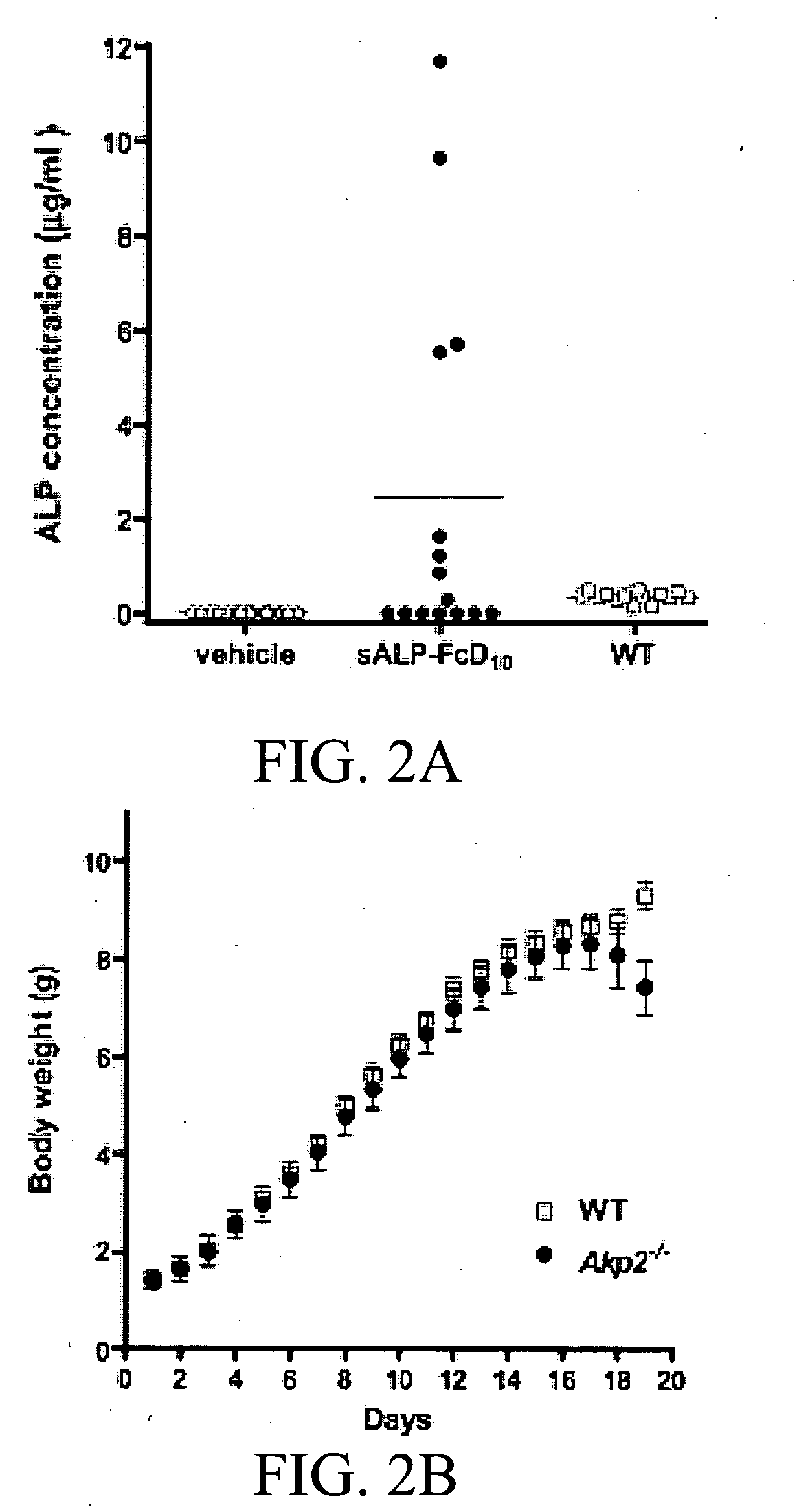
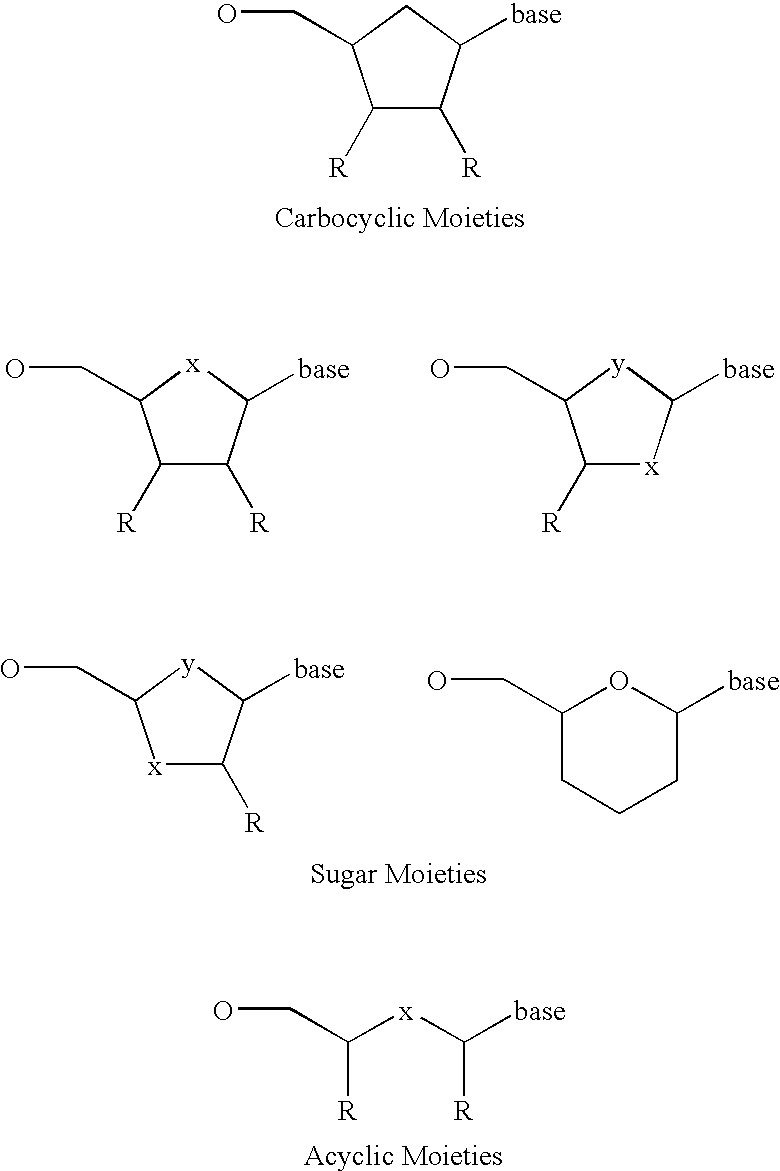
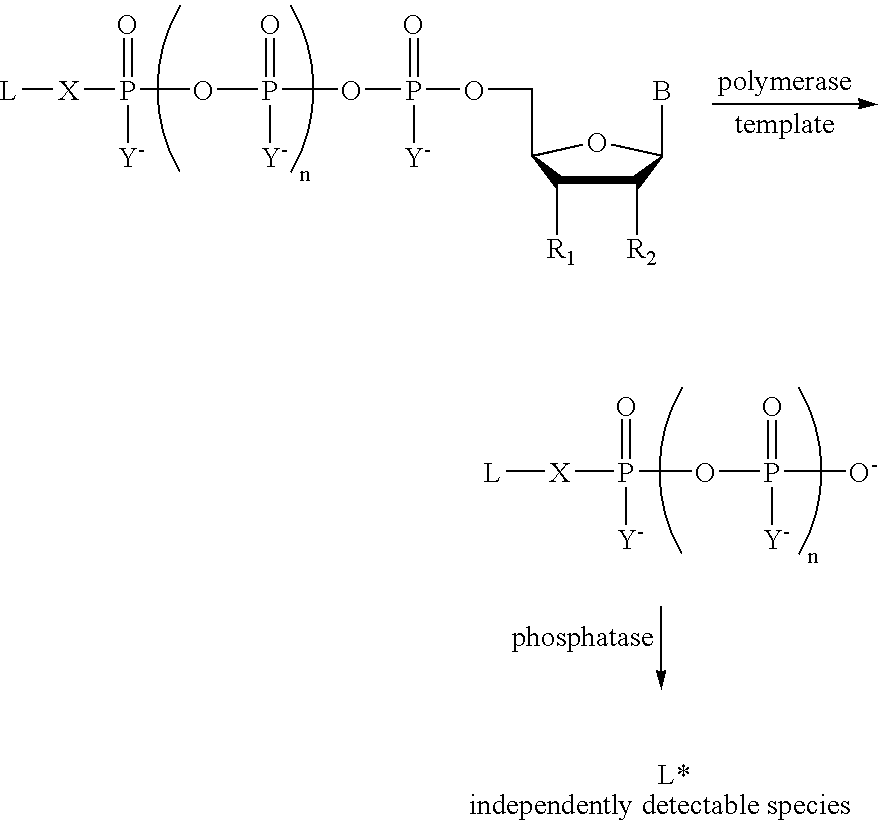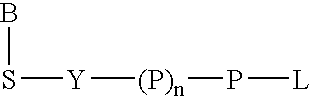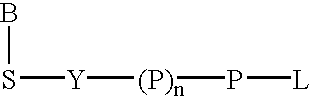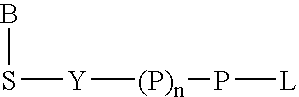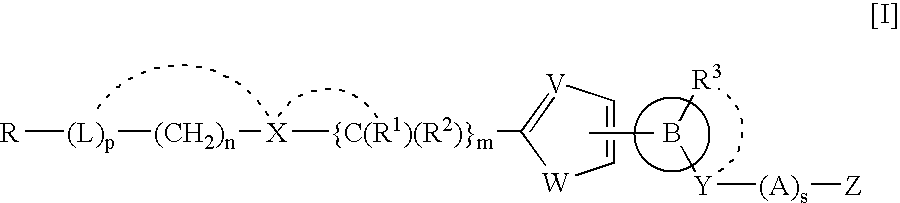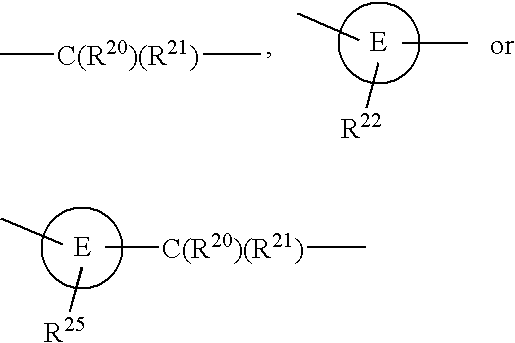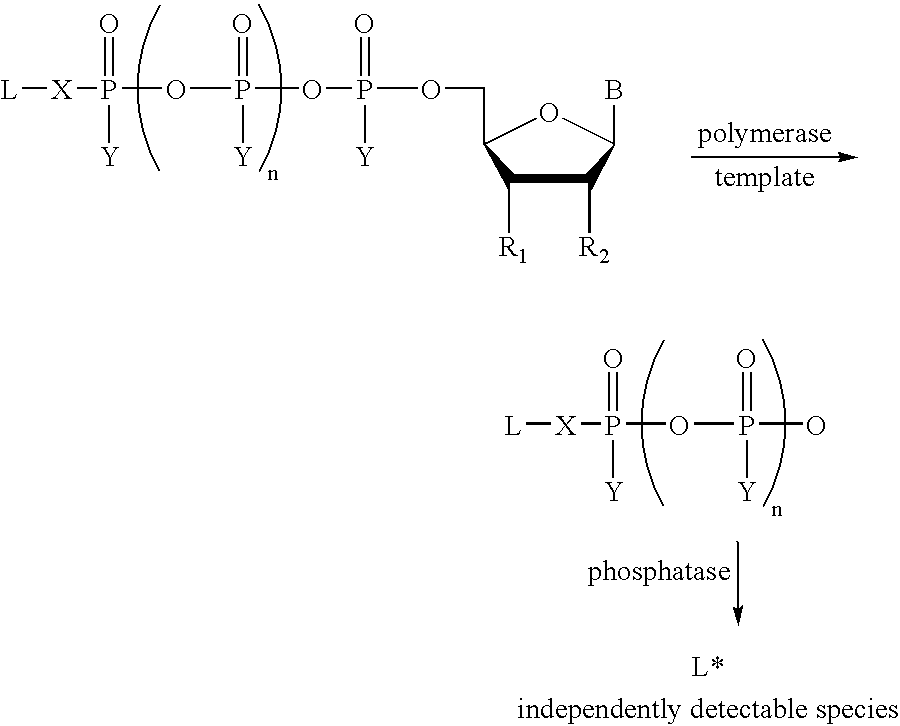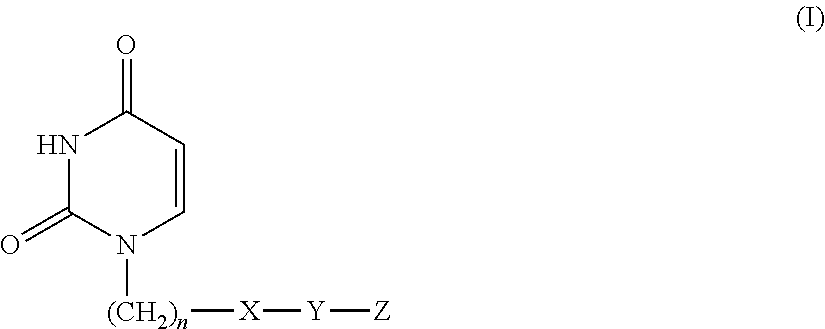Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
1517 results about "Phosphatase" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A phosphatase is an enzyme that uses water to cleave a phosphoric acid monoester into a phosphate ion and an alcohol. Because a phosphatase enzyme catalyzes the hydrolysis of its substrate, it is a subcategory of hydrolases. Phosphatase enzymes are essential to many biological functions, because phosphorylation (e.g. by protein kinases) and dephosphorylation (by phosphatases) serve diverse roles in cellular regulation and signaling. Whereas phosphatases remove phosphate groups from molecules, kinases catalyze the transfer of phosphate groups to molecules from ATP. Together, kinases and phosphatases direct a form of post-translational modification that is essential to the cell's regulatory network. Phosphatase enzymes are not to be confused with phosphorylase enzymes, which catalyze the transfer of a phosphate group from hydrogen phosphate to an acceptor. Due to their prevalence in cellular regulation, phosphatases are an area of interest for pharmaceutical research.
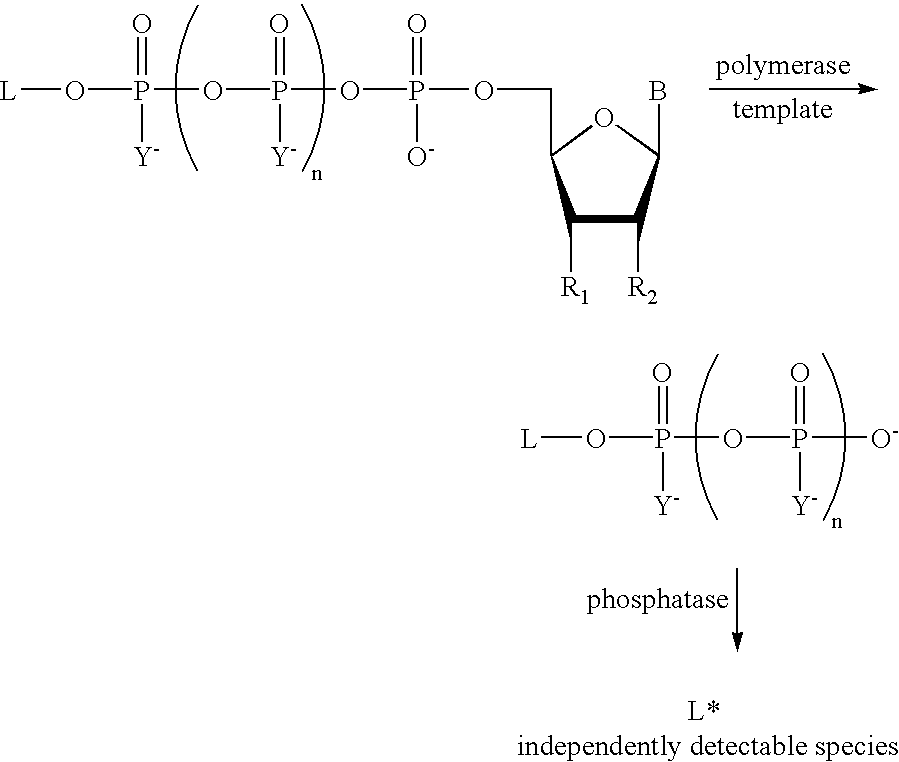
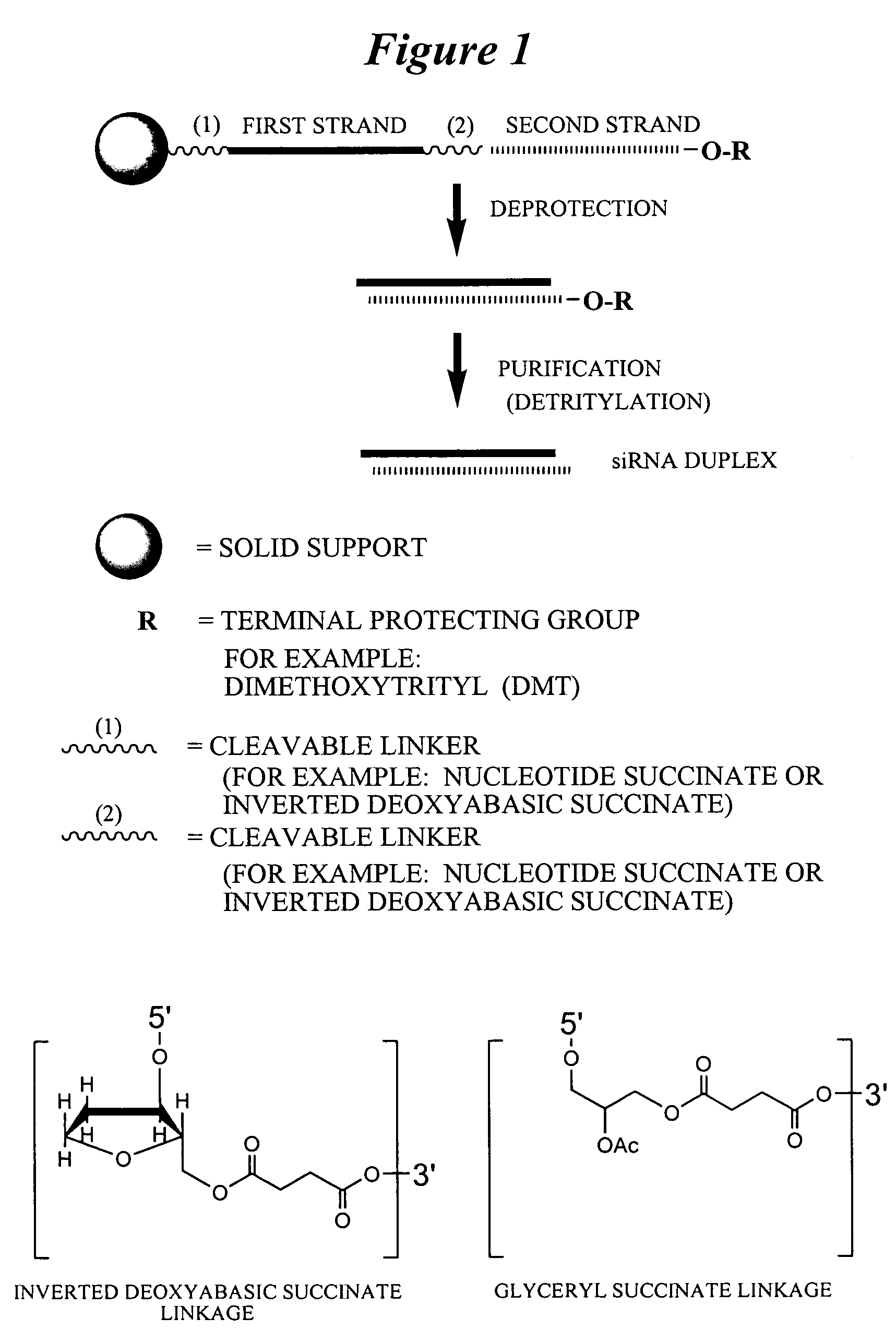
Labeled nucleoside polyphosphates
InactiveUS7041812B2Sugar derivativesMaterial analysis by observing effect on chemical indicatorNucleic acid detectionFluorescence
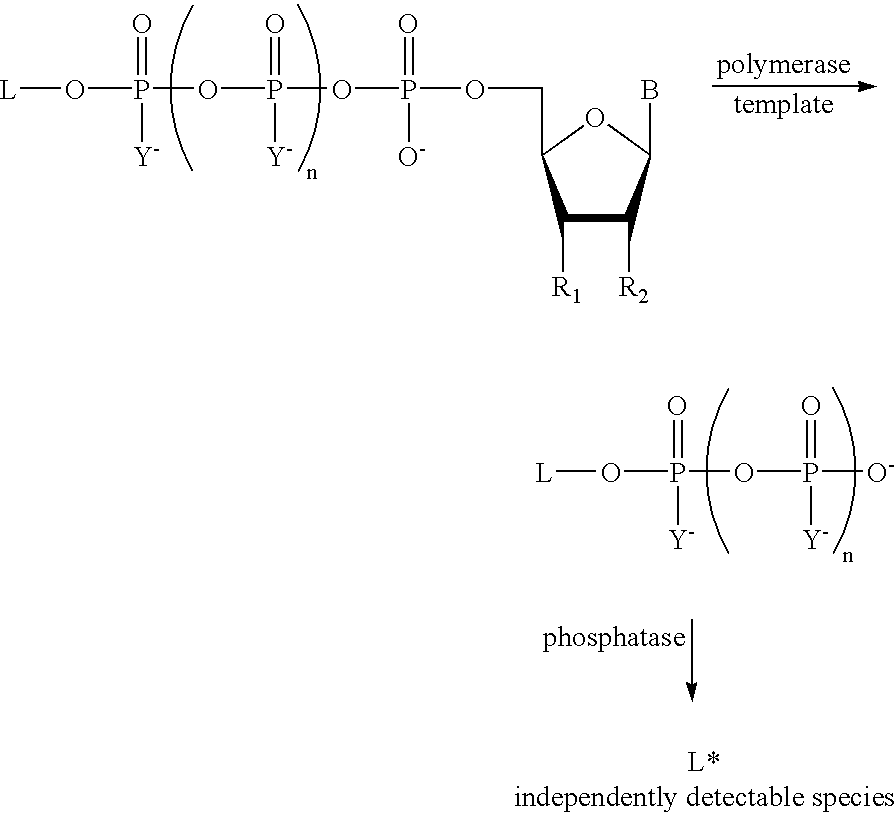
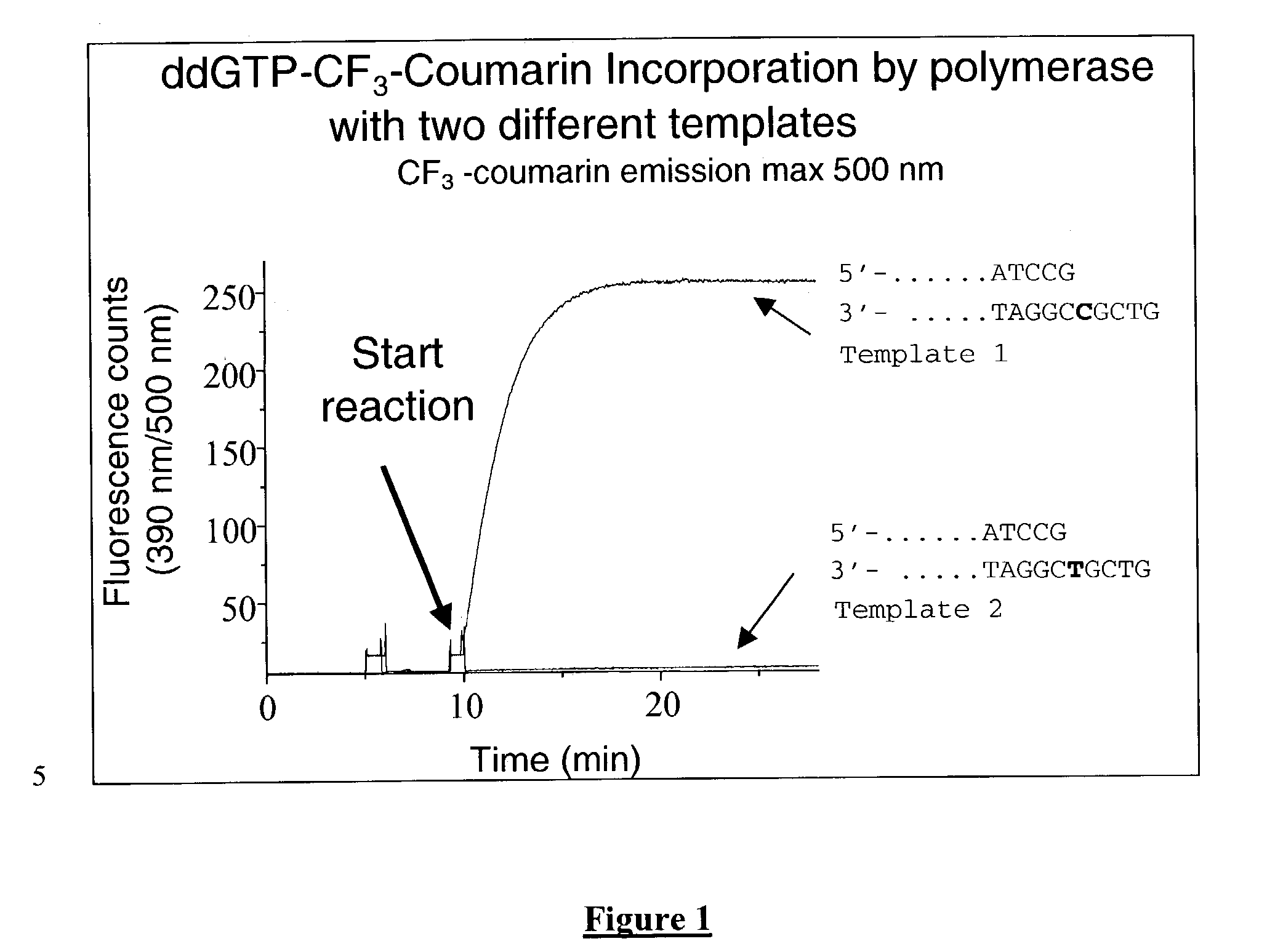
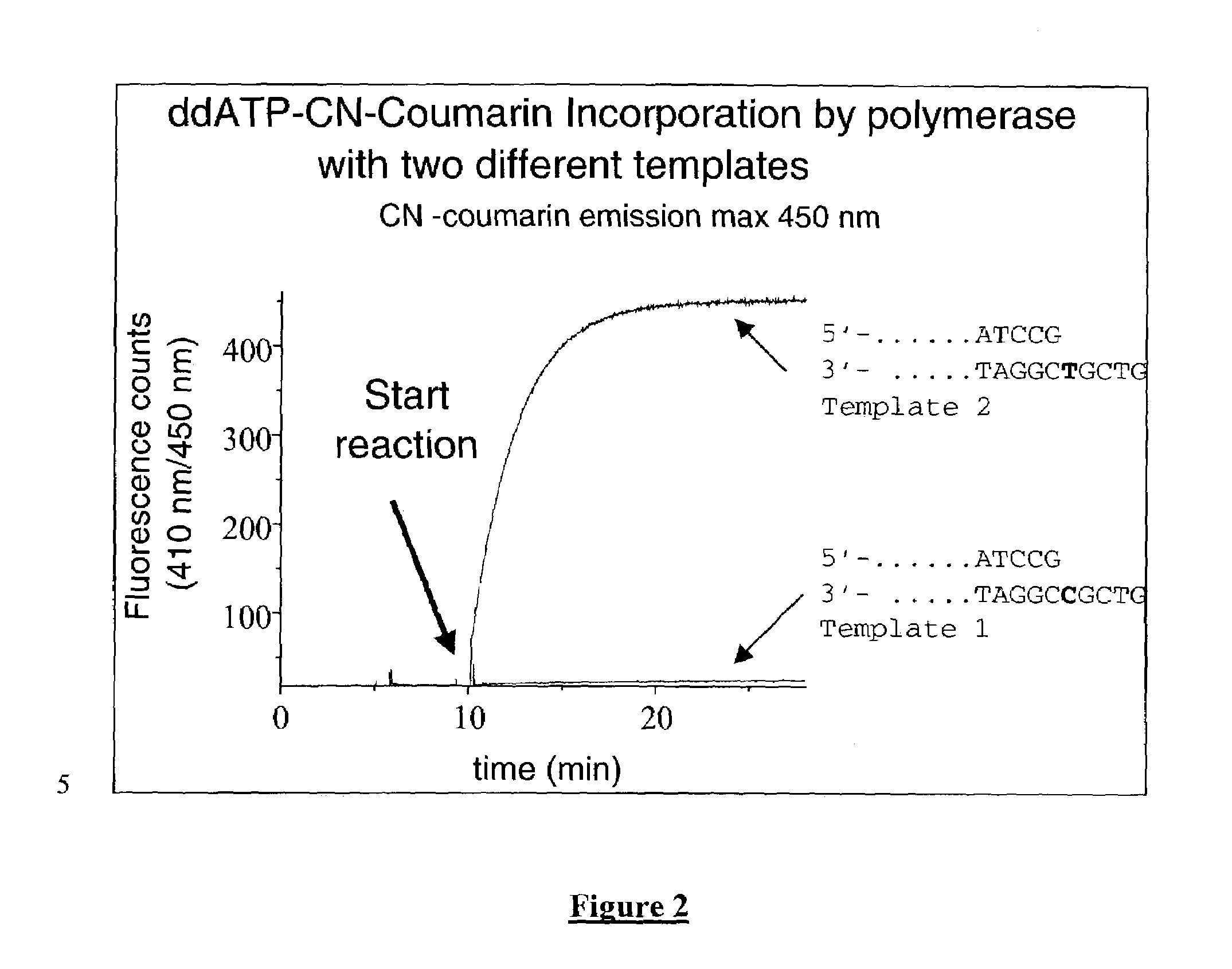
The present invention describes new compositions of matter in the form of labeled nucleoside polyphosphates with four or more phosphates. In addition compositions of nucleoside polyphosphates with four or more phosphates that are substrates for nucleic acid polymerases with enhanced substrate properties and methods of using these nucleoside polyphosphates for nucleic acid detection, characterization and quantification are described. The compositions provided by this invention include nucleoside polyphosphate, dideoxynucleoside polyphosphate, or deoxynucleoside polyphosphate analogues which have colorimetric, chemiluminescent, or fluorescent moieties, mass tags or an electrochemical