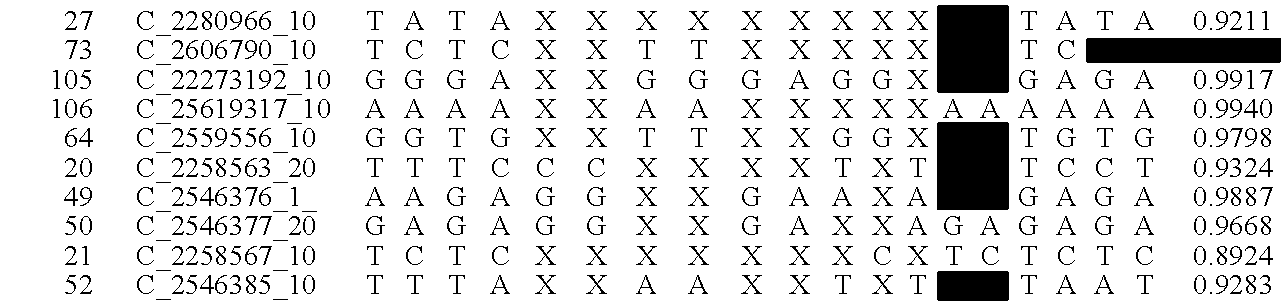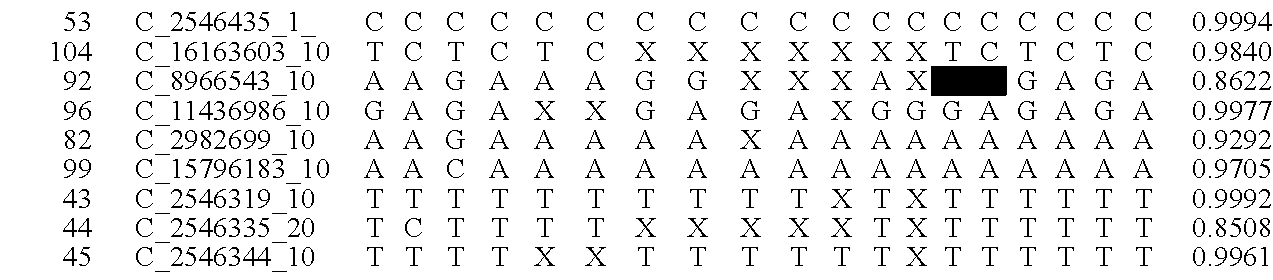Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
2002results about "Molecular structures" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
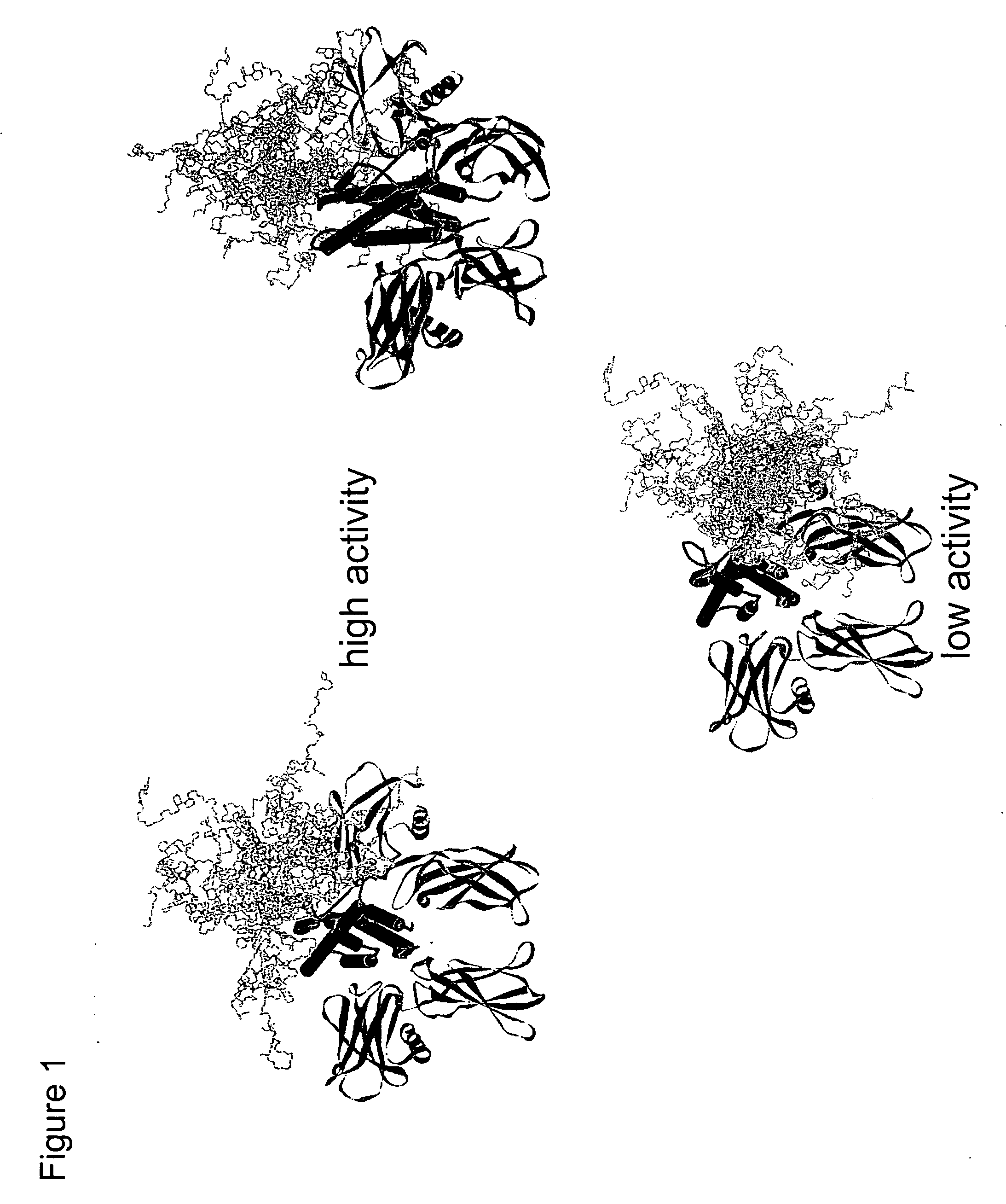
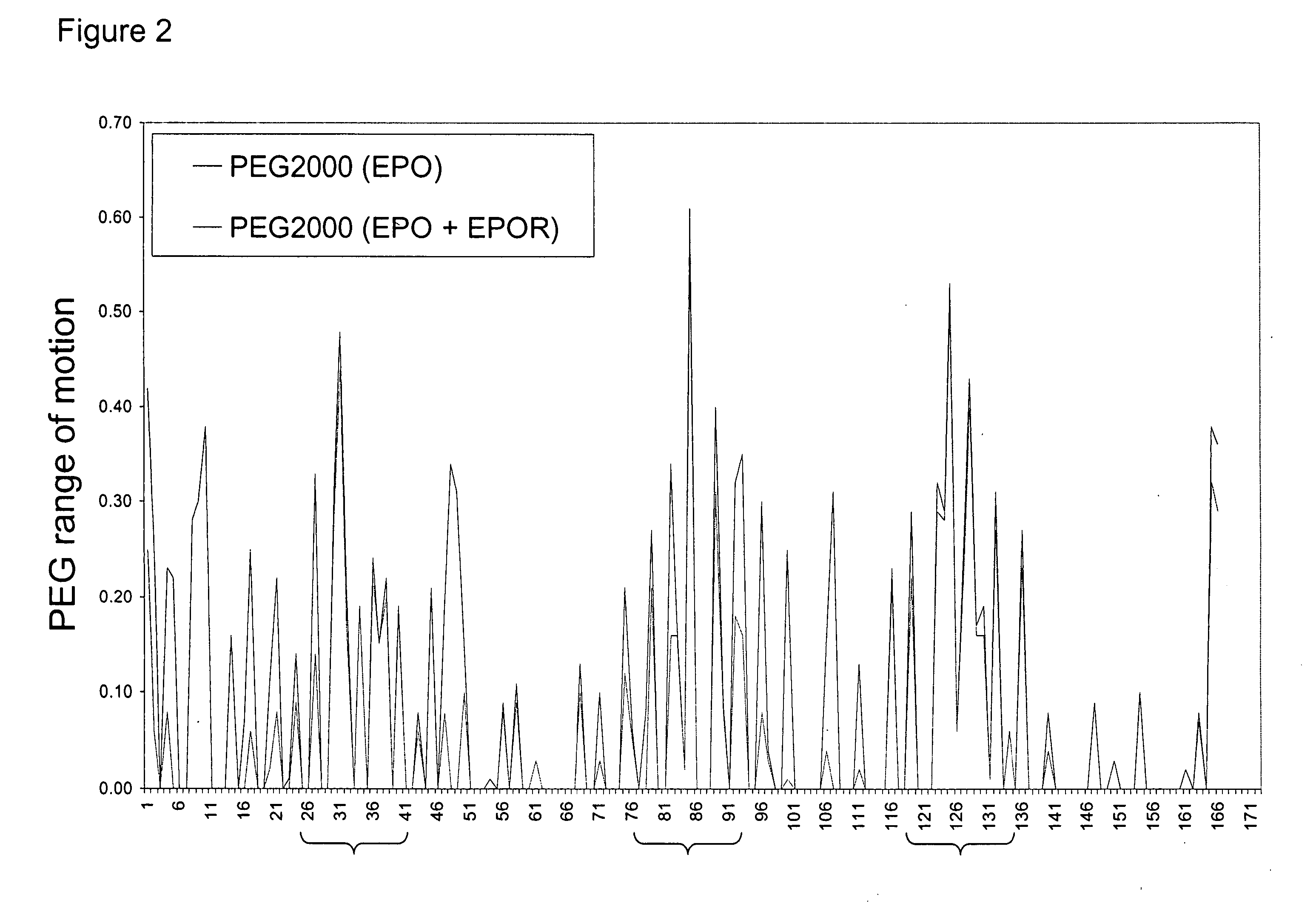
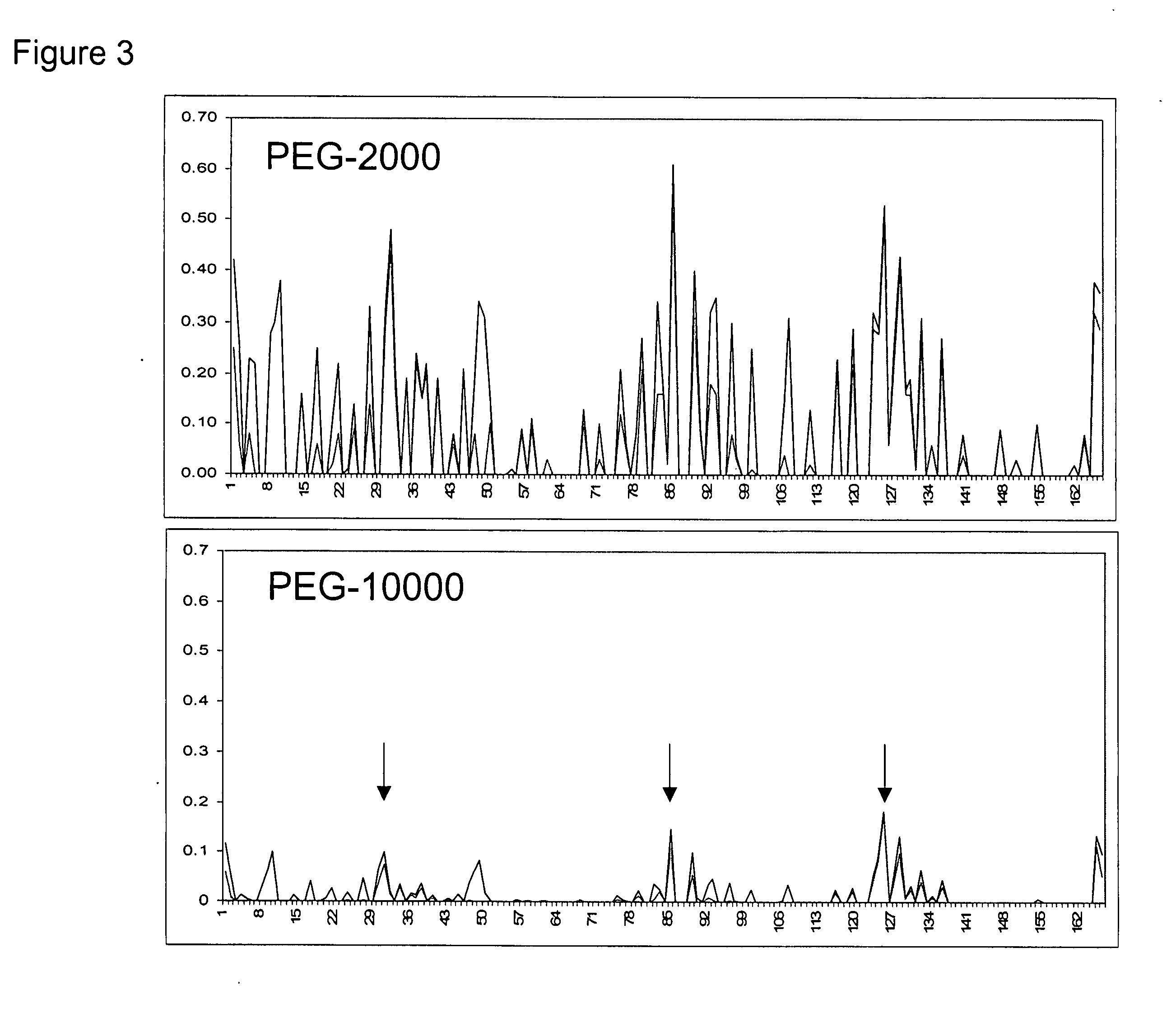
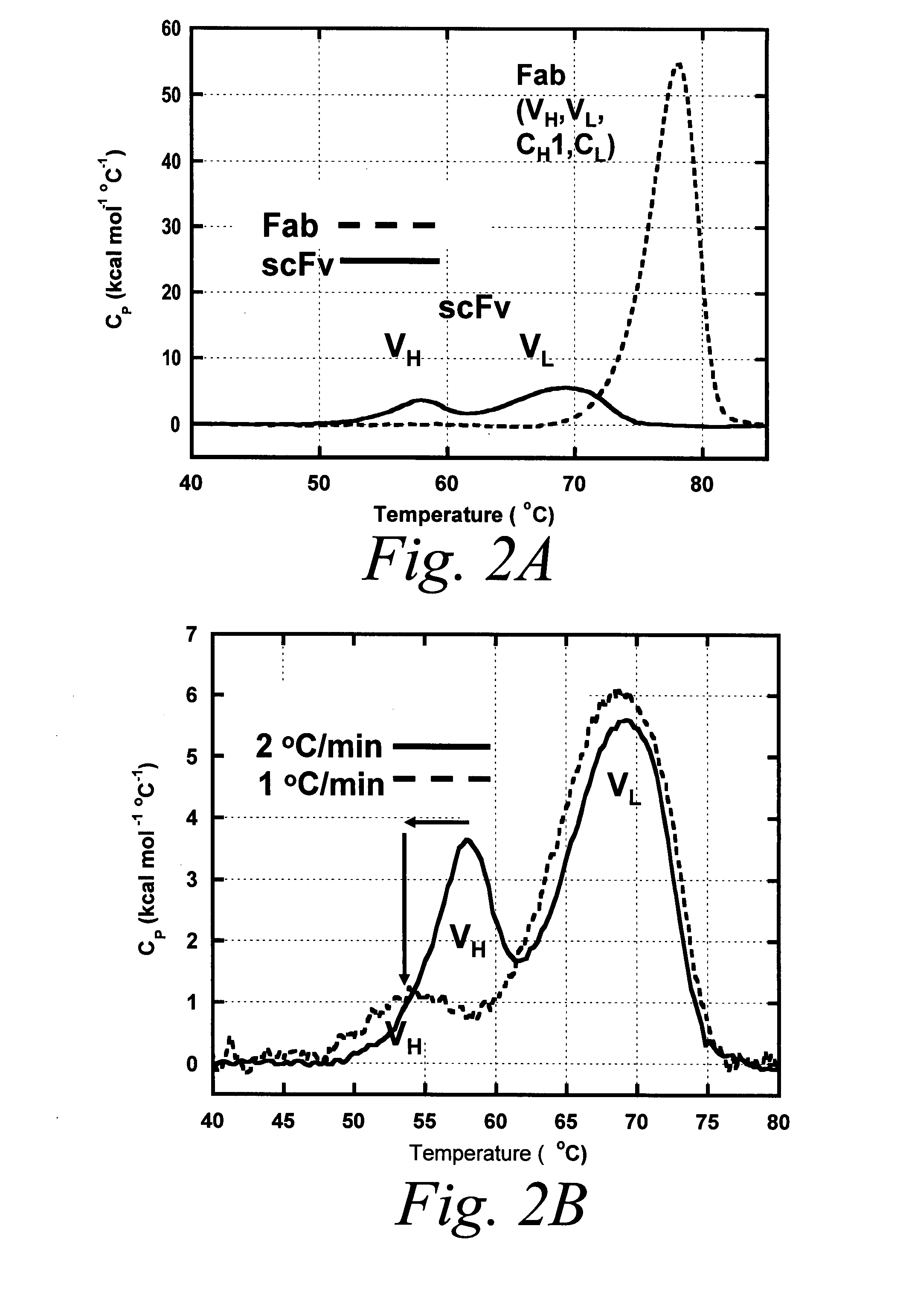
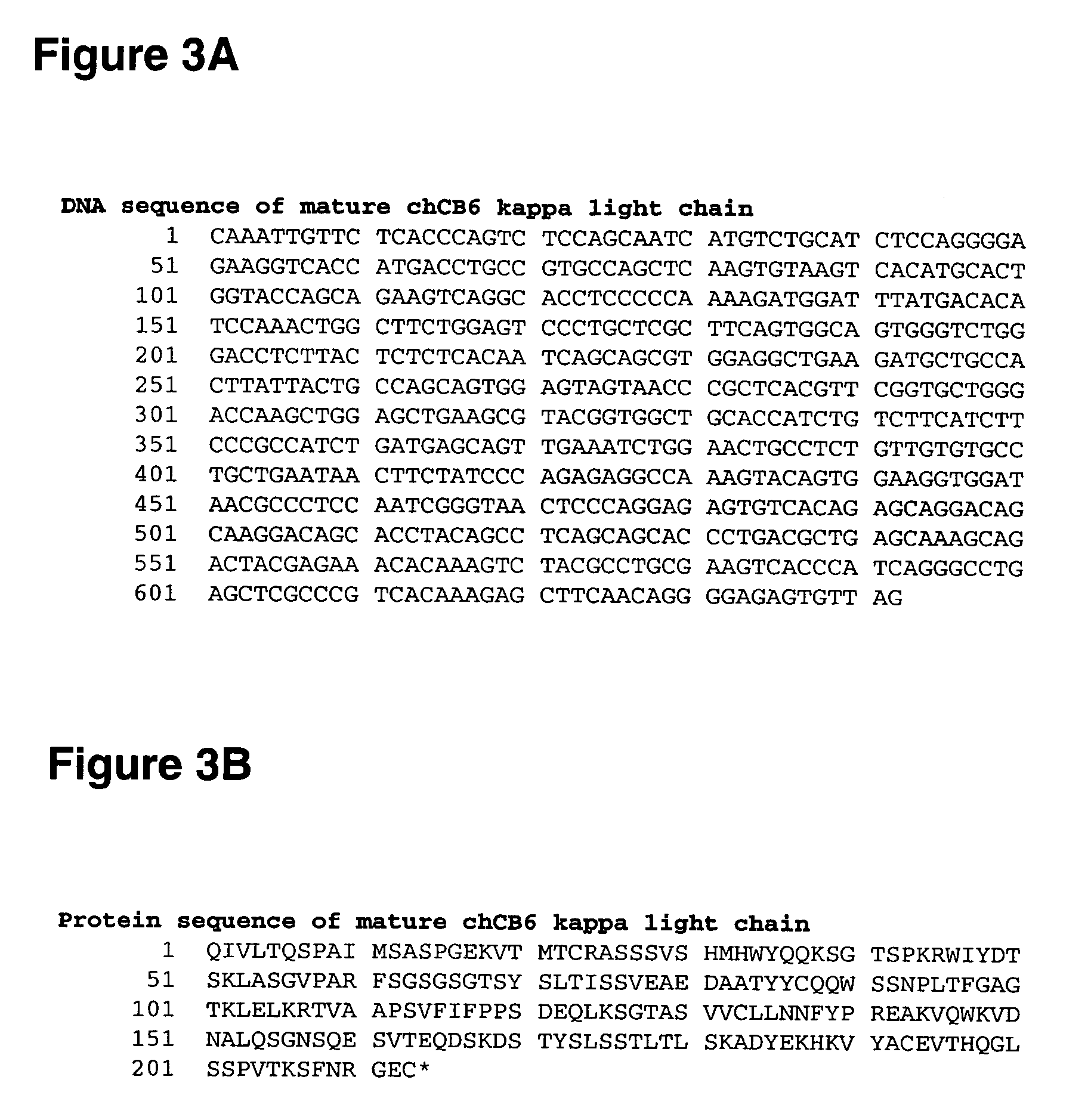
Methods for rational pegylation of proteins
The present invention relates to the use of simulation technology to rationally optimize the locations and sizes of attached polymeric moieties for modification of therapeutic proteins and the proteins generated from this method.
