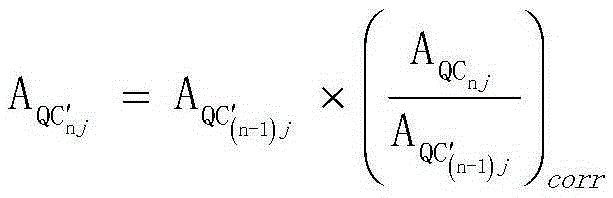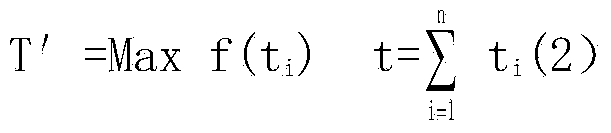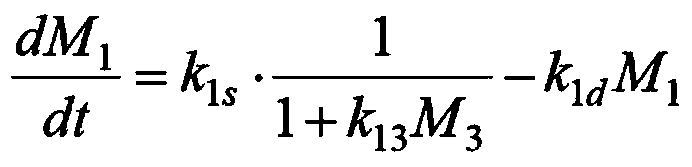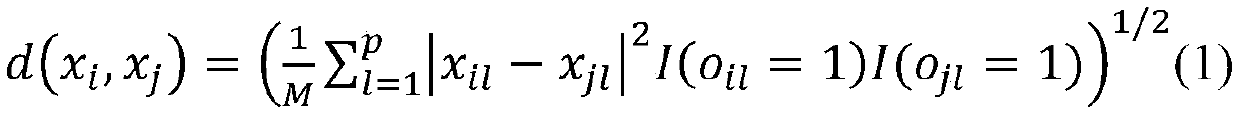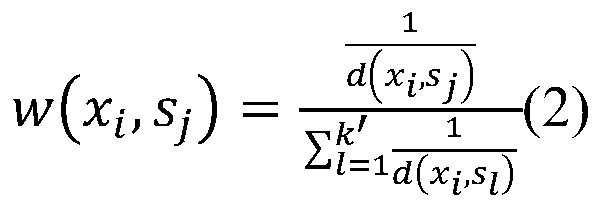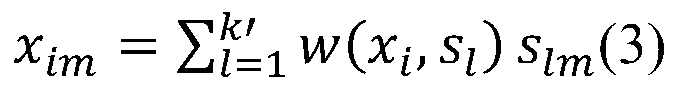Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
63 results about "Metabolomics data" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
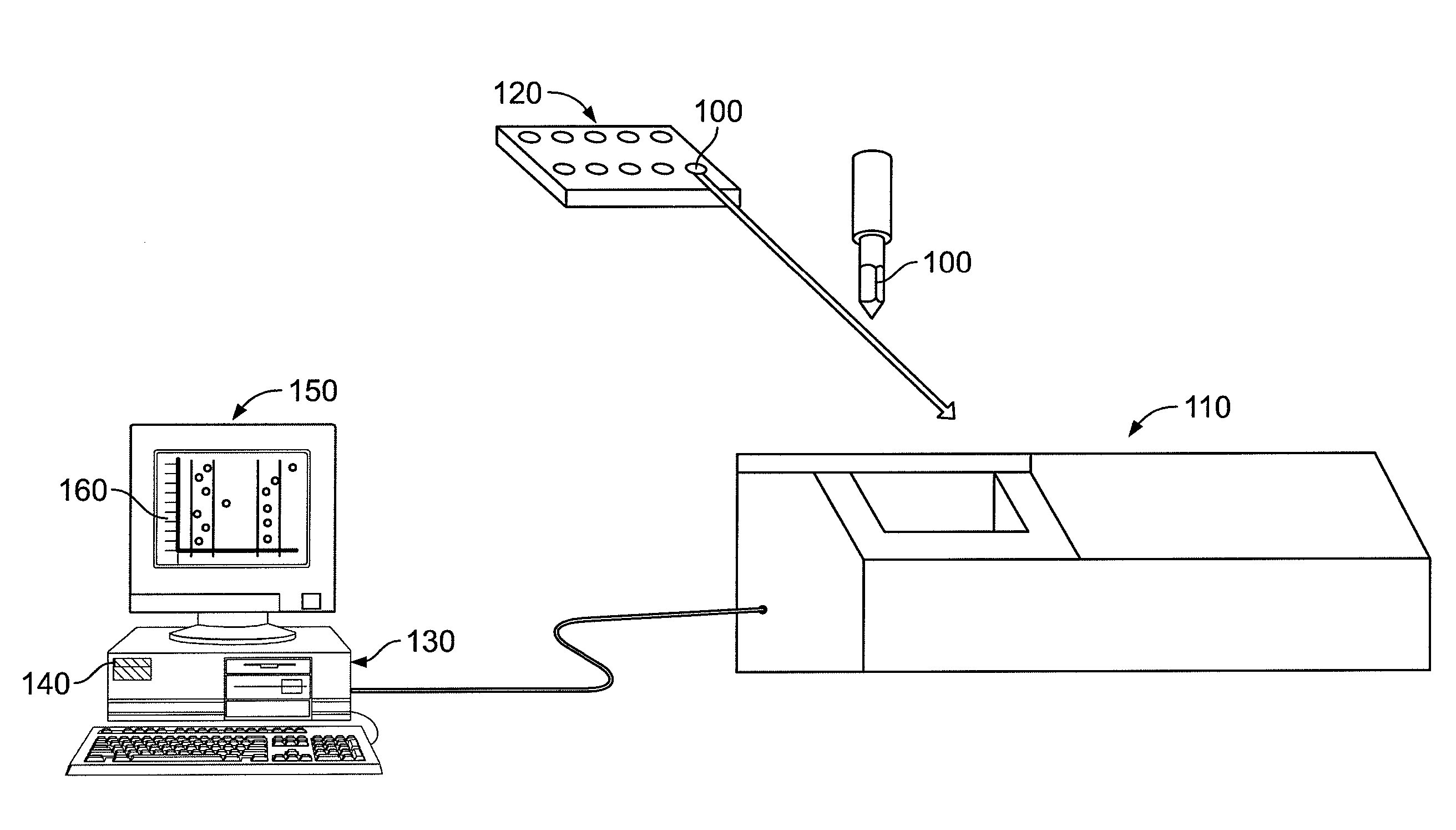
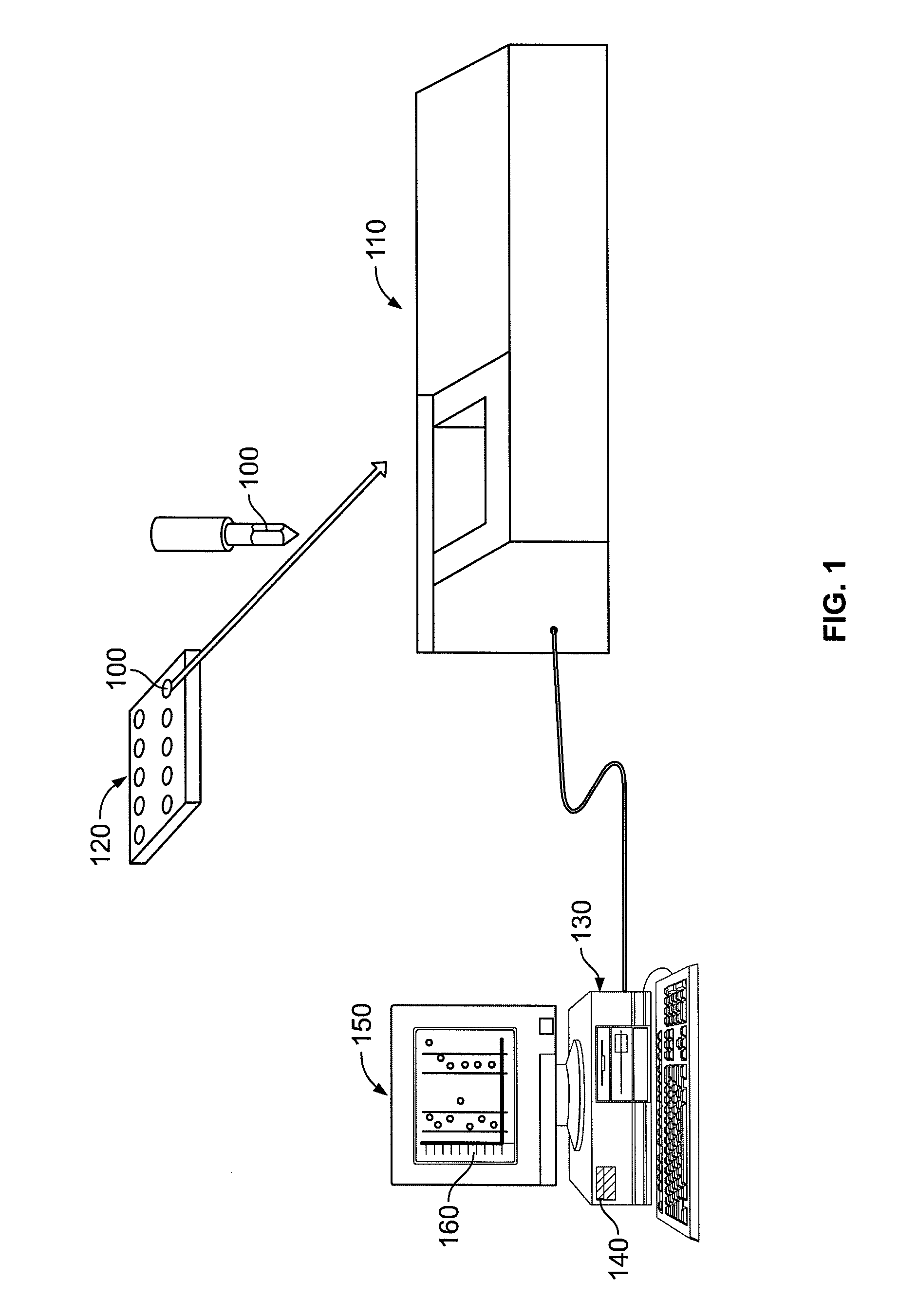
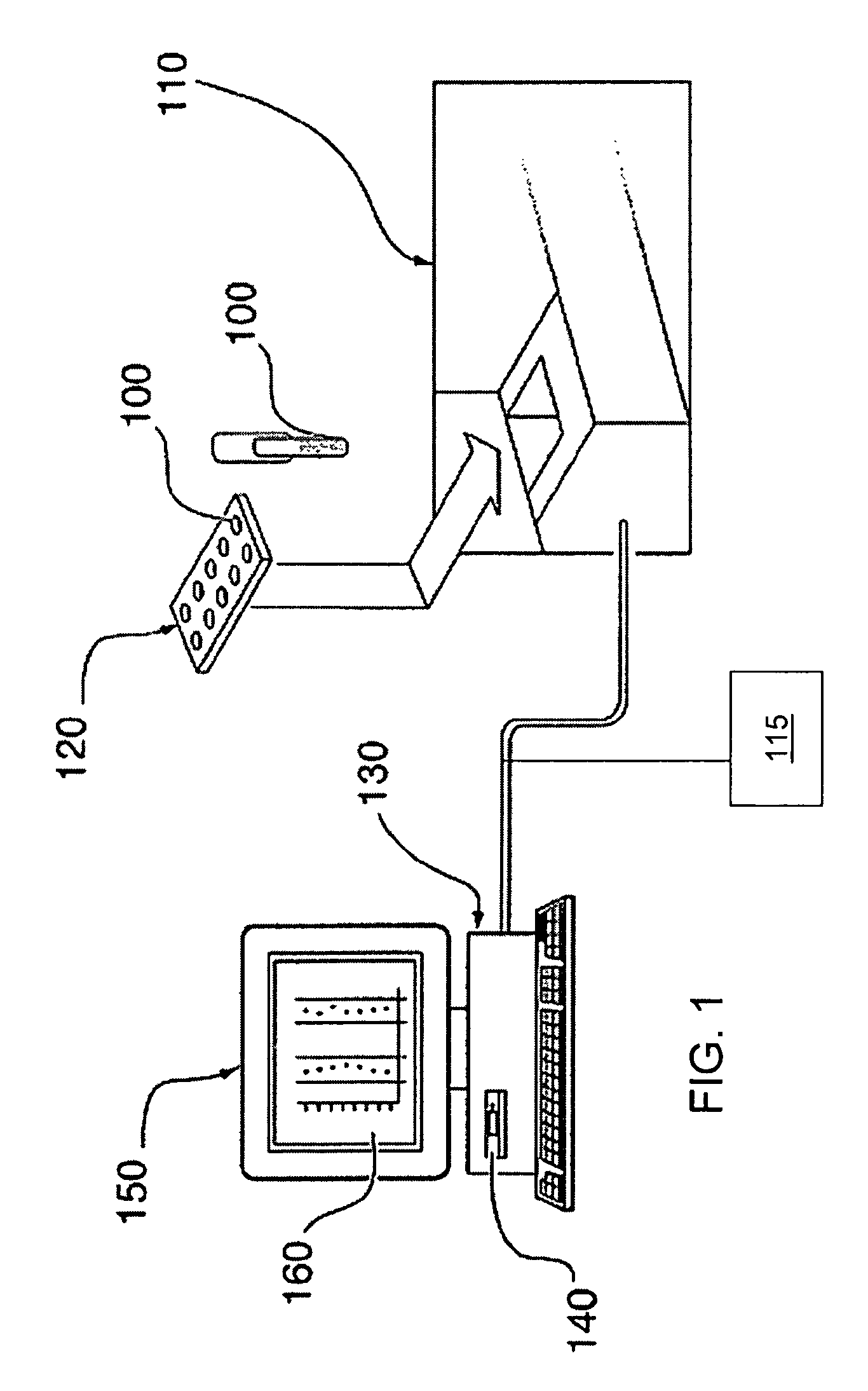
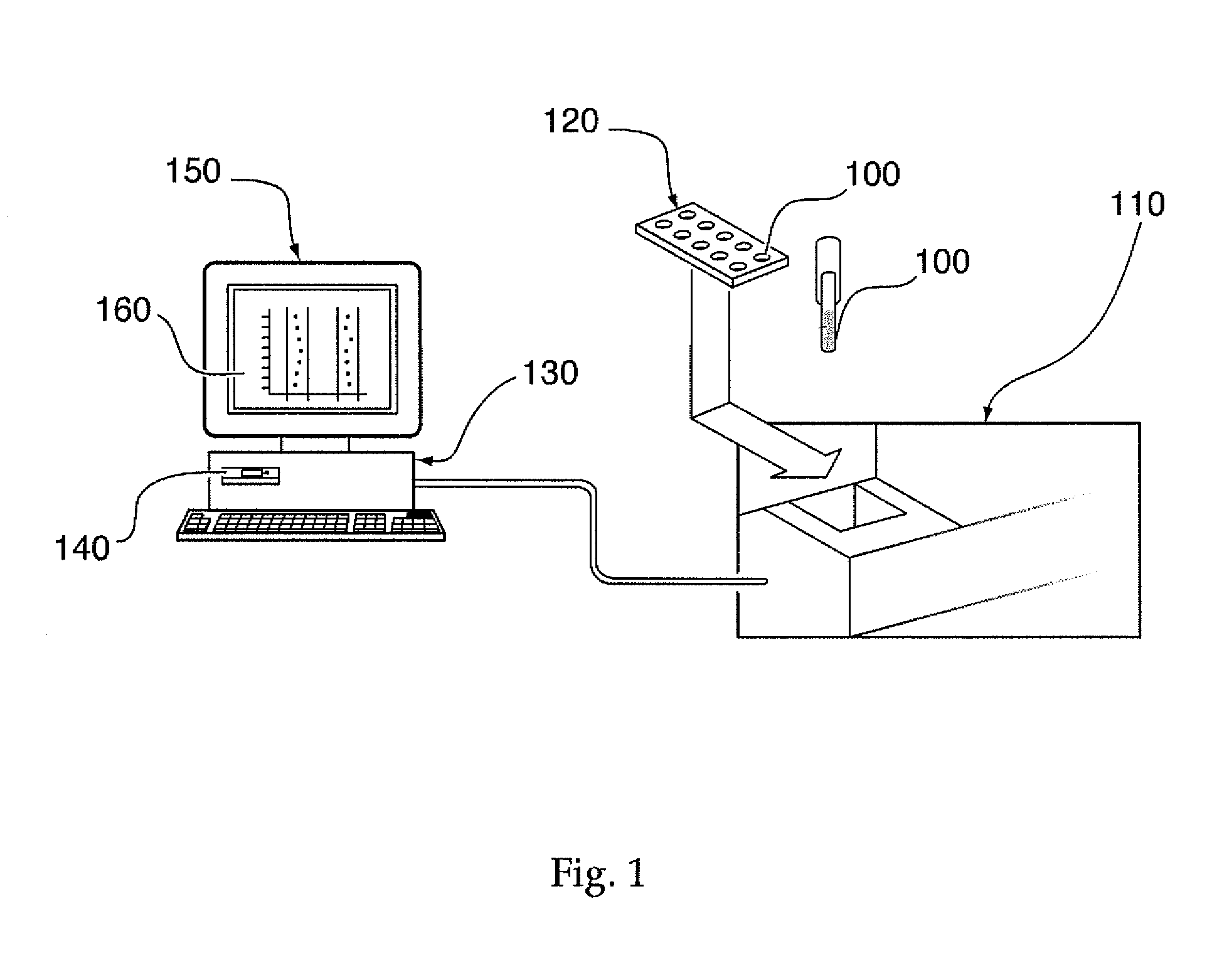
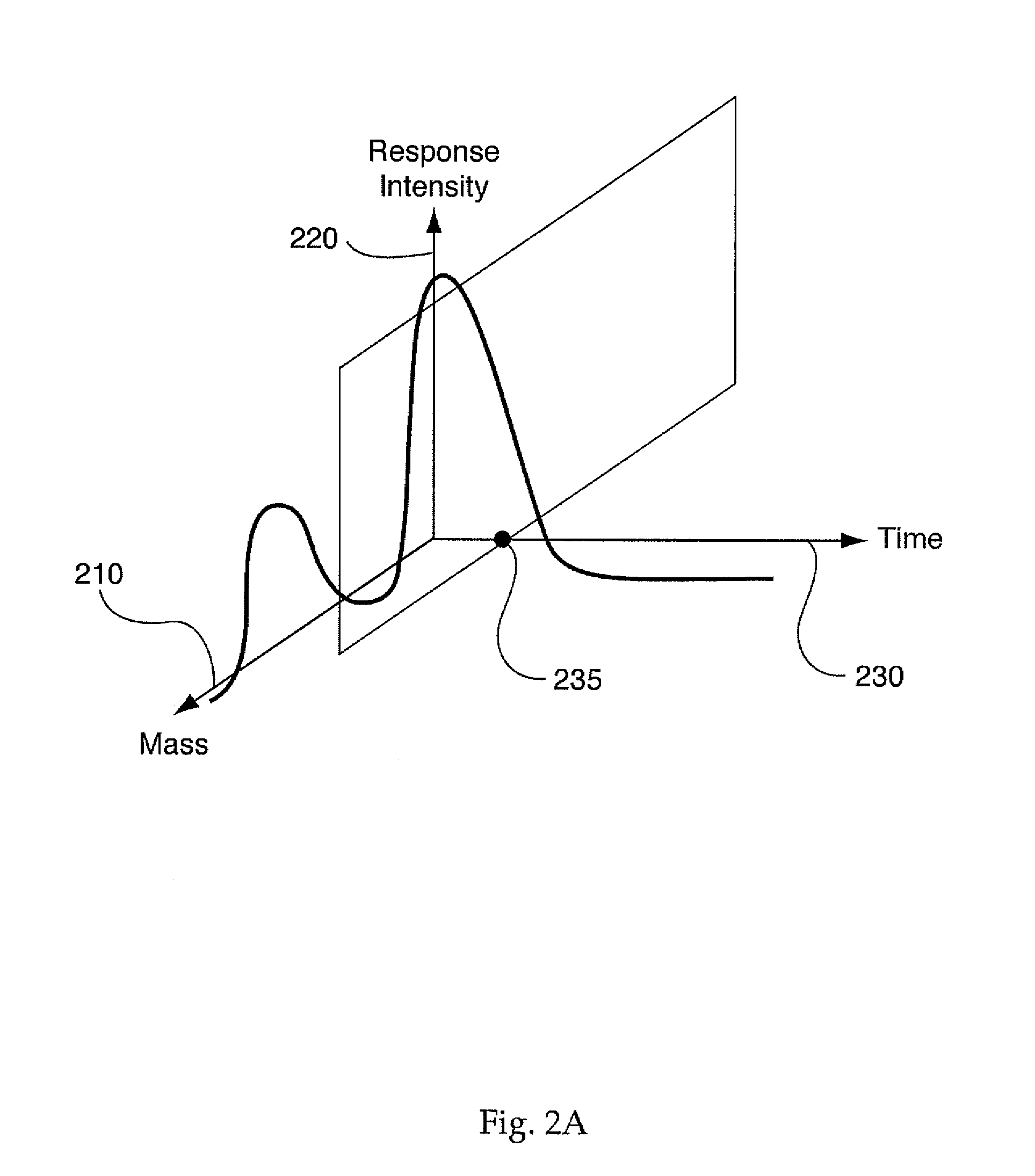
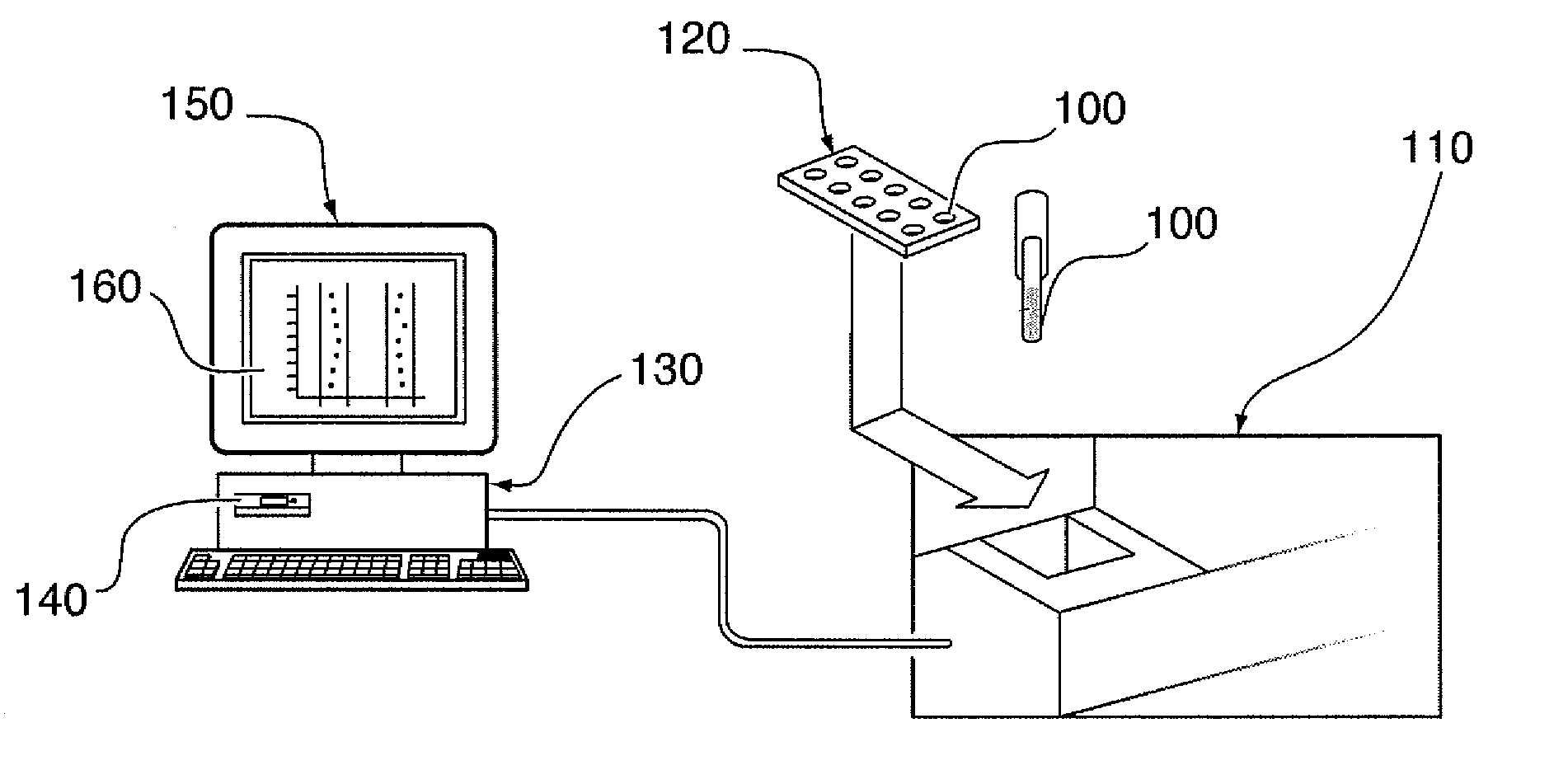
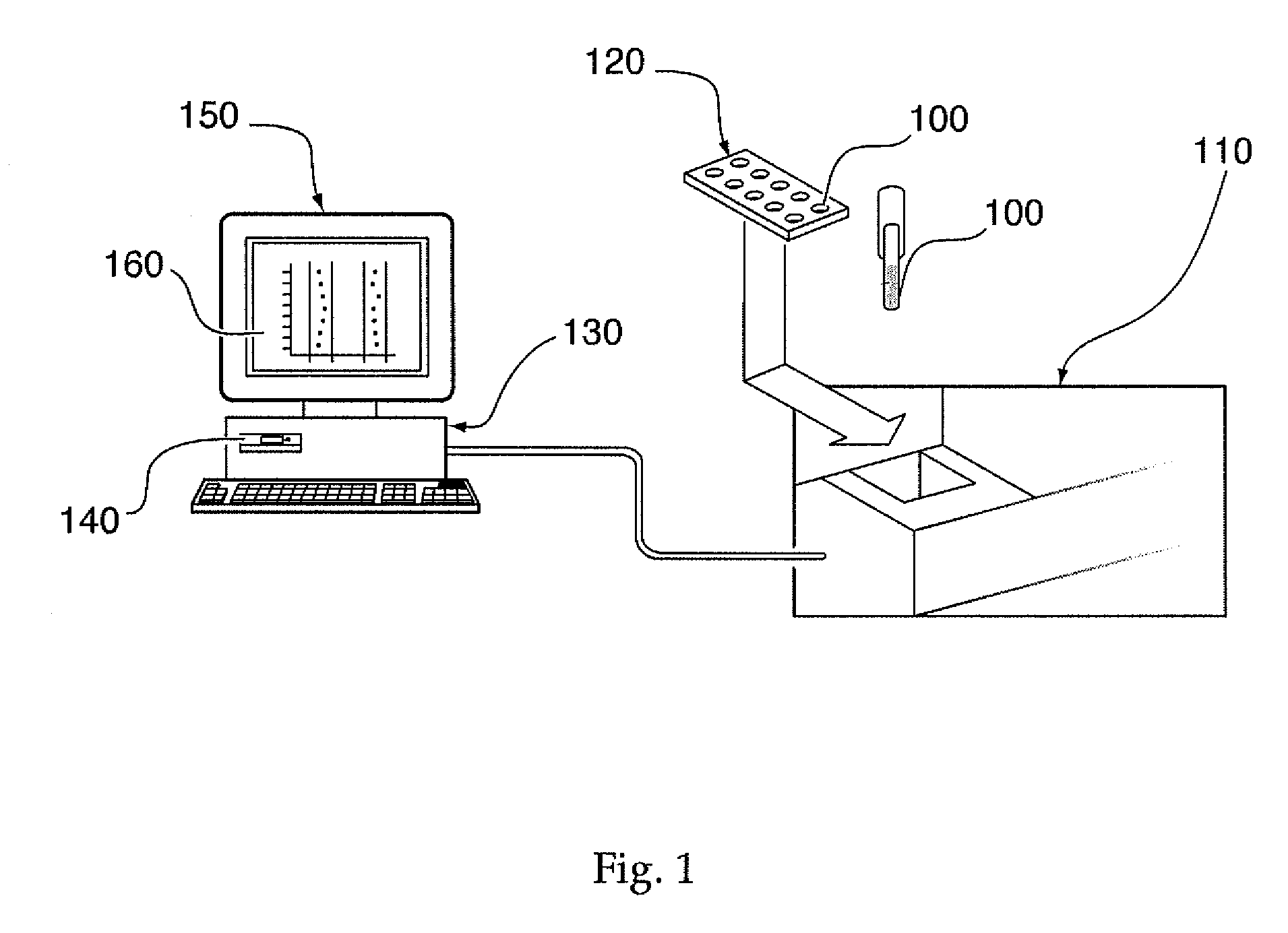
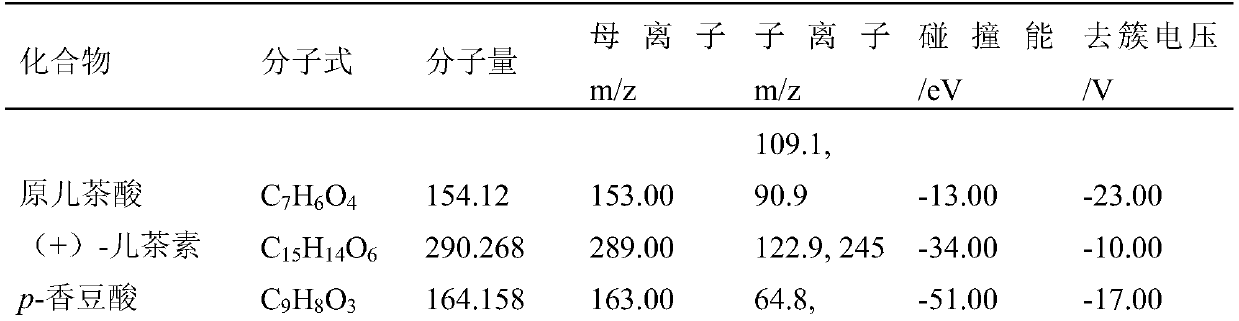
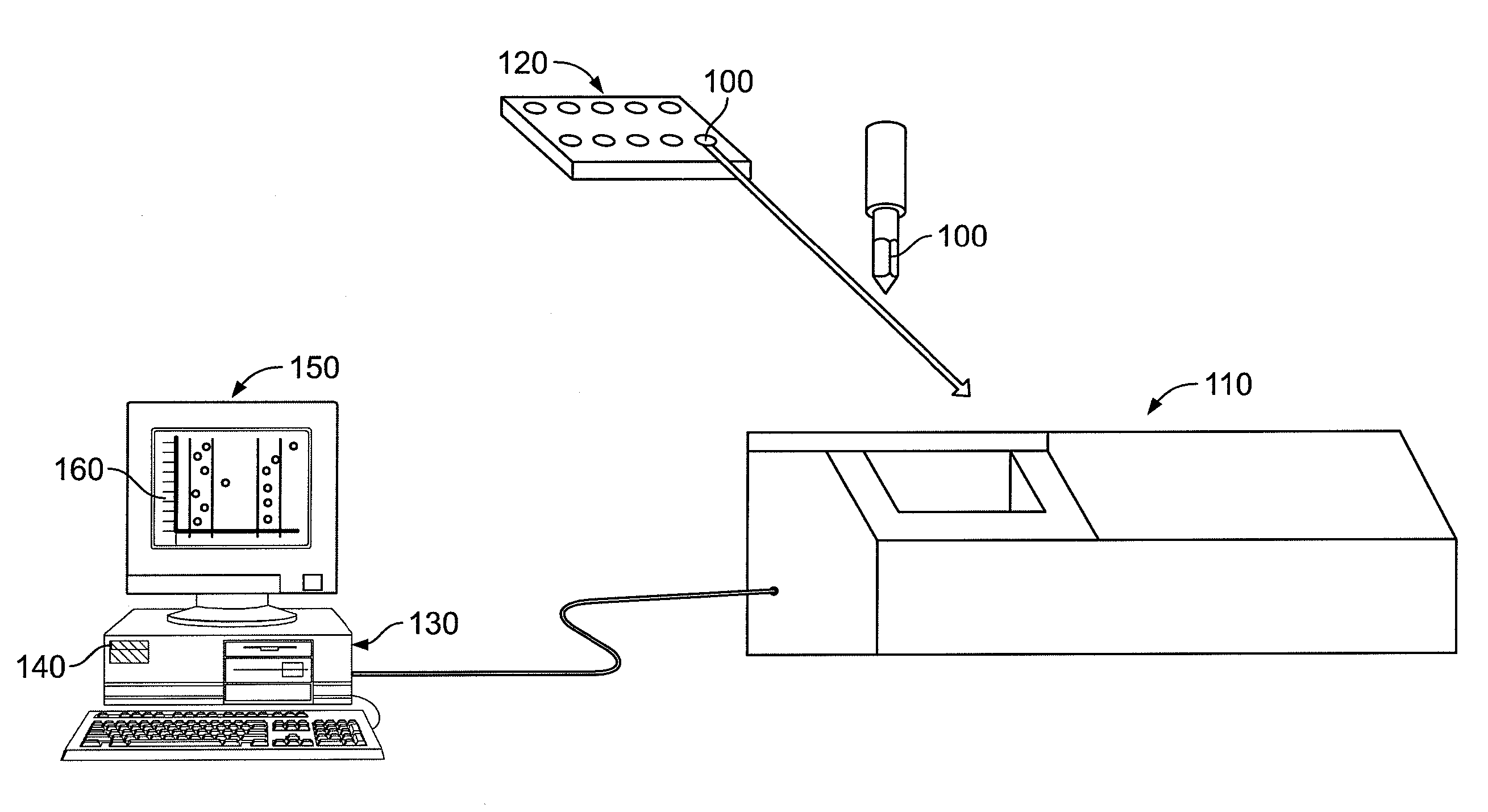
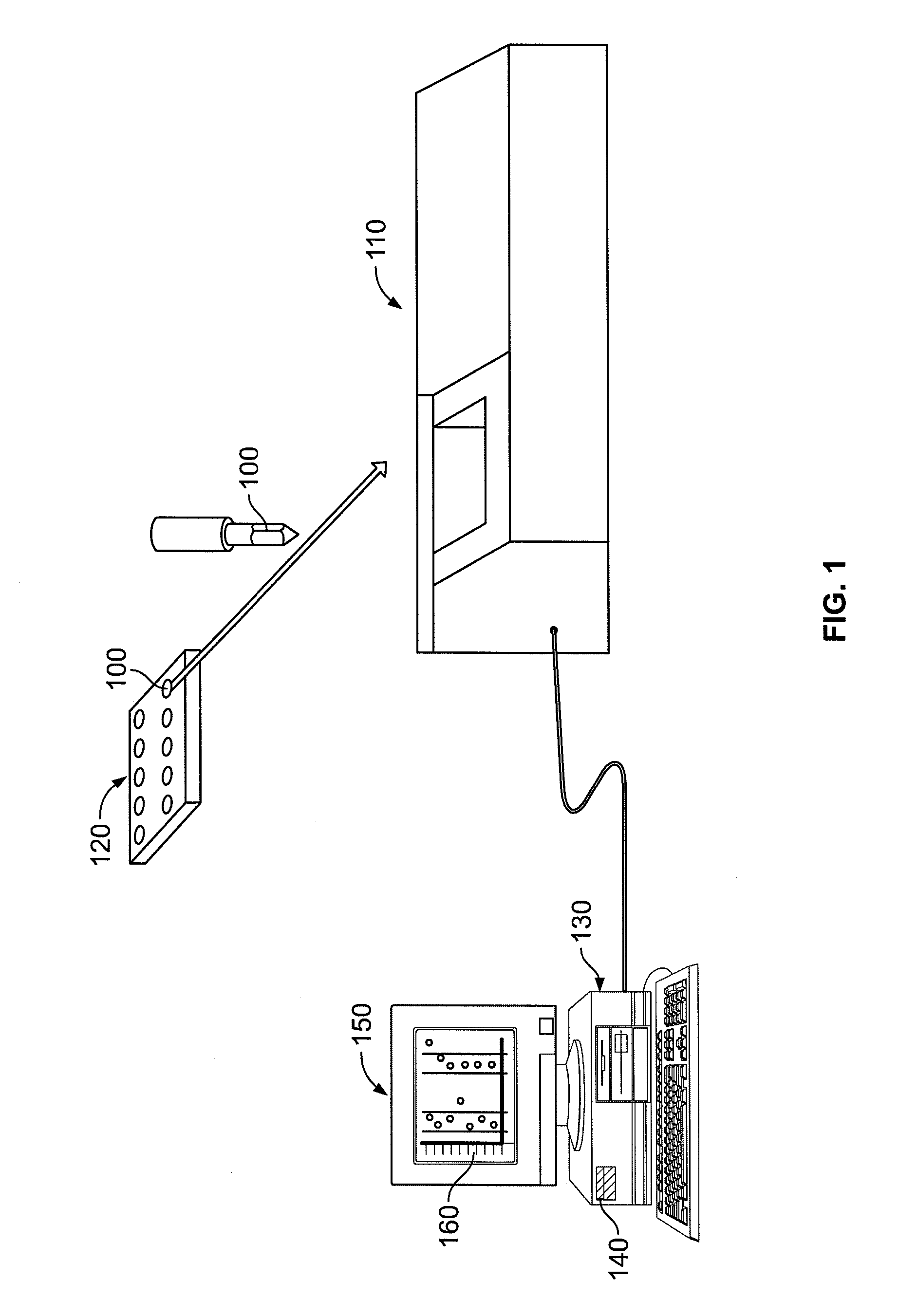
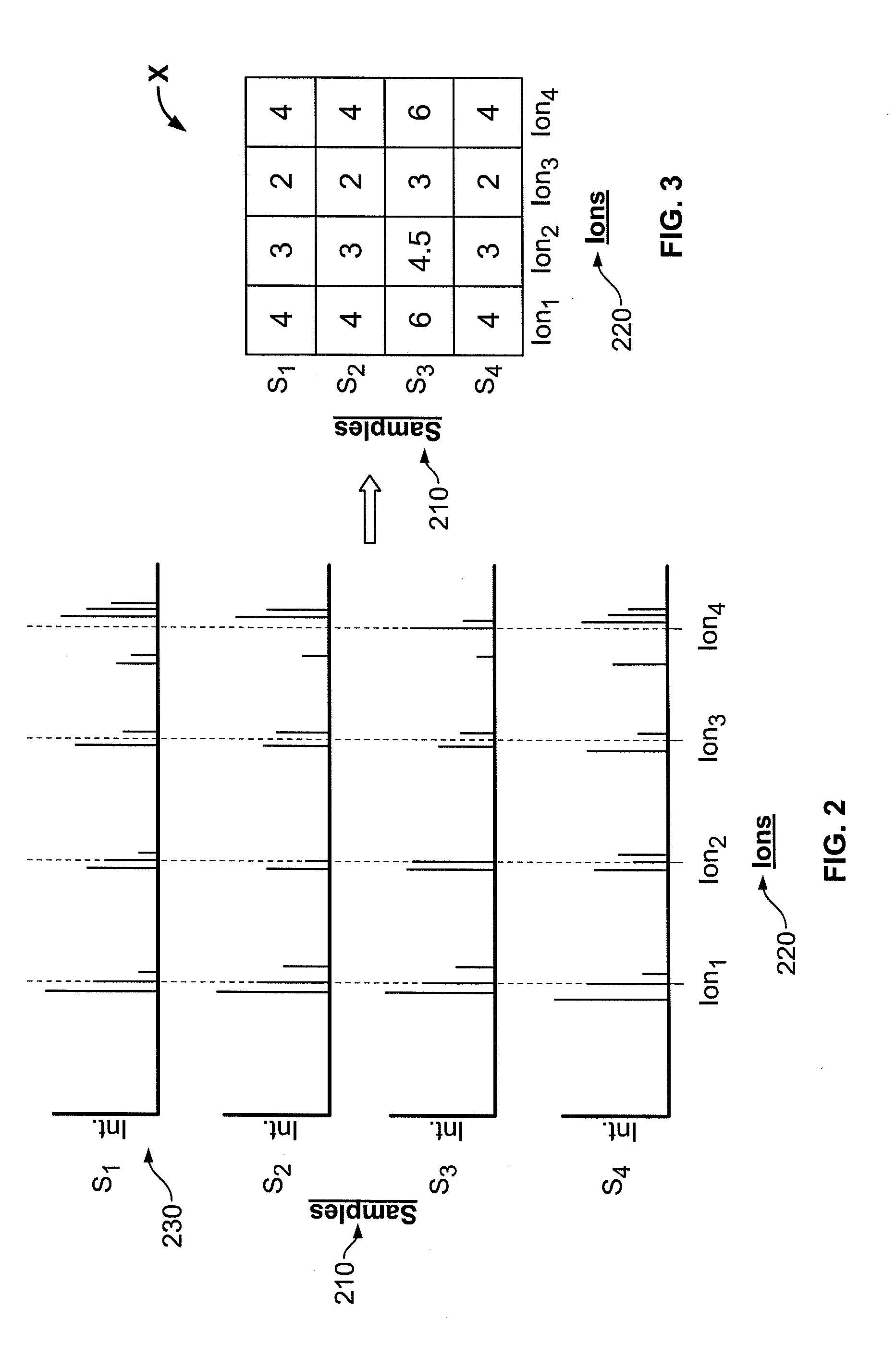
System, method, and computer program product for analyzing spectrometry data to identify and quantify individual components in a sample