tags attached to the terminal-phosphate. When a nucleic acid polymerase uses this analogue as a substrate, an enzyme-activatable label would be present on the inorganic polyphosphate by-product of phosphoryl transfer. Removal of the polyphosphate product of phosphoryl transfer via phosphate or polyphosphate transferring enzyme leads to a detectable change in the label attached thereon. When the polymerase assay is performed in the presence of a phosphatase, there is provided a convenient method for real-time monitoring of DNA or RNA synthesis and detection of a target nucleic acid.
Owner:GLOBAL LIFE SCI SOLUTIONS USA LLC
Single nucleotide amplification and detection by polymerase
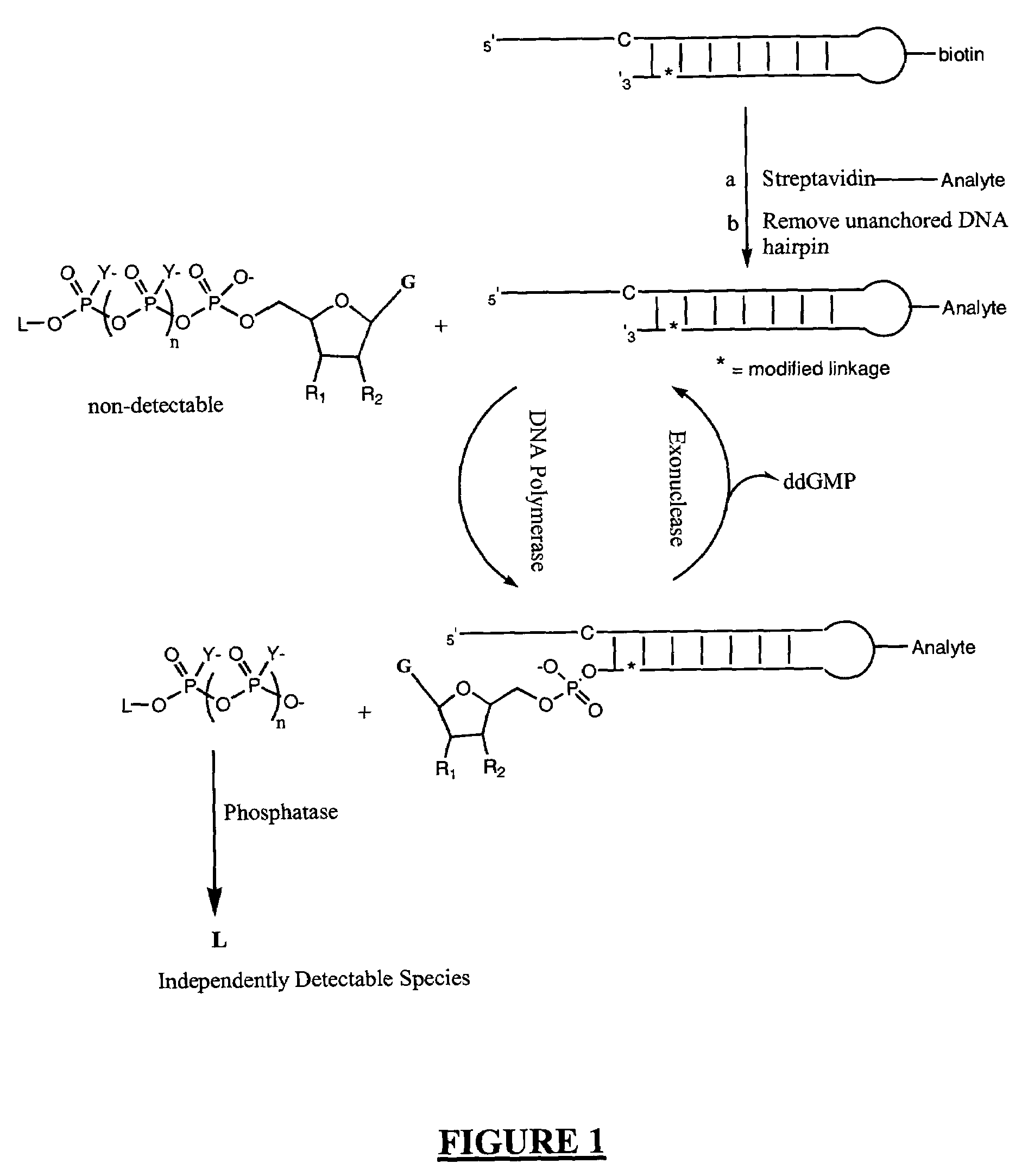
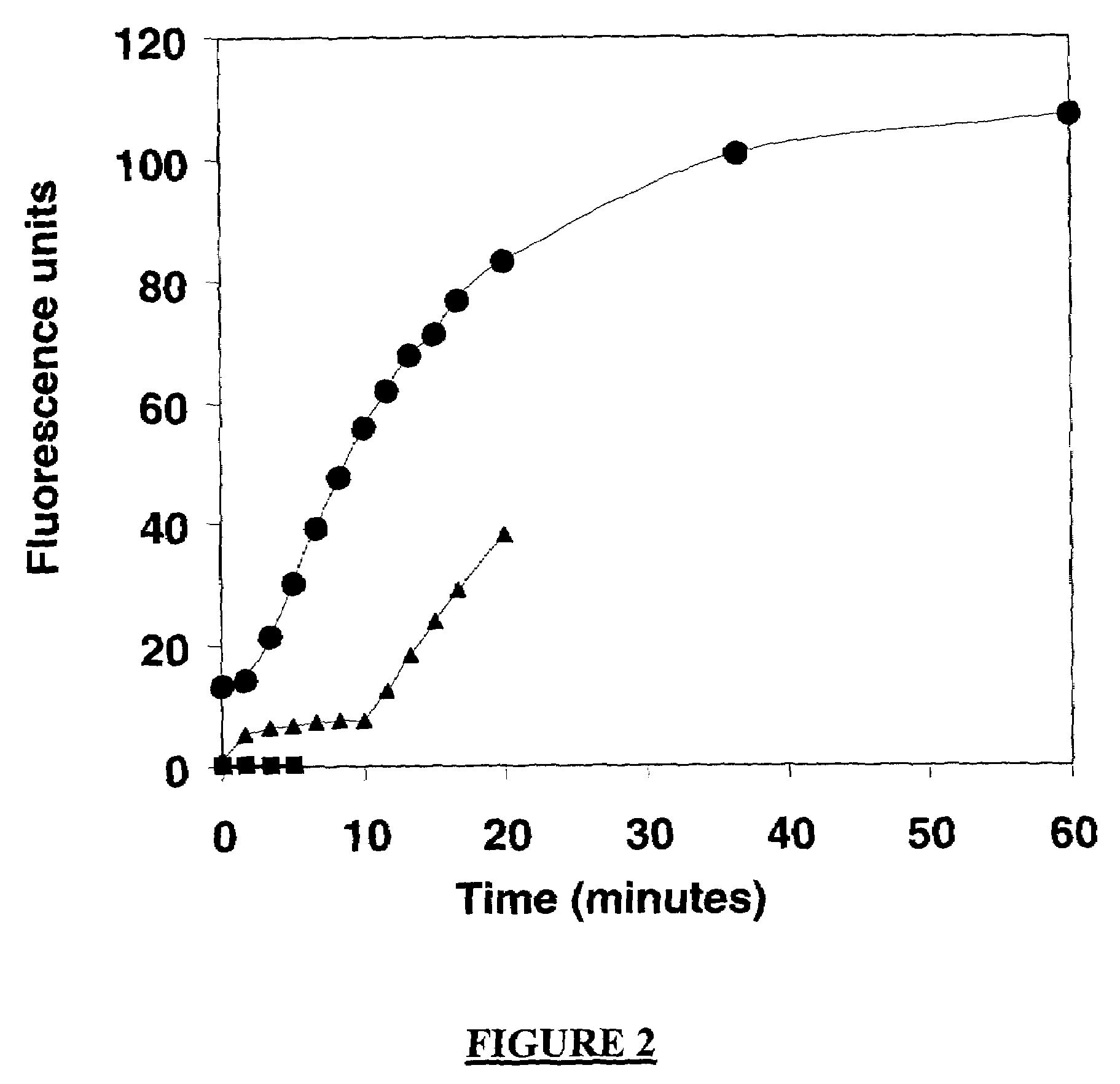
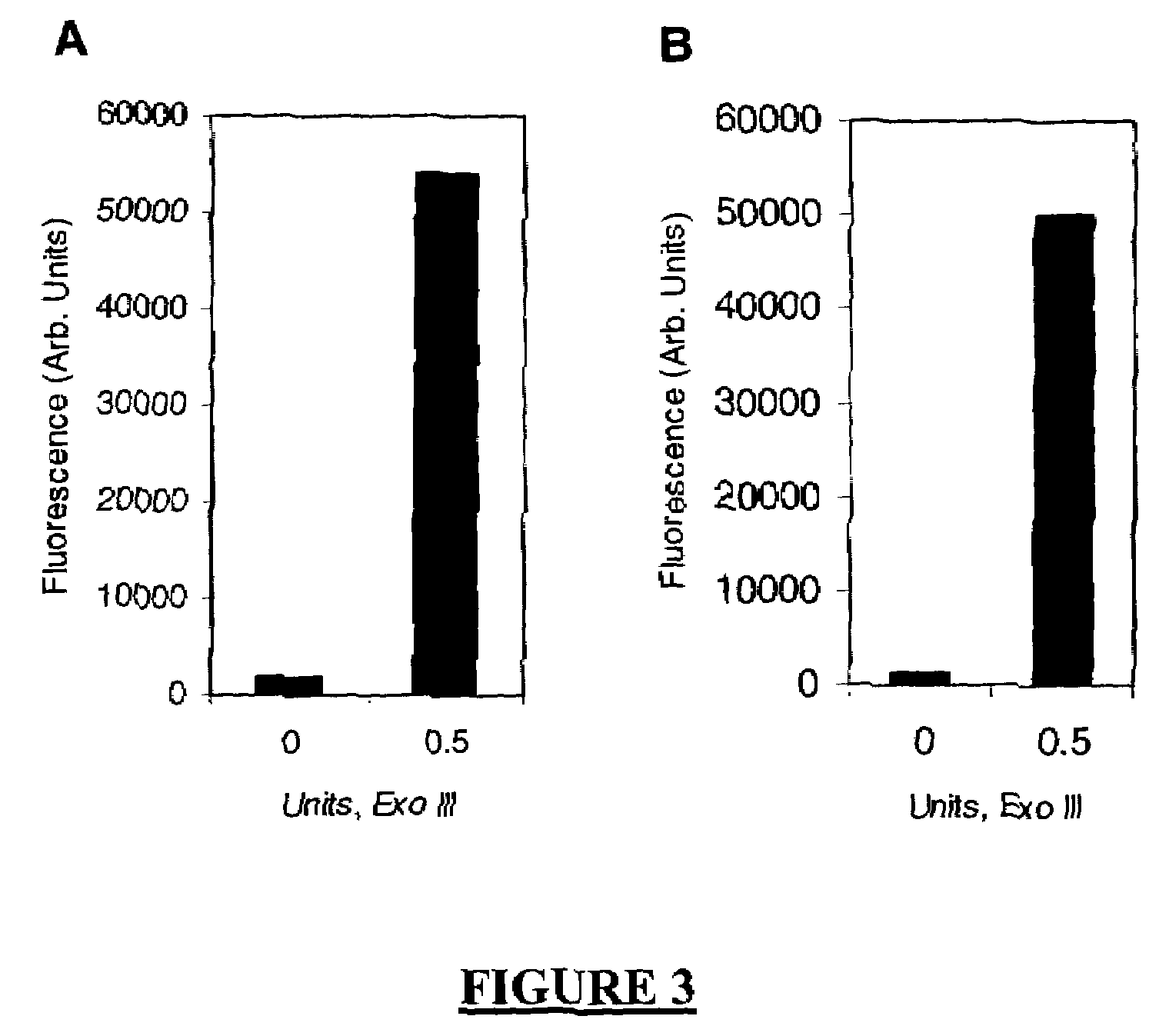
A method of characterizing a nucleic acid sample is provided that includes the steps of: (a) conducting a DNA polymerase reaction that includes the reaction of a template, a non-hydrolyzable primer, at least one terminal phosphate-labeled nucleotide, DNA polymerase, and an enzyme having 3′→5′ exonuclease activity which reaction results in the production of labeled polyphosphate; (b) permitting the labeled polyphosphate to react with a phosphatase to produce a detectable species characteristic of the sample; (c) detecting the detectable species; and (d) characterizing the nucleic acid sample based on the detection.
Owner:GLOBAL LIFE SCI SOLUTIONS USA LLC
Tissue-nonspecific alkaline phosphatase (TNAP) activators and uses thereof
Disclosed herein are tissue-nonspecific alkaline phosphatase (TNAP) activators and uses thereof for promoting bone mineral deposition.
Owner:SANFORD BURNHAM MEDICAL RES INST
Analyte detection
A method of characterizing an analyte sample is provided that includes the steps of: (a) anchoring the analyte to a nucleic acid template of known sequence; (b) conducting a DNA polymerase reaction that includes the reaction of a template, a non-hydrolyzable primer, at least one terminal phosphate-labeled nucleotide, DNA polymerase, and an enzyme having 3'->5' exonuclease activity which reaction results in the production of labeled polyphosphate; (c) permitting the labeled polyphosphate to react with a phosphatase to produce a detectable species characteristic of the sample; (d) detecting the detectable species. The method may include the step of characterizing the nucleic acid sample based on the detection. Also provided are methods of analyzing multiple analytes in a sample, and kits for characterizing analyte samples.
