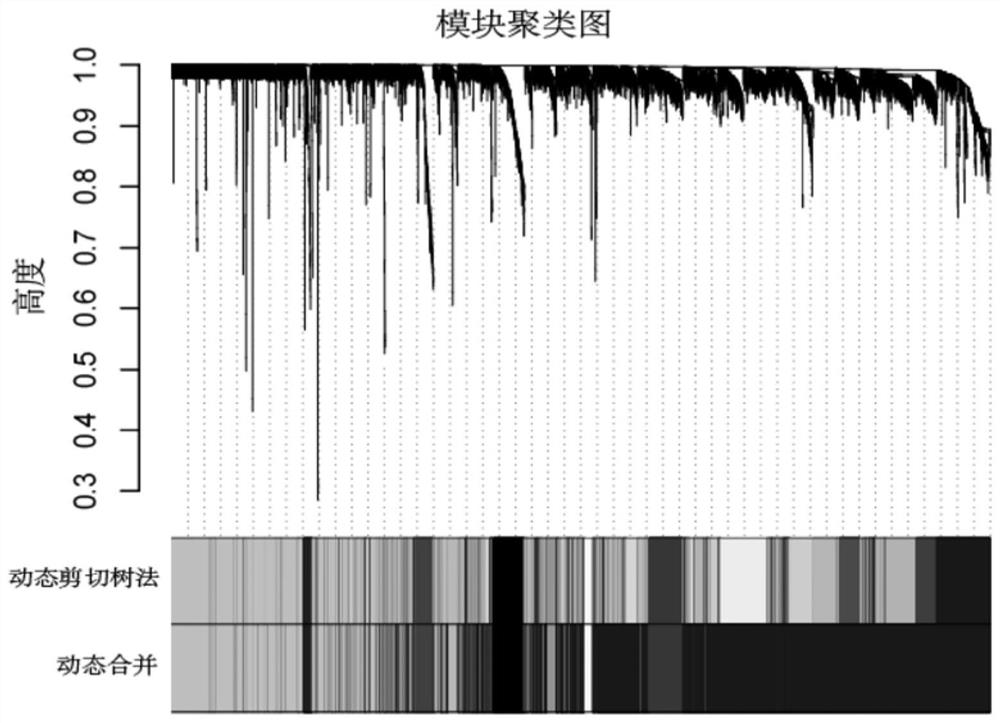
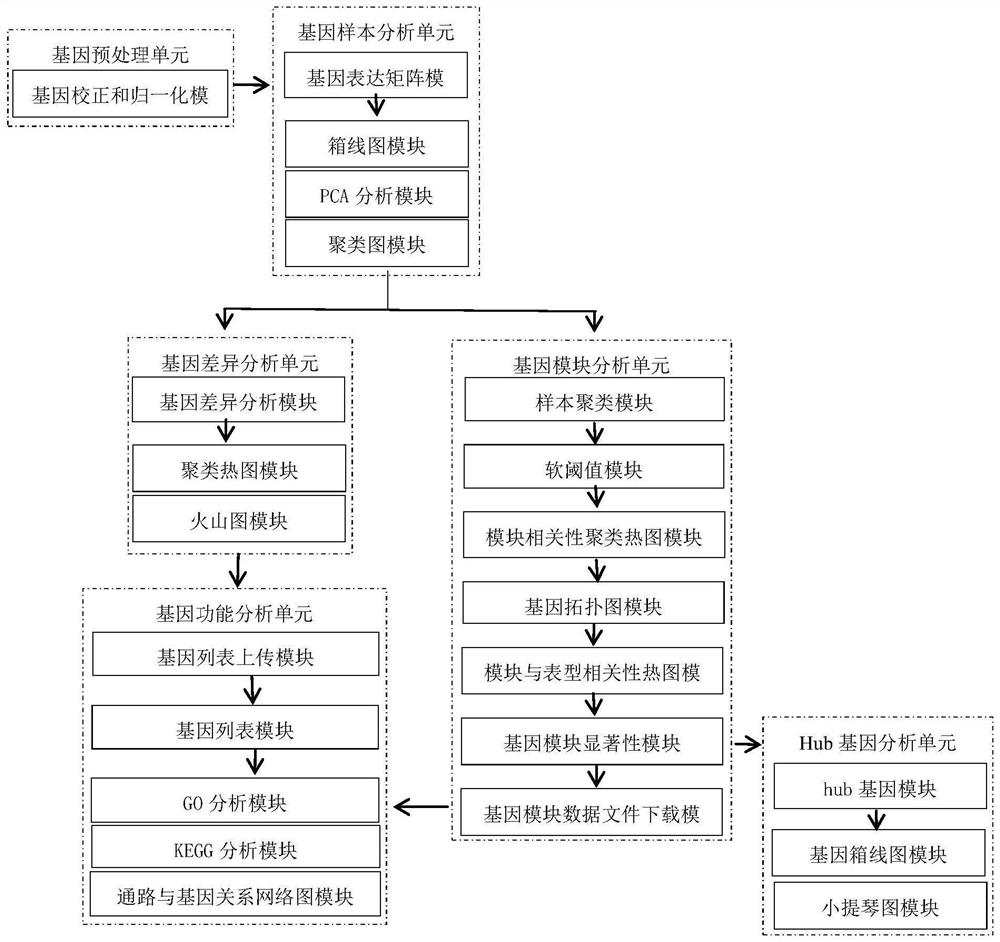
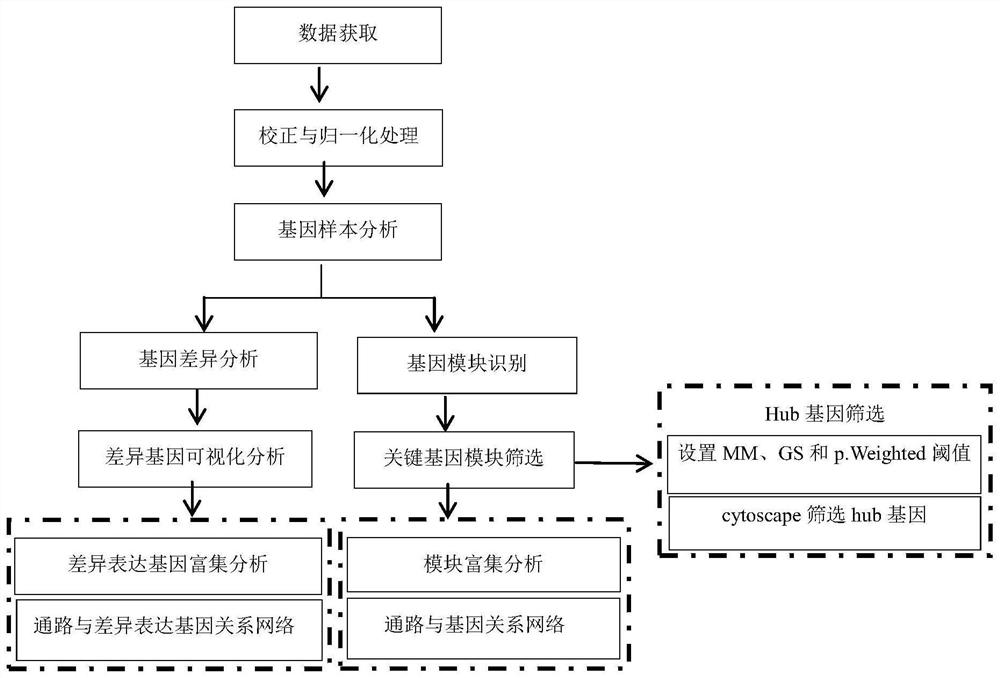
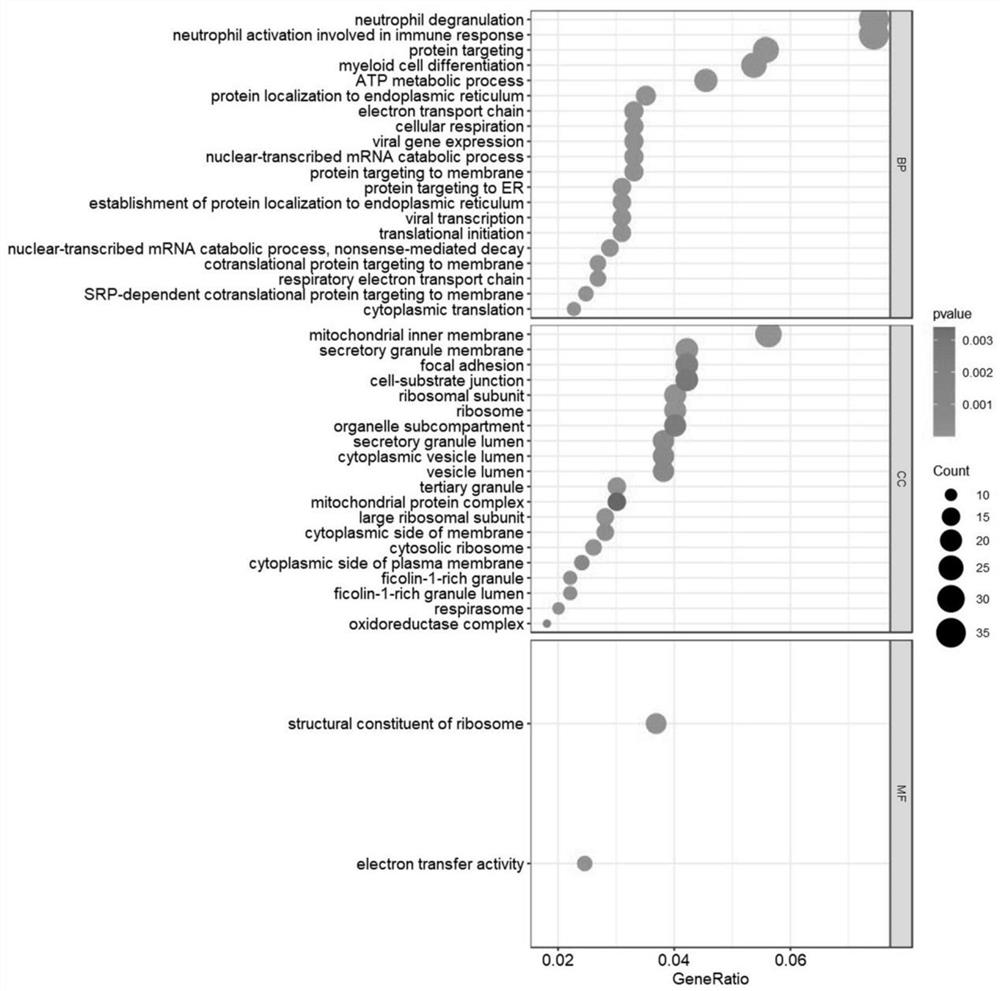
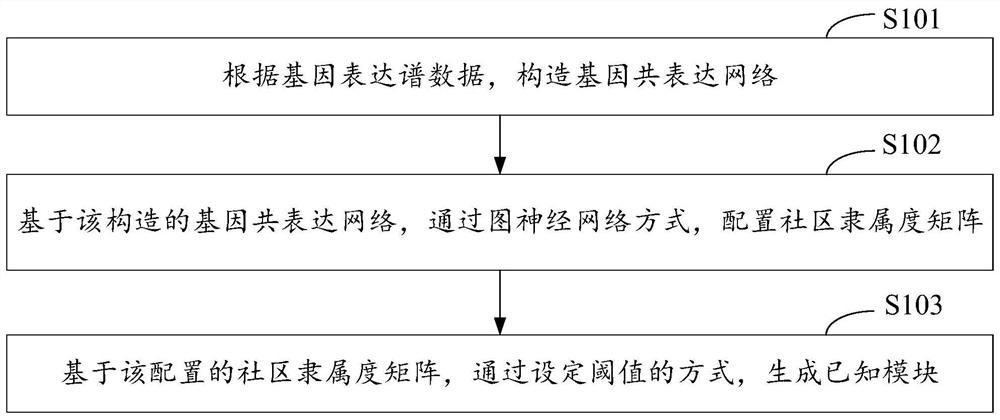
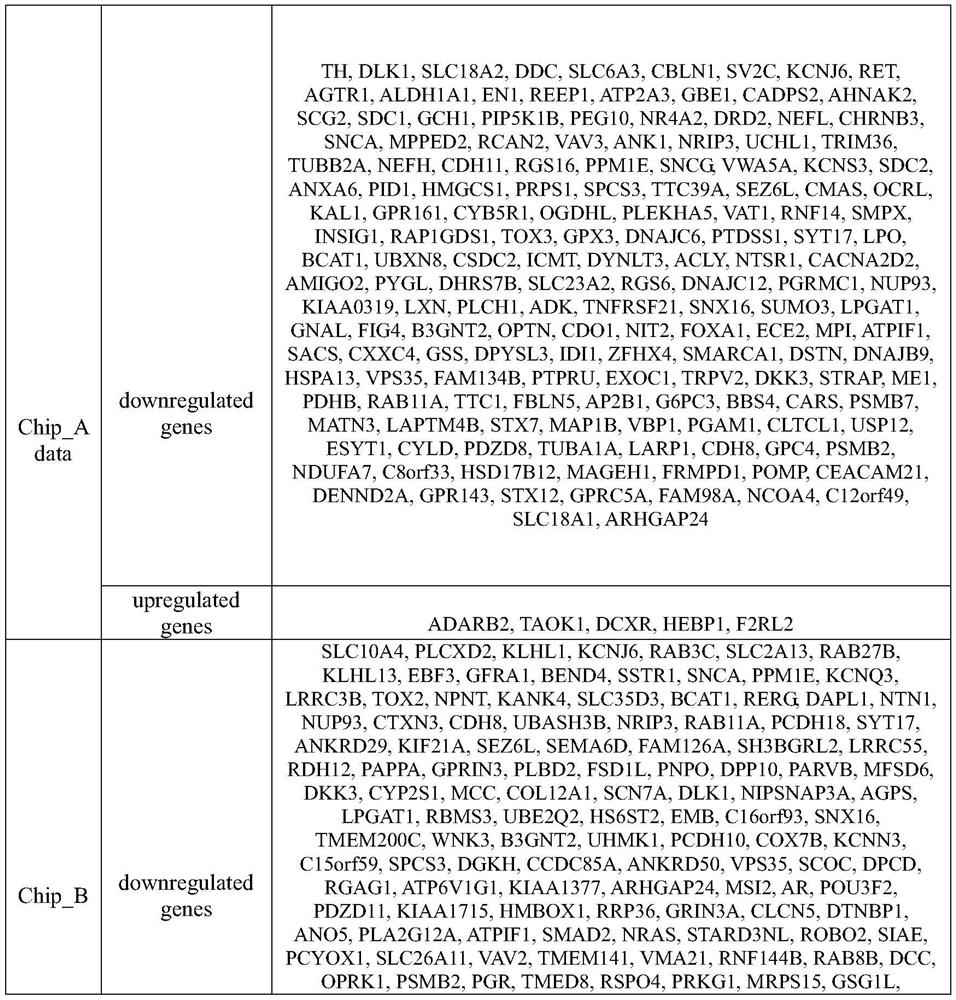
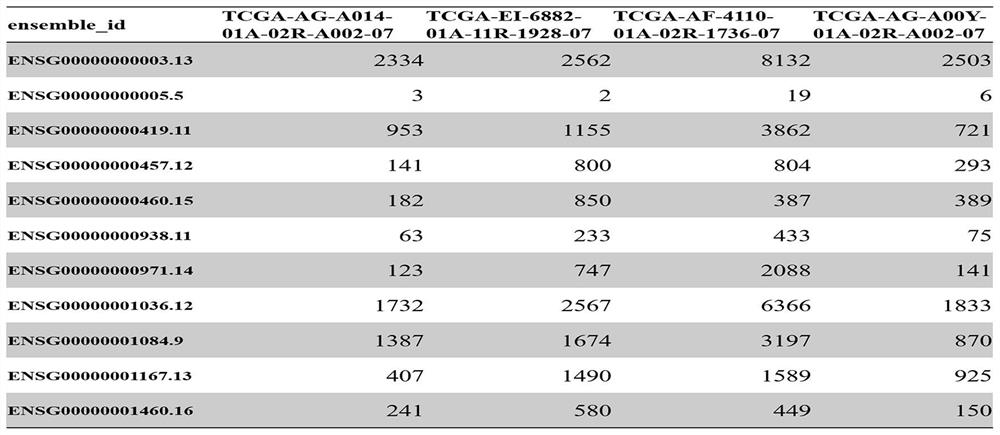
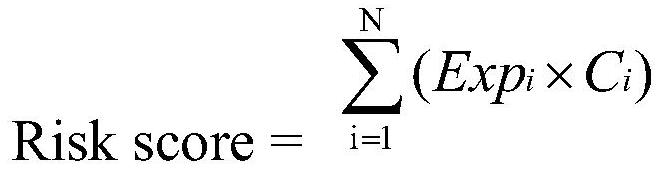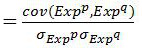Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
32 results about "Gene co-expression network" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
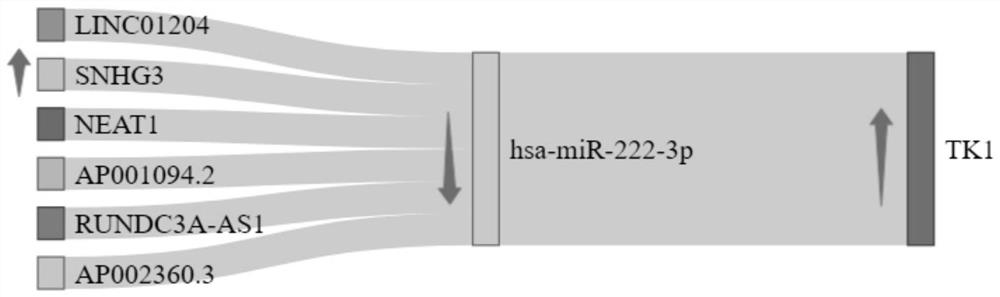
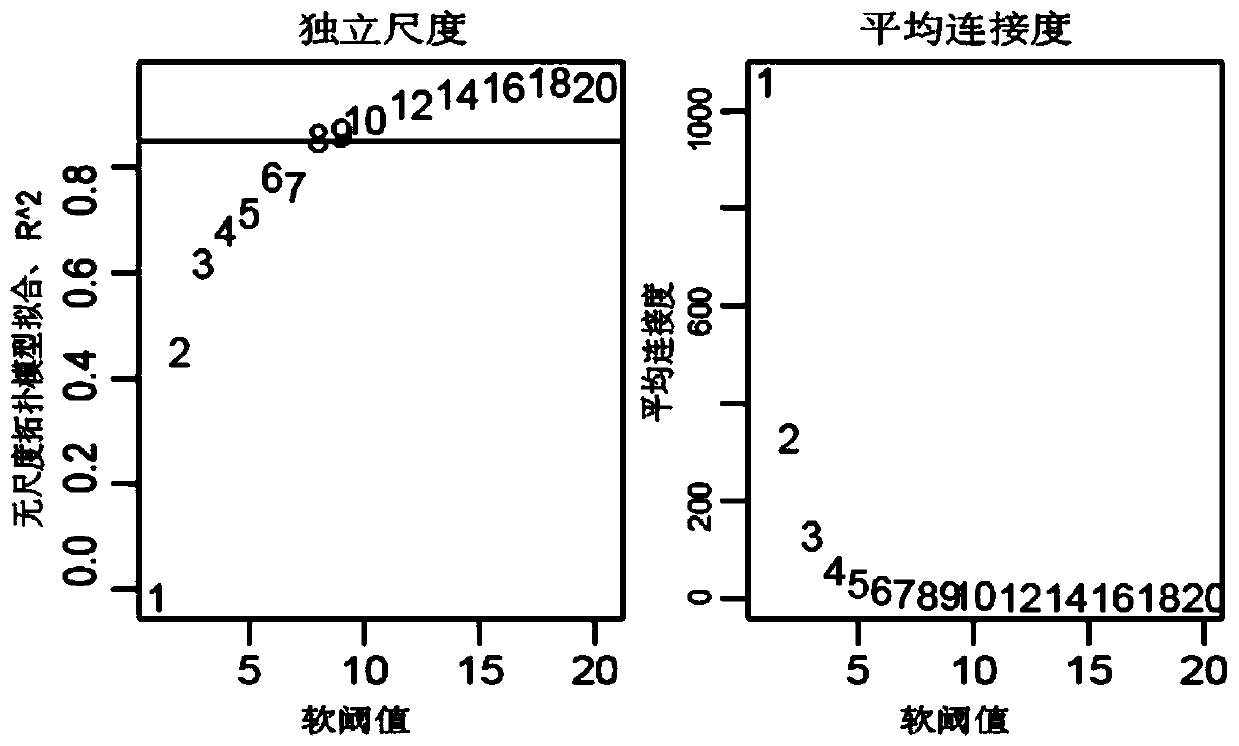
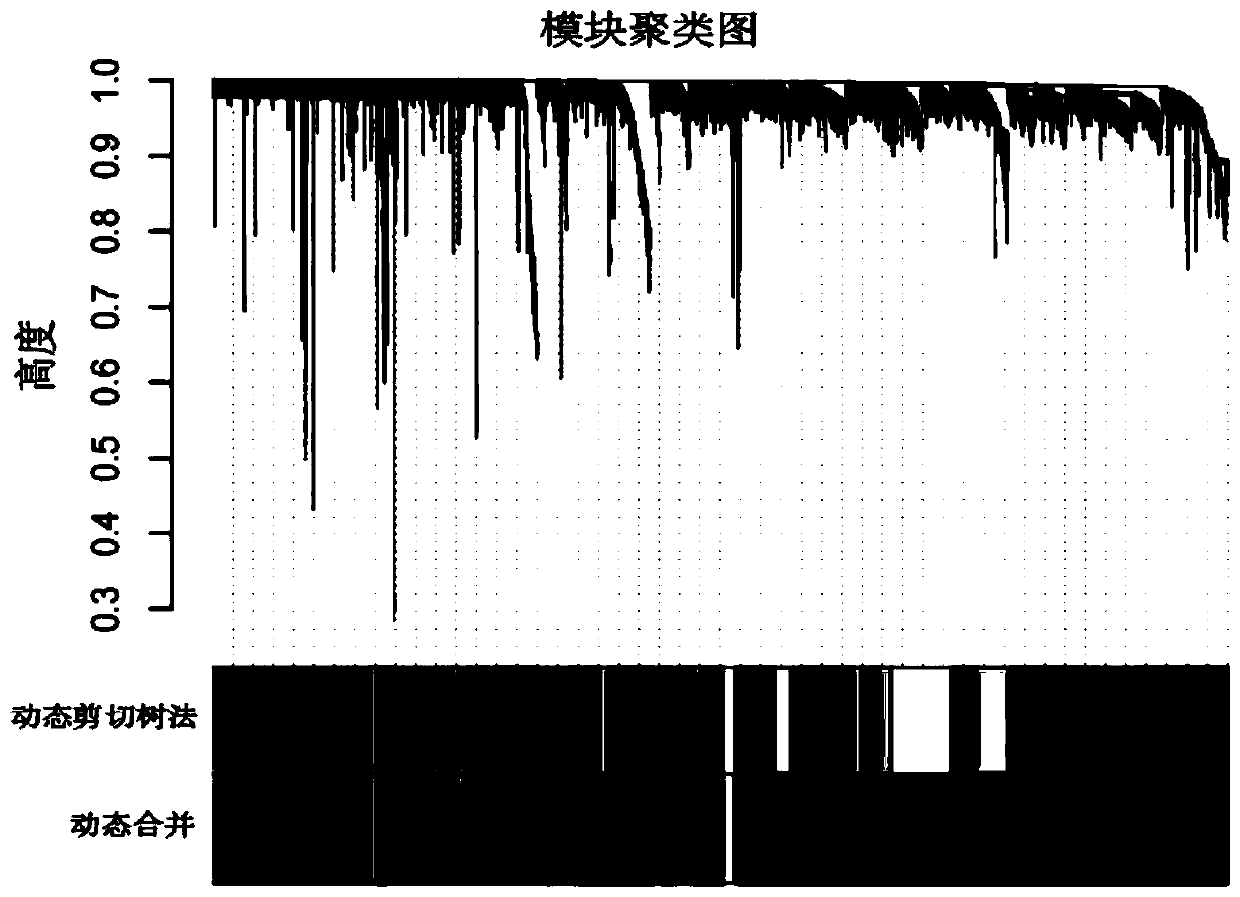
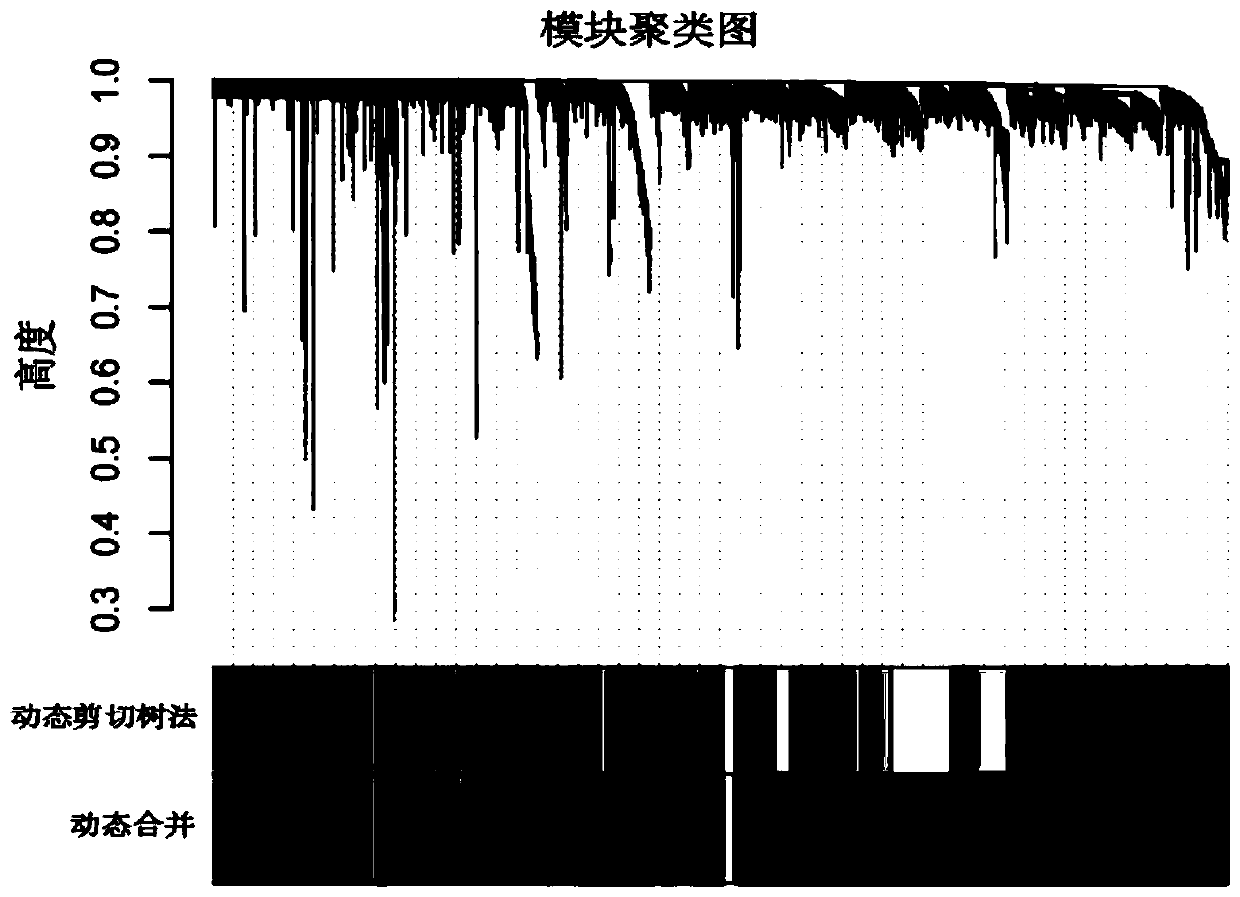
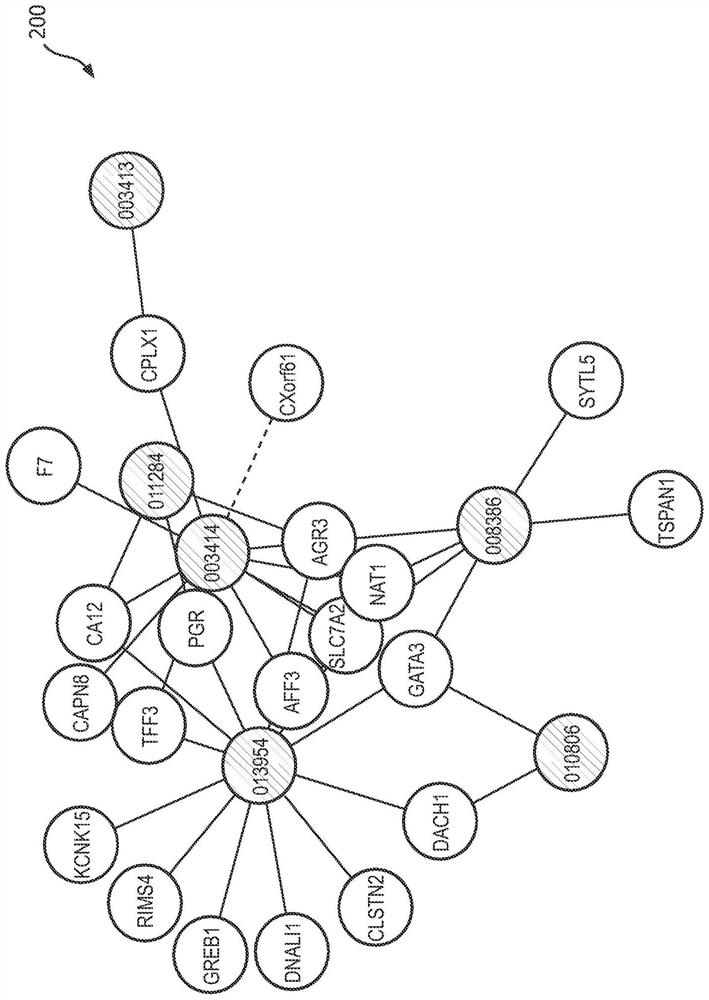
A gene co-expression network (GCN) is an undirected graph, where each node corresponds to a gene, and a pair of nodes is connected with an edge if there is a significant co-expression relationship between them. Having gene expression profiles of a number of genes for several samples or experimental conditions, a gene co-expression network can be constructed by looking for pairs of genes which show a similar expression pattern across samples, since the transcript levels of two co-expressed genes rise and fall together across samples. Gene co-expression networks are of biological interest since co-expressed genes are controlled by the same transcriptional regulatory program, functionally related, or members of the same pathway or protein complex.
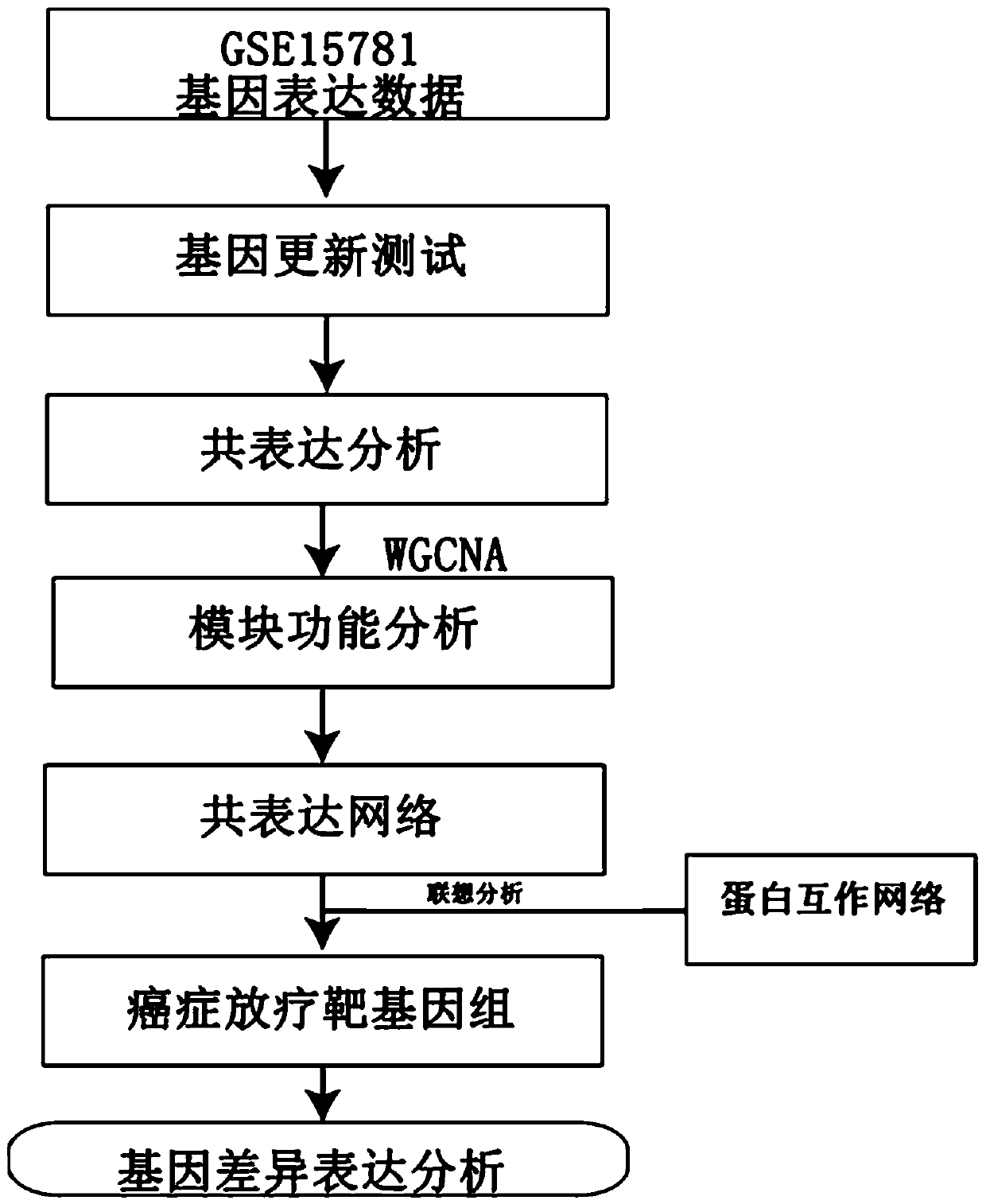
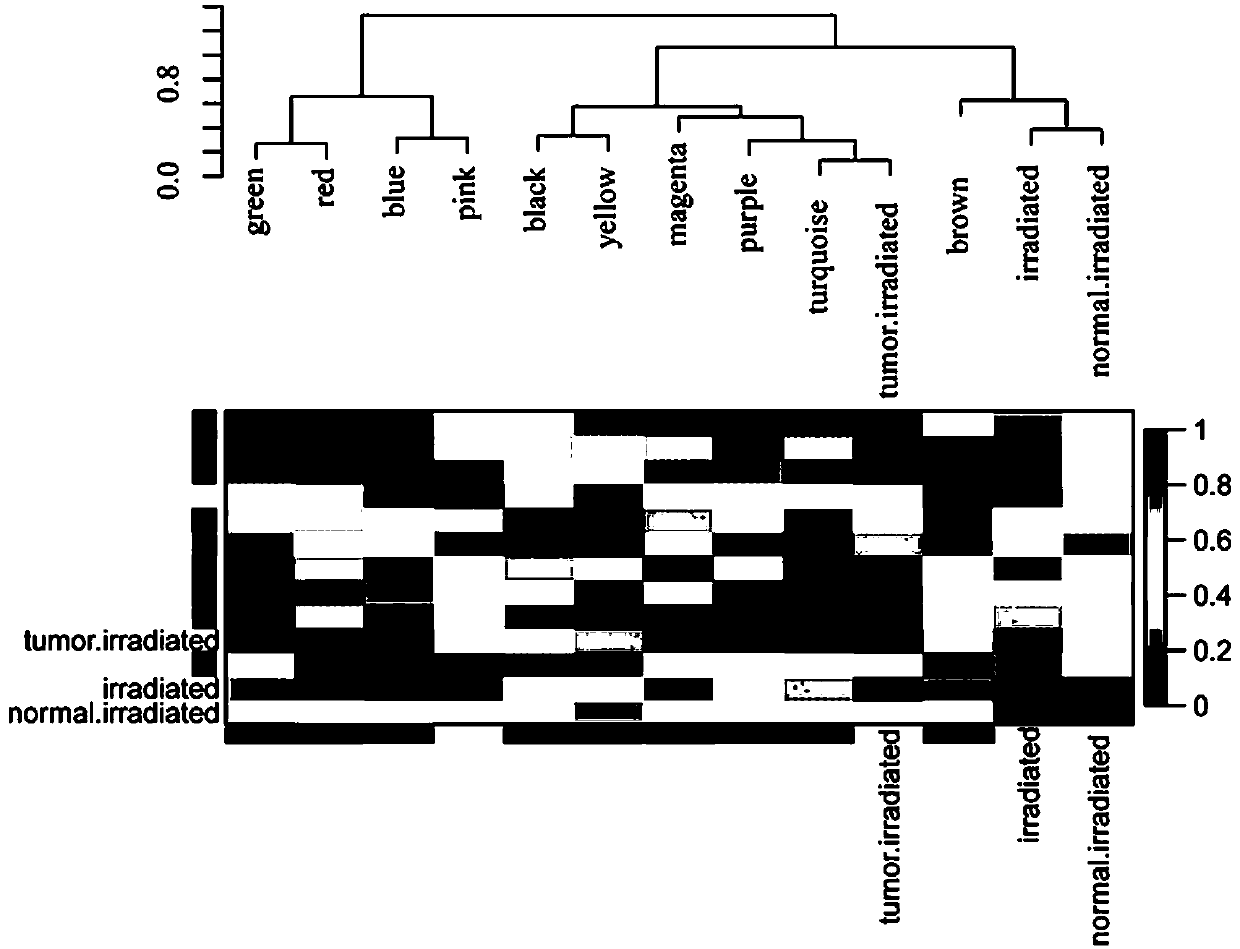
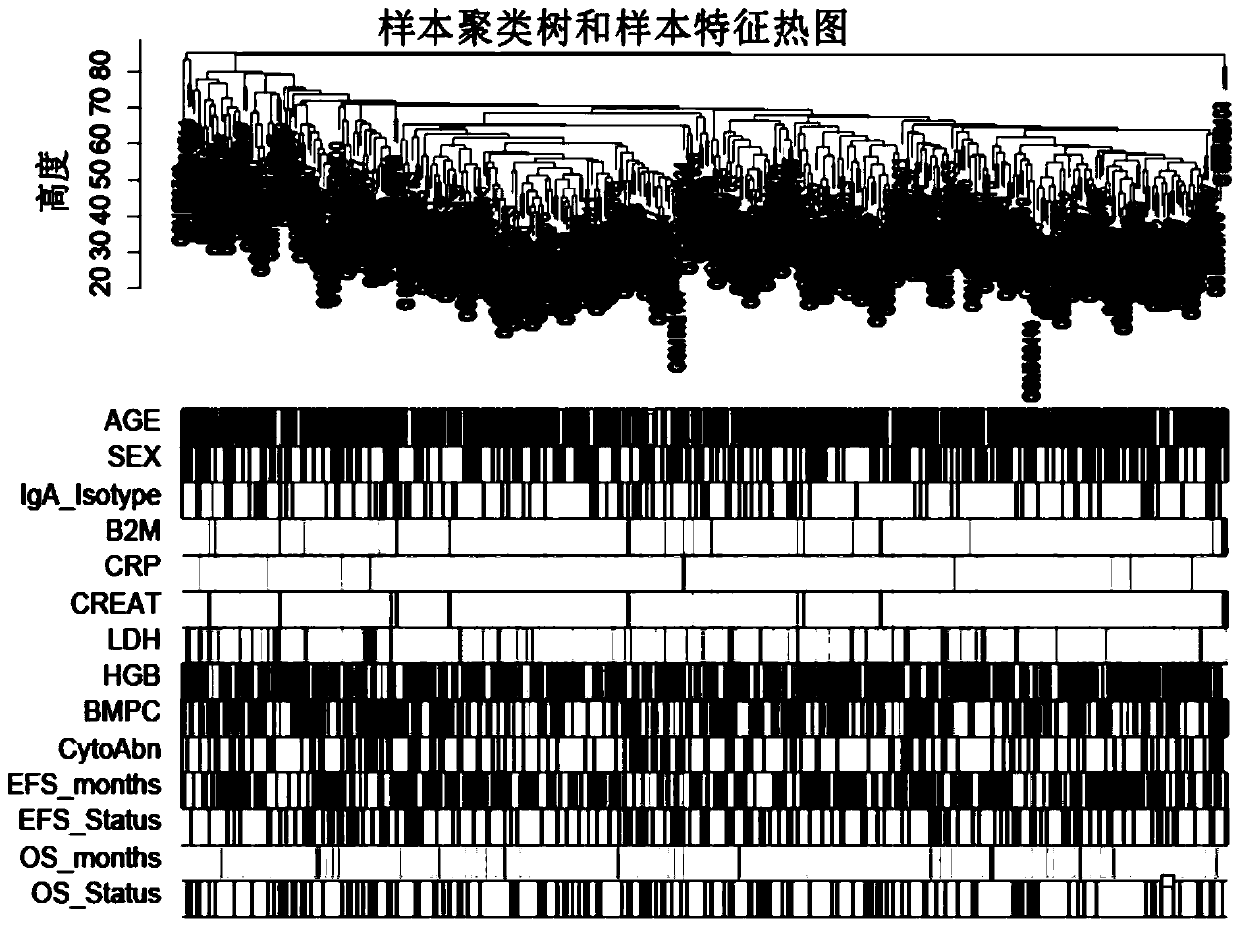
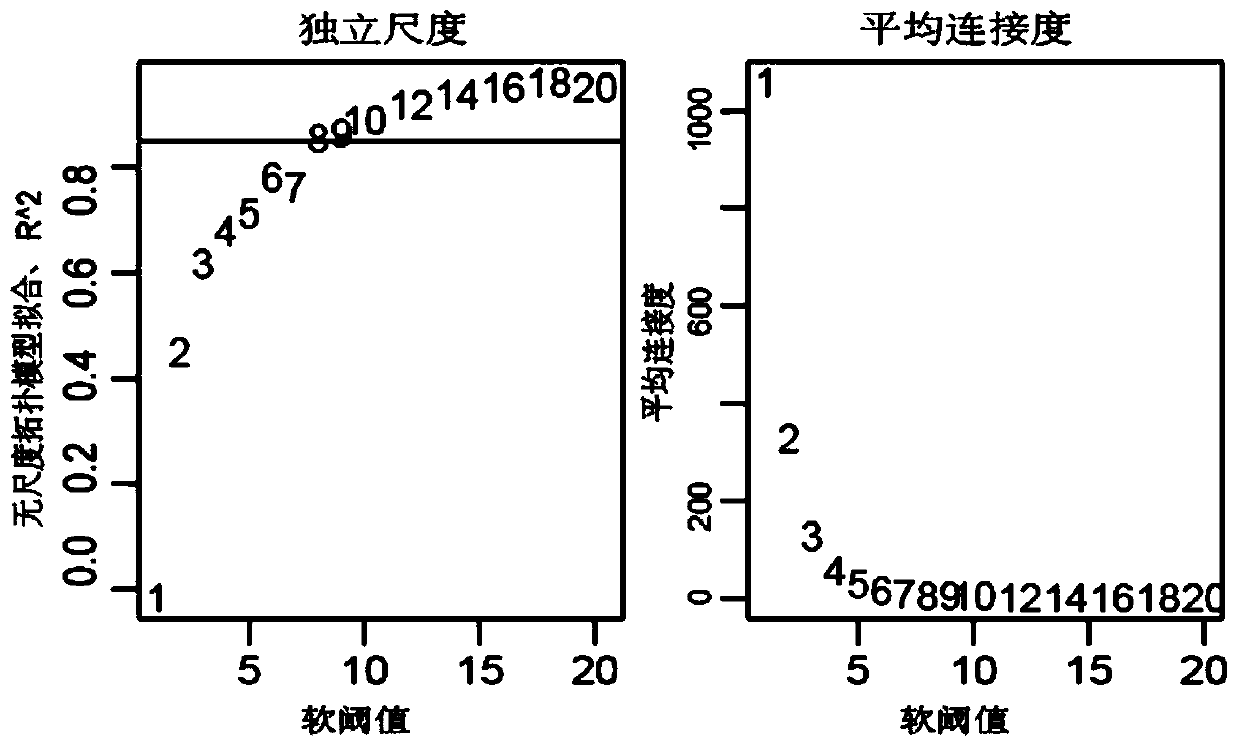
Method for mining radiotherapy specific genes of colorectal cancer by using weight gene co-expression network
ActiveCN109872772AReduce mortalitySolve clinical problemsBiostatisticsProteomicsSurvival prognosisMortality rate
The invention relates to a weight gene co-expression network, and specifically relates to a method for mining radiotherapy specific genes of colorectal cancer by using a weight gene co-expression network. The method for mining radiotherapy specific genes of colorectal cancer by using a weight gene co-expression network provides a new way to mine the specific response genes of colorectal cancer byusing the weight gene