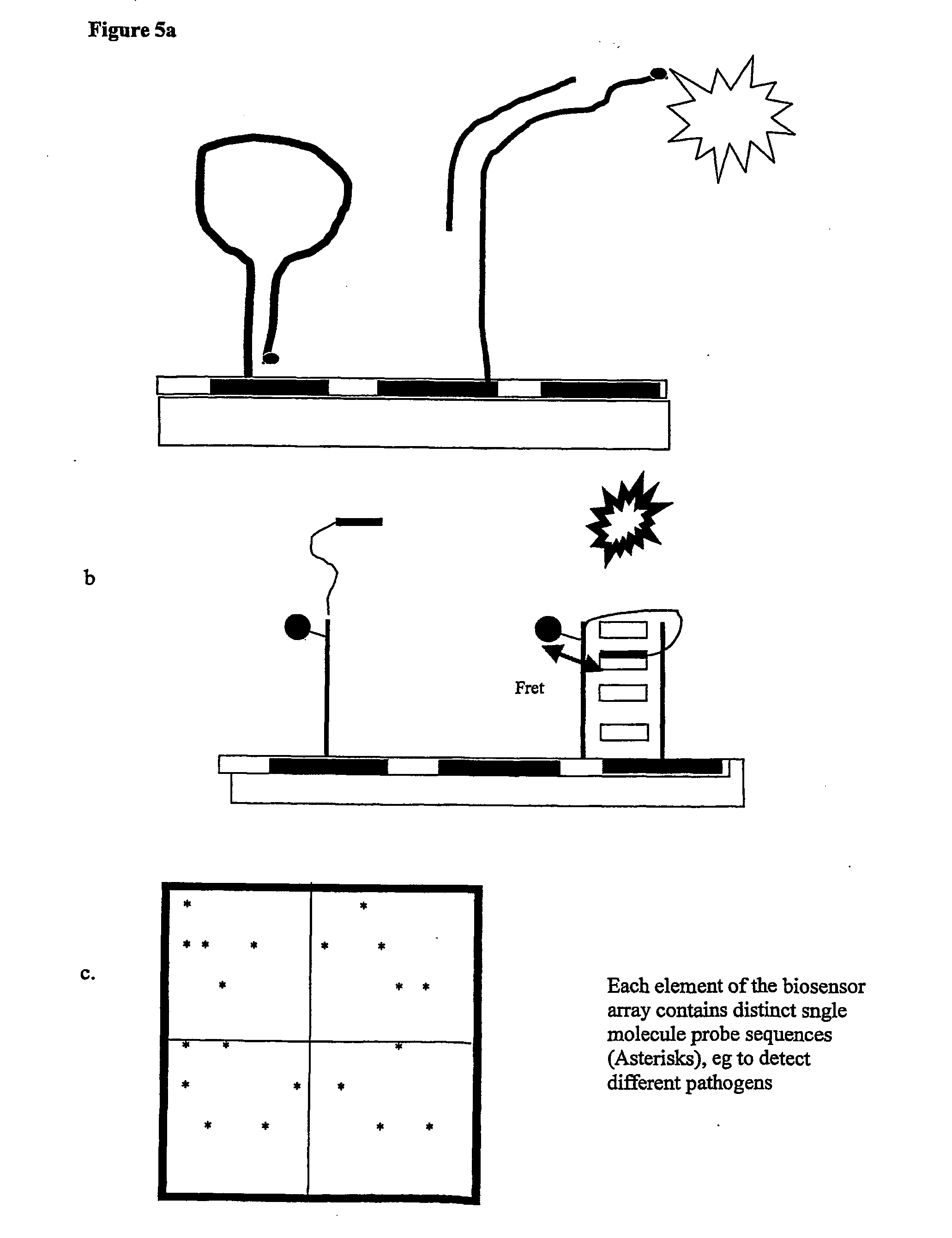
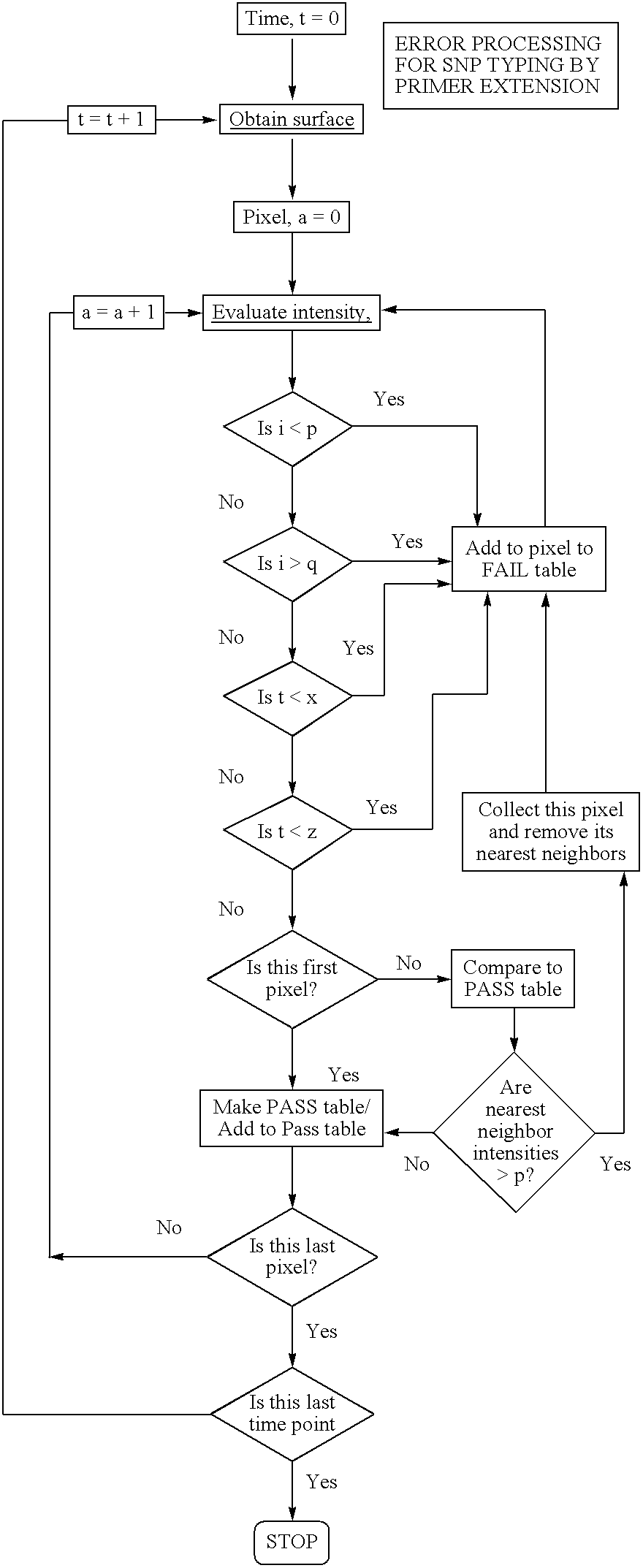
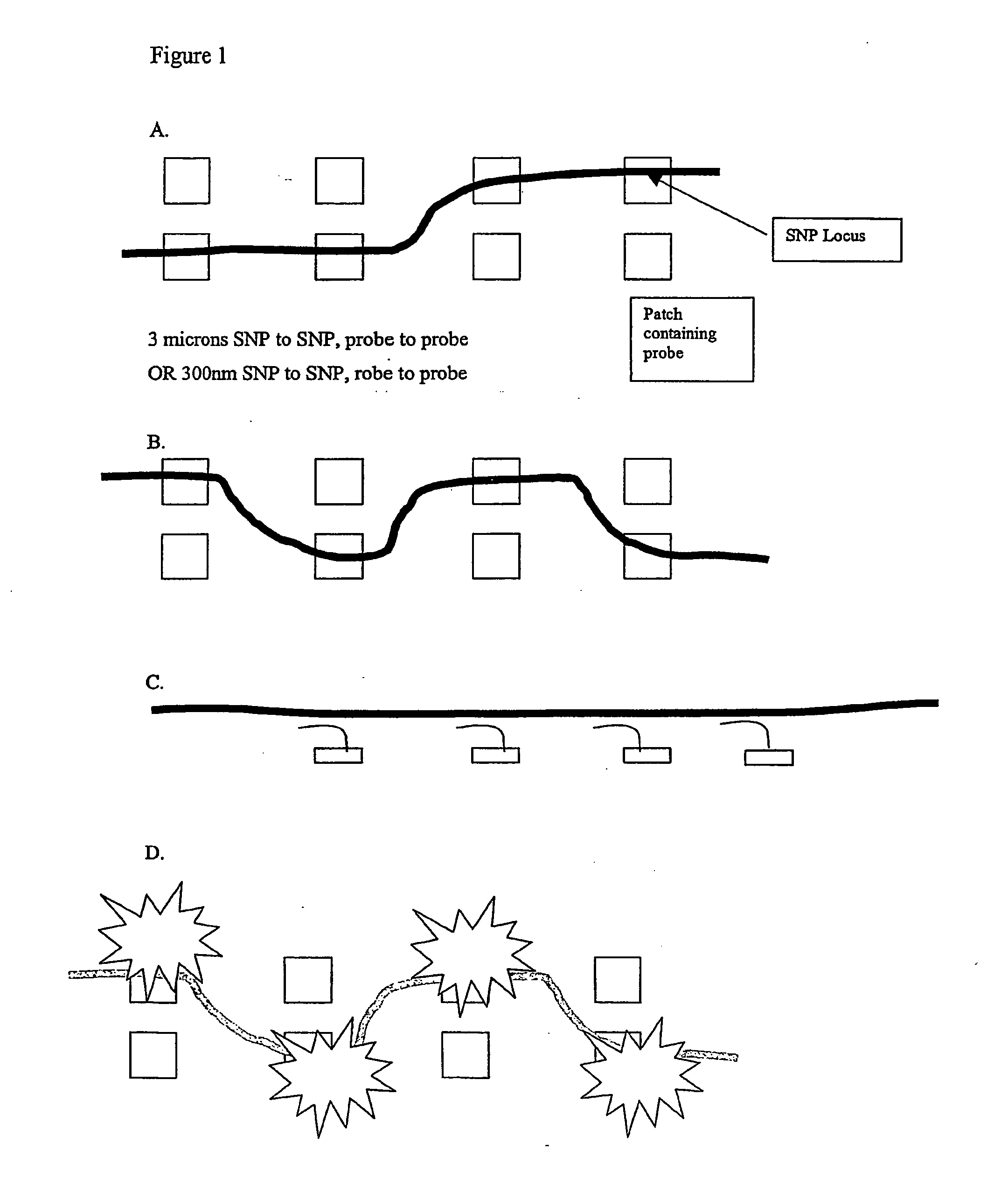
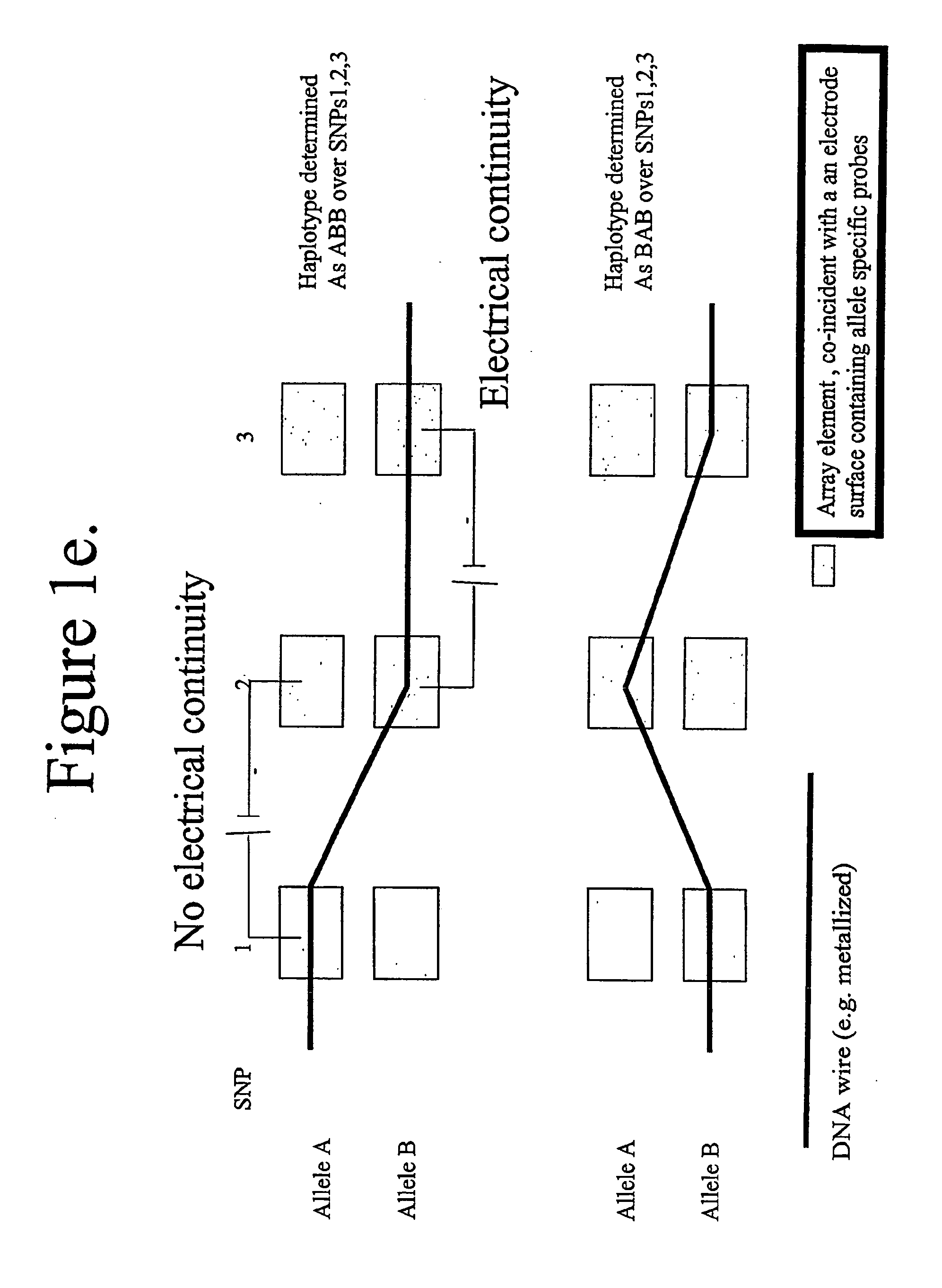
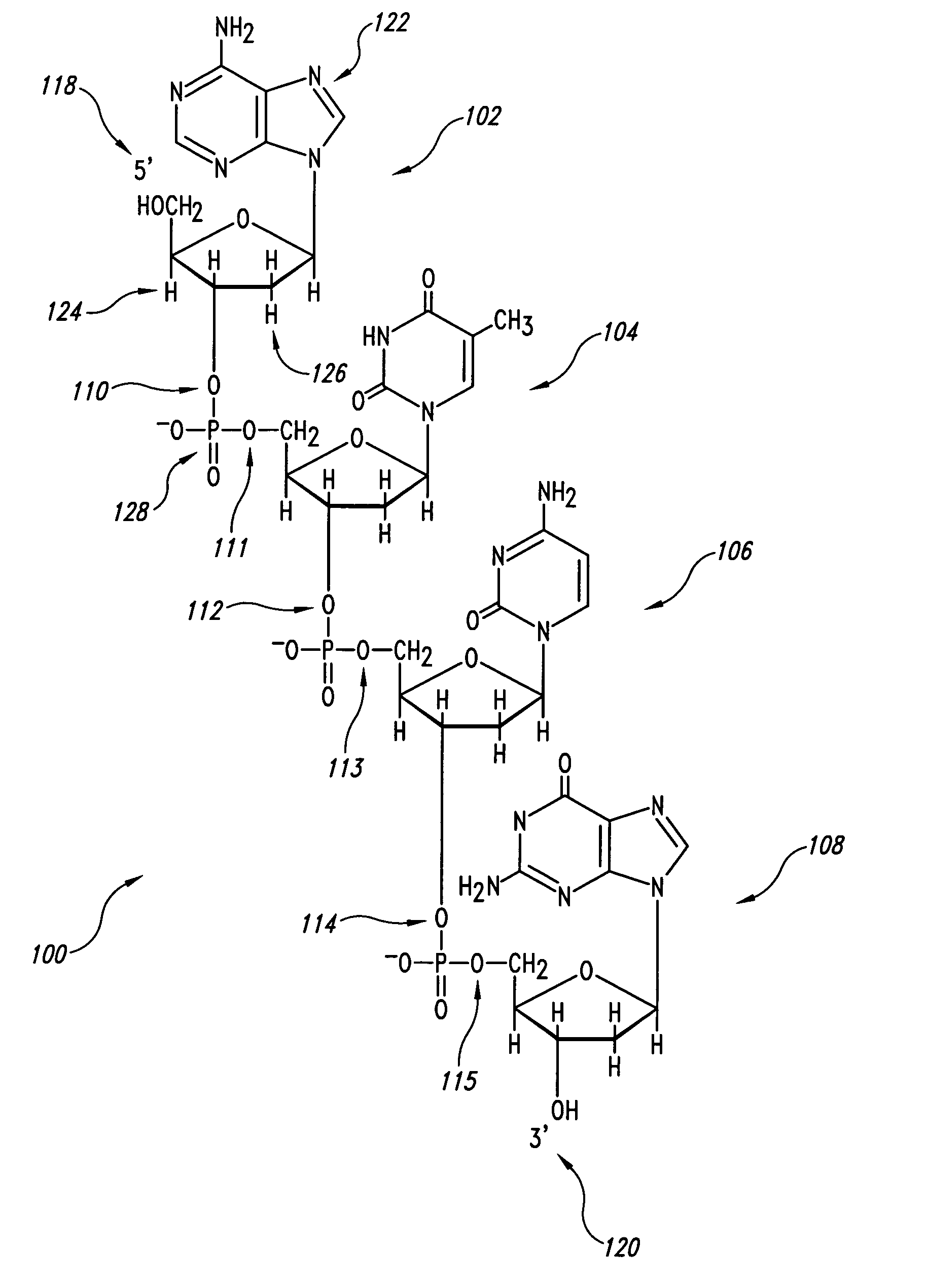
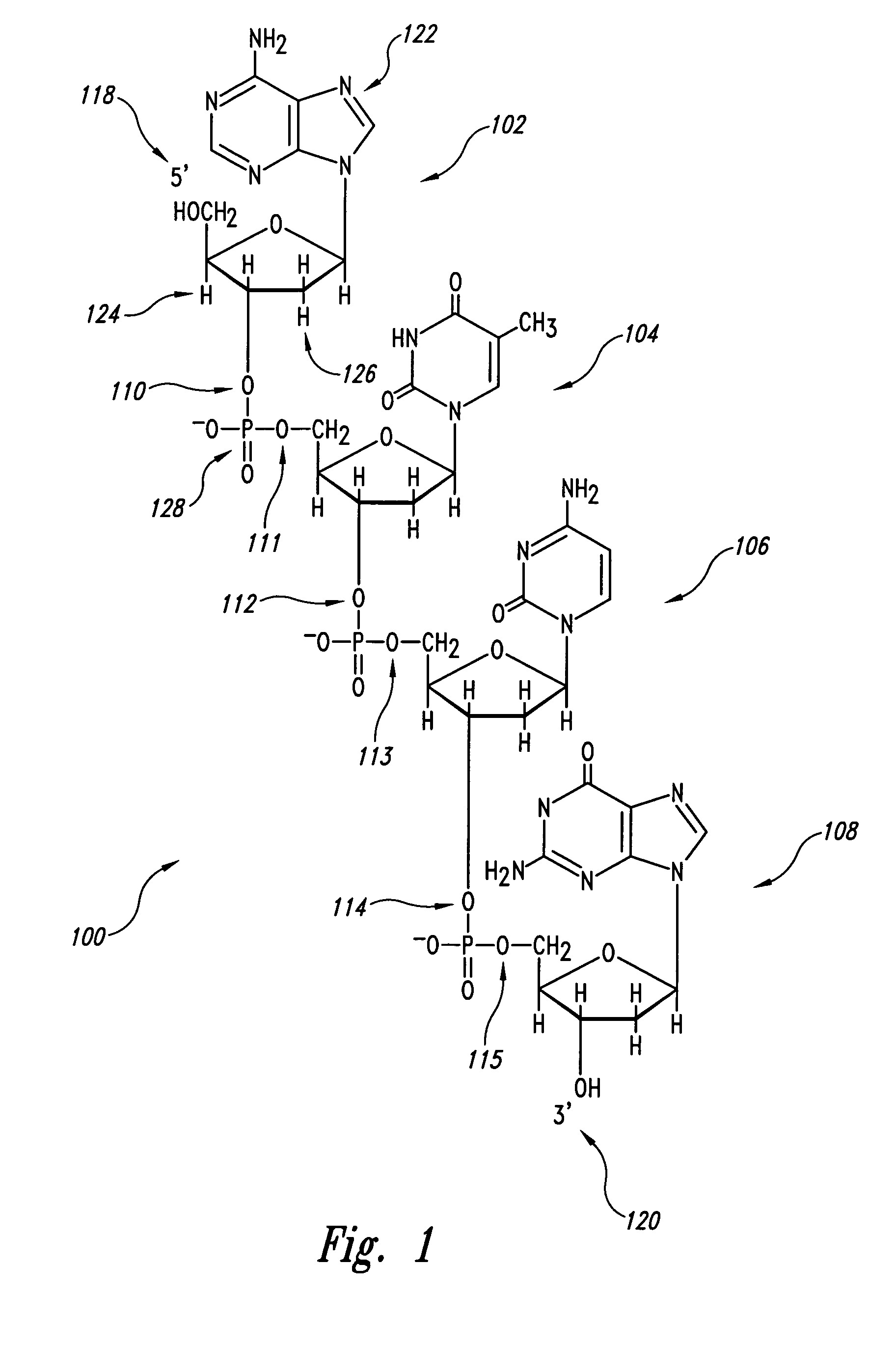
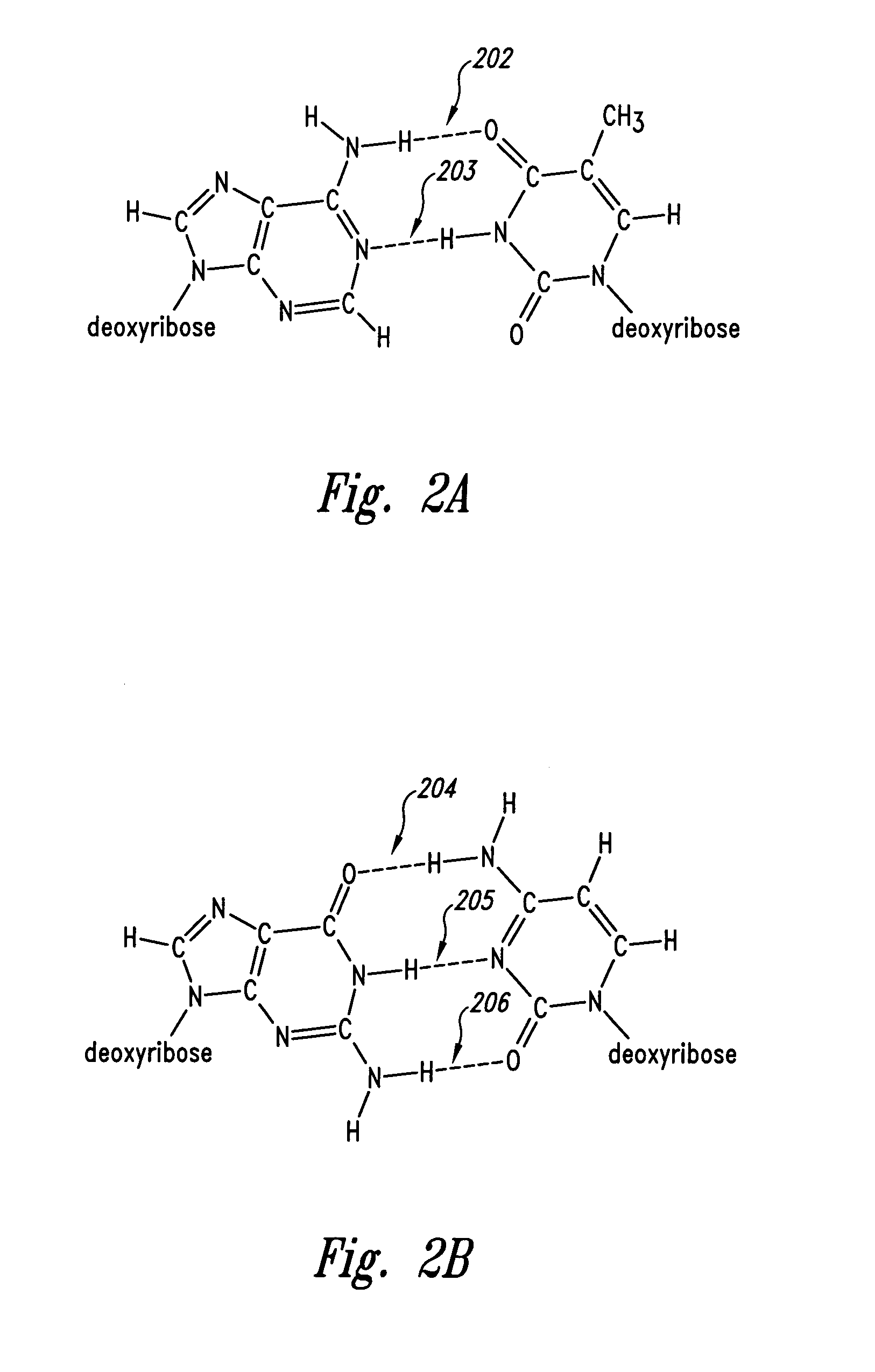
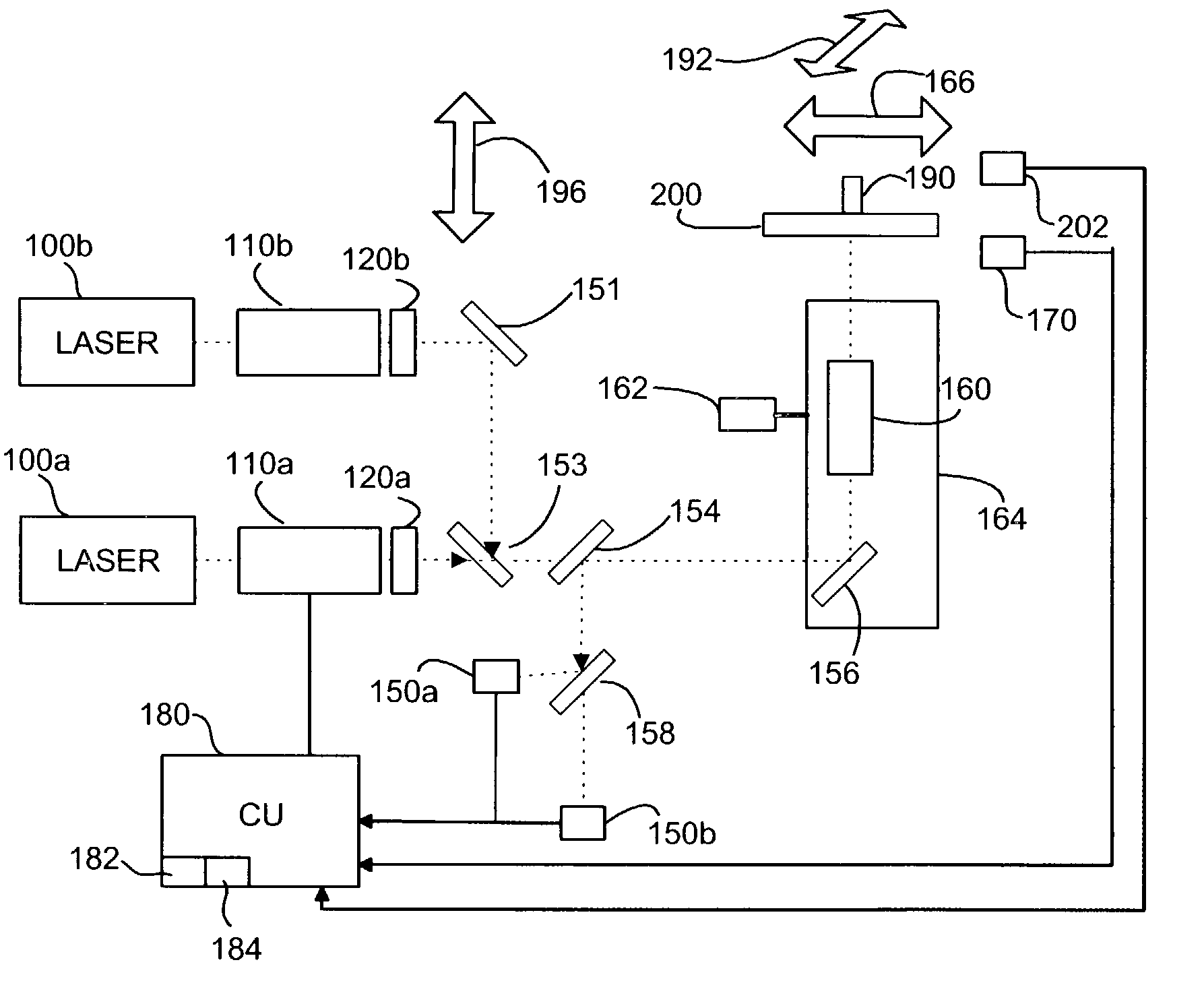
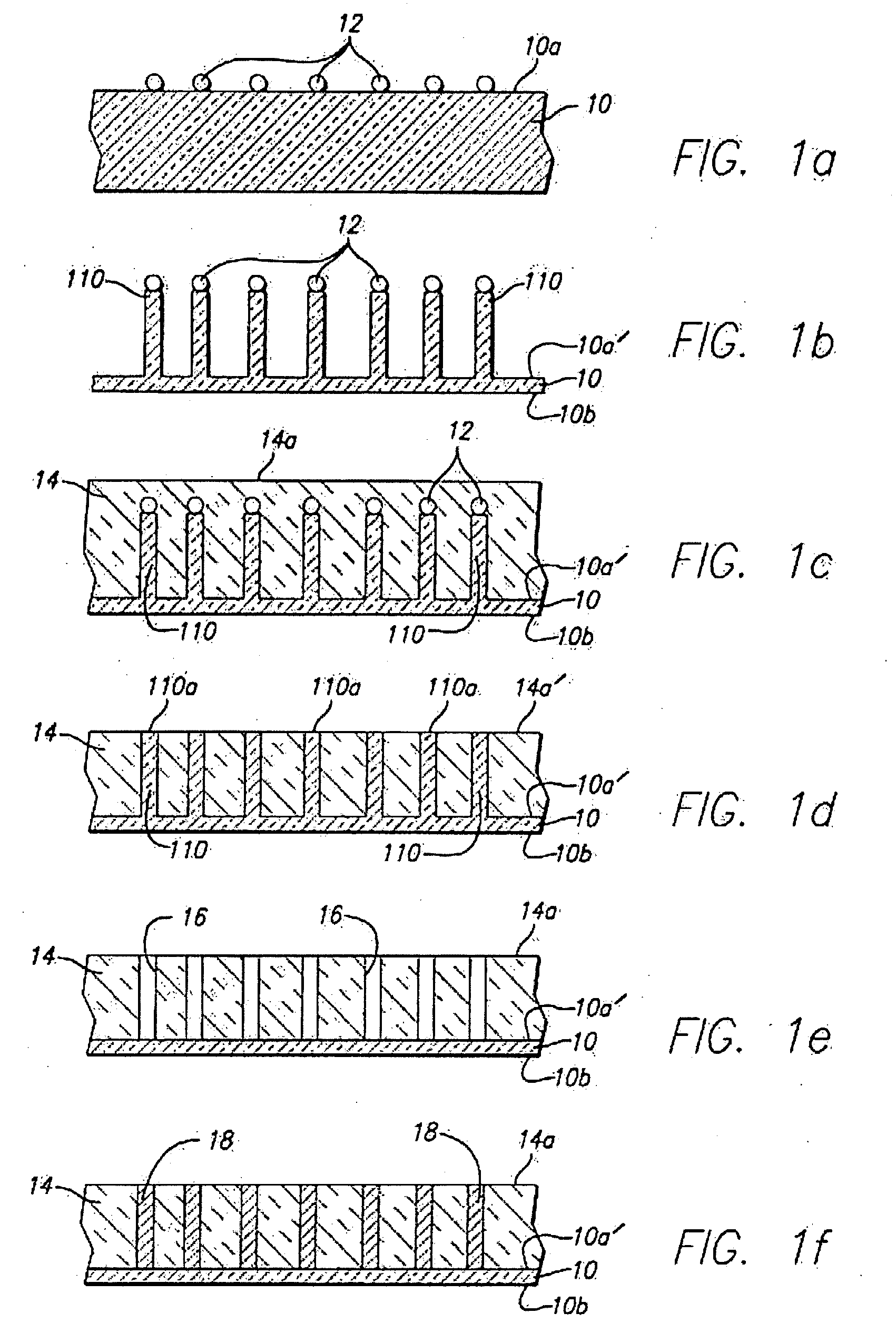
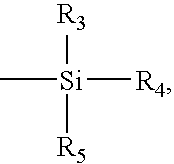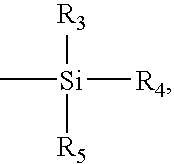Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
65 results about "Molecular array" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
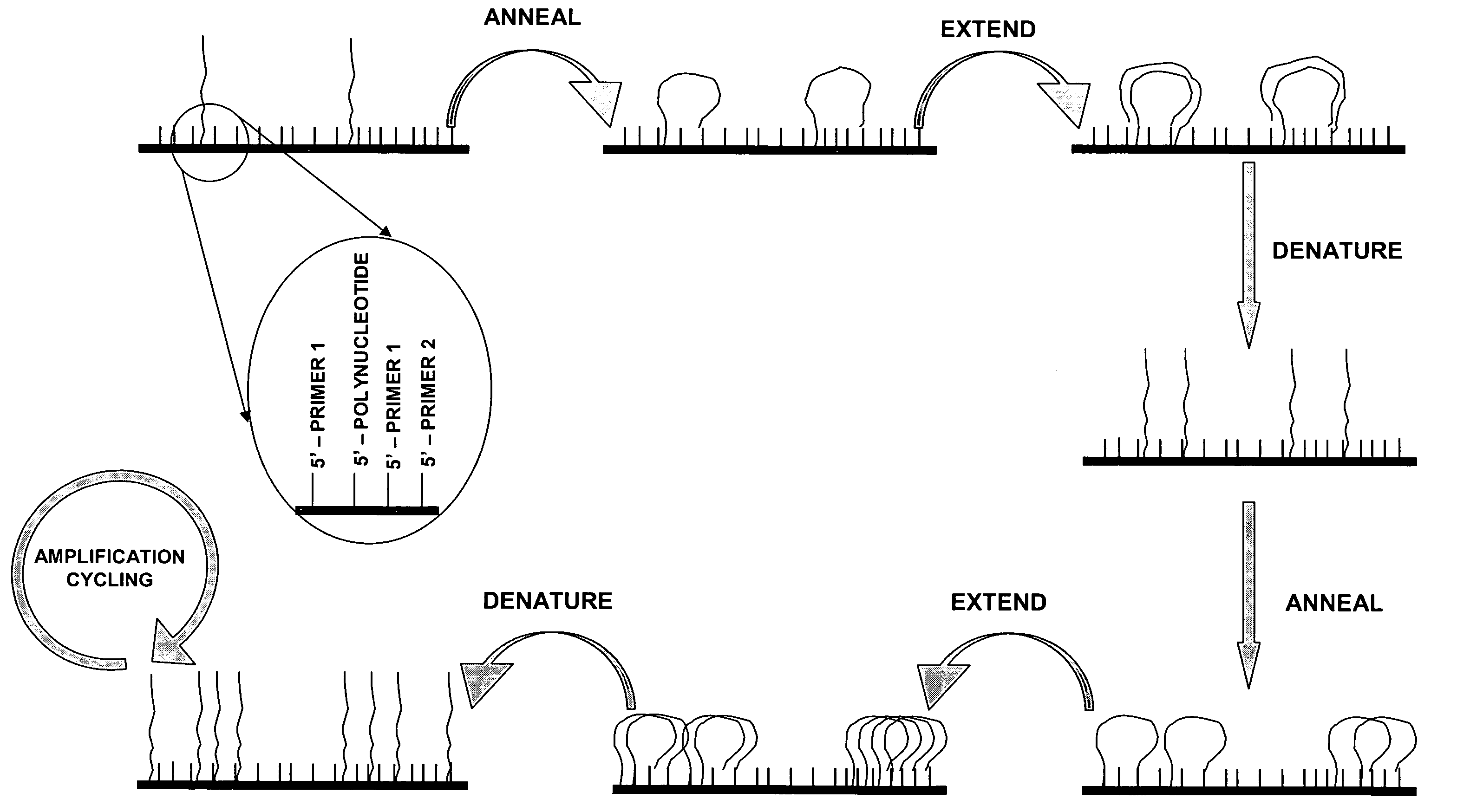
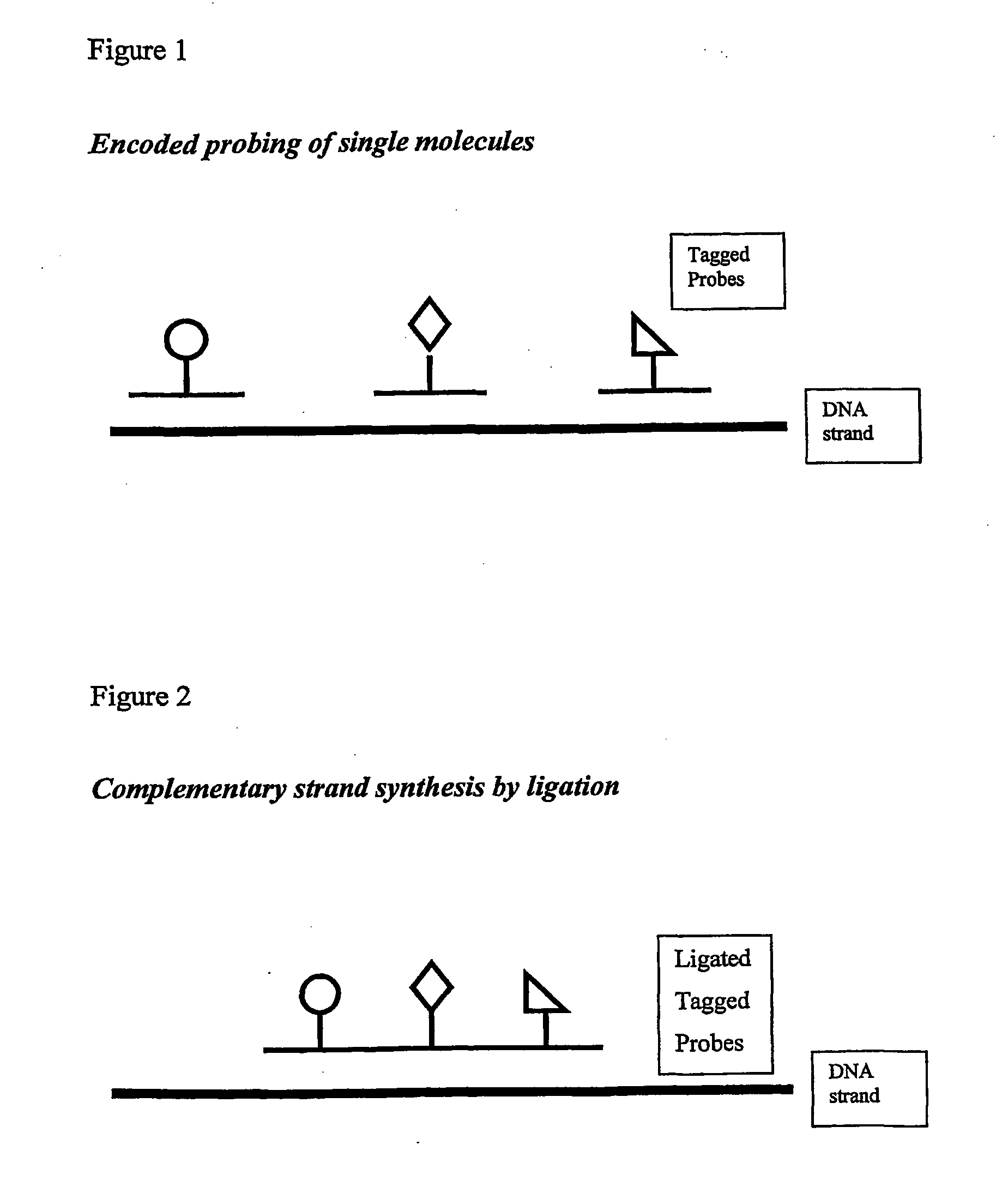
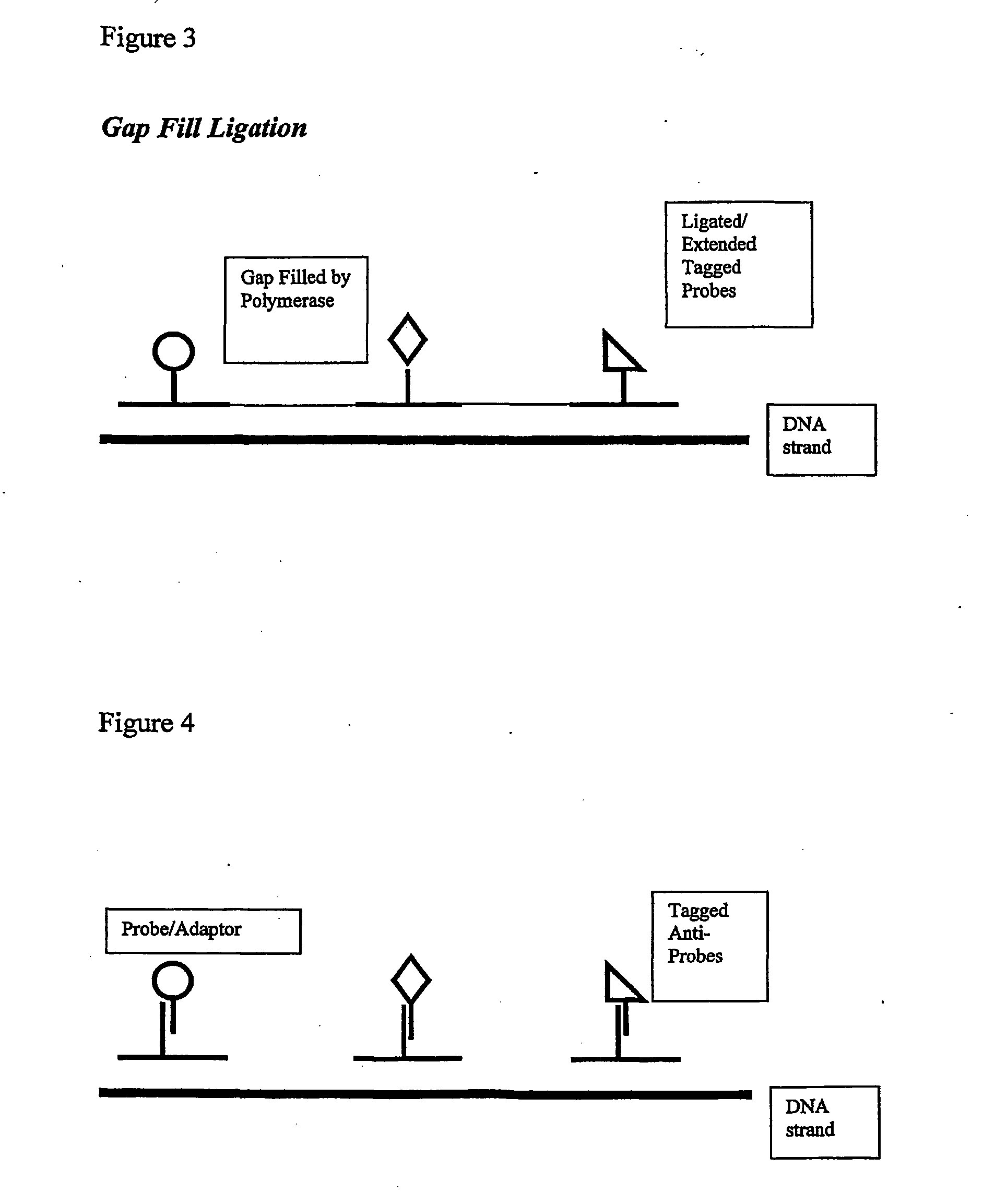
