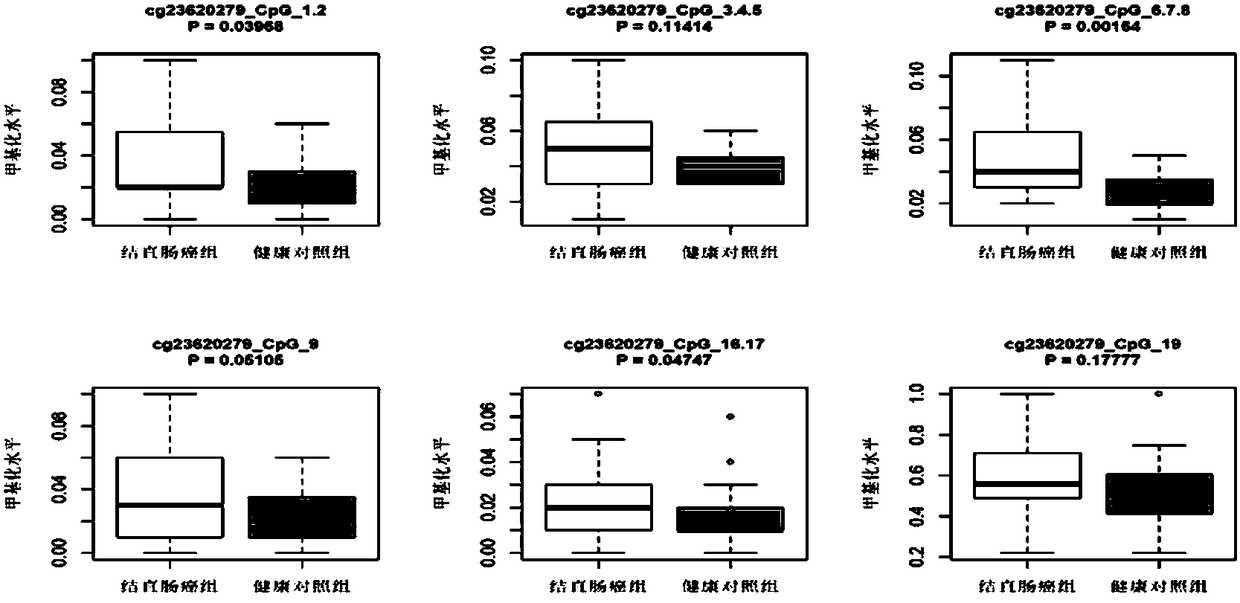
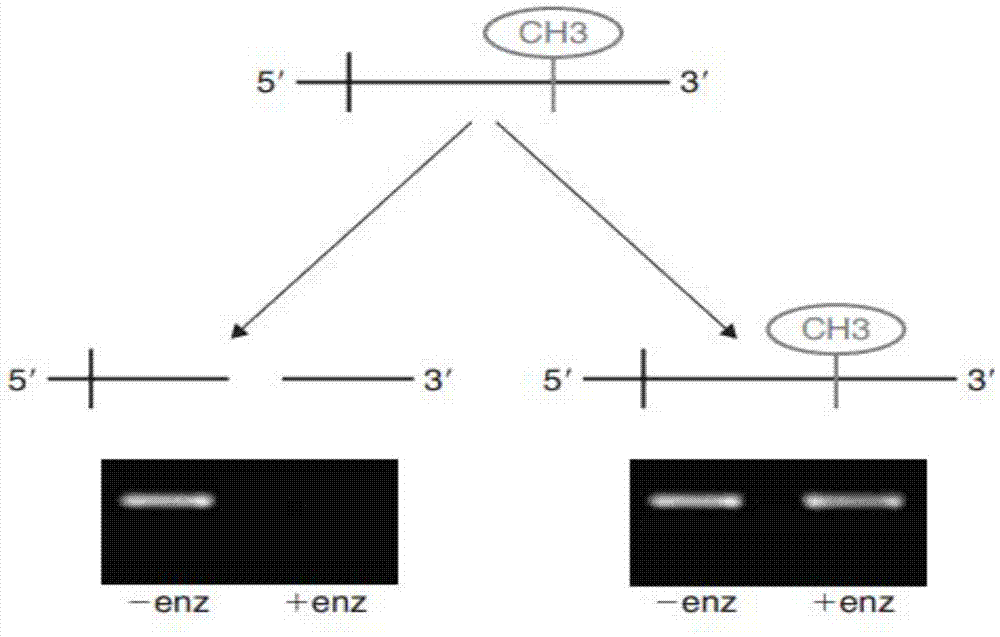
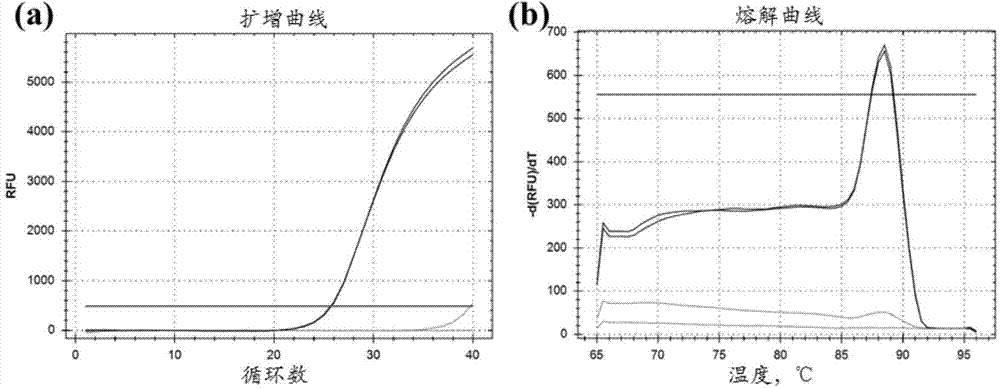
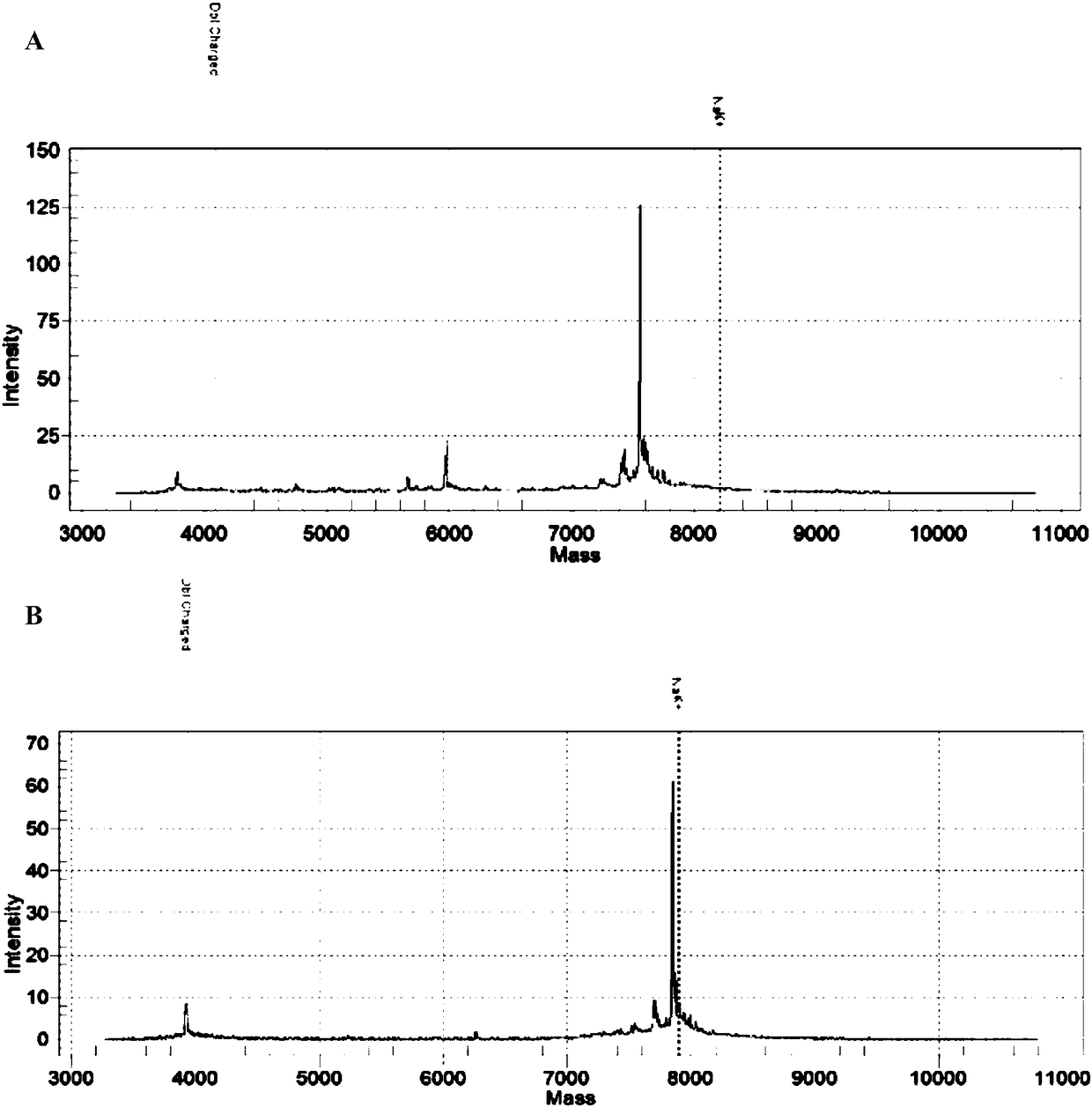
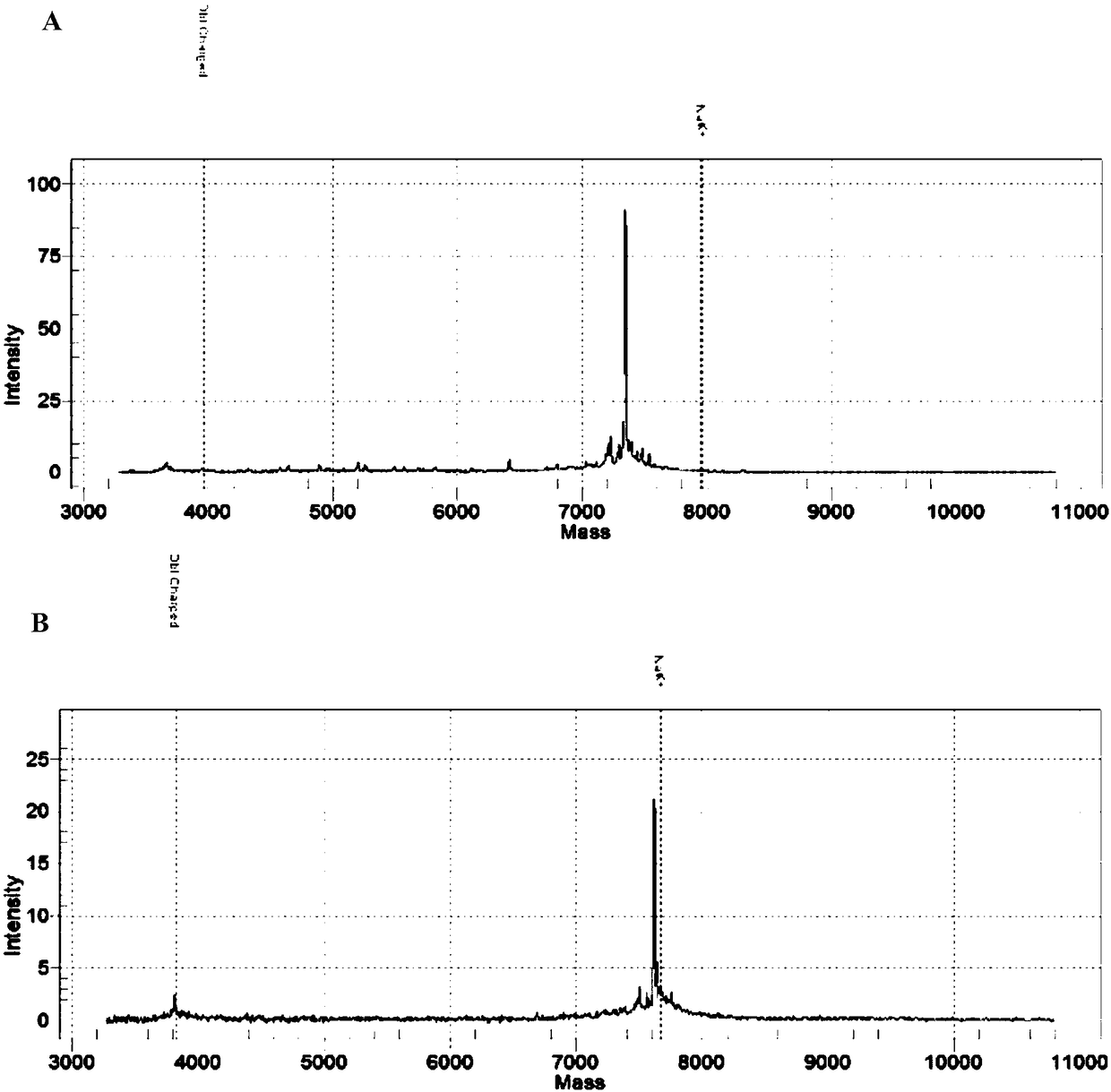
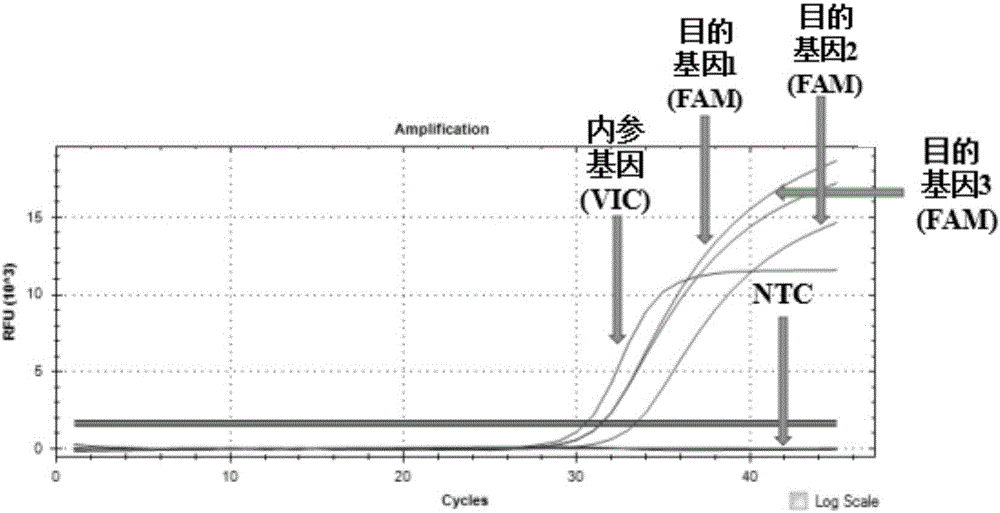
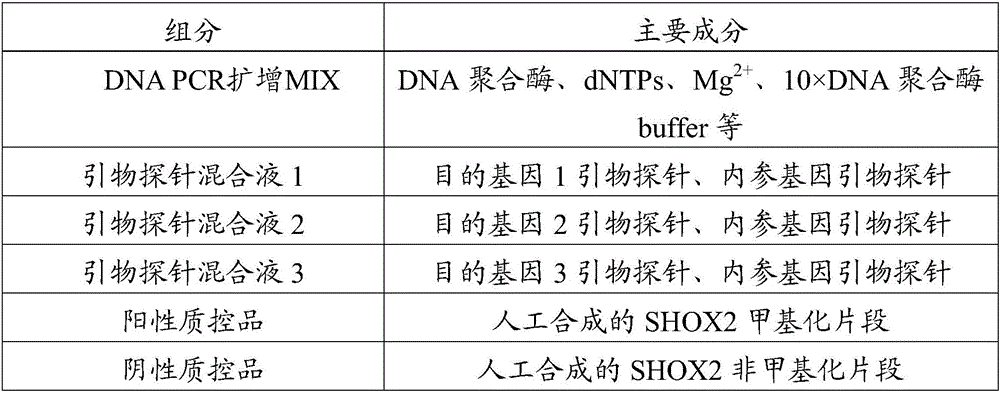
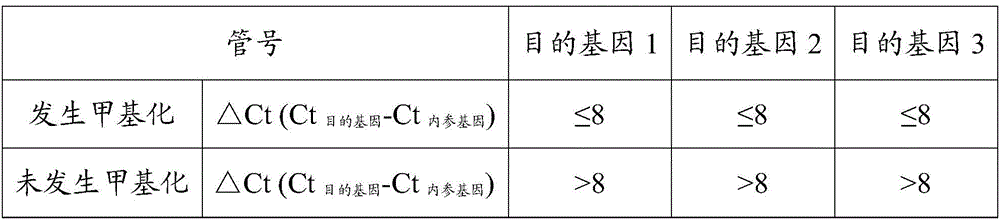
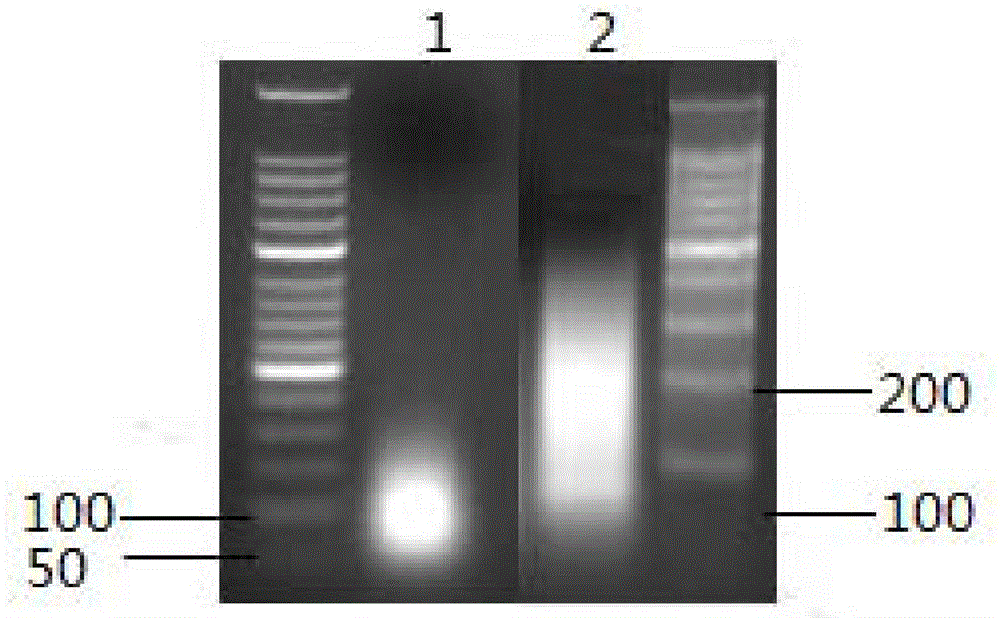
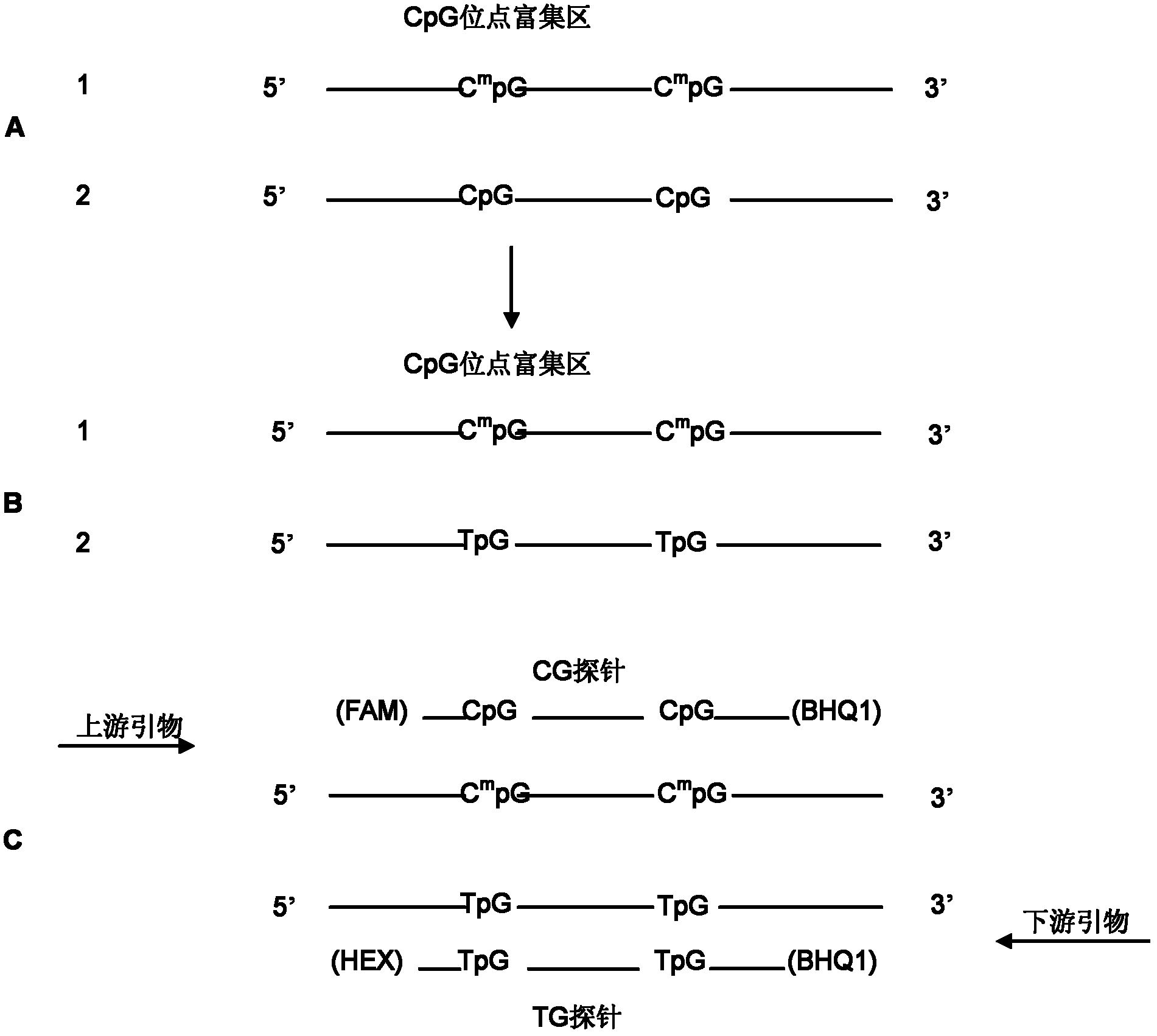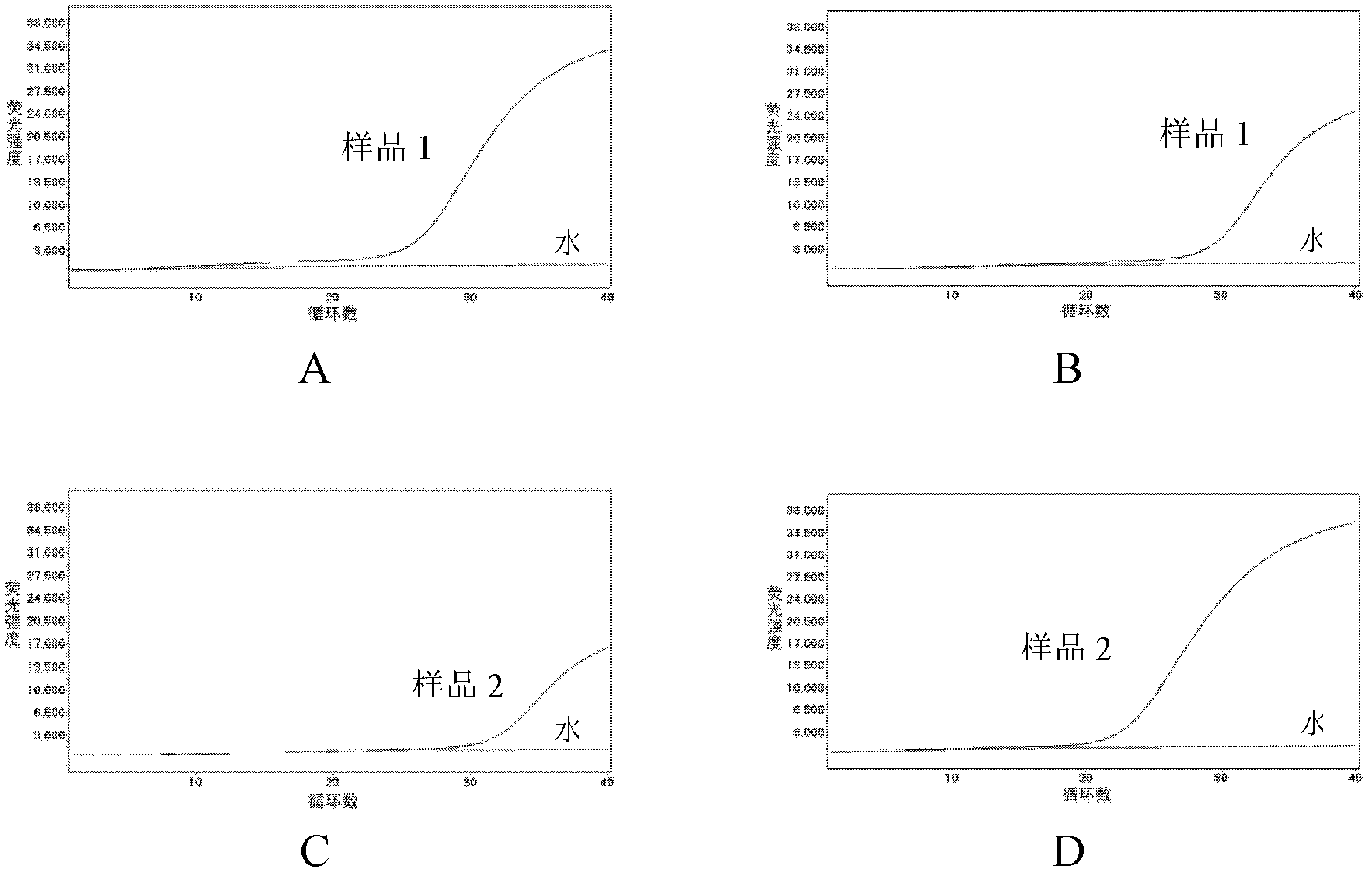Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
291 results about "Methylation Site" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A sequence in DNA that can be recognized by a specific methylase enzyme and that contains the specific base target for methyl transfer.
