Metabonomics network marker identification method based on horizontal relation
A network marker and metabolomics technology, applied in the field of biological data analysis, can solve problems such as sample change disturbance, achieve the effect of good discrimination ability and effective data processing means
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0028] Example: Screening of potential network markers for breast cancer subtype discrimination based on human metabolism.
[0029] (1) Human Metabolic Breast Cancer Data
[0030] The human metabolic breast cancer dataset used in this example is a public dataset (Jan Budczies, Scarlet F. Berit M.Müller, et al.Comparative metabolomics of estrogen receptor positive and estrogen receptor negative breast cancer: alterations inglutamine and beta-alanine metabolism[J].Journal of Proteomics,2013,94:279-288), including 162 qualitative metabolites There are two categories: estrogen receptor negative (ER-) and estrogen receptor positive (ER+). The data is divided into a training set and a test set. Among them, the training set contains 41 ER- samples and 143 ER+ samples. The test set contains 26 ER- samples and 61 ER+ samples.
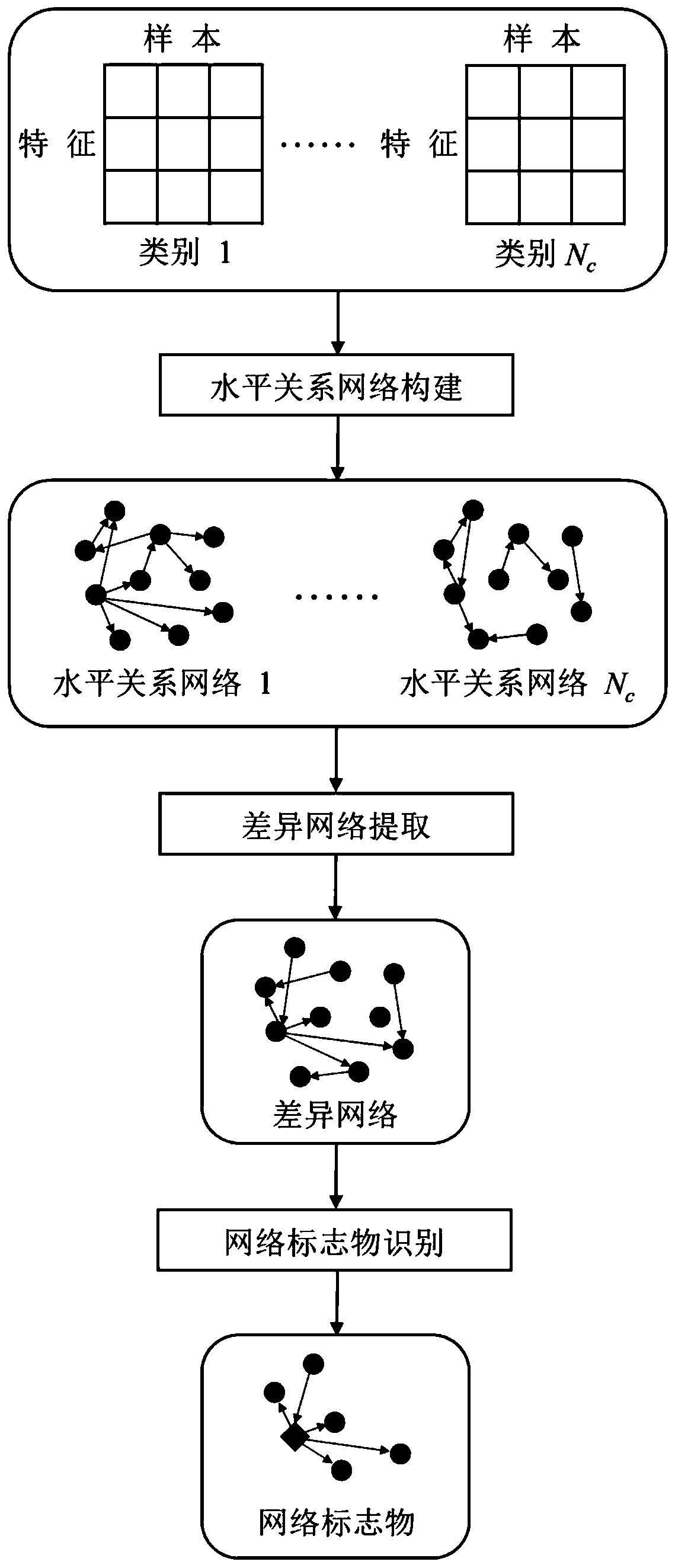
[0031] (2) Build a horizontal relationship network on each type of sample in the training set
[0032] (2.1) Build a horizontal relationship network on ER...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com