Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
43 results about "Host factors" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
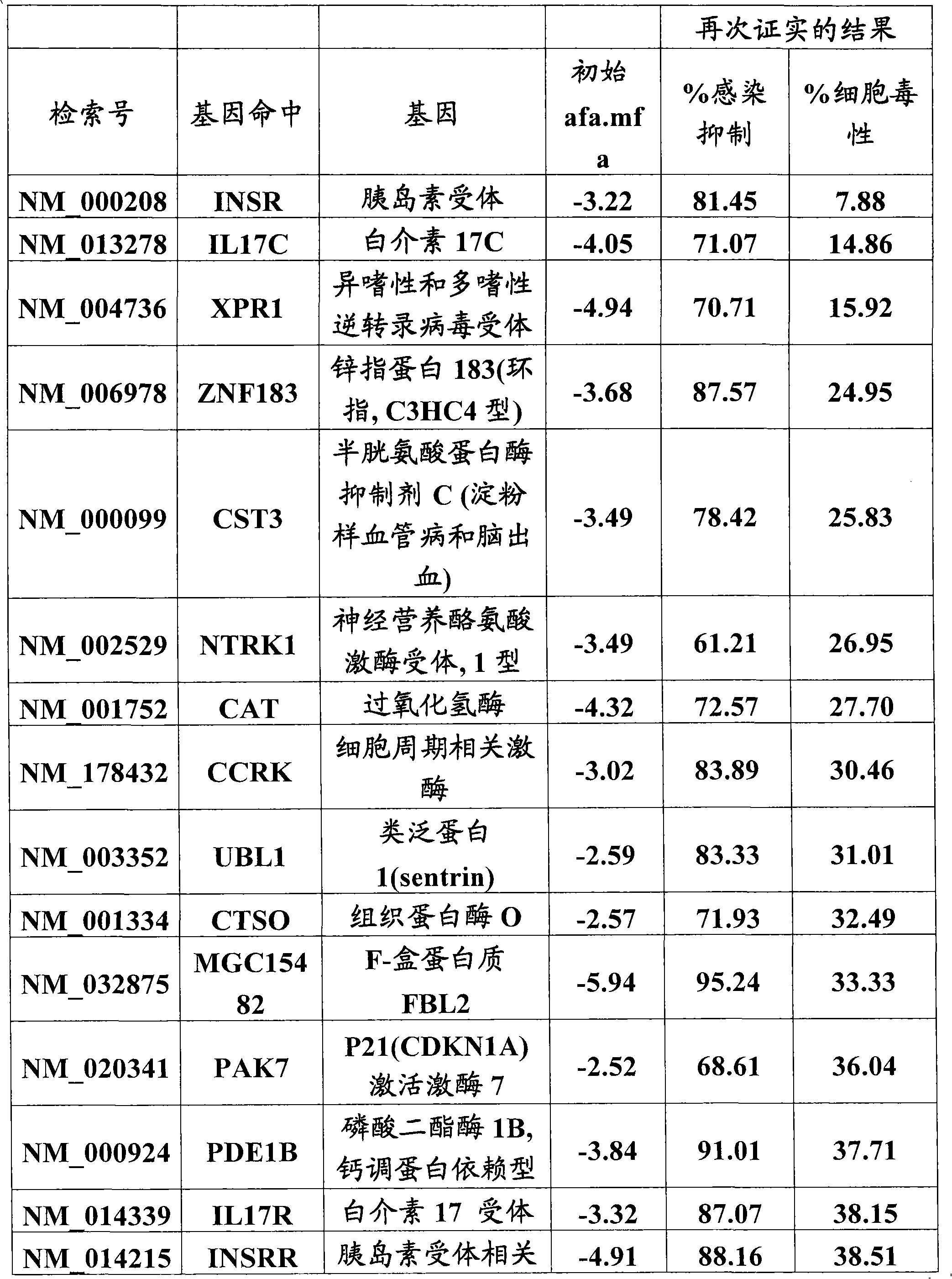
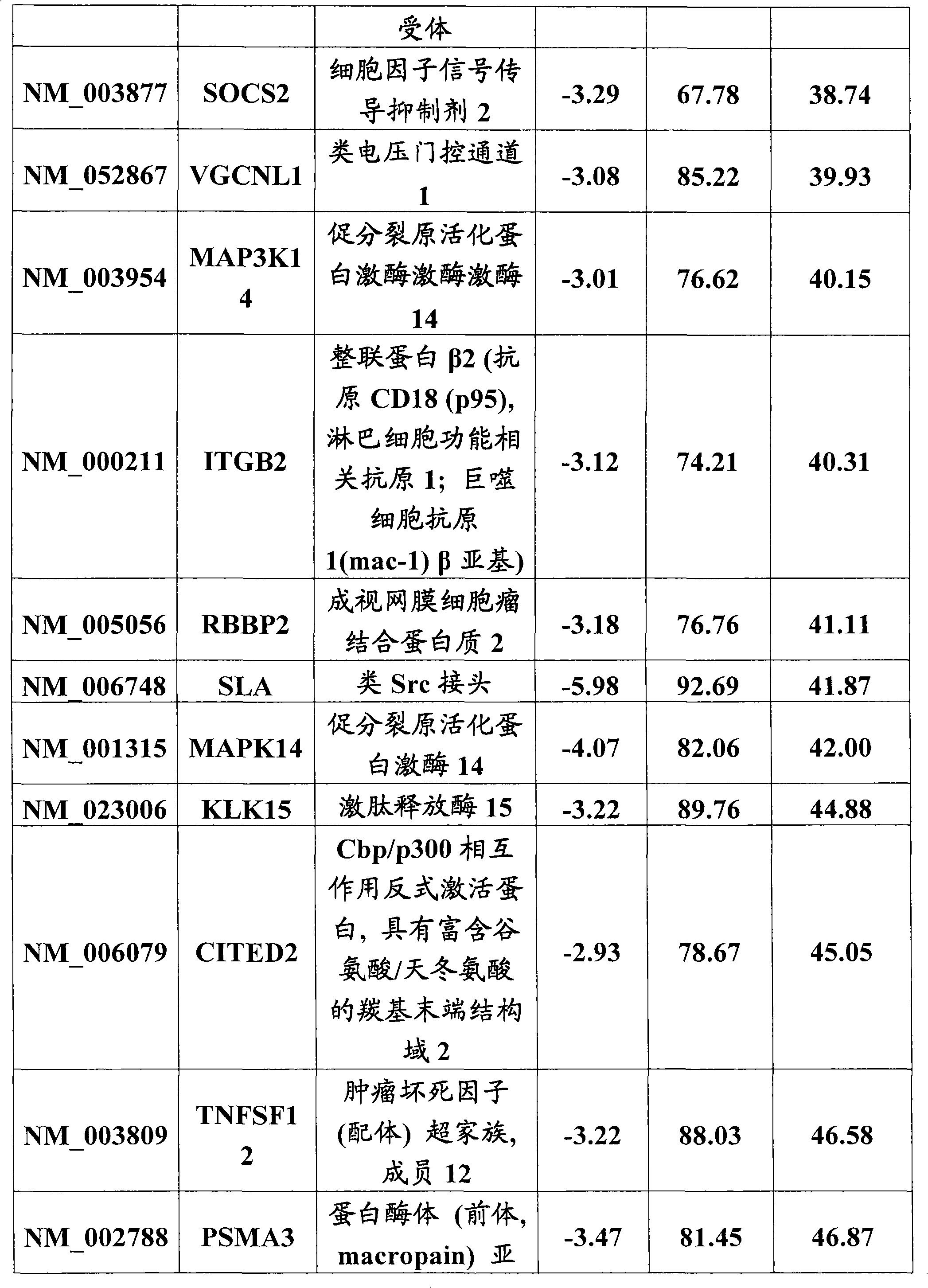
A host factor is considered to be one of the intrinsic qualities of a person that helps determine that person's susceptibility to disease. Host factors may be thought of as risk factors for disease, but they form part of the patient's own physical or psychological constitution.
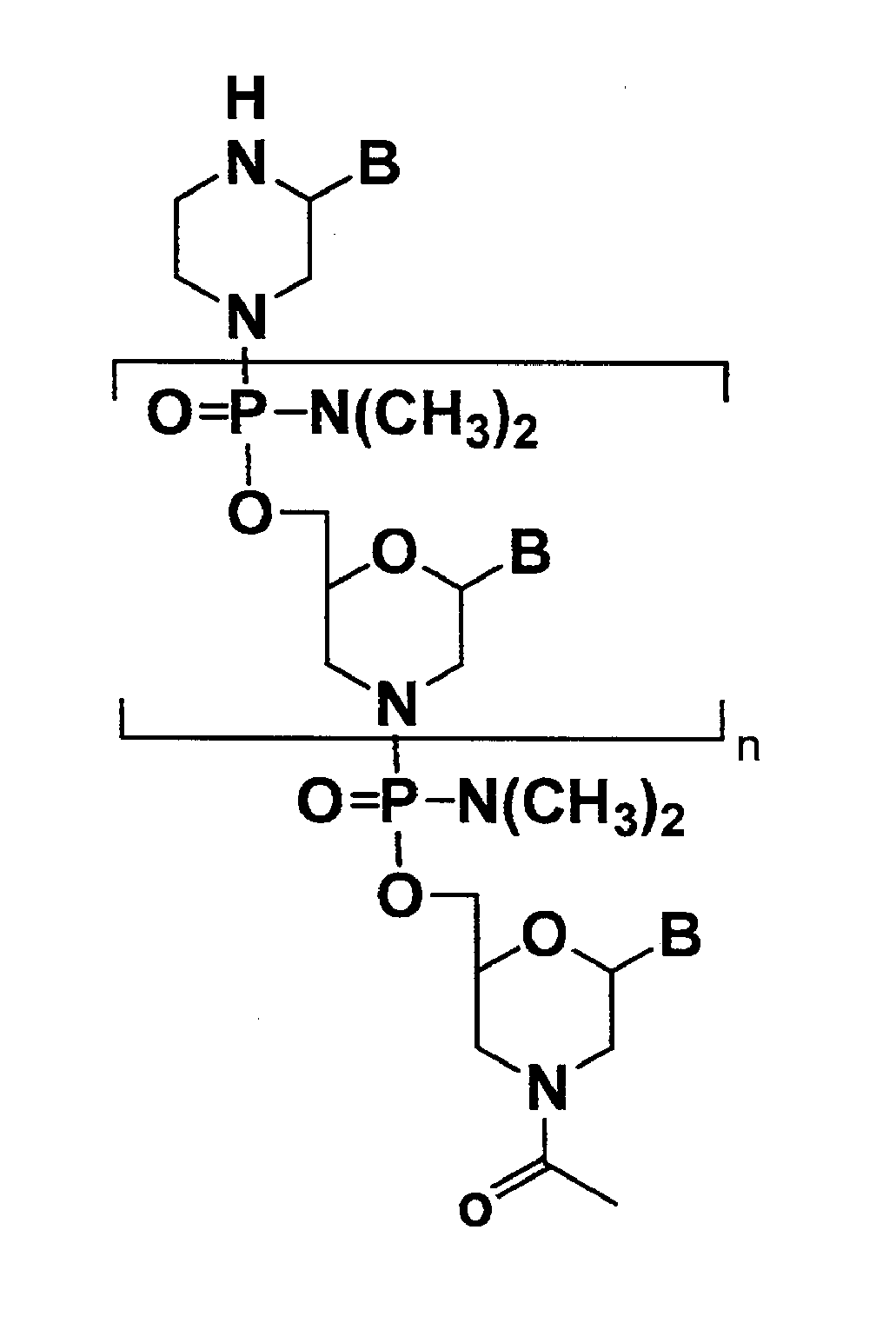
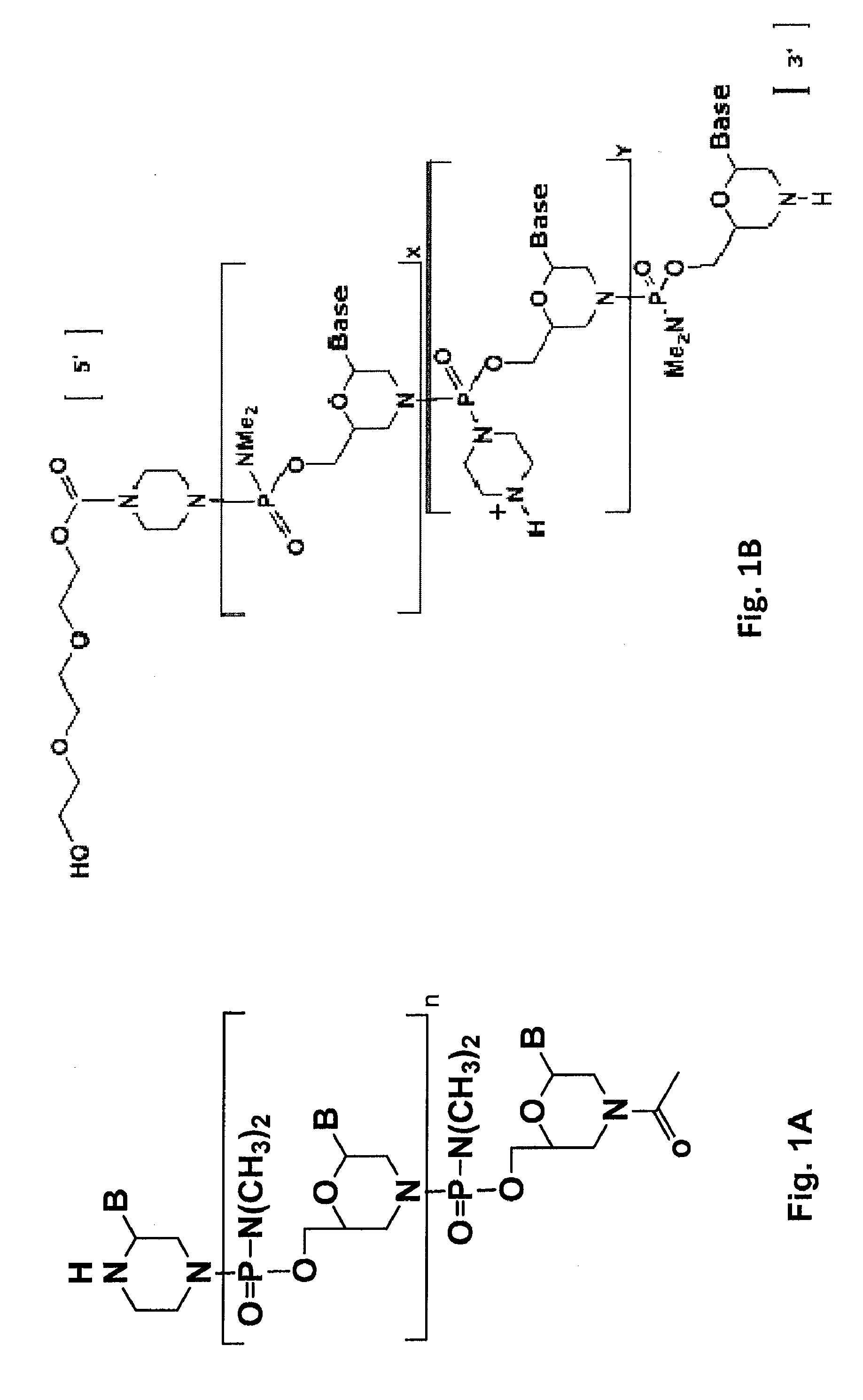
Antisense antibacterial compounds and methods
Antibacterial antisense compounds and methods of their use in treating a Mycobacterium tuberculosis infection in a mammalian host are disclosed. The compounds include an antisense oligonucleotide conjugated to a carrier peptide that significantly enhances the antibacterial activity of the oligonucleotide. The antisense oligonucleotides contain 10-20 nucleotide bases and have a targeting nucleic acid sequence complementary to a target sequence containing or within 20 bases, in a downstream direction, of the translational start codon of a bacterial mRNA that encodes a bacterial protein essential for bacterial replication, where the compound binds to a target mRNA with a Tm of between 45° to 60° C. The carrier peptide is an arginine-rich peptide containing between 6 and 14 amino acids. Antisense compounds that target host factor genes that facilitate Mycobacterium tuberculosis infection are also provided, as are methods of using these compounds to treat Mycobacterium tuberculosis infections, alone or in combination with other therapies.
Owner:AVI BIOPHARMA
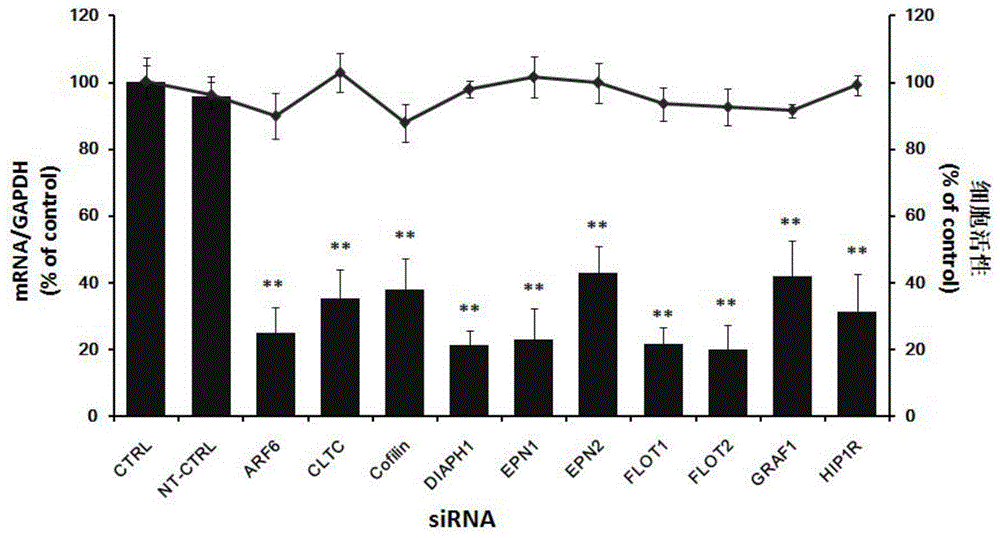
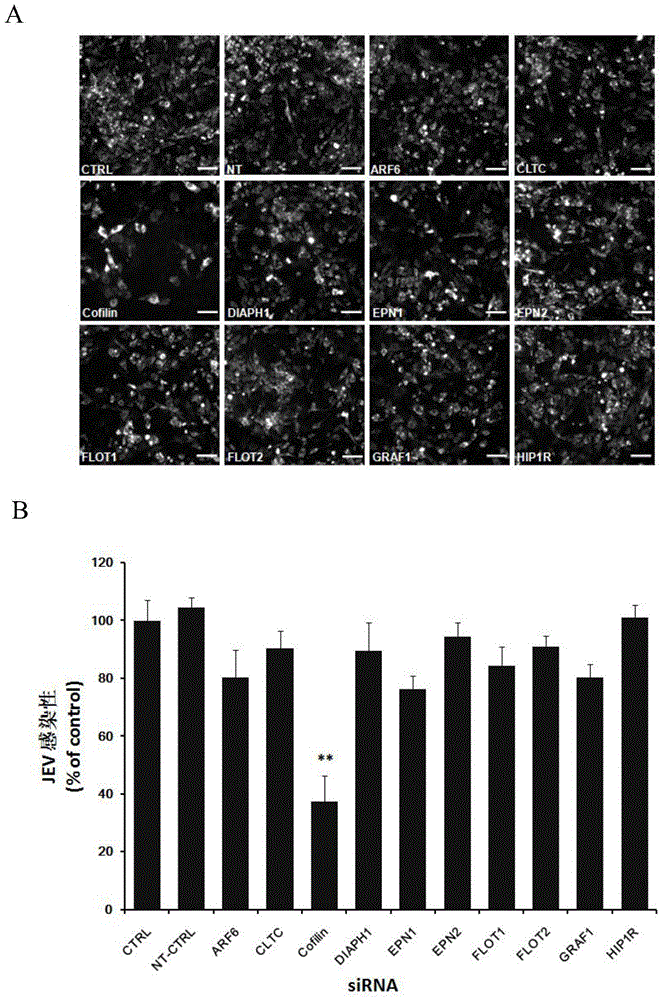
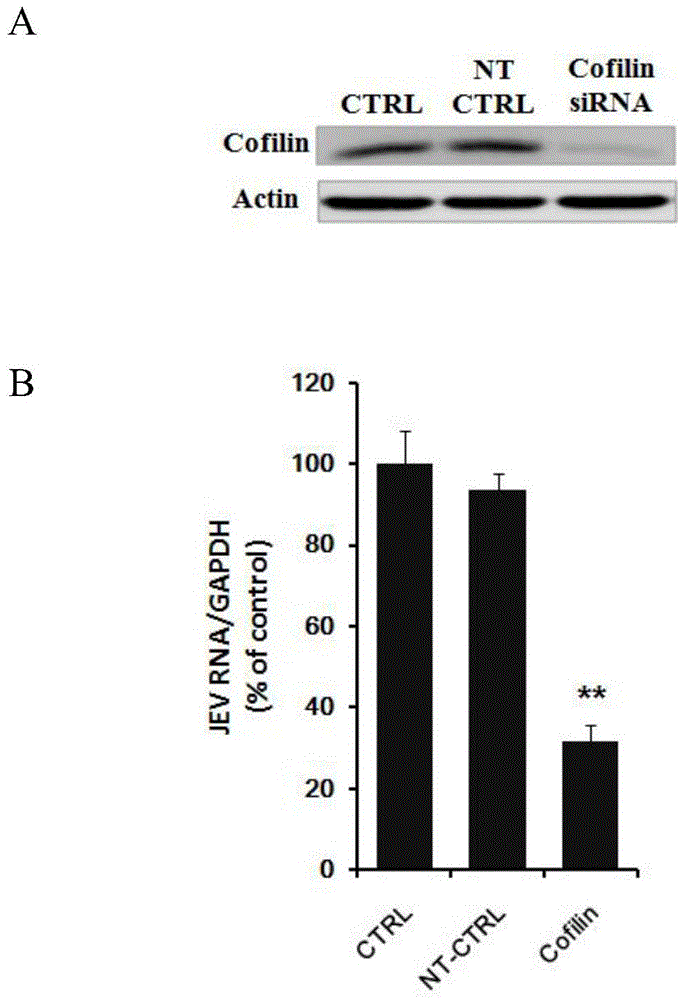
Application of Cofilin in preventing and treating Japanese encephalitis virus infection
The invention relates to the technical field of biomedicine, in particular to a novel target resistant to Japanese encephalitis virus (JEV) infection and application. Human neuroblastoma cells (SK-N-SH) are taken as target cells, RNA technology is adopted to reduce expression of host protein of the target cells to find host factors capable of effectively inhibiting JEV-infected human neuroblastoma cells (SK-N-SH) so as to protect functions of a nerve system and prevent virus from infecting a central nerve system to cause inflammation. The invention provides application of Cofilin in preventing and treating JEV infection, and experiments find that Cofilin plays an important role in JEV-infected SK-N-SH, and JEV infection can be inhibited remarkably by reducing expression of Cofilin. The invention further provides application of Cofilin in preparing drug for preventing or treating JEV infection.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
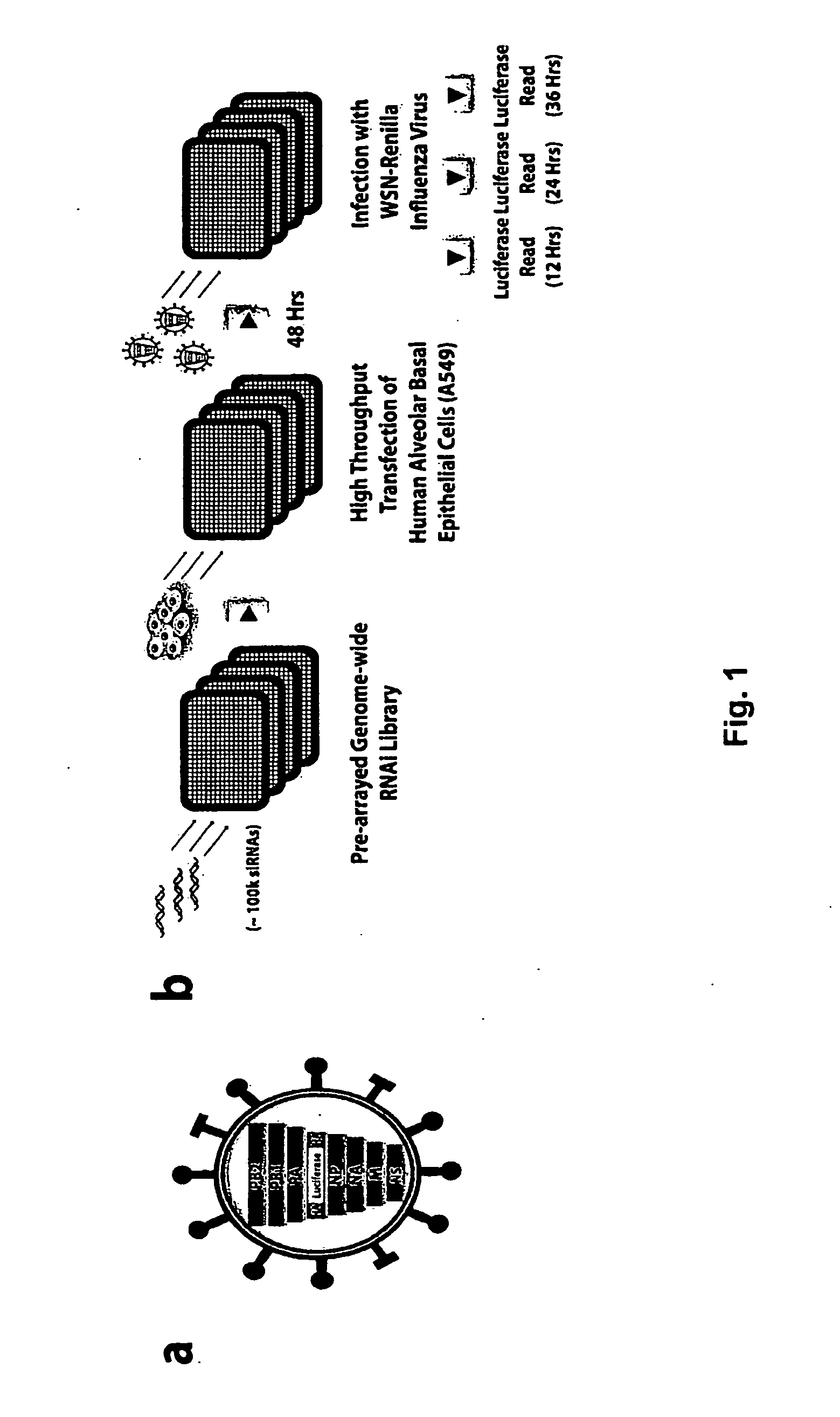
Compositions and Methods for Inhibiting Human Host Factors Required for Influenza Virus Replication
ActiveUS20130090367A1Reduction and amelioration of severityShorten the construction periodBiocideAmide active ingredientsDiseaseHost factors
This application relates to the modulation of host cell factors required for influenza virus replication. The application relates to compounds, including nucleic acid compounds (such as, e.g., small interfering RNAs (siRNAs)) and small molecules, that target human host cell factors involved in influenza virus replication, and the use of such compounds for modulating influenza virus replication and as antiviral agents. The application also relates to methods of treating an influenza virus infection and methods of treating or preventing a symptom or disease associated with influenza virus infection, comprising administering to a subject a composition comprising a compound, such as a nucleic acid compound (e.g., an siRNA) or small molecule, that targets a human host cell factor involved in influenza virus replication.
