Thermostable or thermoactive dna polymerase with attenuated 3'-5' exonuclease activity
An exonuclease and thermostability technology, applied in the field of DNA polymerase, can solve the problems of time consumption and high price
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
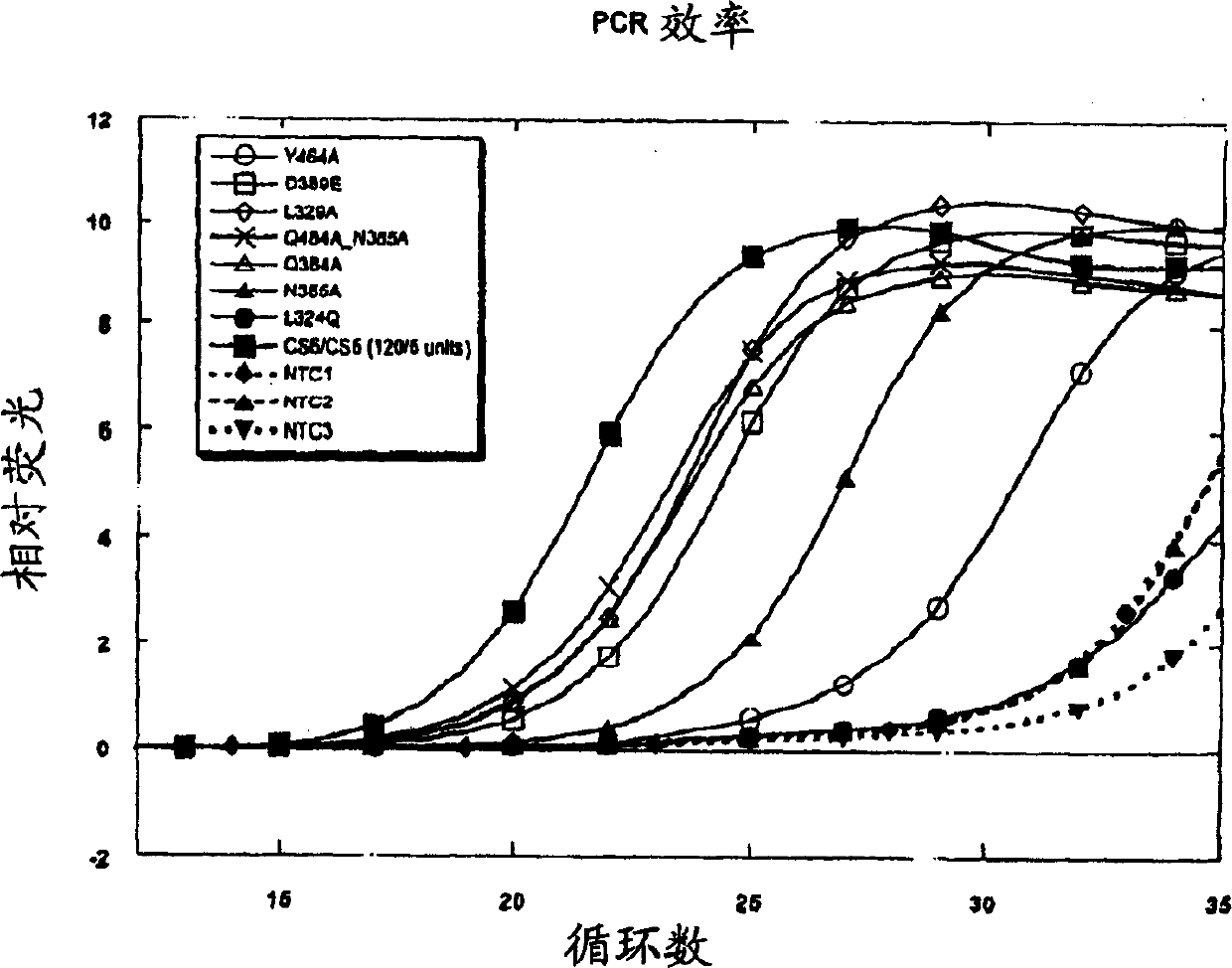
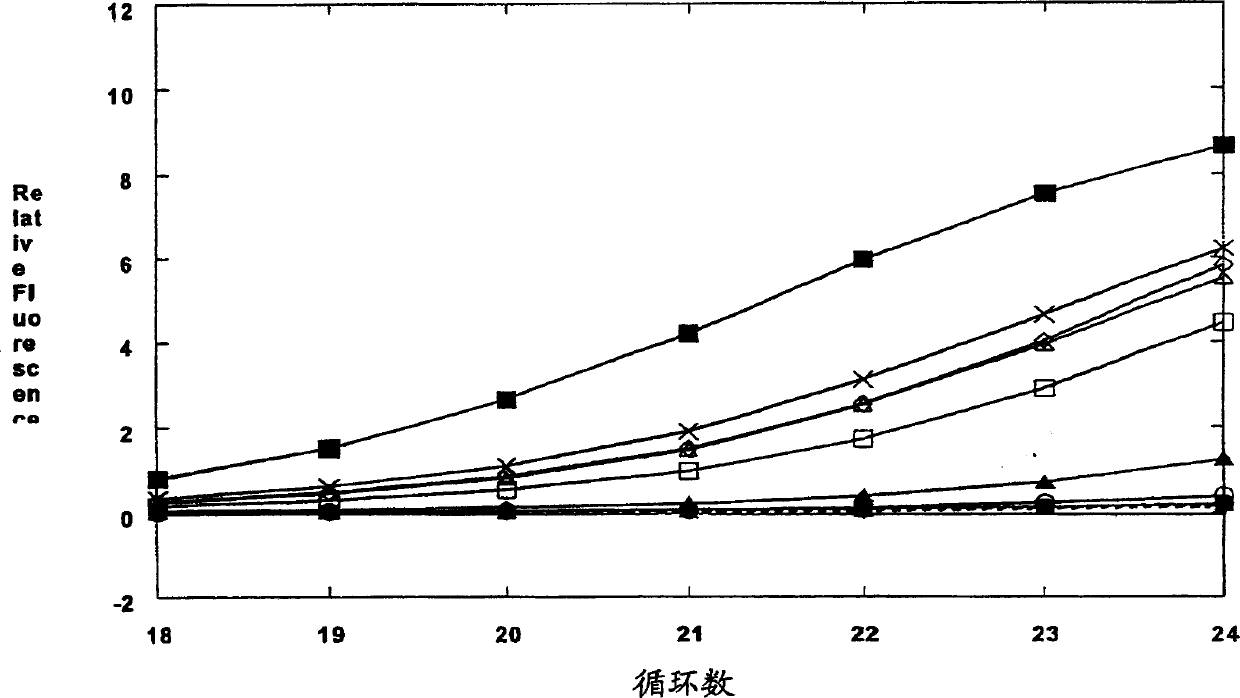
[0699] Example 1: Mutations of pCS6 and pCS5
[0700] This example describes site-directed mutagenesis of the 3'-5' exonuclease domain of a thermostable and heat-activated DNA polymerase.
[0701] In the first round of mutagenesis, site-directed mutagenesis was used to change the residues of thermostable DNA polymerase. These residues were either known to have an important role in coordinating divalent metal cations, the high-fidelity domain of the Klenow fragment The homologous residues in the active site are either located between these homologous residues. See Derbyshire et al., 1995, Methods In Enzymology 262:363-85, which is incorporated herein by reference in its entirety. The plasmid pCS6 encoding the mutant chimeric thermostable DNA polymerase CS6 is used, as shown in Figure 5. CS6 is a mutant of the chimeric thermostable DNA polymerase CS5, as shown in Figure 4. The N-terminal part of CS5 is 291 residues from the N-terminal of Thermus species Z05 (Thermus species Z05) DNA...
Embodiment 2
[0717] Example 2: Production and purification of mutant thermostable DNA polymerase
