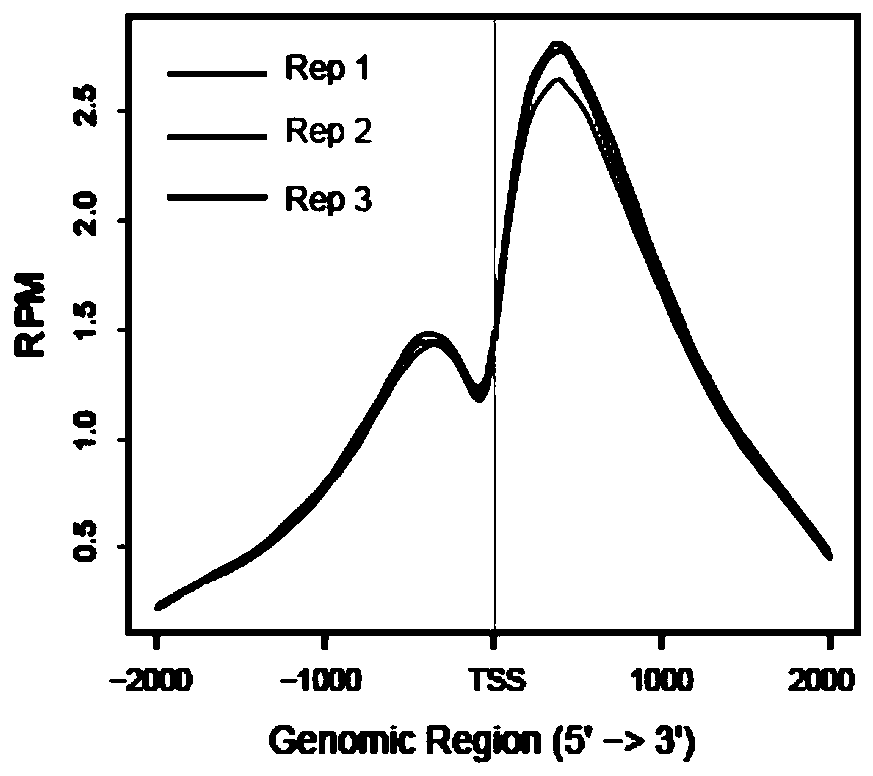
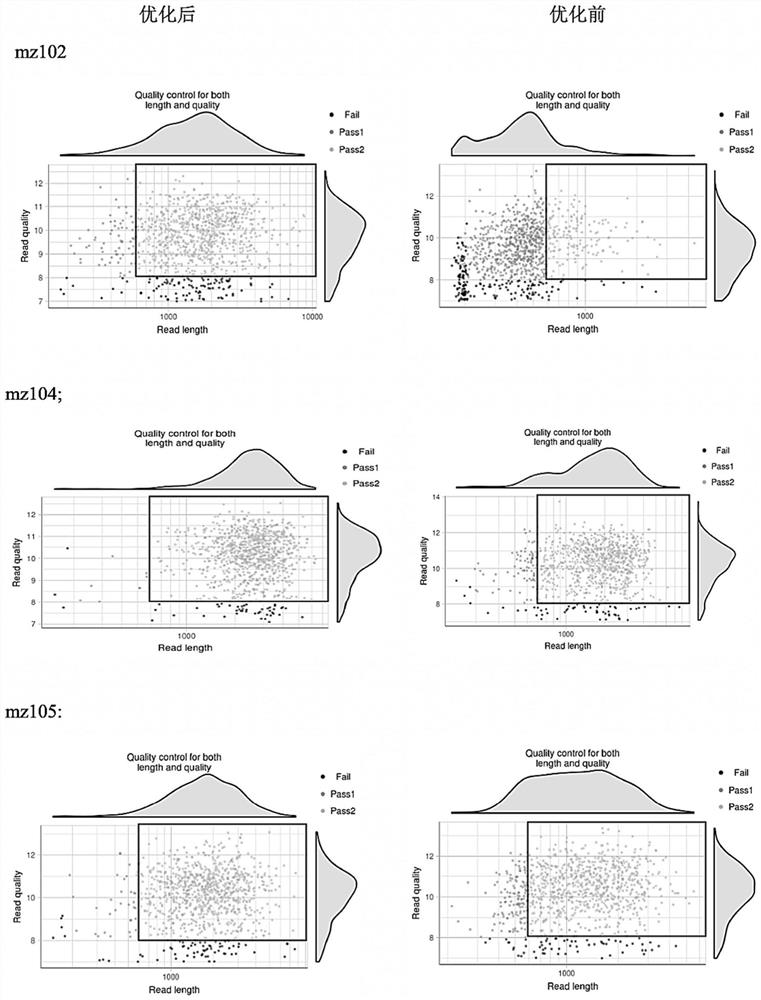
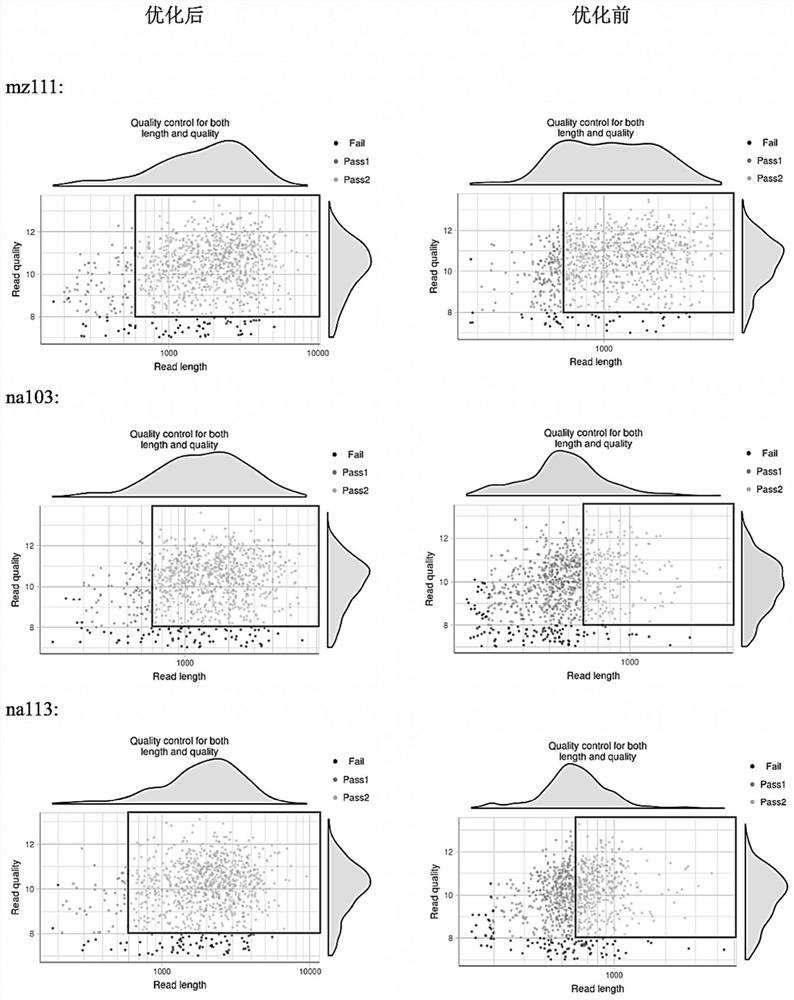
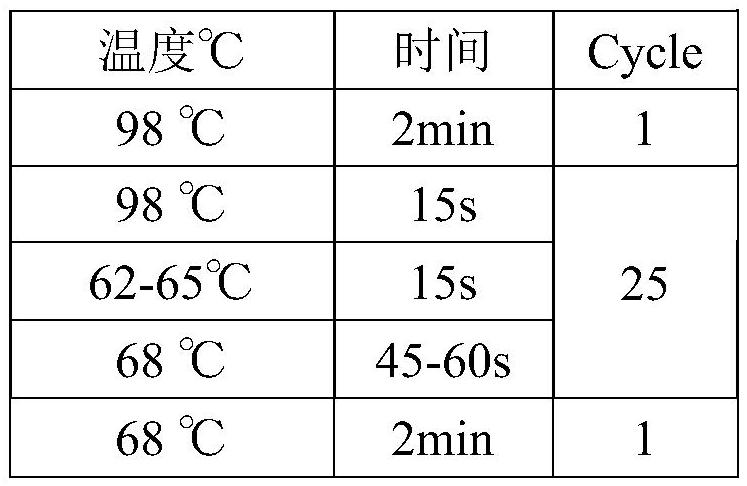
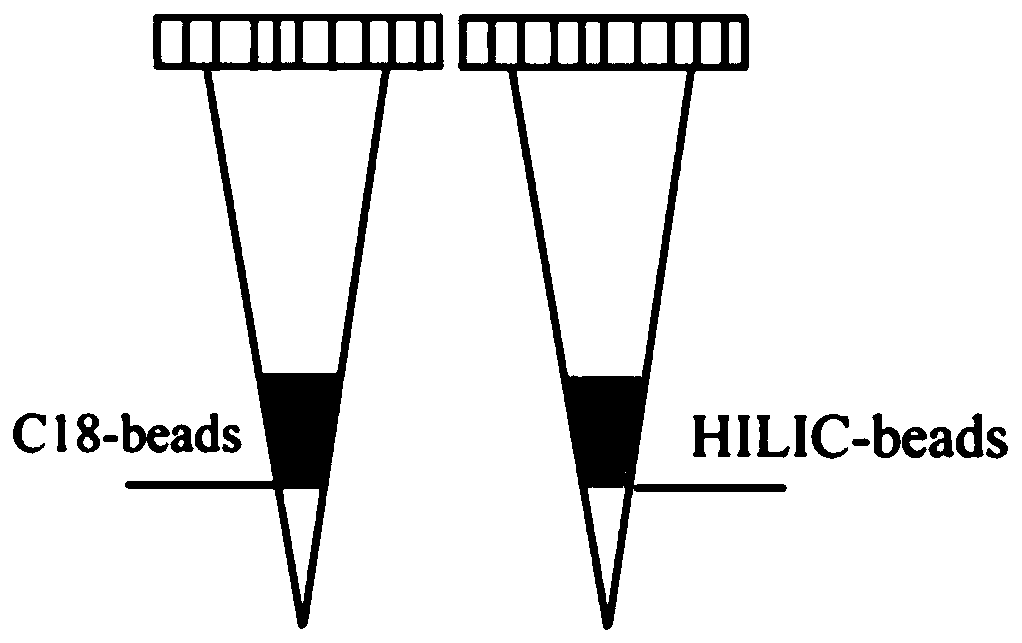
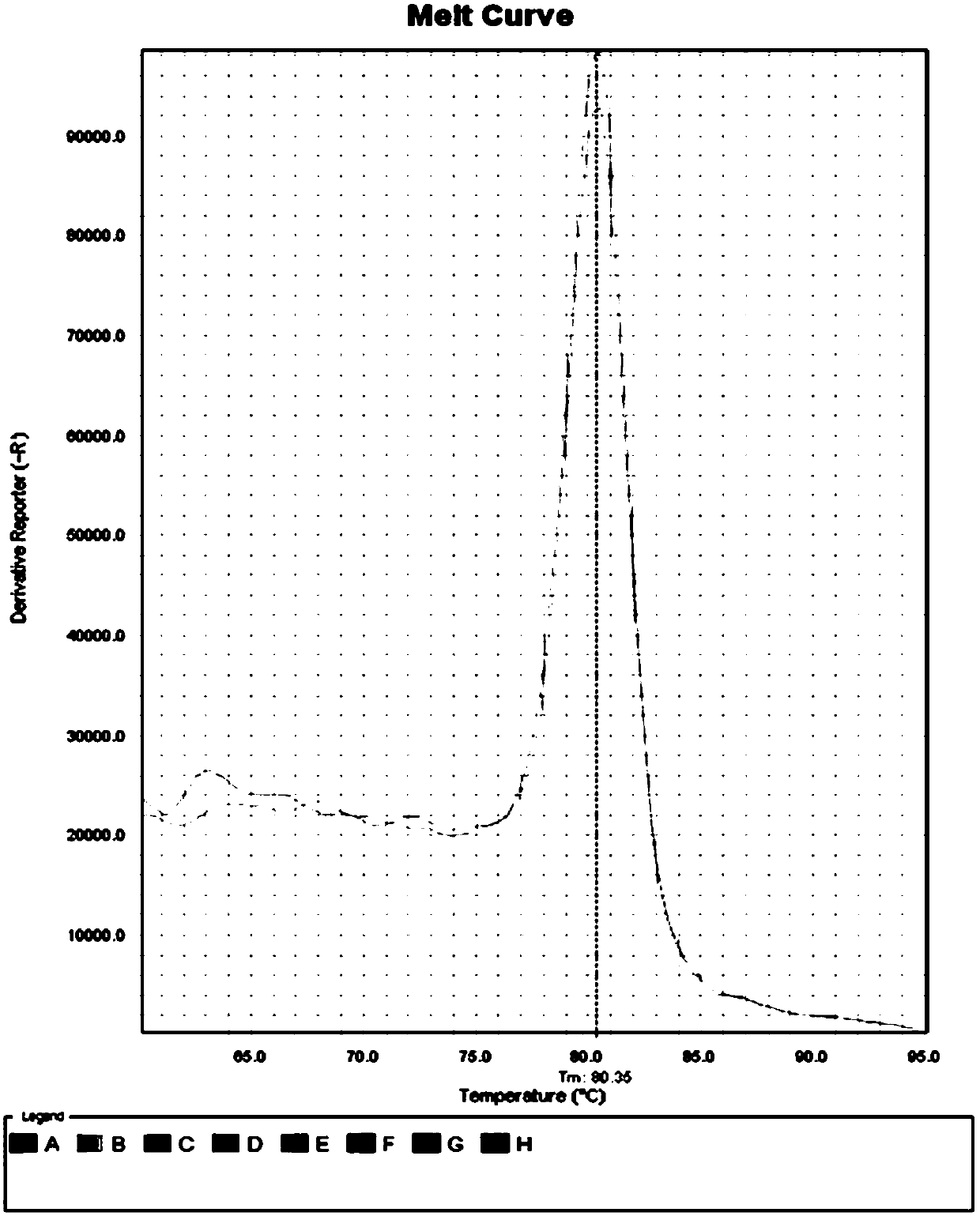
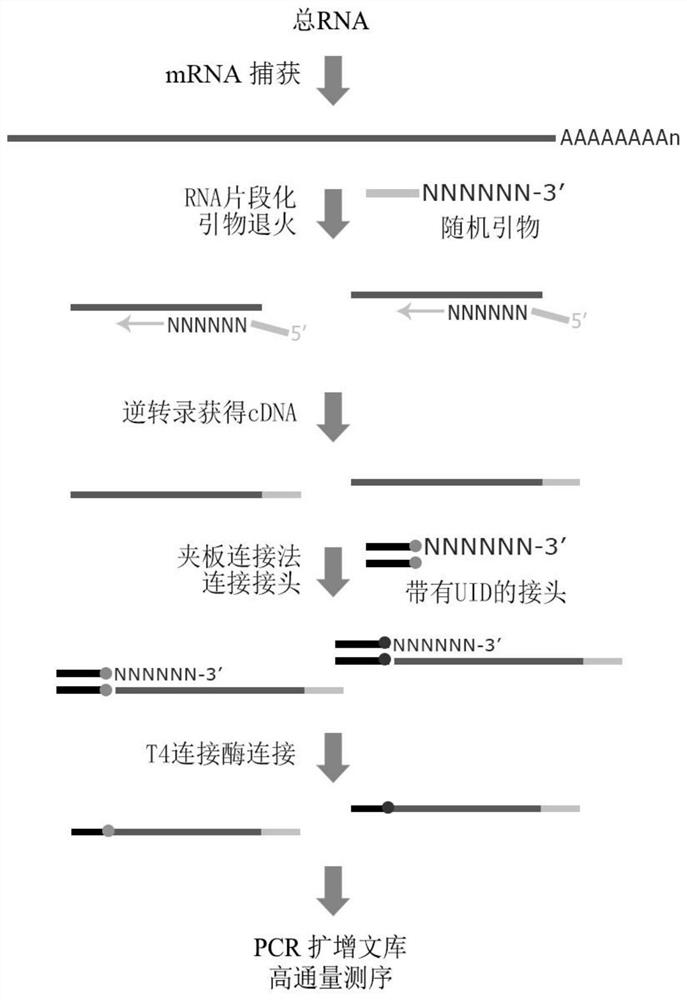
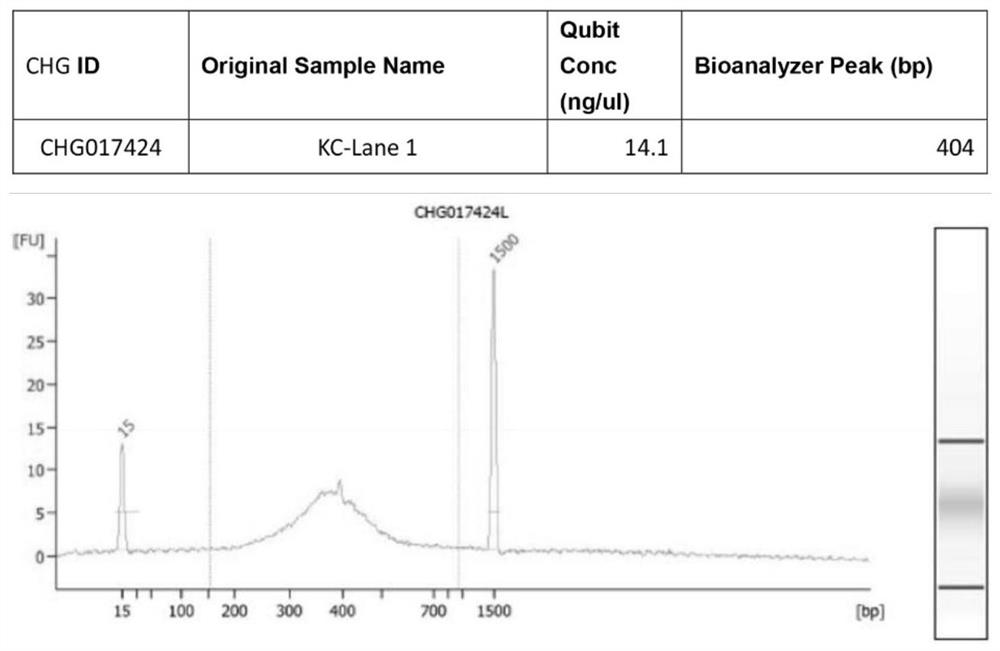
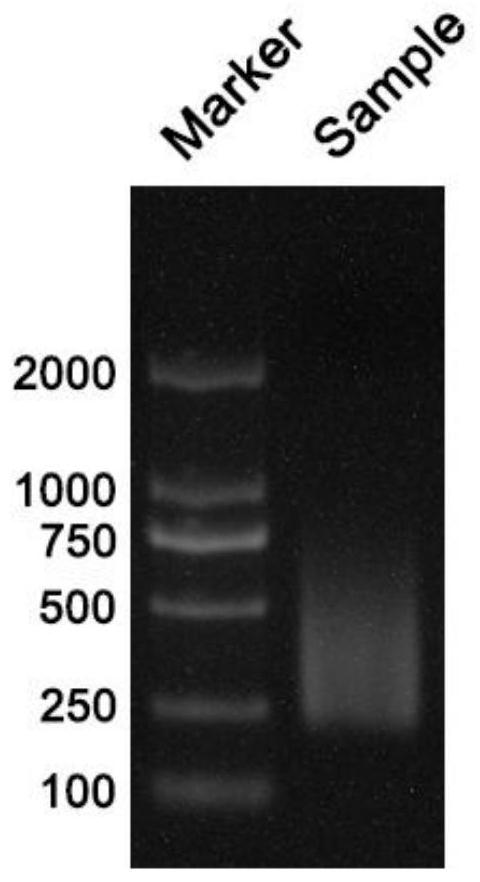
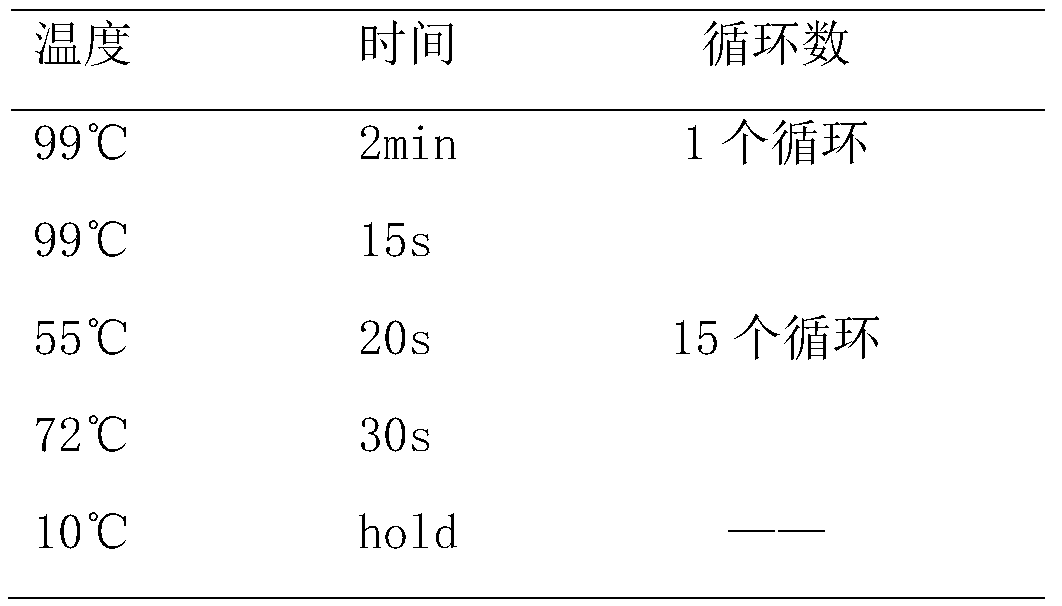
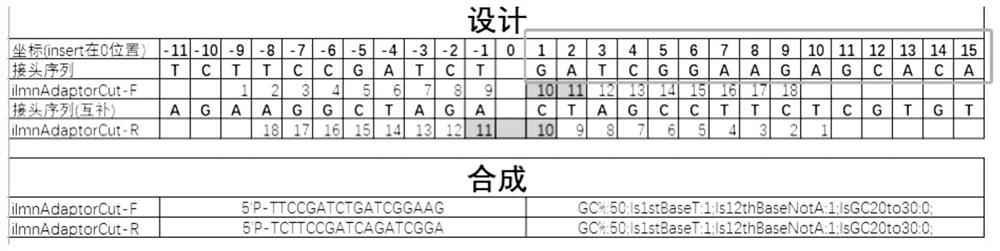
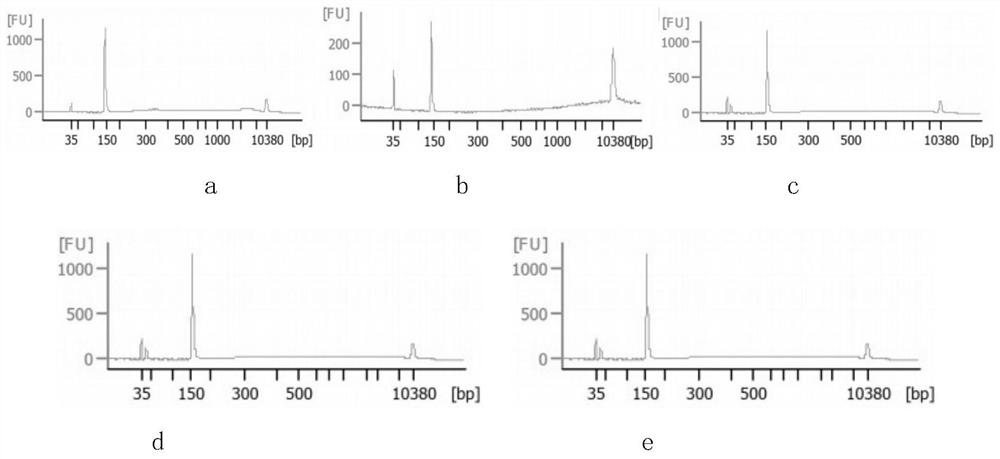
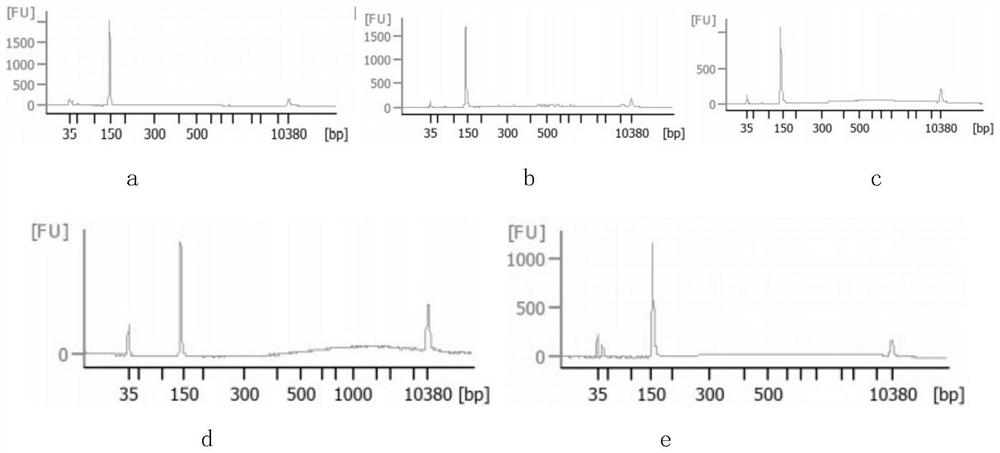
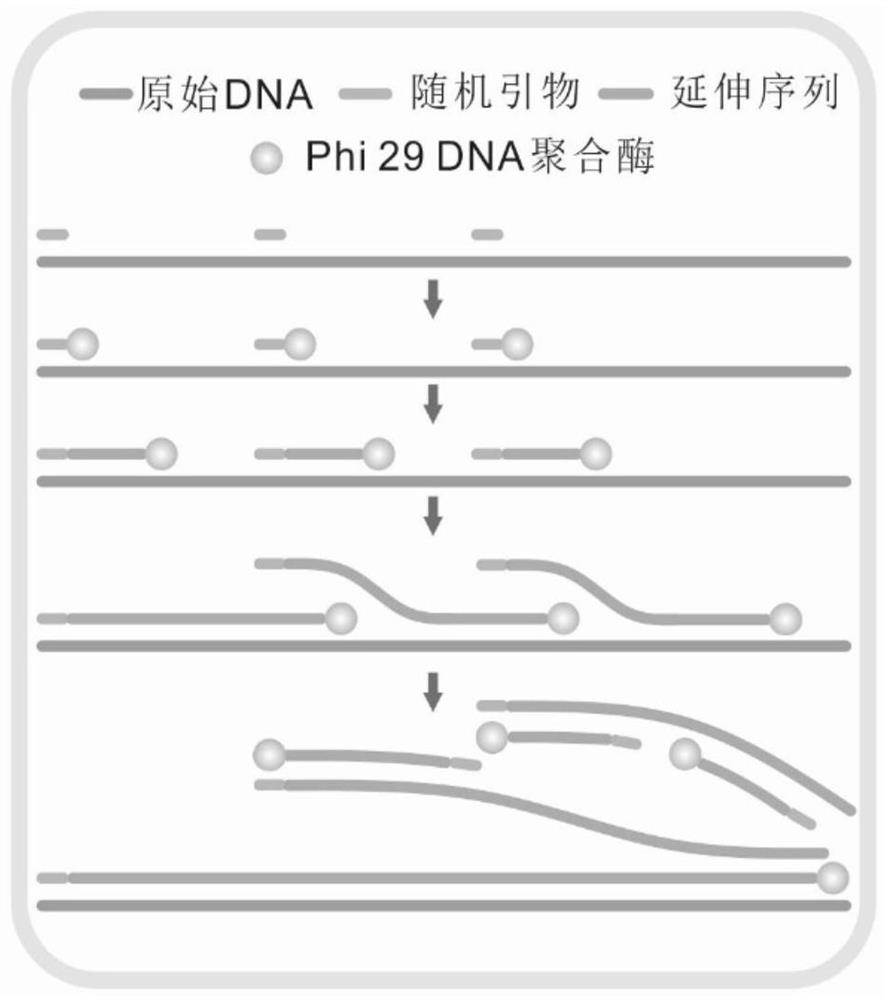
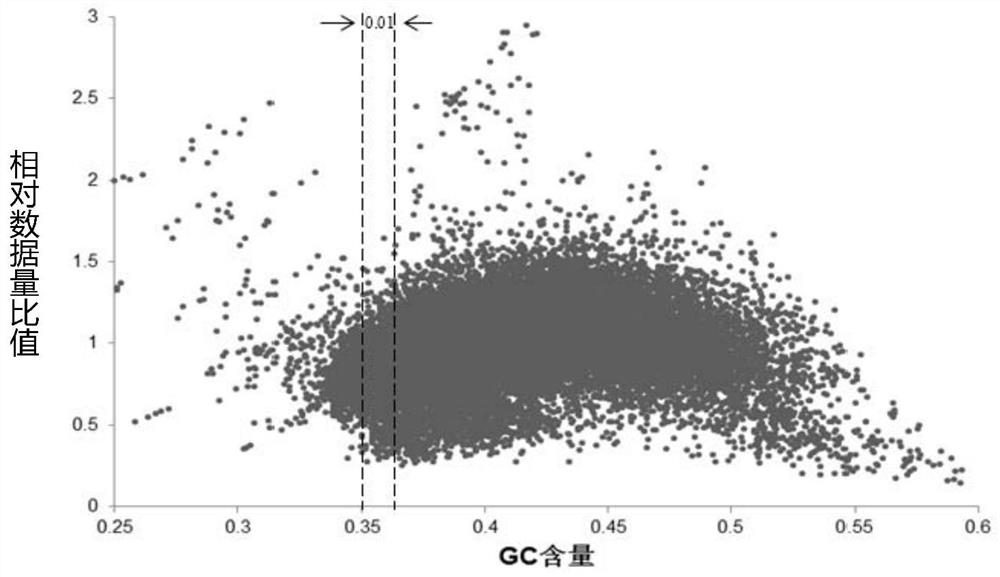
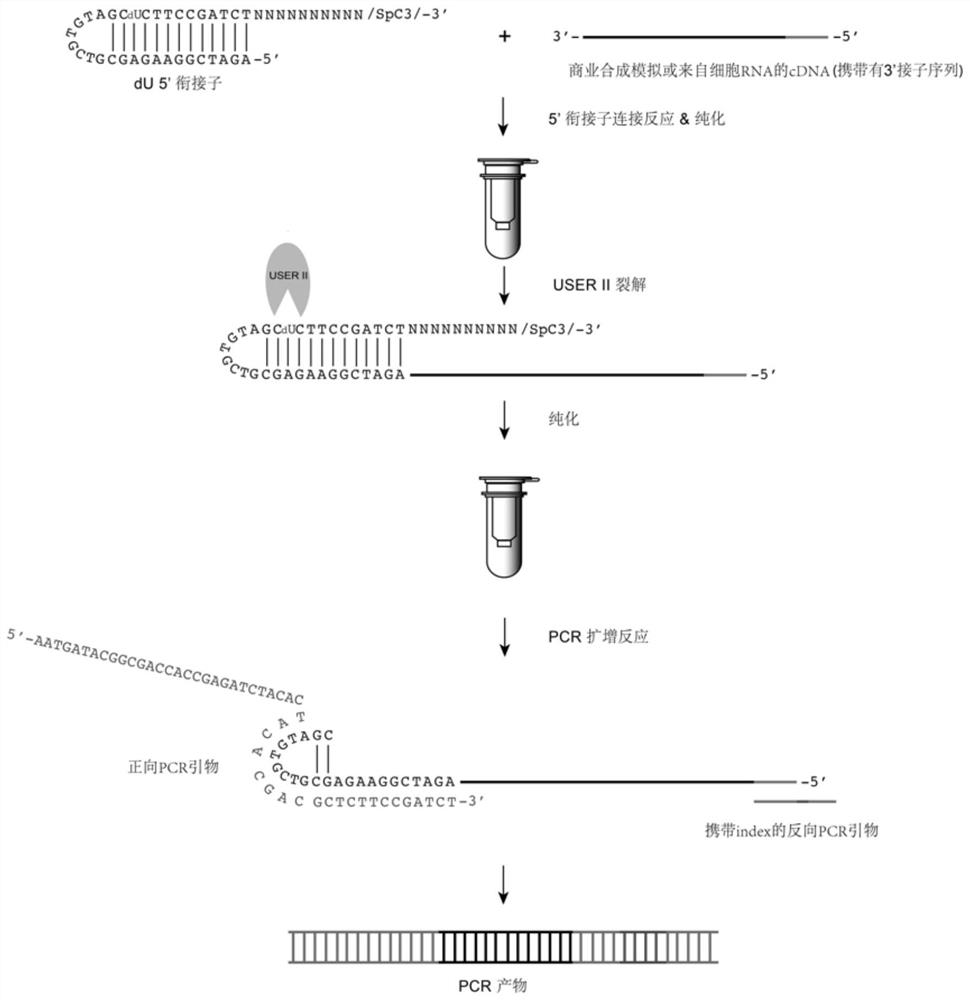
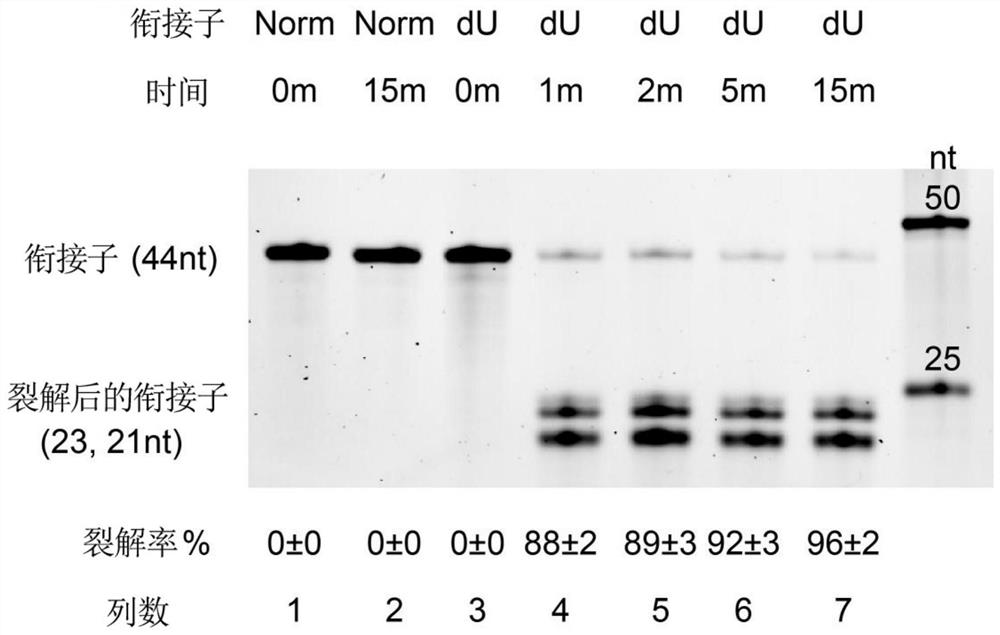
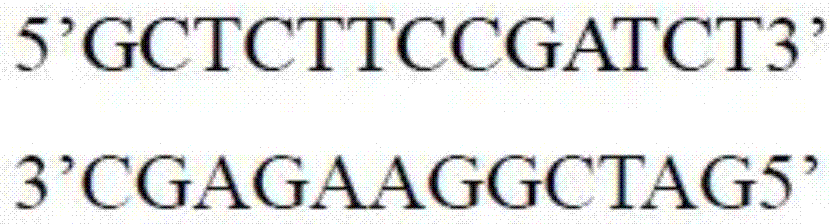Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
46results about How to "Reduce the starting amount" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Sequencing library building method and reagent based on molecular inverse probe
ActiveCN105714383AReduce errorsSimple processMicrobiological testing/measurementLibrary creationExonuclease IReagent
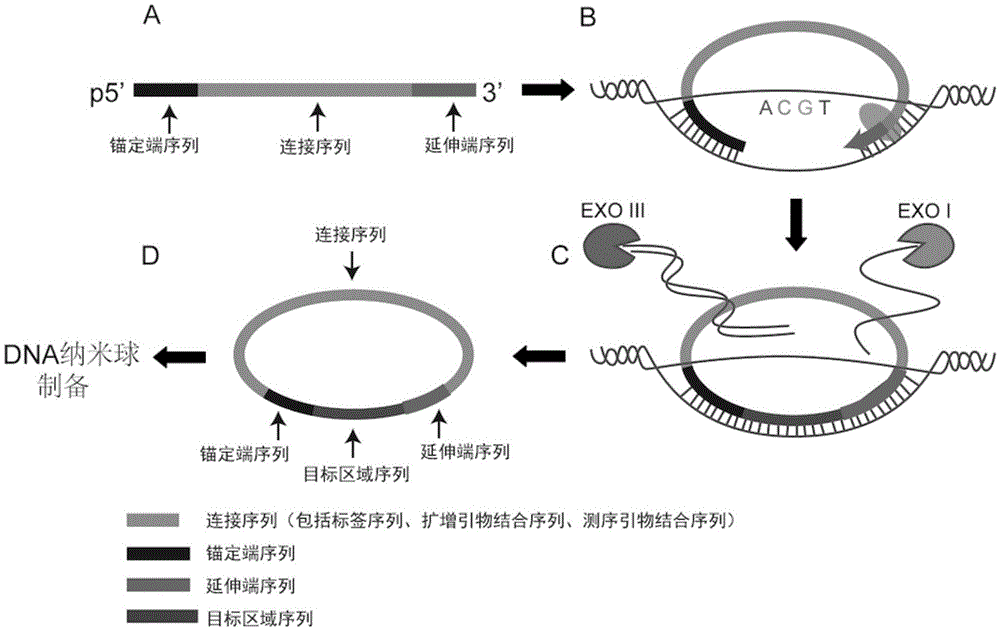
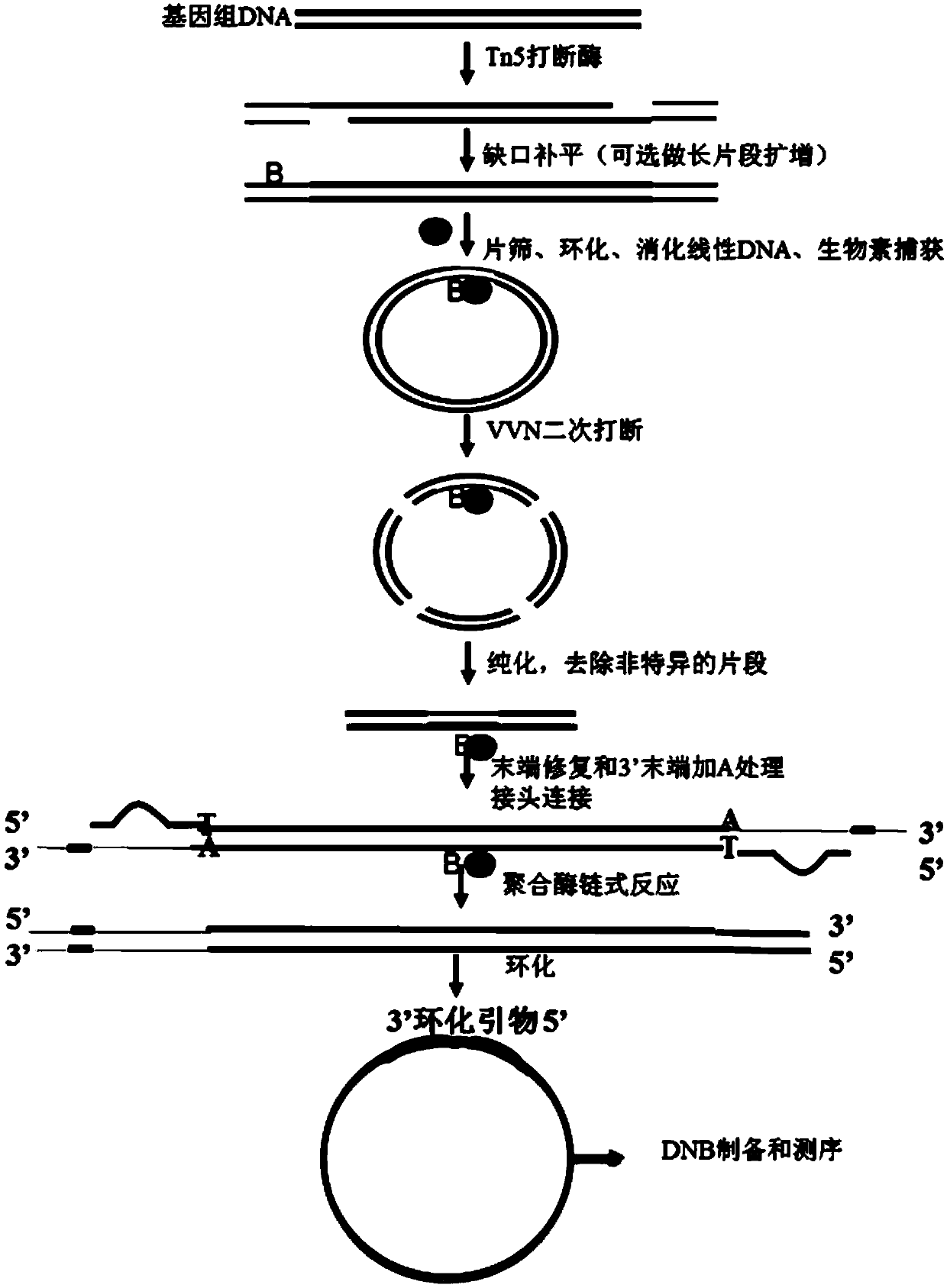
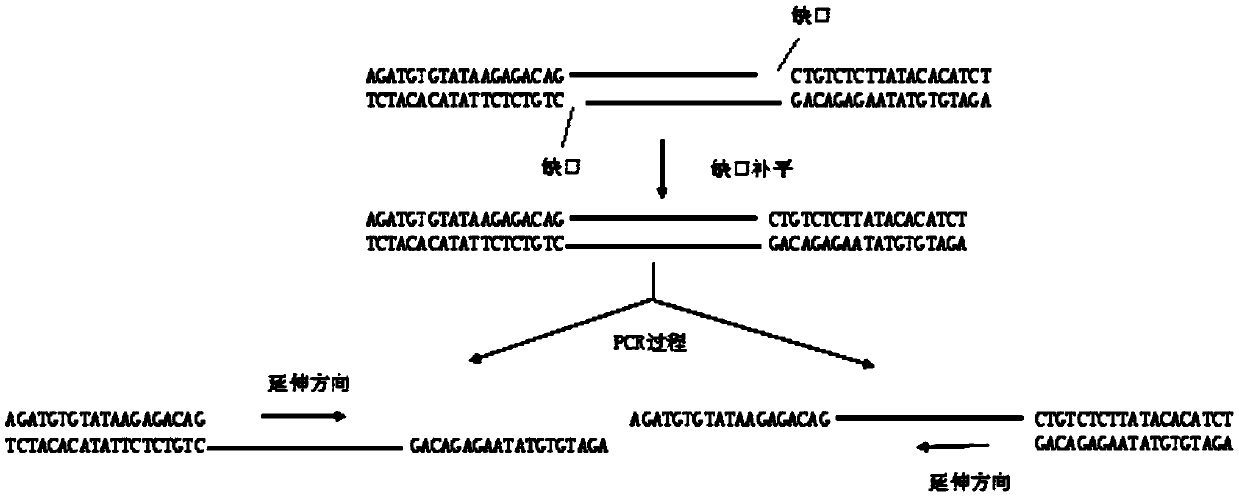
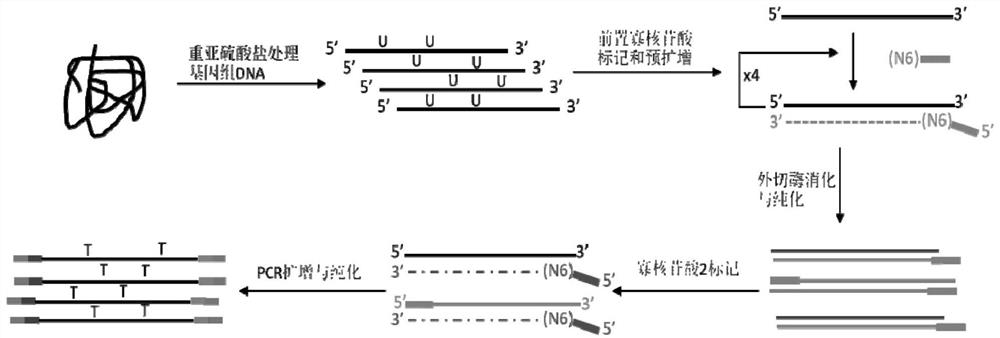
