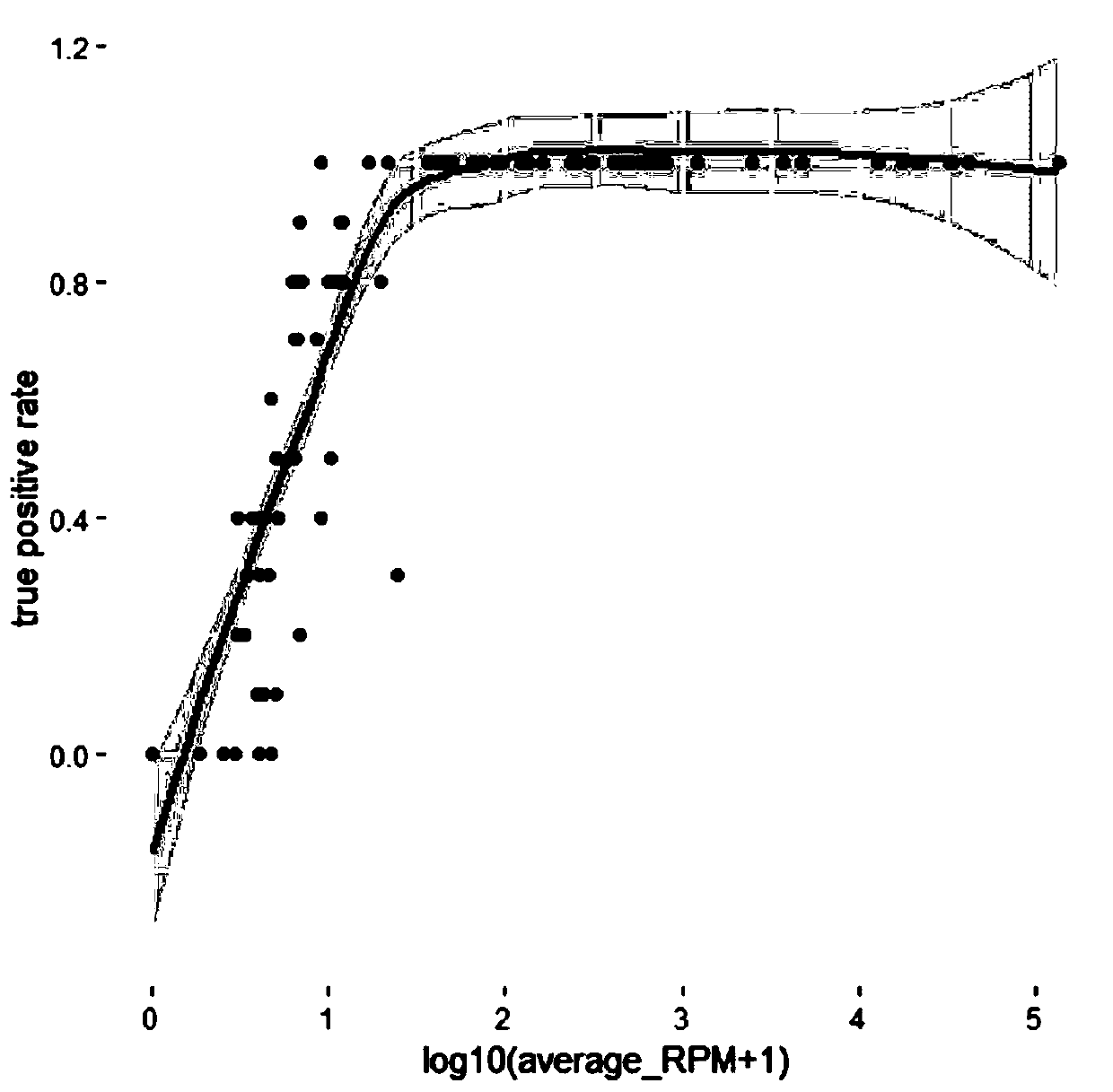
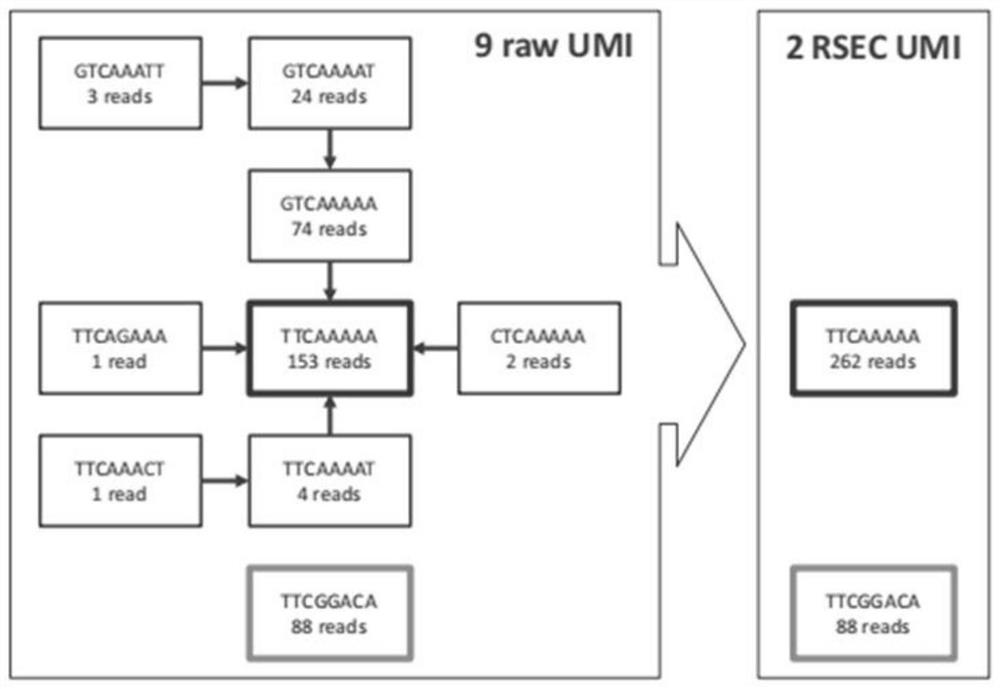
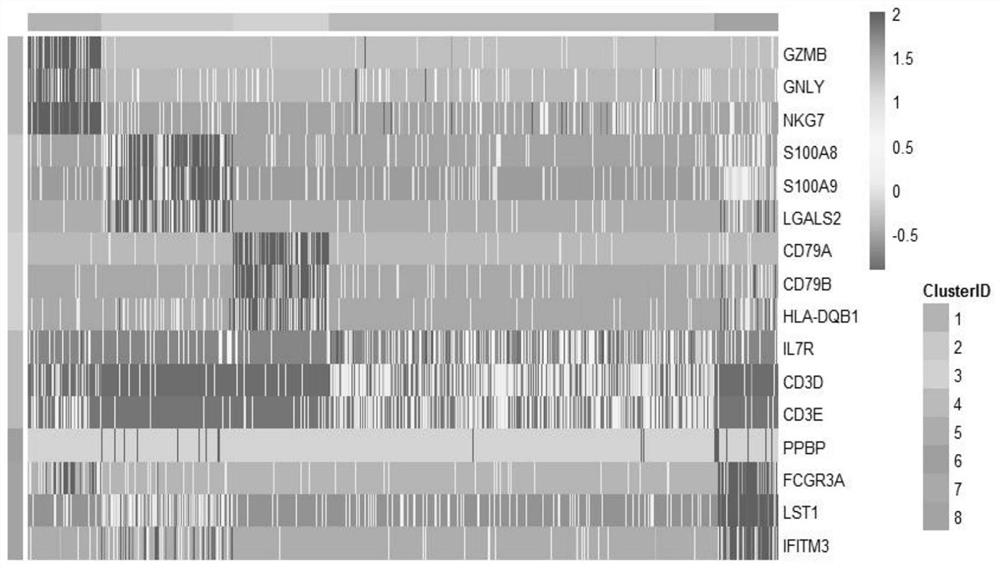
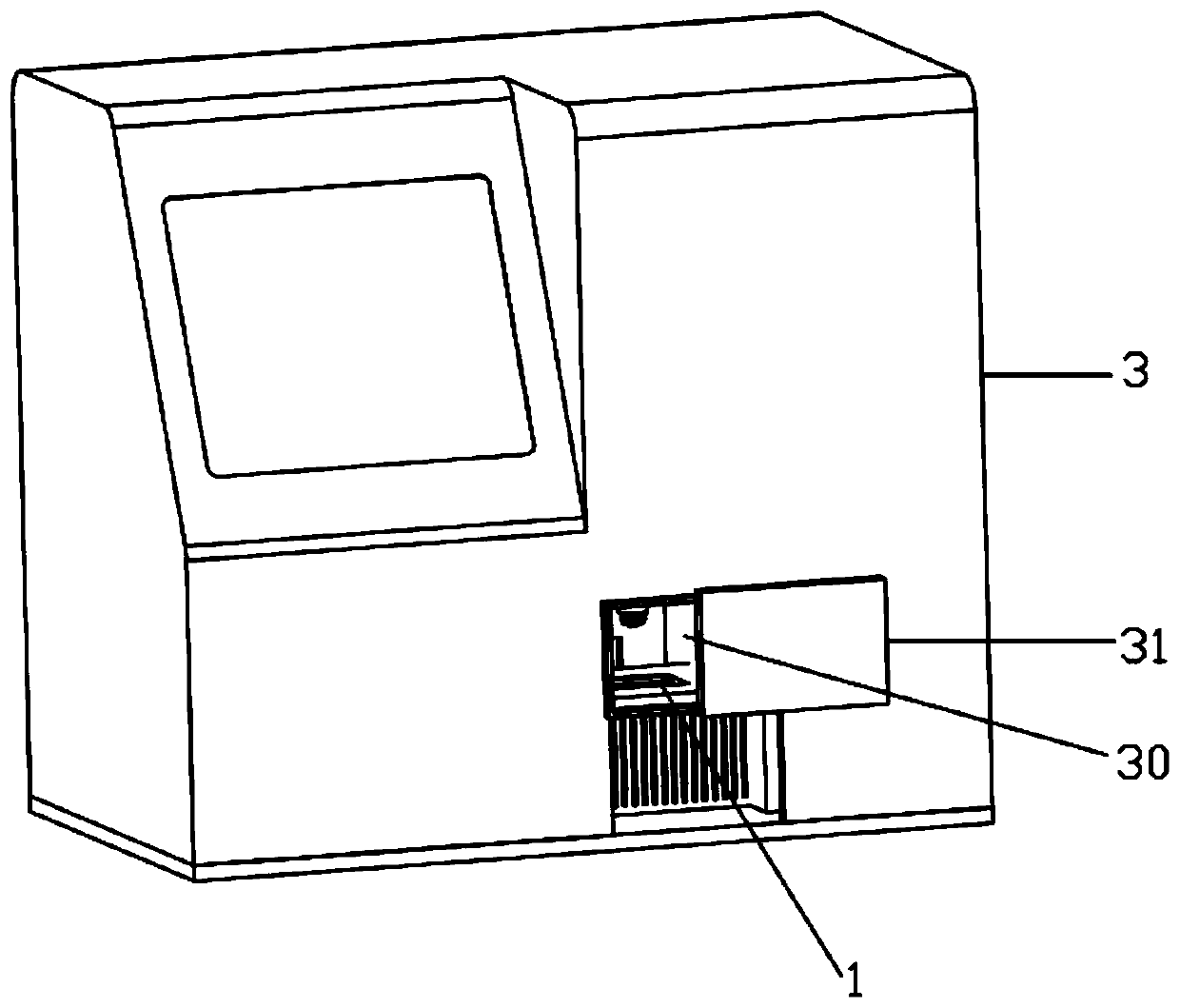
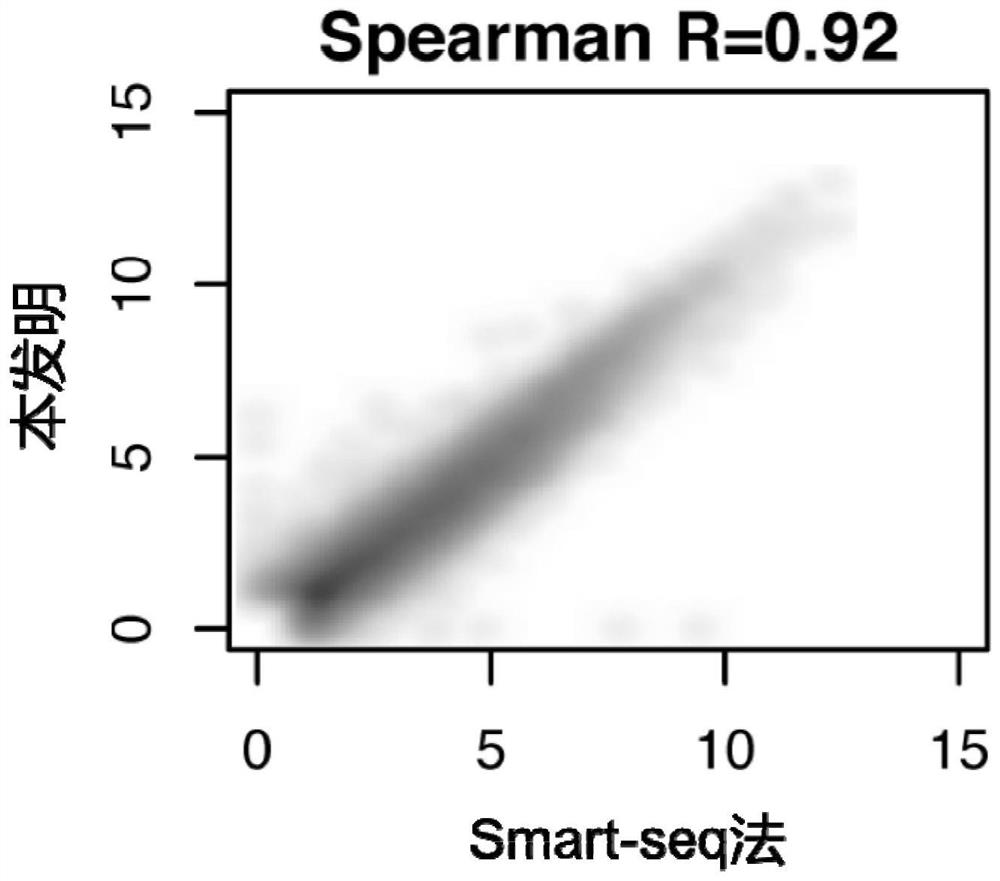
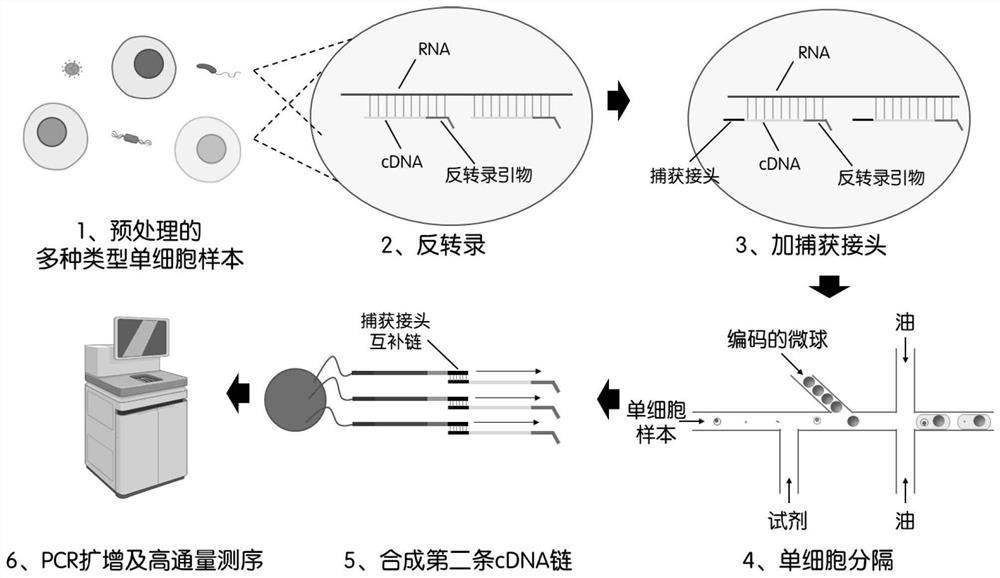
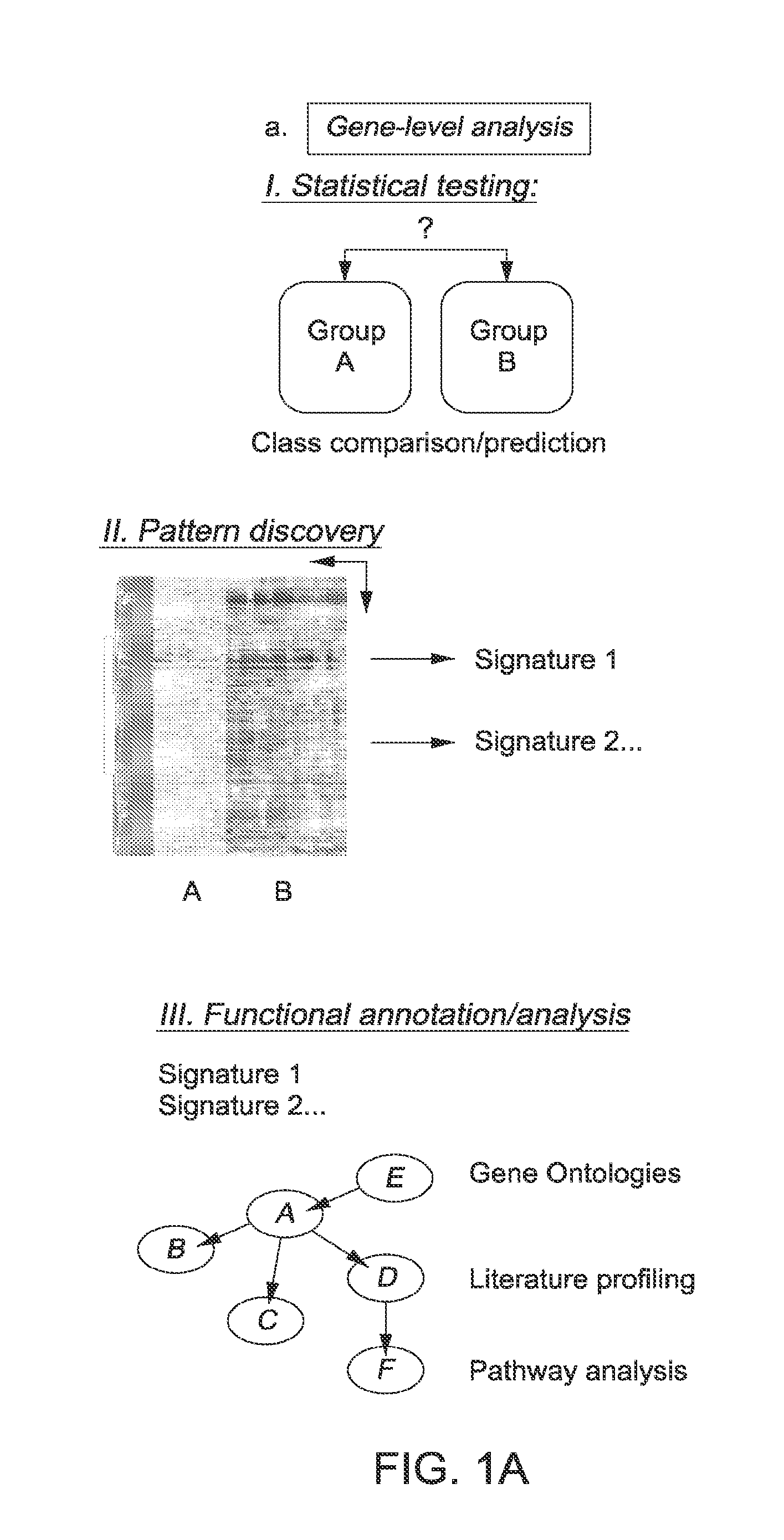
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
78 results about "Cellular transcription" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Internet-Based Tools for Teaching Transcription and Translation. Transcription is the process of making an RNA copy of a gene sequence. This copy, called a messenger RNA (mRNA) molecule, leaves the cell nucleus and enters the cytoplasm, where it directs the synthesis of the protein, which it encodes.
Method for the determination of cellular transcriptional regulation
InactiveUS20060134639A1Rapidly and reliably detectingRapidly and reliably and quantifyingBioreactor/fermenter combinationsBiological substance pretreatmentsSignal onTranscriptional regulation
The present invention relates to a new method for determining the RNAi mediated transcriptional regulation of a cell by the determination of a pattern of at least 3 miRNA detected simultaneously and quantified in the same cell extract. The determination of a pattern of miRNA comprises the steps of: (i) providing an array onto which are fixed capture probes, said capture probes being arranged on pre-determined locations and reflecting the genomic or transcriptional matter of a cell; (ii) isolating a miRNA pool potentially present from a cell; (iii) elongating or ligating said miRNAs into labeled capture probes, (iv) contacting said labeled polynucleotides with the array under conditions allowing hybridization of the labeled polynucleotides to complementary capture probes present on the array; (v) detecting and quantifying a signal present on the specific locations of the array, wherein the detection of a pattern of at least 3 signals on the array reflects the pattern of miRNAs being involved in the RNAi mediated cellular transcriptional regulation.
Owner:EPPENDORF ARRAY TECH SA
Construction method of single cell transcriptome sequencing library and application of construction method
InactiveCN104032377AReduce sample lossImprove stabilityMicrobiological testing/measurementLibrary creationTranscriptome SequencingThroughput
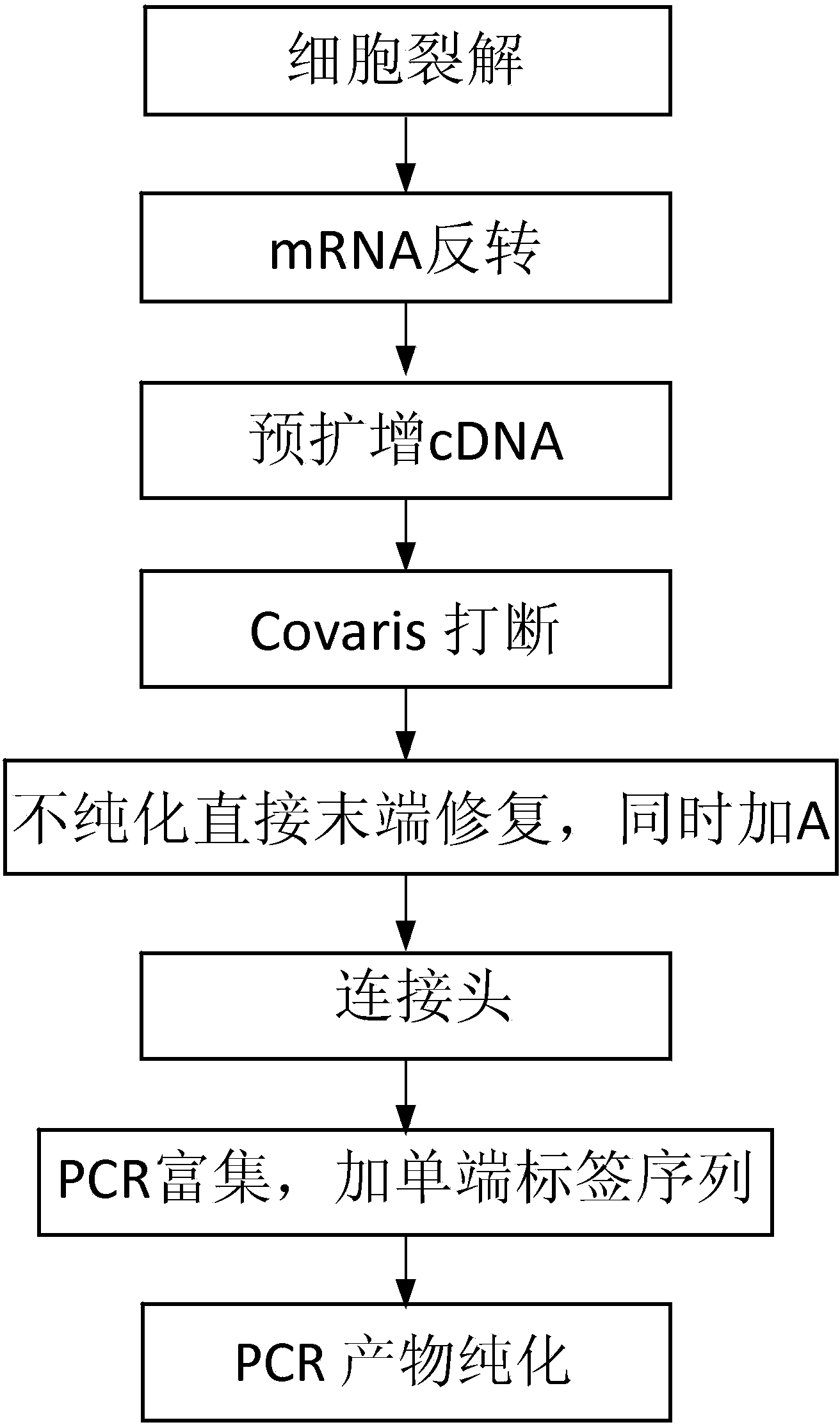
