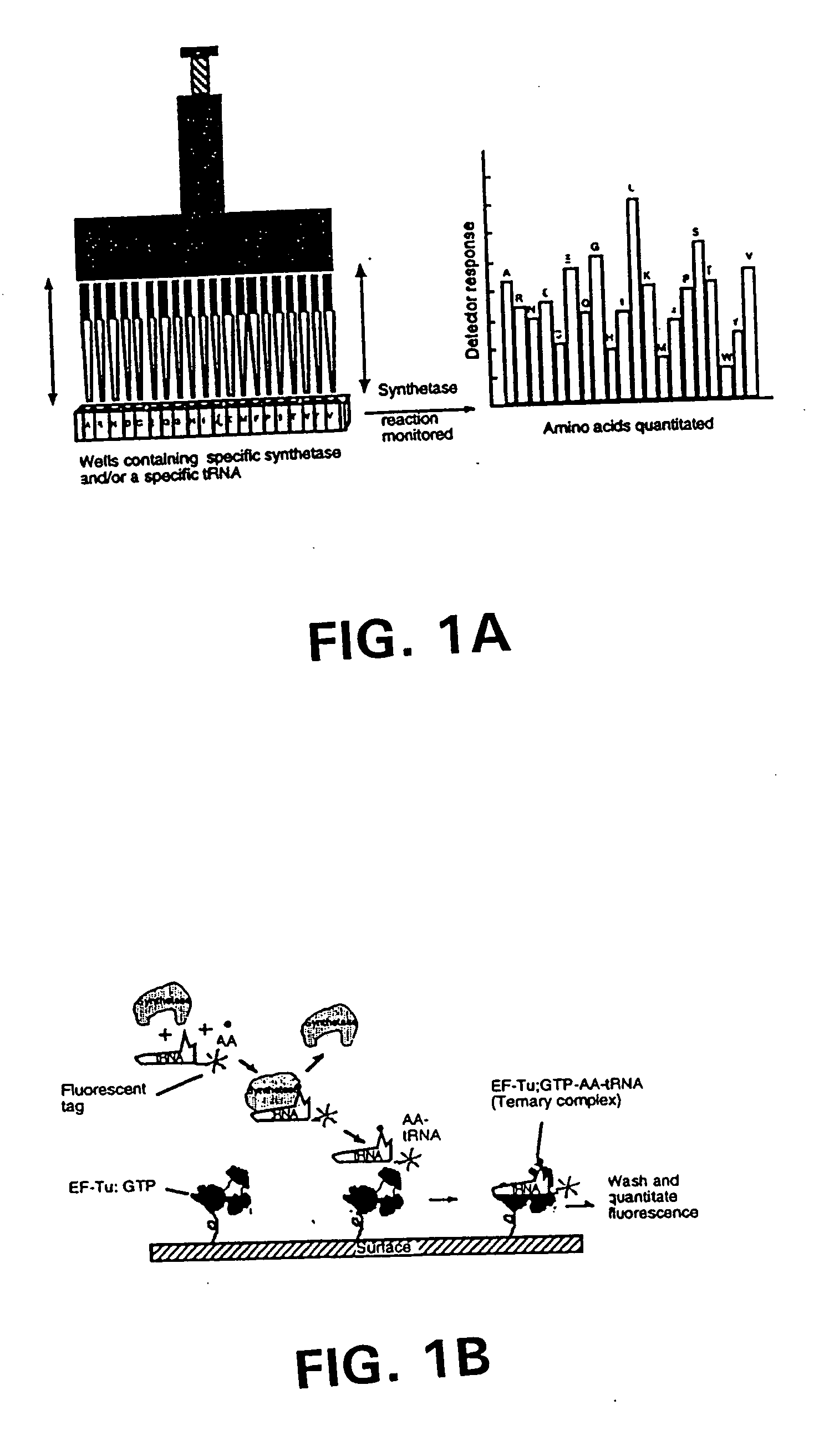
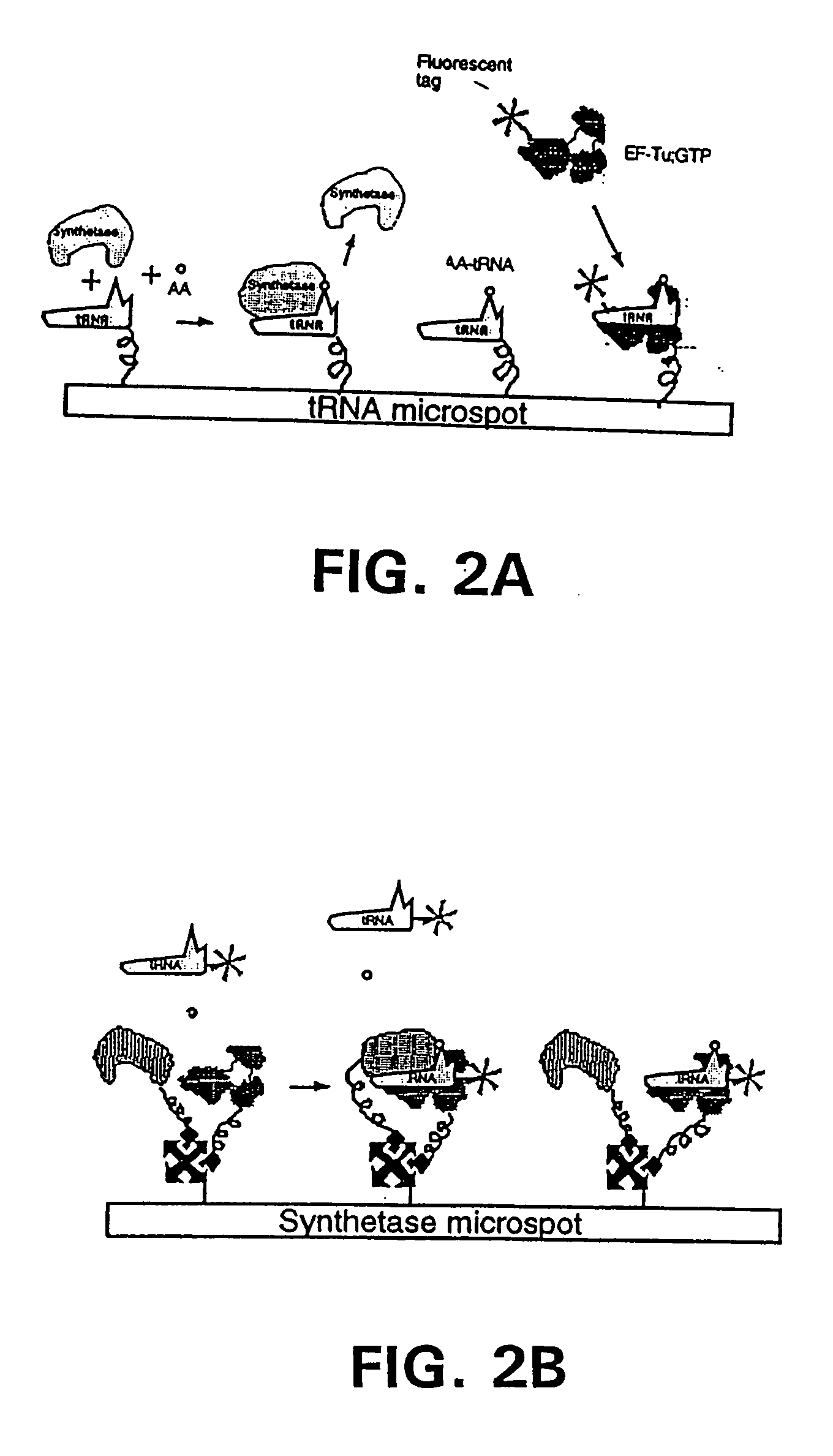
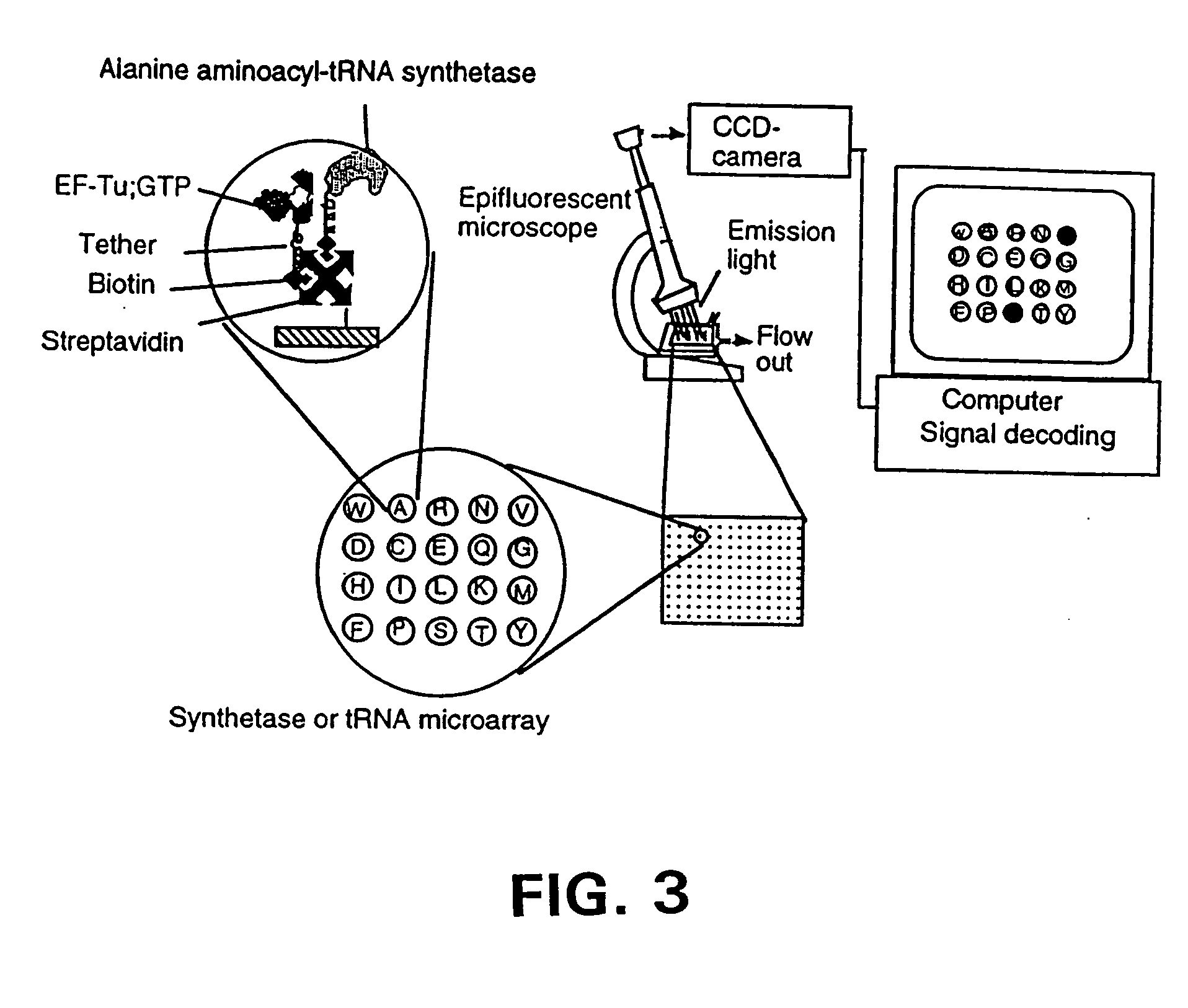
Methods, compositions, kits, and apparatus are provided wherein the aminoacyl-tRNA synthetase system is used to analyze amino acids. The method allows very small devices for quantitative or semi-quantitative analysis of the amino acids in samples or in sequential or complete proteolytic digestions. The methods can be readily applied to the detection and / or quantitation of one or more primary amino acids by using cognate aminoacyl-tRNA synthetase and cognate tRNA. The basis of the method is that each of the 20 synthetases and / or a tRNA specific for a different amino acid is separated spatially or differentially labeled. The reactions catalyzed by all 20 synthetases may be monitored simultaneously, or nearly simultaneously, or in parallel. Each separately positioned synthetase or tRNA will signal its cognate amino acid. The synthetase reactions can be monitored using continuous spectroscopic assays. Alternatively, since elongation factor Tu:GTP (EF-Tu:GTP) specifically binds all AA-tRNAs, the aminoacylation reactions catalyzed by the synthetases can be monitored using ligand assays. Microarrays and microsensors for amino acid analysis are provided. Additionally, amino acid analysis devices are integrated with protease digestions to produce miniaturized enzymatic sequenators capable of generating either N- or C-terminal sequence and composition data for a protein or peptide. The possibility of parallel processing of many samples in an automated manner is discussed.
Owner:NANOBIODYNAMICS
Method and system for rapid biomolecular recognition of amino acids and protein sequencing
InactiveUS20050164264A1Quick analysisBioreactor/fermenter combinationsBiological substance pretreatmentsProtein Sequence DeterminationDigestion
Methods, compositions, kits, and apparatus are provided wherein the aminoacyl-tRNA synthetase system is used to analyze amino acids. The method allows very small devices for quantitative or semi-quantitative analysis of the amino acids in samples or in sequential or complete proteolytic digestions. The methods can be readily applied to the detection and / or quantitation of one or more primary amino acids by using cognate aminoacyl-tRNA synthetase and cognate tRNA. The basis of the method is that each of the 20 synthetases and / or a tRNA specific for a different amino acid is separated spatially or differentially labeled. The reactions catalyzed by all 20 synthetases may be monitored simultaneously, or nearly simultaneously, or in parallel. Each separately positioned synthetase or tRNA will signal its cognate amino acid. The synthetase reactions can be monitored using continuous spectroscopic assays. Alternatively, since elongation factor Tu:GTP (EF-Tu:GTP) specifically binds all AA−tRNAs, the aminoacylation reactions catalyzed by the synthetases can be monitored using ligand assays. Microarrays and microsensors for amino acid analysis are provided. Additionally, amino acid analysis devices are integrated with protease digestions to produce miniaturized enzymatic sequenators capable of generating either N- or C-terminal sequence and composition data for a protein or peptide. The possibility of parallel processing of many samples in an automated manner is discussed.
