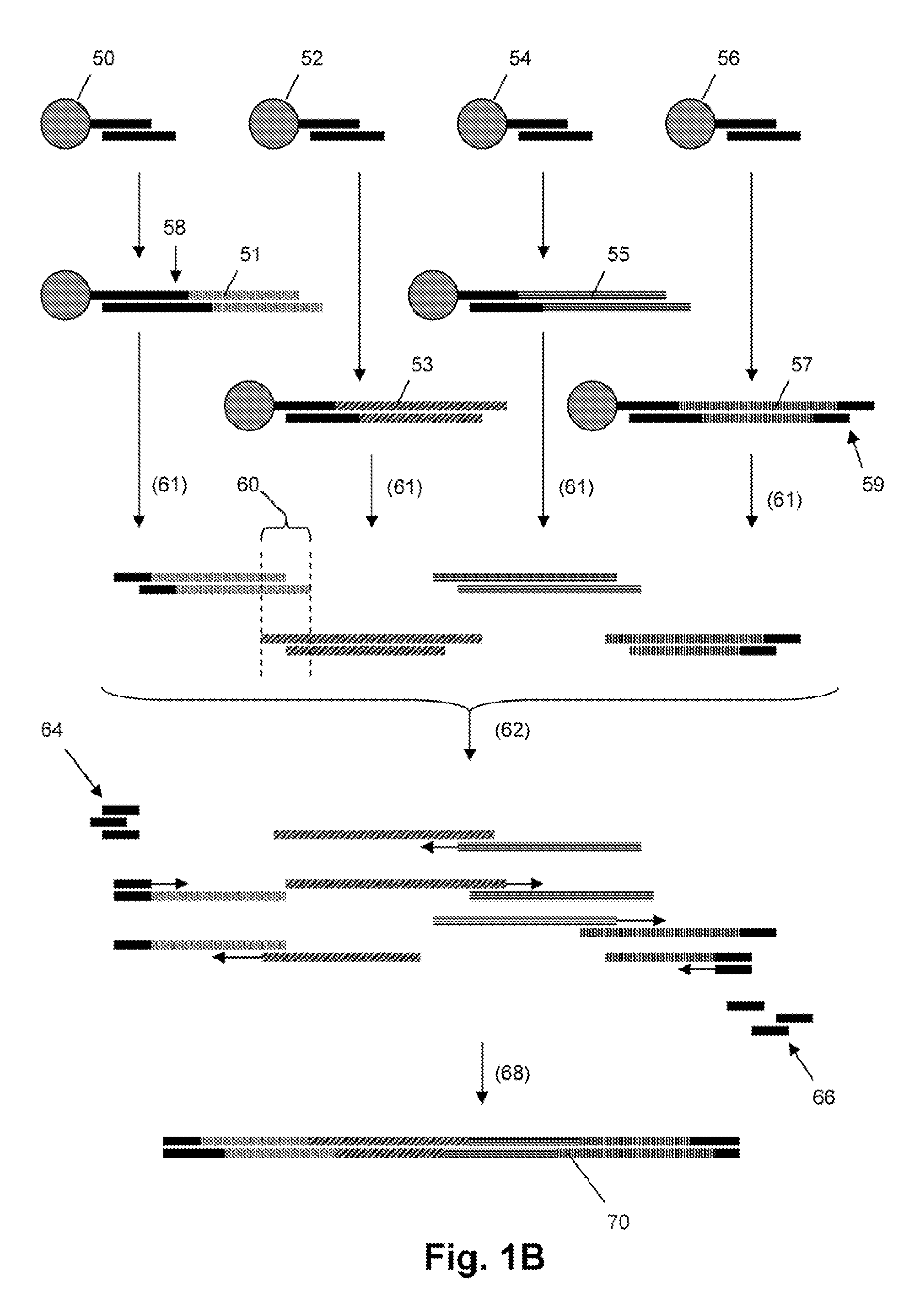
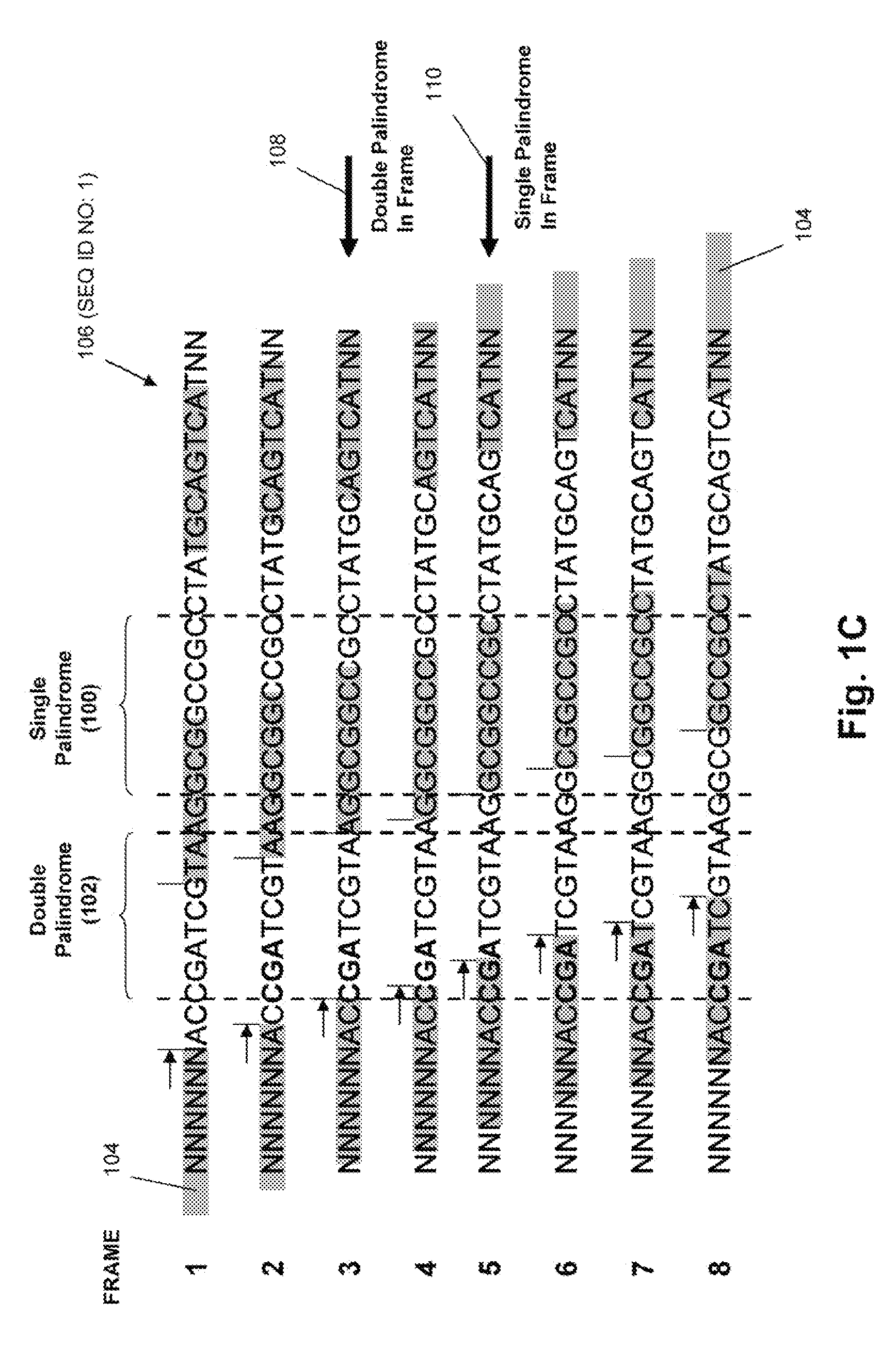
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
45 results about "Palindromic sequence" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
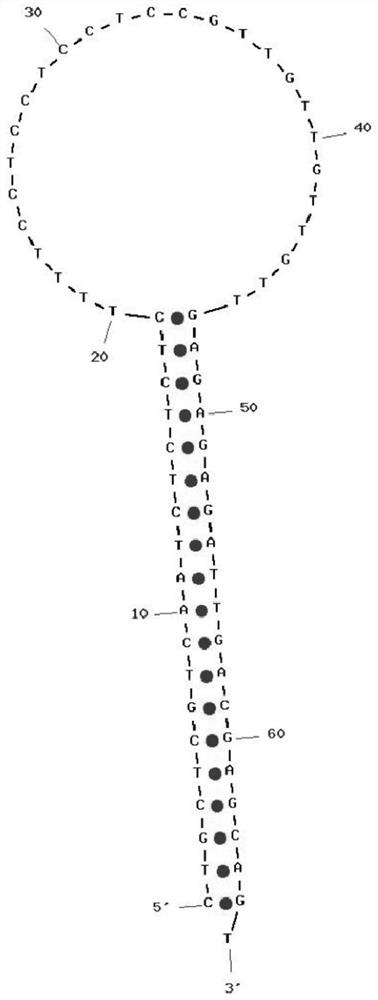
A palindromic sequence is a nucleic acid sequence in a double-stranded DNA or RNA molecule wherein reading in a certain direction (e.g. 5' to 3') on one strand matches the sequence reading in the same direction (e.g. 5' to 3') on the complementary strand. This definition of palindrome thus depends on complementary strands being palindromic of each other.
