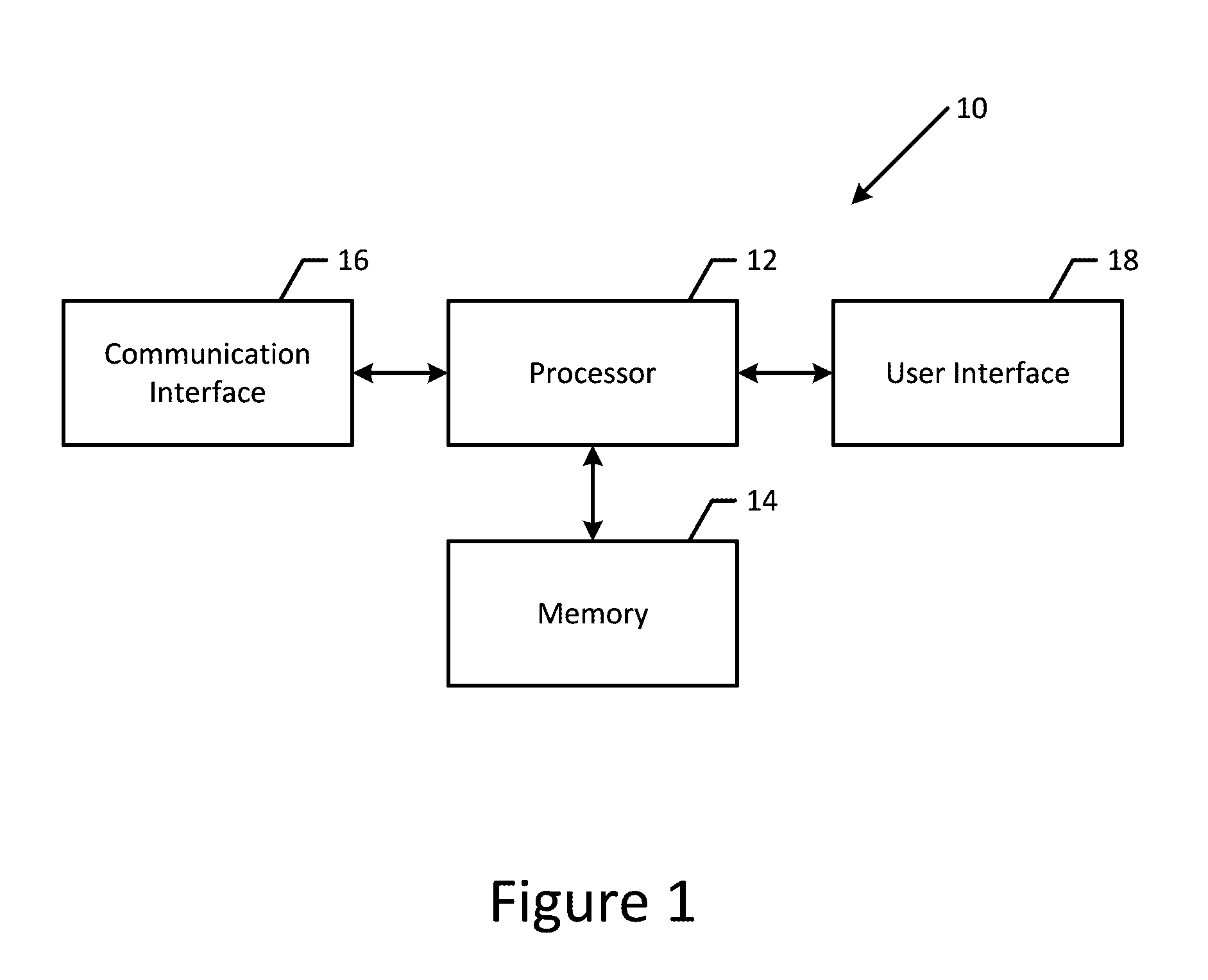
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
117 results about "Spatial search" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Method and system for extracting and classifying geolocation information utilizing electronic social media
InactiveUS20130086072A1DistanceReduce “ noisy ” dataDigital data information retrievalDigital data processing detailsData ingestionGeotargeting
Methods, systems and processor-readable media for extracting and classifying location information utilizing social media messages and / or data thereof. The social media messages can be sampled from a social media database and the messages filtered based on a heuristic rule. A geolocation entity from the unstructured social media messages can be extracted utilizing a geolocation entity extracting module. The messages with the geoentities can be uploaded onto a crowd sourcing platform to manually annotate the messages with a label. A text classification model can be built and learned from the label utilizing a machine learning algorithm and the messages can be classified by a location classifier in order to extract the user location. The user location can then be transformed into a geocode so that a spatial search can be enabled and the distance between the locations can be easily calculated.
Owner:XEROX CORP
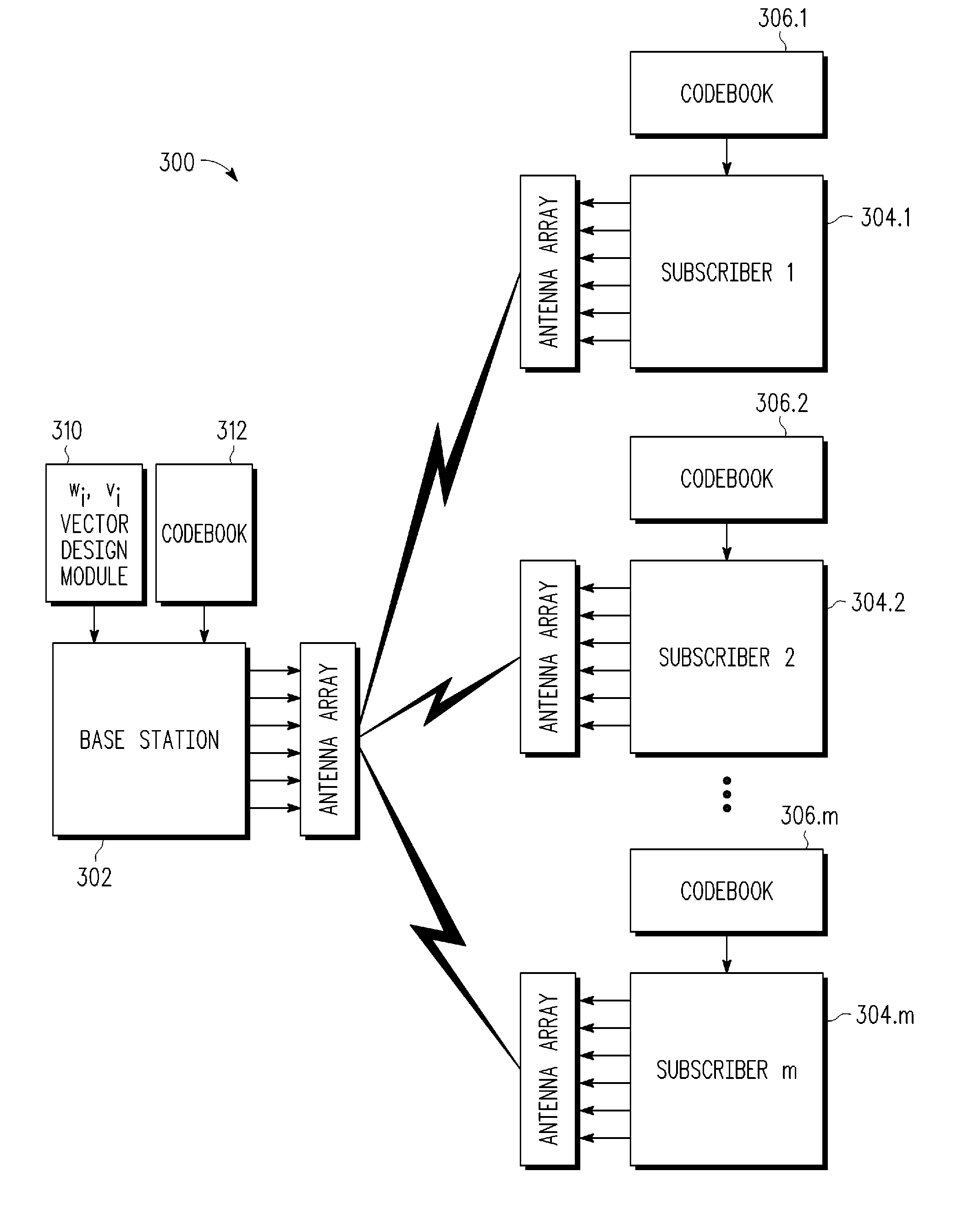
Methods for optimal collaborative MIMO-SDMA
ActiveUS20080076370A1Diversity/multi-antenna systemsSecret communicationCommunications systemTrade offs
A system and method for collaboratively designing optimized beamforming vectors is disclosed for a wireless multiple input, multiple output (MIMO) space division multiple access (SDMA) communication system to optimize an aggregate SNR performance metric across the different users, thereby permitting the flexibility to trade off computational requirements and size of control information exchanged with performance. Using adaptive vector space search methods, the space of all receive beamformers is searched to find the set which maximizes either the sum or product of SNRs of the users.
Owner:APPLE INC
Method for segmenting different objects in three-dimensional scene
InactiveCN101877128AEliminate occlusionEliminate the effects ofImage analysisPattern recognitionData set
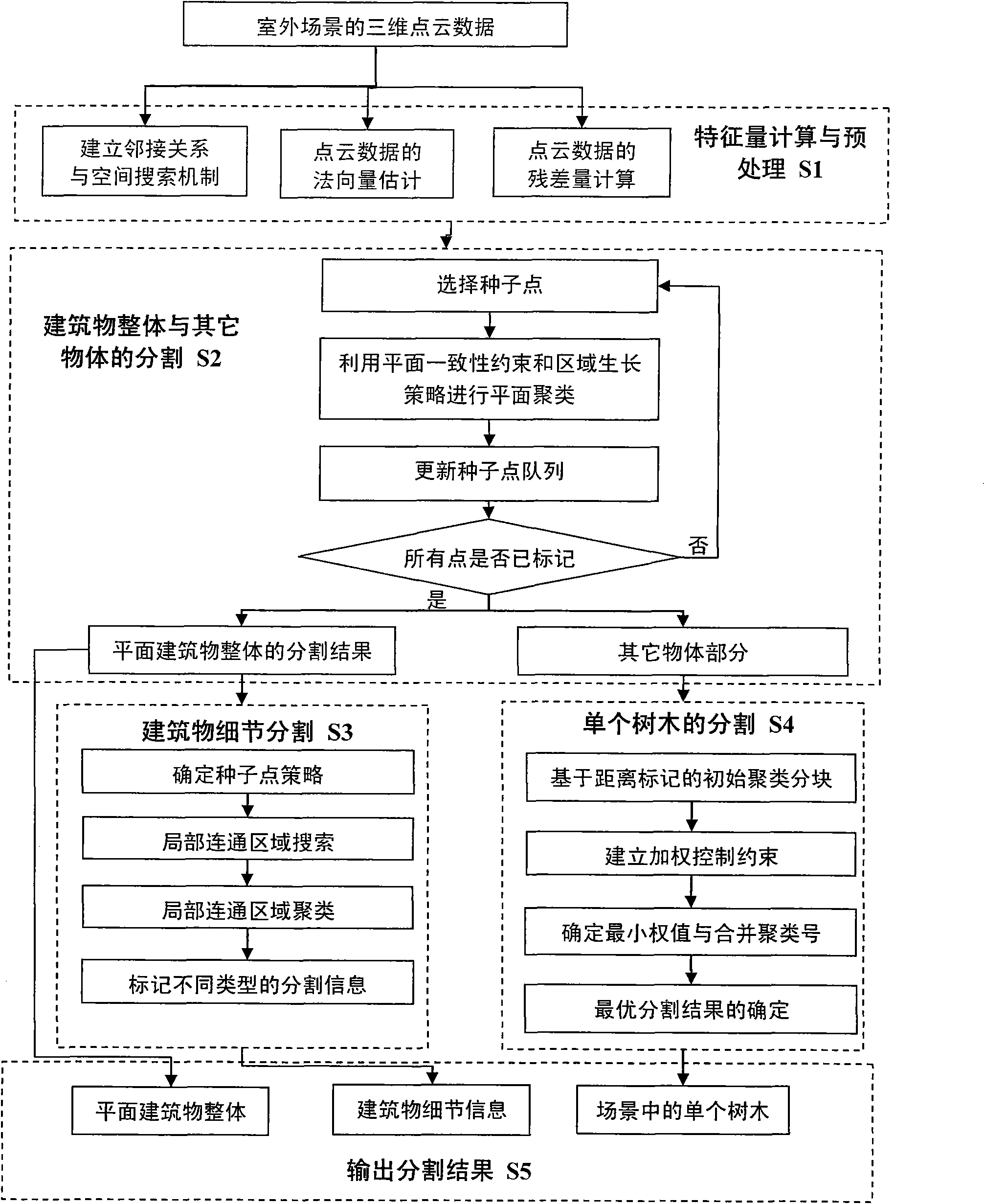
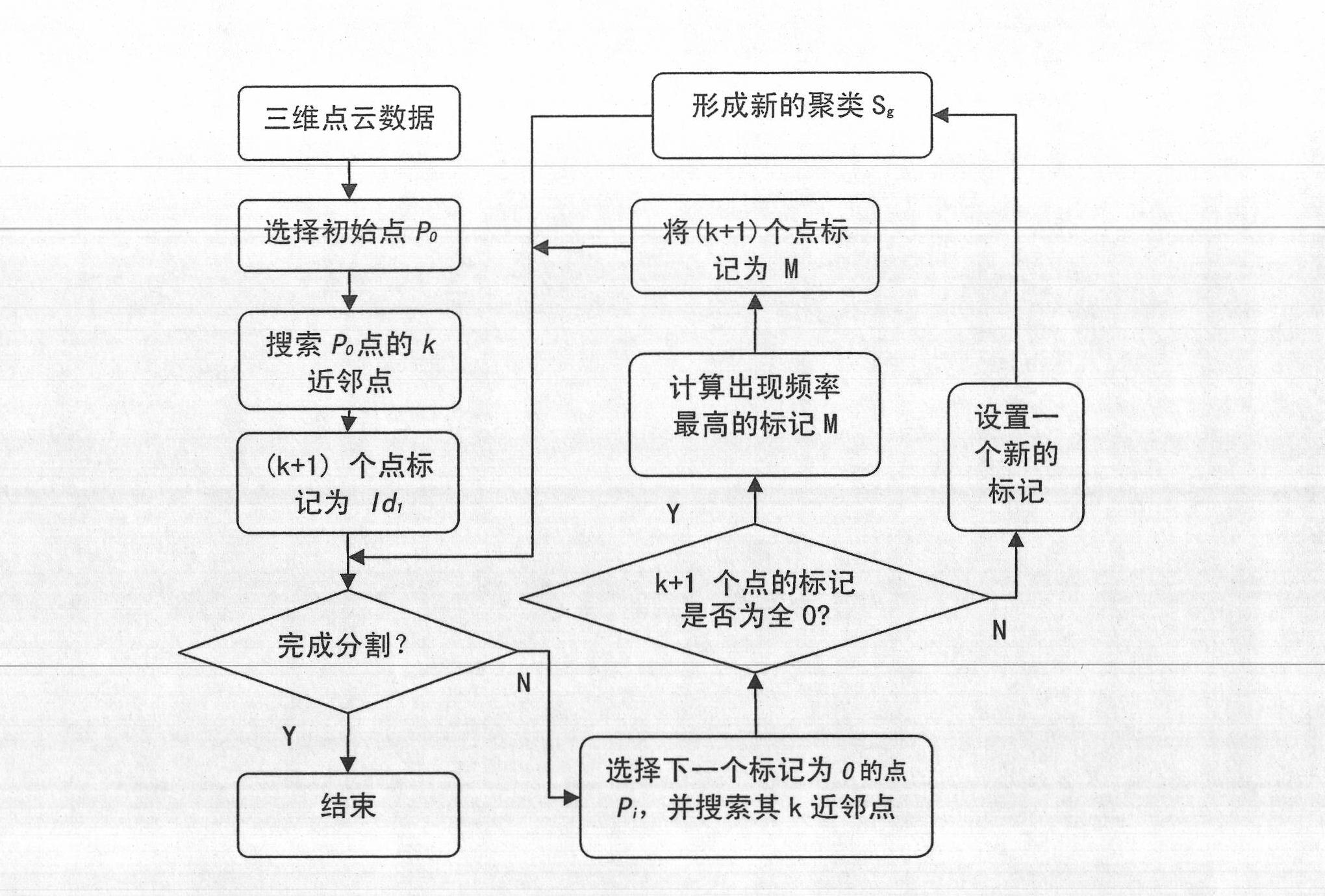
The invention discloses a method for segmenting different objects in a three-dimensional scene. The method comprises the following steps of: establishing adjacency relations and a spatial searching mechanism of point cloud data to estimate a normal vector and a residual of each point for the outdoor scene three-dimensional point cloud data acquired by laser scanning; determining the point with the minimum residual as a seed point and performing plane clustering by using a plane consistency restrictive condition and a region growing strategy to form the state that the entire plane building is segmented from other objects in the three-dimensional scene; establishing locally connected search for a plane building region for the segmented entire building part, and clustering the points with connectivity in the same plane by using different seed point rules to realize the detailed segmentation of the plane of the building; and constructing distance label-based initial cluster blocks for the other segmented objects and establishing weighting control restriction for cluster merging to realize the optimal segmentation result of trees. Tests on a plurality of data sets show that the method can be used for effectively segmenting the trees and buildings in the three-dimensional scene.
Owner:INST OF AUTOMATION CHINESE ACAD OF SCI +1
Auditory localization method
InactiveCN1832633AHigh precisionImprove estimation performanceFrequency/directions obtaining arrangementsSpatial positioningSound sources
Owner:HUAWEI TECH CO LTD +1
Map-centric map matching method and apparatus
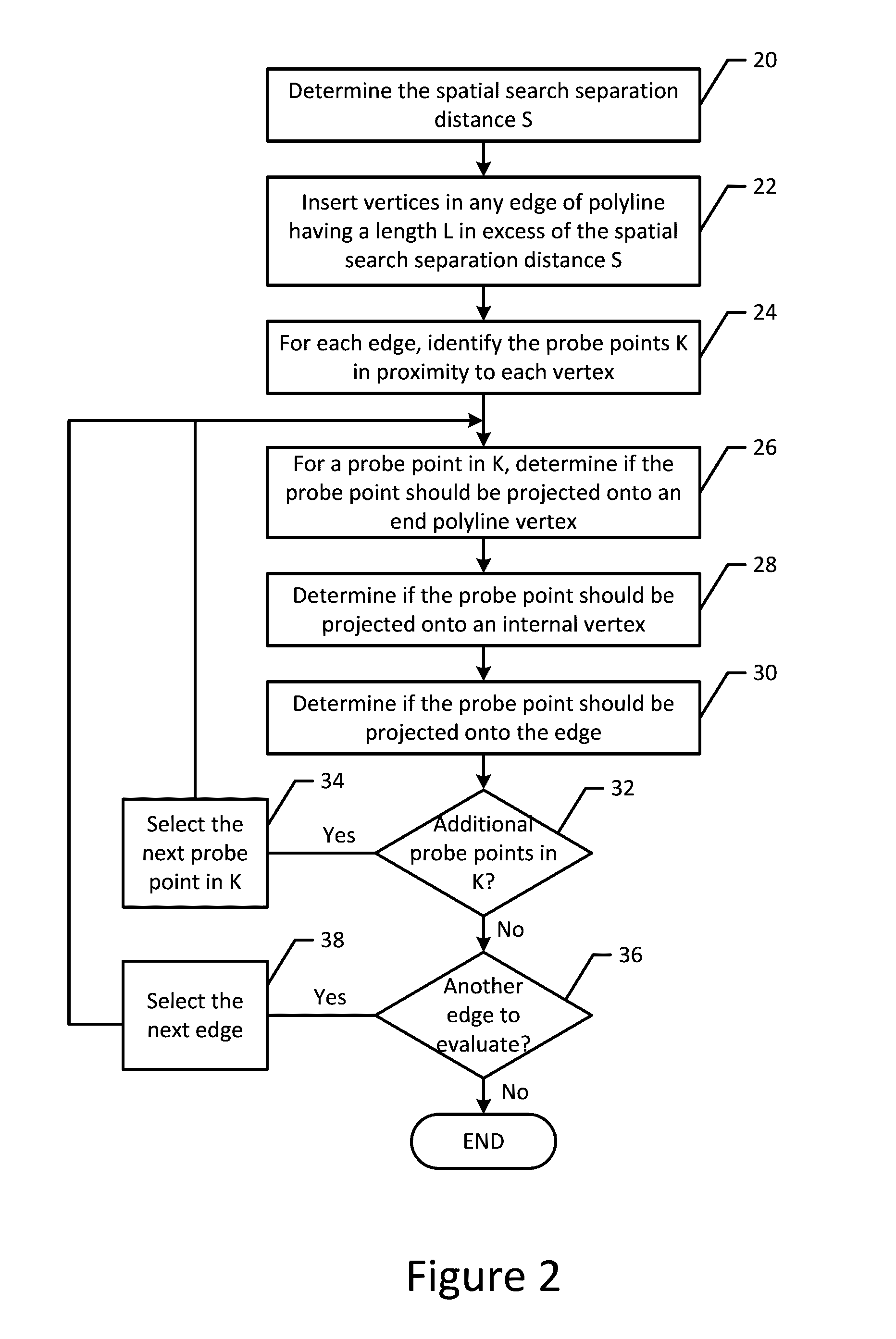
ActiveUS20160377440A1Less resourcesReduce in quantityInstruments for road network navigationRoad vehicles traffic controlGuidelineComputer science
A method, apparatus and computer program product are provided to process probe data in accordance with a map-centric map matching technique. In the context of a method, a plurality of vertices are defined along a polyline representative of a road segment such that the polyline includes one or more edges. Each edge extends between a pair of neighboring vertices. For each vertex of a respective edge, spatial searches are conducted to identify each probe point within a region about a respective vertex. For each probe point identified within a region about a respective vertex, a determination is made as to whether the probe point satisfies a projection criteria in order for the probe point to be projected onto the edge or one of the neighboring vertices between which the edge extends.