The invention discloses a construction method of a single cell transcriptome sequencing library and application of the construction method. The construction method includes the following steps of preparing a cDNA sample from a single cell and conducting fragmentized library construction on the cDNA sample to obtain the transcriptome sequencing library of the single cell, wherein fragmentation is conducted on the cDNA sample with a physical breaking method in the step of constructing the fragmentized library, and the step for purifying the fragmentized cDNA sample is not needed. According to the method, fragmentation is conducted on the sample of the single cell with the physical breaking method, and therefore fragments obtained through breaking are small and relatively uniform, and the peak width of the constructed single cell transcriptome sequencing library is concentrated; the purification step is not needed for the fragmentized sample, sample loss is reduced, construction of the single cell transcriptome sequencing library of the micro-sample can be achieved, stability of output of data volume can be improved, and therefore the reliability of large-scale and high-throughput sequencing of the single cell transcriptome sequencing library is improved.
Owner:BEIJING NOVOGENE TECH CO LTD
Functional nucleic acids and methods
InactiveUS20100087336A1High affinityIncrease stringencyMicroorganismsTissue cultureAntisense nucleic acidNucleic Acid Probes
The present invention relates to methods of generating amounts of selective nucleic acids. The present invention further relates to selective nucleic acids incorporated within non-coding nucleic acids, capable of binding to or altering a target molecule. Selective nucleic acids may generally refer to, but are not limited to, deoxyribonucleic acids (DNAs), ribonucleic acids (RNAs), artificially modified nucleic acids, combinations or modifications thereof. Selective nucleic acids may also generally refer to, but are not limited to, nucleic acid aptamers, aptazymes, ribozymes, deoxyribozymes, nucleic acid probes, small interfering RNAs (siRNAs), micro RNAs (miRNAs), short hairpin RNAs (shRNAs), antisense nucleic acids, diagnostic probes or probe libraries, aptamer inhibitors, precursors of any of the above and / or combinations or modifications thereof. In one aspect, a method for generating amounts of selective nucleic acids includes incorporating a selective nucleic acid sequence into a carrier nucleic acid. In general, the carrier nucleic acid may be transcribed by a cell into a product nucleic acid which may carry an incorporated selective nucleic acid sequence.