co-expression network, and provides a new basis for predicting the survival prognosis of patients with colorectal cancer. The radiotherapy of the target gene discovered by the method of the present invention can improve the survival prognosis of patients with colorectal cancer to some extent, can reduce the mortality of the patients with colorectal cancer, can solve the actual clinical problems, and can provide a better choice for the majority of patients.
Owner:辽宁省肿瘤医院
Method and system for predicting cancer
ActiveCN111899882AEfficient forecastingImprove accuracyMedical data miningHealth-index calculationMedicineData mining
The invention discloses a method and system for predicting cancer. The method comprises the following steps: carrying out difference analysis on the gene expression profile data of a cancer patient and a normal person to obtain difference genes; analyzing the gene expression profile data of the cancer patient and the normal person based on weighted gene co-expression network analysis to obtain a hub gene; processing the gene expression profile data of the difference genes through a variational auto-encoder algorithm to obtain dimension reduction data; and with the gene expression profile dataof the hub gene and the dimension reduction data as classification features of a cancer classifier of a preset type, conducting accurate classification of cancer patients and normal people through thecancer classifier. According to the method and the system for predicting cancer, the gene expression profile data of the hub gene obtained by weighted gene co-expression network analysis and the dimension reduction data processed by the variational auto-encoder are jointly used as the classification characteristics of the cancer classifier, so the accuracy of the cancer classifier is effectivelyimproved, and the purpose of efficiently predicting cancer is achieved.
Owner:UNIV OF SCI & TECH BEIJING
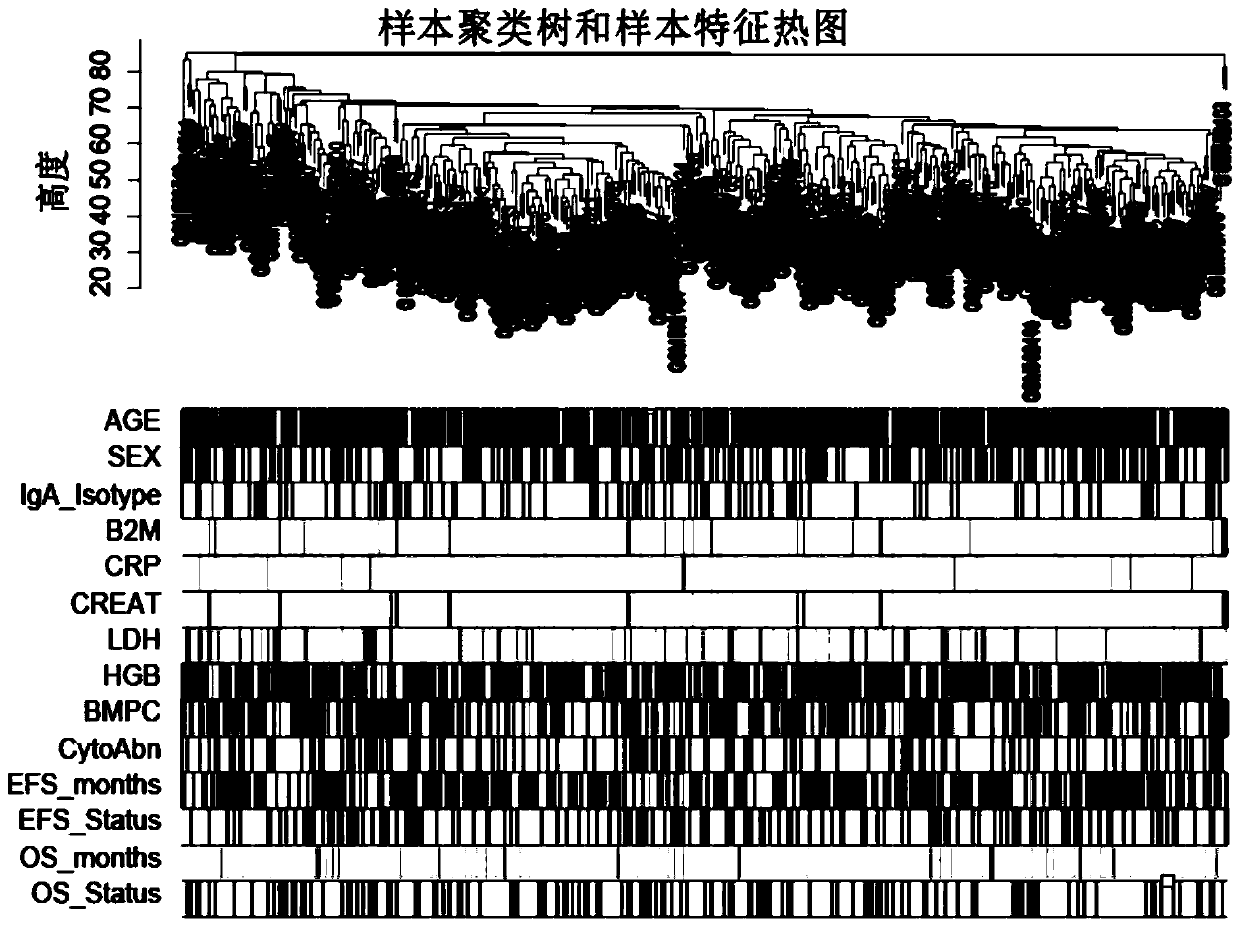
Novel comprehensive risk scoring method for multiple myeloma
ActiveCN110232974AImprove forecast accuracyMedical data miningHealth-index calculationPearson Correlation TestGeo database
