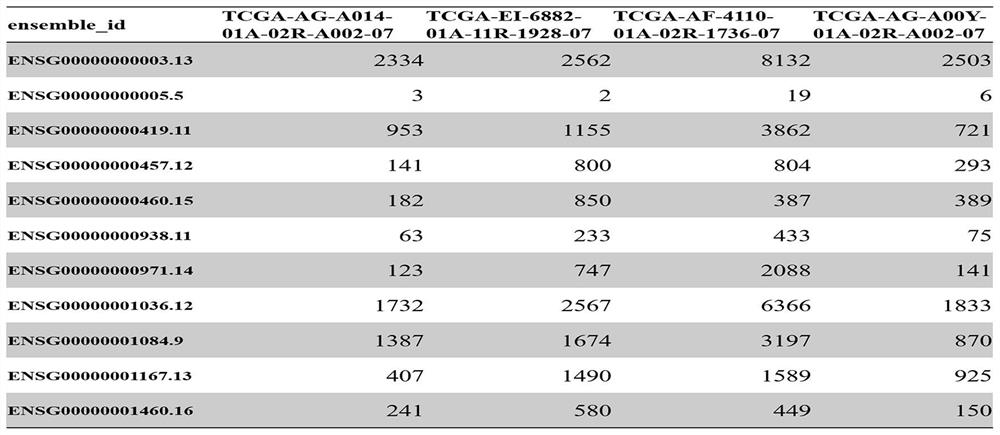
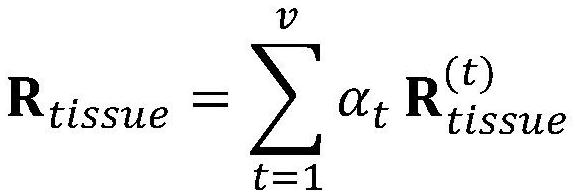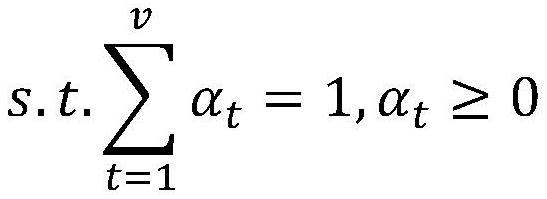Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
30 results about "Coexpression network" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Method for mining radiotherapy specific genes of colorectal cancer by using weight gene co-expression network
ActiveCN109872772AReduce mortalitySolve clinical problemsBiostatisticsProteomicsSurvival prognosisMortality rate
The invention relates to a weight gene co-expression network, and specifically relates to a method for mining radiotherapy specific genes of colorectal cancer by using a weight gene co-expression network. The method for mining radiotherapy specific genes of colorectal cancer by using a weight gene co-expression network provides a new way to mine the specific response genes of colorectal cancer byusing the weight gene co-expression network, and provides a new basis for predicting the survival prognosis of patients with colorectal cancer. The radiotherapy of the target gene discovered by the method of the present invention can improve the survival prognosis of patients with colorectal cancer to some extent, can reduce the mortality of the patients with colorectal cancer, can solve the actual clinical problems, and can provide a better choice for the majority of patients.
Owner:辽宁省肿瘤医院
Identification of esophageal cancer related characteristic pathways and construction method of early diagnosis model
ActiveCN109841280ALimit random fluctuationsFine molecular mechanismHealth-index calculationMedical automated diagnosisPathway analysisWilms' tumor
The invention belongs to the technical field of tumor diagnosis, and particularly relates to identification of esophageal cancer related characteristic pathways of and a construction method of an early diagnosis model. The identification and constructing method includes the steps: expression spectrum preprocessing, differential expression gene extraction, sample cluster analysis, gene cluster analysis, specific gene set functional pathway analysis, comparison of pathway aberration scores, comparative analysis of functional differences, construction of esophageal cancer specific co-expression networks, characteristic selection of genes, prediction of deep learning models, and the like. The method of the invention divides genes into different groups according to expression similarity and functional consistency of the genes, and analyzes the genes in the form of gene collection, thus being able to avoid the disadvantages of high false positive rate, big random error and unstable result inthe traditional method, and also being able to more specifically identify the function significantly associated with esophageal cancer.
Owner:THE FIRST AFFILIATED HOSPITAL OF ZHENGZHOU UNIV
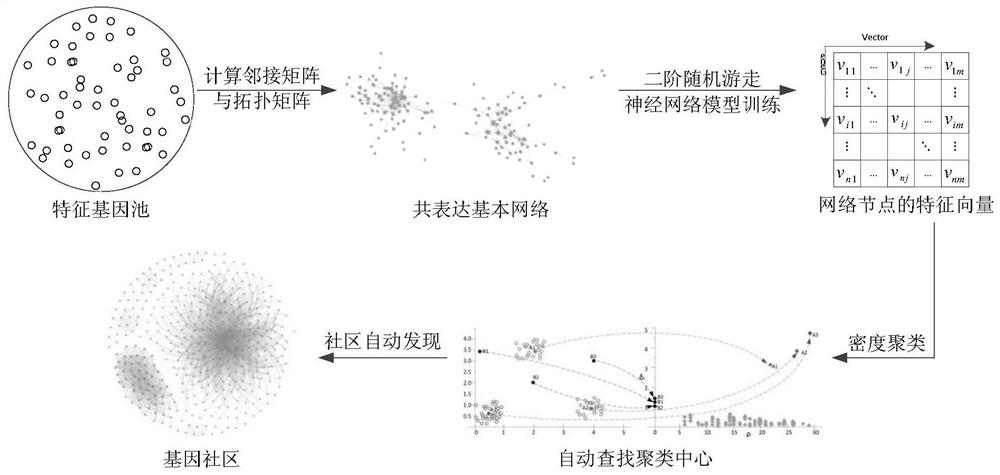
Cancer targeted marker mapping method based on coexpression network
The invention discloses a cancer targeted marker mapping method based on a coexpression network. The method comprises the following steps: 1) building a coexpression basic network, according to gene expression data of a feature gene, calculating an adjacent matrix and a topology matrix; 2) extracting features of the coexpression basic network, namely converting each gene node of the topology network to a feature vector which is used as a feature value of the network; 3) training a neural network model, according to a migration sequence, executing the training of neutral network model parameters; and 4) executing the cancer targeted marker mapping, according to a clustering center self-adaptive algorithm based on a density peak, executing the automatic discovery of a targeted gene community. The provided method has the good universality and precision, and is capable of realizing the target gene mapping by using the building of the coexpression basic network, the extraction of the node feature vector and the automatic discovery of the gene community.
Owner:ZHEJIANG UNIV OF TECH
Intramuscular fat related lncRNA (long non-coding Ribonucleic Acid) and application thereof
ActiveCN108103206ASpeed up entryMicrobiological testing/measurementDNA/RNA fragmentationDiseaseIntramuscular fat
The invention discloses intramuscular fat related lncRNA (long non-coding Ribonucleic Acid) and application thereof. The invention finds out XLOC_004398 related to intramuscular fat of pork; a targetgene NAP1L3 for differentially expressing the lncRNA is analyzed and predicated through co-expression network analysis and a trans regulation effect and is verified. The invention provides certain evidences for breeding high-meat-quality livestock and poultry varieties and treating and preventing fat metabolism related diseases, and a new target is explored.
Owner:INST OF ANIMAL SCI OF CHINESE ACAD OF AGRI SCI
Gene expression time series data classification method based on visibility graph algorithm
ActiveCN108846261AImprove applicabilityGood precisionSpecial data processing applicationsNODALFeature vector
The invention discloses a gene expression time series data classification method based on a visibility graph algorithm. The method comprises the steps of (1) constructing a basic network, selecting adata strip according to preprocessed gene expression time series data, constructing a visibility graph and a connection graph by using the visibility graph algorithm, and determining a basic structureof a co-expression network, (2) extracting relevant traditional features according to the obtained basic network, (3) obtaining a feature vector of each gene node in the basic network by using second-order random walk and neural network model learning, and (4) integrating features of the basic network and using different strategies based on the obtained features of the basic network through a density clustering algorithm to complete the classification of gene expression time series data. The invention provides a method for realizing the gene expression time series data classification by usingvisibility graph foundation network construction, node feature vector extraction and the density clustering algorithm, and the method has good precision and practical performance.
Owner:ZHEJIANG UNIV OF TECH
Method for screening quantitative character candidate genes
ActiveCN109830261AEfficient screeningExpedited screeningComponent separationMaterial analysis by optical meansMolecular breedingCandidate Gene Association Study
The invention belongs to the field of molecular breeding, and mainly relates to a method for screening quantitative character candidate genes. The method includes the steps: extracting genes of quantitative character related intervals (QTL interval and GWAS interval) as candidate gene sets; collecting known genes related to the characters as bait genes; carrying out gene co-expression analysis orweight co-expression analysis on the transcriptome data of a set of samples; constructing a co-expression network; and selecting genes with strong connectivity to the bait genes as candidate genes. Compared with previous analysis methods, the method for screening quantitative character candidate genes can quickly and effectively screen candidate genes in the relevant range of quantitative characters, can narrow the screening range of candidate genes, can predict the interaction between genes, can overcome the defect that traditional fine positioning is large in workload and cumbersome, and canaccelerate the molecular breeding process of species.