Owner:BIOTEX +1
Method for establishing single cell transcriptome sequencing library and application of method
InactiveCN104389026AReduced amplificationIncrease the amount of effective dataMicrobiological testing/measurementLibrary creationSingle cell transcriptomeTranscriptome Sequencing
The invention discloses a method for establishing a single cell transcriptome sequencing library and application of the method. The method comprises the following steps: performing reverse transcription on RNA in a single cell, thus obtaining cDNA; performing pre-amplification on cDNA by using an amplification primer, thus obtaining amplified cDNA; and performing fragmentation library construction on the amplified cDNA, thus obtaining a transcriptome sequencing library of the single cell, wherein dTTP in the amplification primer is substituted by dUTP. According to the method disclosed by the invention, as the dTTP in the pre-amplification primer is substituted by dUTP, fragments containing the pre-amplification primer in a jointed fragment can be interrupted in an enzymic digestion step after the step of joint connection, and are removed in later high-temperature pre-denaturation and denaturation steps; furthermore, the amplification of fragments with the pre-amplification primer can be reduced, the ratio that the proportion of data with pre-amplification primer pollution in the obtained sequencing data is greatly reduced, and the effective data amount of the obtained data is greatly increased.
Owner:BEIJING NOVOGENE TECH CO LTD
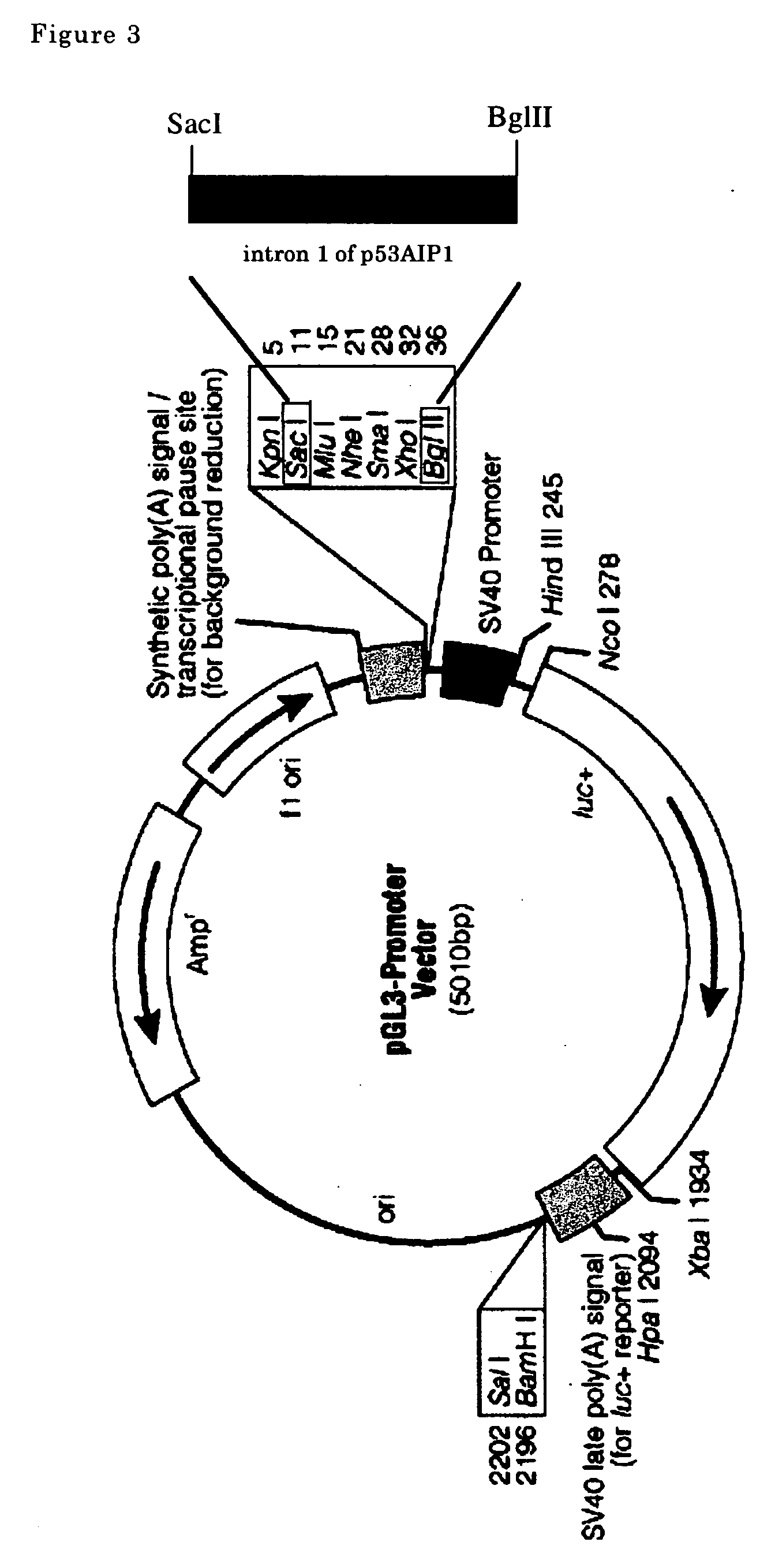
Transcriptional factor inducing apoptosis in cancer cell
A new method for treating cancer by inducing apoptosis exclusively in cancer cells and killing them is provided. The present invention relates to a transcriptional factor, comprising p53 or a mutated type p53, wherein one or more amino acids are deleted, substituted or added with respect to the amino sequence of p53, and clathrin heavy chains and having an activity to induce apoptosis of cancer cells. The transcriptional factor enhances the transcriptional activity of p53AIP1 promoter and induces apoptosis of cancer cells.
Owner:JAPAN SCI & TECH CORP
METHODS AND COMPOSITIONS FOR cDNA SYNTHESIS AND SINGLE-CELL TRANSCRIPTOME PROFILING USING TEMPLATE SWITCHING REACTION
ActiveUS20160258016A1Increase the lengthHigh yieldMicrobiological testing/measurementFermentationCDNA libraryCost effectiveness
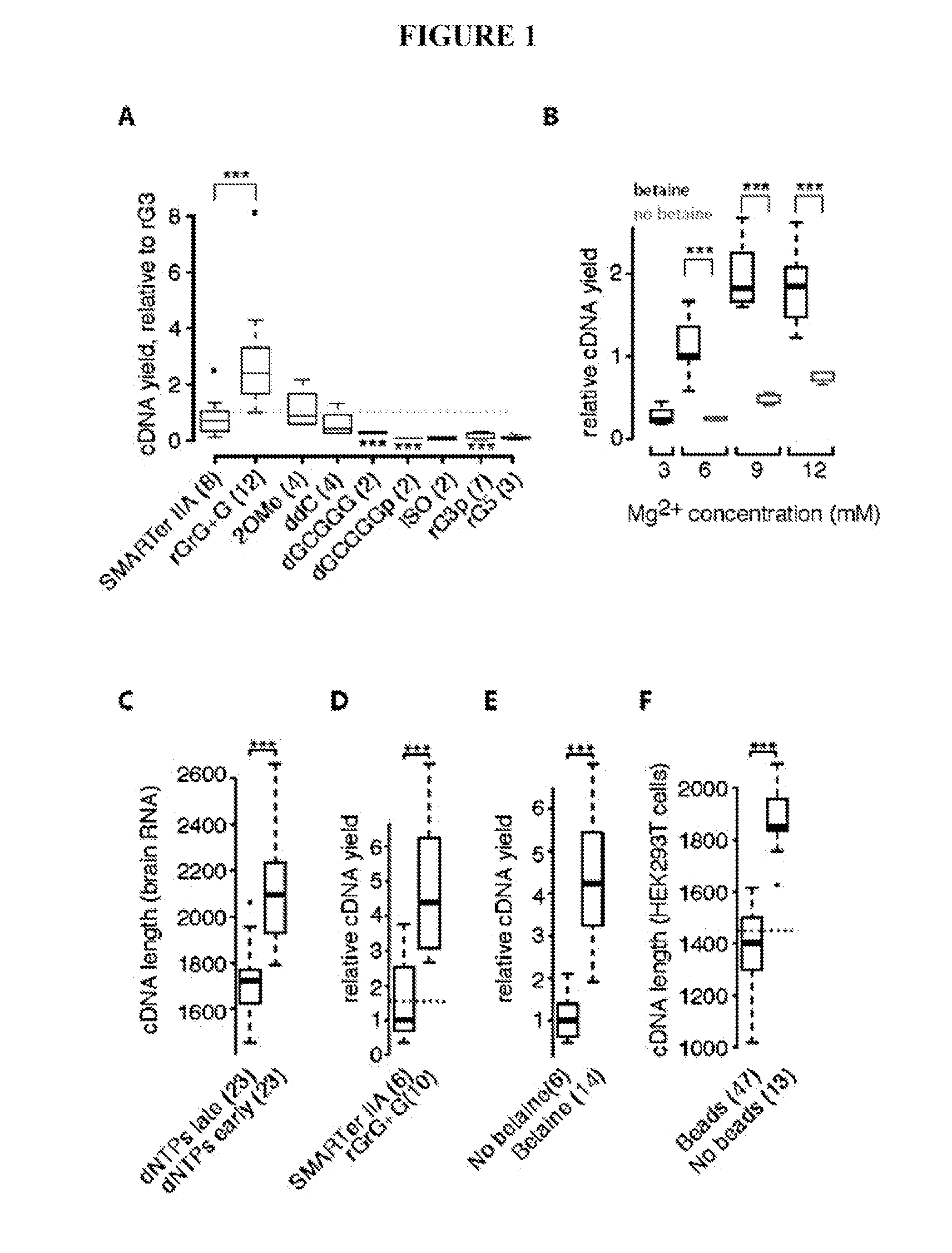
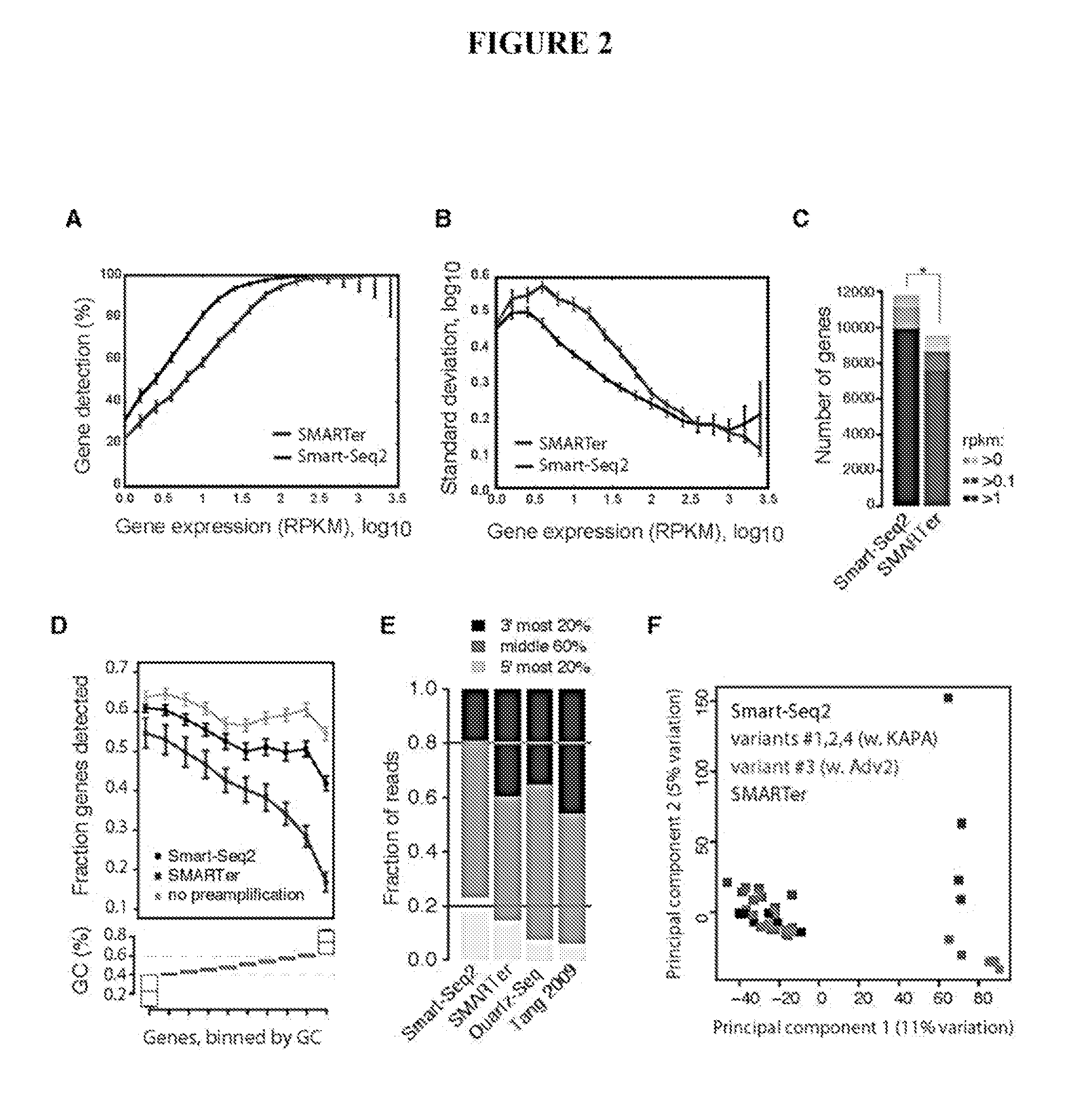
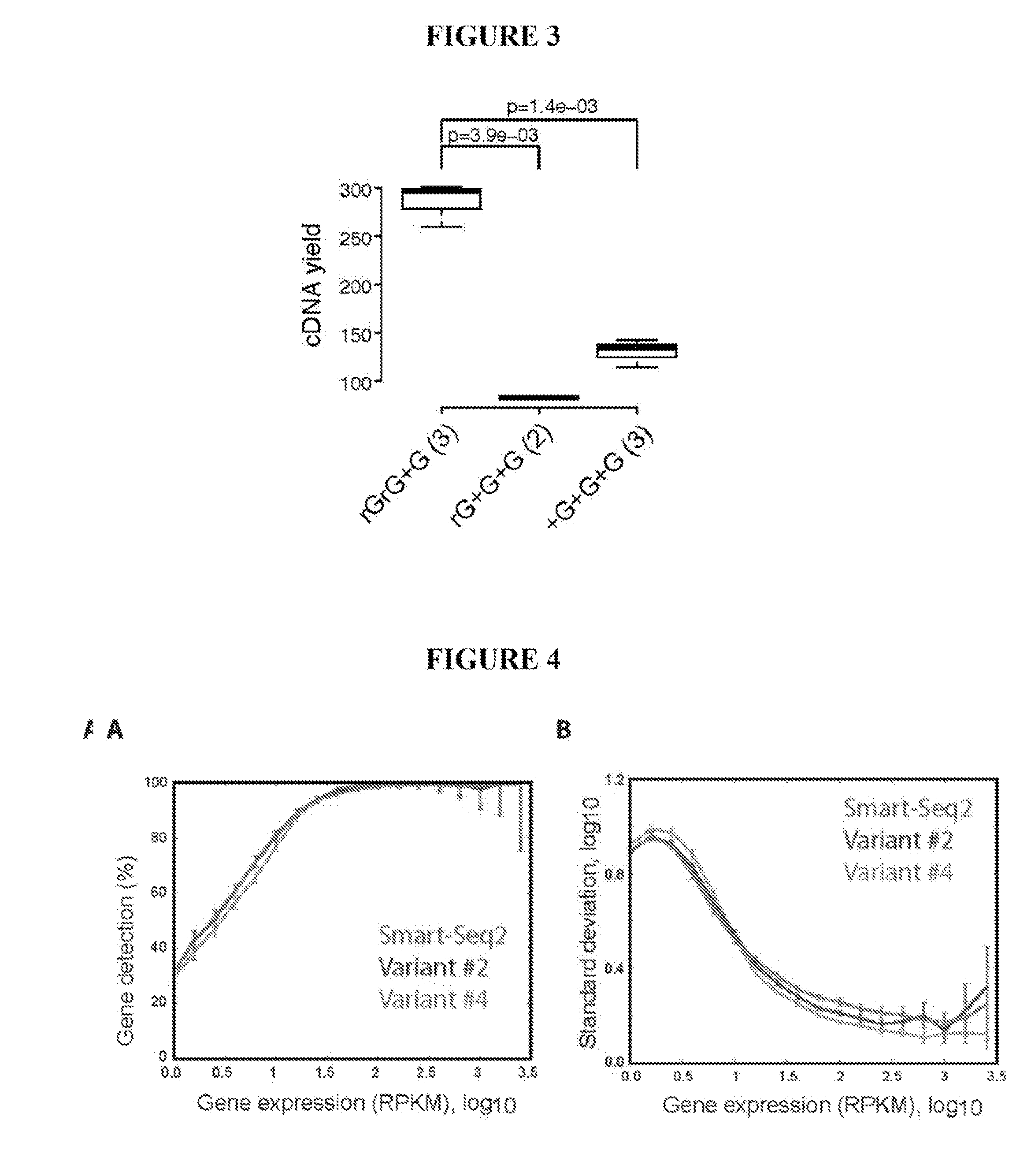
This application discloses methods for cDN′A synthesis with improved reverse transcription, template switching and preamplification to increase both yield and average length of cDNA libraries generated from individual cells. The new methods include exchanging a single nucleoside residue for a locked nucleic acid (INA) at the TSO 3′ end, using a methyl group donor, and / or a MgCb concentration higher than conventionally used. Single-cell transcriptome analyses incorporating these differences have full-length coverage, improved sensitivity and accuracy, have less bias and are more amendable to cost-effective automation. The invention also provides cDNA molecules comprising a locked nucleic acid at the 3′-end, compositions and cDNA libraries comprising these cDNA molecules, and methods for single-cell transcriptome profiling.