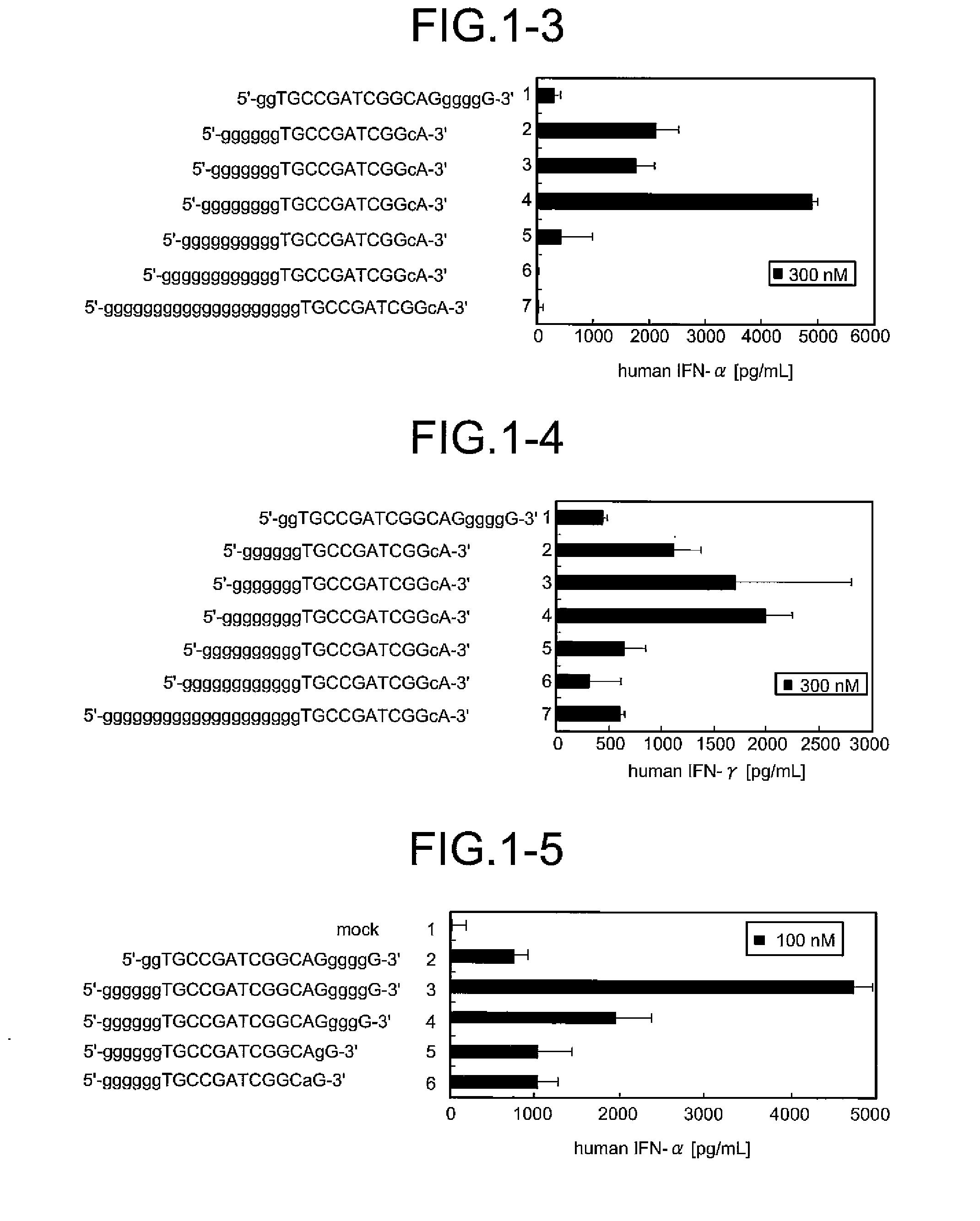
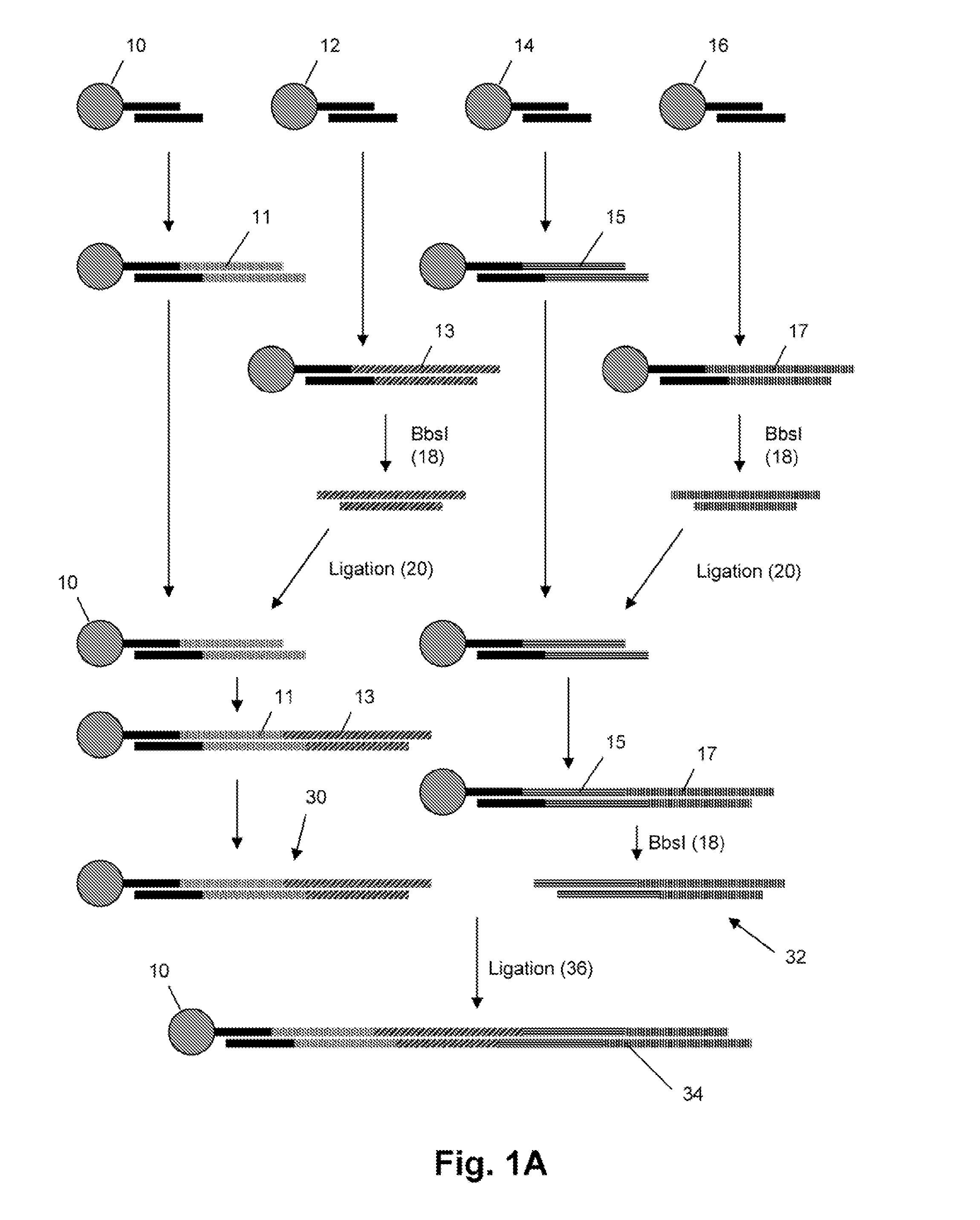
Immunostimulatory oligonucleotides and uses thereof
Oligonucleotides containing the non-palindromic sequence motif:X1X2X3X4X5X6X7X8,wherein X1 is C,T,G or A (preferably T or C); wherein X2 is C,T,G or A; wherein X7 is C,T,G or A (preferably G); at least three, and preferably all, of X3, X4, X5, X6 and X8 are T; and with the proviso that, in the motif, a C does not precede a G (in other terms, the nucleic acid motif does not consist of a CpG oligonucleotide), that modulate the immune response of animals of the order Primate, including humans, are disclosed. This immune modulation is characterized by stimulation of proliferation, differentiation, cytokine production and antibody production on B-cells and cell differentiation on plasmacytoid dendritic cells.
Owner:DAVID HORN LLC
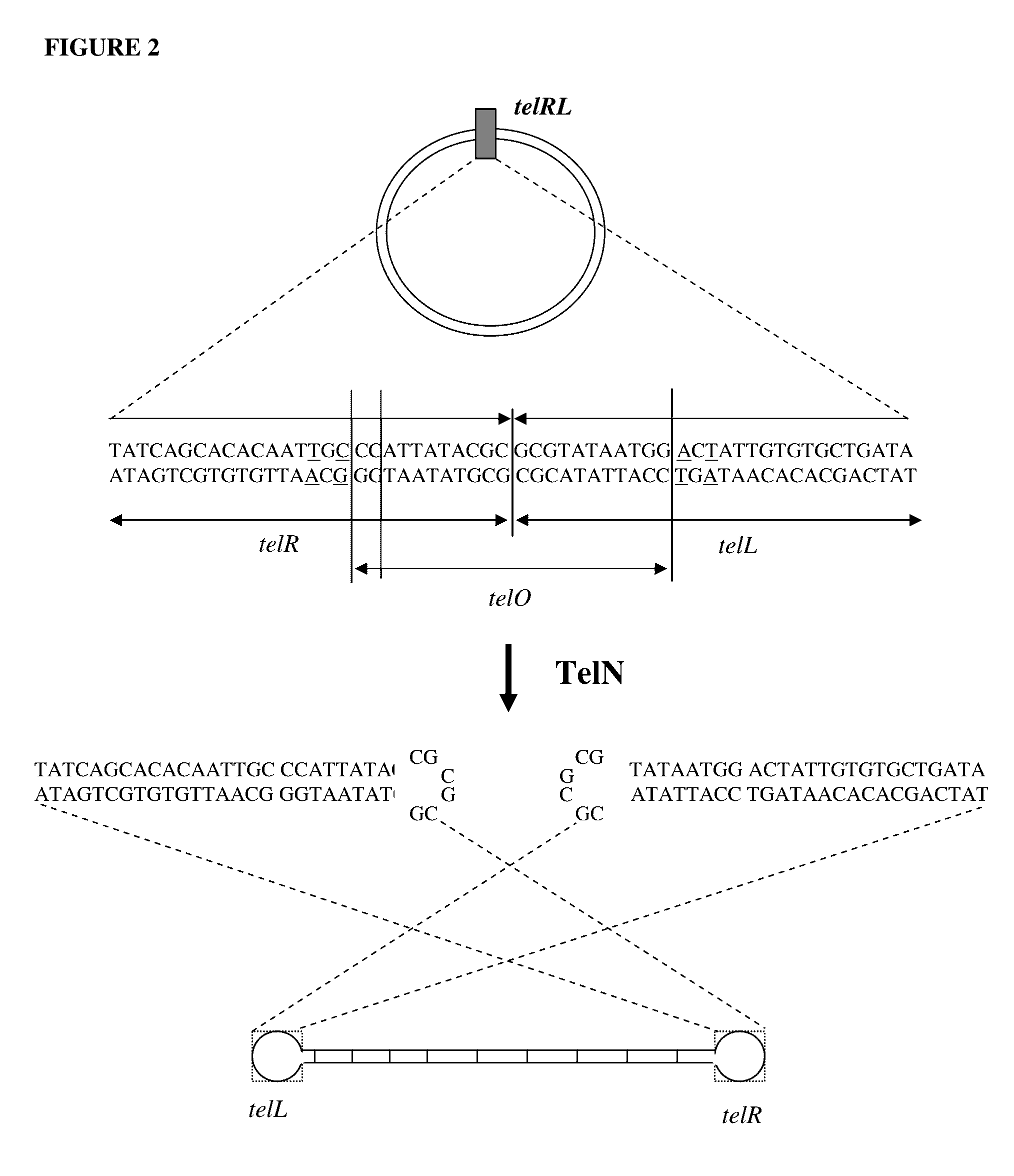
Production of closed linear DNA using a palindromic sequence
A primer for the amplification of a DNA template comprising a protelomerase target sequence, particularly for production of closed linear DNA, which primer is capable of specifically binding to a palindromic sequence within a protelomerase target sequence and priming amplification in both directions.
Owner:TOUCHLIGHT IP LTD
Production of closed linear DNA using a palindromic sequence
A primer for the amplification of a DNA template comprising a protelomerase target sequence, particularly for production of closed linear DNA, which primer is capable of specifically binding to a palindromic sequence within a protelomerase target sequence and priming amplification in both directions.