Owner:XENCOR
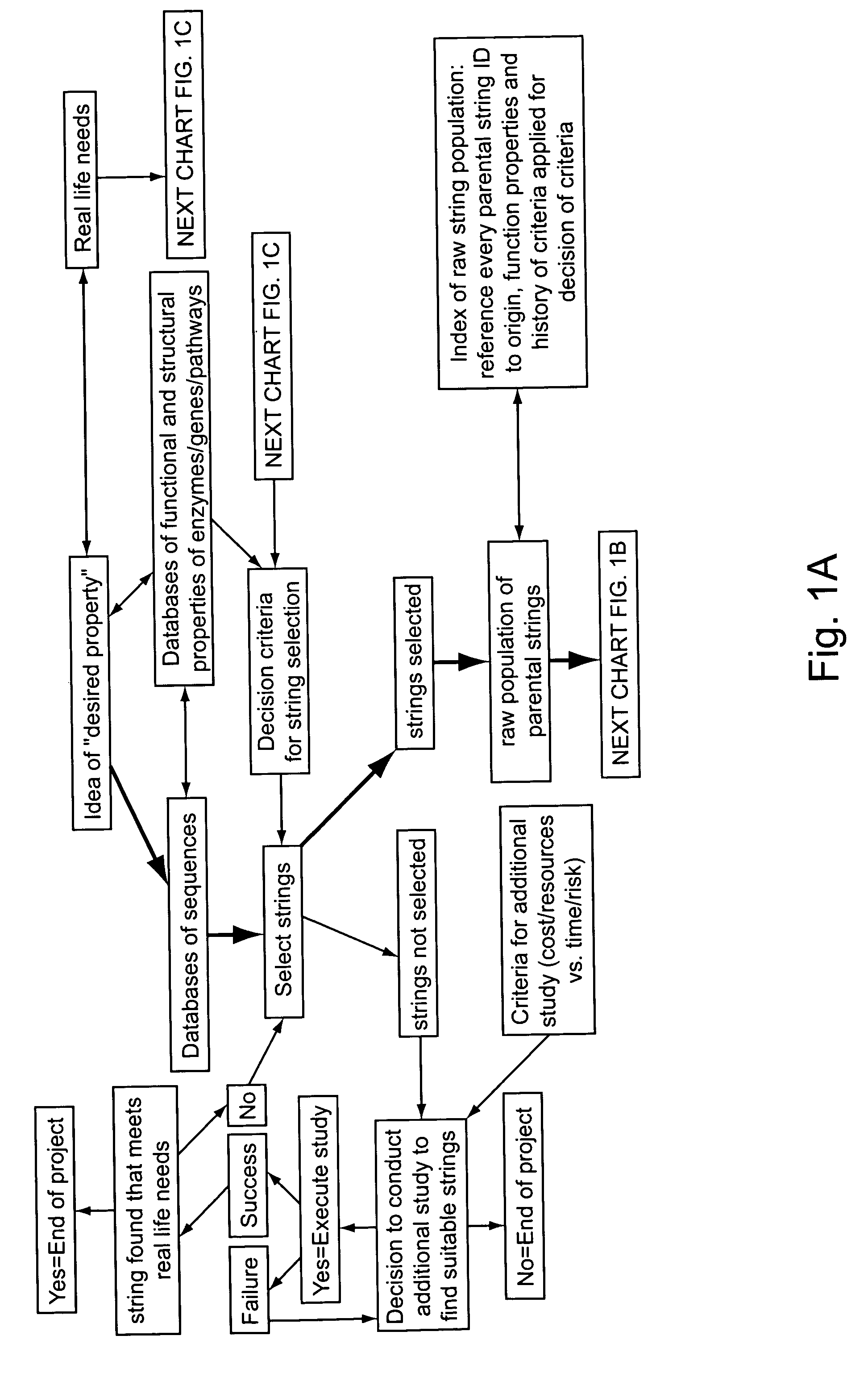
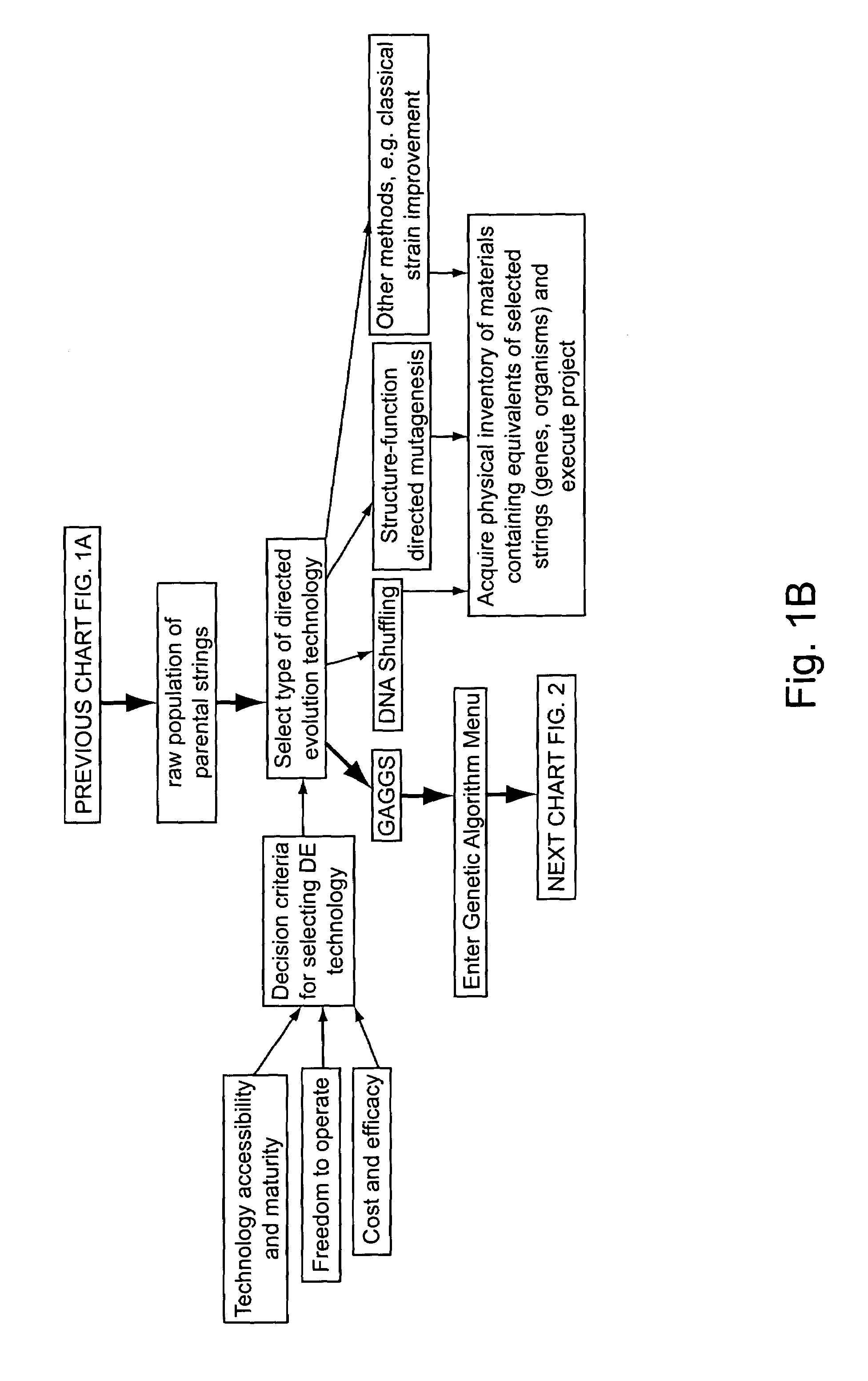
Methods for making character strings, polynucleotides and polypeptides having desired characteristics
InactiveUS7024312B1Simplifies overall synthesis strategyLow levelPeptide/protein ingredientsBiostatisticsPolynucleotideIn silico
“In silico” nucleic acid recombination methods, related integrated systems utilizing genetic operators and libraries made by in silico shuffling methods are provided.
Owner:CODEXIS MAYFLOWER HLDG LLC
Stabilized polypeptide compositions
ActiveUS20080050370A1Improved polypeptide compositionImprove methodPeptide/protein ingredientsAntibody mimetics/scaffoldsCrystallographyPolypeptide composition
Owner:BIOGEN MA INC
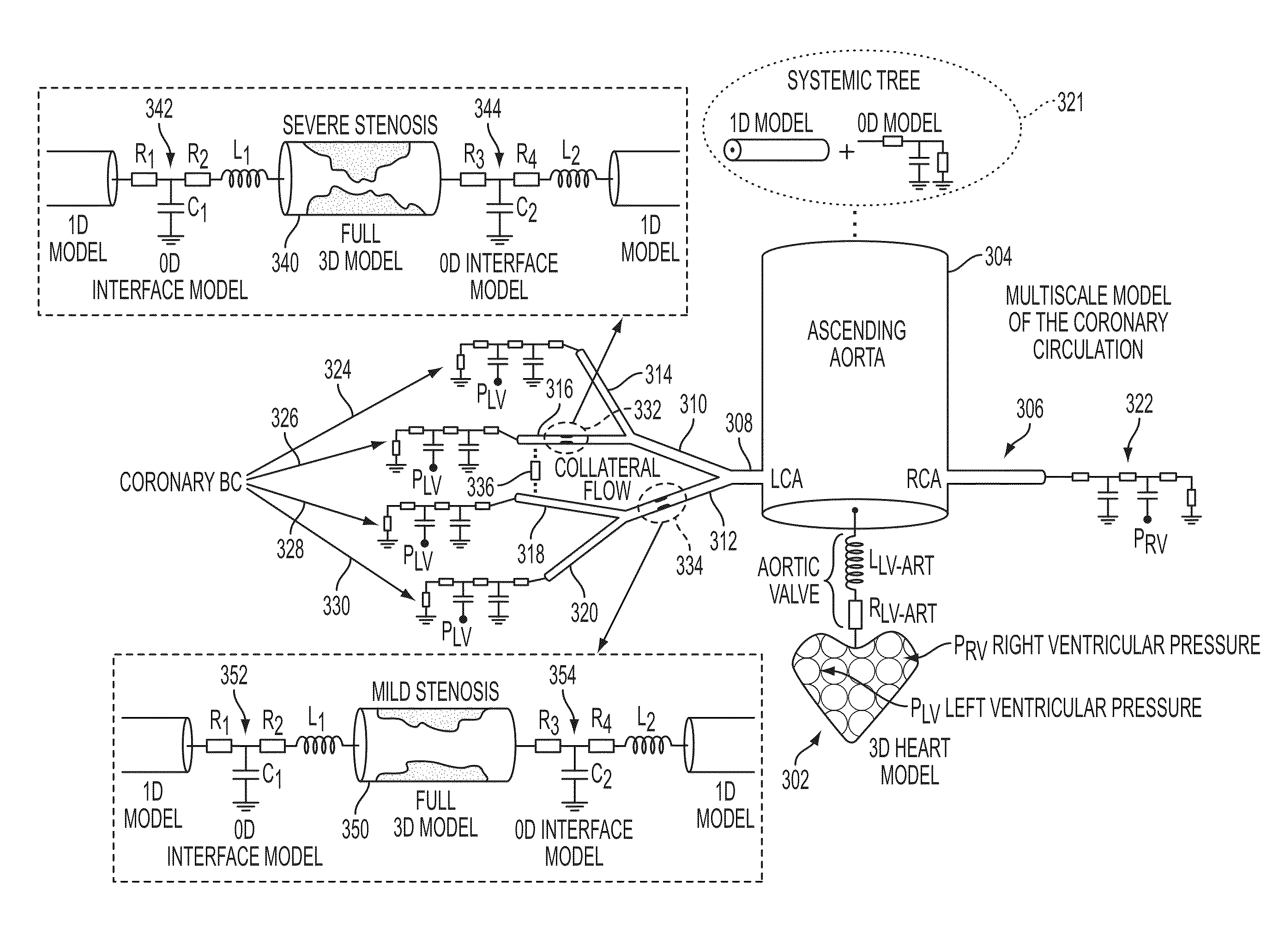
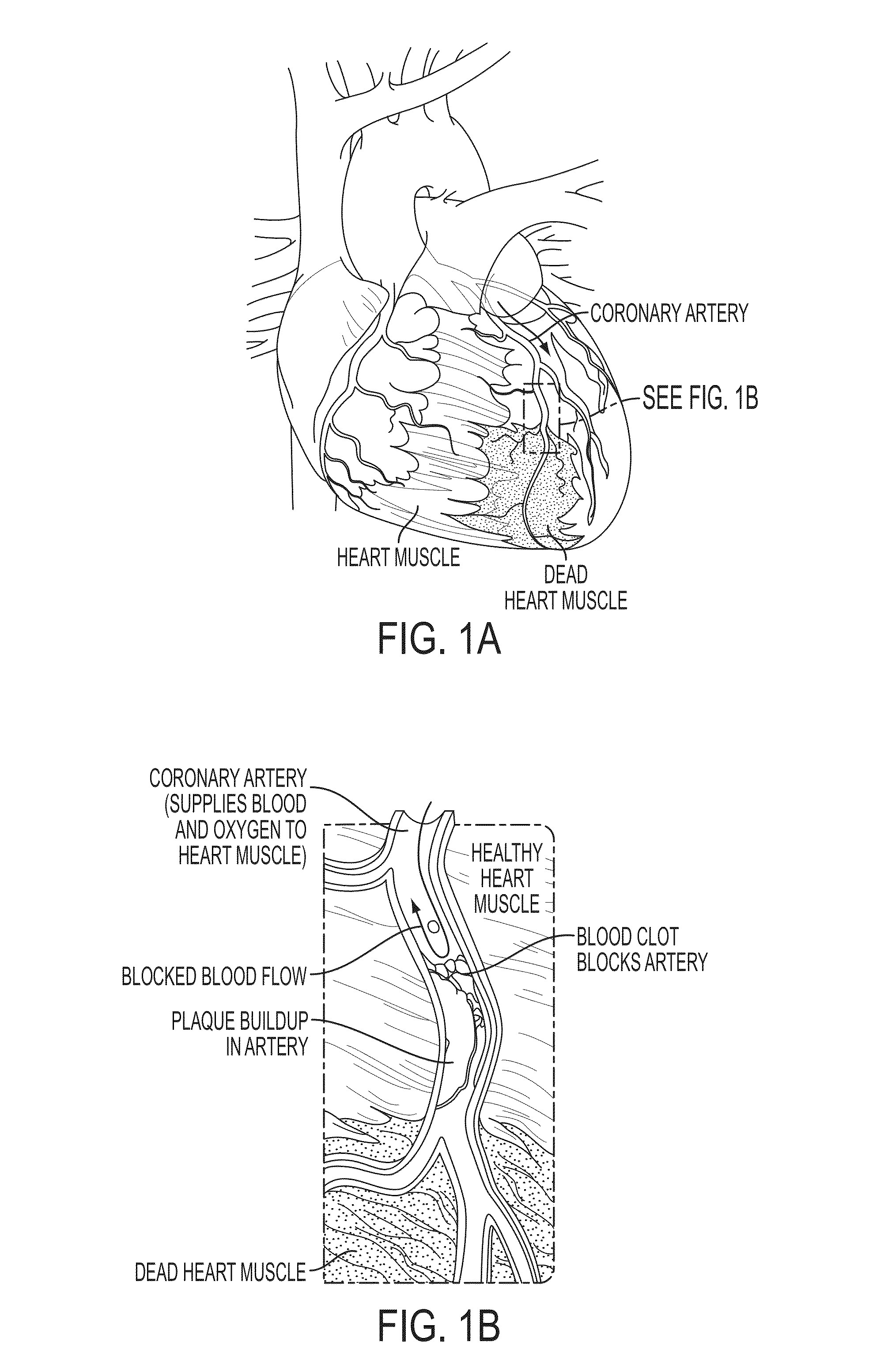
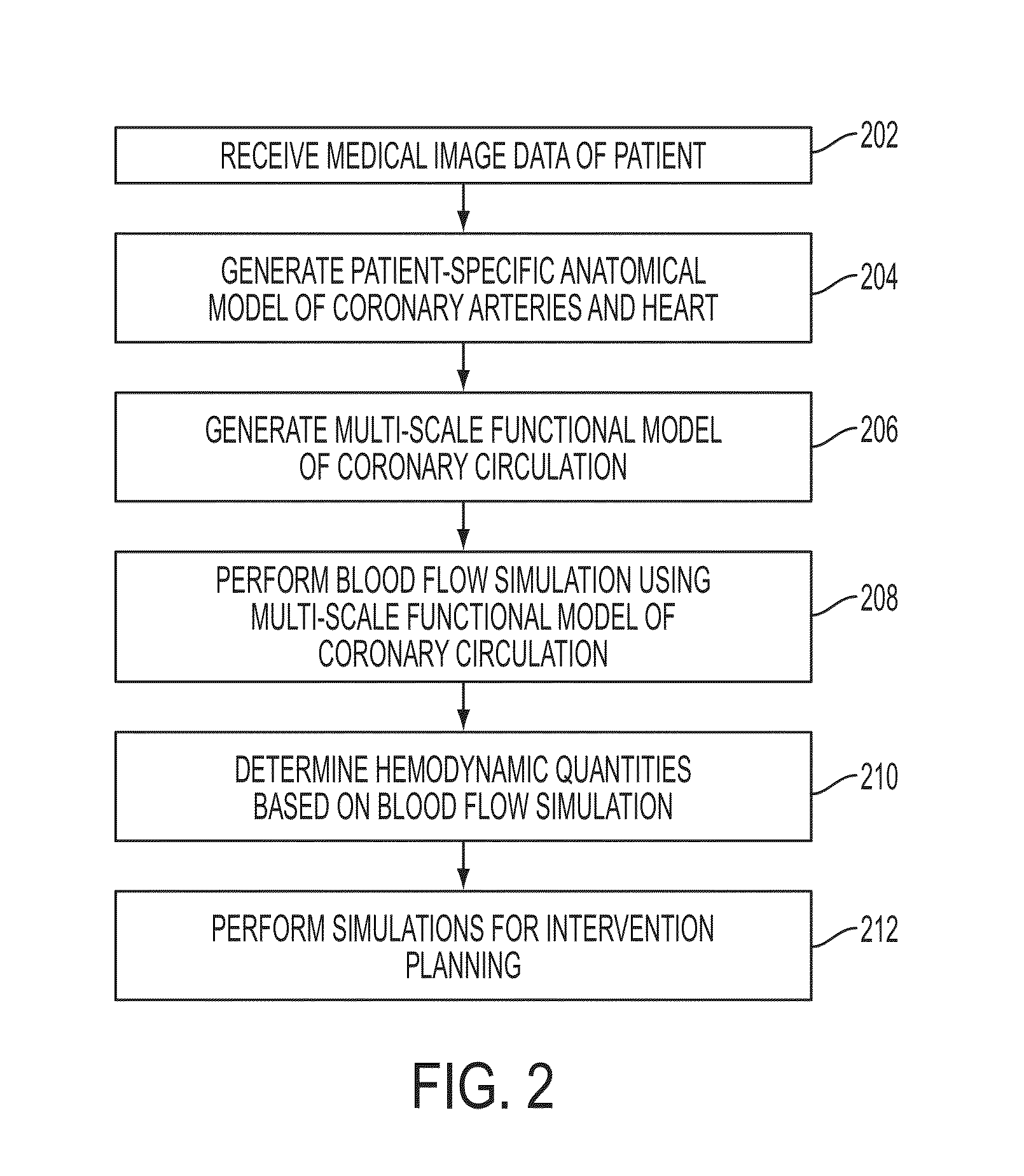
Method and System for Multi-Scale Anatomical and Functional Modeling of Coronary Circulation
ActiveUS20130132054A1Improve predictive performanceImprove clinical managementChemical property predictionChemical structure searchCoronary arteriesIntervention planning
A method and system for multi-scale anatomical and functional modeling of coronary circulation is disclosed. A patient-specific anatomical model of coronary arteries and the heart is generated from medical image data of a patient. A multi-scale functional model of coronary circulation is generated based on the patient-specific anatomical model. Blood flow is simulated in at least one stenosis region of at least one coronary artery using the multi-scale function model of coronary circulation. Hemodynamic quantities, such as fractional flow reserve (FFR), are computed to determine a functional assessment of the stenosis, and virtual intervention simulations are performed using the multi-scale function model of coronary circulation for decision support and intervention planning.
