Method for preparing chromatin co-immunoprecipitation library by using trace of clinical puncture sample and application of method
A co-immunoprecipitation and chromatin technology, applied in the biological field, can solve the problems of low success rate, cumbersome steps, and complicated chromatin co-immunoprecipitation experimental steps.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] Example 1. Preparation method of chromatin immunoprecipitation library
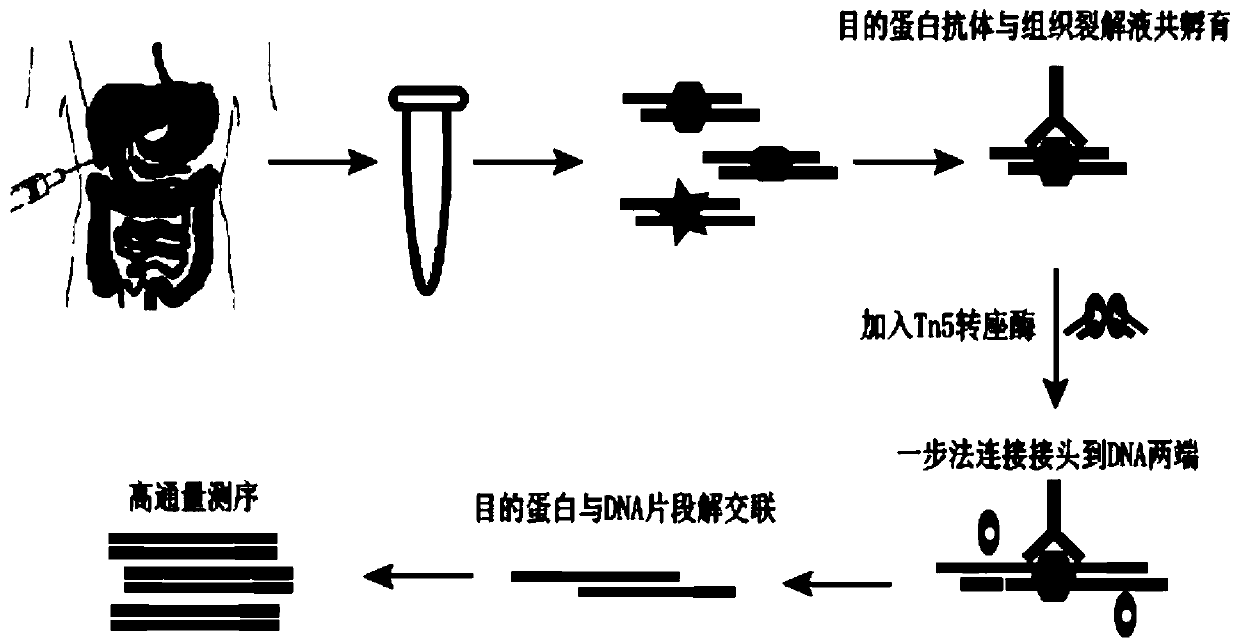
[0079] The flow chart of the rapid co-immunoprecipitation based on the Tn5 transposition system of the present invention is as follows figure 1 shown. These include clinical puncture sample collection, formaldehyde cross-linking, tissue homogenization, cell lysis, ultrasonic interruption of DNA fragments, target protein immunoprecipitation, Tn5 transposition system to break the DNA fragments bound by the target protein and one-step ligation of library adapters, Protein digestion and decompression of DNA fragments bound by cross-linked target proteins, enrichment of DNA fragments bound by target proteins and amplification. Specific steps are as follows:
[0080] 1. Put the clinical puncture sample tissue into a 1.5ml centrifuge tube, add 500μl pre-cooled PBS solution (pH 7.4) to wash the tissue, use a desktop centrifuge at 4°C and 500g for 10 minutes, remove the PBS solution, and get Washed tissu...
Embodiment 2
[0099] Example 2, Application of the preparation method of chromatin immunoprecipitation library
[0100] 1. Clinical puncture samples
[0101] Liver biopsy samples from 3 different individuals (from the Second Affiliated Hospital of Nanchang University, with informed consent from all individuals) were used as clinical biopsy samples, and the initial volumes of clinical biopsy samples were 7.7 mg, 11.4 mg, and 5.5 mg, respectively.
