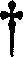Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
30 results about "Proofreading" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
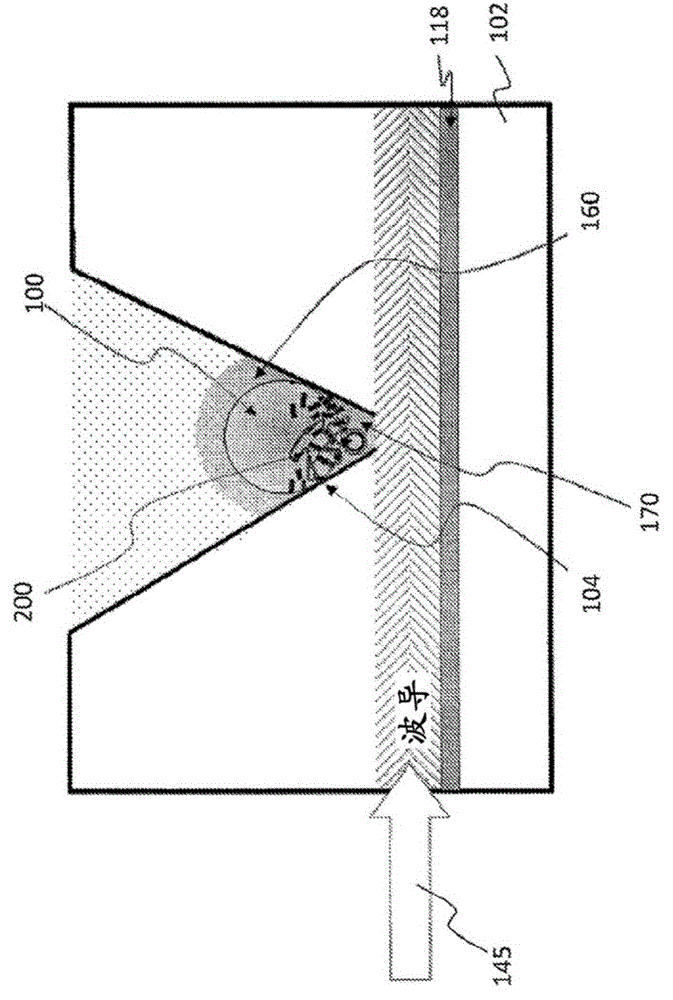
The term proofreading is used in genetics to refer to the error-correcting processes, first proposed by John Hopfield and Jacques Ninio, involved in DNA replication, immune system specificity, enzyme-substrate recognition among many other processes that require enhanced specificity. The proofreading mechanisms of Hopfield and Ninio are non-equilibrium active processes that consume ATP to enhance specificity of various biochemical reactions.
DNA (deoxyribonucleic acid) barcoding based molecular identification method of leech
ActiveCN103898235AImprove accuracyHigh sensitivityMicrobiological testing/measurementMolecular identificationWhitmania pigra
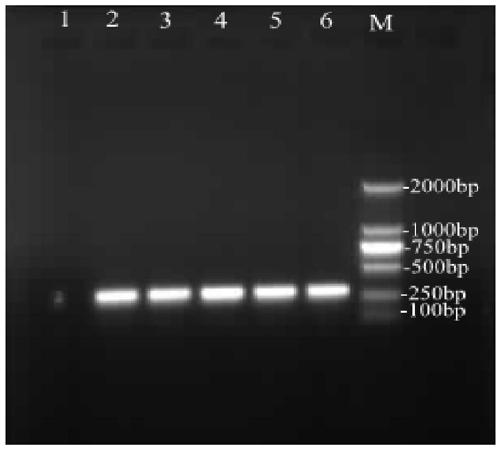
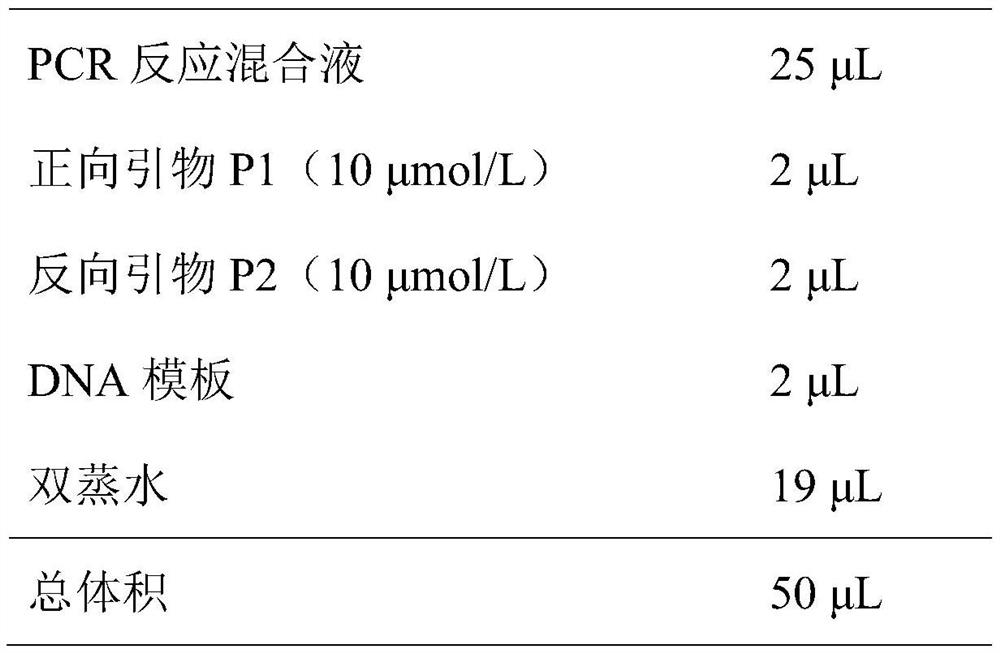
The invention discloses a DNA (deoxyribonucleic acid) barcoding based molecular identification method and CO I sequence of a leech (whitmania pigra). Polymerase chain reaction (PCR) is carried out by using the method to amplify a CO I gene of the leech, a PCR product is sent to a sequencing company to be sequenced after being verified by agarose gel, the sequencing results are subjected to manual proofreading and sequence splicing and are compared with public sequences, and the tissue to be tested can be judged to be the source of whitmania pigra if the homology of the sequencing results and a gene sequence SEQ ID NO.1 is above 99%. The whitmania pigra variety is quickly and accurately identified by adopting the reliable DNA molecular identification technology, thus improving the accuracy and the reliability and ensuring the safety of the leech as a medicine.
Owner:MUDANJIANG YOUBO PHARMA CO LTD
A DNA bar code standard gene sequence for Taiwan Lasiohelea
ActiveCN102690829AFacilitate the realization of molecular identificationShorten identification timeMicrobiological testing/measurementFermentationRapid identificationLasiohelea
The invention discloses a DNA bar code standard gene sequence for Taiwan Lasiohelea, which is characterized in that a bar code standard detection gene is COI gene, possessing gene sequence SEQ ID NO.1. Manual proofreading and sequence assembly are carried out to the sequencing result and Blast similarity search is carried out in NCBI to ensure that the obtained sequence is the target sequence. Species identification of Taiwan Lasiohelea is realized through morphological identification, thereby result reliability is guaranteed. The DNA bar code standard gene sequence for Taiwan Lasiohelea provided in the invention is in favor of realizing rapid identification of Taiwan Lasiohelea, and reduces identification time.
Owner:FUJIAN INT TRAVEL HEALTH CARE CENT
Compositions for DNA amplification, synthesis, and mutagenesis
InactiveUS20060246462A1Product yield is lowReduced target-length capabilityHydrolasesMicrobiological testing/measurementNucleotideA-DNA
This invention provides compositions comprising a thermostable non-proofreading DNA polymerase, a thermostable proofreading DNA polymerase, and a factor that substantially inhibits the incorporation of undesired nucleotides or analogs thereof into a DNA polymer. The compositions may further comprise a buffer that enhances a polymerization reaction involving DNA polymerases. The invention also provides various methods of amplifying, synthesizing, or mutagenizing nucleic acids of interest using these novel compositions. Kits that comprise the compositions are also provided for amplifying, synthesizing, and mutagenizing nucleic acids.
Owner:STRATAGENE INC US
DNA bar code standard detection fragment for identifying vespa soror and application of detection fragment
ActiveCN111635949ASolve the difficult problem of rapid and accurate identificationShorten identification timeMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationMedicinal herbs
The invention discloses a DNA bar code standard detection fragment for identifying vespa soror and application of the detection fragment, and belongs to the field of biological species identification.The nucleotide sequence of the DNA bar code standard detection fragment is as shown in SEQ ID No. 1, and the invention also provides a method for molecular identification of insect medicinal materialvespa soror by using the gene sequence and a PCR amplification technology. A PCR product is verified through agarose gel electrophoresis and then subjected to sequencing, a sequencing result is subjected to manual proofreading and sequence splicing and is compared with a COI gene sequence SEQ ID NO.1 disclosed by the invention, and therefore whether a sample to be detected is vespa soror or not is judged. According to the DNA molecular identification technology, morphological identification does not need to be carried out on insect medicinal material scorpions, quick, accurate and reliable identification on fresh or dry medicinal materials, whole worms or part of worms, pupae or larvae or adults is achieved, and the safety of taking the vespa soror as a medicinal material is guaranteed.
Owner:DALI UNIV
Method for analyzing plant endophyte colonies
The invention discloses a method for analyzing plant endophyte colonies. In the method, a primer pair used for analyzing the plant endophyte colonies is provided, the target sequence of the primer pair is the overall length or part of the region from V3 to V4 of bacteria 16SrDNA; there are differences between the last basic group at the 3' end of one primer in the primer pair and the plant mitochondria 18S rDNA, and the number of the difference sites between the overall length of the primer and the plant mitochondria 18S rDNA is equal to or larger than 3; there are differences between the lastbasic group at the 3' end of the other primer and the plant chloroplast 16S rDNA, and the number of the difference sites between the overall length of the primer and the plant mitochondria 18S rDNA is equal to or larger than 3. The two primers 322F-1 and 796R are designed, the DNA polymerase with hot-start activity but without 3' to 5' exonuclease proofreading activity is selected to achieve theaim that the pollution of the host plant chloroplast 16S rDNA and the pollution of the host plant mitochondria 18S rDNA are avoided at the same time, and therefore, a pure bacteria colony 16S ampliconlibrary is obtained.
Owner:INST OF MICROBIOLOGY - CHINESE ACAD OF SCI
Curcama kwangsiensis DNA barcode standard gene sequence
InactiveCN103695536AFacilitate the realization of molecular identificationAchieve molecular identificationMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationDNA barcoding
The invention discloses a curcama kwangsiensis DNA barcode standard gene sequence. The curcama kwangsiensis DNA barcode standard gene sequence is characterized in that the barcode standard detection gene is an ITS gene and has the gene sequence shown in the formula of SEQ ID NO.1. The sequencing result is subjected to manual proofreading, sequence splicing and Blast similarity searching in NCBI so that it is ensured that the obtained sequence is a required sequence. Curcama kwangsiensis species identification is realized based on morphology and molecular identification so that result reliability is guaranteed. The curcama kwangsiensis DNA barcode standard gene sequence is conducive to fast identification of curcama kwangsiensis, shortens identification time and improves identification accuracy.
Owner:GUILIN MEDICAL UNIVERSITY
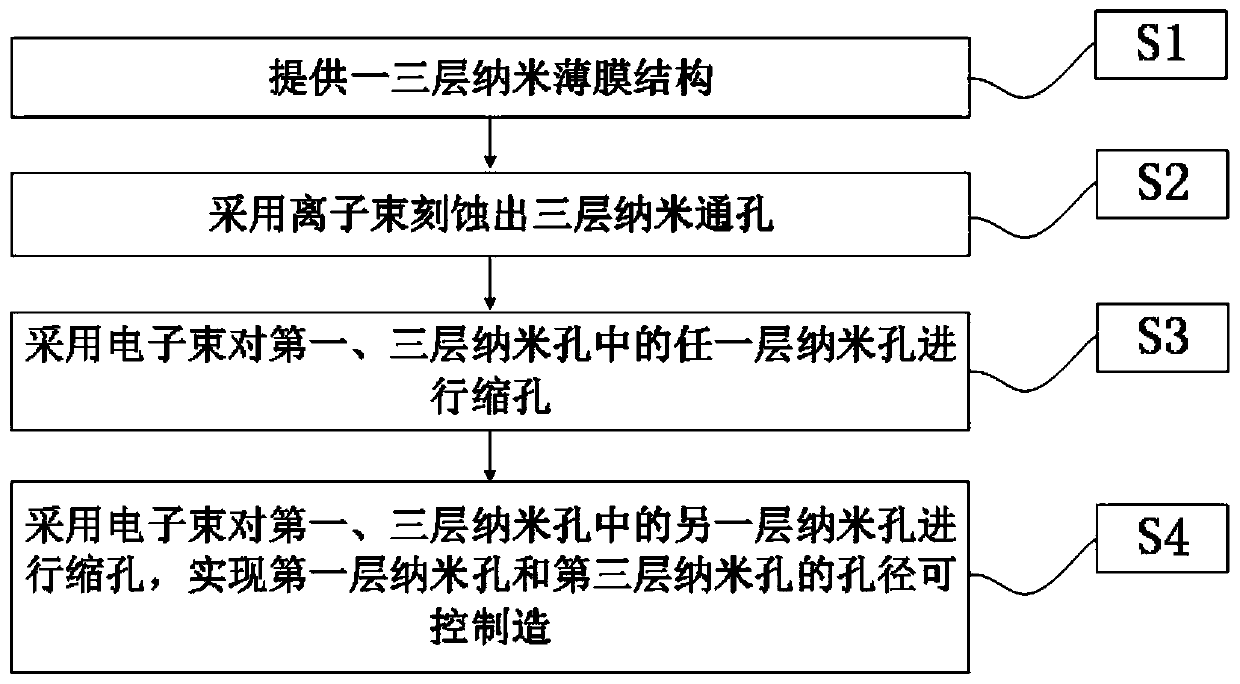
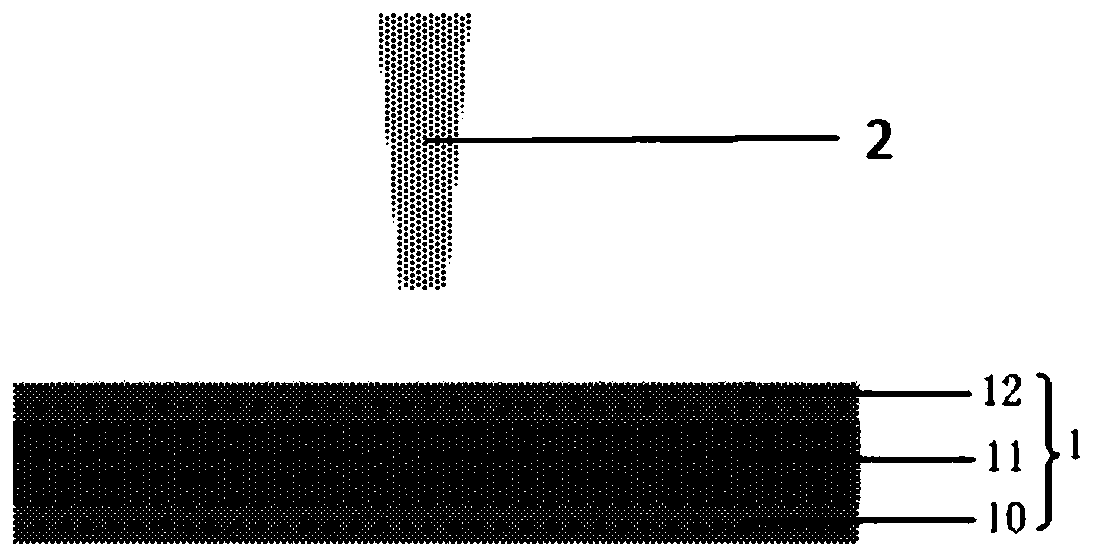
Size-controllable three-layered nanopore structure and preparation method and application thereof
PendingCN109811046AGood size controlRealize dynamic school pairingMicrobiological testing/measurementNanomedicineImage resolutionKinetic proofreading
The invention discloses a size-controllable three-layered nanopore structure and a preparation method and application thereof. The preparation method comprises the following steps: (S1) by etching a three-layered nano-film structure, a three-layered through nanopore is obtained; (S2) an electron beam is adopted to shrink the three-layered through nanopore in S1, so that a three-layered nanopore structure is obtained; and the scanning area of the electron beam is 300nm multiplied by 300nm to 20 Mu m multiplied by 20 Mu m. The invention integrates an ion beam with an electron beam to prepare thethree-layered nanopore structure with a smaller nanopore aperture, and moreover, the size controllability of the nanopore can be realized. The invention can prepare the three-layered nanopore structure with a thinner film and a nanopore aperture less than 10nm, and the three-layered nanopore structure can realize the mechanical proofreading of DNA molecules and increase the vertical resolution and horizontal resolution of base sequence recognition.