Owner:NANOBIODYNAMICS
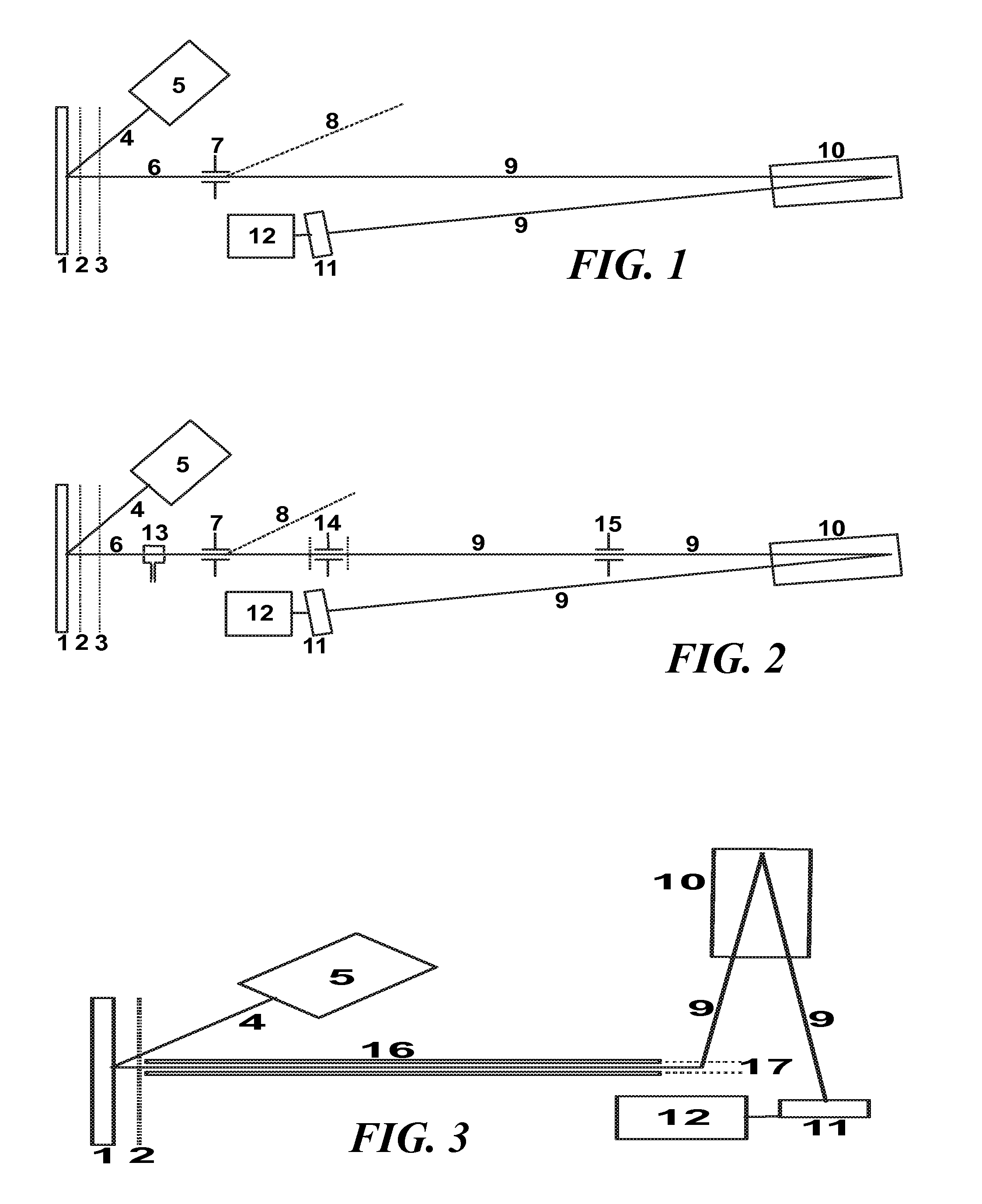
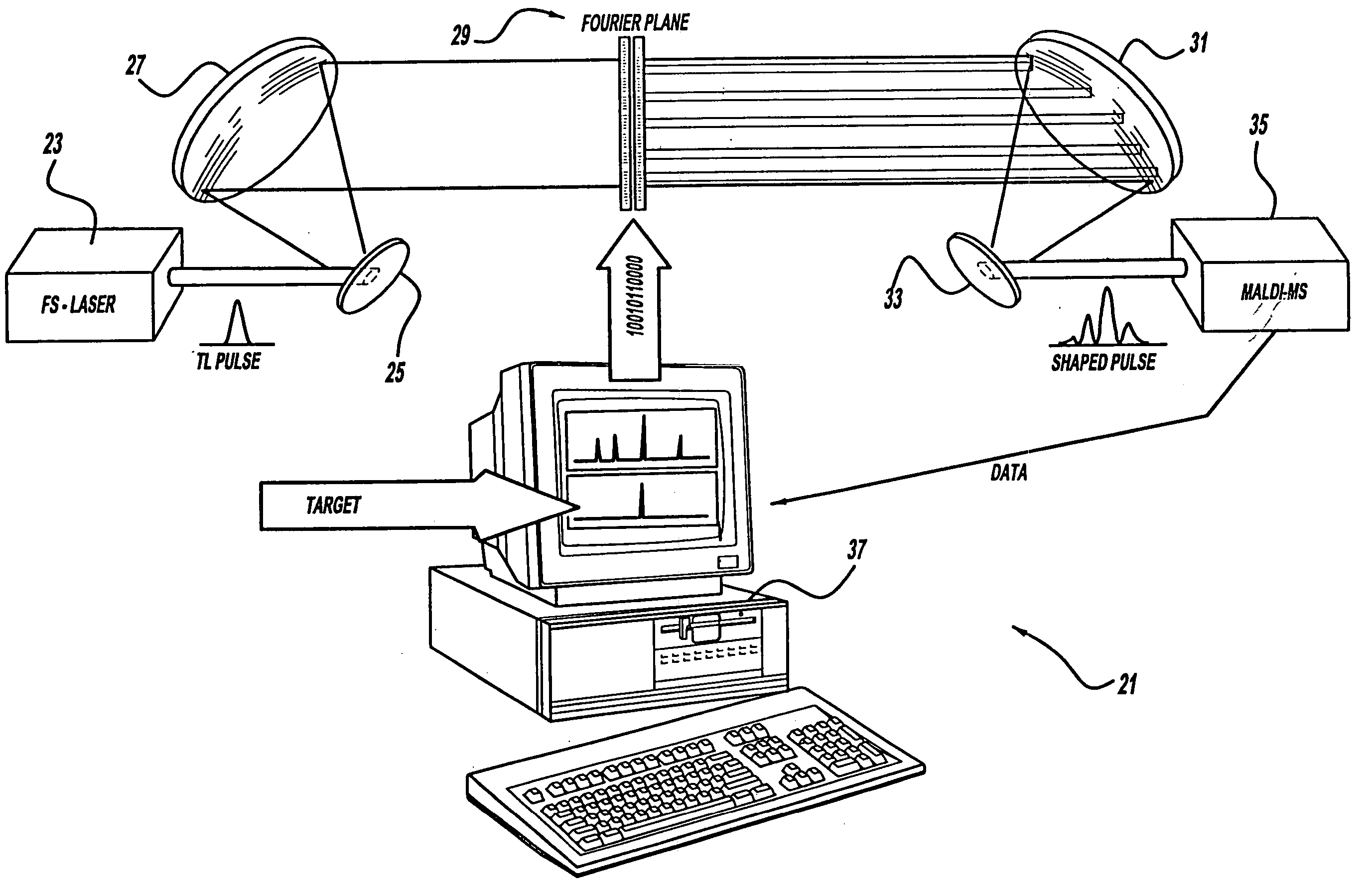
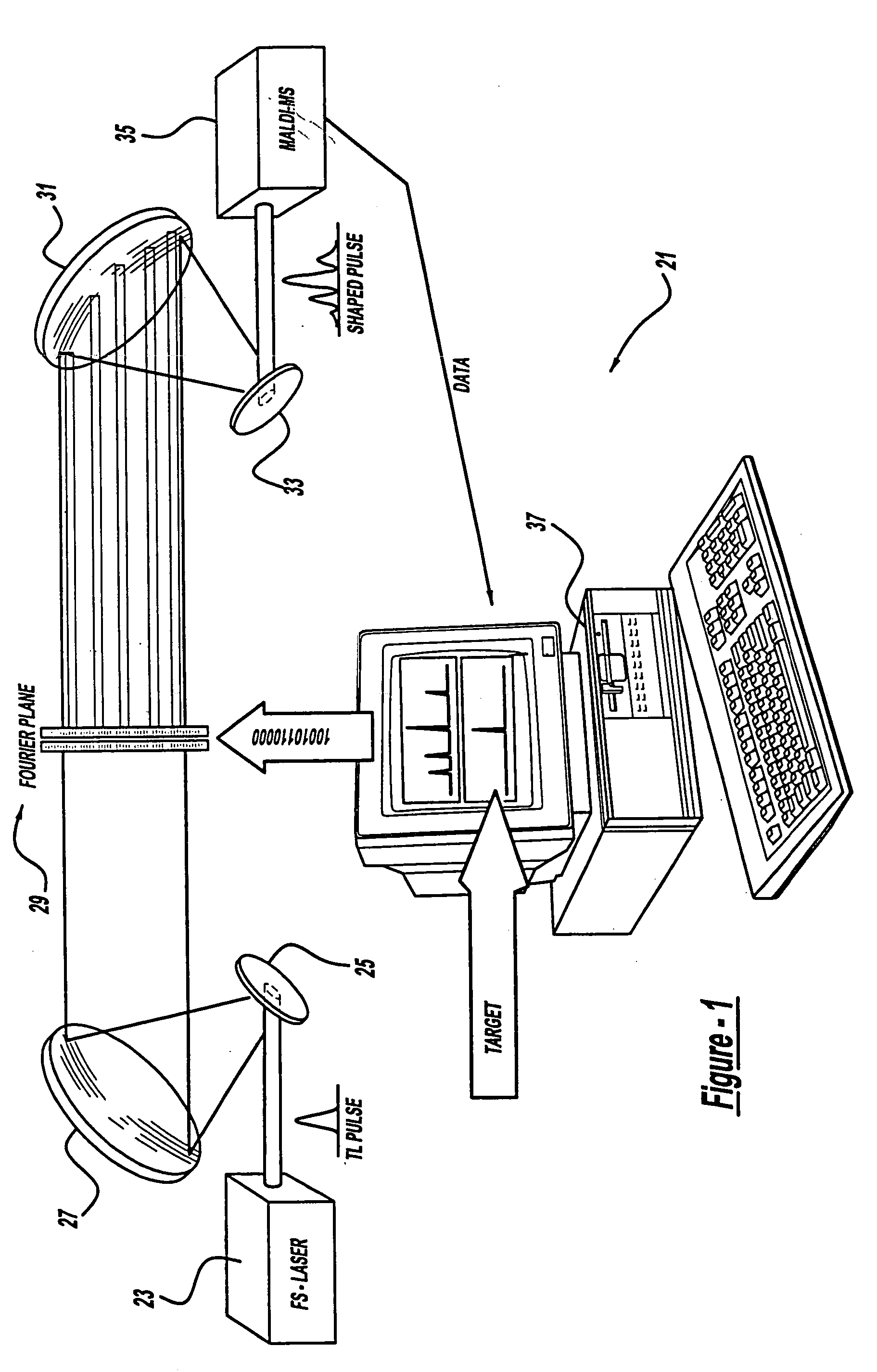
Control system and apparatus for use with laser excitation or ionization
InactiveUS7105811B2Way fastMaximize sensitivityLaser detailsIon sources/gunsPhotodynamic therapyControl system
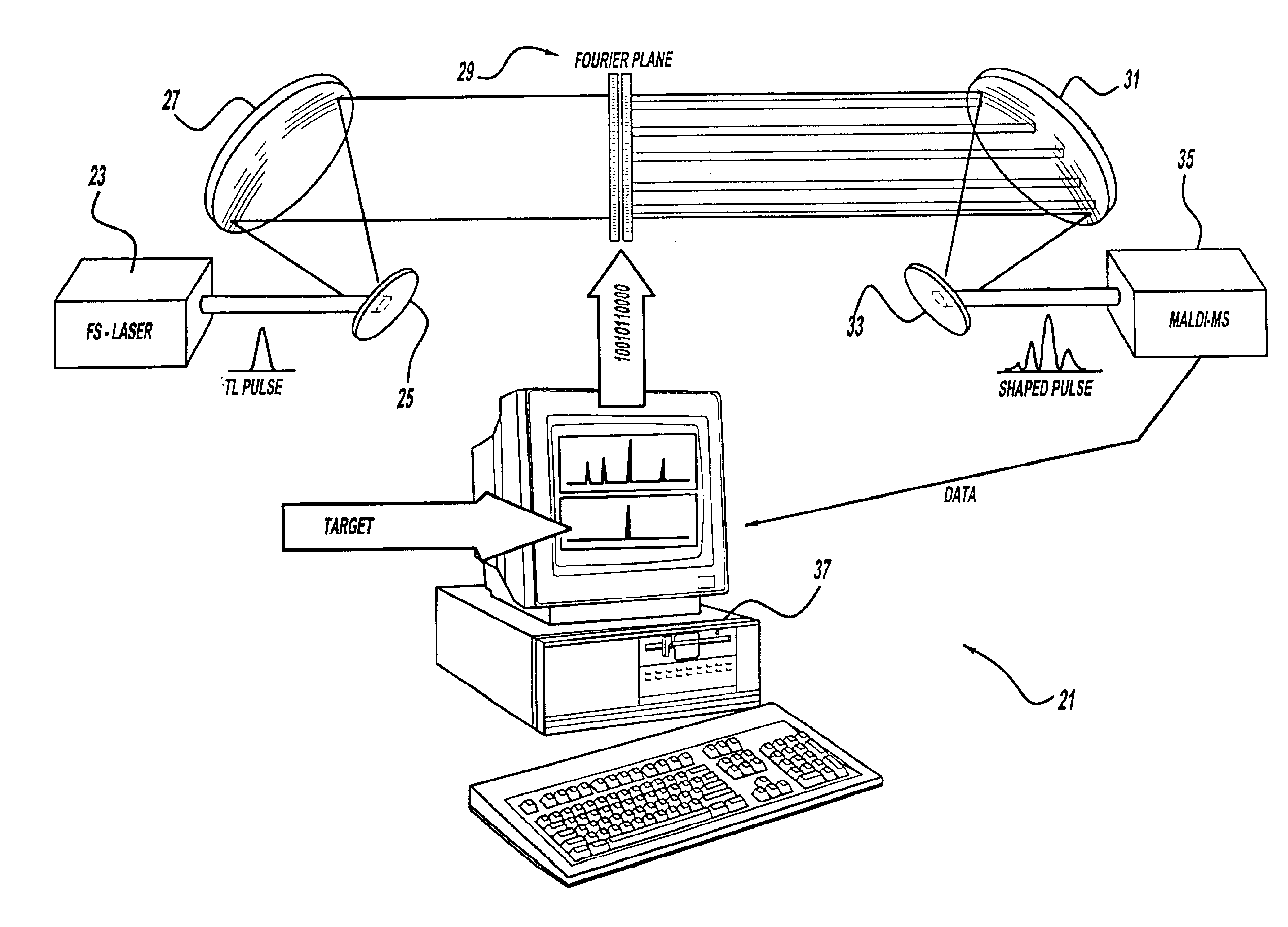
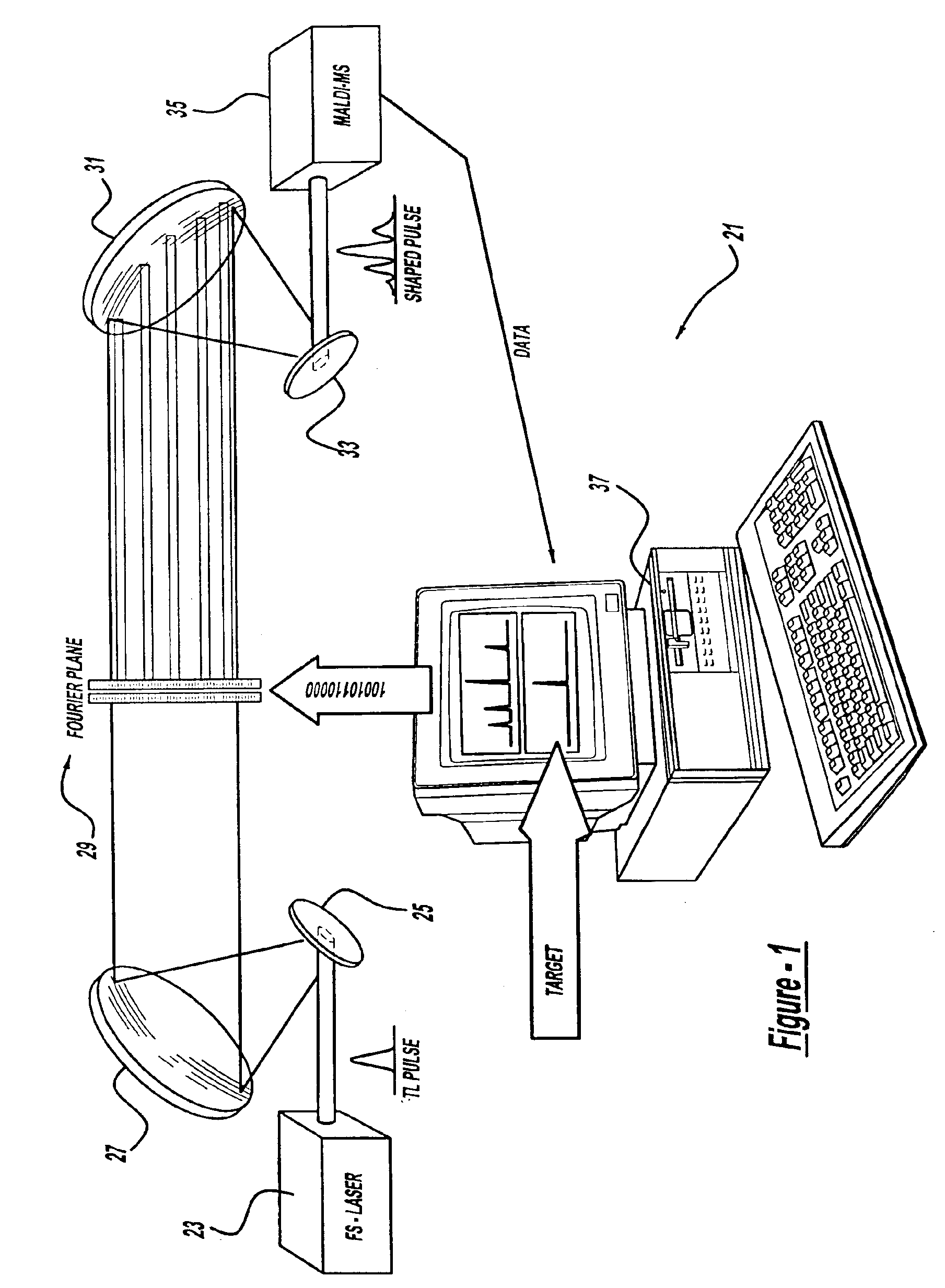
A control system and apparatus for use with laser ionization is provided. In another aspect of the present invention, the apparatus includes a laser, pulse shaper, detection device and control system. A further aspect of the present invention employs a femtosecond laser and a mass spectrometer. In yet other aspects of the present invention, the control system and apparatus are used in MALDI, chemical bond cleaving, protein sequencing, photodynamic therapy, optical coherence tomography and optical communications processes.
