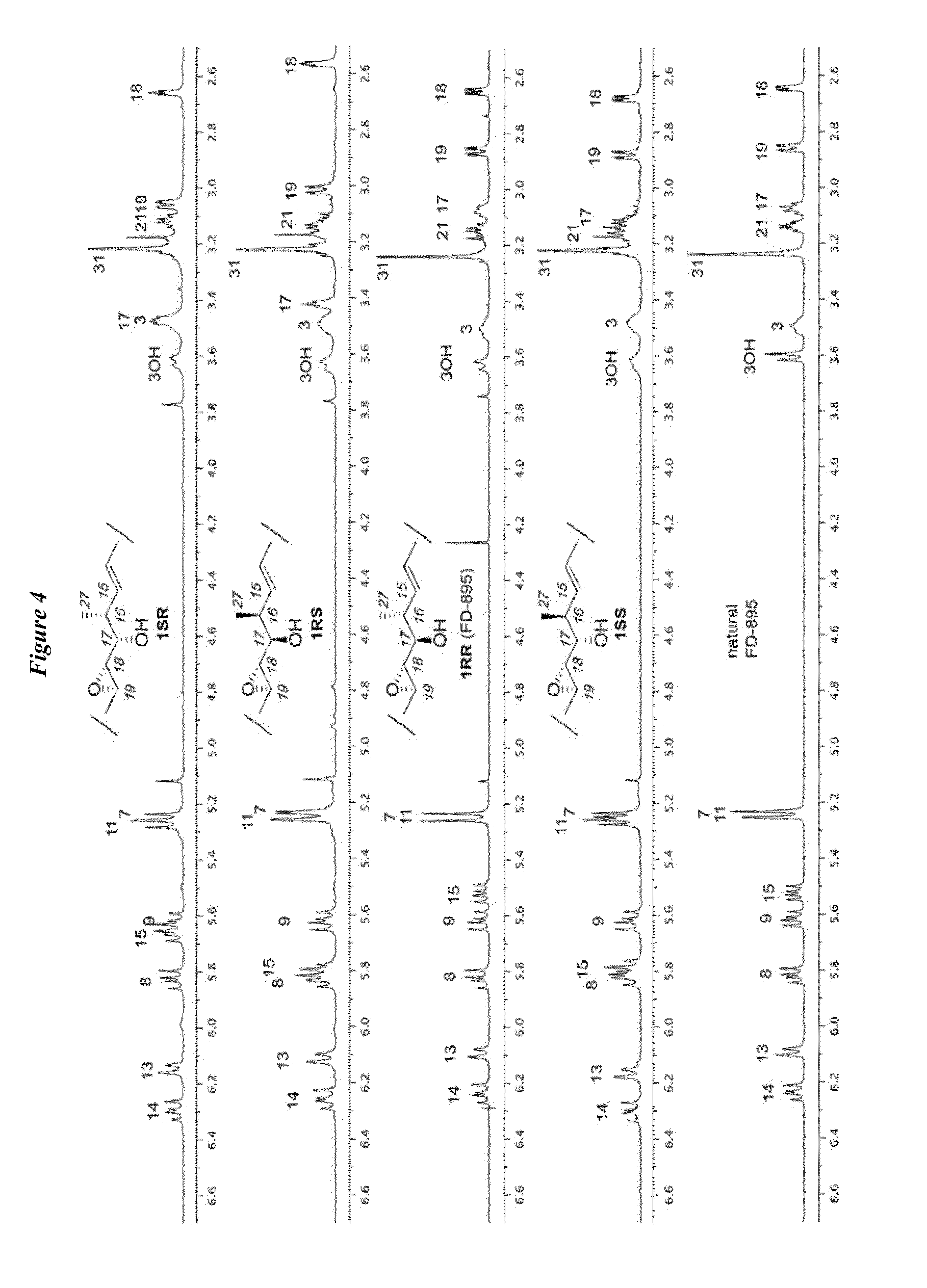
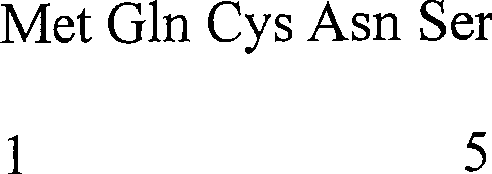Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
57 results about "Spliceosome complex" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A spliceosome is a large and complex molecular machine found primarily within the splicing speckles of the cell nucleus of eukaryotic cells. The spliceosome is assembled from snRNAs and SR protein. The spliceosome removes introns from a transcribed pre-mRNA, a type of primary transcript.
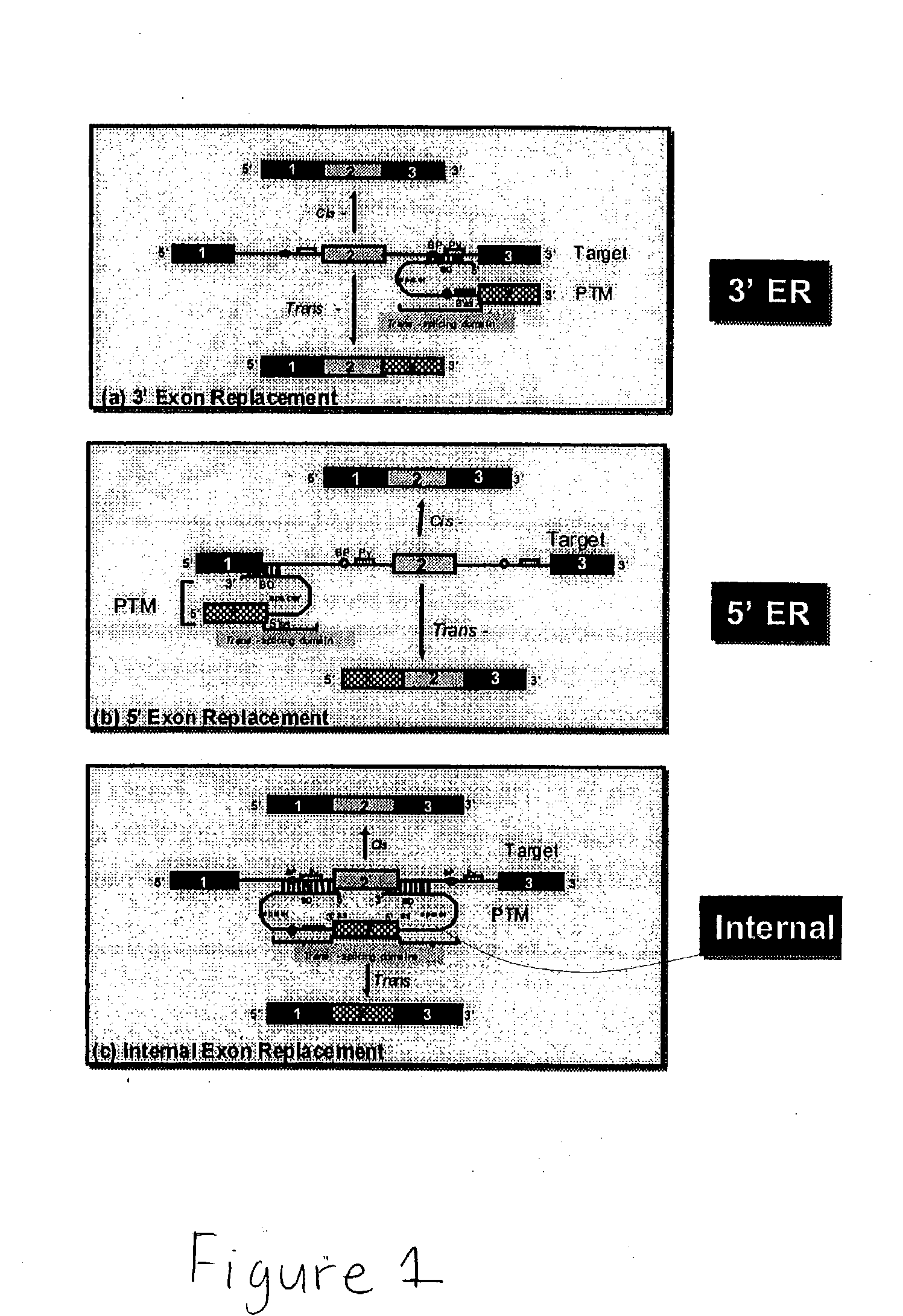
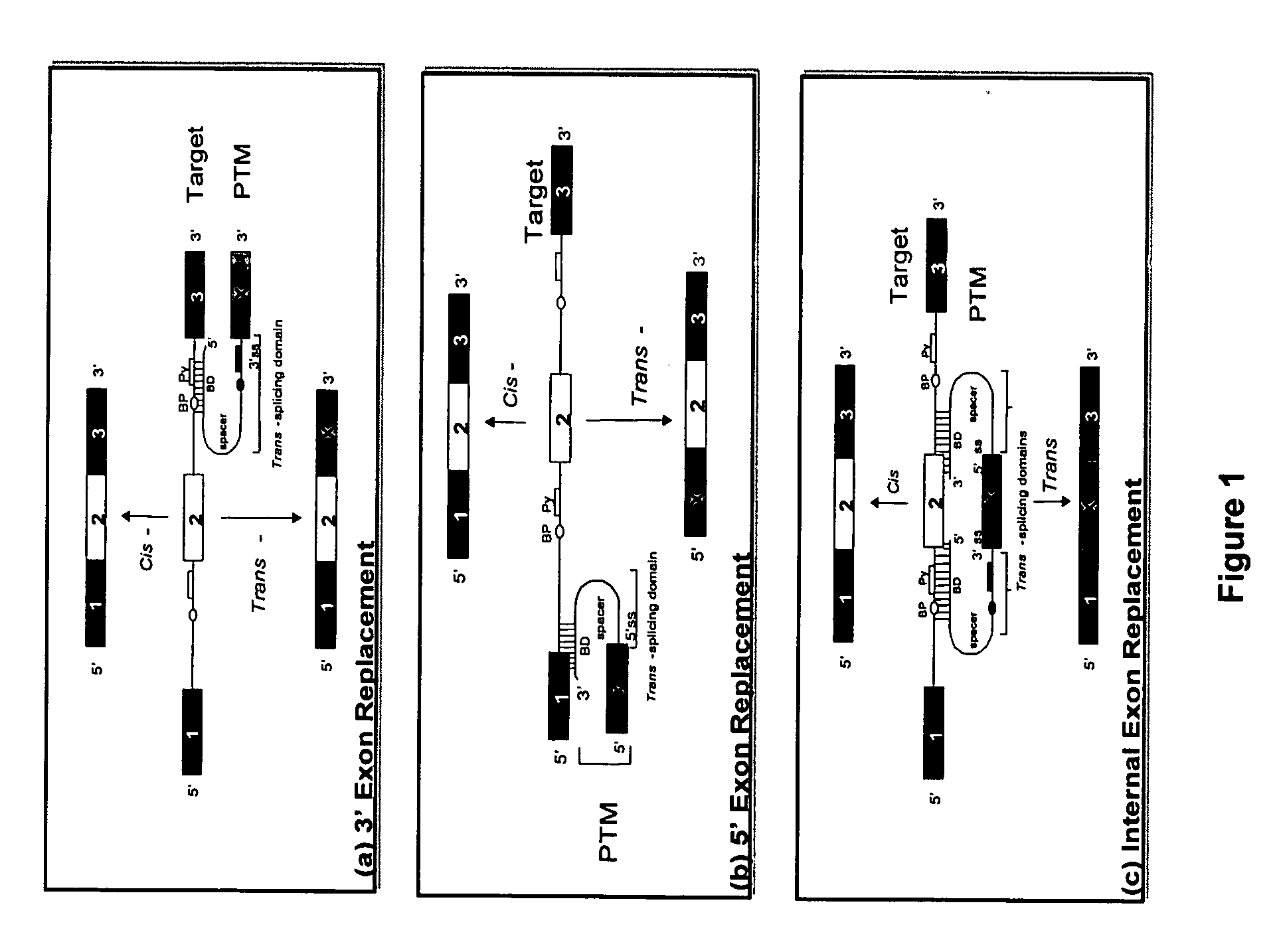
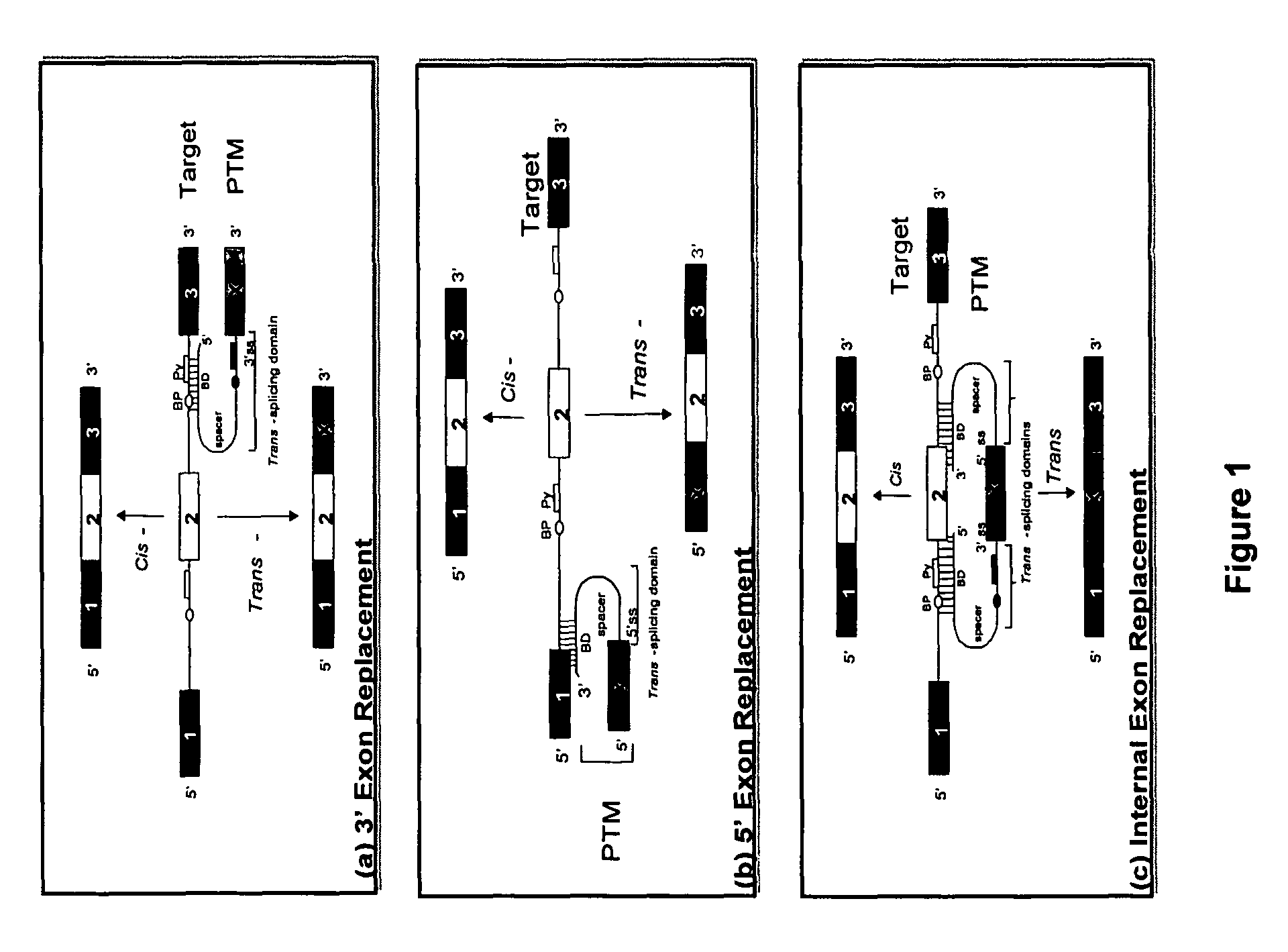
Methods and compositions for use in spliceosome mediated RNA trans-splicing
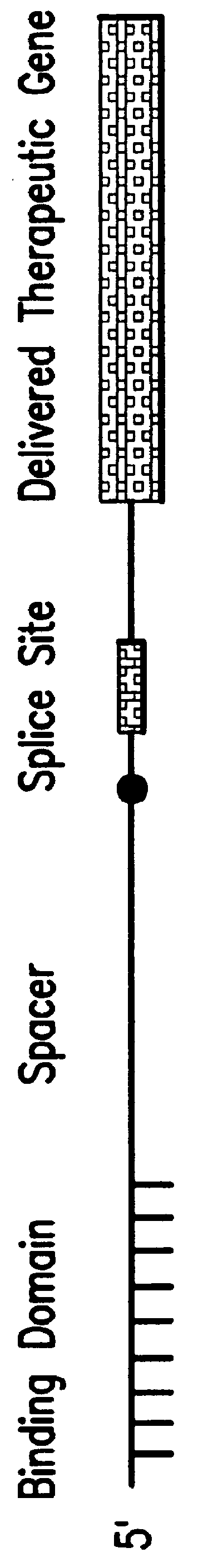
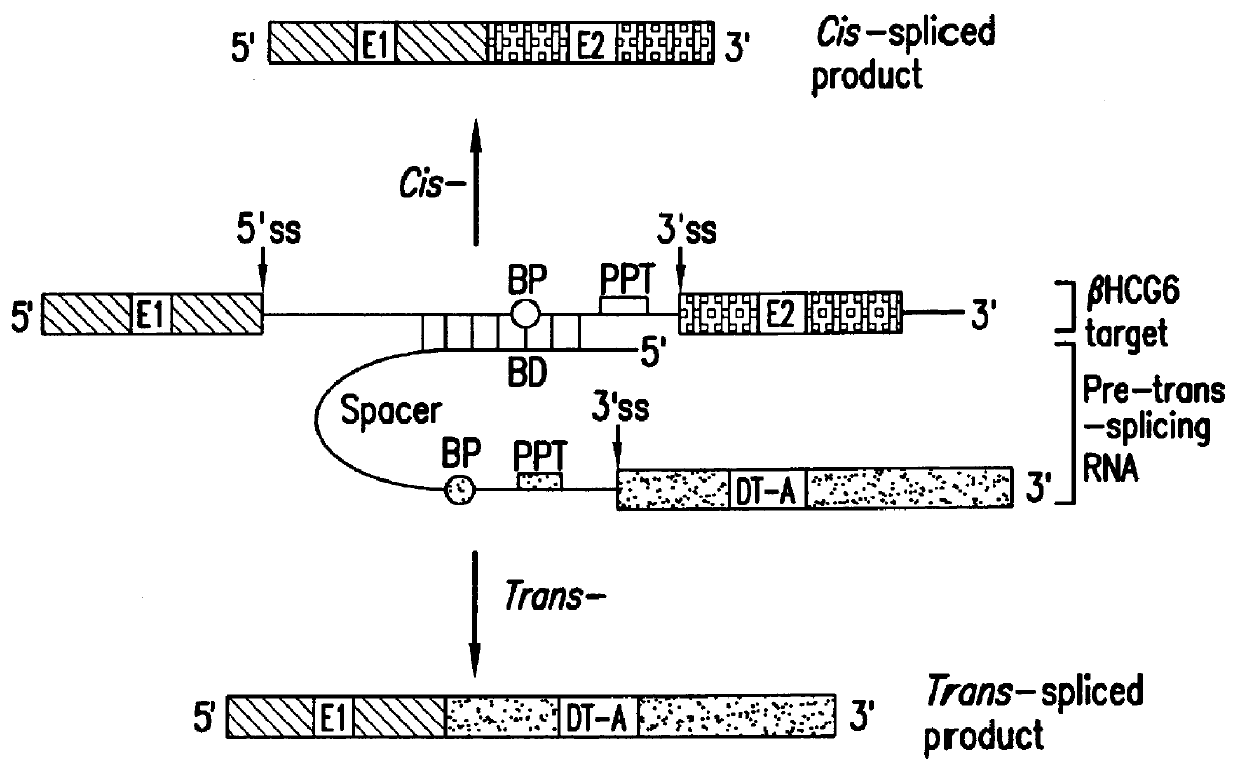
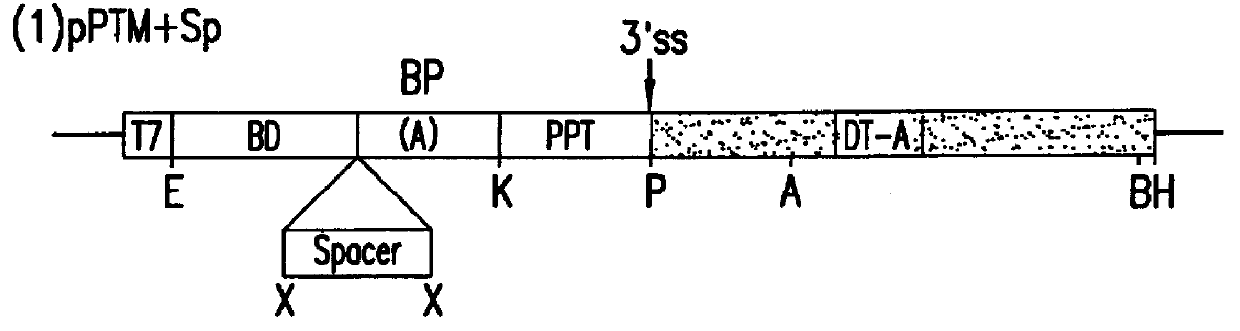
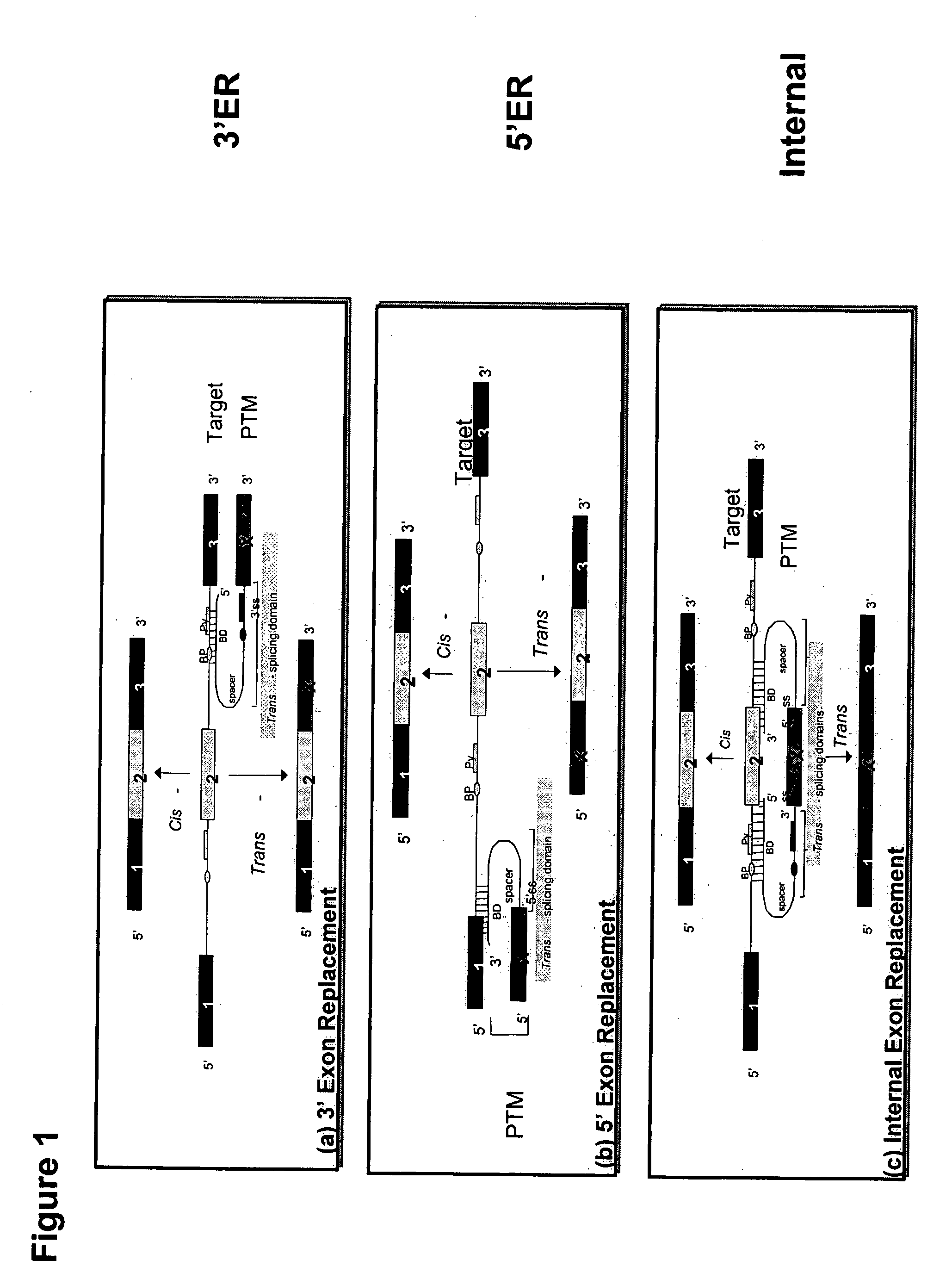
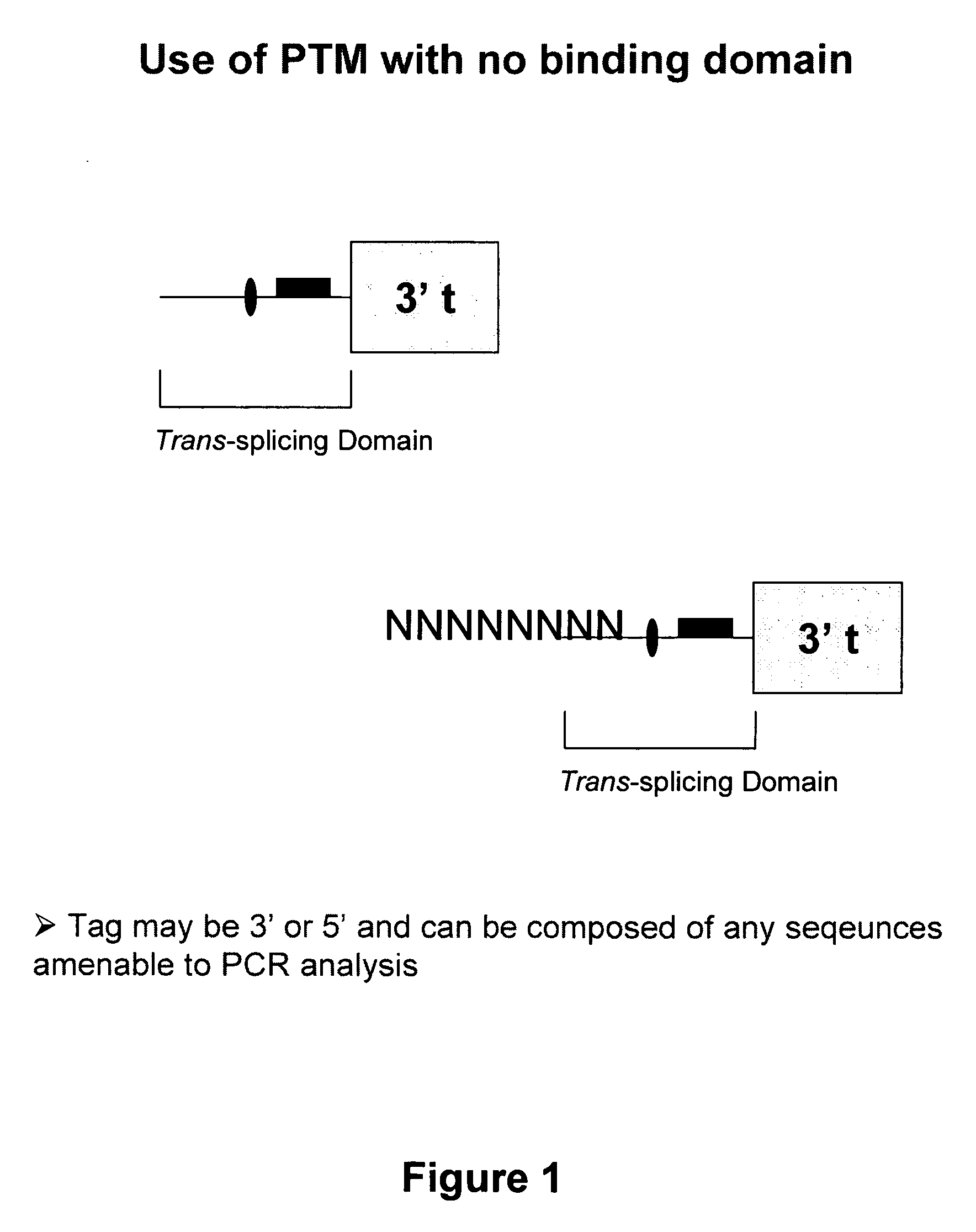
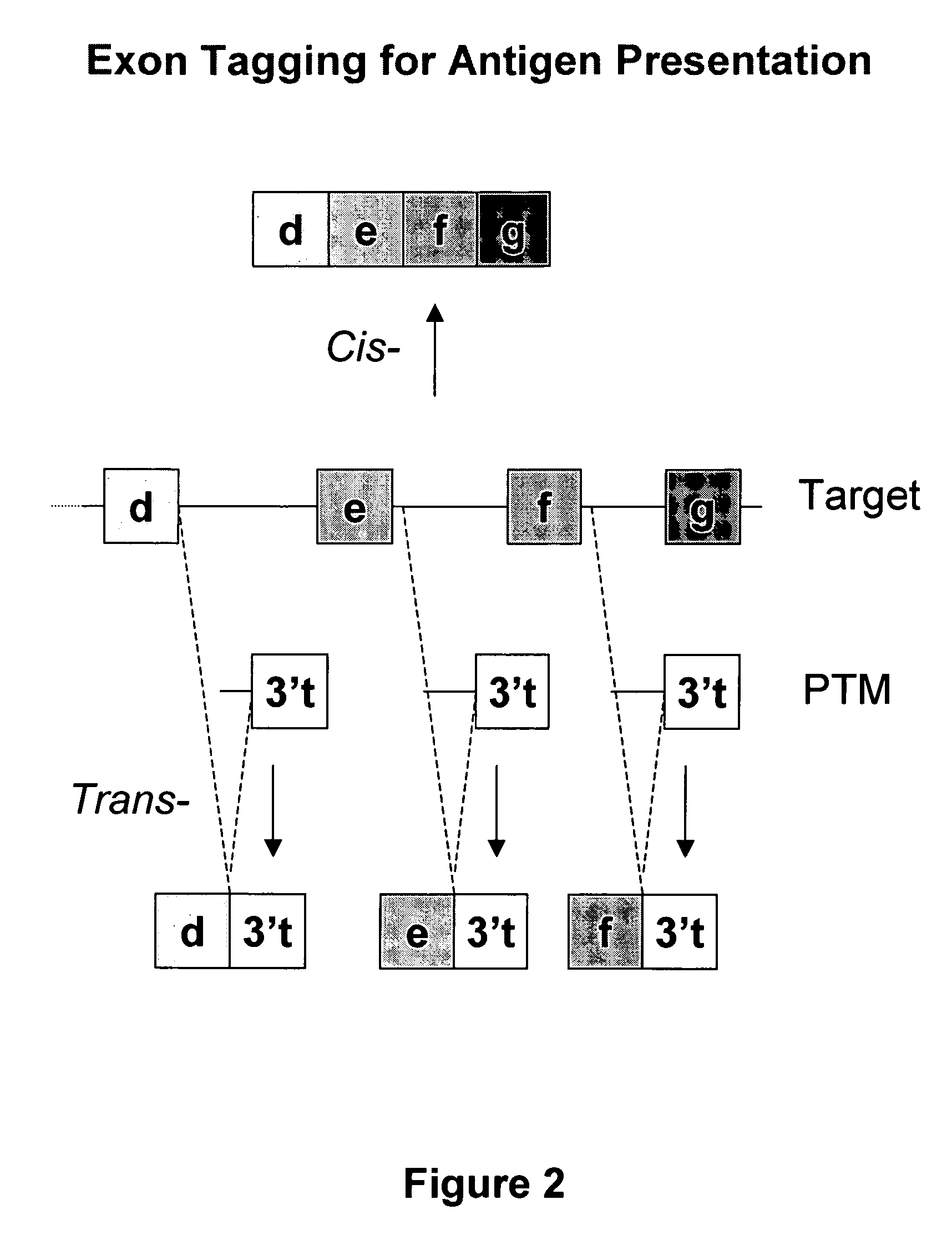
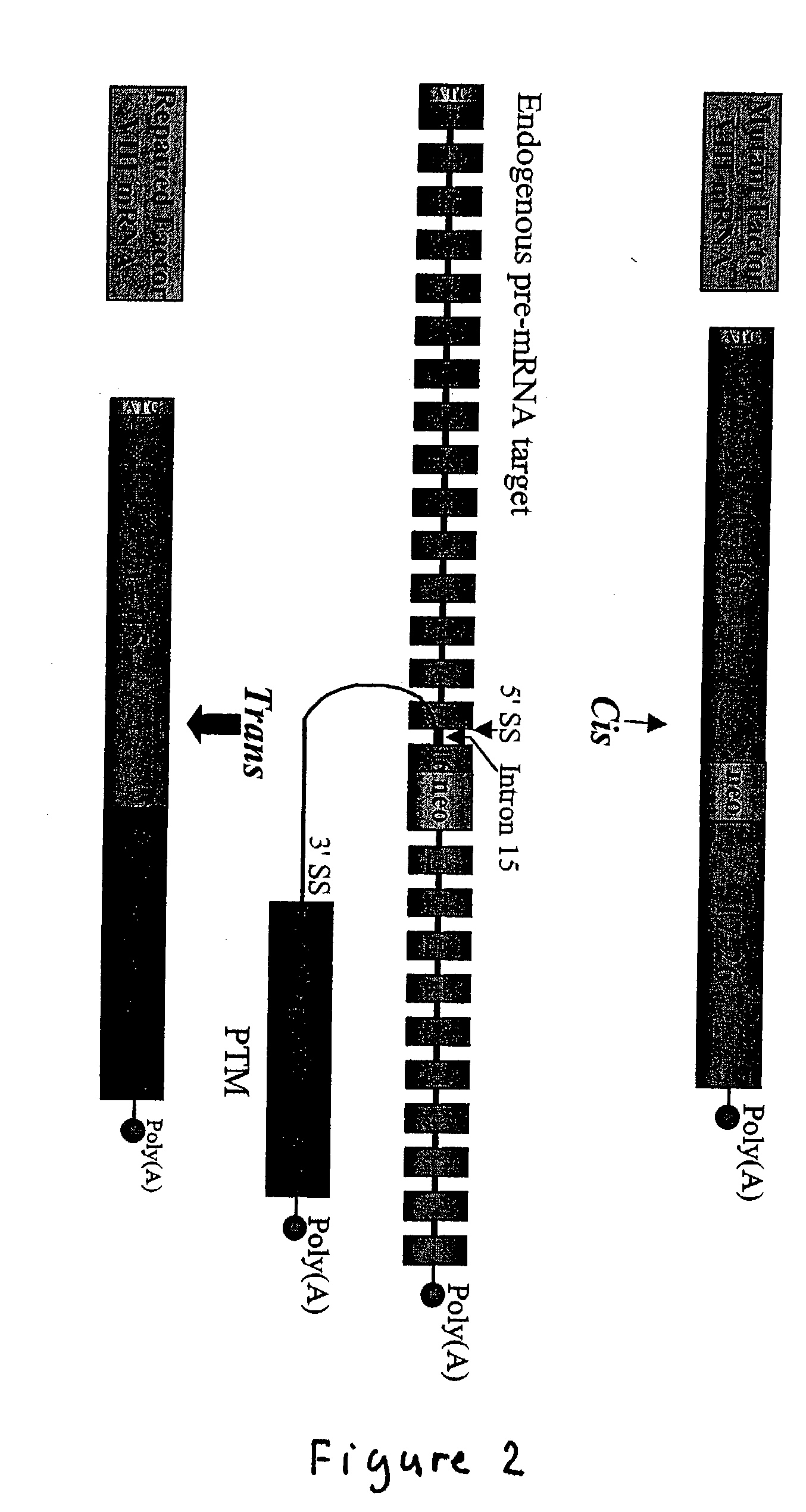
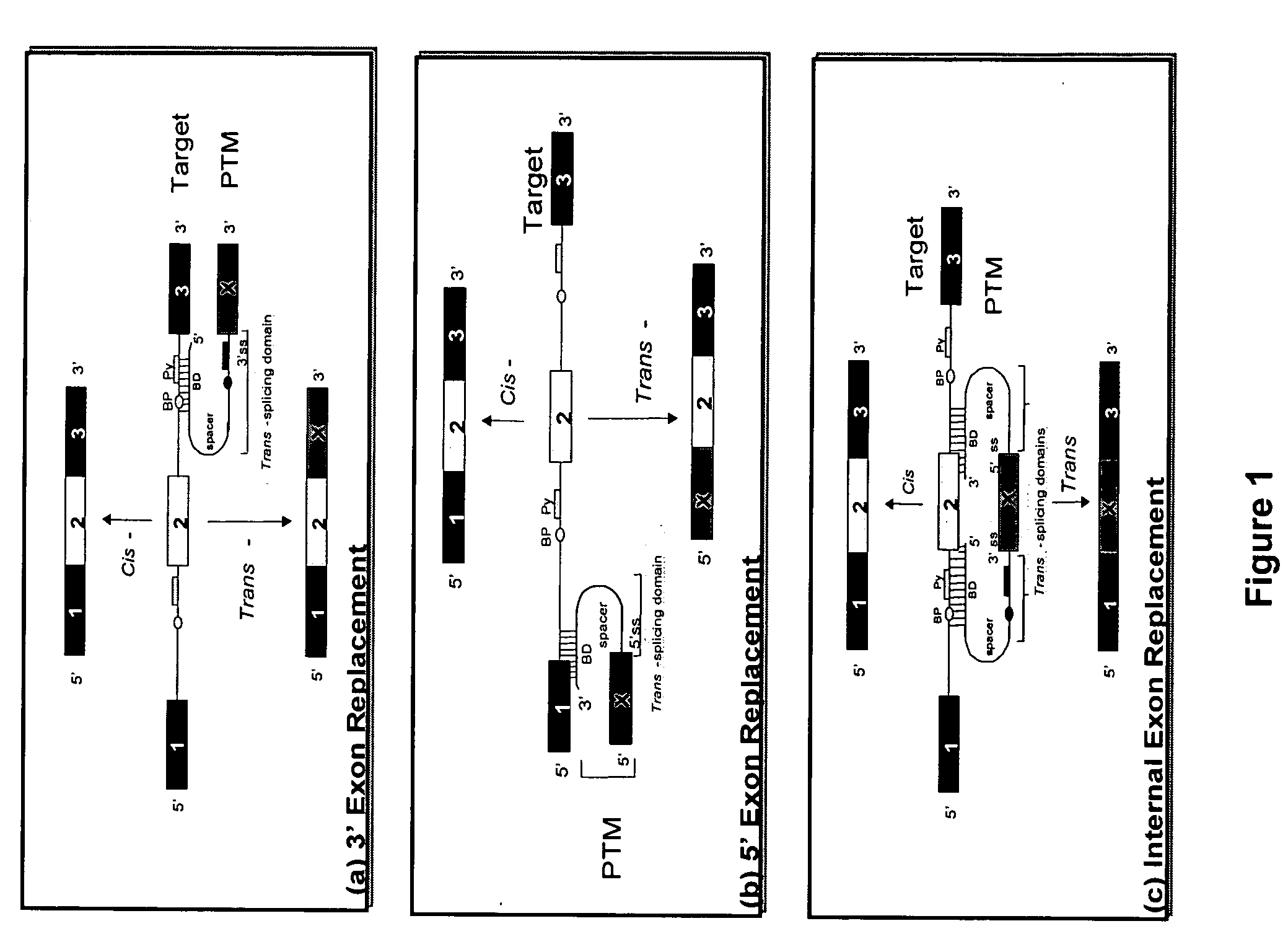
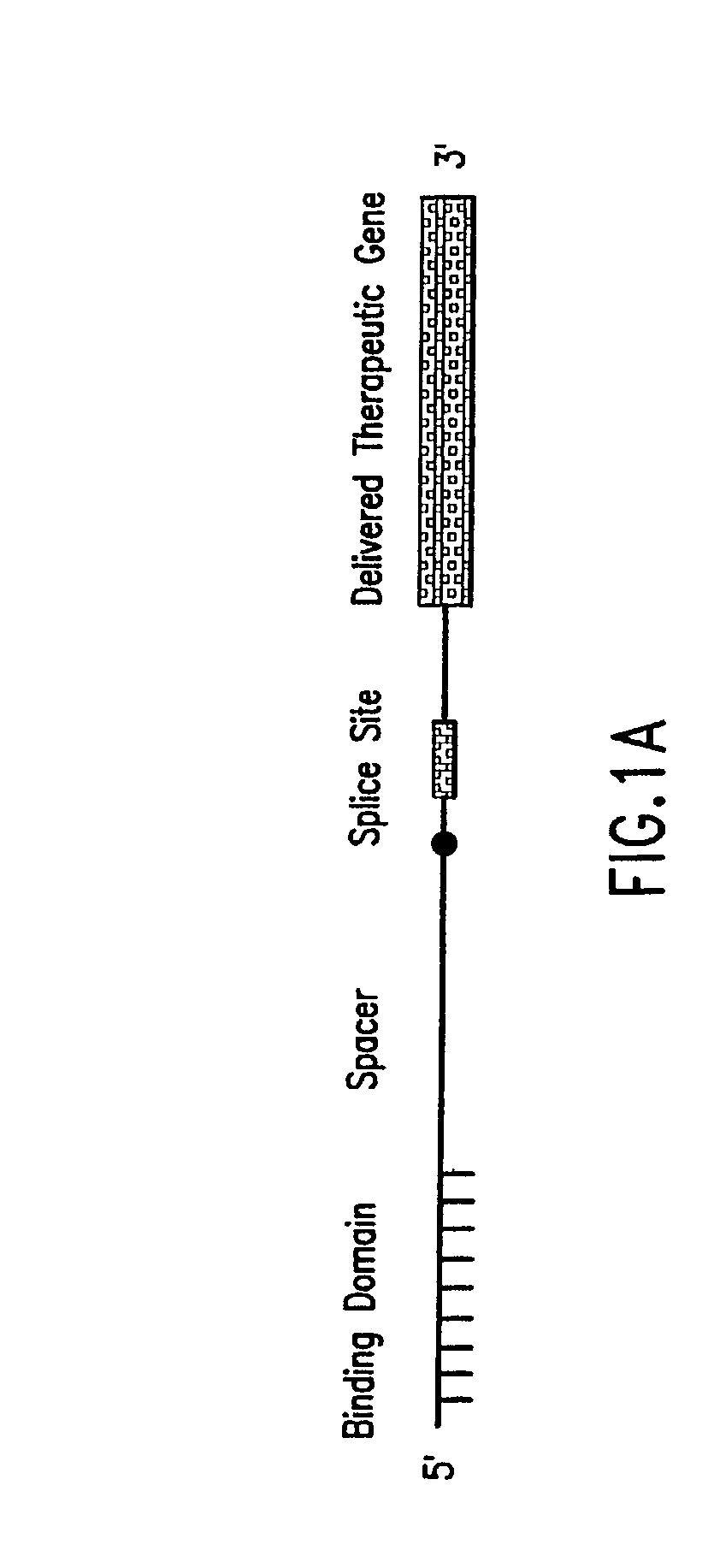
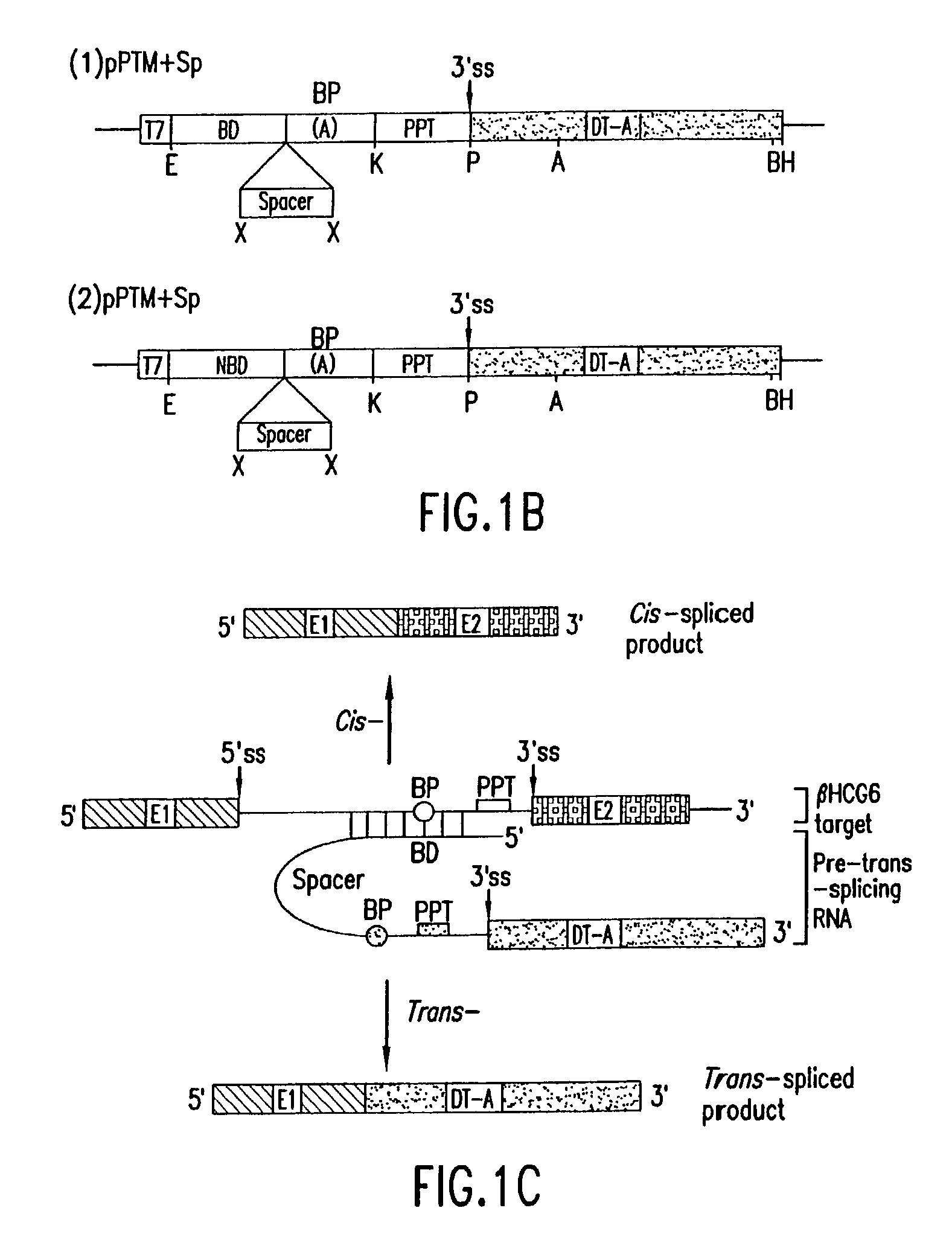
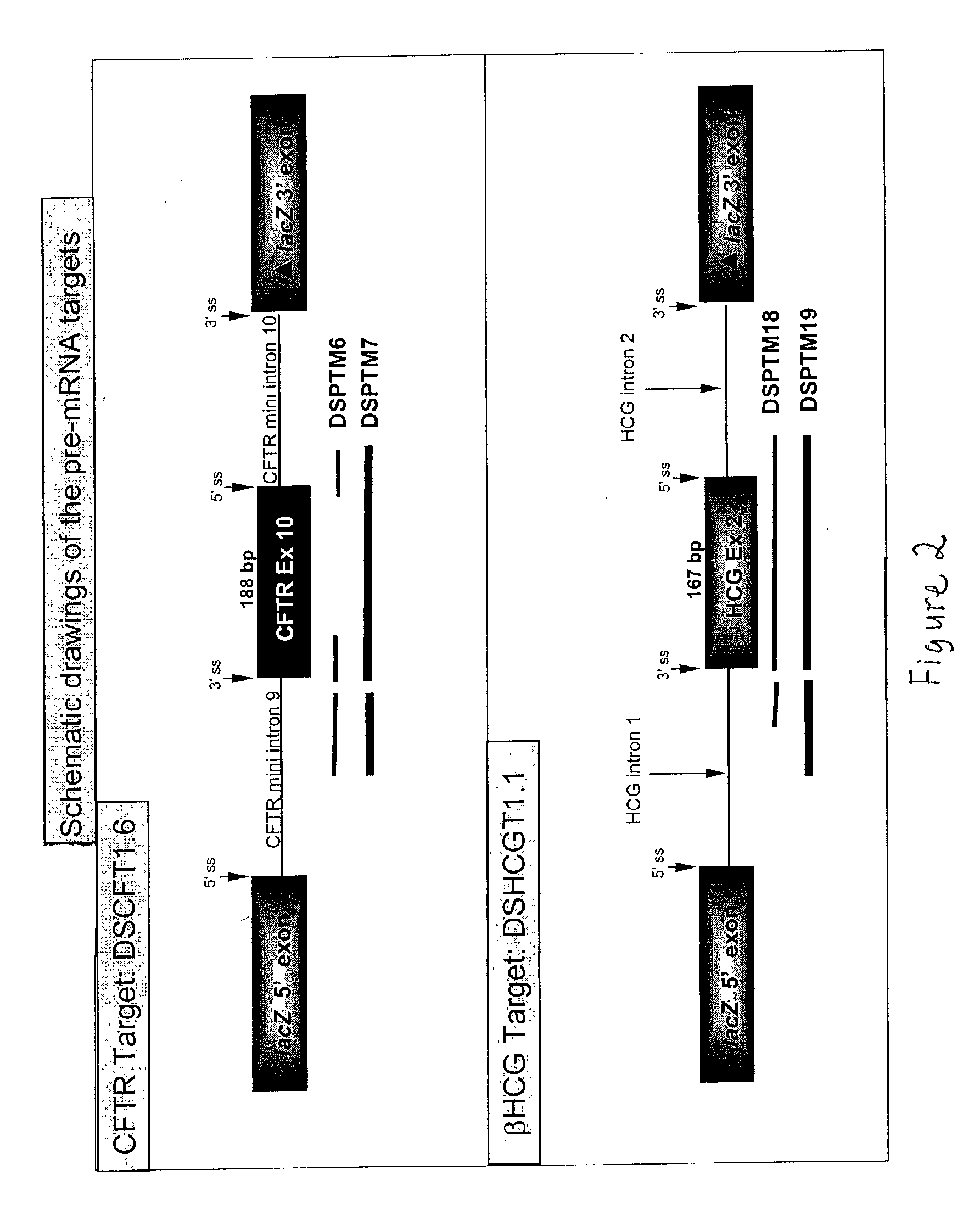
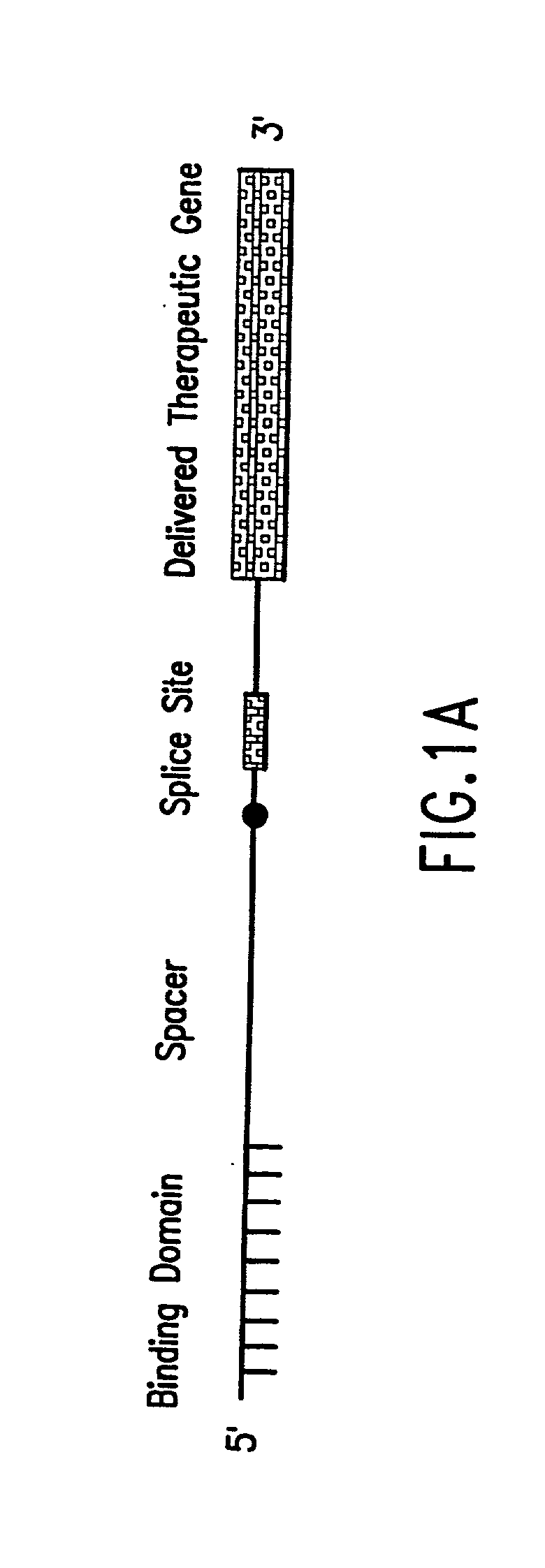
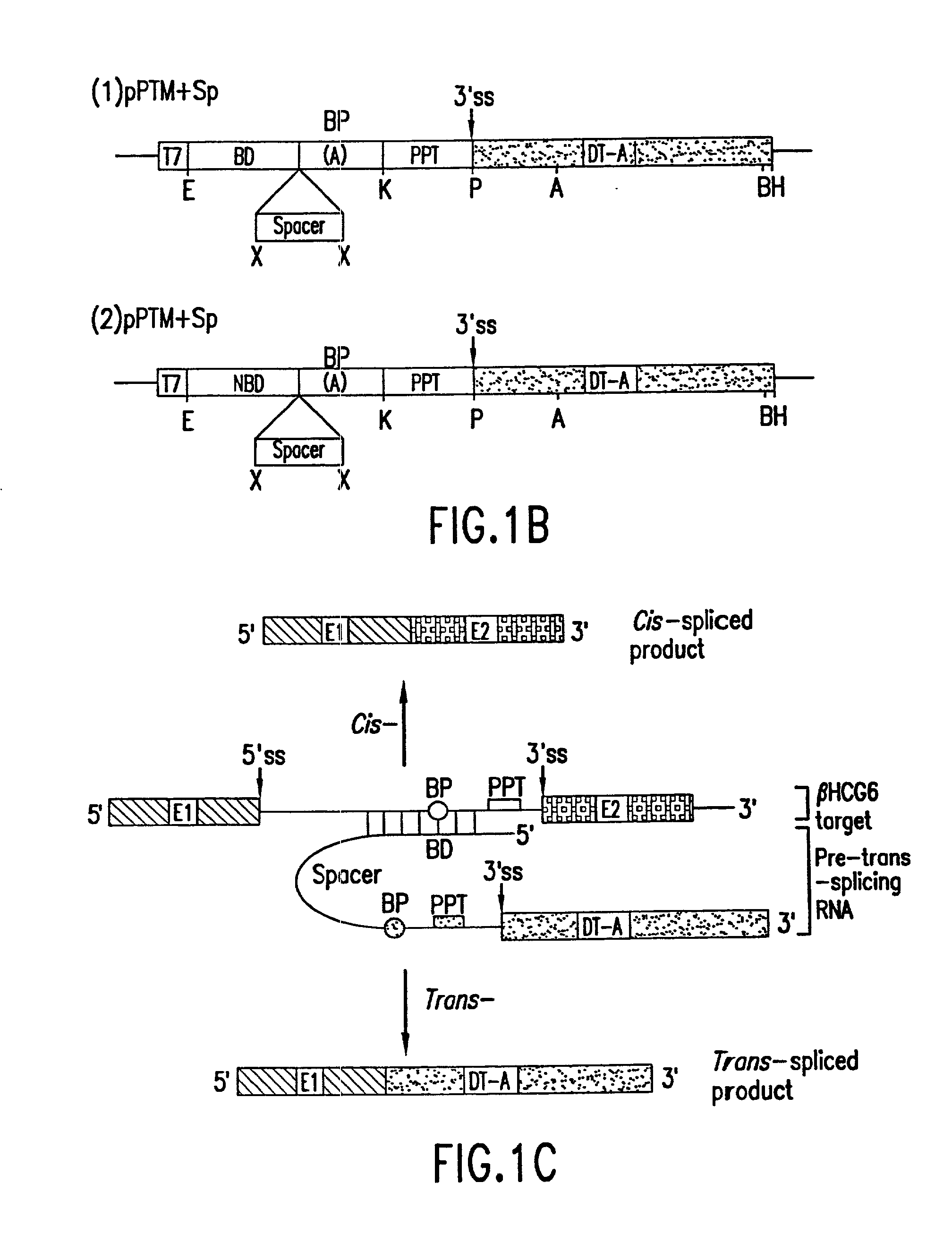
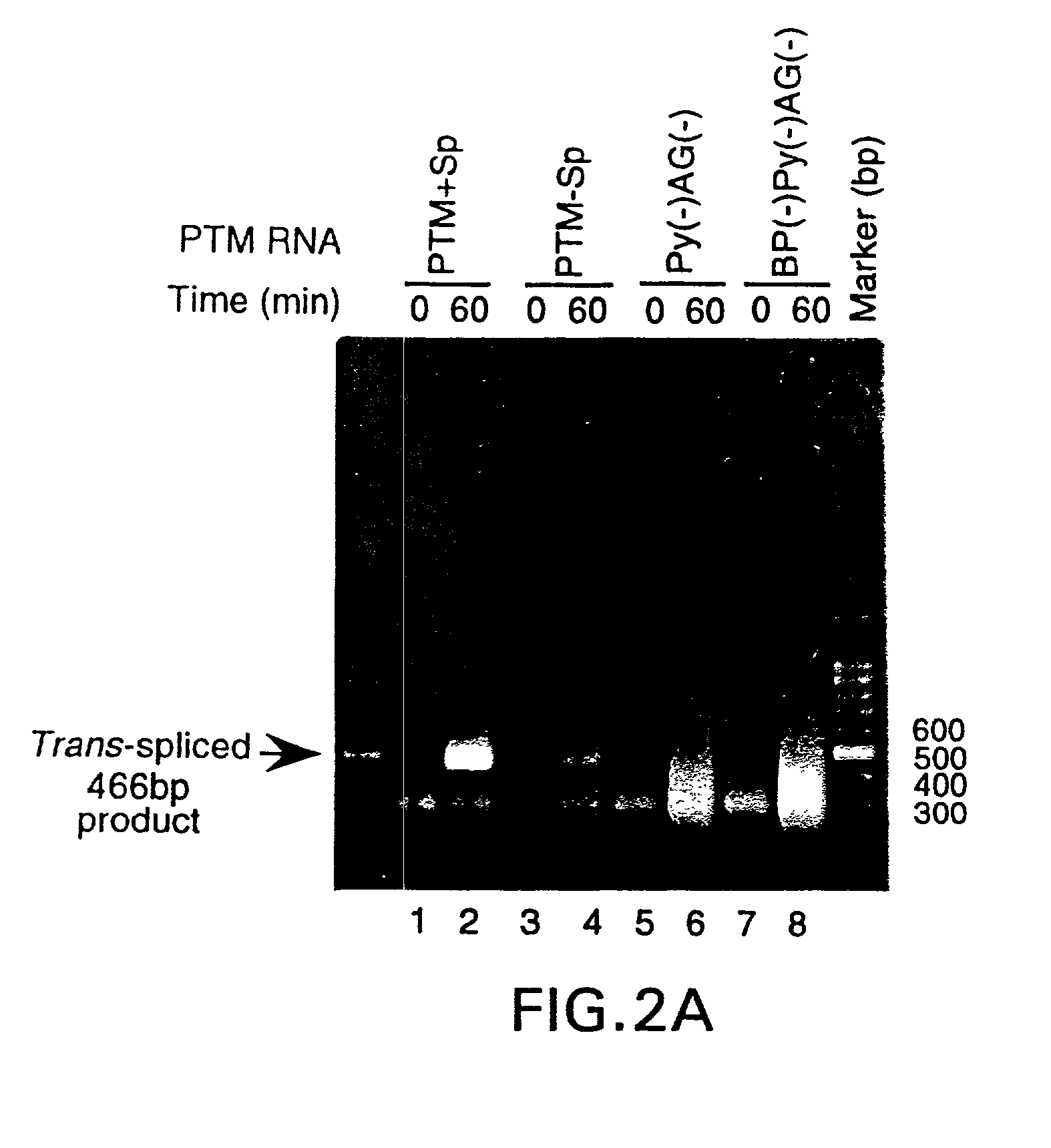
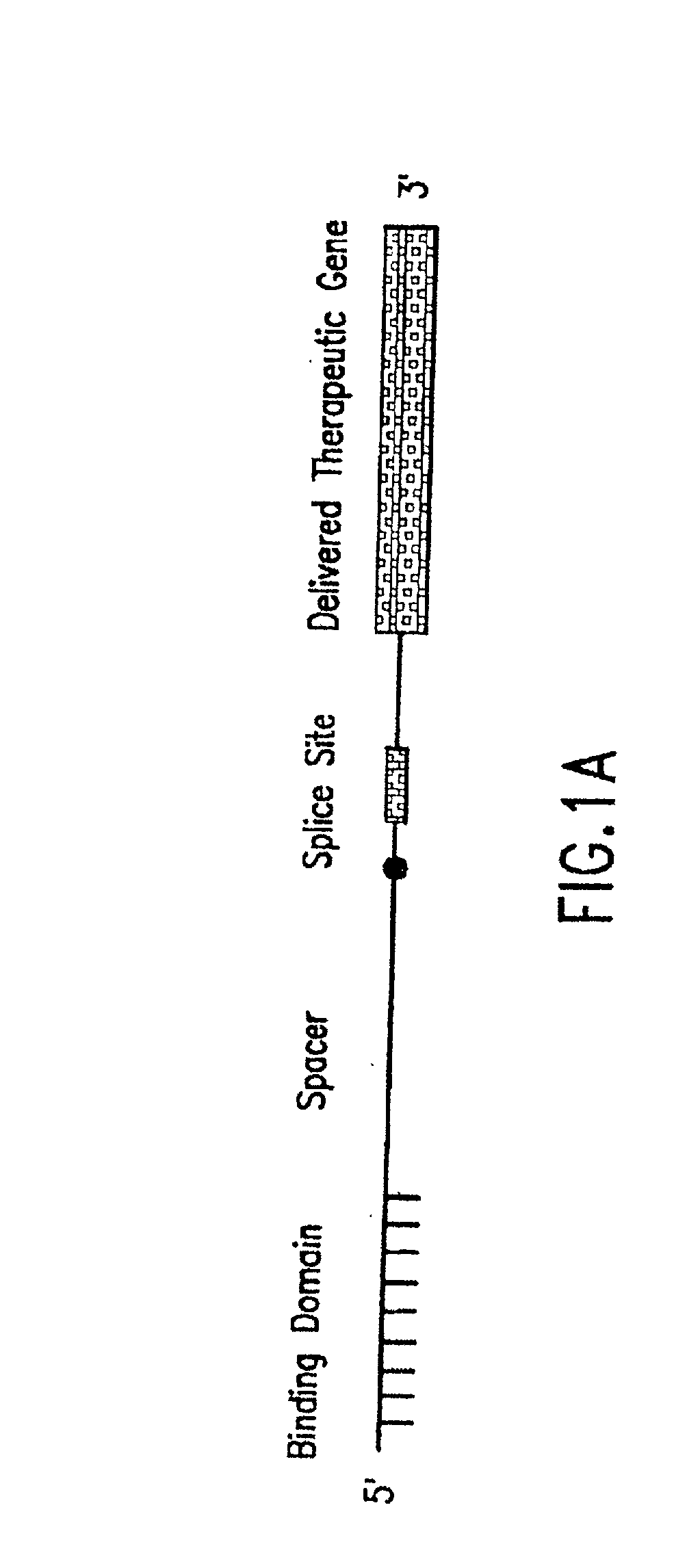
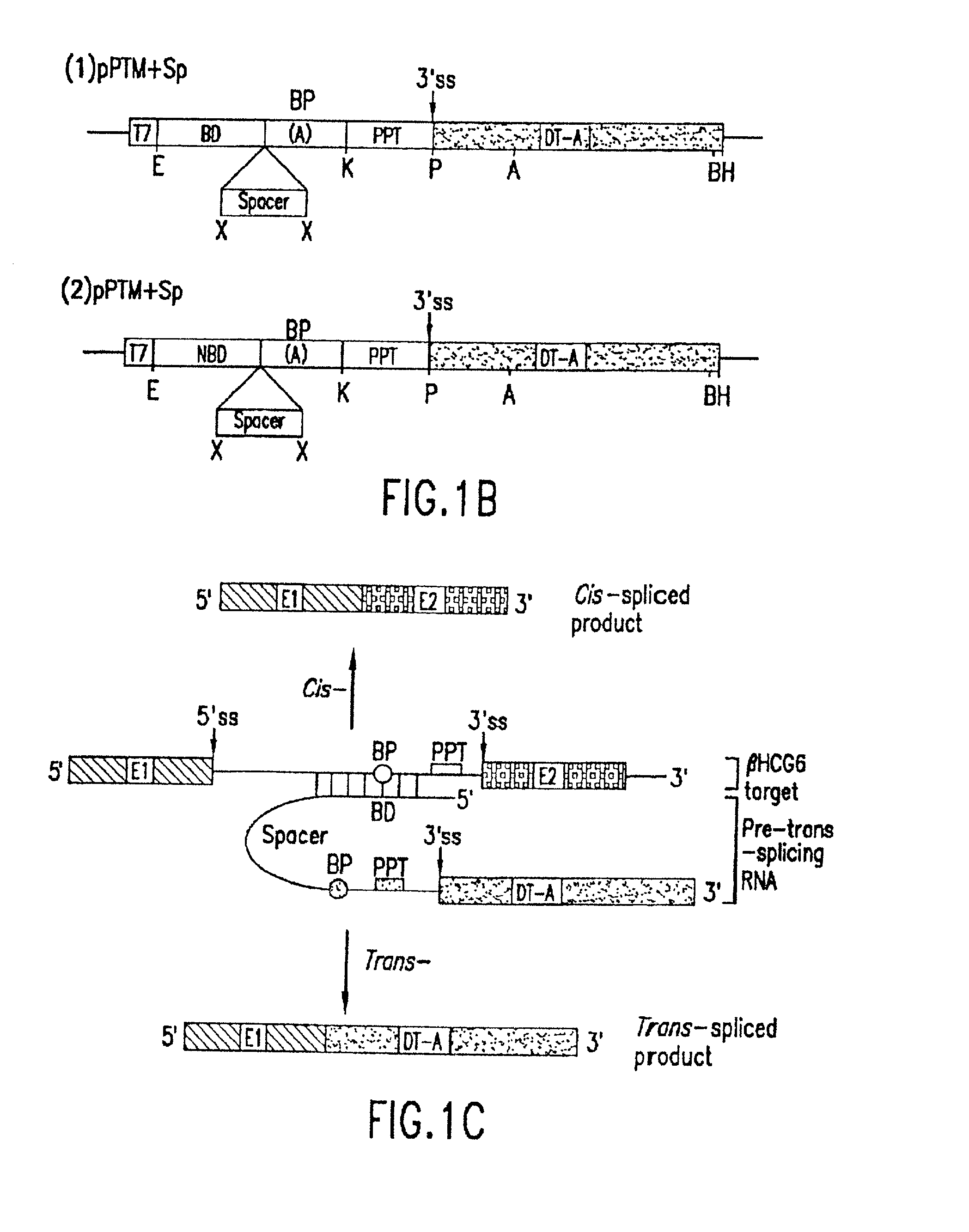