Owner:GLOBAL LIFE SCI SOLUTIONS USA LLC
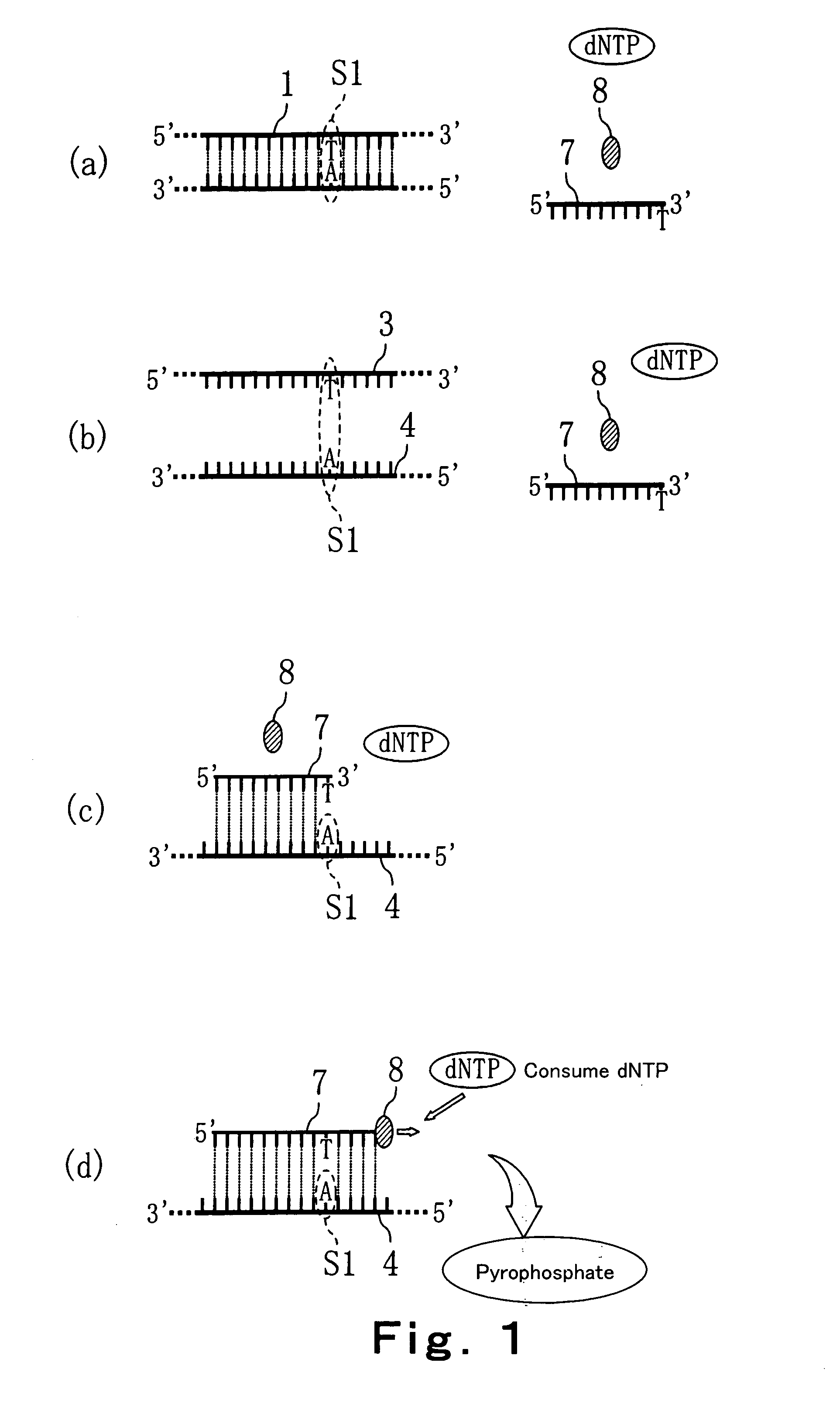
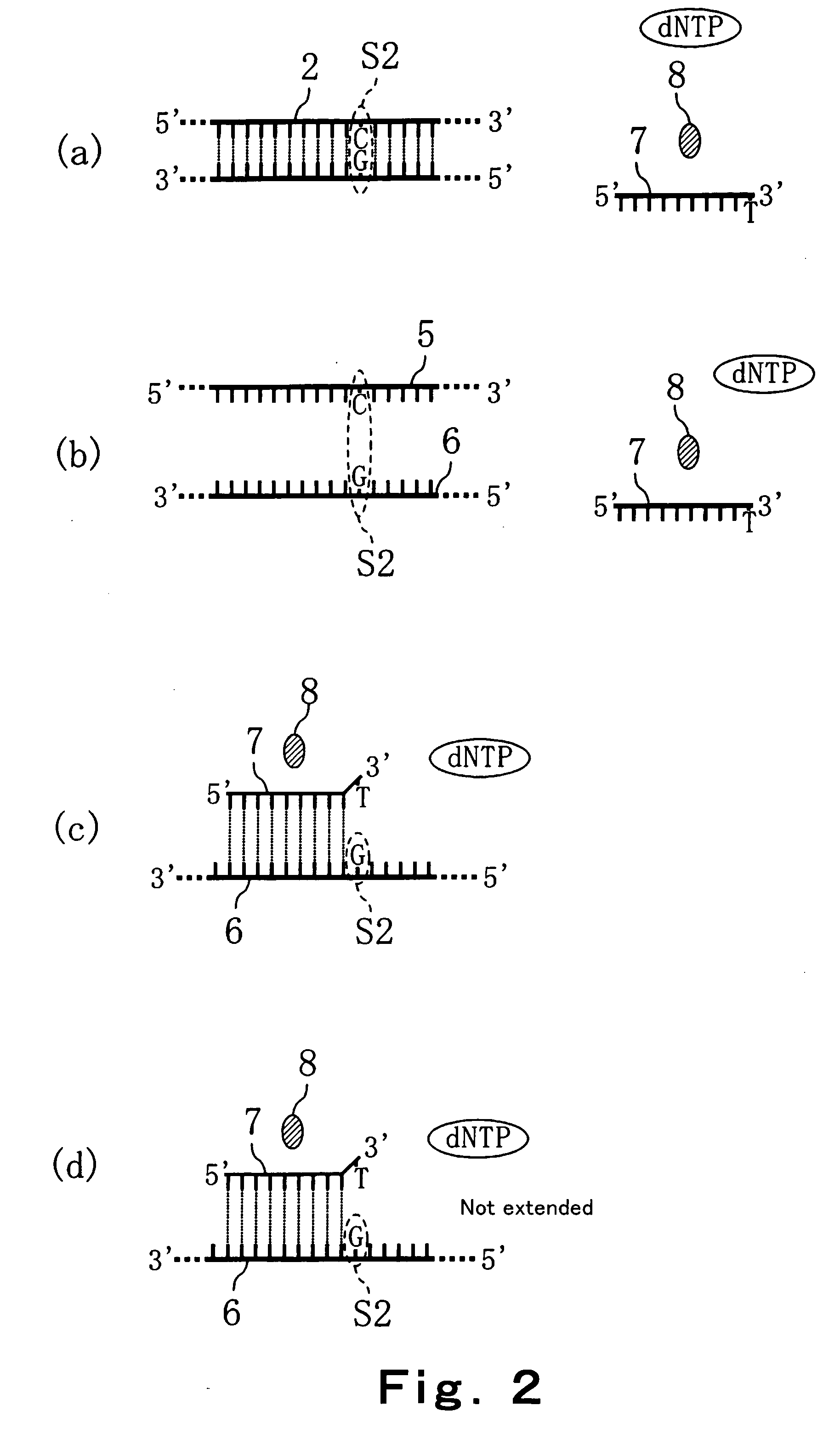
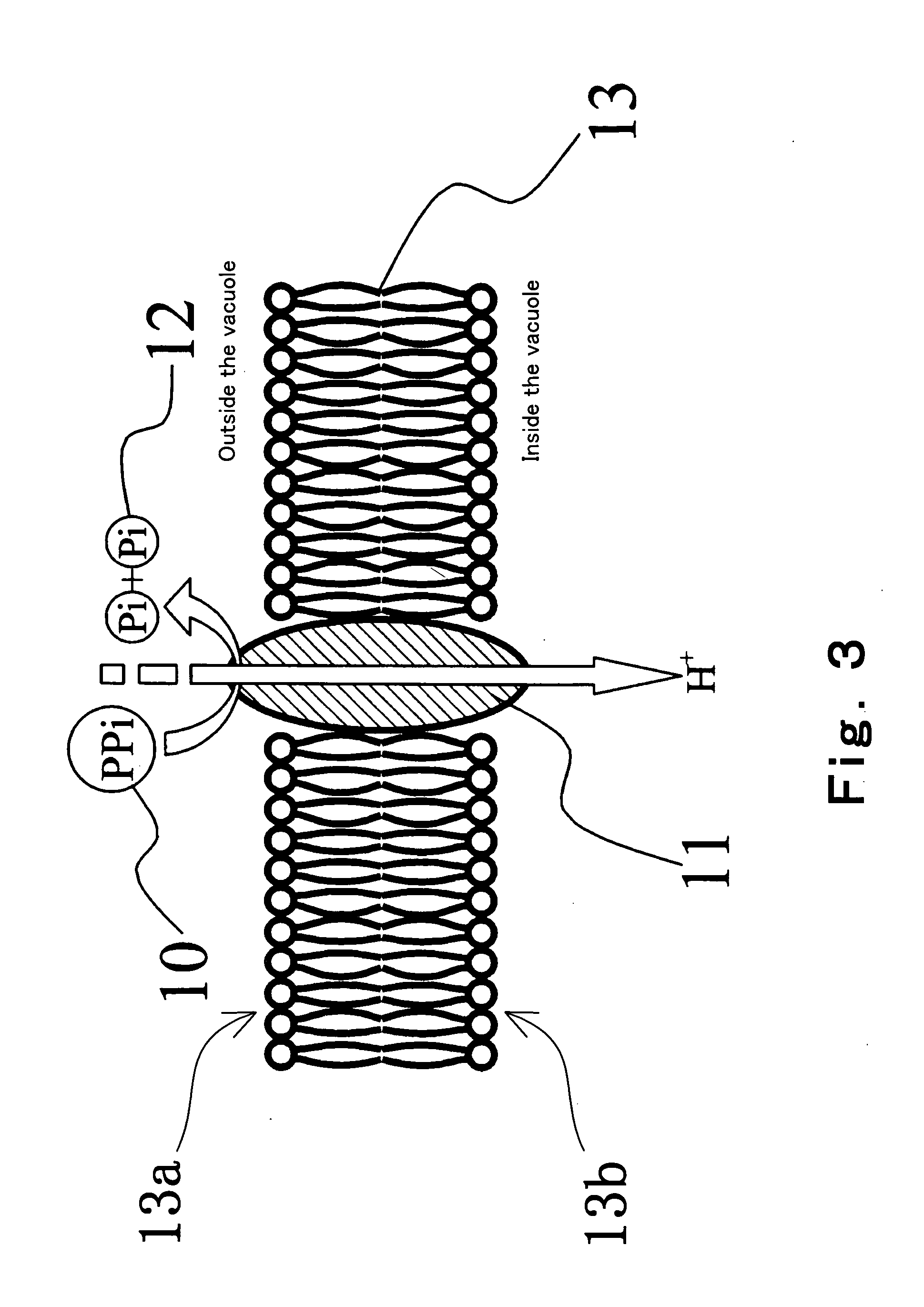
Method of detecting primer extension reaction, method of discriminating base type, device for discriminating base type, device for detecting pyrophosphate, method of detecting nucleic acid and tip for introducing sample solution
InactiveUS20050032075A1Decrease of H+ concentrationImprove concentrationMicrobiological testing/measurementMaterial analysisBase JNucleotide
Convenient techniques for discriminating the base type in a base sequence of a nucleic acid are provided. The technique includes the step (a) of preparing a sample solution containing a nucleic acid, a primer having a base sequence that includes a complementary binding region which complementarily binds to the nucleic acid, and a nucleotide; the step (b) of allowing the sample solution to stand under a condition to cause an extension reaction of the primer, and producing pyrophosphate when the extension reaction is caused; the step (c) of bringing the sample solution into contact with the front face of a H+ hardly permeable membrane having H+-pyrophosphatase, which penetrates from front to back of the membrane, of which active site that hydrolyzes pyrophosphate being exposed to the front face; the step (d) of measuring the H+ concentration of at least either one of the solution at the front face side of the H+ hardly permeable membrane or the solution at the back face side of the H+ hardly permeable membrane, in a state where the H+-pyrophosphatase is immersed in the solution; the step (e) of detecting the extension reaction on the basis of the result of measurement in the step (d) ; and the step (f) of discriminating the base type in the base sequence of the nucleic acid on the basis of the result of detection in the step (e).
Owner:PANASONIC CORP
Labeled nucleoside polyphosphates
InactiveUS20030124576A1Sugar derivativesMaterial analysis by observing effect on chemical indicatorNucleic acid detectionFluorescence
The present invention describes new compositions of matter in the form of labeled nucleoside polyphosphates with four or more phosphates. In addition compositions of nucleoside polyphosphates with four or more phosphates that are substrates for nucleic acid polymerases with enhanced substrate properties and methods of using these nucleoside polyphosphates for nucleic acid detection, charcterization and quantification are described. The compositions provided by this invention include nucleoside polyphosphate, dideoxynucleoside polyphosphate, or deoxynucleoside polyphosphate analogues which have calorimetric, chemiluminescent, or fluorescent moieties, mass tags or an electrochemical tags attached to the terminal-phosphate. When a nucleic acid polymerase uses this analogue as a substrate, an enzyme-activatable label would be present on the inorganic polyphosphate by-product of phosphoryl transfer. Cleavage of the polyphosphate product of phosphoryl transfer via phosphatase leads to a detectable change in the label attached thereon. When the polymerase assay is performed in the presence of a phosphatase, there is provided a convenient method for real-time monitoring of DNA or RNA synthesis and detection of a target nucleic acid.
Owner:GLOBAL LIFE SCI SOLUTIONS USA LLC
Terminal-phosphate-labeled nucleotides and methods of use
The present invention describes methods of detecting a nucleic acid in a sample, based on the use of terminal-phosphate-labeled nucleotides as substrates for nucleic acid polymerases. The methods provided by this invention utilize a nucleoside polyphosphate, dideoxynucleoside polyphosphate, or deoxynucleoside polyphosphate analogue which has a colorimetric dye, chemiluminescent, or fluorescent moiety, a mass tag or an electrochemical tag attached to the terminal-phosphate. When a nucleic acid polymerase uses this analogue as a substrate, an enzyme-activatable label would be present on the inorganic polyphosphate by-product of phosphoryl transfer. Cleavage of the polyphosphate product of phosphoryl transfer via phosphatase leads to a detectable change in the label attached thereon. When the polymerase assay is performed in the presence of a phosphatase, there is provided a convenient method for real-time monitoring of DNA or RNA synthesis and detection of a target nucleic acid.
Owner:GLOBAL LIFE SCI SOLUTIONS USA LLC
Terminal-phosphate-labeled nucleotides and methods of use
The present invention relates to improved methods of detecting a target using a labeled substrate or substrate analog. The methods comprise reacting the substrate or substrate analog in an enzyme-catalyzed reaction which produces a labeled moiety with independently detectable signal only when such substrate or substrate analog reacts. The present invention, in particular, describes methods of detecting a nucleic acid in a sample, based on the use of terminal-phosphate-labeled nucleotides as substrates for nucleic acid polymerases. The methods provided by this invention utilize a nucleoside polyphosphate, dideoxynucleoside polyphosphate, or deoxynucleoside polyphosphate analogue which has a colorimetric dye, chemiluminescent, or fluorescent moiety, a mass tag or an electrochemical tag attached to the terminal-phosphate. When a nucleic acid polymerase uses this analogue as a substrate, an enzyme-activatable label would be present on the inorganic polyphosphate by-product of phosphoryl transfer. Cleavage of the polyphosphate product of phosphoryl transfer via phosphatase leads to a detectable change in the label attached thereon. When the polymerase assay is performed in the presence of a phosphatase, there is provided a convenient method for real-time monitoring of DNA or RNA synthesis and detection of a target nucleic acid.
Owner:GLOBAL LIFE SCI SOLUTIONS USA LLC
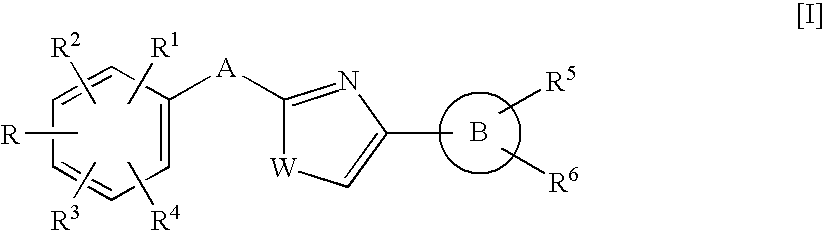
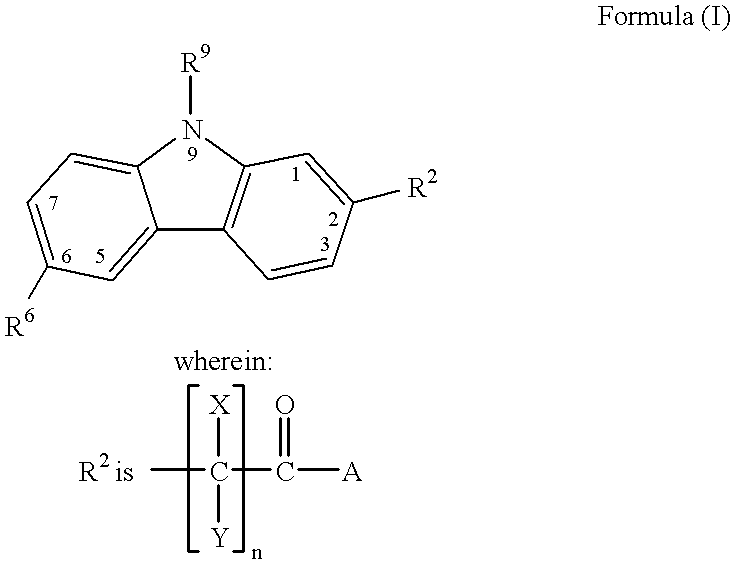
Heteroaromatic pentacyclic compound and medicinal use thereof
A 5-membered heteroaromatic ring compound represented by the formula [I]wherein V is CH or N; W is S or O; R1 and R2 are each H etc.; X is —N(R4)—, —O—, —S—, —SO2—N(R5)—, —CO—N(R7)— etc.; L is wherein R20, R21, R22 and R25 are each H etc.; E is aryl or heteroaromatic ring group; R is —COOH etc.; B is aryl etc.; R3 is H etc.; Y is —C(R13)(R14)—N(R12)—C(R13)(R14)—O—, —N(R11)—, —O— etc.; A is alkylene; and Z is aryl etc., a prodrug thereof and a pharmaceutically acceptable salt thereof. The compound [I] has a superior protein tyrosine phosphatase 1B inhibitory activity and is useful as a therapeutic agent for diabetes, diabetic complications, hyperlipidemia, obesity and the like.
Owner:JAPAN TOBACCO INC
Allele specific primer extension
InactiveUS20040048301A1Rapid rise in fluorescenceEnhanced signalSugar derivativesMicrobiological testing/measurementPhosphateNucleotide
A method of characterizing a nucleic acid sample is provided that includes the steps of: (a) conducting a DNA polymerase reaction that includes the reaction of a template, an allele specific primer, at least one terminal phosphate-labeled nucleotide, DNA polymerase, and optionally an enzyme having 3'->5' exonuclease activity when the primer is non-hydrolyzable, which reaction results in the production of labeled polyphosphate; (b) permitting the labeled polyphosphate to react with a phosphatase to produce a detectable species; (c) detecting the detectable species; and (d) characterizing the nucleic acid sample based on such detection.
Owner:GLOBAL LIFE SCI SOLUTIONS USA LLC
Sample collection and bioluminescent analysis system
ActiveUS20140004548A1Promote useReduce power consumptionBioreactor/fermenter combinationsBiological substance pretreatmentsPhoton detectionPhoton counter
Methods and apparatus for evaluating the quality of an environment or process by measuring light emitted from a bioluminescent sample containing ATP, ADP, or alkaline phosphatase. The apparatus comprises a sample collection and analysis system used to collect a sample, mix reagents, react the sample, and collect it in a measurement chamber. The system includes an instrument having a photon detection assembly for use with the sample testing device and one or more probe assemblies that optically cooperate with the instrument. The instrument includes a dark chamber with a reflective interior surface which may be concave or preferably spherical, and a photon detection sensor such as a multi-pixel photon counter sensor. A substantially transparent portion of the probe assembly, and liquid contained therein, focus bioluminescence toward the photon detection sensor.