Owner:BOARD OF TRUSTEES OPERATING MICHIGAN STATE UNIV
Methods and device for analyte characterization
ActiveUS8278055B2Microbiological testing/measurementBiological testingAnalyteProtein Sequence Determination
Owner:INTEL CORP
Methods and device for analyte characterization
ActiveUS20050282229A1Microbiological testing/measurementBiological testingAnalyteProtein Sequence Determination
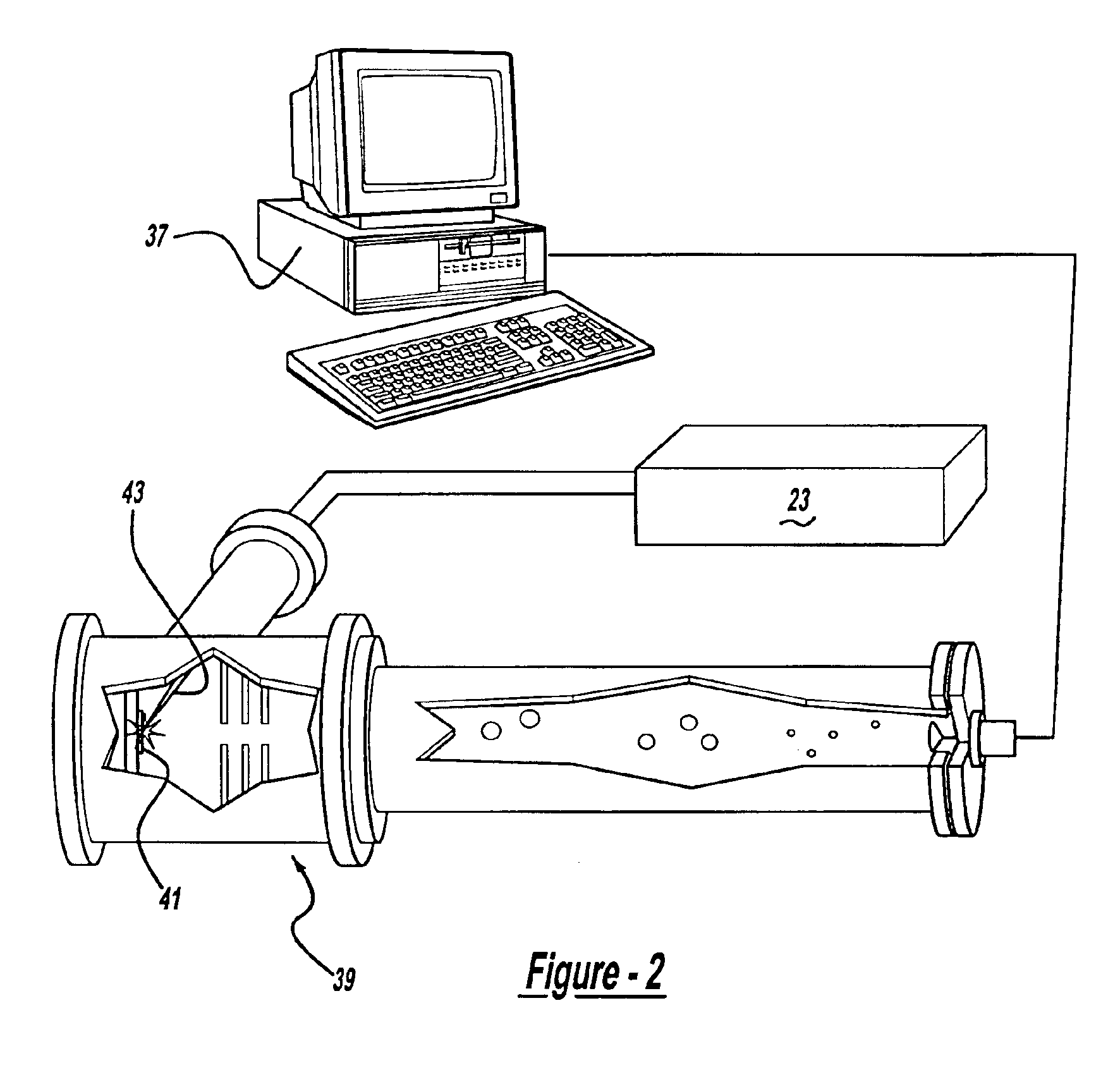
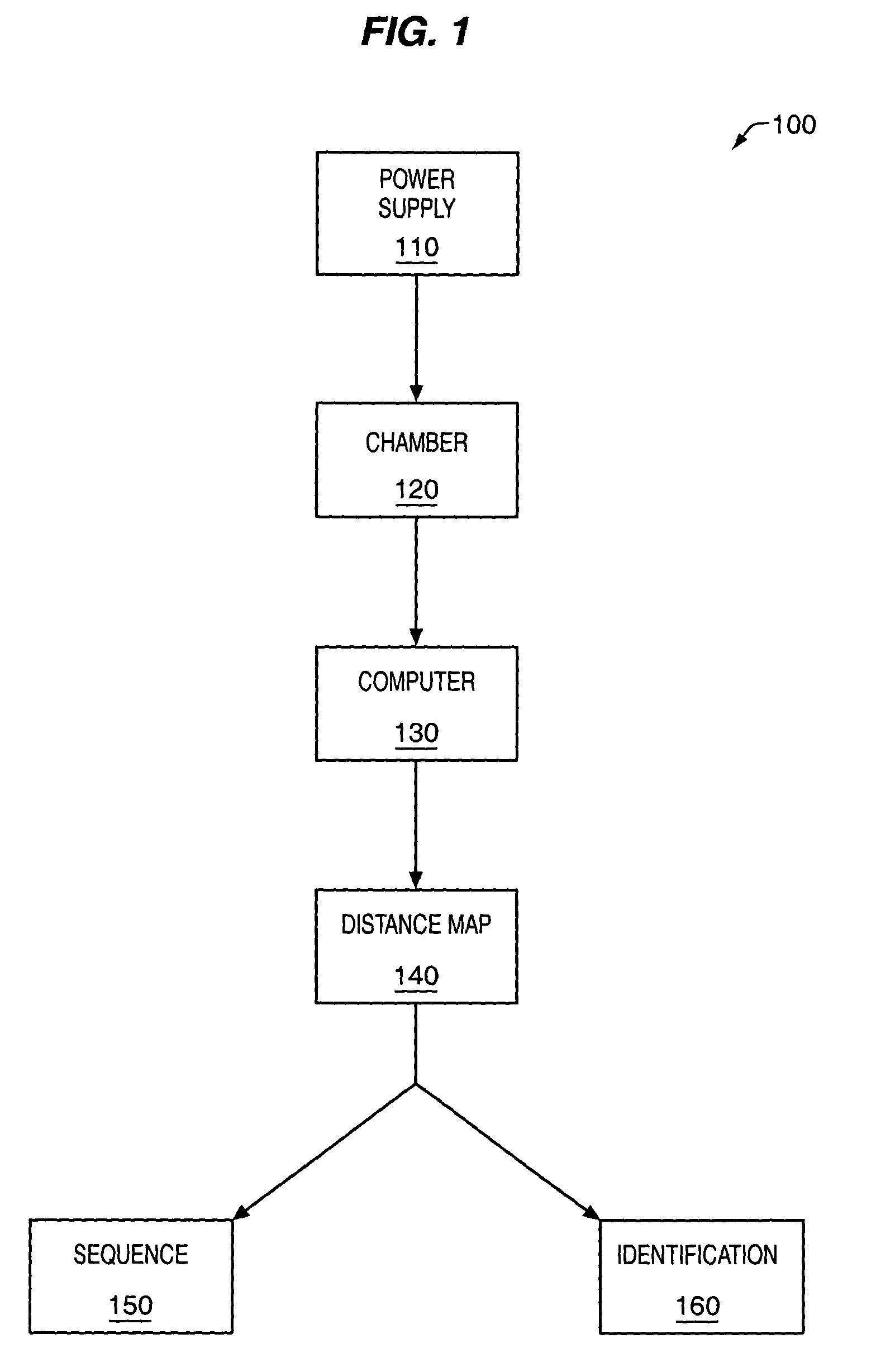
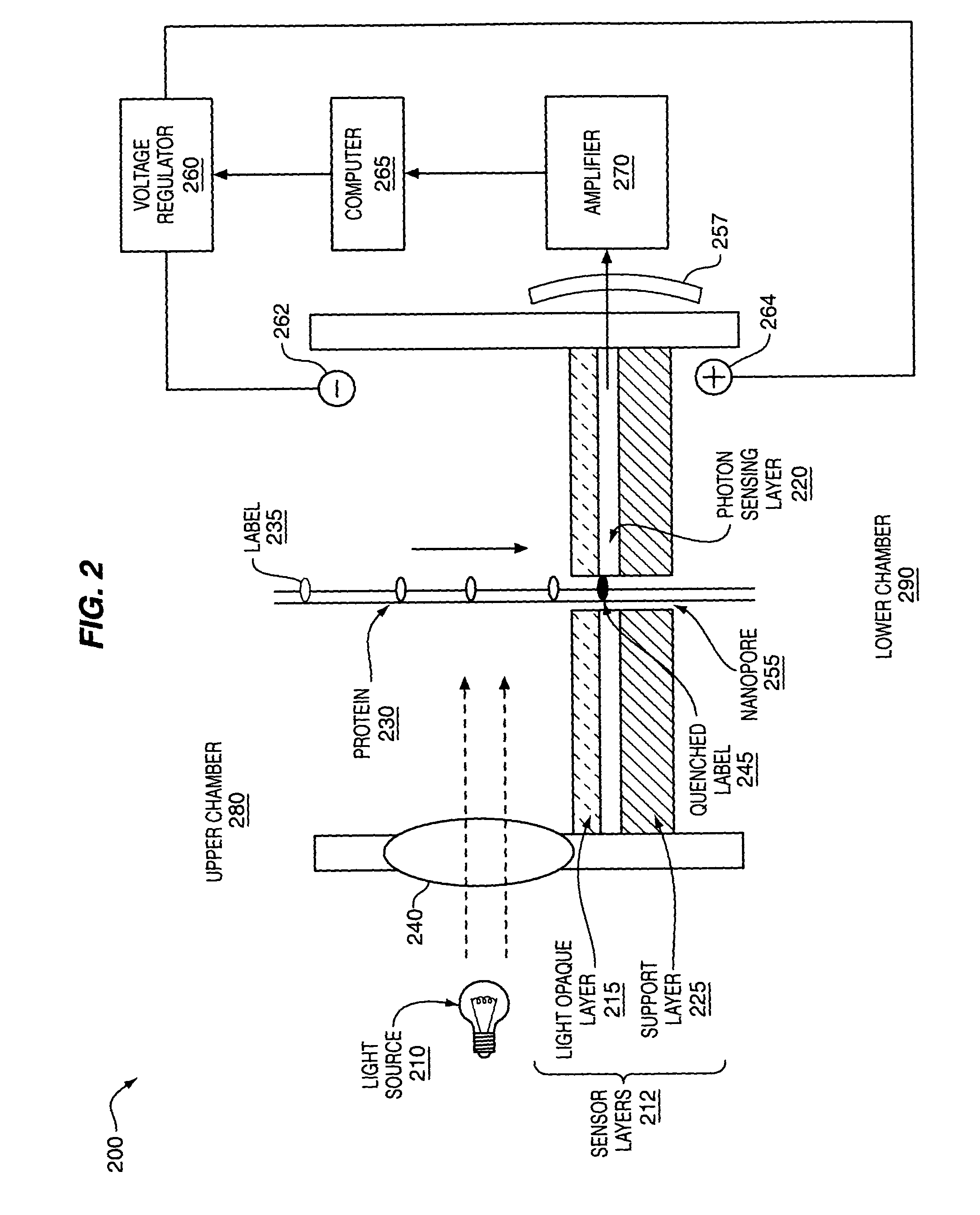
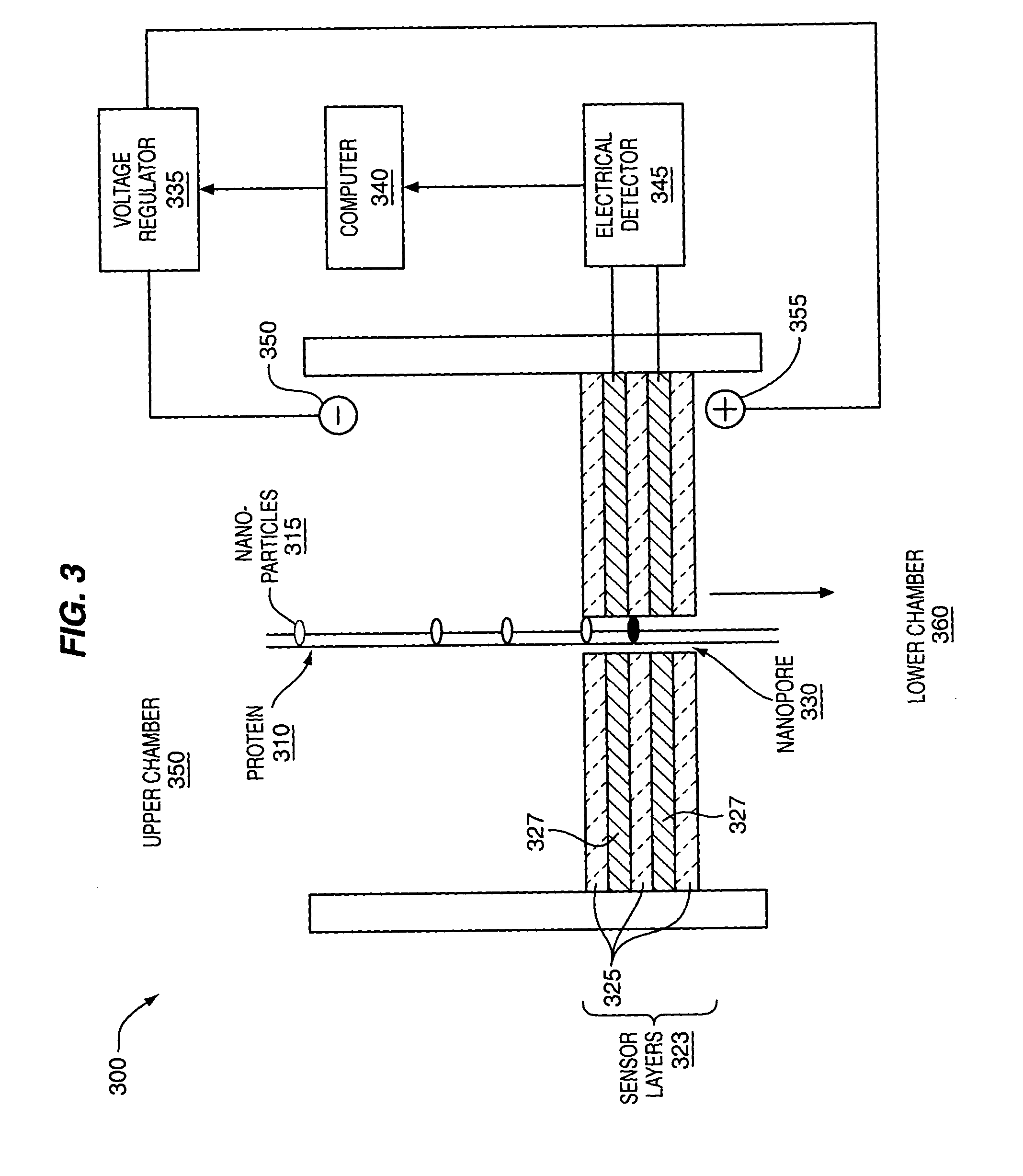
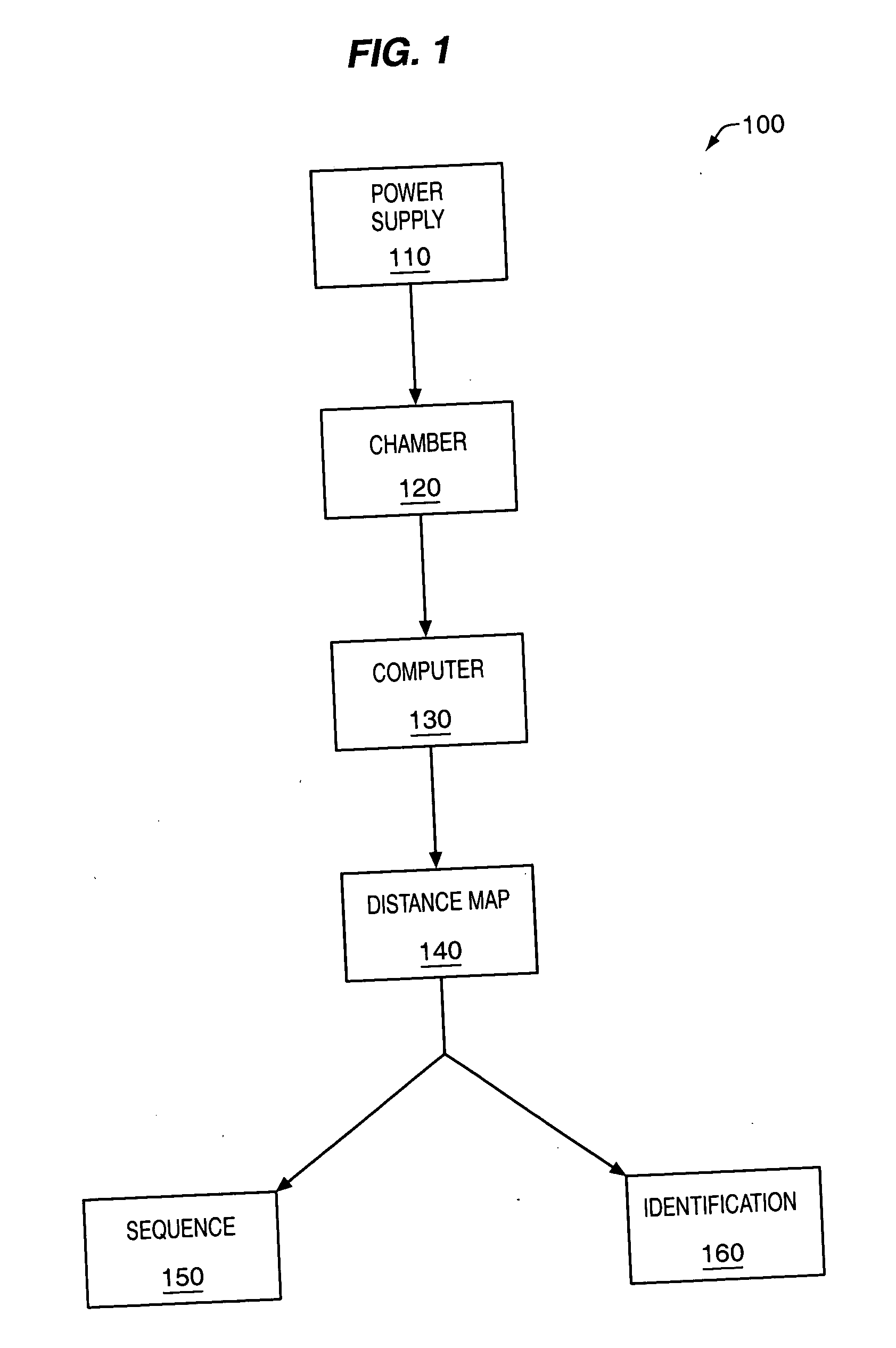
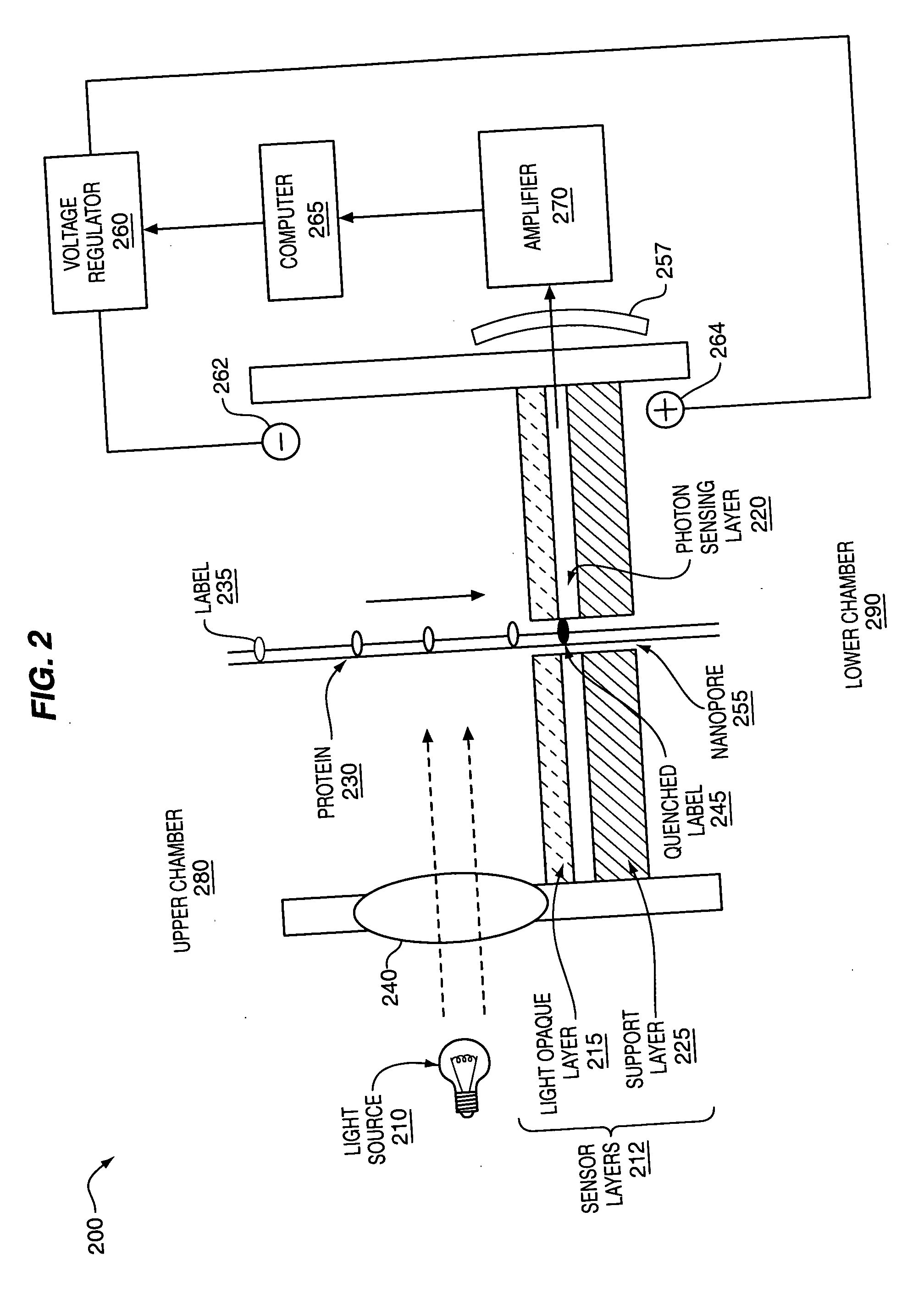
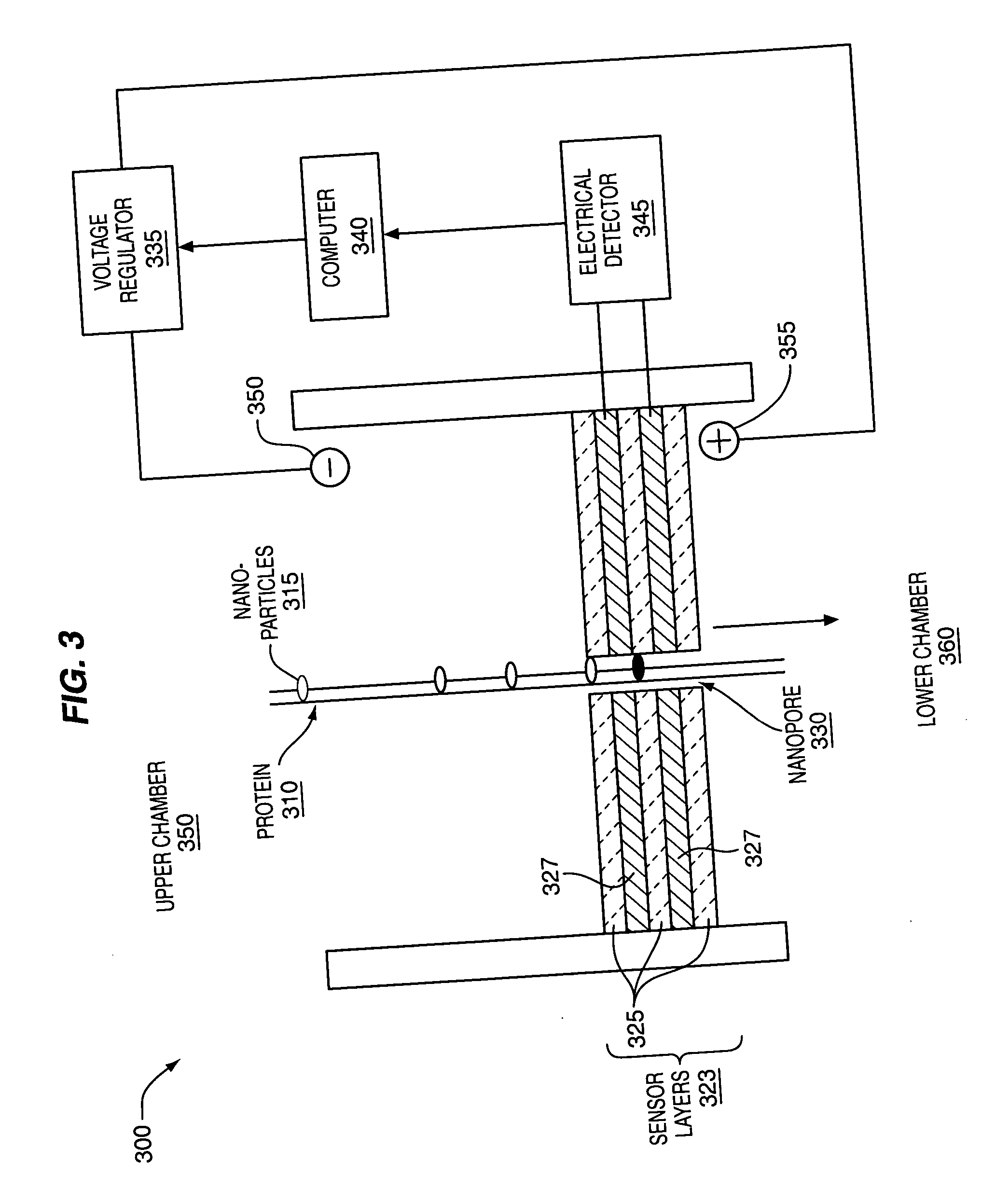
The methods and apparatus, disclosed herein are of use for sequencing and / or identifying proteins, polypeptides and / or peptides. Proteins containing labeled amino acid residues may be synthesized and passed through nanopores. A detector operably coupled to a nanopore may detect labeled amino acid residues as they pass through the nanopore. Distance maps for each type of labeled amino acid residue may be compiled. The distance maps may be used to sequence and / or identify the protein. Apparatus of use for protein sequencing and / or identification is also disclosed herein. In alternative methods, other types of analytes may be analyzed by the same techniques.