The invention discloses a sequencing library building method and reagent based on a molecular inverse probe. The method includes: using the molecular inverse probe and denatured nucleic acid to perform annealing hybridization, wherein the molecular inverse probe comprises the anchoring sequence of a 5' end, the extension sequence of a 3' end and the sequencing connector sequence between the anchoring sequence and the extension sequence, and the anchoring sequence and the extension sequence are respectively inversely complementary with sequences at two ends of a target area; using the target area as a template, and performing polymerization reaction starting from the extension sequence of the 3' end to generate the supplementary sequence of the target area; connecting the anchoring sequence of the 5' end with the supplementary sequence of the target area to form cyclic annular nucleic acid molecules; using exonuclease to digest non-cyclized linear molecules to obtain single-chain cyclic annular molecules containing the target area; sequencing the obtained single-chain cyclic annular molecules so as to achieve capture sequencing of the target area. The sequencing library building method has the advantages that the building of a single-chain cyclic annular library and target area capture are integrated, and library building process and cycle are shortened greatly.
Owner:MGI TECH CO LTD
Construction method of lung cancer polygenic variation library
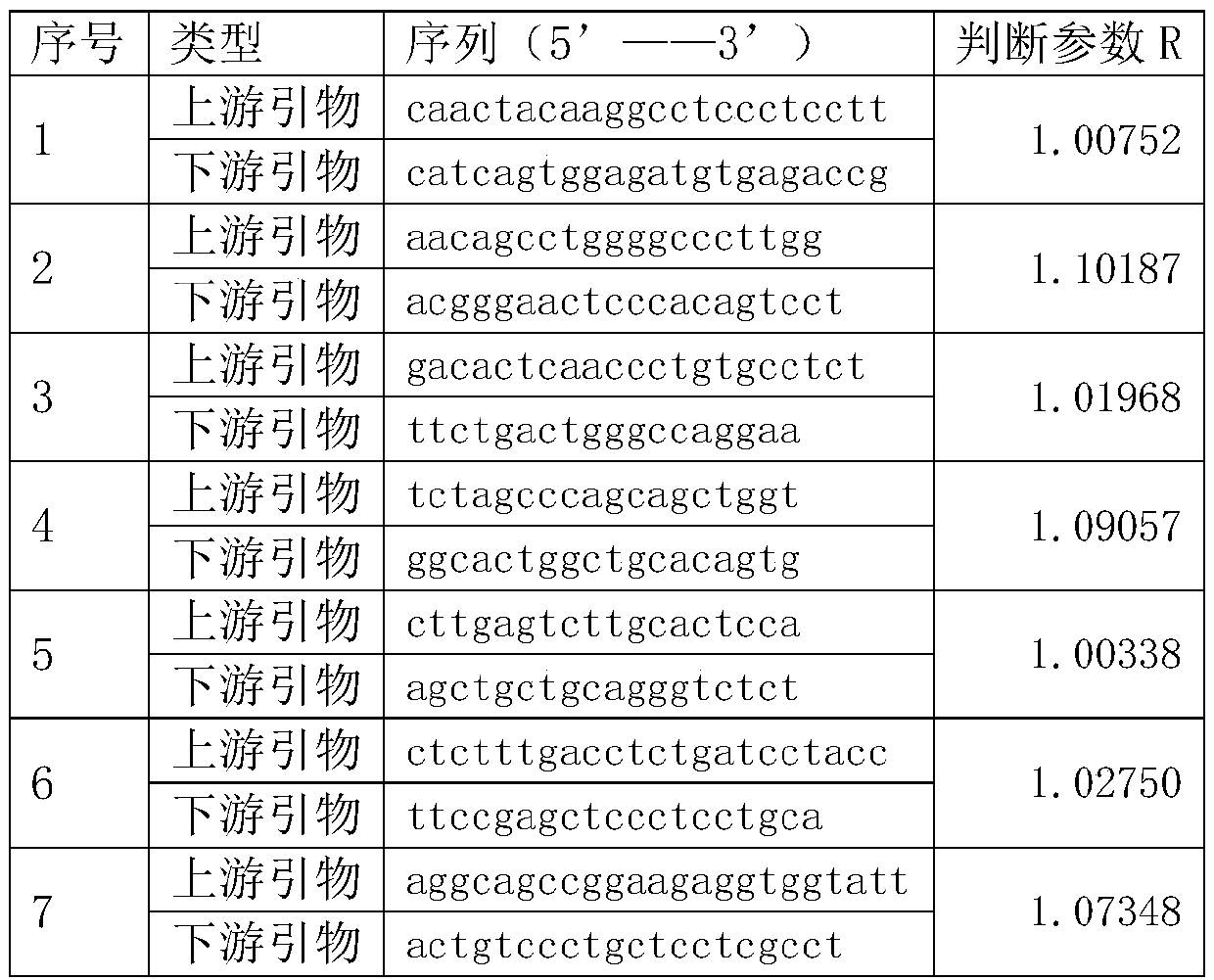
ActiveCN106757379AImprove throughputLow costNucleotide librariesMicrobiological testing/measurementLung cancerBiology