The invention provides a comprehensive risk scoring method for multiple myeloma, comprising the following steps: S1, obtaining a gene expression profile GSE24080 of an MM patient from a GEO database,and pre-treating genes in the gene expression profile GSE24080 to obtain the first 25% of the 5413 genes with the largest variance of expression value; S2, subjecting the 5413 genes to WGCNA gene co-expression network analysis to identify co-expressed functional modules; S3, evaluating the correlation between the functional modules and clinical information through Pearson correlation test to determine the most significant modules; S4, performing univariate survival analysis on the genes in the most significant modules by using a Cox proportional hazard model, and screening out a ten-gene scoring model composed of the 10 best genes through LASSO regression; and S5, setting the rule that each factor scores 1 point when the ten-gene scoring model or serum [beta]2M or LDH is higher than a cut-off value and otherwise 0, and establishing a comprehensive risk scoring system.
Owner:THE FIRST AFFILIATED HOSPITAL OF FUJIAN MEDICAL UNIV
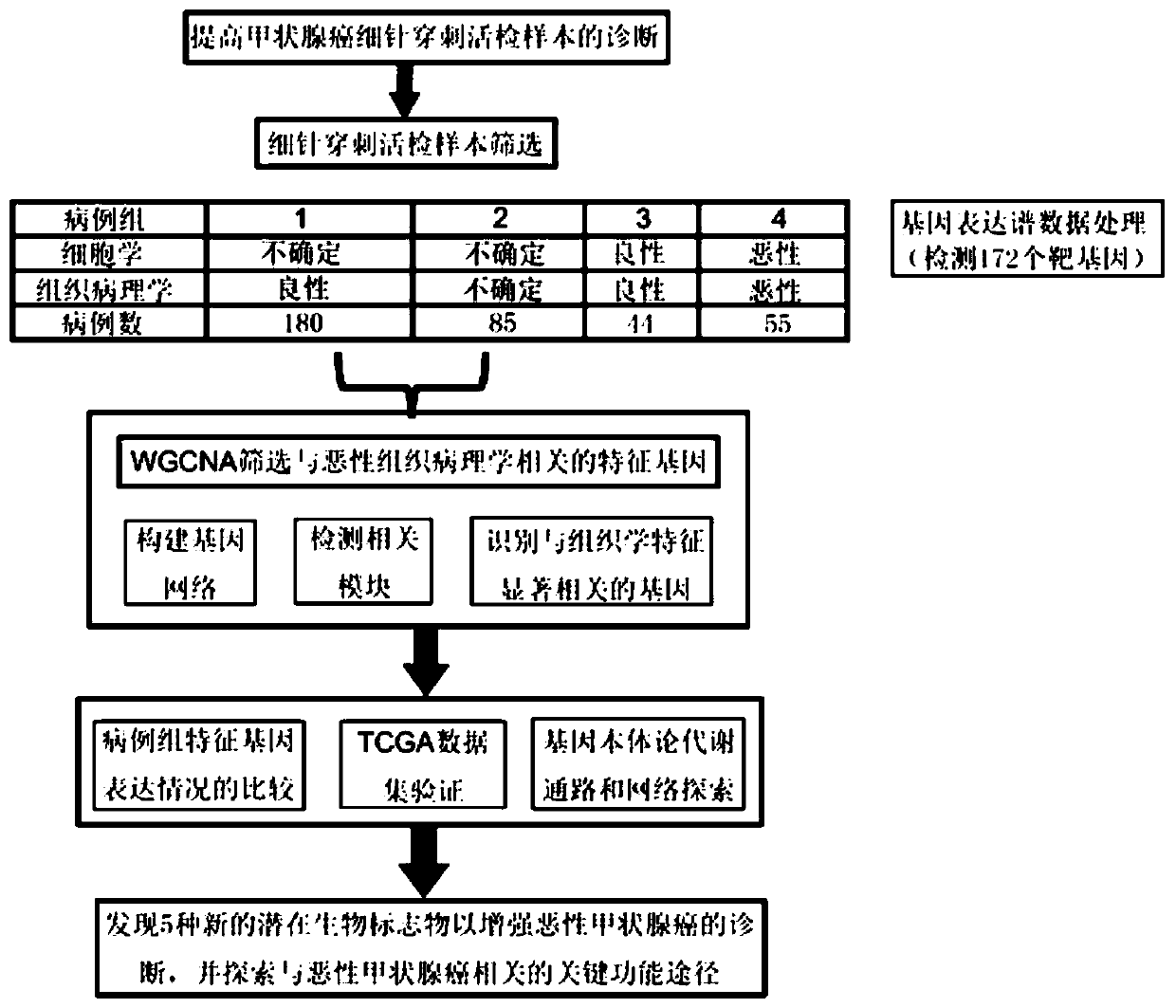
Method for screening potential biomarkers of gastric cancer based on weighted gene co-expression network analysis, and application thereof
ActiveCN109872776AShorten the timeImprove accuracyHealth-index calculationHybridisationIndividualized treatmentScreening method
The invention relates to the biomedicine field, and specifically relates to a method for screening potential biomarkers of gastric cancer based on weighted gene co-expression network analysis, and application thereof. The method for screening potential biomarkers of gastric cancer adopts weighted gene co-expression network analysis (WGCNA) and KEGG pathway and GO enrichment analysis, and other analysis methods. Weighted Gene Co-Expression Network Analysis (WGCNA) is an efficient and comprehensive method for high-dimensional data analysis, and the accuracy and validity in analyzing gene chip data of the WGCNA has been confirmed. The potential biomarker screened by the method of the invention is FERMT2. The method for screening potential biomarkers of gastric cancer based on weighted gene co-expression network analysis provides a new direction for the diagnosis, treatment and prognosis of gastric cancer, and promotes the development of individualized treatment.