ActiveUS7561975B2Molecular entity identificationParticle separator tubesCorrelation functionMetabolomics data
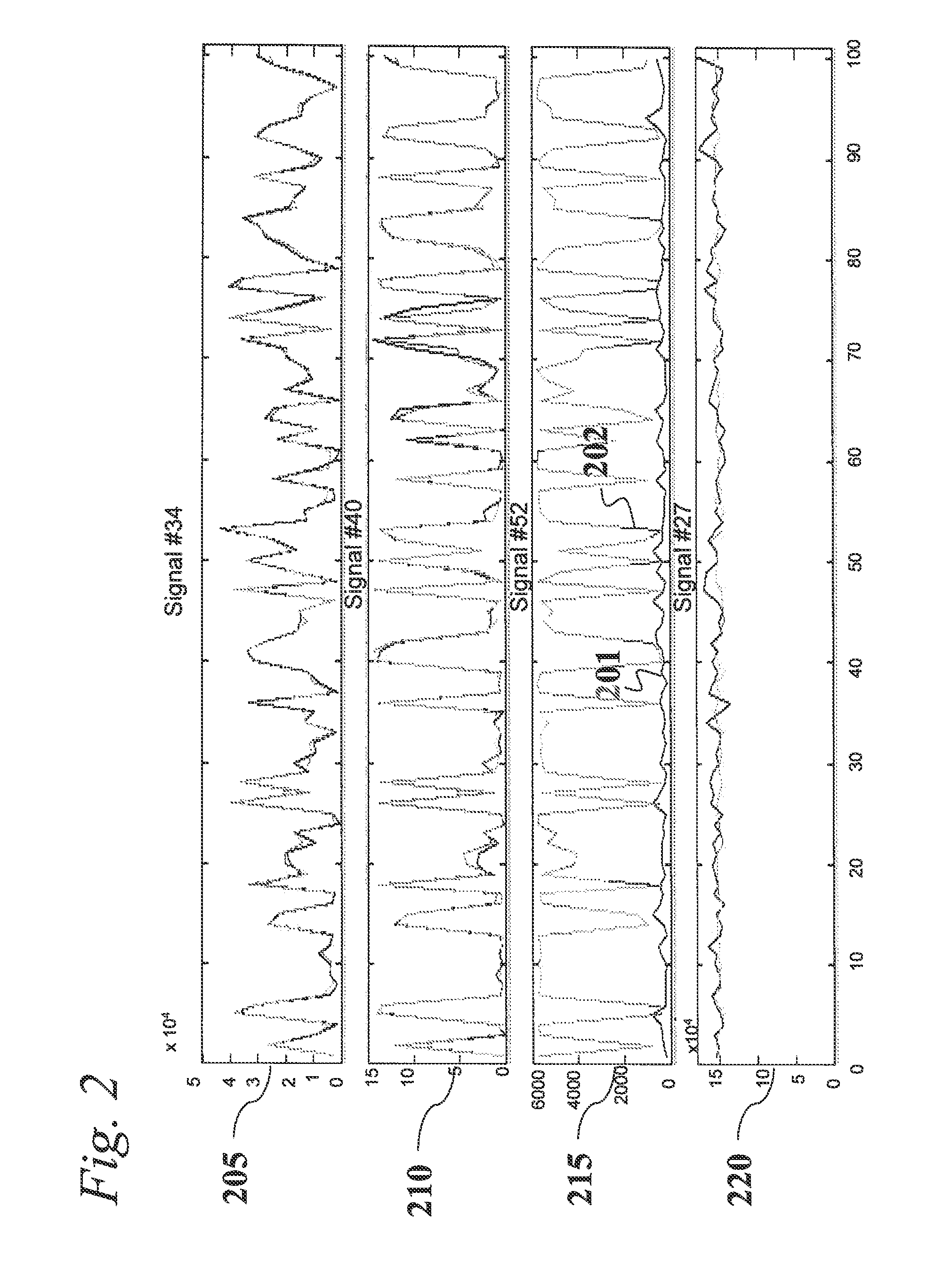
A system is provided for analyzing metabolomics data received from an analytical device across a group of samples. The system automatically receives a data matrix corresponding to each of the samples, wherein the data matrix includes rows corresponding to each of the samples and columns corresponding to a group of ions present in the respective samples. A processor is provided for determining a characteristic value corresponding to at least one of a group of components present in the samples, wherein the components are made up of at least a portion of the group of ions, using at least one of a correlation function and a factorization function. A user interface is in communication with the processor for displaying a visual indication of the characteristic value such that a user may receive a visual indication of the types of components present in the samples.