Owner:SALK INST FOR BIOLOGICAL STUDIES +2
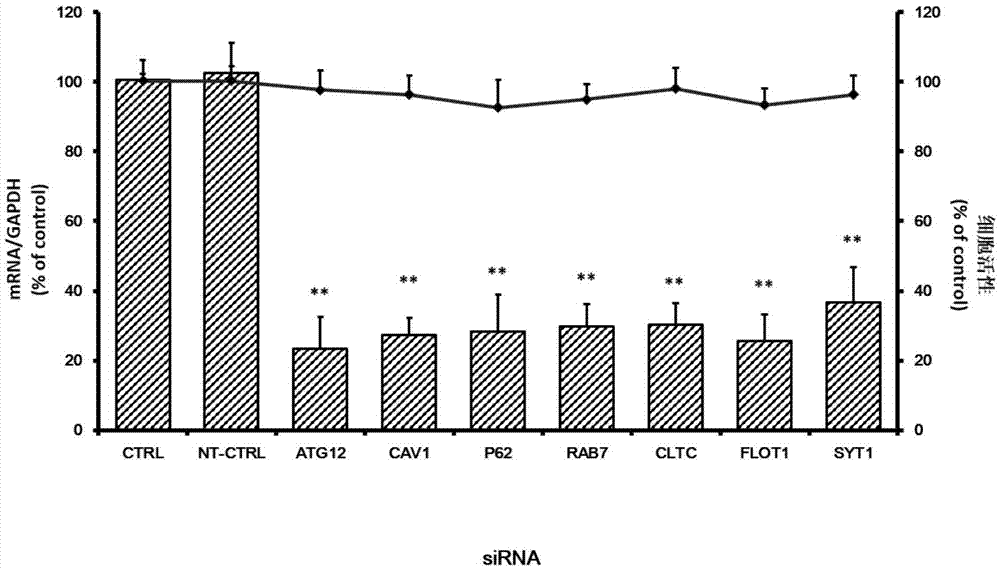
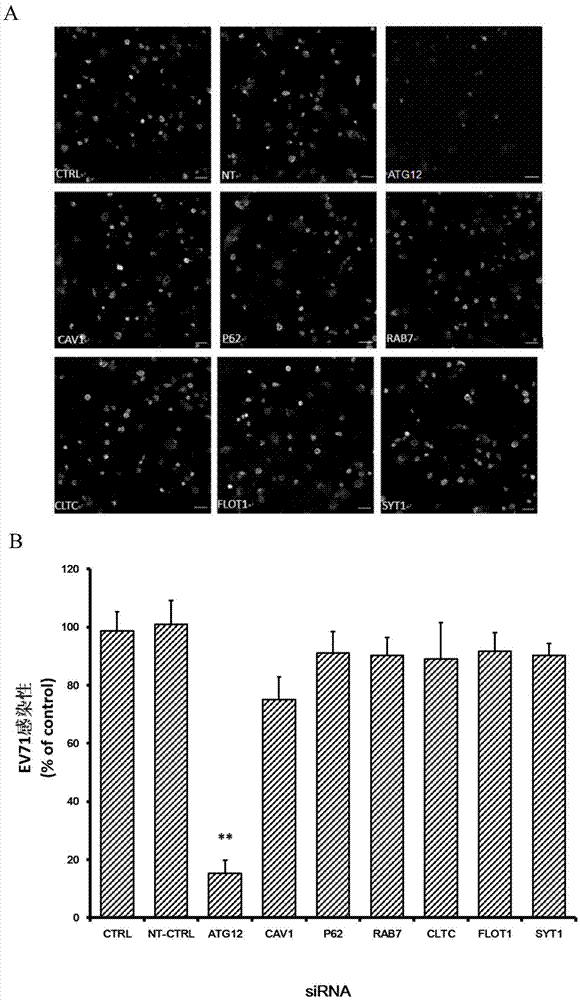
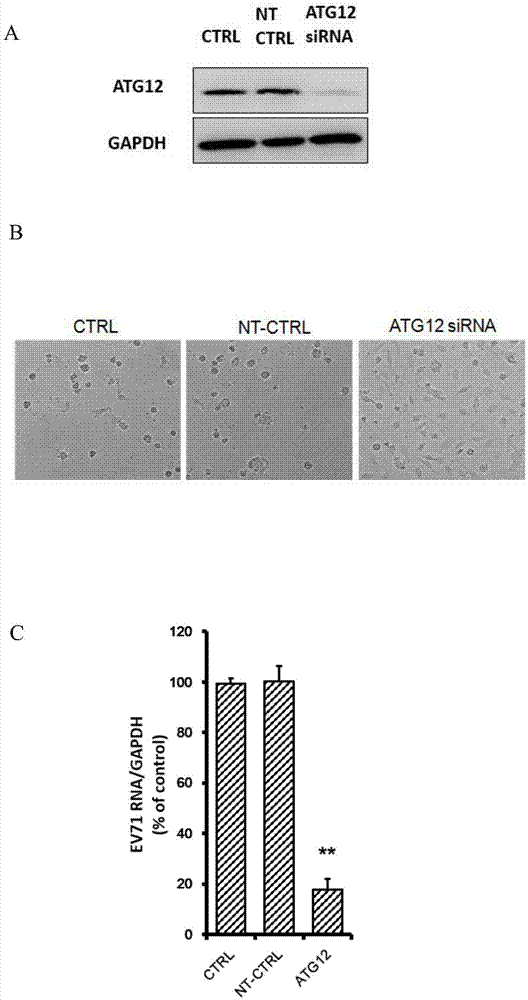
Applications of autophagy-related protein 12 in preventing and treating enterovirus 71 infection
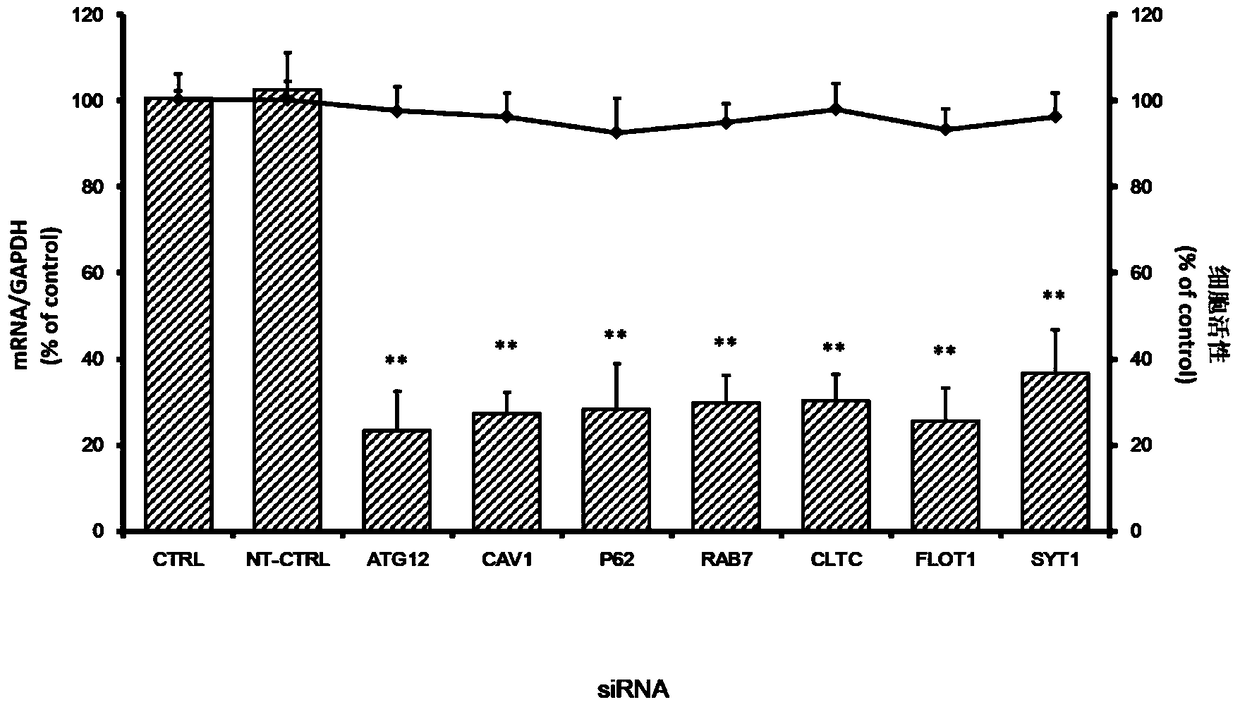
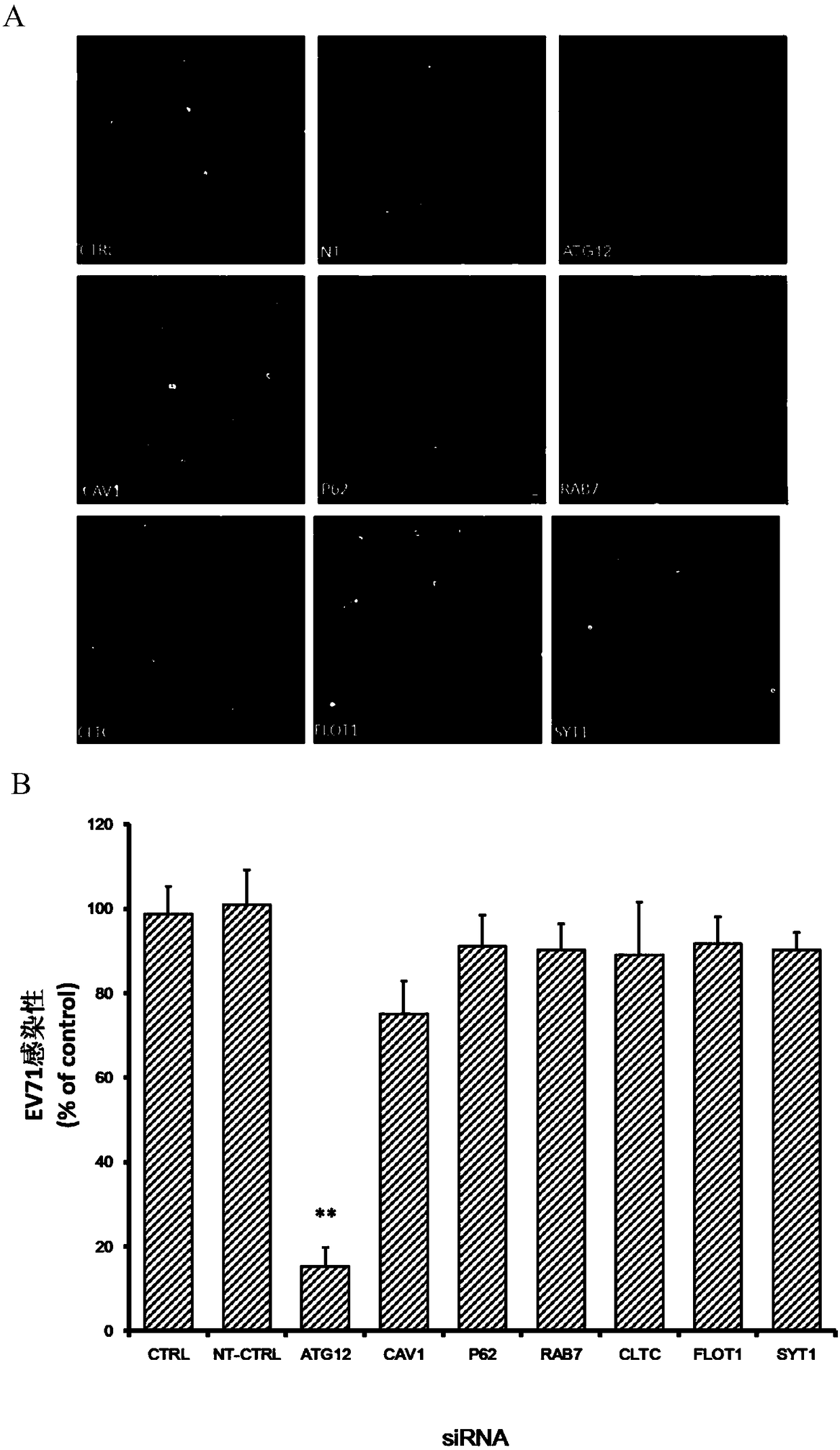
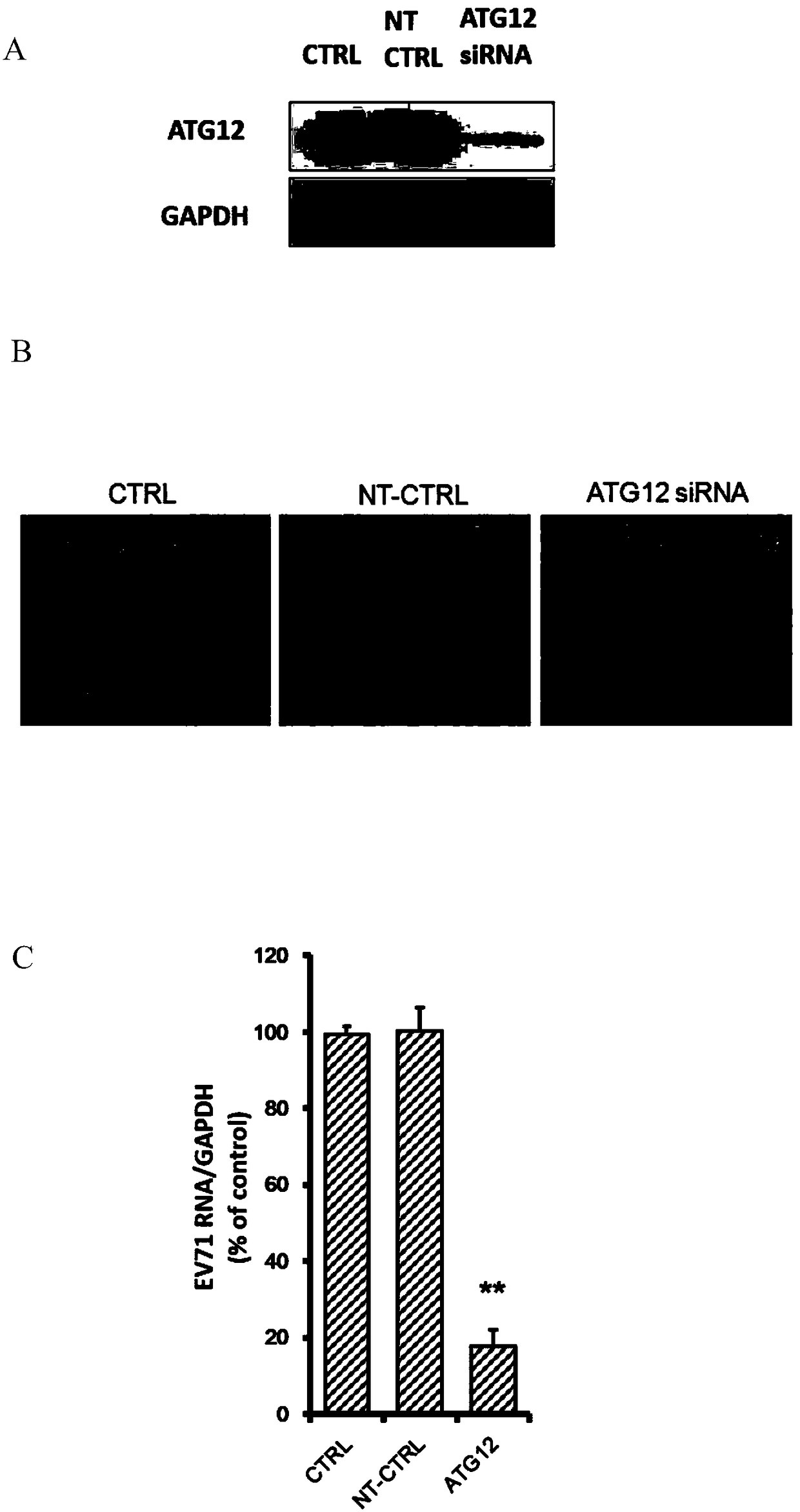
The invention belongs to the technical field of biomedicine, and relates to a novel target used for preventing enterovirus 71 infection, and applications thereof. According to the invention, Human Brain Microvascular Endothelial Cell (HBMEC) is taken as the target cell, and RNA interference technology is adopted for down-regulation of expression of target cell host protein so as to search host factors capable of inhibiting EV71 infection of HBMEC effectively, protect the functions of the blood brain barrier, and prevent central nervous system infection caused by penetrating of the blood brain barrier by virus. It is fond by experiments that the autophagy-related protein 12 (ATG12) possesses significant importance in infection of HBMEC by EV71, and down-regulation of expression of ATG12, and is capable of inhibiting EV71 infection obviously. The invention also provides applications of ATG12 in preparing drugs used for preventing or treating EV71 infection.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
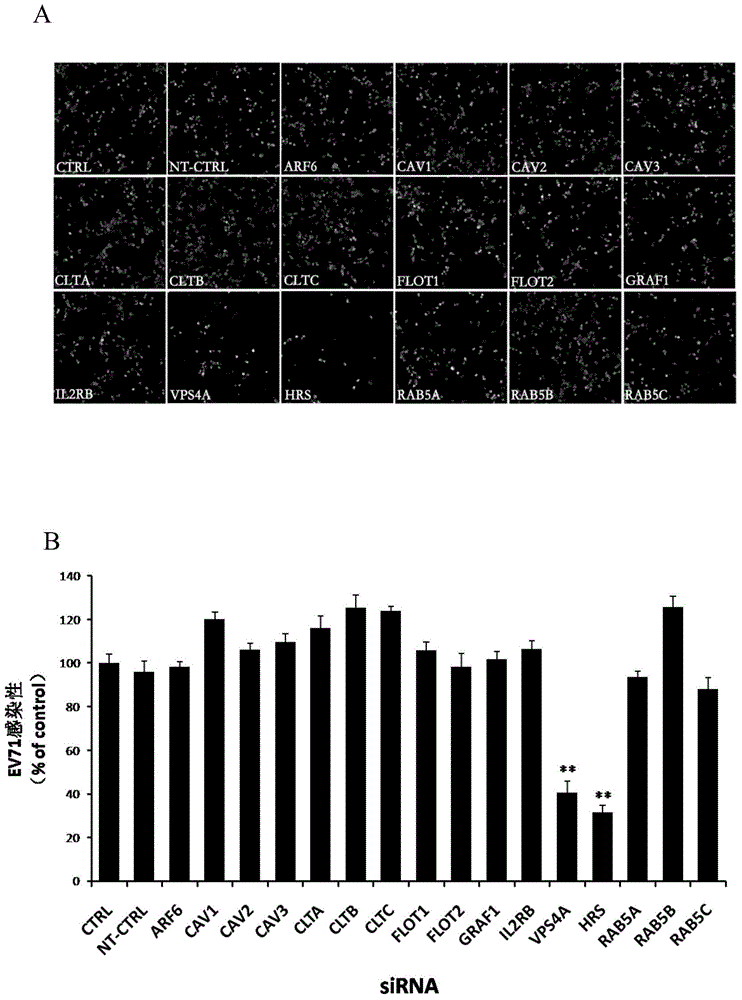
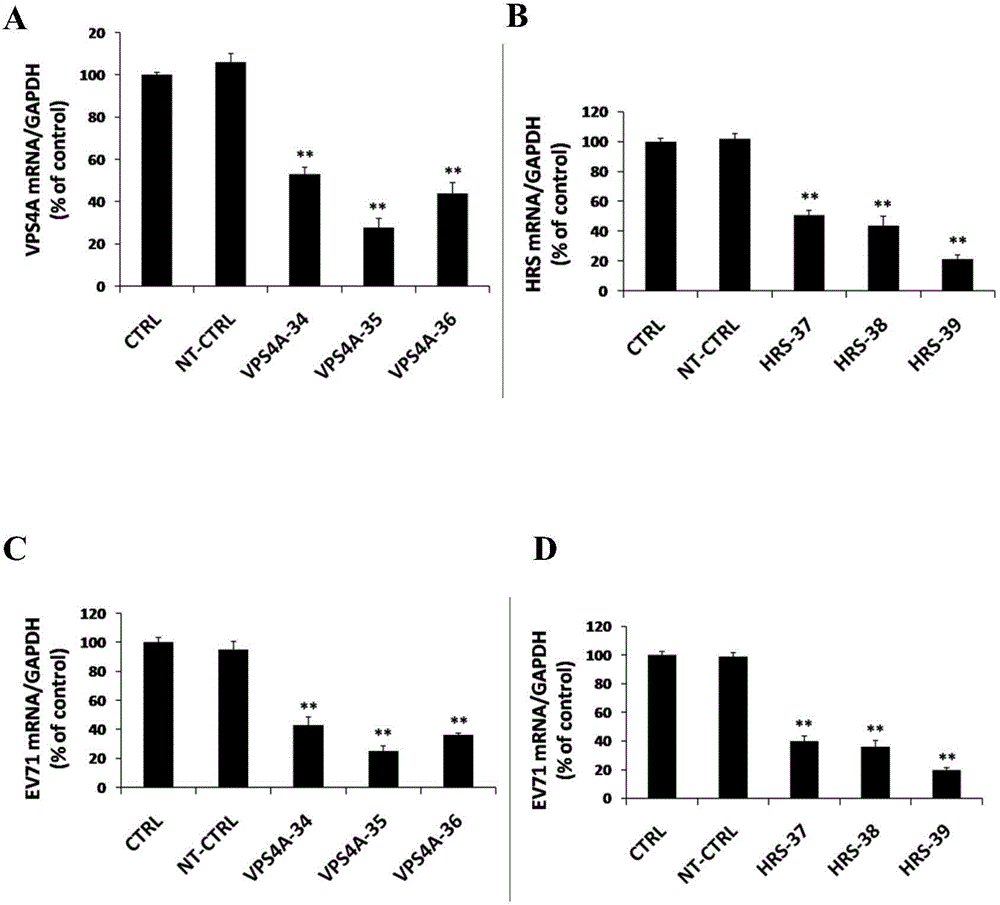
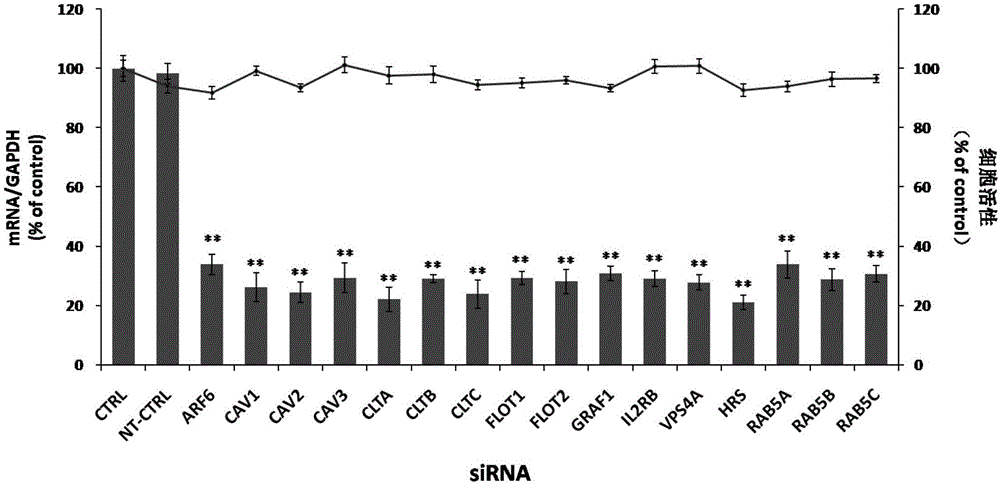
Applications of hepatocyte growth factor-regulated tyropsine kinasesubstrate in preparation of medicines preventing enterovirus 71-type infection
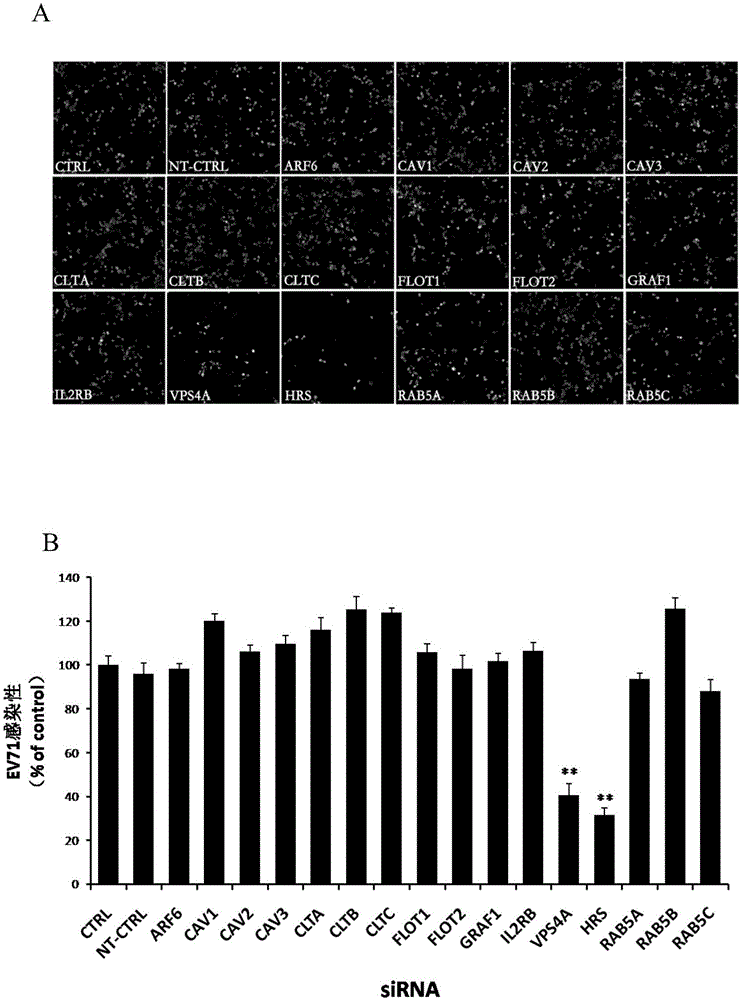
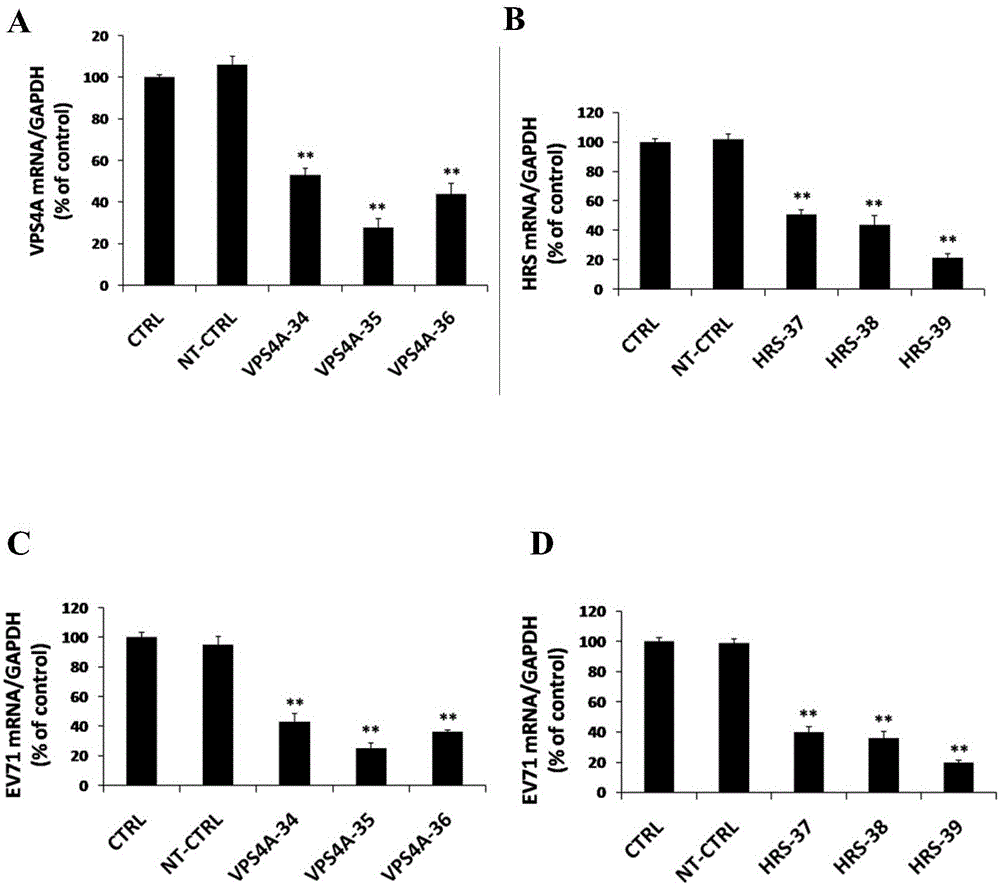