Owner:LUDWIG INST FOR CANCER RES
Compositions and methods for regulating mRNA transcription and translation
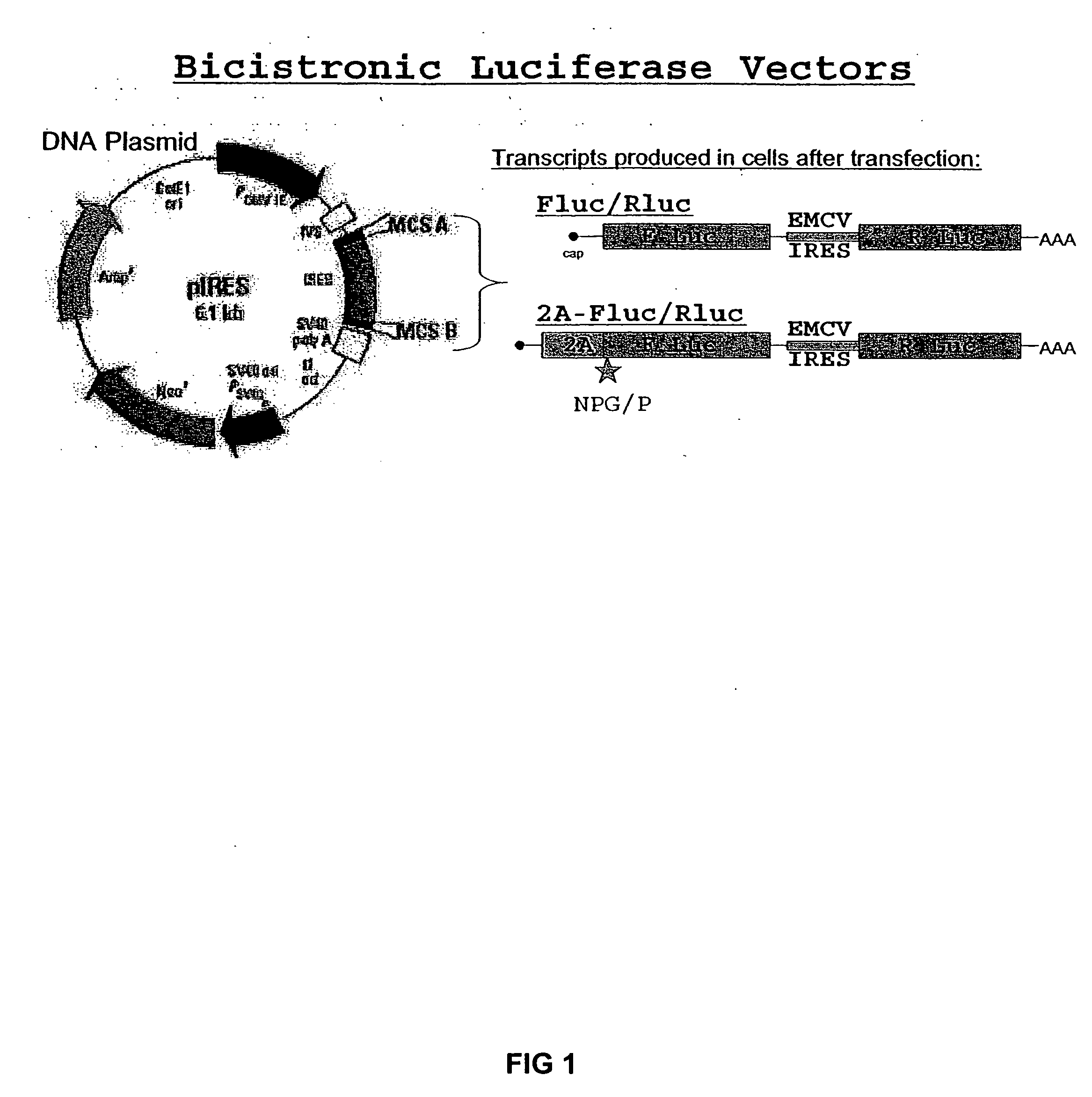
InactiveUS20050019808A1Significant changeSsRNA viruses positive-senseSugar derivativesCell freeInternal ribosome entry site
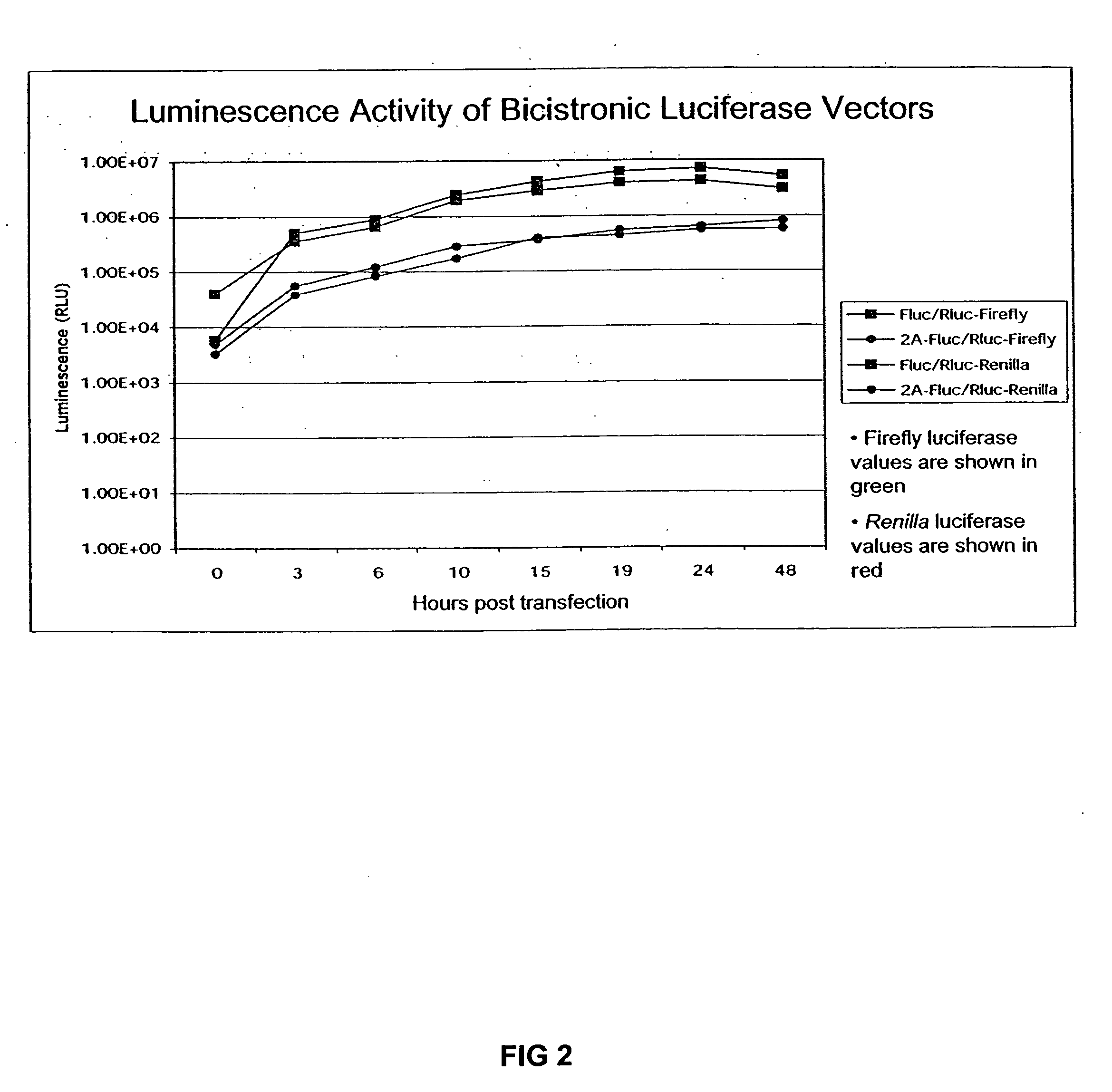
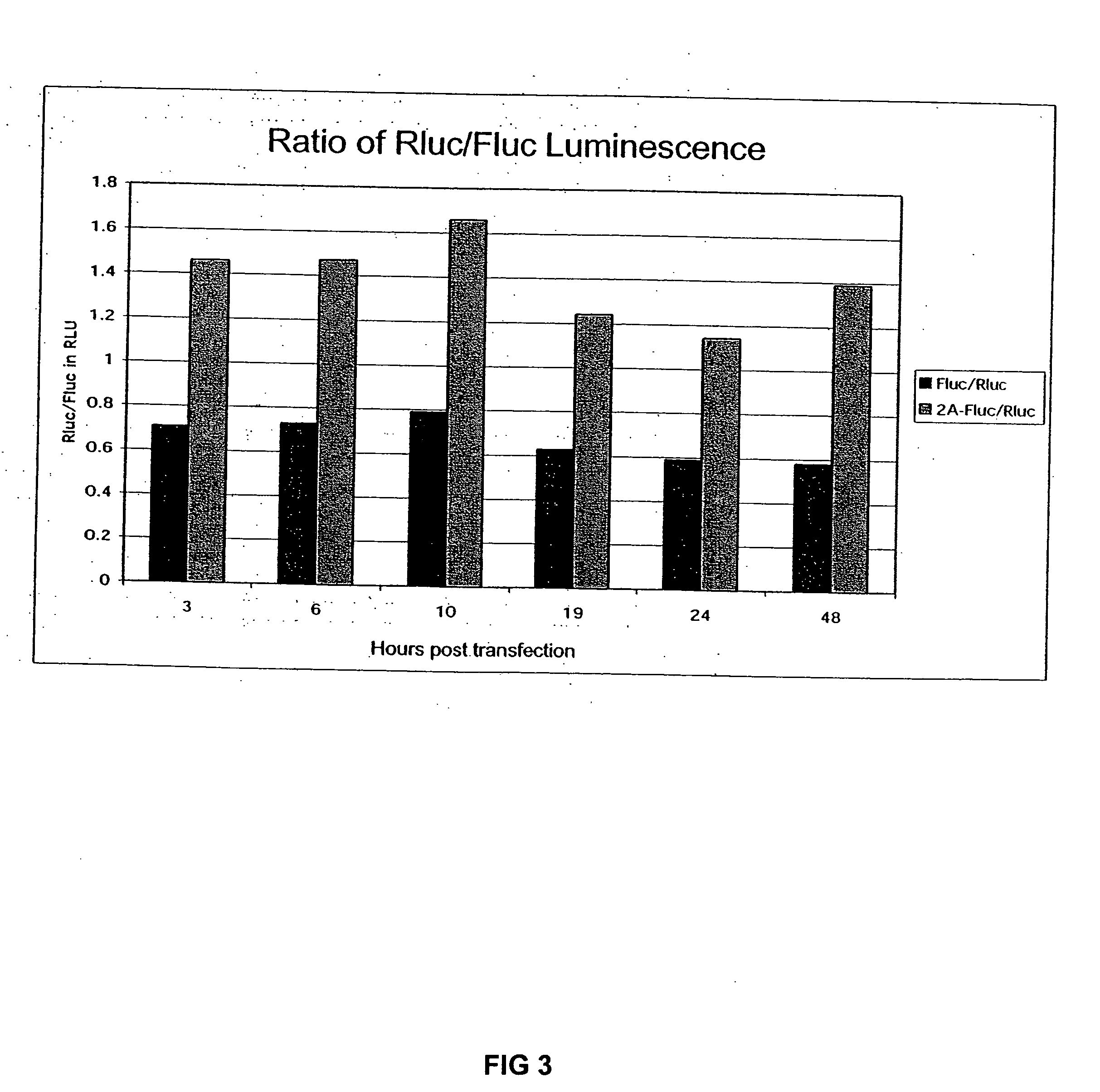
The invention relates to compositions, specifically novel nucleic acid constructs encoding a cardiovirus 2A polypeptide operably linked to suitable promoters. Also, disclosed are methods whereby the nucleic acid constructs are introduced into cells or cell free systems to regulate cellular mRNA transcription and cap-dependent or internal ribosomal entry site (IRES)-dependent mRNA translation.
