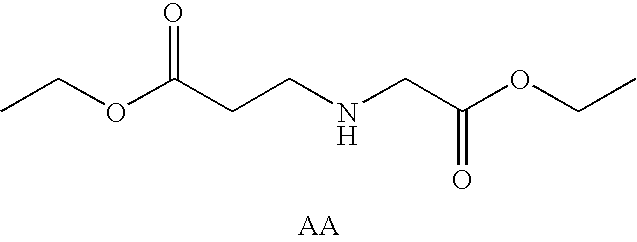
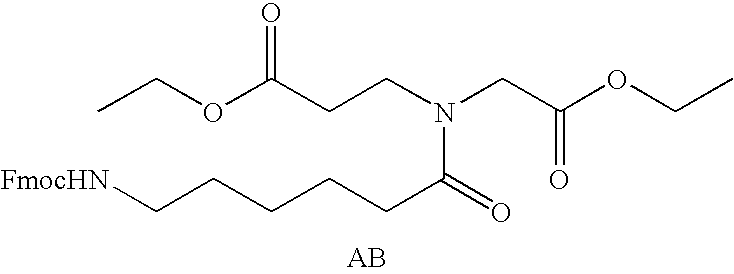
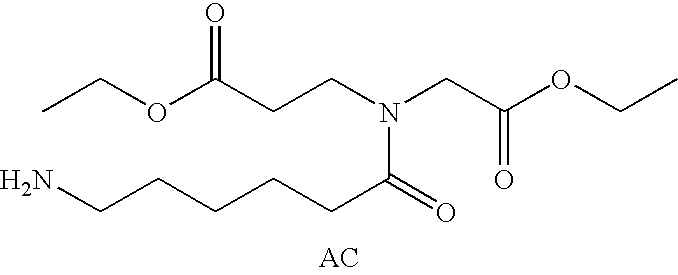
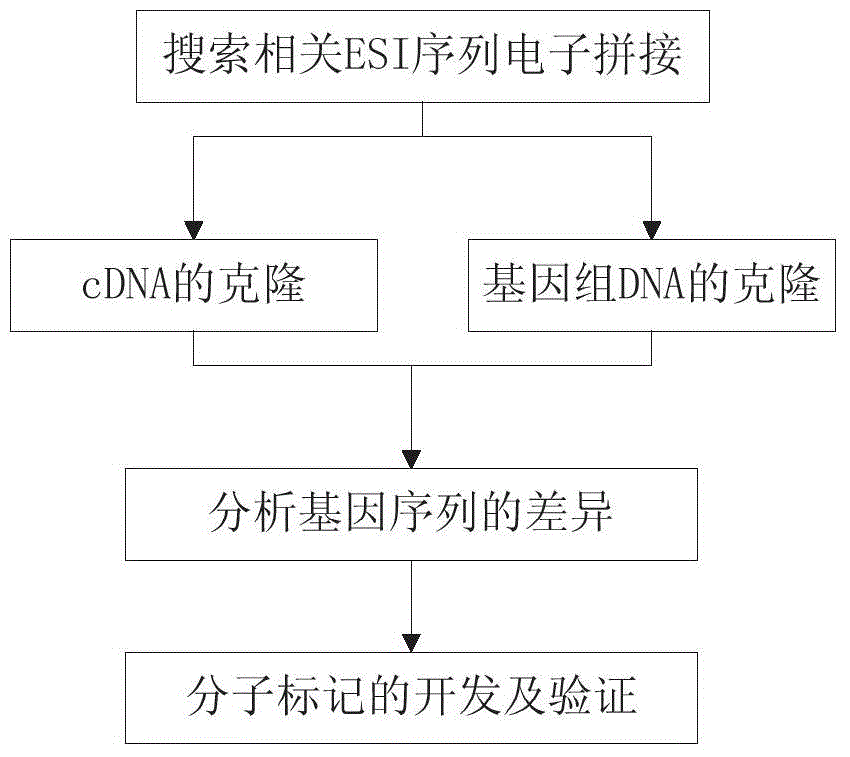
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
443 results about "Versus gene" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Compositions and methods for inhibiting expression of Eg5 gene
The invention relates to a double-stranded ribonucleic acid (dsRNA) for inhibiting the expression of the Eg5 gene (Eg5 gene), comprising an antisense strand having a nucleotide sequence which is less that 30 nucleotides in length, generally 19-25 nucleotides in length, and which is substantially complementary to at least a part of the Eg5 gene. The invention also relates to a pharmaceutical composition comprising the dsRNA together with a pharmaceutically acceptable carrier; methods for treating diseases caused by Eg5 expression and the expression of the Eg5 gene using the pharmaceutical composition; and methods for inhibiting the expression of the Eg5 gene in a cell.
Owner:ALNYLAM PHARMA INC
Modulation of gene expression by oligomers targeted to chromosomal DNA
Synthesis of a target transcript of a gene is selectively increased in a mammalian cell by contacting the cell with a polynucleotide oligomer of 12-28 bases complementary to a region within a target promoter of the gene under conditions whereby the oligomer selectively increases synthesis of the target transcript.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Methods for detection and quantification of analytes in complex mixtures
InactiveUS20070166708A1Sugar derivativesMaterial analysis by observing effect on chemical indicatorAnalyteNucleic Acid Probes
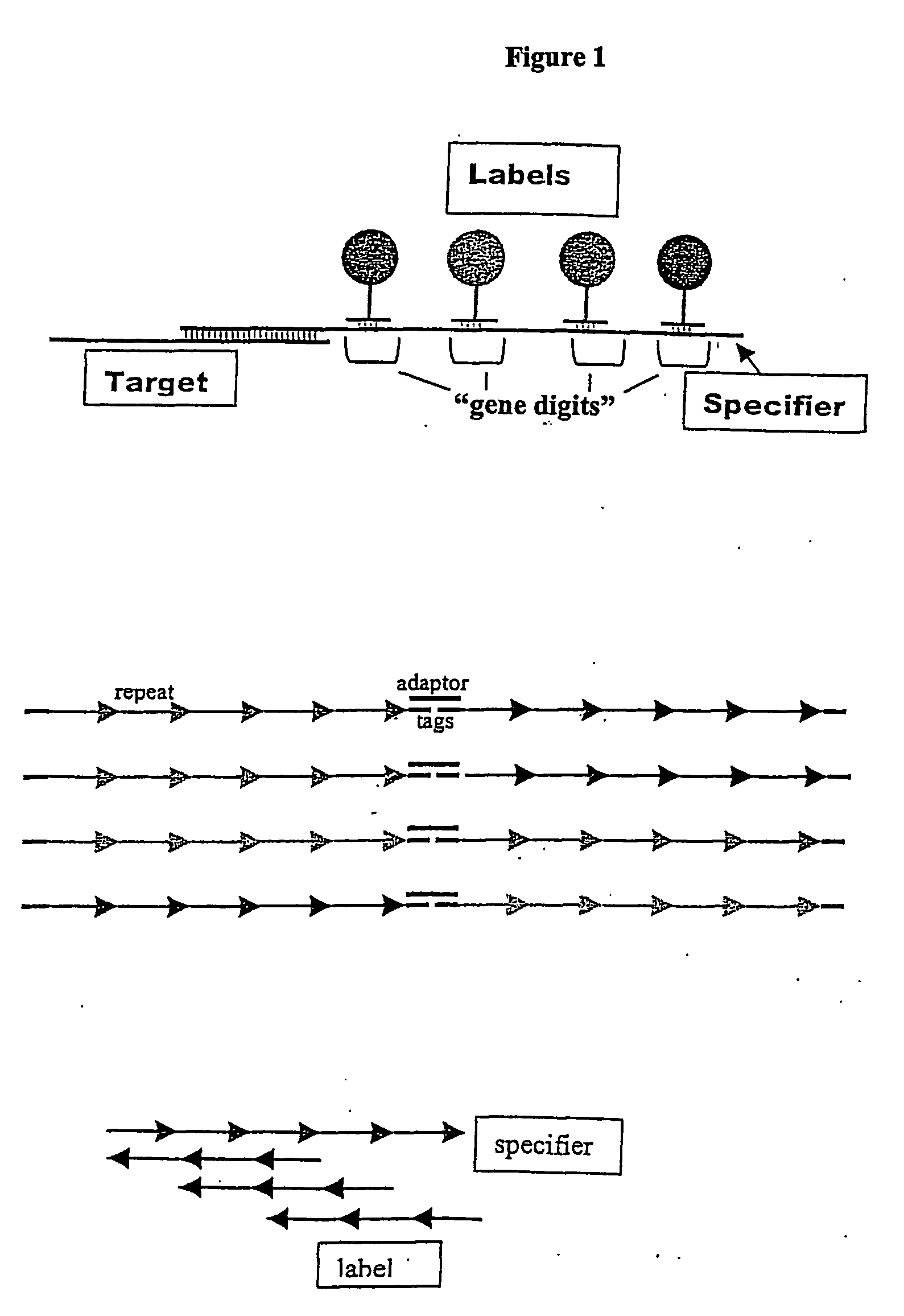
The invention provides a diverse population of uniquely labeled probes, containing about thirty or more target specific nucleic acid probes each attached to a unique label bound to a nucleic acid. Also provided is a method of producing a population of uniquely labeled nucleic acid probes. The method consists of (a) synthesizing a population of target specific nucleic acid probes each having a different specifier; (b) synthesizing a corresponding population of anti-genedigits each having a unique label, the population having a diversity sufficient to uniquely hybridize to genedigits within the specifiers, and (c) hybridizing the populations of target nucleic acid probes to the anti-genedigits, to produce a population in which each of the target specific probes is uniquely labeled. Also provided is a method of detecting a nucleic acid analyte. The method consists of (a) contacting a mixture of nucleic acid analytes under conditions sufficient for hybridization with a plurality of target specific nucleic acid probes each having a different specifier; (b) contacting the mixture under conditions sufficient for hybridization with a corresponding plurality of anti-genedigits each having a unique label, the plurality of anti-genedigits having a diversity sufficient to uniquely hybridize to genedigits within the specifiers, and (c) uniquely detecting a hybridized complex between one or more analytes in the mixture, a target specific probe, and an anti-genedigit.
