Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
50 results about "12s rrna" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
In humans, 12S is encoded by the MT-RNR1 gene and is 959 nucleotides long. MT-RNR1 is one of the 37 genes contained in animal mitochondria genomes. Their 2 rRNA, 22 tRNA and 13 mRNA genes are very useful in phylogenetic studies, in particular the 12S and 16S rRNAs. The 12S rRNA is the mitochondrial homologue of the prokaryotic 16S...
Method for quickly identifying categories of meat and dried meat products of five domestic animals
The invention relates to a method for identifying categories of meat and dried meat products of five domestic animals (namely a pig, a goat, an ox, a buffalo and a yak), which is based on genetic variation of a 440bp gene segment of a chondriosome 12S rRNA gene, adopts a PCR-restriction fragment length polymorphism (PCR-RFLP) technique, and belongs to the field of biological high technology. The identification method of the invention comprises the following main steps: genomic DNA extraction of a sample, PCR amplification, restriction endonuclease digestion, endonuclease product detection and sample identification. The identification method of the invention has the characteristics of quickness, simpleness, economy and accuracy, which can be applied to massive detection of fresh meat and commercialized dried meat samples. The establishment of the method has important significance in the aspects of animal food inspection and quarantine, business fraud prevention and food backtracking system creation.
Owner:KUNMING INST OF ZOOLOGY CHINESE ACAD OF SCI
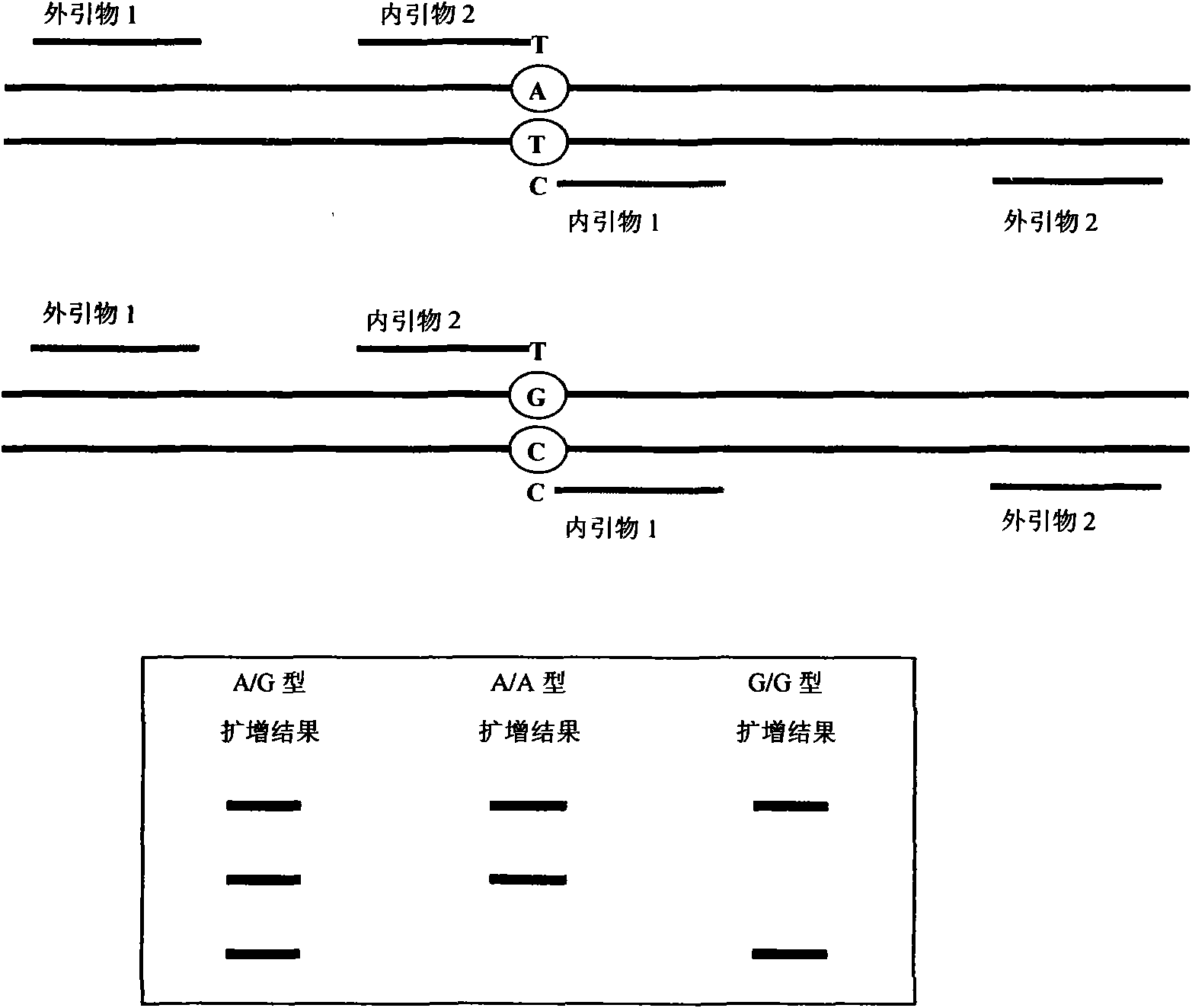
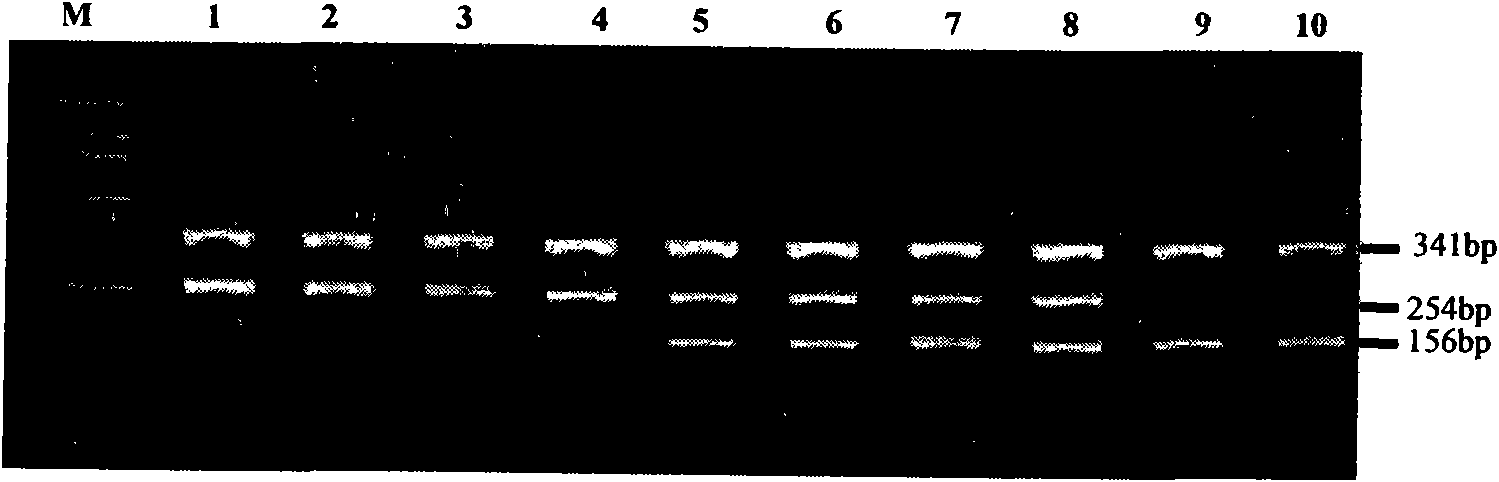
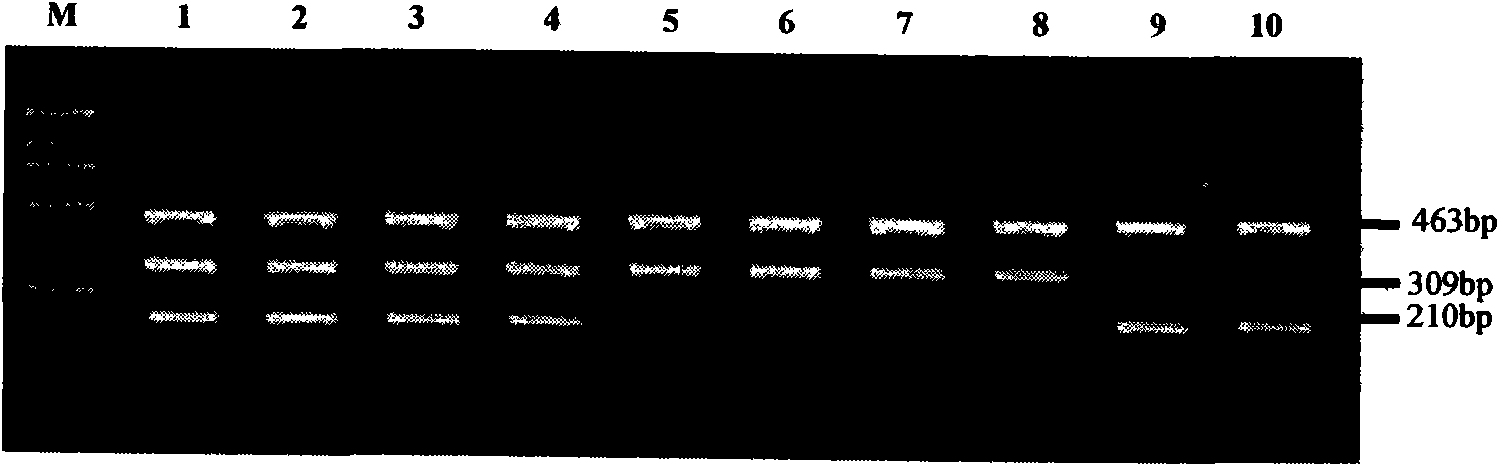
Method for detecting 20 mutation sites of deaf genes
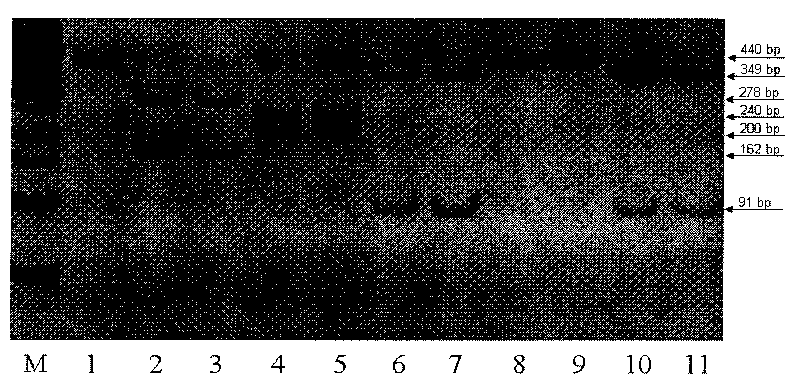
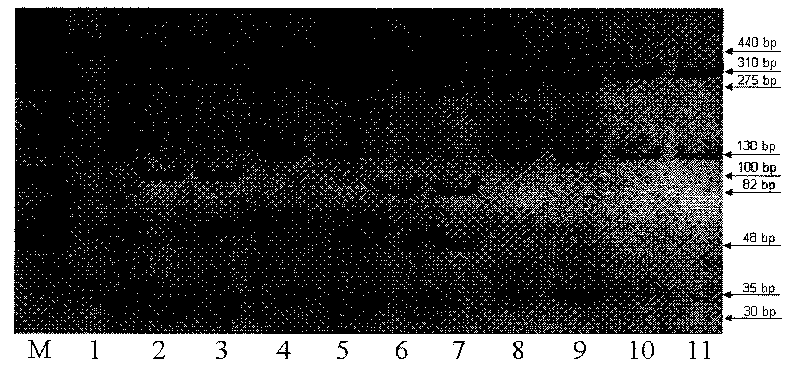
The invention relates to a method for detecting 20 hot mutation sites of four deaf genes such as GJB2, SLC26A4, GJB3 and 12S rRNA in a clinical sample, designing amplimers and single base extension primers of different sites aiming at abnormal SNP gene sites of the four genes by a matrix-assistant laser desorption / ionization flight time mass spectrum detection method and then carrying out mass spectrometry of a specific SNP site genotype.