High-throughput methods for detecting DNA methylation
InactiveUS6605432B1Bioreactor/fermenter combinationsBiological substance pretreatmentsDNA methylationHybridization probe
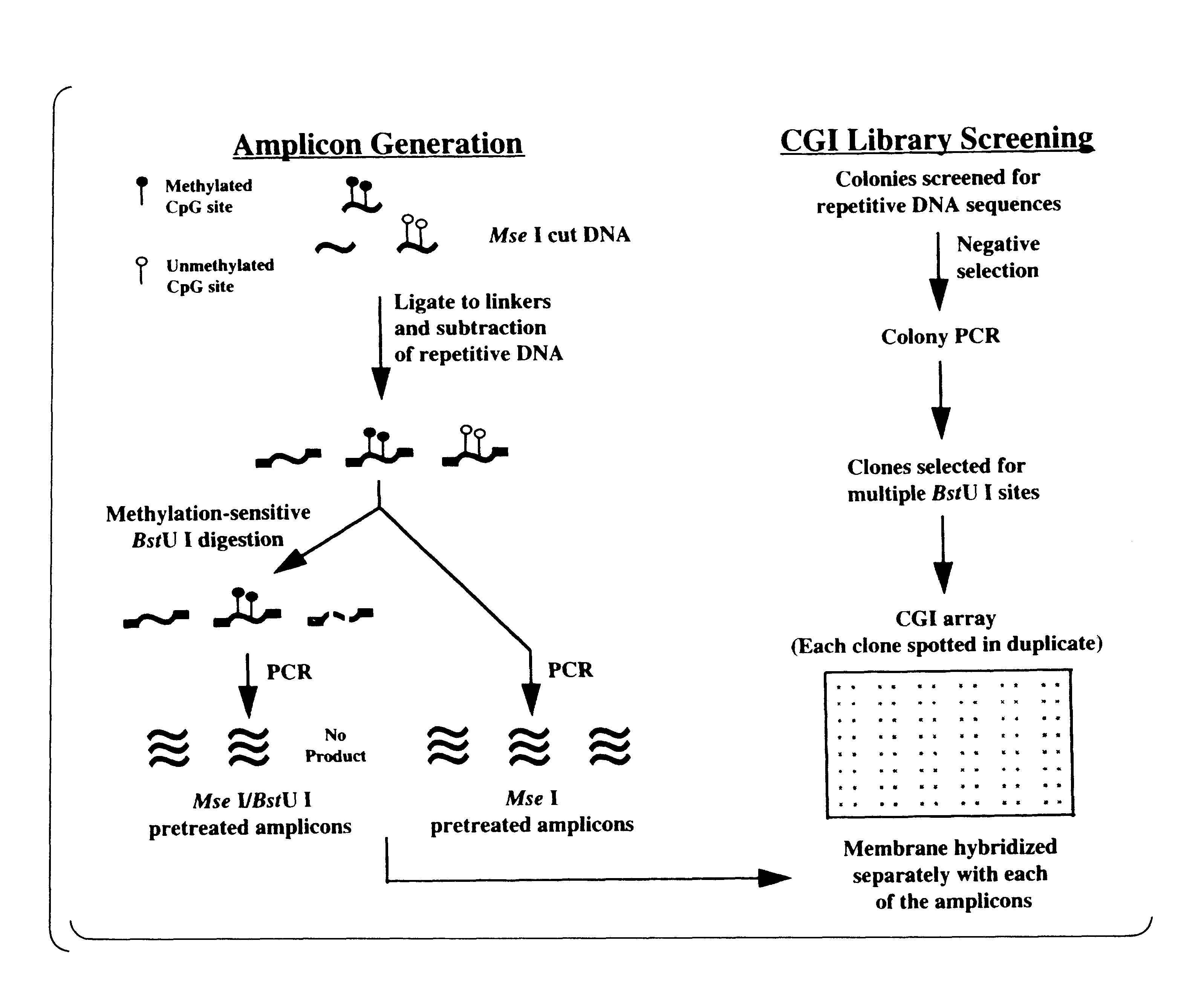
The present invention provides a method of hybridization, differential methylation hybridization (DMH) for high throughput methylation analysis of multiple CpG island loci. DMH utilizes nucleic acid probes prepared from a cell sample to screen numerous CpG dinucleotide rich fragments affixed on a screening array. Positive hybridization signals indicate the presence of methylated sites. Methods of preparing the hybridization probes and screening array are also provided.
Owner:UNIVERSITY OF MISSOURI
Determination of methylated DNA
InactiveUS20070231800A1Microbiological testing/measurementFermentationNucleic Acid ProbesNucleic acid hybridisation
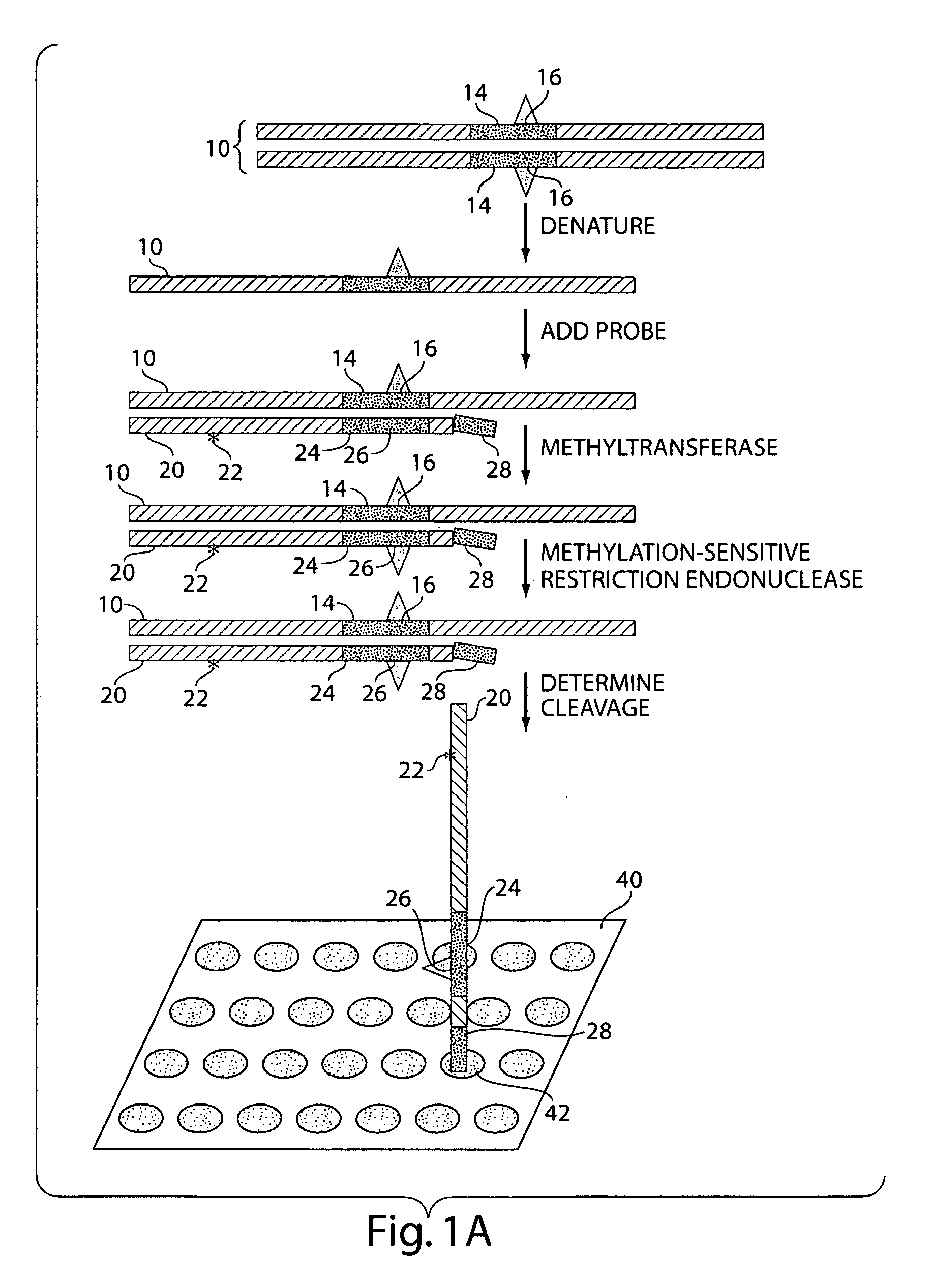
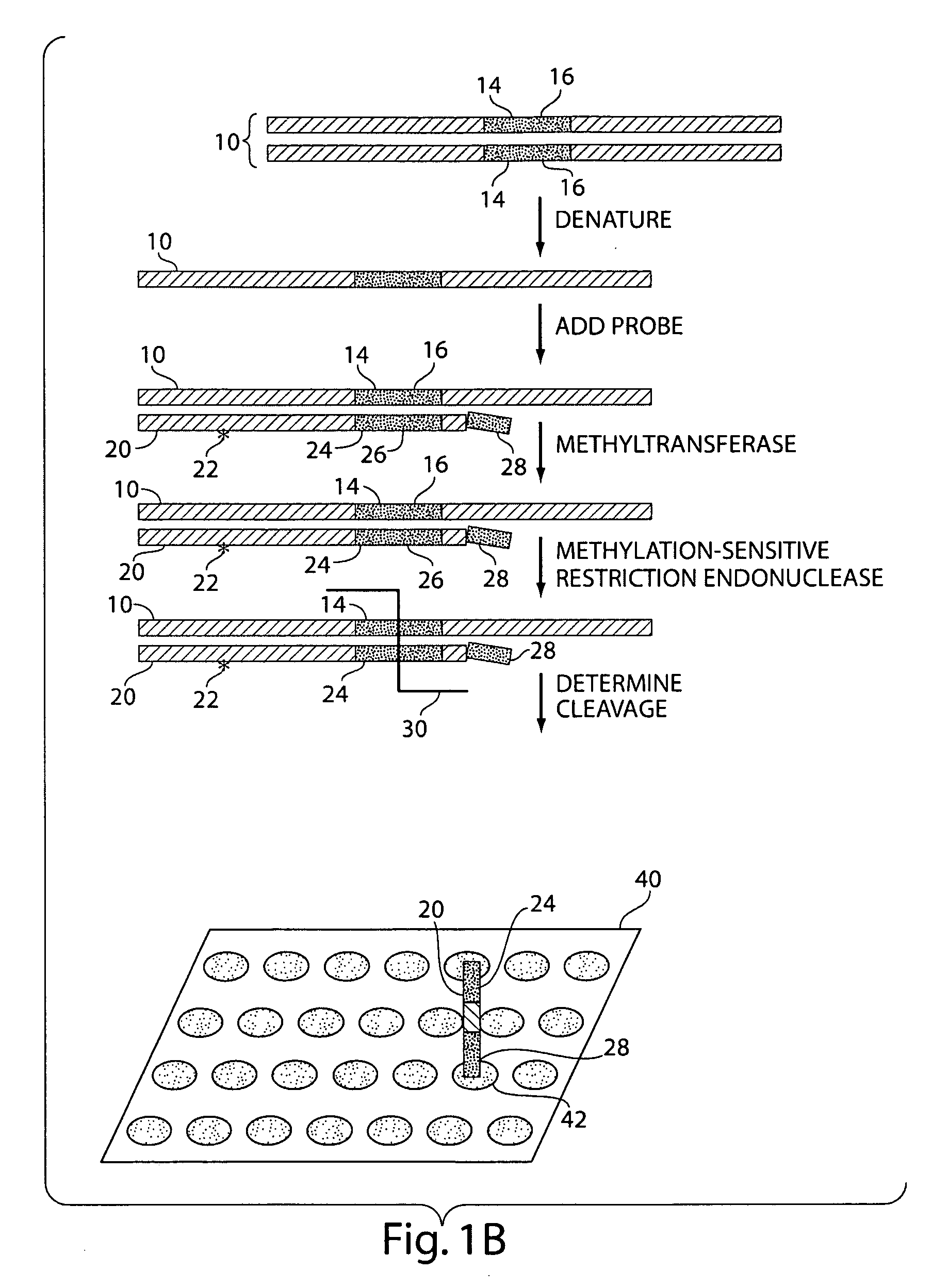
The present invention generally relates to the determination of the state of one or more locations within a nucleic acid and, in particular, to the determination of the methylation state of one or more methylation sites within a nucleic acid such as DNA. In one aspect of the invention, a nucleic acid, such as DNA, that is suspected of being methylated is exposed to a nucleic acid probe able to hybridize the nucleic acid at or near the methylation site. After hybridization, the nucleic acid-probe hybrid is exposed to a methylation-sensitive restriction endonuclease able to bind at or near the methylation site. The restriction endonuclease is not able to cleave the nucleic acid-probe hybrid if the DNA is methylated at the methylation site, but is able to cleave the nucleic acid-probe hybrid if the nucleic acid is not methylated at the methylation site. Determination of the cleavage state of the probe can thus be used to determine the state of the methylation site.
Owner:AGILENT TECH INC
High-throughput methods for detecting DNA methylation
InactiveUS20030129602A1Bioreactor/fermenter combinationsBiological substance pretreatmentsDNA methylationHybridization probe
The present invention provides a method of hybridization, differential methylation hybridization (DMH) for high throughput methylation analysis of multiple CpG island loci. DMH utilizes nucleic acid probes prepared from a cell sample to screen numerous CpG dinucleotide rich fragments affixed on a screening array. Positive hybridization signals indicate the presence of methylated sites. Methods of preparing the hybridization probes and screening array are also provided.