Owner:INSTITUTE FOR SYSTEMS BIOLOGY
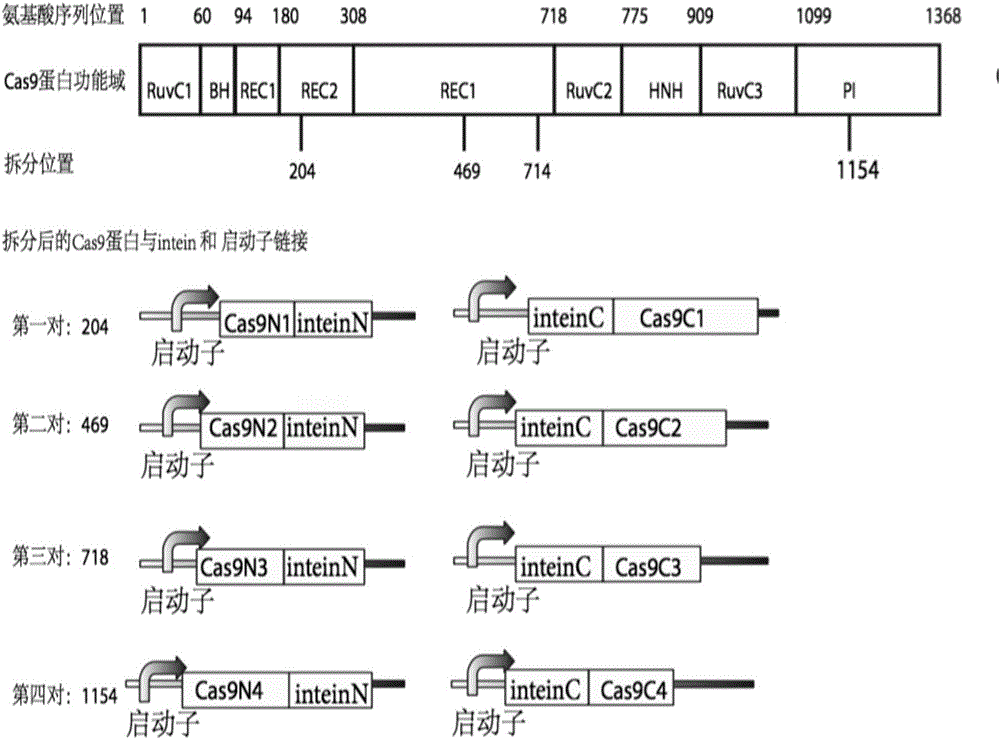
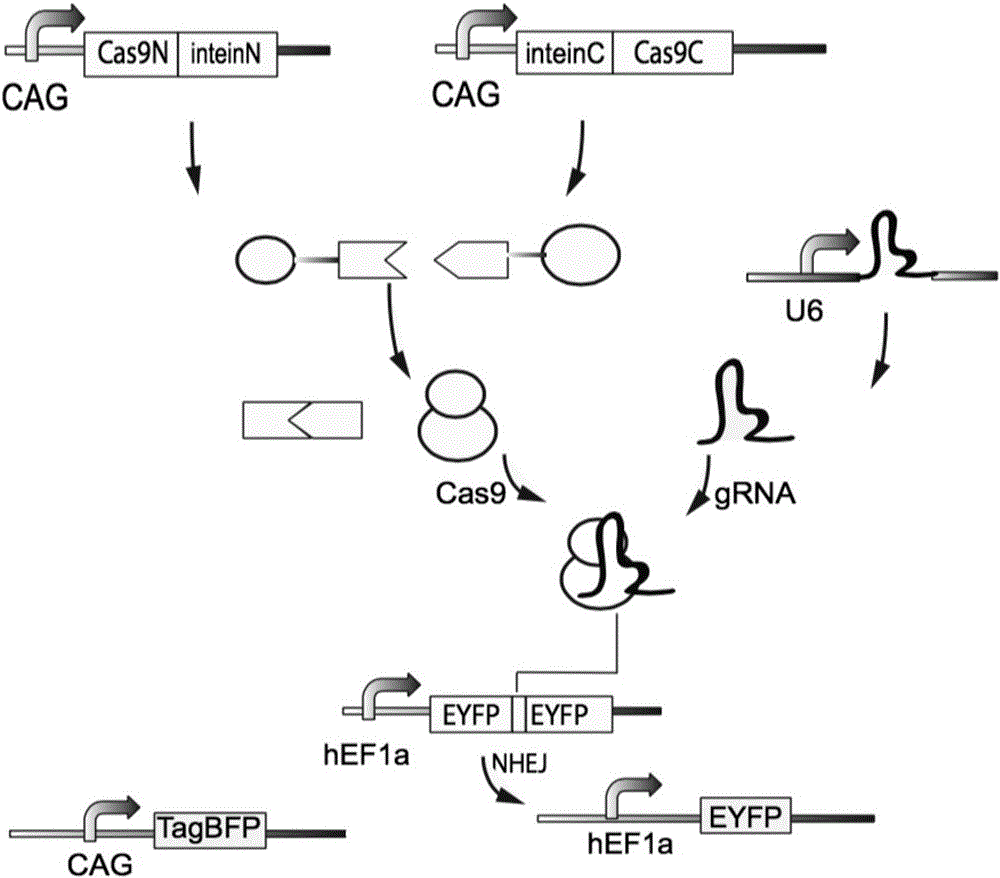
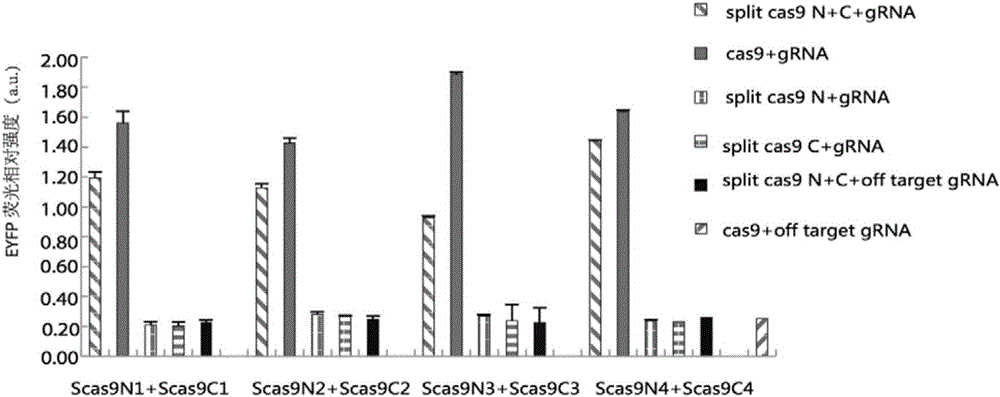
Method for carrying out gene editing and expression regulation by utilizing Cas splitting system
Owner:TSINGHUA UNIV
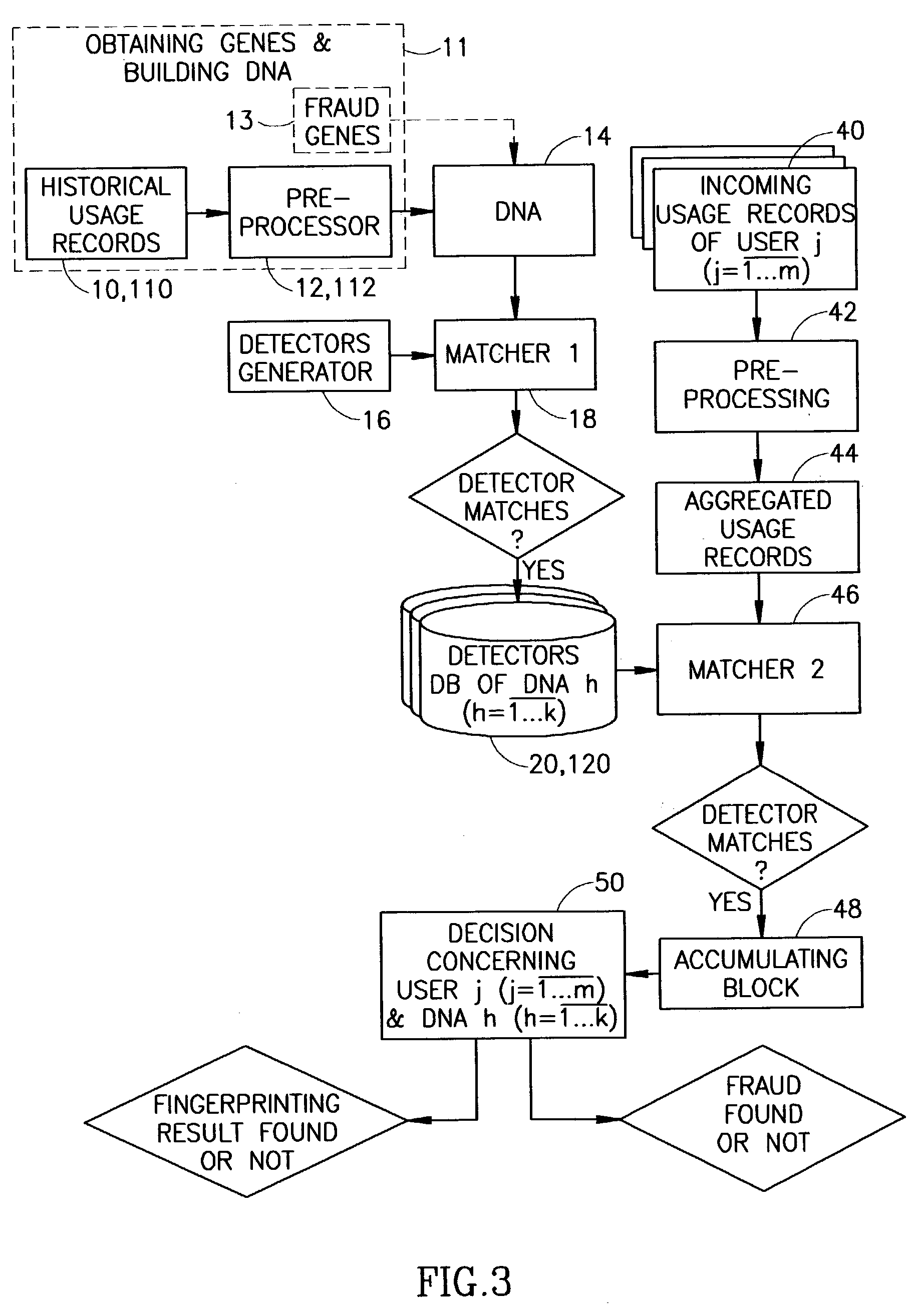
Method for detecting a behavior of interest in telecommunication networks
ActiveUS20030110385A1Digital data processing detailsUser identity/authority verificationTelecom networkVersus gene
The application describes a method for detecting a behavior of interest in a surrounding including at least one telecommunication network, by using approach of Immune Engineering. The method comprises steps of analyzing the behavior of interest by building a characterizing data string called DNA which comprises two or more data sub-strings characterizing fragments of the behavior of interest and called genes. Further, there are selected two or more data fragments called detectors and being substantially close to the genes, and the detectors are applied to identify similar to them data fragments if appearing in information concerning real time activity in the surrounding. Based on the identification results, the method detects whether the behavior of interest is present in the newly incoming information.
Owner:AMDOCS DEV LTD
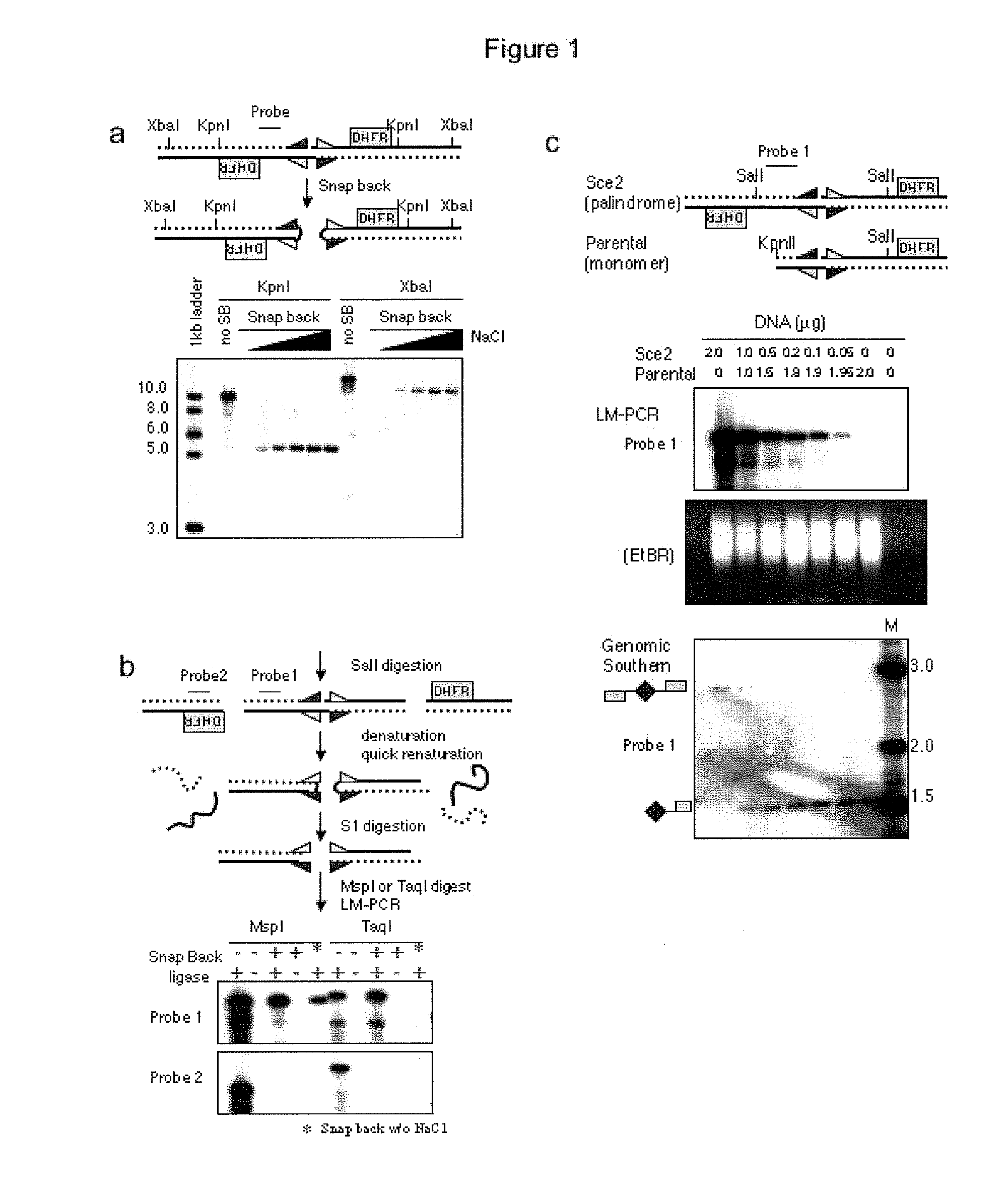
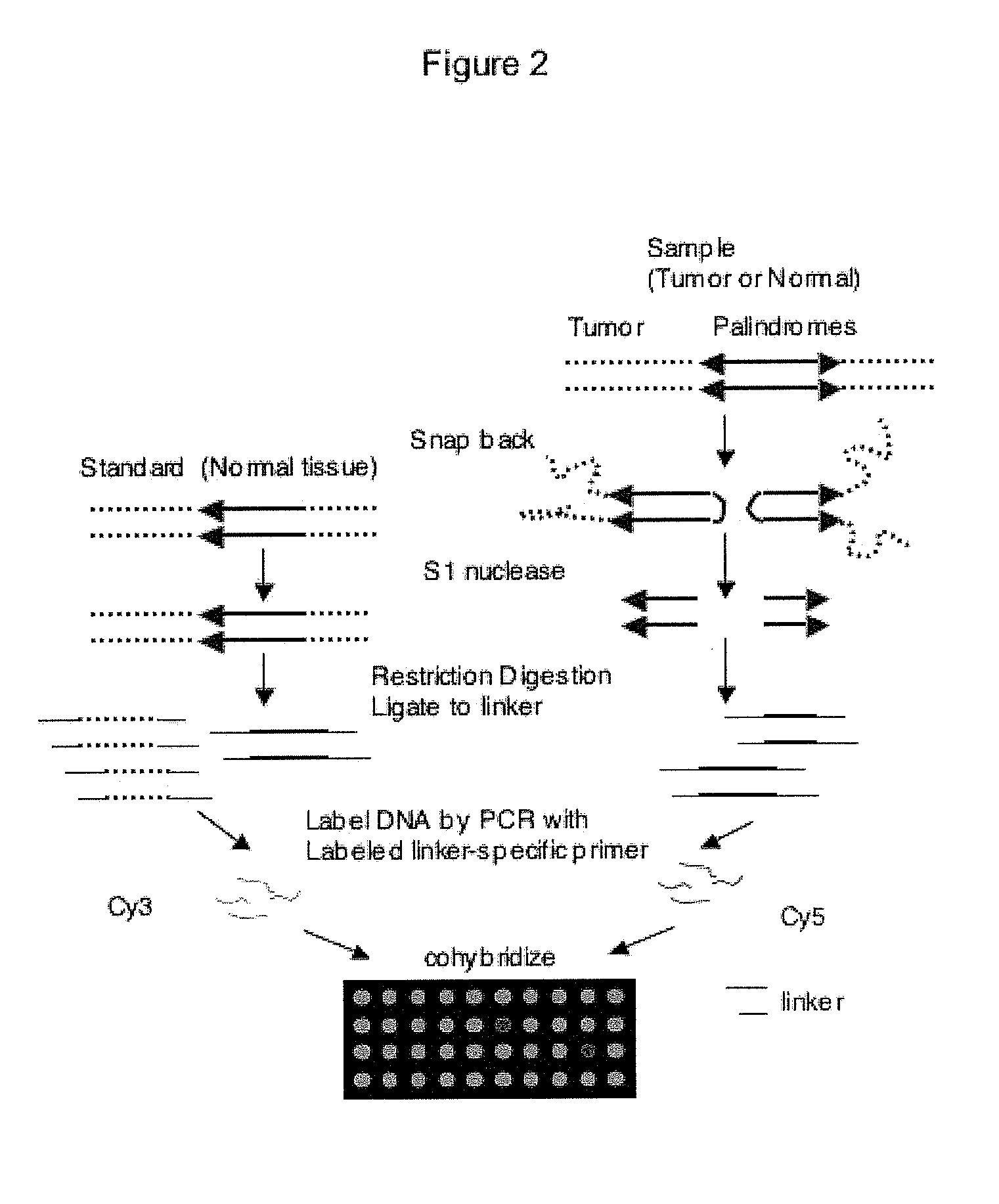
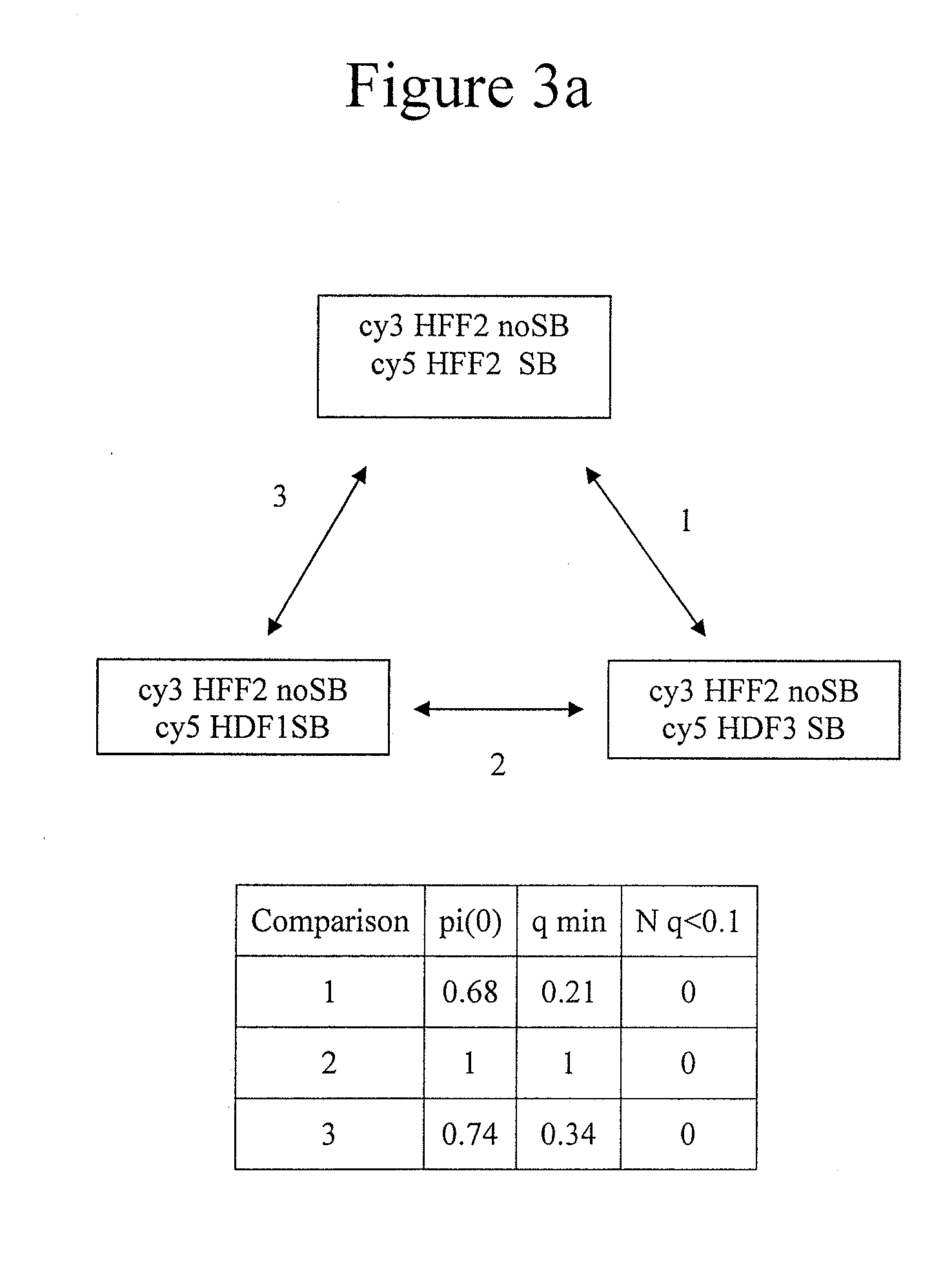
Genome-wide analysis of palindrome formation and DNA methylation
The present disclosure provides methods for detecting the genome-wide presence of methylated DNA and palindrome formation. The present disclosure also provides methods for specific enrichment of methylated DNA or DNA having a DNA palindrome. These methods have demonstrated that somatic palindromes and methylated DNA occur frequently and are widespread in human cancers. Individual tumor types have a characteristic non-random distribution of palindromes in their genome and a small subset of the palindromic loci are associate with gene amplification. The disclosed method can be used to define the plurality of genomic DNA palindromes and regions having methylated DNA associated with various tumor types and can provide methods for the classification of tumors, and the diagnosis, early detection of cancer as well as the monitoring of disease recurrence and assessment of residual disease.
