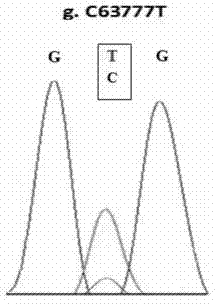
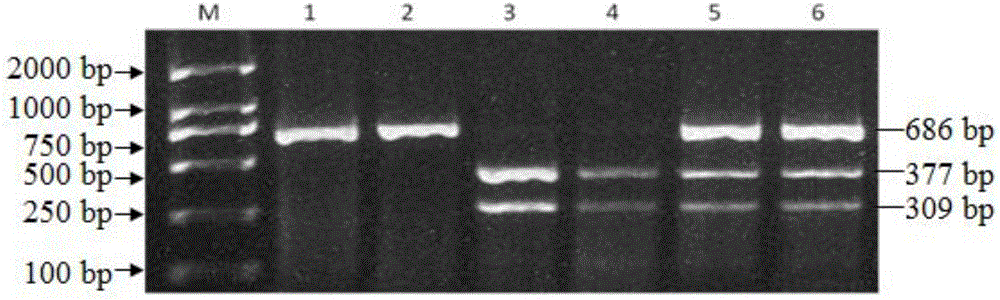
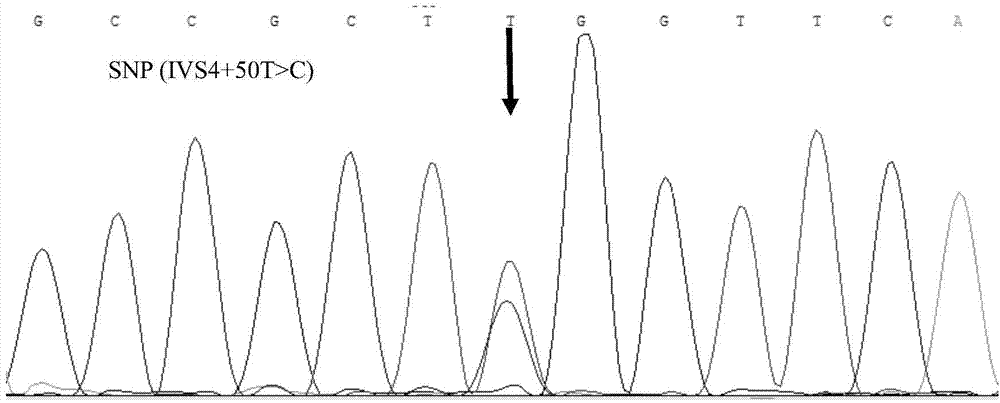
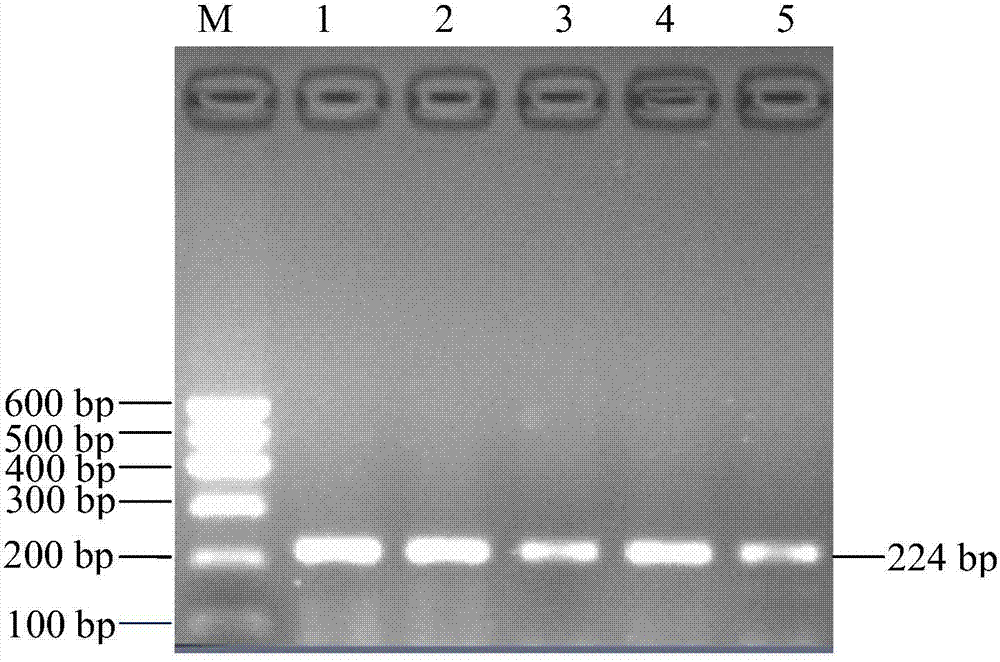
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
113results about How to "Excellent genetic resources" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
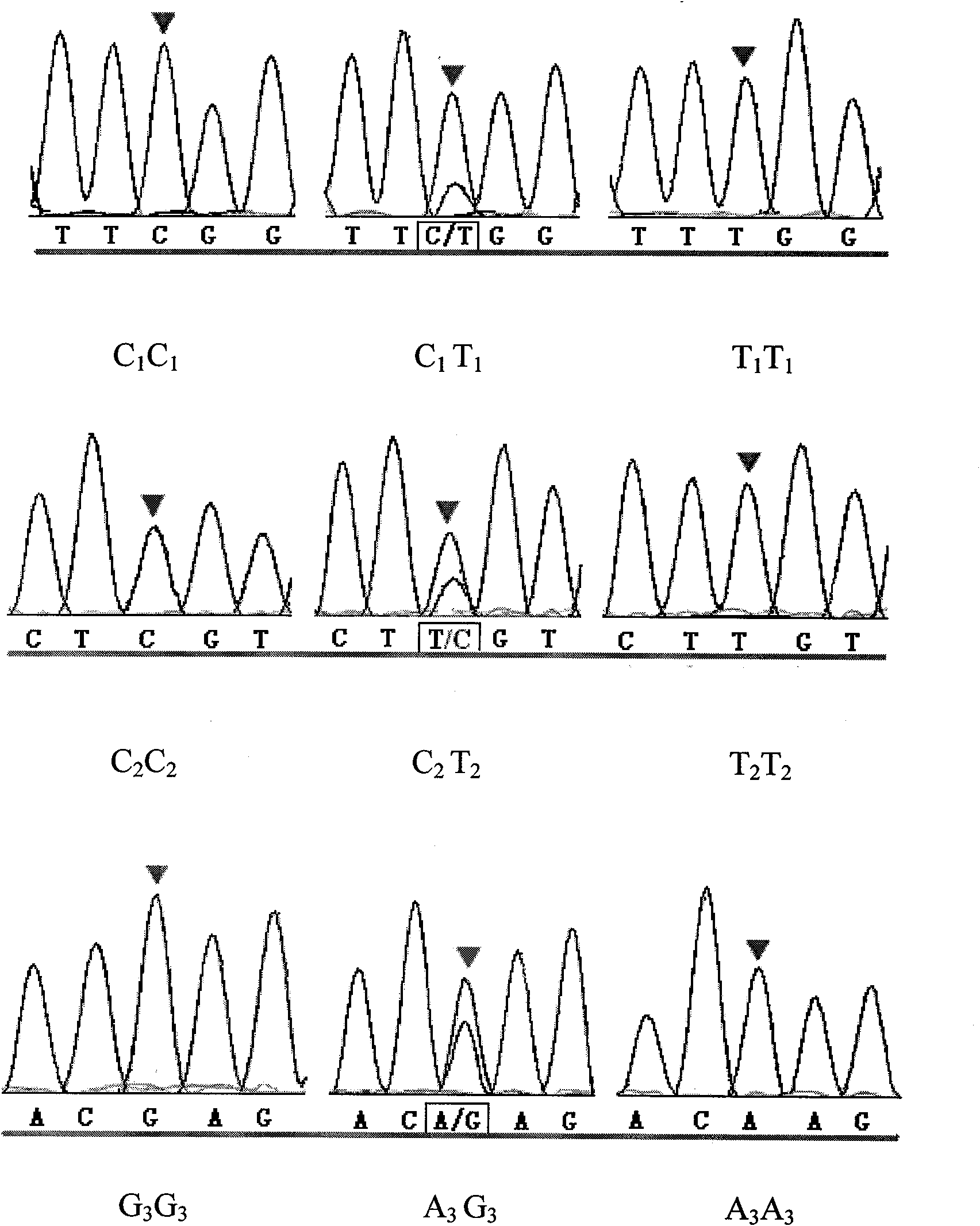
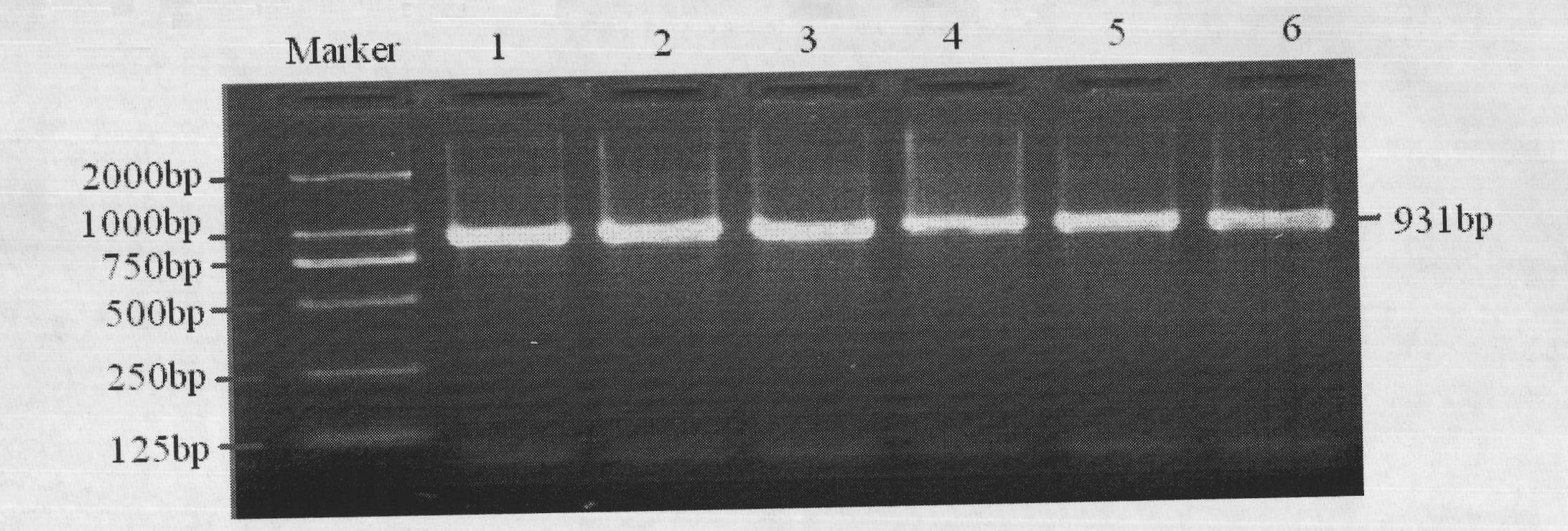
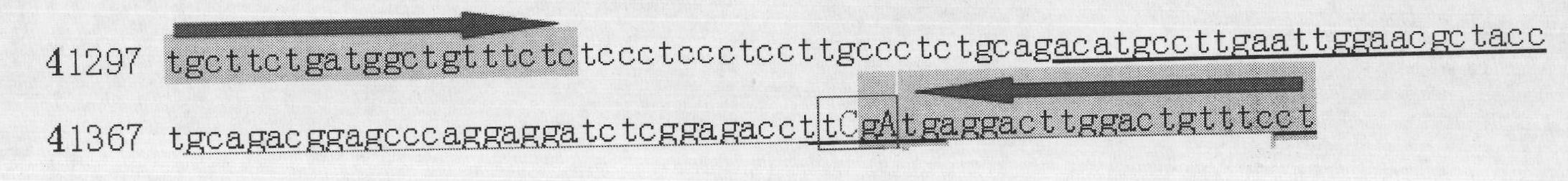
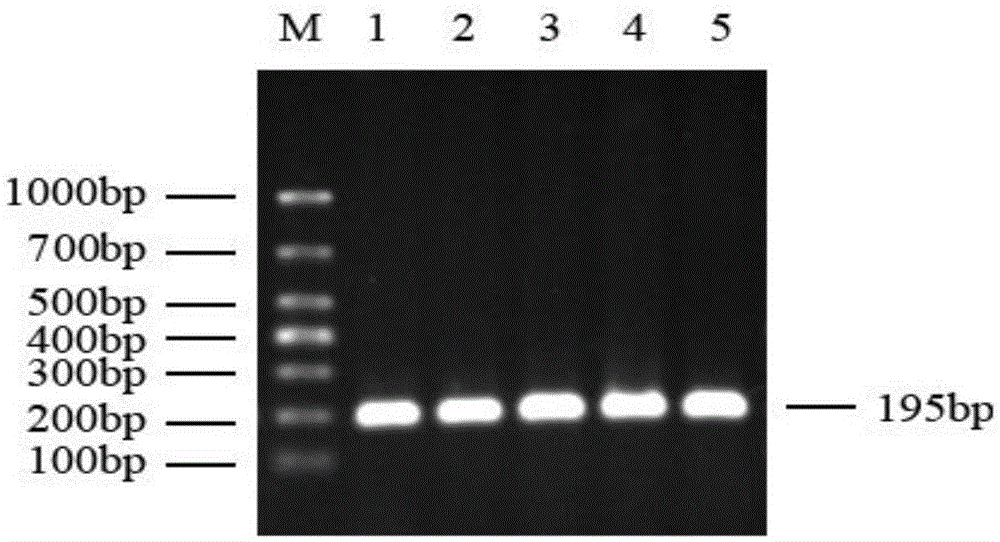
Method for quickly detecting cattle MLLT10 gene CNV marker and application of method
ActiveCN109943647AAccurate identificationLow costMicrobiological testing/measurementMarker-assisted selectionPcr ctpp
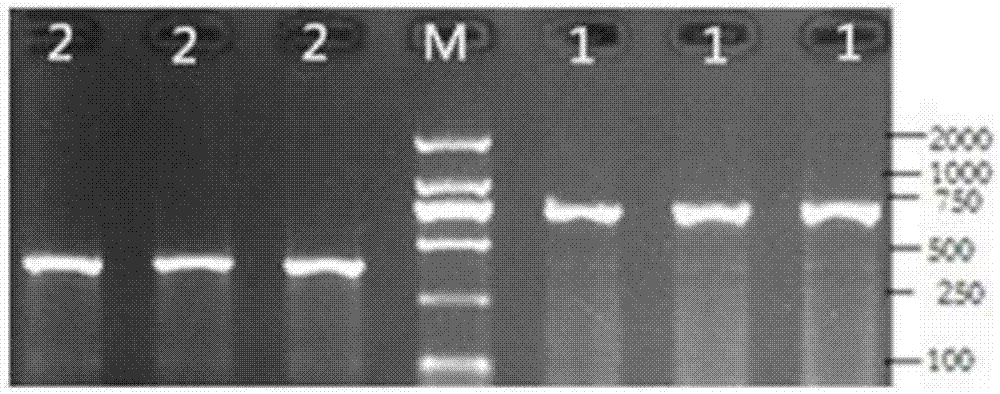
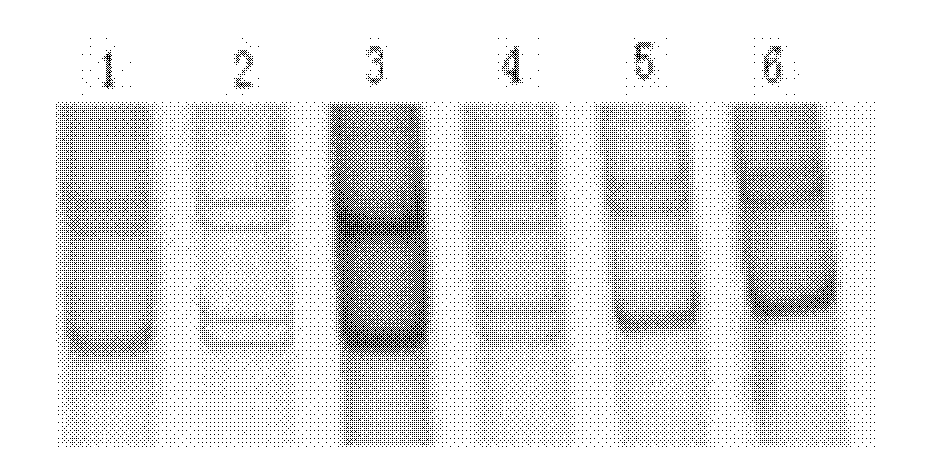
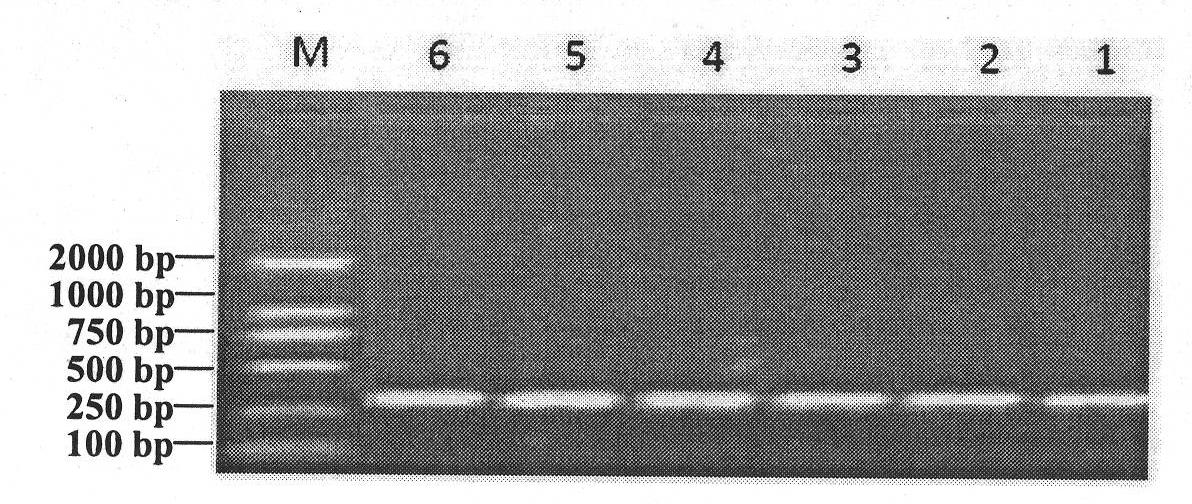
The invention discloses a method for quickly detecting a cattle MLLT10 gene CNV marker and application of the method. Based on the real-time fluorescent quantitation PCR technology, cattle genome DNAserves as a template, the cattle MLLT10 gene copy number variation area is amplified by using specific PCR primers, and partial fragments of the cattle BTF3 gene are amplified to serve as an internalreference, and finally, by using the 2-delta delta Ct method, the individual copy number variation type is computed and determined. The method lays a basis for the relationship between the cattle MLLT10 gene copy number variation and the growth trait, the detection method is simple and fast, the method can be used for accelerating cattle marker-assisted selection breeding work, and application andpopularization are facilitated.