[0718] The plasmid obtained from the above ligation contains the λP LThe polymerase gene under the control of a promoter. The plasmid was used to transform a host cell (DG116, ATCC#53606) containing the cI857 (thermally unstable) lambda repressor. The DG116 cells transformed with the plasmid containing the wild-type or mutant polymerase gene were used to inoculate 10ml of standard shake flask medium (SFM) supplemented with ampicillin (100μg / ml); composed of glucose and vitamin B 1 , Casein hydrolysate and minimal medium), grow overnight at 30°C and 250rpm. Use 5ml overnight culture to inoculate 450ml SFM+Ampicillin medium and incubate at 30℃, 250rpm shaker until the culture OD 600 Reach 0.6-0.8. The culture was then transferred to 37°C and grown overnight at 250 rpm to obtain temperature-induced expression of polymerase. See, for example, U.S. Patent Nos. 5,079,352, 5,420,029, and 5,618,711. The ...
Embodiment 3
[0720] Example 3: Standard analysis of 3'-5' exonuclease activity
[0721] The following examples provide a standard analysis method for measuring the thermostability or heat-activated DNA polymerase 3'-5' exonuclease activity and the definition of 1 unit of 3'-5' exonuclease activity.
[0722] In order to compare the 3'-5' exonuclease activity of the CS5 mutant derivatives, the degradation of the carboxyfluorescein (FAM)-labeled single-stranded oligonucleotide substrate NJS40 was measured.
[0723] NJS40: FAM-GCGCTAGGGCTGGCAAGTGTAGCGGTCAC (SEQID NO: 80)
[0724] Standard analysis method (40μl system) in 50mM Tricine, pH8.3, 25mM KOAc, 5% w / v DMSO, 0.5mM Mn(OAc) 2 And 5% (volume ratio) enzyme storage buffer component (in the reaction solution: 1mM Tris, pH8.0, 0.005mM EDTA, 0.05mM DTT, 5mM KCl, 0.025% Tween 20, 2.5% glycerol) containing 4pmol (100 nM) EAM labeled single-stranded oligonucleotide NJS40. By adding NJS40 and Mn(OAc) 2 The reaction was initiated and incubated at 63°C f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com