Owner:UNIVERSITY OF MISSOURI
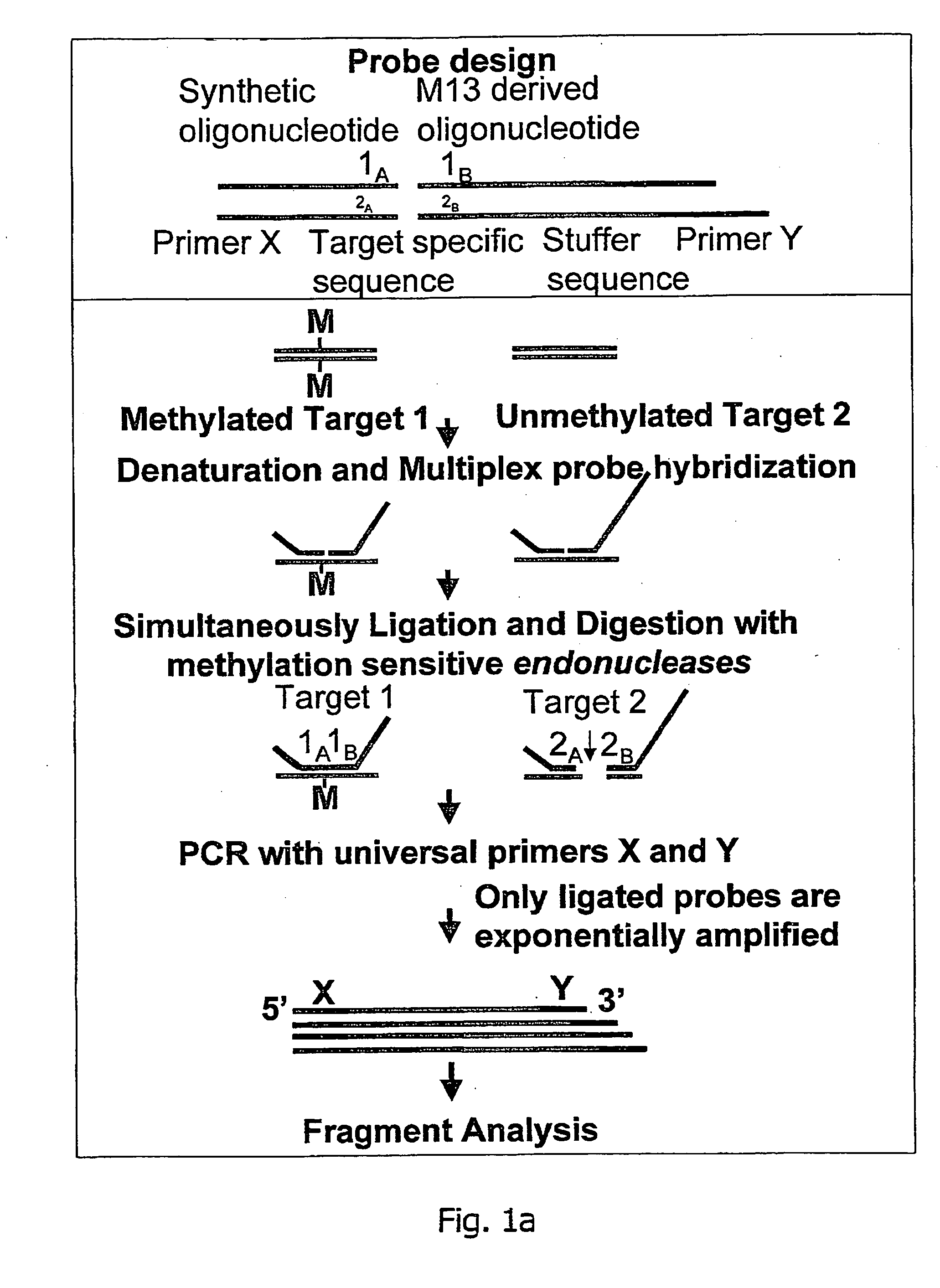
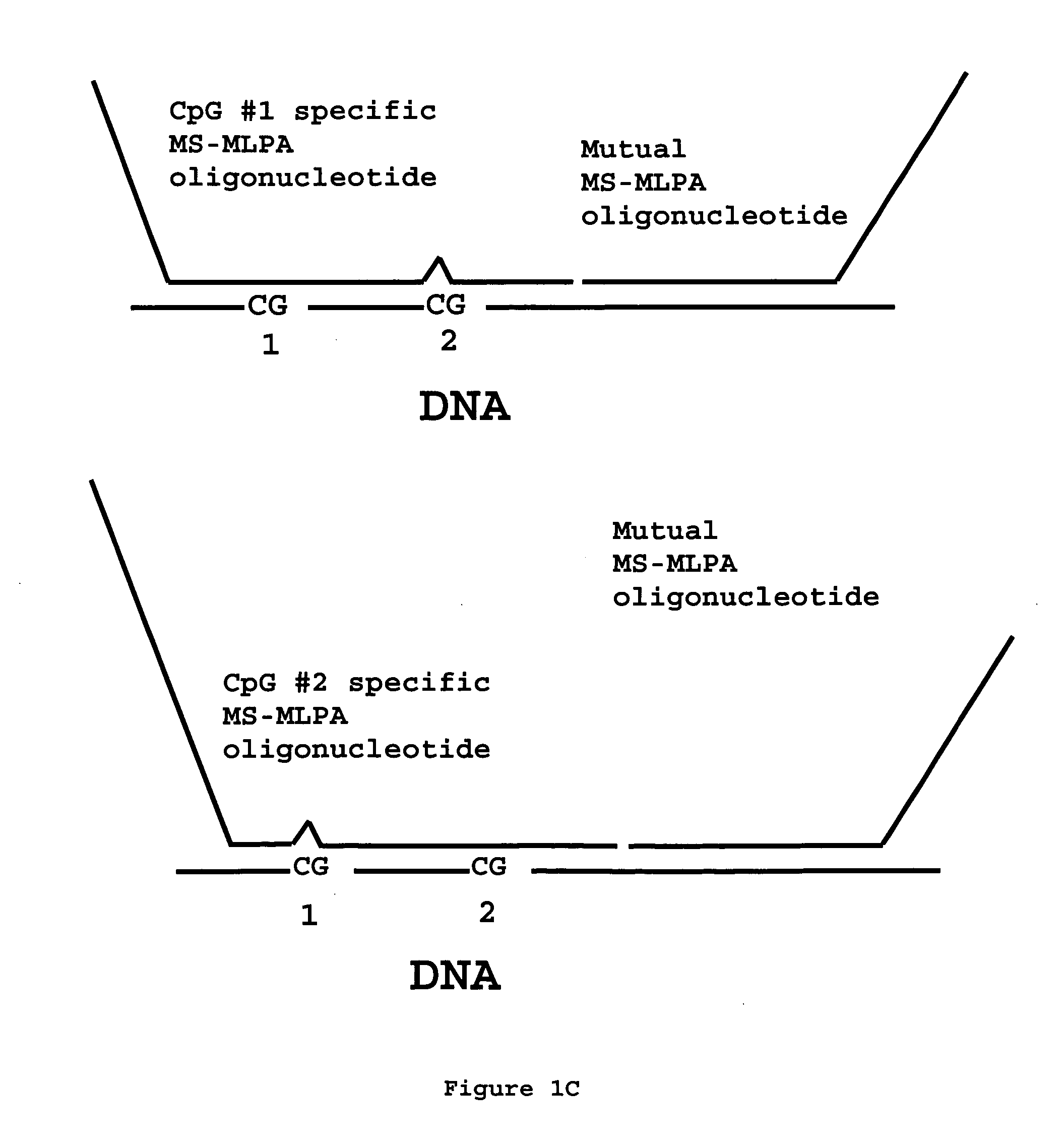
Methylation specific multiplex ligation-dependent probe amplification (MS-MLPA)
InactiveUS20070092883A1Rapid and easy to applyMicrobiological testing/measurementFermentationMultiplex ligation-dependent probe amplificationNucleic acid sequencing
An improved multiplex ligation-dependent amplification method is disclosed for detecting the presence of specific methylated sites in a single stranded target nucleic acid, while simultaneously, the quantification of the target nucleic acid sequence can be performed, using a plurality of probe sets of at least two probes, each of which includes a target specific region and non-complementary region containing a primer binding site. At least one of the probes further includes the sequence of one of the strands of a double stranded recognition site of a methylation sensitive restriction enzyme. The probes belonging to the same set are ligated together when hybridised to the target nucleic acid sequence, the hybrid is subjected to digestion by the methylation sensitive restriction enzyme, resulting in non-methylated recognition sites being cleaved. The probes of the uncleaved (methylated) hybrid are subsequently amplified by a suitable primer set.
Owner:DE LUWE HOEK OCTROOIEN
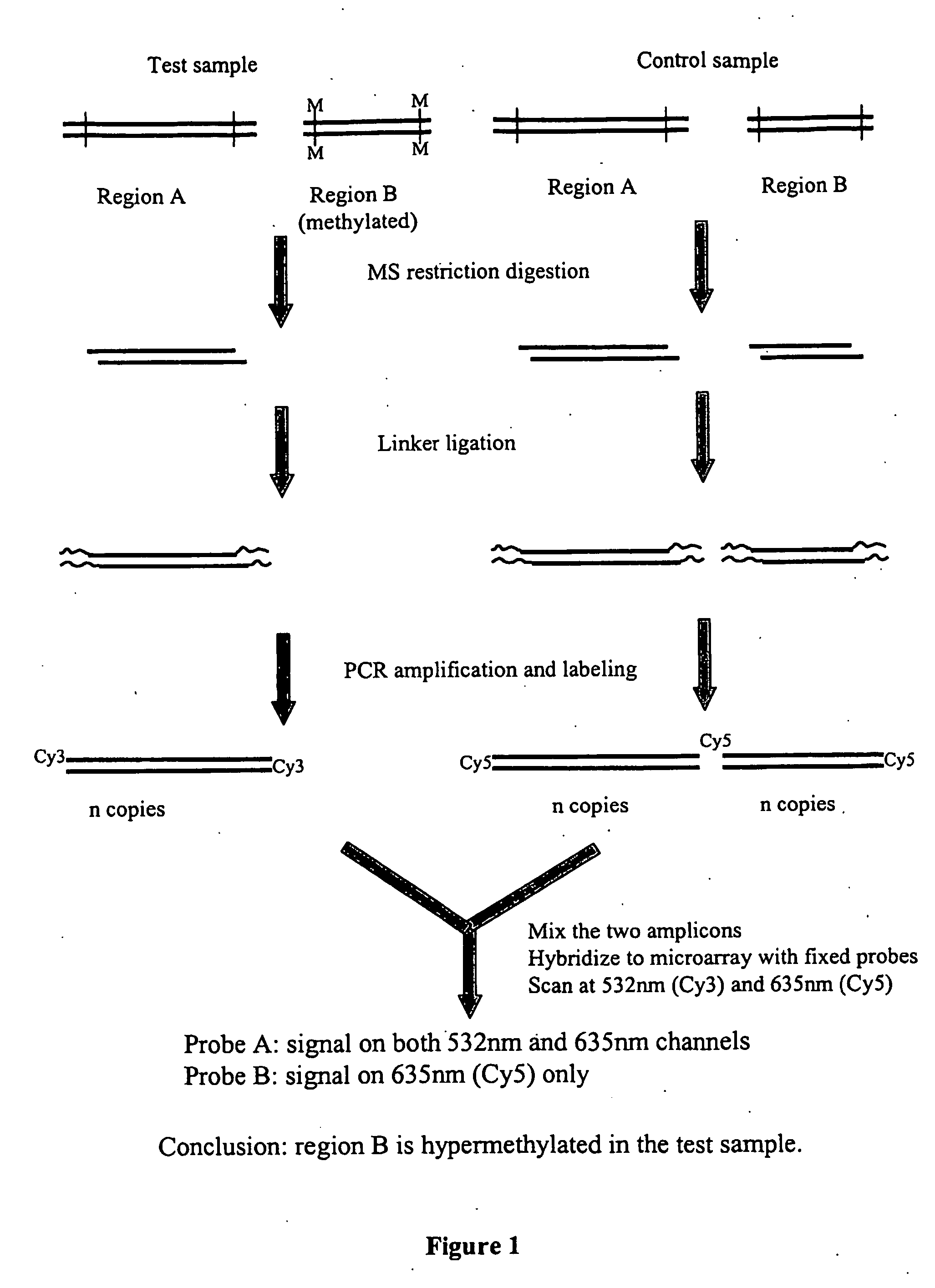
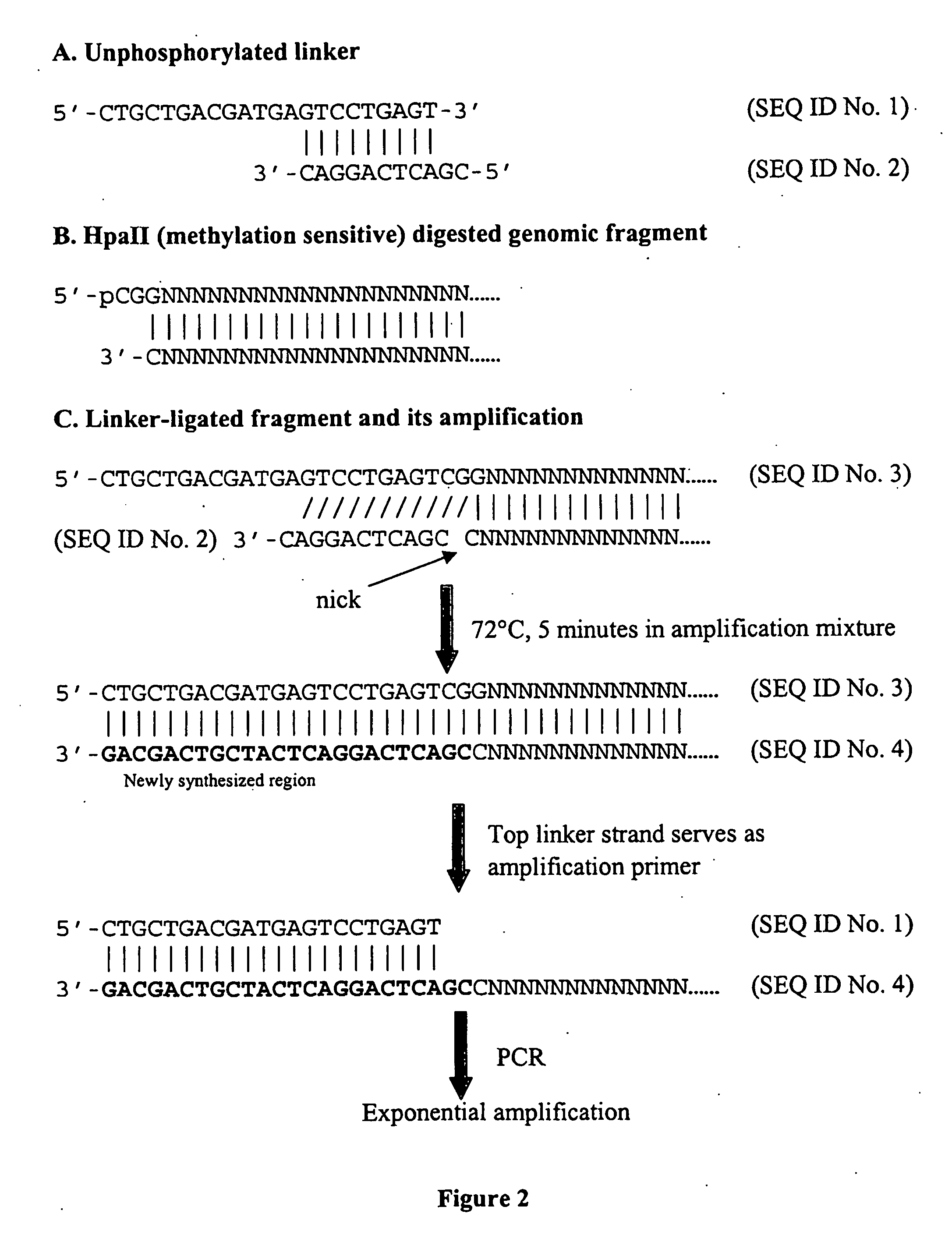
Analysis of methylation using nucleic acid arrays
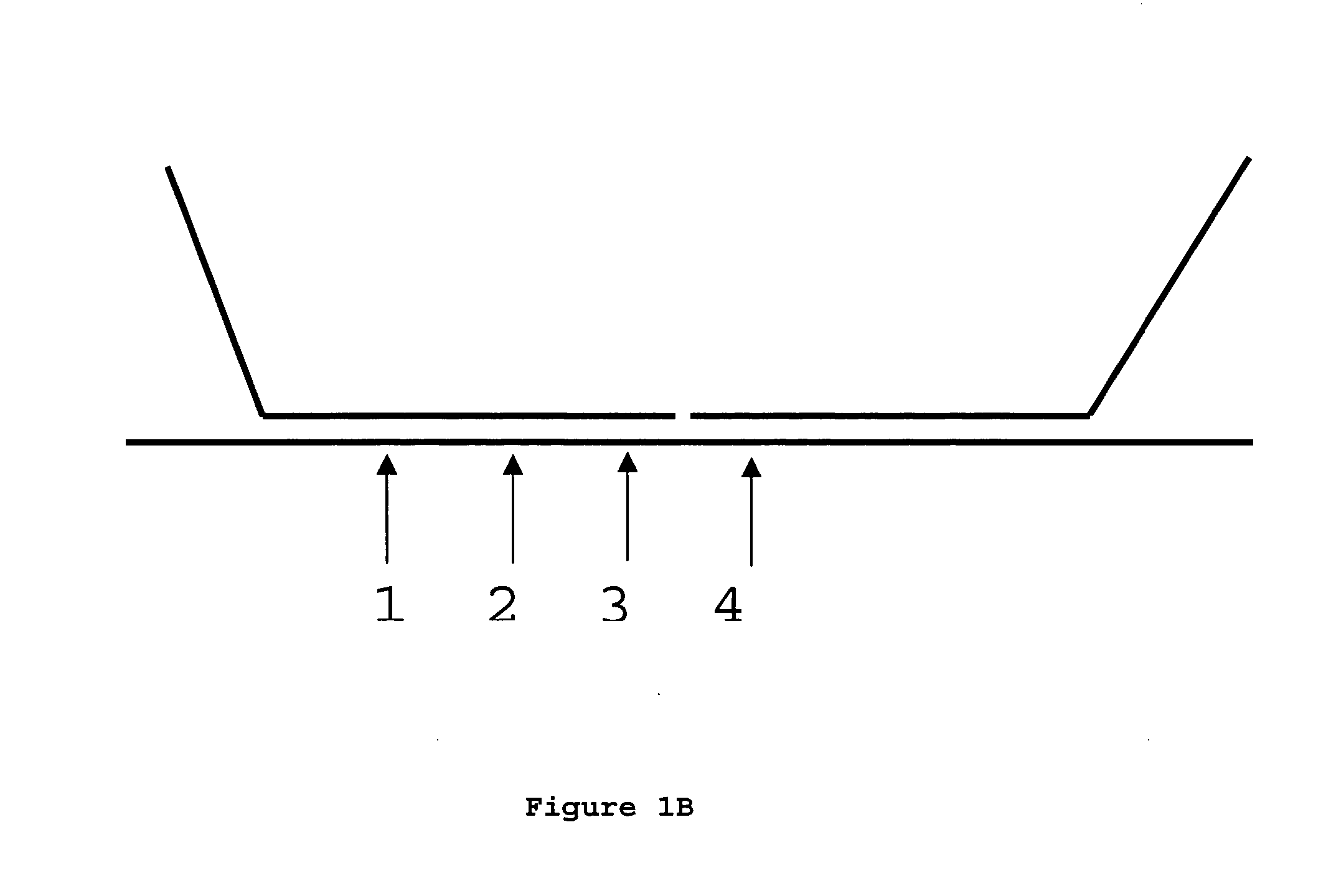
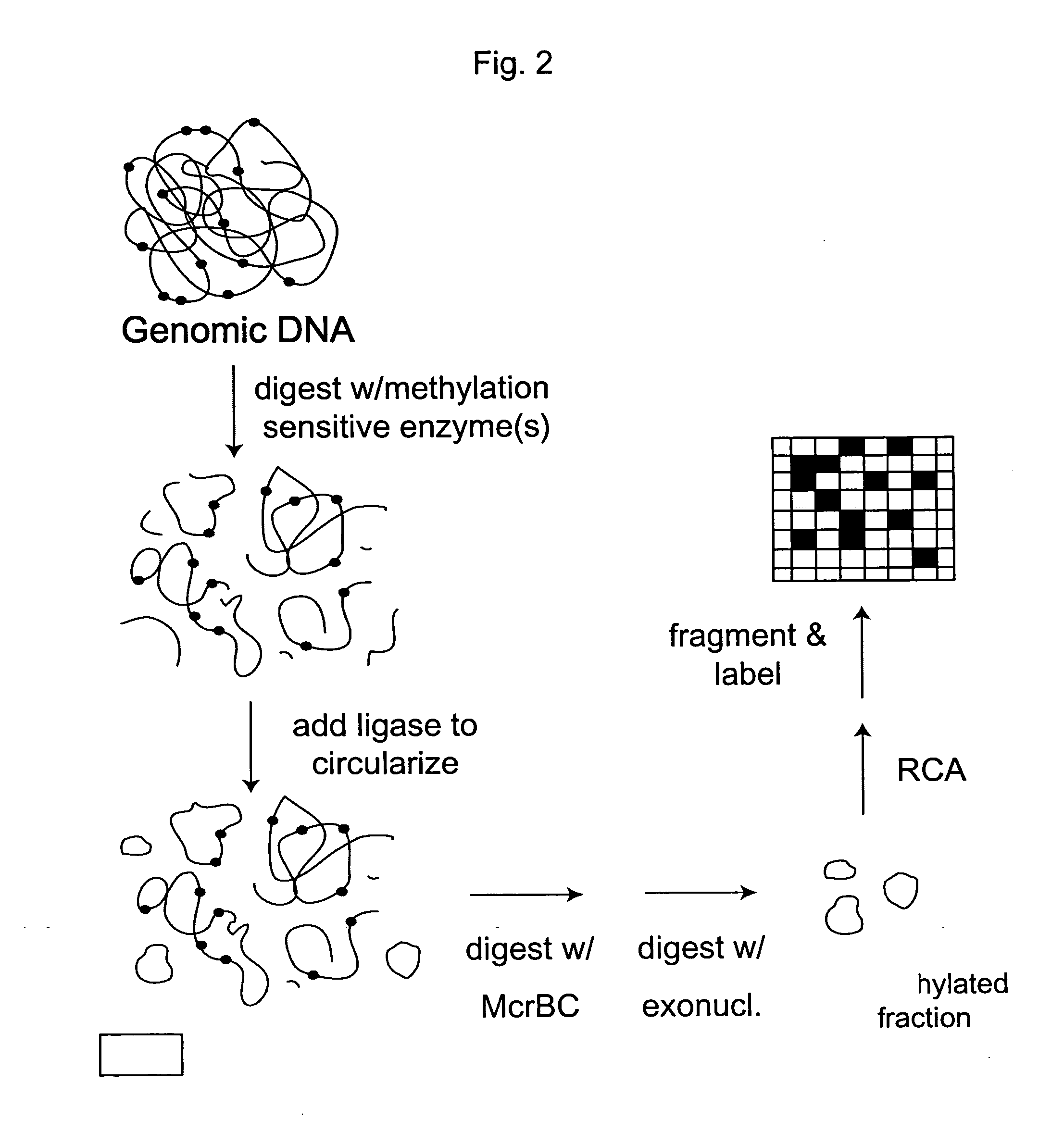
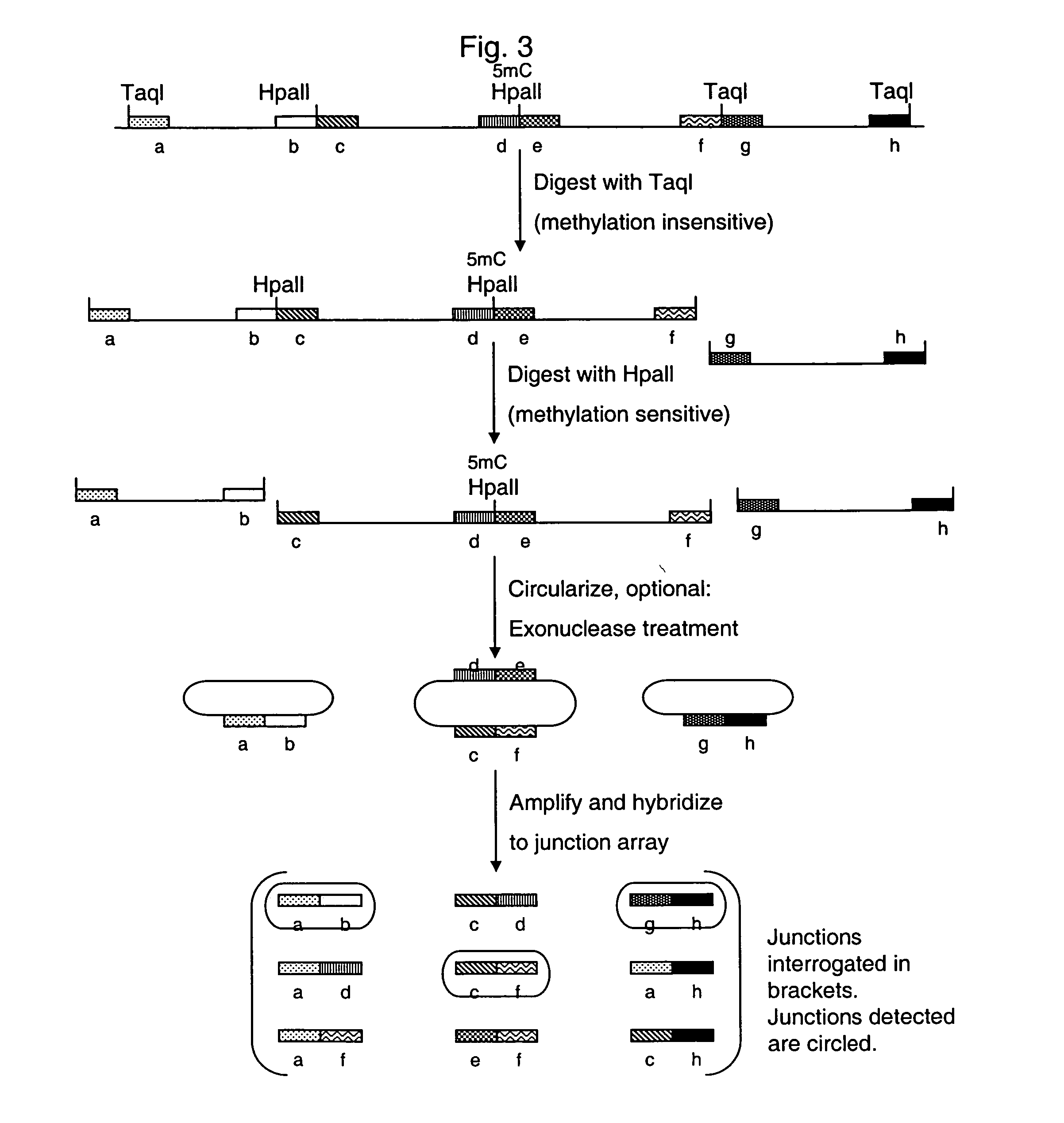
Methods of analyzing DNA to determine the methylation status of a plurality of cytosines are disclosed. In one aspect genomic DNA is fragmented, fragments are circularized, the circles are treated with a methylation sensitive enzyme to enrich for circles with methylated sites or with a methylation dependent enzyme to enrich for circles with unmethylated sites, and the circles are amplified. The amplified product is fragmented, labeled and hybridized to an array of probes. The array of probes may be a tiling array or an array of junction probes. The hybridization pattern is analyzed to determine methylation status of cytosines.