Owner:NORTHWEST A & F UNIV
Method for detecting CNV (copy number variation) mark of MAPK10 gene of Nanyang cattle and application thereof
ActiveCN105506111AExcellent genetic resourcesFacilitates marker-assisted selectionMicrobiological testing/measurementReference genesMarker-assisted selection
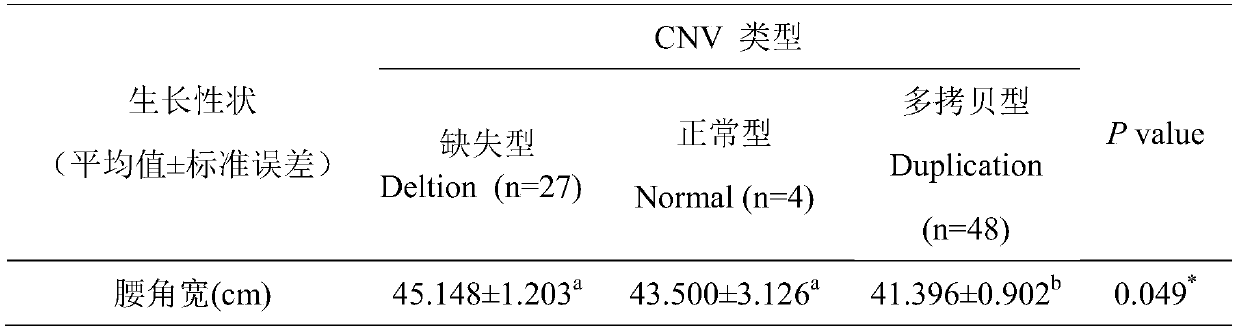
The invention discloses a method for detecting a CNV (copy number variation) mark of a MAPK10 gene of Nanyang cattle and an application thereof. The method comprises the following steps: respectively amplifying the CNV region and reference gene BTF3 of the MAPK10 gene of Nanyang cattle through a real-time fluorescence quantification PCR method by taking blood genome DNA of Nanyang cattle as a template; dividing the result into insert type, deficient type and normal type according to log22-delta delta Ct; and identifying CNV of the MAPK10 gene of Nanyang cattle. Analysis correlated to Nanyang cattle production data indicates that the growth and development of Nanyang cattle is remarkably influenced by different copy numbers of MAPK10, wherein the birth weight, 18-month-aged body height and 24-month-aged body weight and daily gain of a normal type individual body are remarkably higher than those of the insert type and deficient individual bodies. The CNV mark closely related to the production traits of Nanyang cattle is detected on DNA level, and can be used as an important candidate molecular mark for marker assisted selection of the growth traits of Nanyang cattle of China.
Owner:NORTHWEST A & F UNIV
Single nucleotide polymorphism of SCD genes in dairy goat and detection method thereof
InactiveCN101705290AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementElectrophoresisMolecular breeding
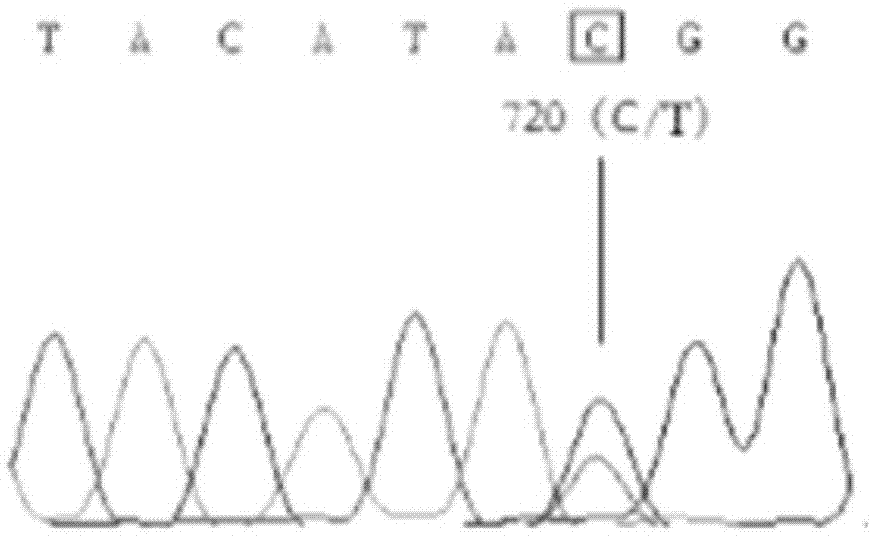
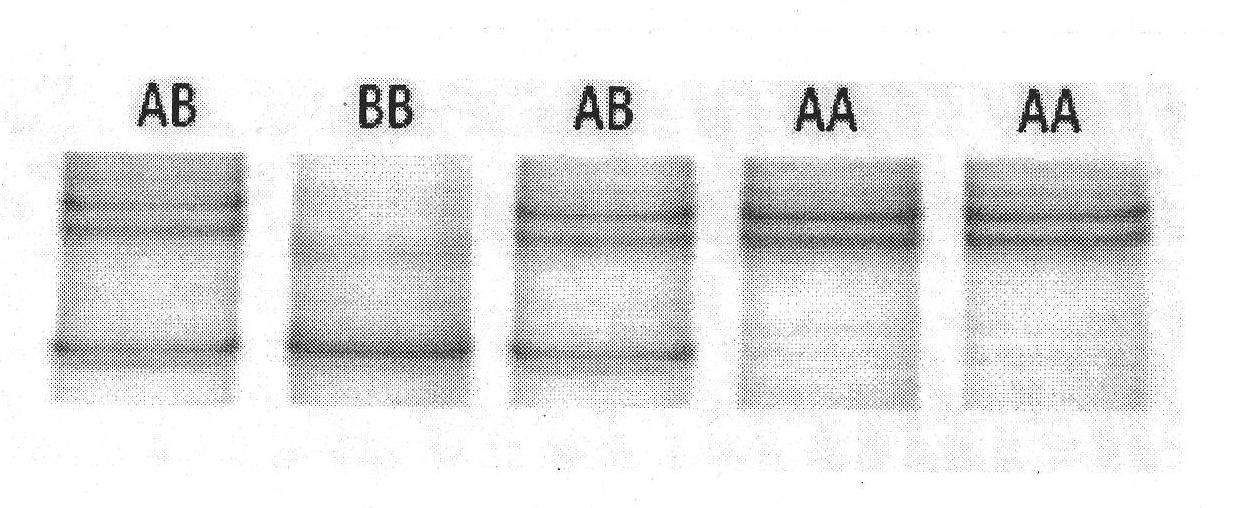
The invention discloses single nucleotide polymorphism of SCD genes in dairy goat and a detection method thereof. The single nucleotide polymorphism of the genes comprises G / A single nucleotide polymorphism which is the 5001st in the SCD genes in the dairy goat. The detection method comprises the following steps: taking the DNA of the whole genome of the dairy goat to be detected containing the SCD genes as a template and a primer pair P as a primer to carry out PCR amplification on the SCD genes in the dairy goat; after using restriction enzyme NdeI to digest the PCR amplified product, carrying out agarose gel electrophoresis on amplified fragments after restriction enzyme; and identifying the 5001bp base polymorphism of the SCD genes in the dairy goat according to the electrophoresis results. The method screens and detects the molecular genetic markers closely related to the milk production traits of the dairy goat on the DNA level to be used for assisted selection and molecular breeding of the dairy goat and speed up find breeding of the dairy goat.
Owner:NORTHWEST A & F UNIV
Soybean cyst nematode resistance gene and application thereof
InactiveCN102206651AEffective guidanceEffectively guide conventional breedingFungiBacteriaGlycineAgricultural science
The invention relates to a soybean cyst nematode resistance gene and application thereof. The cDNA (complementary deoxyribonucleic acid) fragment (SRC-J-6) provided by the invention is derived from soybean (Glycine max (L.) Merrill.) variety L-10, and has a nucleotide sequence shown in 1) a nucleotide sequence shown in SEQ ID NO. 1; 2) a DNA (deoxyribonucleic acid) molecule capable of hybridizingwith the nucleotide sequence shown in SEQ ID NO. 1 and being expressed under strict conditions; 3) an mRNA (messenger ribonucleic acid) molecule, which has homology of more than 90% with the nucleotide sequence shown in 1) and can be expressed; or 4) a DNA (deoxyribonucleic acid) molecule, which has homology of more than 90% with the nucleotide sequence shown in 1) and can be expressed. After theplant tissues are transformed by the gene disclosed by the invention, the resistance to cyst nematode of the transgenic soybean is significantly improved. The soybean cyst nematode resistance gene provided by the invention has important significance, not only can effectively guide conventional breeding, can also provide excellent genetic resources for the transgenic breeding of soybean and has broad application prospects in the soybean production.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