Owner:SOUTHWEST UNIV
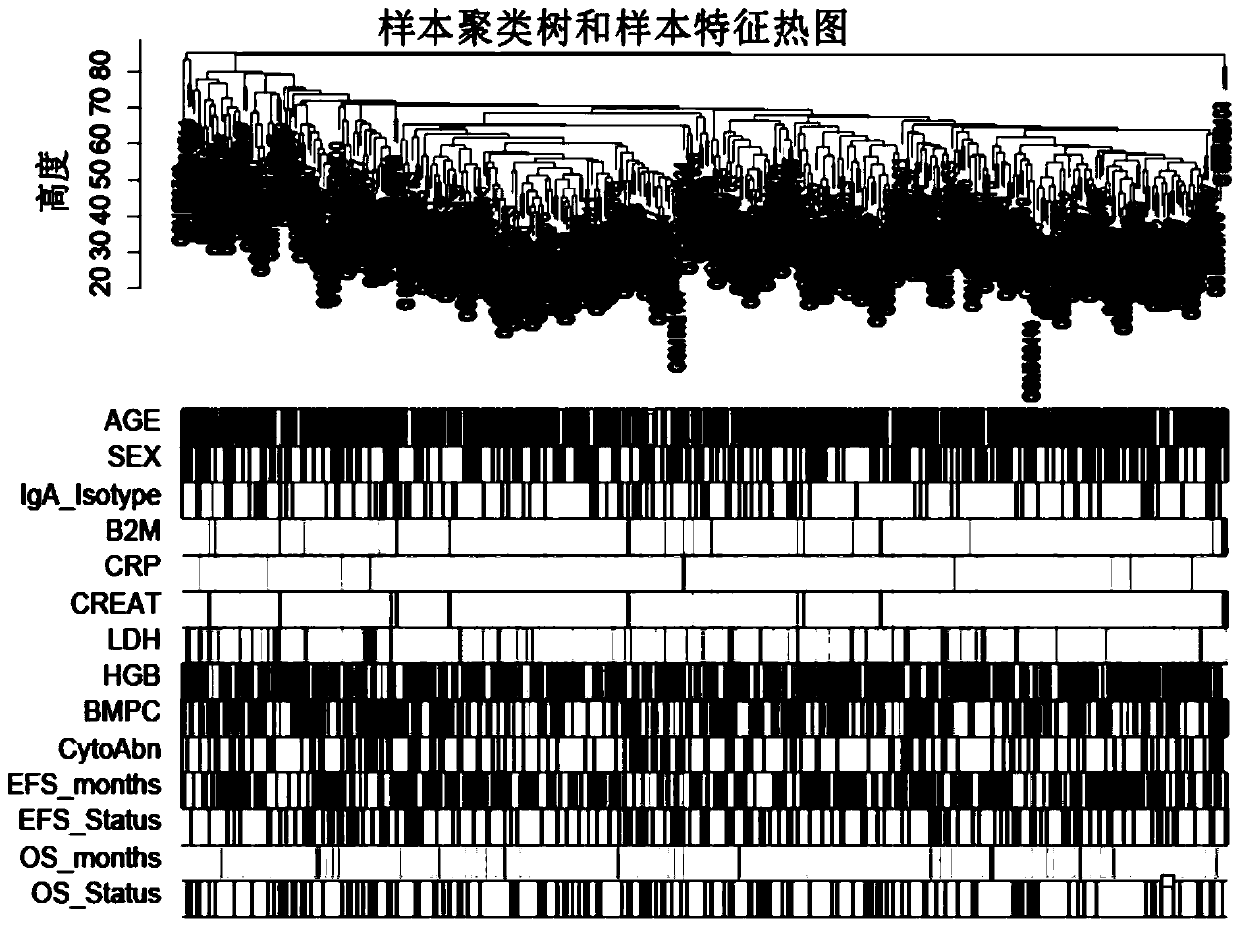
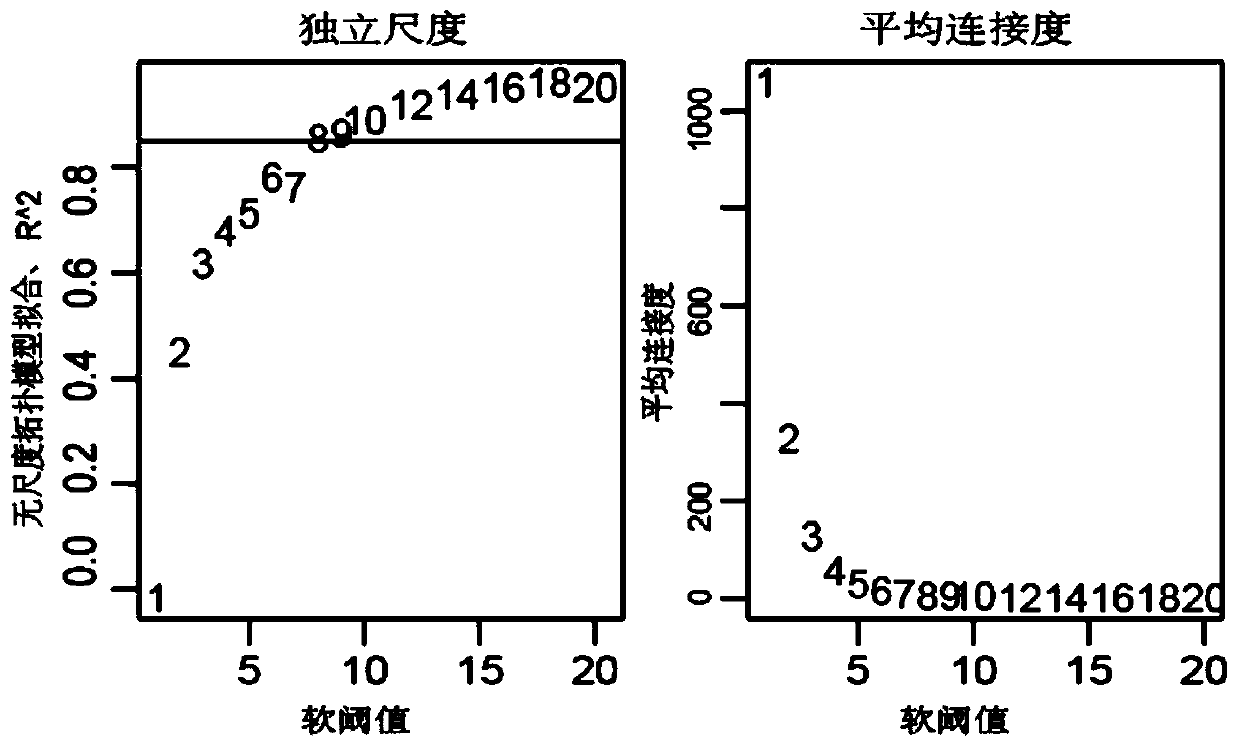
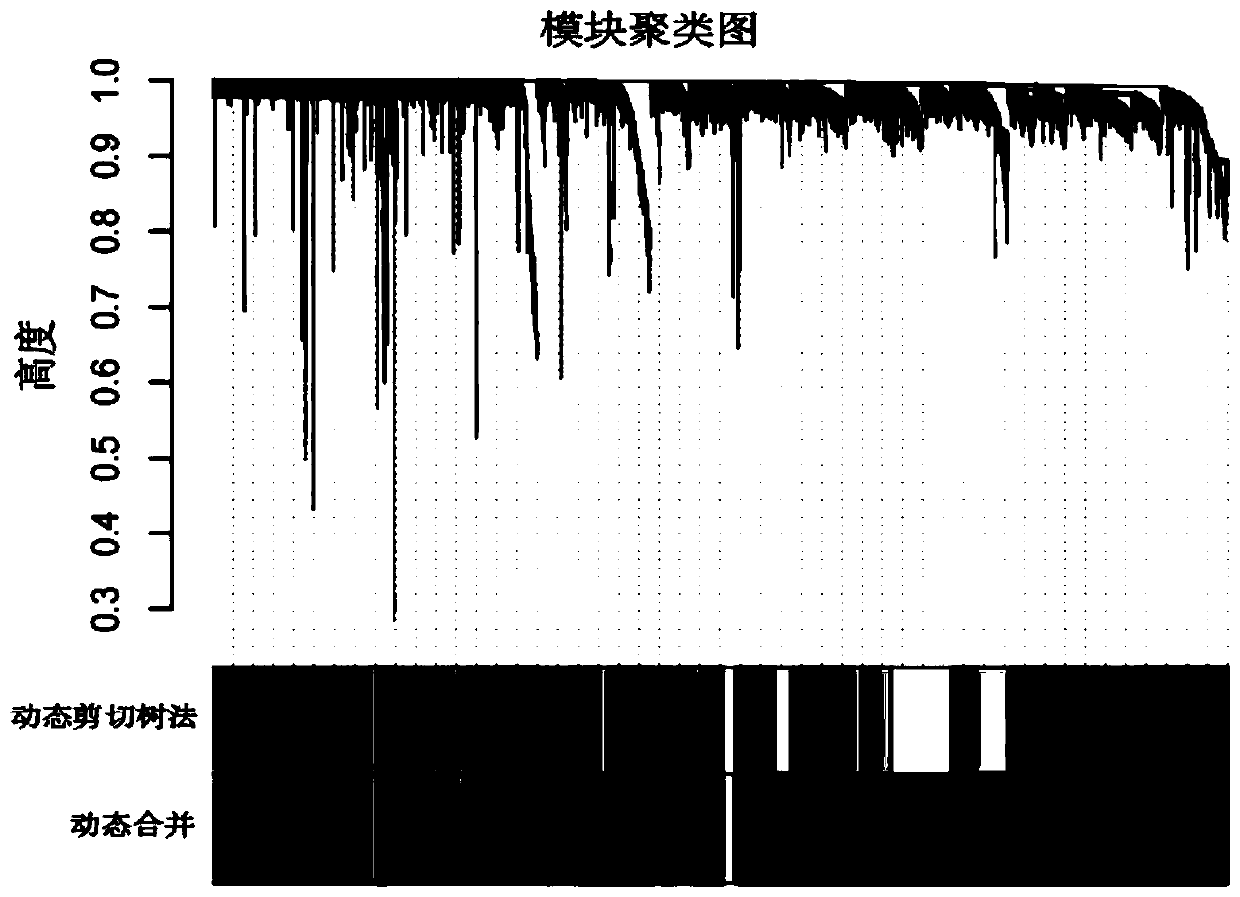
Novel comprehensive risk scoring method for multiple myeloma
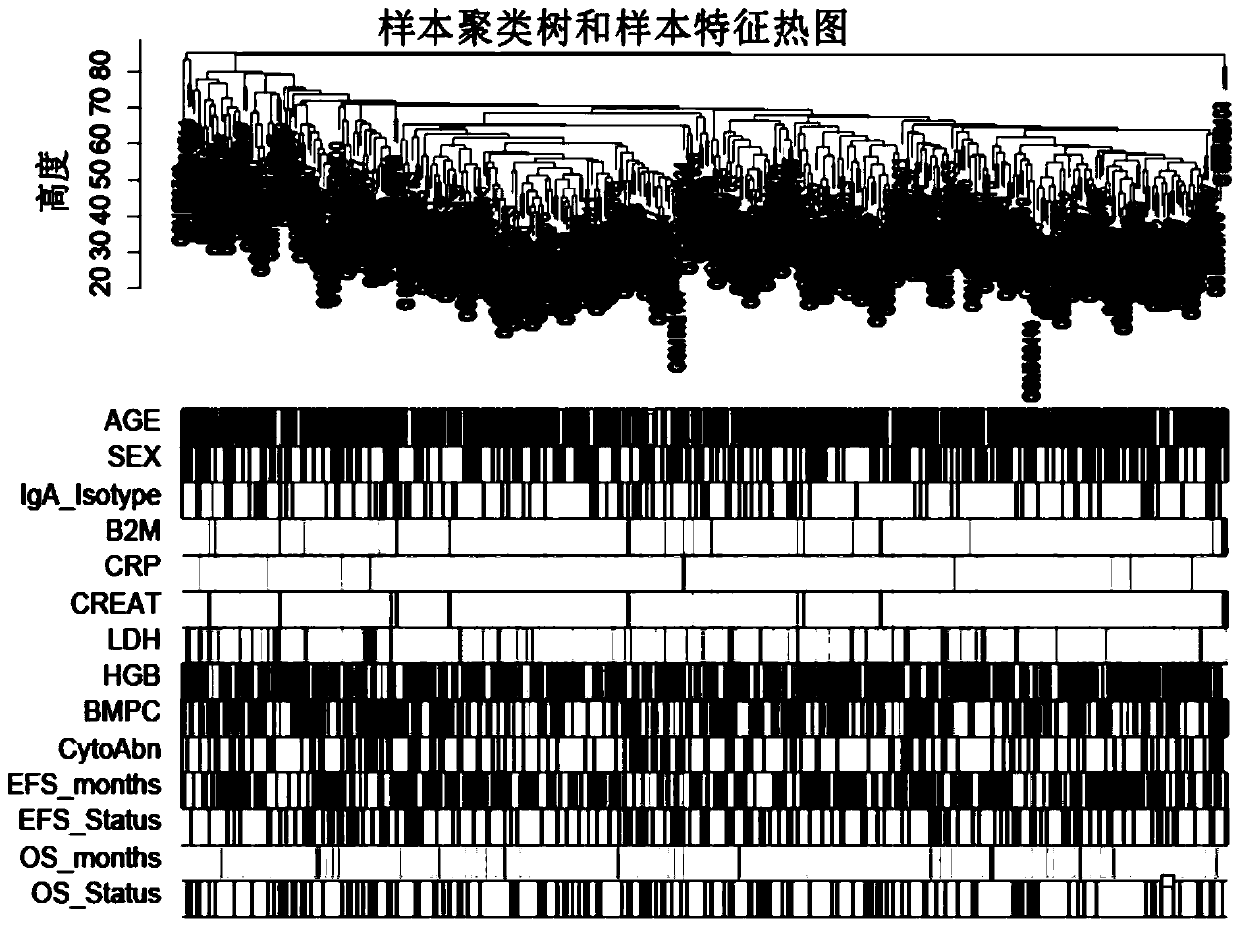
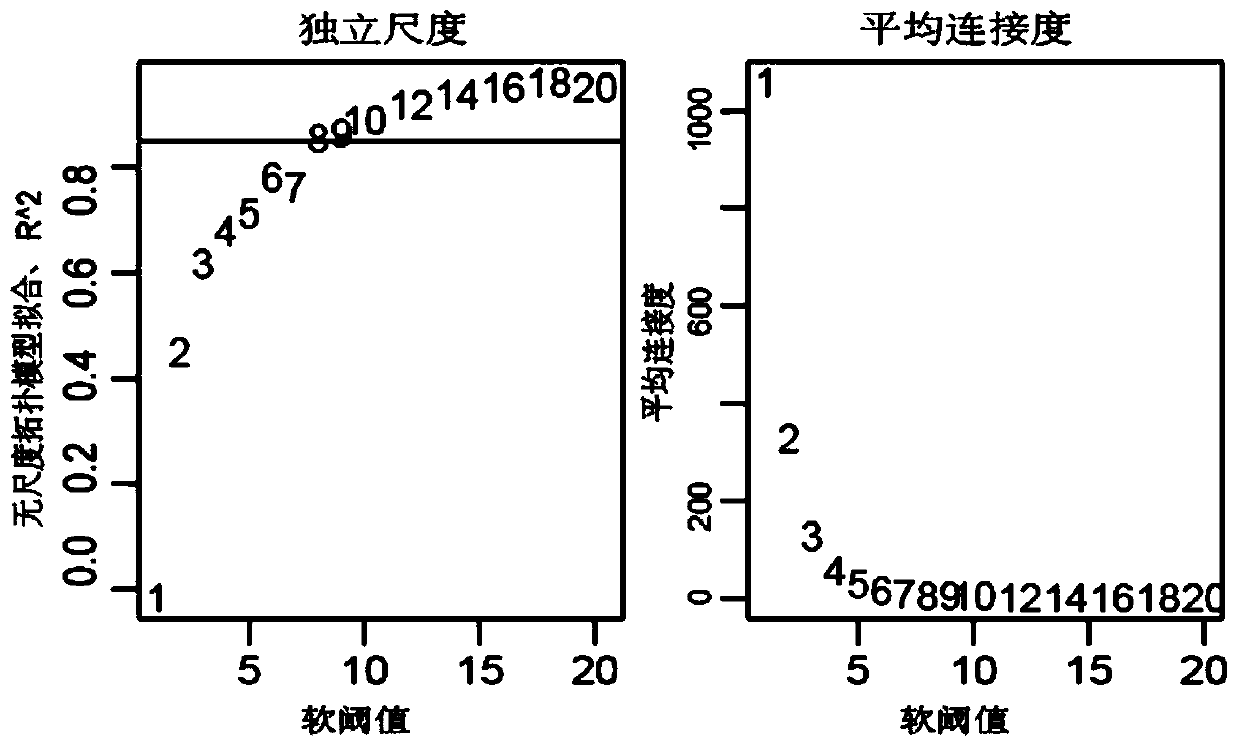
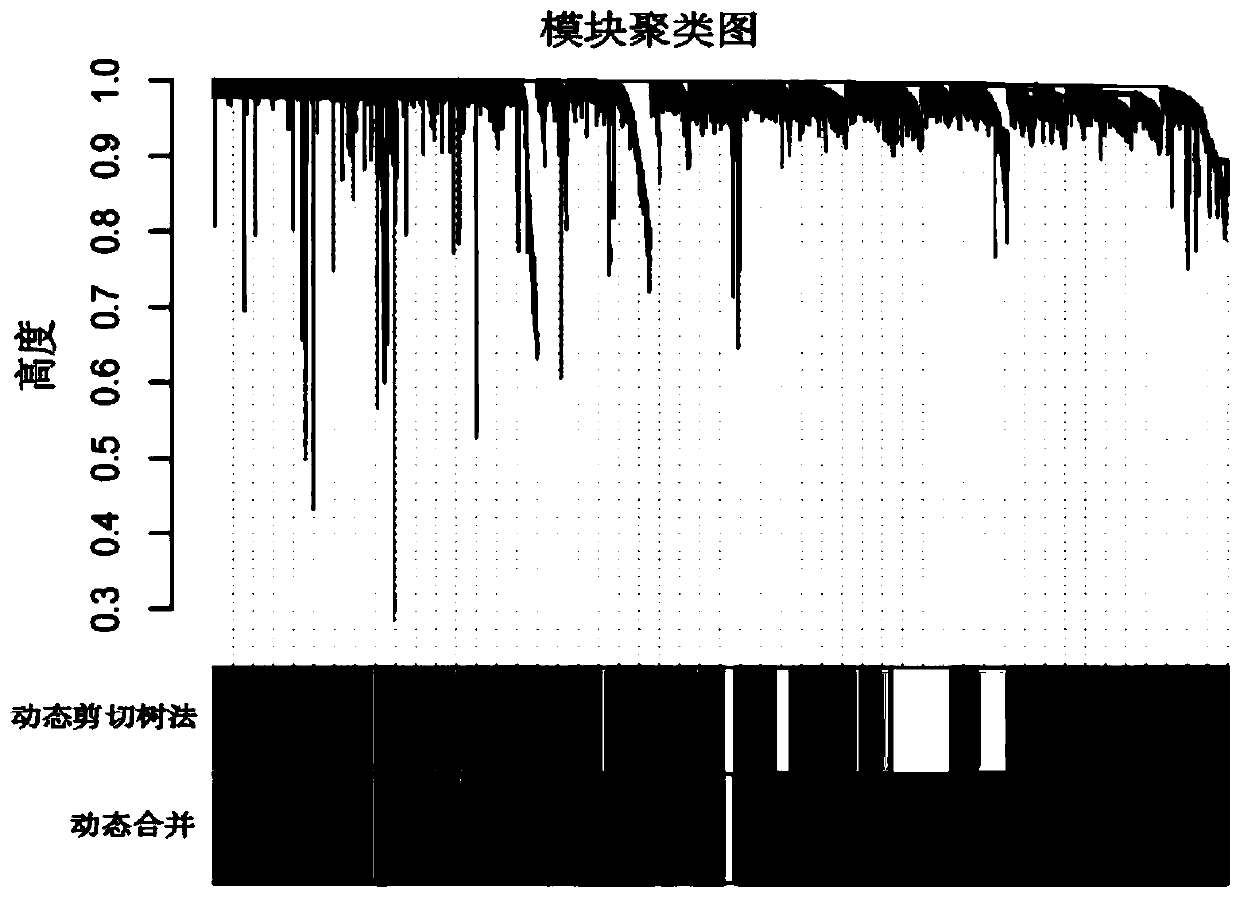
ActiveCN110232974AImprove forecast accuracyMedical data miningHealth-index calculationPearson Correlation TestGeo database
The invention provides a comprehensive risk scoring method for multiple myeloma, comprising the following steps: S1, obtaining a gene expression profile GSE24080 of an MM patient from a GEO database,and pre-treating genes in the gene expression profile GSE24080 to obtain the first 25% of the 5413 genes with the largest variance of expression value; S2, subjecting the 5413 genes to WGCNA gene co-expression network analysis to identify co-expressed functional modules; S3, evaluating the correlation between the functional modules and clinical information through Pearson correlation test to determine the most significant modules; S4, performing univariate survival analysis on the genes in the most significant modules by using a Cox proportional hazard model, and screening out a ten-gene scoring model composed of the 10 best genes through LASSO regression; and S5, setting the rule that each factor scores 1 point when the ten-gene scoring model or serum [beta]2M or LDH is higher than a cut-off value and otherwise 0, and establishing a comprehensive risk scoring system.
Owner:THE FIRST AFFILIATED HOSPITAL OF FUJIAN MEDICAL UNIV
Multi-omics data combined analysis method
The invention discloses a multi-omics data combined analysis method. The multi-omics data combined analysis method comprises the following steps of (A), performing coexpression network analysis on each single-omics index data in to-be-analyzed multi-omics data, and finding a respective expression model; and (b), according to the overlapping relation between respective expression modules of different omics data, screening interaction modules which are remarkably correlated in the to-be-analyzed multi-omics data. The multi-omics data combined analysis method according to the invention is not restricted by the number of omics data, and random multiple groups can be utilized. Furthermore the method does not depend on a data source. The index data (such as gene expression magnitude, apparent methylation degree, SNP mutation rate) which can measure the corresponding omics can be used as input data.