Owner:GUANGZHOU DARUI BIOTECH +1
Deafness susceptibility gene screen test kit
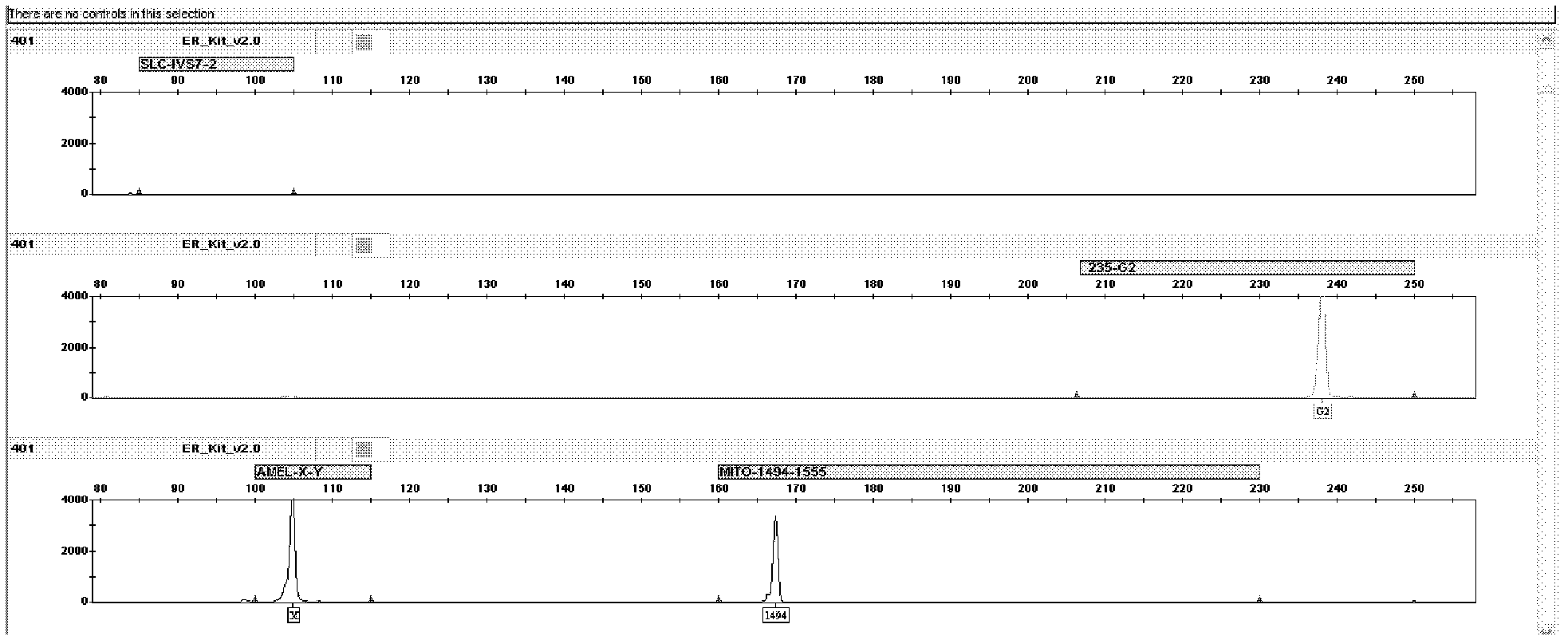
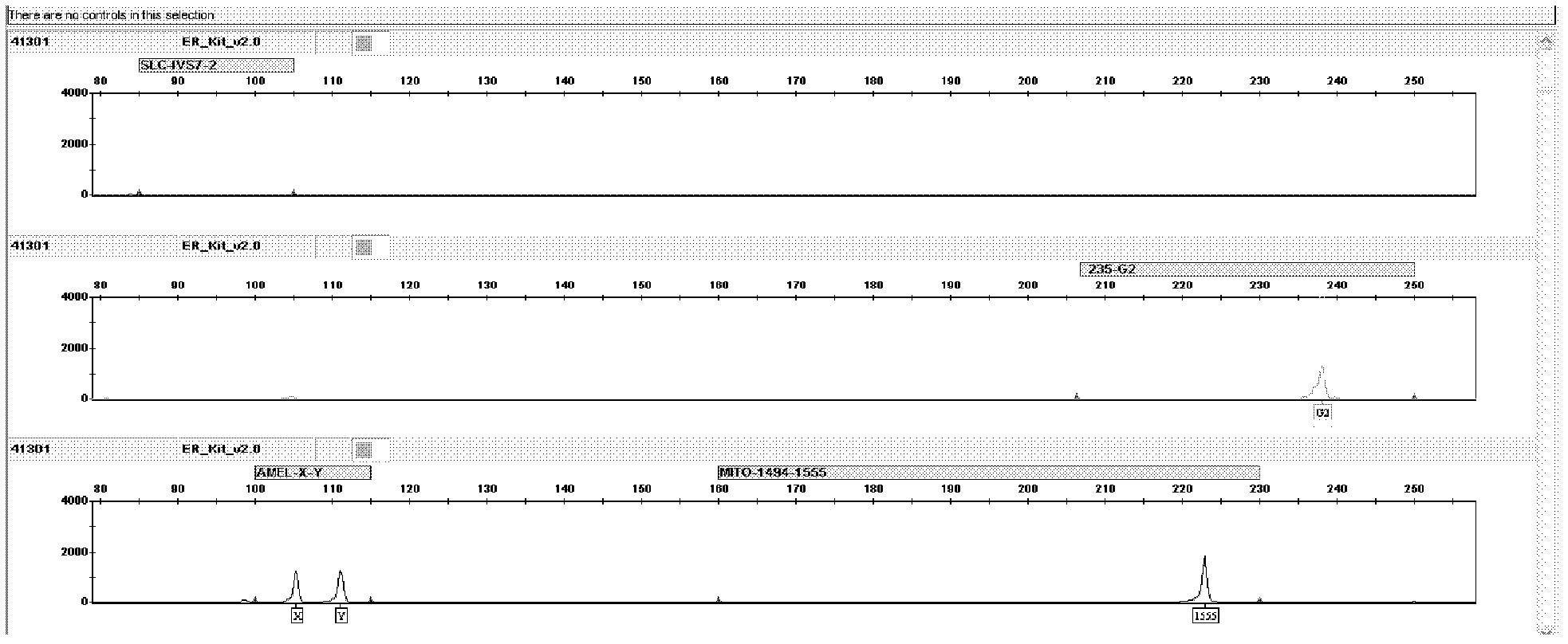
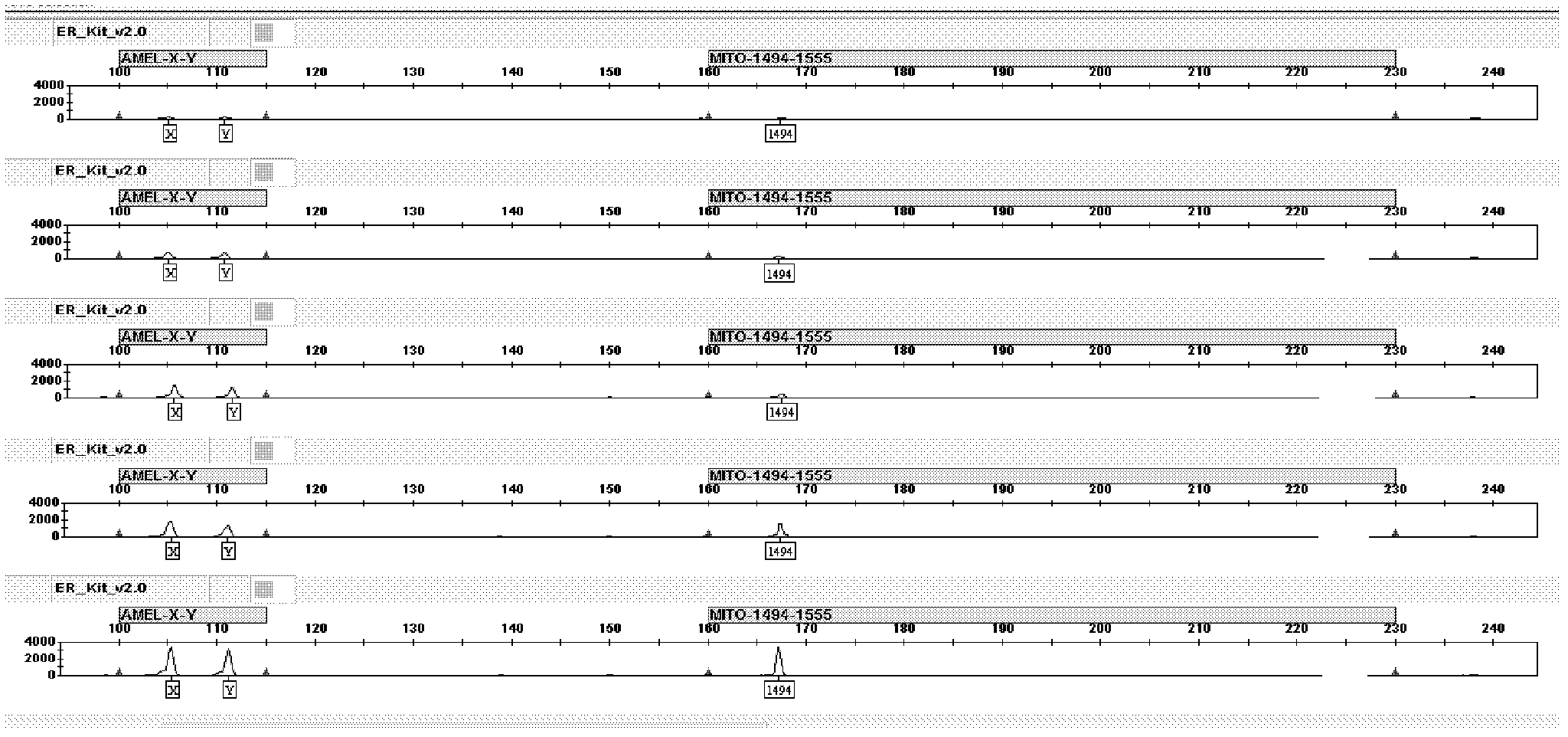
The invention relates to a hereditable deafness susceptibility gene screen test kit. The kit uses mutation from C to T of 1,494 locus of a 12s rRNA gene, mutation from A to G of 1,555 locus of the 12srRNA gene, mutation from A to G of IVS7(-2) locus of an SLC26A4 gene and C(+ / -) of 235 locus of a GJB2 gene as detecting objects, designs and optimizes a set of specific primers respectively againsteach locus to be tested according to the PCR technical principle of a tetra-primer amplification refractory mutation system, amplifies the whole set of the primers in the same reaction tube, and performs primary multiple PCR amplification and primary gel electrophoresis on the four reaction tubes to obtain gene types of four loci simultaneously.
Owner:GENERAL HOSPITAL OF PLA
Kit for detecting hereditary hearing loss
ActiveCN102373265AMicrobiological testing/measurementDNA/RNA fragmentationWild typeMolecular diagnostics
The invention brings forward a kit for detecting hereditary hearing loss. The kit provided by the invention comprises a primer combination and a probe combination of nine hereditary hearing loss polymorphic sites on GJB2(Cx26) gene, SLC26A4(PDS) gene and 12S rRNA(MTRNR1) gene, and can be used to accurately determine the wild type, homozygous mutation or heterzygosity of related sites and realize molecular diagnosis of hearing loss. With the application of the kit provided by the invention, detection can be carried out by cheap equipment or macroscopic observation. The primer combination, reagent panel and kit provided by the invention can be used for the screening of hereditary hearing loss to realize early detection, early prevention and prenatal guidance. The invention is of great commercial value and social significance.