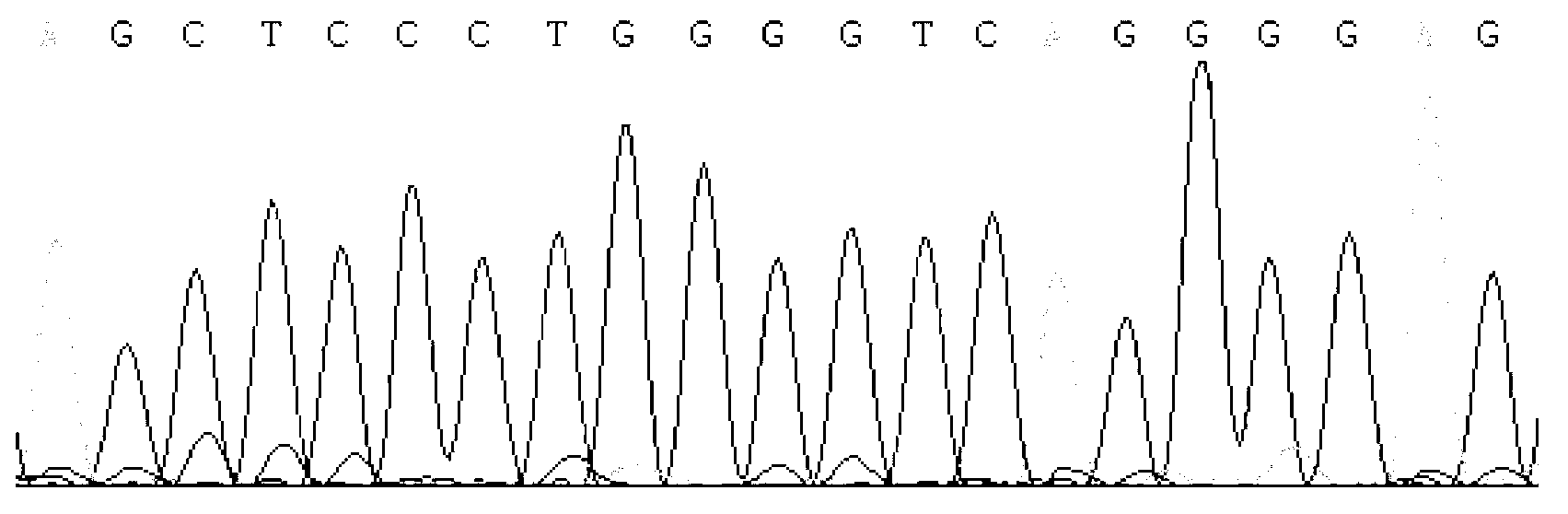
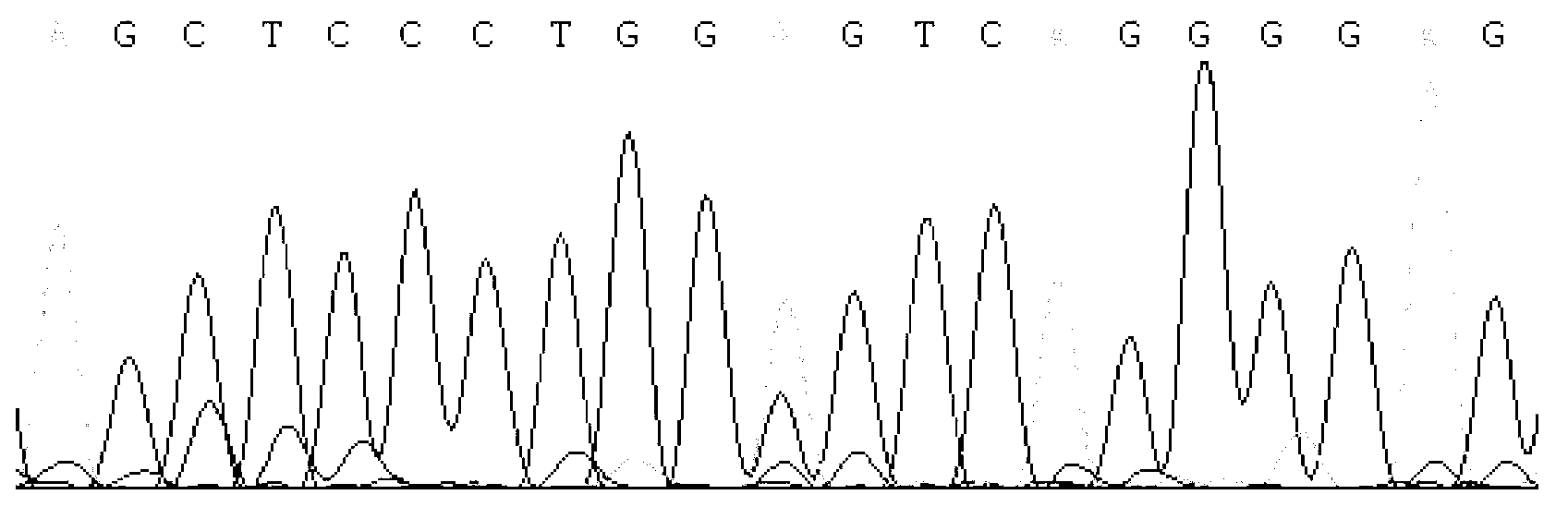
Detecting method and kit of cattle IGF2 (Insulin-like Growth Factor 2) gene mononucleotide polymorphism
InactiveCN103290123AAccurate detectionQuick checkMicrobiological testing/measurementMolecular geneticsRestriction fragment length polymorphism
The invention discloses a detecting method and a kit of cattle IGF2 (Insulin-like Growth Factor 2) gene mononucleotide polymorphism. The detecting method comprises the following steps of: with IGF2-gene-containing cattle whole genome DNA (deoxyribonucleic acid) to be detected as a template, carrying out PCR (Polymerase Chain Reaction) amplification on the cattle IGF2 gene; and detecting and quickly parting the gene polymorphism through DNA sequencing and polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technology. The detecting method is used for screening and detecting a molecular genetic marker closely related to cattle growth traits on DNA level. The detecting method disclosed by the invention can be used for molecular marker-assisted selection of cattle growth traits so as to quickly create cattle population with excellent genetic resources. The detecting method disclosed by the invention is simple, low in cost, direct and reliable in detecting result and fit for screening and diagnosing the cattle IGF2 gene mutation sites in large scale.
Owner:NORTHWEST A & F UNIV
Single nucleotide polymorphic locus of milk goat PITX1 gene, and detection method and application of single nucleotide polymorphic locus
InactiveCN102649956AAccurate detectionLow costMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionMarker-assisted selection
The invention relates to a single nucleotide polymorphic locus of a milk goat PITX1 gene, and a detection method and application of the single nucleotide polymorphic locus, and belongs to the technical field of animal molecular genetic breeding. The gene single nucleotide polymorphic locus comprises the single nucleotide polymorphic locus of the milk goat PITX1 gene with the 201st position is G or A, wherein the genotype is GG, GA and AA. The detection method comprises the following steps: performing polymerase chain reaction (PCR) amplification on the milk PITX1 gene by using the DNA sequence of the PITX1 gene as a template and a primer pair P as a primer; performing enzyme digestion on the PCR product by using restriction endonuclease MspI; and detecting the enzyme-digested product through agarose gel electrophoresis to identify the single nucleotide polymorphic locus of the milk goat PITX1 gene. Through the adoption of the method, the milk goat species with excellent genetic resources can be established quickly in marker assisted selection (MAS) breeding of the milk goat. The method has the advantages of simplicity and quickness in operation, low cost and high detection precision.
Owner:NORTHWEST A & F UNIV
Method for detecting single-nucleotide polymorphism of goat ATBF1 gene and application of goat ATBF1 gene
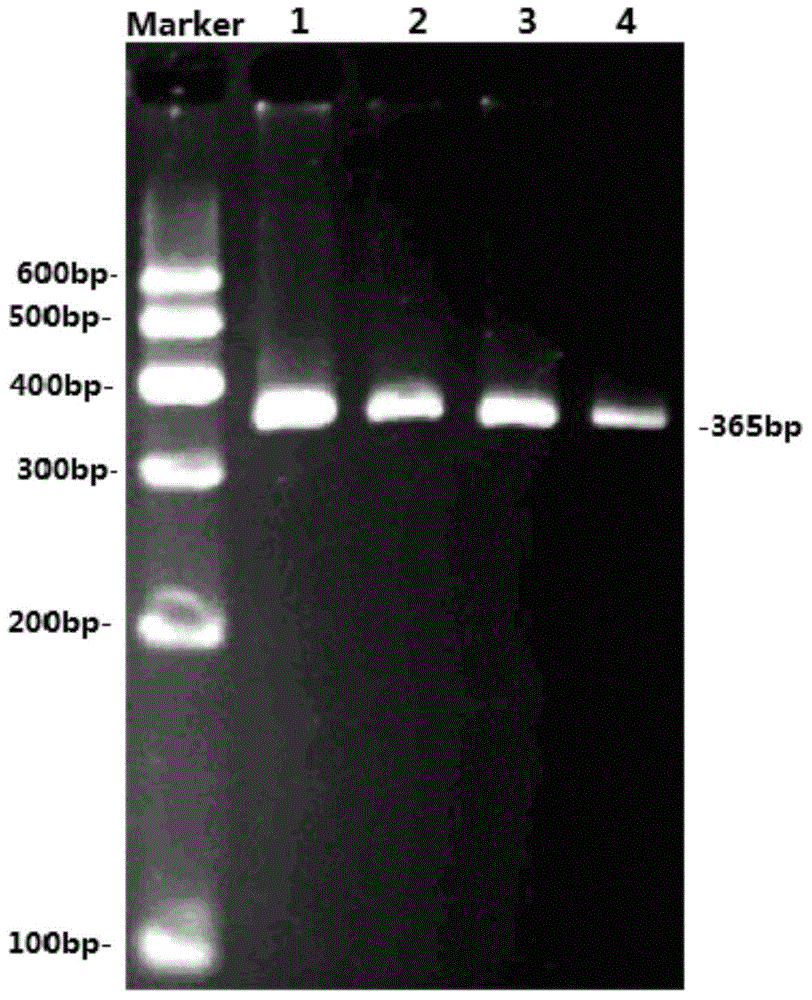
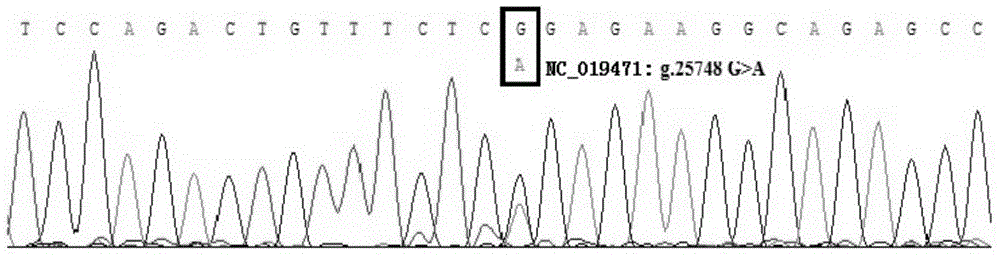
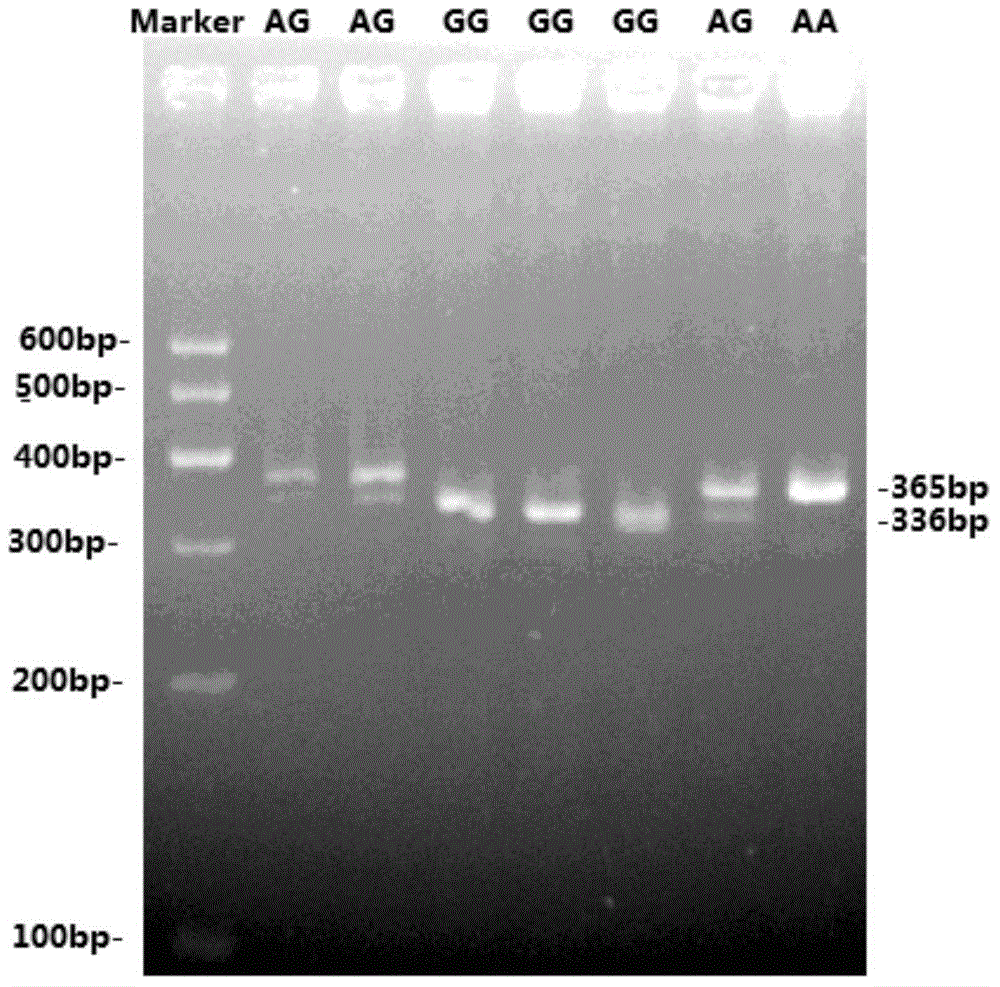
InactiveCN104878099AEasy to detectAccurate detectionMicrobiological testing/measurementDNA/RNA fragmentationElectrophoresisPcr ctpp
The invention discloses a method for detecting the single-nucleotide polymorphism of a goat ATBF1 gene and the application of the goat ATBF1 gene. According to the method, the goat whole-genome DNA to be tested is taken as a template, the goat ATBF1 gene is amplified by use of the PCR technology, the PCR product is digested by use of a restriction enzyme MspI or HpaII, next, the agarose gel electrophoresis is performed, and the genotype of No.25748nt or No.163517nt SNP of the goat ATBF1 gene is identified according to the electrophoresis result; different genotypes of the No.25748nt and No.163517nt SNPs of the ATBF1 gene have remarkable relevance with the growth and development traits of the goat, and therefore, the ATBF1 gene is advantageous for quickly establishing a good goat genetic resource population.