Owner:辽宁省肿瘤医院
Construction method of prostatic cancer prognosis significant correlation ceRNA regulation and control network
ActiveCN112837744AEasy to digExcavate accuratelyEpidemiological alert systemsInstrumentsDiseasePharmaceutical drug
The invention discloses a construction method of a prostatic cancer prognosis significant correlation ceRNA regulation network, which comprises the following steps: integrating prostatic cancer gene expression data sets of TCGA and GTEx databases, respectively carrying out differential gene expression analysis and weighted gene co-expression network analysis, and screening key module genes of differential expression; then acquiring the key gene is obtained through WGCNA analysis and Cox proportional risk regression analysis; subjecting the obtained key gene, miRNA and lncRNA to interaction relationship to construct the ceRNA network, and performing evaluation according to the expression trend of the lncRNA, miRNA and hub gene in PRAD. According to the invention, the survival analysis is combined to obtain the ceRNA regulation network significantly related to PRAD prognosis, such that the early diagnosis of the disease and the precise treatment of the targeted drug design can be conveniently performed.
Owner:NANJING UNIV OF POSTS & TELECOMM
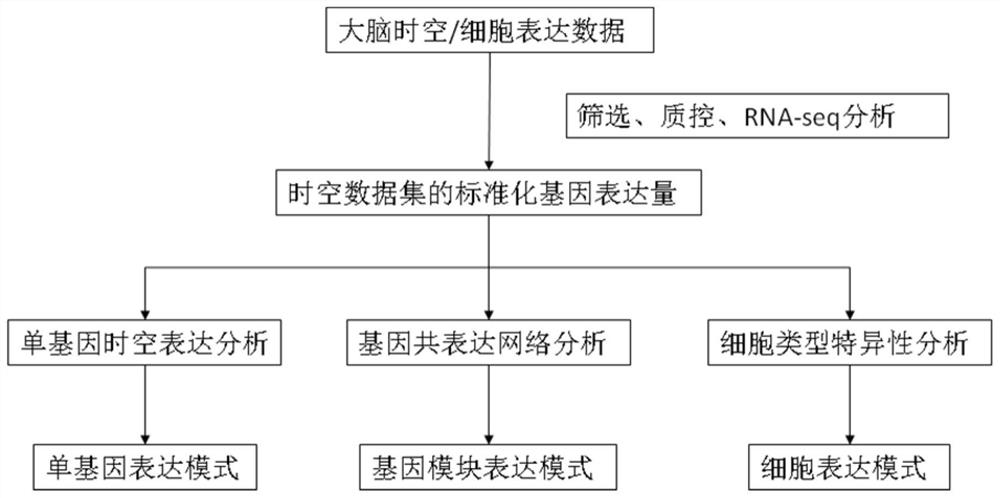
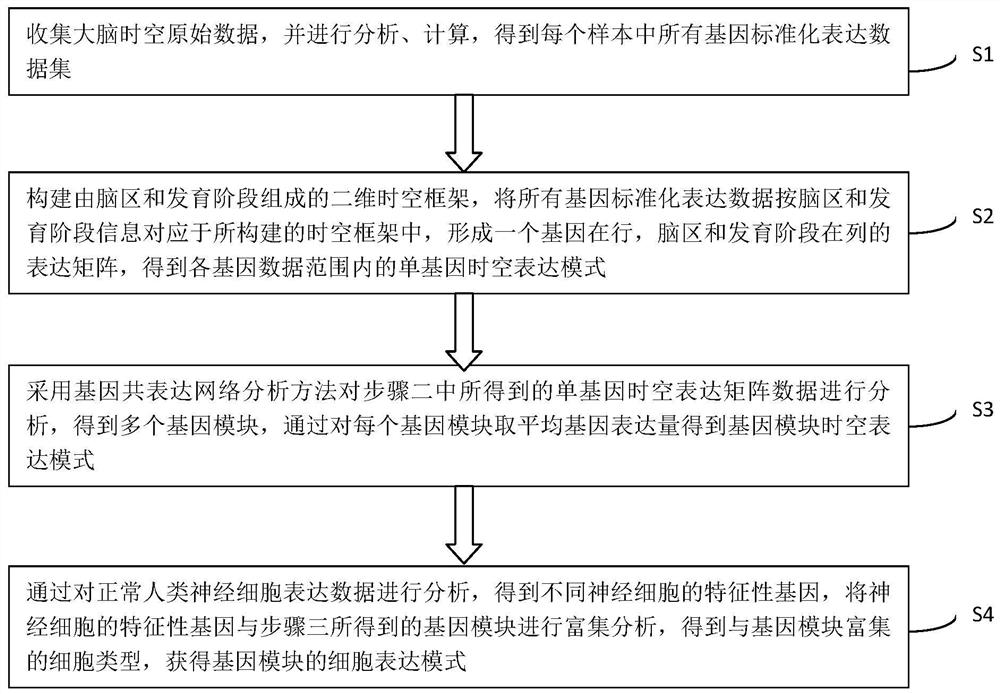
Method for establishing human brain gene expression space-time norm
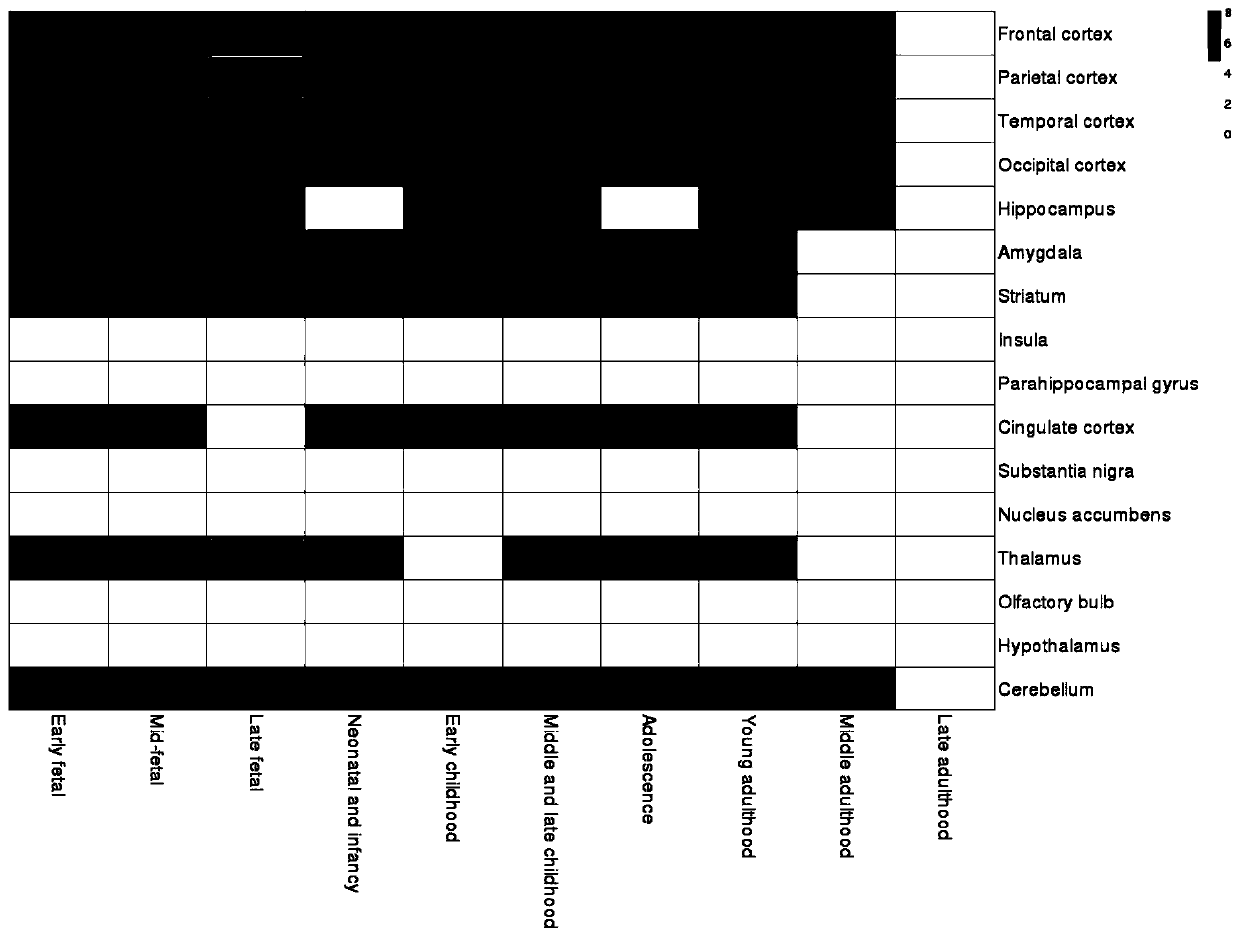
ActiveCN110349625AConvenient supplementEasy to compareBiostatisticsHybridisationData setOriginal data
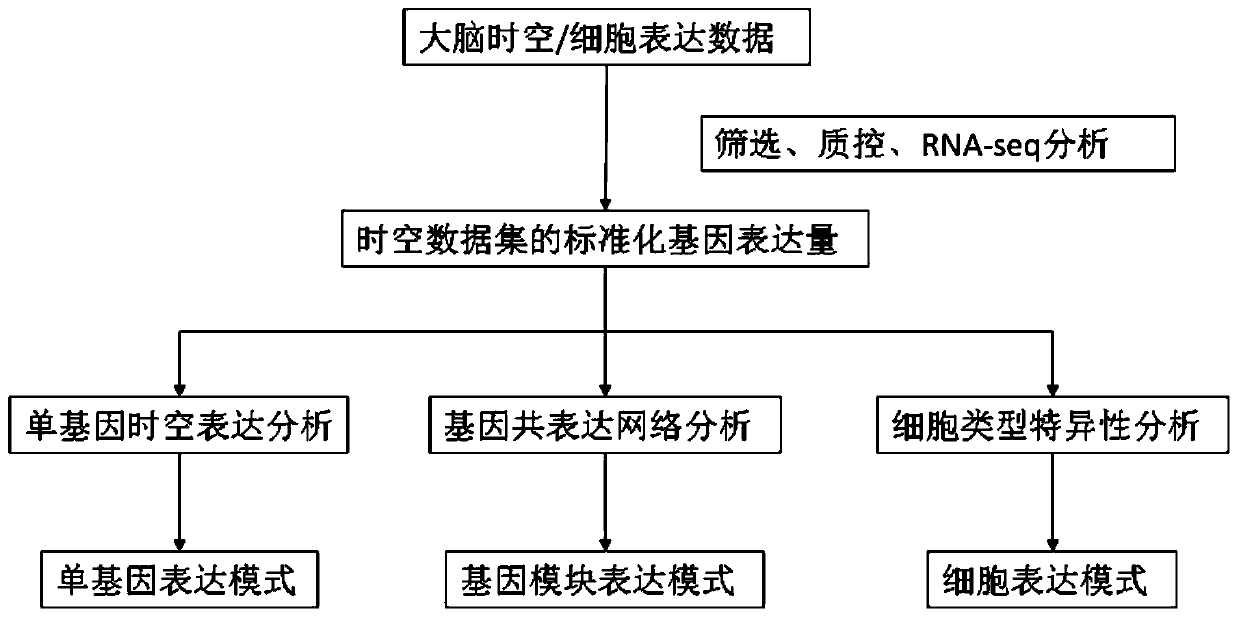
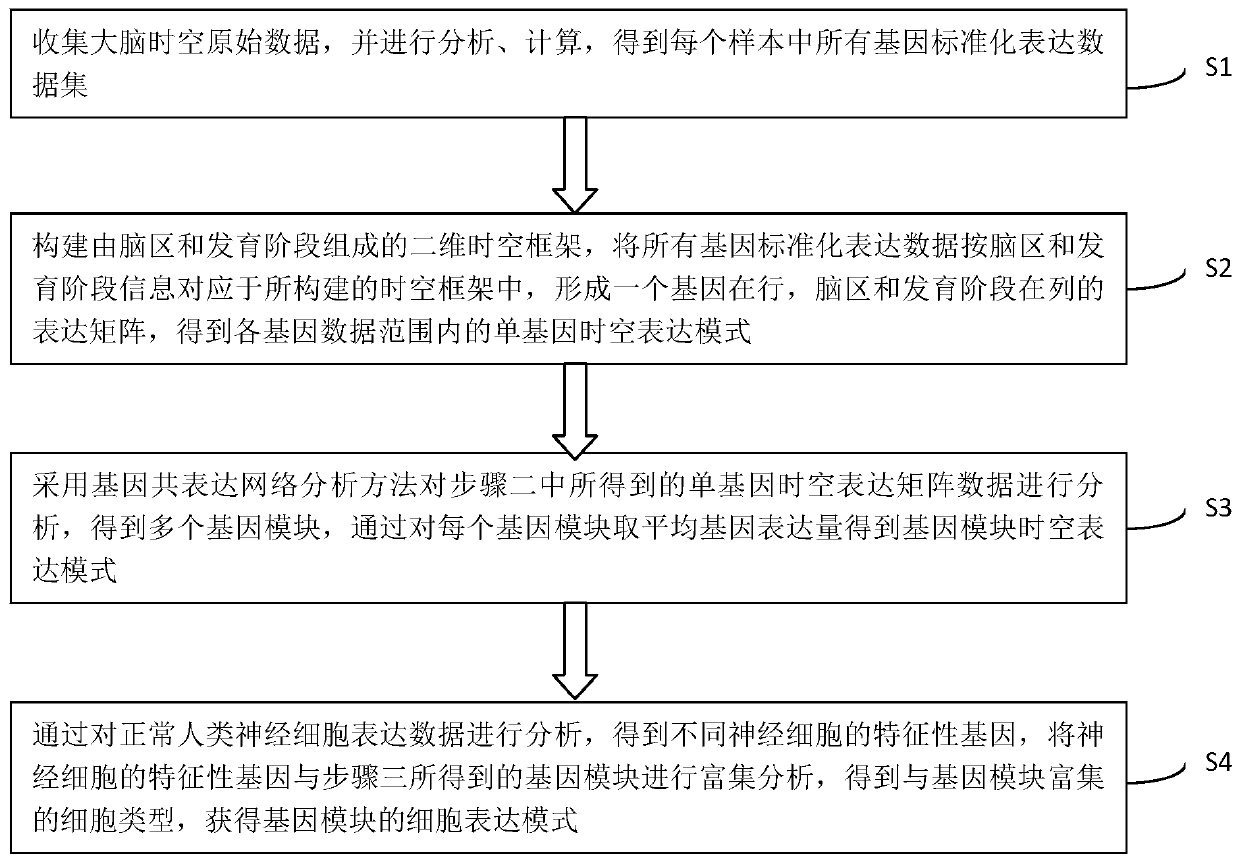
The invention discloses a method for establishing a human brain gene expression space-time norm. The method comprises the following steps: collecting brain space-time original data to obtain a gene standardized expression data set; constructing a two-dimensional space-time framework of a brain region and a development stage, and enabling all gene standardized expression data to correspond to the constructed space-time framework according to the brain region and development stage information to obtain a single-gene space-time expression mode; analyzing the obtained single-gene space-time expression matrix data by adopting a gene co-expression network analysis method to obtain a plurality of gene modules, and averaging the gene expression quantities of the gene modules to obtain a gene module space-time expression mode; analyzing the normal human nerve cell expression data, and performing the enrichment analysis of characteristic genes of nerve cells and the obtained gene modules to obtain a cell expression mode of the gene module. According to the method, all reference data sets are finally presented in the same two-dimensional space-time framework, so that the data sets from different sources are integrated, and researchers can intuitively supplement and compare the content of the space-time framework.
Owner:INST OF PSYCHOLOGY CHINESE ACADEMY OF SCI
Specific gene markers for TMT (thyroid malignant tumor) and application of specific gene markers
PendingCN110283907AAvoid selectivityAvoid lossMicrobiological testing/measurementDNA/RNA fragmentationClinico pathologicalMolecular diagnostics
The invention relates to the field of medical detection, in particular to specific gene markers for TMT (thyroid malignant tumor) and an application of the specific gene markers. On the basis of a weighted gene coexpression network analysis method and by combination with clinical cases, the TMT is specifically correlated with abnormal expression of 17 gene markers and can be taken as a specific gene detection marker set. By means of verification of a large number of independent clinicopathological samples, specific mutation sites of 5 specific genes are further determined as molecular diagnosis markers. The related gene diagnosis marker set has important clinical application value for early molecular accurate diagnosis of the TMT, can be used for preparing a molecular diagnosis kit for detecting malignant thyroid nodules which are indeterminate according to clinical aspiration biopsy cytology, and particularly has important clinical application value for early molecular accurate diagnosis of patients with an uncertain FNAB (fine needle aspiration biopsy) cytology result of malignant thyroid nodules.