Owner:TOUCHLIGHT IP LTD
Immunostimulatory oligonucleotides and use thereof in pharmaceuticals
InactiveUS20090263413A1High therapeutic effectExcellent immunostimulatory activityOrganic active ingredientsSugar derivativesNucleotideCytokine
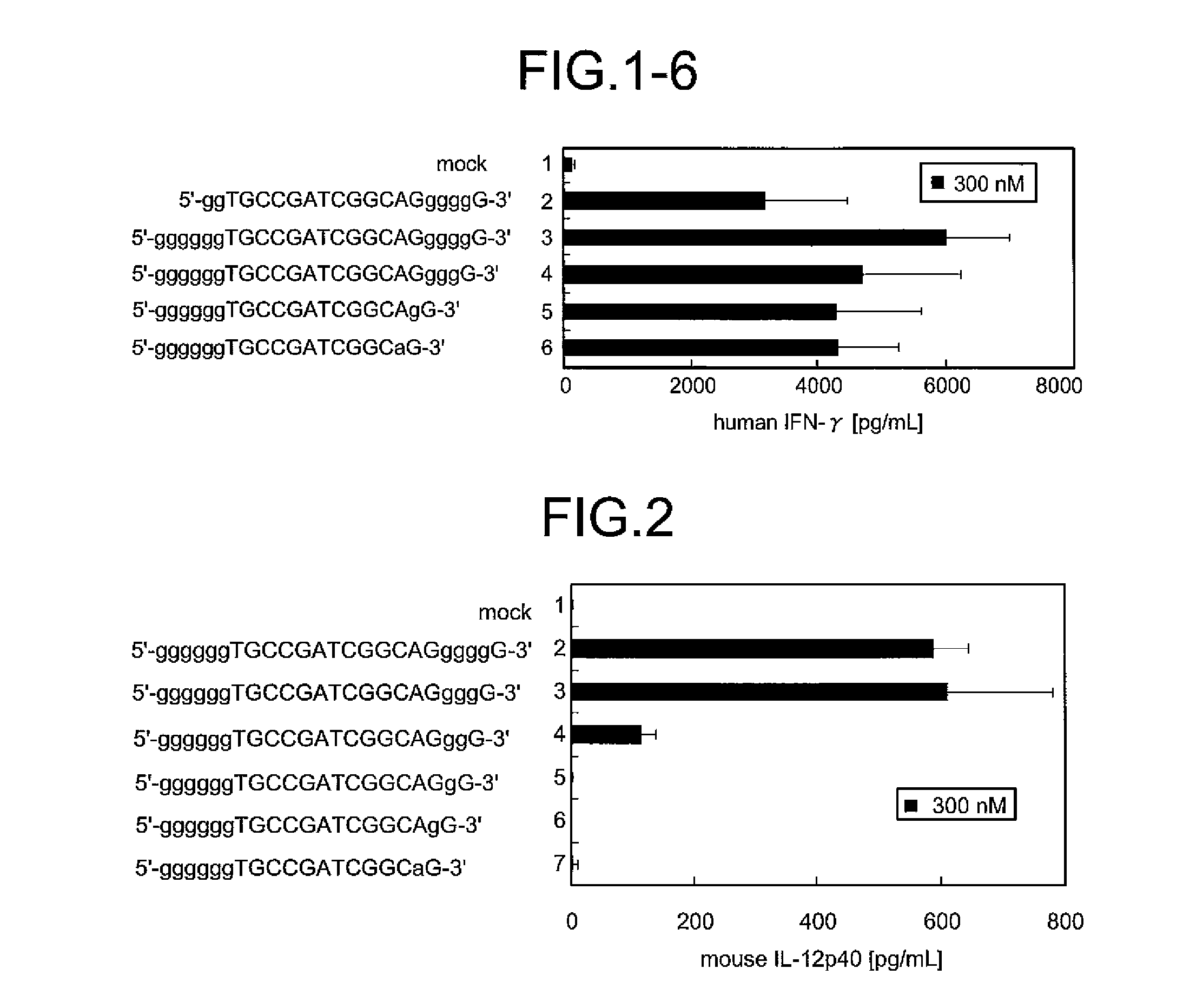
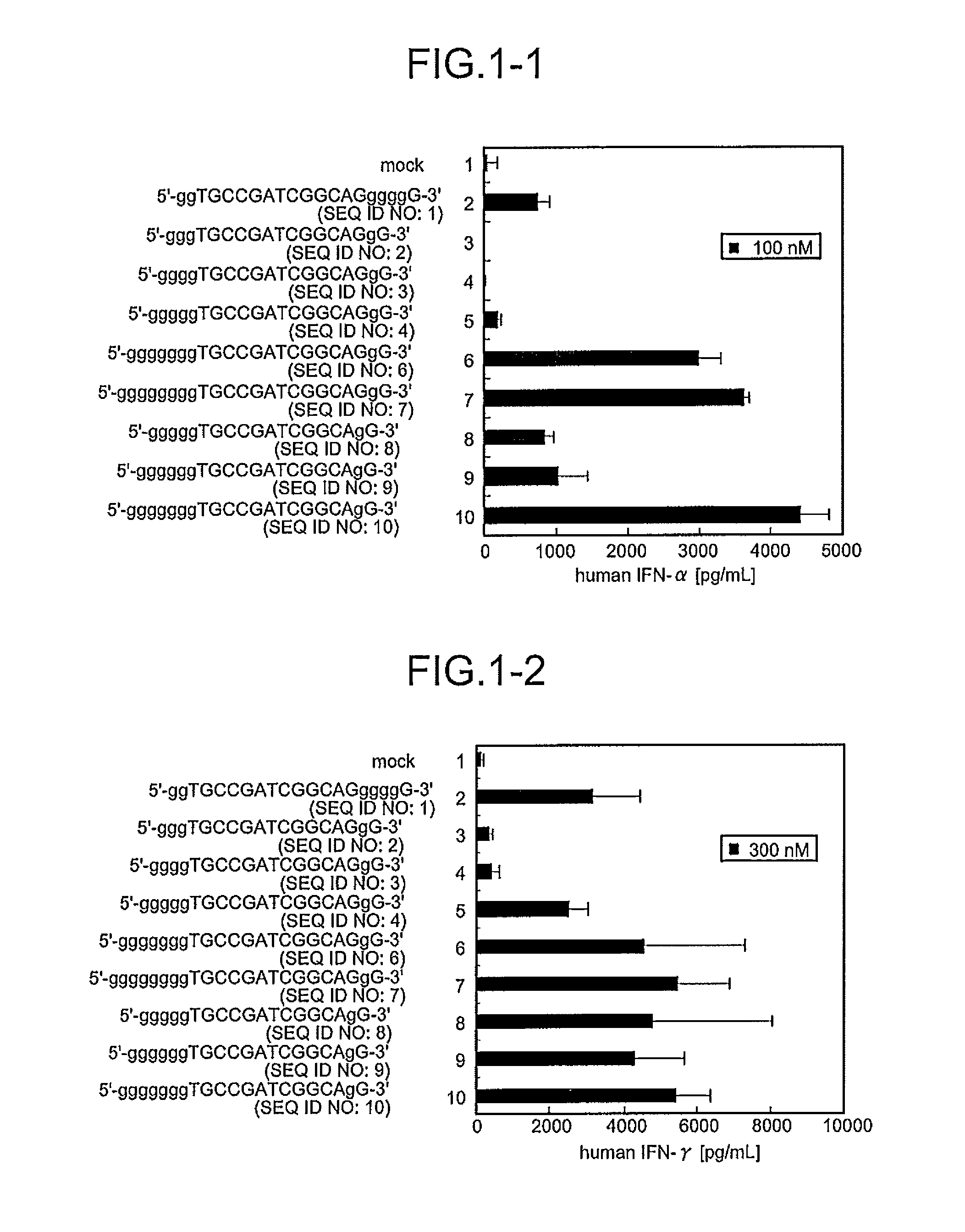
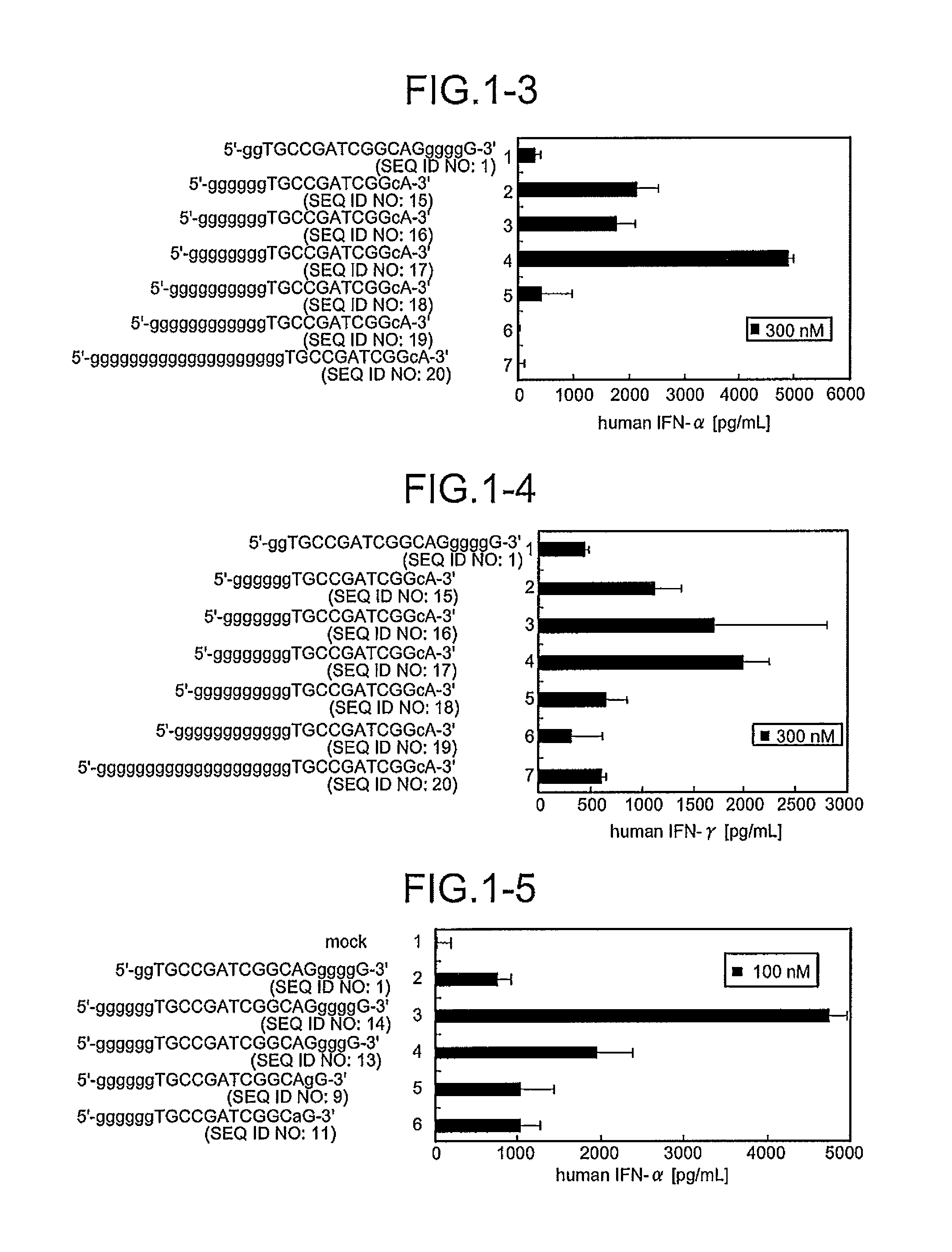
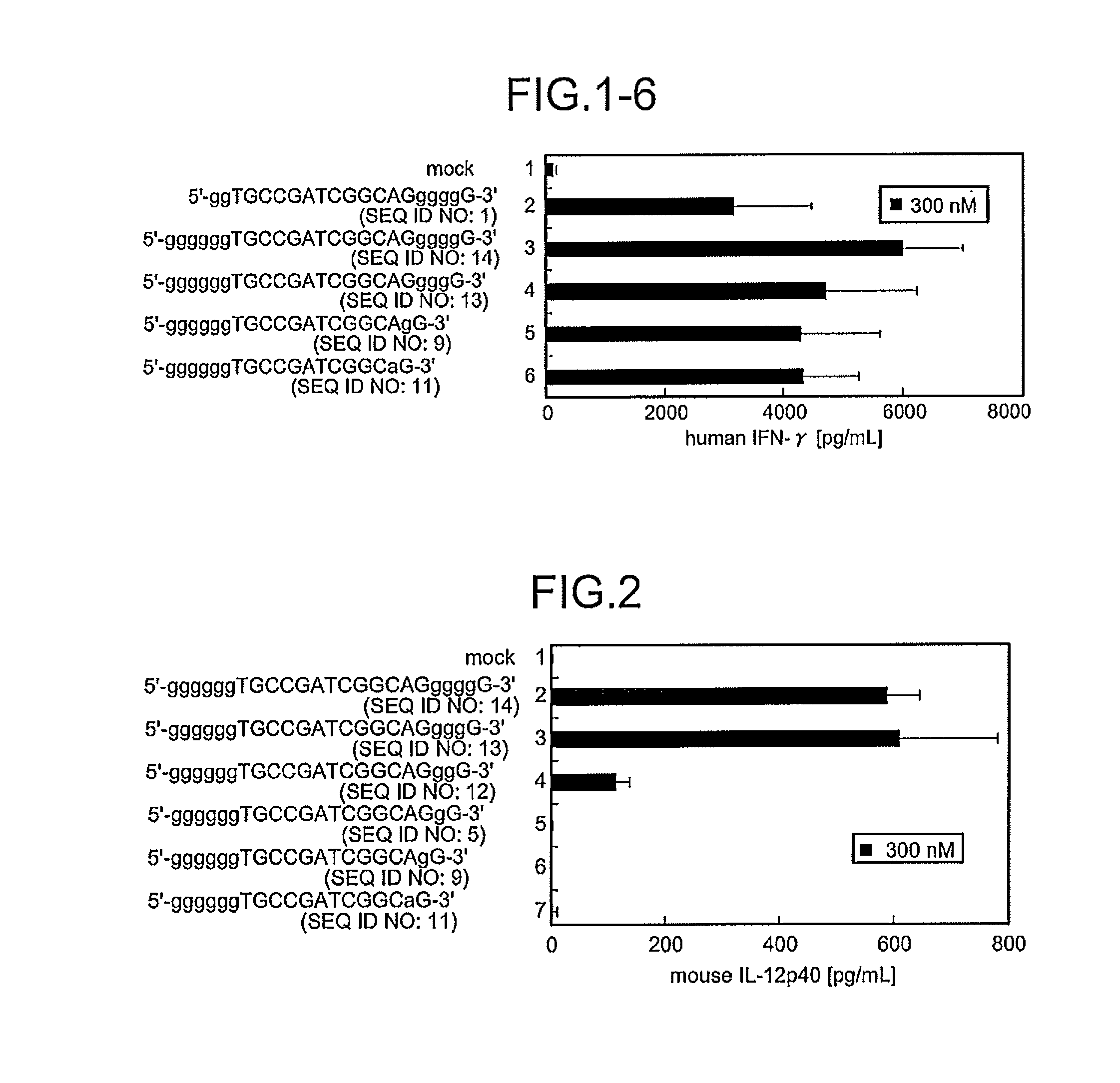
A novel immunostimulatory oligonucleotide by which an IFN-inducing activity is enhanced and an inflammatory cytokine-inducing activity is reduced, and a pharmaceutical containing the same, and an application thereof are provided. That is, the present invention provides the immunostimulatory oligonucleotide composed of a base sequence represented by a formula: 5′-(G)MPXCGYQ(G)N-3′ (X and Y are mutually independent and represent an arbitrary sequence which has a length of 0 to 10 nucleotides and does not contain 4 or more consecutive G residues, and a length of X+Y is 6 to 20 nucleotides; XCGY contains a palindrome sequence having a length of at least 8 nucleotides and has a length of 8 to 22 nucleotides; P and Q are mutually independent and represent one nucleotide other than G; M represents an integer of 6 to 10 and N represents an integer of 0 to 3) wherein a full length thereof is 16 to 37 nucleotides (except for an oligonucleotide composed of a basesequence represented by SEQ ID NO:5), the pharmaceutical application thereof.
Owner:TORAY IND INC +1
Quantitative analysis method aiming at DNA methylation monitoring
InactiveCN104062334ASimple preparationHigh sensitivityMaterial electrochemical variablesAdenosineA-DNA
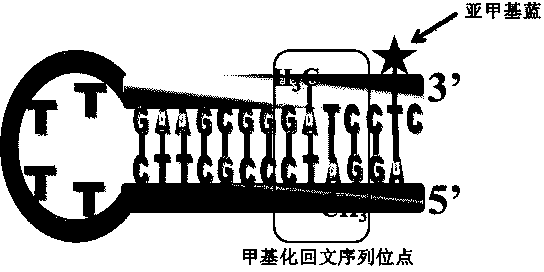
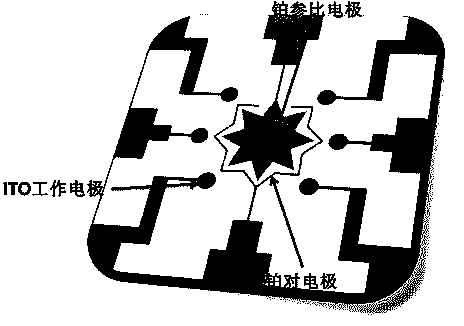
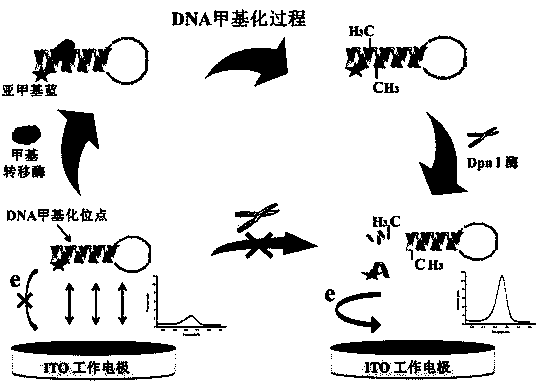
The invention discloses a quantitative analysis method aiming at DNA methylation monitoring. The quantitative analysis method comprises the following steps: (1) designing a hairpin type DNA secondary structure according to a DNA methylation palindromic sequence, and modifying a methylene blue electroactive material