Owner:HERE GLOBAL BV
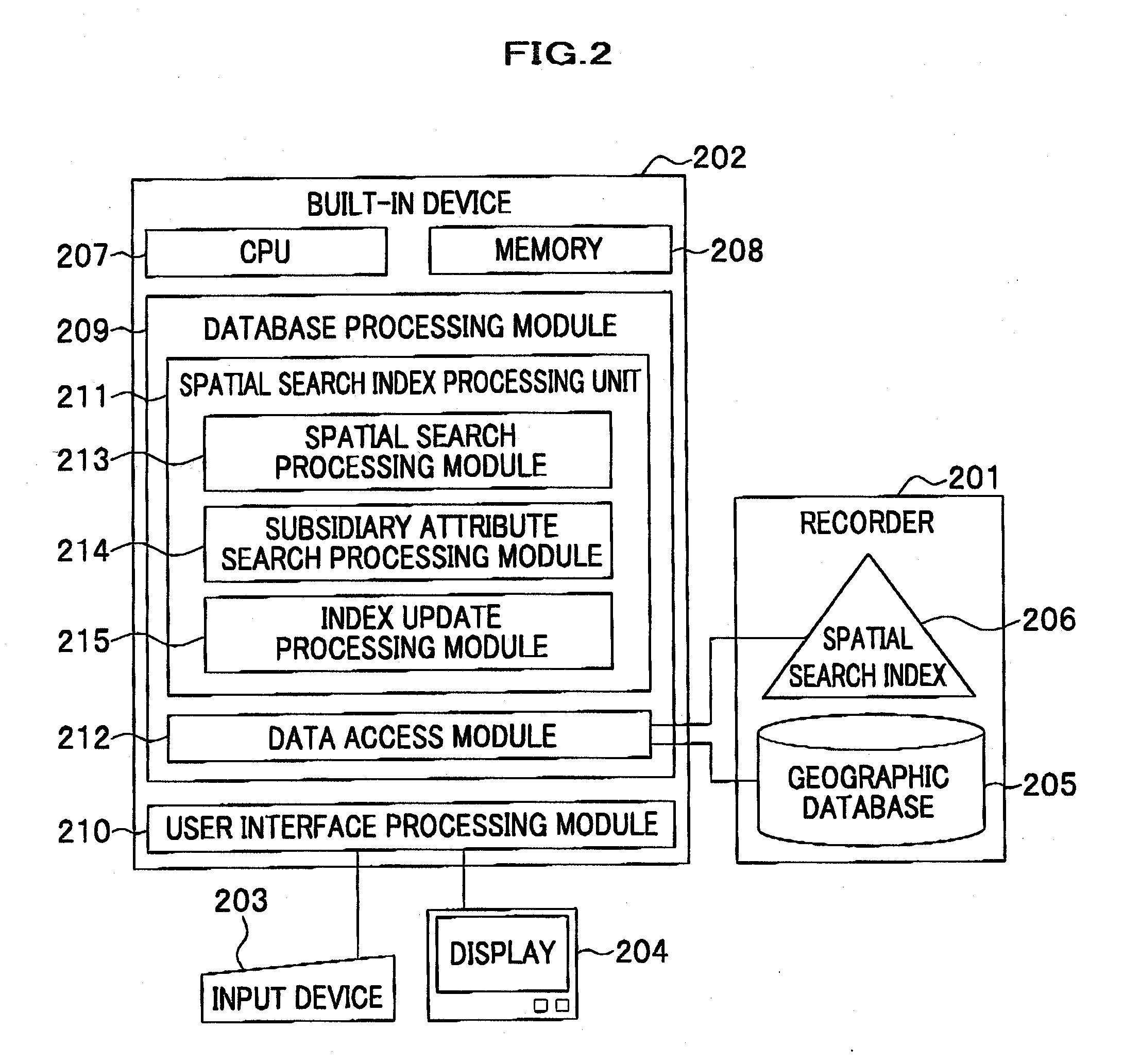
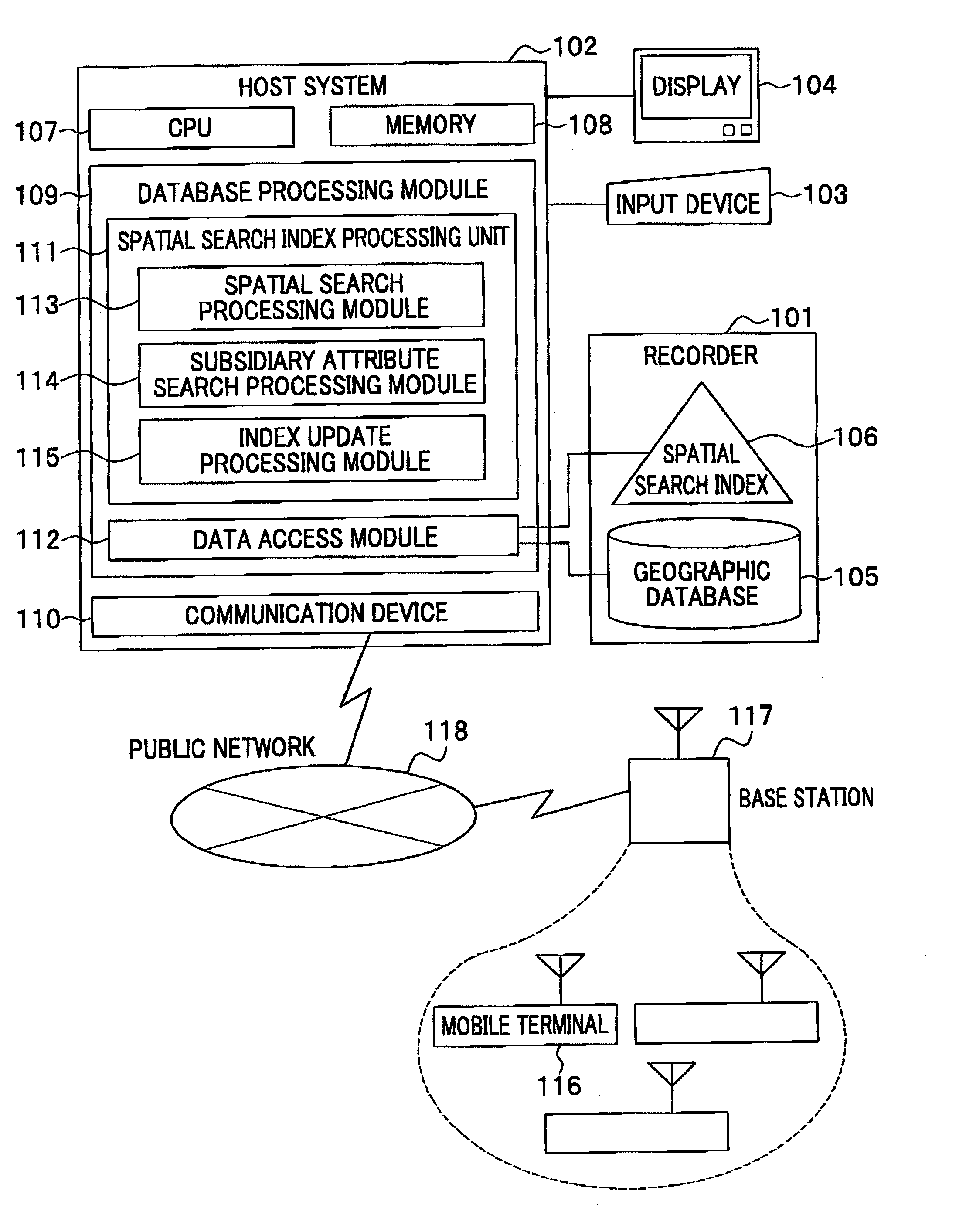
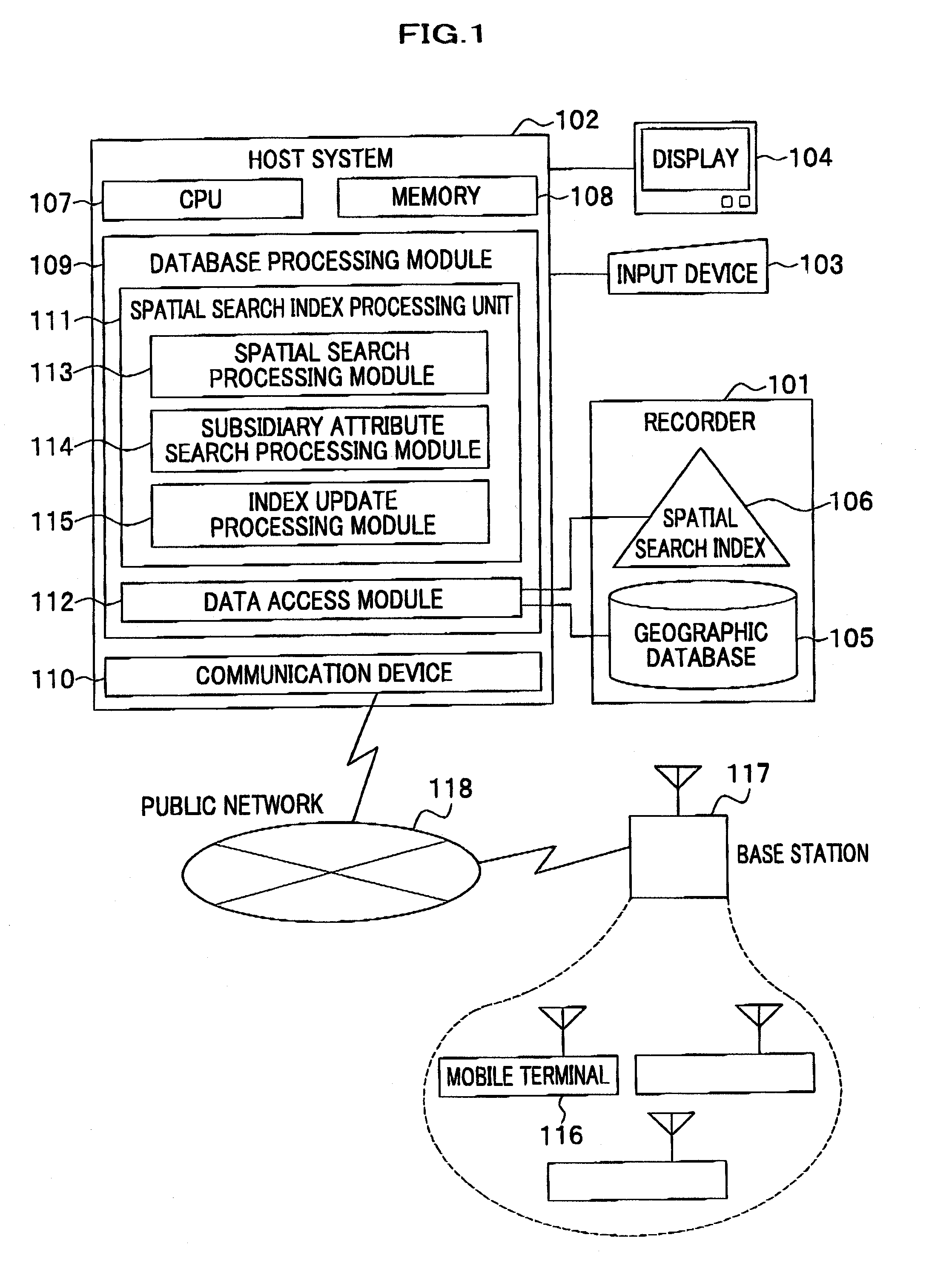
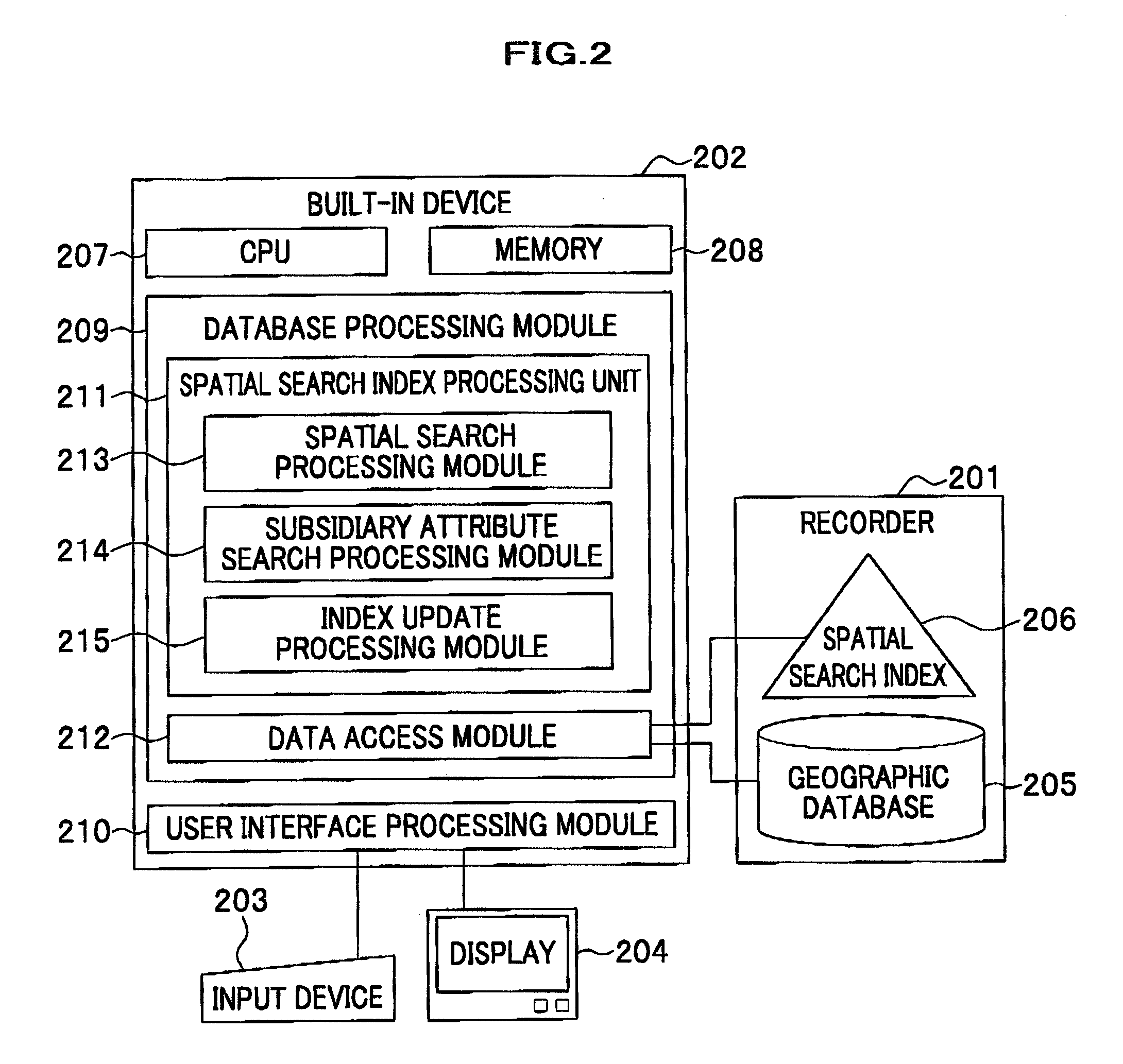
Method and system for data processing with spatial search
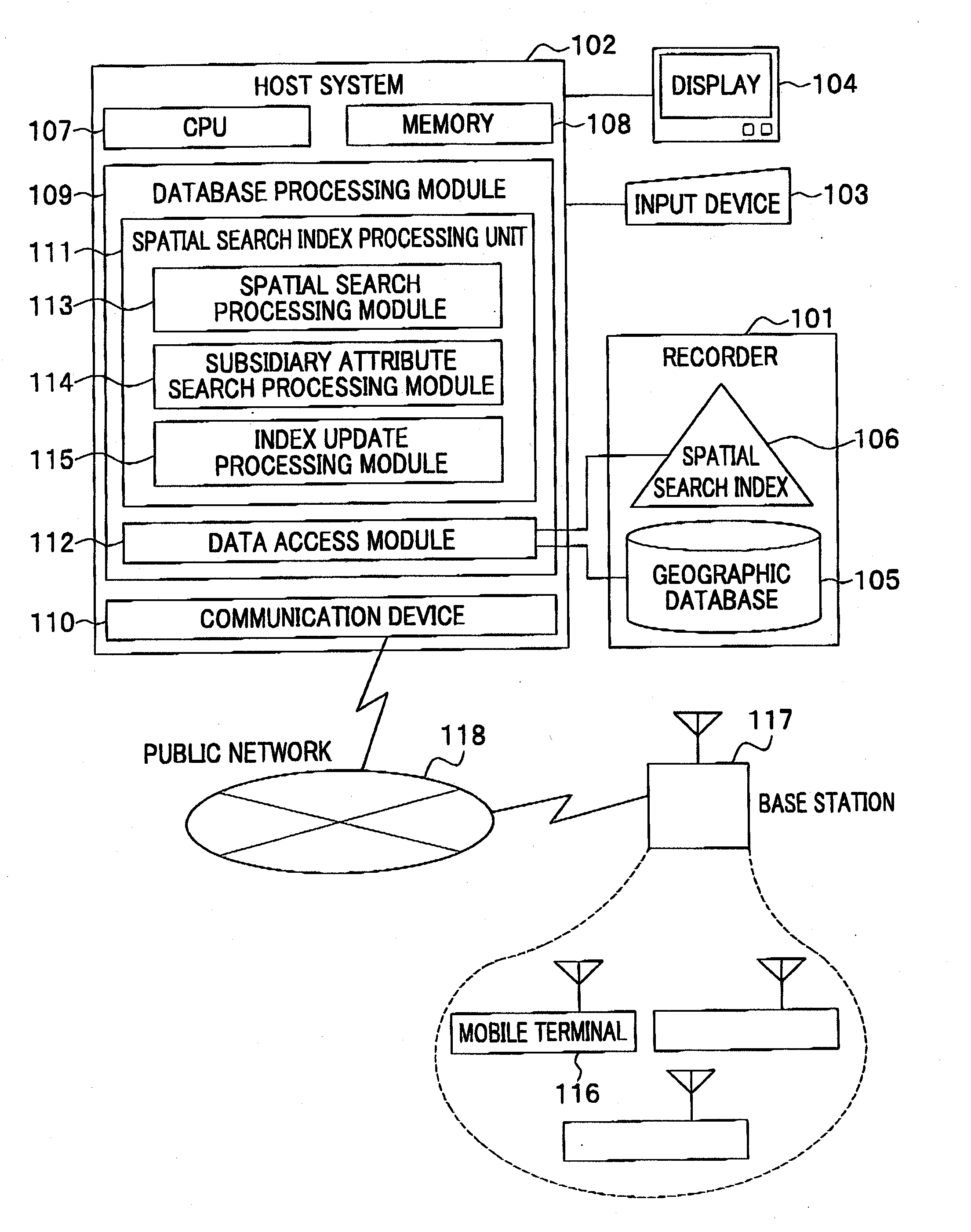
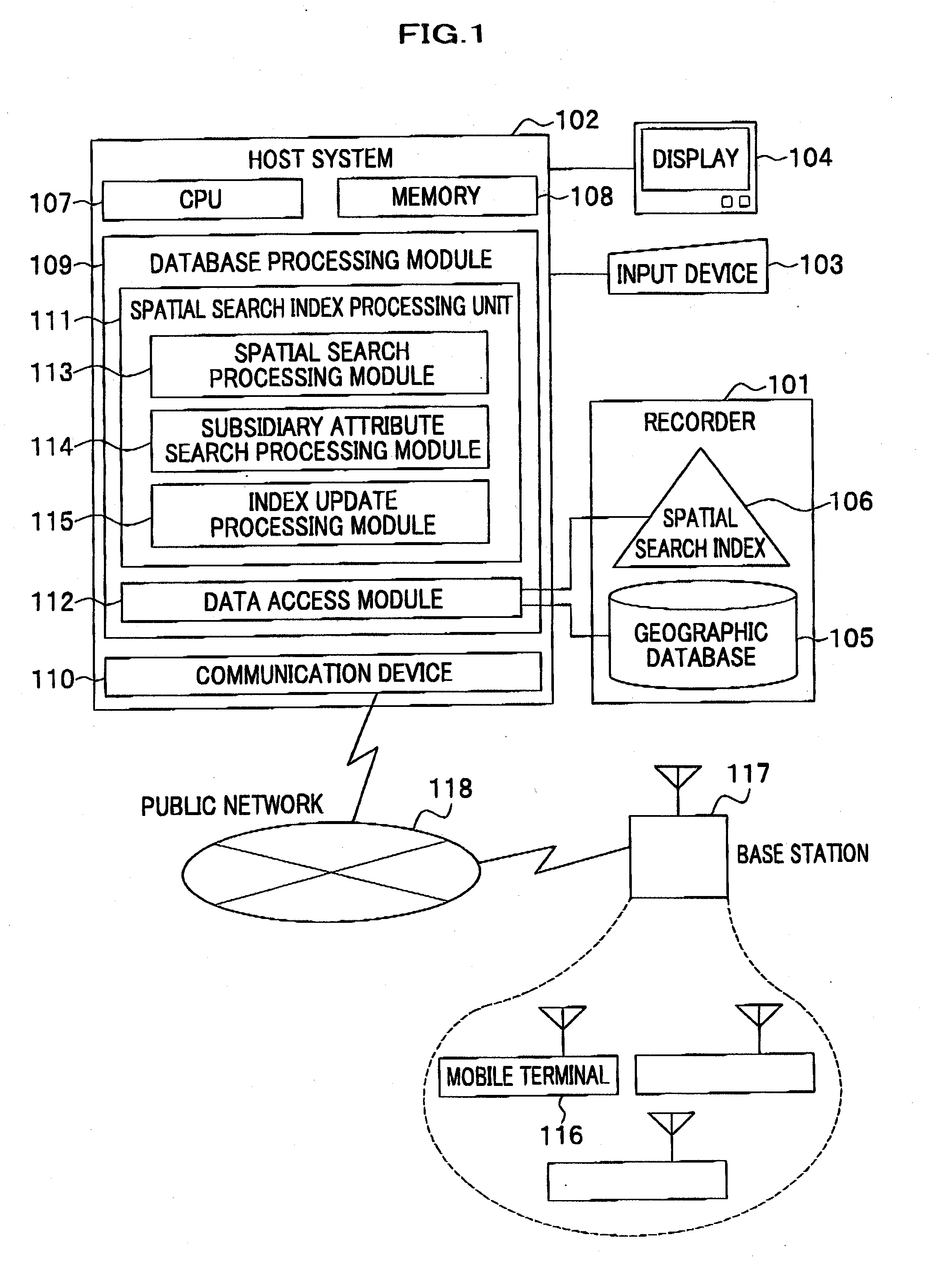
ActiveUS20080016037A1Increase speedSpatial search processingInstruments for road network navigationDigital data processing detailsData processing systemData mining
A method for processing data performed in a data processing system having a storage unit is provided which includes: creating a spatial index comprising a leaf containing location information and attribute information with respect to a plurality of objects to be searched, and storing the spatial index in the storage unit; and referring to the spatial index stored in the storage unit in response to an input of a search request including location information and attribute information for searching the object to be searched, and extracting the object to be searched that agrees with the search request. With this configuration, a spatial search with subsidiary condition search can be processed at high speed, without merging results of both searches.
Owner:HITACHI SOFTWARE ENG
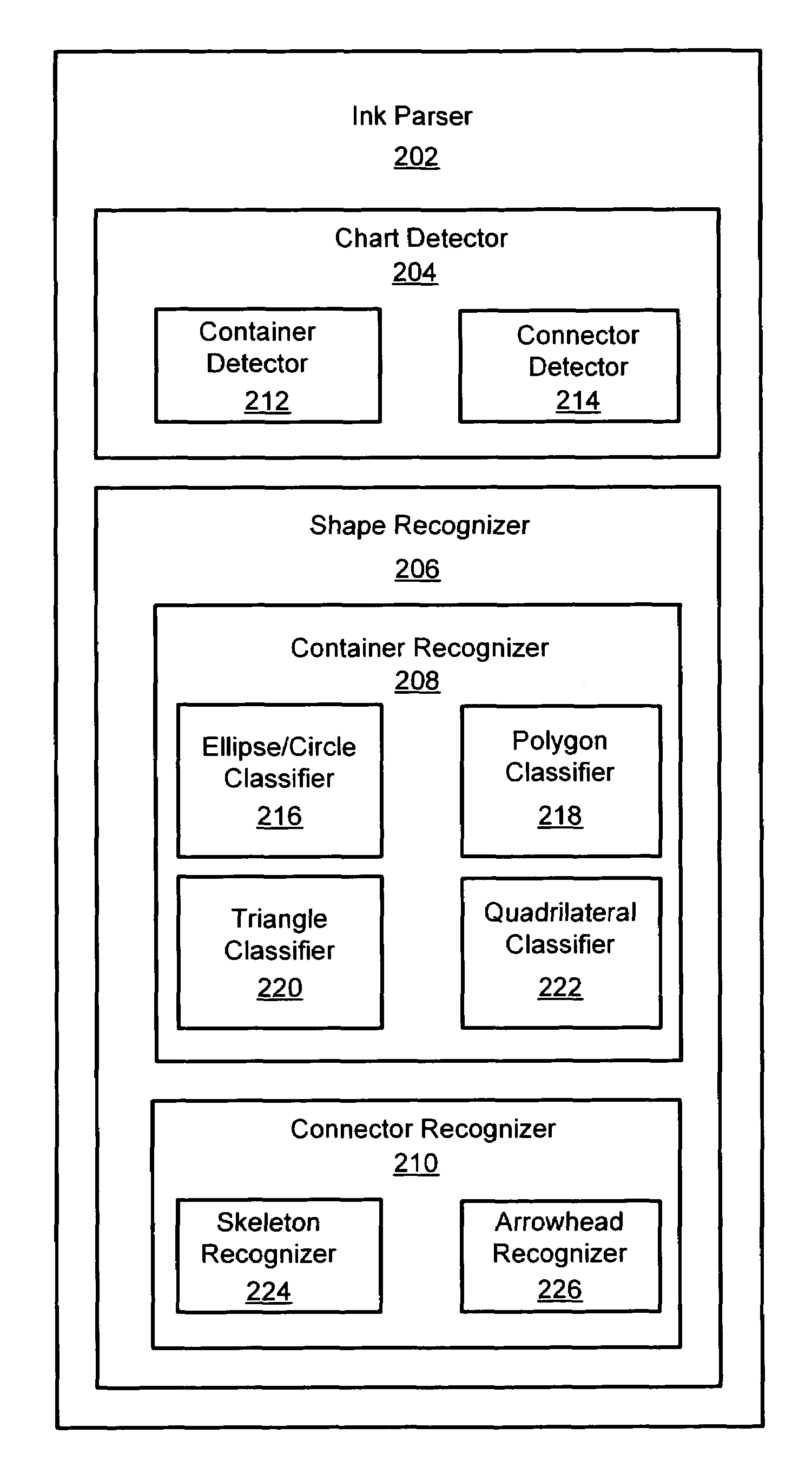
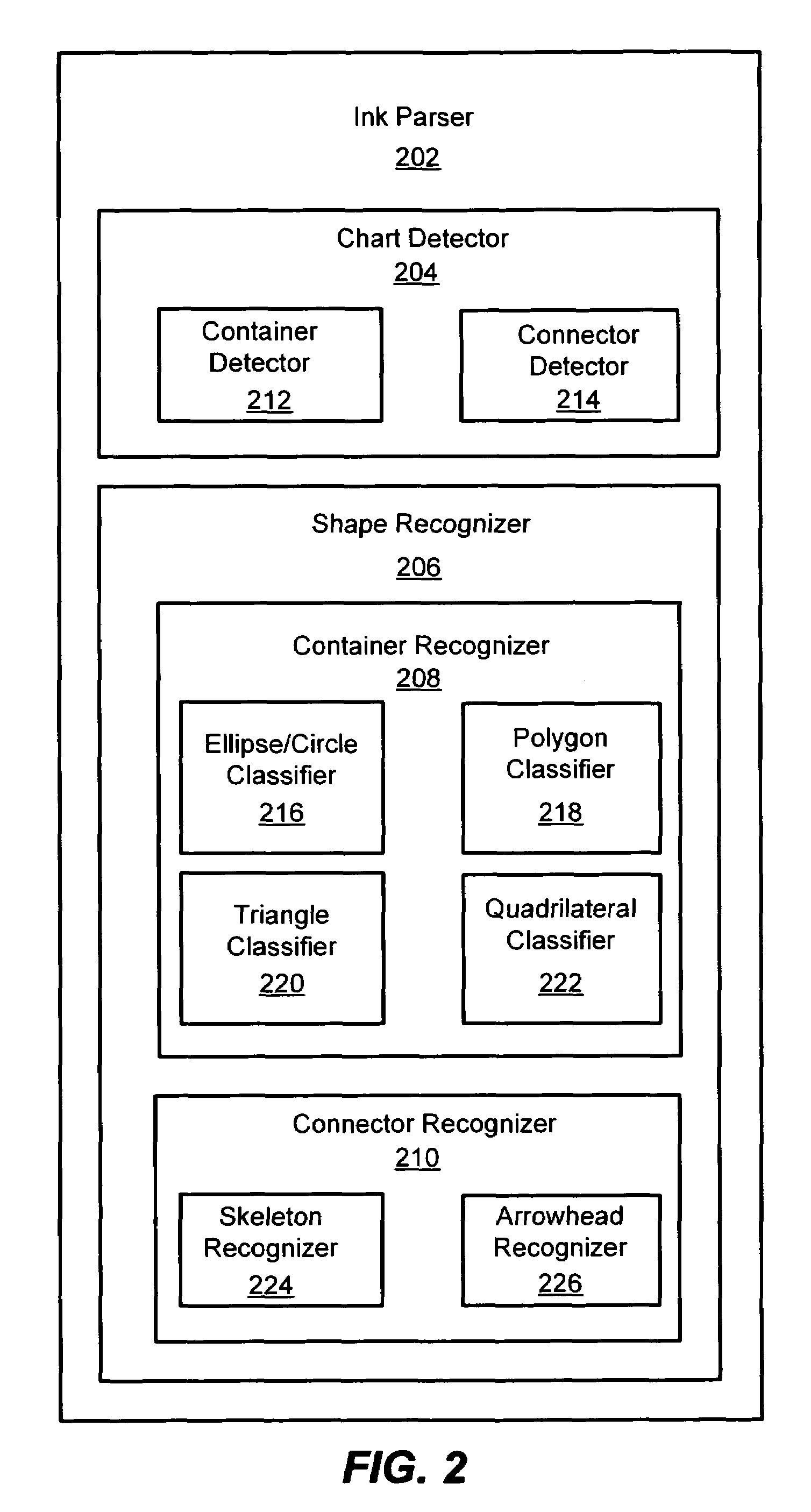
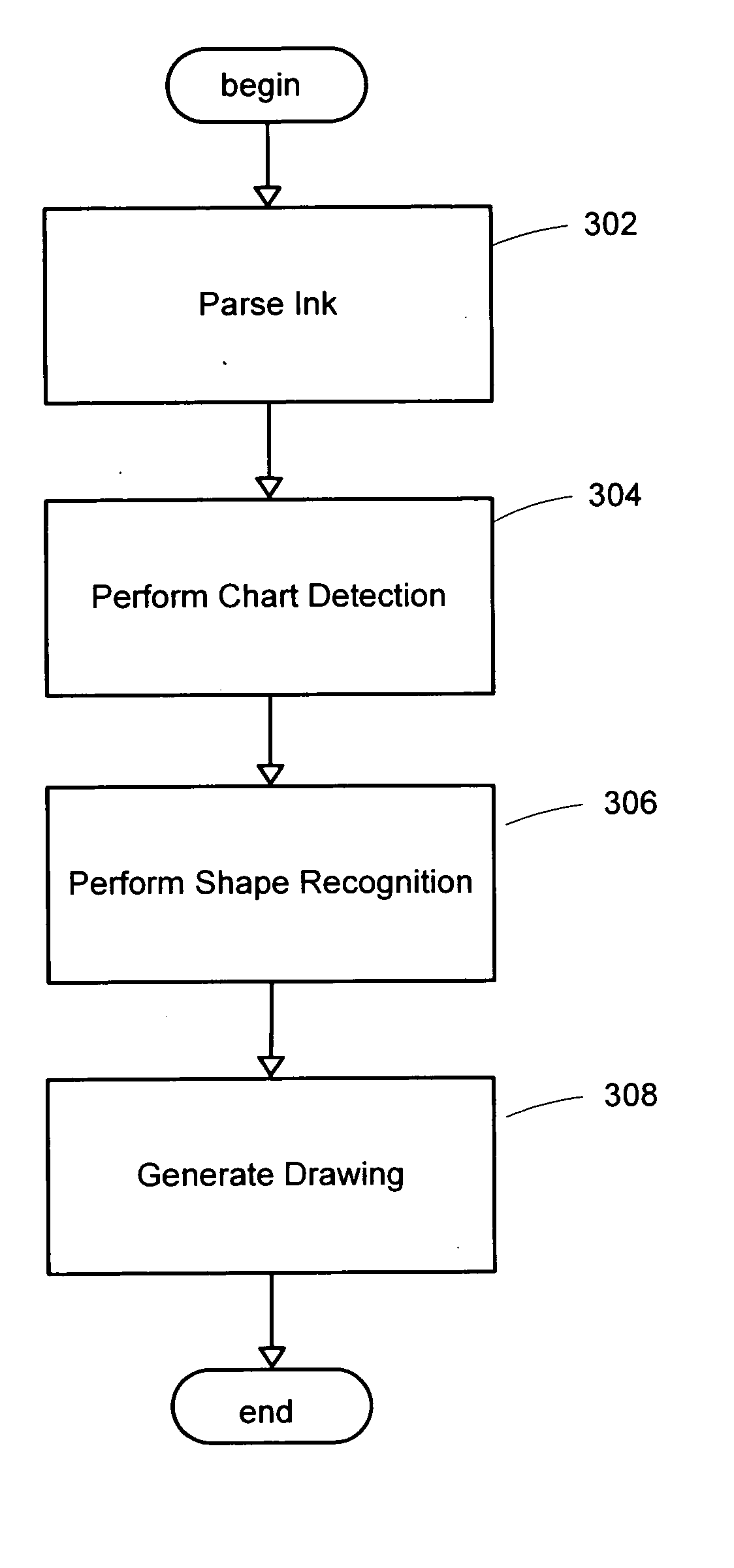
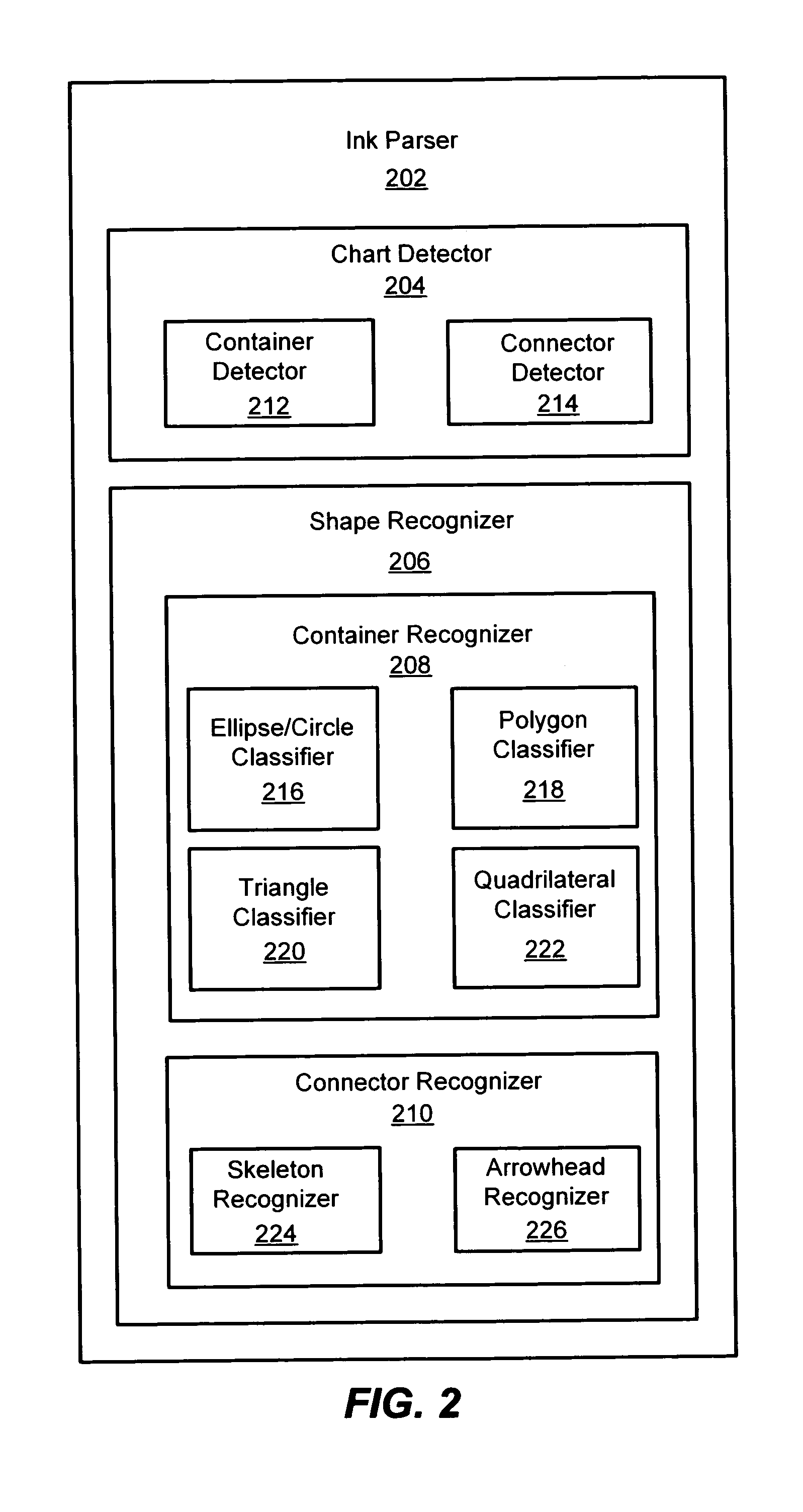
System and method for detecting a hand-drawn object in ink input
Owner:MICROSOFT TECH LICENSING LLC
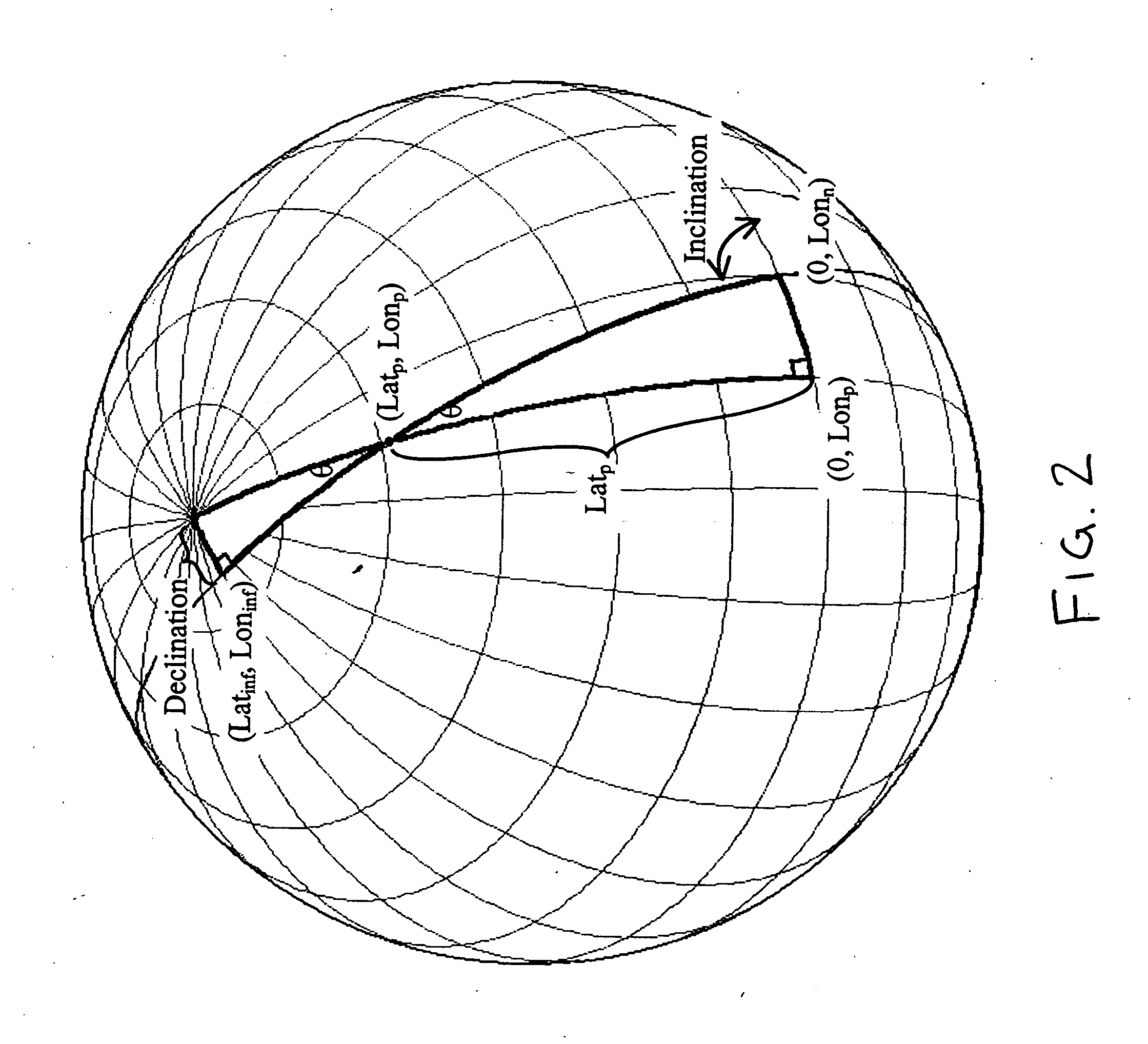
Backtrack orbit search algorithm
InactiveUS20060122776A1LevelRestrict levelInstruments for road network navigationCosmonautic vehiclesSensing dataLongitude
A method of searching an inventory of satellite remote sensing data for data granules, scenes, or images that cover a specified area of interest whereby each data granule is indexed to the relevant ascending equatorial crossing of the satellite. Representative points in the area of interest are traced backwards along the orbital track to determine where the satellite must have crossed the equator on the ascending pass in order for the sensor to have seen the area of interest. Spatial search of the data is thereby reduced to a simple search on a range of crossing longitudes.
Owner:KNOWLES KENNETH W +1
Multidimensional spatial searching for identifying substantially similar data fields
ActiveUS20170177743A1Digital data information retrievalNon-redundant fault processingMultidimensional scalingMatch algorithms
Owner:ORACLE INT CORP
System and method for detecting a hand-drawn object in ink input
A system and method for detection of hand-drawn objects in ink input is provided. A detector may detect a drawing such as a diagram or chart from ink input by detecting closed containers and / or unclosed connectors in the drawing. An efficient grid-based approach may be used for fitting the ink strokes into an image grid with an appropriate size. A flood-fill algorithm may be used to detect the containers and connectors. A time order search may also be performed after a spatial search to handle overlapping of drawing strokes. Finally, content detection may be performed for each detected container. Once any containers and their associated content have been detected in the image grid, connector detection may be performed. By using the present invention, a user may draw diagrams and flow charts freely and without restrictions on the hand-drawn input.
