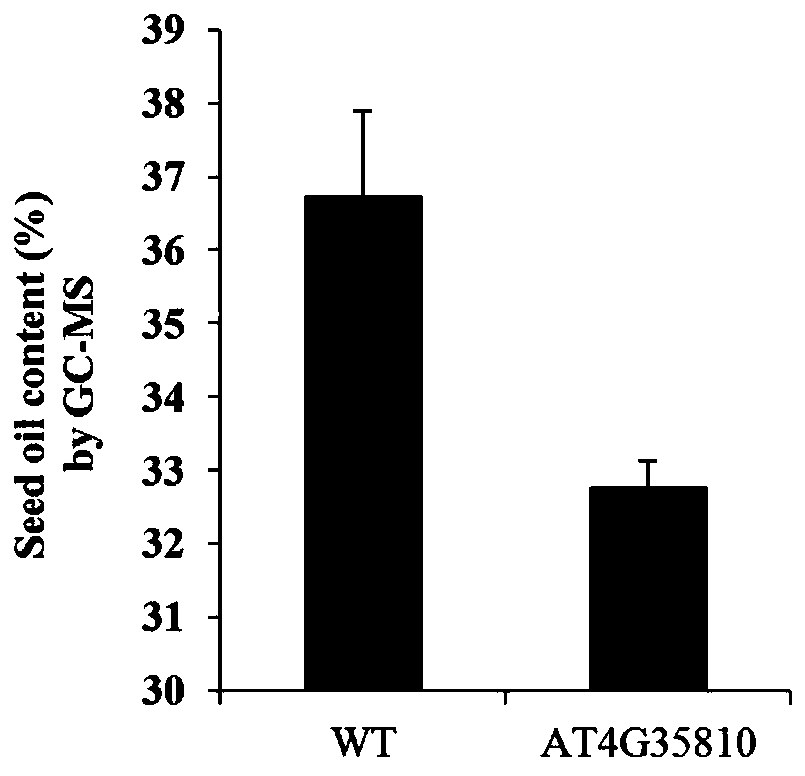
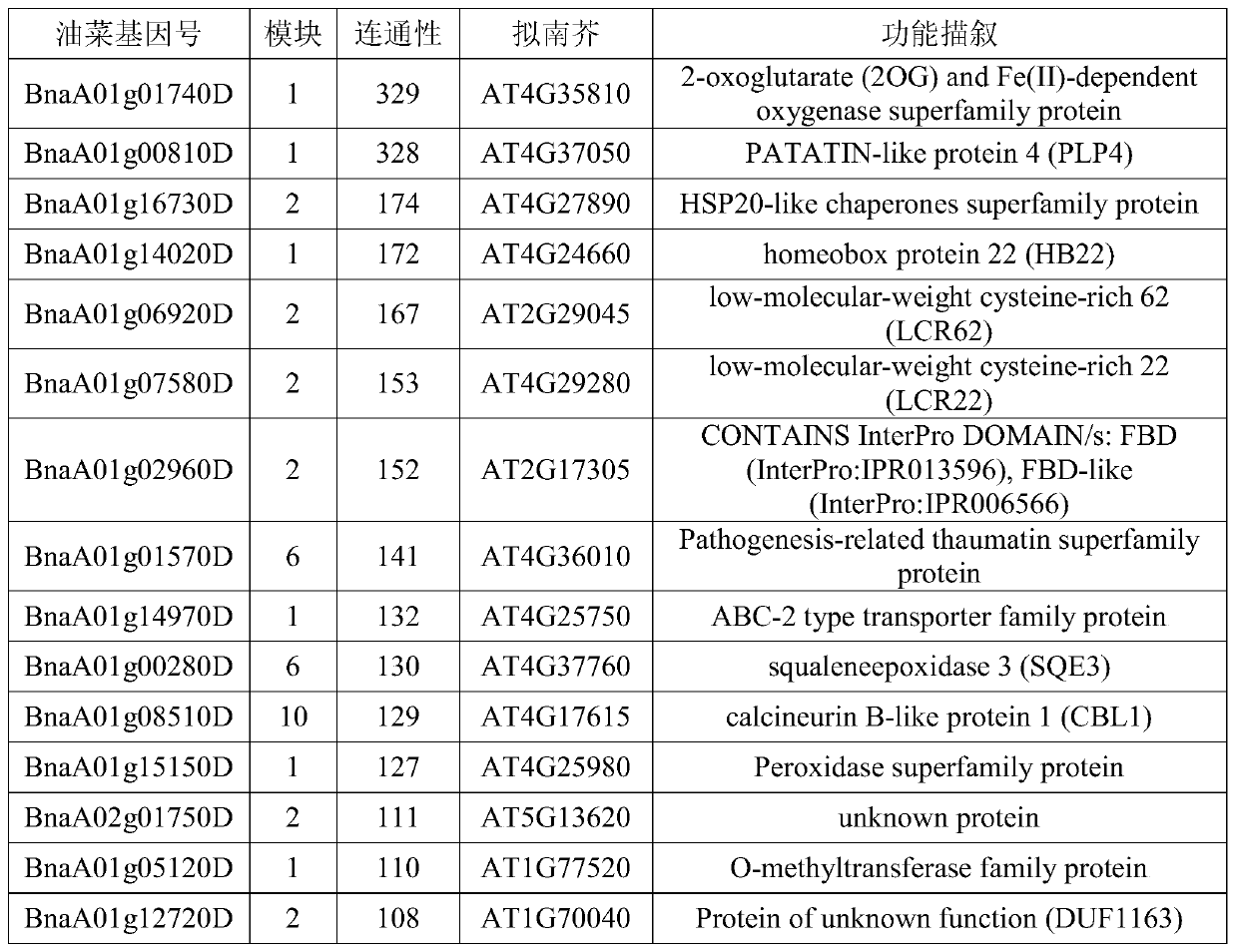
Method for screening quantitative character candidate genes
A technology for candidate genes and quantitative traits, applied in the fields of genomics, measurement devices, material separation, etc., can solve the problems of increased workload, false positive ratio, and high noise in co-expression analysis of candidate gene identification, so as to speed up the screening process, The effect of noise reduction and efficient screening
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] A method for rapidly screening candidate genes for quantitative traits of rapeseed oil is constructed through the following process:
[0024] 1. Natural population resequencing for GWAS analysis of oil content and fatty acid content
[0025] In this example, taking 157 materials with wide genetic sources and large differences in seed oil content and fatty acid content as examples, a natural population was constructed, and the method for obtaining candidate genes related to seed oil content was described in detail, as follows:
[0026] (1) Population planting and phenotype acquisition
[0027] Taking 157 materials with different genetic sources, seed oil content and fatty acid content as research materials, they were continuously planted in the field for 5 years (2012-2016). Each material was planted in 3 rows (24 plants in total), and randomized block design. The sowing period of Beibei in Chongqing is mid-to-late September, and field management is carried out by conv...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com