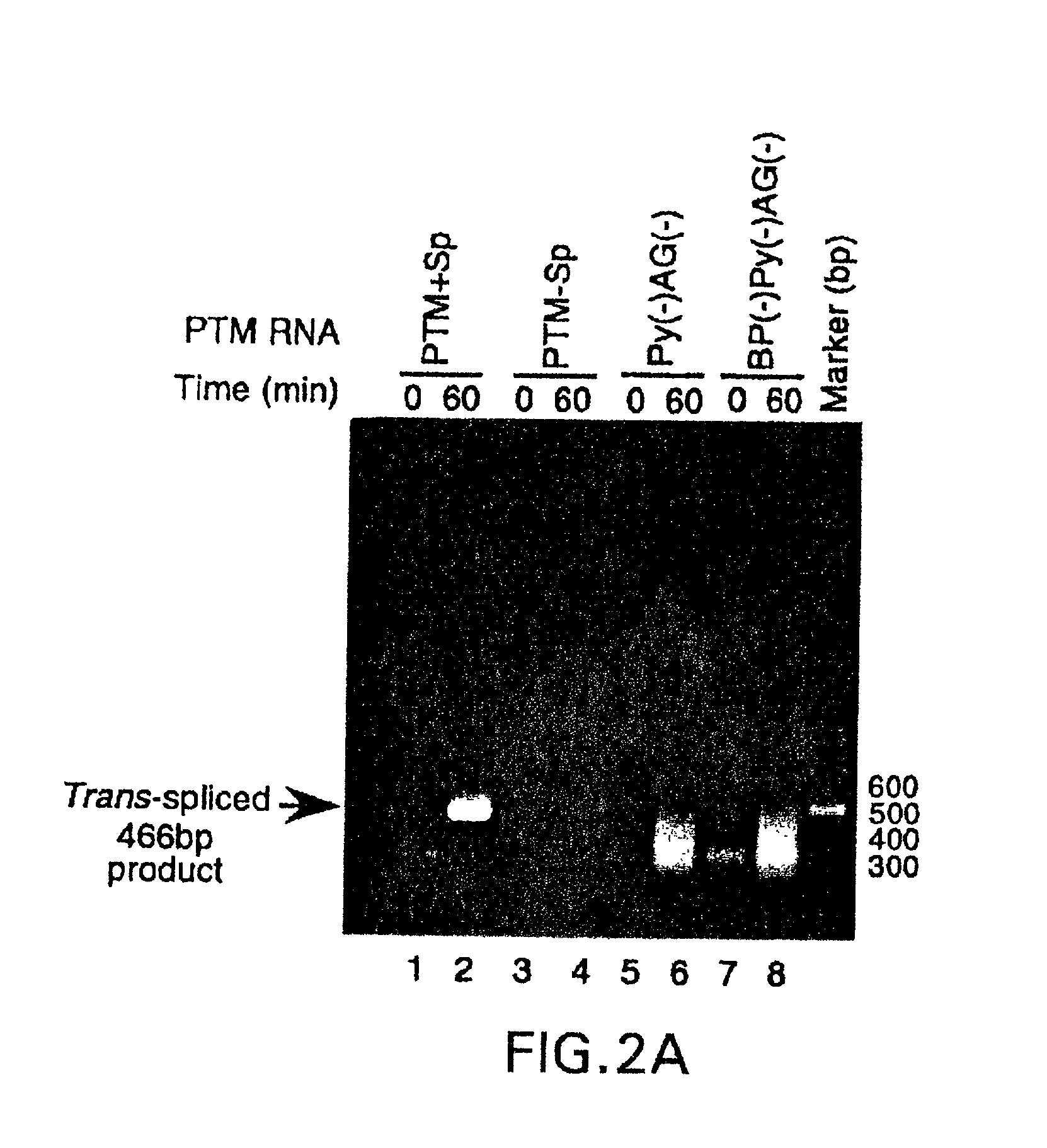
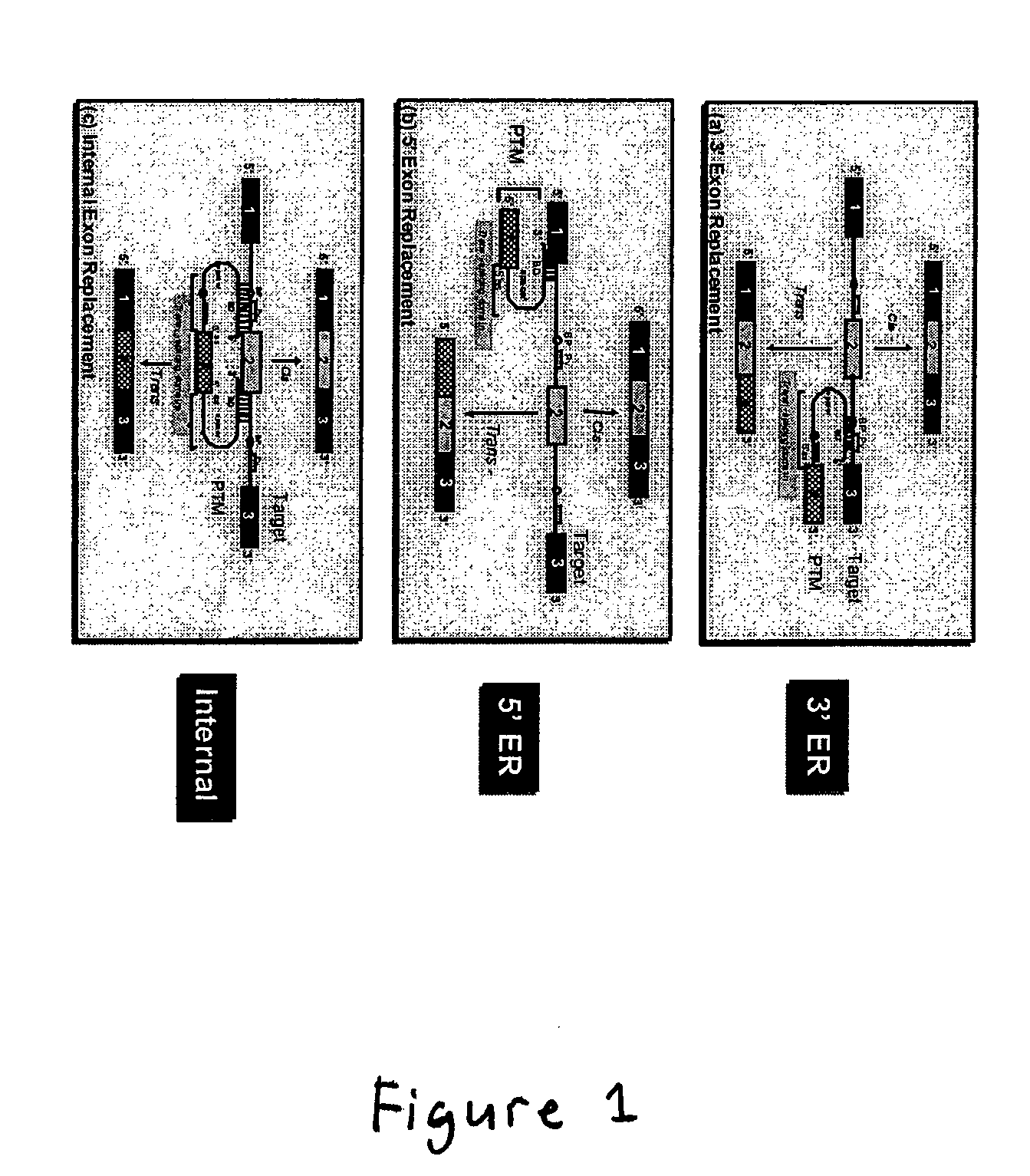
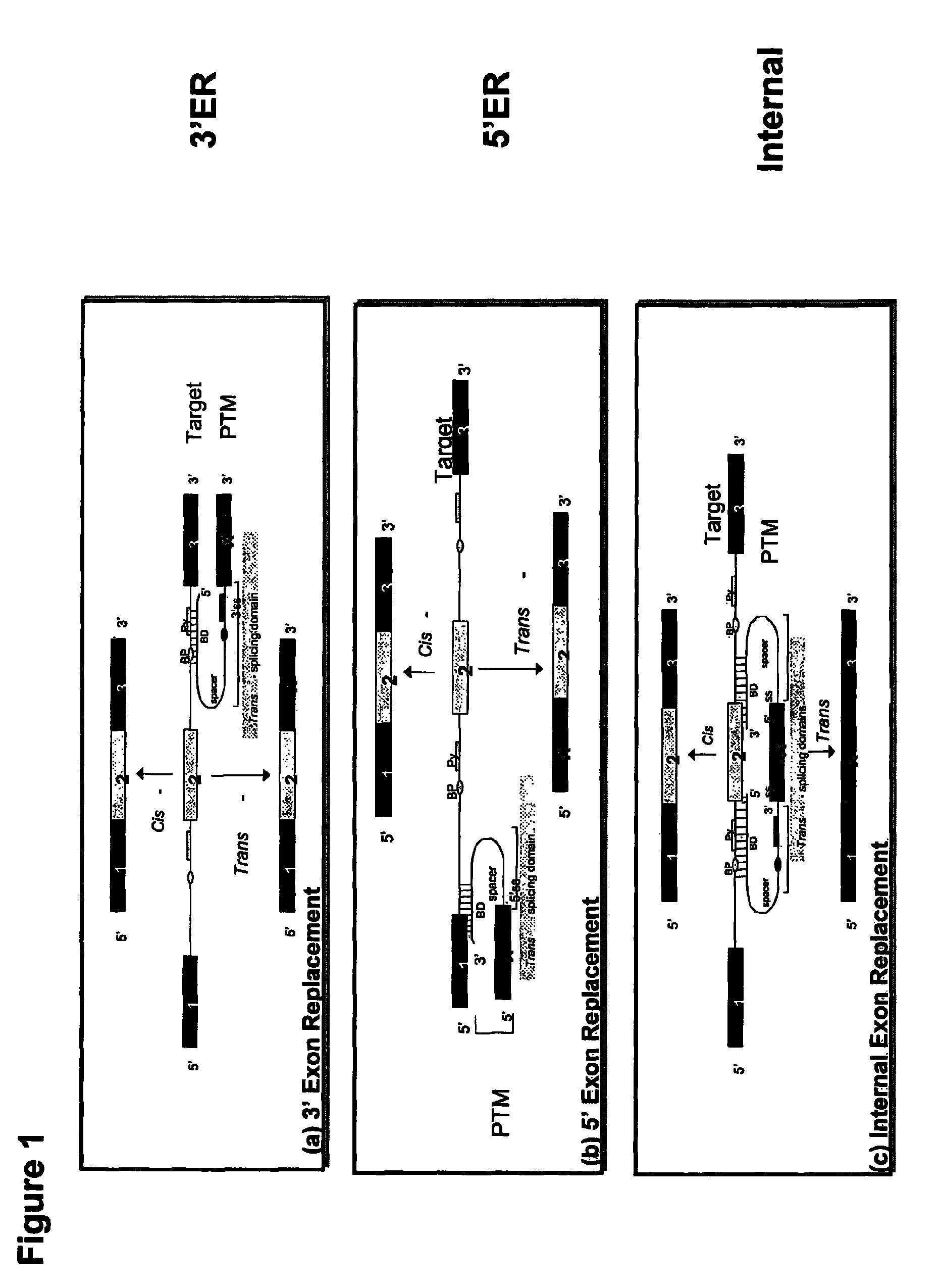
The molecules and methods of the present invention provide a means for in vivo production of a trans-spliced molecule in a selected subset of cells. The pre-trans-splicing molecules of the invention are substrates for a trans-splicing reaction between the pre-trans-splicing molecules and a pre-mRNA which is uniquely expressed in the specific target cells. The in vivo trans-splicing reaction provides a novel mRNA which is functional as mRNA or encodes a protein to be expressed in the target cells. The expression product of the mRNA is a protein of therapeutic value to the cell or host organism a toxin which causes killing of the specific cells or a novel protein not normally present in such cells. The invention further provides PTMs that have been genetically engineered for the identification of exon / intron boundaries of pre-mRNA molecules using an exon tagging method. The PTMs of the invention can also be designed to result in the production of chimeric RNA encoding for peptide affinity purification tags which can be used to purify and identify proteins expressed in a specific cell type.
Owner:INTRONN HLDG +1
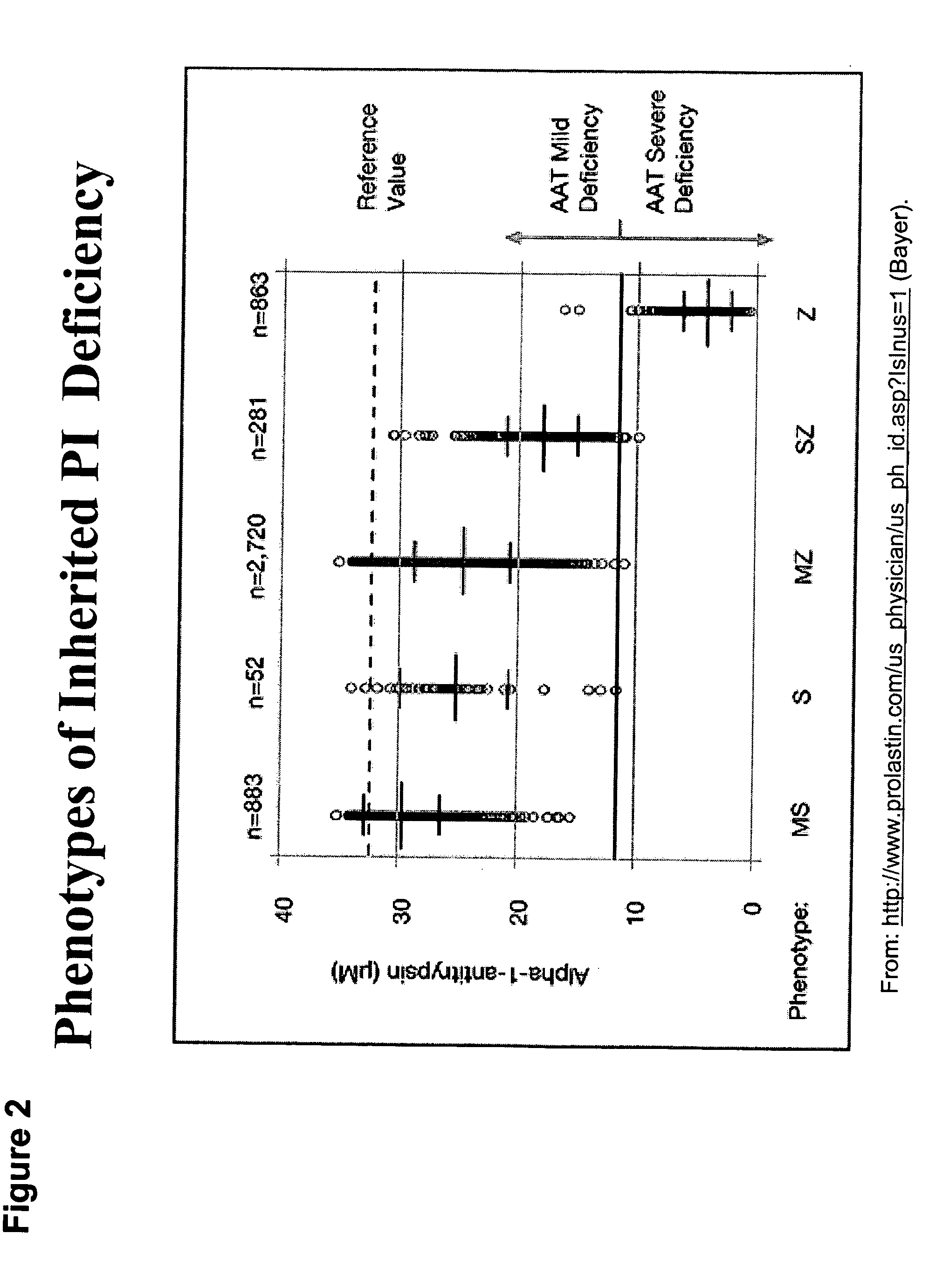
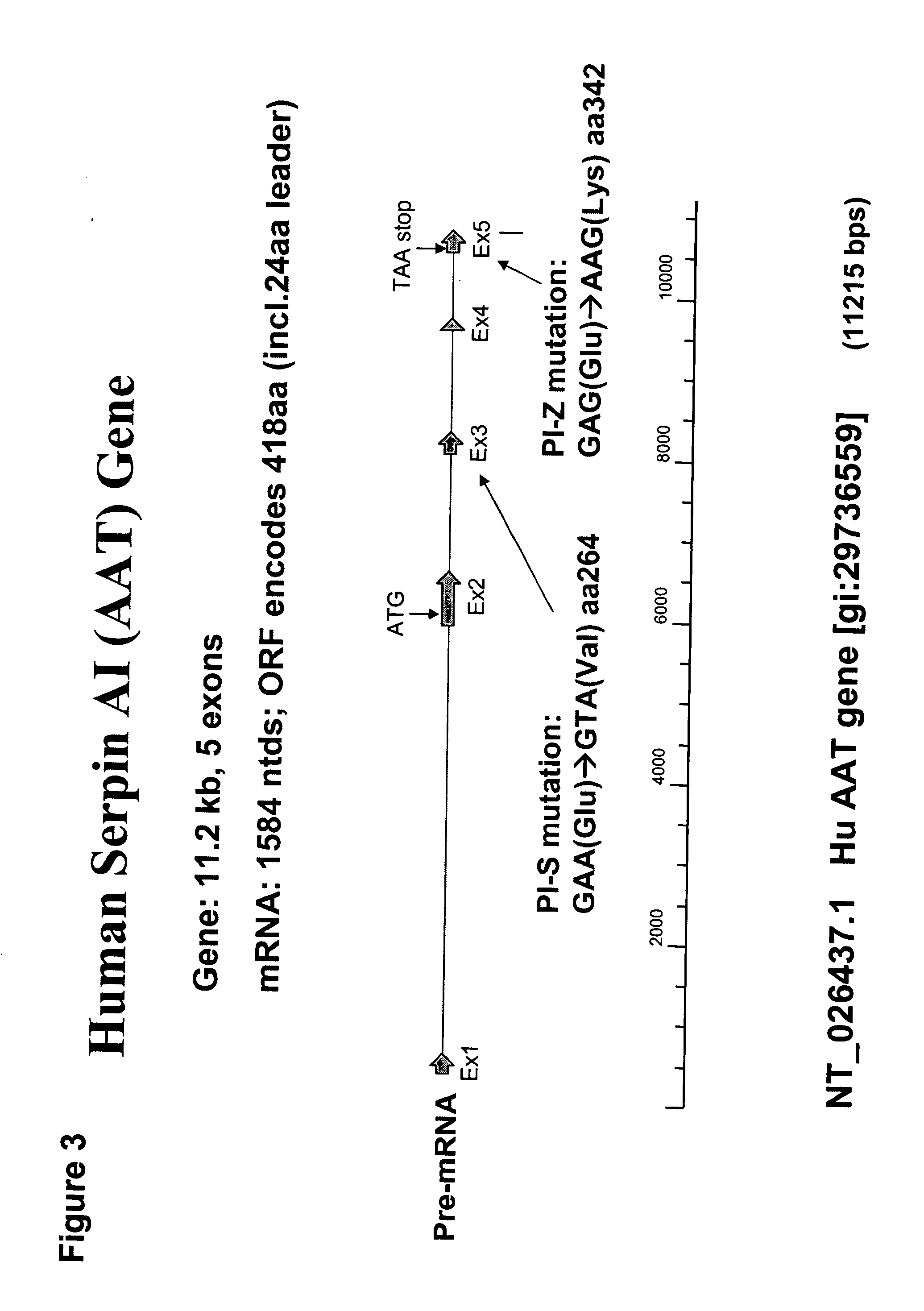
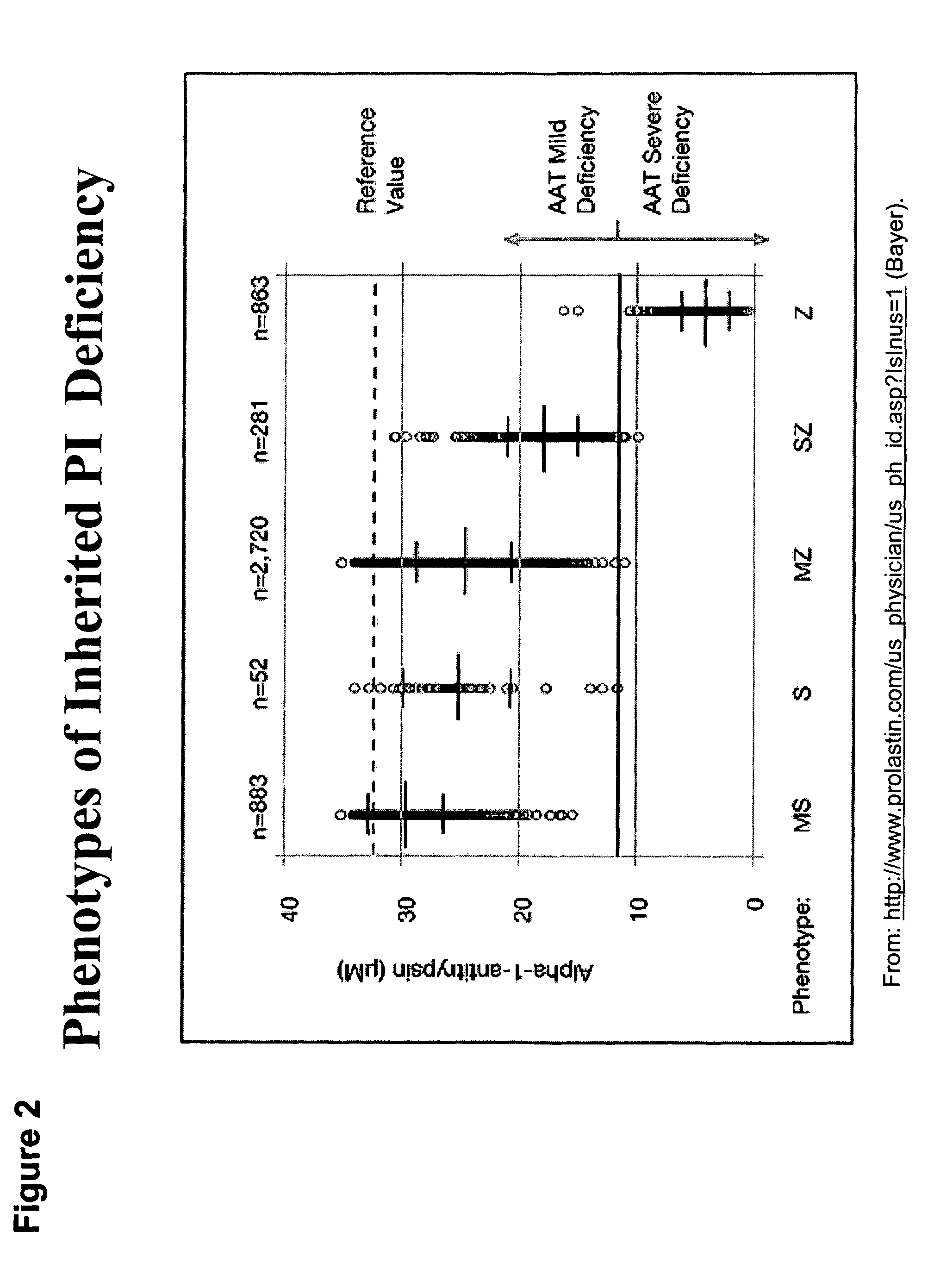