Owner:MICROSOFT TECH LICENSING LLC
Parcelized geographic data medium with internal spatial indices and method and system for use and formation thereof
InactiveUS7266560B2Instruments for road network navigationData processing applicationsInterior spaceNetwork packet
A navigable map database, stored on a computer-readable medium and used with a navigation application program, includes data which are spatially parcelized into a plurality of parcels. Associated with each of the plurality of parcels is a first index which associates the area represented by the data in the parcel with a plurality of sub-areas formed of the area. Also associated with each of the parcels is a second index associating each of the data in the parcel with at least one of the sub-areas. Further disclosed is a method for producing a navigable map database which is parcelized into a plurality of parcels, wherein each of the plurality of parcels includes a first index which associates the area represented by the data in the parcel with a plurality of sub-areas formed of the area and a second index associating each of the data in the parcel with at least one of the sub-areas. Also further disclosed are a program and method for finding data in one or more parcels that match a spatial search criterion using a navigable map database that is parcelized into a plurality of parcels, wherein each of the plurality of parcels includes a first index which associates the area represented by the data in the parcel with a plurality of sub-areas formed of the area and a second index associating each of the data in the parcel with at least one of the sub-areas. The program and method use the first and second indices to identify which of the data in at least one of the plurality of parcels satisfy the spatial search criterion.
Owner:HERE GLOBAL BV
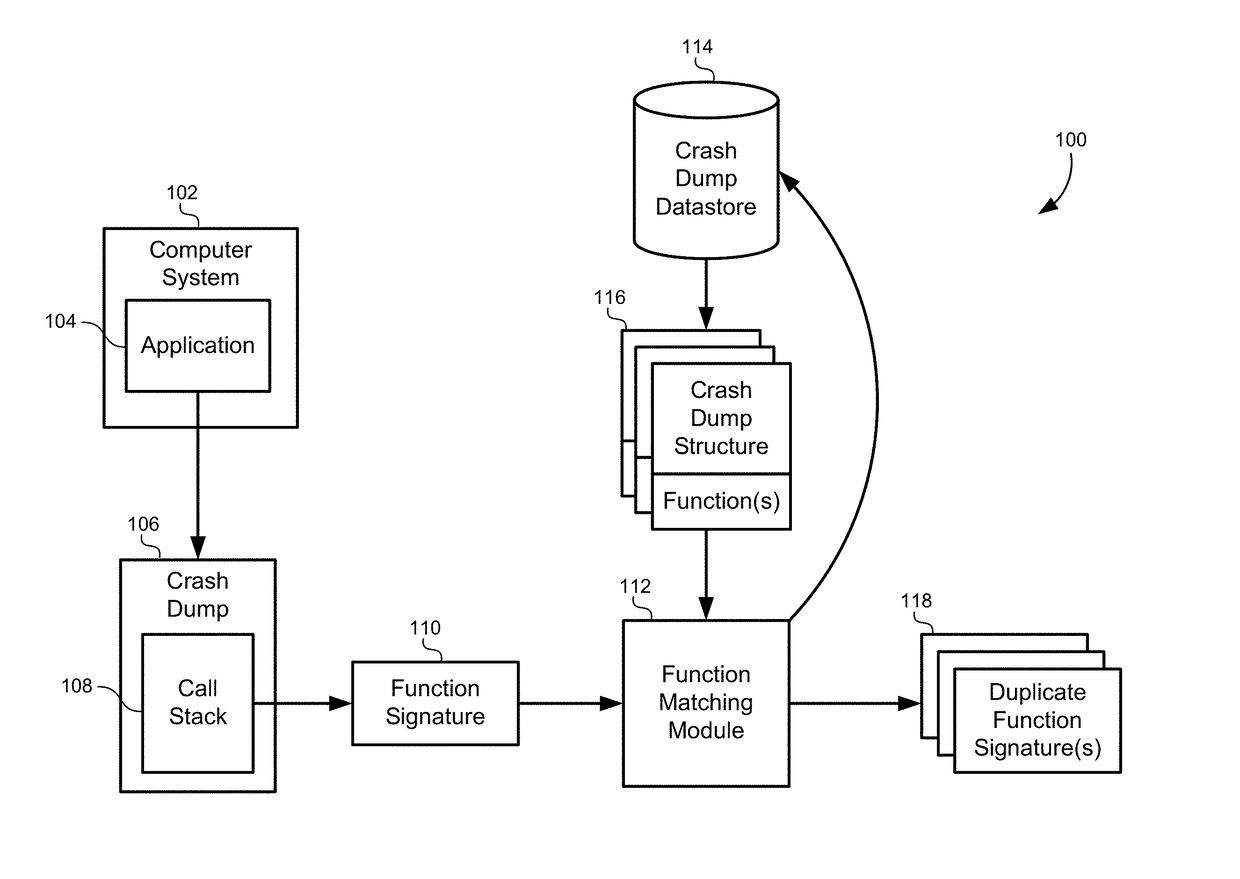
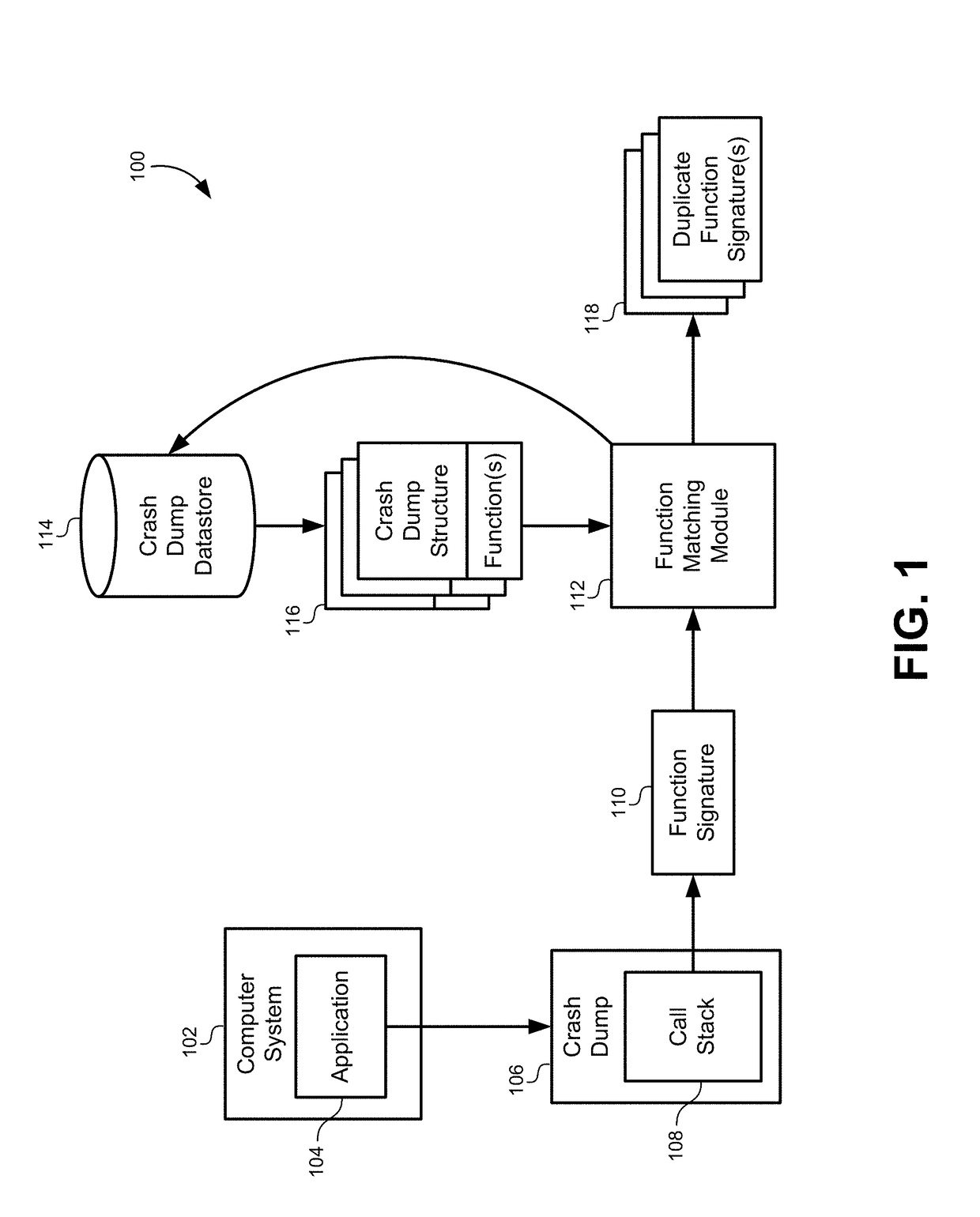
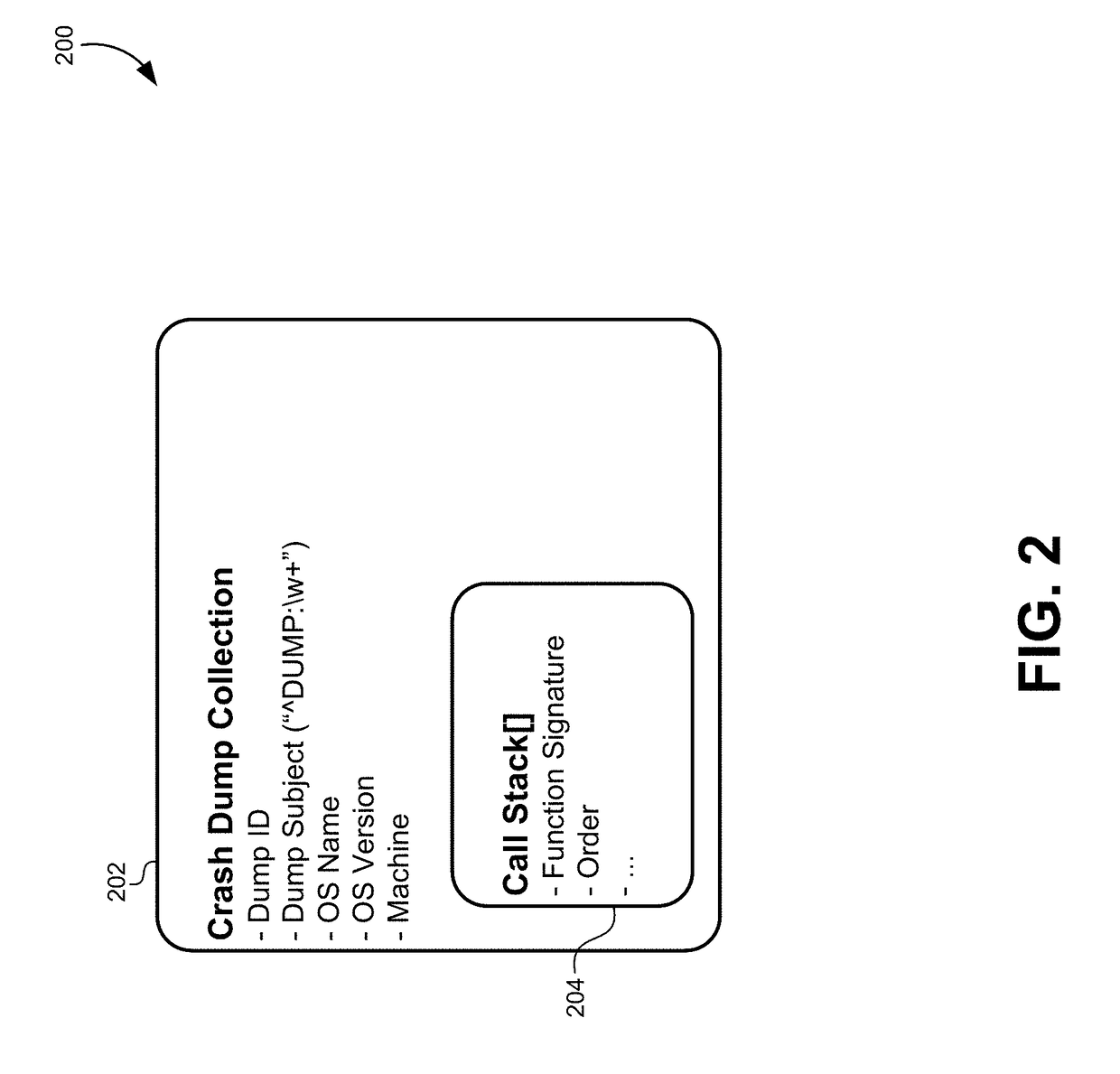
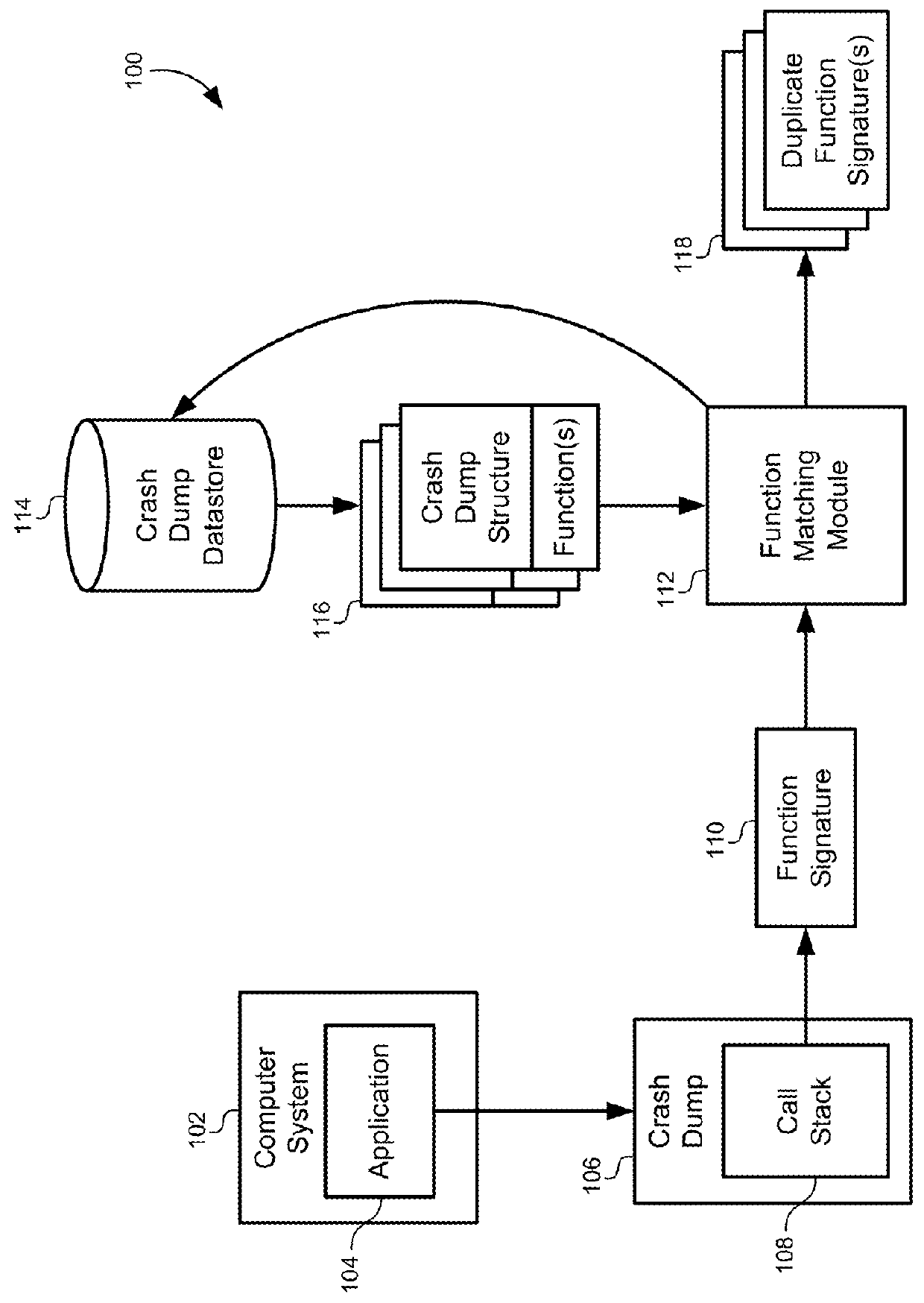
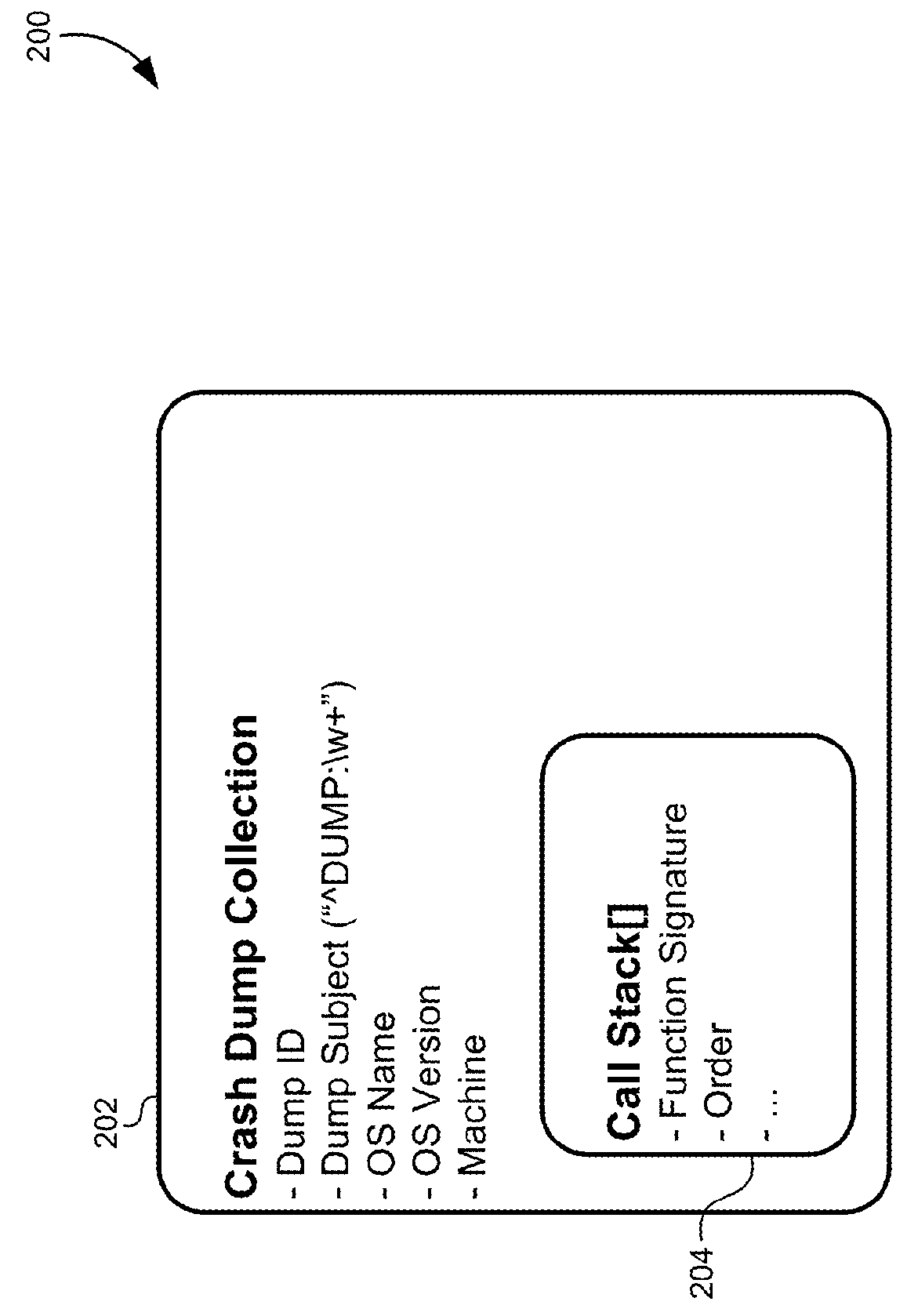
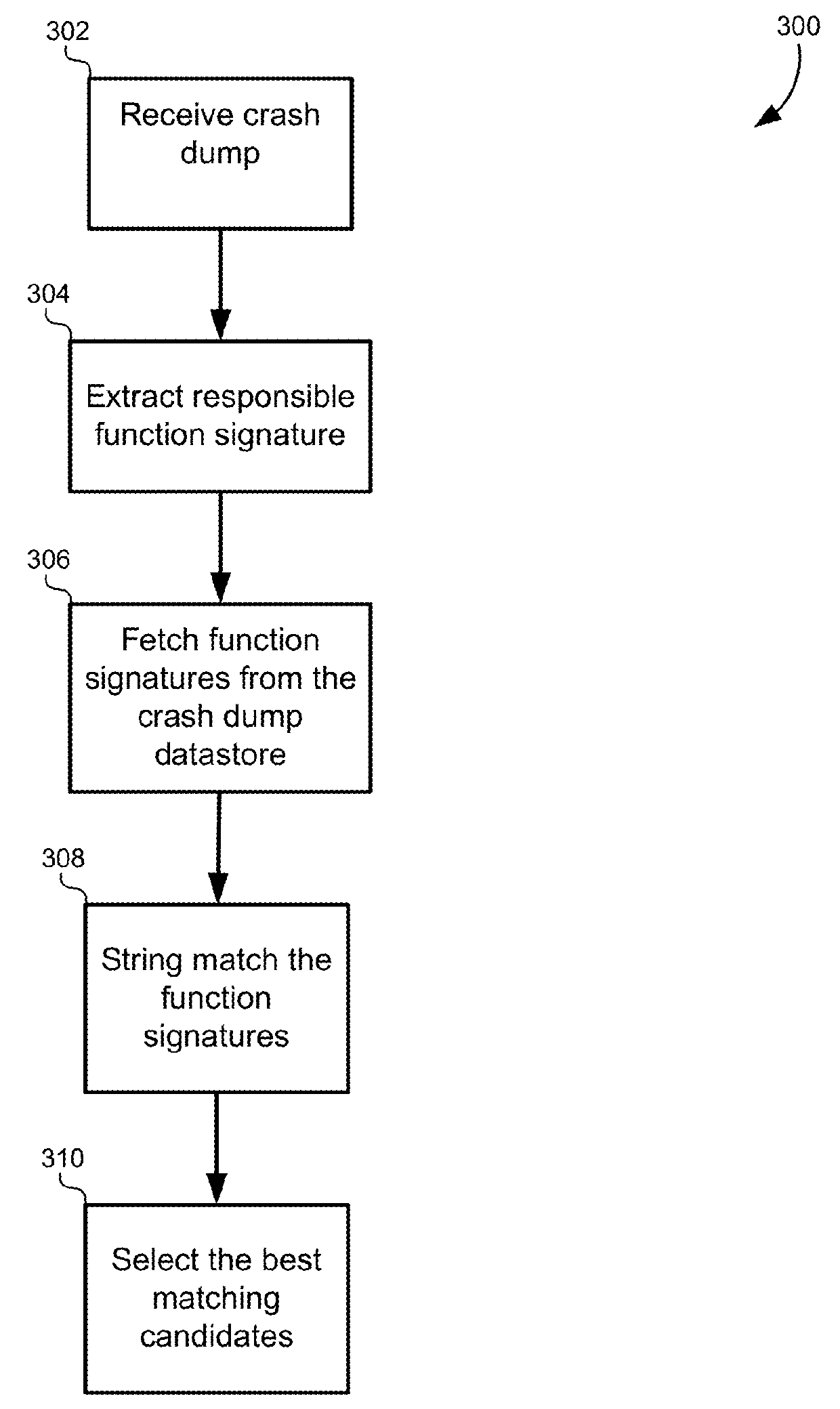
Multidimensional spatial searching for identifying duplicate crash dumps
ActiveUS20160055262A1Digital data information retrievalDigital data processing detailsMultidimensional scalingTheoretical computer science
A method of identifying duplicate crash dumps in a computer system may include receiving a first crash dump caused by an application crash, extracting a first function signature of a function that caused the first crash dump, and searching a datastore of crash dumps for function signatures that substantially match the first function signature. The searching may include performing an approximate string-match between each of the function signatures the first function signature and performing an exact string match between each of the function signatures and the first function signature. The searching may also include combining weighted results of the approximate string-match with weighted results of the exact string match to generate match scores for each of the function signatures, and identifying the function signatures that substantially match the first function signature based on the match scores.