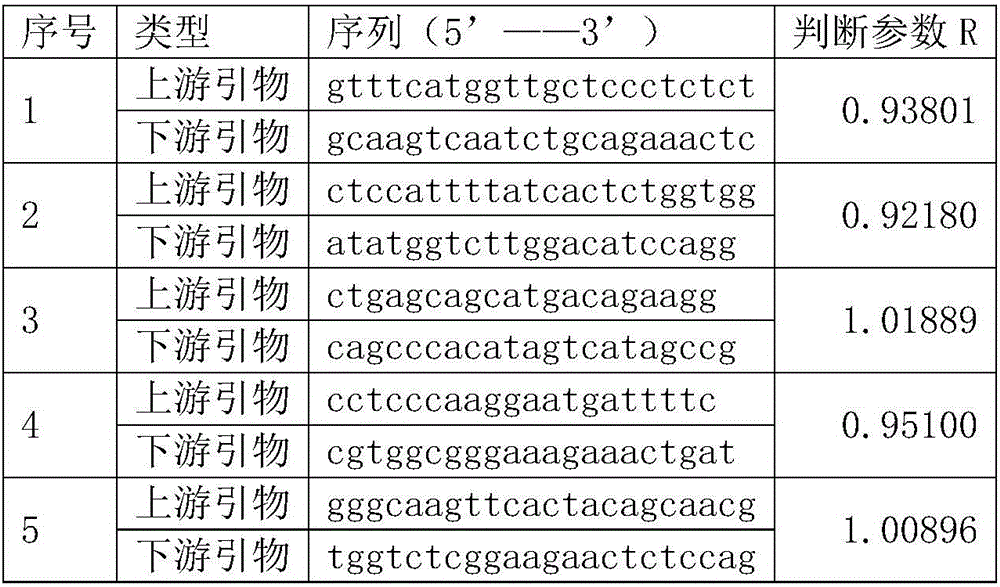
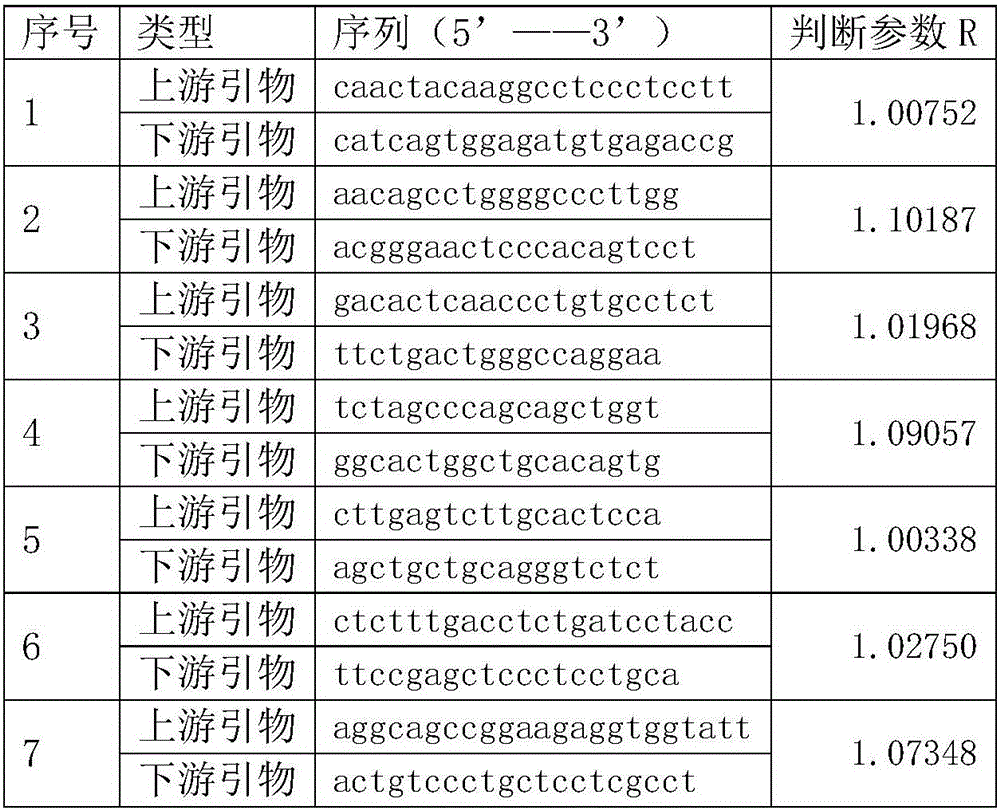
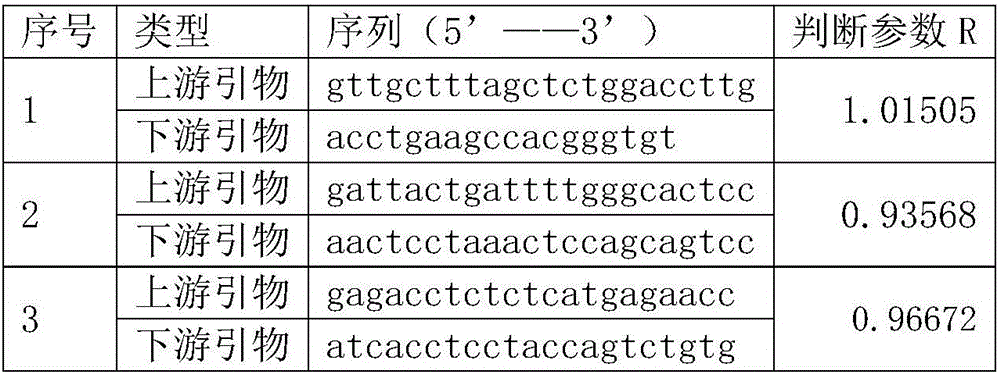
The invention relates to a construction method of a lung cancer polygenic variation library, comprising the steps of A, designing a primer pair capable of detecting variation areas of lung cancer related genes according to these variation areas; B, calculating to obtain judging parameters R of the primer pairs, classifying the primer pairs with the judging parameters R less than or equal to 1 as a first group of primer combination liquids, and the primer pairs with the judging parameters R greater than 1 as a second group of primer combination liquids; C, extracting and purifying sample DNA to be detected; D, subjecting the purified sample to initial PCR (polymerase chain reaction) amplification with the first group of primer combination liquids and the second group of primer combination liquids respectively; E, connecting a linker to a target fragment to obtain a fragment with the linker; F, subjecting a mixed liquid of the first group of primer combination liquids and the second group of primer combination liquids to library PCR amplification to obtain a sequencing library. The library constructed by the construction method has high sequencing throughput, high sensitivity and high specificity, and can detect low-frequency variations of free DNA.