Owner:BIOCONTROL SYST
Polyhydroxyalkanoate production from polyols
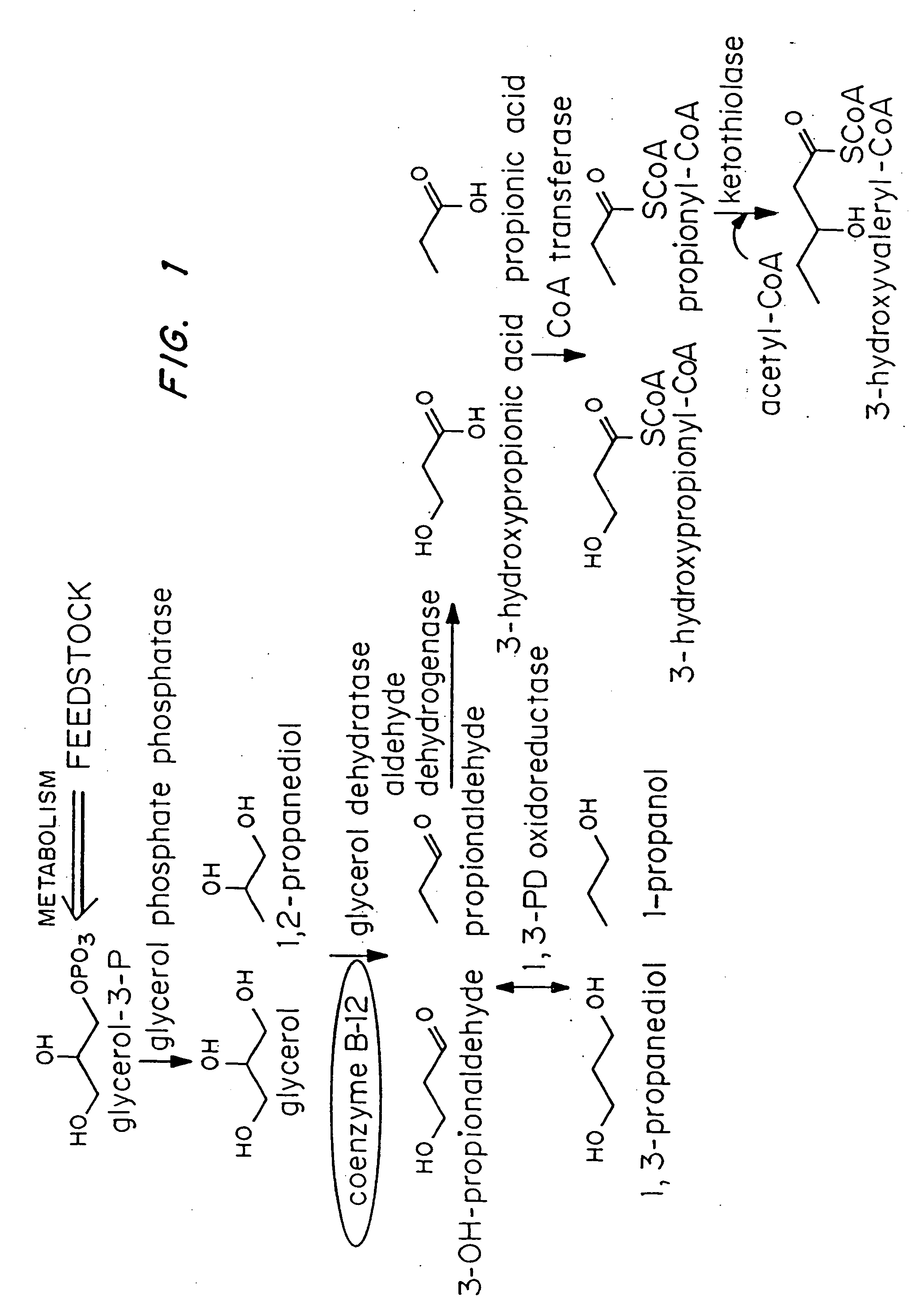
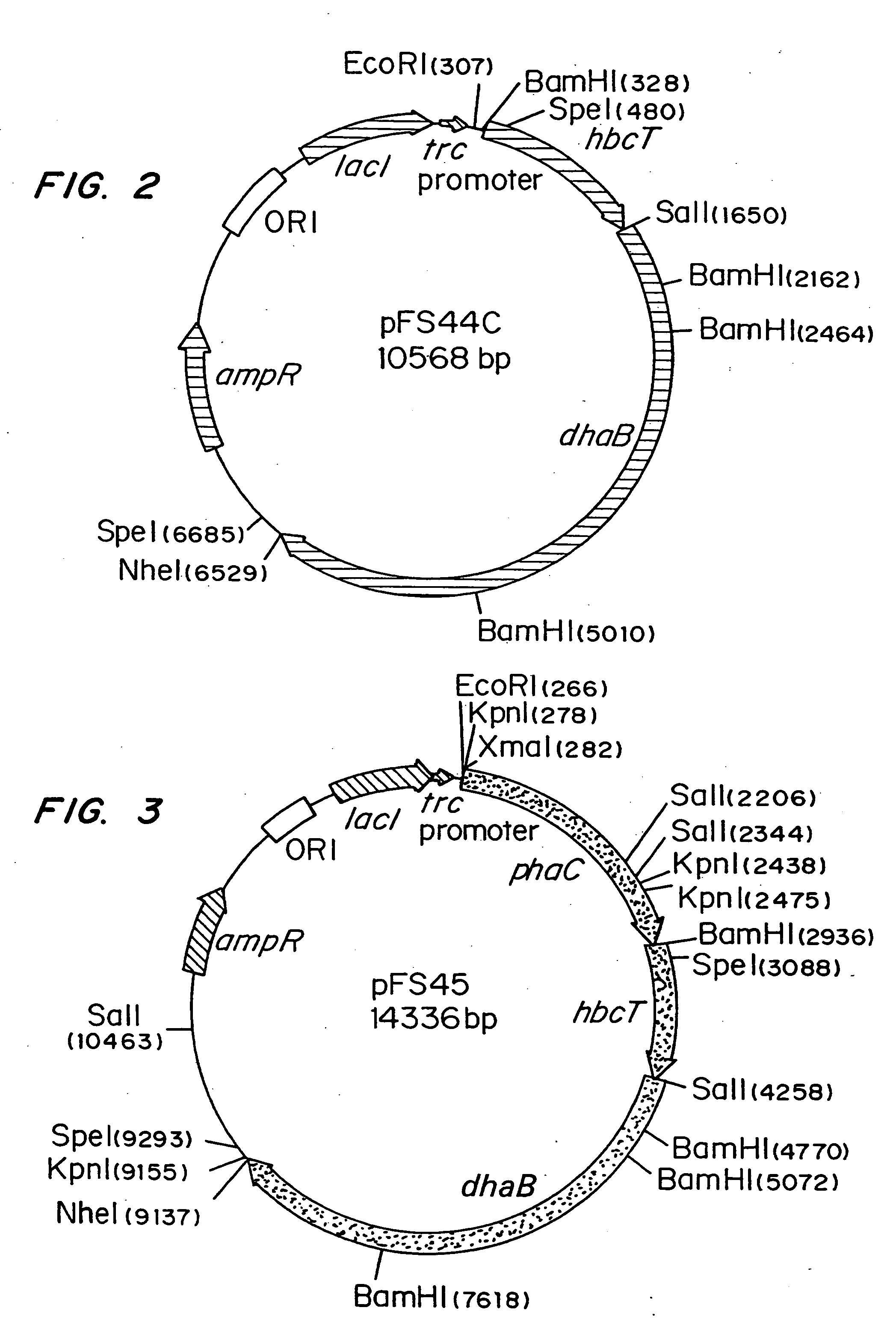
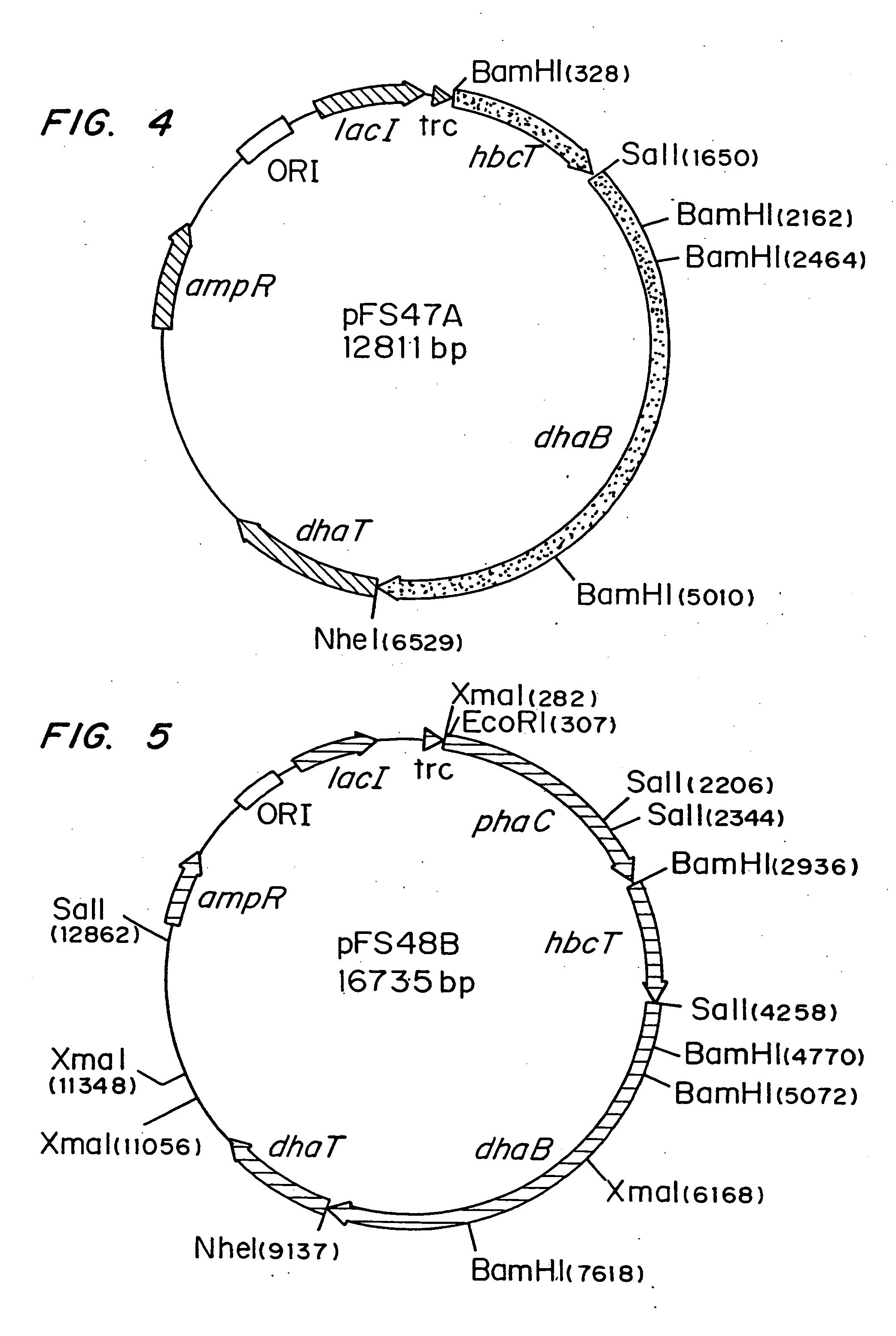
InactiveUS20050239179A1Promote recoveryProduct can be usedBacteriaMicroorganism based processes3-Hydroxypropionic acidPropanoic acid
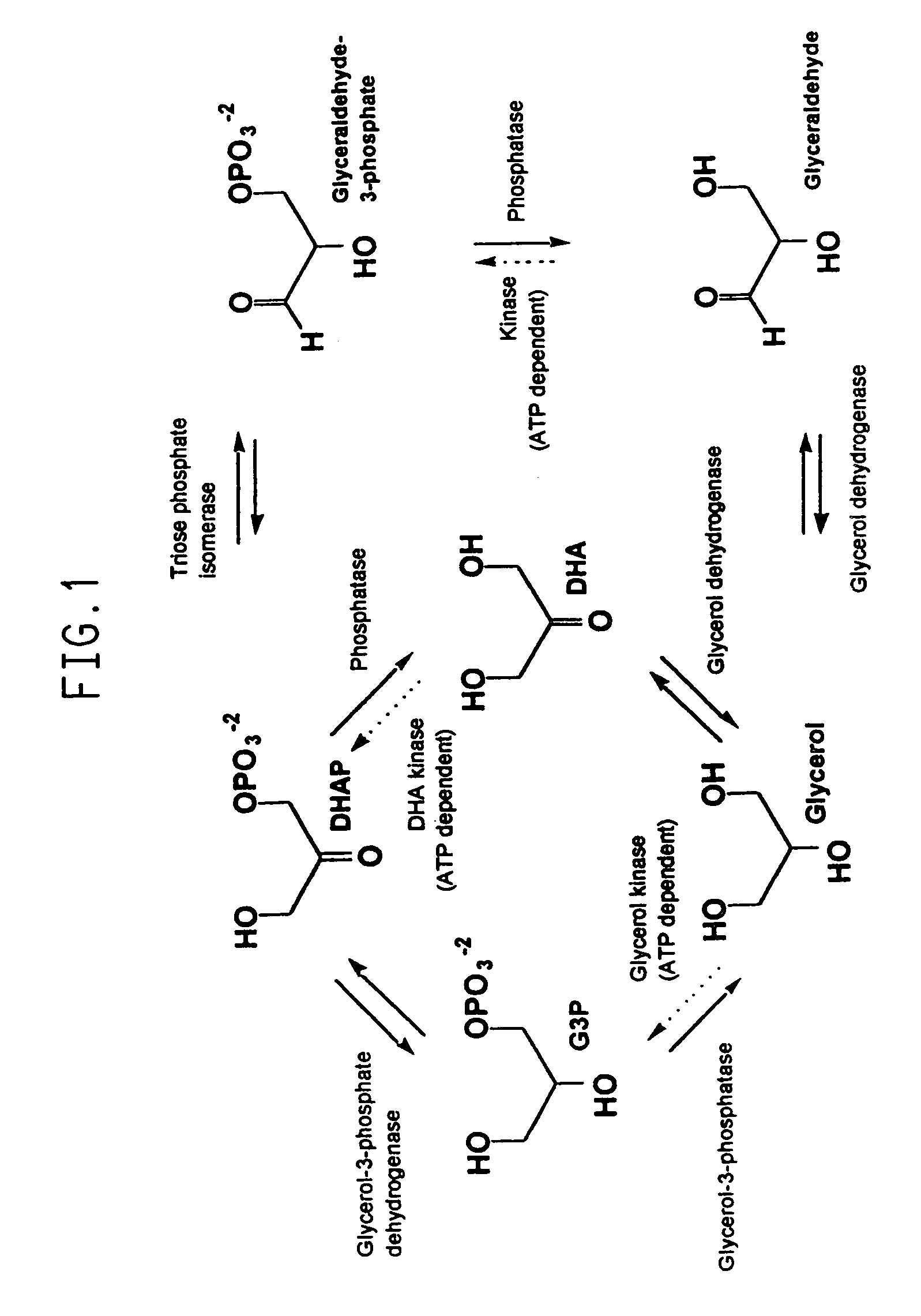
Organisms are provided which express enzymes such as glycerol dehydratase, diol dehydratase, acyl-CoA transferase, acyl-CoA synthetase β-ketothiolase, acetoacetyl-CoA reductase, PHA synthase, glycerol-3-phosphate dehydrogenase and glycerol-3-phosphatase, which are useful for the production of PHAs. In some cases one or more of these genes are native to the host organism and the remainder are provided from transgenes. These organisms produce poly (3-hydroxyalkanoate) homopolymers or co-polymers incorporating 3-hydroxypropionate or 3-hydroxyvalerate monomers wherein the 3-hydroxypropionate and 3-hydroxyvalreate units are derived from the enzyme catalysed conversion of diols. Suitable diols that can be used include 1,2-propanediol, 1,3 propanediol and glycerol. Biochemical pathways for obtaining the glycerol from normal cellular metabolites are also described. The PHA polymers are readily recovered and industrially useful as polymers or as starting materials for a range of chemical intermediates including 1,3-propanediol, 3-hydroxypropionaldehyde, acrylics, malonic acid, esters and amines.
Owner:CJ CHEILJEDANG CORP
Method of dephosphorylating an endotoxin in vivo with alkaline phosphatase
The invention relates to pharmaceutical compositions suitable for treating or curing clinical complications mediated by endotoxin, including sepsis. The compositions contain components suitable for detoxifying endotoxin rendering it less deleterious to mammals such as humans, in particular to patients with reduced host-defence resistance. The invention also relates to pharmaceutical compositions suitable for stimulating bone formation, e.g. for mending broken bone or for prophylaxis or therapy of metabolic bone diseases such as osteoporosis and osteomalacia and pharmaceutical compositions for decreasing or inhibiting undesired bone formation. The pharmaceutical compositions according to the invention are directed at modulating phosphatase activity in vivo.
Owner:UNIVERSITY OF GRONINGEN
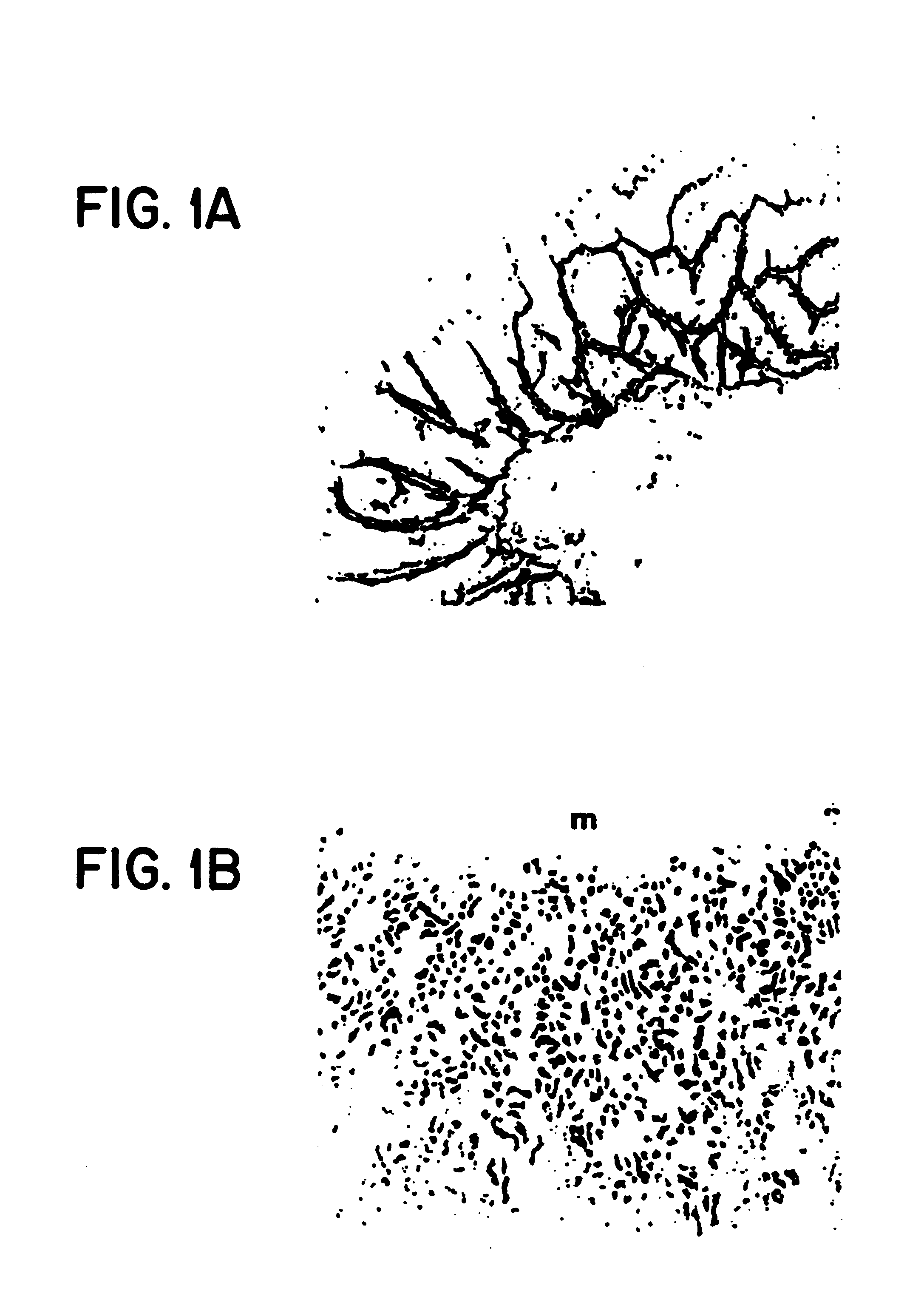
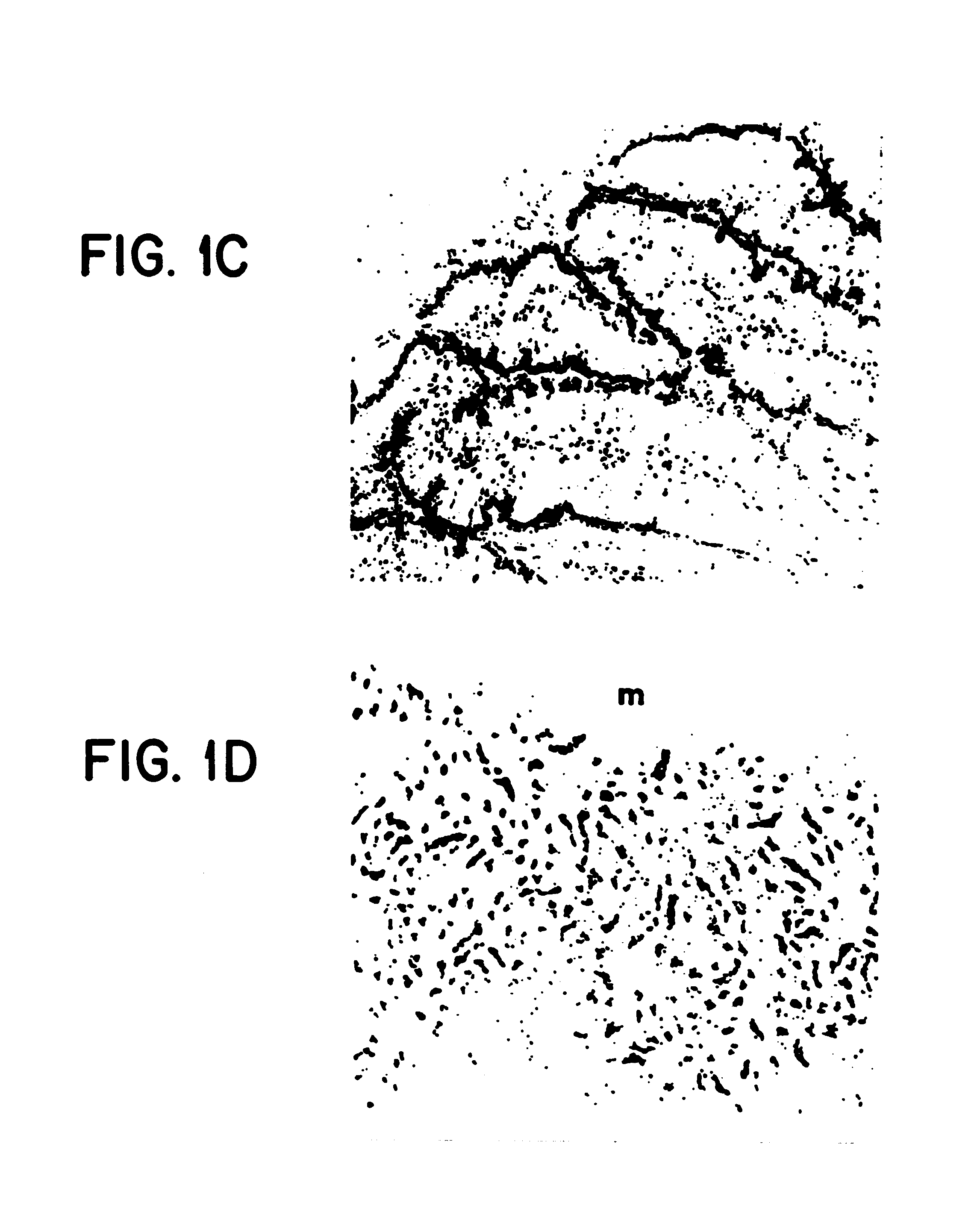
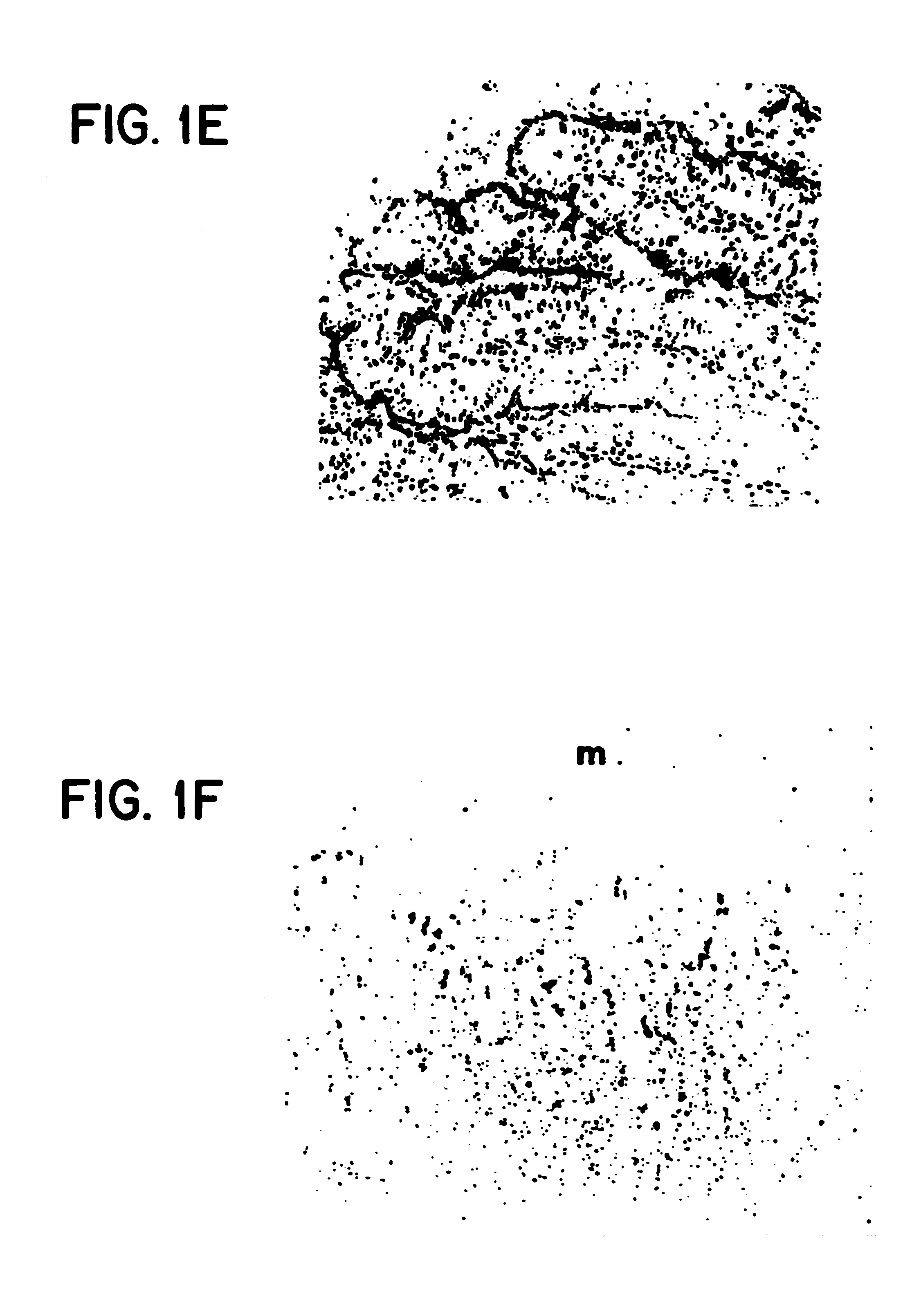
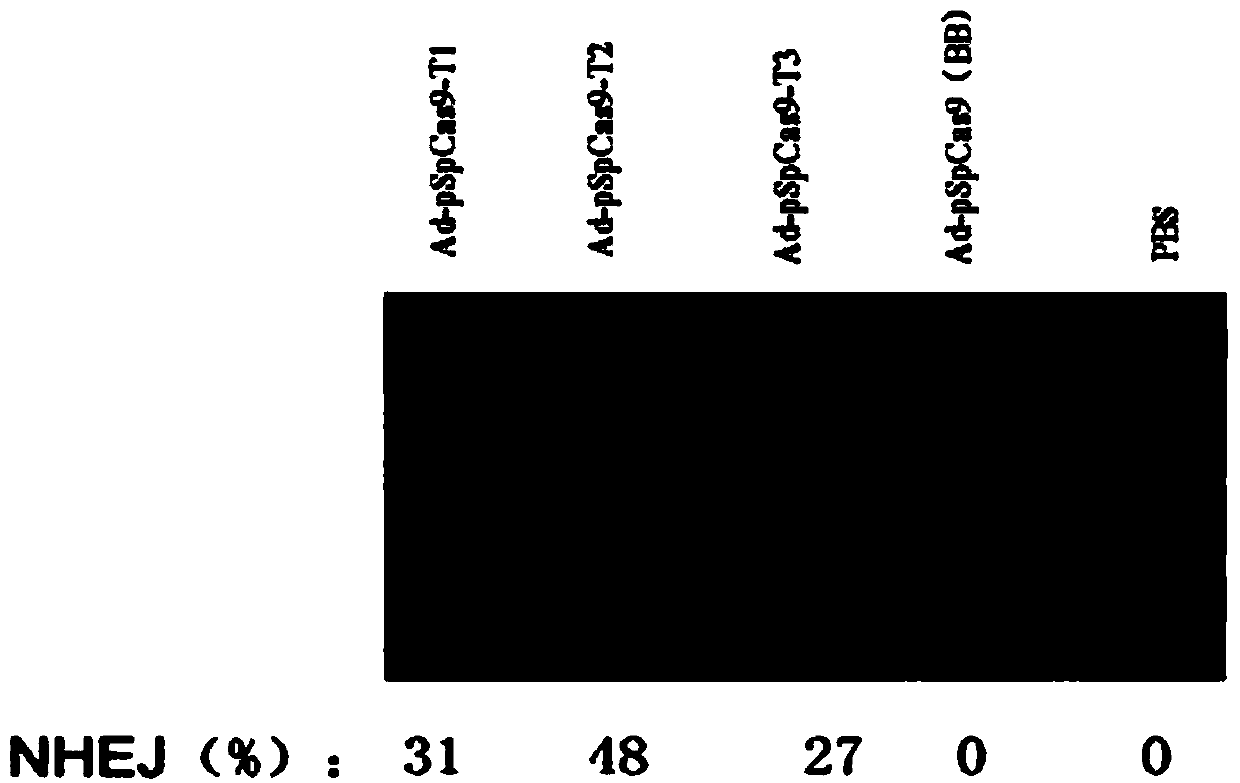
CRISPR/Cas9 system-containing targeted knockout vector and adenovirus and applications thereof
InactiveCN104004778AHigh knockout rateGenetic material ingredientsViruses/bacteriophagesBiotechnologyEnzyme digestion
The invention discloses a CRISPR / Cas9 system-containing targeted knockout vector and adenovirus and applications thereof. The targeted knockout vector is prepared through the following steps: after a pX330 U6-Chimeric_BB-CBh-hSpCas9 plasmid is subjected to enzyme digestion and filling-in by using EcoRI and SacII, connecting the pX330 U6-Chimeric_BB-CBh-hSpCas9 plasmid with a pAdTrack-CMV plasmid subjected to enzyme digestion and filling-in by using BstXI; after the obtained product is linearized by using BbsI, connecting the obtained product to a specific target sequence of a desired gene; and after the obtained object is linearized by using PmeI and dephosphorylated by using CIAP alkaline phosphatase, recombining the obtained product with a pAdEasy-1 plasmid. The targeted knockout vector can mutate gene sequences in target sequence areas, and the mutation rate is high, up to 30.6-45.8%, therefore, the targeted knockout vector can be used for gene site-directed mutation, and lays a foundation for gene therapy.