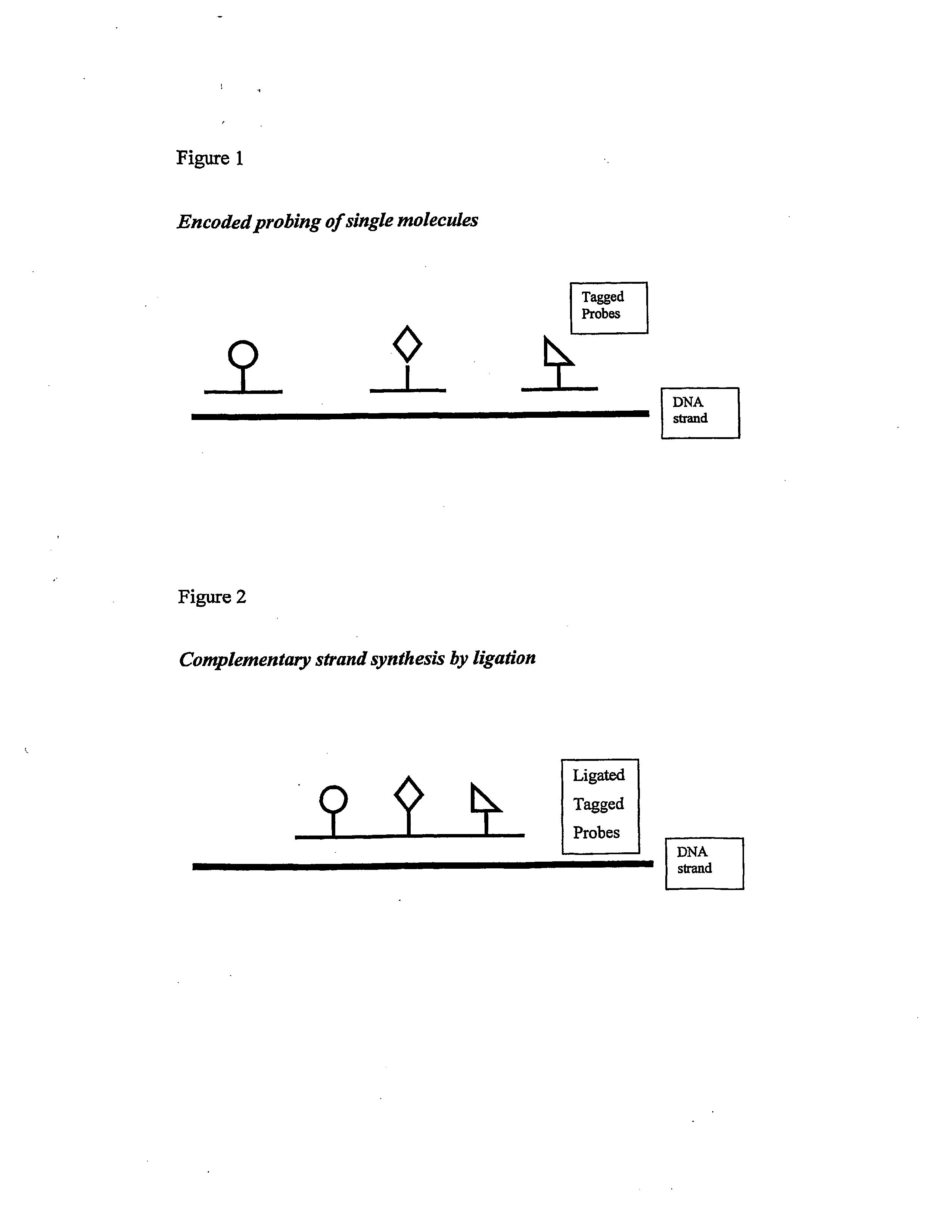
Isothermal methods for creating clonal single molecule arrays
InactiveUS20080009420A1Efficient amplificationIncrease diversityNucleotide librariesMicrobiological testing/measurementMicrobiologyNucleic acid sequencing
The present invention is directed to a method for isothermal amplification of a plurality of different target nucleic acids, wherein the different target nucleic acids are amplified using universal primers and colonies produced thereby can be distinguished from each other. The method, therefore, generates distinct colonies of amplified nucleic acid sequences that can be analyzed by various means to yield information particular to each distinct colony.