Owner:JIANGSU UNIV
Method for screening transcription factors related to postharvest softening of grape fruits
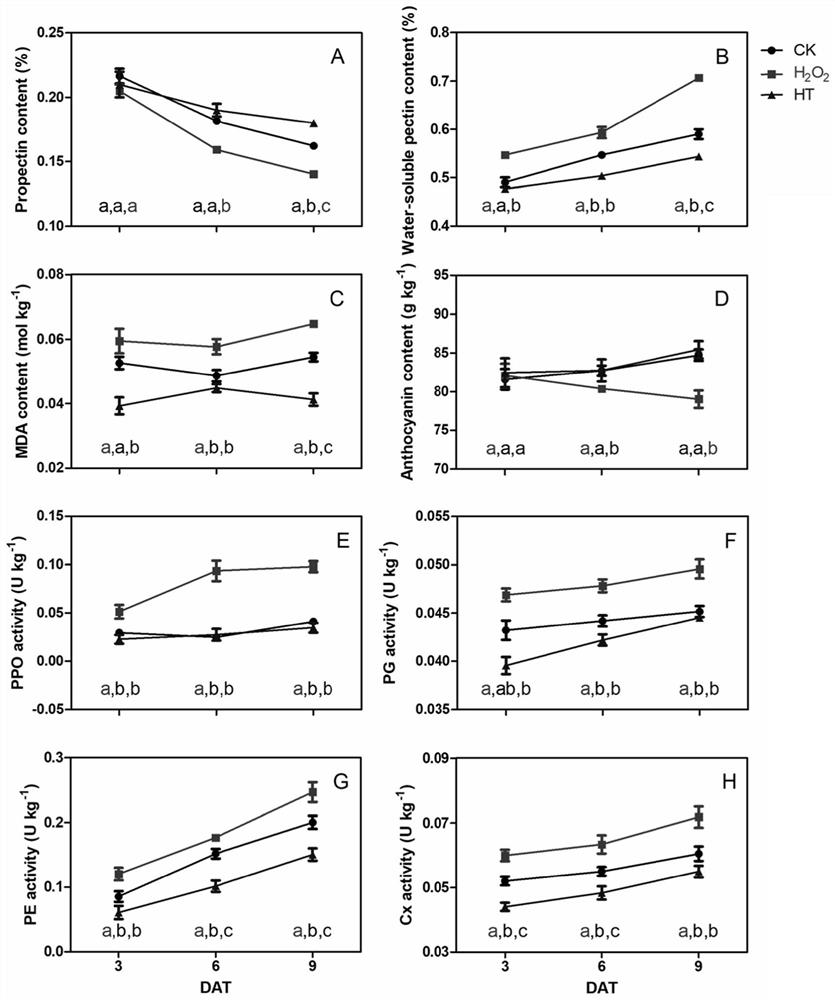
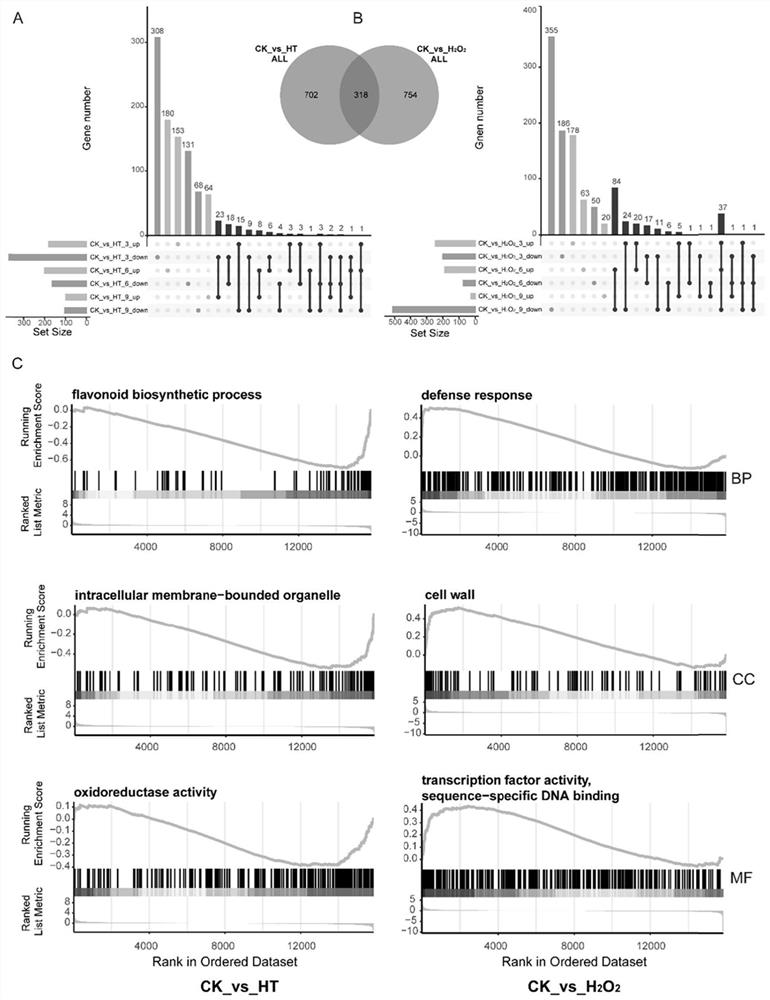
InactiveCN114107456AMicrobiological testing/measurementProteomicsBiotechnologyH2O2 - Hydrogen peroxide
The invention belongs to the technical field of high-throughput sequencing data analysis, and particularly relates to a method for screening transcription factors related to postharvest softening of grape fruits. According to the invention, hypotaurine (HT) and hydrogen peroxide (H2O2) treatment is carried out on harvested 'Kyoho' grapes, through transcriptome sequencing and weighted gene co-expression network analysis (WGCNA), correlation analysis is carried out on differential expression genes obtained through transcriptome analysis and pectin content change, a differential expression gene module closely related to the pectin content change is screened out, and the specific value of the differential expression genes and the specific value of the pectin content change are obtained. And excavating differential expression transcription factors in the softening process of the picked grape fruits. According to the method, the softening character related transcription factors can be effectively screened, and a theoretical reference is provided for researching the softening occurrence mechanism of the picked grape fruits.
Owner:HENAN UNIV OF SCI & TECH
Medicine for improving activity of midbrain substantia nigra dopamine neurons to preventing Parkinson disease
ActiveCN110570903AOvercome limitationsShorten the timeOrganic active ingredientsNervous disorderOrganismNetwork module
The invention provides a medicine for improving the activity of midbrain substantia nigra dopamine neurons to preventing a Parkinson disease. The medicine is fulvestrant. The invention also provides amethod for screening the medicine for improving the activity of midbrain substantia nigra dopamine neurons to preventing the Parkinson disease. A bioinformatics method using a weighted gene co-expression network to analyze and identify a biologically-related differential co-expression module, matches genes closely related to the Parkinson disease to a gene network module so as to obtain a gene co-expression module related to the Parkinson disease, and by a correlation map database and different gene expression profile data between midbrain substantia nigra encephalic region samples in a normal control group from a Parkinson patient, screens out a medicine that can significantly reverse the abnormally expressed genes in the module. The method overcomes the limitation of a single pathway ortarget, and also uses the CMAP database to screen approved clinical medicines, saves time, money, and labor costs, and is more conducive to the development of safe and effective medicines.
Owner:SUN YAT SEN MEMORIAL HOSPITAL SUN YAT SEN UNIV
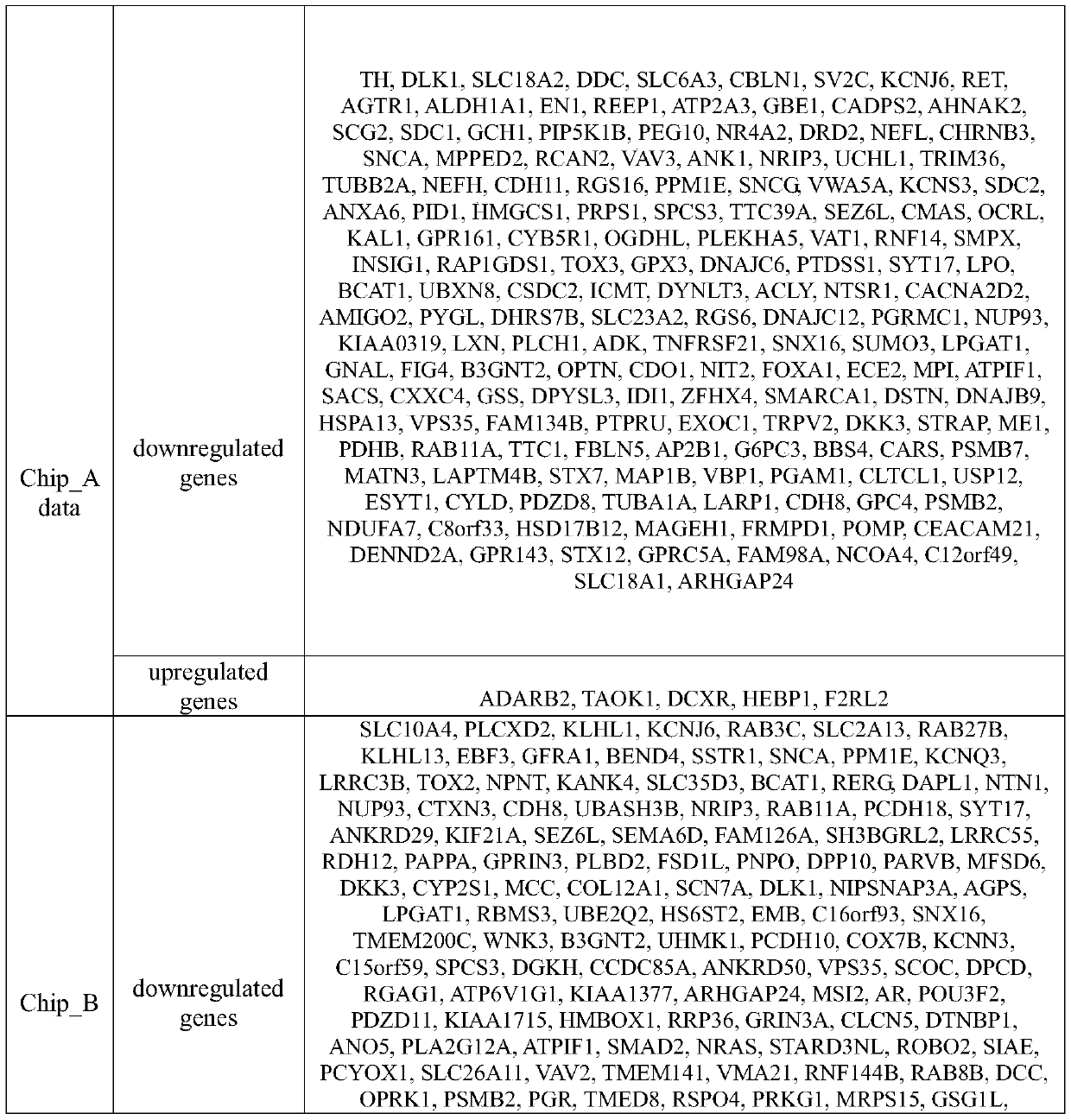
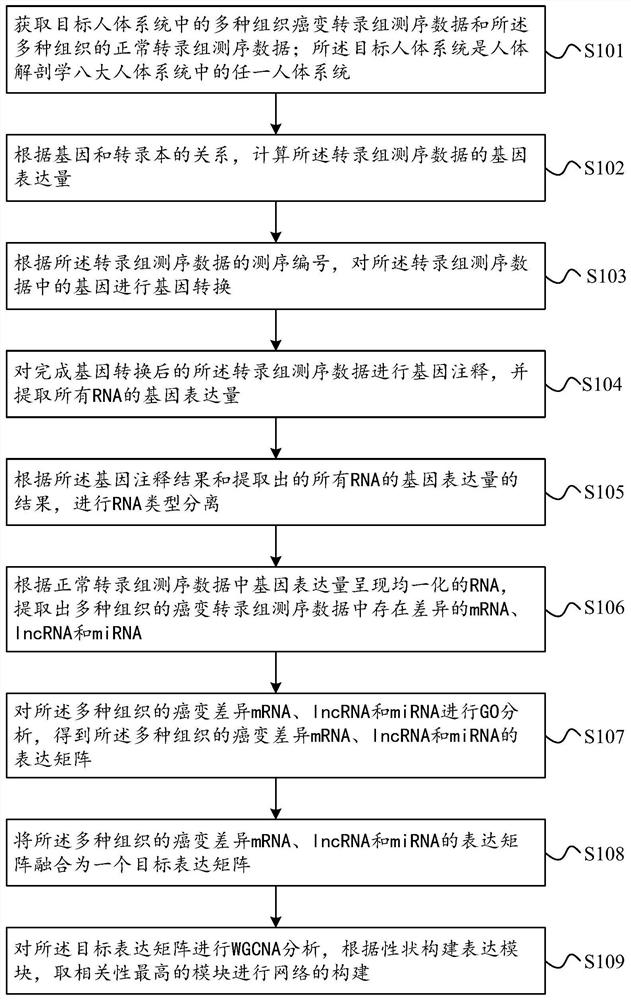
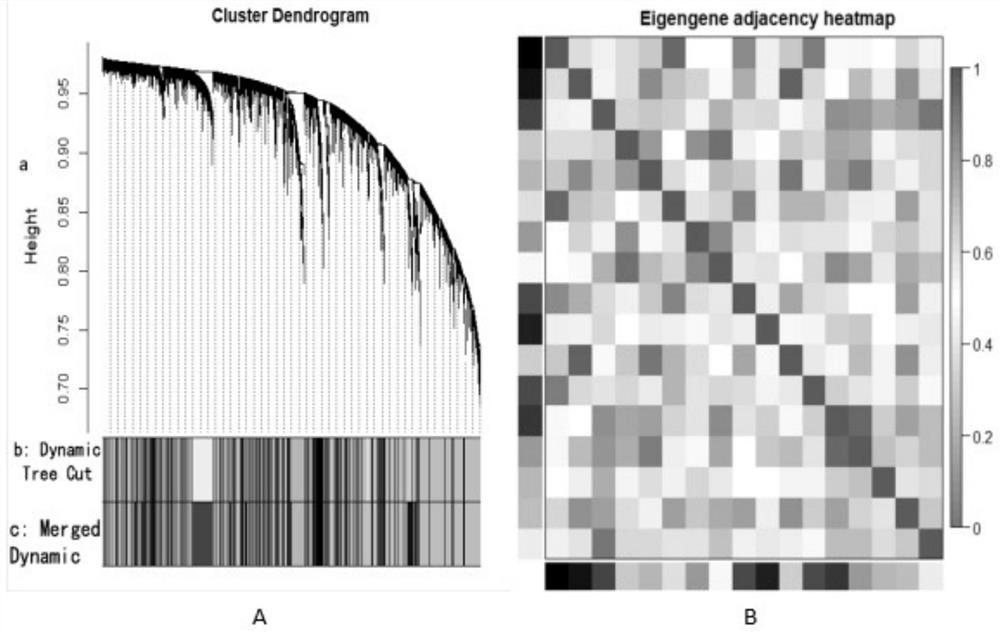
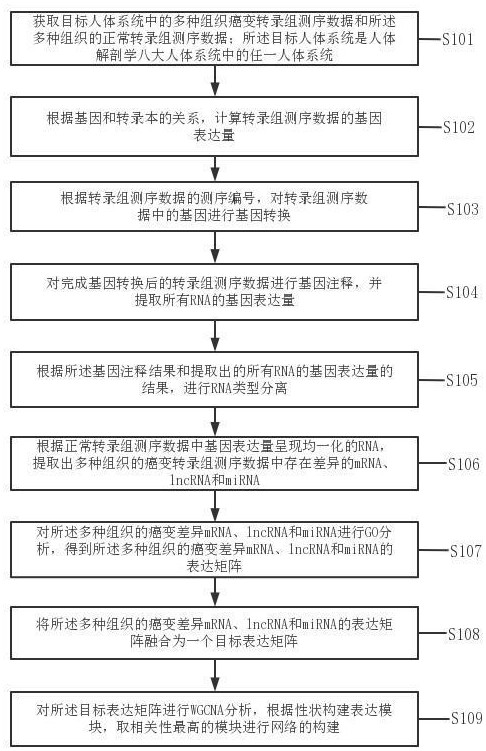
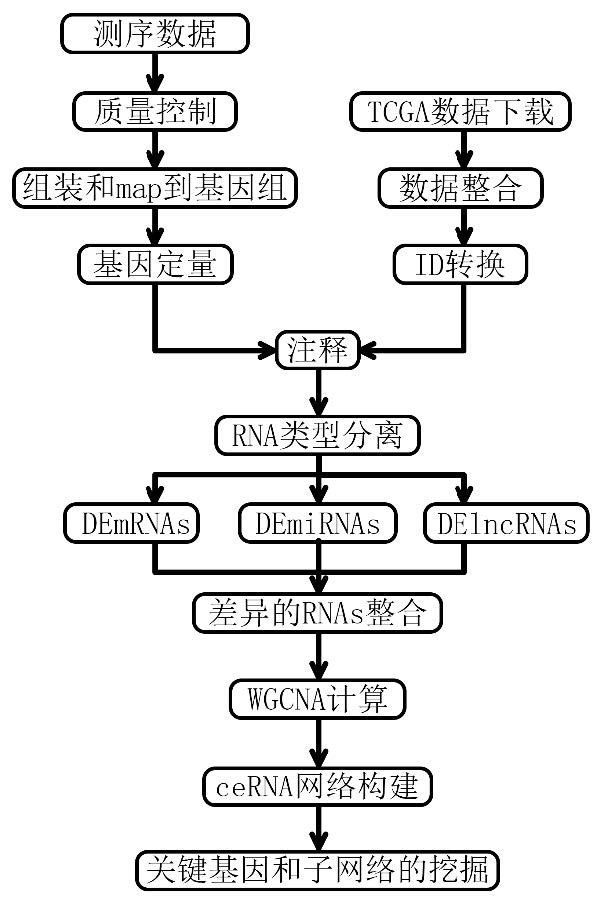
Tumor gene co-expression network construction method and device, equipment and storage medium
The invention provides a tumor gene co-expression network construction method and device, equipment and a storage medium. The method comprises the following steps: acquiring multiple tissue canceration transcriptome sequencing data and normal transcriptome sequencing data of multiple tissues in a target human body system, wherein the target human body system is any one human body system in eight human body systems of human anatomy; extracting mRNA, lncRNA and miRNA with difference in canceration transcriptome sequencing data of various tissues according to a relationship between genes and transcripts and RNA with uniform gene expression quantity in normal transcriptome sequencing data; carrying out GO analysis on canceration difference mRNA, lncRNA and miRNA of the plurality of tissues to obtain a target expression matrix fused with the mRNA, lncRNA and miRNA; and performing WGCNA analysis on the target expression matrix, constructing expression modules according to characters, and performing network construction on the module with the highest correlation. Network construction of various cancers can be realized.