The invention relates to the biomedical technology field, and provides a new target spot for resisting enterovirus 71-type infection and applications. Human colon cancer cells (Caco-2) are employed as target cells, the RNA interference technology is employed to reduce expression of the target cell host proteins, a host factor capable of inhibiting EV71 infection of human colon cancer cells (Caco-2) effectively is sought and the purpose of cutting off EV71 infection from the source (intestinal tract) effectively is achieved. Experiments show that hepatocyte growth factor-regulated tyropsine kinasesubstrate (HRS) plays an important role in inhibiting EV71 infection of Caco-2, the HRS expression is reduced, and EV71 infection can be inhibited obviously. Applications of HRS in preparation of medicines preventing or treating enterovirus 71-type infection are provided.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
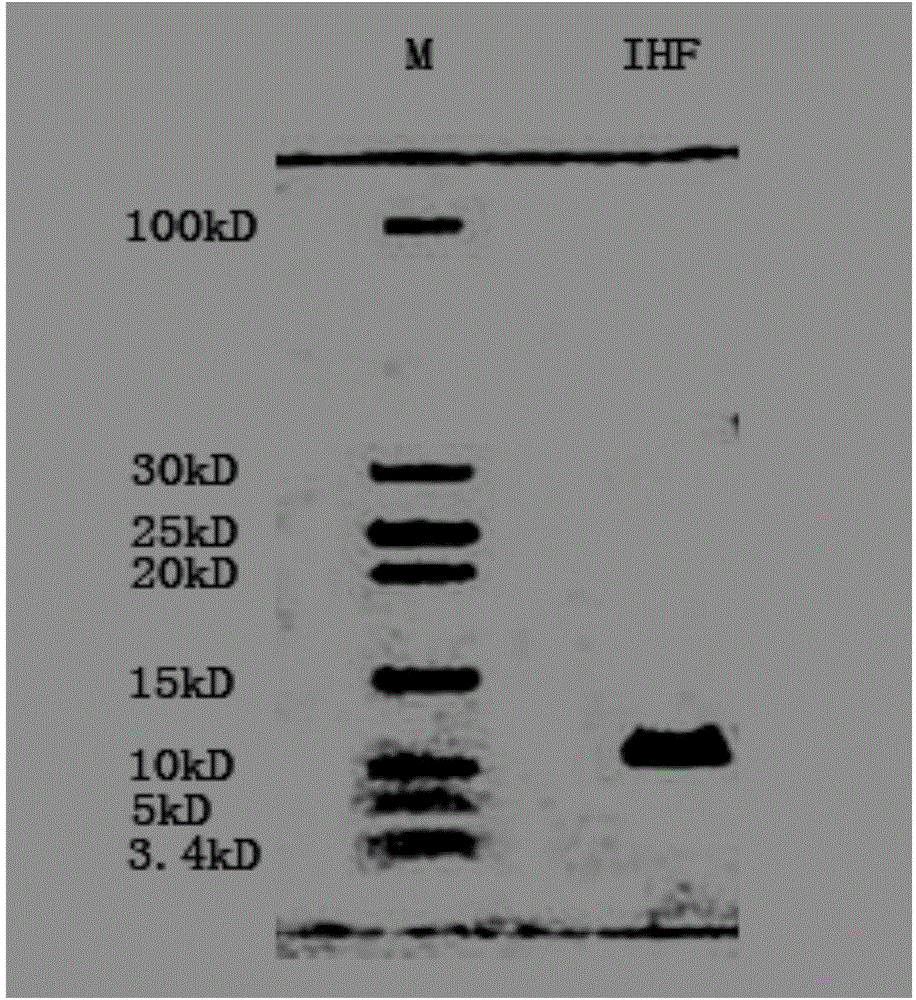
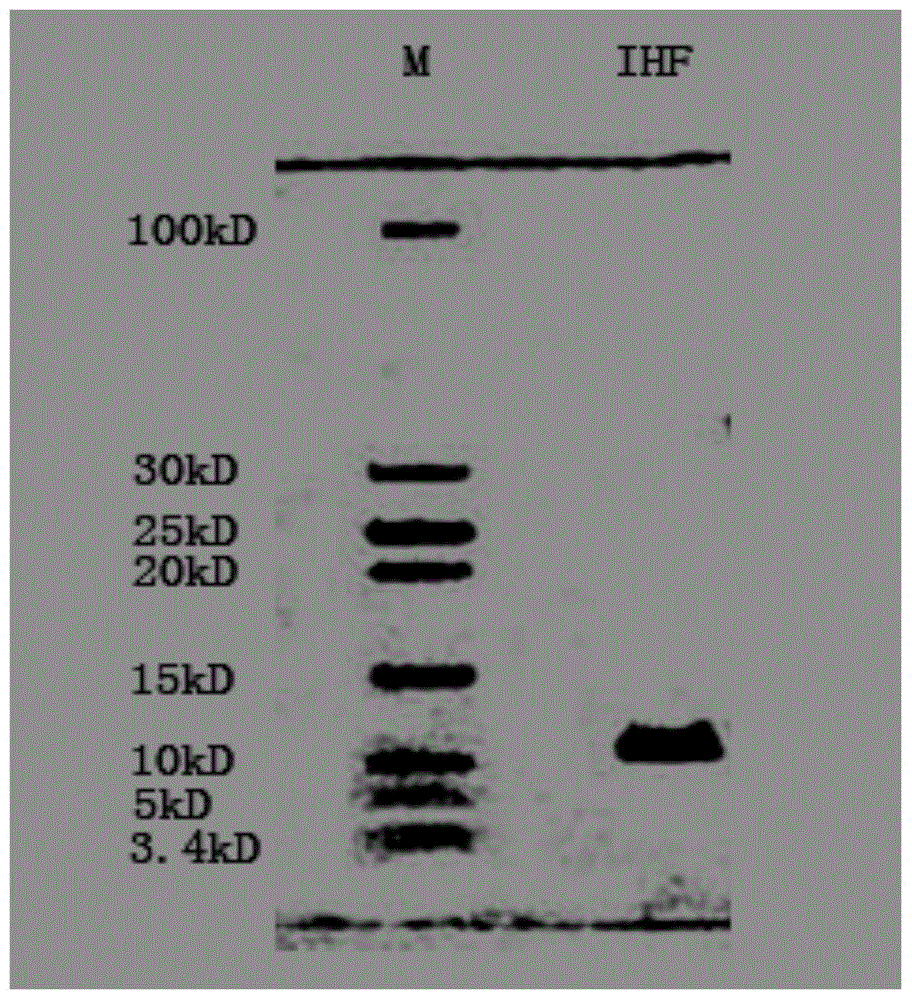
Method for improving PCR amplification sensitivity by using integrated host factor
ActiveCN104593491AHigh sensitivityImprove efficiencyMicrobiological testing/measurementHost factorsOligonucleotide primers
The invention provides a method for improving PCR amplification sensitivity by using an integrated host factor. The method comprises the following steps: adding the integrated host factor IHF into a reaction solution containing deoxyribonucleoside triphosphate, DNA polymerase, an oligonucleotide primer and a nucleic acid fragment which serves as a template, carrying out repeated heat circulation according to three steps of unwinding, annealing and extending of the normal PCR, thereby amplifying a target nucleic acid fragment, wherein the mole ratio of a primer to the integrated host factor IHF is 0.005-1. The method is capable of specifically and efficiently amplifying a target nucleic acid sequence.