Owner:METABOLON
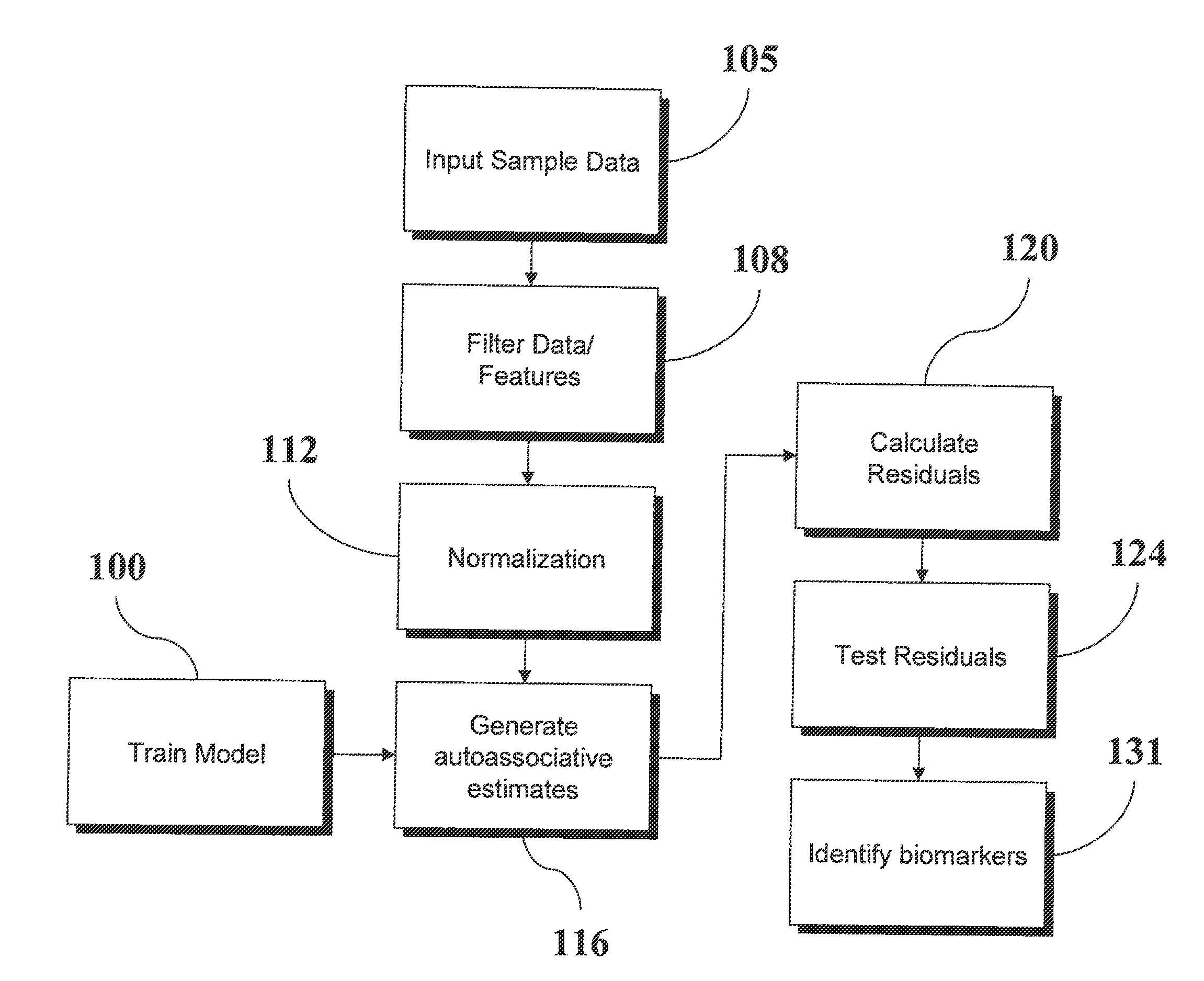
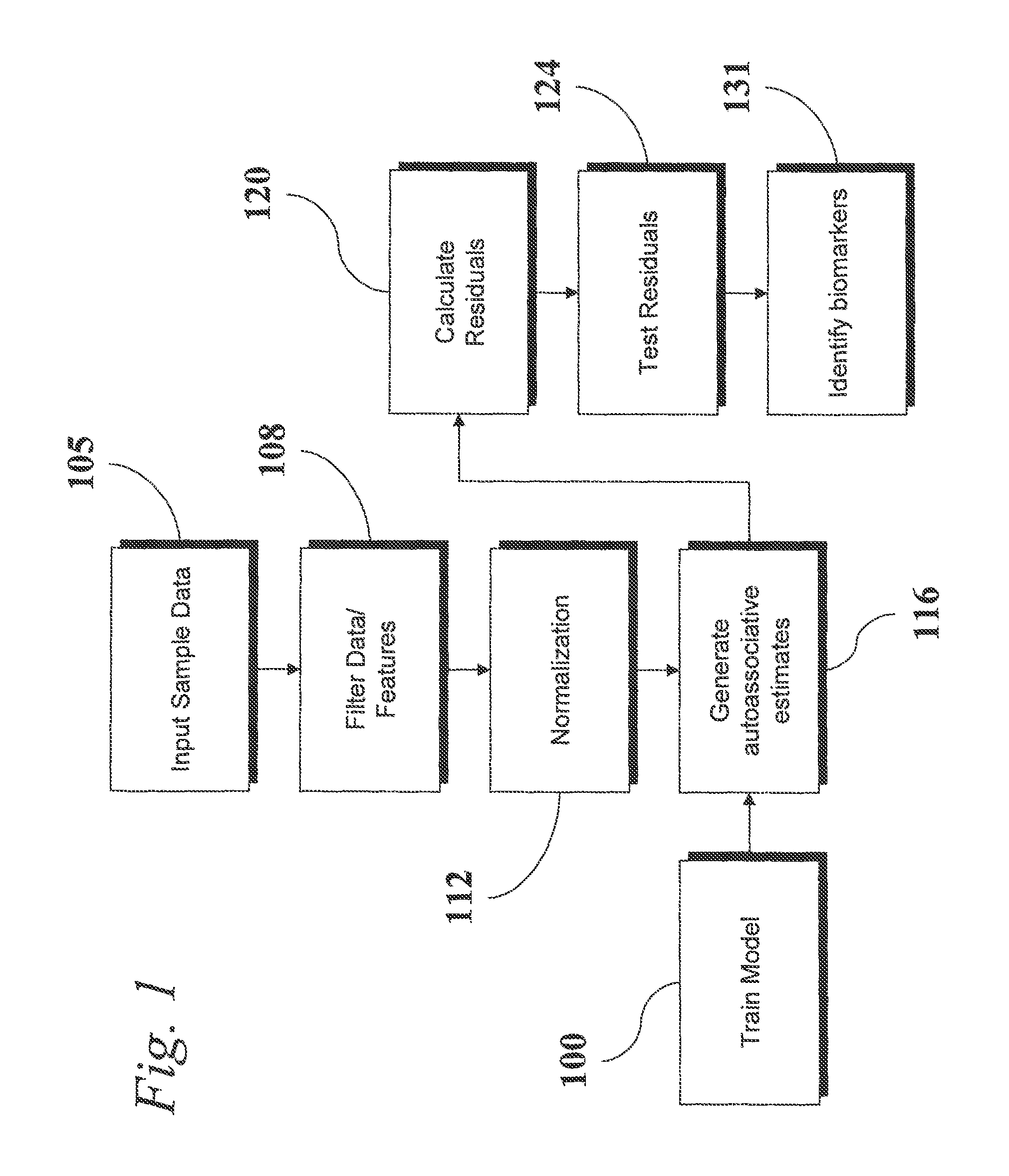
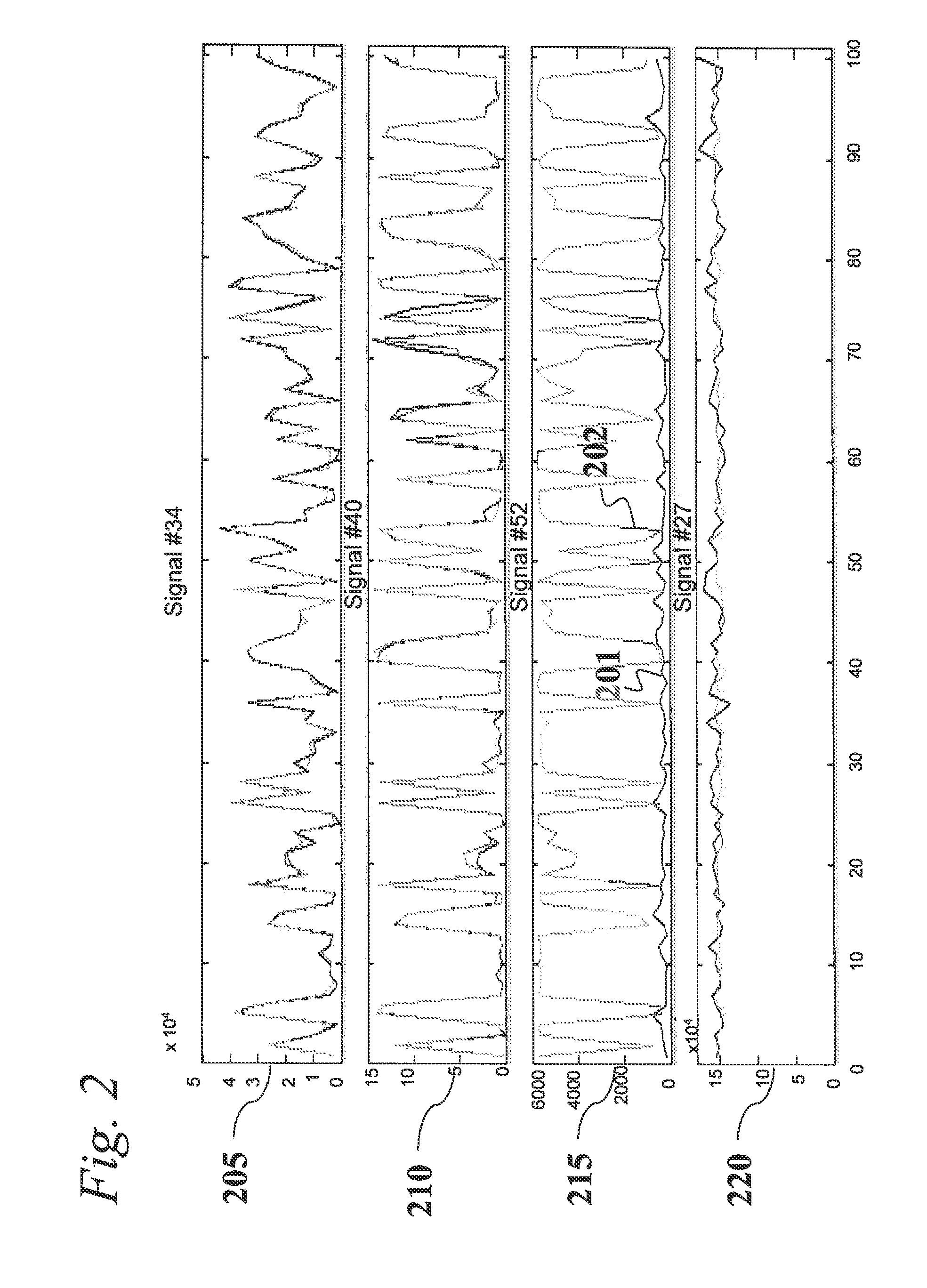
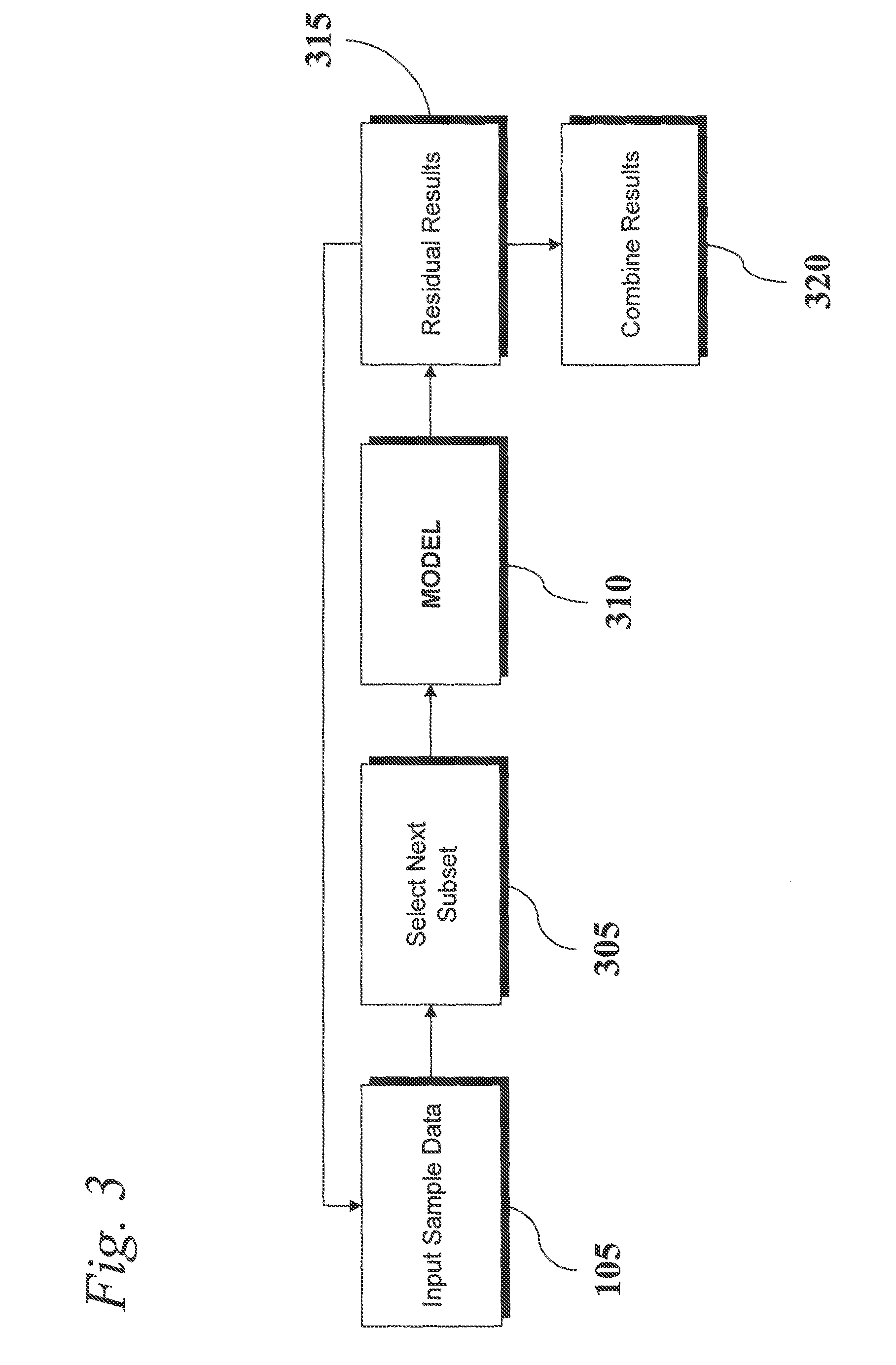
Analysis of Transcriptomic Data Using Similarity Based Modeling
ActiveUS20110093244A1Small footprintFast data processingBiostatisticsComputation using non-denominational number representationBiomarker discoveryOrganism
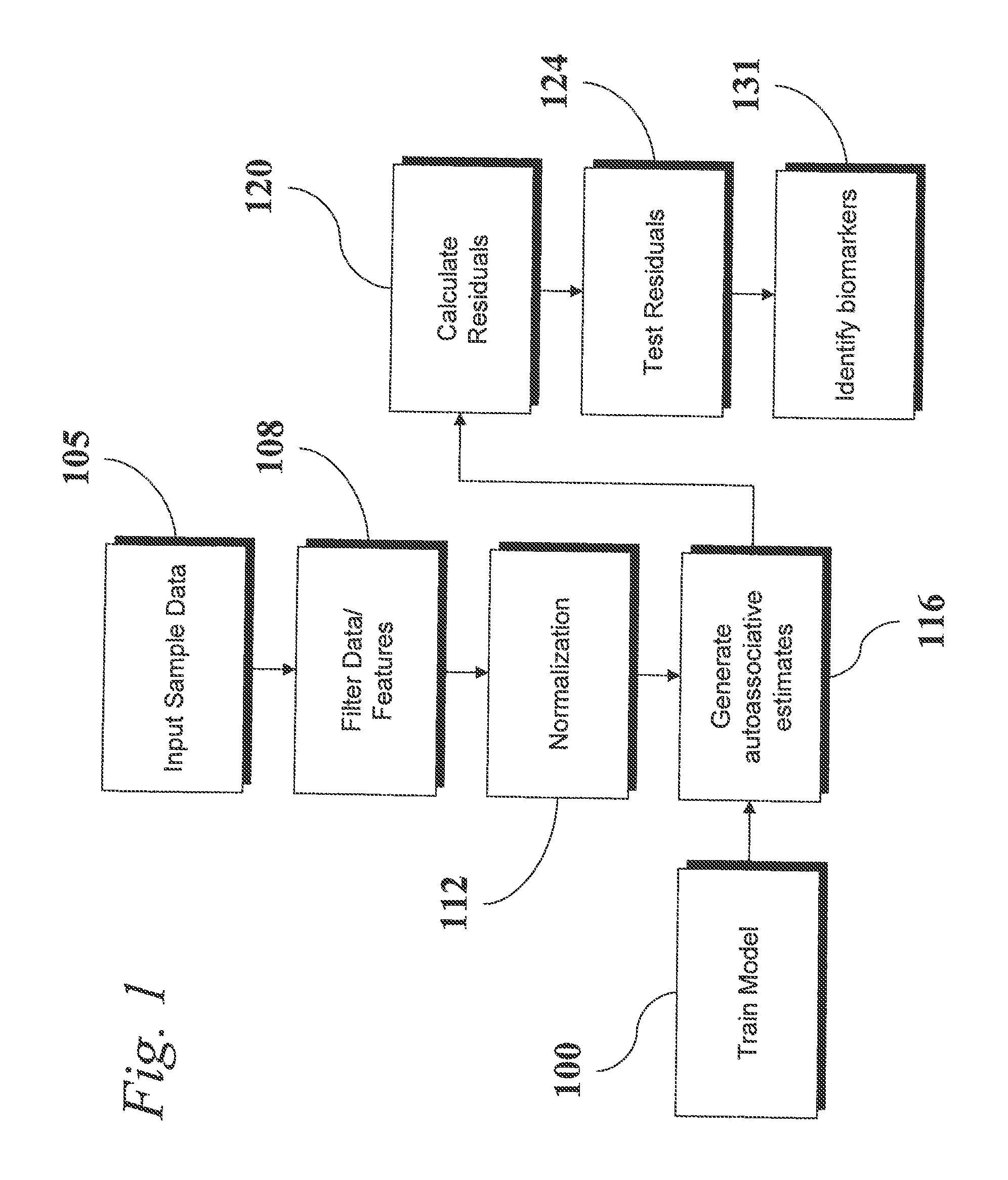
An analytic apparatus and method is provided for diagnosis, prognosis and biomarker discovery using transcriptome data such as mRNA expression levels from microarrays, proteomic data, and metabolomic data. The invention provides for model-based analysis, especially using kernel-based models, and more particularly similarity-based models. Model-derived residuals advantageously provide a unique new tool for insights into disease mechanisms. Localization of models provides for improved model efficacy. The invention is capable of extracting useful information heretofore unavailable by other methods, relating to dynamics in cellular gene regulation, regulatory networks, biological pathways and metabolism.
Owner:PHYSIQ
Method for establishing platform for analyzing and predicting cognitive impairment of diabetes based on metabonomics data
InactiveCN104933277AReduce morbidityAvoid bad consequencesComponent separationSpecial data processing applicationsMetabolitePrincipal component analysis
The present invention discloses a method for establishing a platform for analyzing and predicting the cognitive impairment of diabetes based on metabonomics data. According to the method, a high-performance liquid chromatography-mass spectrometry combined technology is adopted to separate and identify three groups of blood plasma of a healthy person, a diabetic patient who does not suffer from the cognitive impairment and a diabetic patient who suffers from the cognitive impairment; substances obtained by separation are respectively subjected to main component analysis and partial least square discriminant analysis by software; difference metabolites are determined and structures of the difference metabolites