Owner:FRED HUTCHINSON CANCER RES CENT
Diagnosis of diseases associated with gene regulation
InactiveUS20030082609A1Bioreactor/fermenter combinationsOrganic active ingredientsOligonucleotideDisease
The present invention relates to the chemically modified genomic sequences of genes associated with gene regulation, to oligonucleotides and / or PNA-oligomers for detecting the cytosine methylation state of genes associated with gene regulation which are directed against the sequence, as well as to a method for ascertaining genetic and / or epigenetic parameters of genes associated with gene regulation.
Owner:EPIGENOMICS AG
Double-probe gene mutation detecting method based on allele special amplification as well as special chip and kit thereof
ActiveCN101619352AEasy to detectAvoid the hassle of markingMicrobiological testing/measurementGenotypeOligonucleotide
The invention relates to a method for identifying gene mutation types in the field of gene analysis as well as a special chip and a kit thereof. The gene mutation detecting method comprises the following steps: taking a genome to be detected from a human tissue as a template, carrying out multiple allele special PCR amplification by a primer group that is designed aiming at special mutant sites and DNA polymerase without 3'-5' end exonclease activity, then hybridizing the obtained PCR product and an oligonucleotide probe (allele special probe) on the gene chip, and confirming mutation types of all gene sites according to the hybridizing result. The allele special probe is designed aiming at special gene types of gene mutant sites to be detected. The invention can detect gene mutations in comprehensive, systemic and high-flux ways and has light environmental pollution as well as simple and rapid operation compared with PCR-RFLP and a sequencing method.
Owner:CENT SOUTH UNIV
Methods and composition related to in vivo imaging of gene expression
InactiveCN101171337AVectorsCell receptors/surface-antigens/surface-determinantsMethod of imagesIn vivo
Disclosed are nucleic acid sequences encoding a recombinant seven transmembrane G-protein associated receptor (GPCR) amino acid sequence operatively coupled to a tissue-selective promoter sequence. Also disclosed are methods of imaging or treating cells in a subject that involve introducing a nucleic acid encoding a reporter detectable in vivo using non-invasive methods operatively coupled to a tissue-selective promoter or amplified tissue specific promoter to the subject, and subjecting the subject to a non-invasive imaging technique or a therapeutic that selectively interacts with the reporter or gene of interest. For example, the subject may be a subject with cancer, and the therapeutic may be an anti-cancer agent. In particular, the hTERT promoter or amplified hTERT promoter drives expression of a reporter such hSSTR2 and / or hSSTR2 derivative) and these may be linked to a gene of interest. Imaging may be performed by a variety of methods including nuclear medicine.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Constructs for enhancement of gene expression in muscle
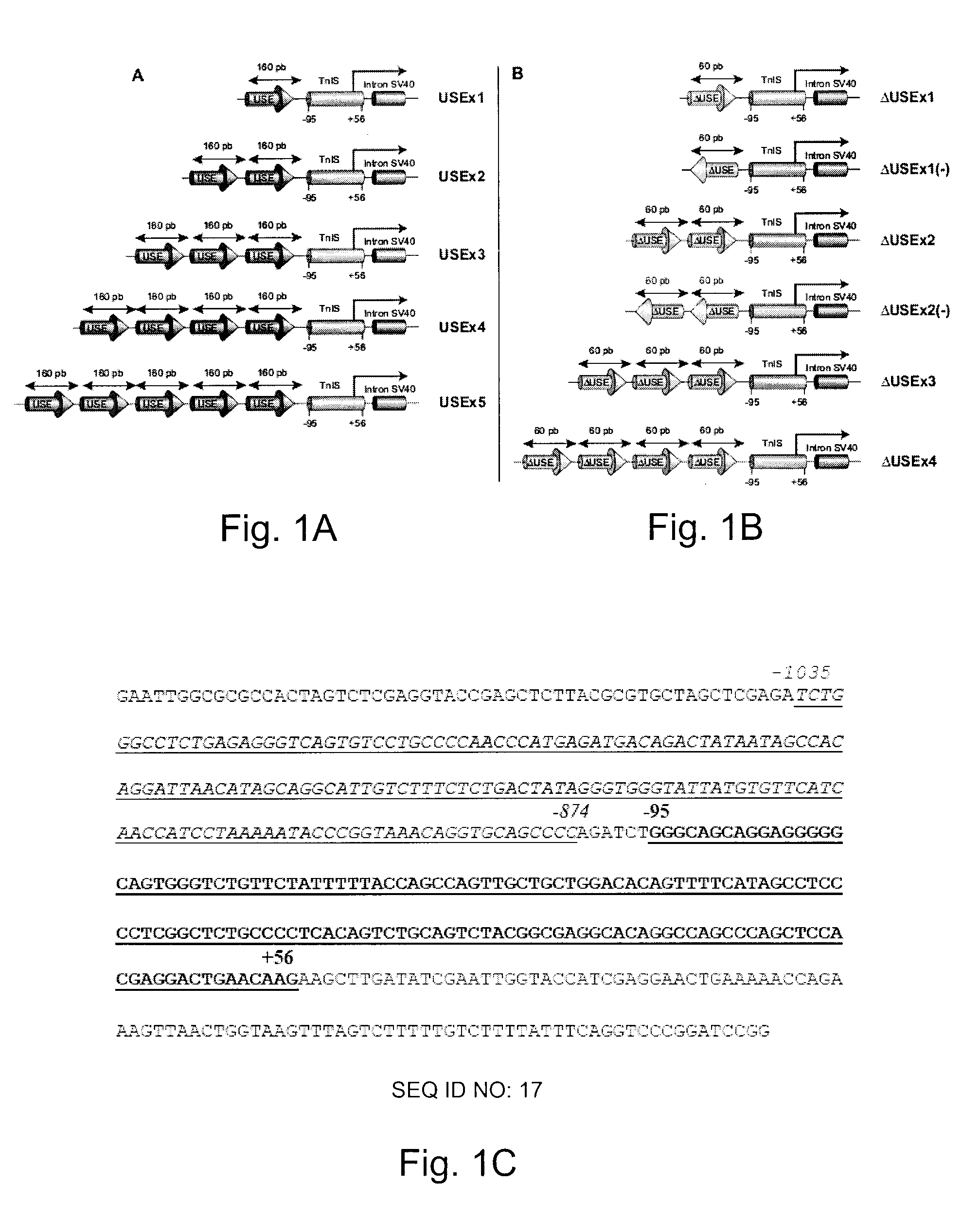
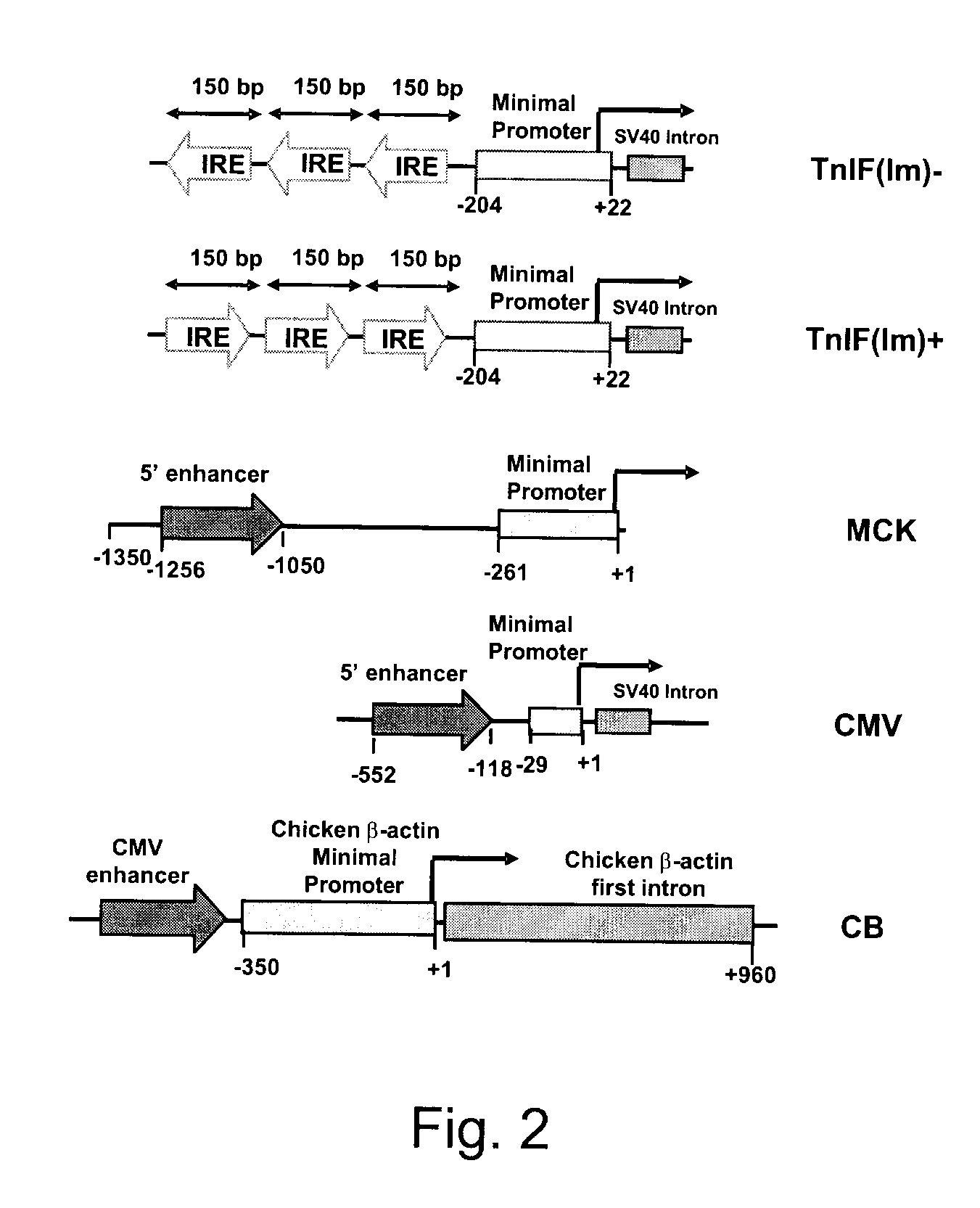
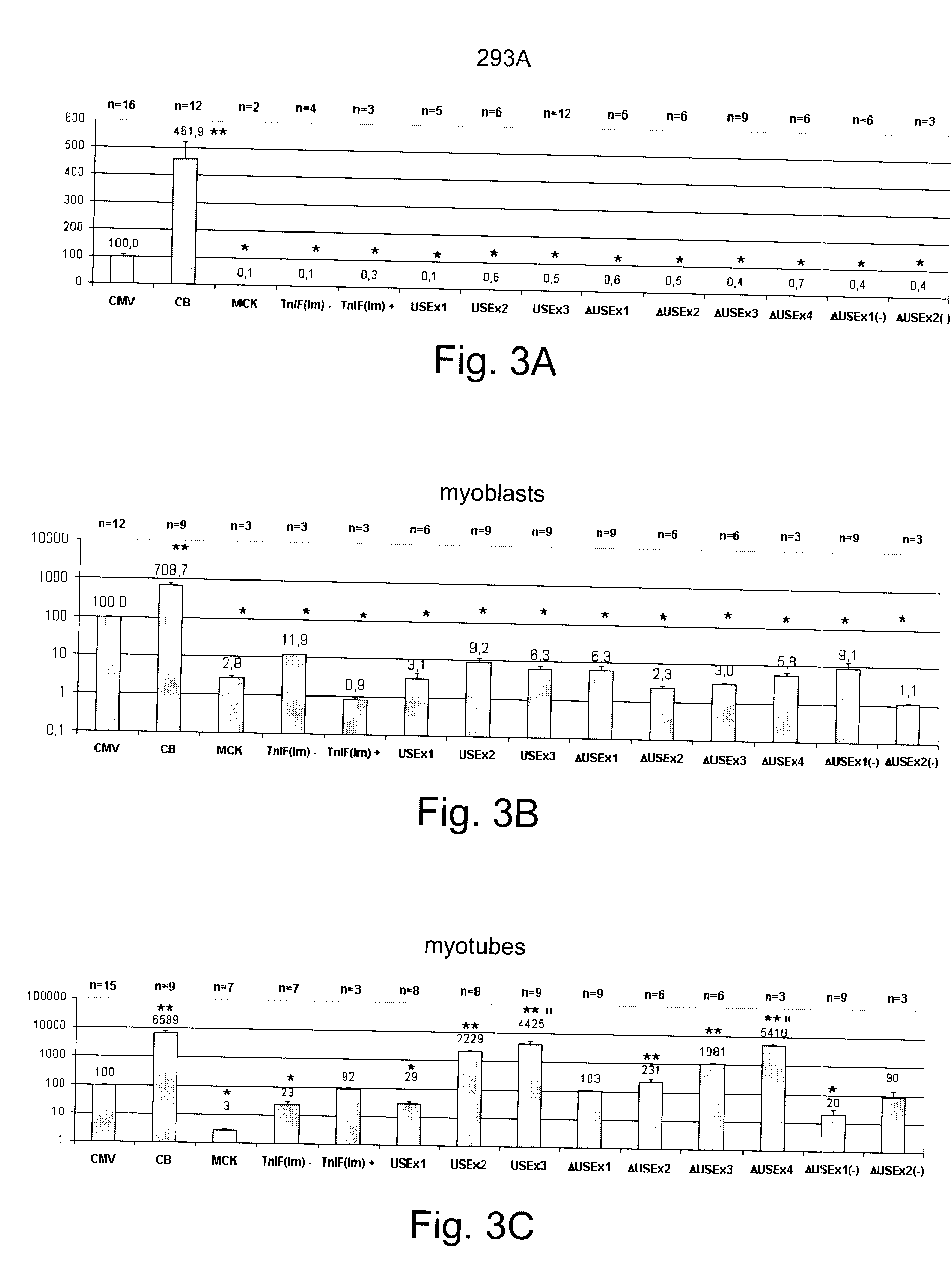
Efficient and muscle-specific gene expression can be obtained with constructs containing two or more copies of USE and / or ΔUSE fused to the minimal promoter of the TnISlow gene. USE is a small (about 160-bp) upstream enhancer of the TnISlow gene that confers slow-twitch muscle fiber specificity. ΔUSE is generated from a 100-bp deletion at the 5′ end of USE. ΔUSE confers expression in slow- and fast-twitch muscle fibers. The strength and relatively small size (less than 600-bp) of these constructs make them useful for gene therapy applications.