Owner:重庆高圣生物医药有限责任公司
Compositions and methods for detection and isolation of phosphorylated molecules
ActiveUS7102005B2Simplifies subsequent analysisSilicon organic compoundsOther chemical processesTernary complexPhosphorylation
Owner:MOLECULAR PROBES
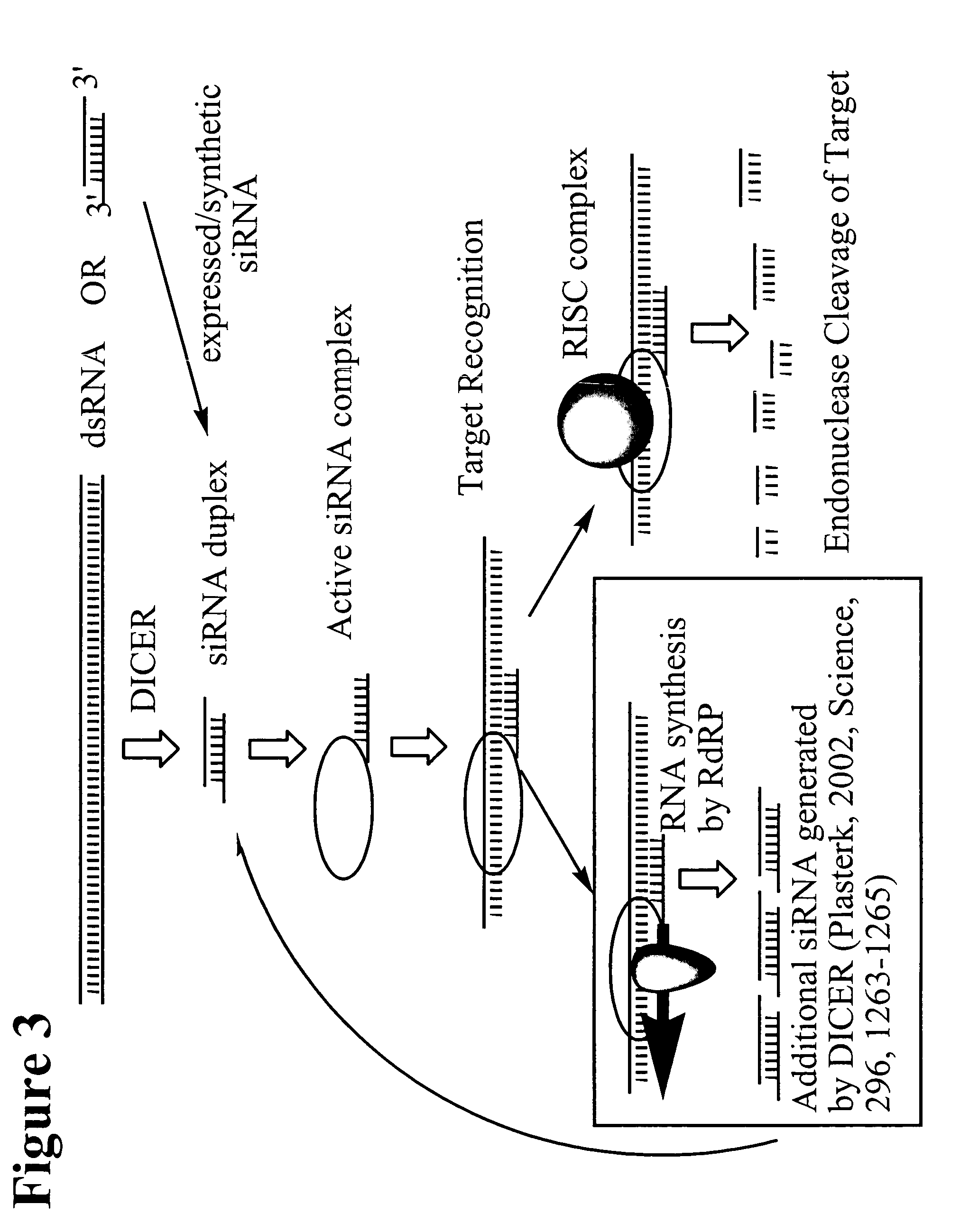
RNA interference mediated inhibition of protein tyrosine phosphatase-1B (PTP-1B) gene expression using short interfering nucleic acid (siNA)
InactiveUS20060025361A1Improves various propertyImprove the immunityCompounds screening/testingSugar derivativesDiseaseTyrosine
This invention relates to compounds, compositions, and methods useful for modulating protein tyrosine phosphatase-1B (PTP-1B) gene expression using short interfering nucleic acid (siNA) molecules. This invention also relates to compounds, compositions, and methods useful for modulating the expression and activity of other genes involved in pathways of PTP-1B gene expression and / or activity by RNA interference (RNAi) using small nucleic acid molecules. In particular, the instant invention features small nucleic acid molecules, such as short interfering nucleic acid (siNA), short interfering RNA (siRNA), double-stranded RNA (dsRNA), micro-RNA (miRNA), and short hairpin RNA (shRNA) molecules and methods used to modulate the expression of PTP-1B genes. Such small nucleic acid molecules are useful, for example, for treating, preventing, inhibiting, or reducing obesity, insulin resistance, diabetes (eg. type II and type I diabetes) in a subject or organism, and for any other disease, trait, or condition that is related to or will respond to the levels of PTP-1B in a cell or tissue, alone or in combination with other treatments or therapies.
Owner:SIRNA THERAPEUTICS INC
Transformed microorganisms and genes useful in the production of glycerol and 1,3-propanediol
Recombinant organisms are provided comprising genes encoding a glycerol-3-phosphate dehydrogenase and / or a glycerol-3-phosphatase activity useful for the production of glycerol from a variety of carbon substrates. The organisms further contain disruptions in the endogenous genes encoding proteins having glycerol kinase and glycerol dehydrogenase activities.
Owner:EI DU PONT DE NEMOURS & CO +1
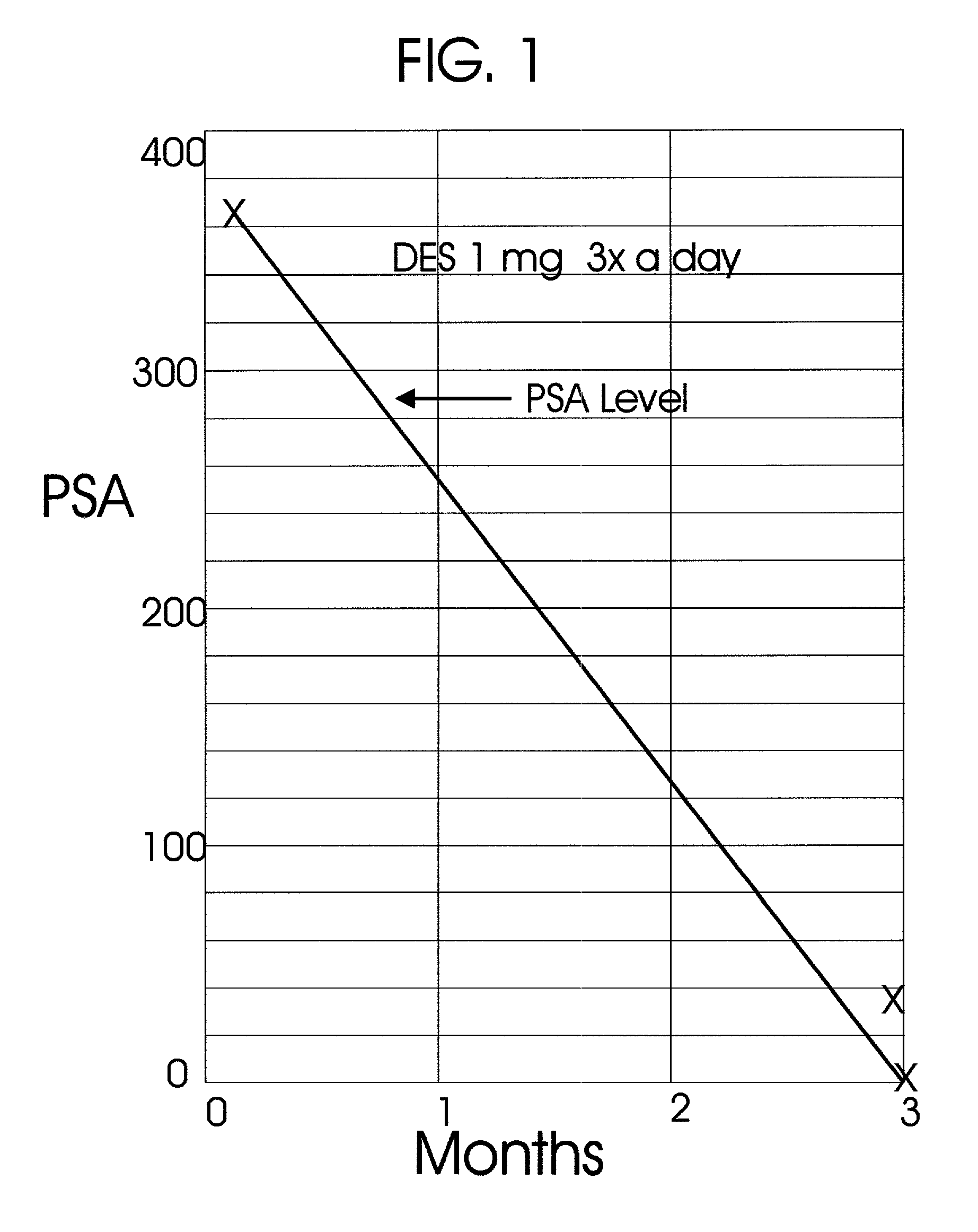
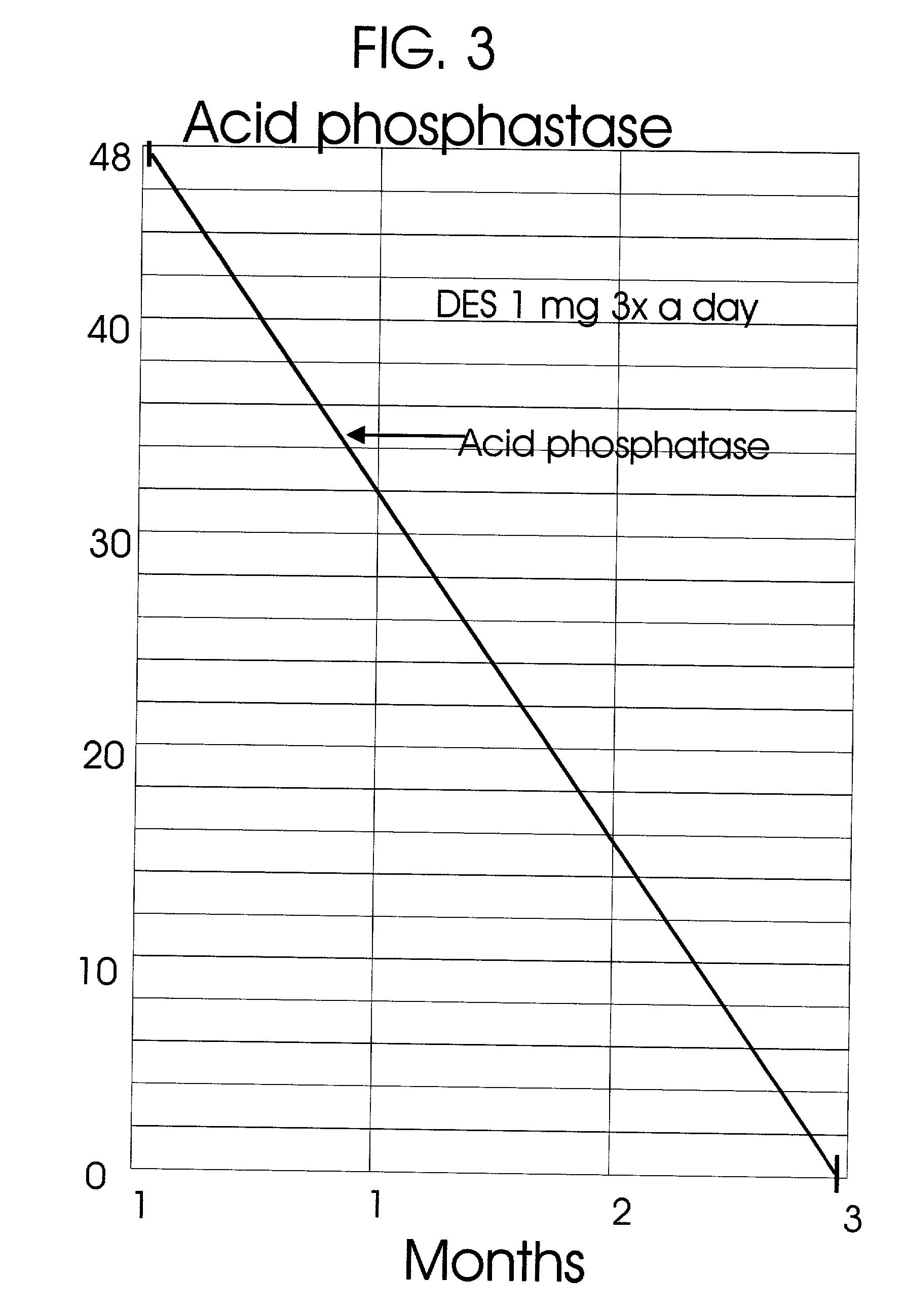
Prostatic hormonal implants treatment of prostate cancer
An improved method and products for the primary hormonal treatment of early stage, low and intermediate risk prostate cancers by prostatic implants of androgen suppressive drugs formulated as fused with a lipoid carrier or encapsulated in microcapsules or in Silastic capsules is provided. Such prostatic implants renders a constant slow-release of their contents to the prostate for extended periods by biodegradation and diffusion. It facilitates higher prostatic and lower systemic concentrations of androgen suppressive hormones. Because of their high prostatic and lower systemic concentrations, tumor control is much improved and the their systemic toxicity is minimized. Tumor control after such primary hormonal implant treatment is followed by clinical examinations and the biochemical tumor control is followed by periodic estimations of serum levels of PSA and acid phosphatase. More complex and expensive surgery or radiation therapy for this group of good prognostic early stage prostate cancer is reserved for those patients failing to this primary hormonal treatment. It will preserve potency more than by surgery or radiation therapy. Furthermore, it would reduce the cost of treatment for early stage prostate cancer significantly. Androgen suppressive hormonal implants to the prostate before, during or after lower dose conventional radiation therapy would also facilitate equal or better cure rates of localized prostate cancer as compared to the more complex and toxic higher dose radiation therapy.
