Method for applying adhesive tail end joints to flanking sequence separation
A technology of flanking sequences and sticky ends, applied in the field of sticky end adapters based on isotailase design, can solve the problems of high cost, incompatibility of connection efficiency and versatility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
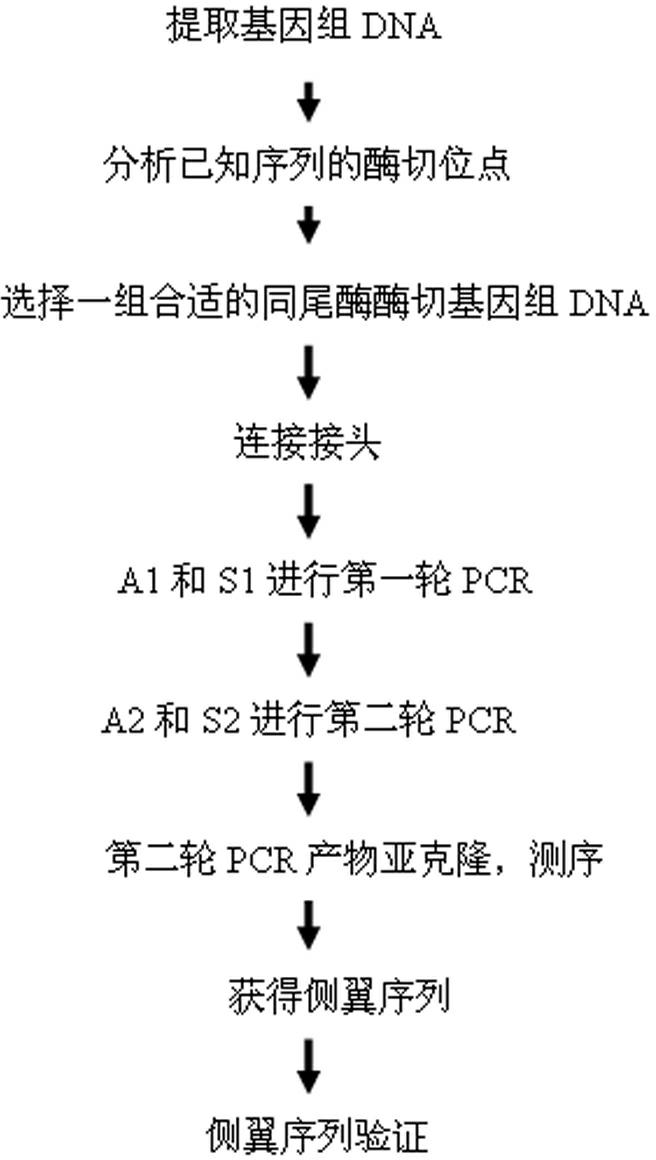
[0055] Example 1: Sticky end adapters are applied to the isolation of flanking sequences of rice pCK vector transgenes to obtain the insertion sites of foreign genes in the rice genome
[0056] The pCK transgene vector has two T-DNA regions, one of which contains the cDNA sequences of two C4 photosynthase genes (PEPC / PPDK), and each target gene is composed of rice Cab (Chlorophyll a / b binding protein, capture chlorophyll a / b-binding protein) promoter and terminated by 35s and NOS terminators, respectively. A T-DNA region contains the hygromycin phospho-transferase (HPT) gene as a selectable marker gene. After the rice was transformed, the transgenic lines without the HPT gene were selected for multigenerational propagation, and the transgene insertion site was analyzed after the homozygosity was stabilized. Proceed as follows:
[0057] 1. Extract the genomic DNA of transgenic rice;
[0058] 2. Analyze the distribution of restriction sites of the pCK vector, select BamHI...
Embodiment 2
[0073] Example 2: Sticky end joints applied to wheat PEBP Isolation of gene flanking sequences to obtain the promoter sequence of the gene
[0074] PEBP The gene encodes phosphatidylethanolamine-binding protein (Phsphatidylethanolamine-binding protein, PEBP), which is a cold-regulated protein that can change the fluidity of the plasma membrane of the cell and is also closely related to ion channels. The currently published wheat genome sequencing map is still in the form of a draft, and the complete wheat genome sequencing map will be completed in the next few years. This draft does not yet provide direct and effective sequences for most genomic studies of wheat. Therefore, we cannot base wheat's PEBP The gene sequence directly finds the sequence of the upstream promoter, and its promoter sequence can only be obtained through the technology of flanking sequence separation.
[0075] This embodiment includes the following steps:
[0076] 1. Extract the genomic DNA of whe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com