Owner:华桥生物工程科技有限公司
A method of screening an HIV-1 activity determinative factor based on high-throughput RNAi
InactiveCN106032550AMicrobiological testing/measurementGenetic engineeringScreening techniquesHost factors
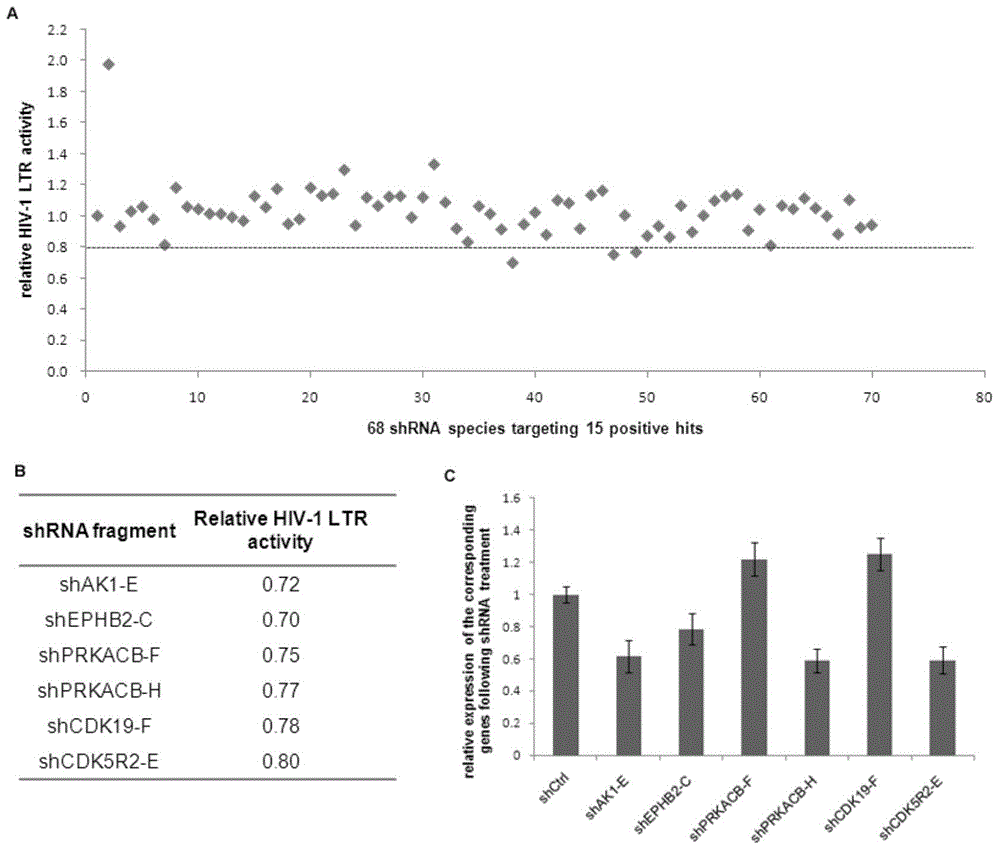
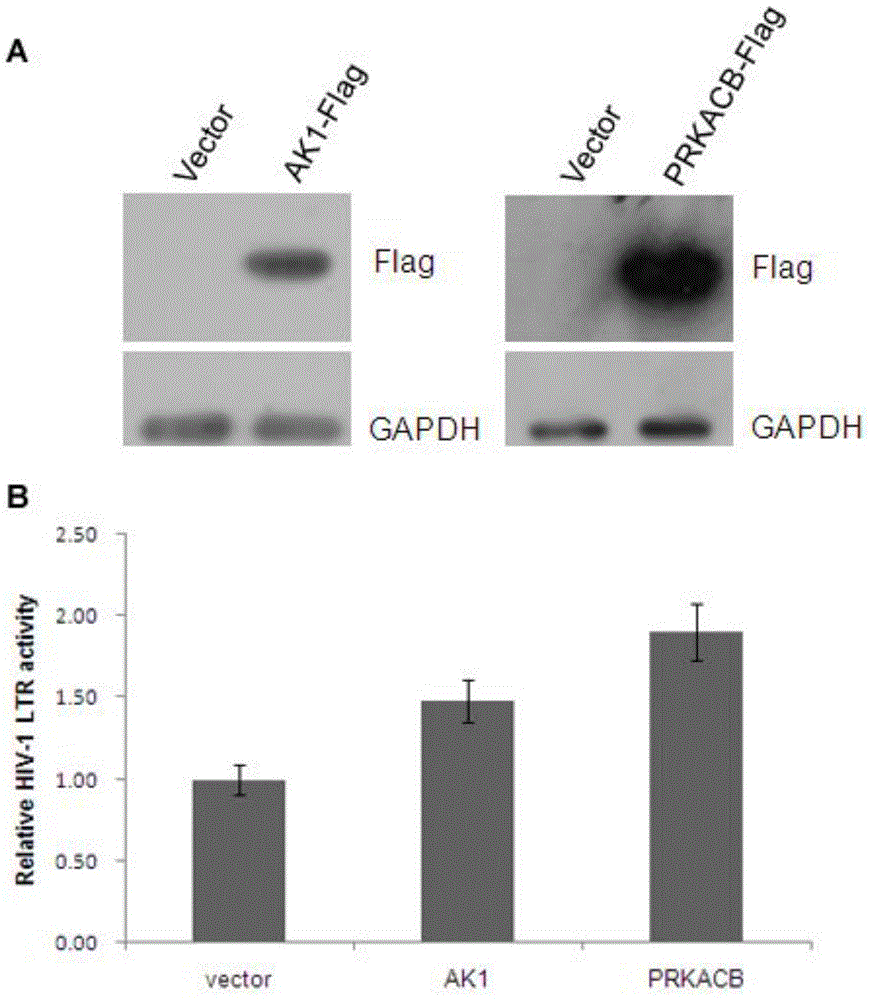
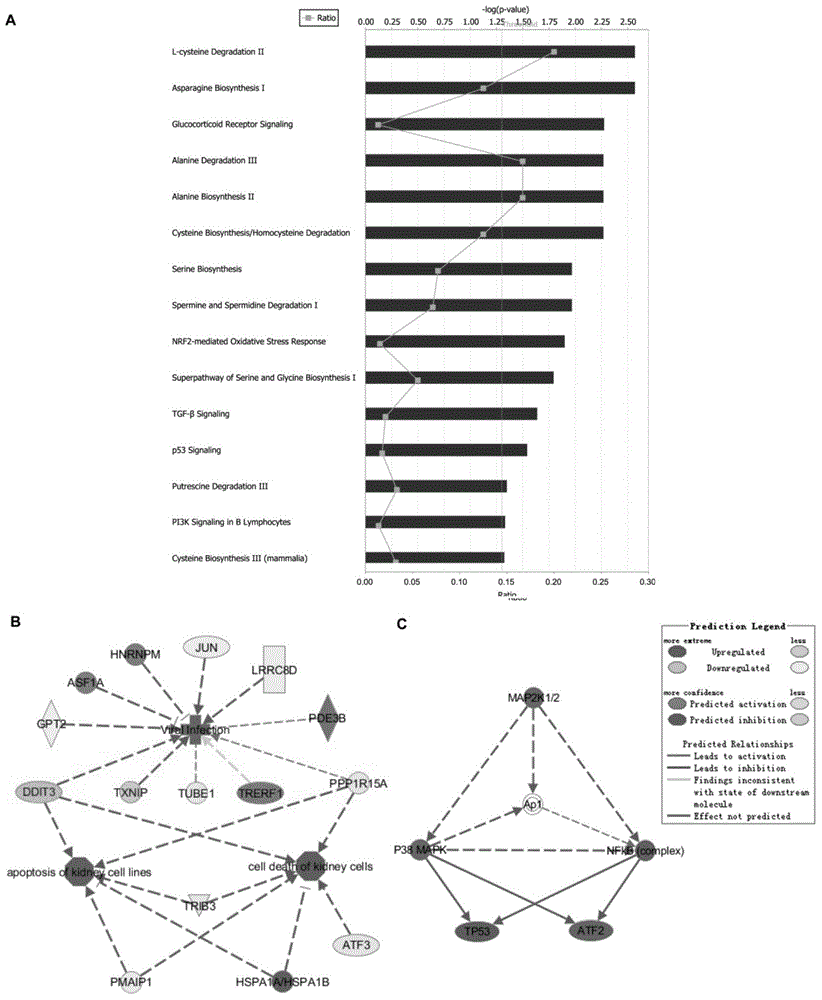
The invention relates to the technical field of molecular biology, and particularly relates to a method of screening an HIV-1 activity determinative factor based on high-throughput RNAi. Aiming at shRNA libraries of 474 kinases, and through a high-throughput RNAi screening technique, HIV-1 LTR-hRluc / HSVTK-hLuc luciferase double reporter plasmids are stably expressed in HEK293T, and kinase-involved HIV-1 LTR regulatory transcriptional activity is obtained by dividing HIV-1 LTR relative transcriptional activity of hRluc expression fluorescence intensity by hLuc fluorescence intensity. The method aims at human kinase genes, performs functional silence screening, searches HIV replication related host factors, and provides valuable reference basis for further understanding interactions between HIV and hosts and for searching HIV-infection resisting potential treatment targets.
Owner:SHANGHAI PUBLIC HEALTH CLINICAL CENT
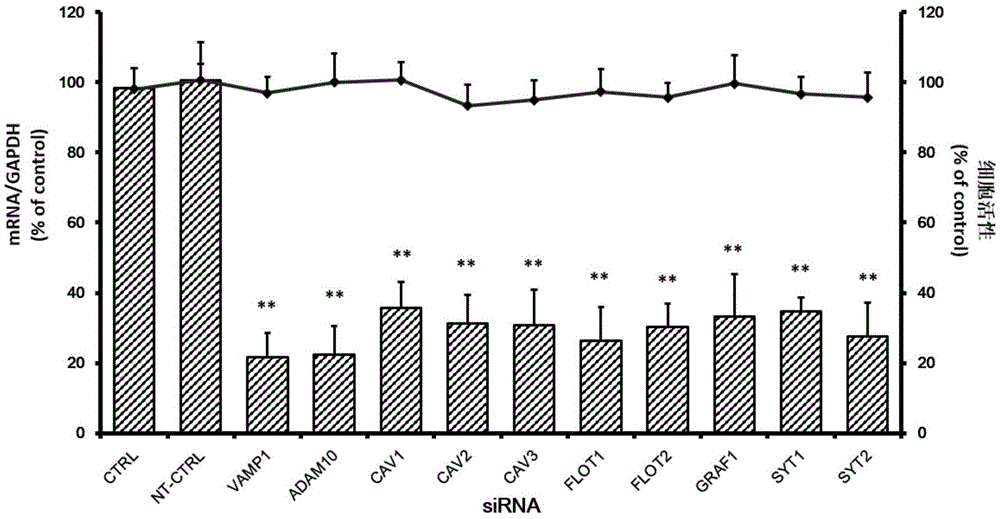
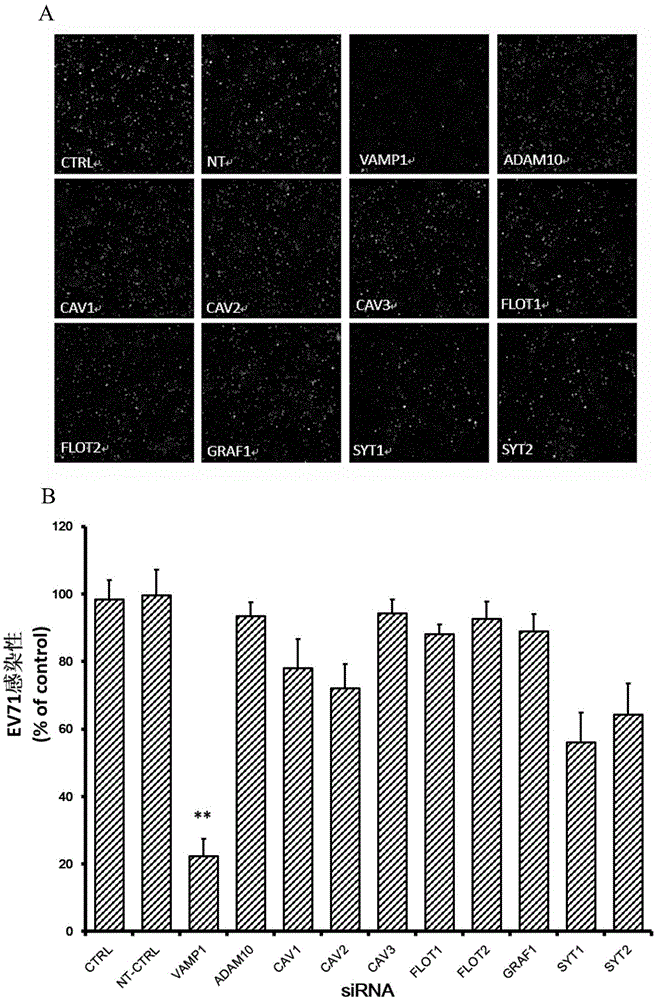
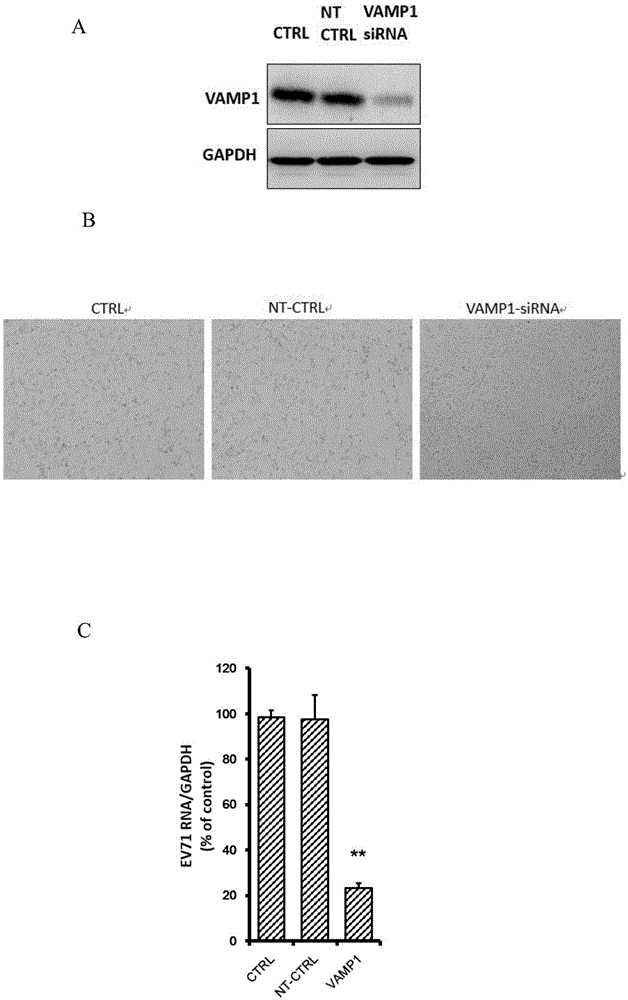
Applications of vesicle-associated membrane protein 1 in prevention and treatment of enterovirus 71 infection
The invention relates to the technical field of biomedicine, and particularly relates to a novel target preventing enterovirus 71 infection and applications thereof. Through adopting a human neuroblastoma cell (SH-SY5Y) as a target cell, and reducing expression of target cell host protein by adopting an RNA interference technique, a host factor effectively inhibiting EV71 infection of the human neuroblastoma cell (SH-SY5Y) is searched so as to prevent central nervous system virus infection and reducing damage to the nerve system. Experiments show that vesicle-associated membrane protein 1 (called as VAMP1 for short) plays an important role in EV71 infection of the human neuroblastoma cell (SH-SY5Y) and EV71 infection can be obviously inhibited by reducing expression of the VAMP1. The invention provides applications of the VAMP1 in preparation of medicines preventing and treating the enterovirus 71 infection.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
Application of vacuolar protein sorting 4A to preparing medicines for preventing or treating enterovirus type 71 infection
The invention provides application of vacuolar protein sorting 4A (VPS4A) to preparing medicines for preventing or treating enterovirus type 71 infection, relates to the technical field of biomedicine, and provides novel enterovirus type 71 infection resisting targets and application. The application includes that expression of host proteins of target cells which are human colon cancer cells (Caco-2) is down-regulated by the aid of an RNA (ribonucleic acid) interference technology, so that host factors capable of effectively inhibiting EV71 (enterovirus type 71)-infected human colon cancer cells (Caco-2) can be searched, and the purpose of effectively eliminating EV71 infection from sources (intestinal tracts) can be achieved. The application has the advantages that as discovered via experiments, important effects can be realized by the vacuolar protein sorting 4homolog A (VPS4A) for the EV71-infected Caco-2, expression of the VPS4A is down-regulated, and accordingly EV71 infection can be obviously inhibited.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
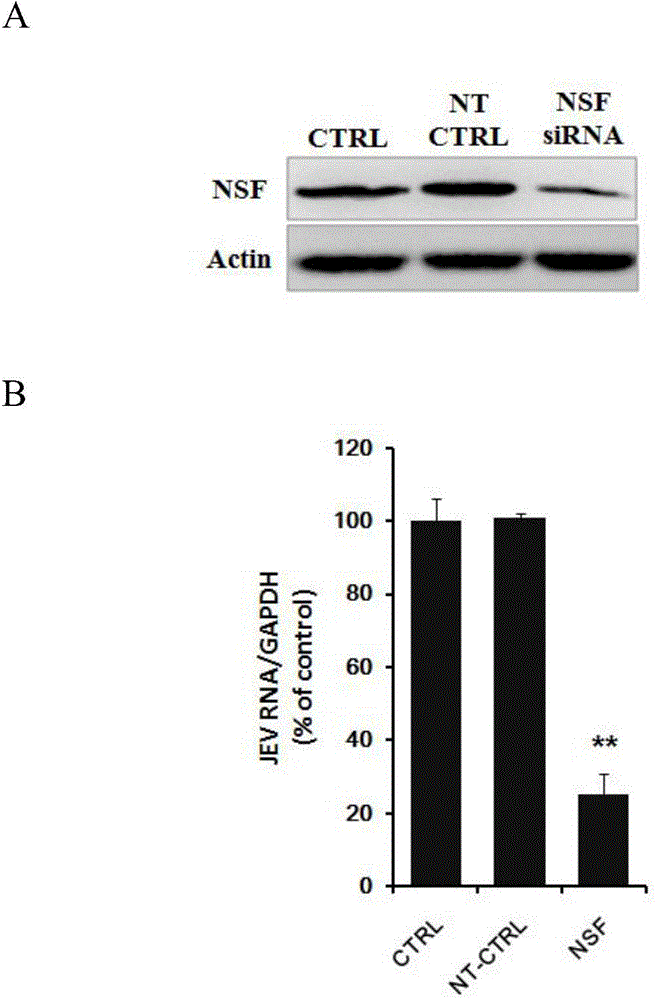
Applications of an N-ethylmaleimide sensitive factor in preventing Japanese encephalitis virus infection
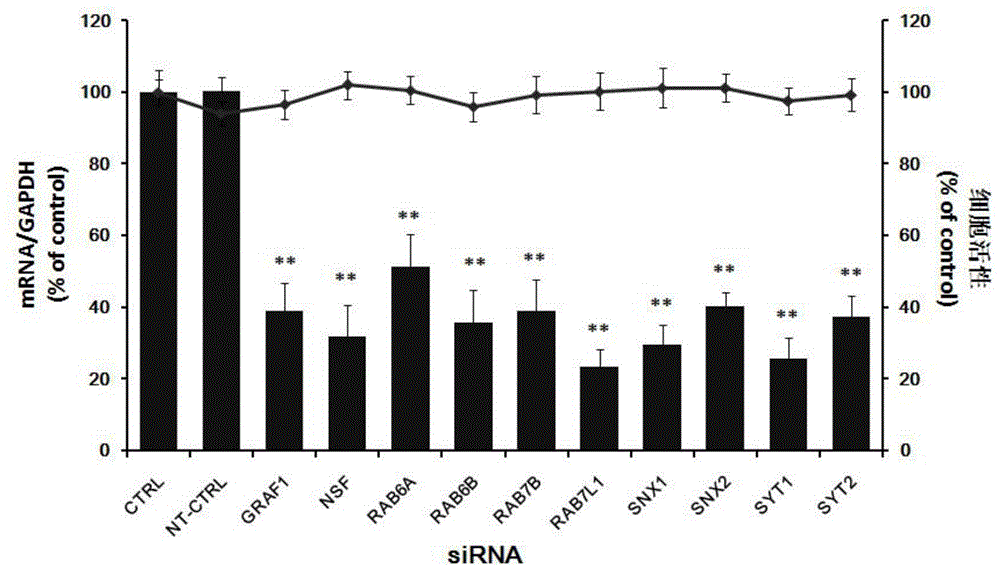
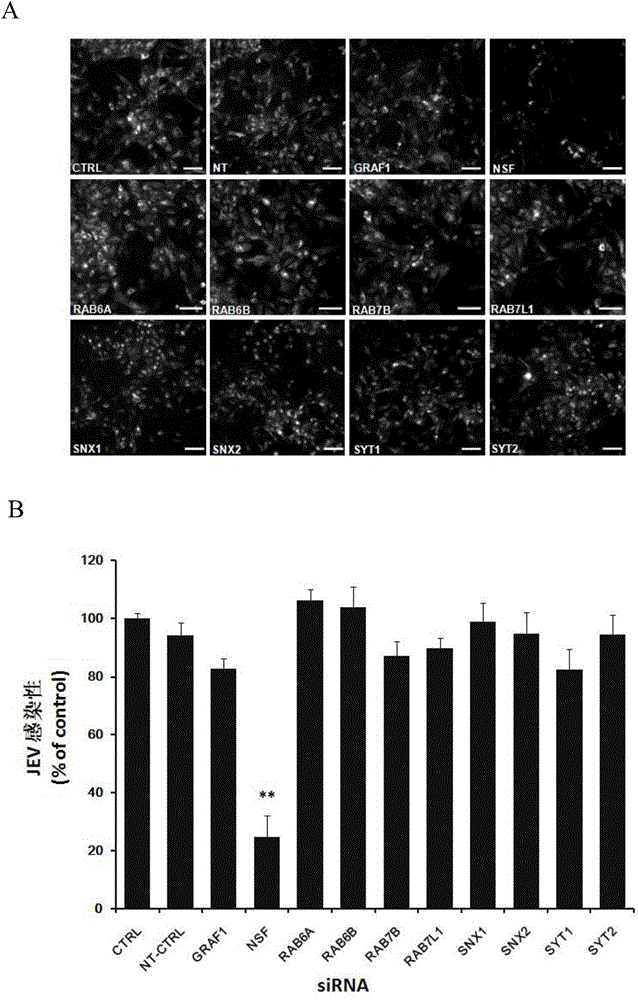
The invention relates to the biomedical technology field, and provides a new target spot for resisting Japanese encephalitis virus infection and applications. Human brain microvascular endothelial cells (HBMEC) are employed as target cells, the RNA interference technology is employed to reduce expression of target cell host proteins, a host factor capable of inhibiting Japanese encephalitis virus infection of human brain microvascular endothelial cells (HBMEC) effectively is sought, the function of protecting the blood brain barrier is achieved, viruses are prevented from going through the blood brain barrier and infecting the central nervous system. Applications of N-ethylmaleimide sensitive factor in preventing Japanese encephalitis virus infection are provided. Experiments show that the N-ethylmaleimide sensitive factor (NSF) plays an important role in inhibiting JEV infection of HBMEC, the NSF expression is reduced, and JEV infection can be inhibited obviously. Applications of NSF in preparation of medicines preventing or treating Japanese encephalitis virus infection are provided.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
Intestinal simulation model-based lactic acid bacteria intestinal probiotic effect measuring method
InactiveCN109439722AOvercoming sampling difficultiesHigh sensitivityBacteriaComponent separationShort-chain fatty acid metabolismBacteroides
The invention discloses an intestinal simulation model-based lactic acid bacteria intestinal probiotic effect measuring method. The intestinal simulation model-based lactic acid bacteria intestinal probiotic effect measuring method comprises the following steps of activating lactobacillus sakei LZ217; adding the activated lactobacillus sakei LZ217 into an intestinal simulation culture medium, meanwhile, setting a control group with no lactobacillus sakei LZ217 added, then adding in processed feces samples for simulating culture to obtain fermentation liquors, performing centrifugal treatment on the fermentation liquors to obtain supernatants; performing gas chromatography analysis on the supernatants to detect short-chain fatty acids. The intestinal simulation model-based lactic acid bacteria intestinal probiotic effect measuring method can be applied to detecting and analyzing the influence of the lactobacillus sakei LZ217 on the intestinal flora change and the short-chain fatty acidmetabolism of the volunteers, and meanwhile, through in vitro simulating fermentation, can overcome the difficulty in sampling intestinal bacteria and eliminate host factors.