Owner:SOLEXA
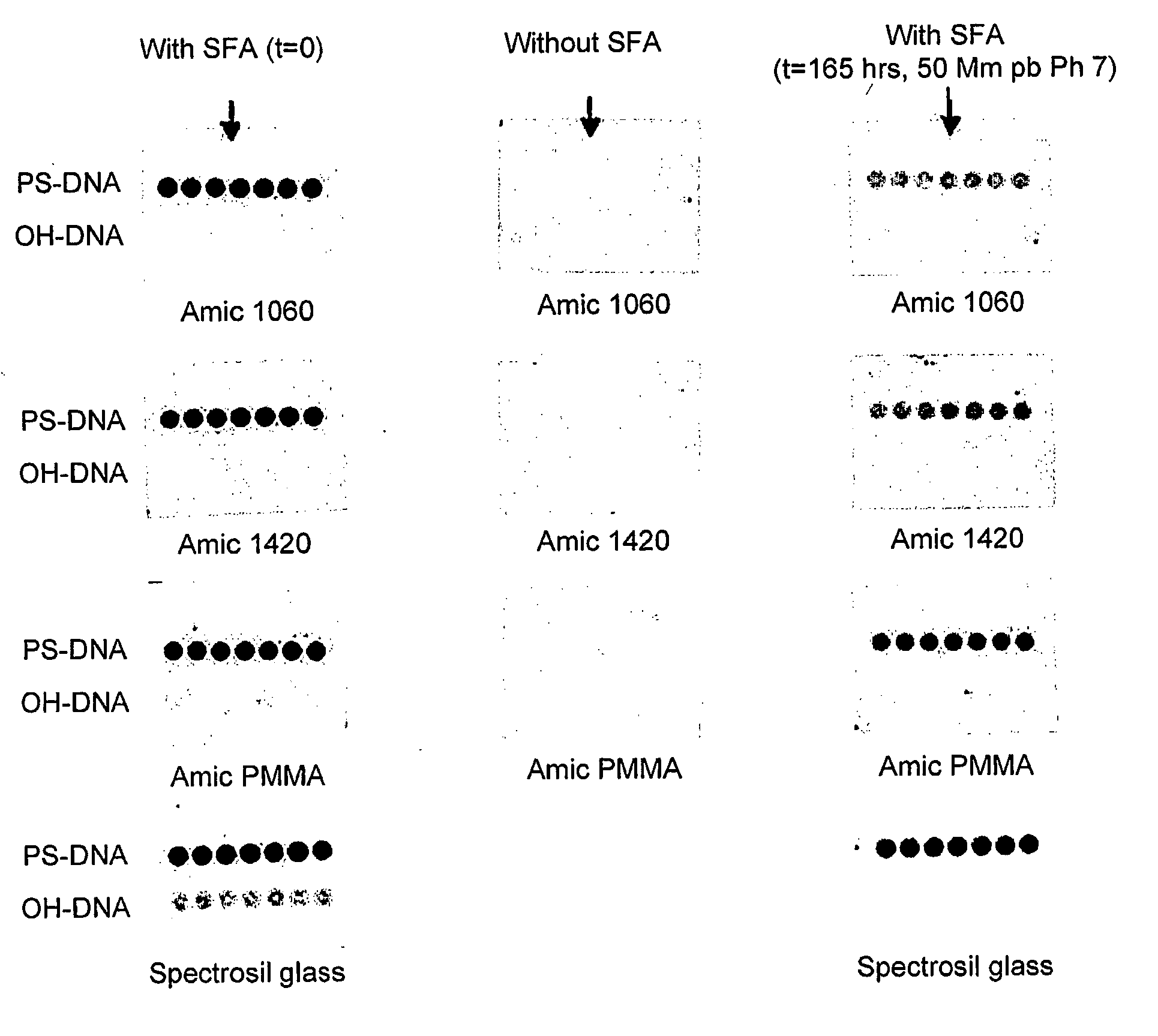
Modified Molecular Arrays
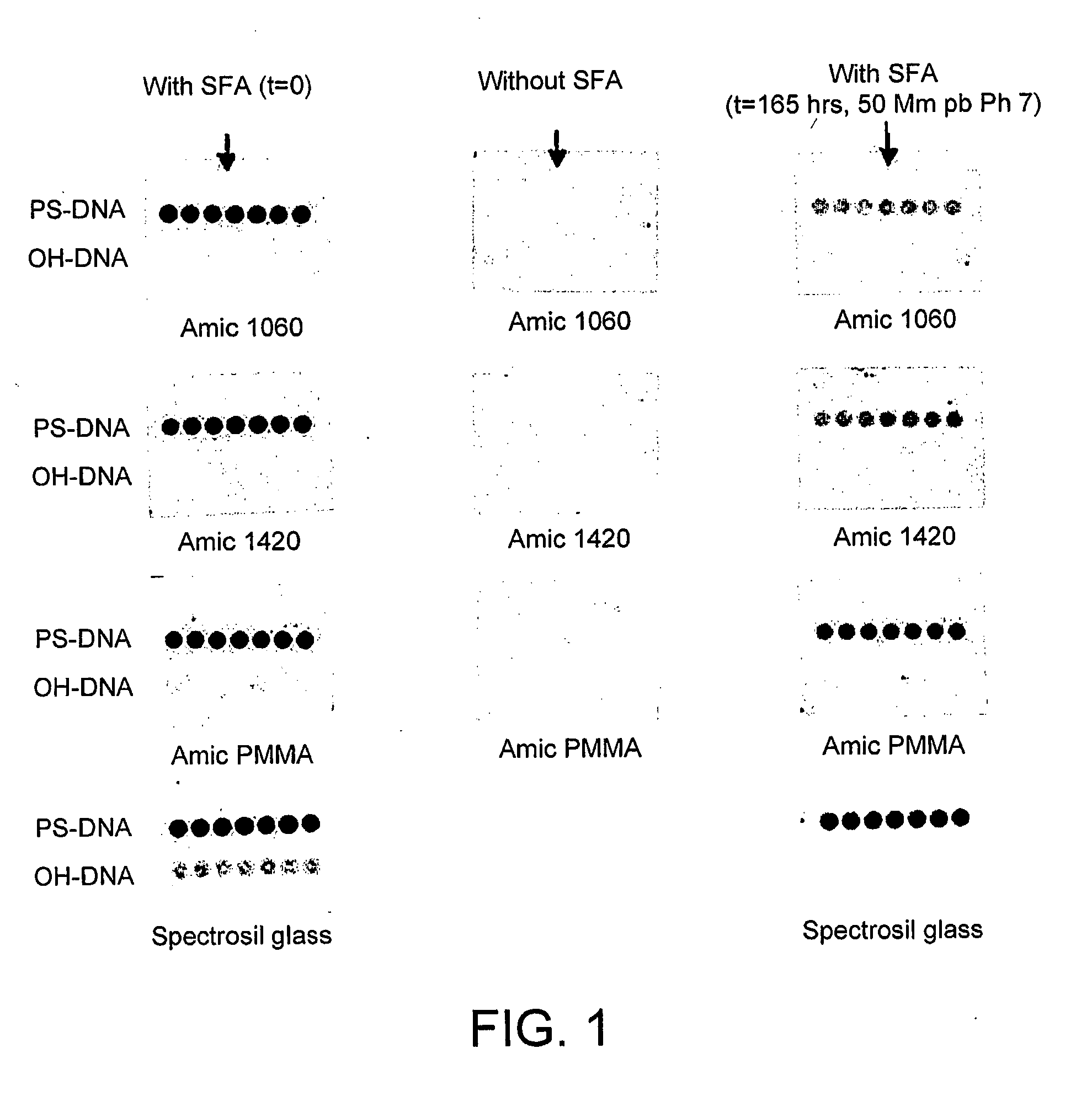
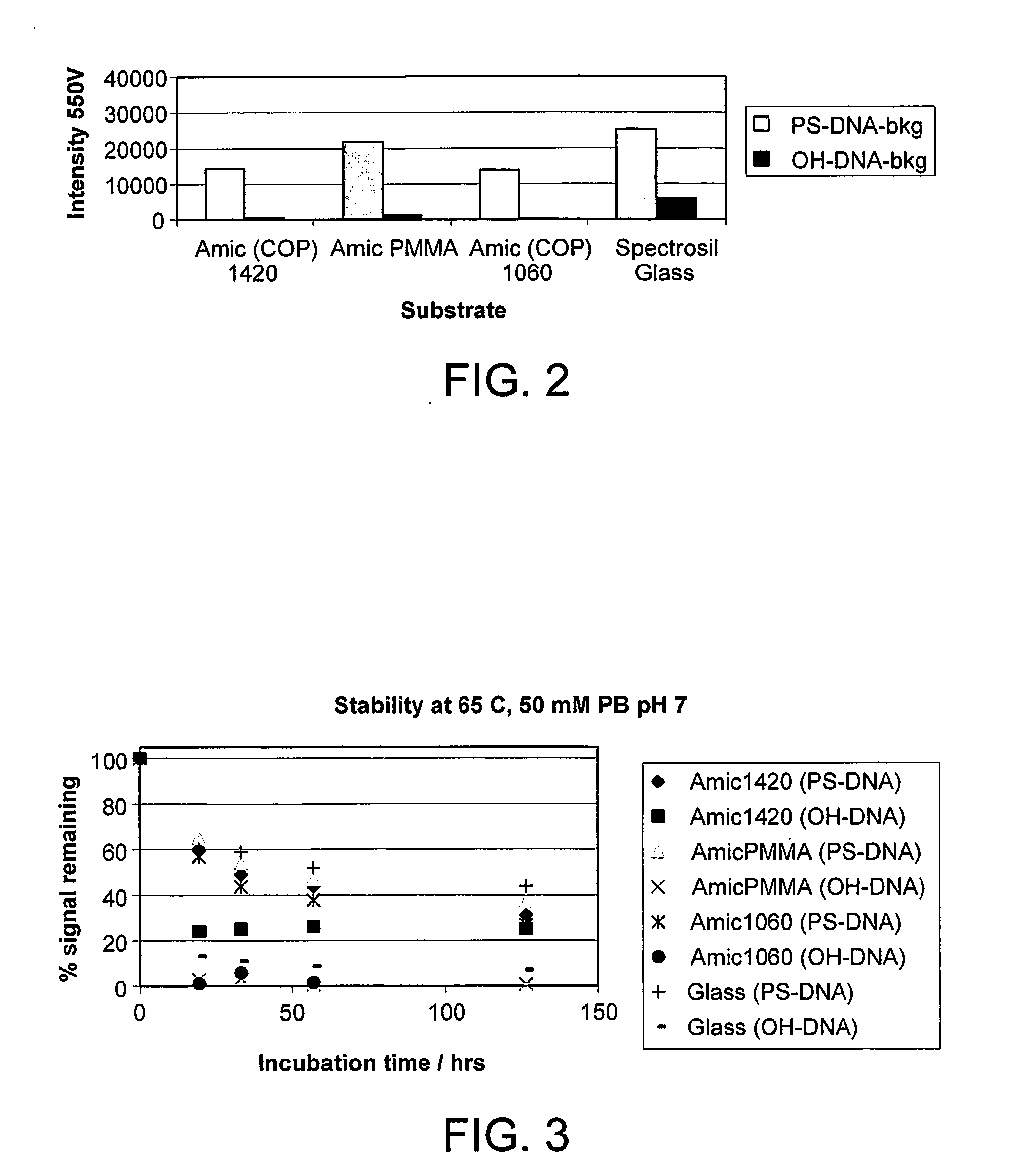
InactiveUS20110059865A1Less reactiveAccelerated programSequential/parallel process reactionsNucleotide librariesMolecular array(Hydroxyethyl)methacrylate
The invention relates to the preparation of a hydrogel surface useful in the formation and manipulation of arrays of molecules, particularly polynucleotides and to the chemical modification of these and other arrays. In particular, the invention relates to a method of preparing a hydrogel immobilised to a solid support comprising polymerising on the support a mixture of a first comonomer which is acrylamide, methacrylamide, hydroxyethyl methacrylate or N-vinyl pyrrolidinone and a second comonomer which is a functionalised acrylamide or acrylate.
Owner:ILLUMINA CAMBRIDGE LTD
Arrays and methods of use
InactiveUS20040248144A1Minimise photobleachingMinimizing transferMaterial nanotechnologyBioreactor/fermenter combinationsMolecular arrayAssay
Methods are provided for producing a molecular array comprising a plurality of molecules immobilised to a solid substrate at a density which allows individual immobilised molecules to be individually resolved, wherein each individual molecule in the array is spatially addressable and the identity of each molecule is known or determined prior to immobilisation. The use of spatially addressable lowdensity molecular arrays in single molecule detection and analysis techniques is also provided. Novel assays and methods are also provided.
Owner:INVITAE CORP
Molecular arrays and single molecule detection
InactiveUS20050244863A1Precision and richness of informationPrecision and richness of and speedBioreactor/fermenter combinationsMaterial nanotechnologyChemical physicsMolecular array
Methods are provided for producing a molecular array comprising a plurality of molecules immobilized to a solid substrate at a density which allows individual immobilized molecules to be individually resolved, wherein each individual molecule in the array is spatially addressable and the identity of each molecule is known or determined prior to immobilisation. The use of spatially addressable low density molecular arrays in single molecule detection techniques is also provided.
Owner:KALIM MIR +2
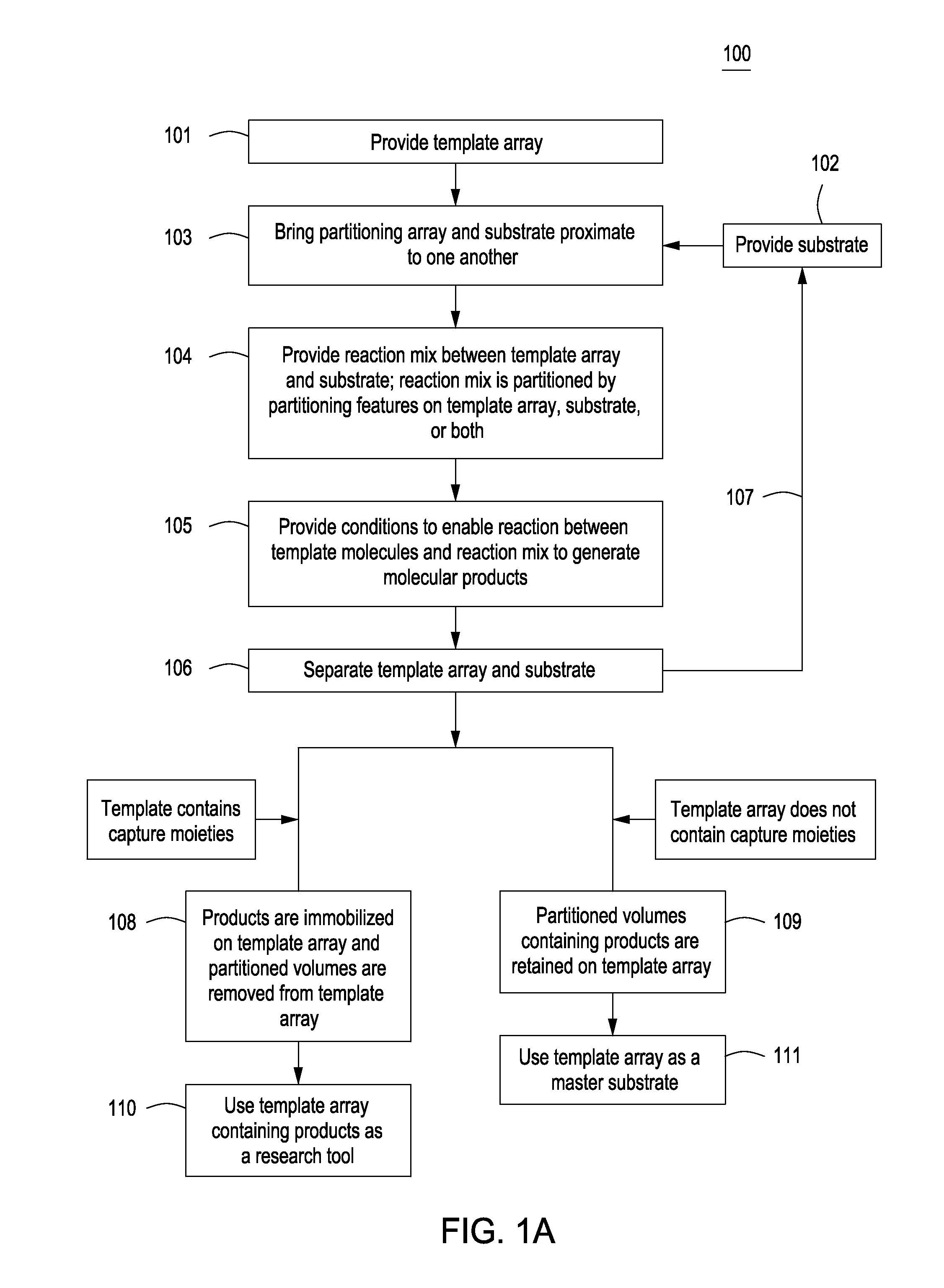
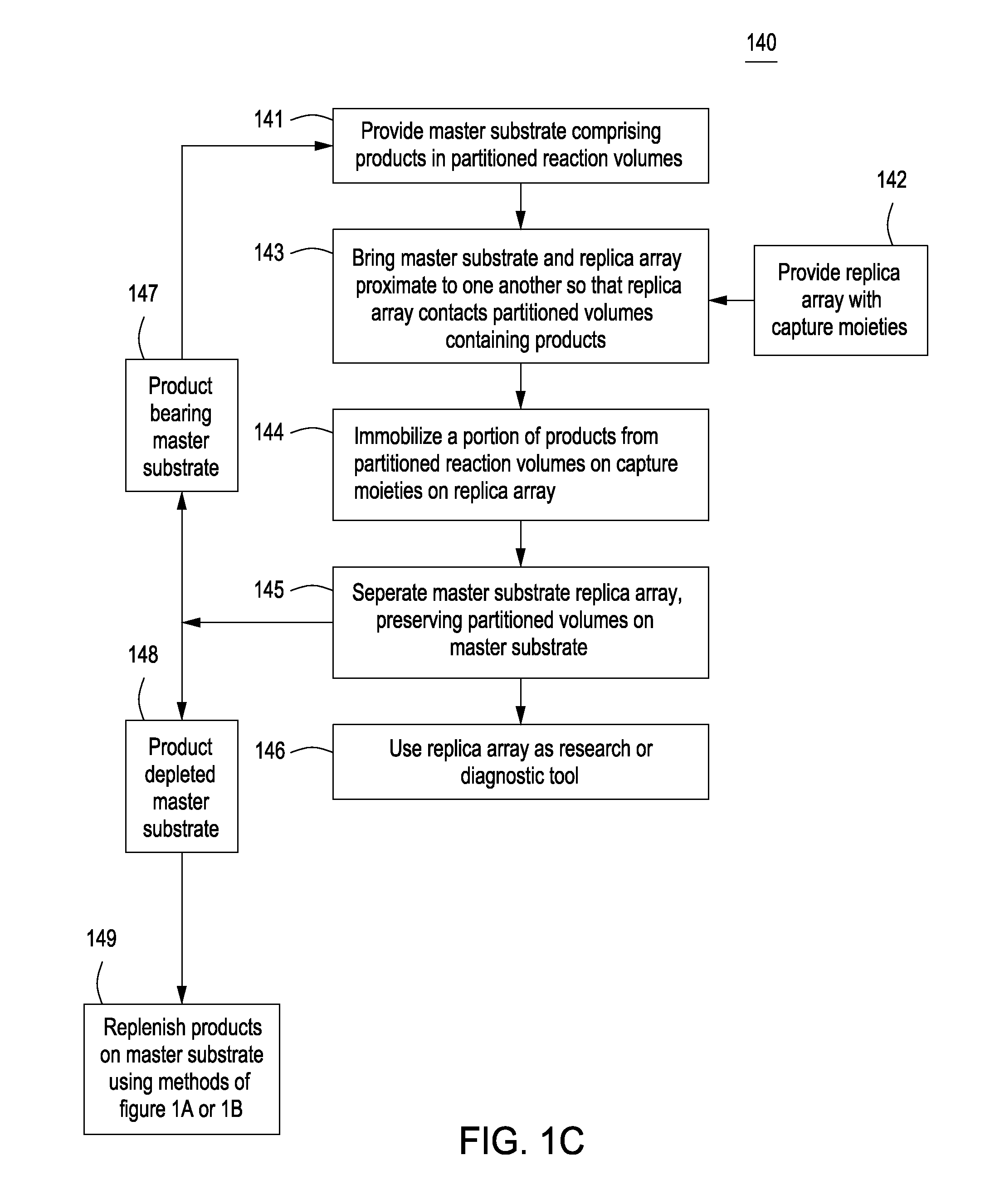
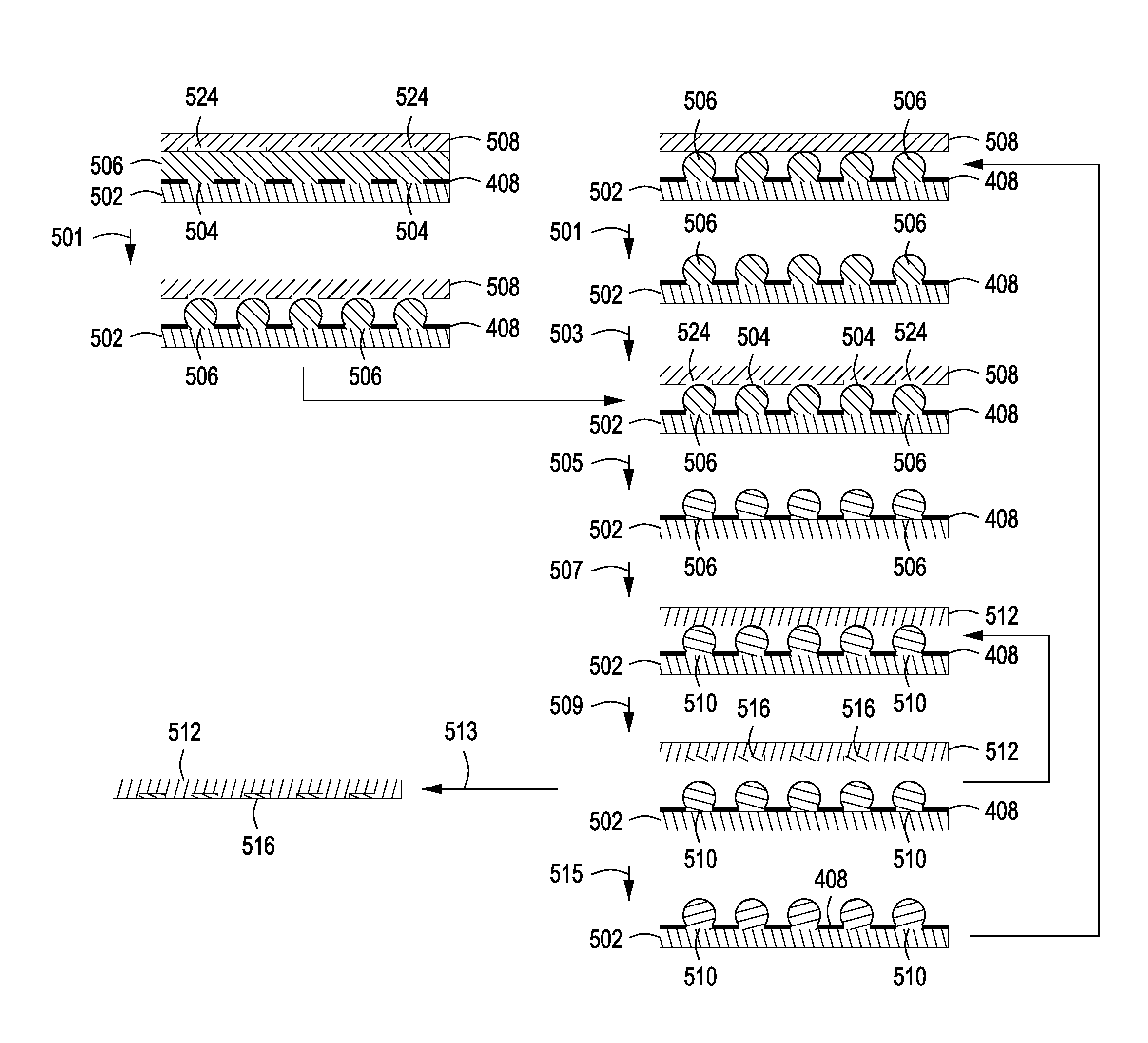
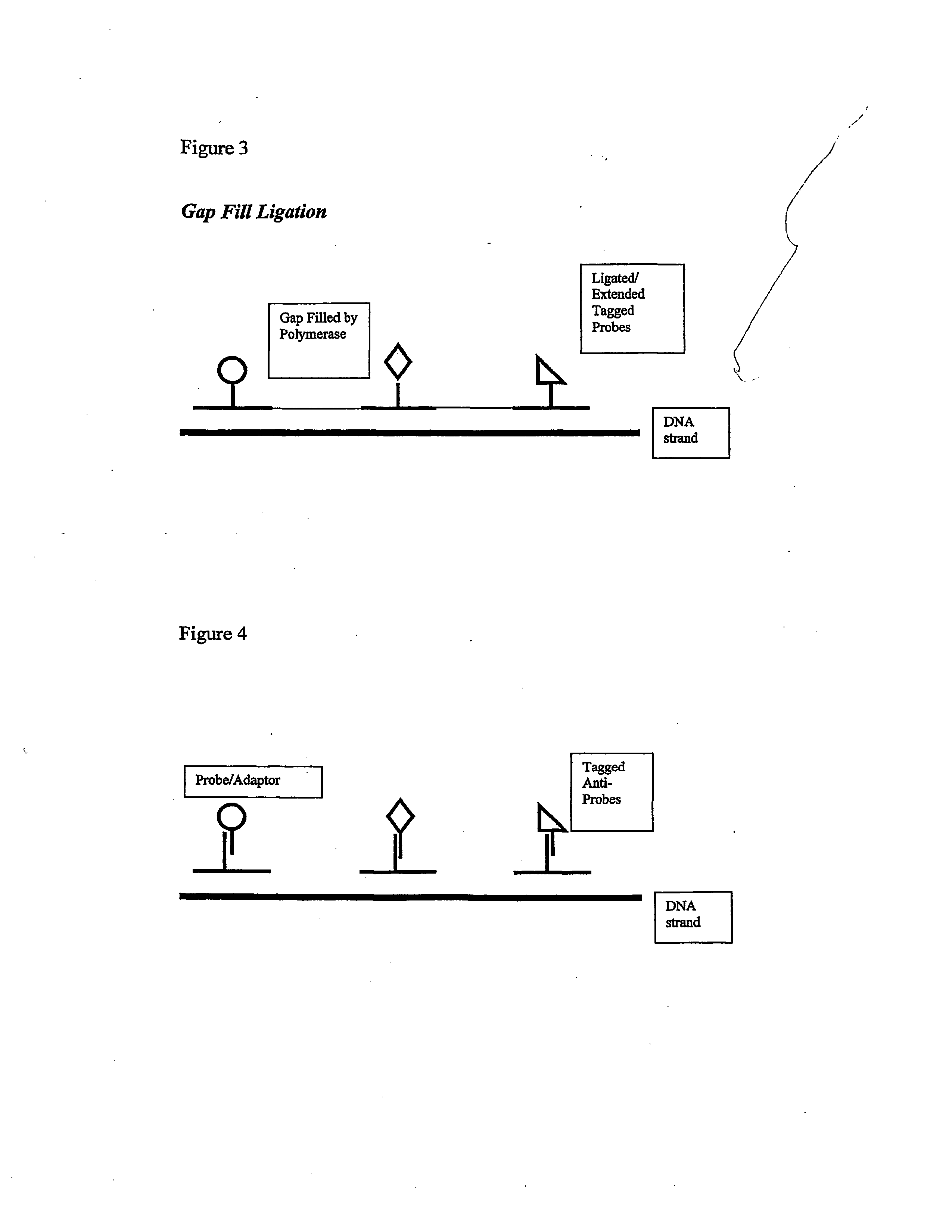
Methods for manufacturing molecular arrays
The methods of the present invention provide methods for manufacturing a master substrate and methods for manufacturing replica arrays from the master substrate. The methods may be used, for example, directly to manufacture or “print” peptide arrays from a DNA array; however, the methods are applicable to a wide range of manufacturing applications for use any time multiple copies of an array needs to be printed.
Owner:PROGNOSYS BIOSCI
Method for growing continuous fiber