Owner:AFFYMETRIX INC
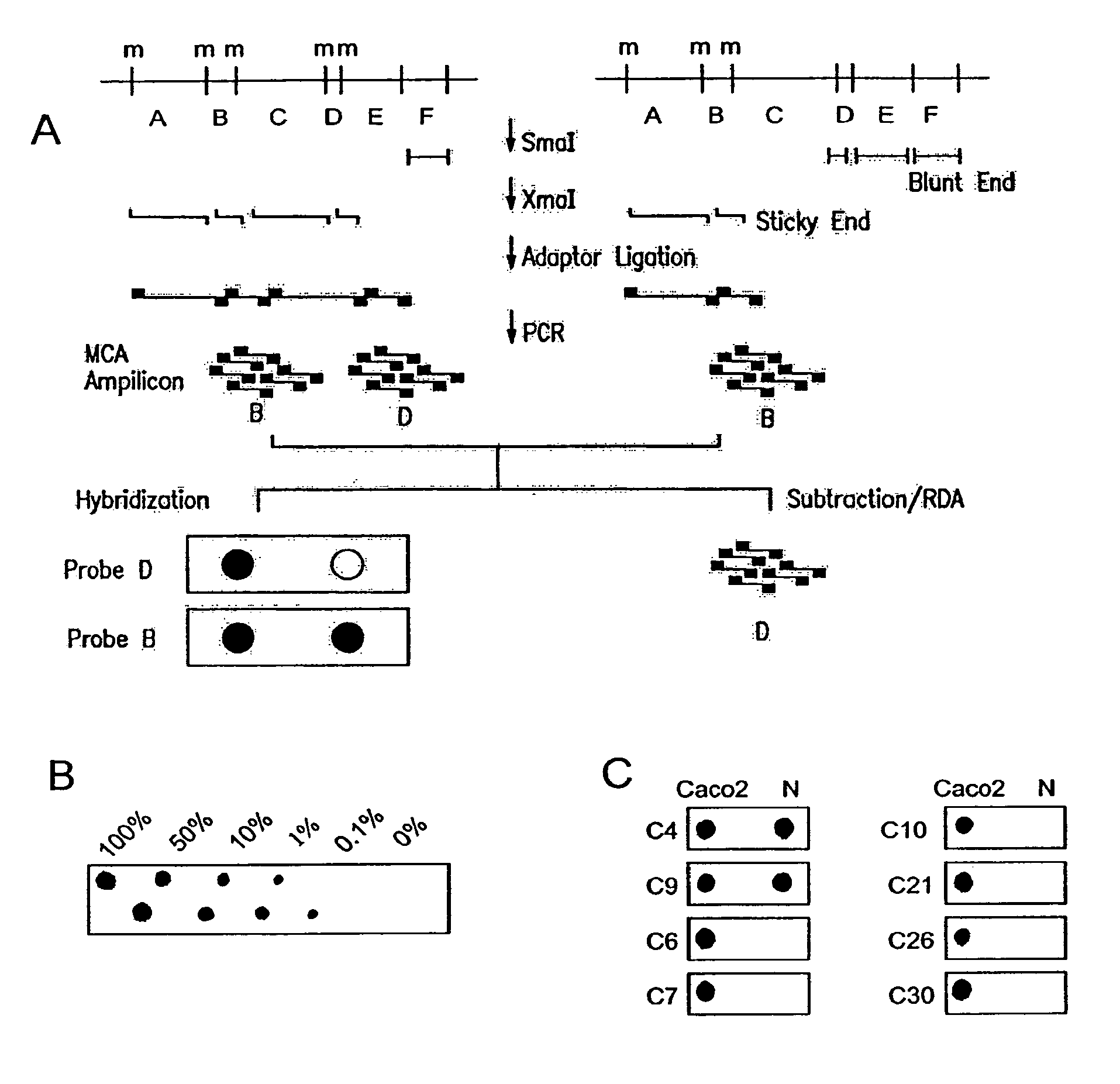
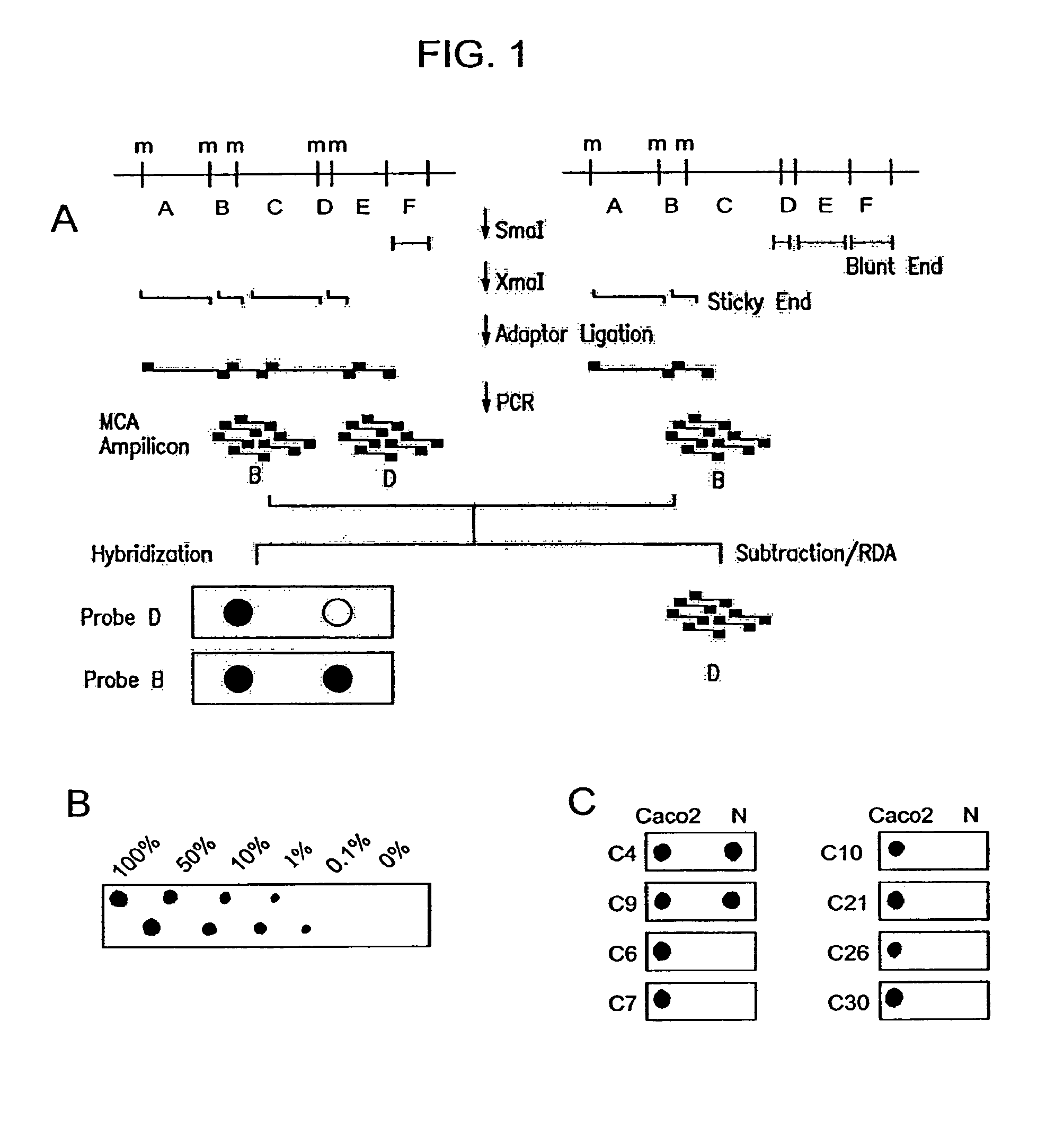
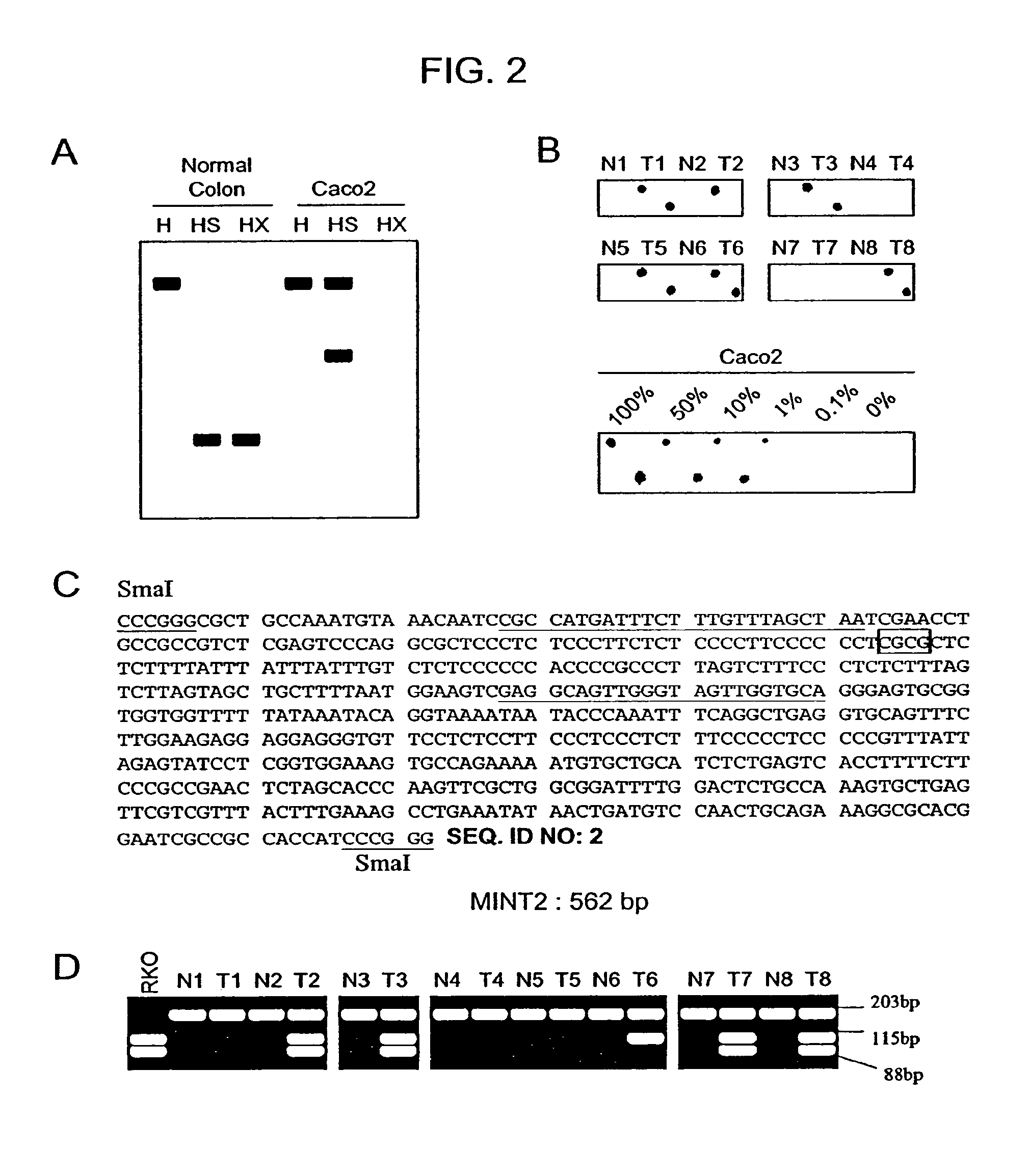
Methylated CpG island amplification (MCA)
The present invention provides a method for identifying a methylated CpG containing nucleic acid by contacting a nucleic acid with a methylation sensitive restriction endonuclease that cleaves unmethylated CpG sites and contacting the sample with an isoschizomer of the methylation sensitive restriction endonuclease, which cleaves both methylated and unmethylated CpG sites. The method also includes amplification of the CpG-containing nucleic acid using CpG-specific oligonucleotide primers A method is also provided for detecting an age associated disorder by identification of a methylated CpG containing nucleic acid. A method is further provided for evaluation the response of a cell to an agent A kit useful for detection of a CpG containing nucleic acid is also provided. Nucleic acid sequences encoding novel methylated clones.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
Method for accurate detection of whole genome methylation sites by utilizing trace genome DNA (deoxyribonucleic acid)
ActiveCN102409408AHigh amplification efficiencyImprove efficiencyMicrobiological testing/measurementLibrary creationFragment sizeExogenous DNA
According to the invention, based on Illumina common tag library sequencing and methylation normal sequencing and combining a common library tag sequencing method, a new method for accurate detection of whole genome methylation sites by utilizing trace genome DNA (deoxyribonucleic acid) is built. According to the method, in the construction of a methylation library by using trace genome DNA, exogenous DNA is innovatively added to carry out bisulfite high-efficiency co-treatment; and at the same time, fragment size selection is not needed, and PCR (polymerase chain reaction) amplification is directly carried out after bisulfite treatment. By the method, the defects that in the common methylation sequencing, samples can not be mixed, the PCR amplification efficiency is low and the trace DNAsample can not be researched are overcome.
Owner:BGI TECH SOLUTIONS
Nucleic acid detection assay
A method for determining the methylation status of a potential methylation site in genomic nucleic acid comprising treating genomic nucleic acid with an agent which modifies cytosine bases but does not modify 5methyl-cytosine bases under conditions to form a. modified nucleic acid template containing a potential methylation site; providing a first clamp containing a first capture sequence complementary to a region flanking one side of the potential methylation site in the modified nucleic acid template, providing a second clamp containing a second capture sequence complementary to a region flanking the other side of the potential methylation site in the modified nucleic acid template; allowing the first clamp and the second clamp to hybridise to the modified nucleic acid template; ligating the hybridised first and second clamps to form a probe spanning the potential methylation site in the modified nucleic acid template; digesting the modified nucleic acid template to obtain the probe; and detecting the probe and determining the methylation status of the potential methylation site in the modified genomic nucleic acid.
Owner:HUMAN GENETIC SIGNATURES PTY LTD
Method for determining the methylation pattern of a polynucleic acid
InactiveUS20060204988A1Reduce complexityEffective and powerfulMicrobiological testing/measurementFermentationBase JNucleotide
Particular aspects relate to a method for determining the methylation pattern of a polynucleic acid, comprising: a) preparing a solution comprising a mixture of fragments of the polynucleic acid; b) coupling the fragments with a substance being detectable with a detection method; c) contacting a solution comprising the fragments of b) with a DNA microarray having a plurality of different immobilized oligonucleotides, each comprising at least one methylation site, at respectively assigned different locations thereon, the contacing under conditions affording hybridization of fragments with correlated immobilized oligonucleotides under defined stringency, and wherein the immobilized oligonucleotides have a length of less than 200 bases; d) optionally performing a a washing step; and e) detecting, using the physical detection method, such immobilized nucleic acids to which solution fragments are hybridized and / or to which solution fragments are not hybridized.
Owner:EPIGENOMICS AG
Prediction method for protein post-translational modification methylation loci
InactiveCN105893787AImprove throughputImprove accuracyProteomicsGenomicsLife activityProtein methylation
The invention discloses a prediction method for protein post-translational modification methylation loci, and belongs to the field of bioinformatics. Protein methylation modification participates in cell functions and many life activities of cell processes, and recognition of protein methylation modification loci has very important significance in understanding of the life activities of cells. The prediction method combines with sequence information, evolutionary information and physical and chemical properties to conduct feature coding on a protein methylation sequence, an information gain optimization feature method is adopted and combines with a support vector machine to construct a prediction model, and it is shown through independent testing results that the prediction method has a good prediction property on the protein methylation loci; meanwhile, a network prediction platform is developed and used for conducting online prediction on the protein methylation loci.
