A method for analyzing plant endophytic bacterial flora
A technology of endogenous bacteria and flora in plants, applied in biochemical equipment and methods, measurement/testing of microorganisms, DNA/RNA fragments, etc., can solve problems such as interference diversity analysis, host DNA contamination, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Embodiment 1, analyze the primer design and synthesis of plant endophytic bacterial flora
[0046] The present invention designs to avoid host plant chloroplast 16S rDNA and mitochondrial 18S rDNA, and designs primer pairs according to the V3-V4 region of bacterial 16S rDNA as follows:
[0047] The last base at the 3' end of one primer is different from the plant mitochondrial 18S rDNA, and the difference between the full length of the primer and the plant mitochondrial 18S rDNA is greater than or equal to 3; the last base at the 3' end of the other primer is different from the plant chloroplast 16S rDNA difference, and the difference between the full length of the primer and the plant mitochondrial 18SrDNA is greater than or equal to 3.
[0048] According to the above design principles, the following primer pairs were designed and synthesized:
[0049] 322F-1: 5'ACGGHCCARACTCCTACGGAA3' (SEQ ID NO: 1)
[0050] 796R: 5'CTACCMGGGTATCTAATCCKG3' (SEQ ID NO: 2);
Embodiment 2
[0051] Embodiment 2, establishment of the method for analyzing plant endophytic bacterial flora
[0052] 1. PCR amplification
[0053] Using the total DNA of healthy rice leaves collected on September 12, 2016 in the rice experimental field of Fujian Agriculture and Forestry University as a template, using the primers 322F-1 and 796R of Example 1, and having hot start activity and no 3'→5' exonuclease Correction activity of DNA polymerase Platinum Taq DNA polymerase (Invitrogen, USA) was used for PCR amplification.
[0054] The reaction system of above-mentioned PCR amplification is as follows:
[0055]
[0056] In the above reaction system, the final concentration of each primer is 0.2 μM; the final concentration of Taq enzyme is 2U / rxn; MgCl 2 The final concentration was 1.5mM.
[0057] Above-mentioned PCR reaction procedure is as follows:
[0058]
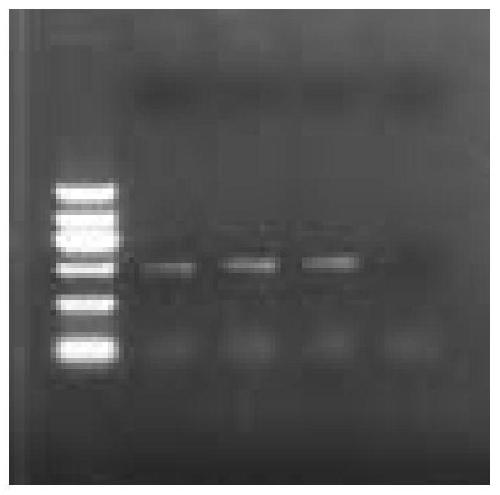
[0059] The result is as figure 1 As shown, Marker: DNA Marker II (TIANGEN), 1, 2, 3: three replicate samples, 4: empt...
Embodiment 3
[0071] Example 3, High-throughput sequencing method for analyzing plant endophytic bacterial flora
[0072] 1. PCR amplification
[0073] 1. Amplification primers
[0074] Six random bases were added to the 5' ends of the primers 322F-1 and 796R designed and synthesized in Example 1 to distinguish the Barcodes of different samples. Each sample has a different primer Barcode combination, which is used for sample identification of subsequent high-throughput sequencing. The Barcode sequence information is provided by Beijing Novogene Technology Co., Ltd., and the amplification products are also provided by Beijing Novogene Technology Co., Ltd. Subsequent library construction and on-machine sequencing were carried out; 4 amplification replicates were set for PCR of each sample, and a total of 200 μl of the final amplification product was obtained, with a total of 4 samples.
[0075] 2. Amplification
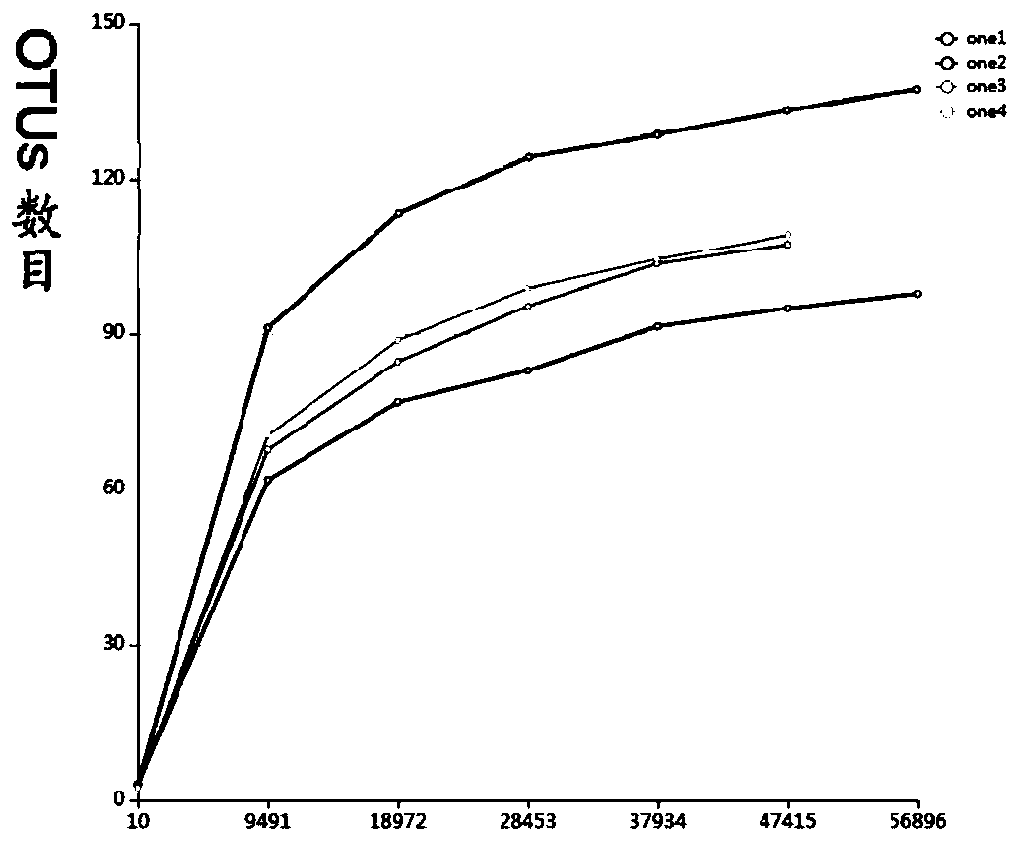
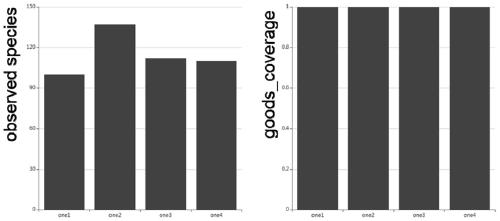
[0076] Total DNA was extracted from rice leaf samples collected in Kaifeng, H...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com