This invention relates generally to a method for growing carbon fiber from single-wall carbon nanotube (SWNT) molecular arrays. In one embodiment, the present invention involves a macroscopic molecular array of at least about 106 tubular carbon molecules in generally parallel orientation and having substantially similar lengths in the range of from about 50 to about 500 nanometers. The hemispheric fullerene cap is removed from the upper ends of the tubular carbon molecules in the array. The upper ends of the tubular carbon molecules in the array are then contacted with a catalytic metal. A gaseous source of carbon is supplied to the end of the array while localized energy is applied to the end of the array in order to heat the end to a temperature in the range of about 500° C. to about 1300° C. The growing carbon fiber is continuously recovered.
Owner:RICE UNIV
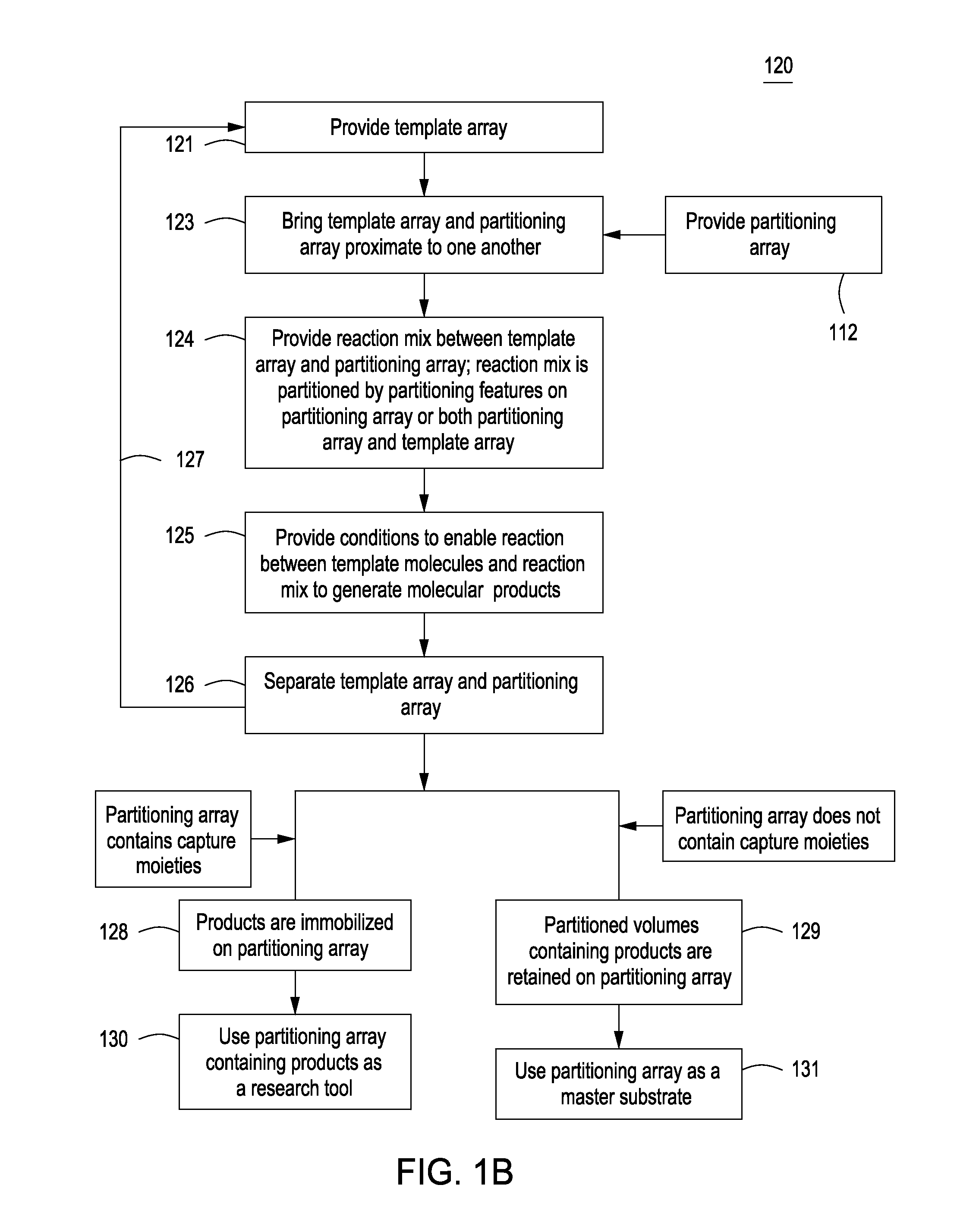
Methods for manufacturing molecular arrays
ActiveUS20130096033A1Sequential/parallel process reactionsNucleotide librariesMolecular arrayPeptide array
The methods of the present invention provide methods for manufacturing a master substrate and methods for manufacturing replica arrays from the master substrate. The methods may be used, for example, directly to manufacture or “print” peptide arrays from a DNA array; however, the methods are applicable to a wide range of manufacturing applications for use any time multiple copies of an array needs to be printed.
Owner:PROGNOSYS BIOSCI
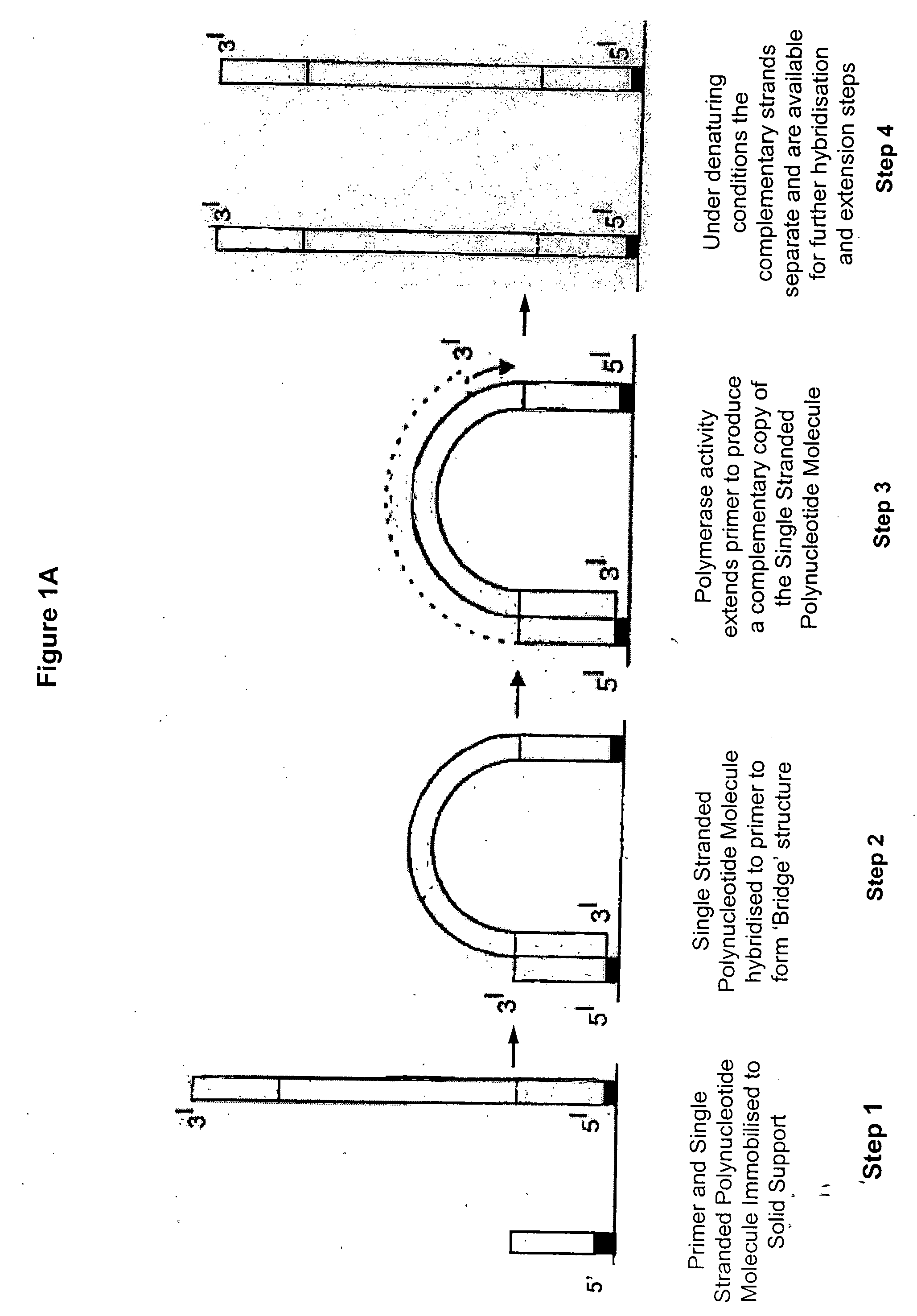
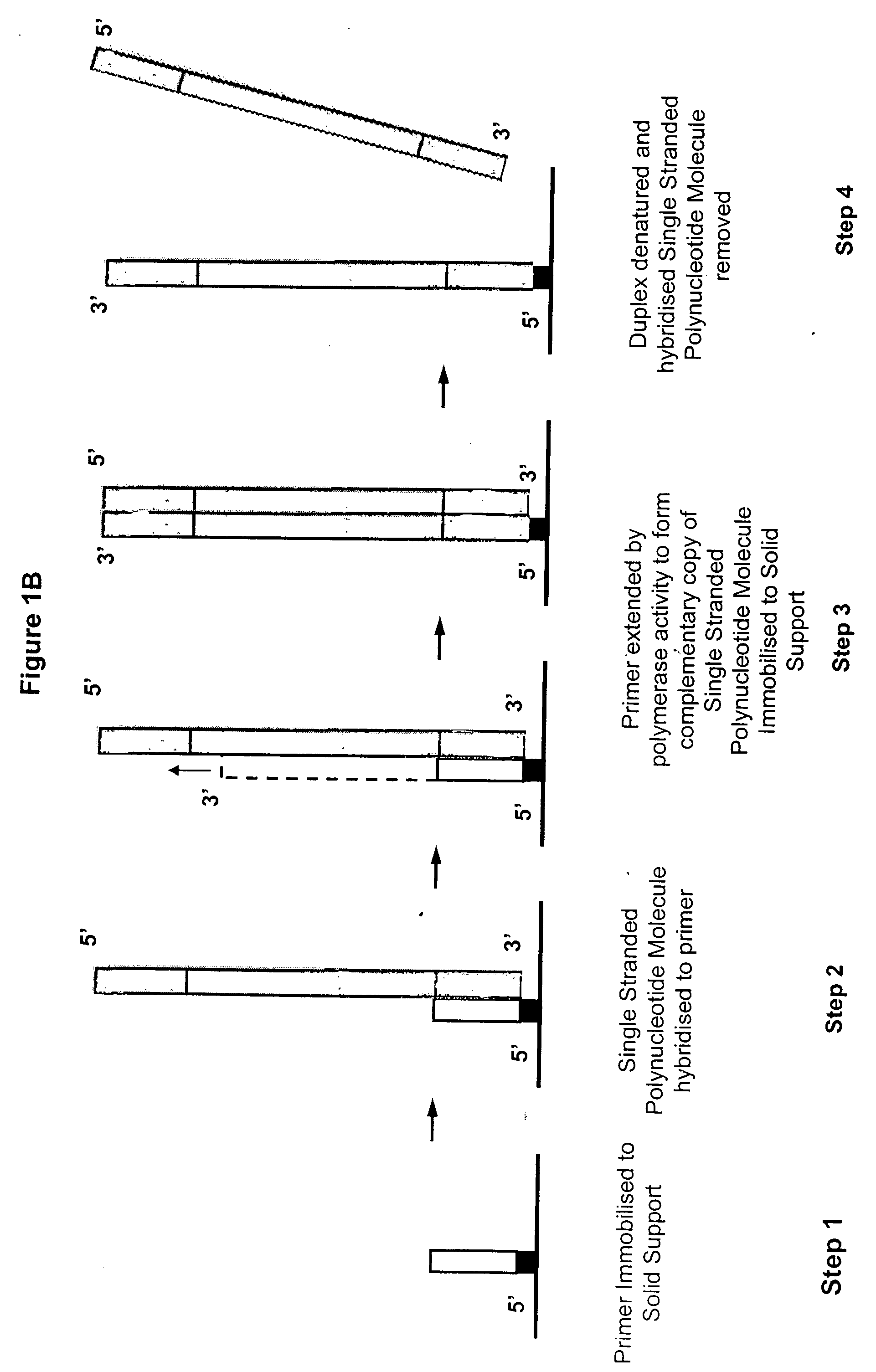
Synthesis of spatially addressed molecular arrays
InactiveUS7384737B2Maximal diversityApproach is usefulBioreactor/fermenter combinationsSequential/parallel process reactionsMolecular arrayPolynucleotide
Owner:SOLEXA
Arrays with cleavable linkers
InactiveUS20070213278A1Treating and preventing HIV infectionAntibacterial agentsBiocideGlycanTest sample
Owner:THE SCRIPPS RES INST
Method and system for measuring a molecular array background signal from a continuous background region of specified size
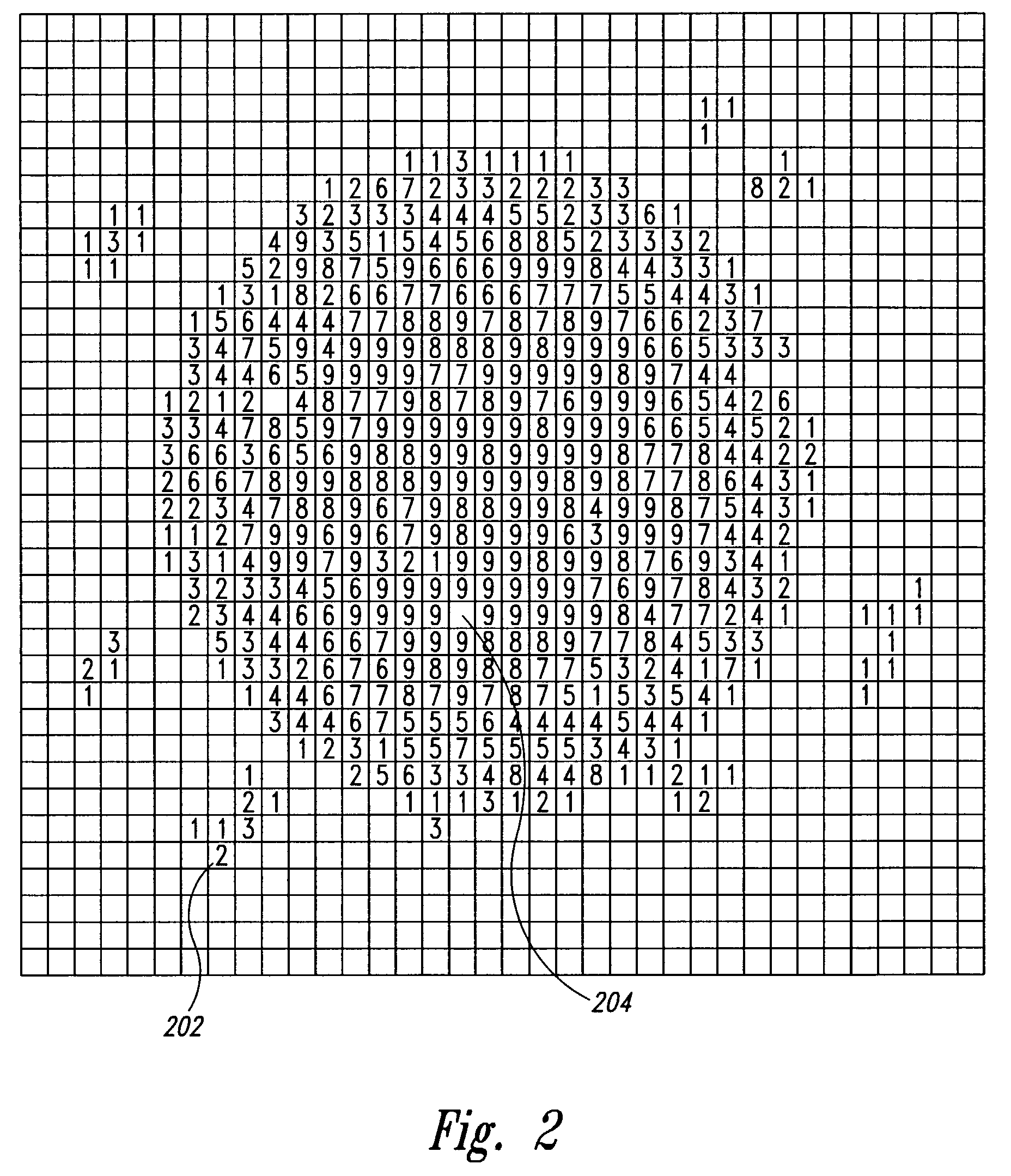
InactiveUS7221785B2Efficiently determinedGenerate efficientlyImage enhancementImage analysisData setMolecular array
A method and system for estimating the background signal over an arbitrarily-sized region of a scanned image of a molecular array, including a background region surrounding the ROI corresponding to the feature. A bit mask is generated, based on a molecular-array, feature-based data set that includes pixel-based intensities and a list of features, including feature coordinates and feature ROI radii, to indicate those pixels in the scanned image of the molecular array corresponding to background, and those pixels in the scanned image of the molecular array corresponding to features and ROIs. An integrated intensity for a background region of arbitrary size and shape can be efficiently determined by selecting pixels within the background region that are indicated to be background pixels in the bit mask. By selecting background anuli of sufficient size to overlap with the background anuli of neighboring features, a relatively continuous function of background-signal-verusus-position can be obtained across the surface of a molecular array in order to examine non-local, background-signal-related phenomena.