Owner:ZHEJIANG GONGSHANG UNIVERSITY
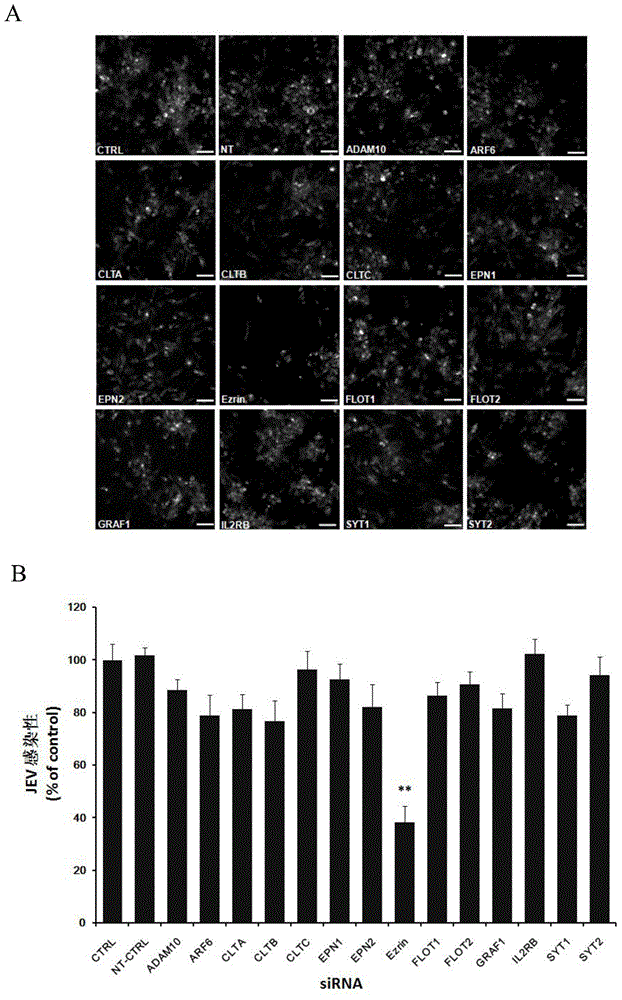
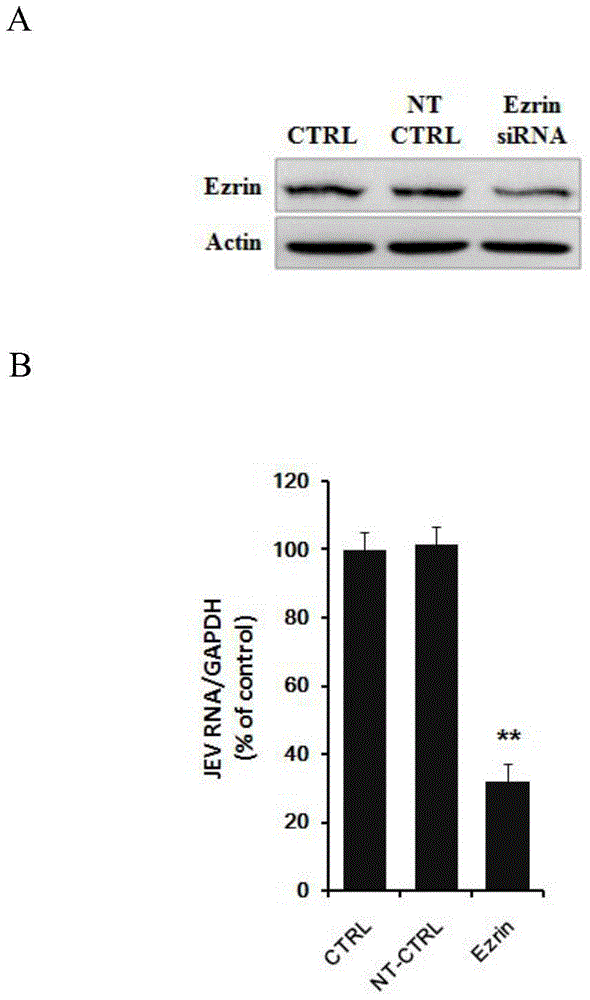
Application of Ezrin in preventing and treating Japanese encephalitis virus infection
ActiveCN105311646AOrganic active ingredientsGenetic material ingredientsVascular endotheliumBlood vessel
The invention relates to the technical field of biomedicine, in particular to a novel target resistant to Japanese encephalitis virus (JEV) infection and application. Human brain microvascular endothelial cells (HBMEC) are taken as target cells, RNA interference technology is adopted to reduce expression of target cell host protein to to find host factors capable of effectively inhibiting JEV-infected HBMECs so as to protect functions of a blood brain barrier and prevent virus from penetrating the blood brain barrier to infect a central nerve system. The invention provides application of Ezrin in preventing and treating JEV infection. Experiments find that Ezrin plays an important role in JEV-infected HBMECs, and JEV infection can be inhibited remarkably by reducing expression of Ezrin. The invention further provides application of Ezrin in preparing drug for preventing or treating JEV infection.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
Preparation method and application of anti-red-spot grouper neuronecrosis virus (tcm) embryonic stem cells
ActiveCN109295046AInhibitionHas a clearing effectMutant preparationEmbryonic cellsHost factorsViral infection
The invention belongs to the technical field of biology, and particularly relates to a preparation method and application of an anti-Epinephelus akaara nervous necrosis virus medaka haploid embryonicstem cell. Medaka haploid embryonic stem cells (HX1 cells) are induced by Ethylmtrosourea (ENU)), and then the anti-Epinephelus akaara nervous necrosis virus medaka haploid embryonic stem cell G1 canbe screened; the anti-Epinephelus akaara nervous necrosis virus medaka haploid embryonic stem cell G1is preserved in china center for type culture collection on January 17 th, 2018, the preservation address is wuhan university in china, and the preservation number is CCTCC NO: C201832. The anti-Epinephelus akaara nervous necrosis virus medaka haploid embryonic stem cell G1 provides an experiment basis and a theoretical basis for further identifying host factors participating in virus infection, and provides a new way for genetic breeding of antiviral fishes.
Owner:SUN YAT SEN UNIV
Application of protein tyrosine kinase FYN proto-oncogene in controlling enterovirus 71 infections
ActiveCN105288654AAvoid infectionDoes not affect physiological functionOrganic active ingredientsGenetic material ingredientsProtein-Tyrosine KinasesVascular endothelium
The invention relates to the technical field of biomedicine, in particular to a novel target resistant to enterovirus 71 infections and application thereof. Brain microvascular endothelial cells (HBMEC) are used as target cells, the expression of host proteins of the target cells are down-regulated by using the RNA interference technology, and thus host factors effective in inhibiting EV71 infections for brain microvascular endothelial cells (HBMEC) are thus found, the blood brain barrier function is protected, and the virus is prevented from penetrating the blood brain barrier to infect the central nervous system. Experiments show that protein tyrosine kinase FYN proto-oncogene (FYN proto-oncogene, Src family tyrosine kinase) molecules play an important role in the HBMEC infection by EV71, and EV71 infections can be significantly inhibited by down-regulating the expression of FYN. The invention provides application of the FYN in the preparation of drugs for preventing or treating enterovirus 71 infections.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
Applications of calcium-and inte-grin-binding protein 1 in preventing and treating enterovirus 71 infection
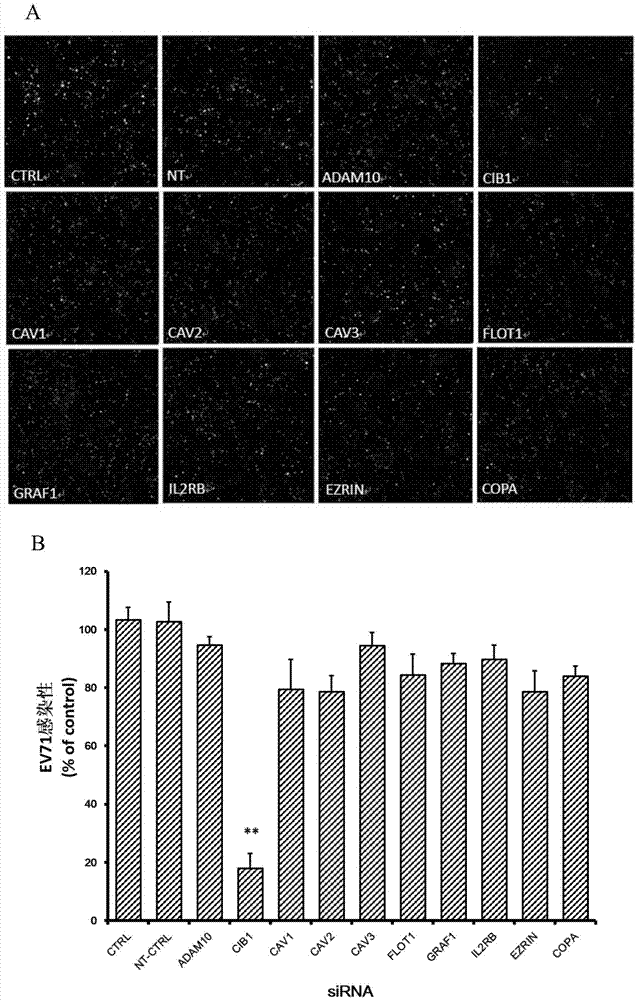
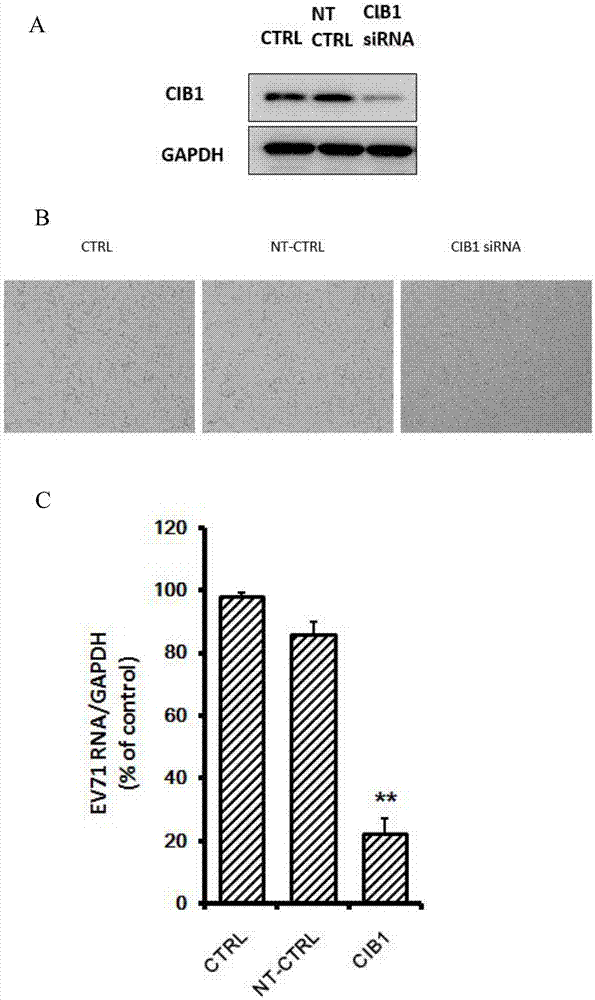
The invention belongs to the technical field of biomedicine, and relates to a novel target used for preventing enterovirus 71 infection, and applications thereof. According to the invention, SH-SY5Y is taken as the target cell, and RNA interference technology is adopted for down-regulation of expression of target cell host protein so as to search host factors capable of inhibiting EV71 infection of SH-SY5Y cells effectively and protect the functions of central nervous systems. It is fond by experiments that calcium-and inte-grin-binding protein, CIB1 molecular possesses significant importance in EV71 infection of SH-SY5Y, and down-regulation of expression of CIB1 is capable of inhibiting EV71 infection obviously. The invention also provides applications of CIB1 in preparing drugs used for preventing or treating EV71 infection.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
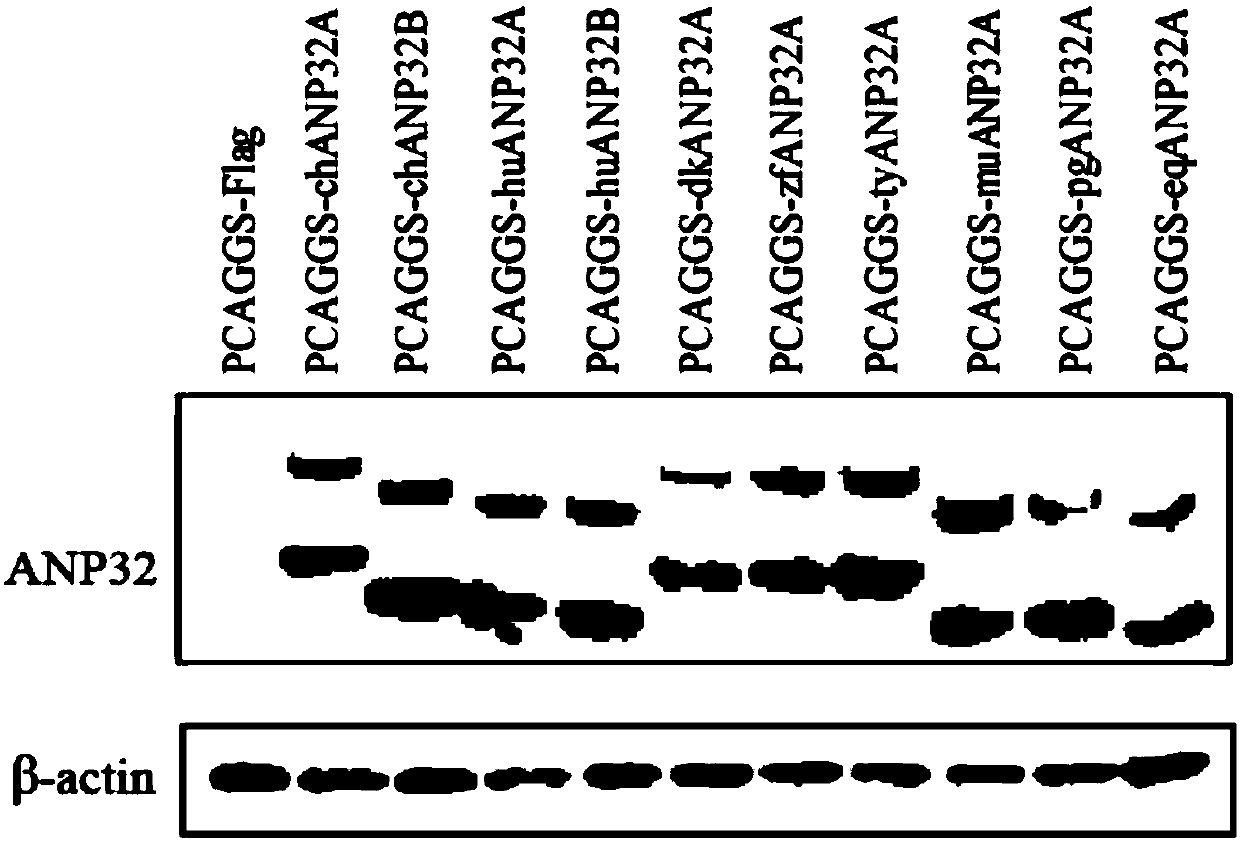
Application of ANP32 protein to maintain influenza virus polymerase activity in host
The invention provides recombination sequence information of a key host factor ANP32A / B necessary for replication of an influenza virus in a host. More specifically, the invention relates to a 129-130motif and a 149 site of host factor ANP32A / B protein, which are key active sites for their promotion of influenza virus replication and are potential targeting sites for anti-influenza drugs.
Owner:HARBIN VETERINARY RES INST CHINESE ACADEMY OF AGRI SCI
RNAi (RNA interference)-based transgenic novel watermelon material construction method
InactiveCN107267553AFermentationVector-based foreign material introductionPlant virusCotyledon plant
The invention relates to an RNAi (RNA interference) vector construction method and an application thereof in watermelon genetic transformation, relates to the technical field of plant virus control and belongs to the application technology in biological and modern agriculture technique fields. The mediated virus infection function is destroyed by acting on host factors and thus watermelons obtain antiviral activity, which has not been reported yet at present. An RNAi vector is constructed and genetically transformed to watermelons, the constructionmethod is characterized in that watermelon eIF4E and a homological isomer eIF(iso)4E gene thereof are cloned and connected, 4E-iso sense and antisense fragments are obtained and connected to two sides of an intron sequence, and a 4E-iso hairpin structure fragment is obtained; the 4E-iso sense fragment, the 4E-iso antisense fragment and the hairpin structure fragment are inserted into a pCAMBIA1301 vector through T4 connection and homologous recombination, and the RNAi vector is obtained; target gene fragments are transformed into watermelon cotyledon explants through agrobacterium tumefaciens-medicated transformation, and transgenic watermelons with the target gene fragments are obtained through screening cultivation. The method is mainly used for providing a novel material and a novel technology for watermelon virus and disease control.