Owner:SHENZHEN POLYTECHNIC +1
Novel screening method for multiple myeloma prognosis gene
ActiveCN110223733AStrong correlationData visualisationHybridisationClinical informationScreening method
The invention provides a novel screening method for a multiple myeloma prognosis gene based on a gene coexpression network. The method comprises the following steps of S1, acquiring a gene expressionspectrum GSE24080 of an MM patient from a GEO database, preprocessing the gene in the gene expression spectrum GSE24080, and obtaining 5413 genes which have the front 25% highest expression value variances; S2, performing WGCNA gene coexpression network analysis on the 5413 genes for identifying a coexpression functional module; and S3, evaluating correlation between the functional module and clinical information through Pearson related examination, thereby determining a most substantial module.
Owner:THE FIRST AFFILIATED HOSPITAL OF FUJIAN MEDICAL UNIV
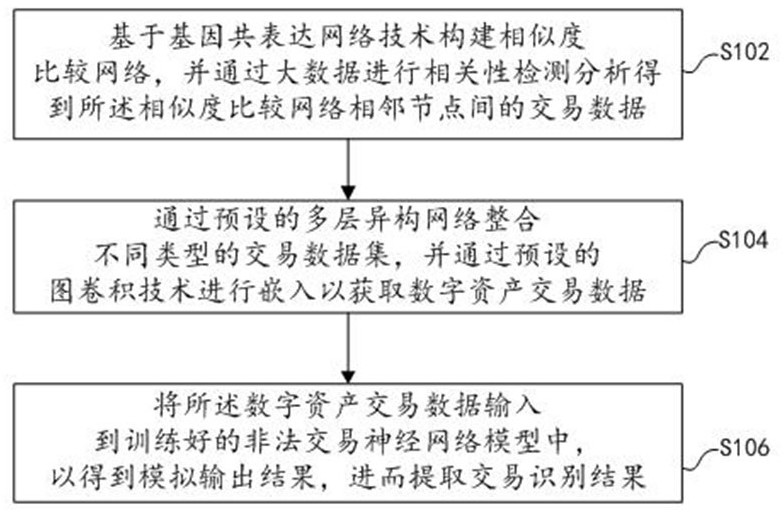
Digital currency identification method and system based on multi-omics technology, and storage medium
InactiveCN113469816AImprove accuracyAccurate trackingFinanceNeural learning methodsDigital currencyNerve network
The invention discloses a digital currency identification method and system based on a multi-omics technology and a storage medium, and the method comprises the steps: constructing a similarity comparison network based on a gene co-expression network technology, and carrying out the correlation detection and analysis through big data, and obtaining the transaction data between adjacent nodes of the similarity comparison network; integrating different types of transaction data sets through a preset multi-layer heterogeneous network, and performing embedding through a preset graph convolution technology to obtain digital asset transaction data; and inputting the digital asset transaction data into a trained illegal transaction neural network model to obtain an analog output result, and extracting a transaction identification result. According to the method, the type of the transaction account and the specific transaction process are determined through multi-omics research, the problem account can be accurately tracked through integration analysis of multi-level and high-throughput omics data, and the method is rapid, efficient and high in accuracy; and meanwhile, the calculation efficiency and the use ductility can be greatly improved by utilizing distributed machine learning.
Owner:浙江中科华知科技股份有限公司
Multiple Myeloma Comprehensive Risk Score
ActiveCN110232974BImprove forecast accuracyMedical data miningHealth-index calculationNetwork analyticsSynexpression
The present invention provides a multiple myeloma comprehensive risk scoring method, comprising the following steps: S1, obtaining the gene expression profile GSE24080 of multiple myeloma patients from the GEO database, and predicting the genes in the gene expression profile GSE24080 processing, to obtain the top 25% of the 5413 genes with the largest variance in expression values; S2, to perform WGCNA gene co-expression network analysis on the 5413 genes to identify co-expressed functional modules; The correlation between the modules and clinical information was evaluated to determine the most significant modules; S4, using the Cox proportional hazards model to perform univariate survival analysis on the genes in the most significant modules, and screened out the 10 most significant modules by LASSO regression The ten-gene scoring model with the best gene composition; S5, when the ten-gene scoring model or serum β2M or LDH is higher than the cut-off value, the score of each factor is 1, otherwise it is 0, and a comprehensive risk scoring system is established.
Owner:THE FIRST AFFILIATED HOSPITAL OF FUJIAN MEDICAL UNIV
System for detecting hypertension-related genes and analyzing gene functions
PendingCN114566222AReduce research costsEasy to implementData visualisationBiostatisticsSynexpressionDifferentially expressed genes
The invention discloses a system for detecting hypertension related genes and analyzing gene functions. Comprising a gene preprocessing unit used for preprocessing hypertension gene expression data to obtain a gene expression matrix; the gene sample analysis unit is used for analyzing a sample distribution condition based on the gene expression matrix; the gene difference analysis unit is used for screening differential genes with significant changes on the basis of the gene expression matrix to obtain a gene difference matrix; the gene module analysis unit is used for determining gene modules and screening key modules through a weighted gene co-expression network method based on the gene expression matrix; the Hub gene analysis unit is used for screening Hub genes in the key genes; and the gene function analysis unit is used for carrying out function enrichment analysis and network analysis of a pathway and gene relationship on the differentially expressed gene or the gene module. The method can assist scientific researchers who are not familiar with hypertension gene related databases and analysis processes in accurately and effectively finding genes and pathways related to complex diseases such as hypertension.
Owner:QINGHAI NORMAL UNIV
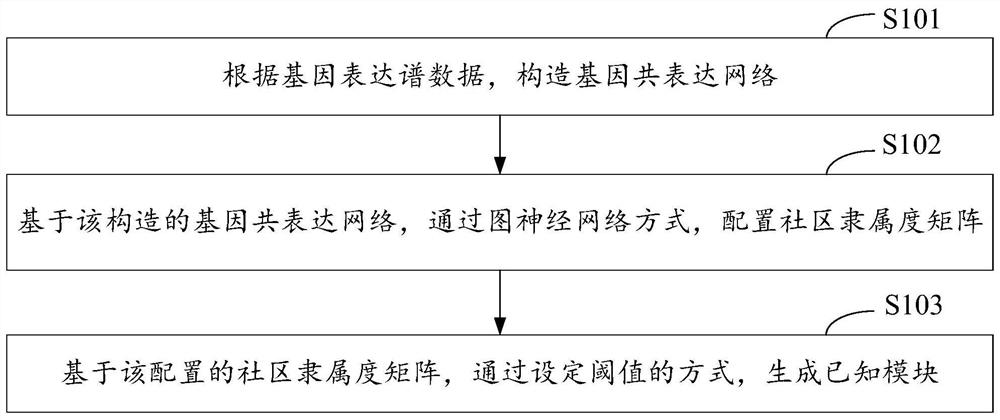
Gene module mining method, device, and computer equipment based on graph neural network
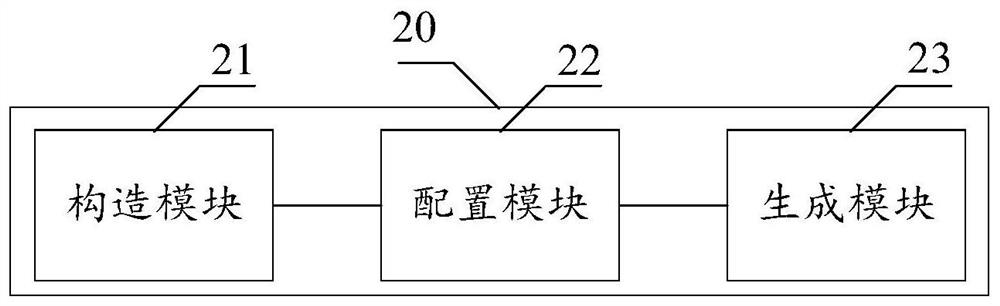
ActiveCN113611366BScale upImprove timing performanceBiostatisticsCharacter and pattern recognitionEngineeringGraph neural networks
The invention discloses a gene module mining method, device and computer equipment based on a graph neural network. Wherein, the method includes: constructing a gene co-expression network according to the gene expression profile data, and based on the constructed gene co-expression network, configuring a community membership matrix through a graph neural network, and a community membership matrix based on the configuration , generate known modules by setting thresholds. Through the above method, it is possible to configure the community affiliation matrix through graph neural network representation learning, and then generate known modules by setting thresholds, so that multiple genes with dense connections can be assigned to the gene module mining results. different modules.
Owner:HARBIN INST OF TECH SHENZHEN GRADUATE SCHOOL
A screening method for multiple myeloma prognostic genes
The present invention provides a method for screening multiple myeloma prognosis genes based on a gene co-expression network, comprising the following steps: S1, obtaining the gene expression profile GSE24080 of MM patients from the GEO database, and analyzing the gene expression profile GSE24080 The genes in are preprocessed to obtain the top 25% of the 5413 genes with the largest expression value variance; S2, WGCNA gene co-expression network analysis is performed on the 5413 genes to identify co-expressed functional modules; S3, through Pearson correlation The test evaluates the correlation between the functional modules and clinical information to identify the most significant modules.
Owner:THE FIRST AFFILIATED HOSPITAL OF FUJIAN MEDICAL UNIV
A drug for improving the activity of dopamine neurons in the substantia nigra of the midbrain and preventing and treating Parkinson's disease
ActiveCN110570903BIncrease the number ofRejuvenate neuronsOrganic active ingredientsNervous disorderFulvestrantBio informatics
Owner:SUN YAT SEN MEMORIAL HOSPITAL SUN YAT SEN UNIV
Application of sodium phenylbutyrate in preparation of medicine for preventing or treating Parkinson's disease
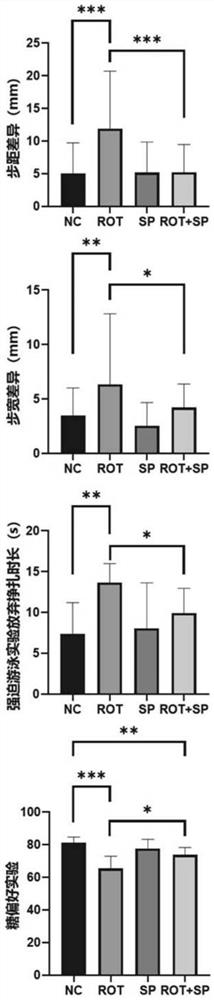
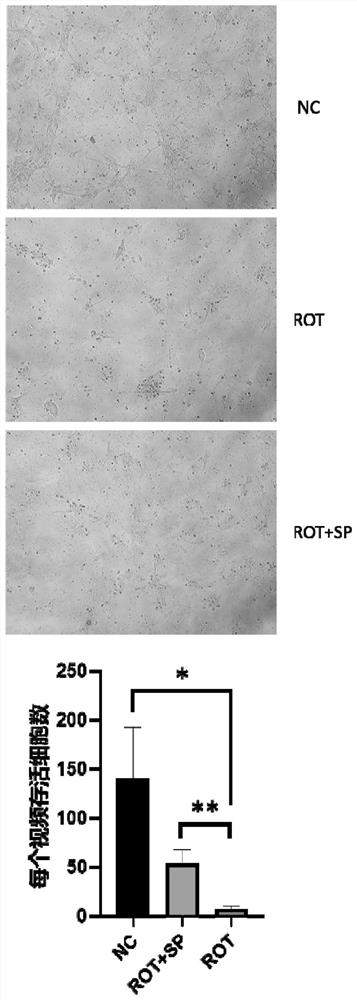
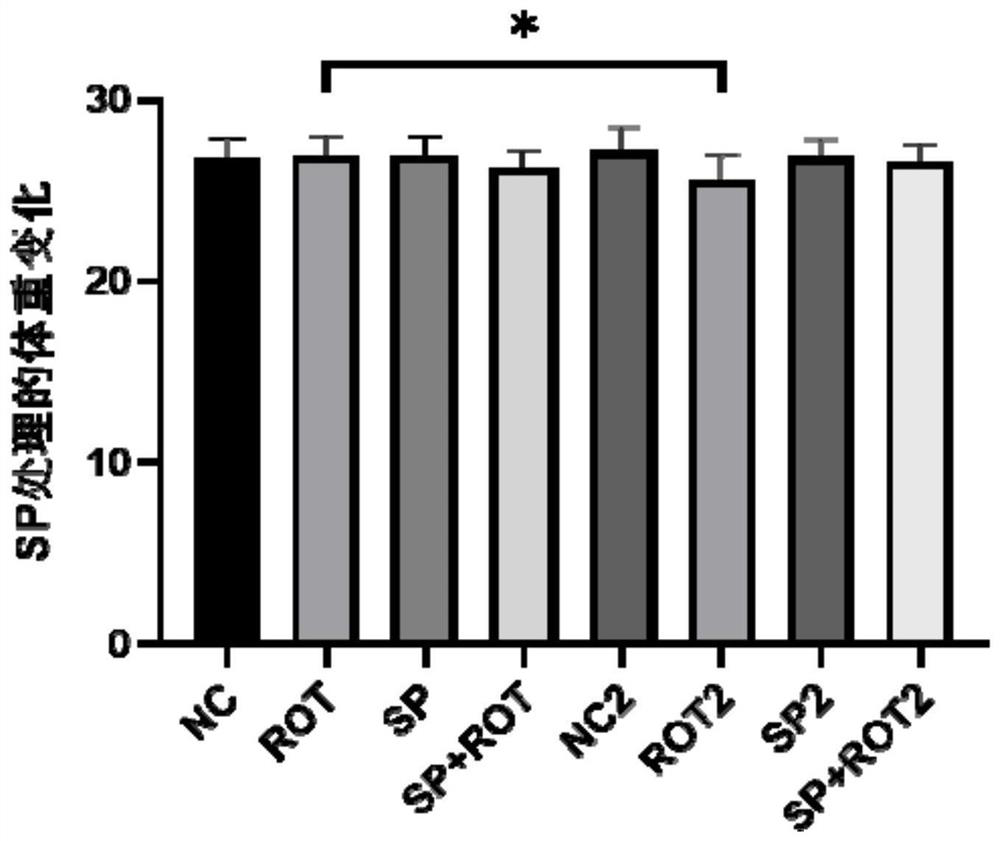
PendingCN113332267APrevent synaptic damagePrevent diseaseOrganic active ingredientsNervous disorderAbnormal expressionAnimal behavior
The invention relates to the field of biological pharmacy, in particular to application of sodium phenylbutyrate (SP) in preparation of a medicine for preventing or treating Parkinson's disease. The gene co-expression network is explored by integrating multiple omics data, and the medicine SP capable of obviously reversing abnormally expressed genes in the module is obtained. Animal experiments show that the medicine can improve animal behaviors and prevent the development of Parkinson's disease. Therefore, the SP has the effect of preventing the development of the Parkinson's disease and the effect of improving the treatment of the Parkinson's disease.
Owner:SUN YAT SEN MEMORIAL HOSPITAL SUN YAT SEN UNIV
A method and system for predicting cancer
ActiveCN111899882BEfficient forecastingImprove accuracyMedical data miningHealth-index calculationSynexpressionAutoencoder
The invention discloses a method and system for predicting cancer. The method includes: performing differential analysis on the gene expression spectrum data of cancer patients and normal people to obtain differential genes; The human gene expression profile data is analyzed to obtain the hub gene; and the gene expression profile data of the differential gene is processed by the variational autoencoder algorithm to obtain the dimensionality reduction data; the gene expression profile data of the hub gene and the dimensionality reduction data Together as the classification features of the preset type of cancer classifier, the precise classification of cancer patients and normal people can be realized through the cancer classifier. The method and system for predicting cancer of the present invention use the gene expression profile data of pivot genes obtained by weighted gene co-expression network analysis and the dimensionality reduction data processed by variational autoencoders as the classification features of the cancer classifier, thereby effectively improving the The accuracy of the cancer classifier achieves the goal of efficiently predicting cancer.
Owner:UNIV OF SCI & TECH BEIJING
Tumor gene co-expression network construction method, device, equipment and storage medium
The present application provides a method, device, equipment and storage medium for constructing a tumor gene co-expression network. The method includes: acquiring the cancerous transcriptome sequencing data of various tissues in the target human body system and the normal transcriptome sequencing data of the various tissues; the target human body system is any one of the eight major human body systems of human anatomy ; According to the relationship between genes and transcripts, as well as RNAs with uniform gene expression in normal transcriptome sequencing data, differential mRNAs, lncRNAs and miRNAs are extracted from cancer transcriptome sequencing data of various tissues; Carcinogenic differential mRNA, lncRNA, and miRNA of various tissues were analyzed by GO, and a target expression matrix was obtained; the target expression matrix was analyzed by WGCNA, expression modules were constructed according to traits, and the most relevant modules were selected for network construction. It can realize network construction for various cancers.