Owner:NANCHANG UNIV
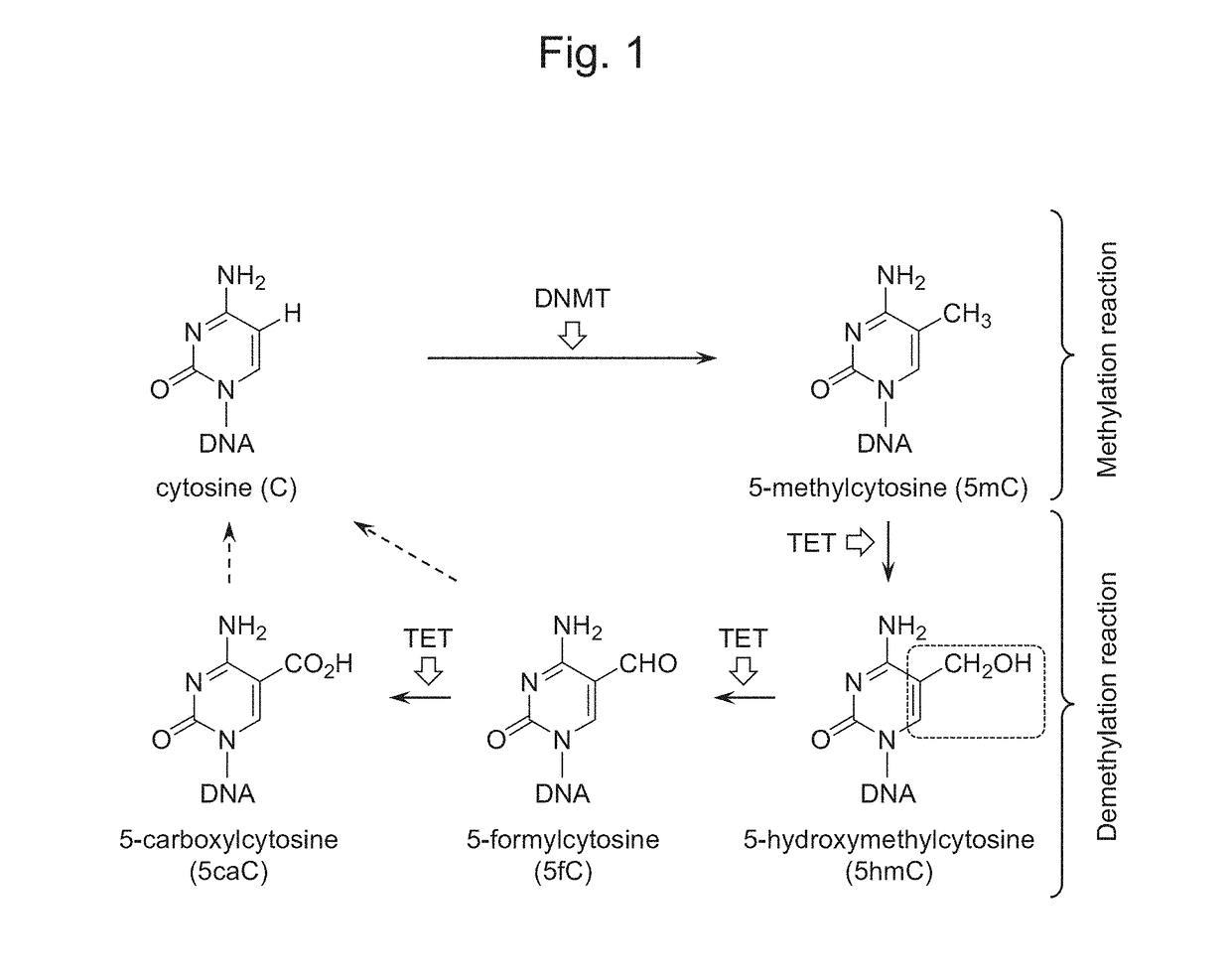
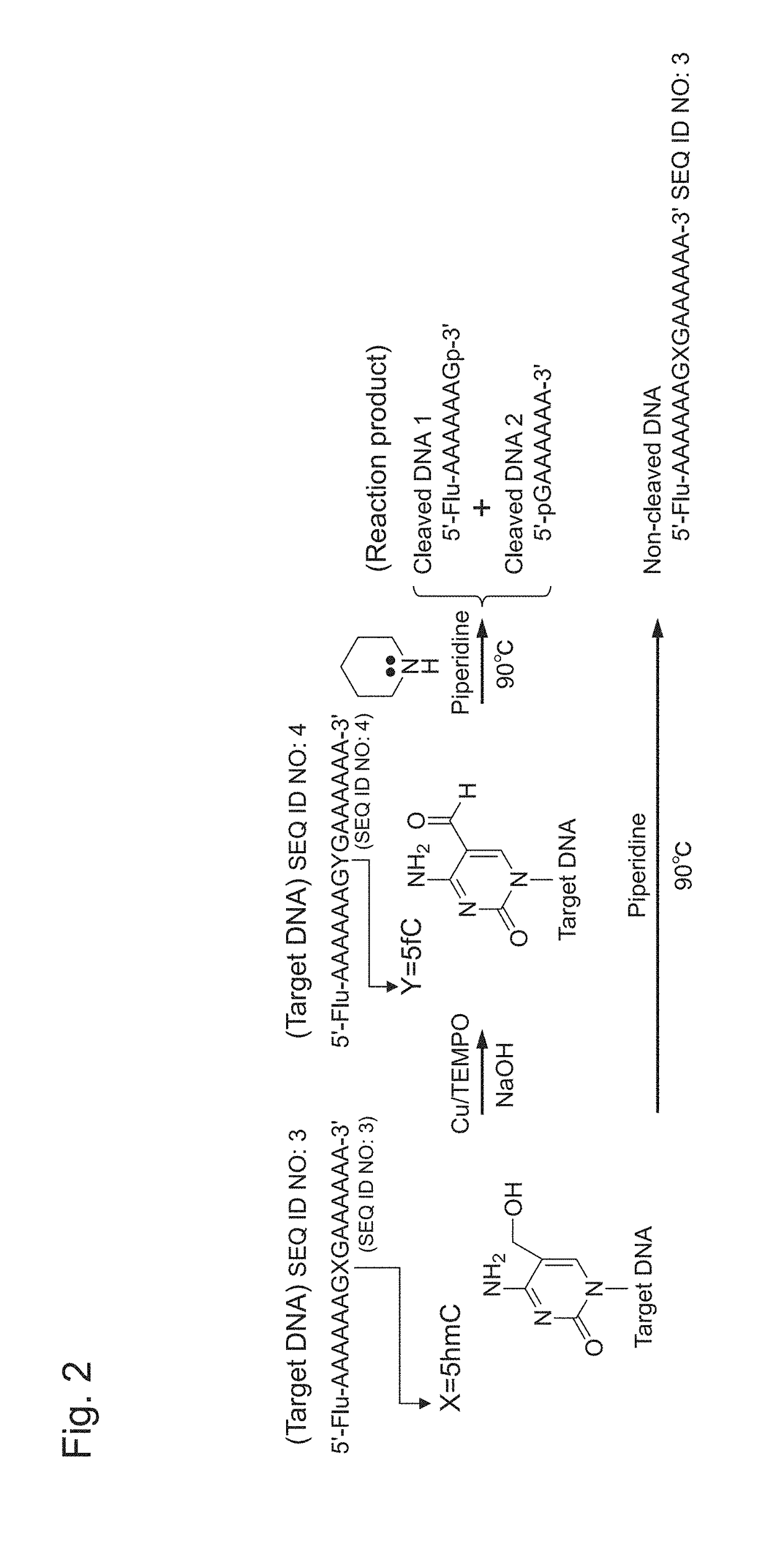
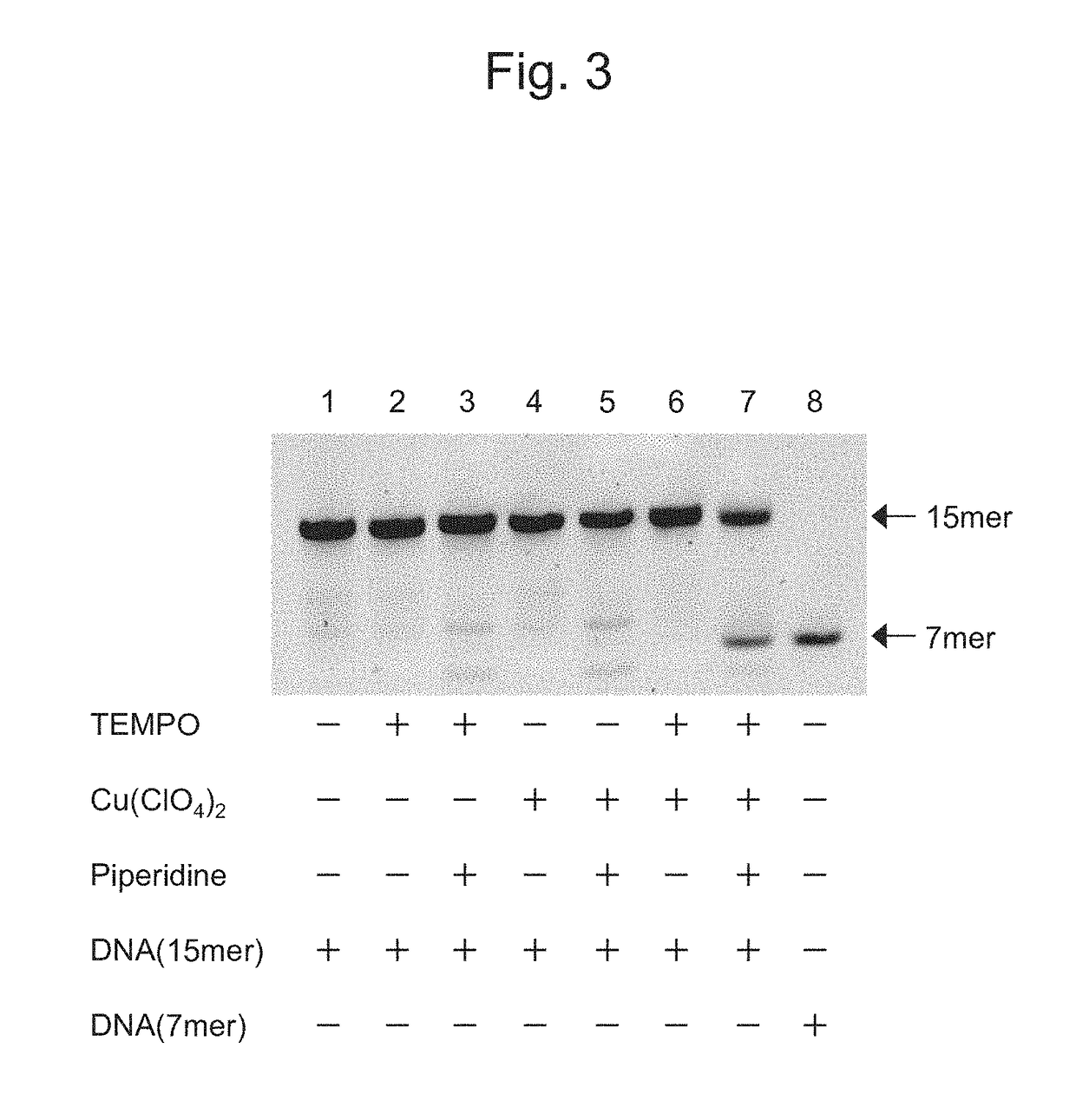
Oxidizing agent for 5-hydroxymethylcytosine and method for analyzing 5-hydroxymethylcytosine
InactiveUS20180251815A1Improve accuracyOrganic chemistryMicrobiological testing/measurementNitroxyl radicalsSide reaction
The purpose of the present invention is to develop and provide a less expensive and novel oxidizing agent for 5-hydroxymethylcytosine, which can selectively oxidize 5-hydroxymethylcytosine on DNA into 5-formylcytosine while preventing the DNA structure from destabilization and suppressing side reactions, and a method for detecting a demethylation site at a high accuracy in demethylation of DNA with the use of the aforesaid oxidizing agent.Provided is an oxidizing agent for 5-hydroxymethylcytosine that comprises a nitroxyl radical molecule and a copper salt or a copper complex, and / or a nitroxyl radical molecule-copper complex.
Owner:THE UNIV OF TOKYO
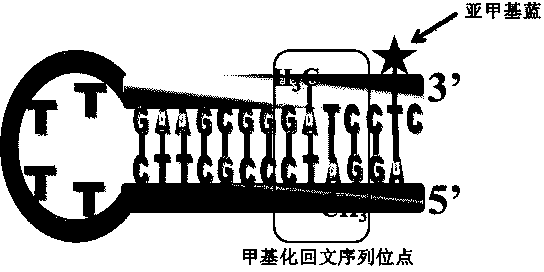
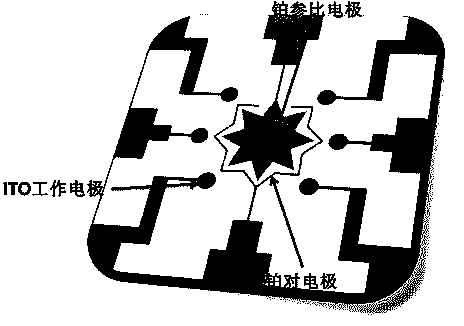
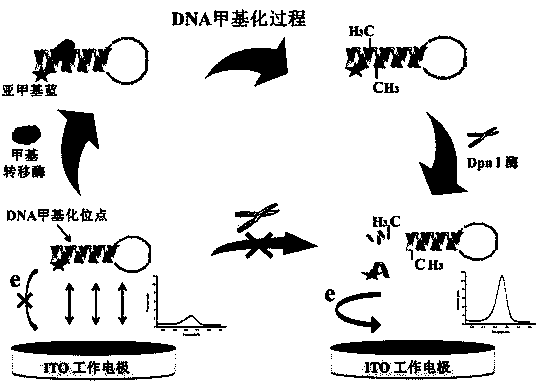
Quantitative analysis method aiming at DNA methylation monitoring
InactiveCN104062334ASimple preparationHigh sensitivityMaterial electrochemical variablesAdenosineA-DNA
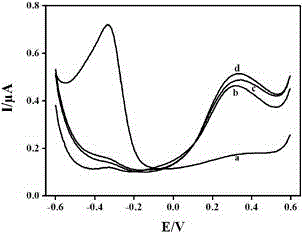
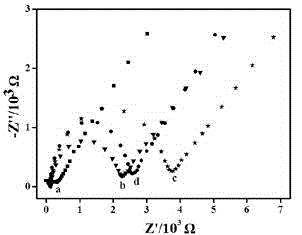
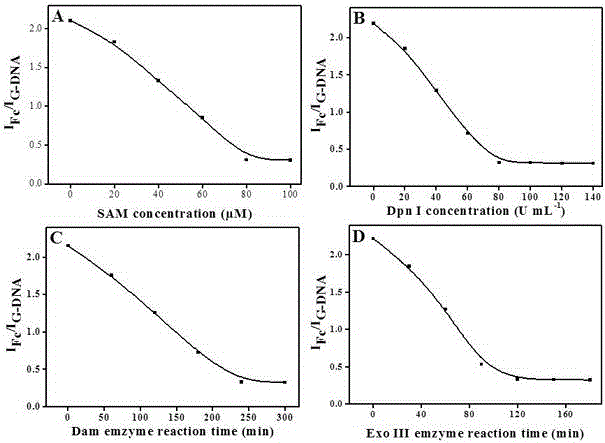
The invention discloses a quantitative analysis method aiming at DNA methylation monitoring. The quantitative analysis method comprises the following steps: (1) designing a hairpin type DNA secondary structure according to a DNA methylation palindromic sequence, and modifying a methylene blue electroactive material on the structure; (2) reacting hairpin type DNA with transmethylase and methylation adenosine to synthesize a methylated hairpin type DNA; (3) performing specific enzymolysis on a DNA methylation site through DpnI enzyme to release DNA fragments connected with methylene blue; (4) performing sample monitoring through an ITO micro-electrode chip with negative electricity, and acquiring electrochemical signals through a multi-pass electric signal reading device; (5) recording a monitoring result; and (6) adding methylation suppression chemicals into a DNA methylation process to screen the DNA methylation suppression chemicals. The quantitative analysis method has the advantages of simplicity in operation, portability, low cost and the like, and an electrode does not need complicated modification.
Owner:FUZHOU UNIV
Assays for dna methylation changes
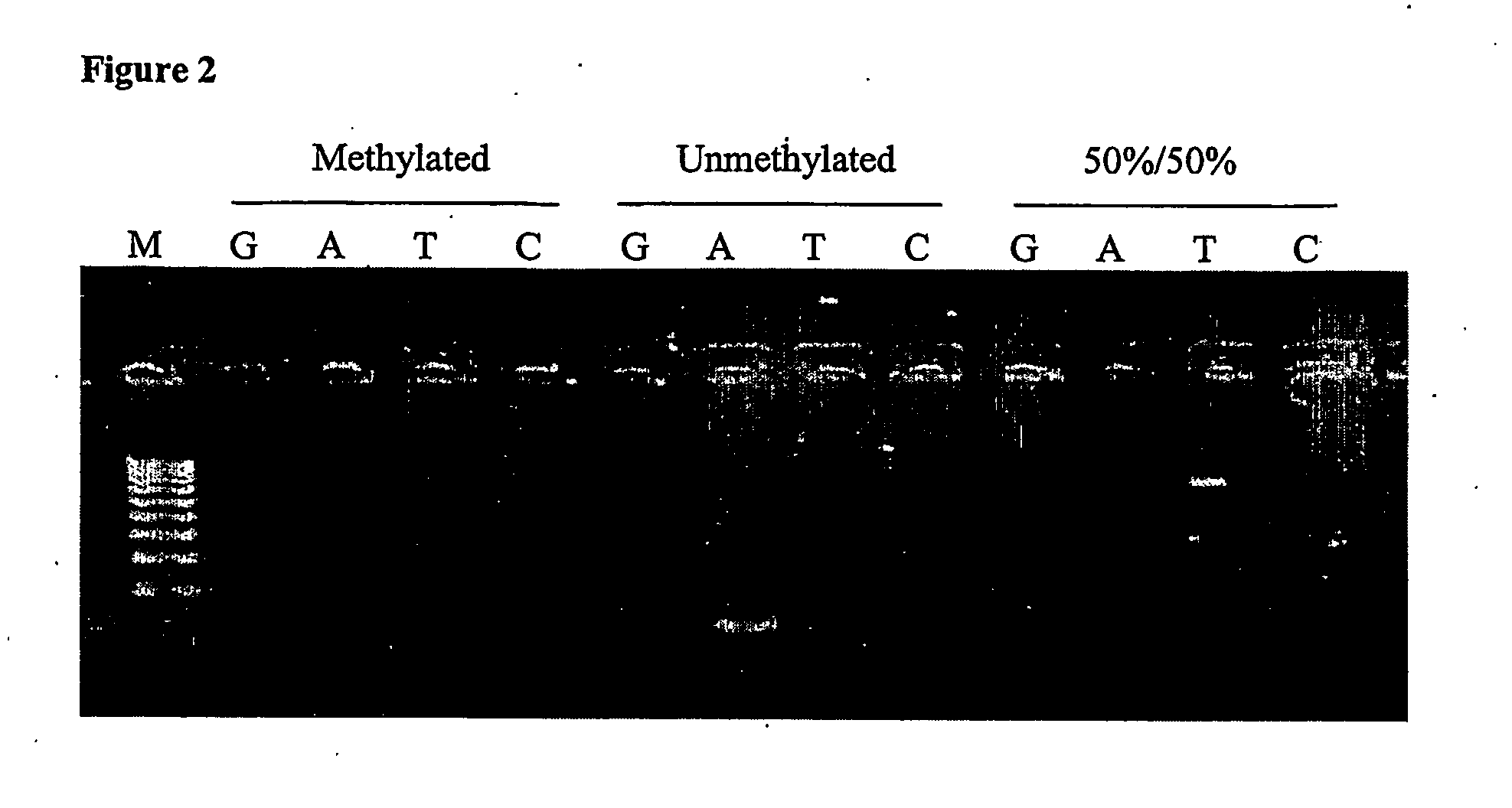
A new technique to determine the extent of DNA methylation entails generating DNA fragments of a test sample by cleaving at methylation sites that are not methylated while sparing methylation sites in the DNA that are methylated. This approach provides enhanced sensitivity to differences of even a single methylated cytosine, within or outside of a CpG island, and yet it can be employed to ascertain the methylation status of a region of DNA comprising a CpG island, a DNA region comprising one or more CpG-containing islands, or even a large variety of DNA regions.
Owner:EPIGENX PHARMA
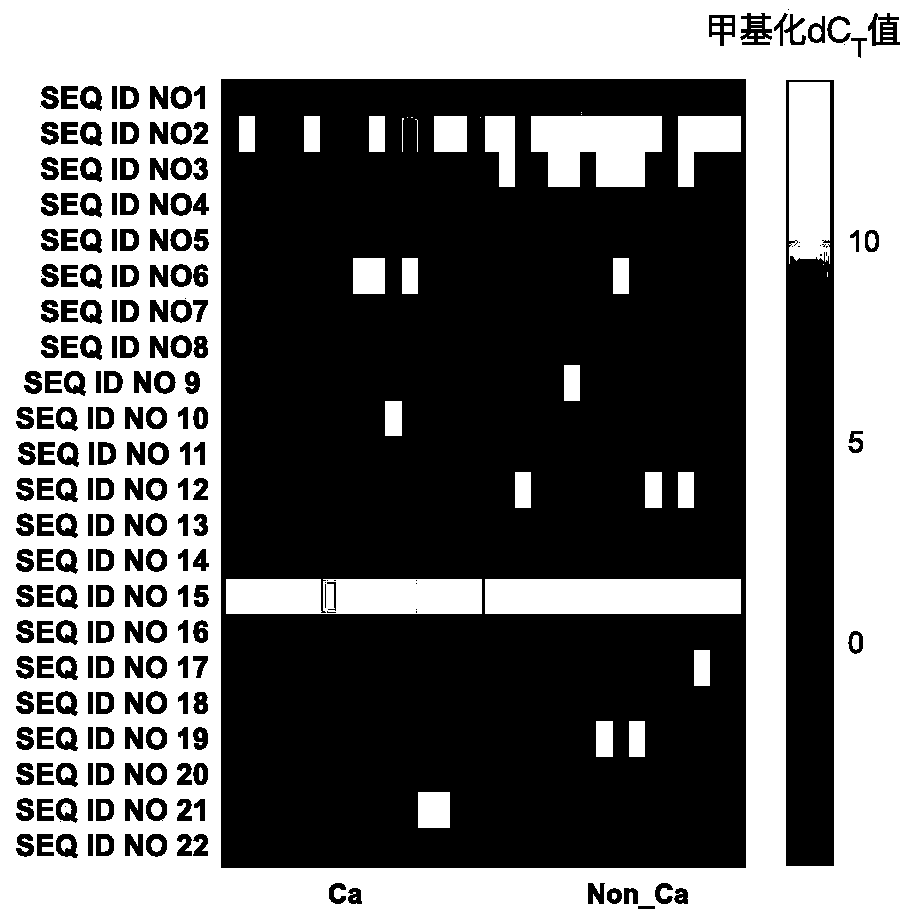
DNA methylation biomarker composition, detection method and kit
ActiveCN110872631AHigh sensitivityReact component optimizationMicrobiological testing/measurementDNA/RNA fragmentationDNA methylationMedicine
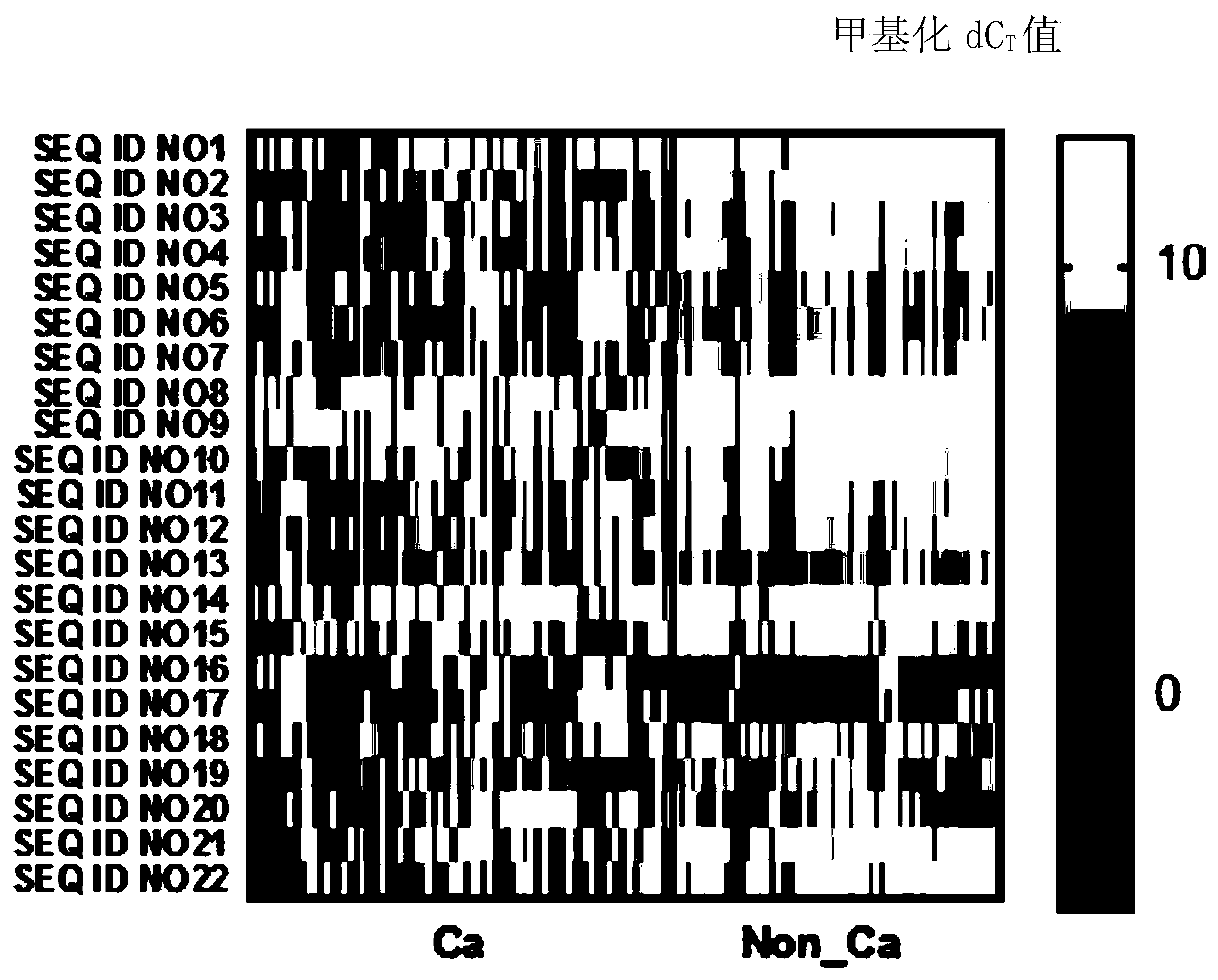
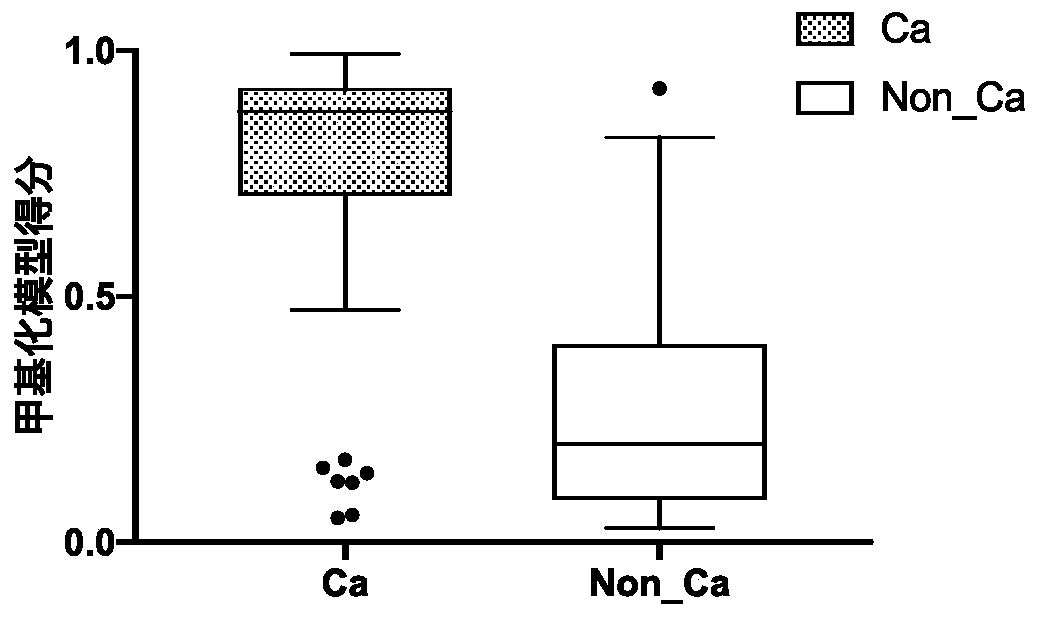
The invention relates to a DNA methylation biomarker composition for bladder cancer detection. The biomarker composition comprises any two or more of sequences shown in the formulas of SEQ ID NO.1 toSEQ ID NO.22, or any two or more of completely complementary sequences shown in the formulas of SEQ ID NO.1 to SEQ ID NO.22. The invention also relates to a detection kit for the methylation biomarkercomposition. The bladder cancer occurrence is discriminated and analyzed by using the composition in the co-methylation state of a plurality of specific methylation regions, the specific methylationcomposition has high sensitivity for discriminating the bladder cancer occurrence, and the detection method is simple, convenient and feasible. The primer pair composition of the kit overcomes the defect of false positive results caused by single methylation site detection mismatch in primer sequence design, and considers the interaction between multiple methylation biomarker primer and probe paircomposition.