Owner:上海赛安生物医药科技股份有限公司
Group of thyroid cancer markers and application thereof
ActiveCN110878358AIn-depth analysis of molecular level informationWide coverageMicrobiological testing/measurementDNA/RNA fragmentationOncologyMolecular typing
The invention provides a group of thyroid cancer markers and an application thereof. The markers comprise thyroid cancer molecular typing related genes, targeted medication related genes, chemotherapymedication related genes, operation prompt related genes, prognosis related genes and thyroid cancer heredity related genes. According to the invention, aiming at all exon regions of 44 genes relatedto thyroid molecular typing, targeted medication, chemotherapy medication, operation prompt, prognosis and heredity, high-depth sequencing is carried out; meanwhile, various variation types like SNV / Indel and gene fusion of genes are analyzed; and molecular level information of thyroid nodules and thyroid cancer is deeply analyzed. Aiming at a capture probe designed for the 44 genes, 558 targeting target regions are covered; the probe comprises 3633 sequences with a total size of 254.096 Kbp; and a prepared kit is wide in coverage, high in cost performance and high in effectiveness, can provide a reference basis for further molecular typing, medication prompt, genetic risk assessment and the like of a thyroid patient, and is applicable to clinical popularization and application.
Owner:SHANGHAI BIOTECAN PHARMA +1
Nucleic acid library construction method, nucleic acid library obtained through the method, and application thereof
PendingCN111379031AReduce the starting amountNucleotide librariesMicrobiological testing/measurementMagnetic beadSingle strand
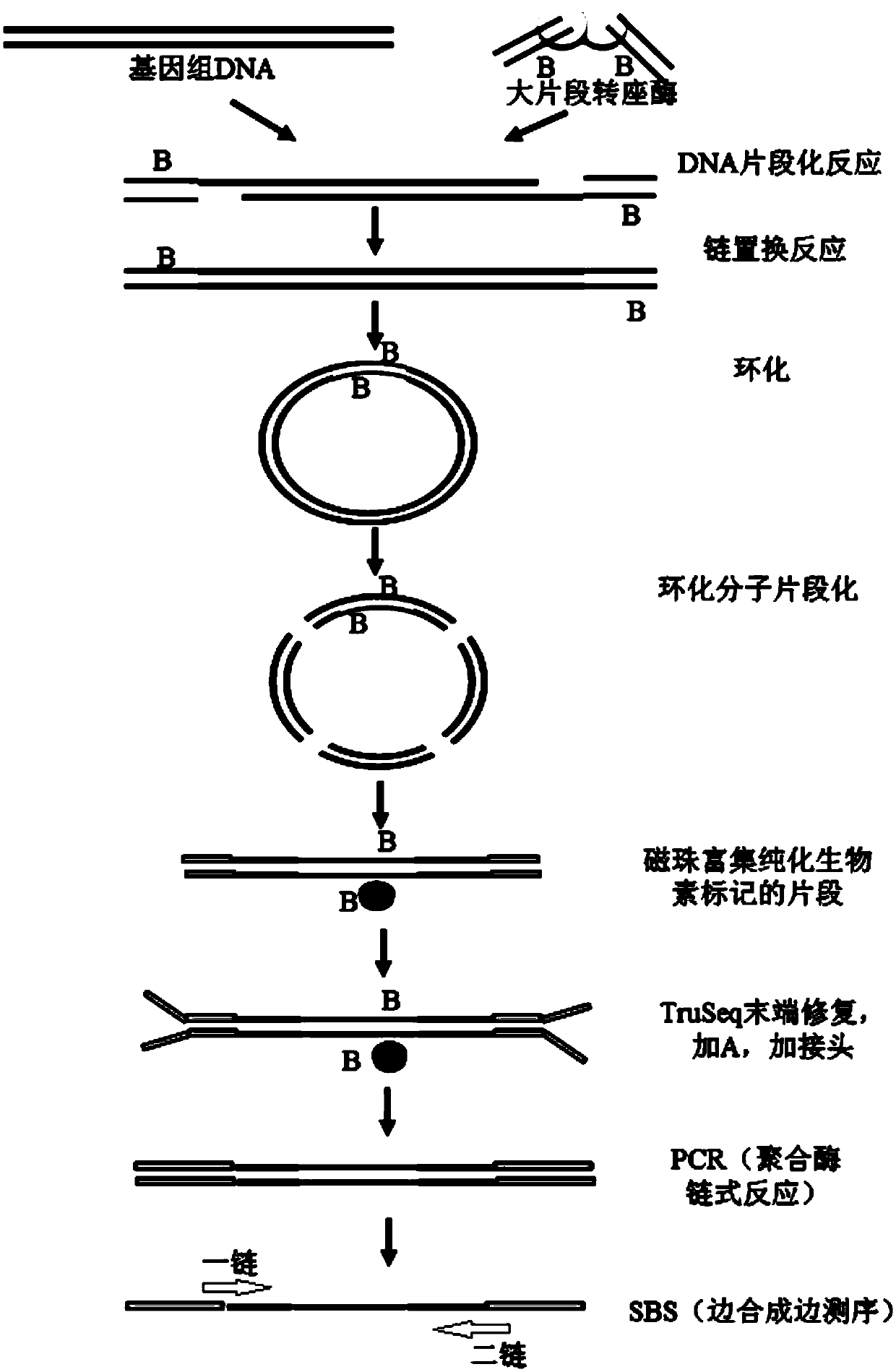
The invention discloses a nucleic acid library construction method, the nucleic acid library obtained through the method, and application thereof. The method comprises the steps: breaking the nucleicacid into fragments through transposase, meanwhile, connecting transposon sequences onto the two ends of the broken fragments, wherein the transposon sequences carry biotin labels; filling a gap between the transposon sequence and the broken segment through strand displacement reaction to obtain a complete double-stranded nucleic acid segment; cyclizing the double-stranded nucleic acid fragment obtained in the previous step into a cyclic nucleic acid fragment; combining streptomycin affinity magnetic beads with the cyclic nucleic acid fragments with biotin labels; breaking cyclic nucleic acidfragments combined on streptomycin affinity magnetic beads by using a breaking enzyme, and carrying out magnetic separation to obtain fragments combined on the streptomycin affinity magnetic beads. The method aims at a large-fragment library, meets the requirement of low initial quantity, can thoroughly get rid of expensive equipment required by physical interruption, can construct a single-chaincyclic library according to a preferred scheme, and enables the library to be used for next-generation sequencing.
Owner:MGI TECH CO LTD
Library establishing method for unicellular RRBS sequencing and application thereof
InactiveCN107475779AReduce the starting amountAvoid lostMicrobiological testing/measurementLibrary creationGenomic DNALinear amplification
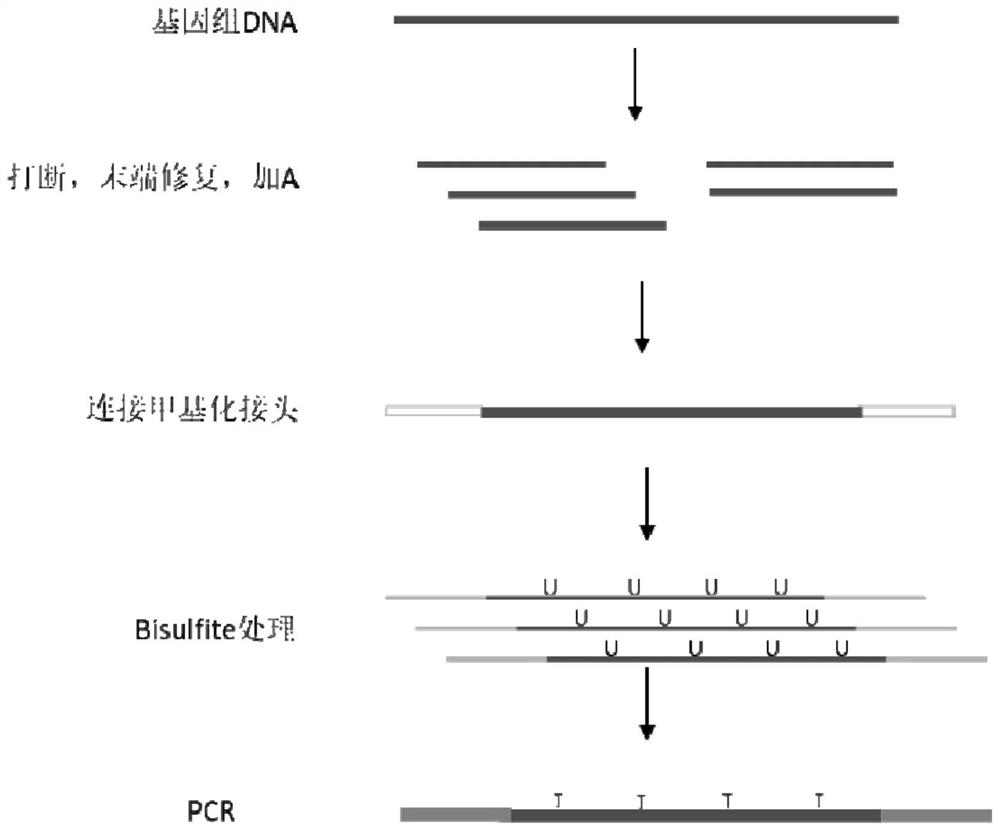
The invention relates to the technical field of molecular biologics and specifically discloses a library establishing method for unicellular RRBS sequencing and an application thereof. The library establishing method for unicellular RRBS sequencing, disclosed by the invention, comprises the following steps: (1) performing digestion, terminal filling, A and joint adding and bisulfate conversion on genomic DNA of a sample in turn; (2) performing linear amplification on the genomic DNA converted in the step (1); (3) performing index amplification on the amplicon subjected to linear amplification in the step (2), wherein the amplicon of the index amplification is used for sequencing the library. The DNA required by the sequencing according to the method is low in initial mass; methylation sequencing can be performed on the genome of a single cell; the method contains an effective library segment control step; the segment of the end product is controlled according to the linear amplification step; complex operation is avoided while the coverage rate is increased.
Owner:上海美吉医学检验有限公司
Absolute quantitative transcriptome library construction method based on specific recognition sequence
ActiveCN110396516AReduce the cost of building a libraryImprove the speed of database constructionMicrobiological testing/measurementLibrary creationReverse transcriptaseRNA library
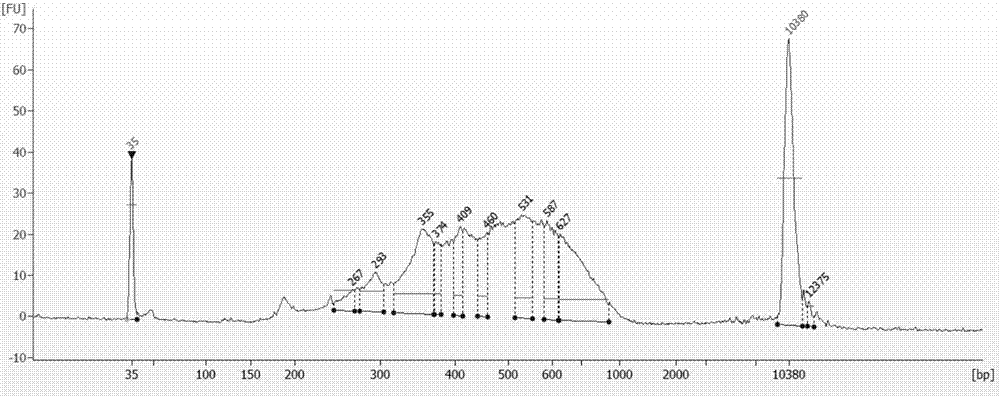
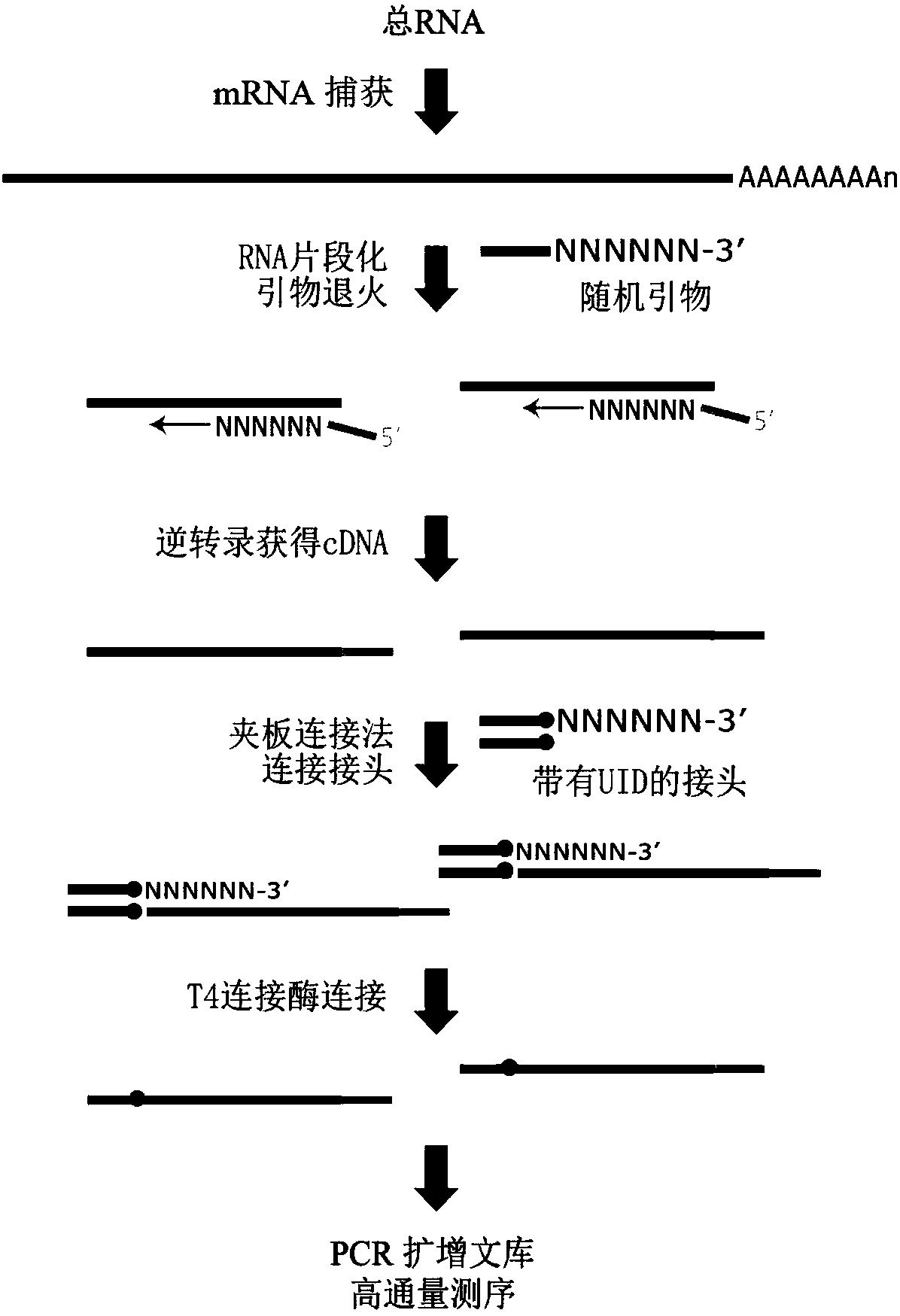
The invention discloses an absolute quantitative transcriptome library construction method based on a specific recognition sequence. Fragmented mRNA is used as a template, a first cDNA chain is synthesized under the action of reverse transcriptase by a primer pool with a general joint sequence, a library construction joint with a special recognition UID sequence is added to cDNA 3' end through anenzymatic reaction, so that each cDNA carries a unique sequence label; and finally, PCR amplification is performed by the general library construction joint to obtain an RNA library. The method constructs the RNA library on the basis of single-stranded cDNA by a splint connection method for the first time, at the same time, a UID sequence is used to precisely reduce the cDNA composition before PCRamplification and can realize precise quantification of transcripts. Library construction is performed by the single-stranded cDNA as a raw material, the step of second strand synthesis is omitted, the template loss rate is reduced, the cost and time are saved, and the defect that the prior art can relatively quantify transcripts is thoroughly solved.
Owner:武汉康测科技有限公司
Construction method of blood platelet nucleic acid library for gene detection and kit
PendingCN108949909AIncrease the amount of informationReduce the starting amountMicrobiological testing/measurementSpecial data processing applicationsCDNA libraryPlatelet
The invention discloses a construction method of a blood platelet nucleic acid library for gene detection and a kit. A nucleic acid capturing probe comprises 5' end biotin modification, an amplification primer sequence P1, a sequencing connector sequence P5, a sample label sequence, a single-molecule label sequence and an Oligo (dT) sequence in sequence from 5'. The invention further provides thekit comprising the nucleic acid capturing probe and the construction method for the blood platelet nucleic acid library by utilizing the nucleic acid capturing probe. According to the construction method disclosed by the invention, the starting amount of blood platelets is greatly reduced, the blood platelets can be separated from less whole blood, and trace amplification and library constructionare directly carried out, so that requirements of liquid biopsy are met. Moreover, samples of different examinees can be mixed in the same reaction system, so that the flux of detection is improved; and the construction method and the kit have important clinical significance and practical value.