Owner:INTEL CORP
Protein sequencing with MALDI mass spectrometry
ActiveUS8581179B2Small amount of calculationIncrease productionTime-of-flight spectrometersSamples introduction/extractionDesorptionProtein Sequence Determination
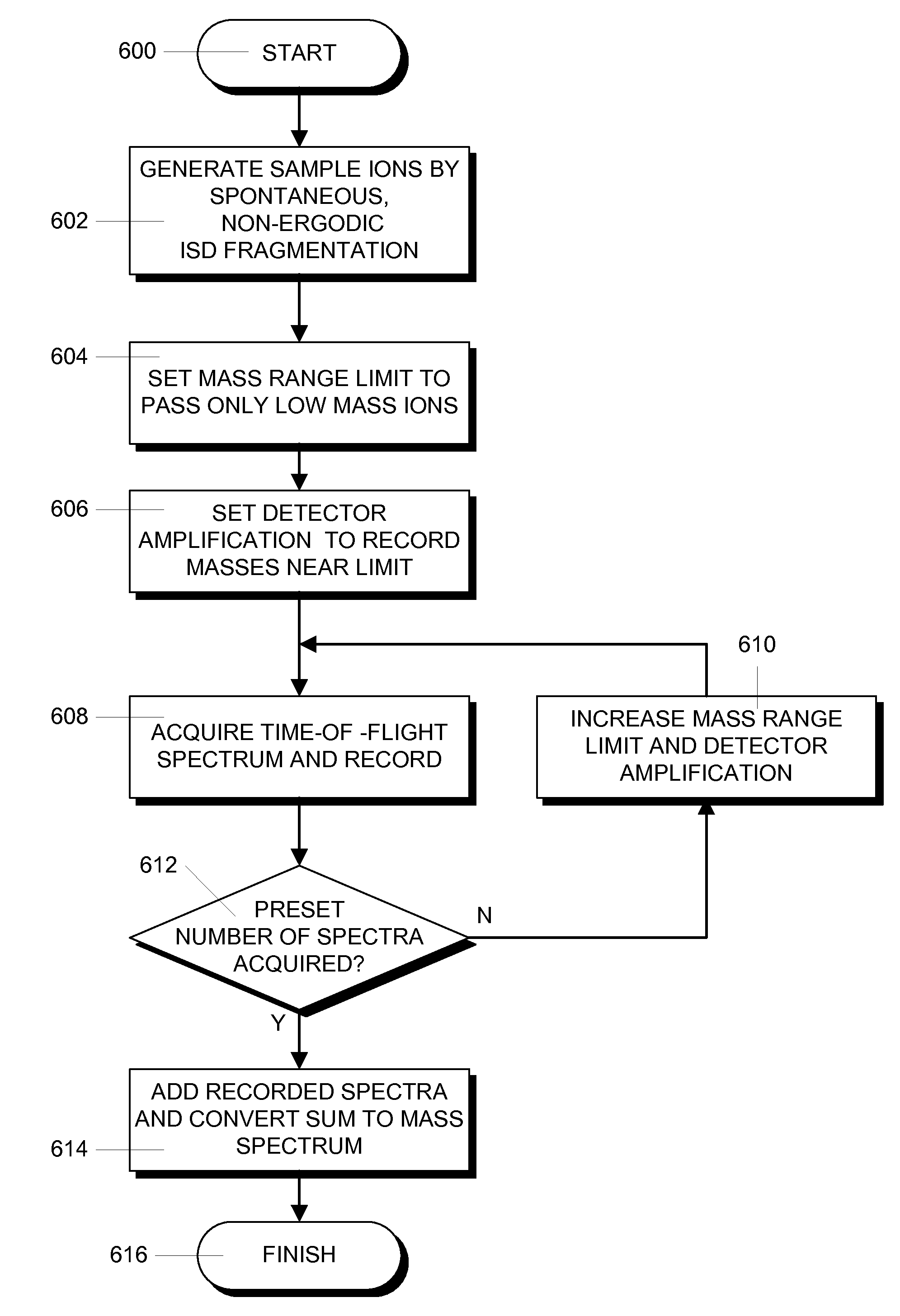
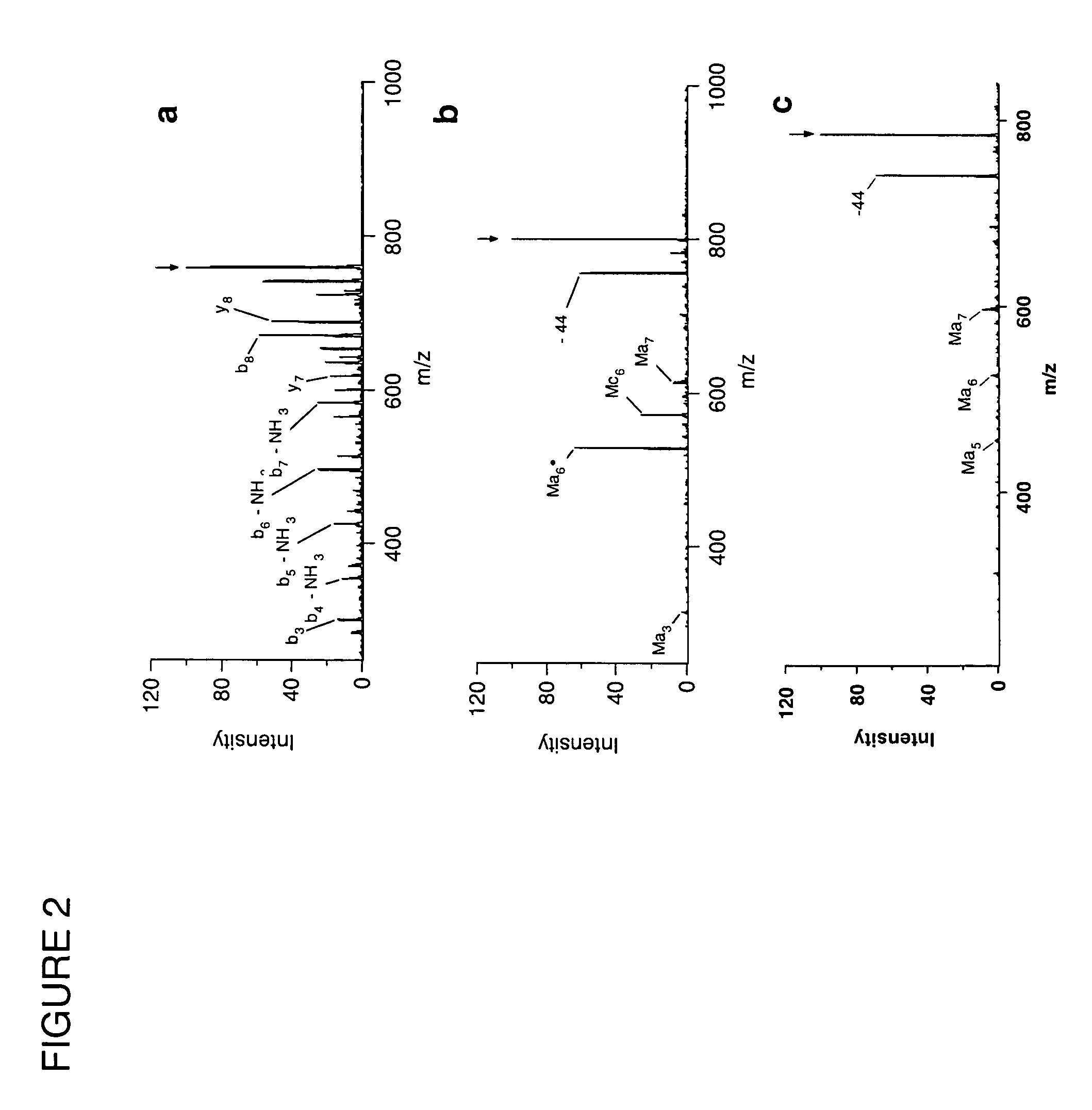
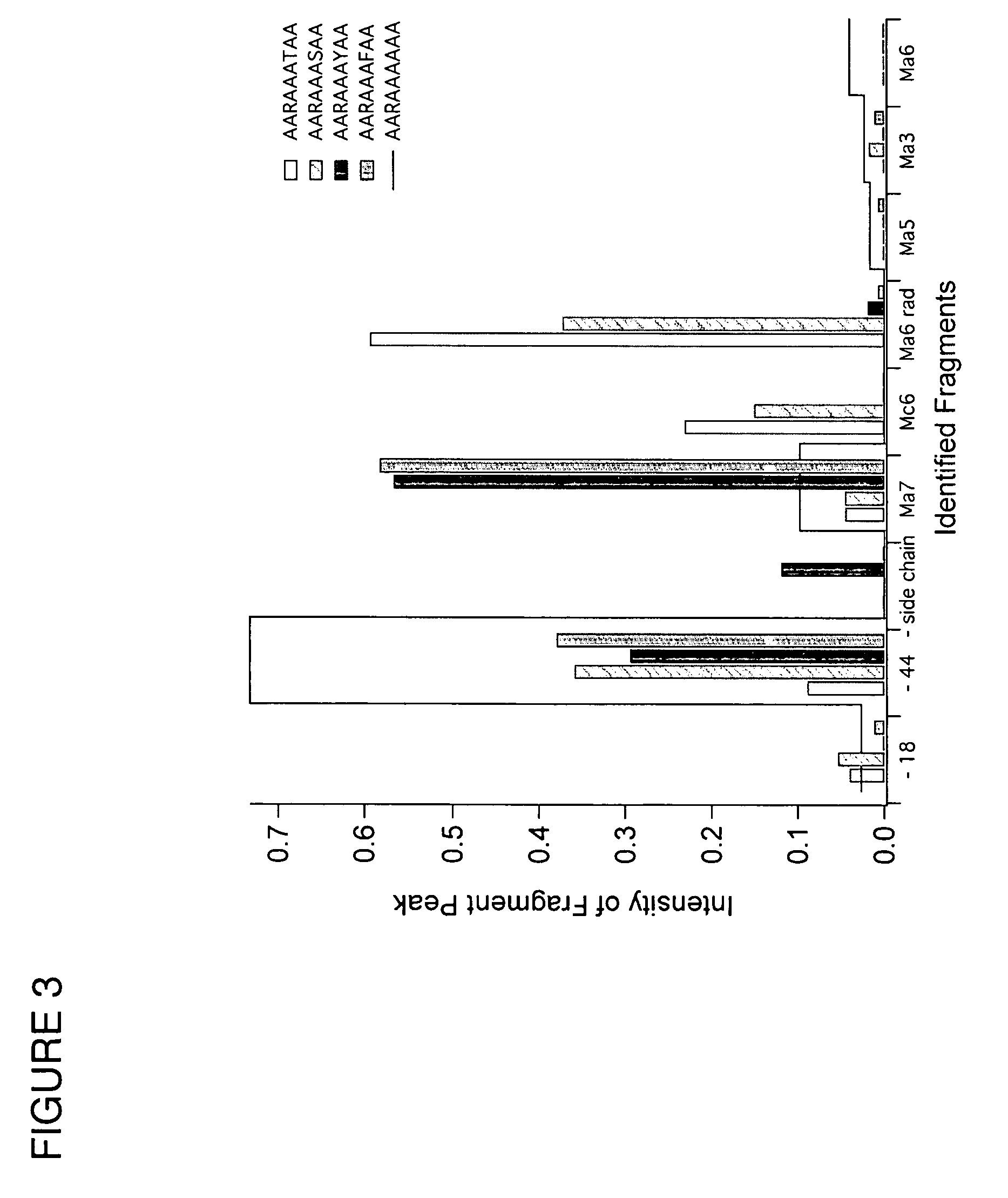
In a mass spectrometer, sample ions are produced by using matrix assisted laser desorption with a matrix substance that supports spontaneous, non-ergodic ISD fragmentation and a laser light source with nanosecond light pulses and a multiple spot beam profile. A plurality of individual time-of-flight spectra are recorded from the resulting ions in such a way that amplification of ion signals in the mass spectrometer detector is initially reduced so that only ions with masses near a mass range limit are initially recorded. During the repeated acquisitions of the individual time-of-flight spectra, both the detector amplification and the mass range limit are increased. By these methods, it is possible to evaluate c and z fragment ions in lower mass ranges and to directly read N-terminal sequences from near terminus up to 80 amino acids and beyond, and C-terminal sequences up to more than 60 amino acids.
Owner:BRUKER DALTONIK GMBH & CO KG
Control system and apparatus for use with laser excitation and ionization
InactiveUS20050232317A1Way fastMaximize sensitivityLaser detailsIon sources/gunsProtein Sequence DeterminationOptical communication
A control system and apparatus for use with laser ionization is provided. In another aspect of the present invention, the apparatus includes a laser, pulse shaper, detection device and control system. A further aspect of the present invention employs a femtosecond laser and a mass spectrometer in yet other aspects of the present invention, the control system and apparatus are used in MALDI, chemical bond cleaving, protein sequencing, photodynamic therapy, optical coherence tomography and optical communications processes.
Owner:BOARD OF TRUSTEES OPERATING MICHIGAN STATE UNIV
Single molecule protein sequencing
InactiveUS20150185199A1Minimal ambiguityError minimizationBioreactor/fermenter combinationsBiological substance pretreatmentsProtein insertionProtein Sequence Determination
The invention provides a device for determining the type of protein in a liquid, the device comprising (a) an immobilized ATP dependent protease based molecular transporter machine configured to guide a protein that is functionalized with labels through a detection area of a detector, (b) said detector, configured to detect a signal as function of the labels of the labelled amino acids, (c) a processor unit, configured to identify from the detector signal a sequence of amino acids of the functionalized protein, wherein the processor unit is further configured to compare the identified sequence of amino acids with the occurrence of such sequence in a database of proteins and to identify the type of protein.
Owner:TECH UNIV DELFT
Protein/Peptide Sequencing By Chemical Degradation in the Gas Phase
ActiveUS20080248585A1The parameters are accurate and reliableHigh performance resultIsotope separationRadio frequency spectrometersGas phaseMass Spectrometry-Mass Spectrometry
A fast and sensitive method and device for protein sequencing are disclosed. The method uses a combination of Edman degradation chemistry and mass spectrometry to sequence proteins and polypeptides. A peptide degradation reaction is performed on a polypeptide or protein ion reactant in the gas phase. The reaction yields a first ion product corresponding to a first amino acid residue of the polypeptide or protein reactant and a polypeptide or protein fragment ion. The mass-to-charge ratio for the first ion product, or the polypeptide or protein fragment ion, or both, is then determined. The first amino acid residue of the polypeptide or protein reactant is then identified from the mass-to-charge ratio so determined.