Owner:BGI TECH SOLUTIONS
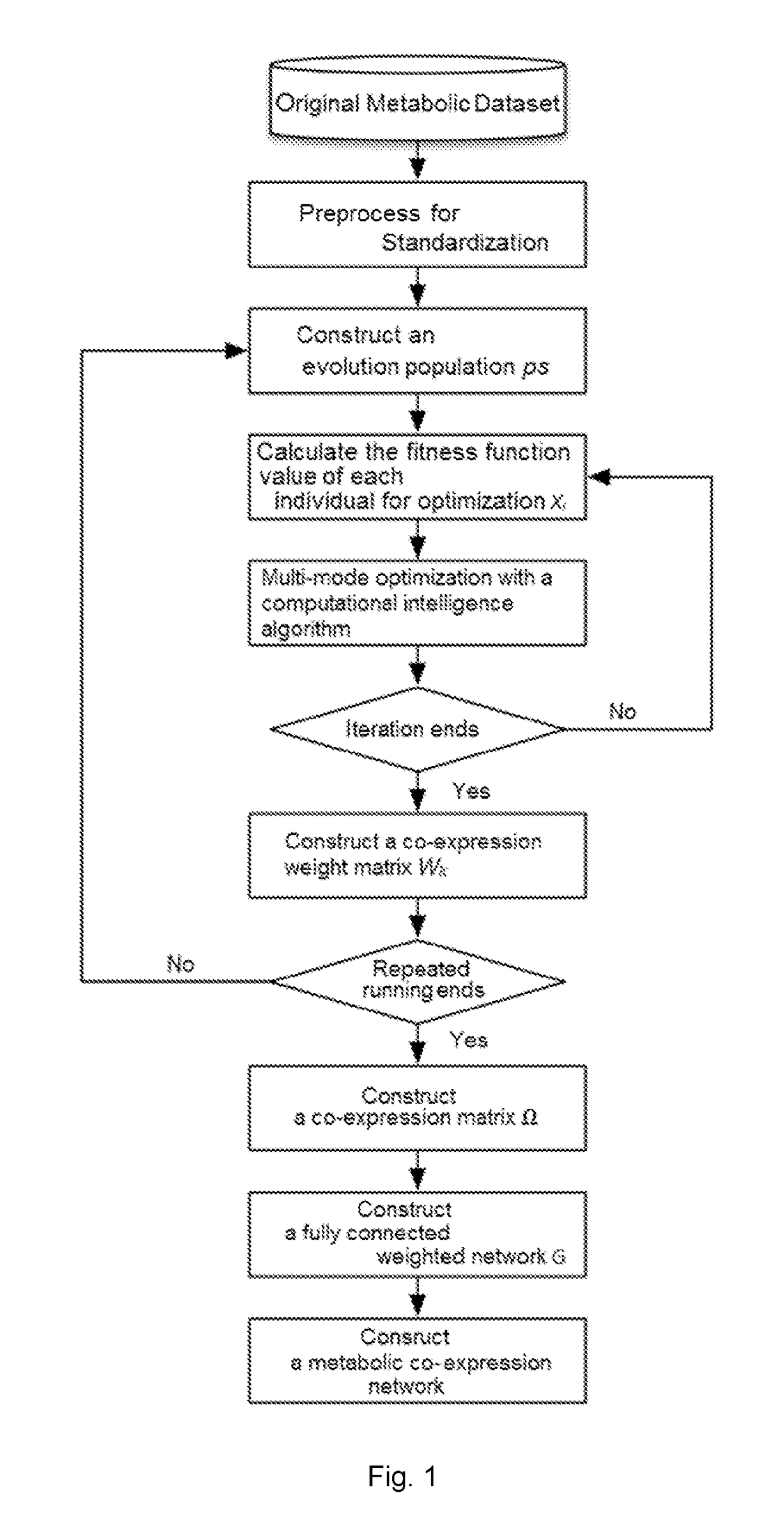
Construction method for heuristic metabolic co-expression network and the system thereof
InactiveUS20170212980A1Low accuracyReduced stabilityBiostatisticsBiomolecular computersProbit modelMultivariate mutual information
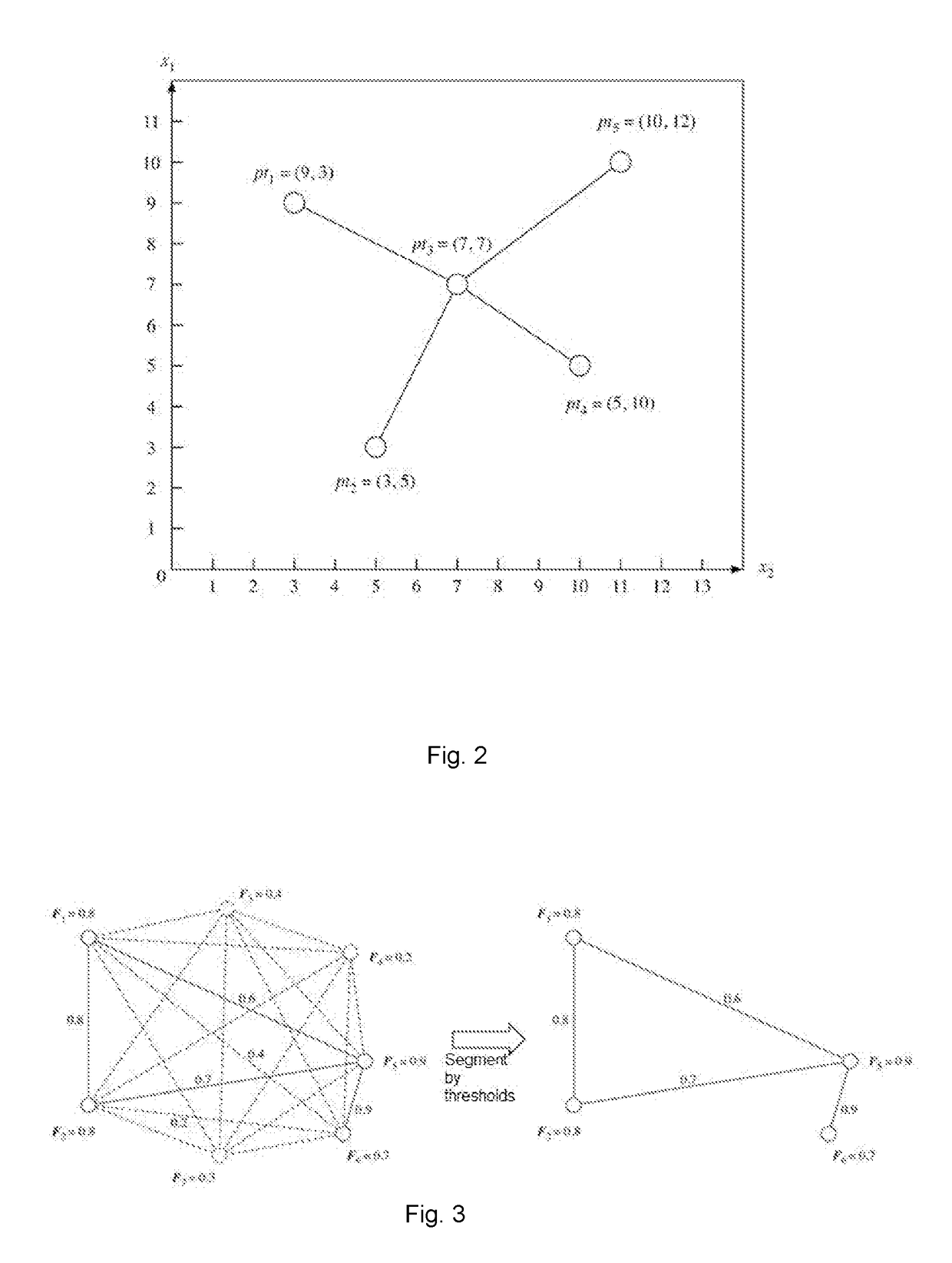
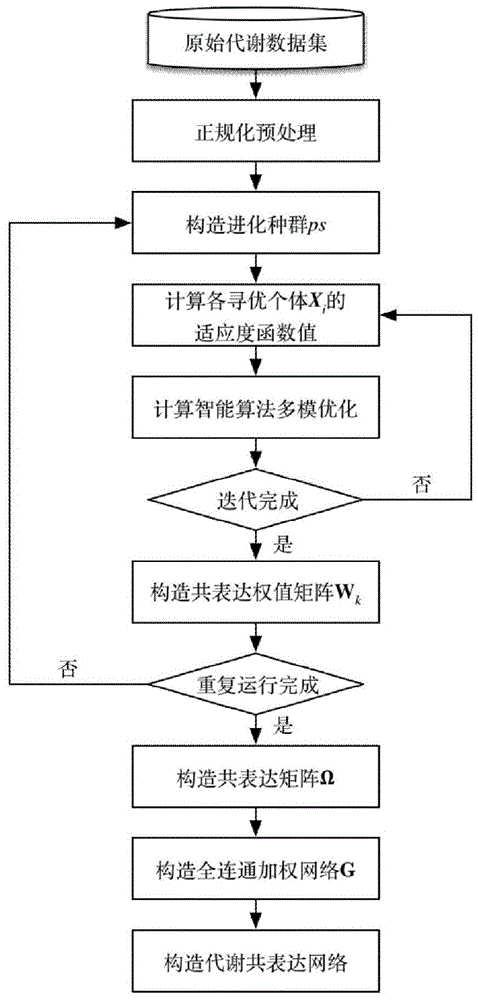
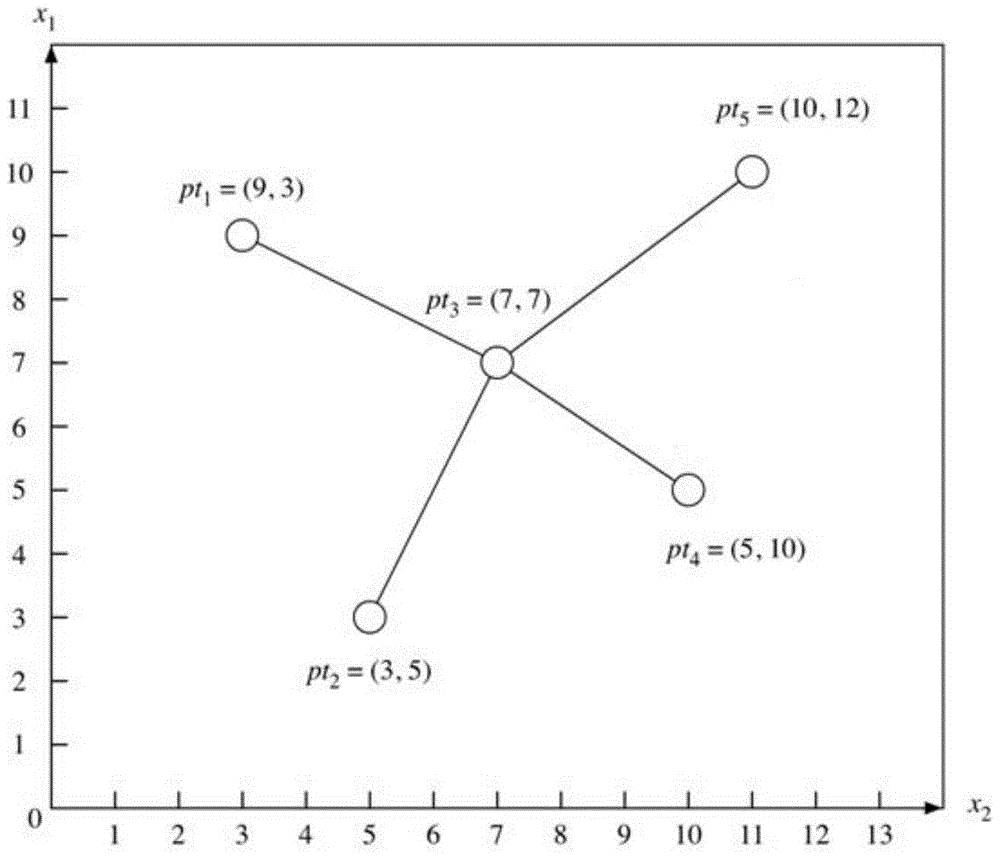
The present invention discloses a construction method for heuristic metabolic co-expression network and the system thereof. Based on the max-dependent criteria, the present invention treats the characterized multivariate mutual information of a plurality of metabolites as mutual function value, and applies an optimization searching for the best feature subset, with a heuristics computational intelligence multimodal optimization algorithm. And by running the optimization process in a plurality of times, combining and studying the results in each time running, a co-expression network structure is built. Finally, a threshold for segmentations is calculated through probability models, and an exact and stable metabolic co-expression network is obtained.
Owner:SHENZHEN UNIV
Construction method and system of heuristic metabolic co-expression network
The invention discloses a construction method and system of a heuristic metabolic co-expression network. According to the most dependent criterion that mutual information of multiple metabolite features serves as a fitness function value, a heuristic intelligent multimode optimization algorithm is used to search optimization from an optimal feature subset. The optimization process is carried out for multiple times, and results of the multiple times are combined and learned to construct the co-expression network structure. A probability model is used to calculate a segmentation threshold, and the accurate and stable metabolic co-expression network is obtained.