Owner:NAT RES COUNCIL OF CANADA
Construction of EP153R and EP402R gene co-expression recombinant adenovirus vector and adenovirus packaging method
InactiveCN109652449ADirect infectionEnhance immune responseViral antigen ingredientsVirus peptidesMultiple cloning siteCloning Site
The invention provides construction of an EP153R and EP402R gene co-expression recombinant adenovirus vector and a adenovirus packaging method, and belongs to the technical field of genetic engineering. Based on a pShuttle-CMV eukaryotic expression vector, an EP153R gene and EP402R gene overexpression adenovirus vector pAD-Shuttle-CMV-CTLA4-EP153R-EP402R is introduced with CTLA4, EP153R and EP402Rgenes at multiple cloning sites. The invention also provides recombinant adenovirus for co-expressing EP153R and EP402R genes, and the adenovirus packaging process is achieved by utilizing the constructed pAD-Shuttle-CMV-CTLA4-EP153R-EP402R vector to obtain the adenovirus capable of directly infecting animal or eukaryotic cells, thereby achieving the purpose of co-expressing EP153R and EP402R genes in eukaryotic cells, and laying a foundation for researching the adenovirus vaccine for achieving the co-expression of EP153R and EP402R genes.
Owner:YANGZHOU UNIV
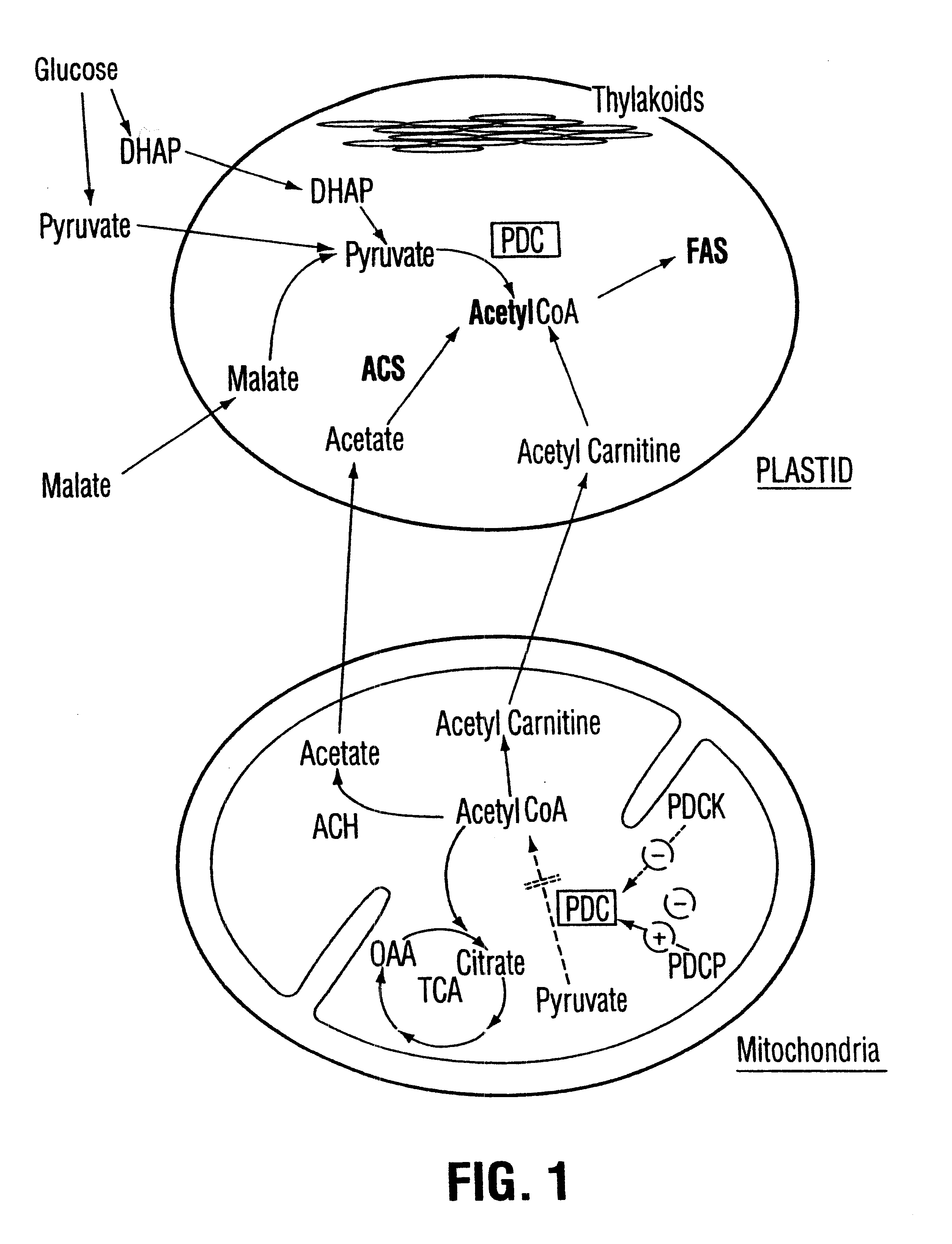
Plant pyruvate dehydrogenase kinase gene
InactiveUS6500670B1High activityImprove availabilitySugar derivativesClimate change adaptationBrassicaceaePlanting seed
The present invention relates to the isolation, purification, characterization and use of a mitochondrial pyruvate dehydrogenase kinase (PDHK) gene [SEQ ID NO:1](pYA5; ATCC No 209562) from the Brassicaceae (specifically Arabidopsis thaliana). The invention includes isolated and purified DNA of the stated sequence and relates to methods of regulating fatty acid synthesis, seed oil content, seed size / weight, flowering time, vegetative growth, respiration rate and generation time using the gene and to tissues and plants transformed with the gene. The invention also relates to transgenic plants, plant tissues and plant seeds having a genome containing an introduced DNA sequence of SEQ ID NO:1; or a part of SEQ ID NO:1 characterized in that said sequence has been introduced into an antisense or sense orientation, and a method of producing such plants and plant seeds. The invention also relates to substantially homologous DNA sequences from plants encoding proteins with deduced amino acid sequences of 25% or greater identity, and 50% or greater similarity, isolated and / or characterized by known methods using the sequence information of SEQ ID NO:1, and to parts of reduced length that are still able to function as inhibitors of gene expression by use in an anti-sense, co-suppression or other gene silencing technologies.
Owner:NAT RES COUNCIL OF CANADA
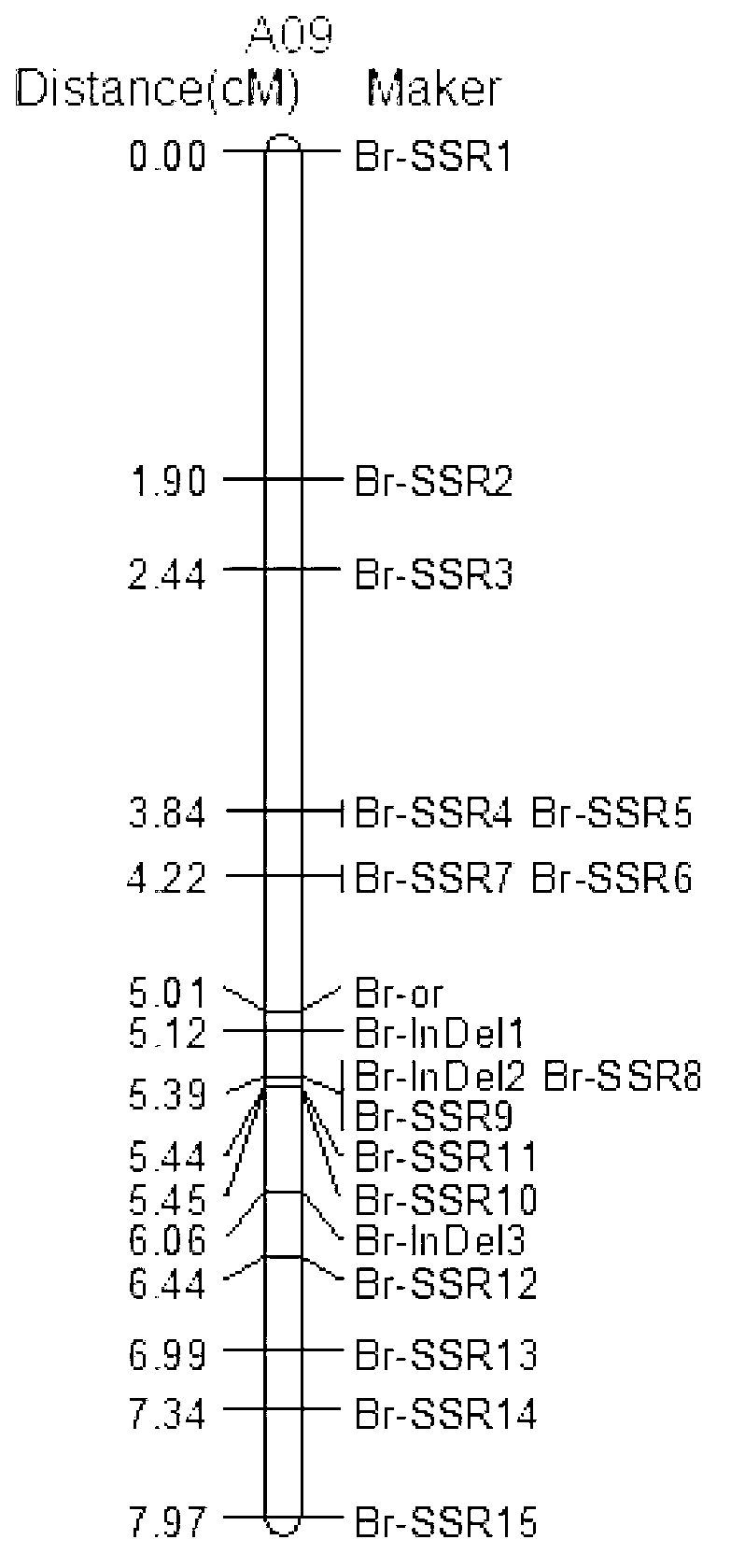
SSR (Simples sequence repeats) and InDel (insertion/deletion) molecular marker primer linked with brassica campestris orange head gene Br-or, and application thereof
InactiveCN103103184AEasy to detectStable amplificationMicrobiological testing/measurementDNA/RNA fragmentationBrassicaAgricultural science
The invention discloses an SSR (simples sequence repeats) and InDel (insertion / deletion) molecular marker primer linked with brassica campestris orange head gene Br-or and an application of the molecular marker primer. The marker primer is obtained by the steps of: based on the genome DNA (deoxyribonucleic acid) of orange brassica campestris and normal white brassica campestris and F2S4 separation group as a template, carrying out polymorphism primer screening to orange head and white head single strain DNA mixed tank in F2S4 by using 480 pairs of SSR and InDel primer pairs, then analyzing single strain of F2S4 group, and screening the molecular marker linked with the brassica campestris orange head gene Br-or to finally obtain fifteen Br-SSR and three Br-InDel molecular markers linked with the Br-or gene, wherein a molecular genetic map of the brassica campestris orange head gene Br-or is created on the ninth link group (A09). According to the link analysis, the genetic distance of two markers most tightly linked with both sides of the Br-or gene are 0.11cM and 0.79cM; and the molecular marker primer has the advantages of being convenient for detection, stable in amplification, and high in repeatability and accuracy, and thus a basis is provided to the molecule assistant selective breeding of brassica campestris orange head character and the breeding process is accelerated.