Owner:SIEMENS HEALTHCARE GMBH +1
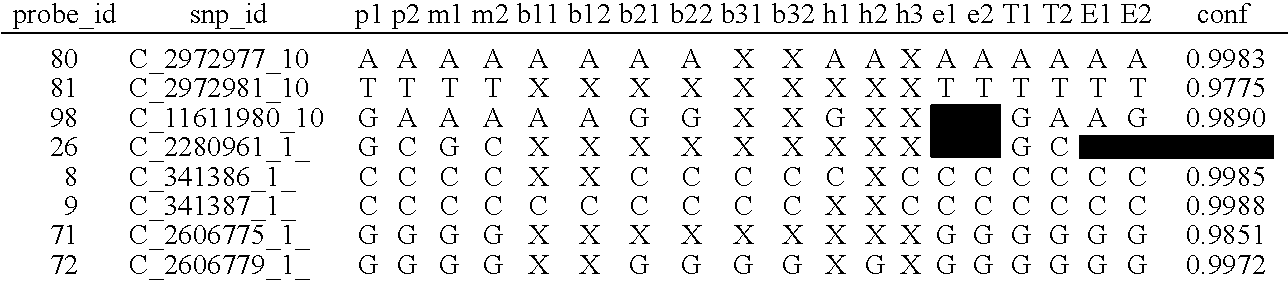
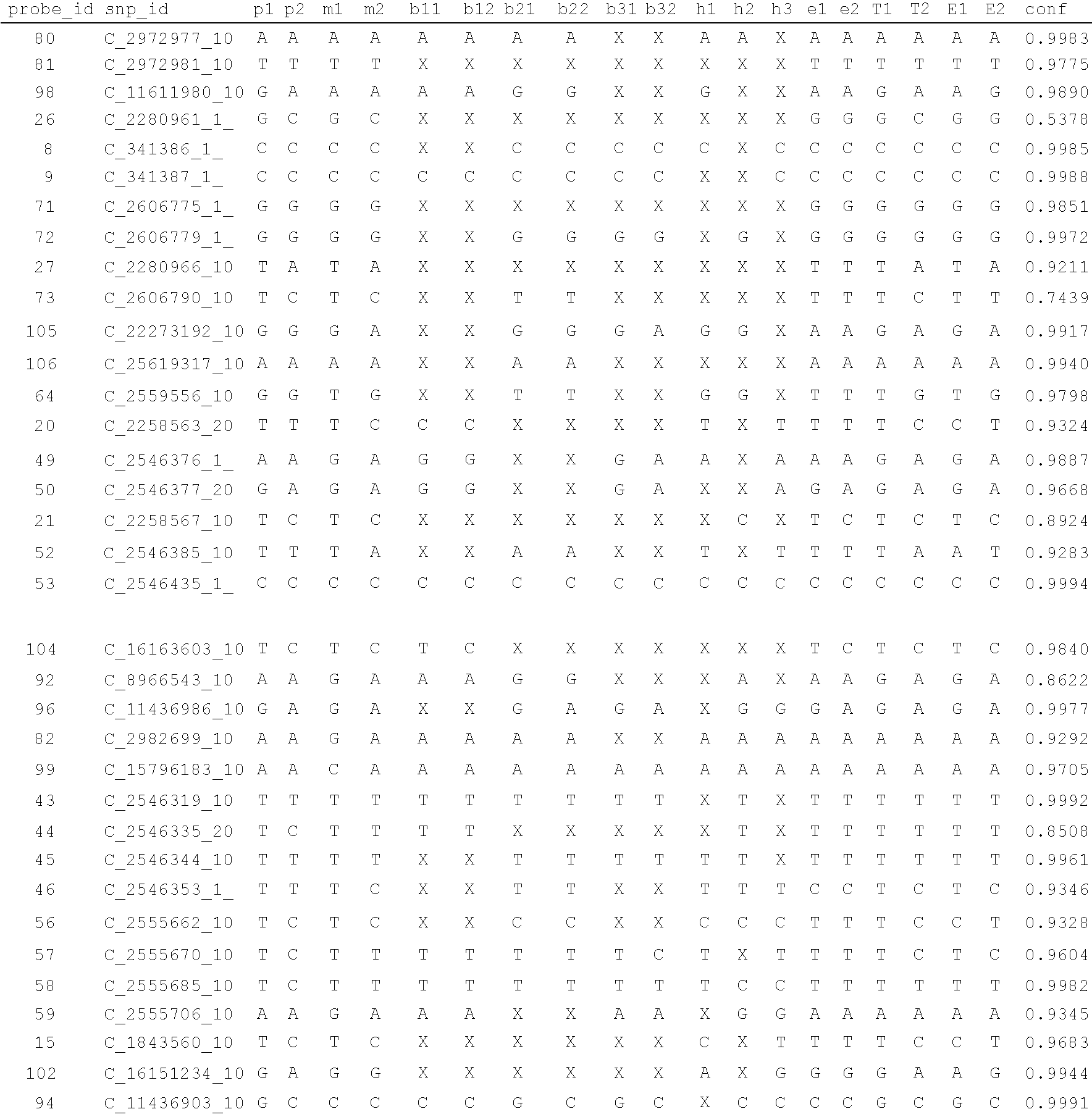
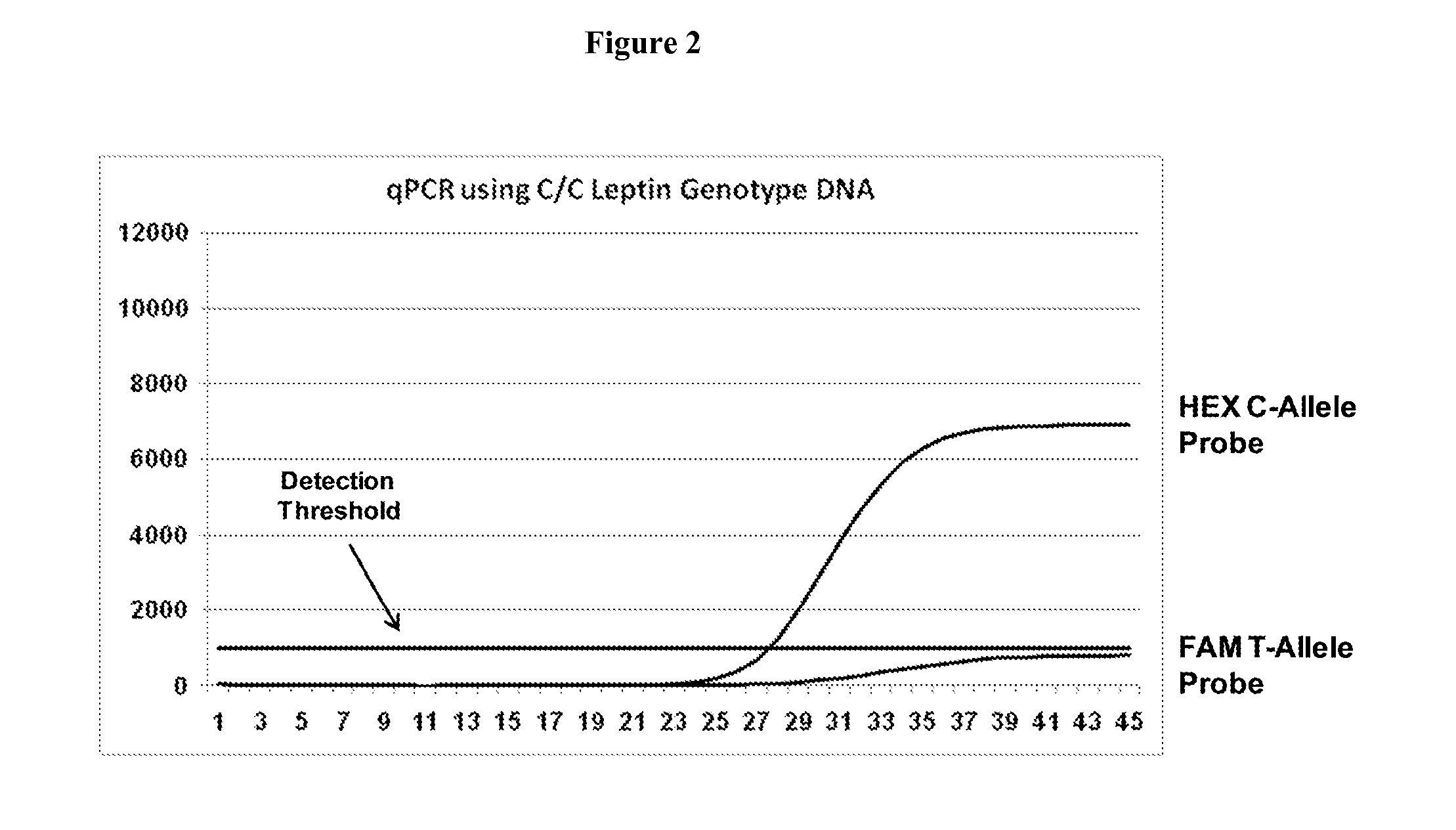
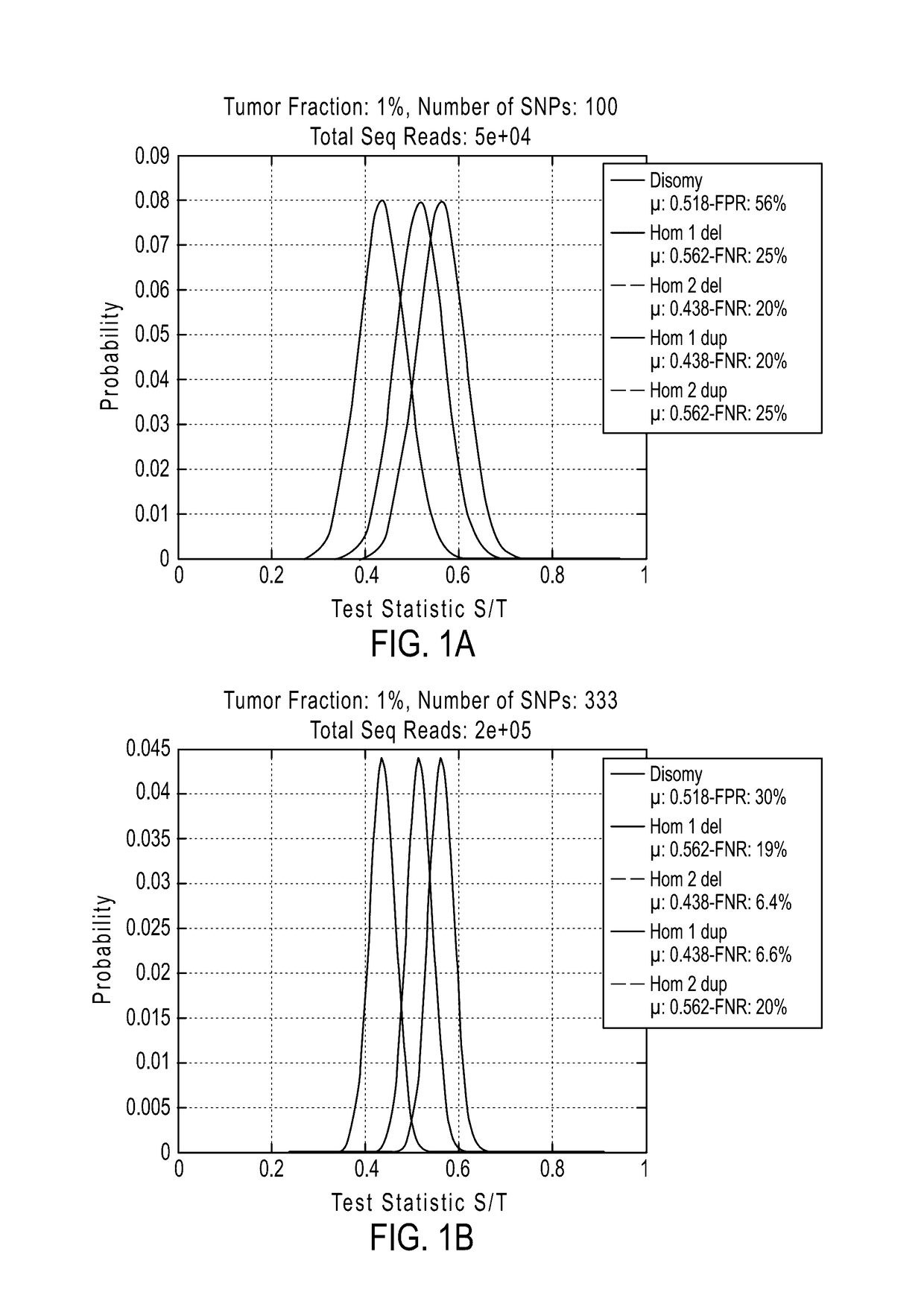
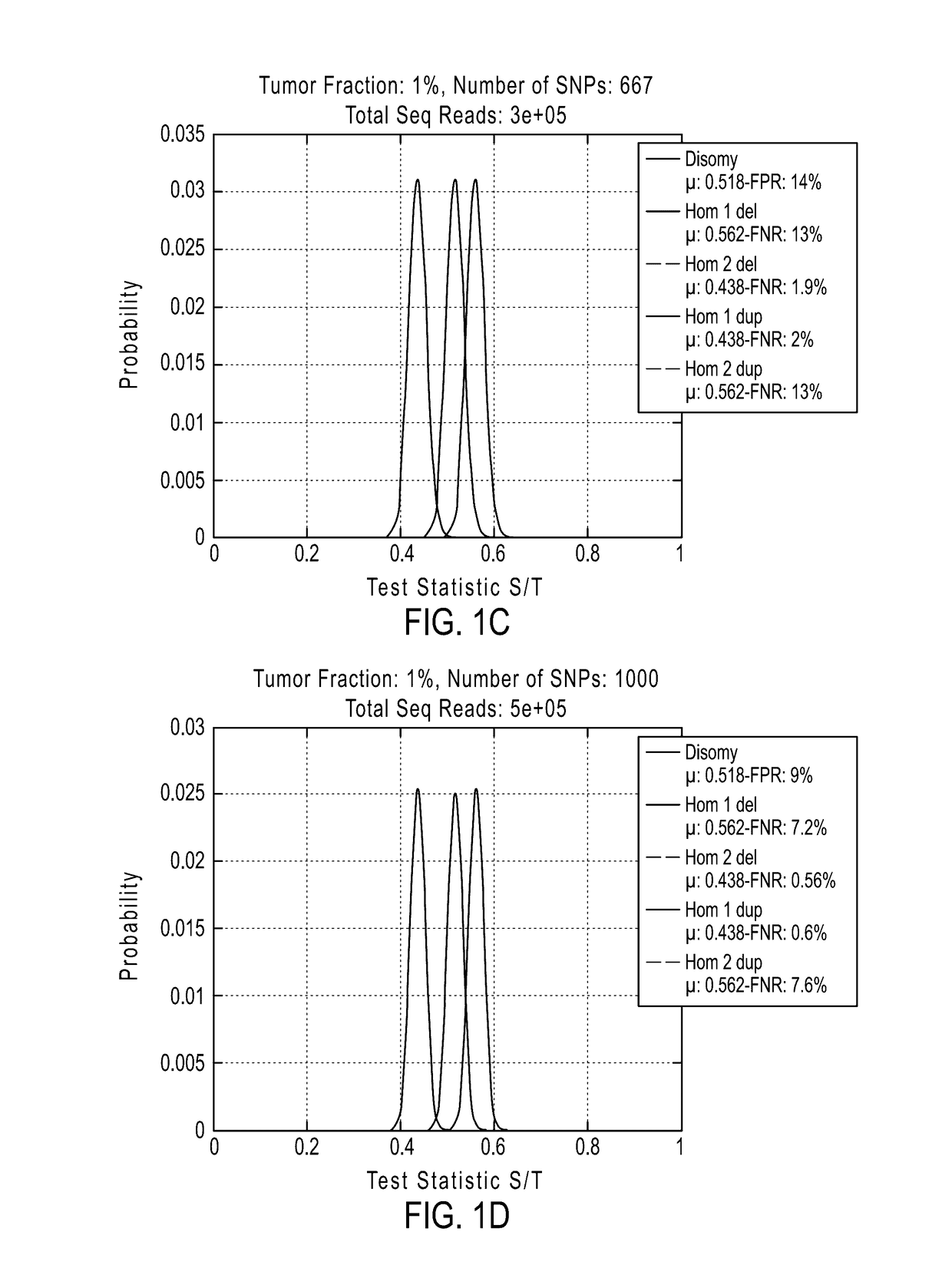
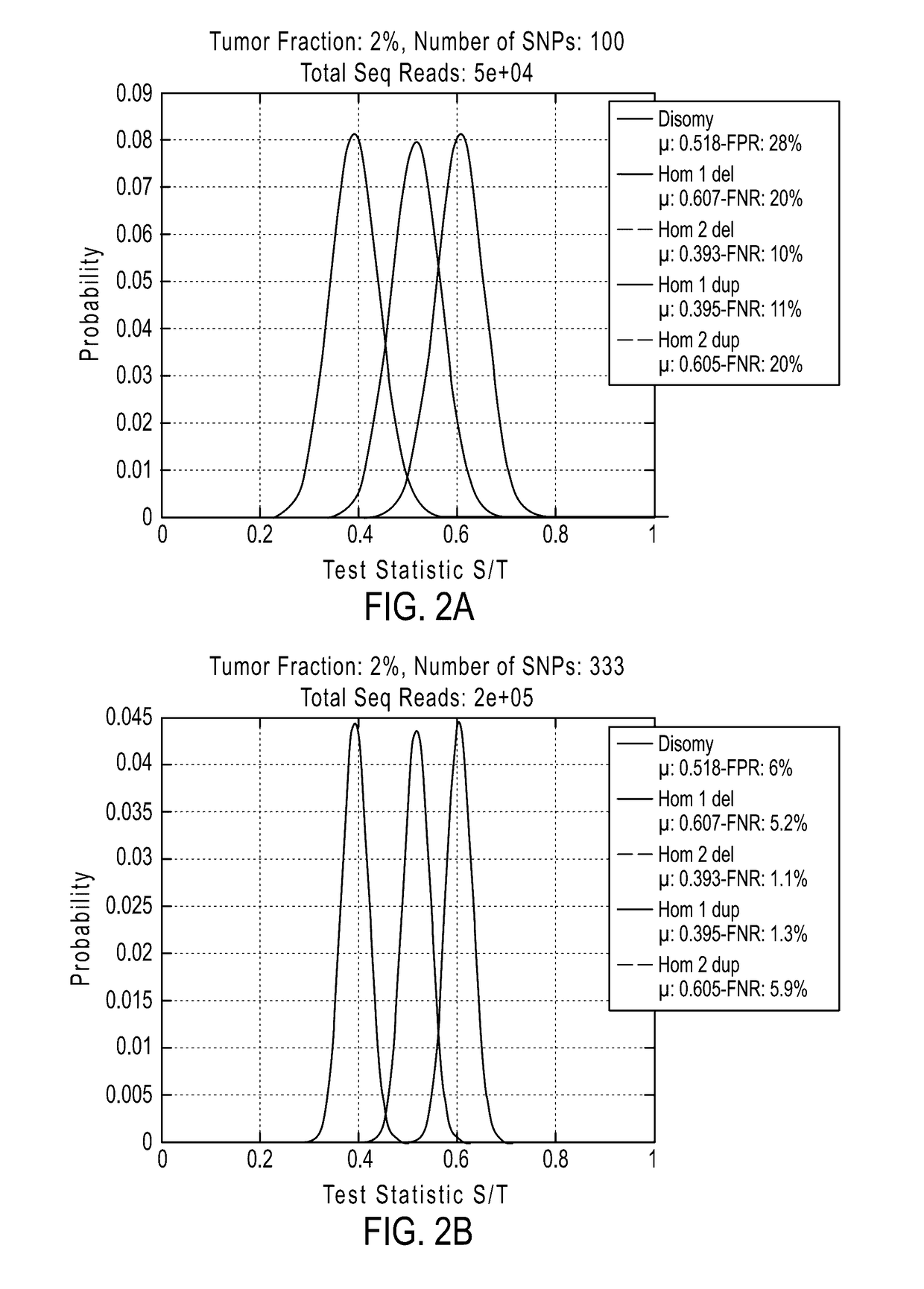
System and method for cleaning noisy genetic data and determining chromosome copy number
ActiveUS20080243398A1Significant resultImprove fidelityData processing applicationsMicrobiological testing/measurementGenetic MaterialsEmbryo
Disclosed herein is a system and method for increasing the fidelity of measured genetic data, for making allele calls, and for determining the state of aneuploidy, in one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Genetic material from the target individual is acquired, amplified and the genetic data is measured using known methods. Poorly or incorrectly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related individuals. In accordance with one embodiment of the invention, incomplete genetic data from an embryonic cell are reconstructed at a plurality of loci using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without haploid genetic data from one or both parents. In another embodiment of the invention, the chromosome copy number can be determined from the measured genetic data of a single or small number of cells, with or without genetic information from one or both parents. In another embodiment of the invention, these determinations are made for the purpose of embryo selection in the context of in-vitro fertilization. In another embodiment of the invention, the genetic data can be reconstructed for the purposes of making phenotypic predictions.
Owner:NATERA
Protein design automation for protein libraries
The invention relates to the use of protein design automation (P DA) to generate computationally prescreened secondary libraries of proteins, and to methods and compositions utilizing the libraries.