Owner:GUANGDONG UNIV OF TECH
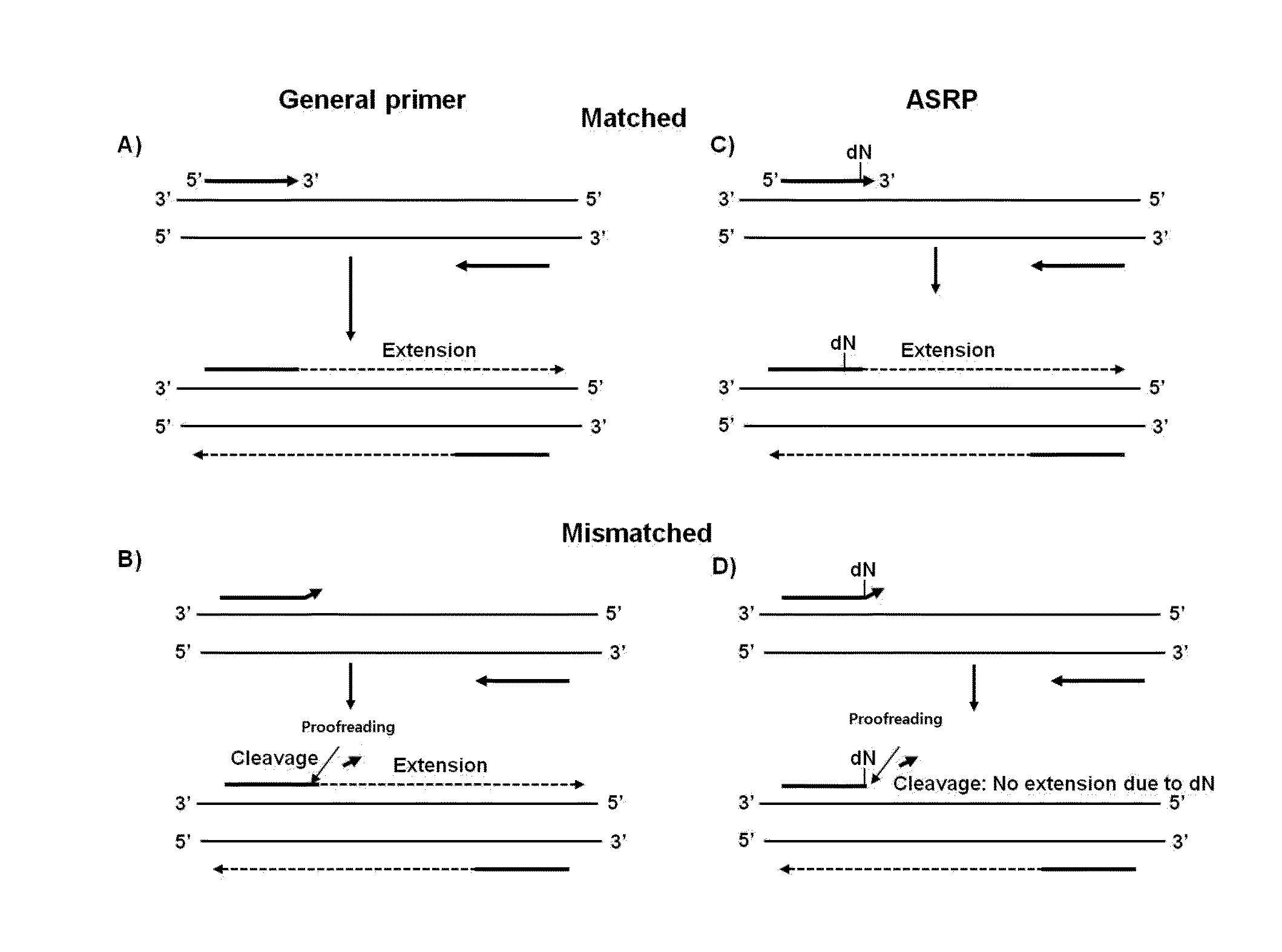
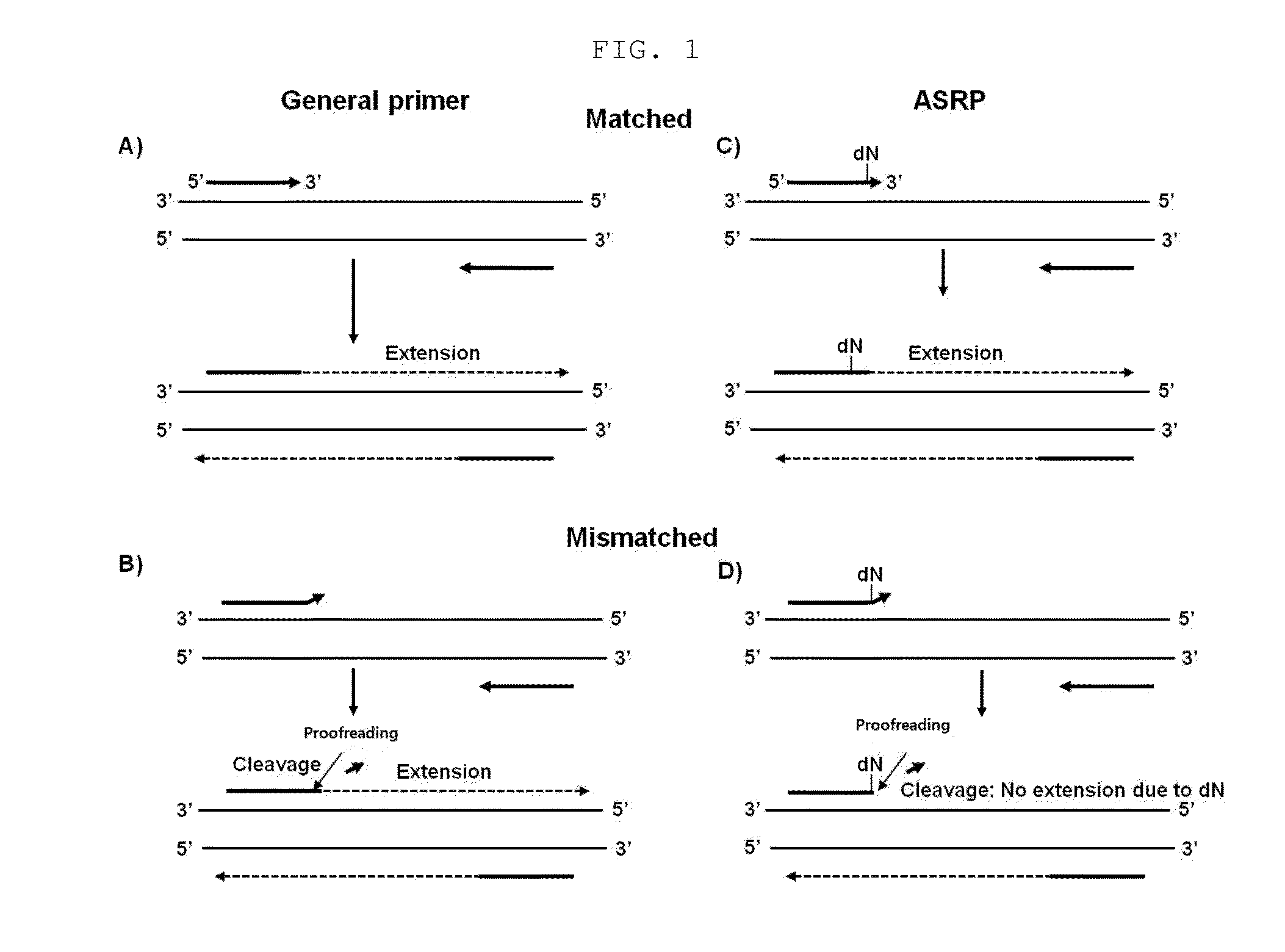
Nucleic acid amplification method using allele-specific reactive primer
ActiveUS20160194695A1Microbiological testing/measurementEnzymology/microbiology apparatusPolymerase LA-DNA
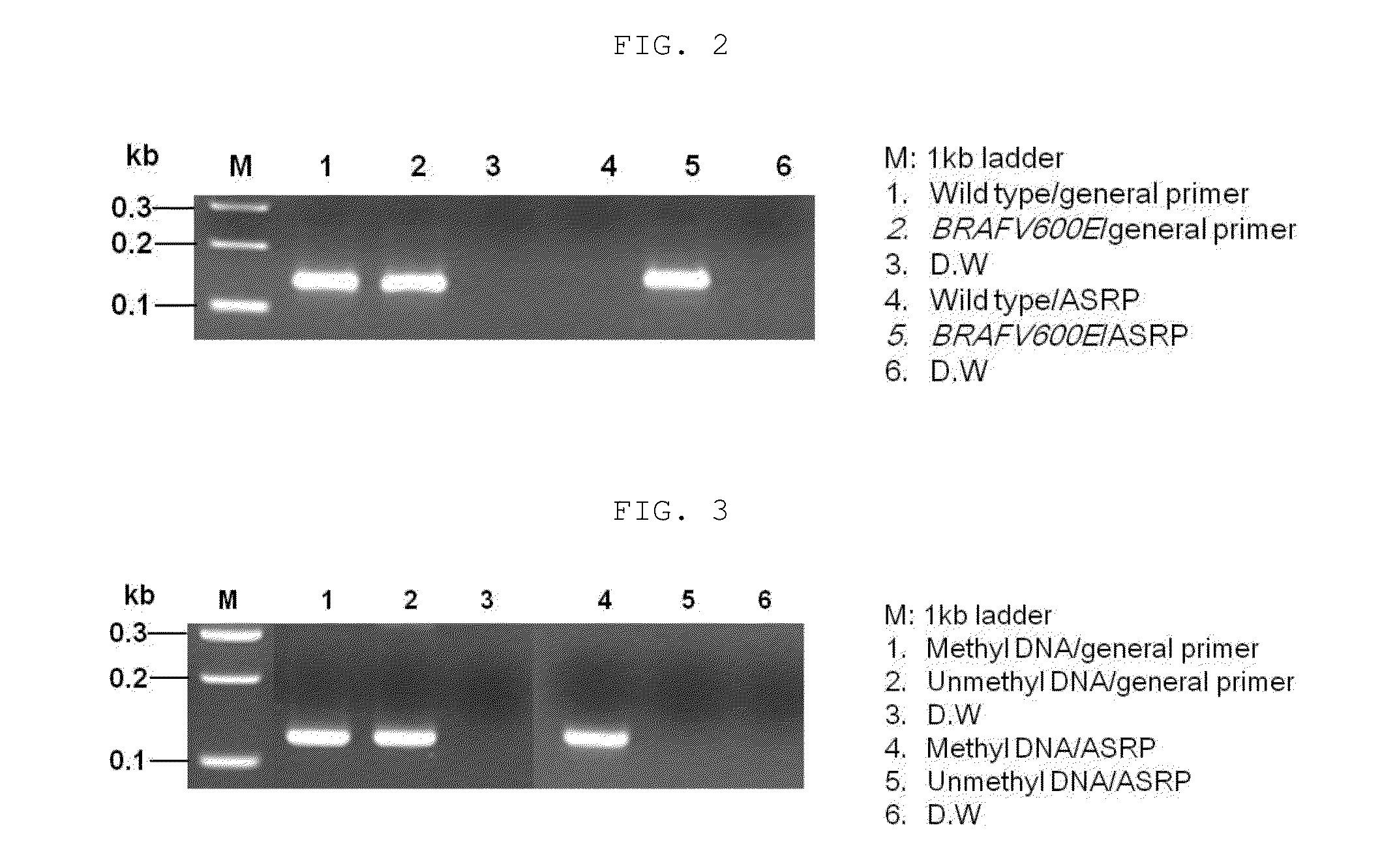
The present invention relates to a method for amplifying nucleic acid using an allele-specific reactive primer (ASRP) designed to solve the problems of conventional allele-specific PCR, and more particularly, to method for detecting a target nucleic acid, which comprises amplifying the target nucleic acid in the presence of a DNA polymerase having proofreading activity, and an allele-specific reactive primer (ASRP) which comprises i) a nucleotide sequence complementary to the target nucleic acid and ii) one or more modified nucleotide(s) located in a region from a nucleotide located right before of a non-complementary nucleotide to 5′ direction of the primer, which will be removed by the proofreading activity of the DNA polymerase when a non-complementary nucleotide is present at the 3′ end, to a nucleotide located at the 5′ end of the primer, in order for the nucleotide in the allele-specific reactive primer cannot act as a primer for a polymerase reaction.The detection method using the allele-specific reactive primer (ASRP) according to the present invention is a technique having a very high specificity of amplification due to the characteristics of the ASRP and a proofreading DNA polymerase. The detection method can effectively detect mutations (point mutation, insertion, deletion, etc.) including a single nucleotide polymorphism (SNP), and can also be used to detect methylation of CpG after bisulfite treatment or to amplify and detect a target DNA starting with a desired nucleotide sequence from a DNA library.