Owner:NORTHWEST A & F UNIV
Qinchuan cattle FoxO1 gene mononucleotide polymorphism molecular marker detection method and application
InactiveCN103233001AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementDNA/RNA fragmentationNucleotideElectrophoresis
The invention discloses cattle FoxO1 gene mononucleotide polymorphism and a detection method thereof. The gene mononucleotide polymorphism comprises that: with a Qinchuan cattle whole genome DNA comprising the FoxO1 gene as a template, and with a primer pair P as primers, the Qinchuan cattle FoxO1 gene is subjected to PCR amplification; a restriction endonuclease HhaI is used for digesting the PCR amplification product, and the digested segment is subjected to agarose gel electrophoresis; and according to the electrophoresis result, base polymorphism on the 178132 site of the Qinchuan cattle FoxO1 gene is identified. The FoxO1 gene is important for animal muscle fat traits and growth traits. With the method, the molecular genetic marker closely related to Qinchuan cattle growth traits is screened and detected on a DNA level. The method is used in Qinchuan cattle assisted selection and molecular breeding, and assists in accelerating Qinchuan cattle breeding speed.
Owner:NORTHWEST A & F UNIV
Method for detecting single nucleotide polymorphism (SNP) of cattle MGAT2 gene
InactiveCN101921848AAccurate detectionLow costMicrobiological testing/measurementNucleotideGel electrophoresis
The invention discloses a method for detecting the single nucleotide polymorphism (SNP) of a cattle MGAT2 gene. The method comprises the following steps of: carrying out PCR (Polymerase Chain Reaction) amplification on the cattle MGAT2 gene by using the DNA containing the cattle MGAT2 gene of a cattle gene genome to be detected as a template and a primer pair p as a primer; digesting a PCR amplification product by using a restriction enzyme HaeIII and then carrying out agarose gel electrophoresis (AGE) on an amplified fragment after enzyme cutting; and identifying the SNP of the 495th site of the cattle MGAT2 gene according to an AGE result. Because the functions of the MGAT2 gene relate to the properties of weight, daily gain growth, and the like, the detection method lays a foundation for establishing a relation between the SNP of the MGAT2 gene and the growth properties so as to facilitate the mark assisted selection of the growth properties for cattle beef in China for quickly establishing cattle populations with good genetic resources.
Owner:NORTHWEST A & F UNIV
Detection method for single nucleotide polymorphism of STMN1 gene of Beijing duck and molecular markers thereof
InactiveCN102816759AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementAnimal husbandryElectrophoresisExon
The invention discloses single nucleotide polymorphism of the STMN1 gene of a Beijing duck and a detection method thereof. The detection method for the single nucleotide polymorphism of the gene comprises the following steps: with whole genome DNA of a to-be-detected Beijing duck containing the STMN1 gene as a template and the primer pair P as primers, carrying out PCR amplification on the STMN1 gene of the Beijing duck; digesting a PCR amplification product with restriction endonuclease and then carrying out agarose gel electrophoresis on amplified segment after digestion; and identifying polymorphism of the 48th nucleotide base of exon 4 of the STMN1 gene of the Beijing duck according to results of electrophoresis. According to the invention, molecular genetic markers closely related to economic characters of the Beijing duck are screened and detected at the level of DNA, which provides molecular bases for rapid selective breeding of the Beijing duck.
Owner:XUZHOU NORMAL UNIVERSITY
Method for selecting molecular marker for goat yeaning traits
InactiveCN101899526AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementBiotechnologyPolymerase L
The invention discloses a method for selecting a molecular marker for goat yeaning traits, which comprises the following steps of: taking a goat genome DNA sequence as a template, amplifying KITL gene introns 1 and 6 by using primers P1 and P2 under a PCR condition in the presence of TaqDNA polymerase, buffer environment, Mg2+ and dNTPs respectively, and judging the size of a destination fragment according to an agarose gel electrophoresis result; digesting the PCR amplification product of the primer P1 by using restriction enzyme CviAII, and then detecting the enzyme digested amplification fragment by using polyacrylamide gel electrophoresis, wherein the amplification product of the primer P1 has mutation of two basic groups; detecting the PCR amplification product of the primer P2 by adopting the polyacrylamide gel electrophoresis, wherein the amplification product of the primer P2 has mutation of one basic group; and then performing gene analysis and gene frequency analysis on the amplification products of the primers P1 and P2, and performing association analysis between the amplification products and the yeaning numbers of Guanzhong dairy goats, western Saanen dairy goats and Boer goats, wherein the analysis results show that an SNPs site of the KITL gene, detected by the primers P1 and P2, can be taken as the molecular marker for goat yeaning trait selection.
Owner:NORTHWEST A & F UNIV
Detection method of yak FOXO1 gene single nucleotide polymorphism and kit thereof
ActiveCN105441567APrecise positioningSensitive positioningMicrobiological testing/measurementNucleotideA-DNA
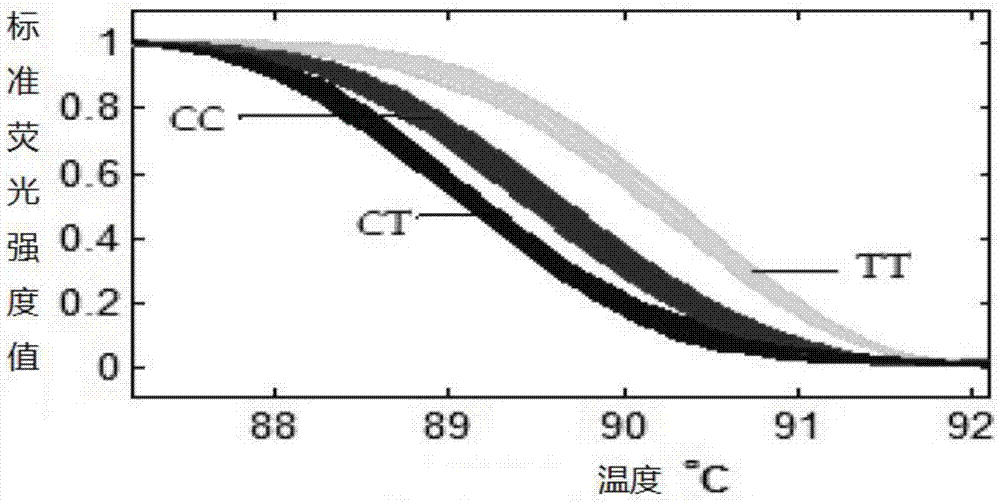
The invention relates to the technical field of gene polymorphism detection, in particular to a detection method of yak FOXO1 gene single nucleotide polymorphism and a kit thereof. The detection method comprises the following steps: 1) preparing a yak FOXO1 gene amplification product; 2) synthesizing a high and low temperature interior label; 3) preparing an HRM-PCR (High Resolution Melt-Polymerase Chain Reaction) amplification product; 4) collecting a fluorescence signal. Compared with an HRM method used for detecting polymorphic sites in the prior art, a gene probe designed by the detection method is more accurate and flexible in positioning, is accurate in parting, does not need PCR post-processing, has high working efficiency and good repeatability and is low in sample quality and quantity requirements, especially, the concentration requirement of a DNA template can be lowered to 0.1ng / mu L, and time and labor are saved when a sample size is large. Meanwhile, the detection kit has the advantages of high sensitiveness, high detection speed and good stability.