Owner:XENCOR
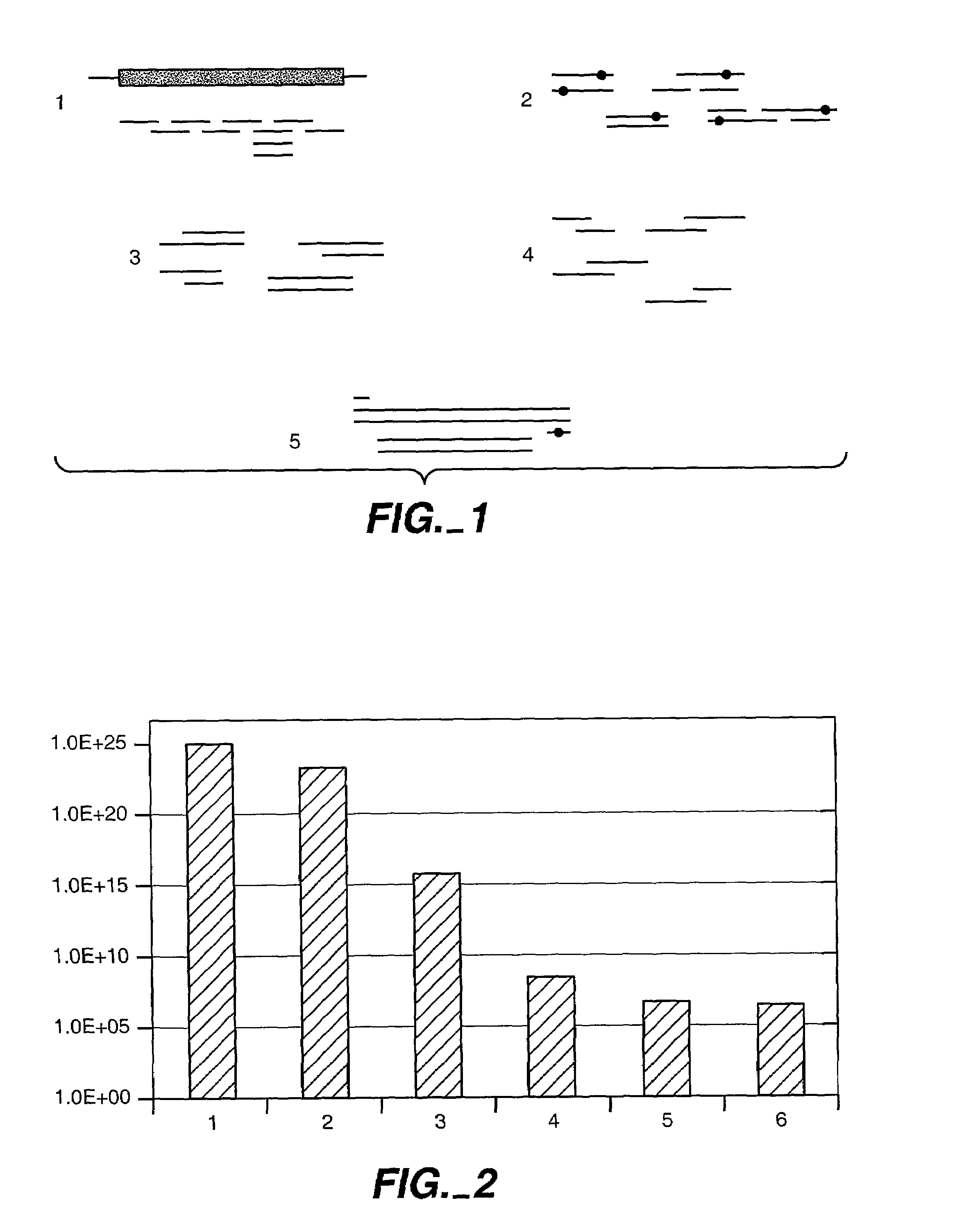
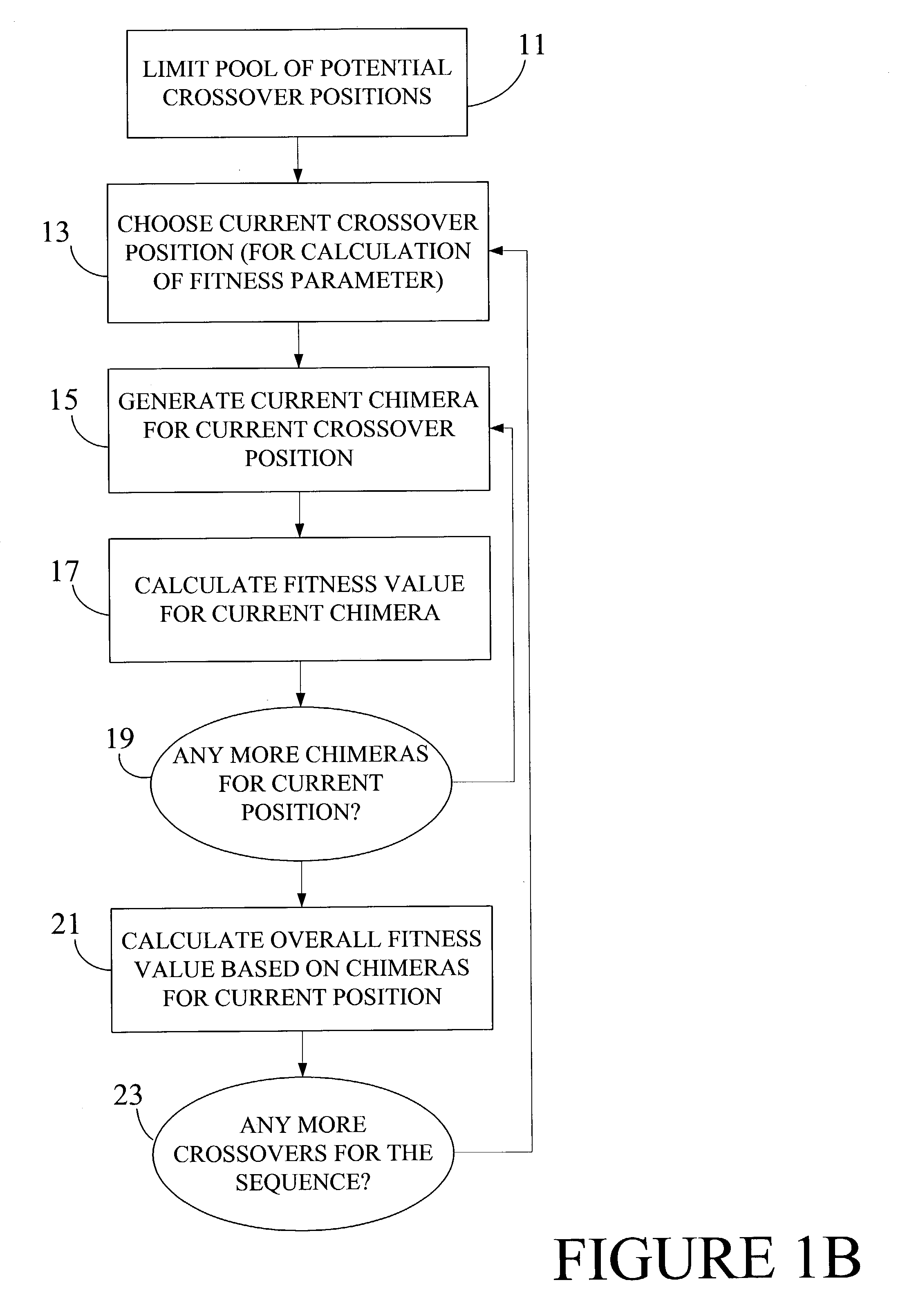
Optimization of crossover points for directed evolution
ActiveUS7620500B2Microbiological testing/measurementAnalogue computers for chemical processesNucleotideNucleotide sequencing
Owner:CODEXIS MAYFLOWER HLDG LLC
Methods for providing bacterial bioagent characterizing information
InactiveUS7217510B2Easy to analyzeMicrobiological testing/measurementEpidemiological alert systemsMedicineInformatics
The present invention relates generally to the field of investigational bioinformatics and more particularly to secondary structure defining databases. The present invention further relates to methods for interrogating a database as a source of molecular masses of known bioagents for comparing against the molecular mass of an unknown or selected bioagent to determine either the identity of the selected bioagent, and / or to determine the origin of the selected bioagent. The identification of the bioagent is important for determining a proper course of treatment and / or irradication of the bioagent in such cases as biological warfare. Furthermore, the determination of the geographic origin of a selected bioagent will facilitate the identification of potential criminal identity.
Owner:IBIS BIOSCI
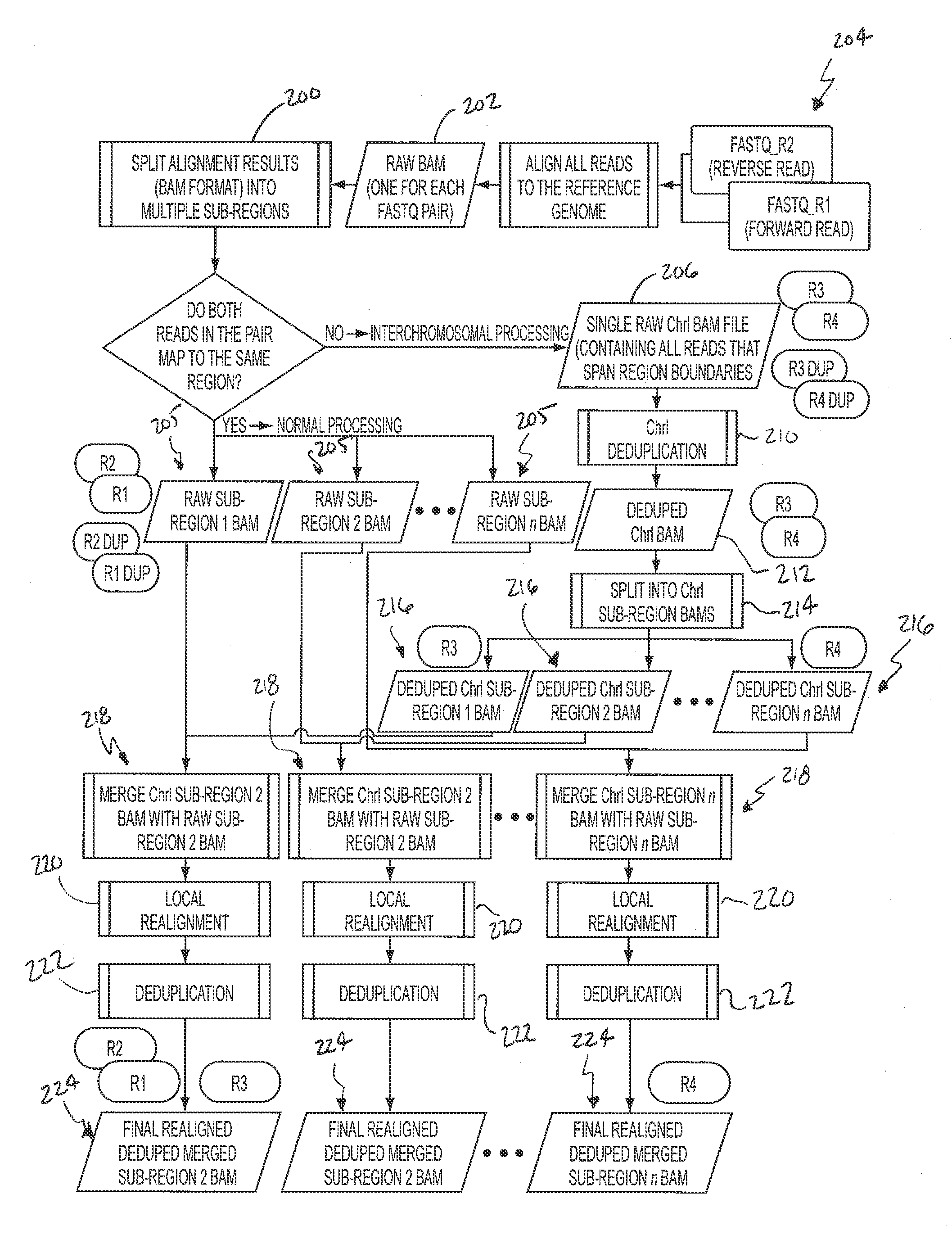
Comprehensive Analysis Pipeline for Discovery of Human Genetic Variation
ActiveUS20130311106A1Shorten analysis timeQuick identificationMicrobiological testing/measurementProteomicsData setProcessing core
Systems and methods for analyzing genetic sequence data involve: (a) obtaining, by a computer system, genetic sequencing data pertaining to a subject; (b) splitting the genetic sequencing data into a plurality of segments; (c) processing the genetic sequencing data such that intra-segment reads, read pairs with both mates mapped to the same data set, are saved to a respective plurality of individual binary alignment map (BAM) files corresponding to that respective segment; (d) processing the genetic sequencing data such that inter-segment reads, read pairs with both mates mapped to different segments, are saved into at least a second BAM file; and (e) processing at least the first plurality of BAM files along parallel processing paths. The plurality of segments may correspond to any given number of genomic subregions and may be selected based upon the number of processing cores used in the parallel processing.
Owner:RES INST AT NATIONWIDE CHILDRENS HOSPITAL
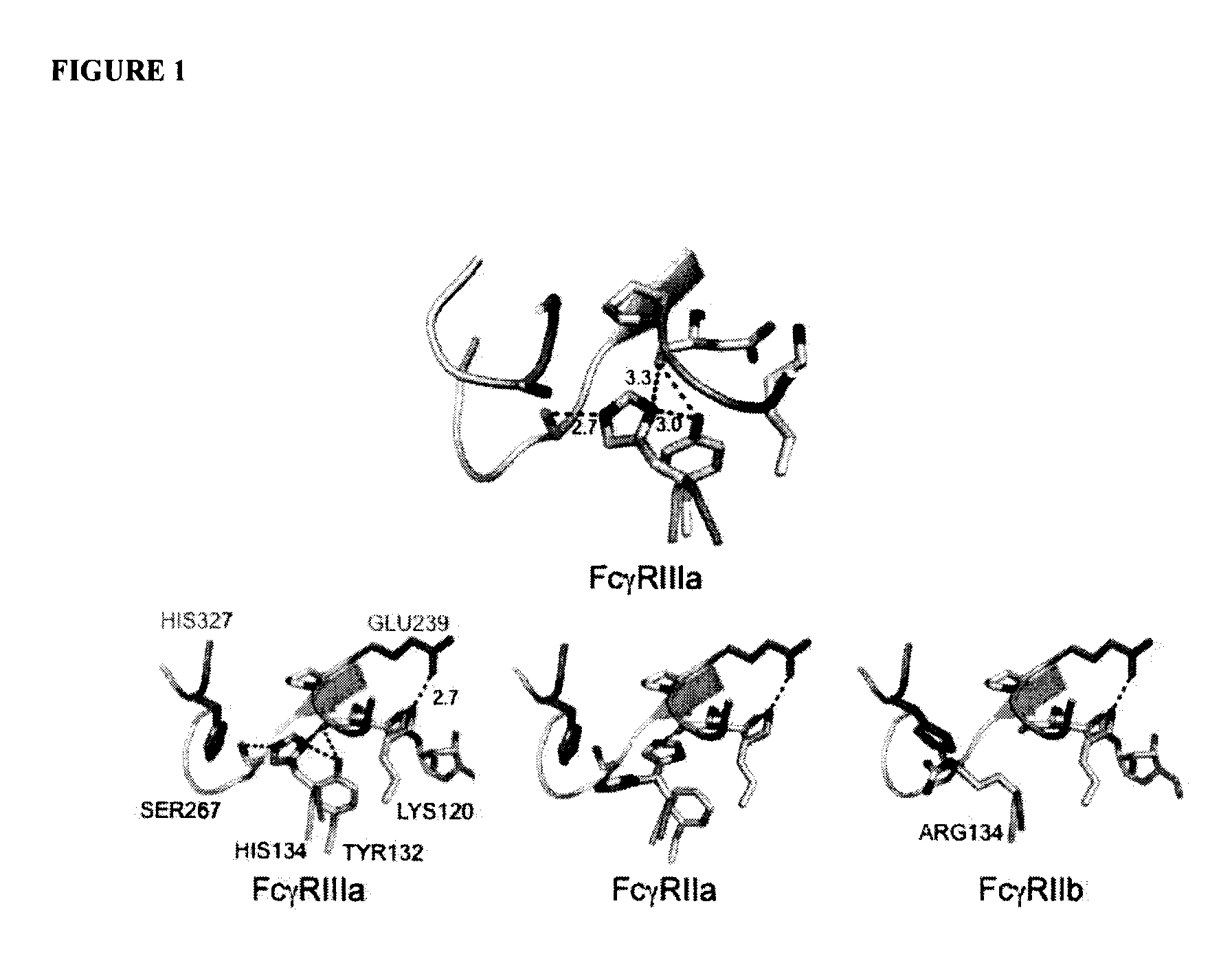
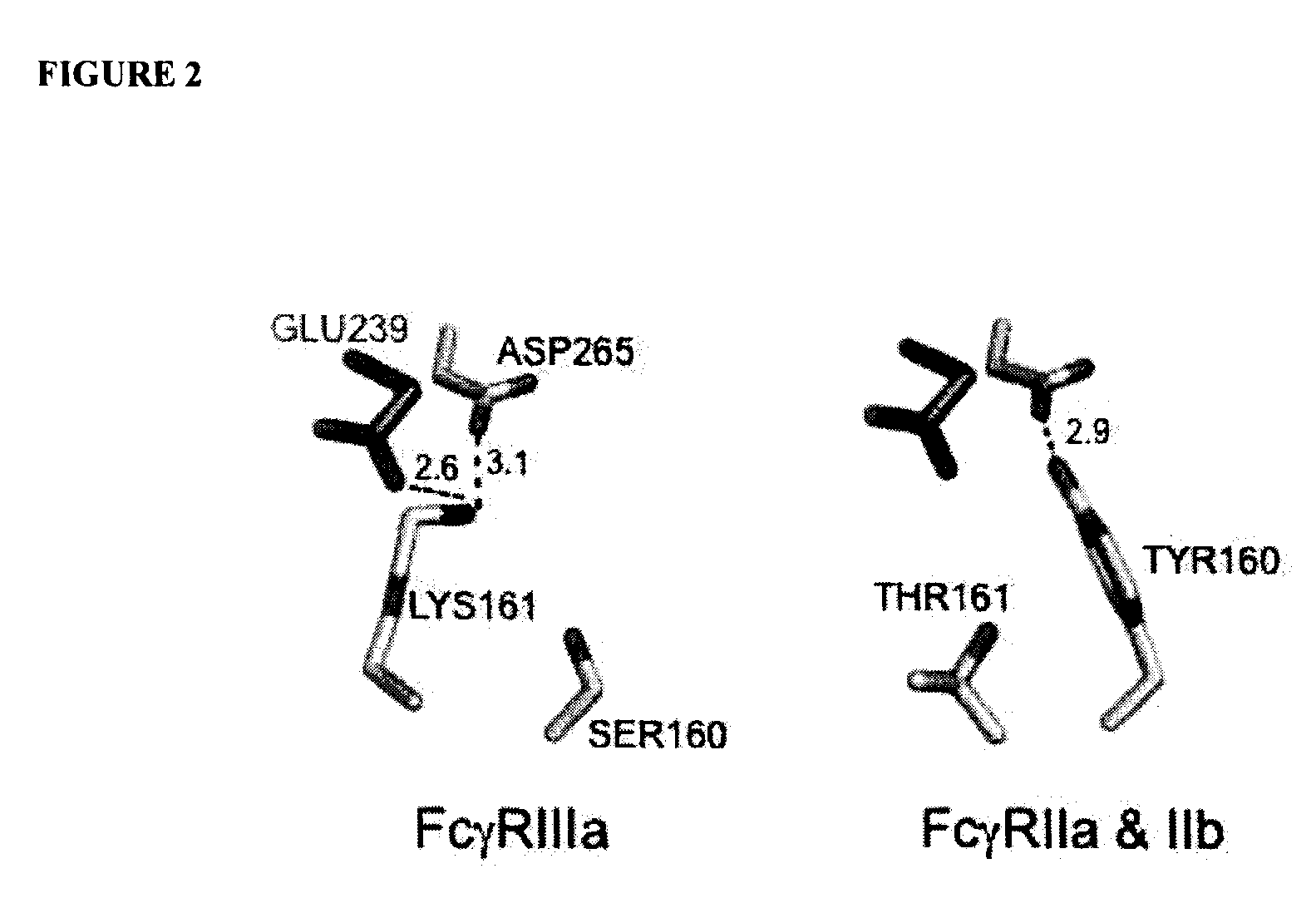
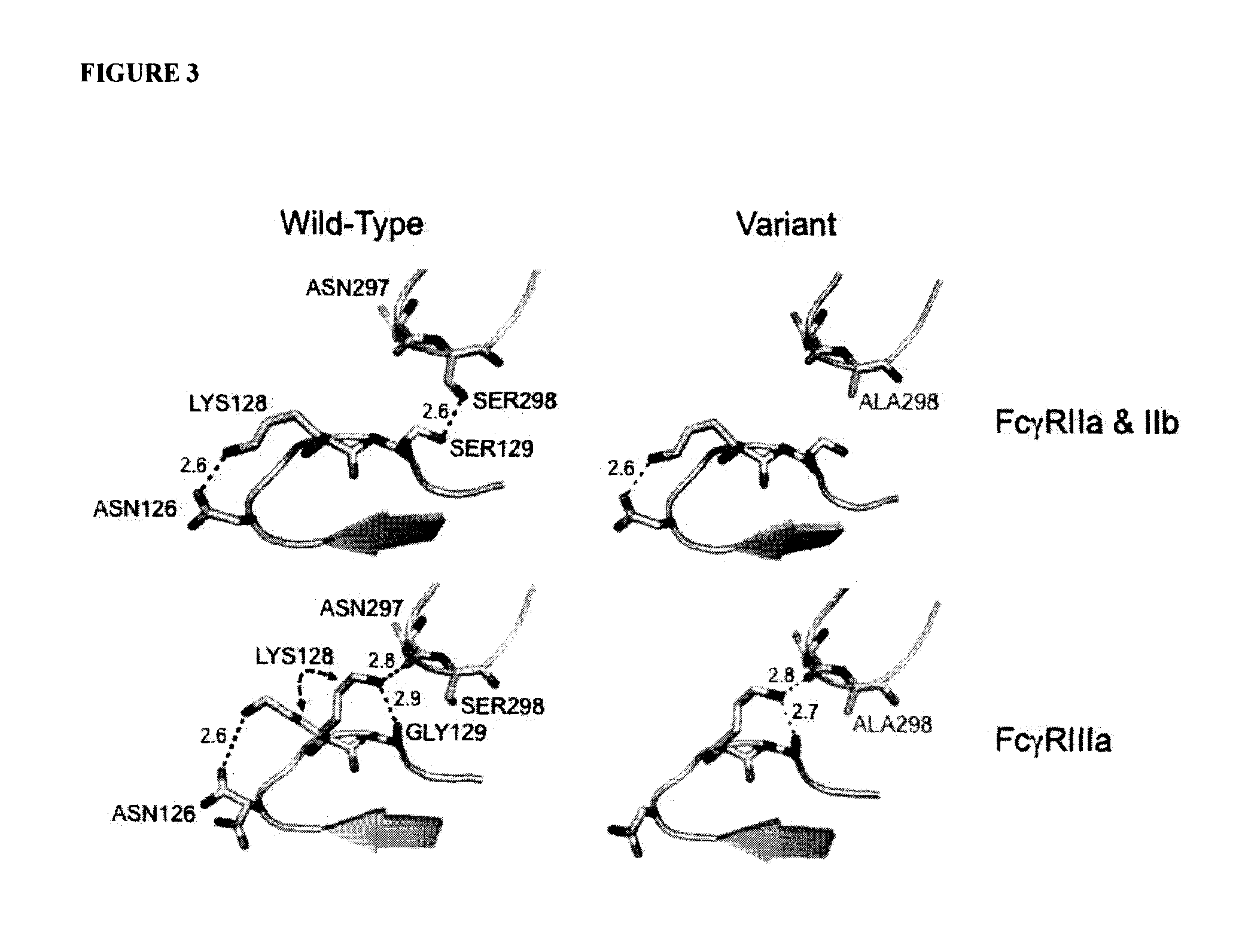
Fcgamma receptor-binding polypeptide variants and methods related thereto
InactiveUS20060275283A1Increase free energyReduced binding affinityFermentationPlant genotype modificationAntibodyMolecular biology
The compositions and methods of the present invention are based, in part, on our discovery that an effector function mediated by an Fc-containing polypeptide can be altered by modifying one or more amino acid residues within the polypeptide (by, for example, electrostatic optimization). The polypeptides that can be generated according to the methods of the invention are highly variable, and they can include antibodies and fusion proteins that contain an Fc region or a biologically active portion thereof.