Owner:GENOMICTREE
Thermostable or thermoactive dna polymerase with attenuated 3'-5' exonuclease activity
Owner:F HOFFMANN LA ROCHE & CO AG
Massively Parallel Enzymatic Synthesis of Nucleic Acid Strands
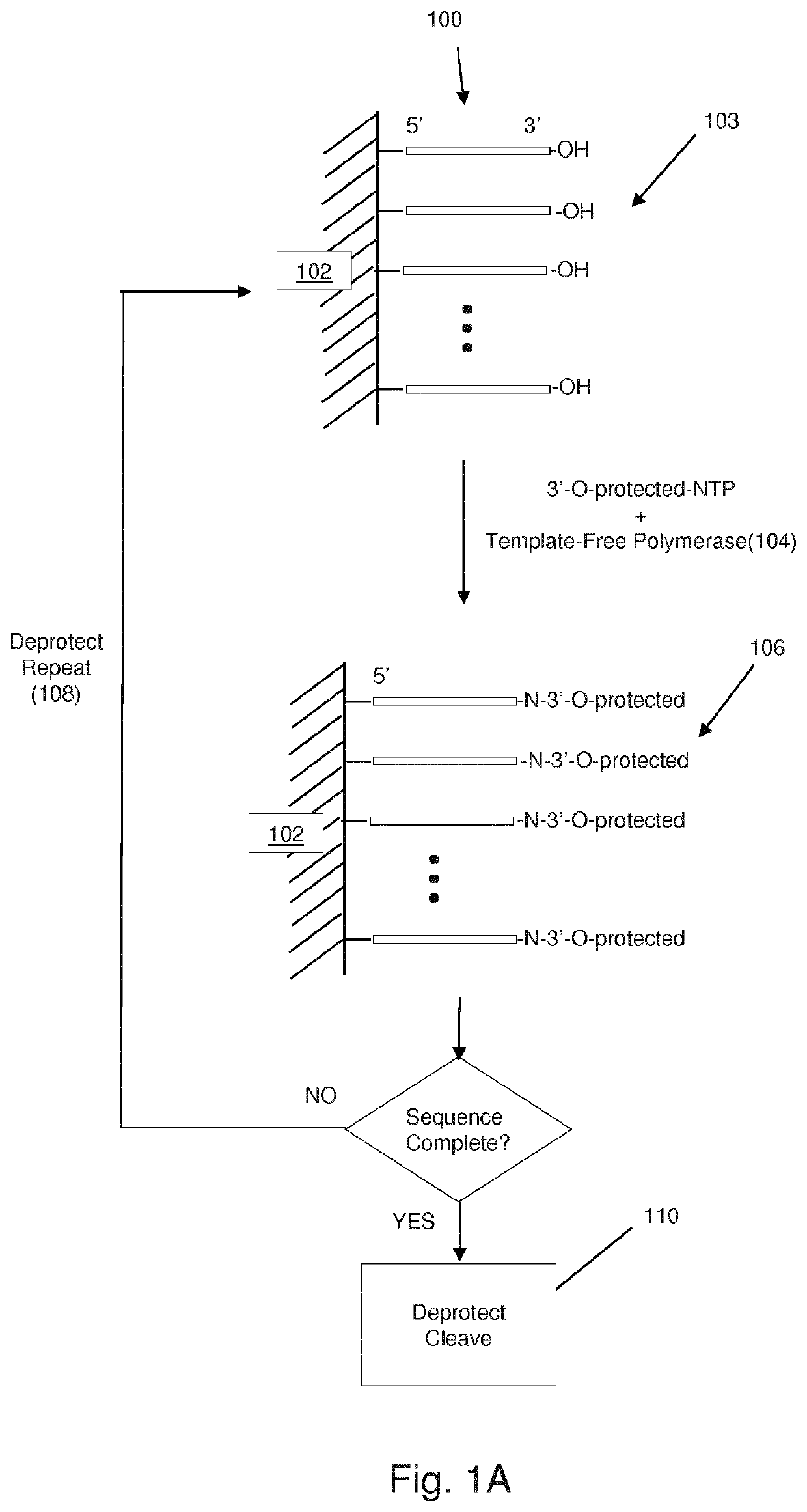
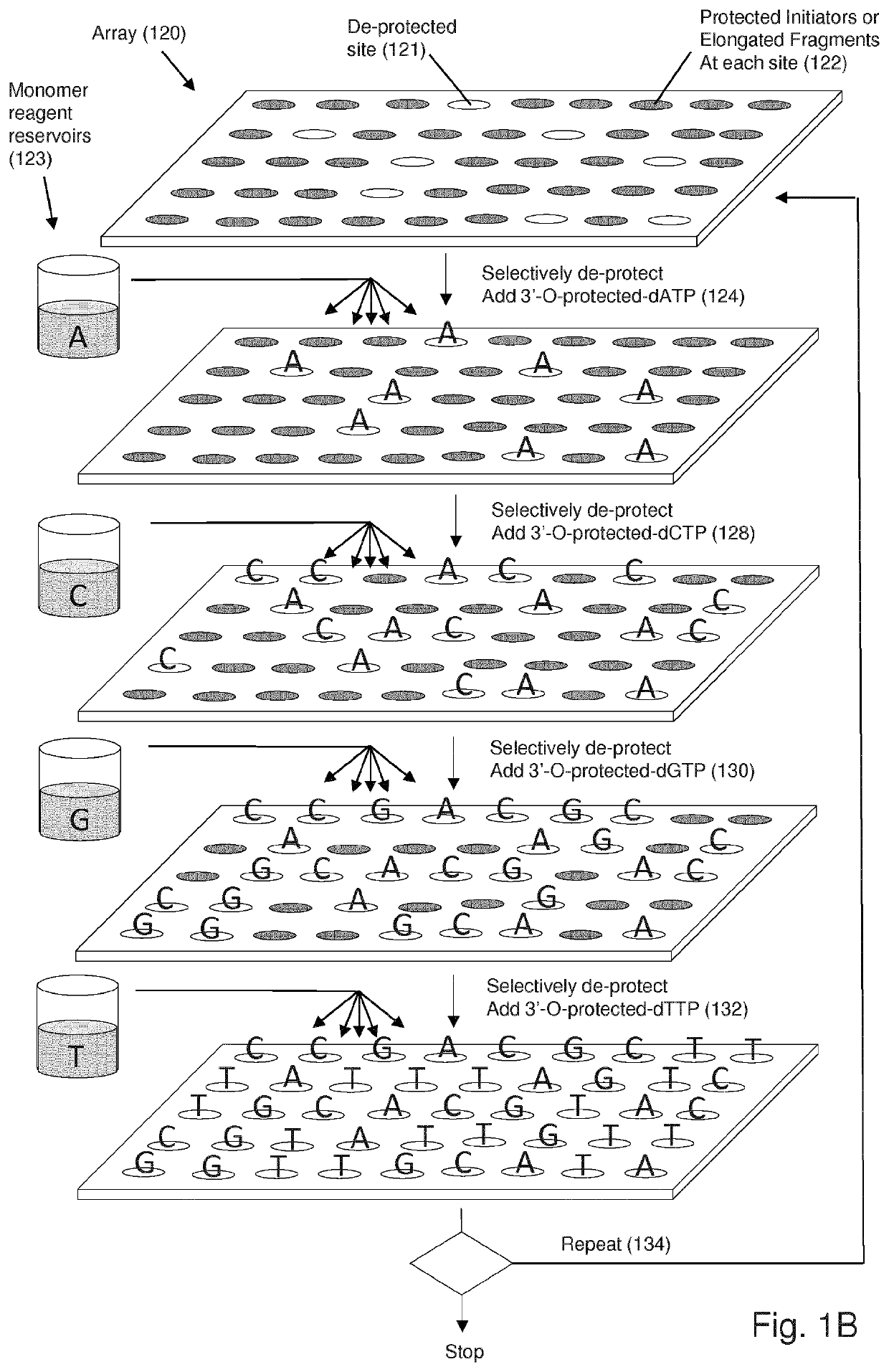
PendingUS20210332351A1Extended storage lifeImprove information integritySequential/parallel process reactionsBioinformaticsEnzymatic synthesisHomopolynucleotide
The invention is directed to methods for massively parallel template-free enzymatic synthesis of a plurality of different polynucleotides of predetermined sequences. In one aspect, methods of the invention employ large scale arrays of reaction sites each associated with at least one working electrode for controlling deprotection and deblocking steps at predetermined user selected sites. In another aspect, the invention provides template-free enzymatic synthesis with proofreading, wherein completed polynucleotides at predetermined reaction sites are sequenced using a sequencing by synthesis technique, particularly employing electrochemically labile blocking groups.
Owner:DNA SCRIPT SAS +1
Pathological picture three-dimensional reconstruction method
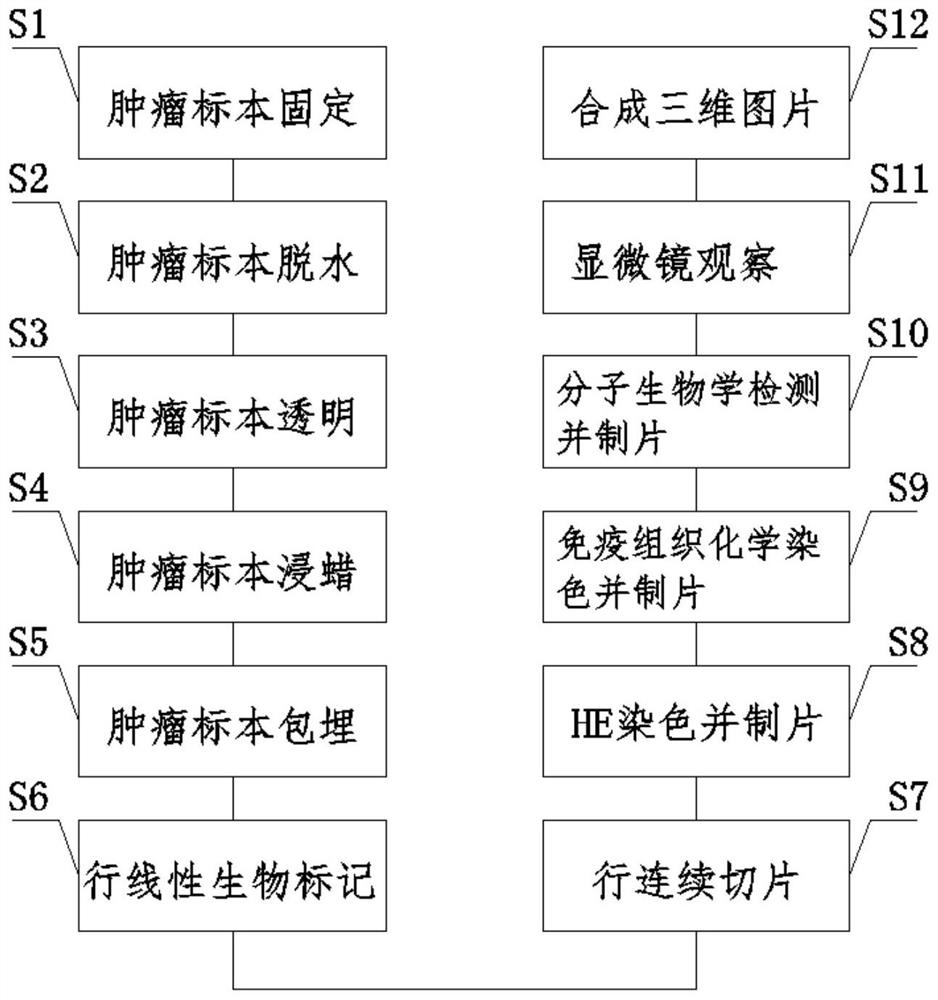
InactiveCN113138110ARealize 3D reconstructionComplete picturePreparing sample for investigationMaterial analysis by optical meansStainingMicroscopic observation
The invention discloses a pathological picture three-dimensional reconstruction method in the technical field of pathological picture display. The method comprises the following steps: S1, fixing a tumor specimen; S2, dehydrating the tumor specimen; S3, transparentizing the tumor specimen; S4, performing wax dipping on the tumor specimen; S5, embedding the tumor specimen; S6, carrying out linear bio-labeling on rows; S7, performing continuous slicing on rows; S8, carrying out HE dyeing and flaking; S9, performing immunohistochemical staining and flaking; S10, performing molecular biological detection and flaking; S11, observing with a microscope; and S12, synthesizing a three-dimensional picture. According to the method, on the basis of the two-dimensional pictures, continuous slicing and accurate positioning of the pathological specimen are carried out, so that after the tumor leaves the body, three-dimensional reconstruction of a plurality of fault pictures and three-dimensional reconstruction of the pathological picture are finally realized through sequential listing, proofreading and accurate positioning of the two-dimensional pictures in a pathological space; pathologists can observe more complete tumor tissue images as much as possible, and more complete and accurate pathology reports are provided for patients and clinicians.
Owner:JILIN UNIV
Biomolecule detection method
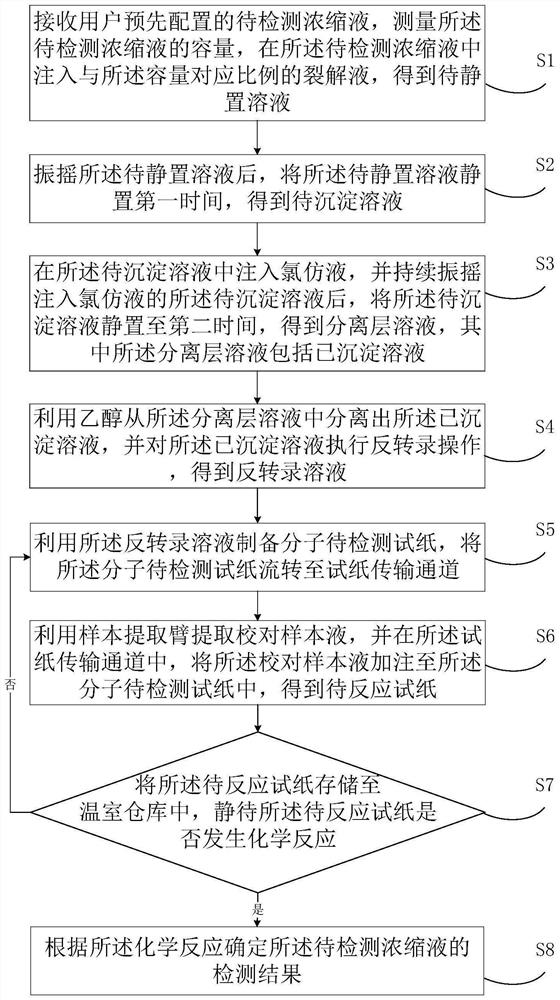
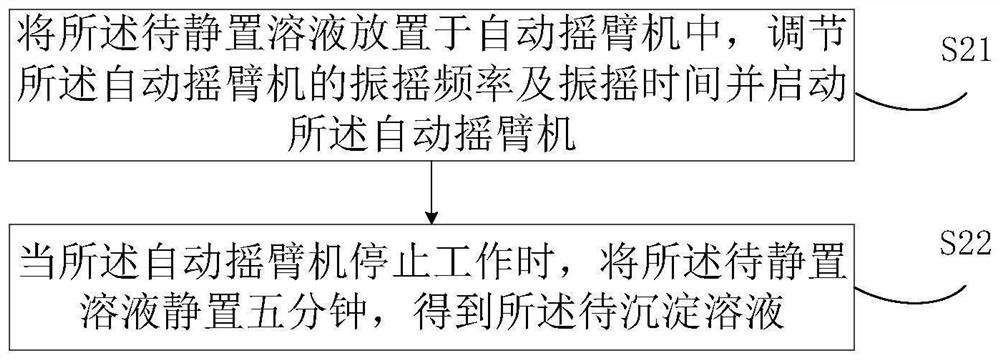
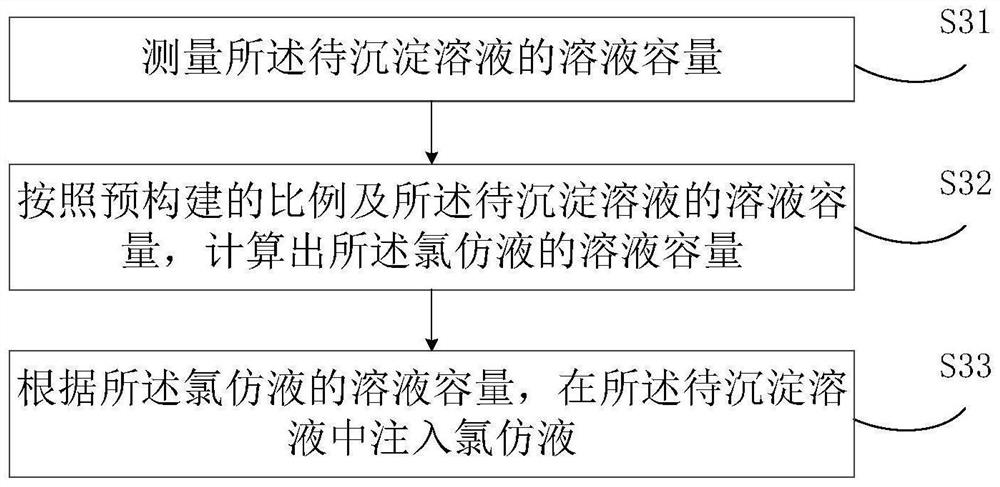
PendingCN114400044ASolve the problem of low detection intelligenceReduce interventionProteomicsGenomicsBio moleculesChemical reaction
The invention relates to the field of biological detection, and discloses a biological molecule detection method which comprises the following steps: separating a precipitated solution from a pre-constructed separation layer solution, performing reverse transcription operation on the precipitated solution to obtain a reverse transcription solution, preparing molecular test paper to be detected by using the reverse transcription solution, and detecting the molecular test paper to be detected. Transferring the to-be-detected molecular test paper to a test paper transmission channel, extracting a proofreading sample liquid by using a sample extraction arm, filling the to-be-detected molecular test paper with the proofreading sample liquid in the test paper transmission channel to obtain to-be-reacted test paper, and storing the to-be-reacted test paper in a greenhouse warehouse. And judging whether the test paper to be reacted has chemical reaction or not, and if the test paper to be reacted has chemical reaction, determining the detection result of the concentrated solution to be detected according to the chemical reaction. The invention further discloses a biomolecule detection method and device, electronic equipment and a storage medium. The problem that the intelligent degree of biomolecule detection is not high can be solved.
Owner:量准(上海)实业有限公司
Mutant DNA polymerases and their genes
The present invention relates to mutant DNA polymerases, their genes and their uses. More specifically, the present invention relates to mutant DNA polymerases which are originally isolated from Thermococcus sp NA1. strain and produced by site-specific mutagenesis, their amino acid sequences, genes encoding said mutant DNA polymerases, their nucleic acids and PCR methods by using thereof. As mutant DNA polymerases according to the present invention have decreased proofreading activity and changed function of inosine sensing effectively compared to wild type DNA polymerase, PCR using primers with specific nucleic acids has made rapid progress. Therefore, the present invention is broadly applicable for PCR in various molecular genetic technologies.