Owner:GANSU AGRI UNIV
Method for detecting single nucleotide polymorphism of goat signal transducer and activator of transcription (STAT3) gene and application of STAT3 gene
InactiveCN103866032ARapid establishment of genetic resources and excellent genetic resourcesExcellent genetic resourcesMicrobiological testing/measurementPopulationPolymerase chain reaction
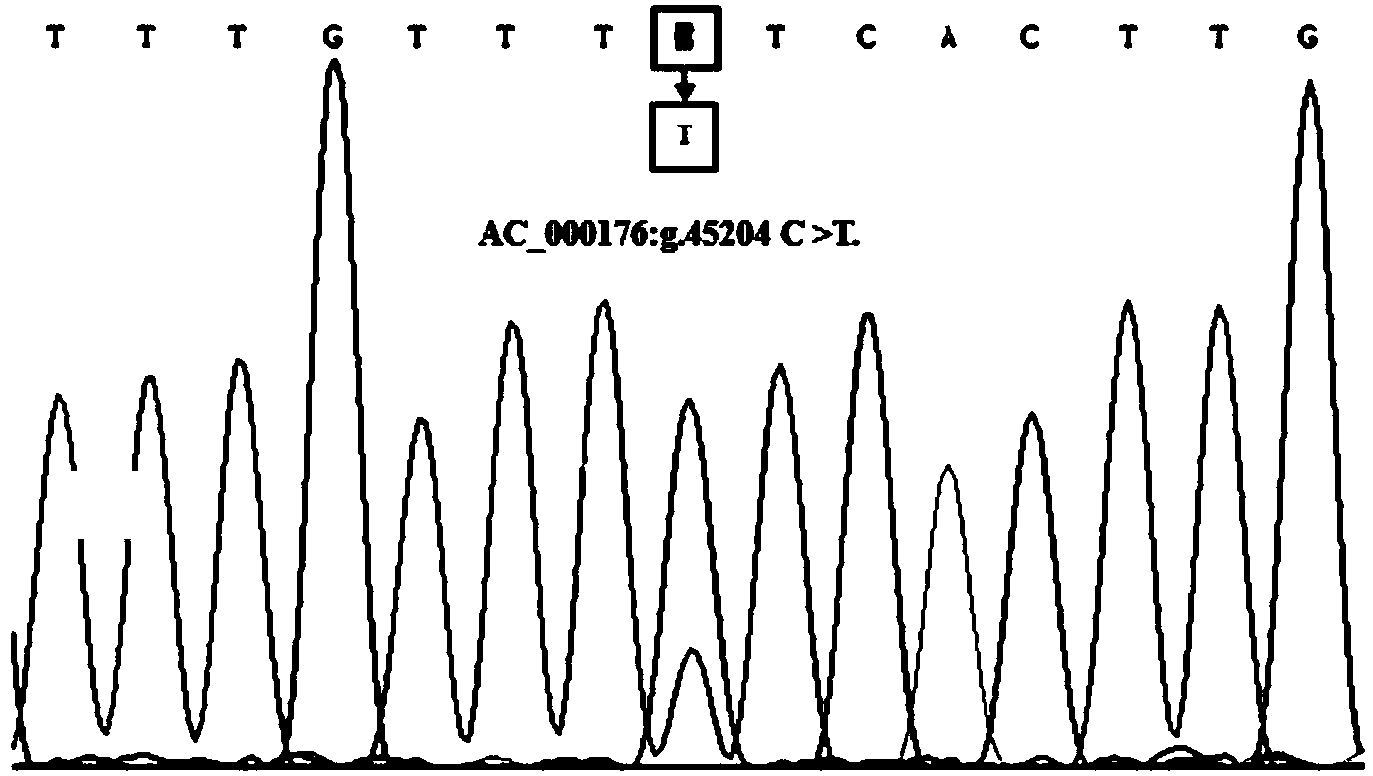
The invention discloses a method for detecting single nucleotide polymorphism of a goat signal transducer and activator of transcription (STAT3) gene and an application of the STAT3 gene. The method comprises the following steps: with a to-be-detected goat whole genome deoxyribonucleic acid (DNA) containing the STAT3 gene as a template and a primer pair P1, P2 or P3 as a primer, carrying out polymerase chain reaction (PCR) amplification on the goat STAT3 gene; digesting a PCR amplification product by using restriction enzyme DdeI, and then carrying out agarose gel electrophoresis on a digested amplification segment; and identifying the single nucleotide polymorphism of the 45204th site, the 62058th site or the 62230th site of the goat STAT3 gene according to the agarose gel electrophoresis result. Three sites can be used as molecular markers for improving the height of the xinong sannen diary goat, wherein one site is used as the molecular marker for improving cannon circumference of the Hainan black goat, and one site is used as the marker for improving the body length index of the Hainan black goat. Therefore, marker assisted selection (MAS) of growth characteristics of the native goats in China is facilitated, and quick building of goat populations with excellent genetic resources is also facilitated.
Owner:NORTHWEST A & F UNIV
Method for detecting Qinchuan cattle Wnt7a gene single nucleotide polymorphism, and application thereof
InactiveCN103320429AAccurate detectionLow costMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionMarker-assisted selection
The invention discloses a method for detecting Qinchuan cattle Wnt7a gene single nucleotide polymorphism (SNP). According to the invention, genome DNA of a Chinese local cattle breed Qinchuan cattle is adopted as a template; Wnt7a gene sequence is subjected to PCR amplification by using Taq DNA polymerase, Buffer (a buffer solution), Mg<2+>, dNTPs and primers M; a PCR product is subjected to enzyme digestion by using restriction endonuclease Alw44I; a fragment is subjected to separation and detection through agarose gel electrophoresis, and Qinchuan cattle Wnt7a gene single nucleotide polymorphism is finally determined. The Wnt7a gene function is related to growth traits such as weight and body length, such that the detection method provided by the invention establishes a basis for establishing a relationship between the SNP of the Wnt7a gene and growth traits. Therefore, marker-assisted selection of meat-purpose growth traits of Chinese Qinchuan cattle is facilitated, and a Qinchuan cattle population with excellent genetic resource can be rapidly established.
Owner:NORTHWEST A & F UNIV
Method for detecting single nucleotide polymorphism of sheep KITLG gene and application of method
ActiveCN106498078AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementKITLG GeneLitter
The invention discloses a method for detecting single nucleotide polymorphism of a sheep KITLG gene and application of the method. The method comprises the following steps: carrying out PCR amplification on fragments of the sheep KITLG gene in a manner of taking a whole-genome DNA of sheep to be detected as a template and taking a primer pair P as primers; digesting a PCR amplification product with restriction endonuclease SspI, and then, carrying out agarose gel electrophoresis on the amplified fragments subjected to digestion; identifying the polymorphism of a 47569-th base of the sheep KITLG gene according to electrophoresis results. The mutant site of the base is closely associated with reproduction traits, i.e., litter size of the sheep, so that the method is a method for detecting molecular genetic markers, which are closely related to the reproduction traits of the sheep, from a DNA level, and can be applied to the assisted selection and molecular breeding of the sheep, and thus, the good-variety breeding speed of the sheep is accelerated.
Owner:天津奥群羊业研究院有限公司
Molecular marking method of using neuroendocrine factor genes to select kidding characters
InactiveCN101906480AImprove selection efficiencySpeed up breeding progressMicrobiological testing/measurementSECRETOR FACTORGenomic DNA
The invention discloses a molecular marking method of using neuroendocrine factor genes to select kidding characters. The method comprises the following steps of: using a goat genomic DNA sequence as a template; amplifying Kisspeptin gene intron 2 by using a primer P1 under the PCR condition in the presence of TaqDNA polymerase, buffering environment, Mg2+, dNTPs, and then judging the size of the target fragment by agarose gel electrophoresis; detecting the PCR application product of the primer P1 by polyacrylamide gel electrophoresis, finding that the amplification SNPs of the primer P1 has one-base mutation and 8bp-base deletion, and then carrying out genotyping and gene frequency analysis on the amplification product of the primer P1, and the correlation analysis with the kidding characters of Guanzhong diary goat and Xinong Saanen dairy goat. Shown by experiments, the SNPs of the Kisspeptin gene intron 2 detected by the primer P1can be used as molecular marker of the selection of kidding characters.
Owner:NORTHWEST A & F UNIV
Single nucleotide polymorphism of GHRHR genes in dairy goat and detection method thereof
InactiveCN101705289AAccurate estimateImprove selection efficiencyMicrobiological testing/measurementEnzymeMolecular breeding
The invention discloses single nucleotide polymorphism of GHRHR genes in dairy goat and a detection method thereof. The single nucleotide polymorphism of the genes comprises C / G base polymorphism which is the 231st in the sequence table of the GHRHR gene in the dairy goat shown in SEQ ID NO.1. The detection method comprises the following steps: taking the DNA of the whole genome of the dairy goat to be detected containing the GHRHR genes as a template and a primer pair P as a primer to carry out PCR amplification on the GHRHR genes in the dairy goat; after using restriction enzyme MspI to digest the PCR amplified product, carrying out agarose gel electrophoresis on amplified fragments after restriction enzyme; and identifying the polymorphism of the genes according to the electrophoresis results. The method screens and detects the molecular genetic markers closely related to the milk production traits of the dairy goat on the DNA level to be used for assisted selection and molecular breeding of the dairy goat and speed up find breeding of the dairy goat.
Owner:NORTHWEST A & F UNIV
Molecular breeding method based on single nucleotide polymorphism of cattle Angpt18 genes
InactiveCN105671189AImprove growth traitsIncrease varietyMicrobiological testing/measurementMolecular breedingMetapopulation
The invention discloses a molecular breeding method based on the single nucleotide polymorphism of cattle Angpt18 genes.The molecular breeding method includes the steps that to-be-detected cattle whole genome DNA with the Angpt18 genes serves as a template, primer pairs P serve as primers, the cattle Angpt18 genes are subjected to PCR amplification, and PCR amplification products are obtained; the PCR amplification products are digested with restriction enzyme HinfI to obtain enzyme-digested product fragments, and the enzyme-digested product fragments are subjected to polyacrylamide gel electrophoresis; the polyacrylamide-gel-electrophoresis result shows that the 308th bits of the cattle Angpt18 genes have the single nucleotide polymorphism; meanwhile, the detecting result of the single nucleotide polymorphism of the cattle Angpt18 genes and the growth characteristics of a cattle are analyzed in a relevance mode, suitable genotypes are used for improving molecular breeding of the growth characteristics of the cattle, and cattle populations with the excellent genetic resources can be rapidly established.