Owner:SHENZHEN UNIV +4
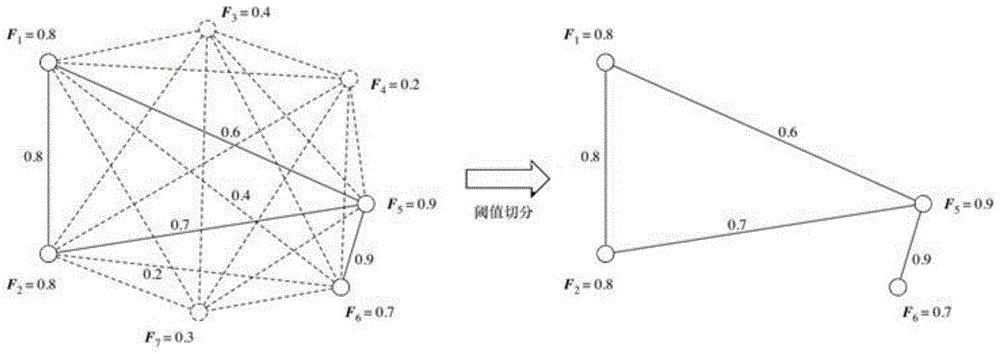
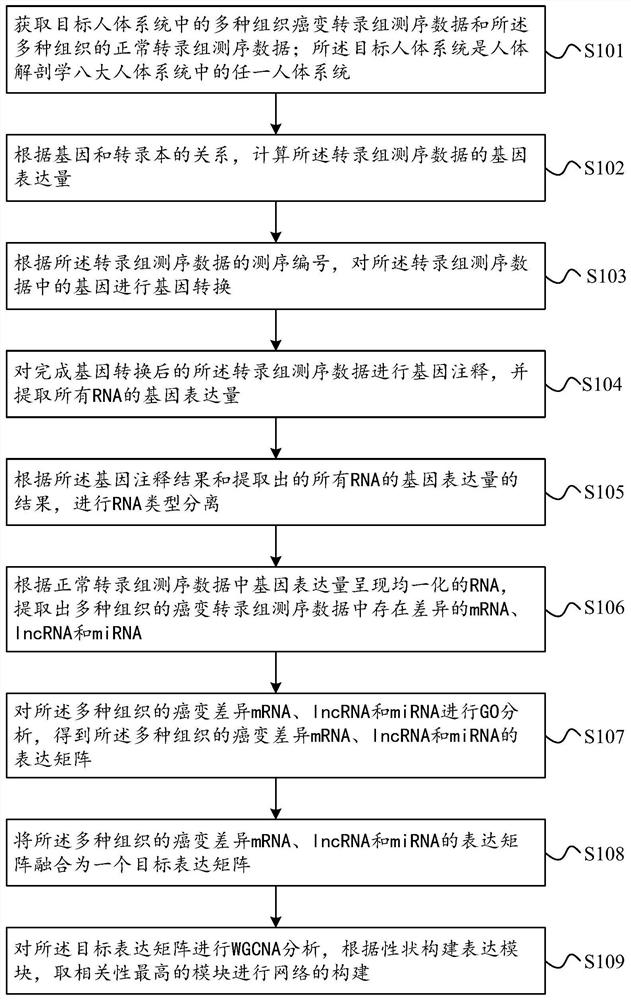
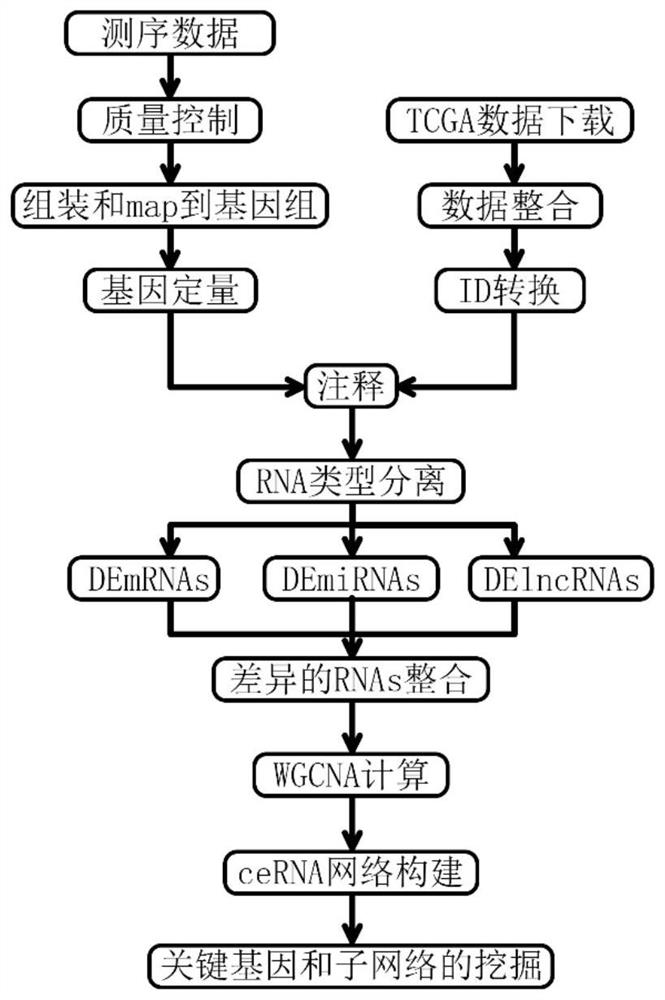
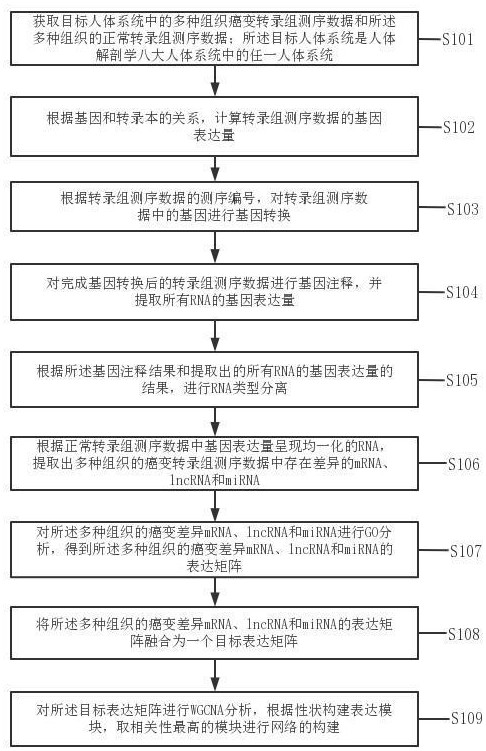
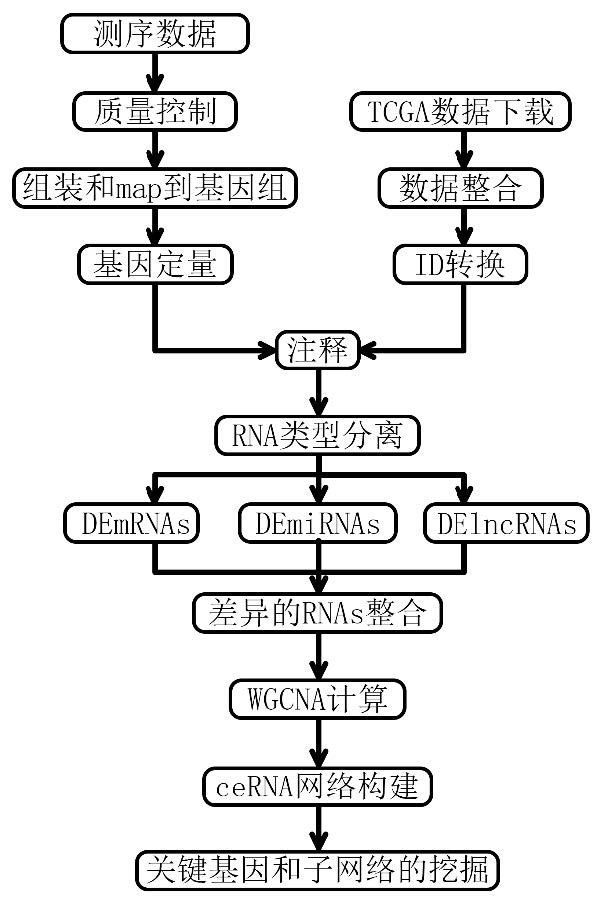
Tumor gene co-expression network construction method and device, equipment and storage medium
The invention provides a tumor gene co-expression network construction method and device, equipment and a storage medium. The method comprises the following steps: acquiring multiple tissue canceration transcriptome sequencing data and normal transcriptome sequencing data of multiple tissues in a target human body system, wherein the target human body system is any one human body system in eight human body systems of human anatomy; extracting mRNA, lncRNA and miRNA with difference in canceration transcriptome sequencing data of various tissues according to a relationship between genes and transcripts and RNA with uniform gene expression quantity in normal transcriptome sequencing data; carrying out GO analysis on canceration difference mRNA, lncRNA and miRNA of the plurality of tissues to obtain a target expression matrix fused with the mRNA, lncRNA and miRNA; and performing WGCNA analysis on the target expression matrix, constructing expression modules according to characters, and performing network construction on the module with the highest correlation. Network construction of various cancers can be realized.
Owner:SHENZHEN POLYTECHNIC +1
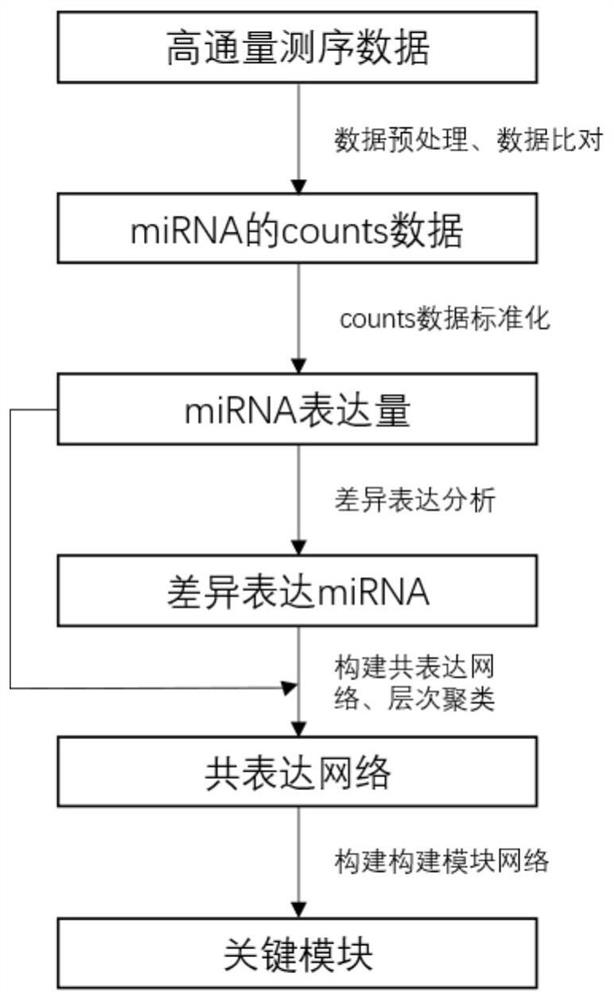
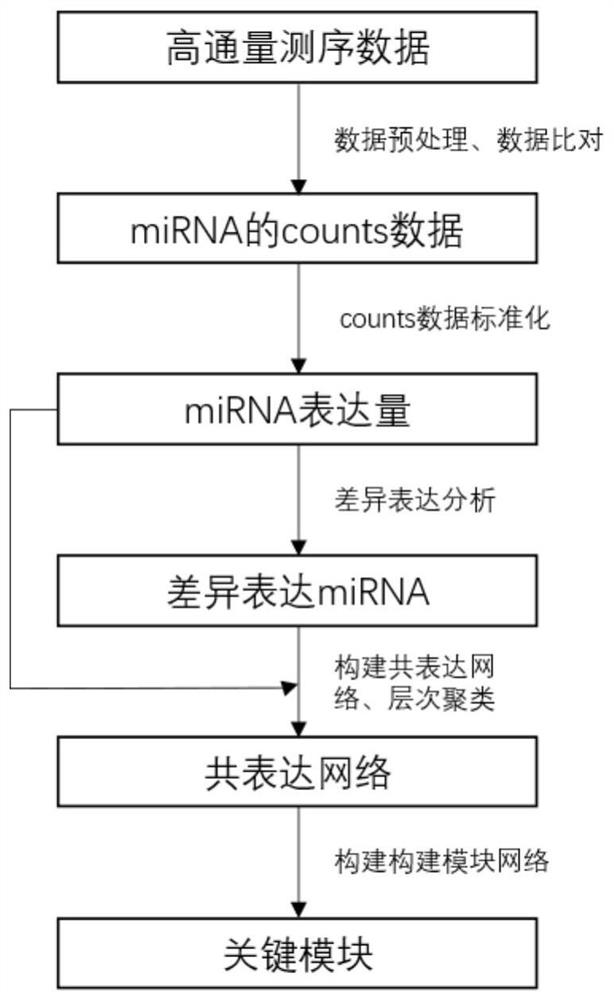
Parkinson disease evolution key module identification method based on miRNA sequencing data
The invention provides a Parkinson's disease key module identification method based on miRNA (micro Ribonucleic Acid) sequencing data. The method comprises the following steps: firstly, preprocessing high-throughput sequencing data; then, grouping the samples according to different stages of PD diseases, and carrying out differential expression analysis; then, carrying out hierarchical clustering according to correlation coefficients between differential expression miRNAs, and constructing a co-expression network and a module; and finally, constructing a module network and performing identifying to obtain a PD key module. By means of the method, key module identification in the PD evolution process can be carried out, the PD stage of the current patient is judged according to the key module, and help is provided for doctors to find early PD patients.