Owner:XIAMEN LIFEINT TECH CO LTD
Construction method of ovarian cancer susceptibility gene mutation library
ActiveCN106498082AReduce morbidityReduce mortalityNucleotide librariesMicrobiological testing/measurementOvarian cancerSusceptibility gene
The invention relates to a construction method of an ovarian cancer susceptibility gene mutation library. The construction method comprises the following steps: A, according to mutation regions of related genes of ovarian cancer, designing primer pairs capable of detecting the mutation regions; B, calculating to obtain judgment parameters R of all the primer pairs; classifying the primer pairs with the judgment parameters R smaller than or equal to 1 into a first group of primer combined solution, and classifying the primer pairs with the judgment parameters R more than 1 into a second group of primer combined solution; C, extracting and purifying DNA (Deoxyribonucleic Acid) of a sample to be detected; D, carrying out initial PCR (Polymerase Chain Reaction) amplification on the purified sample through adopting the first group of primer combined solution and the second group of primer combined solution respectively; E, carrying out joint connection on target segments to obtain segments with joints; and F, carrying out library PCR amplification by adopting a mixed solution of the first group of primer combined solution and the second group of primer combined solution to obtain a sequencing library. According to the construction method of the ovarian cancer susceptibility gene mutation library, provided by the invention, the sequencing flux of the constructed library is high, the sensitivity is high and the specificity is high; the constructed library can be used for low-frequency mutation of free DNA.
Owner:菁良科技(深圳)有限公司
Method for separating mixed samples by SNP detection technology
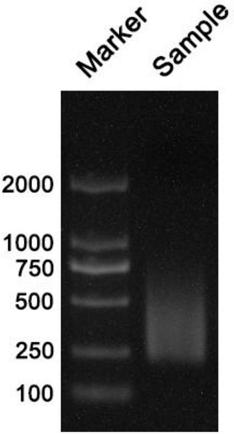
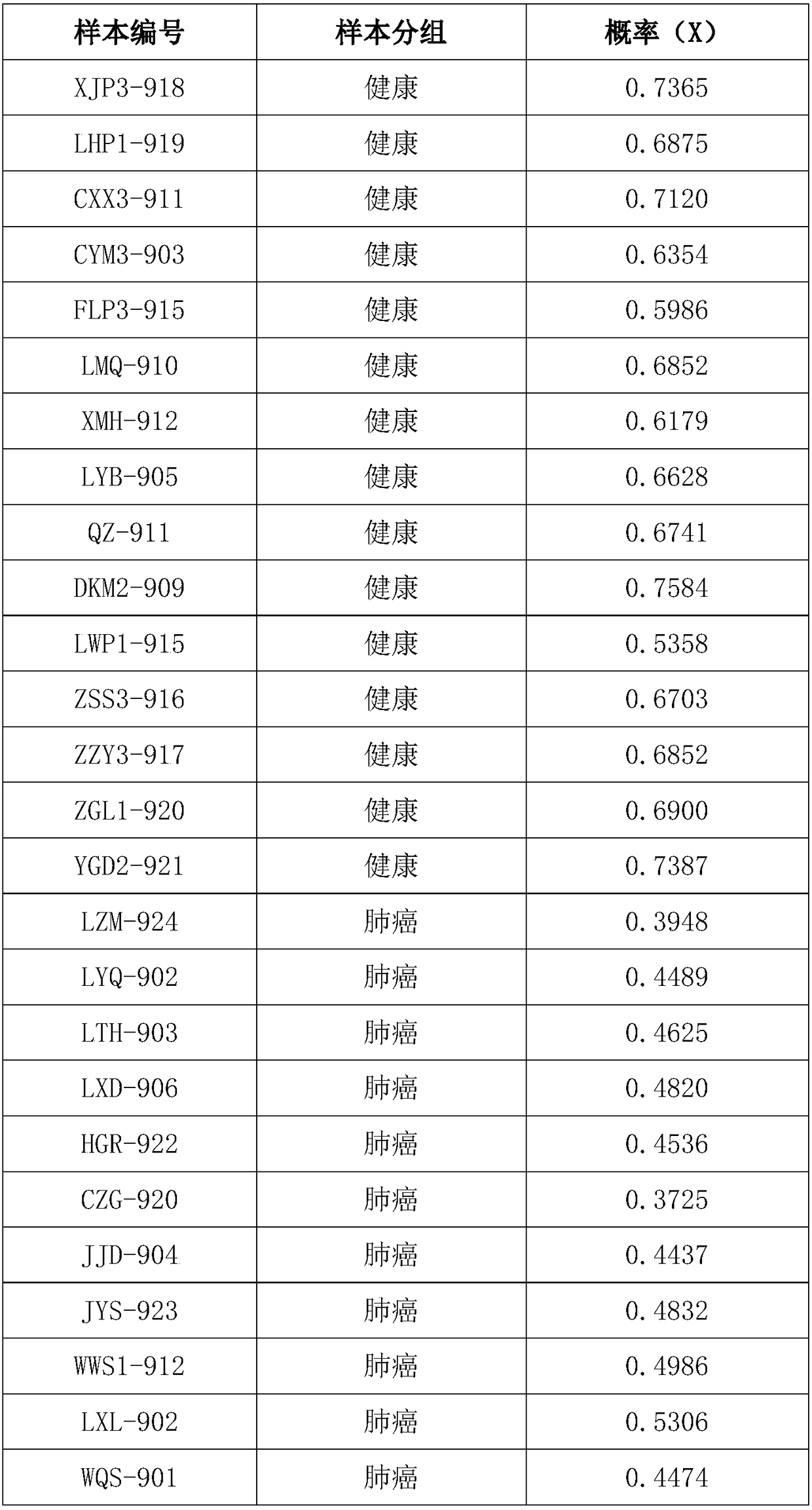
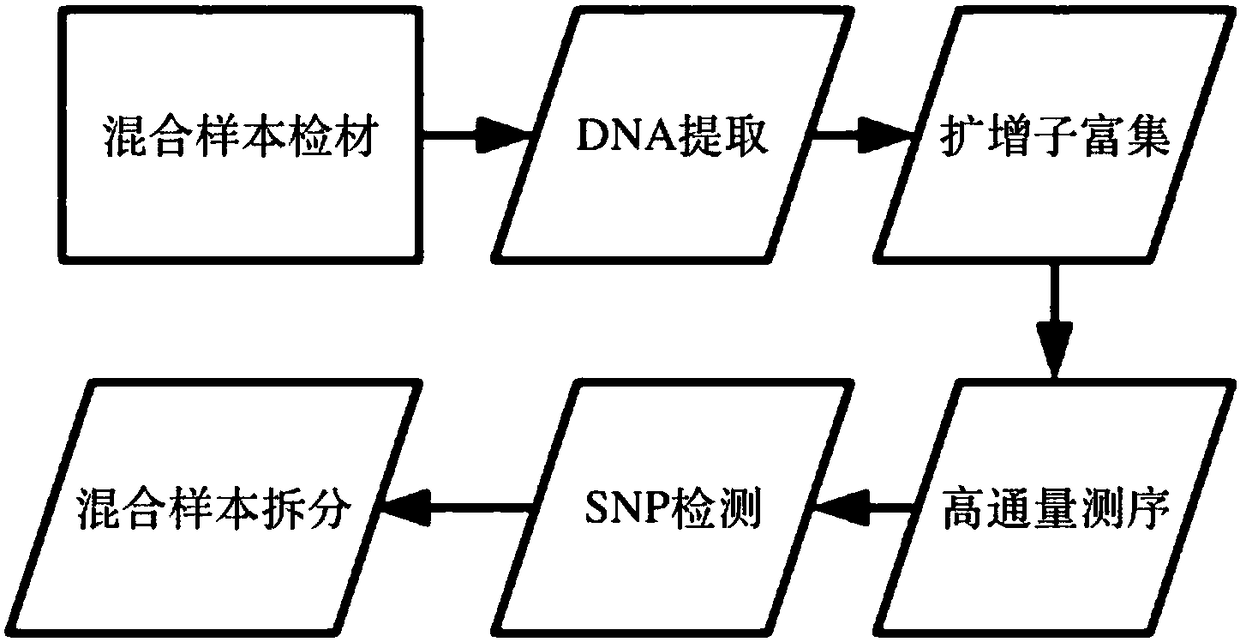
ActiveCN109280696AReduce the starting amountHigh detection sensitivityMicrobiological testing/measurementNucleotideDNA fragmentation
The invention discloses a method for separating mixed samples by an SNP detection technology. The method uses a high-throughput gene sequencing platform (NGS) to detect multiple single nucleotide polymorphisms (SNPs) by amplicon sequencing, which can solve the problems that a mixed sample of two persons cannot be separated happened in criminal evidence identification, civil judicial identificationand clinic. The method includes the following technical points: 1) in order to ensure that amplification efficiency of amplicons is uniform and stable, the 219 amplicons are obtained to achieve simultaneous amplification in the same tube after strict condition design and screening; 2) in order to ensure successful amplification of a degraded sample (the length of a DNA fragment being 160 bp), theSNPs are selected as biomarkers, wherein the SNPs are located in the middle of the 140 bp amplicons; and 3) in order to distinguish whether the sample is a sample of one person or a mixed sample of two persons, a mixing proportion of the mixed sample and SNPs genotype sets of the mixed sample of two persons are further separated.
Owner:安塞斯(北京)生物技术有限公司 +1
Construction method and application of microbial diversity library
InactiveCN107904668AReduce the starting amountWiden the variety of applicationsMicrobiological testing/measurementLibrary creationMicroorganismMicrobiology
The invention relates to the technical field of molecular biology and particularly relates to a construction method and an application of a microbial diversity library. According to the novel construction method of the microbial diversity library, the microbial diversity library of a metagenome is constructed by virtue of single chain linkage and a special label primer joint; the scheme is applicable to conventional samples, trace samples, FFPE samples and less than 150 cell groups, and the application range of microbial diversity research is extended; and by introducing the single chain linkage and a special label primer, different target populations fragments can be enriched in a small deviation manner, so that the richness of species and the authenticity of abundance of different species are guaranteed. Besides, the construction method further has the advantages that the initial mass is low, and the same sample can be subjected to multi-species authentication.
Owner:SHANGHAI MAJORBIO BIO PHARM TECH
Method for treating single-stranded DNA, and applications thereof
ActiveCN110791813AIncrease profitIncrease the number ofMicrobiological testing/measurementLibrary creationSingle strandDouble strand
The invention discloses a method for treating single-stranded DNA, and applications thereof, and relates to the field of gene sequencing, particularly to a method for constructing a library based on single-stranded DNA. The method comprises: linking a first linker to the 3' tail end of single-stranded DNA, wherein the first linker is linked to binding agent, and comprises two partially complementary strands, the two partially complementary strands are respectively a first strand and a second strand, and the second strand has 5-10 basic groups more than the first strand; and adsorbing the single-stranded DNA linked to the first linker by using an adsorbent so as to remove the second strand, so that the double-stranded DNA containing the single-stranded DNA is obtained. According to the invention, the gene library constructed by the method has advantages of high molecular utilization rate, low initial quantity of samples for library construction and low minimum detection limit.
Owner:BGI GUANGZHOU MEDICAL LAB CO LTD +2
Colorectal cancer susceptibility gene mutation library constructing method
ActiveCN106757378AReduce morbidityReduce mortalityNucleotide librariesMicrobiological testing/measurementFhit geneSusceptibility gene
The invention relates to a colorectal cancer susceptibility gene mutation library constructing method. The colorectal cancer susceptibility gene mutation library constructing method comprises the following steps: A, designing primer pairs capable of detecting a mutation region according to the mutation region related to a colorectal cancer; B, calculating to obtain judgment parameters R of the various primer pairs, classifying the primer pairs of which the values of the judgment parameters R are smaller than or equal to 1 into a first group of primer combination liquid, and classifying the primer pairs of which the values of the judgment parameters R are greater than 1 into a second group of primer combination liquid; C, extracting and purifying DNA of a sample to be detected; D, carrying out initial PCR amplification on the purified sample by using the first group of primer combination liquid and the second group of primer combination liquid respectively; E, connecting target fragments via joints to obtain fragments with joints; and F, carrying out library PCR amplification by using mixed liquid of the first group of primer combination liquid and the second group of primer combination liquid to obtain a sequencing library. The library constructed by the colorectal cancer susceptibility gene mutation library constructing method is high in sequencing flux, high in sensitivity and high in specificity, and can be used for detecting free DNA low frequency mutation.
Owner:深圳市艾伟迪生物科技有限公司
Method for preparing chromatin co-immunoprecipitation library by using trace of clinical puncture sample and application of method
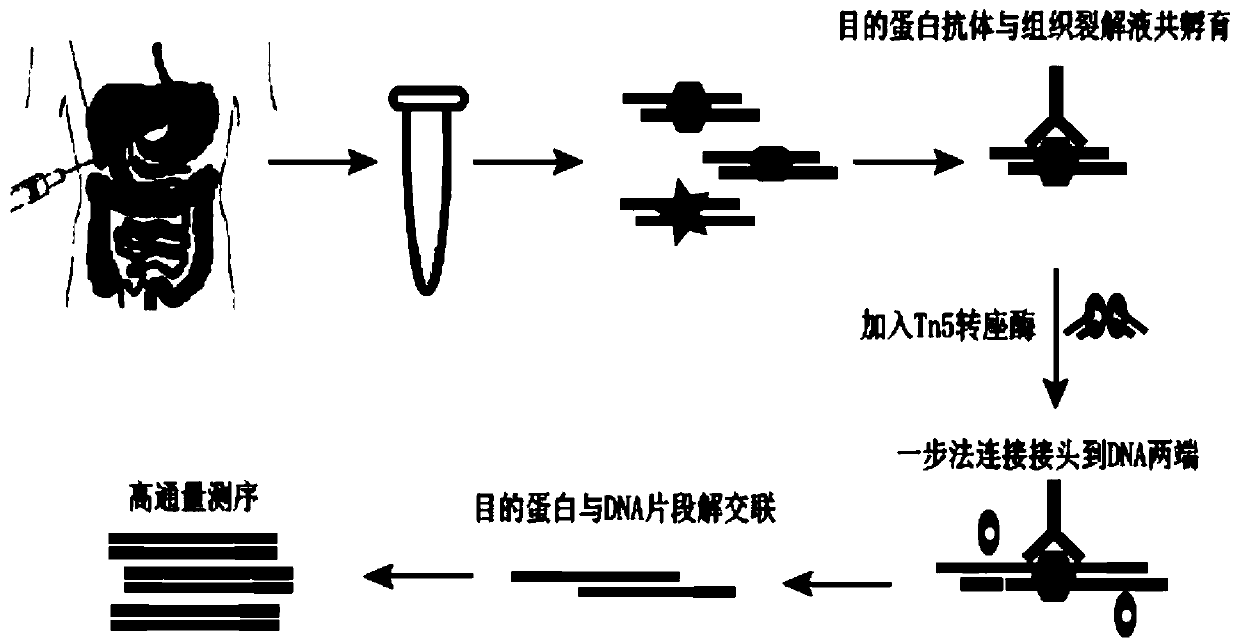
InactiveCN111440843AShort cycleLow costMicrobiological testing/measurementLibrary creationTn5 transposaseTissue sample
The invention discloses a method for preparing a chromatin co-immunoprecipitation library by using a trace clinical puncture sample and an application of the method. The invention successfully develops the method for preparing the chromatin co-immunoprecipitation sequencing library based on trace of clinical puncture sample by utilizing high-sensitivity Tn5 transposase, and ChIP-seq library construction can be effectively and rapidly carried out on the trace of puncture tissue sample. Compared with a traditional ChIP-seq library building mode, the method has the advantages that the initial sample amount is greatly reduced (5-50 mg), the library building time is effectively shortened (about 1.5 days), and the requirements of the sequencing market on trace samples and large-scale sample processing speed are met.
Owner:INSITUTE OF BIOPHYSICS CHINESE ACADEMY OF SCIENCES
Breast cancer susceptibility gene variable library construction method
ActiveCN106754878AReduce morbidityReduce mortalityMicrobiological testing/measurementLibrary creationBreast cancer susceptibility genesTrue positive rate
The invention relates to a breast cancer susceptibility gene variable library construction method. The construction method comprises the following steps that 1, according to variation regions of a breast cancer-related gene, primer pairs capable of detecting the variation regions are designed; 2, judgment parameters R of the primer pairs are calculated, the primer pairs of which values of the judgment parameters are smaller than or equal to 1 are classified into a first primer combination solution, and the primer pairs of which values of the judgment parameters are larger than 1 are classified into a second primer combination solution; 3, a to-be-detected DNA sample is extracted and purified; 4, initial PCR amplification is conducted on the purified sample by adopting the first primer combination solution and the second primer combination solution separately; 5, linker connection is conducted on a target fragment to obtain a fragment with a linker; 6, library PCR amplification is conducted through a mixed solution of the first primer combination solution and the second primer combination solution to obtain a sequencing library. The library constructed through the breast cancer susceptibility gene variable library construction method is high in sequencing flux, sensitivity and specificity and can be used for detecting low-frequency mutation of free DNA.
Owner:南京艾迪康医学检验所有限公司
FFPE DNA library building kit, use thereof and library building method
InactiveCN107937986AImprove efficiencyReduce the starting amountMicrobiological testing/measurementLibrary creationA-DNABuffer solution
The invention relates to an FFPE DNA library building kit, use thereof and a library building method. The kit includes combination of specific joint connection buffer solution, a DNA ligase solution and joint sequences. The kit provided by the invention can maximumly enhance the connection efficiency and PCR amplification efficiency in FFPE DNA library building, thus lowering the initial amount ofDNA library building, reducing the amplification cycle number, and meeting the demand of probe captured pre-amplification product quantity, lowering repetition generated in the sequencing process, and improving data utilization.
Owner:深圳裕策生物科技有限公司
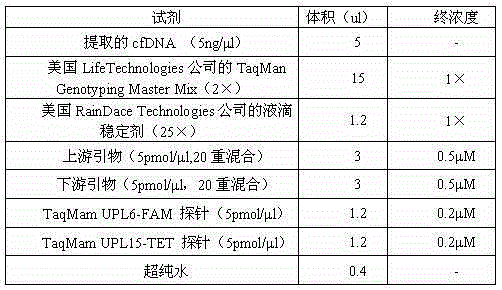
Noninvasive prenatal screening 2, 1-trisomy syndrome kit
ActiveCN105132572AReasonable primer designPrevent birthMicrobiological testing/measurementTrisomyTrisomy X syndrome
The invention discloses a noninvasive prenatal screening 2, 1-trisomy syndrome kit, and relates to the technical field of a nucleic acid determination or inspection method. The noninvasive prenatal screening 2, 1-trisomy syndrome kit comprises a PCR reaction solution, wherein the PCR reaction solution contains a TaqMam UPL6-FAM probe and a TaqMam UPL15-TET probe, the TaqMam UPL6-FAM probe and the TaqMam UPL15-TET probe are respectively used for detecting No.18 and No.21 chromosomes, 20 pairs of primers are respectively designed for the No.21 and No.18 chromosomes, gene sequences of the primers are SEQ ID NO.1 to 40 and SEQ ID NO.41-80; and gene sequences of the TaqMam UPL6-FAM probe and the TaqMam UPL15-TET probe are SEQ ID NO.81 and SEQ ID NO.82. By adopting the noninvasive prenatal screening 2, 1-trisomy syndrome kit, the noninvasive prenatal screening of 21-trisomy syndrome, and the accuracy is high.
