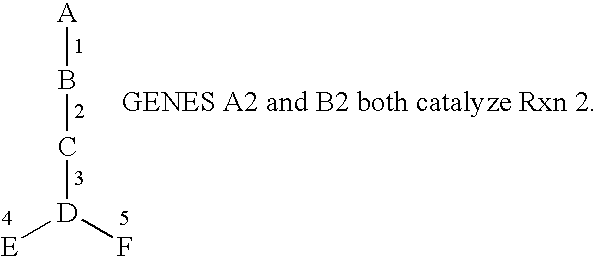Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
136 results about "Biological pathway" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A biological pathway is a series of interactions among molecules in a cell that leads to a certain product or a change in a cell. Such a pathway can trigger the assembly of new molecules, such as a fat or protein. Pathways can also turn genes on and off, or spur a cell to move. Some of the most common biological pathways are involved in metabolism, the regulation of gene expression and the transmission of signals. Pathways play a key role in advanced studies of genomics.
Methods for testing biological network models
InactiveUS6132969AModel is bulkyLow costMicrobiological testing/measurementLibrary screeningBiological pathwayNetwork model
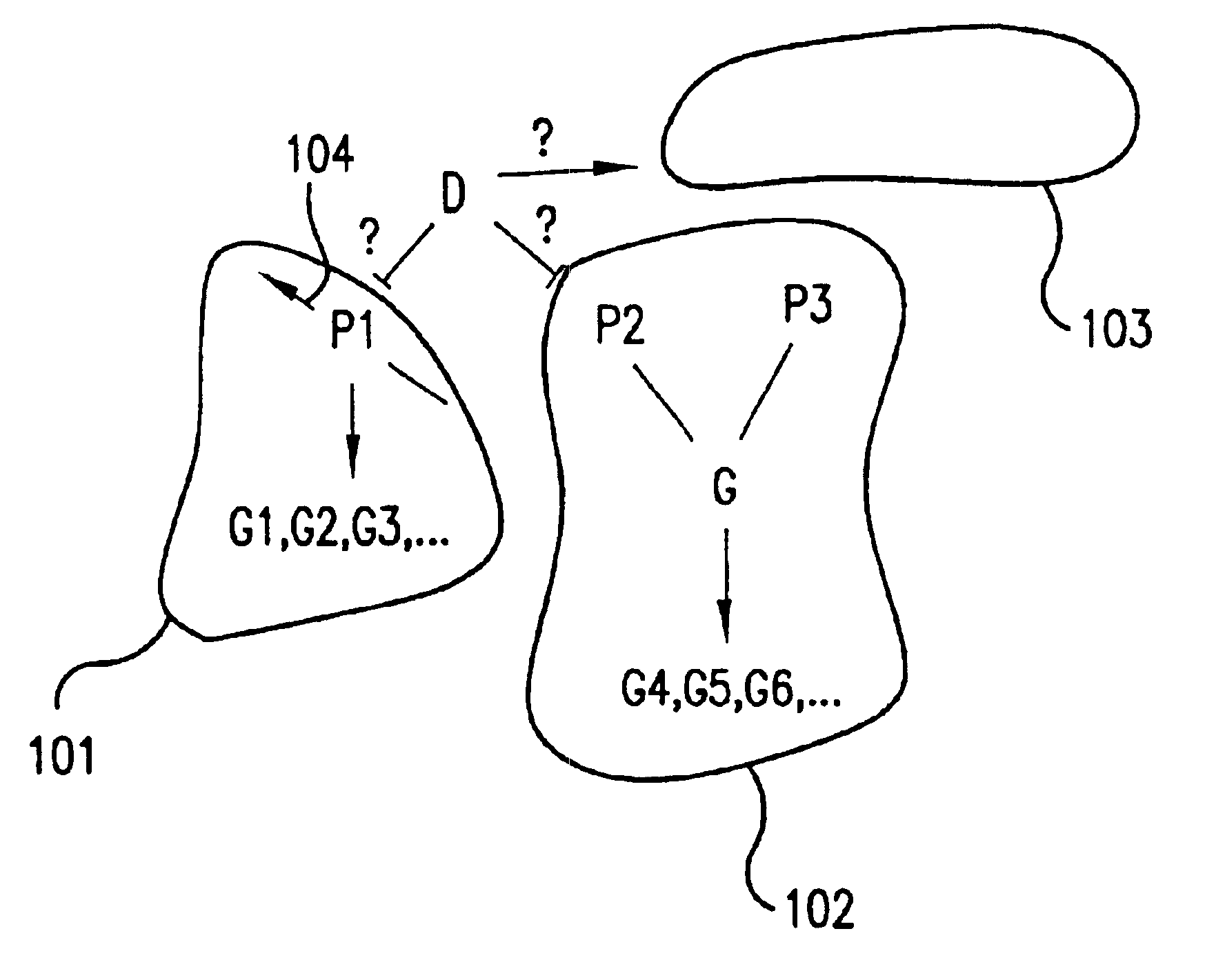
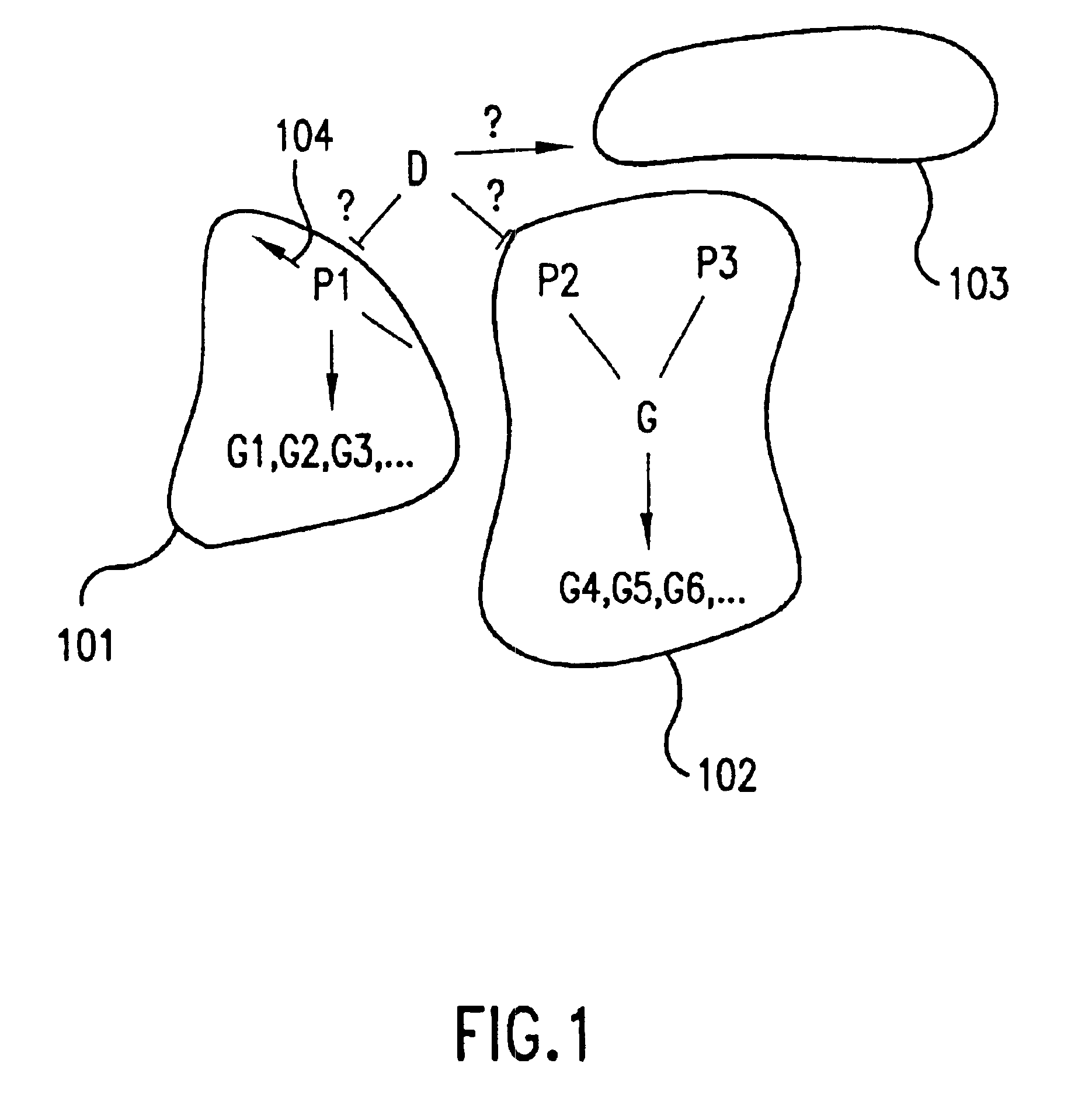
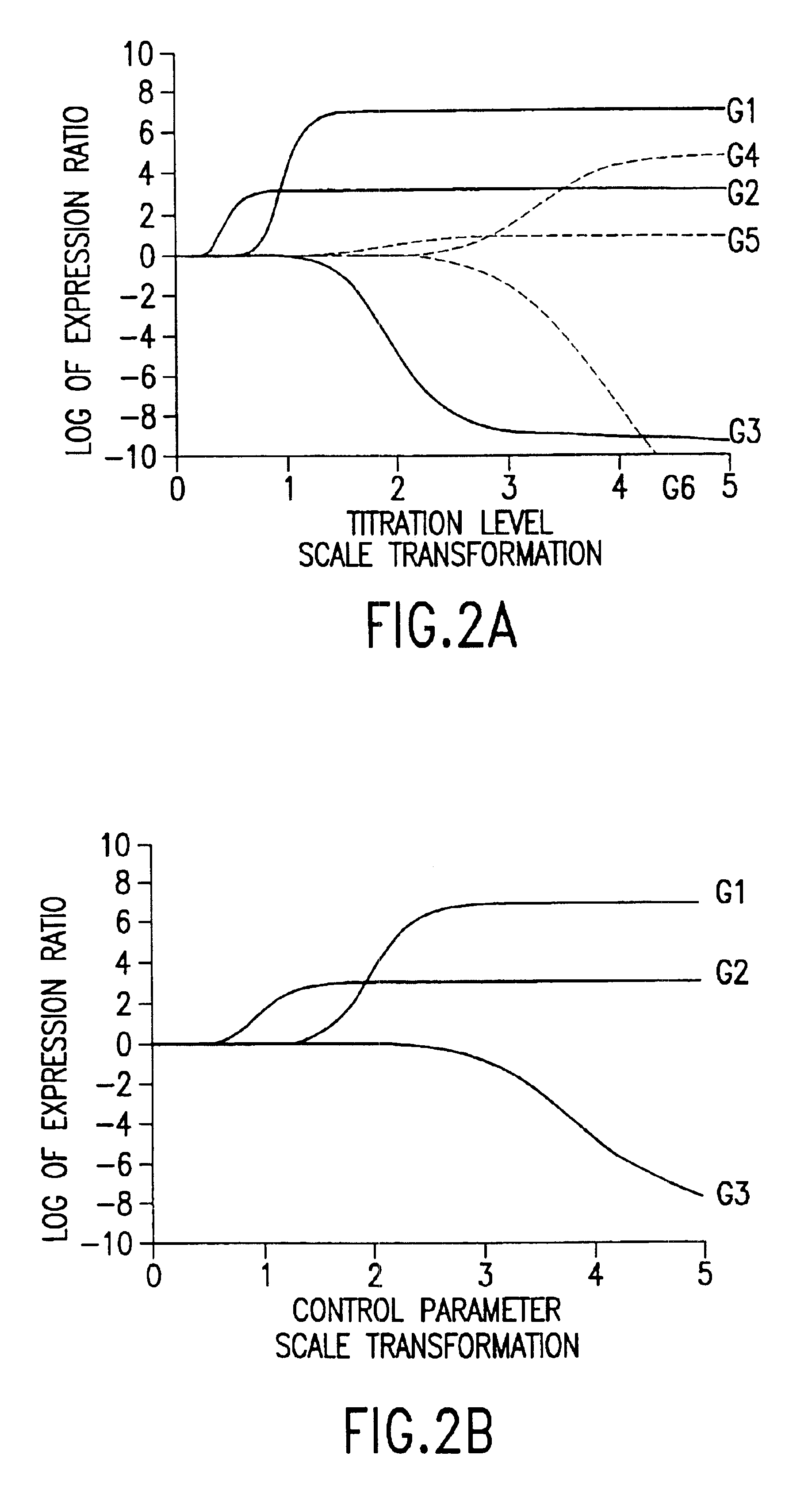
The present invention provides methods and systems for testing and confirming how well a network model represents a biological pathway in a biological system. The network model comprises a network of logical operators relating input cellular constituents (e.g., mRNA or protein abundances) to output classes of cellular constituents, which are affected by the pathway in the biological system. The methods of this invention provide, first, for choosing complete and efficient experiments for testing the network model which compare relative changes in the biological system in response to perturbations of the network. The methods also provide for determining an overall goodness of fit of the network model to biological system by: predicting from the network model how output classes behave in response to the chosen experiments, finding measures of relative change of cellular constituents actually observed in the chosen experiments, finding goodnesses of fit of each observed cellular constituent