Owner:NORTHWESTERN POLYTECHNICAL UNIV
Novel screening method for multiple myeloma prognosis gene
ActiveCN110223733AStrong correlationData visualisationHybridisationClinical informationScreening method
The invention provides a novel screening method for a multiple myeloma prognosis gene based on a gene coexpression network. The method comprises the following steps of S1, acquiring a gene expressionspectrum GSE24080 of an MM patient from a GEO database, preprocessing the gene in the gene expression spectrum GSE24080, and obtaining 5413 genes which have the front 25% highest expression value variances; S2, performing WGCNA gene coexpression network analysis on the 5413 genes for identifying a coexpression functional module; and S3, evaluating correlation between the functional module and clinical information through Pearson related examination, thereby determining a most substantial module.
Owner:THE FIRST AFFILIATED HOSPITAL OF FUJIAN MEDICAL UNIV
Method for predicting functions of unknown genes of maize based on dynamic correlation between gene expression level and characters
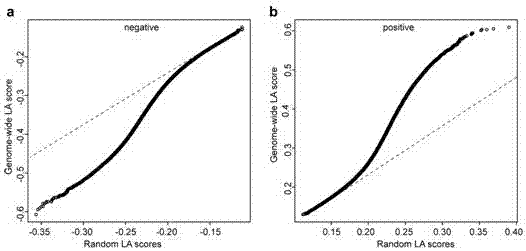
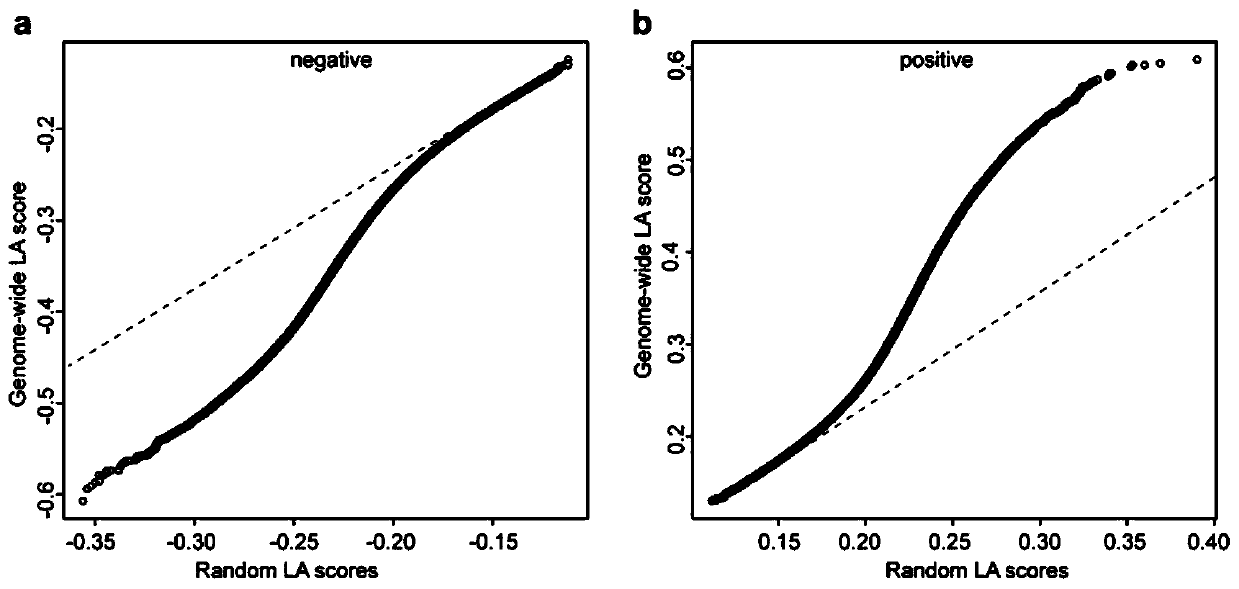
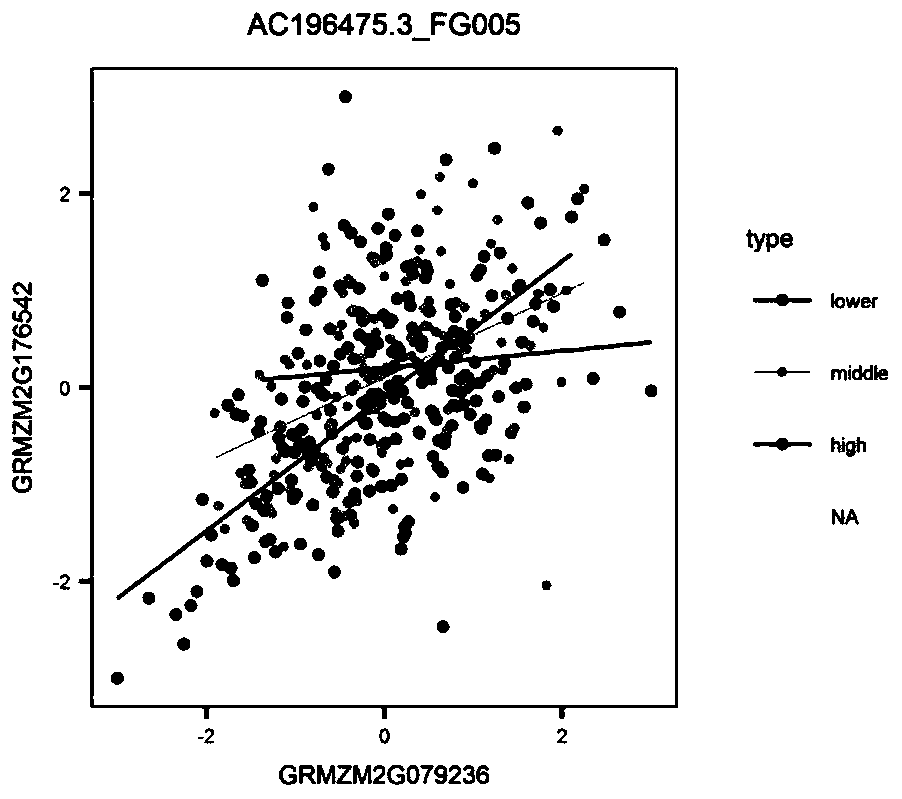
ActiveCN107058525AIdea innovationMicrobiological testing/measurementHybridisationCorrelation analysisGene expression level
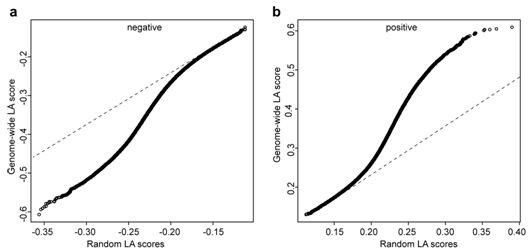
The invention belongs to the field of plant molecular biotechnology and gene engineering and particularly relates to a method for predicting functions of unknown genes of maize based on dynamic correlation between gene expression level and characters. The method comprises the following steps: firstly collecting grain transcripts obtained after pollinating maize inbred lines for 15 days, and sequencing to obtain gene expression level data; building a dynamic correlation analysis LA model; evaluating significance of LA; excavating dynamic correlation of gene expression patterns of whole genomes of the maize; and carrying out function annotation of genes of significant LA results and predicting functions of the genes. The method is capable of predicting the functions of the unknown genes by taking the phenomenon of dynamic correlation between co-expression patterns of gene pairs in the maize grains as sally port; compared with the conventional co-expression network construction, the dynamic correlation analysis is capable of rapidly finding regulation genes for regulating the co-expression patterns.
Owner:UNIV OF JINAN
Method for predicting unknown corn gene functions on the basis of oil content-related genes and oil content dynamic correlation
The invention belongs to the field of plant molecular biotechnology and genetic engineering and relates to a method for predicting unknown corn gene functions, in particular to a method for predictingunknown corn gene functions on the basis of oil content-related genes and oil content dynamic correlation. The method comprises the following steps: firstly, collecting the grain transcript sequencing 15 days after pollination of a corn selfing line to obtain gene expression quantity data; collecting gene data associated with the oil content of the corn kernels; collecting grain oil content dataof an associated group formed by the corn selfing line; establishing a dynamic association analysis LA model; carrying out LA significance evaluation; excavating a gene for regulating and controllingthe dynamic correlation between the oil content associated gene and the oil content within the whole genome range; and performing function annotation on the gene with the obvious LA result, and predicting the function of an unknown gene. According to the method, the phenomenon that genes in corn kernels are dynamically associated with a co-expression mode is taken as a breakthrough. The function of an unknown gene is predicted. Compared with traditional co-expression network construction, the regulation gene for regulating the co-expression mode can be quickly found through dynamic correlationanalysis.
