Method for detecting CNV (copy number variation) mark of MAPK10 gene of Nanyang cattle and application thereof
A gene and marker technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as the influence of Nanyang cattle growth traits and gene function that have not been seen, and achieve the effect of simple operation and accurate and reliable method.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
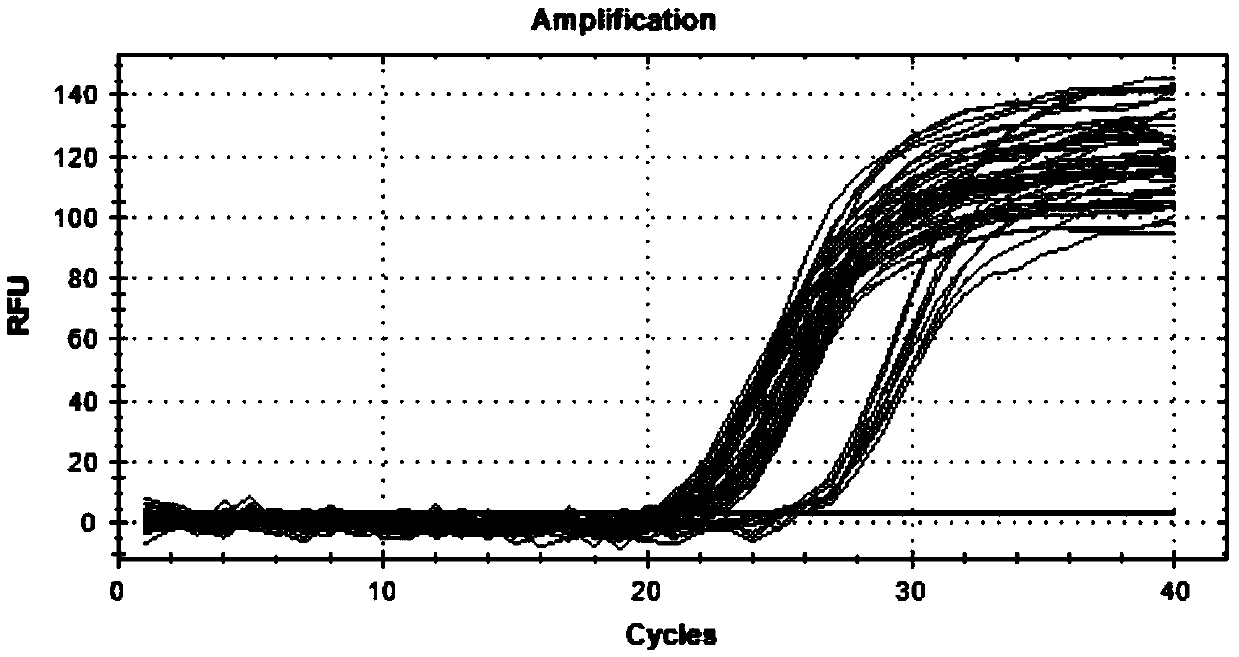
[0032] The present invention utilizes QPCR to detect the copy number variation of the MAPK10 gene of Nanyang cattle. The present invention will be described in detail below in conjunction with the accompanying drawings and examples, which are explanations rather than limitations of the present invention.
[0033] A CNV marker detected based on QPCR technology involved in the present invention refers to the copy number variation of the bovine MAPK10 gene candidate region (Chr6: 102708523-102710298). Specifically, the CNV marker is located in the 610515bp-612290bp region of the sequence shown in the bovine MAPK10 gene (GenBankAccessionNo.AC_000163), according to the log 2 2 -ΔΔCt Divide the results into three categories: Log 2 2 -ΔΔCt >0.5 is plug-in type; Log 2 2 -ΔΔCt 2 2 -ΔΔCt ≤|±0.5| is the normal type.
[0034] 1. Nanyang Cattle Sample Collection
[0035] In the present invention, the local Nanyang cattle breed in China is used as the detection object, and blood samp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com