Owner:KOREA OCEAN RES & DEV INST
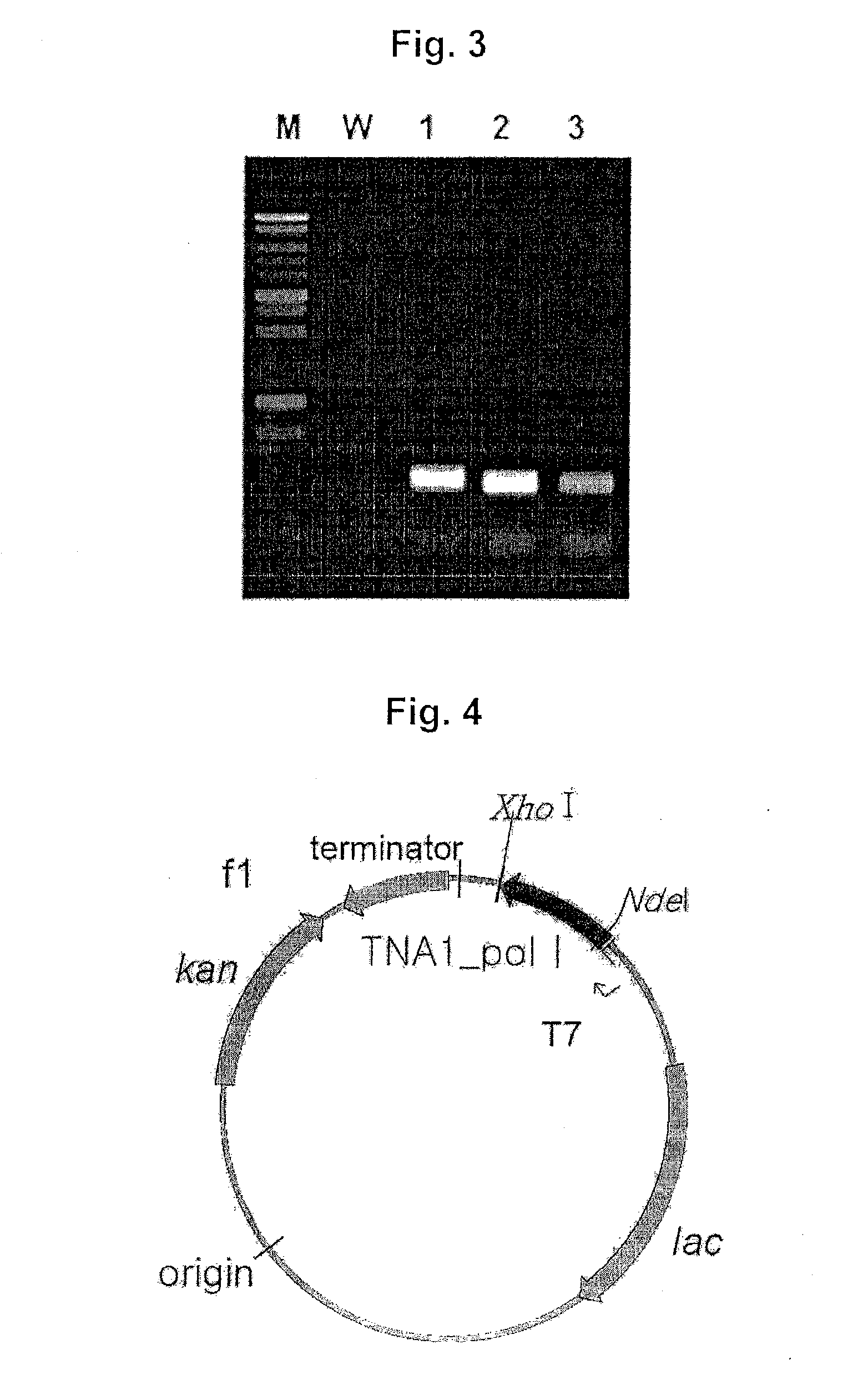
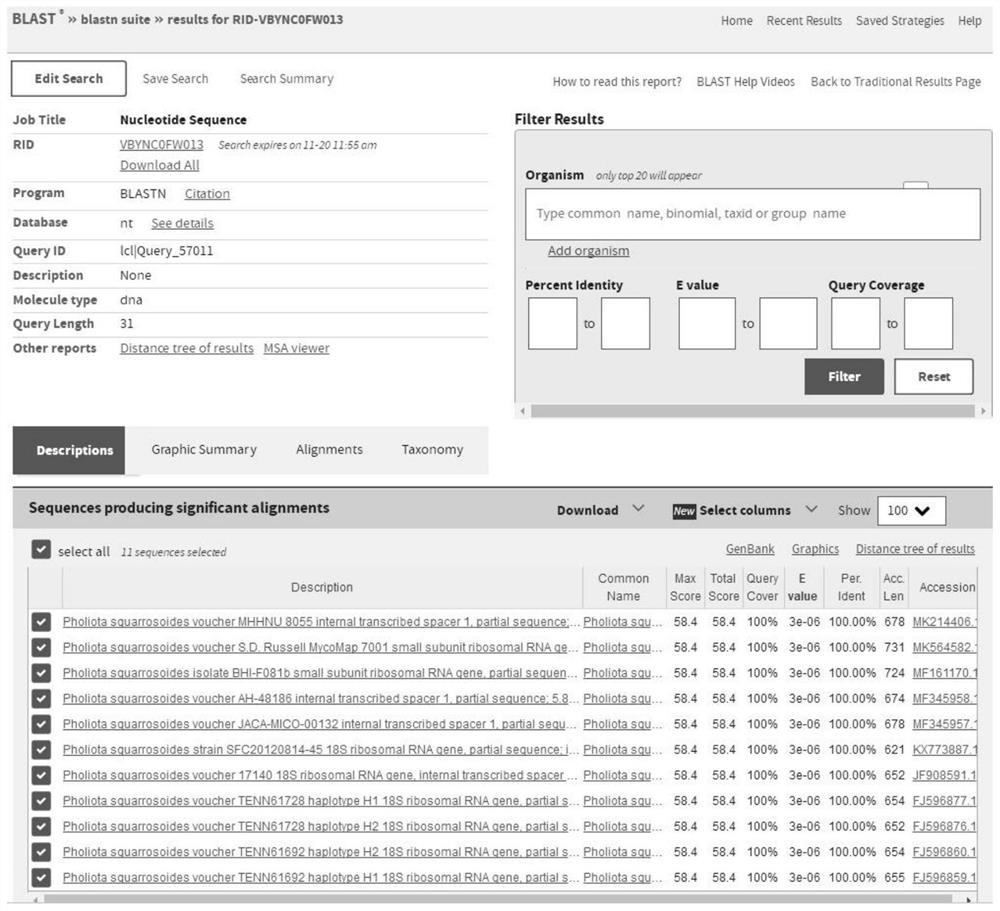
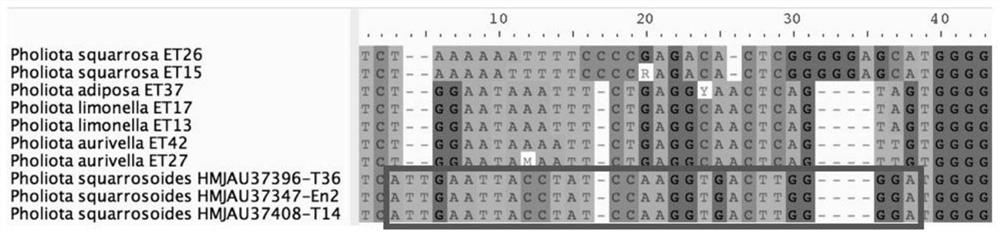
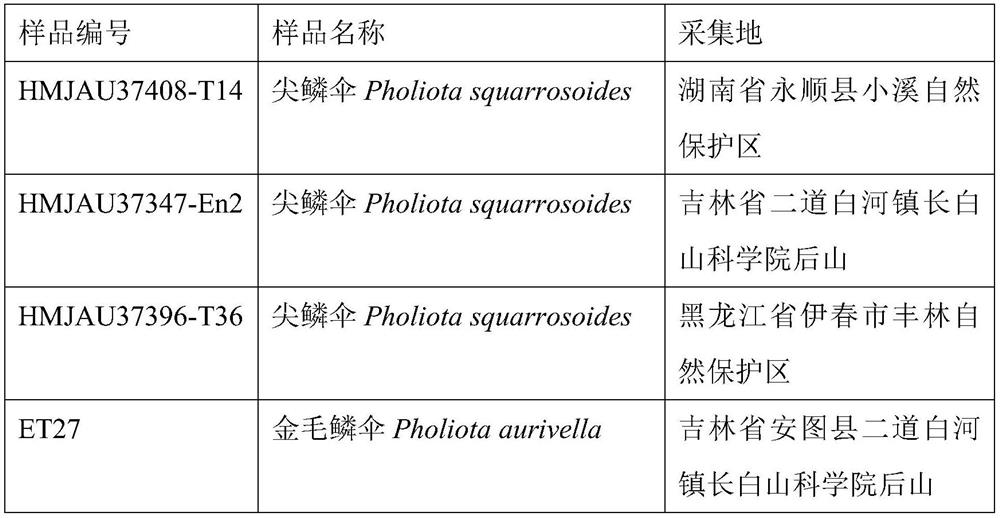
Specific gene of pholiota adipose and application of specific gene
ActiveCN112725490ARapid and Accurate AmplificationSimple and fast operationMicrobiological testing/measurementDNA/RNA fragmentationPholiotaEdible mushroom
The invention relates to a specific gene of pholiota adipose and an application of the specific gene, a short sequence of the specific gene is shown as SEQ ID NO.1: ATTGAATTACCTATCCAAGGTGACTTGGGGA, and a method for identifying pholiota adipose molecules by using the specific gene comprises the following steps: (1) extracting genome DNA from a to-be-detected sample tissue; (2) by taking the genome DNA extracted in the step (1) as a template, carrying out polymerase chain reaction by using a pholiota adipose specific primer to obtain an amplified DNA product; (3) sequencing the DNA product obtained by amplification in the step (2); and (4) splicing and proofreading sequencing results to obtain an amplified fragment, detecting whether the specific gene short sequence SEQ ID NO.1 exists in the amplified fragment or not, and identifying the sample as pholiota adipose if the specific gene short sequence SEQ ID NO.1 exists in the amplified fragment. The specific short sequence and the method provided by the invention can be used for rapidly, accurately and accurately identifying the poison mushroom pholiota adiposa from the easily confused edible mushrooms, and the specific short sequence is easy to amplify, simple and convenient to operate and wide in applicability.
Owner:JILIN AGRICULTURAL UNIV
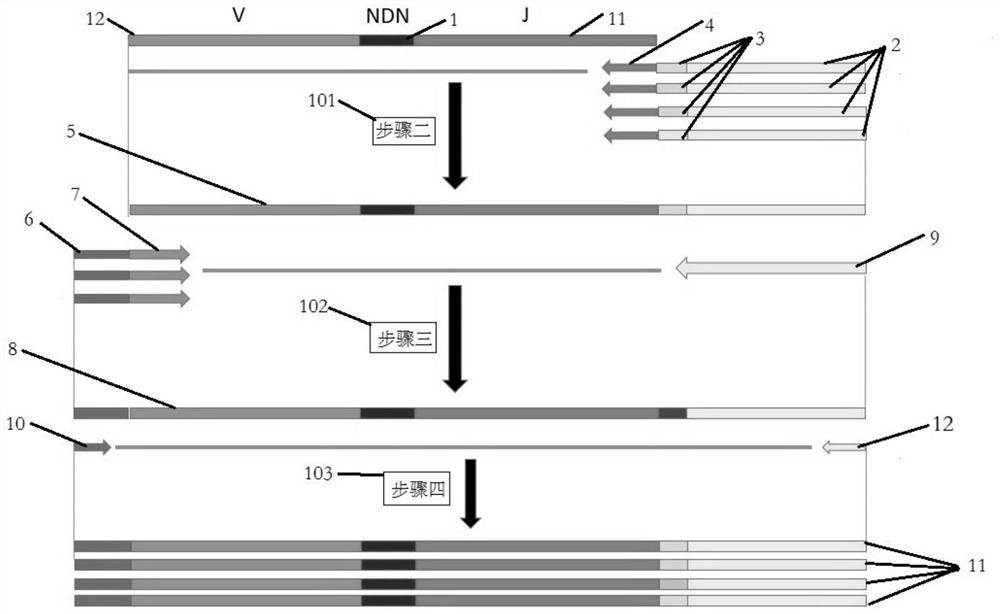
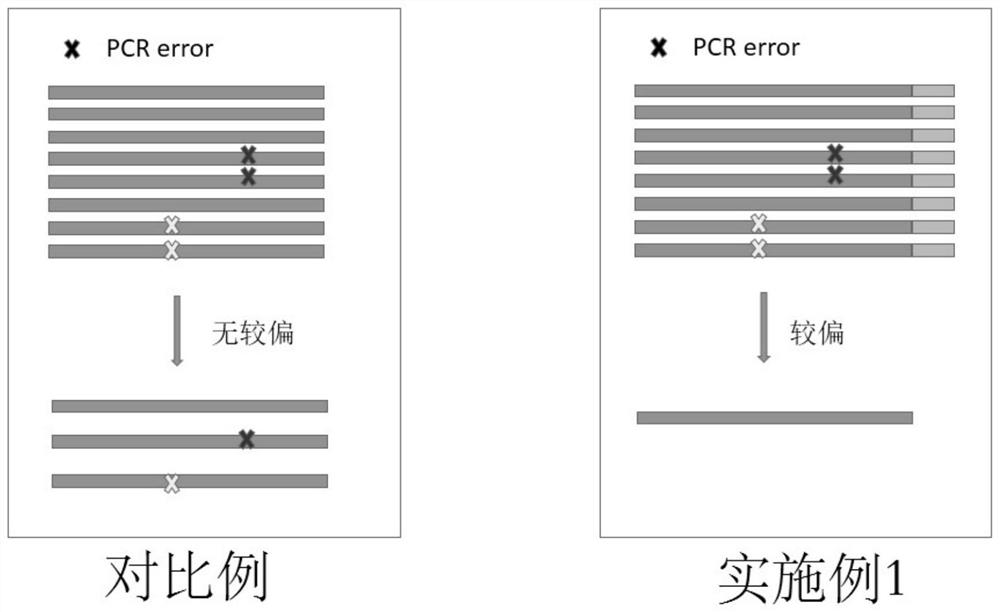
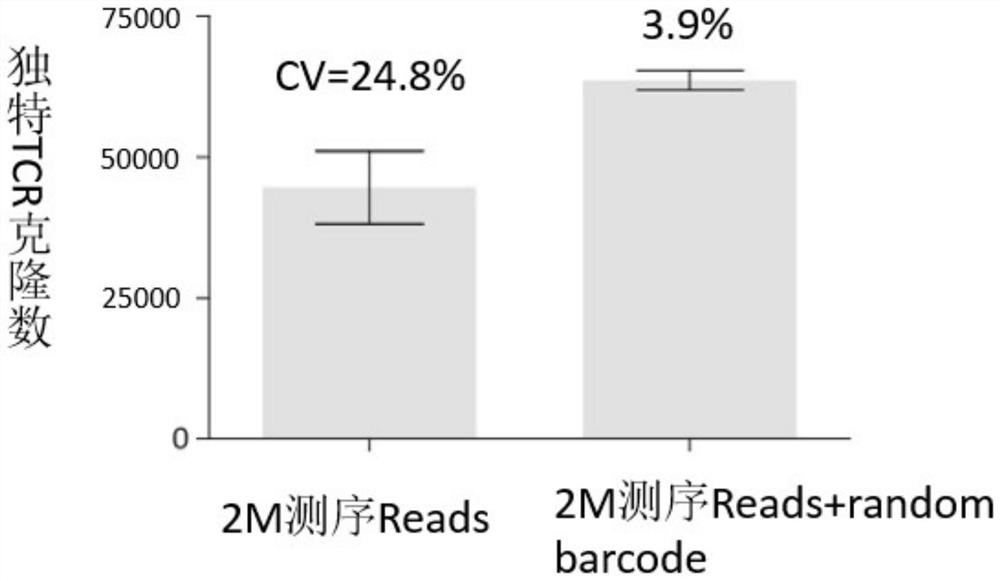
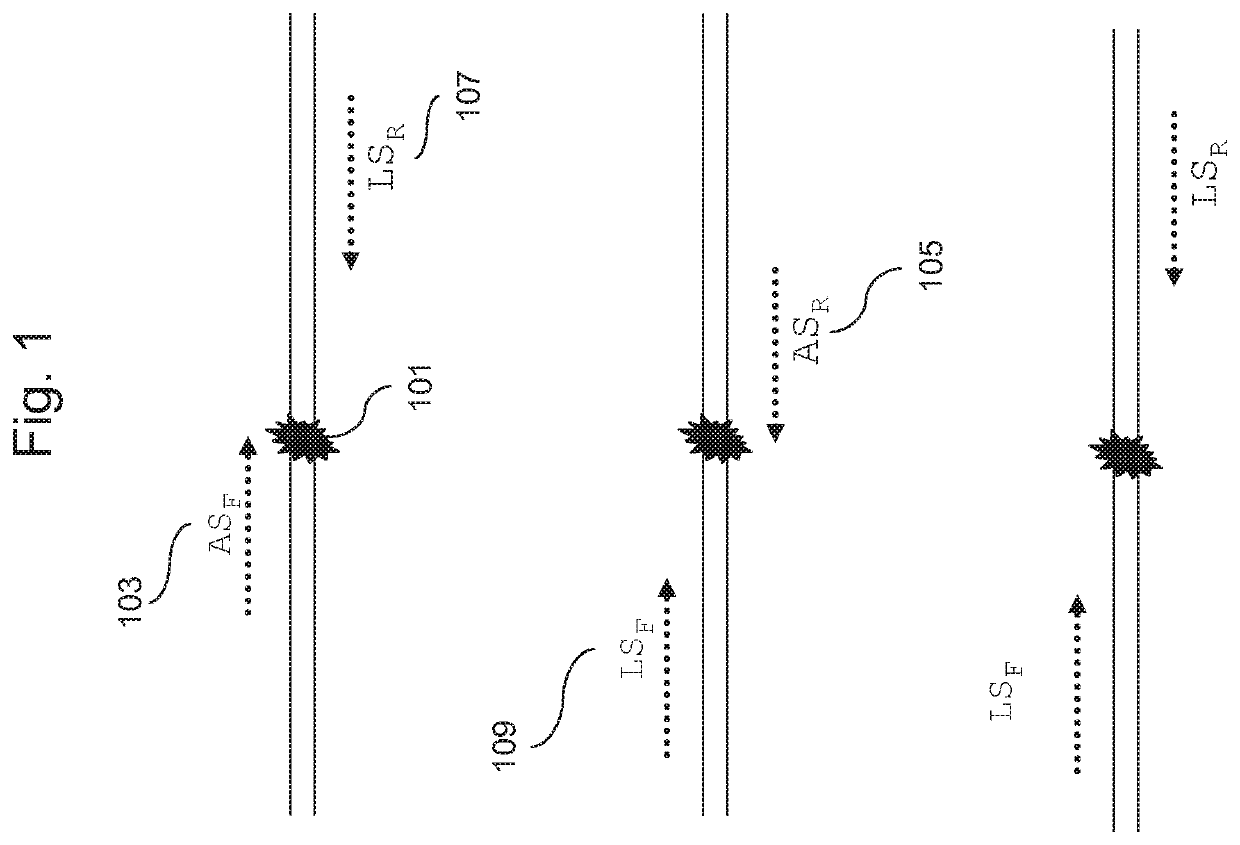
Sequencing method for constructing variable region sequence library kit and variable region sequence
The invention belongs to the field of biotechnology, and in particular relates to a kit for constructing a variable region sequence library and a sequencing method for the variable region sequence. The invention discloses a kit for constructing a variable region sequence library, comprising the following primers: a first primer group, a first adapter primer and a second primer group; the first primer group includes coding sequences that specifically recognize all subtypes of the J region The first primer; the first primer includes the nucleotide sequence of the coding sequence of the specific recognition J region connected in sequence, the proofreading random segment and the first adapter; the second primer group includes the specific recognition of the V region connected in sequence The second primers for the coding sequences of all subtypes; the second primers include a nucleotide sequence that specifically recognizes the coding sequences of the V region and a second linker. The invention is used to solve the technical defects of low amplification efficiency, bias in amplification and easy loss of sequence information of the variable region sequence library existing in the current variable region sequence library technology for constructing TCR or BCR.