Owner:AGILENT TECH INC
Method and system for molecular array scanner calibration
InactiveUS6929951B2Bioreactor/fermenter combinationsBiological substance pretreatmentsMolecular arrayConcentration ratio
A method and system for calibrating molecular arrays to a reference molecular array, and for subsequently calibrating the molecular arrays to maintain a constant signal-intensity-to-label-concentration ratio. In the first step of the two-step calibration method, a reference array coated with the fluorophore or chromophore used to label probe molecules is employed, while in the second step of the two-step method, a reference array coated with a stable dye is employed.
Owner:AGILENT TECH INC
Method for synthesizing a specific, surface-bound polymer uniformly over an element of a molecular array
InactiveUS6936472B2Reduce synthesisMaterial nanotechnologyLibrary screeningMolecular arrayComputational chemistry
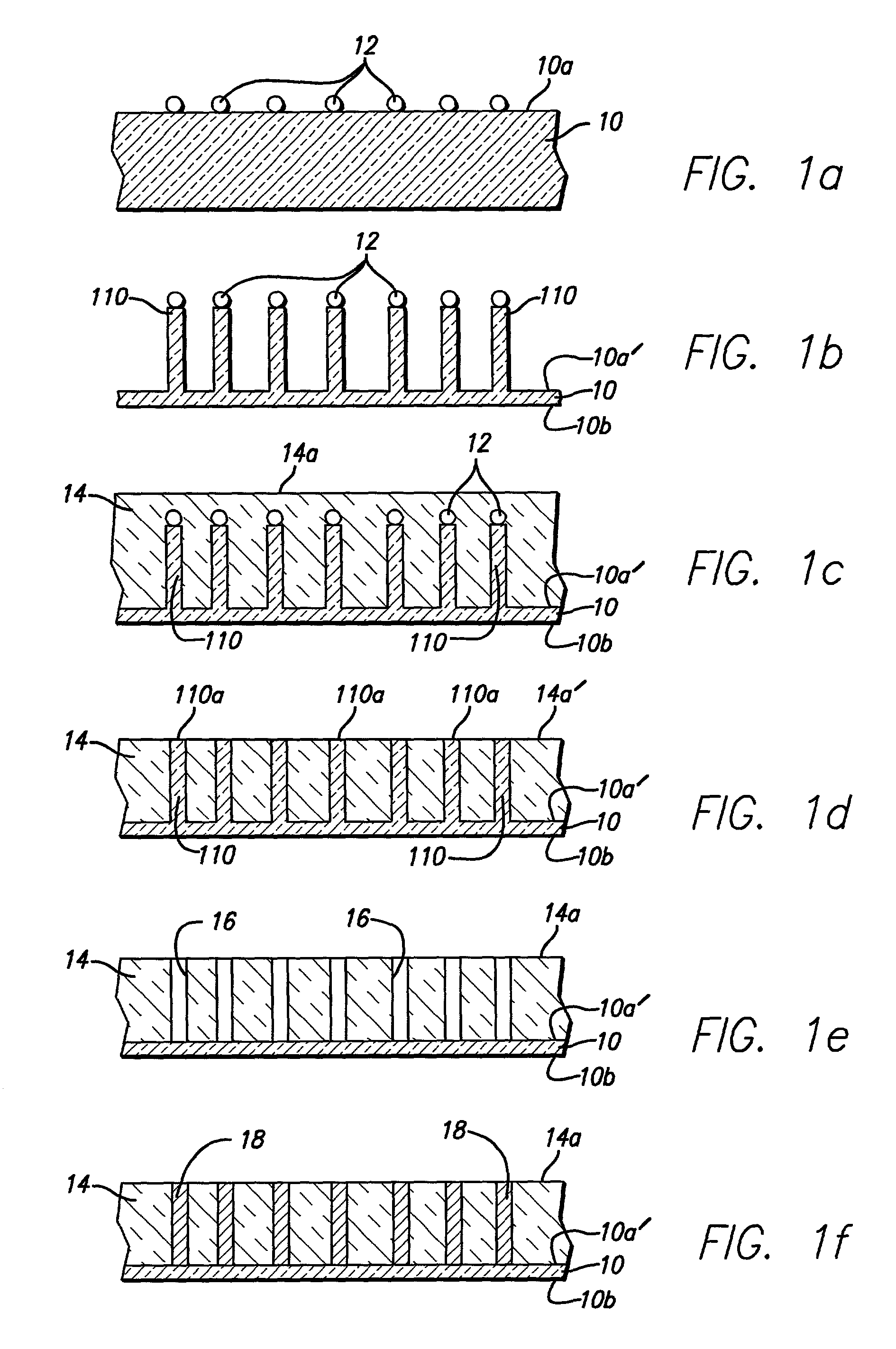
A method for specifically and uniformly synthesizing desired polymers within molecular array elements. Droplets containing a reactive monomer are successively applied to the elements of a molecular array in order to synthesize a substrate-bound polymer. Application of an initial droplet, having a first volume, defines the position and size of a molecular array element. Subsequent droplets are applied, to add successive reactive monomers to growing nascent polymers within the molecular array element, with covering volumes so that, even when application of the subsequent droplets is misregistered, the entire surfaces of the elements of the molecular array are exposed to the subsequently applied droplets. Following application of initial droplets, the surface of the molecular array is exposed to a solution containing a very efficient capping agent in order to chemically cap any unreacted nascent growing polymers and any unreacted substrate molecules.
Owner:AGILENT TECH INC
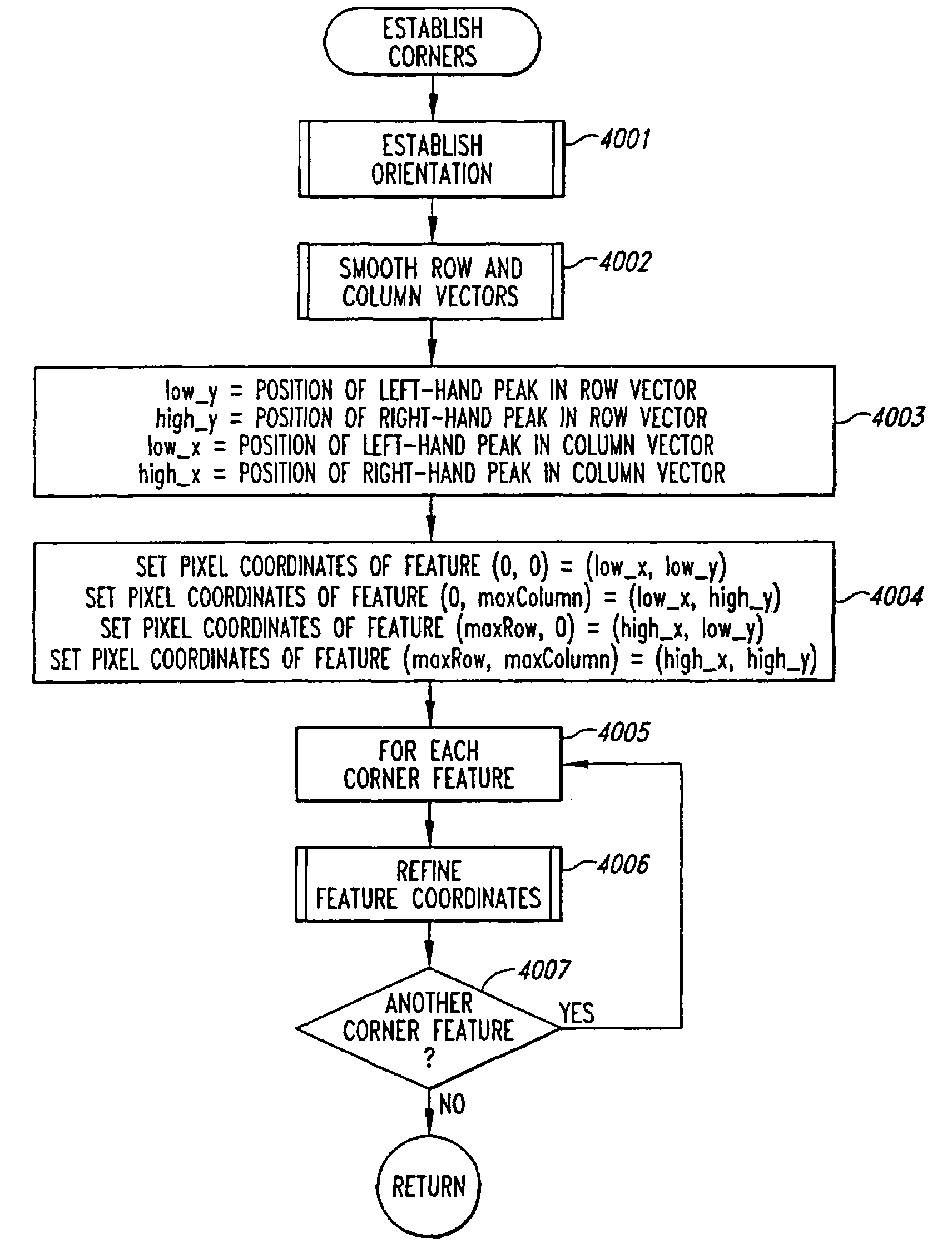
Method and system for extracting data from surface array deposited features
InactiveUS7006927B2Easy to processImprove signal-to-noise ratioImage enhancementRadiation pyrometryMolecular arrayConfidence interval
A method and system for extracting data signals from a scanned image resulting from optical, radiometric, or other types of analysis of a molecular array. The positions of corner features are first located. Then, an initial feature coordinate grid is determined from the positions of the corner features. A refined feature coordinate grid is then calculated based on the positions of strong features, and is used to identify the positions of weak features and the positions of the local background regions surrounding all features. Finally, signal intensity values are extracted from the features and their respective local background regions in the scanned image, and background-subtracted signal intensity values, background-subtracted and normalized signal intensity ratios, and variability information and confidence intervals are determined based on the extracted values.
Owner:AGILENT TECH INC
Method of forming one or more nanopores for aligning molecules for molecular electronics
InactiveUS20030116531A1Less sensitiveDecorative surface effectsNanoinformaticsMolecular arrayMolecular electronics
A technique is provided for forming a molecule or an array of molecules having a defined orientation relative to the substrate or for forming a mold for deposition of a material therein. The array of molecules is formed by dispersing them in an array of small, aligned holes (nanopores), or mold, in a substrate. Typically, the material in which the nanopores are formed is insulating. The underlying substrate may be either conducting or insulating. For electronic device applications, the substrate is, in general, electrically conducting and may be exposed at the bottom of the pores so that one end of the molecule in the nanopore makes electrical contact to the substrate. A substrate such as a single-crystal silicon wafer is especially convenient because many of the process steps to form the molecular array can use techniques well developed for semiconductor device and integrated-circuit fabrication.
Owner:HEWLETT PACKARD DEV CO LP
Arrays comprising pre-labeled biological molecules and methods for making and using these arrays
InactiveUS20050032060A1Bioreactor/fermenter combinationsPeptide librariesMolecular arrayComparative genomic hybridization
The invention provides arrays comprising a plurality of biological molecules, wherein each biological molecule is immobilized to a discrete and known spot on a substrate surface to form an array of biological molecules, and each biological molecule comprises a detectable label. The invention provides a multiplexed system for performing comparative genomic hybridization (CGH) using an array of the invention. The invention also provides methods of making and using these arrays, including a method of comparing copy numbers of unique nucleic acid sequences in a sample.
Owner:PERKINELMER HEALTH SCIENCES INC
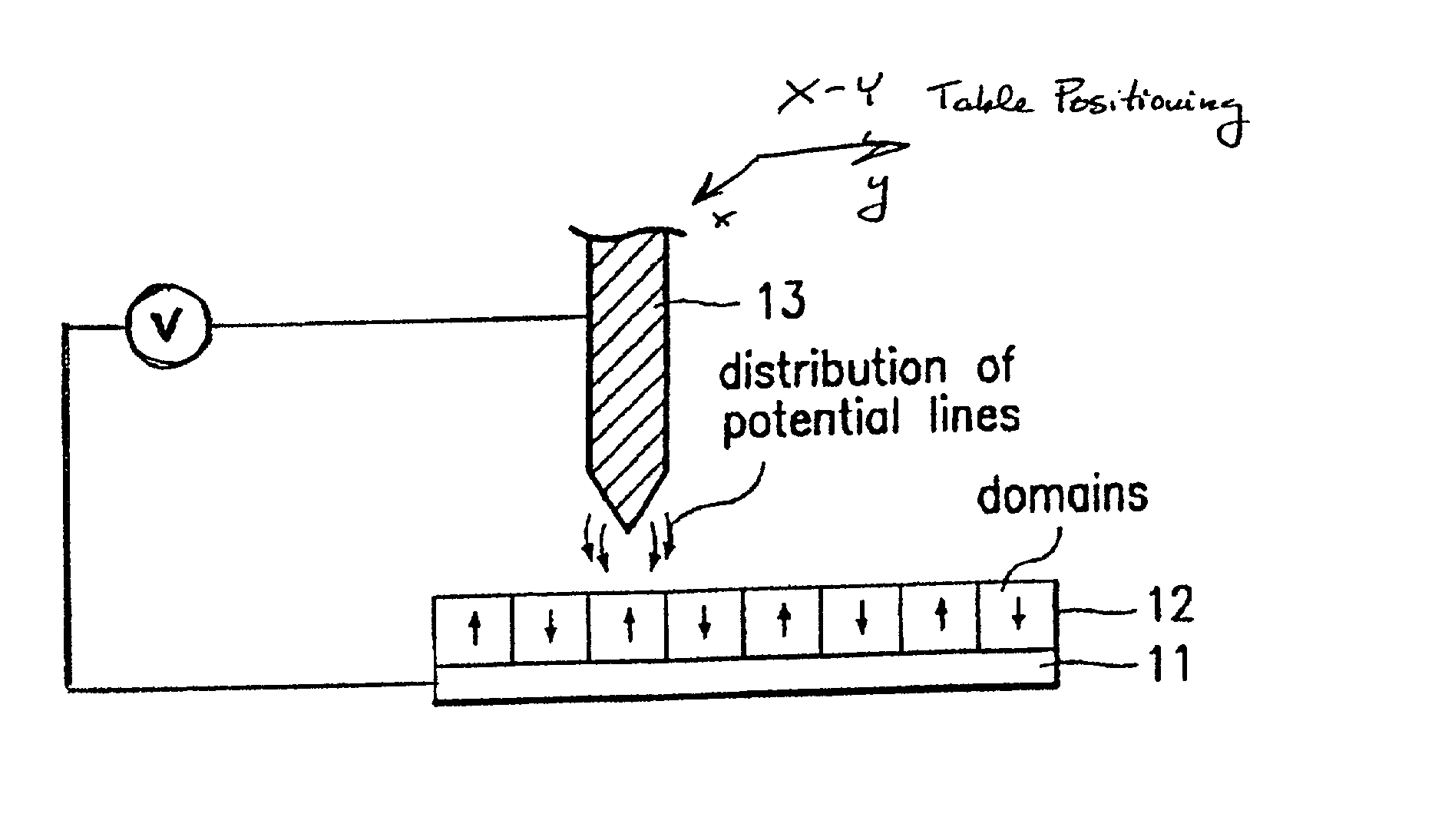
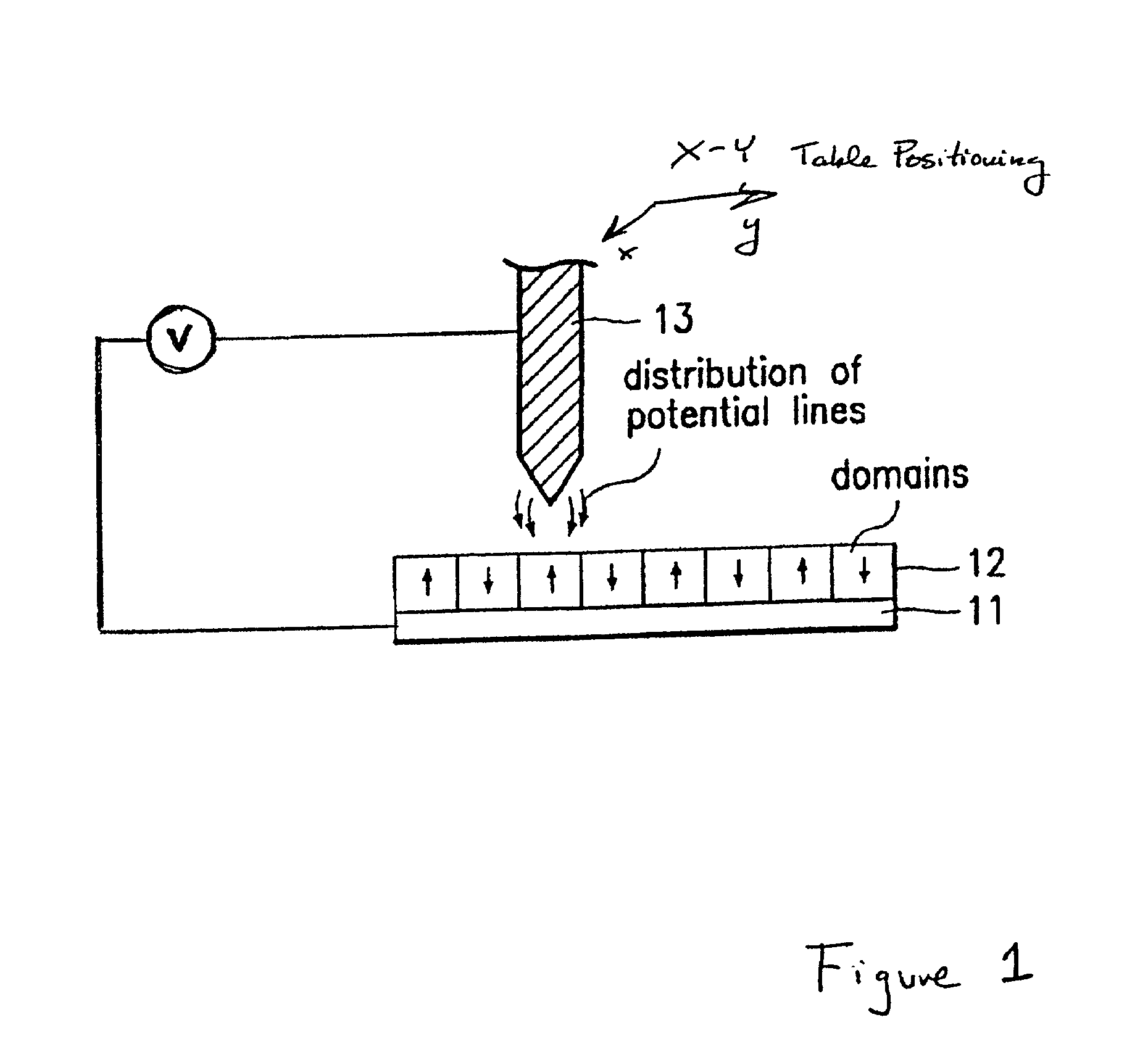
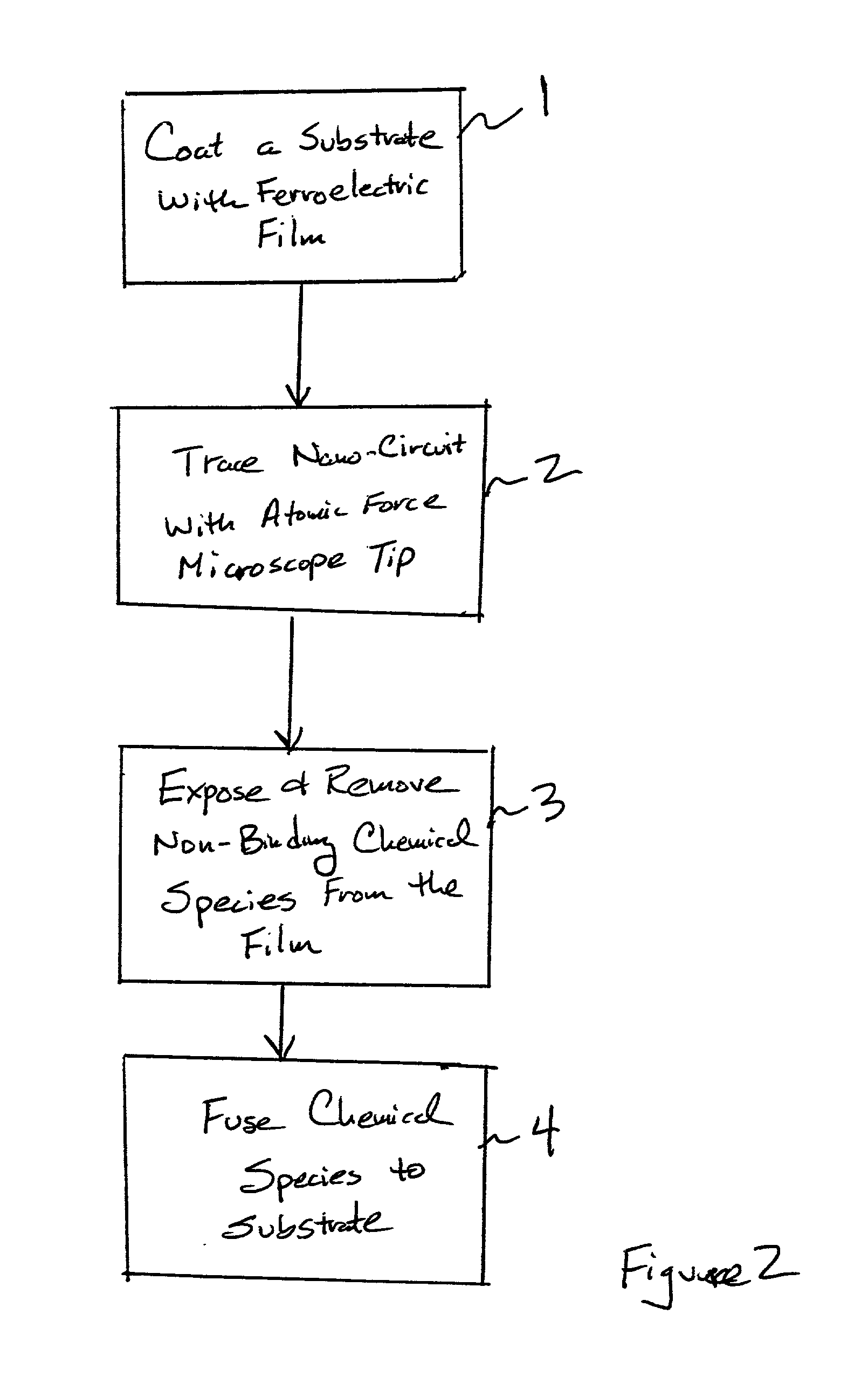
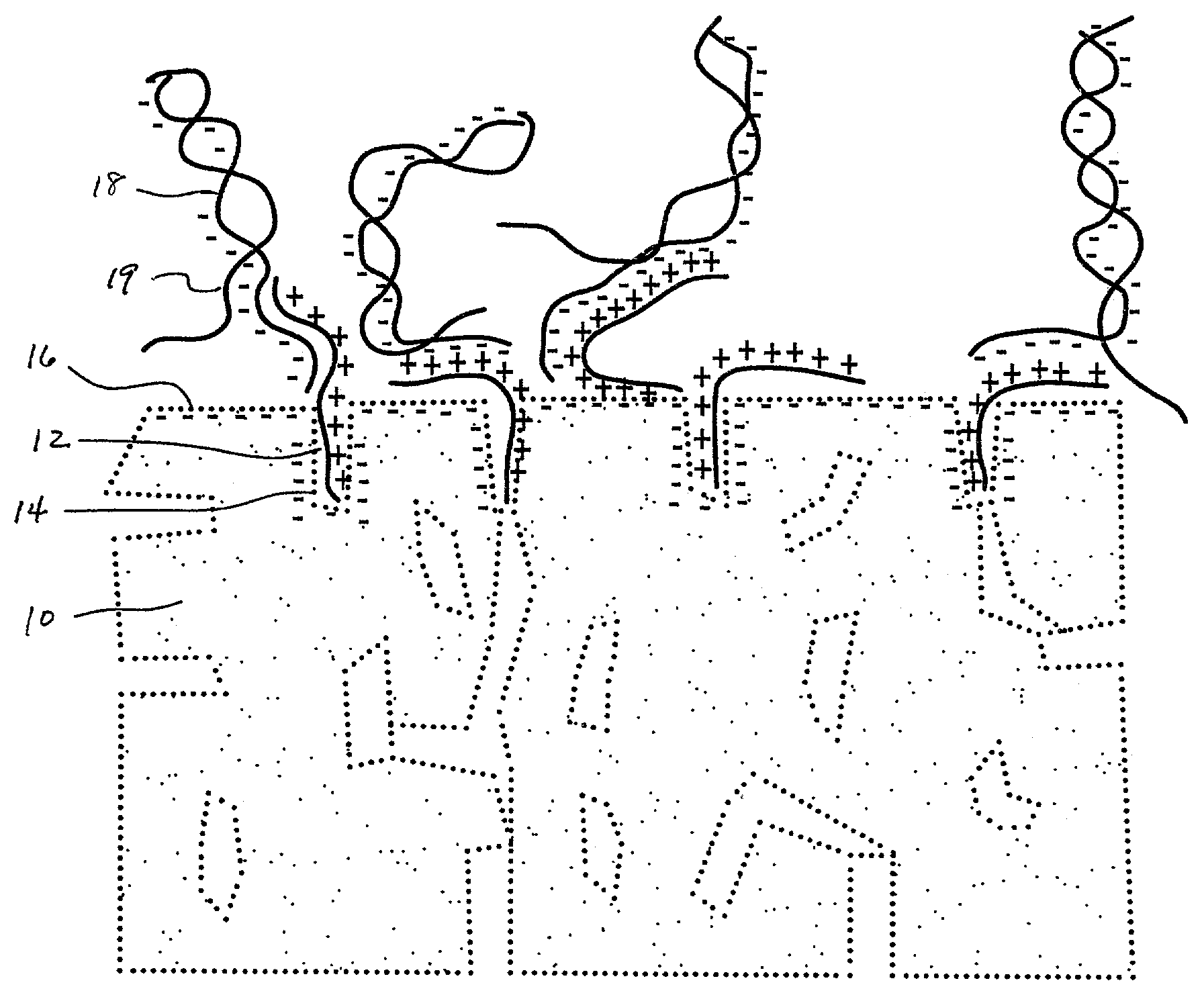
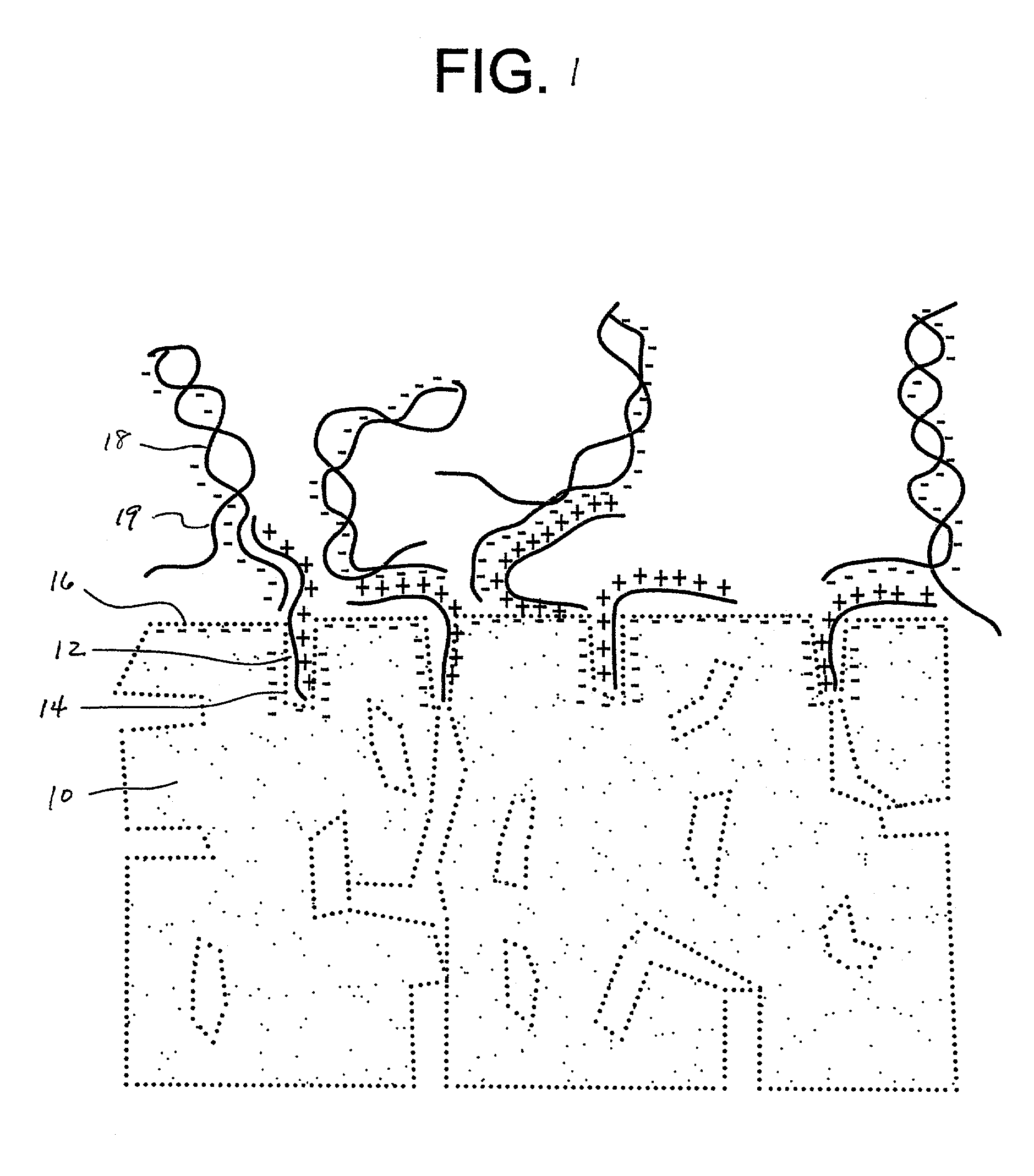
Assembling arrays of small particles using an atomic force microscope to define ferroelectric domains
InactiveUS20020118369A1Nanostructure manufactureMaterial analysis using wave/particle radiationMagnetic force microscopeChemical species
A method of assembling arrays of small particles or molecules using an atomic force microscope to define ferroelectric domains includes depositing a ferroelectric thin film upon a substrate forming workpiece, then using an atomic force microscope having a conductive, tip for generating a pattern on this thin film to define desired nano-circuit patterns. Next, exposure of this thin film to a solution containing chemical species which selectively adsorb or accumulate under the influence of electrophoretic forces in selected regions of this thin film.
Owner:IBM CORP
Porous substrates for DNA arrays
