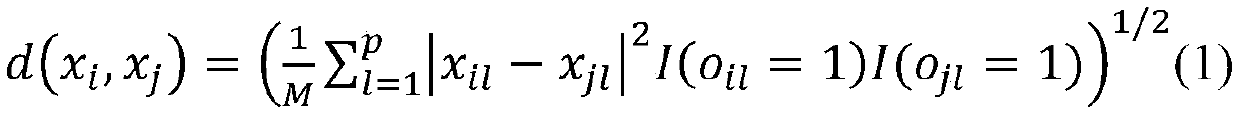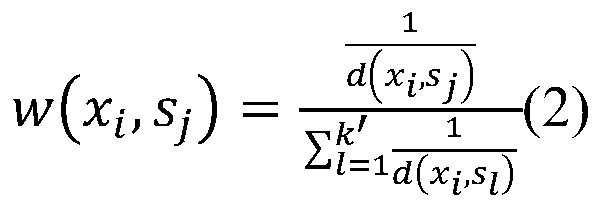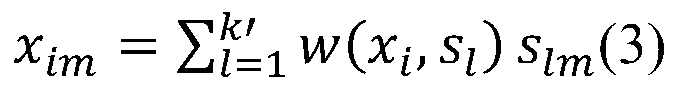Metabolomics data missing value filling method based on neighboring stability
A technology of metabolomics data and filling method, which is applied in the field of missing value filling of metabolomics data, considering the missing type of metabolite missing value, and can solve the problem of unsatisfactory filling effect of non-random missing type data.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0028]The specific implementation of the method will be further described on the simulated data below in conjunction with the technical solution. The simulated data is only limited to explain the present invention for easy understanding, but not to limit the present invention.
[0029] In table 1, be the simulated data of the present invention, x i Indicates the i-th sample, the data contains 10 samples, m 1 ~ m 5 Indicates 5 metabolites in the data and NaN indicates missing values in the data.
[0030] Table 1: Simulation data
[0031]
[0032] The data in Table 1 contains 4 missing values, which are x 13 ,x 52 ,x 84 ,x 93 . Below with x 13 Take as an example to specify.
[0033] (1) Use the formula (1) to calculate the sample x 1 The distance d between other samples, get: d(x 1 ,x 2 )=1.94,d(x 1 ,x 3 )=1.73,d(x 1 ,x 4 )=3.39,d(x 1 ,x 5 )=3.46,d(x 1 ,x 6 )=4.12,d(x 1 ,x 7 )=2.29,d(x 1 ,x 8 )=2.71,d(x 1 ,x 9 )=2.74,d(x 1 ,x 10 ) = 3.16. Let k=...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com