Owner:BIOGEN IDEC MA INC
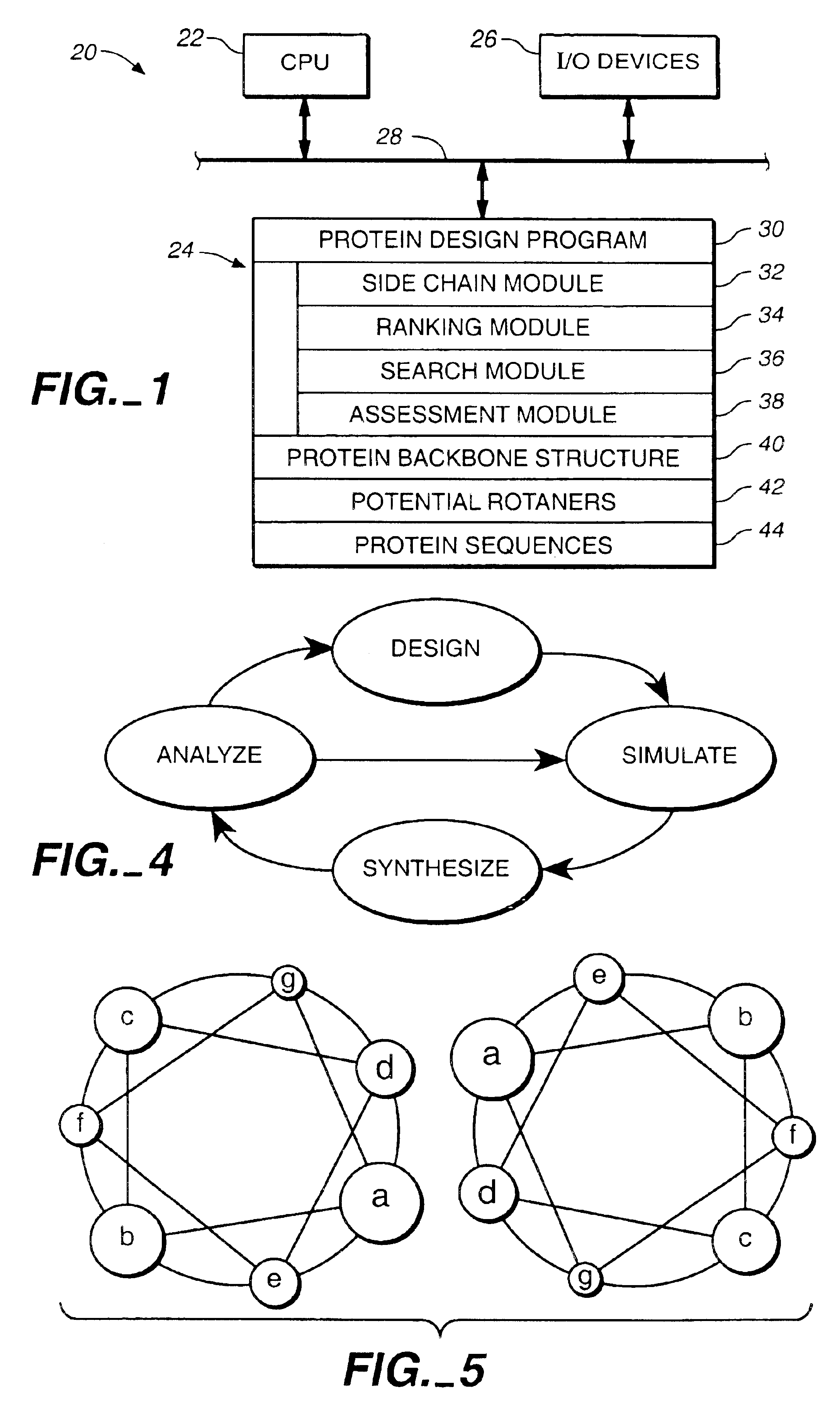
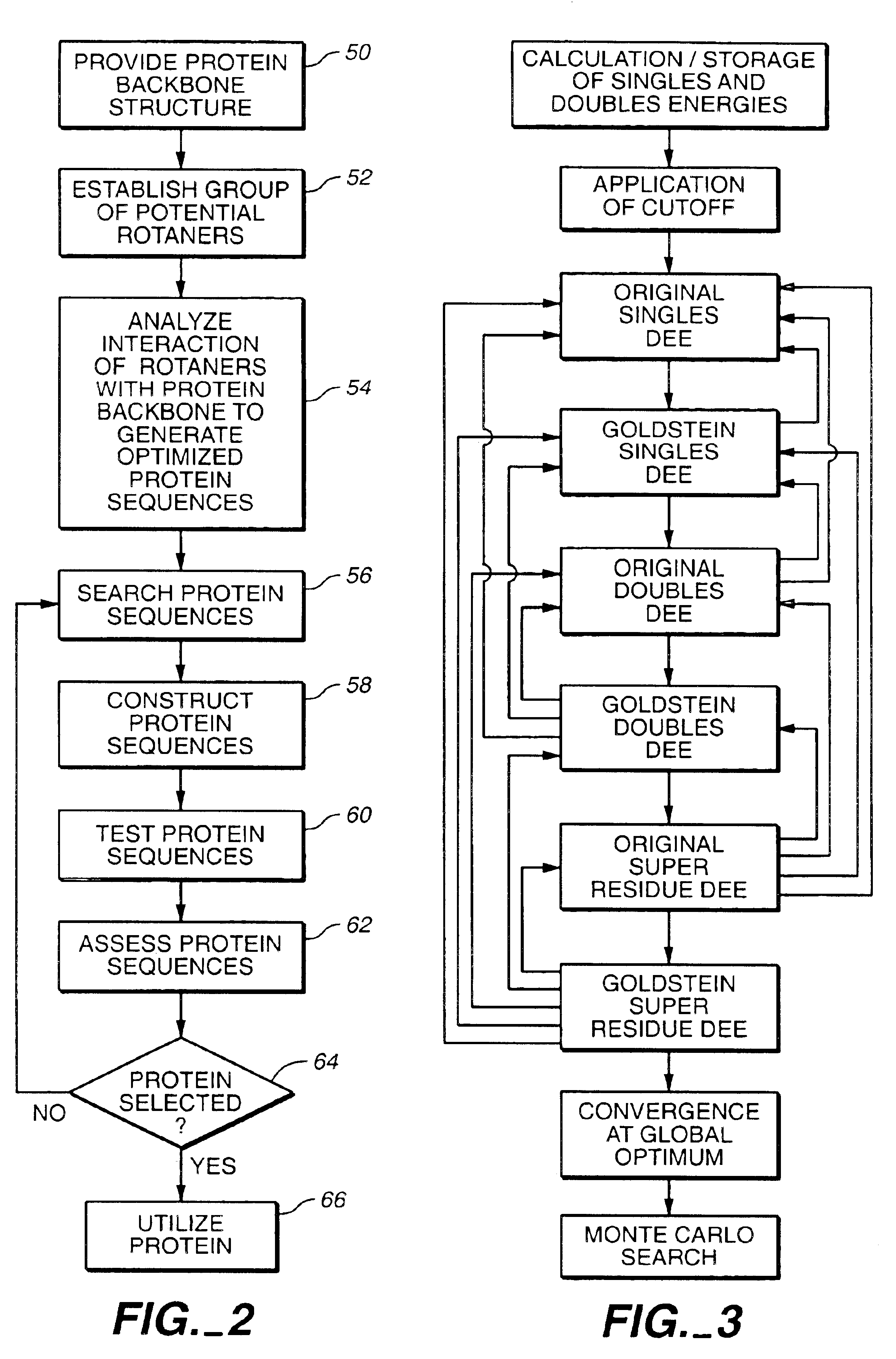
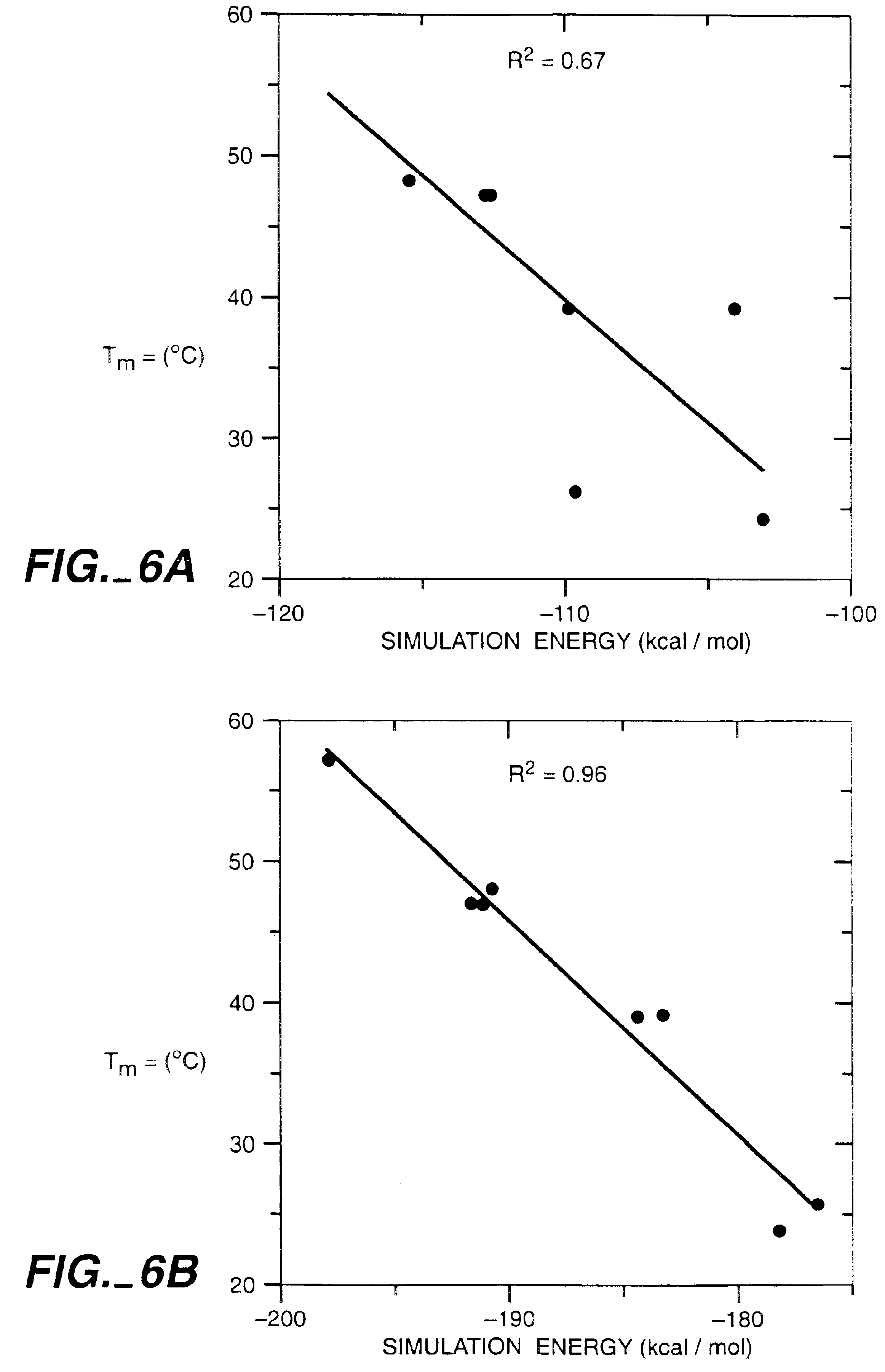
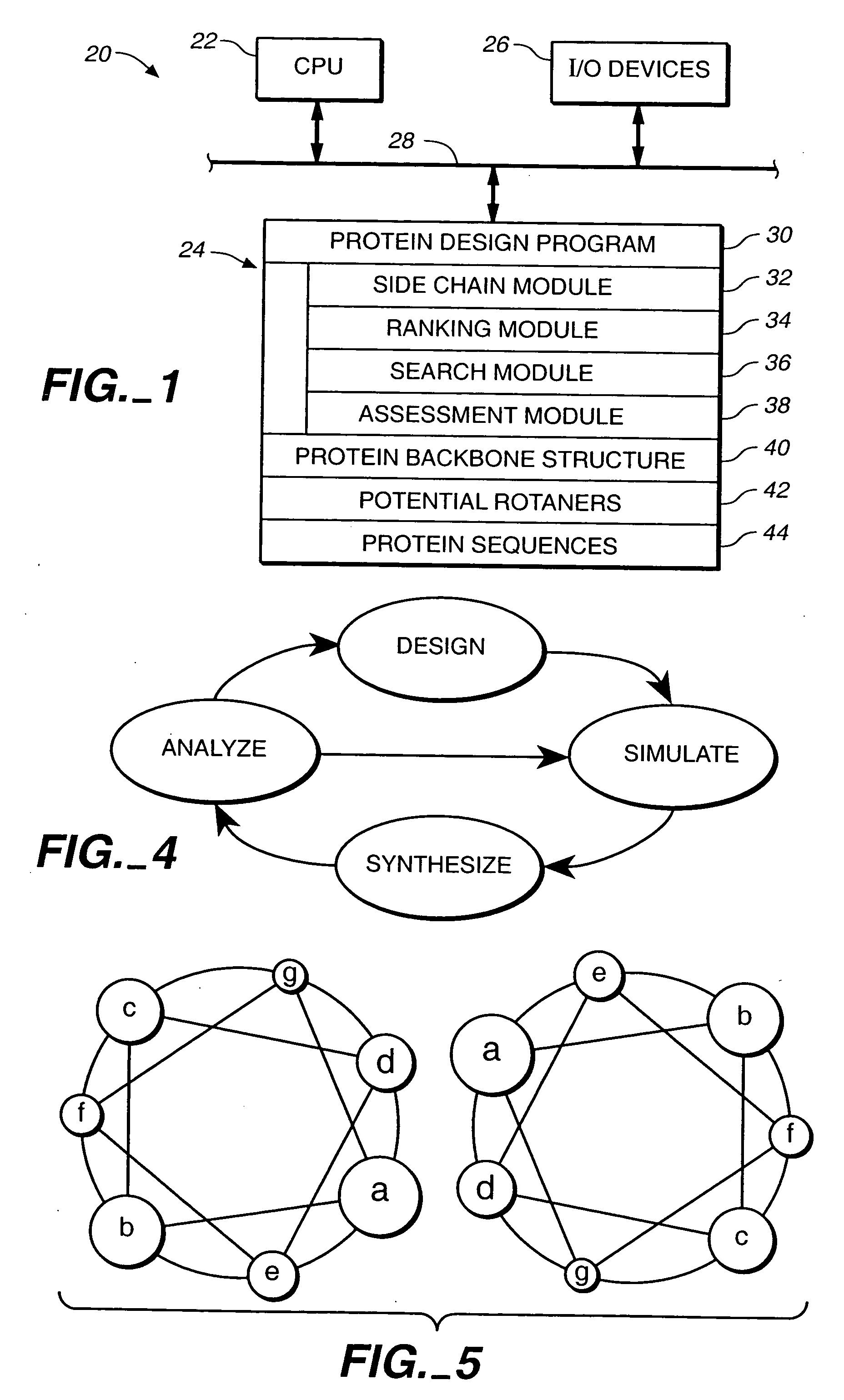
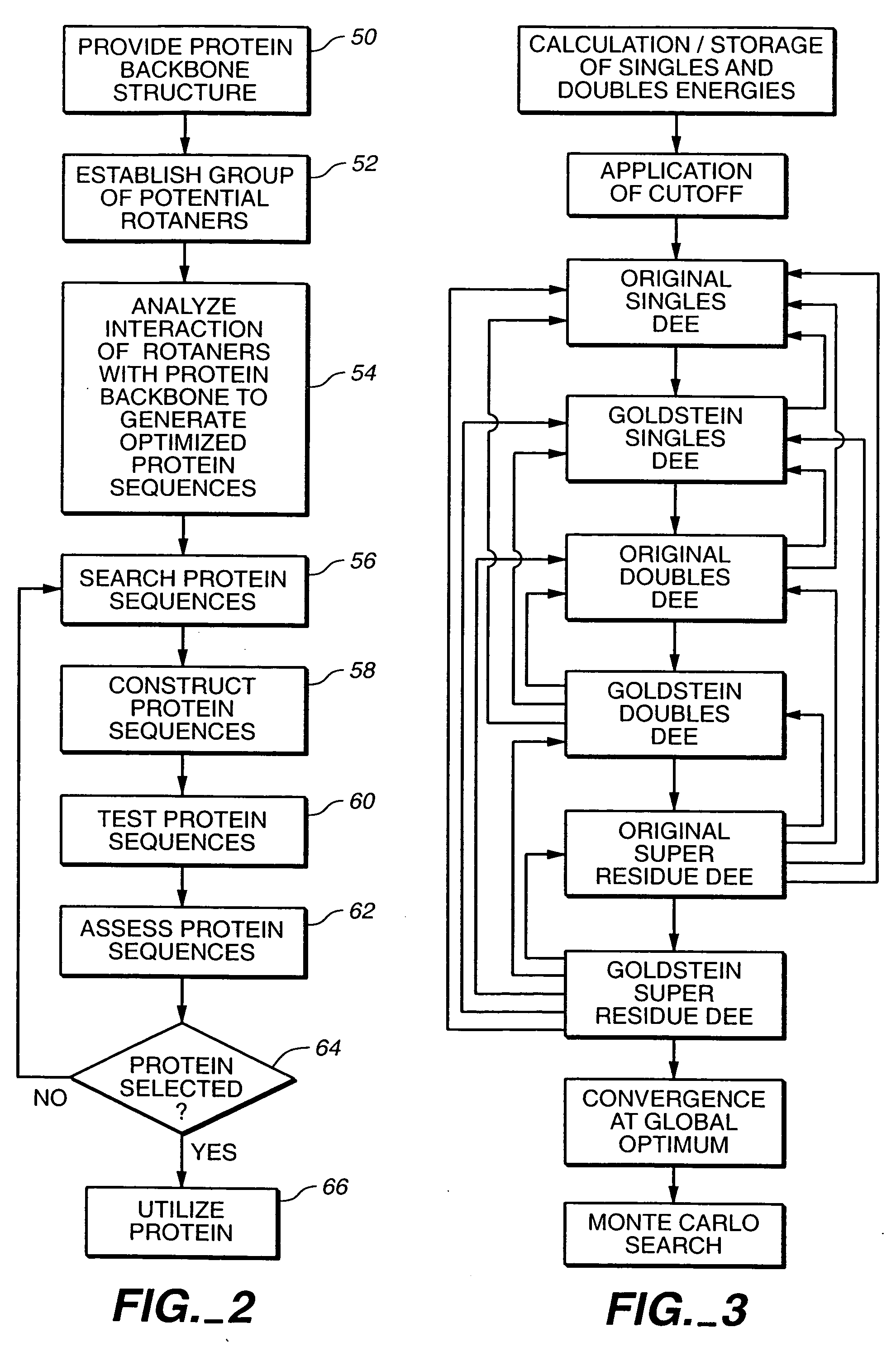
Apparatus and method for automated protein design
The present invention relates to apparatus and methods for quantitative protein design and optimization.