[0102] 2. Preparation of chromatin immunoprecipitation library
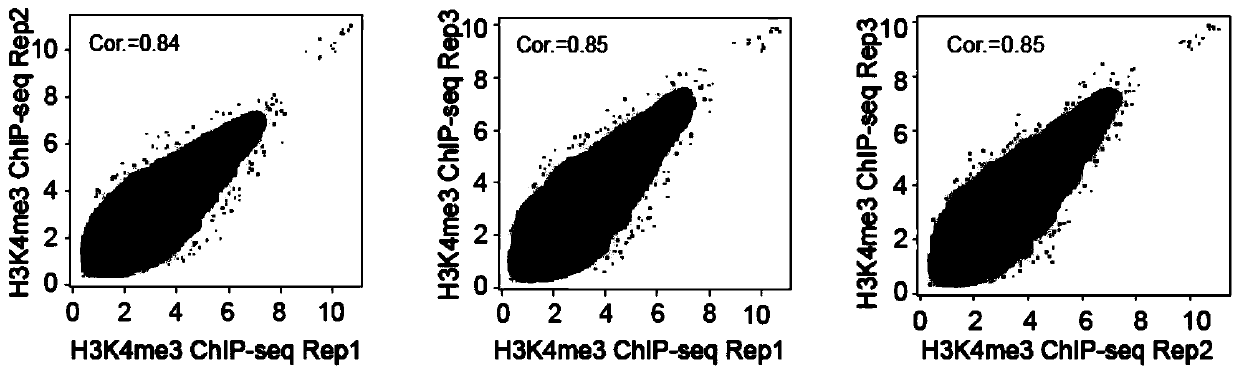
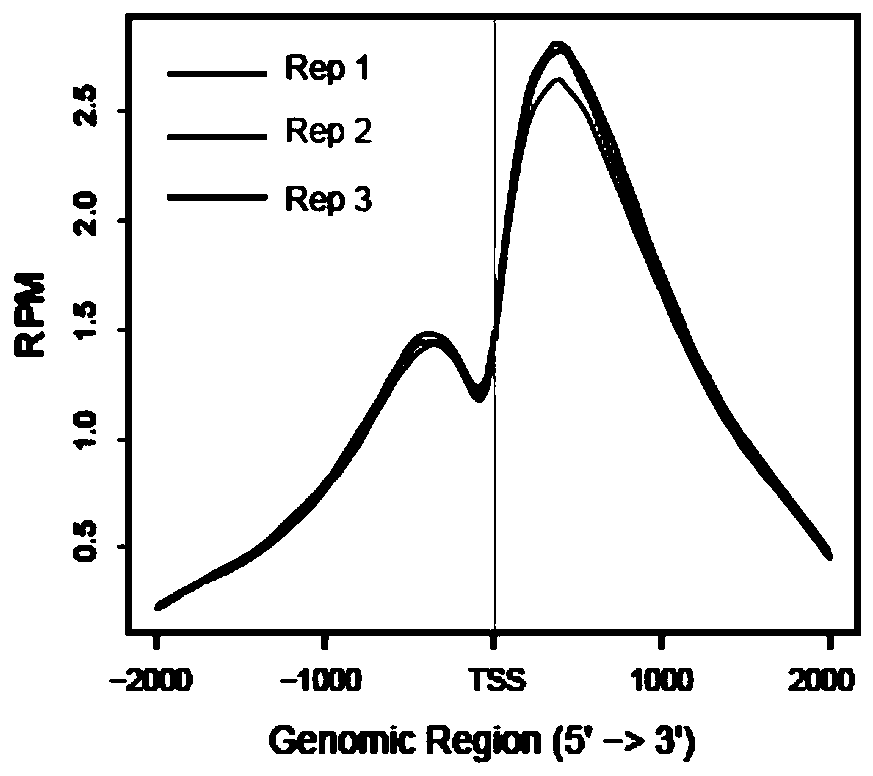
[0103] Based on the clinical biopsy samples in step 1, a chromatin immunoprecipitation library was constructed according to the method in Example 1. Wherein, the target protein is H3K4me3, and the specific target protein antibody in step 6 is H3K4me3 antibody (ActiveMotif). The sequences of the upstream and downstream primers in step 12 are as follows (NNNNNNNNNN is the library Index sequence):
[0104] S501:
[0105] AATGATACGGCGACCACCGAGATCTACANNNNNNTCGTCGGCAGCGTC;
[0106] N7...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com