Owner:BOAO BIOLOGICAL CO LTD +1
Marine fish mitochondrion 12S rRNA (ribosomal ribonucleic acid) gene amplification primer and design and amplification method thereof
InactiveCN102912012AAmplify high efficiency and specificityImprove developmentMicrobiological testing/measurementDNA preparationBiotechnologyNucleotide
The invention belongs to the field of marine fish mitochondrial genome research, and particularly relates to a marine fish mitochondrion 12S rRNA (ribosomal ribonucleic acid) gene amplification primer. The primer is composed of two single-chain oligonucleotide chains, wherein a light chain primer is a nucleotide sequence shown as SEQ ID NO.1, and a heavy chain primer is a nucleotide sequence shown as SEQ ID NO.2. The invention also provides a design method of the amplification primer and a method for amplifying a marine fish DNA (deoxyribonucleic acid) solution by using the amplification primer. The invention can efficiently and specifically amplify multiple marine fish mitochondrion 12S rRNA genes and can be used for the analytical study on the system evolution of different taxonomic categories of fishes, thereby providing a powerful tool for fish species identification, germplasm resource survey and system evolution research.
Owner:ZHEJIANG OCEAN UNIV
High variation zone amplication primer of rockfish mitochondrial genome and its design method
InactiveCN1900318AAccurate identificationMicrobiological testing/measurementConserved sequenceLong pcr
The present invention relates to one pair of PCR primers named as G-dloop, and provides one pair of amplification primers capable of amplifying the high variation zone of rockfish mitochondrial genome in high efficiency and its design method. The amplification primers consist of two single strand oligonucleotides. Through logging-on Genebank to search vertebrate mitochondrion DNA cytochrome b gene and the high conservation area of 16S rRNA gene sequence and homologous comparison, one pair of amplification primers is obtained. Through long PCR amplification, the target segments of 32 varieties of rockfish are obtained and sequenced. The obtained sequences are compared by means of using homologous comparison software Clustal X 1.83 to fine the conservation sequences in the cytochrome b5' ends and the 12S rRNA 3' ends of the mitochondrial genomes of the 32 varieties of rockfish, and the said amplification primers are designed based on the degeneracy principle.
Owner:XIAMEN UNIV
Library building kit for detecting hereditary deafness genes and application
InactiveCN107287314AReduce testing costsEnable high-throughput detectionMicrobiological testing/measurementLibrary creationMultiplexMedicine
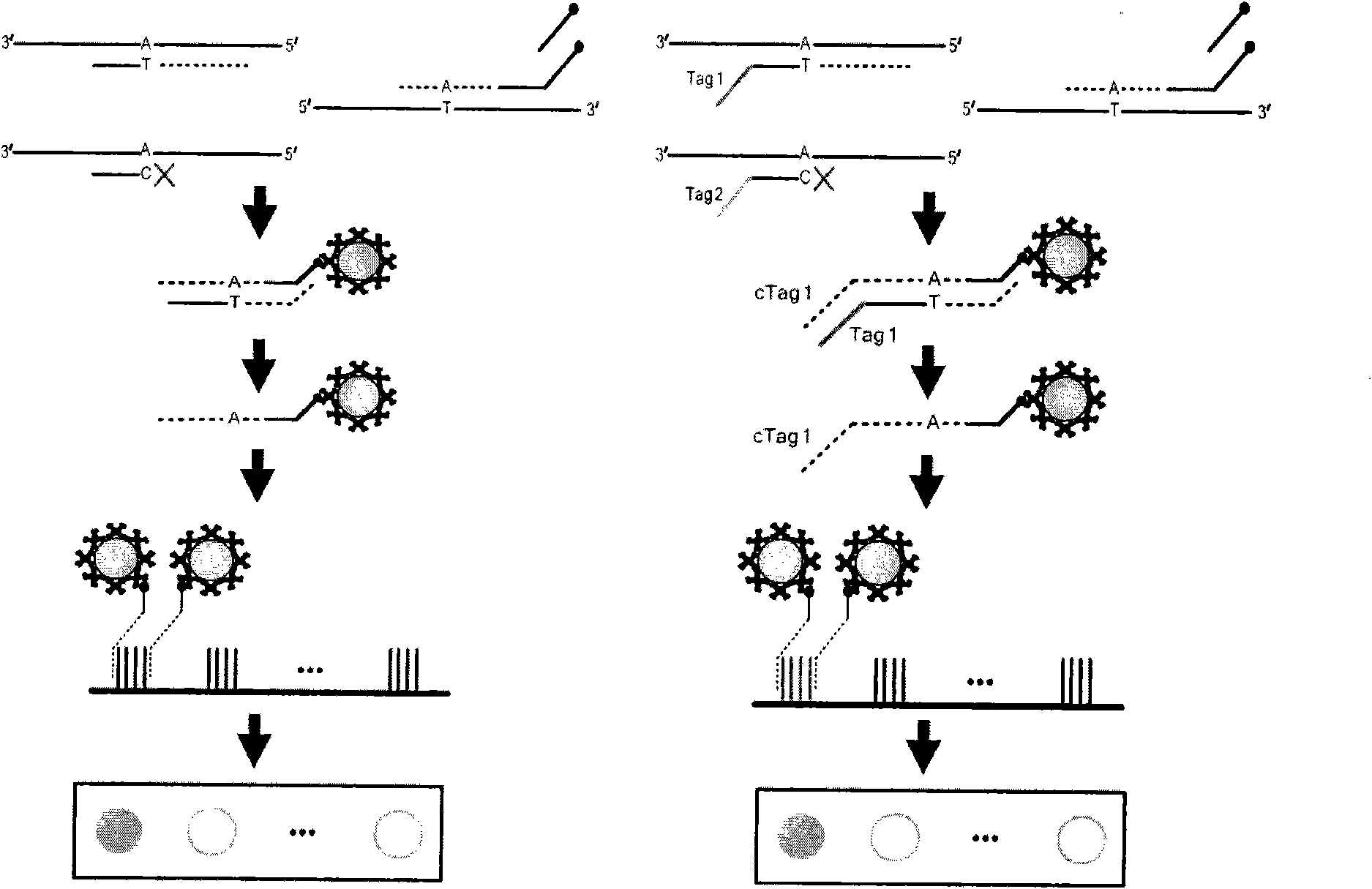
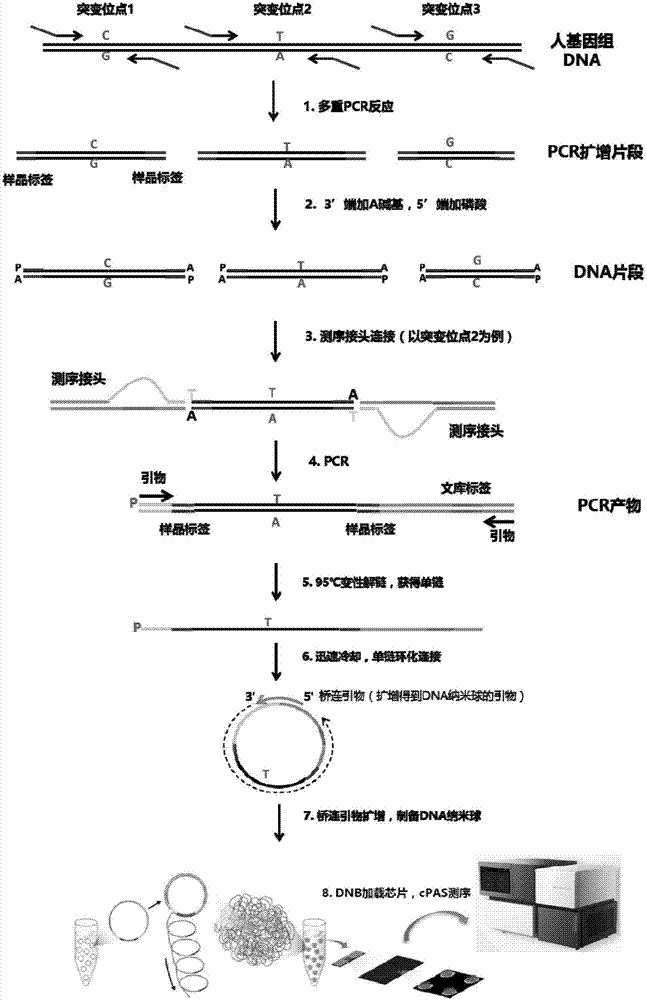
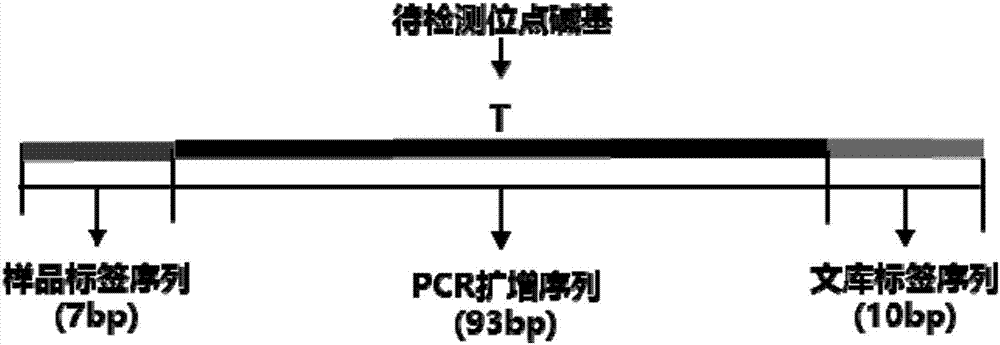
The invention relates to a kit for building a library for detecting hereditary deafness genes and its application. The kit includes an amplification primer set for amplifying hereditary deafness genes; Primers designed for the polymorphism of the gene and the mutation site of the 12s rRNA gene. The present invention adopts multiplex PCR technology, which can detect more than 20 mutation sites of deafness genes in a single reaction; in conjunction with the "double label" system, the DNA amplification product of each sample has two sets of independent label sequences, so The amplification products of all samples can be mixed and sequenced simultaneously to achieve high-throughput detection, thereby greatly reducing the detection cost of a single sample.
Owner:MGI TECH CO LTD +1
Reagent kit for extracting DNA in Chinese alligator chorion film and use method thereof
InactiveCN101368179AHigh purityIncrease concentrationMicrobiological testing/measurementDNA preparationHigh concentrationChinese alligator
The invention discloses a kit for extracting DNA in the egg shell membrane of Yangtze alligator and a use method thereof, and the kit comprises the following reagents: a reagent A, a reagent B, a reagent C, a reagent D, a reagent E and a reagent F. The template DNA which is extracted by the kit has good purity, and the kit can be used for gene amplification sequencing and typing. In the amplification of the 12S rRNA gene, the mitochondrial D-loop region and the Cyt b gene, all the extracted shell membrane DNA templates can be successfully amplified to obtain the fragment products with expected identical sizes, high concentration and high specificity. Microsatellite DNA marker specificity primers are utilized to obtain typing data through the PCR amplification to be used for the identification in disputed paternity of Yangtze alligator. In addition, the method also can be applied to the extraction of other ovipara such as the turtles, the birds and the like. The obtained template DNA can be widely applied to the studies of the ovipara on the fields such as population genetic analysis, molecular evolution, evolution of relationship among individuals and the like.