to an output class with which the cellular constituent has the strongest correlation, and determining an overall goodness of fit of the network model from the individual goodnesses of fit of each observed cellular constituent. Additionally, these methods provide for testing the significance of the overall goodness of fit according to a nonparametric statistical test using an empirically determined distribution of possible goodnesses of fit. This invention also provides for computer systems for carrying out the computational steps of these methods.
Owner:MICROSOFT TECH LICENSING LLC
Transcriptional regulatory elements of biological pathways tools, and methods
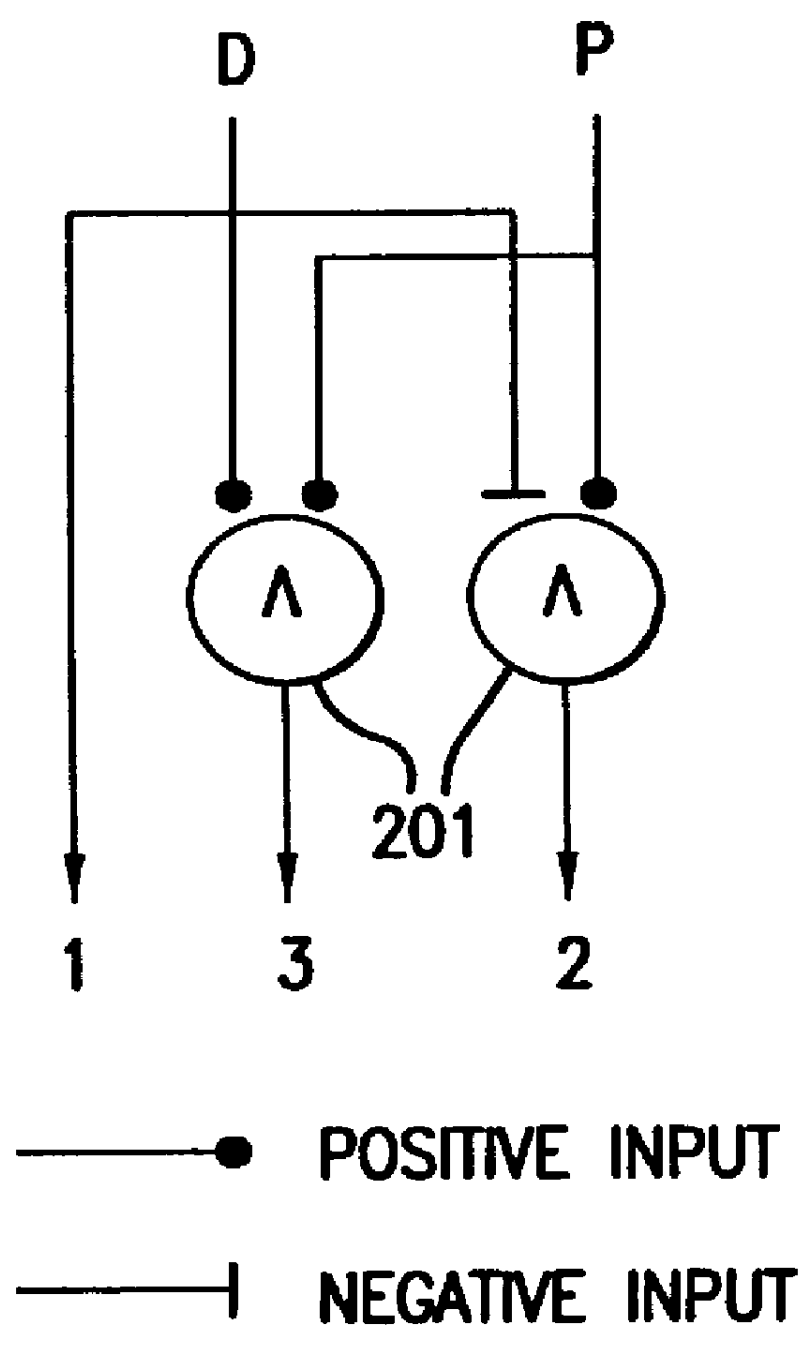
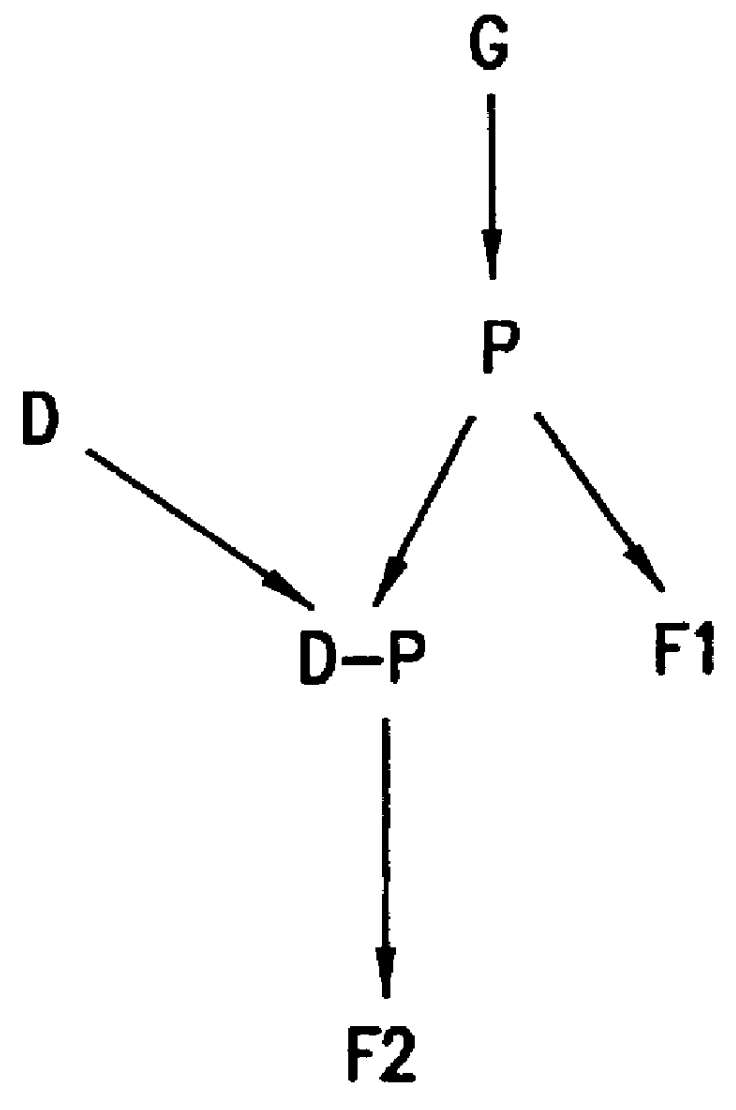
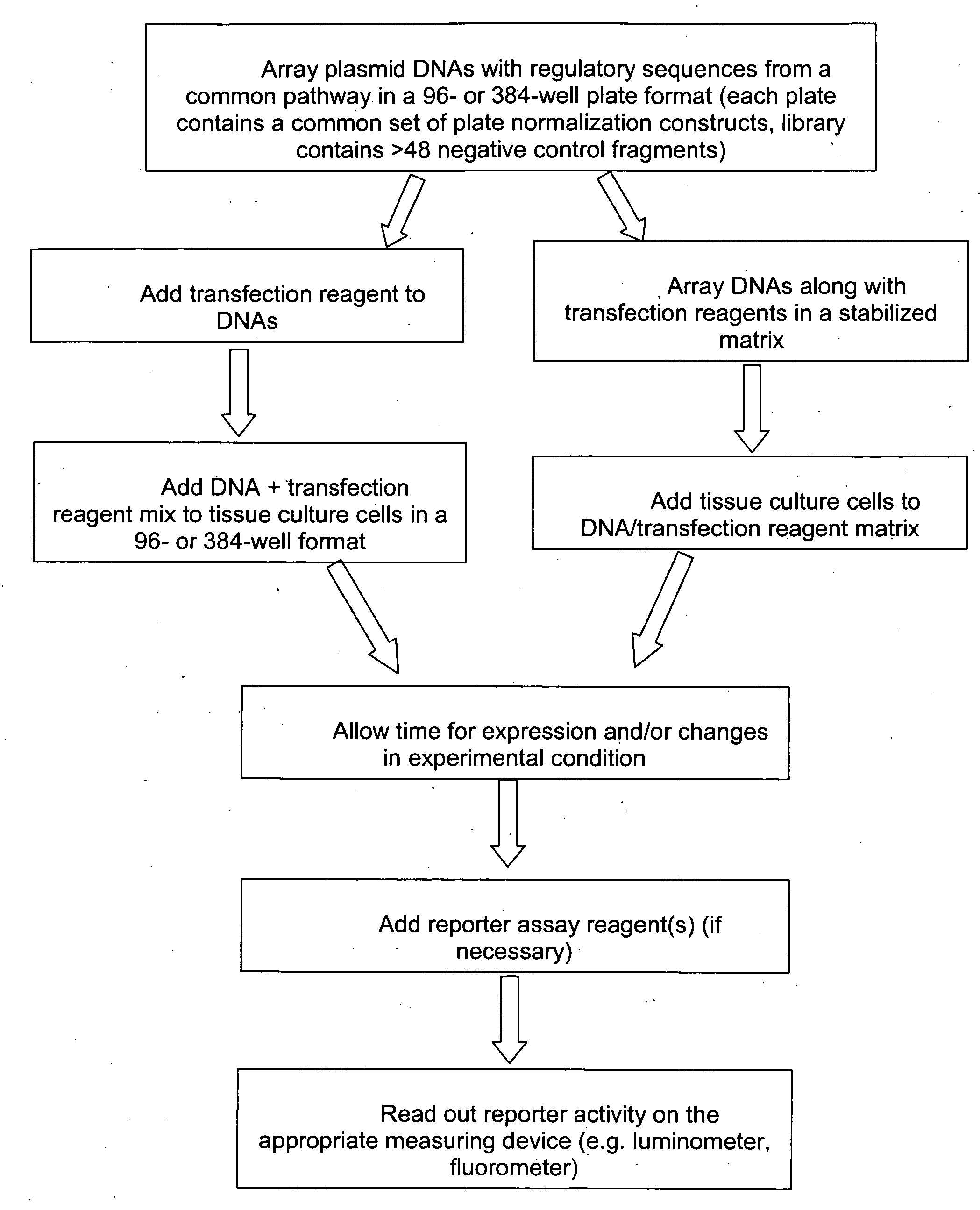
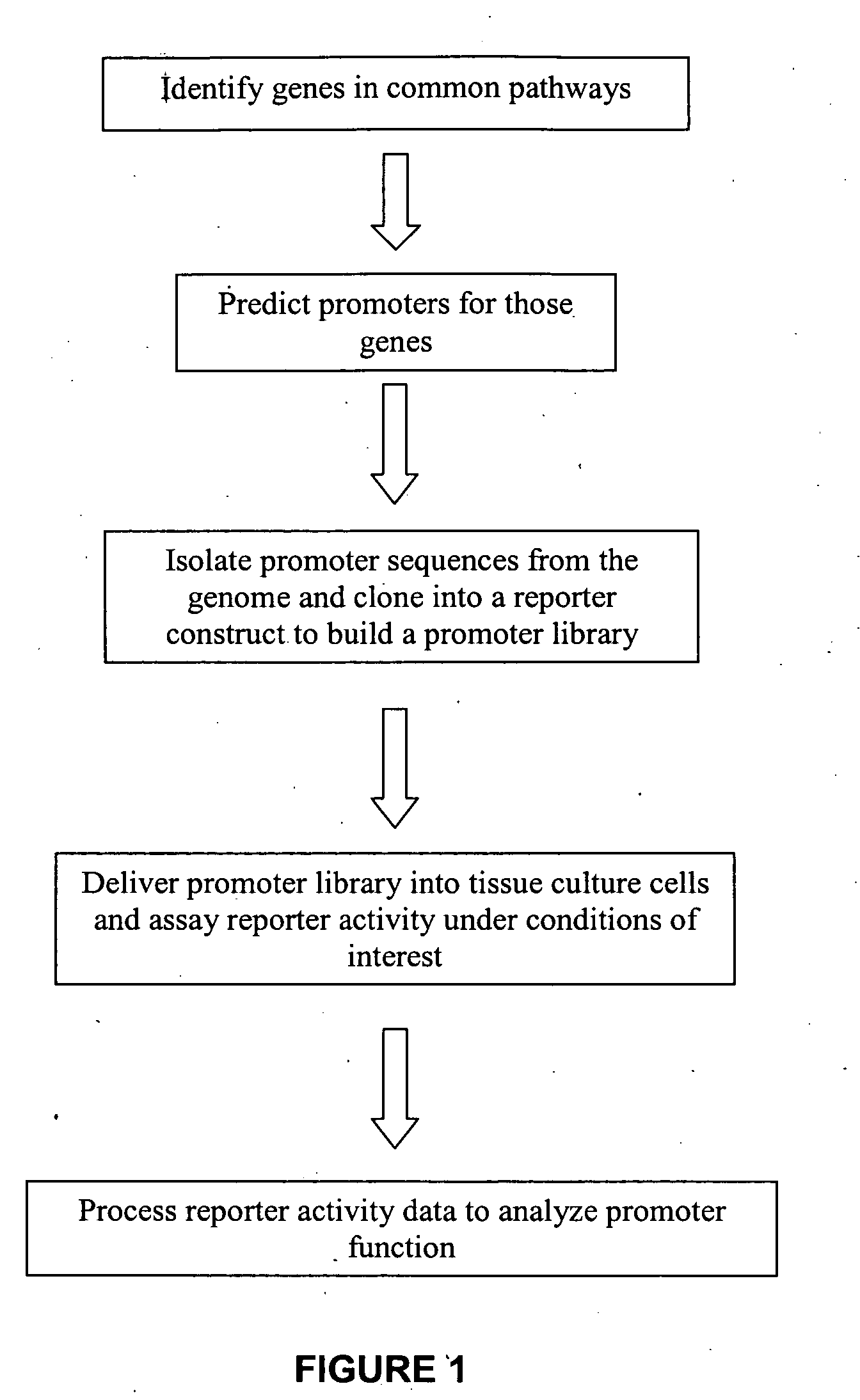
InactiveUS20090018031A1Determine effectNucleotide librariesLibrary screeningHuman DNA sequencingRegulation of gene expression
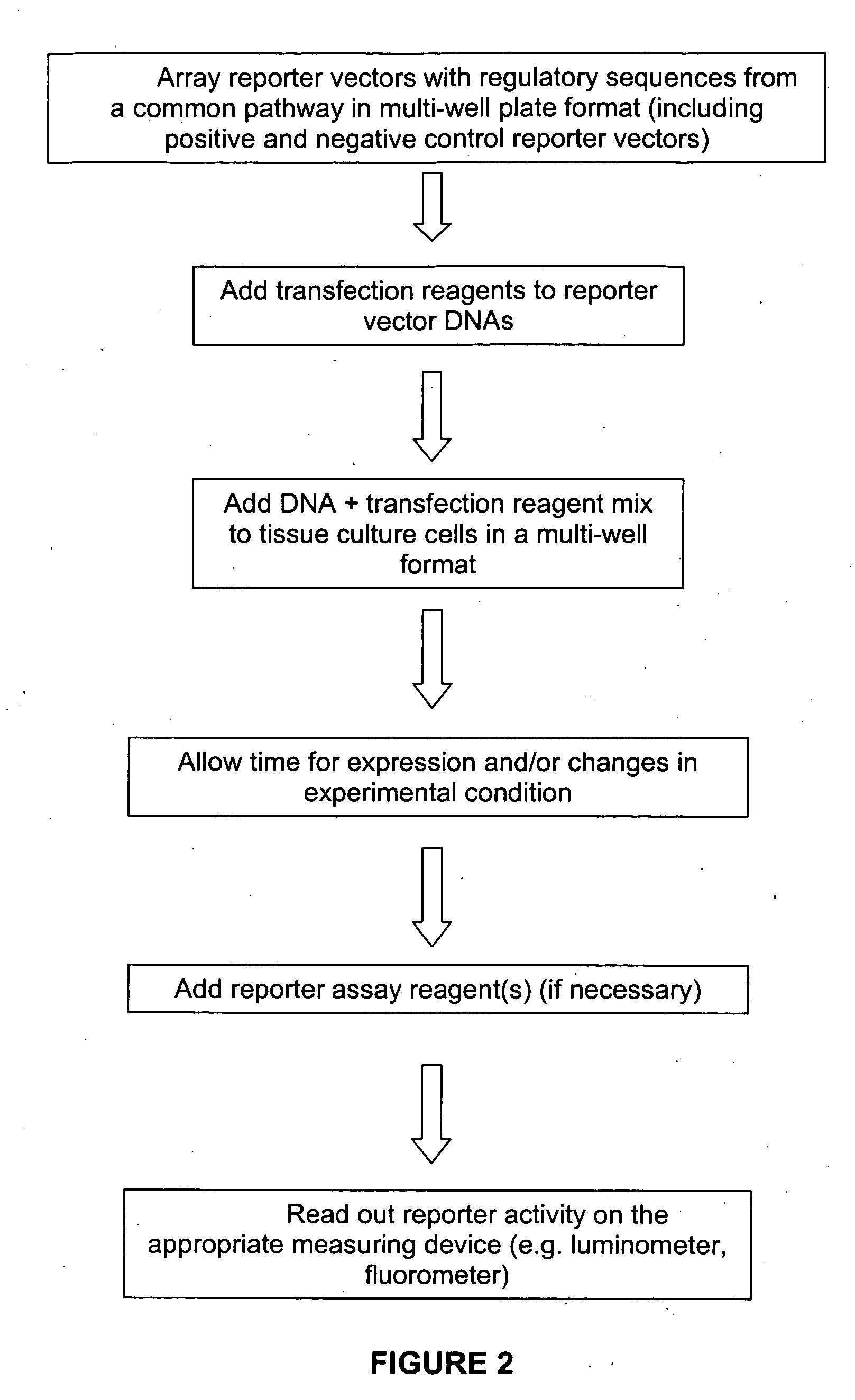
The present invention provides compositions, kits, assemblies, libraries, arrays, and high throughput methods for large scale structural and functional characterization of gene expression regulatory elements in a genome of an organism, especially in a human genome, that are part of a common pathway. In one aspect of the invention, an array of expression constructs is provided, each of the expression constructs comprising: a nucleic acid segment operably linked with a reporter sequence in an expression vector such that expression of the reporter sequence is under the transcriptional control of the nucleic acid segment. The present invention can have a wide variety of applications such as in personalized medicine, pharmacogenomics, and correlation of polymorphisms with phenotypic traits.
Owner:SWITCHGEAR GENOMICS
Analysis of transcriptomic data using similarity based modeling