InactiveUS6994972B2Increase positive chargeEasy to attachImmobilised enzymesBioreactor/fermenter combinationsPorous substrateMolecular array
A planar, rigid substrate made from a porous, inorganic material coated with cationic polymer molecules for attachment of an array of biomolecules, such as DNA, RNA, oligonucleotides, peptides, and proteins. The substrate has a top surface with about at least 200 to about 200,000 times greater surface area than that of a comparable, non-porous substrate. The cationic polymer molecules are anchored on the top surface and in the pores of the porous material. In high-density applications, an array of polynucleotides of a known, predetermined sequence is attached to this cationic polymer layer, such that each of the polynucleotide is attached to a different localized area on the top surface. The top surface has a surface area for attaching biomolecules of approximately 387,500 cm2 / cm2 of area (˜7.5 million cm2 / 1×3 inch piece of substrate). Each pore of the plurality of pores in the top surface of the substrate has a pore radius of between about 40 Å to about 75 Å. Not only does the cationic coating in and over the pores of the substrate greatly increase the overall positive charge on the substrate surface, but also given the size of the pores provides binding sites to which biomolecules can better attach.
Owner:CORNING INC
Arrays and Methods of Use
InactiveUS20130172216A1Low densityQuantitative precisionMaterial nanotechnologyMicrobiological testing/measurementArray data structureMolecular array
Owner:INVITAE CORP
Method and system for extracting data from surface array deposited features
InactiveUS7330606B2Superior in pointHigh positioning accuracyImage enhancementImage analysisMolecular arrayData signal
A method for evaluating an orientation of a molecular array having features arranged in a pattern. An image of the molecular array is obtained by scanning the molecular array to determine data signals emanating from discrete positions on a surface of the molecular array. An actual result of a function on pixels of the image which pixels lie in a second pattern, is calculated. This actual result is compared with an expected result which would be obtained if the second pattern had a predetermined orientation on the array. Array orientation can then be evaluated based on the result.
Owner:AGILENT TECH INC
Polymer coatings
ActiveCN104508060AMicrobiological testing/measurementPretreated surfacesMolecular arrayPolymer coatings
The present disclosure relates to polymer coatings covalently attached to the surface of a substrate and the preparation of the polymer coatings, such as poly(N-(5-azidoacetamidylpentyl)acrylamide-co-acrylamide) (PAZAM), in the formation and manipulation of substrates, such as molecular arrays and flow cells. The present disclosure also relates to methods of preparing a substrate surface by using beads coated with a covalently attached polymer, such as PAZAM, and the method of determining a nucleotide sequence of a polynucleotide attached to a substrate surface described herein.
Owner:ILLUMINA INC
Method and system for molecular array scanner calibration
InactiveUS20060019398A1Color/spectral properties measurementsBiological testingMolecular arrayConcentration ratio
A method and system for calibrating molecular arrays to a reference molecular array, and for subsequently calibrating the molecular arrays to maintain a constant signal-intensity-to-label-concentration ratio. In the first step of the two-step calibration method, a reference array coated with the fluorophore or chromophore used to label probe molecules is employed, while in the second step of the two-step method, a reference array coated with a stable dye is employed.
Owner:CORSON JOHN F +3
Method and system for automated outlying feature and outlying feature background detection during processing of data scanned from a molecular array
InactiveUS6993172B2Non-uniform signal intensityCharacter and pattern recognitionColor/spectral properties measurementsPattern recognitionMolecular array
A method and system for employing pixel-based, signal-intensity data contained within areas of a scanned image of a molecular array corresponding to features and feature backgrounds in order to determine whether or not the features or feature backgrounds have non-uniform signal intensities and are thus outlier features and outlier feature backgrounds. A calculated, estimated variance for the signal intensities within a feature or feature background is compared to a maximum allowable variance calculated for the feature or feature background based on a signal intensity variance model. When the experimental variance is less than or equal to the maximum allowable variance, the feature or feature background is considered to have acceptable signal-intensity uniformity. Otherwise, the feature or feature background is flagged as an outlier feature or outlier feature background.
Owner:AGILENT TECH INC
Method for improved focus control in molecular array scanning by using a symmetrical filter to determine in-focus-distance
InactiveUS7067783B2High-frequency noiseRadiation pyrometryParticle separator tubesMolecular arrayAutomated method
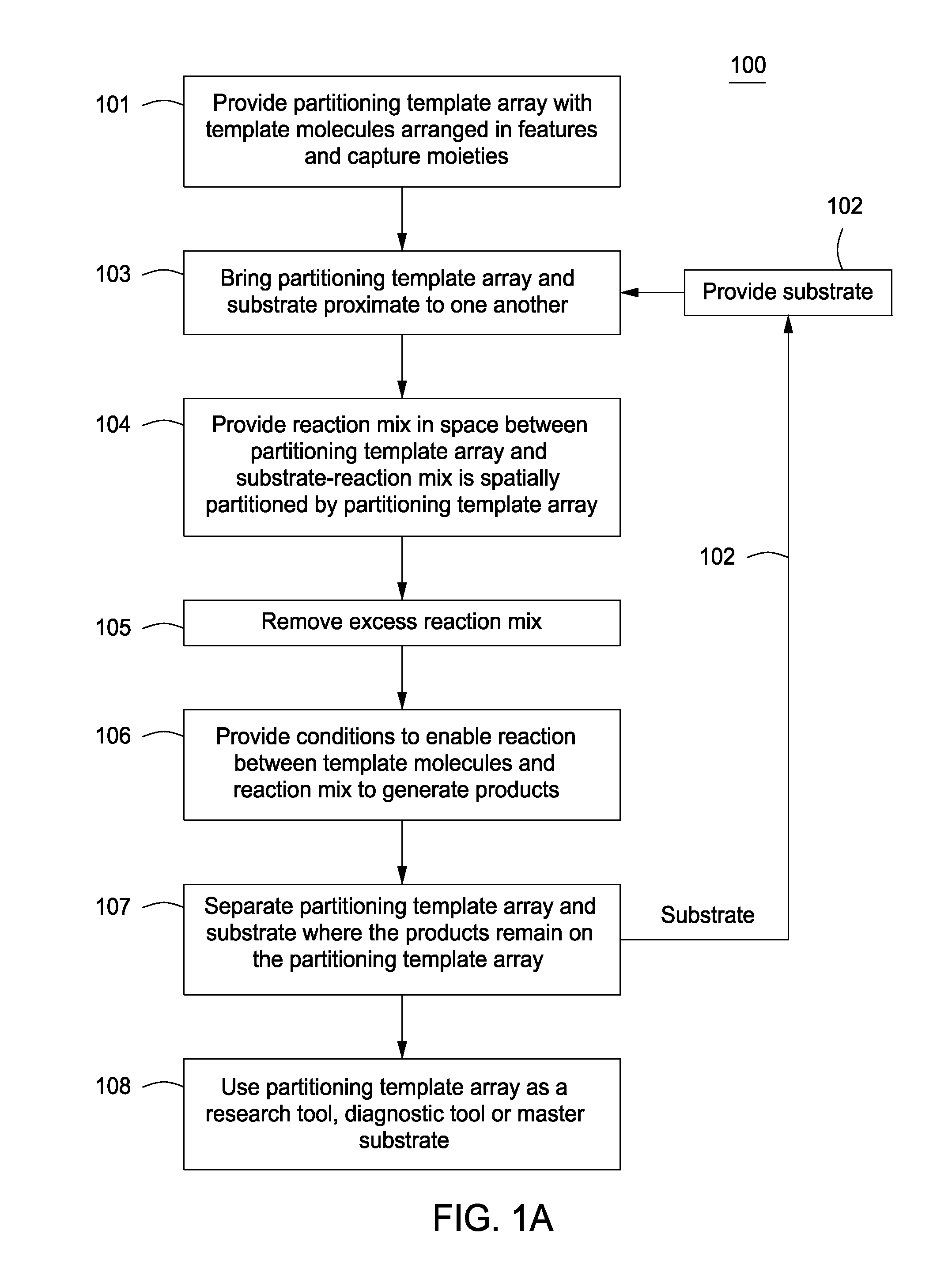
Automated methods and systems for determining an in-focus-distance for a position on the surface of a molecular array substrate using a molecular array scanner are provided. A signal from a first position of an array substrate is detected and noise is filtered out of the detected signal using a symmetrical filter to produce an in-focus-distance. In one embodiment, the in-focus-distance is utilized as an estimated in-focus-distance at a second position of the array substrate. The method finds use in maintaining the focus of a light source while scanning the array by the scanner. Also provided are methods of assaying a sample using the methods and systems of the invention, and kits for performing the invention. The subject invention finds use in a variety of different applications, including both genomics and proteomics applications.
