Characteristic extraction and classification method and device for DNA (Deoxyribo-Nucleic Acid) binding protein sequence information
A technology that combines protein and sequence information, applied in the field of computer biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0038] In order to make the objectives, technical solutions, and advantages of the present invention clearer, the present invention will be further described in detail below with reference to the accompanying drawings and embodiments, but the embodiments of the present invention are not limited thereto.
[0039] Examples of the method for extracting and classifying DNA-binding protein sequence information of the present invention:
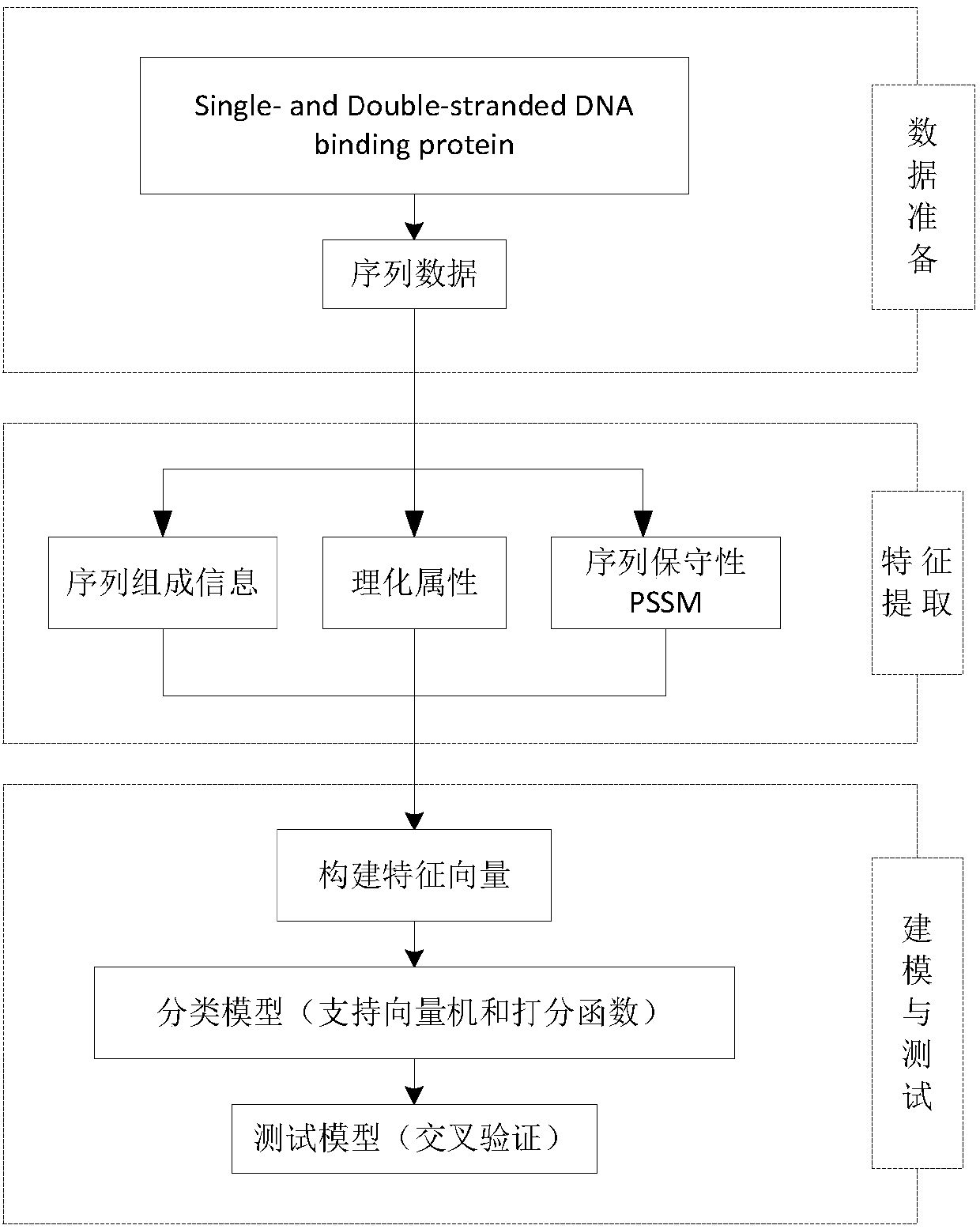
[0040] The overall scheme of the present invention is as figure 1 Shown.
[0041] The commonly used structure databases of DNA binding proteins include: Protein Data Bank (PDB) NDB, ProNuc, and structure classification databases SCOP and CATH; commonly used sequence databases include: SWISS-PROT, PIR, OWL, NRL3D, TrEMBL, etc. Among them, the protein database was established in 1971 by Walter Hamilton of Brookhaven National Laboratory. It is now managed by the Structural Biology Cooperative Research Association (RCSB) and has become an international organi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com