InactiveUS20060293859A1Classification capability can be extendedSmall and fast computing footprintBiostatisticsBiological testingBiomarker discoveryModelling analysis
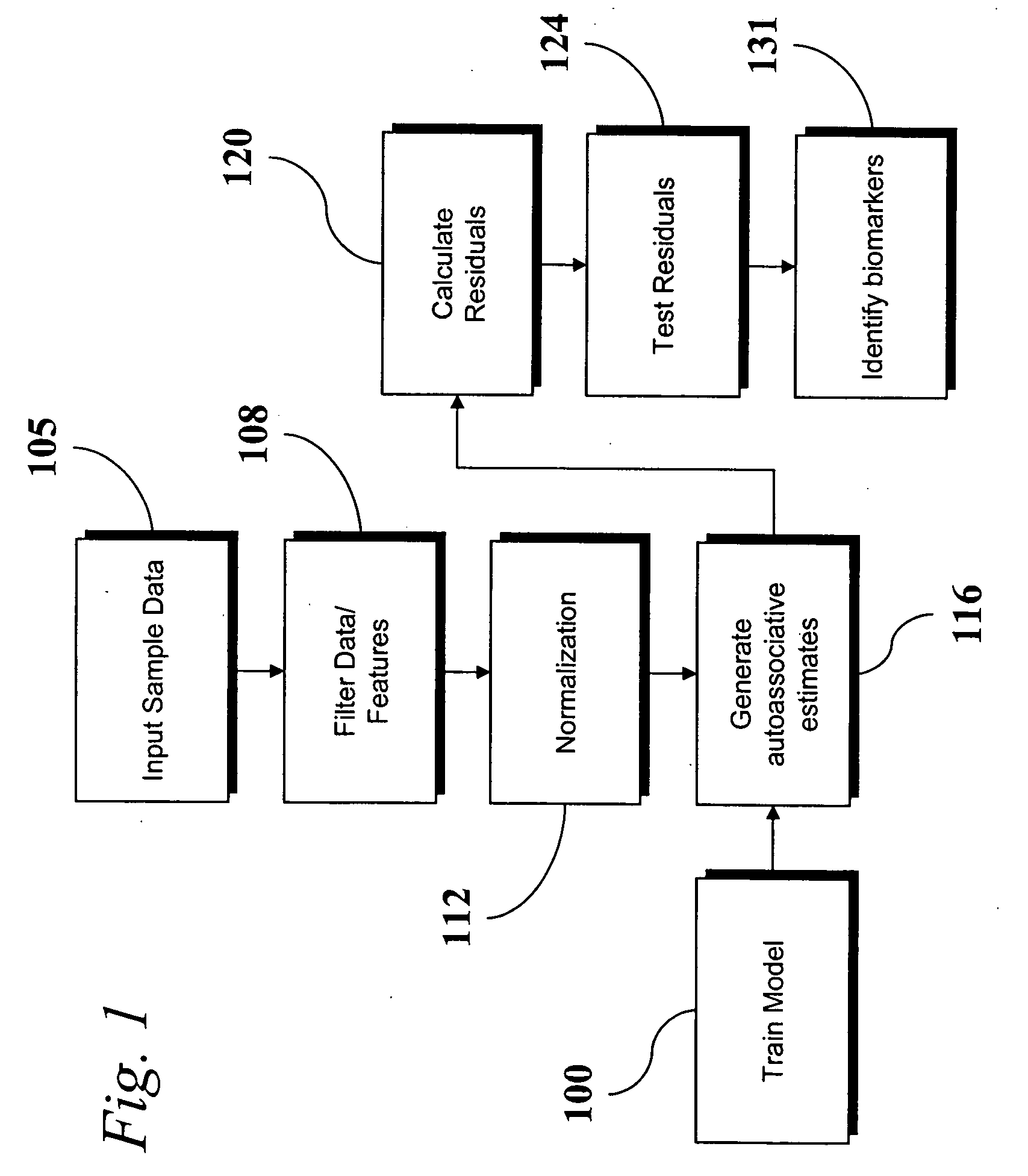
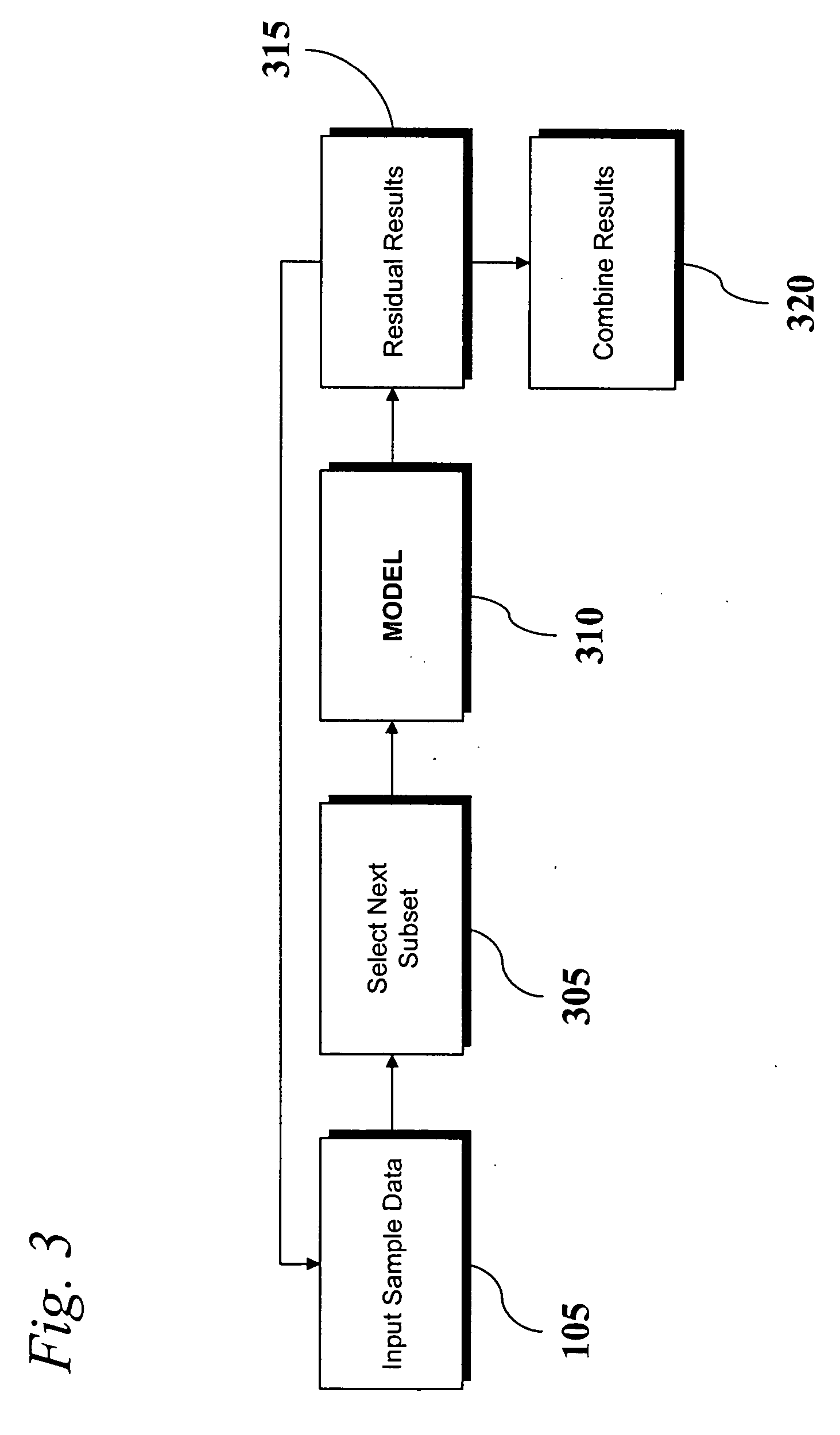
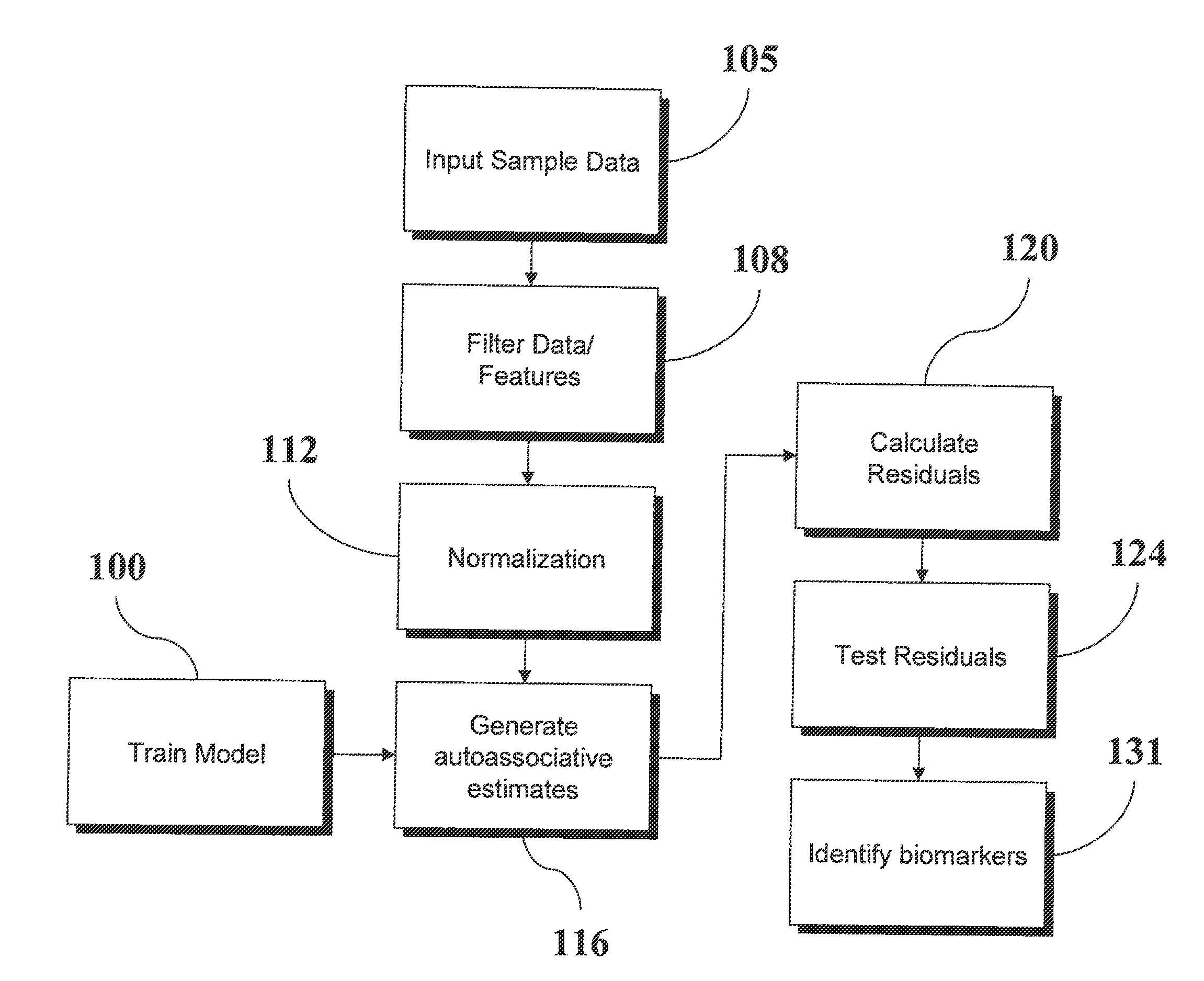
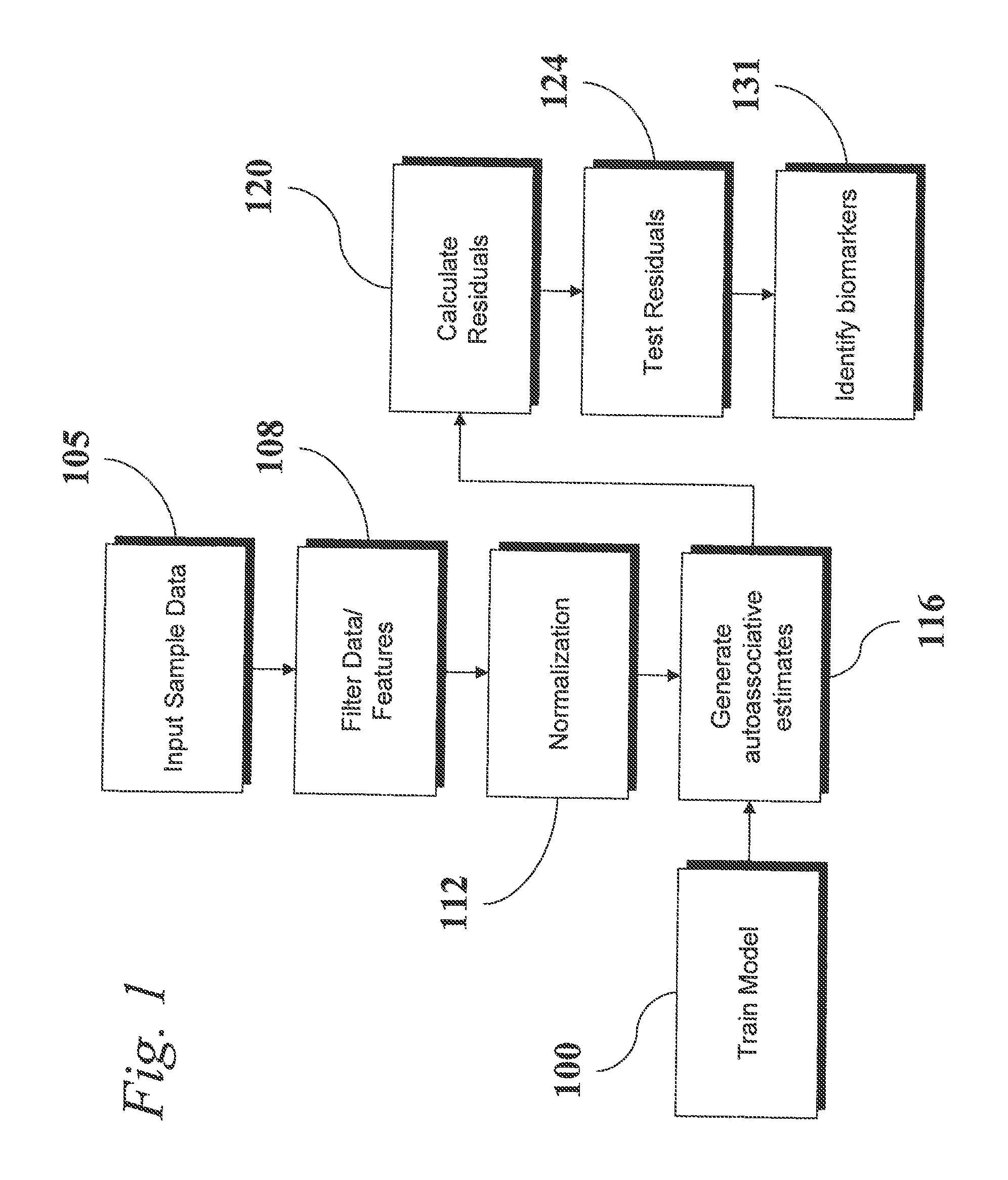
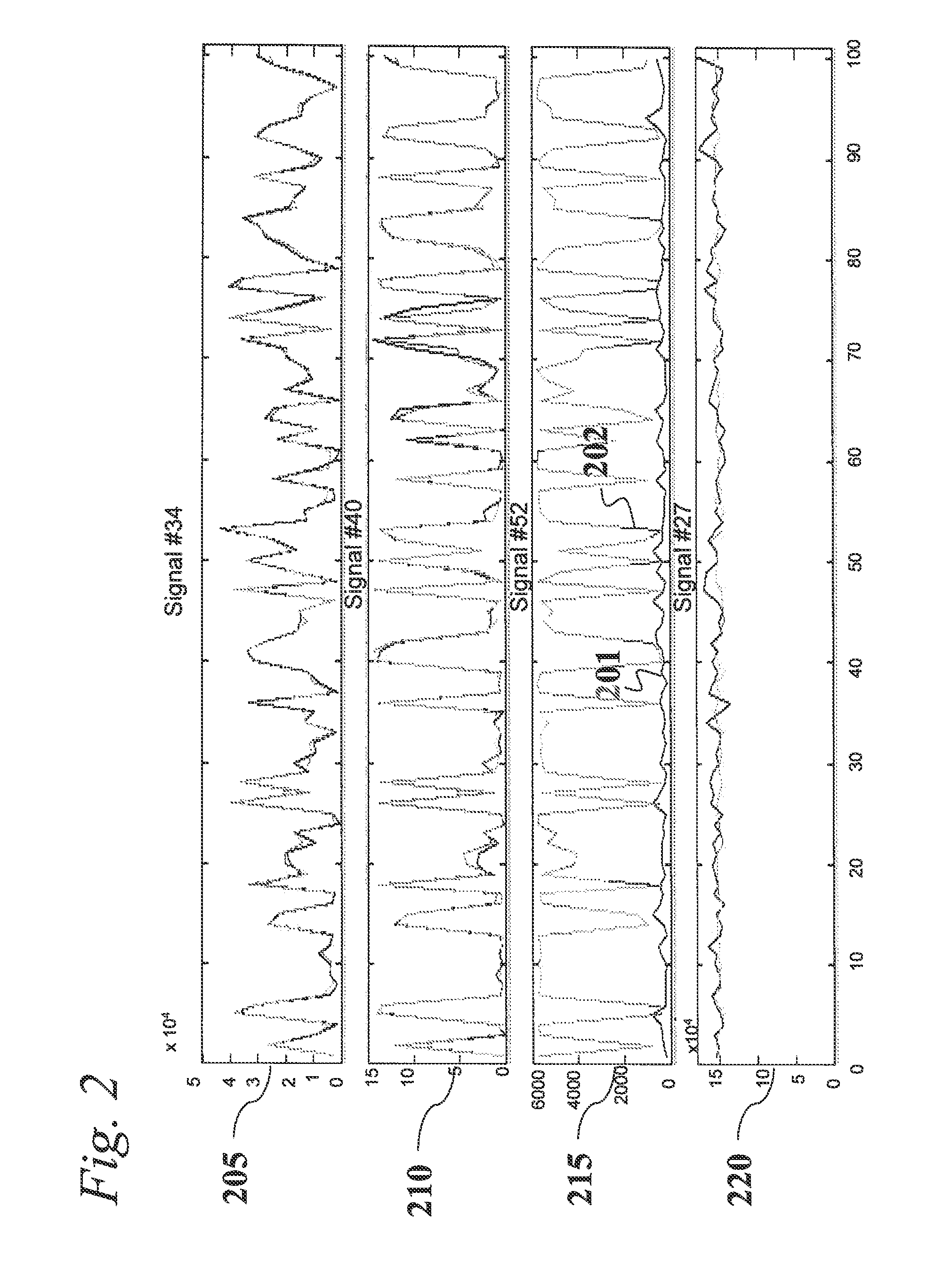
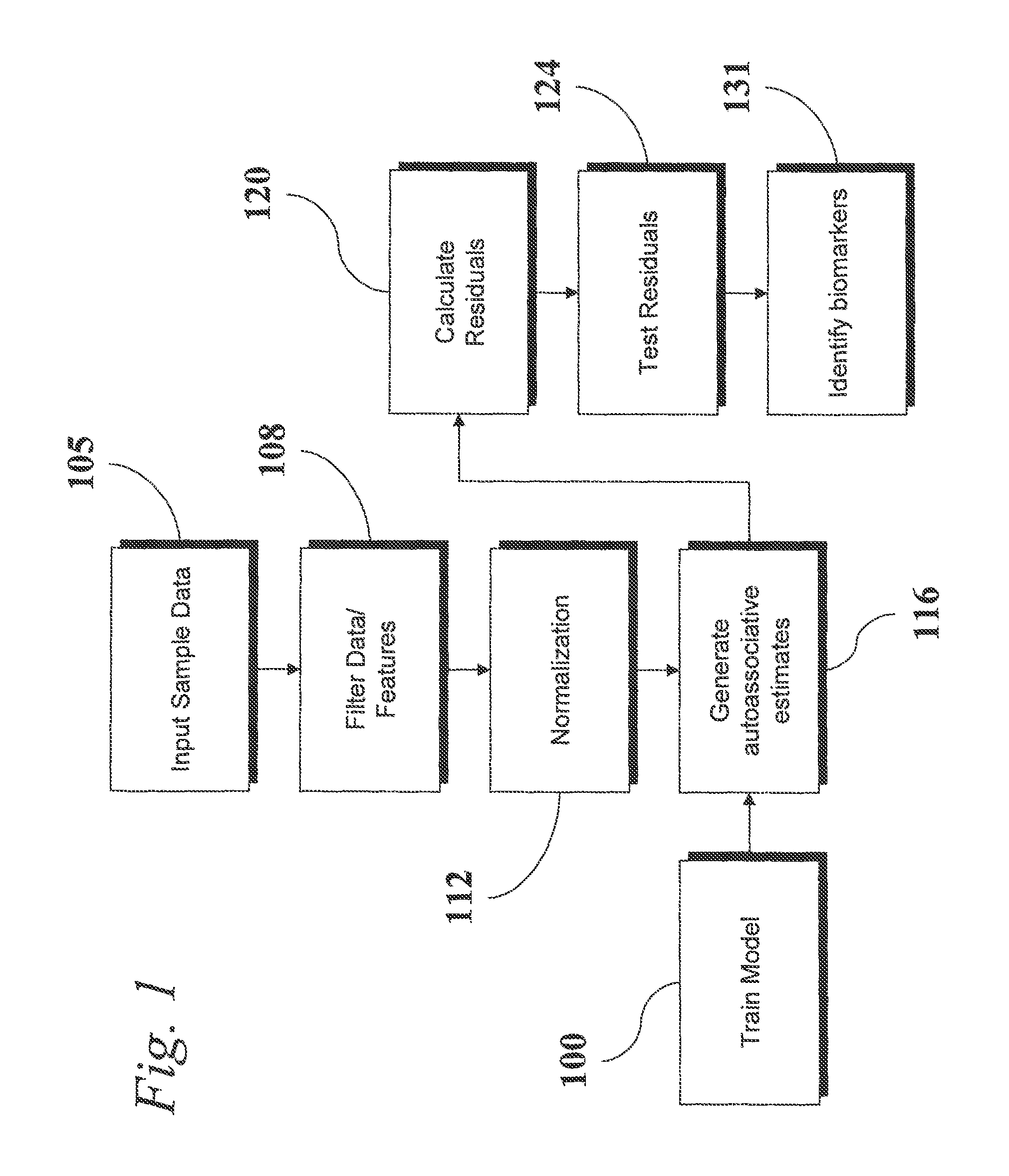
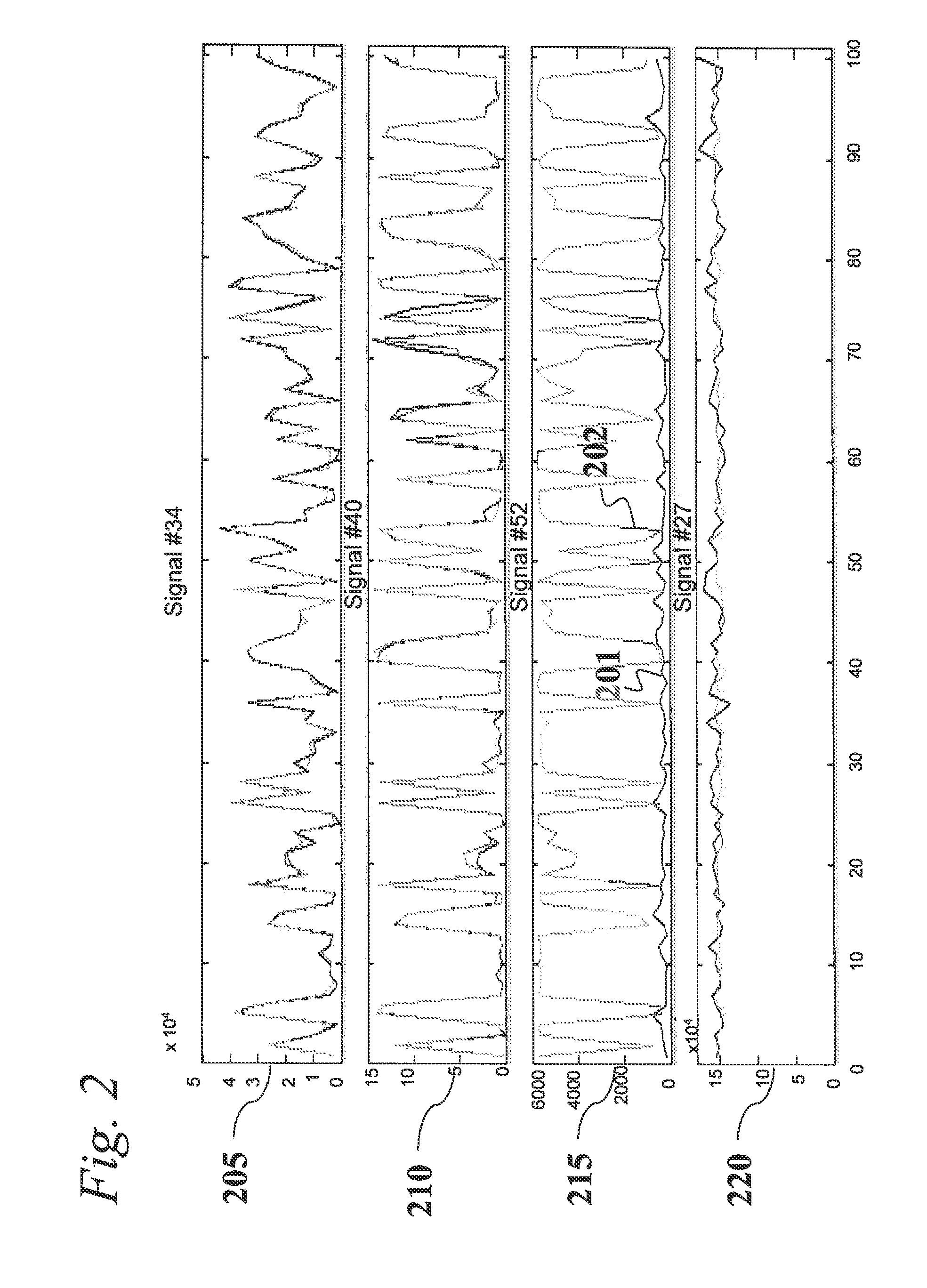
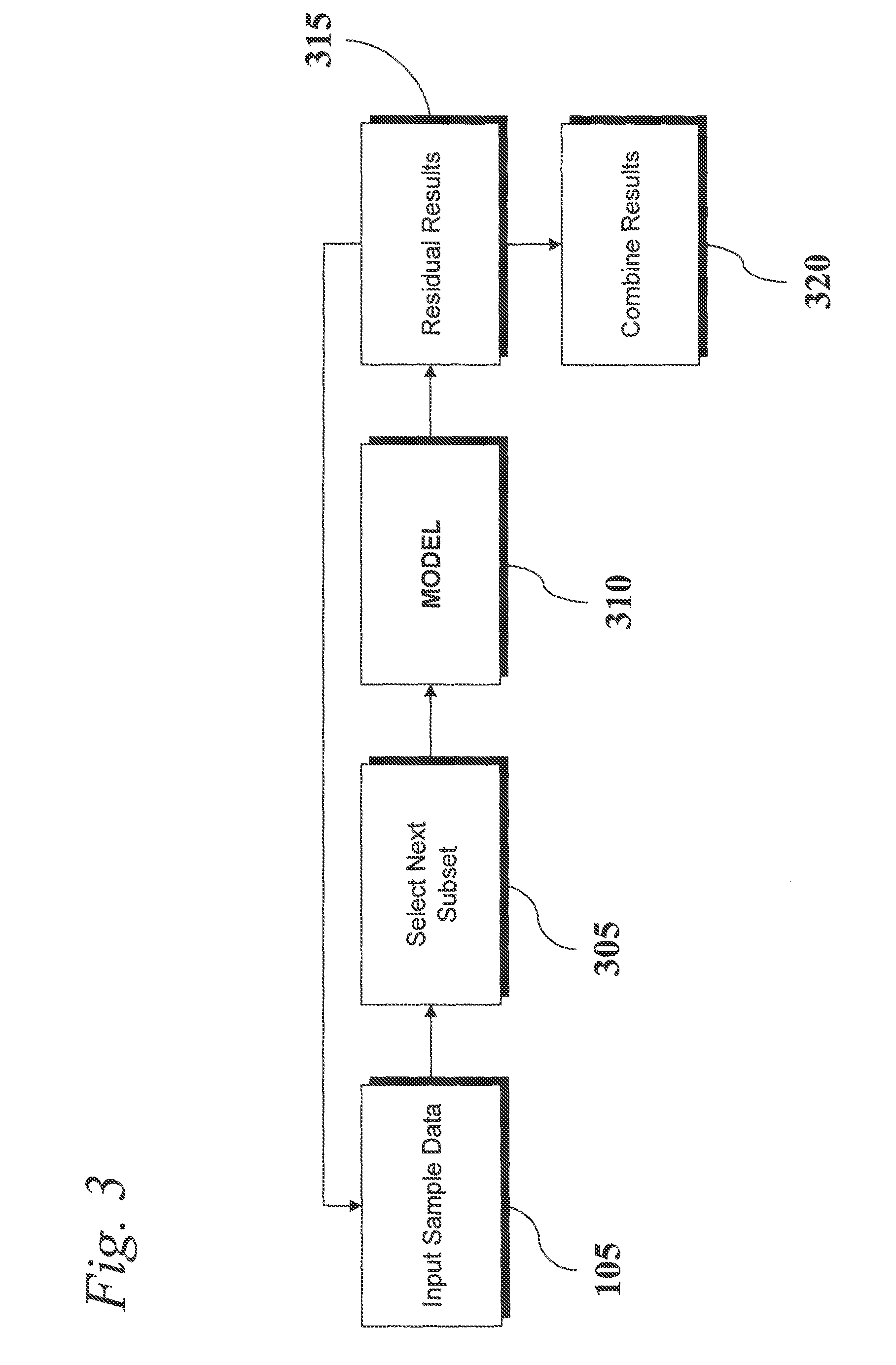
An analytic apparatus and method is provided for diagnosis, prognosis and biomarker discovery using transcriptome data such as mRNA expression levels from microarrays, proteomic data, and metabolomic data. The invention provides for model-based analysis, especially using kernel-based models, and more particularly similarity-based models. Model-derived residuals advantageously provide a unique new tool for insights into disease mechanisms. Localization of models provides for improved model efficacy. The invention is capable of extracting useful information heretofore unavailable by other methods, relating to dynamics in cellular gene regulation, regulatory networks, biological pathways and metabolism.
Owner:VENTURE GAIN L L C
Apparatus and method for electromagnetic treatment
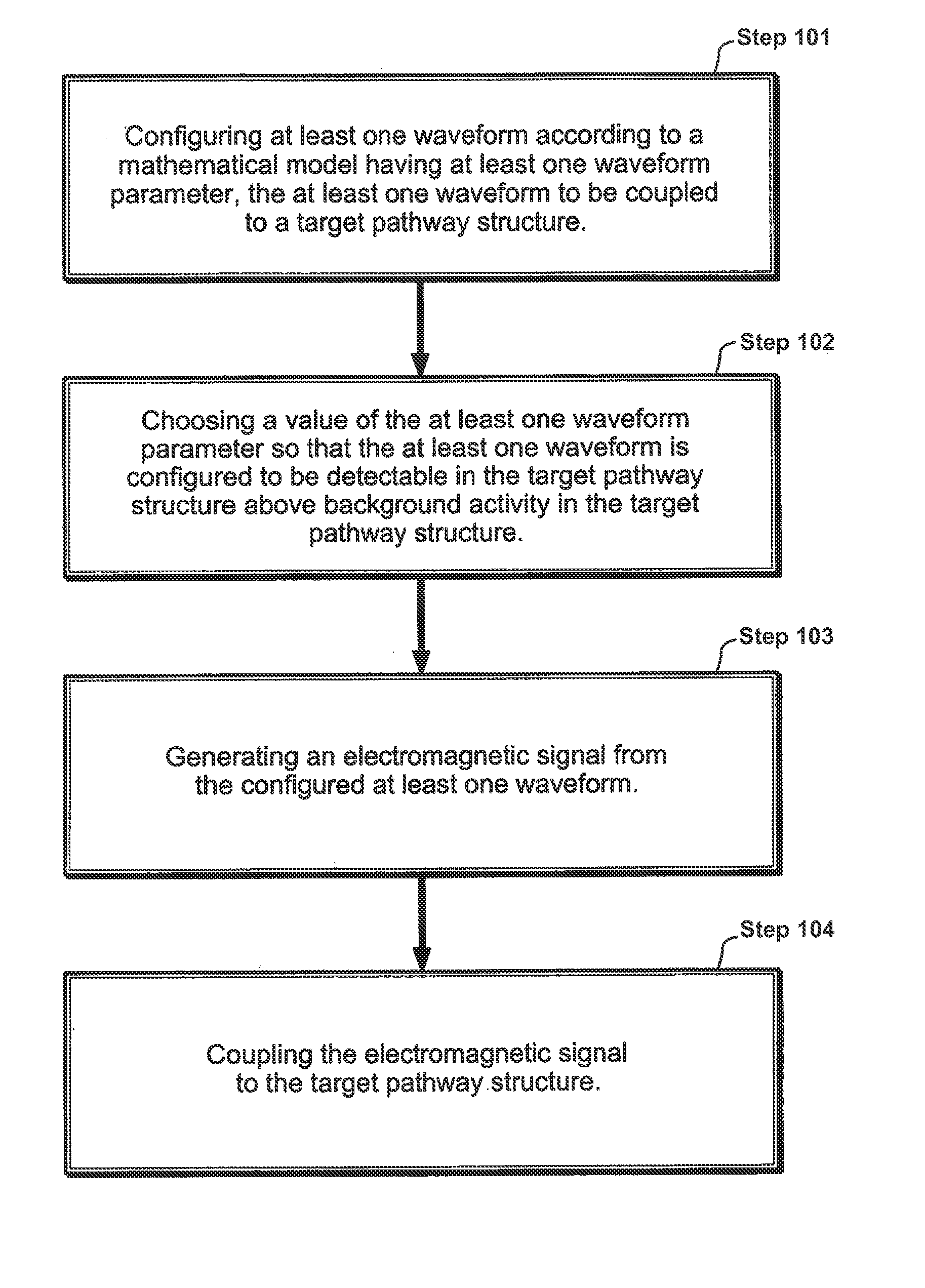
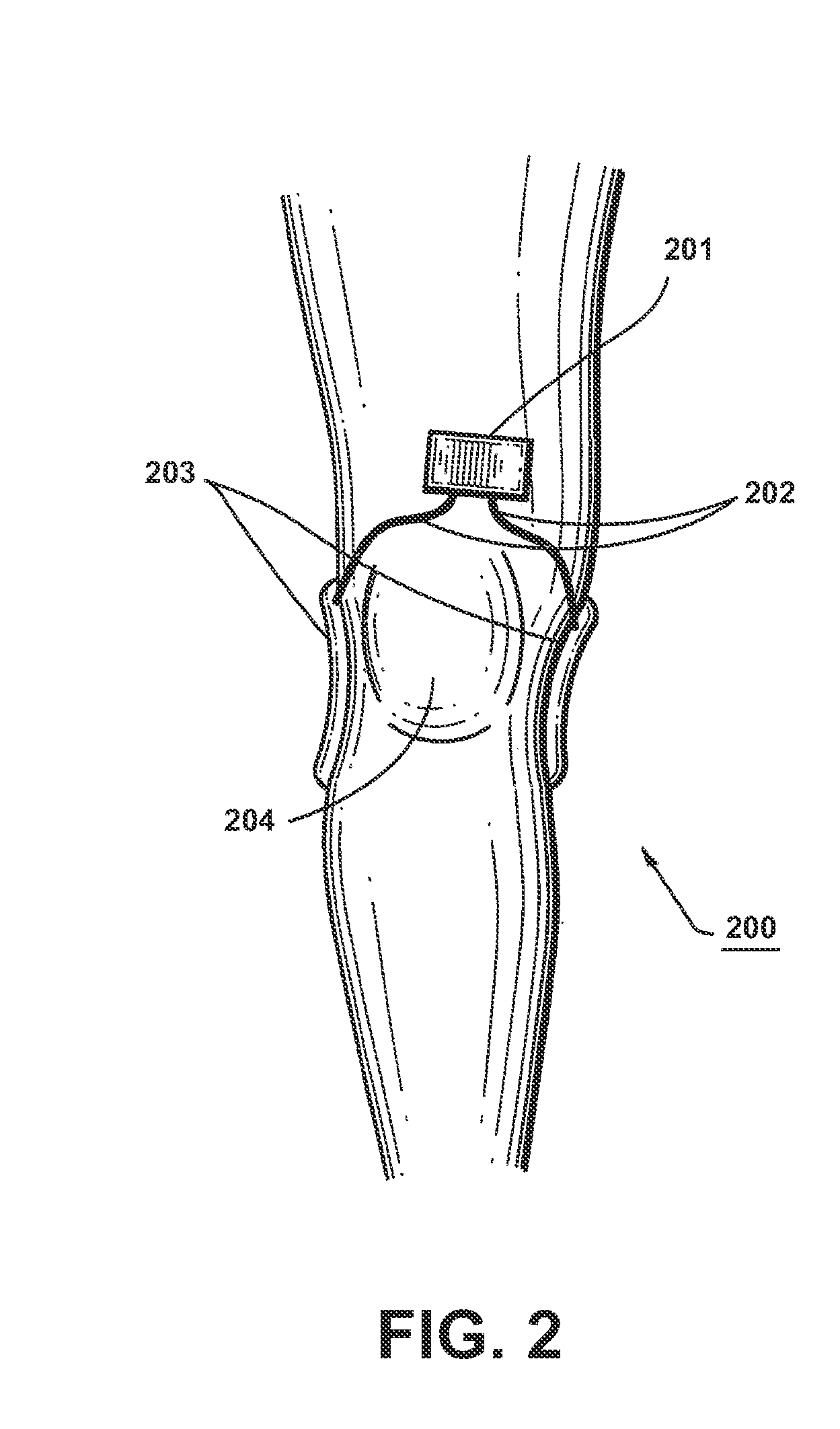
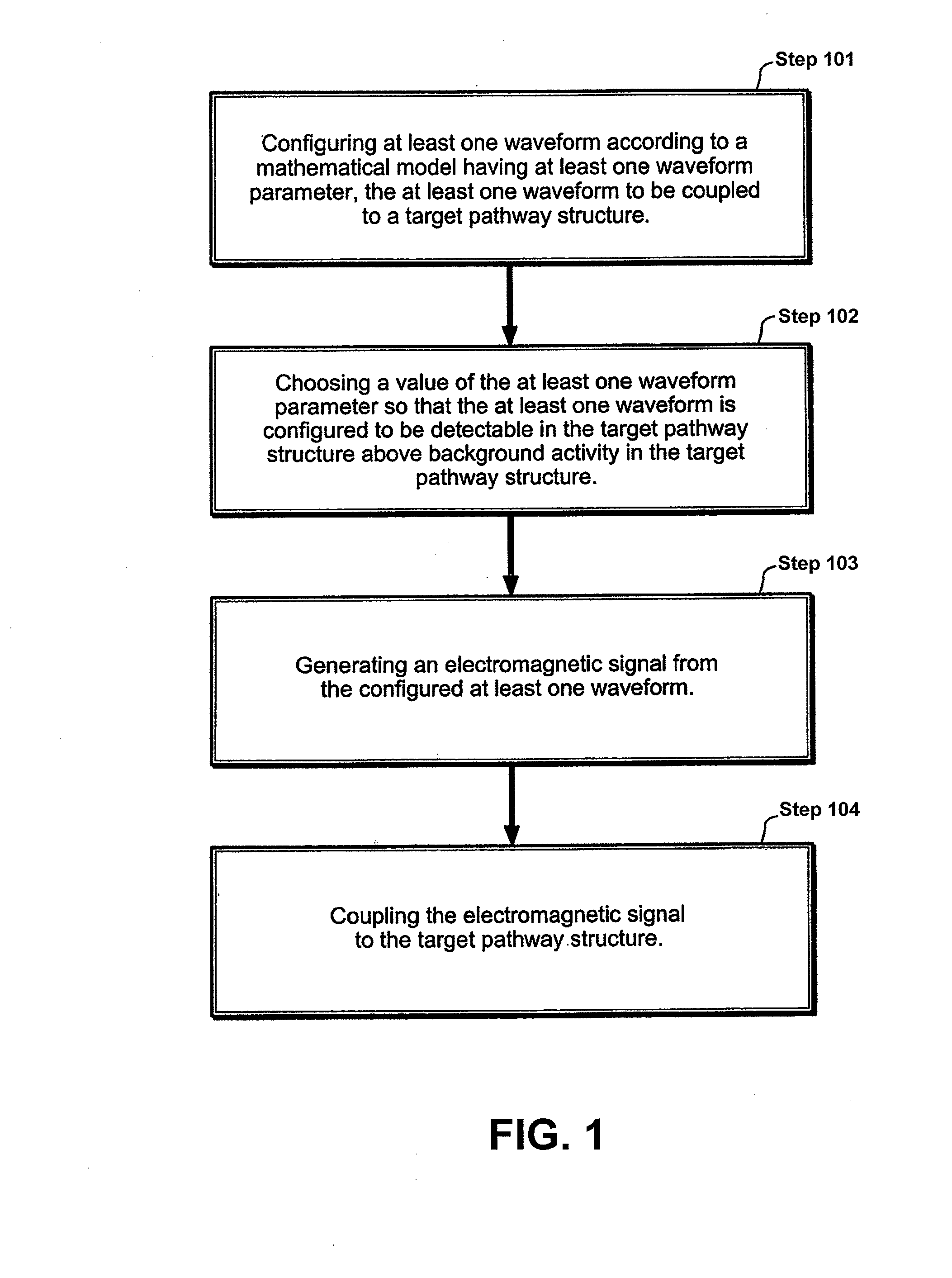
InactiveUS20110112352A1Enhanced transmitted dosimetryReduced Power RequirementsElectrotherapyMagnetotherapy using coils/electromagnetsMedicineBiological pathway
Described herein are electromagnetic treatment devices for treatment of tissue. In particular, described herein are lightweight, wearable, low-energy variations that are specifically configured to specifically and sufficiently apply energy within a specific bandpass of frequencies of a target biological pathway, such as the binding of Calcium to Calmodulin, and thereby regulate the pathway. Methods and systems for treating biological tissue are also described.