Owner:WISCONSIN ALUMNI RES FOUND
Protein sequencing methods and reagents
ActiveUS20180299460A1Facilitates chemical cleavage of N-terminalOrganic chemistryLibrary member identificationProtein Sequence DeterminationComputational chemistry
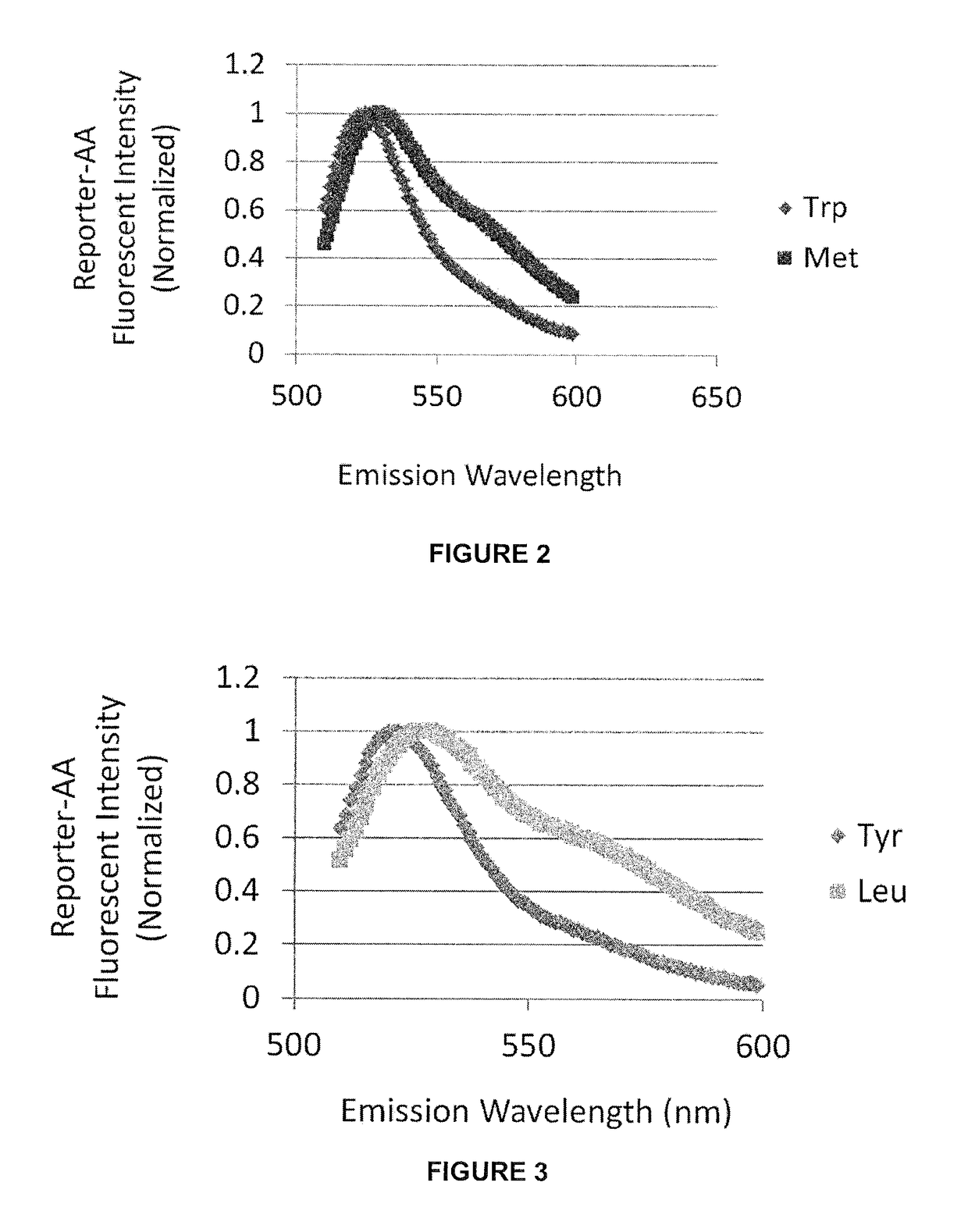
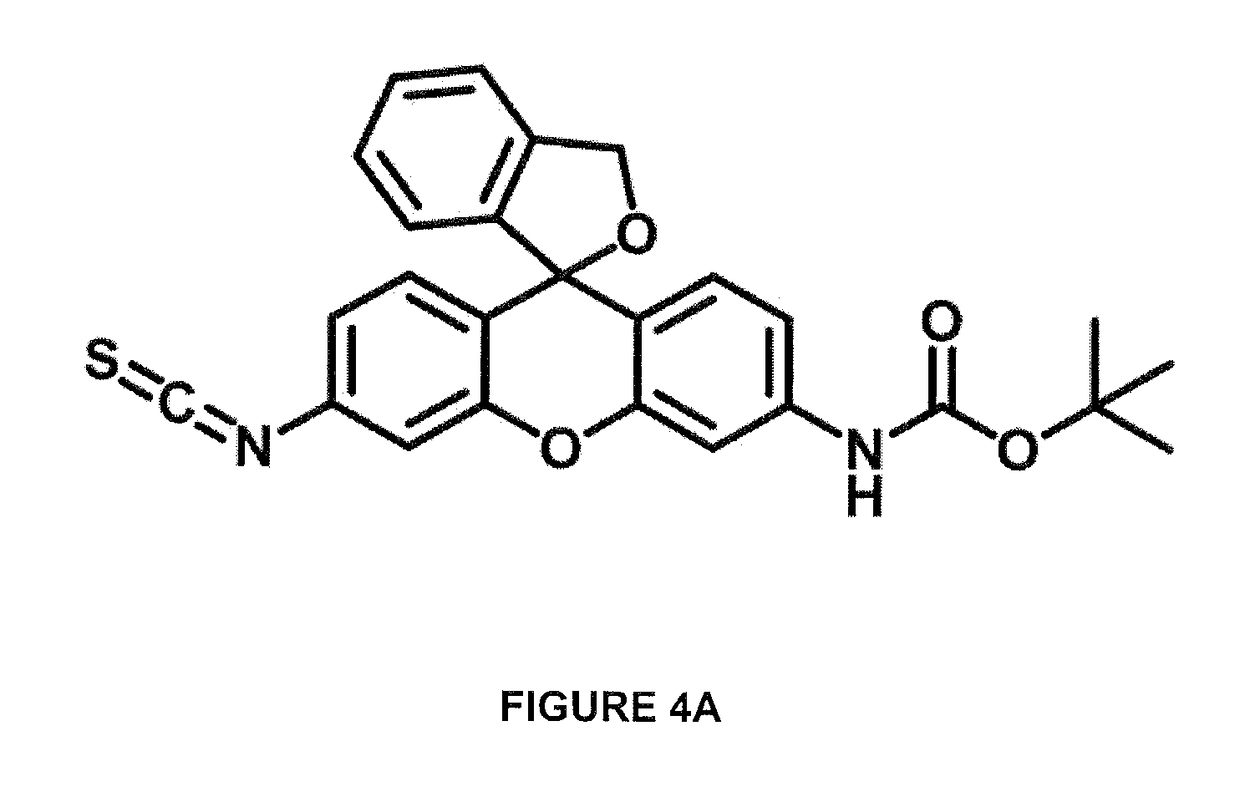
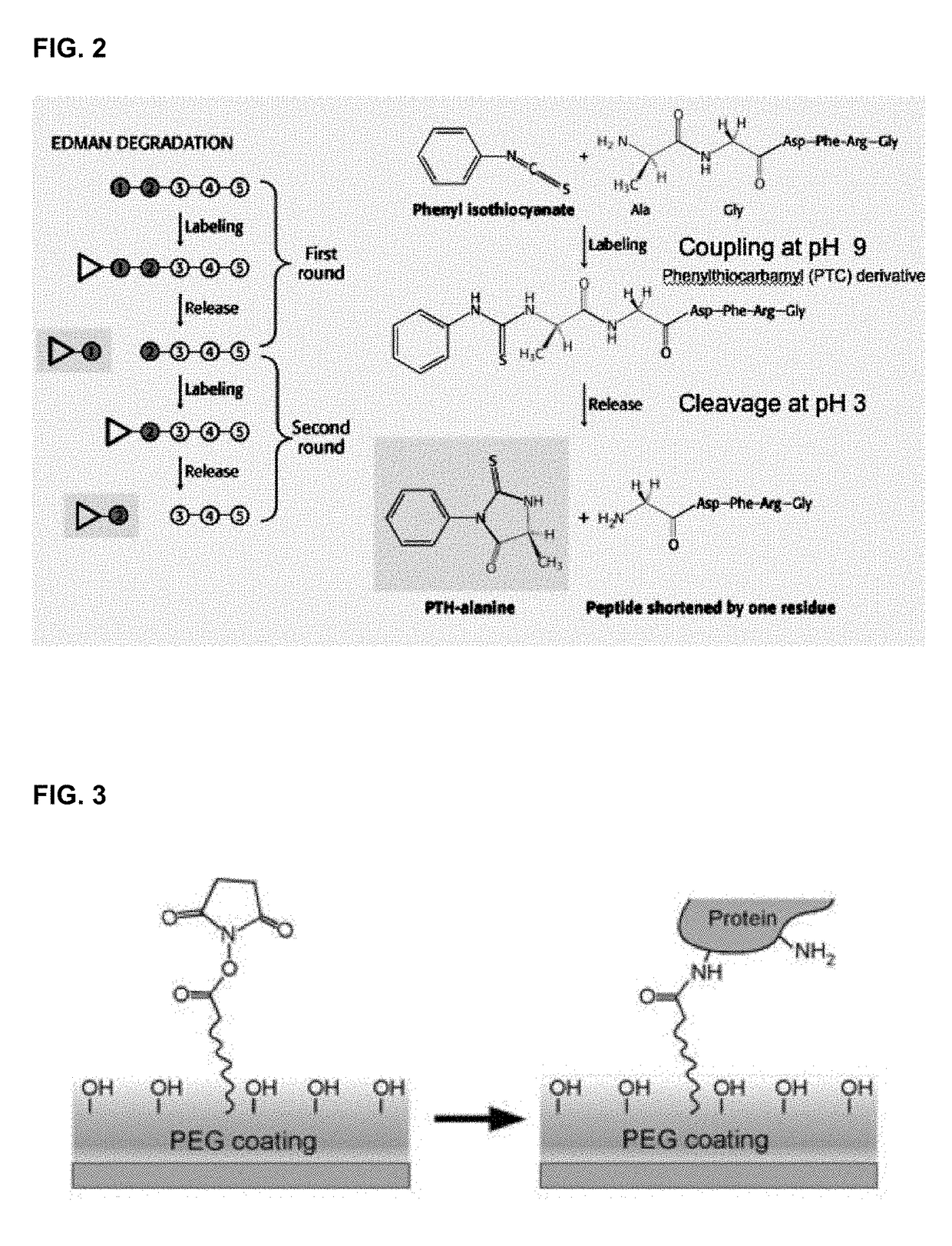
Described are optical methods and reagents for sequencing polypeptides. A probe that exhibits different spectral properties when conjugated to different N-terminal amino acids is conjugated to the N-terminal amino acid of a polypeptide. Sequentially detecting one or more spectral properties of the probe conjugated to the N-terminal amino acid and cleaving the N-terminal amino acid produces sequence information of the polypeptide. The use of super-resolution microscopy allows for the massively parallel sequencing of individual polypeptide molecules in situ such as within a cell. Also described are probes comprising hydroxymethyl rhodamine green, an isothiocyanate group and a protecting group.
Owner:THE GOVERNINIG COUNCIL OF THE UNIV OF TORANTO
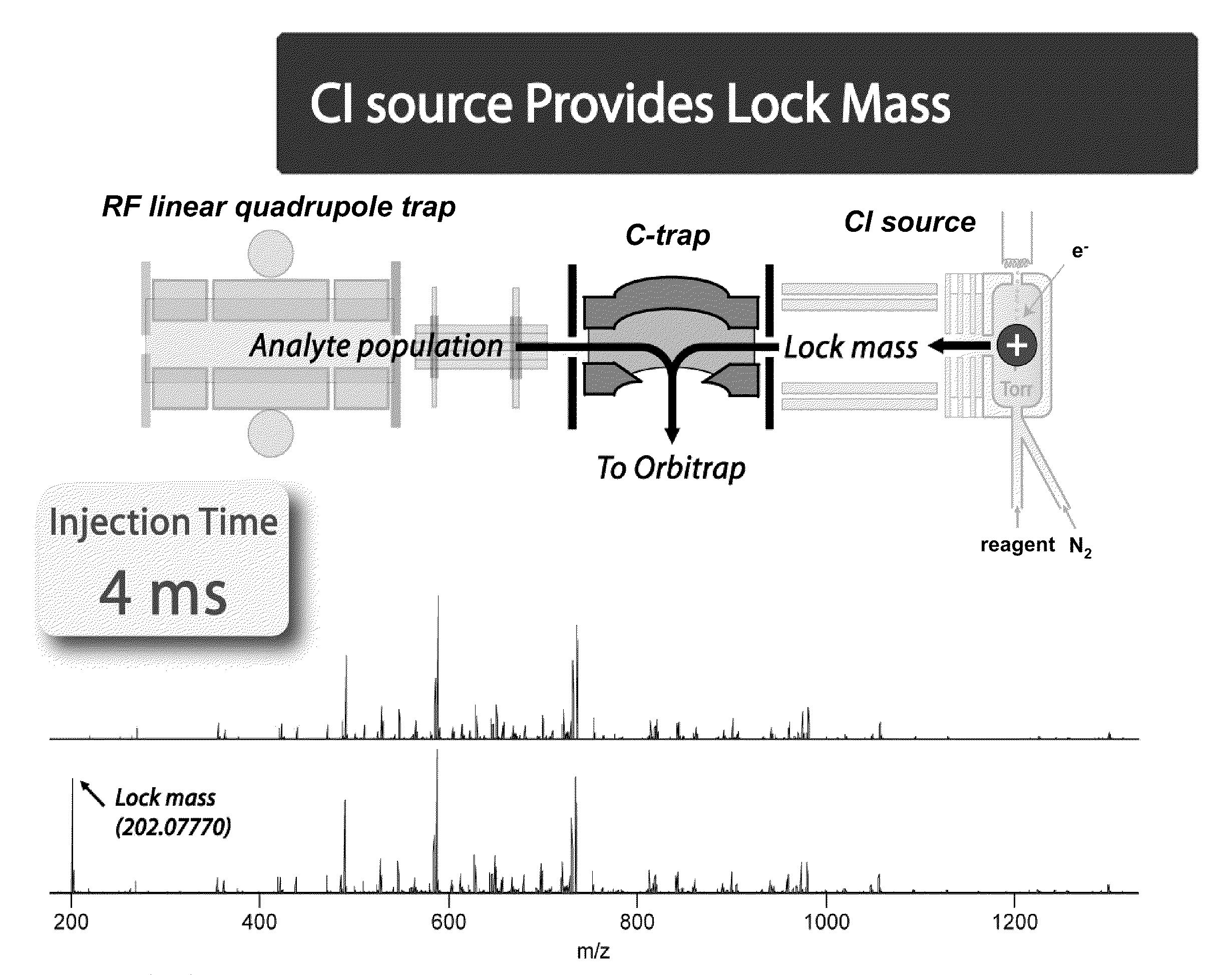
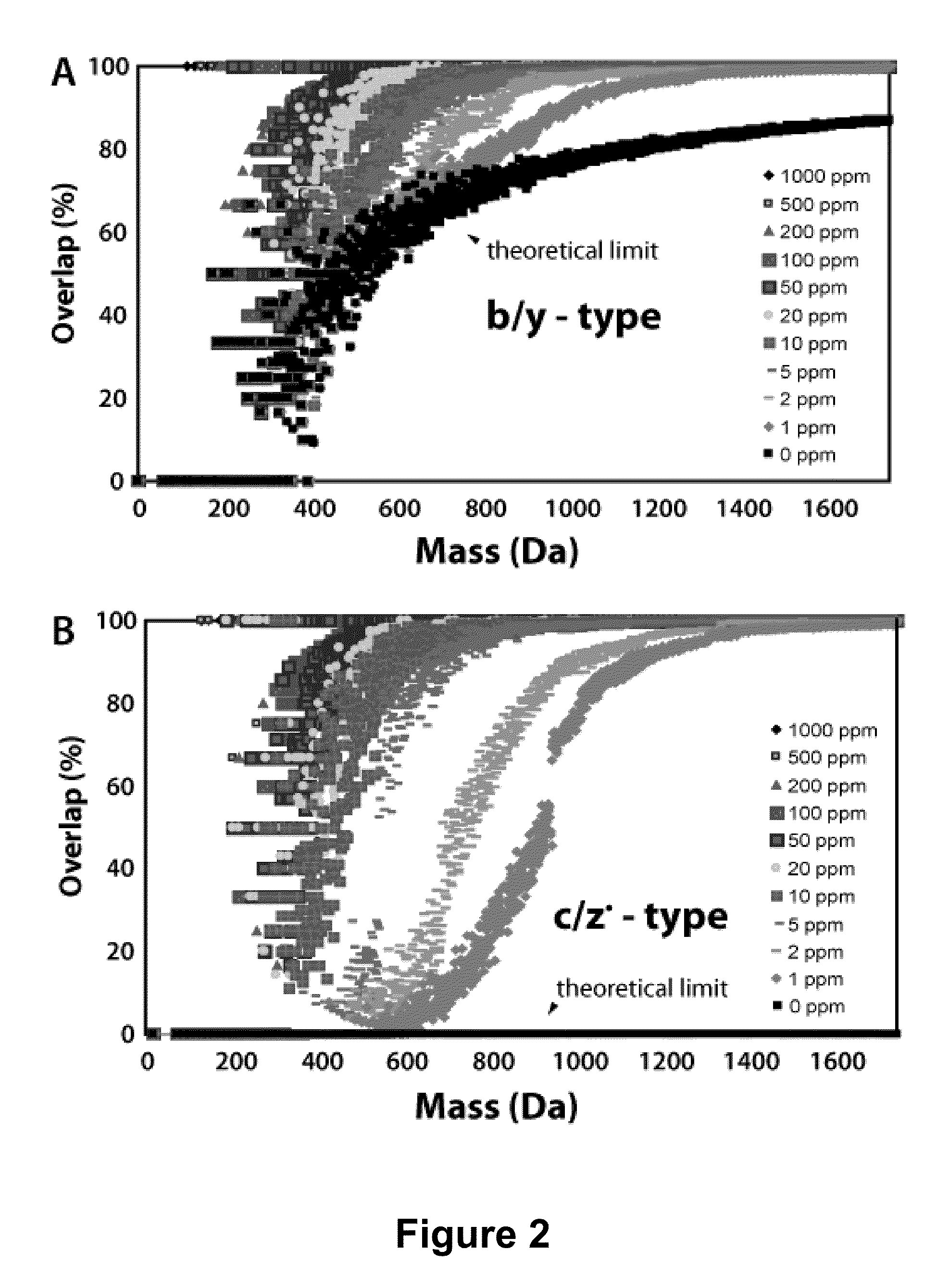
Methods for Processing Tandem Mass Spectral Data for Protein Sequence Analysis
ActiveUS20090173878A1Reliably and uniquely identifying and classifyingThe composition is uniqueParticle separator tubesCalibration apparatusFrequency spectrumProtein sequence analysis
Various mass spectroscopy-based methods are provided to improve protein sequencing by detecting z-type product ions generated from the protein. A polypeptide is introduced to a mass spectrometer, and in particular c- and z-type product ions that are generated by selectively fragmenting the polypeptide. The z-type product ions are distinguished from the c-type product ions and the mass-to-charge ratio of at least a portion of the z-type product ions are determined. From the mass of the z-type product ions, a putative chemical composition is identified for at least a portion of the z-type product ions, c-type product ions, or both, which is used to determine polypeptide compositions. Further provided are various methods for reducing spectral noise, instrument calibration and database searching and verification.