Owner:GUANGZHOU HOMEY HEALTH TECH
Methods for genotyping
ActiveUS11306351B2Reduce complexityMicrobiological testing/measurementAllele-specific oligonucleotideA-DNA
Owner:AFFYMETRIX INC
A set of bac markers for distinguishing 13 pairs of chromosomes in sesame
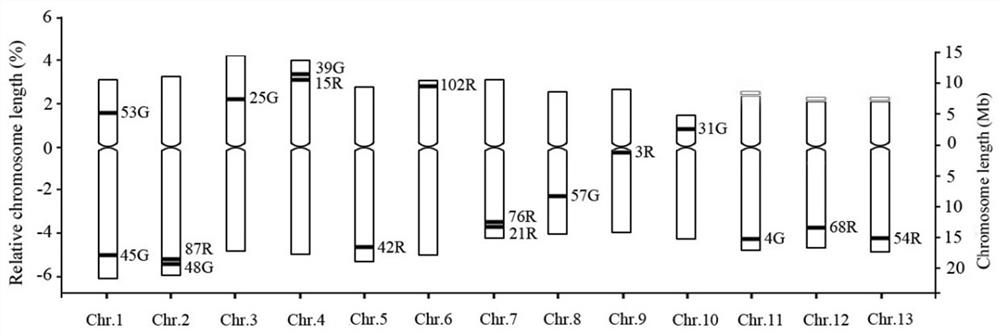
ActiveCN107541553BStable hybridization signalStable and reliable hybridization resultsMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyZoology
The invention belongs to the technical field of sesame genomes, and specifically relates to a set of BAC marker patent applications for distinguishing 13 pairs of chromosomes in sesame. The group of BAC markers includes a total of 17 BAC information. The basic information of the BAC markers is shown in the table below, and the base sequence information of the paired ends of the BAC is shown in SEQ ID NO.1~34. The 17 BAC markers provided by this application are all single specific sites on the chromosome, with site specificity and uniqueness; using the differentiation method provided by this application, each chromosome of sesame can be accurately identified and determined, which can be used for sesame high The research on the construction of density cytogenetic map and proofreading of sesame genome has important theoretical significance and scientific value, which can provide theoretical support and lay an application foundation for subsequent sesame genomics research and sesame breeding.
Owner:HENAN SESAME RES CENT HENAN ACADEMY OF AGRI SCI
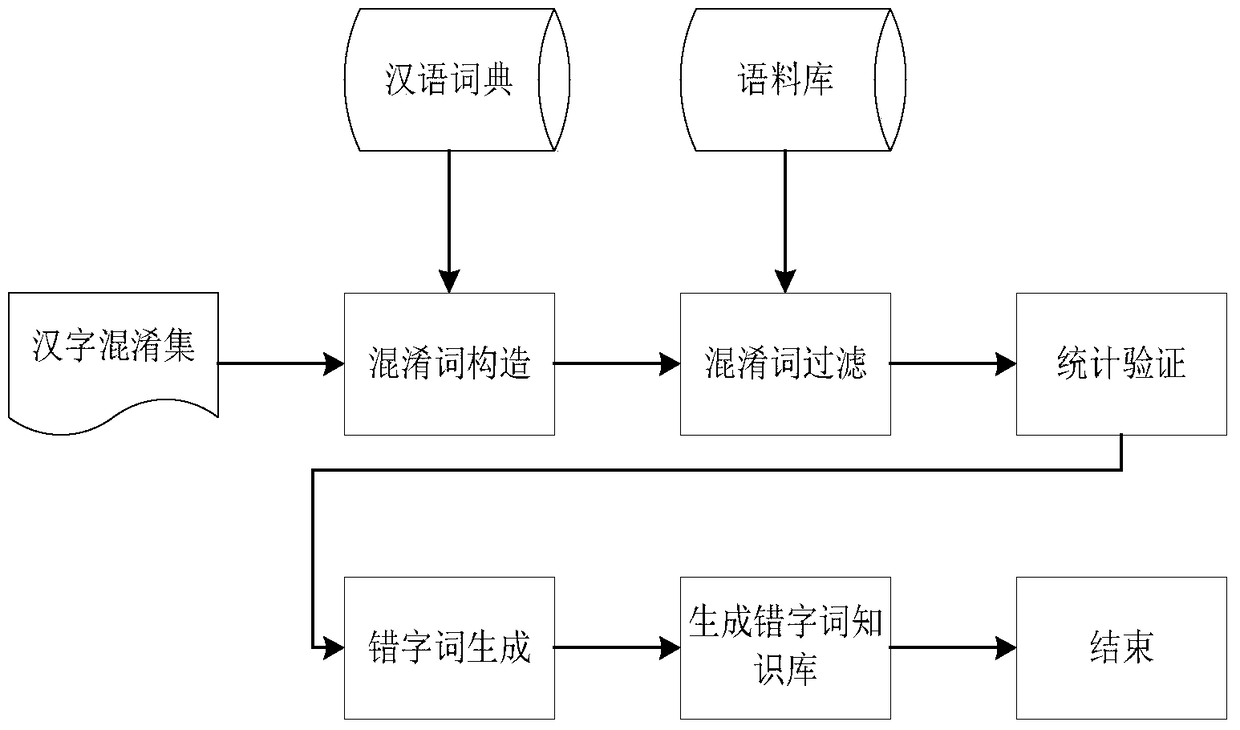
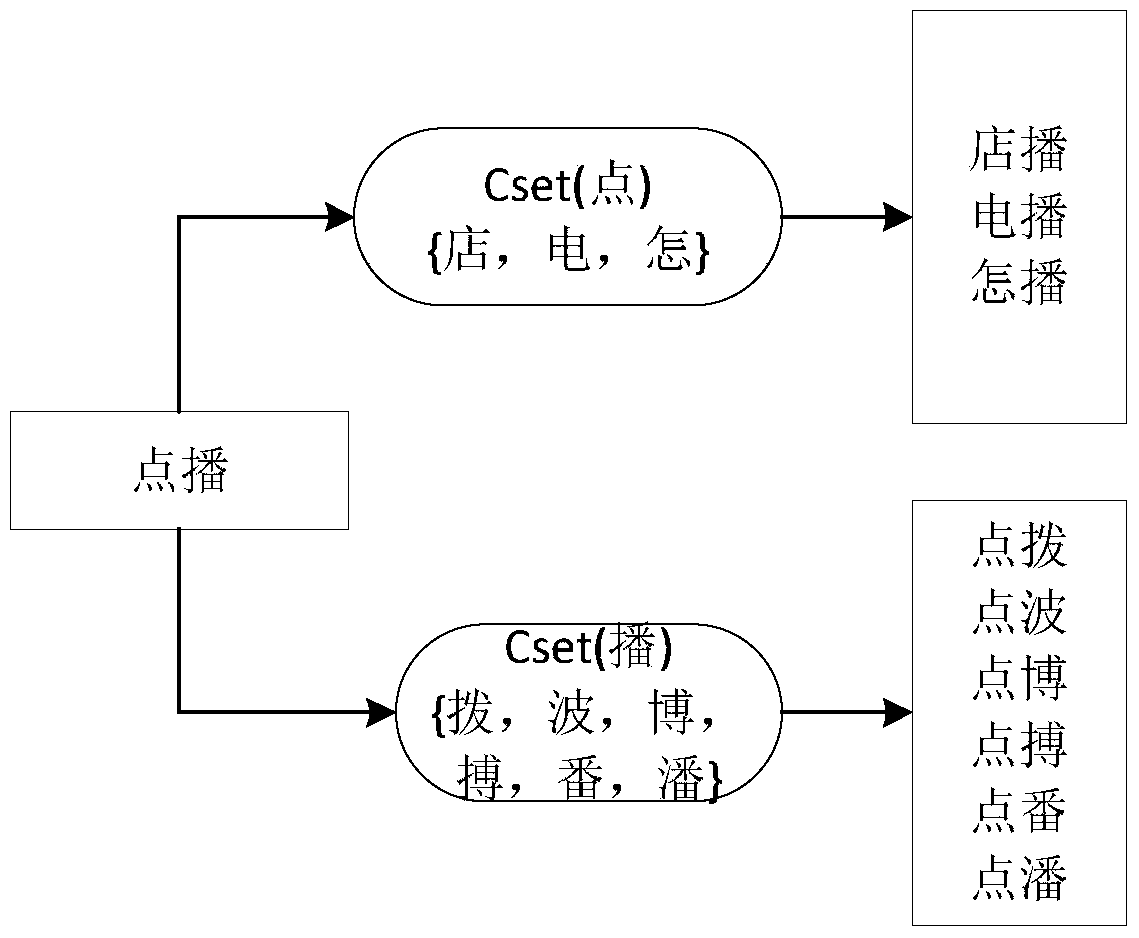
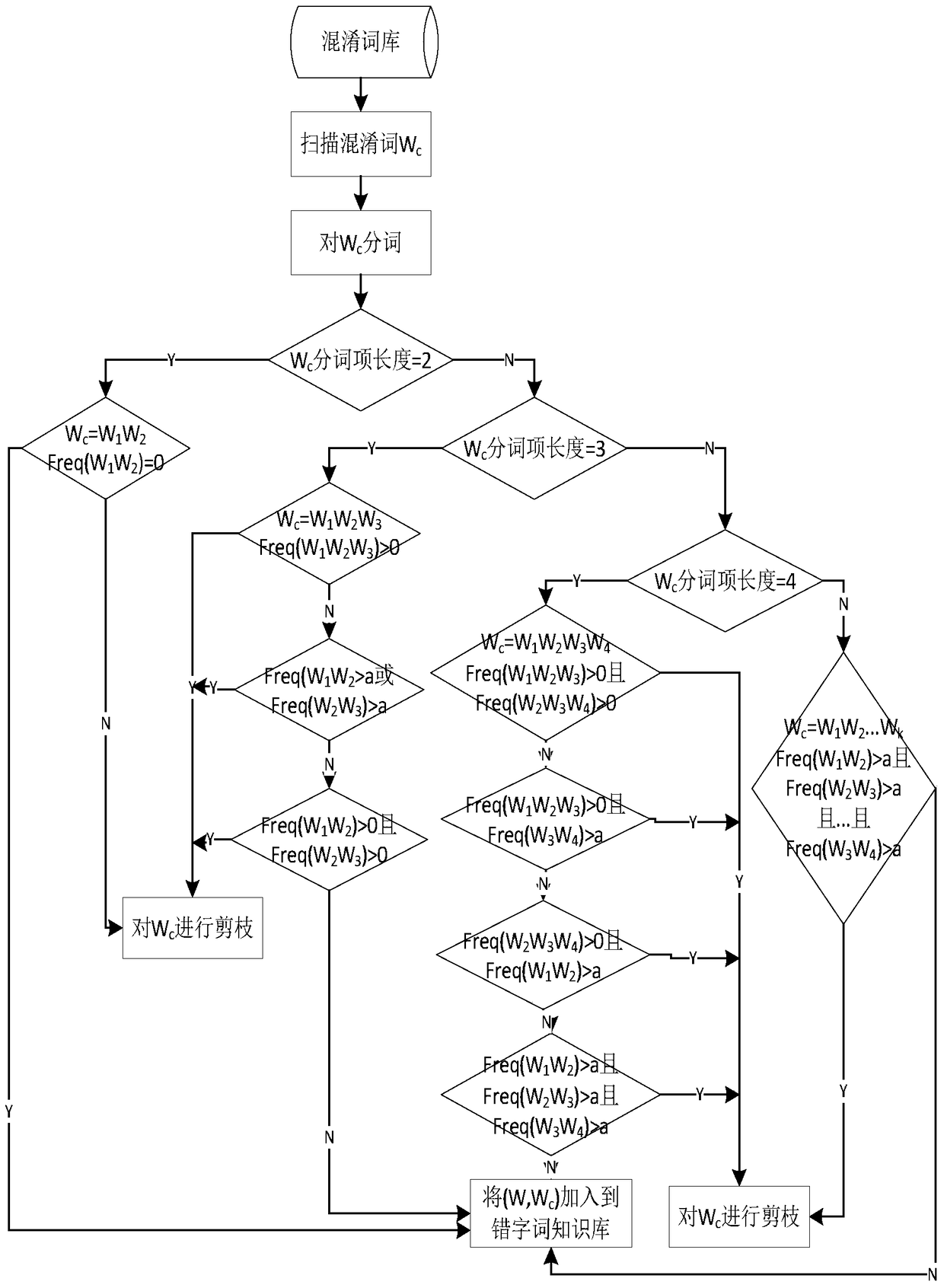
A method for generating typo word knowledge based on Chinese character confusion set
ActiveCN105573979BEnsure comprehensivenessGuaranteed accuracyNatural language data processingSpecial data processing applicationsNatural language processingChinese characters
The invention discloses a method for generating knowledge of wrongly written words based on a Chinese character confusion set. The method first uses a correct word dictionary and a Chinese character confusion set to generate a confusion word set; prunes the generated confusion word set through corpus and rules to complete preliminary filtering ; Then use the forward maximum matching word segmentation to segment the confused words in the confused word set after the initial filtering, and use statistical knowledge to verify the confused words according to the pre-set typo word judgment rules, and finally generate typo word knowledge. The method of the present invention solves the problems of low efficiency and large labor load of the existing manual proofreading, and automatically proofreads and corrects errors by using the knowledge of typos and words obtained by the method of the present invention, which improves the error correction quality and error correction of the automatic proofreading of Chinese texts speed.