Owner:RIO GRANDE NEUROSCI
Method and systems for high throughput single cell genetic manipulation
InactiveUS20180312873A1Strong specificityUptake of the reagent into the cell is facilitatedMicroencapsulation basedBiological pathwayDrug target
Provided herein are methods and systems for introducing nucleic acid manipulation agents into single cells. Such high throughput delivery of nucleic acid manipulation reagents into single cells and subsequent genetic manipulation of such cells allow for large scale genetic analysis that can be useful, for example, for the study of biological pathways and drug target discovery.
Owner:10X GENOMICS
Using knowledge pattern search and learning for selecting microorganisms
InactiveUS20080090736A1Recover clean energyMost efficientMicrobiological testing/measurementLibrary screeningScreening methodClean energy
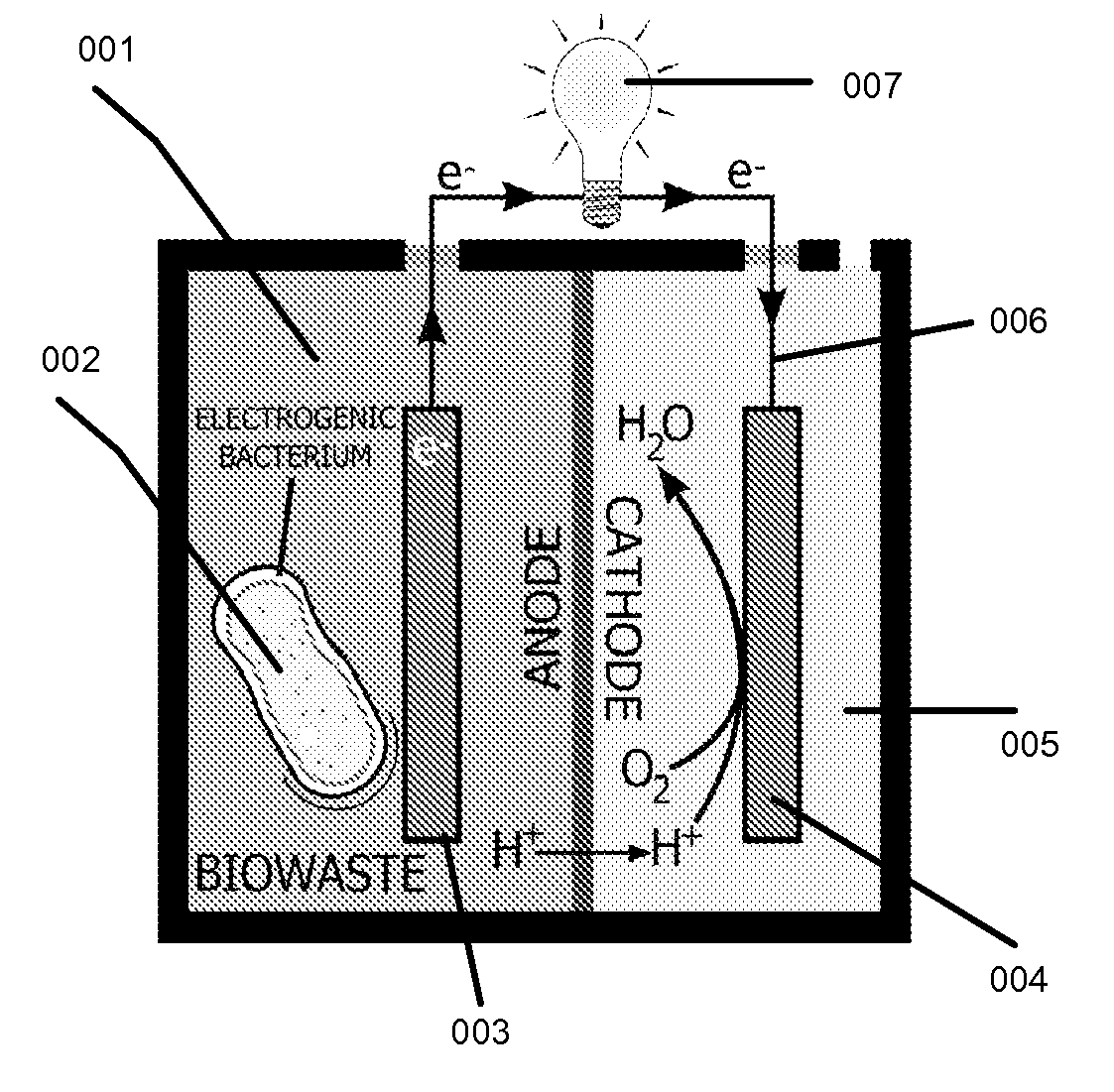
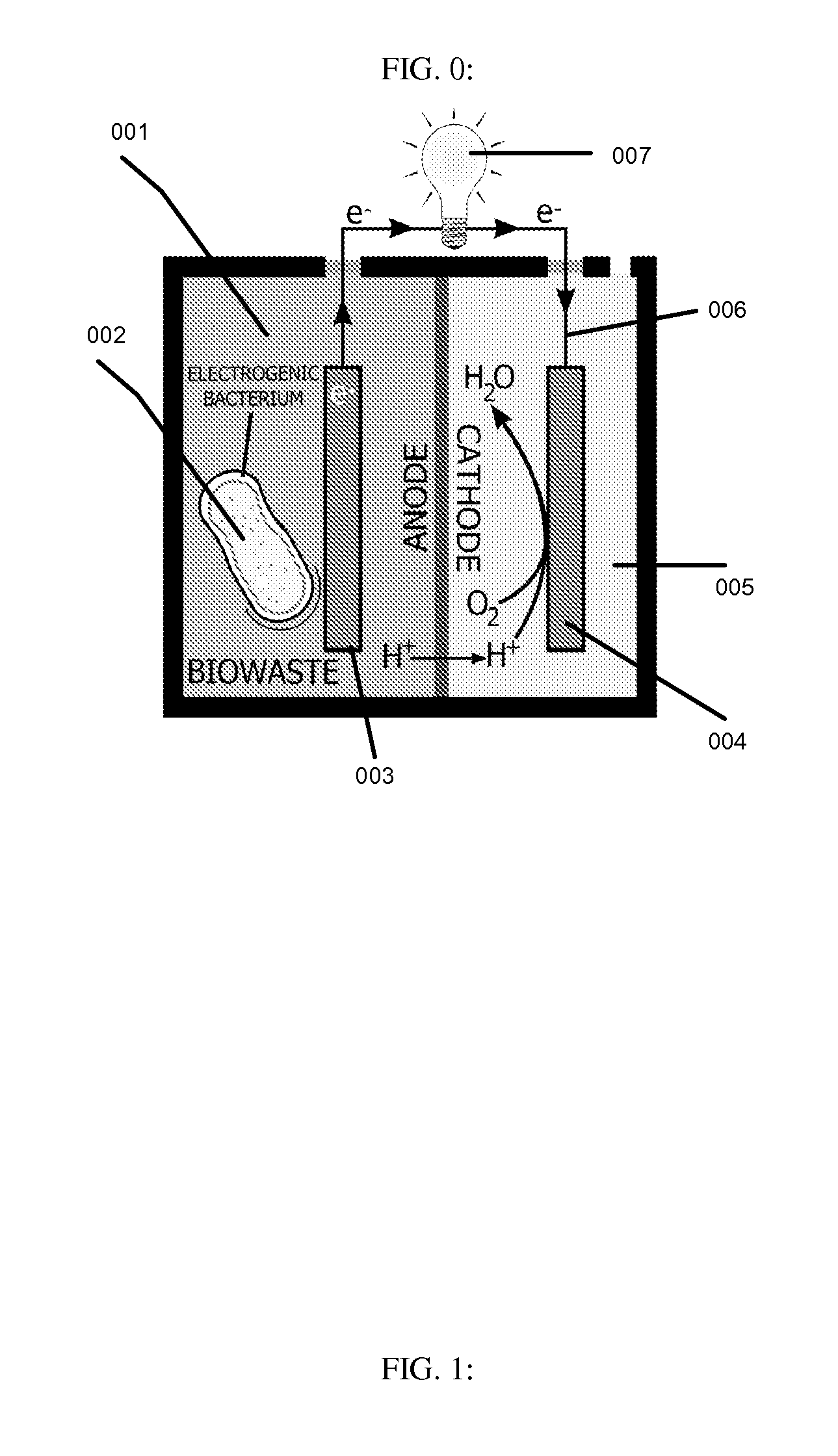
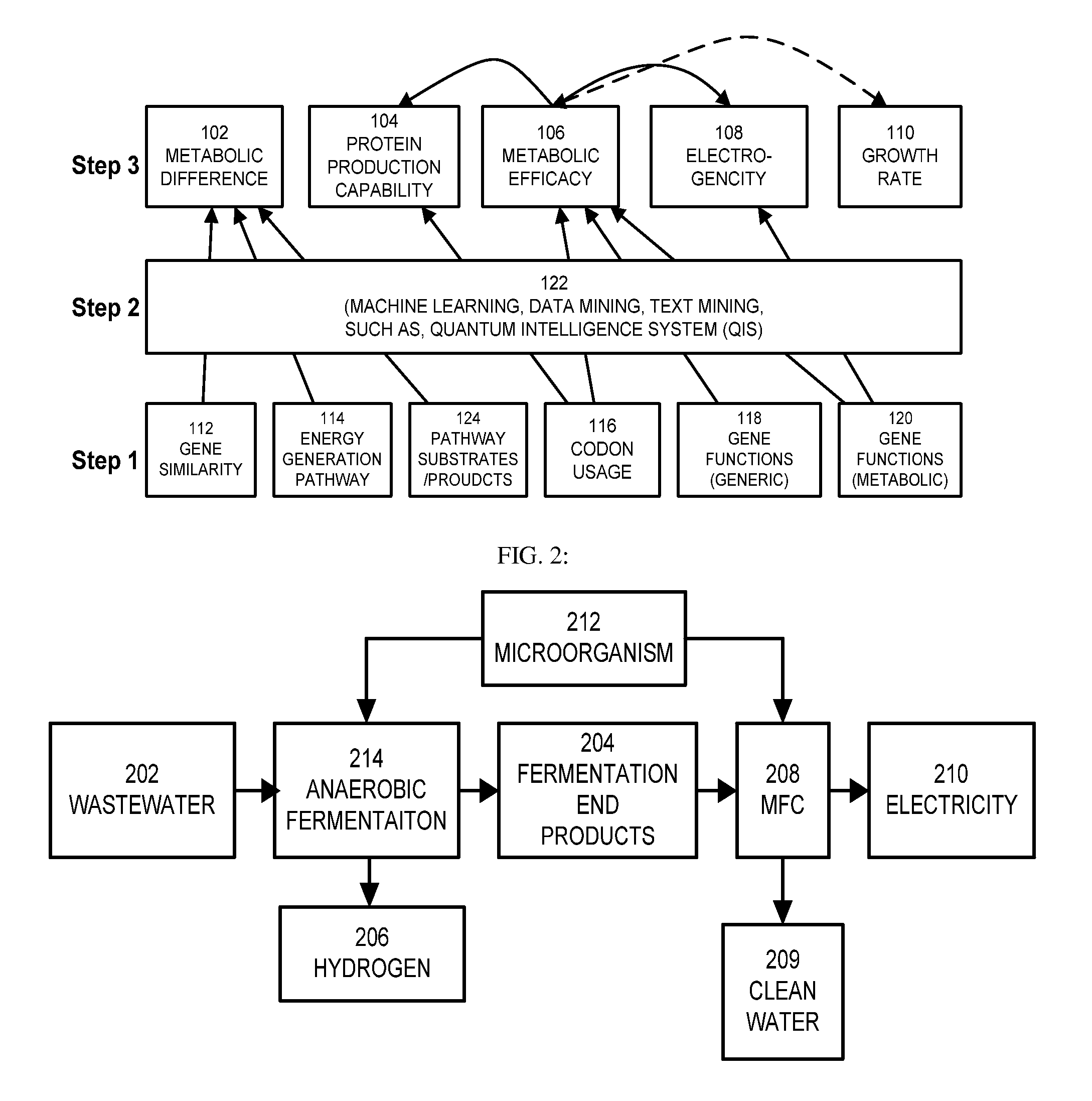
This invention is to use knowledge pattern learning and search system for selecting microorganisms to produce useful materials and to generate clean energy from wastes, wastewaters, biomass or from other inexpensive sources. The method starts with an in silico screening platform which involves multiple steps. First, the organisms' profiles are compiled by linking the massive genetic and chemical fingerprints in the metabolic and energy-generating biological pathways (e.g. codon usages, gene distributions in function categories, etc.) to the organisms' biological behaviors. Second, a machine learning and pattern recognition system is used to group the organism population into characteristic groups based on the profiles. Lastly, one or a group of microorganisms are selected based on profile match scores calculated from a defined metabolic efficiency measure, which, in term, is a prediction of a desired capability in real life based on an organism's profile. In the example of recovering clean energy from treating wastewaters from food process industries, domestic or municipal wastes, animal or meat-packing wastes, microorganisms' metabolic capabilities to digest organic matter and generate clean energy are assessed using the invention, and the most effective organisms in terms of waste reduction and energy generation are selected based on the content of a biowaste input and a desired clean energy output. By selecting a microorganism or consortia of multiply microorganisms using this method, one can clean the water and also directly generate electricity from Microbial Fuel Cells (MFC), or hydrogen, methane or other biogases from microorganism fermentation. In addition, using similar screening method, clean hydrogen can be recovered first from an anaerobic fermentation process accompanying the wastewater treatment, and the end products from the fermentation process can be fed into a Microbial Fuel Cell (MFC) process to generate clean electricity and at the same time treat the wastewater. The invention can be used to first select the hydrogenic microorganisms to efficiently generate hydrogen and to select electrogenic organisms to convert the wastes into electricity. This method can be used for converting wastes to one or more forms of renewable energies.