on the structure; (2) reacting hairpin type DNA with transmethylase and methylation adenosine to synthesize a methylated hairpin type DNA; (3) performing specific enzymolysis on a DNA methylation site through DpnI enzyme to release DNA fragments connected with methylene blue; (4) performing sample monitoring through an ITO micro-electrode chip with negative electricity, and acquiring electrochemical signals through a multi-pass electric signal reading device; (5) recording a monitoring result; and (6) adding methylation suppression chemicals into a DNA methylation process to screen the DNA methylation suppression chemicals. The quantitative analysis method has the advantages of simplicity in operation, portability, low cost and the like, and an electrode does not need complicated modification.
Owner:FUZHOU UNIV
Immunostimulatory oligonucleotides and use thereof in pharmaceuticals
InactiveUS8592566B2High therapeutic effectHigh activityOrganic active ingredientsSugar derivativesNucleotideCytokine
A novel immunostimulatory oligonucleotide by which an IFN-inducing activity is enhanced and an inflammatory cytokine-inducing activity is reduced, and a pharmaceutical containing the same, and an application thereof are provided. That is, the present invention provides the immunostimulatory oligonucleotide composed of a base sequence represented by a formula: 5′-(G)MPXCGYQ(G)N-3′ (SEQ ID NO: 118) (X and Y are mutually independent and represent an arbitrary sequence which has a length of 0 to 10 nucleotides and does not contain 4 or more consecutive G residues, and a length of X+Y is 6 to 20 nucleotides; XCGY contains a palindrome sequence having a length of at least 8 nucleotides and has a length of 8 to 22 nucleotides; P and Q are mutually independent and represent one nucleotide other than G; M represents an integer of 6 to 10 and N represents an integer of 0 to 3) wherein a full length thereof is 16 to 37 nucleotides (except for an oligonucleotide composed of a base sequence represented by SEQ ID NO:5), the pharmaceutical application thereof.