Owner:苏州定一智能技术有限公司
High-fidelity polymerase with preference for gapped DNA and use thereof
ActiveUS20220017880A1High activityMicrobiological testing/measurementTransferasesForward primerDNA polymerase I
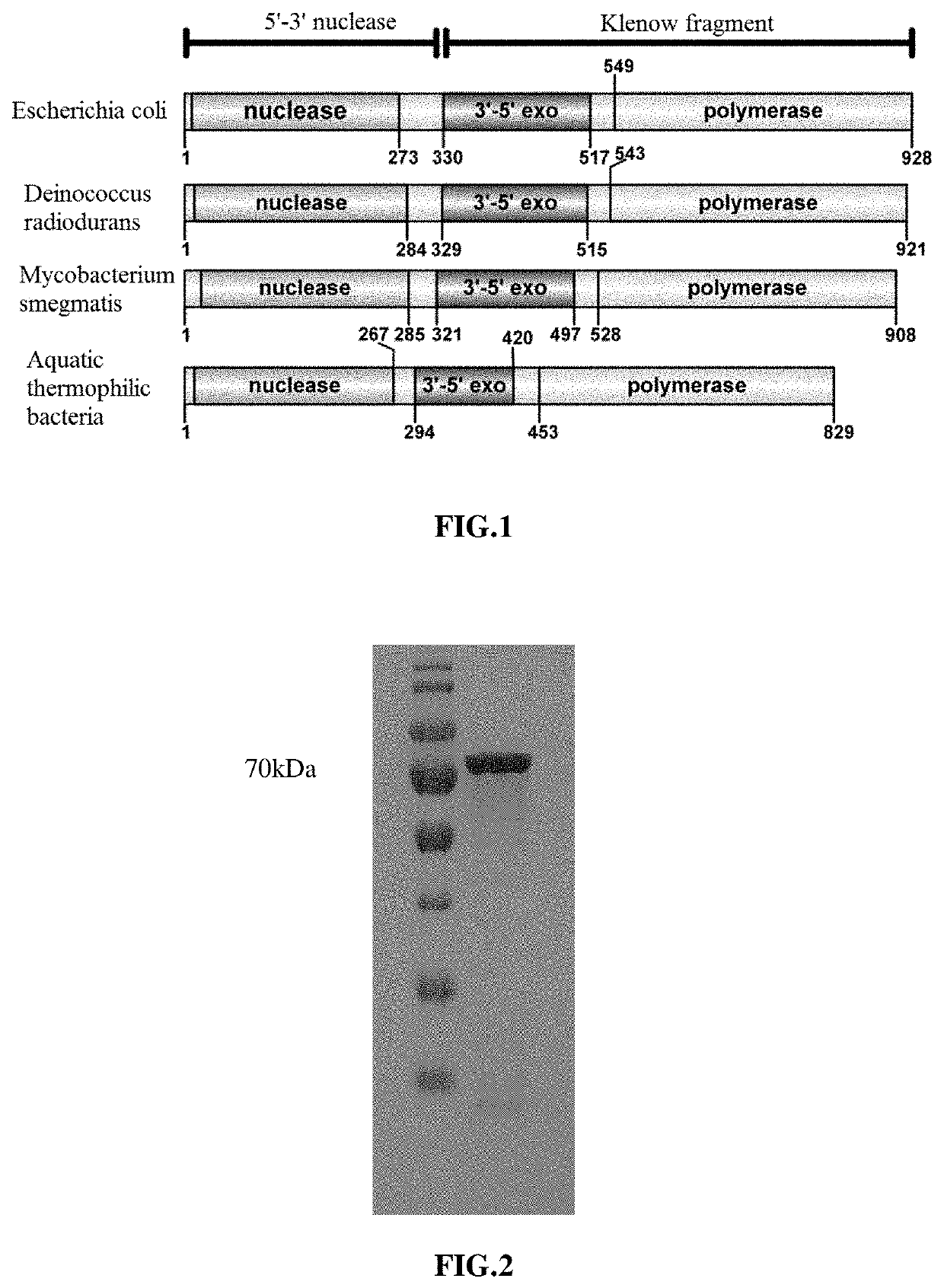
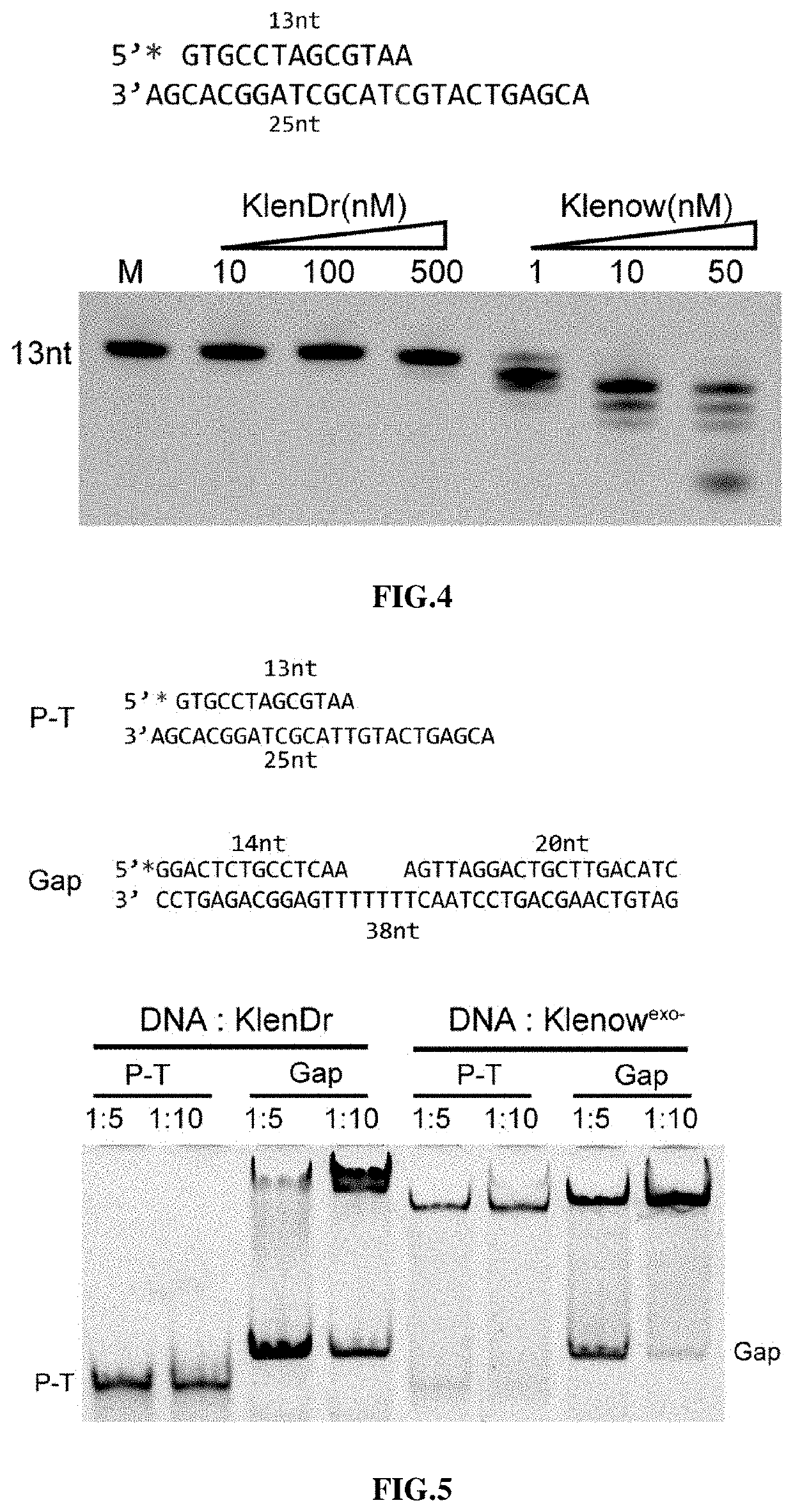
The disclosure provides a high-fidelity polymerase with preference for gapped DNA and use thereof. The Klenow fragment (KlenDr) derived from Deinococcus radiodurans DNA polymerase I, which has the high-fidelity polymerization characteristics, is independent of 3′-5′ proofreading exonuclease activity, has the preference for binding gapped DNA, and is different from the existing commercial high-fidelity polymerase. Due to the specific affinity of KlenDr to gapped DNA substrate, the 3′ end of the forward primer will not be cut off, and the downstream nucleotide chain is rarely replaced.
Owner:ZHEJIANG UNIV
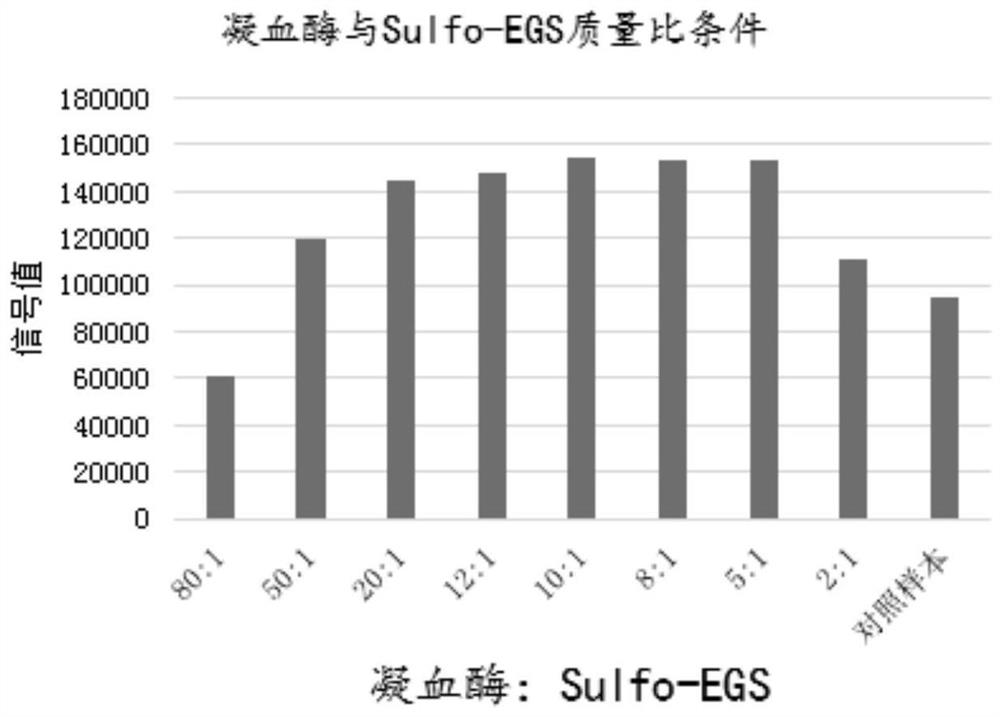
Thrombin-antithrombin III compound determination kit as well as preparation method and application thereof
PendingCN114705870AGood signal valueSolve available puzzlesChemiluminescene/bioluminescenceBiological material analysisAntigenMedicine
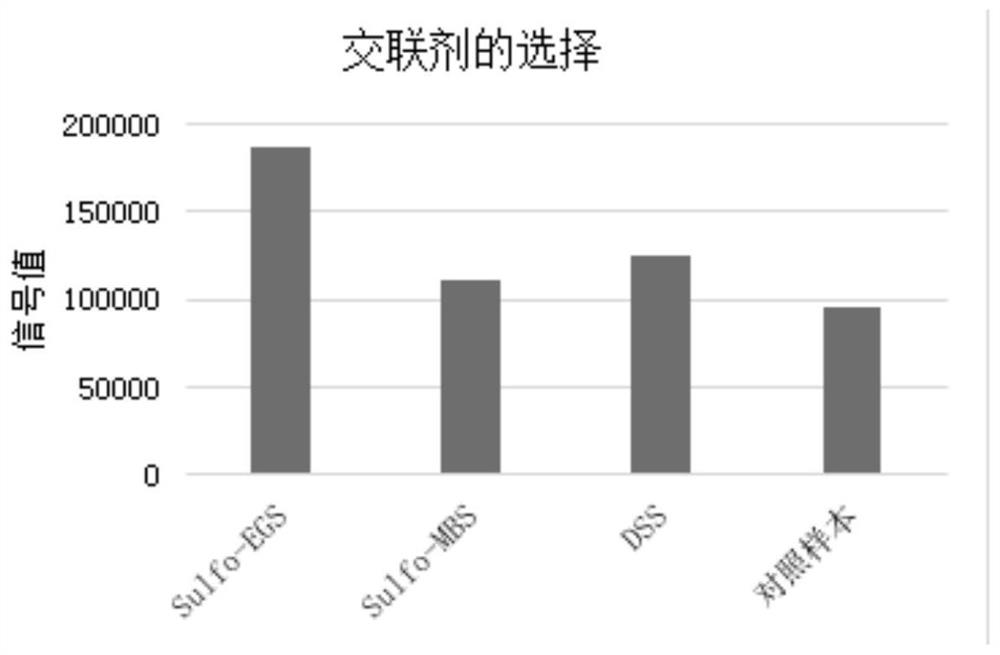
The invention relates to a thrombin-antithrombin III compound antigen compound, a preparation method thereof and a thrombin-antithrombin III compound determination kit, and the thrombin-antithrombin III compound is prepared by cross-linking thrombin with a cross-linking agent Sulfo-EGS and then cross-linking with antithrombin III. The thrombin-antithrombin III compound determination kit is a chemiluminescence method, takes a thrombin-antithrombin III compound as a proofreading product, and has the advantages of high sensitivity, good stability and strong specificity, and is not easy to interfere. According to the detection method of the thrombin-antithrombin III compound, the probabilities that the dosage of a detection sample is small and the detection is faster are achieved, and a better choice is provided for clinical TAT quantification.
Owner:GUANGZHOU WONDFO BIOTECH
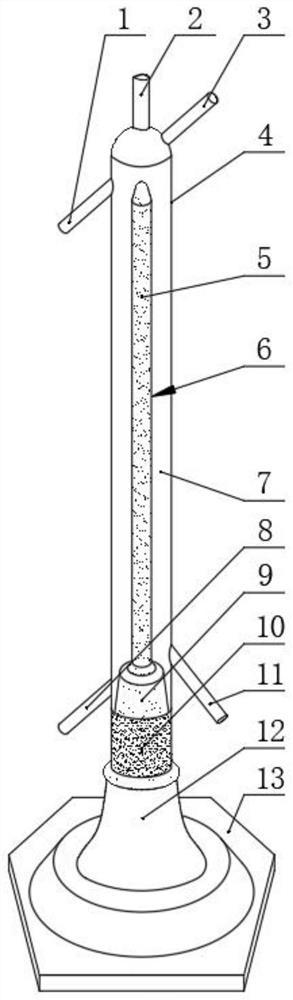
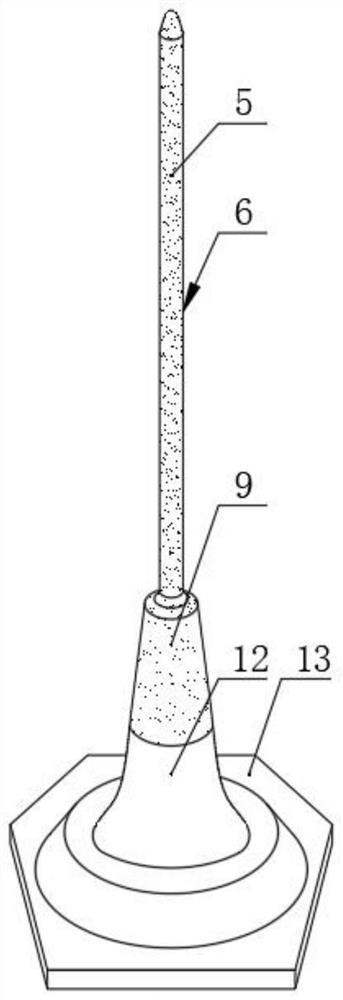
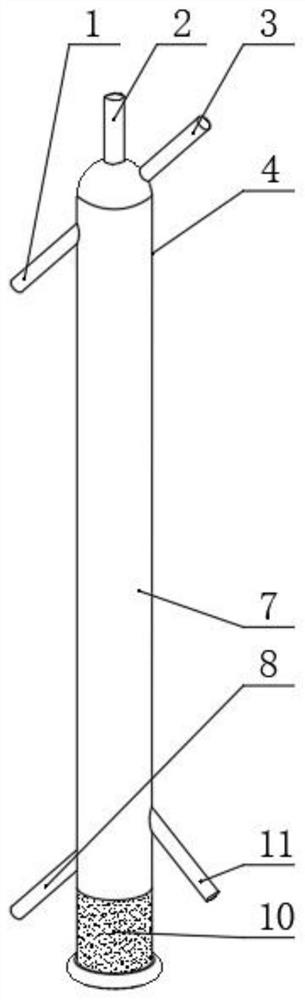
Mercury reduction reaction gas-liquid separation device for high-precision mercury isotope determination and use method
PendingCN114088805AEasy to cleanCleaning will not affectMaterial analysis by electric/magnetic meansSurface reactionChloride
The invention relates to a mercury reduction reaction gas-liquid separation device for high-precision mercury isotope determination, the device comprises an outer sleeve and a reaction tube, the upper end of the outer sleeve is provided with a reactant outlet, a proofreading sample inlet and a feed inlet which are communicated with the inner cavity of the outer sleeve, and the lower end of the outer sleeve is provided with a gas inlet; a frosted surface is arranged on the outer surface of the reaction tube, and a connecting part is arranged at the lower end of the reaction tube and used for being in sealed connection with the lower end opening of the outer sleeve. According to the invention, as the vertical height of the frosted surface is large, mercury and stannous chloride can fully react, and gaseous zero-valent mercury can be fully generated and output along with gas flow, so that the deviation between the gaseous zero-valent mercury and the proofreading sample is reduced, and the accuracy of subsequent isotope detection is ensured. In addition, the outer sleeve and the reaction tube are very easy to separate, the outer sleeve and the reaction tube are separated and respectively cleaned after the reaction is completed, the frosted surface of the reaction tube is easy to clean, and the next reaction is not influenced.