Owner:WISCONSIN ALUMNI RES FOUND
Transcriptome sequencing method
InactiveCN104711340AHigh technical repeatabilityRich researchMicrobiological testing/measurementEthylene HomopolymersTranscriptome Sequencing
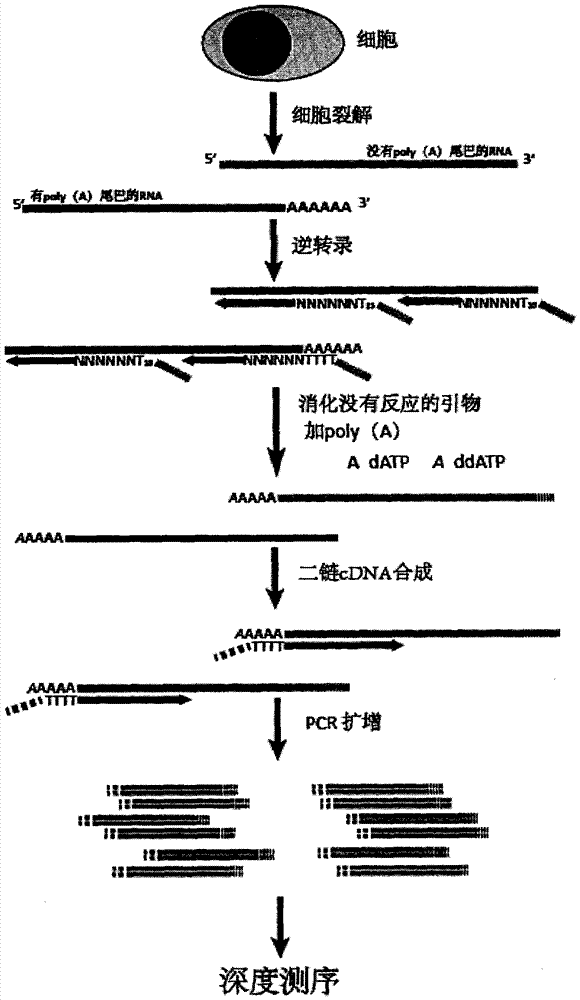
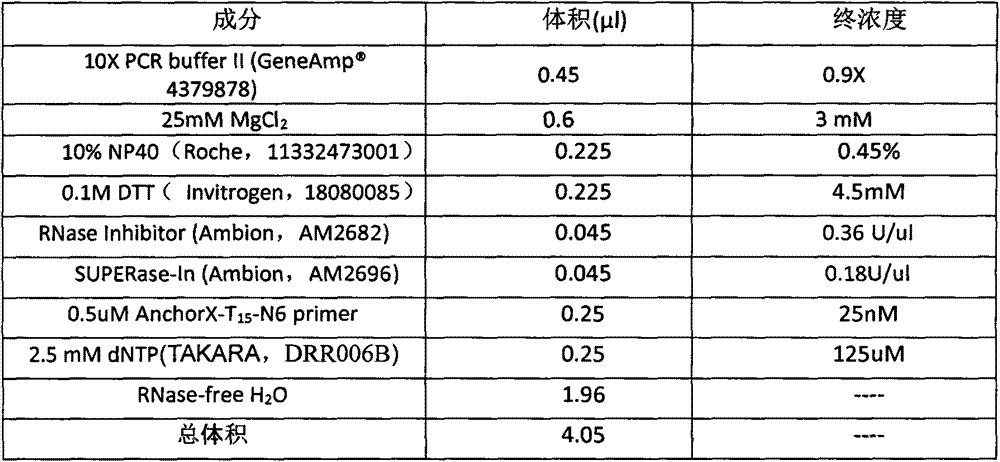
The present invention provides a transcriptome sequencing method, wherein primers used during a reverse transcription process from RNA to cDNA comprise a random sequence and a specific sequence, the random sequence is positioned at the 3' terminal of the specific sequence, bases from 0 to 5 exist between the random sequence and the specific sequence, the random sequence comprises 4-12 random bases, the specific sequence is a homopolymer of 5-30 bases, and the base of the specific sequence is any one selected from A, G, C and T. According to the present invention, with the design on the reverse transcription primers, the RNA having the poly (A) tail can be sequenced while the RNA with no poly (A) tail can be sequenced, the rRNA removal step is not required during the cell transcriptome sequencing, and the method is especially suitable for the sequencing on the transcriptome having the low initial RNA amount and the low cell number.
Owner:PEKING UNIV
High throughput transcriptome analysis
InactiveUS20170137806A1Extending lengthMicrobiological testing/measurementDNA preparationNucleotideSingle strand
Kits and methods for single cell or multiple cell transcriptome analysis are provided. An adapter polynucleotide is disclosed which comprises a double-stranded DNA portion of 15 base pairs and no more than 100 base pairs with a 3′single stranded overhang of at least 3 bases and no more than 10 bases, wherein said double stranded DNA portion is at the 5′ end of the polynucleotide and wherein the sequence of said 3′ single stranded overhang is selected from the group consisting of SEQ ID NOs: 1-8 and 9, wherein the 5′ end of the strand of said double-stranded DNA which is devoid of said 3′ single stranded overhang comprises a free phosphate.
Owner:YEDA RES & DEV CO LTD
Second-generation sequencing technology based microbe unicell transcriptome analysis method
ActiveCN104561244AEnabling Whole Transcriptome AnalysisRealize physiological function analysisMicrobiological testing/measurementTotal rnaLinear amplification
The invention discloses a second-generation sequencing technology based microbe unicell transcriptome analysis method. The analysis method comprises the following steps: (1) separating microbe unicells; (2) extracting total RNA; (3) conducting linear amplification on total RNA; (4) constructing an RNA sequencing library; (5) analyzing the RNA sequencing library. By adopting the analysis method, analysis on gene expression difference among different microbe cells can be realized, and physiological function analysis of the unicell level on microbes which cannot be cultivated in extreme environments can be realized; furthermore, the analysis method can be suitable for analysis on eukaryotic unicells, and has quite wide applications in the fields of important biological and medical researches and the like, such as tumor generation and evolution, and stem cell induction and development.
Owner:TIANJIN UNIV
Attenuation of encephalitogenic alphavirus and uses thereof
InactiveUS20100015179A1SsRNA viruses positive-senseViral antigen ingredientsUltrasound attenuationCytopathic effect
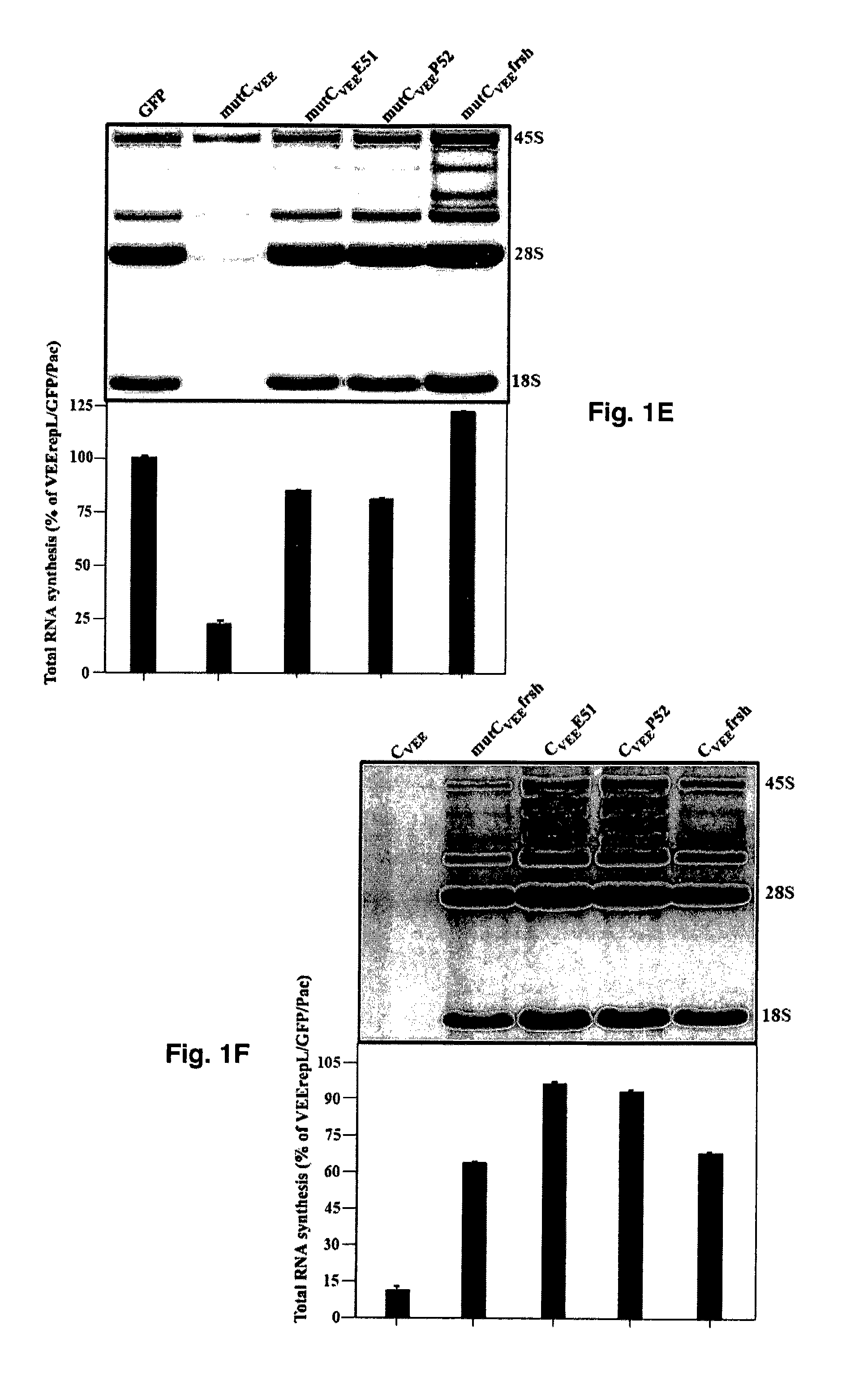
The present invention is drawn to generating attenuated and less cytopathic forms of New World alphaviruses that can be used in immunogenic compositions as vaccines against both Old and New World alphaviruses. In this regard, the present invention discloses that the N-terminal, ˜35-aa-long peptide of VEEV, EEEV and, most likely, of WEEV capsid proteins plays the most critical role in the downregulation of cellular transcription and development of cytopathic effect. The identified, VEEV-specific peptide, CVEE30-68, includes two domains with distinguished functions. The integrity of both domains determines not only the intracellular distribution of CVEE, but is also essential for direct capsid function in the inhibition of transcription. The replacement of the N-terminal fragment of CVEE by its SINV-specific counterpart in VEEV TC-83 genome does not affect virus replication in vitro, but makes it less cytopathic and more attenuated in vivo.
Owner:FROLOV ILYA V +2
High-throughput single-cell transcriptome sequencing method and kit
PendingCN110684829AReduce mutual contaminationIncrease the proportionMicrobiological testing/measurementGenomicsSingle cell transcriptome
The invention discloses a high-throughput single-cell transcriptome sequencing method and a kit. The high-throughput single-cell transcriptome sequencing method includes the steps that a droplet generation system is adopted to encapsulate a single cell and labeled microbeads in a droplet, and reverse transcription is performed in the droplet. According to the high-throughput single-cell transcriptome sequencing method, a throughput of 9,000 cells which is equivalent to 10 x genomics can be achieved, after the droplet is demulsified, the microbeads are less contaminated with each other, and theproportion of valid data is increased. According to the high-throughput single-cell transcriptome sequencing method, reverse transcription is performed in the droplet, fewer reagents are required, and the cost is low. In the preferred scheme of the high-throughput single-cell transcriptome sequencing method, a SMART template conversion technology is adopted in reverse transcription, and productscan be directly used for subsequent Tn5 library construction and BGISeq-500 platform sequencing without library conversion; and multiple amplification and introduction of deviations are avoided, a droplet-based microfluidics platform is used in conjunction with a BGISeq-500 sequencing platform, and large-scale single-cell sequencing is simplified and facilitated.
