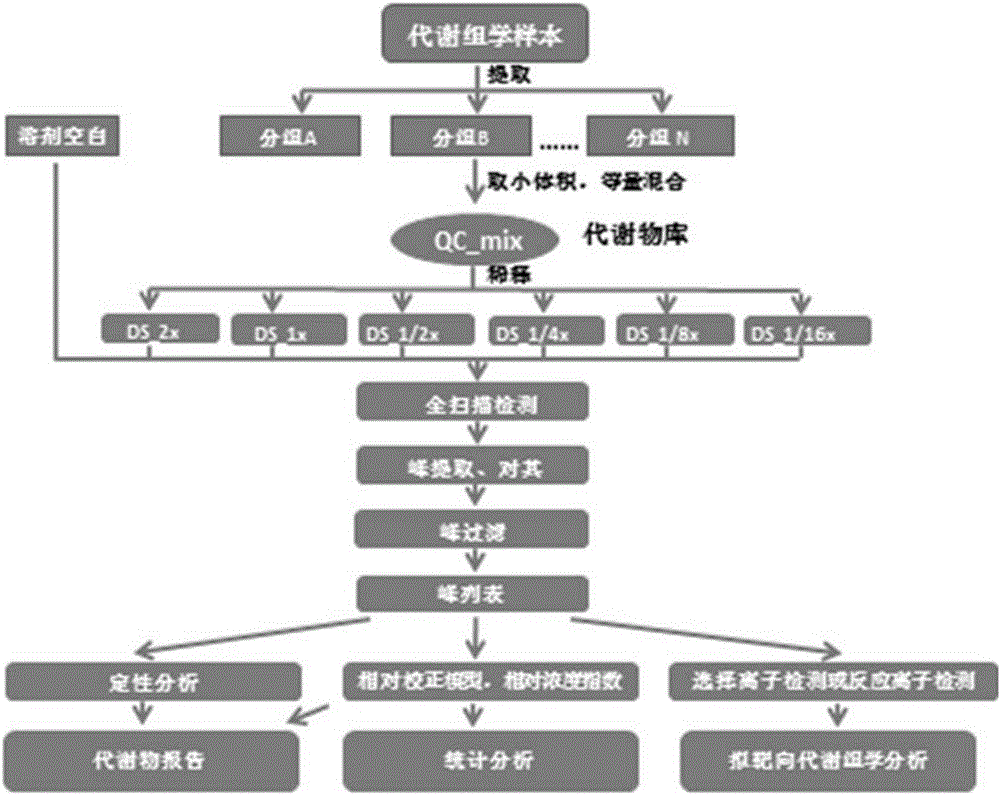
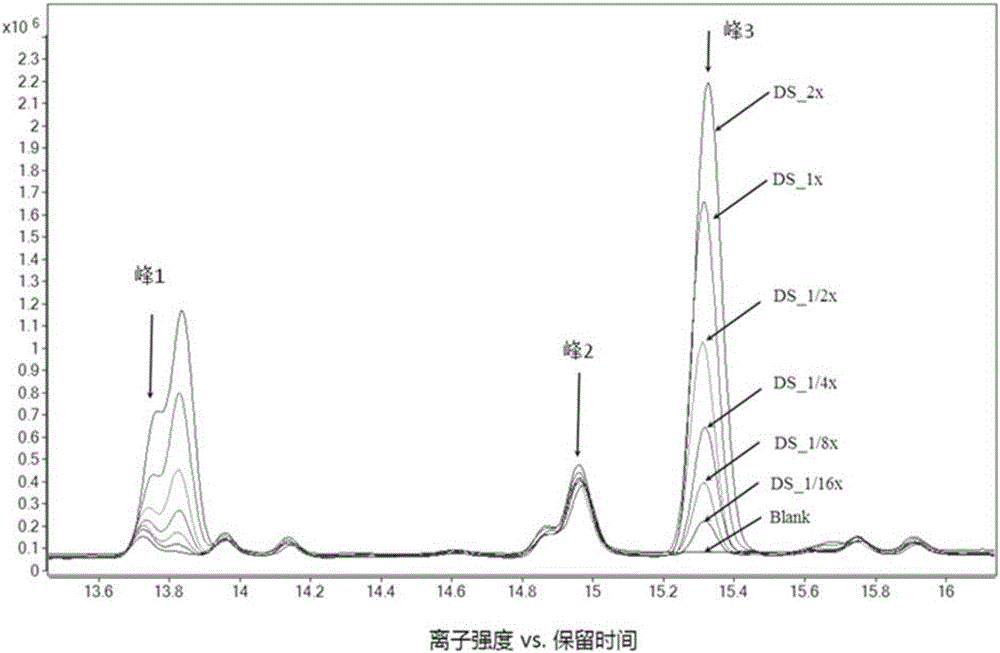
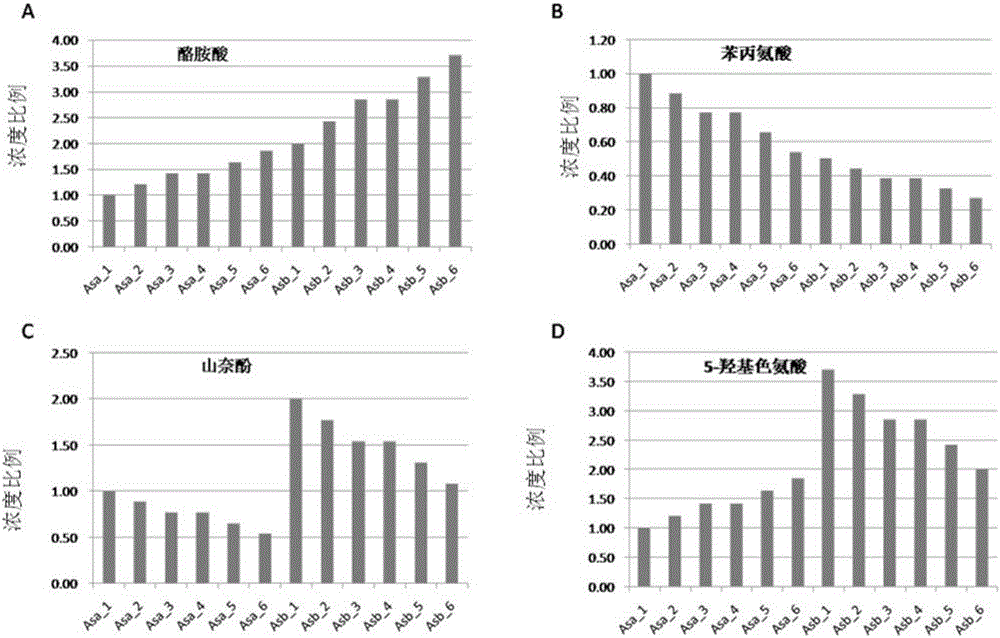
Metabolism group method for distinguishing false positive mass spectra peak signals and quantificationally correcting mass spectra peak area
A technology of mass spectrometry and metabolites, applied in the biological field, can solve problems such as high cost, inappropriate non-targeted metabolomics experiments, and high technical requirements, and achieve reliable data, elimination of false positive mass spectrometry peak signals, and low cost effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] Example 1. The present invention is used to distinguish false positive mass spectrum peak signals and quantitatively correct the metabolomics method of mass spectrum peak area
[0079] One, the method for eliminating false positive mass spectrum peaks in the detection of metabolites, comprising the steps:
[0080] 1. Prepare QC_mix samples, solvent blank samples, QC_mix samples with different dilution ratios, QC_mix samples with different concentration ratios, and metabolite samples of multiple samples to be tested;
[0081] The QC_mix sample is to mix the metabolite solutions of multiple samples to be tested to obtain a QC_mix sample;
[0082] The metabolite solution is composed of metabolites and organic solvents or obtained by extracting metabolites of the sample to be tested with an organic solvent, and is used as a metabolite sample of the sample to be tested;
[0083] Solvent blank samples consisted of internal standard and organic solvent;
[0084] QC_mix sampl...
Embodiment 2
[0131] Embodiment 2, artificial simulation sample verification method of the present invention reduces the effect of false positive mass spectrum peak
[0132] In order to verify the validity, feasibility and reliability of the method, artificial samples (Artificial Samples, AS) composed of 20 standard products were configured in the experiment. figure 1 It is a flowchart of the present invention.
[0133] Table 1 is the artificial simulation sample and concentration (mmol) that 20 kinds of standard substances are composed
[0134]
[0135]
[0136] *: Asa: artificial samples group A, artificial simulated sample group A, Asa_1,_2,,,_6 simulated 6 biological replicates.
[0137] ^: Asb: artificial samples group B, artificial simulation sample group B, Asb_1,_2,,,_6 simulated 6 biological repeats.
[0138] Concentration unit is mmol
[0139] Standards: Tyrosine (T2900000), Phenylalanine (147966), Kaempferol (96353), 3-Indolebutyric Acid (57310), Taxifolin (78666), 5-Hyd...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com