Owner:ANCHORDX MEDICAL CO LTD
Methylation Biomarkers for Diagnosis of Prostate Cancer
InactiveUS20140094380A1Increase of methylationMicrobiological testing/measurementLibrary screeningOncologyBiomarker (petroleum)
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV +1
Primer and kit for detecting septin9 gene methylation
InactiveCN106244724AEasy to detectEliminate distractionsMicrobiological testing/measurementDNA/RNA fragmentationForward primerReference genes
The invention discloses a primer and a kit for detecting septin9 gene methylation. The primer comprises a target gene primer pair Septin9 forward primer F and an Septin9 reverse primer R. The kit comprises the primer and optimally further comprises a reference gene primer, a TaqMan-MGB probe and two Blocker primers. The kit is simple and convenient to operate, is easy to judge and is low in requirement for instruments; the whole PCR process is fully closed; the cross contamination possibility is avoided; the result is more accurate. Compared with the method of detecting by adopting PCR specific primer, the method of identifying the methylated site by utilizing the probe has higher flexibility and accuracy. Two Blocker primers are utilized to strictly control the amplification of the non-specific stripe, so that the kit has higher detection sensitivity and specificity and is especially fit for early screening of free DNA of colorectal cancer plasma.
Owner:北京鑫诺美迪基因检测技术有限公司
Methods of modulating smyd3 for treatment of cancer
InactiveUS20090191181A1Lower methylation levelsRelieve symptomsCompound screeningApoptosis detectionBladder cancerHepatocellular carcinoma
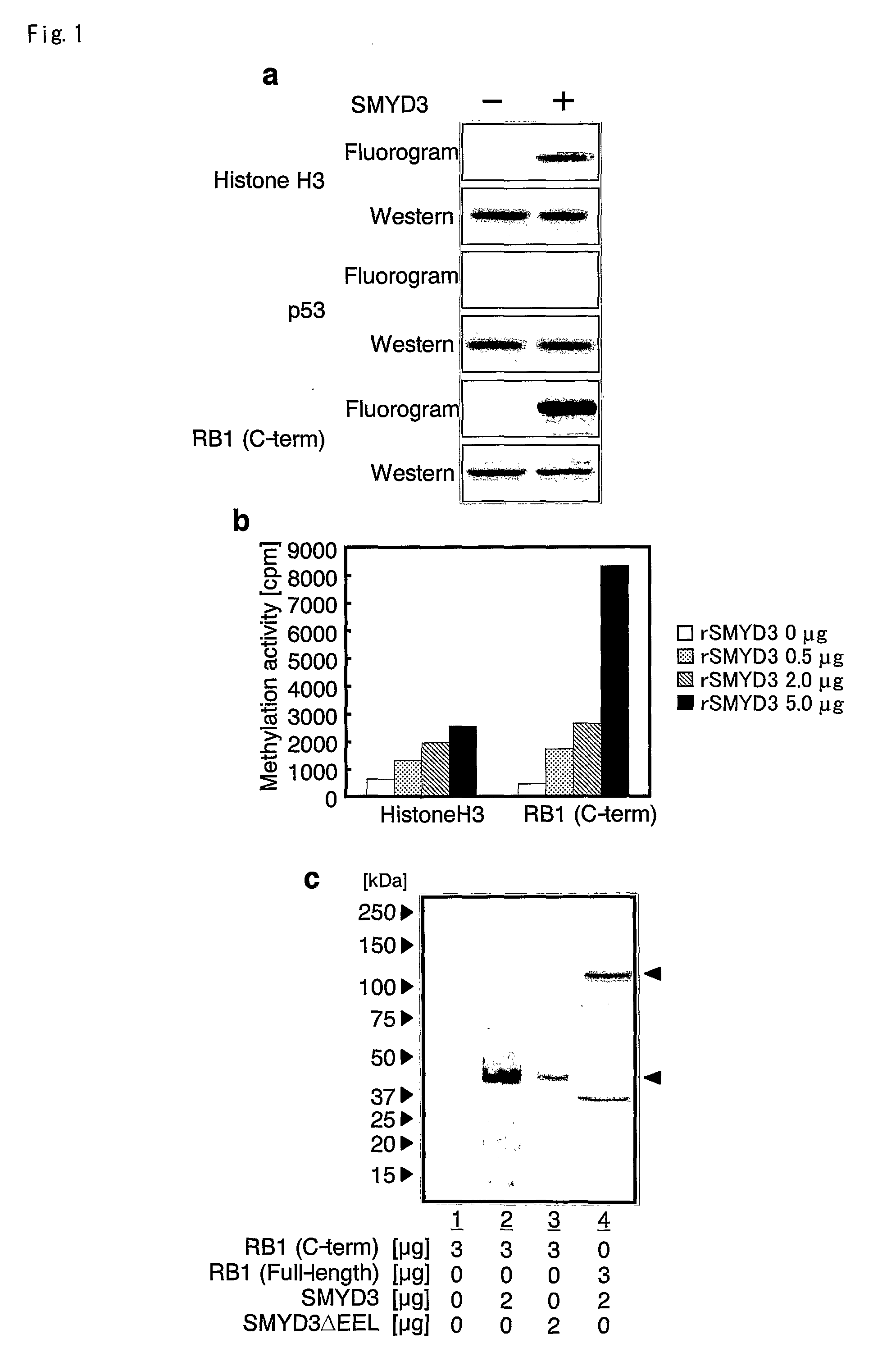
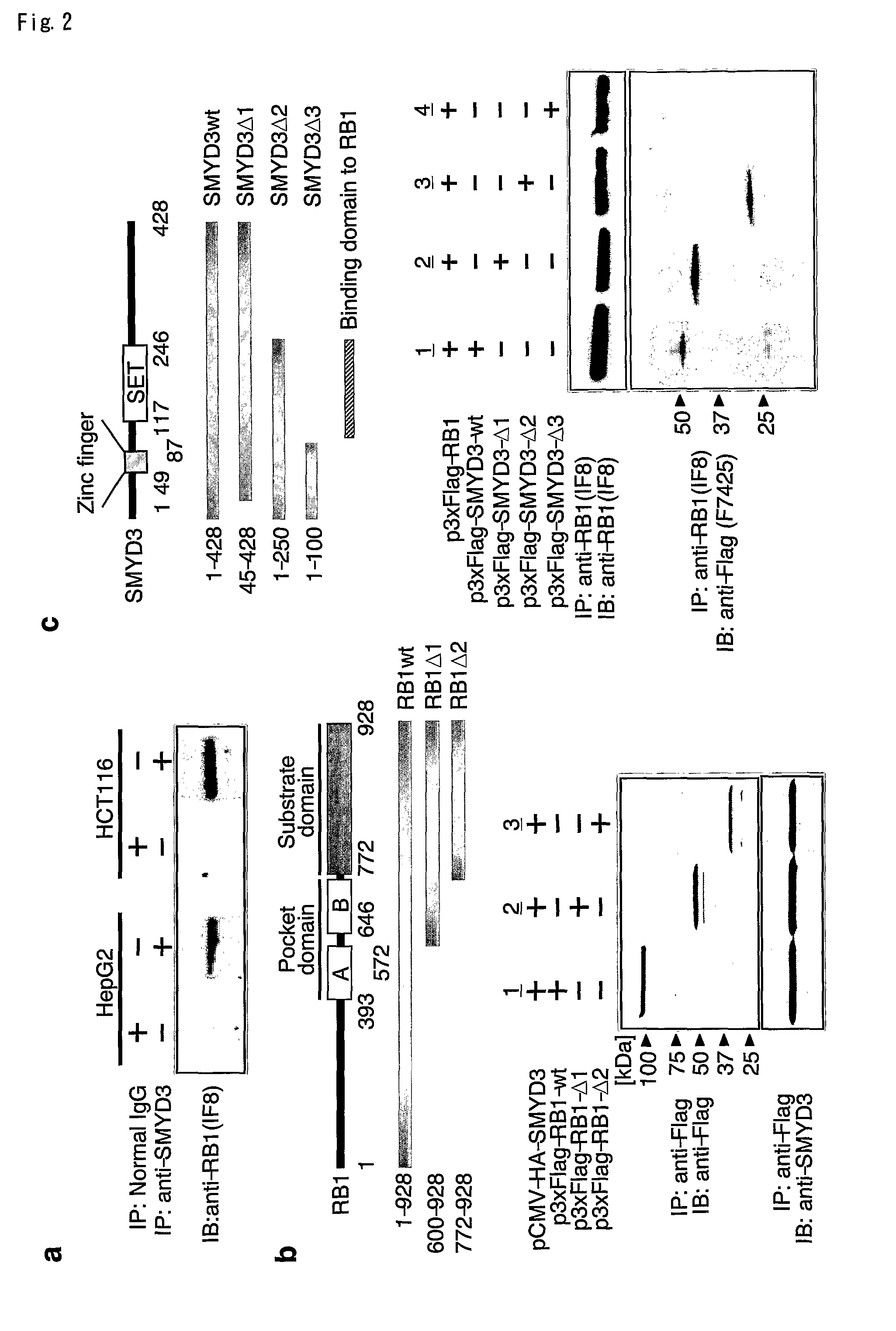
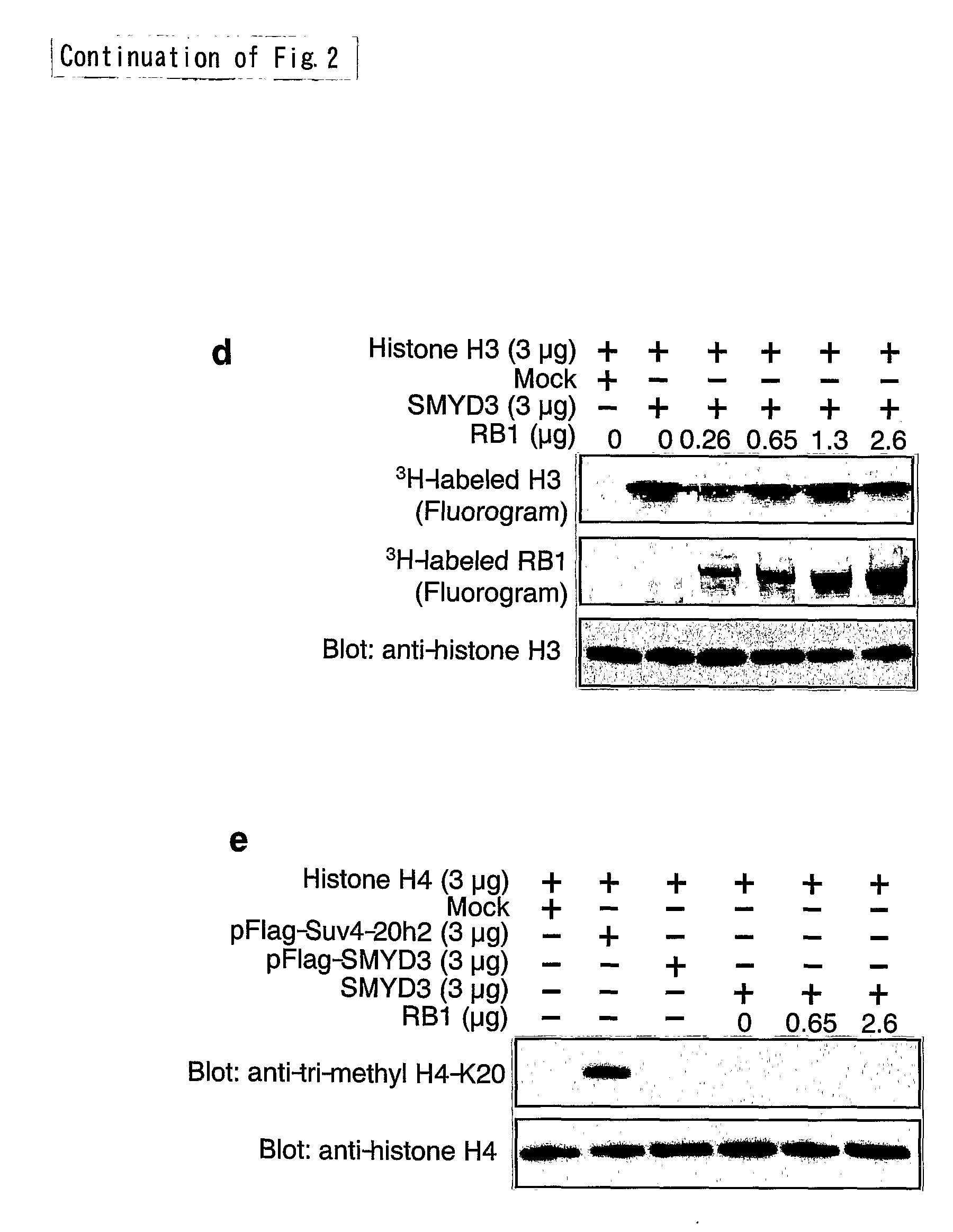
The present invention features a method for determining the methyltransferase activity of a polypeptide and screening for modulators of methyltransferase activity, more particularly for modulators of the methylation of retinoblastoma by SMYD3. The invention further provides a method or pharmaceutical composition for prevention or treating of colorectal cancer, hepatocellular carcinoma, bladder cancer and / or breast cancer using a modulator so identified. N-terminal truncated forms of SMYD3 (alias ZNFN3A1) have higher methylating activity. Lys 824 is a preferred methylation site on the RB1 protein for SMYD3.
Owner:ONCOTHERAPY SCI INC
Method and kit for quantitatively detecting methylation degree of DNA specific site
ActiveCN102321745AGuaranteed accuracyEliminate distractionsMicrobiological testing/measurementCpG siteA-DNA
The invention provides a method and kit for quantitatively detecting the methylation degree of a DNA specific site. The method provided by the invention comprises the following steps: 1) designing a PCR primer in accordance with a DNA sequence to be detected, and using the PCR primer for amplifying the DNA to be detected; 2) designing a pair of TaqMan probes in accordance with a CpG site-enriched region in the PCR product obtained by amplification in the step 1), wherein one probe is a CG probe designed in accordance with the DNA of which the CpG site is completely methylated, and the other probe is a TG probe designed in accordance with the DNA of which the CpG site is completely non-methylated and is used for specifically combining the PCR product and detecting the PCR product; and 3) carrying out real-time quantitative PCR based on the primer designed in the step 1) and the probes designed in the step 2), thus detecting the methylation degree of the DNA to be detected. By using the method, a specific methylated site can be detected, and the methylation states of other sites can not affect the detection result. The kit provided by the invention can be used for detecting the methylation degree of a DNA specific site.
Owner:BIOCHAIN BEIJING SCI & TECH
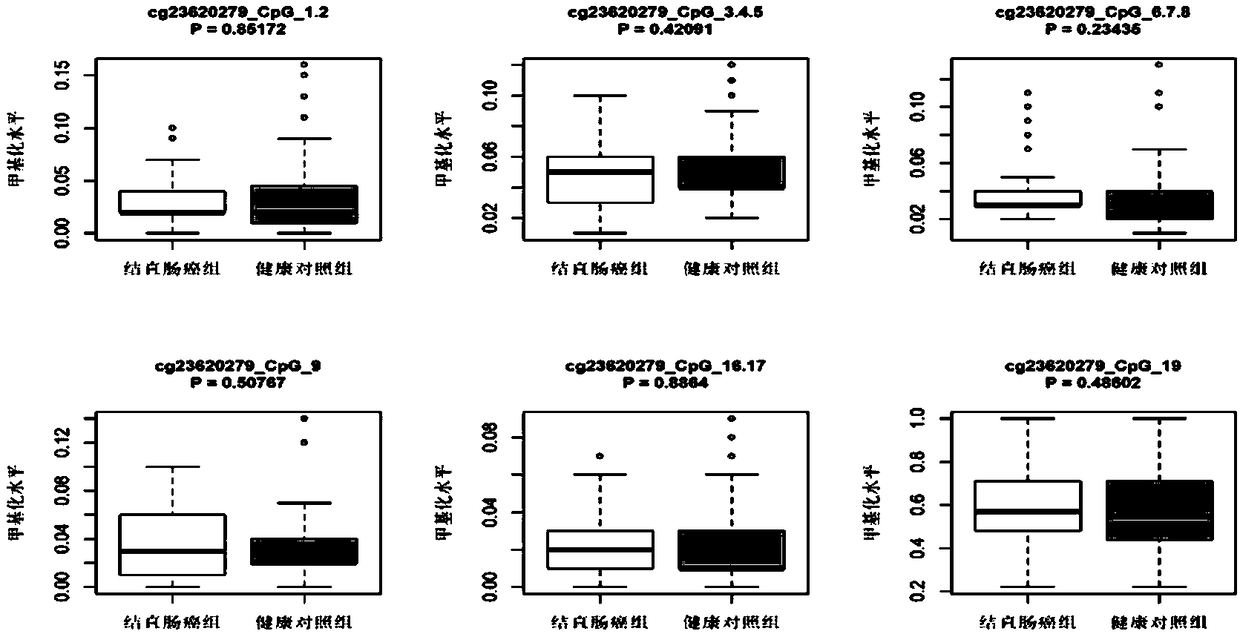
Method, marker, and kit used for colorectal cancer diagnosis, screening, and risk prediction
ActiveCN108315418AEasy to detectLow costMicrobiological testing/measurementDNA/RNA fragmentationCpG siteComputer module
The invention discloses a DNA methylation marker used for diagnosis, screening, and prediction of male colorectal cancer. The DNA methylation marker is obtained via methylation of a combination of thefirst groups, or the first group with the second group, or the third groups, (1), +22, +27, and +29; (2), -24,-18, +89, and +92; and (3) +136; of three CpG sites in the front and back sequence of gene RPS24 transcription start site. The sequence number is based on sequence number in SEQ ID NO.1. The 143th site transcription start site A is recorded as +1. The invention also discloses a probe, a method, and a kit used for detecting the DNA methylation marker, and a computer module used for prediction of male colorectal cancer risk using the data of the DNA methylation sites.
Owner:太科航天智能康养技术(深圳)有限责任公司
Kit and method for detecting methylation level of liver cancer risk gene TSPYL5
ActiveCN104762301AImprove detection efficiencyImprove throughputMicrobiological testing/measurementDNA/RNA fragmentationMedicineOncology
The invention discloses a kit and a method for detecting the methylation level of liver cancer risk gene TSPYL5, and belongs to the field of biotechnologies. The kit for detecting the methylation level of the TSPYL5 comprises a primer pair A used for an MSRE-qPCR method or a primer pair B used for a BSP method, wherein the primer sequence is shown in SEQ ID NO. 1-22. According to the kit and the method disclosed by the invention, the relevance between the methylation of the TSPYL5 gene and liver cancer occurrence is found, and the methylation loci of the TSPYL5 are authenticated; the methylation method for the TSPYL5 by virtue of fluorescent quantitative PCR is established by combining specific methylation sensitive restriction endonuclease with the specific primers; the methylation level of the TSPYL5 is detected through the kit and the method based on MSRE-qPCR, the sensitivity is up to 83.9%, and the specificity is up to 93.25%.
