Quantitative analysis method aiming at DNA methylation monitoring
A quantitative analysis and methylation technology, applied in the direction of material electrochemical variables, etc., can solve the problems of large consumption of monomer samples and complicated detection methods, and achieve low cost, reduced time-consuming experiments, and low consumption Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1 Fabrication of ITO Microelectrode Chips
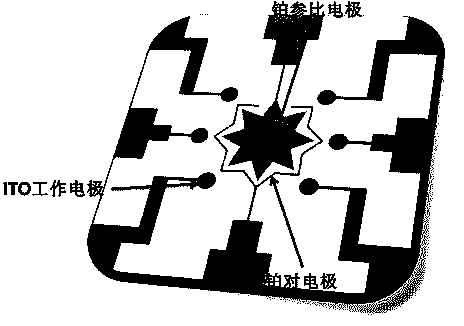
[0028] Use CorelDraw12 software to draw the two-dimensional structure of the chip and make a mask. Use soft lithography technology to transfer the pattern to the ITO glass plate, develop and remove the exposure layer, and then place the chip in the acid etching solution (HCl:H2O:NHO3=50:50:3) for wet etching for 45 minutes, namely The ITO working electrode structure of the chip can be obtained, and the remaining photoresist is removed. Photoetching the reference electrode and counter electrode pattern on the ITO glass plate again, after developing and removing the photoresist of the exposure layer, metal sputtering and sputtering metal platinum layer, the platinum reference electrode and counter electrode structure of the chip can be obtained. And remove the remaining photoresist. Rinse the microchip with secondary water and blow dry with nitrogen. Soak the prepared ITO microchip in Alconox cleaning solution (10 ...
Embodiment 2
[0029] Example 2 Monitoring of DNA methylation process
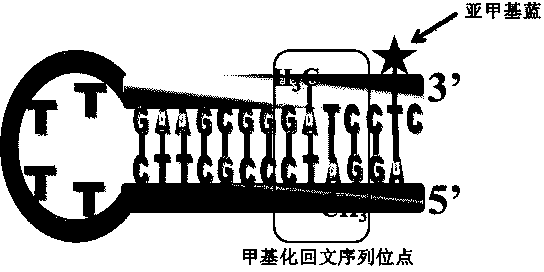
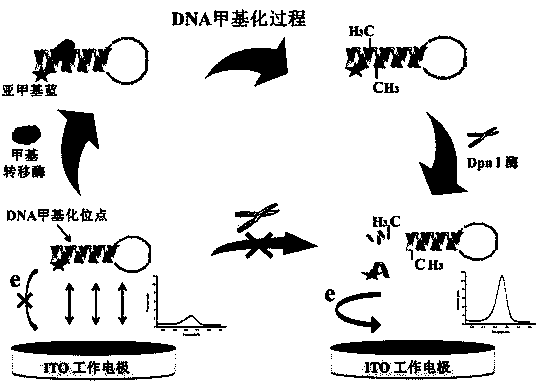
[0030] with 10 mM Tris-HCl buffer (containing 10 mM MgCl 2 , 50 mM NaCl, and pH 7.5) as a solvent mix 1 μM hairpin DNA, 160 μM SAM (methylated adenosine), 1 mM DTT (dithiothreitol), 50 U / mL Dam MTase (methyltransferase) and 20 U / mL of Dpn I enzyme. React at a constant temperature of 37°C, take out 2μL samples every 10 minutes and drop them on the working unit of the ITO microelectrode chip, use the electrochemical workstation to collect the DPV differential pulse electrical signal, the level of the DPV electrical signal indicates DNA methyl The level of DNA methylation can be monitored in real time. image 3 It is a schematic diagram of the principle of the mixed reaction, that is, the hairpin DNA will first co-react with SAM and Dam MTase to convert into methylated hairpin DNA, and the generated DNA methylation site will be cleaved by Dpn I enzyme , so that the fragmented nucleotide chain with methylene blue is close...
Embodiment 3
[0031] Example 3 Screening of DNA Methylation Inhibiting Drugs
[0032] with 10 mM Tris-HCl buffer (containing 10 mM MgCl 2, 50 mM NaCl, and pH 7.5) as a solvent mix 1 μM hairpin DNA, 160 μM SAM (methylated adenosine), 1 mM DTT (dithiothreitol), 50 U / mL Dam MTase (methyltransferase) and drug candidates. React in a water bath at 37°C for 2 hours to complete DNA methylation. Then add (20 U / mL) Dpn I enzyme into the sample and react for another 2 hours (37°C). Finally, 2 μL of sample was taken out and dropped onto the working unit of the ITO microelectrode chip, and the electrochemical workstation was used to collect the DPV differential pulse electrical signal. If the DPV electrical signal is strong, it means that the inhibitory effect of the drug on DNA methylation is not obvious; otherwise, better DNA methylation inhibitory drugs can be screened. According to the designed protocol, DNA probes (sequence: 5'-AG GATC CCGC TTCT TTTG AAGC GGGA TCCTC-3') were used to detect amox...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com