are identified; and the obvious difference metabolites are selected to form the platform for analyzing and predicting the cognitive impairment of diabetes. The cognitive impairment of diabetes can be timely analyzed and predicted according to the blood plasma of diabetic patients, can be timely observed and timely treated; the morbidity of the cognitive impairment of diabetes can be reduced; and a series of family and social problems caused by the cognitive impairment of diabetes are avoided.
Owner:THE SECOND HOSPITAL OF DALIAN MEDICAL UNIV
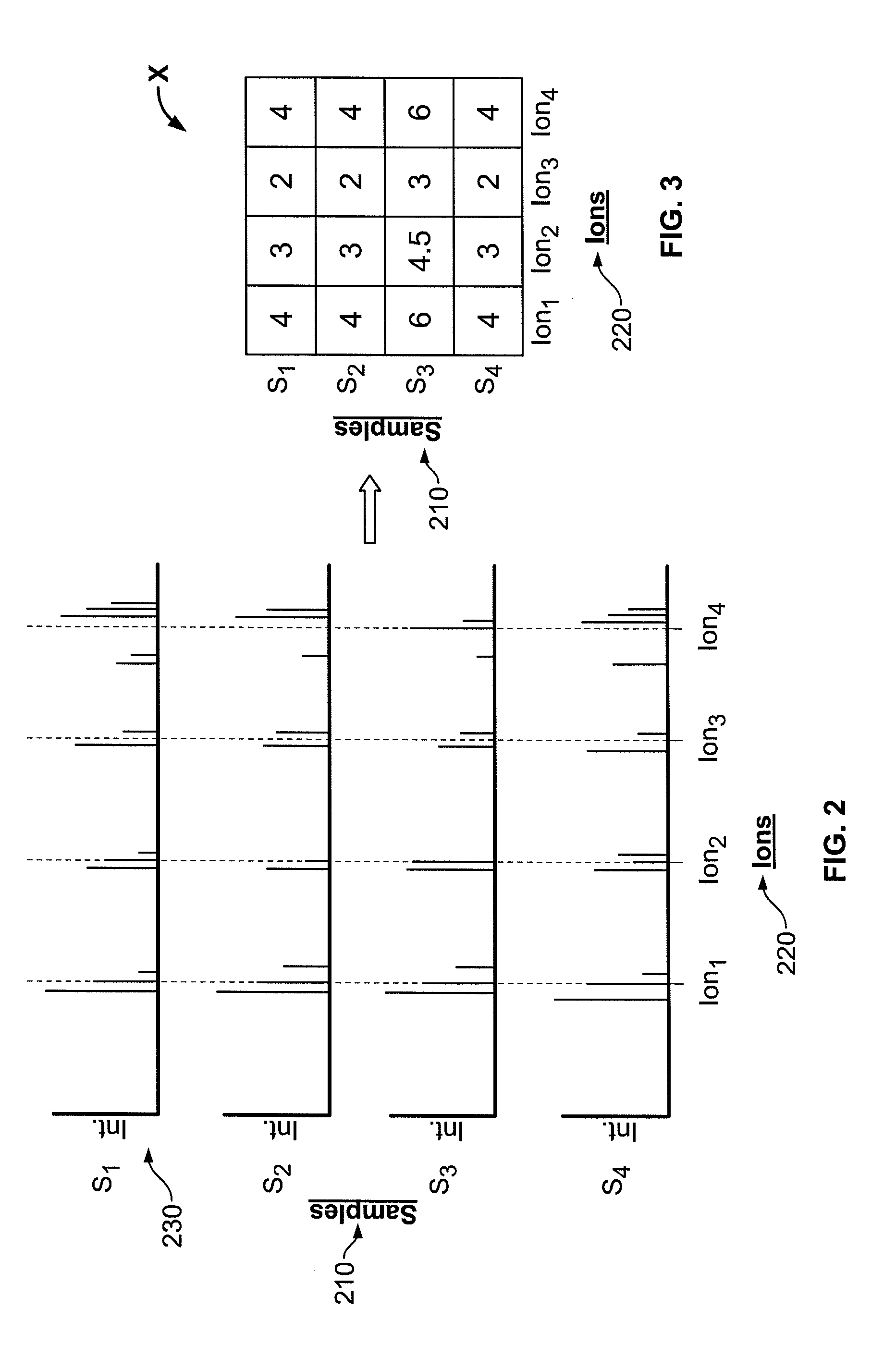
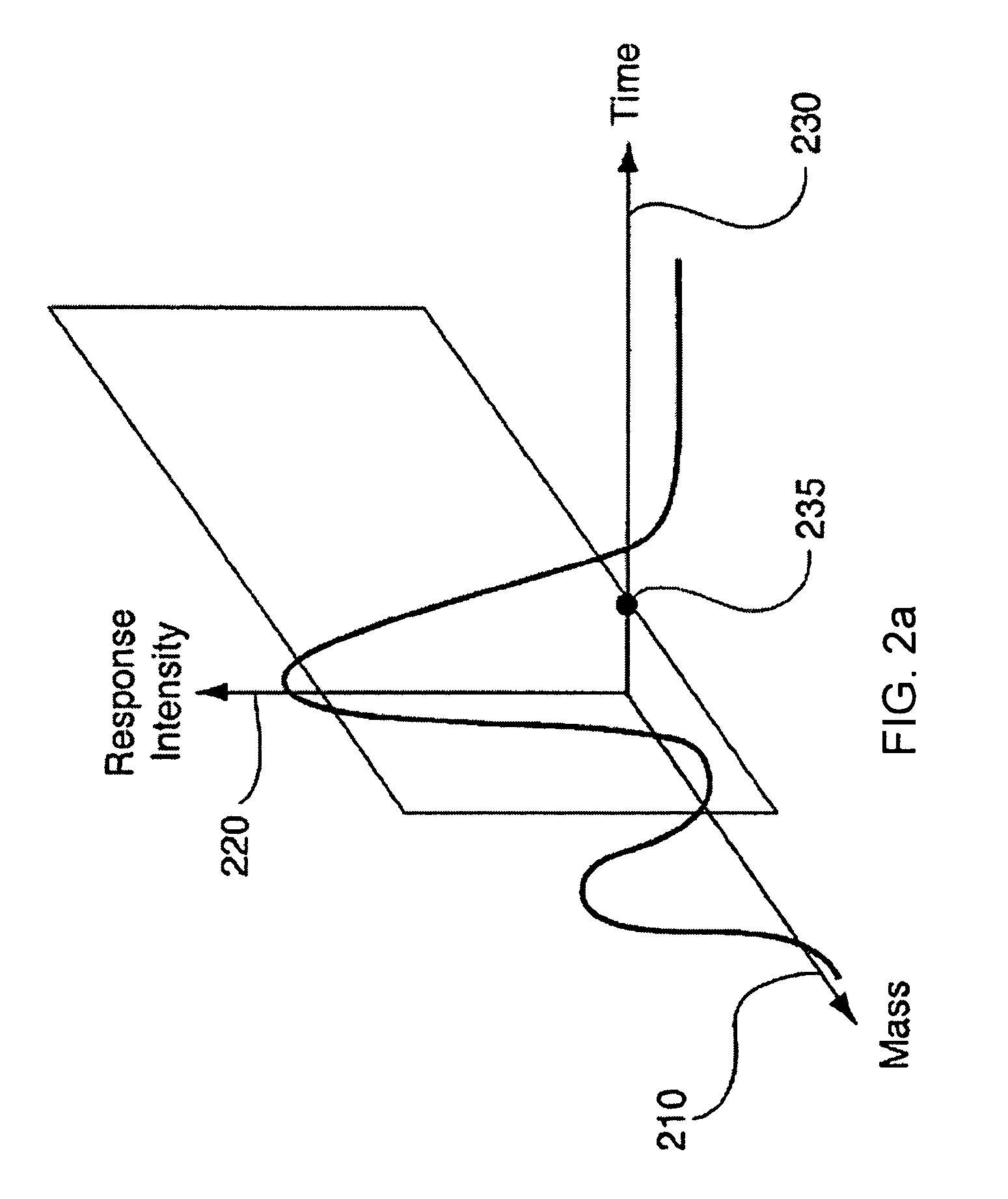
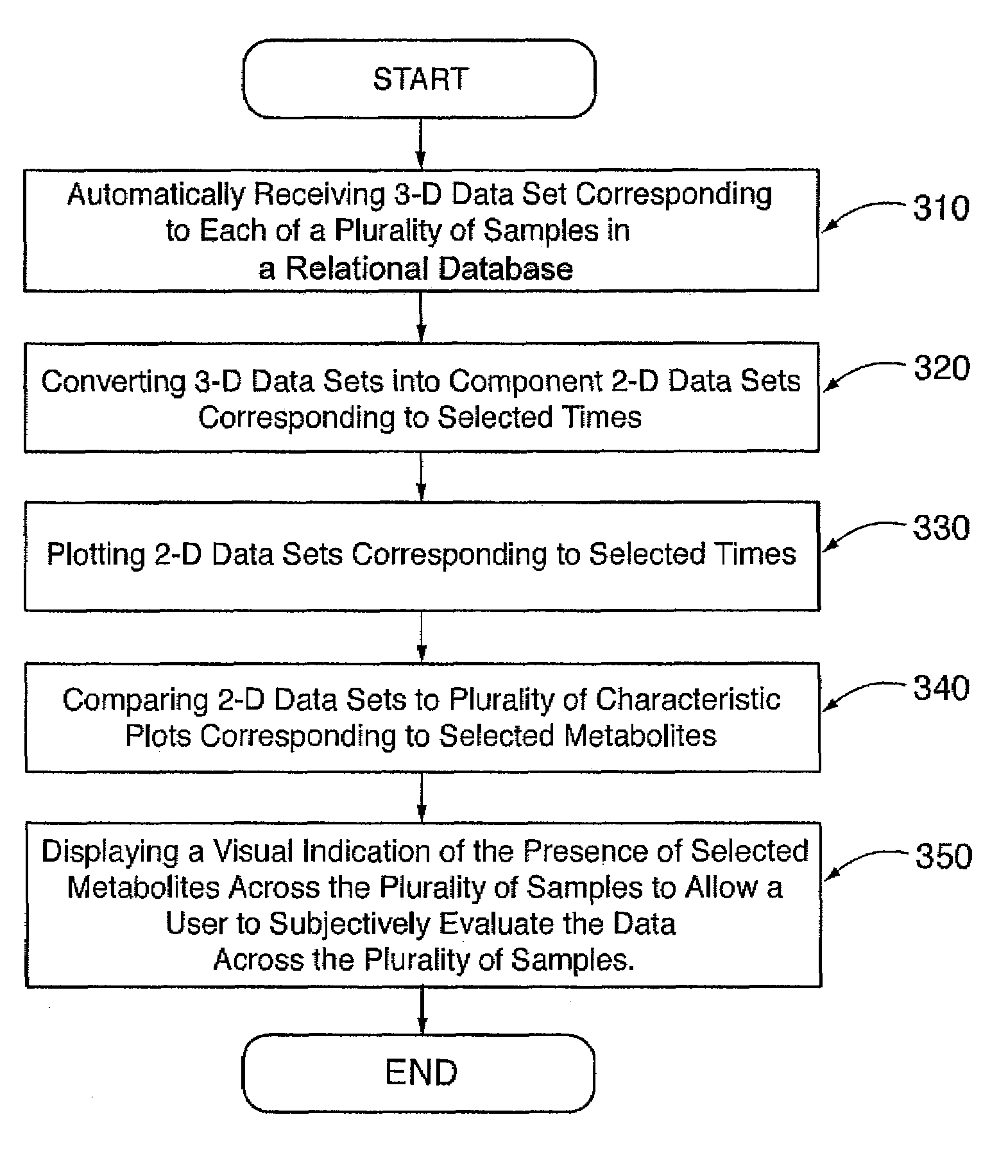
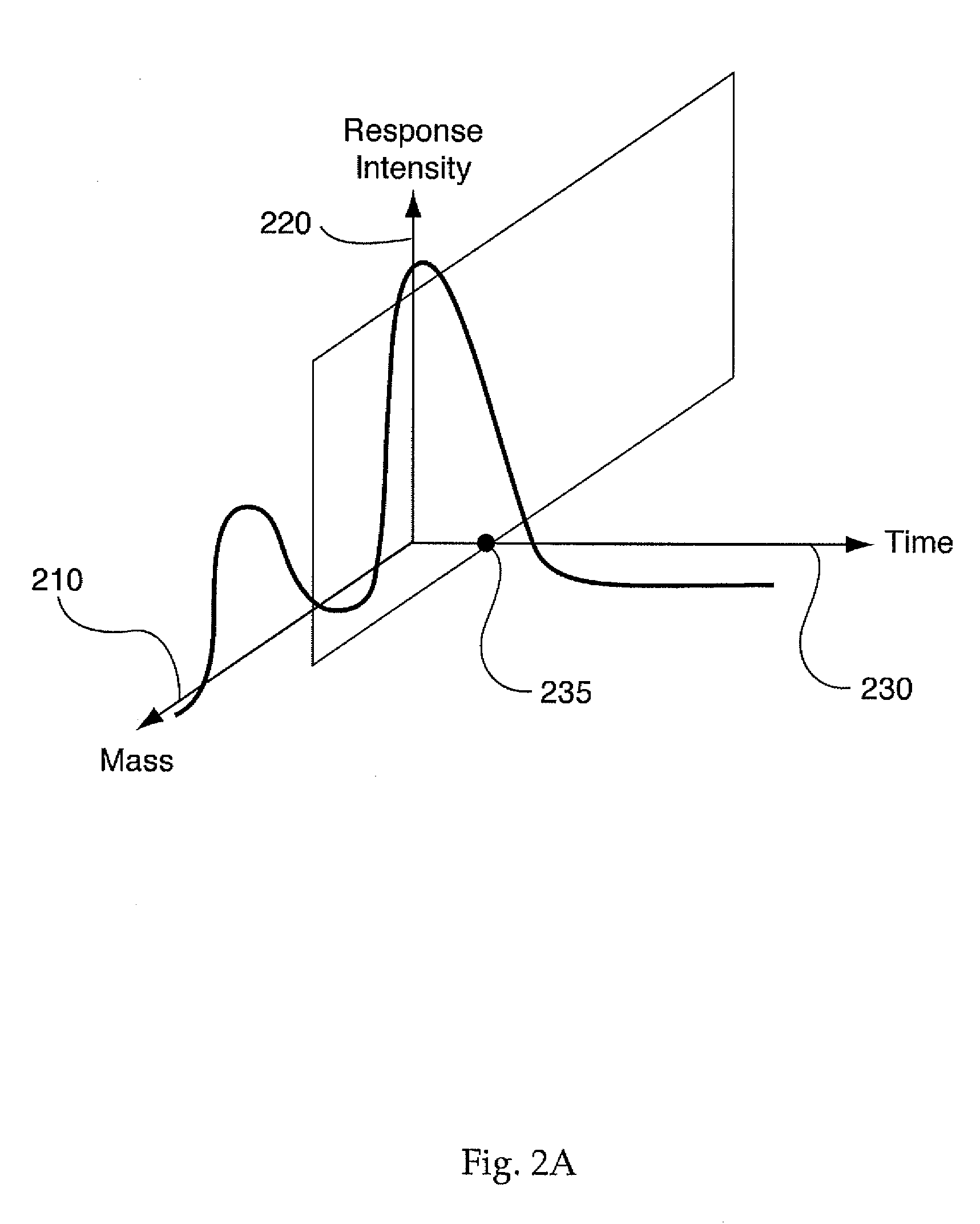
System and Method for Analyzing Metabolomic Data
ActiveUS20090093971A1Easy to determineEasy to compareMolecular entity identificationData visualisationMetaboliteData set
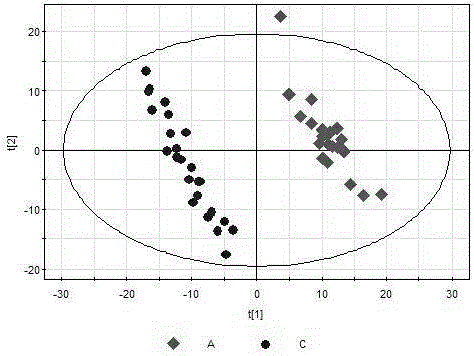
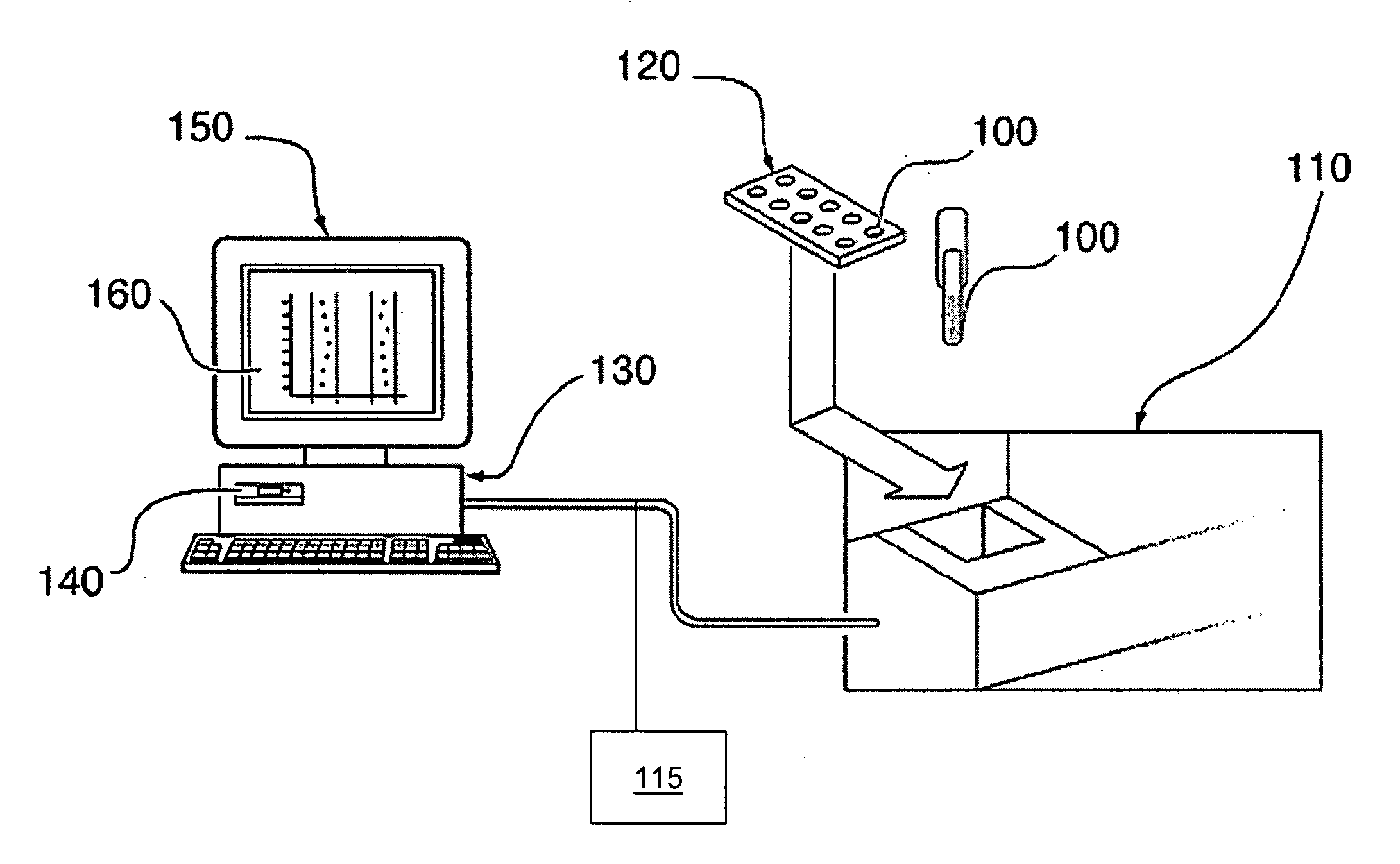
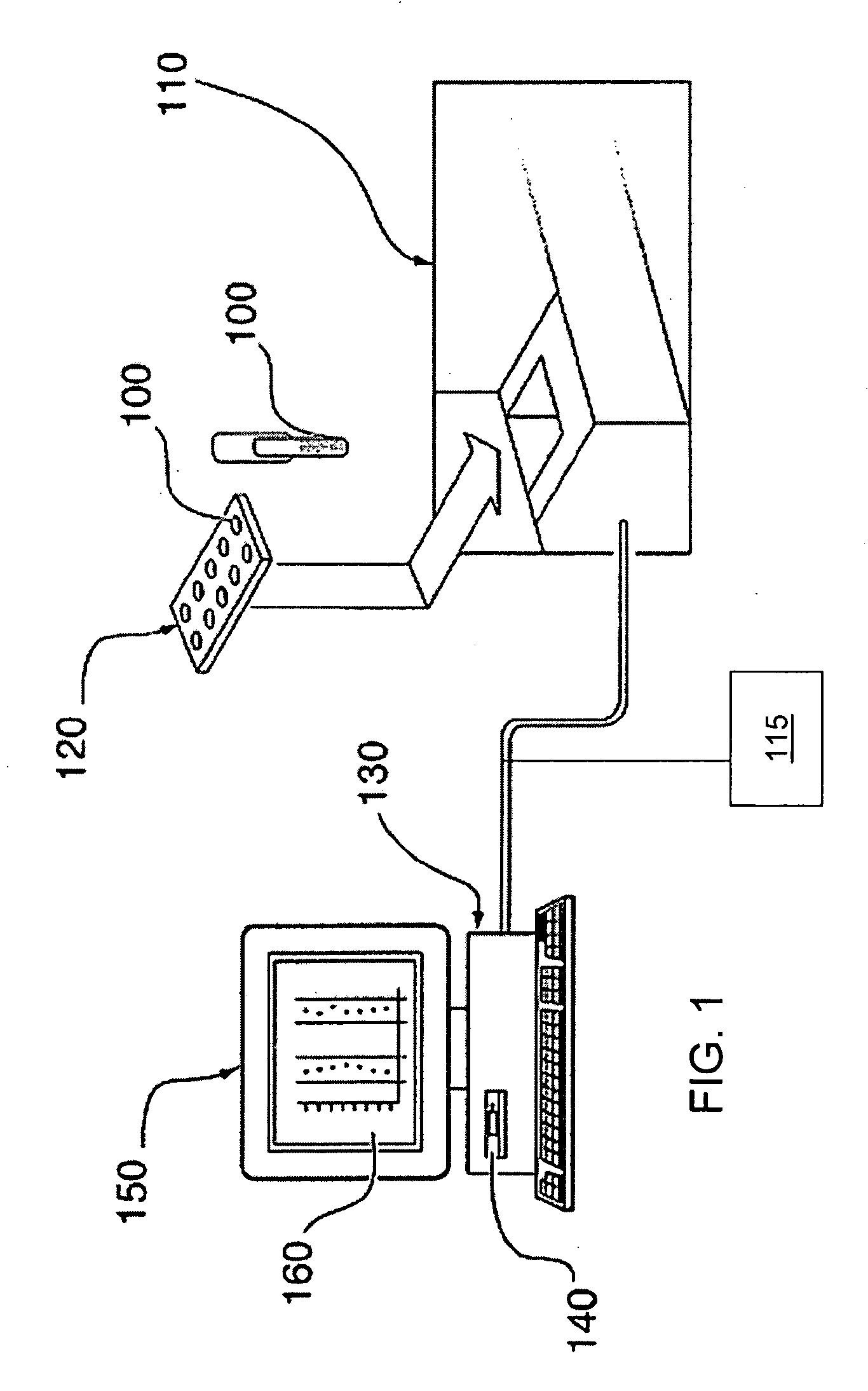
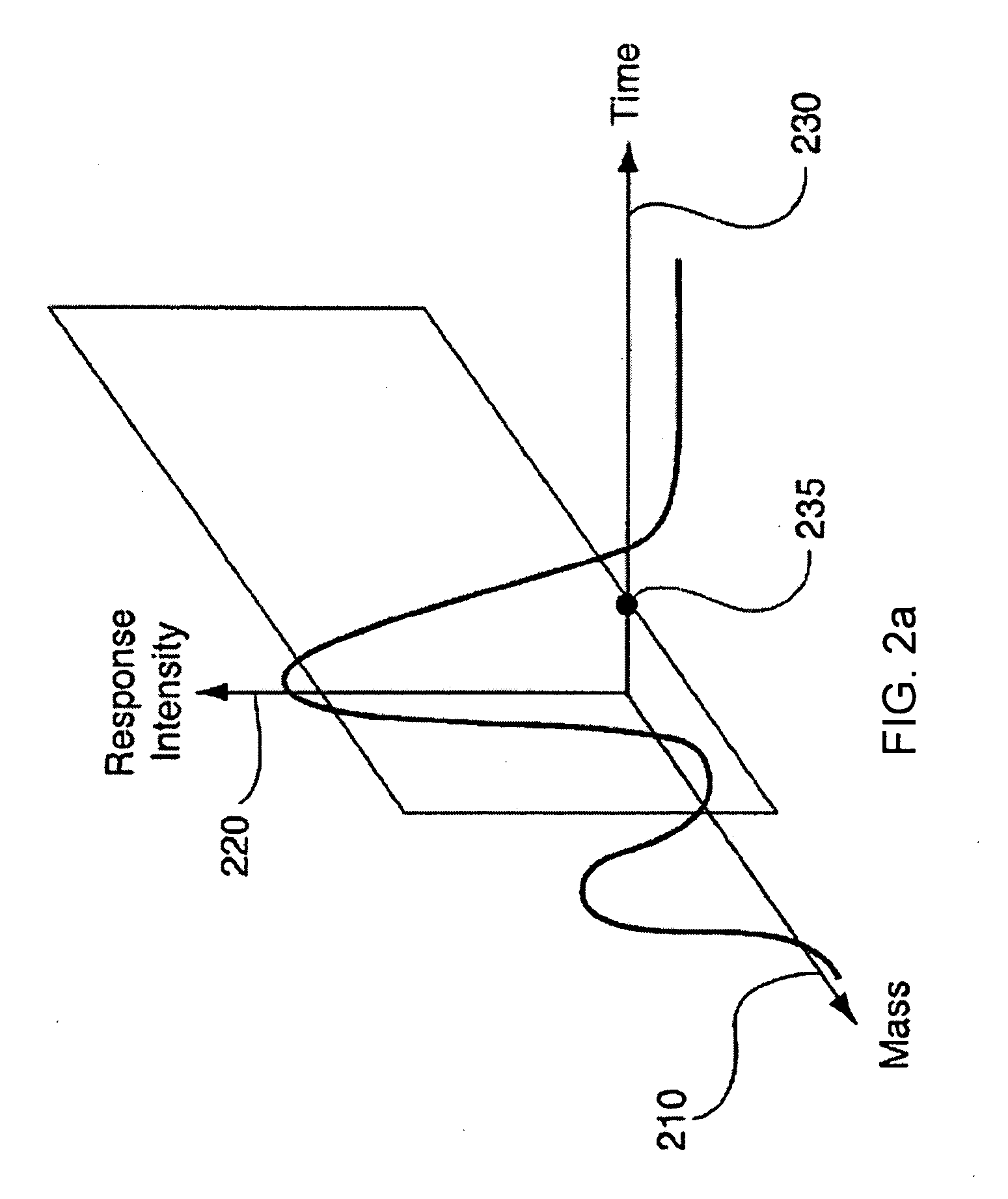
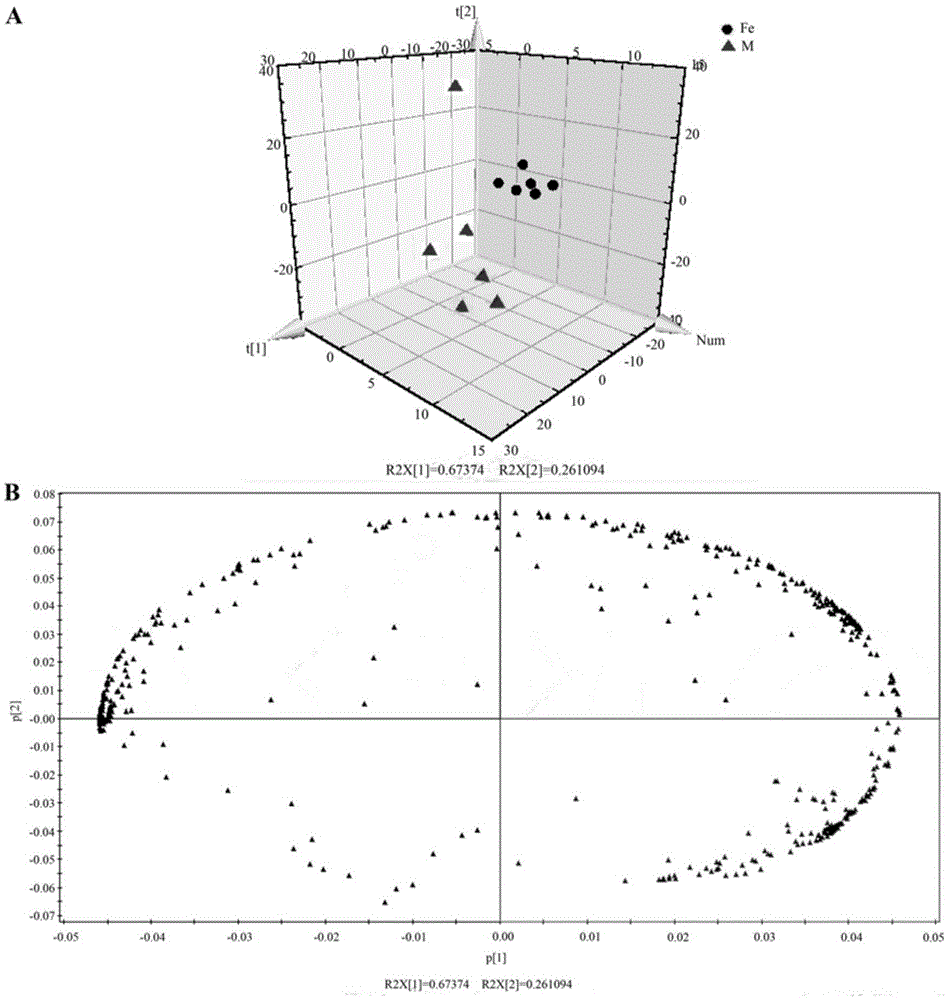
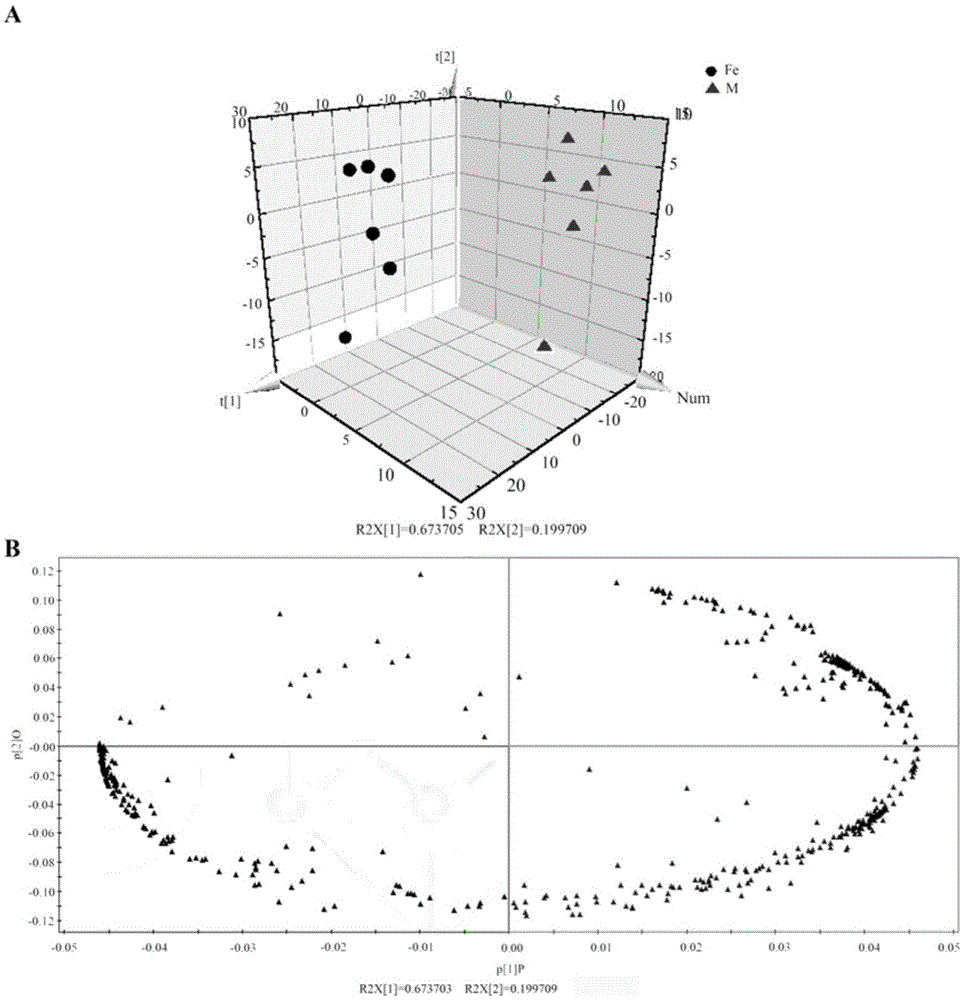
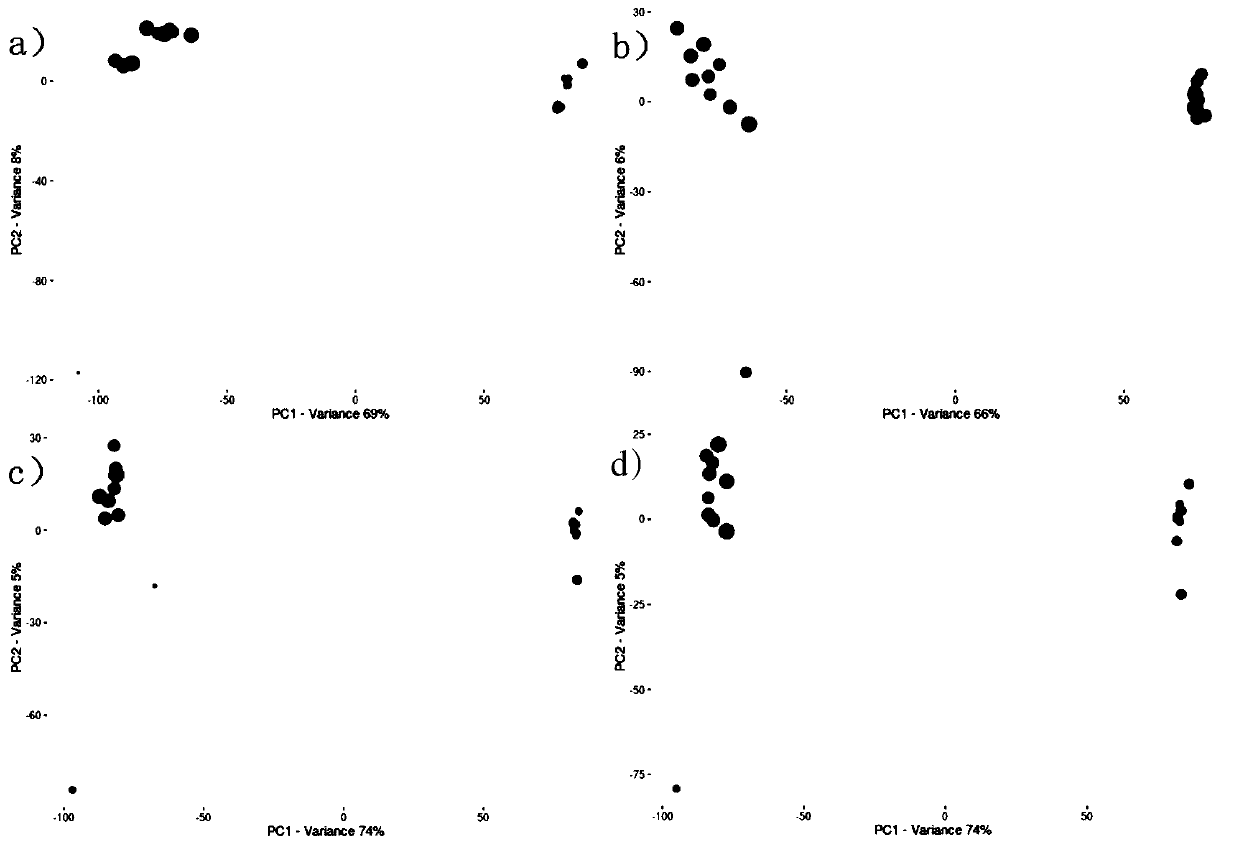
The present invention generates a visual display of metabolomic data compiled by a database and associated processor. More particularly, the present invention provides a database for automatically receiving a three-dimensional spectrometry data set for a group of samples. The present invention also provides a processor device for manipulating the data sets to produce plots that are directly comparable to a plurality of characteristic plots corresponding to a plurality of selected metabolites. Furthermore, the processor device may generate a visual display indicating the presence of the selected metabolites across the group of samples. Thus, the present invention enables a user to analyze a series of complex data sets in a visual display that may indicate the presence of the selected metabolites across the group of samples. Furthermore, the visual display generated by embodiments of the present invention also expedites the subjective analysis of the spectrometry data sets.