Owner:TORAY IND INC +1
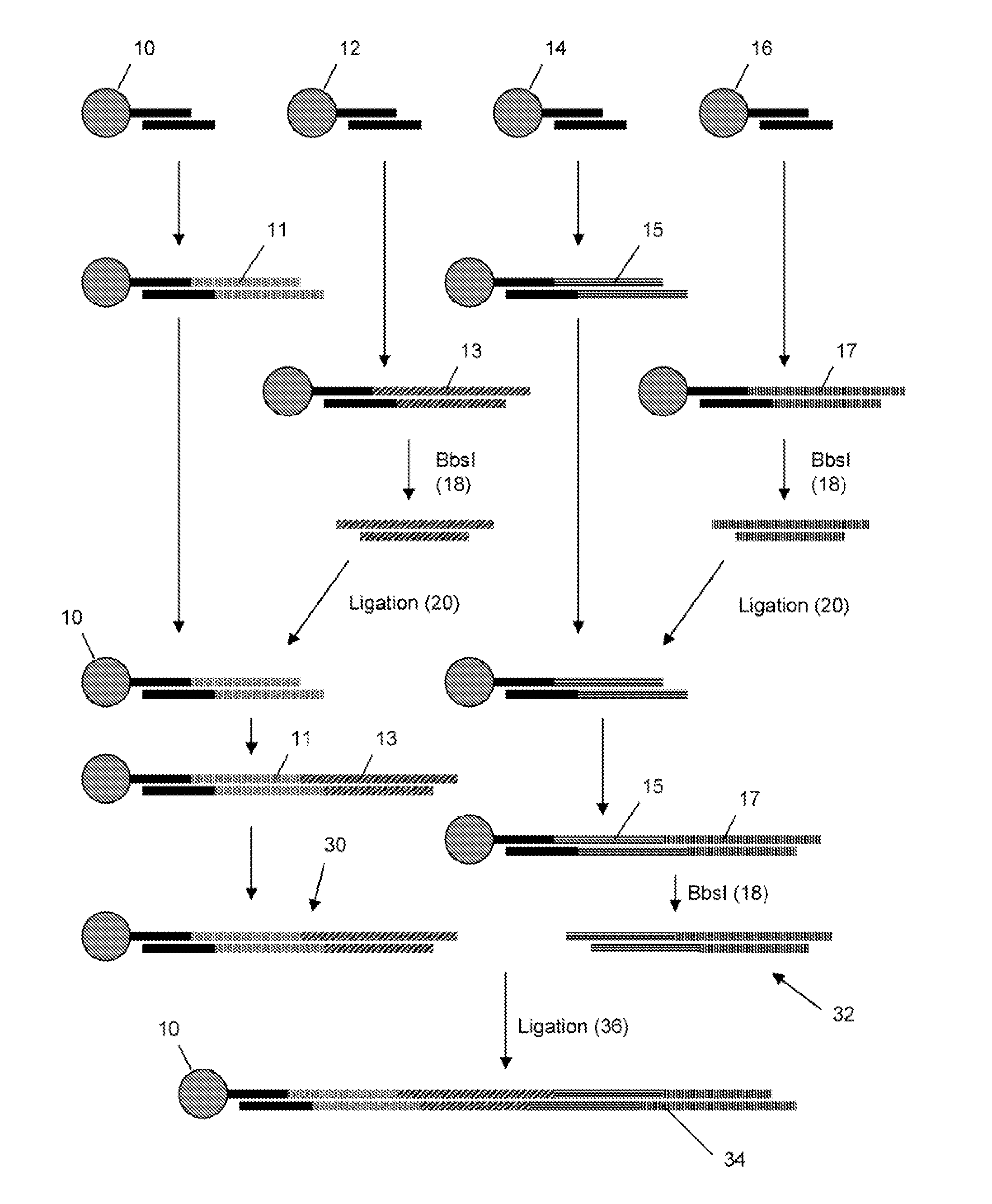
Gene synthesis by convergent assembly of oligonucleotide subsets
InactiveUS20110287490A1Facilitated releaseEasy to separateBioreactor/fermenter combinationsBiological substance pretreatmentsNucleotidePolynucleotide
The invention provides a system and method for synthesizing polynucleotides by solid phase assembly oligonucleotide precursors, in accordance with the method, a polynucleotide is partitioned into an ordered set of subunits, wherein each subunit is assembled in a single reaction from a subset of oligonucleotide precursors that uniquely anneal together to produce the subunit. The subunits are then assembled to form the desired polynucleotide. An important feature of the invention is the selection of subunits that are free of undesired sequence elements, such as palindromes, repetitive sequences, and the like, which would result in more than one subunit product alter ligating a pool of oligonucleotide precursors.
Owner:BRITISH COLUMBIA CANCER AGENCY BRANCH
Method and kit for constructing sequencing library
InactiveCN104313699ALow costIncrease flexibilityLibrary creationEnzyme digestionRecognition sequence
The invention provides a method and a kit for constructing a sequencing library. The method comprises the following steps: performing enzyme digestion on a genome DNA of a sample to be tested to obtain an enzyme digestion fragment with sticky terminals; adding P1 connectors at the two ends of the enzyme digestion fragment to obtain a fragment with the P1 connectors; fragmenting the fragment with the P1 connectors to obtain fragments with the target sizes; adding P2 connectors at the two ends of the fragments with the target sizes to obtain a sequencing library, wherein the genome DNA comprises a recognition sequence which can be recognized by an enzyme; the recognition sequence comprises a palindromic sequence consisting of basic groups at the two ends of the recognition sequence, and a variable sequence in the middle of the palindromic sequence; the variable sequence comprises a one or more basic groups; the sticky terminals comprise one or more basic groups in the variable sequence. Through enzyme digestion on the enzyme of the variable sequence in the recognition sequence, different capturing tag numbers can be achieved through the types and the number of the basic groups in the fixing connectors, so that the flexibility is improved.
Owner:天津诺禾致源生物信息科技有限公司
Asymmetrical hairpin probe and application thereof
InactiveCN102864214AOvercome the problem of large steric hindranceSolve pollutionMicrobiological testing/measurementMicroorganism based processesNucleotideBiology
The invention relates to an asymmetrical hairpin probe and an application thereof, and belongs to the biological detection field. The asymmetrical hairpin probe consists of a target nucleotide distinguished sequence and a palindromic sequence; under the condition of no target sequence, the asymmetrical hairpin probe is under the metastable hairpin structure; once a target sequence to be detected is added, chain-type polymerization reaction free from the participation of enzyme is triggered; the target nucleic acid distinguished sequence part of the asymmetrical hairpin probe is combined with a corresponding target nucleic acid locus; rest probes are pairwise renatured into parts of double-chain probe of which the two ends are both provided with the palindromic sequence; and during molecular recognition hybridization, a detection signal is substantially enhanced so as to quickly detect the target sequence to be detected. According to the chain reaction technology of the asymmetrical hairpin probe, the high specificity of molecular hybridization and high efficiency of chain type polymerization are artfully combined, and free energy released by DNA (deoxyribose nucleic acid) polymerization is used for driving molecular cascade amplification. The asymmetrical hairpin probe has the advantages of simple reaction system and high detection efficiency, and the laboratory pollution problem existing in the traditional technology, such as PCR (polymerase chain reaction) and the like, can be effectively avoided.
Owner:THE FIRST AFFILIATED HOSPITAL OF THIRD MILITARY MEDICAL UNIVERSITY OF PLA
Immunostimulatory oligonucleotides and uses thereof
Oligonucleotides containing the non-palindromic sequence motif: X1X2X3X4X5X6X7X8, wherein X1 is C,T,G or A (preferably T or C); wherein X2 is C,T,G or A; wherein X7 is C,T,G or A (preferably G); at least three, and preferably all, of X3, X4, X5, X6 and X8 are T; and with the proviso that, in the motif, a C does not precede a G (in other terms, the nucleic acid motif does not consist of a CpG oligonucleotide), that modulate the immune response of animals of the order Primate, including humans, are disclosed. This immune modulation is characterized by stimulation of proliferation, differentiation, cytokine production and antibody production on B-cells and cell differentiation on plasmacytoid dendritic cells.
Owner:DAVID HORN LLC
Method for cultivating high-TYLCV (tomato yellow leaf curl virus)-resistant tomatoes
ActiveCN104313051AShorten the breeding cycleGenetic engineeringFermentationTomato yellow leaf curl virusBiology
The invention discloses a method for cultivating a high-TYLCV (tomato yellow leaf curl virus)-resistant tomatoes. The method comprises the following steps: selecting fragments of AV1 gene, AC1 gene and AC3 gene in a TYLCV genome for gomphosis to obtain polygenes chimera AV1-AC1-AV3, and constructing an interference vector RNAi of a palindromic sequence; carrying out transient expression of dsRNA on tomato cotyledon by virtue of agrobacterium-mediated transformation to obtain a transgenic tomato for transforming the polygenes chimera AV1-AC1-AV3; and if the transgenic tomato shows TYLCV resistance after system infection of the TYLC, is not observed with obvious symptoms and has high-TYLCV resistance, screening out the high-TYLCV-resistant tomato. By utilizing the method, the high-TYLCV-resistant tomato strains can be quickly screened out and cultivated in 2 months, the cultivation period is greatly shortened, and a technical support is provided for preventing and curing TYLCV.
Owner:SHANXI AGRI UNIV
EMSA method, probe thereof and preparation method of probe
ActiveCN105506150AOvercoming developing errorsNo false negativesMicrobiological testing/measurementBiological testingSingle strandNucleotide sequencing
The invention discloses an EMSA method, a probe thereof and a preparation method of the probe. The EMSA probe comprises a first strip chain, and the first strip chain comprises a transcription factor combination sequence and joint sequences located at the 5' end and the 3' end of the transcription factor combination sequence. The preparation method of the EMSA probe comprises the following steps of synthesizing a template strand and a primer which is labeled through biotin and modified through LNA, wherein the template strand contains the transcription factor combination sequence and the joint sequences located at the 5' end and the 3' end of the transcription factor combination sequence, each joint sequence is a palindromic sequence, and the nucleotide sequence of the primer is the same as the nucleotide sequence of each joint sequence; adopting the primer for conducting PCR amplification with the template strand as the template and synthesizing the EMSA probe . The EMSA probe can be used for conducting EMSA. According to the EMSA method, the probe thereof and the preparation method of the probe, the double-stranded probe is prepared from a single-strained probe through PCR, false negativeness due to incomplete renaturation is avoided, and developing errors caused by the single-stranded free probe are further overcome.