Owner:WISCONSIN ALUMNI RES FOUND
Methods for processing tandem mass spectral data for protein sequence analysis
ActiveUS8278115B2Reliably and uniquely identifying and classifyingThe composition is uniqueParticle separator tubesIsotope separationSequence analysisFrequency spectrum
Various mass spectroscopy-based methods are provided to improve protein sequencing by detecting z-type product ions generated from the protein. A polypeptide is introduced to a mass spectrometer, and in particular c- and z-type product ions that are generated by selectively fragmenting the polypeptide. The z-type product ions are distinguished from the c-type product ions and the mass-to-charge ratio of at least a portion of the z-type product ions are determined. From the mass of the z-type product ions, a putative chemical composition is identified for at least a portion of the z-type product ions, c-type product ions, or both, which is used to determine polypeptide compositions. Further provided are various methods for reducing spectral noise, instrument calibration and database searching and verification.
Owner:WISCONSIN ALUMNI RES FOUND
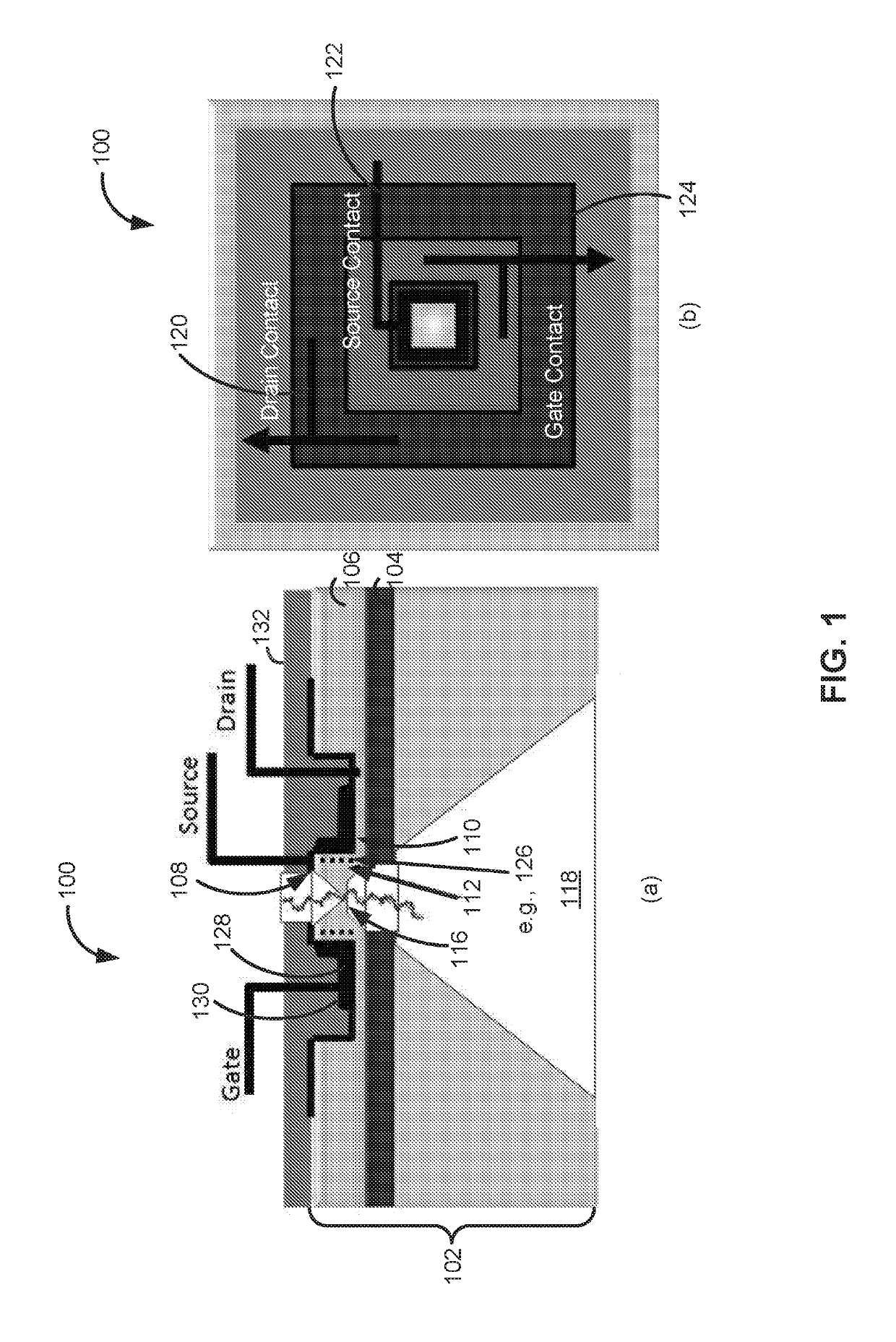
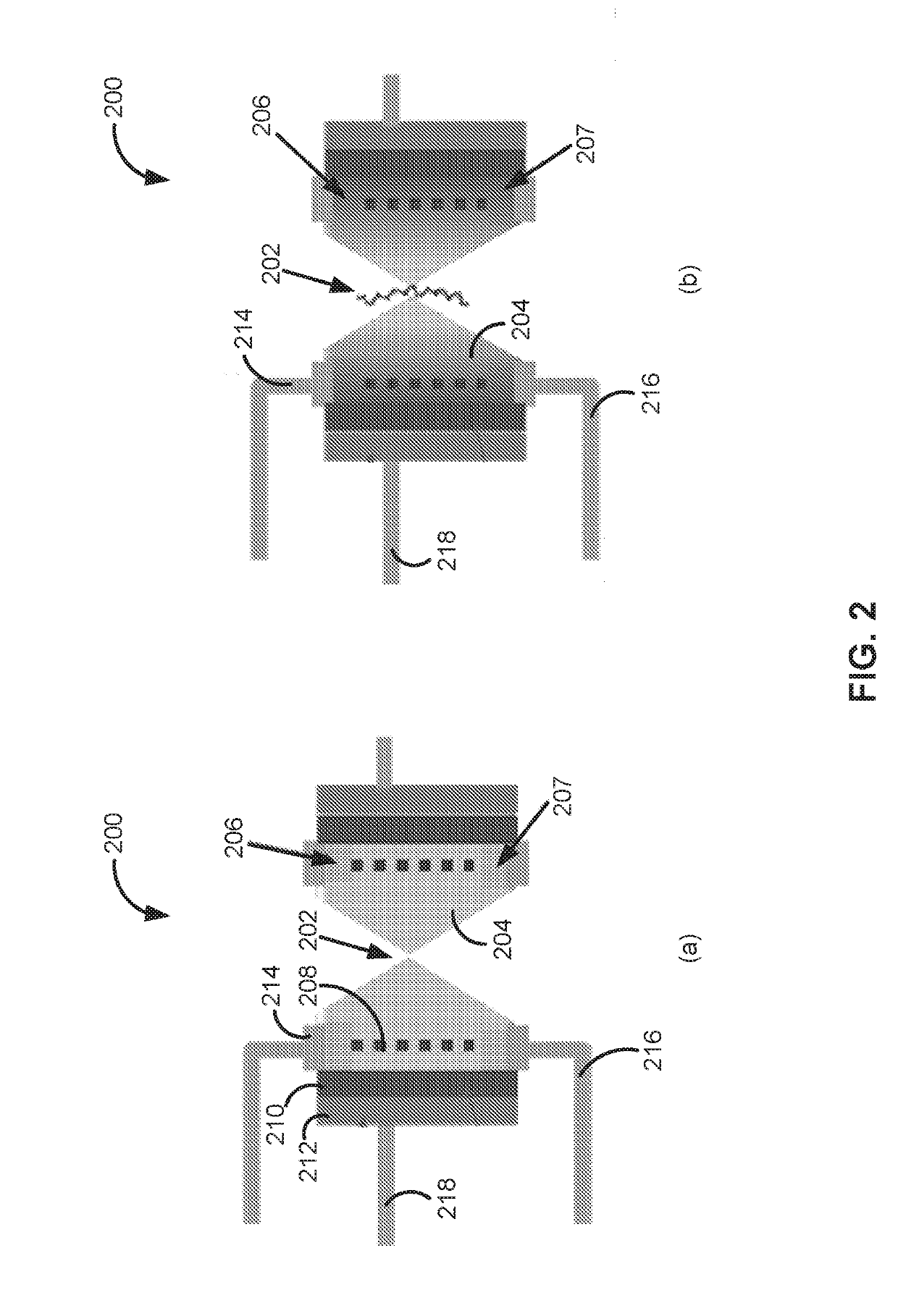
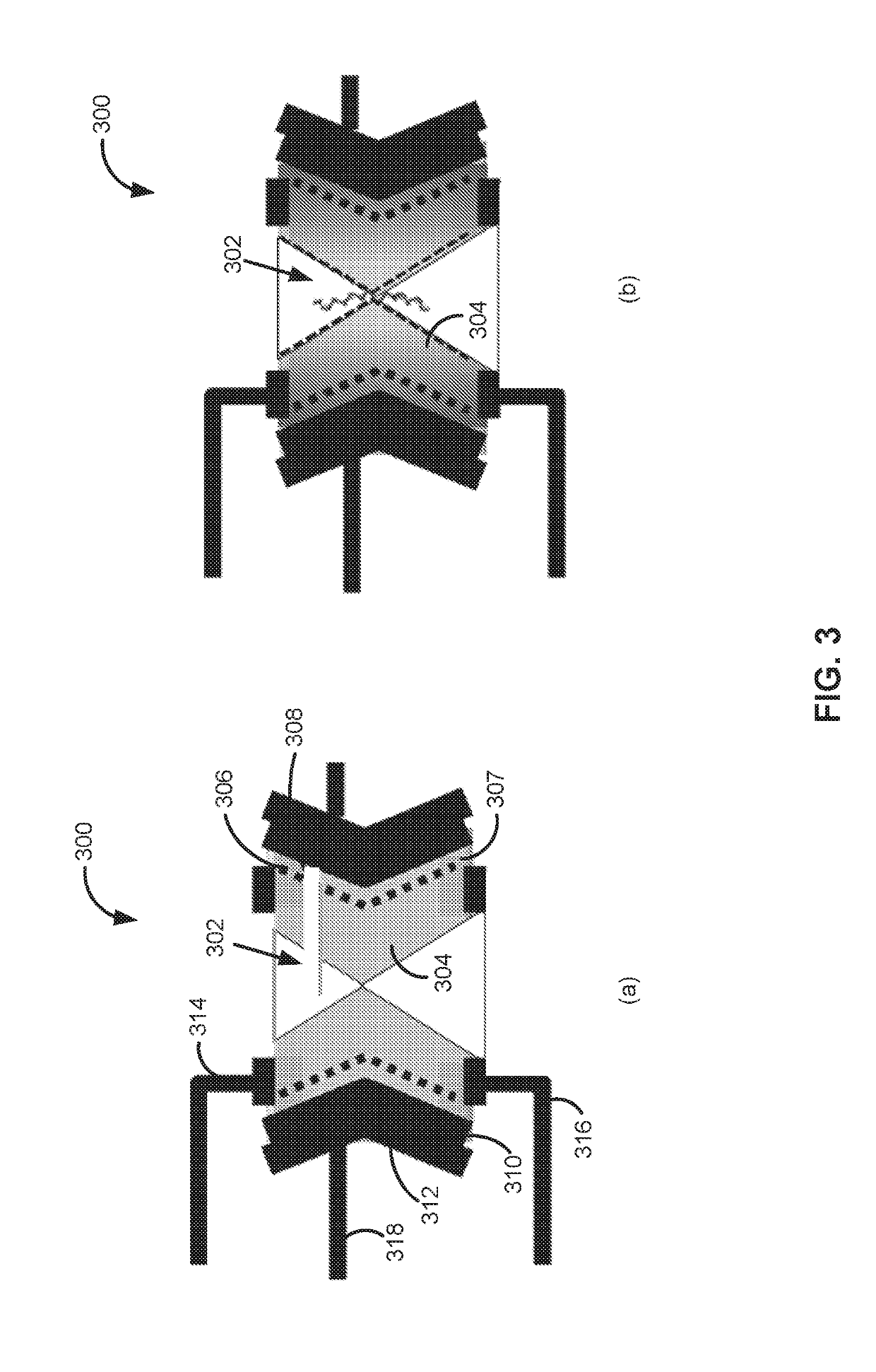
Field effect transistor, device including the transistor, and methods of forming and using same
ActiveUS20150060952A1Microbiological testing/measurementMaterial analysis by electric/magnetic meansGenomic sequencingOrganic field-effect transistor
The present disclosure provides an improved field effect transistor and device that can be used to sense and characterize a variety of materials. The field effect transistor and / or device including the transistor may be used for a variety of applications, including genome sequencing, protein sequencing, biomolecular sequencing, and detection of ions, molecules, chemicals, biomolecules, metal atoms, polymers, nanoparticles and the like.
Owner:INANOBIO
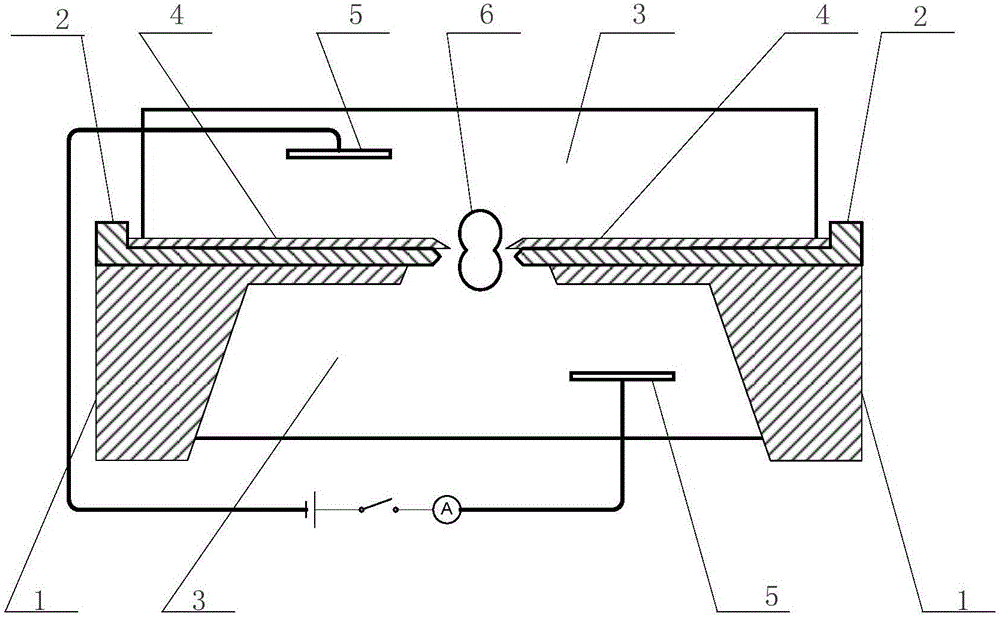
Single-layer mica sheet and preparation method and application of nanopore electronic device thereof
ActiveCN105883838AAchieve single layer cleavage effectRealize identificationBioreactor/fermenter combinationsBiological substance pretreatmentsProtein moleculesHigh energy
The invention discloses a single-layer mica sheet and a preparation method and application of a nanopore electronic device thereof. The method comprises the steps that an ultrathin single mica layer is prepared and coats a proper supporting substrate, and a high-energy focusing electronic beam or an ion beam is used for preparing nanopores of the specification being 1-5 nm in the single-layer mica sheet to obtain a solid-state nanopore electronic device based on the single-layer mica sheet. Single-stranded nucleic acid or protein molecules are driven by an electric field to penetrate through the nanopores in the electronic device, and a basic group on the single-stranded nucleic acid penetrating through the pores or amino acid on the single-stranded protein is recognized by detecting changes of via-pore current in real time. A high throughput parallel molecular device can be prepared from the electronic device, and rapid and ultralow cost nucleic acid / protein sequencing can be achieved.