Owner:ORACLE INT CORP
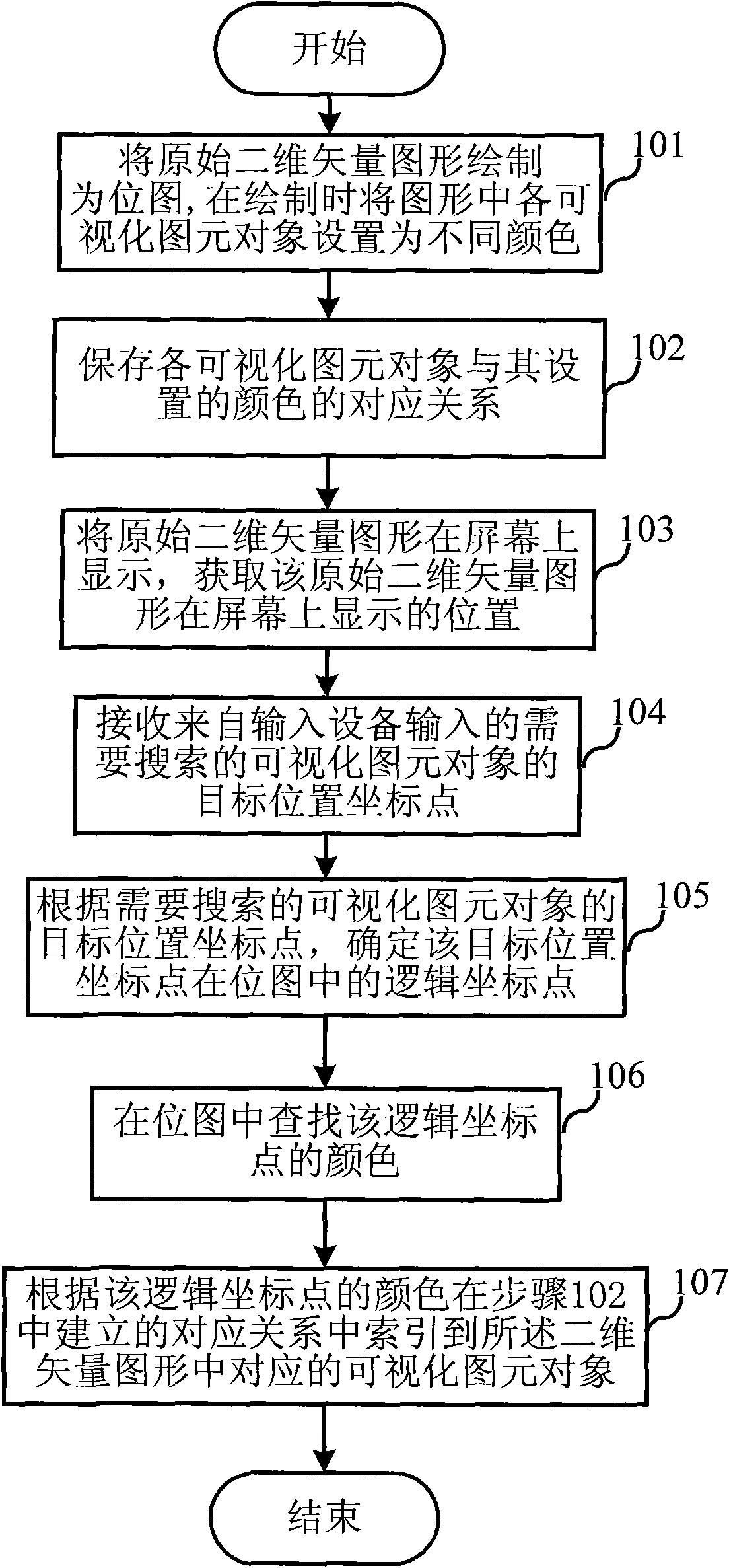
Spatial index method of two-dimension vector graphics and device thereof
ActiveCN101582077AFast search and locationRapid positioningGeometric image transformationSpecial data processing applicationsComputer visionLinear search
The invention relates to a spatial index method of two-dimension vector graphics and a device thereof. The spatial index method comprises the following steps: step one: the two-dimension vector graphics is drawn into bitmap, pixels in bitmap corresponding to visual primitive objects of the two-dimension vector graphics are set in different colors, and corresponding relationship between each visual primitive object and set colors thereof is stored; step two: corresponding pixel color of coordinate point of target position in bitmap is determined after the coordinate point of target position of the visual primitive object to be indexed which is input by an input equipment is received; step three: the determined color of the coordinate point of target position in step two is indexed into the corresponding visual primitive object of the two-dimension vector graphics. Compared with the prior art, the invention transfers complicated spatial search in wide range to linear search, and avoids complex mathematical operation in mapping progress, thus realizing rapid position of search target.
Owner:ASAT CHINA TECH
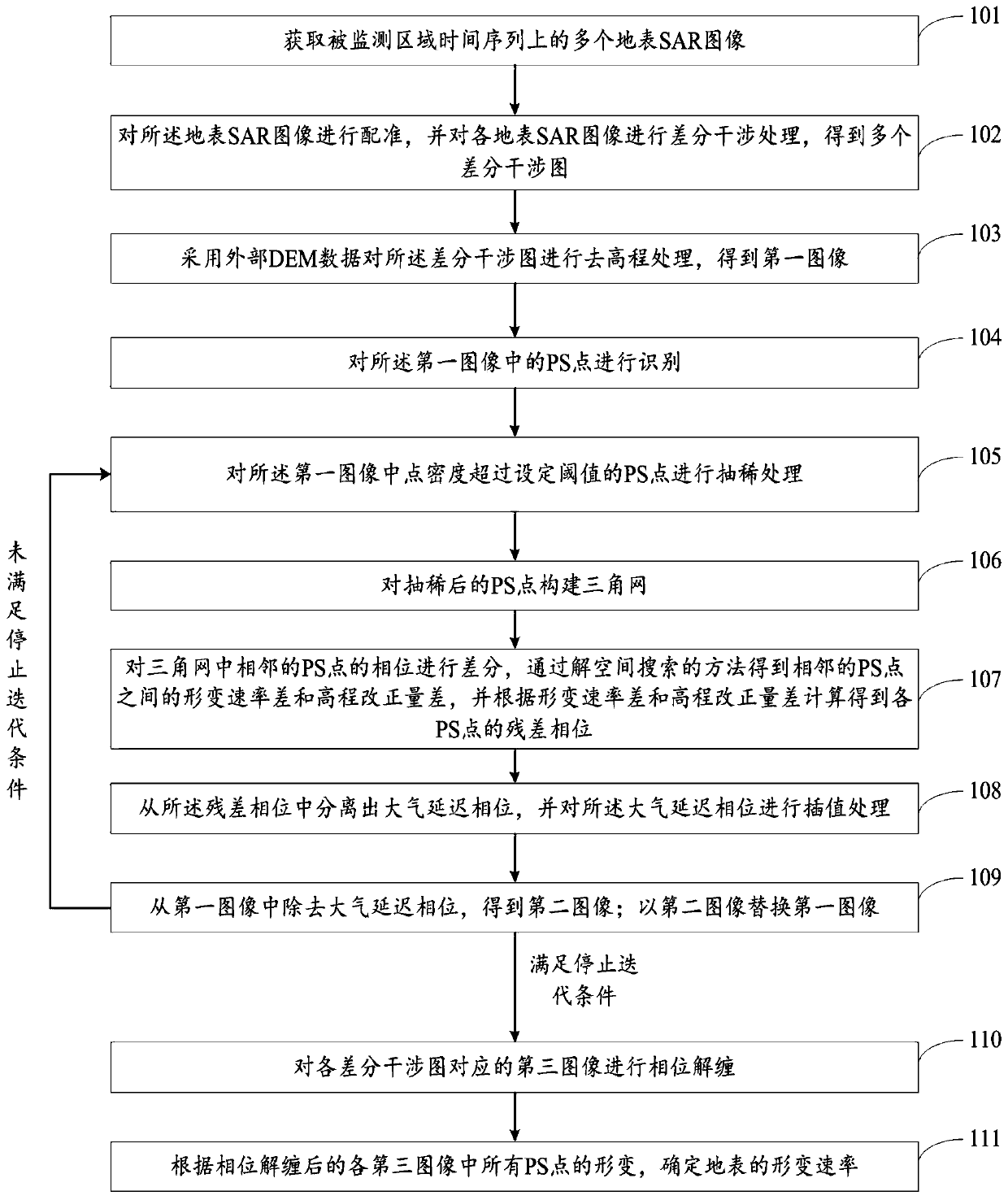
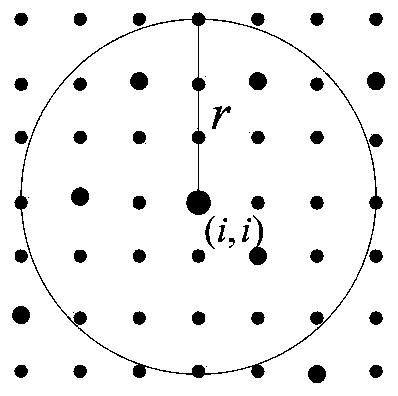
Time sequence InSAR deformation monitoring method and system based on high resolution
ActiveCN111059998AShorten the timeEasy to handleWave based measurement systemsElectrical/magnetic solid deformation measurementImaging processingDeformation monitoring
The invention discloses a time sequence InSAR deformation monitoring method and system based on high resolution. The method comprises the steps of carrying out registration, differential interferenceprocessing and elevation removal processing on a surface SAR image; identifying PS points in the image; thinning the PS points of which the point density exceeds a set threshold value; constructing atriangulation network for the thinned PS points; differentiating the phases of the adjacent PS points in the triangulation network, obtaining a deformation rate difference and an elevation correctiondifference between the adjacent PS points through a solution space search method, and calculating the residual phase of each PS point according to the deformation rate difference and the elevation correction difference; separating an atmospheric delay phase from the residual phase, and carrying out interpolation processing on the atmospheric delay phase; removing the atmospheric delay phase from the differential interferogram, repeating the above steps for iteration, obtaining an image after the atmospheric delay phase is removed for many times, carrying out the phase unwrapping of the image,carrying out analysis on all PS points, and determining the deformation rate of the ground surface. According to the invention, the image processing rate is improved.
Owner:CHINA UNIV OF GEOSCIENCES (BEIJING) +1
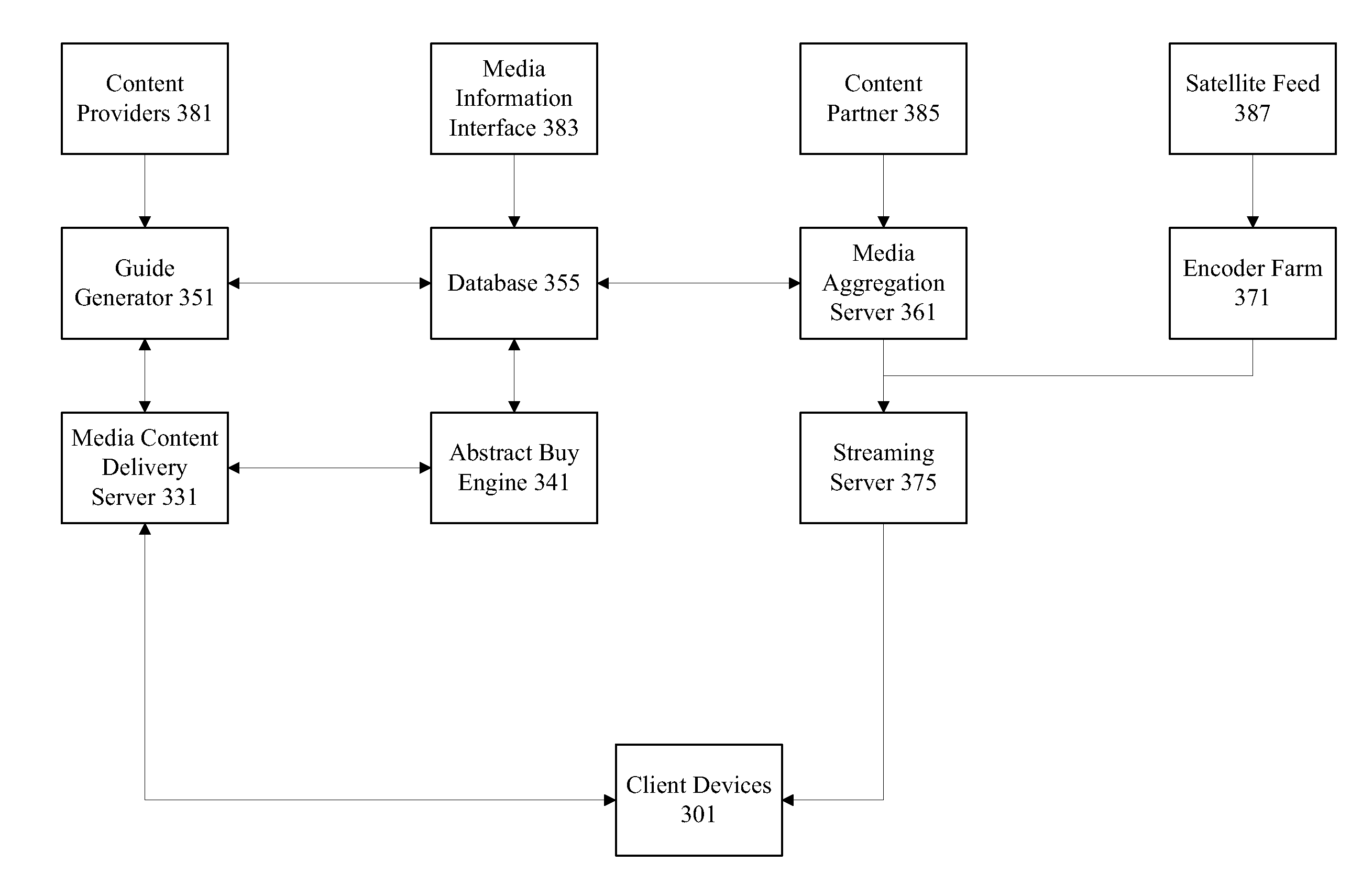
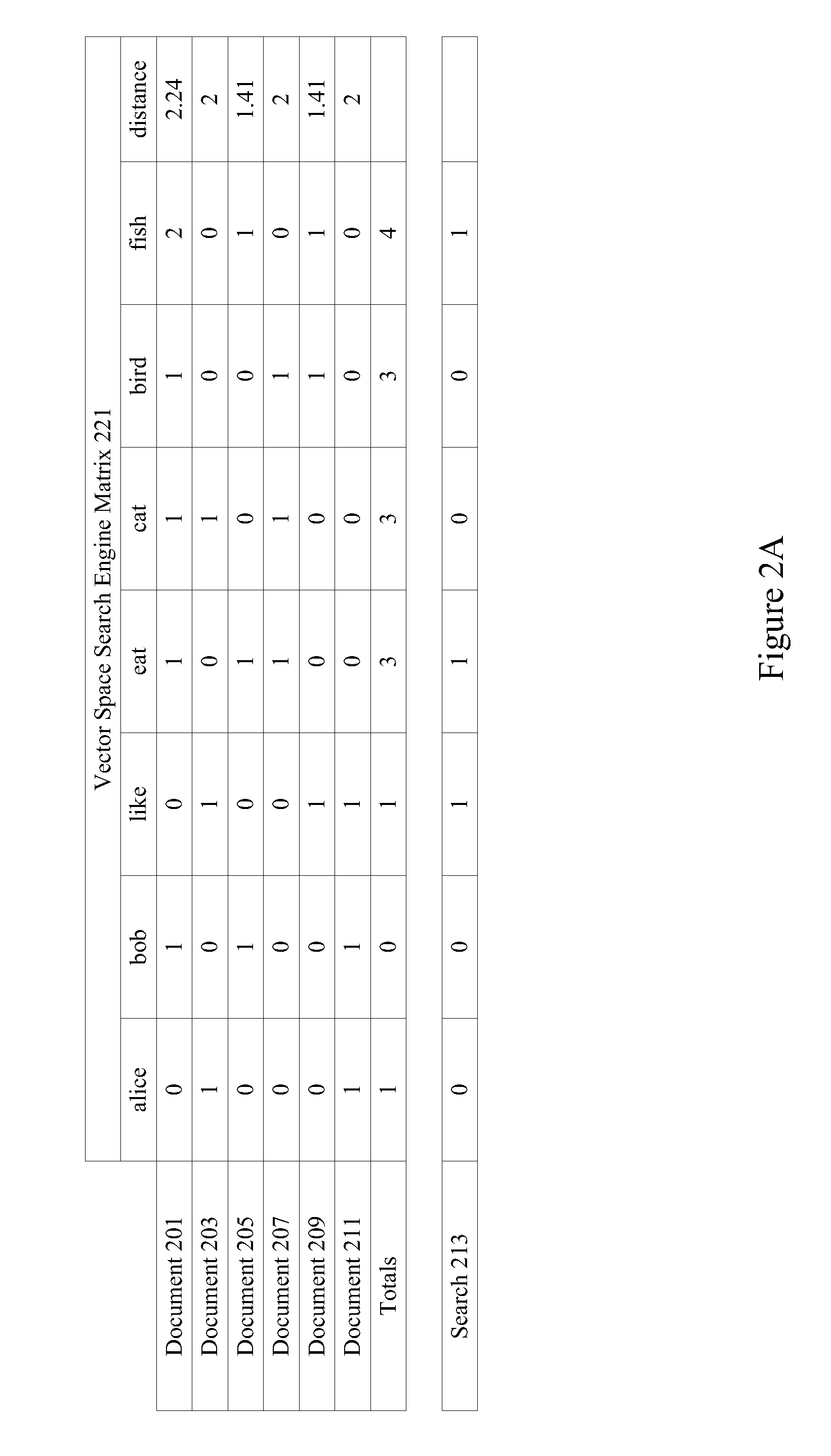
Retrieval and display of related content using text stream data feeds
ActiveUS20110202515A1Data processing applicationsDigital data processing detailsData feedText stream
Mechanisms are provided for retrieving and presenting related content using text stream data feeds. Text stream data feeds such as caption information associated with media content or conversations associated with social networking applications are aggregated and used to retrieve related media content, text documents, and advertisements. Text stream data feeds that a user is exposed to may indicate that the user is interested or at least primed for particular types of related content. In particular examples, an inverse vector space search engine is used to determine particular pieces of related content and categories of interest. Post filtering may also be applied to the results.