Owner:MGI TECH CO LTD
Attenuation of encephalitogenic alphavirus and uses thereof
The present invention is drawn to generating attenuated and less cytopathic forms of New World alphaviruses that can be used in immunogenic compositions as vaccines against both Old and New World alphaviruses. In this regard, the present invention discloses that the N-terminal, ˜35-aa-long peptide of VEEV, EEEV and, most likely, of WEEV capsid proteins plays the most critical role in the downregulation of cellular transcription and development of cytopathic effect. The identified, VEEV-specific peptide, CVEE30-68, includes two domains with distinguished functions. The integrity of both domains determines not only the intracellular distribution of CVEE, but is also essential for direct capsid function in the inhibition of transcription. The replacement of the N-terminal fragment of CVEE by its SINV-specific counterpart in VEEV TC-83 genome does not affect virus replication in vitro, but makes it less cytopathic and more attenuated in vivo.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
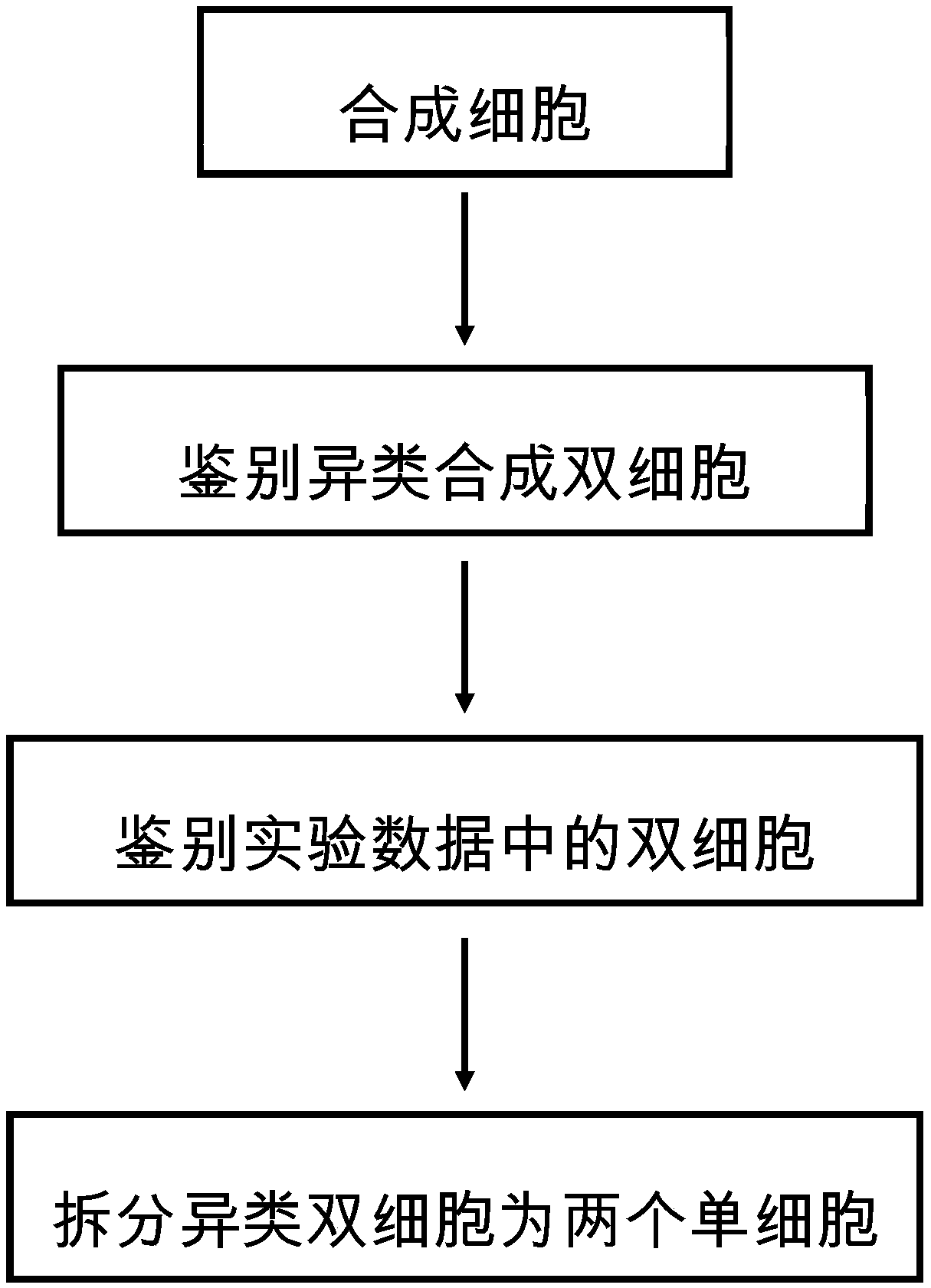
Method for analyzing double cells in single-cell transcriptome data
ActiveCN111292807AImprove the efficiency of detecting the number of cellsBiostatisticsSequence analysisSingle cell transcriptomeCell type
The invention discloses a method for analyzing double cells in single cell transcriptome data, and belongs to the technical field of single cell transcriptome sequencing. Through computer simulation,a cell expression profile is synthesized on the basis of experimental data, and double cells in the experimental data are identified by identifying and synthesizing heterogeneous double cells. The method does not depend on the existing knowledge of cell types and does not depend on the hypothesis of the total expression quantity of the cells, and the full-automatic double-cell identification can be realized on the basis of computer simulation.
Owner:SINGLERON NANJING BIOTECHNOLOGIES LTD
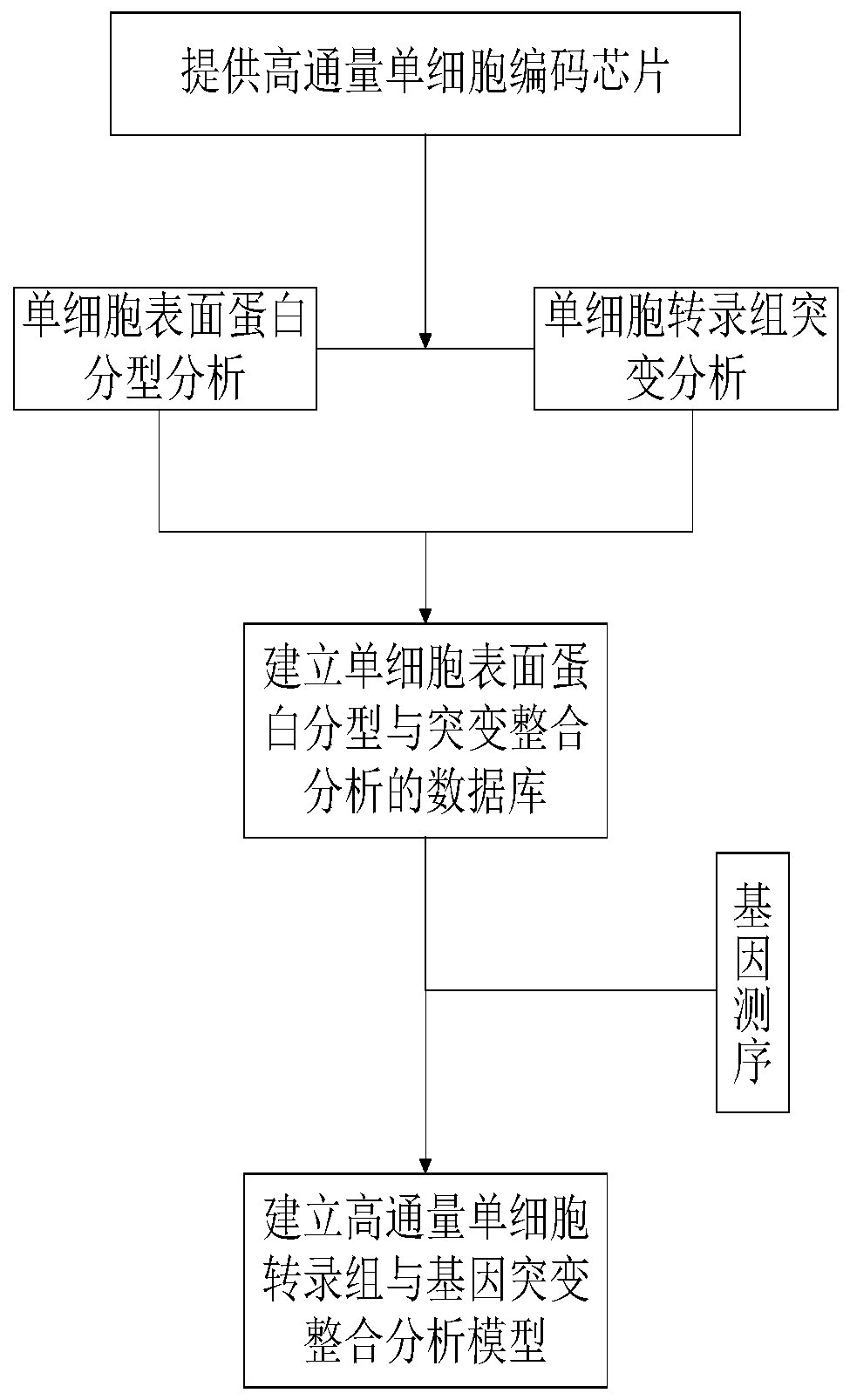
High-throughput single-cell transcriptome and gene mutation integration analysis method
InactiveCN110577983AAchieve a comprehensive understandingAchieve comprehensive characterizationMicrobiological testing/measurementSingle cell transcriptomeCell Surface Proteins
The invention discloses a high-throughput single-cell transcriptome and gene mutation integration analysis method. The high-throughput single-cell transcriptome and gene mutation integration analysismethod comprises the following steps that (1) a high-throughput single-cell encoding chip is provided; (2) single-cell surface protein parting analysis is conducted; (3) single-cell transcriptome mutation analysis is conducted; (4) a database for single-cell surface protein parting and mutation integration analysis is established; and (5) a high-throughput single-cell transcriptome and gene mutation integration analysis model is established. According to the high-throughput single-cell transcriptome and gene mutation integration analysis method, by designing the single-cell encoding chip withtriple encoding technologies of microporous spatial coordinates, cell nucleic acid labels and molecular nucleic acid labels and by combining the modes of single-cell surface protein parting, single-cell transcriptome mutation analysis and gene sequencing, gene mutation information, transcriptome information and protein expression information of single cells can correspond one by one, the completedatabase for high-throughput single-cell transcriptome and gene mutation integration analysis is formed, and the multi-omics integration analysis model is obtained.
Owner:SUZHOU INST OF BIOMEDICAL ENG & TECH CHINESE ACADEMY OF SCI
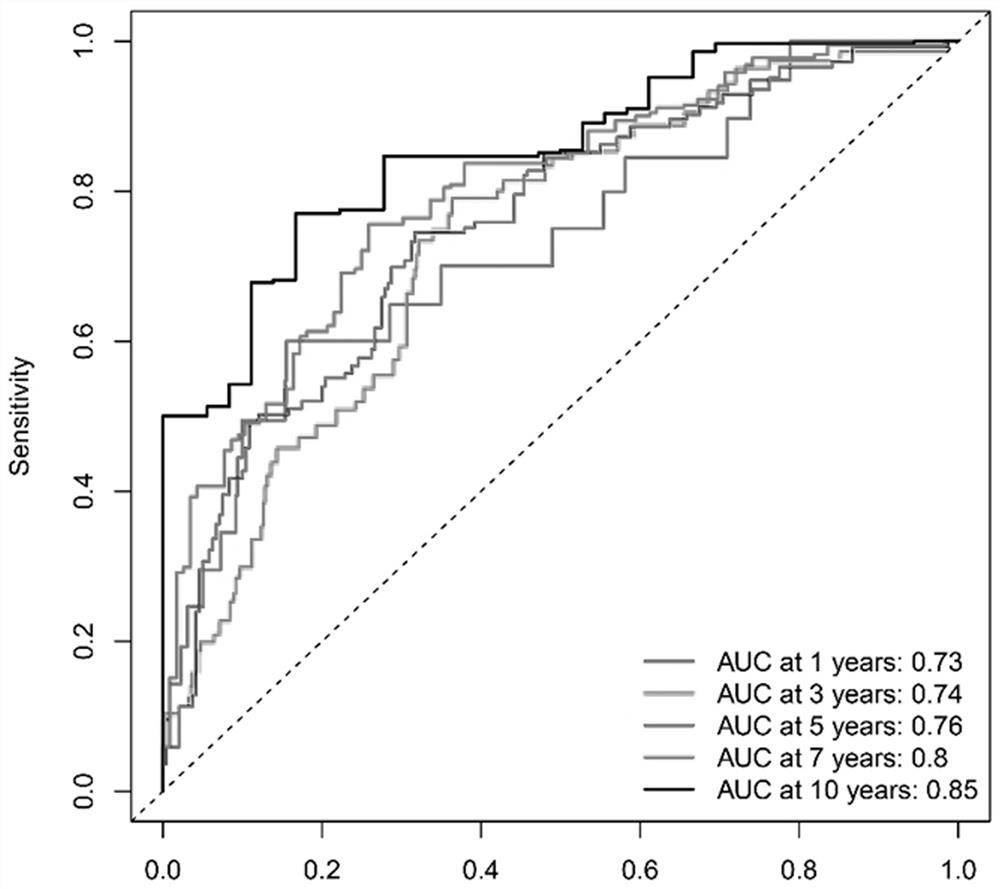
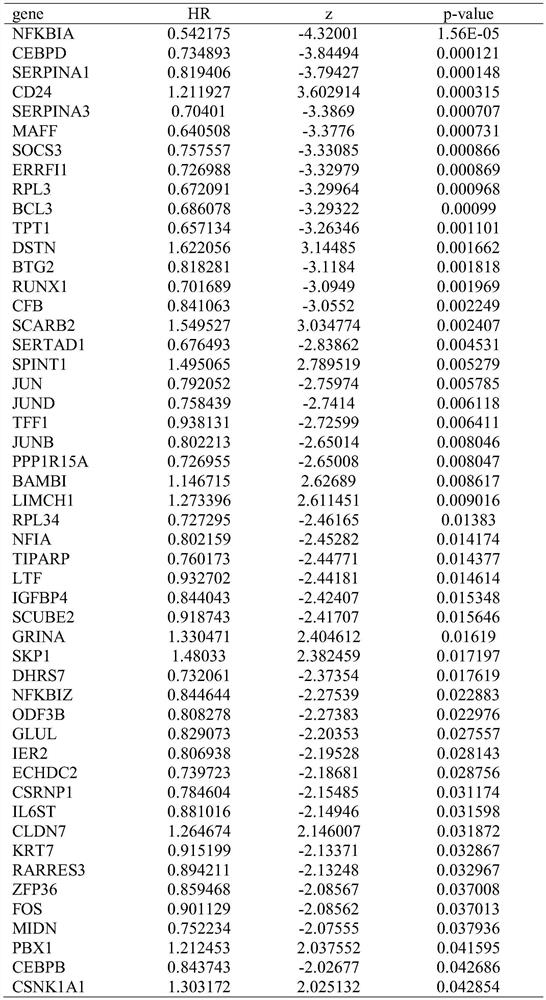
Breast cancer patient recurrence risk 20 gene prediction model based on breast cancer single cell transcriptome sequencing analysis
PendingCN112481378AAccurate judgmentPrecision therapyMicrobiological testing/measurementSequence analysisSingle cell transcriptomeAdjuvant therapy
The invention relates to the field of gene detection technologies and biomedicine, in particular to a breast cancer patient recurrence risk 20 gene prediction model based on breast cancer single-celltranscriptome sequencing analysis and an establishment method and application thereof. The model is composed of 20 genes of CEBPD, SERPINA1, CD24, ERRFI1, BCL3, DSTN, BTG2, SERTAD1, SPINT1, BAMBI, LIMCH1, NFIA, SKP1, DHRS7, ODF3B, KRT7, ZFP36, CEBPB, BHLHE40 and UGDH. The breast cancer patient recurrence risk 20 gene prediction model provided by the invention provides more accurate judgment for long-term prognosis of a breast cancer patient, and provides a basis for selection of a postoperative adjuvant therapy scheme of the patient, so that individualized accurate treatment is realized, and meanwhile, a new research perspective is provided for research of breast cancer tumor stem cells.