Owner:QUANTUM INTELLIGENCE
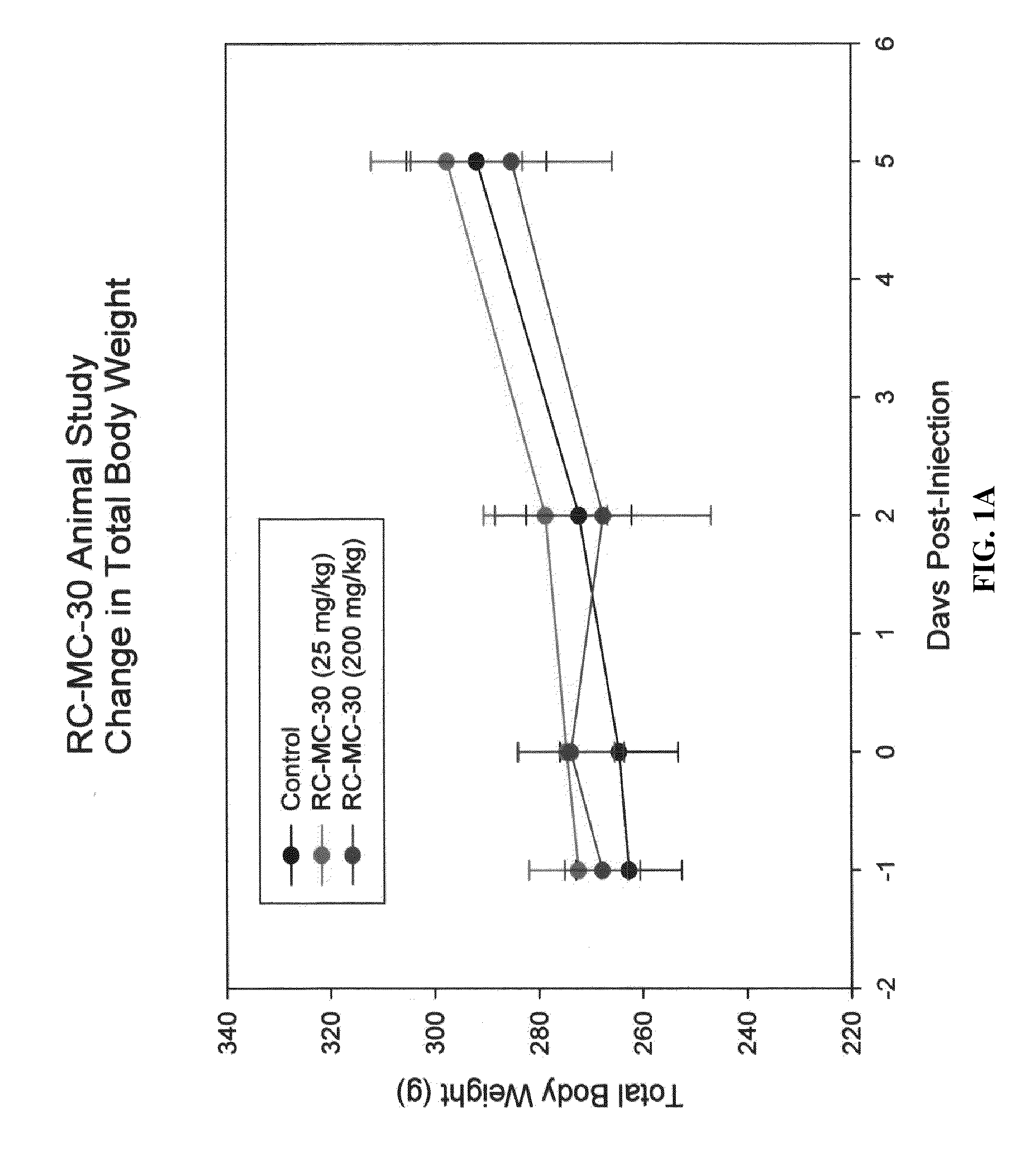
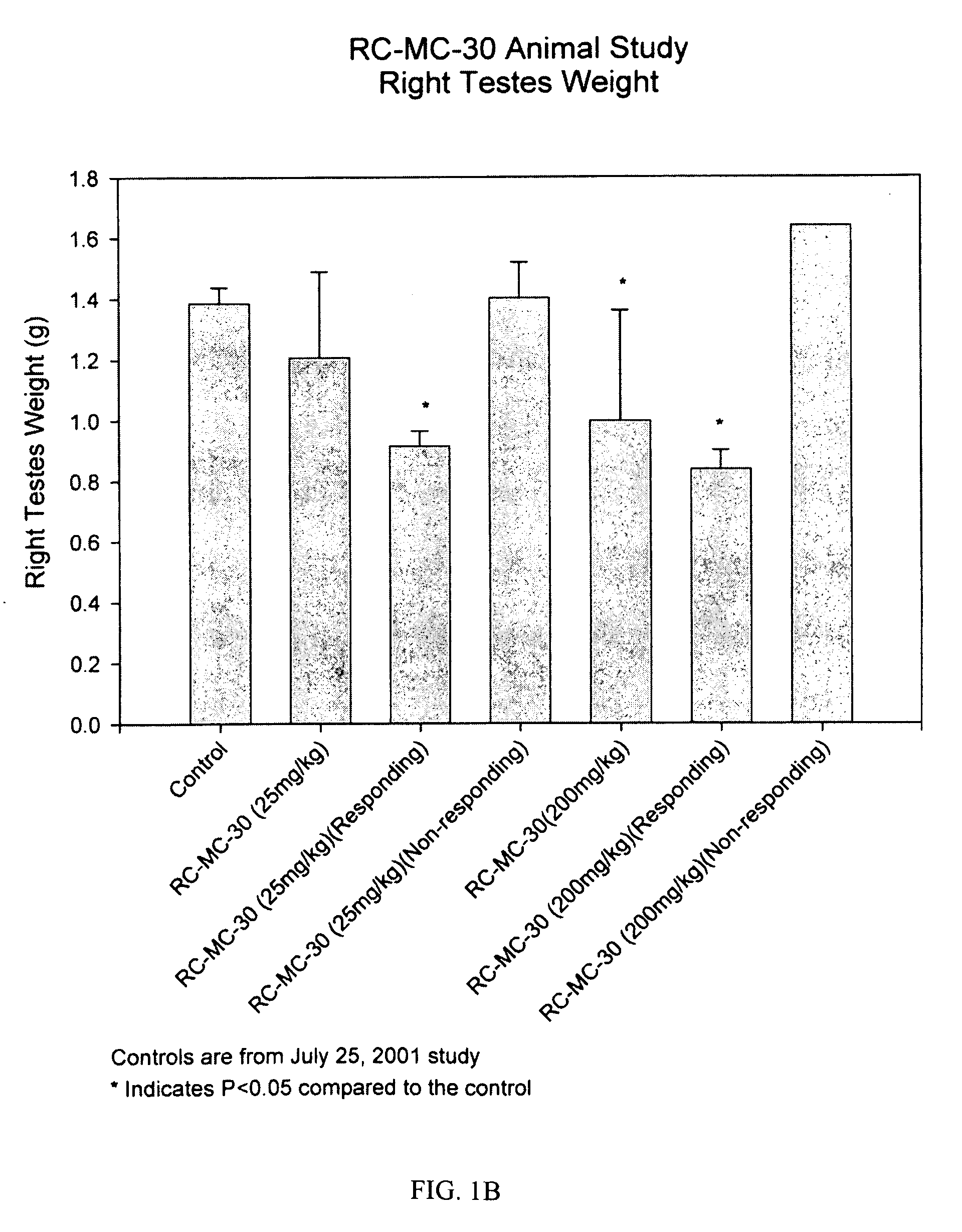
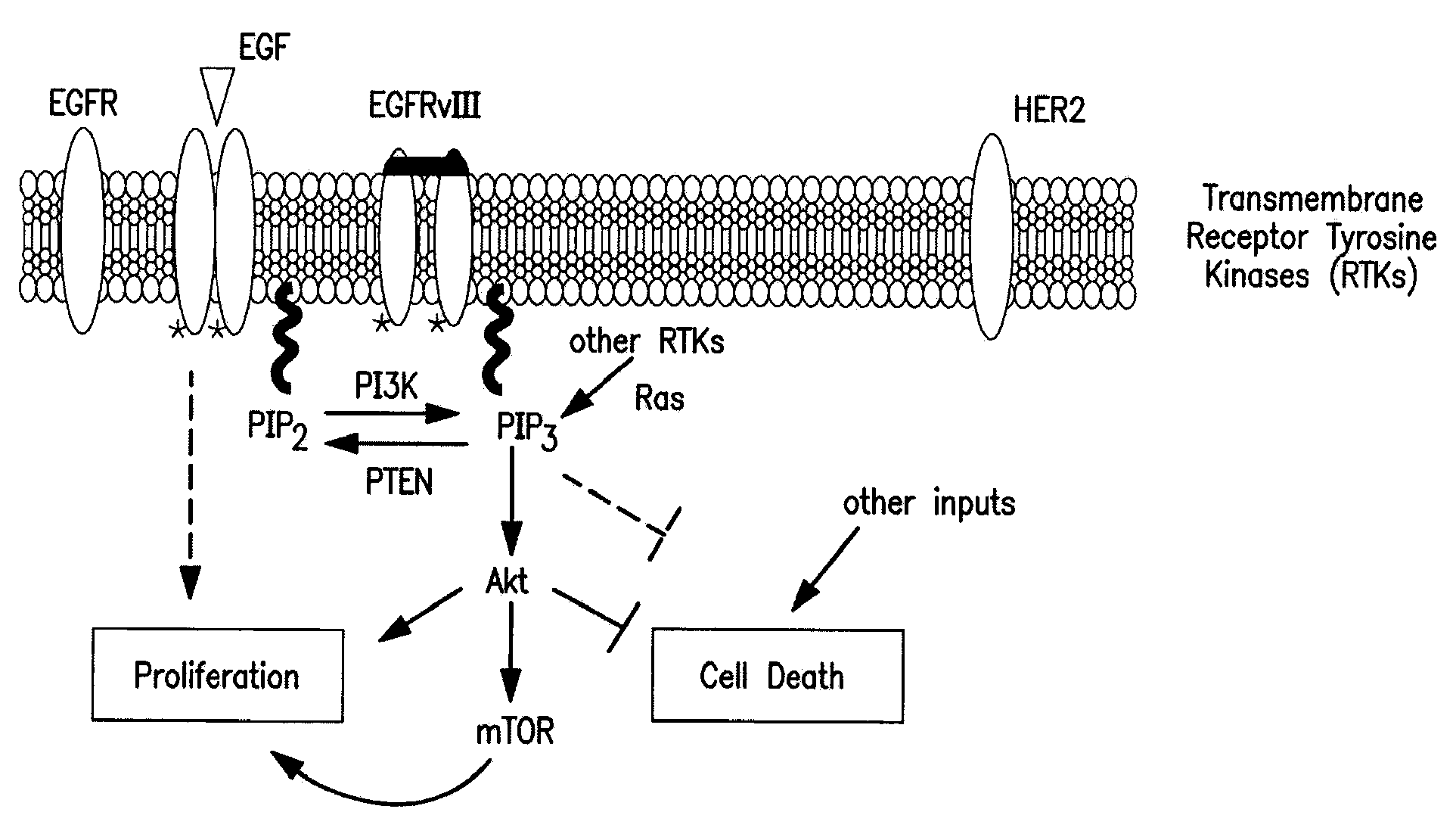
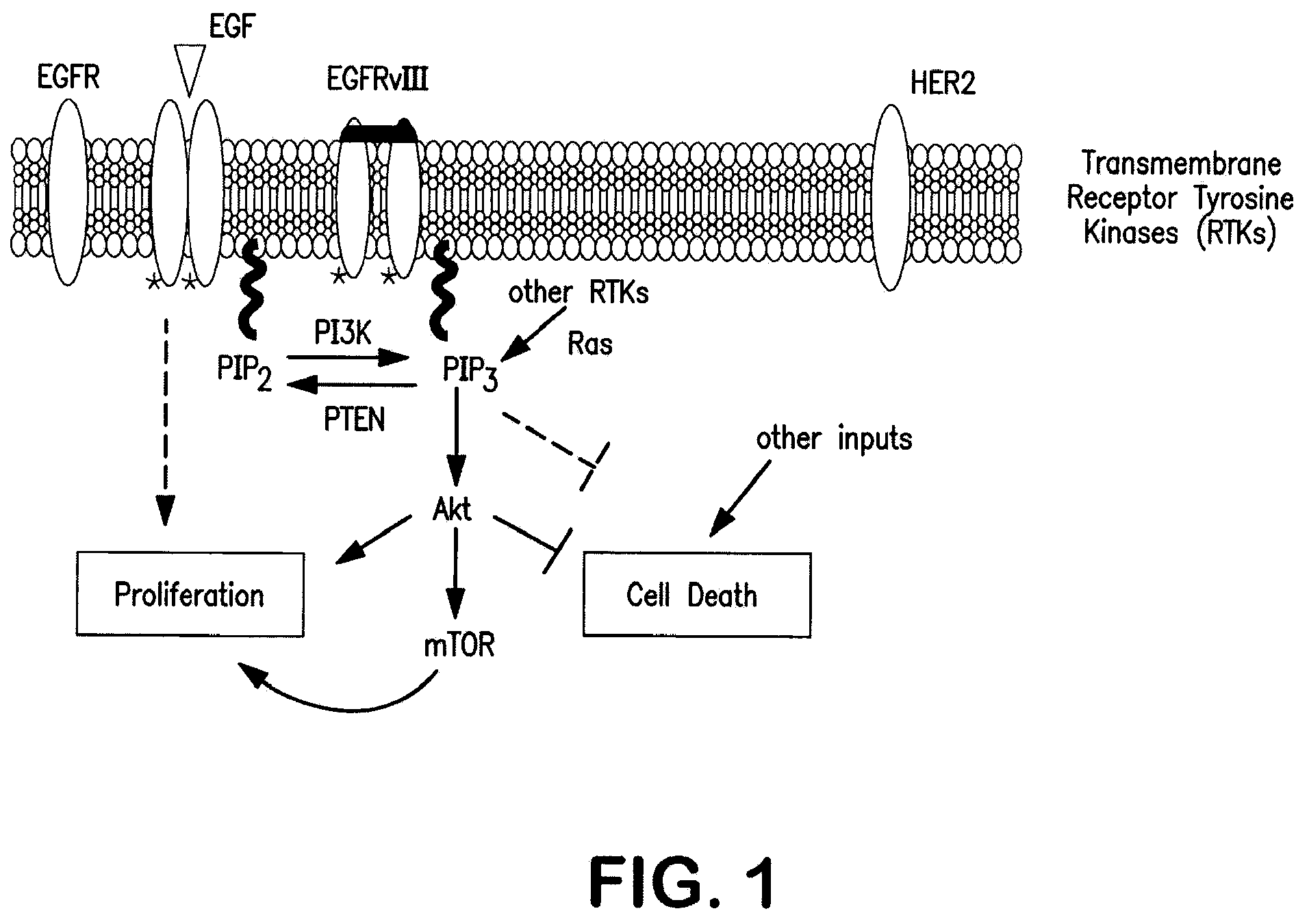
Method for treating cancer using a combination of chk1 and atr inhibitors
ActiveUS20150359797A1Surprising synergistic effectBiocideHeavy metal active ingredientsProtein targetMedicine
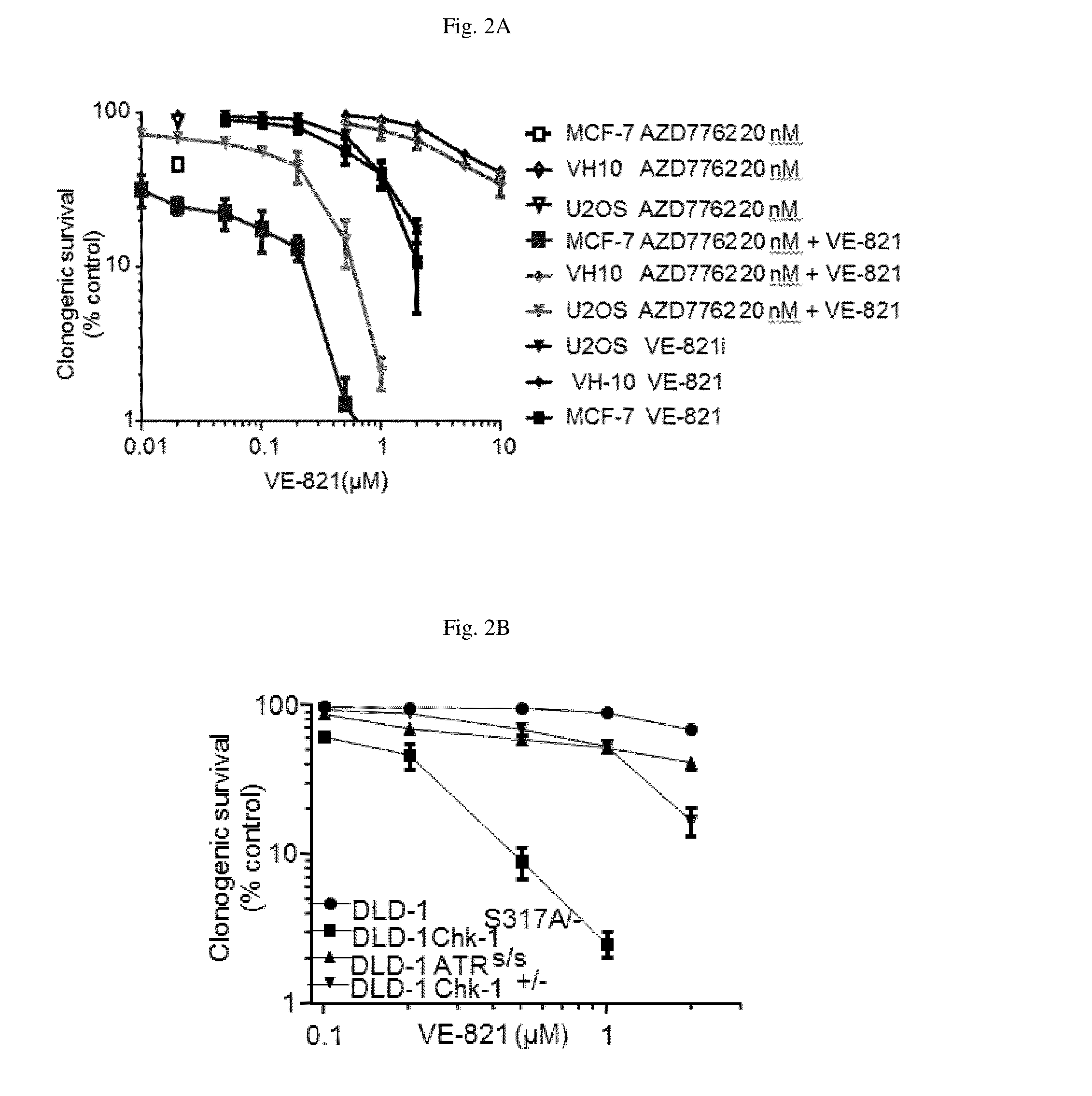
The present invention relates to methods of treating cancer in a patient by administering a compound useful as an inhibitor of ATR protein kinase in combination with a compound useful as an inhibitor of Chk1 protein kinase. The aforementioned combination displays a surprising synergistic effect in treating cancer despite the targeted protein kinases being within the same biological pathway. Moreover, the present invention also relates to methods of treating cancer by administering a compound useful as an inhibitor of ATR protein kinase; administering a compound useful as an inhibitor of Chk1 protein kinase; as well as administering a DNA damaging agent to a patient.The compounds utilized in this invention are represented by formula I and formula II:wherein the variables are as defined herein.
Owner:VERTEX PHARMA INC
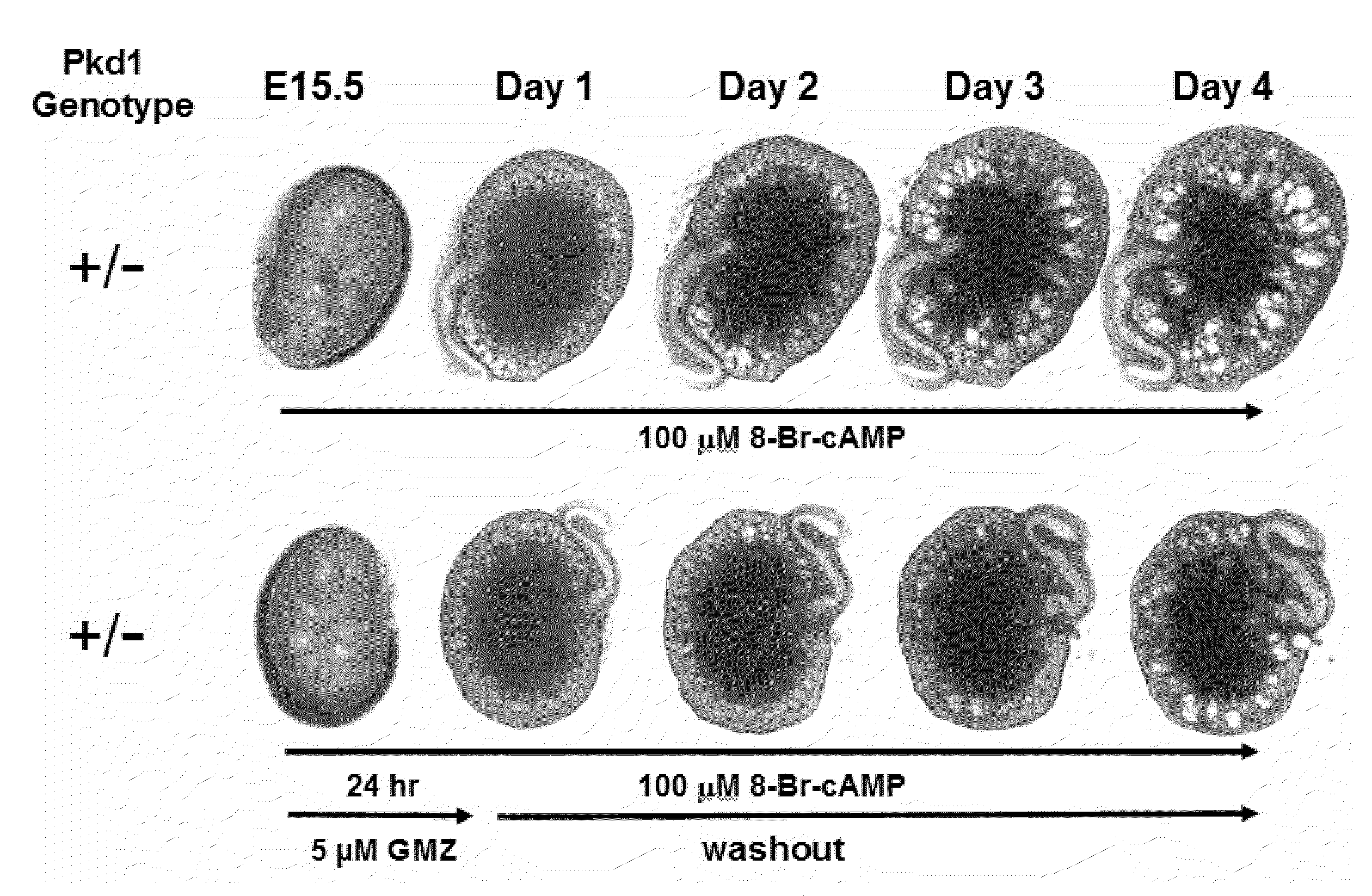
Lonidamine analogues and treatment of polycystic kidney disease
Lonidamine derivatives can be useful in methods of treating, inhibiting, and / or preventing polycystic kidney disease (PKD). Accordingly, lonidamine derivatives can be administered in a therapeutically effective amount for inhibiting, and / or preventing polycystic kidney disease (PKD) in the subject. This can include administering a therapeutically effective amount of the lonidamine derivatives for inhibiting CFTR and / or Hsp90 or biological pathway thereof. Also, the method can include administering the lonidamine derivatives in a therapeutically effective amount for inhibiting ErbB2, Src, Raf-1, B-Raf, MEK, Cdk4, NKCC1, or combinations thereof. For example, the therapeutically effective amount of the lonidamine derivatives can be configured so as to provide a concentration in or adjacent to a kidney cell of about 0.25 uM or more or less.
Owner:UNIVERSITY OF KANSAS
Apparatus and method for electromagnetic treatment
InactiveUS20130274540A1Enhanced transmitted dosimetryReduced Power RequirementsElectrotherapyMagnetotherapy using coils/electromagnetsEnergy variationMedicine
Described herein are electromagnetic treatment devices for treatment of tissue. In particular, described herein are lightweight, wearable, low-energy variations that are specifically configured to specifically and sufficiently apply energy within a specific bandpass of frequencies of a target biological pathway, such as the binding of Calcium to Calmodulin, and thereby regulate the pathway. Methods and systems for treating biological tissue are also described.
Owner:RIO GRANDE NEUROSCI
Methods for Enhancing the Quality of Life of a Senior Animal
Methods for enhancing the quality of life of a senior or super senior animal by feeding the animal a composition comprising at least one omega-3 polyunsaturated fatty acid and various combinations of amino acids, minerals, and antioxidants in amounts effective to enhance alertness, improve vitality, protect cartilage, maintain muscle mass, enhance digestibility, and improve skin and pelage quality. Changes in expression of genes associated with several biological pathways induced in an animal by feeding it said composition are consistent with an enhanced quality of life.
Owner:HILLS PET NUTRITION INC
Analysis of Transcriptomic Data Using Similarity Based Modeling
ActiveUS20110093244A1Small footprintFast data processingBiostatisticsComputation using non-denominational number representationBiomarker discoveryOrganism
An analytic apparatus and method is provided for diagnosis, prognosis and biomarker discovery using transcriptome data such as mRNA expression levels from microarrays, proteomic data, and metabolomic data. The invention provides for model-based analysis, especially using kernel-based models, and more particularly similarity-based models. Model-derived residuals advantageously provide a unique new tool for insights into disease mechanisms. Localization of models provides for improved model efficacy. The invention is capable of extracting useful information heretofore unavailable by other methods, relating to dynamics in cellular gene regulation, regulatory networks, biological pathways and metabolism.
Owner:PHYSIQ
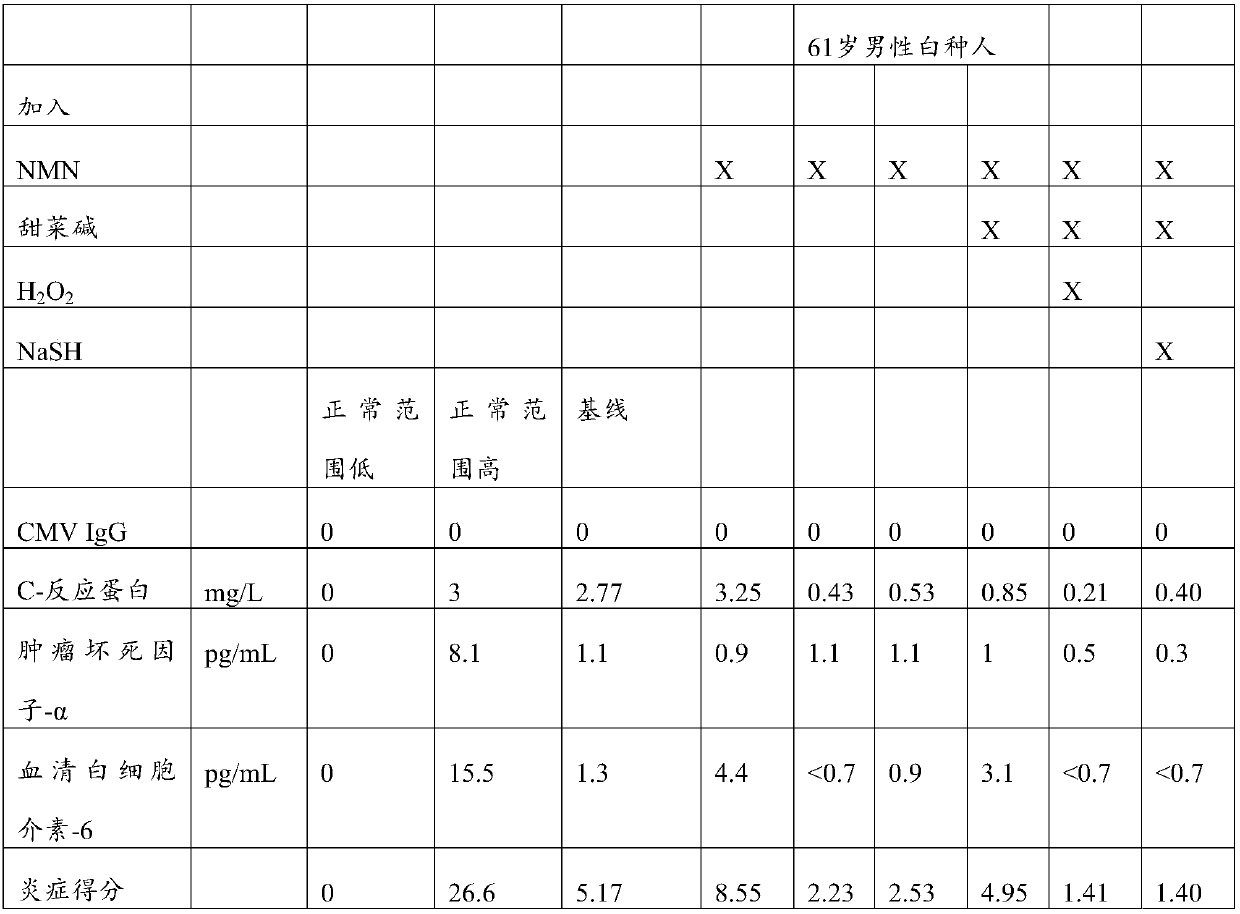
Resetting biological pathways for defending against and repairing deterioration from human aging
PendingCN108290059ADispersion deliveryHydroxy compound active ingredientsS-Adenosyl-l-methionineNicotinamide riboside
Compositions for addressing one or more of the effects of aging are described. The compositions comprise a first component comprising repair system activator(s) such as nicotinamide adenine dinucleotide (NAD+), nicotinamide mononucleotide (NMN), nicotinamide riboside (NR), nicotinic acid adenine mononucleotide (NaMN), nicotinic acid adenine dinucleotide (NaAD), nicotinic acid riboside (NAR), 1-methylnicotinamide (MNM), cyclic adenosine monophosphate (cAMP) and combinations thereof; a second component comprising methyl donor(s), such as S-5'-adenosyl-L-methionine (SAM), methionine, betaine, choline, folate, vitamin B12, or combinations thereof; and a third component comprising antioxidant defense activators, such as H2O2, N2S, NaSH, Na2S, and several others, including combinations thereof.Methods of administering the disclosed compositions or separate formulations of repair system activator, methyl donors, and antioxidant defense activators are also disclosed.
