Subgroup-specific co-expression network identification method
A technology of co-expression and co-expression of genes, applied in the field of identification of subgroup-specific co-expression networks, which can solve the problems of sharp increase in memory and time consumption, less drastic changes in regulatory genes, and inapplicability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Example 1. Establishment of Subgroup-Specific Co-expression Network Identification Method
[0059] 1. Cells are divided into subpopulations based on gene expression
[0060] Obtain all gene expression amounts in all cells to be tested from database or detect (the present invention measures expression amount with UMI quantity); According to different expression amounts (UMI quantity) use Monocle2 (this method is recorded in following document: Trapnell C, Cacchiarelli D, Grimsby J, et al.The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells[J].Naturebiotechnology,2014,32(4):381.) clustered all the tested cells into different cell subpopulations .
[0061] 2. Establishment of identification method for cell subgroup-specific co-expression network
[0062] 1. Normalize the original UMI quantity of the gene
[0063] The original UMI (Uniquemolecular identifiers, UMI is the measurement method of expression) of each gene ...
Embodiment 2
[0101] Example 2, Application of Subgroup-Specific Co-expression Network Identification Method
[0102] 1. Cells are divided into subpopulations based on gene expression
[0103] The source is 4,000 cells of healthy human peripheral blood mononuclear cells (PBMC) published on the official website of 10x genomics as the cells to be tested, and the genetic data comes from:
[0104] https: / / support.10xgenomics.com / single-cell-gene-expression / datasets / 2.1.0 / pbmc4k.
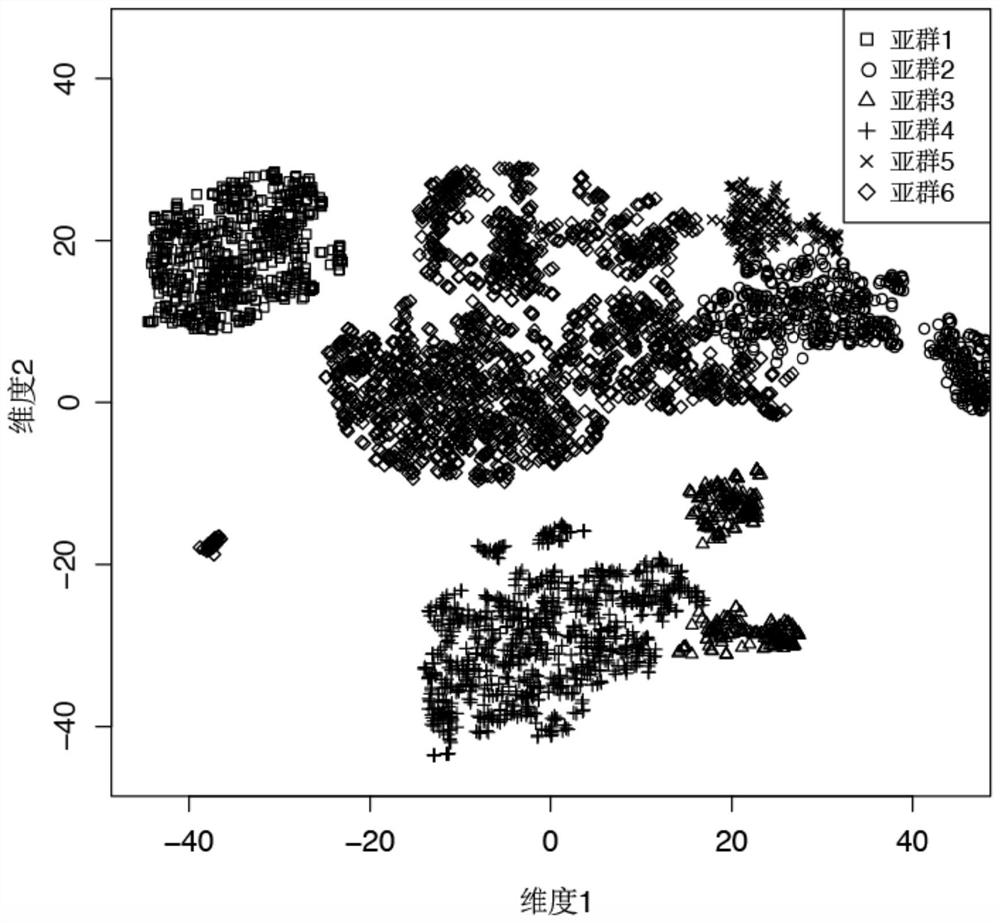
[0105] After clustering the gene expression data of the 4000 peripheral blood mononuclear cells (PBMC) of the above-mentioned healthy people with the Monocle2 method, they were divided into six cell subgroups according to the gene expression level in the cells, as shown in figure 1 shown.
[0106] 2. Establishment of identification method for cell subgroup-specific co-expression network
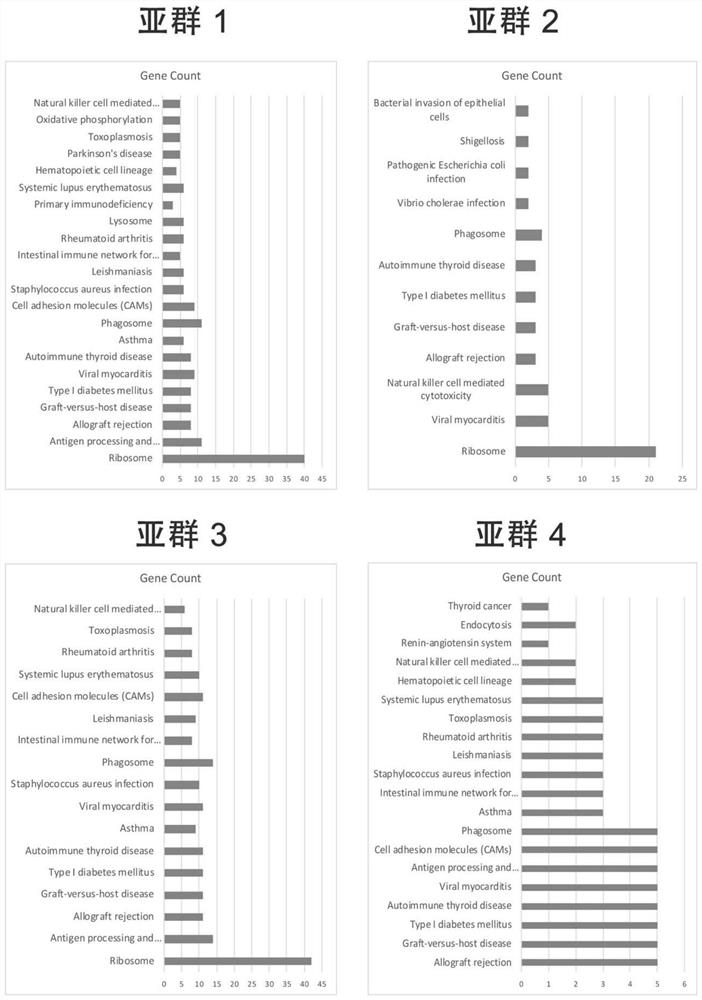
[0107] According to the second step in the method of Example 1, six cell subpopulation-specific co-expression networks were obtain...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com