Bio-metabonomics data processing method, analysis method, device and application
A technology for metabolomics data and processing methods, applied in measurement devices, analytical materials, scientific instruments, etc., which can solve problems such as difficulty in complementarity, increased data processing time and difficulty, and poor repeatability of metabolite detection.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
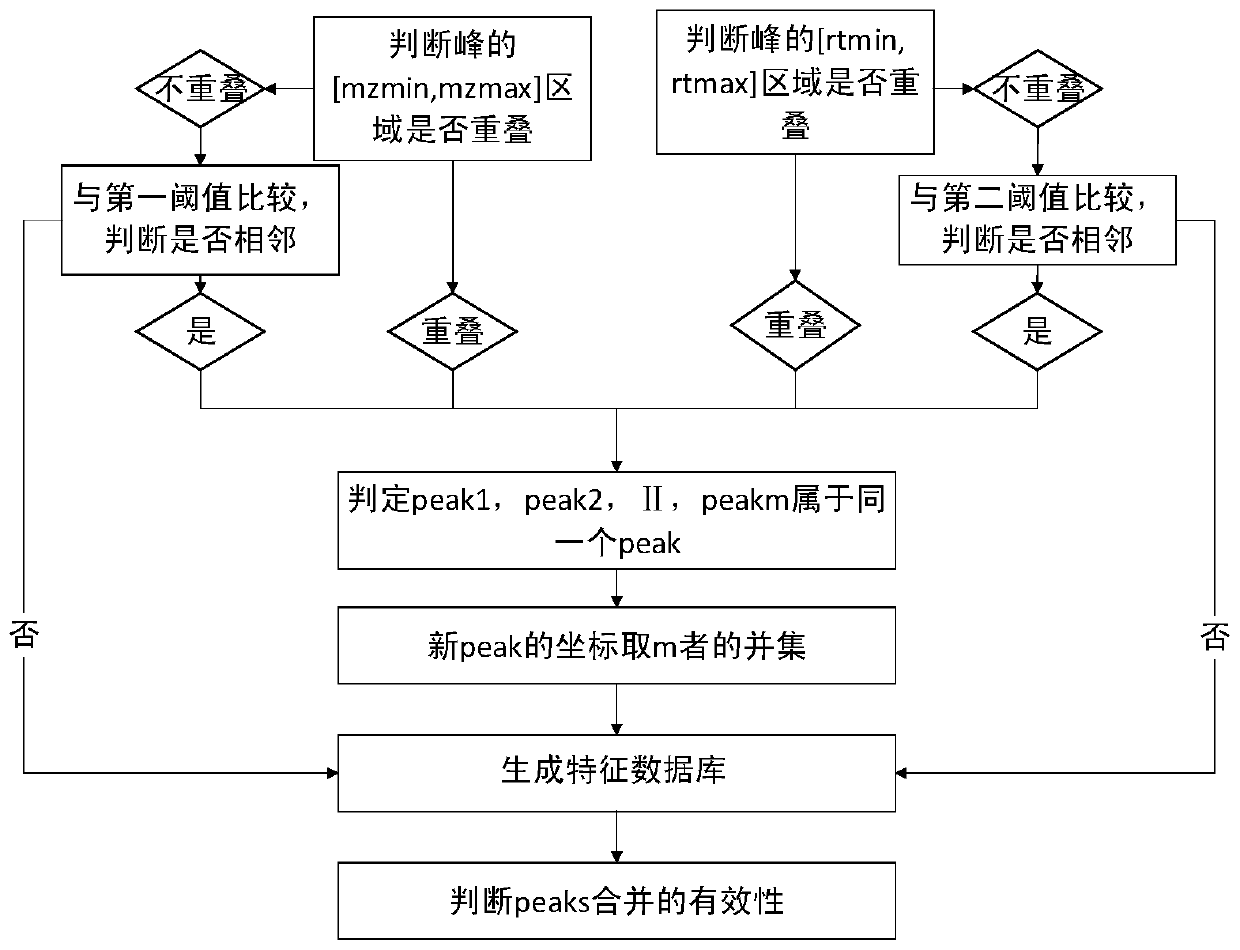
[0107] According to a typical implementation of the present invention, the step of qualitatively identifying metabolites through secondary mass spectrometry data information includes:
[0108] S21, obtaining the mass-to-charge ratio data of each standard compound;
[0109] S22, arbitrarily selecting a feature value in the feature database obtained after the biological metabolomics data processing, and finding all the mass-to-charge ratio data of the secondary mass spectrum corresponding to the feature value, according to all the mass-to-charge ratio data of the secondary mass spectrum, Find the set of standard compounds that match it;
[0110] S23, taking all the mass-to-charge ratio data corresponding to a characteristic value selected in S22 as one side, and taking the mass-to-charge ratio data of the matched standard compound found in S22 as the other side, and performing a process on both Similarity scoring, calculating point integrals, and characterizing metabolites base...