Owner:乔尔胡伊赞加
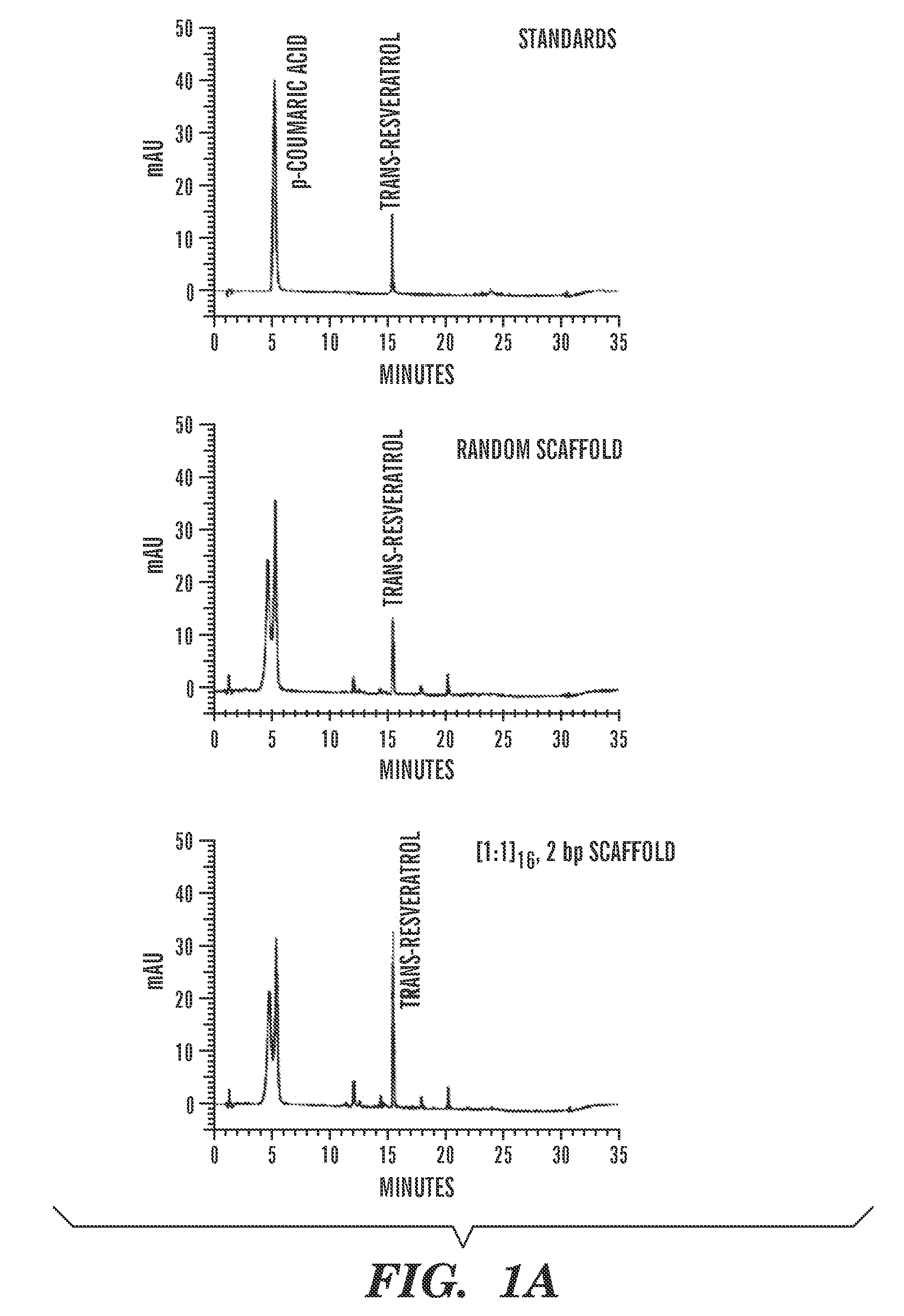
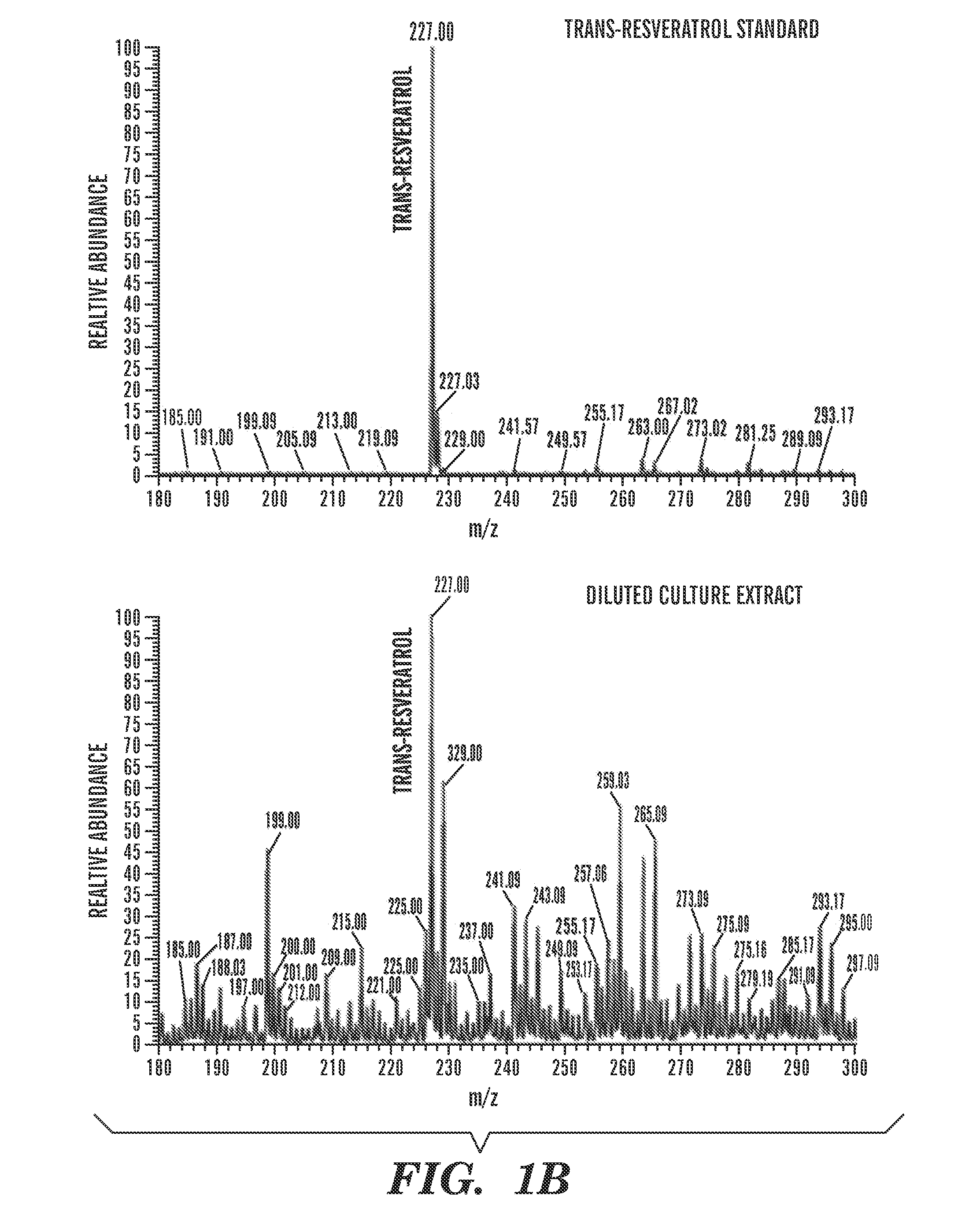
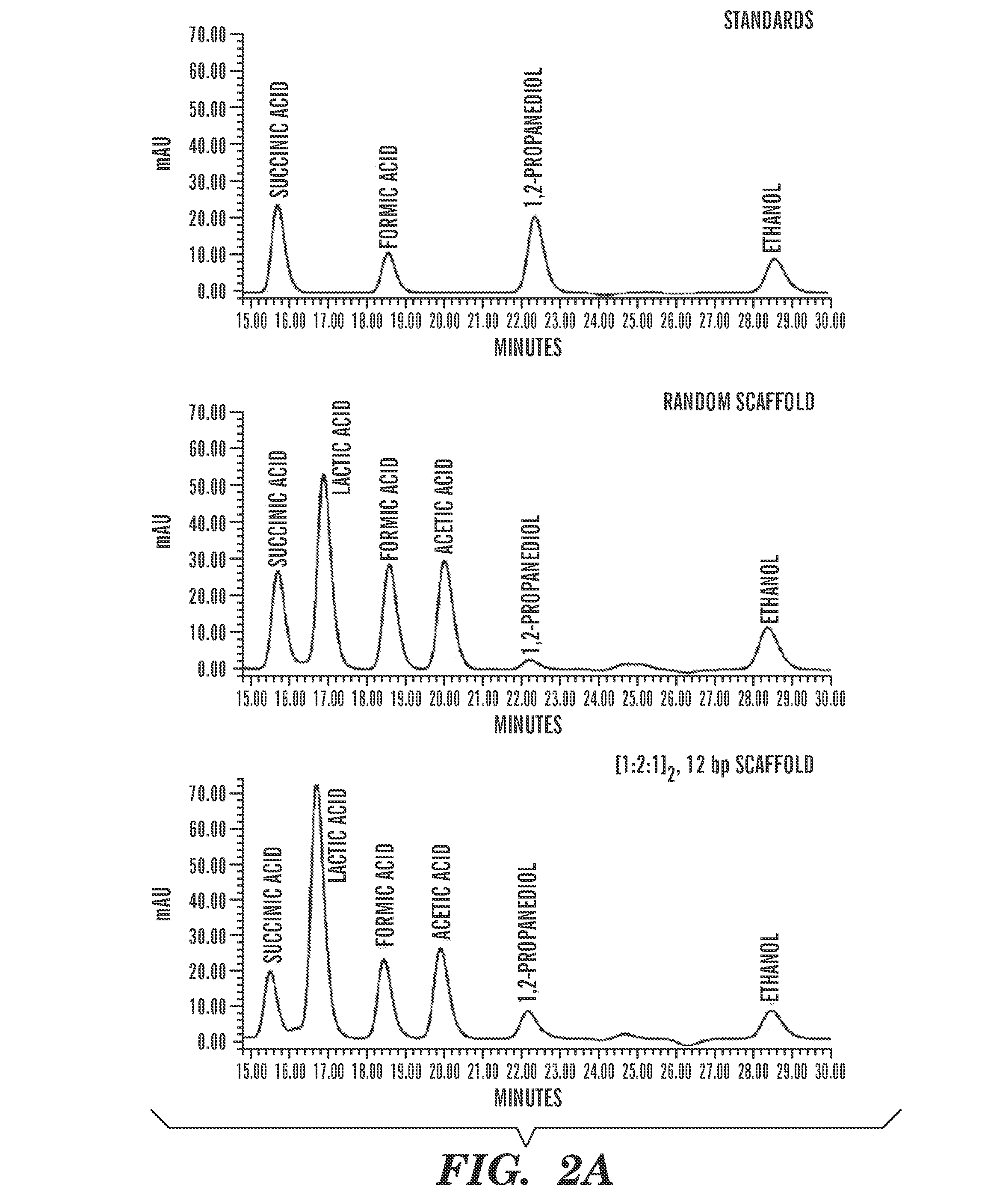
Constructs and methods for the assembly of biological pathways
The present invention is directed to a synthetic nucleic acid scaffold comprising one or more subunits, each subunit comprising two or more different protein-binding sequences coupled together. The present invention further relates to systems and methods for assembling a synthetic biological pathway and producing a biological pathway product or a precursor product using the synthetic nucleic acid scaffold.
Owner:CORNELL UNIVERSITY
Methods for enhancing the quality of life of a senior animal
Methods for enhancing the quality of life of a senior or super senior animal by feeding the animal a composition comprising at least one omega-3 polyunsaturated fatty acid and various combinations of amino acids, minerals, and antioxidants in amounts effective to enhance alertness, improve vitality, protect cartilage, maintain muscle mass, enhance digestibility, and improve skin and pelage quality. Changes in expression of genes associated with several biological pathways induced in an animal by feeding it said composition are consistent with an enhanced quality of life.
Owner:HILLS PET NUTRITION INC
An integrated method and system for identifying functional patient-specific somatic aberations using multi-omic cancer profiles
A system and method for determining the functional impact of somatic mutations and genomic aberrations on downstream cellular processes by integrating multi-omics measurements in cancer samples with community-curated biological pathways are disclosed. The method comprises the steps of extracting biological pathway information from well-curated biological pathway sources, using the pathway information to generate an upstream regulatory parent sub-network tree for each gene of interest, integrating measurement-based omic data for both cancer and normal samples to determine a nonlinear function for each gene expression level based on the gene's epigenetic information and regulatory network status, using the nonlinear function to predict gene expression levels and compare activation and consistency scores with inputted patient- specific gene expression data, and using the patient-specific gene expression predictions to identify significant deviations and inconsistencies in gene expressionlevels from expected levels in individual patient samples to identify potential biomarkers in providing predictive information in relation to cancer and cancer treatment.
Owner:KONINKLJIJKE PHILIPS NV +1
Method for analyzing biological networks
ActiveUS8068994B2Microbiological testing/measurementAnalogue computers for chemical processesDiseaseBiological pathway
Significance of biological pathway in disease state is predicted by (a) providing expression level data for a plurality of biomolecules differentially expressed in a disease state, compared with same biomolecules expressed in a non-diseased state: (b) determining presence probability of the biomolecules in disease state; (c) determining effect of each biomolecule from the plurality of biomolecules on the expression of different downstream biomolecules within pathway to provide perturbation factor for each biomolecule in the pathway; (d) combining statistical significance of differentially expressed biomolecules present in the disease state, with a sum of perturbation factors for all of the biomolecules, generating an impact factor; (e) calculating statistical significance of impact factor based upon determined probability of having statistical significant presence of differentially expressed biomolecules in step (b) and the sum of perturbation factors in step (c); and (f) outputting statistical significance of impact factor for the pathway relevant to the disease.
Owner:WAYNE STATE UNIV
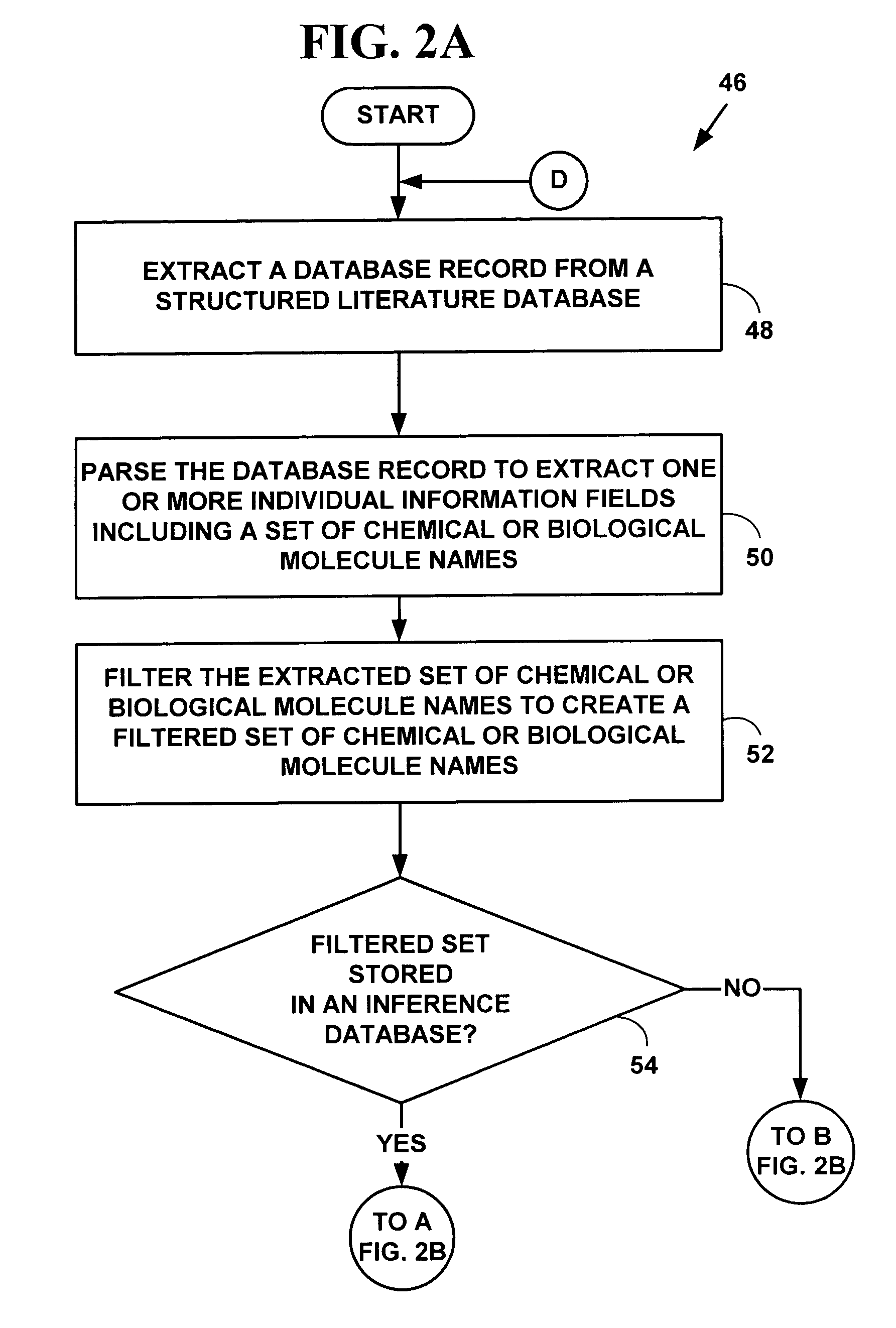
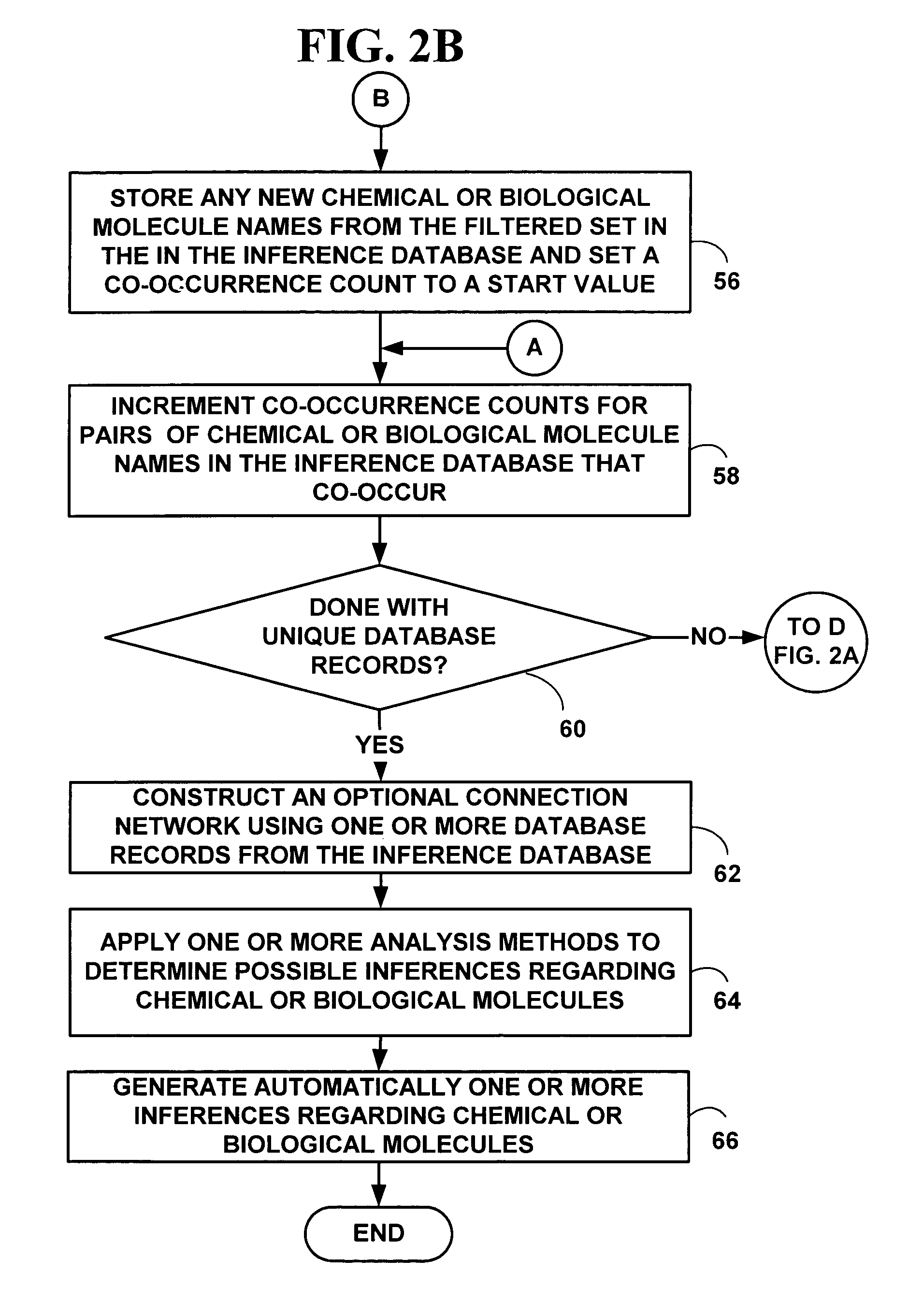
Method and system for automated inference creation of physico-chemical interaction knowledge from databases of co-occurrence data
InactiveUS7356416B2Easy to understandThe result is moreData processing applicationsDigital data processing detailsDiseaseCo-occurrence
Methods and system for automated inference of physico-chemical interaction knowledge from databases of term co-occurrence data. The co-occurrence data includes co-occurrences between chemical or biological molecules or co-occurrences between chemical or biological molecules and biological processes. Likelihood statistics are determined and applied to decide if co-occurrence data reflecting physico-chemical interactions is non-trivial. A next node or an unknown target representing chemical or biological molecules in a biological pathway is selected based on co-occurrence values. The method and system may be used to further facilitate a user's understanding of biological functions, such as cell functions, to design experiments more intelligently and to analyze experimental results more thoroughly. Specifically, the present invention may help drug discovery scientists select better targets for pharmaceutical intervention in the hope of curing diseases. The method and system may also help facilitate the abstraction of knowledge from information for biological experimental data and provide new bioinformatic techniques.