Owner:AGILENT TECH INC
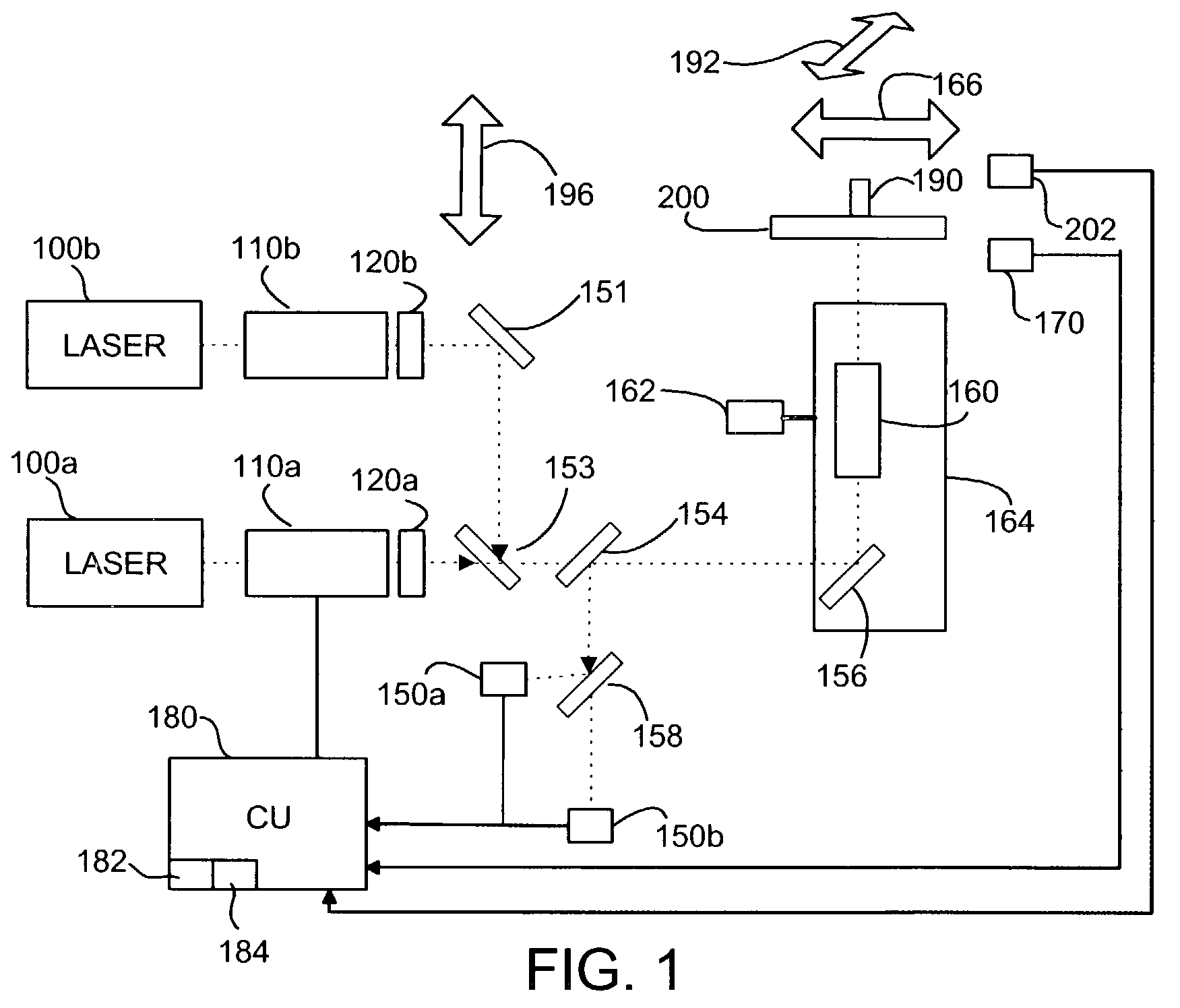
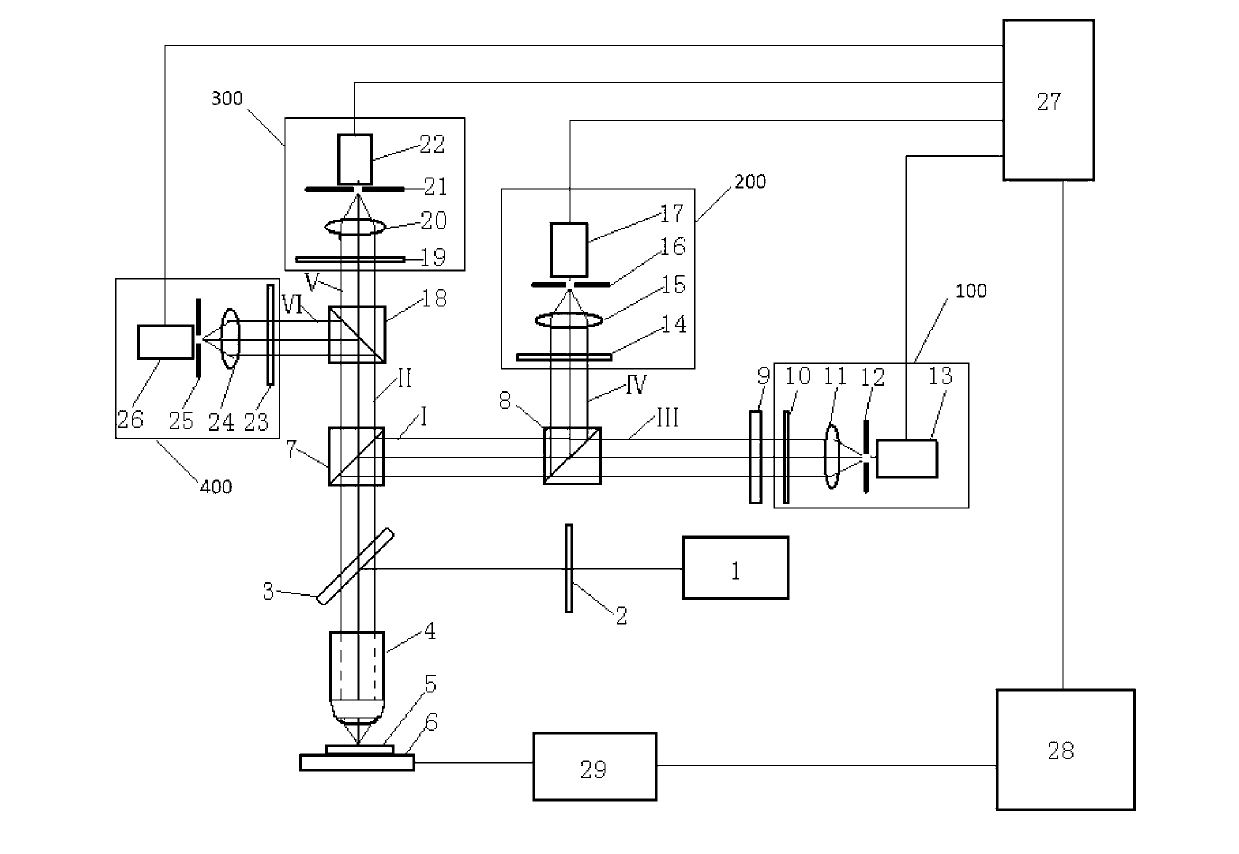
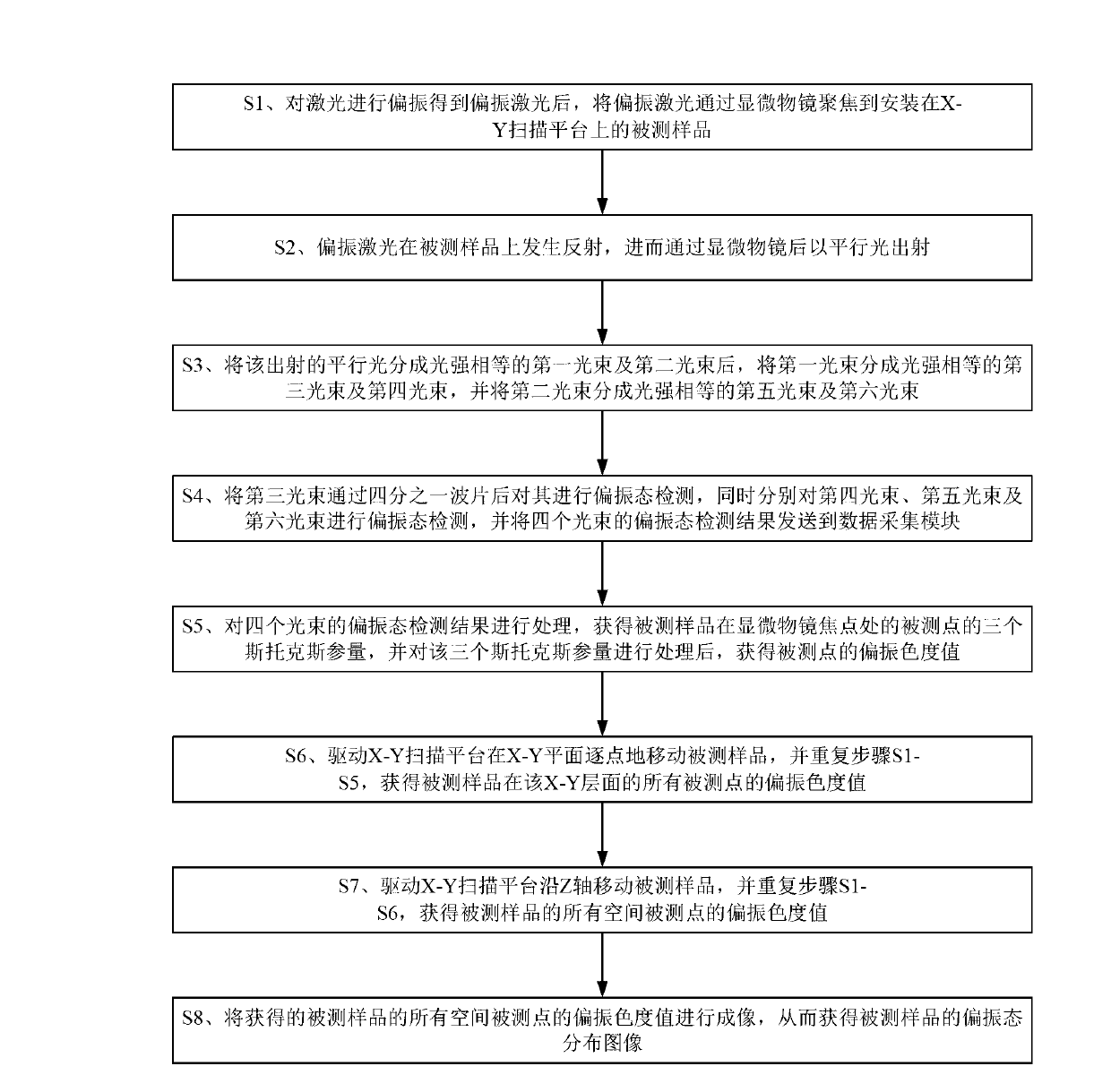
Polarization state tomography micro-imaging device and method thereof
ActiveCN103134756AAccurate reflectionReflection realPolarisation-affecting propertiesMicro imagingBeam splitter
The invention discloses a polarization state tomography micro-imaging device and a method thereof. The device comprises a laser, a polarization generator, a semi-reflecting semitransparent mirror, a microscope objective, a first beam splitter, a second beam splitter, a third beam splitter, a quarter-wave plate, a first Stokes system, a second Stokes system, a third Stokes system, a fourth Stokes system, a data acquiring module, a computer, a drive module and an X-Y scanning platform used for installing a tested sample. The device and the method allow the omni-directional polarization information of the tested sample to be obtained, the polarization states of an object at different levels to be detected and micro-imaged, the stress size and direction, the substance structure, the molecular array orientation, the refractive index, the internal stress distribution, the surface conductivity distribution, the surface roughness and the like of the internal of the object at different levels to be obtained, and the information of the tested sample to be truly and accurately reflected, and can be widely used in the polarization state measurement field.
Owner:SOUTH CHINA NORMAL UNIVERSITY
Organic field effect tube using single molecular layer as oriented transfer layer and its preparing method
InactiveCN1684286AIncreased anisotropyImprove performanceSolid-state devicesSemiconductor/solid-state device manufacturingMolecular arraySilanes
This invention provides an OFET device and its preparation method including an orientation transfer layer for transferring orientation efficiency, which can increase the carrier mobility of the OFET since the orientation transfer layer is the insulation one. Said one is a mono-film formed by processing the surface with silane coupler which can optimize the orientation result of the orientation layer and transfer it to the organic layer. This invention can improve molecular arrays of the organic molecular layer by transferring the orientation result to improve the carrier mobility of the OFET devices applying said orientation layer.
Owner:FUDAN UNIV
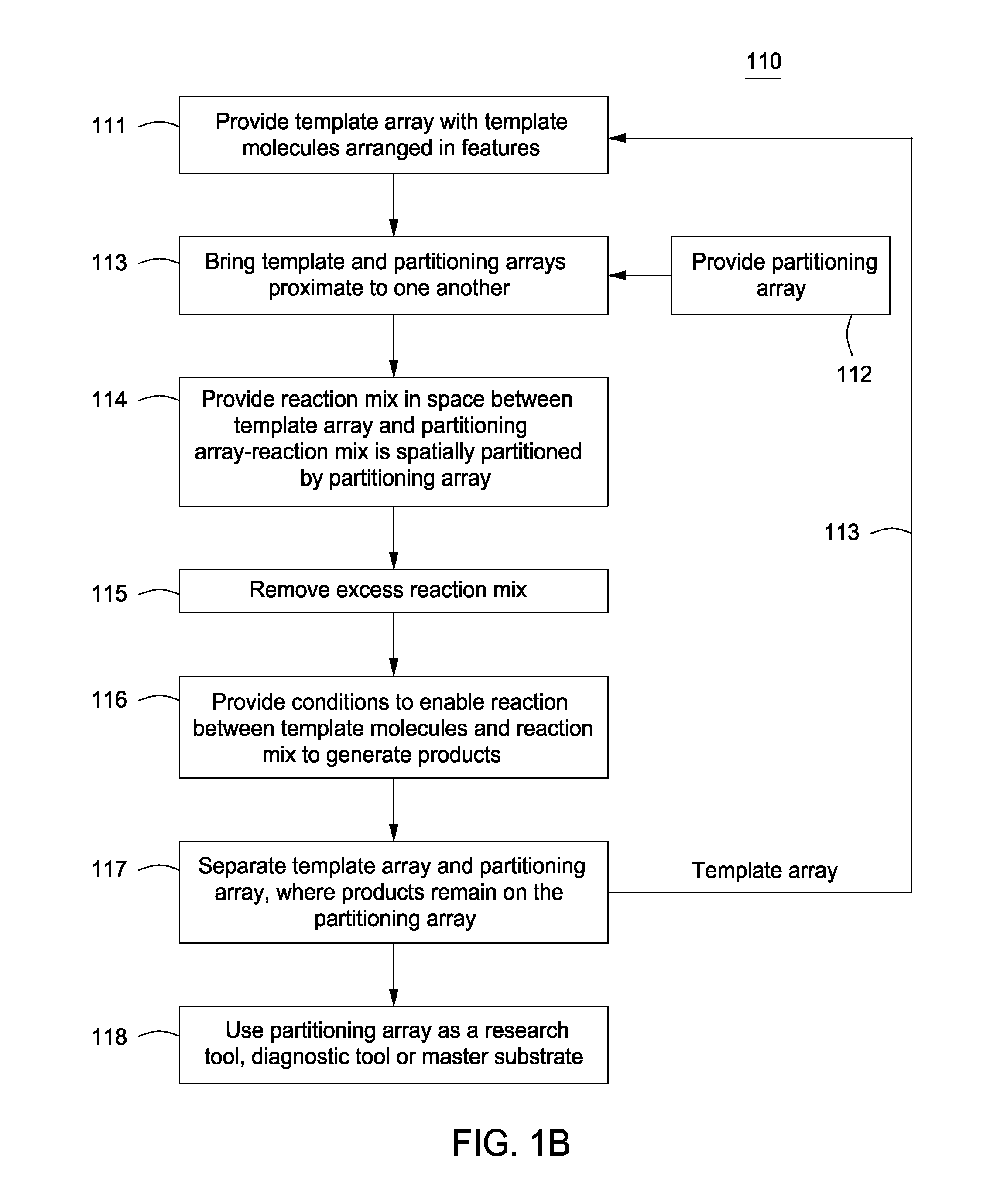
Array-based biomolecule analysis
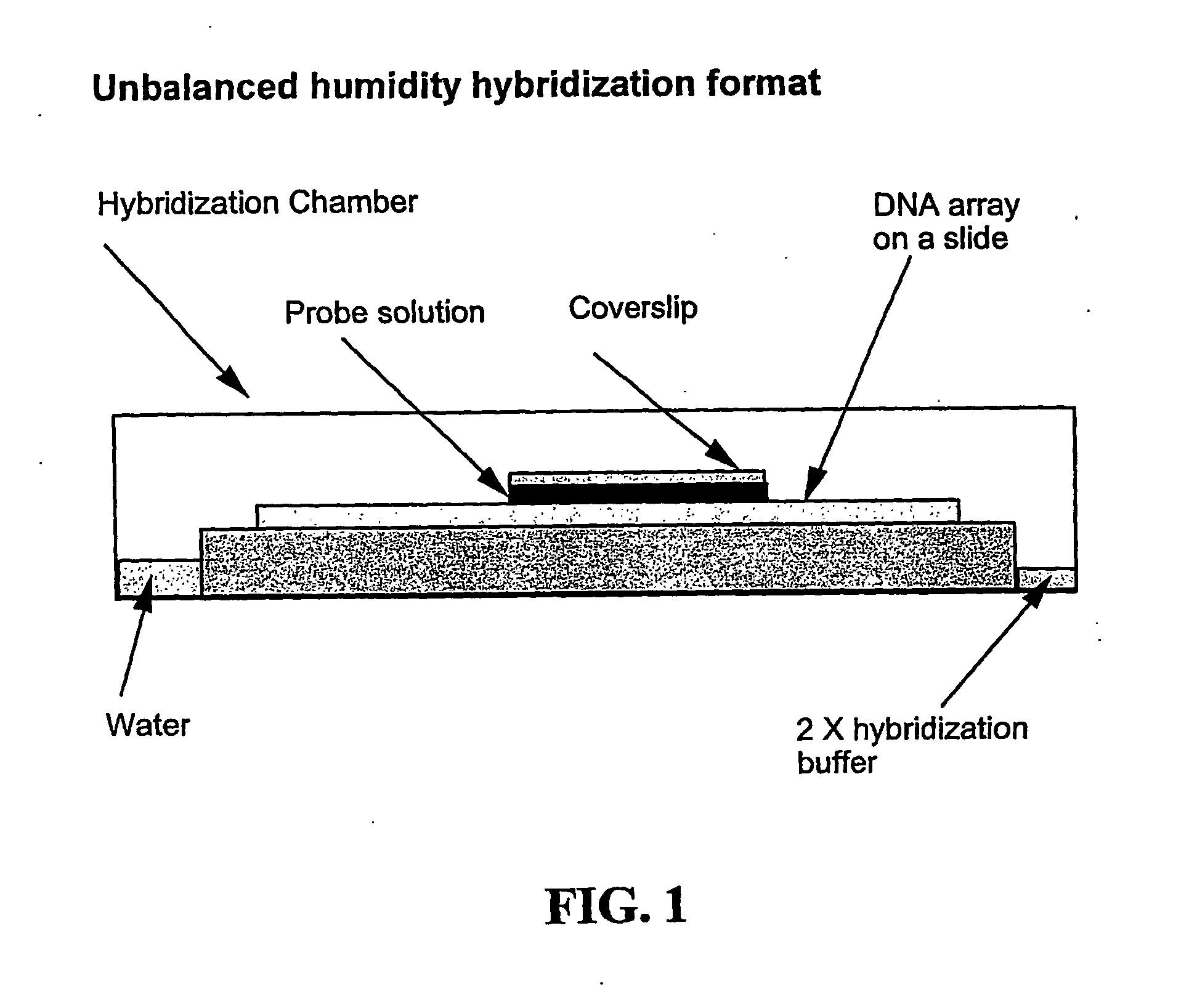
InactiveUS20050019942A1Preserving post-translational modificationDifference can be detectedSamplingMaterial analysis by electric/magnetic meansPredictabilityGel electrophoresis
Separation of macromolecules by one-dimensional or two-dimensional methods, such as gel electrophoresis, produces an array of macromolecules, which can be transferred to a support, thereby producing the same array as on the gel. In the case of one-dimensional gel electrophoresis, because of the regular spacing of the gel lanes and the predictable direction of migration of the macromolecules, the positions of the macromolecule spots or bands in the array can be predicted to be at least within the area of the support corresponding to the lanes of the gel. Where the molecular weight of a macromolecule of interest is known, molecular weight markers can be used to determine where the macromolecule band is on the support, even if the macromolecule is not stained in the gel or on the support. Assays that reveal characteristics of the macromolecule can be carried out by spotting reagents onto the support in a series of microspots of small volume in a line which intersects the macromolecule band, and which corresponds to the line of the direction of migration of the macromolecules on the gel. Appropriate detection methods can be applied, depending on the reagent, to see the results. The steps for locating the bands of macromolecules, applying reagents, and detecting the effect of the reagent on the macromolecule can be automated in an appropriate instrument.
Owner:SHIMADZU CORP
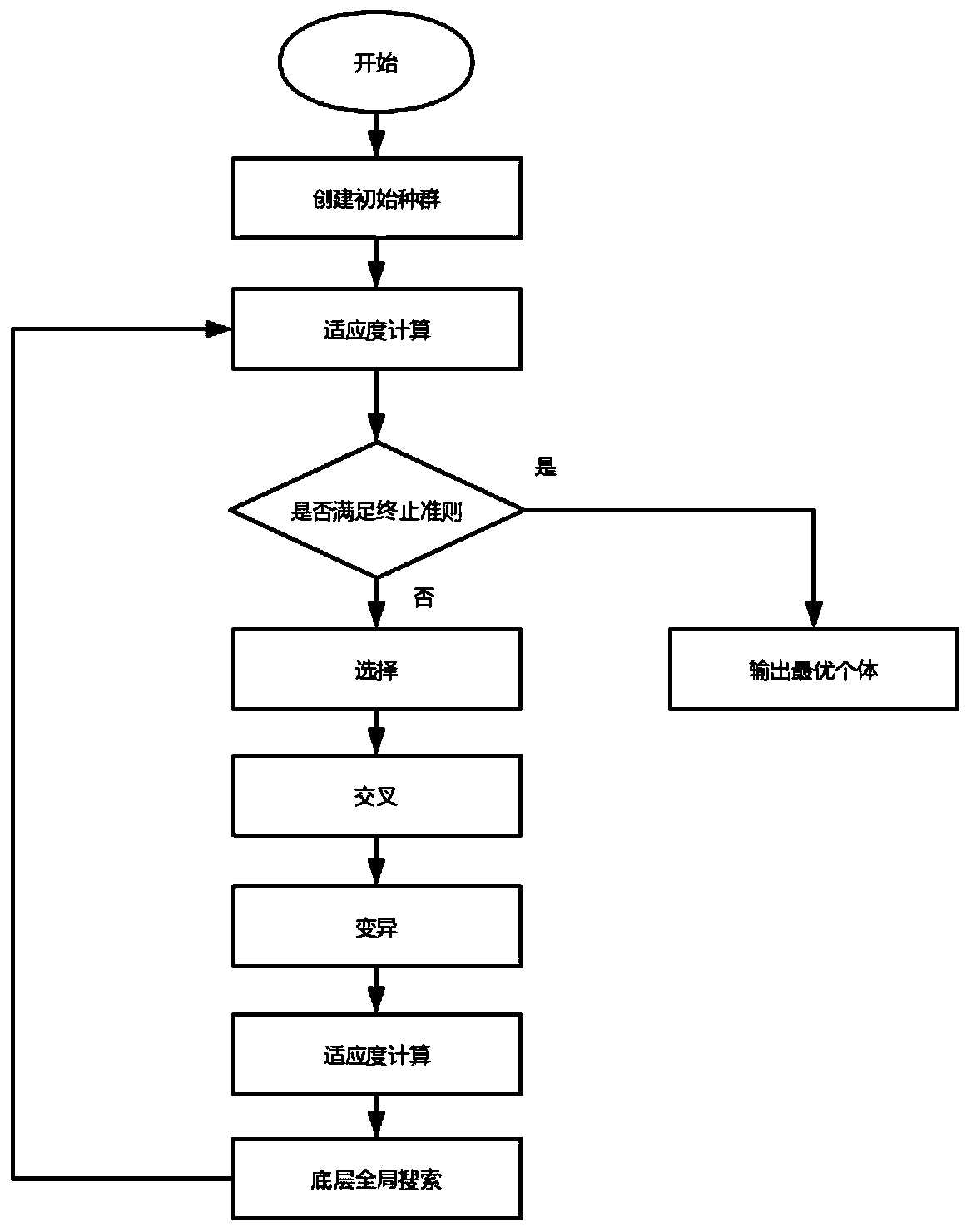
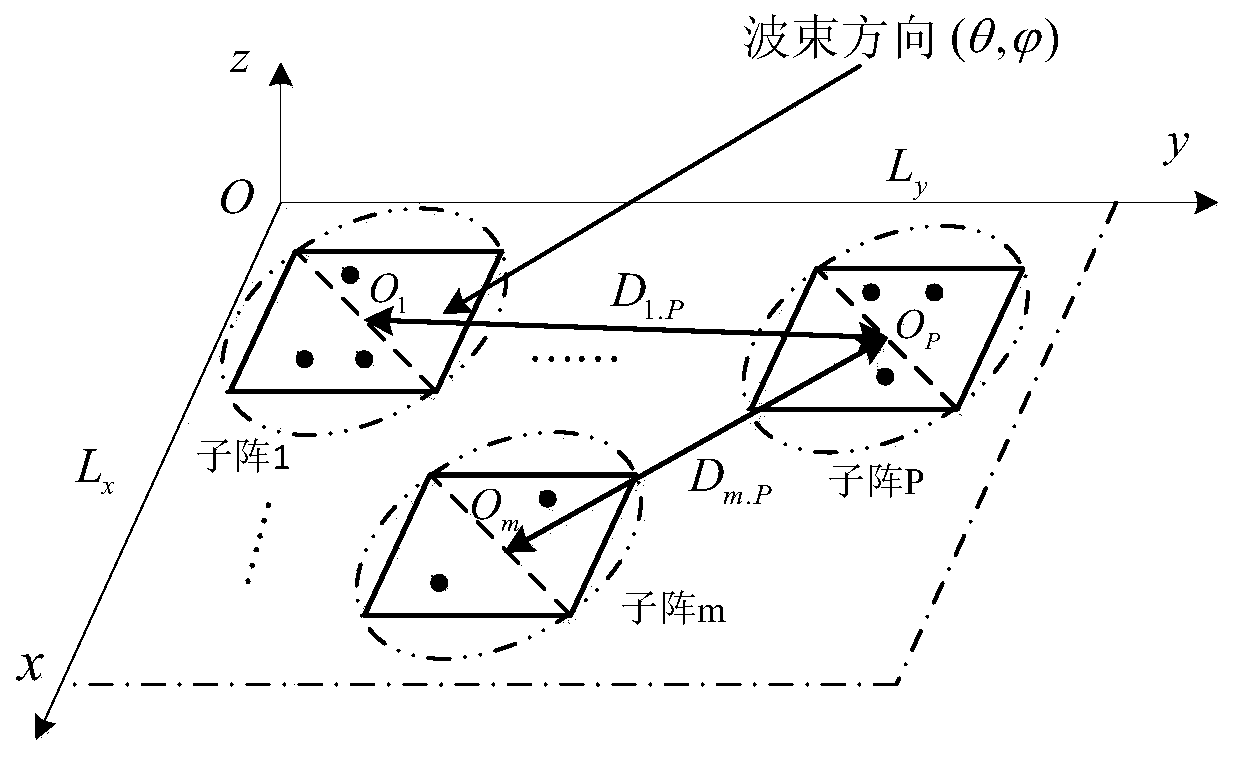
Novel planar molecular array antenna array comprehensive arrangement method based on improved genetic algorithm
ActiveCN111353605AReduce peak sidelobe levelsIncrease the distributionAntenna arraysGenetic algorithmsLocal optimumMolecular array
The invention relates to an antenna array arrangement optimization technology, in particular to an optimization arrangement technology of a sub-array-level antenna array. According to the method, an improved genetic algorithm is proposed firstly, compared with the problem that a traditional genetic algorithm is prone to falling into local optimum, the step of underlying global search is added, andthe optimization performance of the genetic algorithm is improved. Besides, for the problem of planar molecular array arrangement, whether sub-arrays are overlapped or not is difficult to judge; theinvention provides a simple and feasible judgment method, the effect of the method in planar molecular array arrangement is verified by adopting an improved genetic algorithm, and compared with a traditional array arrangement method, the new method effectively improves the degree of freedom of subarray distribution and further improves the array arrangement optimization performance.