Owner:TIVO CORP
Millimeter wave beam tracking and beam sweeping
Aspects of mmWave beam tracking and beam sweeping are described, for example, spatial searching operations, directional beam forming, complex channel measurement operations, and adaptive power savings. Some aspects include using priori information for mmWave beam tracking and beam sweeping. Some aspects include using priori information to modify a superset of beam criteria to obtain a subset of beam criteria, select a spatial region according to the subset of beam criteria, and initiate a spatial searching operation within the spatial region for establishing a communication link. Some aspects include obtaining complex channel measurements of beams and combining the measurements with priori information to determine a beam for use in a communication link. Some aspects include providing signals from Nr over K1 input / output (IO) links and receiving signals over K1 IO
Owner:INTEL CORP
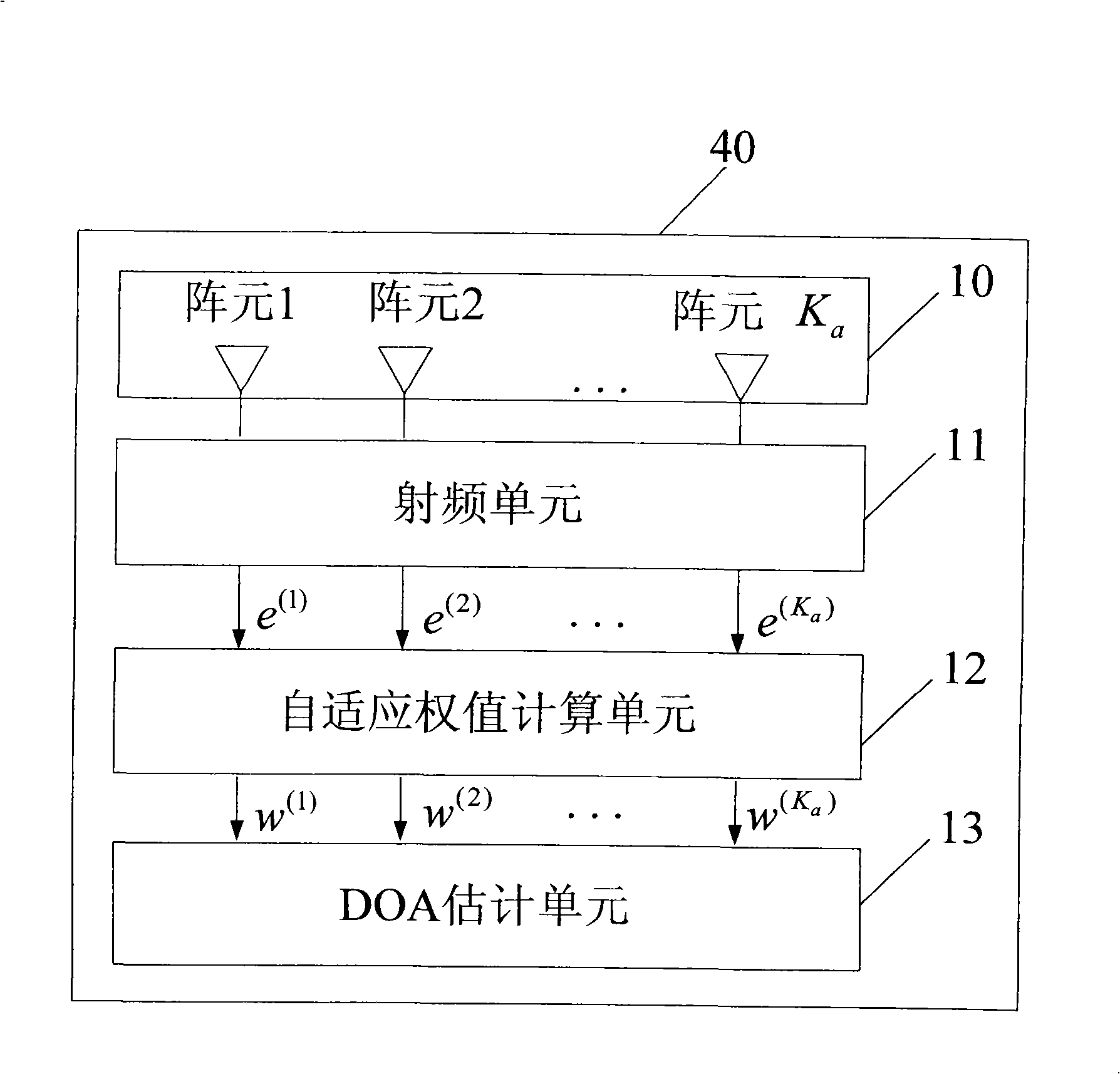
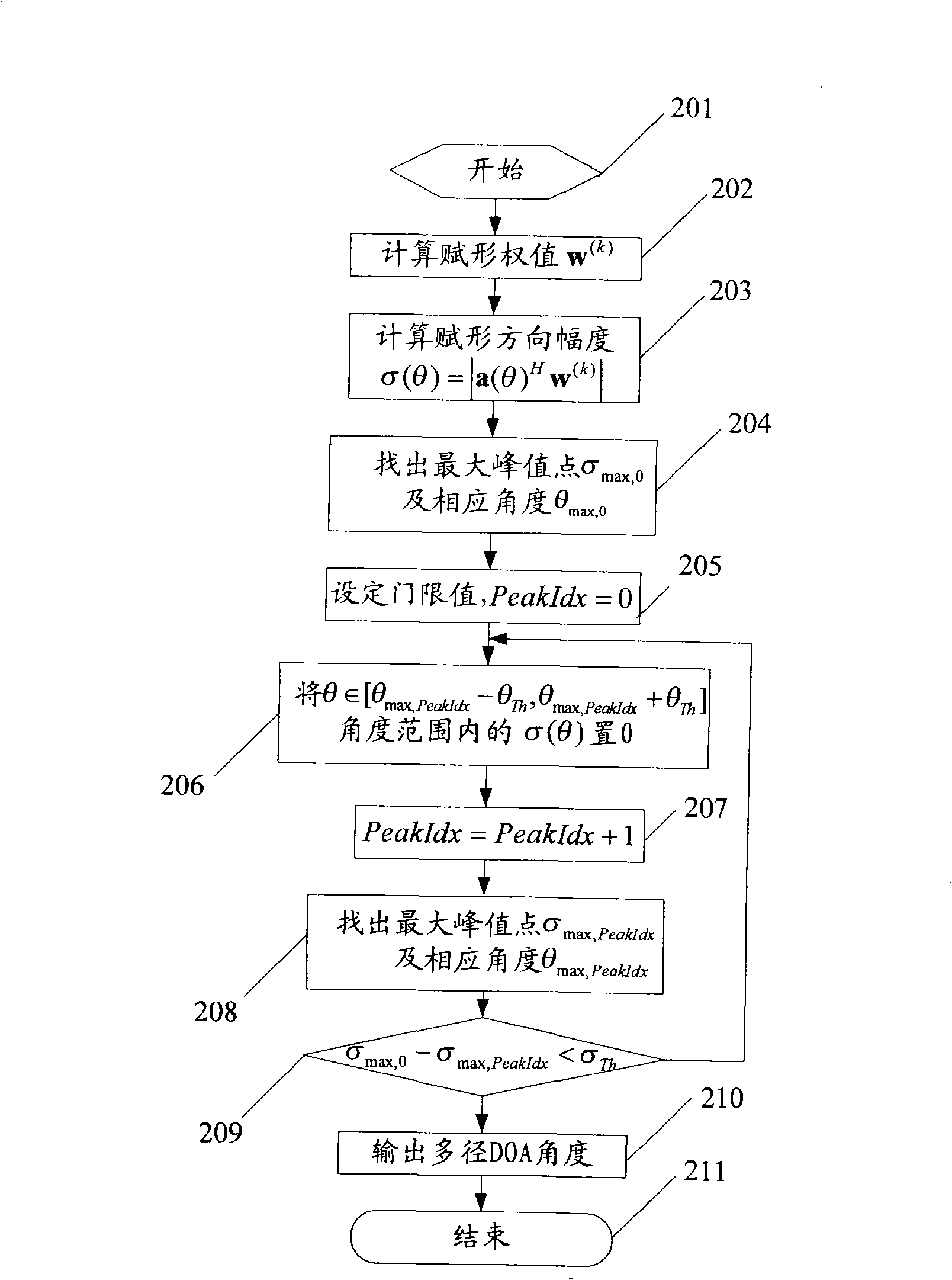
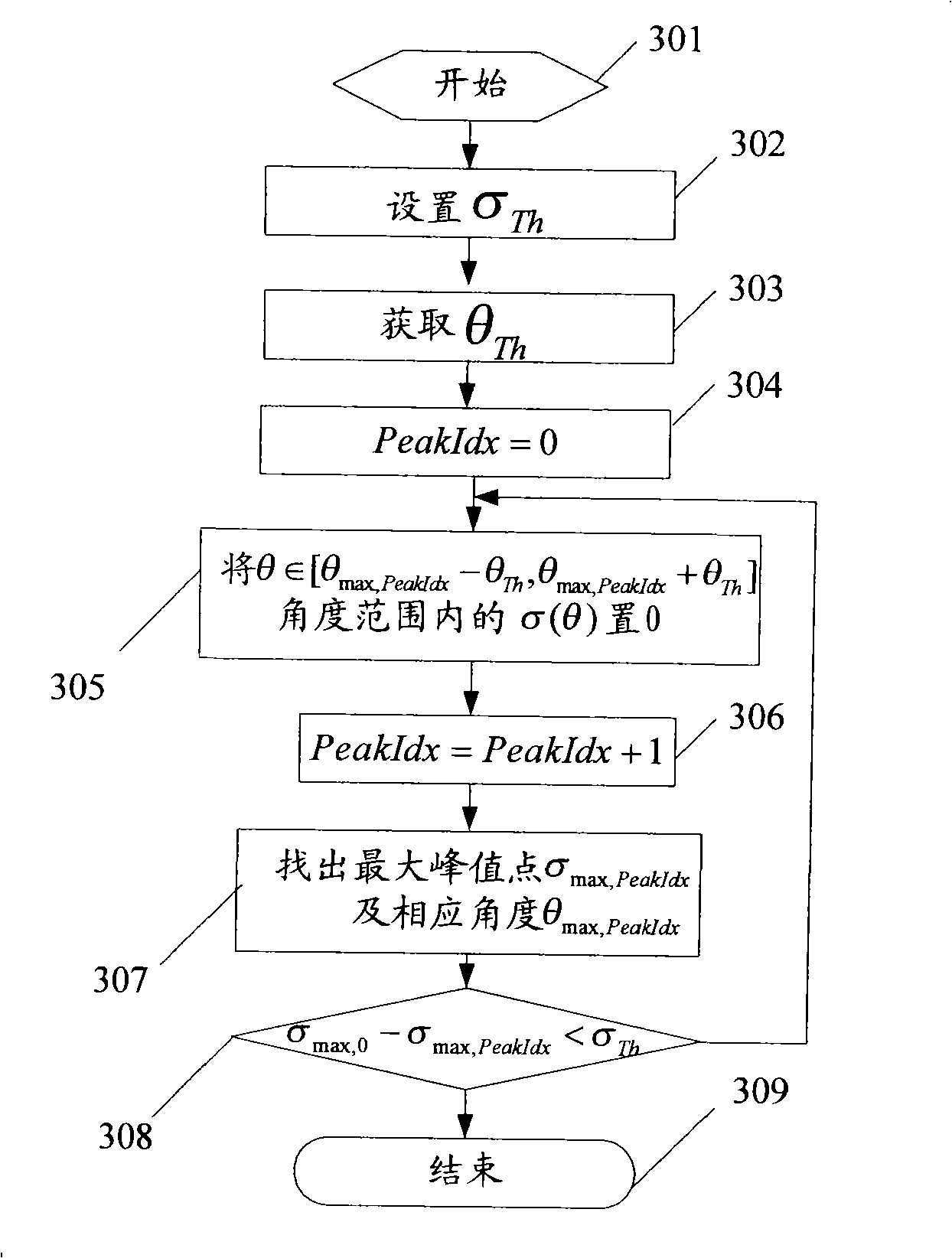
Method and device for calculating arrival direction in an intelligent antenna system
InactiveCN101330304ASolve the acquisition problemSpatial transmit diversityRadio transmission for post communicationWave formSmart antenna
The invention relates to a method for calculating arrival direction in a smart antenna system. The method comprises the following steps: step one, an adaptive weigh value calculating unit figures out a user wave-forming weight value by using the reference algorithm of non-arrival direction; step two, an arrival direction estimating unit figures out a forming direction amplitude in a whole angle scope by adopting a space searching method; step 3, the arrival direction estimating unit finds out a maximal peak point and an angle corresponding to the maximal peak point from the forming direction amplitude. The method can directly acquire a DOA and a multidiameter DOA according to the wave-forming, thereby solving the DOA acquisition problems of a non-DOA reference method.
Owner:ZTE CORP
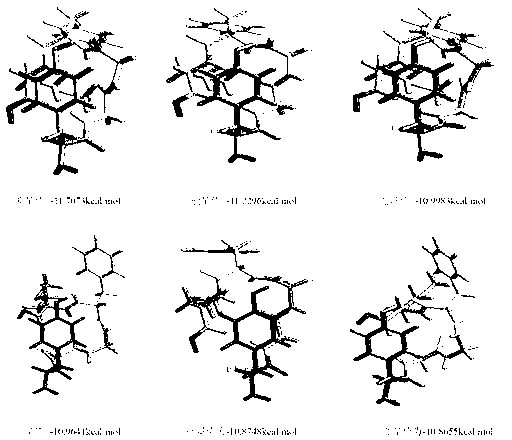
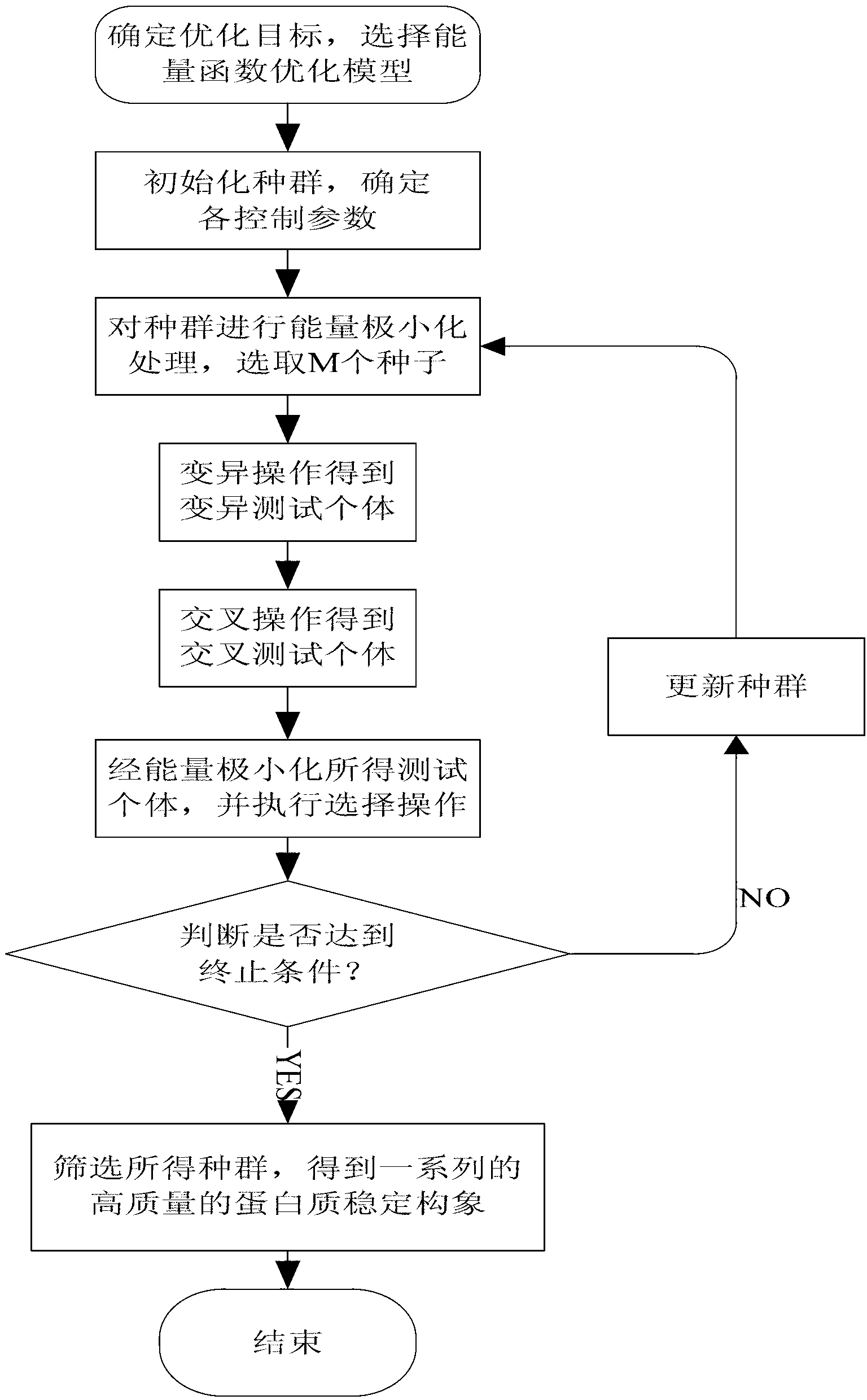
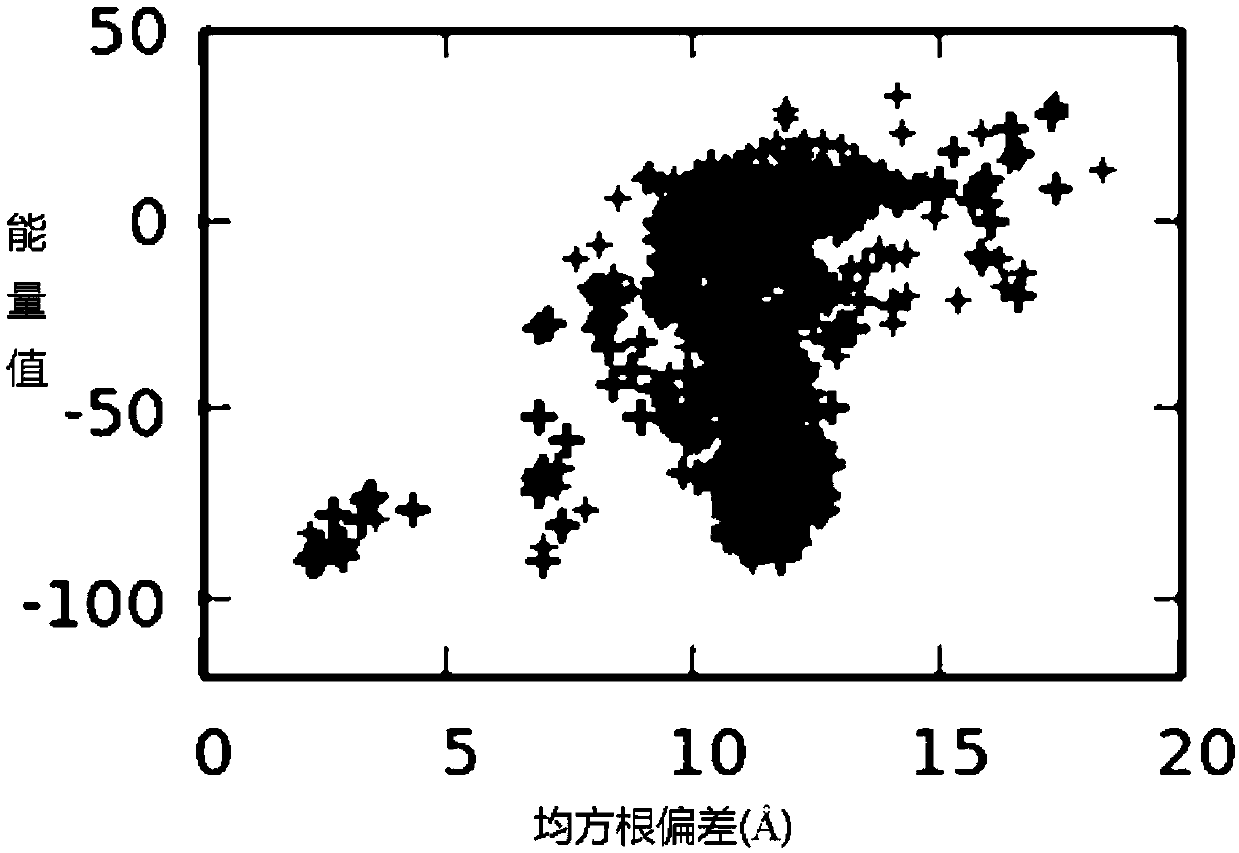
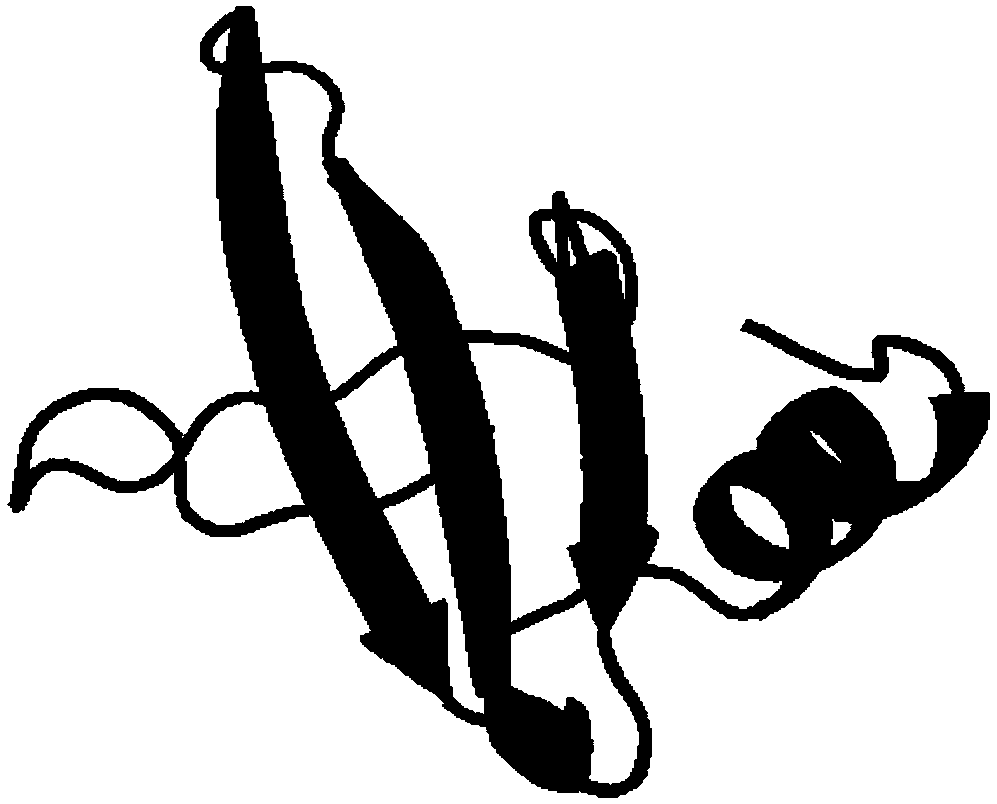
Spatial search method for multi-modal protein conformations
ActiveCN103077226AFast convergencePrevent falling intoBiological modelsEnergy efficient computingDiseaseProtein structure
The invention provides a spatial search method for multi-modal protein conformations. According to the spatial search method, the thoughts of a spatial locality principle and an assembly process are integrated based on a crowding differential evolution algorithm, and the protein conformations obtained through experiments are processed by adopting an energy minimization process. Due to the spatial locality principle, the convergence rate of the algorithm is increased, and the problems of local convergence and modal diversity in multi-modal optimization are effectively solved; in the assembly process, different crossover strategies are randomly selected, so that better segments in the conformations are prevented from being damaged by the algorithm, and the diversity of the multi-modal protein conformations is improved; and due to the energy minimization process, the complexity of space solution of the protein conformations is reduced, and the search space of feasible zones of the protein conformations is effectively shortened. According to the spatial search method, enkephalin is taken as an example, not only is the well-known most global stable structure of the enkephalin obtained, but also a series of high-quality local stable structure are obtained, so that the problem of multi-gene and multi-target paths of diseases which cannot be solved by the traditional single-target-directed single-modal research method is solved, and the requirements of computer-aided drug design on multi-modal protein structures at the present stage are met.
Owner:ZHEJIANG UNIV OF TECH
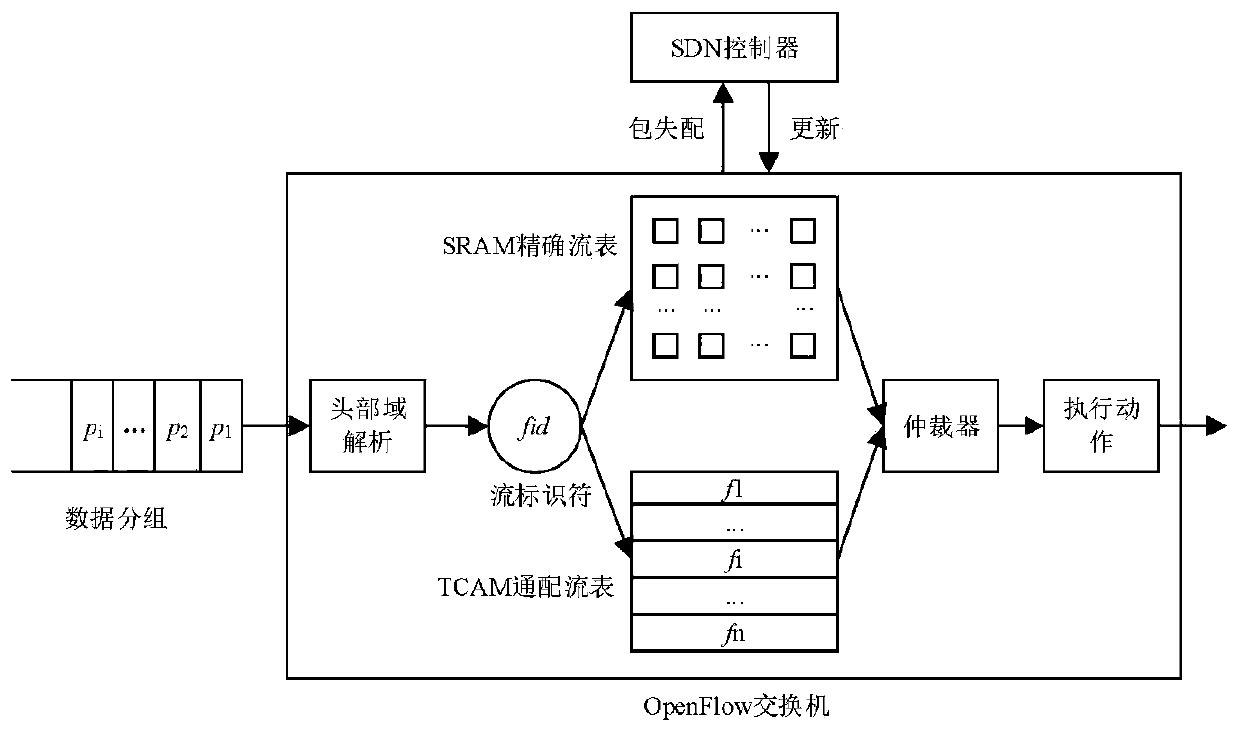
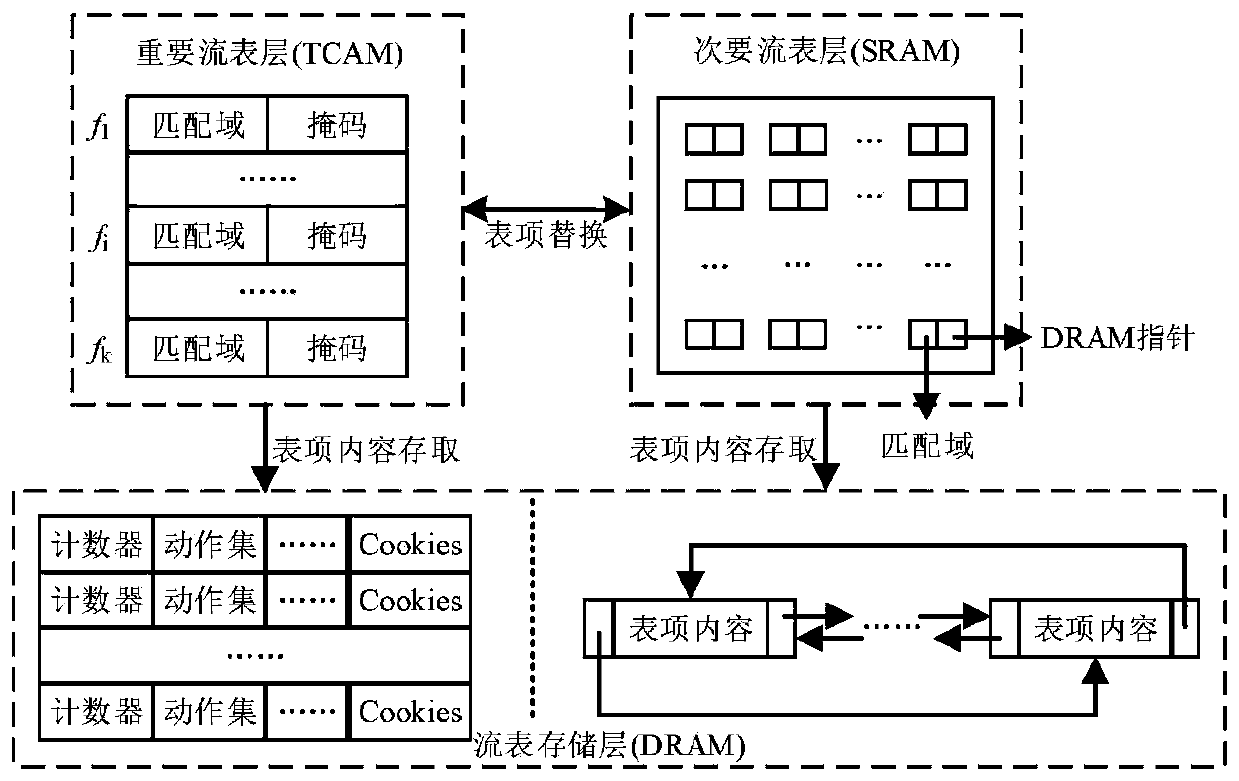
QoS (Quality of Service)-aware OpenFlow flow table hierarchical storage architecture and application
ActiveCN111131084AGuaranteed packet forwarding performanceReduce the average flow table lookup timeData switching networksHigh level techniquesQuality of serviceData pack
The invention discloses a QoS (Quality of Service)-aware OpenFlow flow table hierarchical storage architecture and application, and the architecture provided by the invention stores a high-priority flow in a TCAM (Ternary Content Addressable Memory), so that the grouping search performance of the high-priority flow is ensured; a Cuckoo hash structure is adopted to design an accurate flow cache mechanism and store low-priority and non-priority active accurate flows, so that data packets in the accurate flows directly hit caches, corresponding SRAM flow table entries are quickly found, the average flow table search time is remarkably shortened, and the performance of the OpenFlow switch is improved. According to the method, corresponding flow table searching speeds are provided for network flows with different priorities, wherein network packets in a high-priority flow directly hit a TCAM flow table, and corresponding flow table entries are quickly found; a hit Cuckoo cache is searched by a network packet in the low-priority flows, and a corresponding SRAM flow table entry is directly positioned; most of the groups in the priority-free flows can directly hit the Cuckoo cache, and a tuple space search method needs to be further adopted for searching the SRAM flow table for a small part of the groups, and therefore the overall searching efficiency is high.