Owner:CELLOMICS
Method for estimation of information flow in biological networks
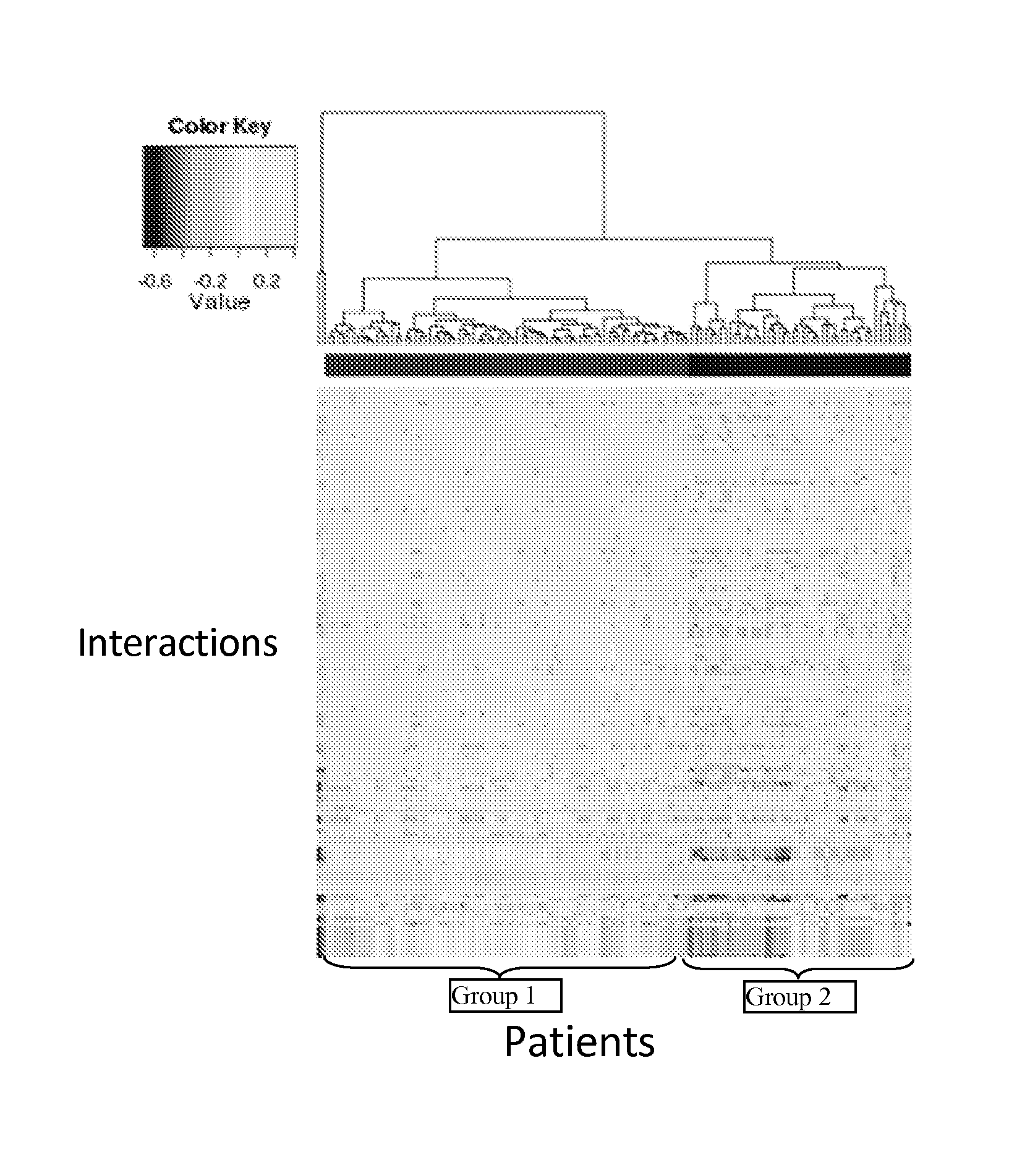
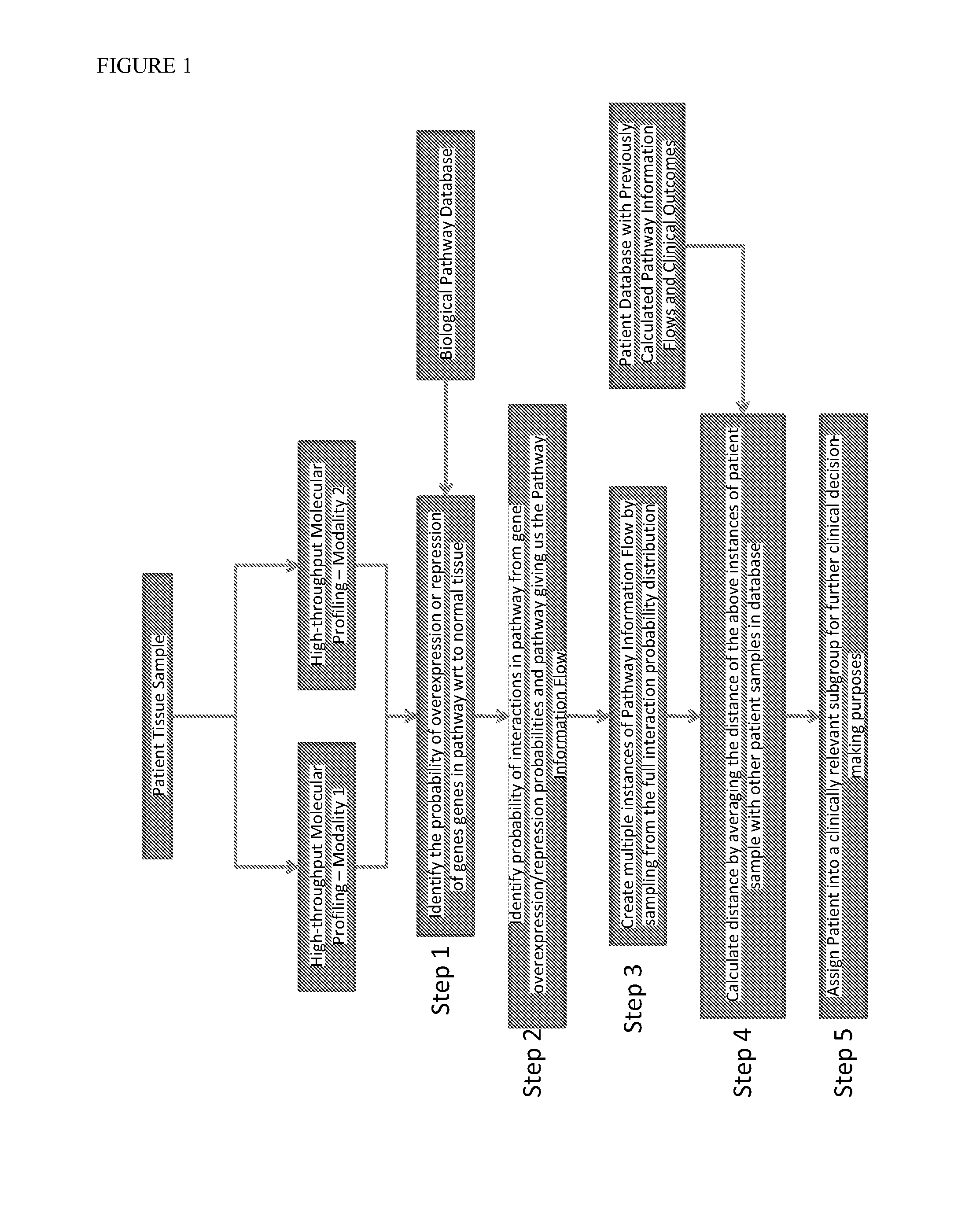
InactiveUS20140040264A1Stratify patients into clinically relevant groups very accuratelyAccurate layeringDigital data processing detailsBiostatisticsPatient databaseNetwork information flow
The present invention relates to a method for stratifying a patient into a clinically relevant group comprising the identification of the probability of an alteration within one or more sets of molecular data from a patient sample in comparison to a database of molecular data of known phenotypes, the inference of the activity of a biological network on the basis of the probabilities, the identification of a network information flow probability for the patient via the probability of interactions in the network, the creation of multiple instances of network information flow for the patient sample and the calculation of the distance of the patient from other subjects in a patient database using multiple instances of the network information flow. The invention further relates to a biomedical marker or group of biomedical markers associated with a high likelihood of responsiveness of a subject to a cancer therapy wherein the biomedical marker or group of biomedical markers comprises altered biological pathway markers, as well as to an assay for detecting, diagnosing, graduating, monitoring or prognosticating a medical condition, or for detecting, diagnosing, monitoring or prognosticating the responsiveness of a subject to a therapy against said medical condition, in particular ovarian cancer. Furthermore, a corresponding clinical decision support system is provided.
Owner:KONINKLJIJKE PHILIPS NV
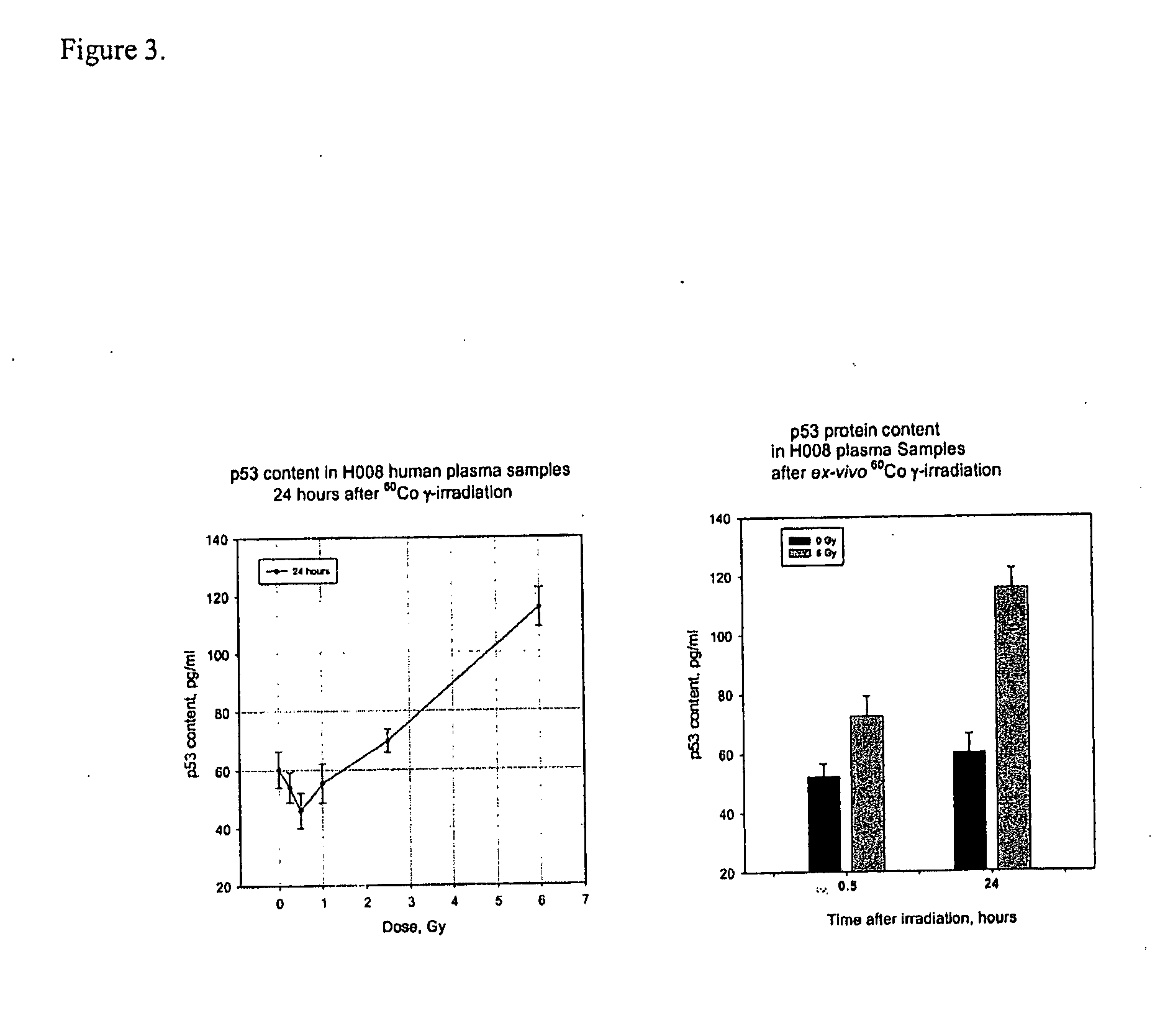
Biomarker panels for assessing radiation injury and exposure
Methods and kits are provided for assessing radiation injury and exposure in a subject. The methods comprise measuring the levels of at least two (2) protein biomarkers from different biological pathways and correlating the levels with an assessment of radiation injury and exposure. Additional use of peripheral blood cell counts and serum enzyme biomarkers, evaluated in the early time frame after a suspected radiation exposure, and use of integrated multiple parameter triage tools to enhance radiation exposure discrimination and assessment are also provided. The information obtained from such methods can be used by a clinician to accurately assess the extent of radiation injury / exposure in the subject, and thus will provide a valuable tool for determining treatment protocols on a subject by subject basis.
Owner:THE HENRY M JACKSON FOUND FOR THE ADVANCEMENT OF MILITARY MEDICINE INC
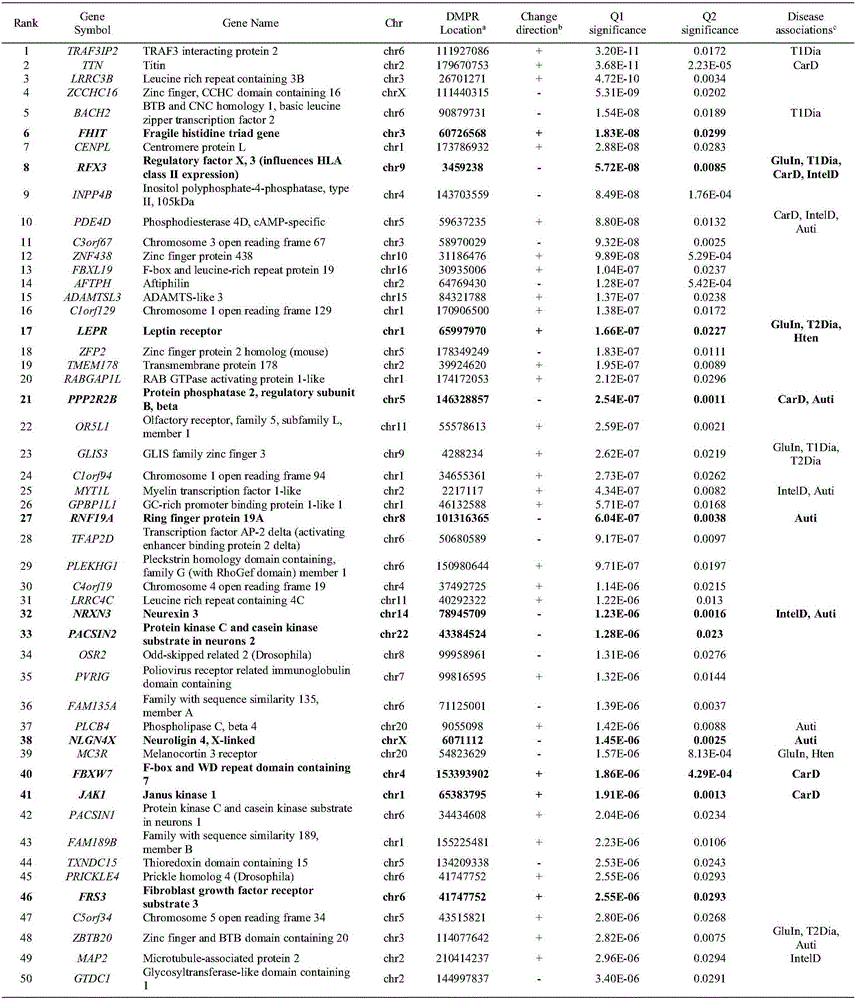
Gene function association network-based method for discovering chronic disease mechanism and early warning intervention policy thereof
ActiveCN106126893AShow validityEpigenetic far-reachingHealth-index calculationProteomicsDisease riskCrowds
The present invention discloses a gene function association network-based method for discovering a chronic disease mechanism and an early warning intervention policy thereof. The method can be applied to the field of related research and application of biology, medicine and pharmacy. Epigenetic changes of the high-risk groups surveyed and discovered according to epidemiology before disease attack are detected, the epigenetic changes before the disease is associated with indicator gene changes of the disease through gene function network analysis, then a long-term molecular mechanism contributing to the disease risk is discovered, and risk early warning for the group, risky diagnosis for an individual, and indicators and methods for preventive intervention are provided. According to the method, a long-term steady biological path or biological function change may leave a mark on related gene epigenetic inheritance, and a chronic disease biological path or biological function level mechanism may be found through analysis of the synergistic effect of epigenetic differences.
Owner:ZHEJIANG UNIV
Method for determining the effects of external stimuli on biological pathways in living cells
InactiveUS20080280777A1Microbiological testing/measurementLibrary screeningInformation processingImaging analysis
The present invention describes methods for carrying out experiments on living cells, including making measurements of the operation transcriptional regulatory processes and indicators of the kinds of processes operating in the cell in response to external stimuli. Image analysis allows for gathering data concerning the flow of information through a cell's genomic regulatory network as it is executing a programmatic change in its activities as a function of said stimuli. The method also allows collection of data of the results of the information-processing in the cell by observing the decisions the cell makes when modulating cellular process activities.
Owner:TRANSLATIONAL GENOMICS RESEARCH INSTITUTE
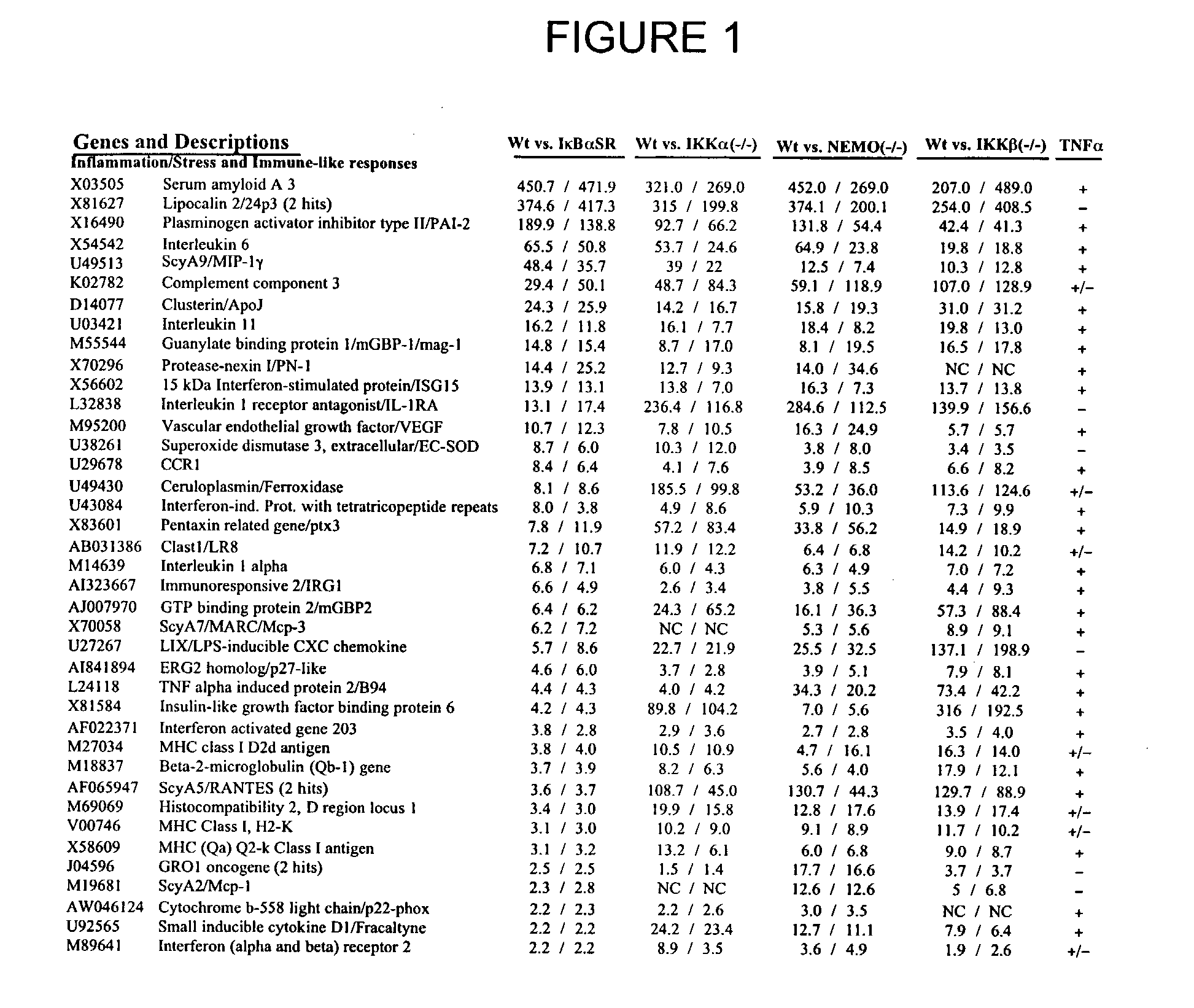
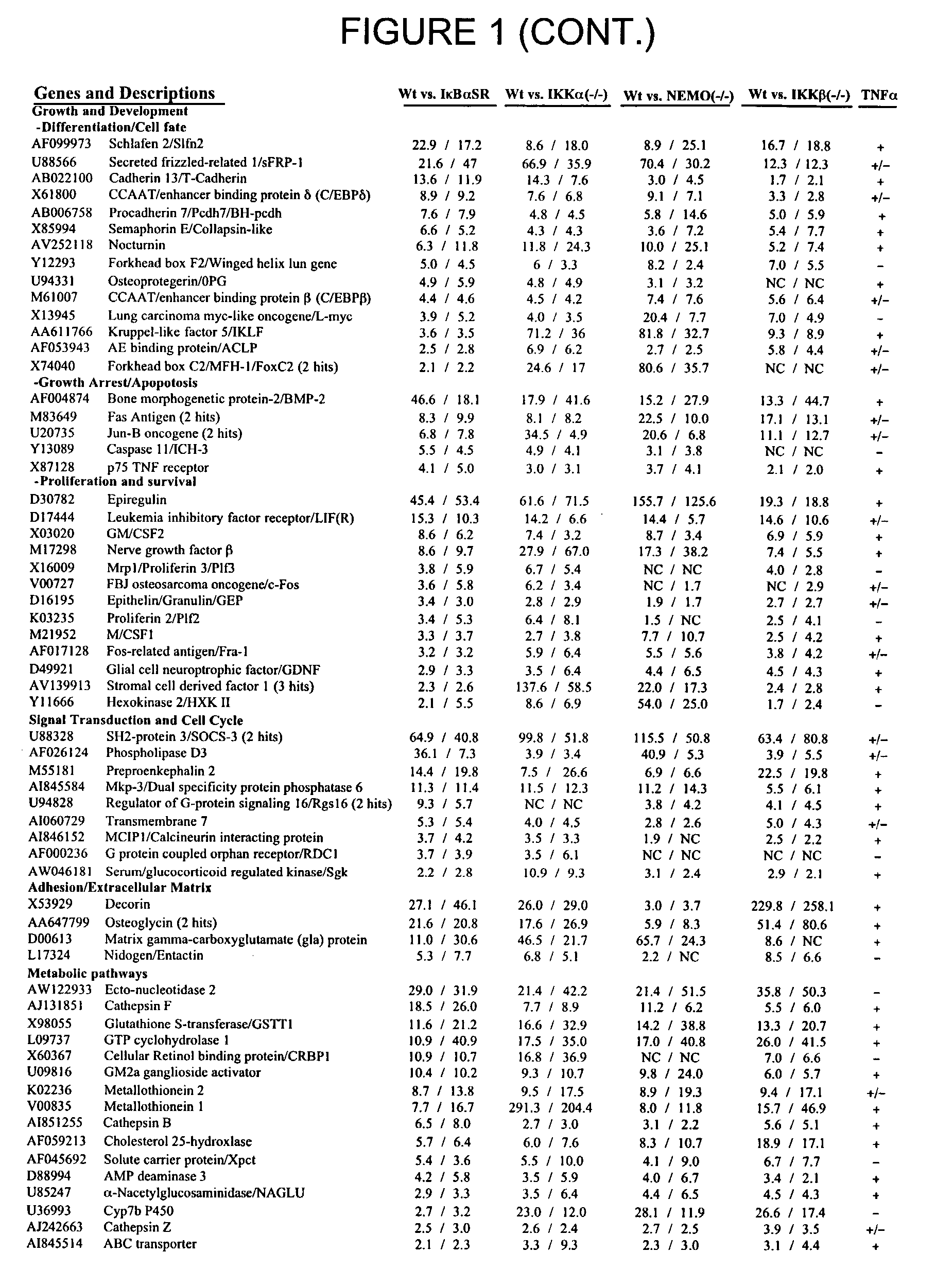
Methods for the Identification of IKKalpha Function and other Genes Useful for Treatment of Inflammatory Diseases
InactiveUS20070128648A1Nervous disorderPeptide/protein ingredientsBiological pathwayGene expression level
The invention provides a method for identifying genes involved in the NF-κB pathway comprised of the steps of determining the level of expression of a gene in an experimental sample obtained from the cells having deficient levels of a component of the NF-κB pathway; determining the level of expression of said gene in a control sample obtained from wild type cells having levels of a component of a biological pathway; selecting genes having a level of expression that are modulated in said experimental sample relative to said wild type sample. The invention also provides a method of treating inflammatory related diseases by modulating the activity of IKKα.