Owner:UNIV OF ELECTRONICS SCI & TECH OF CHINA
Single molecule detection and quantification of nucleic acids with single base specificity
InactiveUS20200123592A1Microbiological testing/measurementFluorescence/phosphorescenceMolecular arrayAssay
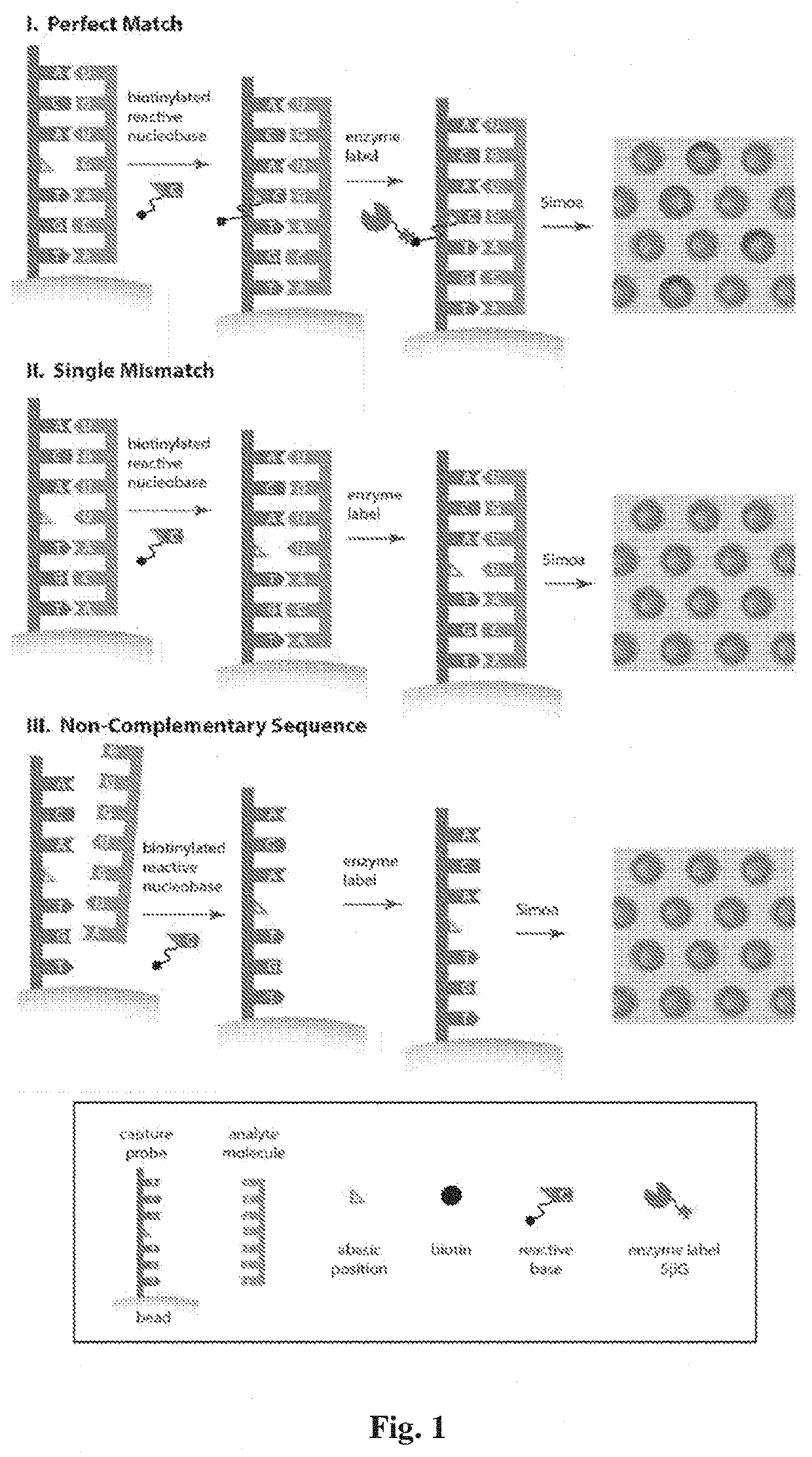
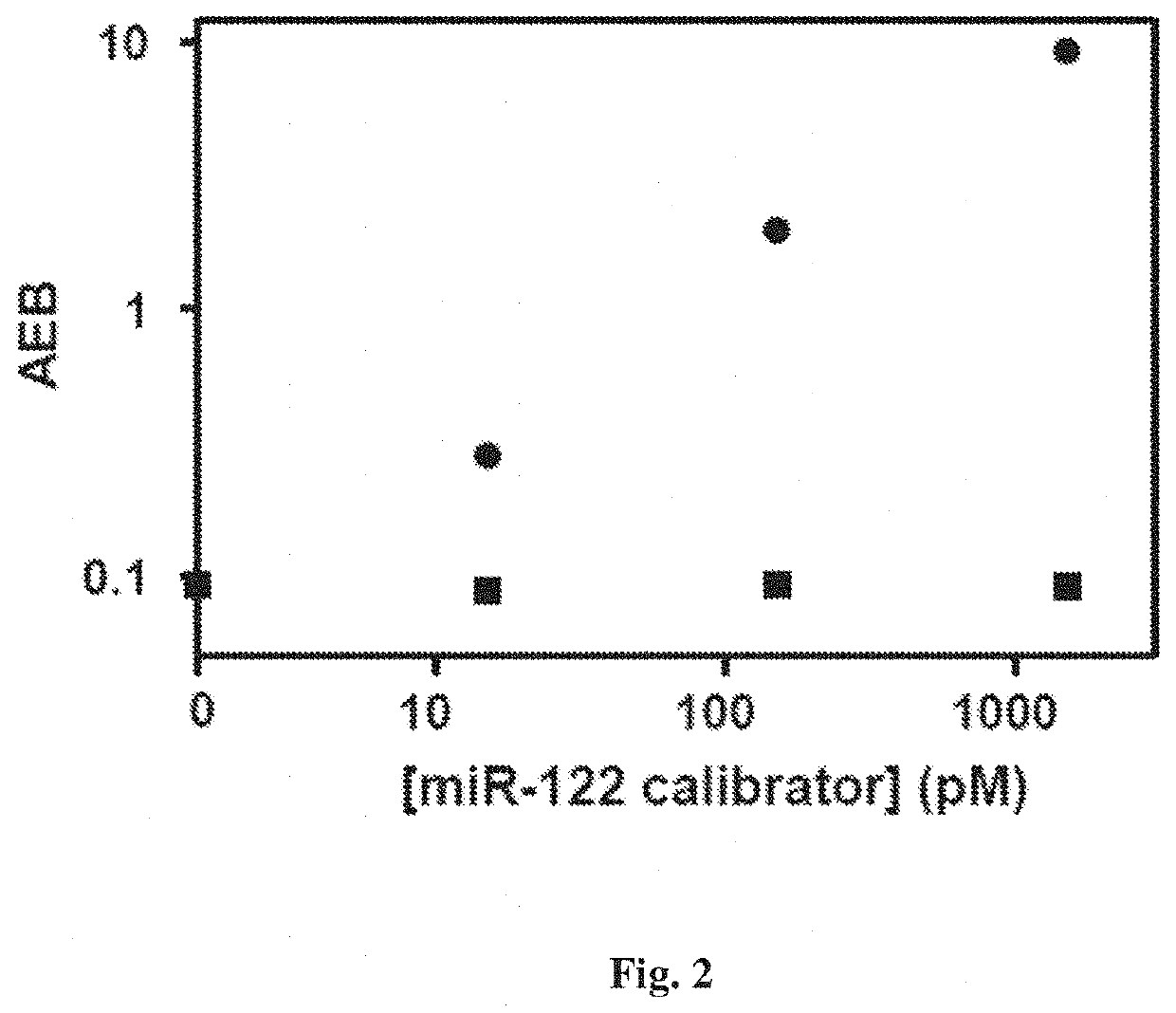
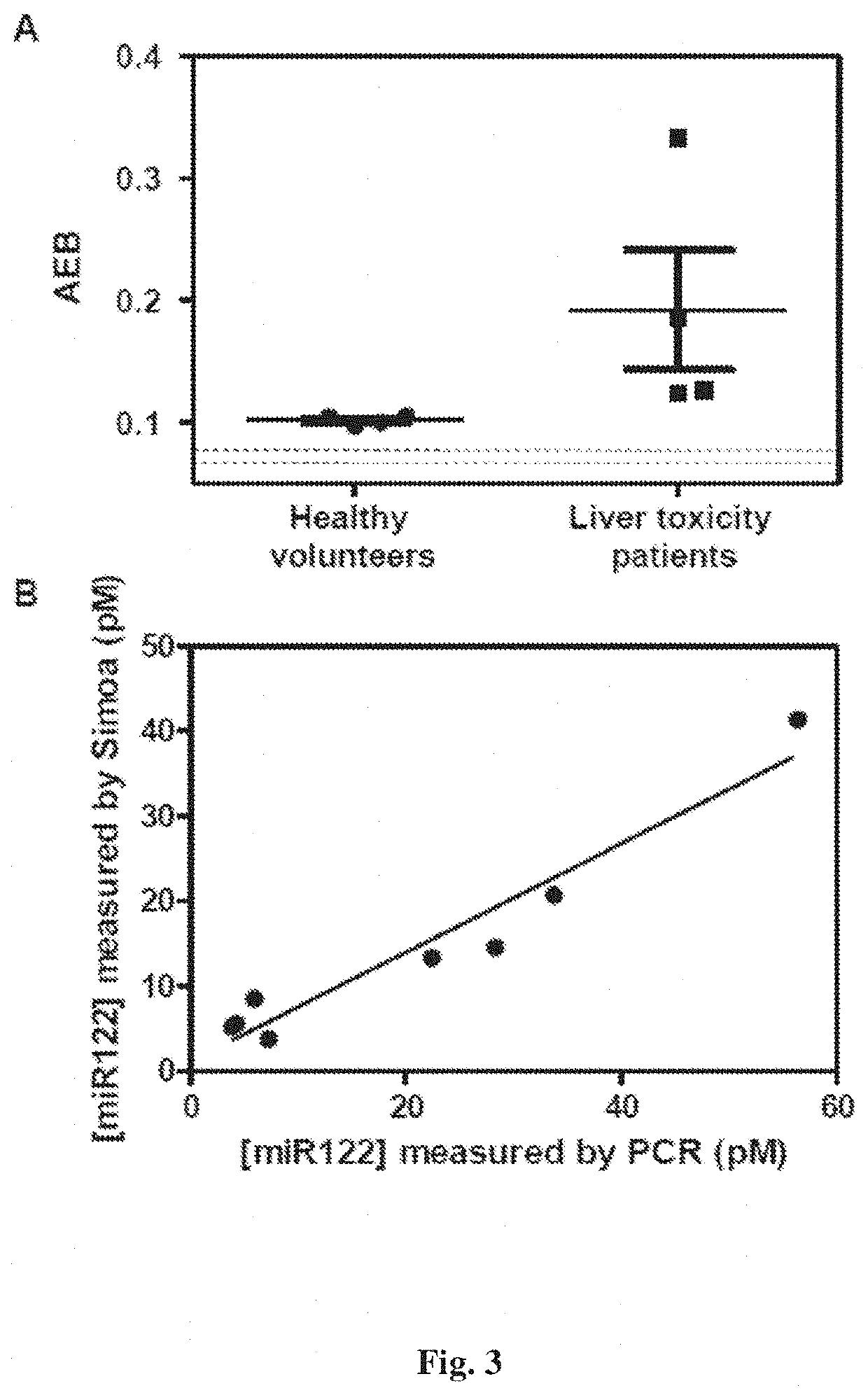
Methods for detecting nucleic acids, particularly small nucleic acids such as microRNAs, involving the use of a peptide nucleic acid (PNA) probe that lacks a base and a labelled modified base corresponding to the omitted base in the PNA probe. A complex containing the target nucleic acid, the PNA probe, and the modified base can be determined using a single molecule array assay.
Owner:QUANTERIX CORP +1
Method of forming one or more nanopores for aligning molecules for molecular electronics
InactiveUS20080203055A1Paper/cardboard articlesDecorative surface effectsMolecular arrayMolecular electronics
A technique is provided for forming a molecule or an array of molecules having a defined orientation relative to the substrate or for forming a mold for deposition of a material therein. The array of molecules is formed by dispersing them in an array of small, aligned holes (nanopores), or mold, in a substrate. Typically, the material in which the nanopores are formed is insulating. The underlying substrate may be either conducting or insulating. For electronic device applications, the substrate is, in general, electrically conducting and may be exposed at the bottom of the pores so that one end of the molecule in the nanopore makes electrical contact to the substrate. A substrate such as a single-crystal silicon wafer is especially convenient because many of the process steps to form the molecular array can use techniques well developed for semiconductor device and integrated-circuit fabrication.
Owner:HEWLETT PACKARD DEV CO LP
Exponential pattern recognition based cellular targeting, compositions, methods and anticancer applications
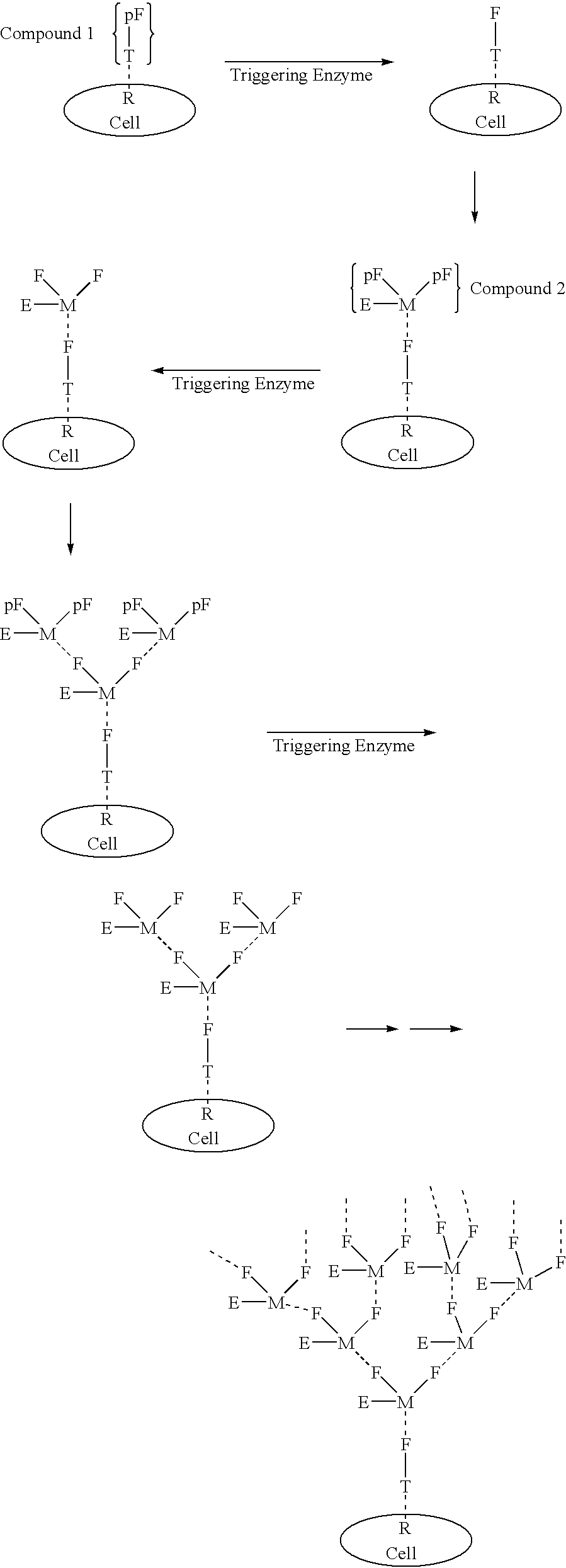
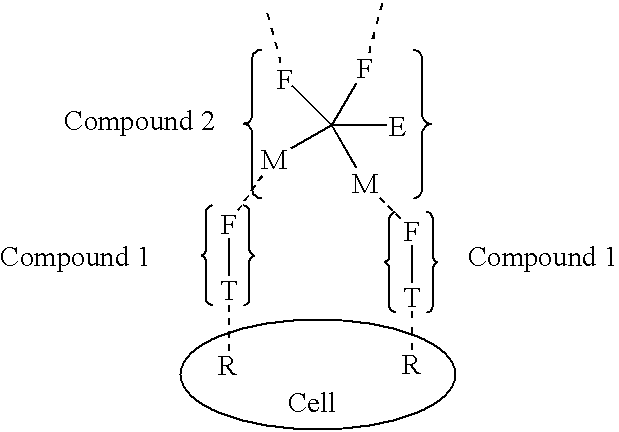
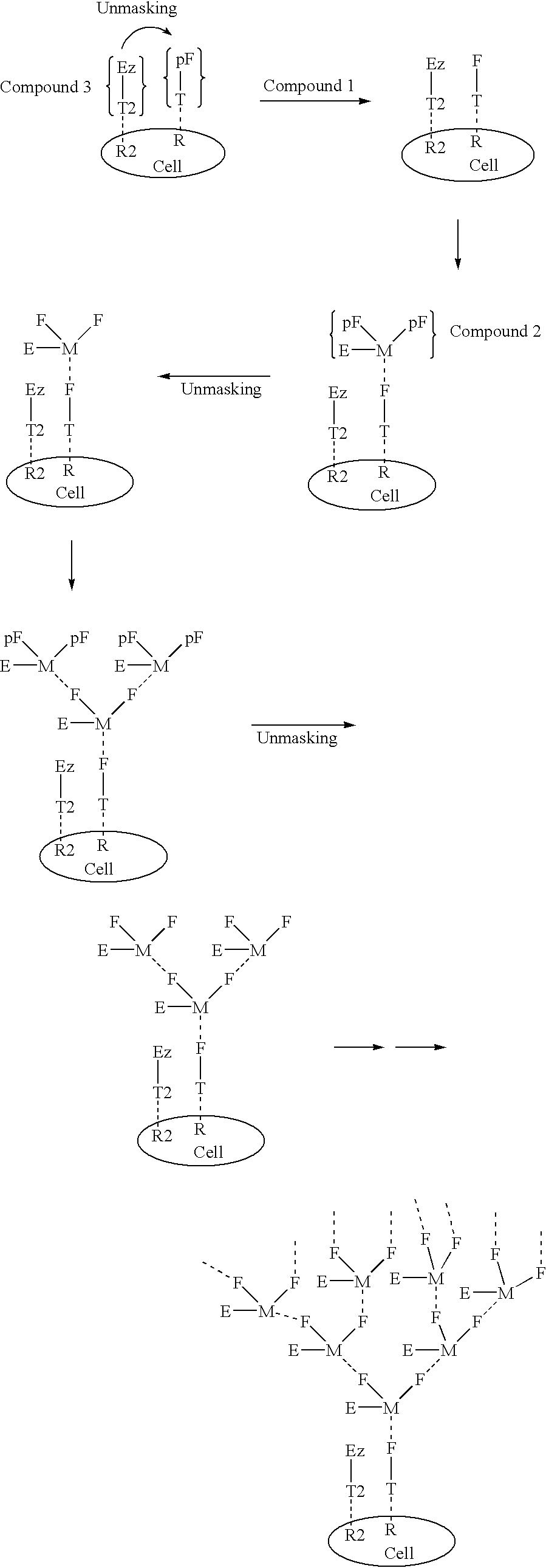
InactiveUS20030031677A1High target affinityLow cytotoxicityAntibody ingredientsPharmaceutical non-active ingredientsEnzymeTherapeutic intent
The present invention relates to the compositions, methods, and applications of a new approach to pattern recognition based targeting by which an exponential amplification of effector response can be specifically obtained at a targeted cells. The purpose of this invention is to enable the selective delivery of large quantities of an array of effector molecules to target cells for diagnostic or therapeutic purposes. The invention is comprised of two components designated as "Compound 1" and "Compound 2": Compound 1 is comprised of a cell binding agent and a masked female adaptor. Compound 2 is comprised of a male ligand, an effector agent, and two or more masked female receptors. The male ligand is selected to bind with high affinity to the female adaptor. Compound 1 can bind with high affinity to the target cell and the female receptor can then be unmasked by an enzyme enriched at the tumor cell. The male ligand of Compound 2 can then bind to the unmasked female adaptor bound to the target cell. The masked female adaptor on the bound Compound 2 can then be specifically unmasked. One receptor has in effect become two. Two new molecules of Compound 2 can bind to the unmasked adaptors receptors. After unmasking two receptors in effect become four. The process can continue in an explosive exponential like fashion resulting in enormous amplification of the number of effector molecules specifically deposited at the target cell.
Owner:DRUG INNOVATION & DESIGN
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com