Owner:CHANGSHA NORMAL UNIV
Access control method of spatial database
ActiveCN103870548AImprove authorization query efficiencyGuaranteed response speedDigital data protectionSpecial data processing applicationsData miningSpatial database
The invention discloses an access control method of a spatial database. The access control method comprises the following steps of realizing authorization of the spatial database by adding access rules into an R+ tree, searching the R+ tree added with the access rules according to an access request, obtaining a spatial entity from the spatial database according to the searched result, further cutting the spatial entity according to the access request and the corresponding access rules, and returning the cut spatial entity to a user. The access control method disclosed by the invention not only supports the authorization of the spatial entity, but also supports the authorization of fine-grit splitting of the entities; the access control rules can only cover one part of the spatial entity, simultaneously, the twice queries such as judgment of the access rules and the spatial search are combined into once query, and in once query, the result of query of the user is determined and corresponding information is returned, so that the query efficiency of authorization is improved and the response speed in accessing spatial vector data with permission limitation is ensured.
Owner:ZHEJIANG UNIV
Conformation diversity policy-based protein three-dimensional structure prediction method
ActiveCN108647486AEnhanced Spatial SearchIncrease diversitySpecial data processing applicationsMolecular structuresProtein structureGenetic algorithm
The invention discloses a conformation diversity policy-based protein three-dimensional structure prediction method. Under a framework of a genetic algorithm, a Rosetta standard protocol is adopted; aloop region of a conformation is operated by utilizing a genetic operator, so that spatial search for the loop region is enhanced; and a secondary protein structure information-based diversity indexis introduced, and an energy value of the conformation serves as a selection standard of population update, so that the negative influence caused by an inaccurate energy function is reduced while thepopulation diversity is enhanced, and a near-native conformation with higher precision is predicted. The conformation diversity policy-based protein three-dimensional structure prediction method provided by the invention is relatively high in prediction precision.
Owner:ZHEJIANG UNIV OF TECH
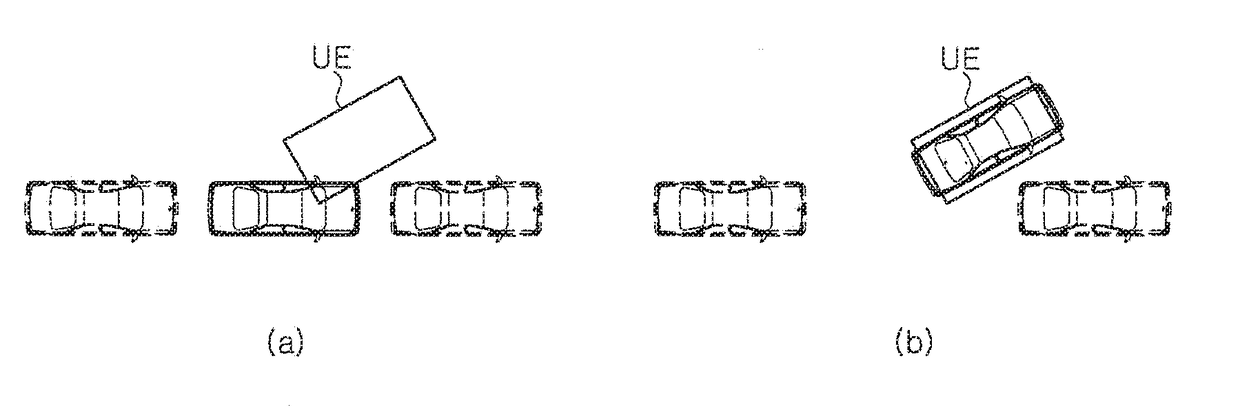
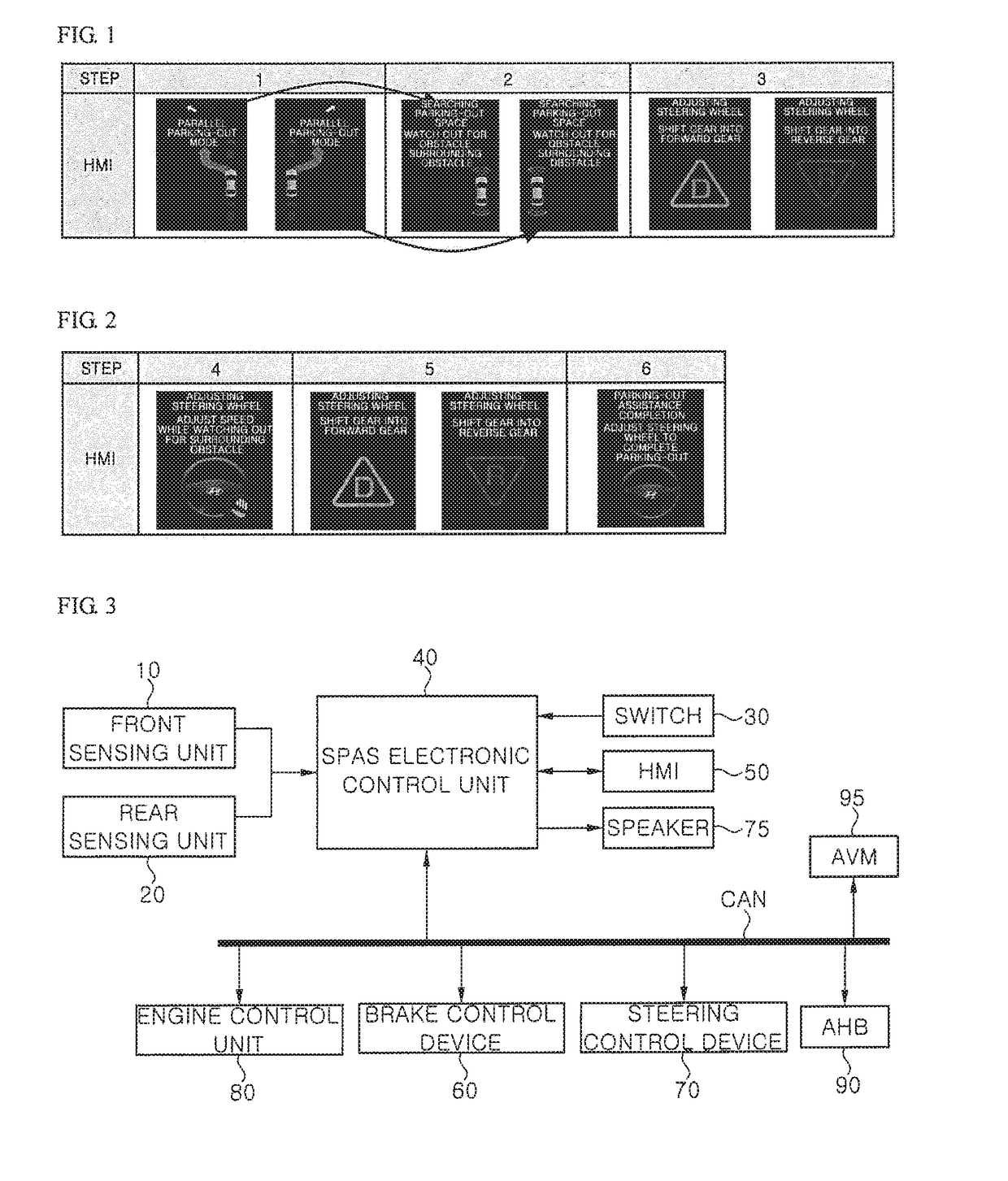
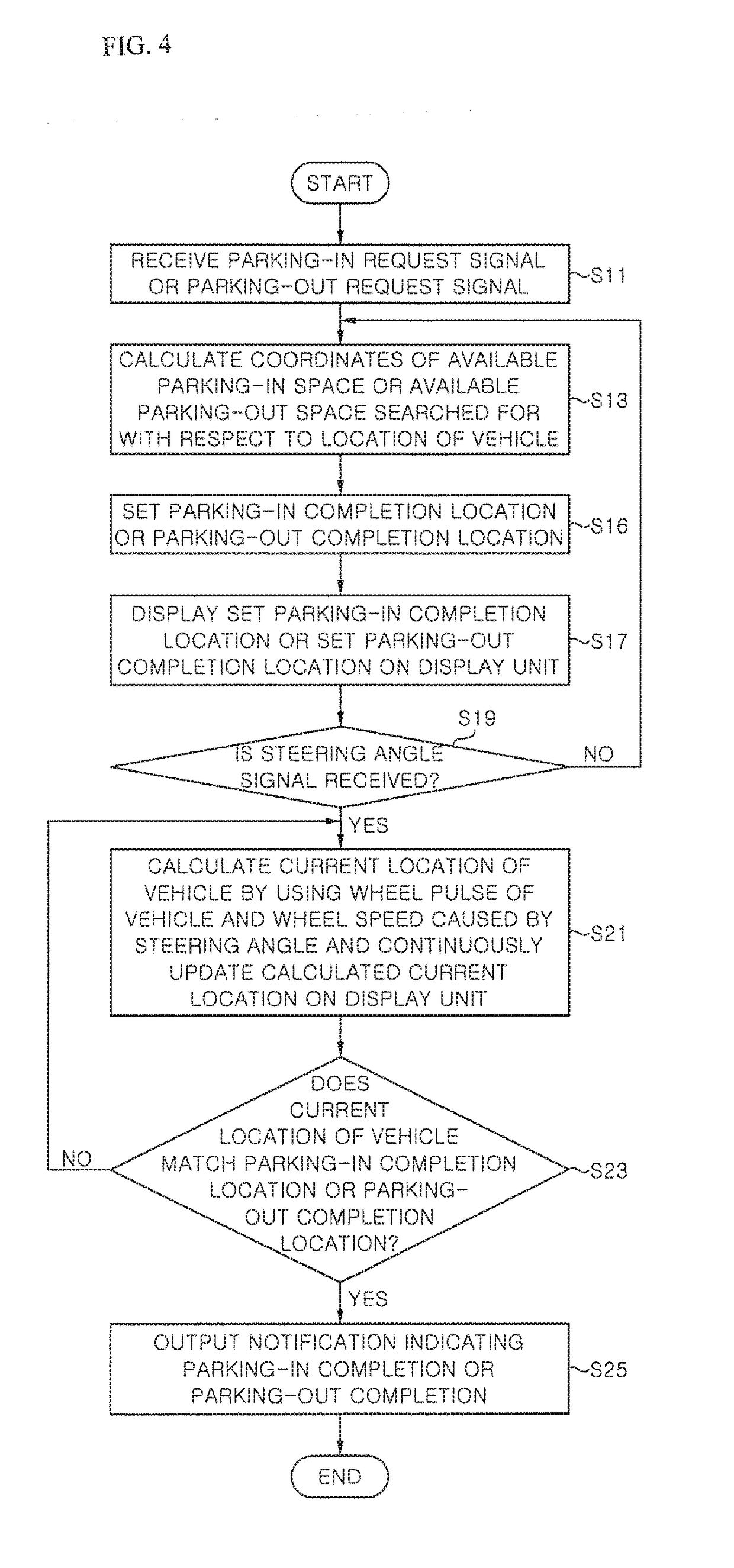
Driving assistance device
ActiveUS20170297624A1Pedestrian/occupant safety arrangementIndication of parksing free spacesAutomatic steeringEngineering
Provided is a driving assistance device allowing a driver to recognize a path of a vehicle by outputting a location of a vehicle and a location of a target space to the driver, the location of the target space being searched for with respect to the location of the vehicle. The driving assistance device includes: a space searching unit configured to search for a target space around a vehicle with respect to the vehicle; a space information acquisition unit configured to acquire a location of the vehicle and a location of the target space; and an output unit configured to output the location of the vehicle and the location of the target space to a driver, wherein, when an automatic steering function is operated, the output unit updates and outputs a current location of the vehicle on a certain cycle, preferably, in real time until the location of the vehicle matches the location of the target space.
Owner:HL KLEMOVE CORP
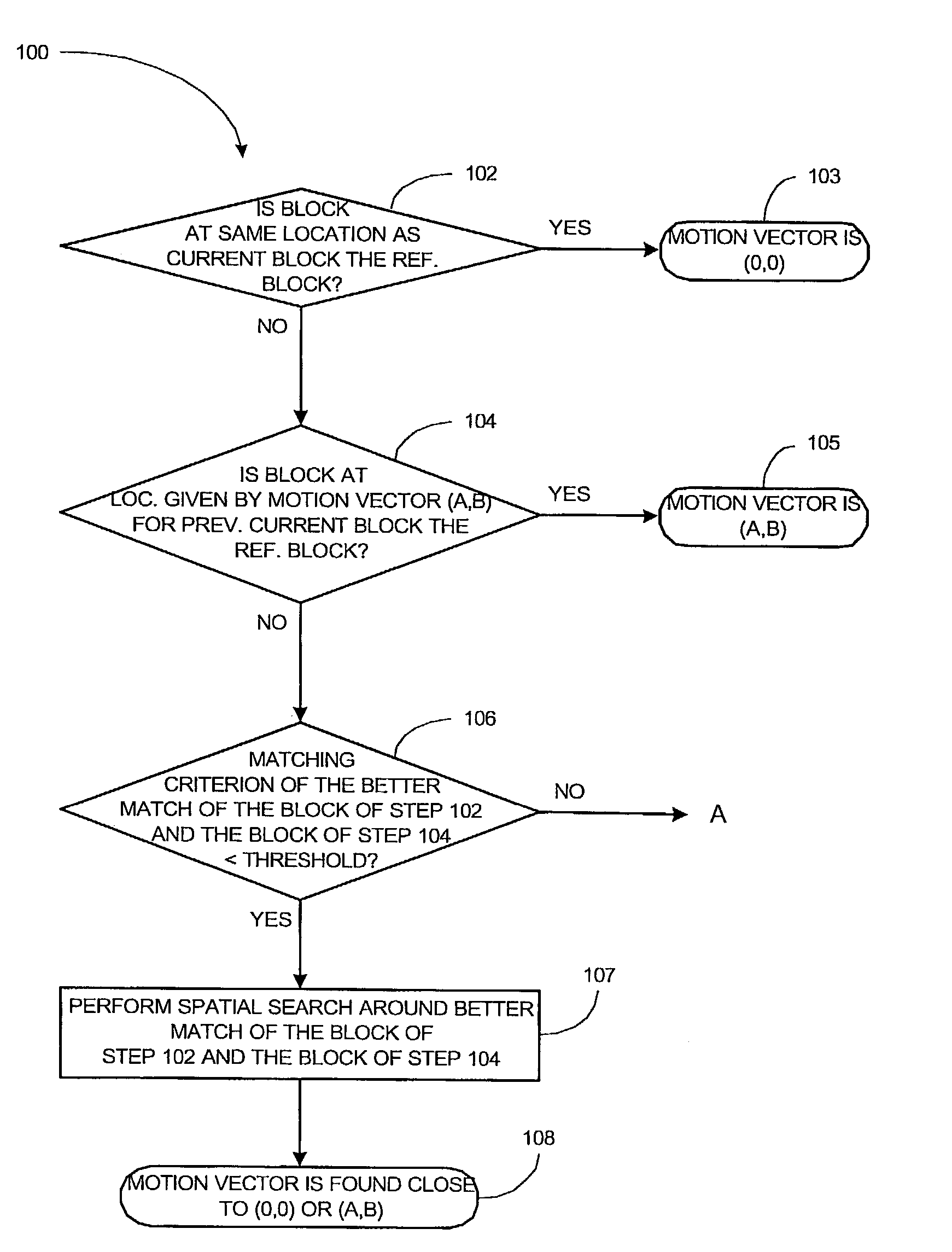
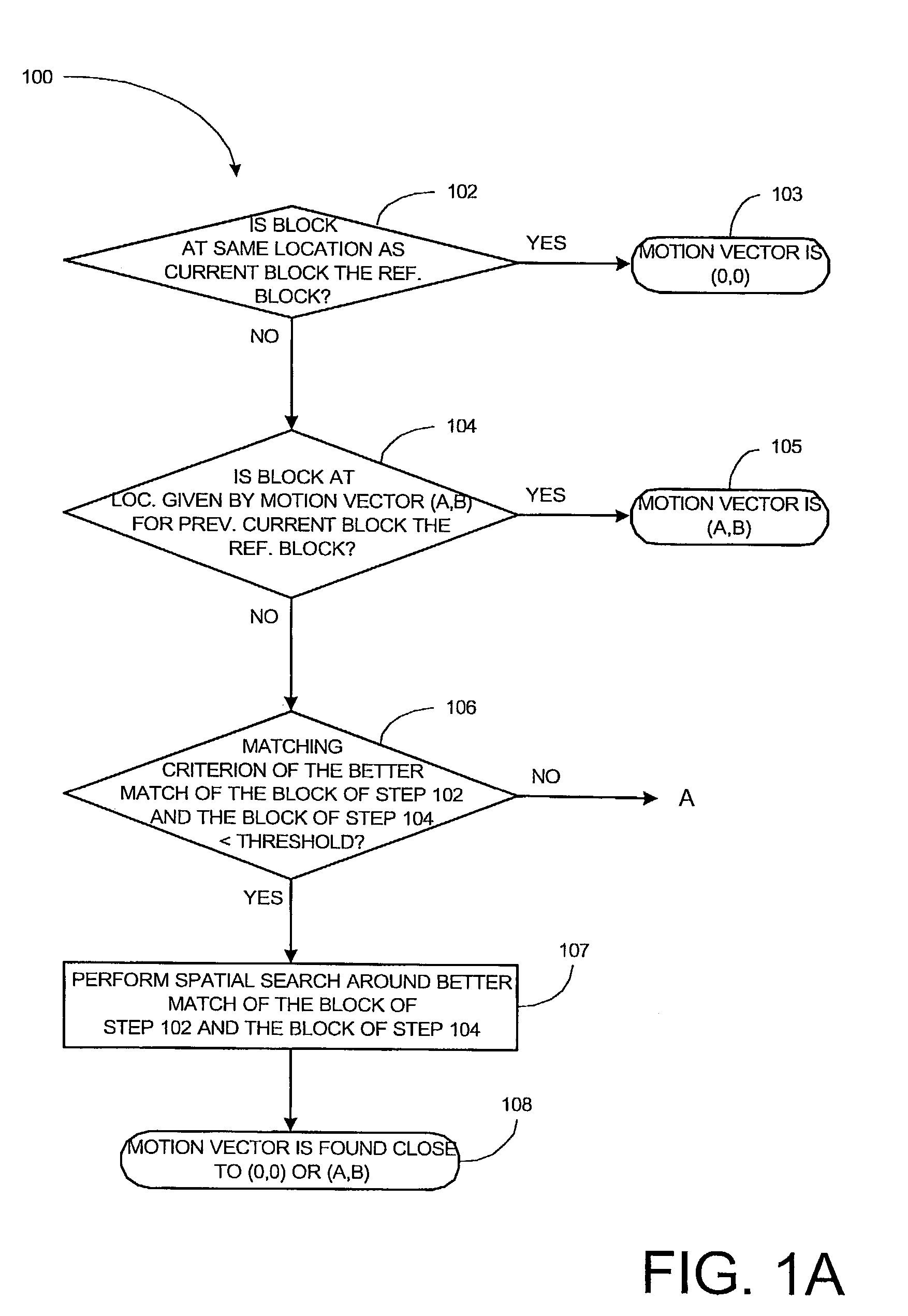
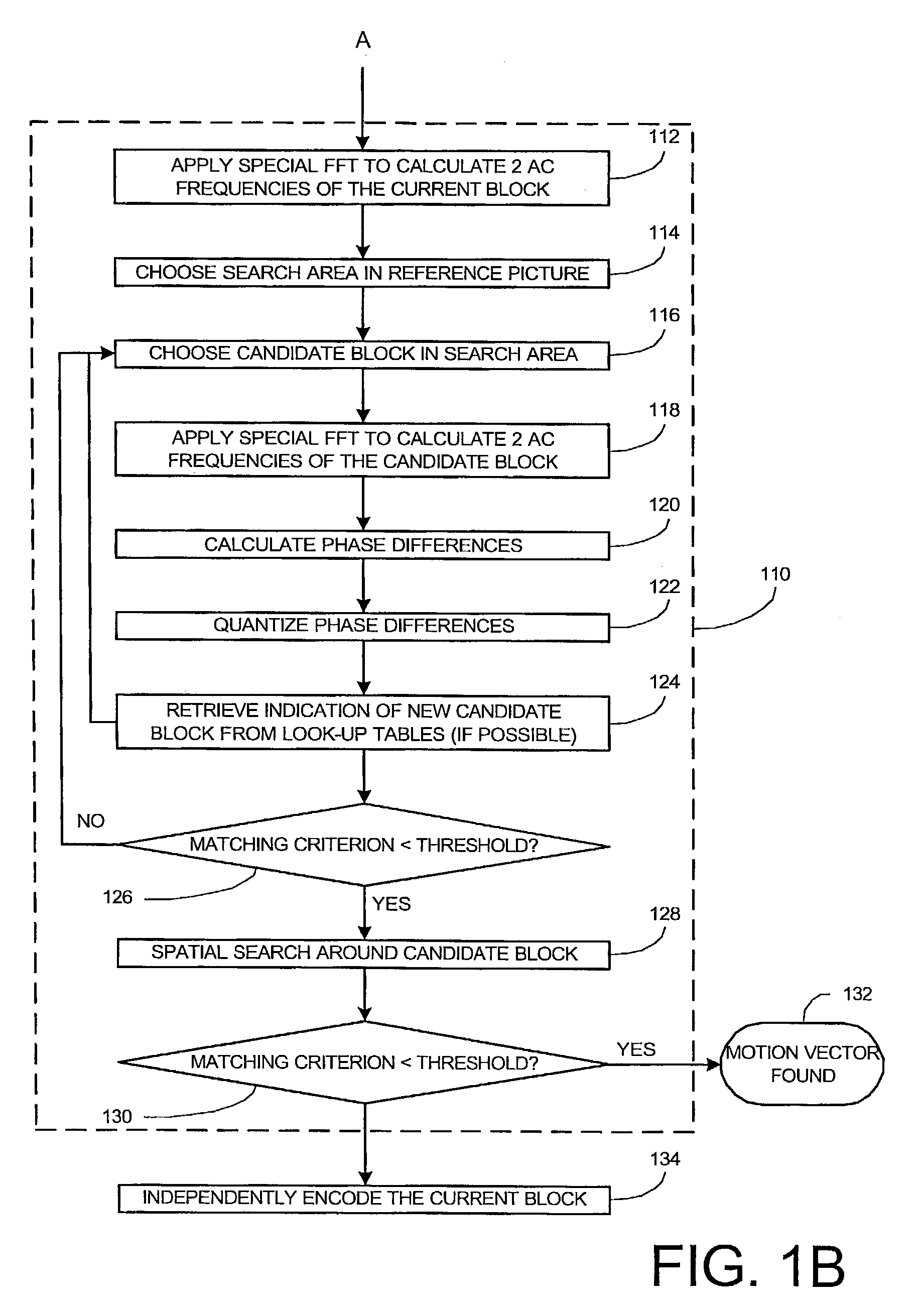
Method and apparatus for motion estimation in a sequence of digital images
ActiveUS7280594B2Color television with pulse code modulationColor television with bandwidth reductionPhase differenceMotion vector
In some embodiments of the present invention, low frequency components for a current block in a current picture and a candidate block in a reference picture are obtained. For each of the low frequency components, quantized phase differences between the current block and the candidate block are calculated. Using the quantized phase differences as an index, coordinates of a global maximum of a correlation function corresponding to the quantized phase differences are retrieved from the table. The coordinates represent a motion vector. A refined candidate block is determined from a spatial search in the vicinity of a candidate block in the reference picture from which the motion vector originates when the motion vector terminates at the current block. The refined candidate block is selected as a reference block for the current block if a matching criterion between the refined candidate block and the current block satisfies a predetermined criterion.
Owner:PARTHUSCEVA
Method and system for data processing with spatial search
ActiveUS7668817B2Spatial search processingIncrease speedInstruments for road network navigationDigital data processing detailsData processing systemData mining
A method for processing data performed in a data processing system having a storage unit is provided which includes: creating a spatial index comprising a leaf containing location information and attribute information with respect to a plurality of objects to be searched, and storing the spatial index in the storage unit; and referring to the spatial index stored in the storage unit in response to an input of a search request including location information and attribute information for searching the object to be searched, and extracting the object to be searched that agrees with the search request. With this configuration, a spatial search with subsidiary condition search can be processed at high speed, without merging results of both searches.