Owner:METABOLON
Method for identifying varieties of dendrobium huoshanense and dendrobium officinale based on metabonomics
ActiveCN105548404AEasy to prepareAvoid one-sidednessComponent separationEndogenous metabolismMetabolite
The invention discloses a method for identifying the varieties of dendrobium huoshanense and dendrobium officinale based on metabonomics. The method comprises the following specific steps: (1) collecting dendrobium samples; (2) preparing dendrobium metabonomics samples; (3) detecting metabolites of the dendrobium samples; and (4) performing metabonomics data processing and analysis. The method provided by the invention has the following advantages: a lot of endogenous metabolite information of dendrobium is acquired by means of metabonomics; among the endogenous metabolites, the characteristic metabolites for identification of dendrobium huoshanense and dendrobium officinale are selected out, and then such two dendrobium varieties are distinguished from each other on the whole; the technical mode is advanced and the experimental result is reliable; and a good means is provided for dendrobium variety identification and dendrobium quality assessment.
Owner:ANHUI AGRICULTURAL UNIVERSITY
System and method for analyzing metabolomic data
ActiveUS7949475B2Easy to determineEasy to compareMolecular entity identificationData visualisationData setMetabolite
The present invention generates a visual display of metabolomic data compiled by a database and associated processor. More particularly, the present invention provides a database for automatically receiving a three-dimensional spectrometry data set for a group of samples. The present invention also provides a processor device for manipulating the data sets to produce plots that are directly comparable to a plurality of characteristic plots corresponding to a plurality of selected metabolites. Furthermore, the processor device may generate a visual display indicating the presence of the selected metabolites across the group of samples. Thus, the present invention enables a user to analyze a series of complex data sets in a visual display that may indicate the presence of the selected metabolites across the group of samples. Furthermore, the visual display generated by embodiments of the present invention also expedites the subjective analysis of the spectrometry data sets.
Owner:METABOLON
System, method, and computer program product using a database in a computing system to compile and compare metabolomic data obtained from a plurality of samples
ActiveUS7433787B2Reduce the burden onOvercome problemsData visualisationDiagnostic recording/measuringData setMetabolite
The present invention generates a visual display of metabolomic data compiled by a database and associated processor. More particularly, the present invention provides a database for automatically receiving a three-dimensional spectrometry data set for a group of samples. The present invention also provides a processor device for manipulating the data sets to produce plots that are directly comparable to a plurality of characteristic plots corresponding to a plurality of selected metabolites. Furthermore, the processor device may generate a visual display indicating the presence of the selected metabolites across the group of samples. Thus, the present invention enables a user to analyze a series of complex data sets in a visual display that may indicate the presence of the selected metabolites across the group of samples. Furthermore, the visual display generated by embodiments of the present invention also expedites the subjective analysis of the spectrometry data sets.
Owner:METABOLON
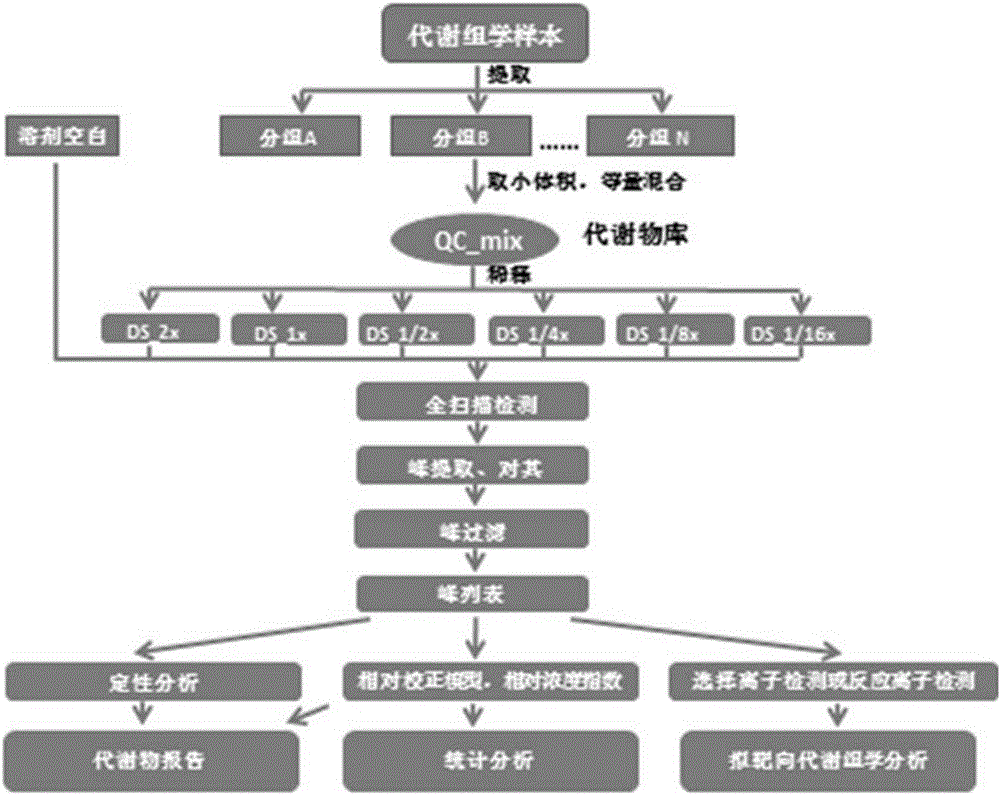
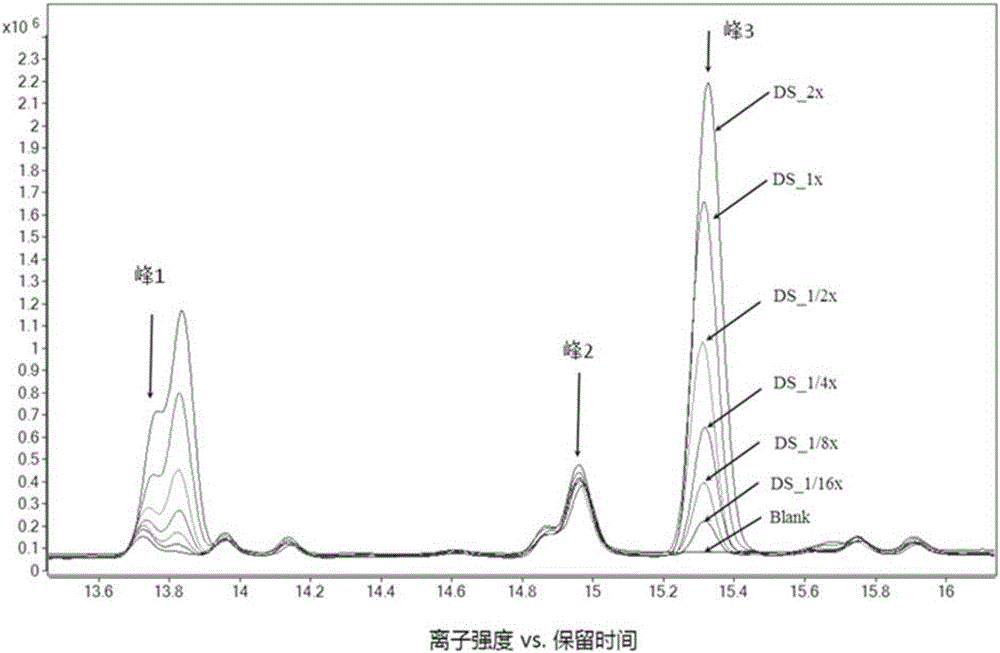
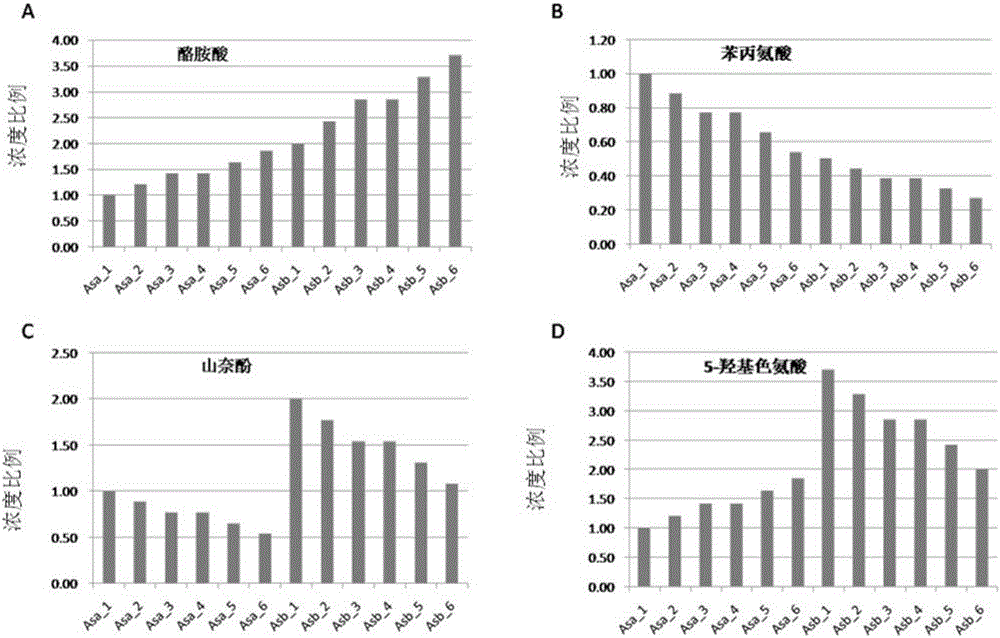
Metabolism group method for distinguishing false positive mass spectra peak signals and quantificationally correcting mass spectra peak area
ActiveCN106018600AEasy to filterSimple Metabolomics StudyComponent separationGroup studySample dilution
The invention discloses a metabolism group method for distinguishing false positive mass spectra peak signals and quantificationally correcting mass spectra peak area. The invention provides a metabolism group study method; by the method, the biological source and non-biological source mass spectra peak signals can be effectively distinguished; the mass spectra peak signals are quantificationally evaluated; the mass spectra peaks with poor quantification performance are excluded; through QC sample dilution, a relative content correction model is built; the mass spectra peak area is corrected. The method has the maximum characteristics that the false positive mass spectra signals can be effectively eliminated, so that the metabolism group data becomes reliable; the real biomarkers can be favorably screened. The method can aim at plant, animal and microbe samples; the method can also be suitable for the mass-spectra-platform-based metabolism group analysis of GC-MS, LC-MS and CE-MS.
Owner:INST OF BOTANY CHINESE ACAD OF SCI
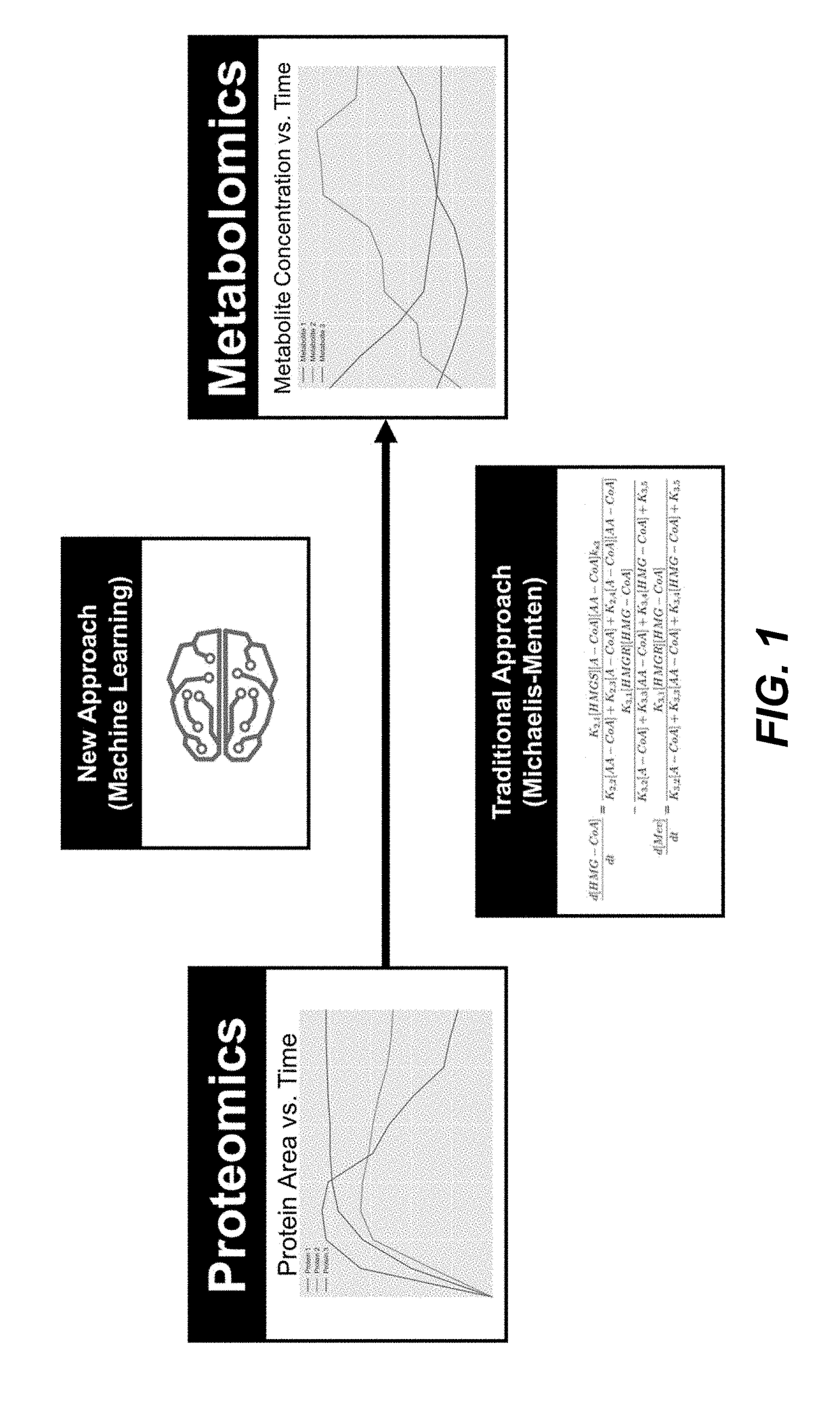
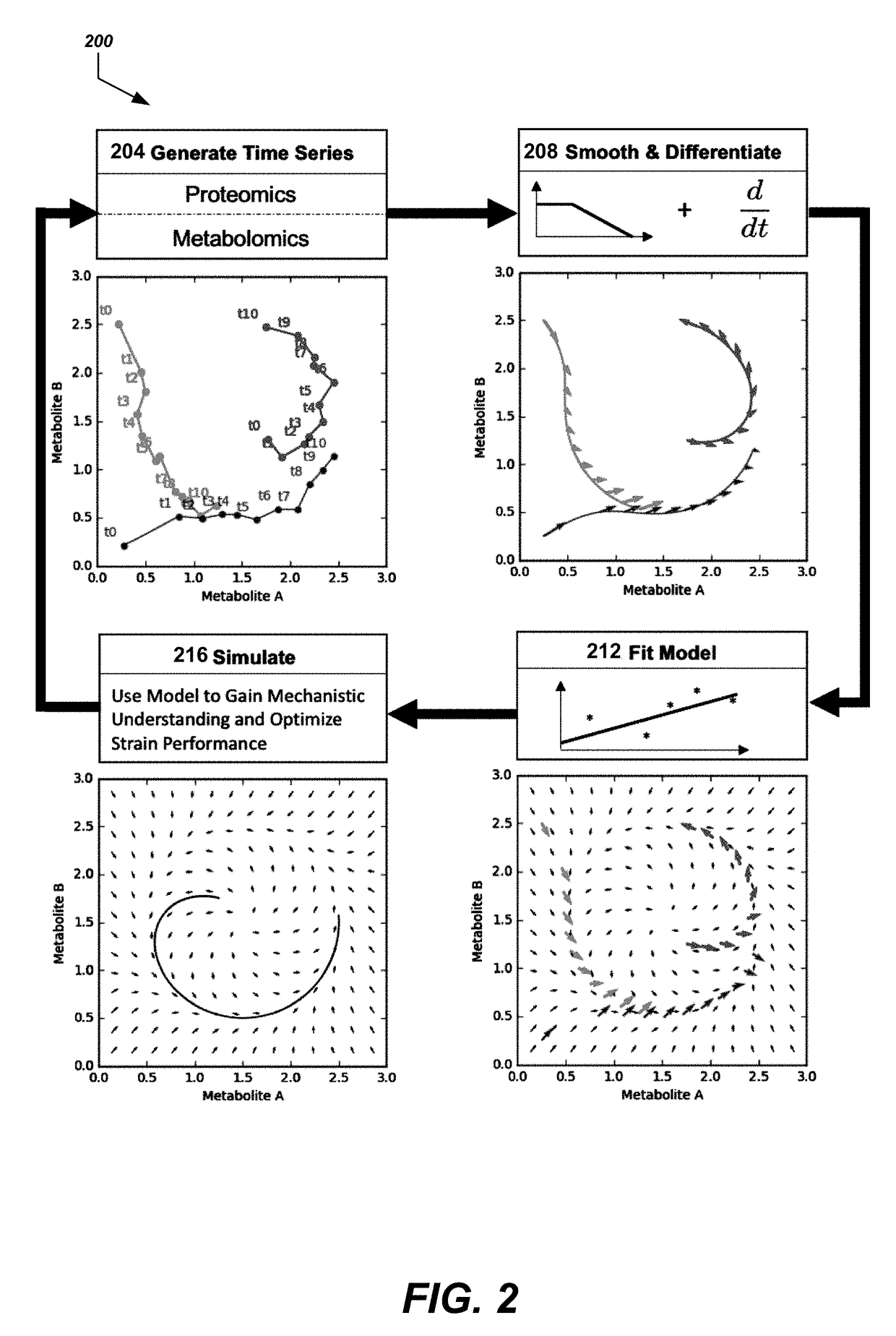
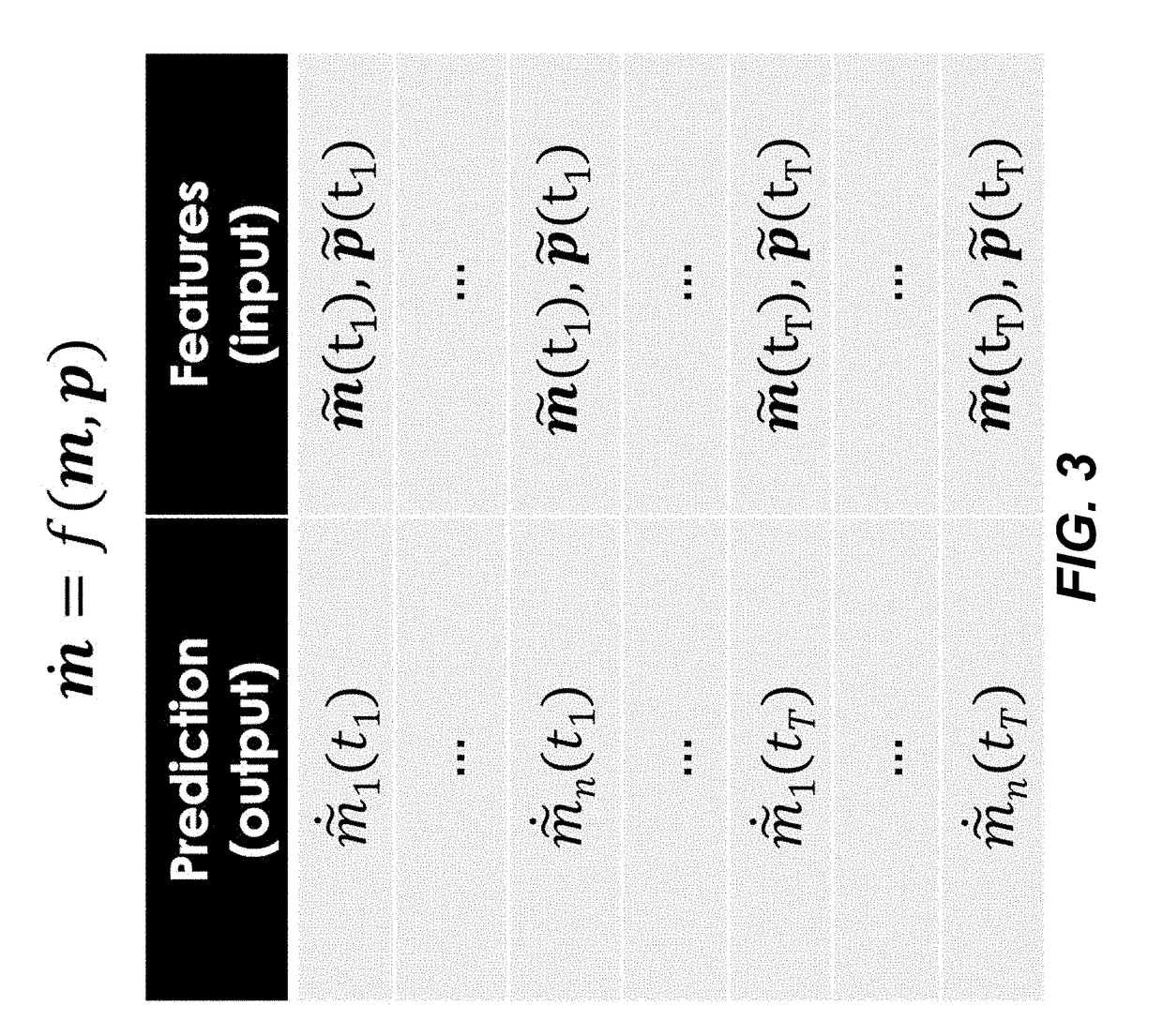
Simulating the metabolic pathway dynamics of an organism
Disclosed herein are systems and methods for determining metabolic pathway dynamics using time series multiomics data. In one example, after receiving time series multiomics data comprising time-series metabolomics data associated a metabolic pathway and time-series proteomics data associated with the metabolic pathway, derivatives of the time series multiomics data can be determined. A machine learning model, representing a metabolic pathway dynamics model, can be trained using the time series multiomics data and the derivatives of the time series multiomics data, wherein the metabolic pathway dynamics model relates the time-series metabolomics data and time-series proteomics data to the derivatives of the time series multiomics data. The method can include simulating a virtual strain of the organism using the metabolic pathway dynamics model.