Owner:SHENGJING HOSPITAL OF CHINA MEDICAL UNIV
Modulation of cellular transcription factor activity
InactiveUS6610650B1Inhibit bindingAntibacterial agentsBiocideTranscription factor activityDouble strand
Owner:NEILSEN PETER E
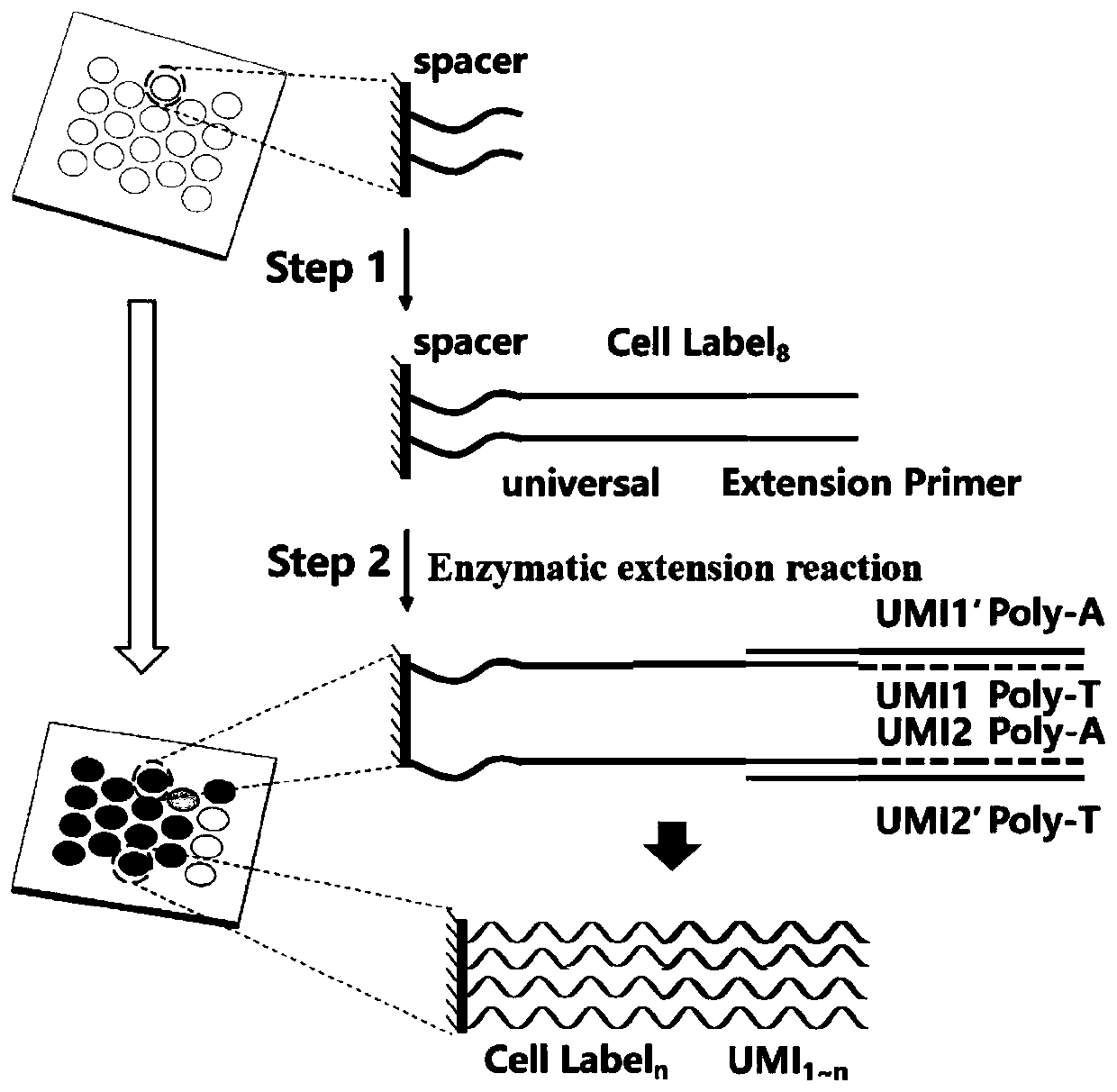
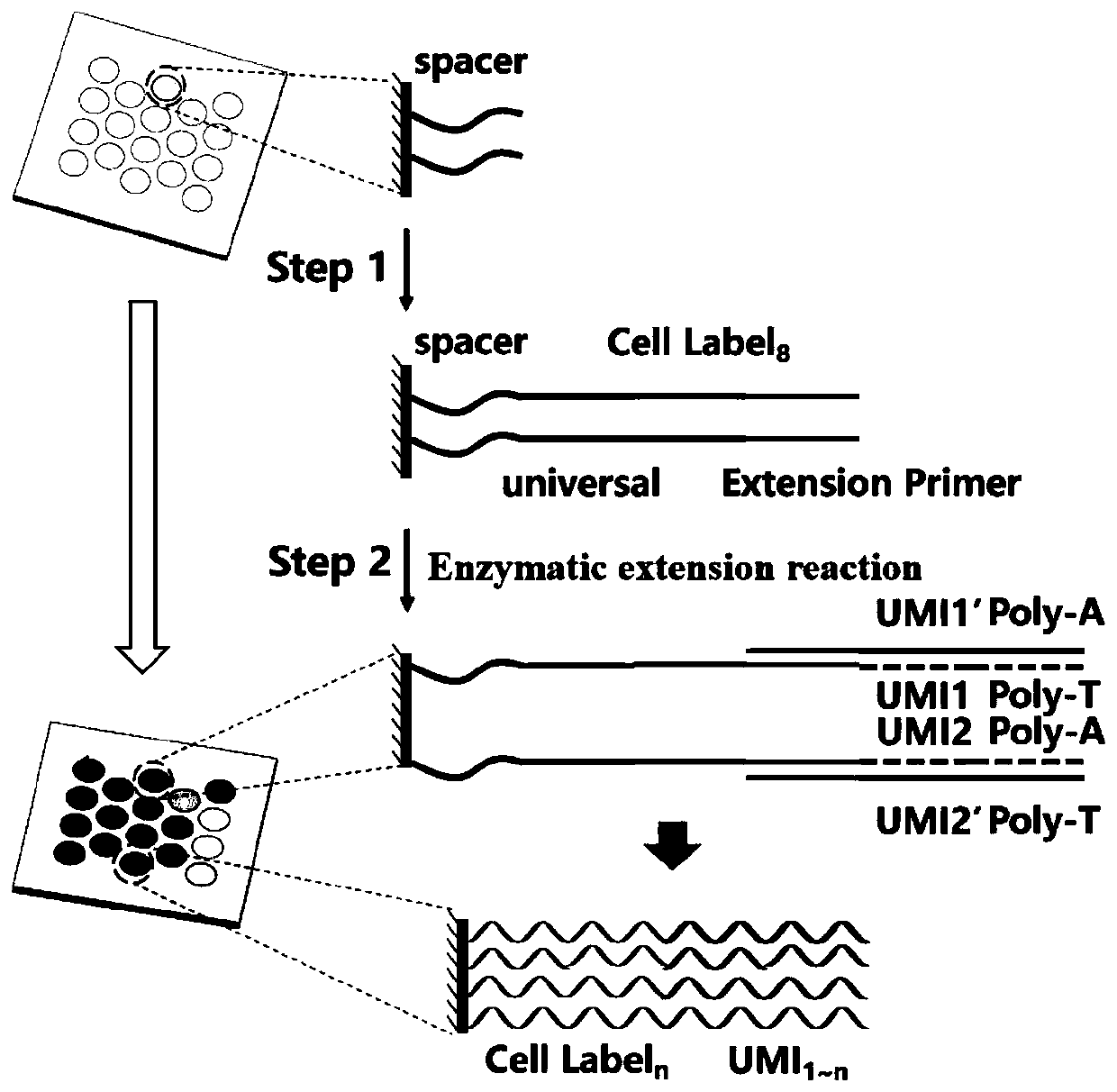
High-throughput single-cell transcriptome and gene mutation integration analysis coding chip
InactiveCN110577982AMicrobiological testing/measurementSingle cell transcriptomeNucleic acid sequencing
The invention discloses a high-throughput single-cell transcriptome and gene mutation integration analysis coding chip. According to the chip, a substrate is provided with a plurality of microholes, the micropores have a size and a shape which can only accommodate a single cell in a micropore, each micropore has a unique spatial coordinate encoding, the micropores are internally modified with a plurality of known nucleic acid sequences, the nucleic acid sequences successively include Spacer sequences, universal primer sequences, cell labels, molecular labels, and Ploy T, wherein the universalprimer sequences are used as primer-binding regions when a PCR is augmented, the cell labels are used for labeling cells from which the RNA originated, and the molecular labels are used for labeling combinative RNA. The invention provides the chip capable of being used for high-throughput single-cell transcriptomes and gene mutation integration analysis. By adopting a triple coding technique of microporous space coordinates, cell nucleic acid labels and molecular nucleic acid labels, gene mutation, the transcriptomes and protein expression information of a single cell can be matched one by one.
Owner:SUZHOU INST OF BIOMEDICAL ENG & TECH CHINESE ACADEMY OF SCI
HEXIM1 as a suppressor of HIV replication and cardiac hypertrophy
Cellular transcription is modulated by increasing or decreasing the amount of active HEXIM1 in the cell. The methods are applied to the treatment of HIV infection and cardiac hypertrophy. Assays using reconstituted 7SK:P-TEFb snRNP screen for agents that modulate HEXIM1-P-TEFb binding.
Owner:RGT UNIV OF CALIFORNIA
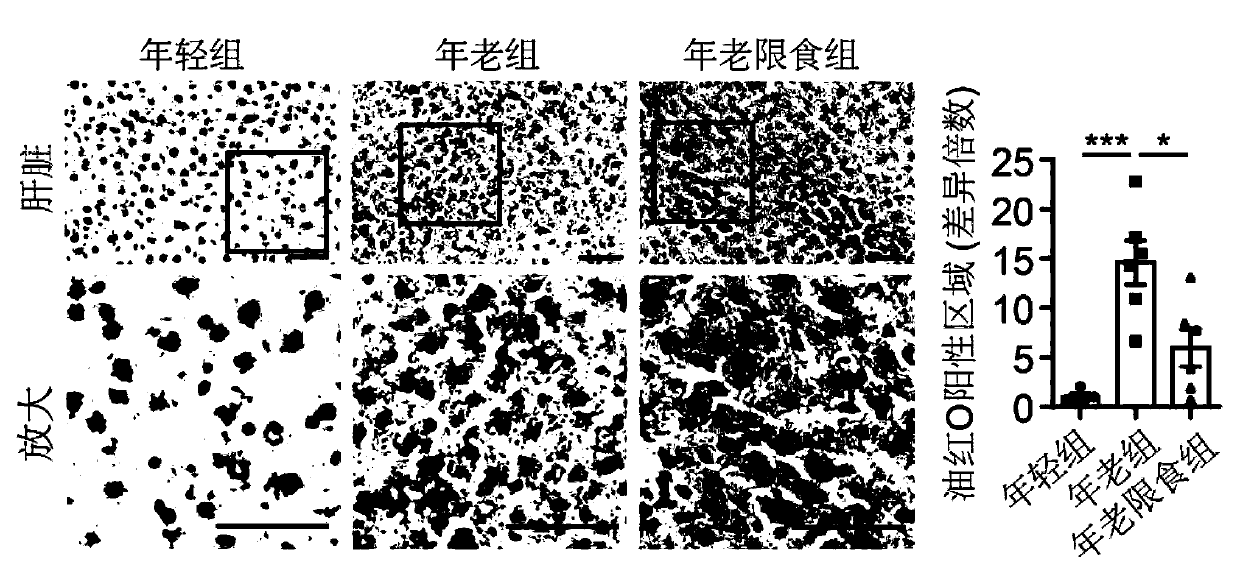
Application of senescence markers and caloric restrictions of multiple tissue organs and cell types in delaying senescence of organism
ActiveCN111218515AMicrobiological testing/measurementBiological material analysisBiotechnologySingle cell transcriptome
The invention discloses an application of senescence markers and caloric restrictions of multiple tissue organs and cell types in delaying senescence of organism. According to the invention, integration analysis of a multi-tissue single-cell transcriptome is carried out by taking a rat as a model so as to research the regulation and control effects of senescence and calorie limitation on a mammalsingle-cell level, and 160000 single cells in seven tissues of a young diet group, an old group and an old diet limitation group are systematically analyzed; based on a high-throughput single-cell transcriptome sequencing technology, a first mammal aging and CR multi-tissue organ high-throughput single-cell transcriptome map is established, the effects of senescence and CR on different tissues andcell types are comprehensively and systematically evaluated in the aspects of cell type composition, cell and tissue specificity differential expression gene, core regulation and control transcription factors, cell-cell communication networks and the like, the complex processes of senescence and CR are revealed, and a basis is provided for systematically researching the molecular regulation and control mechanisms of senescence and CR in the future.