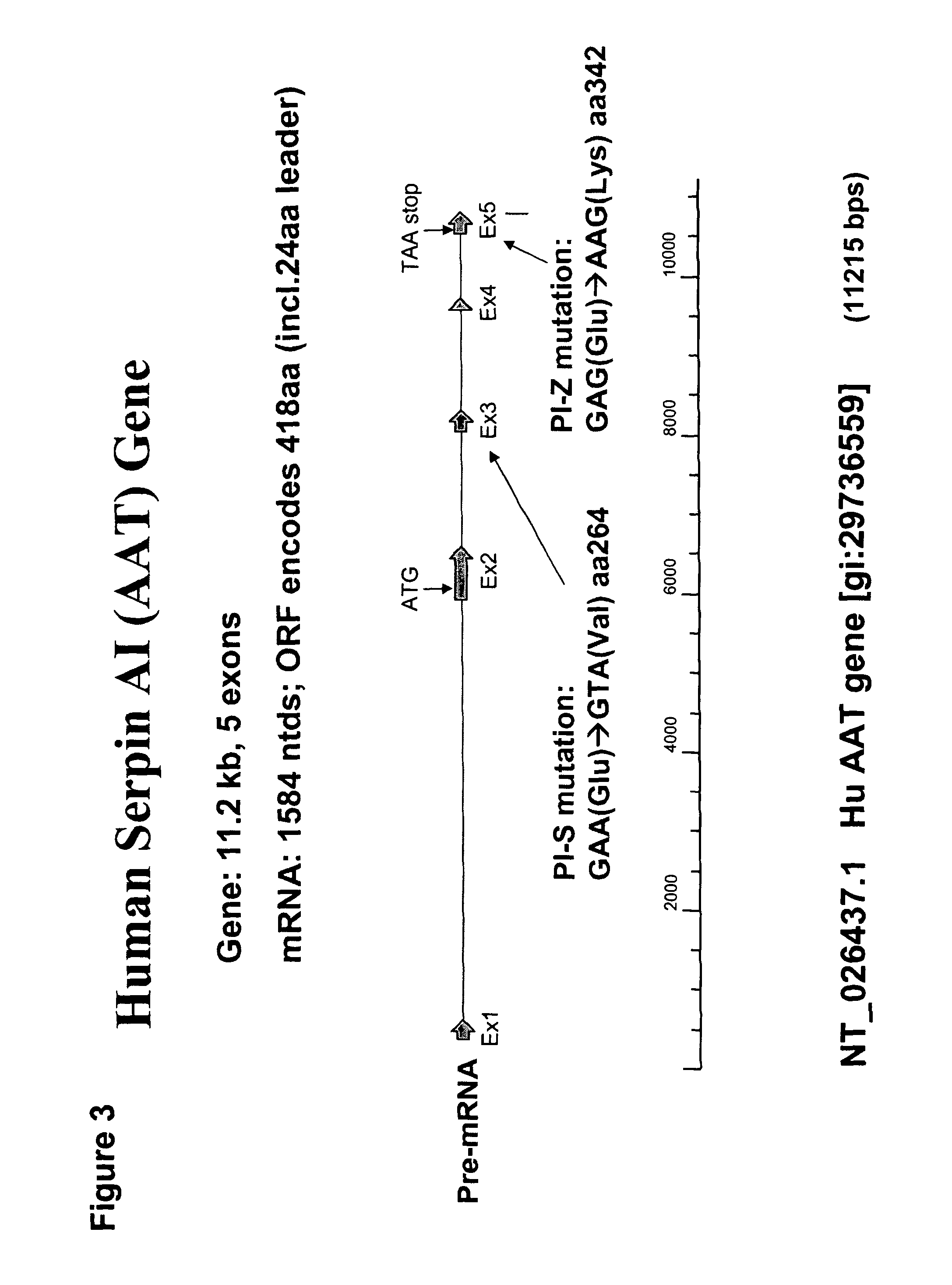
Correction of alpha-1-antitrypsin genetic defects using spliceosome mediated RNA trans splicing
InactiveUS20060234247A1Reduce lungReduce liver pathologySugar derivativesMicrobiological testing/measurementDiseaseRNA Trans-Splicing
The present invention provides methods and compositions for generating novel nucleic acid molecules through targeted spliceosomal mediated RNA trans-splicing. The compositions of the invention include pre-trans-splicing molecules (PTMs) designed to interact with a SERPINA1 target precursor messenger RNA molecule (target pre-mRNA) and mediate a trans-splicing reaction resulting in the generation of a novel chimeric RNA molecule (chimeric RNA). In particular, the PTMs of the present invention include those genetically engineered to interact with SERPINA1 target pre-mRNA so as to result in correction of SERPINA1 genetic defects responsible for AAT deficiency. The PTMs of the invention may also comprise sequences that are processed out of the PTM to yield duplex siRNA molecules directed specifically to mutant SERPIN A1 mRNAs. Such duplexed siRNAs are designed to reduce the accumulation of toxic AAT protein in liver cells. The methods and compositions of the present invention can be used in gene therapy for correction of SERPINA1 disorders such as AAT deficiency.