Owner:RGT UNIV OF CALIFORNIA
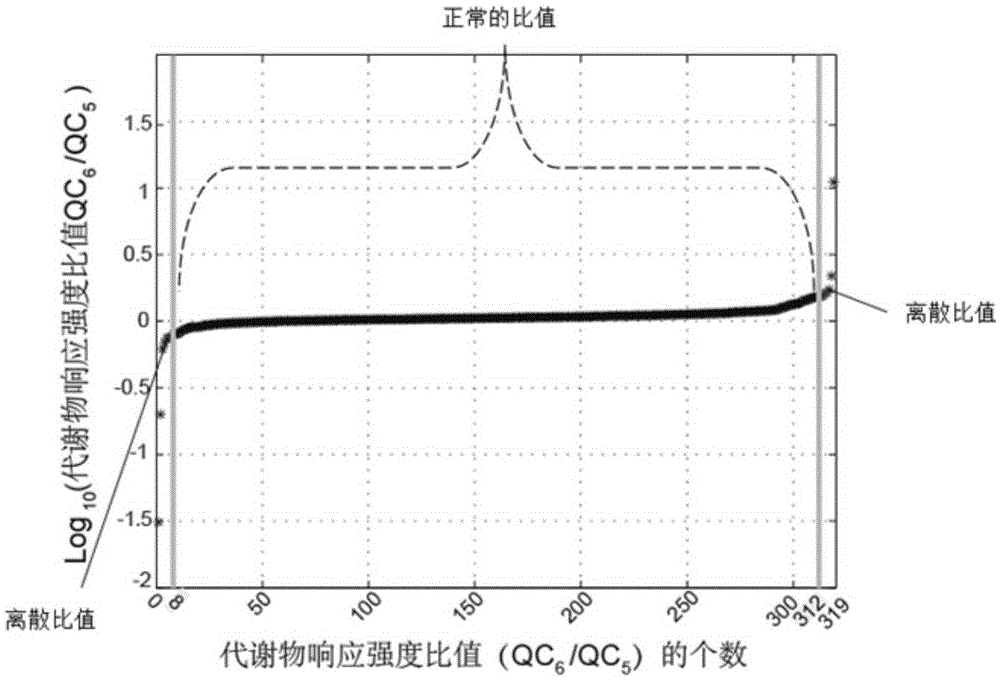
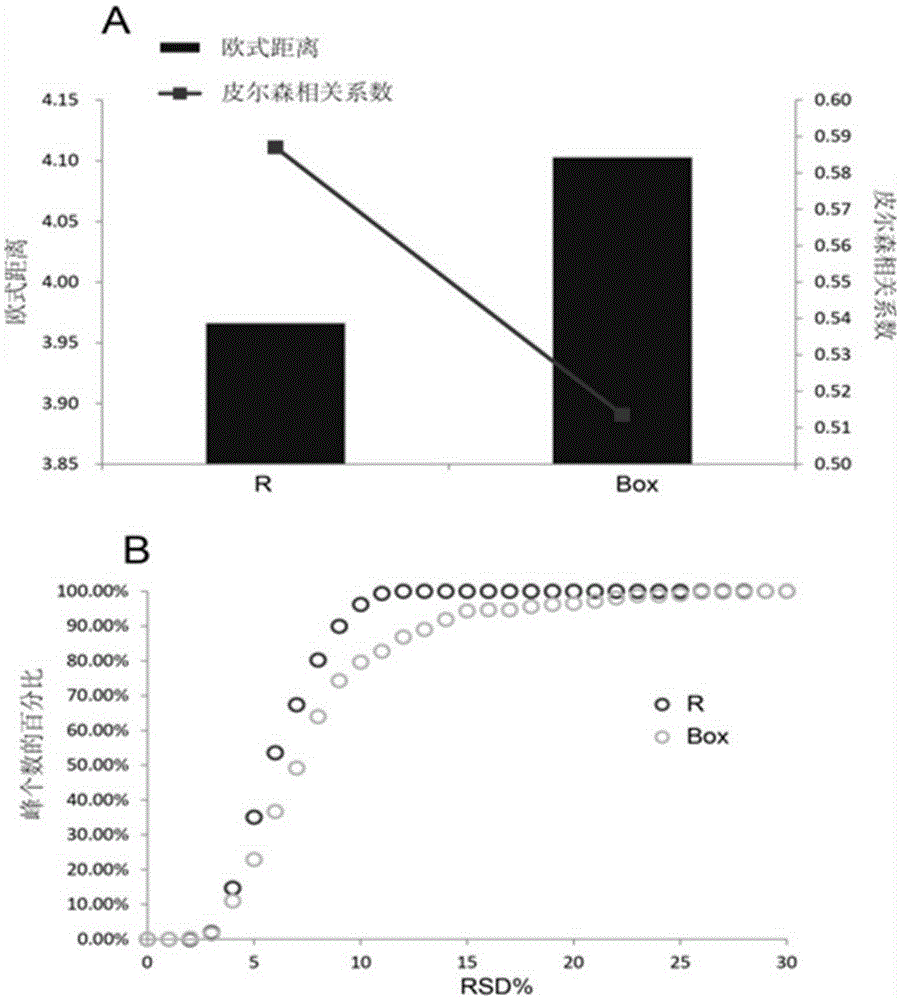
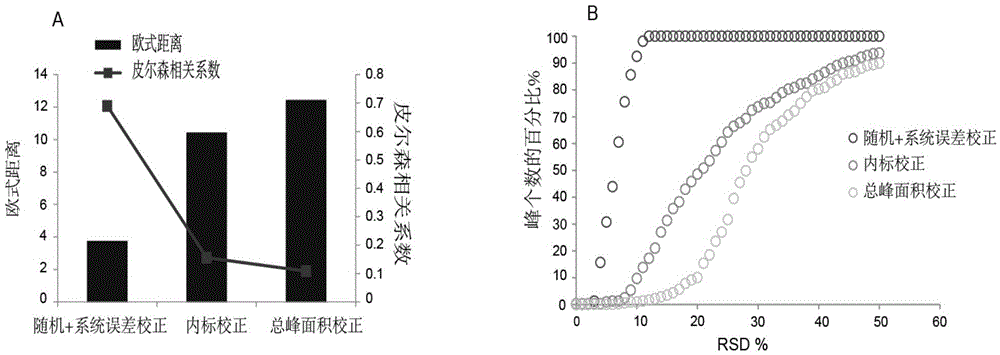
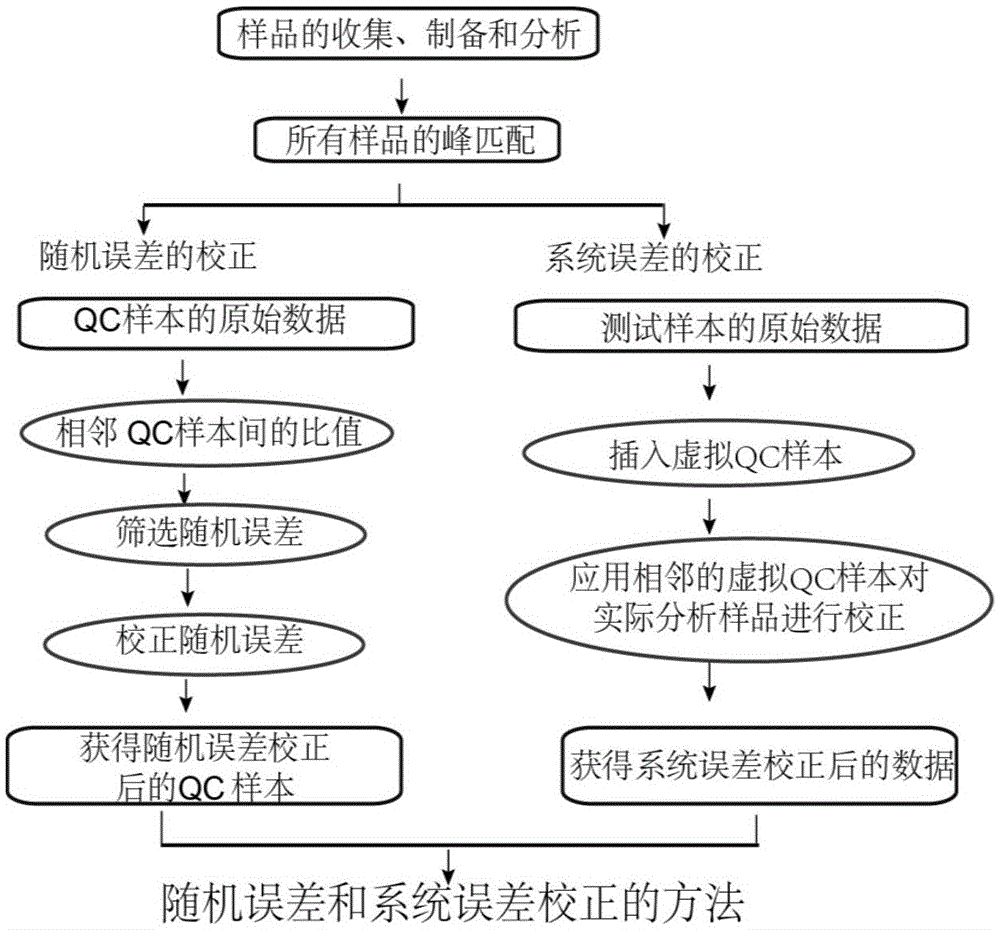
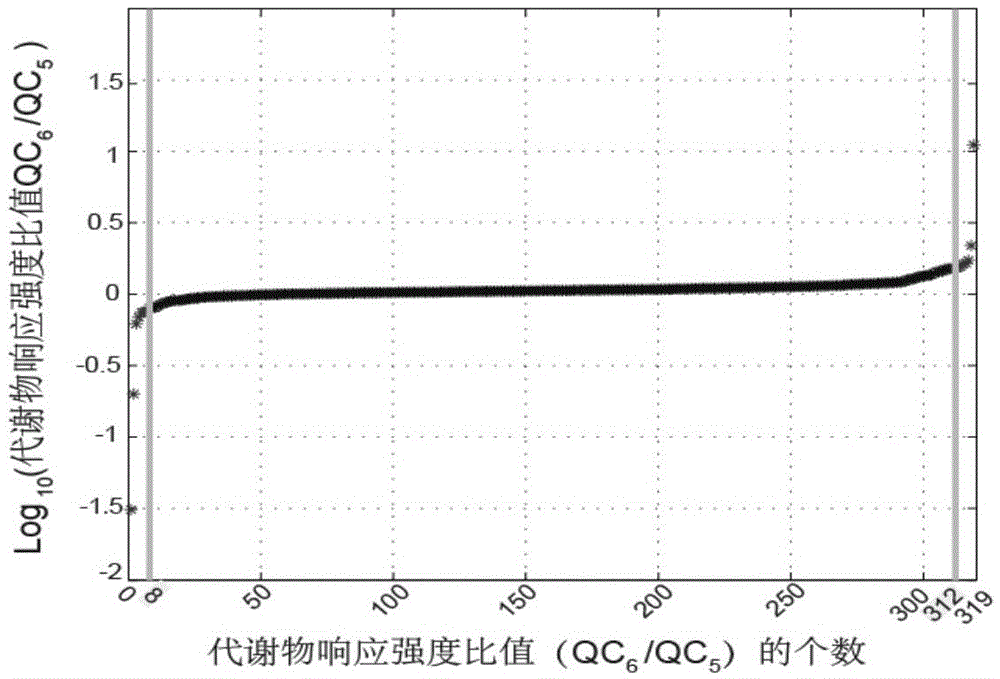
Screening and calibrating method of metabolomic data random errors
InactiveCN105424827AEffective correctionAccurate correctionComponent separationQuality controlMetabolomics data
The invention discloses a screening and calibrating method of metabolomic data random errors. The method comprises the steps that a sample is analyzed through a chromatograph-mass spectrometer to obtain a metabolome profile, the specific value of response intensities of metabolites in every two adjacent quality control (QC) samples is calculated, the specific values are sorted from small to large, 5% of the total number of the specific values are screened to serve as discrete points, the discrete points accounting for 5% of the total number of the specific values are averagely distributed to the two ends of the sorted specific values, and therefore a model is established to screen the random errors in metabolomic data; then a linear fitting model of the specific values is utilized to calibrate the random errors. According to the screening and calibrating method of the metabolomic data random errors, the core is that the model is established through the response intensities of the metabolites in every two adjacent QC samples to screen and calibrate the random errors; the random errors in the metabolomic data can be screened and calibrated efficiently and accurately, and the quality of the metabolomic data can be improved.
Owner:DALIAN UNIV OF TECH
Analysis of transcriptomic data using similarity based modeling
ActiveUS8515680B2Small footprintFast data processingSugar derivativesDigital data processing detailsBiomarker discoveryModelling analysis
An analytic apparatus and method is provided for diagnosis, prognosis and biomarker discovery using transcriptome data such as mRNA expression levels from microarrays, proteomic data, and metabolomic data. The invention provides for model-based analysis, especially using kernel-based models, and more particularly similarity-based models. Model-derived residuals advantageously provide a unique new tool for insights into disease mechanisms. Localization of models provides for improved model efficacy. The invention is capable of extracting useful information heretofore unavailable by other methods, relating to dynamics in cellular gene regulation, regulatory networks, biological pathways and metabolism.
Owner:PROLAIO INC
Method for analyzing change rule of metabolites during fermentation of cephalosporium acremonium
ActiveCN102262136AQualitatively accurateAchieve the same effect as absolute quantificationComponent separationMetabolitePrincipal component analysis
The invention discloses a method for analyzing a change rule of metabolites during fermentation of cephalosporium acremonium. The method comprises the following steps of: sampling and centrifuging before fermentation, during fermentation and after fermentation of cephalosporium acremonium generated by cephalosporin C, mixing phosphate buffer solution into sediment uniformly and centrifuging; quenching by using liquid nitrogen and grinding; adding powdered cells into methanol aqueous solution and butanedioic acid aqueous solution which is marked by deuterium and is used as an internal standard substance, and mixing uniformly; freezing and melting, centrifugally collecting the supernate and drying; performing oximation reaction; performing silanization reaction; determining components and relative content of a sample by using a gas chromatography-flying time mass spectrometer; analyzing principal components of preprocessed data by using software so as to analyze metabolism difference of the samples; performing cluster analysis on the preprocessed data by using the software so as to analyze the metabolism difference of metabolites among the samples; determining the nature of the metabolites to classify the metabolites; and analyzing the change rule of all types of the metabolites during the fermentation so as to comprehensively disclose metabolism information of the cephalosporium acremonium according to a large amount of metabolism physiome data.
Owner:TIANJIN UNIV
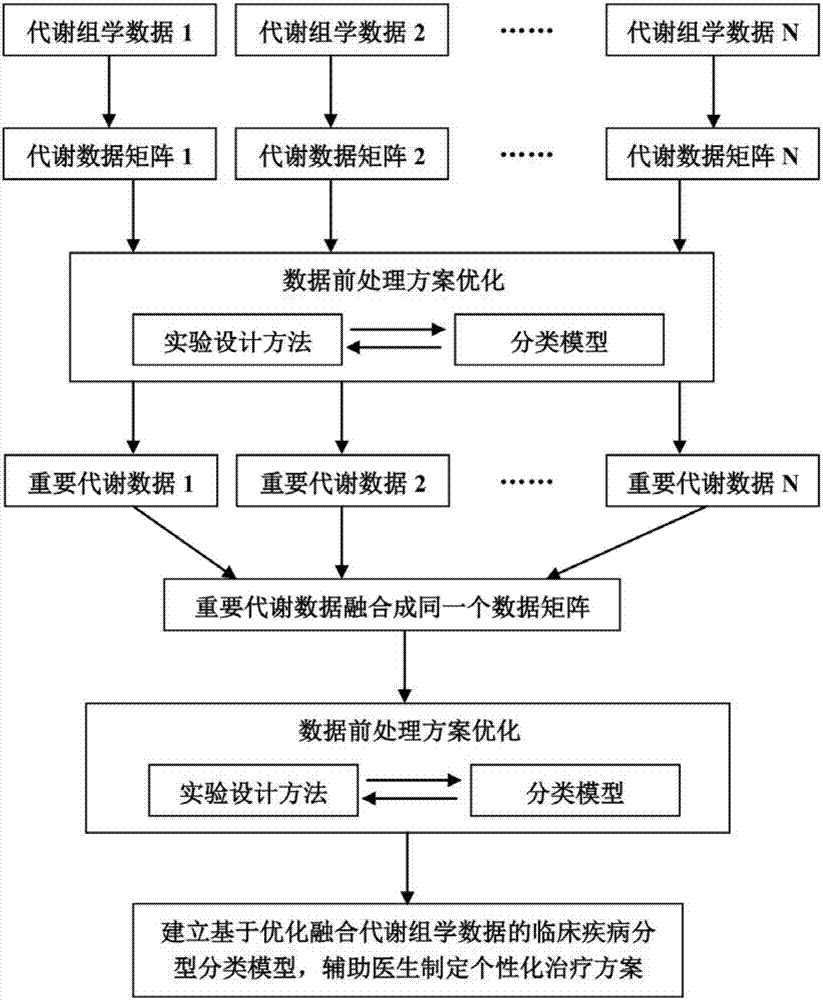
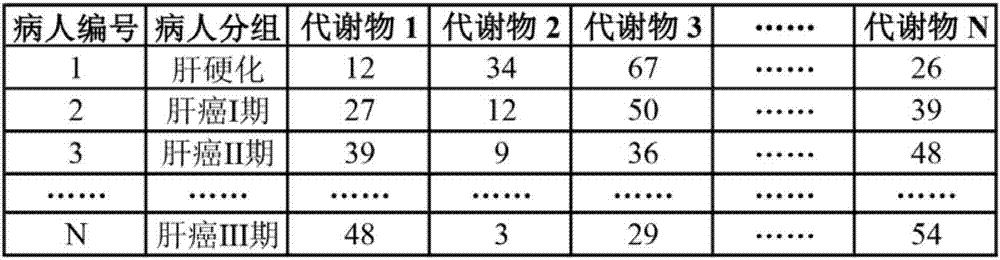
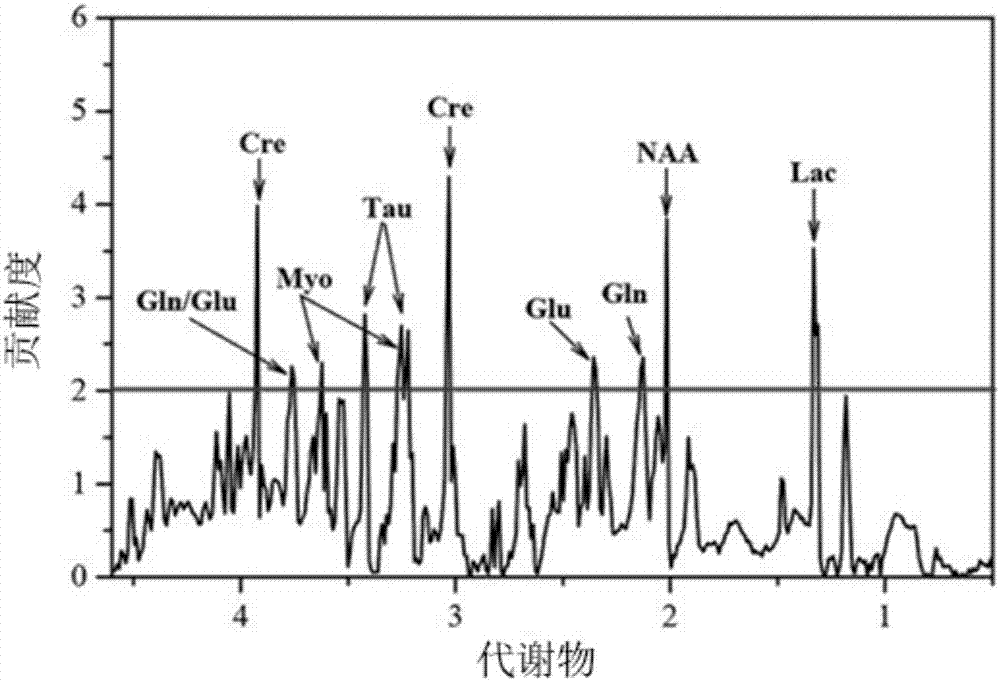
Metabonomics data fusion optimization processing method
ActiveCN107133448AImprove classificationImprove classification accuracyChemical processes analysis/designChemical machine learningPersonalizationDisease
The invention discloses a metabonomics data fusion optimization processing method. The method comprises the following steps of 1, converting metabonomics data of many patients of different sources into metabonomics data matrixes separately, 2, using an experiment design method to optimize a metabonomics data optimal preprocessing method with different sources separately, then through the optimized data optimal preprocessing method to process the metabonomics data matrixes in the step 1, and through the combination of a classification model, finding metabolite data with great contribution; 3, converting the metabolite data with the great contribution found in the step 2 into an important metabolic data matrix; 4, using the experiment design method to optimize the data optimal preprocessing method of the important metabolic data matrix, through the optimized data optimal preprocessing method, processing the important metabolic data matrix obtained in the step 3, and through the combination of the classification model, typing and classifying different patients. According to the metabonomics data fusion optimization processing method, the accuracy for typing and classifying diseases is improved, and a doctor can be assisted in more precisely laying out a personalized treatment scheme for a patient.
Owner:WENZHOU MEDICAL UNIV
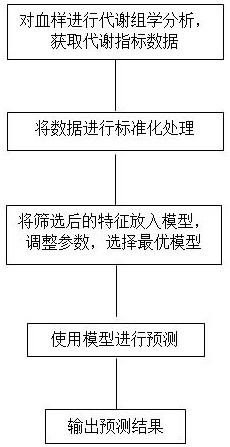
Machine learning diabetes onset risk prediction method and application
InactiveCN112786204AImprove stabilityImprove predictive performanceMedical data miningHealth-index calculationDiseaseDiabetes mellitus
The invention provides a machine learning diabetes onset risk prediction method. The method comprises the following steps: a data acquisition module for acquiring metabonomics data; a data preprocessing module for preprocessing the acquired data; a machine learning module for constructing a prediction model based on a machine learning algorithm and metabonomics data for the purpose of diabetes risk prediction; and a display output module for testing the obtained to-be-predicted sample and outputting a prediction result. If the prediction result is 1, the diabetes risk exists, and if the prediction result is 0, the diabetes risk does not exist. According to the embodiment of the invention, the diabetes risk prediction model is constructed by combining metabonomics characteristics based on technologies mainly based on the random forest and the support vector machine algorithm. The method can be used for improving decision-making efficiency, guiding non-medical personnel to carry out disease risk detection or assisting clinical decision-making, and achieving the purposes of three-level prevention of diseases and promotion and development of health of the whole people.
Owner:TIANJIN MEDICAL UNIV
System, method, and computer program product using a database in a computing system to compile and compare metabolomic data obtained from a plurality of samples
ActiveUS20070032969A1Reduce the burden onOvercome problemsData visualisationElectrical measurementsMetaboliteData set
The present invention generates a visual display of metabolomic data compiled by a database and associated processor. More particularly, the present invention provides a database for automatically receiving a three-dimensional spectrometry data set for a group of samples. The present invention also provides a processor device for manipulating the data sets to produce plots that are directly comparable to a plurality of characteristic plots corresponding to a plurality of selected metabolites. Furthermore, the processor device may generate a visual display indicating the presence of the selected metabolites across the group of samples. Thus, the present invention enables a user to analyze a series of complex data sets in a visual display that may indicate the presence of the selected metabolites across the group of samples. Furthermore, the visual display generated by embodiments of the present invention also expedites the subjective analysis of the spectrometry data sets.
Owner:METABOLON
Analysis method of cecal metabolome in broiler chicken
PendingCN109521113AReduce contentClear thinkingComponent separationBiotechnologyGas chromatography–mass spectrometry
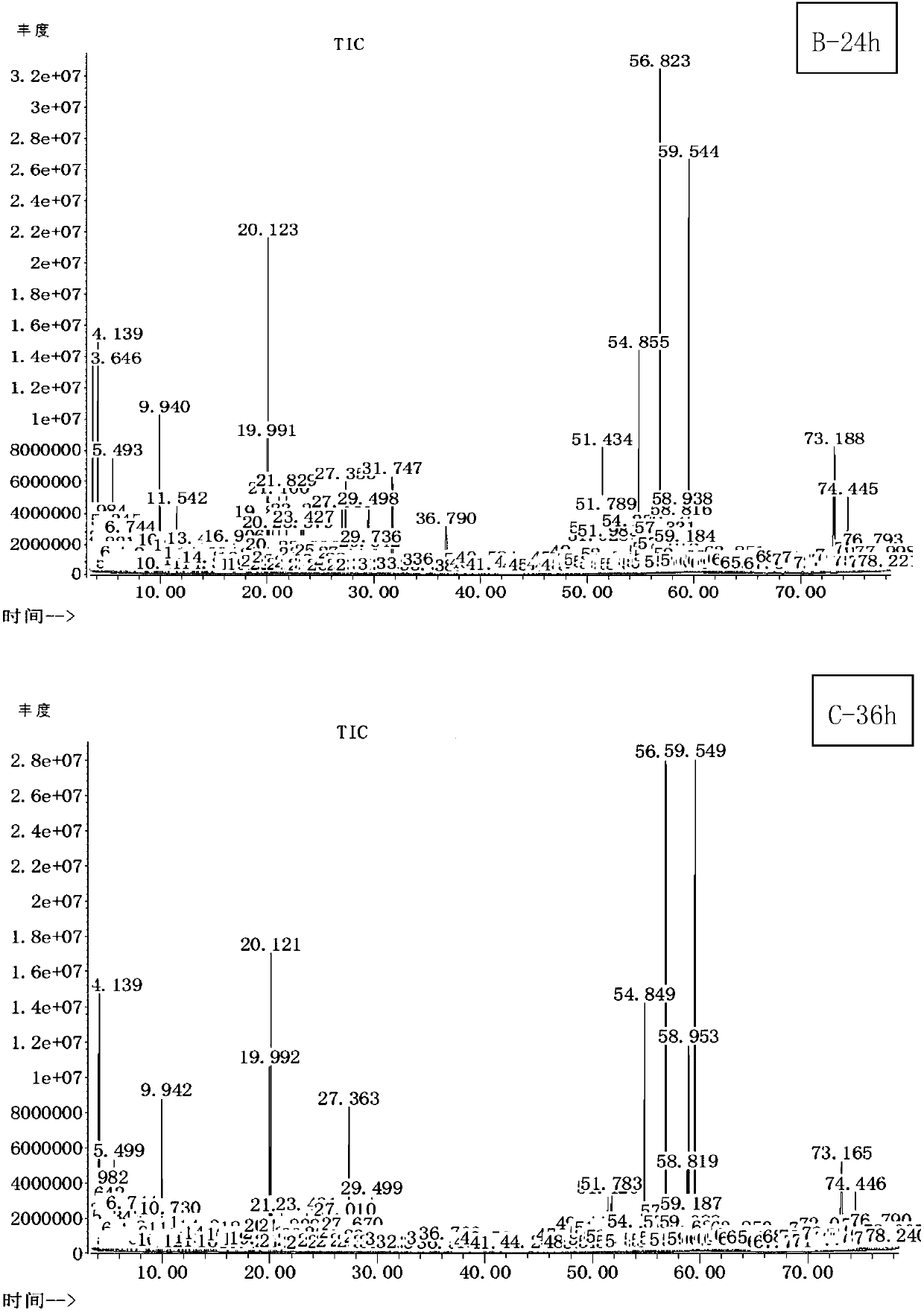
The invention relates to the field of biological analysis methods, and in particular to an analysis method of cecal metabolome in a broiler chicken. The change of metabolites and metabolic pathways ofcecal contents after the broiler chicken eats a yeast culture produced at different fermentation time is detected by jointly using a gas chromatography-mass spectrometry, thereby determining the influence of the yeast culture on the cecal metabolic regulation and the metabolites of the broiler chicken. Metabolomics data show that yeast culture is used for feeding the broiler chicken, the cecal differential metabolites mainly involve in amino acid metabolism and cholesterol metabolism, and participates in fat, vitamin digestion and absorption processes.