Owner:CALIFORNIA INST OF TECH
System and method for cleaning noisy genetic data and determining chromosome copy number
ActiveUS8515679B2Improve fidelityData processing applicationsMicrobiological testing/measurementGenetic MaterialsEmbryo
Disclosed herein is a system and method for increasing the fidelity of measured genetic data, for making allele calls, and for determining the state of aneuploidy, in one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Genetic material from the target individual is acquired, amplified and the genetic data is measured using known methods. Poorly or incorrectly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related individuals. In accordance with one embodiment of the invention, incomplete genetic data from an embryonic cell are reconstructed at a plurality of loci using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without haploid genetic data from one or both parents. In another embodiment of the invention, the chromosome copy number can be determined from the measured genetic data of a single or small number of cells, with or without genetic information from one or both parents. In another embodiment of the invention, these determinations are made for the purpose of embryo selection in the context of in-vitro fertilization. In another embodiment of the invention, the genetic data can be reconstructed for the purposes of making phenotypic predictions.
Owner:NATERA
Stabilized polypeptide compositions
InactiveUS20090048122A1Improve efficiencyImprove effectivenessAntibody mimetics/scaffoldsLibrary screeningPolypeptide compositionOrganic chemistry
Owner:BIOGEN MA INC
Scar-less multi-part DNA assembly design automation
The present invention provides a method of a method of designing an implementation of a DNA assembly. In an exemplary embodiment, the method includes (1) receiving a list of DNA sequence fragments to be assembled together and an order in which to assemble the DNA sequence fragments, (2) designing DNA oligonucleotides (oligos) for each of the DNA sequence fragments, and (3) creating a plan for adding flanking homology sequences to each of the DNA oligos. In an exemplary embodiment, the method includes (1) receiving a list of DNA sequence fragments to be assembled together and an order in which to assemble the DNA sequence fragments, (2) designing DNA oligonucleotides (oligos) for each of the DNA sequence fragments, and (3) creating a plan for adding optimized overhang sequences to each of the DNA oligos.
Owner:RGT UNIV OF CALIFORNIA
Novel proteins with altered immunogenicity
The present invention provides methods for combining computational methods for modulating protein immunogenicity with computational methods for identifying sequences with desired structural and functional properties. More specifically, the methods of the present invention may be used to identify modifications that increase or decrease the immunogenicity of a protein by affecting antigen uptake, MHC binding, T-cell binding, or antibody binding, while retaining or enhancing functional properties.
Owner:XENCOR
System and method for cleaning noisy genetic data and determining chromosome copy number
ActiveUS20140032128A1Significant resultImprove fidelityMicrobiological testing/measurementBiostatisticsDiploid cellsEmbryo
Disclosed herein is a system and method for increasing the fidelity of measured genetic data, for making allele calls, and for determining the state of aneuploidy, in one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Poorly or incorrectly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related individuals. In accordance with one embodiment, incomplete genetic data from an embryonic cell are reconstructed at a plurality of loci using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without haploid genetic data from one or both parents. In another embodiment, the chromosome copy number can be determined from the measured genetic data, with or without genetic information from one or both parents.
Owner:NATERA
Inhibitors of GSK-3 and crystal structures of GSK-3beta protein and protein complexes
Owner:VERTEX PHARMA INC
Antibodies with Enhanced or Suppressed Effector Function
ActiveUS20130089541A1Inhibition is effectiveStrong specificityAntipyreticAnalgesicsFc receptorFc(alpha) receptor
Rationally designed antibodies and polypeptides that comprise multiple Fc region amino acid substitutions that synergistically provide enhanced selectivity and binding affinity to a target Fc receptor are provided. The polypeptides are mutated at multiple positions to make them more effective when incorporated in antibody therapeutics than those having wild-type Fc components.
Owner:ZYMEWORKS INC
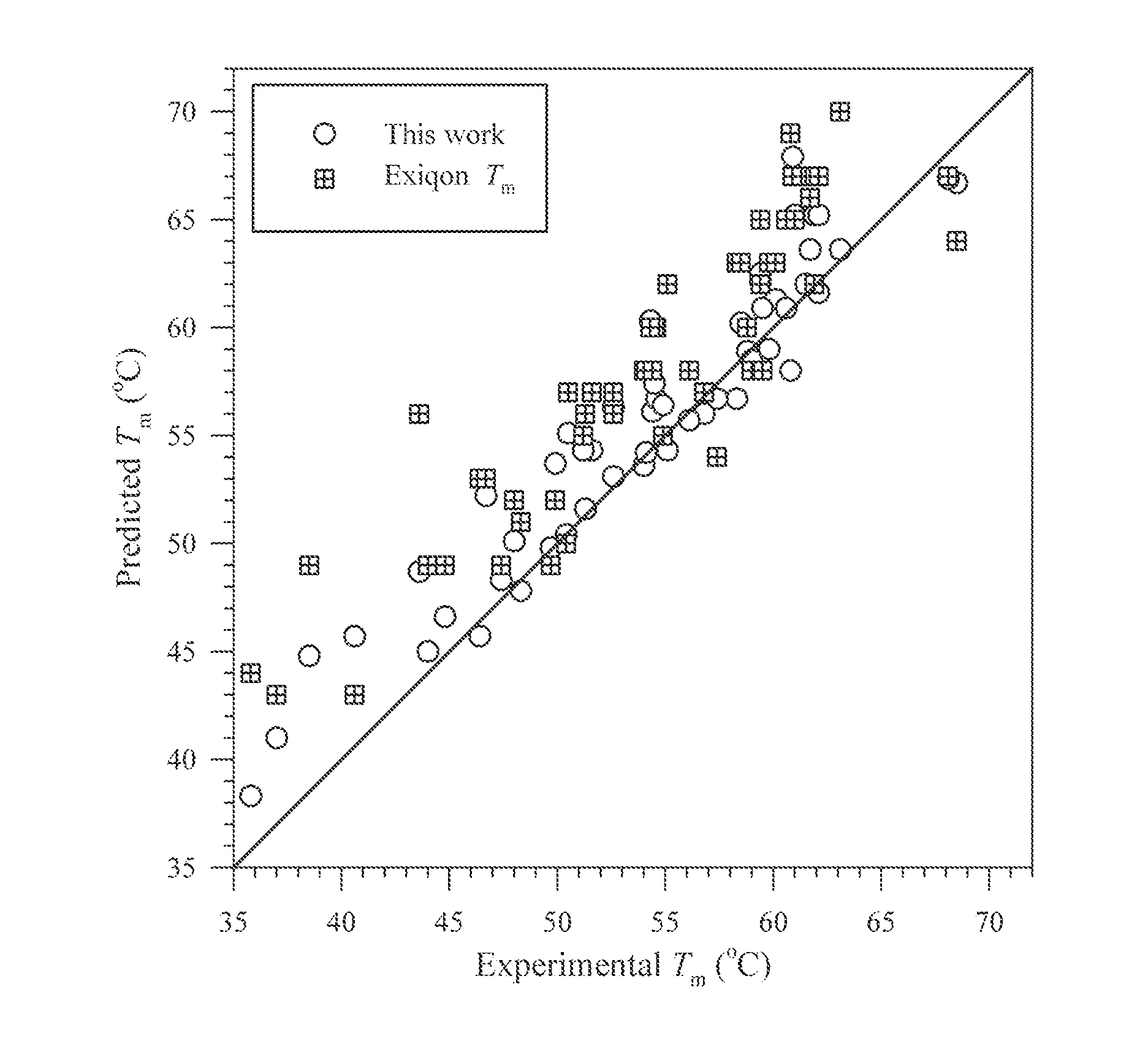
Methods for Predicting Stability and Melting Temperatures of Nucleic Acid Duplexes
ActiveUS20120029891A1Microbiological testing/measurementThermometers using physical/chemical changesOligomerLocked nucleic acid
The present invention provides methods that more accurately predict melting temperatures for duplex oligomers. The invented methods predict the Tm of chimeric duplexes containing various amounts of locked nucleic acid modifications in oligonucleotide strands.
Owner:INTEGRATED DNA TECHNOLOGIES
Apparatus and method for automated protein design
The present invention relates to apparatus and methods for quantitative protein design and optimization.
Owner:MAYO STEPHEN +4
Interaction predicting device
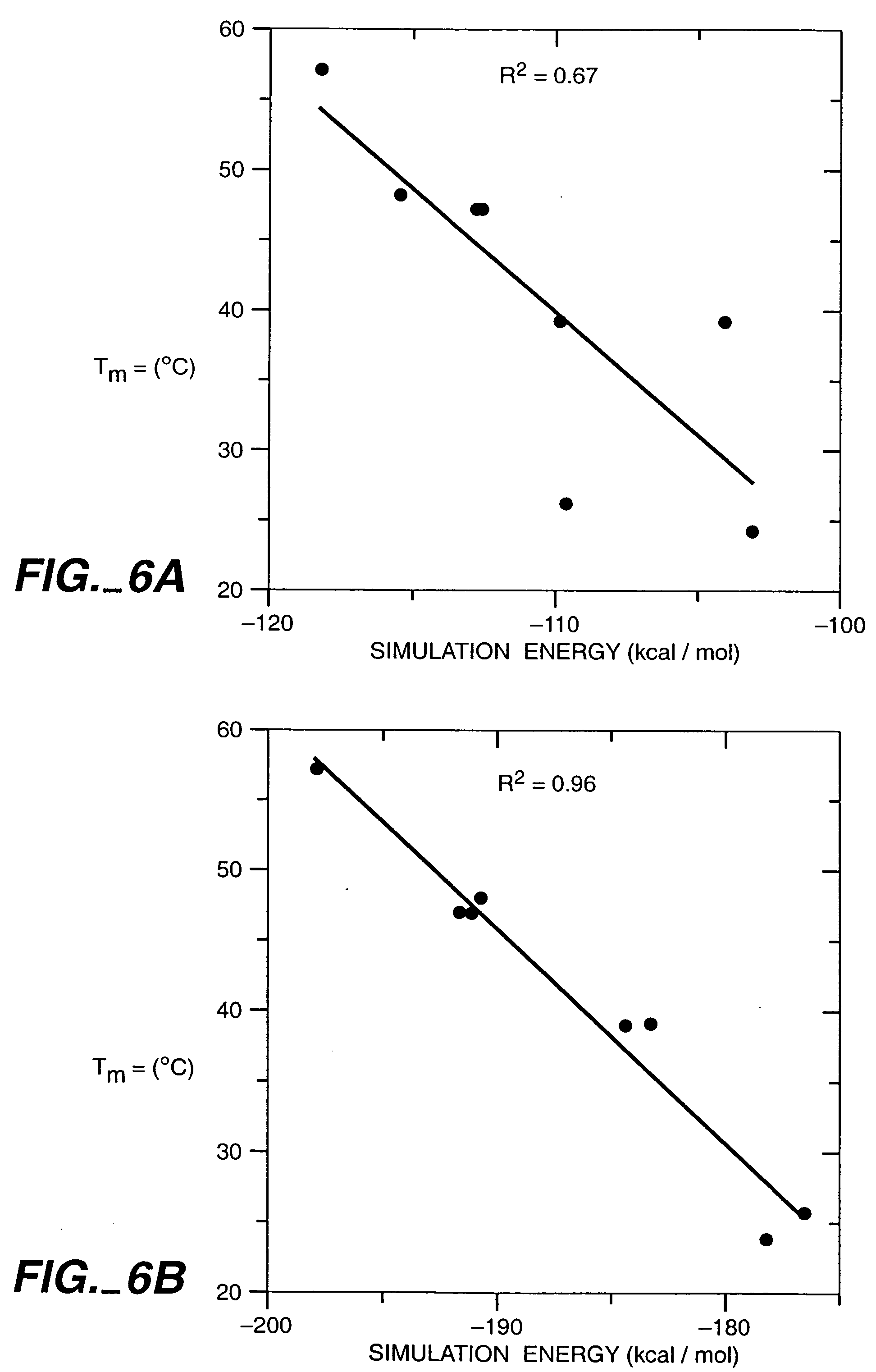
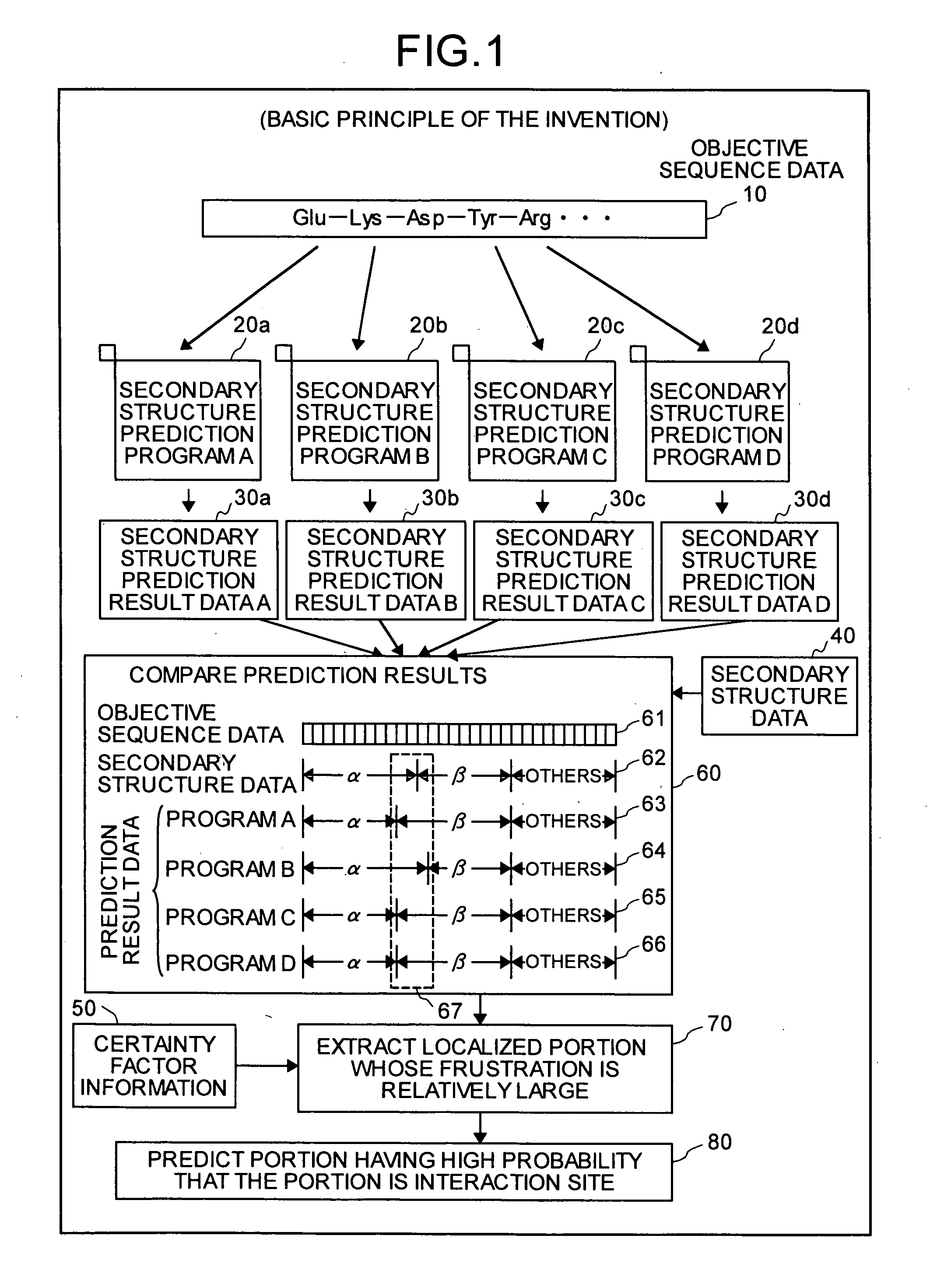
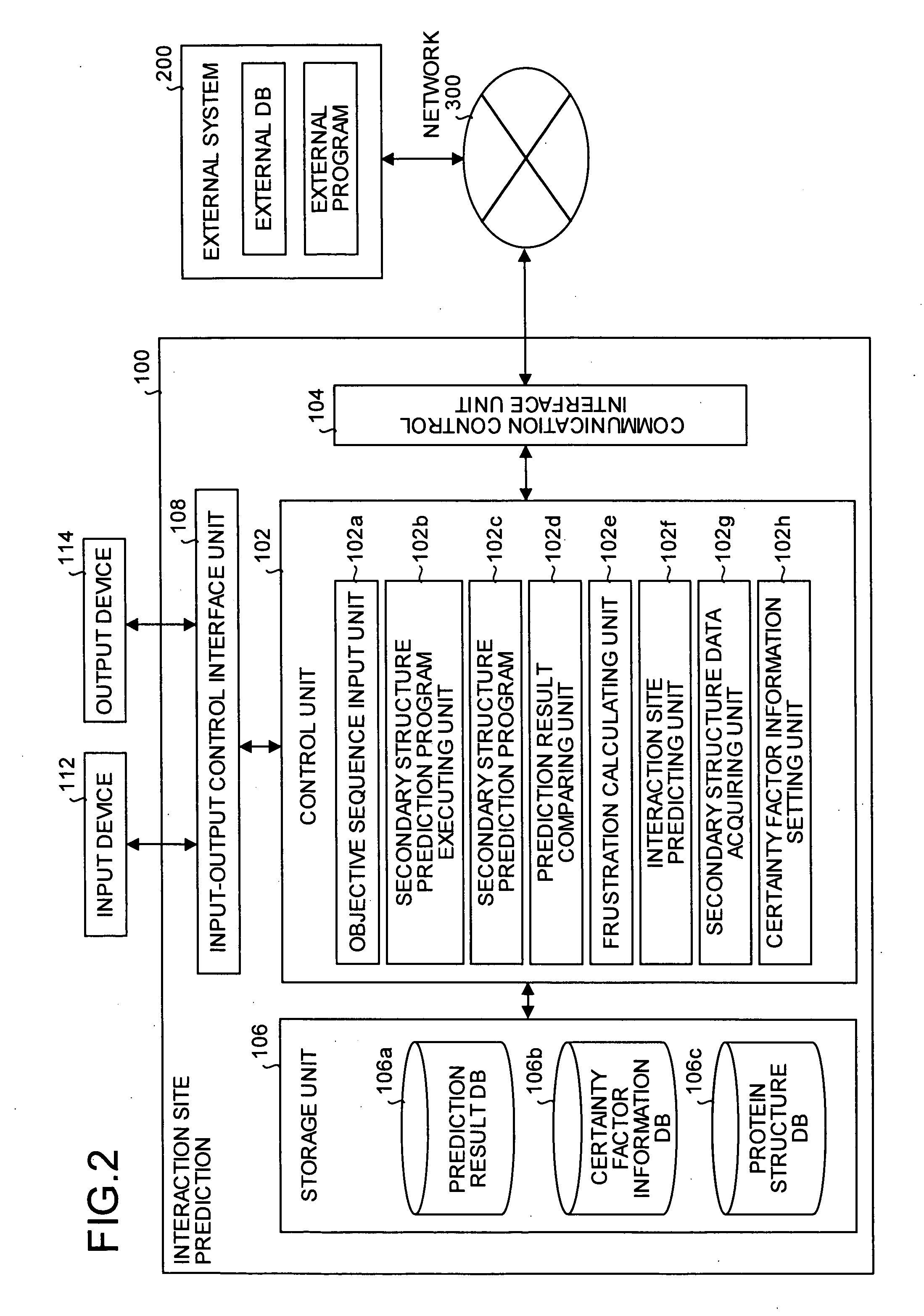
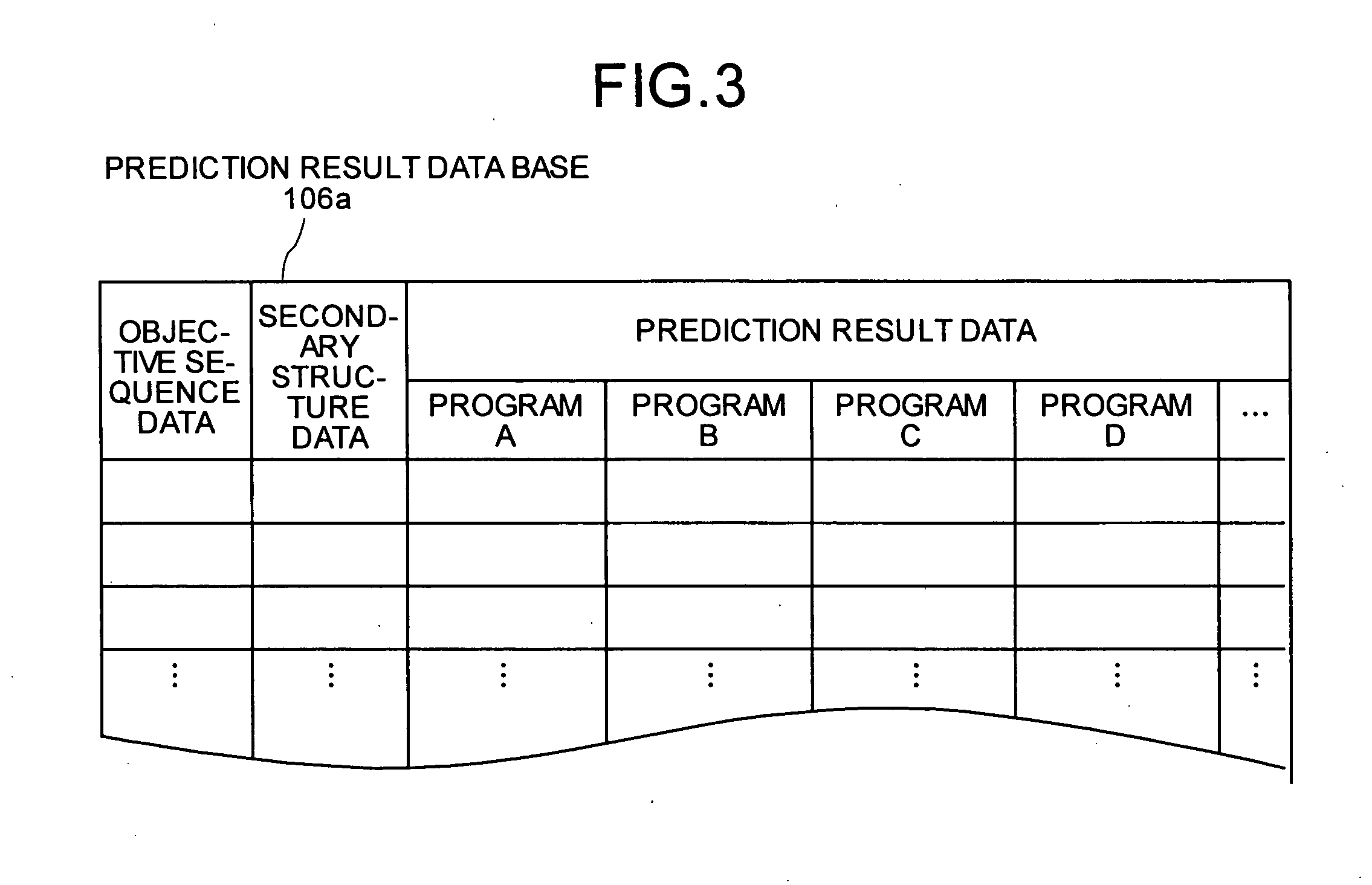
InactiveUS20050130224A1Predict interactionAnalogue computers for chemical processesPeptide preparation methodsFrustrationInteraction site
Objective sequence data (10) which is primary sequence information on an objective protein is entered in an interaction site predicting device by the user. A secondary structure prediction simulation is executed on the objective sequence data (10) entered for secondary structure prediction programs (20a to 20d) that predict a secondary structure of a protein from primary sequence information of the protein. Results of secondary structure prediction (30a to 30d) from the respective secondary structure prediction programs (20a to 20d) are compared (60). Based on the comparison result, frustration of a local portion in the primary sequence information of the objective protein is calculated (70). An interaction site of the objective protein is predicted from the calculated frustration of the local portion (80).