Owner:邯郸市康业生物科技有限公司
Detection method for mitochondrial copy number and sequence variation
InactiveCN110387410AReduce pollutionReduce the starting amountMicrobiological testing/measurementDNA/RNA fragmentationDiseaseInformation analysis
The invention discloses a detection method for mitochondrial copy number and sequence variation. Based on a BGI-Seq series sequencing platform, a high-flux, low-cost and automated mitochondrial variation detection technology is established, and the technology comprises mitochondrial whole-genome DNA separation and genome housekeeping gene amplification, library construction and sequencing, and supporting information analysis algorithm development. The technology can simultaneously achieve mitochondrial copy number variation and sequence variation detection, and is high in flux, small in difference between batches, high in accuracy, low in cost, short in used time, and capable of fully meeting the need of automated production. The technology can be applied to mtDNA somaclonal variation research and detection, diagnosis of mitochondrion related diseases, research of the role, played in the aging process, of mitochondria, tumor cell metabolic activity related research and the like, and has good prospects.
Owner:SHENZHEN HUADA GENE INST
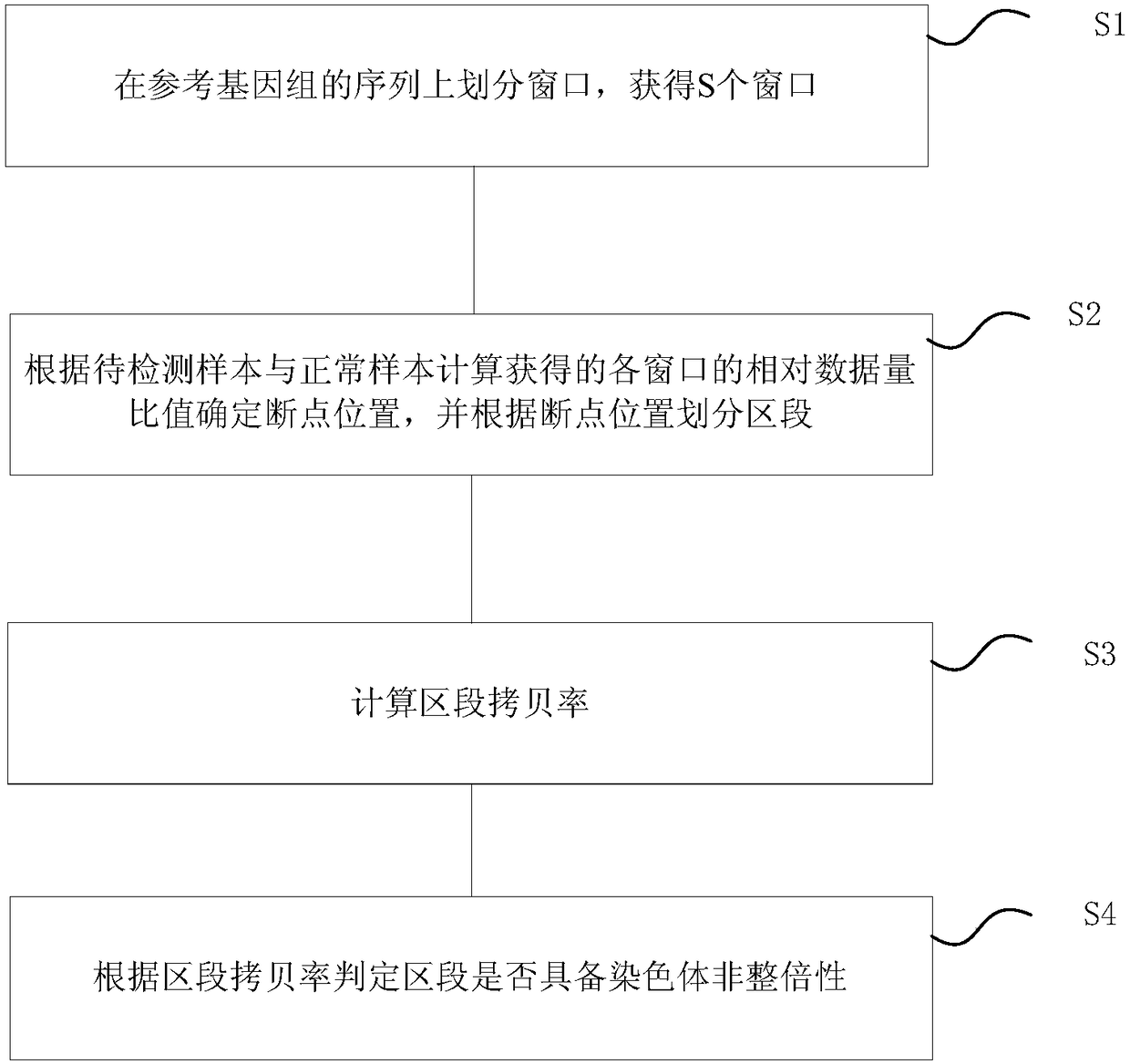
Chromosome aneuploidy detection system suitable for single cells and application
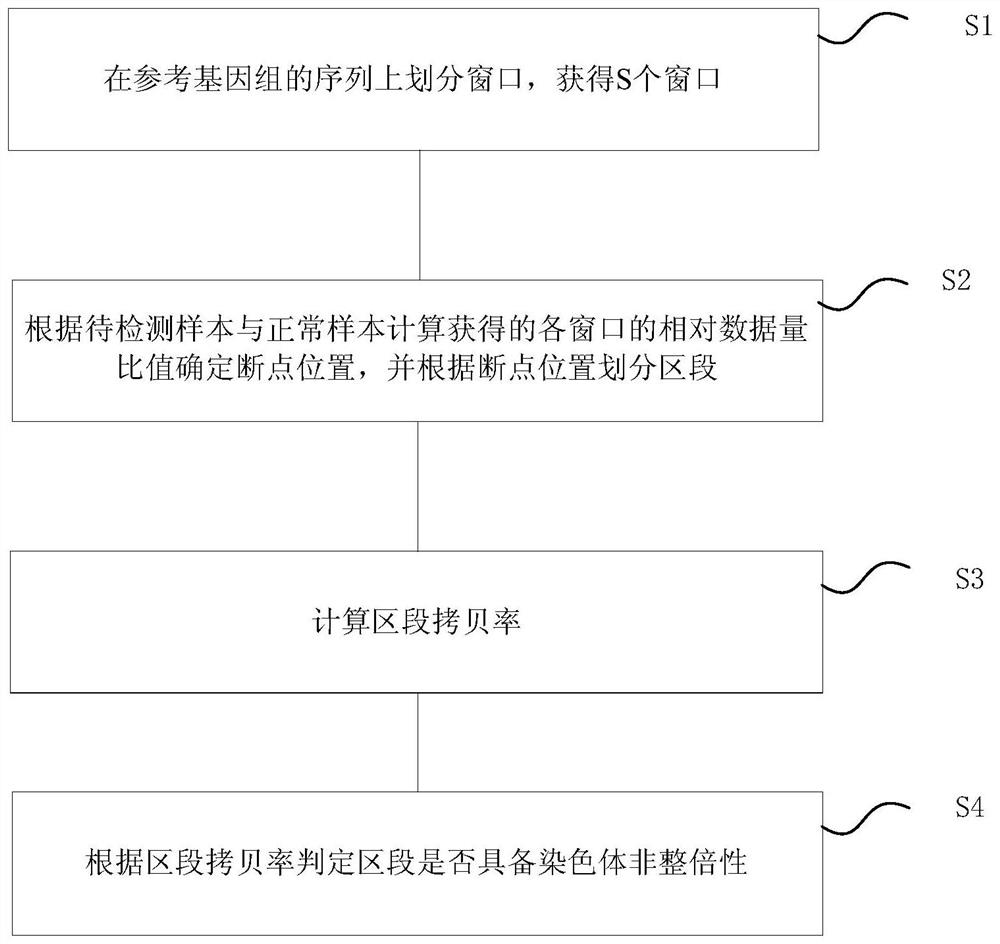
ActiveCN108363903AReduce the starting amountEnables detection of aneuploidyMicrobiological testing/measurementBiostatisticsComputer moduleGene engineering
The invention belongs to the technical field of gene engineering, and particularly relates to a chromosome aneuploidy detection system suitable for single cells and an application. The detection system comprises a window division module, a section division module, a section copy rate calculation module and a section aneuploidy judgment module. The detection system uses at most 700k reads / samples and can detect more than 5Mb chromosome deletion / duplication for the samples; the accuracy can reach 99.9%; the sequencing depth is reduced; and the cost is lowered.
Owner:UNIMED BIOTECH SHANGHAI CO LTD
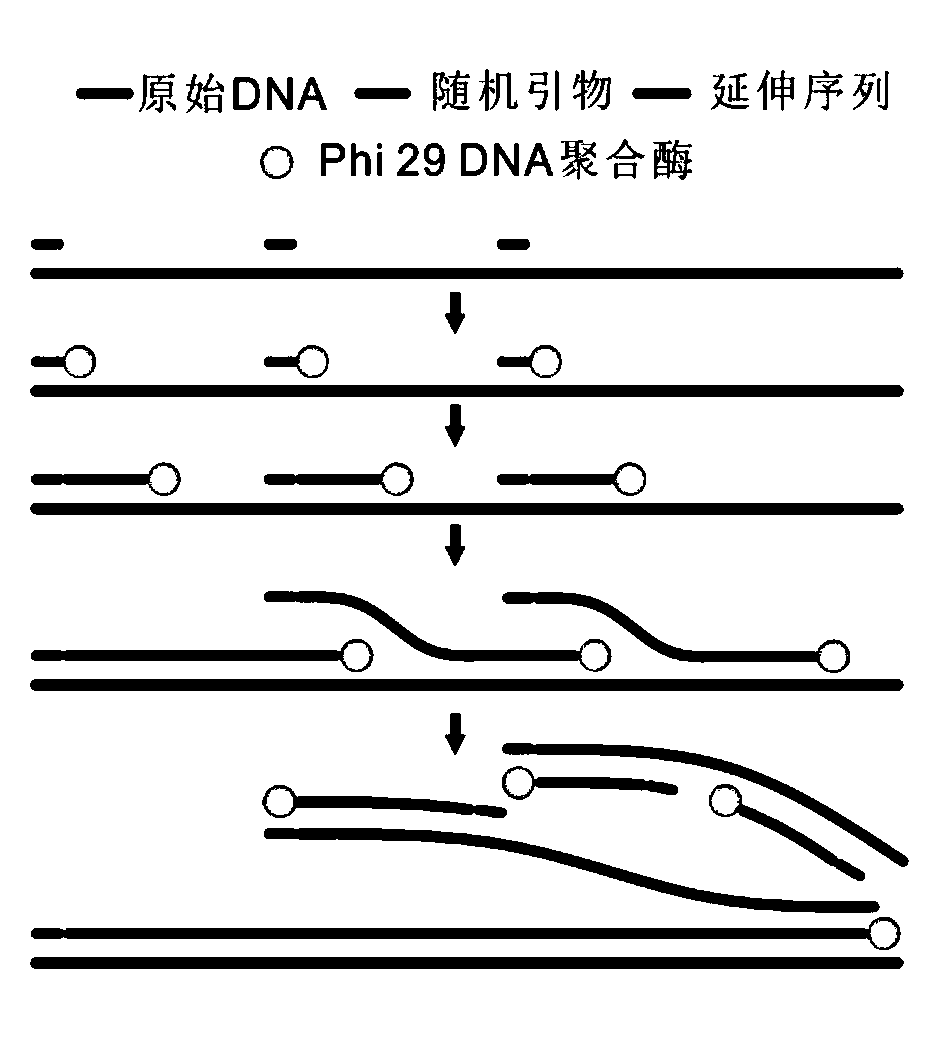
Novel low-initial-quantity DNA methylation library building method
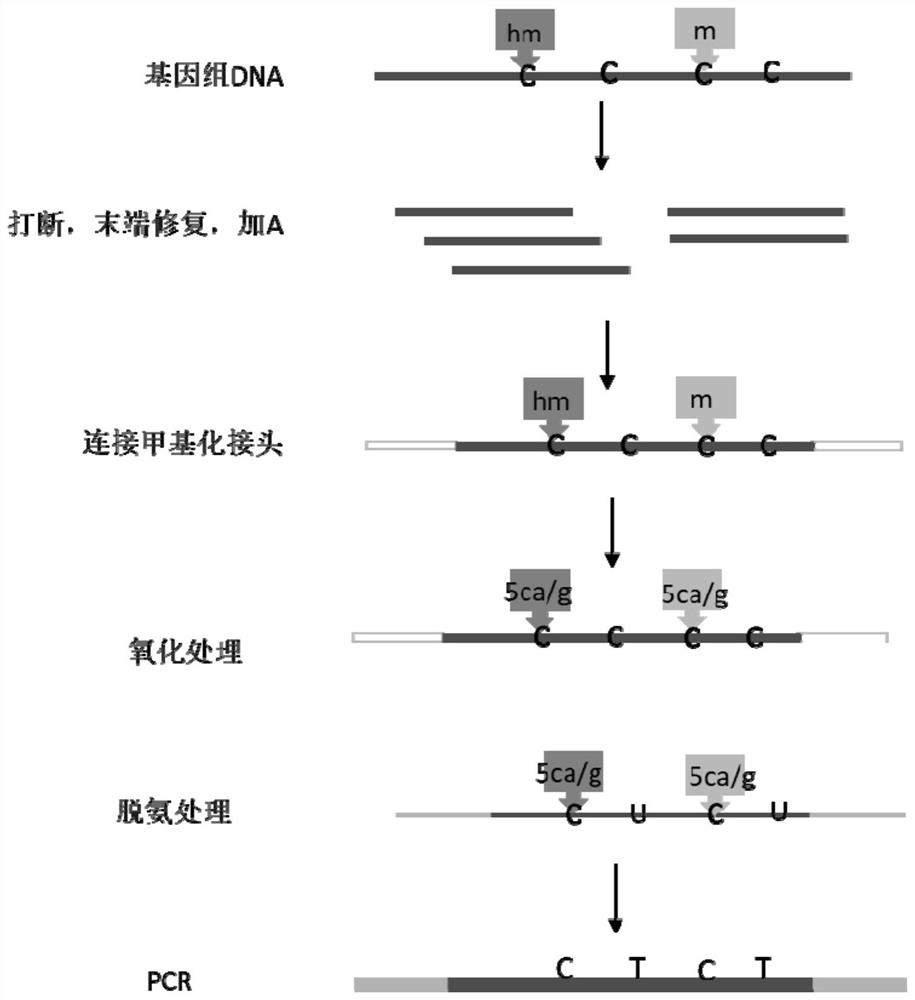
PendingCN113088562AReduce the starting amountAvoid damageMicrobiological testing/measurementLibrary creationDNA methylationCytosine deaminase
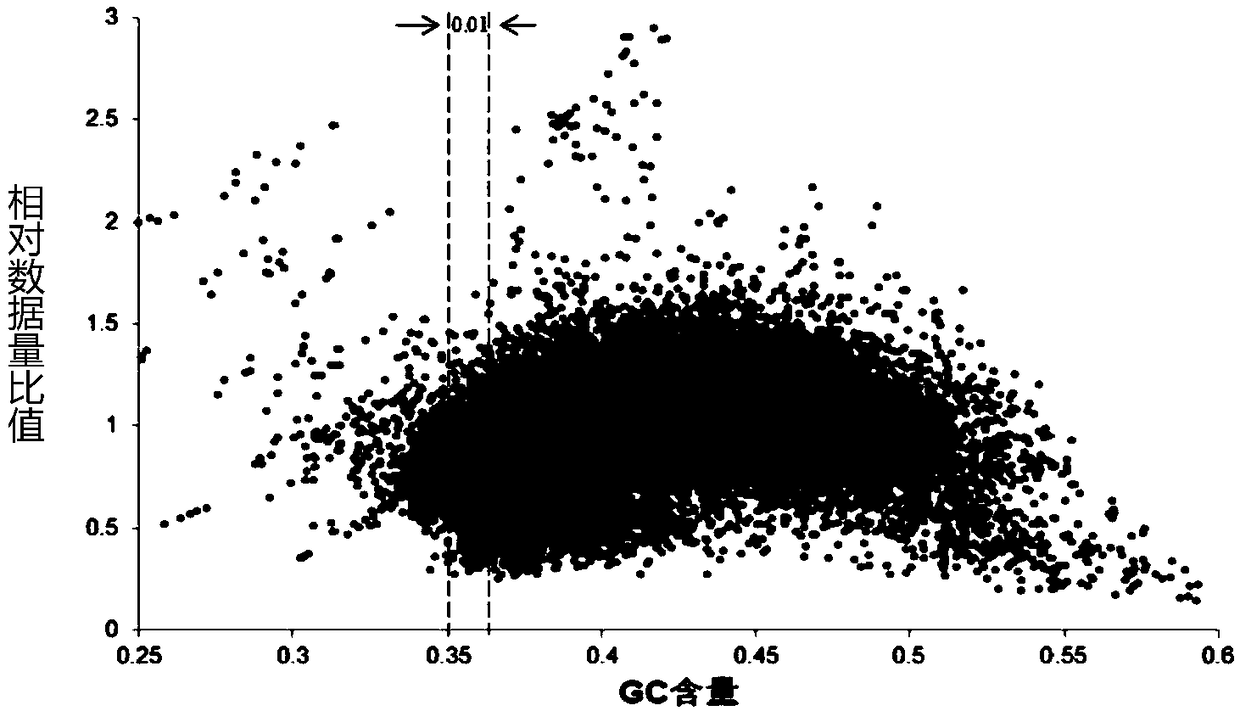
The invention discloses a novel low-initial-quantity DNA methylation library building method. The invention provides a method for preparing a methylation sequencing library, which sequentially comprises the following steps of: (1) taking genome DNA, and carrying out oxidative deamination treatment; (2) carrying out whole genome amplification; and (3) constructing a sequencing library. In the step (1), the oxidative deamination treatment is carried out by adopting an enzyme chemical method, the oxidation treatment is carried out by adopting dioxygenase (TET2), and the deamination treatment is carried out by adopting cytosine deaminase (APOBEC). The purpose of whole genome amplification is to convert DNA at an ng level into DNA at a mu g level. The method provided by the invention has the following beneficial effects: genome DNA methylation treatment with extremely low initial quantity can be realized, and the initial quantity can be as low as a single cell level; the operation steps are simple; the damage to DNA is minimum; and the phenomena of unbalanced GC base distribution, low coverage, high duplication rate and the like of the existing trace methylation library establishment are improved.