Owner:JILIN AGRICULTURAL UNIV
Metabonomics network marker identification method based on horizontal relation
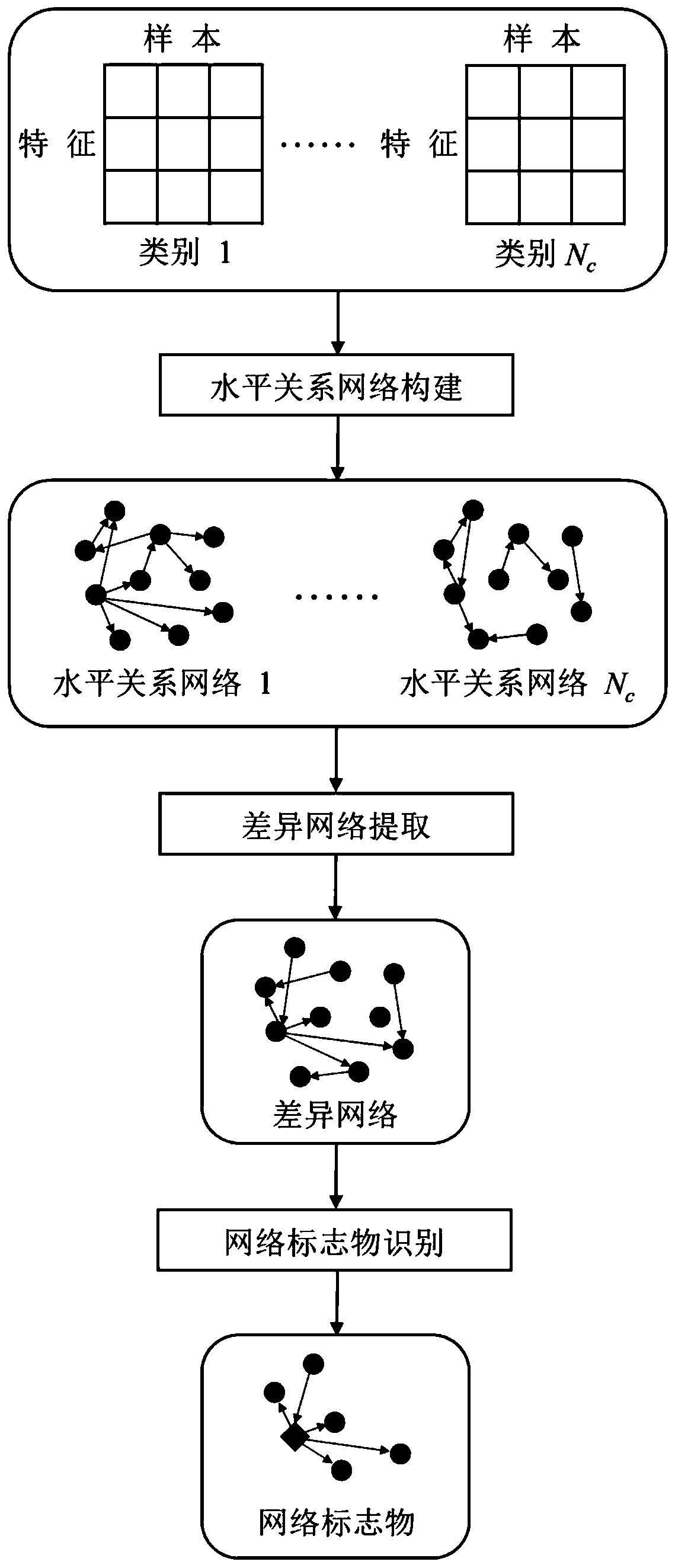
ActiveCN110322930AEfficient miningEfficient Data Processing MeansBiostatisticsSystems biologyNetwork connectionMetabolomics data
The invention provides a metabonomics network marker identification method based on a horizontal relation, belongs to the technical field of biological data analysis, and relates to a metabonomics data analysis method DNB-HC for screening potential network markers of complex diseases. The horizontal relation between the features is defined by using probability scores and is used for measuring thesize relation of the relative expression levels of a pair of metabolic features in the same sample, and the robustness of the horizontal relation is determined through a random disturbance test, so that the network connection edge is determined. Besides, the network markers are identified by using a difference network analysis method, and the screened network metabolic markers have relatively gooddistinguishing capability, so that a practical and effective data processing means can be provided for researching a disease occurrence and development mechanism and diagnosing diseases.
Owner:DALIAN UNIV OF TECH
Evaluation method of food quality based on metabolomics data fusion and artificial neural network and application thereof
The invention discloses an evaluation method of food quality and application thereof. The evaluation method of the food quality is performed based on metabolomics and an artificial neural network model; the artificial neural network model comprises an input layer including multiple input parameters, wherein the input parameters contain a content parameter of at least one food metabolite, and the content parameter of the food metabolite is acquired based on the metabolomics data fusion; a hidden layer; and an output layer including multiple output parameters, wherein the output parameters contain at least one food sense organ score. Through the method disclosed by the invention, a metabolomics key index of the food is used as the input layer, and the sense organ score of the food is used asthe output layer so as to establish a quality evaluation model of the food based on the artificial neural network; the mathematical formula or weight is unnecessary, and the method is simple; the evaluation method is objective, comprehensive, scientific and easy to apply and popularize.
Owner:CHINESE ACAD OF INSPECTION & QUARANTINE
System, method, and computer program product for analyzing spectrometry data to identify and quantify individual components in a sample
ActiveUS20070288174A1Molecular entity identificationParticle separator tubesCorrelation functionMetabolomics data
A system is provided for analyzing metabolomics data received from an analytical device across a group of samples. The system automatically receives a data matrix corresponding to each of the samples, wherein the data matrix includes rows corresponding to each of the samples and columns corresponding to a group of ions present in the respective samples. A processor is provided for determining a characteristic value corresponding to at least one of a group of components present in the samples, wherein the components are made up of at least a portion of the group of ions, using at least one of a correlation function and a factorization function. A user interface is in communication with the processor for displaying a visual indication of the characteristic value such that a user may receive a visual indication of the types of components present in the samples.
Owner:METABOLON
Universal correction method of large-scale metabonimics data
The invention discloses a universal correction method of large-scale metabonimics data. The method comprises the following steps: analyzing samples through adopting a chromatograph-mass spectrometer to obtain a metabolic profiling, calculating a ratio of the response intensities of metabolites in every two adjacent quality control samples (QC), sequencing the obtained ratios from small to large, screening the ratios accounting for 5% of the total quantity of the ratios as discrete points, averagely distributing the 5% discrete points to two ends of the sequenced ratios in order to establish a model and screen the random error in the metabonimics data, ad correcting the random error through using a linear fitting model of the ratios; and constructing a virtual QC technology by using a linear regression model in order to realize the system error correction of a large-scale metabonimics data set. The method has the advantages of highly-efficient and accurate correction of the random error and the system error of the large-scale metabonimics data, realization of integration of multi-batch and different device metabonimics data.
Owner:DALIAN INST OF CHEM PHYSICS CHINESE ACAD OF SCI
Metabolome molecule variable comprehensive screening technology
InactiveCN109856307AEasy to classifyStrong application valueComponent separationMetabolomics dataMetabolome
The invention provides a metabolome molecule variable comprehensive screening technology, and belongs to the technical field of metabonomics data analysis. According to the technology, a plurality ofpossible sample distribution modes on molecular pair variables are considered; the distinguishing capability of the molecular pair variables is comprehensively evaluated; indexes as same as the molecular pair variables are used; single-variable evaluation processes are organically fused; scores of all single variables and pair variables are sorted; and the pair variables with the highest scores and the distribution modes of the pair variables or the single variables are selected to carry out subsequent target metabolic analysis. According to the core technology of a method, based on actual characteristics of metabonomics, multi-angle analysis and comprehensive evaluation are carried out on the plurality of possible sample distribution modes on the pair variables, and the variables with rich information are excavated; and the selected molecular variables and molecular pair variables are subjected to classification testing, and the classification performance is excellent, so that the technology provides the practical and effective method for early-stage analysis processing of metabonomics data, and has relatively high application values.
Owner:DALIAN UNIV OF TECH
Machine learning diabetic retinopathy onset risk prediction method and application
InactiveCN112786203ASmall amount of calculationReduce demandMedical data miningHealth-index calculationDiseaseDiabetes retinopathy
The invention provides a machine learning diabetic retinopathy onset risk prediction method. The method comprises the following steps: a data acquisition module for acquiring metabonomics data; a data preprocessing module for preprocessing the acquired data; a machine learning module for constructing a prediction model based on a machine learning algorithm and metabonomics data in order to predict the risk of diabetic retinopathy; a display output module for testing the obtained to-be-predicted sample and outputting a prediction result, wherein if the prediction result is 1, the diabetic retinopathy risk exists, and if the prediction result is 0, the diabetic retinopathy risk does not exist. By applying the embodiment of the invention, the diabetic retinopathy risk prediction model is constructed by combining metabonomics characteristics on the basis of technologies mainly based on the random forest and the support vector machine algorithm. The method can be used for improving decision-making efficiency, guiding non-medical personnel to carry out disease risk detection or assisting clinical decision-making, and achieving the purposes of three-level prevention of diseases and promotion and development of health of the whole people.
Owner:TIANJIN MEDICAL UNIV
Metabonomics database establishment method and application of Chinese wolfberry tissues
ActiveCN111798937ARapid identificationGood qualityCheminformatics data warehousingCheminformatics programming languagesBiotechnologyMetabolite
The invention provides a metabonomics database establishment method and application of Chinese wolfberry tissues. The metabonomics database establishment method comprises the following steps: collecting leaves and fruit tissues of different varieties of Chinese wolfberry, summarizing the leaves of different varieties, summarizing the fruits of different varieties, respectively carrying out pretreatment, and adding an extraction reagent for extraction to obtain a leaf extracting solution and a fruit extracting solution; carrying out LC-MS / MS (Liquid Chromatography-Mass Spectrometry / Mass Spectrometry) detection on the extracting solution obtained in the step S1 and carrying out data analysis; analyzing the chemical structure of each metabolite and positioning an ion chromatographic peak; andbuilding a database. The method disclosed by the invention has the beneficial effects that (1) through mutual cooperation of a pretreatment method, a mass spectrometry method and a chromatographic method, a large amount of substances of the Chinese wolfberry leaves and the Chinese wolfberry fruits are rapidly identified, and a comprehensive metabolite database is constructed; (2) the metabolite database provides convenience for subsequent qualitative and quantitative analysis of the Chinese wolfberry metabolites; and (3) detection and determination of specific metabolites of the Chinese wolfberry leaves and the Chinese wolfberry fruits, so that a material basis is provided for subsequent metabolism research and development of Chinese wolfberry.
Owner:WOLFBERRY ENG RES INST NINGXIA ACADEMY OF AGRI & FORESTRY SCI
Model construction method for researching black sea bream immune mechanism enhancement by selenized glycosaminoglycan based on liver metabonomics
ActiveCN110346466AImprove throughputHigh sensitivityComponent separationMass spectrometric analysisBiomarker (petroleum)Organism
A model construction method for researching black sea bream immune mechanism enhancement by selenized glycosaminoglycan based on liver metabonomics relates to the field of biomedical analysis, and comprises the following steps: 1) uniformly dividing young black sea bream into an experimental group and a blank group; 2) adding selenized glycosaminoglycan into the feed of the experimental group; 3)carrying out hunger treatment on the young black sea bream in the experimental group and the blank group, anesthetizing, taking out liver tissues, and storing for later use; 4) preparing a liver tissue sample, preparing a quality control sample in the same time, and using ultra-high performance liquid chromatography-time-of-flight mass spectrometry technology for treatment; and (5) analyzing and processing the collected liver metabonomics data of the black sea breams, identifying and screening out liver metabonomics biomarkers for enhancing the immune mechanism of the black sea breams relatedto the selenide glycosaminoglycan, and constructing and analyzing metabolic pathways pointed by the liver metabonomics biomarkers. The construction method is good in systematicness, high in accuracy,low in cost and simple to operate.
Owner:舟山出入境检验检疫局综合技术服务中心
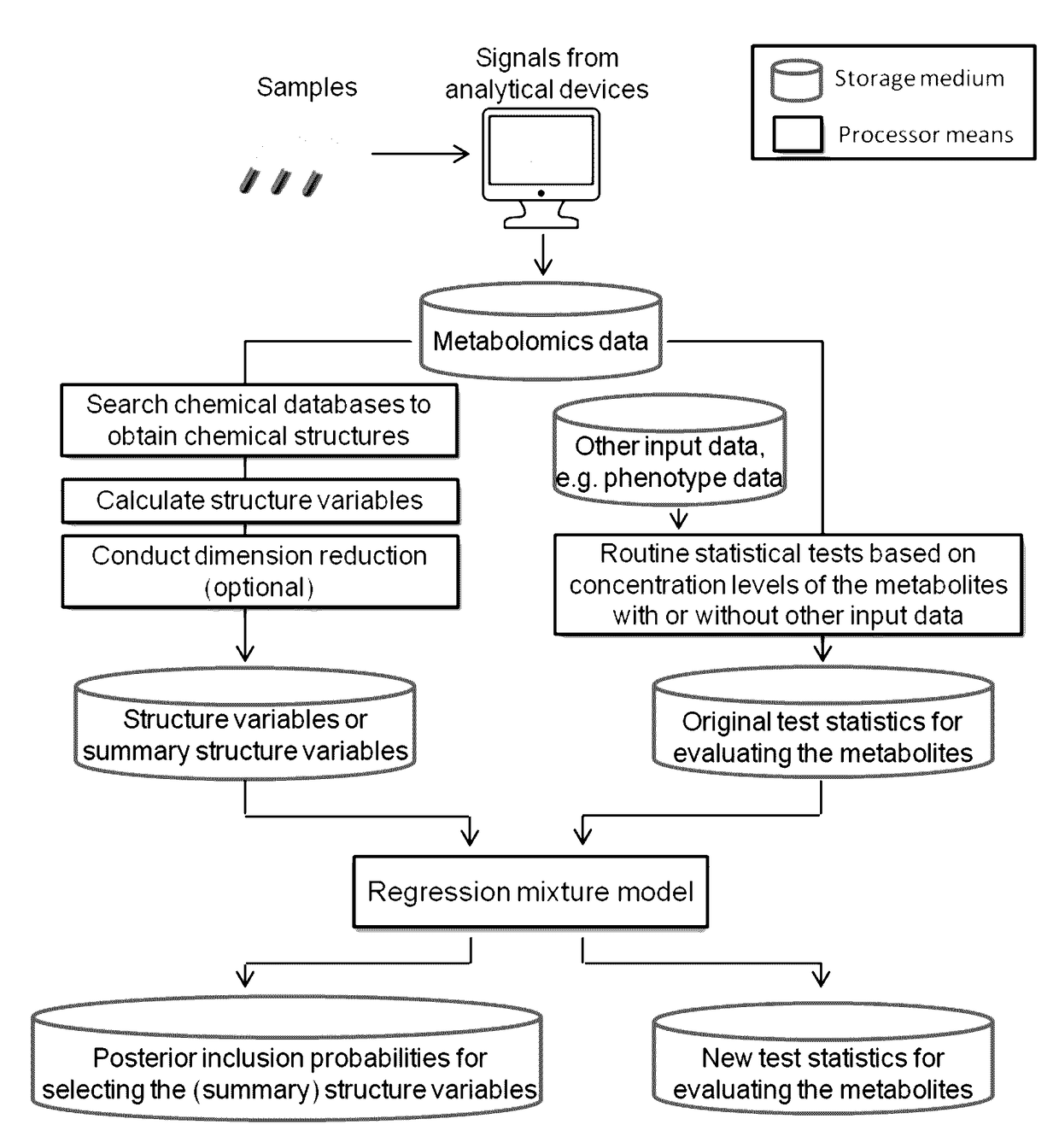
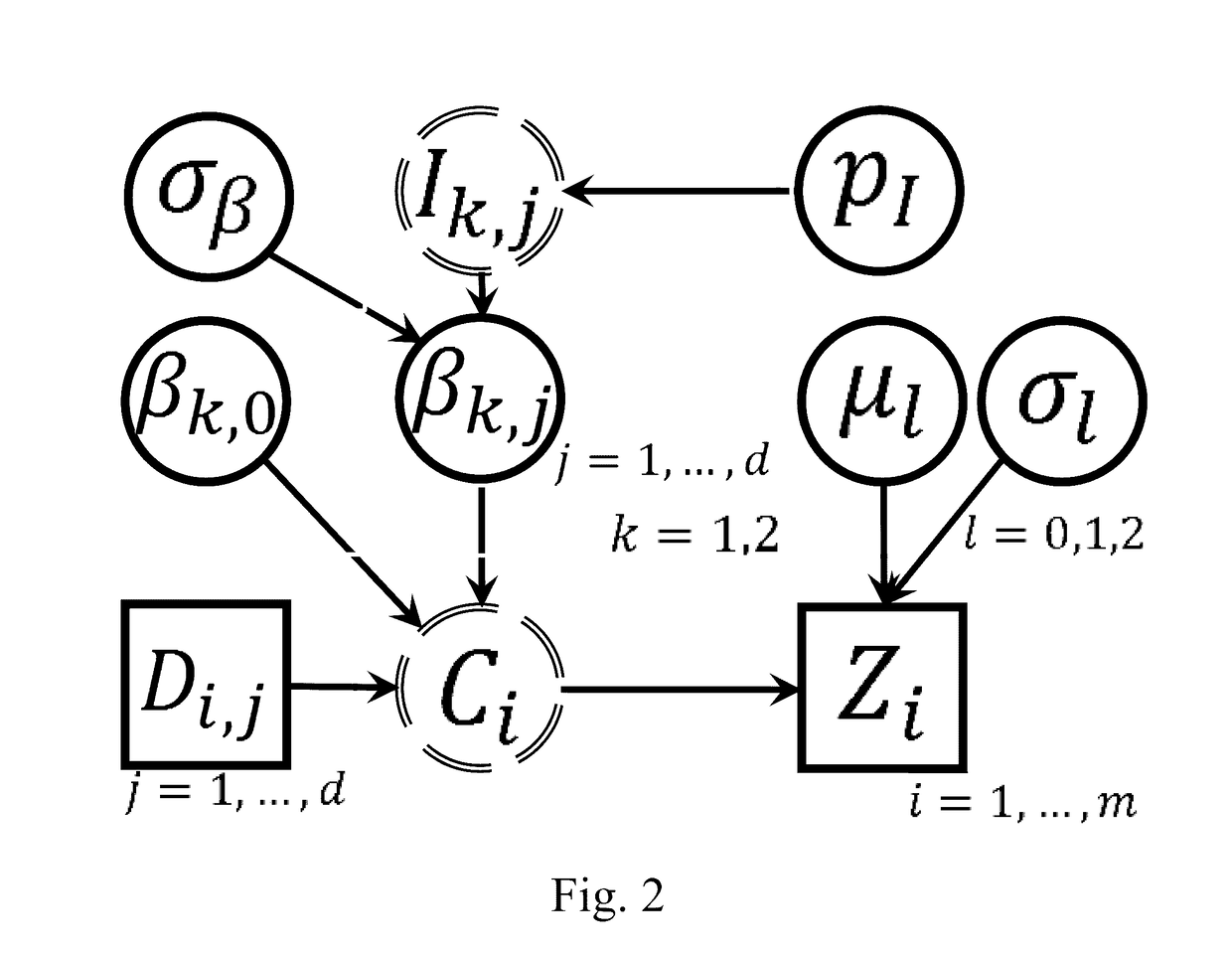
Chemical structure-informed metabolomics data analysis
ActiveUS9646139B1Improve performanceImprove biological activityMolecular entity identificationChemical structure searchChemical structureMetabolite
This invention relates to statistically significant methods for metabolomics data analysis that incorporate the structure information of metabolites. Understanding of disease pathogenesis and drug effects, as well as prediction of variation in drug response can be achieved by analyzing quantitative data measuring metabolomics biomarker profiles from biological samples. This invention is to boost the statistical power of analyzing metabolomics data. The comprising methods may include retrieving information of metabolites' chemical structures, converting them into structural data, and integrating the structural data into analysis of metabolite concentration data to improve the evaluation of metabolites and to better identify metabolomics signatures.
Owner:ZHU HONGJIE +1
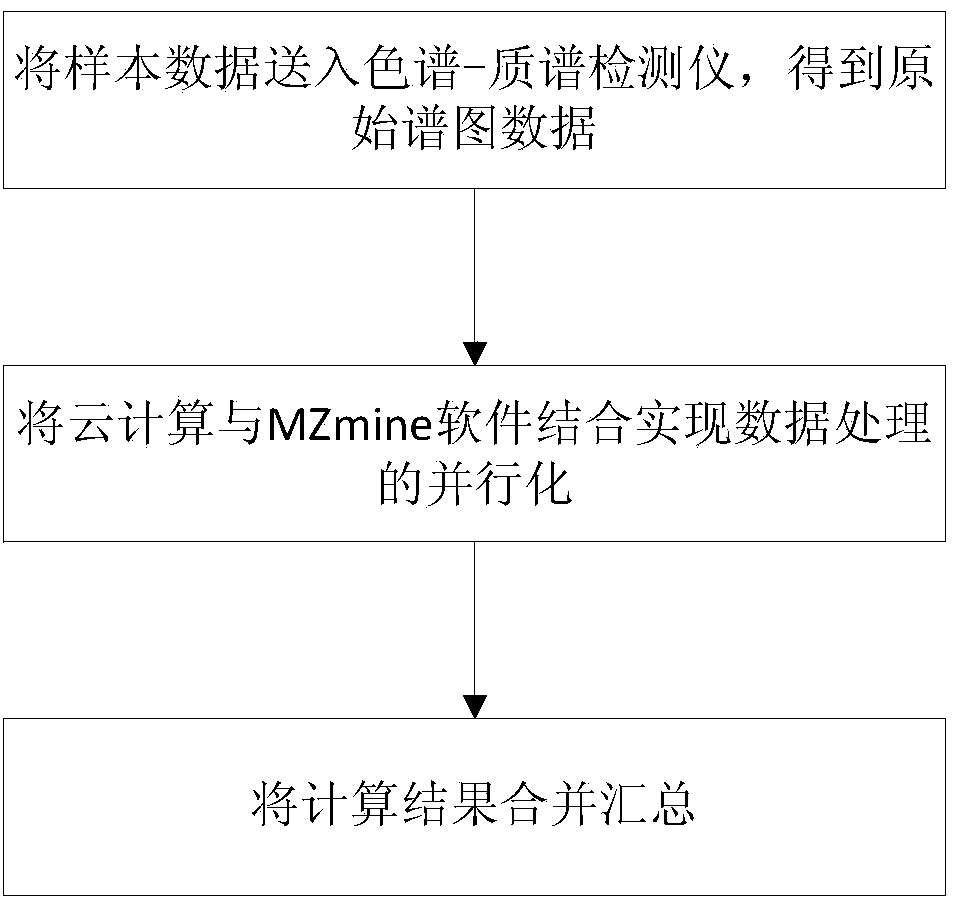
Parallel computing based metabonomics data processing method
InactiveCN104111819AConfiguration highSave energyConcurrent instruction executionSpecific program execution arrangementsRetention timeOriginal data
The invention discloses a parallel computing based metabonomics data processing method. The parallel computing based metabonomics data processing method includes the steps: (1) feeding a sample into a chromatography-mass spectrometry detector, so that original data displayed in a spectrogram form and sample component retention time are obtained; (2) achieving data processing parallelization by means of combining parallel computing with MZmine software: performing task decomposition of the original data obtained from the step (1) according to spectral peaks and the sample component retention time, and enabling a plurality of servers configured with the MZmine software to perform parallel computing for sub-tasks obtained from task decomposition, so that a sample component data table is obtained; (3) merging and summarizing computing results. By the parallel computing based metabonomics data processing method, data processing can be accelerated and computing cost can be saved.