Owner:GUANGZHOU BIOSENSE BIOSCI
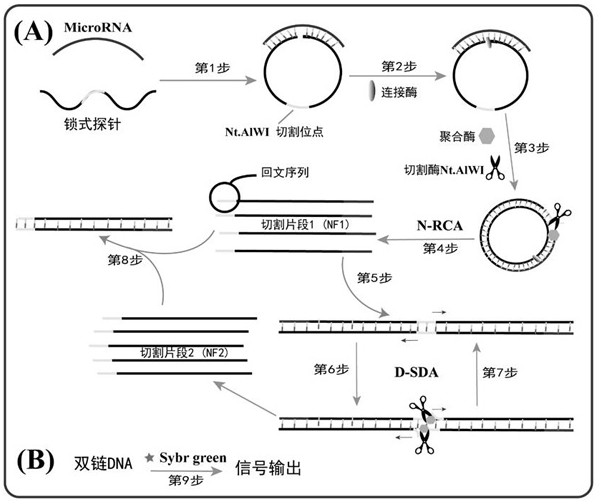
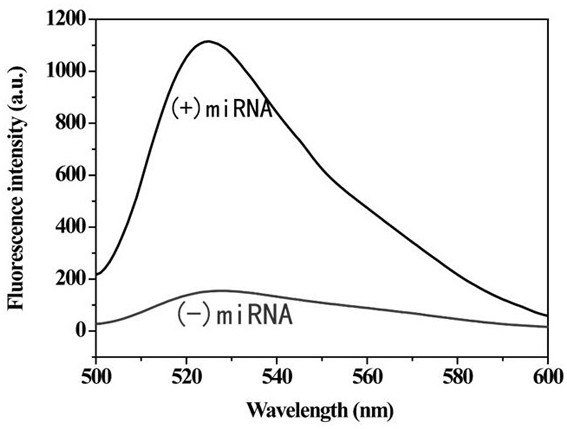
Method for detecting tumor biomarker by using palindrome lock-type probe
InactiveCN108642137AHigh sensitivityStrong specificityMicrobiological testing/measurementTumor BiomarkersBiomarker (petroleum)
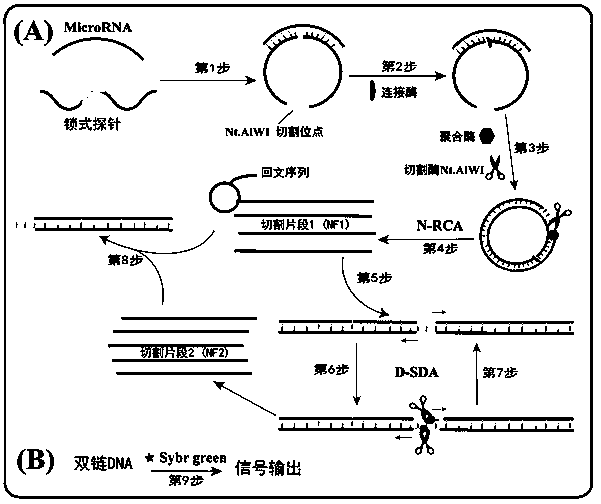
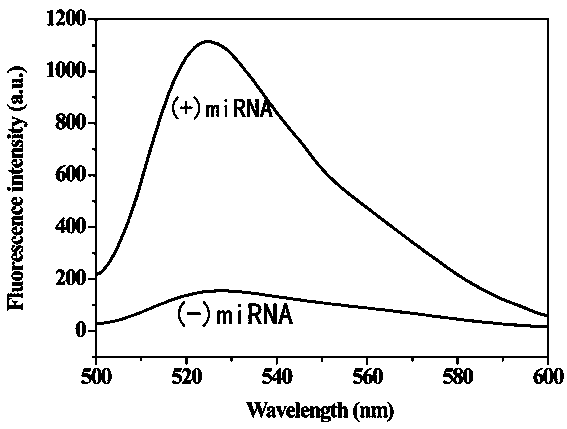
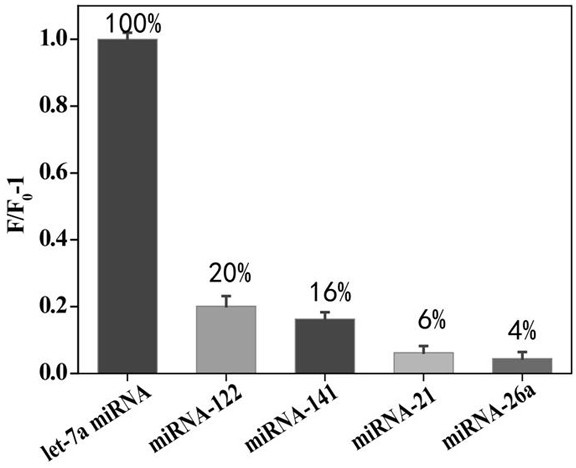
The invention provides a method for detecting a tumor biomarker by using a palindrome lock-type probe. A palindrome-sequence non-fluorescent mark lock-type RCA probe is designed. The probe is composedof a target identification part and a locus cutting part. The target identification part and target miRNA can completely complement each other and is located at a 3' end and a 5' end of the lock-typeprobe; a semi-identification locus of restriction enzyme Nt.AlWI is fused with a palindrome alkali segment. The probe is combined with nicking enzyme, an N-RCA concept is provided, a double-directionchain replacement reaction (D-SDA) is conducted for combination, and the probe is used for detecting the tumor marker miRNA. By means of the method, let-7a miRNA can be efficiently detected through amplification, and the probe is economical and practical and is expected to be applied to development of clinical diagnostic kits.
Owner:FUZHOU UNIV
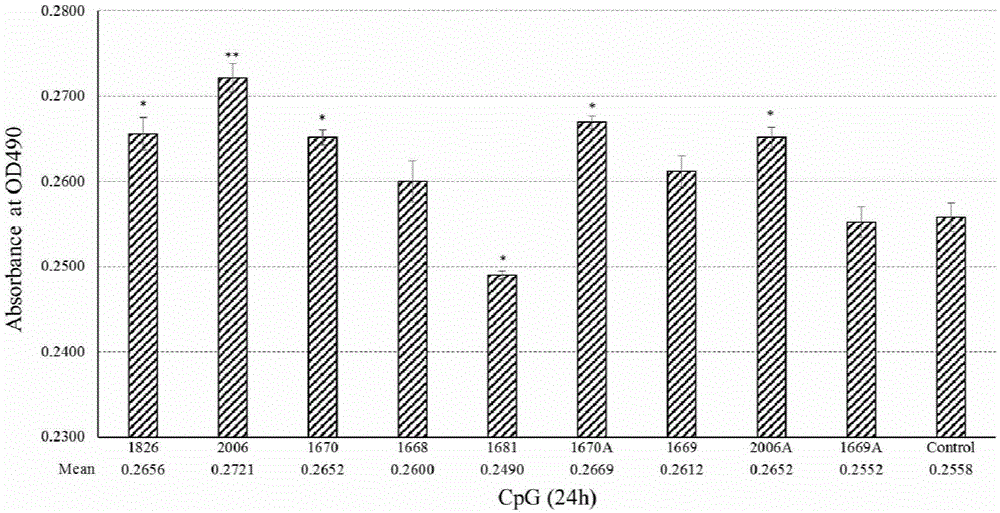
CpG ODN sequence with grass carp immune enhancement activity and use thereof
ActiveCN106256905AEnhance immune responseStrong immune responseOrganic active ingredientsGenetic material ingredientsAquatic animalNucleotide
The invention belongs to the field of genetic engineering of aquatic animals, and relates to a CpG ODN sequence with immune enhancing activity on grass carp and application thereof. Disclosed is a CpG ODN sequence that has immune enhancing activity on grass carp, the sequence is the nucleotide sequence shown in SEQ ID NO: 1, it contains the oligodeoxynucleotide sequence of the CpG motif, and the sequence contains three 'AACGTT' repeats, and a 'TCGA' palindromic sequence. Through preliminary screening of grass carp head kidney mononuclear cells and grass carp kidney cell lines, the enhancement effect of CpG on the immune activity of grass carp was verified, and the CpG with the strongest immune enhancement activity on grass carp cells was screened out through its activation effect on genes related to signaling pathways ODN sequence. Through the challenge test, it was verified again that the CpG ODN sequence has a strong immune enhancing effect. The sequence can be prepared into a grass carp immune enhancer, an immune adjuvant or a vaccine, and can be popularized and applied in the prevention of grass carp diseases.
Owner:HUAZHONG AGRI UNIV
Pseudo attP site in swine genome and application thereof
InactiveCN102796736ADoes not affect expressionDoes not affect normal physiological functionVector-based foreign material introductionForeign genetic material cellsBiotechnologyWild type
The invention discloses a pseudo attP site in the swine genome and applications thereof, belonging to the technical field of biotechnology and biomedical engineering. According to the invention, a pseudo attP site is isolated from the swine genome, and the pseudo attP site has a sequence similarity of 40% with the wild type attP site and has the characteristic of palindromic sequence. The pseudo attP site can be recognized by Streptomycete bacteriophage phi C 31 intergrase, and thereby exogenous genes having attP site can be mediated to integrate in the site in relatively high efficiency. According to protein content tests, the expression level of EGFP gene maintained by the pseudo attP site is 50 times that by the genome without exogenous genes. The invention provides a new technical scheme for targeted integration and high efficiency expression for swine transgene, and a new strategy for the research of swine functional genome, so the invention has important science value and broad application prospect.
Owner:INST OF ANIMAL SCI & VETERINARY HUBEI ACADEMY OF AGRI SCI
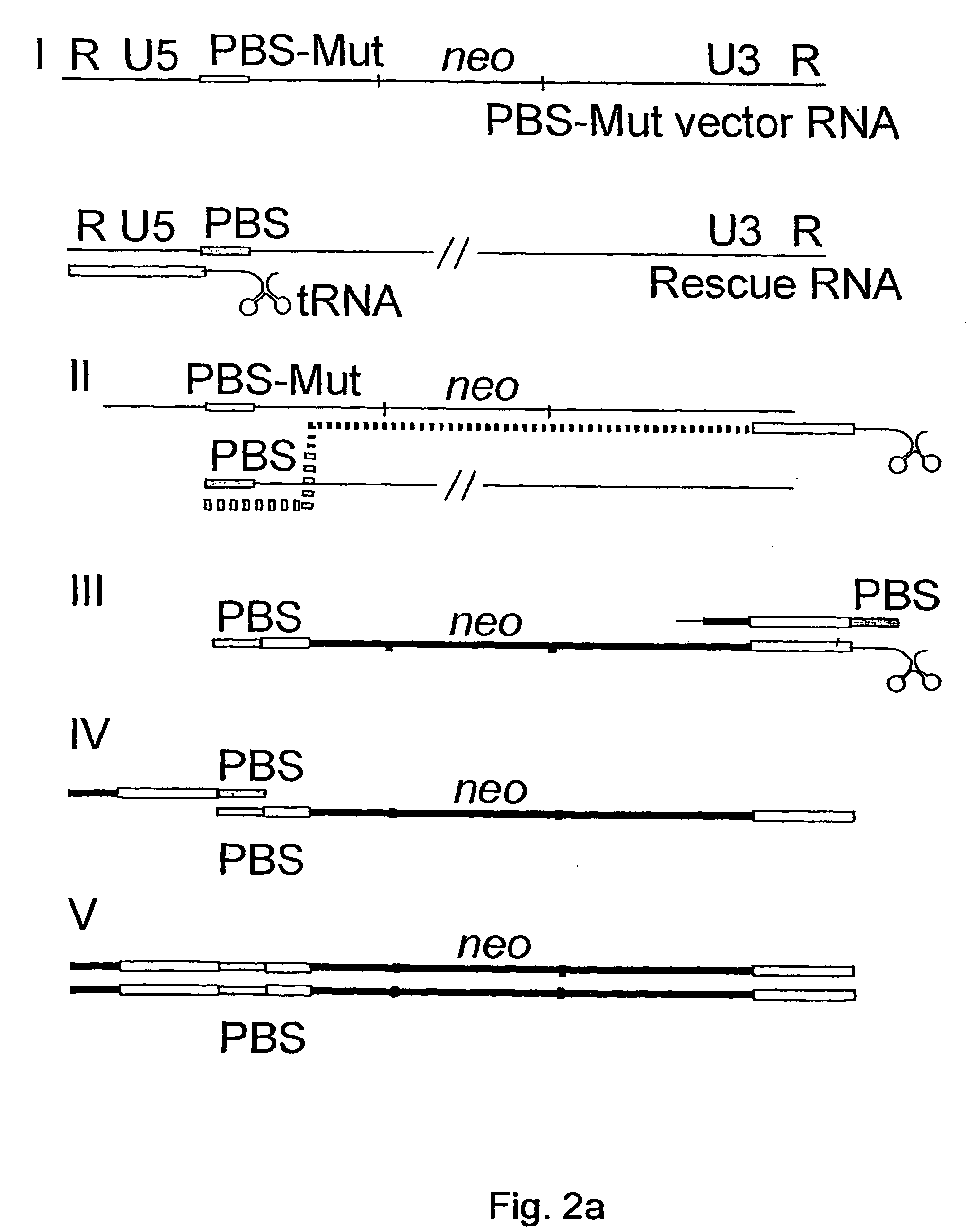
Vectors for gene therapy
InactiveUS20040248083A1Improve securityDecreasing frequency of recombinationFungiBacteriaDimerMedicine
The present invention relates to improved vectors useful in gene therapy which improvement particularly resides in improved safety of such vectors. The improvement is achieved by incorporating sequences into the gene therapy vector which promote dimer formation of transcripts derived from said vector. Such sequences are in a preferred embodiment self-complementary palindromic or nonpalindromic sequences.