Owner:WUHAN UNIV
Method and kit for screening target region of methylation PCR detection and application of target region
ActiveCN108410980AComprehensive forecastReliable predictionMicrobiological testing/measurementDNA/RNA fragmentationTrue positive rateTranscriptome Sequencing
The invention discloses a method and a kit for screening a target region of methylation PCR detection and an application of the target region. The method comprises the following steps: (1) acquiring ato-be-analyzed tumor methylation chip and corresponding transcriptome sequencing data from a database; (2) calculating methylation degree values of a normal group and a cancer group, and screening methylation sites, which keep obvious differences, in the various groups; (3) in combination with analysis on a transcriptome sequencing expression profile, calculating related coefficients, and screening negatively related sites; and (4) associating the methylation candidate sites which are acquired in the step (3) with related disclosed documents, screening sites which are supported and reported by many documents, keep a high inter-group methylation difference group and are in expression amount negative relation, and on the basis of a regression algorithm, acquiring a site set, namely the target region, achieving the optimal sensitivity and specificity. According to the method, through comprehensive analysis of the database, transcriptome sequencing and the documents and in combination with multiple data filtering and the regression algorithm, the target region of methylation PCR detection can be obtained sensitively and specifically.
Owner:ENVELOPE HEALTH BIOTECHNOLOGY CO LTD
Kit for detecting methylation of lung cancer-associated gene SHOX2 (short stature homebox2)
InactiveCN106011292AHigh sensitivityImprove featuresMicrobiological testing/measurementMethylation SiteMgb probe
The invention discloses a kit for detecting methylation of a lung cancer-associated gene SHOX2 (short stature homebox2). Methylated loci of target genes in a free DNA (deoxyribonucleic acid) are detected, and the kit comprises primers corresponding to three methylated target genes and an internal reference gene primer, and preferably further comprises MGB (minor groove binder) probes corresponding to methylated loci of the three target genes, an MGB probe corresponding to an internal reference gene and MGB probes corresponding to the three methylated target genes. The detection loci comprise promoter areas of the genes and coding areas of the genes, and multi-area detection is favorable for improving the methylation accuracy and specificity of the SHOX2. The methylated loci are preferably detected more accurately by specifically combining the MGB probes and a methylation sequence. The kit is convenient to operate and easy to read, and apparatus requirements are low; a complete sealing form is adopted for the whole PCR (polymerase chain reaction) process, so that the probability of cross infection is avoided, and a result is more accurate. The kit with high detection sensitivity is applied to early screening of lung cancer.
Owner:北京鑫诺美迪基因检测技术有限公司
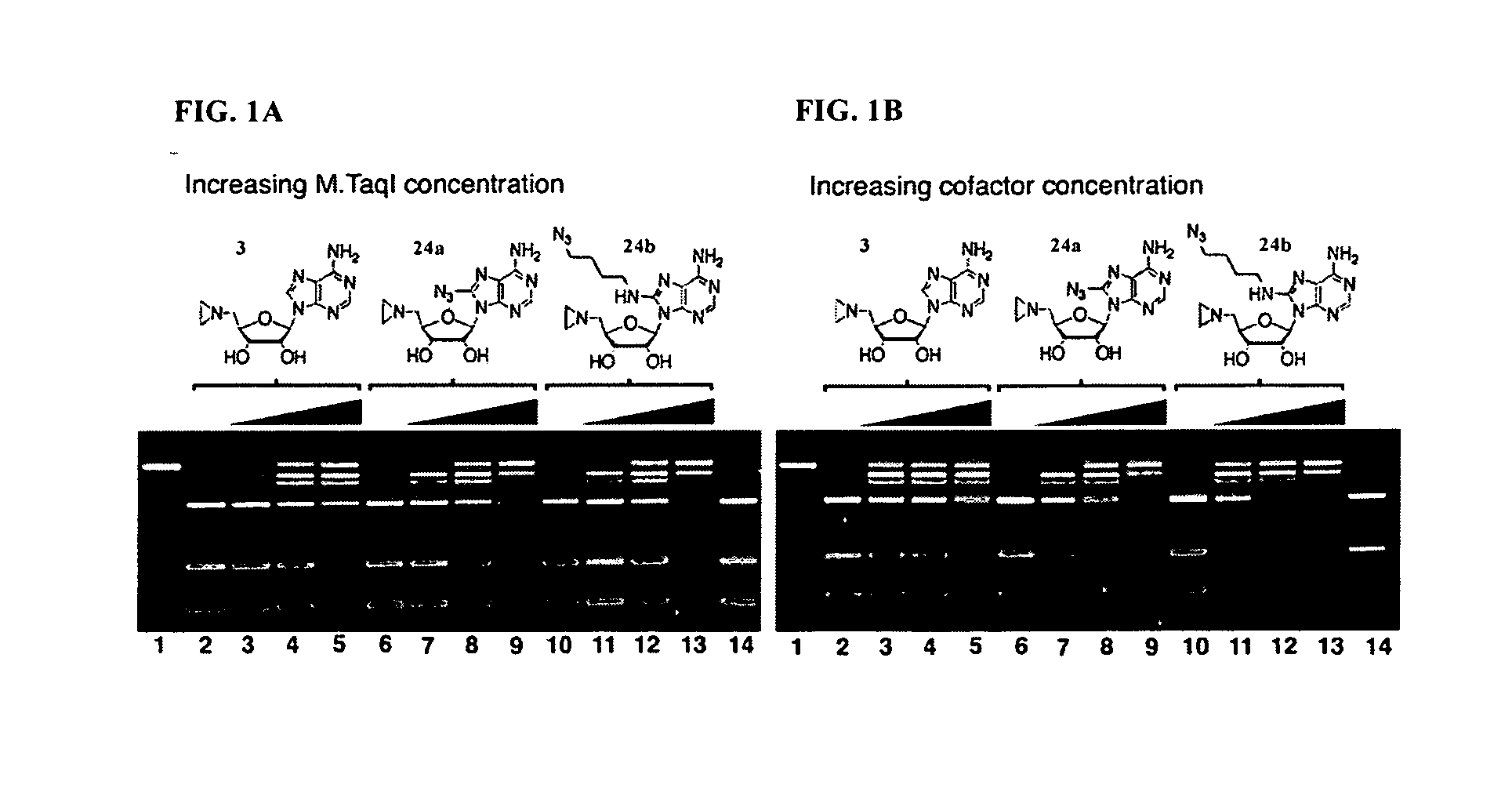
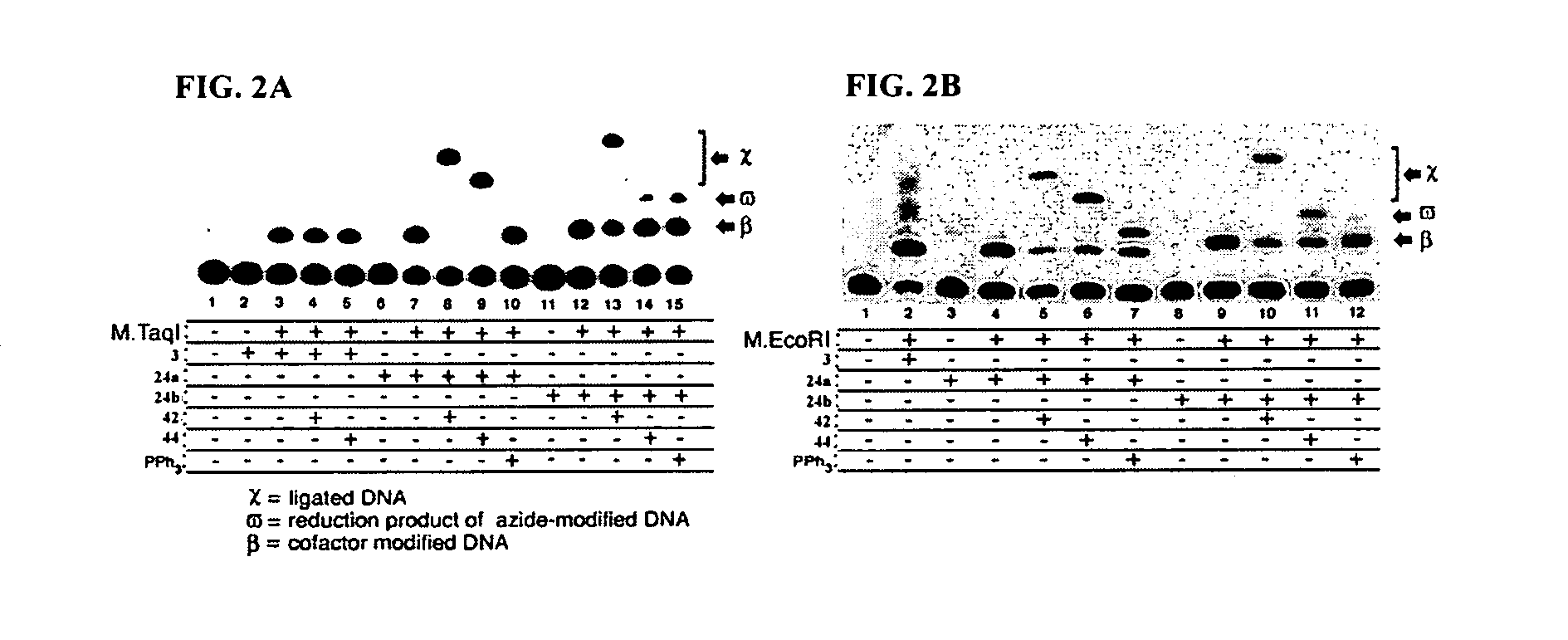
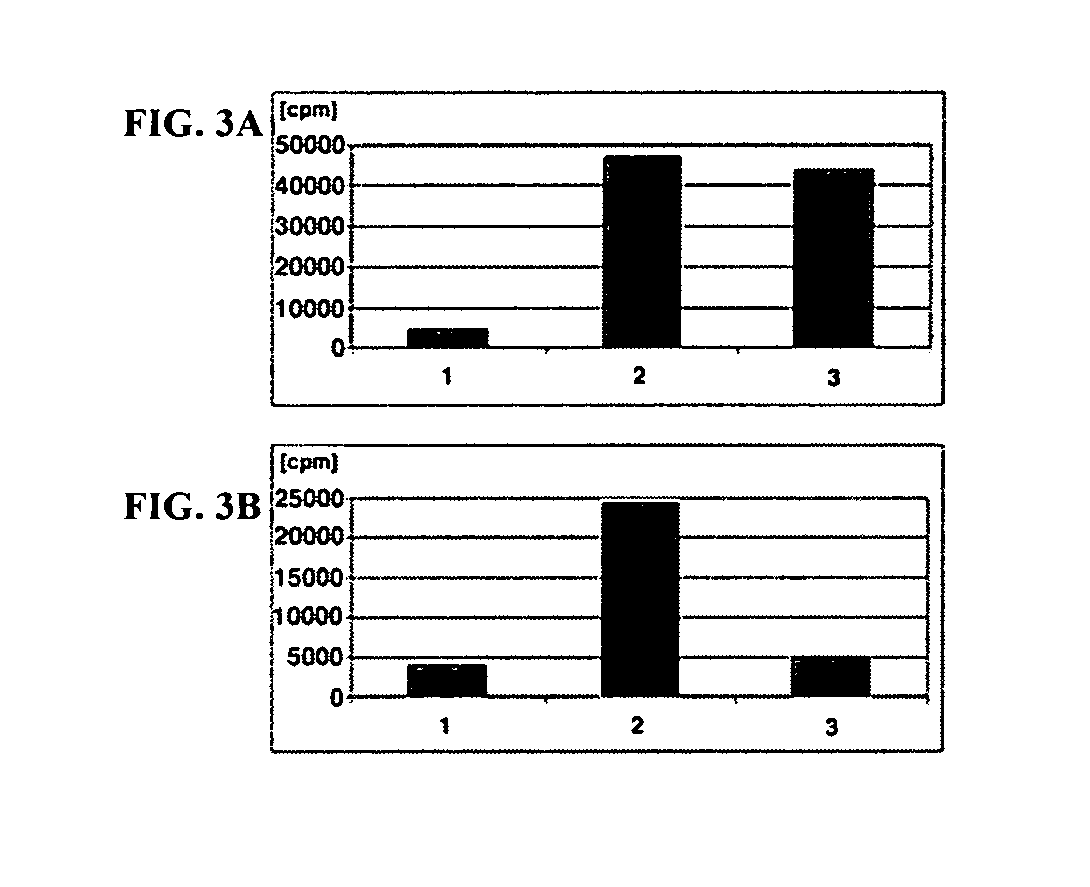
Synthetic cofactor analogs of S-adenosylmethionine as ligatable probes of biological methylation and methods for their use
InactiveUS20070161007A1Sugar derivativesMicrobiological testing/measurementS-Adenosyl methionineS-Adenosyl-l-methionine
The present invention discloses compounds and methods used to specifically target substrates of methylation by S-adenosyl-L-methionine (SAM)-dependent methyltransferases. The substrates can be peptides, single stranded nucleic acids or double stranded nucleic acids, including RNA, DNA and PNA or phospholipids. The compounds disclosed are SAM analogs that are ligated to a methylation site by the methyltransferase. Also disclosed, are reacting groups that are ligatable to the cofactor analogs and can also be used as detectable labels. The reacting group can be used to cleave the substrate providing a methylation footprint. The invention can be used clinically to determine methylation state of a gene or gene promoter such as those involved in imprinting and transcription. In some preferred embodiments, the invention includes a kit, which can include one or more suitable SAM analogs and may include one or more detectable labels. In other preferred embodiments, the invention includes a pharmaceutical composition.
Owner:WISCONSIN ALUMNI RES FOUND
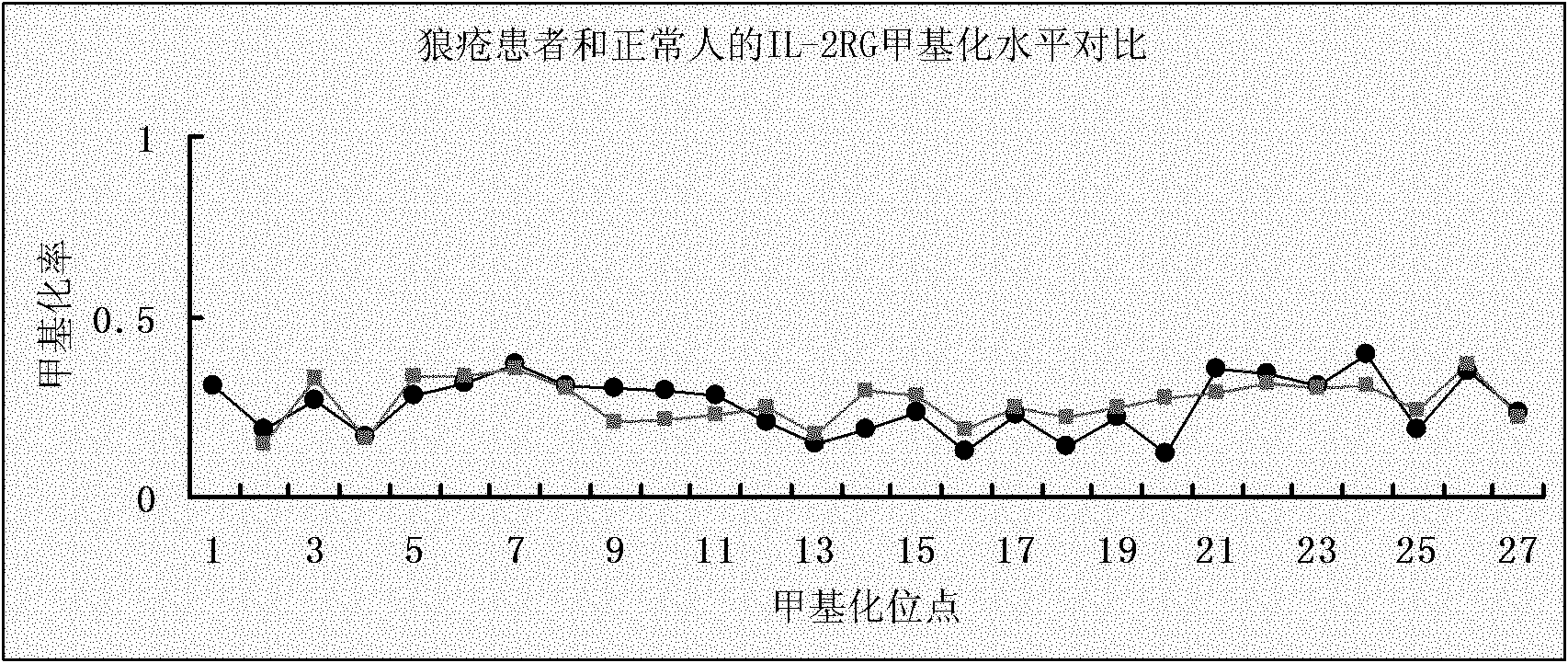
Methylated marker IL-2RG in systemic lupus erythematosus (SLE) whole blood genome and application thereof
InactiveCN102140511AImprove diagnostic efficiencyReduce diagnostic costsMicrobiological testing/measurementCurative effectDisease activity
The invention discloses a methylated marker IL-2RG in a systemic lupus erythematosus (SLE) whole blood genome and application thereof. The sequence of the marker is shown as SEQ ID NO:1. The methylated marker IL-2RG in the SLE whole blood genome is applied to the preparation of a reagent for diagnosing SLE, in particular to a kit for diagnosing SLE. By detecting the methylation level of the methylated marker IL-2RG, a specific methylation locus with a great meaning and a remarkable difference is counted and taken as a basis for early diagnosis of SLE. By adopting the methylated marker, an accurate, cheap and quick clinical tool for early diagnosis of SLE, the disease activity, the curative effect and the prognostic evaluation is provided.
Owner:CENT SOUTH UNIV
Nucleic acid detection assay
A method for determining the methylation status of a potential methylation site in genomic nucleic acid comprising treating genomic nucleic acid with an agent which modifies cytosine bases but does not modify 5methyl-cytosine bases under conditions to form a. modified nucleic acid template containing a potential methylation site; providing a first clamp containing a first capture sequence complementary to a region flanking one side of the potential methylation site in the modified nucleic acid template, providing a second clamp containing a second capture sequence complementary to a region flanking the other side of the potential methylation site in the modified nucleic acid template; allowing the first clamp and the second clamp to hybridise to the modified nucleic acid template; ligating the hybridised first and second clamps to form a probe spanning the potential methylation site in the modified nucleic acid template; digesting the modified nucleic acid template to obtain the probe; and detecting the probe and determining the methylation status of the potential methylation site in the modified genomic nucleic acid.
Owner:HUMAN GENETIC SIGNATURES PTY LTD
Construction method of RNA methylation sequencing library
The invention provides a construction method of an RNA methylation sequencing library. The construction method comprises the following steps: (S1) carrying out fragmentation on an RNA sample, so as to obtain fragmented RNA; (S2) carrying out co-immunoprecipitation on the fragmented RNA by virtue of an RNA methylation specific antibody, so as to obtain precipitates; (S3) digesting the precipitates by virtue of protease, so as to obtain a digestion product; (S4) extracting RNA from the digestion product, and carrying out reverse transcription on extracted RNA, so as to obtain cDNA; and (S5) constructing a sequencing library by virtue of the cDNA, so as to obtain the RNA methylation sequencing library. The digestion of protease is stronger than the competitive elution of analogues, so that the yield of RNA can be increased, the noise generated due to the introduction of methylation site analogues is avoided, the problem of high noise of constructed libraries in the prior art is solved, the library construction quality is improved, and therefore, the utilization rate of valid data output by the sequencing library is increased.