Owner:INST OF ZOOLOGY CHINESE ACAD OF SCI
Single cell transcript isomer sequencing analysis method and kit
PendingCN110643692AIncreased sensitivityHigh detectionMicrobiological testing/measurementResearch strategiesTranscript isoforms
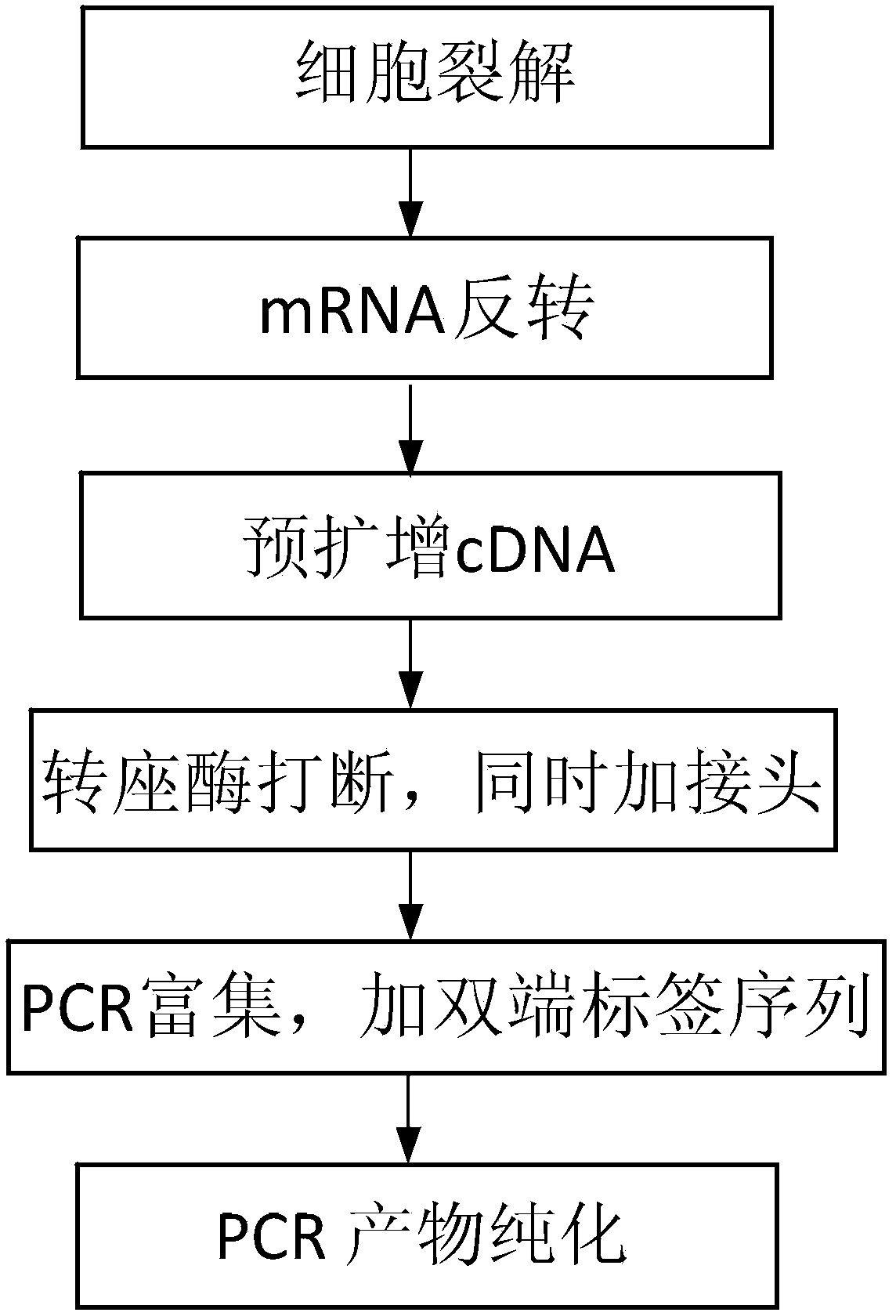
The invention discloses a single cell transcript isomer sequencing analysis method and a kit. By adding tag sequences to the two tail ends of a single mRNA, after trapping and enriching are performed,the whole transcriptome is sequenced, and by accurately judging the head and tail structures of different transcript isomers, full-length transcripts can be judged. Existing technical problems can besolved by the new research strategy. The single cell transcript isomer sequencing analysis method and the kit have the following beneficial effects that 1, the sensitivity of the detection of the full-length transcripts is improved, specifically, due to the targeted detection of the head and tail areas of the transcripts, signals are mostly concentrated at the end edges of the transcripts, so that the detection of the full-length transcripts is higher; 2, simple operation is achieved, and the cost low, specifically, the method is simple to operate, widely applicable and the cost is low; and 3, the accuracy of the detection of the transcripts is improved, specifically, higher accuracy can be achieved by combining the tag sequences of the tail end sequences and the optimization of a data analysis algorithm.
Owner:ZHONGSHAN OPHTHALMIC CENT SUN YAT SEN UNIV
Differential gene analysis method based on BD single cell transcriptome and proteome sequencing data
PendingCN113470743ACharacter and pattern recognitionProteomicsProtein regulationSingle cell transcriptome
The invention discloses a differential gene analysis method based on BD single cell transcriptome and proteome sequencing data, and belongs to the technical field of transcriptome data analysis. In order to solve the problem that no complete data analysis method can combine single cell transcriptome and proteome sequencing data for analysis in the prior art, differential gene analysis of cell subset classification in transcription and protein levels is carried out through a single cell transcriptome and proteome sequencing technology, cell characteristics are represented, and a biological mechanism transcribed to a protein regulation expression process is obtained, and a more accurate and richer BD single cell transcriptome and proteome sequencing data combined analysis method is provided. According to the analysis method, deep understanding of differential expression can be obtained, key mRNA or protein regulated after transcription is excavated, some important regulatory pathways are found and verified, and the analysis method can be applied to the field of tumors.
Owner:哈尔滨星云医学检验所有限公司
Integrated device for integrated analysis of high-throughput single-cell transcriptome and gene mutation
ActiveCN110951580AIntegrated Analysis RealizationAmplifyBioreactor/fermenter combinationsBiological substance pretreatmentsGenes mutationSingle cell transcriptome
The invention discloses an integrated device for integrated analysis of high-throughput single-cell transcriptome and gene mutation. The integrated device includes a high-throughput single-cell codingchip and an integrated analysis device body, wherein the integrated analysis device body includes a housing and a temperature-control thermal cycle module, a fluorescence imaging module and a data storage analysis module which are disposed in the housing, and the fluorescence imaging module includes a light source component, a microscope objective, a fluorescence splitting component and an imaging detector. One-to-one correspondence of gene mutation, transcriptome and protein expression information of single cells can be achieved through the designed high-throughput single-cell coding chip which is provided with triple-encoding functions of micropore spatial coordinates, cellular nucleic acid tags and molecular nucleic acid tags, and PCR amplification can be realized through the temperature-control thermal cycle module; and the fluorescence imaging module is used for collecting fluorescence images of a sample, and the fluorescence images are stored and analyzed through the data storage analysis module, so that integrated analysis of single-cell transcriptome and gene mutation is achieved.
Owner:SUZHOU INST OF BIOMEDICAL ENG & TECH CHINESE ACADEMY OF SCI
Multi-omics method for single cell transcriptome and translation group combined sequencing
ActiveCN112831552AQuality improvementMicrobiological testing/measurementCDNA librarySingle cell transcriptome
The invention provides a method for simultaneously carrying out transcriptome sequencing and transscriptome sequencing, which is characterized by comprising the following steps: carrying out first-step lysis treatment on cells and a lysis buffer solution to obtain a first lysis solution; after uniformly mixing and centrifuging, taking a part of the first lysis solution in the supernate as a transcriptome template, and taking the rest of the supernate and the lysis buffer solution to carry out second lysis treatment so as to obtain a second lysis solution; centrifuging the second lysate, and collecting supernate as a translation group template; obtaining a transcriptome cDNA library by using the transcriptome template, and sequencing the transcriptome cDNA library; and extracting a ribosome protected RNA fragment from the translation group template, and carrying out library establishment and sequencing on the ribosome protected RNA fragment. According to the method, high-quality transcriptome and translation group data in a whole genome range can be obtained at the same time, and the method is particularly suitable for single cells, so that possibility is provided for knowing collaborative regulation of single cell transcription and translation levels.
Owner:TSINGHUA UNIV
Single cell transcriptome sequencing method and application thereof
PendingCN114507711AWide range of typesHigh detection sensitivityMicrobiological testing/measurementLibrary creationSingle cell transcriptomeSingle cell suspension
The invention provides a single-cell transcriptome sequencing method and application thereof. The method comprises the following steps: preparing a to-be-detected cell sample into a single-cell suspension, and fixing the single-cell suspension with a stationary liquid; and performing an in-situ reverse transcription reaction on the RNA of the fixed single cell by using a reverse transcription primer to synthesize a cDNA first chain.
Owner:ZHEJIANG UNIV
High-throughput single-cell transcriptome sequencing method and kit
PendingCN113026110AReduce mutual contaminationImprove capture efficiencyMicrobiological testing/measurementLibrary creationHigh cellSingle cell transcriptome
The invention discloses a high-throughput single cell transcriptome sequencing method. The method comprises the following steps: capturing and packaging a single cell by using a micro-fluidic chip, carrying out RAN reverse transcription, pre-amplifying cDNA, carrying out primary amplification, carrying out secondary amplification, performing fragmenting, building a library, performing sequencing, conducting analyzing and the like. The sequencing method disclosed by the invention has the advantages of high cell capture efficiency, high flux (up to 10,000 cells), high flexibility ( wherein 1-8 samples can be prepared at the same time), low cross contamination, low double-package rate, simplicity in operation, low cost and the like, and has wide application prospects in various fields such as environment, infectious diseases, rejection after organ transplantation, immunotherapy and the like.
Owner:SUZHOU GENO TRUTH BIOTECHNOLOGY CO LTD
Method of amplifying single cell transcriptome
PendingCN111406114AMicrobiological testing/measurementDNA preparationSingle cell transcriptomeBarcode
The present disclosure provides methods for amplifying RNA by using a combination of reverse transcription and amplification cycles based on multiple annealing and cyclization. Primers are used such that the resulting amplicons comprise a first cell-specific barcode sequence, a second cell-specific barcode sequence, and a unique molecular identifier barcode sequence.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Module-level analysis of peripheral blood leukocyte transcriptional profiles
InactiveUS20140179807A1Easy to explainDifficult to interpretBiocideMicrobiological testing/measurementIndividual geneGene expression
The present invention includes an apparatus, system and method for the development and use of transcriptional modules by obtaining individual gene expression levels from cells obtained from one or more patients with a disease or condition; recording the expression value for each gene in a table that is divided into clusters; iteratively selecting gene expression values for one or more transcriptional modules by: selecting for the module the genes from each cluster that match in every disease or condition; removing the selected genes from the analysis; and repeating the process of gene expression value selection for genes that cluster in a sub-fraction of the diseases or conditions; and iteratively repeating the generation of modules.
Owner:BAYLOR RES INST
HEXIM1 as a suppressor of HIV replication and cardiac hypertrophy
Cellular transcription is modulated by increasing or decreasing the amount of active HEXIM1 in the cell. The methods are applied to the treatment of HIV infection and cardiac hypertrophy. Assays using reconstituted 7SK:P-TEFb snRNP screen for agents that modulate HEXIM1-P-TEFb binding.
Owner:RGT UNIV OF CALIFORNIA
Construction method and application method of lung cancer prognosis model and electronic equipment
PendingCN112735592AMedical simulationHealth-index calculationWhite blood cellCandidate Gene Association Study
The invention discloses a construction method and an application method of a lung cancer prognosis model, electronic equipment and a storage medium. The construction method comprises the following steps: acquiring peripheral blood leukocyte transcription spectrum expression data of a plurality of lung cancer patients and a plurality of reference people; screening candidate genes based on the transcriptional profile expression data; constructing a risk scoring model based on the candidate genes; wherein a lung cancer prognosis model comprises a risk scoring model; screening candidate genes based on the transcriptional profile expression data, including: performing similarity analysis on the transcriptional profile expression data to determine a plurality of lung cancer subgroups; respectively carrying out differential expression gene analysis on the plurality of lung cancer subgroups and a plurality of reference people, and determining respective differential expression genes of the plurality of lung cancer subgroups; comparing the expression states of the differentially expressed genes of the plurality of lung cancer subgroups, and selecting the lung cancer subgroup with more immunosuppressive genes in a high expression state from the plurality of lung cancer subgroups as a candidate lung cancer subgroup; and screening candidate genes from the differentially expressed genes of the candidate lung cancer subgroups.
Owner:CANCER INST & HOSPITAL CHINESE ACADEMY OF MEDICAL SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com