Owner:CELESTAR LEXICO SCI
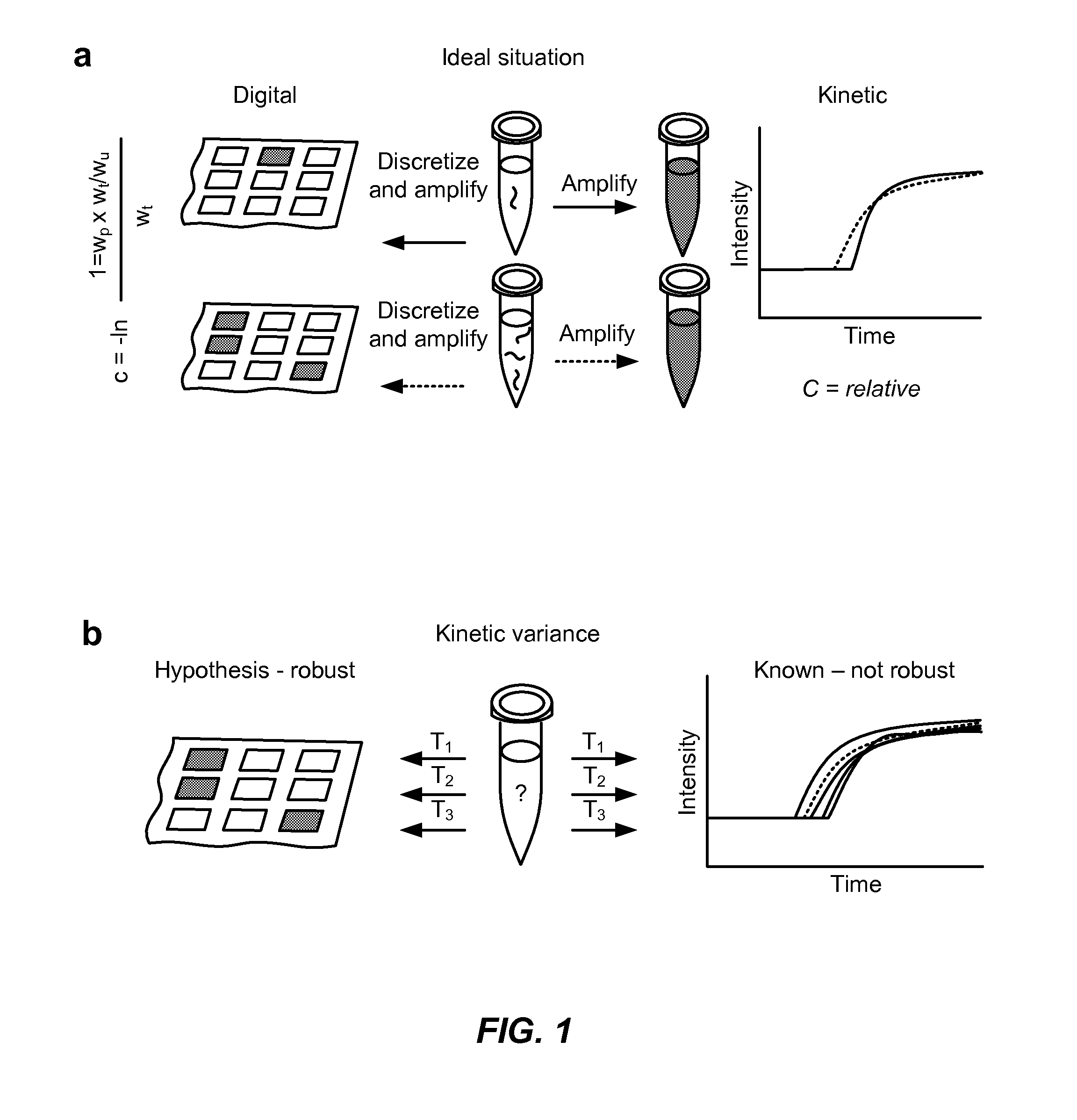
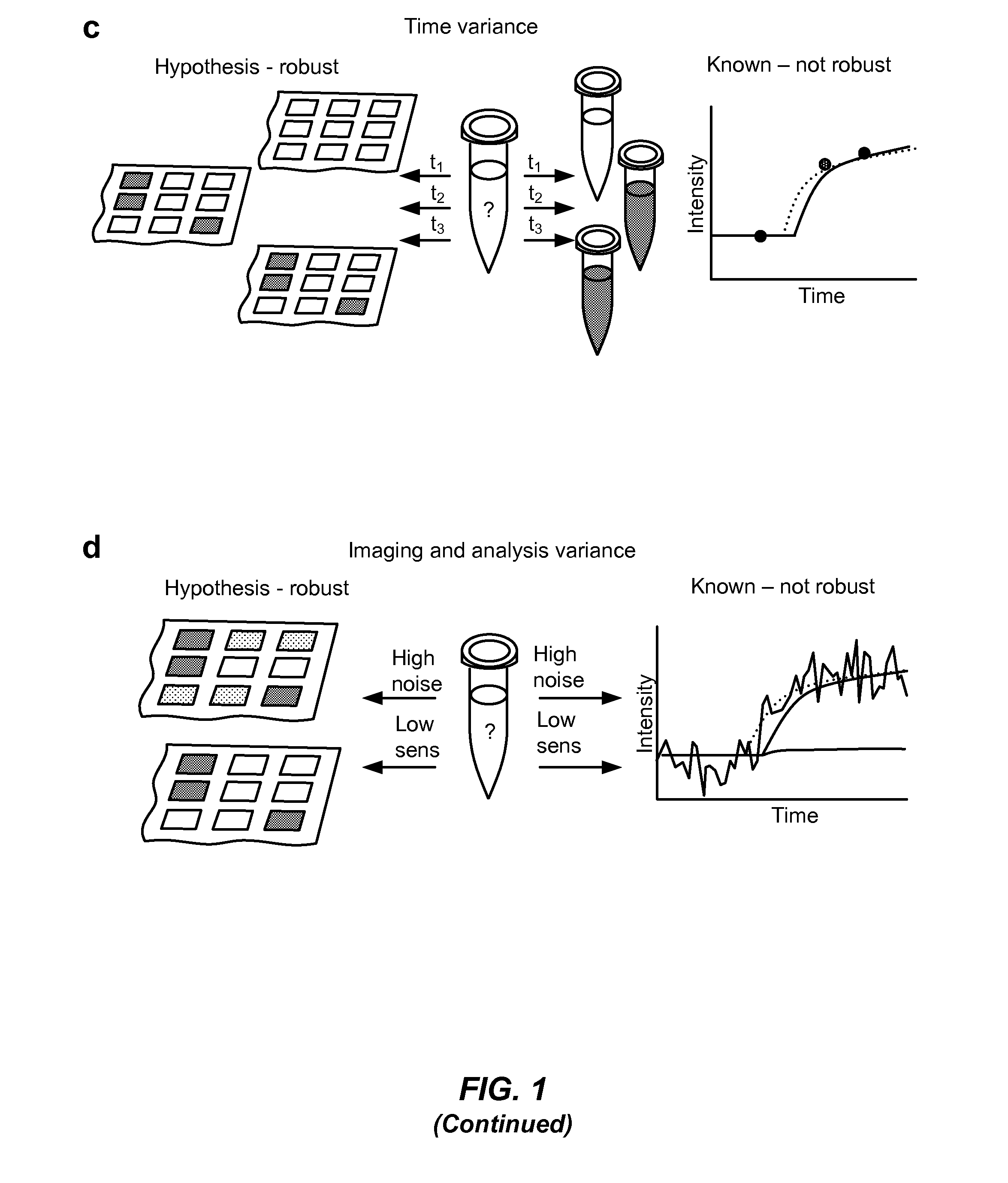
Detecting mutations and ploidy in chromosomal segments
ActiveUS20170107576A1Reduce sensitivityIncreased susceptibilityMicrobiological testing/measurementBiostatisticsNucleotideBiology
The invention provides methods, systems, and computer readable medium for detecting ploidy of chromosome segments or entire chromosomes, for detecting single nucleotide variants and for detecting both ploidy of chromosome segments and single nucleotide variants. In some aspects, the invention provides methods, systems, and computer readable medium for detecting cancer or a chromosomal abnormality in a gestating fetus.
Owner:NATERA
Cytokine receptor
InactiveUS20070032640A1Enhanced interactionEnhanced signalAntibacterial agentsOrganic active ingredientsMedicineScreening method
A crystalline composition comprising a crystal of the IL-6 receptor I chain is provided. Also provided are methods of using the crystal and related structural information to screen for and design compounds that interact with IL-6R, or variants thereof. Also provided arc methods of modulating an IL-6 receptor comprising contacting the IL-6 receptor with a compound identified by the screening method of the invention.
Owner:COMMONWEALTH SCI & IND RES ORG +2
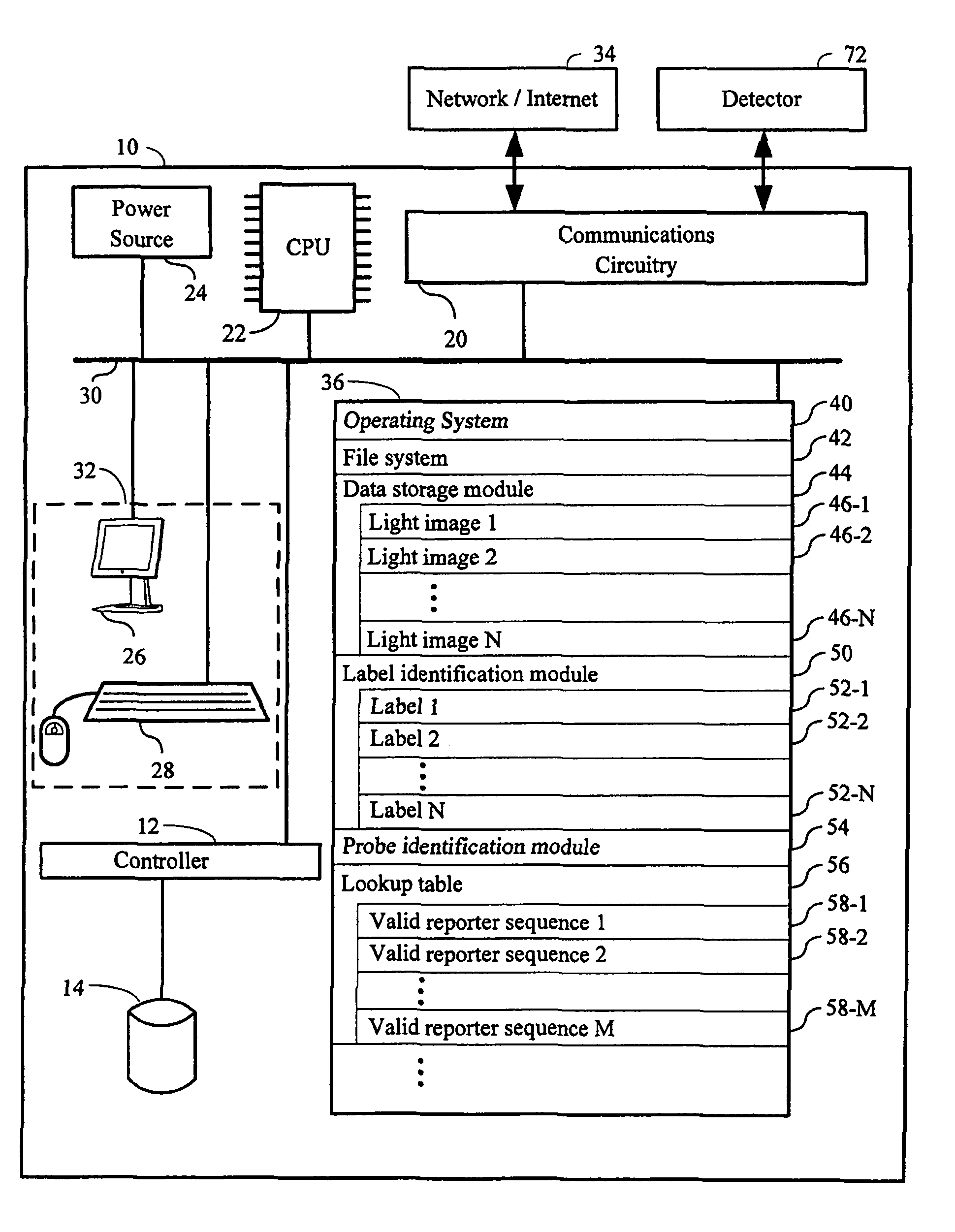
Systems and methods for analyzing nanoreporters
Methods, computers, and computer program products for detecting the presence of a probe within a sample overlayed on a substrate are provided. The probe comprises a plurality of spatially arranged labels. A data storage module stores a plurality of light images, where each light image has light from the sample at a corresponding wavelength range in a plurality of different wavelength ranges. A label identification module identifies a plurality of labels in the plurality of light images that are proximate to each other on the substrate. A spatial order of the plurality of labels determines a string sequence of the plurality of labels. A probe identification module determines whether the string sequence of the plurality of labels comprises a valid reporter sequence.
Owner:NANOSTRING TECH INC
System and method for cleaning noisy genetic data and determining chromosome copy number
ActiveUS10083273B2Improve fidelityData processing applicationsMicrobiological testing/measurementDiploid cellsEmbryo
Disclosed herein is a system and method for increasing the fidelity of measured genetic data, for making allele calls, and for determining the state of aneuploidy, in one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Poorly or incorrectly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related individuals. In accordance with one embodiment, incomplete genetic data from an embryonic cell are reconstructed at a plurality of loci using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without haploid genetic data from one or both parents. In another embodiment, the chromosome copy number can be determined from the measured genetic data, with or without genetic information from one or both parents.
Owner:NATERA
Ligand-directed covalent modification of protein
InactiveUS20110269244A1Easily assumeMolecular designTransferasesCovalent modificationPharmaceutical formulation
The present invention relates to enzyme inhibitors. More specifically, the present invention relates to ligand-directed covalent modification of proteins; method of designing same; pharmaceutical formulation of same; and method of use.
Owner:CELGENE CAR LLC
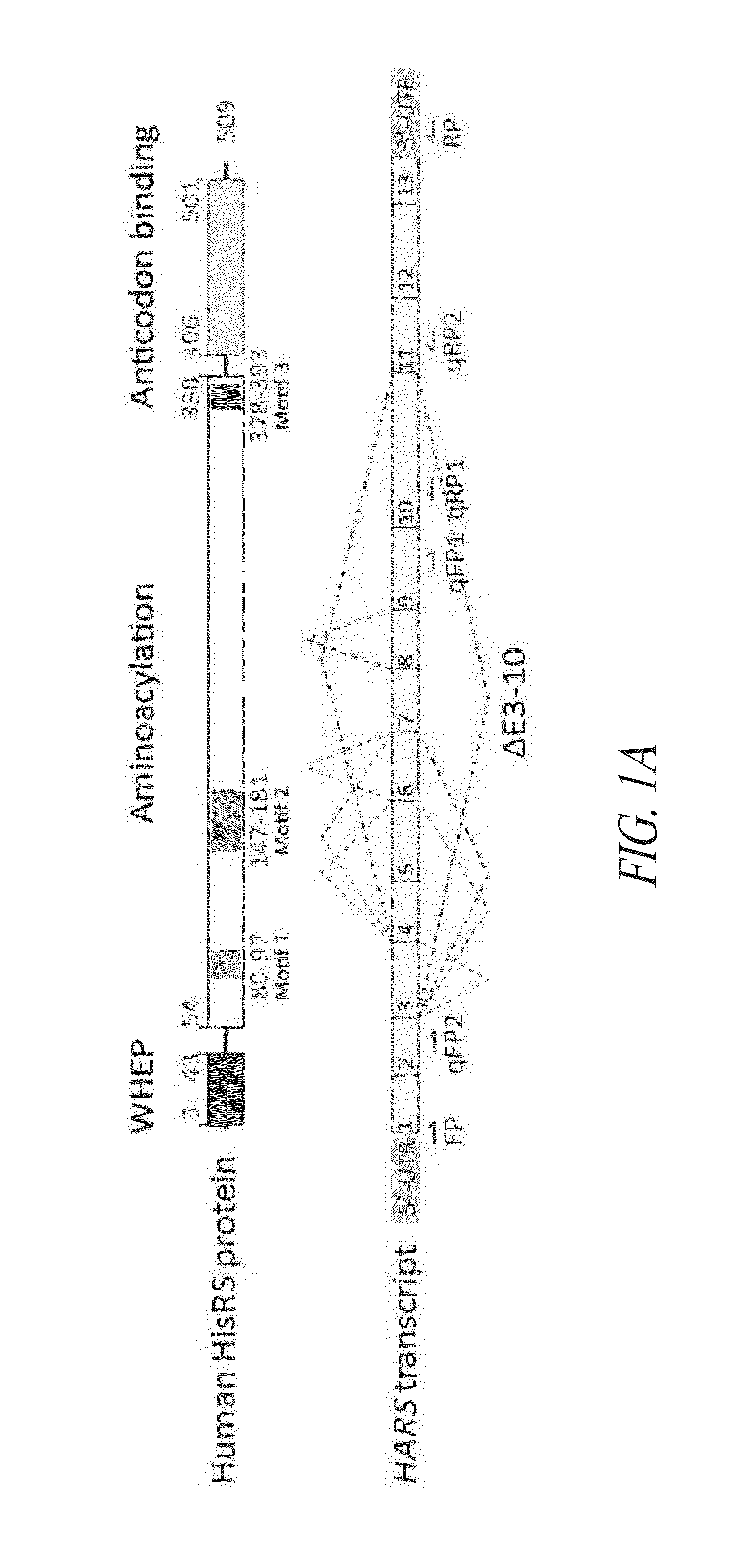
Structures of human histidyl-trna synthetase and methods of use
InactiveUS20140066321A1Improve homogeneityLibrary screeningAnalogue computers for chemical processesMedicineX-ray
Provided are histidyl-tRNA synthetase variant polypeptides, X-ray crystallographic and NMR spectroscopy structures of HRS polypeptides, and related compositions and methods for therapy and drug discovery.