Owner:HITACHI SOFTWARE ENG
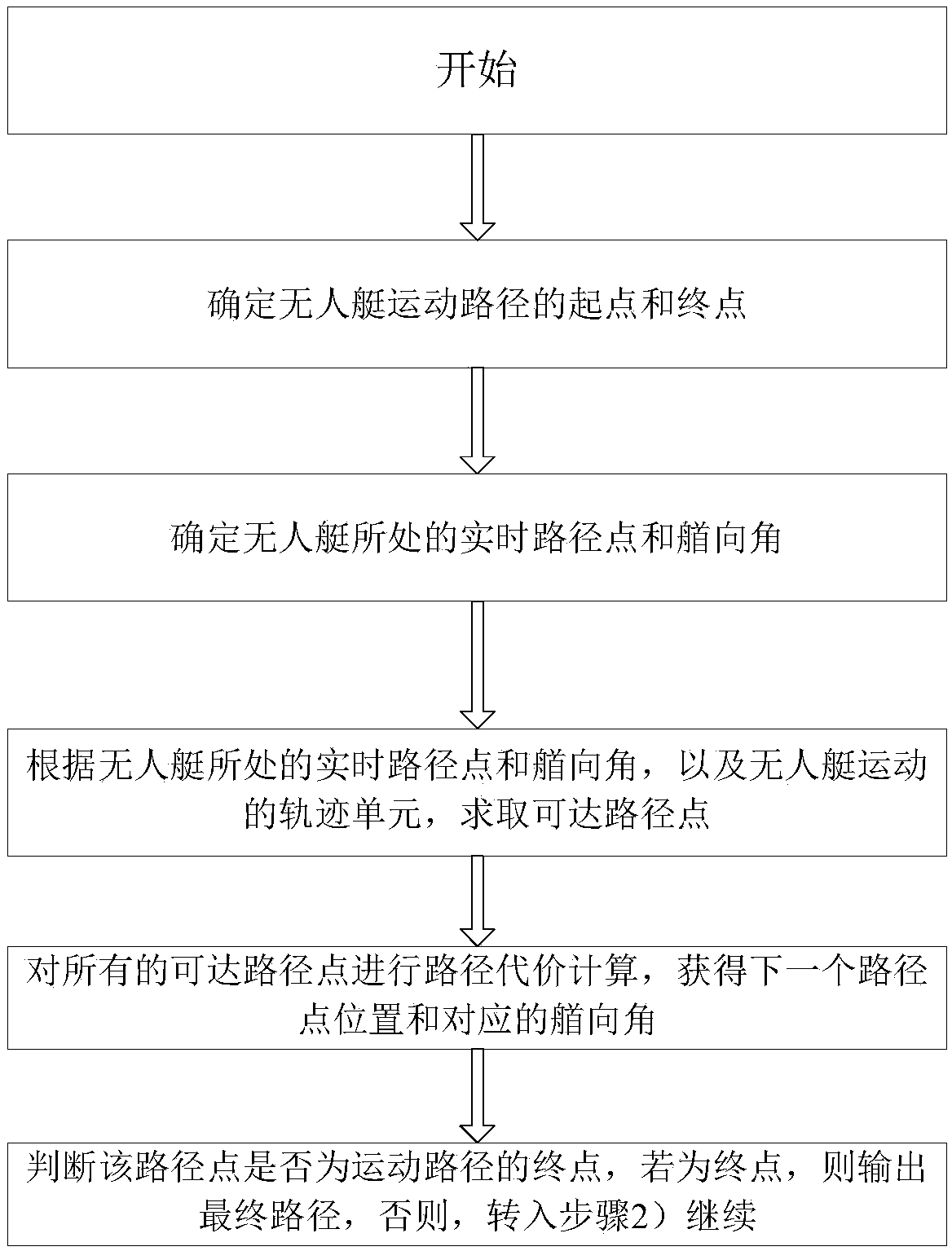
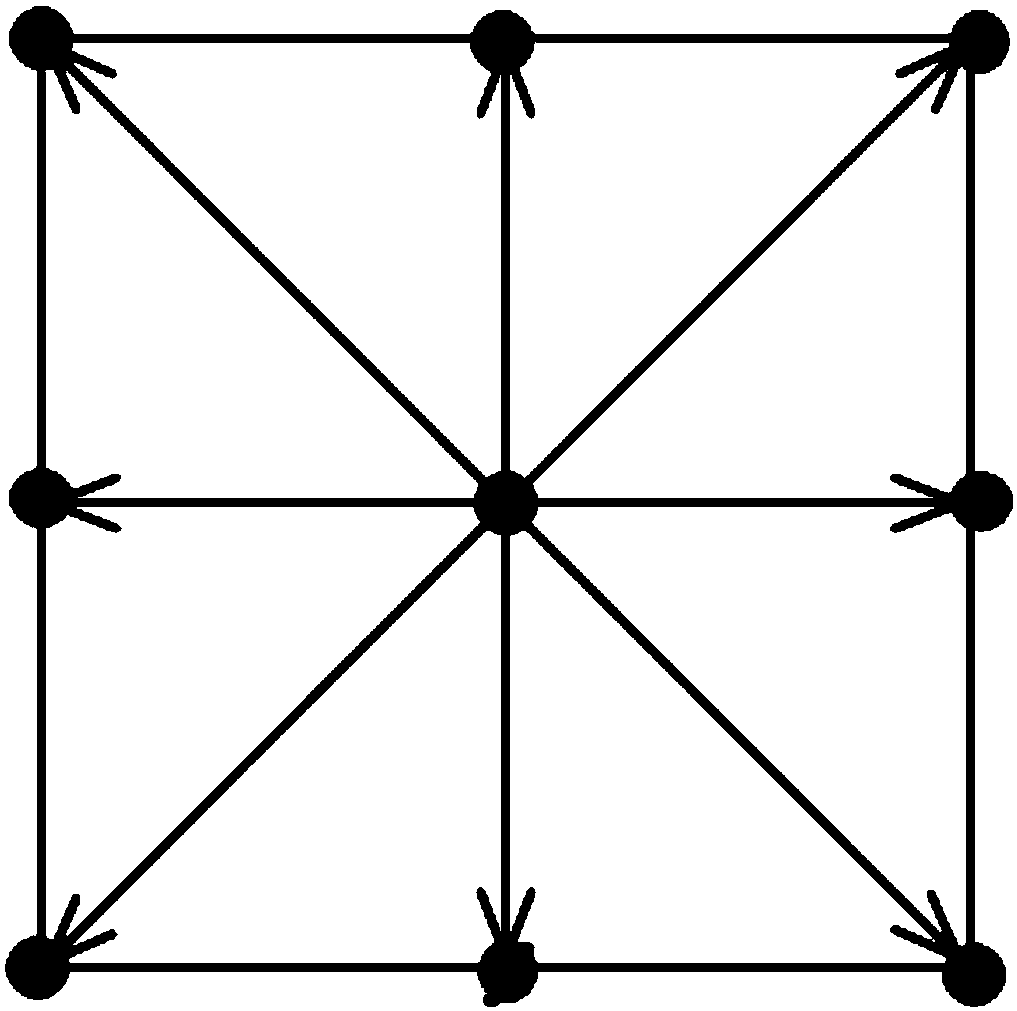
USV obstacle avoidance method based on circular trajectory unit
ActiveCN111123903ASolve discretizationResolve continuityPosition/course control in two dimensionsSimulationObstacle avoidance
The invention discloses an obstacle avoidance method based on a circular trajectory unit. According to the method, dynamic characteristics of an unmanned surface vehicle (USV) can be well combined with an obstacle avoidance process. Firstly, an arc grid allows candidate heading and path points of the USV to change continuously on an arc, and the candidate heading and path points have good correlation with a continuous motion curve; and secondly, the problem of complete space coverage is solved by utilizing a circular grid tree search structure. The trajectory unit adds trajectory constraints into a spatial search process, and the combination of a motion curve and obstacle avoidance is realized. And finally, the problems of discretization of the trajectory and maintenance of the continuityof a final path through a standardization rule are solved. According to the method, a new motion planning obstacle avoidance method is provided, static and dynamic obstacles can be well avoided by applying the method, the avoidance path not only conforms to the dynamic characteristics of the unmanned surface vehicle, but also provides a steering instruction for the operation of the unmanned surface vehicle, so that precise motion planning of the unmanned surface vehicle is realized in an assisted manner.
Owner:WUHAN UNIV OF TECH
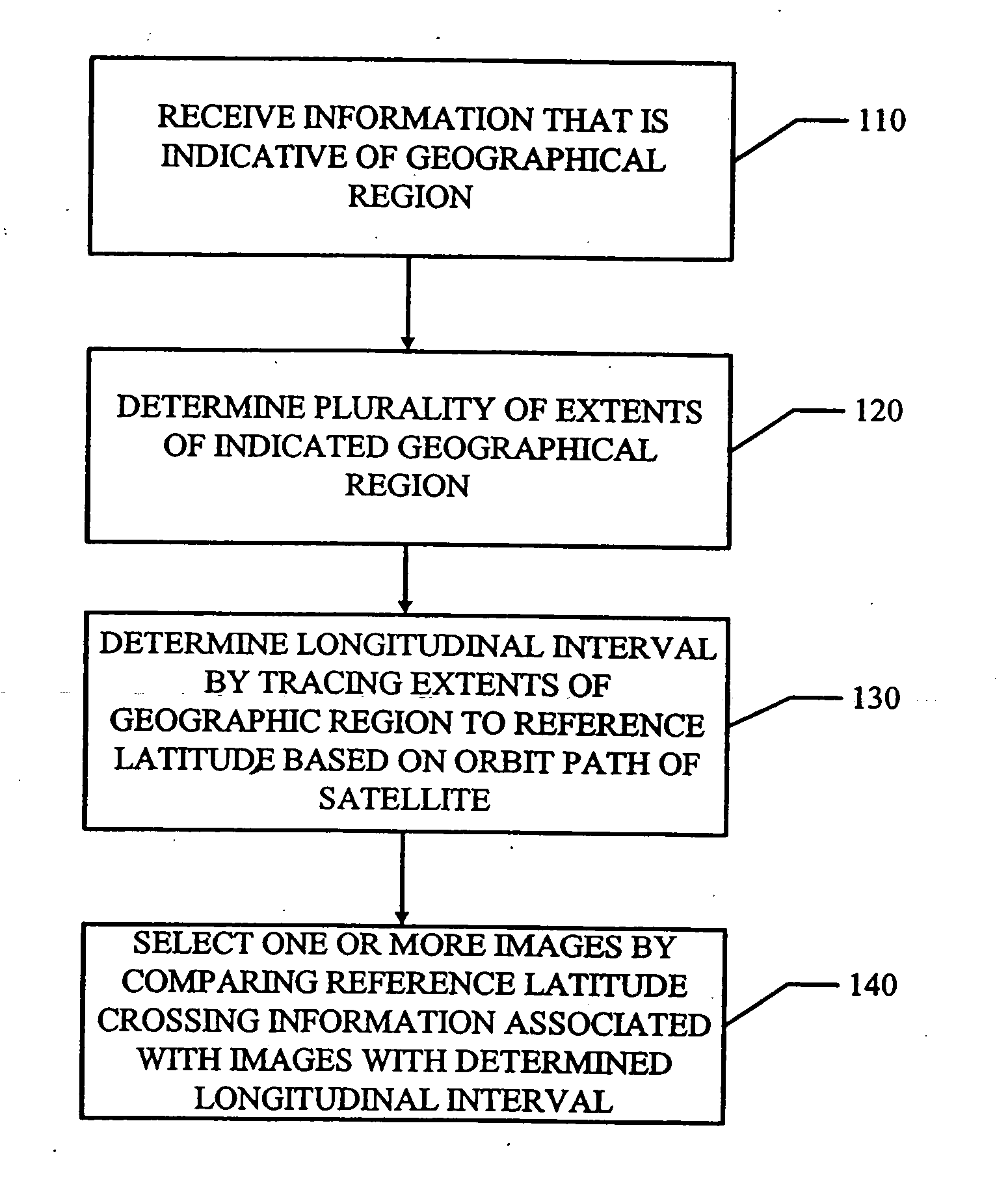
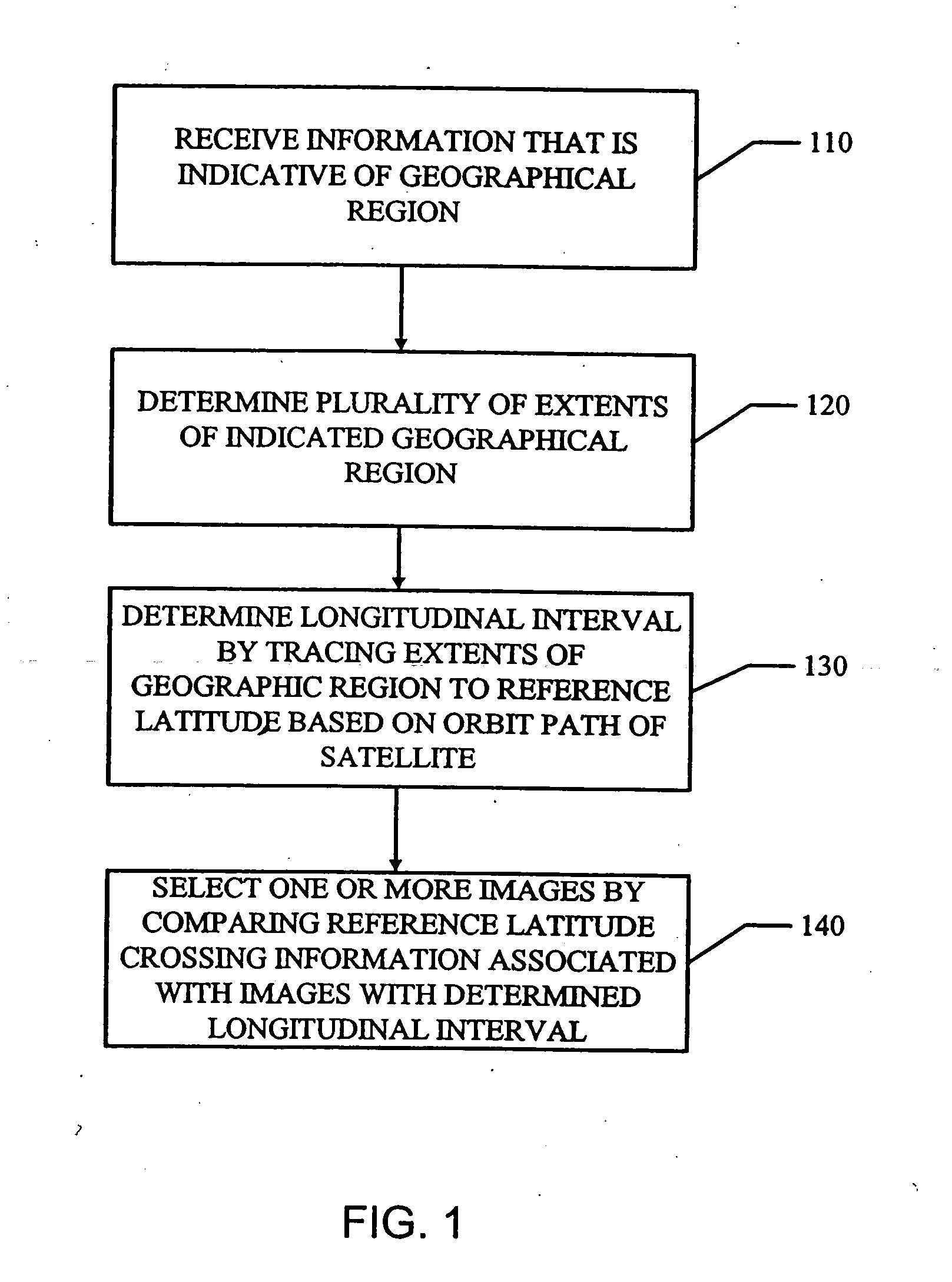
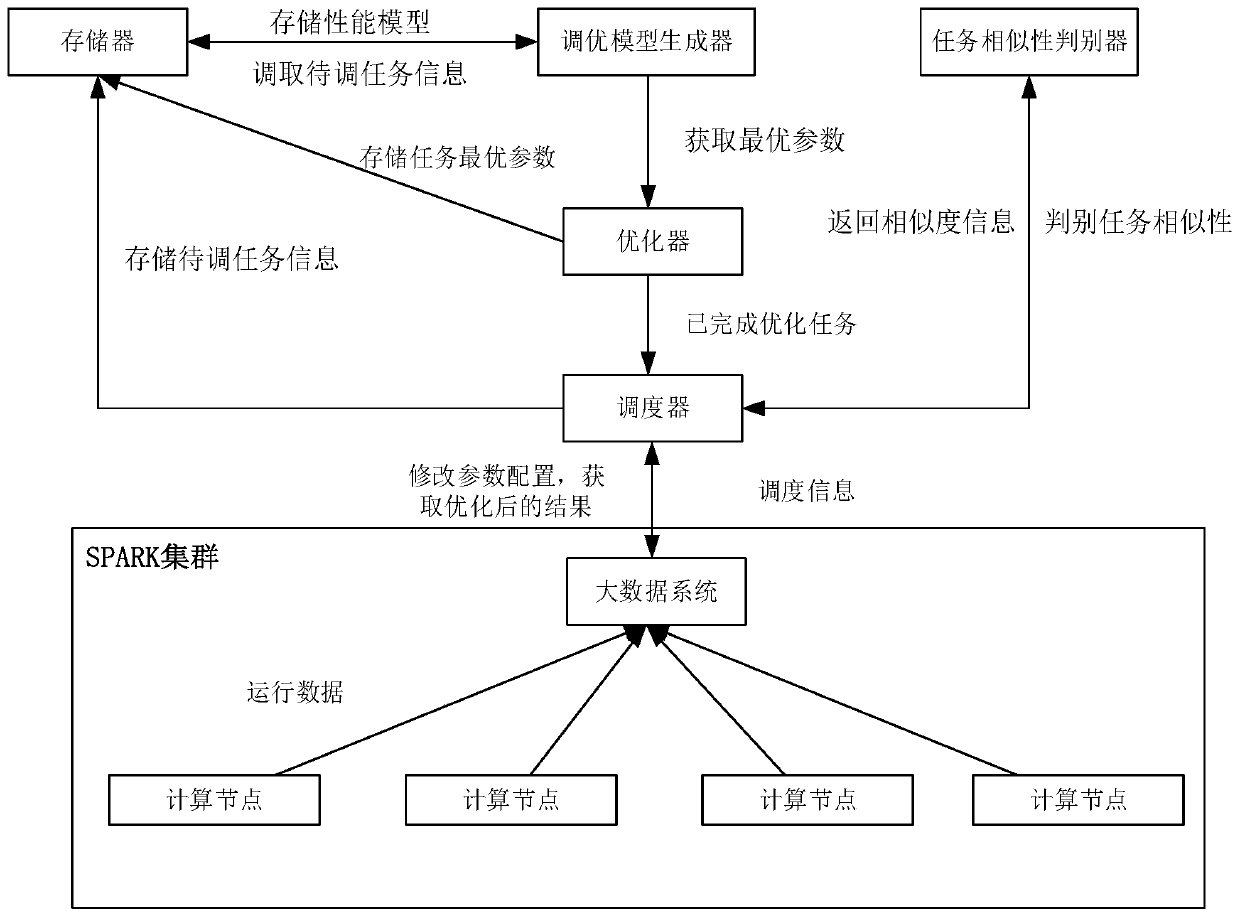
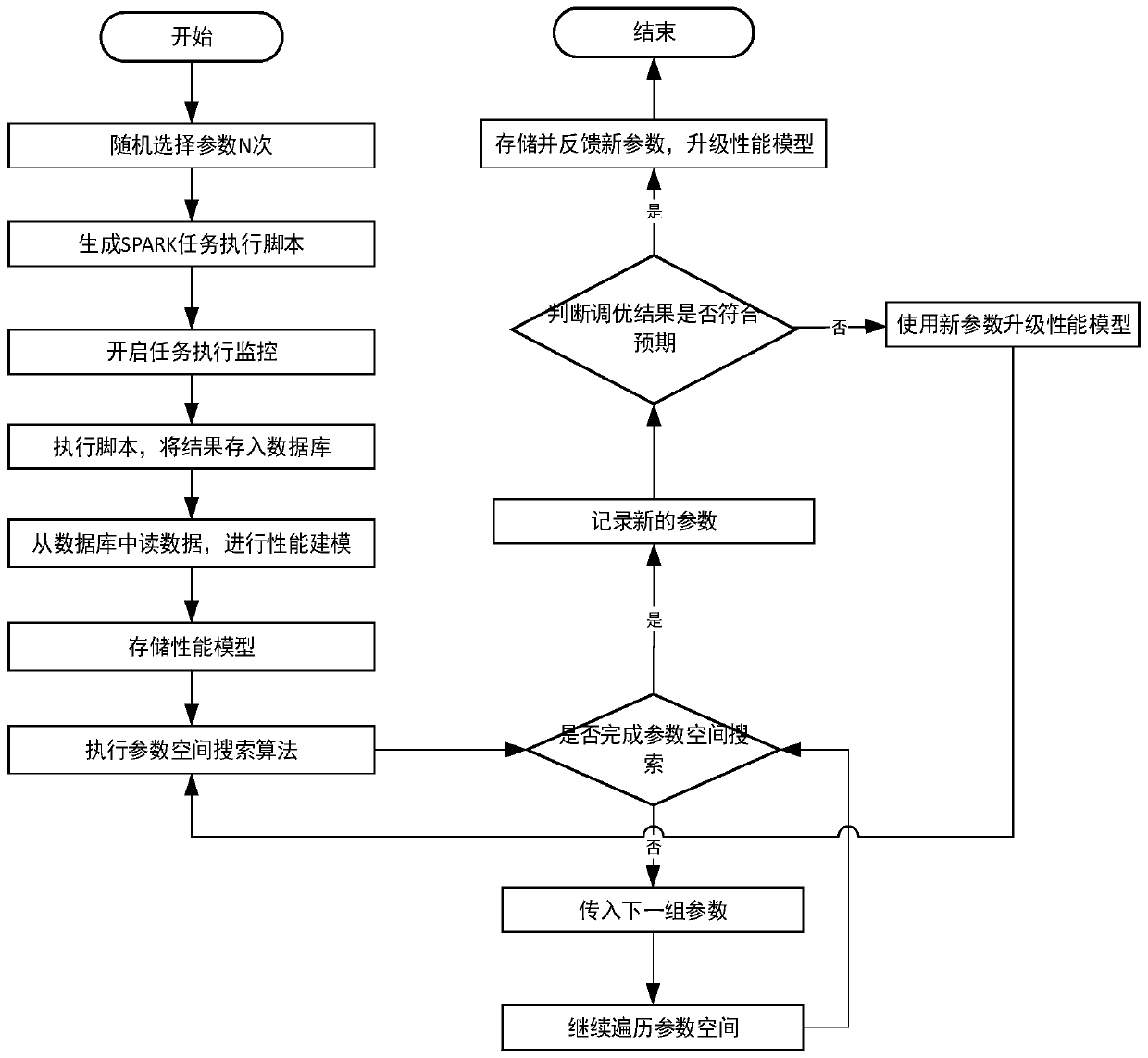
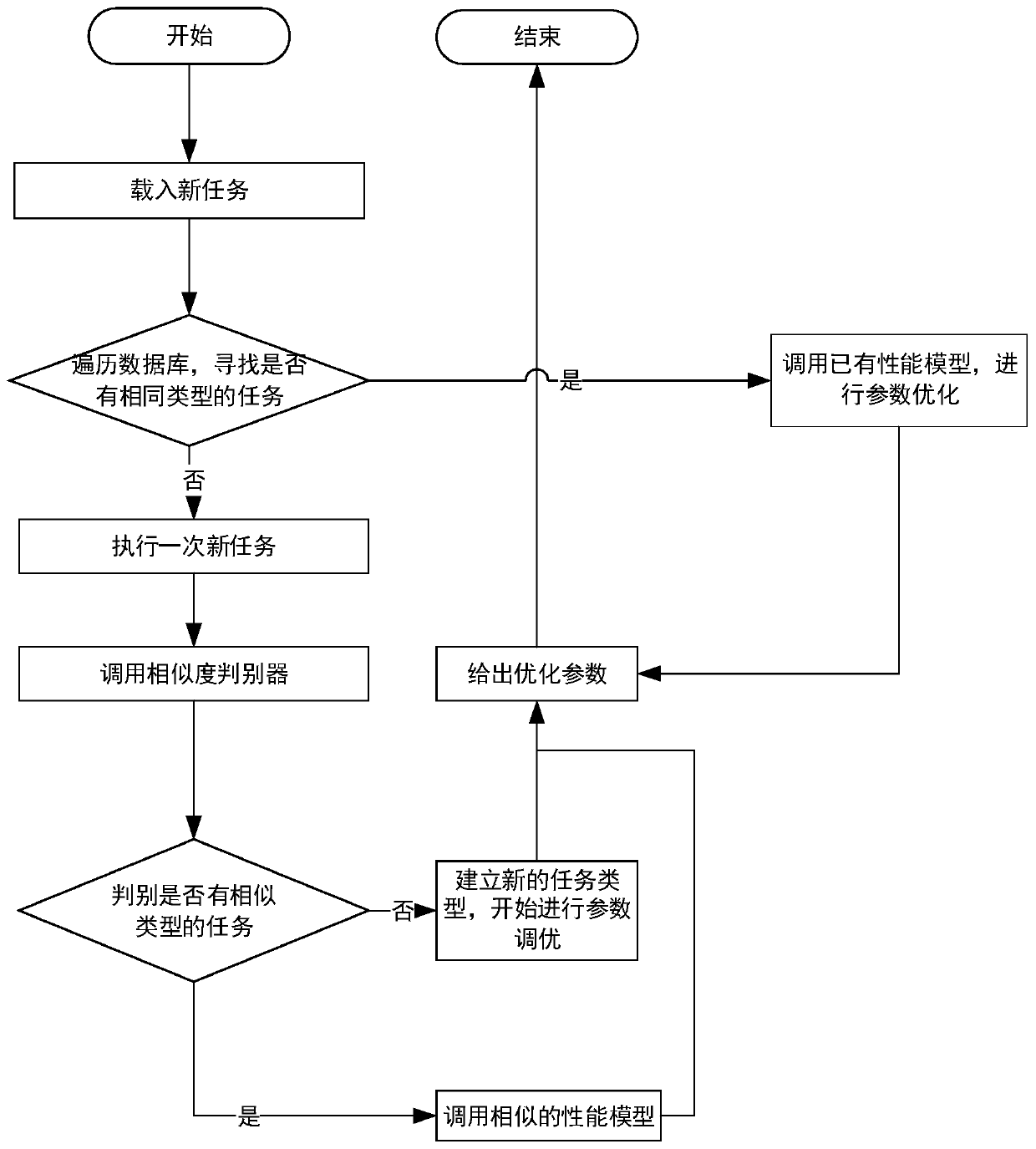
SPARK parameter automatic adjustment and optimization method based on cost model
ActiveCN110727506AReduce workloadReduced performance impactProgram initiation/switchingTask analysisComputer science
The invention provides a Spark parameter automatic adjustment and optimization method based on a cost model. The method comprises the following steps: 1, constructing a cost-based performance model byobtaining configuration of task execution and corresponding cost information, and obtaining optimal configuration in a given parameter space; 2, for tasks of unknown types, using default parameters for one-time operation, and giving a reference value of optimal configuration by judging the similarity of the tasks. According to the invention, a cost-based performance model is provided for solvingproblems possibly existing in current configuration parameter adjustment and optimization. A performance model is generated by analyzing Spark historical tasks, optimization parameters are obtained through a parameter space search algorithm. Meanwhile, the model is continuously upgraded and adjusted along with running of new tasks, the accuracy of the model is improved, and parameter reference values are provided after the model runs once for tasks of unknown types.
