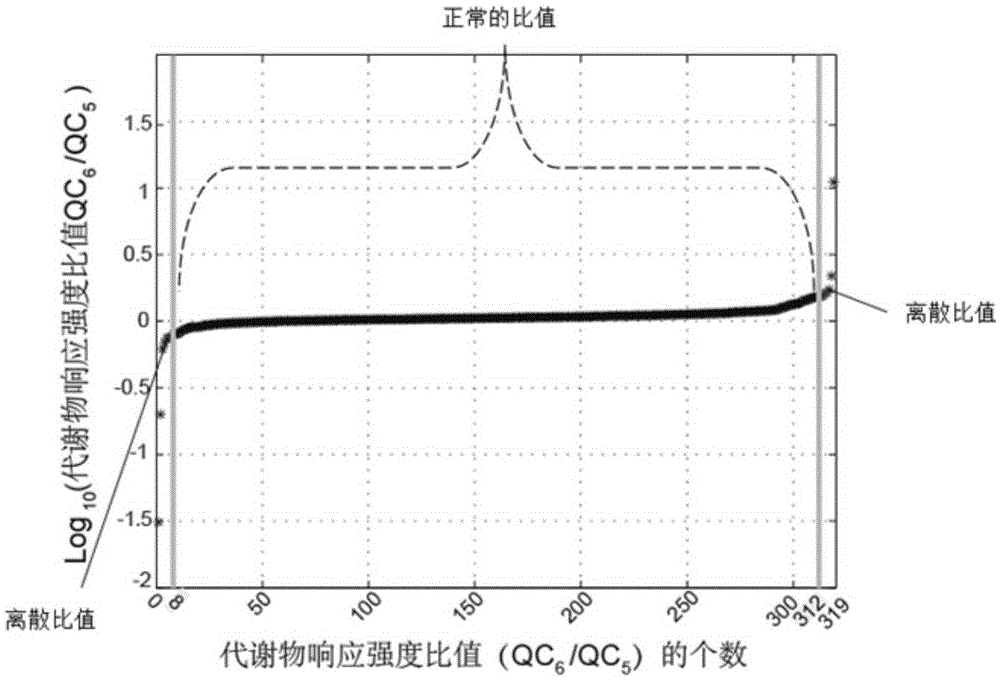
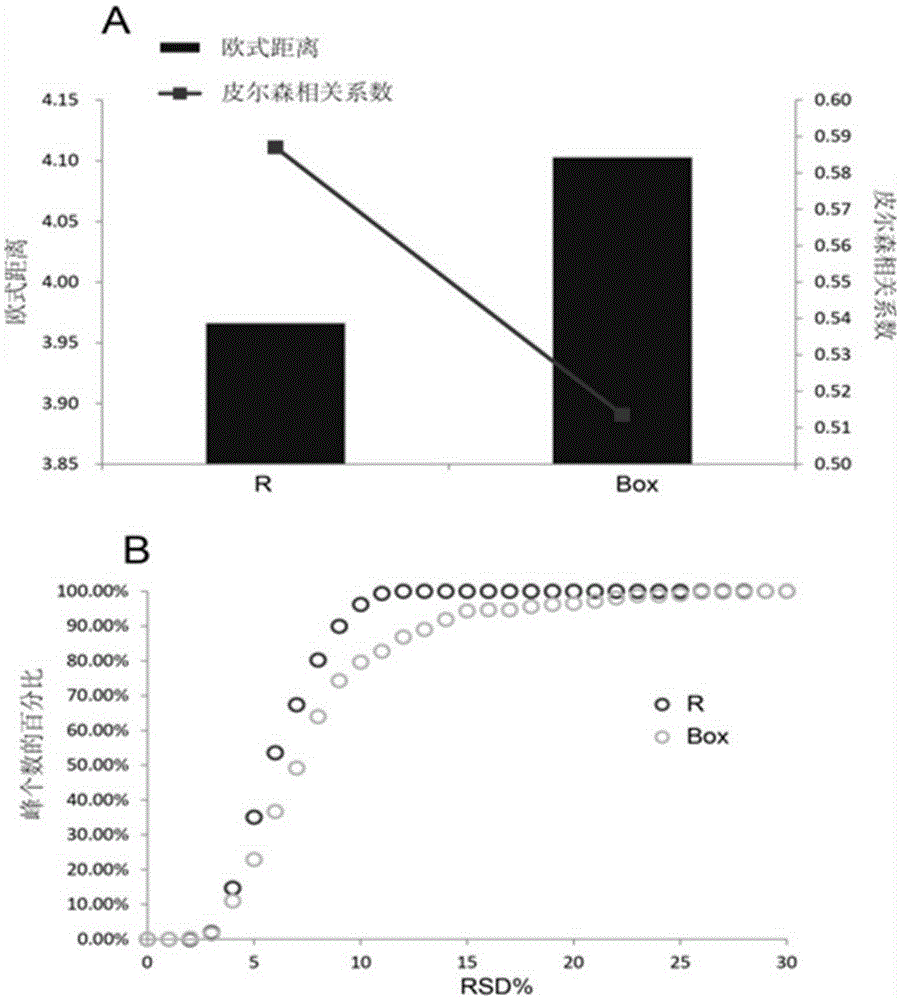
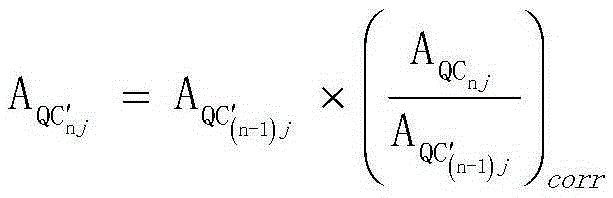Screening and calibrating method of metabolomic data random errors
A metabolomics data and random error technology, which is applied in the field of analytical chemistry and metabolomics, can solve problems such as interference with experimental results and random errors, and achieve obvious correction effects, quality improvement, and high throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0023] 1 sample
[0024] In this example, fresh tobacco leaves were used as samples. A total of 447 fresh tobacco leaves were collected from three production areas in Henan, Yunnan and Guizhou. They were stored in liquid nitrogen at -196°C, transported, ground under liquid nitrogen, freeze-dried at low temperature, and stored in a refrigerator at -80°C. Weigh 0.5g of each tobacco powder sample, mix them evenly, and generate a new sample, ie, a quality control (QC) sample. QC samples can be used to establish gas chromatography-mass spectrometry (GC-MS) quasi-target metabolomics methods, evaluate the repeatability of analytical methods, and correct errors in metabolomic data of actual samples.
[0025] 2. Random error screening and correction method:
[0026] 2.1 GC-MS metabolomics analysis
[0027] (1) Sample pretreatment: the fresh tobacco leaf samples were taken out from the -80°C refrigerator, placed in a 4°C refrigerator overnight, and then placed at room temperature for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com