Owner:XINYANG NORMAL UNIVERSITY
Method for detecting single nucleotide polymorphism of sheep PCNP (PEST-Containing Nuclear Protein) gene by using PCR-RFLP (Polymerase Chain Reaction-Restriction Fragment Length Polymorphism) and application of method
ActiveCN106755371AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementMolecular geneticsBiology
The invention discloses a method for detecting single nucleotide polymorphism of a sheep PCNP (PEST-Containing Nuclear Protein) gene by using PCR-RFLP (Polymerase Chain Reaction-Restriction Fragment Length Polymorphism) and application of the method. The method comprises the following steps: carrying out PCR amplification on a sheep PCNP gene segment by taking a sheep whole genome DNA to be measured as a template and taking a primer pair P as a primer; after a PCR amplification product is digested by a restriction enzyme SPhI, carrying out agarose gel electrophoresis on the digested amplified segment; authenticating the single nucleotide polymorphism of the 5019th site of the sheep PCNP gene according to an electrophoresis result. As a base mutation site is closely associated with the kidding number of sheep reproduction traits, the method is a method for detecting molecular genetic markers closely associated with the sheep reproduction traits at the DNA level, and can be used for auxiliary selection and molecular breeding of sheep and increasing the improved variety breeding speed of the sheep.
Owner:甘肃润牧生物工程有限责任公司
RFLP method and kit for detecting SNP locus of MEF2C gene of cattle
InactiveCN104498610AExcellent genetic resourcesEasy to operateMicrobiological testing/measurementMarker-assisted selectionMEF2C Gene
The invention discloses a RFLP method and a kit for detecting an SNP locus of an MEF2C gene of cattle. The method comprises the following steps: by taking cattle genome DNA or DNA containing MEF2C gene sequences as a template, performing PCR amplification, digesting PCR amplification products by using restriction endonuclease Hind III, performing agarose gel electrophoresis on digested segments, and identifying the single nucleotide polymorphism of the NEF2C gene of cattle. As the MEF2C gene plays a significant role in the muscle development process and is tightly related to cattle growth and meat quality property, and SNP is related to multiple significant economic properties of cattle, the detection method disclosed by the invention can be applied to auxiliary selection of cattle meat growth marks, is simple and rapid to operate, low in cost and high in detection precision, and has good advantages in establishing cattle population with excellent genetic resources.
Owner:NORTHWEST A & F UNIV
Method for detecting single nucleotide polymorphism of cattle krupple-like factor (KLF) 7 gene
InactiveCN101899500AExcellent genetic resourcesAccurately estimate breeding valuesMicrobiological testing/measurementEnzyme digestionNucleotide
The invention discloses a method for fast detecting the single nucleotide polymorphism (SNP) of a cattle krupple-like factor (KLF) 7 gene. The method comprises the following steps of: performing polymerase chain reaction (PCR) amplification on the cattle KLF7 gene by taking a whole genome DNA comprising a KLF7 gene of cattle to be detected as a template and the mixture of primer pairs P1, P2, P3 and the like in a mol ratio as a primer; after digesting a PCR amplification product by using a restriction enzyme TaqI, performing agarose gel electrophoresis on an amplified fragment subjected to enzyme digestion; and according to the result of the agarose gel electrophoresis, identifying the SNP of the 41041st, 42025th and 42075th positions of the cattle KLF7 gene. In the method, the SNPs of the KLF7 gene are subjected to genotyping and gene frequency analysis, and the growth traits of Nanyang cattle in different growth stages are subjected to trait association analysis. The results show that the SNP locus for the KLF7 gene, particularly P2, can serve as a molecular marker selected in the early stage (6 month age and 12 month age) of the growth traits of the cattle.
Owner:NORTHWEST A & F UNIV
Detection method for single nucleotide polymorphism of cattle PPARbeta gene and molecular breeding method
ActiveCN105543362AExcellent genetic resourcesMicrobiological testing/measurementPopulationAgricultural science
The invention discloses a detection method for single nucleotide polymorphism of a cattle PPARbeta gene. The detection method comprises the steps that PCR amplification is performed on the cattle PPARbeta gene by taking to-be-detected cattle whole genome DNA containing the PPARbeta gene as a template and taking a primer pair (P) as primers; a PCR amplification product is digested with a restriction enzyme (Pvu II), and unmodified polyacrylamide gel electrophoresis is performed on segments obtained through enzyme digestion; the polymorphism of the 71616 single nucleotide of the cattle PPARbeta gene is identified according to an electrophoresis result. The invention further relates to a method for constructing a cattle molecular breeding index system by means of the single nucleotide polymorphism of the PPARbeta gene. Due to the fact that the PPARbeta gene has the important biological function, the gene single nucleotide polymorphism has the significant influence on the weight, the withers height and the ischium end width of a cattle in the early stage and can be used for cattle beef and molecular marker-assisted selection of growth characteristics, and then a cattle population which has excellent genetic resources can be quickly constructed.
Owner:XINYANG NORMAL UNIVERSITY
Method for detecting single nucleotide polymorphisms of cattle 17HSDB8 gene
InactiveCN104278083AUnique technical effectImprove molecular markersMicrobiological testing/measurementNucleotideWeight gain
The invention discloses a method for detecting the single nucleotide polymorphisms of a cattle 17HSDB8 gene. The method comprises the following steps: by taking a 17HSDB8 gene-containing cattle genome DNA to be detected as a template and P as a primer, amplifying the cattle 17HSDB8 gene by using a PCR method, then digesting a PCR amplification product by using restriction endonuclease Sma I, then performing polyacrylamide gel electrophoresis to the enzyme-digested segments, and identifying the single nucleotide polymorphisms at 153th locus of the cattle 17HSDB8 gene according to the electrophoresis result. The 17HSDB8 gene has important biological functions on the growth and development characters, and a basis is established for association of the SNP of the gene with the growth character. The single nucleotide polymorphisms of the cattle 17HSDB8 gene has remarkable effect on the early-stage body weight, standing height, body length, chest girth and daily weight gain of a cattle, and can be used as the molecular marker-assisted selection of cattle beef and growth characters so as to further fast establish a cattle population with excellent genetic resources.
Owner:XINYANG NORMAL UNIVERSITY
Boar StAR gene 5-bp repetitive deletion polymorphism detection method and application
InactiveCN108410997ALow costAccurate detectionMicrobiological testing/measurementDNA/RNA fragmentationBOARGenotype
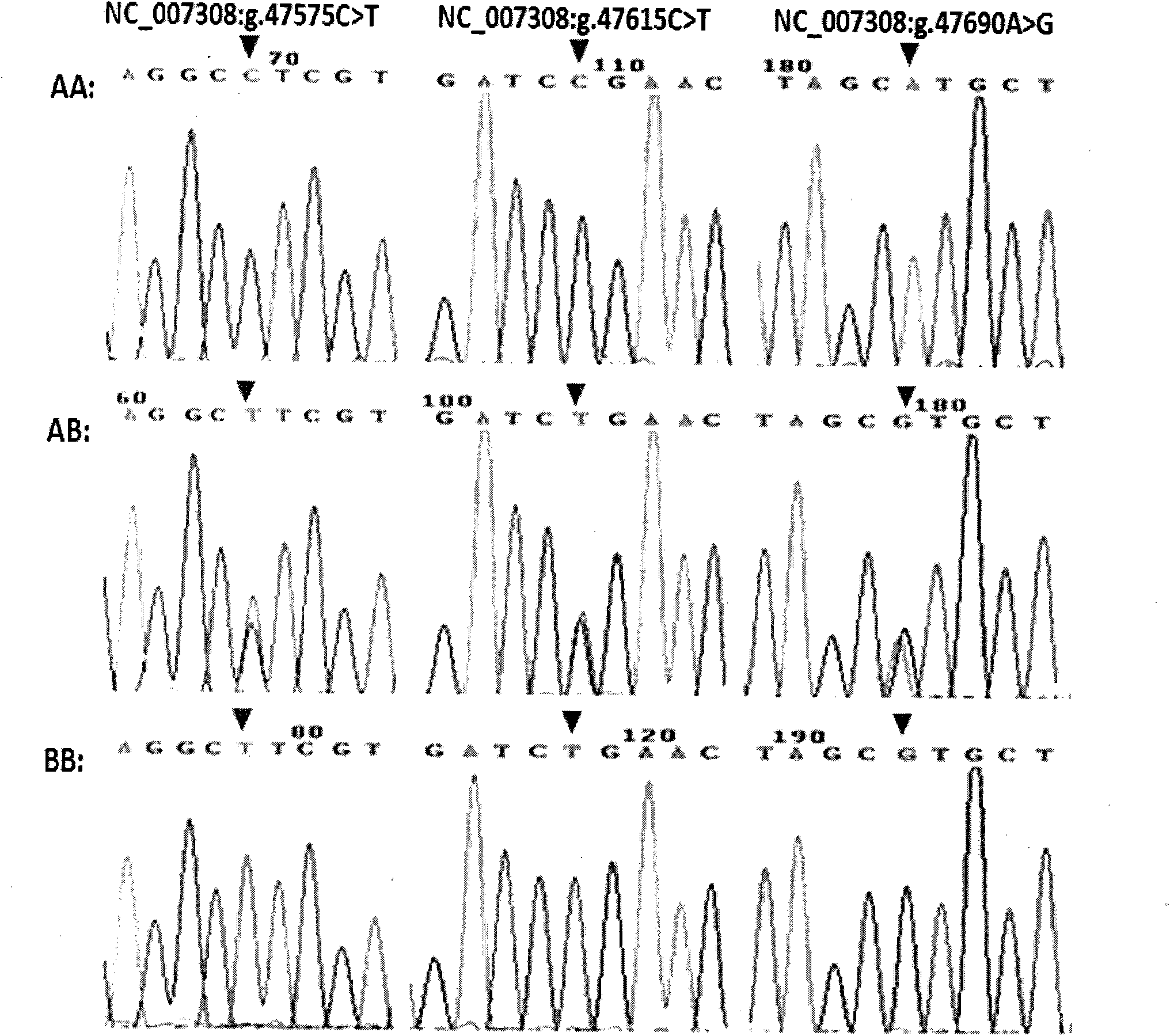
The invention discloses a boar StAR gene 5-bp repetitive deletion polymorphism detection method and application. The method includes: taking to-be-detected boar whole-genome DNA as a template to amplify a StAR gene segment by means of PCR (polymerase chain reaction); performing agarose gel electrophoresis, and authenticating 5-bp repetitive deletion polymorphism of StAR gene at NC_010457.5:g.5524-5528 locus according to an electrophoresis result. Different genotypes of StAR gene 5-bp repetitive deletion polymorphism are remarkably associated with reproductive traits including testicle long axis length, testicle short axis length, testicle weight and the like of large white boars at ages of 15 days and 40 days. By a boar StAR gene insertion / deletion method, application in marker assisted selective breeding of boar reproductive traits can be realized, and boar genetic resource groups excellent in reproductive performance can be quickly established.