Owner:VIRXSYS
Use of polypeptides micro-molecule in preparing medicine for preventing and treating ischemic cerebrovascular disease
ActiveCN101361961AImprove protectionHigh transparencyTripeptide ingredientsCardiovascular disorderDiseaseBrain protection
The invention relates to the technical field of biological medicines, which is an application of a small polypeptide molecule in preparing a drug for preventing ischemic cerebrovascular disease. The research of the invention finds out that polypeptide bioactive molecules and polypeptide spliceosomes extracted from the secretion of amoeba histolytica have the pharmacological action of preventing ischemic cerebrovascular disease. The pentapeptide and tripeptide are proved to be capable of remarkably reducing the brain damage to the mouse caused by ischemia-reperfusion and remarkably improving the neuro behavioral of the mouse after ischemia by the results of pharmacological experiment on mouse models with focal cerebral ischemia. Compared with the kindred protein drugs, the small polypeptide molecule has strong brain protection effects; in addition, molecular weight is small, thereby being easy to permeate blood-brain barrier and having remarkable advantage of brain permeability. Therefore, the small polypeptide molecule provided has good application prospect in preparing the drug for preventing the ischemic cerebrovascular disease.
Owner:GUILIN EIGHT PLUS ONE PHARMA CO LTD
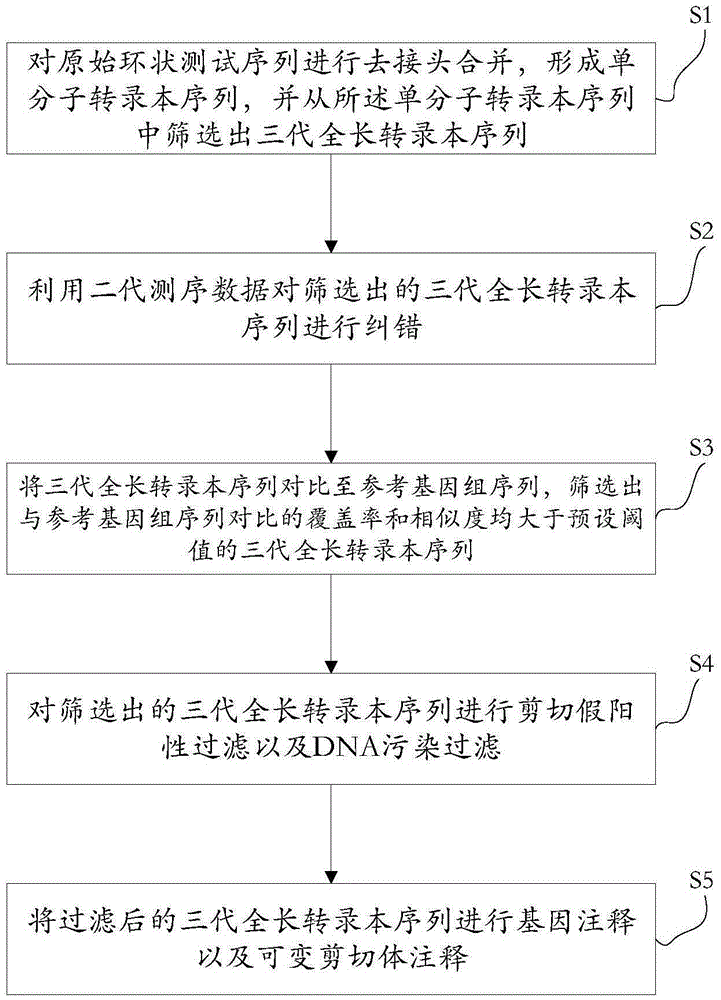
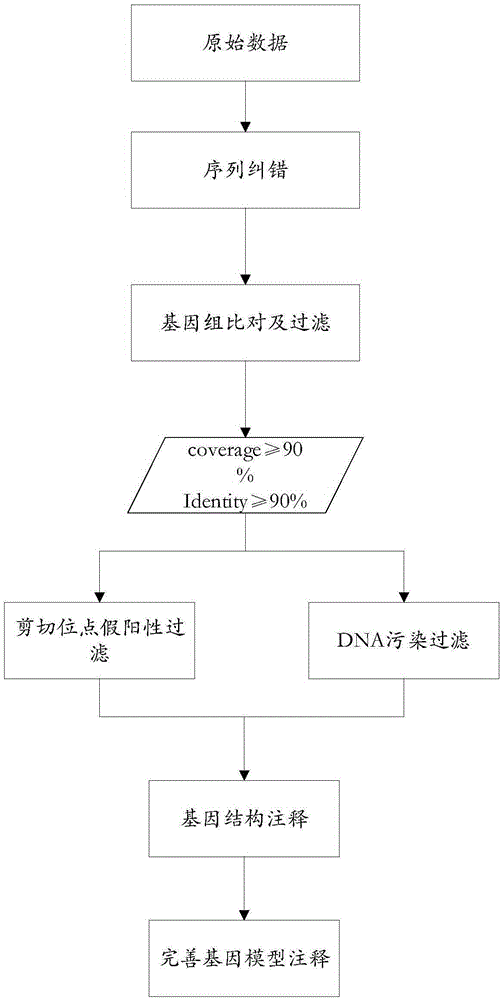
Method for detecting variable spliceosome in third generation full-length transcriptome
ActiveCN105389481AEfficient access to shear structuresPerfect commentSequence analysisSpecial data processing applicationsReference genome sequenceGene model
The invention discloses a method for detecting a variable spliceosome in a third generation full-length transcriptome. The method comprises the following steps: merging original annular test sequences with joints removed to form a monomolecular transcript sequence, and screening a third generation full-length transcript sequence; comparing the third generation full-length transcript sequence with a reference genome sequence, and screening a third generation full-length transcript sequence having coverage and similarity with the reference genome sequence larger than preset thresholds; carrying out splicing false positive filtration and DNA contamination filtration on the screened third generation full-length transcript sequence; and carrying out gene annotation and variable spliceosome annotation on the filtered third generation full-length transcript sequence. An overlong read length of a third generation sequencing technology mentioned in the method disclosed by the invention is large enough to cover most RNA, the third generation full-length transcript sequence can be obtained by SMRT sequencing transcriptomes without being assembled, and a splicing structure of a gene can be effectively obtained by third generation transcriptome sequencing, and more perfect gene model annotation can be constructed.
Owner:嘉兴菲沙基因信息有限公司
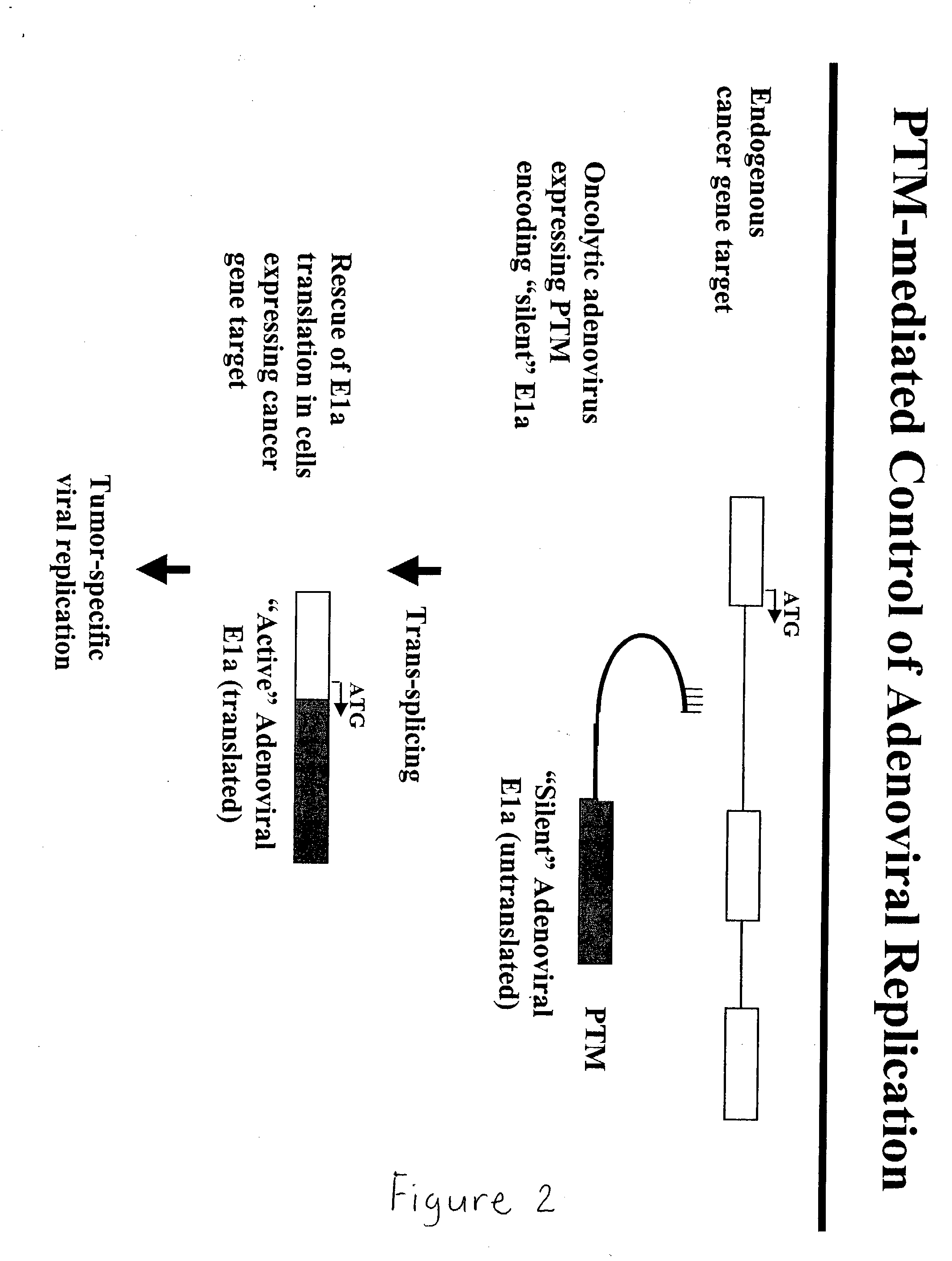
Use of spliceosome mediated RNA trans-splicing to confer cell selective replication to adenoviruses
The present invention provides methods and compositions for conferring tumor selective cell death on cancer cells expressing specific target precursor messenger RNA molecules (cancer cell selective target pre-mRNAs). The compositions of the invention include conditionally replicative adenoviruses that have been genetically engineered to express one or more pre-trans-splicing molecules (PTMs) designed to interact with one or more cancer cell target pre-mRNA and mediate a trans-splicing reaction resulting in the generation of novel chimeric RNA molecules (chimeric RNA) capable of encoding adenovirus specific protein(s). Adenovirus specific proteins include those proteins complementing an essential activity necessary for replication of a defective adenovirus. The methods and compositions of the invention may be used to target a lytic adenovirus infection to cancer cells thereby providing a method for selective destruction of cancer cells. In addition, the adenoviruses of the invention may be engineered to encode PTMs designed to interact with target pre-mRNAs encoded by infectious agents within a cell, thereby targeting selective destruction of cells infected with such agents.
Owner:VIRXSYS
Targeted oligonucleotides
Methods and compositions are provided for oligonucleotides that bind targets of interest. The targets include cells and microvesicles, such as those derived from various diseases. The oligonucleotides can be used for diagnostic and therapeutic purposes. The target of the oligonucleotides can be a member of a ribonucleoprotein or spliceosomal complex such heterologous nuclear ribonucleoprotein U (hnRNP U).
Owner:CARIS SCIENCE INC
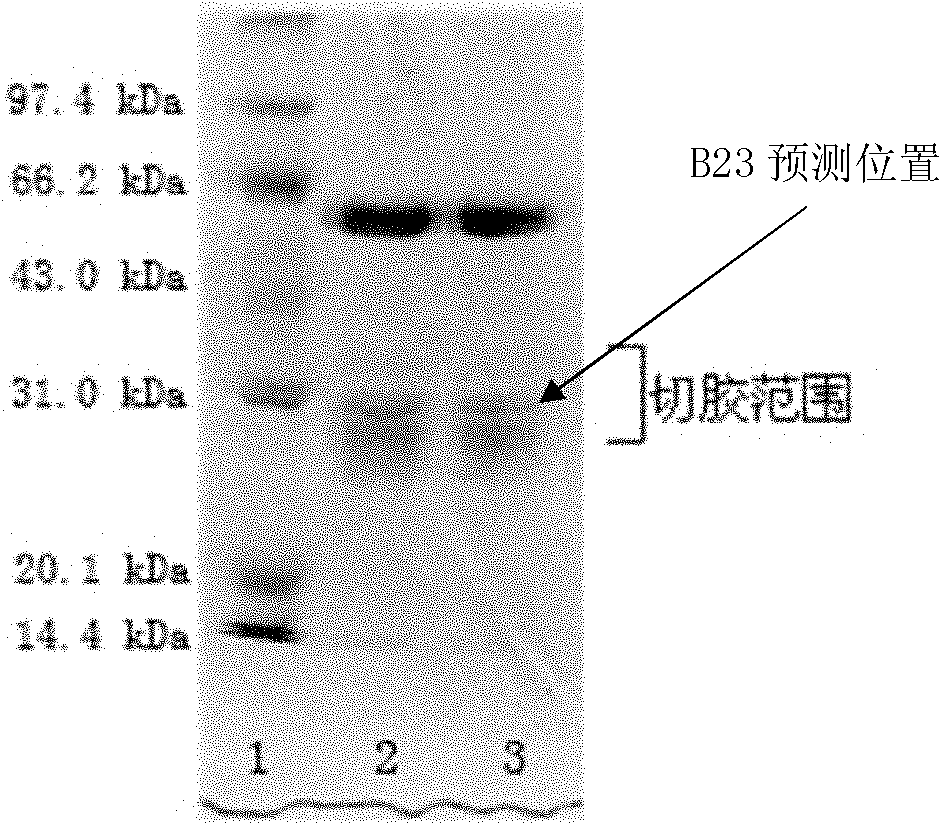
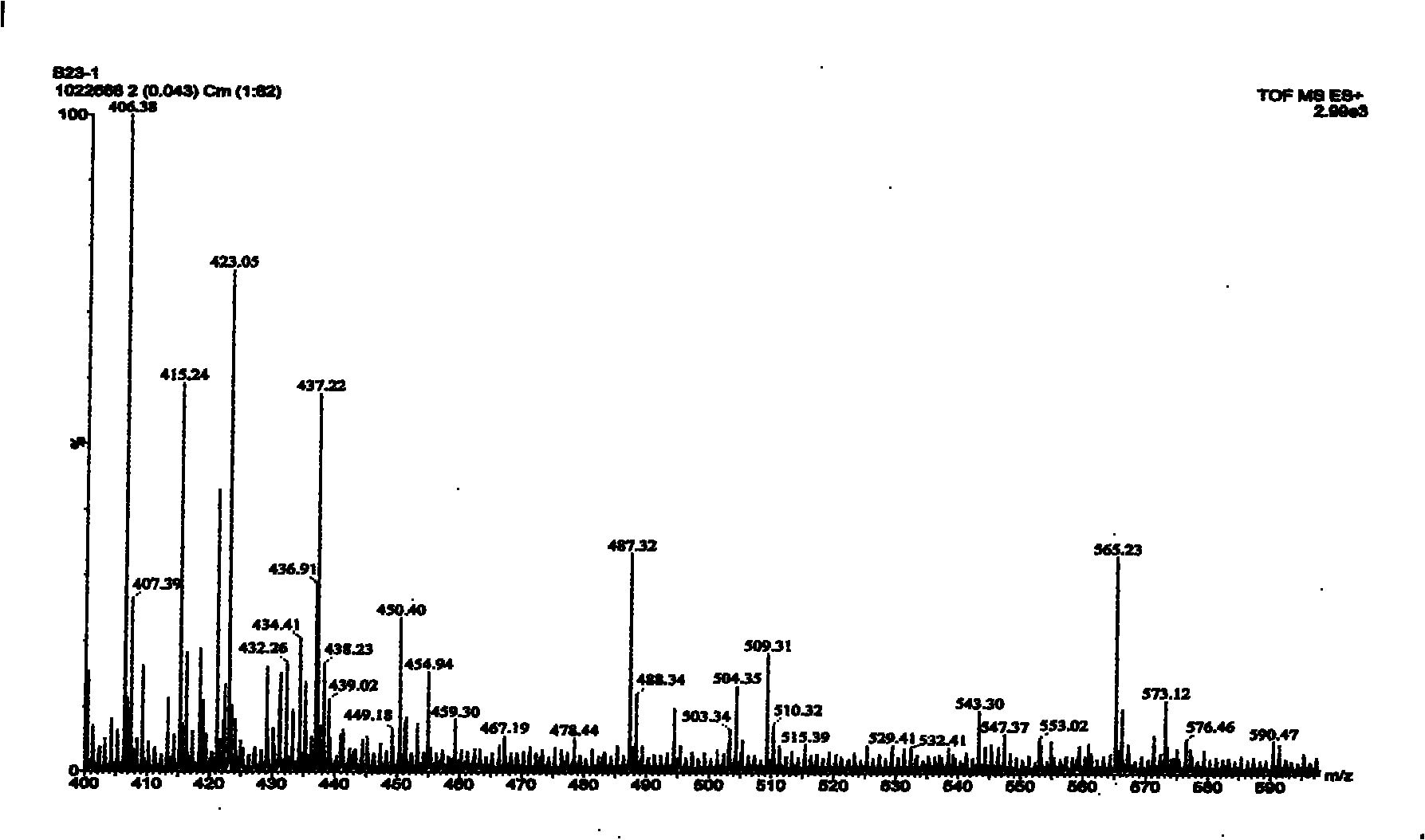
Mass spectra identification method of nucleophosmin variable spliceosome and gastric cancer diagnostic reagent kit
ActiveCN102156164AReliable determinationGood repeatabilityComponent separationMaterial analysis by electric/magnetic meansSpliceosome complexMass Spectrometry-Mass Spectrometry
The invention provides a mass spectrometry identification method of nucleophosmin variable spliceosome and a gastric cancer diagnostic reagent kit. The method is combined by a Glu-C enzyme cutting method and a Q-TOF (quadrupole time-of flight) mass spectra method, and can be applied to the clinical diagnosis and treatment evaluation of the gastric cancer; and by the method disclosed by the invention, the rapid and reliable identification result can be acquired as compared with the traditional methods.
Owner:BEIJING 3S CENTURY TECH CORP
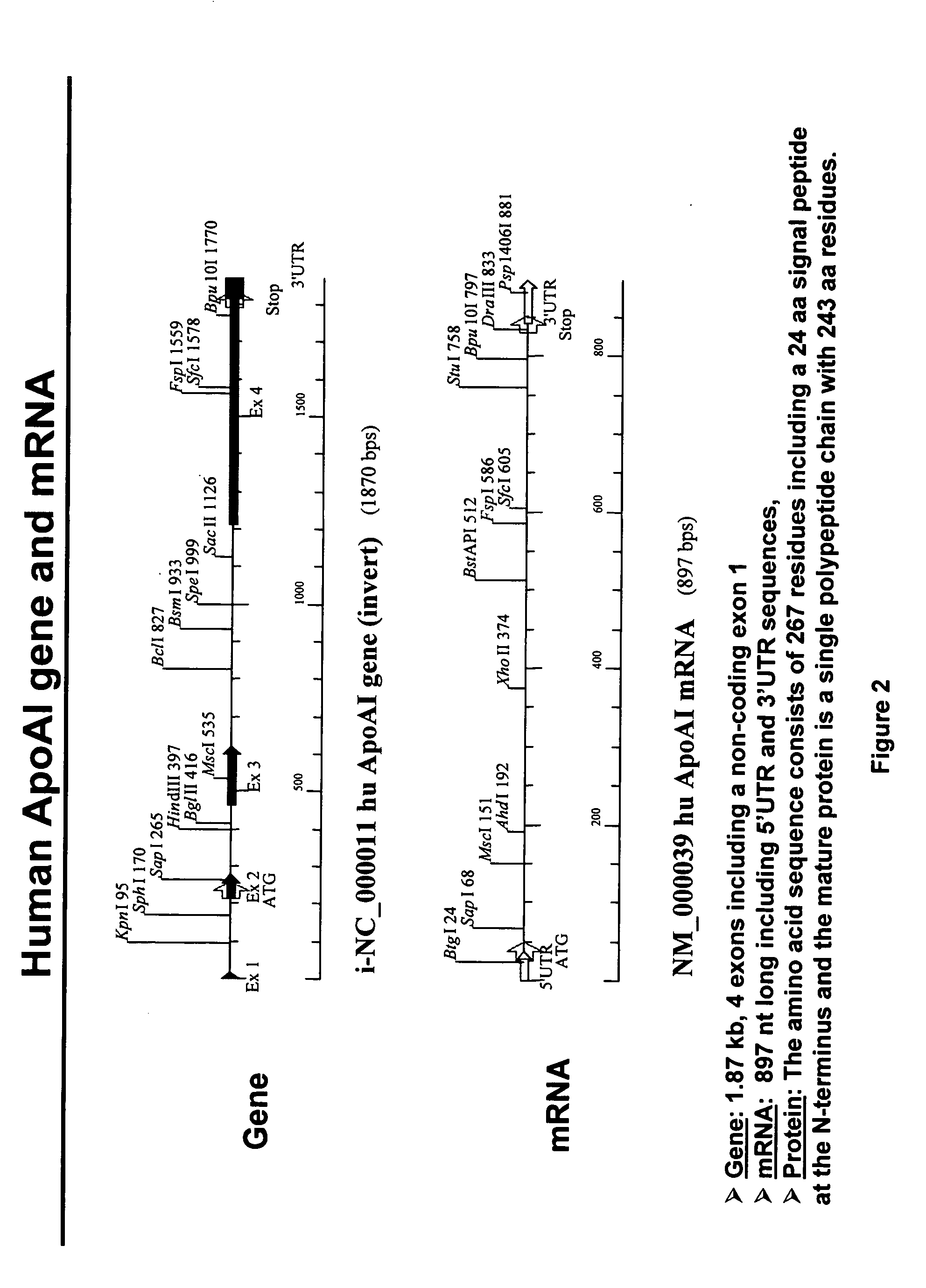
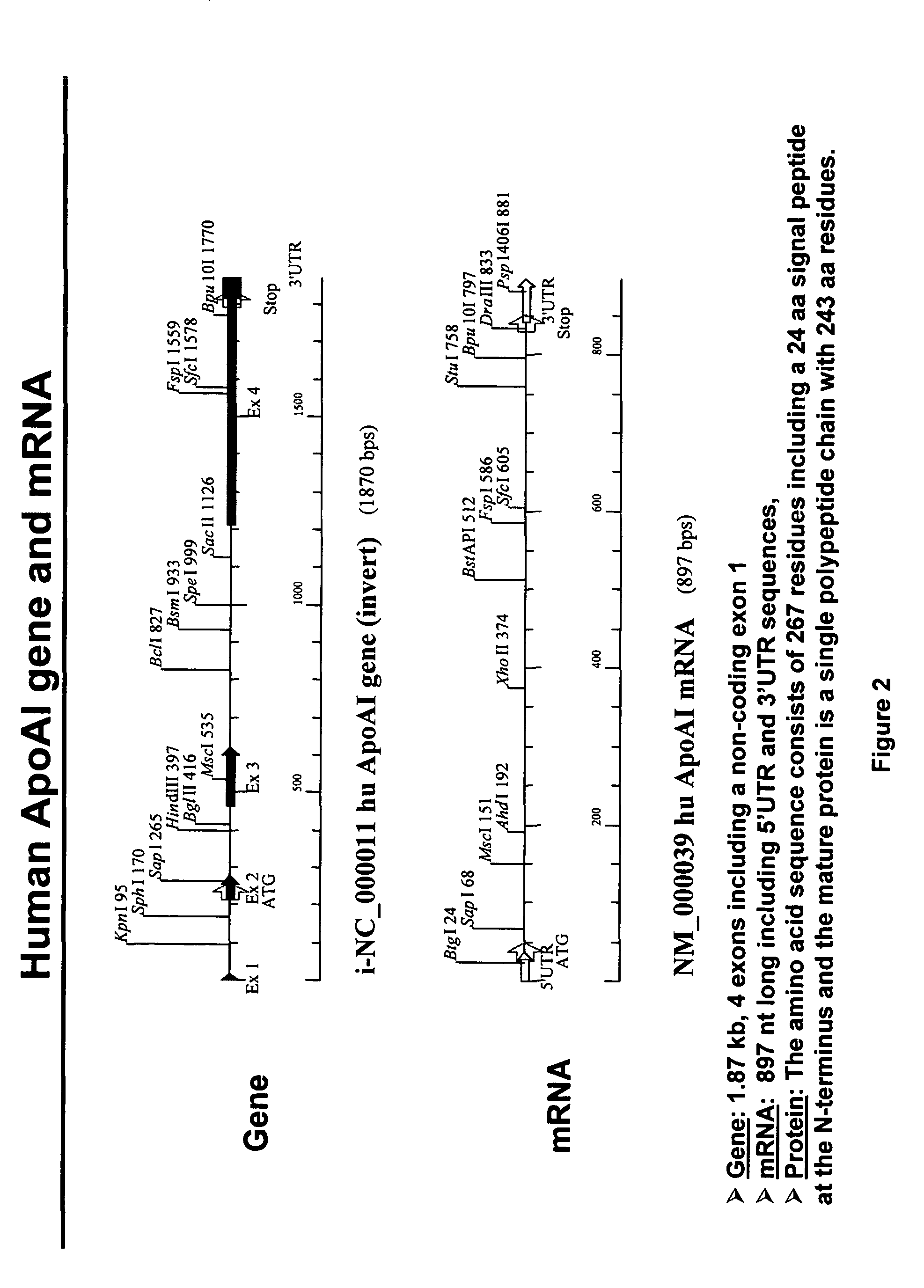
Expression of apoAI and variants thereof using spliceosome mediated RNA trans-splicing
InactiveUS20060194317A1Improving biological half lifeFunction increaseSplicing alterationAntibody mimetics/scaffoldsRNA Trans-SplicingMessenger RNA
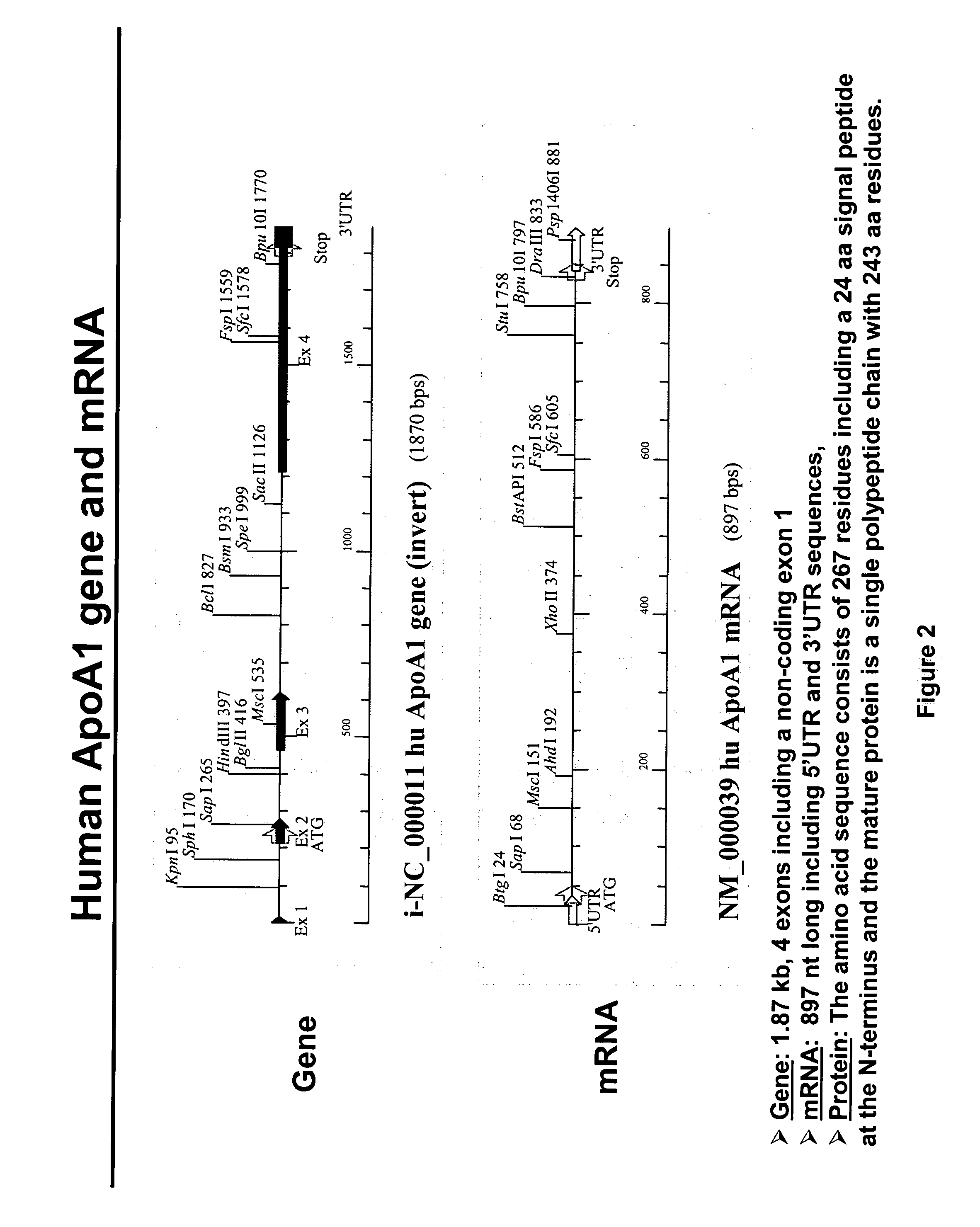
Methods and compositions for generating novel nucleic acid molecules through targeted spliceosome mediated RNA trans-splicing that result in expression of a apoAI protein, an apoAI variant, the preferred embodiment referred to herein as the apoAI Milano variant, a pre-pro-apoAI or an analogue of apoAI. The methods and compositions include pre-trans-splicing molecules (PTMs) designed to interact with a target precursor messenger RNA molecule (target pre-mRNA) and mediate a trans-splicing reaction resulting in the generation of a novel chimeric RNA molecule (chimeric RNA) capable of encoding apoAI, the apoAI Milano variant, or an analogue of apoAI. The expression of this apoAI protein results in protection against vascular disorders resulting from plaque build up, i.e., atherosclerosis, strokes and heart attacks.