Owner:NORTHWEST A & F UNIV
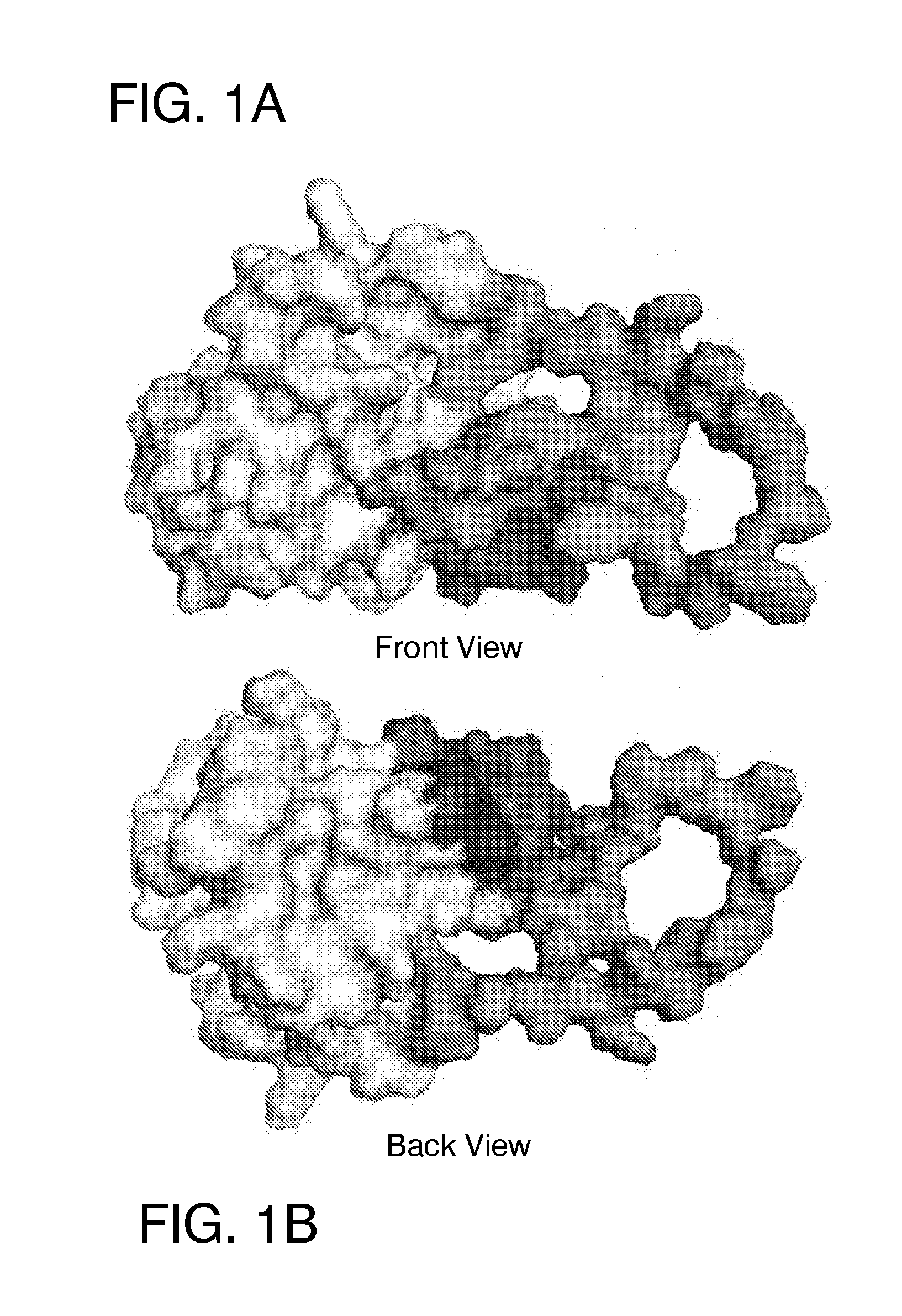
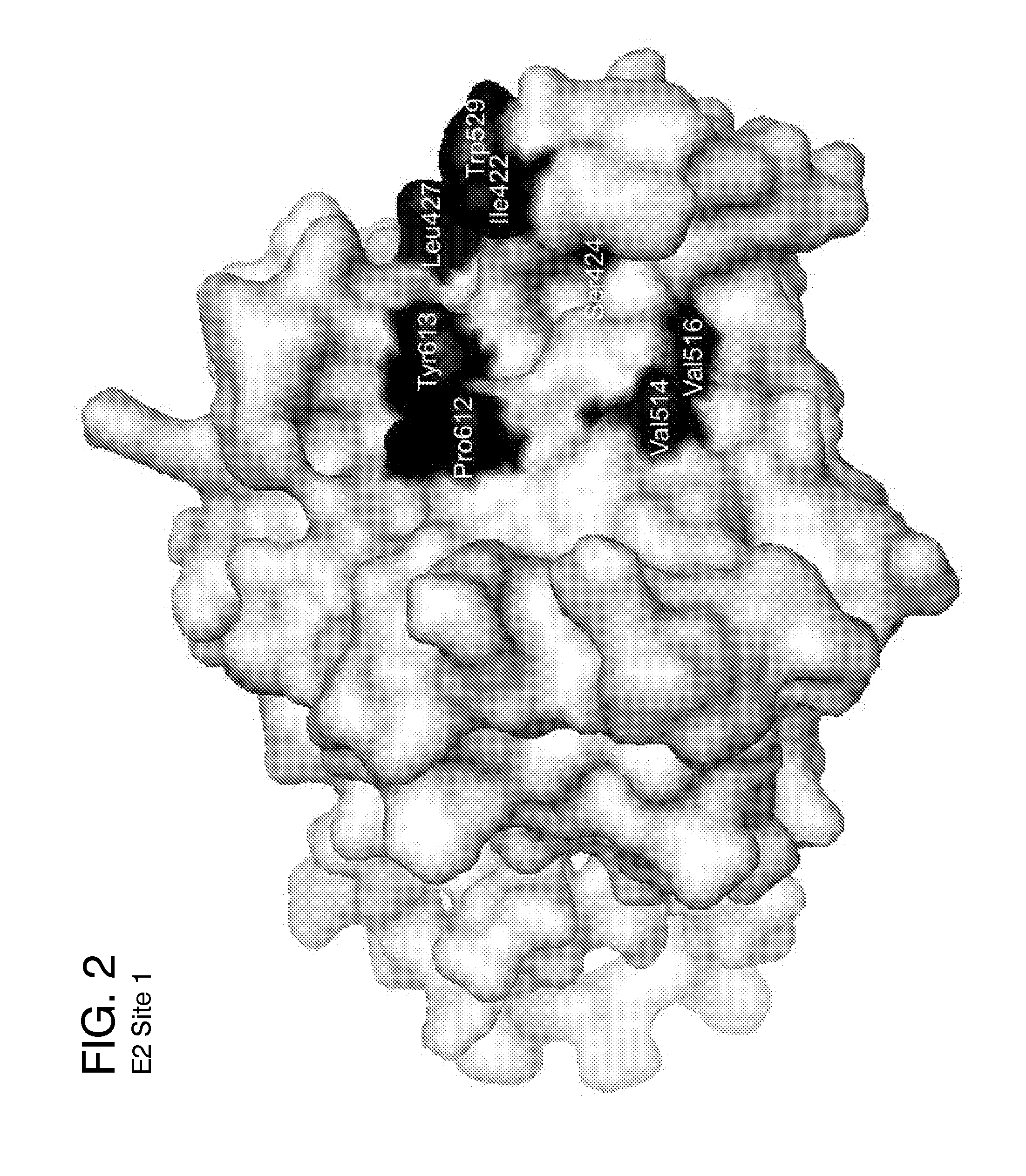
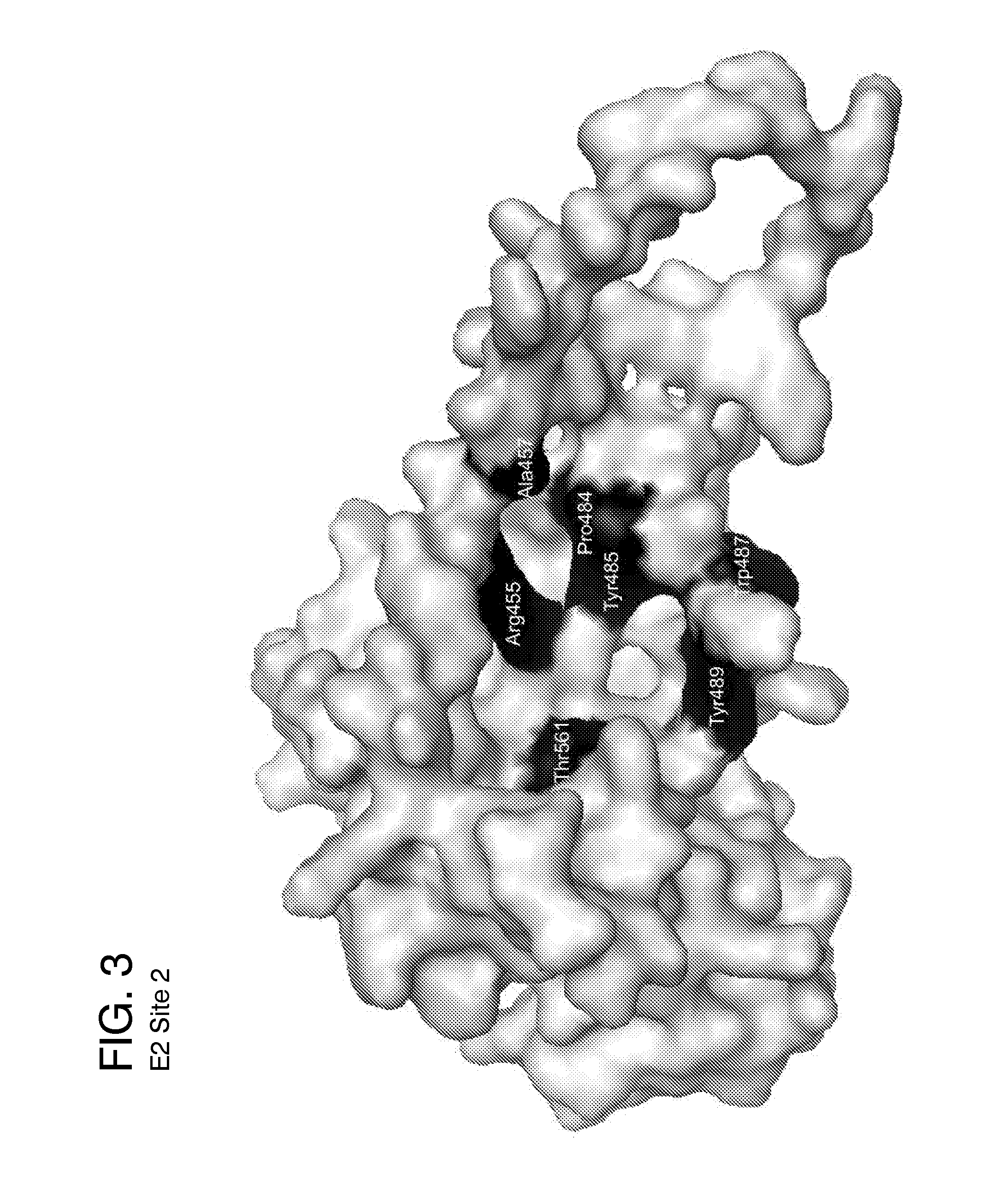
Ligands that target hepatitis c virus e2 protein
InactiveUS20160361311A1Reduce severityPharmaceutical non-active ingredientsHeterocyclic compound active ingredientsPolymerase LInterferon alpha
Hepatitis C Virus (HCV) infects 200 million individuals worldwide. Although several FDA approved drugs targeting the HCV serine protease and polymerase have shown promising results, there is a need for better drugs that are effective in treating a broader range of HCV genotypes and subtypes without being used in combination with interferon and / or ribavirin. Recently, the crystal structure of the core of the HCV E2 protein (E2c) has been determined, providing structural information that can now be used to target the E2 protein and develop drugs that disrupt the early stages of HCV infection by blocking E2's interaction with different host factors. By targeting sites containing conserved E2 amino acids in the CD81 binding site on HCV E2, one might also be able to develop drugs that block HCV infection in a genotype-independent manner. Using the E2c structure as a template, a structural model of the E2 protein core (residues 421-645) was developed that includes the three amino acid segments that are not present in the E2c crystal structure. Blind docking of a diverse library of 1715 small molecules to this model led to the identification of a set of 34 ligands predicted to bind near conserved amino acid residues involved in the HCV E2:CD81 interaction. Surface plasmon resonance was used to screen the ligand set for binding to recombinant E2 protein, and the best binders were subsequently tested to identify compounds that inhibit the infection of hepatocytes by HCV. One compound, 281816, blocked E2 binding to CD81 and inhibited hepatocyte infection by HCV genotypes 1a, 1b, 2a, 2b, 4a and 6a with IC50's ranging from 2.2 μM to 4.6 μM. Methods are described for preventing or treating HCV infection using small molecule inhibitors such as 281816 that target E2 and disrupt its interactions.
Owner:AMERICAN UNIV OF CAIRO
Methods and compositions for integration-defective lentiviral vectors
The present invention provides an integration-defective lentiviral vector based on a parental lentivirus and related methods, the integration-defective lentiviral vector including one or more of the following: (a) a mutation, deletion or other modification of one or more binding sites for a host factor involved in gene silencing; (b) an addition of one or more binding sites for a transcription activator, which can be natural (such as but not limited to ubiquitous and / or tissue / cell type specific) including but not limited to SP1 NFkB, or synthetic including but not limited to binding sites for tetracycline regulated trans activators tTA, rtTA, tT65, and / or rtT65; (c) one or more nucleic acid sequences from a SV40 genome, wherein the one or more sequences are obtained from a region of the SV40 genome upstream to the SV40 poly-adenylation signal; (d) a shRNA expression cassette, which encodes a shRNA directed to a host gene involved in epigenetic silencing and / or in DNA repair pathways; or (e) any combination of (a), (b), (c) and (d), wherein as compared to the parental lentivirus, the integration-defective lentiviral vector resists gene silencing.
Owner:THE UNIV OF NORTH CAROLINA AT CHAPEL HILL
Methods and compositions for inhibiting HIV infection
This invention provides novel HIV-interacting host factors. The invention also provides methods of using the HIV-interacting host factors to screen for compounds that inhibit HIV infection. The methods comprise first screening test compounds for modulators of an HIV-interacting host factor disclosed herein, and then further screening the identified modulating compounds for ability to inhibit HIV infection. The invention further provides methods and pharmaceutical compositions for treating diseases and conditions associated with HIV infection.
Owner:IRM
Application of dynamin 1 in prevention and treatment on enterovirus type 71 infection
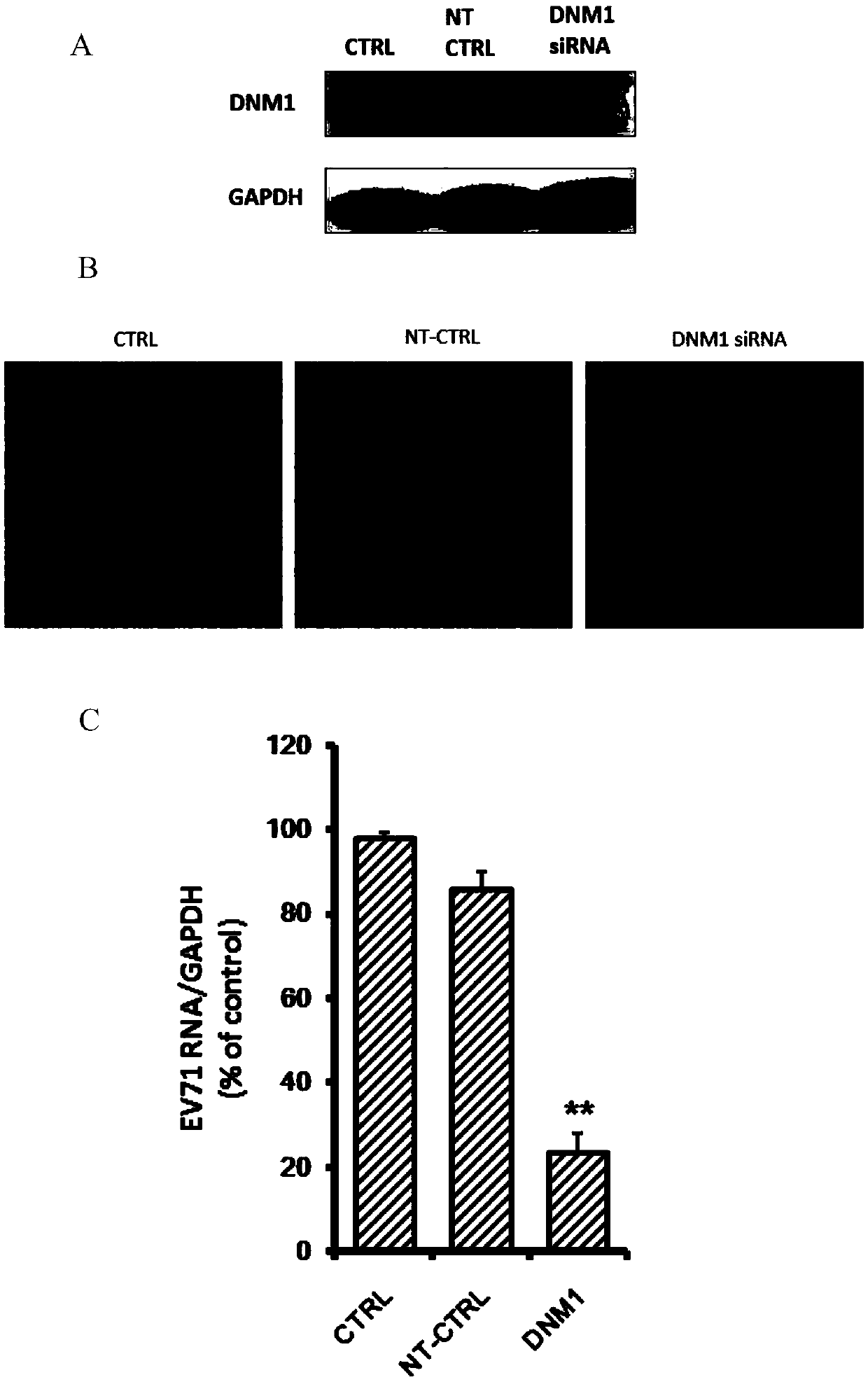
ActiveCN105497918AAvoid infectionDoes not affect physiological functionOrganic active ingredientsGenetic material ingredientsNervous systemNeuroblastic Tumor
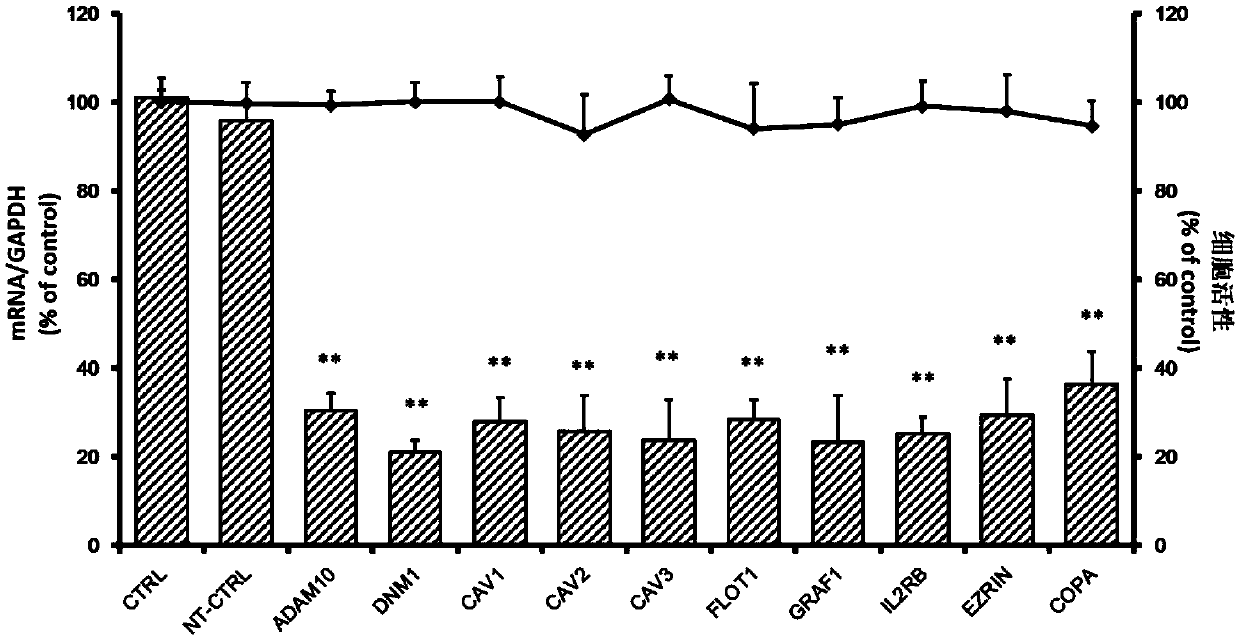
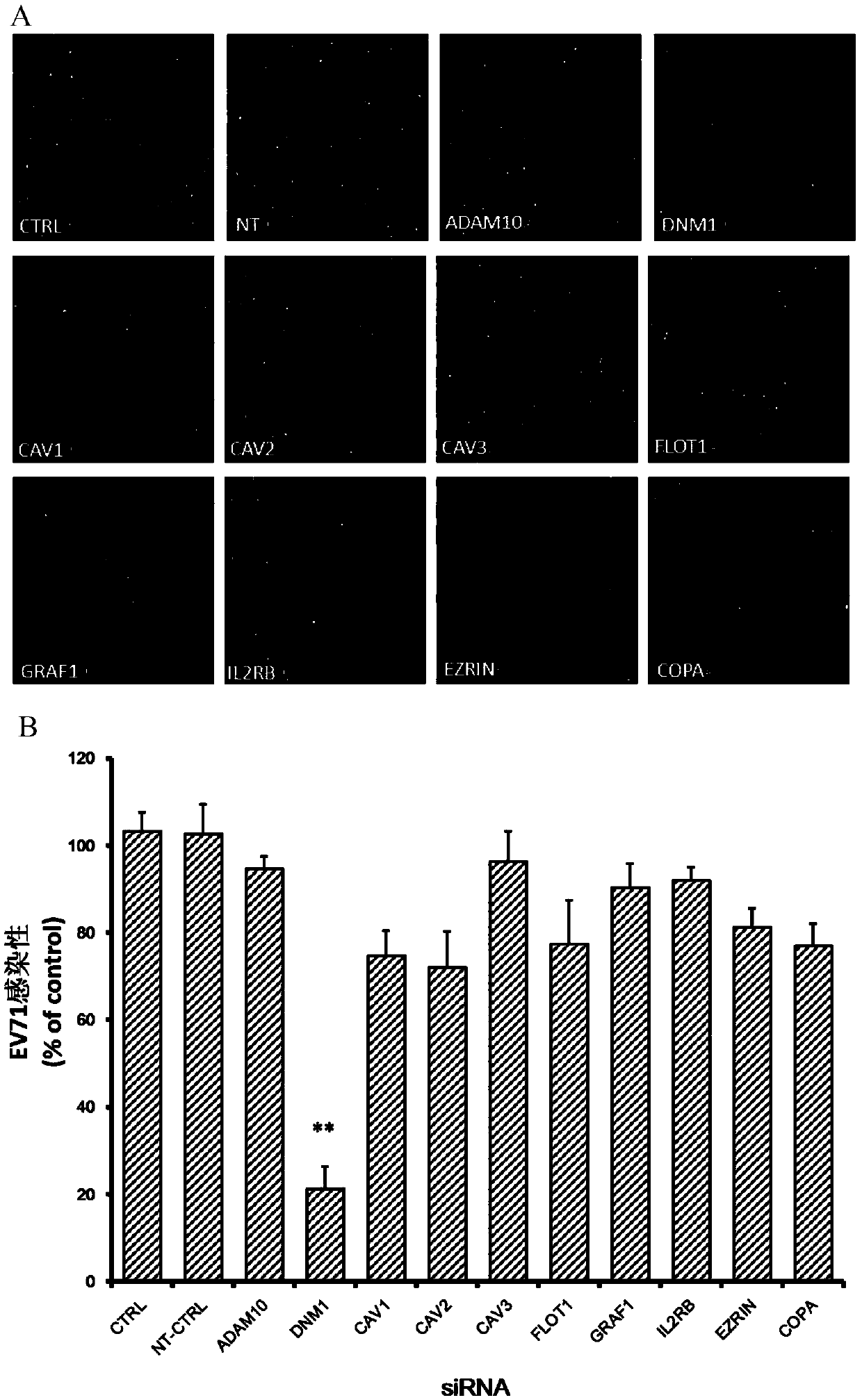
The invention relates to the technical field of biomedicine, and provides a new target for resisting entervirus type 71 infection and application. A human neuroblastoma cell (SH-SY5Y) is taken as a target cell, expression of host protein of the target cell is down-regulated by adopting an RNA interference technique to look for a host factor which can effectively inhibit the EV71-infected human neuroblastoma cell (SH-SY5Y), and therefore the function of a central nervous system is protected. Through experiments, it is found that a dynamin 1 (NDM1) plays an important role on the EV71-infected SH-SY5Y, and EV71 infection can be obviously inhibited by down-regulating expression of the DNM1. The invention provides application of the DNM1 in preparation of medicine for preventing or treating the entervirus type 71 infection.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
Methods and compositions for integration defective lentiviral vectors
The present invention provides an integration-defective lentiviral vector based on a parental lentivirus and related methods, the integration-defective lentiviral vector including one or more of the following: (a) a mutation, deletion or other modification of one or more binding sites for a host factor involved in gene silencing; (b) an addition of one or more binding sites for a transcription activator, which can be natural (such as but not limited to ubiquitous and / or tissue / cell type specific) including but not limited to SP1 NFkB, or synthetic including but not limited to binding sites for tetracycline regulated trans activators tTA, rtTA, tT65, and / or rtT65; (c) one or more nucleic acid sequences from a SV40 genome, wherein the one or more sequences are obtained from a region of the SV40 genome upstream to the SV40 poly-adenylation signal; (d) a shRNA expression cassette, which encodes a shRNA directed to a host gene involved in epigenetic silencing and / or in DNA repair pathways; or (e) any combination of (a), (b), (c) and (d), wherein as compared to the parental lentivirus, the integration-defective lentiviral vector resists gene silencing.