Owner:SAHADEVAN VELAYUDHAN
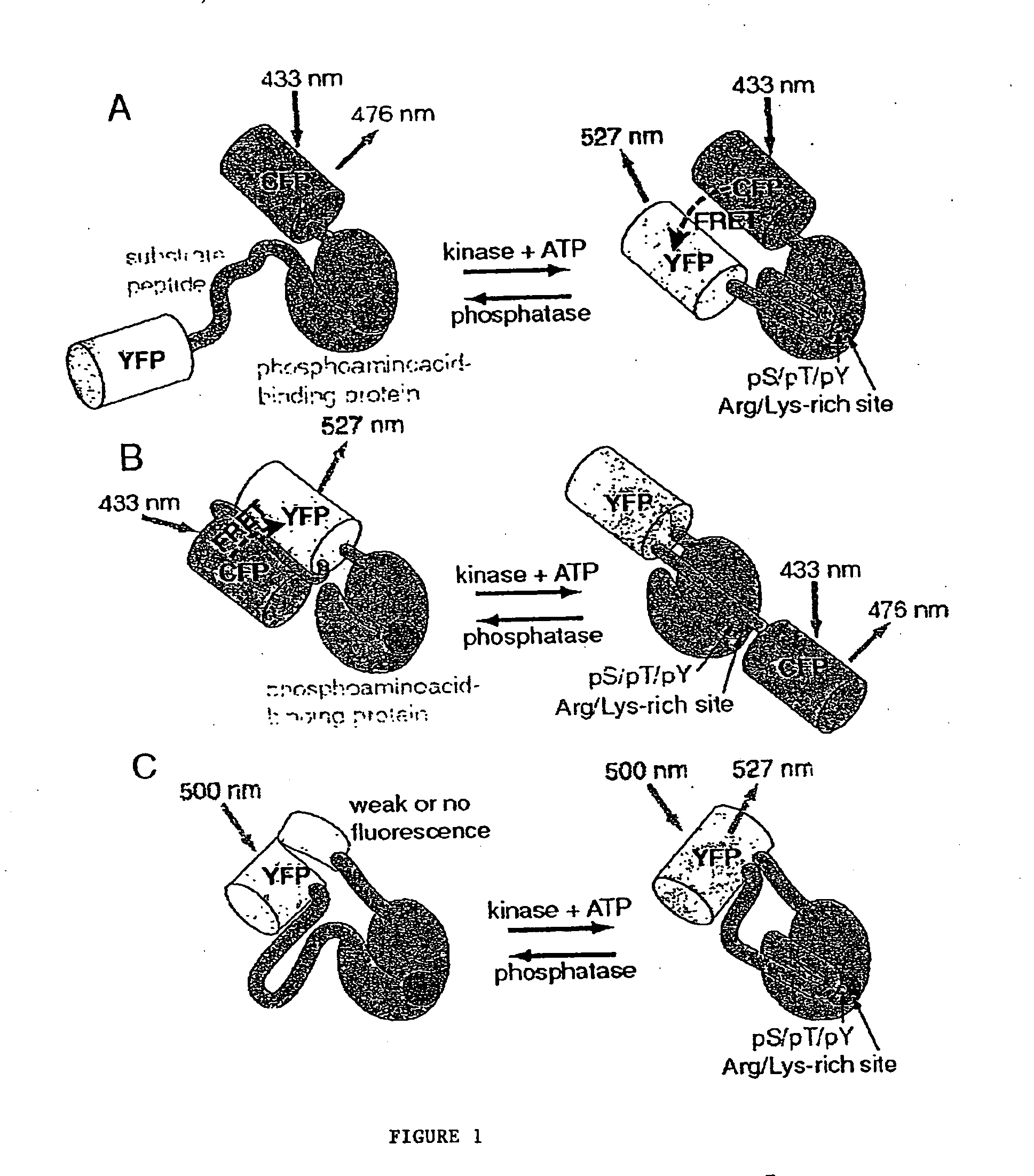
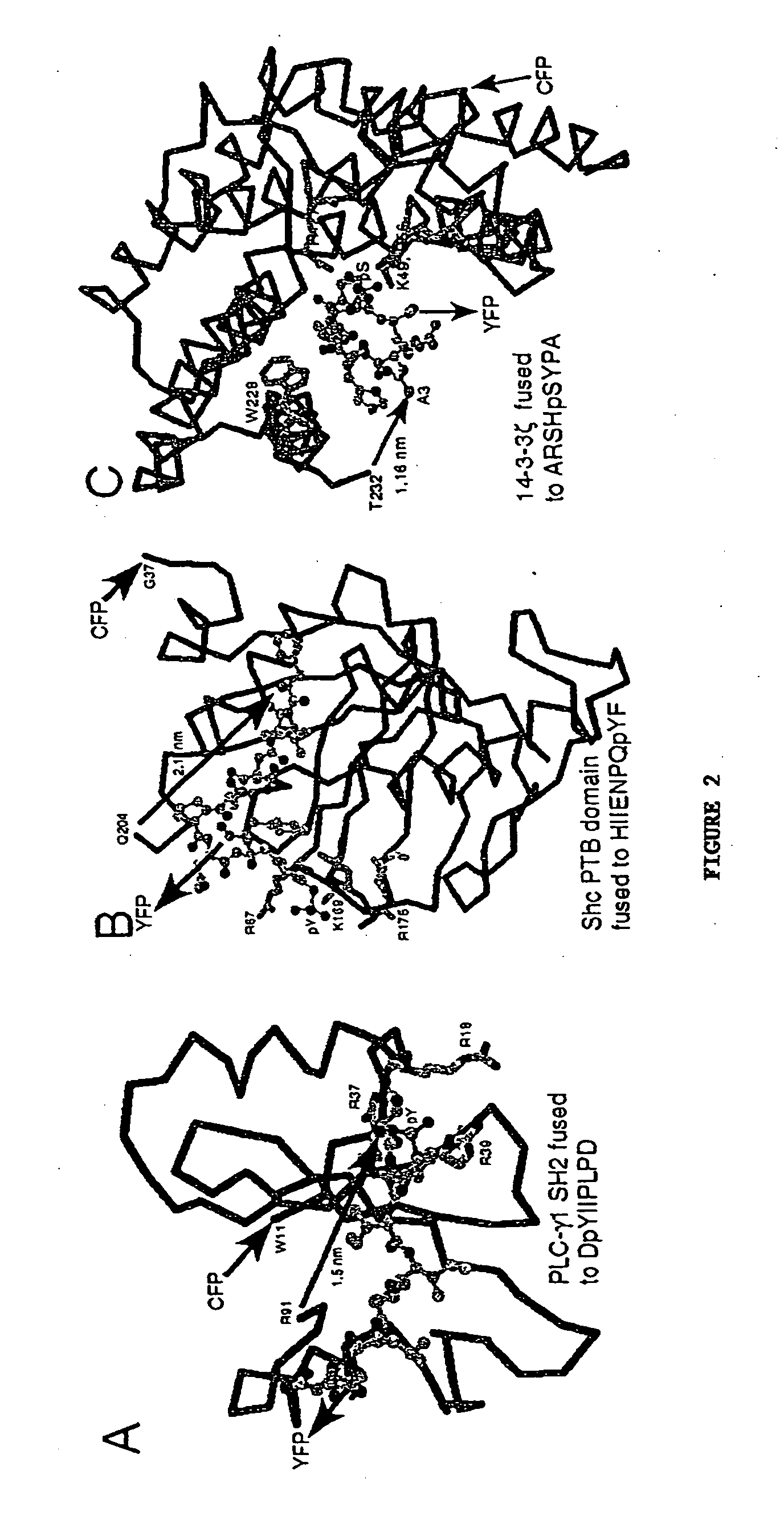
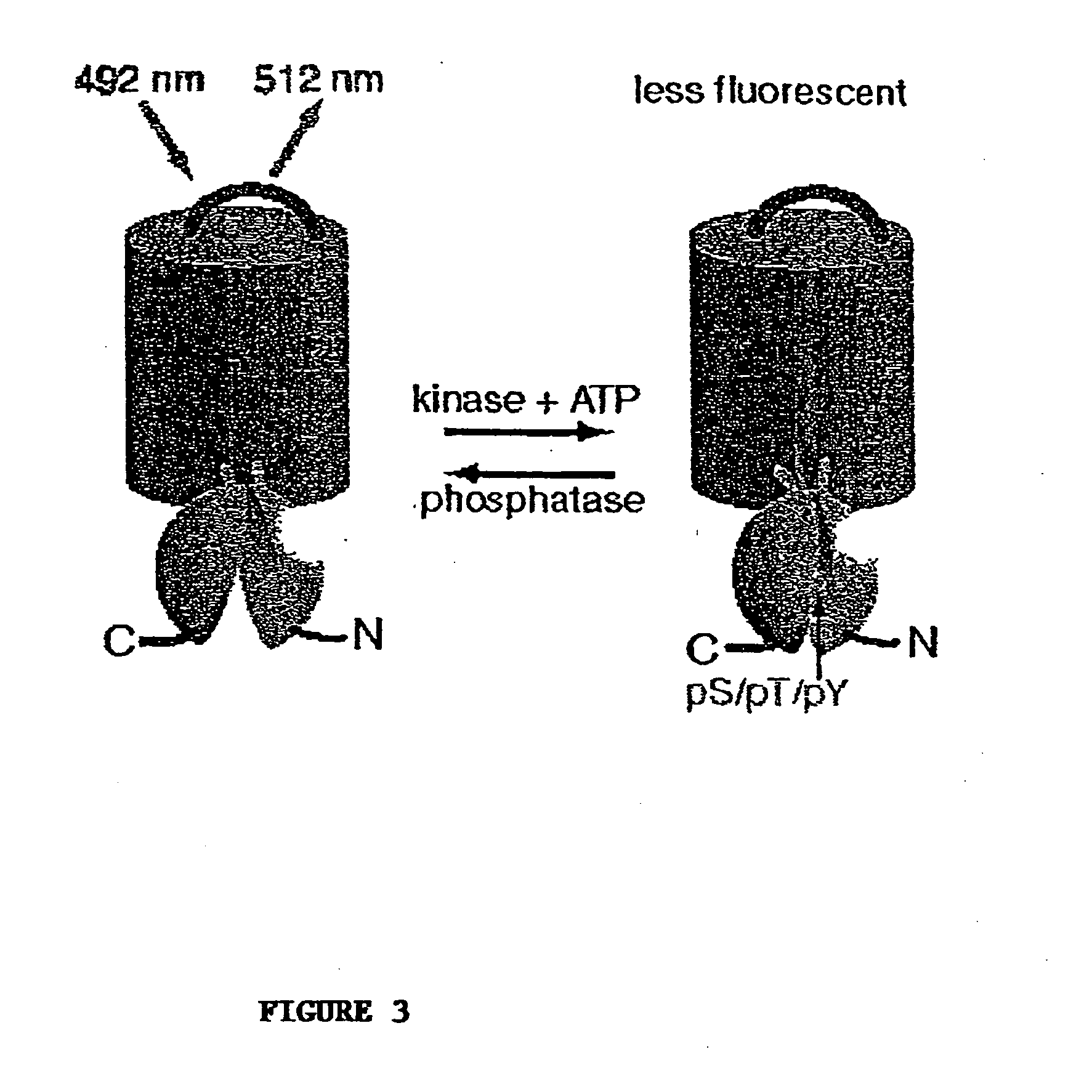
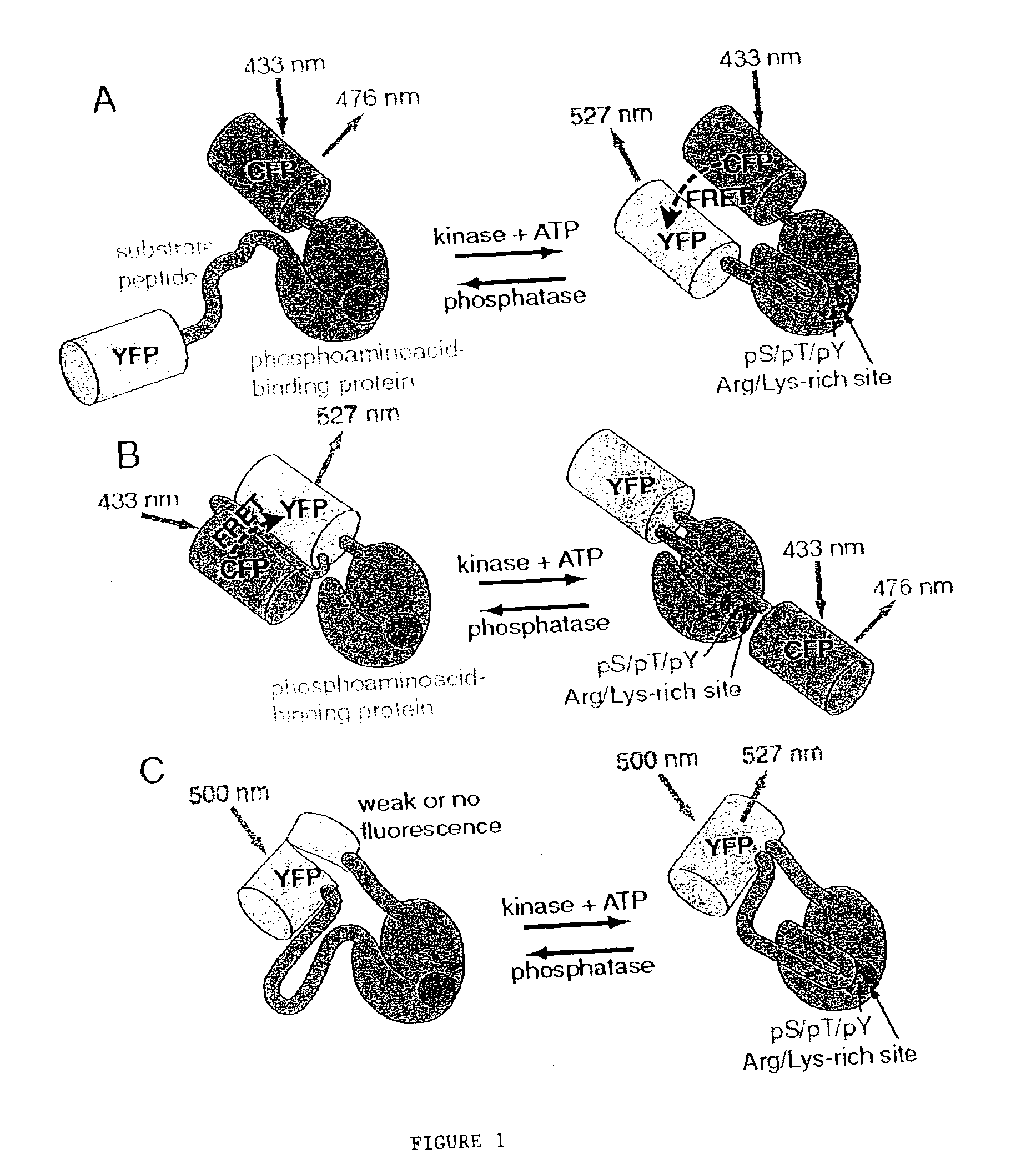
Emission ratiometric indicators of phosphorylation by C-kinase
InactiveUS20050026234A1Facilitate its translocationPeptide/protein ingredientsAntibody mimetics/scaffoldsNucleotideFluorophore
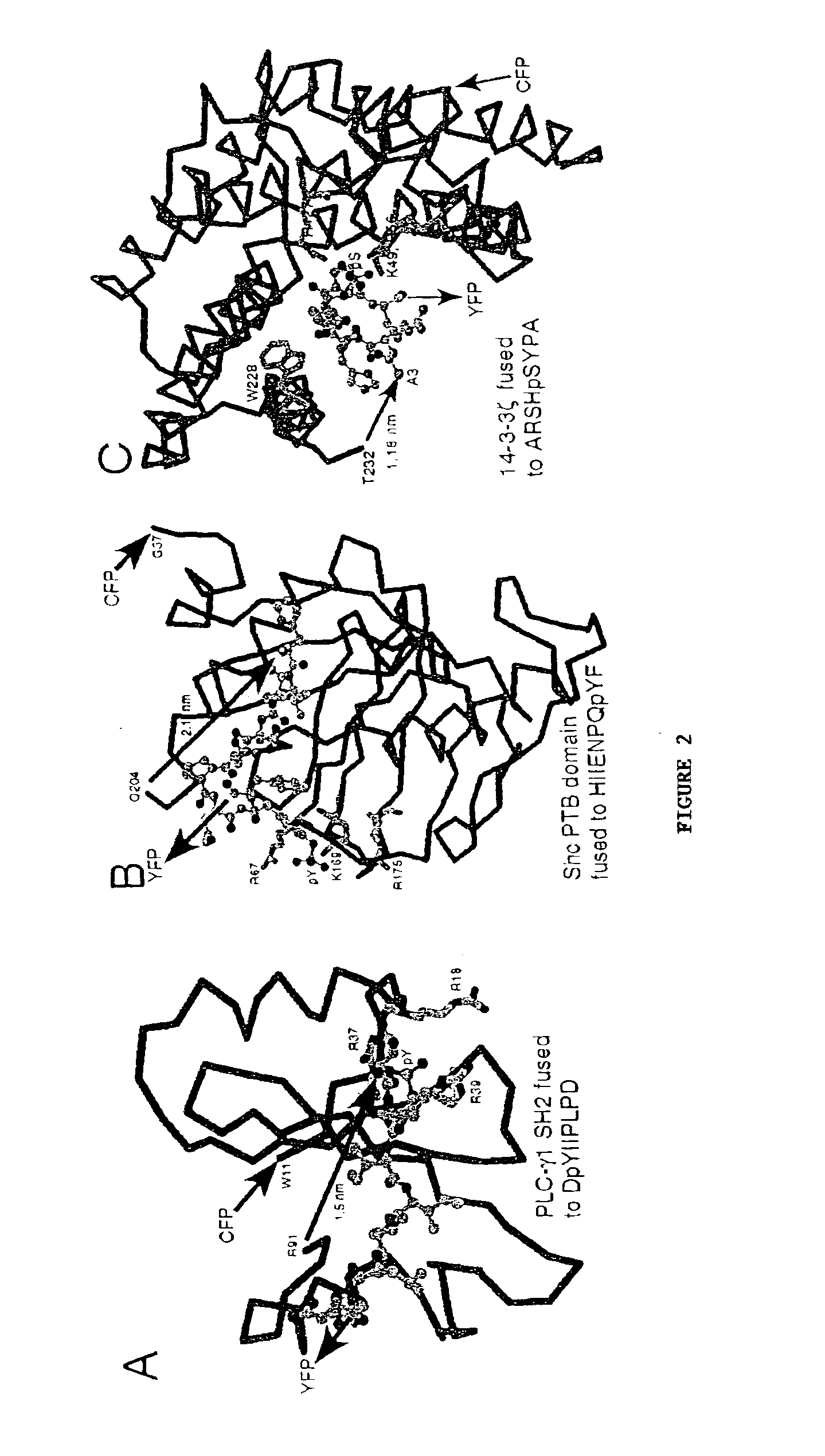
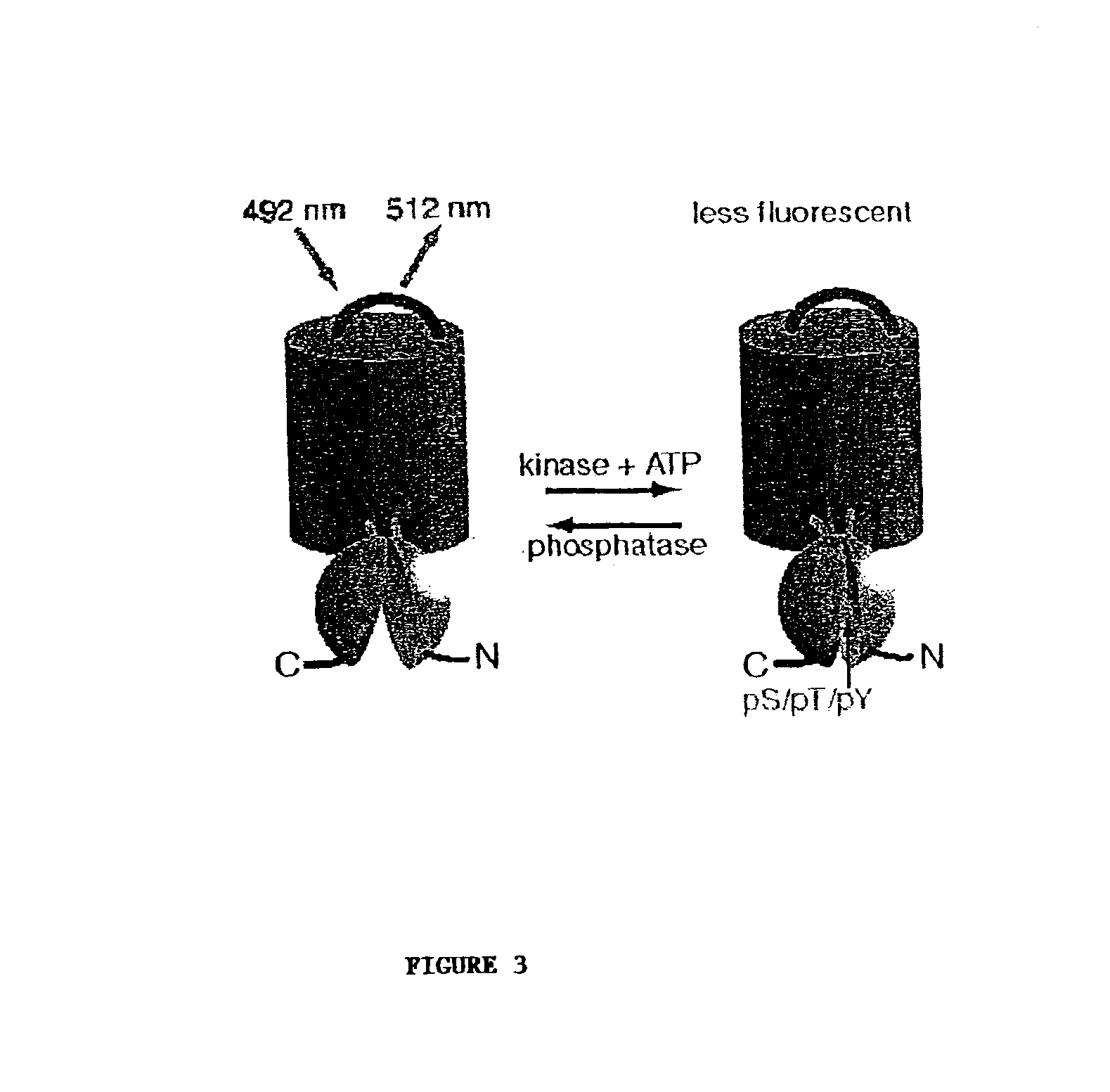
A chimeric phosphorylation indicator (CPI) as provided herein can contain a donor molecule, a phosphorylatable domain, a phosphoaminoacid binding domain (PAABD), and an acceptor molecule. Where the phosphorylatable domain is phosphorylatable by protein kinase C (PKC), the CPI is a c-kinase activity reporter (CKAR). Donor and acceptor molecules may be, independently, fluorescent proteins such as non-oligomerizing fluorescent proteins. A CPI can contain a phosphorylatable polypeptide and a fluorescent protein; the phosphorylatable polypeptide may be contained within the sequence of the fluorescent protein, or the fluorescent protein may be contained within the sequence of the phosphorylatable polypeptide. The spatiotemporal properties of the PKC signal pathway may be tested with CKAR, calcium-sensing fluorophores and FRET-based translocation assays. Polynucleotides encoding such CPIs, and kits containing the indicators and / or the polynucleotides, are provided. A method of using the chimeric phosphorylation indicators to detect a kinase or phosphatase in a sample is provided.
Owner:RGT UNIV OF CALIFORNIA +1
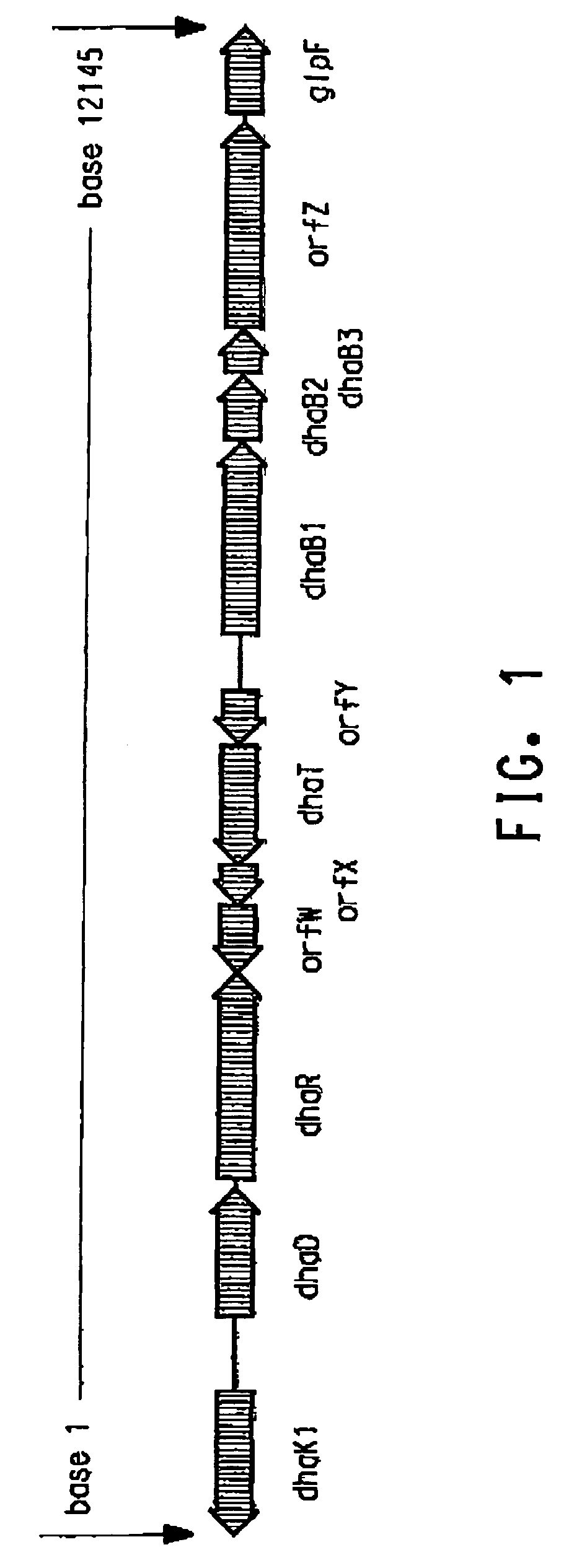
Process for the biological production of 1,3-propanediol with high titer
The present invention provides an improved method for the biological production of 1,3-propanediol from a fermentable carbon source in a single microorganism. In one aspect of the present invention, an improved process for the conversion of glucose to 1,3-propanediol is achieved by the use of an E. coli transformed with the Klebsiella pneumoniae dha regulon genes dhaR, orfY, dhaT, orfX, orfW, dhaB1, dhaB2, dhaB3, and orfZ, all these genes arranged in the same genetic organization as found in wild type Klebsiella pneumoniae. In another aspect of the present invention, an improved process for the production of 1,3-propanediol from glucose using a recombinant E. coli containing genes encoding a G3PDH, a G3P phosphatase, a dehydratase, and a dehydratase reactivation factor compared to an identical process using a recombinant E. coli containing genes encoding a G3PDH, a G3P phosphatase, a dehydratase, a dehydratase reactivation factor and a 1,3-propanediol oxidoreductase (dhaT). The dramatically improved process relies on the presence in E. coli of a gene encoding a non-specific catalytic activity sufficient to convert 3-hydroxypropionaldehyde to 1,3-propanediol.
Owner:DUPONT US HLDG LLC
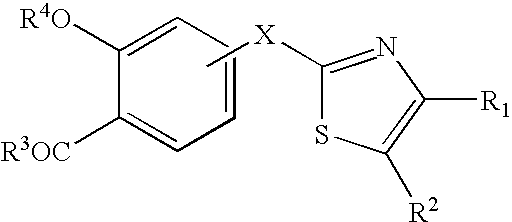
Azole compound and medicinal use thereof
The present invention relates to an azole compound represented by the formula [I]wherein W is S or O; R is —COOR7, —X1-A1-COOR7 (R7 is H, alkyl) or tetrazolyl; R1, R2, R3 and R4 are H and the like; A is —(CH2)m—X— (X is —N(R8)—, —C(R9)(R10)—, —CO— or —CO—N(R8)—); B is aryl or aromatic heterocyclic group; R5 is H and the like; R6 is —(Y)s1-(A2)s-Z (Y is —O—, —S(O)t—, —N(R13)—, —N(R14)—CO—, —N(R14)—SO2—, —SO2—N(R14)— and the like, A2 is alkylene, and Z is cycloalkyl, aryl, aromatic heterocyclic group, indanyl, piperazinyl, a prodrug thereof or a pharmaceutically acceptable salt thereof. The compound [I] of the present invention has a protein tyrosine phosphatase 1B inhibitory activity, and is useful as a therapeutic agent for diabetes, a therapeutic drug for diabetic complications or a therapeutic drug for hyperlipidemia.