Owner:VIRXSYS
Use of spliceosome mediated RNA trans-splicing for immunotherapy
InactiveUS20060094110A1Stimulate immune responseGood health benefitsSugar derivativesTissue cultureRNA Trans-SplicingMessenger RNA
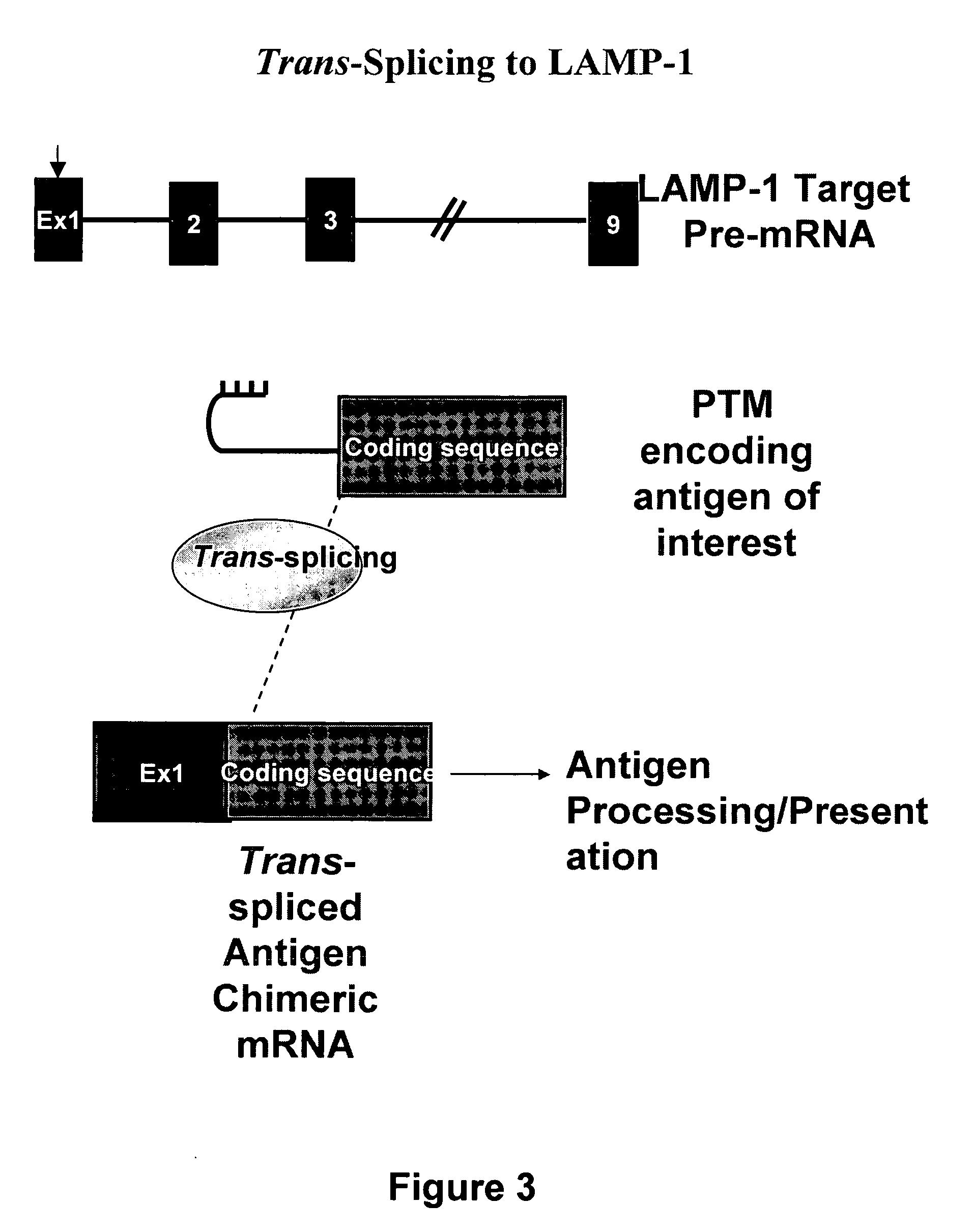
Methods and compositions for generating novel nucleic acid molecules through targeted spliceosomal mediated trans-splicing that result in expression of an immunogenic polypeptide. The invention includes pre-trans-splicing molecules (PTMs) designed to interact with a target precursor messenger RNA molecule (target pre-mRNA) and mediate a trans-splicing reaction resulting in the generation of a novel chimeric RNA molecule (chimeric RNA) capable of encoding the immunogenic polypeptide, recombinant vector systems capable of expressing the PTMs of the invention, and cells expressing said PTMs. The target pre-mRNA are those encoding proteins that function in antigen uptake, antigen presentation and chaperoning. The methods of the invention encompass contacting the PTMs of the invention with a target pre-mRNA, under conditions in which a portion of the PTM is trans-spliced to a portion of the target pre-mRNA to form a chimeric mRNA molecule capable of encoding an immunogenic polypeptide.
Owner:VIRXSYS
Correction of factor VIII genetic defects using spliceosome mediated RNA trans splicing
InactiveUS20040126774A1Splicing alterationMicrobiological testing/measurementRNA Trans-SplicingVector system
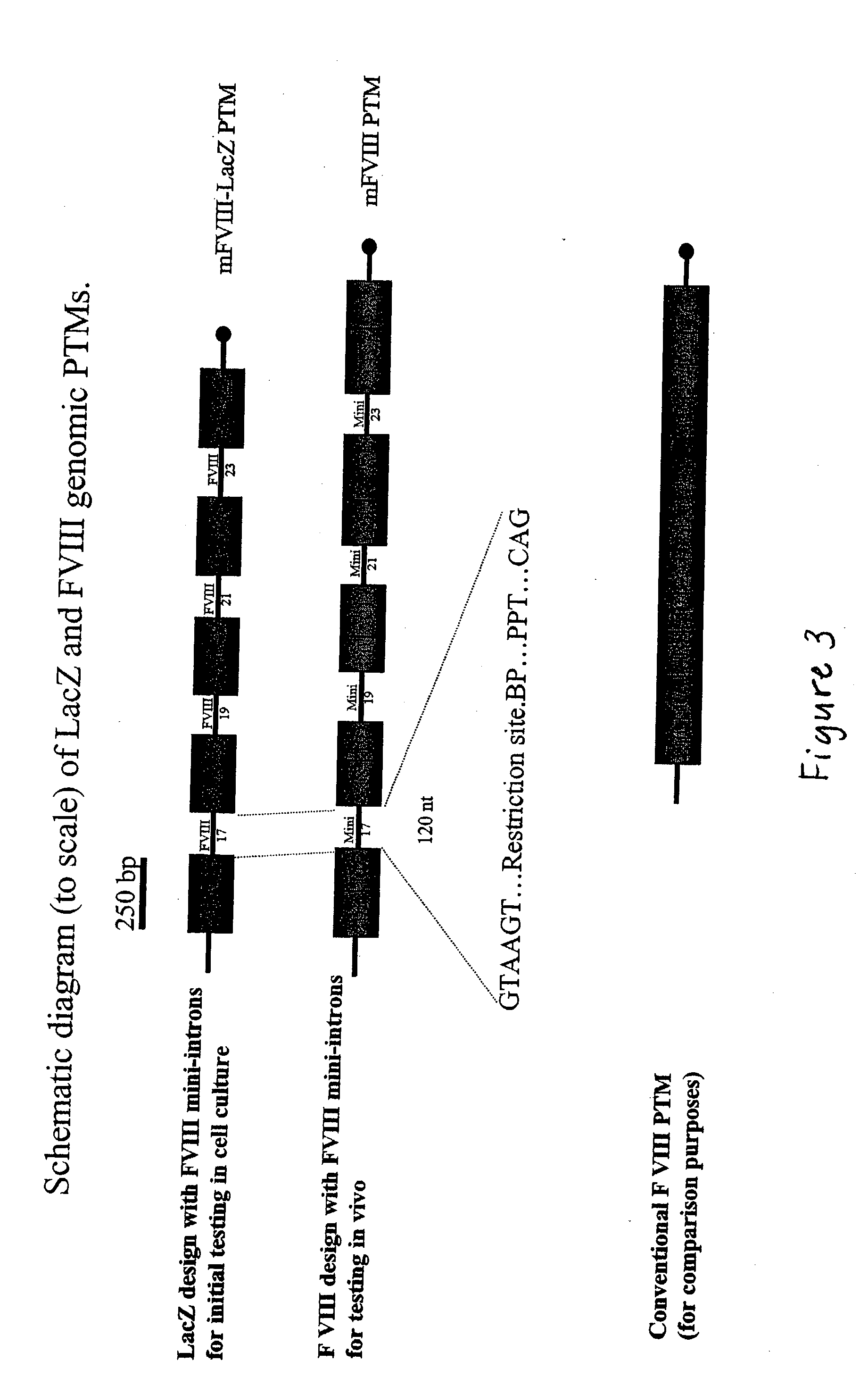
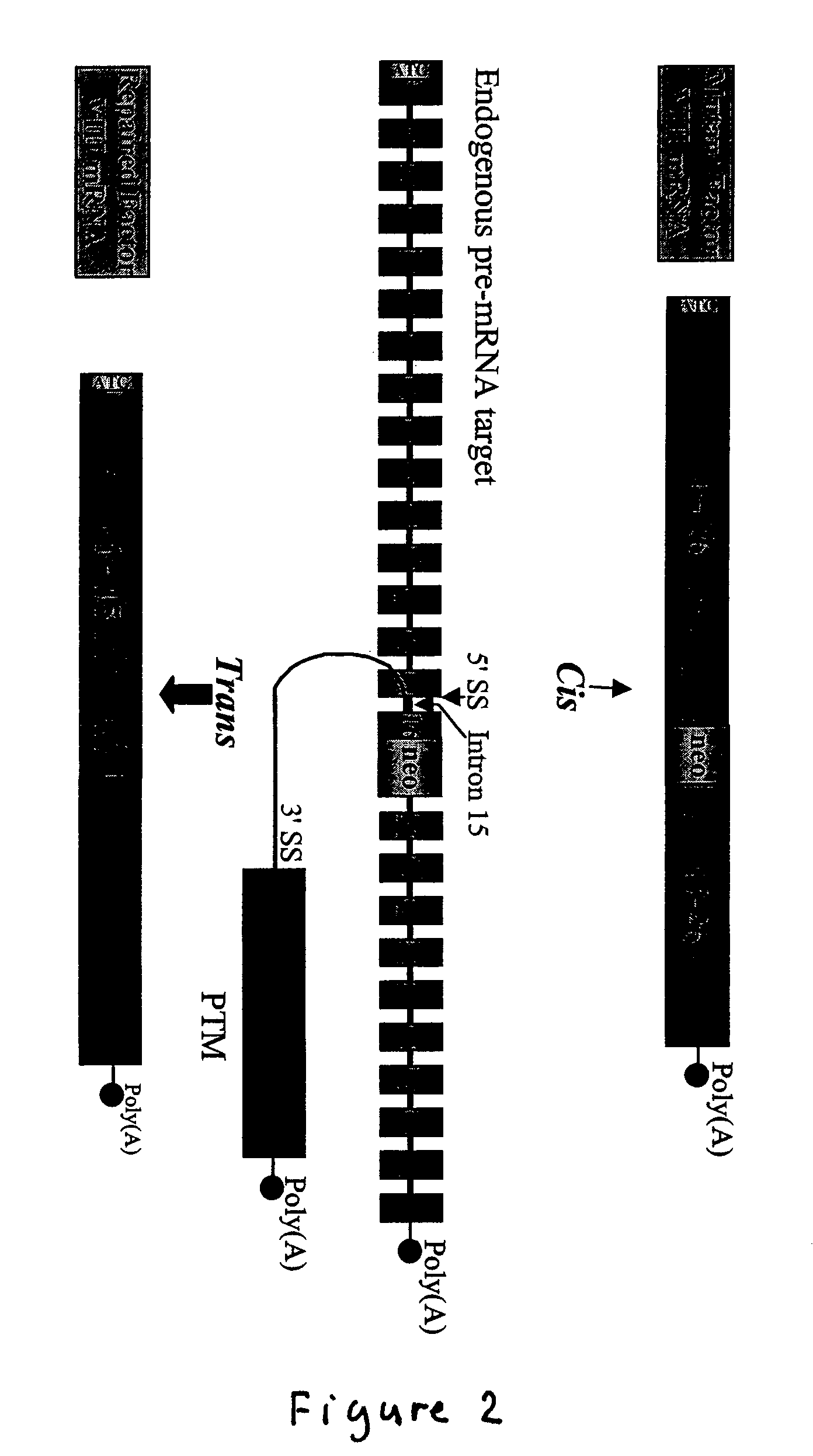
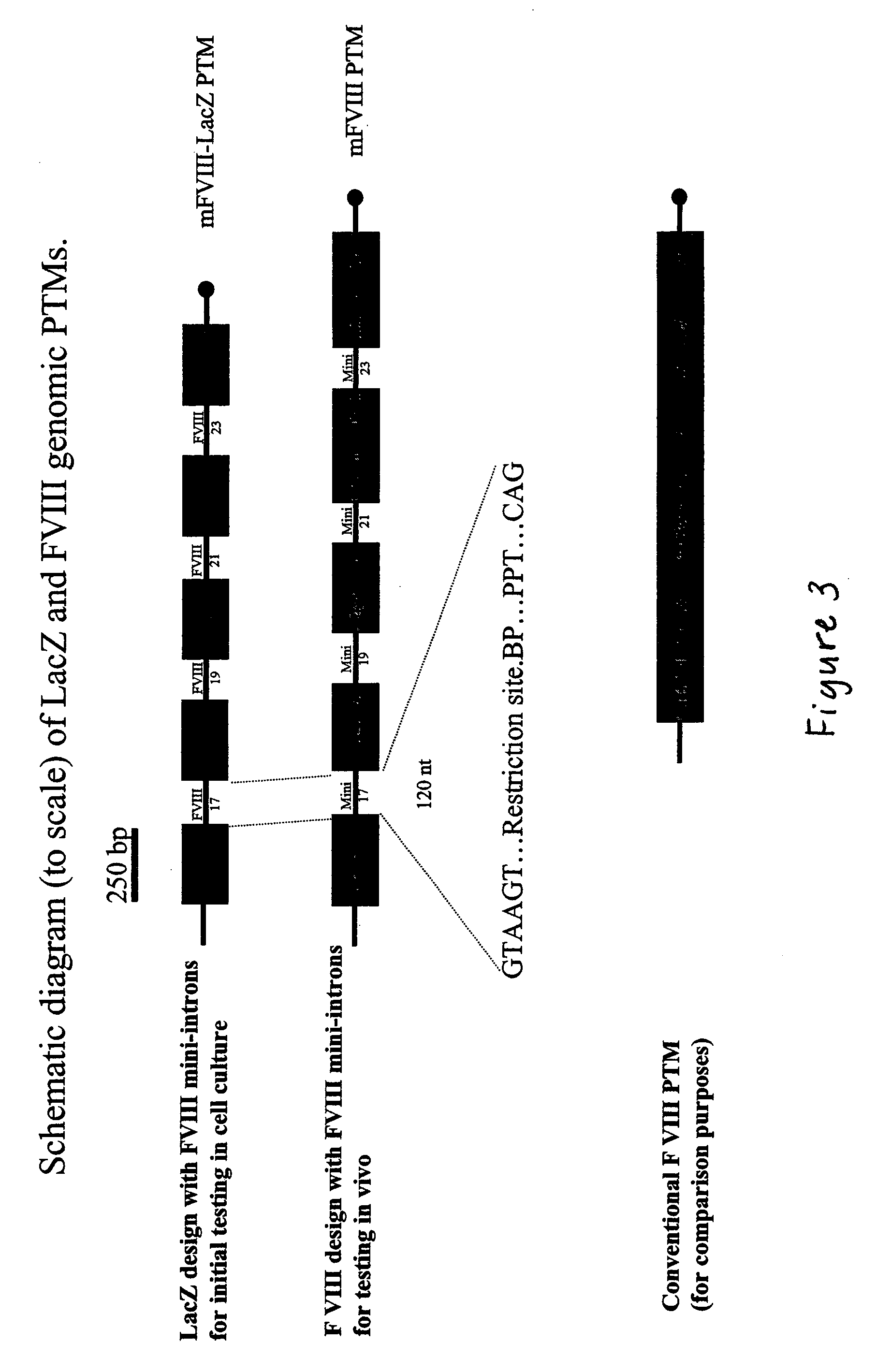
The present invention provides methods and compositions for generating novel nucleic acid molecules through targeted spliceosomal mediated trans-splicing. The compositions of the invention include pre-trans-splicing molecules (PTMs) designed to interact with a target precursor messenger RNA molecule (target pre-mRNA) and mediate a trans-splicing reaction resulting in the generation of a novel chimeric RNA molecule (chimeric RNA). In particular, the PTMs of the present invention are genetically engineered to interact with factor VIII (FVIII) target pre-mRNA so as to result in correction of clotting FVIII genetic defects responsible for hemophilia A. The compositions of the invention further include recombinant vector systems capable of expressing the PTMs of the invention and cells expressing said PTMs. The methods of the invention encompass contacting the PTMs of the invention with a FVIII target pre-mRNA under conditions in which a portion of the PTM is trans-spliced to a portion of the target pre-mRNA to form a RNA molecule wherein the genetic defect in the FVIII gene has been corrected. The methods and compositions of the present invention can be used in gene therapy for correction of FVIII disorders such as hemophilia A.
Owner:VIRXSYS
Expression of apoA-1 and variants thereof using spliceosome mediated RNA trans-splicing
InactiveUS20060177933A1Reduce buildReduce usageSplicing alterationApolipeptidesRNA Trans-SplicingMessenger RNA
The present invention provides methods and compositions for generating novel nucleic acid molecules through targeted spliceosome mediated RNA trans-splicing that result in expression of an apoA-1 variant, the preferred embodiment referred to herein as the apoA-1 Milano variant. The compositions of the invention include pre-trans-splicing molecules (PTMs) designed to interact with a target precursor messenger RNA molecule (target pre-mRNA) and mediate a trans-splicing reaction resulting in the generation of a novel chimeric RNA molecule (chimeric RNA) capable of encoding the apoA-1 Milano variant. The expression of this variant protein results in protection against vascular disorders resulting from plaque build up, i.e., strokes and heart attacks. In particular, the PTMs of the present invention include those genetically engineered to interact with the apoA-1 target pre-mRNA so as to result in expression of the apoA-1 Milano variant. In addition, the PTMs of the invention include those genetically engineered to interact with the apoB or albumin or other specific target pre-mRNAs so as to result in expression of an apoB / apoA-1 and / or alb / apoA-1 wild type or Milano fusion protein thereby reducing apoB expression and simultaneously produce ApoA-1 function.
Owner:VIRXSYS
Methods and compositions for use in spliceosome mediated RNA trans-splicing
InactiveUS20020193580A1Improve abilitiesEfficient processVectorsSugar derivativesTrans-splicingSpliceosome complex
The present invention provides methods and compositions for delivery of synthetic pre-trans-splicing molecules (synthetic PTMs) into a target cell. The compositions of the invention include synthetic pre-trans-splicing molecules (PTMs) with enhanced stability against chemical and enzymatic degradation. The synthetic PTMs are designed to interact with a natural target precursor messenger RNA molecule (target pre-mRNA) and mediate a trans-splicing reaction resulting in the generation of a novel chimeric RNA molecule (chimeric RNA).
Owner:VIRXSYS
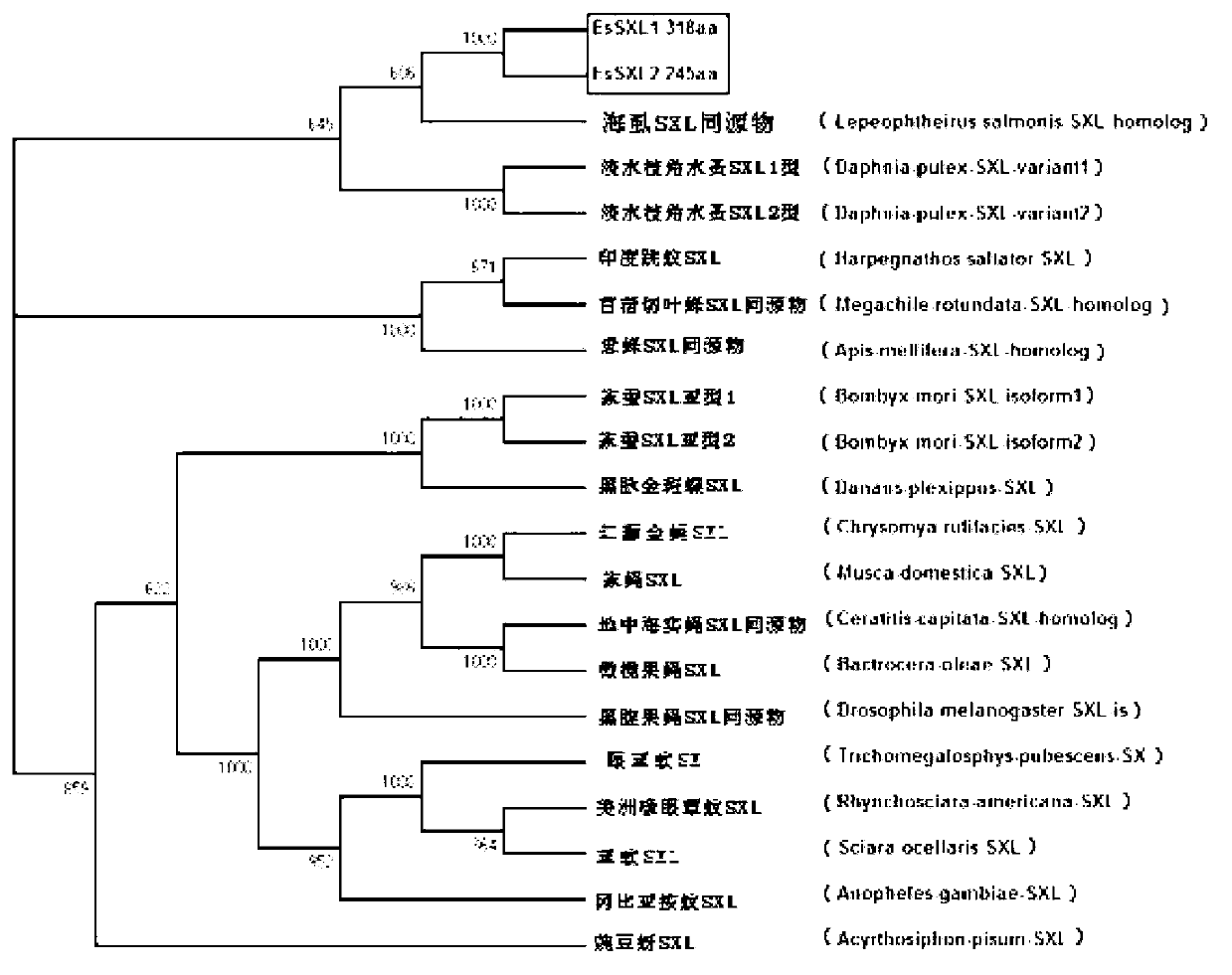
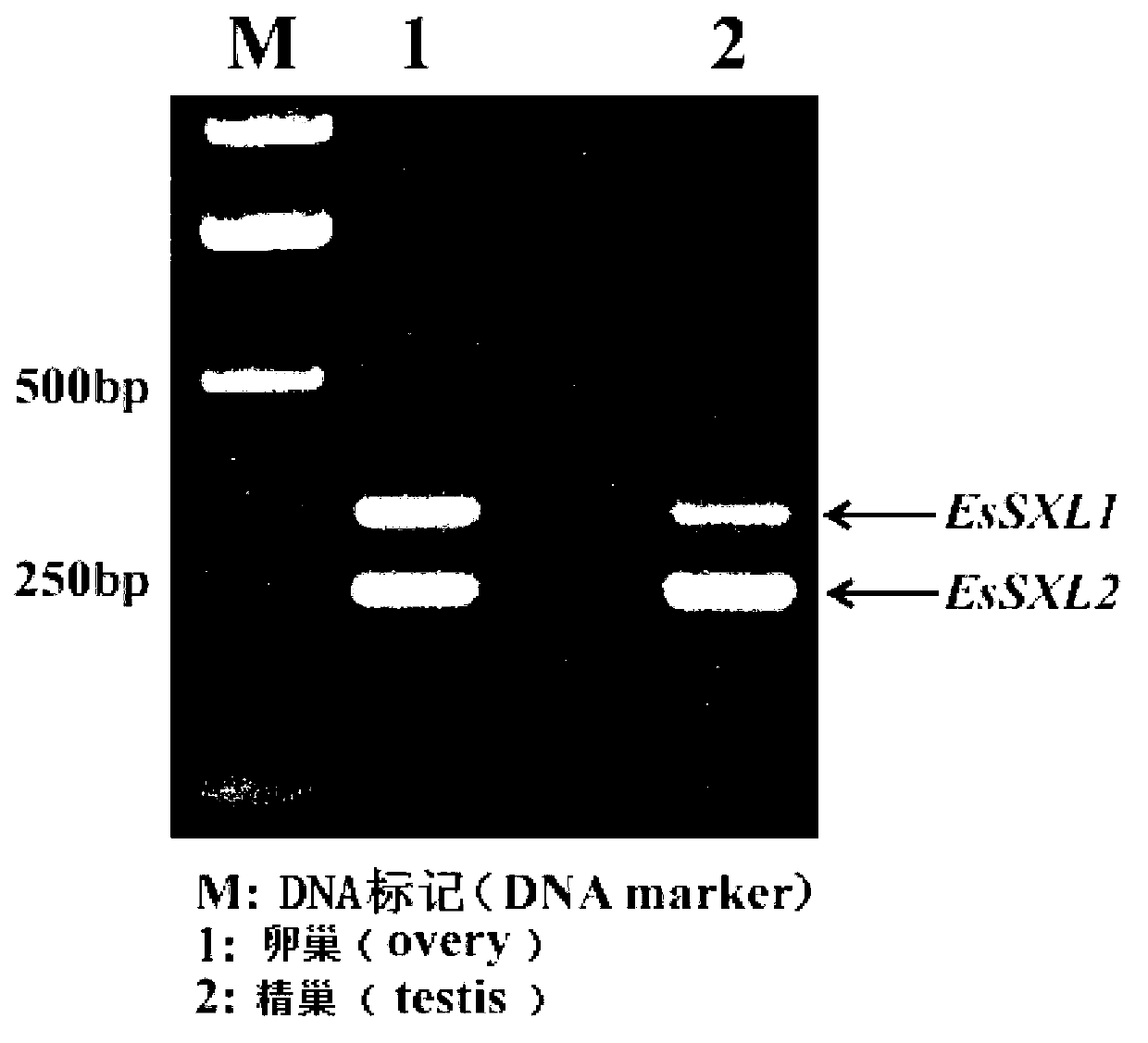
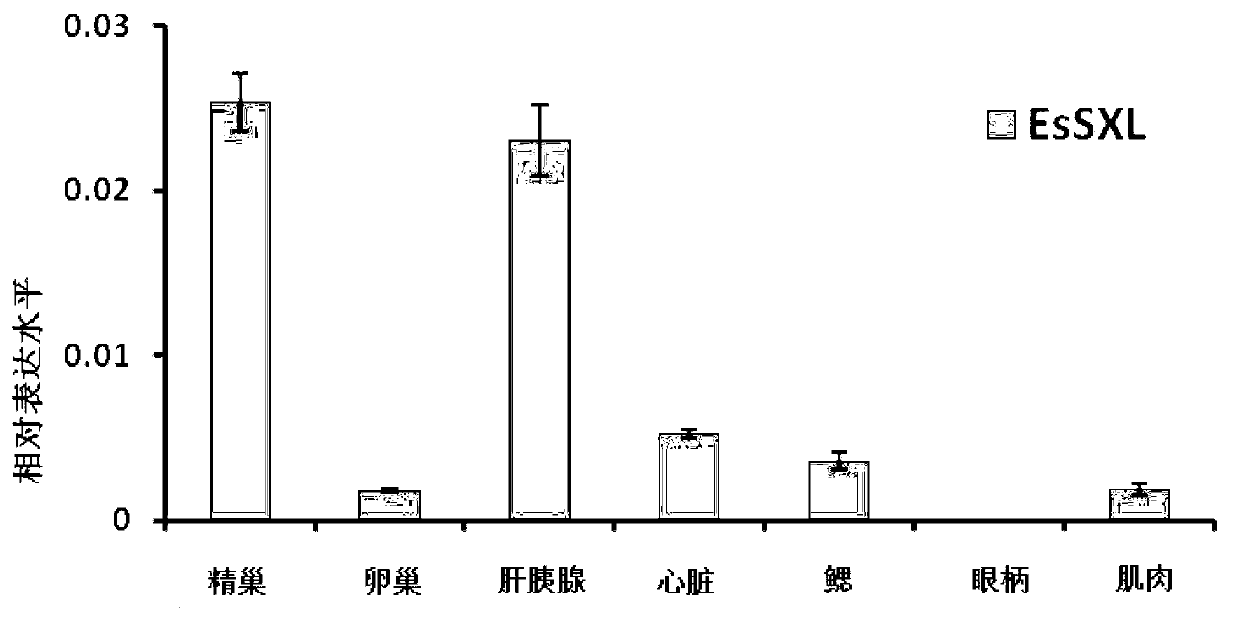
Eriocheir sinensis EsSXL gene, amplification primer group thereof and amplification method
The invention discloses an Eriocheir sinensis EsSXL gene, an amplification primer group thereof and an amplification method. A full-length cDNA (complementary Deoxyribose Nucleic Acid) sequence of the EsSXL gene is cloned from an Eriocheir sinensis testis tissue by using the primer group provided by the invention, and the EsSXL gene comprises two variable spliceosomes EsSXL1 and EsSXL2, and lays an important foundation for the research of the important function of the function of the Eriocheir sinensis EsSXL gene in a sex differentiation process of the Eriocheir sinensis EsSXL gene.
