A method for detecting tumor biomarkers using palindromic lock probes
A technology of biomarkers and padlock probes, which is applied in the field of detection of tumor biomarkers using palindromic padlock probes, can solve the problems of high temperature control and insufficient sensitivity, and achieve high sensitivity and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] In the verification process of this invention, 20 microliters is used as the reaction system, and 200 microliters is used as an example for the spectroscopic measurement. The sequence used is as follows:
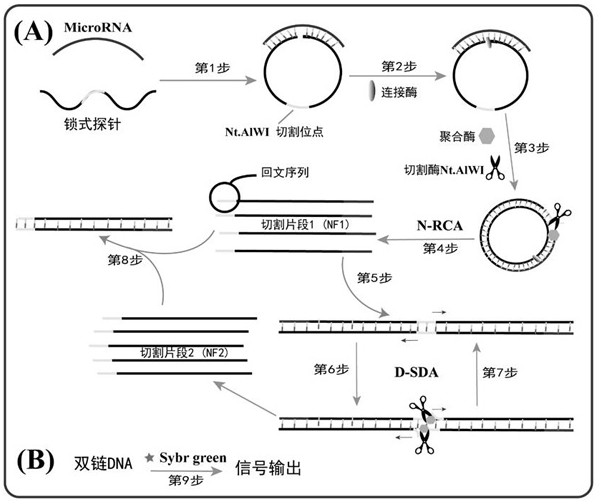
[0024] 2. Feasibility study of palindromic lock probes for miRNA detection:
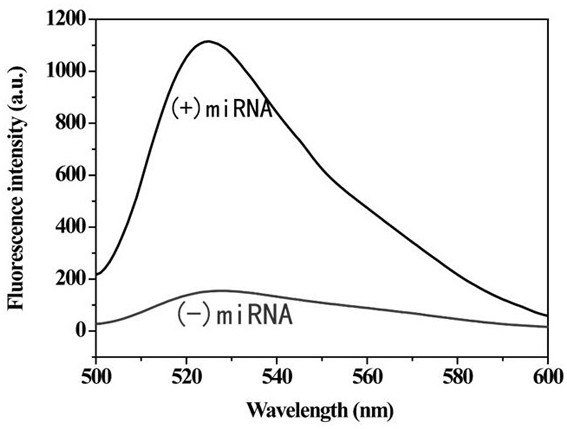
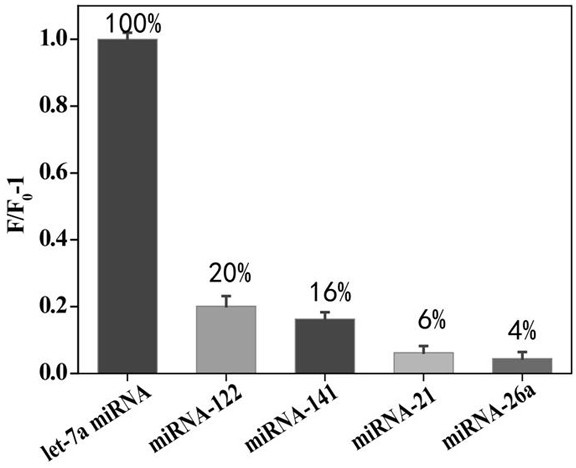
[0025] In order to prove the feasibility of this invention, we set up the experimental group (with target miRNA) and control group (without target miRNA) respectively. First, the palindromic lock probe (final concentration 1 μM) and miRNA (final concentration 1 μM) were added to T4 ligase buffer at a ratio of 1:1, and ligated at 350 U / μL T4 DNA Under the action of the enzyme, incubate at 16°C for 2 hours to allow the padlock probe to undergo a ligation reaction.
[0026] Next, add 10 mM 2 μL dNTPs, 3 U phi29 polymerase, 5 UNt.AlWI cutting enzyme, 0.5 μL 100×BSA, 2 μL 10×phi29 buffer solution and 4.7 μL secondary water to 10 μL of the above mixture, Incubate the reaction at 37°C for 2 ho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com