Owner:JAPAN TOBACCO INC
Nucleic acids encoding kinase and phosphatase enzymes, expression vectors and cells containing same
InactiveUS7517970B2Good quality crystalImprove crystallizationSugar derivativesHydrolasesActive enzymeSystematic variation
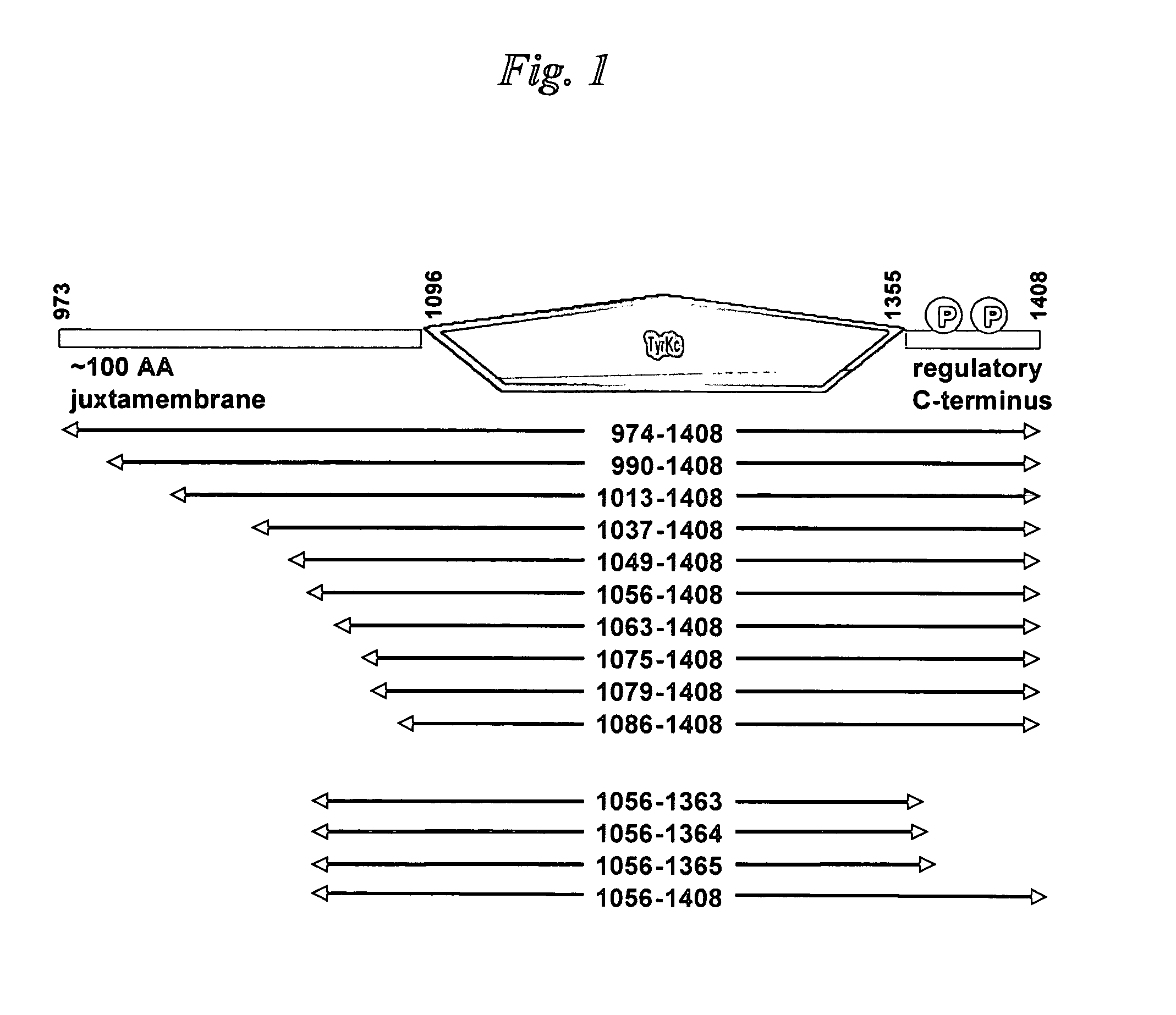
Methods for expressing active enzymes are described that involve co-expressing a first enzyme with a second enzyme that has an enzymatic activity that reverses a modification on the first enzyme and / or for identification of soluble and / or active catalytic domains by systematic variation of fragment lengths around catalytic domain boundaries.
Owner:PLEXXIKON INC
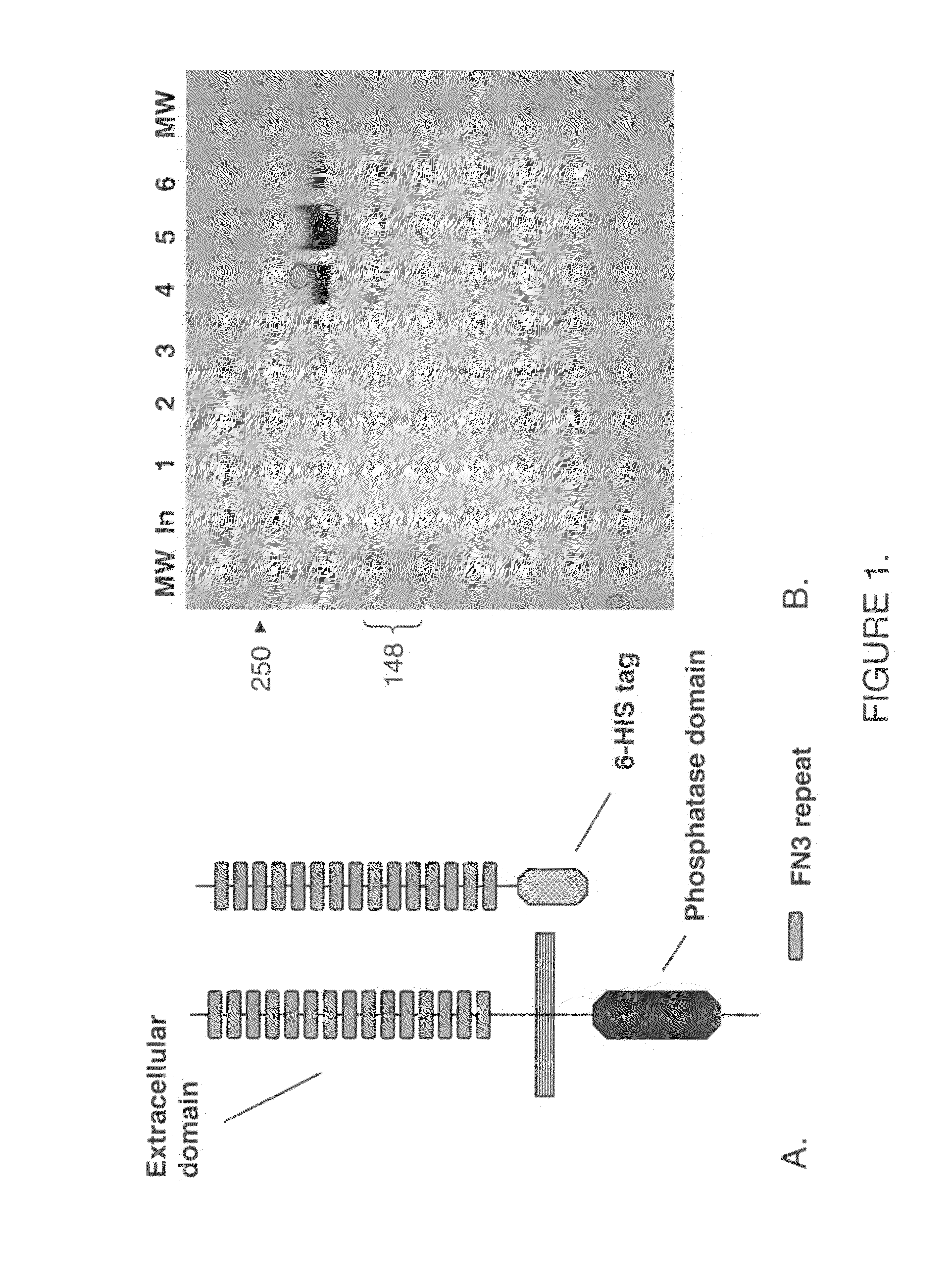
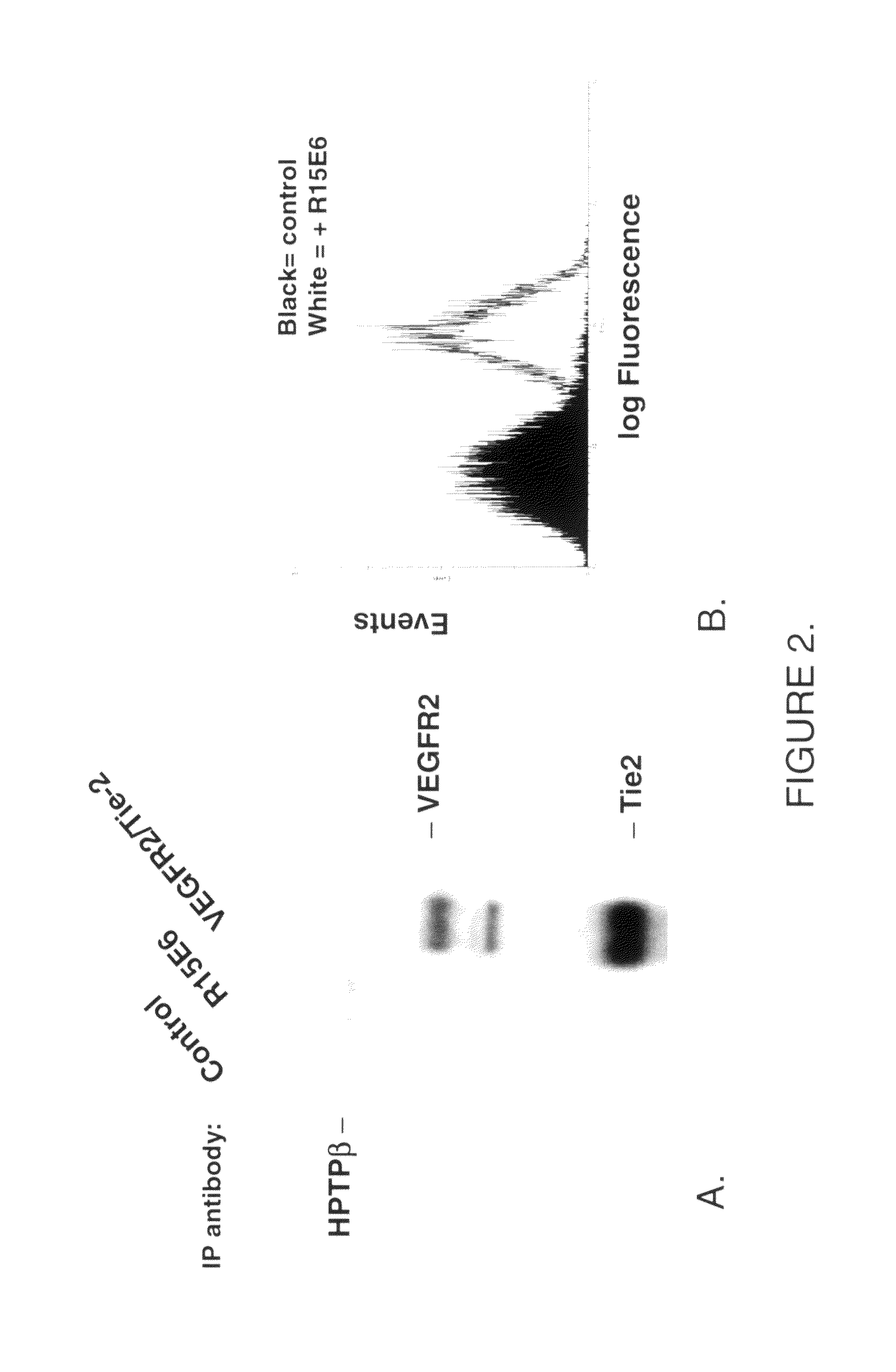
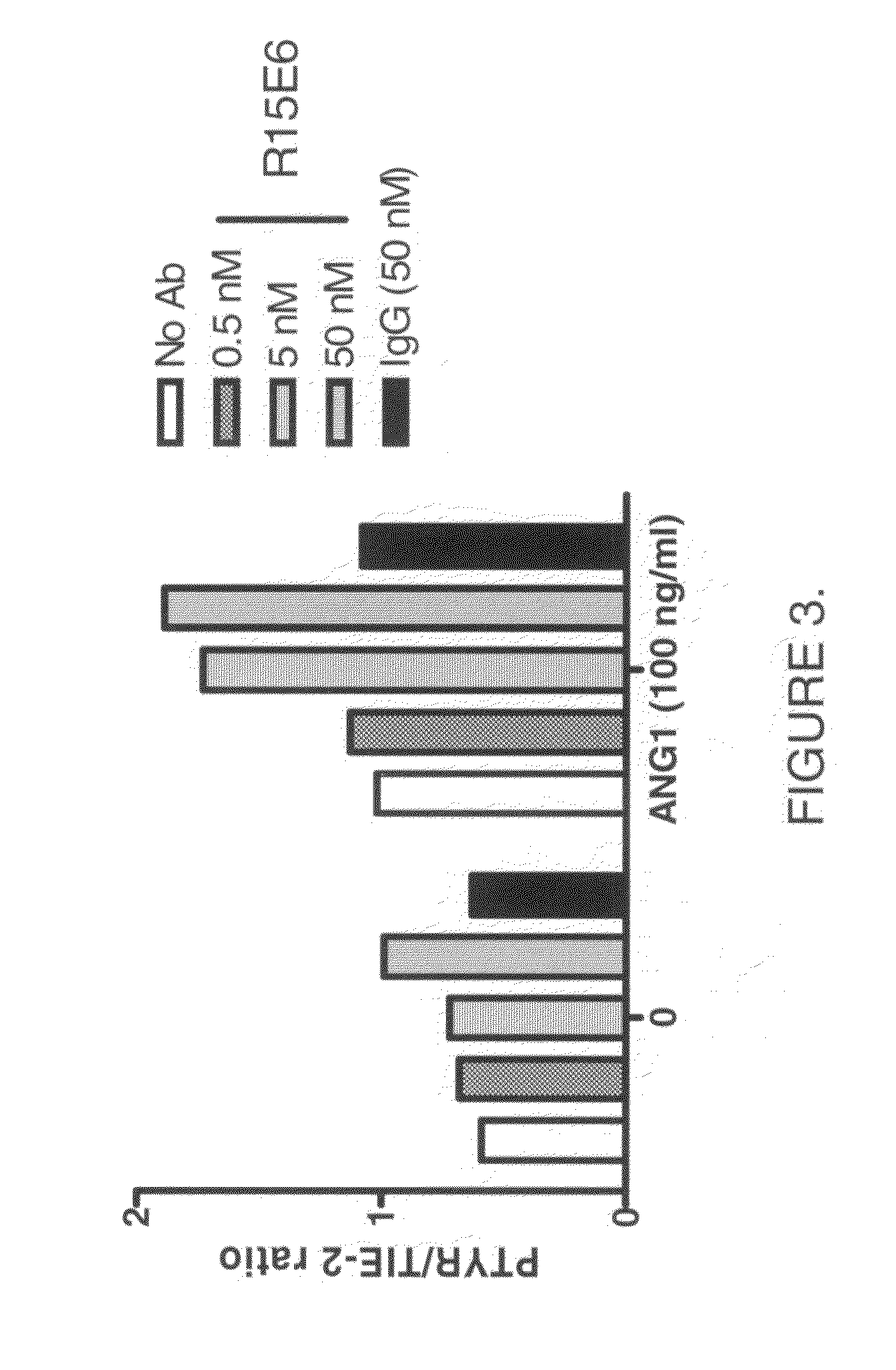
Antibodies that bind human protein tyrosine phosphatase beta (HPTPβ)
Owner:EYEPOINT PHARMA INC
Analyte detection
A method of characterizing an analyte sample is provided that includes the steps of: (a) anchoring the analyte to a nucleic acid template of known sequence; (b) conducting a DNA polymerase reaction that includes the reaction of a template, a non-hydrolyzable primer, at least one terminal phosphate-labeled nucleotide, DNA polymerase, and an enzyme having 3′→5′ exonuclease activity which reaction results in the production of labeled polyphosphate; (c) permitting the labeled polyphosphate to react with a phosphatase to produce a detectable species characteristic of the sample; (d) detecting the detectable species. The method may include the step of characterizing the nucleic acid sample based on the detection. Also provided are methods of analyzing multiple analytes in a sample, and kits for characterizing analyte samples.
Owner:GLOBAL LIFE SCI SOLUTIONS USA LLC
Novel uracil compound or salt thereof having human deoxyuridine triphosphatase inhibitory activity
ActiveUS20110082163A1Potent human dUTPase inhibitory activityAntibacterial agentsBiocideDeoxyuridinePharmaceutical Substances
Provided is a uracil compound or a salt thereof, which has potent human dUTPase inhibitory activity and is useful as, for example, an antitumor drug.A uracil compound represented by the general formula (I) or a salt thereof:wherein n represents an integer of 1 to 3; X represents a bond, an oxygen atom, a sulfur atom, or the like; Y represents a linear or branched alkylene group having 1 to 8 carbon atoms, or the like; and Z represents —SO2NR1R2 or —NR3SO2—R4, wherein R1 and R2 each represent an alkyl group having 1 to 6 carbon atoms, an aralkyl group which is optionally substituted, or the like; R3 represents an alkyl group having 1 to 6 carbon atoms, or the like; and R4 represents an aromatic hydrocarbon group, an unsaturated heterocyclic group, or the like.
Owner:TAIHO PHARMA CO LTD
Treating or preventing the early stages of degeneration of articular cartilage or subchondral bone in mammals using carprofen and derivatives
Treating or preventing the early stages of degeneration of articular cartilage or subchondral bone in the affected joint of a mammal is accomplished by administering a chondroprotective compound of Formula (I):where A is hydroxy, (C1-C4)alkoxy, amino, hydroxy-amino, mono-(C1-C2)alkylamino, di-(C1-C2)alkylamino; X and Y are independently H or (C1-C2)alkyl; and n is 1 or 2; R6 is halogen, (C1-C3)alkyl, trifluoromethyl, or nitro; R9 is H; (C1-C2)alkyl; phenyl or phenyl-(C1-C2)alkyl, where phenyl is optionally mono-substituted by fluoro or chloro; -C(=O)-R, where R is (C1-C2)alkyl or phenyl, optionally mono-substituted by fluoro or chloro; or -C(=O)-O-R', where R1 is (C1-C2)alkyl.This treatment ameliorates, diminishes, actively treats, reverses or prevents any injury, damage or loss of articular cartilage or subchondral bone subsequent to said early stage of said degeneration. Whether or not a mammal needs such treatment is determined by whether or not it exhibits a statistically significant deviation from normal standard values in synovial fluid or membrane from the affected joint, with respect to at least five of the following substances: increased interleukin-1 beta (IL-1beta); increased tumor necrosis factor alpha (TNFalpha); increased ratio of IL-1beta to IL-1 receptor antagonist protein (IRAP); increased expression of p55 TNF receptors (p55 TNF-R); increased interleukin-6 (IL-6); increased leukemia inhibitory factor (LIF); decreased insulin-like growth factor-1 (IGF-1); decreased transforming growth factor beta (TGFbeta); decreased platelet-derived growth factor (PDGF); decreased basic fibroblast growth factor (b-FGF); increased keratan sulfate; increased stromelysin; increased ratio of stromelysin to tissue inhibitor of metalloproteases (TIMP); increased osteocalcin; increased alkaline phosphatase; increased cAMP responsive to hormone challenge; increased urokinase plasminogen activator (uPA); increased cartilage oligomeric matrix protein; and increased collagenase.
Owner:PFIZER INC +1
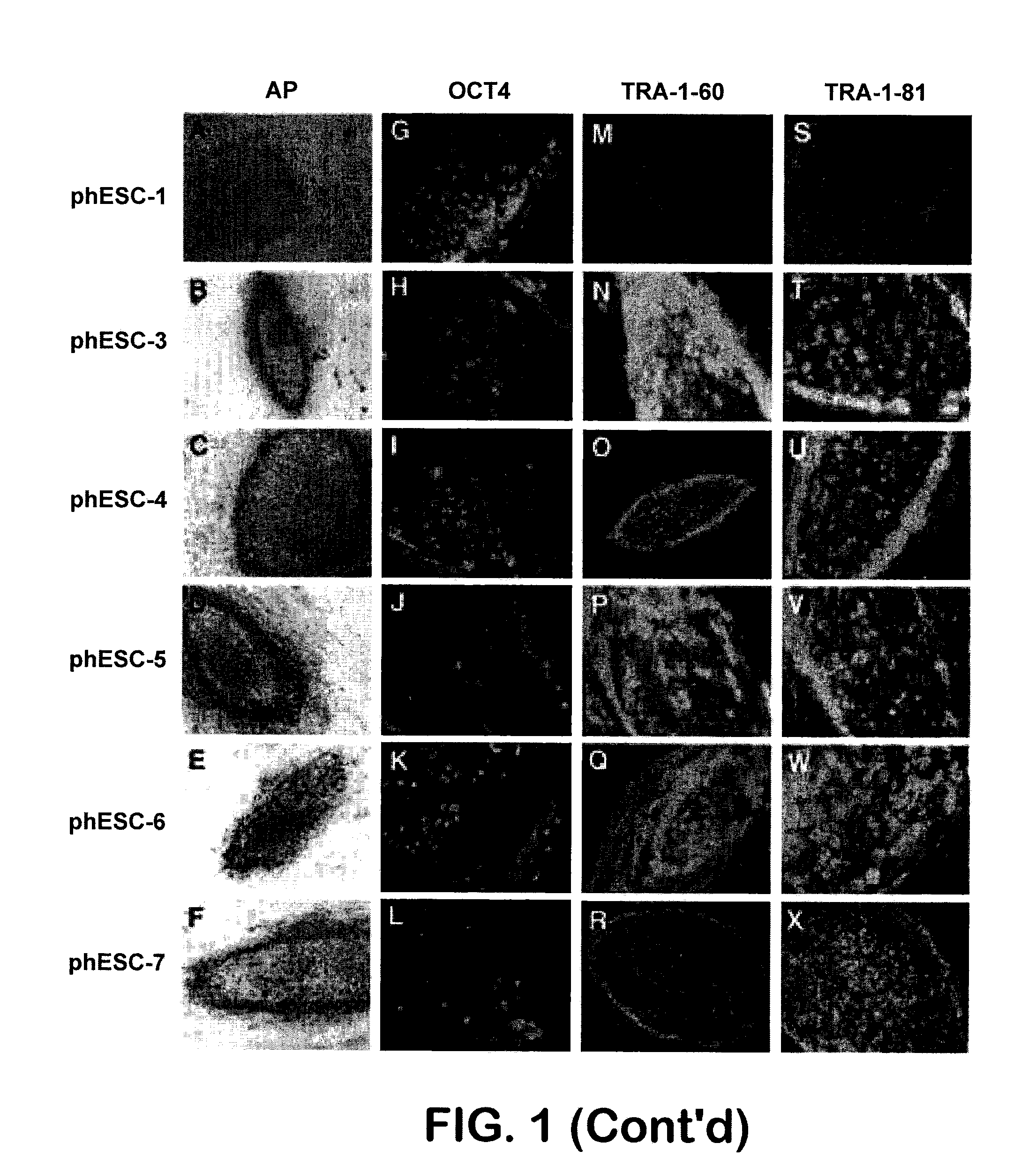
Patient-specific stem cell lines derived from human parthenogenetic blastocysts
InactiveUS20080299091A1Increase alkaline phosphatase activityHigh levelBiocideSenses disorderTelomeraseOocyte donor
Methods are disclosed for generating HLA homozygous parthenogenetic human stem cell (hpSC-Hhom) lines from both HLA homozygous and HLA heterozygous donors. These hpSC-Hhom lines demonstrate typical human embryonic stem cell morphology, expressing appropriate stem cell markers and possessing high levels of alkaline phosphatase and telomerase activity. Additionally, injection of these cell lines into immunodeficient animals leads to teratoma formation. Furthermore, in the case of HLA heterozygous donors, the hpSC-Hhom lines inherit the haplotype from only one of the donor's parents. SNP data analysis suggests that hpSC-Hhom lines derived from HLA heterozygous oocyte donors are homozygous throughout the genome as assessed by single-nucleotide polymorphism (SNP) analysis. The protocol as disclosed minimizes the use of animal-derived components, which makes the stem cells more practical for clinical application.