Owner:ANHUI NORMAL UNIV
Method and special kit for detecting gene multi-mutant site
ActiveCN101597638AStrong specificityInstrument requirements are simpleMicrobiological testing/measurementMT-RNR1Melting temperature
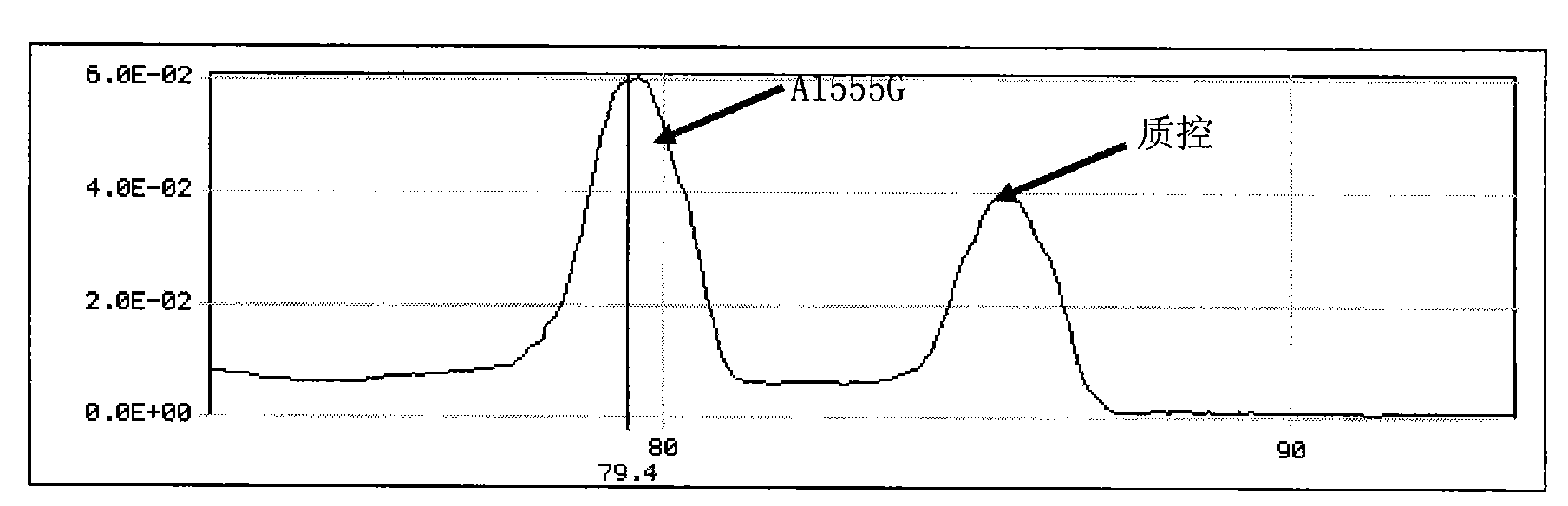
The invention discloses a method for detecting gene multi-mutant site and a special kit thereof. The method for detecting the gene multi-mutant site is a method for detecting the A-to-G mutation of a 1555 site and / or the C-to-T mutation of a 1494 site of a 12S rRNA gene of plastosome, three primers are adopted, and the sequences of the three primers are respectively a sequence 1, a sequence 2 and a sequence 3 in a sequence table. The method comprises the following steps: PCR amplification is carried out by using the three primers; the melting curve of a PCR amplification product is detected; if a melting peak with the melting temperature of 79.4 DEG C appears, point mutation exists at the 1555 site of the 12S rRNA gene of the human plastomsome, and the mutant type is A to G; if a melting peak with the melting temperature of 76.6 DEG C appears, point mutation exists at the 1494 site of the 12S rRNA gene of the human plastomsome, and the mutant type is C to T. The invention also discloses a special kit for detecting the gene multi-mutant site.
Owner:CAPITALBIO CORP +1
Method for identifying yak meat and cattle meat
ActiveCN106755341AAccurate detectionEasy to operateMicrobiological testing/measurementGenomeMelt temperature
The invention provides a method for identifying yak meat and cattle meat by adopting an HRM method. The method comprises the following steps: (1) selecting different loci of the mitochondrial 12S rRNA gene of the yak and the mitochondrial 12S rRNA gene of the cattle, and designing a marking primer and a marking probe according to the selected different loci; (2) extracting the whole genome DNA of a to-be-detected sample; (3) by taking the whole genome DNA of the to-be-detected sample obtained in the step (2) as a template, adding with the marking primer and the marking probe in the step (1), and carrying out asymmetrical PCR amplification, thus obtaining a PCR amplification product; and (4) detecting the melting temperature of the PCR amplification product obtained in the step (3) by adopting an HRM method, and drawing a melting curve, wherein if the melting peak is within 55-60 DEG C, the to-be-detected sample is detected to be the yak meat, and if the melting peak is within 65-70 DEG C, the to-be-detected sample is detected to be the cattle meat.
Owner:LANZHOU INST OF ANIMAL SCI & VETERINARY PHARMA OF CAAS
Method for conducting real-time fluorescence identification on edible bird's nest product through PCR
The invention relates to the technical field of chemical detection and discloses a method for conducting real-time fluorescence identification on an edible bird's nest product through PCR. The method comprises the following steps: 1, extracting DNA of the edible bird's nest product to be detected; 2, conducting real-time fluorescence PCR amplification on esculent swift constituents, wherein firstly, 12.5 microliters of 2*PCR Master Mix, 0.5-1 microliter of an upstream primer, 0.5-1 microliter of a downstream primer, 0.5-1 microliter of Taqman probes and 1-2 microliters of the edible bird's nest product to be detected with the concentration being 50 ng / microliter are added into a PCR reaction tube, supplementing double distilled water to be 25 microliters, and conducting uniform mixing; secondly, putting the PCR reaction tube into a fluorescent quantitative PCR instrument to complete PCR amplification; 3, analyzing the amplification result with real-time fluorescence PCR instrument analysis software. The main principle of the method is that an edible bird's nest is mainly from saliva of esculent swifts and contains DNA of the esculent swifts, and realness of the edible bird's nest is identified by detecting the 12S rRNA gene of the esculent swifts in the edible bird's nest.
Owner:英格尔检测技术服务(上海)有限公司 +1
Kit for jointly detecting four deafness predisposing genes and application thereof
InactiveCN102534030AAvoid false positivesMicrobiological testing/measurementFluorescenceLNA nucleoside
The invention discloses a fluorescent detection kit for detecting four deafness predisposing genes simultaneously. The kit comprises reagents before amplification and reagents after amplification, wherein the reagents before amplification comprise a polymerase chain reaction (PCR) buffer solution, a reaction mixture of MgCl2 and deoxyribonucleoside triphosphates (DNTPs), Taq DNA polymerase, ultrapure water, and a primer mixture for amplifying loci of GJB2235delC, 12S rRNA 1555A>G mutation, 1494C>T mutation, IVS7-2A>G and Amelogenin at high specificity; and the reagents after amplification comprise a genotyping standard and an internal standard. Deafness gene loci of the GJB2235delC, 12S rRNA 1555A>G mutation, 1494C>T mutation, IVS7-2A>G and Amelogenin are simultaneously detected at high sensitivity and high specificity by combining a fluorescent labeling technology, a linolenic acid (LNA) nucleoside monomer doping-primer modification technology and a capillary electrophoresis technology for the first time, manpower and material resources and time are greatly saved, and pollution due to multi-step operation is prevented.
Owner:上海芯鑫生物科技有限公司
Deafness pathogenic gene detection kit utilizing time-of-flight mass spectrometry
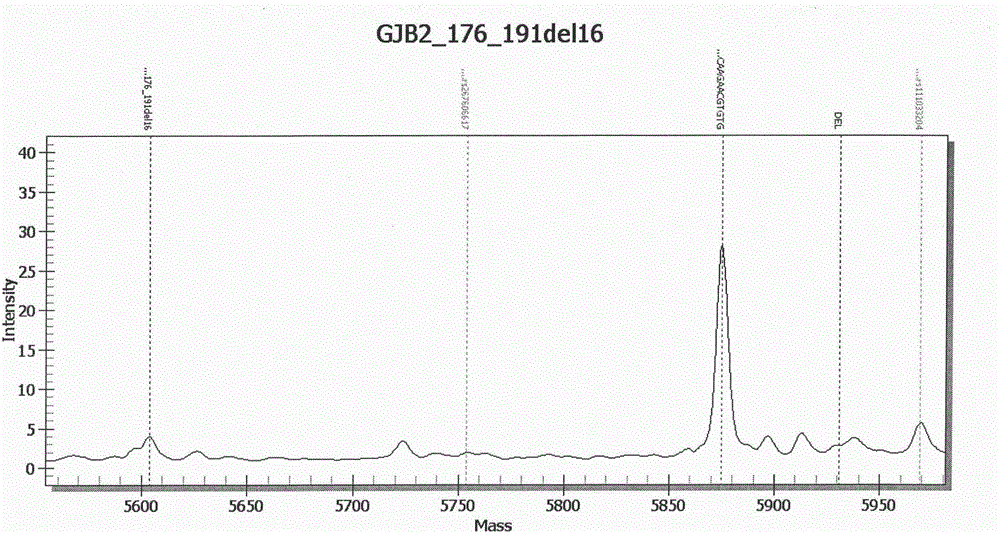
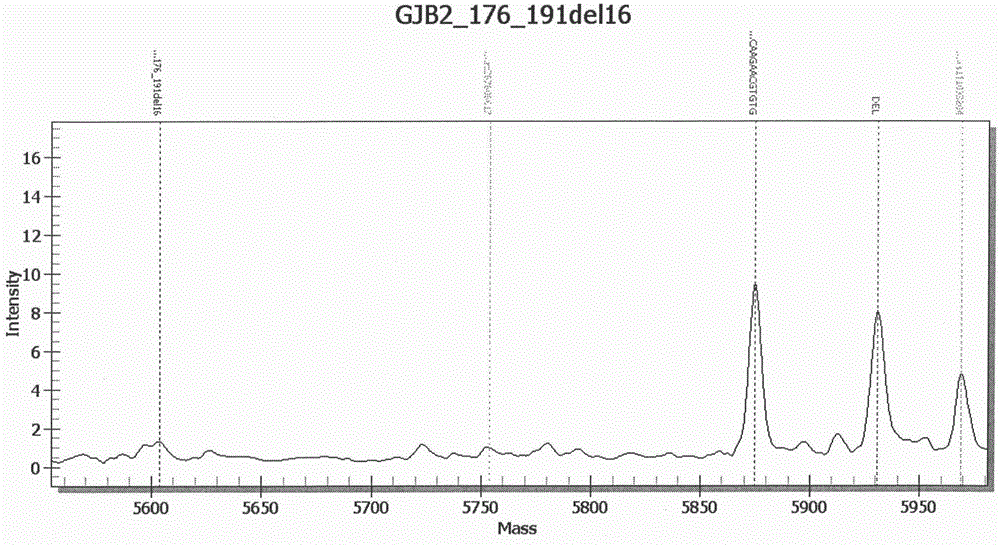
InactiveCN107974497AResolve incompatibilitiesMany mutation detection sitesMicrobiological testing/measurementTotal DeafnessSlc26a4 gene
The invention provides a deafness pathogenic gene detection kit utilizing a time-of-flight mass spectrometry. The kit comprises a reagent for detecting at least following 26 mutation sites of 12S rRNA(ribosomal Ribonucleic Acid), GJB2 and SLC26A4 genes: 12S rRNAm.1494-C is greater than T, 12S rRNAm.1555Ais greater thanG, GJB2c.35delG, GJB2c.257C is greater than G, GJB2c.427C is greater than T, GJB2c.176del16, GJB2c.9G is greater than A, GJB2c.235delC, GJB2c.299-300delAT, SLC26A4IVS4+2T is greater than C, SLC26A4c.1673A is greater than T, SLC26A4c.1520delT, SLC26A4c.2027T is greater than A, SLC26A4c.1975G is greater than C, SLC26A4c.1226G is greater than A, SLC26A4c.1318A is greater than T, SLC26A4c.1229C is greater than T, SLC26A4c.281C is greater than T, SLC26A4c.2168A is greater than G,SLC26A4IVS7-2A is greater than G, SLC26A4c.1174A is greater than T, SLC26A4c.235C is greater than T, SLC26A4c.1340delA, SLC26A4_c.589G is greater than A, SLC26A4c.916-917insG and SLC26A4c.IVS15+5G isgreater than A. The kit provided by the invention has the advantages of more mutation detection sites, high throughput, simplicity in operation and short period, high accuracy, high stability and lowcost.