Owner:FRESHWATER FISHERIES RES CENT OF CHINESE ACAD OF FISHERY SCI
Methods and compositions for use in spliceosome mediated RNA trans-splicing
InactiveUS20030027250A1Inhibition of translationPeptide/protein ingredientsGenetic material ingredientsRNA Trans-SplicingIntein
The molecules and methods of the present invention provide a means for in vivo production of a trans-spliced molecule in a selected subset of cells. The pre-trans-splicing molecules of the invention are substrates for a trans-splicing reaction between the pre-trans-splicing molecules and a pre-mRNA which is uniquely expressed in the specific target cells. The in vivo trans-splicing reaction provides a novel mRNA which is functional as mRNA or encodes a protein to be expressed in the target cells. The expression product of the mRNA is a protein of therapeutic value to the cell or host organism a toxin which causes killing of the specific cells or a novel protein not normally present in such cells. The invention further provides PTMs that have been genetically engineered for the identification of exon / intron boundaries of pre-mRNA molecules using an exon tagging method. The PTMs of the invention can also be designed to result in the production of chimeric RNA encoding for peptide affinity purification tags which can be used to purify and identify proteins expressed in a specific cell type.
Owner:VIRXSYS
Methods and compositions for use in spliceosome mediated RNA trans-splicing
InactiveUS20030148937A1Inhibition of translationFactor VIIPeptide/protein ingredientsRNA Trans-SplicingIntein
The molecules and methods of the present invention provide a means for in vivo production of a trans-spliced molecule in a selected subset of cells. The pre-trans-splicing molecules of the invention are substrates for a trans-splicing reaction between the pre-trans-splicing molecules and a pre-mRNA which is uniquely expressed in the specific target cells. The in vivo trans-splicing reaction provides a novel mRNA which is functional as mRNA or encodes a protein to be expressed in the target cells. The expression product of the mRNA is a protein of therapeutic value to the cell or host organism a toxin which causes killing of the specific cells or a novel protein not normally present in such cells. The invention further provides PTMs that have been genetically engineered for the identification of exon / intron boundaries of pre-mRNA molecules using an exon tagging method. The PTMs of the invention can also be designed to result in the production of chimeric RNA encoding for peptide affinity purification tags which can be used to purify and identify proteins expressed in a specific cell type.
Owner:VIRXSYS
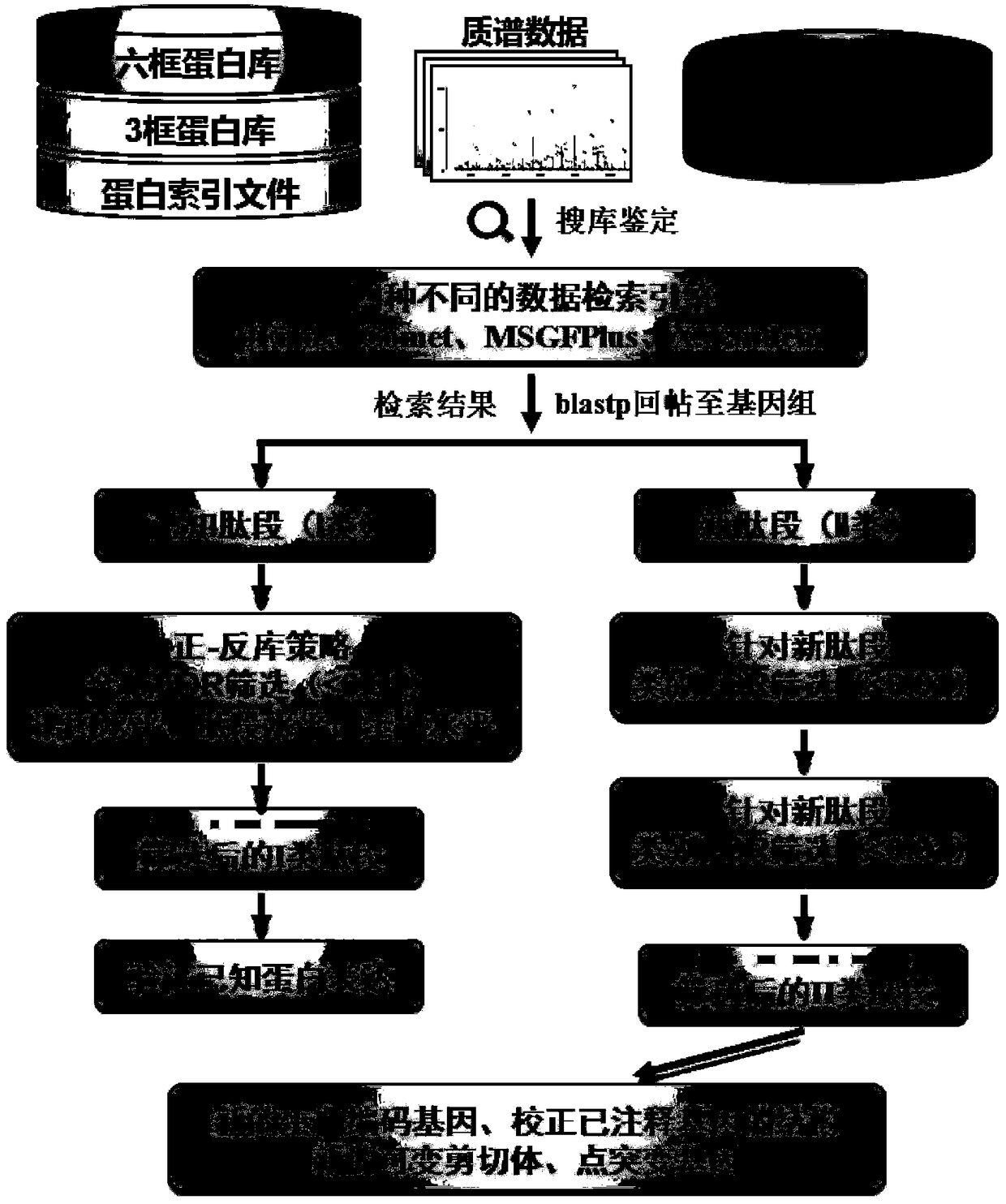
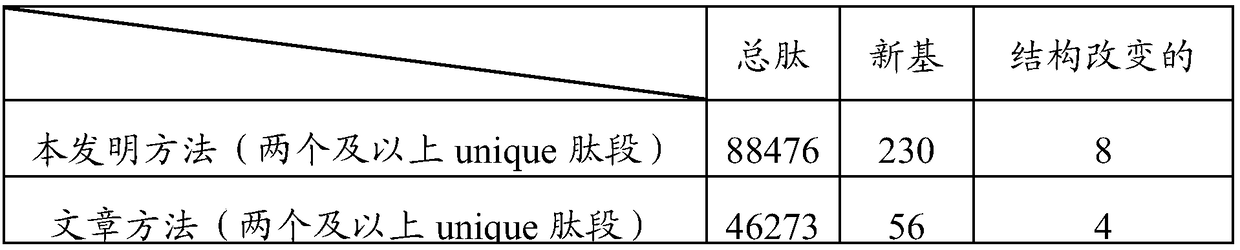
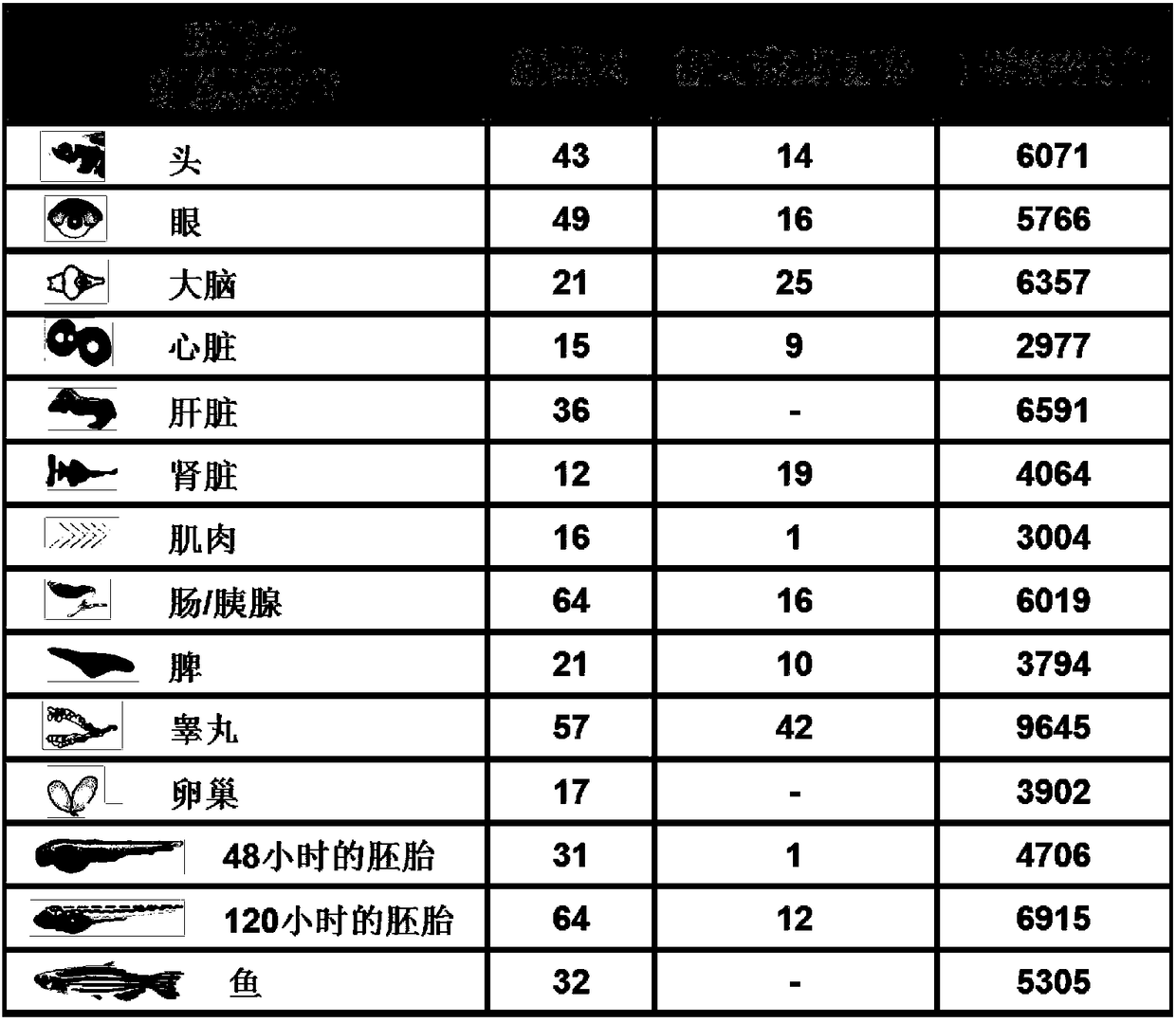
Method for rapidly analyzing eukaryotic protein genomic data
ActiveCN108920898ARapid Identification AnalysisIncrease coverageSpecial data processing applicationsPositive controlScreening method
The invention provides a method for rapidly analyzing eukaryotic protein genomic data, and belongs to the technical field of protein genomic data analysis methods. According to the method for rapidlyanalyzing eukaryotic protein genomic data, II-type credible peptide fragments are obtained by adoption of a prokaryote multi-group data arrangement method and a screening method; and three different genome replying methods for the aims of predicting new genes, variant spliceosomes and point mutation genes and correcting structures of annotated genes are designed. The method provided by the invention is suitable for any sequenced eukaryon, and through a variant spliceosome and point mutation gene prediction method, the coverage degree of authentication is improved; by adoption of different relatively strict false positive control strategies, the credibility of the authentication is improved; and through predicting and correcting original mass spectrometric data, final new genes, variant spliceosome and point mutation genes, annotated gene structure series are analyzed, so that rapid authentication and analysis of eukaryotic mass spectrometric data are really realized.
Owner:INST OF AQUATIC LIFE ACAD SINICA
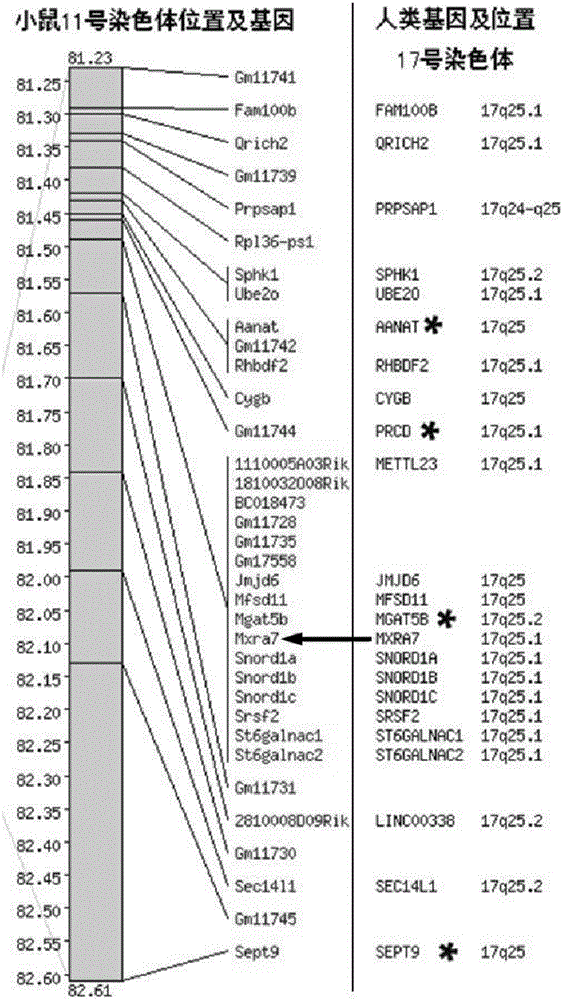
Tumor marker and application thereof
ActiveCN105753960AStrong specificityMicrobiological testing/measurementBiological material analysisSpliceosome complexTreatment strategy
The invention discloses a tumor marker and an application thereof. The tumor marker comprises a spliceosome 1 or an amino acid sequence SEQ ID NO.1 thereof, a spliceosome 2 or an amino acid sequence SEQ ID NO.2, and a spliceosome 3 or an amino acid sequence SEQ ID NO.3. (1) Three different spliceosomes of MXRA 7 show different change patterns in different tumors, the gene can be expressed in different tumors, and the MXRA 7 has different expression patterns in different stages of the tumors or during the growth and metastasis of the tumors, so that the occurrence, growth, metastasis and the like of different tumors can be determined according to the transcription or expression level of the MXRA7; (2) the MXRA 7 can be used as a target spot of tumor treatment, and a new tumor treatment strategy is provided by regulating and controlling the expression process of the MXRA 7; and (3) the marker disclosed by the invention is strong in specificity, and is suitable for multiple types of tumors.
Owner:THE FIRST AFFILIATED HOSPITAL OF SOOCHOW UNIV
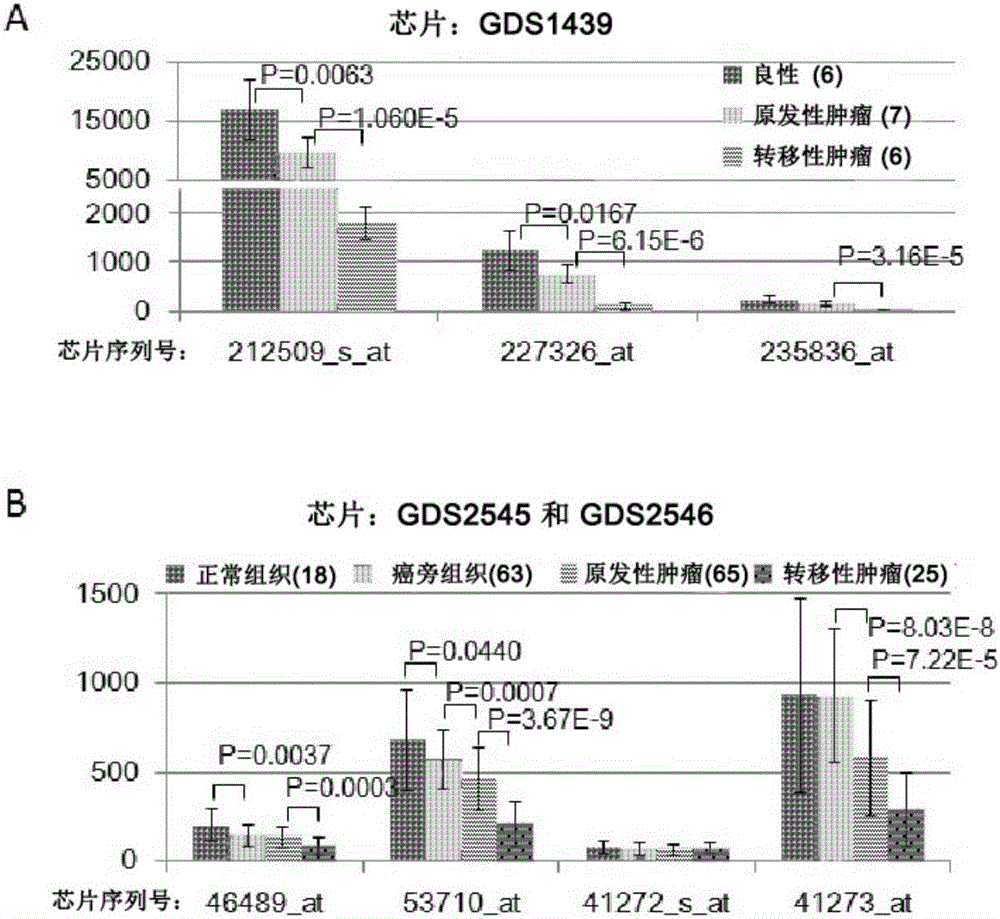
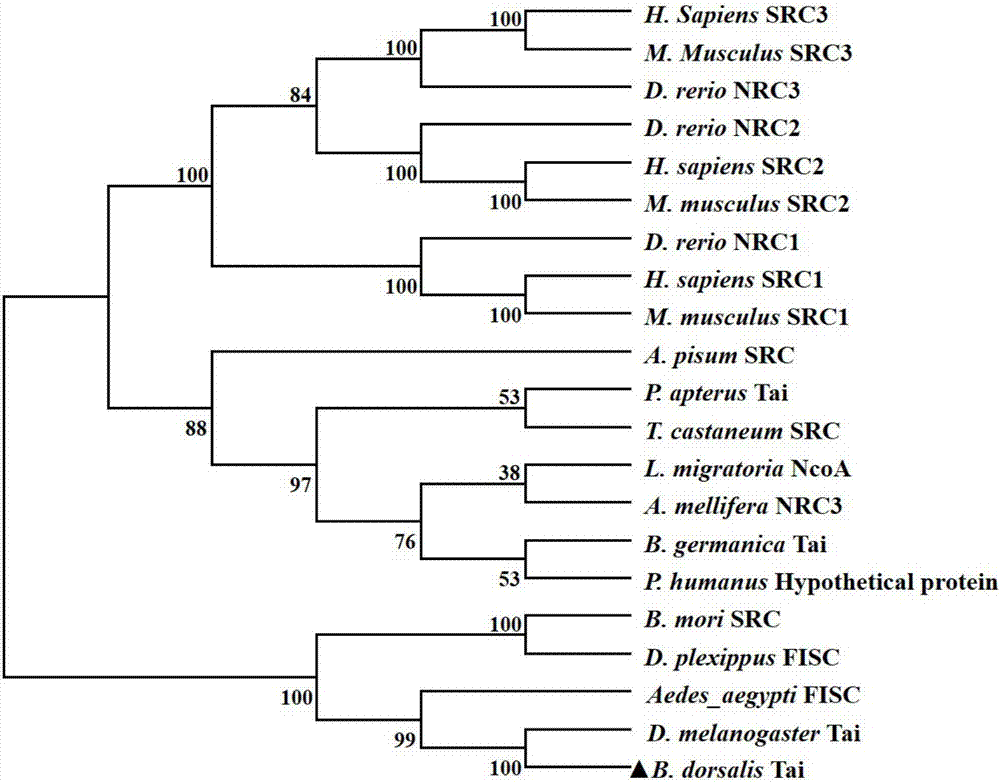
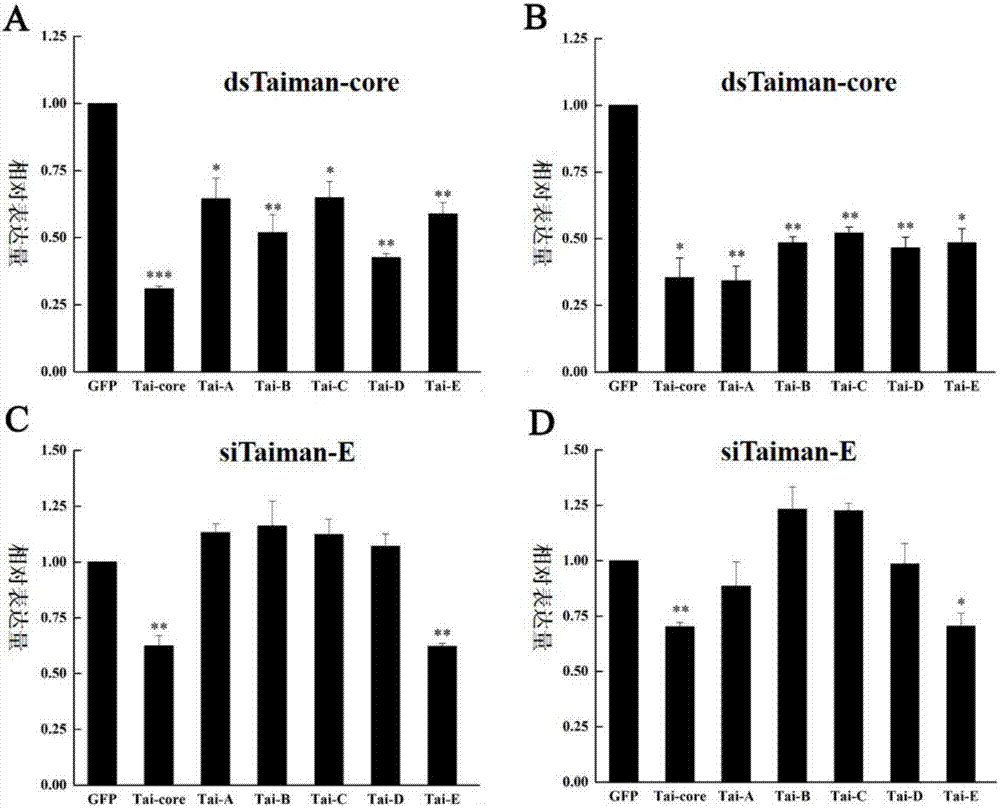
A bactrocera dorsalis Taiman gene, a detecting method of variable spliceosomes thereof and dsRNA of the gene
InactiveCN106916824ASolving the dsRNA problemSilent efficiency is highMicrobiological testing/measurementPeptidesSpliceosome complexNucleotide sequencing
A bactrocera dorsalis Taiman gene is disclosed. The nucleotide sequence of the gene is shown as SEQ ID NO:11 or 12 or 13 or 14. Segmented amplification primer pairs for amplifying the full-length bactrocera dorsalis Taiman gene are also disclosed. Sequences of the primers are shown as SEQ ID NO:1 to SEQ ID NO:10 in order. The invention also discloses dsRNA of the gene, a synthetic method thereof and applications of the dsRNA. The invention discloses a method of subjecting the gene to qRT-PCR detection. Sequences of primers used in qPCR are shown as SEQ ID NO:27 and SEQ ID NO:28. The invention also discloses methods of performing qRT-PCR detection on the Taiman gene shown as the SEQ ID NO:11, 12, 13 and 14 respectively and primer sequences are shown as SEQ ID NO:29 to SEQ ID NO:36 respectively.