Owner:INT STEM CELL CORP
Emission ratiometric indicators of phosphorylation
InactiveUS6900304B2Facilitate its translocationUltrasonic/sonic/infrasonic diagnosticsFungiPhosphoamino AcidsNucleotide
A chimeric phosphorylation indicator is provided. A chimeric phosphorylation indicator can contain a donor molecule, a phosphorylatable domain, a phosphoaminoacid binding domain (PAABD), and an acceptor molecule. A chimeric phosphorylation indicator also can contain a phosphorylatable polypeptide and a fluorescent protein, wherein the phosphorylatable polypeptide is contained within the sequence of the fluorescent protein, or wherein the fluorescent protein is contained within the sequence of the phosphorylatable polypeptide. Also provided are polynucleotides encoding such chimeric phosphorylation indicators, as well as kits containing the indicators or the polynucleotides. In addition, a method of using the chimeric phosphorylation indicators to detect a kinase or phosphatase in a sample is provided.
Owner:RGT UNIV OF CALIFORNIA
Chemiluminescence quantitative detection kit for procalcitonin, and preparation method and detection method thereof
ActiveCN103901203ALow cross-reactivityImprove accuracyChemiluminescene/bioluminescenceBiotin-streptavidin complexN-Hydroxysuccinimide
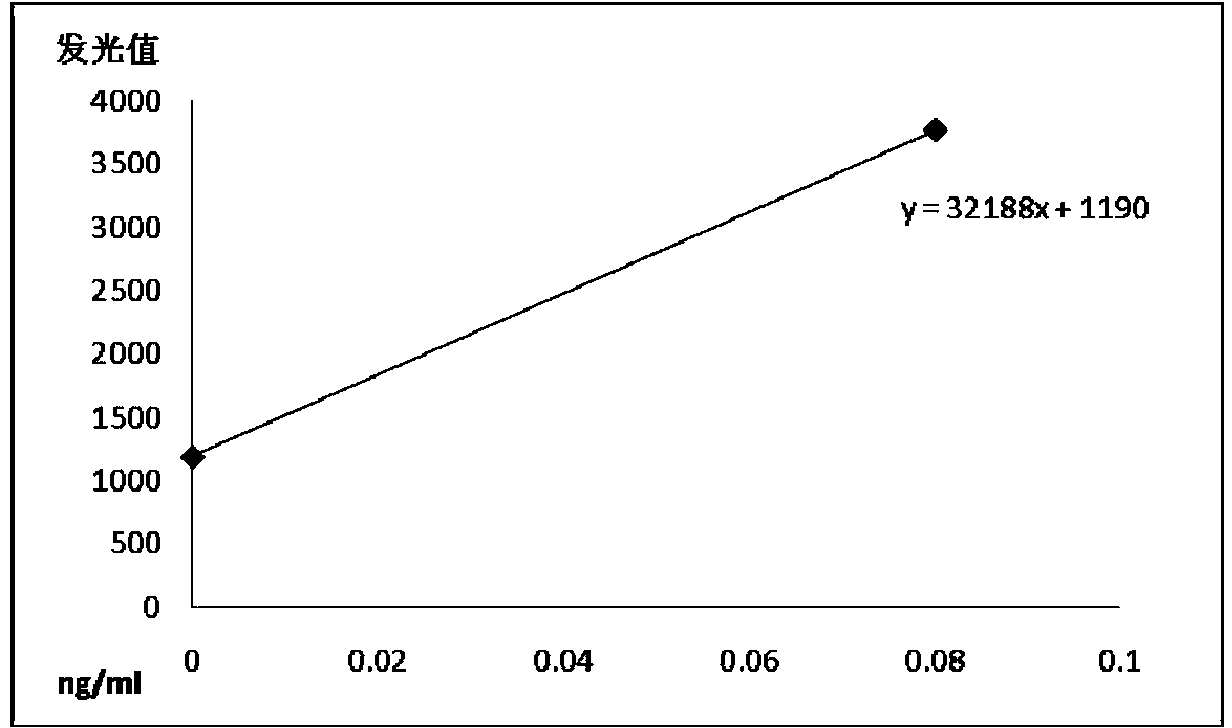
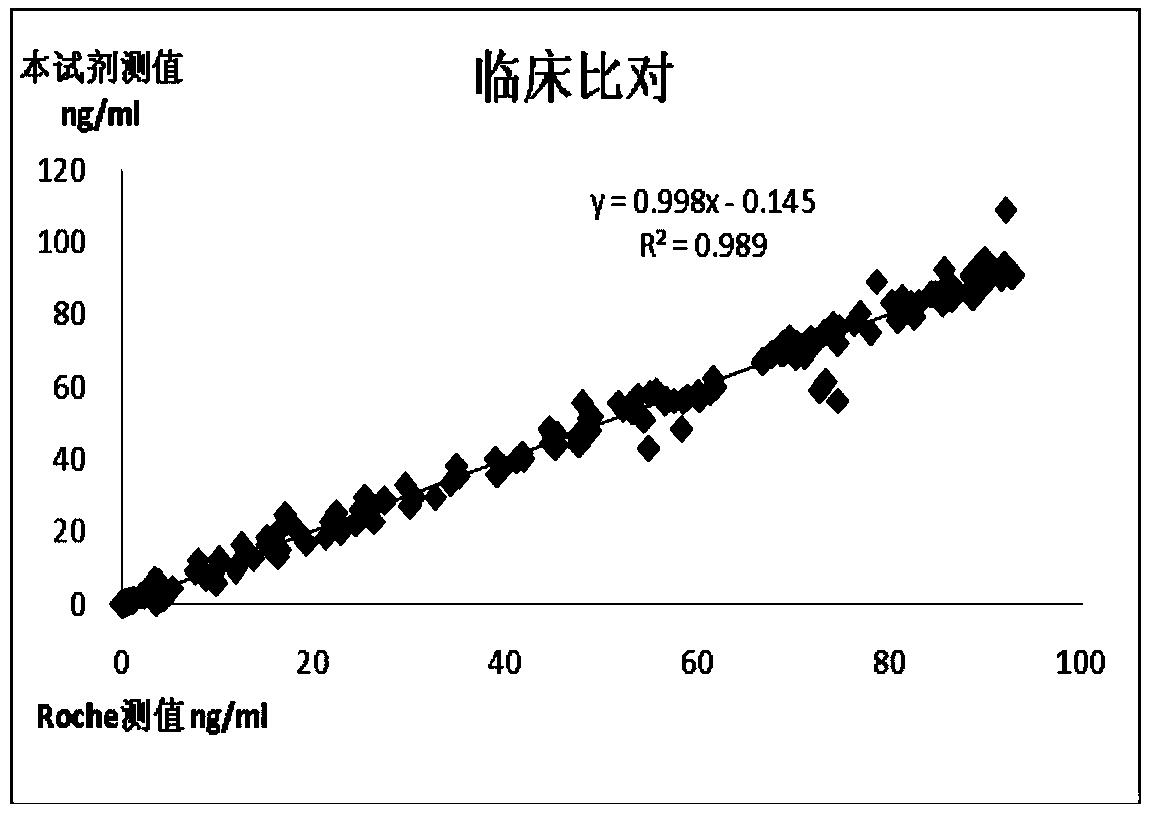
The invention relates to a chemiluminescence quantitative detection kit for procalcitonin, and a preparation method and detection method thereof. The kit comprises a procalcitonin series standard substance, a magnetic separation reagent (magnetic particle suspension coupled with streptavidin), a first reagent (anti-procalcitonin monoclonal antibody solution containing biotin N-hydroxysuccinimide ester label) and a second reagent (anti-procalcitonin monoclonal antibody solution containing alkaline phosphatase label). The sensitivity of the kit prepared from the magnetic particle suspension coupled with streptavidin, anti-procalcitonin monoclonal antibody solution containing biotin N-hydroxysuccinimide ester label and anti-procalcitonin monoclonal antibody solution containing alkaline phosphatase label is up to 0.008ng / ml; and the kit has the advantages of high accuracy, high precision, no need of prediluting the sample, and wide detection range, and is simple and time-saving to operate.
Owner:SUZHOU HAOOUBO BIOPHARML
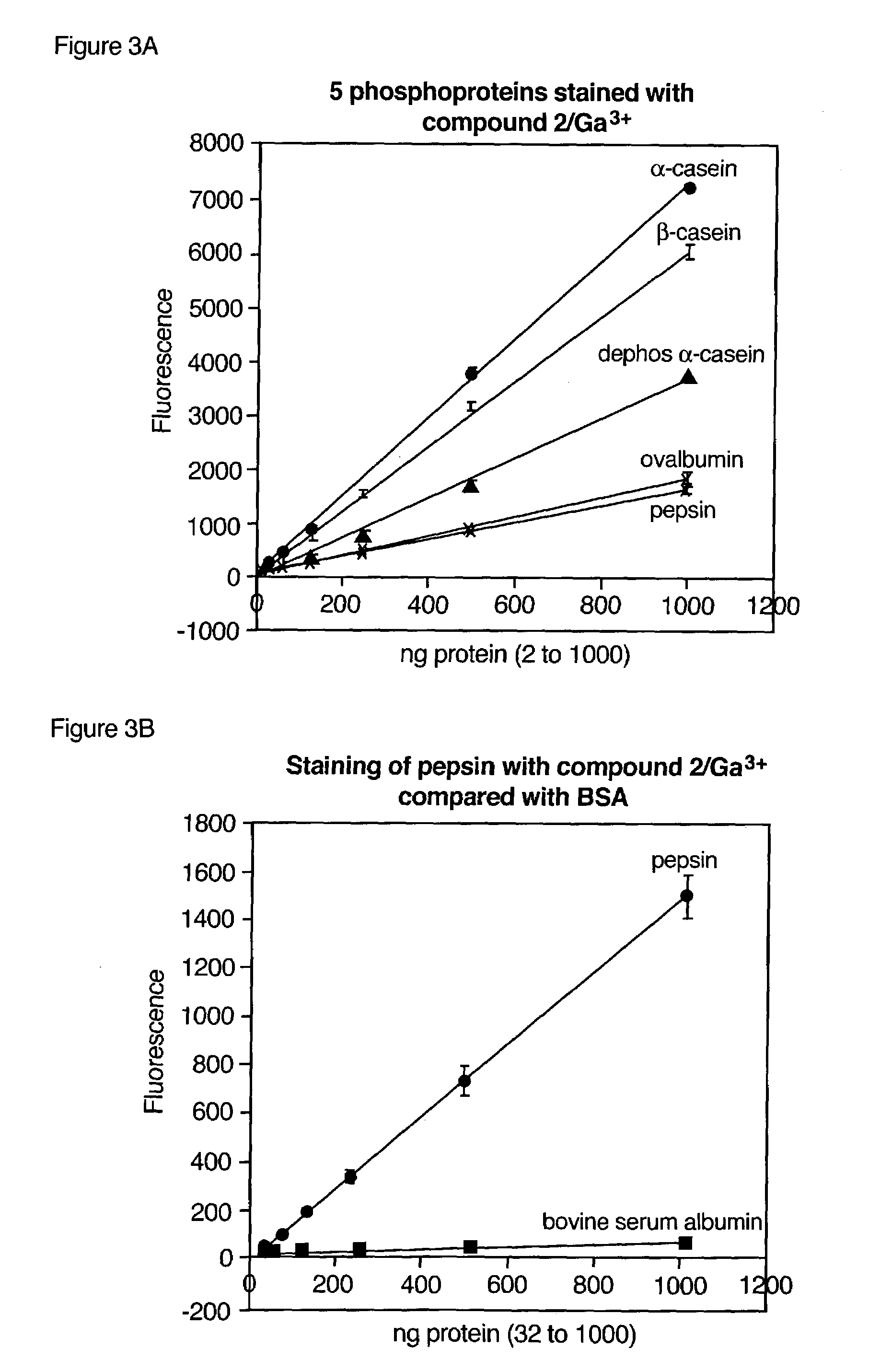
Compositions and methods for detection and isolation of phosphorylated molecules
ActiveUS20050014197A1Simplifies subsequent analysisSugar derivativesPeptide/protein ingredientsTernary complexPhosphorylation
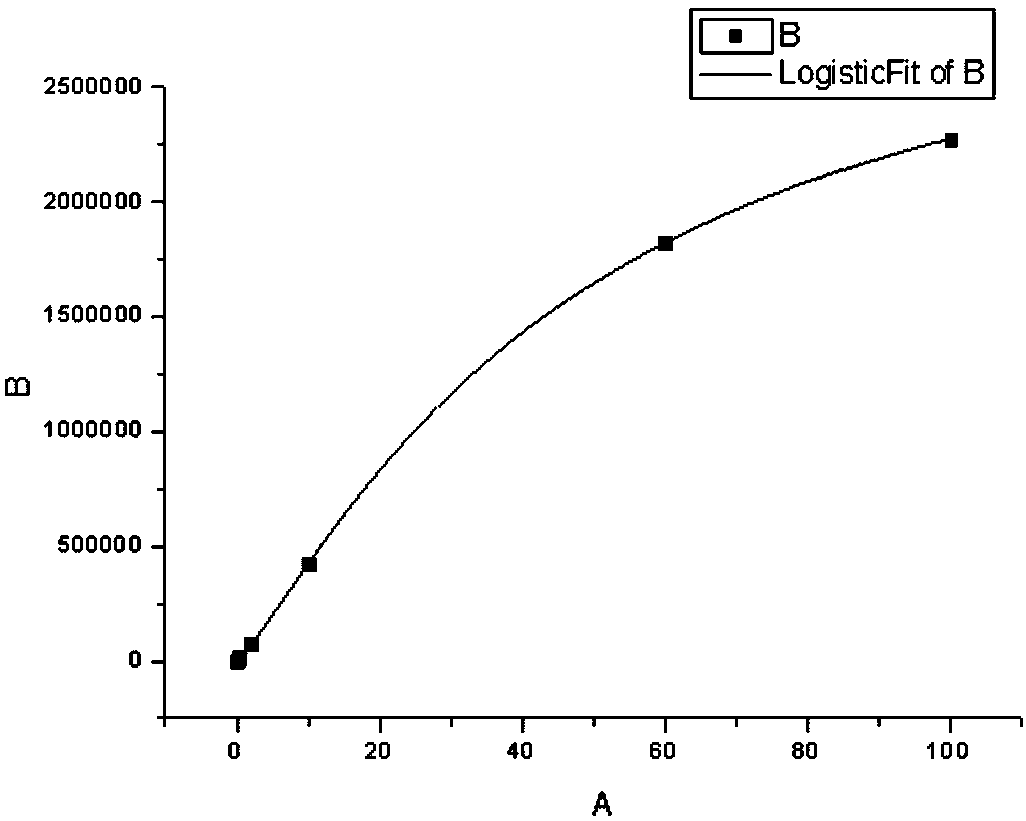
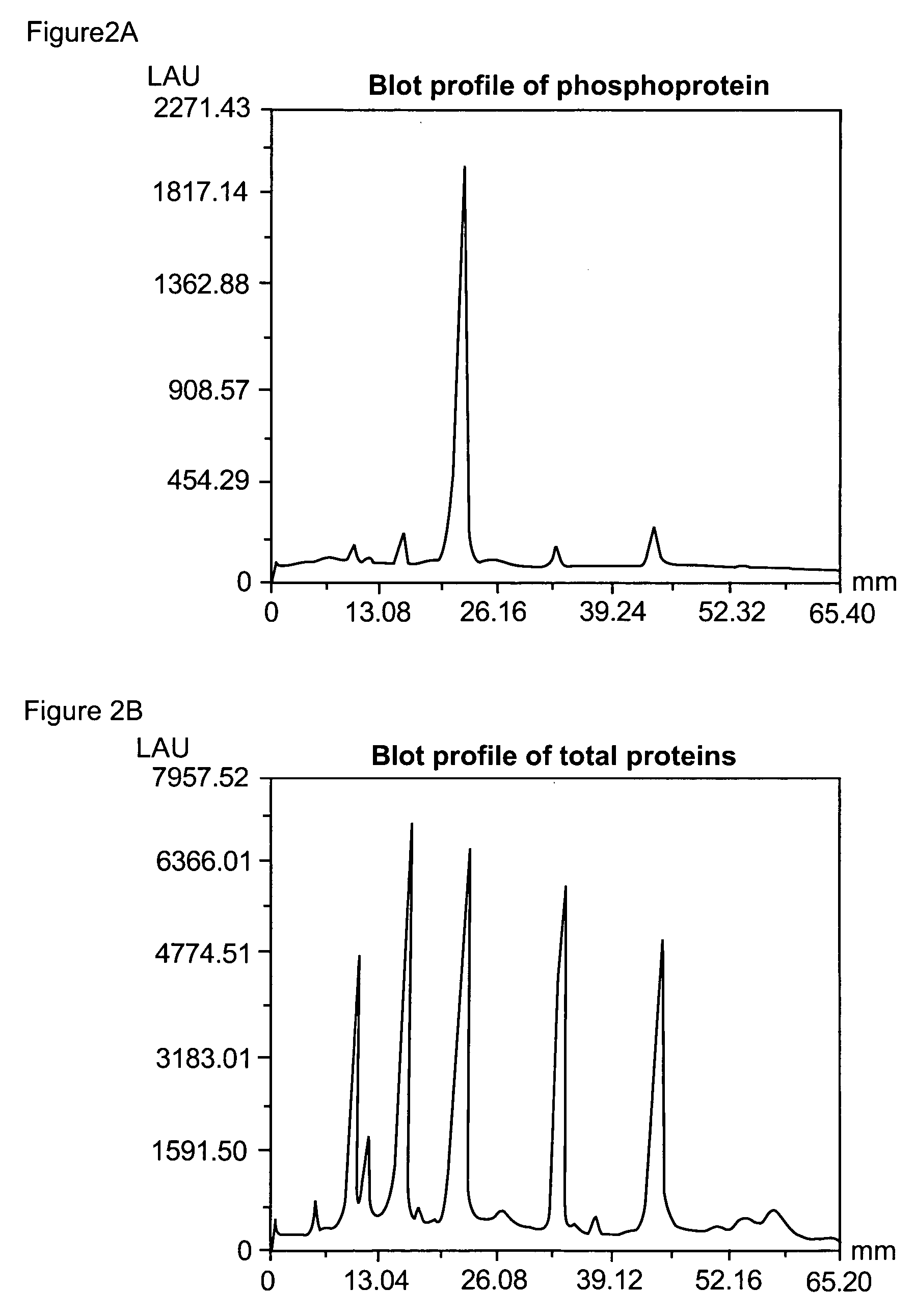
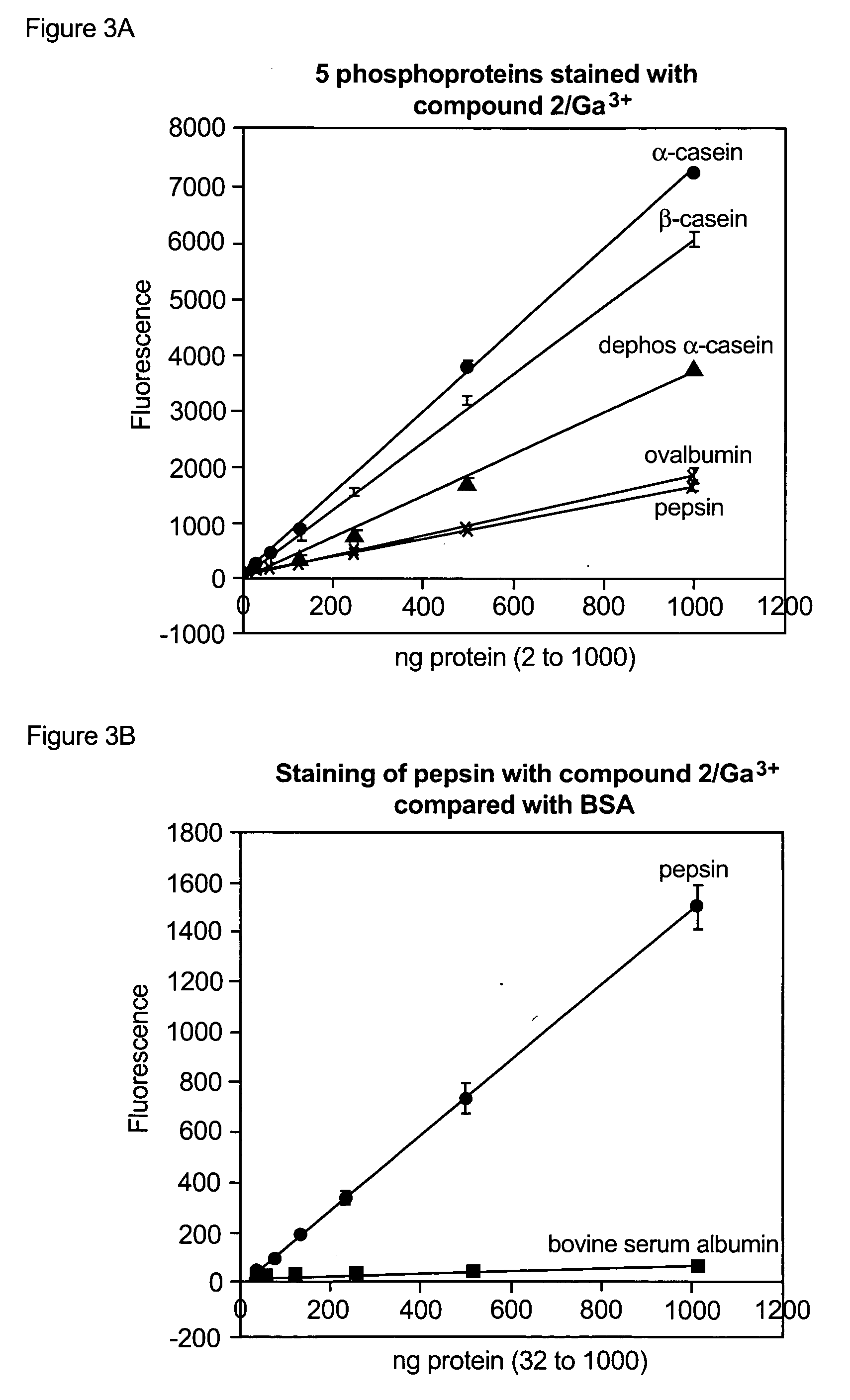
The present invention relates to phosphate-binding compounds that find use in binding, detecting and isolating phosphorylated target molecules including the subsequent identification of target molecules that interact with phosphorylated target molecules or molecules capable of being phosphorylated. A binding solution is provide that comprises a phosphate-binding compound, an acid and a metal ion wherein the metal ion simultaneously interacts with an exposed phosphate group on a target molecule and the metal chelating moiety of the phosphate-binding compound forming a bridge between the phosphate-binding compound and a phosphorylated target molecule resulting in a ternary complex. The binding solution of the present invention finds use in binding and detecting immobilized and solubilized phosphorylated target molecules, isolation of phosphorylated target molecules from a complex mixture and aiding in proteomic analysis wherein kinase and phosphatase substrates and enzymes can be identified.
Owner:MOLECULAR PROBES
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com