Owner:AARHUS UNIV
PCR primer and application thereof in DNA fragment connection
ActiveCN111748611AImprove effective utilizationReduce Sequencing Redundant DataMicrobiological testing/measurementDNA preparationNucleotideDouble strand
The invention provides a PCR primer and an application thereof in DNA fragment connection. The PCR primer comprises a palindrome sequence segment and any PCR primer sequence segment, the 3'end of thepalindrome sequence segment is connected with the 5'end of the any PCR primer sequence segment, the nucleotide of the 3' end of the palindrome sequence segment is U, and the any PCR primer sequence segment is complementarily paired with the 3'end of a pre-amplified DNA template. The primer provided by the embodiment of the invention can be used for constructing a sequencing library; in the construction process, the primer provided by the embodiment of the invention is used as a PCR amplification primer based on DNA, the two ends of an obtained amplification product DNA double strand have a palindrome sequence structure, and the 3'end of the palindrome sequence structure has nucleotide U, so that a foundation is laid for subsequent interconnection of different amplification product double strand DNAs.
Owner:BGI TECH SOLUTIONS
Repetitive extragenic palindromic sequence for improving exogenous gene mRNA and application thereof
ActiveCN108531496AHigh expressionAlter the secondary structureVectorsBacteriaEscherichia coliProtein target
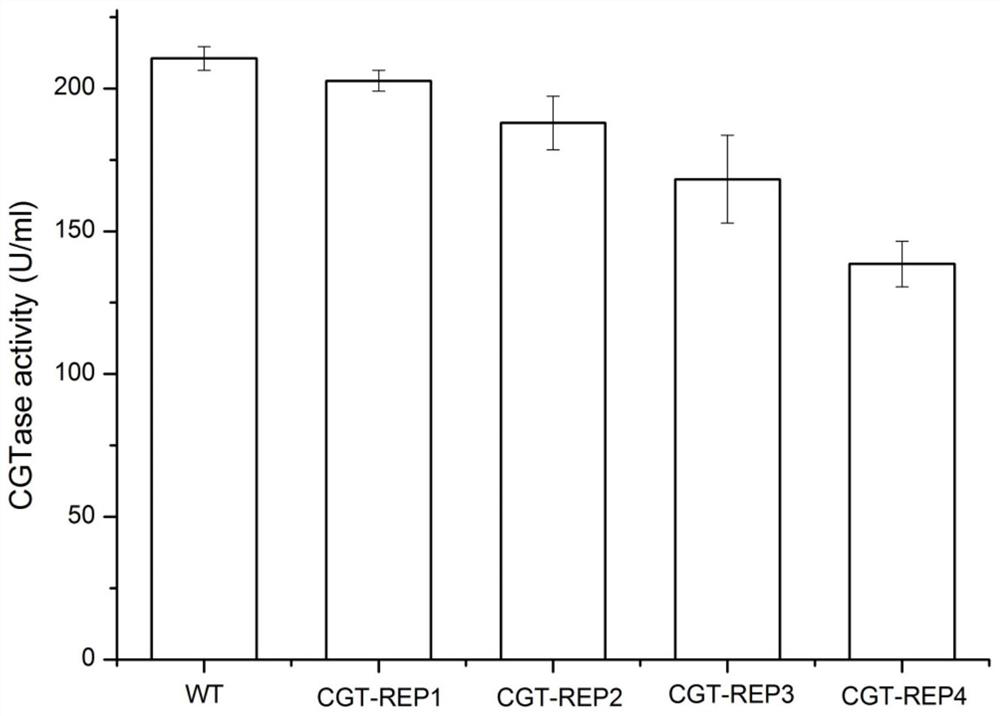
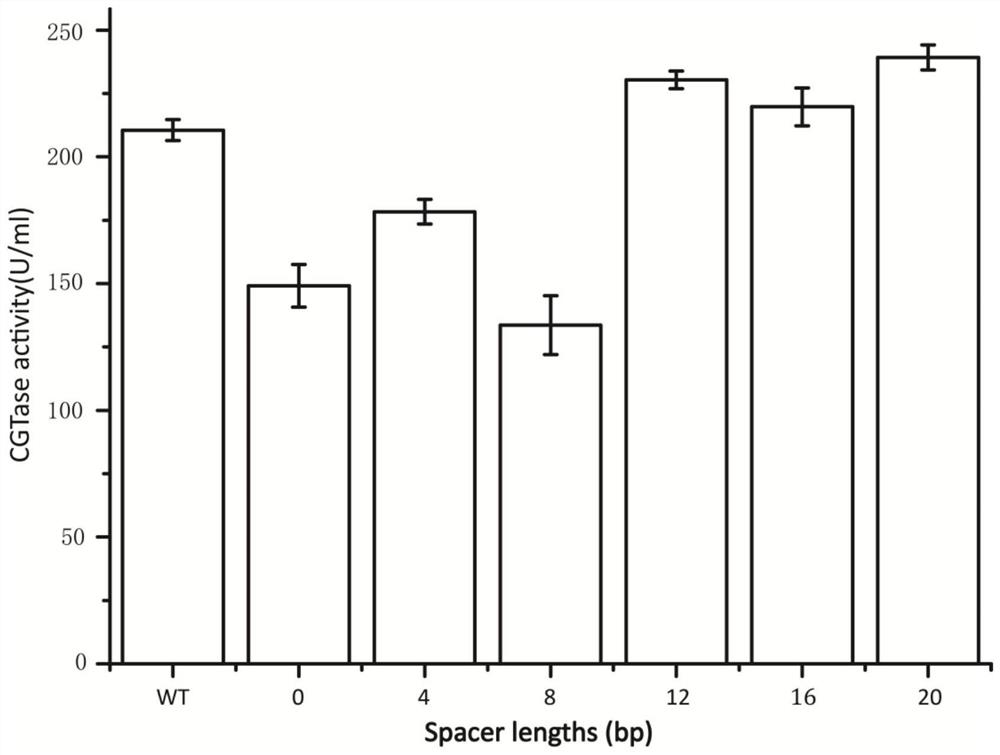
Belonging to the technical field of genetic engineering, the invention discloses a repetitive extragenic palindromic sequence for improving exogenous gene mRNA and application thereof. The repetitiveextragenic palindromic sequence and a spacer region sequence thereof are shown as SEQ ID NO, 1, the signal peptide can improve the stability of mRNA by changing the secondary structure, so that the extracellular enzyme activity of the target protein cyclodextrin glycosyltransferase can be improved by 14%, the recombinant escherichia coli modified by the signal peptide has strengthened extracellular protein production capacity, therefore the repetitive extragenic palindromic sequence can be used for escherichia coli to express exogenous protein.
Owner:JIANGNAN UNIV
Gene synthesis by convergent assembly of oligonucleotide subsets
InactiveUS9068209B2Facilitated releaseEasy to separateBioreactor/fermenter combinationsBiological substance pretreatmentsNucleotidePolynucleotide
The invention provides a system and method for synthesizing polynucleotides by solid phase assembly oligonucleotide precursors, in accordance with the method, a polynucleotide is partitioned into an ordered set of subunits, wherein each subunit is assembled in a single reaction from a subset of oligonucleotide precursors that uniquely anneal together to produce the subunit. The subunits are then assembled to form the desired polynucleotide. An important feature of the invention is the selection of subunits that are free of undesired sequence elements, such as palindromes, repetitive sequences, and the like, which would result in more than one subunit product alter ligating a pool of oligonucleotide precursors.
Owner:BRITISH COLUMBIA CANCER AGENCY BRANCH
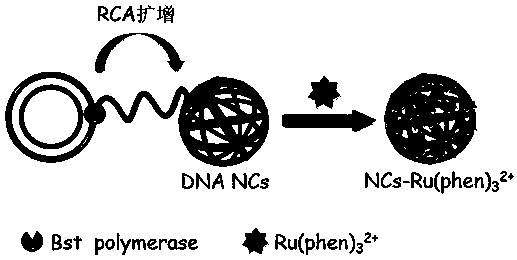
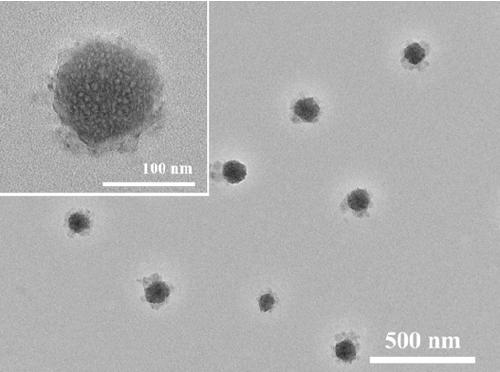
Preparation method of DNA nanowire cluster loaded with phenanthroline ruthenium
InactiveCN110295219AThe synthesis method is simpleEasy to controlMicrobiological testing/measurementNanowireRuthenium
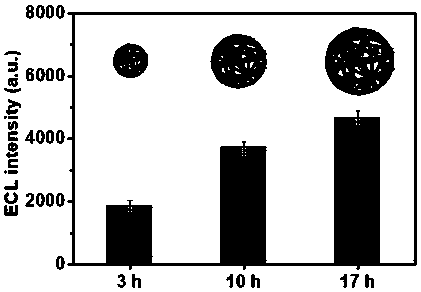
The invention discloses a preparation method of a DNA nanowire cluster loaded with phenanthroline ruthenium. The preparation method comprises the following steps that 1, a rolling circle amplification(RCA) reaction template chain coded by a palindromic sequence and a corresponding primer chain are designed; 2, the RCA template chain is subjected to connection cyclization by using a DNA ligase; 3,an RCA amplification reaction is started, and products of the RCA amplification reaction are self-assembled into the three-dimensional spherical DNA nanowire cluster (DNA NCs); 4, the phenanthrolineruthenium (Ru(phen)32+) is subjected to high-efficiency loading into a double-chain structure of the DNA NCs to prepare an ECL beacon molecule. The method has the advantages of simplicity in operation, low cost and good reproducibility. The three-dimensional spherical DNA NCs can be used for high-efficiency loading of Ru(phen) 32+ as the ECL beacon molecule, and signal amplification is realized; meanwhile, the stability is high, the biocompatibility is good, and the method is expected to be widely applied to the fields of life science, medical clinical detection and the like.
Owner:FUZHOU UNIV
A kind of dna for increasing the amount of foreign gene mRNA and its application
ActiveCN108531496BHigh expressionAlter the secondary structureVectorsBacteriaEscherichia coliProtein target
Belonging to the technical field of genetic engineering, the invention discloses a repetitive extragenic palindromic sequence for improving exogenous gene mRNA and application thereof. The repetitiveextragenic palindromic sequence and a spacer region sequence thereof are shown as SEQ ID NO, 1, the signal peptide can improve the stability of mRNA by changing the secondary structure, so that the extracellular enzyme activity of the target protein cyclodextrin glycosyltransferase can be improved by 14%, the recombinant escherichia coli modified by the signal peptide has strengthened extracellular protein production capacity, therefore the repetitive extragenic palindromic sequence can be used for escherichia coli to express exogenous protein.