Owner:SOUTHWEST UNIV
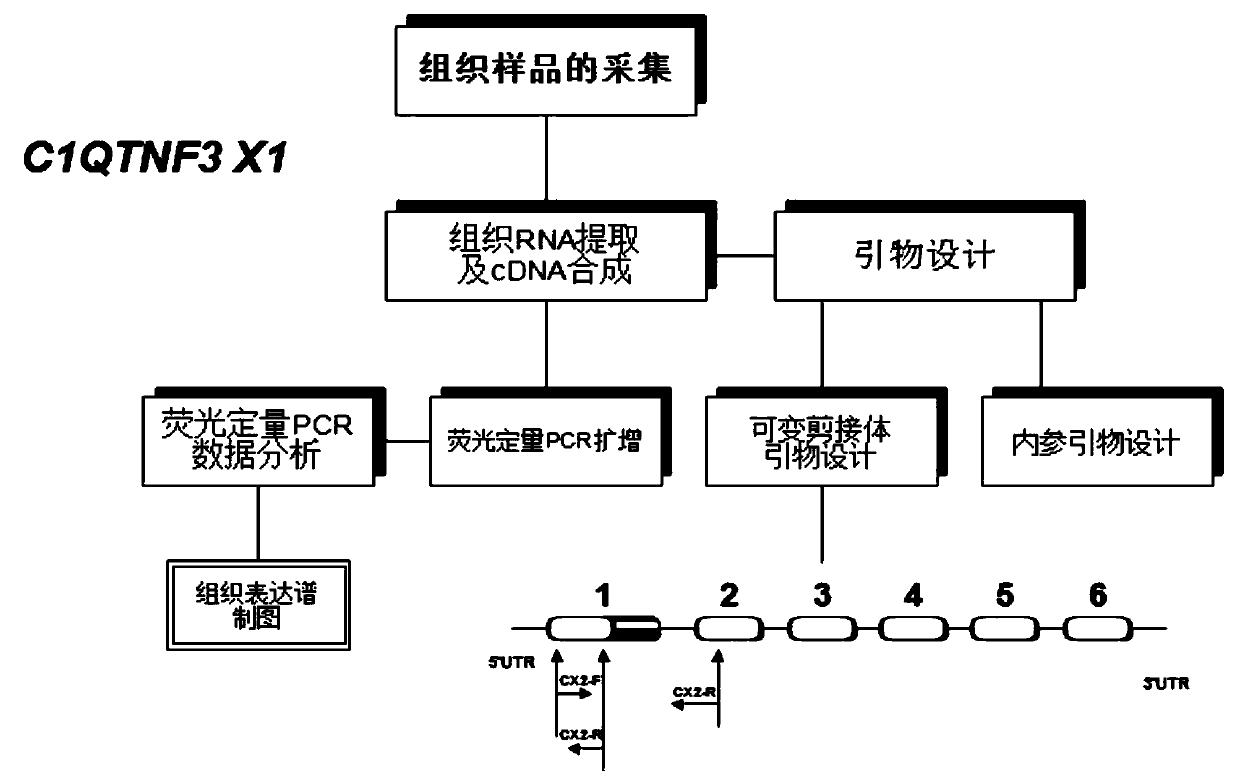
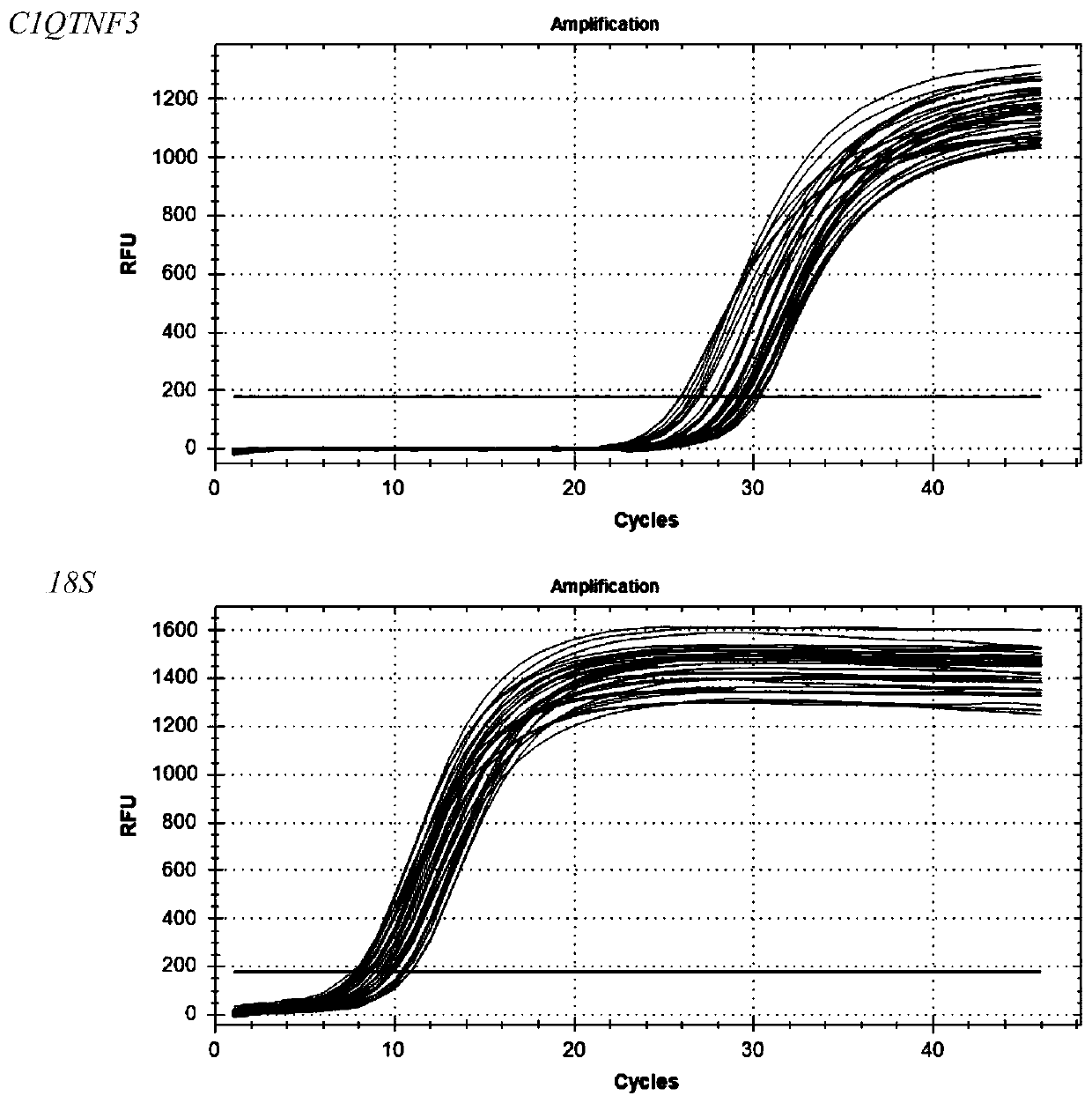
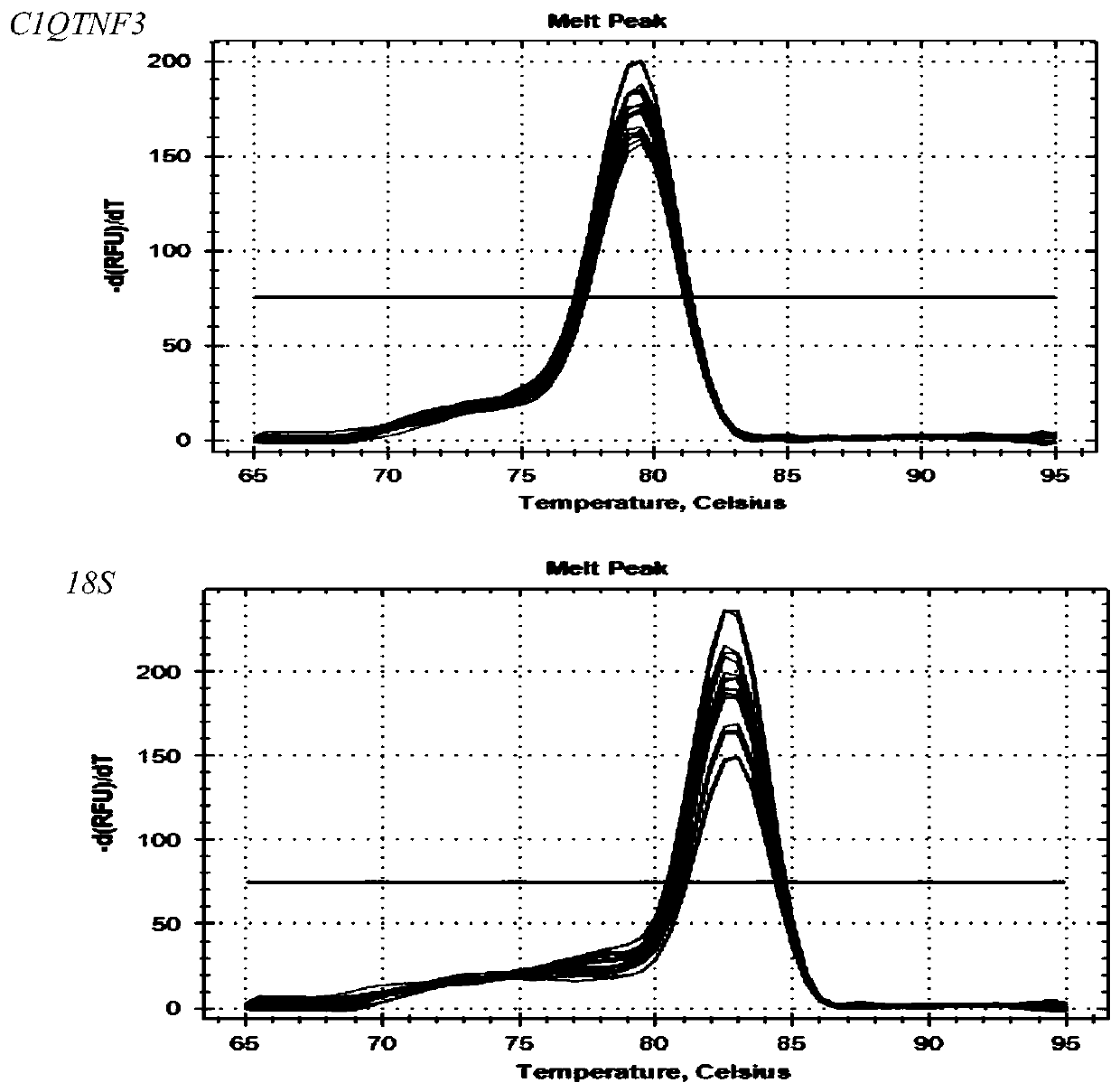
Primer, kit and detection method for detecting C1QTNF3 gene 219bp-deletion variable spliceosome
PendingCN109825560APracticalMicrobiological testing/measurementDNA/RNA fragmentationSpliceosome complexGene
The invention provides a primer, kit and detection method for detecting C1QTNF3 gene 219bp-deletion variable spliceosome. The sequence of the primer for detecting the C1QTNF3 gene 219bp-deletion variable spliceosome is shown in SEQ ID NO:1 and 2. The for detecting C1QTNF3 gene 219bp-deletion variable spliceosome is separated from various splicing forms of the gene , the expression level of the C1QTNF3 gene 219bp-deletion variable spliceosome is quantitatively analyzed, the detection method is accurate, quick, convenient to use and high in practicability.
Owner:SHANXI AGRI UNIV
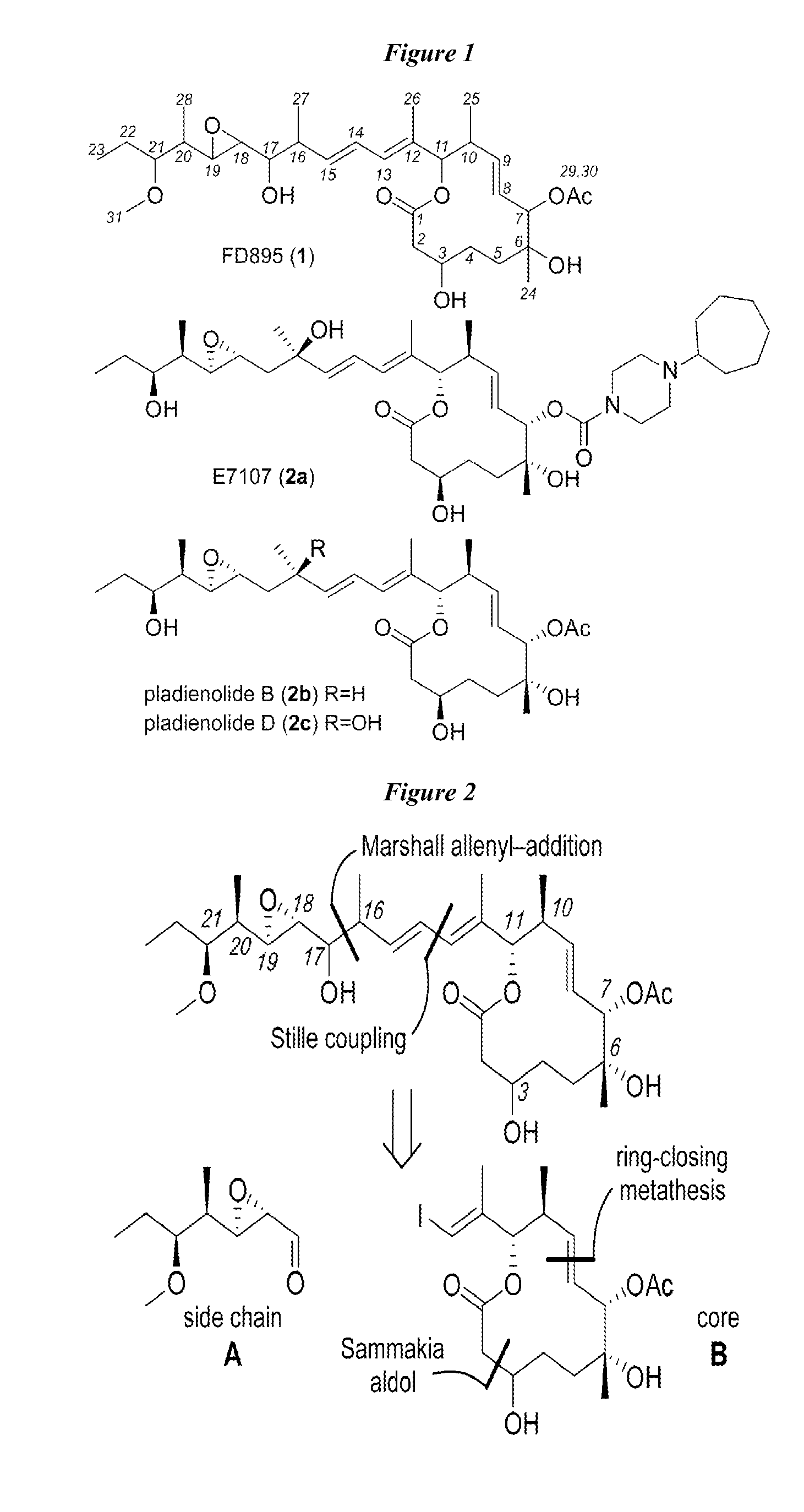
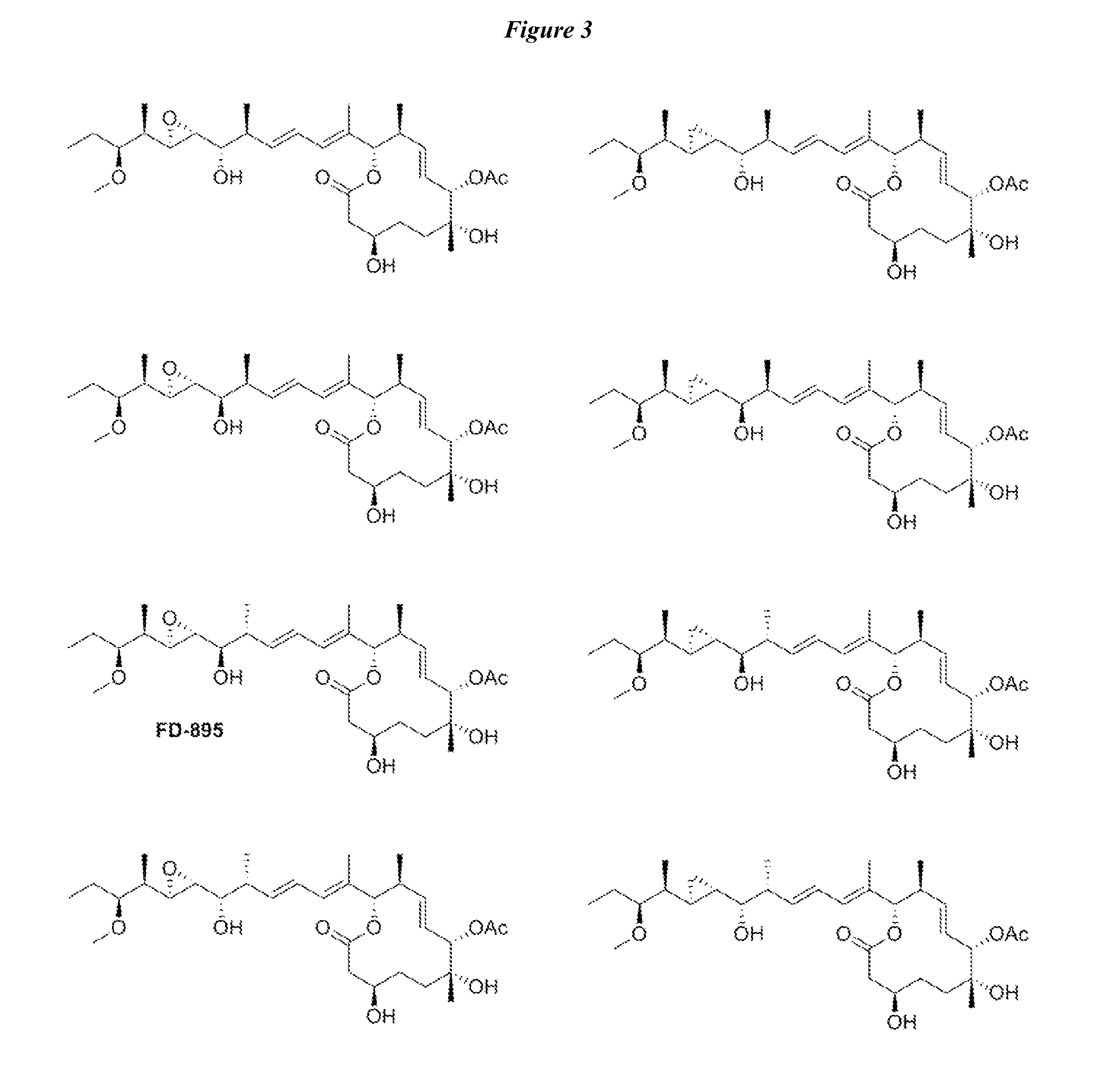
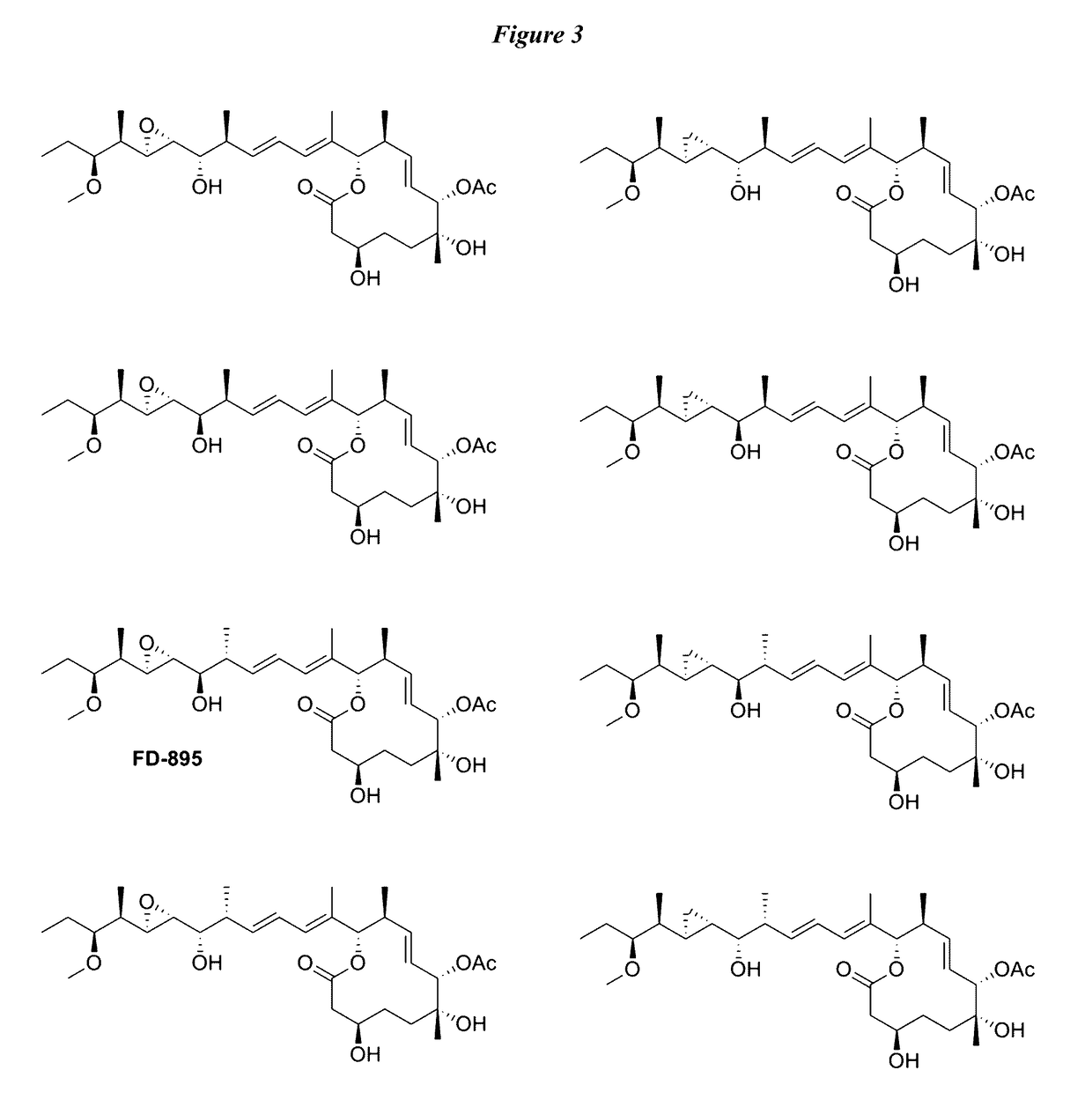
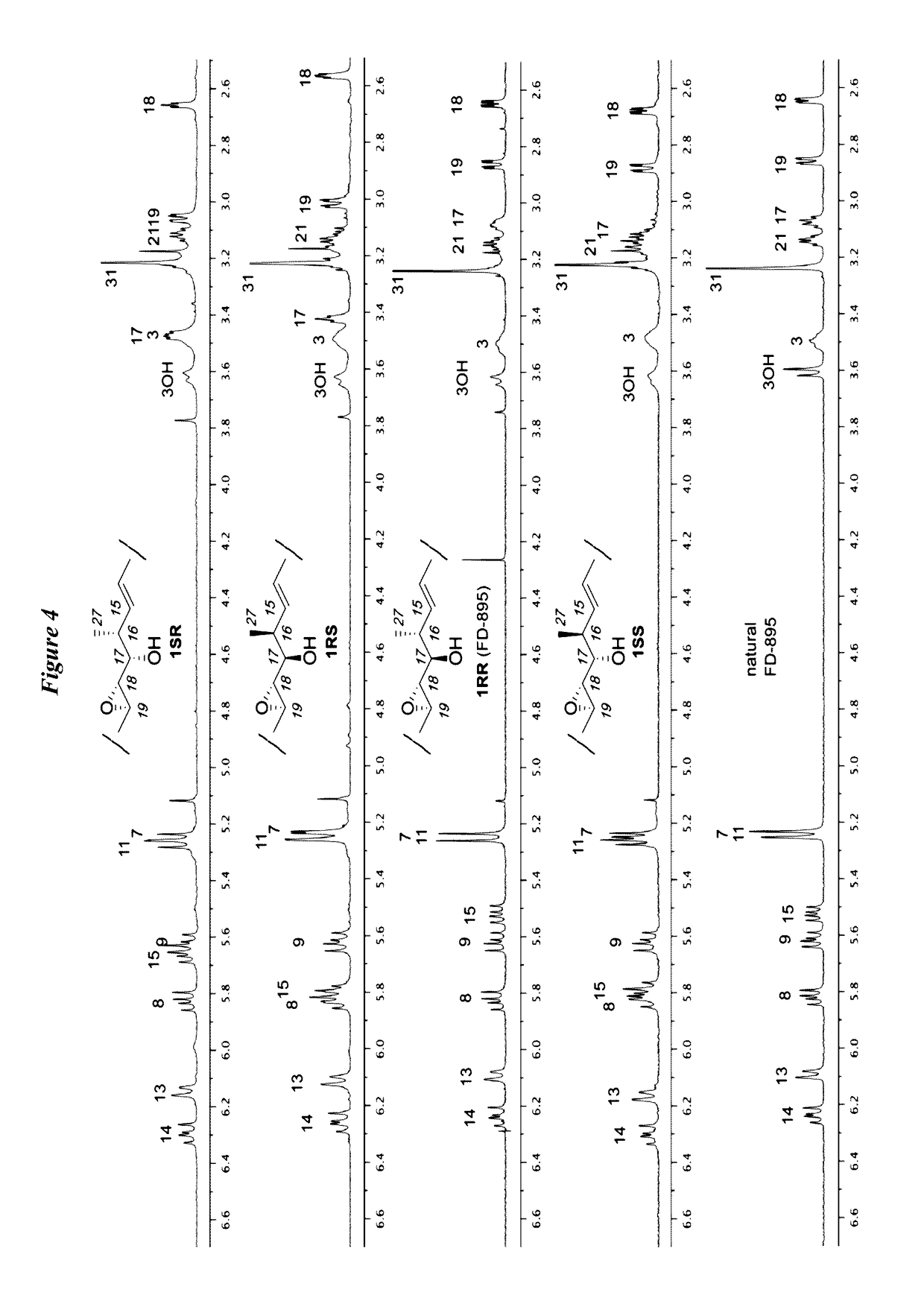
Anti-cancer polyketide compounds
Provided herein, inter alia, are anticancer polyketides. The uses of the polyketides described herein include treatment of cancer, for example, through regulation of the spliceosome and detection of spliceosome inhibition.
Owner:RGT UNIV OF CALIFORNIA
Application of PD-L1 (Programmed Death-Ligand 1) spliceosome as marker B for instructing administration of PD-L1/PD1 immunotherapy
The invention discloses application of a PD-L1 (Programmed Death-Ligand 1) spliceosome B as a marker for instructing administration of PD-L1 / PD1 immunotherapy. Expression of three different variable spliceosomes of PD-L1 can be observed in normal and mutant colorectal cancer cell systems, and eukaryotic expression vectors of the three spliceosomes are established and transfected into tumor cells without or with low PD-L1 expression. Tests further show that in co-culture of a tumor cell system with isoform b over-expression and immune cells, apoptosis of the immune cells is promoted, secretionof immune cell factors is inhibited, inhibition of immunoreactions in bodies of patients can be reminded, and clinical sample testing shows that isoform b of PD-L1 is a monitoring target of poor prognosis of tumor.
Owner:ZHEJIANG UNIV
Spliceosome mediated RNA trans-splicing for correction of factor VIII genetic defects
The present invention provides methods and compositions for generating novel nucleic acid molecules through targeted spliceosomal mediated trans-splicing. The compositions of the invention include pre-trans-splicing molecules (PTMs) designed to interact with a target precursor messenger RNA molecule (target pre-mRNA) and mediate a trans-splicing reaction resulting in the generation of a novel chimeric RNA molecule (chimeric RNA). In particular, the PTMs of the present invention are genetically engineered to interact with factor VIII (FVIII) target pre-mRNA so as to result in correction of clotting FVIII genetic defects responsible for hemophilia A. The compositions of the invention further include recombinant vector systems capable of expressing the PTMs of the invention and cells expressing said PTMs. The methods of the invention encompass contacting the PTMs of the invention with a FVIII target pre-mRNA under conditions in which a portion of the PTM is trans-spliced to a portion of the target pre-mRNA to form a RNA molecule wherein the genetic defect in the FVIII gene has been corrected. The methods and compositions of the present invention can be used in gene therapy for correction of FVIII disorders such as hemophilia A.
Owner:MITCHELL LLOYD G +1
Anti-cancer polyketide compounds
Provided herein, inter alia, are anticancer polyketides. The uses of the polyketides described herein include treatment of cancer, for example, through regulation of the spliceosome and detection of spliceosome inhibition.
Owner:RGT UNIV OF CALIFORNIA
Expression of apoAI and variants thereof using spliceosome mediated RNA trans-splicing
InactiveUS7968334B2Reduce buildReduce usageSplicing alterationAntibody mimetics/scaffoldsRNA Trans-SplicingMessenger RNA
Methods and compositions for generating novel nucleic acid molecules through targeted spliceosome mediated RNA trans-splicing that result in expression of a apoAI protein, an apoAI variant, the preferred embodiment referred to herein as the apoAI Milano variant, a pre-pro-apoAI or an analogue of apoAI. The methods and compositions include pre-trans-splicing molecules (PTMs) designed to interact with a target precursor messenger RNA molecule (target pre-mRNA) and mediate a trans-splicing reaction resulting in the generation of a novel chimeric RNA molecule (chimeric RNA) capable of encoding apoAI, the apoAI Milano variant, or an analogue of apoAI. The expression of this apoAI protein results in protection against vascular disorders resulting from plaque build up, i.e., atherosclerosis, strokes and heart attacks.
Owner:VIRXSYS
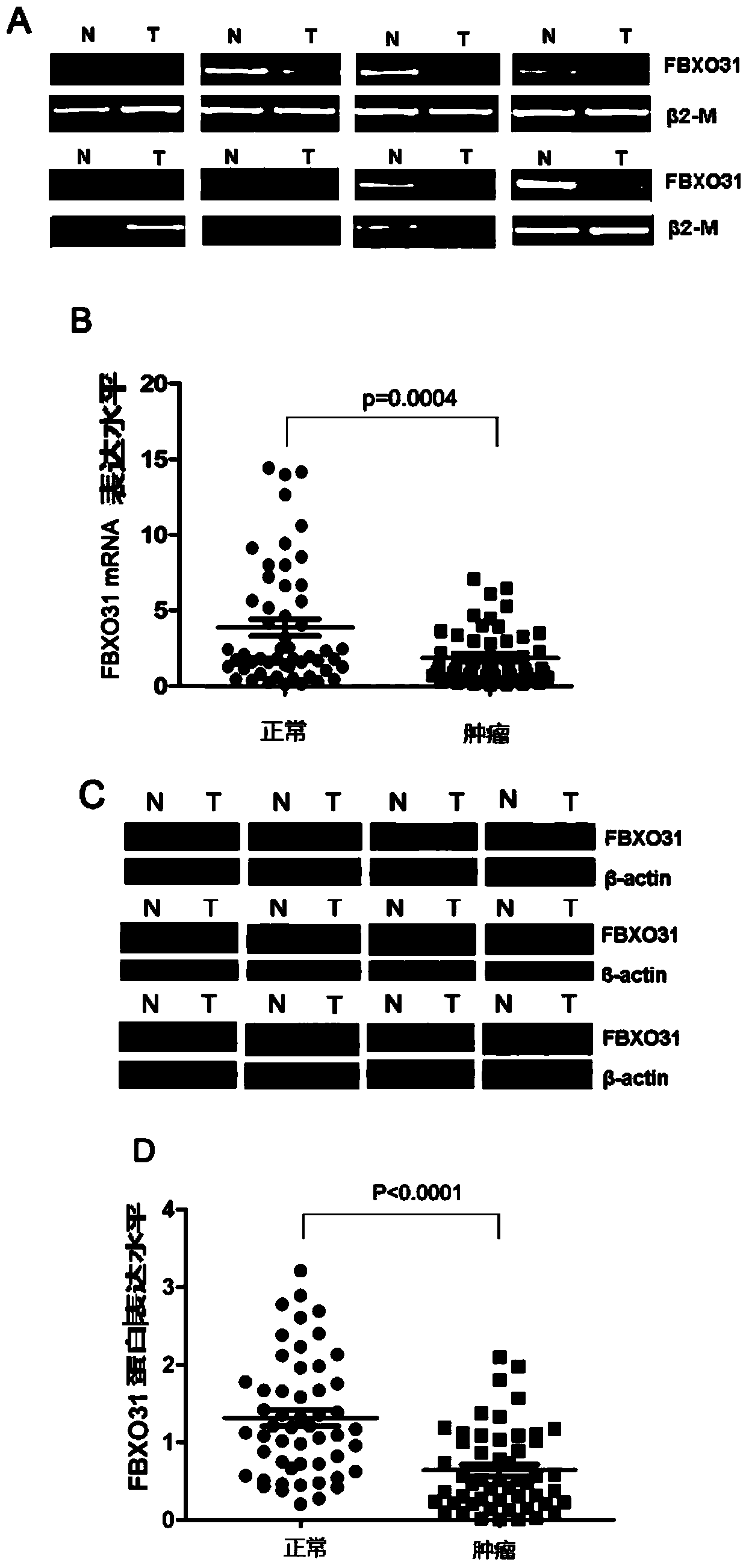
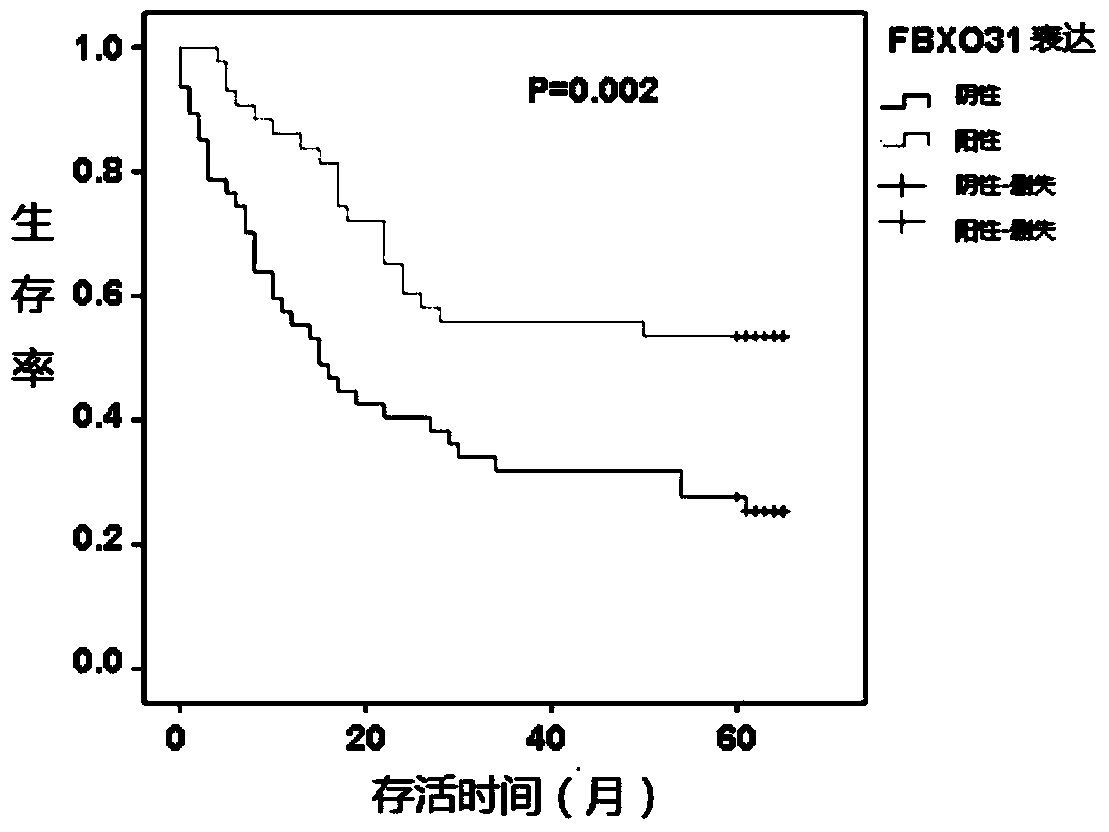
Application of FBXO31 gene and related products in preparing gastric cancer diagnostic reagent
InactiveCN104017879AReduce expressionMicrobiological testing/measurementBiological material analysisSpliceosome complexAntisense oligodeoxynucleotides
The invention relates to application of an FBXO31 gene and related products in preparing a gastric cancer diagnostic reagent. The related products of the FBXO31 gene are one or more of spliceosome of the FBXO31 gene, antisense oligodeoxynucleotide of the FBXO31 gene, mRNA (Messenger Ribonucleic Acid) of the FBXO31 gene, a protein of the FBXO31 gene, a peptide fragment encoded by the FBXO31 gene, an antibody of the protein of the FBXO31 gene, a regulating small molecular RNA related to the FBXO31 gene or an inhibitor of the regulating small molecular RNA related to the FBXO31 gene. The invention provides a novel concept of diagnosing gastric cancer and provides a novel path for researching and developing the gastric cancer diagnostic reagent.