Owner:NORTHWEST A & F UNIV
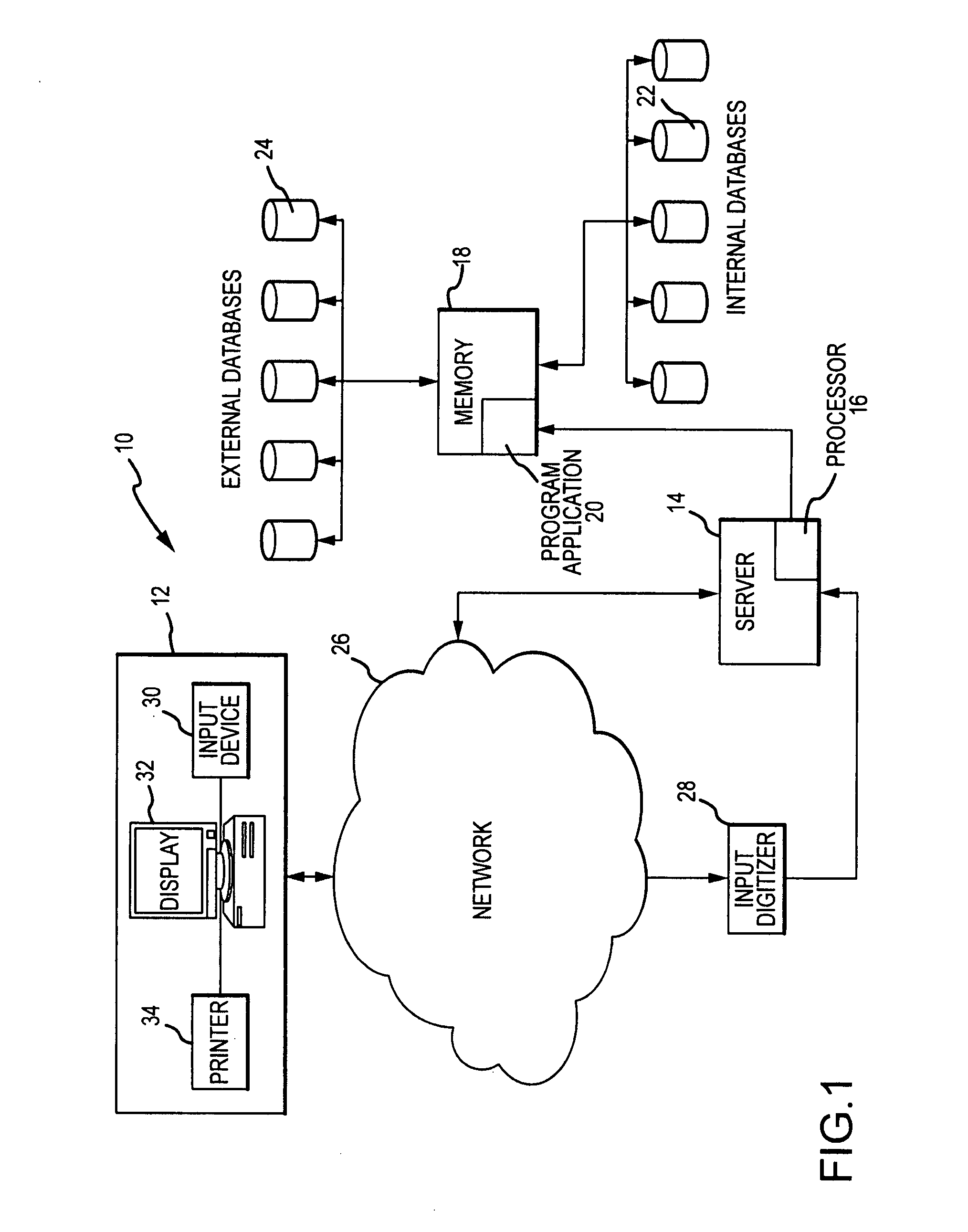
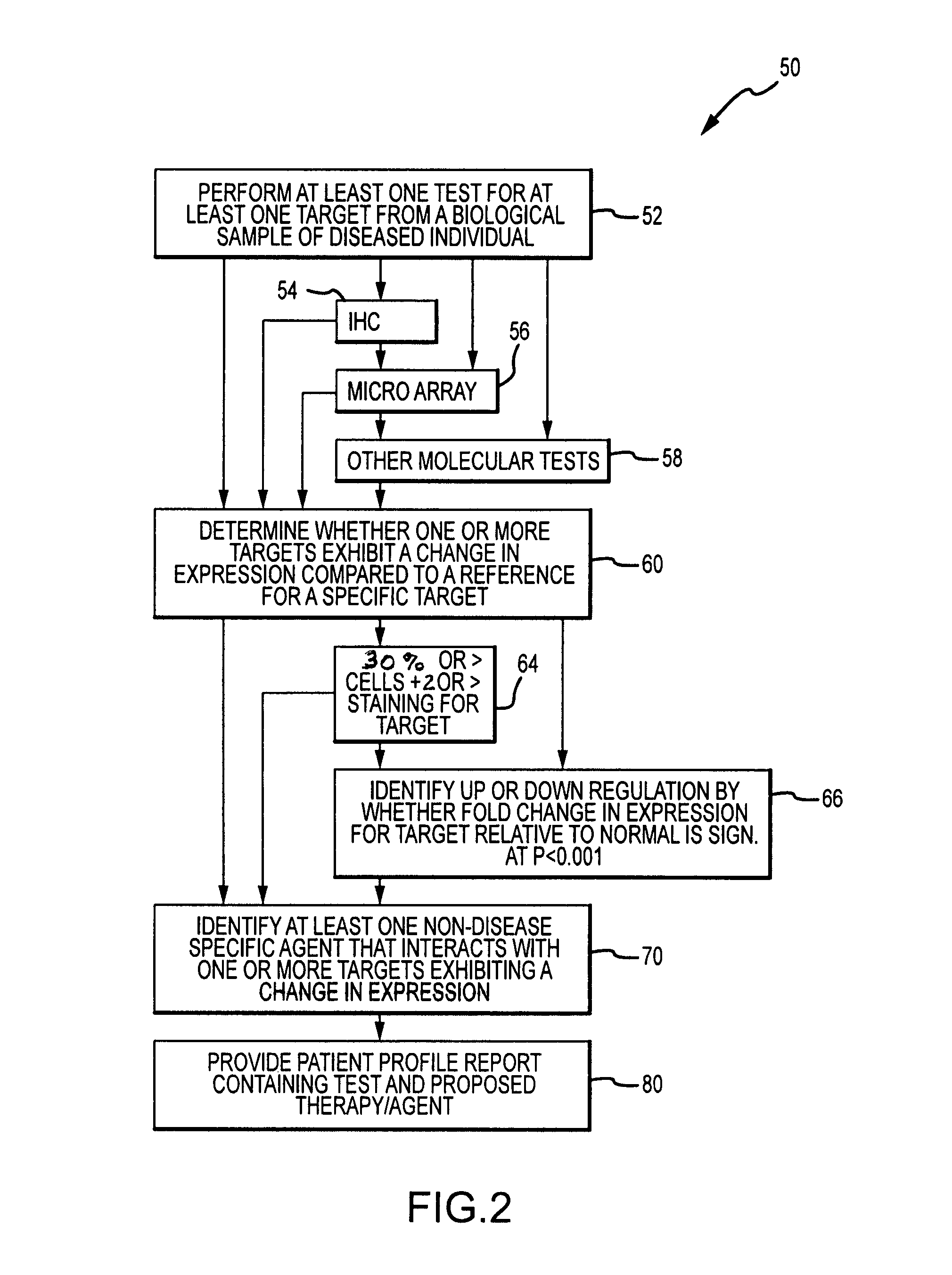
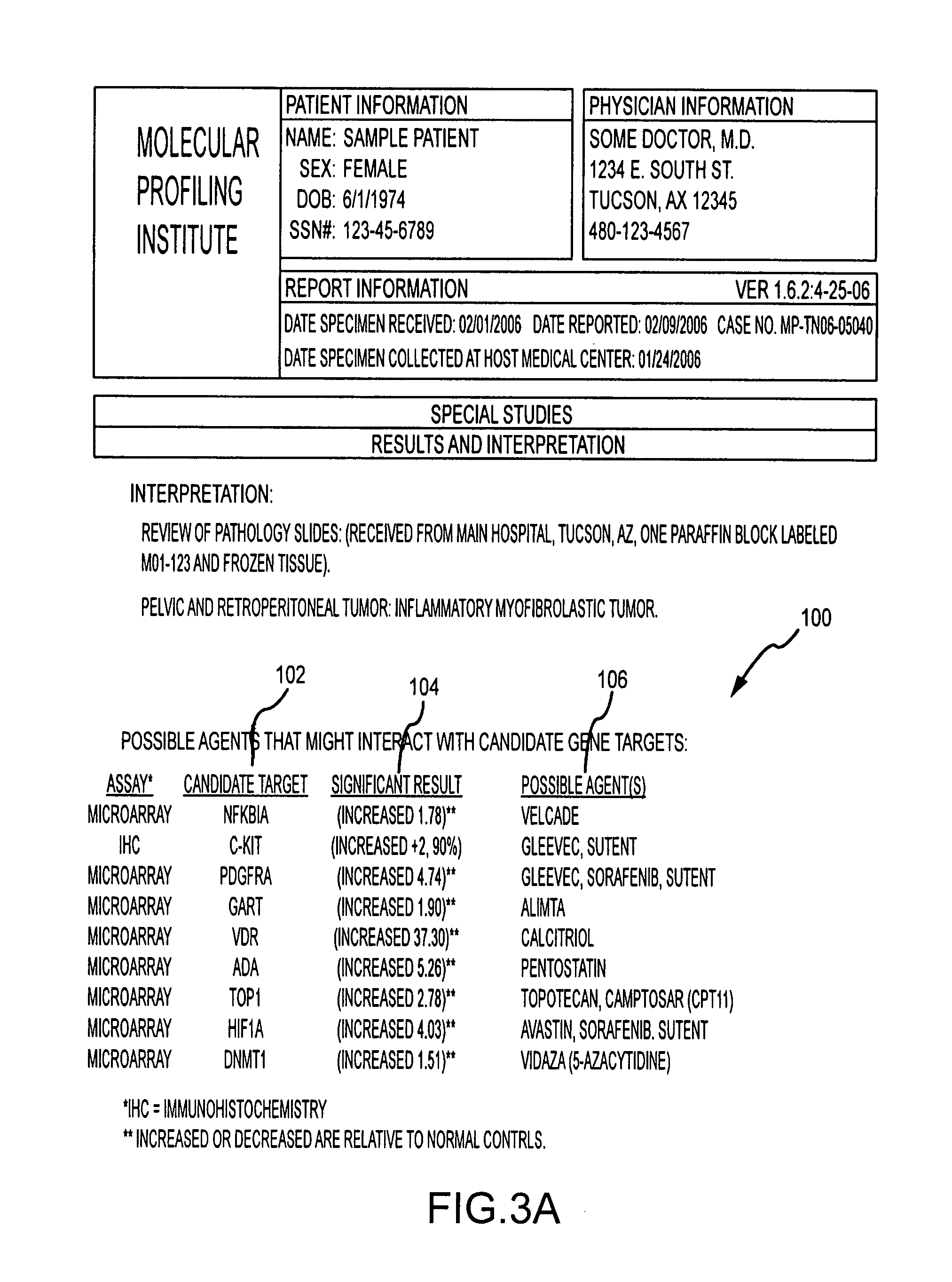
System and method for determining individualized medical intervention for a disease state
ActiveUS8700335B2Physical therapies and activitiesDrug and medicationsDisease specificMedical treatment
Owner:CARIS MPI INC
Method for identifying HBV gene mutation type, special chip and reagent kit
The invention discloses a method for identifying HBV gene mutation type and a chip and a reagent box which are especially used with the method. The method for identifying the HBV gene mutation type takes the detected the genome of the hepatitis B virus as the template and uses the following primer group and a DNA polymerase which has no exonuclease activity from the 3'end to 5' end for implementing multiple PCR magnification; the primer group comprises a universal primer and a or more than one special primers; the special primer 5' end is a bar code sequence and the length of the bar code sequence is 5 to 25 nt; the bar code sequence has a comparatively low homology with the hepatitis B virus; the special primer totally matches with the corresponding segment of the hepatitis B virus comprising wild base group or mutant base group at a mutation point; the gene chip comprises a plurality of probes and the probes are corresponding to the bar code sequence one to one and every probe only contains one bar code sequence.
Owner:BOAO BIOLOGICAL CO LTD +1
Molecular markers co-separated with bruchid resistance genes VrPGIP of vigna radiate and application of molecular markers
InactiveCN105950736ASimple methodFast wayMicrobiological testing/measurementDNA/RNA fragmentationVignaMolecular marker
The invention provides molecular markers co-separated with bruchid resistance genes VrPGIP of vigna radiate. The bruchid resistance genes VrPGIP of the vigna radiate are positioned in regions of 5.558Mb and 5.625Mb of a chromosome 5 of the vigna radiate; the molecular markers co-separated with the genes VrPGIP comprise SSR (Simple Sequence Repeat) markers Vr05-5590 and Vr05-5597; physical distances between the Vr05-5590 and the Vr05-5597 and the bruchid resistance genes VrPGIP of the vigna radiate are 1.5kb and 850bp respectively. The invention also provides application of the molecular markers in mapping of the bruchid resistance genes of the vigna radiate or genetic breeding of the vigna radiate. According to the molecular markers and the application thereof disclosed by the invention, by finely mapping the bruchid resistance genes VrPGIP of the vigna radiate, two SSR markers co-separated with the bruchid resistance genes VrPGIP of the vigna radiate are found; the development of the molecular markers provides a feasible way for assistant breeding of bruchid resistant molecules of the vigna radiate.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
Hybrid network gene screening method based on gene expression data
The invention relates to the field of genetic diagnosis, and particularly relates to a hybrid network gene screening method based on gene expression data. According to the scheme, by introducing the gene expression data, a hybrid network gene screening method model based on the gene expression data is designed, and specific steps how to construct different networks by the gene expression data are given out; and the gene expression data is utilized to carry out more detailed and deep quantization on diseases and genes and a mutual relationship network between the genes so as to reinforce comprehensive capacity of the entire hybrid model on the aspect of disease gene screening.