Owner:ATYR PHARM INC +1
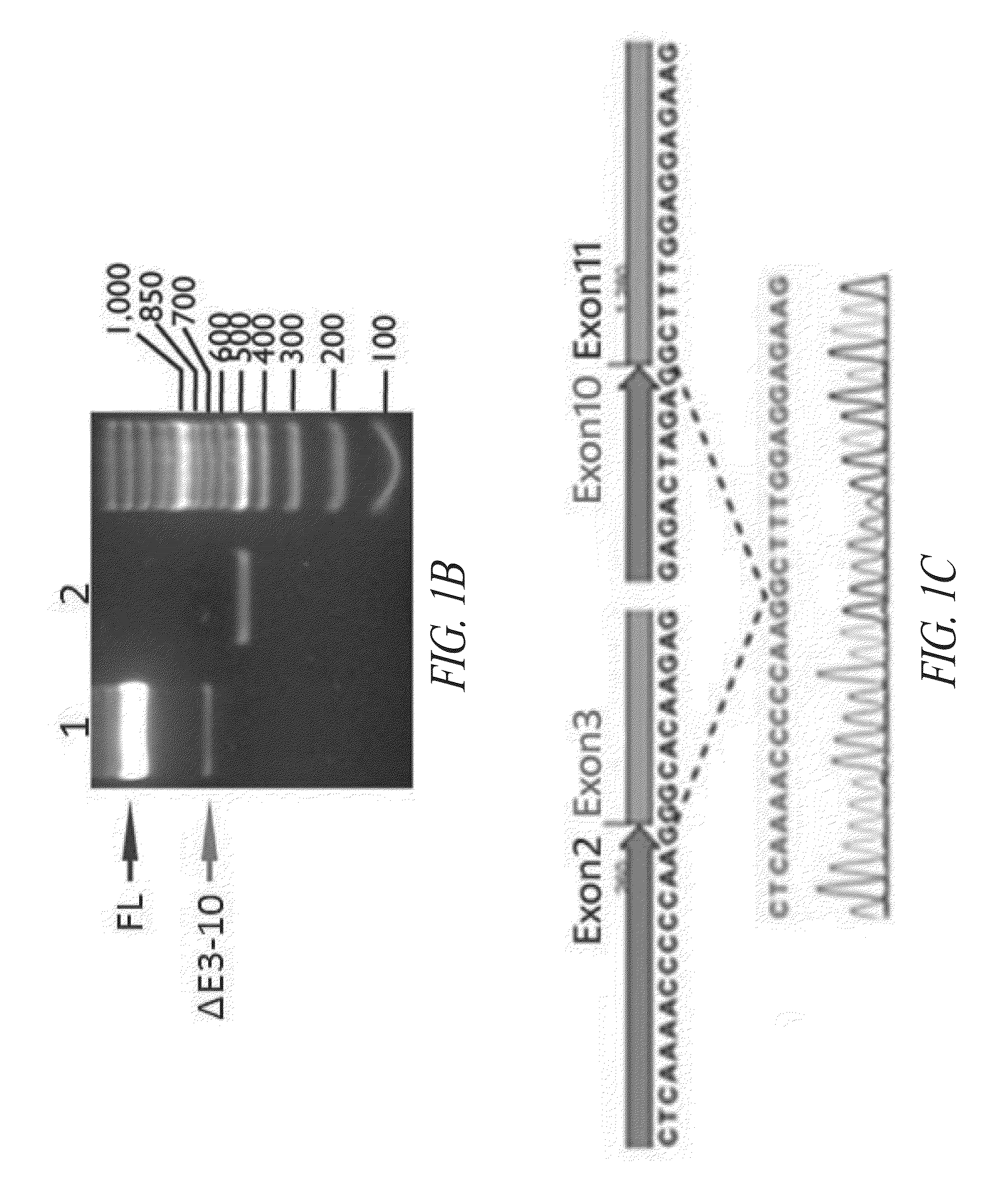
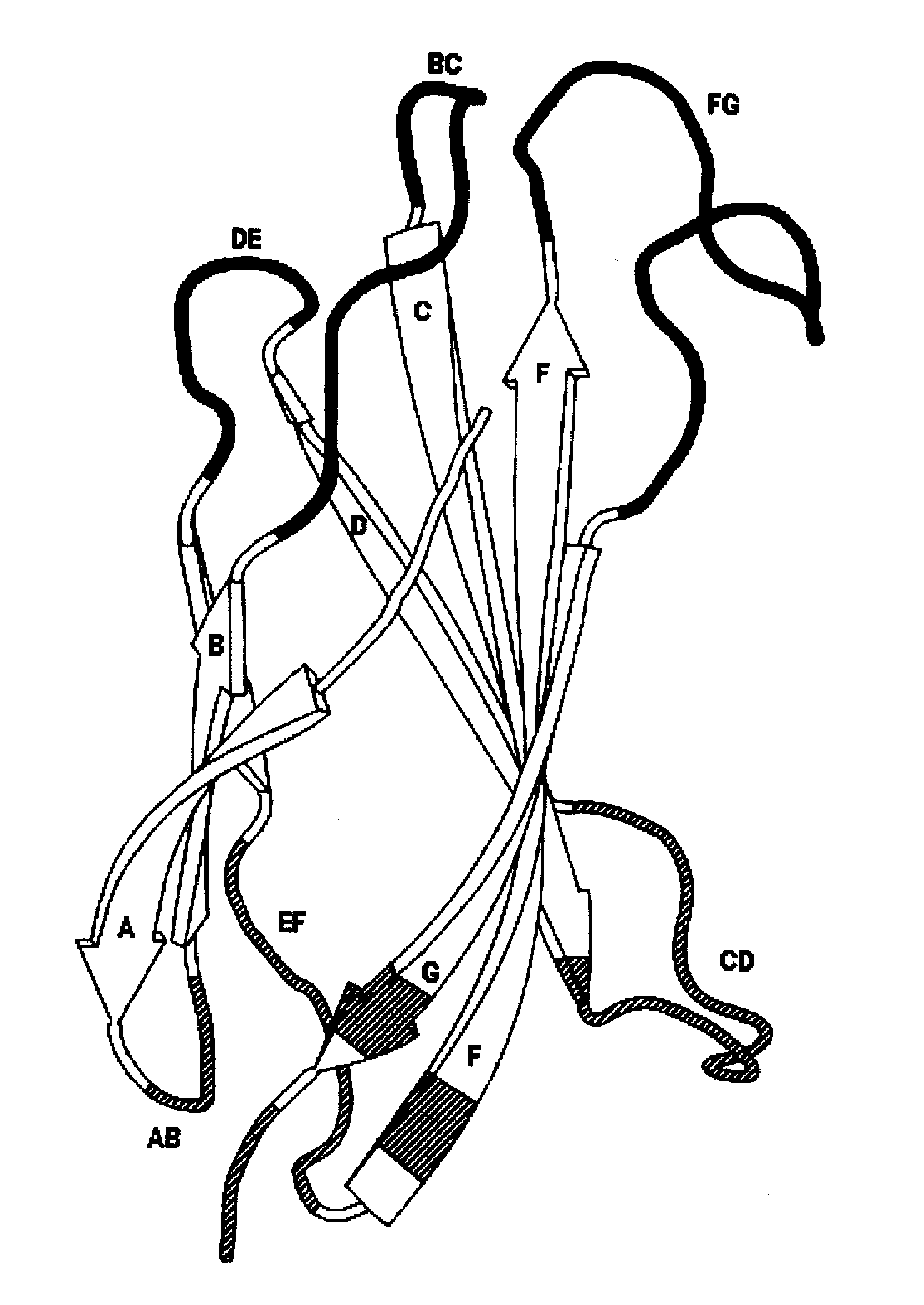
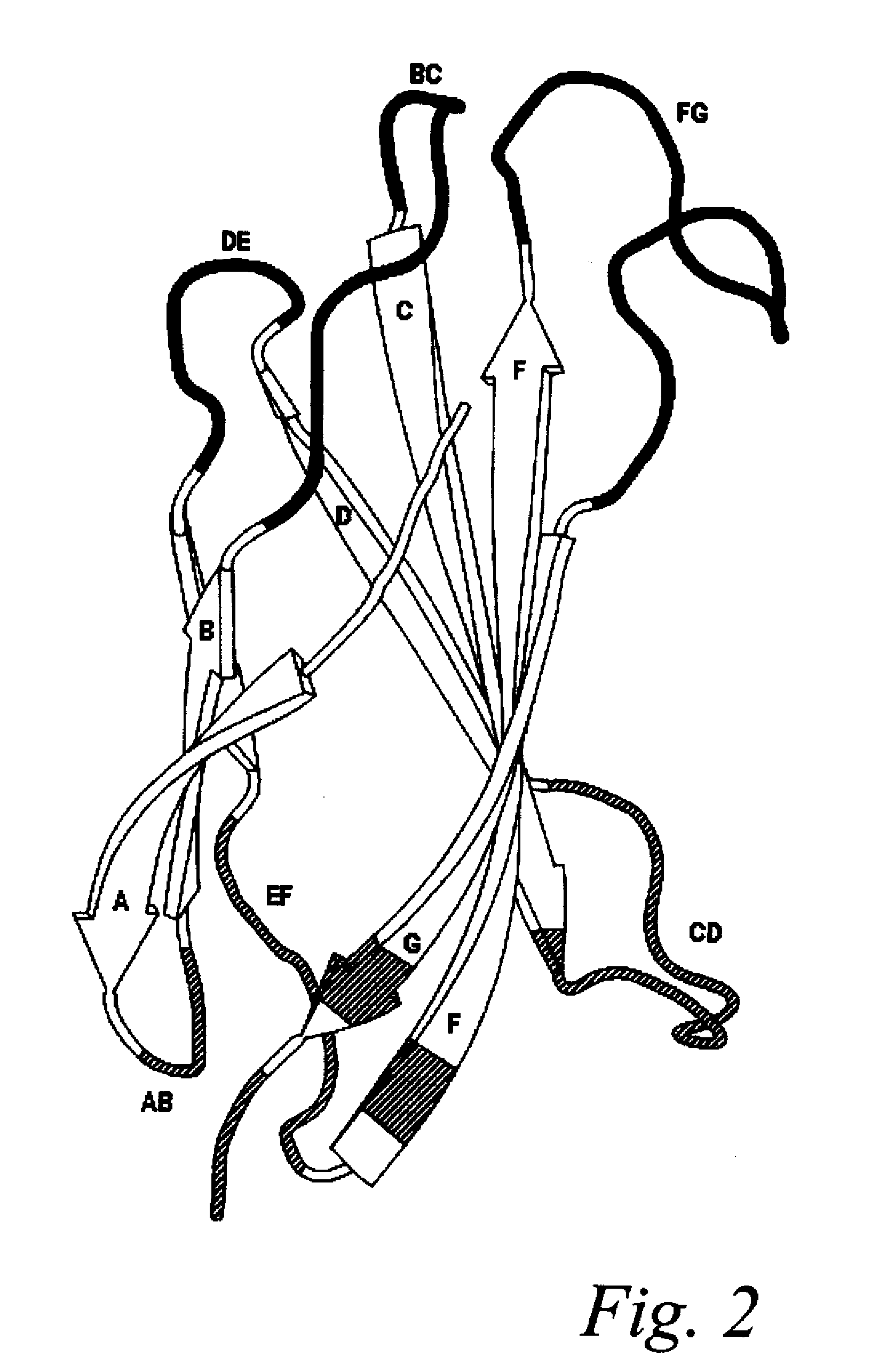
Universal fibronectin type iii bottom-side binding domain libraries
The invention pertains to a natural-variant combinatorial library of fibronectin Type 3 domain (Fn3) polypeptides useful in screening for the presence of one or more polypeptides having a selected binding or enzymatic activity. The library polypeptides include (a) regions A, AB, B, C, CD, D, E, EF, F, and G having wildtype amino acid sequences of a selected native fibronectin Type 3 polypeptide or polypeptides, and (b) loop regions AB, CD, and EF having selected lengths (Bottom Loops). The Fn3 may also have loop regions BC, DE, and FG having wildtype amino acid sequences, having selected lengths, or mutagenized amino acid sequences (Top Loops).
Owner:NOVARTIS AG
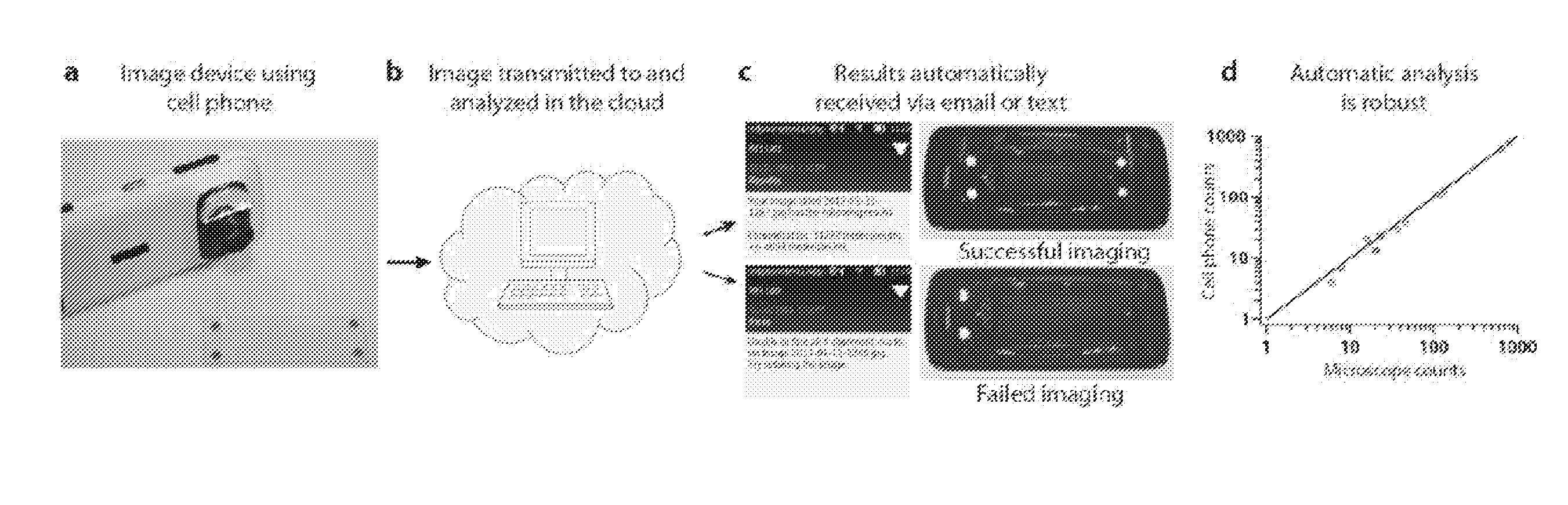
Methods and systems for microfluidics imaging and analysis
InactiveUS20150247190A1Bioreactor/fermenter combinationsBiological substance pretreatmentsBiological bodyMicrofluidics
Owner:CALIFORNIA INST OF TECH
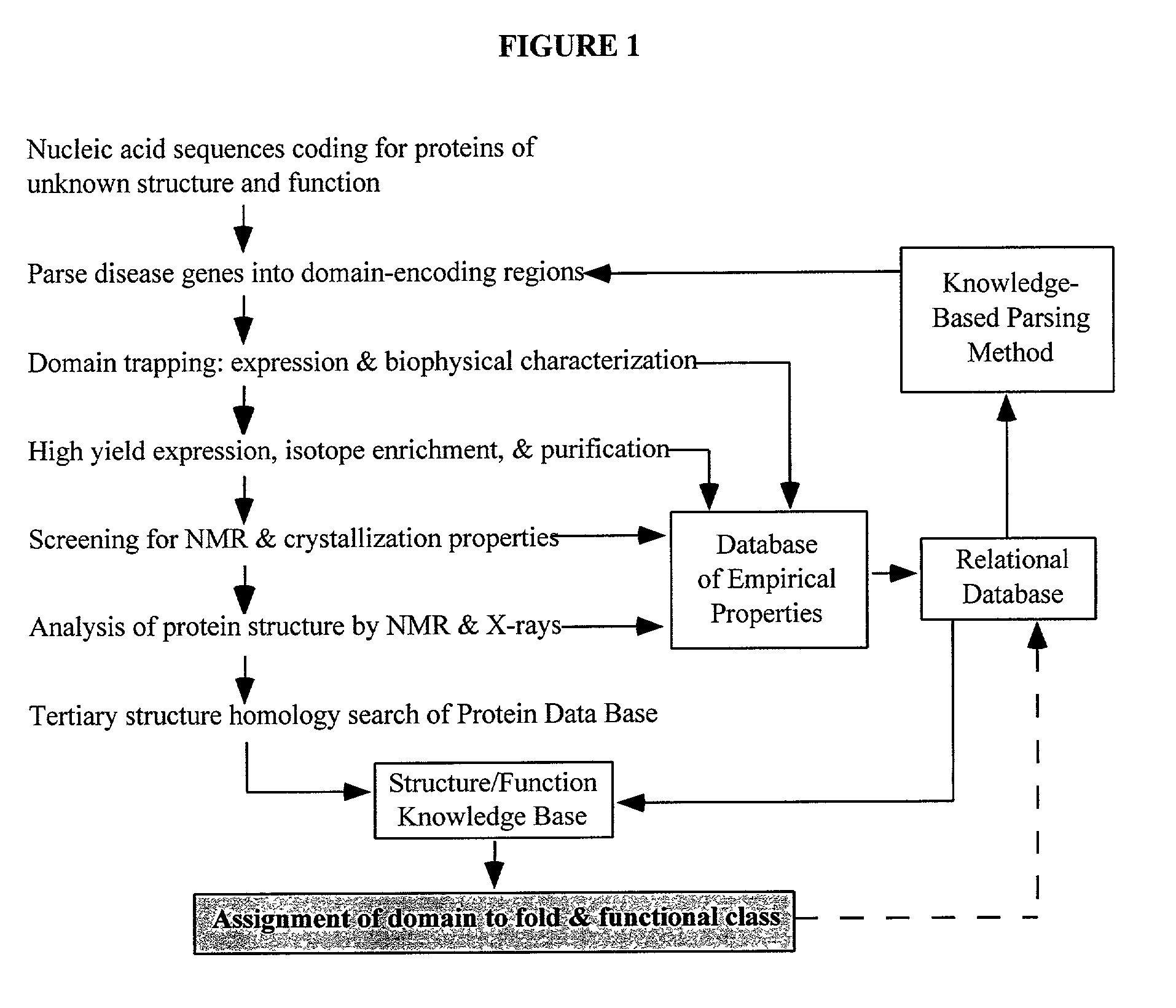
Linking gene sequence to gene function by three dimesional (3D) protein structure determination
InactiveUS20010016314A1Sugar derivativesMicrobiological testing/measurementFunctional profilingProtein structure
The present invention provides a structure-functional analysis engine for the high-throughput determination of the biochemical function of proteins or protein domains of unknown function. The present invention uses bioinformatics, molecular biology and nuclear magnetic resonance tools for the rapid and automated determination of the three-dimensional structures of proteins and protein domains.
Owner:RUTGERS THE STATE UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com