Owner:BGI GENOMICS CO LTD
Macro-gene library building method of nanopore sequencing platform and kit thereof
ActiveCN112029823AQuality improvementReduce sequencing timeMicrobiological testing/measurementLibrary creationEngineeringBioinformatics
The invention relates to a macro-gene library building method of a nanopore sequencing platform and a kit thereof. By optimizing and adjusting end-repair connection, PCR amplification and the like inlibrary building, the library building method and the kit thereof effectively reduce the initial quantity of library building, improve the reading length and quality of sequencing data, shorten the library building and sequencing time, save the sequencing cost, improve the sensitivity of the whole process, meet the clinical requirements for infection detection, and are suitable for popularizationand application.
Owner:JIANGSU SIMCERE MEDICAL DEVICE CO LTD +2
Device for preparing proteomics micro-sample
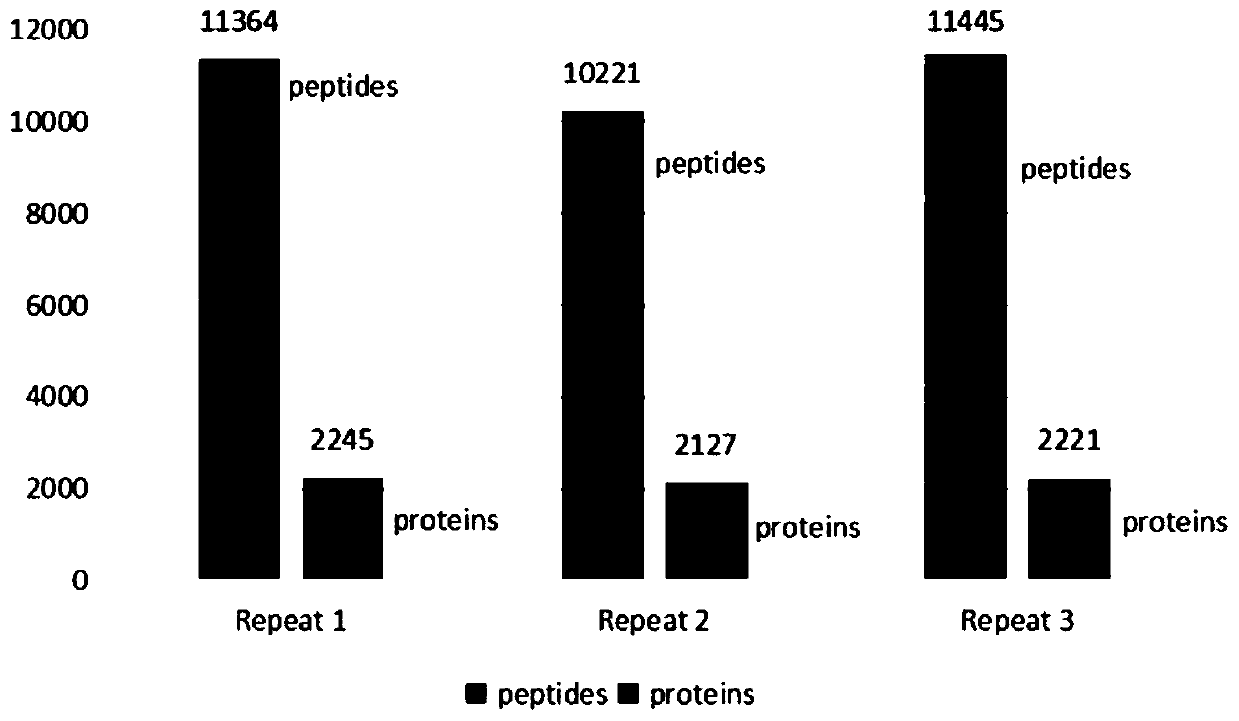
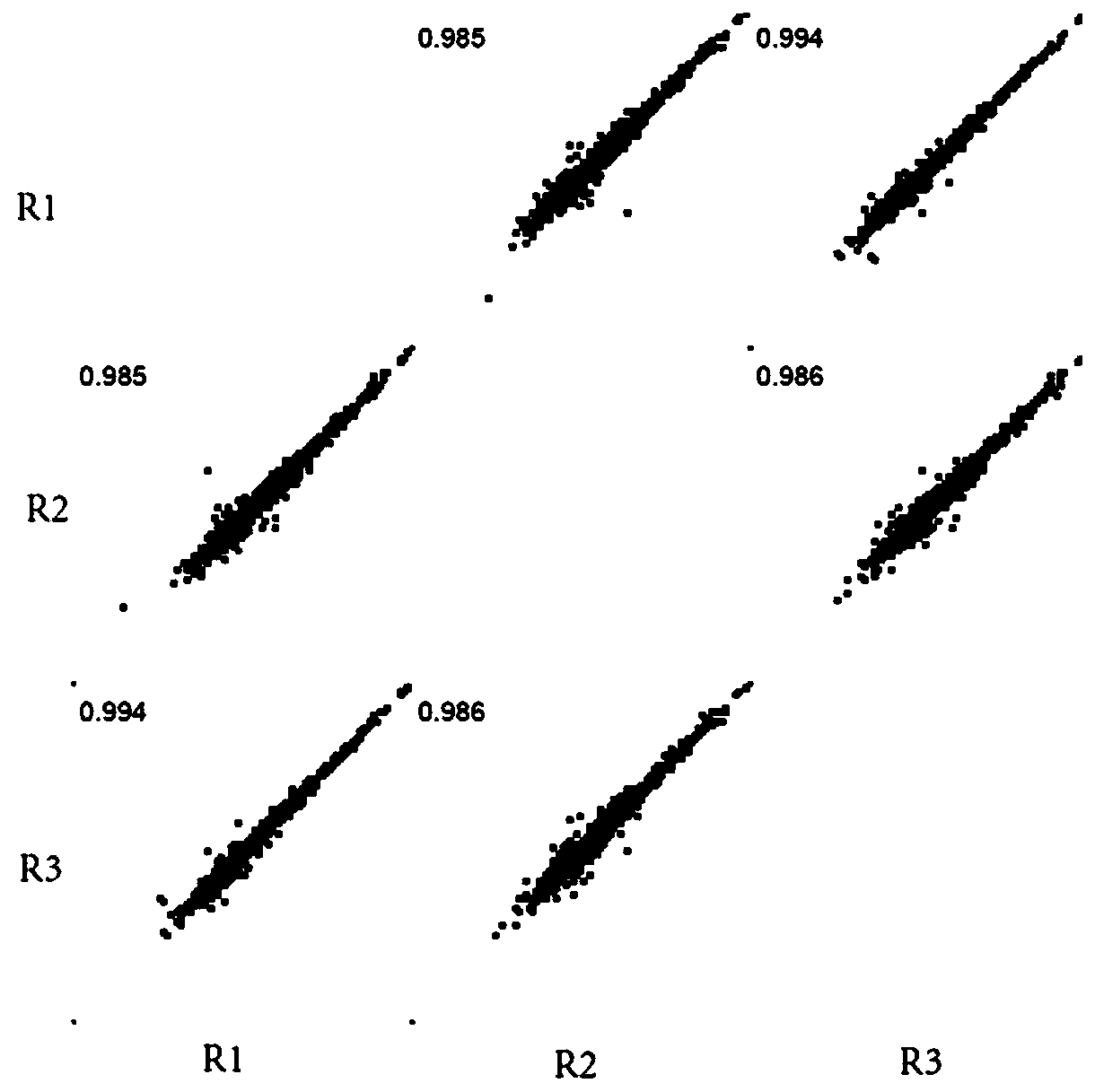
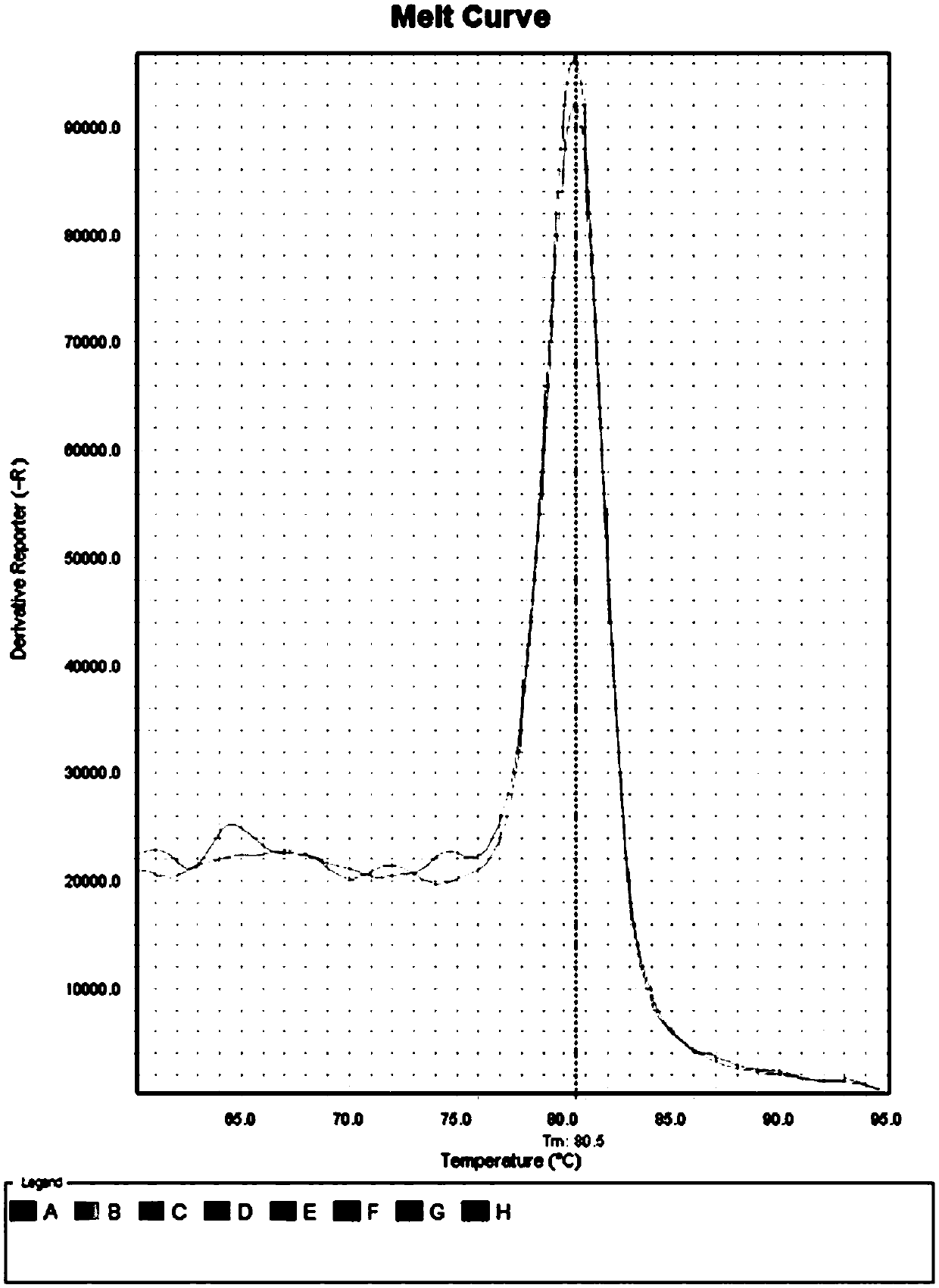
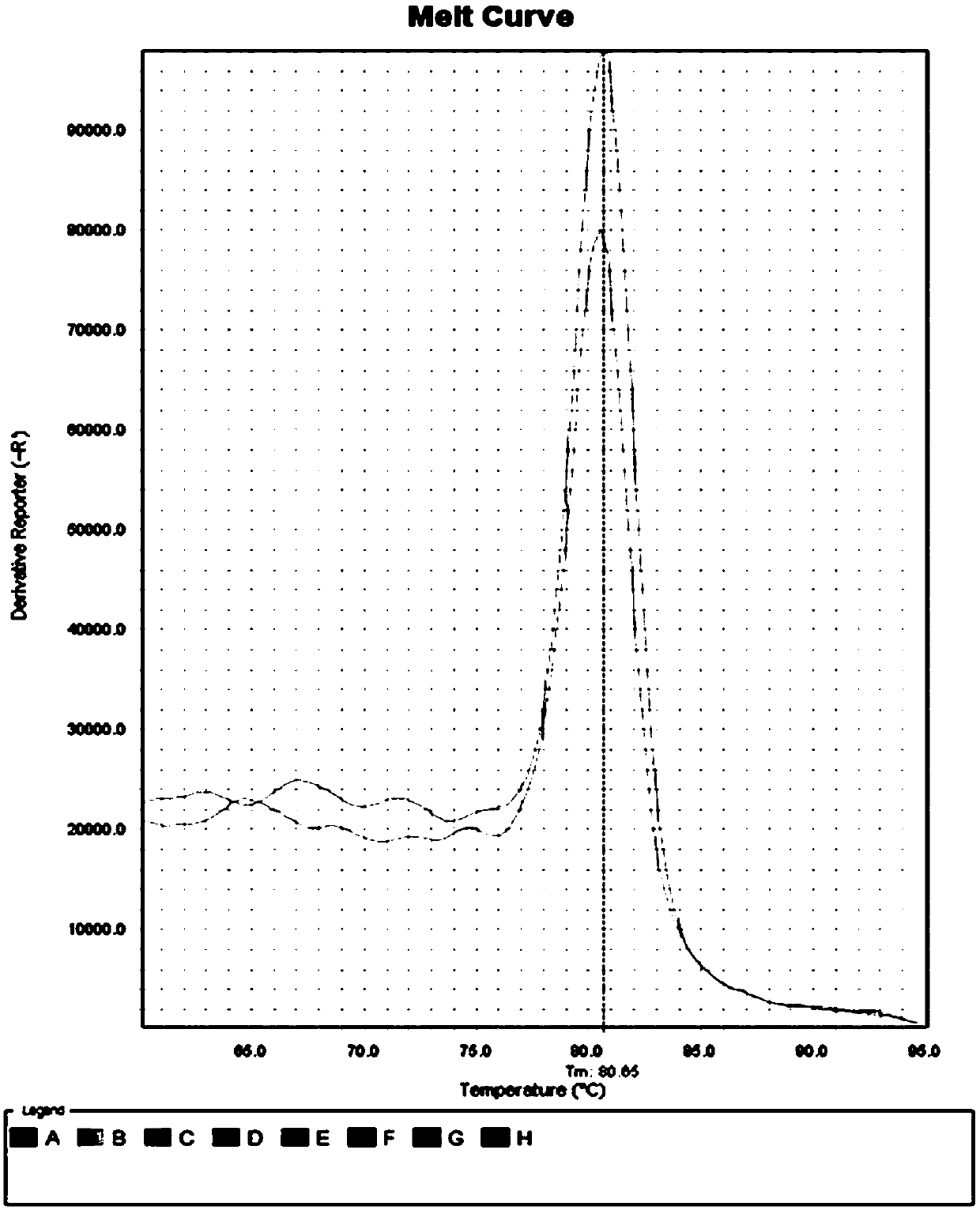
PendingCN111521473AReduce the starting amountMeeting Research NeedsComponent separationPreparing sample for investigationBiotechnologyBiochemistry
The invention discloses a device for preparing a proteomics trace sample. The device is easy to manufacture and low in cost, and the peptide fragment recovery rate and the protein identification amount are superior to those of a conventional method under the condition that the initial protein amount is low. The device realizes seamless combination of one-stop in-situ protein digestion, peptide fragment acquisition, and N-sugar chain release and enrichment. The loss of the sample is reduced to the greatest extent, and the whole-process sample preparation of the micro-sample proteomics is realized.
Owner:ACADEMY OF MILITARY MEDICAL SCI +1
Method for constructing low-initial-quantity PCR-free high-throughput sequencing library
PendingCN109609597AOmit the purification stepOptimize the reaction systemMicrobiological testing/measurementLibrary creationNon invasiveBlood plasma
The present invention relates to the field of bioinformatics, and specifically provides a method for constructing a low-initial-quantity PCR-free high-throughput sequencing library. The method includes the following steps: (1) extracting cfDNA from plasma, carrying out terminal repair and 5' terminal phosphorylation, and purifying and recovering terminal repair products with magnetic beads; (2) adding "A" bases to 3' terminal of the purified terminal repair products to be integrated into one system with a joint connection system, and carrying out a reaction; and (3) purifying joint products with magnetic beads to obtain the sequencing library. The constructing method has the advantages of low cost, short time-consuming, no need of PCR amplification for library construction, maintained stable data output and low requirements for the initial DNA quantity. Only 1-2ng of cfDNA can meet the needs of library construction, and the method can be used for effective sequencing, and has very broad application prospects in high-throughput sequencing, non-invasive diagnosis and other research.
Owner:BEIJING USCI MEDICAL LAB CO LTD
Trace nucleic acid sequencing pretreatment method based on magnetic bead coating and kit
InactiveCN106701993AReduce the number of transfersReduce lossMicrobiological testing/measurementDNA preparationPretreatment methodMagnetic bead
The invention discloses a trace nucleic acid sequencing pretreatment method based on magnetic bead coating and a kit. The trace nucleic acid sequencing pretreatment method based on magnetic bead coating comprises the following steps of: firstly coating DNA (deoxyribonucleic acid) to be treated with magnetic beads, carrying out tail end repairing on the DNA to be treated, which is coated with the magnetic beads, purifying, leaving the magnetic beads, carrying out A addition reaction, purifying again, leaving the magnetic beads, carrying out joining reaction, purifying, discarding the magnetic beads, carrying out PCR (polymerase chain reaction), and purifying by adding the magnetic beads after completing polymerase chain reaction to obtain the treated DNA. The trace nucleic acid sequencing pretreatment method based on magnetic bead coating has the beneficial effects that the frequency of transferring trace nucleic acids in a sequencing pretreatment process is effectively reduced by coating the DNA with the magnetic beads, thereby greatly reducing the loss of the nucleic acids in a sequencing pretreatment process; reaction conditions are mild; meanwhile, the DNA damage is also reduced; the success rate of trace nucleic acid sequencing pretreatment is increased to 90% or above; the method has the characteristics of high success rate, high quality and low initial amount in comparison with a traditional nucleic acid sequencing pretreatment method; and used reagent raw materials are low in cost, simple and easily available.