Owner:PEKING UNIV +1
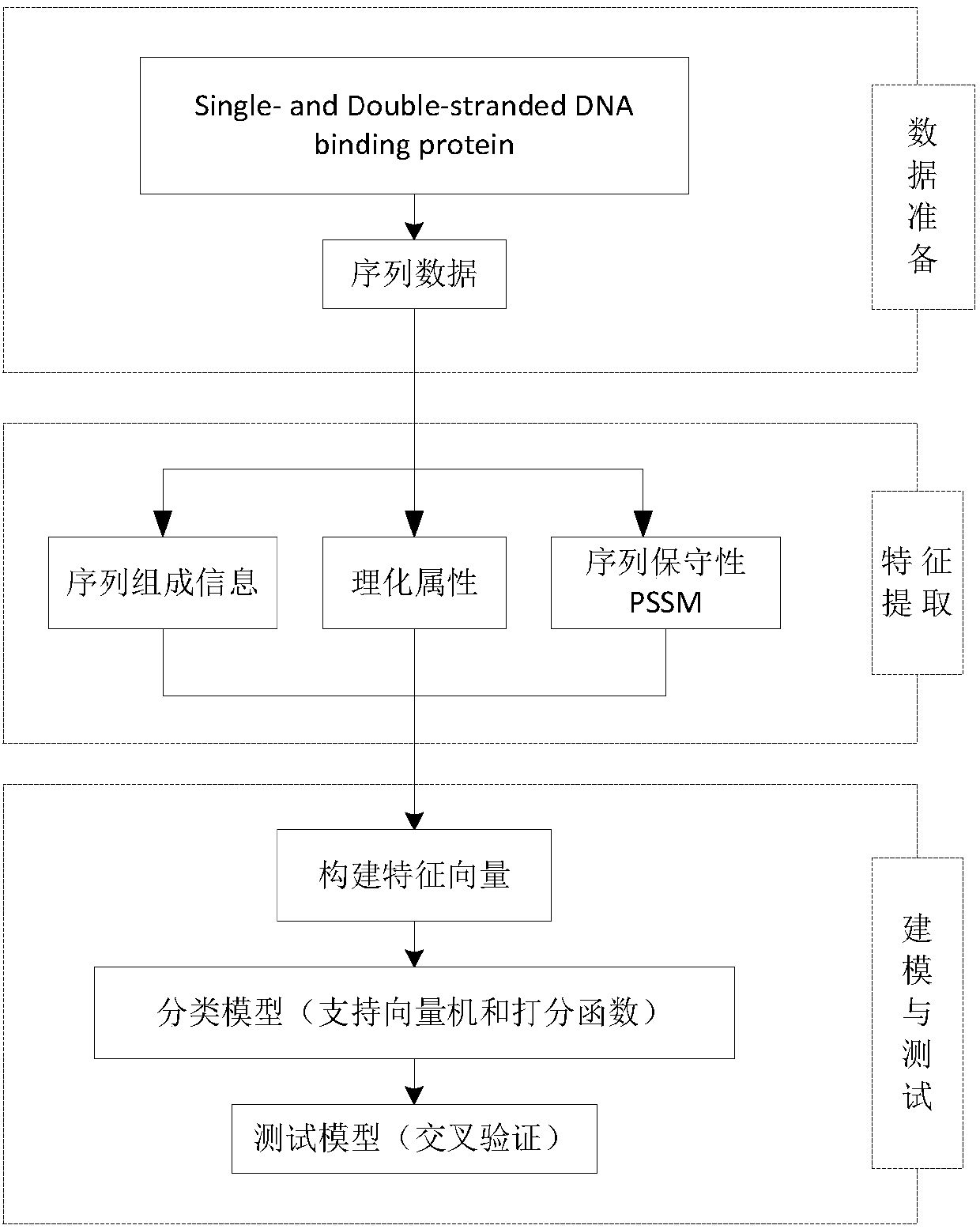
Characteristic extraction and classification method and device for DNA (Deoxyribo-Nucleic Acid) binding protein sequence information
InactiveCN108875310AImplement function annotationsAchieve classification goalsSpecial data processing applicationsData setProtein insertion
The invention relates to a characteristic extraction and classification method and device for DNA (Deoxyribo-Nucleic Acid) binding protein sequence information. The method comprises the following steps that: firstly, carrying out theory argumentation, and analyzing and arranging collected data to obtain a reliable dataset with a biological meaning and a statistical meaning; then, extracting effective protein sequence data characteristic parameters from a complex protein three-dimensional structure as a key link which is a way of converting sequence character information into digital characteristic information, designing a reasonable classification algorithm for extracted characteristic data, and screening the characteristics favorable for classification for realizing target classification;and finally, adopting a reasonable and fair evaluation system, including testing methods, checking means, evaluation index selection and the like, for classification performance. By use of the method, requirements on high-flux protein sequencing function annotation can be met, automated DNA binding protein sequence function annotation can be realized, and meanwhile, the characteristics which areput forward can assist biologists in carrying out experimental analysis and research on the DNA binding protein sequence.
Owner:HENAN NORMAL UNIV
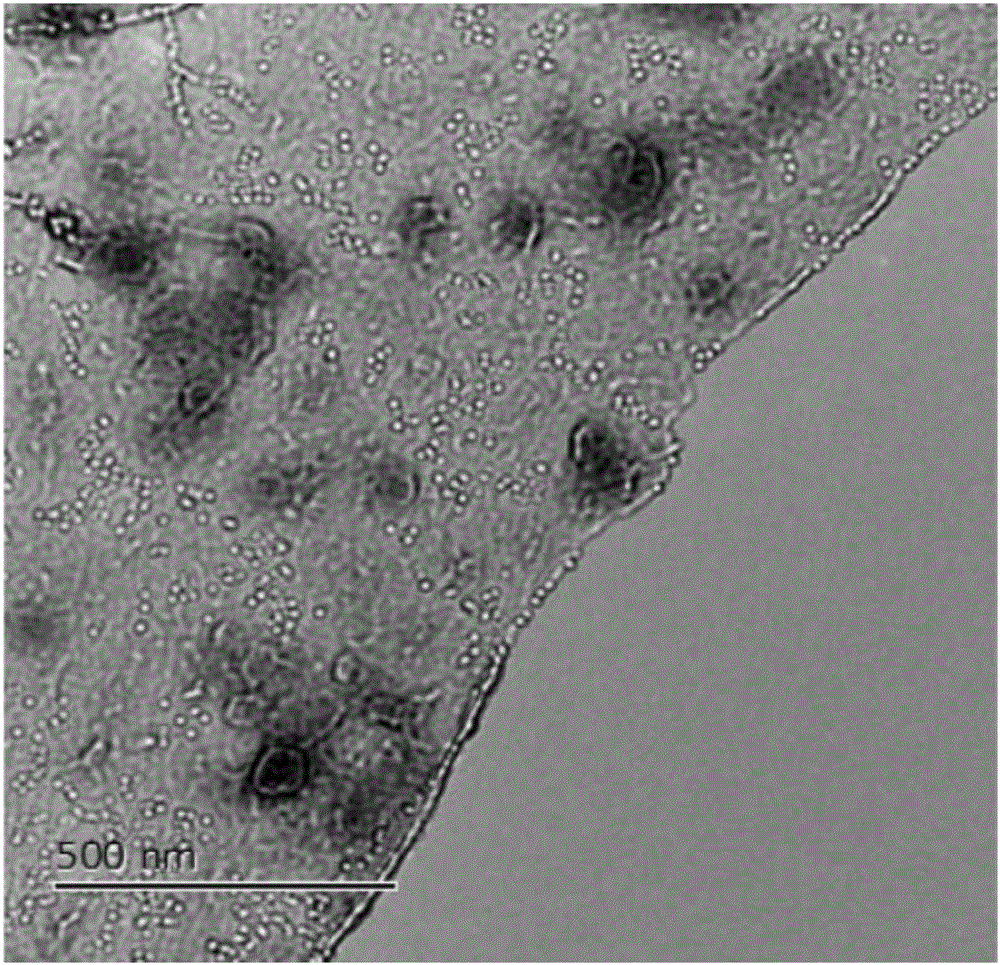
Monodisperse nanolamella-based nanometer single hole, and making method and application thereof
ActiveCN107356580AStable structureEnabling single-molecule Raman detectionMaterial nanotechnologyRaman scatteringEngineeringProtein Sequence Determination
The invention relates to a monodisperse nanolamella-based nanometer single hole, and a making method and an application thereof. The nanometer single hole is positioned in the nanometer tip of a capillary tube, and is formed by a monodisperse nanolamella capping the tip of the capillary tube, and the aperture of the nanometer single hole is 10-100 nm. The metal nanometer single hole has the advantages of simple making method, low cost, small dimension and controllable structure, and can be combined with Raman spectrum as an electrochemical device to realize electrochemical controllable single molecule detection and realize the identification of single bases, so a method is provided for DNA sequencing and even protein sequencing.
Owner:NANJING UNIV
Field effect transistor, device including the transistor, and methods of forming and using same
ActiveUS9341592B2Microbiological testing/measurementMaterial analysis by electric/magnetic meansGenomic sequencingNanoparticle
The present disclosure provides an improved field effect transistor and device that can be used to sense and characterize a variety of materials. The field effect transistor and / or device including the transistor may be used for a variety of applications, including genome sequencing, protein sequencing, biomolecular sequencing, and detection of ions, molecules, chemicals, biomolecules, metal atoms, polymers, nanoparticles and the like.
Owner:INANOBIO
Protein/polypeptide sequencing method adopting Aerolysin nanopores
PendingCN112480204AEasy to identifyAccurately obtain single-molecule sequence informationPeptide preparation methodsMaterial analysisAerolysinProtein molecules
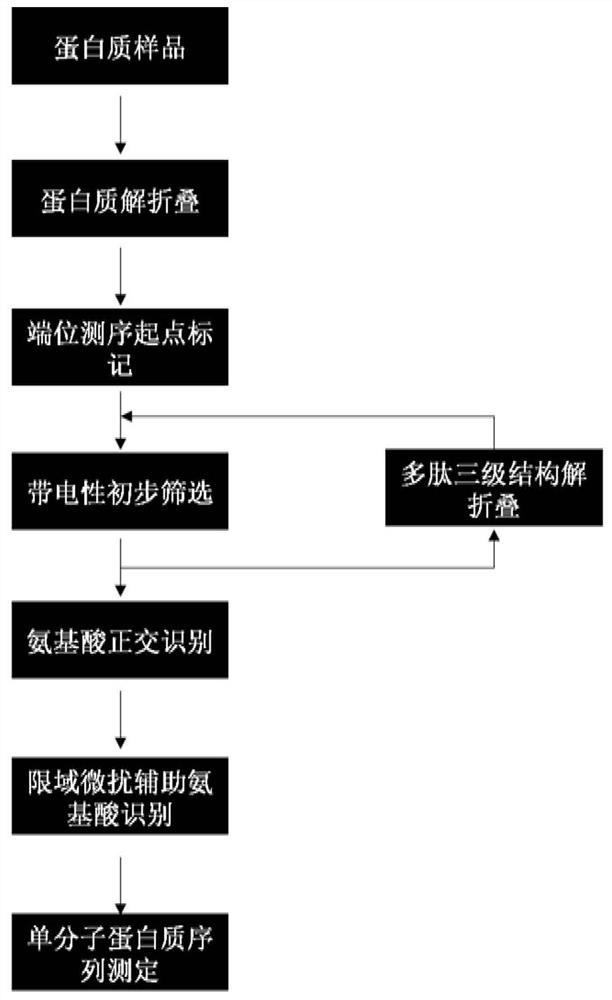
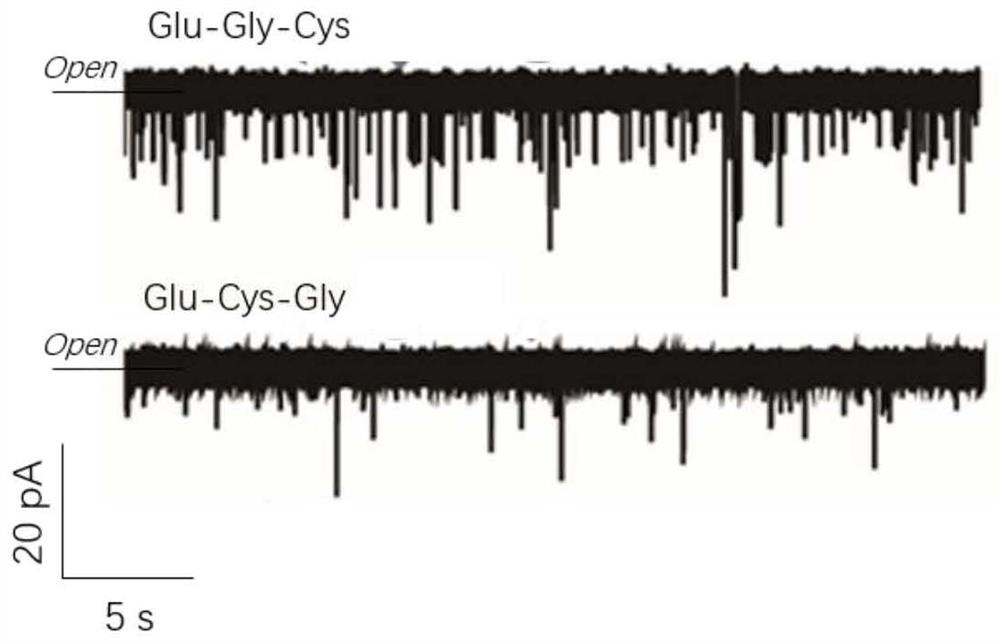
The invention provides a method for protein / polypeptide sequencing based on Aerolysin nanopores. The method realizes specific resolution of natural amino acid and post-translational modification thereof and accurate acquisition of a single-molecule protein sequence, and comprises the following steps: (1) carrying out protein unfolding; (2) marking an end position sequencing starting point; (3) carrying out electrified preliminary screening; (4) unfolding a polypeptide tertiary structure; (5) carrying out amino acid orthogonal recognition; (6) carrying out limited range perturbation assisted amino acid recognition; and (7) carrying out single molecule protein sequence determination. According to the method, an innovative method for accurate measurement of single protein molecule amino acidsequence and post-translational modification is established for sensitive detection of information of 20 amino acid sequences.
Owner:NANJING UNIV
Labidura japonica de haan antimicrobial peptide DEI and application thereof in duck feed
InactiveCN108503702AStrong toleranceGrow fastAccessory food factorsAnimals/human peptidesEscherichia coliPichia pastoris
The invention discloses a labidura japonica de haan antimicrobial peptide DEI and application thereof in duck feed, and belongs to the technical field of bioengineering. The labidura japonica de haanantimicrobial peptide DEI is prepared by acquiring an antimicrobial peptide DEI of labidura japonica de haan, testing an amino acid sequence of the antimicrobial peptide DEI by using an automatic protein sequencing instrument, further by means of genetic engineering, designing a cDNA (complementary deoxyribonucleic acid) fragment of the amino acid sequence, establishing cloning plasmid pUC57-TM, converting escherichia coli DH 5alpha, performing PCR (polymerase chain reaction) and sequencing to carry out further identification, establishing a recombinant expression vector pPICZaC-TM, performinglinearization on the recombinant expression vector, performing electric conversion into pichia pastoris SMD1168, screening to obtain a high-copy recombinant yeast conversion factor, and culturing a single colony of the recombinant yeast conversion factor, thereby obtaining the labidura japonica de haan antimicrobial peptide DEI. The labidura japonica de haan antimicrobial peptide DEI can be applied to duck feed, then the immunity of ducks can be improved, the morbidity of the ducks can be greatly reduced, the survival rate of the ducks can be increased, and thus the feeding cost of duck farmers can be reduced.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
System and method of free radical initiated protein sequencing
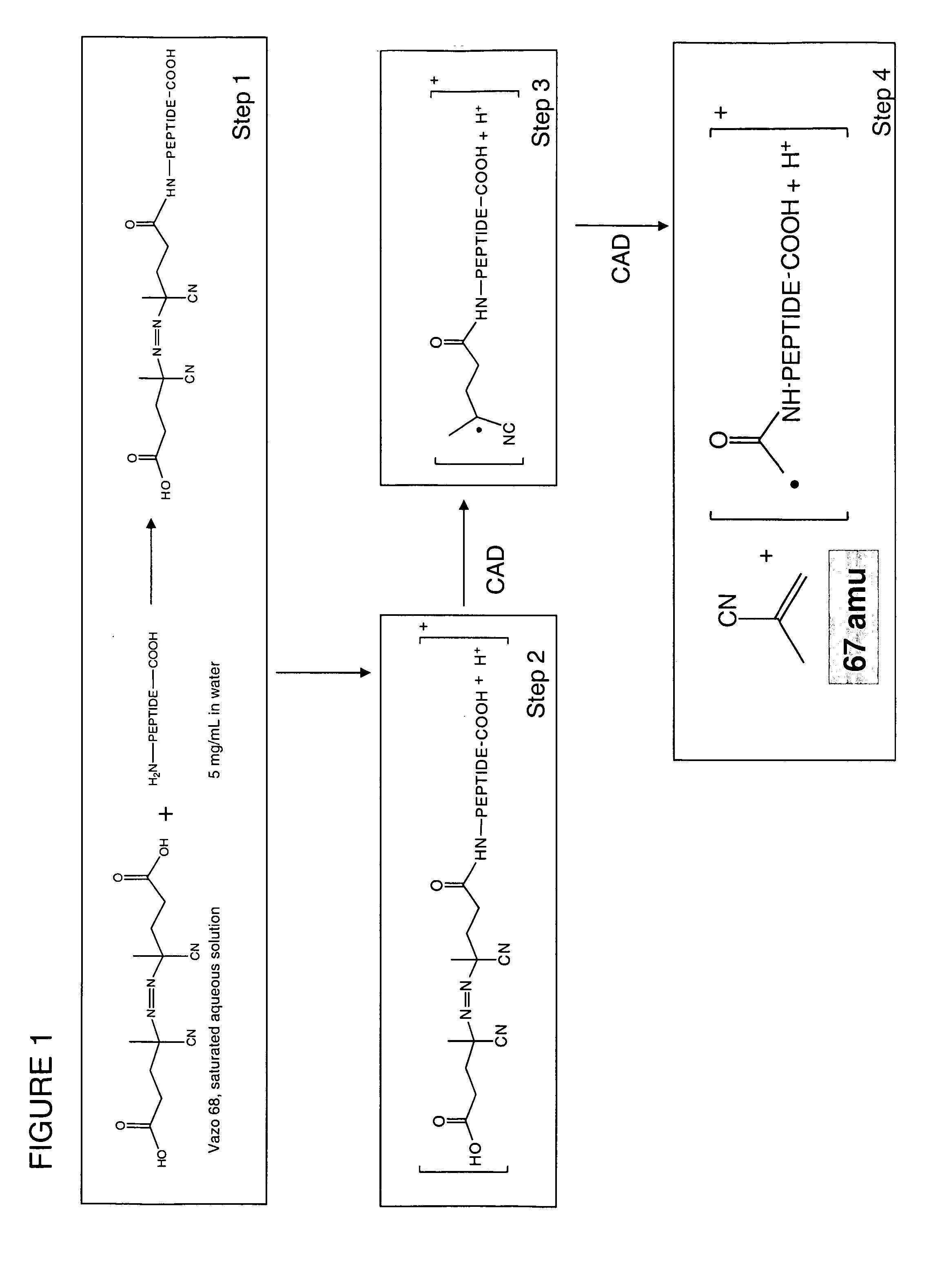
A method for the selective fragmentation of peptides using free radical initiator-peptide conjugates is provided. Free radical initiated peptide sequencing, or FRIPS, consists of covalently attaching a free radical initiator to a peptide and collisionally activating this conjugate. This results in fragment formation. Subsequent collision-activated dissociation further dissociates the fragments, producing a group of ions based on the interaction of the free radical initiator and the target molecule. The methodology is applied to the fundamental study of biologically relevant reactions of free radicals with peptides and proteins in the gas phase, as well as to a completely gas-phase approach to protein sequencing.