Owner:LI JUN +5
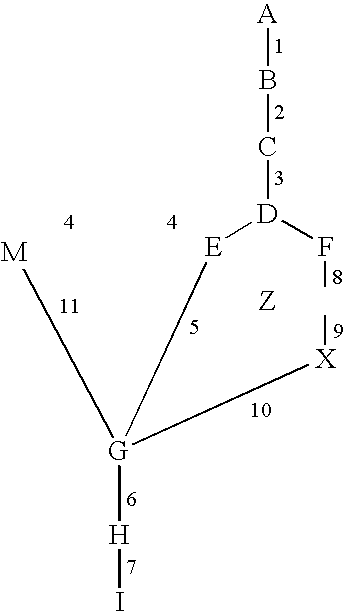
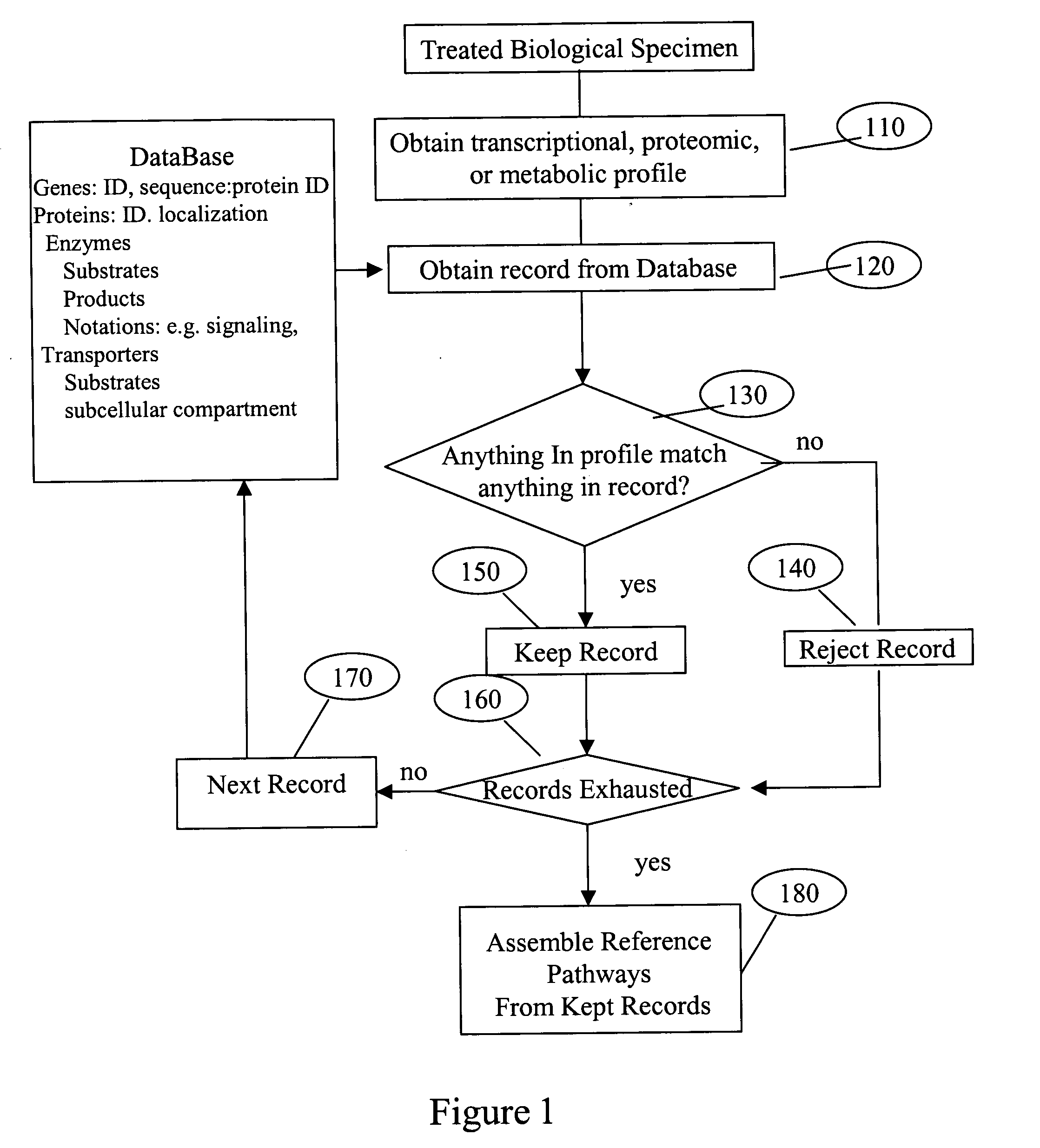
System and process of determining a biological pathway based on a treatment of a biological specimen
InactiveUS20050260615A1Rapid identificationMicrobiological testing/measurementBiological testingMetabolomeOrganism
Provided herein is a system and method for analyzing microarray data (transcriptome profiles), metabolite data (metabolome profiles), protein level data (proteome profiles), or any combination thereof to determine the biochemical pathways affected by a treatment. The system and method can be used to generate biochemical pathway information for any organism for which metabolic profile data can be obtained. The system and method may allow users to query the pathways generated and to filter the results of queries based on the -omic profile data, pathways involving molecules of interest, notions of biochemical importance, milestone molecules, and other factors. The system and method may also be suitable for discovery of regulatory sequences in genes. In an exemplary use, the system and method can be utilized to identify three genes involved in “de novo” ammonia “biosynthesis” that are induced by light, and was able to identify a putative cis-element, GWTTGTGG, that is likely involved in the regulation of those genes.
Owner:NEW YORK UNIV
Methods for the identification of IKKalpha function and other genes useful for treatment of imflammatory diseases
The invention provides a method for identifying genes involved in the NF-kappaB pathway comprised of the steps of determining the level of expression of a gene in an experimental sample obtained from the cells having deficient levels of a component of the NF-kappaB pathway; determining the level of expression of said gene in a control sample obtained from wild type cells having levels of a component of a biological pathway; selecting genes having a level of expression that are modulated in said experimental sample relative to said wild type sample. The invention also provides a method of treating inflammatory related diseases by modulating the activity of IKKalpha.
Owner:BOEHRINGER INGELHEIM PHARMA INC
Information management system for biochemical information
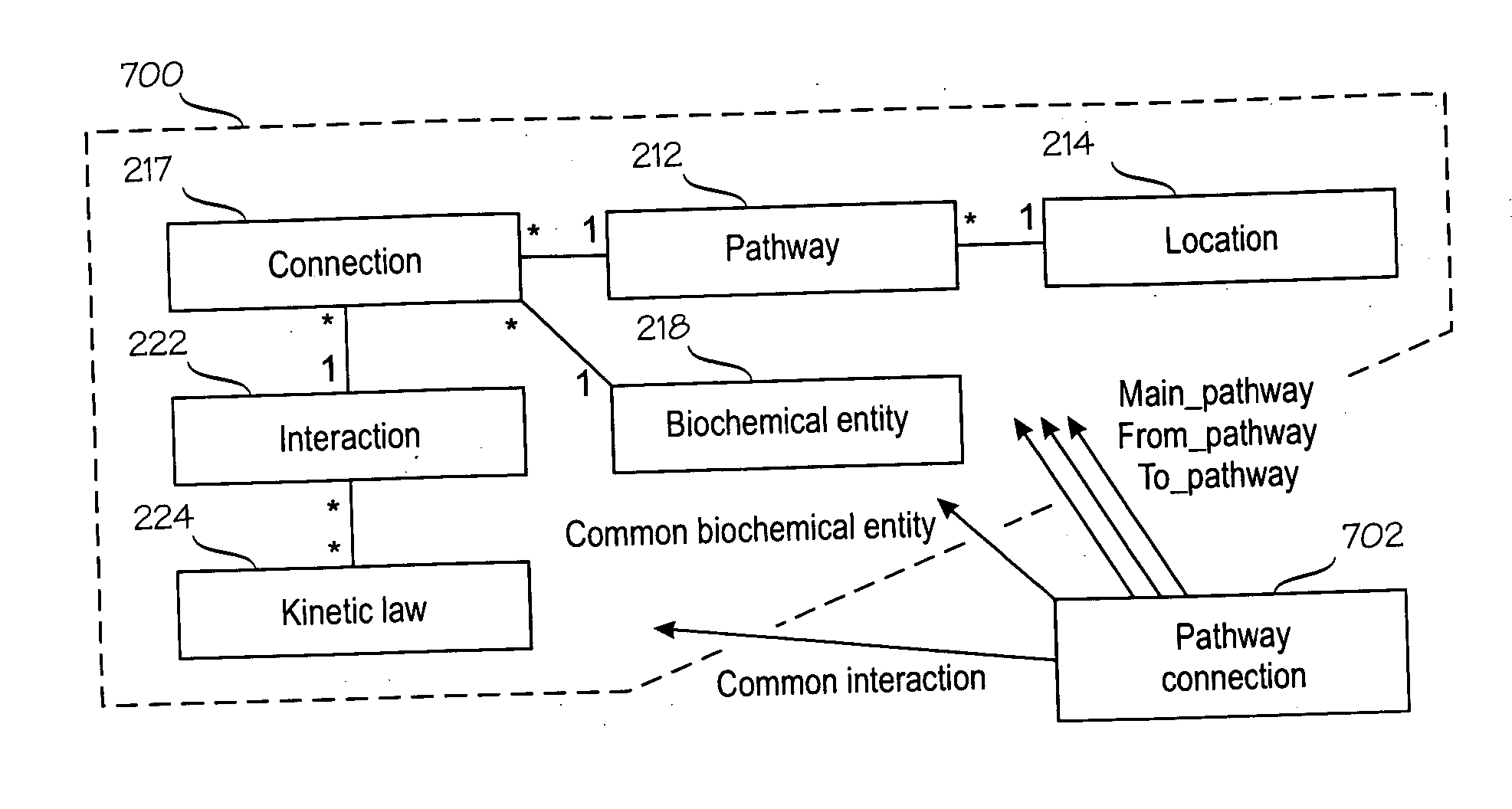
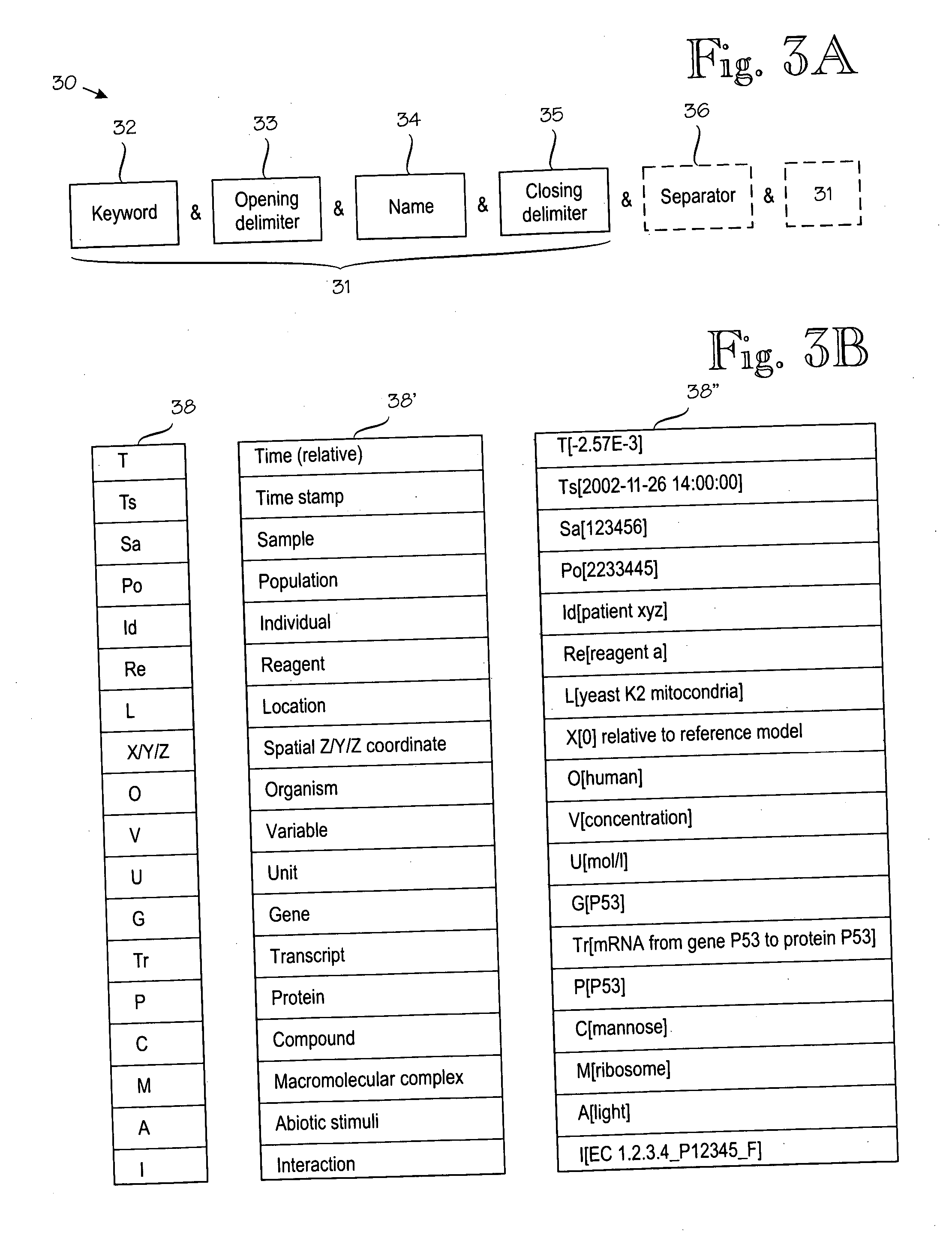
An information management system for managing biological information (200). The information management system comprises by structured descriptions of biological pathways (700) that are formed of at least pathways (212), biochemical entities (218), connections (216) and interactions (222), such that each pathway (212) relates to one or more connections (216); each connection (216) joins one biochemical entity (218) and one interaction (222); and each pathway (212) relates to a specific location (214).
Owner:MEDICEL
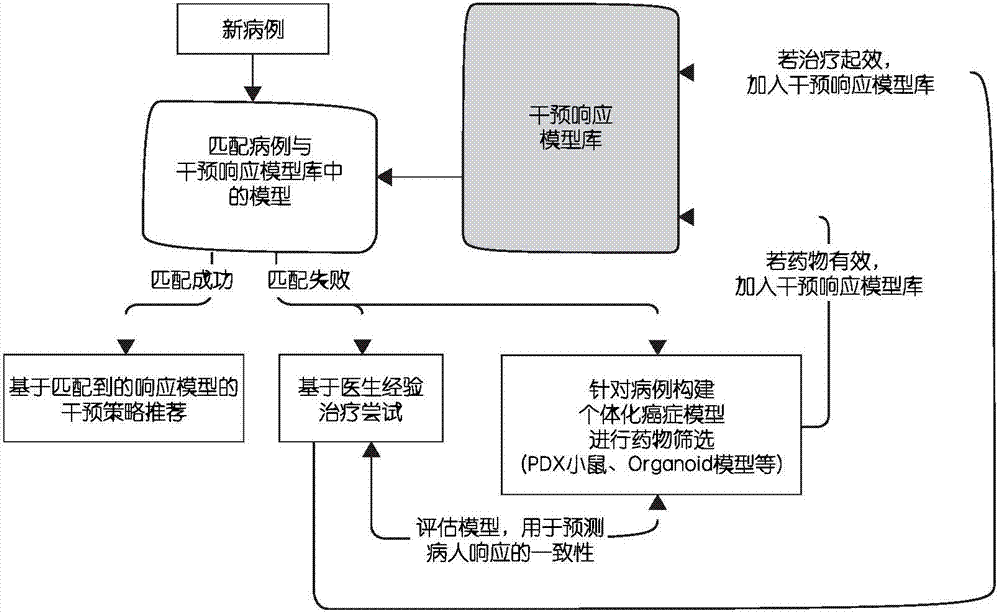
Cancer individuation intervention strategy selection method based on molecular physiome
InactiveCN106897549ASimplify the diagnostic cycleImprove follow-up intervention responseMedical data miningMedical automated diagnosisResponse effectMolecular level
The invention discloses a cancer individuation intervention strategy selection method based on molecular physiome. The method comprises the following steps that: establishing an intervention response model library; when a new case appears, comparing the new case with the pre-administration proteomics difference of a model; and analyzing biological approaches / physiological function differences and the like which may be caused by the difference. A sample data type on which the method depends has high dependency with a physiological level phenotype. Proteomics data related to cell physiology and functions including protein and the like can be transcribed and subjected to epigenetic inheritance. The method is more sensitive to the intervention response effect of diseases, can provide rich indexes to observe molecular level physiological changes caused by diseases, and is suitable for clinic transformation and practical popularization.
Owner:ZHEJIANG UNIV
Analysis of transcriptomic data using similarity based modeling
ActiveUS8515680B2Small footprintFast data processingSugar derivativesDigital data processing detailsBiomarker discoveryModelling analysis
An analytic apparatus and method is provided for diagnosis, prognosis and biomarker discovery using transcriptome data such as mRNA expression levels from microarrays, proteomic data, and metabolomic data. The invention provides for model-based analysis, especially using kernel-based models, and more particularly similarity-based models. Model-derived residuals advantageously provide a unique new tool for insights into disease mechanisms. Localization of models provides for improved model efficacy. The invention is capable of extracting useful information heretofore unavailable by other methods, relating to dynamics in cellular gene regulation, regulatory networks, biological pathways and metabolism.
Owner:PROLAIO INC
Agents, compositions and methods for treating pathologies in which regulating an ache-associated biological pathway is beneficial
The present invention provides agents which are capable of regulating the function of a micro-RNA component which can be used to regulate an AChE-associated biological pathway. In addition, the present invention provides methods and pharmaceutical compositions for the treatment of various pathologies related to AChE-associated biological pathways such as apoptosis, aberrant cholinergic signaling, abnormal hematopoietic proliferation and / or differentiation, cellular stress, exposure to inflammatory response-inducing agents, and / or exposure to organophosphates or other AChE inhibitors.
Owner:YISSUM RES DEV CO OF THE HEBREWUNIVERSITY OF JERUSALEM LTD
Computer systems for identifying pathways of drug action
InactiveUS6859735B1Minimizes valueLess-robust and reliable resultMicrobiological testing/measurementAnalogue computers for chemical processesPharmaceutical drugBiological pathway
The present invention provides methods and computer systems for identifying and representing the biological pathways of drug action on a cell The present invention also provides methods and computer systems for assessing the significance of the identified representation and for verifying that the identified pathways are actual pathway of drug action. The present invention also provides methods and computer systems for drug development based on the methods for identifying biological pathways of drug action, and methods and computer systems for representing the biological pathways involved in the effect of an environmental change upon a cell.
Owner:MICROSOFT TECH LICENSING LLC +1
Gene for constructing 5-aminolevulinic acid C4 biosynthesis pathway in Escherichia coli and construction method thereof
InactiveCN103146694AEasy to synthesizeIncrease productionBacteriaFermentationEscherichia coliSuccinic acid
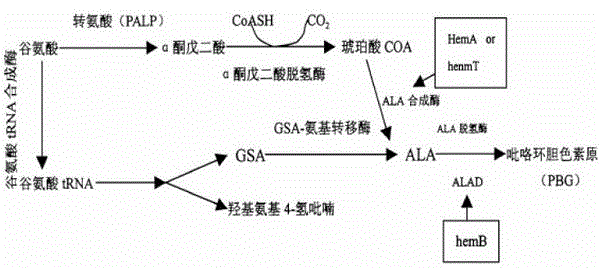
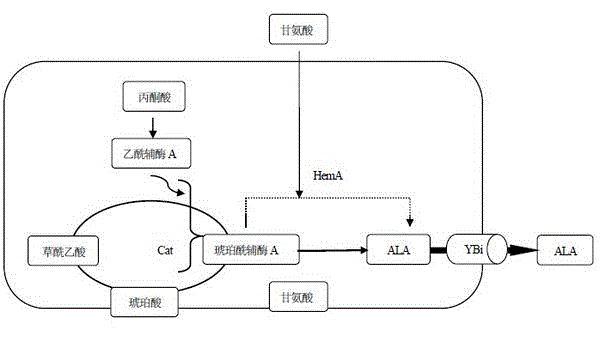
The invention discloses a gene for constructing a 5-aminolevulinic acid C4 biosynthesis pathway in Escherichia coli and a construction method thereof. By analyzing the problems of rate-limiting steps, intermediate supply, product secretion and the like of the 5-aminolevulinic acid C4 biosynthesis pathway, a Rhodobactersphaeroides-derived ALA synthetase gene hemA, a Propionibacterium shermanil-derived coenzyme A transferase gene (Cat) and an Escherichia coli-derived ALA secretor gene (YBi) are cloned; by the aid of a pETDuet-1 co-expression vector, the gene hemA for synthesizing the 5-aminolevulinic acid, the gene Cat for generating the intermediate succinic acid COA for synthesizing the 5-aminolevulinic acid, and the gene Ybi for promoting secretion of the 5-aminolevulinic acid are co-expressed; and the co-expression of the three genes reinforces the biosynthesis of the 5-aminolevulinic acid. The gene with C4 biological pathway enhances the 5-aminolevulinic acid yield of Escherichia coli, and the ALA secreted out of the cells reaches 1124 mg / L.
Owner:HEBEI UNIVERSITY OF SCIENCE AND TECHNOLOGY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com