Owner:TIANJIN UNIV
Input error correction method and device and device for input error correction
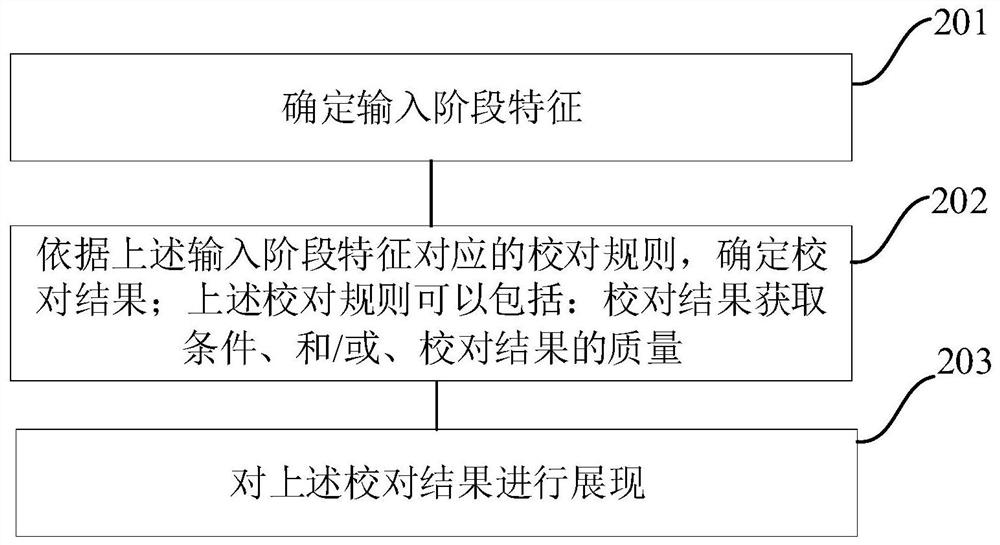
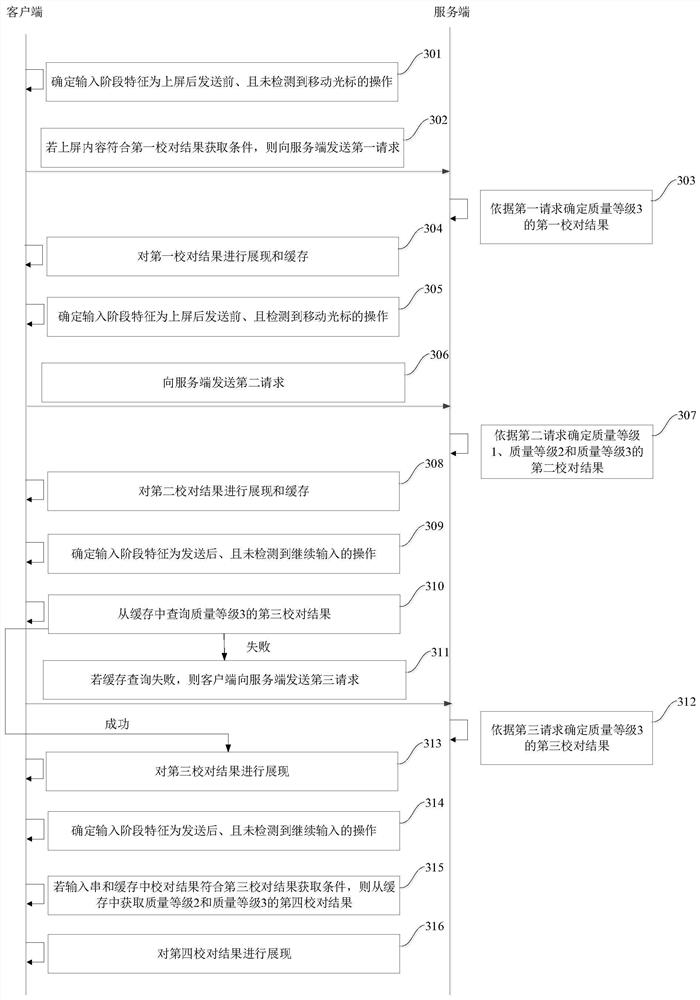
PendingCN113435185AImprove experienceImprove intelligenceNatural language data processingInput/output processes for data processingAlgorithmEngineering
The embodiment of the invention provides an input error correction method and device and a device for input error correction. The method specifically comprises the following steps: determining an input stage feature; determining a proofreading result according to a proofreading rule corresponding to the input stage feature, wherein the proofreading rule comprises a proofreading result acquisition condition and / or the quality of a proofreading result; and displaying the proofreading result. According to the embodiment of the invention, the intelligence of input proofreading and proofreading results can be improved, and a user is helped to use the proofreading results to correct errors, so that the input efficiency and the user experience are improved.
Owner:BEIJING SOGOU TECHNOLOGY DEVELOPMENT CO LTD
Compositions and methods for sequencing nucleic acids
ActiveCN102782159BSequencing errors dropSugar derivativesMicrobiological testing/measurementNucleotidePolymerase L
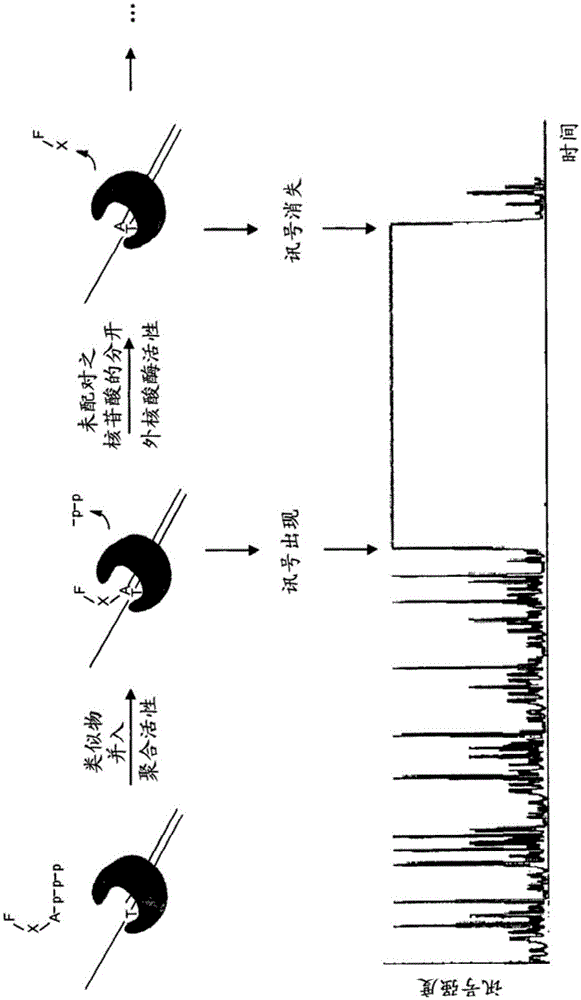
Examples relate to methods of sequencing nucleic acids. Examples include the use of nucleotide analogs with a nucleic acid polymerase enzyme or enzyme complex that includes proofreading activity. Nucleotide analogs can become incorporated into a replicating strand and induce proofreading activity of the polymerase, thus prolonging the duration of the signal associated with nucleotide incorporation, producing more significant sequencing results and increasing the efficiency of nucleic acid sequencing. Correctness.
Owner:IND TECH RES INST
A method for analyzing plant endophytic bacterial flora
The invention discloses a method for analyzing plant endophyte colonies. In the method, a primer pair used for analyzing the plant endophyte colonies is provided, the target sequence of the primer pair is the overall length or part of the region from V3 to V4 of bacteria 16SrDNA; there are differences between the last basic group at the 3' end of one primer in the primer pair and the plant mitochondria 18S rDNA, and the number of the difference sites between the overall length of the primer and the plant mitochondria 18S rDNA is equal to or larger than 3; there are differences between the lastbasic group at the 3' end of the other primer and the plant chloroplast 16S rDNA, and the number of the difference sites between the overall length of the primer and the plant mitochondria 18S rDNA is equal to or larger than 3. The two primers 322F-1 and 796R are designed, the DNA polymerase with hot-start activity but without 3' to 5' exonuclease proofreading activity is selected to achieve theaim that the pollution of the host plant chloroplast 16S rDNA and the pollution of the host plant mitochondria 18S rDNA are avoided at the same time, and therefore, a pure bacteria colony 16S ampliconlibrary is obtained.
Owner:INST OF MICROBIOLOGY - CHINESE ACAD OF SCI
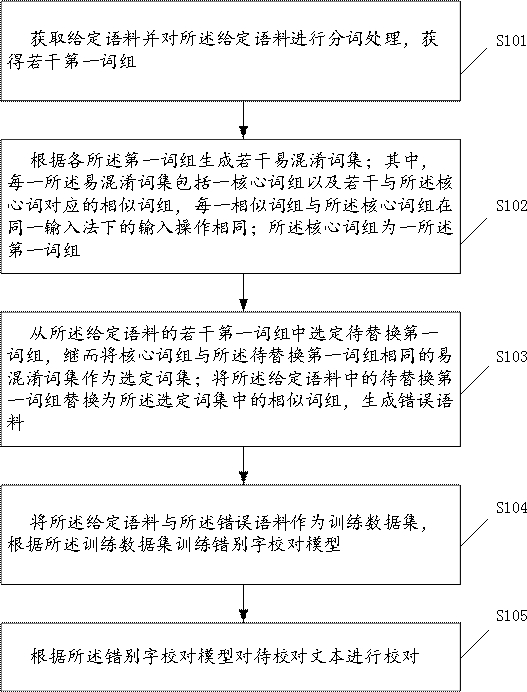
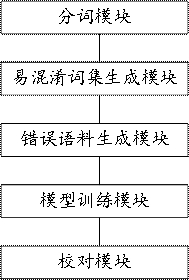
A typo proofreading method and device for automatically generating training data
The invention discloses a method and device for correcting typos that automatically generate training data. The method includes performing word segmentation processing on a given corpus to obtain a number of first phrases; generating a number of confusing word sets according to each first phrase; Select the first phrase to be replaced from several first phrases in the first phrase, and then use the same confusing word set as the core phrase and the first phrase to be replaced as the selected word set; replace the first phrase to be replaced in the given corpus with the selected The similar phrases in the fixed word set generate error corpus; the given corpus and the error corpus are used as training data sets, and the typo proofreading model is trained according to the training data set; the text to be proofread is proofread according to the typo proofreading model. By implementing the present invention, the problems of long time-consuming and low efficiency of manual collection of wrong corpus can be solved in the prior art.
Owner:京华信息科技股份有限公司
A low-abundance gene mutation enrichment method based on the removal of wild-type sequences
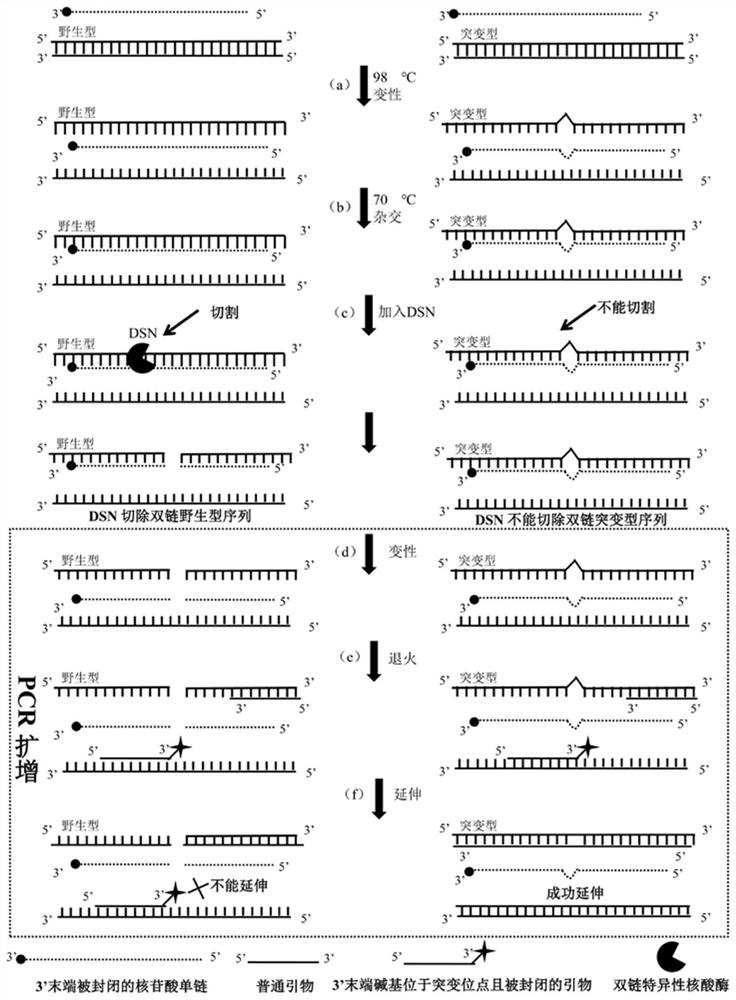
ActiveCN107893109BHigh sensitivityImprove accuracyMicrobiological testing/measurementHeterologousHeteroduplex
The invention discloses a low-abundance gene mutation enrichment method based on the removal of wild-type sequences. The method of the invention processes the sample DNA used as a DNA template before PCR amplification, so that the 3' end is blocked and contains mutations. The nucleotide single strand at the site is used as a hybrid strand to hybridize with the wild-type and mutant sequences, and the double-strand-specific nuclease can specifically remove the homologous double-stranded DNA that is completely complementary to the wild-type sequence formed by the hybrid strand, However, the non-completely complementary heteroduplex DNA formed by the hybrid strand and the mutant sequence cannot be removed. The removed product is amplified by PCR, and the 3' end of a primer used in PCR amplification is located at the mutation site and is blocked. Under the action of a polymerase with 3'-5' proofreading activity, the wild-type sequence cannot However, the mutant sequence can be amplified exponentially, thereby effectively enriching the low-abundance gene mutant sequence and improving the enrichment sensitivity and accuracy. At the same time, the method of the present invention can also simultaneously enrich a variety of mutant sequences so as to improve the amplification efficiency. increase efficiency.