Owner:NORTHWEST A & F UNIV
Method for detecting mononucleotide polymorphism of scalper SREBP1c gene
InactiveCN101875977AAccurate detectionQuick checkMicrobiological testing/measurementEnzyme digestionMarker-assisted selection
The invention discloses a method for detecting mononucleotide polymorphism of a scalper SREBP1c gene, which comprises the following steps of: using a scalper whole genome DNA to be detected containing the SREBP1c gene as a template, using a primer pair P as a primer, and performing PCR amplification on the scalper SREBP1c gene; using a restriction endonuclease Bg1 I to digest a PCR amplification product, and then performing sepharose gel electrophoresis on an amplified fragment after enzyme digestion; and identifying the mononucleotide polymorphism of the 10914th bit of the scalper SREBP1c gene according to the result of the sepharose gel electrophoresis. Because the functions of the SREBP1c gene relate to four important growth traits of body weight, daily gain, body length and chest circumference, the detection method provided by the invention lays a good foundation for establishing a relation between SNP and the growth traits of the SREBP1c gene, and is convenient to be used in a marker assistant selection (MAS) of the growth traits of Chinese scalper beef to quickly establish a scalper population with an excellent genetic resource.
Owner:NORTHWEST A & F UNIV
Method for detecting base mutation polymorphism of goat gonadotropin releasing hormone and growth differentiation factor 9
InactiveCN101845507AAccurately estimate breeding valuesImprove selection efficiencyMicrobiological testing/measurementMaterial analysis by electric/magnetic meansBiotechnologyIntein
The invention discloses a method for detecting the polymorphism of three base mutations of the gene exon 4 of goat gonadotropin releasing hormone and the gene exon 2 of the growth differentiation factor 9. The method comprises the following steps: using the DNA sequence of goat genome as template, using 2 to amplify the primer P1 and P2 under PCR condition in the presence of Taq DNA polymerase, buffer environment, Mg2+ and dNTPs, using agarose gel electrophoresis to judge the size of the target fragments; using polyacrylamide gel electrophoresis to detect the PCR amplified product of the primer P1 and P2 to find that the two amplification sites have three base mutations, then performing genotyping and gene frequency analysis to the SNPs of the two genes, and performing association analysis with the fecundity traits of Boer goat and Xinong Saanen dairy goat. The result shows that the SNPs sites detected by the primer P1 and P2, of the GnRH gene exon 4 (containing partial intron 3 and 3' non-translational region) and the GDF9 gene exon 2, thus the method can be used for molecular marking of fecundity trait selection of goat.
Owner:NORTHWEST A & F UNIV
Method for detecting single nucleotide polymorphisms of AQP9 genes in native Chinese cattle
InactiveCN101985656AExcellent genetic resourcesEasy to detectMicrobiological testing/measurementPopulationGenomic DNA
The invention discloses a method for detecting single nucleotide polymorphisms of AQP9 genes in native Chinese cattle, which is characterized by taking genomic DNA of the cattle to be detected containing the AQP9 genes as the template and the specific primer pair P6 as the primers and amplifying the exons 6 of the AQP9 genes in the cattle by PCR; and carrying out SSCP detection on the segments of the amplified products and sequencing the products. As the functions of the AQP9 genes relate to such growth traits as weight, height, length, chest circumference and the like, the method screens and detects the molecular genetic markers closely related to the growth traits of the cattle on the DNA level so as to be used for marker-assisted selection (MAS) of the growth traits for the Chinese cattle meat and rapidly build the cattle population with excellent genetic resources.
Owner:XUZHOU NORMAL UNIVERSITY
Method for detecting single nucleotide polymorphism of cattle PPARGC1A genes
InactiveCN102888451AEasy to detectAccurate detectionMicrobiological testing/measurementSingle-strand conformation polymorphismGenomic DNA
The invention discloses a method for detecting single nucleotide polymorphism of cattle PPARGC1A genes. The method comprises the steps: taking to-be-detected cattle genomic DNA containing PPARGC1A genes as a formwork and a primer pair P as a primer, conducting PCR (Polymerase Chain Reaction) application to the cattle PPARGC1A genes, and then carrying out agarose gel electrophoresis to detect the size of the amplified product segment; and detecting the SSCP (Single Strand Conformation Polymorphism). The method is used for screening and detecting molecular genetic marker closely related with growth traits of cattle on the DNA level, and therefore, the detection method provided by the invention can be used for molecular marker-assisted selection of Chinese cattle beef and growth traits, so that cattle crowds with excellent genetic resources can be further established.
Owner:NORTHWEST A & F UNIV
Racomitrium canescens protein phosphatase 2C gene RcPP2C as well as encoded protein and application thereof
ActiveCN107746851AImprove stress resistanceExcellent genetic resourcesHydrolasesFermentationDrought resistanceMolecular breeding
The invention relates to a racomitrium canescens protein phosphatase 2C gene RcPP2C as well as encoded protein and an application thereof. The invention aims at providing the racomitrium canescens protein phosphatase 2C gene RcPP2C as well as the encoded protein and the application thereof. The sequence of the racomitrium canescens protein phosphatase 2C gene RcPP2C is shown as SEQ ID No: 1 in a sequence list, and an amino acid sequence is shown as SEQ ID No.2 in the sequence list; and the gene is applied to the improvement of plant drought resistance. The RcPP2C gene is related to drought resistance in a cell signal transduction pathway; the gene, when transformed into a plant, can improve the drought resistance of the plant and the gene can lay a foundation for breeding a novel drought-resistant plant variety and enhancing the drought resistance of the plant; therefore, the gene has important significance. The gene provided by the invention is applied to the field of molecular breeding.
Owner:QIQIHAR UNIVERSITY
Method for rapidly detecting polymorphism of sugarcane sucrose phosphate synthase B (SPSB) gene and application of method
InactiveCN104561365APrimer specificityImprove accuracyMicrobiological testing/measurementSucroseMarker-assisted selection
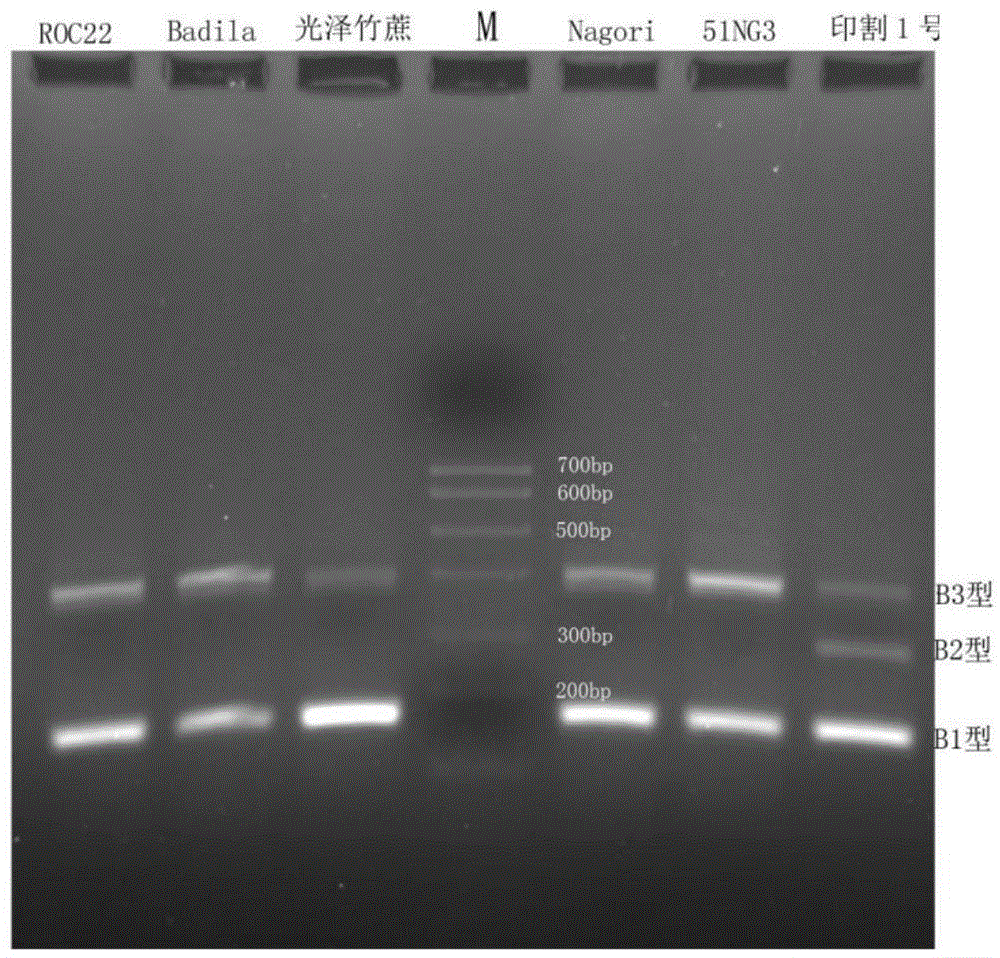
The invention discloses a method for rapidly detecting the polymorphism of a sugarcane sucrose phosphate synthase B (SPSB) gene. The method comprises the steps of designing a pair of primers to carry out PCR amplification by taking the whole-genome DNA of saccharum robustum B.51NG3, saccharum species ROC22, saccharum.officinarum L.Badila, saccharum sinense R. lustrous bamboocane, saccharum barberi J.Nagori or saccharum spontaneum L.yinge No.1 as a template; and carrying out agarose gel detection to name the SPSB genotype of a fragment with the size of 175bp as type B1, name the SPSB genotype of a fragment with the size of 294bp as type B2 and name the SPSB genotype of a fragment with the size of 421bp as type B3. The invention provides a DNA level identifying method for saccharum sugar trait marker-assisted selection, and the DNA level identifying method is beneficial to tracking of different haplotypes of SPSB genes in filial generations of saccharum and has important significance for rapidly establishing saccharum germplasm with excellent heredity.
Owner:SUGARCANE RES INST OF YUNNAN ACADEMY OF AGRI SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com