Owner:SHENZHEN POLYTECHNIC +1
Early pathogenic factor detection method based on gene co-expression network propagation analysis
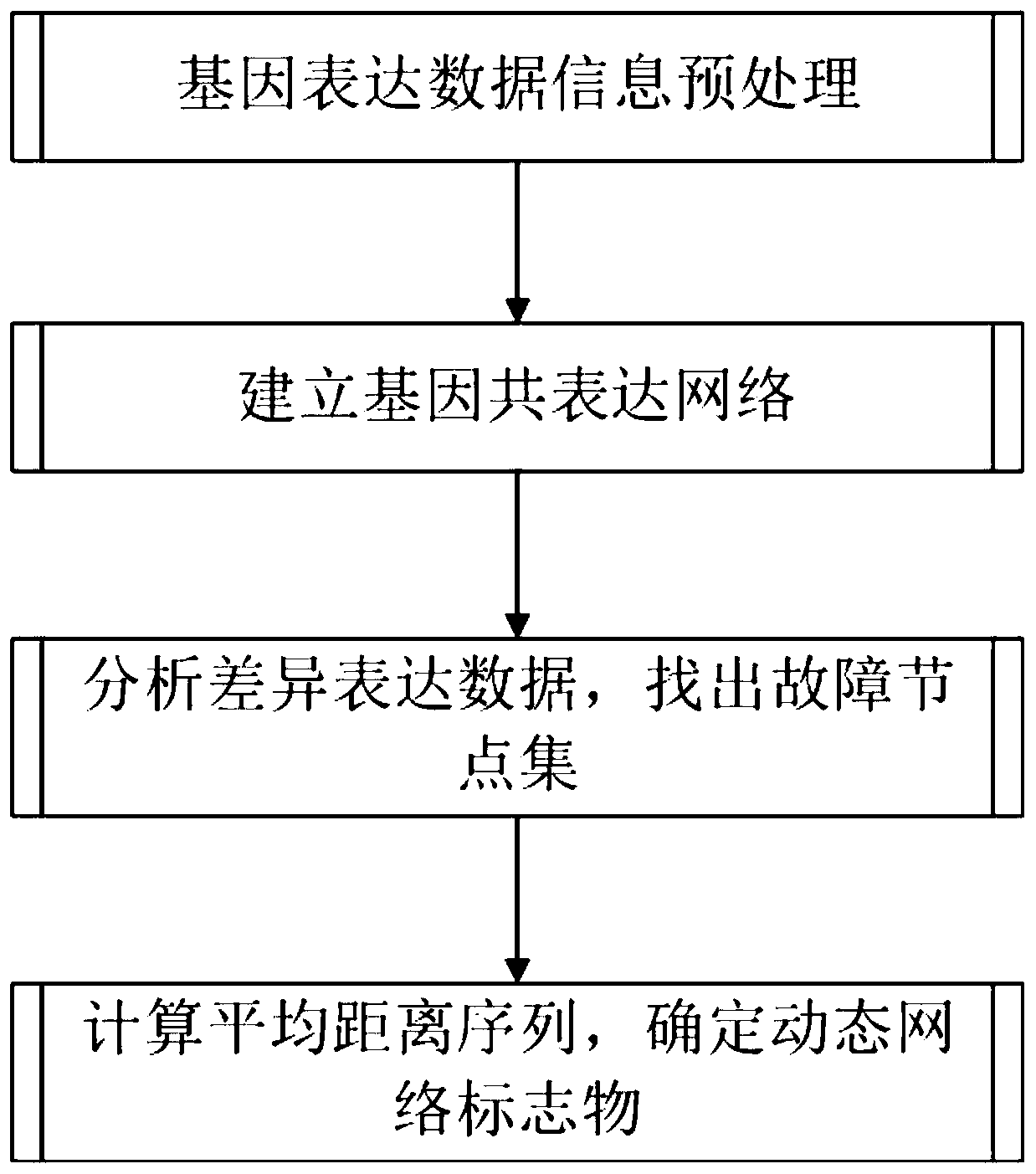
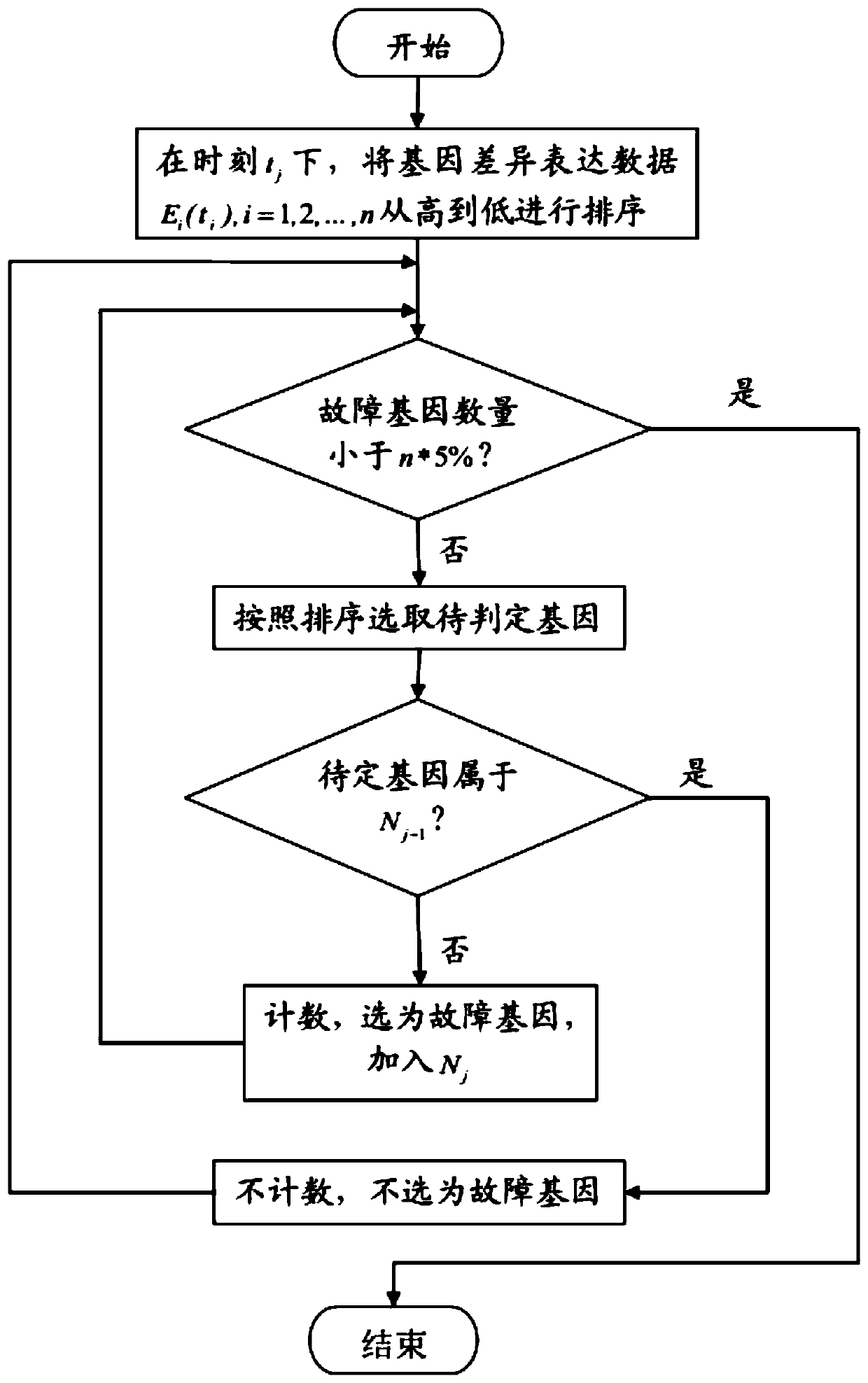
The invention provides an early-stage pathogenic factor detection method based on gene co-expression network spreading analysis. The method comprises the following steps that: 1) obtaining data from the true gene expression of the case group and the control group of a certain disease, and preprocessing the gene expression data of a certain disease; 2) through correlation analysis, determining whether two genes have a coexpression relationship or not so as to establish a gene coexpression network; 3) finding a fault node set from the differential expression soring of the gene; and 4) calculating an average distance of the nodes of a fault node subset and a prepared fault spreading center, finding an incremental average distance sequence, and determining a dynamic network biomarker. The method aims at the detection problem of the early-stage pathogenic factor of a complex disease to put forward the early-stage pathogenic factor detection method based on gene co-expression network spreading analysis from the perspective of a system in order to make up the deficiencies of a traditional molecular biomarker and a statistic network biomarker, the early-stage pathogenic factor of a disease can be effectively found, and a contribution is made for accurate medical treatment.
Owner:BEIHANG UNIV
Molecular classification and application of glioma based on cdc20 gene co-expression network
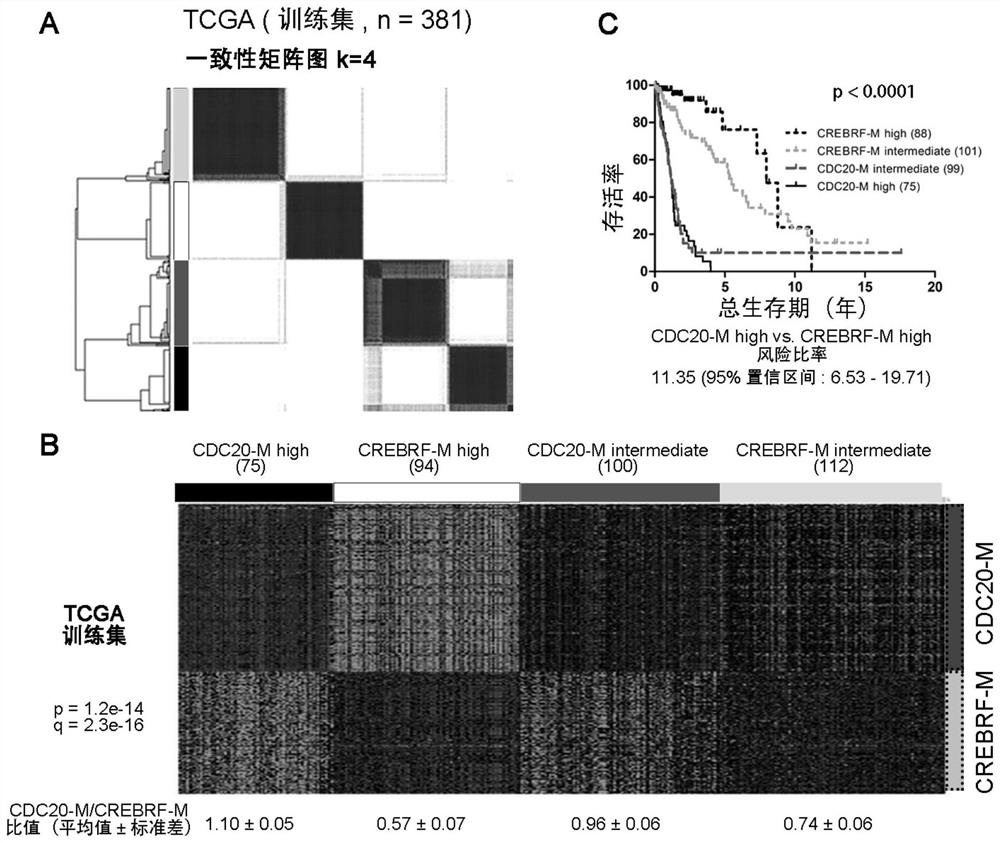
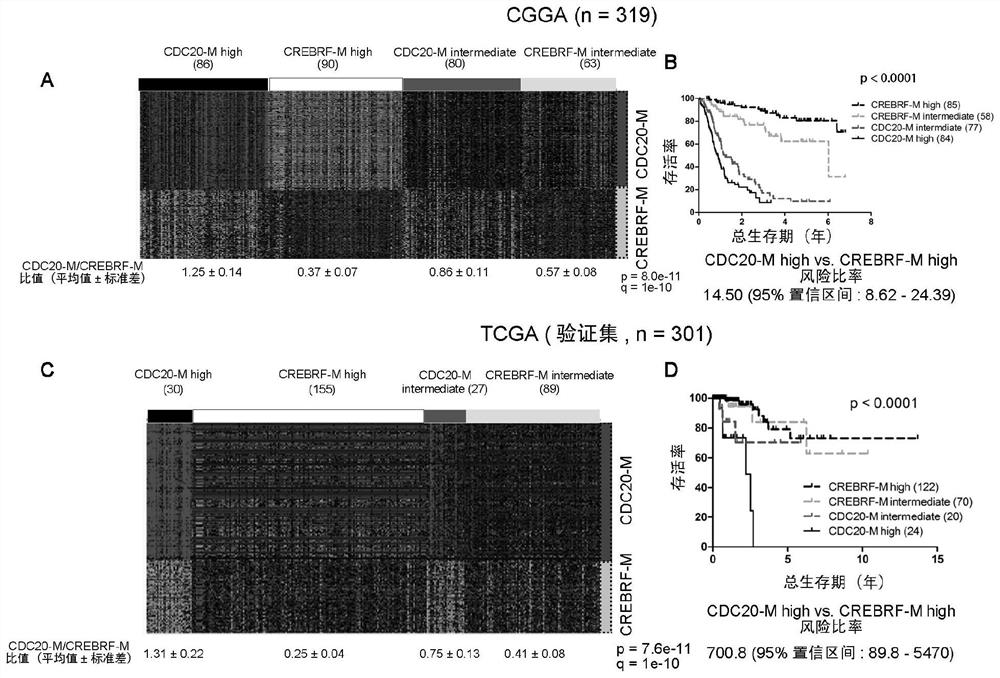
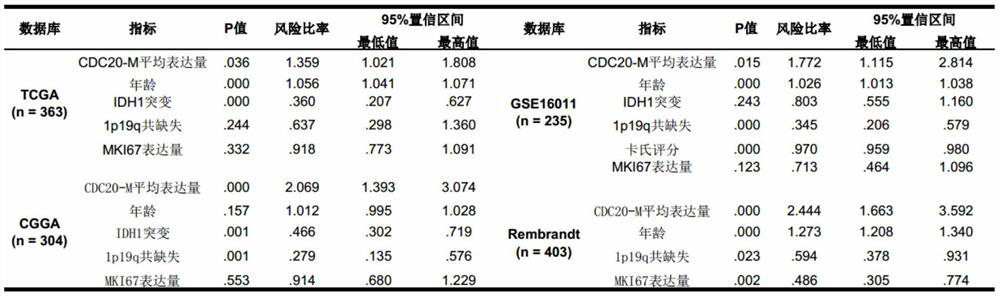
The invention discloses glioma molecular typing based on CDC20 gene co-expression network and application and provides application of various gene expression matters in CDC20-M gene group and CREBRF-Mgene group for detecting glioma patients in the preparation of products for estimating prognosis risk of the glioma patients; the CDC20-M gene group is composed of 139 genes; the CREBRF-M gene groupis composed of 120 genes. It is verified herein that CDC20-M is a sensitive marker for poor prognosis for gliomas.
Owner:BEIJING NORMAL UNIVERSITY
Novel method for constructing nomogram of multiple myeloma
ActiveCN110197701APredicted survivalHelps quantitatively predict survivalHealth-index calculationData visualisationNewly diagnosedNomogram
The invention provides a method for constructing a nomogram of multiple myeloma based on ten gene characteristics, serum [beta]2 microglobulin and LDH. The method comprises the following steps: S1, obtaining a gene expression profile GSE24080 of an MM patient from a GEO database, and pre-treating genes in the gene expression profile GSE24080 to obtain the first 25% of the 5413 genes with the largest variance in expression values; S2, subjecting the 5413 genes to WGCNA gene co-expression network analysis to identify function modules of co-expression; S3, evaluating the correlation between the functional modules and clinical information by Pearson correlation test to determine the most significant module; S4, performing univariate survival analysis on the genes in the most significant modules using a Cox proportional hazard model, and screening a score model consisting of 10 best genes by LASSO regression; and S5, establishing a novel nomogram based on the scoring model, serum [beta]2M and high LDH to predict 3-year OS and 5-year OS of newly diagnosed MM patients.