Owner:HARBIN INST OF TECH SHENZHEN GRADUATE SCHOOL
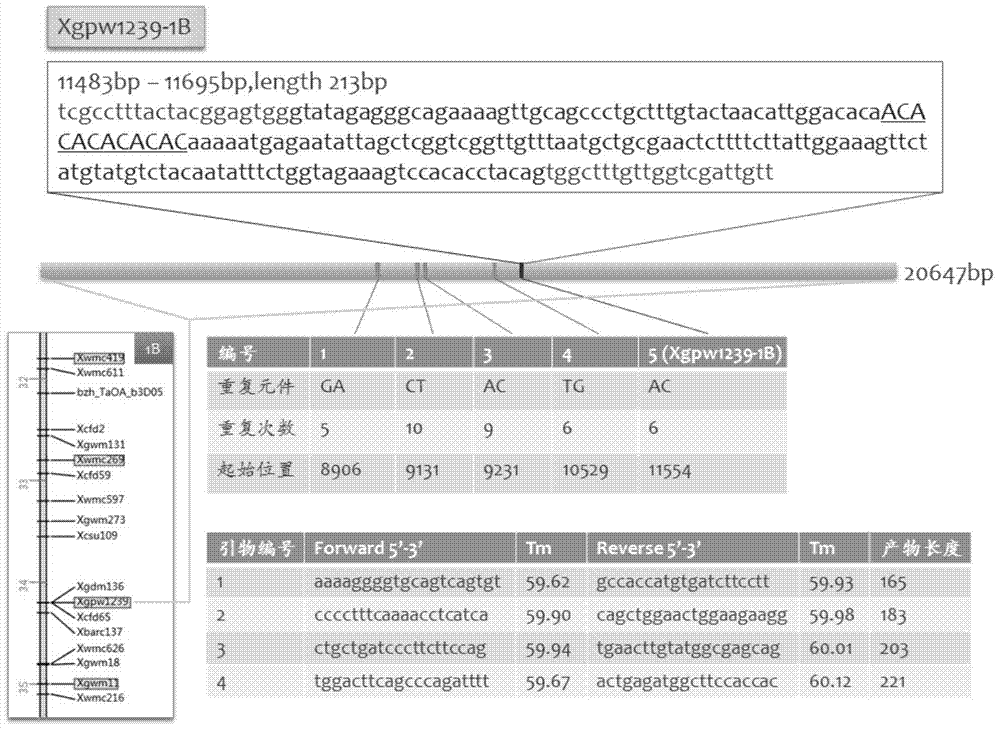
Design method of SSR label primer and wheat SSR label primers
InactiveCN103571833AIncrease the number ofFast encryptionMicrobiological testing/measurementSpecial data processing applicationsContigTest group
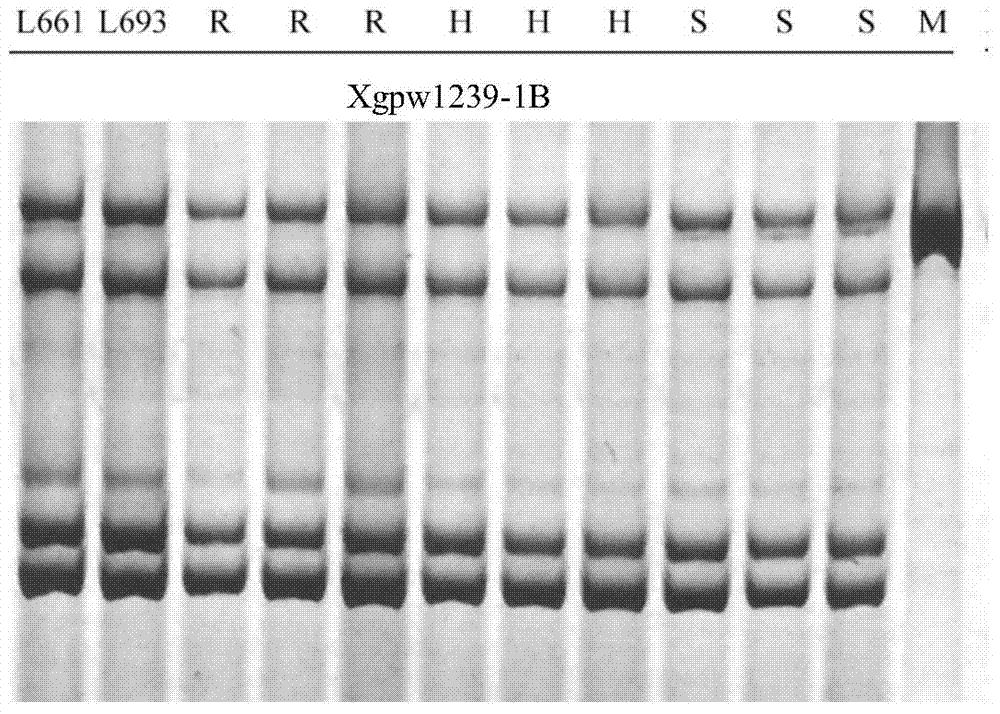
The invention discloses a design method of an SSR label primer. In order to overcome the defect of insufficient polymorphism of the conventional SSR label primer, the invention provides a method for designing a novel SSR label primer based on a draft sequence of a genome. The design method comprises the following steps: firstly selecting an SSR label primer with a known site as a starting SSR label primer; secondly, comparing the starting SSR label primer with the draft sequence of a chromosome to which the starting SSR label primer belongs, and finding a contig in a comparison result; thirdly, searching an SSR sequence in the contig as a finishing SSR sequence; finally, designing the SSR label primer based on the finishing SSR sequence. The invention provides 14 pairs of novel SSR label primers related to wheat stripe rust resistance, and a method for establishing a wheat genetic map with L693*L661 and L661*L693F2 single plants as a mapping population, wherein five pairs of the SSR label primers generate genetic polymorphism in a genetic test group. According to the design method, the number of the SSR label primers with known sites can be quickly increased and the polymorphism of the SSR label is increased; the genetic map can be quickly encrypted through combination with initial gene localization.
Owner:SICHUAN AGRI UNIV
Maize cytoplasmic male sterility (CMS) c-type restorer rf4 gene, molecular markers and their use
ActiveUS20120090047A1Increased seed yieldLow costSugar derivativesMicrobiological testing/measurementHybrid seedCandidate Gene Association Study
This disclosure concerns high-resolution mapping and candidate gene cloning of Rf4, a maize restorer of fertility gene that restores fertility to C-type cytoplasmic male sterility. The disclosure also relates to molecular markers that are tightly-linked to, or reside within, the Rf4 gene. In some embodiments, methods are provided whereby hybrid seeds may be produced from crosses of a male plant comprising nucleic acid molecular markers that are linked to or that reside within the Rf4 gene and a female plant carrying C-type CMS.
Owner:CORTEVA AGRISCIENCE LLC
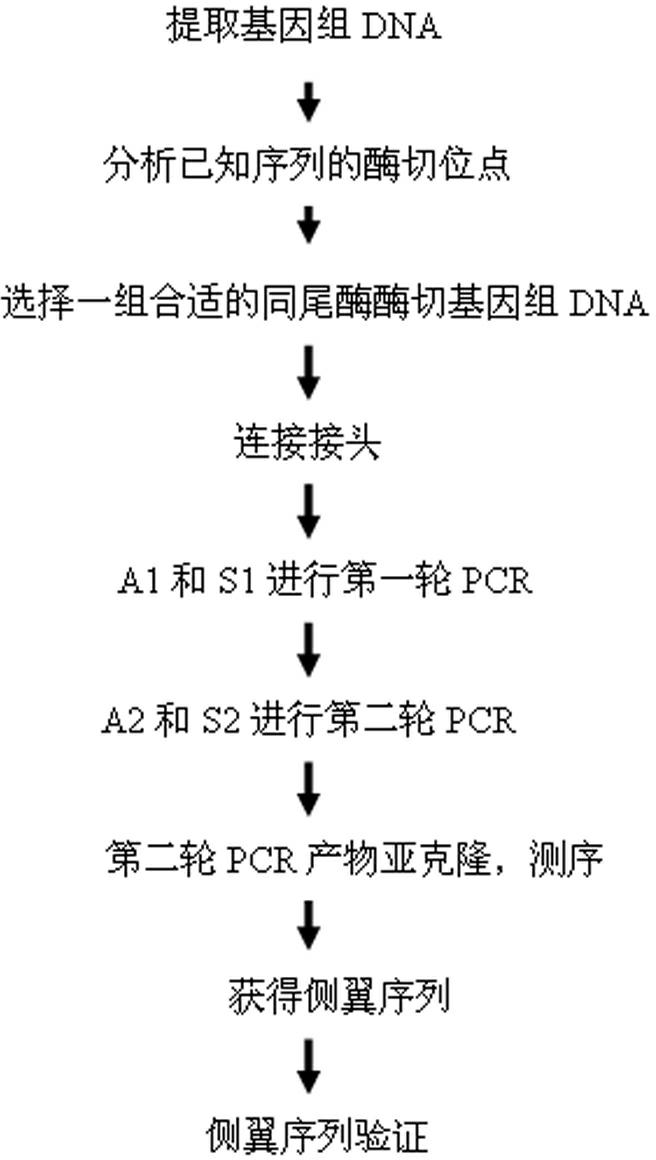
Method for applying adhesive tail end joints to flanking sequence separation
A method for applying adhesive tail end joints to flanking sequence separation includes following steps: 1) extracting a genome DNA (deoxyribonucleic acid) to be detected; 2) selecting isocaudarner and designing and synthesizing corresponding joints; 3) digesting the genome by the isocaudarner to form DNA segments with identical adhesive tail ends; 4) detecting digestion quality in an electrophoresis manner, quantifying the concentration of the DNA segments, connecting the joints with digestion products of the genome, and forming each DNA segment with 'the joint, the digestion segment and the other joint' sequentially; 5) purifying the DNA segments with 'the joints, the digestion segments and the other joints' sequentially, and performing PCR (polymerase chain reaction); 6) recycling the segments obtained from the step 3) and sequencing; 7) analyzing a sequencing result and obtaining an unknown flanking sequence of a known sequence; and 8) checking the unknown flanking sequence. The method for flanking sequence separation is high in connection efficiency and generality and low in cost.
Owner:HUNAN HYBRID RICE RES CENT
High-oleic-acid-content peanut molecular marker, assistant selection back cross breeding method and application of back cross breeding method
ActiveCN103468678ASimple methodEasy to operateMicrobiological testing/measurementPlant genotype modificationBiotechnologyForward primer
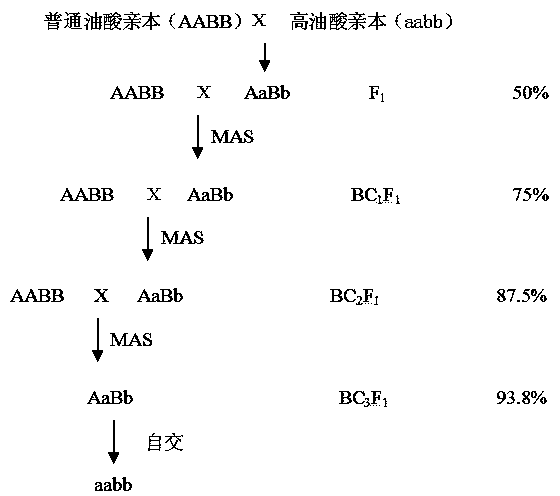
The invention relates to a high-oleic-acid-content molecular marker, a back cross breeding method for carrying out assistant selection by utilizing the molecular marker, and an application of the back cross breeding method. Sequences of variant specific primers of the molecular marker are as follows: a forward primer MITE-INS-F, and a reverse primer MITE-INS-R; the sequences of non-variant specific primers are as follows: a forward primer WILD-F, and a reverse primer WILD-R. The back cross breeding method utilizing the molecular maker comprises the following steps: carrying out hybridization on a peanut parent of a gene type AABB and a parent of a gene type aabb, then carrying out cross breeding; amplifying an FAD2A gene segment by utilizing one pair of primers in a genome, and selecting a single plant containing a genotype Aa; carrying out PCR (polymerase chain reaction) detection by utilizing a primer, at a site ahFAD2B, of a peanut as a marker, and selecting a single plant containing a genotype Bb; carrying out back crossing on the single plant which is dominant in two markers, namely the single plant containing the genotype AaBb, and the parent AABB until a high-oleic-acid-content single plant which is of a genotype aabb and is consistent with a recurrent parent in other economical characters is selected from later generations, and then carrying out self-crossing and amplifying propagation. The high-oleic-acid-content molecular marker has the advantages of being simple in method, low in cost and stable in result; when the high-oleic-acid-content molecular marker is used for carrying out the back cross breeding, the accuracy of character selection can be obviously improved, the breeding cost is reduced, and the breeding efficiency is improved.