Owner:宿州洛奇医学检验实验室有限公司
An Absolute Quantitative Transcriptome Library Construction Method Based on Unique Recognition Sequences
ActiveCN110396516BReduce the cost of building a libraryImprove the speed of database constructionMicrobiological testing/measurementLibrary creationReverse transcriptaseRNA library
The invention discloses a method for constructing an absolute quantitative transcriptome library based on a unique recognition sequence. Using the fragmented mRNA as a template, the first cDNA strand is synthesized under the action of reverse transcriptase using a primer pool with a universal linker sequence, and a unique recognition UID sequence is added to the 3' end of the synthesized cDNA by an enzymatic reaction. Build a library linker so that each cDNA has a unique sequence tag; finally, use a universal library linker to perform PCR amplification to obtain an RNA library. For the first time, the present invention uses the splint ligation method to construct an RNA library based on single-stranded cDNA. At the same time, the UID sequence is used to accurately restore the cDNA composition before PCR amplification, which can realize accurate quantification of transcripts; the present invention uses single-stranded cDNA as a raw material for library construction. The step of second-strand synthesis is omitted, the template loss rate is reduced, cost and time are saved, and the existing technology can only relatively quantify transcripts.
Owner:武汉康测科技有限公司
Method for constructing breast cancer susceptibility gene variation library
ActiveCN106754878BReduce morbidityReduce mortalityMicrobiological testing/measurementLibrary creationBreast cancer susceptibility genesMedicine
The invention relates to a breast cancer susceptibility gene variable library construction method. The construction method comprises the following steps that 1, according to variation regions of a breast cancer-related gene, primer pairs capable of detecting the variation regions are designed; 2, judgment parameters R of the primer pairs are calculated, the primer pairs of which values of the judgment parameters are smaller than or equal to 1 are classified into a first primer combination solution, and the primer pairs of which values of the judgment parameters are larger than 1 are classified into a second primer combination solution; 3, a to-be-detected DNA sample is extracted and purified; 4, initial PCR amplification is conducted on the purified sample by adopting the first primer combination solution and the second primer combination solution separately; 5, linker connection is conducted on a target fragment to obtain a fragment with a linker; 6, library PCR amplification is conducted through a mixed solution of the first primer combination solution and the second primer combination solution to obtain a sequencing library. The library constructed through the breast cancer susceptibility gene variable library construction method is high in sequencing flux, sensitivity and specificity and can be used for detecting low-frequency mutation of free DNA.
Owner:南京艾迪康医学检验所有限公司
Method for eliminating self-connector of sequencing library and application
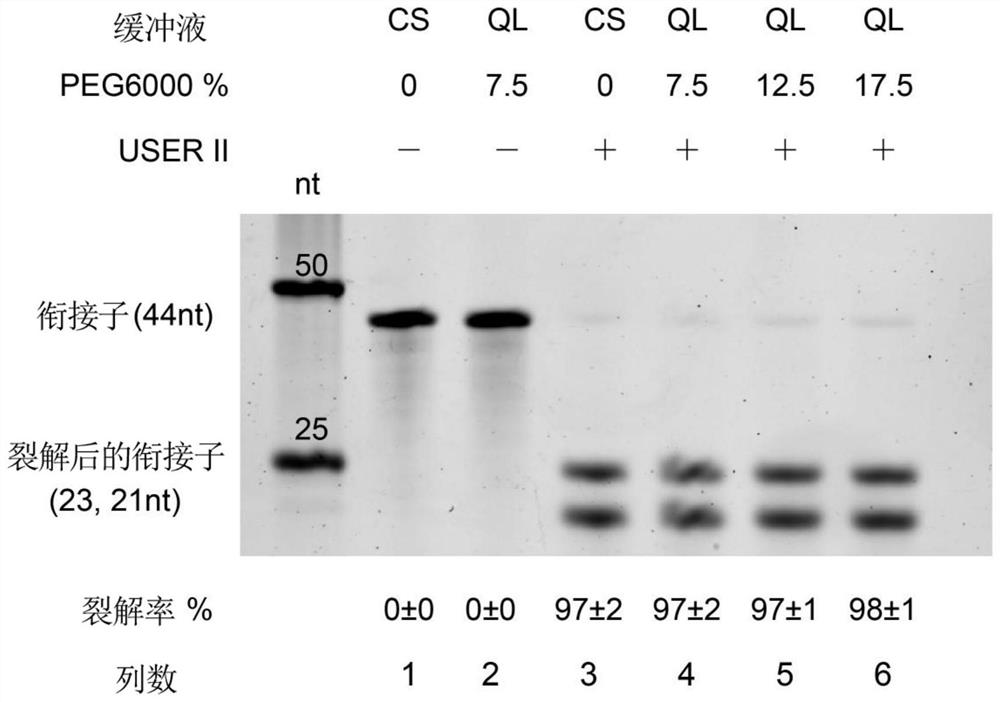
PendingCN113817804AEfficient removalReduce the ratioMicrobiological testing/measurementLibrary creationEngineeringData efficiency
The invention relates to a method for eliminating a self-connector of a sequencing library and an application. The method comprises the following steps that a short sequence guide DNA is designed by taking a sequencing connector self-connecting library sequence as a target sequence, double-strand breakage of a dsDNA library molecule is realized by combining with Argonaute endonuclease, and the self-connector is cut from the dsDNA library molecule, so as to prevent the self-connector from being amplified in a subsequent PCR reaction. According to the method for eliminating the self-connector of the sequencing library, the proportion of self-connectors of the library can be remarkably reduced, the sequencing clean reads can be increased, and the data efficiency can be improved.
Owner:上海金匙医学检验实验室有限公司
Method for constructing sequencing library based on single-stranded dna molecule and its application
ActiveCN105463585BEfficient constructionEfficiently obtainedNucleotide librariesMicrobiological testing/measurementSingle strand dnaDouble stranded
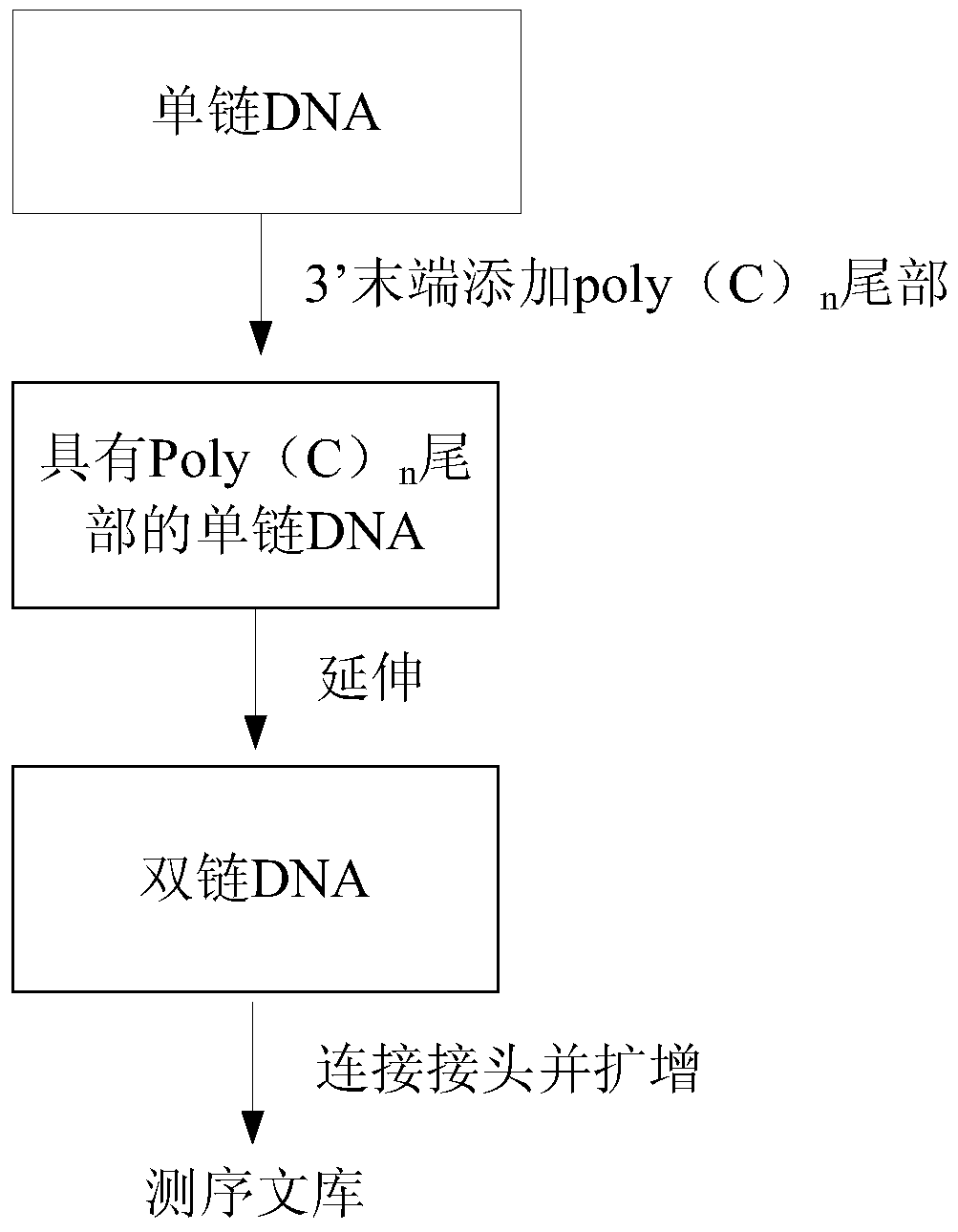
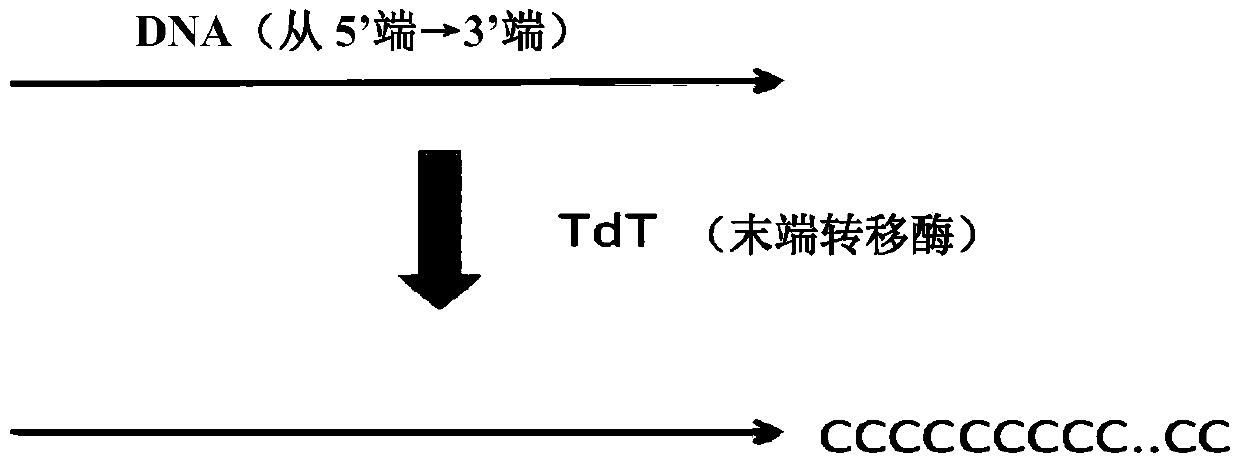
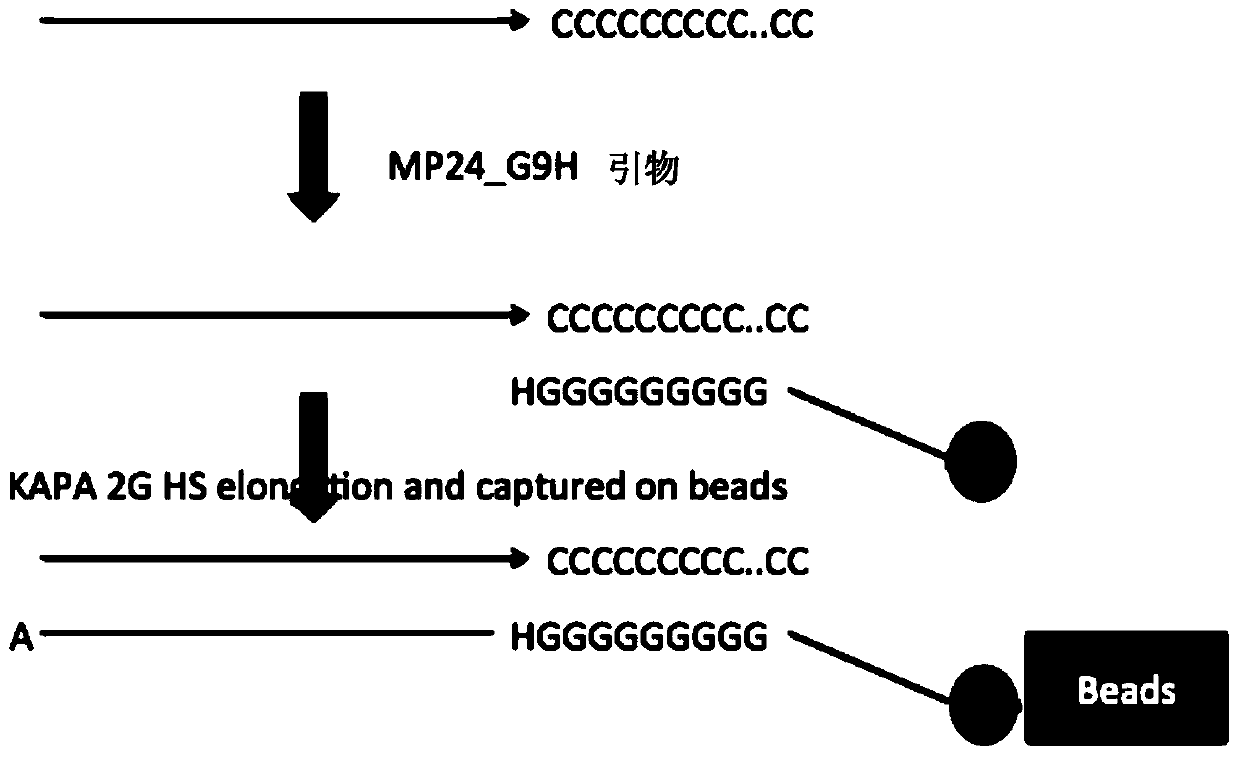
The invention discloses a method for constructing a sequencing library based on a single-stranded DNA molecule and its application, wherein the method for constructing a sequencing library includes: (1) forming a poly(C)n tail at the 3' end of the single-stranded DNA molecule, wherein n represents The number of base C; (2) Utilize the extension primer, based on the single-stranded DNA molecule with Poly(C)n tail, obtain the double-stranded DNA molecule, wherein, the extension primer includes H(G)m at its 3' end unit, H is base A, base T or base C, and m is the number of base G; and (3) connecting a linker at the end of the double-stranded DNA molecule away from the H(G)m unit, and connecting the The resulting ligation products are amplified, and the amplified products constitute the sequencing library. The method can effectively construct a sequencing library of single-stranded DNA molecules, especially a sequencing library of micro samples.
Owner:TSINGHUA UNIV +1
A chromosome aneuploidy detection system suitable for single cells and its application
ActiveCN108363903BReduce the starting amountEnables detection of aneuploidyMicrobiological testing/measurementBiostatisticsGenetic engineeringChromatosome
The invention belongs to the technical field of genetic engineering, and in particular relates to a single-cell chromosome aneuploidy detection system and its application. The detection system includes a window division module, a segment division module, a segment copy rate calculation module, and a segment aneuploidy judgment module. The detection system uses up to 700k reads / sample, and can detect chromosome deletion / duplication of more than 5Mb in the sample, with an accuracy rate of up to 99.9%, reducing the depth of sequencing and reducing costs.
Owner:UNIMED BIOTECH SHANGHAI CO LTD
DU5 adaptor, application thereof and cDNA library constructed by dU5 adaptor
PendingCN114763546AReduce formationReduce the number of cyclesMicrobiological testing/measurementLibrary linkersCDNA libraryDimer
The invention provides a dU5'adapter, application thereof and a cDNA library constructed by the dU5 'adapter. The structure of the dU5'adapter is 5 '-p-S1-S2-S3-S4-SpC3-3', S1, S2, S3 and S4 are gene segments which are sequentially connected through chemical bonds, and p represents phosphorylation modification performed at the 5'end; s1 is a first stem; s2 is a ring part; s3 is a second stem part and contains one deoxidized uracil dU; the 5 '-> 3' sequence of S1 and the 3 '-> 5' sequence of S3 are complementary, S4 is a guide part (N) m, N is any one selected from A, T, G and C basic groups, m is 10, and SpC3 represents C3 interval modification performed at the 3'terminus. The dU5'adapter can reduce formation of adapter-primer dimers, improve the PCR amplification efficiency, shorten the PCR period and improve the library quality, and a new important platform is provided for constructing a cDNA library for wide biological application.
Owner:香港城市大学深圳研究院
Method for constructing colorectal cancer susceptibility gene variation library
ActiveCN106757378BReduce morbidityReduce mortalityNucleotide librariesMicrobiological testing/measurementIntestinal CancerFhit gene
The invention relates to a colorectal cancer susceptibility gene mutation library constructing method. The colorectal cancer susceptibility gene mutation library constructing method comprises the following steps: A, designing primer pairs capable of detecting a mutation region according to the mutation region related to a colorectal cancer; B, calculating to obtain judgment parameters R of the various primer pairs, classifying the primer pairs of which the values of the judgment parameters R are smaller than or equal to 1 into a first group of primer combination liquid, and classifying the primer pairs of which the values of the judgment parameters R are greater than 1 into a second group of primer combination liquid; C, extracting and purifying DNA of a sample to be detected; D, carrying out initial PCR amplification on the purified sample by using the first group of primer combination liquid and the second group of primer combination liquid respectively; E, connecting target fragments via joints to obtain fragments with joints; and F, carrying out library PCR amplification by using mixed liquid of the first group of primer combination liquid and the second group of primer combination liquid to obtain a sequencing library. The library constructed by the colorectal cancer susceptibility gene mutation library constructing method is high in sequencing flux, high in sensitivity and high in specificity, and can be used for detecting free DNA low frequency mutation.
Owner:深圳市艾伟迪生物科技有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com