Owner:SHANDONG NORMAL UNIV
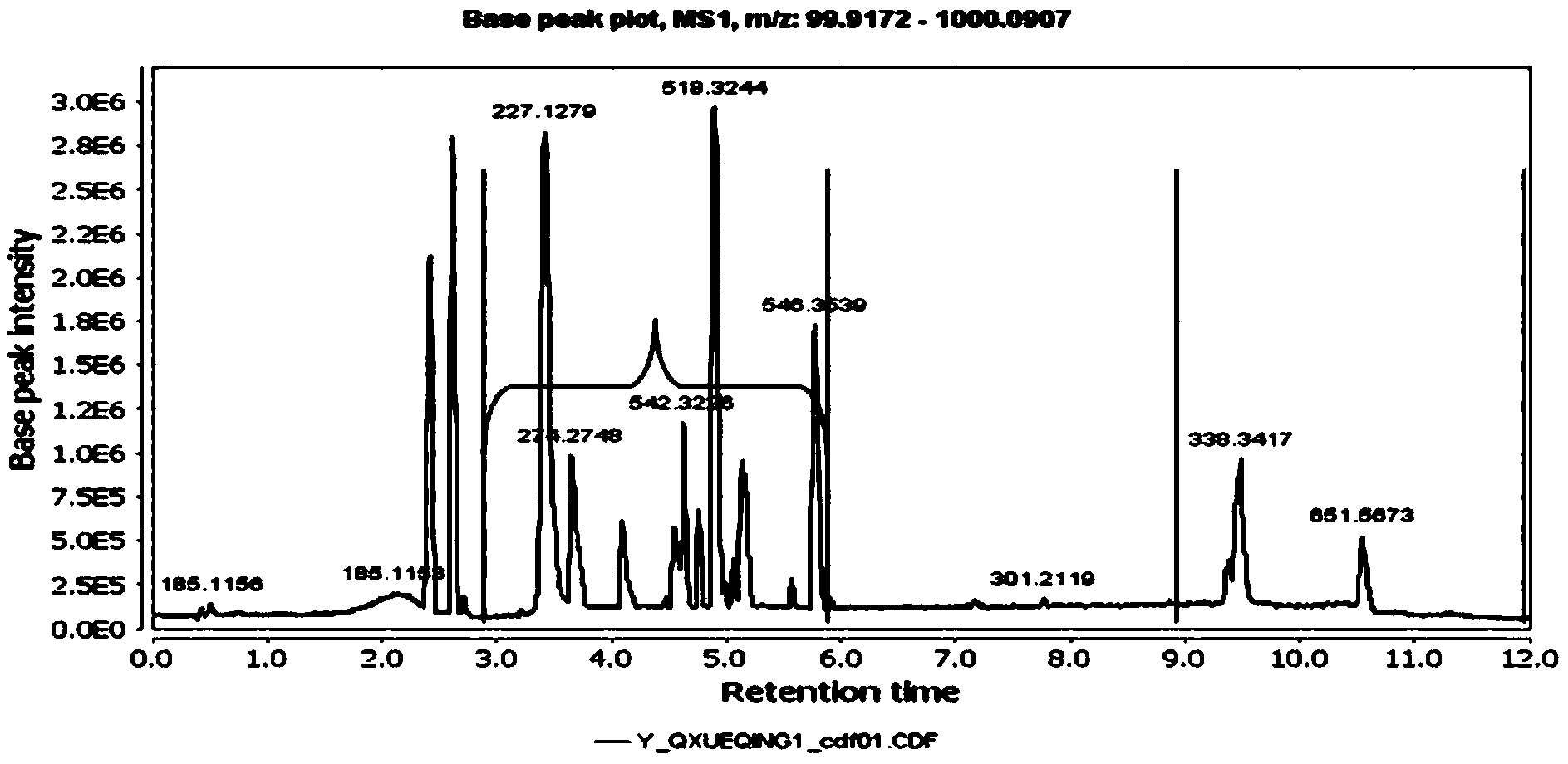
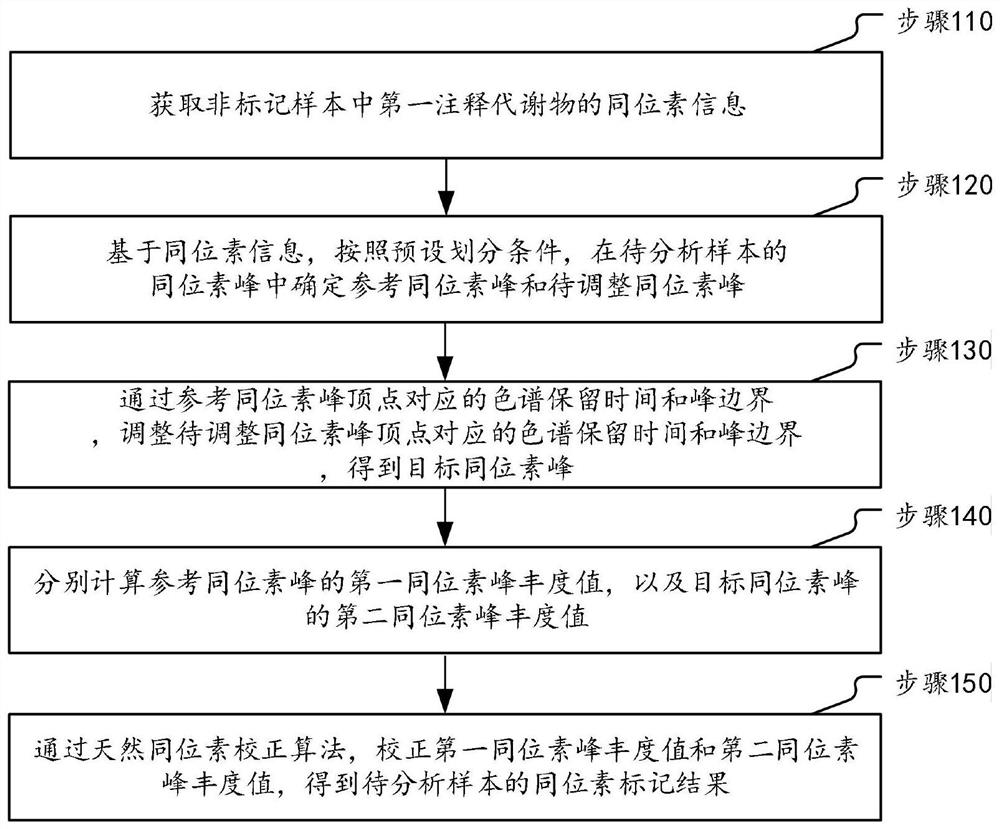
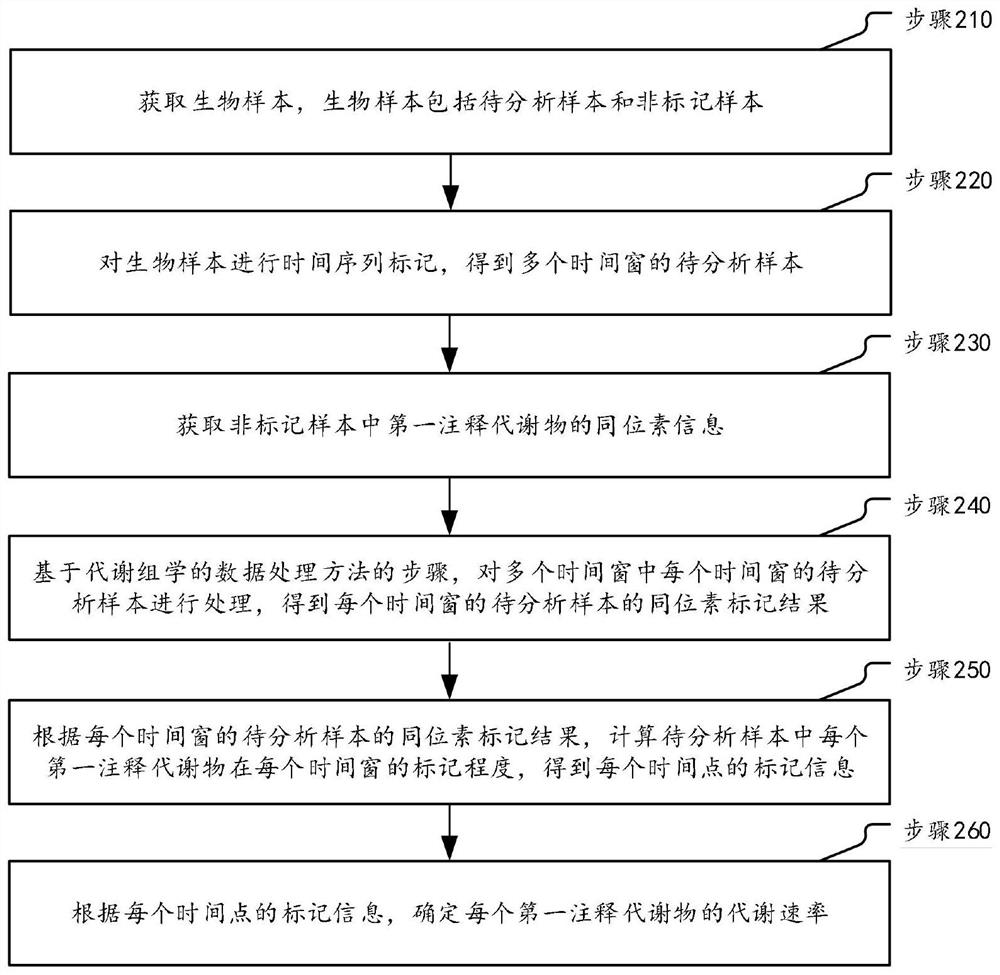
Metabonomics data processing method and device based on stable isotope labeling
ActiveCN114295766AImprove accuracyComponent separationCharacter and pattern recognitionStable Isotope LabelingIsotopic labeling
The invention discloses a metabonomics data processing method and device based on stable isotope labeling. The method specifically comprises the following steps: acquiring isotope information of a first annotation metabolite in a non-labeled sample; based on the isotope information, determining a reference isotope peak and a to-be-adjusted isotope peak in the isotope peaks of the to-be-analyzed sample according to a preset division condition; according to the chromatographic retention time and the peak boundary corresponding to the peak top point of the reference isotope, adjusting the chromatographic retention time and the peak boundary corresponding to the peak top point of the isotope to be adjusted to obtain a target isotope peak; respectively calculating a first isotope peak abundance value of the reference isotope peak and a second isotope peak abundance value of the target isotope peak; and correcting the first isotope peak abundance value and the second isotope peak abundance value through a natural isotope correction algorithm to obtain an isotope labeling result of the to-be-analyzed sample. Therefore, the marked condition of each metabolite can be accurately obtained, so that comprehensive dynamic metabolism analysis on the metabolome in the organism is realized.
Owner:SHANGHAI INST OF ORGANIC CHEM CHINESE ACAD OF SCI
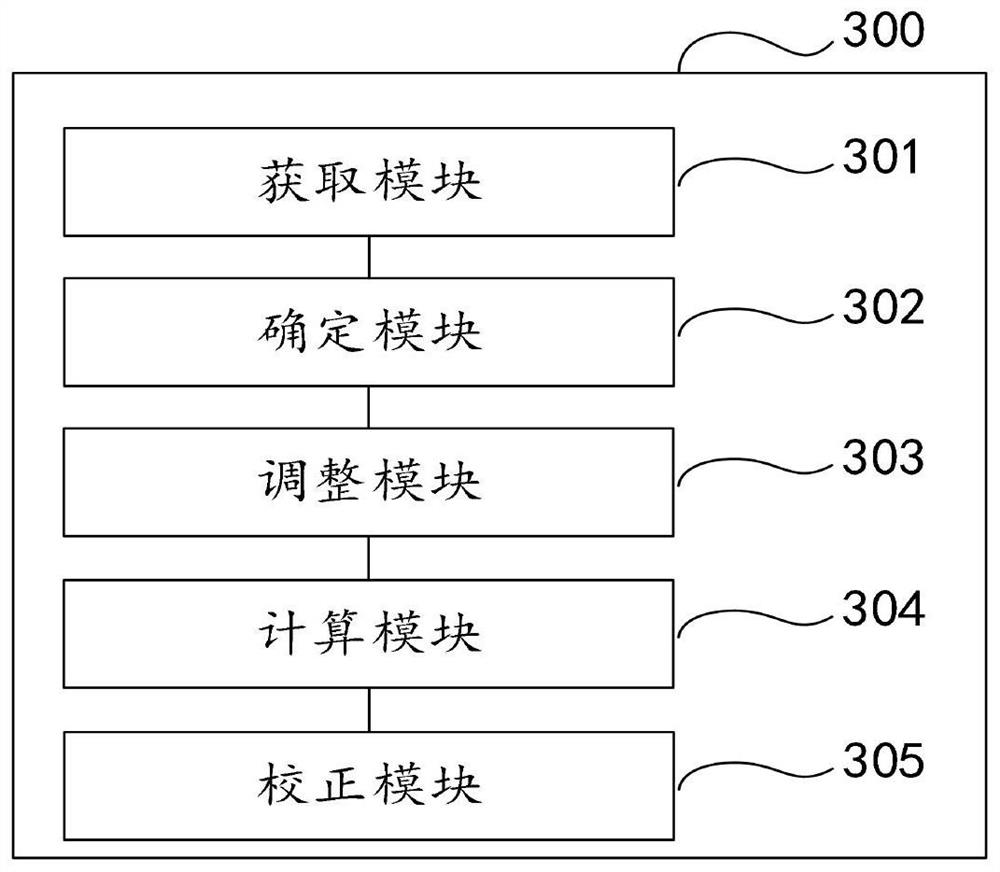
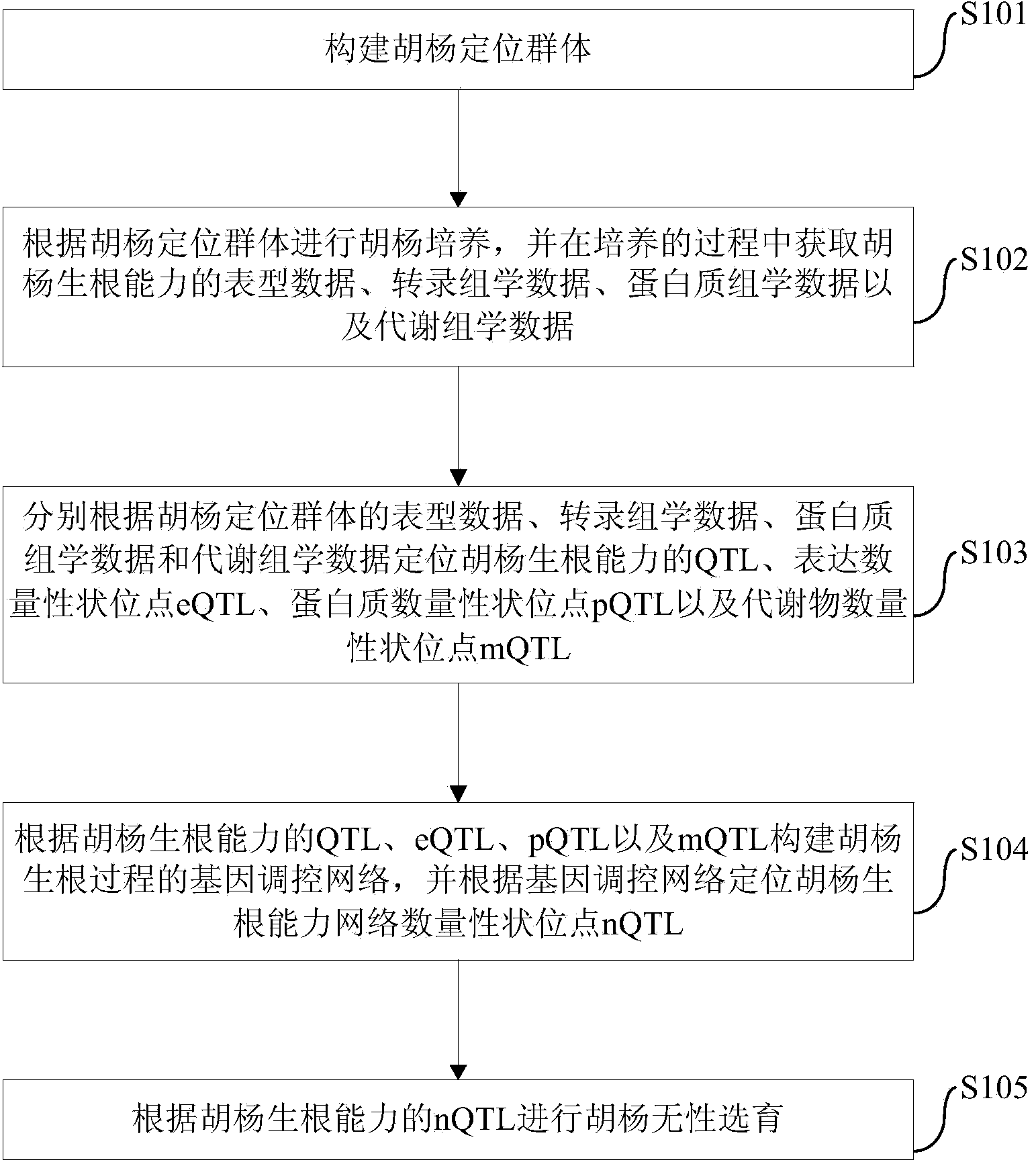
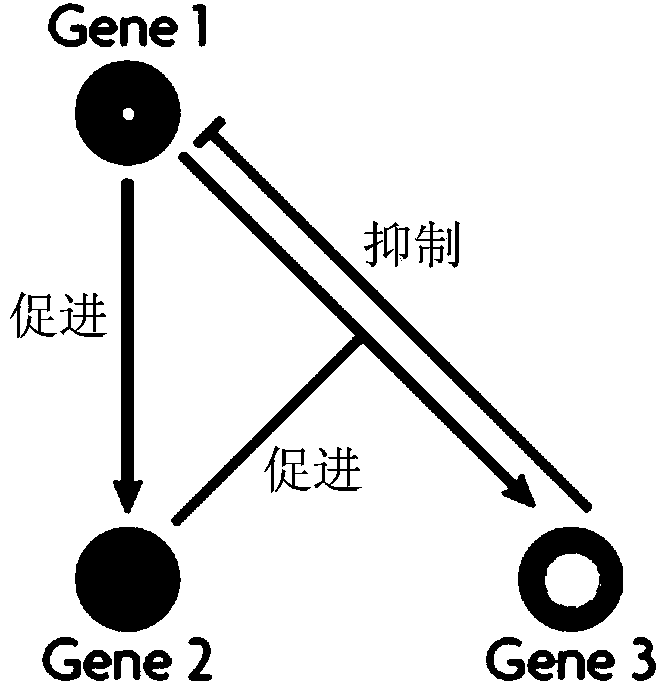
Populus diversifolia sexless breeding method based on gene regulatory network
ActiveCN104361262AOvercoming the problem that shape separation is seriously detrimental to screeningHigh reproductive rateSpecial data processing applicationsPopulus diversifoliaMetabolomics data
The invention discloses a populus diversifolia sexless breeding method based on a gene regulatory network. The method comprises the following steps: forming a populus diversifolia positioning group; conducting explant culture according to the populus diversifolia positioning group, and obtaining phenotype data, transcriptomics data, proteomics data and metabonomics data of the populus diversifolia rooting ability according to the populus diversifolia positioning group; positioning QTL, eQTL, pQTL and mQTL of the populus diversifolia rooting ability respectively according to the phenotype data, the transcriptomics data, the proteomics data and the metabonomics data of the populus diversifolia positioning group; forming the gene regulatory network of the populus diversifolia rooting process according to the QTL, the eQTL, the pQTL and the mQTL of the populus diversifolia rooting ability, and positioning network quantitative trait loci (nQTL) of the populus diversifolia rooting ability according to the gene regulatory network; conducting populus diversifolia sexless breeding according to the nQTL of the populus diversifolia rooting ability. According to the method of the embodiment, the populus diversifolia single plant with strong rooting ability can be effectively screened out, the reproductive rate of the populus diversifolia is improved, and the stress resistant sexless breeding of the populus diversifolia is greatly promoted.
Owner:BEIJING FORESTRY UNIVERSITY
Metabolomics data missing value filling method based on neighboring stability
ActiveCN110097920AEasy to fillMaterial analysis by electric/magnetic meansSystems biologyMetaboliteMetabolomics data
The invention provides a metabolomics data missing value filling method based on neighboring stability, wherein the method belongs to the field of metabolomics data analysis technology. The core technology of the method is metering stability of content of k most neighboring samples of a sample containing a missing metabolite in the corresponding metabolite, and filling different types of missing values by means of different strategies. The method has relatively high filling effect to the metabolomics data which contain the missing value and has an important significance to subsequent data analysis, metabolic marker selection, etc.
Owner:DALIAN UNIV OF TECH
Bio-metabonomics data processing method, analysis method, device and application
InactiveCN111157664AAchieve integrationAchieve correctionComponent separationReference sampleGas liquid chromatographic
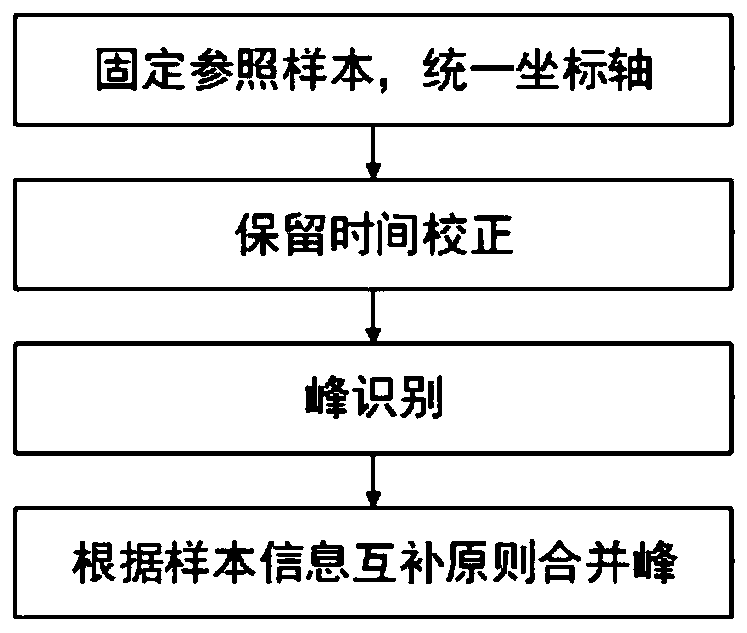
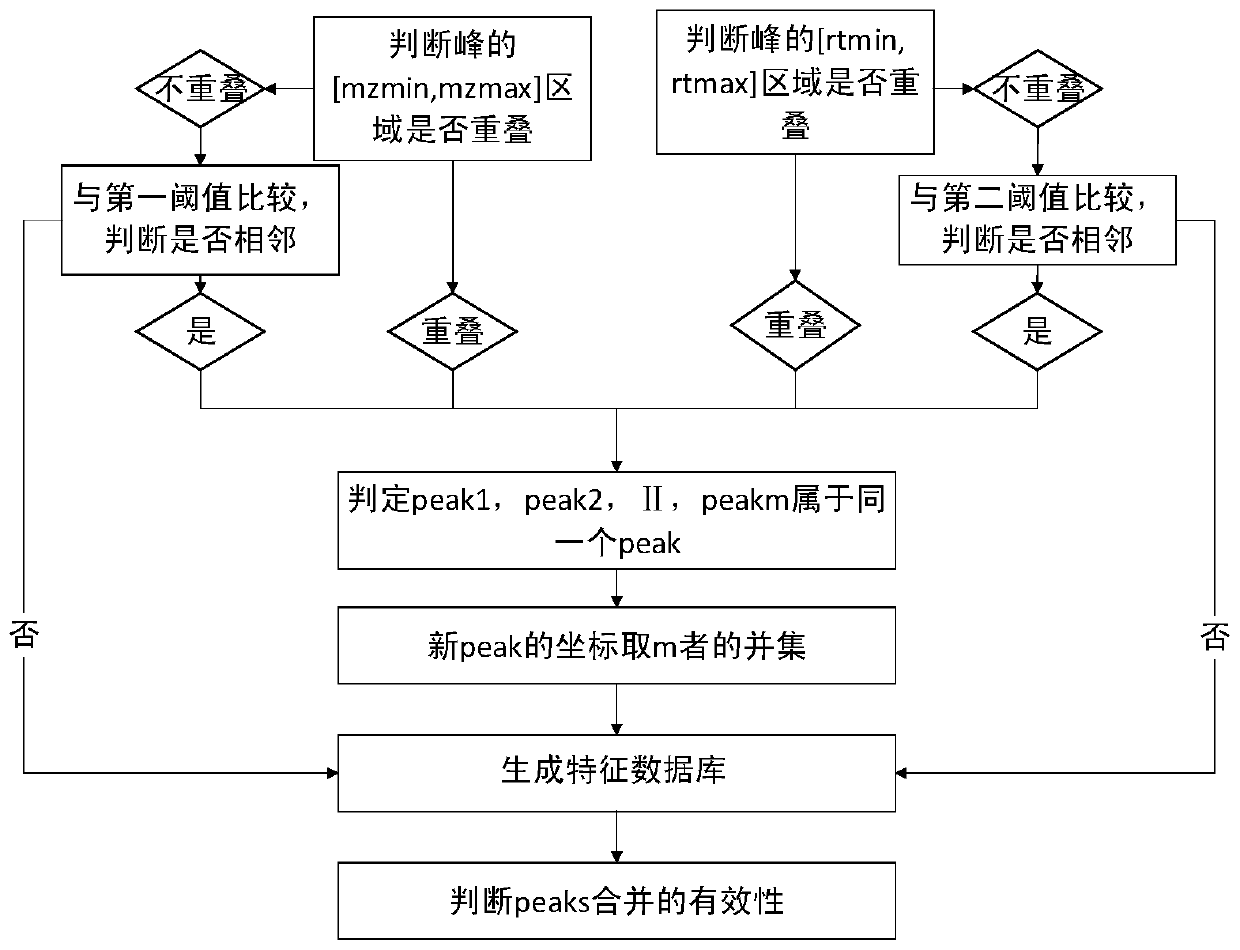
The invention discloses a bio-metabonomics data processing method, a biometabonomics data analysis method, a biometabonomics data processing device, a biometabonomics data analysis device and application. The biological metabonomics data processing method comprises the step of integrating liquid chromatography-mass spectrometry data or gas chromatography-mass spectrometry data of a plurality of biological samples to form a characteristic database, the integration step comprises the following steps: S11, randomly selecting one sample in a plurality of biological samples as a reference sample, and correcting the time axes of other samples one by one according to the time axis of the reference sample; S12, for each corrected sample, carrying out peak identification processing of primary massspectrum ion peaks one by one to obtain a plurality of identification characteristic peaks; and S13, combining the plurality of identification characteristic peaks according to a sample information complementation principle to obtain a characteristic database of the plurality of biological samples. According to the technical scheme, super-large-scale metabonomics data integration can be achieved,data correction and data integration of batches or a single sample can be achieved, and the method is not affected by detection batches.
Owner:SHENZHEN ICARBONX DIGITAL LIFE MANAGEMENT CO LTD +2
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com