Owner:JIANGNAN UNIV
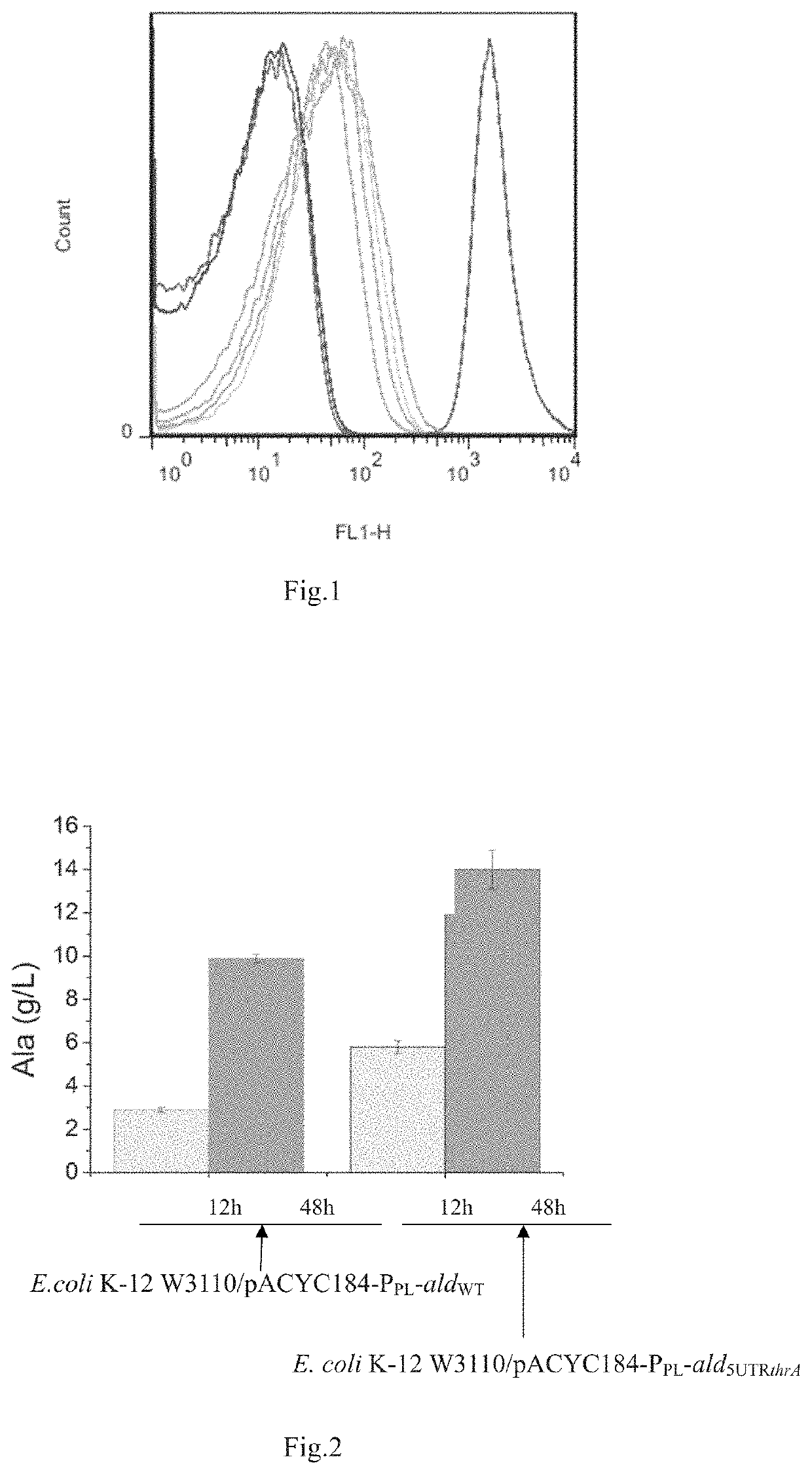
Method for modifying amino acid attenuator and use of same in production
ActiveUS11492616B2Improve expression levelIncrease productionFermentationVector-based foreign material introductionTrp operonThreonine
The present invention discloses a method for modifying an amino acid attenuator, a class of amino acid attenuator mutants, engineered bacteria created on the basis of the amino acid attenuator mutants, and use of the engineered bacteria. The present invention protects a method for relieving the attenuation regulation of an amino acid operon gene, which is modification of the amino acid operon gene by: removing a gene coding for a leader peptide and an anterior reverse complementary palindromic sequence in the terminator stem-loop structure, and maintaining a posterior reverse complementary palindromic sequence in the terminator. The amino acid operon particularly can be histidine operon, tryptophan operon, phenylalanine operon, alanine operon, threonine operon and etc. The present invention can be used for the production of amino acids and derivatives thereof in fermentation by bacteria, providing a novel method for improving the production of amino acids in fermentation.
Owner:INST OF MICROBIOLOGY - CHINESE ACAD OF SCI
pcr primers and their application in ligation of dna fragments
ActiveCN111748611BImprove effective utilizationReduce Sequencing Redundant DataMicrobiological testing/measurementDNA preparationNucleotideDouble strand
The invention provides a PCR primer and an application thereof in DNA fragment connection. The PCR primer comprises a palindrome sequence segment and any PCR primer sequence segment, the 3'end of thepalindrome sequence segment is connected with the 5'end of the any PCR primer sequence segment, the nucleotide of the 3' end of the palindrome sequence segment is U, and the any PCR primer sequence segment is complementarily paired with the 3'end of a pre-amplified DNA template. The primer provided by the embodiment of the invention can be used for constructing a sequencing library; in the construction process, the primer provided by the embodiment of the invention is used as a PCR amplification primer based on DNA, the two ends of an obtained amplification product DNA double strand have a palindrome sequence structure, and the 3'end of the palindrome sequence structure has nucleotide U, so that a foundation is laid for subsequent interconnection of different amplification product double strand DNAs.
Owner:BGI TECH SOLUTIONS
Method for cultivating transgenic rice based on RNAi binary vector
InactiveCN107446948AHigh suppression efficiencyObvious characteristics of salt-alkali resistanceVector-based foreign material introductionAngiosperms/flowering plantsGenetically modified riceTransgenic lines
The invention relates to a method for cultivating transgenic rice based on an RNAi binary vector, and belongs to the technical field of rice genetic engineering. Selection and cloning of target fragments, construction of palindromic sequences of target fragments, selection of cloning systems and construction of RNAi recombinant plasmids, Agrobacterium transformation of RNAi recombinant plasmids, Agrobacterium-mediated transformation and rice tissue culture, detection of transgenic rice, transgenic Stress resistance analysis of plants. The invention utilizes RNAi technology to effectively reduce the expression of rice Csn5 gene, can be used to breed rice transgenic strains with certain resistance, and provides technical support for crop variety improvement and application in agricultural molecular breeding.
Owner:JILIN BUSINESS & TECH COLLEGE
DNA molecular weight standard fragment amplification single-chain primer, amplification method and DNA molecular weight standard preparation method
ActiveCN113249438AAvoid the problem of easy template residueEfficient purificationMicrobiological testing/measurementDNA/RNA fragmentationSingle strandA-DNA
Owner:北京擎科生物科技股份有限公司
Nucleic acid aptamers targeting lymphocyte activation gene 3 (lag-3) and uses thereof
PendingUS20210115447A1Improve anti-tumor activityHigh affinityImmunological disordersAntineoplastic agentsAptamerNucleotide
Nucleic acid aptamers capable of binding to lymphocyte activation gene 3 (LAG-3) and uses thereof for modulating immune responses. Such aptamers may comprise a G-rich motif, for example, GX1GGGX2GGTX3A (SEQ ID No: 1), in which each of X1 and X2 are independently G, C, or absent, and X3 is T or C, or L-(G)n-L′, in which n is an integer of 5-9 inclusive, and L and L′ are nucleotide segments having complementary sequences. Also provided herein are multimeric nucleic acid aptamers containing a backbone moiety, which comprises a palindromic sequence.
Owner:ONENESS BIOTECH
A method for detecting tumor biomarkers using palindromic lock probes
InactiveCN108642137BHigh sensitivityStrong specificityMicrobiological testing/measurementEnzyme bindingDouble strand
Owner:FUZHOU UNIV
Nucleic acid aptamers targeting lymphocyte activation gene 3 (lag-3) and uses thereof
Nucleic acid aptamers capable of binding to lymphocyte activation gene 3 (LAG-3) and uses thereof for modulating immune responses. Such aptamers may comprise a G-rich motif, for example, GX1GGGX2GGTX3A (SEQ ID No: 1), in which each of X1 and X2 are independently G, C, or absent, and X3 is T or C, or L- (G)n-L', in which n is an integer of 5-9 inclusive, and L and L'are nucleotide segments having complementary sequences. Also provided herein are multimeric nucleic acid aptamers containing a backbone moiety, which comprises a palindromic sequence.
Owner:ONENESS BIOTECH
Quality control method based on pacbio full-length transcriptome sequencing data
ActiveCN109817277BReduce the ratioImprove accuracyProteomicsGenomicsTranscriptome SequencingData mining
The present invention provides a quality control method based on PacBio full-length transcriptome sequencing data, including steps: 1) using the IsoSeq analysis process to obtain high-quality and low-quality consistent full-length sequences from the original PacBio full-length transcriptome sequencing data; 2) Correct low-quality consensus full-length sequences based on Illumina sequencing data, and filter sequences that still fail to meet high-quality standards; 3) Merge high-quality and low-quality consistent full-length sequences that meet the conditions after correction, and filter according to the following criteria : Remove overly long sequences generated by sequence chimerism; remove consensus full-length sequences that have palindromic sequences in their own alignment results; remove sequences that can be aligned to multiple positions by other consistent full-length sequences. The chimeric sequences that may exist in the consensus full-length sequence are filtered through multiple criteria, reducing the proportion of false positive results in the final transcriptome and improving the accuracy of subsequent transcriptome-related analysis results.
Owner:BIOMARKER TECH
Nanoparticle based on cisplatin and DNA coordination self-assembly, and preparation method and application of nanoparticle
ActiveCN111514163AImprove solubilityGood biocompatibilityInorganic active ingredientsPharmaceutical non-active ingredientsBiocompatibilityDrug loading dose
The invention provides a nanoparticle based on cisplatin and DNA coordination self-assembly, and a preparation method and application thereof. The preparation method comprises the steps: dissolving cisplatin with normal saline under a high-temperature condition to prepare a cisplatin solution; then mixing DNA palindromic sequences with the cisplatin solution, and conducting a constant-temperaturereaction in a PCR instrument; and after annealing, supplementing an equal amount of DNA palindromic sequences, and continuously carrying out the constant-temperature reaction in the PCR instrument, soas to obtain nanoparticles with uniform shape and size. The DNA palindromic sequence is non-toxic and has good biocompatibility, self-crosslinking growth can occur through sequence design, and a hydrogel layer can be formed on the shell of the nanoparticle to improve the stability of the nanoparticle; meanwhile, cisplatin can be coated at the core of the nanoparticle by the DNA palindromic sequences to finally form the nanoparticle which is good in water solubility and relatively stable; the nanoparticle is simple in production process, environment-friendly in production process and relatively high in drug loading capacity, and has a wide application prospect in the field of drug delivery.
Owner:SHANGHAI JIAO TONG UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com