Owner:CHONGQING UNIV OF POSTS & TELECOMM
Thermostable reverse transcriptase
ActiveUS20170327818A1Extensive and accurate copyingExtensive and accurate copying of any RNA populationTransferasesMaterial analysis by electric/magnetic meansReverse transcriptaseRNA Sequence
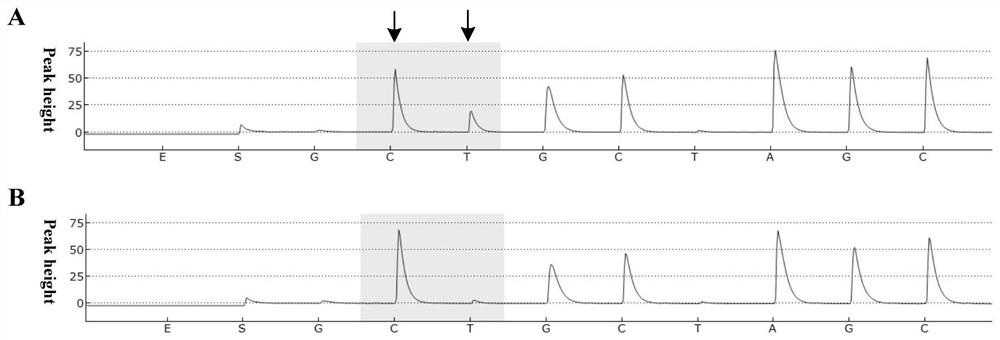
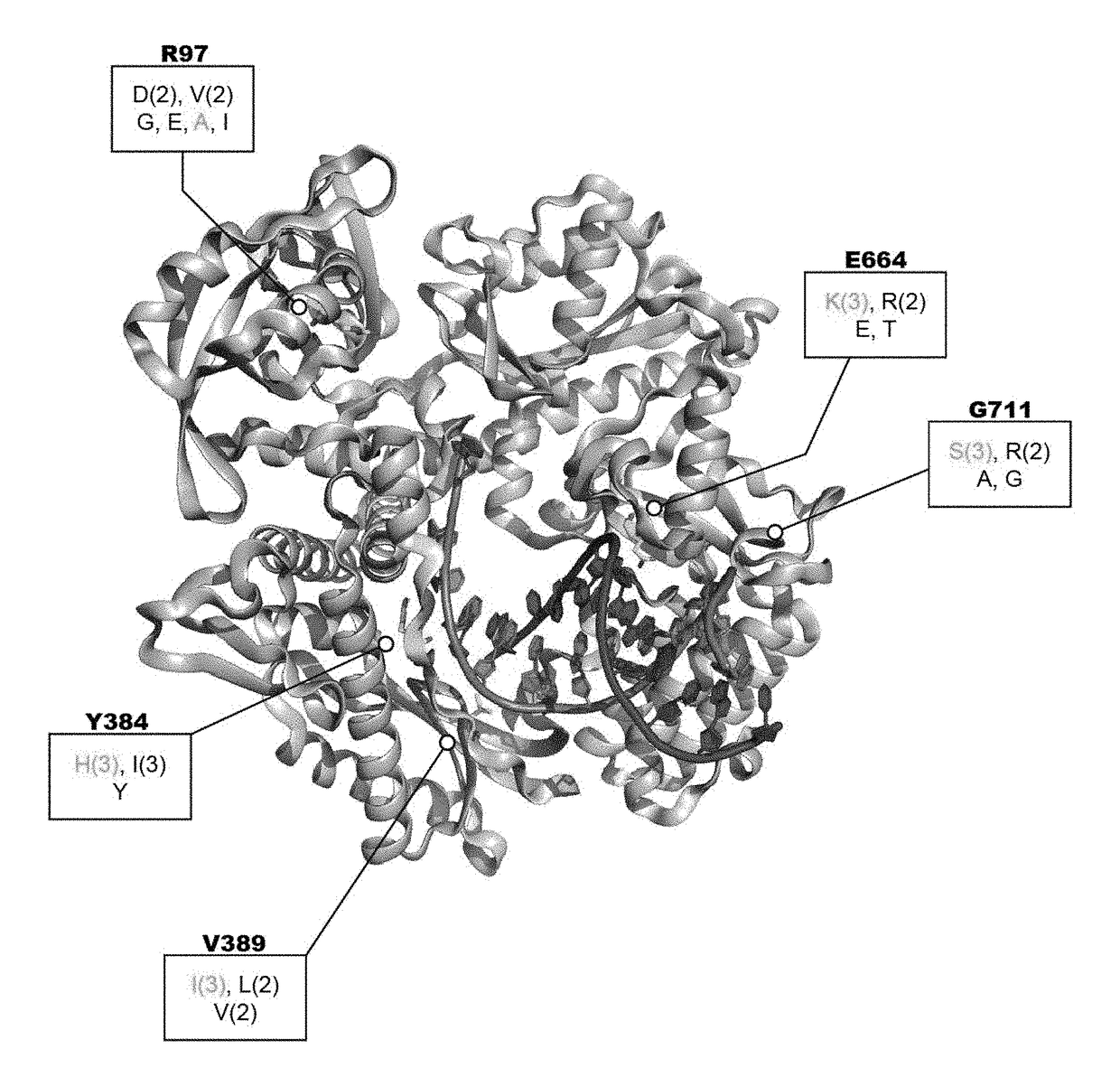
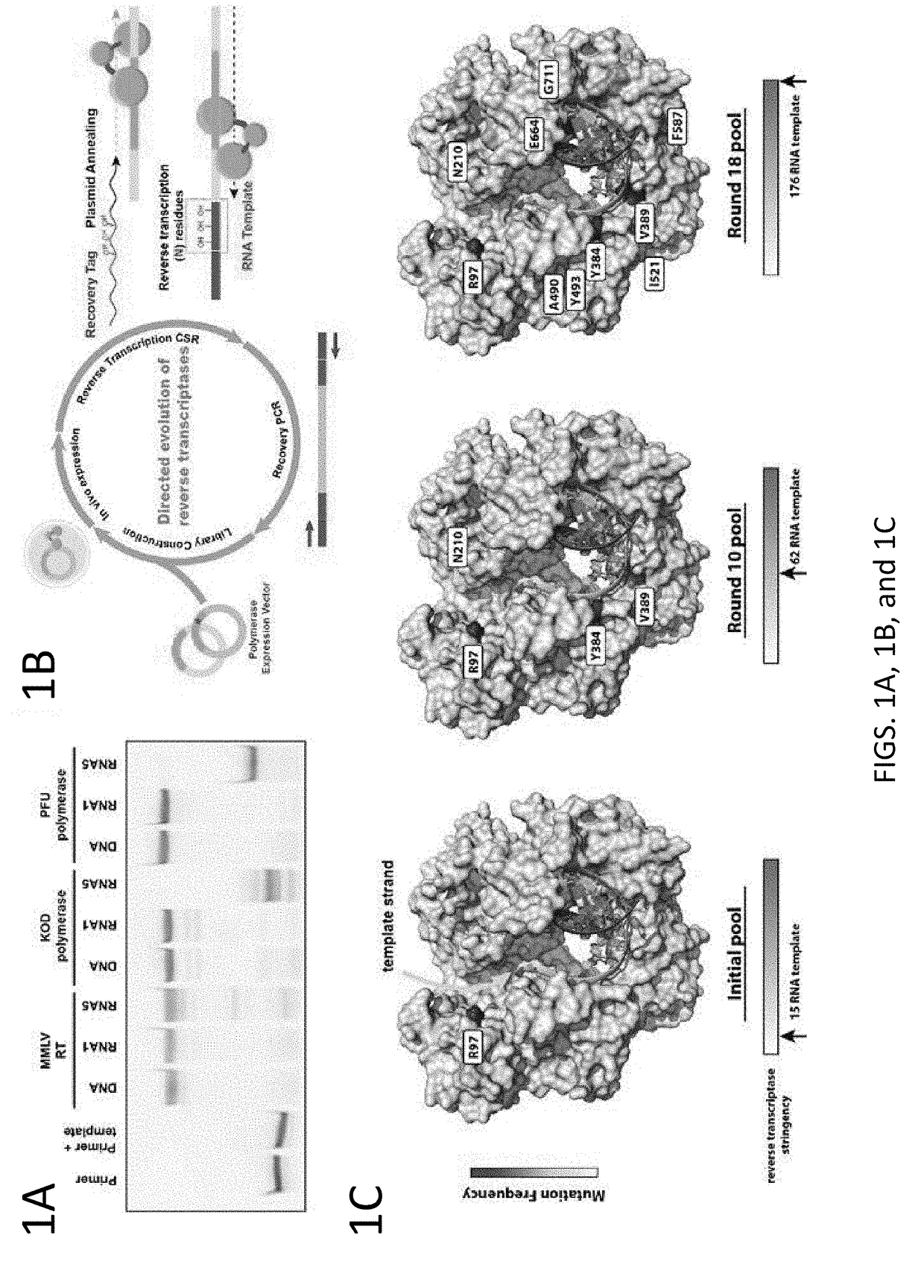
Embodiments of the disclosure concern methods and compositions related to generation and / or use of proofreading reverse transcriptases, including those that are thermophilic or hyperthermophilic. The disclosure encompasses specific recombinant polymerases and their use. In some embodiments, the polymerases are utilized for RNA sequencing in the absence of generation of a cDNA intermediate.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
DNA barcoding standard detection fragment for identifying vespa mandarinia and application thereof
InactiveCN111647667ARapid identificationAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationMedicinal herbs
The invention discloses a DNA barcoding standard detection fragment for identifying vespa mandarinia and application thereof, and belongs to the field of biological species identification. The invention discloses a discloses a DNA barcoding standard detection gene sequence as shown in SEQ ID NO.1, and a key technology for molecular identification of the insect medicinal material vespa mandarinia by using the CO II gene sequence and a PCR amplification technology. A PCR product is verified through agarose gel electrophoresis and then subjected to sequencing, a sequencing result is subjected tomanual proofreading and sequence splicing, the spliced sequence is compared with the CO II gene sequence disclosed by the invention, and if the homology is 98% or above, the invention can judge that the insect medicinal material to be identified is the vespa mandarinia source. The DNA molecular identification technology provided by the invention realizes rapid, accurate and reliable identificationon fresh or dry medicinal materials, whole worms or part of worms, pupae or larvae or adults, and ensures the safety of the vespa mandarinia as a medicinal material.
Owner:DALI UNIV
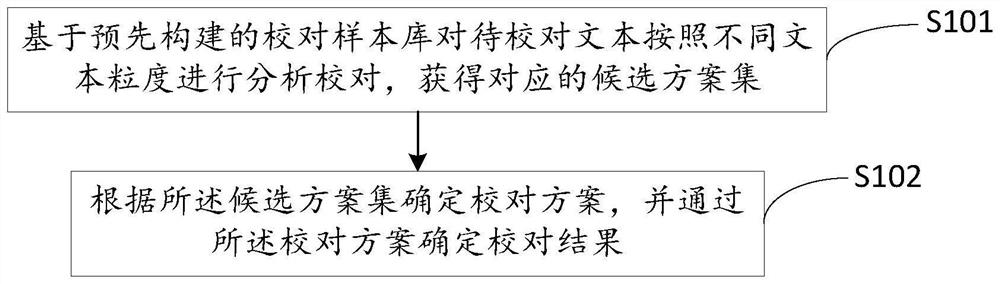
Transcription text proofreading method and storage medium
ActiveCN112836493AImprove accuracyImprove rationalityNatural language data processingText database queryingAlgorithmGranularity
The invention discloses a transcription text proofreading method and a storage medium. The method comprises the steps: carrying out the analysis and proofreading of a to-be-proofreading text according to different text granularities based on a pre-constructed proofreading sample library, and obtaining a corresponding candidate scheme set; and determining a proofreading scheme according to the candidate scheme set, and determining a proofreading result through the proofreading scheme. The method comprises the following steps: analyzing and proofreading a to-be-proofreading text according to different text granularities on the basis of a pre-constructed proofreading sample library to obtain a corresponding candidate scheme set; and determining the proofreading scheme according to the candidate scheme set, so that the proofreading scheme is determined from different text granularities, and the accuracy of the transliteration text and the reasonability of semantics are improved.
Owner:NAT COMP NETWORK & INFORMATION SECURITY MANAGEMENT CENT
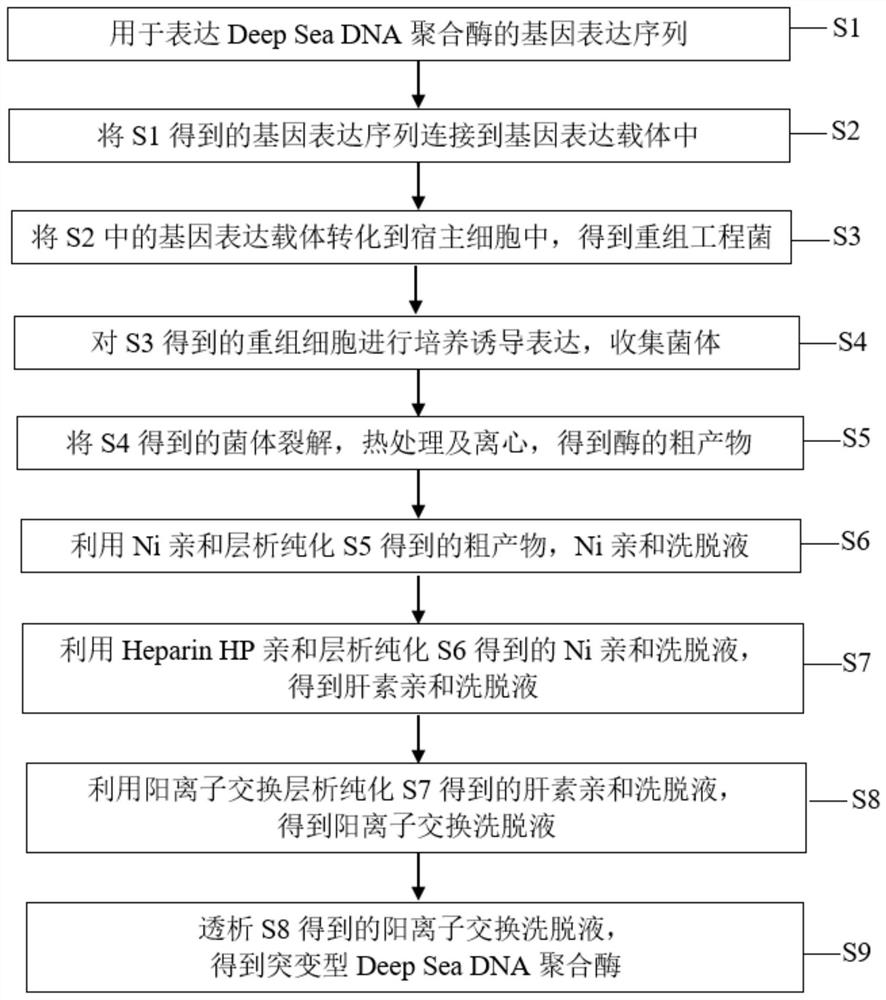
A kind of dna polymerase and its preparation method and application
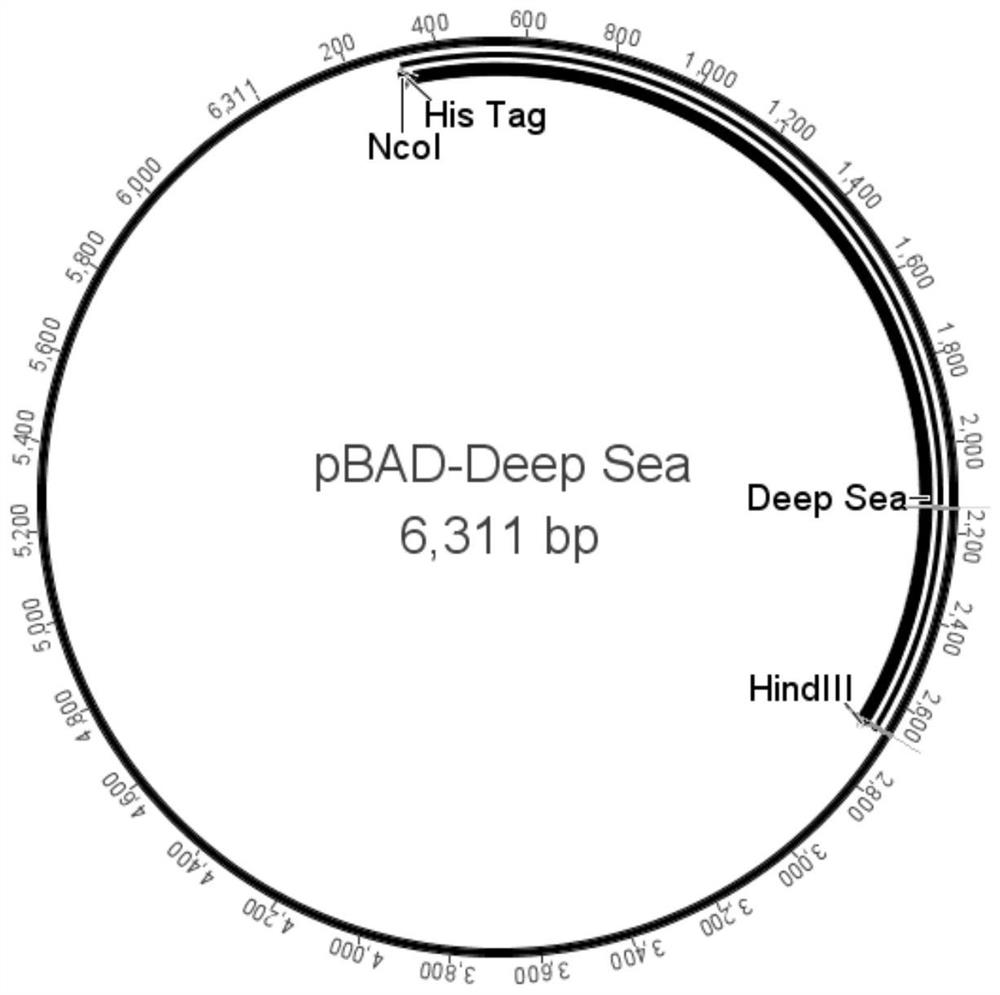
The present invention provides a kind of DNA polymerase and its preparation method and application, described DNA polymerase has any one in the aminoacid sequence shown in (I), (II) or (III): (I) SEQ ID NO:1 The amino acid sequence shown; (II) a polypeptide having ≥98% homology with the amino acid sequence shown in SEQ ID NO:1; (III) the amino acid sequence shown in SEQ ID NO:1 after 1 to 20 amino acid residues The amino acid sequence obtained by substituting, deleting or adding groups, and the obtained amino acid sequence has DNA polymerase activity. The Deep Sea DNA polymerase of the present invention removes part of the 3'→5' exonuclease proofreading activity of the wild-type DNA polymerase, has 5 times higher fidelity than Taq enzyme, good stability and high activity, and is suitable for For the amplification of difficult templates containing high GC sequences and cyclic sequences, it can be used as a tool enzyme for third-generation sequencing library construction.
Owner:莫纳(武汉)生物科技有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com