Owner:BEIJING NOVOGENE TECH CO LTD
Method for mining methylation pattern by whole genome data
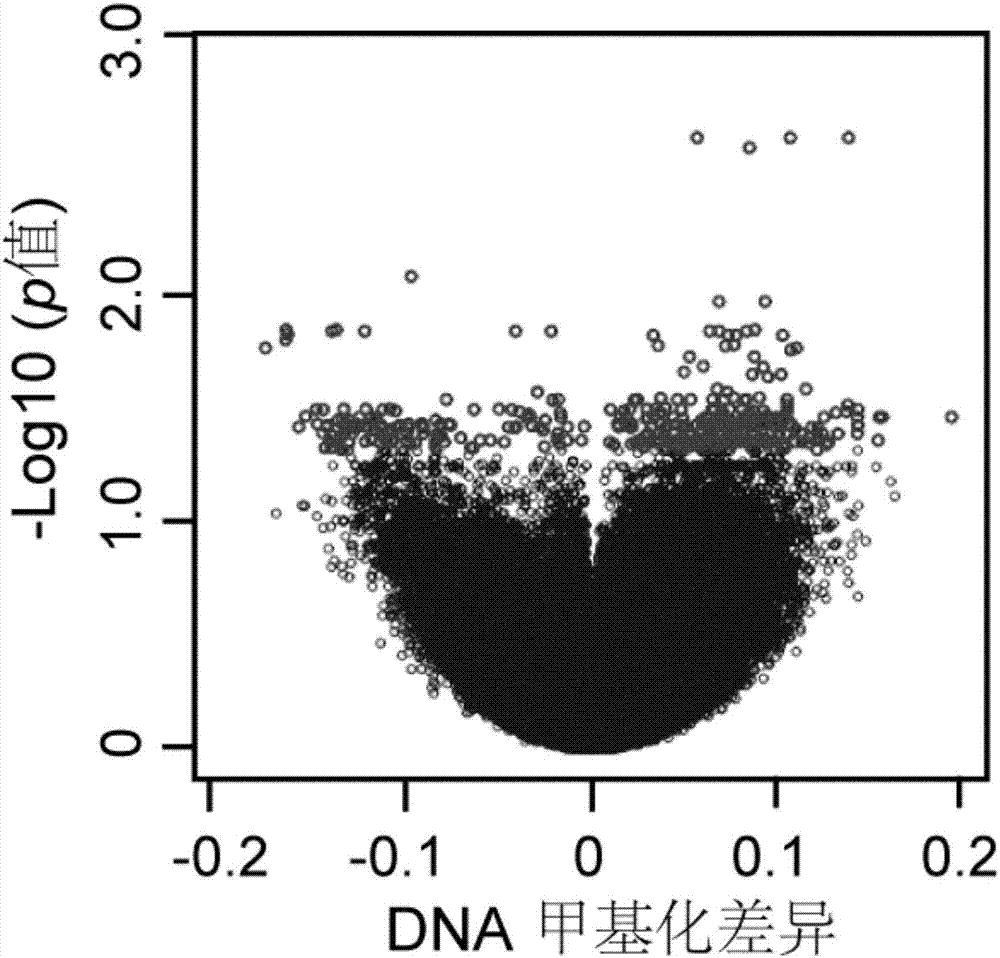
InactiveCN107301330AAddress distribution requirementsProof of validityBiostatisticsHybridisationDifferential MethylationMethylation Site
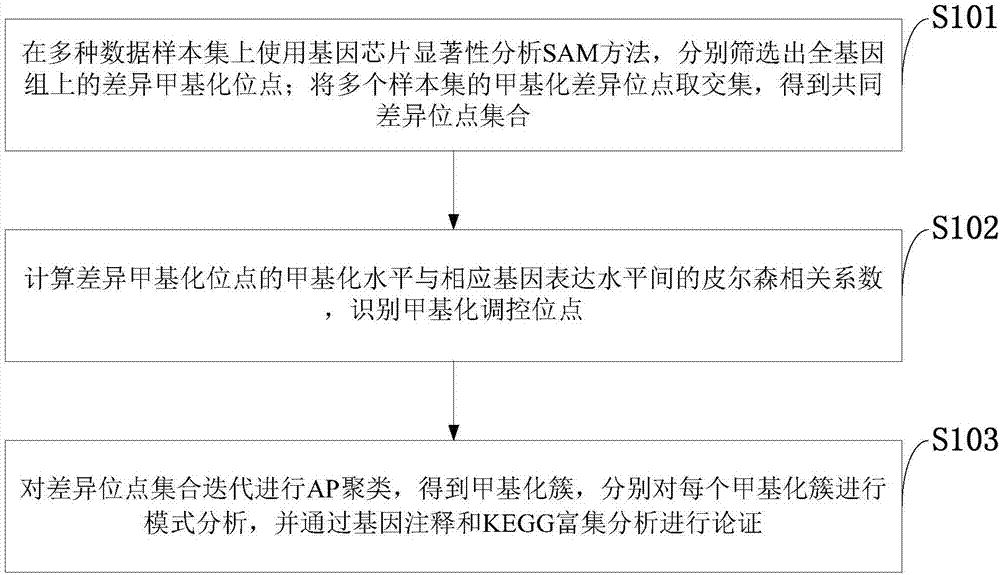
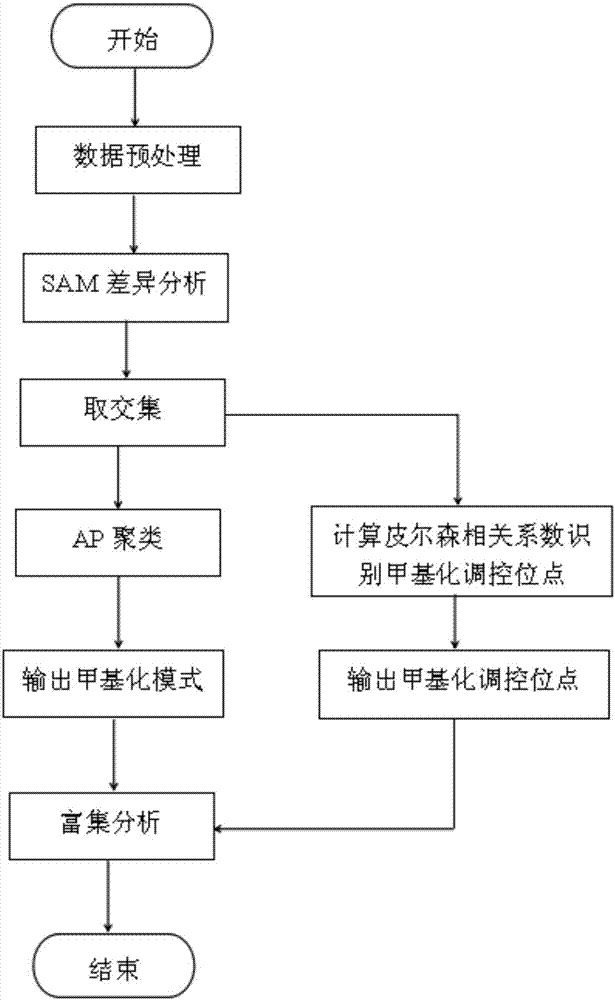
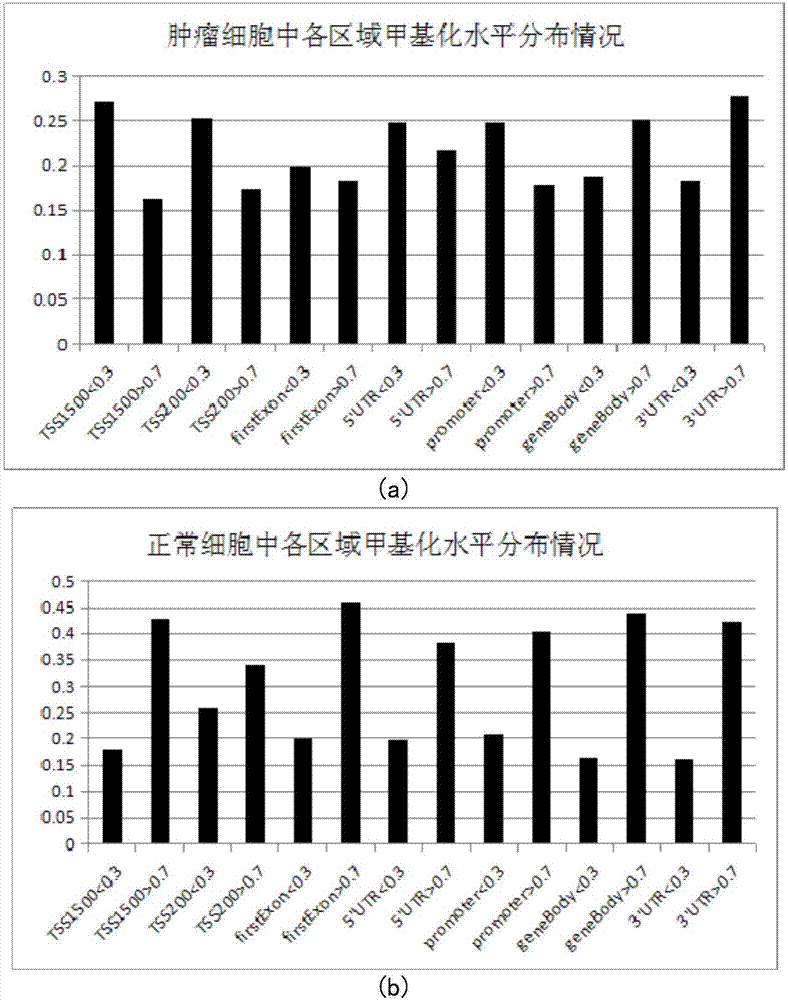
The invention belongs to the technical field of data processing of bioinformatics, and discloses a method for mining the methylation pattern by whole genome data. The method comprises the following steps that: using a SAM (Significance Analysis of Microarrays) method in various data sample sets to independently screen differential methylation sites on a whole genome; taking an intersection from the differential methylation sites of a plurality of sample sets to obtain a common difference site set; calculating a Pearson correlation coefficient between the methylation levels of the differential methylation sites and a corresponding gene expression level, and identifying a methylation regulation and control site; and carrying out AP (Affinity Propagation) clustering on the differential methylation site set to obtain a methylation cluster, independently carrying out pattern analysis on each methylation cluster, and carrying out demonstration through gene annotation and KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis. By use of the method, references can be provided by aiming at a demethylation medicine, different types of diseases have generality on a methylation pattern, and the method has a practical and clinic meaning for researching a relationship between the methylation pattern and the disease from a perspective of the whole genome.
Owner:XIDIAN UNIV
Double electrical signal and DNA circulating amplification technique-based Dam methyltransferase activity detection method
InactiveCN104833712AHigh sensitivityGood choiceMaterial electrochemical variablesEndonuclease IIISingle strand
The invention discloses a double electrical signal and DNA circulating amplification technique-based Dam methyltransferase activity detection method, and belongs to the technical field of electrochemical sensing. After methylation of hairpin-shaped DNA containing methylation sites under the effect of Dam methyltransferase and cutting by DpnI endonuclease, double-stranded DNA is formed by hybridization of released single stranded DNA and protruding parts of dicyclopentadienyl iron-DNA on the electrode surface, and then is cut into DNA fragments by endonuclease III, so that dicyclopentadienyl iron leaves away from the electrode surface so as to reduce the current, current of G-four chain body-heme complex formed by the the single stranded DNA on the electrode surface and heme is enhanced, and Dam methyltransferase activity sensitive detection can be realized by proportion of current intensity of the two.
Owner:NANCHANG UNIV
Human tumor-related methylation site as well as screening method and use thereof
ActiveCN107190076AEarly diagnosisMicrobiological testing/measurementDNA/RNA fragmentationHuman tumorScreening method
The invention provides a human tumor-related methylation site as well as a screening method and use thereof. The human tumor-related methylation site is selected from at least one methylation site on a human 18th chromosome, a human third chromosome and a human second chromosome; the change of methylation site, caused by BMI, on chromosomes is related to a human endometrial cancer; prognosis on a human tumor can be performed by detecting the methylation site changes on the chromosomes; the human tumor-related methylation site can be used as a potential prognosis marker of the tumor, and especially can be used as a prognosis marker of the human endometrial cancer.
Owner:SUZHOU INST OF BIOMEDICAL ENG & TECH CHINESE ACADEMY OF SCI +1
Application of specific methylation sites as breast cancer molecular subtype diagnosing markers
InactiveCN108676879AMicrobiological testing/measurementDNA/RNA fragmentationDiseaseBreast cancer subtype
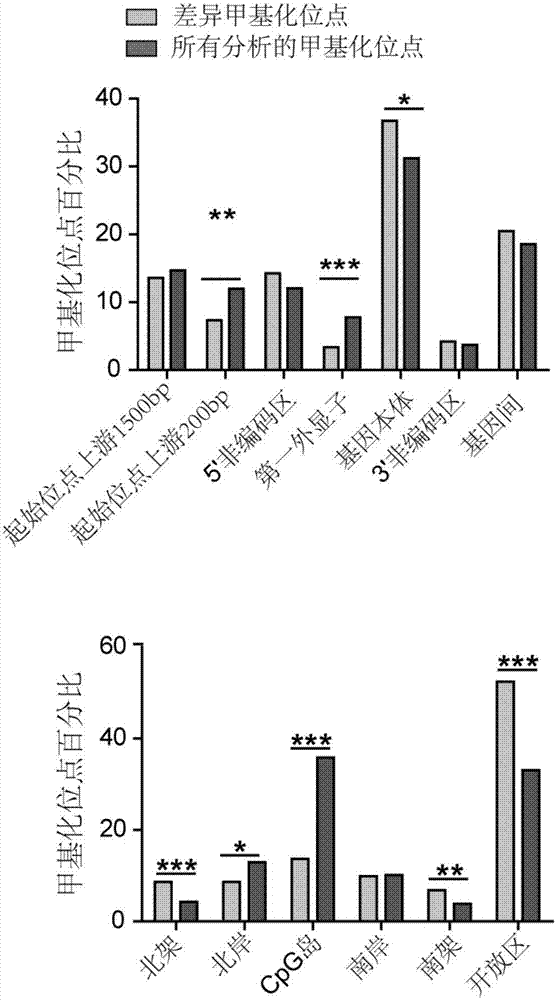
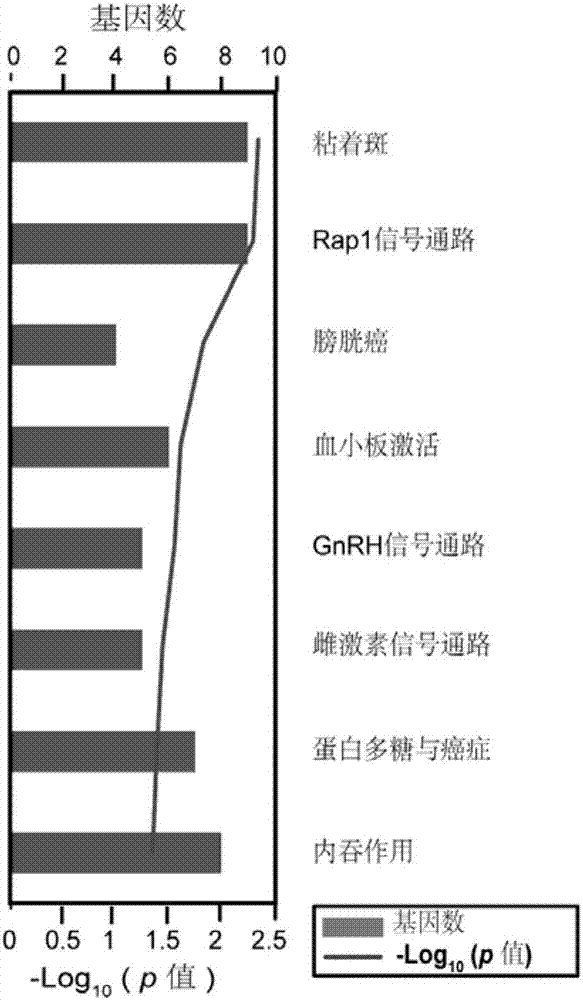
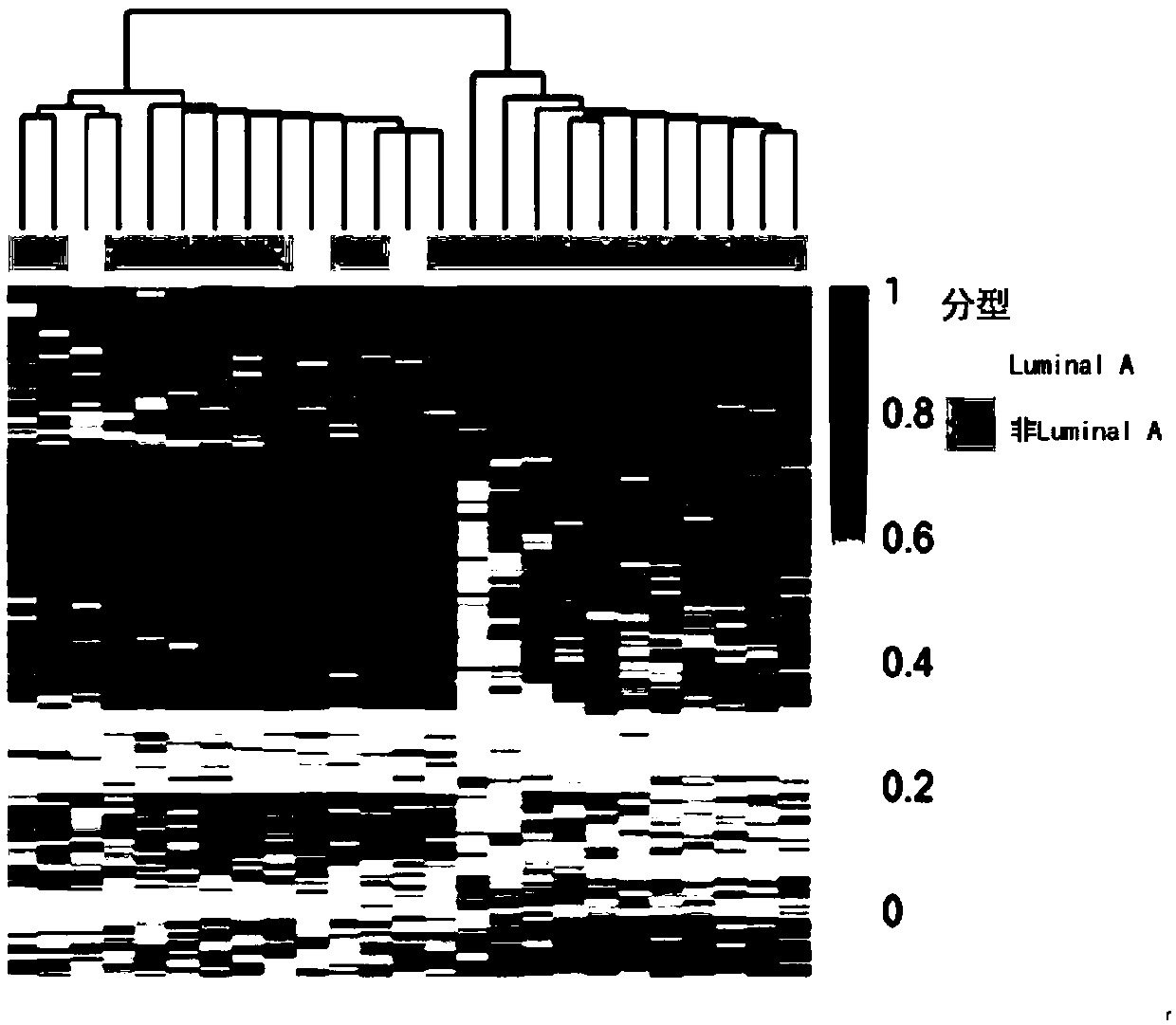
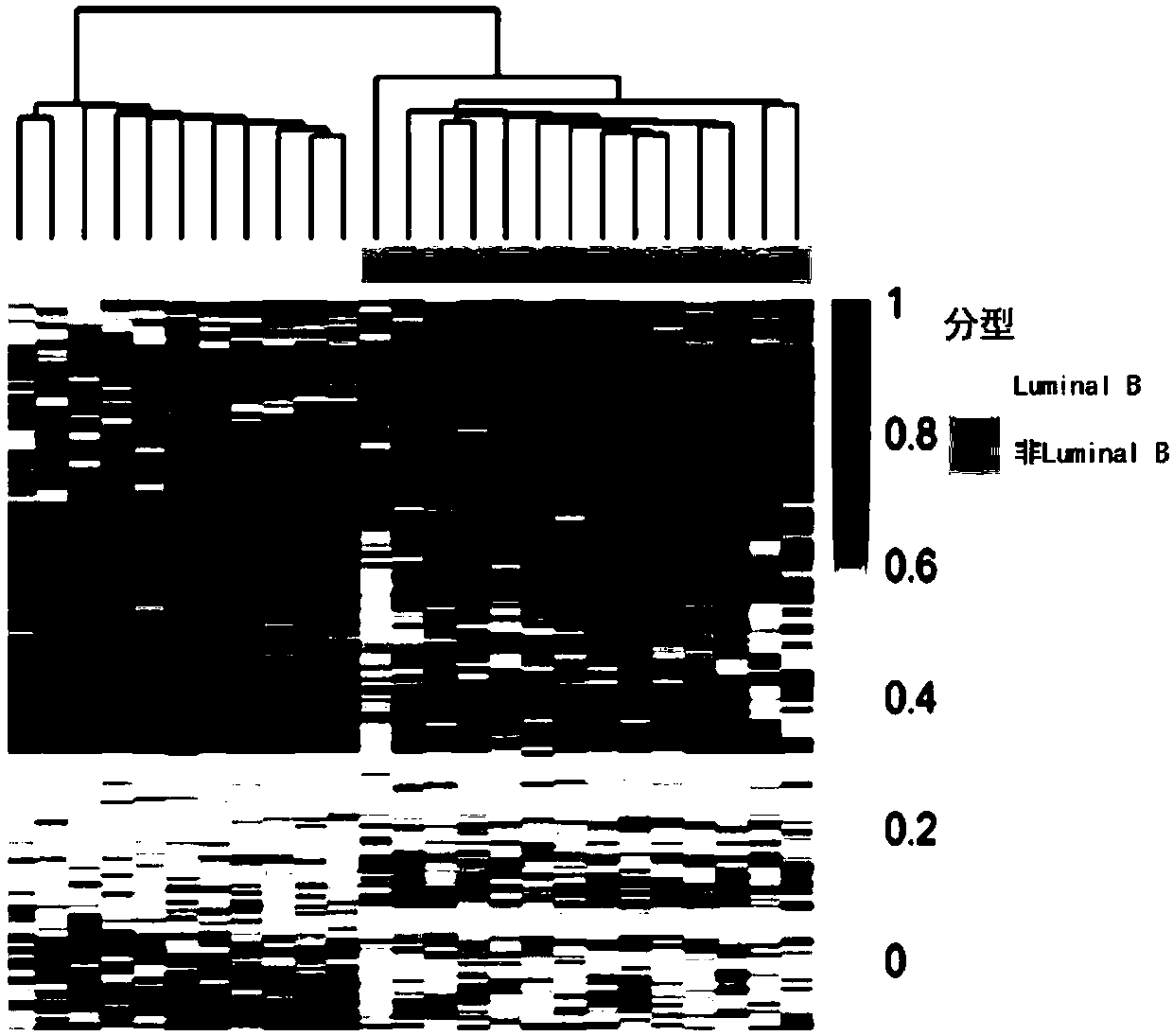
The invention discloses application of specific methylation sites as breast cancer molecular subtype diagnosing markers. The invention establishes a method and a standard for screening disease-relatedchromosomal methylation sites, and screens 40 methylation sites related to breast cancer diagnosis by using breast cancer patients as cases. The screened specific methylation sites as the breast cancer molecular subtype diagnosing markers can be applied to non-invasive detection of molecular breast cancer subtypes and diagnosis of other diseases. A kit is used for one or more of the following applications: diagnosis of the breast cancer molecular subtypes, assessment of breast cancer risks, assessment of a breast cancer treating effect, screening of breast cancer treating medicines and the like.
Owner:BEIJING INST OF GENOMICS CHINESE ACAD OF SCI CHINA NAT CENT FOR BIOINFORMATION
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com