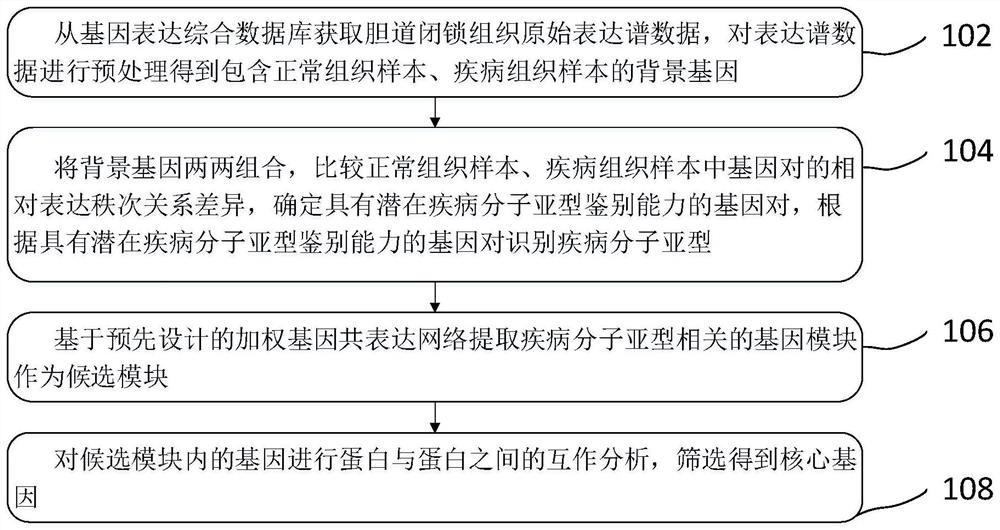
Owner:SHANGHAI TISSUEBANK MEDICAL LAB CO LTD
Application of meclofenoxate hydrochloride in preparation of medicine for preventing or treating Parkinson's disease
PendingCN113332271APrevent synaptic damagePrevent diseaseOrganic active ingredientsNervous disorderPharmaceutical SubstancesSynexpression
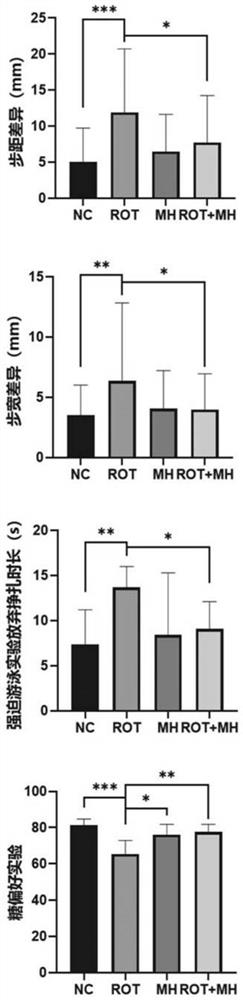
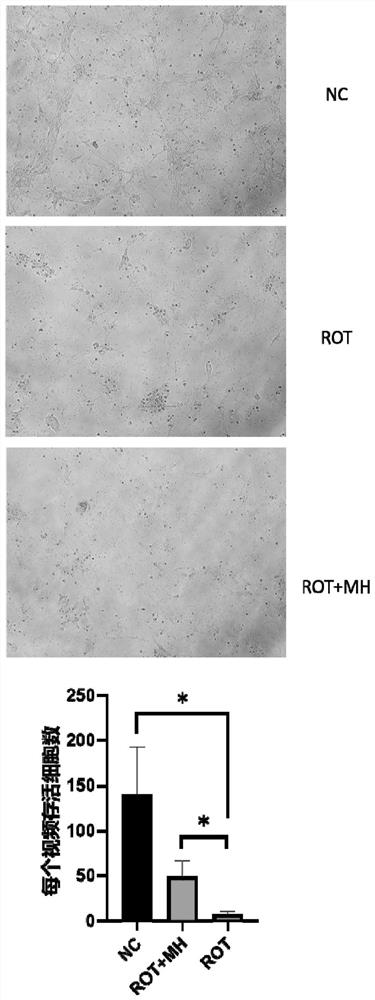
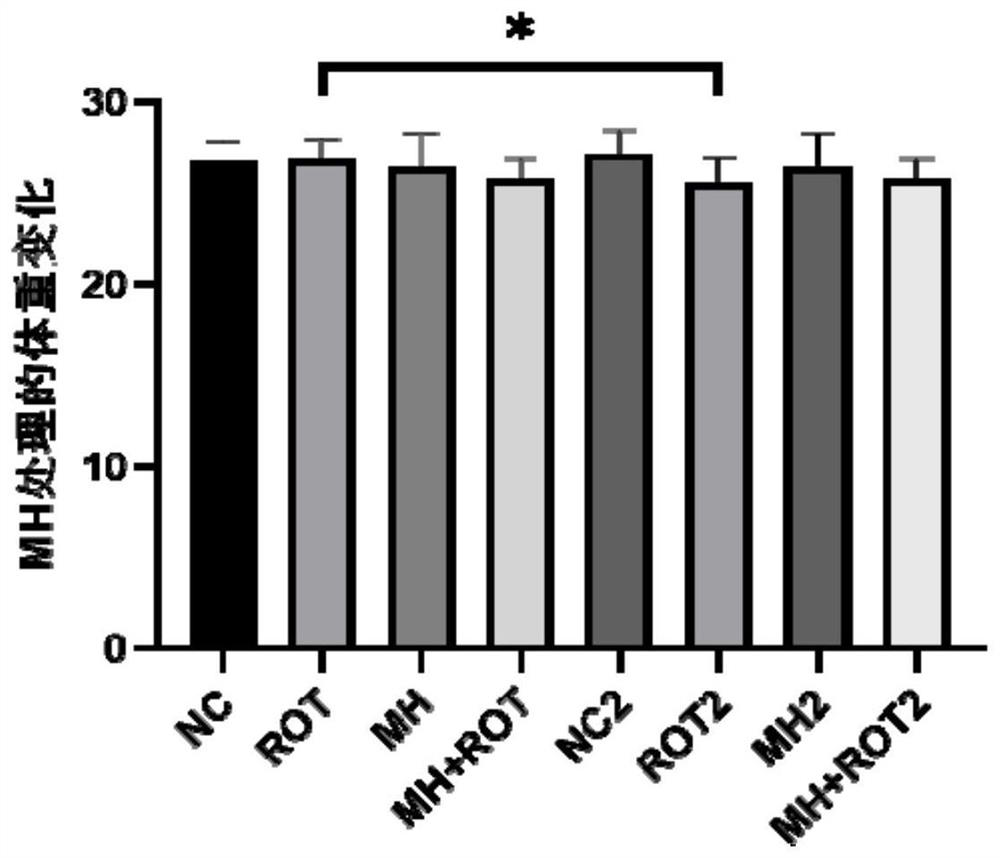
The invention relates to the field of biological pharmacy, in particular to application of meclofenoxate hydrochloride (MH) in preparation of a medicine for preventing or treating Parkinson's disease. By integrating multi-omics data to explore a gene co-expression network and applying cell and animal experiments to verify, the meclofenoxate hydrochloride can obviously prevent synaptic injury caused by mitochondrial function decline in the Parkinson's disease, can effectively reduce mitochondrial membrane injury and reduce mitochondrial edema, and can effectively enable the expression of synapsis specific proteins to rise; it is indicated that the Parkinson's disease characterized by neuronal synaptic lesion can be prevented, dopamine activity is improved, Parkinson's disease lesion is prevented, and behavioral symptoms of the Parkinson's disease is improved.
Owner:SUN YAT SEN MEMORIAL HOSPITAL SUN YAT SEN UNIV
Biliary atresia potential molecular subtype and recognition method of core gene of biliary atresia potential molecular subtype
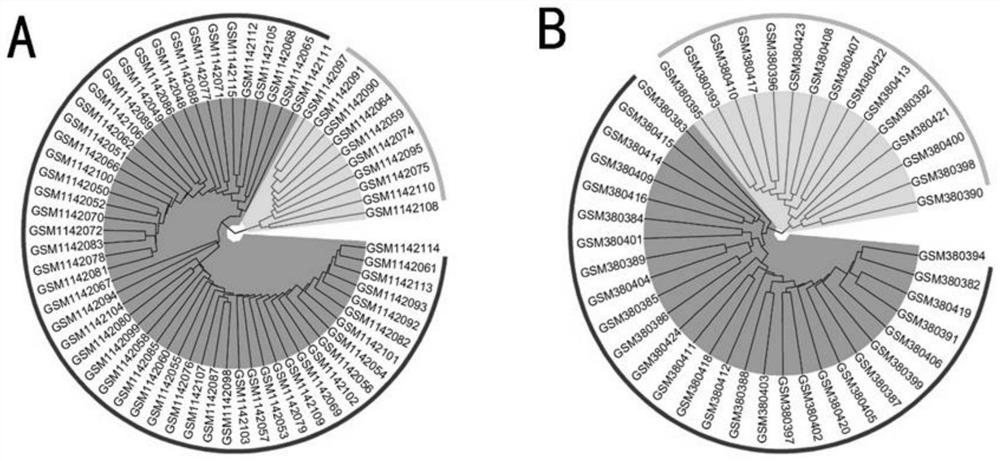
PendingCN114783515AOvercome the weakness of incomparable expression valuesOvercome incomparable weaknessesCharacter and pattern recognitionMedical automated diagnosisCore geneMedicine
The invention relates to a biliary atresia potential molecular subtype and core gene identification method, which comprises the following steps: based on a relative expression rank relationship among genes in a sample, mining potential gene pairs with subtype identification capability, and identifying and verifying the molecular subtype through clustering analysis. And then analyzing and identifying gene modules related to BA subtypes by using a weighted gene co-expression network, and screening core genes related to the subtypes. The biliary tract atresia subtype key gene identification method gets rid of limitation of gene expression absolute values, overcomes the disadvantage that expression values among different laboratories are incomparable, eliminates the influence of group difference on results to the greatest extent, and can be applied to single individual samples by the biliary tract atresia subtype key genes identified by the biliary tract atresia subtype key gene identification method.
Owner:GANNAN MEDICAL UNIV
Gene module mining method and device based on graph neural network, and computer equipment
ActiveCN113611366AScale upImprove timing performanceBiostatisticsCharacter and pattern recognitionEngineeringGraph neural networks
Owner:HARBIN INST OF TECH SHENZHEN GRADUATE SCHOOL
Methods and systems for generating non-coding-coding gene co-expression networks
ActiveCN107111689BMicrobiological testing/measurementData visualisationComputer scienceCoexpression network
A method for identifying coexpressed coding and noncoding genes is disclosed. The method may comprise: receiving a gene sequence; mapping the gene sequence to known coding and non-coding genes; correlating the mapped genes; and generating a co-expression network. A system for generating a co-expression network and providing the co-expression network to a user on a display is disclosed. The system may include memory, one or more processors, one or more databases, and a display.
Owner:KONINKLJIJKE PHILIPS NV
GPU (Graphics Processing Unit)-based rapid optimization operation gene co-expression network method
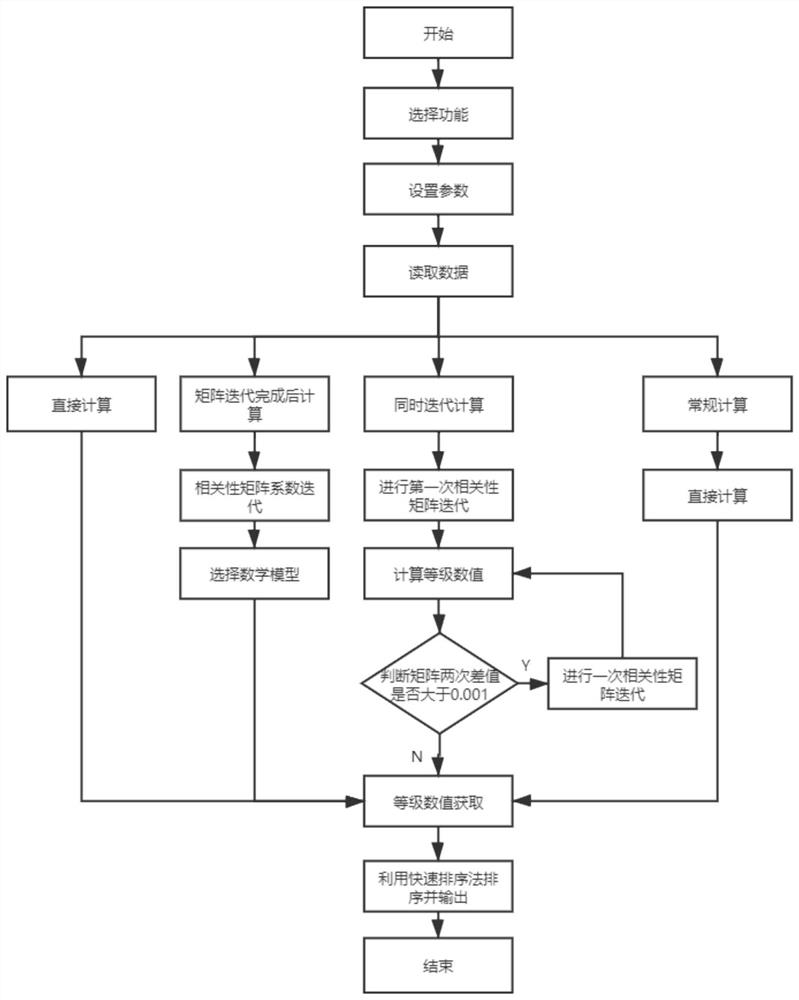
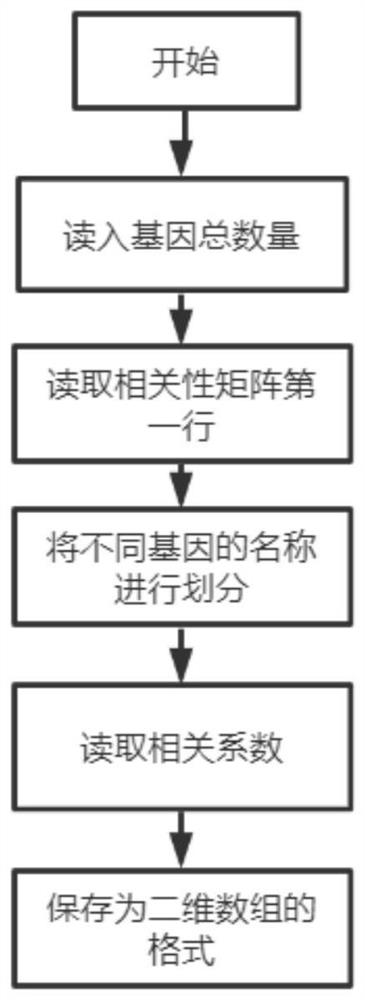
PendingCN113920003AFacilitate follow-up researchSimple environmentDesign optimisation/simulationProcessor architectures/configurationThe InternetEngineering
The invention discloses a GPU (Graphics Processing Unit)-based rapid optimization operation gene co-expression network method, which belongs to the field of internet and comprises the following steps: 1, establishing a data reading module, a correlation matrix coefficient iteration module and a grade numerical value iteration module, providing computing power by using a GPU, and constructing the gene co-expression network; and 2, reading related data of the input file by using a data reading module. An iteration idea is introduced, and a new value is recursively calculated by using an old value, so that the new value can adapt to gene co-expression data under different conditions, and then a weighted association network is constructed and a result with more biological significance is obtained; and the GPU has multiple cores and is applied to gene co-expression network analysis to process a gene co-expression matrix, so that the analysis period can be greatly shortened, and the analysis waiting time can be shortened. In this way, needed hardware and software environments are simple, and installation is convenient.
Owner:NANJING UNIV OF POSTS & TELECOMM
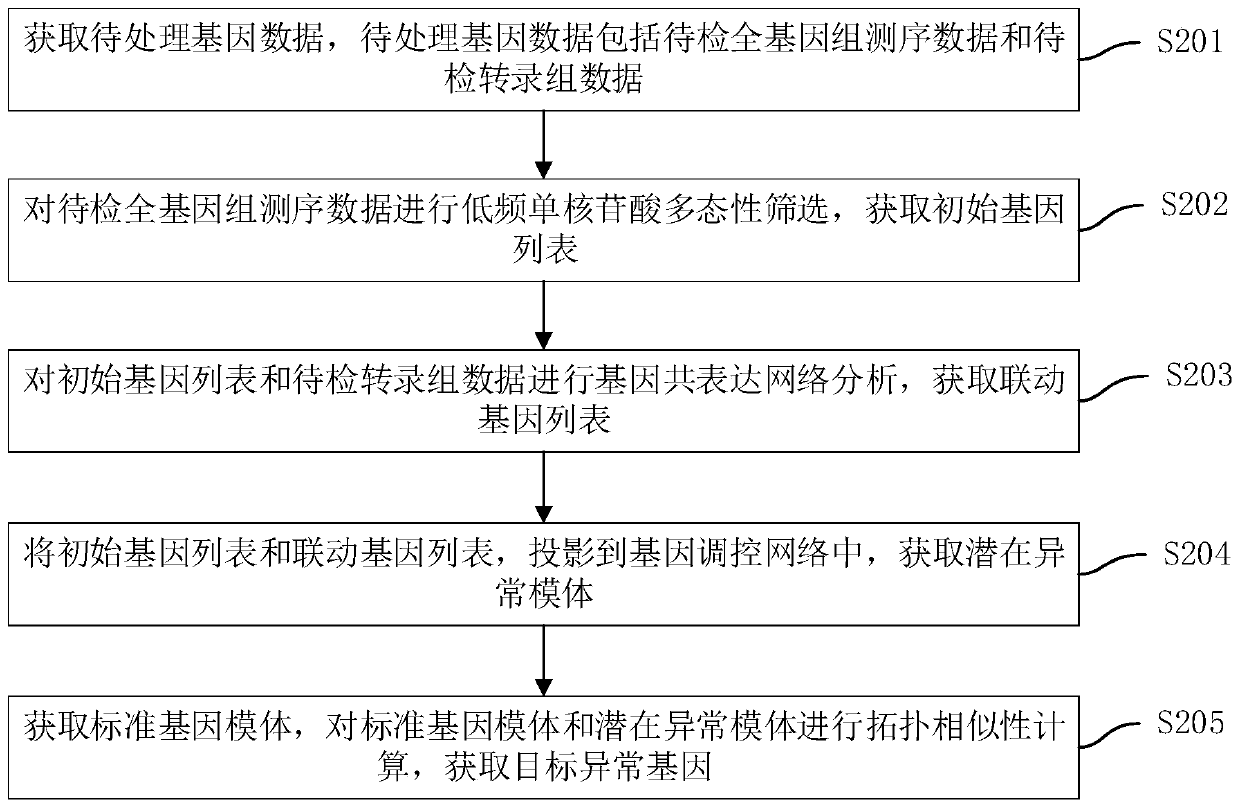
Gene positioning method and device, computer equipment and storage medium
PendingCN110910958AImprove processing efficiencyImprove positioning efficiencySequence analysisHybridisationNucleotideSynexpression
The invention discloses a gene positioning method and device, computer equipment and a storage medium. The gene positioning method comprises the following steps: acquiring to-be-processed gene data, wherein the to-be-processed gene data comprises to-be-detected whole genome sequencing data and to-be-detected transcriptome data; carrying out low-frequency mononucleotide polymorphism screening on the whole to-be-detected genome sequencing data to obtain an initial gene list; performing gene co-expression network analysis on the initial gene list and the to-be-detected transcriptome data to obtain a linkage gene list; projecting the initial gene list and the linkage gene list into a gene regulation and control network to obtain a potential abnormal motif; obtaining a standard gene motif, andperforming topological similarity calculation on the standard gene motif and the potential abnormal motif to obtain a target abnormal gene. The gene positioning method provided by the invention can realize rapid and accurate positioning of abnormal genes.
Owner:PING AN TECH (SHENZHEN) CO LTD
A method for establishing spatiotemporal norms of human brain gene expression
ActiveCN110349625BConvenient supplementEasy to compareBiostatisticsHybridisationDevelopmental stageData set
The invention discloses a method for establishing a spatio-temporal norm of gene expression in the human brain, collecting brain spatio-temporal raw data to obtain a gene standardized expression data set; constructing a two-dimensional spatio-temporal framework of brain regions and developmental stages, and dividing all gene standardized expression data into brain regions Corresponding to the constructed spatiotemporal framework and developmental stage information, the spatiotemporal expression pattern of single gene is obtained; the gene coexpression network analysis method is used to analyze the obtained single gene spatiotemporal expression matrix data, and multiple gene modules are obtained and the average gene expression of each gene is obtained. Quantitatively obtain the spatiotemporal expression pattern of gene modules; through the analysis of normal human nerve cell expression data, the characteristic genes of nerve cells and the obtained gene modules are enriched and analyzed to obtain the cell expression patterns of gene modules. The present invention finally presents all reference data sets in the same two-dimensional space-time frame, which not only synthesizes data sets from different sources, but also facilitates researchers to intuitively supplement and compare the contents of the space-time frame.
Owner:INST OF PSYCHOLOGY CHINESE ACADEMY OF SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com