Embodiment 1
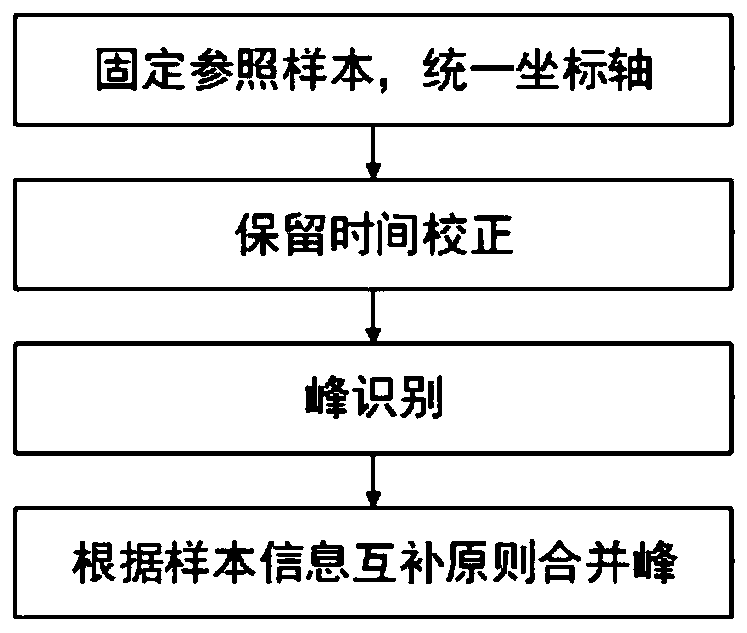
[0165] 1. Build the features database
[0166] 1) Determine a unified coordinate axis to make the samples comparable in time. Select a fixed sample from all samples as a reference, and the time axes of other samples are corrected according to this sample, which is reference.xml.
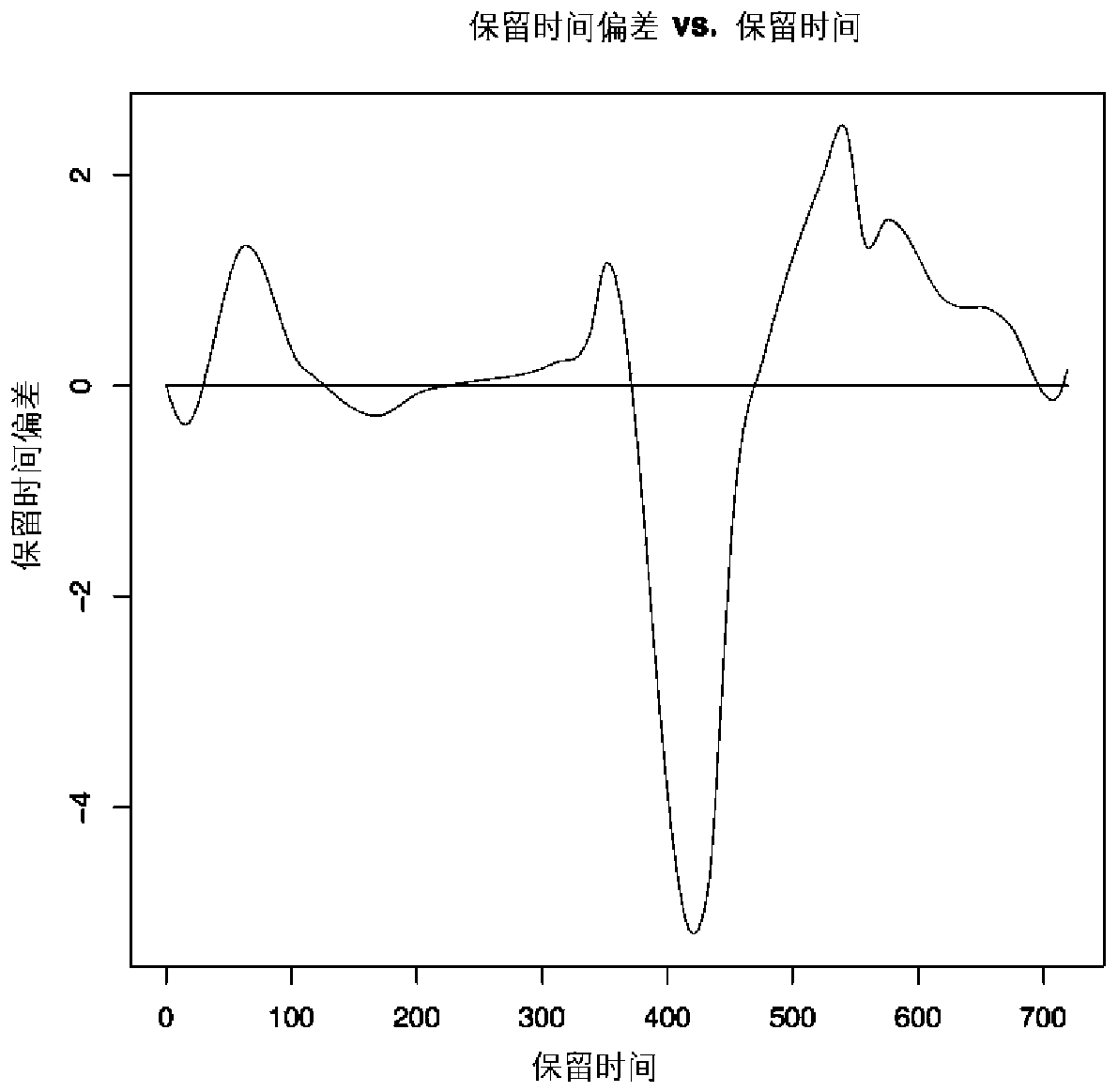
[0167] 2) The new sample is first corrected for retention time (RT). In this step, the Obiwarp algorithm is used to correct the retention time (RT) of the primary mass spectrometry data (MS1) and secondary mass spectrometry data (MS2). In this embodiment, a sample retention time is corrected as image 3 As shown (note: the horizontal axis is the retention time RT (unit: s), the vertical axis is the time when the sample retention time deviates from the reference sample (unit: s), also known as the retention time deviation. The horizontal line is the reference sample (reference), Curves are other samples.
[0168] 3) On the corrected time axis, use the CentWave algorithm to perform peak identificati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com