Owner:国家卫生健康委科学技术研究所
A fluorescence detection kit for detecting deafness susceptibility gene 12S rRNA 1555A>G and application thereof
InactiveCN102660636AAvoid false positivesMicrobiological testing/measurementFluorescenceLNA nucleoside
The present invention discloses a fluorescence detection kit for detecting deafness susceptibility gene 12S rRNA 1555A>G, comprising a pre-amplification reagent and a post-amplification reagent, wherein the reagent before amplification includes a PCR buffer solution, a reaction mixture of MgC12 and dNTPs, a Taq enzyme, an ultra pure water, and a primer mixture for high- specificity amplification of 12S rRNA 1555A>G and Amelogenin loci; the post-amplification reagent includes a genotyping standard and an internal standard. The detection kit, with the loci of 12S rRNA 1555A>G and Amelogenin as detection objects, can screen out individuals having mutation at the above loci through the amplification of the deafness susceptibility gene locus, detection by capillary electrophoresis, and comparison between the detection object and the genotyping standard. The detection kit is of great significance to detection of deafness susceptibility gene and greater significance to deafness gene screening of the neonate. The detection kit is the first in the field of deafness gene screening to comprehensively combine fluorescence labeling technology, LNA nucleoside monomer incorporation-primer modification technology and capillary electrophoresis technology to realize detection of the deafness susceptibility gene locus 12S rRNA 1555A>G with high sensitivity and specificity.
Owner:北京科聆金仪生物技术有限公司
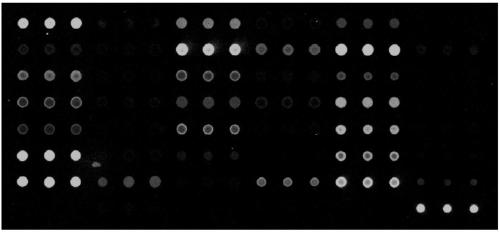
Hereditary deafness related gene detection chip kit
ActiveCN109504753AImprove accuracyOvercome the defects that easily cause missed inspectionsMicrobiological testing/measurementMutantLeak detection
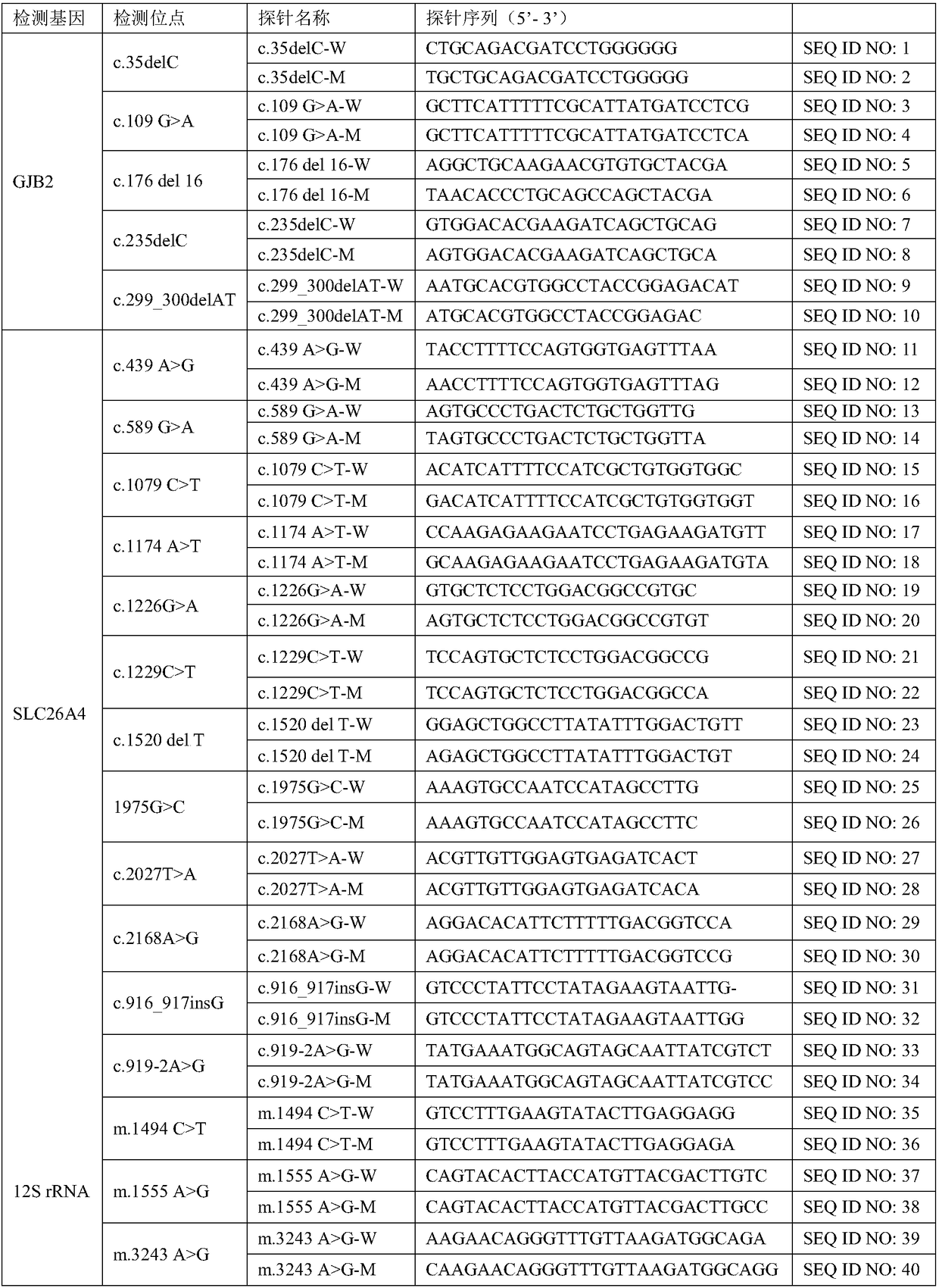
The invention discloses a hereditary deafness related gene detection chip kit, which comprises a primer and a specific probe for detecting 20 site variation on three hotspot genes of hereditary deafness, wherein 20 sites comprise 5 mutant types on GJB2, 12 mutant types on SLC26A4 and 3 mutant types on 12S rRNA (MTRNR1, belonging to a chondriogene); the sequence of the specific probe is shown as SEQ ID NO: 1 to SEQ ID NO: 40; the sequence of the specific probe is shown as SEQ ID NO: 41 to SEQ ID NO: 60. The invention also discloses a concrete detection method. The kit is designed especially forthe hotspot mutation sites of the hereditary deafness gene of people groups of southern China; the detection accurate rate of the hereditary deafness of people groups of southern China can be improved; the kit overcomes the defects that the existing kit is mainly designed for the sites of people groups of Northern China, and the detection on the people groups of southern China is liable to leak detection.
Owner:上海伯豪生物技术有限公司
Deafness predisposing gene 12S rRNA (ribosomal ribonucleic acid) 1494C>T fluorescence detection kit and application thereof
InactiveCN102586433AIncrease the Tm valueShorten the lengthMicrobiological testing/measurementFluorescenceInternal standard
The invention discloses a fluorescence detection kit for detecting deafness predisposing gene 12S rRNA (ribosomal ribonucleic acid) 1494C>T. The kit comprises a reagent before amplification and a reagent after amplification, wherein the reagent before amplification comprises a reaction mixture of a PCR (polymerase chain reaction) buffer solution, MgCl2 and dNTPs (deoxyribonucleoside triphosphate), Taq enzyme, ultrapure water, and a primer mixture for high-specific amplification of 12SrRNA1494C>T and amelogenin locus; and the reagent after amplification comprises an allelic ladder and an internal standard. According to the invention, 12SrRNA1494C>T and amelogenin locus are used as detecting objects, so that the fluorescence detection kit has important meaning to detection of deafness predisposing genes, particularly screening of newborn deafness genes.
Owner:万戈江
Drug-induced deafness gene multichannel fluorescence PCR (polymerase chain reaction) detection kit
InactiveCN104711366AImprove effectivenessTest effectivenessMicrobiological testing/measurementFluorescenceContamination
The invention discloses a drug-induced deafness gene multichannel fluorescence PCR (polymerase chain reaction) detection kit. Two pairs of specific primers and four TaqMan probes are designed according to four sites on the drug-induced deafness related gene mtDNA 12S rRNA: 1555A>G, 1494C>T, 1095T>C and 827A>G, and a pair of specific primers and probes are designed by using the housekeeping gene beta-actin as the internal reference to monitor the effectiveness of the sample template and reaction system and avoid the appearance of false negative; and meanwhile, the setting of the negative and positive quality control ensures the accuracy of the detection result. The kit implements simultaneous detection on four mutant sites of the drug-induced deafness gene in one reaction tube by a multichannel fluorescence PCR process for the first time. The method is simple to operate, has the advantages of high speed, high accuracy, high flux and low cost, avoids the cross contamination due to shut tube operation, improves the purposiveness and screening range of deafness screening, effectively prevents the drug-induced deafness, and is easy for popularization and application.
Owner:JINAN YING SHENG BIOTECH
Probes for myctophid fish and a method for developing the same
The DNA probes produced by molecular cloning and the characterization of specific gene region sequences is provided, these can be used as genetic markers for the genes such as Cytochrome b (cyt b); Mitochondrial control region (D-Loop); Inter Transcribed Spacers (ITS2) and Rhodopsin (ROD), 12S rRNA and 16S rRNA in mesopelagic lantern fishes which are found in the mesopelagic zones of the oceans where the photic regime is of dim light and associate themselves with the oxygen minimum layer, it also includes the recombinant DNA techniques for the preparation of specific gene probes and sequences of species specific primers of lantern fishes, novel gene probes and novel oligonucleotides for amplification of myctophid genes are disclosed.
