Method for selecting molecular marker for goat yeaning traits
A technology of molecular markers and goats, which is applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of expensive detection, non-existence, cumbersome operation, etc., to speed up breeding progress, accurately estimate, and improve selection efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
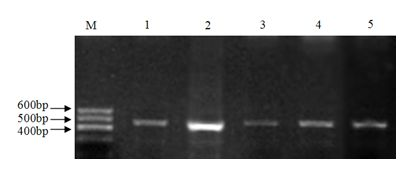
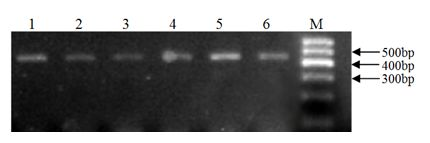
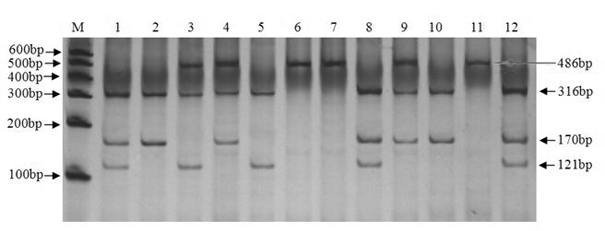
[0037] A. PCR amplification of intron 1 and 6 of KITL gene and detection of its polymorphism
[0038] 1. Collection and processing of goat blood samples
[0039] Take 5 mL of goat blood sample, add 0.2 μL of ACD (2.4 g of citric acid; 6.6 g of trisodium citrate; 7.35 g of glucose; dilute to 50 mL, autoclave) for anticoagulation, slowly invert 3 times and put it in an ice box, Store at -80°C for later use.
[0040] In this example, a total of 681 ewe blood samples from 3 goat populations were used, specifically: 290 blood samples of Xinong Saneng sheep, collected from Qianyang Saneng sheep breeding farm in Shaanxi Province; 193 blood samples of Guanzhong dairy goats, collected from Shaanxi Zhouzhi Green New Century Biological Co., Ltd.; 198 Boer goat blood samples were collected from the Boer goat breeding farm in Linyou County, Shaanxi.
[0041] 2. Extraction and purification of genomic DNA from blood samples
[0042] 1) Thaw the frozen blood sample at room temperatur...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com