Owner:HENAN ACAD OF AGRI SCI
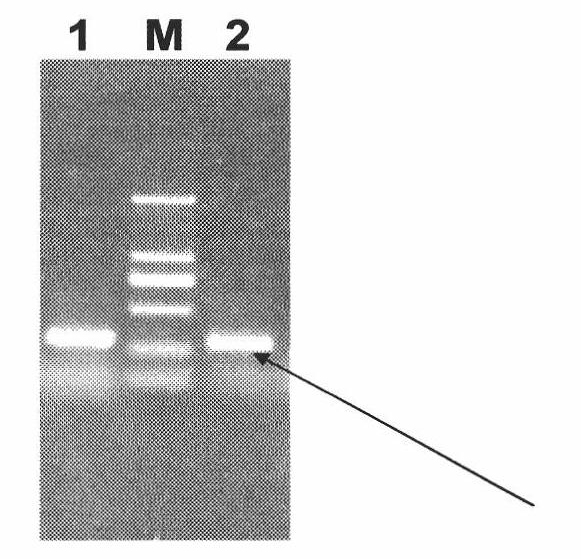
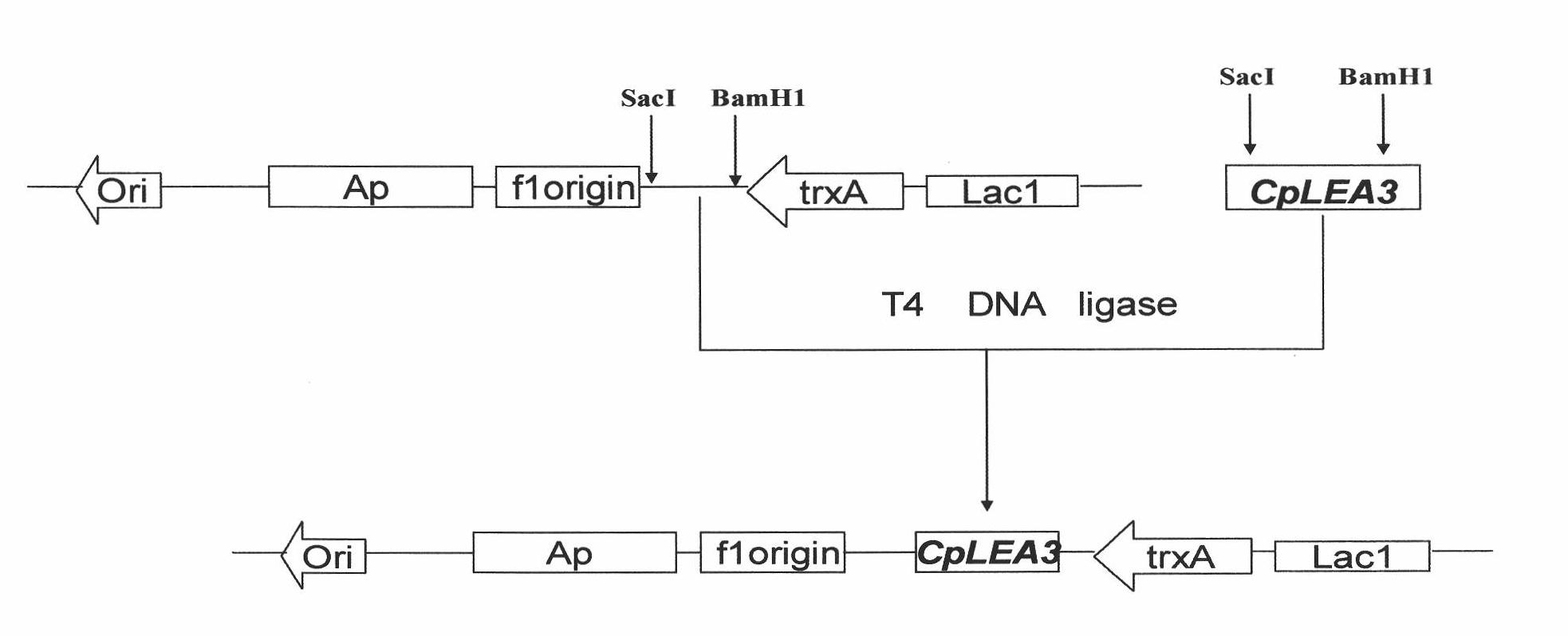
Gene of wintersweet late embryogenesis abundant protein (LEA) and low-temperature resistant application thereof
InactiveCN102010869AImprove the ability to resist low temperatureImprove low temperature resistanceBacteriaMicroorganism based processesLate embryogenesis abundant proteinsExpression vector
The invention relates to a gene of wintersweet late embryogenesis abundant protein (LEA) and low-temperature resistant application thereof, belonging to the field of molecular biology and gene engineering. The invention provides a gene of wintersweet LEA and protein and also provides an expression vector and a host cell corresponding to the gene. The invention has the advantage that the application of the gene of the wintersweet LEA can improve the low-temperature resistance of a strain.
Owner:JILIN UNIV
Mycoplasma hyopneumoniae fusion gene and application
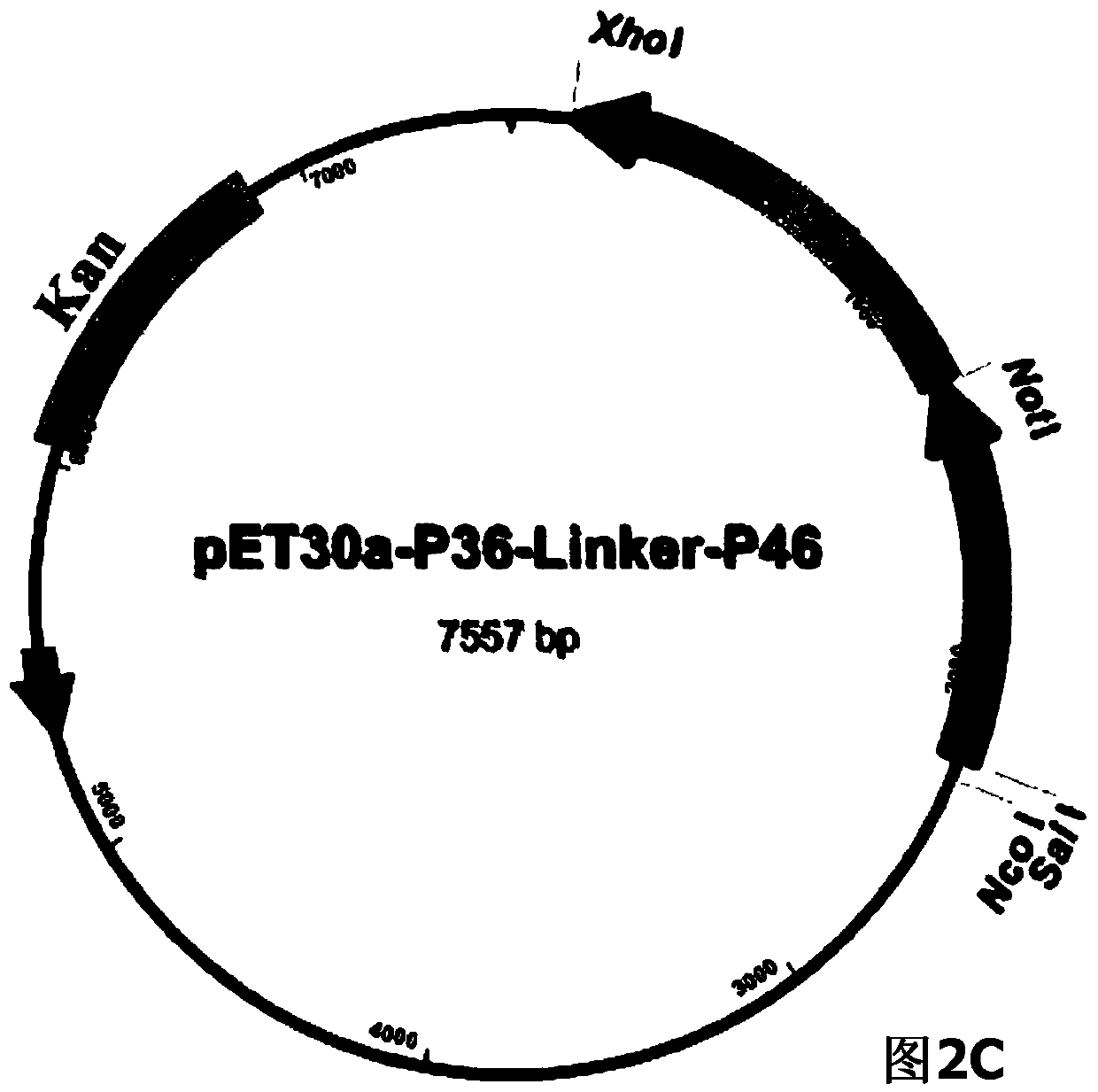
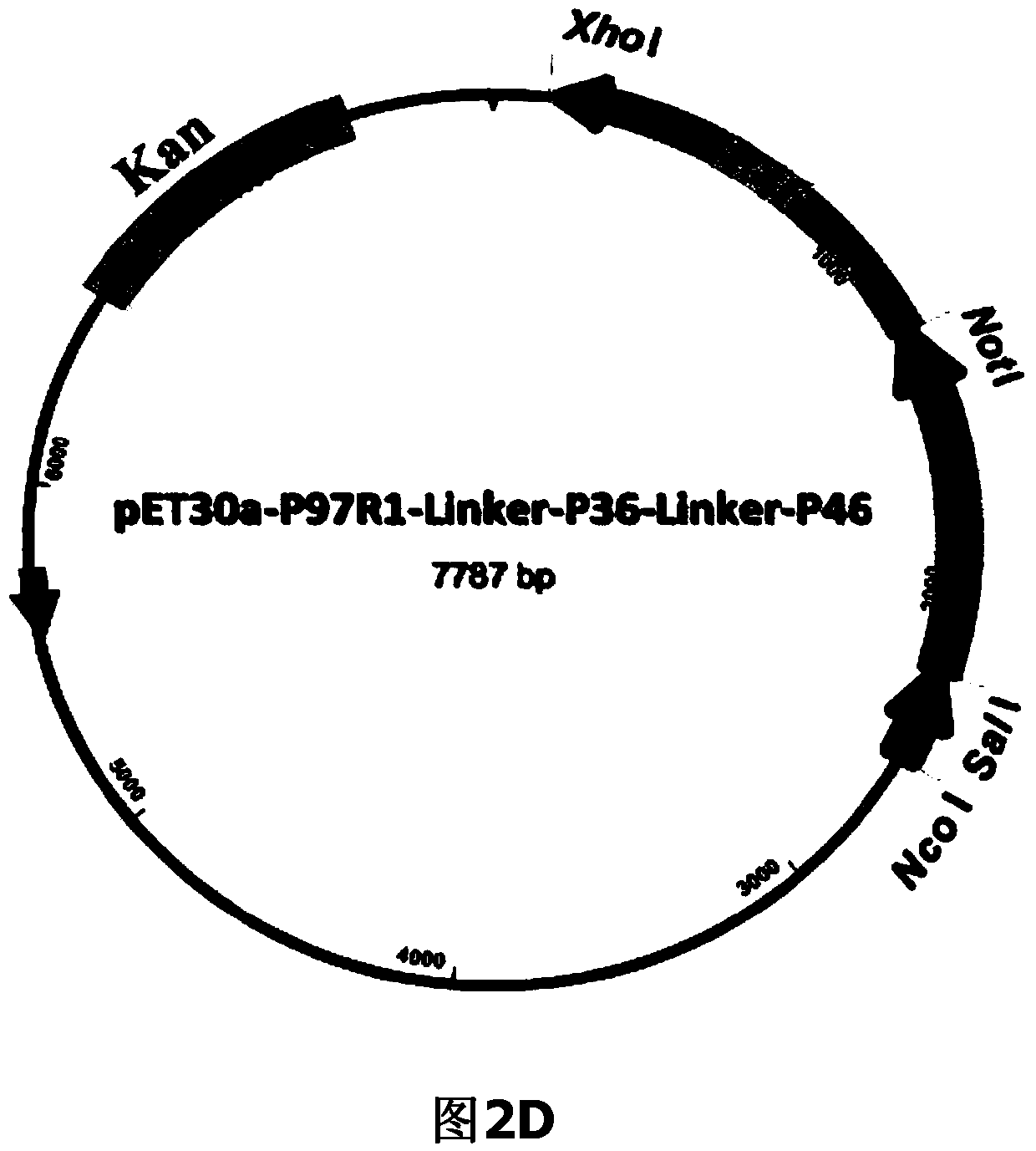
ActiveCN104293816AImproving immunogenicityHigh expressionAntibacterial agentsBacterial antigen ingredientsEscherichia coliNucleotide
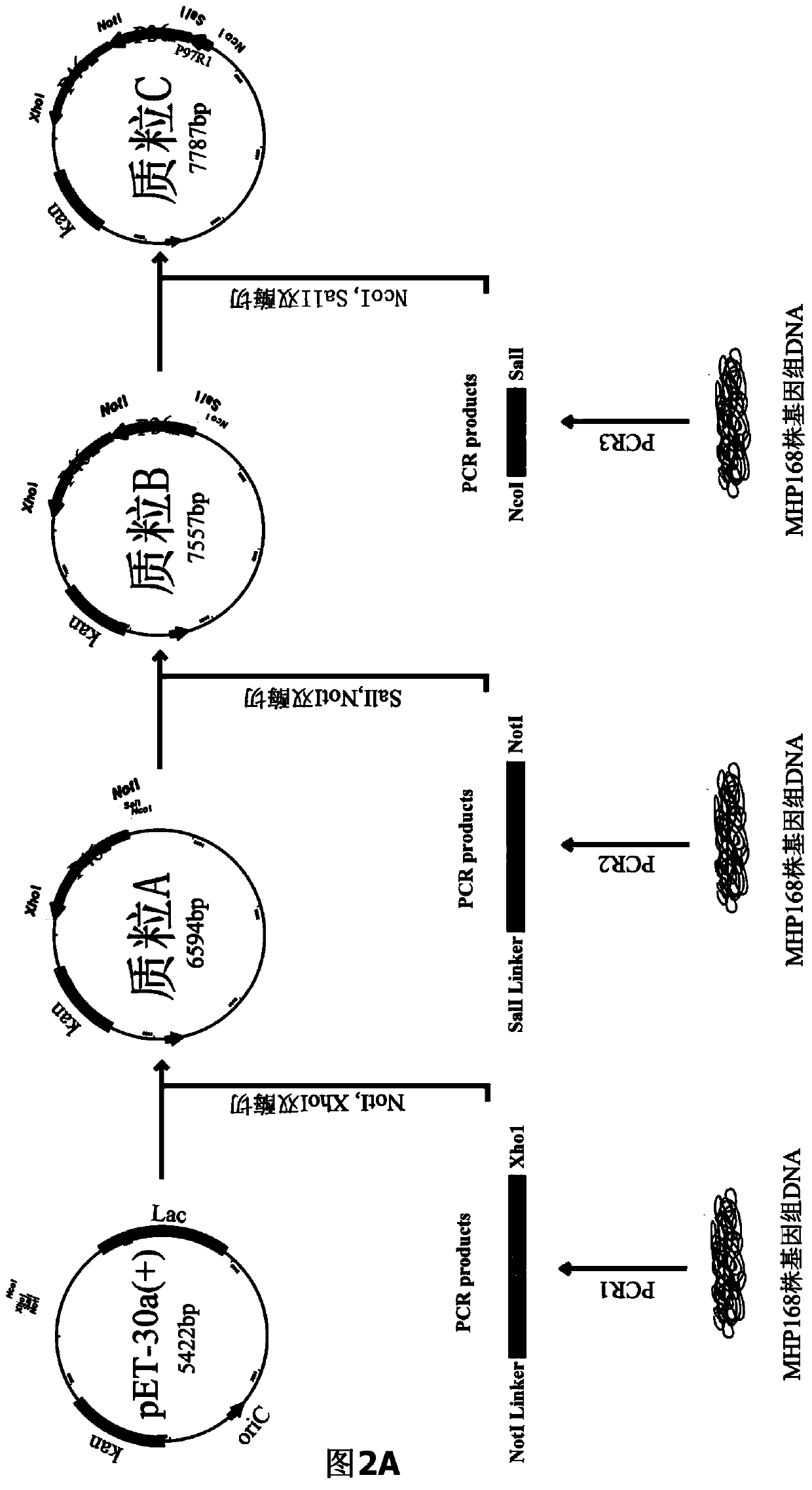
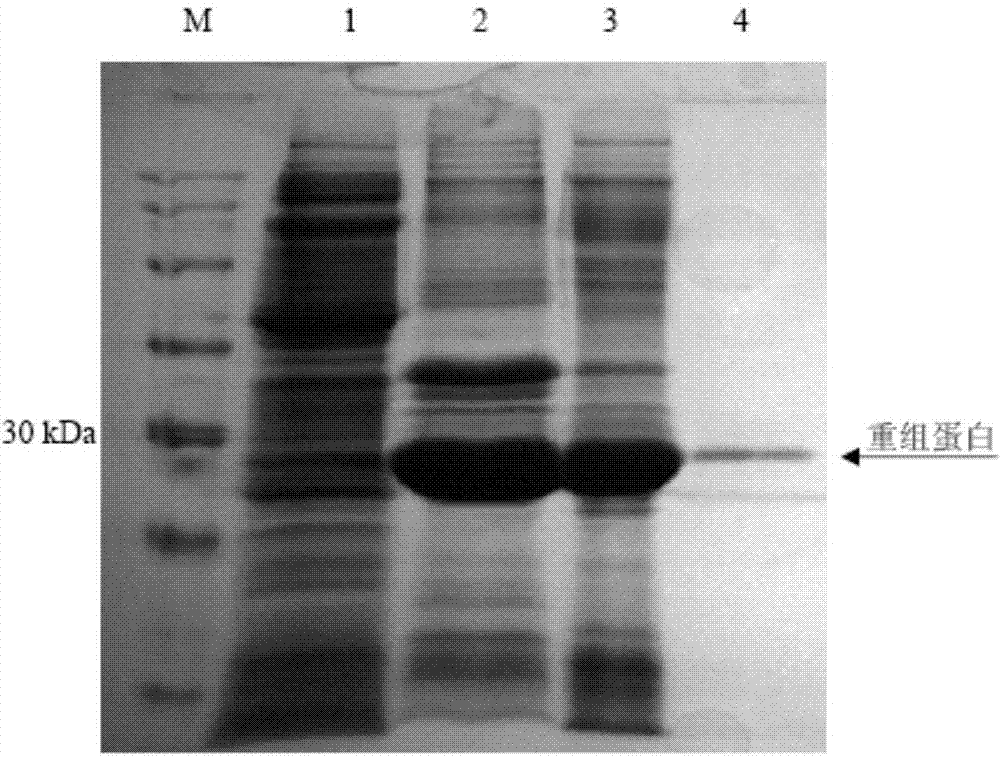
The invention belongs to the field of animal gene engineering, and in particular relates to a synthetic fusion gene for expressing mycoplasma hyopneumoniae and application. The fusion gene is characterized in that a mycoplasma hyopneumoniae P97R1 gene is connected in series with a P36 gene through a Linker, the termination codon of the P36 gene is deleted, the P46 gene with signal peptide removed is fused at the C end of the P36 gene through Linker, and then the fusion gene P97R1-P36-P46 is obtained, wherein the nucleotide sequence of the fusion gene is shown as SEQ ID NO: 1. The fusion gene is included in a prokaryotic expression plasmid, and escherichia coli BL21 / pET30a-P97R1-Linker-P36-Linker-P46 containing the plasmid is collected in CCTCC with the collection number CCTCC NO: M2014269. The invention further discloses immune efficacy evaluation of the fusion gene and application of the fusion gene in a novel vaccine.