Owner:THE UNIV OF NORTH CAROLINA AT CHAPEL HILL
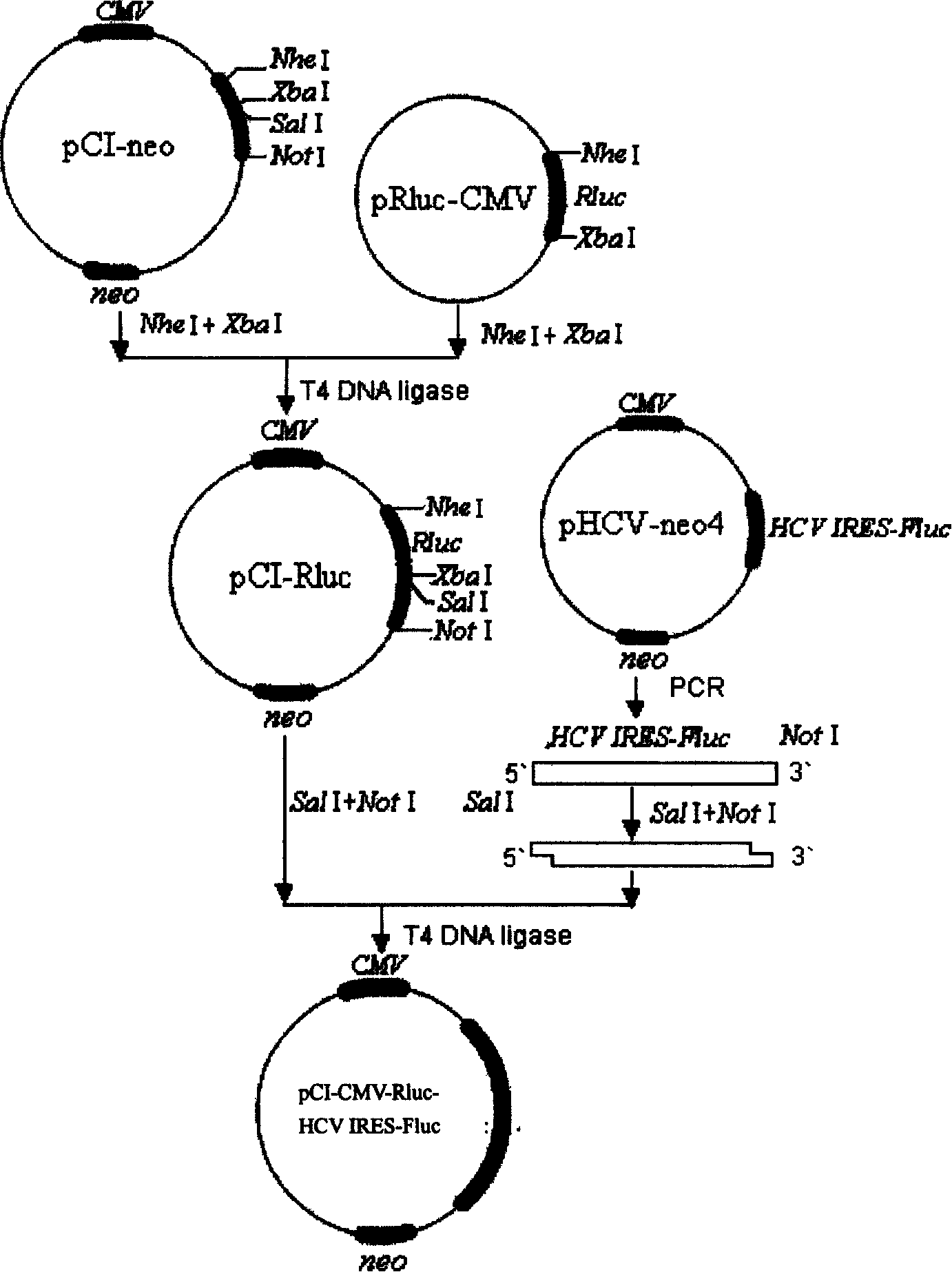
HCV transgene mouse model and its construction method and application
InactiveCN100410381CAvoid errorsCutting costsFermentationVector-based foreign material introductionHost factorsMedicine use
A HCV transgenic mouse model is prepared through configuring the bicistronic expression carrier including sea pensy leuciferinase gene, glowworm leuciferinase gene and the HCV IRES sequence between them, and transfecting it into mouse body to obtain said model. It can be used for screening and evaluating the HCV-resistant medicines using HCV IRES as target, such as antisense oligonucleotide, specific ribozyme, siRNA, etc, and researching the affection of host factors to HCV IRES.
Owner:FIELD OPERATION BLOOD TRANSFUSION INST OF PLA SCI ACAD OF MILITARY
Tomato RNA virus host factor and genes encoding same and use thereof
The present invention discloses one kind of tomato RNA virus host factor and its coding gene and application in antiviral breeding of plant. The tomato RNA virus host factor is protein possessing one of the following amino acid residue sequences: 1) SEQ ID No. 1 in the sequence list; and 2) the amino acid residue sequence of SEQ ID No. 1 in the sequence list and through the substitution, deletion or addition of 1-10 amino acid residues and supporting virus copying protein. Silencing the protein coding gene inside plant body via transgenic antisense gene segment or RNA interfering can obtain antiviral plant. The tomato RNA virus host factor gene ToTOM3 and the method of inhibiting the expression of the gene has practical significance and wide application foreground in culturing and breeding antiviral plant.
Owner:GUANGXI UNIV
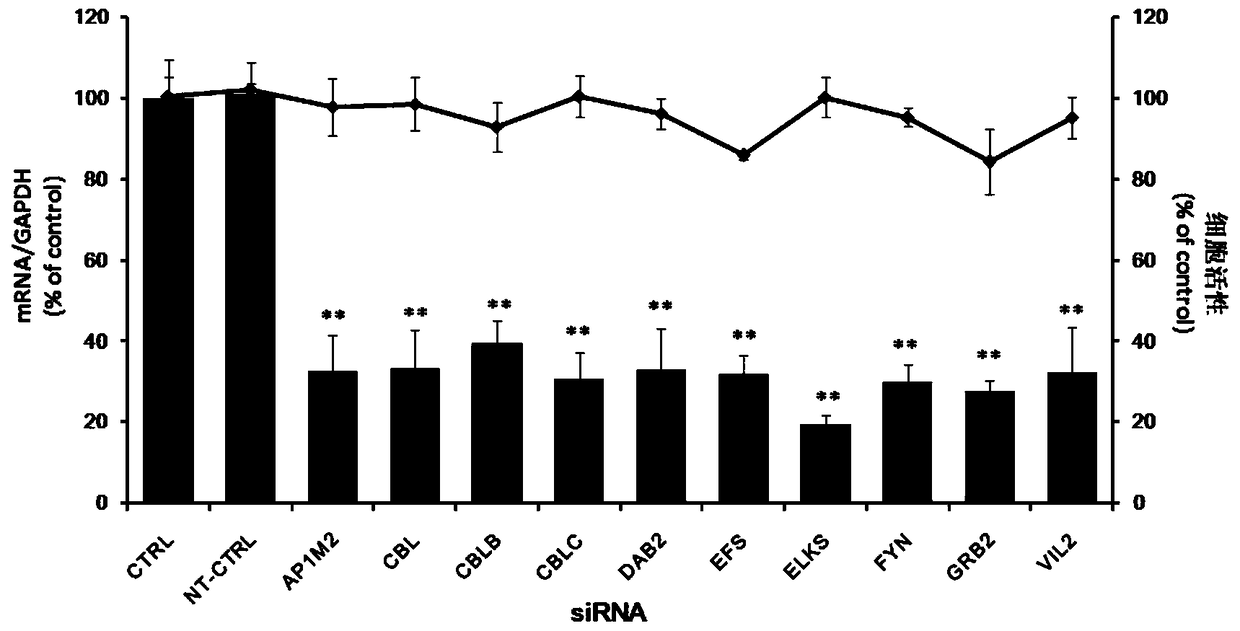
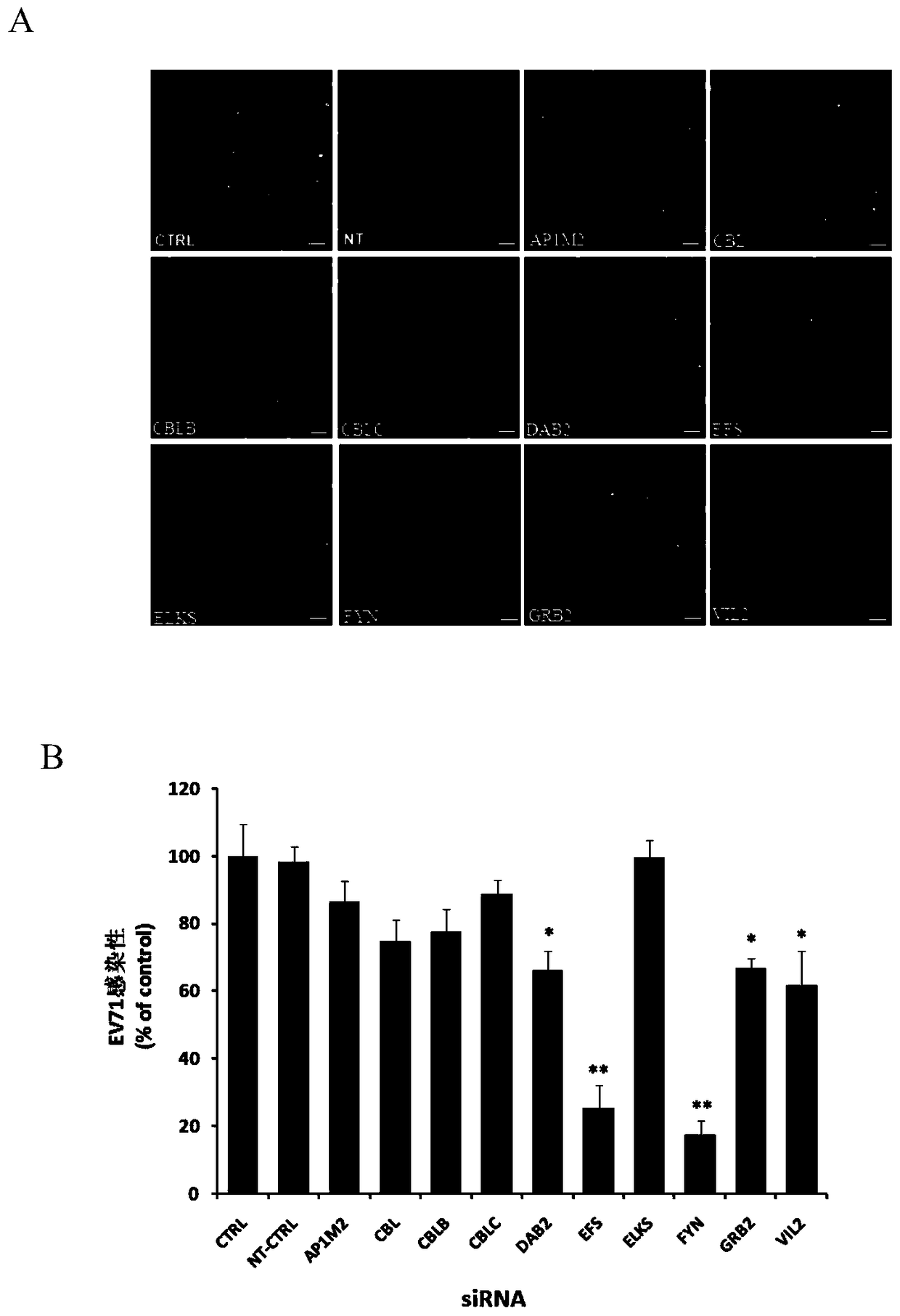
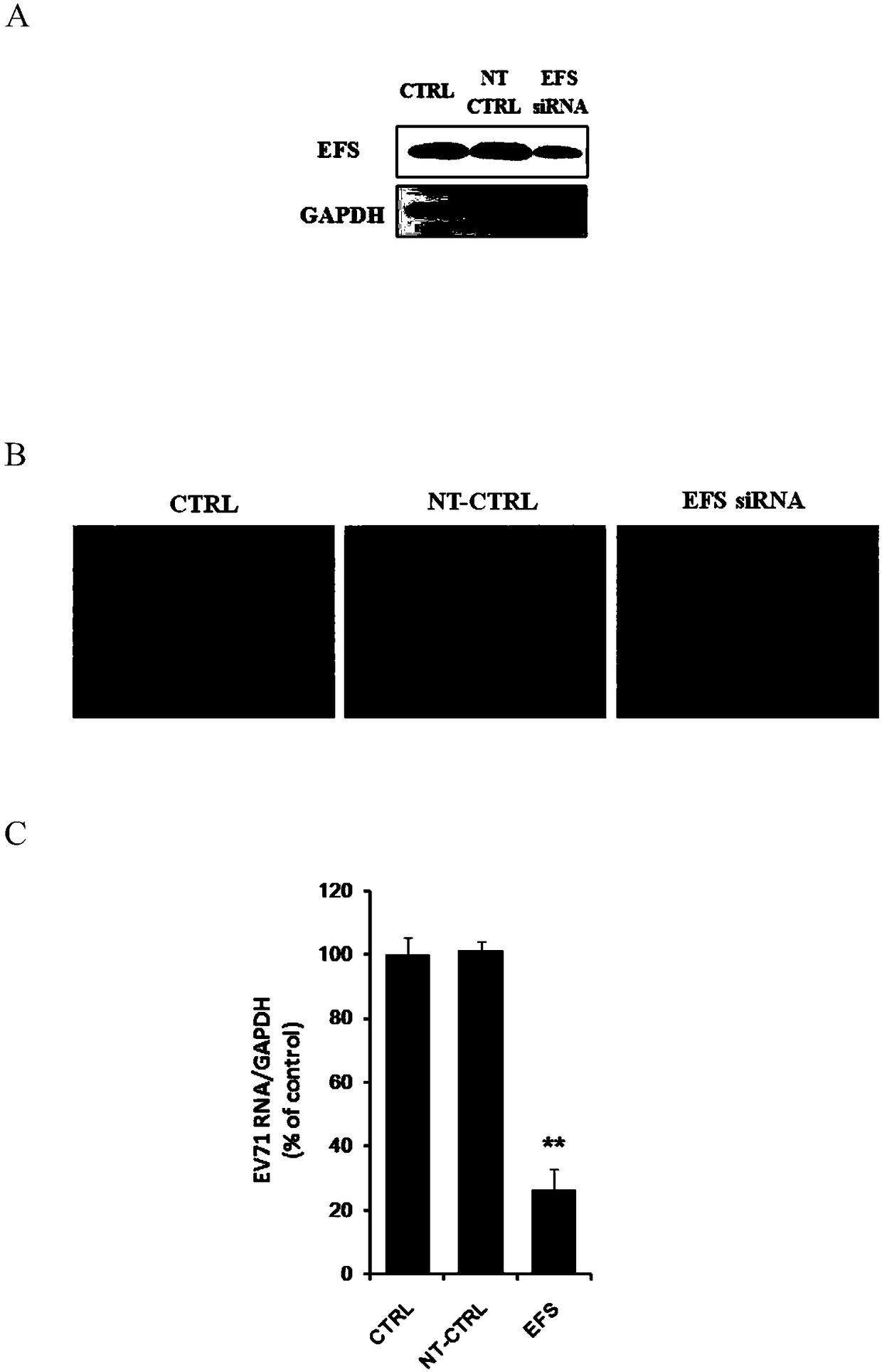
Application of embryonic fyn-related substrate efs in the prevention and treatment of enterovirus 71 infection
ActiveCN105311645BAvoid infectionDoes not affect physiological functionOrganic active ingredientsMicrobiological testing/measurementVascular endotheliumEmbryo
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
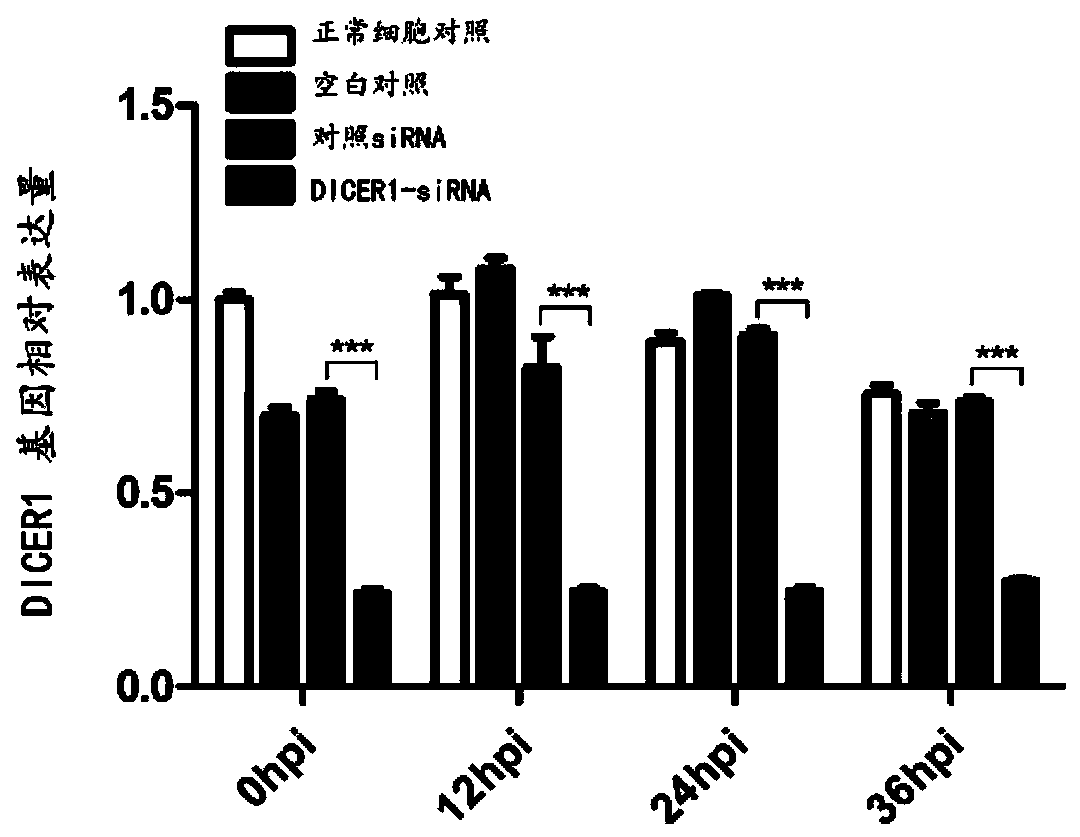
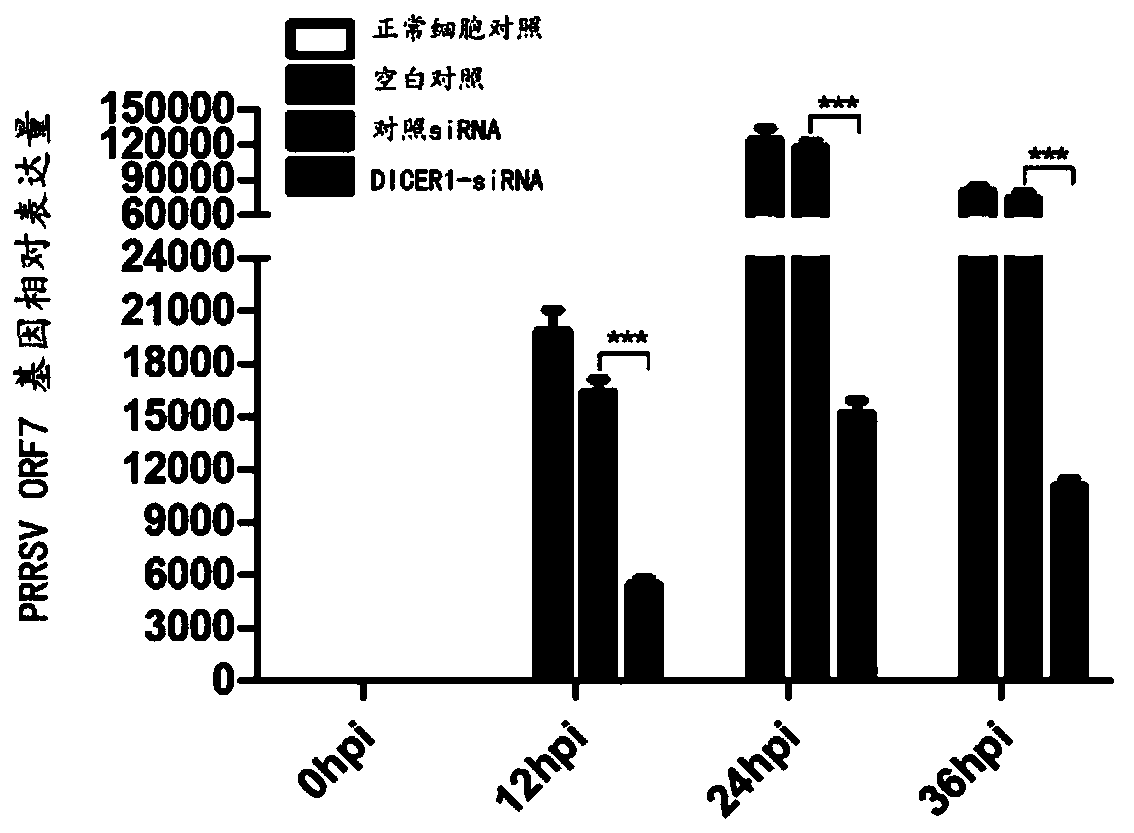
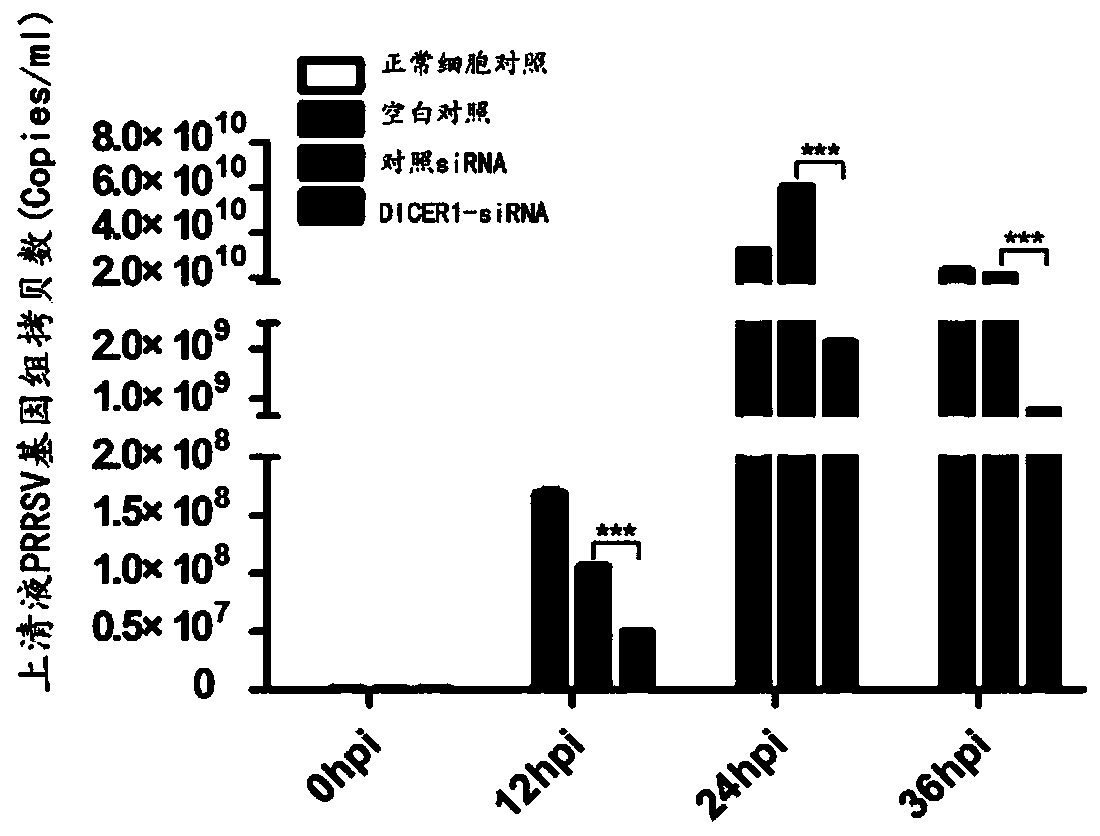
Application of DICER1 gene and siRNA thereof
The invention discloses an application of a DICER1 gene and siRNA thereof in the influence of PRRSV duplication. si-RNA is used to knock-down DICER1, then the influence of DICER1 on a PRRSV infected host cell is detected by qRT-PCR and TCID50; after the host factor DICER1 is specifically knocked-down, the duplication and proliferation of PRRSV are influenced; the specific siRNA of the host factorDICER1 can be used to develop drugs for preventing and treating PRRSV; and corresponding gene DICER1 can be applied to research on PRRSV transgenic pigs.