Owner:UNIV OF JINAN
A method for predicting the function of unknown genes in maize based on the dynamic correlation between gene expression and traits
ActiveCN107058525BMicrobiological testing/measurementHybridisationCorrelation analysisExpression gene
Owner:UNIV OF JINAN
Tumor gene co-expression network construction method, device, equipment and storage medium
The present application provides a method, device, equipment and storage medium for constructing a tumor gene co-expression network. The method includes: acquiring the cancerous transcriptome sequencing data of various tissues in the target human body system and the normal transcriptome sequencing data of the various tissues; the target human body system is any one of the eight major human body systems of human anatomy ; According to the relationship between genes and transcripts, as well as RNAs with uniform gene expression in normal transcriptome sequencing data, differential mRNAs, lncRNAs and miRNAs are extracted from cancer transcriptome sequencing data of various tissues; Carcinogenic differential mRNA, lncRNA, and miRNA of various tissues were analyzed by GO, and a target expression matrix was obtained; the target expression matrix was analyzed by WGCNA, expression modules were constructed according to traits, and the most relevant modules were selected for network construction. It can realize network construction for various cancers.
Owner:SHENZHEN POLYTECHNIC +1
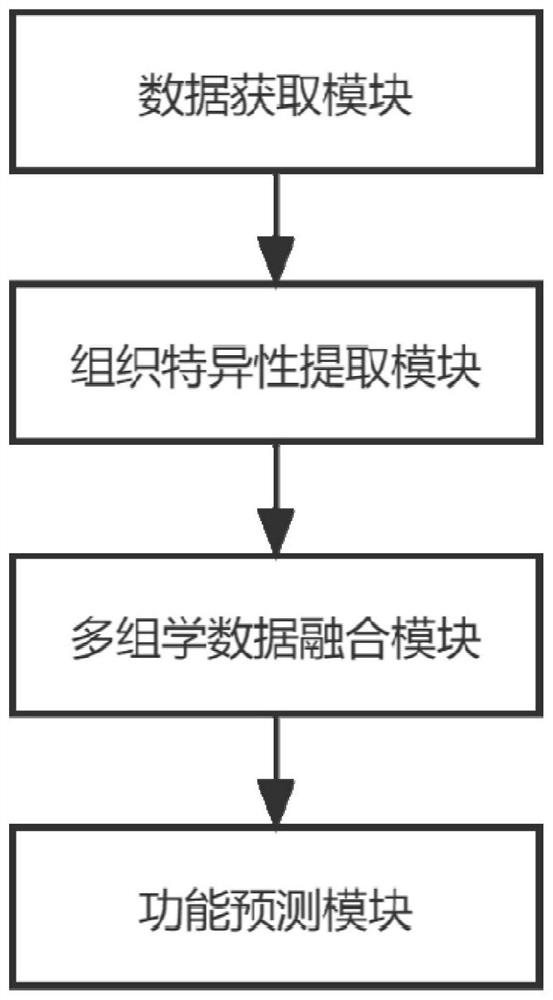
Corn variable splicing isomer function prediction system based on tissue specificity
PendingCN114863992ATake advantage ofImprove function prediction effectBiostatisticsSystems biologyNonnegative matrix factorisationGenetics
The invention provides a corn variable splicing isomer function prediction system based on tissue specificity, which is characterized in that isoform co-expression networks of a plurality of tissues are constructed by processing expression data of isoform on each tissue, and the isoform co-expression networks of each tissue are integrated through self-adaptive weight, so that the function of the corn variable splicing isomer is predicted. A high-quality isoform organization specific association network can be obtained; according to the method, an isoform sequence similarity network is constructed by using isoform sequence data, and the isoform sequence similarity network is fused with an isoform organization specificity association network, so that a better isoform function association network can be obtained; multi-example learning is performed through non-negative matrix factorization, and meanwhile, the non-negative matrix factorization is guided by using an isoform function association network, so that more accurate and more comprehensive function prediction of isoform is realized.
Owner:SHANDONG UNIV
Novel method for constructing nomogram of multiple myeloma
ActiveCN110197701APredicted survivalHelps quantitatively predict survivalHealth-index calculationData visualisationNewly diagnosedNomogram
The invention provides a method for constructing a nomogram of multiple myeloma based on ten gene characteristics, serum [beta]2 microglobulin and LDH. The method comprises the following steps: S1, obtaining a gene expression profile GSE24080 of an MM patient from a GEO database, and pre-treating genes in the gene expression profile GSE24080 to obtain the first 25% of the 5413 genes with the largest variance in expression values; S2, subjecting the 5413 genes to WGCNA gene co-expression network analysis to identify function modules of co-expression; S3, evaluating the correlation between the functional modules and clinical information by Pearson correlation test to determine the most significant module; S4, performing univariate survival analysis on the genes in the most significant modules using a Cox proportional hazard model, and screening a score model consisting of 10 best genes by LASSO regression; and S5, establishing a novel nomogram based on the scoring model, serum [beta]2M and high LDH to predict 3-year OS and 5-year OS of newly diagnosed MM patients.
Owner:SHANGHAI TISSUEBANK MEDICAL LAB CO LTD
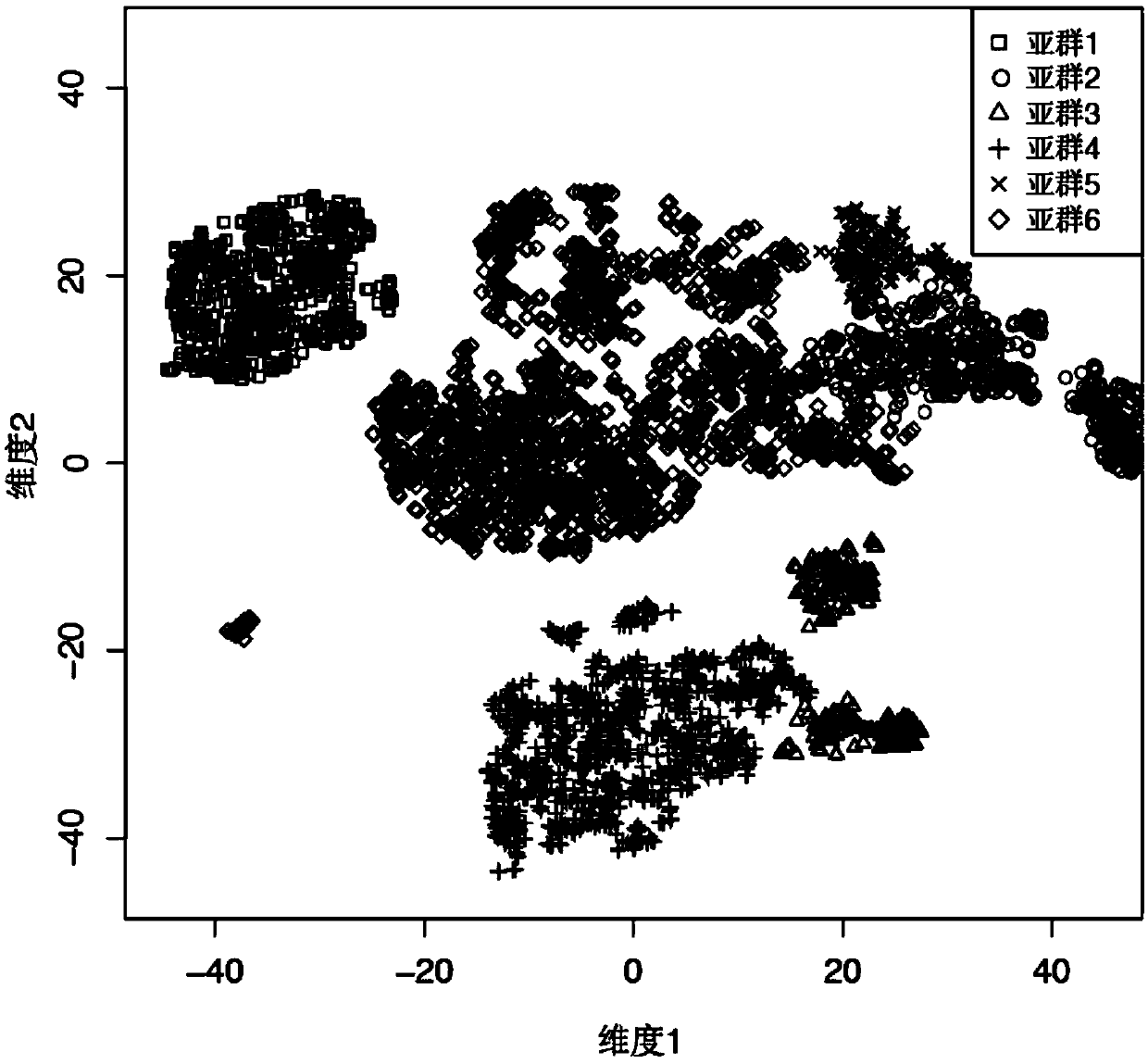
Subgroup specific co-expression network identification method
ActiveCN110706744AUnderstand functionEfficient screeningProteomicsGenomicsExpression geneCell subpopulations
The invention discloses a subgroup specific co-expression network identification method. The invention provides a method for identifying a specific co-expression network of a subgroup of a sample to be detected. The method comprises the following steps of: 1) carrying out homogenization treatment on an original UMI number value of each gene of each sample in the subgroup of the sample to be detected; 2) filtering an original UMI homogenization result; 3) calculating the weighted median correlation bicor of every two filtered genes; 4) filtering the weighted median correlation result to obtaina strong co-expression gene relationship pair; 5) subtracting the shared strong co-expression gene relationship pair appearing in all cells in the subgroup from the filtered strong co-expression generelationship pair of the subgroup of the cells to be detected to obtain a specific co-expressed gene pair of the subgroup of the cells; and constructing a specific gene co-expression network of the subgroup of the sample to be detected by using the specific co-expression gene pair of the subgroup of the cells. The method for the subgroups is more targeted and more sensitive.
Owner:BGI GENOMICS CO LTD
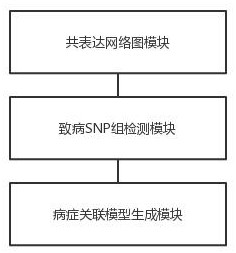
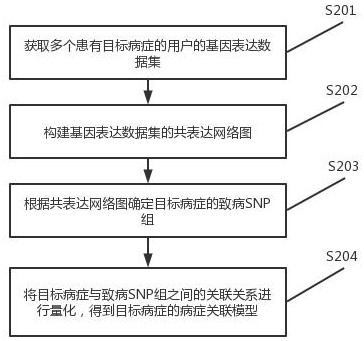
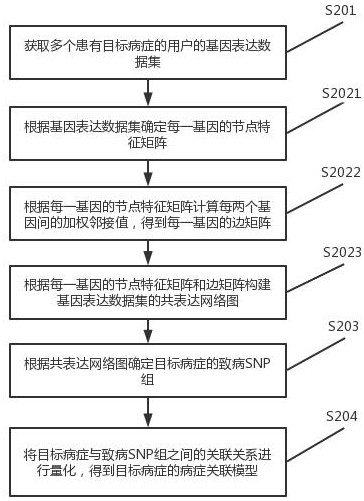
Disease correlation analysis system and method based on gene big data
The invention discloses a disease correlation analysis system and method based on gene big data, and relates to the technical field of biological information. Comprising a co-expression network diagram module, a pathogenic SNP group detection module and a disease association model generation module, the co-expression network diagram module obtains gene expression data sets of a plurality of users suffering from target diseases; constructing a co-expression network diagram of the gene expression data set; the pathogenic SNP group detection module determines a pathogenic SNP group of the target disease according to the co-expression network diagram; and the disease association model generation module quantifies the association relationship between the target disease and the pathogenic SNP group to obtain a disease association model of the target disease. The co-expression network diagram is constructed through the gene expression data set, and then the pathogenic SNP group of the target disease is determined, so that the relationship between the target disease and the pathogenic SNP group can be quantified, and the disease relevance between the gene expression and the disease is enhanced.