Owner:CALIFORNIA INST OF TECH
Protein sequencing method and reagents
InactiveUS20170212126A1Biological material analysisDepsipeptidesAcid derivativeProtein Sequence Determination
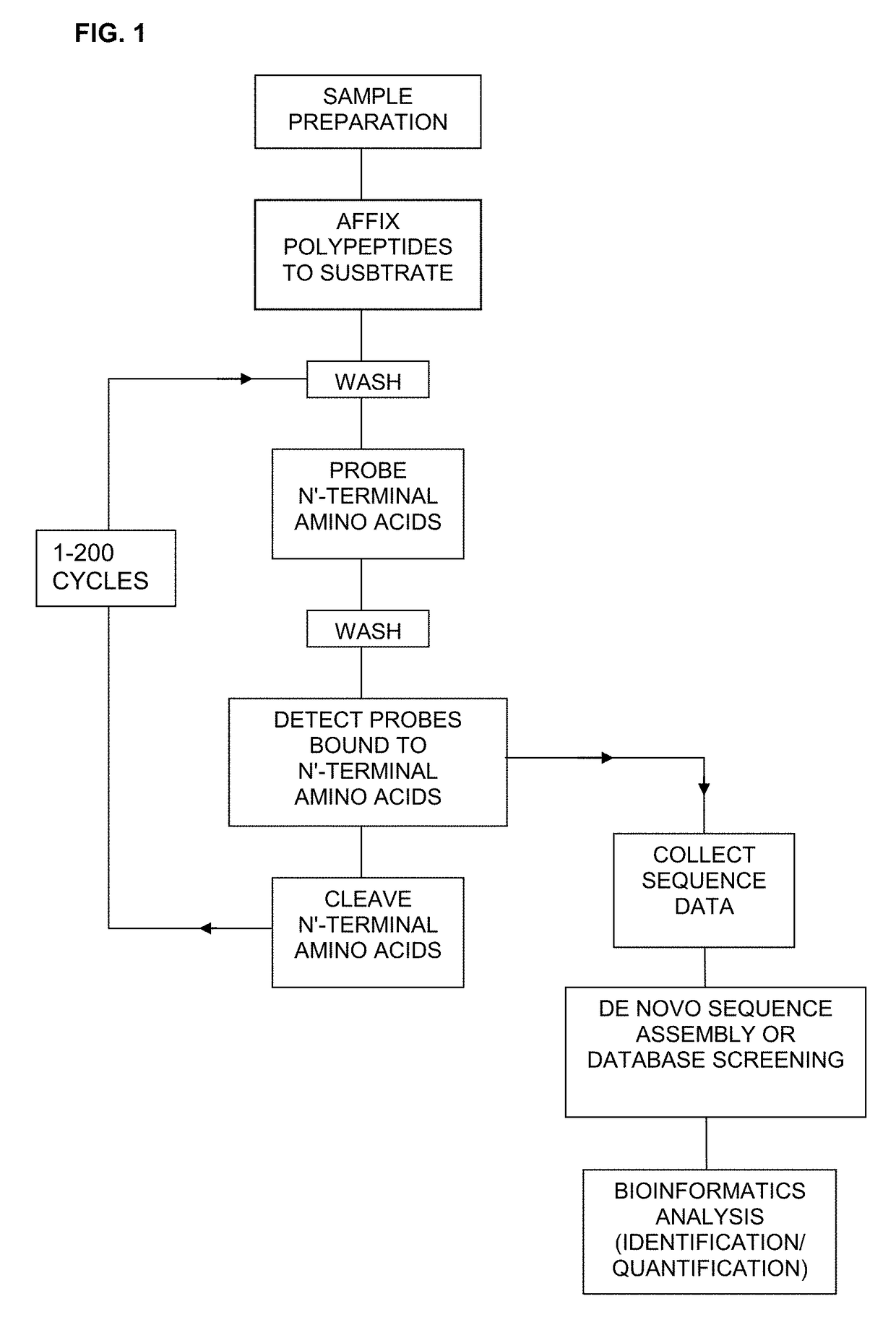
The invention describes methods and reagents useful for sequencing polypeptide molecules. The method comprises affixing a polypeptide to a substrate and contacting the polypeptide with a plurality of probes. Each probe selectively binds to an N-terminal amino acid or an N-terminal amino acid derivative. Probes bound to the polypeptide molecule are then identified before cleaving the N-terminal amino acid or N-terminal amino acid derivative of the polypeptide. Also provided are methods for the sequencing a plurality of polypeptide molecules in a sample and probes specific for N-terminal amino acids or N-terminal amino acid derivatives.
Owner:THE GOVERNING COUNCIL OF THE UNIV OF TORONTO
Method for Isolating and Purifying Immuno-Modulating Polypeptide from Cow Placenta
InactiveUS20080319163A1High purityPeptide/protein ingredientsMammal material medical ingredientsAnion-exchange chromatographyProtein Sequence Determination
The present invention provides a method for isolating and purifying immuno-modulating polypeptide from cow placenta, which is characterized by using the steps of anion-exchange chromatography, gel exclusion chromatography and reverse-phase high performance liquid chromatography to isolate and purify immuno-modulating polypeptide from cow placenta, identifying its activity of stimulating lymphocyte proliferation in vitro by MTT method, then determining its molecular weight by MALDI-TOF-MS, its isoelectric point by CIEF and its amino acid sequence with analyzer for protein sequencing. Since the obtained immuno-modulating polypeptide by the method according to the present invention has more than 90% purity, its bioactivity can reach medicinal standards.
Owner:SHI JIA ZHUANG SANLU GRP
Nanopore sensor, structure and device including the sensor, and methods of forming and using same
PendingUS20190145950A1Increase interaction distanceImprove accuracyMaterial analysis by electric/magnetic meansLaboratory glasswaresGenomic sequencingNanoparticle
The present disclosure provides an improved device that can be used to sense and characterize a variety of materials. The device may be used for a variety of applications, including genome sequencing, protein sequencing, biomolecular sequencing, and detection of ions, molecules, chemicals, biomolecules, metal atoms, polymers, nanoparticles and the like.
Owner:TAKULAPALLI BHARATH
Amplitude-modulated laser
PendingUS20210218218A1Fluorescence enhancementExtension of timeFluorescence/phosphorescenceSolid state laser constructional detailsFluorescenceEngineering
Systems and methods are described for producing an amplitude-modulated laser pulse train. The laser pulse train can be used to cause fluorescence in materials at which the pulse trains are directed. The parameters of the laser pulse train are selected to increase fluorescence relative to a constant-amplitude laser pulse train. The amplitude-modulated laser pulse trains produced using the teachings of this invention can be used to enable detection of specific molecules in applications such as gene or protein sequencing.
Owner:QUANTUM SI
Single molecule peptide sequencing
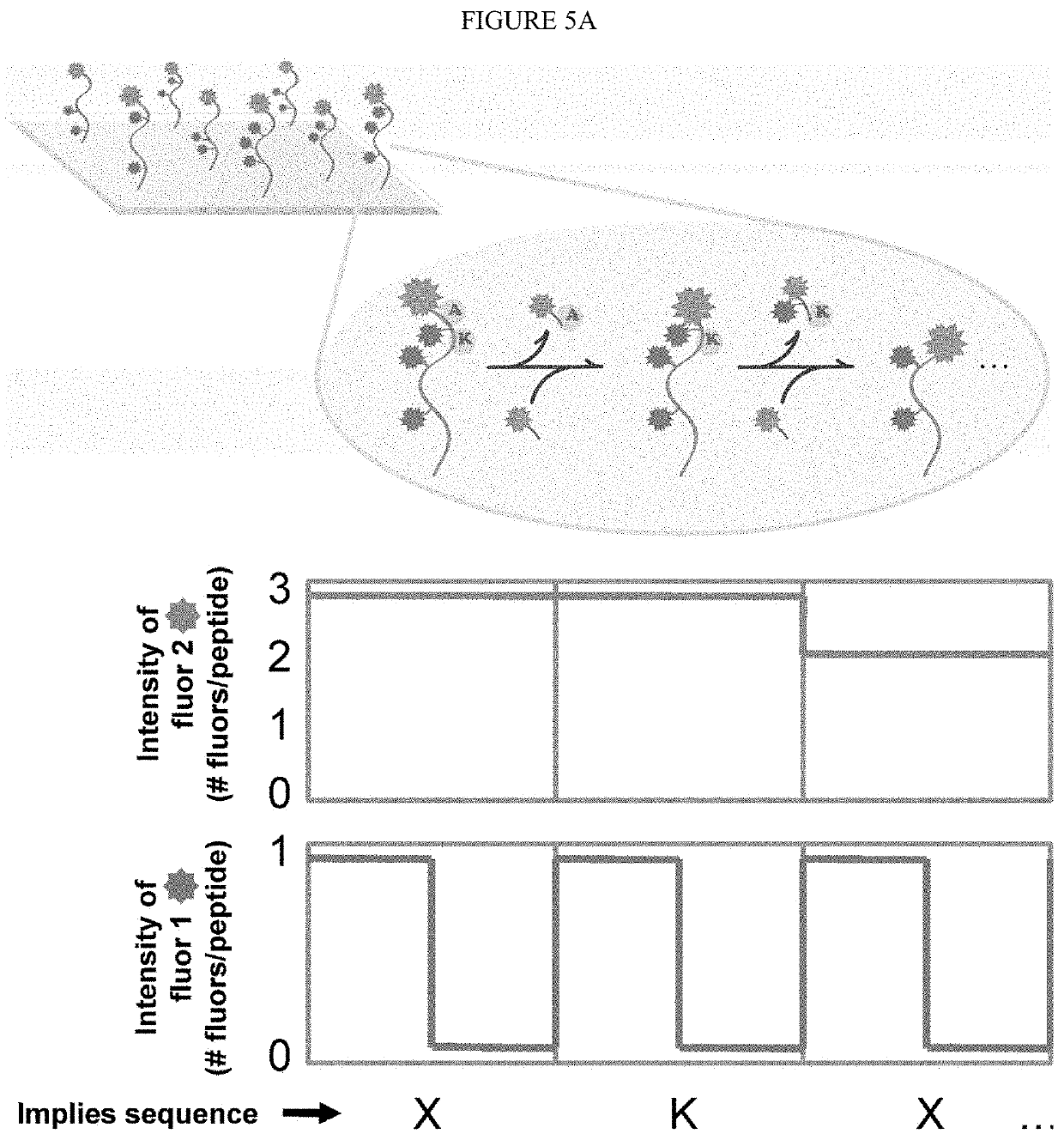
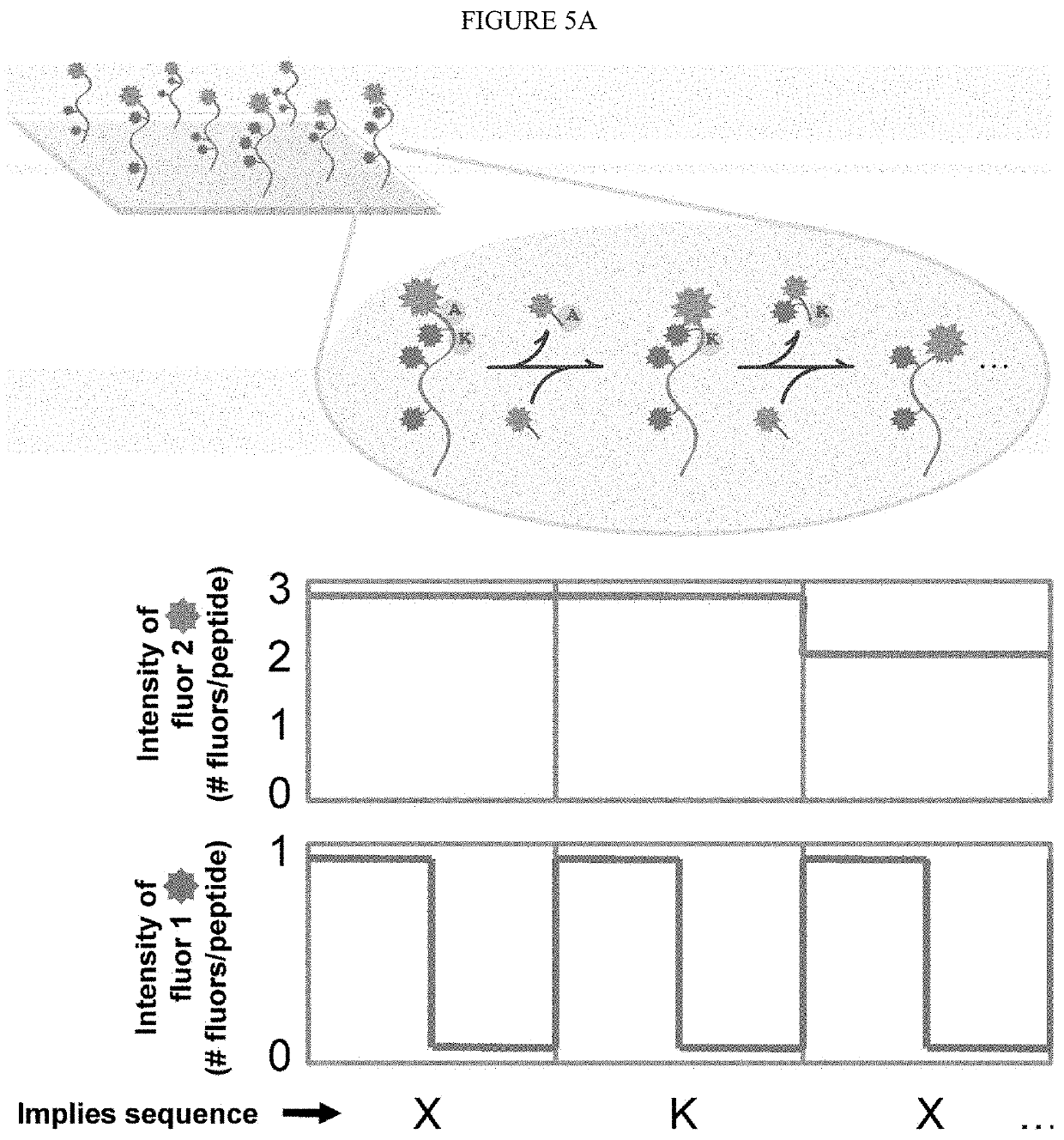
ActiveUS20200018768A1OmicsCarrier-bound/immobilised peptidesLarge-Scale SequencingProtein Sequence Determination
Identifying proteins and peptides, and more specifically large-scale sequencing of single peptides in a mixture of diverse peptides at the single molecule level is an unmet challenge in the field of protein sequencing. Herein are methods for identifying amino acids in peptides, including peptides comprising unnatural amino acids. In one embodiment, the N-terminal amino acid is labeled with a first label and an internal amino acid is labeled with a second label. In some embodiments, the labels are fluorescent labels. In other embodiments, the internal amino acid is Lysine. In other embodiments, amino acids in peptides are identified based on the fluorescent signature for each peptide at the single molecule level.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Single molecule peptide sequencing
ActiveUS20220091130A1OmicsCarrier-bound/immobilised peptidesLarge-Scale SequencingProtein Sequence Determination
Identifying proteins and peptides, and more specifically large-scale sequencing of single peptides in a mixture of diverse peptides at the single molecule level is an unmet challenge in the field of protein sequencing. Herein are methods for identifying amino acids in peptides, including peptides comprising unnatural amino acids. In one embodiment, the N-terminal amino acid is labeled with a first label and an internal amino acid is labeled with a second label. In some embodiments, the labels are fluorescent labels. In other embodiments, the internal amino acid is Lysine. In other embodiments, amino acids in peptides are identified based on the fluorescent signature for each peptide at the single molecule level.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Method for identifying transcription factor binding element in cotton fiber development period by DNA affinity protein sequencing
PendingCN112646867AFast and accurate identificationStrong practical valueMicrobiological testing/measurementProteomicsBiotechnologyProtein target
The invention relates to a method for identifying a transcription factor binding element in a cotton fiber development period by DNA affinity protein sequencing. The method comprises the following steps of: 1) extracting cotton fiber genome DNA, randomly breaking into fragments of about 200bp, and constructing an Index-DNA library with a sequencing joint by using the broken DNA; 2) performing prokaryotic expression on a cotton transcription factor protein with a label, and performing immunoblotting to confirm the expressed target protein; 3) incubating the target protein and the Index-DNA library in vitro to fully combine the target protein and the Index-DNA library, and extracting Index-DNA specifically bound with the protein; and 4) performing amplification and sequencing, matching the obtained data to a cotton genome, and identifying and confirming the sequence of the obtained binding element after the obtained data corresponds to a DHS site in the cotton fiber development period. The method provided by the invention can quickly and accurately identify the downstream binding site of the cotton transcription factor without genetic transformation, and has the advantages of being fast, accurate, economical, labor-saving and the like.
Owner:ANYANG INST OF TECH +1
Nano single hole based on capillary tip, preparation method and application thereof
ActiveCN113155808ARealize dynamic Raman detectionStable structureRaman scatteringNanosensorsProtein moleculesCapillary Tubing
The invention relates to a nano single hole based on a capillary tip, a preparation method and application thereof. The nano single hole is characterized in that the inner side wall of a capillary tip is provided with a deposition layer, the deposition layer extends out of a capillary to form a hollow conical structure, and a nano single hole is located at the tip of the conical structure. According to the invention, the nano single hole is simple in preparation method, low in cost, small in size and controllable in structure, and can be used as an electrochemical device to be combined with Raman spectroscopy; and bias voltage is applied to the two ends of a nanotube, so that the detection of a single molecule with a small size by dynamic Raman is realized, the detection of a single protein molecule based on current and Raman spectrum is realized, and a new method is provided for DNA sequencing and protein sequencing.
Owner:NANJING UNIV
Amino acid configuration analysis method and N-polypeptide terminal sequence sequencing method of polymyxin B
ActiveCN111505142ARapid purificationRapid purification and separationComponent separationFluid phaseMass Spectrometry-Mass Spectrometry
The invention discloses an amino acid configuration analysis method and an N-polypeptide end sequence sequencing method of polymyxin B. A liquid phase purification method of a polymyxin B component isestablished, and polymyxin B1, polymyxin B2 and polymyxin B1-I can be rapidly purified and separated from the polymyxin B component. A preparation method and a liquid phase purification method of thepolymyxin B enzymolysis component are established, and the enzymolysis components of polymyxin B1, polymyxin B2 and polymyxin B1-I can be rapidly purified and separated; polymyxin B component hydrolysis-amino acid chiral derivation-liquid chromatography-mass spectrometry are adopted to determine the amino acid configuration; the N-polypeptide terminal sequence is determined by combining enzymolysis of the polymyxin B component, liquid chromatography purification and protein sequencing, so that a research basis is provided for strictly controlling the quality of polymyxin B.
Owner:SHANGHAI INST FOR FOOD & DRUG CONTROL
Single molecule peptide sequencing methods
ActiveUS20200400677A1Microbiological testing/measurementChemiluminescene/bioluminescenceProtein Sequence DeterminationPeptide sequencing
The invention, in part, includes methods of single molecule protein sequencing that include using weak binding spectra in the amino acid identification.
Owner:MASSACHUSETTS INST OF TECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com