Owner:HUAZHONG AGRI UNIV
Methods and compositions relating to gene silencing
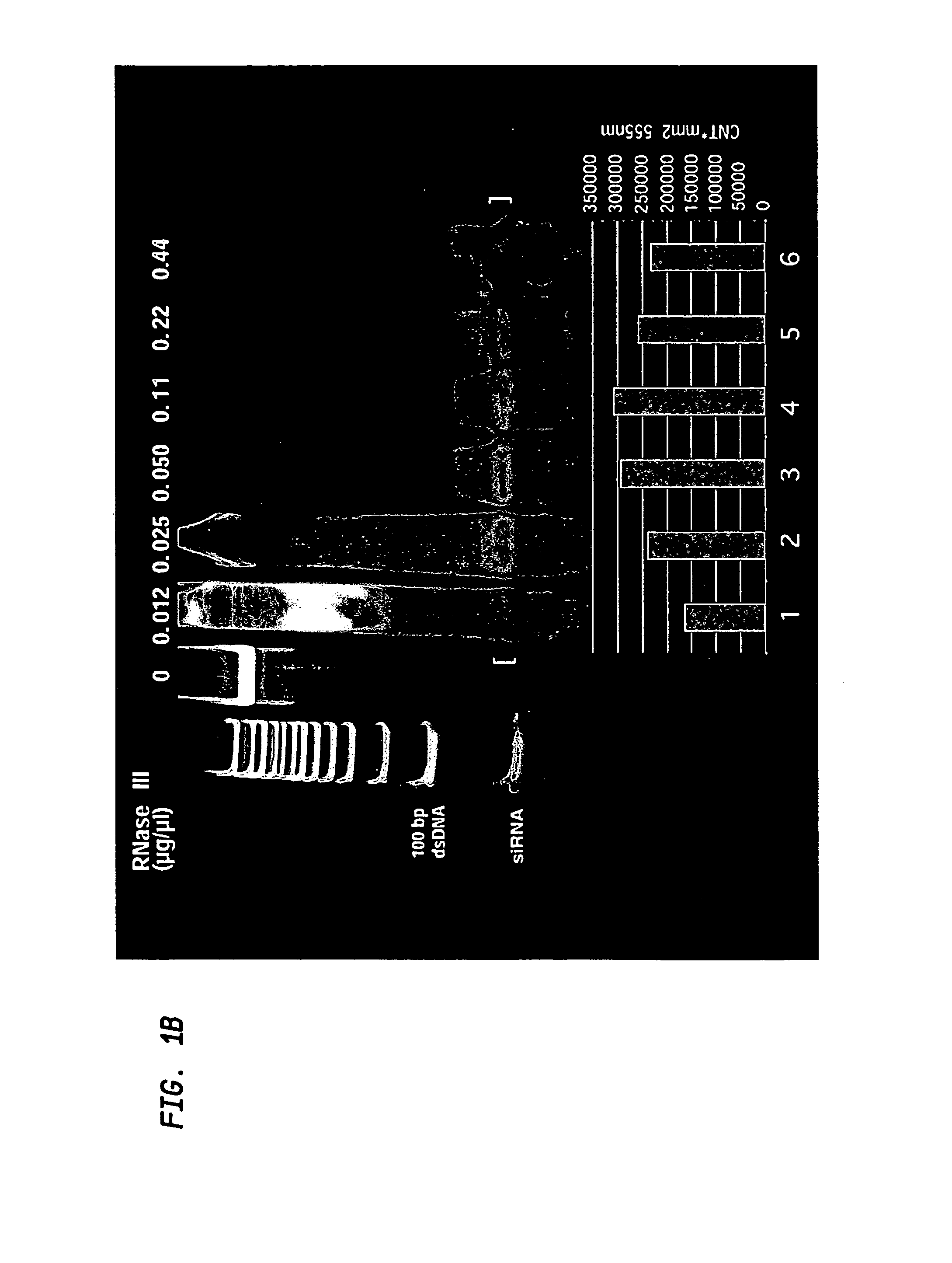
InactiveUS7700758B2Reduce expressionImprove featuresSugar derivativesHydrolasesHeterologousNucleotide
A method for obtaining a mixture of heterogenous short double-stranded RNA molecules suitable for use in gene silencing (hsiRNA) by subjecting large double-stranded RNA to enzymatic cleavage under specified conditions. The resulting mixture consistently includes enhanced representation of fragments having a size of 21-22 nucleotides absent any fractionation step. The fragments contain sequences that collectively span the entire length of the large double-stranded RNA from which they are derived. Double-stranded RNA with sequences that individually represent segments of a target mRNA may be analyzed using the methods described herein to identify the most active subset of hsiRNA fragments or individual siRNA fragments for achieving gene silencing for any gene or transcribed sequences. A method is additionally provided for preparing and cloning DNA encoding selected siRNA, hsiRNA mixtures or hairpin sequences to provide a continuous supply of a gene silencing reagent derived from any long double-stranded RNA.
Owner:NEW ENGLAND BIOLABS
Method for detecting Xanthomonas oryzae pv.oryzae, Xoo, X.oryzae pv. Oryzicola, Xoox, X.axonopodis pv. Citri, Xac, and X.campestris pv.campestris, Xcc
InactiveCN101921843ANot easy to polluteHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyMicrobiology
The invention discloses a method for detecting Xanthomonas oryzae pv.oryzae, Xoo, X.oryzae pv. Oryzicola, Xoox, X.axonopodis pv. Citri, Xac, and X.campestris pv.campestris, Xcc. The method comprises the following steps: carrying out PCR (polymerase chain reaction) amplification on the DNA template of a sample to be detected by using two pairs of primers at the same time; then, hybridizing the PCR products with the specific probe arranged on a gene chip; and judging whether the sample to be detected is with Xanthomonas oryzae pv.oryzae, Xoo, X.oryzae pv. Oryzicola, Xoox, X.axonopodis pv. Citri, Xac, and X.campestris pv.campestris, Xcc according to the fact whether the hybridization result is positive or not. Therefore, the method of the invention is capable of detecting four bacteria by one time with high detection speed and has the advantages of direct-viewing and clear detection result and easy judgment.
Owner:SHENZHEN AUDAQUE DATA TECH +1
Stichopus japonicas BPI gene, encoded protein, cloning method of stichopus japonicas BPI gene, and method for constructing recombinant stichopus japonicas BPI genetically engineered bacterium
ActiveCN104745595AEfficient killingAntibacterial agentsPeptide/protein ingredientsBPI proteinVibrio parahaemolyticus
The invention discloses a stichopus japonicas BPI gene, an encoded protein, a cloning method of the stichopus japonicas BPI gene, and a method for constructing a recombinant stichopus japonicas BPI genetically engineered bacterium. The stichopus japonicas BPI gene is characterized in that a stichopus japonicas BPI gene sequence is as shown in SEQ ID NO.1; the cloning method comprises the step of designing a nested primer of RACE according to an expressed sequence tag EST sequence which is homologous to the BPI gene; a full-length gene is expanded by employing an RACE technique; a stichopus japonicas BPI protein sequence is as shown in SEQ ID NO.2; an N-terminal protein structure domain sequence is as shown in SEQ ID NO.3; an N-terminal structure domain of the stichopus japonicas BPI protein is amplified by employing primers which respectively comprise BamH I sites and Not I sites; the cloned target gene is inserted into a vector to obtain recombinant plasmids; and the recombinant plasmids are subjected to induced expression, and purification and renaturation, so as to obtain the genetically engineered bacterium. The stichopus japonicas BPI gene has the advantage of having obvious sterilization effect on vibrio parahaemolyticus, vibrio harveyi and micrococcus luteus.
Owner:NINGBO UNIV
Construction and application of vector used for developing high-expression cell strain
ActiveCN103525867AReduce formation rateEasy to getVector-based foreign material introductionForeign genetic material cellsVersus geneBiochemistry
The invention relates to a pSGS vector and application of the vector to development of a high-expression cell strain. The construction of the vector comprises the following steps: based on a neomycin resistance screening gene NEO, through combined strategy of weakening screening gene expression and reinforcing target gene expression, constructing an expression vector pSNEO, and in view of the amplification function of a GS system, through designing a primer, replacing the hairpin structure sequence and the NEO gene of the vector pSNEO with a hairpin structure sequence and a GS gene to obtain a universal vector pSGS for screening a high-expression and stable-transfection cell strain. Through the pSGS vector constructured by the invention, the construction of a stable and high-expression cell strain of the target gene can be finished conveniently and quickly, and a new tool is provided for preparation of macromolecular recombinant protein.
Owner:BEIJING DONGFANG BIOTECH
Gene related to chlorophyll content of leaf and grain weight of wheat as well as Indel marker and application thereof
InactiveCN105132440ASynergisticGrain weight reductionMicrobiological testing/measurementFermentationAgricultural scienceGrain weight
The invention relates to a gene TaBAS1-2B related to the chlorophyll content of a leaf and the grain weight of wheat and an Indel marker of the gene. According to the gene, the gene TaBAS1-2B related to the chlorophyll content of the leaf and the grain weight of the wheat is separated and cloned from wheat for the first time, the Indel marker with polymorphism is developed between two varieties of Jing 411 and Hongmangchun 21 according to a cloned TaBAS1-2B gene sequence, the effect on phenotype is analyzed in 194 natural wheat varieties, and finally, a functional marker which is separated together with the gene TaBAS1-2B and is closely related to the chlorophyll content of the leaf and the grain weight of the wheat is developed. The Indel marker can be used for respectively explaining 8.2% and 4.4% of phenotypic variation of the chlorophyll content of a flag leaf and the thousand seed weight after wheat anthesis, wherein a strip of which the molecular weight is 494bp (Seq ID No.3) has a synergistic effect on the increase of the grain weight, and a strip of which the molecular weight is 493bp (Seq ID No.4) has a synergistic effect on the reduction of the grain weight. The development of the molecular marker provides a practical way for assistant breeding of high-yield wheat molecules.
Owner:ANHUI AGRICULTURAL UNIVERSITY
Short-chain dehydrogenase and mutant thereof as well as preparation of gene and application of short-chain dehydrogenase
The invention belongs to the technical field of biology, and relates to wild type short-chain dehydrogenase LfSDR1 and a gene of mutant enzyme of the short-chain dehydrogenase, construction of a recombinant expression vector containing the gene, preparation of recombinant expression transformant, recombinase and a preparation method of the recombinase. Through mutation of the wild type short-chaindehydrogenase LfSDR1, the mutant enzyme having two stereoselectivities (R or S stereoselectivities) is obtained, and catalytic asymmetrical reduction on prochiral ketone is carried out by utilizing the mutant enzyme, such as asymmetrical reduction of catalytic 1-phenyl ethanone compounds. Compared with the wild type short-chain dehydrogenase, the mutant disclosed by the invention has the advantages that the stereoselectivities is improved or reversed, a substrate is catalyzed so as to obtain chiral alcohol having two types (R or S type), such as optical pure (R)- or (S)-2-chloro-1-phenylethanol; moreover, the mutant has a good application value in the field of preparation of chiral alcohol.
Owner:SHENYANG PHARMA UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com