Owner:COUNCIL OF SCI & IND RES
Method for analyzing phyletic lineage of scallop
InactiveUS20020081593A1Efficient analysisQuick analysisSugar derivativesMicrobiological testing/measurementPatinopecten yessoensisCoding region
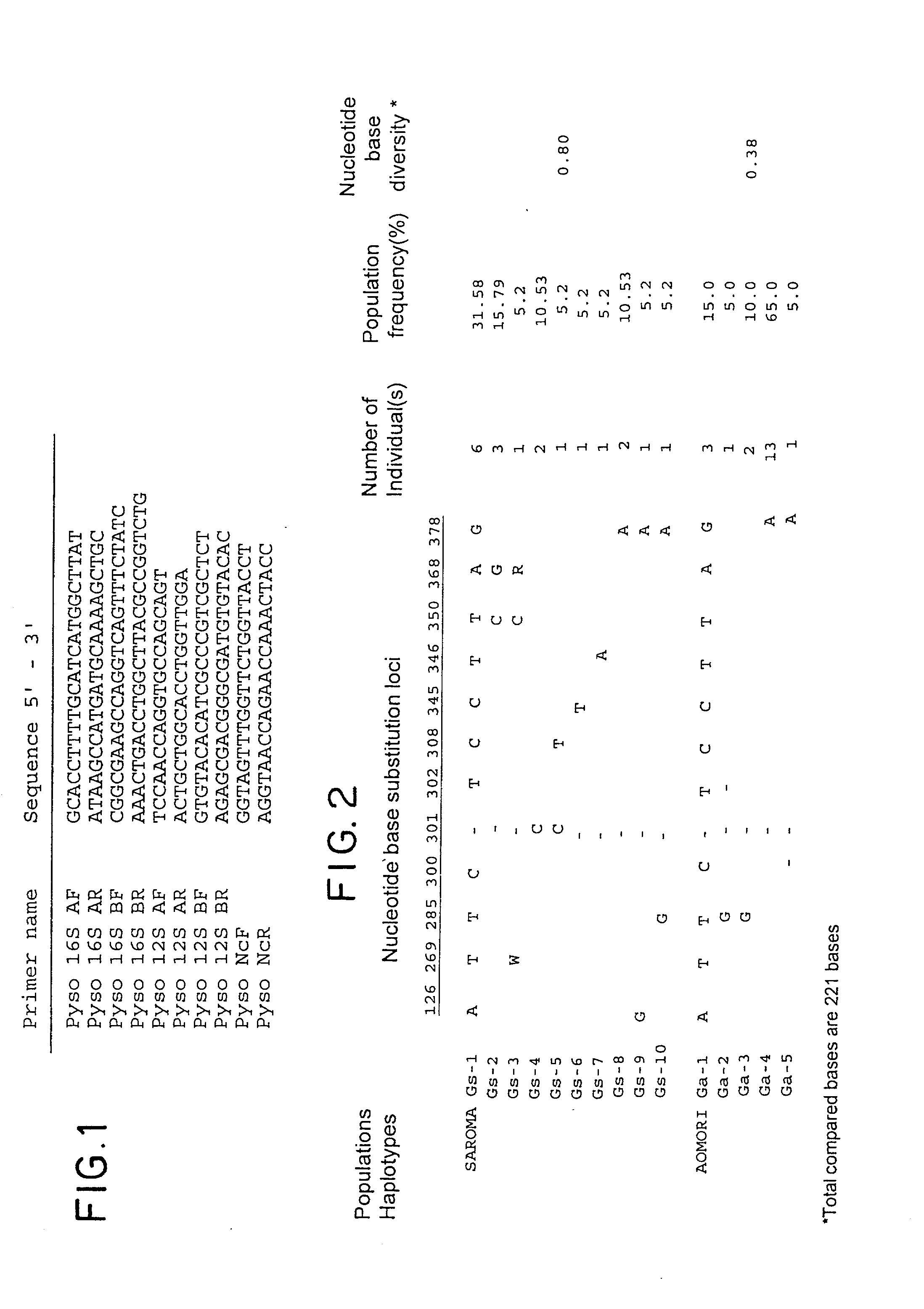
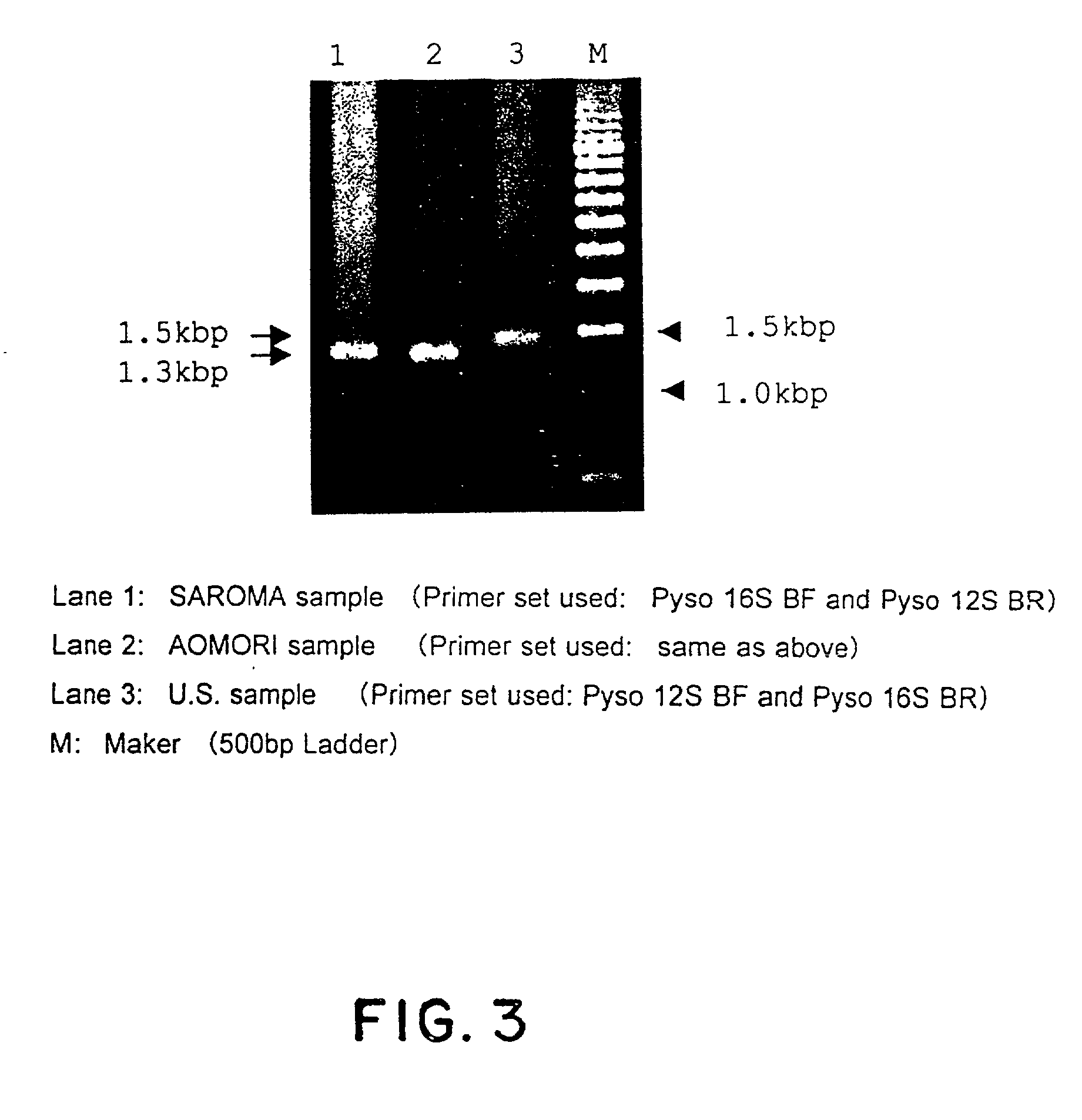
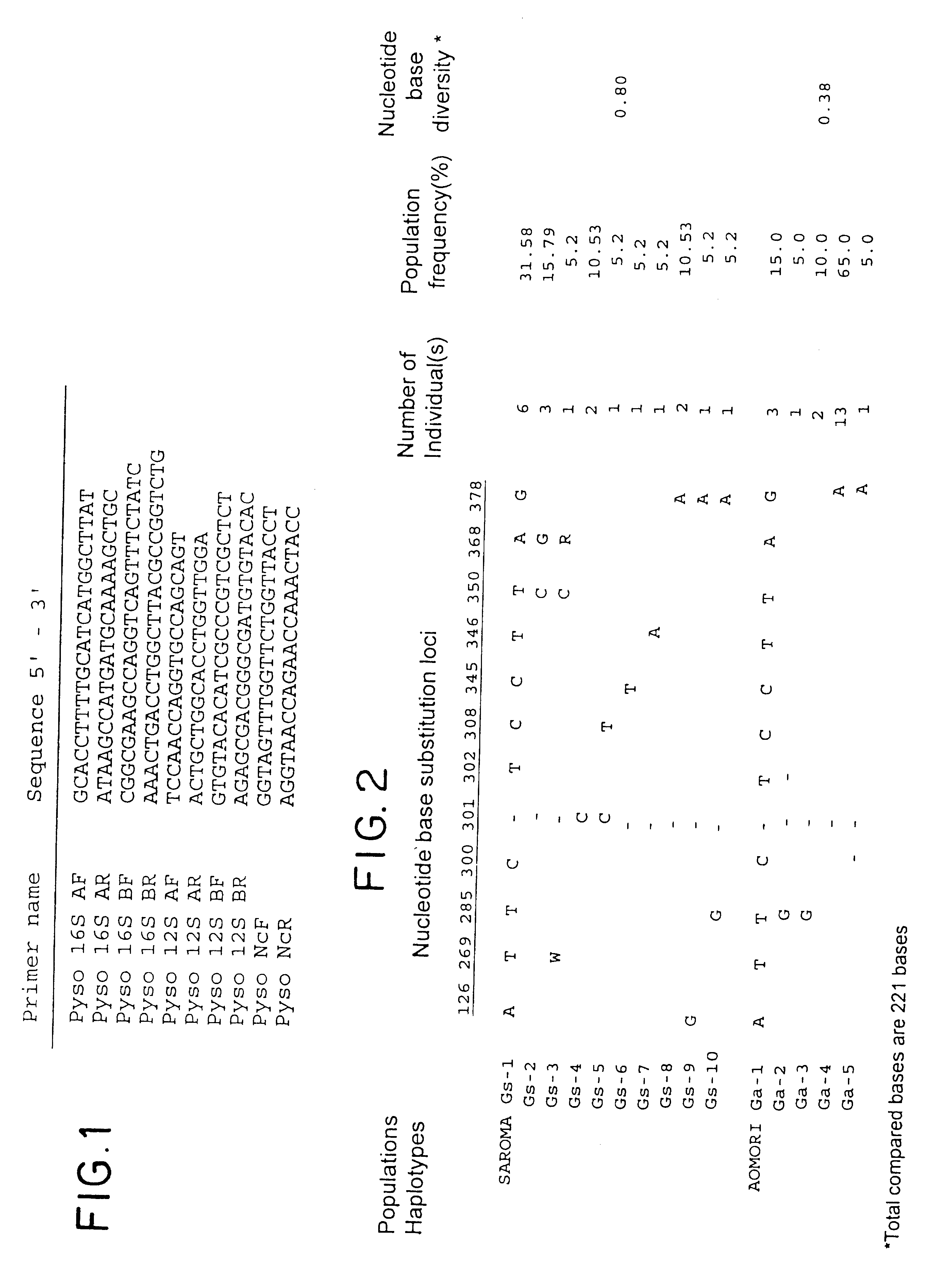
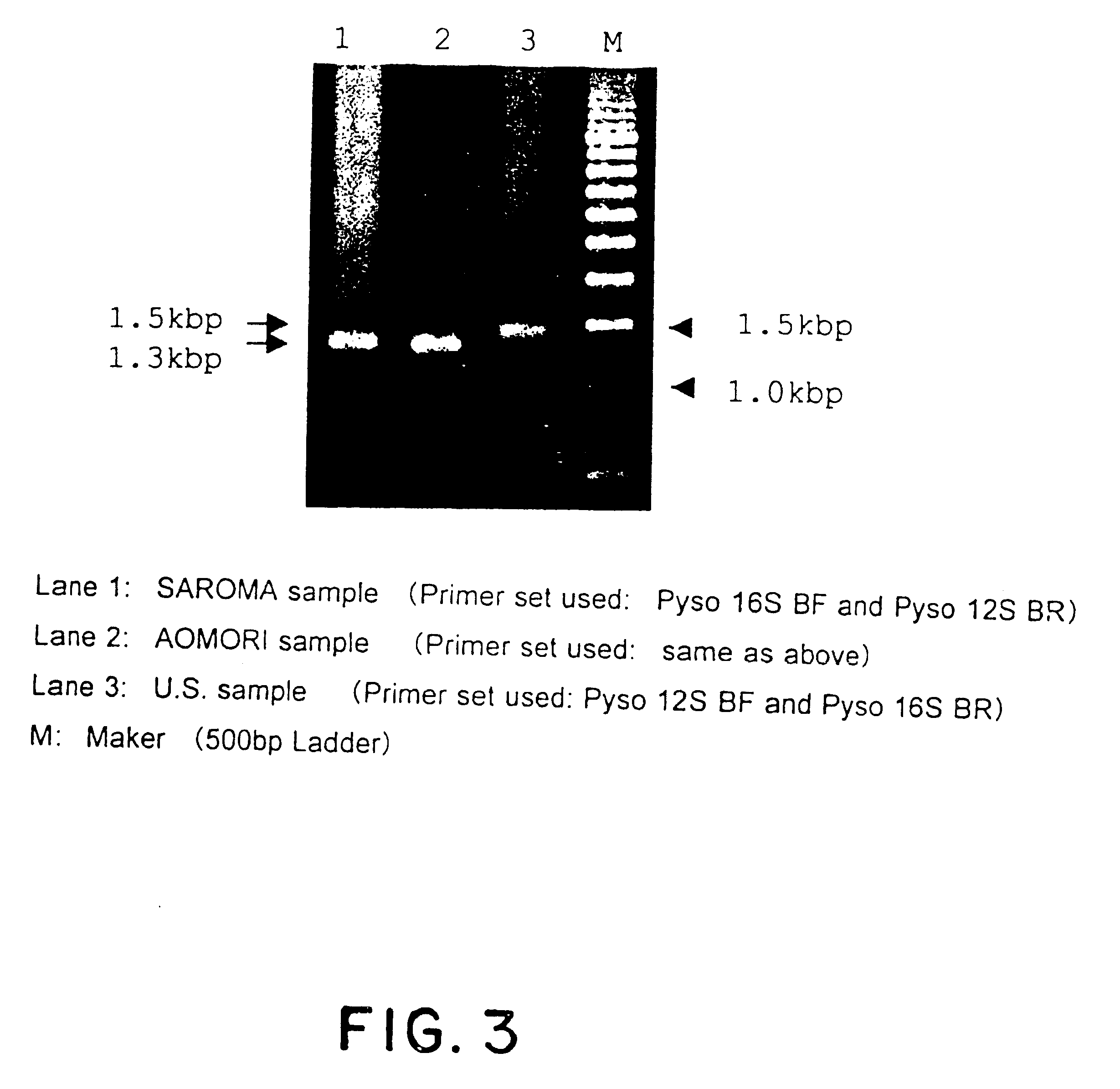
Method for analyzing a phyletic lineage of scallop, which comprises the steps of sequencing a mitochondrial DNA of the scallop to determine a nucleotide base sequence of the mitochondrial DNA, wherein the nucleotide base sequence includes a non-coding region containing a nucleotide base substitution locus which shows a sequence polymorphism indicative of a particular lineage of the scallop. Mitochondrial DNA of the scallop is amplified by PCR, using a suitable primer set designed on the basis of nucleotide base sequences conserved among known shellfish mitochondrial 16S rRNA and 12S rRNA genes, followed by sequencing to determine a non-coding region therein. In such non-coding region, a nucleotide base substitution locus is located, whereby a particular lineage of the scallop is determined. Based on those steps, a Japanese scallop, Patinopecten yessoensis, is analyzed as to the nucleotide base sequence and lineage thereof.