Owner:SHANDONG UNIV
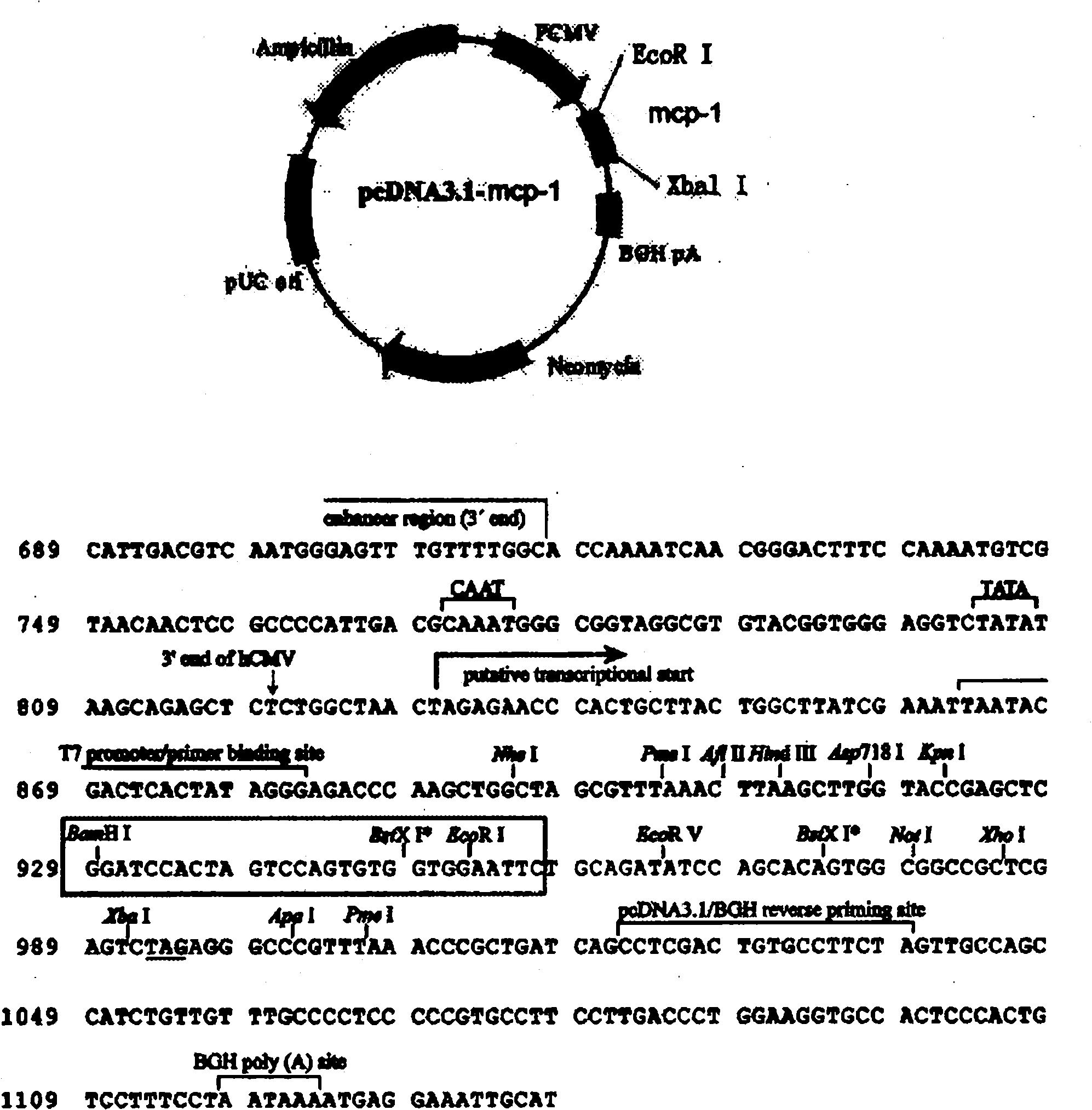
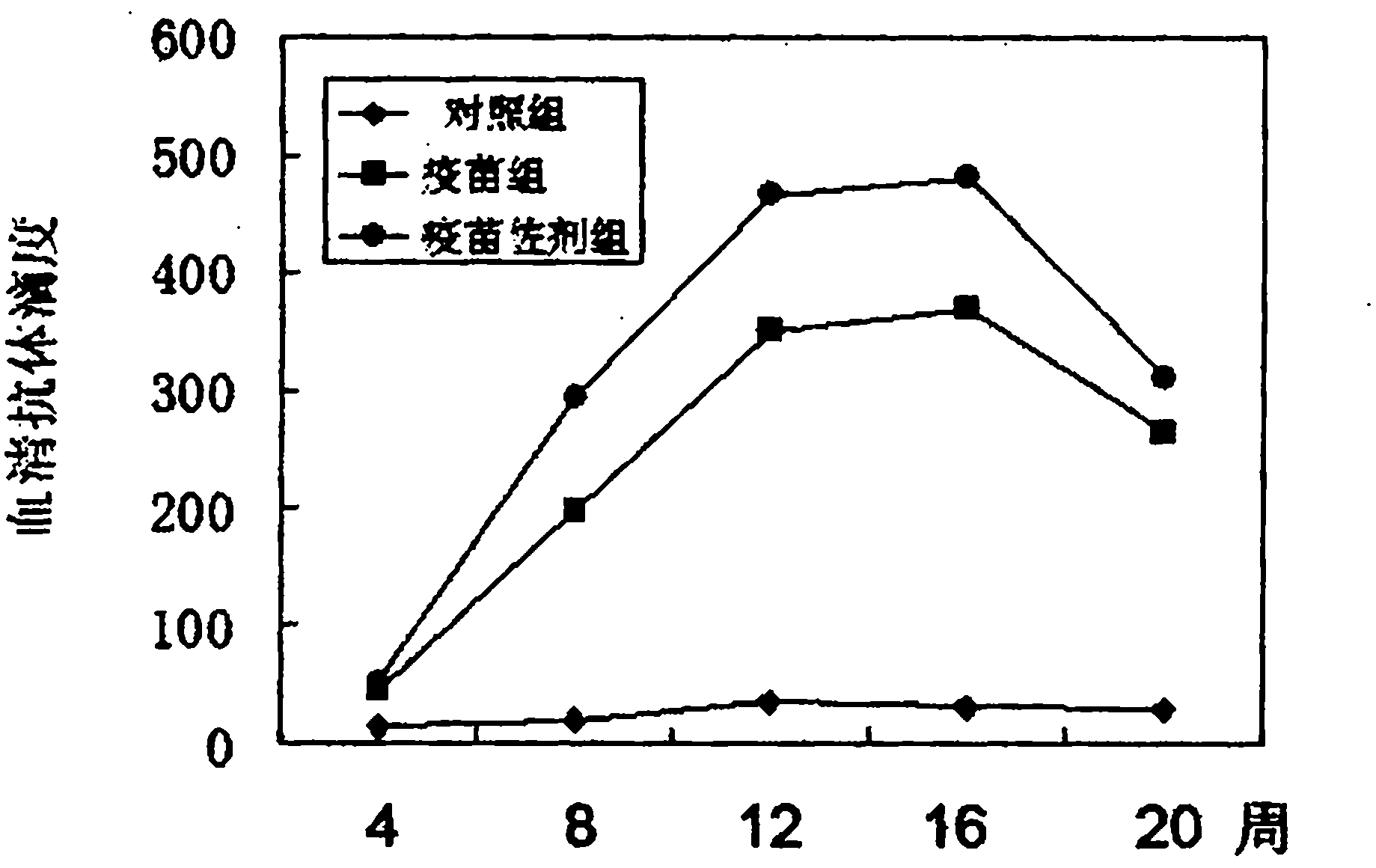
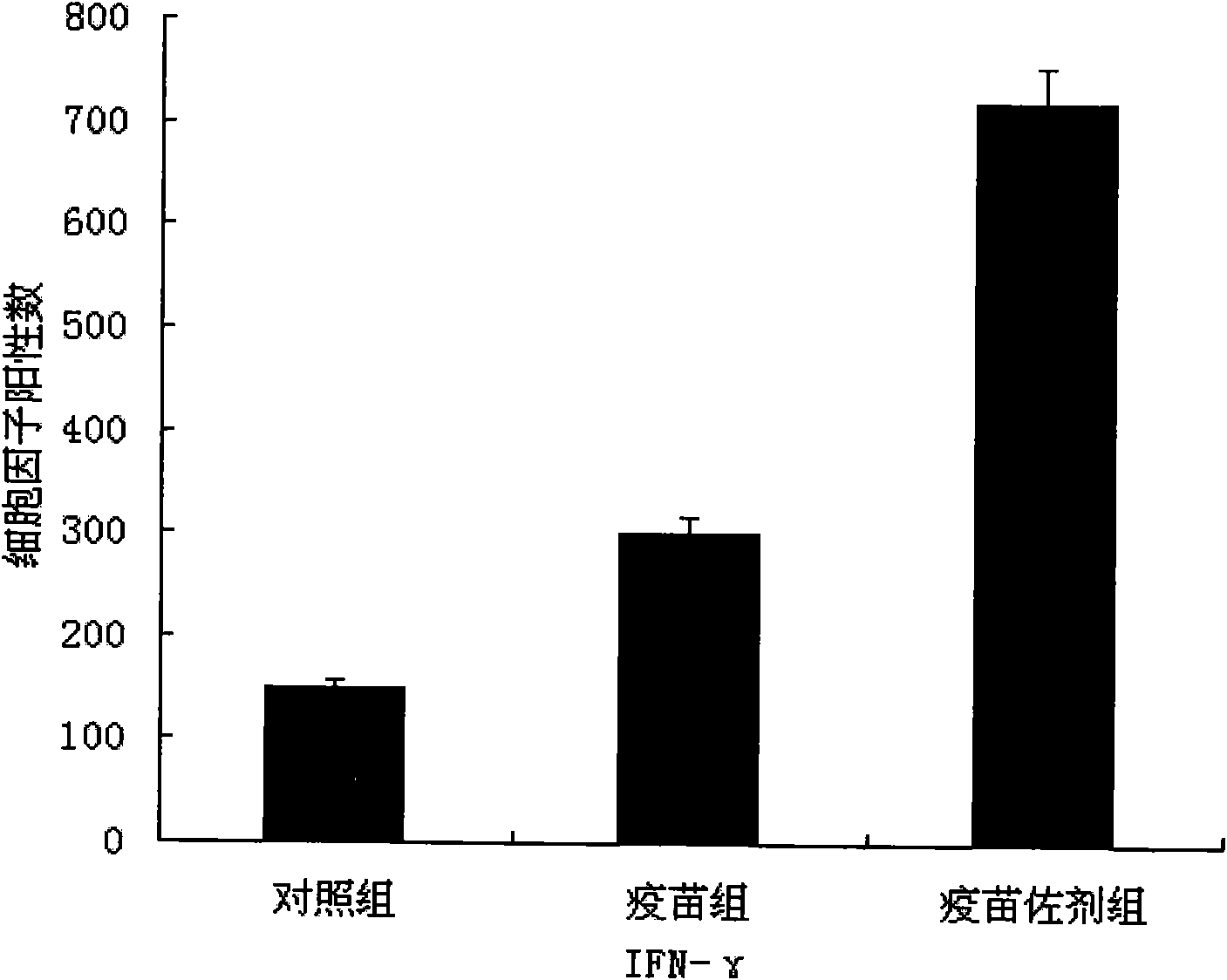
Nucleic acid vaccine adjuvant designed based on MCP-1 and construction method thereof
InactiveCN102038951AImprove expression levelDoes not affect antigen expressionAntibody medical ingredientsFermentationCD8Spleen
The invention relates to the technical field of medical biology and discloses a plasmid for enhancing cellular immune effect of a nucleic acid vaccine, which is designed based on monocyte chemoattractant protein-1 (MCP-1). An expressed sequence of a novel MCP-1 spliceosome molecule is inserted into a pcDNA-3.1 plasmid to form a pcDNA-3.1-MCP-1 plasmid, the plasmid can efficiently express the novel MCP-1 spliceosome molecule, and the expression ability of the co-transfected nucleic acid vaccine is not influenced. In an in vivo test, the plasmid and the nucleic acid vaccine are injected simultaneously to have the capacity of effectively inducing cellular immune response, namely antigenic specificity spleen interferon (IFN)-gamma positive cells are increased and target-killing capacity of spleen CD8 positive cells is improved. Therefore, the plasmid can be added into the conventional nucleic acid vaccine preparation for mixed injection so as to obviously accelerate the nucleic acid vaccine-induced cellular immune response, and the plasmid serves as a novel cellular immune adjuvant.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
Correction of alpha-1-antitrypsin genetic defects using spliceosome mediated RNA trans splicing
InactiveUS8053232B2Reduce lung and liver pathologyReduce accumulationSugar derivativesMicrobiological testing/measurementRNA Trans-SplicingMessenger RNA
The present invention provides methods and compositions for generating novel nucleic acid molecules through targeted spliceosomal mediated RNA trans-splicing. The compositions of the invention include pre-trans-splicing molecules (PTMs) designed to interact with a SERPINA1 target precursor messenger RNA molecule (target pre-mRNA) and mediate a trans-splicing reaction resulting in the generation of a novel chimeric RNA molecule (chimeric RNA). In particular, the PTMs of the present invention include those genetically engineered to interact with SERPINA1 target pre-mRNA so as to result in correction of SERPINA1 genetic defects responsible for AAT deficiency. The PTMs of the invention may also comprise sequences that are processed out of the PTM to yield duplex siRNA molecules directed specifically to mutant SERPIN A1 mRNAs. Such duplexed siRNAs are designed to reduce the accumulation of toxic AAT protein in liver cells. The methods and compositions of the present invention can be used in gene therapy for correction of SERPINA1 disorders such as AAT deficiency.
Owner:VIRXSYS
Wear-resistant cam rotor
PendingCN108825490AReduce wearReduce frictionRotary piston pumpsRotary piston liquid enginesSpliceosome complexWear resistant
The invention relates to a cam rotor, specifically to a wear-resistant cam rotor. The wear-resistant cam rotor comprises a rotor body equipped with several convex sides. A spindle shaft is arranged atthe center of the rotor body. Engagement bodies are separately arranged on the convex sides. By separately arranging the engagement bodies on the convex sides, when abrasion of a wear resistant stickon the engagement body is large, the engagement bodies can be dismounted, and a brand-new wear resistant stick is placed into a groove, and then a spliceosome A and a spliceosome B are connected andfixed. By arranging the wear resistant stick on the top of the curved surface of the engagement body, friction between the wear resistant stick and a pump cavity can be reduced so as to reduce abrasion of the wear resistant stick. By arranging an elastic device at the bottom of the wear resistant stick, the wear resistant stick can greatly fit the pump cavity. By arranging a waterproof plate and agroove, a fluid can be prevented from entering the inside through a seam; and by arranging a deflector hole on the engagement body, a fluid can be thrown out through the deflector hole by centrifugalforce of the rotor.
Owner:奥戈恩(广州)泵业有限公司
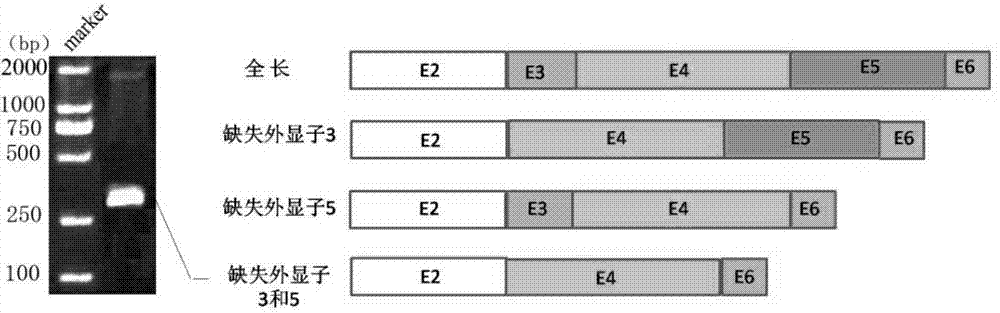
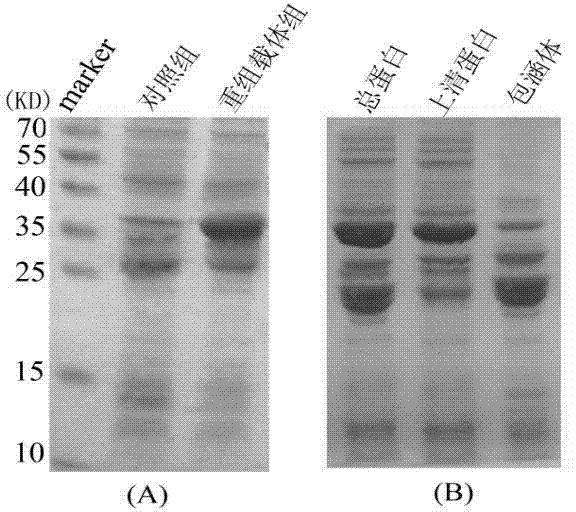
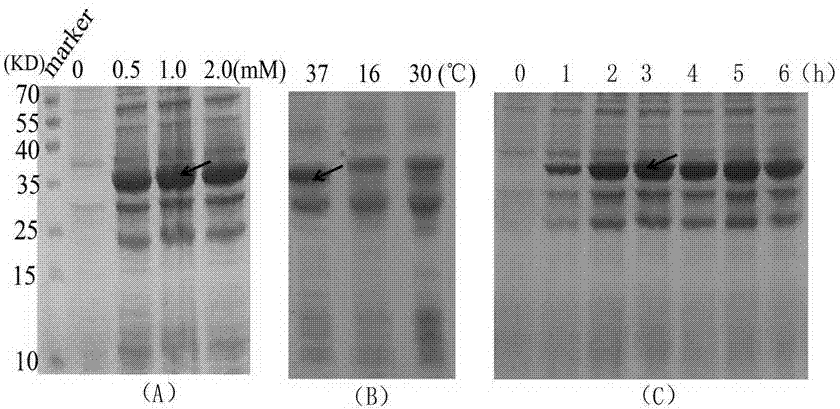
Method for expressing and purifying simultaneous exons 3 and 5 deficient alpha synuclein alternative spliceosome protein
ActiveCN104711280AImprove parallelismEasy to operateFermentationAnimals/human peptidesSpliceosome complexCytotoxicity
The invention provides a method for expressing and purifying a simultaneous exons 3 and 5 deficient alpha synuclein alternative spliceosome protein belongs to the technical field of biology, and a method for expressing the simultaneously exons 3 and 5 deficient alpha synuclein alternative spliceosome protein in an E.coli body and purifying the protein. The method comprises the steps of obtaining of the cDNA of the simultaneous exons 3 and 5 deficient alpha synuclein, vector construction, induced expression of protein, GST-alpha-syndelta3delta5 purification and GST label removal. The method can be used for realizing the in-vitro expression of the simultaneous exons 3 and 5 deficient alpha synuclein alternative spliceosome protein; as a result, a foundation is laid for the in-vitro study of the gathering characteristic, cytotoxicity and transfer characteristic of the protein and the construction of an animal model of the simultaneous exons 3 and 5 deficient alpha synuclein alternative spliceosome protein, and a method is provided for deep studying the correlation of the protein to the pathogenic mechanism of Parkinson disease.
Owner:INST OF MEDICAL BIOLOGY CHINESE ACAD OF MEDICAL SCI
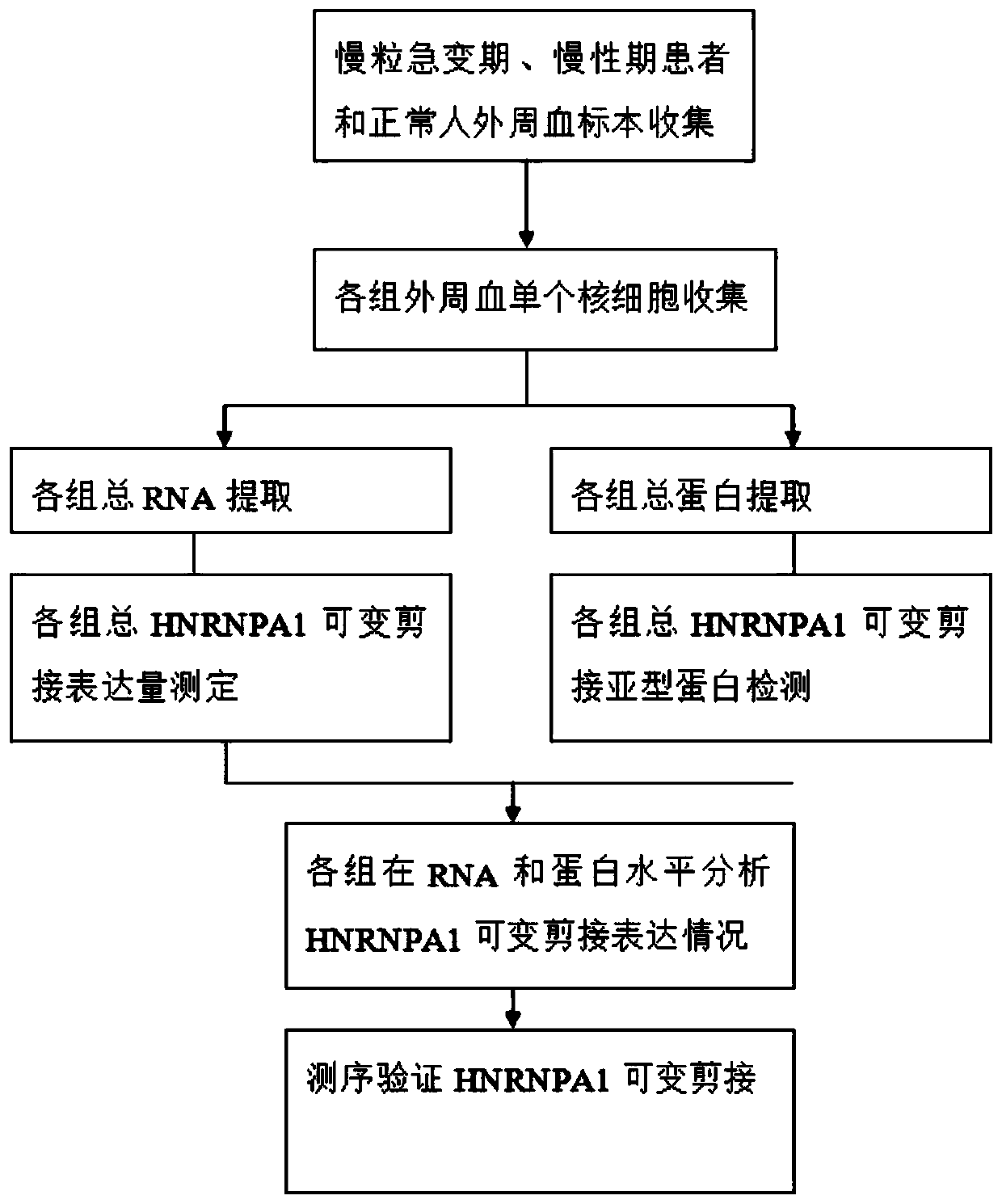
Reagent for diagnosing chronic myeloid leukemia blast crisis based on hnRNPA1 splicing variant and kit
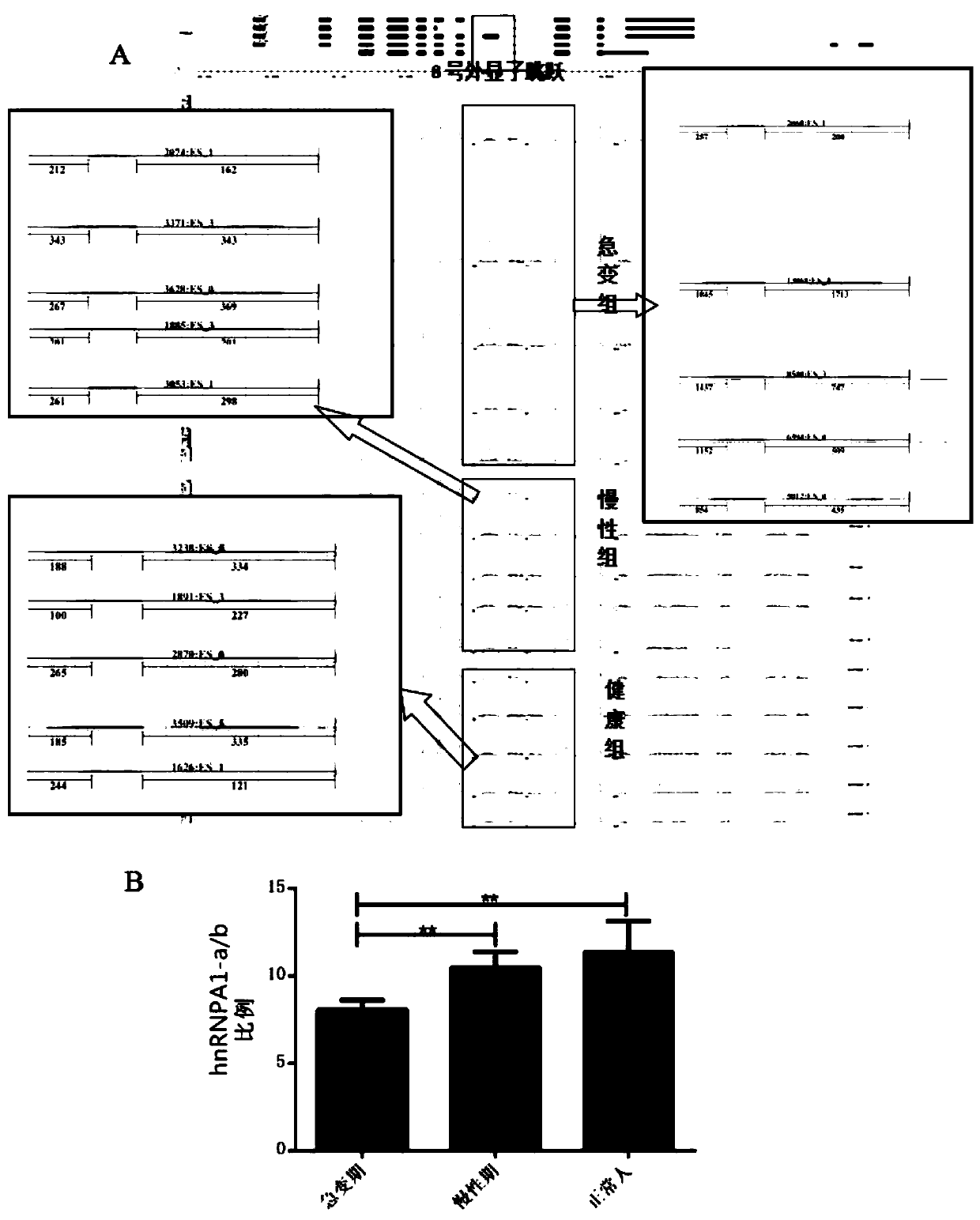
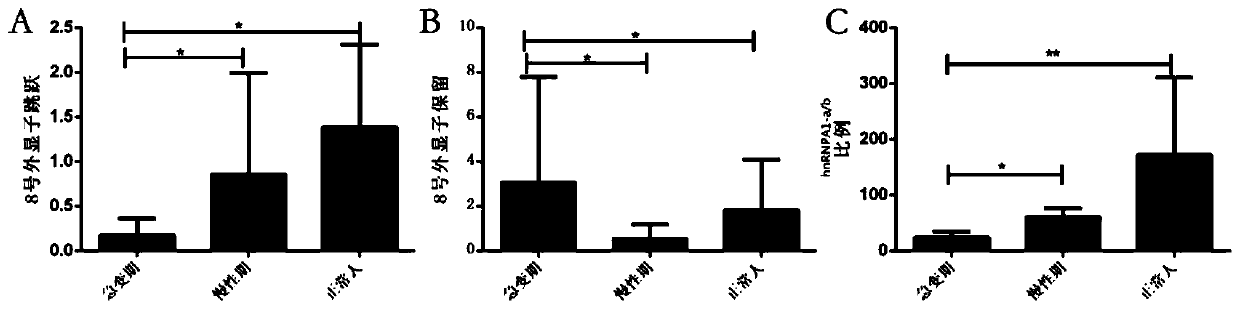
ActiveCN109880908AHigh sensitivityImprove featuresMicrobiological testing/measurementMaterial analysisBlast CrisisSpliceosome complex
The invention discloses application of No.8 exon skip / retained spliceosome (hnRNPA1-a and hnRNPA1-b) of hnRNPA1 in preparing a reagent for diagnosing chronic myeloid leukemia blast crisis or in a kitcontaining the diagnostic reagent. The reagent has higher sensitivity and specificity, can observe the chronic myeloid leukemia blast crisis more simply, accurately and early so as to timely take treatment means, and has higher clinical application value.
Owner:THE SECOND AFFILIATED HOSPITAL TO NANCHANG UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com