A method and system for predicting cancer
A cancer and preset type technology, applied in the fields of bioinformatics and computational biology, can solve problems such as poor classification results, and achieve the effects of improving accuracy, efficient classification, and efficient prediction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
no. 1 example
[0045] This embodiment provides a method for predicting cancer, which can be implemented by an electronic device, and the electronic device can be a terminal or a server. The execution flow of this method is as follows figure 1 shown, including the following steps:
[0046] S101, performing difference analysis on gene expression profile data between cancer patients and normal people, and obtaining differential genes between cancer patients and normal people;
[0047] It should be noted that the data basis of this example is the expression profile data of differential genes between cancer patients and normal people. For this reason, this example uses the limma package in R language to realize the gene expression profile data of cancer patients and normal people. Differential expression analysis. limma (Linear Models for Microarray Data) is a robust T-test method based on empirical Bayesian, which has been implemented in the limma package of Bioconductor. The limma method is ...
no. 2 example
[0100] This embodiment provides a system for predicting cancer, and the system for predicting cancer includes the following modules:
[0101] The differential gene acquisition module is used for differential analysis of gene expression profile data between cancer patients and normal people, and obtains differential genes between cancer patients and normal people;
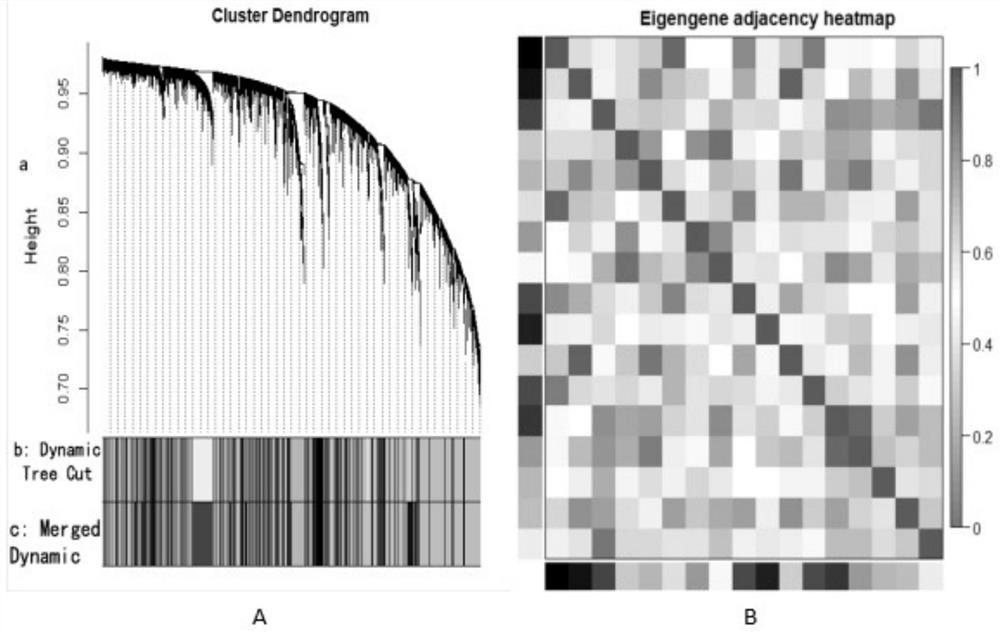
[0102] The feature data acquisition module is used to analyze the gene expression profile data of cancer patients and normal people based on weighted gene co-expression network analysis to obtain hub genes; Process the gene expression profile data of differential genes to obtain dimensionality reduction data;
[0103] The classification module is used to use the gene expression profile data and dimensionality reduction data of the hub genes acquired by the feature data acquisition module as the classification features of the preset type of cancer classifier, so as to realize the relationship between cancer patients ...
no. 3 example
[0106] This embodiment provides an electronic device, which includes a processor and a memory; at least one instruction is stored in the memory, and the instruction is loaded and executed by the processor, so as to implement the method of the first embodiment.
[0107] The electronic device may have relatively large differences due to different configurations or performances, and may include one or more processors (central processing units, CPU) and one or more memories, wherein at least one instruction is stored in the memory, so The above instructions are loaded by the processor and perform the following steps:
[0108] S101, performing difference analysis on gene expression profile data between cancer patients and normal people, and obtaining differential genes between cancer patients and normal people;
[0109] S102, analyze the gene expression profile data of cancer patients and normal people based on weighted gene co-expression network analysis to obtain hub genes; and p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com