Owner:NORTHWEST A & F UNIV
Compounds and methods for altering RSV replication rate
ActiveUS10219492B2SsRNA viruses negative-senseCarbohydrate active ingredientsHost factorsInfection risk
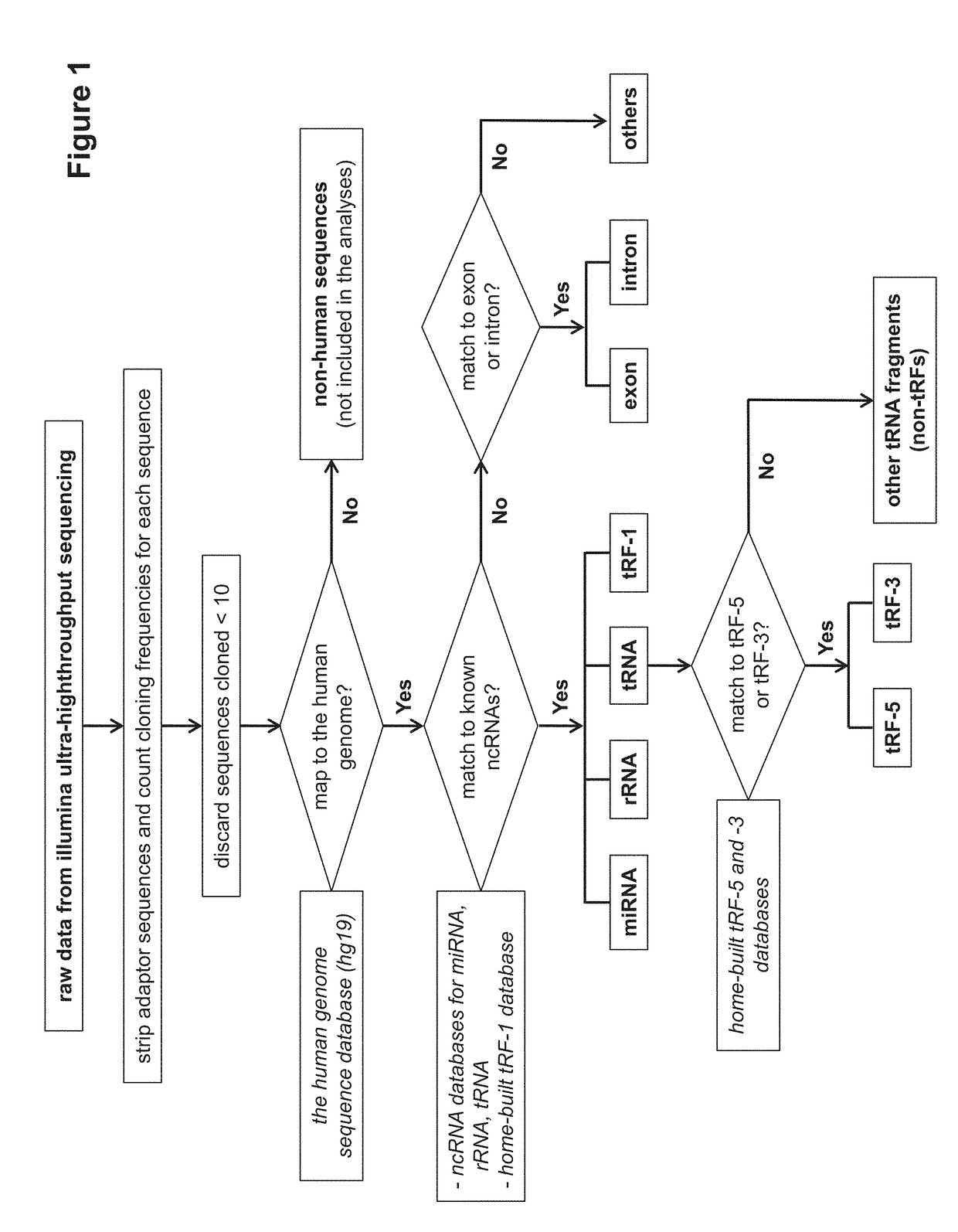
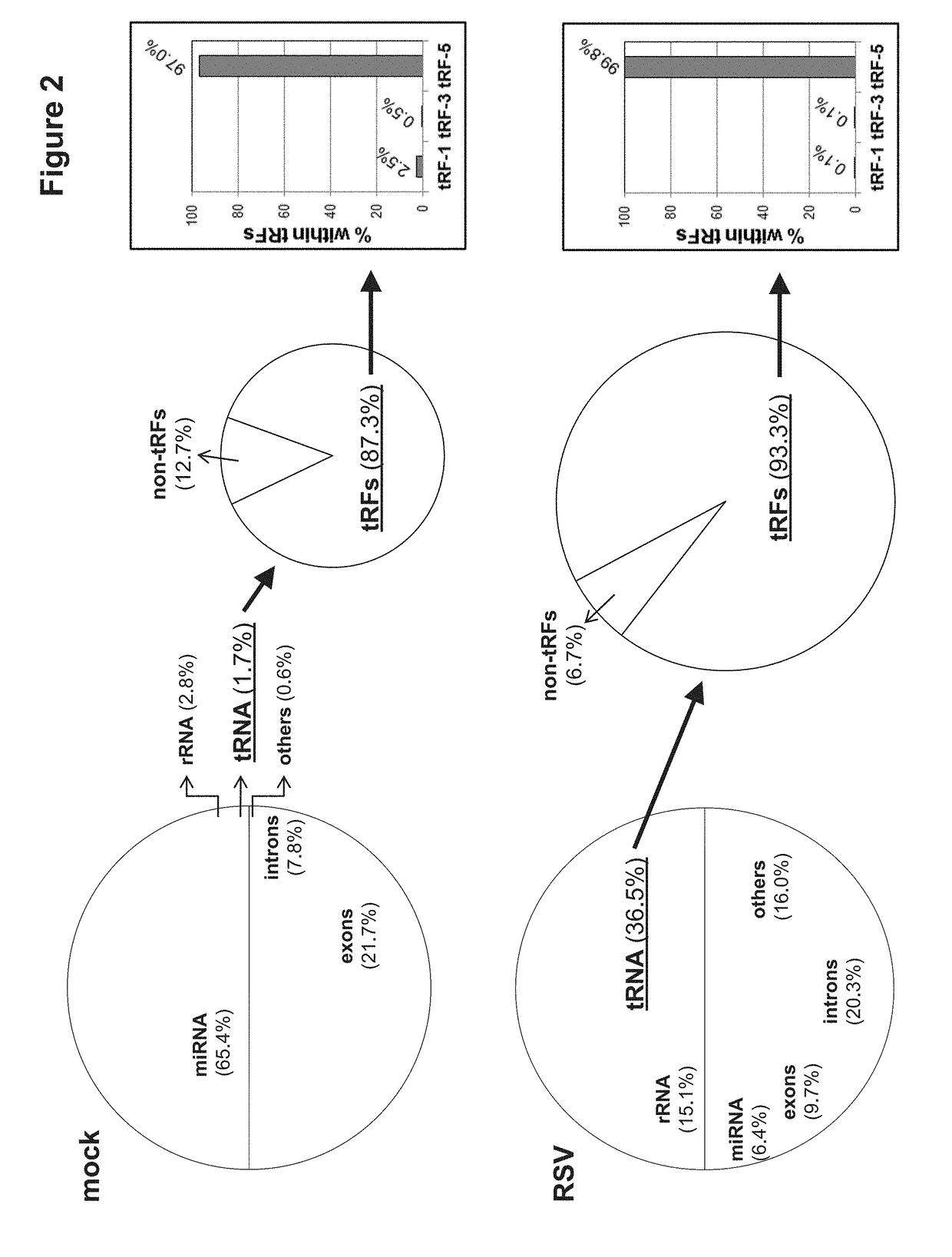
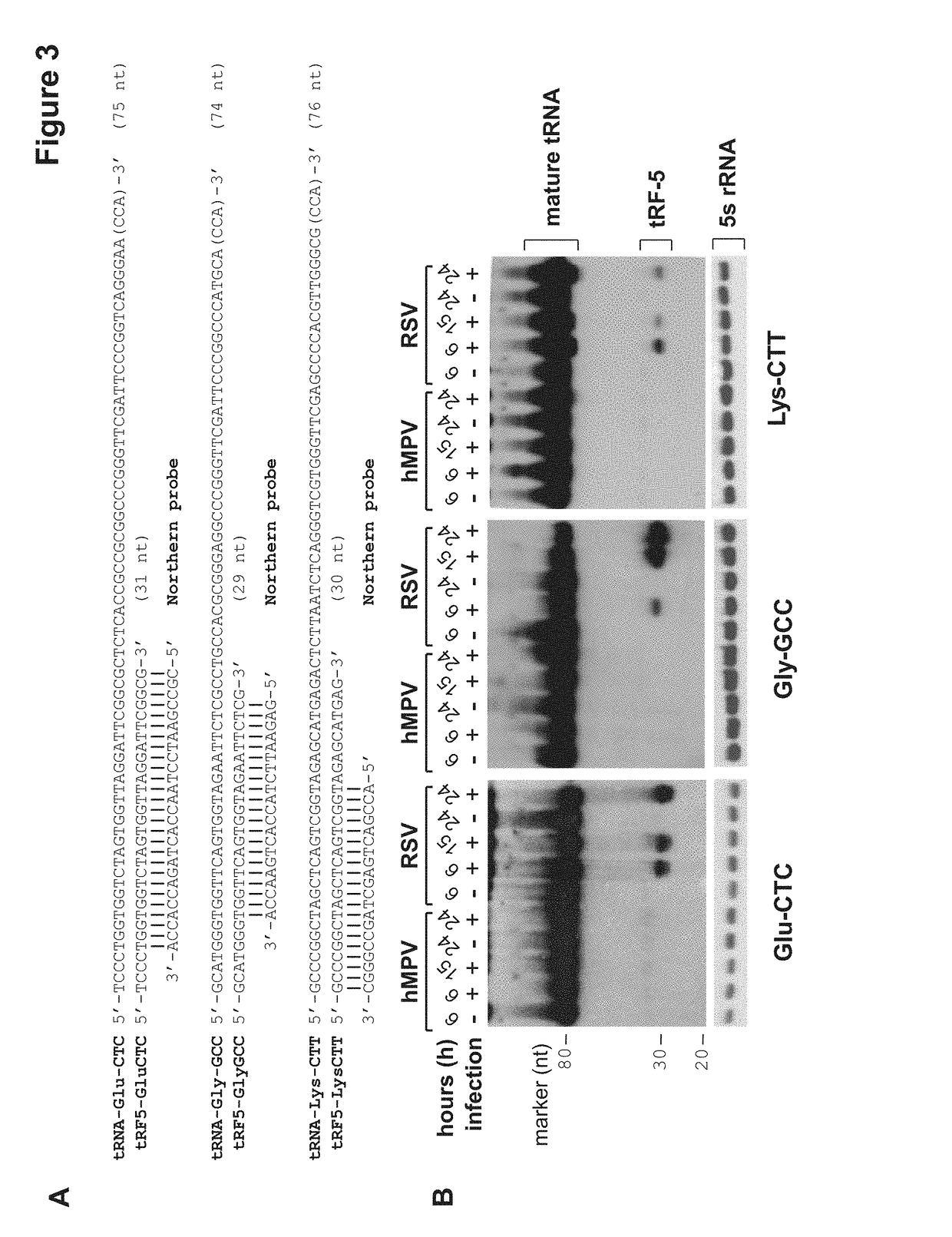
Provided herein are methods for altering respiratory syncytial virus (RSV) replication in a cell using oligonucleotides derived from tRNAs, also referred to as tRFs (tRNA-derived RNA Fragments). The oligonucleotides may be used to decrease or increase replication of RSV. Also provided herein are methods for treating a subject having or at risk of having an RSV infection, and animal models for evaluating viral and host factors in RSV pathogenesis.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
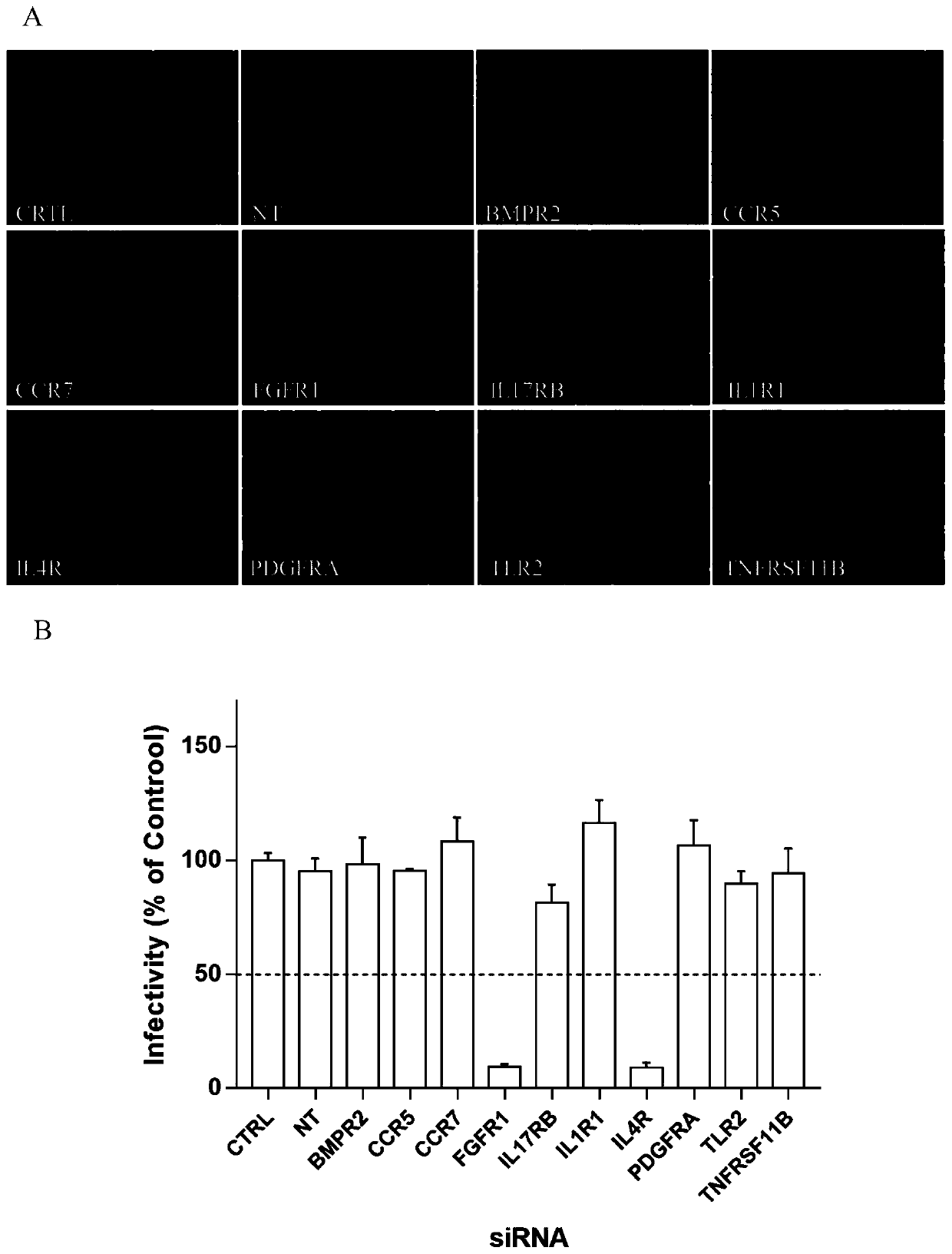
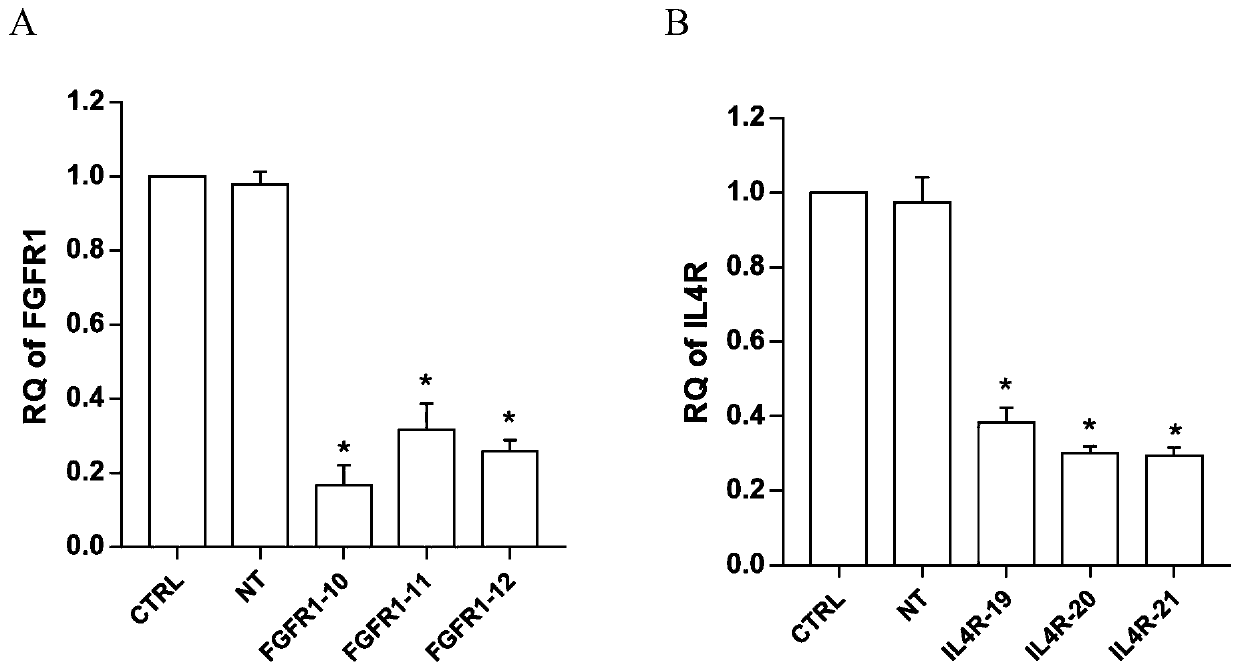
Application of fibroblast growth factor receptor 1 to preparation of medicines for preventing and treating enterovirus 71 type infection
ActiveCN110585432AAvoid infectionDoes not affect normal physiological functionAntiviralsPharmaceutical active ingredientsVirus typeBrainstem encephalitis
The invention relates to the technical field of biomedical science, in particular to a new target for resisting enterovirus 71 type infection, and an application. Human colon cancer cells (Caco-2) areused as target cells, an RNA interference technique is adopted for reducing expression of a target cell membrane receptor, a host factor which can effectively restrain EV71 infected human colon cancer cells is sought, and the purpose of blocking EV71 infection from a source (intestinal tract) is achieved. Through experiment, the inventor of the invention finds that the fibroblast growth factor receptor 1 (FGFR1) exerts important effects in EV71 infected Caco-2 cells, so that when the expression of the FGFR1 is reduced, the EV71 infection can be obviously restrained. The invention provides anapplication of the FGFR1 to preparation of medicines for preventing or treating enterovirus 71 type infection, and a new target and a treatment scheme are provided for clinical prevention and treatment of diseases of hand-foot-mouth diseases, meningitis, brainstem encephalitis or poliomyelitis and the like caused by the EV71 infection.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
A method for improving the sensitivity of pcr amplification by integrating host factors
ActiveCN104593491BHigh sensitivityImprove efficiencyMicrobiological testing/measurementHost factorsNucleic acid sequencing
The invention provides a method for improving PCR amplification sensitivity by using an integrated host factor. The method comprises the following steps: adding the integrated host factor IHF into a reaction solution containing deoxyribonucleoside triphosphate, DNA polymerase, an oligonucleotide primer and a nucleic acid fragment which serves as a template, carrying out repeated heat circulation according to three steps of unwinding, annealing and extending of the normal PCR, thereby amplifying a target nucleic acid fragment, wherein the mole ratio of a primer to the integrated host factor IHF is 0.005-1. The method is capable of specifically and efficiently amplifying a target nucleic acid sequence.
Owner:华桥生物工程科技有限公司
Application of autophagy-related protein 12 in the prevention and treatment of enterovirus 71 infection
The invention belongs to the technical field of biomedicine, and relates to a novel target used for preventing enterovirus 71 infection, and applications thereof. According to the invention, Human Brain Microvascular Endothelial Cell (HBMEC) is taken as the target cell, and RNA interference technology is adopted for down-regulation of expression of target cell host protein so as to search host factors capable of inhibiting EV71 infection of HBMEC effectively, protect the functions of the blood brain barrier, and prevent central nervous system infection caused by penetrating of the blood brain barrier by virus. It is fond by experiments that the autophagy-related protein 12 (ATG12) possesses significant importance in infection of HBMEC by EV71, and down-regulation of expression of ATG12, and is capable of inhibiting EV71 infection obviously. The invention also provides applications of ATG12 in preparing drugs used for preventing or treating EV71 infection.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com