Owner:HOKKAIDO FEDERATION OF FISHERIES COOP ASSOCS
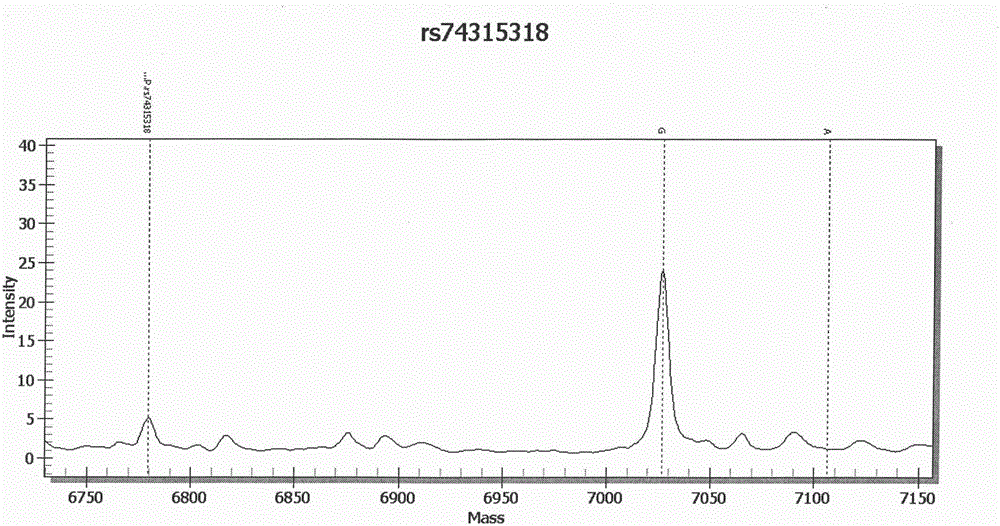
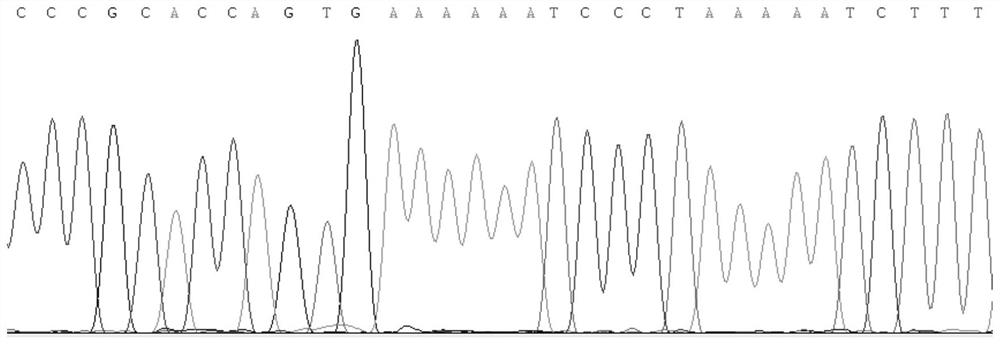
Method and kit for realizing sequencing-based typing of mitochondria 12S rRNA genome full-length sequence
ActiveCN105400874AImprove recognition resultsEasy to interpret resultsMicrobiological testing/measurementMT-RNR1Typing methods
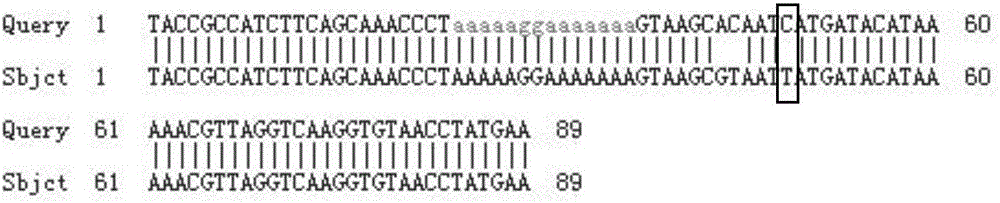
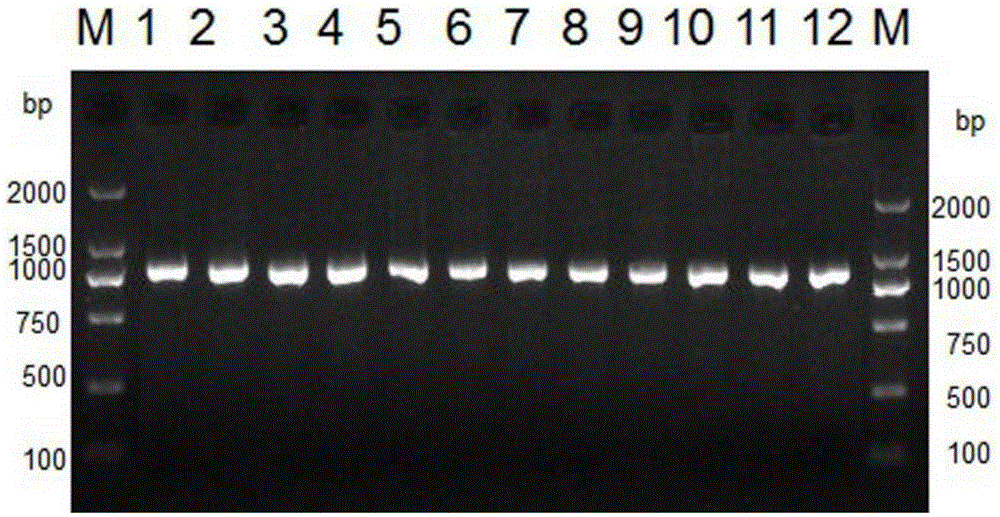
The invention belongs to the field of biological technical detection, and discloses a method and a kit for realizing sequencing-based typing of a mitochondria 12S rRNA genome full-length sequence. The method comprises the following steps: carrying out PCR amplification reaction on a typing target area, namely a 954bp genome full-length sequence, by adopting a pair of PCR amplification primers, and carrying out bi-directional sequencing reaction on the amplification product through four forward and reverse sequencing primers. The invention establishes the sequencing-based typing method for the 12S rRNA genome full-length sequence which is accurate, stable and reliable. The obtained base sequence peak map contains no background signals and impure peaks, and is easy for identification and result determination. The method can carry out sequencing-based typing on the 12S rRNA genome full-length sequence, so that the equivocal result appearing in the sequencing-based typing is avoided; the method is suitable for the typing of the 12S rRNA genes, and the basic and applied study works on the aspects of population genetics, evolutiology, disease association and the like of the 12S rRNA genes.
Owner:SHENZHEN CITY BAOAN DISTRICT MATERNAL & CHILD HEALTH HOSPITAL
High variation zone amplication primer of rockfish mitochondrial genome and its design method
The present invention relates to one pair of PCR primers named as G-dloop, and provides one pair of amplification primers capable of amplifying the high variation zone of rockfish mitochondrial genome in high efficiency and its design method. The amplification primers consist of two single strand oligonucleotides. Through logging-on Genebank to search vertebrate mitochondrion DNA cytochrome b gene and the high conservation area of 16S rRNA gene sequence and homologous comparison, one pair of amplification primers is obtained. Through long PCR amplification, the target segments of 32 varietiesof rockfish are obtained and sequenced. The obtained sequences are compared by means of using homologous comparison software Clustal X 1.83 to fine the conservation sequences in the cytochrome b5' ends and the 12S rRNA 3' ends of the mitochondrial genomes of the 32 varieties of rockfish, and the said amplification primers are designed based on the degeneracy principle.
Owner:XIAMEN UNIV
Kit and method for identifying Eospalax species
PendingCN113430281AEasy to determineStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationConserved sequenceGenetics
The invention provides a kit for identifying Eospalax species. The kit comprises reagents for detecting the following 14 SNP sites of a species to be detected: the 37 site of a conserved motif sequence shown in SEQ ID NO: 5 in a 12S rRNA gene, the 18 site of a conserved motif sequence shown in SEQ ID NO: 6, the 33<rd> site of a conserved motif sequence shown in SEQ ID NO: 7, the 7 site of a conserved motif sequence shown in SEQ ID NO: 8, the 47 site of a conserved motif sequence shown in SEQ ID NO: 11, the 1 site and the 2<nd> site of a conserved motif sequence shown in SEQ ID NO: 13, the 4 site of a conserved motif sequence shown in SEQ ID NO: 14, and the 25 site of a conserved motif sequence shown in SEQ ID NO: 15; and the 33<rd> site of a conserved motif sequence shown in SEQ ID NO: 9 in a 16S rRNA gene, the 34 site of a conserved motif sequence shown in SEQ ID NO: 10, the 35 site of a conserved motif sequence shown in SEQ ID NO: 12, and the 22<nd> site and the 31 site of a conserved motif sequence shown in SEQ ID NO: 16. The kit can simply and accurately identify the species of individuals in Eospalax, solves the problem of identifying the Eospalax species through morphology, and has excellent application prospects.