Owner:BEIHANG UNIV
Group fraud identification method and device based on spatial search algorithm
ActiveCN111127062AQuick searchEfficient identificationEnergy efficient computingMarketingData miningSearch computing
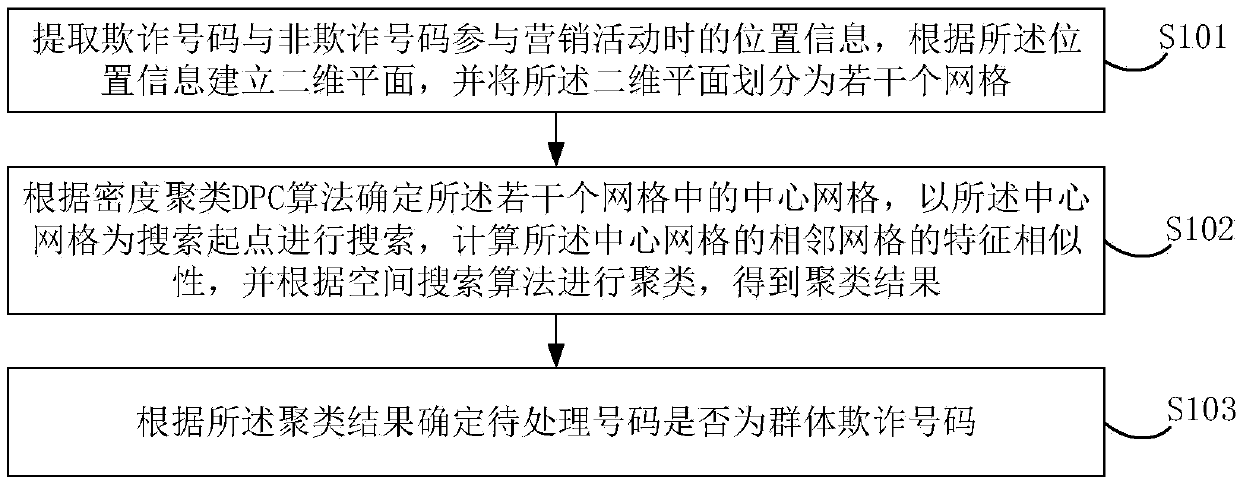
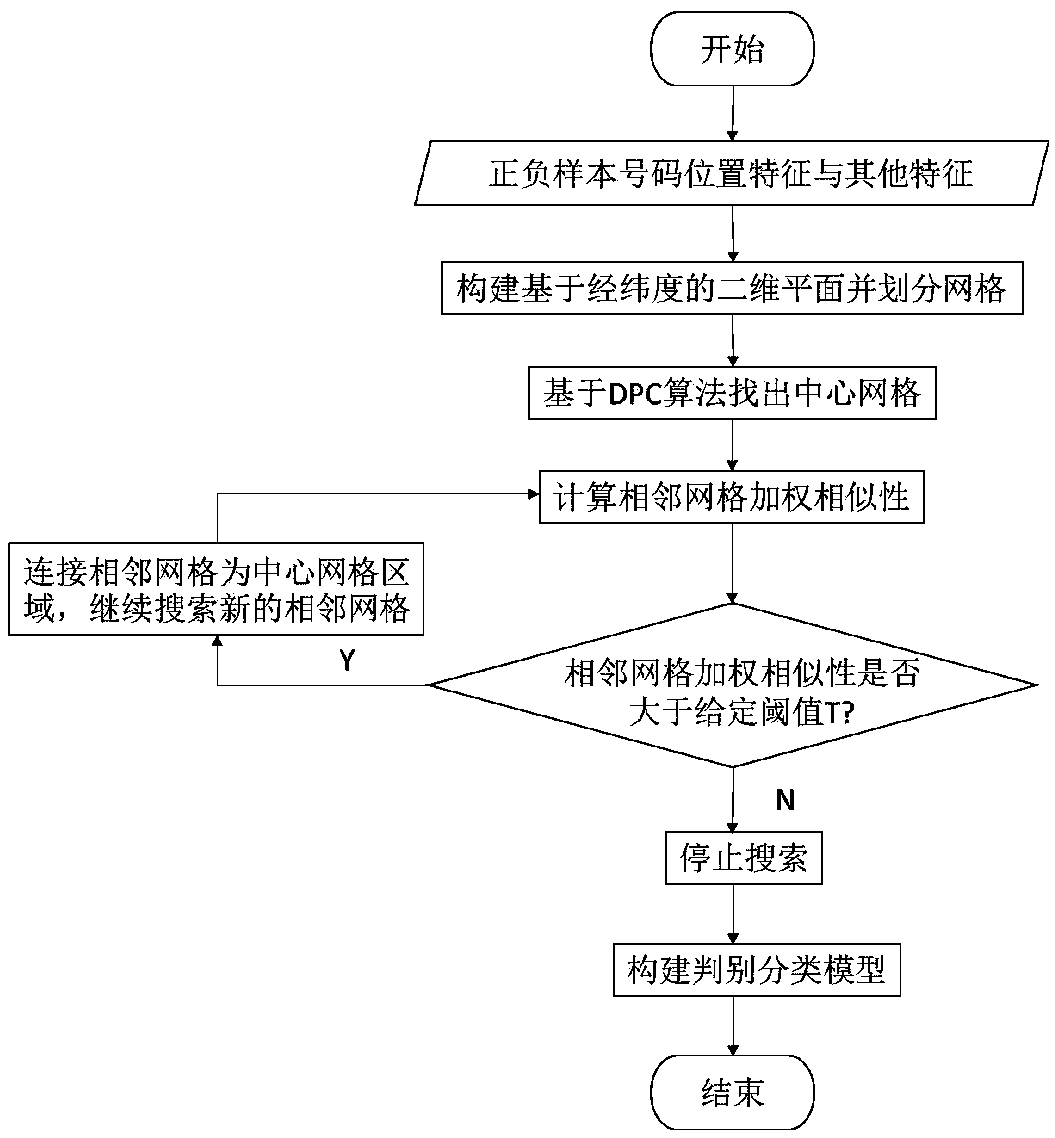
The embodiment of the invention discloses a group fraud identification method and device based on a spatial search algorithm, and the method comprises the steps: extracting the position information ofa fraud number and a non-fraud number during the participation of a marketing activity, building a two-dimensional plane according to the position information, and dividing the two-dimensional planeinto a plurality of grids; determining a central grid in the plurality of grids according to a DPC algorithm, searching by taking the central grid as a search starting point, calculating feature similarity of adjacent grids of the central grid, and clustering according to a spatial search algorithm to obtain a clustering result; and determining whether the to-be-processed number is a group fraudulent number according to the clustering result. The user is projected into a two-dimensional plane through the geographic position; grids are divided, the grids with high similarity with the grids withthe high fraud probability are quickly searched, group fraud numbers are found, fraud risks are directly calculated from group behavior characteristics, fraud user groups can be efficiently and accurately recognized, major fraud group risks are found, and enterprises are helped to effectively recognize fraud behaviors.
Owner:CHINA MOBILE GRP GUANGDONG CO LTD +1
VLSI (Very Large Scale Integration) layout design method for solving given border constraint
InactiveCN106777849ARestricted movementReduce the temperatureCAD circuit designSpecial data processing applicationsRandomizationVery-large-scale integration
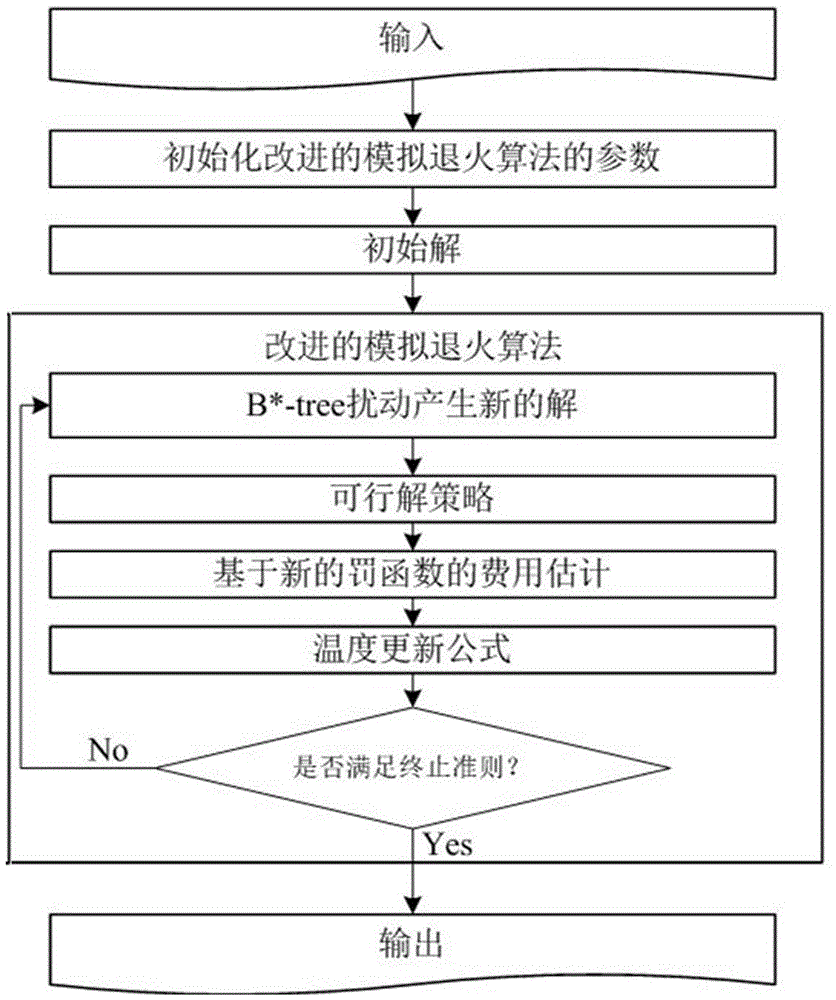
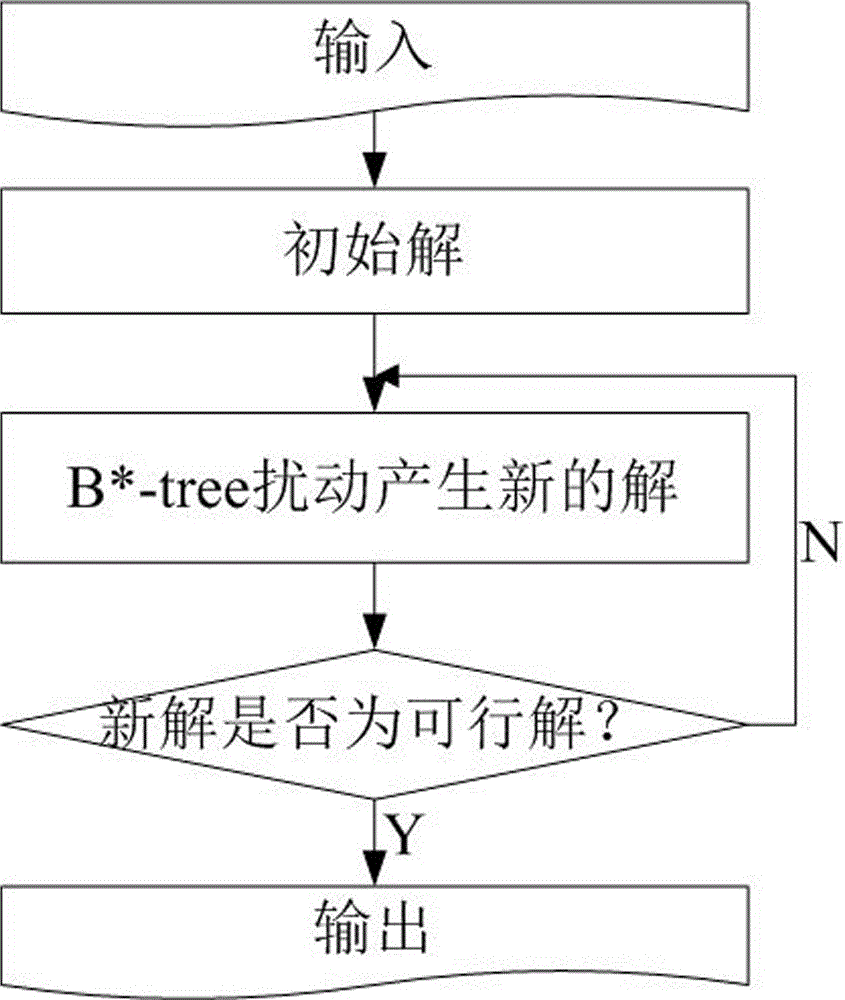
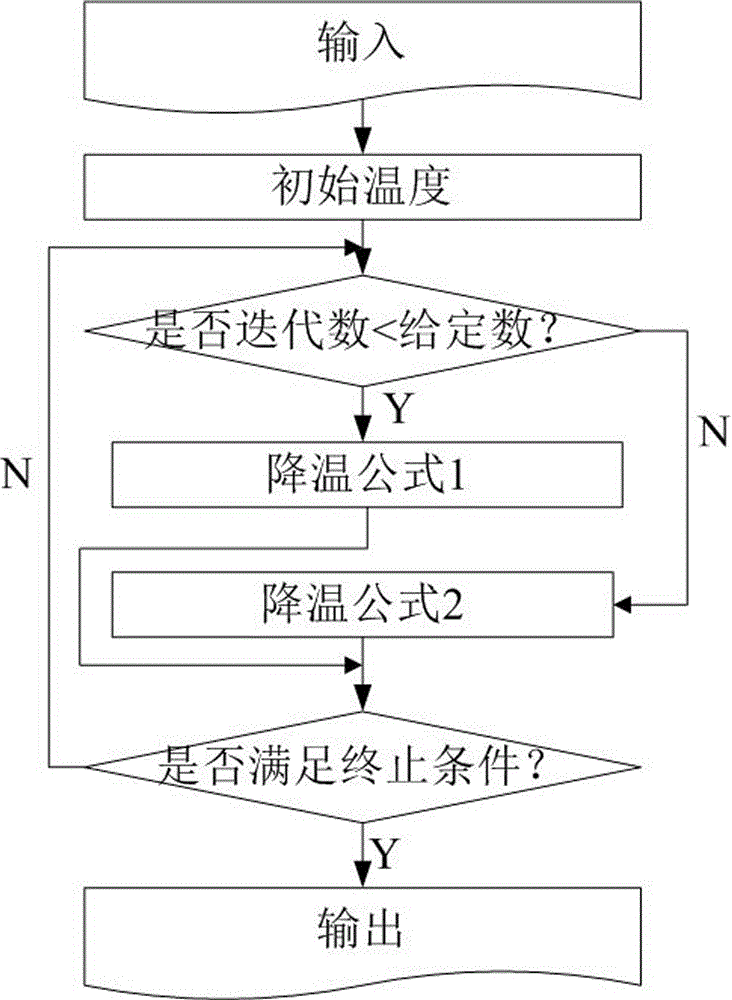
The invention relates to a VLSI (Very Large Scale Integration) layout design method for solving a given border constraint. By adopting a strategy of randomization, a random initial layout is used as an initial solution of a hybrid simulated annealing algorithm. The border constraint is proposed as a violating penalty function of a target function. The violating penalty function introduces a length function considering each module over a given border based on an area function considering a minimum rectangle enclosing candidate layout results over the given border. Based on the hybrid simulated annealing algorithm, a feasible solution strategy is introduced. Solutions produced via a series B*-tree disturbance are feasible solutions, so that better solutions can be obtained from the algorithm. The optimal solution is searched by introducing the hybrid simulated annealing algorithm, and a new temperature update formula is adopted in the algorithm. The new temperature update formula shortens the time for solving the spatial search phase, and more time is used for searching the better solutions in the temperature rise phase, so that the probability of finding the optimal solution is improved.
Owner:FUZHOU UNIV
Visual question and answer method based on network structure search
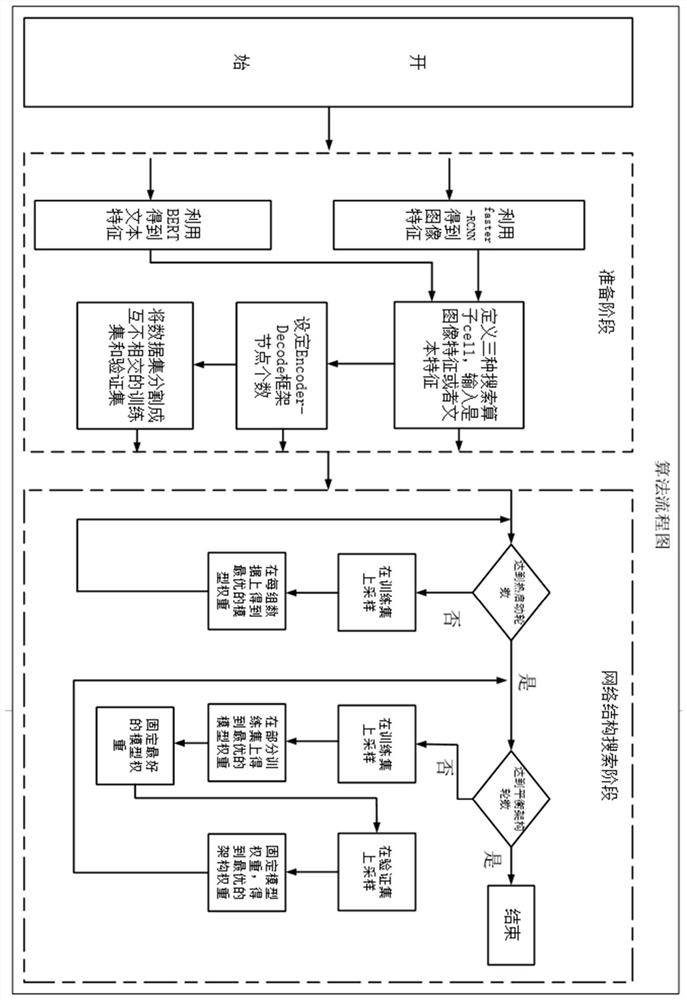
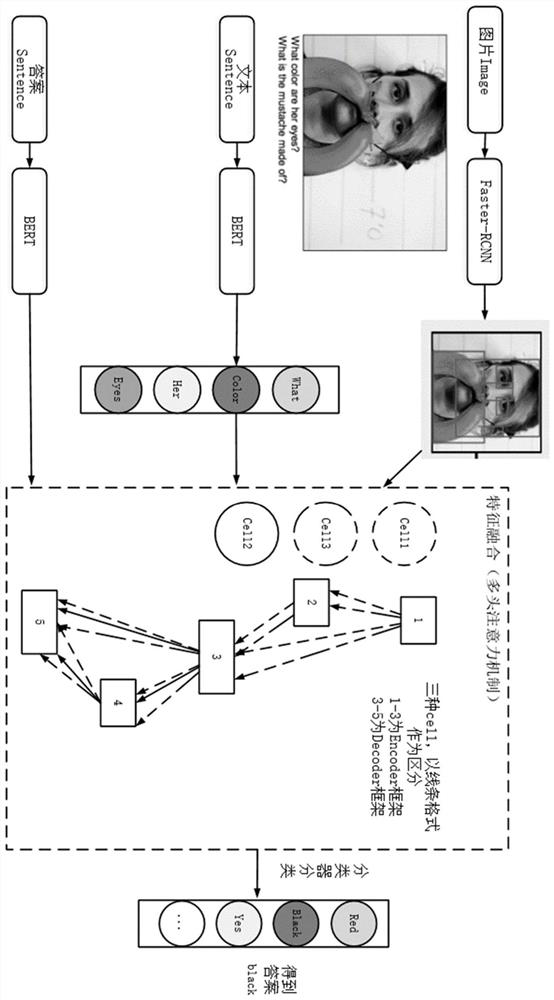
ActiveCN113282721ANeural architecturesText database queryingFeature extractionClassification methods
The invention provides a visual question and answer method based on network structure search. The method comprises the following steps: performing feature extraction on an original picture by adopting a first artificial neural network model; performing feature extraction on the text information by adopting a second artificial neural network model; a to-be-searched network structure framework being a coding-decoder framework, defining three search operators for framework network search, and the input of the search operators being image features or text features extracted based on the original picture or / and the text information; searching the architecture weight of the network structure and the operation weight of an operator by using a gradient-based alternative optimization strategy; and enabling the search network to output candidate word vectors according to a multi-classification method, and selecting the word vector with the maximum probability as an answer to be output. The visual question and answer method based on network structure search has the beneficial effect that a better effect can be searched in a larger space.
Owner:NANJING UNIV
Deep learning operator automatic optimization system and method based on Shenwei processor
PendingCN110929850AEfficient manual optimization technologyEfficient automatic optimization technologyNeural architecturesNeural learning methodsCode generationAlgorithm
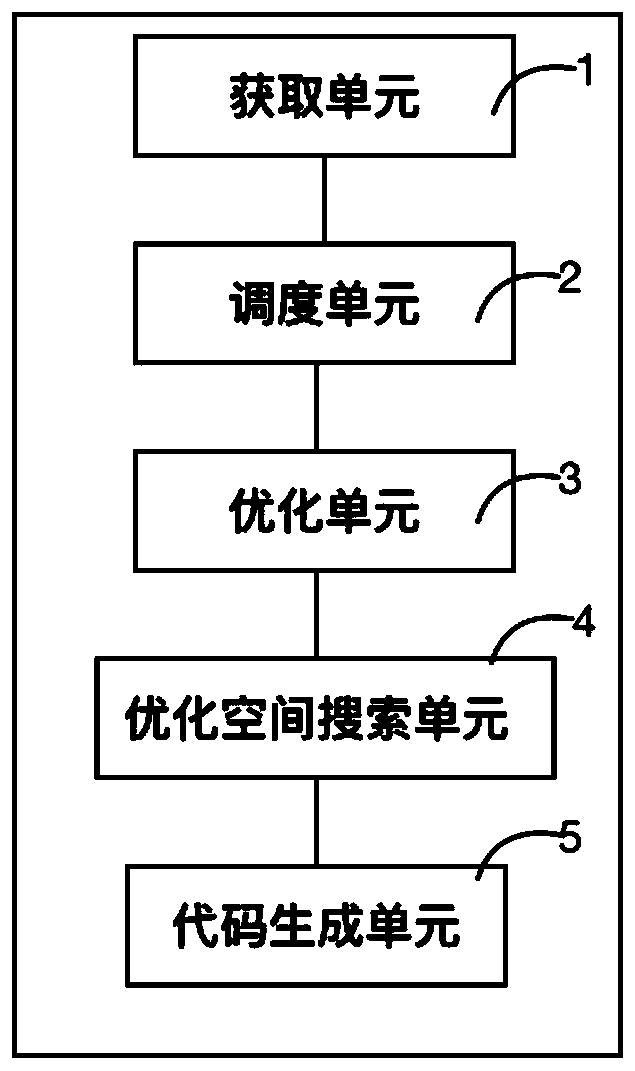
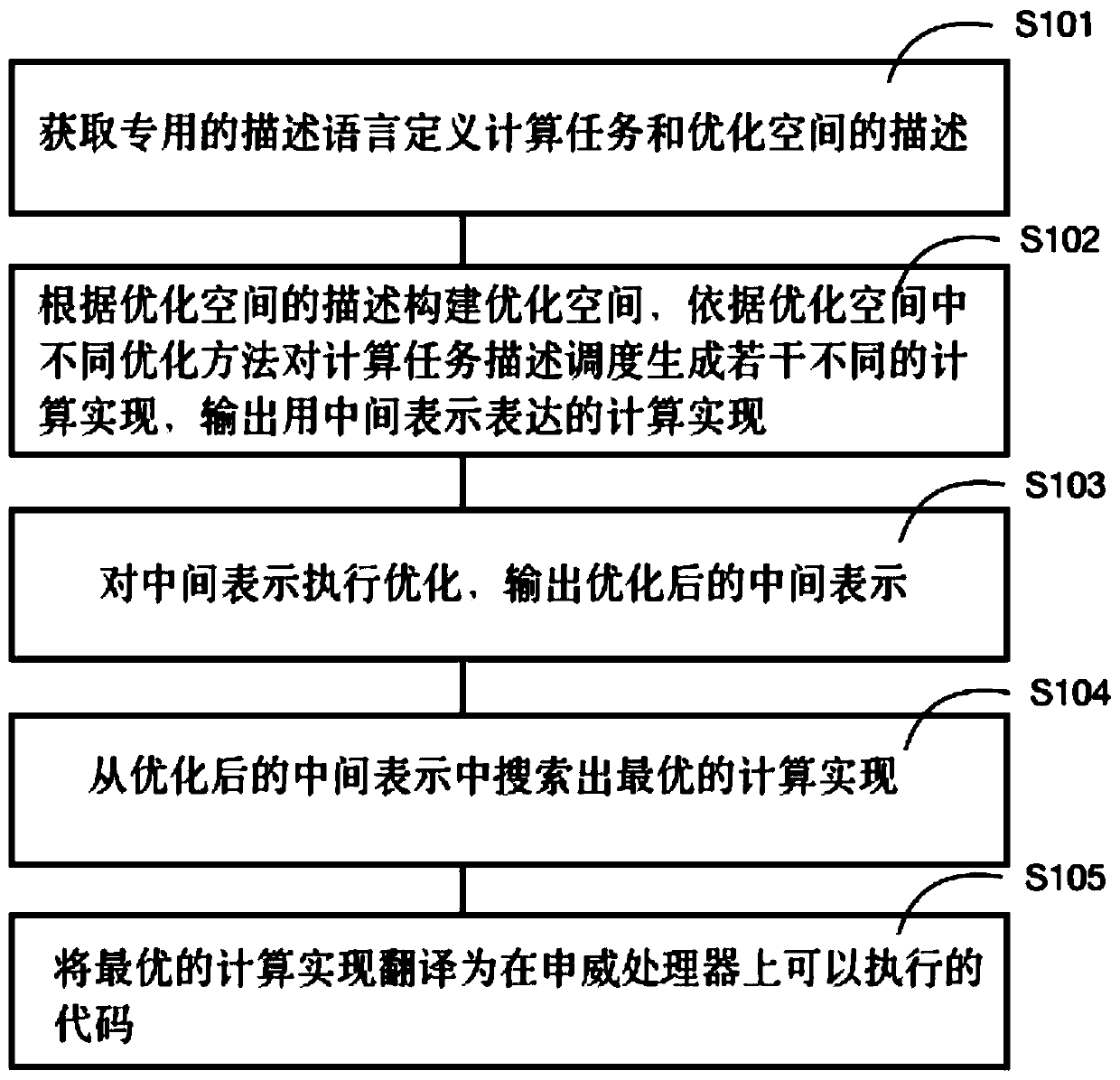
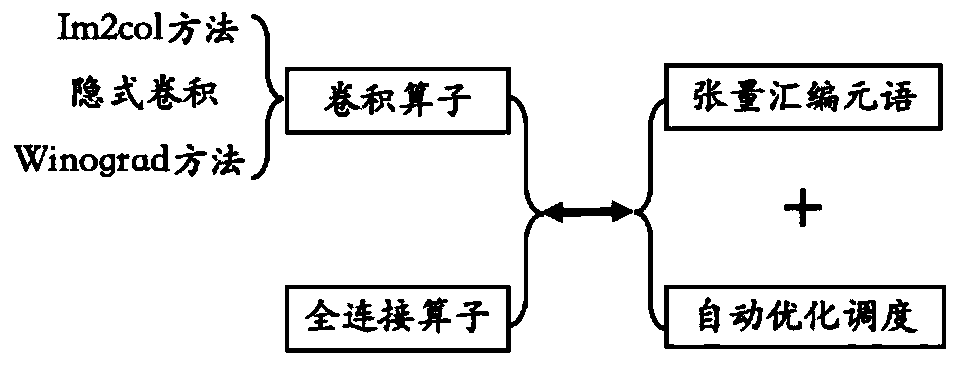
The invention provides a deep learning operator automatic optimization system based on an Shenwei processor, and the system comprises an obtaining unit which obtains the description of a special description language definition calculation task and an optimization space; a scheduling unit which constructs an optimization space according to the description of the optimization space, schedules the calculation task description according to different optimization methods in the optimization space to generate a plurality of different calculation implementations, and outputs the calculation implementations expressed by intermediate representation; an optimization unit which is used for receiving the intermediate representation, optimizing the intermediate representation and outputting the optimized intermediate representation; an optimized space searching unit which is used for searching the optimal calculation implementation from the optimized intermediate representation; and a code generation unit which is used for translating the optimal calculation implementation into codes which can be executed on the Shenwei processor. The method can solve the problems of low optimization performance, difficult transplantation and large optimization time overhead in the prior art, is more efficient than a manual optimization technology and an automatic optimization technology, and can be conveniently transplanted to other mechanisms for use.
Owner:NAT SUPERCOMPUTING WUXI CENT
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com