Owner:广州盛安医学检验有限公司
A co-expression network-based method for mapping cancer target markers
ActiveCN107992720BImprove applicabilityGood precisionMolecular designBiostatisticsCancer targetingNetwork model
A cancer target marker mapping method based on a co-expression network, comprising the following steps: 1) constructing a co-expression basic network, and calculating an adjacency matrix and a topology matrix according to gene expression data of characteristic genes; 2) extracting the characteristics of the co-expression basic network , that is to convert each gene node of the topological network into an eigenvector as the eigenvalue of the network; 3) train the neural network model, and train the parameters of the neural network model according to the walk sequence; 4) conduct cancer target marker mapping, according to the An adaptive algorithm for cluster centers of density peaks for automatic discovery of targeted gene communities. The invention provides a method with good universality and precision, which adopts co-expression basic network construction, node feature vector extraction and gene community automatic discovery to realize target gene mapping.
Owner:ZHEJIANG UNIV OF TECH
Subgroup-specific co-expression network identification method
ActiveCN110706744BUnderstand functionEfficient screeningProteomicsGenomicsExpression geneCell subpopulations
The invention discloses a subgroup-specific co-expression network identification method. The invention provides a method for identifying the specific co-expression network of a subgroup of samples to be tested, comprising the following steps: 1) processing the original UMI number values of each gene in each sample in the subgroup of samples to be tested to be normalized; 2) converting the original UMI Filter the homogenization results, 3) calculate the pairwise weighted median correlation bicor of the filtered genes; 4) filter the weighted median correlation results to obtain strong co-expression gene relationship pairs; 5) filter the cell subgroups to be tested Strongly co-expressed gene relationship pairs, subtracting the shared strong co-expressed gene relationship pairs in all cells in the subgroup, that is, to obtain the specific co-expressed gene pairs of the cell subgroup; and then using the cell subgroup Group-specific co-expressed gene pairs were used to construct a subgroup-specific gene co-expression network of the sample to be tested. The method for subgroups of the present invention is more targeted and more sensitive.
Owner:BGI GENOMICS CO LTD
Identification method of key modules in Parkinson's disease evolution based on miRNA sequencing data
ActiveCN113035279BBiostatisticsSequence analysisDifferentially expressed mirnasDifferential expression analysis
Owner:NORTHWESTERN POLYTECHNICAL UNIV
A method for predicting spindle assembly checkpoint abnormalities in triple-negative breast cancer using lncRNA-mRNA co-expression network
The present invention relates to a method for predicting the abnormality of triple-negative breast cancer spindle assembly checkpoints by using lncRNA-mRNA co-expression network, comprising the following steps: experimental grouping, RNA extraction, differential expression analysis of lncRNA chip data, and lncRNA target gene analysis and Control network construction. The method of the present invention using lncRNA-mRNA co-expression network to predict spindle assembly checkpoint abnormality in triple-negative breast cancer provides a new molecular marker for triple-negative breast cancer by predicting spindle assembly checkpoint abnormality through lnRNA-mRNA co-expression network A new approach has been established, which provides a reference for predicting the prognosis of triple-negative breast cancer.
Owner:范志民
Using miRNA-gene co-expression networks to predict abnormal gap junctional communication in anthracycline-treated cardiomyocytes
The invention relates to a method for predicting anthracycline drug cardiomyocyte gap junction communication abnormality using miRNA-gene co-expression network, comprising the following steps: Step 1: Experimental grouping; Step 2: RNA extraction; Step 3: miRNA-gene chip data Differential expression analysis; step 4: miRNA target gene analysis and regulatory network construction. The present invention uses the miRNA-gene co-expression network to predict the abnormality of anthracycline drug cardiomyocyte gap junction communication, and predicts the abnormal cardiomyocyte gap junction communication through the miRNA-gene co-expression network, which is a new molecular marker for discovering anthracycline drug cardiotoxicity A new approach is provided, which provides a reference for predicting its prognosis.
Owner:JINZHOU MEDICAL UNIV
Methods and systems for generating non-coding-coding gene co-expression networks
ActiveCN107111689BMicrobiological testing/measurementData visualisationComputer scienceCoexpression network
A method for identifying coexpressed coding and noncoding genes is disclosed. The method may comprise: receiving a gene sequence; mapping the gene sequence to known coding and non-coding genes; correlating the mapped genes; and generating a co-expression network. A system for generating a co-expression network and providing the co-expression network to a user on a display is disclosed. The system may include memory, one or more processors, one or more databases, and a display.
Owner:KONINKLJIJKE PHILIPS NV
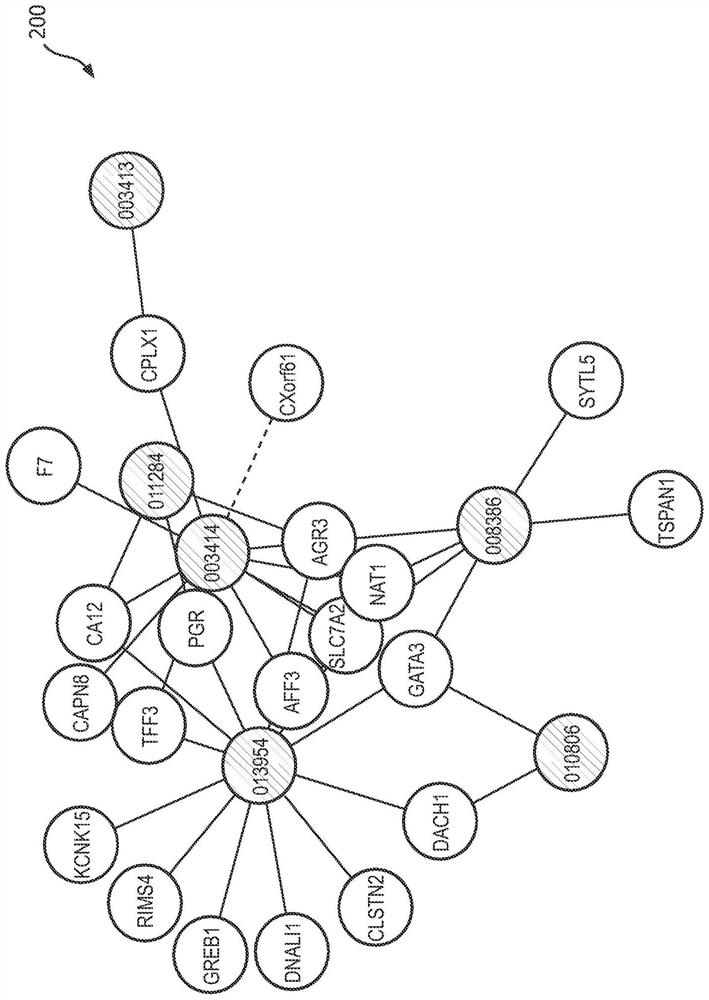
Research method for key genes of intracranial aneurysm formation and rupture
The invention relates to the field of bioinformatics, in particular to a research method of key genes of intracranial aneurysm formation and rupture, which comprises the following steps: S1, chip data acquisition and chip data pretreatment; s2, screening key genes; s3, gene function enrichment analysis and immune microenvironment analysis, on the basis of screening of key genes, a method combining differential expression gene analysis and WGCNA is adopted, on the basis, the key genes obtained after intersection of results of the two methods are subjected to protein interaction network analysis, the gene with the most core of IAs formation and rupture is screened out, and the key genes with the core of IAs formation and rupture are screened out. The method realizes identification of potential key genes causing formation and rupture of IAs through a weight gene co-expression network analysis method.
Owner:AFFILIATED HUSN HOSPITAL OF FUDAN UNIV
A method for joint analysis of multi-omics data
The invention discloses a method for joint analysis of multi-omics data. The method for joint analysis of multi-omics data provided by the present invention includes the following steps: (A) performing co-expression network analysis on each single-omics index data in the multi-omics data to be analyzed, and finding respective expression modules; B) According to the overlapping relationship between the respective expression modules of different omics data, screen out the significantly correlated interaction modules among the multi-omics data to be analyzed. The multi-omics data joint analysis method provided by the present invention is not limited by the number of omics data, and theoretically can be any number of groups; at the same time, it does not depend on the source of the input data, as long as it is the index data that can measure the corresponding omics ( Such as gene expression value, apparent degree of methylation, SNP mutation rate, etc.), can be used as input data.
Owner:BGI TECH SOLUTIONS
Biological network clustering method and system based on high-order structure
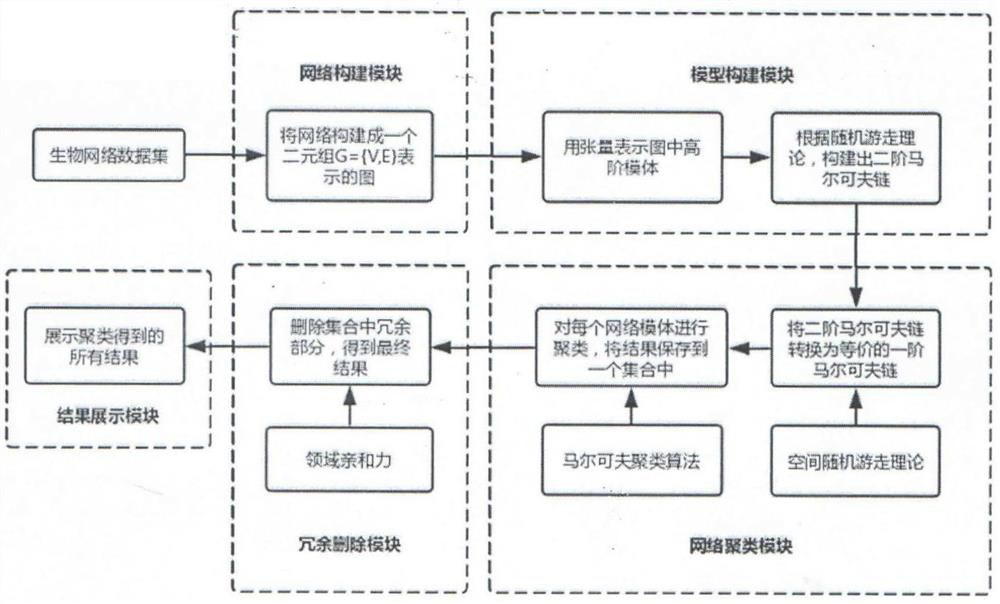
PendingCN113470737AEfficient clusteringHigh precisionBiostatisticsInstrumentsEngineeringSynexpression
The invention relates to a biological network clustering method and system based on a high-order structure. The system comprises a network construction module, a model construction module, a network clustering module, a redundancy deletion module and a result display module. Rich high-order structure information in a biological network is utilized to identify functional modules in the biological network, and clustering analysis can be performed on various types of network motifs by combining the advantages of a high-order Markov random process. The method has excellent performance; based onthe clustering result of the high-order structure information, a new thought is provided for biological network analysis, such as recognition of overlapping protein complexes and inference of new signal paths; and meanwhile, abundant organization structures presented in the biological network are disclosed. The biological network clustering method and system directly act on biological networks such as a protein interaction network and a gene co-expression network, are high in effect accuracy, and are a very reliable biological network clustering method and system.
Owner:XINJIANG TECHN INST OF PHYSICS & CHEM CHINESE ACAD OF SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com