Owner:CHINA ACAD OF SCI NORTHWEST HIGHLAND BIOLOGY INST
Molecular biological method for rapidly identifying two leuciscus waleckii species distributed in same area of Sinkiang
InactiveCN109251968AIncrease successImprove accuracyMicrobiological testing/measurementMT-RNR1Leuciscus
The invention discloses a molecular biological method for rapidly identifying two leuciscus waleckii species distributed in the same area of Sinkiang. The molecular biological method comprises the following steps: (1) extracting genomes of leuciscus waleckii tissue by adopting an improved phenol-chloroform method; (2) designing primers of mitochondria 12S rRNA genes; (3) by adopting DNAs of the two fish species as templates, carrying out PCR amplification; (4) carrying out 1% agarose gel electrophoresis detection on the PCR products, selecting the PCR products with clear stripes, and carryingout sequencing; and (5) carrying out sequence contrast. The molecular biological method has the beneficial effects that the molecular biological method is scientific and effective, is simple and rapid, is accurate and feasible, is strong in specificity, and is high in reliability, leuciscus waleckii does not need to be killed, amplification can be carried out on the two species at once, and thus the operation is efficient, rapid and convenient.
Owner:ZHEJIANG OCEAN UNIV
Method for identifying western Henan black pigs by using 12S rRNA gene mutation sites
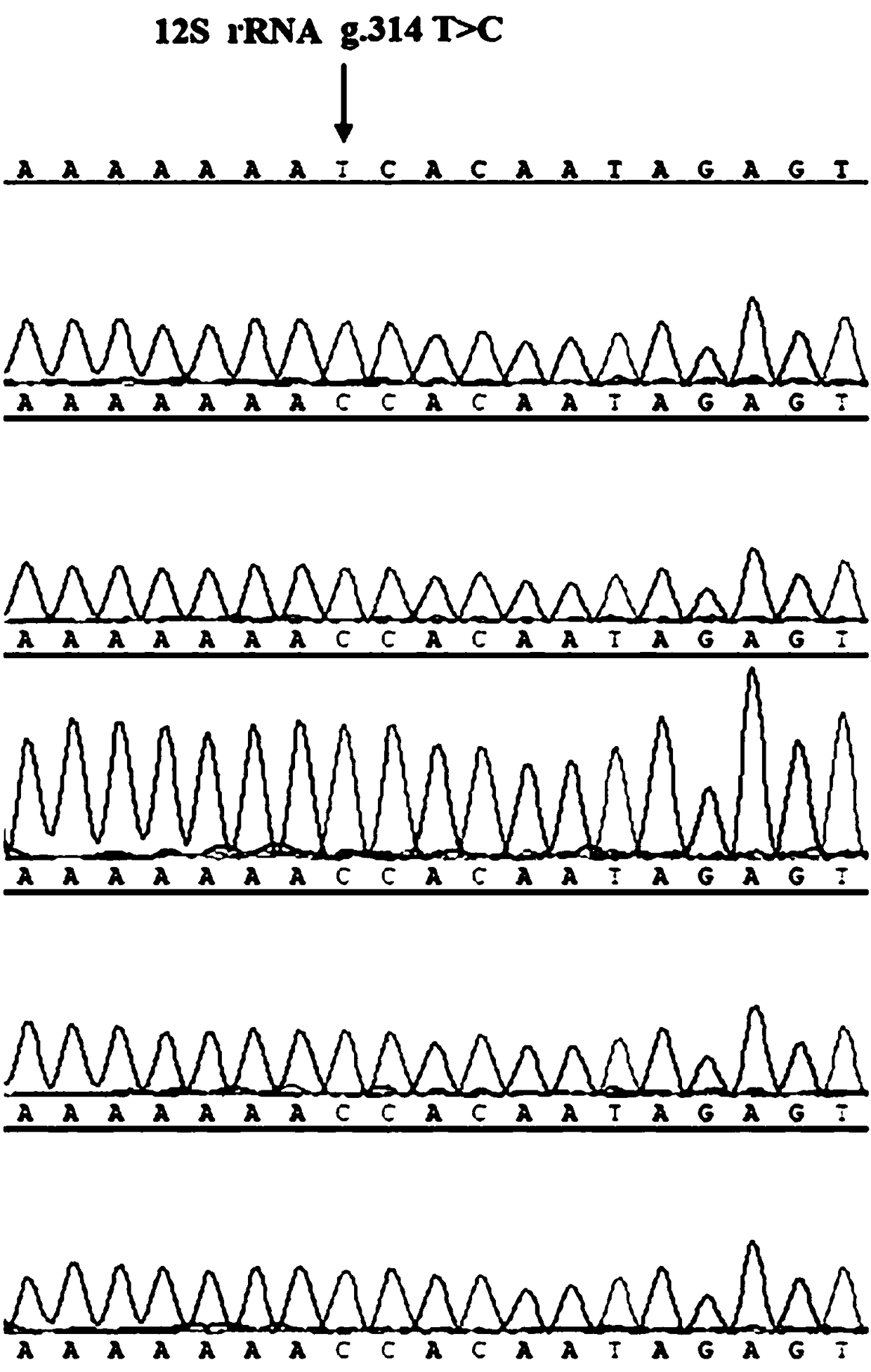
ActiveCN108130363ARapid identificationAccurate identificationMicrobiological testing/measurementAnimal GeneticsElectrophoresis
The invention relates to the field of animal genetics, in particular to a method for identifying western Henan black pigs by using 12S rRNA gene mutation sites. The method comprises the steps of downloading the 12S rRNA gene in a reference sequence in NCBI; respectively designing a PCR primer at the upstream part and the downstream part of the 314 site basic group of the 12S rRNA gene; extractingDNA of a sample to be tested; performing PCR amplification by using the DNA of the sample to be tested as a template and the PCR primers in the step (2) as primers; performing electrophoresis gel running (agarose gel electrophoresis) identification on the PCR product, and recovering a target product; comparing the target product subjected to sequencing with the 12S rRNA gene of the western Henan black pigs to complete the identification of the western Henan black pigs. The method solves the problem of distinguishing among the black pigs in the same region, and also solves the problems that theexisting pork market is disordered, the defective pork is used as high-quality pork, and an effective detection method does not exist.
Owner:HENAN AGRICULTURAL UNIVERSITY
Kit and method for identifying zokor
PendingCN113637771ALow sequencing length requirementReliable resultsMicrobiological testing/measurementDNA/RNA fragmentationZokorConserved sequence
The invention provides a kit for identifying zokor. The kit comprises a reagent for detecting any one or more of the following three SNP sites of a species to be detected: the 33 <rd> site of a conserved motif sequence shown in SEQ ID NO: 5 in a 12S rRNA gene, the 7 site of a conserved motif sequence shown in SEQ ID NO: 6, and the 33 <rd> site of a conserved motif sequence shown in SEQ ID NO: 7 in a 16S rRNA gene. The invention provides the kit for simply, conveniently and accurately identifying the zokor species and a method for identifying the zokor, the problem of identifying the zokor species through morphology is solved, and the kit has excellent application prospects in zokor species identification.
Owner:CHINA ACAD OF SCI NORTHWEST HIGHLAND BIOLOGY INST
Method for analyzing phyletic lineage of scallop
InactiveUS6514705B2High precision and reliabilityEfficient analysisSugar derivativesMicrobiological testing/measurementPatinopecten yessoensisCoding region
Method for analyzing a phyletic lineage of scallop, which comprises the steps of sequencing a mitochondrial DNA of the scallop to determine a nucleotide base sequence of the mitochondrial DNA, wherein the nucleotide base sequence includes a non-coding region containing a nucleotide base substitution locus which shows a sequence polymorphism indicative of a particular lineage of the scallop. Mitochondrial DNA of the scallop is amplified by PCR, using a suitable primer set designed on the basis of nucleotide base sequences conserved among known shellfish mitochondrial 16S rRNA and 12S rRNA genes, followed by sequencing to determine a non-coding region therein. In such non-coding region, a nucleotide base substitution locus is located, whereby a particular lineage of the scallop is determined. Based on those steps, a Japanese scallop, Patinopecten yessoensis, is analyzed as to the nucleotide base sequence and lineage thereof.
Owner:HOKKAIDO FEDERATION OF FISHERIES COOP ASSOCS
Method for screening Cobb broiler DNA barcode and application of DNA barcode
ActiveCN110144406ABreakout cycleBreakout feeMicrobiological testing/measurementDNA/RNA fragmentationDNA barcodingScreening method
The invention discloses an application of a Cobb broiler DNA barcode 'TCTACGGGC' in screening Cobb broilers, and the Cobb broiler DNA barcode is obtained by the screening method. SNP sites and haplotype sequences specific on 12S rRNA are screened from mitochondrial genes by a PCR direct sequencing method by use of bioinformatic analysis software, one DNA barcode sequence specific to the Cobb broilers is obtained by large-group verification, and the DNA barcode breaks through the defects of long detection period and high cost of traditional microsatellite molecular markers, can rapidly and conveniently detect whether the blood lineage of the Cobb broilers is mixed in local varieties and can be applied to verification analysis of Cobb broiler purebreds.
Owner:GUANGXI UNIV
Kit and method for identifying myospalax psilurus
PendingCN113637769ALow sequencing length requirementReliable resultsMicrobiological testing/measurementDNA/RNA fragmentationConserved sequenceGenetics
The invention provides a kit for identifying myospalax psilurus. The kit comprises a reagent for detecting any one or more of the following three SNP sites of species to be detected: the 21st site of a conserved motif sequence shown as SEQ ID NO: 5 in a 12S rRNA gene, the 4th site of the conserved motif sequence shown as SEQ ID NO: 6, and the 25th site of the conserved motif sequence as shown in SEQ ID NO: 7 in the 16S rRNA gene. The invention provides the kit for simply, conveniently and accurately identifying the species of the myospalax psilurus and the method for identifying the myospalax psilurus, the problem of identifying the species of the myospalax psilurus through forms is solved, and the kit and the method have excellent application prospects in species identification of the myospalax psilurus.
Owner:QINGHAI UNIVERSITY +1
Aminoglycosides related 12s rRNA gene mutation site detection kit
InactiveCN108588213AIncreased sensitivityStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationPolymorphism DetectionAminoglycoside Drugs
The invention relates to an aminoglycosides related 12s rRNA gene mutation site detection kit, in particular to a detection primer composition, a kit and a method for an aminoglycosides related 2s rRNA gene polymorphism mutation site. The primer composition comprises polymorphism detection primers and internal control gene detection primers, wherein sequences of the polymorphism detection primersare as follows: a first polymorphism primer: 5'-TACGCATTTA TATAGAGGATA-3'; a second polymorphism primer: 5'-TACGCATTTA TATAGAGGACG-3'; a third polymorphism primer: 5'-GTGCACTTGG ACGAACCAGA-3'; a fourth polymorphism primer: 5'-GCGTACACAC CGCCCGTCAAC-3'; a fifth polymorphism primer: 5'-GCGTACACAC CGCCCGTCAGT-3'; a sixth polymorphism primer: 5'-TAAATGCGTA GGGGTTTTAG-3'; sequences of the internal control gene detection primers are as follows: a first internal control primer: 5'-AGCAAGCAGG AGTATGACG-3'; a second internal control primer: 5'-GAAAGGGTGT AACGCAACT-3'. Compared to the prior art, the aminoglycosides related 12s rRNA gene mutation site detection kit has the advantages of high sensitivity, high specificity, short time consumption and the like in the process of detecting 12s rRNA gene mutation.
Owner:宁波美丽人生医药生物科技发展有限公司
Deafness susceptibility gene screen test kit
Owner:GENERAL HOSPITAL OF PLA
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com